
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol., 12 July 2023
Sec. Microbial Physiology and Metabolism
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1179607
This article is part of the Research TopicHydrogenase: Structure, Function, Maturation, and ApplicationView all 12 articles
Hydrogen is considered one of the key enablers of the transition towards a sustainable and net-zero carbon economy. When produced from renewable sources, hydrogen can be used as a clean and carbon-free energy carrier, as well as improve the sustainability of a wide range of industrial processes. Photobiological hydrogen production is considered one of the most promising technologies, avoiding the need for renewable electricity and rare earth metal elements, the demands for which are greatly increasing due to the current simultaneous electrification and decarbonization goals. Photobiological hydrogen production employs photosynthetic microorganisms to harvest solar energy and split water into molecular oxygen and hydrogen gas, unlocking the long-pursued target of solar energy storage. However, photobiological hydrogen production has to-date been constrained by several limitations. This review aims to discuss the current state-of-the art regarding hydrogenase-driven photobiological hydrogen production. Emphasis is placed on engineering strategies for the expression of improved, non-native, hydrogenases or photosynthesis re-engineering, as well as their combination as one of the most promising pathways to develop viable large-scale hydrogen green cell factories. Herein we provide an overview of the current knowledge and technological gaps curbing the development of photobiological hydrogenase-driven hydrogen production, as well as summarizing the recent advances and future prospects regarding the expression of non-native hydrogenases in cyanobacteria and green algae with an emphasis on [FeFe] hydrogenases.
Sunlight is the most abundant and widespread energy source around the globe. Every year the sun irradiates Earth’s surface with more than 7,000 times the current global energy consumption. On average 173 PW of energy strikes Earth’s surface at any time, providing a virtually unlimited energy source that can be harvested locally. The conversion of solar energy into chemical energy, e.g., in the form of hydrogen (H2), greatly facilitates storage, distribution and applicability in a wide-range of settings. Presently H2 is mainly produced via steam reforming (yielding so-called “grey” hydrogen), but truly fossil-free hydrogen production is increasing. Current fossil-free processes rely on electrolyzers driven by carbon-free electricity. Albeit efficient these technologies feature several limitations, e.g., high demand of electricity and scarce rare earth metals requirements. Renewable energies (which can yield green and yellow hydrogen) have become cost-competitive and their expected growth trends are astonishing [estimated annual growth rates of 8.4% for renewables (Novus-Light-Technologies, 2023)] during the next decade. Similarly, nuclear power (pink hydrogen) is expected to increase by 5.7% (Bussiness-Research-Company, 2023) in the same time period. Still, electricity demands are expected to increase significantly, up to 50%, during the electrification process towards a net-zero carbon emissions economy (IEA, 2021; Exxon-Mobil, 2022). It follows that electrochemical hydrogen production is likely to be limited to contexts where there is a frequent surplus of clean electricity generation. Consequently, novel hydrogen production technologies are required for global large-scale production, ideally integrated within other industrially relevant processes.
Biotechnological hydrogen gas production provides an inexpensive and scalable alternative technology for H2 production. Indeed, our drive towards a hydrogen society is pre-dated by evolution, which has developed an efficient hydrogen economy involving biological machineries capable of hydrogen production without the requirement of rare-earth metals and avoiding heavy electricity consumption. Waste-to-hydrogen has been intensively explored as a sustainable alternative to conventional hydrogen production from fossil fuels. However, the demand of high-cost pre-treatment methods, the use of inconsistent feedstock as well as low energy conversion efficiencies are noteworthy constrictions of this technology (Lui et al., 2020). Also, the competition with well-established biogas production processes and the overall reduced energy content in waste limits its application for large-scale H2 production. Instead, oxygenic photosynthetic organisms stand out as ideal platforms for biological H2 production as they are capable of direct solar-to-hydrogen (STH) energy conversion.
