
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 26 June 2023
Sec. Extreme Microbiology
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1193955
This article is part of the Research Topic Extremozymes: Characteristics, Structure, Protein Engineering and Applications View all 8 articles
TrLipE is a thermophilic lipase that has potential commercial applications because of its catalytic ability under extreme conditions. Consistent with most lipases, the lid of TrLipE is located over the catalytic pocket, controls the substrate channel to the active center, and regulates the substrate specificity, activity, and stability of the enzyme through conformational changes. TrLipE from Thermomicrobium roseum has potential industrial applications, which is hindered by its weak enzymatic activity. Here, 18 chimeras (TrL1-TrL18) were reconstructed by N-terminal lid swapping between TrLipE and structurally similar enzymes. The results showed that the chimeras had a similar pH range and optimum pH as wild TrLipE but a narrower temperature range of 40–80°C, and TrL17 and the other chimeras showed lower optimum temperatures of 70°C and 60°C, respectively. In addition, the half-lives of the chimeras were lower than those of TrLipE under optimum temperature conditions. Molecular dynamics simulations indicated that chimeras had high RMSD, RMSF, and B-factor values. When p-nitrophenol esters with different chains were used as substrates, compared with TrLipE, most of the chimeras had a low Km and high kcat value. The chimeras TrL2, TrL3, TrL17, and TrL18 could specifically catalyze the substrate 4-nitrophenyl benzoate, with TrL17 showing the highest kcat/Km value of 363.88 ± 15.83 L⋅min–1⋅mmol–1. Mutants were then designed by investigating the binding free energies of TrL17 and 4-nitrophenyl benzoate. The results indicated that single, double, and triple substitution variants (M89W and I206N; E33W/I206M and M89W/I206M; and M89W/I206M/L21I and M89W/I206N/L21I, respectively) presented approximately 2- to 3-fold faster catalysis of 4-nitrophenyl benzoate than the wild TrL17. Our observations will facilitate the development of the properties and industrial applications of TrLipE.
Lipases (EC 3.1.1.3) are well-known biotechnologically relevant biocatalysts of the α/β-hydrolase superfamily that act on carboxylic ester bonds and catalyze a variety of chemical reactions, including hydrolysis, transesterification, esterification, alcoholysis, aminolysis, and acidolysis, in aqueous and organic solvents (Sarmah et al., 2017; Casas-Godoy et al., 2018; Vivek et al., 2022). Based on their thermostability, organic solvent tolerance, and excellent catalytic ability under extreme conditions, lipases have been widely used in the food (Asmat et al., 2019; Cipolatti et al., 2020), detergent, leather (Verma et al., 2021), pharmaceutical, and biodiesel industries (Cavalcante et al., 2020; Zhong et al., 2020; Almeida et al., 2021). Lipases have a highly conserved catalytic triad consisting of nucleophilic serine or cysteine, aspartic or glutamic acid, and histidine (Anobom et al., 2014; Gutiérrez-Domínguez et al., 2022; Liu et al., 2022). Most lipases display a very similar fold in which mixed β-sheets are surrounded by α-helices and a domain named lid that controls the substrate channel to the active pocket (Mohd Din et al., 2021; Xing et al., 2021; Cui et al., 2022).
Most lipases are widely considered to exert interfacial activation, and the lid domain located above the catalytic pocket is responsible for this phenomenon because the lid switches between open and closed conformations at the interface to correspond to the active and inactive states of the lipase, respectively (Jan et al., 2017; Maiangwa et al., 2017; Soni et al., 2019; Wang et al., 2021). However, CALB from Candida antarctica with a small lid has also been reported to display almost no interfacial activation in the presence of short-chain substrates (Stauch et al., 2015). The lid plays an important role in most lipases because it not only regulates the activity of the enzyme but also contributes to its catalytic specificity, thermostability, and binding of the substrate. In recent years, considerable research interest has focused on the role of lids in enzymes. Tang et al. (2015) reported that the T66L/D70N mutant of lipase (PEL) from Penicillium expansum displayed a 136.4-fold increase in p-nitrophenol palmitate activity by replacing the amino acids of the N-terminal hinge of the lid. Soni et al. (2019) engineered a monoglyceride lipase (TON-LPL) from Thermococcus onnurineus into triglyceride lipase by lid swapping. The values of t1/2 at 60°C and Tm were increased by introducing a disulfide bridge into the lid hinge of a lipase from Rhizopus chinensis (Yu et al., 2012).
The optimal enzyme-substrate complex exhibits good binding affinity and a relatively low binding free energy (Anuar et al., 2021). Therefore, a variety of computational programs have been developed to calculate the binding energy of protein and ligand complexes as a protein mutation design strategy to improve enzyme catalytic efficiency and stereoselectivity (Kumari and Gupta, 2013; Zhang et al., 2013; Bai et al., 2022).
TrLipE from T. roseum possesses remarkable enzymatic properties, such as excellent thermostability, organic solvent tolerance, and pH tolerance; thus, it has great potential for industrial application. However, its weak enzymatic activity hinders the application of TrLipE in the above-mentioned fields. In this study, 18 chimeras were successfully constructed by lid swapping between TrLipE and structurally similar enzymes. Different from other studies on lid, our study changed the binding ability of chimeras to different substrates by exchanging the complete lid domains, and thus changing their specificity. In addition, to further improve the enzymatic activity of the chimera, mutation sites were designed based on strategies to reduce the binding energy of the protein–ligand docking complex. This study provides valuable insights into the rational design of TrLipE for further industrial applications.
Escherichia coli JM109 and BL21 were used for cloning and TrLipE expression, respectively, and stored at −80°C in our laboratory. The ClonExpress II One-Step Cloning Kit was purchased from Vazyme Biotech Co., Ltd. (Nanjing, China). The substrates (p-nitrophenyl fatty acid esters) were purchased from Sigma-Aldrich (USA). A BCA protein quantification kit was purchased from Tiangen Biotech Co., Ltd. (Beijing, China). A Mag-Beads His-Tag Protein Purification Kit was purchased from Sangon Biotech (Shanghai, China). Other analytical reagents and biological materials were provided by local suppliers.
Standard manipulation techniques have been used for the construction and isolation of plasmid DNA and transformation of E. coli (Kay et al., 2016). Briefly, the DNA fragment containing the TrLipE gene sequence without a lid and the sequence of plasmid pTIG, and other 18 lid sequences fragments from 18 different lipases similar to TrLipE were obtained by polymerase chain reaction (PCR) using the primers sequence in Supplementary Table 1. Then, the expression plasmids for the chimeras were obtained by homologous recombination of the above fragments, and the expression plasmids for the chimeras were then transformed in E. coli JM109 for amplification and preservation. In addition, the primers in Supplementary Table 2 were designed on the recombinant plasmid pTIG-TrL17 for site-directed mutagenesis, and a Mut Express MultiS Fast Mutagenesis Kit V2 (Vazyme) was used for the designed substitutions according to the manufacturer’s recommendations. After PCR, the mixture was digested with DpnI to remove the template (pTIG-TrL17). The plasmids containing mutated sites were then transformed in E. coli JM109 for amplification and preservation. After verifying the sequence, the chimeric and mutant expression plasmids were transformed into E. coli BL21. Recombinant cells were cultured, harvested, and disrupted by sonication where the program was that the power was set at 39%, work for 1 s, stop for 2 s, and the total time was 15 min. The Mag-Beads His-Tag Protein Purification kit was used for purification of TrLipE, chimeras, and mutants, as described by Fang et al. (2021), and then the pure enzyme was centrifuged (4,000 rpm 30 min) by ultrafiltration to remove imidazole, and the buffer was replaced with Tris-HCL (50 mmol⋅L–1, pH 8.0).
The TrLipE and chimera enzymatic activity was determined using emulsified p-nitrophenol esters as the substrates (Sha et al., 2013; Fang et al., 2022). The substrates were dissolved in dimethyl sulfoxide at a concentration of 15 mmol⋅L–1, and the buffer was composed of 50 mM Tris-HCl containing 0.55 g⋅L–1 Arabic gum (sigma) and 1.2 g⋅L–1 sodium deoxycholate (sigma) (pH 8.0). The reaction was processed in a final volume of 500 μL, which contained 20 μL enzyme at a concentration of 0.3 mg/mL and 480 μL of substrate mixture, in which the substrates were mixed with the buffer at a ratio of 1:100. The absorbance value at OD410nm was determined. Mixture without the enzyme was applied as the blank. One enzyme unit (μmol⋅mg–1⋅min–1) was defined as the amount of enzyme that released 1 μmol of p-nitrophenol per minute under the assay conditions. All results were obtained from experiments performed in triplicate, and the variance was calculated.
