
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 17 April 2023
Sec. Microbiotechnology
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1181967
Numerous microorganisms and other invertebrates that are able to degrade polyethylene (PE) have been reported. However, studies on PE biodegradation are still limited due to its extreme stability and the lack of explicit insights into the mechanisms and efficient enzymes involved in its metabolism by microorganisms. In this review, current studies of PE biodegradation, including the fundamental stages, important microorganisms and enzymes, and functional microbial consortia, were examined. Considering the bottlenecks in the construction of PE-degrading consortia, a combination of top-down and bottom-up approaches is proposed to identify the mechanisms and metabolites of PE degradation, related enzymes, and efficient synthetic microbial consortia. In addition, the exploration of the plastisphere based on omics tools is proposed as a future principal research direction for the construction of synthetic microbial consortia for PE degradation. Combining chemical and biological upcycling processes for PE waste could be widely applied in various fields to promote a sustainable environment.
Plastic pollution remediation is always a global environmental protection issue. As the most commonly used material, plastic products are, however, increasingly in demand and used in all fields globally along with socio-economic development due to their excellent characteristics such as durability, low cost and convenience, and it is predicted that the global production of plastic products will be over 26 billion tons by 2050 (Plastics Europe, 2021; Zeenat et al., 2021; Liu et al., 2022b). Approximately 80% of the world’s 400 million tons of plastic waste is dumped in landfills or discarded directly into the environment (Tejaswini et al., 2022). And the most common types of plastic waste are divided into two categories according to their thermal properties: thermoplastics and thermosetting plastics (Canopoli et al., 2018). Thermoplastics are types of linear-chain polymer compounds that have plasticity at a certain temperature, the most common of which are polyethylene (PE), polyvinyl chloride (PVC), polypropylene (PP), and polystyrene (PS). On the other hand, thermosetting plastics, such as polyurethane (PUR), cannot be melted due to irreversible thermal chemical changes (Amobonye et al., 2021). The backbones of thermosetting polymers are highly crosslinked by heteroatoms, making it easy for fracture to occur at ester or amide bonds. In contrast, the junctions of the primary chains in thermoplastics, which mostly comprise carbon atoms, render them more resistant to deterioration (Zheng et al., 2005). The majority of plastic waste is sent to landfill or discharged directly into the environment where it undergoes a very sluggish natural degradation process, which for PE specifically, the most inert polyolefin plastic, the half-lives were estimated to vary from decades to centuries (Chamas et al., 2020).
As the primary C–C-chain polymer (Figure 1), PE is currently the most extensively used plastic type, accounting for approximately 38% of the market share (Danso et al., 2019). PE is a thermoplastic made of ethylene that has been through high-pressure polymerization (Kumar Sen and Raut, 2015). According to different densities, branching degrees, and the availability of surface functional groups, PE can be divided into low-density polyethylene (LDPE), high-density polyethylene (HDPE), linear low-density polyethylene (LLDPE), etc. (Restrepo-Flórez et al., 2014). PE is widely utilized in cling film, commercial plastic bags, pharmaceutical and food packaging films, and other consumer and manufacturing industries, because of its desirable properties, such as non-toxicity, tastelessness, high tensile strength, low permeability, and durability (Shah et al., 2008; Liu et al., 2021). Due to its large molecular weight, stable chemical structure, high hydrophobicity, crystallinity, and limited number of functional groups required for biodegradation, PE is the plastic polymer most resistant to degradation, this means that polyethylene debris has lingered in the marine and other ecosystems for two decades and is extremely resistant to degradation (Kyaw et al., 2012; Dey et al., 2020; Krause et al., 2020). In addition, PE is the most prevalent municipal solid waste (MSW), and accounts for a high proportion of plastic waste in the environment (Zhou et al., 2014). Therefore, understanding the mechanisms of PE degradation and devising effective and environmentally friendly methods for PE degradation could provide ideas and data to help to mitigate our ever-worsening plastic pollution on a massive scale.
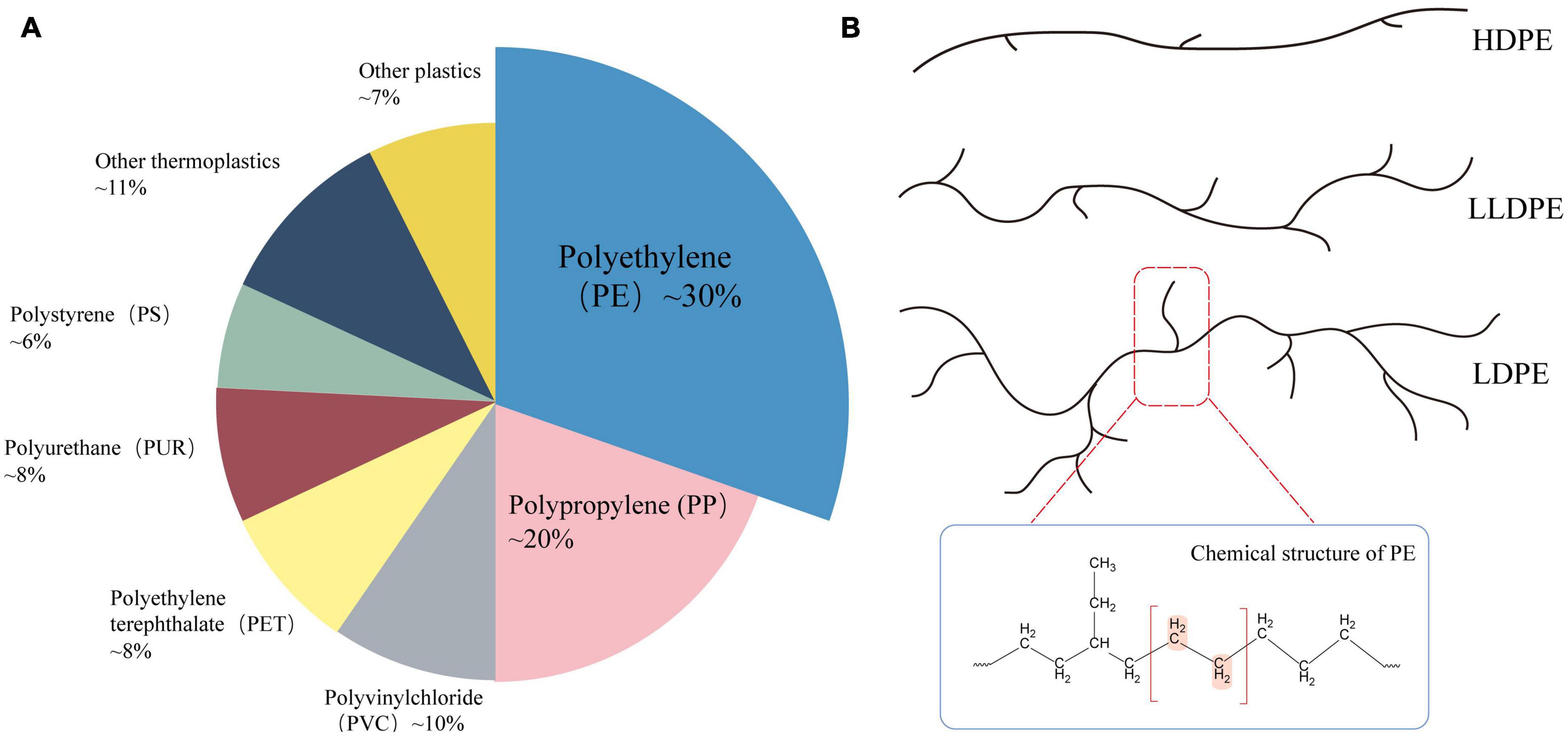
Figure 1. (A) Market share distribution of plastic types in 2021 (Plastics Europe, 2021); (B) common types of polyethylene (PE) and their molecular structure.
Current research indicates that the decomposition of plastic waste, such as PE, in landfills and other ecosystems is predominantly accomplished via physicochemical (abiotic) degradation and biodegradation (Dimassi et al., 2022). Physical degradation, such as cracking, embrittlement, and spalling, typically modifies the primary structures of polymers; whereas chemical degradation primarily modifies the molecular structures of polymers, such as breaking bonds or the oxidation of long polymer chains to produce compounds with low molecular weights (Andrady, 2011; Gewert et al., 2015; Ali et al., 2021). Mechanical degradation (including tidal forces, waves, and erosion), photo-oxidation (such as ultraviolet rays), thermal oxidation (such as incineration, pyrolysis, and gasification), and chemical hydrolysis processes (such as those involving acids, alkalis, and other organic solvents) are the primary physicochemical degradation mechanisms for PE and other plastic waste in the environment (Kyrikou and Briassoulis, 2007). Among them, photodegradation and thermo-oxidative degradation are the most common mechanisms of PE degradation in the environment (Canopoli et al., 2020). Through a variety of physical and chemical processes, large-sized plastic waste is broken down into plastic debris and micro/nanoplastics, which are then more easily digested and consumed by microorganisms (Taghavi et al., 2021b; Singh Jadaun et al., 2022). Researchers have become increasingly interested in the biodegradation of plastic in recent years due to the high efficiency, availability, and eco-friendliness of this approach (Khatoon et al., 2017). Deepening our understanding of the natural microbial communities that arise in the plastisphere may lead to novel approaches for the development of PE-degrading consortia. The key to the construction of efficient, stable, and controllable consortia is the design, which needs to ensure that the microorganisms in these communities interact synergistically. Certain technologies, such as next-generation sequencing technologies like metagenomic sequencing, can be used to determine the microbial community structure of the plastisphere (Gilmore et al., 2019; San León and Nogales, 2022).
This study focused on PE, the most prevalent plastic polymer. We briefly examined its characteristics and biodegradation mechanisms, as well as the current research gaps and recent advances in PE biodegradation. Then, we explored the advantages of using microbial functional consortia and their potential for PE pollution remediation compared to single strains. Lastly, we emphatically investigated the prospect of combining the plastisphere concept and metagenomics to inform the construction of functional microbial consortia.
Polyethylene (PE) biodegradation occurs via bacteria, fungi, algae, and other microorganisms that adhere to the plastic’s surface and consume it for growth and reproduction (Taghavi et al., 2021a). Microorganisms face a considerable challenge with PE due to its high hydrophobicity, large molecular size, and lack of reactive functional groups in its polymer backbone (Cowan et al., 2022). Numerous microorganisms (including bacteria, fungi, actinomycetes, and even some algae) that are capable of degrading PE polymers in various ecosystems (including oceans, soil, farmland, animal manure, compost, landfills, sewage, etc.) have been discovered using culture-dependent or culture-independent techniques (Dey et al., 2020). Several microbial enzymes associated with plastic biodegradation, particularly those involved in PE biodegradation, such as laccase, manganese peroxidase, lignin peroxidase, and alkane hydroxylase (AH), have also been found to play crucial roles (Wei and Zimmermann, 2017). It is worth noting that laccase and peroxidase (manganese peroxidase and lignin peroxidase) are also key enzymes in lignin degradation (Kavitha and Bhuvaneswari, 2021). Lignin as a complex amorphous aromatic biopolymer, which is connected by carbon–carbon bonds and ether bonds. Additionally, lignin and PE have partial similarities in terms of their structural and physicochemical properties, as they both have the characteristics of high hydrophobicity and high molecular weight. Thus, the enzymes required for the biodegradation of lignin, as a recalcitrant natural polymer, are similar to those required for the degradation of PE in that their degradation relies on the involvement of oxidoreductases to catalyze oxidation and generate free radicals to drive depolymerization (Kumar and Chandra, 2020; Daly et al., 2021).
The microbial degradation of plastic polymers, including PE, generally involves five basic stages (Figure 2): microbial colonization, biodeterioration, biofragmentation, assimilation, and mineralization (Gu, 2003).
(i) The initial phase involves the colonization of microorganisms on the surface of the plastic material. During this period, extracellular polysaccharides or biosurfactants allow microbes to cluster more easily and cling to the hydrophobic surface of the plastic material (Kavitha and Bhuvaneswari, 2021; Ji et al., 2022). The formation of biofilms on polymer surfaces not only is conducive to the colonization of microorganisms, but also severely damages the surfaces, enabling the addition of functional groups.
(ii) Typically, the microorganisms that colonize the surface of a plastic material are the cause of biodeterioration. Through the production of enzymes (such as lipase or dehydrogenase) or other secretions, these colonizing microbes penetrate polymer materials and subsequently generate interstices and cracks (Thakur et al., 2018; Rana et al., 2022). Additionally, some filamentous microorganisms (such as actinomycetes and molds) can deteriorate plastic matrices via their filamentous network structures (Sánchez, 2020). Moreover, the production of acids and alkalis during microbial metabolic processes may, in some cases, intensify the erosion of polymer surfaces (Ali et al., 2021).
(iii) Biofragmentation is the lytic process by which microorganisms convert macromolecular polymers into oligomers, dimers, or monomers via the action of enzymes or free radicals (Ghatge et al., 2020). Since polymers with macromolecular structures cannot be directly absorbed or utilized by microorganisms, they have to be disintegrated into smaller products that are suited to cell absorption and subsequent metabolism outside the cell (Lucas et al., 2008). PE, the main C–C-chain polymer with a high molecular weight, can only be supplied to microbial cells for further metabolism after being broken into oligomers of 10–50 carbon atoms (Krueger et al., 2015).
(iv) When polymers have been degraded into oligomers with low molecular weights (< 600 Da), microbes can easily absorb these oligomers via assimilation (Du et al., 2021). Finally, the polymers are mineralized under aerobic or anaerobic conditions through a range of metabolic pathways and to produce metabolites such as CO2, CH4, and H2O (Pathak and Navneet, 2017).
Several studies have found that bacteria can inhabit plastic-polluted environments (soil, water, etc.) by forming biofilms on the surfaces of discarded plastic debris, meaning that these microbial communities may be able to colonize plastic surfaces more effectively (Basili et al., 2020; Vaksmaa et al., 2021). The colonizing bacteria secrete the corresponding plastic-degrading enzymes, which catalyze the decomposition of the polymers into oligomers, dimers, and monomers through a combination of enzymes and finally mineralize them into other products, such as CO2 and H2O. Recent research has shown that Bacillus (Samanta et al., 2020), Enterobacter (Ren et al., 2019; Sarker et al., 2020), Brevibacterium (Hadad et al., 2005; Dwicania et al., 2019b), Pseudomonas (Hou et al., 2022), Arthrobacter (Balasubramanian et al., 2010), Acinetobacter (Montazer et al., 2018; Zhang et al., 2023), and other prevalent bacterial genera have the ability to degrade PE (Table 1). Notably, Bacillus spp. and Pseudomonas spp. also play key roles in the degradation of other plastic polymers, including polyethylene terephthalate (PET), PS, PP, and PUR (Auta et al., 2018; Roberts et al., 2020; Ganesh Kumar et al., 2021; Roy et al., 2021). In addition, bacteria that survive in extreme conditions, such as those that are thermophilic, acidophilic/alkaline, halophilic, radiation-resistant, etc., have significant potential for degrading PE (Atanasova et al., 2021). Mouafo Tamnou et al. (2021) utilized Pseudomonas aeruginosa to degrade PE under the conditions of an acidic environment (pH = 5) and a temperature of 44°C, and the weight loss rate of PE reached 6.25% after 30 days of culture. Hadad et al. (2005) recovered a thermophilic bacterial strain (Brevibacillus borstelensis 707) from soil and demonstrated that it reduced the weight of LDPE by 11%. Although previous studies have revealed many bacteria with PE-degradation functions, these bacteria have been determined to largely belong to the genera Bacillus and Pseudomonas; however, with the development of isolation and cultivation technologies, additional PE-degrading bacterial resources may still be discovered.
