
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 11 May 2023
Sec. Virology
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1186869
Although the natural hosts of avian influenza viruses (AIVs) are wild birds, multiple subtypes of AIVs have established epidemics in numerous mammals due to their cross-species spillover. Replication and evolution in intermedia mammalian hosts may facilitate AIV adaptation in humans. Because of their large population and intimacy with humans, dogs could act as such an intermedia host. To monitor the epidemiology of canine influenza viruses (CIVs) in Liaoning, China, we performed three surveillances in November 2018, March 2019, and April 2019. Five H3N2 and seven novel H3N6 CIVs had been isolated. Since the N6 neuraminidase (NA) genes were clustered with the H5N6 AIV, there is a high possibility that these H3N6 CIVs were generated from a H3N2 CIVs and H5N6 AIVs reassortment case. In addition, the H3N6 CIV showed increased mammalian adaptation ability compared to all the H3N2 strains in both in vitro and in vivo studies. Even though isolated 3 months later, the March 2019 isolated H3N2 viruses replicated more efficiently than the November 2018 isolated viruses. Our study indicated that H3 CIVs were undergoing an evolution process, through both genetic mutations and gene reassortment, at an incredible speed.
Influenza A virus (IAV), a member of the family Orthomyxoviridae, contains eight single-stranded negative-sense RNA segments that encode more than 15 viral proteins (Liu et al., 2020). A total of 18 hemagglutinin (HA) subtypes and 11 neuraminidase (NA) subtypes have been identified in IAV, except H17N10 and H18N11, all of which emerge in avian strains. The natural reservoirs of IAVs are migratory waterfowl, but certain IAV lineages may infect diverse species, including domestic poultry, humans, and other mammals (Krammer et al., 2018).
The H3N2 influenza virus is a major cause of human epidemics annually. H3N2 viruses are also commonly detected in other animals, where they undergo adaptive evolution that facilitates cross-species transmission. The H3N2 avian influenza virus (AIV) can infect and transmit among mammals, posing a potential pandemic threat (Guan et al., 2019; Zhang et al., 2021). H3N2 AIV entered the canine population and formed the canine influenza virus (CIV) lineage in around 2006. Since then, H3N2 CIVs have been isolated in Thailand (Bunpapong et al., 2014) and other areas of Southeast Asia (Sun et al., 2017). In 2015, H3N2 CIVs emerged in the United States (Watson et al., 2017). A new antigenic clade of H3N2 CIV emerged in China in 2016 (Lyu et al., 2019) and became dominant since then (Li et al., 2021).
Dogs can act as a “mixing vessel” of the influenza virus, because they express both human-like and avian-like receptors (Wasik et al., 2017). A large number of studies have shown that dogs could be naturally infected by different origins of influenza viruses, such as human H1N1 and H3N2, swine H1N1, avian H5N1, H5N2, H6N1, H9N2, and H10N8 viruses (Lin et al., 2015; Sun et al., 2017). When multiple influenza virus strains co-infect a dog, a novel reassortant could be generated by gene segments exchanging between influenza viruses. Novel CIVs had been derived in this way from the recombination between H3N2 CIV and the 2009 pandemic H1N1 virus (pdm/09) and swine H3N2 and swine H1N1 viruses (Song et al., 2012; Chen et al., 2018). Given the closeness of dogs and humans, the epidemic and evolutionary statuses of CIVs deserve attention.
Liaoning Province has the largest pet dog breeding and trading base in China. Numerous dogs are distributed daily from Liaoning to other provinces of China. In this study, we investigated the epidemic situation of CIVs and analyzed the biological properties of different isolates in Liaoning.
Mice experiments were approved for use by the Animal Experimentation and Laboratory Animal Welfare Committee of Shenyang Agricultural University (No. 202106009, approved on 8 March 2021).
Active CIV surveillance studies were conducted from November 2018 to April 2019. Dog nasal swab samples were collected from a large-scale animal shelter in Liaoning Province, China. The swab samples were separately put into 1 mL of phosphate-buffered saline (PBS) containing penicillin (2,000 U/mL) and streptomycin (2,000 μg/mL). Viral RNA was extracted from each sample using the TIAN amp Virus DNA/RNA Kit (TIANGEN Biotech, Beijing, China). Reverse transcription-PCR (RT-PCR) was performed using primers targeting the influenza virus matrix (M) gene (the primer sequences are available upon request). The PCR-positive samples were inoculated into 10-day-old specific pathogen-free embryonated chicken eggs (Harbin Weike Biotechnology Development Company, Harbin, China). The inoculated eggs were incubated at 37°C for 72 h. Then, the allantoic fluid was collected for the hemagglutination test, and the positive ones were reserved at −80°C as viral stock.
The isolated viruses were applied to whole-genome sequencing and phylogenetic analysis. The viral RNA was extracted using the TIAN amp Virus DNA/RNA Kit (TIANGEN Biotech, Beijing, China). The RT-PCR test was carried out with primers for each viral gene (the primer sequences are available upon request). Viral genomes were sequenced by Comate Bioscience Company Limited (Changchun, Jilin, China). For phylogenetic analysis, reference sequences of canine, avian, swine, equine, mink, and human influenza viruses were downloaded from three main influenza virus databases: the NCBI influenza virus database, the GISAID influenza database, and the Influenza Research database (Supplementary Table S1). The unrooted tree was constructed using the maximum-likelihood (ML) method with MEGA X (https://www.megasoftware.net) with 1,000 bootstrapreplicates.
To evaluate the replicative ability of the CIVs in vitro, Madin–Darby canine kidney (MDCK) cells were infected with 102 50% tissue culture infectious doses (TCID50) of each virus with volumes of 100 μl. The infected cells were incubated at 37°C for 72 h with Dulbecco's modified Eagle's medium containing 1 μg/mL of tolylsulfonyl phenylalanyl chloromethyl ketone (TPCK)-treated trypsin. The supernatant was collected every 12 h and titrated in MDCK cells.
To evaluate the replicative ability of the CIVs in vivo, eight 6-week-old and three 4-week-old female BALB/c mice (Changsheng Biotechnology, Liaoning, China) were inoculated intranasally with each CIV as described previously (Shi et al., 2017). Each mouse was infected intranasally with 106 50% egg infectious doses (EID50) of each virus with volumes of 50 μL. Three mice from each group were euthanized on day 3 post-infection (p.i.), and their nasal turbinates, lungs, brains, spleens, and kidneys were collected for viral titration. The remaining five mice were observed for clinical symptoms. Their body weights were measured every day until day 14 p.i. Five 6-week-old mice were infected intranasally with a 50 μl volume of PBS as the mock groups.
Receptor-binding property is a key determinant of the mammalian adaption of the influenza virus (Gao et al., 2009; Zhang et al., 2012). Hemagglutination assays using resialylated chicken red blood cells (cRBCs) and sheep red blood cells (sRBCs) were performed to analyze the receptor-binding property of the CIVs. The surface of cRBCs expresses both human-like α2,6 and avian-like α2,3 sialic acid receptors (Ito et al., 1997), while sRBCs only contain avian-like α2,3 sialic acid receptors (Medeiros et al., 2001). The α2,3 sialic acids were removed from the cRBCs' surface by incubating with α2,3-sialidase (Takara, Dalian, Liaoning, China). The desialylation cRBCs were generated by incubating with Vibrio cholerae neuraminidase (VCNA, Roche, San Francisco, CA, United States). The desialylation cRBCs were used as the negative control. The A/Sichuan/1/2009(H1N1) and A/chicken/Hebei/3/2013(H5N2), which bind to α2,6-cRBCs and α2,3-cRBCs, respectively (Shi et al., 2017), were used as controls.