Following the enzyme activity assay, the optimum temperature and pH were determined within a temperature range of 40–80°C and a pH range of 5.5–11 using buffers with different pH values, including phosphate buffer (pH = 5.5–7.5), Tris-HCl buffer (pH = 7.5–9.0) and glycine-sodium hydroxide buffer (pH = 9.0–11.0). The maximum enzymatic activity was defined as the control (100%).
For determining the half-life, the pure enzyme solution was placed at constant temperatures of 60, 70, and 80°C for TrL17 and 50, 60, and 70°C for the other chimeras, and then samples were collected at intervals of 1 h for chimeras and 3 h for TrLipE to determine the remaining enzyme activity as described above. As mentioned above, enzymatic activity at 0 h was defined as the control (100%).
Substrate specificity was determined using different p-nitrophenyl esters, including p-NP acetate (C2), p-NP butyrate (C4), p-NP hexanoate (C6), p-NP caprylate (C8), p-NP decanoate (C10), p-NP laurate (C12), p-NP palmitate (C16), and p-NP benzoate. The Km value was calculated using the Non-linear Curve Fit, and the kcat and kcat/Km values were calculated according to the molecular weight and concentration of the enzyme. For each substrate, the reaction was determined at 60°C. All results were obtained from experiments performed in triplicate, and the variance was calculated.
After modeling with Alphafold2, molecular dynamics (MD) simulations of chimeras and TrLipE were performed as described by Fang et al. (2021), Kay et al. (2016). Briefly, the Gromacs 2018.4 package (Tambunan et al., 2014) containing the Amber14SB (Maier et al., 2015) all-atom force field was used under constant temperature, pressure, and periodic boundary conditions. MD runs were performed for 100 ns with a time step of 2 fs at 300 K. The molecular docking of TrL17 with 4-nitrophenyl benzoate was carried out using Schrödinger software (Godara et al., 2022), and the docking results were converted into PDB format using PyMOL. Discovery Studio 2019b (DS) was used to analyze the docking and virtual mutations (Gao et al., 2018).
The amino acid sequence of TrLipE was submitted to the PDB database for alignment, and 18 crystal structures with 20–60% similarity to TrLipE were selected to reconstruct chimeras. Table 1 shows the lid sequence range used to swap, the PDB code of these 18 crystal structures, and the names of the chimeric enzymes correspond to different lids. After recombination between TrLipE and the lids of the above-mentioned structures, the gene sequences were optimized and synthesized E. coli. After expression and purification according to the above method, the pure enzyme was further verified by SDS-PAGE (12% separating gel). The results indicated that these chimeras could be expressed efficiently in E. coli BL21 and showed a band with the calculated molecular mass range of approximately 35–37 kDa because of the different sizes of the lid regions. Furthermore, the chimeras displayed different expression levels than TrLipE. The reason for this phenomenon was that the proportion of hydrophilic amino acids in the lid region changed after lid swapping (Supplementary Table 3).
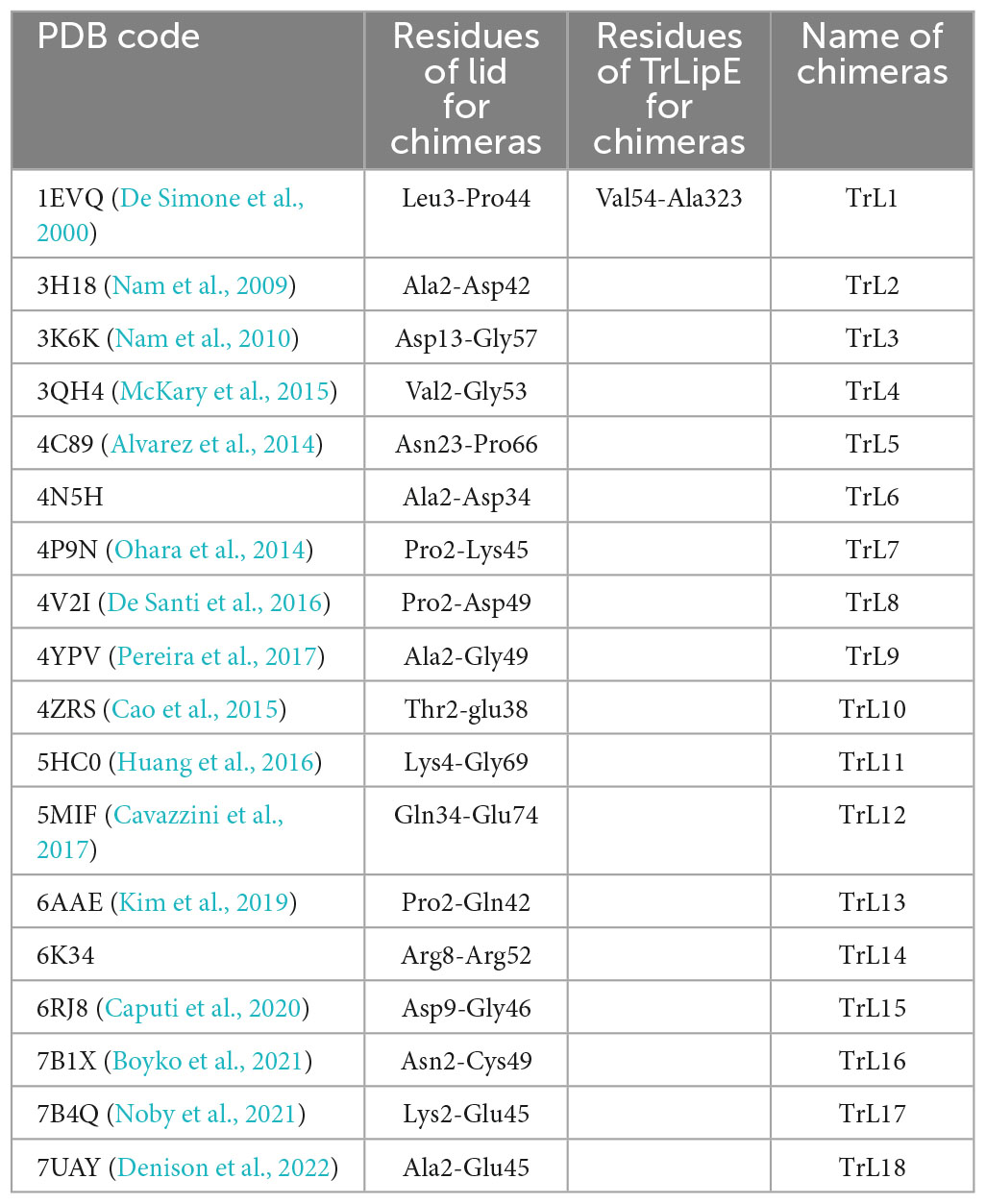
Table 1. The PDB code of 18 crystal structures, lid sequences range used to swap and the names of chimeric enzymes correspond to different lid.
Figure 1 illustrates that the chimeras had a low optimal temperature of 70°C for TrL17 and 60°C for the other chimeras, which was lower than that of TrLipE with an optimal temperature of 85°C (Kay et al., 2016). The half-life of 18 chimeras was in the range of 5–9 h at the optimal temperature, which was also lower than that of TrLipE (>12 h) shown in Supplementary Table 4. Although the half-lives of chimeras increased when the temperature was 10°C lower than the optimal temperature, the same phenomenon of slightly lower thermostability of the chimeras relative to the wild-type TrLipE was observed. The common feature shared by chimeras and TrLipE was that they nearly completely lost their enzymatic activity after 3 h when the temperature was 10°C higher than the optimal temperature. Therefore, the above results indicate that the lid affects the thermostability of the chimeras, which has also been confirmed by other studies. Yu et al. (2014) found that lid swapping has a negative effect on the thermostability of chimeras when studying the conversion of lipase to esterase. Coincidentally, Huang et al. (2019) indicated an obvious decrease in Tm and half-life values of chimera (AFLB-CALBlid) obtained by lid swapping. As reported by Khan et al. (2017), the thermostability of lipases can be altered by modifications in their lid domains because thermophilic lipases generally have a large and rigid lid structure. Nevertheless, these chimeras are less thermostable than TrLipE and superior to other reported lipases (Yang et al., 2015, 2016; Li et al., 2022).