Cyanobacteria and green algae exhibit biological features that make them highly suitable for the engineering of hydrogenase based STH systems. They feature a highly specialized light harvesting machinery coupled to a complex photosynthetic electron transport chain (PETC). Briefly, protons (H+) and electrons are extracted from the water splitting reaction at photosystem II (PSII). Subsequently, H+ reduction to hydrogen can be performed by redirecting the high potential electrons via the PETC towards photosystem I (PSI) and then transferring them to a hydrogenase. Indeed, most cyanobacteria and green algae feature at least one native hydrogenase and certain strains are capable of native hydrogen production under specific conditions. Commonly, hydrogenases in cyanobacteria are used as a mechanism to recover energy from the H2 produced as a byproduct of nitrogenase in nitrogen fixing strains (uptake hydrogenases). Additionally, hydrogenases also act as photoprotective electron valves during periods when metabolic electron sinks are inactive or cannot cope with a sudden increase in the reducing power supplied from photosynthetic apparatus (bidirectional hydrogenases). Important differences between both organisms are found in the light harvesting systems and internal cellular organization. While eukaryotic green algae harbor light harvesting complexes similar to plants, cyanobacteria are prokaryotes and feature phycobilisomes containing specific light harvesting phycobiliproteins not present in green algae. Still, both exhibit photosynthetic efficiencies and biomass production rates significantly higher than any plants. Previous reviews (Khanna and Lindblad, 2015; Kosourov et al., 2021; Touloupakis et al., 2021; Li et al., 2022; Redding et al., 2022) provide a broad picture of the STH paradigm, reviewing the most relevant photobiological H2 production strategies reported to date.
Photobiological H2 production can occur via two main mechanisms, employing either nitrogenases or hydrogenases. The former relies in co-occurring H2 production during N2-fixation. Moreover, nitrogenase H2 production requires both ATP and reducing equivalents. Therefore, the maximum efficiency of nitrogenases is significantly lower than hydrogenase-based systems, which only require reducing equivalents from a compatible electron donor. Additionally, some hydrogenases have turnover numbers up to 1,000-fold greater than those of nitrogenases (Hallenbeck and Benemann, 2002). Thus, hydrogenases are undoubtedly better catalysts for the purpose of biotechnological STH conversion owing to its higher catalytic rates and more efficient energy utilization. Historically, hydrogenase derived H2 production has attracted less attention due to a range of limitations, including the bidirectional nature and catalytic bias of the enzymes, and an apparent trade-off between turnover rates and O2 tolerance. These perceived issues, in combination with an earlier lack of tools for extensive engineering of photosynthetic organisms and limited information about hydrogenase structure(s) and mechanism(s) has hindered the development of hydrogenase-driven STH production. However, during the last years, a number of multiple factors has unlocked the possibility of further development of hydrogenase based photobiological H2 production.
Firstly, a deeper understanding of structural and functional principles of hydrogenases has been obtained. In addition, several novel hydrogenases have been discovered or engineered, exhibiting highly interesting features for biotechnological utilization. Secondly, a wider understanding of photosynthetic metabolism, cell physiology and regulation has been acquired during the last decades, enabling the development of more sophisticated genetic engineering and synthetic biology tools. In combination with a growing interest in photosynthetic microorganisms within the scientific community and industry, this has unlocked the possibility to further engineer photosynthetic organisms for photobiological hydrogenase-driven H2 production. Current electrochemical STH technologies, feature energy conversion efficiencies ranging between 10 and 20% and production costs about 6 to 8 € · kg H2−1 (Grimm et al., 2020). Maximum theoretical estimations for non-engineered photobiological hydrogen production indicate that efficiencies close to 10% could be achieved, being on-par with current electrochemical technologies (Kruse et al., 2005). We expect that further developments combining the different strategies summarized here will allow a dramatic increase of efficiency relative to current approaches. Depending on the obtainable STH conversion efficiencies and further bioprocess design, our production costs estimations may be as low as 5 € · kg H2−1, making photobiological H2 production potentially cheaper than current STH electrochemical methods.
In the present review we provide a summary of the, in our opinion, most relevant achievements in the context of hydrogenase-based photobiological H2 production systems, highlighting how these efforts are bringing us closer towards the development of cost-effective photobiological hydrogen production technologies.
Hydrogenases are an evolutionary convergent enzyme family of oxidoreductases that catalyze the reduction of protons into dihydrogen as well as the reverse oxidation of dihydrogen into two protons and two electrons. They are found in all domains of life, predominately present in Archaea and Bacteria and rarely in Eukarya (Vignais and Billoud, 2007; Greening et al., 2016). Hydrogenases are classified into [NiFe], [FeFe], and [Fe] hydrogenases based on their unique organometallic cofactors that facilitate reversible redox reactions (Lubitz et al., 2014; Berggren et al., 2022). [Fe] hydrogenases show comparably low catalytic rates and differ substantially from [NiFe] and [FeFe] hydrogenases in their reaction mechanism and substrates (Vogt et al., 2008). Therefore, they are not considered suitable for a photosynthesis-coupled hydrogen production and will not be discussed further. [NiFe] and [FeFe] hydrogenases on the other hand can achieve hydrogen turnover in vivo by using ferredoxin and NAD(P)H/NAD(P)+ (among others) as redox partner which are closely linked to PSI and the photosynthetic machinery (Appel et al., 2020; Lupacchini et al., 2021) However, we note that the they are extremely diverse enzyme families, reflecting large variations in activity between different hydrogenases (Land et al., 2020; Berggren et al., 2022).
