- 1Department of Animal Science, University of California, Davis, Davis, CA, United States
- 2Western Institute for Food Safety and Security, University of California, Davis, Davis, CA, United States
- 3Department of Population Health and Reproduction, School of Veterinary Medicine, University of California, Davis, Davis, CA, United States
- 4California Department of Public Health, Richmond, CA, United States
Shrimp is one of the most consumed seafood products globally. Antimicrobial drugs play an integral role in disease mitigation in aquaculture settings, but their prevalent use raises public health concerns on the emergence and spread of antimicrobial resistant microorganisms. Vibrio spp., as the most common causative agents of seafood-borne infections in humans, and Enterococcus spp., as an indicator organism, are focal bacteria of interest for the monitoring of antimicrobial resistance (AMR) in seafood. In this study, 400 samples of retail shrimp were collected from randomly selected grocery stores in the Greater Sacramento, California, area between September 2019 and June 2020. The prevalence of Vibrio spp. and Enterococcus spp. was 60.25% (241/400) and 89.75% (359/400), respectively. Subsamples of Vibrio (n = 110) and Enterococcus (n = 110) isolates were subjected to antimicrobial susceptibility testing (AST). Vibrio isolates had high phenotypic resistance to ampicillin (52/110, 47.27%) and cefoxitin (39/110, 35.45%). Enterococcus were most frequently resistant to lincomycin (106/110, 96.36%), quinupristin-dalfopristin (96/110, 87.27%), ciprofloxacin (93/110, 84.55%), linezolid (86/110, 78.18%), and erythromycin (58/110, 52.73%). For both Vibrio and Enterococcus, no significant associations were observed between multidrug resistance (MDR, resistance to ≥3 drug classes) in isolates from farm raised and wild caught shrimp (p > 0.05) and in isolates of domestic and imported origin (p > 0.05). Whole genome sequencing (WGS) of a subset of Vibrio isolates (n = 42) speciated isolates as primarily V. metschnikovii (24/42; 57.14%) and V. parahaemolyticus (12/42; 28.57%), and detected 27 unique antimicrobial resistance genes (ARGs) across these isolates, most commonly qnrVC6 (19.05%, 8/42), dfrA31 (11.90%, 5/42), dfrA6 (9.5%, 4/42), qnrVC1 (9.5%, 4/42). Additionally, WGS predicted phenotypic resistance in Vibrio isolates with an overall sensitivity of 11.54% and specificity of 96.05%. This study provides insights on the prevalence and distribution of AMR in Vibrio spp. and Enterococcus spp. from retail shrimp in California which are important for food safety and public health and exemplifies the value of surveillance in monitoring the spread of AMR and its genetic determinants.
1. Introduction
Shrimp is the most popular seafood in the United States (U.S.) and is vaunted by the United States Department of Agriculture (USDA) as a healthy protein due to its nutrient density, lower unhealthy fat content relative to red meat and poultry, and lower levels of methylmercury compared to many other seafoods [U.S. Department of Agriculture and U.S. Department of Health and Human Services (USDA and USDHHS), 2020; National Fisheries Institute Media, 2022]. Annual per-capita consumption of shrimp in the U.S. approached five pounds in 2020, despite nearly 90% of Americans eating less than the recommended quantity of shrimp [U.S. Department of Agriculture and U.S. Department of Health and Human Services (USDA and USDHHS), 2020; National Marine Fisheries Service, 2022; Thomsen et al., 2022]. Still, the U.S. is one of the largest importers in the global shrimp industry which was valued at USD 24.7 billion in 2022 and is still growing at an accelerating rate [National Fisheries Institute Media, 2022; Food and Agriculture organization of the United Nations (FAO), 2022a].
Shrimp aquaculture, which already outproduces wild shrimp fisheries three times over, is the fastest growing animal food sector in the world [Kumar et al., 2016; Golder et al., 2022; Food and Agriculture Organization of the United Nations (FAO), 2022b]. The majority of shrimp production occurs in countries in Asia (primarily China, Thailand, Indonesia, and India) and South and Central America (especially Ecuador) [Food and Agriculture Organization of the United Nations (FAO), 2022b]. Farm raised shrimp are highly susceptible to infectious disease due to high stocking densities and their decreased capacity for adaptive immunity relative to vertebrates (Ali et al., 2018; Thornber et al., 2020; Lanz-Mendoza and Contreras-Garduño, 2022). Outbreaks can endanger entire harvests without quick and aggressive treatment. Traditional vaccination, which has gained traction in finfish aquaculture, is not an option for shrimp, while new DNA-based vaccination methods have limited and poorly understood efficacy (Gudding and Van Muiswinkel, 2013; Chang et al., 2018). Consequentially, bacterial infectious diseases in farmed shrimp are almost always treated with antimicrobial agents. Since the scale of the industry and the risks posed by those diseases are so great, substantial volumes of antimicrobial drugs are used in shrimp production. From previous reports, 2.7% of all global antimicrobial usage of any type is attributable to shrimp aquaculture (Schar et al., 2020; Thornber et al., 2020).
The most common form of antimicrobial use in shrimp farming is feed-mediated metaphylaxis after the detection of an infection (Thornber et al., 2020). Prophylactic use of antimicrobials in shrimp aquaculture was once commonplace and is an ongoing practice, especially in hatchery settings, but has generally declined through the 21st century (Holmström et al., 2003; Zhang et al., 2011; Smith, 2012; Thornber et al., 2020). While antimicrobials are the first line treatment against pathogens in food production and clinical contexts, their use and misuse can increase selective pressures that lead to antimicrobial resistance (AMR) in bacteria [Centers for Disease Control and Prevention (CDC), 2019a]. Shrimp-associated bacteria are thus at high risk of developing AMR, which reduces the ability to treat infections that compromise animal welfare, human health, and industry. Moreover, since antimicrobial resistance genes (ARGs) can spread via horizontal gene transfer, even non-pathogenic bacteria – or those that are pathogenic for different hosts – that develop resistance can spread resistance to bacteria of greater public health concern (Lulijwa et al., 2020). Monitoring the prevalence and patterns of AMR in shrimp is therefore critical to evaluate food safety and public health risks.
Recently, a study conducted through the National Antimicrobial Resistance Monitoring System (NARMS) detected Vibrio spp. and Enterococcus spp. as the most prevalent Gram-negative and Gram-positive bacteria in retail seafood samples, respectively, and highlighted them as good candidates for tracking AMR (Tate et al., 2022). Vibrio spp. are cosmopolitan and are normal flora in the coastal and estuarine habitats of wild shrimp, but certain species are also the most common seafood-borne pathogens in humans (Costa et al., 2015a; Stratev et al., 2023) and the main pathogens for shrimp (El-Far et al., 2015; Amatul-Samahah et al., 2020). Enterococcus spp. are primarily commensal bacteria in the environment and animal gut microbiomes, but are often employed as indicator organisms for the monitoring of antimicrobial resistance since they can readily acquire ARGs conferring resistance to a large diversity of antimicrobials and transfer them to other bacteria, including pathogens (Byappanahalli et al., 2012; Chajęcka-Wierzchowska et al., 2016; Çardak et al., 2022). The objective of this study was to evaluate the prevalence and antimicrobial resistance profiles of Vibrio spp. and Enterococcus spp. in retail shrimp from grocery stores in the greater Sacramento area in California.
2. Materials and methods
2.1. Sample collection
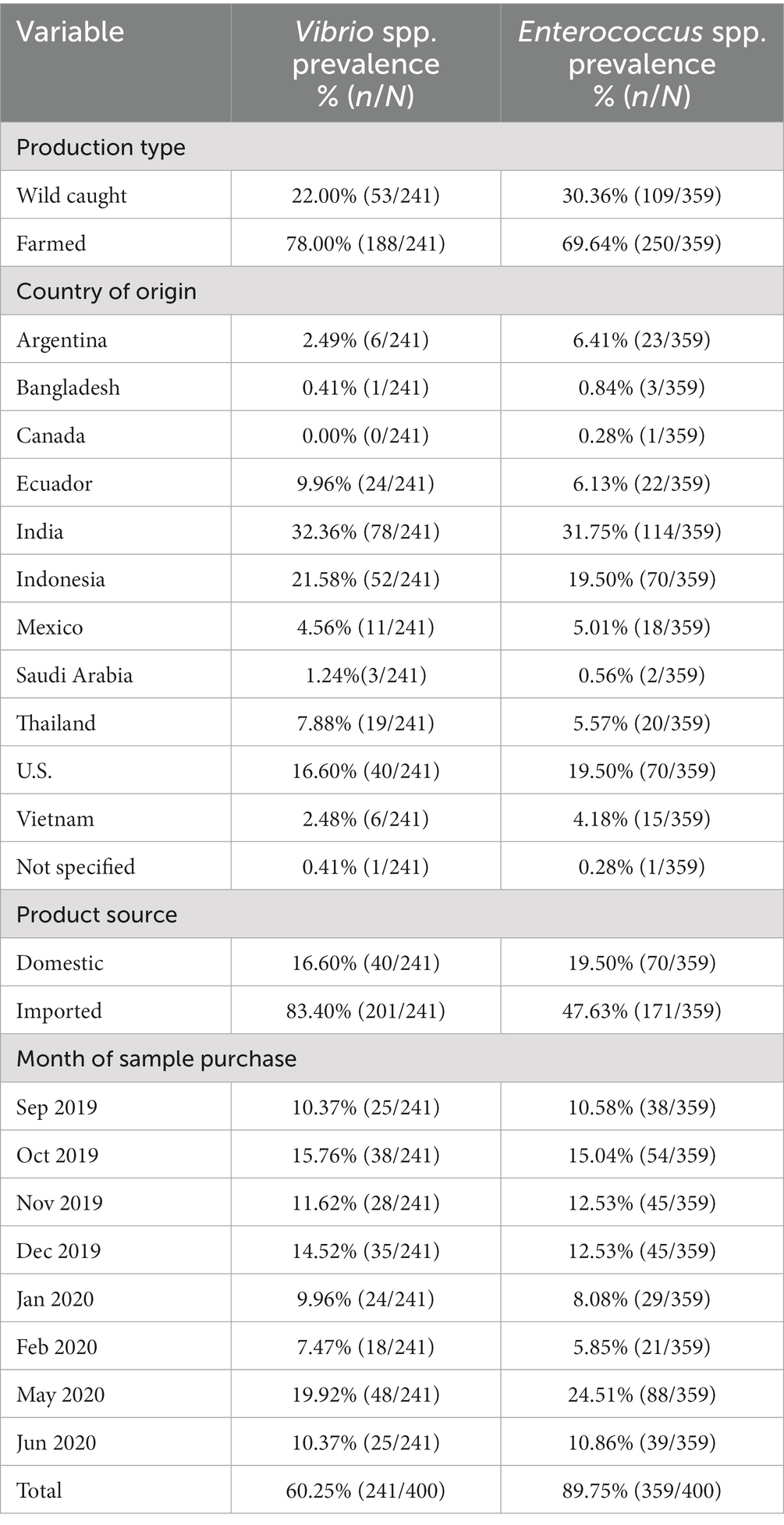
A total of 400 shrimp samples, either prepackaged or in half-pound packages from bulk seafood counters, were collected from grocery stores in the Greater Sacramento area in California over four seasonal periods between September 2019 and June 2020. A list of 100 grocery stores was randomly selected among those located in Sacramento zip codes according to Google Maps. During each sampling event, stores were randomly selected from this pool. Samples were randomly selected from chilled retail displays in the manner they were presented to consumers. Along with the samples, metadata including production type (farm raised or wild caught), country of origin, species and size of shrimp, store handling method, sold forms (fresh, frozen, or previously frozen), and time of collection were collected during sampling. Samples were kept on ice during transport, refrigerated upon receipt at the laboratory, and processed within 72 h of collection.
2.2. Sample processing, bacterial isolation, and confirmation
Samples were processed using the NARMS seafood pilot laboratory protocol (FDA, 2021). Briefly, two aliquots of 25 g from each shrimp sample were placed into two sterile stomacher bags, one containing 225 mL of alkaline peptone water (APW) and another with buffered peptone water (BPW). Samples were homogenized in a Neutec Masticator Paddle Blender (Neutec Group, Inc., Farmingdale, NY, United States) for 2 min at 230 RPM and incubated at 35°C for 24 ± 2 h. Subsequently, overnight APW and BPW enrichments were streaked onto thiosulfate-citrate-bile salts-sucrose (TCBS) (BD Difco, Detroit, MI, United States) and Enterococcosel (BD BBL, Franklin Lakes, NJ, United States) agars, respectively, and incubated at 35°C for 18–24 h for identification of Vibrio spp. and Enterococcus spp., respectively. One colony with positive colony morphology (yellow or green to blue-green colonies being characteristic of Vibrio, and beige colonies with strong black halos being characteristic of Enterococcus) was selected from each plate and streaked to purity on blood agar plates. Presumptive positives for Vibrio were confirmed to genus level by PCR using the forward primer: 5’-GGC GTA AAG CGC ATG CAG GT-3′; and the reverse primer: 5′-GAA ATT CTA CCC CCC TCT ACA G-3′, as previously described in Thompson et al. (2004). Enterococcus were confirmed with Gram-staining for identification of Gram-positive cocci and biochemical tests (catalase negative and PYR positive) using BD BBL DrySlide™ PYR kits and following methods previously described by Aryal (2016).
