- 1Botany and Microbiology Department, Faculty of Science, Alexandria University, Alexandria, Egypt
- 2Nucleic Acid Research Department, Genetic Engineering and Biotechnology Research Institute, the City of Scientific Research and Technological Applications, Alexandria, Egypt
Introduction: Lactose intolerance is a widespread problem that affects people of many different races all over the world. The following pharmacological supplements can improve the lives of those who suffer from this issue.
Methods: This work focused on lactase producer isolation and statistical design (Plackett–Burman, and BOX–Behnken) to maximize the effectiveness of environmental factors. A lactase-producing bacterium was chosen from a discovery of 100 strains in soil that had previously been polluted with dairy products. Plackett-Burman investigated fifteen variables.
Results: The most critical variables that lead to increased lactase synthesis are glucose, peptone, and magnesium sulfate (MgSO4). The ideal process conditions for the creation of lactase yield among the stated variables were then determined using a BOX-Benken design. To establish a polynomial quadratic relationship between the three variables and lactase activity, the Box–Behnken design level was used. The EXCEL-solver nonlinear optimization technique was used to predict the best form for lactase production. The ideal temperature and pH levels have been determined, both before and after the lactase purification process, to achieve the highest performance of isolated lactase.
Conclusion: According to this study, Bacillus licheniformis is a perfect supply of the lactase enzyme (β -Galactosidase), It can be used as a product to assist people who have health issues due to lactose intolerance.
1. Introduction
Enzymes are active proteins that function as biochemical catalysts (Piumetti and Illanes, 2022). These are biomolecules that are required for both the synthesis and breakdown reactions of living organisms (Aggarwal et al., 2021). The enzymes known as β-galactosidases (EC 3.2.1.23), sometimes known as lactases, hydrolyze lactose (the most prevalent milk sugar) into glucose and galactose (Liburdi and Marco, 2022). Although these enzymes are accessible in a range of biological systems, such as plants or microorganisms, they are currently only available commercially from yeasts, molds, and bacteria (Saqib et al., 2017; Martarello et al., 2019). Although these enzymes from various bacteria have varied features, their specificity is fundamentally the same (Ramos and Malcata, 2011; Kocabaş et al., 2021). Each chain of the four polypeptides in the tetramer of β-galactosidase has 1,023 amino acids, and together they form five different structural domains (Sedzro et al., 2018; Swentowsky, 2021). One of these domains is the jellyroll barrel, while the others are fibronectin, b-sandwich, and a central domain with a TIM-type barrel that also serves as the active site. Tetramer subunits make up the core domain, which is catalytically active (Wang et al., 2021). The catalytic site is disabled when a tetramer is broken up into dimers. Those enzymes are active proteins and their function as biocatalysts were discovered by Cao et al. (2021). Similarly, the β-galactosidase amino acid sequence was done by Poch et al. (1992) and the structure was determined by Jacobson et al. (1994). This enzyme’s amino-terminal sequence contains a-peptide that engages in a-complementation and aids in the formation of the subunit interaction (Aydin and Coin, 2021; Xu et al., 2021). Numerous organisms, including fungi, plants, yeast, and bacteria, may produce lactase (Shaima'a and Rashid, 2017). In the industrial sector, bacterial strains offer a lot of promise for large-scale manufacturing. Lactase generated by bacteria was frequently used to hydrolyze lactose because of its ease of fermentation, high activity, and good stability (Movahedpour et al., 2022). β-Galactosidase is used in a variety of industries (Ureta et al., 2021). β-galactosidase is utilized to handle whey disposal concerns on a commercial scale in addition to manufacturing lactose-free goods for lactose-intolerant patients (Paulo, 2018). To avoid the problem of hygroscopic lactose crystallizing in food, β -galactosidase is employed to hydrolyze the lactose in frozen, concentrated sweets (Portnoy and Barbano, 2021). This treatment lowers the lactose level of milk so that lactose-intolerant people can drink it. Lack of lactase, a digestive enzyme that prevents the body from hydrolyzing lactose in meals, is the primary cause of lactose intolerance. About 75% of people worldwide have lactose intolerance, which significantly lowers their quality of life (Venkateswarulu et al., 2017; Peele et al., 2018). Probiotic supplements may be beneficial for those with lactase deficiency. The dairy industry used lactase-producing bacterial strains to cure milk-based products (Venkateswarulu et al., 2017). Stomach pain and abdominal distension, abdominal colic, diarrhea, and nausea are all signs of lactose intolerance (Shrestha et al., 2021). Lactase deficiency can be classified as primary, congenital, or secondary. Primary lactase insufficiency affects adults aged 2 to 20. A more prevalent variety is primary lactase deficiency, which is caused by a decrease in β-galactosidase production along the small intestine’s brush boundaries (lactase). The second kind of lactase deficit is a birth defect in lactase production, which is brought on by a genetic defect and is defined by patients having either little or nonexistent lactase enzyme at all. The third kind, sometimes referred to as secondary lactase deficit, is when there are inadequate levels of this enzyme as a result of a GI tract issue (Paulo, 2018; Szilagyi et al., 2019; Muzaffar et al., 2021).
The current study’s major purpose is to develop exploitation tactics for the eventual advantage of extracellular bacterial lactase production. The goal of this study was to uncover a novel approach to lactase manufacturing in a newly discovered lactase producer. Furthermore, the utilization of mathematical models was the main emphasis of the current investigation to optimize the growth conditions that result in the best lactase productivity from a newly discovered Bacillus licheniformis ALSZ2. To our knowledge, lag phase bacterial lactase production has not yet been documented in the literature; the current study’s findings are the first report of Egyptian lactase manufacturing that is significantly cost-effective.
2. Materials and method
2.1. Isolation and screening of lactase producers
Samples of diverse dairy products (12) (milk, Romy cheese, and karish cheese) were collected in a sterile dry container. For bacterial isolation and colony purification, an LB medium that was diluted to one-tenth of medium strength was used. For lactase production, 100 purified bacterial isolates were screened. Isolation of lactase-producing bacteria was achieved using an LB broth medium fortified with lactose substrate (5%) and 1% X-gal. Blue color after growth refers to lactase production.
2.1.1. Rapid plate assay method
Qualitative identification of lactase producers was achieved using an LB agar medium containing 1 mM of IPTG (substrate of lactase) and X-gal (1%). The solidified agar plates were incubated in an inverted position at 37°C for 24 h. Isolated colonies that are blue (positive lactase producer) were subcultured in nutrient broth and incubated at 37°C and preserved for further studies (Petassi et al., 2020).
2.2. Molecular identification of the most potent producer
Fresh bacterial cells are used for DNA extraction using GeneJET Genomic DNA Purification Kit. Using standard primers (F: AGAGTTTGATCMTGGCTCAG and R: TACGGYACCTTGTTACGACTT) (Alkhafaje et al., 2019) intended to magnify a 1,500 base pair portion of the 16S rDNA region (Kim and Chun, 2014; Soliman et al., 2018), the 16S rDNA was amplified using PCR (polymerase chain reaction). The condition of the cyclical reaction was 4 min at 95°C and then 40 cycles of 40 s at 94°C, 50 s at 55°C, and 50 s at 72°C, monitored by an additional 10 min at 72°C. PCR reactions were run on an agarose gel, and the remaining mixture was purified for sequence (Alkhafaje et al., 2022).
2.2.1. Phylogenetic analysis
To evaluate the DNA similarity of the obtained 16SrDNA sequence [by using a 3,130 X DNA Sequencer (Genetic Analyzer, Applied Biosystems, Hitachi, Japan)], phylogenetic analysis was conducted using the BLAST tool.1 Mega7 software was used to accomplish molecular phylogeny and multiple sequence alignment present in the database (NCBI). A neighbor-joining (NJ) tree and a maximum parsimony (MP) tree using bootstrapping were made using this alignment (Hassan et al., 2018; Shaaban et al., 2022).
2.3. Enzyme assay
The culture media were prepared, inoculated with lactase producers, and incubated for 24 h at 37°C. At the end of the incubation period, the fermented culture was centrifuged at 5000 rpm for 10 min at room temperature (at 25°C). 0.8 mL of the cell-free extract (crude enzyme) was incubated with 1.2 mL of 5% lactose at 37°C for 30 min. Enzyme lactase hydrolyses lactose into glucose and galactose, and this reaction was terminated by boiling the crude enzyme extract for 10 min. Glucose was then measured using a Glucose kit (Biosystem) at 540 nm (Amamcharla and Metzger, 2011). The quantity of enzyme necessary to yield 1 μg of glucose every minute under typical test circumstances was considered to be a single lactase unit.
2.4. Optimization of lactase producer using experimental design
2.4.1. Plackett-Burman screening design
Using the Plackett-Burman experimental design, the effects of various medium compositions on the production of the lactase enzyme were investigated. Constructed using a Plackett-Burman matrix (Supplementary Table S1) (Ali et al., 2013; Sorour et al., 2020), a two-level factorial design was used to examine fifteen variables at two levels, −1 for the low value and + 1 for the high value. This permits the analysis of n − 1 variables with the fewest number of tests. To determine the relative significance of 15 elements or variables (MgSO4, Glucose, NaNO3, CaCl2, CuSO4, MnSO4, ZnSO4, FeSO4, KCL, NaHPO4.12H2O, KH2PO4, K2HPO4, yeast extract (YE), Beef extract, and Peptone), an experimental design which includes a set of 20 experiments (trials) was used. The first-order model served as the basis for the Plackett-Burman design: Y = o + iXi, where Y represents the response (enzyme activity), o represents the model intercept, I represents the linear coefficient, and Xi represents the degree of an independent variable. Developing the free Essential Experimental Design program, data analysis, coefficient determination, and polynomial model reduction was performed on the lactase enzyme statistics.