Figure 1. The optimal temperature of chimeras. The chimeras had a low optimal temperature of 70°C for TrL17 and 60°C for the other chimeras, which was lower than that of TrLipE with an optimal temperature of 85°C.
To further evaluate the effect of the lid area on thermostability, MD simulations were performed at 300 K. The Root Mean Square Deviation (RMSD), Root Mean Square Fluctuation (RMSF), and B-factor values were analyzed, which represent the flexibility of the protein structure. Higher values indicate greater flexibility of the protein structure and vice versa, so they can characterize the thermostability of the protein. As shown in Figure 2, the results clearly demonstrated that chimeras had higher RMSD, RMSF, and B-factor values than TrLipE, which indicated that chimeras had greater flexibility and poorer thermostability than wild-type TrLipE. However, although the chimeras had higher average RMSD (0.174 ± 0.052 nm), RMSF (0.126 ± 0.037 nm), and B-factor (53.13 ± 3.18) values than TrLipE, the values for TrL17 were lower than those of other chimeras (Supplementary Figure 1), thus confirming the optimal temperature results mentioned above.
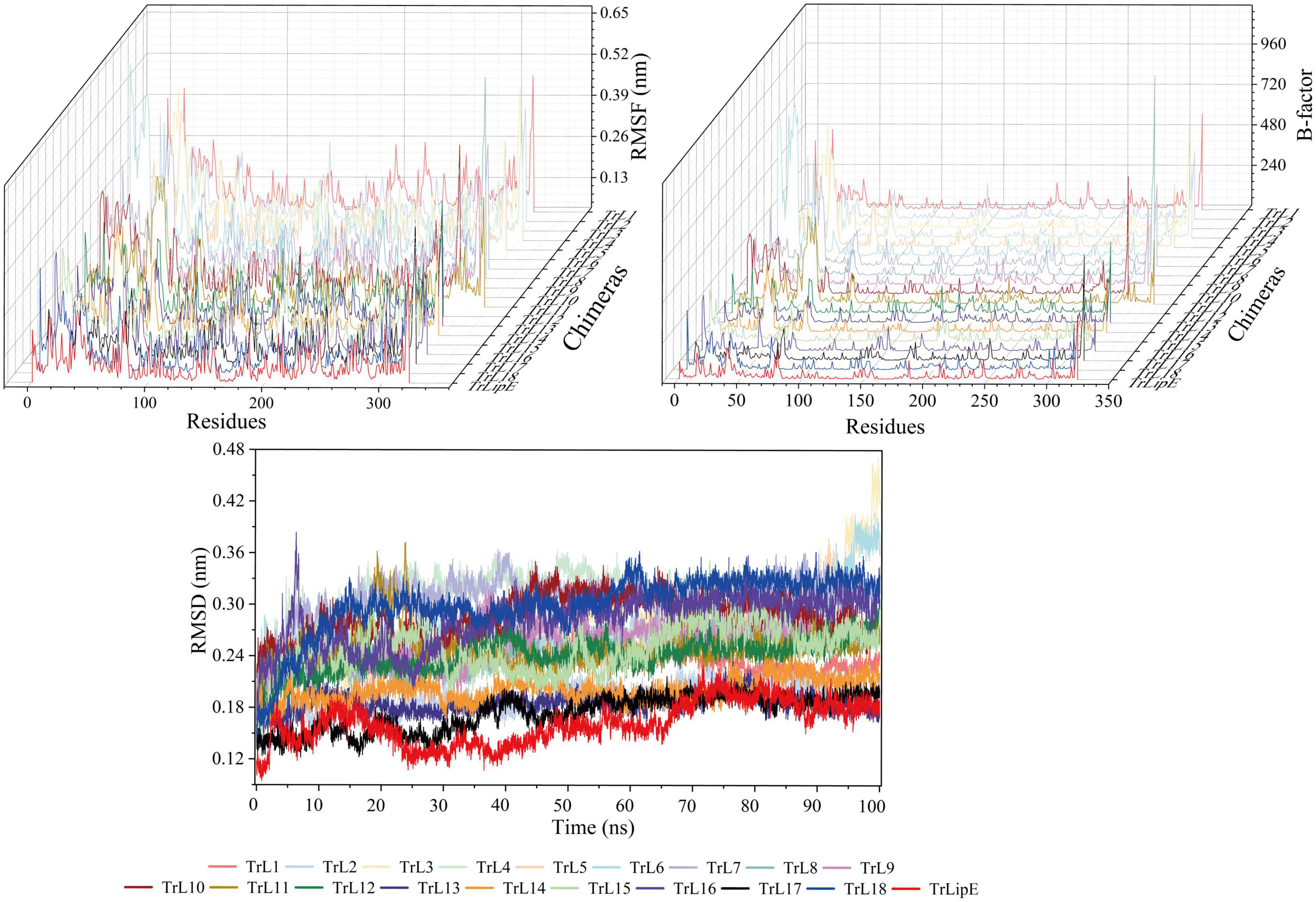
Figure 2. The MD simulation result of TrLipE and chimeras. MD simulations were performed at 300 K, and the chimeras had higher RMSD, RMSF, and B-factor values than TrLipE.
Compared to the thermostability of chimeras, the optimum pH did not change significantly. Figure 3 illustrates the optimal pH of the chimeras and TrLipE. The results showed that lid swapping had a weak effect on the pH of the chimeras. The optimal pH for the chimeras was 8–8.5, and this value was slightly lower than the pH of 8.5–9 for TrLipE, which was similar to that of most alkaline lipases (Melani et al., 2019).
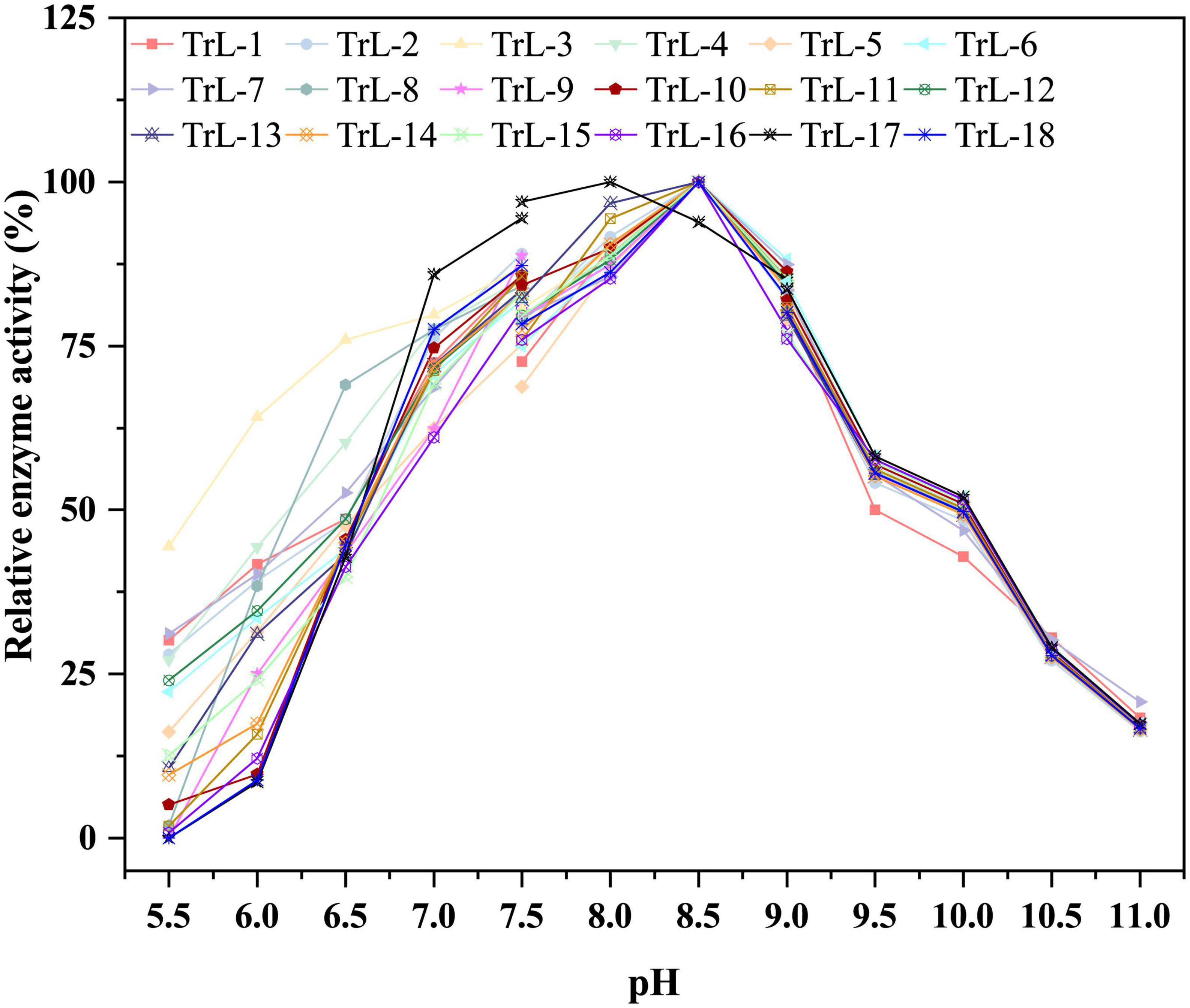
Figure 3. The optimal pH of chimeras. The optimal pH for the chimeras was 8–8.5, and this value was slightly lower than the pH of 8.5–9 for TrLipE.