Statistical analysis between different groups was performed using a one-way analysis of variance (ANOVA) test via the GraphPad Prism version 8.0 (Graph Pad Software Inc., CA, USA). A difference with a value of p < 0.05 was considered statistically significant, while p < 0.01 was considered highly statistically significant.
Three active CIV surveillance studies were conducted on 26 November 2018, 2 March 2019, and 30 April 2019. A total of 534 nasal swab samples were collected from dogs kept in a large-scale animal shelter in Liaoning Province that houses multiple kinds of animals including dogs, cats, swine, and birds. The swab samples were tested for the influenza virus M gene by RT-PCR. In total, 60 samples were positive. The CIV-positive rate in this animal shelter was 11.24% (Table 1). The positive samples were inoculated into embryonated specific-pathogen-free eggs for CIV isolation, which resulted in 12 CIVs being isolated.
The genome sequence of the 12 CIVs was determined by using Sanger sequencing, which revealed that of the 12 isolates, five were H3N2 (isolated on 26 November 2018 and 2 March 2019) and seven were H3N6 (isolated on 30 April 2019) CIVs (Table 2). Their nucleotide sequences have been deposited in GenBank (Accession Number: MZ323741-MZ323748, MZ323750-MZ323757, MZ323776-MZ323807, and MZ323813-MZ323860). Phylogenetic trees were constructed with MEGA X software using the maximum-likelihood (ML) method with 1000 bootstrap replications based on the sequences of the open reading frames. Except for the NA genes of the H3N6 viruses, each genome segment of the H3N2 and H3N6 viruses shared high genetic similarity (99.3%−100%) at the nucleotide level and grouped together in each phylogenetic tree. All HA genes clustered with the antigenic variants that emerged in 2016 (Figure 1). The N2 NA genes clustered with H3N2 CIVs were isolated in China, South Korea, and North America after 2016 (Figure 2). The six internal genes of all 12 isolates, the basic polymerase 2 (PB2), basic polymerase1 (PB1), acidic polymerase (PA), nucleoprotein (NP), matrix (M), and non-structural protein (NS) genes also clustered into H3N2 CIVs groups that were isolated after 2016 (Figures 3–8).

Figure 1. Phylogenetic analysis of the HA genes of H3N2 and H3N6 CIVs. It is based on nucleotide positions 30–1,730 for HA. Isolates in this study are colored red. The scale bar indicates the number of nucleotide substitutions per site.
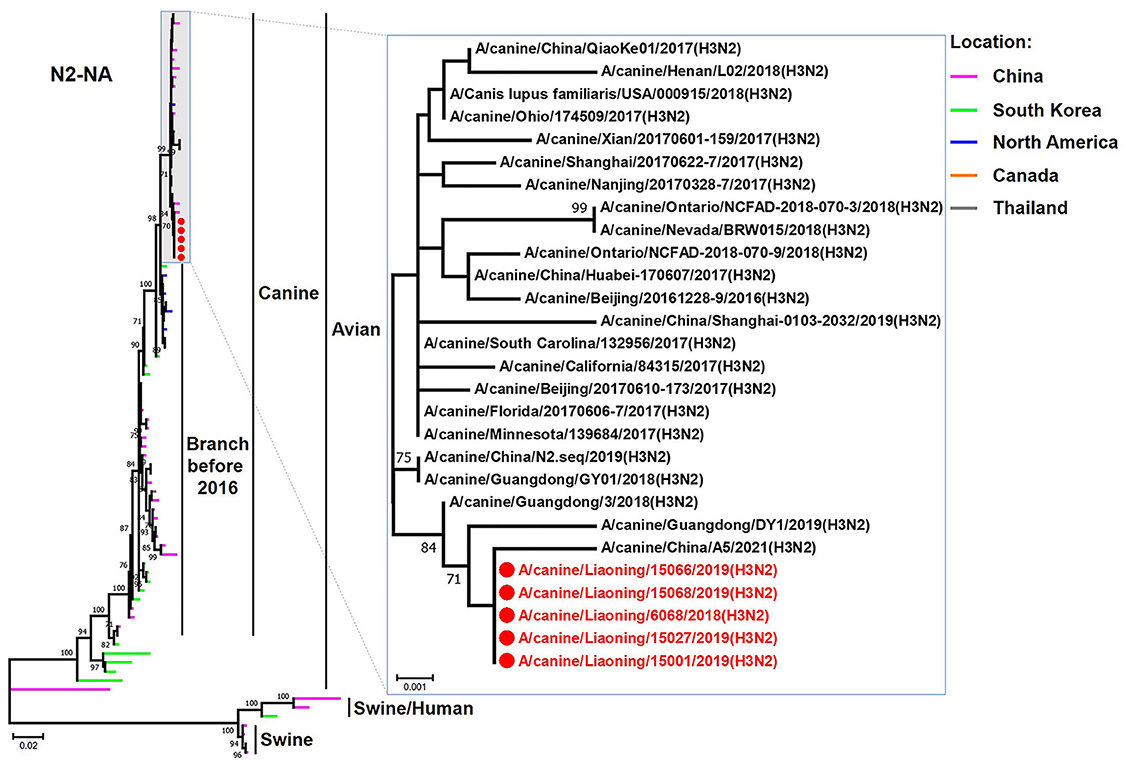
Figure 2. Phylogenetic analysis of the NA genes of H3N2 CIVs. It is based on nucleotide positions 20–1,429 for NA. Isolates in this study are colored red. The scale bar indicates the number of nucleotide substitutions per site.
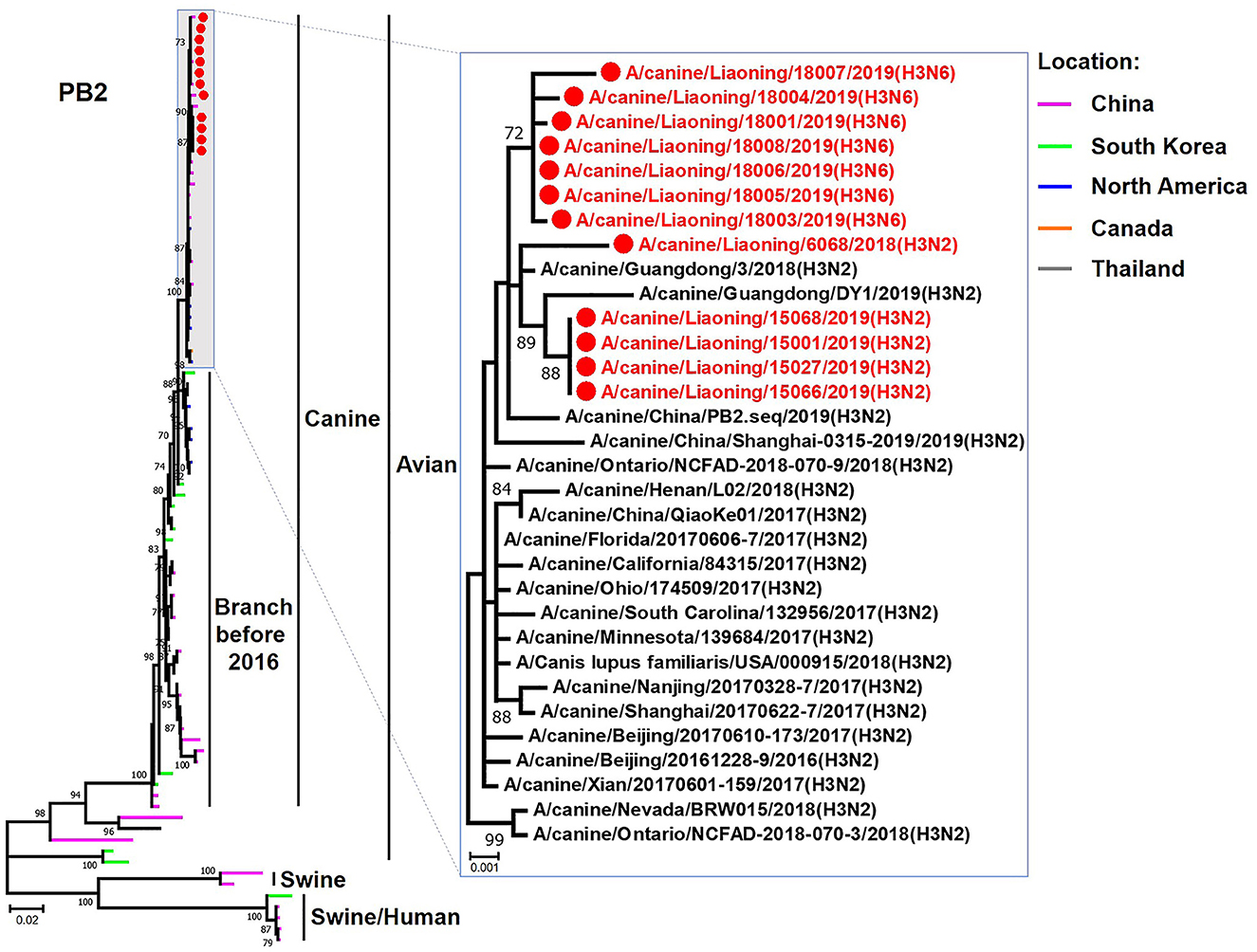
Figure 3. Phylogenetic analysis of the PB2 genes of H3N2 and H3N6 CIVs. It is based on nucleotide positions 28–2,307 for PB2. Isolates in this study are colored red. The scale bar indicates the number of nucleotide substitutions per site.
