- 1College of Plant Protection, Hebei Agricultural University, Baoding, China
- 2Graduate School of Chinese Academy of Agricultural Sciences, Beijing, China
The gut microbiota is essential for the growth and development of insects, and the intestinal immune system plays a critical role in regulating the homeostasis of intestinal microorganisms and their interactions with pathogenic bacteria. Infection with Bacillus thuringiensis (Bt) can disrupt the gut microbiota of insects, but the regulatory factors governing the interaction between Bt and gut bacteria are not well understood. Uracil secreted by exogenous pathogenic bacteria can activate DUOX-mediated reactive oxygen species (ROS) production, which helps maintain intestinal microbial homeostasis and immune balance. To elucidate the regulatory genes involved in the interaction between Bt and gut microbiota, we investigate the effects of uracil derived from Bt on gut microbiota, and host immunity using a uracil deficient Bt strain (Bt GS57△pyrE) obtained by homologous recombination. We analyze the biological characteristics of the uracil deficient strain and found that the deletion of uracil in Bt GS57 strain changed the diversity of gut bacteria in Spodoptera exigua, as investigated using Illumina HiSeq sequencing. Furthermore, qRT-PCR analysis showed that compared with Bt GS57 (control), the expression of the SeDuox gene and the level of ROS were significantly decreased after feeding with Bt GS57△pyrE. Adding uracil to Bt GS57△pyrE restored the expression level of DUOX and ROS to a higher level. Additionally, we observed that PGRP-SA, attacin, defensin and ceropin genes were significant different in the midgut of S. exigua infected by Bt GS57 and Bt GS57△pyrE, with a trend of increasing first and then decreasing. These results suggest that uracil regulates and activates the DUOX-ROS system, affects the expression of antimicrobial peptide genes, and disturb intestinal microbial homeostasis. We preliminarily speculate that uracil is a key factor in the interaction between Bt and gut microbiota, and these findings provide a theoretical basis for clarifying the interaction between Bt, host, and intestinal microorganisms, as well as for gaining new insights into the insecticidal mechanism of B. thuringiensis in insects.
Introduction
Insects harbor complex microbial communities in their gut, which form a diverse microbial ecosystem that plays a critical role in the host’s physiology and resistance against pathogens (Shin et al., 2011; Kamareddine et al., 2018; Bai et al., 2021). The insect immune response is responsible for maintaining gut microbial homeostasis and plays a crucial role in regulating the interactions between commensal and pathogenic bacteria (Wu et al., 2016; Gao et al., 2020). Reactive oxygen species (ROS) and antimicrobial peptides (AMPs) are important components of the insect intestinal immunity that regulate pathogen growth and protect commensal bacteria (Bae et al., 2010; Ryu et al., 2010). ROS are primarily produced by dual oxidases (DUOX), while the immune-deficiency pathways generate AMPs (Ha et al., 2005, 2009; Zeng et al., 2022).
Recent studies have revealed that intestinal microorganisms can impact the insecticidal activity of pathogenic bacteriaused to control pests. For instance, elimination of indigenous midgut bacteria by oral administration of antibiotics has been shown to reduce the efficacy of Bacillus thuringiensis (Bt) against Lymantria dispar larvae (Broderick et al., 2006). Additionally, in some insects, the entry of Bt into the insect leads to the transformation of beneficial bacteria into pathogenic bacteria, thereby affecting the sensitivity of insects to Bt (Broderick et al., 2009). However, Raymond et al. demonstrated that antibiotics have an effect on the activity of Bt, and a series of experiments have shown that midgut bacteria are not essential for Bt to exert its insecticidal activity (Raymond et al., 2009). Entomopathogenic fungi have also been shown to downregulate DUOX expression in the midgut, leading to an imbalance in the intestinal flora that can cause the opportunistic pathogen Serratia marcescens to proliferate and transfer to the insect’s hemocoel, ultimately promoting mosquito death (Wei et al., 2017).
Free nucleobases are not normally present intraor extracellularly in bacteria, However, uracil was observed in Escherichia coli and the release of uracil is likely influenced by various environmental conditions (Rinas et al., 1995). In Pseudomonas aeruginosa, uracil auxotrophic mutant strains decreased virulence and biofilm formation (Ueda et al., 2009), such uracil-modulated bacterial responses were speculated that potentially threatening to host fitness (Attila et al., 2009). It has been shown that pathogenic bacteria can release uracil, which activate DUOX-dependent gut immunity, that plays an essential role in host protection against pathogen infection (Lee et al., 2013, 2015). Uracil released by allochthonous bacteria, but not by symbiotic resident bacteria, helps the host to distinguish intestinal symbiotic bacteria from pathogenic bacteria, thereby maintaining intestinal microbial homeostasis and immune balance (Lee et al., 2013). The car and pyr gene (such as carA, carB, pyrB, pyrC, pyrD, pyrE) related to uracil synthesis, and Orotate phosphoribosyl transferase (OPRTase) encoded by pyrE is a key enzyme in pyrimidine biosynthesis (Ueda et al., 2009). pyrE gene mutants are resistant to 5-FOA but are uracil auxotrophs, and the selection system utilizing these properties has been successfully established (Ueda et al., 2009; Akasaka et al., 2013).
In our previous work, we investigated the effect of Bt on the gut microbiota diversity of the Spodoptera exigua (Li et al., 2022). However, the regulatory genes of Bt that interact with insect gut microbiota and the molecular mechanism of host immune regulation were not clear. In this study, we obtained an uracil auxotrophic strain, Bt GS57△pyrE, and clarified the role of uracil in activating the DUOX-ROS immune system and maintaining the stability of intestinal microbes. Our findings provide a theoretical basis for studying new insecticidal mechanisms of Bt.
Materials and methods
Insect rearing
Spodoptera exigua larvae were reared on an artificial diet following the method described previously, the main components are wheat germ powder, soybeans, and so on (Ren et al., 2013), and maintained at 26 ± 1°C with a relative humidity of 70 ± 10%.
Bacterial strains, plasmids and growth conditions
The highly virulent strain of Bt GS57, known to be effective against S. exigua larvae, was maintained in our laboratory (Li et al., 2022). Competent Bt GS57 cells were prepared in 3.7% brain heart infusion medium, as per literature (Xu et al., 2020). Non-methylated plasmid DNA for transformation into B. thuringiensis cells was generated using E. coli. All E. coli strains were grown in LB medium. The pMAD and pSTK plasmids used in this study were obtained from Institute of Plant Protection, Chinese Academy of Agricultural Sciences.
Bt GS57△pyrE strain construction
B. thuringiensis is an important biocontrol agent against various insect pests, including S. exigua. The pyrE gene plays a crucial role in regulating uracil synthesis in bacteria (Akasaka et al., 2013). To investigate the role of uracil synthesis in Bt virulence, a Bt GS57△pyrE mutant strain was constructed. In this study, the location of pyrE gene in the Bt GS57 genome was determined (Chromosome1:263694:264326), the 997-bp fragment upstream of pyrE (pyrE fragment A, pyrEA) and the 993-bp fragment downstream of pyrE (pyrE fragment B, pyrEB) were amplified from Bt GS57 genomic DNA with pyrE-1F/EU-RX and pyrE-1R/EU-FX primers, respectively. A 1.4 kb fragment of kanamycin resistance gene was amplified using EPK-F/R as primers and pSTK plasmid as templates. pyrEA, kan, and pyrEB were ligated together using overlapping PCR with primers pyrE-1F/R. The resulting fragment was inserted into the BamH I-Sal I restriction sites of pMAD plasmids to construct the recombinant plasmid vector pMAD△pyrE, which was identified using PCR and DNA sequencing with pMAD-F and pMAD-R primers. The pMAD△pyrE plasmid was electroporated into competent Bt GS57 cells, and transformants were selected on LB agar plates based on erythromycin and kanamycin resistance. The pyrE gene deletion in the Bt GS57 was accomplished using homologous recombination as described previously (Janes and Stibitz, 2006; Liu et al., 2021). The Bt GS57△pyrE mutant was selected on LB agar plates containing kanamycin resistance without erythromycin and verified using PCR with primers pyrE-JF/R.