2.4.2. Box-Benken design
To discover the kind of response surface in the experiment and to decide the greatest circumstances for enzyme synthesis, a Box-Benken design (Ali et al., 2013; Sorour et al., 2020; Tamer et al., 2021) was adopted. The factorial design, which includes thirteen trials, was constructed to look into the most critical parameters that influence enzyme synthesis. Each variable was examined at three different levels, with low, moderate, and high values, respectively, denoted by −1, 0, and + 1 (Supplementary Table S2). A second-order polynomial equation was developed to determine the appropriate location and to connect the relationship between the independent components and the response for three parameters. The equation was:
Where Y is the expected response, 0 is the design constant, X1, X2, and X3 are the independent factors, 12–13 and 23 are cross-product constants, 11–22 and 33 are quadratic constants, and 12–13 and 23 are coefficients of the cross product, respectively, and 11–22 and 33 are quadratic coefficients, and 12–13 and 23 are cross product coefficients, respectively. The experimental data were subjected to a regression analysis using Microsoft Excel 2010. The constant of determination, R2, was employed to explain the polynomial model equation’s grade of fit (Ali et al., 2013; Sorour et al., 2020). The tests were repeated three times, with the average results presented.
2.5. Purification of lactase enzyme
The crude enzyme was purified using the column chromatography (gel filtration) technique. Multiple columns with different pour sizes were applied to detect the molecular weight and extract the target enzyme (Amicon system). The system combines downstream sample concentration and buffer exchange with affinity-based spin column purification. The tool eliminates the requirement for numerous centrifugation processes and has a large reservoir that can hold a variety of sample quantities. For simultaneous elution, concentration, and extremely effective diafiltration (>99%) in a single spin, the included Amicon® Ultra filter (Philippines - Merck Millipore Division, an affiliate of Merck KGaA, Darmstadt, Germany) was attached. According to the protein cut-off different filters were used (UFC8010, UFC8020, UFC8030, UFC8050, UFC8070, and UFC8100 for 10, 20, 30, 50, 70, and 100 KD respectively). The concentrated protein fraction was very carefully layered on top of the pre-equilibrated and stabilized falcon tubes. The tubes were then centrifuged under cooling (4°C) for 20 min at 8000 rpm, and the upper layer was transferred to the tube with the cut-off (10, 20, 30, 50, 70, and 100 KD) as presented in the supplementary file. Protein content was estimated in each of the filter cut-offs, using the bovine serum albumin (BSA) standard curve The enzyme activity of each of the resulting various molecular weight proteins was then determined (Patel et al., 2017).
2.5.1. Characterization of purified and crude lactase
2.5.1.1. Determination of optimum temperature
At pH 7, temperatures between 30–60°C were found to be the ideal temperature for the crude and purified enzyme activity.
2.5.1.2. Obtaining the ideal pH for enzyme activity
In a trial to test the effect of pH level on crude and purified enzyme activities one at a time, different buffers, namely sodium phosphate buffer, Tris–HCl buffer, and potassium phosphate buffer with varied pH ranges (5.8–8), (6–8.5), and (5.8–8) respectively, were used.
2.6. The antibiotics sensitivity of ALSZ2
Seventeen antibiotic disks were tested, namely amikacin (AMK, 30 μg), amoxicillin-clavulanic acid (AMC, 20 μg/10 μg), ampicillin (AMP, 10 μg), aztreonam (ATM, 30 μg), cefepime (FEP, 30 μg), ceftazidime (CAZ, 30 μg), chloramphenicol (CHL, 30 μg), ciprofloxacin (CIP, 5 μg), clindamycin (CLI, 2 μg), clotrimazole (CC, 10 μg), erythromycin (ERY, 15 μg), gentamicin (GEN, 10 μg), imipenem (IPM, 10 μg), kanamycin (KAN, 30 μg), ticarcillin-clavulanic acid (TIM, 75 μg/10 μg), trimethoprim (TMP, 5 μg) and vancomycin (VAN, 30 μg). The prepared plates were incubated for 24 h at 37°C. The inhibition zone for each antibiotic was measured with a ruler. The indicator of antibiotic sensitivity has traditionally been the existence of a clear zone surrounding the disk. The diameter of the inhibitory zone was used to calculate the results of antibiotic sensitivity tests.
3. Results
In the beginning, many bacteria were isolated from different sources of dairy products, and then the isolates were compared to produce the required enzyme and determine the best one in terms of productivity.
3.1. Isolation and screening of lactase (β-galactosidase) producers
In a screening process for lactase enzyme-producing bacteria, 100 isolates were collected from dairy product samples and examined for enzyme production. Lactase-producing bacteria were screened qualitatively on X-Gal agar plates (Figure 1A) and nutrient broth media containing X-Gal (Figure 1B) at 37°C. Thirteen out of 100 bacterial isolates showed a variance in enzyme activity. Isolate ALSZ2 was the most promising lactase producer and was related for further work.
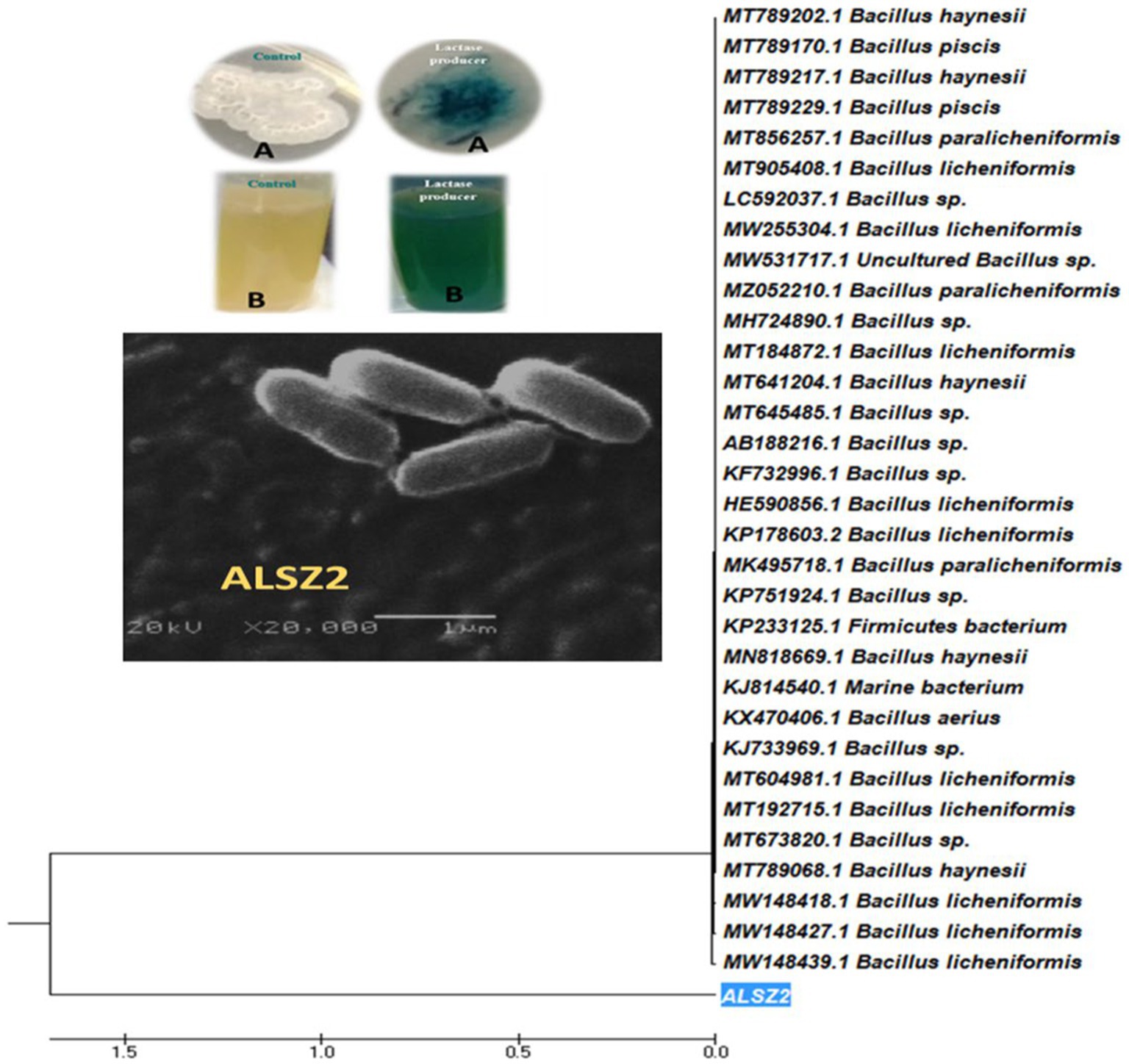
Figure 1. Phylogenetic tree of Bacillus licheniformis ALSZ2 based on 16S RNA sequence assessments. Detection of lactase production using colorimetric methods: (A) shows the negative control and positive lactase producer using solid media and (B) shows the negative control and positive lactase producer using liquid media.