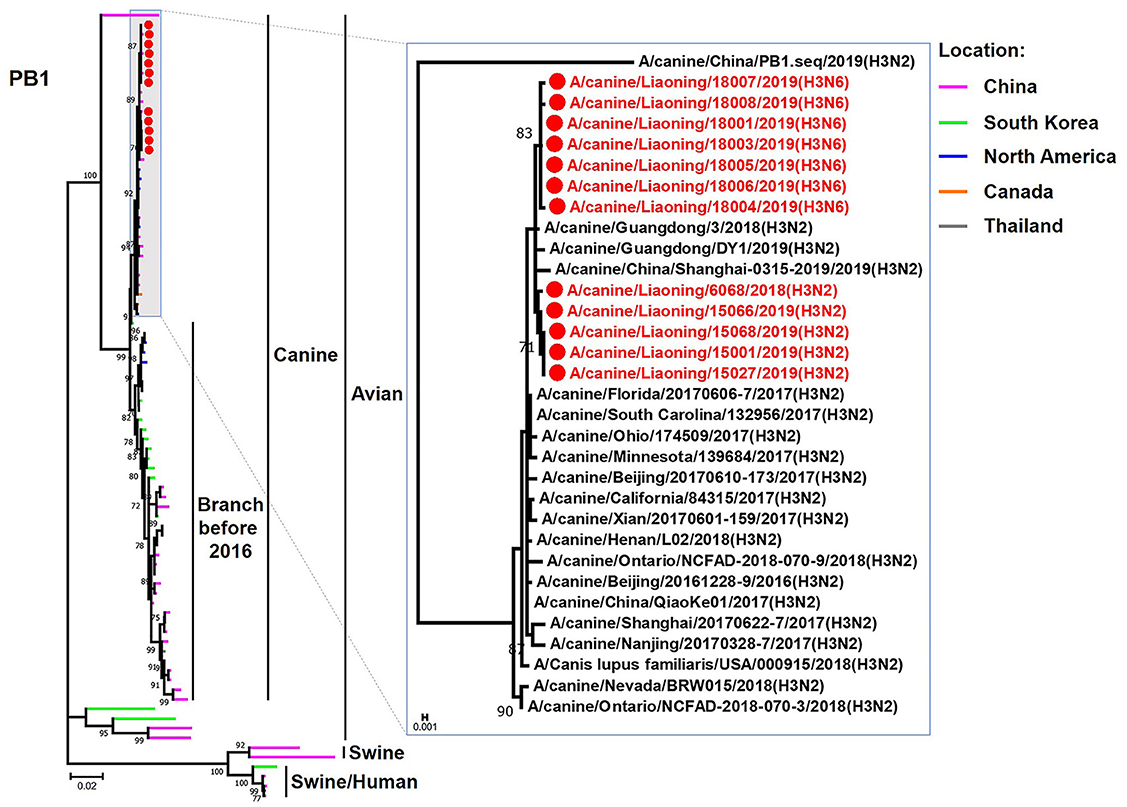
Figure 4. Phylogenetic analysis of the PB1 genes of H3N2 and H3N6 CIVs. It is based on nucleotide positions 25–2,298 for PB1. Isolates in this study are colored red. The scale bar indicates the number of nucleotide substitutions per site.
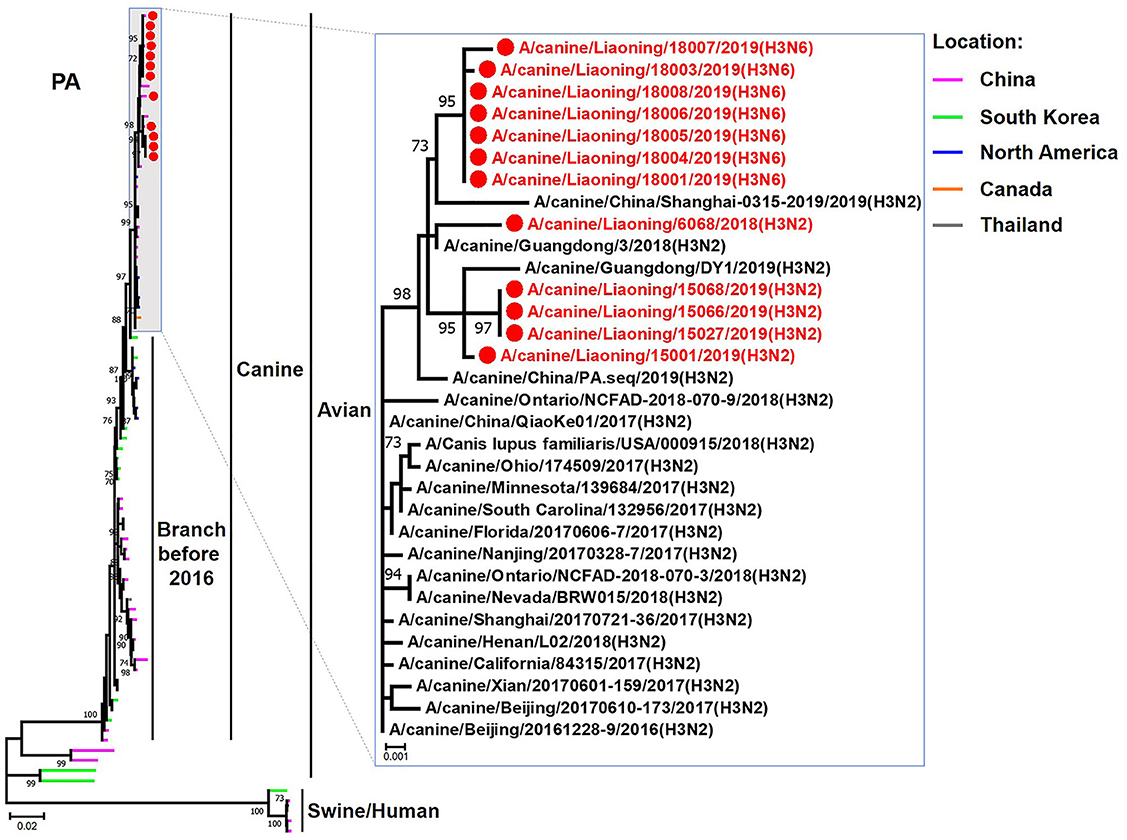
Figure 5. Phylogenetic analysis of the PA genes of H3N2 and H3N6 CIVs. It is based on nucleotide positions 25–2,175 for PA. Isolates in this study are colored red. The scale bar indicates the number of nucleotide substitutions per site.