Actinomycetes, as a special subgroup of prokaryotes that can form branching hyphae and conidia, are abundant in soil and other habitats (Barka et al., 2016). As highly efficient polymer-degrading agents, Actinomycetes have been extensively studied in the degradation of natural polymers, such as lignocellulose (Tan et al., 2022) and natural rubber (Gibu et al., 2020). Regarding the degradation of polymeric polymers by Actinomycetes, current research primarily focuses on PET degradation. In the breakdown of PET by Actinomycetes, many enzymes that can hydrolyze polyester have been discovered (Wei et al., 2014; Charnock, 2021). Actinomycetes also secrete related enzymes during the degradation of PE. In one study, Santo et al. (2013) discovered that the extracellular laccase produced by the actinomycete Rhodococcus ruber C208 during PE degradation had greater PE-degradation activity when copper was present. Similar to other bacteria, the actinomycete colonization process is highly impacted by biofilm development. Actinomycetes are able to produce extracellular polymeric compounds, such as mucopolysaccharides rich in N-acetylglucosamine, which may improve their adhesion to plastic surfaces for later microbial action (Singh and Sedhuraman, 2015). The actinomycetes that have been identified in recent studies as having the ability to degrade PE are predominantly members of the genera Rhodococcus (Santo et al., 2013), Streptomyces (Soleimani et al., 2021), and Nocardia (Table 1; Koutny et al., 2006).
Fungi play indispensable roles in decomposing recalcitrant substrates and can make recalcitrant polymers, such as plastic, parts of their food web structures, driving the carbon cycle and nutrient regeneration in terrestrial ecosystems (Zeghal et al., 2021). The primary benefit of using fungi over single bacterial strains that can degrade plastic is that fungi grow more broadly in soil and can penetrate polymer surfaces using mycelia, making it easier for them to interact with plastic polymers and subsequently degrade plastic films (Sánchez, 2020). Additionally, fungi can secrete hydrophobic proteins that are capable of adhering to extremely hydrophobic polymer surfaces. These proteins enhance the development of aerial mycelia in filamentous fungi and the adherence of mycelia to hydrophobic polymer surfaces. Due to the high surface activity and strong adhesion properties of these proteins, fungal mycelia can grow more readily than bacteria on hydrophobic plastic surfaces (Piscitelli et al., 2017; Sánchez, 2020). Moreover, the enzymes required for PE degradation, such as laccase, lignin peroxidase, etc., are also found in the fungi-mediated PE breakdown process (Temporiti et al., 2022). Fungi have been shown to be capable of degrading PE, especially strains that belong to the genera Aspergillus, Fusarium, and Penicillium (Taghavi et al., 2021a), among others (Table 2).
The involvement of enzymes is essential for the microbial degradation of plastic. The molecular weights of plastic polymers are too high for direct uptake by microorganisms; therefore, extracellular enzymes are produced in advance to depolymerize polymers with high molecular weights into oligomers, dimers, monomers, etc., which can be ingested by microbial cells (John, 2019).
As the molecular structures of different plastic polymers vary, identification and further investigation of the specific enzymes for the degradation of different polymers is ongoing. For instance, in the biodegradation of polyethylene terephthalate (PET), it has been found that PETase (an aromatic polyester enzyme) from Ideonella sakaiensis 201-F6 can metabolize PET into bis-hydroxyethyl terephthalate (BHET), monohydroxyethyl terephthalate (MHET), terephthalic acid (TPA), and other intermediates, while MHETase (an auxiliary enzyme) further acts on MHET intermediates to convert them into terephthalic acid and ethylene glycol. Both enzymes are found in I. sakaiensis secretions and may work cooperatively to degrade PET (Austin et al., 2018). Magnin et al. (2019) demonstrated that a mixture of two enzymes [an efficient amidase (E4143) that hydrolyzes the PUR bonds of molecules with low molar masses and an esterase (E3576) that hydrolyzes aqueous polyester PUR dispersions] could be used to treat thermoplastic polyurethanes, resulting in a synergistic increase in the hydrolysis of PUR bonds.
Polyethylene (PE) has an extremely stable C–C backbone that is more resistant to hydrolysis than those of PET and PUR, which have chemical bonds in their backbones that can be attacked by hydrolases (Yeom et al., 2022). For this reason, certain enzymes and their related mechanisms are still inadequate for PE degradation. As mentioned in the previous section “2.3 Fungi involved in PE degradation”, the main fungal enzymes that have been discovered to be involved in PE degradation are laccase, manganese peroxidase (MnP), and lignin peroxidase (LiP). Gao et al. (2022) analyzed the genomics and transcriptomics of the isolated marine fungus Alternaria alternata FB1 with the potential for PE degradation and revealed significant upregulation of the genes that encode 153 enzymes potentially associated with PE degradation, including laccase, peroxidase, and hydroxylase, which further confirmed the ability of some of these enzymes to degrade PE. Laccase is also a potential functional enzyme for PE-degrading bacteria such as Bacillus, Psychrobacter, etc. (Liu et al., 2022c; Zhang A. et al., 2022). Jeon and Kim (2015) discovered an AH system composed of alkane monooxygenase, rubredoxin, and rubredoxin reductase in P. aeruginosa E7. The alkane monooxygenase had LMWPE (low-molecular-weight polyethylene) degradation activity, while the rubredoxin and rubredoxin reductase were indirectly involved in PE degradation through the transfer of relevant electrons. They also identified two alkane monooxygenases (AlkB1 and AlkB2) in P. aeruginosa and found that AlkB2 displayed higher transcriptional activity in the presence of LMWPE, thus indicating that AlkB2 was more efficient at degrading LMWPE than AlkB1 (Jeon and Kim, 2016).
Alkane hydroxylases (AHs) are key enzymes in the degradation of alkane compounds such as petroleum, and the genes encoding them have recently been identified in the genomes of a variety of PE-degrading species (Nie et al., 2014; Khalil et al., 2018; Hou et al., 2022). AHs harbor considerable diversity and can differ in terms of substrate ranges and degradation pathways (Ji et al., 2013). Polyethylene polymers with macromolecular weights can be depolymerized to form polymers with low molecular weights, as well as other alkanes, alcohols, and fatty acids with different chain lengths, via a range of abiotic processes (UV irradiation, heat treatment, oxidation, etc.) or enzymatic processes (laccase, peroxidase, etc.) (Zhang Y. et al., 2022). The ends of low-molecular-weight polyethylene can be oxidized by AHs through oxidation processes, including terminal oxidation, subterminal oxidation, etc. As a group of AHs, cytochrome P450 (CYPs) are also candidate enzymes for PE degradation (in addition to the AlkB family that was mentioned previously) and are also key enzymes for PE degradation by PE-degrading bacteria, such as Rhodococcus spp. and Bacillus spp. (Zampolli et al., 2021; Liu et al., 2022c). The alkanes of different chain lengths that are depolymerized by PE are oxidized by AHs to form alcohols, which are subsequently reacted with a series of cascade enzymes [such as alcohol dehydrogenase (Adh), aldehyde dehydrogenase (Aldh), Baeyer–Villiger monooxygenase (BVMO), and esterase, depending on the hydroxylation terminus] to form alkanoic acids that eventually enter the metabolic pathways of organisms and are consumed (Figure 3; Yeom et al., 2022).
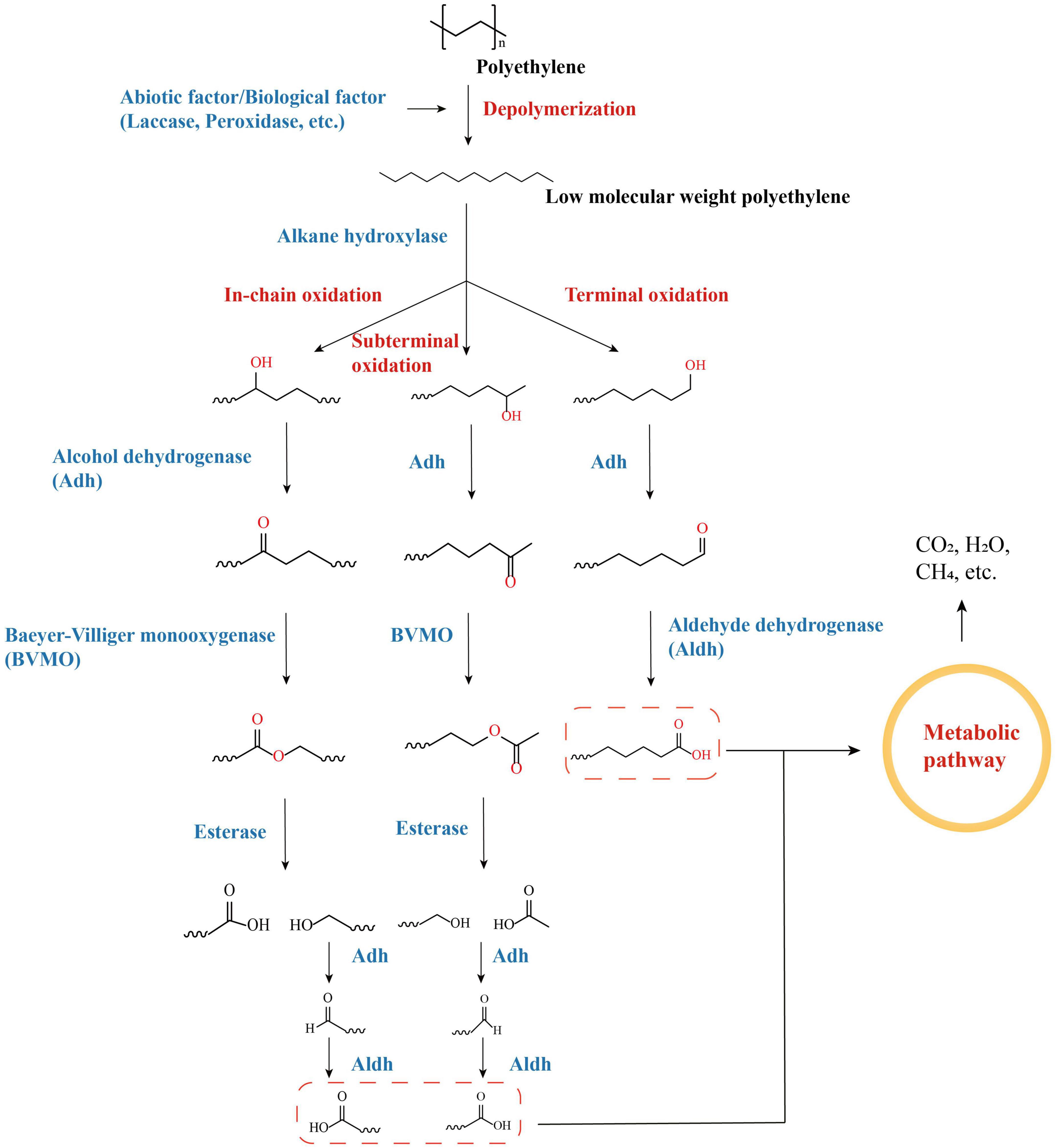
Figure 3. Degradation pathways of PE-derived long-chain linear alkanes by oxidation with alkane hydroxylases (AHs).
Microbial consortia, as efficient pollutant-degrading agents, have been applied to various environmental remediation fields, such as the degradation of agricultural and forestry residues [cellulose (Wang et al., 2010; Zhao et al., 2017; Du et al., 2018), hemicellulose (Wang et al., 2013; Tabanag et al., 2018; Zhang et al., 2018; Zheng et al., 2020), lignin (Kim et al., 2019; Lu et al., 2019; Zheng et al., 2020; Mendes et al., 2021), etc.], sewage treatment (Halat-Las et al., 2018; Feng et al., 2021), the degradation of persistent organic pollutants (POPs) (Arulazhagan et al., 2010; Li et al., 2021; Xu et al., 2021), and oil pollution treatment (Wu et al., 2017). In recent years, the potential for using microbial consortia in the biodegradation of plastic pollution has been investigated by an increasing number of relevant researchers.
Although many strains capable of degrading PE have been isolated and identified, it takes an incredibly long time for single strains/enzymes to degrade PE and the rate of degradation is exceedingly inefficient due to some of the durability characteristics of PE (Kotova et al., 2021). In addition, studies on the PE degradation products (PE-DPs) that are generated in these biodegradation processes are still in the preliminary stages. Some studies have used gas chromatograph-mass spectrometer (GC-MS) to conduct qualitative analyses and toxicity tests on PE-DPs (Shahnawaz et al., 2016). Studies have confirmed that PE-DPs are mostly composed of fatty acids, plasticizers, benzene, and alcohol; however, it is unclear whether PE-DPs subsequently interfere with microbial growth and metabolic activities (Sangale et al., 2019a). Consequently, these unknown and complicated byproducts may prevent single strains from degrading PE in the latter stages of fermentation. The stable and complicated coexistence of microbial communities arises via evolutionary changes in ecological and biological systems that have occurred over long periods of metabolic activity in the natural environment (Escalante et al., 2015). However, single strains gradually lose their naturally occurring microbial interdependence after isolation and purification, resulting in some attributes being reduced or lost (Ding et al., 2019).
Compared to single strains, microbial consortia are more efficient, robust, and controllable (Qian et al., 2020). (i) Firstly, the biodegradation of plastic polymers is a complex process driven by collaboration between various enzymes and metabolic pathways (Jaiswal et al., 2020). Microbial consortia have more abundant enzyme systems and metabolic activities than single strains (Zhang N. et al., 2022). (ii) Moreover, microbial consortia also create new microenvironments for strains, which may activate some metabolic pathways that are dormant under single-strain culture conditions (Qian et al., 2020). (iii) And then, microbial consortia generally adopt intercellular communication modes, such as quorum sensing (QS), to minimize competition between strains or synergistically regulate gene expression (Duncker et al., 2021). (iv) Concurrently, microbial consortia can execute the division of labor by partitioning metabolic pathways, thereby minimizing the accumulation of byproducts and the metabolic burden of individual strains (Jagmann and Philipp, 2014). (v) From the aspect of stability, microbial consortia that are composed of multifunctional microbes are more robust to environmental perturbations during biodegradation than single-strain cultures (Xu and Yu, 2021). (vi) It is important to note that despite the fact that some strains that are quite prevalent in microbial communities lack degrading capabilities, they may be of indispensable importance in the formation of biofilms on the surface of plastic debris (Giri et al., 2020; Gao and Sun, 2021). (vii) From a long-term perspective, microbial consortia can also effectively use carbon sources and access an expanded spectrum of substrates (Xu and Yu, 2021). Although the majority of plastic debris in the environment consists of mixed plastic, single strains can only target specific substrate polymers. In contrast, multifunctional consortia that are composed of diverse microorganisms can cope with plastic waste containing various types of polymers more effectively due to their vast biodegradation potential (Skariyachan et al., 2022).