Efficient (photo-)biological hydrogen production is dependent on successful expression of active and robust hydrogenases. The oxygen (O2) sensitivity of most [NiFe] and [FeFe] hydrogenases represent a particular challenge for coupling in vivo hydrogen production to the photosynthetic electron transport, as PS II produces molecular oxygen during the water oxidation. There are critical differences between hydrogenases in their reactivity towards O2, but the terminology of oxygen tolerance and oxygen sensitivity can lead to misconceptions.
In the following, we will distinguish between oxygen-sensitive and oxygen-insensitive hydrogenases. Oxygen-insensitive enzymes are defined as functional under atmospheric oxygen concentration. Conversely, oxygen-sensitive hydrogenases are inhibited by oxygen, but can be further subdivided into oxygen-sensitive tolerant and oxygen-sensitive intolerant hydrogenases based on the reversibility of their oxygen-induced inhibition. It should be noted that these definitions are different as compared to the general hydrogenase literature, where the term O2 tolerant is commonly used to describe what we would refer to as O2 insensitive hydrogenases (see, e.g., Lubitz et al., 2014). Most [NiFe] hydrogenases reported to-date are oxygen-sensitive tolerant enzymes, i.e., they display reversible oxygen-induced inhibition. A few [NiFe] hydrogenases even stand out as oxygen-insensitive as their function under atmospheric oxygen concentrations is not impaired (Buhrke et al., 2005; Leroux et al., 2008; Liebgott et al., 2010; Schäfer et al., 2013; Grinter et al., 2023). In contrast, the majority of [FeFe] hydrogenases are oxygen-sensitive intolerant enzymes as the active site is irreversibly damaged when exposed to trace amounts of oxygen. Still, there are also reports of oxygen-sensitive tolerant [FeFe] hydrogenases (Morra et al., 2016; Corrigan et al., 2020; Winkler et al., 2021). Within these broad classifications, the kinetics of the inactivation and reactivation vary between enzymes (see for example Cracknell et al., 2008; Goldet et al., 2009; Lukey et al., 2011; Kubas et al., 2017; Koo and Swartz, 2018).
Most [NiFe] hydrogenases are dimeric enzymes, consisting of a “small” and a “large” subunit of which the latter one contains the active site with the catalytic [NiFe] cofactor. The small subunit harbors usually a chain of three FeS-clusters, denoted as proximal, medial and distal depending on their relative position to the [NiFe] active site, which ensures electron transport between protein surface and NiFe center. The medial cluster is often [3Fe-4S] while the proximal and distal clusters are [4Fe-4S] (Volbeda et al., 1995, 1996). Exceptions to this FeS-cluster arrangement have been observed, and include the soluble bidirectional cytoplasmatic [NiFe] hydrogenases found in different aerobic microorganisms such as the HoxFUYH complex from Synechocystis PCC 6803. The NAD(P)+-reducing [NiFe] hydrogenase HoxFUYH is a multi-subunit complex with HoxH as catalytic (or “large”) subunit. The accessory subunits HoxFUY build a complex electron relay where HoxY serves as the immediate electron transfer partner of HoxH, and can be considered equivalent to the small subunit, but features only a single [4Fe-4S] cluster (Carrieri et al., 2011). Other reported exceptions with changes to the electron relay clusters have been shown to influence oxygen tolerance and catalytic bias as further outlined below. [FeFe] hydrogenases often show substantially higher turnover rates than [NiFe] hydrogenases at the expense of being irreversibly inactivated by trace amounts of molecular oxygen (Goldet et al., 2009). [FeFe] hydrogenases are generally functional as monomers but can consist of several domains (Calusinska et al., 2010; Land et al., 2020). The H-domain is essential for the function as it contains the active site (H-cluster) which comprises a canonical [4Fe-4S] cluster ([4Fe-4S]H) that is linked to the di-iron metal center via a bridging cysteine. [FeFe] hydrogenases commonly carry additional domains with FeS-clusters in the N- or C-terminus of the H-domain (Meyer, 2007). The modularity of these so-called F-domains ensures that they can provide the enzyme with different functions, e.g., ensuring efficient intra- or intermolecular electron transport between the highly conserved active site and redox partners (Gauquelin et al., 2018).