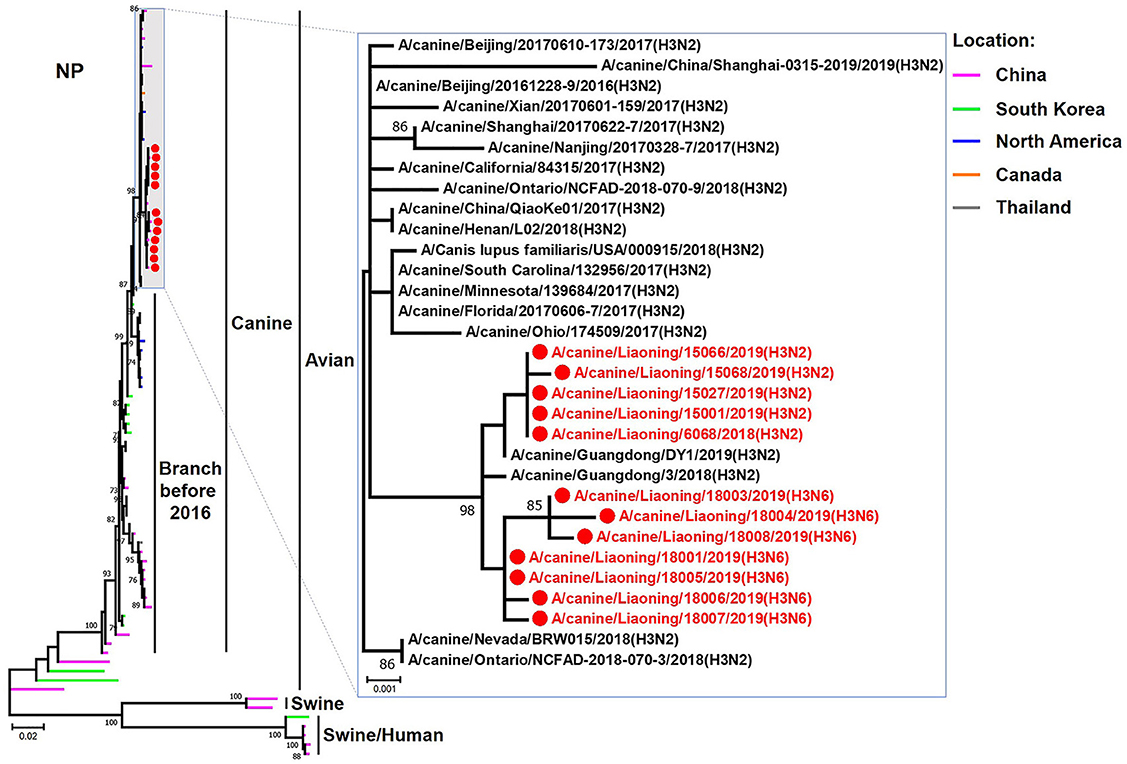
Figure 6. Phylogenetic analysis of the NP genes of H3N2 and H3N6 CIVs. It is based on nucleotide positions 46–1,542 for NP. Isolates in this study are colored red. The scale bar indicates the number of nucleotide substitutions per site.
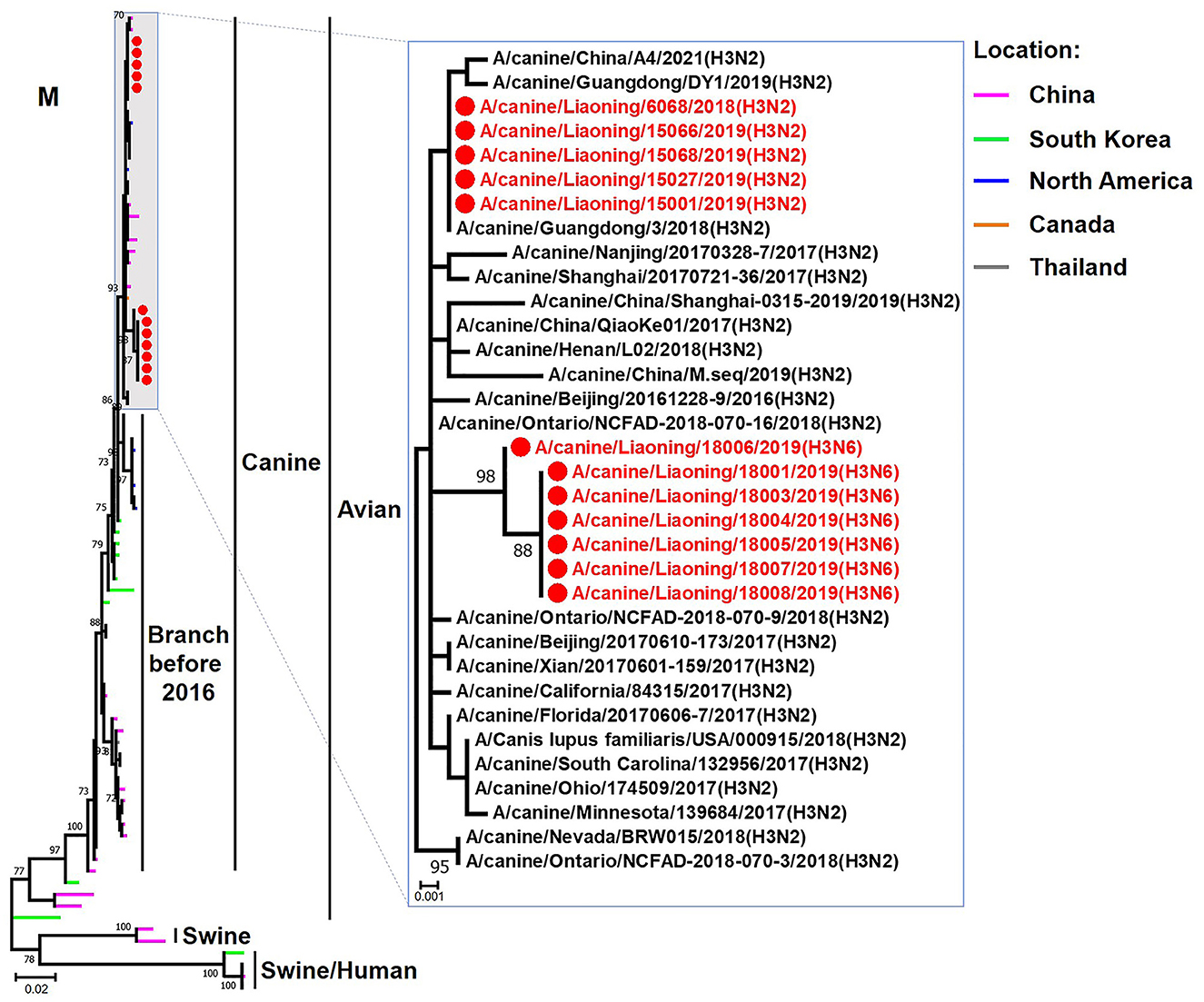
Figure 7. Phylogenetic analysis of the M genes of H3N2 and H3N6 CIVs. It is based on nucleotide positions 26–1,007 for M. Isolates in this study are colored red. The scale bar indicates the number of nucleotide substitutions per site.