Current research on the development of synthetic microbial consortia has generally followed two traditional approaches: “top-down” and “bottom-up” (Liang et al., 2022). Top-down techniques involve modification of environmental factors and continual enrichment and serial dilution of natural microbial communities derived from the environment, from which key microbial populations can be identified and stable PE -degrading consortia can ultimately be obtained (Lin, 2022). On the other hand, bottom-up techniques combine isolated strains and/or engineered microorganisms from the same or different sources, depending on their characteristics, and optimize them via appropriate adjustments to construct the desired functional synthetic consortia (Ding et al., 2019).
Recently, some researchers have attempted to construct microbial consortia/co-cultures by mixing single strains to achieve the more efficient degradation of PE, and part investigations on applications of microbial consortia in PE degradation are summarized in Table 3. Park and Kim (2019) combined Bacillus sp. and Paenibacillus sp. strains that were isolated from landfill sediments and had PE -degradation capabilities to construct a mixed bacterial consortium that, after 60 days of incubation, degraded up to 14.7% of PE microparticles. Skariyachan et al. (2016, 2021) isolated PE degrading strains from a plastic waste disposal area and cow manure, respectively, and then constructed PE degrading microbial consortia that achieved higher PE degradation compared to single strains as well as discovering that PE films lost more weight after treatment with the consortia than PE pellets, which is probably owing to the large surface size and thinness of plastic films giving better conditions for the colonization of microbes. Notably, Esmaeili et al. (2013) demonstrated the ability of fungal–bacterial cross-kingdom consortia to co-degrade LDPE films by burying PE films in soil inoculated with Aspergillus and Lysinibacillus strains. The advantage of using fungal–bacterial cross-kingdom consortia is that fungal hyphae may represent media for the bacterial colonization of soil that can facilitate the migration of bacterial strains beyond what can be accomplished individually. These studies have formed the basis for investigations into positive interactions between bacteria and fungi that have high potential for the bioremediation of plastic pollutants (Massot et al., 2022). Interestingly, Lou et al. (2022) and Ruiz Barrionuevo et al. (2022) studied the gut microbial communities of PE-fed wax worms and their larvae suggesting that the gut microbes of invertebrates and the constructed microbial consortia are capable of consuming and degrading plastic polymers, thus providing a new resource for the development of functional microbial consortia that have the capacity to degrade PE and other types of plastics. In addition, hydrocarbon-degrading bacteria (HCB) are prominent members of natural microbial communities derived from the plastisphere, given the long-term exposure to plastics, suggesting that PE enrichment cultures of microbial communities derived from the plastisphere will provide new potential for PE-degrading microbial communities (Harrison et al., 2014; Delacuvellerie et al., 2022a; Wang et al., 2022). For example, Joshi et al. (2022) screened potential LDPE-degrading marine bacterial communities associated with the plastisphere from plastic debris collected in coastal environments and then isolated, identified, and evaluated potential marine bacterial strains. Finally, they formulated an associated bacterial consortium that reduced the weight of LDPE by 47.07 ± 6.67% within 120 days.
The current studies on the construction of functional PE degradation consortia have focused on “bottom-up” construction seeking to incorporate previously isolated strains with significant efficiency of PE degradation aiming to improve the efficiency of biodegradation significantly. Unfortunately, the lack of a clear understanding of the key functional genes, naturally occurring microbial symbioses, and metabolic mechanisms of isolated PE -degrading strains currently restrict the construction of microbial consortia through a variety of limitations, including the existence of antagonistic effects between different strains, the heavy workload associated with arbitrarily attempting different microbial combinations, and the unknown metabolic mechanisms within microbial communities. Significantly, the vast majority of microorganisms in nature cannot be directly isolated in the laboratory using traditional microbial culture technology (Kaeberlein et al., 2002). Consequently, the combination of top-down and bottom-up approaches may be able to compensate for the deficiencies of each. The top-down selective enrichment of natural microbial communities could provide a framework for reconstructing PE -degrading consortia (Gilmore et al., 2019). The application of next-generation sequencing technologies, such as (meta)genomic and (meta)transcriptomic sequencing, could enhance our understanding of microbial diversity, composition, structures, functional characteristics, and metabolic activities of natural microbial communities, thereby enabling the targeted manipulation of synthetic functional consortia structures.
In recent years, genomics, metagenomics, transcriptomics, and proteomics as well as multi omics have been increasingly applied to the study of the microbial ecology of the plastisphere and its plastic-biodegradation mechanisms due to the rapid development of high-throughput sequencing (Gravouil et al., 2017; Zhang Z. et al., 2022; Rüthi et al., 2023). Through metagenomic analysis, the genomic information of microorganisms could be retrieved directly from the landfill plastisphere and used to construct genomic libraries (Culligan and Sleator, 2016). The interpretation of the microbial community structures of the plastisphere could uncover novel genes or enzymes involved in plastic-degradation pathways (Purohit et al., 2020). Pinto et al. (2022) analyzed a metagenome of seawater-derived PE biofilms after 2 years of enrichment with LDPE as a carbon source and found that functional genes gradually became more dominant over time. They also identified numerous microorganisms, metabolic pathways, and genes that could utilize LDPE and related components, including the gene alkb. Hou et al. (2022) discovered that biofilm formation was a distinct metabolic pathway through their genomic analysis of two Pseudomonas strains with potential PE -degradation capacity. Multiple genes that could encode PE -degrading enzymes were found in both strains (Hou et al., 2022). Gravouil et al. (2017) combined transcriptomics and lipidomics to analyze the expression of functional genes associated with R. ruber under PE exposure. Compared to the mannitol-supplemented controls, the PE supplementation resulted in the increased expression of 158 genes in R. ruber, with the most upregulated pathways associated with alkane degradation and fatty-acid β-oxidation. In the recent study, Gao and Sun (2021) reported that they obtained a marine bacterial community that effectively degraded PE and PET by screening a large number of samples collected from plastic waste. They then combined next-generation sequencing techniques to gain insights into the community composition and abundance, as well as three purified bacterial strains that were reconstituted into a synthetic functional bacterial community that was similar to the native microbial community. Transcriptomics was then used to elucidate the degradation mechanisms and the expression of the upregulated genes involved in the associated enzymes.
The integration of multi-omics analysis could not only contribute to the elucidation of PE -degradation pathways, microbial abundance, and community distributions, but also effectively reveal metabolic capacities at community and individual levels (Meyer-Cifuentes et al., 2020). In addition, the real-time multi-omics analysis of synthetic functional consortia could be used to monitor the dynamics and metabolic relationships between the functional consortia and could act as a guide for the later manipulation of population ratios and environmental parameters.
Natural microbial communities that form in ecological environments through long-term diversification, dispersion, selection, and drift have higher levels of complexity and diversity than synthetic synthetic consortia (Nemergut Diana et al., 2013). The symbiotic relationships that are established through interactions, such as reciprocity (commensalism and mutualism) and resource-based competition, help to maintain the diversity and stability of natural microbial communities (Blanchard and Lu, 2015; Liang et al., 2022; Qin et al., 2022). Correspondingly, the plastisphere is formed of natural microbial communities that are tightly attached to the surfaces of plastic debris that have been present in the natural environment for a long time. The structures of the microbial communities in the plastisphere vary depending on environmental factors, polymer type, and exposure time (Kirstein et al., 2019; Zhang S. J. et al., 2022).
The concept of the plastisphere, first proposed by Zettler et al. (2013), refers to diverse microbial communities consisting of heterotrophs, autotrophs, predators, and symbionts, which are attached to the surfaces of waste plastic materials. Through the discovery of pits on the surfaces of plastic waste debris using scanning electron microscope (SEM) techniques, it was hypothesized that the plastisphere, as a naturally occurring microbial ecological phenomenon, would have the potential to metabolize plastic waste. To date, many studies have explored the plastisphere in aquatic ecosystems such as oceans (Delacuvellerie et al., 2022b), lakes (Di Pippo et al., 2022), and rivers (Delacuvellerie et al., 2022a), as well as in soil ecosystems such as farmlands (Wang et al., 2022), mountains (Rüthi et al., 2023), and landfills (MacLean et al., 2021), using techniques such as amplicon sequencing and metagenome sequencing. These studies have analyzed the microbial community structures in the plastisphere and compared them to the microbial community structures in the surrounding environment from the same environmental source (Zhang S. J. et al., 2022; Zhurina et al., 2022). Moreover, the formation of mature and stable biofilms on the surfaces of plastic debris is required for the establishment of the plastisphere and the gradual formation of biofilms reduces the hydrophobicity of plastic and alters its functional groups (Tu et al., 2020; Du et al., 2022). With the formation of biofilms, the richness and diversity of species in the plastisphere gradually declines and the microorganisms involved in biofilm development and plastic degradation eventually come to dominate the microbial communities (Nguyen et al., 2022). In this regard, studies on biofilm formation and structure in the plastisphere may provide new ideas for consortium construction. In the construction of synthetic consortia, the existence of biofilms can stimulate the colonization of functional strains, stabilize the diversity of genotypes, and protect cells from detrimental environmental disturbances (Giri et al., 2020). Furthermore, studies on terrestrial ecosystems, especially landfills, as they are the most contaminated with plastic waste, have lagged significantly behind those on aquatic ecosystems. Since landfills are perennially contaminated with various types of plastic waste, the landfill plastisphere contains a variety of microbial community structures and potentially functional microorganisms that differ from those of marine ecosystems (Kumar et al., 2021). As a result, further research on the terrestrial plastisphere (especially in landfills, petroleum-hydrocarbon-contaminated soils, etc.) may provide new opportunities for the development of as yet unexplored potential plastic-degrading microbes/genes.
In general terms, top-down strategies have been used to selectively enrich the natural microbial communities of the plastisphere and optimize environmental parameters to obtain minimal and efficient plastisphere-derived PE -degrading microbial communities (Díaz-García et al., 2021). The studies to date on the metagenomics of the plastisphere and its functional microbial communities after enrichment have contributed to the identification of core and specific microbial populations and their ecological preferences, which could help to more effectively manipulate the compositions of microbial communities and comprehend the synergistic effects of strains (Zettler et al., 2013; Pinnell and Turner, 2019). Moreover, multi-omics, i.e., the combination of metagenomic, transcriptomic, and proteomic analyses, could be applied to interpret the microbial ecology of the plastisphere at the species, gene, and metabolic levels and help us to better understand the metabolic pathways that are involved in the process of PE degradation (Tiwari et al., 2022). Single strains with potential PE -degradation activities have been isolated from enriched communities and artificially reconstructed into stable and efficient synthetic functional consortia using bottom-up strategy, based on an omics analysis of the original plastisphere and its functional microbial communities, as well as a whole-genome analysis of the single strains (Figure 4; Jia et al., 2016; Liu et al., 2020).
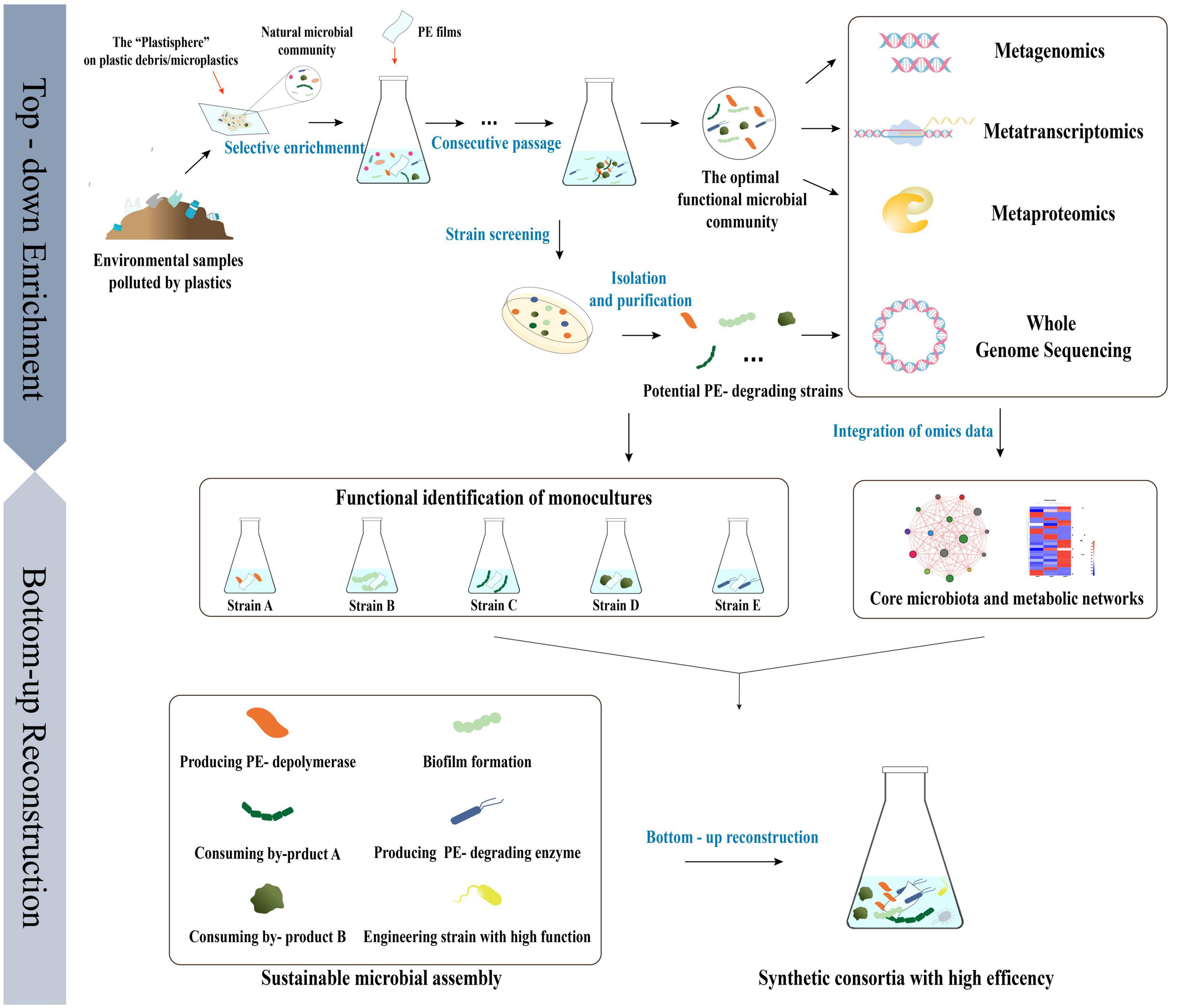
Figure 4. The strategy of “top-down” enrichment and “bottom-up” reconstruction combined to develop efficient PE degrading microbial consortia based on multi-omics analysis of the natural microbial community derived from the “Plastisphere” and the functional microbial community achieved after enrichment.
Despite the isolation and identification of a few functional PE-degrading microbes, the biodegradation process has been relatively slow in the majority of recent studies, with microbial culturing times ranging from 30 to 120 days and only minor degradation observed using characterization techniques such as weight loss or scanning electron microscopy. The most evident limitation of present research is the lack of uniform criteria and reliable methods for precisely quantifying deterioration efficiency. Currently, the most common type of PE substrate that is utilized in investigations is commercial PE. In addition, different studies have used different PE types (powder, particles, pieces, or films), initial weights, pretreatments, measurements, and culture conditions. Therefore, it is crucial to establish standard procedures that can be used to quantify PE degradation in subsequent studies to evaluate the biodegradation ability of microorganisms (Zhang Y. et al., 2022). Commercial PE also contains additives, such as plasticizers, colorants, and antioxidants, which make PE less degradable. It is also difficult to tell whether microorganisms preferentially utilize the additives as preferred carbon sources over the plastic, resulting in false positives. In addition, it is uncertain whether the various additives have negative impacts on the process of microbial biodegradation due to their levels of toxicity. Therefore, it is recommended that further studies eliminate additives and other contaminants using pretreatment processes, such as extraction, prior to investigating biodegradation (Ferreira et al., 2022). Alternatively, the integration of strains with specific degradation additives into synthetic consortia could be effective in both degrading the various additives and resolving PE polymers in synergy with other functional bacteria in the consortia.