To test the enzymatic activity, 4-nitrophenol laurate was used as a substrate. Figure 4A shows the specific enzymatic activities of the chimeras. All chimeras were able to hydrolyze the substrate 4-nitrobenzene laurate, but their activity changed. Chimeras TrL2, TrL5, TrL7, TrL8, TrL9, TrL11, TrL13, TrL14, and TrL17 were increased, with TrL17 and TrL8 representing the best performing chimeras and showing an increase in specific enzyme activity by approximately 20 ± 3.043% and 47 ± 2.361%, respectively. However, the other chimeras showed an opposite trend compared to TrLipE, which might be caused by the different lid structure on the binding ability of substrate 4-nitrophenol laurate. In addition, regarding the substrate specificity of chimeras, TrL2, TrL3, TrL17, and TrL18 could specifically recognize and hydrolyze 4-nitrophenyl benzoate, whereas TrLipE and the other chimeras could not. TrL17 had a specific enzyme activity of 0.5 ± 0.0247 μmol⋅mg–1⋅min–1 p-nitrophenol when 4-nitrophenyl benzoate was used as the substrate at a concentration of 150 μmol⋅L–1, and this activity was higher than that of TrL2, TrL3, and TrL18, as shown in Figure 4B, the specific enzymatic activity of chimera TrL2, TrL3 and TrL18 was only 19.68 ± 0.72%, 18.98 ± 0.70% and 16.41 ± 0.68% of TrL17, respectively. Then, we also used 4-nitrophenyl benzoate analogies (4-nitrophenyl anthranilate and 4-nitrophenyl salicylate) as substrates and found that all chimeras did not hydrolyze these two substrates, which was similar to the findings for TrLipE. As reported by Brocca et al. (2003), it could be concluded that the role of the lid in affecting the enzyme activity and specificity of chimeras might be complex, which has also been demonstrated previously (Akoh et al., 2004; Skjøt et al., 2009; Vagstad et al., 2012).
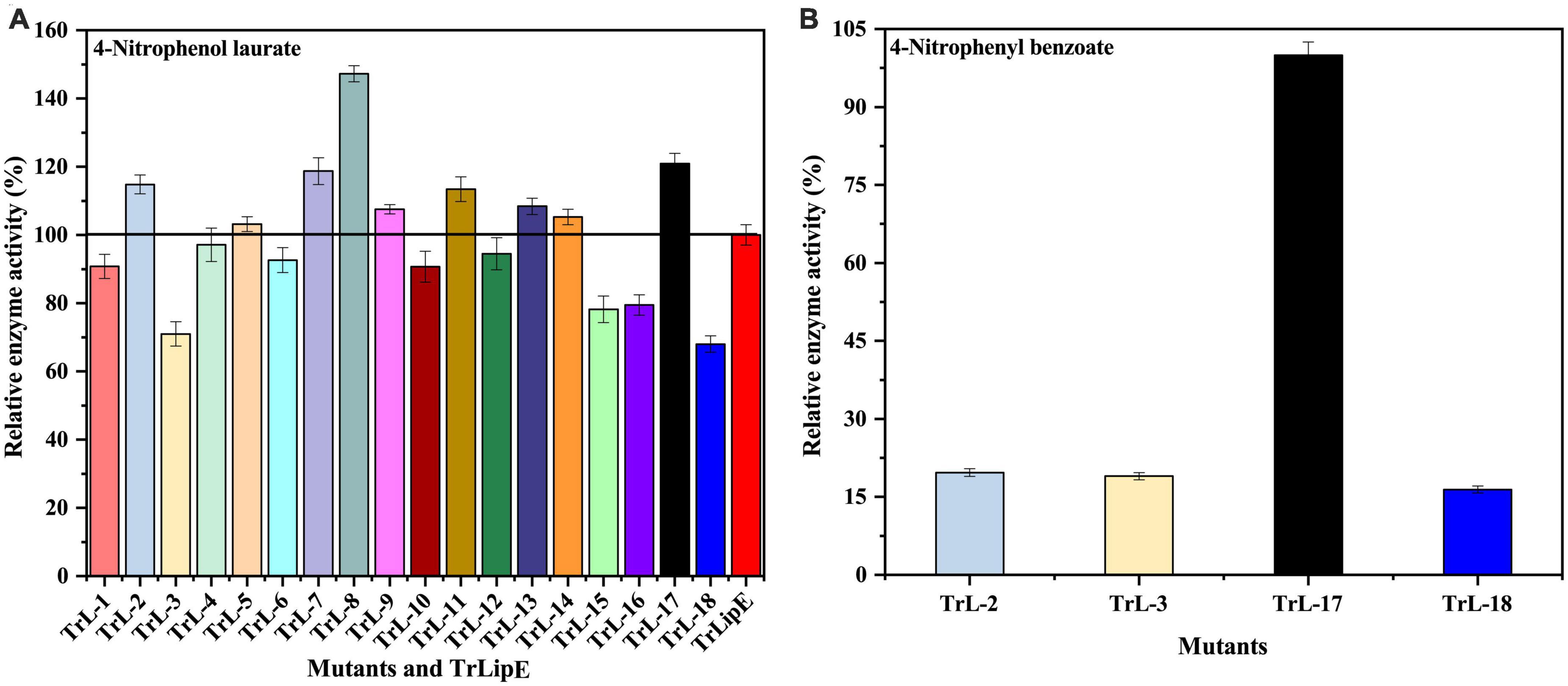
Figure 4. The relative enzyme activity of TrLipE and chimeras. (A) The relative enzyme activity of TrLipE and chimeras for 4-nitrophenol laurate; (B) The relative enzyme activity of chimeras TrL2, TrL3, TrL,17, and TrL18 for 4-nitrophenyl benzoate.
To analyze the changes in the affinity of the chimeras for different substrates, p-NP acetate (C2), p-NP butyrate (C4), p-NP hexanoate (C6), p-NP caprylate (C8), p-NP decanoate (C10), p-NP laurate (C12), p-NP palmitate (C16), and p-NP benzoate were used to determine the kinetic parameters of the chimeras and TrLipE. The values of Km, kcat, and kcat/Km for the chimeras and TrLipE are listed in Tables 2–4, respectively, and the fitted curves are shown in Supplementary Figure 2. The results indicated that the 18 chimeras had different affinities and catalytic abilities for different substrates and showed characteristics similar to those of TrLipE; that is, they showed better affinity for substrates with longer carbon chains. Among the tested substrates, 15 chimeras (except TrL7, TrL13, and TrL16) showed decreased activity for 4-nitrophenol palmitate and had a minimum Km value in the range of 7.46–77.35 μmol⋅L–1, which was lower than the 553.19–861.37 μmol⋅L–1 for 4-nitrophenol acetate. However, despite the good affinity for 4-nitrophenol palmitate, the chimeras TrL1, TrL5, TrL6, TrL8, TrL10, and TrL11 had lower kcat and kcat/Km values (Tables 3, 4) compared to that obtained for the other substrates and chimeras. This may be because high affinity slows the release of the product after lipase hydrolysis. Regarding the other substrates, the chimeras exhibited obvious differences in Km, kcat, and kcat/Km values, suggesting that the lid played an important role in the specificity or selectivity of the lipase (Brocca et al., 2003).