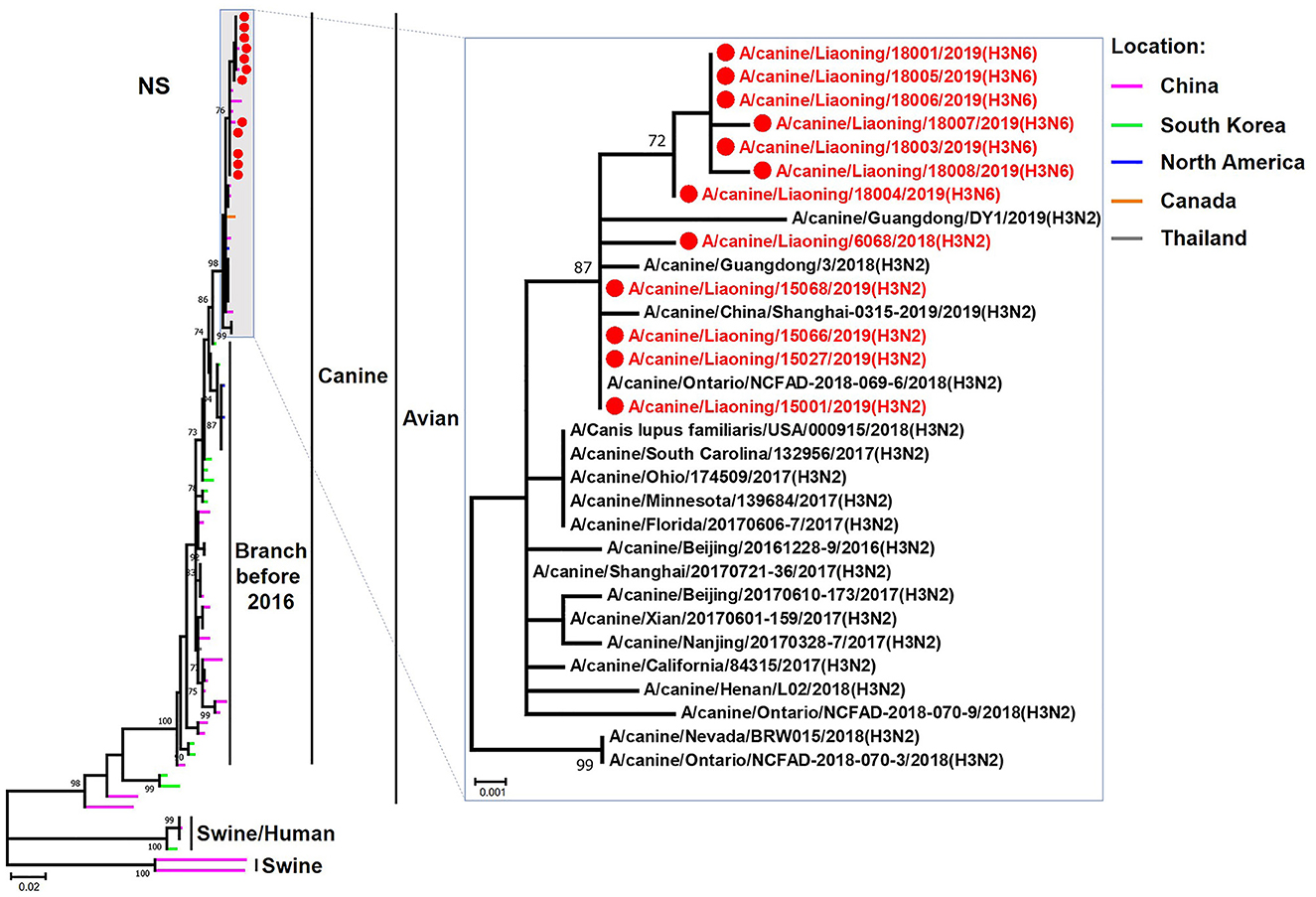
Figure 8. Phylogenetic analysis of the NS genes of H3N2 and H3N6 CIVs. It is based on nucleotide positions 27–864 for NS. Isolates in this study are colored red. The scale bar indicates the number of nucleotide substitutions per site.
The NA genes of the seven H3N6 CIVs shared 99.9%−100% genetic similarity at the nucleotide level and clustered with the H5N6 AIVs that were isolated in China between 2017 and 2018 (Figure 9). According to the genetic analysis results, the H3N6 CIVs represent a novel reassortant between H3N2 CIV and H5N6 AIV, probably generated from a dog co-infection case.
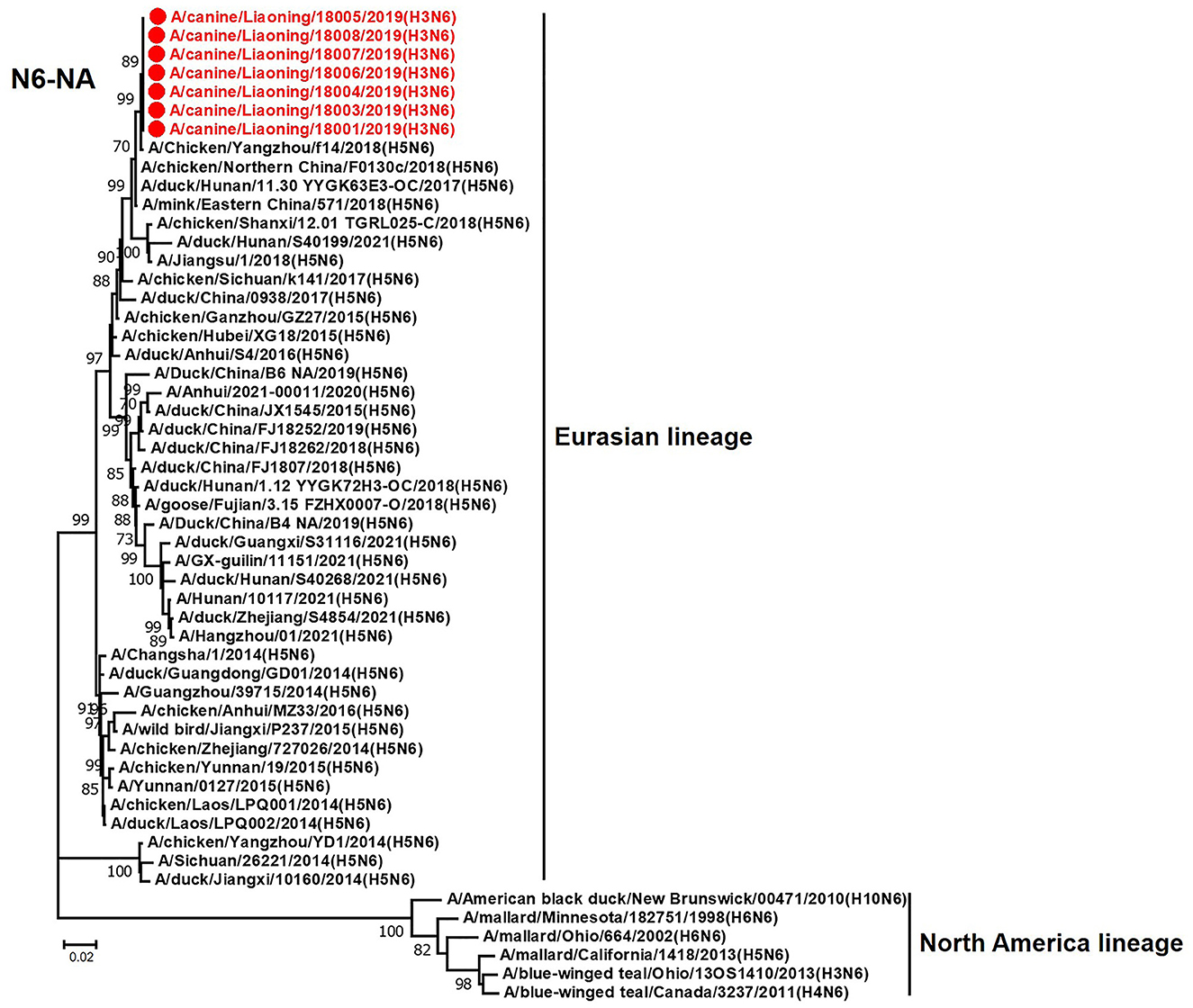
Figure 9. Phylogenetic analysis of the NA genes of H3N6 CIVs. It is based on nucleotide positions 19–1,398 for NA. Isolates in this study are colored red. The scale bar indicates the number of nucleotide substitutions per site.