Moreover, the low enzyme production capacities and simple enzymatic systems of single strains, as well as the lack of discovery of efficient specific enzymes for PE degradation in studies to date, render single strains incapable of effectively tackling the complex PE -degradation process. Therefore, it is suggested that both the exploration of novel efficient PE-degrading enzymes/genes and the investigation of degradation mechanisms be strengthened in future research. Additionally, the combination of genetic engineering and enzyme engineering technologies could help with the engineering of efficient PE-degrading-enzyme-producing bacteria and the enhancement of related enzyme activities. Various abiotic (e.g., temperature, UV, dissolved oxygen, humidity, pH, etc.) and biotic (e.g., microbial diversity and abundance, microbial enzymatic activity, etc.) factors affect the process of microbial PE degradation (Maity et al., 2021). Thus, different factors, including environmental parameters and culture conditions, should be considered to establish the optimal conditions for PE biodegradation.
As the production and properties of byproducts from PE -biodegradation pathways are not currently fully understood, there is an urgent need for additional studies on the properties of these byproducts and their implications for later-stage fermentation and environmental toxicity.
Remarkably, the majority of previous studies have focused on the degradation of large pieces of PE. In contrast, there has been a lack of studies on PE micro/nanoplastics, which are relatively tiny in size and well-hidden in the environment. These micro/nanoplastics are not as easily collected and disposed of as large pieces of plastic and, as they absorb enormous quantities of dangerous compounds during their movements around the environment, they significantly enhance the difficulty of plastic degradation (Liu et al., 2022a). For this reason, there is still a need to further investigate collection methods for and the biodegradation of PE micro/nanoplastics in the environment.
The microbial community structures and associated biofilms of the plastispheres in different ecosystems polluted by plastic waste have not been fully explored, particularly those in terrestrial ecosystems; hence, comprehensive multi-omics studies should be conducted to reveal the biodiversity, biofilm-formation mechanisms, and biodegradation capacity of these plastispheres in the future. Additionally, microbial metabolic engineering based on recombinant DNA or gene-editing technology may be effective to some degree in solving the limitations of PE-degrading single strains, such as poor degradation ability and low enzyme production capacity, thereby enhancing the metabolic activities of PE -degrading strains and significantly enhancing their biological degradation efficiency (Sharma and Shukla, 2022). Engineered microorganisms with high efficiency for degrading PE that are obtained through recombinant DNA technology or gene editing may have potential for future large-scale industrial applications.
Especially remarkable is the fact that, with the severe damage to the environment caused by conventional plastic disposal methods, more economically and environmentally acceptable plastic valorization is now increasingly being proposing and developed to address the ever-increasing volume of plastic waste. The valorization of waste plastics aims to maximize the value of plastic waste by recycling, upcycling, biodegradation, or energy recovery to dispose of the plastic waste in more sustainable ways and/or to transform them into brand new and functional products (Hou et al., 2021; Zhang F. et al., 2022; Table 4). Residual bulk plastic waste can be processed by either mechanical or chemical recycling, which involves manufacturing waste plastics into homogeneous or homologous renewable plastic products or breaking them down into their constituent molecules and applying them to produce other products, such as fuels, chemicals, etc. (Ragaert et al., 2017). Nowadays, the most common and time-efficient method of recovering energy from waste plastics is incineration, whereby part of the heat generated by the incineration process is recovered and applied to heat or electricity generation (Damayanti et al., 2022). Unfortunately, conventional recycling or directly incinerating plastic waste has many shortcomings, namely the labourious processes of collection, sorting and cleaning, the finite recycling times, exorbitant costs, the emission of hazardous residues and the unsuitability for heavily weathered plastic debris or micro/nano plastics (Lomwongsopon and Varrone, 2022). Instead, upcycling of plastic waste via chemical or biological methods into smaller molecules and transforming these molecules into new and valuable products is gradually appearing to be an attractive and more eco-friendly route to valorizing plastics in order to maximize the value of post-consumer plastics while minimizing the amount of plastic waste being directly discarded into the environment (Hou et al., 2021). The current studies on the upcycling of PE and other plastic wastes mainly focuses on the production of plastic wastes into fuels, high-value chemicals and multifunctional materials by chemical reactions including thermochemical catalysis (hydrolysis, oxidation, tandem catalysis, and carbonization), electrocatalysis, and photocatalysis, which will be widely applied in various fields such as energy, chemicals, medicine, construction, and textiles (Zhou et al., 2022).
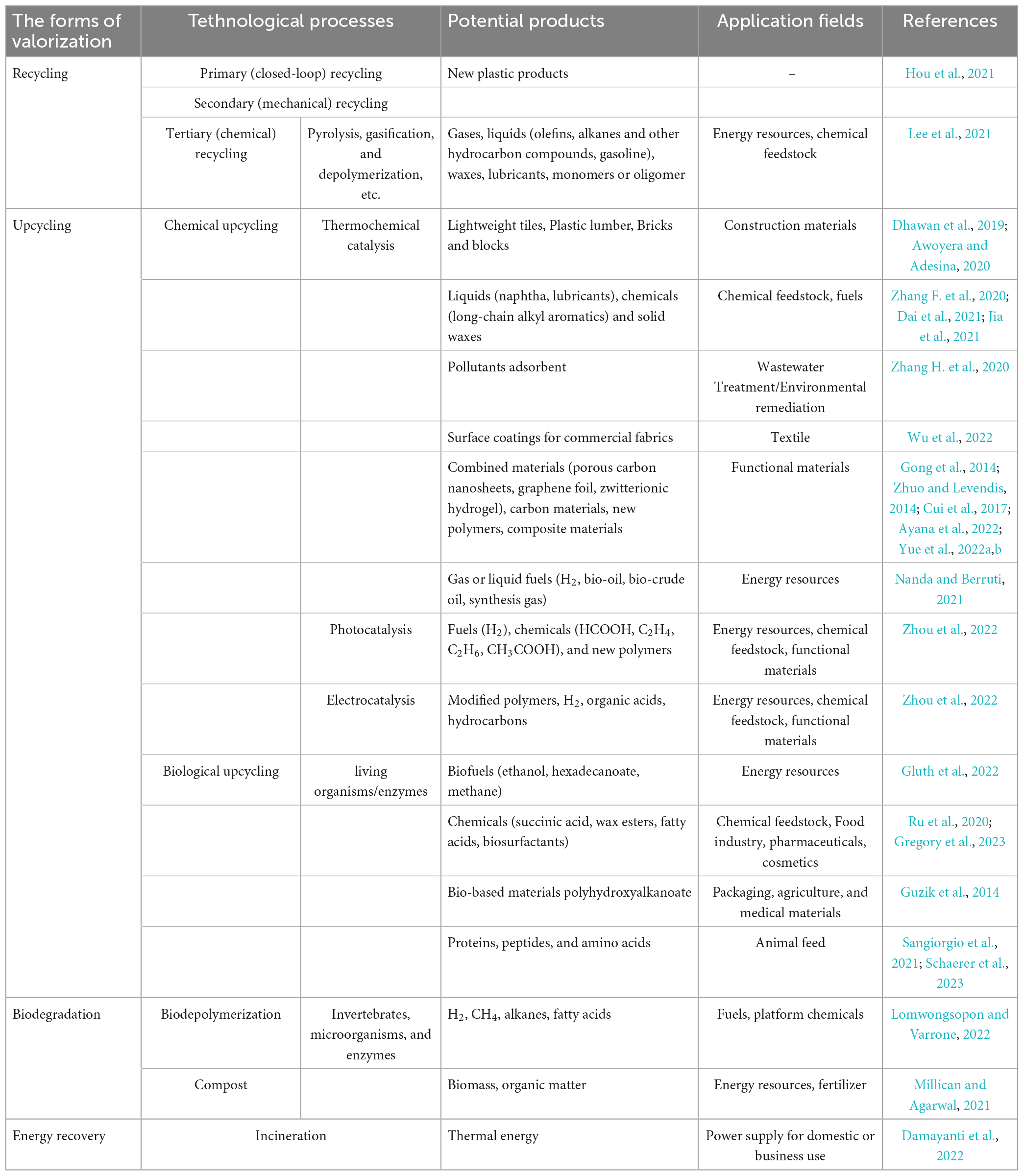
Table 4. Potential products and application area of waste plastic valorized via different strategies and technical routes.
The bio-upcycling of plastic waste is more sustainable and reaction conditions are more moderate than chemical processes. Combining biodegradation and upcycling of plastic waste enables to treat the non-recyclable plastic waste in the environment as well as to overcome the issue of product and by-product treatment during degradation of plastics. For one thing, the by-products and products derived from the biodegradation of PE could be taken directly into fuels or platform chemicals such as hydrogen, methane and various alkanes, fatty acids, etc. For the other hand, the oligomers or monomers obtained from the biodepolymerization of plastic can be biotransformed by engineered microbial cell factories to produce various value-added products (e.g., biofuels, fine chemicals, bio-based materials, biosurfactants, etc.) by upcycling (Guzik et al., 2014; Ru et al., 2020; Hou et al., 2021; Jehanno et al., 2022; Figure 5). Significantly, there is still relatively few studies on the upcycling of plastic waste, especially polyolefins such as PE, by microorganisms or enzymes given that there is limited understanding around the mechanisms of plastic biodegradation. The strategy of integrating microbial consortiums with multi-omics can further suggest a new vision on the bio-upcycling of plastic waste. The application of multi-omics will contribute to the deeper insights on the metabolic mechanisms of plastic biodegradation and the understanding of microbial interactions between microbial communities. The rational design and synthesis of the microbial consortia’s composition and the pathways of microbial metabolism would be more favorable to the promising biotransformation into a larger variety of potential products of plastic biodegradation, thereby extending the range of products that can be produced through upcycling of plastic waste.
Furthermore, the adoption of gene engineering, metabolic engineering and other technologies allow further enhancement of functional strains and optimization of metabolic pathways for more efficient and targeted biotransformation into different products or specific chemicals derived from waste plastics (Sullivan et al., 2022).
The prevalence of plastic waste and micro/nanoplastic pollution has always been a great challenge facing environmental remediation. Traditional plastic disposal methods have not been effective in solving the global plastic waste pollution problem, which is becoming increasingly serious. However, recent studies on plastic biodegradation have led to new perspectives on the sustainable disposal of plastic waste and the discovery of an increasing number of microbes and other invertebrates that are involved in plastic degradation. PE is the most widespread and recalcitrant petroleum-based plastic and the study of its biodegradation has received considerable interest over the years. The microorganisms and other organisms involved in the degradation of PE have been extensively explored in the literature; however, there have been no breakthroughs in the study of PE biodegradation, particularly regarding the mechanisms of PE degradation and the identification of specific PE depolymerases. Recent studies have examined the constraints that single strains encounter when trying to degrade PE, and progressive efforts have been made to combine several strains to develop more effective consortia or co-cultures. Compared to single strains, functional microbial consortia offer many advantages in degrading plastic waste; however, the lack of an understanding of the byproducts and mechanisms of PE degradation has limited the development of artificial functional consortia for PE degradation, which was the focus of this review.
Considering the existing bottlenecks in the construction of PE-degrading consortia and the pros and cons of each strategy, we propose combining top-down and bottom-up approaches for the synthesis of efficient PE-degrading consortia. In this way, the selective, top-down enrichment of natural microbial communities can be applied to optimize community functions, and the enriched communities can undergo omics analyses of their functions and community structures to help us better understand the microbial PE -degradation mechanisms and microbial interactions at play within the communities. The insights into microbial communities achieved using top-down strategies could contribute to a more reasonable and appropriate bottom-up synthesis of functional PE -degrading consortia. The plastisphere is a new ecological niche that spontaneously forms on plastic debris and is closely associated with plastic polymers. Investigating the microorganisms or microbial communities that grow in the plastisphere could uncover principal sources of novel PE-degrading microorganisms/microbiota. The combination of genomic, transcriptomic, metagenomic, and other omics/meta-omics analyses could help to unravel the microbial diversity and community composition of the plastispheres in different ecosystems and the metabolic relationships between microbes, which could also enable the discovery of core microbiota and key pathways/genes for PE degradation and provide a foundation for the subsequent construction of functional consortia.
We close with a discussion of concerns and bottlenecks in the existing studies on PE biodegradation as well as some insights into the topics that should be considered in future research. Due to insufficient study of the mechanisms of PE biodegradation, the construction of effective microbial consortia currently faces numerous obstacles. In addition, the safety and stability of synthetic microbial consortia and the regulation of coexistence between microorganisms could represent challenges for future research due to the dynamic nature of microbial communities, biological variability, and various other uncertainties.
CZ: conceptualization, data curation, and writing-original draft preparation. YM, TL, and F-JJ: data curation and visualization; C-ZJ and H-MO: resources and visualization; H-GL. and LJ: supervision and writing-reviewing and editing. All authors have read and agreed to the published version of the manuscript.