Even though [NiFe] and [FeFe] hydrogenases are not evolutionary related, they share certain key characteristics among their active sites. Both catalytic cofactors are dinuclear, and the metal ions bridged by thiol ligands. In the case of [NiFe] the thiols are derived from cysteine, while [FeFe] hydrogenases are dependent on a 2-aza-propane-dithiolate (adt) ligand that bridges the di-iron metal center. Furthermore, both feature Fe ions ligated by carbon monoxide and cyanide. These strong field ligands keep the Fe ions in a low-spin and low valent state, which enables the heterolytic splitting of H2 or the dihydrogen formation. Considering their unique structures, the assembly and introduction of these organometallic active site cofactors unsurprisingly depend on hydrogenase-type specific maturation enzymes (Lubitz et al., 2014). Cyanobacteria express natively one or two [NiFe] hydrogenases, which either serve as sink for excess electrons from photosynthesis or are closely linked to the hydrogen uptake for nitrogenase activity. They rely on the expression of at least six [NiFe] cofactor maturation enzymes (HypABCDEF) which have been identified in E. coli but are known to be existent in all organisms that express [NiFe] hydrogenases. [FeFe] hydrogenases on the other side are not present in any known cyanobacterial organism. So far [FeFe] hydrogenases and their specific cofactor maturation enzymes (HydEFG) have been exclusively found in algal organisms and non-cyanobacterial prokaryotes (Böck et al., 2006; Khanna and Lindblad, 2015). Consequently, heterologously expressed hydrogenases are only functional if either the expression host expresses natively the specific maturation enzymes or the maturation machinery is co-expressed (King et al., 2006; Wegelius et al., 2021). It was recently shown that maturation enzymes of [FeFe] hydrogenases require an additional auxiliary pathway providing a co-substrate, a glycine cleavage system generating methylated lipoyl-H-protein (Pagnier et al., 2022). However, this co-substrate may usually be regenerated by the native host metabolism.
Several different approaches have been explored to achieve a photosynthesis-coupled hydrogen production. In the most cases, the work focused on coupling hydrogenases to electron carriers or PSI itself. Even though the high catalytic rates of [FeFe] hydrogenases exhibit a considerable potential for such applications the oxygen sensitivity and the irreversible inactivation are major obstacles as mentioned above. Therefore, it is necessary to explore possible ways to create oxygen-insensitive [FeFe] hydrogenases.
In the case of [NiFe] hydrogenases evolution has provided a number of different features identified to provide oxygen tolerance. Primarily these protection strategies appear to focus on the controlled reduction of O2 to avoid reactive oxygen species (ROS) and/or limiting gas access to the O2-sensitive active-site. These features could be considered for the generation of oxygen-insensitive [FeFe] hydrogenase. For several membrane bound oxygen-insensitive (originally referred as oxygen tolerant) [NiFe] hydrogenases an altered FeS cluster has been reported proximal to the di-nuclear metal center. This proximal [4Fe-3S]-6Cys cluster enables the oxygen-insensitive [NiFe] hydrogenases to rapidly transfer two electrons to the active site, ensuring detoxification of O2 through reduction to water (Ogata et al., 2016). However, such an alteration of the [FeFe] active site is likely challenging as the proximal [4Fe-4S]H-cluster is directly involved in the catalytic cycle. However, there are reports of F-clusters improving O2 tolerance (Kubas et al., 2017). Still, we note that any protection mechanism depending on reduction of O2 will result in loss of electrons towards unproductive process. Several studies also explored oxygen tolerance achieved by limiting mass transfer through the putative gas channels (Buhrke et al., 2005; Leroux et al., 2008; Liebgott et al., 2010). Buhrke et al. reported that in the H2-sensing [NiFe] hydrogenase RH from Ralstonia eutropha bulky hydrophobic residues restrict the diffusion of molecular oxygen to the active site. A recent striking example of this mechanism at play is provided by the Huc hydrogenase from Mycobacterium smegmatis, which is capable of performing H2 oxidation under fully aerobic conditions (Grinter et al., 2023). Molecular dynamics experiments of [FeFe] hydrogenases suggested that O2 can only diffuse through hydrophobic gas channels whereas H2 can also diffuse freely through temporary cavities in the flexible protein structure (Cohen et al., 2005). However, engineering approaches which aimed at modulating the gas transfer through the assumed main gas channel have not been successful so far (Lautier et al., 2011). Furthermore, a complete obstruction of the gas channels in [FeFe] hydrogenases is also not assumed beneficial, as the build-up of H2 accumulation in the active site can inhibit the H2 production (Fourmond et al., 2013).