2.3. Antimicrobial susceptibility testing
Antimicrobial susceptibility testing (AST) was conducted on a subset of isolates comprised of 110 Vibrio (110/241) and 110 Enterococcus (110/359) isolates using the broth microdilution method with the NARMS Gram-negative (CMV3AGNF) and Gram-positive (CMV3AGPF) panels, respectively. Isolates were streaked onto selective agar plates (TCBS and Enterococcosel agar for Vibrio and Enterococcus, respectively) and incubated at 35°C for 18–24 h. A colony with typical morphology was then restreaked onto blood agar (Thermo Fisher Scientific, Waltham, MA, United States) and incubated at 35°C for 20–24 h. Pure colonies on fresh overnight blood agar plates were suspended in sterile demineralized water to an optical density (OD) between 0.08 and 0.10 as measured by a spectrophotometer (BioMate 3; ThermoSpectronic, Rochester, NY) at 625 nm. Aliquots of the suspension (20 uL for Vibrio and 10 uL for Enterococcus) were then transferred to 11 mL of cation-adjusted Mueller-Hinton broth (CAMHB; BD Difco, Detroit, MI, United States), and the mixture was vortexed for 5–10 s. Subsequently, 50 uL of the CAMHB mixture was transferred to each well of the AST plate. Additionally, a loopful (10 uL) of the CAMHB suspension was streaked onto a blood agar plate for quality control. AST plates and blood agar plates were then incubated at 35°C for 18–24 h. The minimum inhibitory concentration (MIC) was recorded as the lowest concentration of each drug with fully inhibited growth in the wells and per guidelines from Clinical and Laboratory Standards Institute (CLSI) methods [Clinical and Laboratory Standards Institute (CLSI), 2018].
Resulting MIC values were interpreted as susceptible, resistant, or intermediate based on CDC breakpoints for non-cholera Vibrio [Centers for Disease Control and Prevention (CDC), 2019a] and FDA NARMS breakpoints for Enterococcus [U.S. Food and Drug Administration (FDA), 2019], both of which are based on CLSI breakpoints [Clinical and Laboratory Standards Institute (CLSI), 2017]. Six of the fourteen drugs in the Gram-negative panel for Vibrio isolates (ceftiofur, ceftriaxone, chloramphenicol, nalidixic acid, streptomycin, and sulfisoxazole) have no CLSI or NARMS breakpoints, and were omitted from analysis. The composition of each drug panel and interpretive breakpoints for each drug are listed in Supplementary Table 1. Intermediate results were counted as resistant in the analysis. Multidrug resistance (MDR) was defined as resistance to three or more classes of antimicrobial drugs (Tate et al., 2022).
2.4. Whole genome sequencing (WGS) and identification of antimicrobial resistance genes (ARGs)
Whole genome sequencing was conducted on a subset of Vibrio isolates that exhibited phenotypic resistance (n = 42) at the Food and Drug Laboratory Branch of the California Department of Public Health. The isolates were streaked onto Trypticase Soy Agar with 0.6% Yeast Extract (TSA-YE) and 3% saline for recovery as well as CHROM Vibrio plates for confirmation. A single colony was restreaked on TSA-YE with 3% saline and incubated at 35°C for 18–24 h. Genomic DNA was extracted from bacteria using the DNeasy Blood and Tissue Kit (Qiagen) and quantified using a Qubit fluorometer (Thermo Fisher Scientific). DNA libraries were prepared with Illumina DNA Prep kits (Illumina Inc., San Diego, California). Whole genome sequencing was performed on the Illumina MiSeq DNA sequencing system using the MiSeq reagent kit version 2 (2 × 250-bp paired-end reads) per CDC PulseNet guidelines (PulseNet, n.d.). After the successful completion of the sequencing runs, the FASTQ files along with the corresponding metadata were submitted to the PulseNet for data analysis and uploaded to NCBI. Identification of antimicrobial resistance genes was done with raw reads using the ResFinder database (version 4.1, Center for Genetic Epidemiology, Kongens Lyngby, Denmark) with genes determined as present if sequences met thresholds of 90% identity and 60% minimum length (Zankari et al., 2017; Clausen et al., 2018; Bortolaia et al., 2020).
2.5. Statistical analysis
Descriptive statistics for the prevalence of Vibrio and Enterococcus in shrimp samples, the distribution of resistant patterns among isolates, sample characteristics, and the prevalence of resistance genes were conducted in Microsoft Excel (version 2,207, Redmond, WA, U.S.). Percent of isolates resistant to an antimicrobial agent was determined by dividing the number of isolates with a MIC value classified as resistant based on the appropriate CDC or FDA breakpoint criteria by the total number of isolates.
Prevalence and metadata analyses were performed using R version 4.1.2 (Vienna, Austria). Fisher’s exact test with adjusted p-values was used to evaluate the associations between these demographic factors and multidrug resistance. An α value of 0.05 was used to determine statistical significance for all analyses. Figures were created in R using packages ggplot2, ggtext, and heatmap.3.
Concordance between phenotypic resistance from AST and genotypic resistance from ARGs identified through WGS were evaluated for non-cholera Vibrio isolates (n = 40) for gentamicin, amoxicillin/clavulanic acid 2:1 ratio, cefoxitin, trimethoprim/sulfamethoxazole, azithromycin, ampicillin, and tetracycline as previously described (Lee et al., 2022). Phenotype and genotype were considered concordant when an isolate with phenotypic resistance to a drug in the MIC panel also had ARGs associated with the corresponding drug (true positive, TP), or when an isolate with phenotypic susceptibility to a drug also did not contain any corresponding ARGs (true negative, TN). TP and TN results indicated that in-silico predictions of resistance based on WGS were concordant with phenotypic observations. False negatives (FN) were defined as isolates that exhibited phenotypic resistance but did not harbor any ARGs known to confer resistance to the corresponding drug, and false positives (FP) were defined as isolates that exhibited phenotypic susceptibility to a drug but contained ARGs associated with that drug. FP and FN results indicated discordance between in-silico predictions and observed phenotypic resistance. Sensitivity was calculated as TP/(TP + FN) and specificity was calculated as TN/(TN + FP). Two Vibrio isolates that were speciated as V. cholerae were omitted from concordance analysis because phenotypic resistance was determined based on MIC breakpoints defined specifically for non-cholera Vibrio.
3. Results
3.1. Prevalence of Vibrio spp. and Enterococcus spp. in retail shrimp
The overall prevalence of Vibrio spp. in retail shrimp samples in this study was 60.25% (241/400). Farmed samples (78.44%, 211/269) had higher Vibrio prevalence than wild caught samples (45.80%, 60/131). Vibrio prevalence was also higher in imported samples (71.61%, 227/317) than domestic samples (53.01%, 44/83). Enterococcus spp. were present in 89.75% of all samples (359/400), including 92.94% of farmed samples (250/269), 83.21% of wild caught samples (109/131), 91.17% of imported samples (289/317), and 84.34% of domestically sourced samples (70/83) (Table 1).
3.2. Phenotypic resistance from antimicrobial susceptibility testing
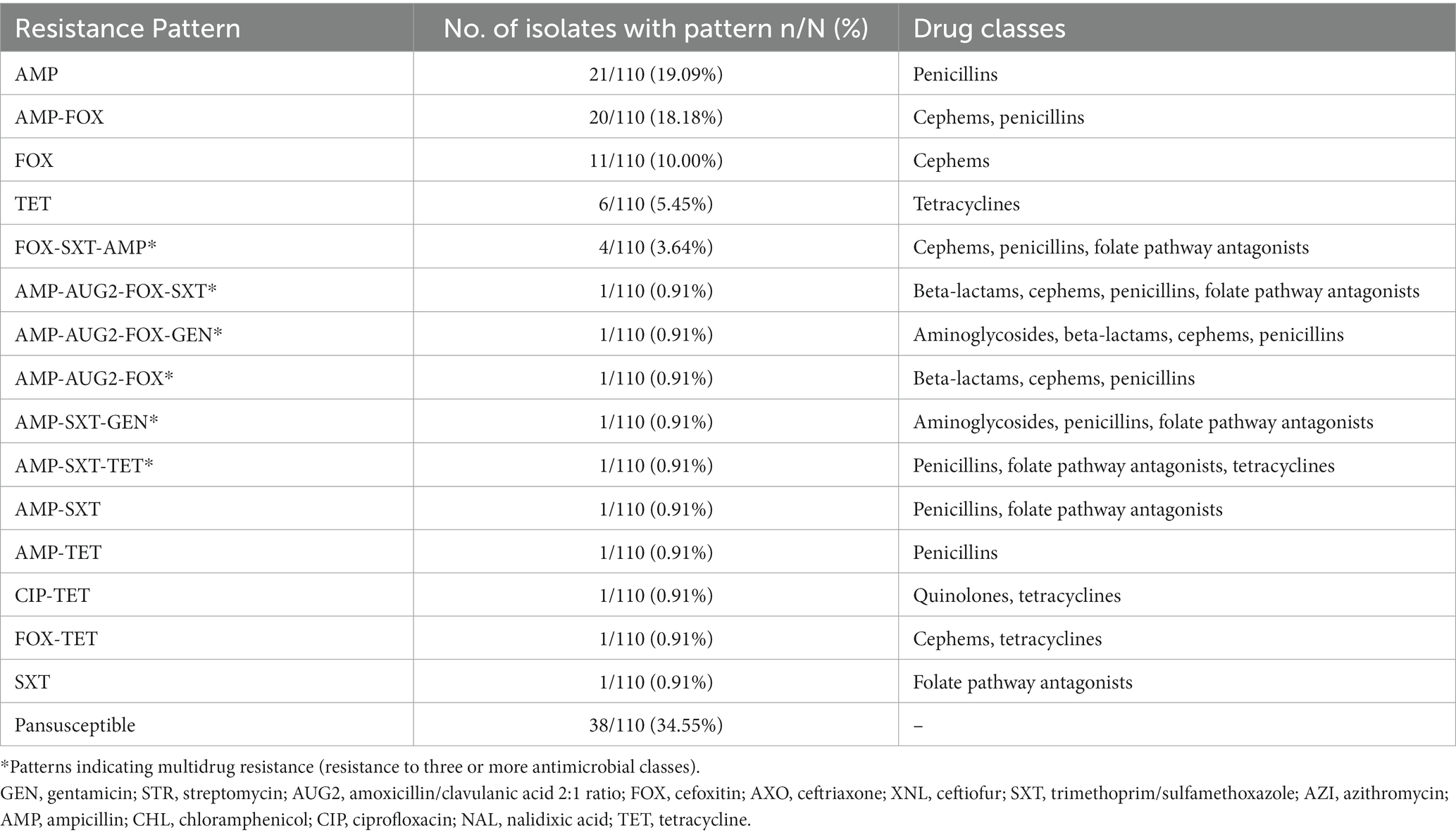
The predominant antimicrobials that the 110 Vibrio isolates tested for phenotypic resistance were resistant to were ampicillin (47.27%, 52/110) and cefoxitin (35.45%, 39/110). Low prevalence of resistance was observed for tetracycline (9.09%, 10/110), trimethoprim-sulfamethoxazole (8.18%, 9/110), amoxicillin/clavulanic acid 2:1 ratio (2.73%, 3/110), gentamicin (1.82%, 2/110), ciprofloxacin (0.91%, 1/110), and azithromycin (0%, 0/110) (Table 2). The number of Vibrio isolates with MIC values below the lowest concentration and above the highest concentration tested for those drugs in the Gram-negative panel are reported in Supplementary Table 2. Multidrug resistance was observed in 8.18% (9/110) of Vibrio isolates. A further 21.82% (24/110) were resistant to two antimicrobial classes, 35.45% (39/110) were resistant to one class, and the remaining 34.55% (38/110) were pansusceptible (Table 3).