This research was funded by the Korea Environment Industry & Technology Institute (KEITI) through a project to develop new, eco-friendly materials and processing technology derived from wildlife, funded by the Korea Ministry of Environment (MOE) (grant number 2021003240004), by the National Research Council of Science and Technology (NST) grant by the Korea government (MSIT) (CAP20023-200), and by the Korea Research Institute of Bioscience and Biotechnology (KRIBB) Research Initiative Program (KGM5252322).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Ali, S. S., Elsamahy, T., Koutra, E., Kornaros, M., El-Sheekh, M., Abdelkarim, E. A., et al. (2021). Degradation of conventional plastic wastes in the environment: A review on current status of knowledge and future perspectives of disposal. Sci. Total Environ. 771:144719. doi: 10.1016/j.scitotenv.2020.144719
Amobonye, A., Bhagwat, P., Singh, S., and Pillai, S. (2021). Plastic biodegradation: Frontline microbes and their enzymes. Sci. Total Environ. 759:143536. doi: 10.1016/j.scitotenv.2020.143536
Andrady, A. L. (2011). Microplastics in the marine environment. Marine Pollut. Bull. 62, 1596–1605. doi: 10.1016/j.marpolbul.2011.05.030
Arulazhagan, P., Vasudevan, N., and Yeom, I. T. (2010). Biodegradation of polycyclic aromatic hydrocarbon by a halotolerant bacterial consortium isolated from marine environment. Int. Journal of Environ. Sci. Technol. 7, 639–652. doi: 10.1007/BF03326174
Atanasova, N., Stoitsova, S., Paunova-Krasteva, T., and Kambourova, M. (2021). Plastic degradation by extremophilic bacteria. Int. J. Mol. Sci. 22:5610. doi: 10.3390/ijms22115610
Austin, H. P., Allen, M. D., Donohoe, B. S., Rorrer, N. A., Kearns, F. L., Silveira, R. L., et al. (2018). Characterization and engineering of a plastic-degrading aromatic polyesterase. Proc. Natl. Acad. Sci. 115, E4350–E4357. doi: 10.1073/pnas.1718804115
Auta, H. S., Emenike, C. U., and Fauziah, S. H. (2017). Screening of Bacillus strains isolated from mangrove ecosystems in Peninsular Malaysia for microplastic degradation. Environ. Pollut. 231, 1552–1559. doi: 10.1016/j.envpol.2017.09.043
Auta, H. S., Emenike, C. U., Jayanthi, B., and Fauziah, S. H. (2018). Growth kinetics and biodeterioration of polypropylene microplastics by Bacillus sp. and Rhodococcus sp. isolated from mangrove sediment. Mar. Pollut. Bull. 127, 15–21. doi: 10.1016/j.marpolbul.2017.11.036
Awoyera, P. O., and Adesina, A. (2020). Plastic wastes to construction products: Status, limitations and future perspective. Case Stud. Constr. Mater. 12:e00330. doi: 10.1016/j.cscm.2020.e00330
Ayana, K. D., De Angelis, M., Schmidt, G., Krause, A., and Ali, A. Y. (2022). Valorization of potential post-consumer polyethylene (PE) plastics waste and ethiopian indigenous highland bamboo (EHB) for wood plastic composite (WPC): Experimental evaluation and characterization. Fibers 10:85. doi: 10.3390/fib10100085
Balasubramanian, V., Natarajan, K., Hemambika, B., Ramesh, N., Sumathi, C. S., Kottaimuthu, R., et al. (2010). High-density polyethylene (HDPE)-degrading potential bacteria from marine ecosystem of Gulf of Mannar, India. Lett. Appl. Microbiol. 51, 205–211. doi: 10.1111/j.1472-765X.2010.02883.x
Bardají, D. K. R., Furlan, J. P. R., and Stehling, E. G. (2019). Isolation of a polyethylene degrading Paenibacillus sp. from a landfill in Brazil. Arch. Microbiol. 201, 699–704. doi: 10.1007/s00203-019-01637-9
Barka, E. A., Vatsa, P., Sanchez, L., Gaveau-Vaillant, N., Jacquard, C., Klenk, H.-P., et al. (2016). Taxonomy, physiology, and natural products of actinobacteria. Microbiol. Mol. Biol. Rev. 80, 1–43. doi: 10.1128/MMBR.00019-15
Basili, M., Quero, G. M., Giovannelli, D., Manini, E., Vignaroli, C., Avio, C. G., et al. (2020). Major role of surrounding environment in shaping biofilm community composition on marine plastic debris. Front. Mar. Sci. 7:262. doi: 10.3389/fmars.2020.00262
Biki, S. P., Mahmud, S., Akhter, S., Rahman, M. J., Rix, J. J., Al Bachchu, M. A., et al. (2021). Polyethylene degradation by Ralstonia sp. strain SKM2 and Bacillus sp. strain SM1 isolated from land fill soil site. Environ. Technol. Innov. 22:101495. doi: 10.1016/j.eti.2021.101495
Blanchard, A. E., and Lu, T. (2015). Bacterial social interactions drive the emergence of differential spatial colony structures. BMC Syst. Biol. 9:59. doi: 10.1186/s12918-015-0188-5
Canopoli, L., Coulon, F., and Wagland, S. T. (2020). Degradation of excavated polyethylene and polypropylene waste from landfill. Sci. Total Environ. 698:134125. doi: 10.1016/j.scitotenv.2019.134125
Canopoli, L., Fidalgo, B., Coulon, F., and Wagland, S. T. (2018). Physico-chemical properties of excavated plastic from landfill mining and current recycling routes. Waste Manage. 76, 55–67. doi: 10.1016/j.wasman.2018.03.043
Chamas, A., Moon, H., Zheng, J., Qiu, Y., Tabassum, T., Jang, J. H., et al. (2020). Degradation rates of plastics in the environment. ACS Sust. Chem. Eng. 8, 3494–3511. doi: 10.1021/acssuschemeng.9b06635
Charnock, C. (2021). Norwegian soils and waters contain mesophilic, plastic-degrading bacteria. Microorganisms 9:94. doi: 10.3390/microorganisms9010094
Cowan, A. R., Costanzo, C. M., Benham, R., Loveridge, E. J., and Moody, S. C. (2022). Fungal bioremediation of polyethylene: Challenges and perspectives. J. Appl. Microbiol. 132, 78–89. doi: 10.1111/jam.15203
Cui, L., Wang, X., Chen, N., Ji, B., and Qu, L. (2017). Trash to treasure: Converting plastic waste into a useful graphene foil. Nanoscale 9, 9089–9094. doi: 10.1039/C7NR03580B
Culligan, E. P., and Sleator, R. D. (2016). Editorial: From genes to species: Novel insights from metagenomics. Front. Microbiol. 7:1181. doi: 10.3389/fmicb.2016.01181
Dai, L., Zhou, N., Lv, Y., Cheng, Y., Wang, Y., Liu, Y., et al. (2021). Chemical upcycling of waste polyolefinic plastics to low-carbon synthetic naphtha for closing the plastic use loop. Sci. Total Environ. 782:146897. doi: 10.1016/j.scitotenv.2021.146897
Daly, P., Cai, F., Kubicek, C. P., Jiang, S., Grujic, M., Rahimi, M. J., et al. (2021). From lignocellulose to plastics: Knowledge transfer on the degradation approaches by fungi. Biotechnol. Adv. 50:107770. doi: 10.1016/j.biotechadv.2021.107770
Damayanti, D., Saputri, D. R., Marpaung, D. S. S., Yusupandi, F., Sanjaya, A., Simbolon, Y. M., et al. (2022). Current prospects for plastic waste treatment. Polymers 14:3133. doi: 10.3390/polym14153133
Danso, D., Chow, J., and Streit Wolfgang, R. (2019). Plastics: Environmental and biotechnological perspectives on microbial degradation. Appl. Environ. Microbiol. 85, e1095–e1019. doi: 10.1128/AEM.01095-19
Delacuvellerie, A., Ballerini, T., Frère, L., Matallana-Surget, S., Dumontet, B., and Wattiez, R. (2022a). From rivers to marine environments: A constantly evolving microbial community within the plastisphere. Mar. Pollut. Bull. 179:113660. doi: 10.1016/j.marpolbul.2022.113660
Delacuvellerie, A., Géron, A., Gobert, S., and Wattiez, R. (2022b). New insights into the functioning and structure of the PE and PP plastispheres from the Mediterranean Sea. Environ. Pollut. 295:118678. doi: 10.1016/j.envpol.2021.118678
Delacuvellerie, A., Cyriaque, V., Gobert, S., Benali, S., and Wattiez, R. (2019). The plastisphere in marine ecosystem hosts potential specific microbial degraders including Alcanivorax borkumensis as a key player for the low-density polyethylene degradation. J. Hazard. Mater. 380:120899. doi: 10.1016/j.jhazmat.2019.120899
Dey, A. S., Bose, H., Mohapatra, B., and Sar, P. (2020). Biodegradation of unpretreated low-density polyethylene (LDPE) by Stenotrophomonas sp. and Achromobacter sp., isolated from waste dumpsite and drilling fluid. Front. Microbiol. 11:603210. doi: 10.3389/fmicb.2020.603210
Dhawan, R., Bisht, B. M. S., Kumar, R., Kumari, S., and Dhawan, S. K. (2019). Recycling of plastic waste into tiles with reduced flammability and improved tensile strength. Process Saf. Environ. Protect. 124, 299–307. doi: 10.1016/j.psep.2019.02.018
Di Pippo, F., Crognale, S., Levantesi, C., Vitanza, L., Sighicelli, M., Pietrelli, L., et al. (2022). Plastisphere in lake waters: Microbial diversity, biofilm structure, and potential implications for freshwater ecosystems. Environ. Pollut. 310:119876. doi: 10.1016/j.envpol.2022.119876
Díaz-García, L., Huang, S., Spröer, C., Sierra-Ramírez, R., Bunk, B., Overmann, J., et al. (2021). Dilution-to-stimulation/extinction method: A combination enrichment strategy to develop a minimal and versatile lignocellulolytic bacterial consortium. Appl. Environ. Microbiol. 87, e2427–e2420. doi: 10.1128/AEM.02427-20
Dimassi, S. N., Hahladakis, J. N., Yahia, M. N. D., Ahmad, M. I., Sayadi, S., and Al-Ghouti, M. A. (2022). Degradation-fragmentation of marine plastic waste and their environmental implications: A critical review. Arab. J. Chem. 15:104262. doi: 10.1016/j.arabjc.2022.104262
Ding, X., Seebeck, T., Feng, Y., Jiang, Y., Davis, G. D., and Chen, F. (2019). Improving CRISPR-Cas9 genome editing efficiency by fusion with chromatin-modulating peptides. Crispr J. 2, 51–63. doi: 10.1089/crispr.2018.0036
Dsouza, G. C., Sheriff, R. S., Ullanat, V., Shrikrishna, A., Joshi, A. V., Hiremath, L., et al. (2021). Fungal biodegradation of low-density polyethylene using consortium of Aspergillus species under controlled conditions. Heliyon 7:e07008. doi: 10.1016/j.heliyon.2021.e07008
Du, H., Xie, Y., and Wang, J. (2021). Microplastic degradation methods and corresponding degradation mechanism: Research status and future perspectives. J. Hazard. Mater. 418:126377. doi: 10.1016/j.jhazmat.2021.126377
Du, R., Guo, L., Li, S., Xie, D., and Yan, J. (2018). Metagenomic DNA extraction of natural cellulose-degrading consortia. Bioenergy Res. 11, 115–122. doi: 10.1007/s12155-017-9882-z
Du, Y., Liu, X., Dong, X., and Yin, Z. (2022). A review on marine plastisphere: Biodiversity, formation, and role in degradation. Comput. Struct. Biotechnol. J. 20, 975–988. doi: 10.1016/j.csbj.2022.02.008
Duncker, K. E., Holmes, Z. A., and You, L. (2021). Engineered microbial consortia: Strategies and applications. Microb. Cell Fact. 20:211. doi: 10.1186/s12934-021-01699-9
Dwicania, E., Rinanti, A., and Fachrul, M. F. (2019b). “Biodegradation of LLDPE plastic by mixed bacteria culture of Pseudomonas aeruginosa and Brevibacterium sp,” in Proceedings of the 4th annual applied science and engineering conference, Bali.
Dwicania, E., Rinanti, A., and Fachrul, M. F. (2019a). Biodegradation of LLDPE plastic by mixed bacteria culture of Pseudomonas aeruginosa and Brevibacterium sp. J. Phys. 1402:022105.
Escalante, A. E., Rebolleda-Gómez, M., Benítez, M., and Travisano, M. (2015). Ecological perspectives on synthetic biology: Insights from microbial population biology. Front. Microbiol 6:143. doi: 10.3389/fmicb.2015.00143
Esmaeili, A., Pourbabaee, A. A., Alikhani, H. A., Shabani, F., and Esmaeili, E. (2013). Biodegradation of low-density polyethylene (LDPE) by mixed culture of Lysinibacillus xylanilyticus and Aspergillus niger in soil. PLoS One 8:e71720. doi: 10.1371/journal.pone.0071720
Feng, S., Gong, L., Zhang, Y., Tong, Y., Zhang, H., Zhu, D., et al. (2021). Bioaugmentation potential evaluation of a bacterial consortium composed of isolated Pseudomonas and Rhodococcus for degrading benzene, toluene and styrene in sludge and sewage. Bioresour. Technol. 320:124329. doi: 10.1016/j.biortech.2020.124329
Ferreira, A. M., Sucena, I., Otero, V., Angelin, E. M., Melo, M. J., and Coutinho, J. A. P. (2022). Pretreatment of plastic waste: Removal of colorants from HDPE using biosolvents. Molecules 27:98. doi: 10.3390/molecules27010098
Gajendiran, A., Krishnamoorthy, S., and Abraham, J. (2016). Microbial degradation of low-density polyethylene (LDPE) by Aspergillus clavatus strain JASK1 isolated from landfill soil. 3 Biotech 6:52. doi: 10.1007/s13205-016-0394-x
Ganesh Kumar, A., Hinduja, M., Sujitha, K., Nivedha Rajan, N., and Dharani, G. (2021). Biodegradation of polystyrene by deep-sea Bacillus paralicheniformis G1 and genome analysis. Sci. Total Environ. 774:145002. doi: 10.1016/j.scitotenv.2021.145002
Gao, R., and Sun, C. (2021). A marine bacterial community capable of degrading poly(ethylene terephthalate) and polyethylene. J. Hazard. Mater. 416:125928. doi: 10.1016/j.jhazmat.2021.125928
Gao, R., Liu, R., and Sun, C. (2022). A marine fungus Alternaria alternata FB1 efficiently degrades polyethylene. J. Hazard. Mater. 431:128617. doi: 10.1016/j.jhazmat.2022.128617
Gewert, B., Plassmann, M. M., and MacLeod, M. (2015). Pathways for degradation of plastic polymers floating in the marine environment. Environ. Sci. 17, 1513–1521. doi: 10.1039/C5EM00207A
Ghatge, S., Yang, Y., Ahn, J.-H., and Hur, H.-G. (2020). Biodegradation of polyethylene: A brief review. Appl. Biol. Chem. 63:27. doi: 10.1186/s13765-020-00511-3
Gibu, N., Arata, T., Kuboki, S., Dao Viet, L., Fukuda, M., Steinbuechel, A., et al. (2020). Characterization of the genes responsible for rubber degradation in Actinoplanes sp. strain OR16. Appl. Microbiol. Biotechnol. 104, 7367–7376. doi: 10.1007/s00253-020-10700-1
Gilan, I., Hadar, Y., and Sivan, A. (2004). Colonization, biofilm formation and biodegradation of polyethylene by a strain of Rhodococcus ruber. Appl. Microbiol. Biotechnol. 65, 97–104. doi: 10.1007/s00253-004-1584-8
Gilmore, S. P., Lankiewicz, T. S., Wilken, S. E., Brown, J. L., Sexton, J. A., Henske, J. K., et al. (2019). Top-down enrichment guides in formation of synthetic microbial consortia for biomass degradation. ACS Synth. Biol. 8, 2174–2185. doi: 10.1021/acssynbio.9b00271
Giri, S., Shitut, S., and Kost, C. (2020). Harnessing ecological and evolutionary principles to guide the design of microbial production consortia. Curr. Opin. Biotechnol. 62, 228–238. doi: 10.1016/j.copbio.2019.12.012
Gluth, A., Xu, Z., Fifield, L. S., and Yang, B. (2022). Advancing biological processing for valorization of plastic wastes. Renew. Sust. Energy Rev. 170:112966. doi: 10.1016/j.rser.2022.112966
Gong, J., Michalkiewicz, B., Chen, X., Mijowska, E., Liu, J., Jiang, Z., et al. (2014). Sustainable conversion of mixed plastics into porous carbon nanosheets with high performances in uptake of carbon dioxide and storage of hydrogen. ACS Sust. Chem. Eng. 2, 2837–2844. doi: 10.1021/sc500603h
Gravouil, K., Ferru-Clément, R., Colas, S., Helye, R., Kadri, L., Bourdeau, L., et al. (2017). Transcriptomics and lipidomics of the environmental strain Rhodococcus ruber point out consumption pathways and potential metabolic bottlenecks for polyethylene degradation. Environ. Sci. Technol. 51, 5172–5181. doi: 10.1021/acs.est.7b00846
Gregory, G. J., Wang, C., Sadula, S., Koval, S., Lobo, R. F., Vlachos, D. G., et al. (2023). Polyethylene valorization by combined chemical catalysis with bioconversion by plastic-enriched microbial consortia. ACS Sust. Chem. Eng. 11, 3494–3505. doi: 10.1021/acssuschemeng.2c07461
Gu, J.-D. (2003). Microbiological deterioration and degradation of synthetic polymeric materials: Recent research advances. Int. Biodeterior. Biodegradation 52, 69–91. doi: 10.1016/S0964-8305(02)00177-4
Gupta, K. K., Sharma, K. K., and Chandra, H. (2022). Micrococcus luteus strain CGK112 isolated from cow dung demonstrated efficient biofilm-forming ability and degradation potential toward high-density polyethylene (HDPE). Arch. Microbiol. 204:402. doi: 10.1007/s00203-022-03023-4
Guzik, M. W., Kenny, S. T., Duane, G. F., Casey, E., Woods, T., Babu, R. P., et al. (2014). Conversion of post consumer polyethylene to the biodegradable polymer polyhydroxyalkanoate. Appl. Microbiol. Biotechnol. 98, 4223–4232. doi: 10.1007/s00253-013-5489-2
Hadad, D., Geresh, S., and Sivan, A. (2005). Biodegradation of polyethylene by the thermophilic bacterium Brevibacillus borstelensis. J. Appl. Microbiol. 98, 1093–1100. doi: 10.1111/j.1365-2672.2005.02553.x
Halat-Las, M., Jedynak, P., Malec, P., Burczyk, J., Vilches, A. I., and Kaszyckl, P. (2018). Application of microbial consortia for biological treatment of post-fermentation effluents from biomethanization of sewage sludge and pig manure. Przem. Chem. 97, 1189–1194.