3.2. Identification of the selected bacterial isolates
The most promising isolate with the code ALSZ2 was identified using PCR-amplified 16S rDNA genes. Molecular identification and gene bank sequence isolate (ALSZ2) was Bacillus genus, with a 96.66 present similarity to B. licheniformis, according to the findings. The Mega 7 program was used to generate a phylogenetic tree, showing that isolate (ALSZ2) is more strongly related to B. licheniformis bacterium (acc.: MW148439.1), shown in Figure 1.
3.3. The antibiotic sensitivity of the selected bacterial isolates
The antibiotics amikacin, amoxicillin-clavulanic acid, ampicillin, aztreonam, cefepime, ceftazidime, chloramphenicol, ciprofloxacin, clindamycin, clotrimazole, erythromycin, gentamicin, imipenem, kanamycin, ticarcillin-clavulanic acid, trimethoprim, and vancomycin were all very sensitive to B. licheniformis ALSZ2. The strain was resistant to ampicillin and cefepime.
3.4. Improving the nutritive necessities upsetting Bacillus licheniformis ALSZ2 lactase production using multifactorial statistical design: Plackett-Burman and Box–Behnken designs
3.4.1. Selection of important variables upsetting lactase production using a Plackett-Burman design
The “two-phase” optimization strategy was used to apply the statistical design. The initial stage was to determine the relative relevance of the various components in the culture media, as well as the levels of variables that have a major impact on lactase synthesis. The trials were then verified to validate the results under precise, optimal experimental settings.
Plackett-Burman design for twenty trials with two concentration levels for fifteen different variables (Supplementary Table S1) was conducted based on the experimental matrix shown in Table 1. The corresponding results were summarized in Table 2. The Plackett-Burman design studies’ results revealed a wide range of variations. To assess variables that affect lactase production, the Plackett-Burman statistical design was employed (B. licheniformis ALSZ2). Calculations and graphical representations of the primary impacts of the investigated factors on lactase activity are shown in Figure 2. A major effect value with a positive sign suggests that a variable’s high concentration is close to its optimal level, whereas the main effect value with a negative sign indicates that a variable’s low concentration is close to its optimal level (Figure 2 and Table 3). It was found that Peptone, MgSO4, and Glucose have the greatest positive effect on the production, followed by ZnSO4, Beef extract, KCl, FeSO4, MnSO4, and CaCl2. Whereas, NaHPO4 has the most negative effect, followed by K2HPO4, CuSO4, yeast extract, KH2PO4, and NaNO3 in order.
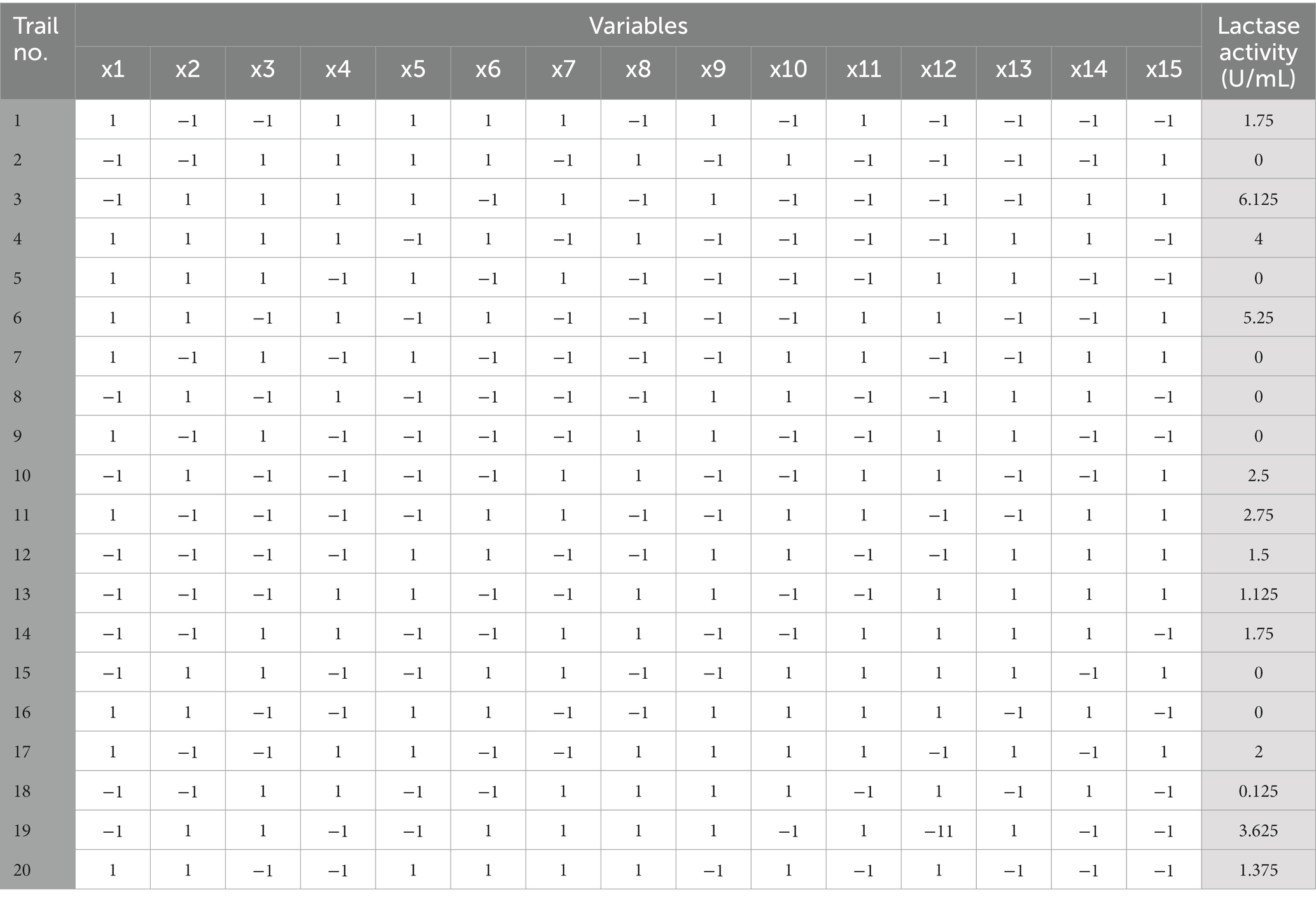
Table 1. Plackett-Burman design matrix for fifteen variables with coded levels for Bacillus licheniformis ALSZ2 lactase optimization.
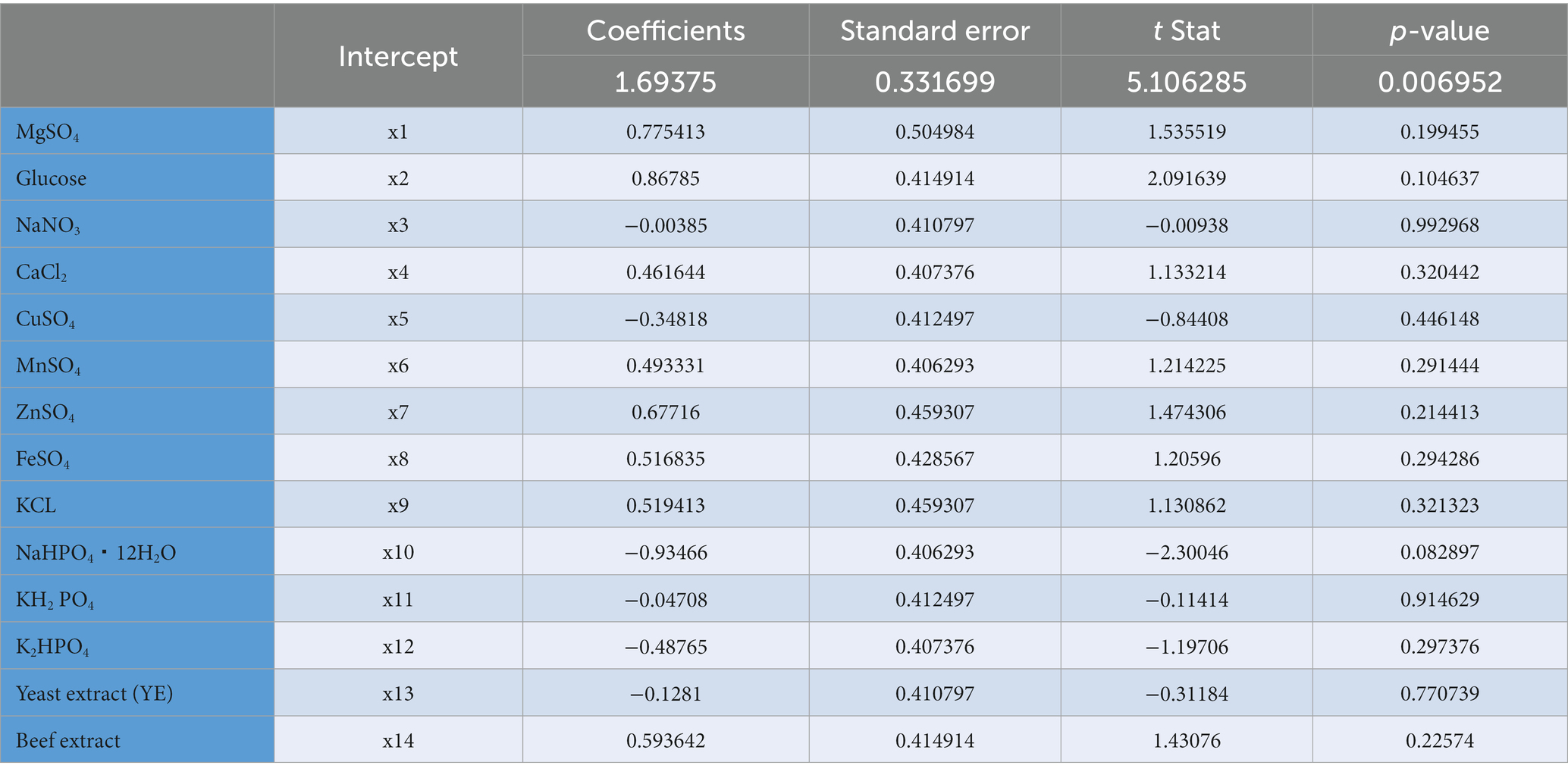
Table 2. Statistical analysis of Plackett–Burman design showing p-values, t stat, standard error, and coefficient values for each variable on B. licheniformis ALSZ2.