Verification of Bt GS57△pyrE mutant strains
Uracil auxotrophs strains are often identified based on their resistance to 5-fluoroorotic acid (5-FOA), an antimetabolite commonly used for selection. To determine the working concentration for Bt GS57, LB broth medium was supplemented with 5-FOA at final concentrations of 0.1 mg/mL, 1.0 mg/mL, and 1.5 mg/mL. Subsequently, a single colony of Bt GS57 was inoculated into each of the media and grown for 24 h. Growth curves of Bt GS57 were generated for each concentration of 5-FOA. To assess the effectiveness of the selected concentration, a single colony of Bt GS57△pyrE and Bt GS57 was inoculated onto LB plates supplemented with the optimal concentration of 5-FOA. Growth of each strain on the plates was compared to determine the concentration at which 5-FOA effectively selects uracil auxotrophic mutants.
Growth characteristics and biological activity assay of Bt GS57△pyrE
The crystal phenotypic and growth characteristics of Bt GS57△pyrE were assessed according to a previously published method (Li et al., 2022). Both Bt GS57△pyrE and Bt GS57 were inoculated into LB medium and grown at 30°C with shaking (200 rpm) for 30 h. The OD600 values were measured every 2 h to monitor the growth of Bt GS57△pyrE, and the results were compared with those of the Bt GS57 strain. The spore crystal mixtures of Bt GS57△pyrE and Bt GS57 were harvested and stained with 0.5% safranin, then observed using a light microscope (Olympus, U-HGLGPS, 100X oil objective) to evaluate their biological activity.
Virulence assays
The Bt GS57△pyrE strain was cultured in 1/2 LB medium at 30°C for 46 h to obtain the spore crystal mixture. The mixture was then suspended in distilled water and diluted to gradient concentrations (0.05, 0.1, 0.2, 0.4, 0.8, 1.6 mg·ml−1). Subsequently, 100 μl of the diluted mixture was overlaid on a 33 mm diameter plastic tube containing artificial diet. The newly hatched larvae and early stage of 4th-instar larvae of S. exigua, which had been starved for 2 h, were then exposed to B. thuringiensis or 20 mM uracil (Sigma-Aldrich, St. Louis, United States) and reared under standard culture conditions, as described in a previous study (Li et al., 2022). The bioassays were repeated three times for each treatment, and mortality was recorded every 24 h. The semi-lethal concentration (LC50) was calculated after 72 h using Probit analysis (SPSS, Chicago, IL, United States).
Quantification of gut bacteria by qPCR
Fourth instar larvae of S. exigua were fed with Bt GS57 or Bt GS57△pyrE strains at the semi-lethal concentration (3 mg cell crystal mixture per mL). The control group was added the sterile water. Genomic DNA from the gut, including contents, was extracted at various time points (0 h, 4 h, 8 h, 12 h and 24 h) using TIANamp Genomic DNA Kit (TIANGEN) following the manufacturer’s instructions. Quantitative PCR (qPCR) was performed using universal bacteria primers (16SF/R) (Table 1) to amplify 16S rRNA fragments with TB Green II Master Mix (TaKaRa Bio Inc. Japan). The expression levels of 16S rRNA were normalized using the actin gene as an endogenous control. All reactions were performed in triplicate, and the data were analyzed using the threshold cycle 2−ΔΔCt method (Livak and Schmittgen, 2001).
Bacterial composition and diversity of gut microbiota in S. exigua infected with Bt
Fourth instar S. exigua larvae were infected with Bt GS57 and Bt GS57△pyrE by spreading cell crystal mixture on the diet surface. The midguts of the infected larvae were dissected at five different time points (0 h, 4 h, 8 h,12 h and 24 h) and three replicates were prepared for each treatment, with 15 larvae per replicate mixed to obtain the samples. The DNA was extracted from the samples and amplified using the 338F/806R primer (Table 1) targeting the V3 and V4 hypervariable region of the bacterial 16S rDNA gene. The Illumina MiSeq platform was used for 16S rDNA gene amplicon sequencing by Beijing Biomarker Technologies company. The high-quality CCS (Circular Consensus Sequencing) sequence were obtained by removing the chimera (UCHIME, version 8.1) and clustering the resulting datasets as operational taxonomic units (OTUs) based on 97% similarity using USEARCH (version 10.0). The alpha diversity indices (ACE and Shannon) and beta-diversity (NMDS) were calculated for each sample, and differences in the microbial community structure between groups were analyzed using BMKCloud.1
Analysis of DUOX and AMPs gene expression
The midguts of fourth instar larvae of S. exigua were collected at 0 h (control), 4 h, 8 h, 12 h and 24 h after Bt GS57 or Bt GS57△pyrE (3 mg/mL) infection, and total RNA was isolated from each sample. First-strand cDNA was synthesized from total RNA using the PrimeScript RT Reagent Kit with gDNA Eraser (Takara, China) following the manufacturer’s instructions. qPCR analysis was performed to analyze the influence of bacterial-derived uracil on immune-related genes. The housekeeping actin gene was used as endogenous control, and each reaction was performed in triplicate. The primers used are shown in Table 1. The results were calculated using the 2−△△Ct method.
Measurement of total ROS activity in vivo
The intestinal tissues of fourth instar larvae of S. exigua were dissected in pre-cooled PBS, ground in 500 μl PBS solution and centrifuged at 4°C and 3,000 g for 5 min. The supernatant was used for ROS levels detection following the instructions of the reactive oxygen species detection kit (Solarbio, China). Samples were incubated with 2′,7′-dichlorodihydrofluorescein diacetate (DCFH-DA) (10 μmol/L) for 30 min at 37°C in the dark and detected using a Tecan Infinite® M200 multiscan spectrum (Swiss) at excitation wavelengths of 488 nm and 525 nm to excite DCF. The fluorescence intensity of the final sample was calibrated with a negative control and three biological replicates were set for each treatment, with 15 larvae in each replicate.
Data analysis
The results of gut bacterial load of S. exigua and the gene expression were tested for significance using Tukey’s HSD one-way ANOVA with SPSS (Statistical Package for the Social Sciences) Statistics 18.0 software. The different letters indicate that the difference is significant among groups at p < 0.05 level (n = 3).
Results
Identification and cloning of pyrE in Bt GS57
The pyrE gene deletion mutant Bt GS57△pyrE was constructed by homologous recombination assay (Figure 1A), pyrE gene was identified in the genome of Bt GS57 (accession number NC_020238.1), located between 263,694 and 264,326 bp. A high amino acid identity of 99.52% was found between pyrE in B. thuringiensis strain GS57 and Bacillus sp. strain SYJ. The upstream and downstream homology arms of the pyrE gene (997-bp & 993-bp) from Bt GS57 strain, and a kanamycin resistance gene fragment (1400-bp) were amplified by PCR, respectively (Figure 1B). The overlapping PCR of the three fragments yielded a fusion fragment of 3,390 bp (Figure 1C).