Han, Y.-N., Wei, M., Han, F., Fang, C., Wang, D., Zhong, Y.-J., et al. (2020). Greater Biofilm Formation and Increased Biodegradation of Polyethylene Film by a Microbial Consortium of Arthrobacter sp. and Streptomyces sp. Microorganisms 8:1979. doi: 10.3390/microorganisms8121979
Harrison, J. P., Schratzberger, M., Sapp, M., and Osborn, A. M. (2014). Rapid bacterial colonization of low-density polyethylene microplastics in coastal sediment microcosms. BMC Microbiol. 14:232. doi: 10.1186/s12866-014-0232-4
Harshvardhan, K., and Jha, B. (2013). Biodegradation of low-density polyethylene by marine bacteria from pelagic waters, Arabian Sea, India. Mar. Pollut. Bull. 77, 100–106. doi: 10.1016/j.marpolbul.2013.10.025
Hou, L., Xi, J., Liu, J., Wang, P., Xu, T., Liu, T., et al. (2022). Biodegradability of polyethylene mulching film by two Pseudomonas bacteria and their potential degradation mechanism. Chemosphere 286:131758. doi: 10.1016/j.chemosphere.2021.131758
Hou, Q., Zhen, M., Qian, H., Nie, Y., Bai, X., Xia, T., et al. (2021). Upcycling and catalytic degradation of plastic wastes. Cell Rep. Phys. Sci. 2:100514. doi: 10.1016/j.xcrp.2021.100514
Jagmann, N., and Philipp, B. (2014). Reprint of design of synthetic microbial communities for biotechnological production processes. J. Biotechnol. 192, 293–301. doi: 10.1016/j.jbiotec.2014.11.005
Jaiswal, S., Sharma, B., and Shukla, P. (2020). Integrated approaches in microbial degradation of plastics. Environ. Technol. Innov. 17:100567. doi: 10.1016/j.eti.2019.100567
Jehanno, C., Alty, J. W., Roosen, M., De Meester, S., Dove, A. P., Chen, E. Y. X., et al. (2022). Critical advances and future opportunities in upcycling commodity polymers. Nature 603, 803–814. doi: 10.1038/s41586-021-04350-0
Jeon, H. J., and Kim, M. N. (2015). Functional analysis of alkane hydroxylase system derived from Pseudomonas aeruginosa E7 for low molecular weight polyethylene biodegradation. Int. Biodeterior. Biodegradation 103, 141–146. doi: 10.1016/j.ibiod.2015.04.024
Jeon, H. J., and Kim, M. N. (2016). Comparison of the functional characterization between alkane monooxygenases for low-molecular-weight polyethylene biodegradation. Int. Biodeterior. Biodegradation 114, 202–208. doi: 10.1016/j.ibiod.2016.06.012
Jeon, J.-M., Park, S.-J., Choi, T.-R., Park, J.-H., Yang, Y.-H., and Yoon, J.-J. (2021). Biodegradation of polyethylene and polypropylene by Lysinibacillus species JJY0216 isolated from soil grove. Polym. Degrad. Stab. 191:109662. doi: 10.1016/j.polymdegradstab.2021.109662
Ji, J., Zhao, T., and Li, F. (2022). Remediation technology towards zero plastic pollution: Recent advance and perspectives. Environ. Pollut. 313:120166. doi: 10.1016/j.envpol.2022.120166
Ji, Y., Mao, G., Wang, Y., and Bartlam, M. (2013). Structural insights into diversity and n-alkane biodegradation mechanisms of alkane hydroxylases. Front. Microbiol. 4:58. doi: 10.3389/fmicb.2013.00058
Jia, C., Xie, S., Zhang, W., Intan, N. N., Sampath, J., Pfaendtner, J., et al. (2021). Deconstruction of high-density polyethylene into liquid hydrocarbon fuels and lubricants by hydrogenolysis over Ru catalyst. Chem Catal. 1, 437–455. doi: 10.1016/j.checat.2021.04.002
Jia, X., Liu, C., Song, H., Ding, M., Du, J., Ma, Q., et al. (2016). Design, analysis and application of synthetic microbial consortia. Synth. Syst. Biotechnol. 1, 109–117. doi: 10.1016/j.synbio.2016.02.001
John, A. G. (2019). “Biological degradation of polymers in the environment,” in Plastics in the environment, ed. G. Alessio (Rijeka: IntechOpen), 5.
Joshi, G., Goswami, P., Verma, P., Prakash, G., Simon, P., Vinithkumar, N. V., et al. (2022). Unraveling the plastic degradation potentials of the plastisphere-associated marine bacterial consortium as a key player for the low-density polyethylene degradation. J. Hazard. Mater. 425:128005. doi: 10.1016/j.jhazmat.2021.128005
Kaeberlein, T., Lewis, K., and Epstein, S. S. (2002). Isolating “uncultivable” microorganisms in pure culture in a simulated natural environment. Science 296, 1127–1129. doi: 10.1126/science.1070633
Kavitha, R., and Bhuvaneswari, V. (2021). Assessment of polyethylene degradation by biosurfactant producing ligninolytic bacterium. Biodegradation 32, 531–549. doi: 10.1007/s10532-021-09949-8
Khalil, A. B., Sivakumar, N., Arslan, M., Saleem, H., and Qarawi, S. (2018). Insights into Brevibacillus borstelensis AK1 through whole genome sequencing: A thermophilic bacterium Isolated from a Hot Spring in Saudi Arabia. Biomed Res. Int. 2018:5862437. doi: 10.1155/2018/5862437
Khan, S., Ali, S. A., and Ali, A. S. (2022). Biodegradation of low density polyethylene (LDPE) by mesophilic fungus ‘Penicillium citrinum’ isolated from soils of plastic waste dump yard, Bhopal, India. Environ. Technol. 43, 1–15. doi: 10.1080/09593330.2022.2027025
Khandare, S. D., Agrawal, D., Mehru, N., and Chaudhary, D. R. (2022). Marine bacterial based enzymatic degradation of low-density polyethylene (LDPE) plastic. J. Environ. Chem. Eng. 10:107437. doi: 10.1016/j.jece.2022.107437
Khatoon, N., Sahar, N. I., Ndu, U., Ali, N., Jamal, A., Ahmed, S., et al. (2017). High-throughput production of peroxidase and its biodegradation potential toward polymeric material. Int. J. Environ. Sci. Technol. 14, 755–766. doi: 10.1007/s13762-016-1182-1
Kim, D., Park, H. J., Nam, S., Kim, S. C., and Lee, H. (2019). Humic substances degradation by a microbial consortium enriched from subarctic tundra soil. Korean J. Microbiol 55, 367–376.
Kirstein, I. V., Wichels, A., Gullans, E., Krohne, G., and Gerdts, G. (2019). The Plastisphere - Uncovering tightly attached plastic “specific” microorganisms. PLoS One 14:e0215859. doi: 10.1371/journal.pone.0215859
Kotova, I. B., Taktarova, Y. V., Tsavkelova, E. A., Egorova, M. A., Bubnov, I. A., Malakhova, D. V., et al. (2021). Microbial degradation of plastics and approaches to make it more efficient. Microbiology 90, 671–701. doi: 10.1134/S0026261721060084
Koutny, M., Sancelme, M., Dabin, C., Pichon, N., Delort, A.-M., and Lemaire, J. (2006). Acquired biodegradability of polyethylenes containing pro-oxidant additives. Polym. Degrad. Stab. 91, 1495–1503. doi: 10.1016/j.polymdegradstab.2005.10.007
Krause, S., Molari, M., Gorb, E. V., Gorb, S. N., Kossel, E., and Haeckel, M. (2020). Persistence of plastic debris and its colonization by bacterial communities after two decades on the abyssal seafloor. Sci. Rep. 10:9484. doi: 10.1038/s41598-020-66361-7
Krueger, M. C., Harms, H., and Schlosser, D. (2015). Prospects for microbiological solutions to environmental pollution with plastics. Appl. Microbiol. Biotechnol. 99, 8857–8874. doi: 10.1007/s00253-015-6879-4
Kumar Sen, S., and Raut, S. (2015). Microbial degradation of low density polyethylene (LDPE): A review. J. Environ. Chem. Eng. 3, 462–473. doi: 10.1016/j.jece.2015.01.003
Kumar, A., and Chandra, R. (2020). Ligninolytic enzymes and its mechanisms for degradation of lignocellulosic waste in environment. Heliyon 6:e03170. doi: 10.1016/j.heliyon.2020.e03170
Kumar, R., Pandit, P., Kumar, D., Patel, Z., Pandya, L., Kumar, M., et al. (2021). Landfill microbiome harbour plastic degrading genes: A metagenomic study of solid waste dumping site of Gujarat, India. Sci. Total Environ. 779:146184. doi: 10.1016/j.scitotenv.2021.146184
Kyaw, B. M., Champakalakshmi, R., Sakharkar, M. K., Lim, C. S., and Sakharkar, K. R. (2012). Biodegradation of Low Density Polythene (LDPE) by Pseudomonas Species. Indian J. Microbiology 52, 411–419. doi: 10.1007/s12088-012-0250-6
Kyrikou, I., and Briassoulis, D. (2007). Biodegradation of agricultural plastic films: A critical review. J. Polym. Environ. 15, 125–150. doi: 10.1007/s10924-007-0053-8
Lee, J., Kwon, E. E., Lam, S. S., Chen, W.-H., Rinklebe, J., and Park, Y.-K. (2021). Chemical recycling of plastic waste via thermocatalytic routes. J. Clean. Prod. 321:128989. doi: 10.1016/j.jclepro.2021.128989
Li, M., Yin, H., Zhu, M., Yu, Y., Lu, G., and Dang, Z. (2021). Co-metabolic and biochar-promoted biodegradation of mixed PAHs by highly efficient microbial consortium QY1. J. Environ. Sci. 107, 65–76. doi: 10.1016/j.jes.2021.02.002
Li, Z., Wei, R., Gao, M., Ren, Y., Yu, B., Nie, K., et al. (2020). Biodegradation of low-density polyethylene by microbulbifer hydrolyticus IRE-31. J. Environ. Manag. 263:110402. doi: 10.1016/j.jenvman.2020.110402
Liang, Y., Ma, A., and Zhuang, G. (2022). Construction of environmental synthetic microbial consortia: Based on engineering and ecological principles. Front. Microbiol. 13:829717. doi: 10.3389/fmicb.2022.829717
Lin, L. (2022). Bottom-up synthetic ecology study of microbial consortia to enhance lignocellulose bioconversion. Biotechnol. Biofuels Bioprod. 15:14. doi: 10.1186/s13068-022-02113-1
Liu, F., Mao, J., Kong, W., Hua, Q., Feng, Y., Bashir, R., et al. (2020). Interaction variability shapes succession of synthetic microbial ecosystems. Nat. Commun. 11:309. doi: 10.1038/s41467-019-13986-6
Liu, T., Xin, Y., Liu, X., Wu, B., and Xiang, M. (2021). [Advances in microbial degradation of plastics]. Sheng Wu Gong Cheng Xue Bao 37, 2688–2702.