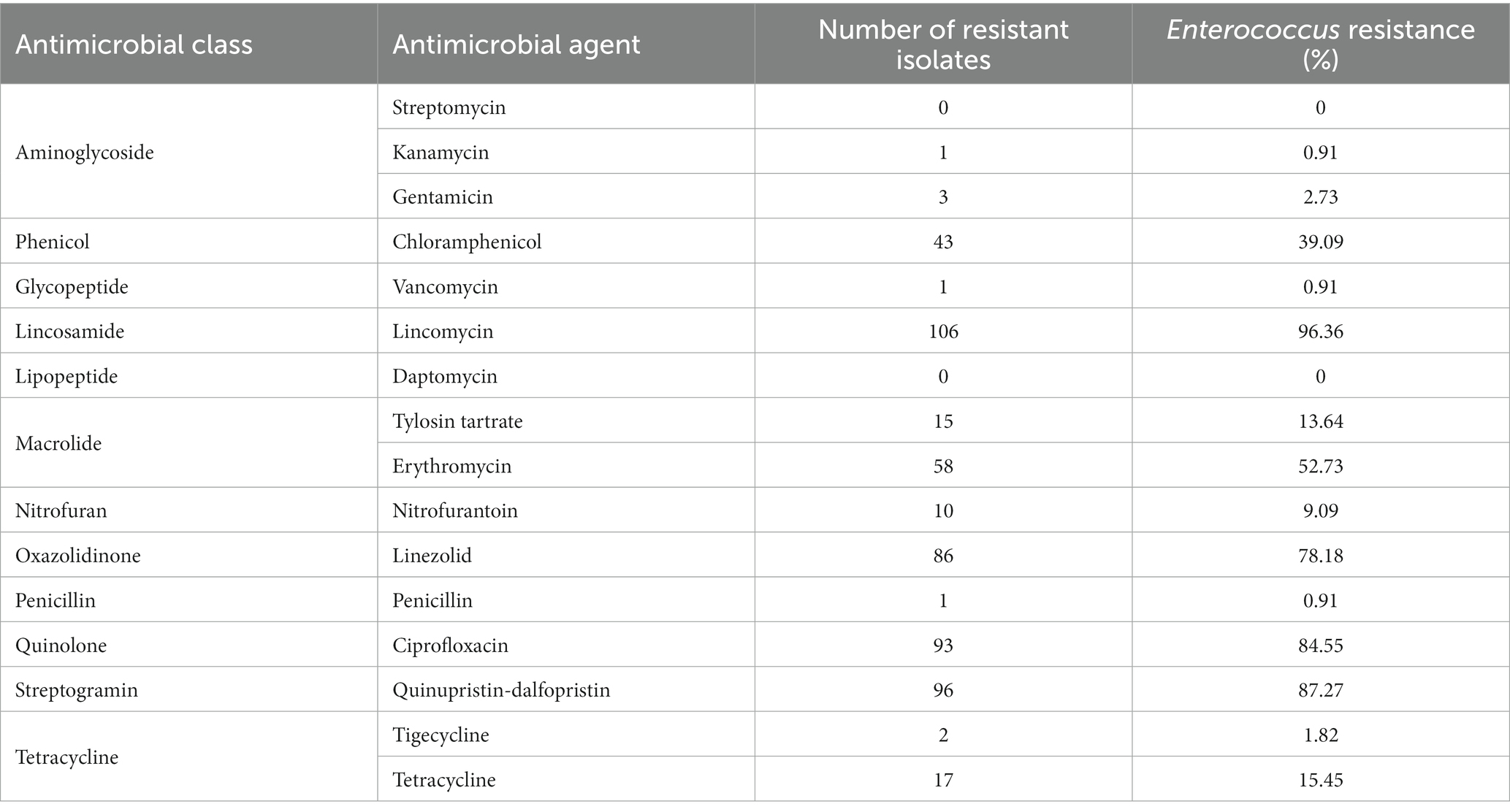
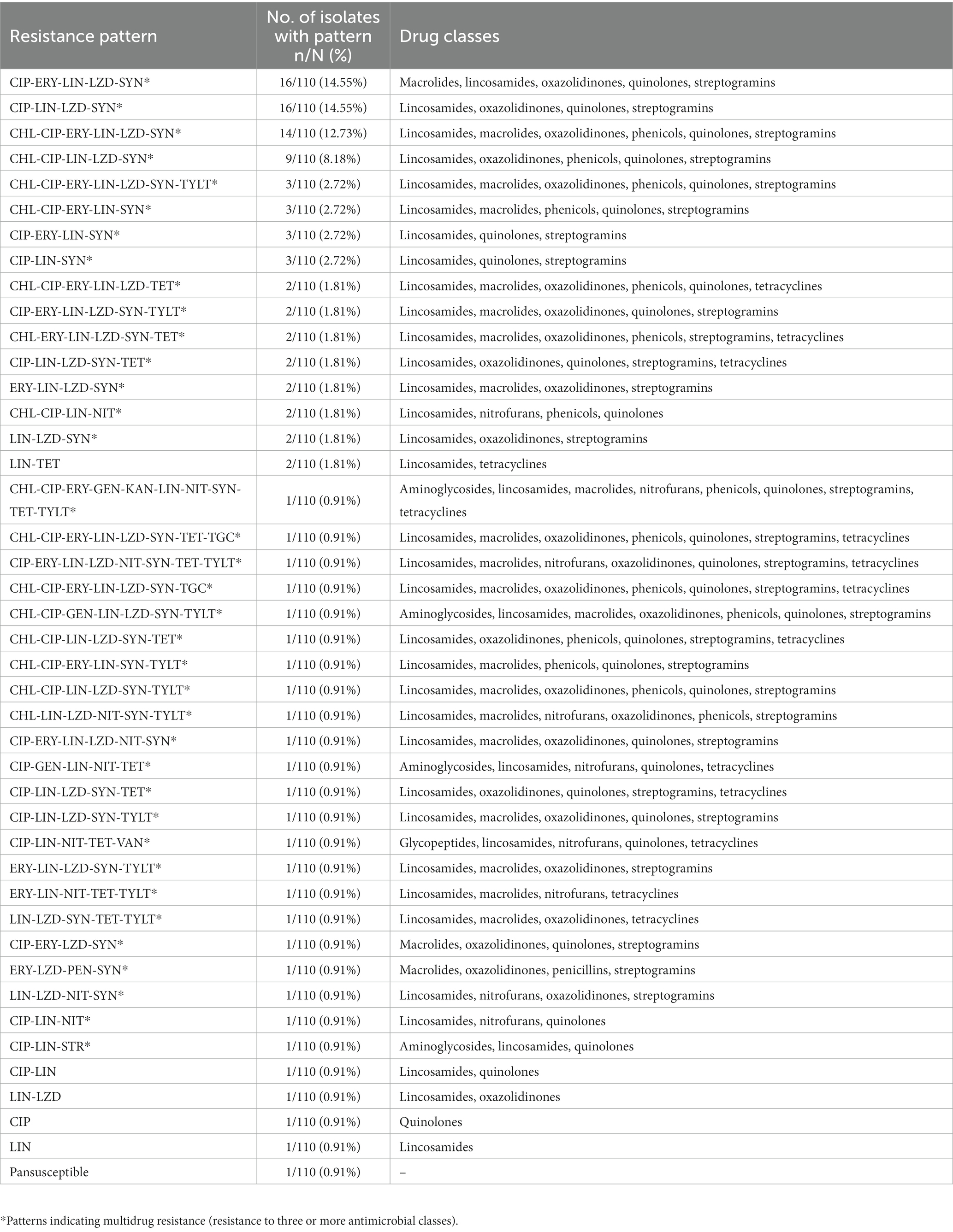
Antimicrobial susceptibility testing of Enterococcus isolates revealed high prevalence of resistance to lincomycin (96.36%, 106/110), quinupristin-dalfopristin (87.27%, 96/110), ciprofloxacin (84.55%, 93/110), linezolid (78.18%, 86/110), erythromycin (52.73%, 58/110), and chloramphenicol (39.09%, 43/110) (Table 4). Only one isolate was pansusceptible to all drugs in the MIC panel and all other isolates exhibited resistance to at least one of these six drugs in addition to various combinations of the other drugs in the panel. Low levels of resistance were found for tetracycline (15.45%, 17/110), tylosin tartrate (13.64%, 15/110), nitrofurantoin (9.09%, 10/110), gentamicin (2.73%, 3/110), tigecycline (1.82%, 2/110), kanamycin (0.91%, 1/110), penicillin (0.91%, 1/110), vancomycin (0.91%, 1/110), daptomycin (0%, 0/110), and streptomycin (0%, 0/110). Of these Enterococcus isolates, 93.64% (103/110) were multidrug resistant, 3.64% (4/110) were resistant to two classes of antimicrobials, 1.82% (2/110) were resistant to one class, and 0.91% (1/110) were pansusceptible. The phenotypic resistance patterns of Enterococcus isolates were diverse, though half exhibited one of four patterns involving chloramphenicol (CHL), ciprofloxacin (CIP), erythromycin (ERY), lincomycin (LIN), linezolid (LZD), and quinupristin-dalfopristin (SYN): CIP-ERY-LIN-LZD-SYN (14.55%, 16/110), CIP-LIN-LZD-SYN (14.55%, 16/110), CHL-CIP-ERY-LIN-LZD-SYN (12.73%, 14/110), and CHL-CIP-LIN-LZD-SYN (8.18%, 9/110) (Table 5).
3.3. Analysis of antimicrobial resistance by shrimp sample metadata
For analysis, origin was collapsed to domestic or imported categories due to small sample size by country. Similarly, season of collection was excluded due to small sample sizes by season. Packaging claims were excluded because few samples included claims about antimicrobial use. All claims that were found came in the form of the Global Seafood Alliance’s Best Aquaculture Practices (BAP) certifications, which mandate veterinary and regulatory oversight of antimicrobial usage and prohibit the use of drugs for growth promotion (Best Aquaculture Practices, 2014). The eight farmed Vibrio isolates from packages with BAP certifications averaged resistance to 1.88 drugs, which was more (p = 0.015) than the 0.96 average of the 73 isolates without the certification. Thirteen Enterococcus isolates were sourced from farmed shrimp samples with BAP certifications, and their average resistance to 5.31 drugs did not significantly differ (p = 0.212) from the average of the 4.79 drug average for the 68 uncertified. Shrimp species was excluded as a variable for analysis because the majority of samples were whiteleg shrimp (57.0%, 228/400) or did not specify species (21.75%, 87/400), and the remainder consisted of nine different species, which limited the ability to make comparisons between samples.
No significant associations were observed between multidrug resistance in Vibrio or Entercoccus isolates and production method (farm raised or wild caught) (Table 6). Vibrio isolates from farm raised shrimp were multidrug resistant 8.64% (7/81) of the time, compared to 6.90% (2/29) for those sourced from wild caught shrimp (p = 1.0). Of Enterococcus isolates from farm raised shrimp samples, 95.1% (77/81) were resistant to at least one of the sixteen antimicrobial agents, compared to 89.66% (26/29) from wild caught shrimp samples (p = 0.377). No significant associations were observed between multidrug resistance and sample origin either (domestic or imported) (Table 6). Domestic and imported Vibrio isolates were multidrug resistant 5.26% (1/19) and 8.79% (8/91) of the time, respectively (p = 1.0). Enterococcus isolates from domestically produced shrimp were multidrug resistant 88.89% (16/18) of the time, while those from imported shrimp were multidrug resistant 94.57% (87/92) of the time (p = 0.321).
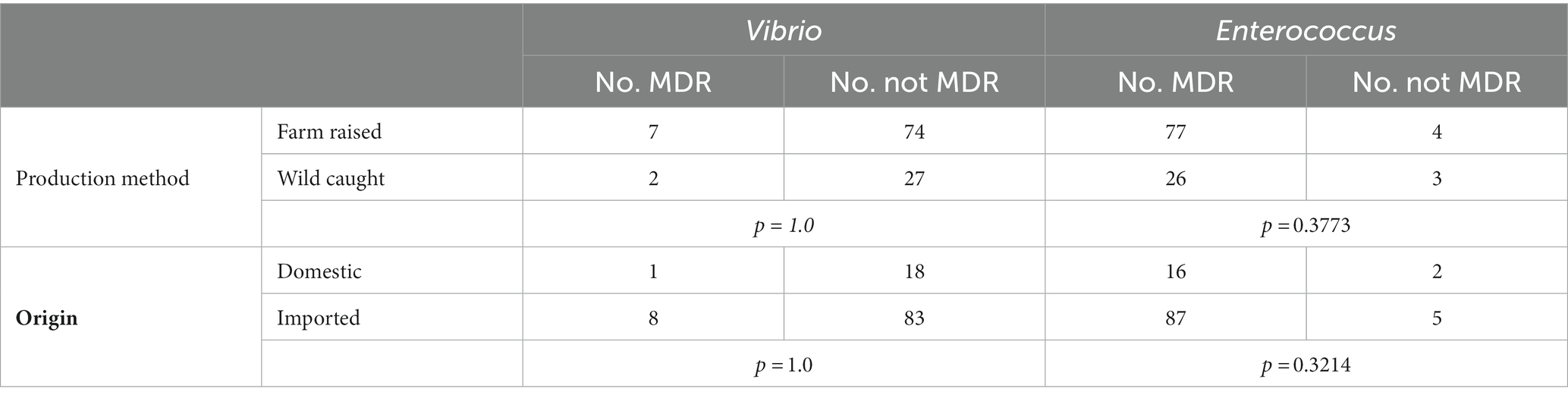
Table 6. Association between multidrug resistant (MDR) Vibrio and Enterococcus from retail shrimp and sample production method and origin.