To evaluate the replicative ability of the CIVs, we selected two H3N2 CIVs, A/Canine/Liaoning/6068/2018 (6068/18) and A/Canine/Liaoning/15001/2019 (15001/19), and one H3N6 virus, A/Canine/Liaoning/18005/2019 (18005/19). The MDCK cells were inoculated with 102 TCID50 of the three CIVs, and the supernatant was collected every 12 h for viral titration. Although isolated within 5 months of each other, the three CIVs showed different biologic properties. In general, the H3N6 virus replicated better than the H3N2 virus. The viral titers of 18005/19 were 10–100 times higher than those of 15001/19 at each timepoint. But 6068/18 the only virus isolated in 2018, did not replicate well in MDCK cells (Figure 10).
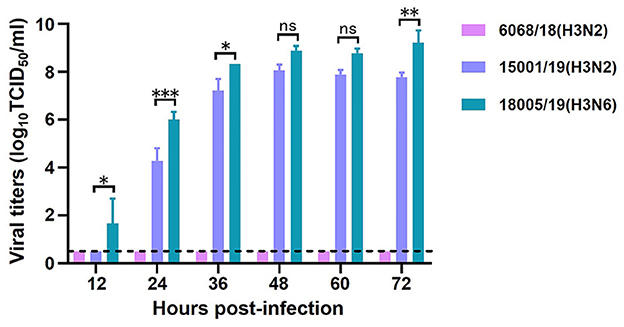
Figure 10. In vitro replication ability of the two H3N2 and one H3N6 CIVs. Growth kinetics in MDCK cells. The means of three repeats are shown, with error bars indicating the SD. Statistical analysis between different groups was performed by using a one-way analysis of variance (ANOVA) test. *(p < 0.05), **(p < 0.01), ***(p < 0.001), and ns (p > 0.05).
We used mice as a model to evaluate the adaptation of the three CIVs in mammalian hosts. Eight 6-week-old BALB/c mice were intranasally inoculated with 106 EID50 of each virus. Three mice from each group were euthanized on day 3 p.i., and their brains, nasal turbinates, lungs, spleens, and kidneys were collected for viral titration. The remaining mice were observed for clinical signs and body weight change until day 14 p.i. No mice exhibited significant symptoms or morbidity during the whole experiment. None of the strains were detected in the brain, spleen, or kidney of the mice (data not shown). The 15001/19 and 18005/19 viruses replicated in both the upper and lower respiratory tract of the mice (Figure 11A), although 15001/19 was only detected in one mouse lung, whereas 18005/19 was detected in the lungs of all three mice. The 6068/18 virus did not replicate well in mice (Figure 11A). Moderate, transient weight loss was observed in each inoculated group during a short period (Figure 11B).
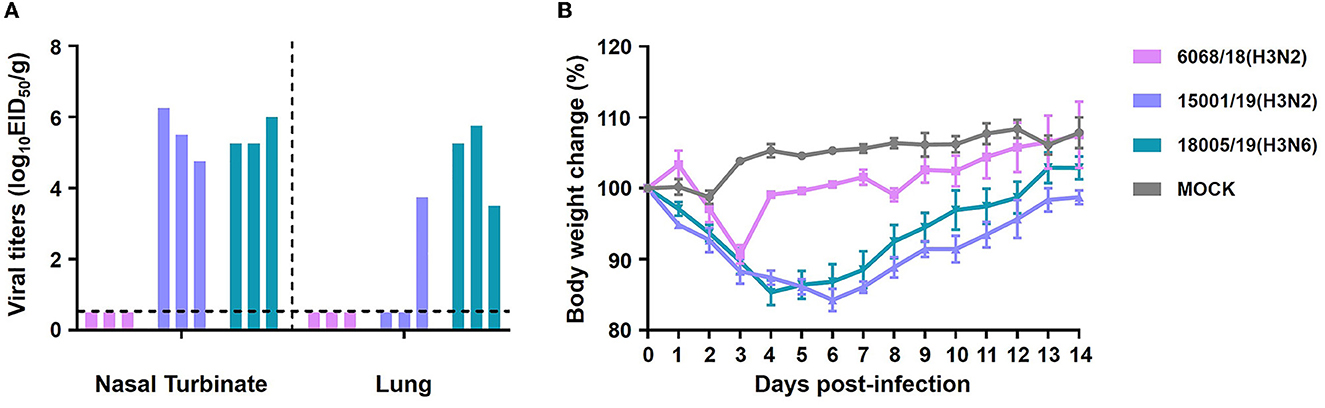
Figure 11. In vivo replication ability of the two H3N2 and one H3N6 CIVs. Replication ability in 6-week-old mice. (A) Mice were euthanized on day 3 p.i., and their nasal turbinates and lungs were collected and titrated for virus infectivity in eggs. (B) Mice were monitored for body weight loss throughout the observation period for 14 days.
We then repeated the replication study in younger animals. Three 4-week-old mice were infected with the three viruses according to the same procedure. Viral titration results indicated that the 15001/19 and 18005/19 viruses replicated more efficiently in the respiratory tracts, and 6068/18 could replicate in the upper respiratory tracts of the younger mice (Figure 12). Both 6068/18 and 15001/19 increased replication ability in younger mice. The immune system development variances of the two age groups of mice might be the reason for the 6868/18 and 15001/19 different replication phenotypes. These findings indicate that 6068/18 isolated on 26 November 2018, had not fully adapted to the mice yet, but 15001/19 had, even though 15001/19 was isolated 3 months after 6068/18.
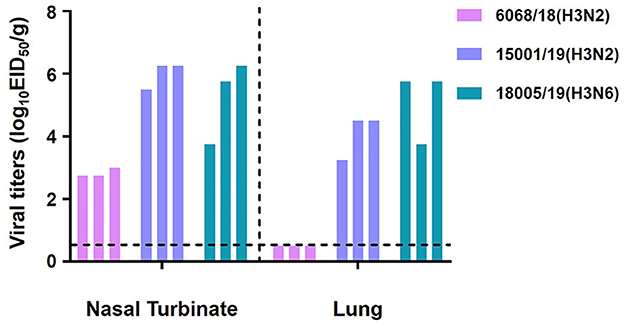
Figure 12. In vivo replication ability of the two H3N2 and one H3N6 CIVs. Replication ability in 4-week-old mice. Mice were euthanized on day 3 p.i., and their nasal turbinates and lungs were collected and titrated for virus infectivity in eggs.