Liu, X., Lei, T., Boré, A., Lou, Z., Abdouraman, B., and Ma, W. (2022b). Evolution of global plastic waste trade flows from 2000 to 2020 and its predicted trade sinks in 2030. J. Clean. Prod. 376:134373. doi: 10.1016/j.jclepro.2022.134373
Liu, X., Zhang, Y., Sun, Q., Liu, Z., Zhao, Y., Fan, A., et al. (2022c). Rapid colonization and biodegradation of untreated commercial polyethylene wrap by a new strain of Bacillus velezensis C5. J. Environ. Manag. 301:113848. doi: 10.1016/j.jenvman.2021.113848
Liu, L., Xu, M., Ye, Y., and Zhang, B. (2022a). On the degradation of (micro)plastics: Degradation methods, influencing factors, environmental impacts. Sci. Total Environ. 806:151312. doi: 10.1016/j.scitotenv.2021.151312
Lomwongsopon, P., and Varrone, C. (2022). Critical review on the progress of plastic bioupcycling technology as a potential solution for sustainable plastic waste management. Polymers 14:4996. doi: 10.3390/polym14224996
Lou, H., Fu, R., Long, T., Fan, B., Guo, C., Li, L., et al. (2022). Biodegradation of polyethylene by Meyerozyma guilliermondii and Serratia marcescens isolated from the gut of waxworms (larvae of Plodia interpunctella). Sci. Total Environ. 853:158604. doi: 10.1016/j.scitotenv.2022.158604
Lu, J., Yang, Z., Xu, W., Shi, X., and Guo, R. (2019). Enrichment of thermophilic and mesophilic microbial consortia for efficient degradation of corn stalk. J. Environ. Sci. 78, 118–126. doi: 10.1016/j.jes.2018.07.010
Lucas, N., Bienaime, C., Belloy, C., Queneudec, M., Silvestre, F., and Nava-Saucedo, J.-E. (2008). Polymer biodegradation: Mechanisms and estimation techniques – A review. Chemosphere 73, 429–442. doi: 10.1016/j.chemosphere.2008.06.064
MacLean, J., Mayanna, S., Benning, L. G., Horn, F., Bartholomäus, A., Wiesner, Y., et al. (2021). The terrestrial plastisphere: Diversity and polymer-colonizing potential of plastic-associated microbial communities in soil. Microorganisms 9:1876. doi: 10.3390/microorganisms9091876
Magnin, A., Pollet, E., Perrin, R., Ullmann, C., Persillon, C., Phalip, V., et al. (2019). Enzymatic recycling of thermoplastic polyurethanes: Synergistic effect of an esterase and an amidase and recovery of building blocks. Waste Manag. 85, 141–150. doi: 10.1016/j.wasman.2018.12.024
Maity, S., Banerjee, S., Biswas, C., Guchhait, R., Chatterjee, A., and Pramanick, K. (2021). Functional interplay between plastic polymers and microbes: A comprehensive review. Biodegradation 32, 487–510. doi: 10.1007/s10532-021-09954-x
Malachová, K., Novotný, Č, Adamus, G., Lotti, N., Rybková, Z., Soccio, M., et al. (2020). Ability of trichoderma hamatum isolated from plastics-polluted environments to attack petroleum-based, synthetic polymer films. Processes 8:467. doi: 10.3390/pr8040467
Maroof, L., Khan, I., Hassan, H., Azam, S., and Khan, W. (2022). Microbial degradation of low density polyethylene by Exiguobacterium sp. strain LM-IK2 isolated from plastic dumped soil. World J. Microbiol. Biotechnol. 38:197. doi: 10.1007/s11274-022-03389-z
Massot, F., Bernard, N., Alvarez, L. M. M., Martorell, M. M., Mac Cormack, W. P., and Ruberto, L. A. M. (2022). Microbial associations for bioremediation. What does “microbial consortia” mean? Appl. Microbiol. Biotechnol. 106, 2283–2297. doi: 10.1007/s00253-022-11864-8
Mendes, I. V., Garcia, M. B., Araujo Bitencourt, A. C., Santana, R. H., Lins, P. d. C, Silveira, R., et al. (2021). Bacterial diversity dynamics in microbial consortia selected for lignin utilization. PLoS One 16:e0255083. doi: 10.1371/journal.pone.0255083
Meyer-Cifuentes, I. E., Werner, J., Jehmlich, N., Will, S. E., Neumann-Schaal, M., and Öztürk, B. (2020). Synergistic biodegradation of aromatic-aliphatic copolyester plastic by a marine microbial consortium. Nat. Commun. 11:5790. doi: 10.1038/s41467-020-19583-2
Millican, J. M., and Agarwal, S. (2021). Plastic pollution: A material problem? Macromolecules 54, 4455–4469. doi: 10.1021/acs.macromol.0c02814
Mohammadi, S., Moussavi, G., and Rezaei, M. (2022). Enhanced peroxidase-mediated biodegradation of polyethylene using the bacterial consortia under H2O2-biostimulation. Polymer 240:124508. doi: 10.1016/j.polymer.2021.124508
Montazer, Z., Habibi-Najafi, M. B., Mohebbi, M., and Oromiehei, A. (2018). Microbial degradation of UV-pretreated low-density polyethylene films by novel polyethylene-degrading bacteria isolated from plastic-dump soil. J. Polym. Environ. 26, 3613–3625. doi: 10.1007/s10924-018-1245-0
Mouafo Tamnou, E. B., Tamsa Arfao, A., Nougang, M. E., Metsopkeng, C. S., Noah Ewoti, O. V., Moungang, L. M., et al. (2021). Biodegradation of polyethylene by the bacterium Pseudomonas aeruginosa in acidic aquatic microcosm and effect of the environmental temperature. Environ. Chall. 3:100056. doi: 10.1016/j.envc.2021.100056
Nag, M., Lahiri, D., Dutta, B., Jadav, G., and Ray, R. R. (2021). Biodegradation of used polyethylene bags by a new marine strain of Alcaligenes faecalis LNDR-1. Environ. Sci. Pollut Res. 28, 41365–41379. doi: 10.1007/s11356-021-13704-0
Nanda, S., and Berruti, F. (2021). Thermochemical conversion of plastic waste to fuels: A review. Environ. Chem. Lett. 19, 123–148. doi: 10.1007/s10311-020-01094-7
Nemergut Diana, R., Schmidt Steven, K., Fukami, T., O’Neill Sean, P., Bilinski Teresa, M., Stanish Lee, F., et al. (2013). Patterns and processes of microbial community assembly. Microbiol. Mol. Biol. Rev. 77, 342–356. doi: 10.1128/MMBR.00051-12
Nguyen, H. T., Choi, W., Kim, E.-J., and Cho, K. (2022). Microbial community niches on microplastics and prioritized environmental factors under various urban riverine conditions. Sci. Total Environ. 849:157781. doi: 10.1016/j.scitotenv.2022.157781
Nie, Y., Chi, C.-Q., Fang, H., Liang, J.-L., Lu, S.-L., Lai, G.-L., et al. (2014). Diverse alkane hydroxylase genes in microorganisms and environments. Sci. Rep. 4:4968. doi: 10.1038/srep04968
Park, S. Y., and Kim, C. G. (2019). Biodegradation of micro-polyethylene particles by bacterial colonization of a mixed microbial consortium isolated from a landfill site. Chemosphere 222, 527–533. doi: 10.1016/j.chemosphere.2019.01.159
Pathak, V. M., and Navneet (2017). Review on the current status of polymer degradation: A microbial approach. Bioresour. Bioprocess. 4:15. doi: 10.1186/s40643-017-0145-9
Pinnell, L. J., and Turner, J. W. (2019). Shotgun metagenomics reveals the benthic microbial community response to plastic and bioplastic in a coastal marine environment. Front. Microbiol. 10:1252. doi: 10.3389/fmicb.2019.01252
Pinto, M., Zhao, Z., Klun, K., Libowitzky, E., and Herndl Gerhard, J. (2022). Microbial consortiums of putative degraders of low-density polyethylene-associated compounds in the Ocean. mSystems 7, e1415–e1421. doi: 10.1128/msystems.01415-21
Piscitelli, A., Cicatiello, P., Gravagnuolo, A. M., Sorrentino, I., Pezzella, C., and Giardina, P. (2017). Applications of functional amyloids from fungi: Surface modification by class i hydrophobins. Biomolecules 7:45. doi: 10.3390/biom7030045
Plastics Europe (2021). Plastics – the facts 2021. Available online at: https://plasticseurope.org/knowledge-hub/plastics-the-facts-2021/ (accessed February 5, 2023).
Purohit, J., Chattopadhyay, A., and Teli, B. (2020). Metagenomic exploration of plastic degrading microbes for biotechnological application. Curr. Genomics 21, 253–270. doi: 10.2174/1389202921999200525155711
Qian, X., Chen, L., Sui, Y., Chen, C., Zhang, W., Zhou, J., et al. (2020). Biotechnological potential and applications of microbial consortia. Biotechnol. Adv. 40:107500. doi: 10.1016/j.biotechadv.2019.107500
Qin, W. F., Xia, Y. J., Xiong, Z. Q., Song, X., Ai, L. Z., and Wang, G. Q. (2022). The intestinal colonization of Lactiplantibacillus plantarum AR113 is influenced by its mucins and intestinal environment. Food Res. Int. 157:111382. doi: 10.1016/j.foodres.2022.111382
Ragaert, K., Delva, L., and Van Geem, K. (2017). Mechanical and chemical recycling of solid plastic waste. Waste Manag. 69, 24–58. doi: 10.1016/j.wasman.2017.07.044
Rana, A. K., Thakur, M. K., Saini, A. K., Mokhta, S. K., Moradi, O., Rydzkowski, T., et al. (2022). Recent developments in microbial degradation of polypropylene: Integrated approaches towards a sustainable environment. Sci. Total Environ. 826:154056. doi: 10.1016/j.scitotenv.2022.154056
Ren, L., Men, L., Zhang, Z., Guan, F., Tian, J., Wang, B., et al. (2019). Biodegradation of polyethylene by Enterobacter sp. D1 from the Guts of Wax Moth Galleria mellonella. Int. J. Environ. Res. Public Health 16:1941. doi: 10.3390/ijerph16111941
Restrepo-Flórez, J.-M., Bassi, A., and Thompson, M. R. (2014). Microbial degradation and deterioration of polyethylene – A review. Int. Biodeterior. Biodegradation 88, 83–90. doi: 10.1016/j.ibiod.2013.12.014
Roberts, C., Edwards, S., Vague, M., León-Zayas, R., Scheffer, H., Chan, G., et al. (2020). Environmental Consortium Containing Pseudomonas and Bacillus Species Synergistically degrades polyethylene terephthalate plastic. mSphere 5, e1151–e1120. doi: 10.1128/mSphere.01151-20
Roy, R., Mukherjee, G., Das Gupta, A., Tribedi, P., and Sil, A. K. (2021). Isolation of a soil bacterium for remediation of polyurethane and low-density polyethylene: A promising tool towards sustainable cleanup of the environment. 3 Biotech 11:29. doi: 10.1007/s13205-020-02592-9
Ru, J., Huo, Y., and Yang, Y. (2020). Microbial degradation and valorization of plastic wastes. Front. Microbiol. 11:442. doi: 10.3389/fmicb.2020.00442
Ruiz Barrionuevo, J. M., Vilanova-Cuevas, B., Alvarez, A., Martín, E., Malizia, A., Galindo-Cardona, A., et al. (2022). The bacterial and fungal gut microbiota of the greater wax moth, Galleria mellonella L. Consuming polyethylene and polystyrene. Front. Microbiol. 13:918861. doi: 10.3389/fmicb.2022.918861
Rüthi, J., Rast, B. M., Qi, W., Perez-Mon, C., Pardi-Comensoli, L., Brunner, I., et al. (2023). The plastisphere microbiome in alpine soils alters the microbial genetic potential for plastic degradation and biogeochemical cycling. J. Hazard. Mater. 441:129941. doi: 10.1016/j.jhazmat.2022.129941
Samanta, S., Datta, D., and Halder, G. (2020). Biodegradation efficacy of soil inherent novel sp. Bacillus tropicus (MK318648) onto low density polyethylene matrix. J. Polym. Res. 27:324. doi: 10.1007/s10965-020-02296-x
San León, D., and Nogales, J. (2022). Toward merging bottom–up and top–down model-based designing of synthetic microbial communities. Curr. Opin. Microbiol. 69:102169. doi: 10.1016/j.mib.2022.102169
Sánchez, C. (2020). Fungal potential for the degradation of petroleum-based polymers: An overview of macro- and microplastics biodegradation. Biotechnol. Adv. 40:107501. doi: 10.1016/j.biotechadv.2019.107501
Sangale, M. K., Shahnawaz, M., and Ade, A. B. (2019a). Gas chromatography-Mass Spectra analysis and deleterious potential of fungal based polythene-degradation products. Sci. Rep. 9:1599. doi: 10.1038/s41598-018-37738-6
Sangale, M. K., Shahnawaz, M., and Ade, A. B. (2019b). Potential of fungi isolated from the dumping sites mangrove rhizosphere soil to degrade polythene. Sci. Rep. 9:5390. doi: 10.1038/s41598-019-41448-y
Sangeetha Devi, R., Rajesh Kannan, V., Nivas, D., Kannan, K., Chandru, S., and Robert Antony, A. (2015). Biodegradation of HDPE by Aspergillus spp. from marine ecosystem of Gulf of Mannar, India. Mar. Pollut. Bull. 96, 32–40. doi: 10.1016/j.marpolbul.2015.05.050
Sangiorgio, P., Verardi, A., Dimatteo, S., Spagnoletta, A., Moliterni, S., and Errico, S. (2021). Tenebrio molitorin the circular economy: A novel approach for plastic valorisation and PHA biological recovery. Environ. Sci. Pollut. Res. 28, 52689–52701. doi: 10.1007/s11356-021-15944-6
Santo, M., Weitsman, R., and Sivan, A. (2013). The role of the copper-binding enzyme – laccase – in the biodegradation of polyethylene by the actinomycete Rhodococcus ruber. Int. Biodeterior. Biodegradation 84, 204–210. doi: 10.1016/j.ibiod.2012.03.001
Sarker, R. K., Chakraborty, P., Paul, P., Chatterjee, A., and Tribedi, P. (2020). Degradation of low-density poly ethylene (LDPE) by Enterobacter cloacae AKS7: A potential step towards sustainable environmental remediation. Arch. Microbiol. 202, 2117–2125. doi: 10.1007/s00203-020-01926-8
Schaerer, L. G., Wu, R., Putman, L. I., Pearce, J. M., Lu, T., Shonnard, D. R., et al. (2023). Killing two birds with one stone: Chemical and biological upcycling of polyethylene terephthalate plastics into food. Trends Biotechnol. 41, 184–196. doi: 10.1016/j.tibtech.2022.06.012
Shah, A. A., Hasan, F., Hameed, A., and Ahmed, S. (2008). Biological degradation of plastics: A comprehensive review. Biotechnol. Adv. 26, 246–265. doi: 10.1016/j.biotechadv.2007.12.005
Shahnawaz, M., Sangale, M. K., and Ade, A. B. (2016). Bacteria-based polythene degradation products: GC-MS analysis and toxicity testing. Environ. Sci. Pollut. Res. 23, 10733–10741. doi: 10.1007/s11356-016-6246-8
Sharma, B., and Shukla, P. (2022). Futuristic avenues of metabolic engineering techniques in bioremediation. Biotechnol. Appl. Biochem. 69, 51–60. doi: 10.1002/bab.2080
Singh Jadaun, J., Bansal, S., Sonthalia, A., Rai, A. K., and Singh, S. P. (2022). Biodegradation of plastics for sustainable environment. Bioresour. Technol. 347:126697. doi: 10.1016/j.biortech.2022.126697
Singh, M. J., and Sedhuraman, P. (2015). Biosurfactant, polythene, plastic, and diesel biodegradation activity of endophytic Nocardiopsis sp. mrinalini9 isolated from Hibiscus rosasinensis leaves. Bioresour. Bioprocess. 2:2. doi: 10.1186/s40643-014-0034-4
Skariyachan, S., Manjunatha, V., Sultana, S., Jois, C., Bai, V., and Vasist, K. S. (2016). Novel bacterial consortia isolated from plastic garbage processing areas demonstrated enhanced degradation for low density polyethylene. Environ. Sci. Pollut. Res. 23, 18307–18319. doi: 10.1007/s11356-016-7000-y