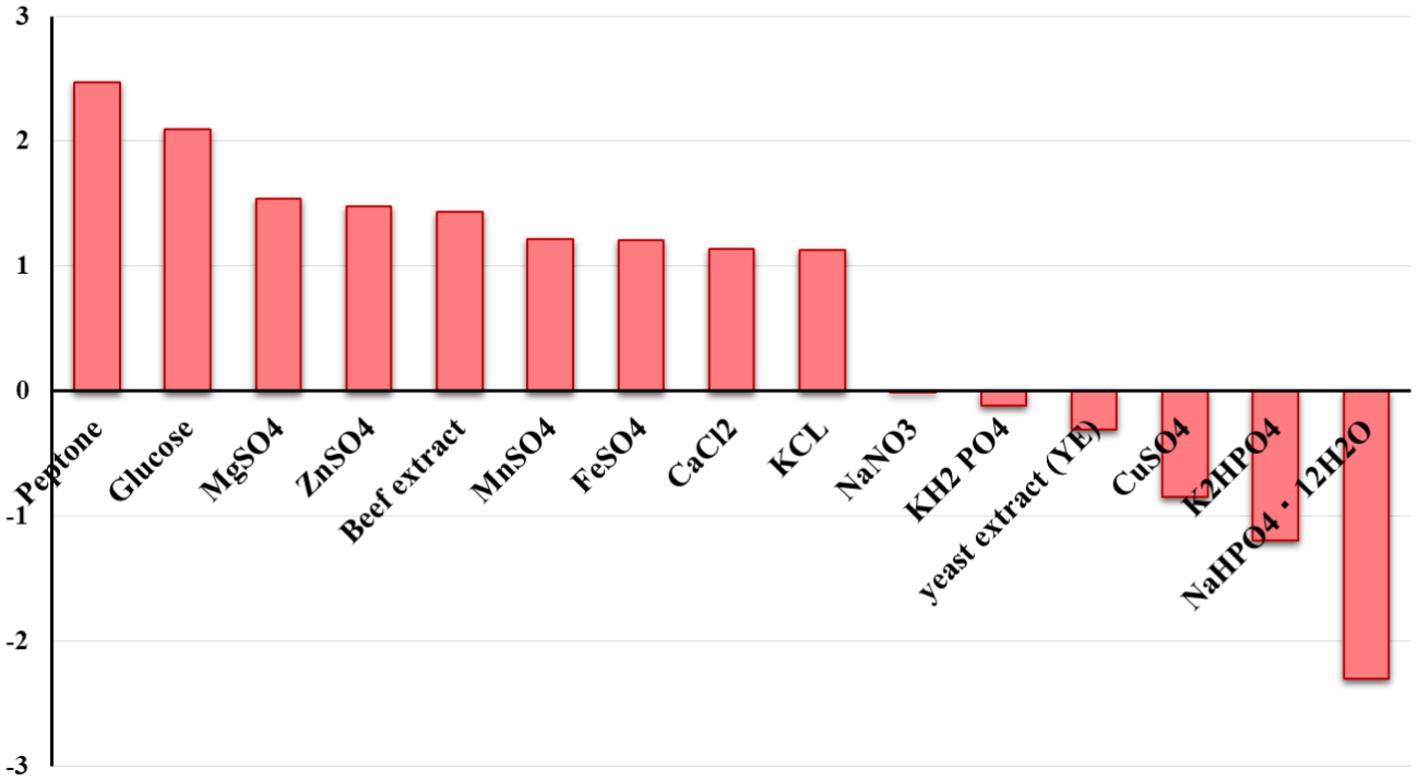
Figure 2. The main effect of different variables on B. licheniformis ALSZ2 lactase production based on Plackett-Burman design results.
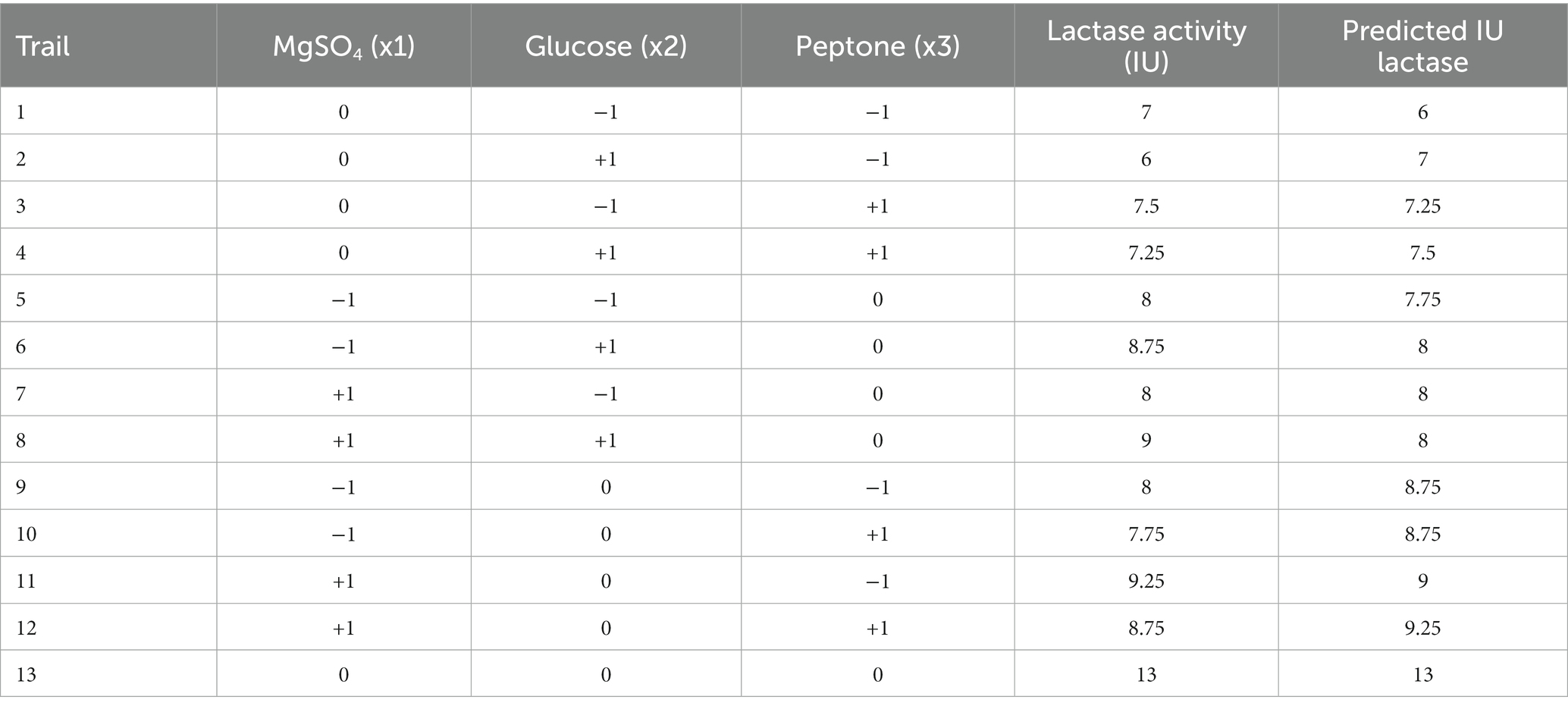
Table 3. Box–Behnken design matrix and results for the three most significant variables that affected B. licheniformis ALSZ2 lactase production.