An interesting result observed for most of the chimeras and TrLipE was the affinity for p-nitrophenyl butyrate (C4). Although the chimeras had a lower Km value and higher kcat and kcat/Km values (Tables 3, 4) for 4-nitrophenol butyrate compared to TrLipE, most chimeras still possessed a poor affinity for this substrate compared to that for 4-nitrophenyl acetate. However, some chimeras (TrL5, TrL6, TrL7, TrL8, TrL9, TrL12, and TrL16) exhibited better affinity for 4-nitrophenol butyrate. This further confirmed that the effect of the lid on lipase specificity is complex.
With respect to substrate specificity, chimeras TrL2, TrL3, TrL17, and TrL18 could specifically hydrolyze 4-nitrophenyl benzoate while TrLipE and other chimeras could not. Although TrL17 had a higher Km value of 60.24 μmol⋅L–1 compared to the other three chimeras (50.03, 36.99, and 44.86 μmol⋅L–1, respectively), TrL17 exhibited higher activity and catalytic capability, with kcat and kcat/Km values of 21.92 min–1 and 363.88 L⋅min–1⋅mmol–1, respectively, which were higher than those of the chimeras TrL2, TrL3, and TrL18. As confirmed by the results of the kinetic study, the lid is involved in the substrate specificity of lipase, which has also been demonstrated by other studies (Vanleeuw et al., 2019).
The binding pocket is an important target for modifying the catalytic activity of enzymes (Ma et al., 2022). Thus, to improve the accessibility and catalytic efficiency of TrL17 for 4-nitrophenyl benzoate, molecular docking simulations were performed using Schrödinger software, which is a powerful simulation tool for evaluating protein–ligand interactions (Anuar et al., 2021). From Supplementary Figure 3, the serine of the active center acts as a nucleophile on the ester bond, as previously reported (Cen et al., 2019), and G84 and G85 participate in the formation of oxygen holes. Furthermore, other interactions, such as C-H bonds, pi-alkyl, and Pi-Pi bonds, were formed between ligands and other amino acids involved in docking. Therefore, based on the results of docking, the “Build and Edit Protein” protocol of DS was applied to perform the mutation and binding energy calculations. Then, 72 single mutations were selected based on the lower binding energy.
Based on the above method of kinetic parameters determination, enzymatic reactions were determined in accordance with assays conditions described above and carried out with 4-nitrophenyl benzoate as the substrate, and the Km, kcat, and catalytic efficiency (kcat/Km) values of the 72 single mutants are summarized in Supplementary Table 5. Most of the mutants reduced the catalytic activity of 4-nitrophenyl benzoate, for example, the catalytic efficiency of the mutant G86L (15.509 L⋅mmol–1⋅min–1) was reduced by 20-fold compared to the wild-type TrL17 (331.39 L⋅mmol–1⋅min–1), and 22 mutants lost their activity directly. In these mutants, such as A158, A156, A254, L256, F257, F284, and G286, residues were mainly distributed around catalytic triplets (S157, D255, and H285). In addition, the mutants of G84 and G85, which are involved in the formation of catalytic tetrahedral structures, also lost their activity. These results suggest that residues in the conserved region play a critical role in enzyme activity (Nezhad et al., 2023).
The heat map shown in Figure 5A demonstrates the ratio of kcat/Km values between single mutants (except for those showing a loss of enzyme activity) and TrL17. The figure clearly shows that the kcat/Km value of 10 mutants at six residue sites were 1.28- to 2.23-fold higher than that of the wild-type TrL17, with mutant M89W (2.23-fold higher) having a highest kcat/Km value of 601.42 L⋅mmol–1⋅min–1 and showing the greatest improvement compared to the other mutations. Compared with that of TrL17, the tryptophan of mutant M89W formed a Pi-Pi interaction with the substrate and the binding energy changed from 0 to −1.94 kcal⋅mol–1, which led to an increase in binding affinity and thus an increase in enzyme activity. The binding energy plays a pivotal role in Km and kcat; therefore, reducing the binding energy of the protein and ligand complex has a positive effect on improving the efficiency of enzyme catalysis (Dutta Banik et al., 2016; Wang et al., 2020). In our study, we also found that reducing the binding energy could improve enzyme activity, although this does not mean that the binding energy could be reduced indefinitely. The results showed that the catalytic efficiency of the mutant increased and then decreased with a decrease in the binding energy when the mutation was carried out at the same site (such as L21). The reason for this phenomenon may be that after the enzyme reaction, the product could not be released quickly owing to the low binding energy. In addition, we did not identify a specific relationship between the binding energy and catalytic efficiency of the mutants from different sites.
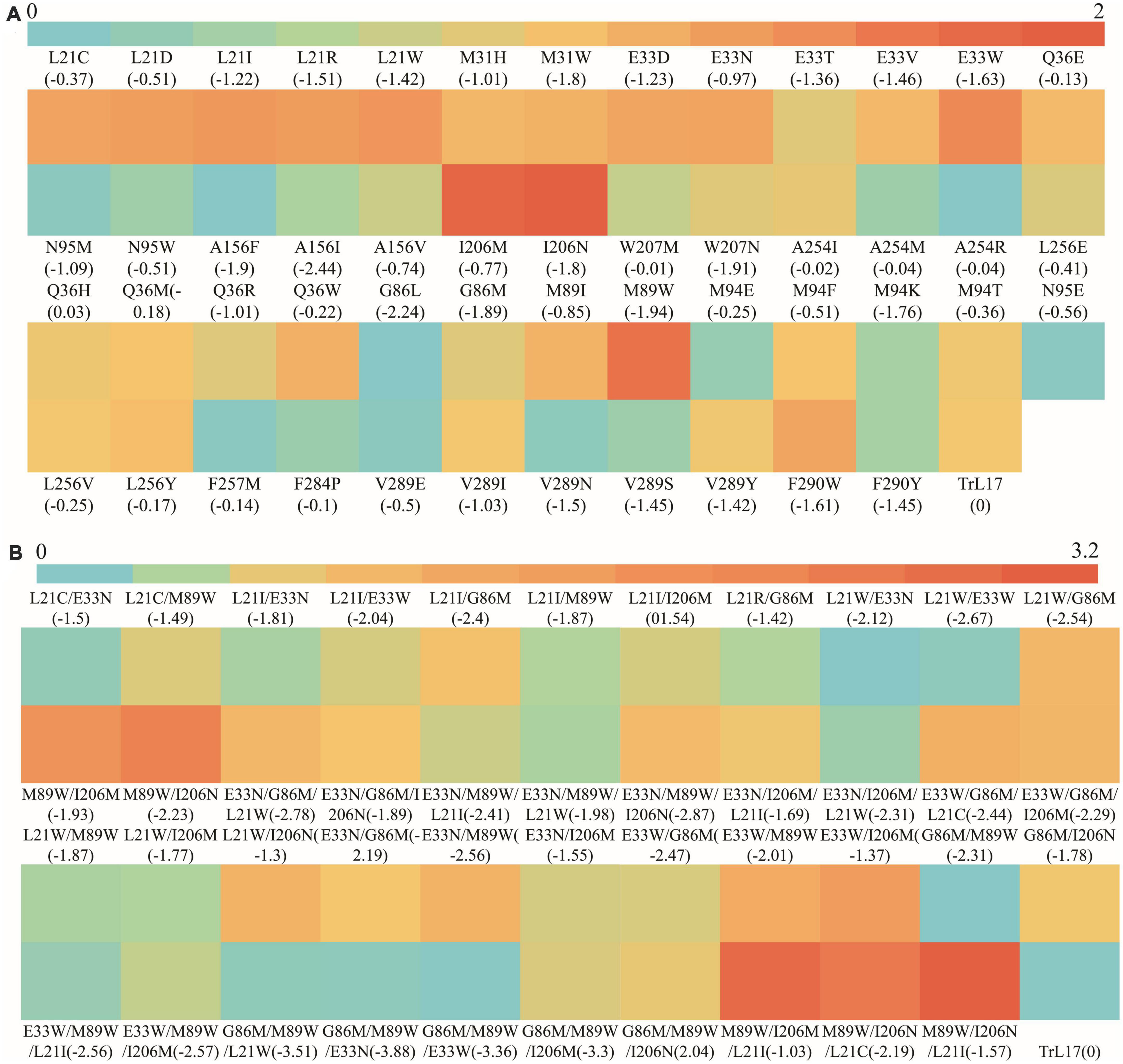
Figure 5. The heat map of the ratio of kcat/Km value between TrL17 and mutants. (A) The ratio of kcat/Km values between single mutants and TrL17; (B) The ratio of kcat/Km values between double, triple mutants, and TrL17.