3.4. Vibrio species identification and metadata trends via whole genome sequencing (WGS)
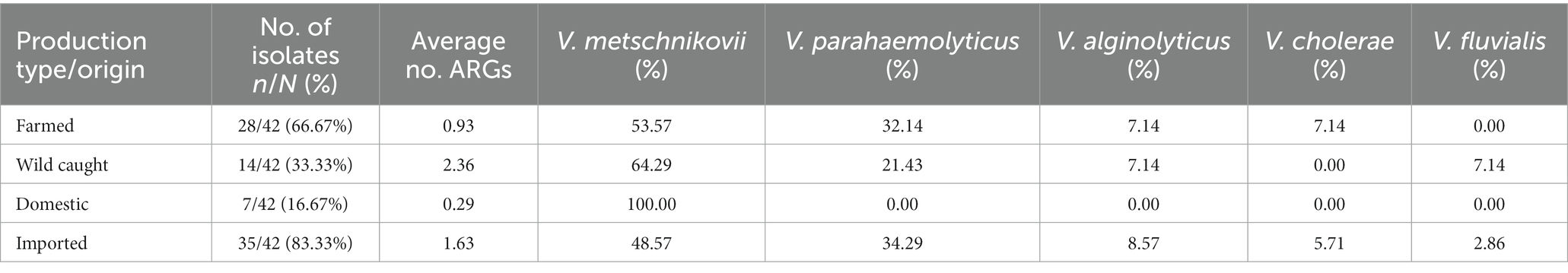
For the 42 Vibrio isolates that underwent whole genome sequencing, the distribution of species and resistance genes alongside metadata characteristics are summarized in Table 7. The most common Vibrio species identified by WGS was V. metschnikovii (24/42; 57.14%), followed by V. parahaemolyticus (12/42; 28.57%), V. alginolyticus (3/42; 7.14%), V. cholerae (2/42; 4.76%), and V. fluvialis (1/42; 2.33%). All seven domestic isolates subjected to WGS were speciated as V. metschnikovii, while the majority of imported isolates were either V. metschnikovii (17/35; 48.57%) or V. parahaemolyticus (12/35; 34.29%). The domestic isolates averaged 0.29 ARGs, compared to 1.63 ARGs on average for imported isolates (Table 8). While the WGS subsample included twice as many isolates sourced from farmed shrimp than wild caught shrimp, the bacterial species compositions within the groups were similar to each other and to the full sample selected for AST.
3.5. Resistance gene identification through WGS
Whole genome sequencing identified 27 unique ARGs from the 42 Vibrio isolates. Among these resistance genes were genes corresponding to two types of aminoglycoside-modifying enzymes (AMEs), phosphotransferases (aph(3′)-Ia, aph(3″)-Ib, and aph(6)-Id) and adenylyltransferases (aph(2″)-Ia). Nine unique blaCARB and one blaVEB ARGs were found, which are associated with resistance to beta-lactam agents including penicillins. Four ARGs associated with folate pathway antagonists were identified, three of which (dfrA1, dfrA6, and dfrA31) are known to confer resistance to trimethoprim and one (sul2) known to confer resistance to sulfamethoxazole. Three ARGs encoding for tetracycline efflux pumps, two pentapeptide genes conferring resistance to quinolones, one chloramphenicol efflux pump gene, and one macrolide inactivation gene were also present in this subsample of Vibrio isolates. Two genes were identified that confer resistance to rifamycins, a drug class not included in the Gram-negative panel used for AST in this study. Cephems were the only class on the panel for which no ARGs were identified. The frequencies at which these ARGs were observed are visualized in Figure 1.
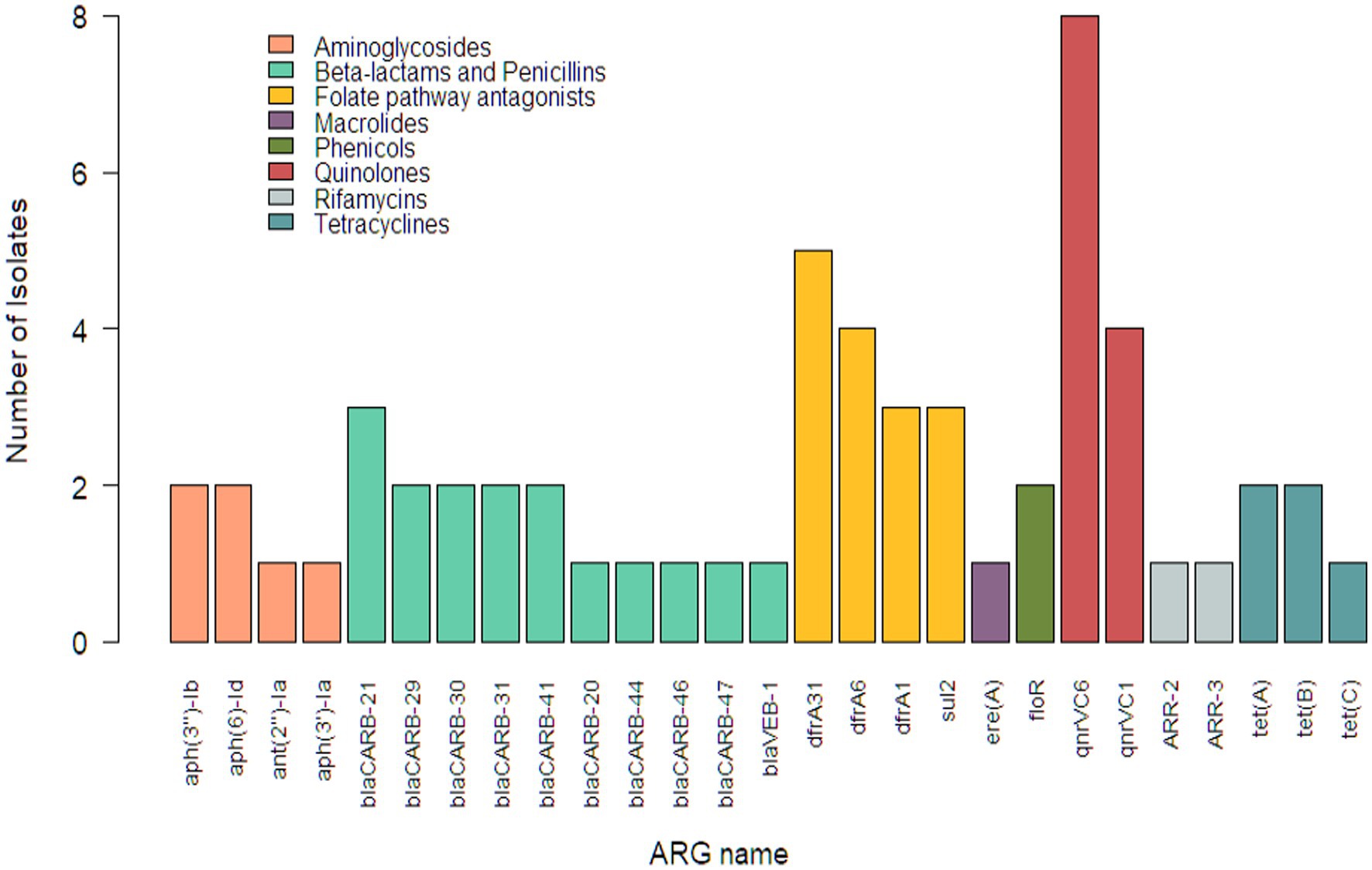
Figure 1. Distribution of antimicrobial resistance genes in Vibrio isolates (n = 42) from retail shrimp.
Vibrio parahaemolyticus isolates in this study had more ARGs than other species, averaging 3.00 ARGs per isolate. They were the only isolates to contain resistance genes associated with beta-lactams including penicillins. Sixteen unique resistance genes were identified in one V. parahaemolyticus isolate from a wild caught shrimp originating from Vietnam. This was also the only isolate with rifamycin and macrolide resistance genes and was one of only two with aminoglycoside resistance genes. All five ARGs identified from V. metschnikovii isolates corresponded to either quinolones or folate pathway antagonists.
Farmed isolates (28/42; 66.67%) contained between zero and five ARGs (mean = 0.93), while wild caught isolates (14/42; 33.33%) contained between zero and sixteen (mean = 2.36). The quinolone ARG qnrVC6 was the predominant resistance gene identified in Vibrio isolates from farmed (4/28; 14.29%), wild caught (4/14; 28.57%), and imported shrimp. Only two ARGs were identified within the seven domestic isolates, qnrVC6 (1/7; 14.29%) and dfrA1 (1/7, 14.29%) (Figure 2).
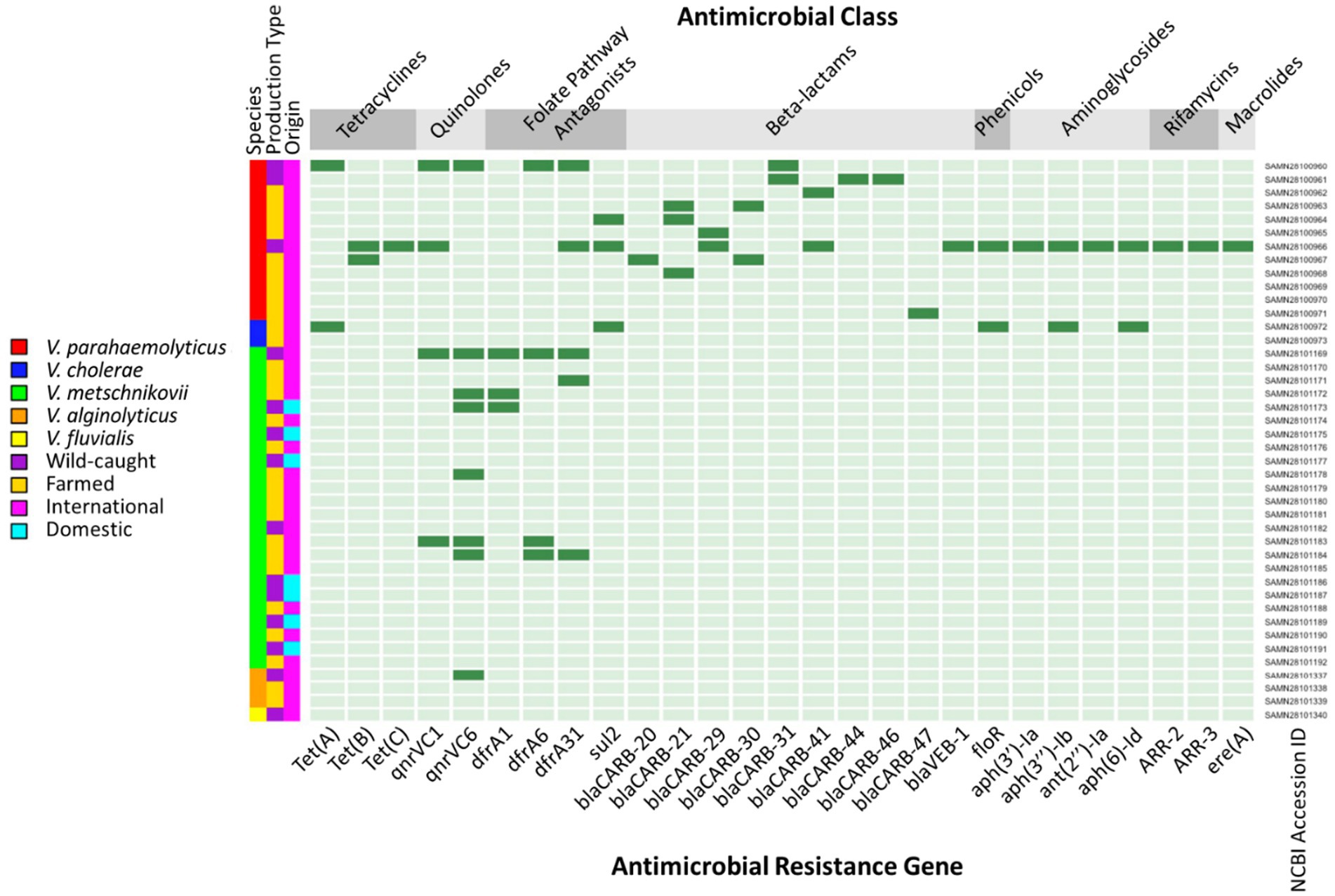
Figure 2. Heatmap of antimicrobial resistance genes (ARGs) identified through whole-genome sequencing in Vibrio isolates from retail shrimp. Dark green indicates the presence of an ARG and light green indicates the absence of an ARG.
3.6. Concordance of phenotypic and genotypic resistance in Vibrio isolates
Comparing phenotypic AMR and resistance genes identified from WGS in 40 non-cholera Vibrio isolates, the overall sensitivity and specificity were determined to be 11.54 and 96.05%, respectively. Discrepancies were observed in all drugs assessed; for each, there was at least one isolate that was categorized as phenotypically resistant but did not harbor any corresponding ARGs. None of the 17 isolates categorized as phenotypically resistant (3 resistant and 14 intermediate isolates) to cefoxitin harbored any associated ARGs. Of the four isolates categorized as phenotypically resistant (3 resistant and 1 intermediate) to trimethoprim/sulfamethoxazole, only one had ARGs associated with both component drugs. The majority of the false positives that contributed to low specificity were for ampicillin (ampicillin associated ARGs were found in 7 of the 15 phenotypically susceptible isolates) (Table 9).