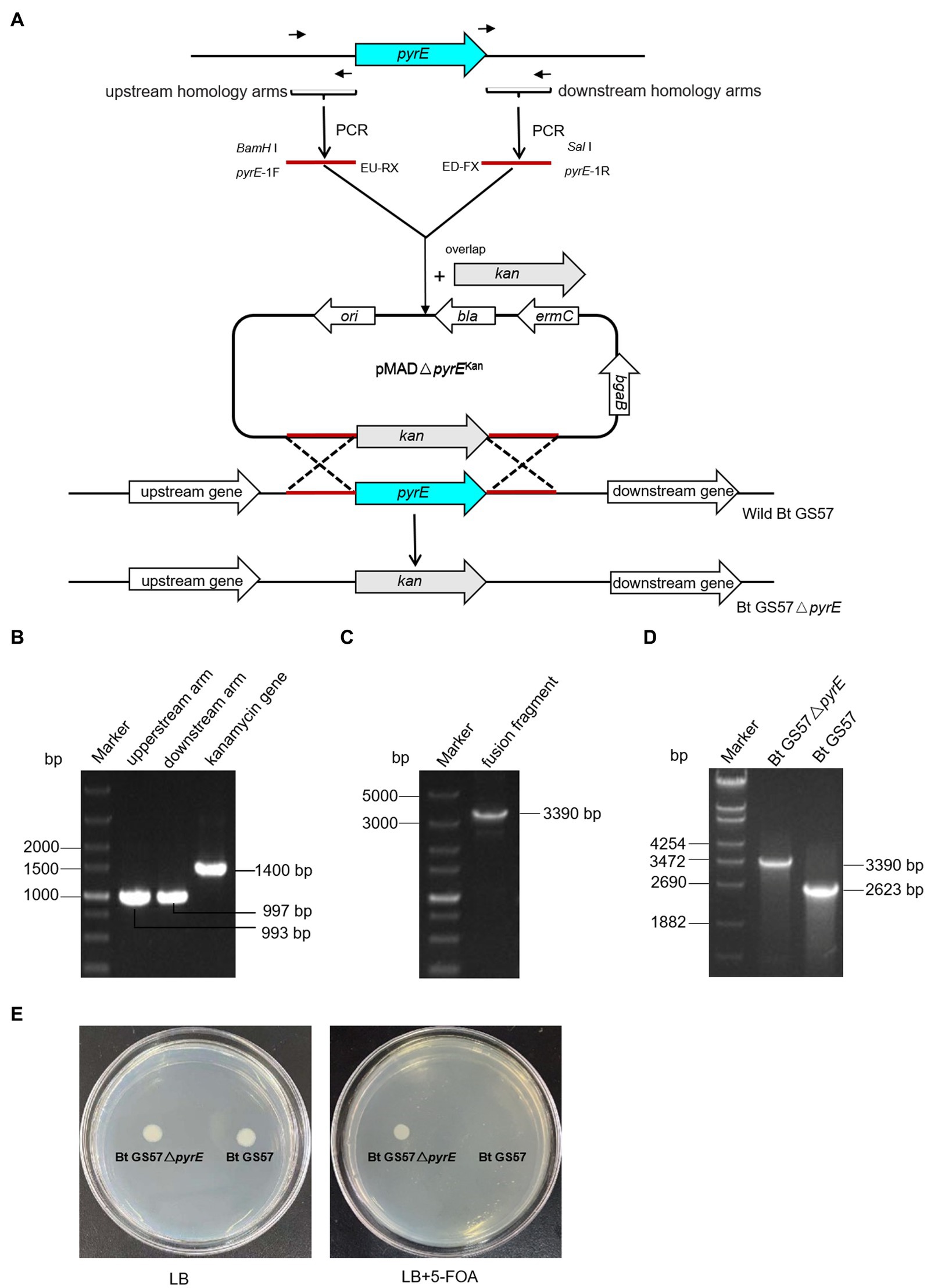
Figure 1. Construction and PCR identification of Bt GS57△pyrE mutant. (A) Bt GS57△pyrE constructed by homologous recombination, the pyrE was exchanged by kanamycin gene as a mutant. (B) PCR amplification of pyrE gene and kanamycin resistance gene. Marker: DL5000 Marker; the upstream homology arms fragment of pyrE gene amplified by pyrE-1F/EU-RX primer; the downstream homology arms fragment of pyrE gene amplified by ED-FX/pyrE-1R primer; the fragment of kanamycin resistance gene amplified by EPK-F/R primer. (C) The fusion fragment of upstream, downstream arms and kanamycin gene amplified by pyrE-1F/pyrE-2R primer. (D) PCR identification of Bt GS57△pyrE mutant. Marker: DL5000 Marker, the lane of Bt GS57△pyrE and Bt GS57 were pyrE fragment amplified by pyrE-JF/pyrE-JR prime. (E) Functional identification of the Bt GS57△ pyrE mutant, Bt GS57△pyrE grows normally on LB plates containing 5-FOA, while Bt GS57 does not.
The purified PCR product was cloned into the pMAD vector and transformed into E. coli SCS110. The recombinant pMAD-pyrE plasmid was then extracted and transformed into wild-type Bt GS57 by electro transformation. After two cycles of homologous recombination, the kanamycin gene replaced the pyrE gene, and the pyrE deletion mutant strain was constructed and verified by PCR (3,390 bp) and DNA sequencing (Figure 1D). Bt GS57△pyrE showed partial resistance to 5-FOA, as it grew in LB medium containing 1.5 mg/mL 5-FOA, while Bt GS57 could not (Figure 1E).
Effects of pyrE deletion on biological characteristics and activity
To investigate the effects of pyrE deletion on biological characteristics and activity, Bt GS57 and Bt GS57△pyrE were cultured in 1/2 LB medium. The growth curves of Bt GS57 and Bt GS57△pyrE were similar, with a lag period of 0–3 h, a logarithmic growth period of 4–22 h, and a stable period (Figure 2A). Both strains produced spores and rhomboidal-shaped crystals at 44 h as assessed by microscope (Figure 2B). The bioassay results of Bt strains against the newly hatched S. exigua larvae showed that there was no significant change in LC50 of Bt GS57△pyrE (0.329 mg·ml−1) (Table 2). We also found that the mortality rate of the 4th-instar larvae of S. exigua after treat with Bt GS57△pyrE was approximately 56.67%, which is slightly lower than the mortality rate of 63.33% of Bt GS57 but is not significant. And the uracil supplement to Bt GS57△pyrE did not significantly changes the mortality rate (Figure 3). These results showed that PyrE deletion did not reduce the susceptibility of S. exigua larvae to Bt GS57.
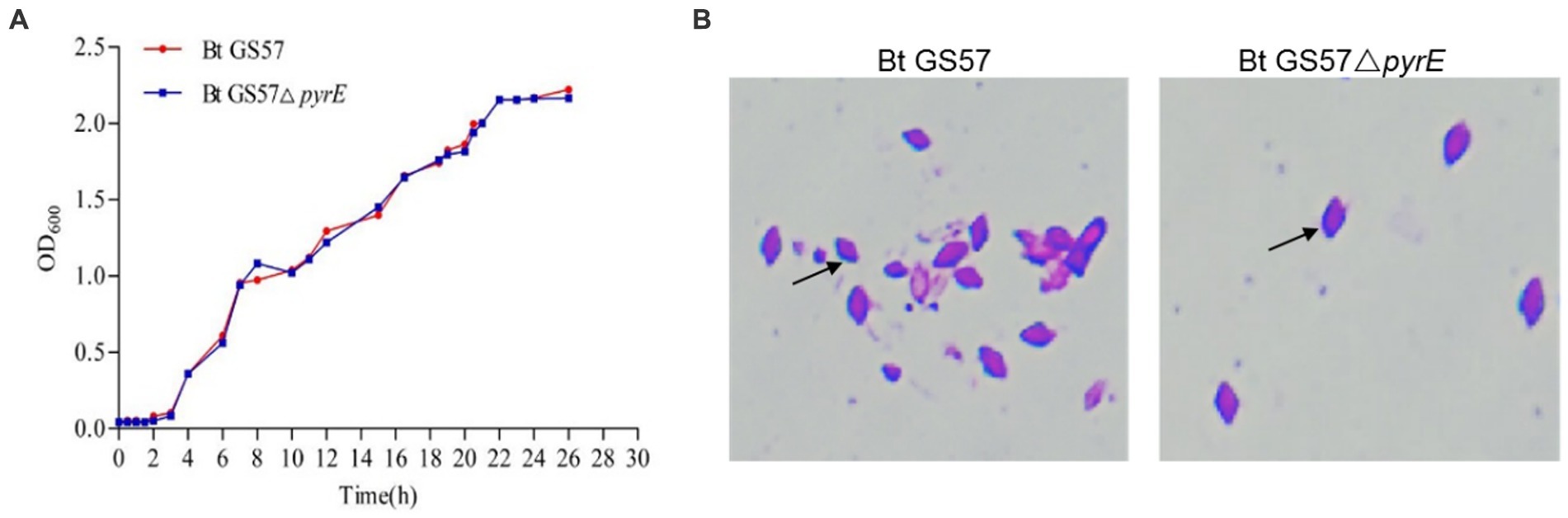
Figure 2. Growth curve and observation of crystal morphology of Bt GS57 and Bt GS57△pyrE. (A) Growth curve of Bt GS57 and Bt GS57△pyrE. (B) Observation of crystals by microscope, and the arrow points to the rhomboidal-shaped crystals.