3.4.1.1. Verification of the model
Variables with negative main effect values were used as (−1) coded values, while variables with positive main effect values were used as (+1) coded values. Supplementary Table S3 shows the lactase activity variations X1, X2, X3, X4, X5, X6, X10, and X19. Considering the data gathered from the outcomes of the Plackett-Burman experiment, the following composition (g/L) is expected to be near to the optimal: Beef extract, 1; MgSO4, 10; Glucose, 75; NaNO3, 30; Yeast extract, 100; CaCl2, 1; CuSO4, 0; MnSO4, 10; ZnSO4, 15; FeSO4, 20; KCl, 1; NaHPO4.12H2O, 10; KH2PO4,10; K2HPO4, 0; Peptone, 25. The medium was reformed to a pH of 7 and the flasks were incubated at 37°C for 24 and 48 h, respectively. A verification experiment was done to estimate the reliability of the Plackett-Burman screening test.
3.4.2. Optimization of medium composition by Box–Behnken design
To validate the precision of the variables identified by the Plackett-Burman design using the Box–Behnken design experimental strategy, Response Surface Methodology was used. The following three tiers of the three crucial factors were looked into: −1, 0, and + 1.
3.4.2.1. Box–Behnken design for Bacillus licheniformis ALSZ2
According to the Box–Behnken design, Supplementary Table S2 shows alternative combinations of the three essential variables. All assemblies had the same concentration of the remaining components as the Plackett-Burman pre-optimized medium. The Box–Behnken experimental design’s results are summarized in Table 3 and the output of the surface plots formula is in (Figure 3). The optimal condition was determined by examining the connection between the individual variables and the lactase activity response. Table 3 shows the design matrix. The design indicated the optimum environmental factors as predictable lactase activity of 13.01754 U/mL, even though 13 U/mL of activity existed under ideal circumstances. As a consequence, the accuracy grade of 99.8% was used to evaluate the power of the perfect matrix under the following ideal circumstances. The medium composition contained g/L: MgSO4, 10.5; Glucose, 75.4491; NaNO3, 30; CaCl2, 1; CuSO4, 0; MnSO4, 10; ZnSO4, 15; FeSO4, 20; KCl, 1; NaHPO4.12H2O, 10; KH2PO4,10; K2HPO4, 0; Yeast extract, 100; Beef extract, 1; Peptone, 25.249 at pH 7, and the bacteria were grown in a rotatory shaker set to 37°C and 200 rpm, for 24 h.
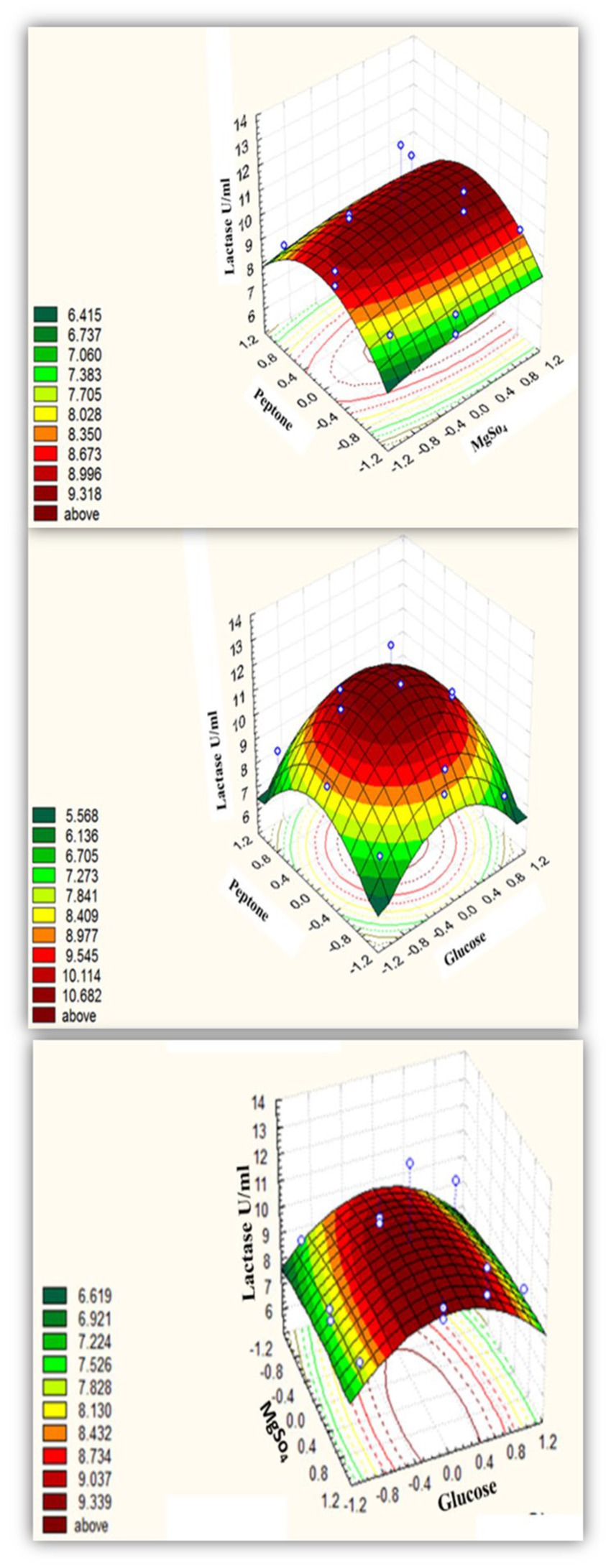
Figure 3. Three-dimensional response surface representing B. licheniformis ALSZ2 lactase enzyme as affected by culture conditions.
3.5. Purification of lactase enzyme
The cell-free extract was subjected to purification using an Amicon system to achieve the best possible result of enzyme activity with different protein molecular weight (MW) cut-offs (100, 50, 30, and 10 kDa). Bacillus licheniformis ALSZ2 lactase was detected in a 30–50 kDa cut-off filter.
3.6. Effect of temperature on lactase activity (crude and purified)
Bacillus licheniformis ALSZ2 lactase (crude & purified) was exposed to seven different temperatures to ascertain the enzyme’s optimum temperature (30, 35, 40, 45, 50, 55, and 60°C Figure 4). Results in Figure 4 revealed that the perfect temperature for maximum enzyme activities (crude & purified) was found to be 35°C with 49 and 46.25 U/mL (30–50 kDa) respectively. Lactase synthesis reached a maximum of 90.05 IU at a temperature of 35°C.
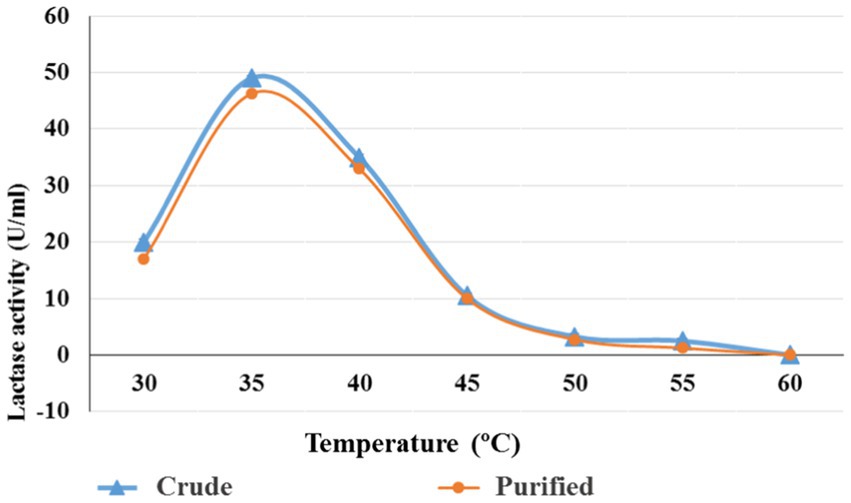
Figure 4. Optimum temperature for crude and purified lactase (30–50 kDa) from B. licheniformis AlSZ2.
3.7. Effect of hydrogen ion concentration on the enzyme activity (crude and purified)
To determine the ideal pH for enzyme activity, three different buffers have been used, namely sodium phosphate, potassium phosphate, and Tris–HCl, each with a distinct pH concentration. Sodium phosphate and potassium phosphate have a pH range of 5.8–8, while Tris–HCl buffer has a pH range of 6–8.5. Results in Figures 5A–C indicated that the optimum enzyme (crude & purified) activities were 7 with all the detected buffers with enzyme activity ranging from 30 & 27.75, 36 & 29, and 20 & 18.25 U/mL, respectively.