To further improve the catalytic efficiency of TrL17 on 4-nitrophenyl benzoate, double and triple substitution variants were designed according to the results of the single substitution. After enzymatic reaction, the Km, kcat, and catalytic efficiency (kcat/Km) values of the 43 mutants (double and triple substitutions) are calculated and shown in Supplementary Table 6. Figure 5B displays the heat map of the ratio of kcat/Km values between the double and triple mutants with TrL17. The results clearly show that increases in substitution sites further reduced the binding ability. For example, the double mutation L21W/E33W (365.98 L⋅mmol–1⋅min–1) and triple mutation E33N/G86M/M89W (365.64 L⋅mmol–1⋅min–1) had the lowest binding energies of −2.67 kcal⋅mol–1 and −3.88 kcal⋅mol–1, respectively; moreover, their catalytic performance on 4-nitrophenyl benzoate was not the best. Among the double mutants, M89W/I206M (833.58 L⋅mmol–1⋅min–1) and M89W/I206N (921.8 L⋅mmol–1⋅min–1) showed excellent performance, and their catalytic efficiencies were 2.51- and 2.78-fold higher than those of TrL17, respectively, and showed corresponding increases compared to those of the single mutation. Among the triple mutations, M89W/I206M/L21I (1019.93 L⋅mmol–1⋅min–1) and M89W/I206N/L21I (1058.89 L⋅mmol–1⋅min–1) were the best, and their catalytic efficiencies were up to 3.08- and 3.20-fold higher than those of TrL17, respectively. In addition, the catalytic efficiency of the other double and triple mutants was also improved compared to that of TrL17. Our study shows that it is feasible to improve the catalytic efficiency of enzymes by reducing the binding energy of protein–ligand complexes.
In this study, 18 chimeras were successfully constructed by lid swapping. Most of the chimeras showed higher expression levels than the wild TrLipE. Meanwhile, the MD simulation results showed that the chimeras had greater flexibility and relatively lower optimal temperatures, such as 70°C for TrL17 and 60°C for the other chimeras, compared with TrLipE, and they also had a poorer half-life than TrL17. Moreover, compared to other reported thermostable lipases, the thermostability of these 18 chimeras was excellent. In addition, they could still withstand a wide pH range, which indicates that they still have great potential for future applications in extreme conditions, such as the detergent industry and biodiesel industry. Among the tested substrates, the specific enzyme activity and affinity of the partial chimeras were higher than those of the wild TrLipE, due to the change of substrate binding ability of chimeras after lid swapping. The chimeras TrL2, TrL3, TrL17, and TrL18 had better characteristics, and they could specifically hydrolyze 4-nitrophenyl benzoate, which cannot be hydrolyzed by TrLipE, with TrL17 showing the best ability to hydrolyze 4-nitrophenyl benzoate. After changing the substrate specificity of the chimera by lid swapping, we further optimized the catalytic center of the chimera by relying on the strategy of reducing the enzyme-substrate binding energy, and finally the catalytic efficiency of chimera TrL17 mutation was up to 3.20-fold faster than that of wild TrL17, which provided certain support for the further application of the chimera TrL17, and also provided valuable insights into the rational design of other lipases.
The original contributions presented in this study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
YF: investigation, formal analysis, and writing—original draft. FL and TY: methodology, validation, and investigation. YS: writing—review and editing. YX: methodology and software. ZG: data curation and visualization. GS: supervision and methodology. LZ: funding acquisition, project administration, and methodology. All authors contributed to the article and approved the submitted version.
This work was supported by the National Key Research and Development Program of China (2018YFA0900300 and 2021YFC2100300).
We greatly acknowledge Prof. Zhihu Zhao from Beijing Institute of Biotechnology (AMMS) for providing the pTIG vector.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1193955/full#supplementary-material
Akoh, C. C., Lee, G. C., and Shaw, J. F. (2004). Protein engineering and applications of Candida rugosa lipase isoforms. Lipids 39, 513–526. doi: 10.1007/s11745-004-1258-7
Almeida, F., Travália, B. M., Gonalves, I. S., and Forte, M. B. S. (2021). Biodiesel production by lipase-catalyzed reactions: Bibliometric analysis and study of trends. Biofuel. Bioprod. Biorefin. 15, 1141–1159.
Alvarez, Y., Esteban-Torres, M., Cortés-Cabrera, A., Gago, F., Acebrón, I., Benavente, R., et al. (2014). Esterase LpEst1 from Lactobacillus plantarum: A novel and atypical member of the αβ hydrolase superfamily of enzymes. PLoS One 9:e92257. doi: 10.1371/journal.pone.0092257
Anobom, C. D., Pinheiro, A. S., De-Andrade, R. A., Aguieiras, E. C., Andrade, G. C., Moura, M. V., et al. (2014). From structure to catalysis: Recent developments in the biotechnological applications of lipases. Biomed Res. Int. 2014:684506.
Anuar, N. F. S. K., Wahab, R. A., Huyop, F., Amran, S. I., Hamid, A. A. A., Halim, K. B. A., et al. (2021). Molecular docking and molecular dynamics simulations of a mutant Acinetobacter haemolyticus alkaline-stable lipase against tributyrin. J. Biomol. Struct. Dyn. 39, 2079–2091. doi: 10.1080/07391102.2020.1743364
Asmat, S., Anwer, A. H., and Husain, Q. (2019). Immobilization of lipase onto novel constructed polydopamine grafted multiwalled carbon nanotube impregnated with magnetic cobalt and its application in synthesis of fruit flavours. Int. J. Biol. Macromol. 140, 484–495. doi: 10.1016/j.ijbiomac.2019.08.086
Bai, Q. Y., Cai, Y. Z., Yang, L. F., Wu, Q. Q., Su, M., and Tang, L. H. (2022). Substitution of L133 with methionine in GXSXG domain significantly changed the activity of Penicillium expansum lipase. Catal. Lett. 152, 2047–2055.
Boyko, K. M., Kryukova, M. V., Petrovskaya, L. E., Kryukova, E. A., Nikolaeva, A. Y., Korzhenevsky, D. A., et al. (2021). Structural and biochemical characterization of a cold-Active PMGL3 Esterase with unusual oligomeric structure. Biomolecules 11:57. doi: 10.3390/biom11010057
Brocca, S., Secundo, F., Ossola, M., Alberghina, L., Carrea, G., and Lotti, M. (2003). Sequence of the lid affects activity and specificity of Candida rugosa lipase isoenzymes. Protein Sci. 12, 2312–2319. doi: 10.1110/ps.0304003
Cao, L. C., Chen, R., Xie, W., and Liu, Y. H. (2015). Enhancing the thermostability of feruloyl esterase EstF27 by directed evolution and the underlying structural basis. J. Agric. Food Chem. 63, 8225–8233. doi: 10.1021/acs.jafc.5b03424
Caputi, L., Franke, J., Bussey, K., Farrow, S. C., Vieira, I. J. C., Stevenson, C. E. M., et al. (2020). Structural basis of cycloaddition in biosynthesis of iboga and aspidosperma alkaloids. Nat. Chem. Biol. 16, 383–386. doi: 10.1038/s41589-019-0460-x
Casas-Godoy, L., Gasteazoro, F., Duquesne, S., Bordes, F., Marty, A., and Sandoval, G. (2018). Lipases: An overview. Methods Mol. Biol. 1835, 3–38.