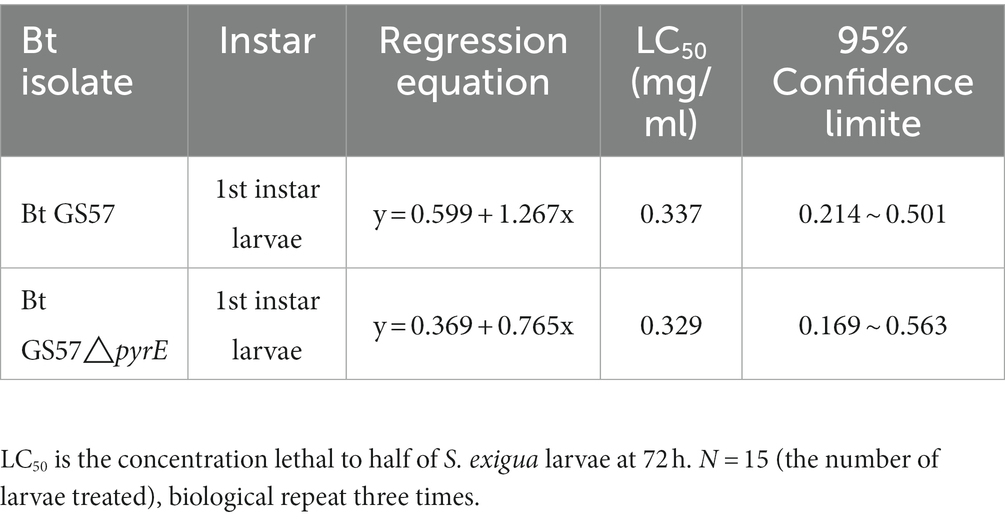
Table 2. Bioassay of Bacillus thuringiensis isolates against the 1st instar larvae of Spodoptera exigua.
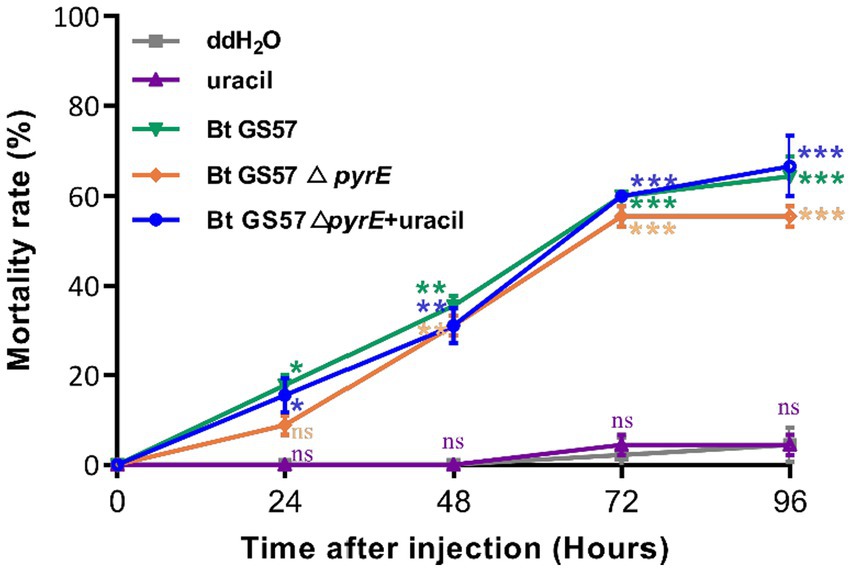
Figure 3. Mortality rate of S. exigua larvae feed with Bt strains at different times. The larvae were exposed to Bt GS57, Bt GS57△pyrE strains (3 mg·ml−1) and Bt GS57△pyrE + 20 mM uracil respectively, the control group was fed with ddH2O and 20 mM uracil (n = 15), repeat three times. The mortality rates of each group were analyzed every 24 h. Data are expressed as the mean ± SD from three independent experiments; asterisks indicate significant differences according to Student’s t-test, compared with the control value (*p < 0.05, **p < 0.01, ***p < 0.001).
Effects of Bt-derived uracil on the gut bacterial load of S. exigua
We investigate the effect of Bt GS57-derived uracil on the gut bacterial load of S. exigua. The gut bacterial loads of S. exigua were assessed at 4 h, 8 h, 12 h and 24 h post infection with Bt GS57△pyrE and Bt GS57, and compared to non-infected controls treated with ddH2O. qPCR analysis revealed that the gut bacterial load was upregulated in S. exigua after infection with both strains at different time intervals, compared to noninfected controls. However, at 8 h post-infection, the uracil deletion strain showed a significantly lower bacterial load than the wild-type strain. At 12 h, when the mutant was supplemented with uracil, there was no significant difference between the uracil deletion strain and the wild-type strain in terms of gut microbial load (Figure 4). These results suggest that Bt infection leads to dysbiosis of gut microbes and uracil is an important regulatory factor, we further investigation of the effect on immune genes.
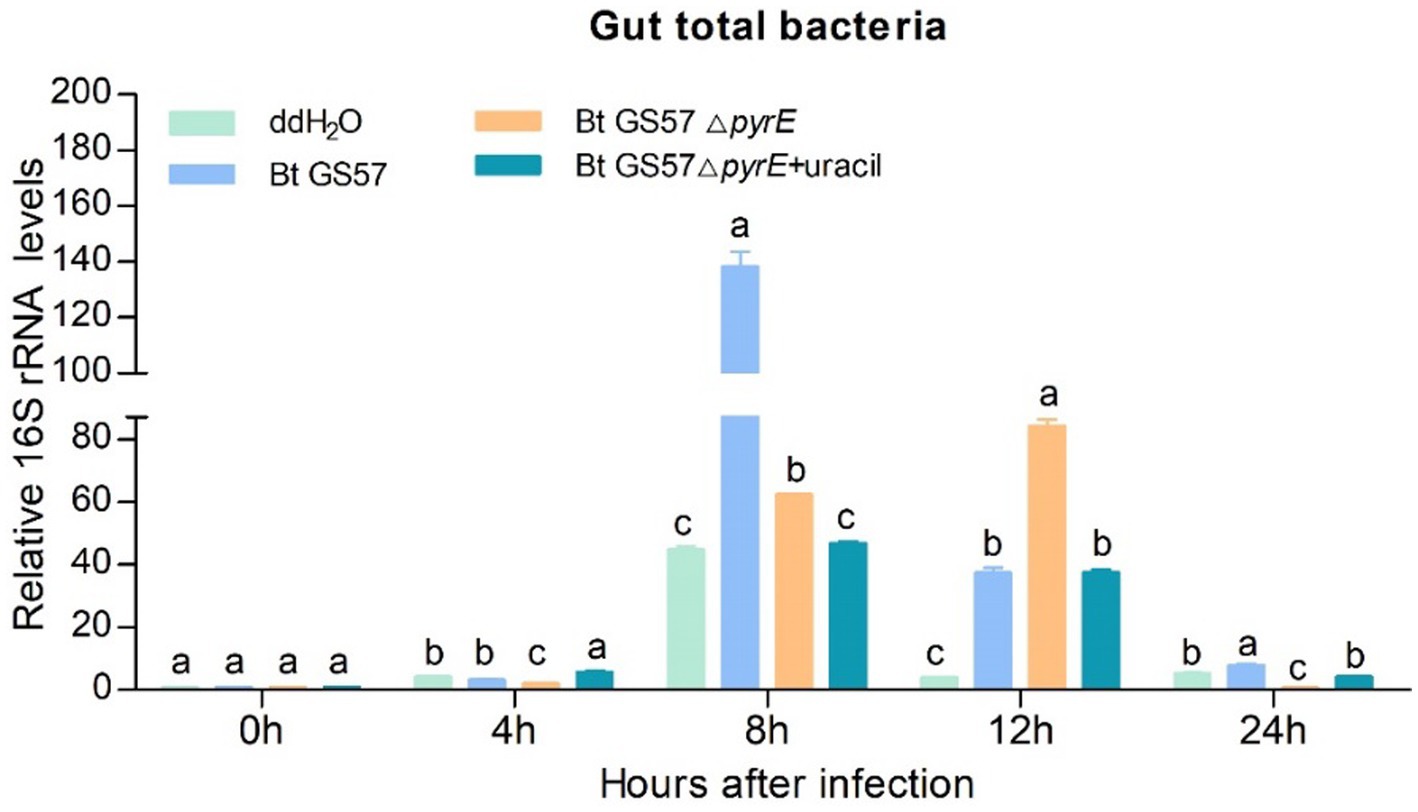
Figure 4. The effect of Bt GS57△pyrE on the bacterial load of the gut of S. exigua. mRNA levels of gut total bacteria in the 4th instar larvae of S. exigua (n = 3) following infection with Bt GS57, Bt GS57ΔpyrE, and Bt GS57ΔpyrE + uracil. Data are representative of three independent experiments (mean + s.e.m.). Significant differences between groups were determined by Tukey’s test as part of a one-way analysis of variance. Different lowercase letters (a, b or c) above bars indicate significant differences in the gut bacteria among groups (p < 0.05).