Skariyachan, S., Patil, A. A., Shankar, A., Manjunath, M., Bachappanavar, N., and Kiran, S. (2018). Enhanced polymer degradation of polyethylene and polypropylene by novel thermophilic consortia of Brevibacillus sps. and Aneurinibacillus sp. screened from waste management landfills and sewage treatment plants. Polym. Degrad. Stab. 149, 52–68. doi: 10.1016/j.polymdegradstab.2018.01.018
Skariyachan, S., Taskeen, N., Kishore, A. P., and Krishna, B. V. (2022). Recent advances in plastic degradation – From microbial consortia-based methods to data sciences and computational biology driven approaches. J. Hazard. Mater. 426:128086. doi: 10.1016/j.jhazmat.2021.128086
Skariyachan, S., Taskeen, N., Kishore, A. P., Krishna, B. V., and Naidu, G. (2021). Novel consortia of Enterobacter and Pseudomonas formulated from cow dung exhibited enhanced biodegradation of polyethylene and polypropylene. J. Environ. Manag. 284:112030. doi: 10.1016/j.jenvman.2021.112030
Soleimani, Z., Gharavi, S., Soudi, M., and Moosavi-Nejad, Z. (2021). A survey of intact low-density polyethylene film biodegradation by terrestrial Actinobacterial species. Int. Microbiol. 24, 65–73. doi: 10.1007/s10123-020-00142-0
Sowmya, H. V., Ramalingappa Krishnappa, M., and Thippeswamy, B. (2014). Degradation of polyethylene by Trichoderma harzianum—SEM, FTIR, and NMR analyses. Environ. Monit. Assess. 186, 6577–6586. doi: 10.1007/s10661-014-3875-6
Spina, F., Tummino, M. L., Poli, A., Prigione, V., Ilieva, V., Cocconcelli, P., et al. (2021). Low density polyethylene degradation by filamentous fungi. Environ. Pollut. 274:116548. doi: 10.1016/j.envpol.2021.116548
Sullivan, K. P., Werner, A. Z., Ramirez, K. J., Ellis, L. D., Bussard, J. R., Black, B. A., et al. (2022). Mixed plastics waste valorization through tandem chemical oxidation and biological funneling. Science 378, 207–211. doi: 10.1126/science.abo4626
Tabanag, I. D. F., Chu, I. M., Wei, Y.-H., and Tsai, S.-L. (2018). Ethanol production from hemicellulose by a consortium of different genetically-modified sacharomyces cerevisiae. J. Taiwan Inst. Chem. Eng. 89, 15–25. doi: 10.1016/j.jtice.2018.04.029
Taghavi, N., Singhal, N., Zhuang, W.-Q., and Baroutian, S. (2021a). Degradation of plastic waste using stimulated and naturally occurring microbial strains. Chemosphere 263:127975. doi: 10.1016/j.chemosphere.2020.127975
Taghavi, N., Udugama, I. A., Zhuang, W.-Q., and Baroutian, S. (2021b). Challenges in biodegradation of non-degradable thermoplastic waste: From environmental impact to operational readiness. Biotechnol. Adv. 49:107731. doi: 10.1016/j.biotechadv.2021.107731
Tan, F., Cheng, J., Zhang, Y., Jiang, X., and Liu, Y. (2022). Genomics analysis and degradation characteristics of lignin by Streptomyces thermocarboxydus strain DF3-3. Biotechnol. Biofuels And Bioprod. 15, 78–78. doi: 10.1186/s13068-022-02175-1
Tejaswini, M. S. S. R., Pathak, P., Ramkrishna, S., and Ganesh, P. S. (2022). A comprehensive review on integrative approach for sustainable management of plastic waste and its associated externalities. Sci. Total Environ. 825:153973. doi: 10.1016/j.scitotenv.2022.153973
Temporiti, M. E., Nicola, L., Nielsen, E., and Tosi, S. (2022). Fungal enzymes involved in plastics biodegradation. Microorganisms 10:1180. doi: 10.3390/microorganisms10061180
Thakur, S., Chaudhary, J., Sharma, B., Verma, A., Tamulevicius, S., and Thakur, V. K. (2018). Sustainability of bioplastics: Opportunities and challenges. Curr. Opin. Green Sust. Chem. 13, 68–75. doi: 10.1016/j.cogsc.2018.04.013
Tiwari, N., Bansal, M., Santhiya, D., and Sharma, J. G. (2022). Insights into microbial diversity on plastisphere by multi-omics. Arch. Microbiol. 204:216. doi: 10.1007/s00203-022-02806-z
Tribedi, P., and Sil, A. K. (2013). Low-density polyethylene degradation by Pseudomonas sp. AKS2 biofilm. Environ. Sci. Pollut. Res. 20, 4146–4153. doi: 10.1007/s11356-012-1378-y
Tu, C., Chen, T., Zhou, Q., Liu, Y., Wei, J., Waniek, J. J., et al. (2020). Biofilm formation and its influences on the properties of microplastics as affected by exposure time and depth in the seawater. Sci. Total Environ. 734:139237. doi: 10.1016/j.scitotenv.2020.139237
Vaksmaa, A., Knittel, K., Asbun, A. A., Goudriaan, M., Ellrott, A., Witte, H. J., et al. (2021). Microbial communities on plastic polymers in the Mediterranean Sea. Front. Microbiol. 12:673553. doi: 10.3389/fmicb.2021.673553
Vimala, P. P., and Mathew, L. (2015). “Biodegradation of polyethylene using Bacillus subtilis,” in Proceedings of the 4th international conference on emerging trends in engineering, science and technology (ICETEST), (Trichur), 2016. doi: 10.1016/j.protcy.2016.05.031
Wang, A., Gao, L., Ren, N., Xu, J., Liu, C., and Lee, D.-J. (2010). Enrichment strategy to select functional consortium from mixed cultures: Consortium from rumen liquor for simultaneous cellulose degradation and hydrogen production. Int. J. Hydrogen Energy 35, 13413–13418. doi: 10.1016/j.ijhydene.2009.11.117
Wang, H., Li, J., Lu, Y., Guo, P., Wang, X., Kazuhiro, M., et al. (2013). Bioconversion of un-pretreated lignocellulosic materials by a microbial consortium XDC-2. Bioresour. Technol. 136, 481–487. doi: 10.1016/j.biortech.2013.03.015
Wang, P., Liu, J., Han, S., Wang, Y., Duan, Y., Liu, T., et al. (2023). Polyethylene mulching film degrading bacteria within the plastisphere: Co-culture of plastic degrading strains screened by bacterial community succession. J. Hazard. Mater. 442:130045. doi: 10.1016/j.jhazmat.2022.130045
Wang, P., Song, T., Bu, J., Zhang, Y., Liu, J., Zhao, J., et al. (2022). Does bacterial community succession within the polyethylene mulching film plastisphere drive biodegradation? Sci. Total Environ. 824:153884. doi: 10.1016/j.scitotenv.2022.153884
Wei, R., and Zimmermann, W. (2017). Microbial enzymes for the recycling of recalcitrant petroleum-based plastics: How far are we? Microb. Biotechnol. 10, 1308–1322. doi: 10.1111/1751-7915.12710
Wei, R., Oeser, T., and Zimmermann, W. (2014). “Chapter Seven – Synthetic polyester-hydrolyzing enzymes from thermophilic actinomycetes,” in Advances in applied microbiology, eds S. Sariaslani and G. M. Gadd (Cambridge, MA: Academic Press), 267–305. doi: 10.1016/B978-0-12-800259-9.00007-X
Wu, M., Li, W., Dick, W. A., Ye, X., Chen, K., Kost, D., et al. (2017). Bioremediation of hydrocarbon degradation in a petroleum-contaminated soil and microbial population and activity determination. Chemosphere 169, 124–130. doi: 10.1016/j.chemosphere.2016.11.059
Wu, Q., Qin, K.-X., Gan, M.-X., Xu, J., Li, Z.-L., and Li, Z.-C. (2022). Recyclable biomass-derived polyethylene-like materials as functional coatings for commercial fabrics: Toward Upcycling of waste textiles. ACS Sust. Chem. Eng. 10, 17187–17197. doi: 10.1021/acssuschemeng.2c05080
Xu, C., and Yu, H. (2021). Insights into constructing a stable and efficient microbial consortium. Chin. J. Chem. Eng. 30, 112–120. doi: 10.1016/j.cjche.2020.12.012
Xu, T., Liu, T., Jiang, D., Yuan, Z., and Jia, X. (2021). Attainment and characterization of a microbial consortium that efficiently degrades biphenyl and related substances. Biochem. Eng. J. 173:108073. doi: 10.1016/j.bej.2021.108073
Yeom, S.-J., Le, T.-K., and Yun, C.-H. (2022). P450-driven plastic-degrading synthetic bacteria. Trends Biotechnol. 40, 166–179. doi: 10.1016/j.tibtech.2021.06.003
Yin, C.-F., Xu, Y., and Zhou, N.-Y. (2020). Biodegradation of polyethylene mulching films by a co-culture of Acinetobacter sp. strain NyZ450 and Bacillus sp. strain NyZ451 isolated from Tenebrio molitor larvae. Int. Biodeterior. Biodegradation 155:105089. doi: 10.1016/j.ibiod.2020.105089
Yue, X.-H., Zhang, F.-S., Wu, L., Zhang, C.-C., and Qian, P. (2022a). Upcycling of blending waste plastics as flexible growing substrate with superabsorbing property. Chem. Eng. J. 435:134622. doi: 10.1016/j.cej.2022.134622
Yue, X.-H., Zhang, F.-S., Zhang, C.-C., and Qian, P. (2022b). Upcycling of blending waste plastics as zwitterionic hydrogel for simultaneous removal of cationic and anionic heavy metals from aqueous system. J. Hazard Mater. 432:128746. doi: 10.1016/j.jhazmat.2022.128746
Zadjelovic, V., Erni-Cassola, G., Obrador-Viel, T., Lester, D., Eley, Y., Gibson, M. I., et al. (2022). A mechanistic understanding of polyethylene biodegradation by the marine bacterium Alcanivorax. J. Hazard. Mater. 436:129278. doi: 10.1016/j.jhazmat.2022.129278
Zampolli, J., Orro, A., Manconi, A., Ami, D., Natalello, A., and Di Gennaro, P. (2021). Transcriptomic analysis of Rhodococcus opacus R7 grown on polyethylene by RNA-seq. Sci. Rep. 11:21311. doi: 10.1038/s41598-021-00525-x
Zeenat Elahi, A., Bukhari, D. A., Shamim, S., and Rehman, A. (2021). Plastics degradation by microbes: A sustainable approach. J. King Saud Univ. Sci. 33:101538. doi: 10.1016/j.jksus.2021.101538
Zeghal, E., Vaksmaa, A., Vielfaure, H., Boekhout, T., and Niemann, H. (2021). The Potential role of marine fungi in plastic degradation – a review. Front. Marine Sci. 8:738877. doi: 10.3389/fmars.2021.738877
Zettler, E. R., Mincer, T. J., and Amaral-Zettler, L. A. (2013). Life in the “plastisphere”: Microbial communities on plastic marine debris. Environ. Sci. Technol. 47, 7137–7146. doi: 10.1021/es401288x
Zhang, A., Hou, Y., Wang, Q., and Wang, Y. (2022). Characteristics and polyethylene biodegradation function of a novel cold-adapted bacterial laccase from Antarctic sea ice psychrophile Psychrobacter sp. NJ228. J. Hazard. Mater. 439:129656. doi: 10.1016/j.jhazmat.2022.129656
Zhang, Y., Pedersen, J. N., Eser, B. E., and Guo, Z. (2022). Biodegradation of polyethylene and polystyrene: From microbial deterioration to enzyme discovery. Biotechnol. Adv. 60:107991. doi: 10.1016/j.biotechadv.2022.107991
Zhang, N., Ding, M., and Yuan, Y. (2022). Current advances in biodegradation of polyolefins. Microorganisms 10:1537. doi: 10.3390/microorganisms10081537
Zhang, Z., Peng, H., Yang, D., Zhang, G., Zhang, J., and Ju, F. (2022). Polyvinyl chloride degradation by a bacterium isolated from the gut of insect larvae. Nat. Commun. 13:5360. doi: 10.1038/s41467-022-32903-y
Zhang, S. J., Zeng, Y.-H., Zhu, J.-M., Cai, Z.-H., and Zhou, J. (2022). The structure and assembly mechanisms of plastisphere microbial community in natural marine environment. J. Hazard. Mater. 421:126780. doi: 10.1016/j.jhazmat.2021.126780
Zhang, F., Wang, F., Wei, X., Yang, Y., Xu, S., Deng, D., et al. (2022). From trash to treasure: Chemical recycling and upcycling of commodity plastic waste to fuels, high-valued chemicals and advanced materials. J. Energy Chem. 69, 369–388. doi: 10.1016/j.jechem.2021.12.052
Zhang, D., Wang, Y., Zhang, C., Zheng, D., Guo, P., and Cui, Z. (2018). Characterization of a thermophilic lignocellulose-degrading microbial consortium with high extracellular xylanase activity. J. Microbiol. Biotechnol. 28, 305–313. doi: 10.4014/jmb.1709.09036
Zhang, F., Zeng, M., Yappert, R. D., Sun, J., Lee, Y.-H., LaPointe, A. M., et al. (2020). Polyethylene upcycling to long-chain alkylaromatics by tandem hydrogenolysis/aromatization. Science 370, 437–441. doi: 10.1126/science.abc5441
Zhang, H., Pap, S., Taggart, M. A., Boyd, K. G., James, N. A., and Gibb, S. W. (2020). A review of the potential utilisation of plastic waste as adsorbent for removal of hazardous priority contaminants from aqueous environments. Environ. Pollut. 258:113698. doi: 10.1016/j.envpol.2019.113698
Zhang, H., Lu, Y., Wu, H., Liu, Q., and Sun, W. (2023). Effect of an Acinetobacter pittobacter on low-density polyethylene. Environ. Sci. Pollut. Res. 30, 10495–10504. doi: 10.1007/s11356-022-22658-w
Zhao, C., Chu, Y., Li, Y., Yang, C., Chen, Y., Wang, X., et al. (2017). High-throughput pyrosequencing used for the discovery of a novel cellulase from a thermophilic cellulose-degrading microbial consortium. Biotechnol. Lett. 39, 123–131. doi: 10.1007/s10529-016-2224-y
Zheng, G., Yin, T., Lu, Z., Boboua, S. Y. B., Li, J., and Zhou, W. (2020). Degradation of rice straw at low temperature using a novel microbial consortium LTF-27 with efficient ability. Bioresour. Technol. 304:123064. doi: 10.1016/j.biortech.2020.123064
Zheng, Y., Yanful, E. K., and Bassi, A. S. (2005). A Review of Plastic Waste Biodegradation. Crit. Rev. Biotechnol. 25, 243–250. doi: 10.1080/07388550500346359
Zhou, C., Fang, W., Xu, W., Cao, A., and Wang, R. (2014). Characteristics and the recovery potential of plastic wastes obtained from landfill mining. J. Clean. Prod. 80, 80–86. doi: 10.1016/j.jclepro.2014.05.083
Zhou, H., Wang, Y., Ren, Y., Li, Z., Kong, X., Shao, M., et al. (2022). Plastic waste valorization by leveraging multidisciplinary catalytic technologies. ACS Catal. 12, 9307–9324. doi: 10.1021/acscatal.2c02775
Zhuo, C., and Levendis, Y. A. (2014). Upcycling waste plastics into carbon nanomaterials: A review. J. Appl. Polym. Sci. 131, 1–14. doi: 10.1002/app.39931
Keywords: polyethylene biodegradation, microbial consortia, plastisphere, microplastics, nanoplastics, omics
Citation: Zhang C, Mu Y, Li T, Jin F-J, Jin C-Z, Oh H-M, Lee H-G and Jin L (2023) Assembly strategies for polyethylene-degrading microbial consortia based on the combination of omics tools and the “Plastisphere”. Front. Microbiol. 14:1181967. doi: 10.3389/fmicb.2023.1181967
Received: 08 March 2023; Accepted: 31 March 2023;
Published: 17 April 2023.
Edited by:
Umer Zeeshan Ijaz, University of Glasgow, United KingdomReviewed by:
Ciara Keating, University of Glasgow, United KingdomCopyright © 2023 Zhang, Mu, Li, Jin, Jin, Oh, Lee and Jin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Long Jin, aXNhY2NraW1AYWx1bW5pLmthaXN0LmFjLmty; Hyung-Gwan Lee, dHJ1c3RpbkBrcmliYi5yZS5rcg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.