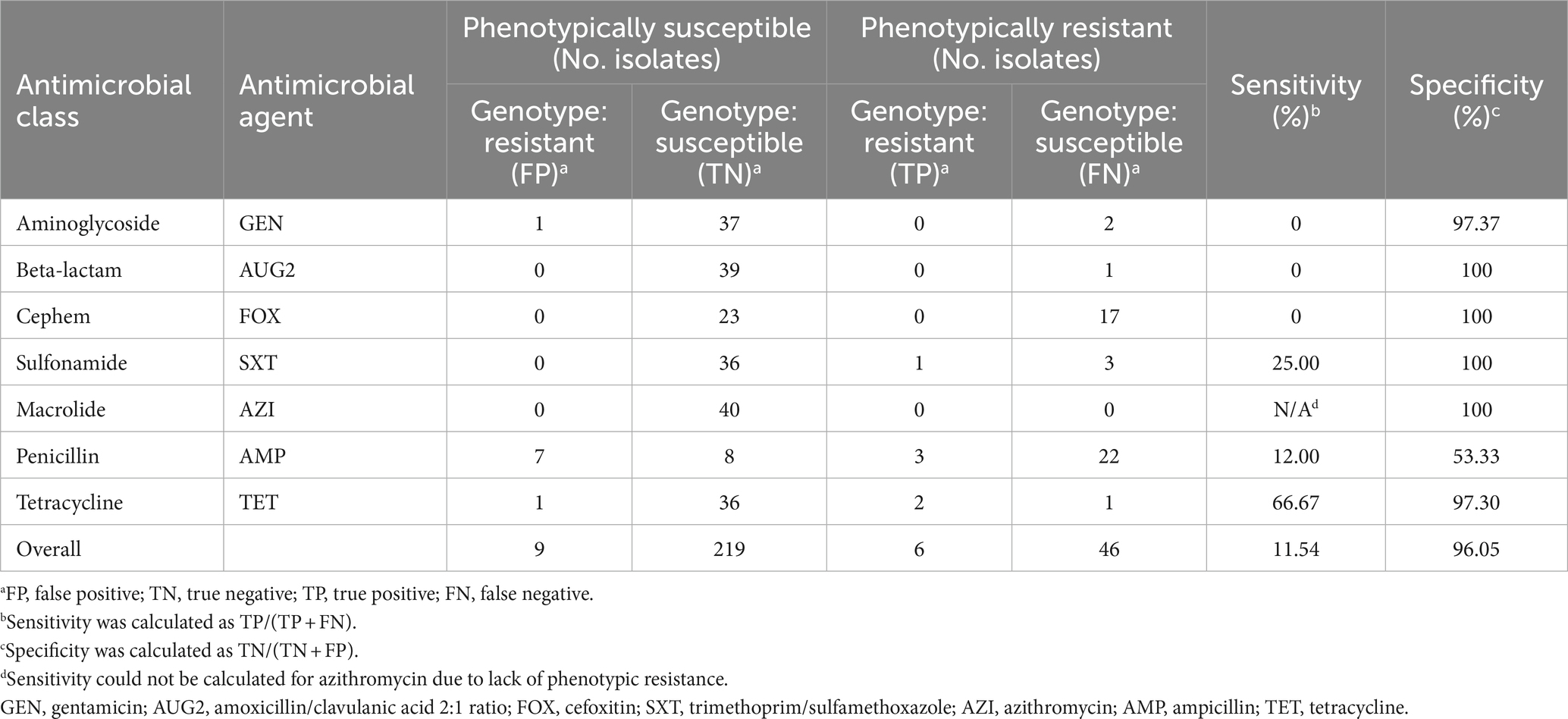
Table 9. Concordance of phenotypic and genotypic resistance of non-cholera Vibrio isolates from retail shrimp (n = 40).
4. Discussion
4.1. Prevalence of Vibrio spp. and Enterococcus spp. in retail shrimp
This study found Vibrio prevalence of 60.25% (241/400) in retail shrimp meat samples of different production types and geographic origins in northern California. Other studies from around the world have reported widely varying prevalences of Vibrio spp. in shrimp samples, ranging from 17.1% in Iran to 88.1% in Mexico and 95.6% in Ecuador (Sperling et al., 2015; Asgarpoor et al., 2018; Guardiola-Avila et al., 2020). The most directly comparable recent assessment to ours was conducted in 2022 by Tate et al. who found 40.85% (290/710) prevalence of Vibrio spp. in United States retail shrimp samples. In this context, the 60.25% Vibrio spp. prevalence we found in our 400 retail shrimp samples is not anomalous.
We observed Enterococcus spp. prevalence to be 89.75% (359/400) in our shrimp samples. Enterococcus spp. are ubiquitous bacteria common in aquatic environments and the overall prevalence in our samples, while high, was in line with expectations. Our prevalence observations were similar to those of other recent studies that measured Enterococcus spp. in samples of retail shrimp meat, which ranged from 58.33% prevalence in shrimp imported to grocery stores in northeastern Poland, to 66% in shrimp samples from American grocery stores, to 84.7% in shrimp imported to Denmark (Chajęcka-Wierzchowska et al., 2016; Ellis-Iversen et al., 2020; Tate et al., 2022).
Whole genome sequencing is a powerful tool in both laboratory and clinical settings and has become popular in the surveillance of foodborne pathogens for its utility in identifying the microbial species present in samples (Grundmann, 2014; Köser et al., 2014). In this study we employed WGS to determine the species of a subset of 42 Vibrio isolates and identify the diversity of ARGs within their genomes. While V. cholerae, as some strains are causative agents of epidemic cholera, and V. parahaemolyticus and V. alginolyticus, as two of the most common causative agents of foodborne illness globally, are the species most associated with human infections that were found among our isolates, all of the species identified are among the twelve known to be associated with human infections (Morris and Acheson, 2003). The most prevalent Vibrio species we isolated was V. metschnikovii (57.14%; 24/42), followed by V. parahaemolyticus (28.57%; 12/42), V. alginolyticus (7.14%; 3/42), V. cholerae (4.76%; 2/42), and V. fluvialis (2.38%; 1/42). By comparison, in a 2011 assessment of Vibrio prevalences in shrimp samples in Baton Rouge, Louisiana, U.S., Wang et al. (2011) observed V. cholerae in 17.8%, V. mimicus in 6.63%, V. parahaemolyticus in 4.57%, and other, unspecified Vibrio species in 21.1% of their samples.
4.2. Phenotypic resistance of Vibrio and Enterococcus
The evaluation of Vibrio resistance was hindered by six drugs without defined CLSI or NARMS breakpoints on the 14-drug NARMS Gram-negative panel. One such drug was nalidixic acid, a quinolone. Quinolones are the most common class of antimicrobial agents used in aquaculture globally (Schar et al., 2020). Chloramphenicol also does not have defined breakpoints despite being highly relevant to aquaculture. It belongs to the third most commonly used class of antimicrobials in aquaculture globally, phenicols, and was the only drug in the panel approved for aquacultural use in the United States (Schar et al., 2020; FDA, 2022a).
The drugs that Vibrio isolates in this study were most commonly resistant to were ampicillin and cefoxitin, which is in line with previous assessments. Ampicillin resistance has been reported in V. parahaemolyticus and V. vulnificus since 1978 and 2001, respectively (Joseph et al., 1978; Zanetti et al., 2001). The observed prevalence of Vibrio resistance to ampicillin in our study (40.91%; 45/110) is very similar to that observed by Akinbowale et al. (2006) in their analysis of Vibrio isolates collected from various aquacultural sources in Australia (40.32%, 25/62), but contrasts starkly with Raissy et al. (2012) who found 97.2% (70/72) ampicillin resistance among Vibrio isolates from wild caught seafood. The prevalence of cefoxitin resistant Vibrio isolates (35.45%) in our study was similar to the findings of García-Aljaro et al. (2014), who found 44% resistance in Vibrio spp. isolates from aquaculture facilities.
Antimicrobial susceptibility testing of Enterococcus isolates revealed resistance prevalence for multiple drugs, namely lincomycin (96.36%), quinupristin-dalfopristin (87.27%), ciprofloxacin (84.55%), linezolid (78.18%), erythromycin (52.73%), and chloramphenicol (39.09%). Interestingly, despite near-ubiquitous resistance to lincomycin among Enterococcus isolates in this assessment, its use in aquaculture has only been reported in China and none of our samples originated from China (Lulijwa et al., 2020). This could suggest exposure to lincomycin residues from non-aquaculture sources; lincomycin is commonly found in the waste streams of terrestrial livestock facilities and could enter surface waterways that feed directly into flow-through aquaculture systems like those that predominate shrimp farming (Boyd et al., 2022; Public Health Agency of Canada, 2022).
Co-selection of AMR, a phenomenon in which selective pressure upon exposure to one antimicrobial agent often results in the acquisition of resistance to other agents, could also have contributed to the high prevalence of lincomycin resistance (Seiler and Berendonk, 2012; Zhang M et al., 2018; Imran et al., 2019). Some heavy metals, notably copper sourced from aquacultural and agricultural pollution, have been shown to cause co-selection of AMR in waterborne bacteria. Seiler and Berendonk (2012) found that exposure to high levels of copper resulted in bacterial resistance to lincomycin, erythromycin, and vancomycin which persisted through the end of their seven-day observation period. It’s possible that the high rates of resistance to lincomycin observed in our assessment could have resulted from inadvertent exposure to metals such as copper.
It is also possible that the use of lincomycin in the countries of origin for these resistant isolates was unreported, or even inadvertent. Accurate tracking of antimicrobial use is a difficult endeavor that often involves non-governmental surveys which depend on the honesty and knowledge of producers, both of which can be unreliable (Shamsuzzaman and Biswas, 2012; Pham et al., 2015). Mislabeling of probiotic products in shrimp and other aquaculture has also introduced unintended and antimicrobial resistant bacteria into farms, which could be another explanation for the observed lincomycin resistance (Noor Uddin et al., 2015; Uma and Rebecca, 2018).
Ciprofloxacin is not used in aquaculture, yet many of our Enterococcus isolates from both farm raised and wild caught shrimp grew uninhibited in its presence (Thornber et al., 2020). Other studies have reported varied levels of ciprofloxacin resistance. Ellis-Iversen et al. (2020) found near-zero resistance to ciprofloxacin in E. faecalis and E. faecium isolates from Asian seafood imported to Denmark, while Igbinosa and Beshiru (2019) found more than 40% ciprofloxacin resistance in Enterococcus isolated from ready-to-eat seafood products. The prevalence observed in our study (84.55%), however, is abnormally high compared to levels of ciprofloxacin resistance in previous studies. Ciprofloxacin is classified as a critically important antimicrobial in human medicine by the World Health Organization (WHO), who cite its frequency of use and unique effectiveness against pathogenic infections as reasons that resistance could pose a significant risk to human health (WHO et al., 2019). While Enterococcus spp. are not among the pathogens of concern in this case, populations with prevalent resistance like those we observed are concerning as potential reservoirs of ARGs that could transfer to more significant pathogens.
Resistance of Enterococcus to linezolid has become increasingly common within the past decade, which is a growing concern in human medicine since it is used as a last resort treatment against vancomycin-resistant Enterococcus infections (Klare et al., 2015). Clinical studies have reported an increase in the rate of linezolid resistance among E. faecium samples over time, including <1% in 2008, >9% in 2014, and >20% in 2021 (Klare et al., 2015 Ma et al., 2021). Although the isolates in this assessment are not human pathogenic E. faecalis, the fact that 78.18% of them exhibited resistance to lincomycin and all have the capability of spreading that trait is notable. The pervasiveness of linezolid resistance in our assessment is also curious because the drug is not applied in aquaculture settings (Lulijwa et al., 2020). It is possible that this resistance could also have been acquired in a co-selection process. Pervasive, acquired resistance to a wide range of antimicrobial drugs in bacteria as adept at ARG transfer as Enterococcus poses a threat to human health.
Intrinsic resistance is a consideration when interpreting Enterococcus MIC results as well. Many Enterococci are known to be intrinsically resistant to aminoglycosides, although few samples were classified as resistant to streptomycin (1.82%), kanamycin (0.91%), or gentamicin (2.73%) in our susceptibility testing (Morrison et al., 1997; Harakeh et al., 2006). Some Enterococcus species also have unique resistances; E. faecalis is intrinsically resistant to streptogramins like quinupristin-dalfopristin, for example, and E. gallinarum and E. casseliflavus are intrinsically resistant to vancomycin (Morrison et al., 1997; Arias and Murray, 2012; Ellis-Iversen et al., 2020). These species-specific traits could not be considered in our assessment, however, since no Enterococcus isolates were identified beyond genus level.
4.3. Multidrug resistance patterns in Vibrio and Enterococcus
Prevalence of MDR in aquaculture-sourced Vibrio has increased in the 21st century (Han et al., 2007; Baker-Austin et al., 2009; Raissy et al., 2012; Shaw et al., 2014; Igbinosa, 2016). However, recent studies have varied substantially in their MDR observations. The prevalence of MDR for our Vibrio isolates (8.18%) was similar to the rates observed in aquacultured shrimp-associated bacteria by Singh et al. (2018) in their study in Punjab, India (8.4%; 10/119), and Helena Rebouças et al. (2011) conducted in northeastern Brazil (12.9%; 4/31). By contrast, Costa et al. (2015b) found that none of their 100 Virbio isolates from farmed shrimp in Brazil were resistant to three or more of the nine drug classes in their MIC panel, while a 2016 analysis of Vibrio spp. sampled from Nigerian aquaculture farms (Igbinosa, 2016) found that 57.49% of isolates (96/167) were resistant to at least three of the eight classes of antimicrobial drugs they tested. While these comparisons are valuable to contextualize the results of this assessment, it should be noted that there is variation in the composition of the drug panels between studies. The studies referenced above feature a similar number and identity of drugs and drug classes to those of our assessment, however. Moreover, the resolution of our findings was hampered by the omission of six drugs from the Gram-negative panel; many other studies assessing Vibrio spp. included multiple drugs within one or more classes, whereas our panel had only one representative per class among interpretable drugs.