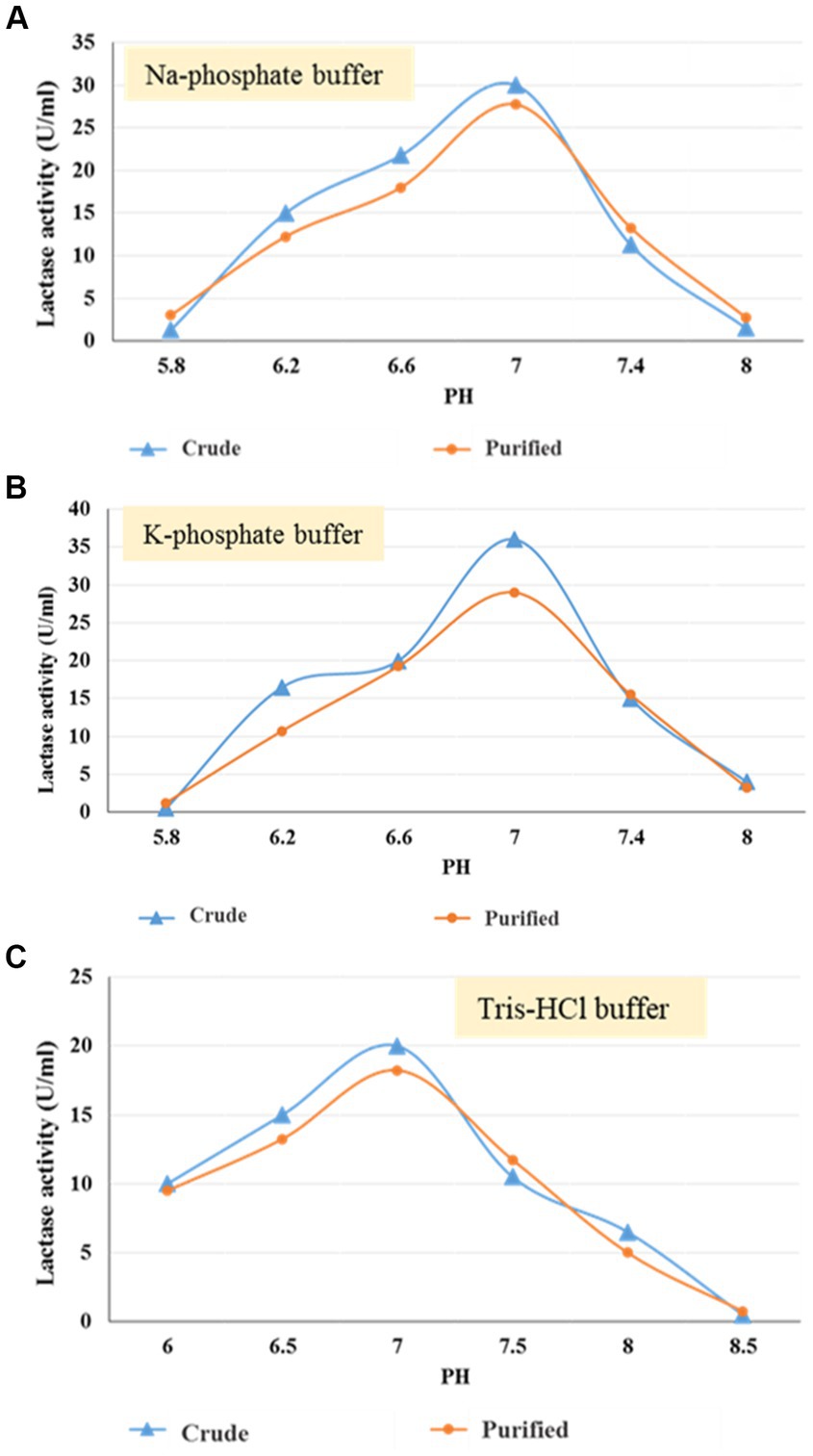
Figure 5. Optimum pH using Na-phosphate (A), K-phosphate (B), and Tris–HCl (C) buffer for crude and purified (30–50 kDa) lactase from B. licheniformis ALSZ2.
3.8. Economic comparison between produced lactase and international products
The need for lactase medical supplements is increasing every day worldwide. The frequency of usage differed significantly between countries. This study provides a promising lactase enzyme product as the initial stage for lactase medical supplements. Comparing AlSZ2 lactase with other products available in the international market, it was found that AlSZ2 lactase is considerably more cost-effective. Many other products are expensive and often unavailable in the Egyptian market. The lactase products’ cost (1,000 U) range from 5.5–8 $, due to the high cost of shipping rather than the real price of the lactase product, whereas 1,000 U of AlSZ2 lactase costs 1.2 $. Therefore, this study is of great importance to cheaply provide lactase, which leads to raising the economic value and aiding in the treatment of a prevalent digestive problem.
4. Discussion
The frequency of Middle Easterners who are lactose intolerant was 70%, with 68 percent of Egyptians affected (Silanikove et al., 2015). Therefore, this study seeks to isolate bacteria capable of producing lactase that can be accessed cheaply.
The amount of carbon in the growth media is critical for bacteria to produce extracellular lactase. Lactase biosynthesis is regulated by carbon availability in various bacteria (Konsoula and Liakopoulou-Kyriakides, 2007; Alazzeh et al., 2009; Maischberger et al., 2010; Akcan, 2018). Based on the experimental matrix, a Plackett-Burman design (Karlapudi et al., 2018; Sorour et al., 2020) was carried out for twenty trials with two concentration levels for fifteen distinct variables (Supplementary Table S1). When using response surface approaches, Plackett and Burman Design is a useful tool for assessing the effects of process factors on yield. It can significantly lower the number of trials that must be repeated in later optimization investigations (Ekpenyong et al., 2017).
In previous research (Manera et al., 2008), lactose was found to be an excellent substrate when used in quantities more than 28.2 g per liter, while Anumukonda and Prabhakar (2010) discovered a 1.5 percent improvement in lactase synthesis in the optimized medium. Another study using solid-state fermentation with Aspergillus terreus NFCCI 1840 found that supplementing the cultured medium with 2.97, 2.88, and 2.67 g per liter of ammonium sulfate, lactose, and magnesium sulfate, respectively, increased lactase production by 2.8 times when compared to the standard basal medium (Al-Jazairi et al., 2015).
Ca2+ and Mg2+ increased enzyme activity in Pediococcus acidilacti, Lactobacillus acidophilus, and Bacillus sp. (Ustok et al., 2010; Carevic et al., 2015; Chanalia et al., 2018). Magnesium is required for the catalytic activity and stability of β -galactosidase. MgSO4 at a concentration of 0.1 percent, Ahmed, et al. (Ahmed et al., 2016) found that Lactobacillus sp. KLSA 22 produced the highest amount of β -galactosidase. Nitrogen sources are the most essential secondary energy molecules for bacterial growth and metabolism. The type of these compounds and the amounts used can either promote or prevent the growth of enzymes (Sharma and Singh, 2014). Sources of nitrogen’s effect on L. caseiMB2’s ability to synthesize galactosidase is important. Peptone supplementation raised the enzyme activity of L. caseiMB2 enzyme production medium to 126.24 IU/mL (specific activity of 315.60 IU/mg). Conversely, urea, sodium chloride, sodium nitrate, ammonium nitrate, and beef extract all showed a reduction in enzyme activity (Heena and Nivedita, 2020).
Numerous separation methods, such as membrane-based separation, ion exchange membrane chromatography, gel permeation chromatography, zinc chloride, protamine sulfate, and ammonium sulfate precipitation, have been investigated for the purification of beta-galactosidase from crude extract (Najer et al., 2022). β - Galactosidases with a range of molecular weights have been discovered in plant sources using the Amicon membrane system with a 100 to 30 kDa cut-off (Aulitto et al., 2021). Five enzymes with molecular weights of 87, 87, 87, 73, and 45 kDa have been recognized (Ajay et al., 2013).
The synthesis of enzymes grew continuously with increasing temperatures up to 35°C and then dropped (Ahmed et al., 2016). Natarajan et al. (2012) found that Bacillus thuringiensis produced the most enzymes at a temperature of 35°C. 37°C was the optimal degree of heating during fermentation, based on how temperature affects the activity of enzymes for the highest β -galactosidase yield (Husain, 2010; Huang et al., 2021).
At pH 7, the greatest enzyme activity was observed (89.94 IU), which was also a good pH for Lactobacillus sp. KLSA 228 growth. However, L. amylophilus GV6 was shown to produce the most lactase when grown at pH 6.513. Lactase from L. delbrueckii spp. ATCC 11842 was found to have the highest activity when pH was 6.8 (Ahmed et al., 2016; Venkateswarulu et al., 2020). Natarajan et al. (2012), on the other hand, found that lactase synthesis was best at a pH of 7.
5. Conclusion
Exploration of bacterial species with unique lactase capacities is critical. As a result, the current study aimed to isolate the most potent extracellular lactase-producing bacteria from various dairy products in various Alexandria locations, as well as increase lactase productivity by a local isolate Bacillus strain ASZ using low-cost materials via a sequential optimization strategy. The findings indicated that the chosen isolate is B. licheniformis ALSZ2, which is employed as a lactase-producing model. Through a two-level Plackett–Burman design, peptone, lactose, and MgSO4 were chosen as investigated variables because of their strong positive influence on lactase efficiency. The lactase yield increased with the initial basal medium, allowing a quadratic polynomial model to be developed that links the relationship between all three factors and lactase production. In comparison to the un-optimized medium, the estimated ideal lactase activity was 13 U/mL, which was four times higher. Following that, the purification of experimental bacterial lactase productivity was done. When compared to comparable lactase products on the worldwide market, AlSZ2 lactase is significantly less expensive.
Data availability statement
The data presented in this study are deposited in the NCBI database under accession number OR535141.