Cavalcante, F., Neto, F. S., Falco, I., Souza, J., and Santos, J. (2020). Opportunities for improving biodiesel production via lipase catalysis. Fuel 288:119577.
Cavazzini, D., Grossi, G., Levati, E., Vallese, F., Montanini, B., Bolchi, A., et al. (2017). A family of archaea-like carboxylesterases preferentially expressed in the symbiotic phase of the mychorrizal fungus Tuber melanosporum. Sci. Rep. 7:7628. doi: 10.1038/s41598-017-08007-9
Cen, Y. X., Singh, W., Arkin, M., Moody, T. S., Huang, M. L., Zhou, J. H., et al. (2019). Artificial cysteine-lipases with high activity and altered catalytic mechanism created by laboratory evolution. Nat. Commun. 10:3198. doi: 10.1038/s41467-019-11155-3
Cipolatti, E. P., Valério, A., Henriques, R. O., Pinto, M., and Pessela, B. C. (2020). Production of new nanobiocatalysts via immobilization of lipase B from C. antarctica on polyurethane nanosupports for application on food and pharmaceutical industries. Int. J. Biol. Macromol. 165, 2957–2963. doi: 10.1016/j.ijbiomac.2020.10.179
Cui, R., Che, X., Li, L., Sun-Waterhouse, D., Wang, J., and Wang, Y. (2022). Engineered lipase from Janibacter sp. with high thermal stability to efficiently produce long-medium-long triacylglycerols. LWT 165:113675.
De Santi, C., Leiros, H. K., Di Scala, A., de Pascale, D., Altermark, B., and Willassen, N. P. (2016). Biochemical characterization and structural analysis of a new cold-active and salt-tolerant esterase from the marine bacterium Thalassospira sp. Extremophiles 20, 323–336. doi: 10.1007/s00792-016-0824-z
De Simone, G., Galdiero, S., Manco, G., Lang, D., Rossi, M., and Pedone, C. (2000). A snapshot of a transition state analogue of a novel thermophilic esterase belonging to the subfamily of mammalian hormone-sensitive lipase. J. Mol. Biol. 303, 761–771. doi: 10.1006/jmbi.2000.4195
Denison, M., Steinke, S. J., Majeed, A., Turro, C., Kocarek, T. A., Sevrioukova, I. F., et al. (2022). Ir(III)-based agents for monitoring the cytochrome P450 3A4 active site occupancy. Inorg. Chem. 61, 13673–13677.
Dutta Banik, S., Nordblad, M., Woodley, J. M., and Peters, G. H. (2016). A correlation between the activity of Candida Antarctica lipase B and differences in binding free energies of organic solvent and substrate. ACS Catal. 6, 6350–6361.
Fang, Y. K., Fan Liu, F., Shi, Y., Ting Yang, T., Yu Xin, Y., Zhenghua Gu, Z. H., et al. (2022). Hotspots and mechanisms of action of the thermostable framework of a microbial thermolipase. ACS Synth. Biol. 11, 3460–3470. doi: 10.1021/acssynbio.2c00360
Fang, Y. K., Zhou, Y. J., Xin, Y., Shi, Y., Guo, Z. T., Li, Y. R., et al. (2021). Preparation and characterization of a novel thermostable lipase from Thermomicrobium roseum. Catal. Sci. Technol. 11, 7386–7397.
Gao, B., Jiang, S. Q., Wang, L. Z., Zhang, L. J., and Wei, D. Z. (2018). Energy and conformation determine the enantioselectivity of enzyme. Biochem. Eng. J. 129, 106–112.
Godara, P., Naik, B., Meghwal, R., Ojha, R., Srivastava, V., Prajapati, V. K., et al. (2022). Rational designing of peptide-ligand conjugates-based immunotherapy for the treatment of complicated malaria. Life Sci. 311:121121. doi: 10.1016/j.lfs.2022.121121
Gutiérrez-Domínguez, D. E., Chí-Manzanero, B., Rodríguez-Argüello, M. M., Todd, J. N. A., Islas-Flores, I., Canseco-Pérez, M. Á, et al. (2022). Identification of a novel lipase with AHSMG pentapeptide in Hypocreales and Glomerellales filamentous fungi. Int. J. Mol. Sci. 23:9367. doi: 10.3390/ijms23169367
Huang, J., Huo, Y. Y., Ji, R., Kuang, S., Ji, C., Xu, X. W., et al. (2016). Structural insights of a hormone sensitive lipase homologue Est22. Sci. Rep. 6:28550. doi: 10.1038/srep28550
Huang, W. Q., Lan, D. M., Popowicz, G. M., Zak, K. M., Zhao, Z. X., Yuan, H., et al. (2019). Structure and characterization of Aspergillus fumigatus lipase B with a unique, oversized regulatory subdomain. FEBS J. 286, 2366–2380. doi: 10.1111/febs.14814
Jan, A. H., Dubreucq, É, and Subileau, M. (2017). Revealing the roles of subdomains in the catalytic behavior of lipases/acyltransferases homologous to CpLIP2 through rational design of chimeric enzymes. Chembiochem 18, 941–950. doi: 10.1002/cbic.201600672
Kay, E. J., Yates, L. E., Terra, V. S., Cuccui, J., and Wren, B. W. (2016). Recombinant expression of Streptococcus pneumoniae capsular polysaccharides in Escherichia coli. Open Biol. 6:150243. doi: 10.1098/rsob.150243
Khan, F. I., Lan, D. M., Durrani, R., Huan, W. Q., Zhao, Z. X., and Wang, Y. H. (2017). The lid domain in lipases: Structural and functional determinant of enzymatic properties. Front. Bioeng. Biotechnol. 5:16. doi: 10.3389/fbioe.2017.00016
Kim, S. H., Kang, P. A., Han, K., Lee, S. W., and Rhee, S. (2019). Crystal structure of chloramphenicol-metabolizing enzyme EstDL136 from a metagenome. PLoS One 14:e0210298. doi: 10.1371/journal.pone.0210298
Kumari, A., and Gupta, R. (2013). Phenylalanine to leucine point mutation in oxyanion hole improved catalytic efficiency of Lip12 from Yarrowia lipolytica. Enzyme Microb. Technol. 53, 386–390. doi: 10.1016/j.enzmictec.2013.08.004
Li, X. J., Li, Q., Zhan, X. X., Zhang, Y. J., Xiong, G. L., and Zheng, J. Y. (2022). Expression and characterization of a thermostable lipase from Thermomyces dupontii. Chem. Papers 76, 2811–2821.
Liu, Y. Q., Xu, W. Z., and Xu, W. (2022). A review on lipase-catalyzed synthesis of geranyl esters as flavor additives for food, pharmaceutical and cosmetic applications. Biotechnol. Adv. 60:108002.
Ma, D. L., Xin, Y., Guo, Z. T., Shi, Y., Zhang, L., Li, Y. R., et al. (2022). Ancestral sequence reconstruction and spatial structure analysis guided alteration of longer-chain substrate catalysis for Thermomicrobium roseum lipase. Enzyme Microb. Technol. 156:109989. doi: 10.1016/j.enzmictec.2022.109989
Maiangwa, J., Ali, M. S. M., Salleh, A. B., Abd Rahman, R. N. Z. R., Normi, Y. M., Shariff, F. M., et al. (2017). Lid opening and conformational stability of T1 Lipase is mediated by increasing chain length polar solvents. PeerJ 5:e3341. doi: 10.7717/peerj.3341
Maier, J. A., Martinez, C., Kasavajhala, K., Wickstrom, L., Hauser, K. E., and Simmerling, C. (2015). ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 11, 3696–3713.
McKary, M. G., Abendroth, J., Edwards, T. E., and Johnson, R. J. (2015). Structural basis for the strict substrate selectivity of the mycobacterial hydrolase LipW. Biochemistry 55, 7099–7111. doi: 10.1021/acs.biochem.6b01057
Melani, N. B., Tambourgi, E. B., and Silveira, E. (2019). Lipases: From production to applications. Sep. Purif. Rev. 49, 143–158.
Mohd Din, M. H., Nair, A., Masomian, M., Mohamad Ali, M. S., and Raja Abd. Rahman, R. N. Z. (2021). Heterologous expression and characterization of plant lipase LIP2 from Elaeis guineensis Jacq. oil palm mesocarp in Escherichia coli. Catalysts 11:244.