We observed 93.64% MDR among Enterococcus isolates. This was driven in large part by pervasive resistance to lincomycin, quinupristin-dalfopristin, ciprofloxacin, linezolid, erythromycin, and chloramphenicol. Other recent studies have reported similar rates of MDR driven by ubiquitous or near-ubiquitous resistance to a subset of drugs. Enany et al. (2022), for instance, found that all of their 72 aquaculture-sourced Enterococcus isolates were resistant to chloramphenicol, macrolides azithromycin, and erythromycin. Further, 91.6% (66/72) of their isolates were resistant to tetracycline, and all exhibited resistance or intermediate resistance to nitrofurantoin. Generally, there has been a growing trend of MDR Enterococcus which is a concern in clinical circles, and the results of this study reinforce that pattern (Klare et al., 2015).
4.4. Multidrug resistance of Vibrio and Enterococcus by shrimp production and origin
No significant association was found between multidrug resistance in isolates sourced from farm raised or wild caught shrimp for Vibrio (p = 1.0) or Enterococcus (p = 0.377). This result was unexpected because shrimp raised in a farm environment are likely to be directly exposed to antimicrobial drugs which would apply selective pressure and presumably result in higher prevalence of resistance. Antimicrobial agents including many of those included on the MIC panels in this assessment have increasingly been found at detectable concentrations in coastal and estuarine ecosystems where wild shrimp are fished, and even diffuse, subinhibitory concentrations have been shown to select for AMR in environmental and shrimp-associated bacteria (Gullberg et al., 2011; Zheng et al., 2021). One other possibility is that the wild caught samples as a group were contaminated between capture and sale in a way that farmed samples were not. Still, our results imply that wild caught shrimp do not pose lower risk than farm raised shrimp for AMR.
There was also no significant association between MDR in isolates sourced from domestically produced or imported shrimp for Vibrio (p = 1.0) or Enterococcus (p = 0.321). All domestic samples were labeled as wild caught at collection, but since no statistical difference was found between farmed and wild caught isolates, that should not affect the interpretation of this result. The similarity between samples of different geographic origins could indicate that the United States’ import monitoring has been successful in holding imported seafood to the same antimicrobial stewardship standards as domestic seafood, or that there is some overlap in the processing or distribution processes that facilitates cross-contamination of bacteria before all shrimp of any origin reach grocery store shelves (FDA, 2022b).
4.5. Distribution of antimicrobial resistance genes in Vibrio and concordance of phenotypic and genotypic antimicrobial resistance
Whole genome sequencing with a subset of 42 Vibrio isolates was performed after antimicrobial susceptibility testing to identify species and ARGs. The WGS revealed that two of the 42 isolates were V. cholerae, and it’s possible others among the non-sequenced isolates were as well. This complicates the interpretation of the results of the MIC analysis, because, as noted in results section 3.6, the breakpoints used to classify the MIC values were specifically defined for non-cholera Vibrio. The confirmed V. cholerae isolates were still considered in the MIC analysis since the identities of those not yet sequenced are unknown. If possible, sequencing isolates before MIC testing would help ensure that this uncertainty does not arise in future assessments. The most common ARGs found in this assessment were qnr genes which encode for pentapeptide repeat proteins and confer reduced susceptibility to quinolones. Resistance to quinolones is primarily mediated by chromosomal quinolone resistance determining region (QRDR) mutations, though, and secondarily by acquired plasmid-mediated quinolone resistance (PMQR) genes like qnrVC1 and qnrVC6 (Zhang Y et al., 2018; Esmaeel et al., 2020; Lee et al., 2022). Still, acquired ARGs impart partial resistance to quinolones on their own and remain dangerous since they are highly transmissible to other organisms and contribute to the selection of resistance-associated chromosomal mutations (Nazik et al., 2011). The raw MIC values for ciprofloxacin and nalidixic acid did not suggest that isolates with one or both of qnrVC1 and qnrVC6 had reduced phenotypic susceptibility to quinolones. Considering only 0.91% of Vibrio isolates were classified as resistant to ciprofloxacin, this likely indicates that few QRDR mutations were present in this subsample.
More unique ARGs were found related to beta-lactam and penicillin resistance than any other antimicrobial classes. Nine of the ten such genes were blaCARB ARGs. The beta-lactamase protein encoded by this class of genes is a major mechanism of resistance to beta-lactam agents in Vibrio spp. and beyond (Potron et al., 2009; Manjusha and Bhat, 2011; Li et al., 2020). The blaCARB ARGs identified in this study are almost exclusively found in V. parahaemolyticus; blaVEB-1, the other beta-lactamase ARG found in one of our isolates, was found in V. parahaemolyticus in this study but has also been previously observed in V. alginolyticus (Alcock et al., 2020).
Genotypic resistance to the trimethoprim/sulfamethoxazole combo agent would require ARGs for both drugs to be present, though only one still confers partial resistance (Suhartono et al., 2016; Das et al., 2020 Ma et al., 2021). Among the seven isolates with one or more allelic variants of the dfrA trimethoprim ARG without any sulfamethoxazole ARGs, there was a trend between number of unique variants and resistance to trimethoprim/sulfamethoxazole (SXT). Of the four isolates with three dfrA variants, two expressed phenotypic resistance. In addition, of the two isolates with two dfrA variants, one expressed intermediate resistance; the one isolate with one dfrA variant was phenotypically susceptible to SXT. Both isolates with sulfamethoxazole-associated sul2 gene and no trimethoprim ARGs were susceptible to the combination agent. The other 32 isolates devoid of folate pathway antagonist ARGs were susceptible to SXT in MIC analysis.
An isolate with sixteen ARGs was the only with both a trimethoprim ARG (dfrA31) and a sulfamethoxazole ARG (sul2) and was phenotypically resistant to SXT. Among the other ARGs this isolate contained was aminoglycoside adenylyltransferase ARG ant(2″)-Id, which is known to confer resistance to gentamicin, though the isolate did not express phenotypic resistance to this drug in MIC testing (Ramirez and Tolmasky, 2010). Two aminoglycoside phosphotransferase ARGs, aph(3″)-Ib and aph(6)-Id, were also among the sixteen. These genes have been observed colocalized with sul2 and other ARGs on RSF1010, an oft-transmitted plasmid (Ramirez and Tolmasky, 2010). Another isolate, an isolate speciated as V. cholerae from a farmed Ecuadorian shrimp, also contained sul2, aph(3″)-Ib and aph(6)-Id. Regardless of whether the RSF1010 plasmid is present in these isolates, the possibility highlights the high transfer potential of the ARGs identified in this study.
Tetracyclines are highly important agents for human and veterinary medicine (WHO et al., 2019). The tetracycline ARGs identified in this study are frequently found in bacterial genomes isolated from aquatic environments like aquaculture ponds and from crustaceans (Schmidt et al., 2001; Dang et al., 2007; Zhang et al., 2009; Kim et al., 2013). Their presence in this study means they could be spread to significant pathogens and complicate treatment in clinical settings.
Analyses of AMR should account for intrinsic resistances in the bacteria of interest (Michelle et al., 2016). It has been shown that V. vulnificus and V. parahaemolyticus are intrinsically resistant to cefoxitin (Elmahdi et al., 2016). The twelve V. parahaemolyticus isolates identified by our WGS, however, did not reflect this pattern: 11/12 (91.67%) were inhibited at cefoxitin concentrations low enough to classify them as susceptible, and the other one (8.33%) was classified as intermediate. Regardless, since not all 110 isolates subjected to antimicrobial susceptibility testing were sequenced, this would not have been accounted for in statistical analyses even if the intrinsic resistance was observed.
Chiou et al. (2015) posited that V. parahaemolyticus intrinsically carries the blaCARB-17 gene which confers resistance to ampicillin, however only two of the twelve isolates identified in our study (16.67%) were phenotypically resistant to ampicillin and one (8.33%) expressed intermediate resistance. By contrast, among the other 28 non-cholera Vibrio isolates, 21 were phenotypically resistant (75.0%) and another was intermediate (3.57%). Further, WGS did not identify blaCARB-17 in any of the 42 isolates, though nine other blaCARB genes were found in V. parahaemolyticus isolates.
We observed a trend in our study that Vibrio from wild caught shrimp harbored a higher number ARGs on average than those from farmed samples. This is an unexpected finding given that there was no significant difference in phenotypic resistance prevalence between these two groups. Domestic isolates in this subset averaged fewer ARGs (0.29) than imported isolates (1.63), though the sample sizes and species compositions of these subgroups were distinct and limited the utility of their comparison. All seven domestic isolates were speciated as V. metschnikovii, whereas the 35 imported isolates had a more representative species distribution. There was also no significant difference between these two groups in phenotypic resistance prevalence.
The identification of ARGs via WGS facilitates in-silico predictions of phenotypic resistance (NIHR Global Health Research Unit on Genomic Surveillance of AMR, 2020; Lee et al., 2022). Phenotypic and genotypic AMR in non-cholera Vibrio isolates in our study correlated with an overall sensitivity of 11.54% and specificity of 96.05%. There are a few explanations for this low sensitivity and imperfect specificity. The largest contributing factor to the low sensitivity in our study is likely the grouping of intermediate isolates with resistant isolates, particularly for cefoxitin where 14 intermediate isolates were categorized as resistant for analysis. The results from our dataset indicate that the treatment of intermediate isolates during analysis has a large impact on the assessment of phenotypic and genotypic concordance. Other explanations include our WGS analysis of ARGs being limited to one database, so it is possible there are undetected and/or unknown AMR genetic determinants present amongst our isolates, in addition to the potential impact of cut-offs for identity and coverage used to determine the presence of ARGs. Lastly, AST and WGS in our study were conducted on separate occasions, so it is possible that plasmid loss occurred at some point, which could further contribute to incongruence of phenotypic and genotypic AMR.
5. Conclusion
The large-scale production and global distribution demands for shrimp results in a food production system that can be conducive to the selection and spread of antimicrobial resistance, prompting the need to better understand the occurrence of AMR in both pathogenic and commensal bacteria from these products. This present study provides food safety and public health insights on the prevalence and distribution of AMR in Vibrio spp. and Enterococcus spp. from retail shrimp in California, and highlights the importance of continued AMR monitoring of seafood products and the value of complementing antimicrobial susceptibility testing with whole-genome sequencing for AMR assessment.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
XL, EA, XY, and KaL: conceptualization. BH, KuL, BB, MG, ZL, and AY: laboratory analysis. BH, KuL, KaL, DK, ZL, and AY: data curation. BH, KaL, DK, and XL: formal analysis. BH, KaL, DK, XL, and XY: methodology. KaL, XL, and XY: supervision. EA, DK, XL, and XY: resources. BH: writing – original draft preparation. KaL, DK, ZL, AY, XL, and XY: writing – review and editing. XL and EA: funding acquisition. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by USDA National Institute of Food and Agriculture Animal Health Formula Funds project no. CALV-AH-395.
Acknowledgments
We thank Jennifer Chase for her guidance and assistance in the laboratory, and Toni Duarte for her feedback throughout the study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1192769/full#supplementary-material
References
Akinbowale, O., Peng, H., and Barton, M. D. (2006). Antimicrobial resistance in bacteria isolated from aquaculture sources in Australia. J. Appl. Microbiol. 100, 1103–1113. doi: 10.1111/j.1365-2672.2006.02812.x
Alcock, B. P., Raphenya, A. R., Lau, T. T. Y., Tsang, K. K., Bouchard, M., Edalatmand, A., et al. (2020). CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 48, D517–D525. doi: 10.1093/nar/gkz935
Ali, H., Rahman, M. M., Rico, A., Jaman, A., Basak, S. K., Islam, M. M., et al. (2018). An assessment of health management practices and occupational health hazards in tiger shrimp (Penaeus monodon) and freshwater prawn (Macrobrachium rosenbergii) aquaculture in Bangladesh. Vet Animal Sci. 5, 10–19. doi: 10.1016/j.vas.2018.01.002
Amatul-Samahah, M. A., Wan Omar, W. H. H., Mohd Ikhsan, N. F., Amal Azmai, M. N., Zamri-Saad, M., and Ina-Salwany, M. Y. (2020). Vaccination trials against vibriosis in shrimp: a review. Aquacult. Rep. 18:100471. doi: 10.1016/j.aqrep.2020.100471
Arias, C. A., and Murray, B. E. (2012). The rise of the Enterococcus: beyond vancomycin resistance. Nat. Rev. Microbiol. 10, 266–278. doi: 10.1038/nrmicro2761
Aryal, S. (2016). PYR test- principle, uses, procedure and result interpretation. Microbiology Info.com. Available at: https://microbiologyinfo.com/pyr-test-principle-uses-procedure-and-result-interpretation/ (Accessed February 20, 2023).