Bt GS57 infection causes dysbiosis of the gut microbiota
To evaluate the impact of Bt GS57 infection on gut microbiota, we analyzed the midgut bacetria communities of Bt GS57-infected and Bt GS57△pyrE-infected S. exigua larvae at different time intervals (0, 4, 8, 12, 24 h) using 16S rRNA gene sequencing (Table 1). We found that the abundance of Firmicutes and Proteobacteria changed dynamically at 8 h and 12 h post-infection5, and that the bacterial diversity decreased significantly in Bt GS57△pyrE larvae compared to Bt GS57-infected larvae (Figure 5A), which Tsuggests that uracil produced by Bt GS57, may play a role in regulating gut microbiota composition. Principal Components Analysis (PCA) further revealed the deletion of uracil resulting in changes of gut microbial communities, which explained 21.28% of the variance in the group difference (Figure 5B). At the genus level, the relative abundances and distributions of bacterial families changed significantly in S. exigua after infection by Bt GS57 and Bt GS57△pyrE (Figure 5C), especially the genus Bacillus and Enterobacter (Supplementary Table S1). The results suggest that bacterial-derived uracil can alter gut bacterial composition and result in dysbiosis of the gut microbiota.
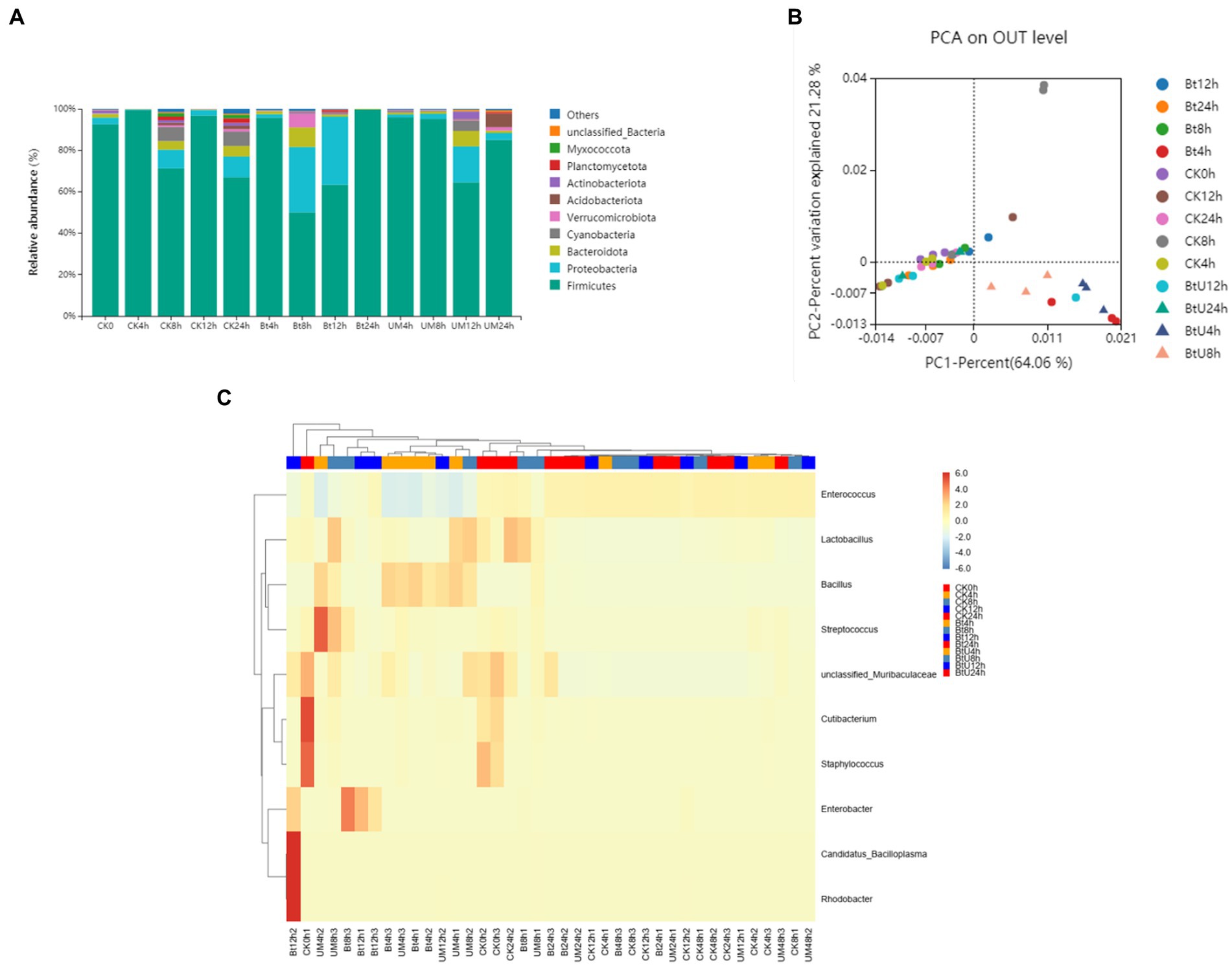
Figure 5. Gut bacterial dynamics in S. exigua after Bt GS57 and Bt GS57△pyrE infection. (A) Relative abundance of bacterial communities at the phylum level in different treatments. (B) PCA analysis of microbial communities according to host developmental stage (ANOSIM test, p = 0.001). (C) Heat map of major taxa at different treatments at the genus level generated by cluster analysis using the average method. Columns were clustered according to the similarity of bacterial abundance profiles. Each row represents an OUT assigned to the genus level. Color gradients represent the abundance variation of different species in the sample. Plotting scale, from red to blue, indicates the decrease in relative abundance of bacteria. Bt, Bt GS57△pyrE; UM, Bt GS57; CK, ddH2O.
Effects of the pyrE deletion on immune gene expression in the midgut
To test whether pyrE deletion could affect uracil deficient Bt strain (Bt GS57△pyrE) midgut immune responses, qPCR analysis was used to examine the effect of uracil on the expression of the DUOX, PGRP-SA, attacin, defensin and ceropin genes. DUOX gene expression and ROS levels was significantly upregulated after infection with Bt GS57, while Bt GS57△pyrE infection resulted in no significant difference compared to the ddH2O control at earlily time (4 h and 8 h), but a significant decrease compared to the wild strain, (Figure 6). Addition of uracil to Bt GS57△pyrE infected larvae restored DUOX and ROS expression to similar levels observed with Bt GS57 infection, indicating that uracil can activate the DUOX-ROS system and promote ROS production.