Nam, K. H., Kim, S. J., Priyadarshi, A., Kim, H. S., and Hwang, K. Y. (2009). The crystal structure of an HSL-homolog EstE5 complex with PMSF reveals a unique configuration that inhibits the nucleophile Ser144 in catalytic triads. Biochem. Biophys. Res. Commun. 389, 247–250. doi: 10.1016/j.bbrc.2009.08.123
Nam, K. H., Park, S. H., Lee, W. H., and Hwang, K. Y. (2010). Biochemical and structural analysis of hormone-sensitive lipase homolog EstE7: Insight into the stabilized dimerization of HSL-homolog proteins. Bull. Korean Chem. Soc. 31, 2627–2632.
Nezhad, N. G., Rahman, R. N. Z. R. A., Yahaya, N. M., Oslan, S. N., Shariff, F. M., and Leow, T. C. (2023). Recent advances in simultaneous thermostability-activity improvement of industrial enzymes through structure modification. Int. J. Biol. Macromol. 232:123440. doi: 10.1016/j.ijbiomac.2023.123440
Noby, N., Auhim, H. S., Winter, S., Worthy, H. L., Embaby, A. M., Saeed, H., et al. (2021). Structure and in silico simulations of a cold-active esterase reveals its prime cold-adaptation mechanism. Open Biol. 11:210182. doi: 10.1098/rsob.210182
Ohara, K., Unno, H., Oshima, Y., Hosoya, M., Fujino, N., Hirooka, K., et al. (2014). Structural insights into the low pH adaptation of a unique carboxylesterase from Ferroplasma: Altering the pH optima of two carboxylesterases. J. Biol. Chem. 289, 24499–24510. doi: 10.1074/jbc.M113.521856
Pereira, M. R., Maester, T. C., Mercaldi, G. F., de Macedo Lemos, E. G., Hyvönen, M., and Balan, A. (2017). From a metagenomic source to a high-resolution structure of a novel alkaline esterase. Appl. Microbiol. Biotechnol. 101, 4935–4949. doi: 10.1007/s00253-017-8226-4
Sarmah, N., Revathi, G., Sheelu, G., Rani, Y. K., Sridhar, S., Mehtab, V., et al. (2017). Recent advances on sources and industrial applications of lipases. Biotechnol. Prog. 34, 5–28.
Sha, C., Yu, X. W., Lin, N. X., Zhang, M., and Xu, Y. (2013). Enhancement of lipase r27RCL production in Pichia pastoris by regulating gene dosage and co-expression with chaperone protein disulfide isomerase. Enzyme Microb. Technol. 53, 438–443. doi: 10.1016/j.enzmictec.2013.09.009
Skjøt, M., De Maria, L., Chatterjee, R., Svendsen, A., Patkar, S. A., Østergaard, P. R., et al. (2009). Understanding the plasticity of the α/β hydrolase fold: Lid swapping on the Candida antarctica lipase B results in chimeras with interesting biocatalytic properties. ChemBioChem 10, 520–527.
Soni, S., Sathe, S. S., Sheth, R. R., Tiwari, P., Vadgama, R. K. N., Odaneth, A. A., et al. (2019). N-terminal domain replacement changes an archaeal monoacylglycerol lipase into a triacylglycerol lipase. Biotechnol. Biofuel. 12, 1–11. doi: 10.1186/s13068-019-1452-5
Stauch, B., Fisher, S. J., and Cianci, M. (2015). Open and closed states of Candida antarctica lipase B: Protonation and the mechanism of interfacial activation. J. Lipid Res. 56, 2348–2358. doi: 10.1194/jlr.M063388
Tambunan, U. S. F., Randy, A., and Parikesit, A. A. (2014). Design of Candida Antarctica lipase B thermostability improvement by introducing extra disulfide bond into the enzyme. OnLine J. Biol. Sci. 14, 108–118.
Tang, L., Su, M., Yan, J., Xie, S., and Zhang, W. (2015). Lid hinge region of Penicillium expansum lipase affects enzyme activity and interfacial activation. Process Biochem. 50, 1218–1223.
Vagstad, A. L., Hill, E. A., Labonte, J. W., and Townsend, C. A. (2012). Characterization of a fungal thioesterase having Claisen cyclase and deacetylase activities in melanin biosynthesis. Chem. Biol. 19, 1525–1534. doi: 10.1016/j.chembiol.2012.10.002
Vanleeuw, E., Winderickx, S., Thevissen, K., Lagrain, B., Dusselier, M., Cammue, B. P. A., et al. (2019). Substrate-specificity of Candida rugosa lipase and its industrial application. ACS Sustain. Chem. Eng. 7, 15828–15844.
Verma, S., Meghwanshi, G. K., and Kumar, R. (2021). Current perspectives for microbial lipases from extremophiles and metagenomics. Biochimie 182, 23–36. doi: 10.1016/j.biochi.2020.12.027
Vivek, K., Sandhia, G. S., and Subramaniyan, S. (2022). Extremophilic lipases for industrial applications: A general review. Biotechnol. Adv. 60:108002.
Wang, L., Marciello, M., Estévez-Gay, M., Soto Rodriguez, P. E. D., Luengo Morato, Y., Iglesias-Fernández, J., et al. (2020). Enzyme conformation influences the performance of lipase-powered nanomotors. Angew. Chem. Int. Ed. Engl. 59, 21080–21087. doi: 10.1002/anie.202008339
Wang, S., Xu, Y., and Yu, X. W. (2021). A phenylalanine dynamic switch controls the interfacial activation of Rhizopus chinensis lipase. Int. J. Biol. Macromol. 173, 1–12. doi: 10.1016/j.ijbiomac.2021.01.086
Xing, Y. N., Tan, J., Wang, Y., and Wang, J. (2021). Enhancing the thermostability of a mono- and diacylglycerol lipase from Malassizia globose by stabilizing a flexible loop in the catalytic pocket. Enzyme Microb. Technol. 149:109849. doi: 10.1016/j.enzmictec.2021.109849
Yang, W., Cao, H., Xu, L., Zhang, H., and Yan, Y. (2015). A novel eurythermic and thermostale lipase LipM from Pseudomonas moraviensis M9 and its application in the partial hydrolysis of algal oil. BMC Biotechnol. 15:94. doi: 10.1186/s12896-015-0214-0
Yang, W., He, Y., Xu, L., Zhang, H., and Yan, Y. (2016). A new extracellular thermo-solventstable lipase from Burkholderia ubonensis SL-4: Identification, characterization and application for biodiesel production. J. Mol. Catal. B Enzym. 126, 76–89.
Yu, X. W., Tan, N. J., Xiao, R., and Xu, Y. (2012). Engineering a disulfide bond in the lid hinge region of Rhizopus chinensis lipase: Increased thermostability and altered acyl chain length specificity. PLoS One 7:e46388. doi: 10.1371/journal.pone.0046388
Yu, X. W., Zhu, S. S., Xiao, R., and Xu, Y. (2014). Conversion of a Rhizopus chinensis lipase into an esterase by lid swapping. J. Lipid Res. 55, 1044–1051. doi: 10.1194/jlr.M043950
Zhang, J. H., Jiang, Y. Y., Lin, Y., Sun, Y. F., Zheng, S. P., and Han, S. Y. (2013). Structure-guided modification of Rhizomucor miehei lipase for production of structured lipids. PLoS One 8:e67892. doi: 10.1371/journal.pone.0067892
Keywords: binding energy, lid swapping, thermophilic lipase, molecular dynamics, substrate specificity
Citation: Fang Y, Liu F, Shi Y, Yang T, Xin Y, Gu Z, Shi G and Zhang L (2023) N-terminal lid swapping contributes to the substrate specificity and activity of thermophilic lipase TrLipE. Front. Microbiol. 14:1193955. doi: 10.3389/fmicb.2023.1193955
Received: 26 March 2023; Accepted: 12 June 2023;
Published: 26 June 2023.
Edited by:
Rajesh K. Sani, South Dakota School of Mines and Technology, United StatesReviewed by:
Muntazir Mushtaq, Shoolini University, IndiaCopyright © 2023 Fang, Liu, Shi, Yang, Xin, Gu, Shi and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liang Zhang, emhhbmdsQGppYW5nbmFuLmVkdS5jbg==; Yu Xin, eXV4aW5AamlhbmduYW4uZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.