Asgarpoor, D., Haghi, F., and Zeighami, H. (2018). Detection and molecular characterization of Vibrio parahaemolyticus in shrimp samples. Open Biotechnol. J. 12, 46–50. doi: 10.2174/1874070701812010046
Baker-Austin, C., McArthur, J. V., Lindell, A. H., Wright, M. S., Tuckfield, R. C., Gooch, J., et al. (2009). Multi-site analysis reveals widespread antibiotic resistance in the marine pathogen Vibrio vulnificus. Microb. Ecol. 57, 151–159. doi: 10.1007/s00248-008-9413-8
Best Aquaculture Practices (2014). Aquaculture Facility Certification – finfish, crustacean, and mollusk hatcheries and nurseries – Version 1
Bortolaia, V., Kaas, R. S., Ruppe, E., Roberts, M. C., Schwarz, S., Cattoir, V., et al. (2020). ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 75, 3491–3500. doi: 10.1093/jac/dkaa345
Boyd, C. E., Davis, R. P., and McNevin, A. A. (2022). Perspectives on the mangrove conundrum, land use, and benefits of yield intensification in farmed shrimp production: a review. J. World Aquacult. Soc. 53, 8–46. doi: 10.1111/jwas.12841
Byappanahalli, M. N., Nevers, M. B., Korajkic, A., Staley, Z. R., and Harwood, V. J. (2012). Enterococci in the environment. Microbiol. Mol. Biol. Rev. 76, 685–706. doi: 10.1128/MMBR.00023-12
Çardak, M., Özmen Toğay, S., Ay, M., Karaalioğlu, O., Erol, Ö., and Bağcı, U. (2022). Antibiotic resistance and virulence genes in Enterococcus species isolated from raw and processed seafood. J. Food Sci. Technol. 59, 2884–2893. doi: 10.1007/s13197-021-05313-z
Centers for Disease Control and Prevention (CDC) (2019a). Antibiotic resistance threats in the United States, 2019. Atlanta, GA: U.S. Department of Health and Human Services, CDC
Centers for Disease Control and Prevention (CDC) (2019b). Antibiotics Tested by NARMS. Available at: https://www.cdc.gov/narms/antibiotics-tested.html (Accessed July 14, 2022).
Chajęcka-Wierzchowska, W., Zadernowska, A., and Łaniewska-Trokenheim, Ł. (2016). Virulence factors, antimicrobial resistance and biofilm formation in Enterococcus spp. isolated from retail shrimps. LWT Food Sci. Technol. 69, 117–122. doi: 10.1016/j.lwt.2016.01.034
Chang, Y.-H., Kumar, R., Ng, T. H., and Wang, H.-C. (2018). What vaccination studies tell us about immunological memory within the innate immune system of cultured shrimp and crayfish. Dev. Comp. Immunol. 80, 53–66. doi: 10.1016/j.dci.2017.03.003
Chiou, J., Li, R., and Chen, S. (2015). CARB-17 family of β-lactamases mediates intrinsic resistance to Penicillins in Vibrio parahaemolyticus. Antimicrob. Agents Chemother. 59, 3593–3595. doi: 10.1128/AAC.00047-15
Clausen, P. T. L. C., Aarestrup, F. M., and Lund, O. (2018). Rapid and precise alignment of raw reads against redundant databases with KMA. BMC Bioinformat. 19:307. doi: 10.1186/s12859-018-2336-6
Clinical and Laboratory Standards Institute (CLSI) (2017). Performance standards for antimicrobial susceptibility testing. CLSI supplement M100 27th Clinical Laboratory Standards Institute Wayne, PA, U.S.
Clinical and Laboratory Standards Institute (CLSI) Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. CLSI document M07 11th (2018). Clinical Laboratory Standards Institute Wayne, PA, U.S.
Costa, R. A., Araújo, R. L., and dos Viera, R. H. S. F. (2015a). Raw tropical oysters as vehicles for multidrug-resistant Vibrio parahaemolyticus. Rev. Inst. Med. Trop. Sao Paulo 57, 193–196. doi: 10.1590/S0036-46652015000300002
Costa, R. A., Araújo, R. L., Souza, O. V., and dos Vieira, R. H. S. F. (2015b). Antibiotic-resistant Vibrios in farmed shrimp. Biomed. Res. Int. 2015:e505914, 1–5. doi: 10.1155/2015/505914
Dang, H., Zhang, X., Song, L., Chang, Y., and Yang, G. (2007). Molecular determination of oxytetracycline-resistant bacteria and their resistance genes from mariculture environments of China. J. Appl. Microbiol. 103, 2580–2592. doi: 10.1111/j.1365-2672.2007.03494.x
Das, B., Verma, J., Kumar, P., Ghosh, A., and Ramamurthy, T. (2020). Antibiotic resistance in Vibrio cholerae: understanding the ecology of resistance genes and mechanisms. Vaccine 38, A83–A92. doi: 10.1016/j.vaccine.2019.06.031
El-Far, S. A. H., Khalil, R. H., Saad, T. T., Tanekhy, M. E., and Abdel-Latif, H. M. R. (2015). Occurrence, characterization and antibiotic resistance patterns of bacterial communities encountered in mass kills of pond cultured Indian prawn (Fenneropenaeus indicus) at Damietta governorate, Egypt. Int. J. Fish. Aquat. Stud. 2, 271–276.
Ellis-Iversen, J., Seyfarth, A. M., Korsgaard, H., Bortolaia, V., Munck, N., and Dalsgaard, A. (2020). Antimicrobial resistant E. coli and enterococci in pangasius fillets and prawns in Danish retail imported from Asia. Food Control 114:106958. doi: 10.1016/j.foodcont.2019.106958
Elmahdi, S., DaSilva, L. V., and Parveen, S. (2016). Antibiotic resistance of Vibrio parahaemolyticus and Vibrio vulnificus in various countries: a review. Food Microbiol. 57, 128–134. doi: 10.1016/j.fm.2016.02.008
Enany, M., Tartor, Y. H., Kishk, R. M., and Ali, E. M. (2022). Occurrence of multidrug-resistant enterococci in fresh water fishes. Suez Canal Vet. Med. J. 27, 91–100. doi: 10.21608/scvmj.2022.125815.1073
Esmaeel, N. E., Gerges, M. A., Hosny, T. A., Ali, A. R., and Gebriel, M. G. (2020). Detection of chromosomal and plasmid-mediated quinolone resistance among Escherichia coli isolated from urinary tract infection cases; Zagazig University Hospitals, Egypt. Infect. Drug Resist. 13, 413–421. doi: 10.2147/IDR.S240013
FDA (2021). Seafood pilot study laboratory protocol. FDA National Antimicrobial Resistance Monitoring System Available at: https://www.fda.gov/media/149957/download
FDA (2022a). Approved Aquaculture Drugs. Available at: https://www.fda.gov/animal-veterinary/aquaculture/approved-aquaculture-drugs (Accessed July 14, 2022).
FDA (2022b). Imported Seafood Safety Program. Available at: https://www.fda.gov/food/importing-food-products-united-states/imported-seafood-safety-program (Accessed July 15, 2022).
Food and Agriculture organization of the United Nations (FAO) (2022a). The state of the world fisheries and aquaculture: towards blue transformation Rome: Food and Agriculture Organization of the United Nations
Food and Agriculture Organization of the United Nations (FAO) FishStatJ: global fishery and aquaculture production statistics Rome: Food and Agriculture Organization of the United Nations (2022b)
García-Aljaro, C., Riera-Heredia, J., and Blanch, A. R. (2014). Antimicrobial resistance and presence of the SXT mobile element in Vibrio spp. isolated from aquaculture facilities. New Microbiol. 37, 339–346.
Golder, H. M., Séon Simon, A. A., Santigosa, E., de Ondarza, M.-B., and Lean, I. J. (2022). Effects of probiotic interventions on production efficiency, survival rate, and immune responses of shrimp: a meta-analysis and meta-regression. Aquaculture 552:737973. doi: 10.1016/j.aquaculture.2022.737973
Grundmann, H. (2014). Towards a global antibiotic resistance surveillance system: a primer for a roadmap. Ups. J. Med. Sci. 119, 87–95. doi: 10.3109/03009734.2014.904458
Guardiola‐Avila, I., Martínez‐Vázquez, V., Juárez‐Rendón, K., Alvarez‐Ainza, M., Paz‐González, A., and Rivera, G. (2020). Prevalence and virulence of Vibrio species isolated from raw shrimp from retail markets in Reynosa, Mexico. Letters in Applied Microbiology. 71, 280–286. doi: 10.1111/lam.13315
Gudding, R., and Van Muiswinkel, W. B. (2013). A history of fish vaccination: science-based disease prevention in aquaculture. Fish Shellfish Immunol. 35, 1683–1688. doi: 10.1016/j.fsi.2013.09.031
Gullberg, E., Cao, S., Berg, O. G., Ilbäck, C., Sandegren, L., Hughes, D., et al. (2011). Selection of resistant bacteria at very low antibiotic concentrations. PLoS Pathog. 7:e1002158. doi: 10.1371/journal.ppat.1002158
Han, F., Walker, R. D., Janes, M. E., Prinyawiwatkul, W., and Ge, B. (2007). Antimicrobial susceptibilities of Vibrio parahaemolyticus and Vibrio vulnificus isolates from Louisiana Gulf and retail raw oysters. Appl. Environ. Microbiol. 73, 7096–7098. doi: 10.1128/AEM.01116-07
Harakeh, S., Yassine, H., Hajjar, S., and El-Fadel, M. (2006). Isolates of Staphylococcus aureus and saprophyticus resistant to antimicrobials isolated from the Lebanese aquatic environment. Mar. Pollut. Bull. 52, 912–919. doi: 10.1016/j.marpolbul.2005.12.008
Helena Rebouças, R., Viana de Sousa, O., Sousa Lima, A., Roger Vasconcelos, F., de Carvalho, P. B., and dos Fernandes Vieira, R. H. S. (2011). Antimicrobial resistance profile of Vibrio species isolated from marine shrimp farming environments (Litopenaeus vannamei) at Ceará, Brazil. Environ. Res. 111, 21–24. doi: 10.1016/j.envres.2010.09.012
Holmström, K., Gräslund, S., Wahlström, A., Poungshompoo, S., Bengtsson, B.-E., and Kautsky, N. (2003). Antibiotic use in shrimp farming and implications for environmental impacts and human health. Int. J. Food Sci. Technol. 38, 255–266. doi: 10.1046/j.1365-2621.2003.00671.x
Igbinosa, E. O. (2016). Detection and antimicrobial resistance of Vibrio isolates in aquaculture environments: implications for public health. Microb. Drug Resist. 22, 238–245. doi: 10.1089/mdr.2015.0169
Igbinosa, E. O., and Beshiru, A. (2019). Antimicrobial resistance, virulence determinants, and biofilm formation of Enterococcus species from ready-to-eat seafood. Front. Microbiol. 10:728. doi: 10.3389/fmicb.2019.00728
Imran, M., Das, K. R., and Naik, M. M. (2019). Co-selection of multi-antibiotic resistance in bacterial pathogens in metal and microplastic contaminated environments: an emerging health threat. Chemosphere 215, 846–857. doi: 10.1016/j.chemosphere.2018.10.114
Joseph, S. W., DeBell, R. M., and Brown, W. P. (1978). In vitro response to chloramphenicol, tetracycline, ampicillin, gentamicin, and beta-lactamase production by halophilic Vibrios from human and environmental sources. Antimicrob. Agents Chemother. 13, 244–248. doi: 10.1128/AAC.13.2.244
Kim, M., Kwon, T.-H., Jung, S.-M., Cho, S.-H., Jin, S. Y., Park, N.-H., et al. (2013). Antibiotic resistance of bacteria isolated from the internal organs of edible snow crabs. PLoS One 8:e70887. doi: 10.1371/journal.pone.0070887
Klare, I., Fleige, C., Geringer, U., Thürmer, A., Bender, J., Mutters, N. T., et al. (2015). Increased frequency of linezolid resistance among clinical Enterococcus faecium isolates from German hospital patients. J. Glob. Antimicrob. Resist. 3, 128–131. doi: 10.1016/j.jgar.2015.02.007
Köser, C. U., Ellington, M. J., and Peacock, S. J. (2014). Whole-genome sequencing to control antimicrobial resistance. Trends Genet. 30, 401–407. doi: 10.1016/j.tig.2014.07.003
Kumar, V., Roy, S., Meena, D. K., and Sarkar, U. K. (2016). Application of probiotics in shrimp aquaculture: importance, mechanisms of action, and methods of administration. Rev. Fish. Sci. Aquacult. 24, 342–368. doi: 10.1080/23308249.2016.1193841
Lanz-Mendoza, H., and Contreras-Garduño, J. (2022). Innate immune memory in invertebrates: concept and potential mechanisms. Develop. Comparat. Immunol. 127:104285. doi: 10.1016/j.dci.2021.104285
Lee, K. Y., Atwill, E. R., Pitesky, M., Huang, A., Lavelle, K., Rickard, M., et al. (2022). Antimicrobial resistance profiles of non-typhoidal Salmonella from retail meat products in California, 2018. Front. Microbiol. 13:835699. doi: 10.3389/fmicb.2022.835699
Li, P., Liu, C., Li, B., and Ma, Q. (2020). Structural analysis of the CARB β-lactamase from Vibrio parahaemolyticus facilitates application of the β-lactam/β-lactamase inhibitor therapy. Biochimie 171–172, 213–222. doi: 10.1016/j.biochi.2020.03.011
Lulijwa, R., Rupia, E. J., and Alfaro, A. C. (2020). Antibiotic use in aquaculture, policies and regulation, health and environmental risks: a review of the top 15 major producers. Rev. Aquac. 12, 640–663. doi: 10.1111/raq.12344
Ma, X., Zhang, F., Bai, B., Lin, Z., Xu, G., Chen, Z., et al. (2021). Linezolid resistance in Enterococcus faecalis associated with urinary tract infections of patients in a tertiary hospitals in China: resistance mechanisms, virulence, and risk factors. Front. Public Health 9:570650. doi: 10.3389/fpubh.2021.570650
Ma, Q., Zhu, C., Yao, M., Yuan, G., and Sun, Y. (2021). Correlation between the sulfamethoxazole-trimethoprim resistance of Shigella flexneri and the sul genes. Medicine 100:e24970. doi: 10.1097/MD.0000000000024970
Manjusha, S., and Bhat, S. (2011). Plasmid associated antibiotic resistance in Vibrio isolated from coastal waters of Kerala. Int. Food Res. J. 18, 1171–1181.