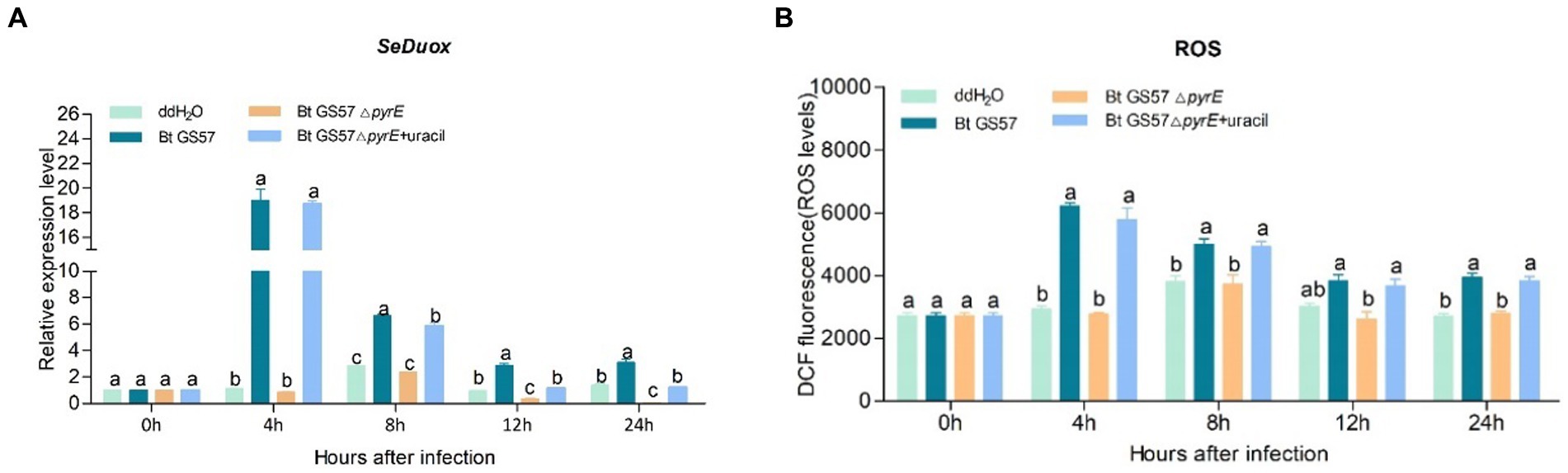
Figure 6. Effects of Bt GS57△pyrE on the expression of DUOX and ROS levels of S. exigua. (A) The expression of SeDuox gene of 4th instar larvae of S. exigua infection with Bt GS57, Bt GS57ΔpyrE, and Bt GS57ΔpyrE + uracil (n = 3). (B) Effects of Bt GS57△pyrE on The ROS level of 4th instar larvae of S. exigua infection with Bt GS57, Bt GS57ΔpyrE, and Bt GS57ΔpyrE + uracil (n = 3). Data are representative of three independent experiments (mean + s.e.m.). Different letters (a, b or c) indicate significant differences in the DUOX gene expression and ROS levels among different group as determined by Tukey’s test as part of a one-way analysis of variance (p < 0.05).
Furthermore, qPCR analysis showed that the expression of PGRP-SA and AMP-related genes (attacin, defensin and ceropin) were significant different in the midgut of S. exigua infected by Bt GS57 and by Bt GS57△pyrE (Figure 7). PGRP-SA, defensin and ceropin was upregulated significantly in the midgut after infection with Bt GS57 and Bt GS57△pyrE at 8 h, while the attacin gene was downregulated which expressed at low levels.
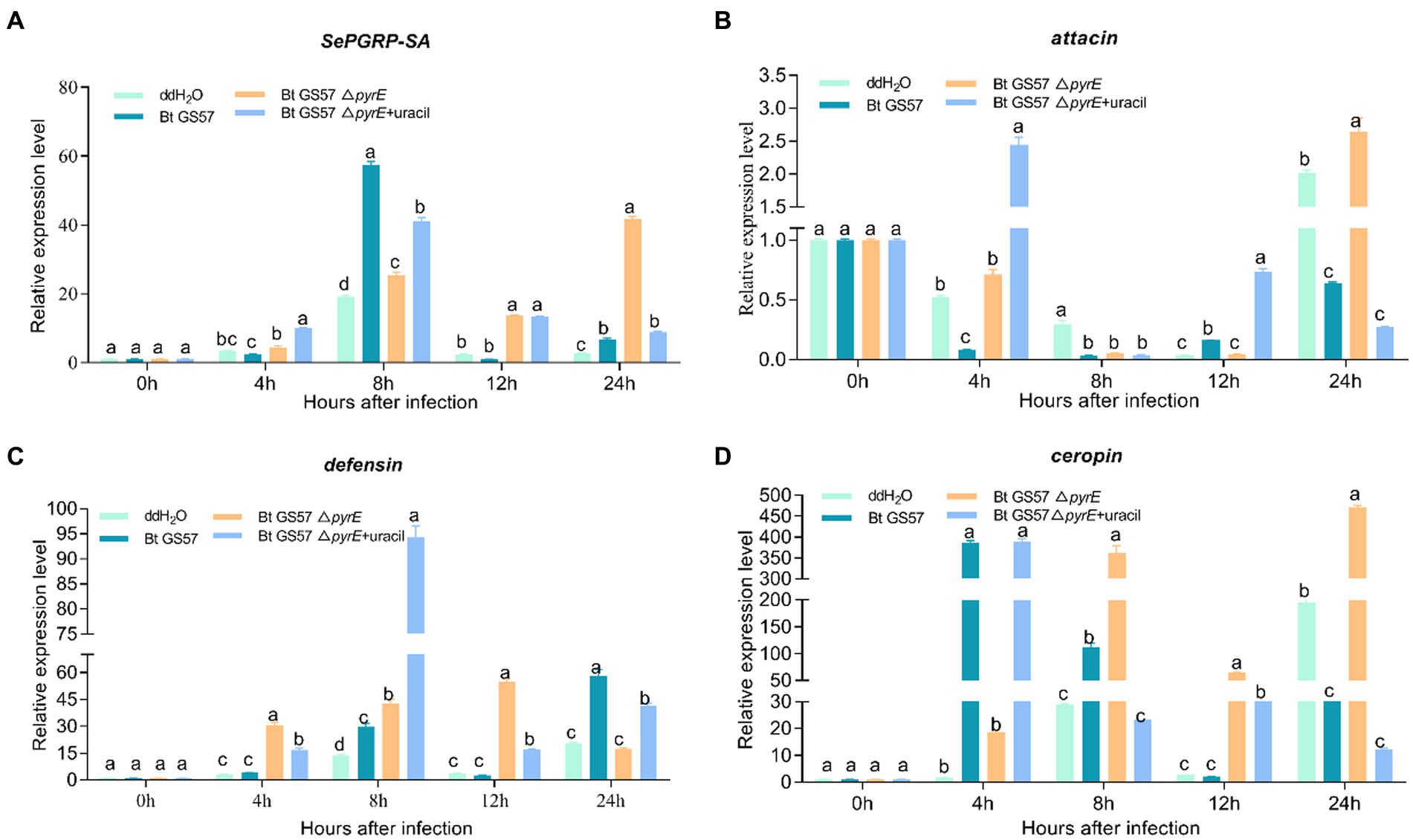
Figure 7. The effect of Bt GS57△pyrE on AMPs genes in the gut of S. exigua. (A–D) mRNA levels of PGRP-SA, attacin, defensin and ceropin in the 4th instar larvae of S. exigua (n = 3) following infection with Bt GS57, Bt GS57ΔpyrE, and Bt GS57ΔpyrE + uracil. Data are representative of three independent experiments (mean + s.e.m.). Significant differences between groups were determined by Tukey’s test as part of a one-way analysis of variance. Different letters (a, b or c) meant there was significant difference in the gene expression level among groups (p < 0.05).