The receptor-binding property is important for influenza virus mammalian adaption. We, therefore, also evaluated the receptor-binding properties of the three CIVs, as described previously (Ito et al., 1997; Medeiros et al., 2001). The assay results demonstrated that all three CIVs bound preferentially to the avian-like receptor, α2,3-linked sialic acid (Figure 13).
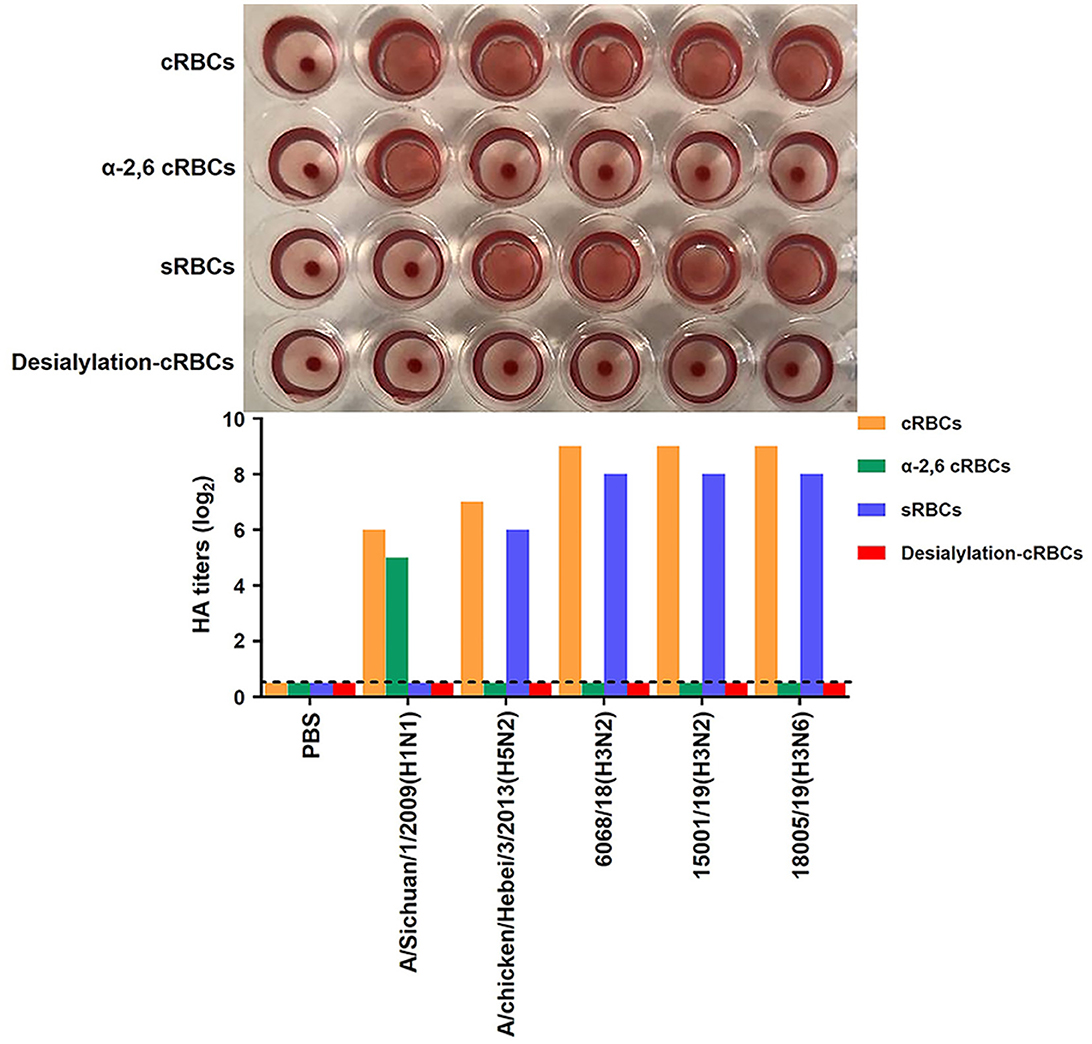
Figure 13. Receptor-binding analysis of the two H3N2 and one H3N6 CIVs. Agglutination activities of the H3N2 and H3N6 CIVs with various red blood cells. Four types of red blood cells: cRBCs (1% chicken red blood cells); sRBCs (1% sheep red blood cells); α-2,6-cRBCs (1% chicken red blood cells treated with α2,3-Sialidase); desialylation-cRBCs (1% chicken red blood cells treated with VCNA). The HA titer showed agglutination activities of 6068/18, 15001/19, and 18005/19 for the four types of red blood cells. PBS was a negative control group, whereas A/Sichuan/1/2009(H1N1) and A/chicken/Hebei/3/2013(H5N2) were the positive groups binding to α-2,6-cRBCs and sRBCs, respectively.
In this study, we performed active CIV surveillance in Liaoning Province for the 2018 to 2019 flu season. Twelve CIVs were isolated, including five H3N2 and seven H3N6 viruses. We systematically analyzed the genetic and biological properties of these CIVs. The phylogenetic analysis of the HA genes of 12 CIVs demonstrated that isolates in this study were all clustered into the antigenic variants emerging in 2016 (Lyu et al., 2019). NA genes of the H3N6 CIVs were clustered into H5N6 AIV groups, which were isolated in China from 2017 to 2018. There is a high possibility that the novel H3N6 CIVs are generated from H3N2 CIV and H5N6 AIV reassortment cases. H5N6 AIVs have circulated in China since 2014, and trans-species infections and reassortant cases have been reported when the H5N6 circulated in domestic poultry (Yu et al., 2015; Gu et al., 2022). Previous studies demonstrated that novel reassortant H3N6 viruses had been isolated from migratory birds and domestic birds, which contained some gene segments from the dominant H5N6 viruses (Li et al., 2020). To our knowledge, this is the first report that avian H5N6 viruses can recombine with H3N2 CIVs.
This study indicated that CIVs are evolving and adapting to mammalian hosts at an incredible speed. The H3N2 CIV isolated in November 2018 showed limited replicative ability in vitro and in vivo, but 3 months later, increased replication was detected in March 2019 H3N2 CIV isolates. Moreover, 1 month later, novel reassortant H3N6 CIVs appeared in April 2019. The H3N6 reassortant possessed enhanced mammalian adaption ability compared with H3N2 CIVs.
Genetic mutation and reassortment were both observed among our CIVs isolates. There were only five amino acid differences between 6068/18 and 15001/19, PB2-A292T, PB2-K578E, PA-I561M, HA-G271D (H3 numbering), and NS1-G142E. The effects of these substitutions on the biological properties of these viruses require further exploration. The alignment of the N6 NA stalk showed that there was an 11 amino acid deletion at positions 58 to 68, which had been identified in H5N6 AIVs. According to the in vitro and in vivo replication study results, it seemed that the H3N2 HA and H5N6 NA genes of the novel CIV are more functionally and optimally cooperative than the HA and NA genes of H3N2 CIVs. Genome reassortment between different subtypes of flu strains is an important driving force for the evolution of influenza viruses and the sources of potential pandemic strains. Three out of four human influenza pandemics of the last century were caused by reassortant viruses (Li and Chen, 2014).
As companion animals, dogs are closely related to humans, and their potential threat to public health deserves high attention. In this study, we found that H3 CIVs were undergoing an evolution process at an incredible speed and provided important information about the evolutionary status of H3 CIVs. Further surveillance and risk analysis of CIVs should be performed.
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
The animal study was reviewed and approved by the mice experiments were approved for such use by the Animal Experimentation and Laboratory Animal Welfare Committee of Shenyang Agricultural University (No. 202106009, approved on 8 March 2021).