Michelle, K., Lihan, S., Felecia, C., Entigu, R., Apun, K., Bilung, L. M., et al. (2016). Antibiotic resistance of diverse bacteria from aquaculture in Borneo. Int. J. Microbiol. 2016, 1–9. doi: 10.1155/2016/2164761
Morris, J. G., and Acheson, D. (2003). Cholera and other types of vibriosis: a story of human pandemics and oysters on the half shell. Clin. Infect. Dis. 37, 272–280. doi: 10.1086/375600
Morrison, D., Woodford, N., and Cookson, B. (1997). Enterococci as emerging pathogens of humans. J. Appl. Microbiol. 83, 89S–99S. doi: 10.1046/j.1365-2672.83.s1.10.x
National Fisheries Institute Media (2022). Top 10 list offers a look back in time. National Fisheries Institute Media. Available at: https://aboutseafood.com/press_release/top-10-list-offers-a-look-back-in-time/ (Accessed May 16, 2022).
National Marine Fisheries Service (2022). Fisheries of the United States, 2020. Available at: https://www.fisheries.noaa.gov/national/sustainable-fisheries/fisheries-united-states
Nazik, H., Bektöre, B., Öngen, B., Ilktaç, M., Özyurt, M., Kuvat, N., et al. (2011). Plasmid-mediated quinolone resistance genes in Escherichia coli urinary isolates from two teaching hospitals in Turkey: coexistence of TEM, SHV, CTX-M and VEB-1 type lactamases. Trop. J. Pharm. Res. 10, 325–333. doi: 10.4314/tjpr.v10i3.9
NIHR Global Health Research Unit on Genomic Surveillance of AMR (2020). Whole-genome sequencing as part of national and international surveillance programmes for antimicrobial resistance: a roadmap. BMJ Glob. Health 5:e002244. doi: 10.1136/bmjgh-2019-002244
Noor Uddin, G. M., Larsen, M. H., Christensen, H., Aarestrup, F. M., Phu, T. M., and Dalsgaard, A. (2015). Identification and antimicrobial resistance of bacteria isolated from probiotic products used in shrimp culture. PLoS One 10:e0132338. doi: 10.1371/journal.pone.0132338
Pham, D. K., Chu, J., Do, N. T., Brose, F., Degand, G., Delahaut, P., et al. (2015). Monitoring antibiotic use and residue in freshwater aquaculture for domestic use in Vietnam. EcoHealth 12, 480–489. doi: 10.1007/s10393-014-1006-z
Potron, A., Poirel, L., Croizé, J., Chanteperdrix, V., and Nordmann, P. (2009). Genetic and biochemical characterization of the first extended-Spectrum CARB-type ß-lactamase, RTG-4, from Acinetobacter baumannii. Antimicrob. Agents Chemother. 53, 3010–3016. doi: 10.1128/AAC.01164-08
Public Health Agency of Canada (2022). Canadian Antimicrobial Resistance Surveillance System Report 2021 Available at: https://www.canada.ca/en/public-health/services/publications/drugs-health-products/canadian-antimicrobial-resistance-surveillance-system-report-2021.html (Accessed February 3, 2023).
PulseNet (n.d.). Laboratory standard operating procedure for whole genome sequencing on MiSeq. Available at: https://www.cdc.gov/pulsenet/ (Accessed April 18, 2023).
Raissy, M., Moumeni, M., Ansari, M., and Rahimi, E. (2012). Antibiotic resistance pattern of some Vibrio strains isolated from seafood. Iran. J. Fish. Sci. 11, 618–626.
Ramirez, M. S., and Tolmasky, M. E. (2010). Aminoglycoside modifying enzymes. Drug Resist. Updat. 13, 151–171. doi: 10.1016/j.drup.2010.08.003
Schar, D., Klein, E. Y., Laxminarayan, R., Gilbert, M., and Van Boeckel, T. P. (2020). Global trends in antimicrobial use in aquaculture. Sci. Rep. 10:21878. doi: 10.1038/s41598-020-78849-3
Schmidt, A. S., Brunn, M. S., Dalsgaard, I., and Larsen, J. L. (2001). Incidence, distribution, and spread of tetracycline resistance determinants and integron-associated antibiotic resistance genes among motile aeromonads from a fish farming environment. Appl. Environ. Microbiol. 67, 5675–5682. doi: 10.1128/AEM.67.12.5675-5682.2001
Seiler, C., and Berendonk, T. (2012). Heavy metal driven co-selection of antibiotic resistance in soil and water bodies impacted by agriculture and aquaculture. Front. Microbiol. 3:399. doi: 10.3389/fmicb.2012.00399
Shamsuzzaman, M., and Biswas, T. K. (2012). Aqua chemicals in shrimp farm: a study from south-west coast of Bangladesh. Egypt. J. Aquat. Res. 38, 275–285. doi: 10.1016/j.ejar.2012.12.008
Shaw, K. S., Rosenberg Goldstein, R. E., He, X., Jacobs, J. M., Crump, B. C., and Sapkota, A. R. (2014). Antimicrobial susceptibility of Vibrio vulnificus and Vibrio parahaemolyticus recovered from recreational and commercial areas of Chesapeake Bay and Maryland Coastal Bays. PLoS One 9:e89616. doi: 10.1371/journal.pone.0089616
Singh, B., Tyagi, A., Billekallu Thammegowda, N. K., and Ansal, M. D. (2018). Prevalence and antimicrobial resistance of vibrios of human health significance in inland saline aquaculture areas. Aquac. Res. 49, 2166–2174. doi: 10.1111/are.13672
Smith, P. (2012). “7 – Antibiotics in aquaculture: reducing their use and maintaining their efficacy” in Infectious disease in aquaculture woodhead publishing series in food science, technology and nutrition. ed. B. Austin (Cambridge, UK: Woodhead Publishing), 161–189.
Sperling, L., Alter, T., and Huehn, S. (2015). Prevalence and antimicrobial resistance of Vibrio spp. in retail and farm shrimps in Ecuador. J. Food Prot. 78, 2089–2092. doi: 10.4315/0362-028X.JFP-15-160
Stratev, D., Fasulkova, R., and Krumova-Valcheva, G. (2023). Incidence, virulence genes and antimicrobial resistance of Vibrio parahaemolyticus isolated from seafood. Microb. Pathog. 177:106050. doi: 10.1016/j.micpath.2023.106050
Suhartono, S., Savin, M., and Gbur, E. E. (2016). Genetic redundancy and persistence of plasmid-mediated trimethoprim/sulfamethoxazole resistant effluent and stream water Escherichia coli. Water Res. 103, 197–204. doi: 10.1016/j.watres.2016.07.035
Tate, H., Ayers, S., Nyirabahizi, E., Li, C., Borenstein, S., Young, S., et al. (2022). Prevalence of antimicrobial resistance in select bacteria from retail seafood—United States, 2019. Front. Microbiol. 13:928509. doi: 10.3389/fmicb.2022.928509
Thompson, J. R., Randa, M. A., Marcelino, L. A., Tomita-Mitchell, A., Lim, E., and Polz, M. F. (2004). Diversity and dynamics of a north atlantic coastal vibrio community. Appl. Environ. Microbiol. 70, 4103–4110. doi: 10.1128/AEM.70.7.4103-4110.2004
Thomsen, S. T., Assunção, R., Afonso, C., Boué, G., Cardoso, C., Cubadda, F., et al. (2022). Human health risk–benefit assessment of fish and other seafood: a scoping review. Crit. Rev. Food Sci. Nutr. 62, 7479–7502. doi: 10.1080/10408398.2021.1915240
Thornber, K., Verner-Jeffreys, D., Hinchliffe, S., Rahman, M. M., Bass, D., and Tyler, C. R. (2020). Evaluating antimicrobial resistance in the global shrimp industry. Rev. Aquac. 12, 966–986. doi: 10.1111/raq.12367
U.S. Department of Agriculture and U.S. Department of Health and Human Services (2020). Dietary Guidelines for Americans, 2020-2025 Available at: https://dietaryguidelines.gov/
U.S. Food and Drug Administration (FDA) (2019). The National Antimicrobial Resistance Monitoring System: NARMS Integrated Report, 2016–2017. Laurel, MD U.S. Food and Drug Administration
Uma, A., and Rebecca, G. (2018). Antibiotic resistance in bacterial isolates from commercial probiotics used in aquaculture. Int. J. Curr. Microbiol. App. Sci. 7, 1737–1743. doi: 10.20546/ijcmas.2018.701.210
Wang, F., Jiang, L., Yang, Q., Han, F., Chen, S., Pu, S., et al. (2011). Prevalence and antimicrobial susceptibility of major foodborne pathogens in imported seafood. J. Food Prot. 74, 1451–1461. doi: 10.4315/0362-028X.JFP-11-146
WHO Critically Important Antimicrobials for Human Medicine 6th (2019). Available at: https://www.who.int/publications/i/item/9789241515528 (Accessed October 12, 2022).
Zanetti, S., Spanu, T., Deriu, A., Romano, L., Sechi, L. A., and Fadda, G. (2001). In vitro susceptibility of Vibrio spp. isolated from the environment. Int. J. Antimicrob. Agents 17, 407–409. doi: 10.1016/S0924-8579(01)00307-7
Zankari, E., Allesøe, R., Joensen, K. G., Cavaco, L. M., Lund, O., and Aarestrup, F. M. (2017). PointFinder: a novel web tool for WGS-based detection of antimicrobial resistance associated with chromosomal point mutations in bacterial pathogens. J. Antimicrob. Chemother. 72, 2764–2768. doi: 10.1093/jac/dkx217
Zhang, M., Chen, L., Ye, C., and Yu, X. (2018). Co-selection of antibiotic resistance via copper shock loading on bacteria from a drinking water bio-filter. Environ. Pollut. 233, 132–141. doi: 10.1016/j.envpol.2017.09.084
Zhang, Y. B., Li, Y., and Sun, X. L. (2011). Antibiotic resistance of bacteria isolated from shrimp hatcheries and cultural ponds on Donghai Island, China. Mar. Pollut. Bull. 62, 2299–2307. doi: 10.1016/j.marpolbul.2011.08.048
Zhang, X.-X., Zhang, T., and Fang, H. H. P. (2009). Antibiotic resistance genes in water environment. Appl. Microbiol. Biotechnol. 82, 397–414. doi: 10.1007/s00253-008-1829-z
Zhang, Y., Zheng, Z., Chan, E. W.-C., Dong, N., Xia, X., and Chen, S. (2018). Molecular characterization of qnrVC genes and their novel alleles in Vibrio spp. isolated from food products in China. Antimicrob. Agents Chemother. 62, e00529–e00518. doi: 10.1128/AAC.00529-18
Zheng, D., Yin, G., Liu, M., Chen, C., Jiang, Y., Hou, L., et al. (2021). A systematic review of antibiotics and antibiotic resistance genes in estuarine and coastal environments. Sci. Total Environ. 777:146009. doi: 10.1016/j.scitotenv.2021.146009
Glossary
Keywords: Vibrio spp., Enterococcus spp., foodborne, shrimp, antimicrobial resistance, whole genome sequencing, resistance genes
Citation: Hirshfeld B, Lavelle K, Lee KY, Atwill ER, Kiang D, Bolkenov B, Gaa M, Li Z, Yu A, Li X and Yang X (2023) Prevalence and antimicrobial resistance profiles of Vibrio spp. and Enterococcus spp. in retail shrimp in Northern California. Front. Microbiol. 14:1192769. doi: 10.3389/fmicb.2023.1192769
Edited by:
Patrick Rik Butaye, Ghent University, BelgiumReviewed by:
Sivalingam Periyasamy, National Research Council (CNR), ItalyYadouleton Anges, Carlos III Health Institute (ISCIII), Spain
Copyright © 2023 Hirshfeld, Lavelle, Lee, Atwill, Kiang, Bolkenov, Gaa, Li, Yu, Li and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xunde Li, eGRsaUB1Y2RhdmlzLmVkdQ==; Xiang Yang, eGNyeWFuZ0B1Y2RhdmlzLmVkdQ==
†These authors have contributed equally to this work
 Brady Hirshfeld
Brady Hirshfeld Kurtis Lavelle
Kurtis Lavelle Katie Yen Lee
Katie Yen Lee Edward Robert Atwill3
Edward Robert Atwill3 Xunde Li
Xunde Li Xiang Yang
Xiang Yang