Discussion
The microbiota in the gut of insects play a vital role in the host’s development, immunity, and metabolism, as well as in the defence against pathogens (Engel and Moran, 2013; Siddiqui et al., 2022; Zhang et al., 2022). The balance between the gut microbial communities and the host’s immune system is critical for maintaining the health of the host. Insects rely on the ROS and AMPs pathway for their immune response, which regulates the intestinal flora and shows a synergistic effect in case of microbial disbalance (Kamareddine et al., 2018; Feng et al., 2020). Previous studies have shown that the joint action of metabolites of B. thuringiensis influences the midgut microbes (Engel and Moran, 2013). Additionally, recent research has demonstrated that uracil produced by pathogenic bacteria can upregulate the expression of the DUOX gene to produce ROS, which helps to resist pathogenic bacterial infection and maintain intestinal microbial homeostasis (Lee et al., 2015). In our previous research, we observed that gut microbiota can accelerate the killing of S. exigua by Bt GS57 (Li et al., 2022), which involves altered community structure and significantly decreased bacteria diversity. Building on this finding, we conducted further research to explore the effect of B. thuringiensis produced uracil on the interaction between intestinal microorganisms and immune genes. We constructed an uracil mutant by deleting the pyre gene in Bt GS57 and confirmed the characteristics of pyrE mutation. The results showed that the pyrE deletion mutant Bt GS57(△pyrE) lost the ability to synthesize uracil and was resistant to 5-FOA (Figure 1). However, there were no significant changes in the biological function and growth characteristics of Bt GS57(△pyrE) compared with the wild strain, and the crystal protein remained bipyramidal (Figure 2). Bioassay results also showed no significant effect on the toxicity of Bt GS57(ΔpyrE) against S. exigua larvae, indicating that the pyrE mutation did not change the strain’s characteristics (Table 2; Figure 3). Uracil, as a bacterial-derived ligand, plays a crucial role in activating the DUOX-dependent gut immune system, and uracil-modulated host signaling pathways are essential for DUOX-dependent ROS generation and host resistance to gut infection (Lee et al., 2015). In this study, we clarified the effect of uracil on activating the DUOX-ROS immune system and maintaining the stability of intestinal microbes. We found that after feeding on Bt GS57△pyrE, the DUOX expression and ROS levels were significantly down-regulated compared to the Bt GS57 (control) (Figure 6). However, after uracil supplementation, the expression of DUOX was significantly increased, inducing ROS generation, which suggests that the uracil secreted from Bt strain is possibly responsible for DUOX activation and could also induce intestinal ROS generation. These results show that uracil mediates the up-regulation of DUOX expression in the midgut, thereby increasing the production of midgut ROS levels.
In addition to ROS production, insects use the induction of AMPs as another defense mechanism to regulate their gut microbiota (Sheehan et al., 2018). The peptidoglycan recognition proteins (PGRPs) family acts as a pattern recognition receptor of the IMD or Toll signaling pathway and synthesizes AMPs, which play an essential role in the innate immunity of insects(Wang et al., 2019). Bt GS57△pyrE also induces an increase in PGRP and antimicrobial peptide-related genes (defensin and ceropin) at 8 h and 12 h compared with the sterile water control, but significantly lower than the wild strain (Figure 7), which showed the uracil also mediates the expression of PGRP and antimicrobial peptide-related genes. These results reveal that bacteria activate uracil-induced gut immune response, maybe act as a critical factor that determine gut microbe diversity and interactions with pathogenic bacteria. During the interaction between B. thuringiensis and gut microbes, the ROS and AMP response have a synergistic effect in resisting pathogenic bacterial infection and maintaining intestinal microbial homeostasis (Wei et al., 2017).
Most studies on the interaction between B. thuringiensis and insect midgut bacteria have focused on gut physiology, bacterial community structure, and composition. However, recent studies have shown that the toxicity of B. thuringiensis is associated with the gut microbiota. An investigation revealed that the diversity of bacterial microbiota in the tolerant Aedes aegypti larvae is reduced after Bt infection (Tetreau et al., 2018). In the Bt-resistant line of Galleria mellonella, Bt infection significantly reduced the diversity and abundance of the gut microbiota, possibly due to secreted antimicrobial factors in the resistant insects (Dubovskiy et al., 2016). The gut bacteria also affect Bt HD-1 insecticidal activity against Plodia interpunctella (Hübner) (Orozco-Flores et al., 2017). Similarly, Bt infection can cause a dynamic change in the gut microbiota of S. exigua (Li et al., 2022), and Bt Cry1Ac protoxin interacts with the gut microbiota to accelerate the mortality of Plutella xylostella larvae. However, the gut microbiota community composition of honeybees did not change significantly after feeding on transgenic Cry1Ah maize pollen (Jiang et al., 2013).
This study aimed to clarify the regulatory genes in the interaction between B. thuringiensis and insect resident midgut microflora. To investigate this, we compared the effect of pyrE deletion mutants of Bt GS57 on gut microbiota with the wild strain. It was found that the deletion of uracil could affect gut homeostasis, leading to an increase in bacteria and changes in bacterial community structure in the midgut of S. exigua larvae. The results suggest that the bacterial-derived uracil may trigger the DUOX-ROS immune response and influence the microbial communities of the gut, providing a better understanding of the interaction of B. thuringiensis-insect microbiota and exploring its insecticidal potential.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: NCBI - PRJNA944960, SRR24031882-SRR24031841.
Author contributions
DZ, HW, and WG conceived the research. DZ, HW, YL, QW, XG, and YJ conducted experiments. DZ and WG contributed material and secured funding. HW and YJ analysed data and conducted statistical analyses. DZ wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the earmarked fund for Modern Agro-industry Technology Research System (CARS-13), the National Natural Science Foundation of China (Grant No. 31471775), the National Key Research and Development Program of China (2017YFD0200401), the Science and Technology Project of Hebei Education Department (QN2020119) and the Foundation of the Graduate School of the Chinese Academy of Agricultural Sciences (CAAS) (1610042022005).
Acknowledgments
The authors would like to express their gratitude to EditSprings (https://www.editsprings.cn) for the expert linguistic services provided.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1182699/full#supplementary-material
Footnotes
References
Akasaka, N., Sakoda, H., Hidese, R., Ishii, Y., and Fujiwara, S. (2013). An efficient method using Gluconacetobacter europaeus to reduce an unfavorable flavor compound, acetoin, in rice vinegar production. Appl. Environ. Microbiol. 79, 7334–7342. doi: 10.1128/AEM.02397-13
Attila, C., Ueda, A., and Wood, T. K. (2009). 5-fluorouracil reduces biofilm formation in Escherichia coli K-12 through global regulator AriR as an antivirulence compound. Appl. Microbiol. Biotechnol. 82, 525–533. doi: 10.1007/s00253-009-1860-8
Bae, Y. S., Choi, M. K., and Lee, W. J. (2010). Dual oxidase in mucosal immunity and host-microbe homeostasis. Trends Immunol. 31, 278–287. doi: 10.1016/j.it.2010.05.003