Author contributions
SA conceived the study and funded the project. Experimental work, data analysis, and primary publication writing were all done in collaboration with AA, ZO, and SA. All authors contributed to the article and approved the submitted version.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1180463/full#supplementary-material
Footnotes
References
Aggarwal, S., Chakravarty, A., and Ikram, S. (2021). A comprehensive review on ofcredible renewable carriers as promising platforms for enzyme immobilization & thereof strategies. Int. J. Biol. Macromol. 167, 962–986. doi: 10.1016/j.ijbiomac.2020.11.052
Ahmed, S., Kattimani, L., Divatar, M., Gajare, S., Shivalee, A., Asma, F., et al. (2016). Optimization of lactase production under submerged fermentation by Lactobacillus sp. KLSA 22. Int. J. Pure App. Biosci 4, 212–220. doi: 10.18782/2320-7051.2321
Ajay, P., Melita, L., and Farhath, K. (2013). Extraction, purification and thermodynamic characterization of almond (Amygdalus communis) β-Galactosidase for the preparation of delactosed milk. Food Technol. Biotechnol. 51, 53–61. Available at: https://hrcak.srce.hr/file/146868
Akcan, N. (2018). Cultural conditions optimization for production of β-galactosidase from Bacillus licheniformis ATCC 12759 under solid-state fermentation. Turkish J. Biochem. 43, 240–247. doi: 10.1515/tjb-2017-0153
Alazzeh, A. Y., Ibrahim, S. A., Song, D., Shahbazi, A., and AbuGhazaleh, A. A. (2009). Carbohydrate and protein sources influence the induction of α-and β-galactosidases in Lactobacillus reuteri. Food Chem. 117, 654–659. doi: 10.1016/j.foodchem.2009.04.065
Ali, S. M., Omar, S. H., and Soliman, N. A. (2013). Co-production of cellulase and xylanase enzymes by thermophilic Bacillus subtilis 276NS. Int. J. Biotechnol. Wellness Indus. 2, 65–74. Available at: https://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.985.9502&rep=rep1&type=pdf
Al-Jazairi, M., Abou-Ghorra, S., Bakri, Y., and Mustafa, M. (2015). Optimization of [Beta]-galactosidase production by response surface methodology using locally isolated Kluyveromyces marxianus. Int. Food Res. J. 22:1361. Available at: https://www.proquest.com/openview/66eb623a60fb5faadca2f12a4e4e5779/1?pq-origsite=gscholar&cbl=816390
Alkhafaje, W. K., Ali, S. M., and Olama, Z. A. (2019). Isolation and molecular differentiation of MDR bacteria isolated from dairy products. Pollut. Res. 38, 959–964. Available at: http://www.envirobiotechjournals.com/article_abstract.php?aid=10075&iid=286&jid=4
Alkhafaje, W. K., Ali, S. M., and Olama, Z. A. (2022). Molecular characterization of virulence factors and microbial resistance of different bacterial isolates in some dairy products. Iraqi Journal of Veterinary Sciences. 36, 333–339. doi: 10.33899/ijvs.2021.130206.1764
Amamcharla, J. K., and Metzger, L. E. (2011). Development of a rapid method for the measurement of lactose in milk using a blood glucose biosensor. J. Dairy Sci. 94, 4800–4809. doi: 10.3168/jds.2011-4416
Anumukonda, P., and Prabhakar, T. (2010). Optimization of bioprocess parameters for the production of β-galactosidase by employing statistical methods. Int. J. Pharm. Biosci 1, 1–9. Available at: http://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.437.7762&rep=rep1&type=pdf
Aulitto, M., Strazzulli, A., Sansone, F., Cozzolino, F., Monti, M., Moracci, M., et al. (2021). Prebiotic properties of Bacillus coagulans MA−13: production of galactoside hydrolyzing enzymes and characterization of the transglycosylation properties of a GH42 β-galactosidase. Microb. Cell Factories 20, 71–17. doi: 10.1186/s12934-021-01553-y
Aydin, Y., and Coin, I. (2021). Biochemical insights into structure and function of arrestins. FEBS J. 288, 2529–2549. doi: 10.1111/febs.15811
Cao, Y., Li, X., and Ge, J. (2021). Enzyme catalyst engineering toward the integration of biocatalysis and chemocatalysis. Trends Biotechnol. 39, 1173–1183. doi: 10.1016/j.tibtech.2021.01.002
Carevic, M., Vukasinovic-Sekulic, M., Grbavcic, S., Stojanovic, M., Mihailovic, M., Dimitrijevic, A., et al. (2015). Optimization of β-galactosidase production from lactic acid bacteria. Hem. Ind 69, 305–312. doi: 10.2298/HEMIND140303044C
Chanalia, P., Gandhi, D., Attri, P., and Dhanda, S. (2018). Purification and characterization of β-galactosidase from probiotic Pediococcus acidilactici and its use in milk lactose hydrolysis and galactooligosaccharide synthesis. Bioorg. Chem. 77, 176–189. doi: 10.1016/j.bioorg.2018.01.006
Ekpenyong, M. G., Antai, S. P., Asitok, A. D., and Ekpo, B. O. (2017). Plackett-Burman design and response surface optimization of medium trace nutrients for Glycolipopeptide biosurfactant production. Iranian Biomed J 21, 249–260. doi: 10.18869/acadpub.ibj.21.4.249
Hassan, S. W., Abd El Latif, H. H., and Ali, S. M. (2018). Production of cold-active lipase by free and immobilized marine Bacillus cereus HSS: application in wastewater treatment. Front. Microbiol. 9:2377. doi: 10.3389/fmicb.2018.02377
Heena, C., and Nivedita, S. (2020). Optimization of Β-Galactosidase produced by a potential lactic acid Bacteria Lactobacillus casei MB2 isolated from traditional dairy product of Himachal Pradesh. Int. J. Curr. Microbiol. App. Sci. 9, 2819–2832. doi: 10.20546/ijcmas.2020.906.341
Huang, C., Feng, Y., Patel, G., Xu, X. Q., Qian, J., Liu, Q., et al. (2021). Production, immobilization and characterization of beta-glucosidase for application in cellulose degradation from a novel Aspergillus versicolor. Int. J. Biol. Macromol. 177, 437–446. doi: 10.1016/j.ijbiomac.2021.02.154
Husain, Q. (2010). β Galactosidases and their potential applications: a review. Crit. Rev. Biotechnol. 30, 41–62. doi: 10.3109/07388550903330497
Jacobson, R. H., Zhang, X. J., DuBose, R. F., and Matthews, B. W. (1994). Three-dimensional structure of β-galactosidase from E. coli. Nature 369, 761–766. doi: 10.1038/369761a0
Karlapudi, A. P., Krupanidhi, S., Reddy, R., Indira, M. M. D. N. B., and Venkateswarulu, T. C. (2018). Plackett-Burman design for screening of process components and their effects on production of lactase by newly isolated Bacillus sp. VUVD101 strain from dairy effluent. Beni-Suef Univ. J. basic Appl. Sci. 7, 543–546. doi: 10.1016/j.bjbas.2018.06.006
Kim, M., and Chun, J. (2014). 16S rRNA gene-based identification of bacteria and archaea using the EzTaxon server. Methods Microbiol. 41, 61–74. doi: 10.1016/bs.mim.2014.08.001
Kocabaş, D. S., Lyne, J., and Ustunol, Z. (2021). Hydrolytic enzymes in the dairy industry: applications, market and future perspectives. Trends Food Sci. Technol. 119, 467–475. doi: 10.1016/j.tifs.2021.12.013
Konsoula, Z., and Liakopoulou-Kyriakides, M. (2007). Co-production of α-amylase and β-galactosidase by Bacillus subtilis in complex organic substrates. Bioresour. Technol. 98, 150–157. doi: 10.1016/j.biortech.2005.11.001
Liburdi, K., and Marco, E. (2022). Galacto-oligosaccharide (GOS) synthesis during enzymatic lactose-free Milk production: state of the art and emerging opportunities. Beverages 8:21. doi: 10.3390/beverages8020021
Maischberger, T., Leitner, E., Nitisinprasert, S., Juajun, O., Yamabhai, M., Nguyen, T. H., et al. (2010). Β-Galactosidase from Lactobacillus pentosus: purification, characterization and formation of galacto-oligosaccharides. Biotechnol. J. 5, 838–847. doi: 10.1002/biot.201000126
Manera, A. P., Ores, J. C., Ribeiro, V. A., Burkert, C. A. V., and Kalil, S. J. (2008). Optimization of the culture medium for the production of β-galactosidase from K. marxianus CCT7082. Food Technol. Biotechnol. 46, 66–72. Available at: https://hrcak.srce.hr/22222
Martarello, R. D. A., Cunha, L., Cardoso, S. L., de Freitas, M. M., Silveira, D., Fonseca-Bazzo, Y. M., et al. (2019). Optimization and partial purification of beta-galactosidase production by Aspergillus Niger isolated from Brazilian soils using soybean residue. AMB Express 9, 81–13. doi: 10.1186/s13568-019-0805-6
Movahedpour, A., Ahmadi, N., Ghalamfarsa, F., Ghesmati, Z., Khalifeh, M., Maleksabet, A., et al. (2022). β-Galactosidase: from its source and applications to its recombinant form. Biotechnol. Appl. Biochem. 69, 612–628. doi: 10.1002/bab.2137