BM performed the experiments, analyzed the data, and wrote the first draft of the manuscript. HL, YF, and DZ collected the canine samples. CF and WG assisted with virus isolation and animal experiments. HC and YZ guided the experiments, conceived the study, analyzed the data, and finalized the manuscript. All authors contributed to the article and approved the submitted version.
This research was financed by the National Key Research and Development Program of China (2021YFD1800200), the National Natural Science Foundation of China (32170539 and 32000357), the Key Research and Development Program of Liaoning (2020JH2/10200035), and the Liao Ning Revitalization Talents Program (XLYC2007114).
We thank Susan Watson for editing this manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1186869/full#supplementary-material
Bunpapong, N., Nonthabenjawan, N., Chaiwong, S., Tangwangvivat, R., Boonyapisitsopa, S., Jairak, W., et al. (2014). Genetic characterization of canine influenza A virus (H3N2) in Thailand. Virus Genes 48, 56–63. doi: 10.1007/s11262-013-0978-z
Chen, Y., Trovão, N. S., Wang, G., Zhao, W., He, P., Zhou, H., et al. (2018). Emergence and evolution of novel reassortant influenza A viruses in canines in southern China. mBio 9, e00909–18. doi: 10.1128/mBio.00909-18
Gao, Y., Zhang, Y., Shinya, K., Deng, G., Jiang, Y., Li, Z., et al. (2009). Identification of amino acids in HA and PB2 critical for the transmission of H5N1 avian influenza viruses in a mammalian host. PLoS Pathog. 5, e1000709. doi: 10.1371/journal.ppat.1000709
Gu, W., Shi, J., Cui, P., Yan, C., Zhang, Y., Wang, C., et al. (2022). Novel H5N6 reassortants bearing the clade 2.3.4.4b HA gene of H5N8 virus have been detected in poultry and caused multiple human infections in China. Emerg. Microbes Infect. 11, 1174–1185. doi: 10.1080/22220222063076
Guan, L., Shi, J., Kong, X., Ma, S., Zhang, Y., Yin, X., et al. (2019). H3N2 avian influenza viruses detected in live poultry markets in China bind to human-type receptors and transmit in guinea pigs and ferrets. Emerg. Microbes Infect. 8, 1280–1290. doi: 10.1080/22220191660590
Ito, T., Suzuki, Y., Mitnaul, L., Vines, A., Kida, H., Kawaoka, Y., et al. (1997). Receptor specificity of influenza A viruses correlates with the agglutination of erythrocytes from different animal species. Virology 227, 493–499. doi: 10.1006/viro.1996.8323
Krammer, F., Smith, G. J. D., Fouchier, R. A. M., Peiris, M., Kedzierska, K., Doherty, P. C., et al. (2018). Influenza. Nat. Rev. Dis. Primers 4, 3. doi: 10.1038/s41572-018-0002-y
Li, C., and Chen, H. (2014). Enhancement of influenza virus transmission by gene reassortment. Curr. Top. Microbiol. Immunol. 385, 185–204. doi: 10.1007/82_2014_389
Li, R., Zhang, T., Xu, J., Chang, J., and Xu, B. (2020). Isolation of two novel reassortant H3N6 avian influenza viruses from long-distance migratory birds in Jiangxi Province, China. Microbiologyopen 9, e1060. doi: 10.1002/mbo3.1060
Li, Y., Zhang, X., Liu, Y., Feng, Y., Wang, T., Ge, Y., et al. (2021). Characterization of canine influenza virus A (H3N2) circulating in dogs in China from 2016 to 2018. Viruses 13, 2279. doi: 10.3390/v13112279
Lin, H. T., Wang, C. H., Chueh, L. L., Su, B. L., and Wang, L. C. (2015). Influenza A(H6N1) virus in dogs, Taiwan. Emerg. Infect. Dis. 21, 2154–2157. doi: 10.3201/eid2112.141229
Liu, Y., Fu, C., Lu, G., Luo, J., Ye, S., Ou, J., et al. (2020). Comparison of Pathogenicity of different infectious doses of H3N2 canine influenza virus in dogs. Front. Vet. Sci. 7, 580301. doi: 10.3389/fvets.2020.580301
Lyu, Y., Song, S., Zhou, L., Bing, G., Wang, Q., Sun, H., et al. (2019). Canine influenza virus A(H3N2) clade with antigenic variation, China, 2016–2017. Emerg. Infect. Dis. 25, 161–165. doi: 10.3201/eid2501.171878
Medeiros, R., Escriou, N., Naffakh, N., Manuguerra, J., and van der Werf, S. (2001). Hemagglutinin residues of recent human A (H3N2) influenza viruses that contribute to the inability to agglutinate chicken erythrocytes. Virology 289, 74–85. doi: 10.1006/viro.2001.1121
Shi, J., Deng, G., Kong, H., Gu, C., Ma, S., Yin, X., et al. (2017). H7N.9 virulent mutants detected in chickens in China pose an increased threat to humans. Cell Res. 27, 1409–1421. doi: 10.1038/cr.2017.129
Song, D., Moon, H. J., An, D. J., Jeoung, H. Y., Kim, H., Yeom, M. J., et al. (2012). A novel reassortant canine H3N1 influenza virus between pandemic H1N1 and canine H3N2 influenza viruses in Korea. J. Gen. Virol. 93, 551–554. doi: 10.1099/vir.0.037739-0
Sun, H., Blackmon, S., Yang, G., Waters, K., Li, T., Tangwangvivat, R., et al. (2017). Zoonotic risk, pathogenesis, and transmission of avian-origin H3N2 canine influenza virus. J. Virol. 91, e00637–17. doi: 10.1128/JVI.00637-17
Wasik, B. R., Barnard, K. N., Ossiboff, R. J., Khedri, Z., Feng, K. H., Yu, H., et al. (2017). Distribution of O-acetylated sialic acids among target host tissues for influenza virus. mSphere 2, e00379–16. doi: 10.1128/mSphere.00379-16
Watson, C. E., Bell, C., and Toohey-Kurth, K. (2017). H3N.2 canine influenza virus infection in a dog. Vet. Pathol. 54, 527–530. doi: 10.1177/0300985816681411
Yu, Z., Gao, X., Wang, T., Li, Y., Li, Y., Xu, Y., et al. (2015). Fatal H5N6 avian influenza virus infection in a domestic cat and wild birds in China. Sci. Rep. 5, 10704. doi: 10.1038/srep10704
Zhang, Y., Zhang, Q., Gao, Y., He, X., Kong, H., Jiang, Y., et al. (2012). Key molecular factors in hemagglutinin and PB2 contribute to efficient transmission of the 2009 H1N1 pandemic influenza virus. J. Virol. 86, 9666–9674. doi: 10.1128/JVI.00958-12
Keywords: canine influenza virus, H3N2, H5N6, reassortant, replicative ability
Citation: Meng B, Li H, Feng C, Guo W, Feng Y, Zhu D, Chen H and Zhang Y (2023) Emergence of a novel reassortant H3N6 canine influenza virus. Front. Microbiol. 14:1186869. doi: 10.3389/fmicb.2023.1186869
Received: 15 March 2023; Accepted: 10 April 2023;
Published: 11 May 2023.
Edited by:
Peirong Jiao, South China Agricultural University, ChinaReviewed by:
Hongliang Chai, Northeast Forestry University, ChinaCopyright © 2023 Meng, Li, Feng, Guo, Feng, Zhu, Chen and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hualan Chen, Y2hlbmh1YWxhbkBjYWFzLmNu; Ying Zhang, eWluZ3poYW5nMThAc3lhdS5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.