Bai, S., Yao, Z., Raza, M. F., Cai, Z., and Zhang, H. (2021). Regulatory mechanisms of microbial homeostasis in insect gut. Insect Sci 28, 286–301. doi: 10.1111/1744-7917.12868
Broderick, N. A., Raffa, K. F., and Handelsman, J. (2006). Midgut bacteria required for Bacillus thuringiensis insecticidal activity. Proc. Natl. Acad. Sci. 103, 15196–15199. doi: 10.1073/pnas.0604865103
Broderick, N. A., Robinson, C. J., McMahon, M. D., Holt, J., Handelsman, J., and Raffa, K. F. (2009). Contributions of gut bacteria to Bacillus thuringiensis-induced mortality vary across a range of Lepidoptera. BMC Biol. 7:11. doi: 10.1186/1741-7007-7-11
Dubovskiy, I. M., Grizanova, E. V., Whitten, M. M., Mukherjee, K., Greig, C., Alikina, T., et al. (2016). Immuno-physiological adaptations confer wax moth galleria mellonella resistance to Bacillus thuringiensis. Virulence 7, 860–870. doi: 10.1080/21505594.2016.1164367
Engel, P., and Moran, N. A. (2013). The gut microbiota of insects - diversity in structure and function. FEMS Microbiol. Rev. 37, 699–735. doi: 10.1111/1574-6976.12025
Feng, M., Fei, S., Xia, J., Labropoulou, V., Swevers, L., and Sun, J. (2020). Antimicrobial peptides as potential antiviral factors in insect antiviral immune response. Front. Immunol. 11:2030. doi: 10.3389/fimmu.2020.02030
Gao, L., Song, X., and Wang, J. (2020). Gut microbiota is essential in PGRP-LA regulated immune protection against plasmodium berghei infection. Parasit. Vectors 13:3. doi: 10.1186/s13071-019-3876-y
Ha, E. M., Lee, K. A., Seo, Y. Y., Kim, S. H., Lim, J. H., Oh, B. H., et al. (2009). Coordination of multiple dual oxidase-regulatory pathways in responses to commensal and infectious microbes in drosophila gut. Nat. Immunol. 10, 949–957. doi: 10.1038/ni.1765
Ha, E.-M., Oh, C.-T., Bae, Y. S., and Lee, W.-J. (2005). A direct role for dual oxidase in Drosophila gut immunity. Science 310, 847–850. doi: 10.1126/science.1117311
Janes, B. K., and Stibitz, S. (2006). Routine markerless gene replacement in Bacillus anthracis. Infect. Immun. 74, 1949–1953. doi: 10.1128/IAI.74.3.1949-1953.2006
Jiang, W.-Y., Geng, L.-L., Dai, P.-L., Lang, Z.-H., Shu, C.-L., Lin, Y., et al. (2013). The influence of Bt-transgenic maize pollen on the bacterial diversity in the Midgut of Chinese honeybees, Apis cerana cerana. J. Integr. Agric. 12, 474–482. doi: 10.1016/s2095-3119(13)60248-8
Kamareddine, L., Robins, W. P., Berkey, C. D., Mekalanos, J. J., and Watnick, P. I. (2018). The Drosophila immune deficiency pathway modulates Enteroendocrine function and host metabolism. Cell Metab. 28, 449–462.e5. doi: 10.1016/j.cmet.2018.05.026
Lee, K. A., Kim, B., Bhin, J., Kim, D. H., You, H., Kim, E. K., et al. (2015). Bacterial uracil modulates Drosophila DUOX-dependent gut immunity via hedgehog-induced signaling endosomes. Cell Host Microbe 17, 191–204. doi: 10.1016/j.chom.2014.12.012
Lee, K. A., Kim, S. H., Kim, E. K., Ha, E. M., You, H., Kim, B., et al. (2013). Bacterial-derived uracil as a modulator of mucosal immunity and gut-microbe homeostasis in Drosophila. Cells 153, 797–811. doi: 10.1016/j.cell.2013.04.009
Li, Y., Zhao, D., Wu, H., Ji, Y., Liu, Z., Guo, X., et al. (2022). Bt GS57 interaction with gut microbiota accelerates Spodoptera exigua mortality. Front. Microbiol. 13:835227. doi: 10.3389/fmicb.2022.835227
Liu, X., Zhang, R., Hou, S., Liu, H., Wang, J., Yu, Q., et al. (2021). Identification and functional characterization of two homologous SpoVS proteins involved in sporulation of Bacillus thuringiensis.Pdf. Microbiol. Spectr 9:e0088121. doi: 10.1128/Spectrum.00881-21
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Orozco-Flores, A. A., Valadez-Lira, J. A., Oppert, B., Gomez-Flores, R., Tamez-Guerra, R., Rodriguez-Padilla, C., et al. (2017). Regulation by gut bacteria of immune response, Bacillus thuringiensis susceptibility and hemolin expression in Plodia interpunctella. J. Insect Physiol. 98, 275–283. doi: 10.1016/j.jinsphys.2017.01.020
Raymond, B., Johnston, P. R., Wright, D. J., Ellis, R. J., Crickmore, N., and Bonsall, M. B. (2009). A mid-gut microbiota is not required for the pathogenicity of Bacillus thuringiensis to diamondback moth larvae. Environ. Microbiol. 11, 2556–2563. doi: 10.1111/j.1462-2920.2009.01980.x
Ren, X. L., Chen, R. R., Zhang, Y., Ma, Y., Cui, J. J., Han, Z. J., et al. (2013). A Spodoptera exigua cadherin serves as a putative receptor for Bacillus thuringiensis Cry1Ca toxin and shows differential enhancement of Cry1Ca and Cry1Ac toxicity. Appl. Environ. Microbiol. 79, 5576–5583. doi: 10.1128/AEM.01519-13
Rinas, U. H. K., Kang, R., Seeger, A., and Schlieker, H. (1995). Entry of Escherichia coli into stationary phase is indicated by endogenous and exogenous accumulation of nucleobases. Appl. Environ. Microbiol. 61, 4147–4151. doi: 10.1128/aem.61.12.4147-4151.1995
Ryu, J. H., Ha, E. M., and Lee, W. J. (2010). Innate immunity and gut-microbe mutualism in Drosophila. Dev. Comp. Immunol. 34, 369–376. doi: 10.1016/j.dci.2009.11.010
Sheehan, G., Garvey, A., Croke, M., and Kavanagh, K. (2018). Innate humoral immune defences in mammals and insects: the same, with differences? Virulence 9, 1625–1639. doi: 10.1080/21505594.2018.1526531
Shin, S. C., Kim, S. H., You, H., Kim, B., Kim, A. C., Lee, K. A., et al. (2011). Drosophila microbiome modulates host developmental and metabolic homeostasis via insulin signaling. Science 334, 670–674. doi: 10.1126/science.1212782
Siddiqui, J. A., Khan, M. M., Bamisile, B. S., Hafeez, M., Qasim, M., Rasheed, M. T., et al. (2022). Role of insect gut microbiota in pesticide degradation: a review. Front. Microbiol. 13:870462. doi: 10.3389/fmicb.2022.870462
Tetreau, G., Grizard, S., Patil, C. D., Tran, F.-H., Tran Van, V., Stalinski, R., et al. (2018). Bacterial microbiota of Aedes aegypti mosquito larvae is altered by intoxication with Bacillus thuringiensis israelensis. Parasit. Vectors 11:121. doi: 10.1186/s13071-018-2741-8
Ueda, A., Attila, C., Whiteley, M., and Wood, T. K. (2009). Uracil influences quorum sensing and biofilm formation in Pseudomonas aeruginosa and fluorouracil is an antagonist. Microb. Biotechnol. 2, 62–74. doi: 10.1111/j.1751-7915.2008.00060.x
Wang, Q., Ren, M., Liu, X., Xia, H., and Chen, K. (2019). Peptidoglycan recognition proteins in insect immunity. Mol. Immunol. 106, 69–76. doi: 10.1016/j.molimm.2018.12.021
Wei, G., Lai, Y., Wang, G., Chen, H., Li, F., and Wang, S. (2017). Insect pathogenic fungus interacts with the gut microbiota to accelerate mosquito mortality. Proc. Natl. Acad. Sci. U. S. A. 114, 5994–5999. doi: 10.1073/pnas.1703546114
Wu, K., Yang, B., Huang, W., Dobens, L., Song, H., and Ling, E. (2016). Gut immunity in Lepidopteran insects. Dev. Comp. Immunol. 64, 65–74. doi: 10.1016/j.dci.2016.02.010
Xu, L., Han, G., Fan, X., Lv, J., Zhang, X., Peng, Q., et al. (2020). Characteristics of the sigK deletion mutant from Bacillus thuringiensis var. israelensis strain Bt-59. Curr. Microbiol. 77, 3422–3429. doi: 10.1007/s00284-020-02150-9
Zeng, T., Jaffar, S., Xu, Y., and Qi, Y. (2022). The intestinal immune defense system in insects. Int. J. Mol. Sci. 23:15132. doi: 10.3390/ijms232315132
Keywords: Bt GS57△pyrE, uracil, gut microbiota, immune gene, Spodoptera exigua
Citation: Zhao D, Wu H, Li Y, Wang Q, Ji Y, Guo X and Guo W (2023) Effects of the pyrE deletion mutant from Bacillus thuringiensis on gut microbiota and immune response of Spodoptera exigua. Front. Microbiol. 14:1182699. doi: 10.3389/fmicb.2023.1182699
Edited by:
Dimitris Mossialos, University of Thessaly, GreeceReviewed by:
Jialin Wang, Central China Normal University, ChinaVipin Rana, University of Maryland, United States
Copyright © 2023 Zhao, Wu, Li, Wang, Ji, Guo and Guo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Guo, Z3Vvd2VpMDVAY2Fhcy5jbg==
†These authors have contributed equally to this work
 Dan Zhao
Dan Zhao Han Wu1,2†
Han Wu1,2† Yazi Li
Yazi Li Yujie Ji
Yujie Ji Wei Guo
Wei Guo