Muzaffar, K., Jan, R., Bhat, N. A., Gani, A., and Shagoo, M. A. (2021). Commercially available probiotics and prebiotics used in human and animal nutrition. Adv. Probiotics, 417–435. doi: 10.1016/B978-0-12-822909-5.00025-3
Najer, A., Blight, J., Ducker, C. B., Gasbarri, M., Brown, J. C., Che, J., et al. (2022). Potent Virustatic polymer–lipid Nanomimics block viral entry and inhibit malaria parasites in vivo. ACS Central Sci. 8, 1238–1257. doi: 10.1021/acscentsci.1c01368
Natarajan, J., Christobell, C., Kumar, D. M., Balakumaran, M. D., Kumar, M. R., and Kalaichelvan, P. T. (2012). Isolation and characterization of b-galactosidase producing Bacillus sp. from dairy effluent. World Appl. Sci. J. 17, 1466–1474. Available at: https://www.semanticscholar.org/paper/Isolation-and-Characterization-of-%CE%B2-Galactosidase-Natarajan-Kumar/d74ae1ebf26c1ac62d865f5b5db8a500dbc76148
Patel, A. K., Singhania, R. R., and Pandey, A. (2017). Production, purification, and application of microbial enzymes. Biotechnology of microbial enzymes. Acade. Press 13–41. doi:doi: 10.1016/B978-0-12-803725-6.00002-9Get rights and content
Paulo, A. J. (2018). Produção, caracterização e purificação parcial de uma nova β-Galactosidade produzida por Cladosporium tenuissimum URM 7803. Universidade Federal de Pernambuco. Brazil
Peele, K. A., Krupanidhi, S., Reddy, E. R., Indira, M., Bobby, M. N., and Venkateswarulu, T. C. (2018). Plackett-Burman design for screening of process components and their effects on production of lactase by newly isolated Bacillus sp. VUVD101 strain from dairy effluent. Beni-Suef Univer. J. Basic Appl. Sci 7, 543–546. doi: 10.1016/j.bjbas.2018.06.006
Petassi, M. T., Hsieh, S. C., and Peters, J. E. (2020). Guide RNA categorization enables target site choice in Tn7-CRISPR-Cas transposons. Cells 183, 1757–1771.e18. doi: 10.1016/j.cell.2020.11.005
Piumetti, M., and Illanes, A. (2022). Enzymes and their function. Mol. Dynamics Complex. Catalysis Biocatalysis, 23–53. doi: 10.1007/978-3-030-88500-7.pdf
Poch, O., L'Hôte, H., Dallery, V., Debeaux, F., Fleer, R., and Sodoyer, R. (1992). Sequence of the Kluyveromyces lactis β-galactosidase: comparison with prokaryotic enzymes and secondary structure analysis. Gene 118, 55–63. doi: 10.1016/0378-1119(92)90248-N
Portnoy, M., and Barbano, D. M. (2021). Lactose: use, measurement, and expression of results. J. Dairy Sci. 104, 8314–8325. doi: 10.3168/jds.2020-18706
Ramos, O.S., and Malcata, F.X. (2011). Comprehensive biotechnology. Industrial biotechnology and commodity products 20.
Saqib, S., Akram, A., Halim, S. A., and Tassaduq, R. (2017). Sources of β-galactosidase and its applications in food industry. Biotech 7, 79–77. doi: 10.1007/s13205-017-0645-5
Sedzro, D. M., Bellah, S. M. F., Akbar, H., and Billah, S. M. S. (2018). Structure, function, application and modification strategy of β–Galactosidase. J Multidis Res Rev. 1, 10–16. Available at: https://www.researchgate.net/publication/331276005
Shaaban, M. T., Abdelhamid, R. M., Zayed, M., and Ali, S. M. (2022). Evaluation of a new antimicrobial agent production (RSMM C3) by using metagenomics approaches from Egyptian marine biotech. Biotechnol. Reports 34:e00706. doi: 10.1016/j.btre.2022.e00706
Shaima'a, R., and Rashid, I. M. (2017). Use and misuse of antimicrobial agents in healthcare settings of Diyala Province. Diyala J. Med. 12, 30–33. Available at: https://djm.uodiyala.edu.iq/index.php/djm/article/view/145
Sharma, S., and Singh, P. (2014). Isolation and characterization of β-Galactosidase enzyme producing microbe and optimization of its enzyme activity under different culture condition. Int. J. Cur. Micro. Appl. Sci. 3, 148–155. Available at: https://www.researchgate.net/publication/306092614_Isolation_and_Characterization_of_beta-Galactosidase_Enzyme_Producing_Microbe_and_Optimization_of_its_Enzyme_Activity_under_different_culture_condition
Shrestha, A., Samuelsson, L., Sharma, P., and Day, L. (2021). Cameron-Smith, D., and Milan, A. M. Comparing response of sheep and cow milk on acute digestive comfort and lactose malabsorption: a randomized controlled trial in female dairy avoiders. Front. Nutr. 8. doi:doi: 10.3389/fnut.2021.603816
Silanikove, N., Leitner, G., and Merin, U. (2015). The interrelationships between lactose intolerance and the modern dairy industry: global perspectives in evolutional and historical backgrounds. Nutrients 7, 7312–7331. doi: 10.3390/nu7095340
Soliman, N., Ali, S., and Omar, S. (2018). Statistical optimization for Thermostable Xylanase production by Geobacillus stearothermophilus NASA267 and access to sugar production from some agricultural waste. Res. J. Biotechnol. 13, 28–37. Available at: https://www.researchgate.net/publication/328870798_Statistical_optimization_for_thermostable_xylanase_production_by_Geobacillus_stearothermophilus_NASA267_and_access_to_sugar_production_from_some_agricultural_waste
Sorour, A. A., Ali, S. M., El-Naggar, M. Y., and Olama, Z. A. (2020). Isolation and optimization of cellulase production by Aspergillus penicillioides 12 ASZ using experimental design. Eco. Env. Cons. 26, S109–S115. Available at: http://www.envirobiotechjournals.com/EEC/Vol26AugSuppl20/EEC-17.pdf
Swentowsky, K. W. (2021). Characterization of two meiotic drive kinesins and loci controlling perenniality in Zea (Doctoral dissertation, University of Georgia) Georgia
Szilagyi, A., Catherine, W., and Mark, G. T. (2019). Lactose intolerance and other related food sensitivities. Lactose, 113–153. doi: 10.1016/B978-0-12-811720-0.00003-9
Tamer, T. M., Eweida, B. Y., Omer, A. M., Soliman, H. M., Ali, S. M., Zaatot, A. A., et al. (2021). Removal of oil spills by novel Amphiphilic chitosan-g-Octanal Schiff Base polymer developed by click grafting technique. J. Saudi Chem. Soc. 25:101369. doi: 10.1016/j.jscs.2021.101369
Ureta, M. M., Martins, G. N., Figueira, O., Pires, P. F., Castilho, P. C., and Gomez-Zavaglia, A. (2021). Recent advances in β-galactosidase and fructosyltransferase immobilization technology. Crit. Rev. Food Sci. Nutr. 61, 2659–2690. doi: 10.1080/10408398.2020.1783639
Ustok, F. I., Tari, C., and Harsa, S. (2010). Biochemical and thermal properties of β-galactosidase enzymes produced by artisanal yoghurt cultures. Food Chemi. 119, 1114–1120. doi: 10.1016/j.foodchem.2009.08.022
Venkateswarulu, T. C., Abraham Peele, K., Krupanidhi, S., Prakash Narayana Reddy, K., Indira, M., Ranga Rao, A., et al. (2020). Biochemical and molecular characterization of lactase producing bacterium isolated from dairy effluent. J. King Saud Univ. Sci. 32, 1581–1585. doi: 10.1016/j.jksus.2019.12.014
Venkateswarulu, T. C., Vidya Prabhakar, K., Bharath Kumar, R., and Krupanidhi, S. (2017). Optimization of variables for lactase production from isolated Bacillus subtilis strainVUVD001 through submerged fermentation. Curr. Trend. Biotechnol. Pharm 11, 371–375. Available at: https://agris.fao.org/agris-search/search.do?recordID=US202100049465
Wang, X., Cheng, M. T., Chen, Z. P., Jiang, Y. L., Ge, Y. S., Xia, R., et al. (2021). Structural and biochemical analyses of the tetrameric carboxypeptidase S9Cfn from Fusobacterium nucleatum. Acta Crystallographica section D. Struct. Biol. 77. doi: 10.1107/S2059798321010810/cb5129sup1.pdf
Keywords: lactase, Plackett-Burman, Box-Benken, characterization, purification
Citation: Amin AA, Olama ZA and Ali SM (2023) Characterization of an isolated lactase enzyme produced by Bacillus licheniformis ALSZ2 as a potential pharmaceutical supplement for lactose intolerance. Front. Microbiol. 14:1180463. doi: 10.3389/fmicb.2023.1180463
Edited by:
Monika Prakash Rai, Amity University, IndiaReviewed by:
Atte Johannes Von Wright, University of Eastern Finland, FinlandSowmyalakshmi Subramanian, McGill University, Canada
Copyright © 2023 Amin, Olama and Ali. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Safaa M. Ali, U2FmYWEubW9oYW1lZGFsaUB5YWhvby5jb20=; U2FsaUBzcnRhY2l0eS5zY2kuZWc=
†ORCID: Safaa M. Ali, https://orcid.org/0000-0001-9850-0174
 Alaa A. Amin1
Alaa A. Amin1 Safaa M. Ali
Safaa M. Ali