The main protection mechanism against O2-induced degradation of the [FeFe] hydrogenase cofactor identified to-date involves binding of thiol ligands at the open Fe-coordination site involved in catalysis. For sulphate-reducing bacteria Desulfovibrio desulfuricans and Desulfovibrio vulgaris Hildenborough, and their respective [FeFe] hydrogenases DdHydAB and DvHydAB, it has been shown that an extrinsic sulfide group can coordinate the substrate binding site. This protected state of the active site causes reversible inactivation but also oxygen-sensitve tolerant characteristics (Roseboom et al., 2006). Chemical treatments of purified [FeFe] hydrogenases with NaS2 can induce this state in vitro under oxidizing conditions (Rodríguez-Maciá et al., 2018). More recently, a similar effect was reported through the binding of exogenous cyanide (Martini et al., 2023). However, in contrast to thiols, cyanide binding appears irreversible thus also causing inactivation of the enzyme. Up to now only one intrinsically oxygen-sensitive tolerant [FeFe] hydrogenase from Clostridium beijerinckii (CbA5H/CbHydA1) has been identified and characterized by Morra et al. (2016). An active site neighboring cysteine enables a reversible inactivation of the [FeFe] hydrogenases by coordinating the substrate binding site of the H-cluster with its thiol sidechain (Corrigan et al., 2020; Winkler et al., 2021). This coordination prevents the reduction of O2 at the H-cluster and therefore also the formation of reactive oxygen species that could irreversibly damage the active site cofactor. Winkler et al. also reported that the interaction of individual distant residues around the loop region of the protective cysteine are crucial for an oxygen-protective conformation. In another recent study it was shown that one amino acid exchange on the surface of CbHydA1 can modulate the transition rates between the inactivated and oxygen-sensitive state (Rutz et al., 2023). With the above-mentioned findings it stands out that protein engineering of characterized [FeFe] hydrogenases is of significant importance to develop more robust hydrogen-producing biocatalysts. In parallel, the identification and characterization of new [FeFe] hydrogenases are needed to explore more beneficial properties such as oxygen sensitivity (Morra, 2022).
Another aspect to consider when using hydrogenases is that they are bidirectional catalysts. However, the activity can be influenced by many parameters, and often the enzymes show a preference for either hydrogen production or oxidation. This kinetic discrepancy between the forward and reverse reaction is referred to as “catalytic bias.” Amino acid side chains in the first coordination sphere of the organometallic cofactor can influence the catalytic bias (Land et al., 2020). Additionally, when present, the FeS clusters in the F-domains of [FeFe] hydrogenase have been shown to introduce a strong catalytic bias (Pandey et al., 2017; Birrell et al., 2021).
Engineering efforts towards efficient photobiological H2 production has focused into two main complementary paths. First, removing native hydrogenases capable of H2 uptake and optimizing the hydrogenase as the key component of the H2 production system. In this context, heterologous, chimeric, or fully synthetic hydrogenases with improved features are over-expressed as a way to enhance hydrogen production. Importantly, enzyme expression also needs to be at high levels, as it has been documented that hydrogen production correlates with the abundance of active hydrogenase enzyme in vivo (Weiner et al., 2018). Secondly, significant efforts have been devoted at re-engineering different sub-systems within the complex bioenergetic machinery of PETC and/or light harvesting systems, aiming to enhance the overall electron supply and efficiently channel it towards the expressed hydrogenase.
During the last decades a deep understanding of cyanobacterial physiology has been acquired, giving rise to the development of an expanding synthetic biology toolbox for sophisticated strain engineering. These factors in combination with its generally faster growth rates, more flexible light harvesting machinery and engineering simplicity when compared to green algae makes them a very promising platform for developing highly efficient green cell H2 producing factories. Table 1 provides a summary of achievements on H2 production when expressing non-native hydrogenases in oxygenic photosynthetic microorganisms.
[FeFe] hydrogenases have been widely considered the most active and promising group for hydrogenase-catalyzed H2 production. Unfortunately, cyanobacteria do not feature native [FeFe] hydrogenases, neither the maturation machinery required for its activation. This limitation has been overcome by the co-expression of an [FeFe] hydrogenase together with its maturation machinery (HydEFG). An [FeFe] hydrogenase from Clostridium acetobutylicum (HydA1) together with the HydEF and HydG maturation machinery from Chlamydomonas reinhardtii were introduced and expressed in the non-N2-fixing unicellular cyanobacterium Synechococcus elongatus PCC 7942 (Ducat et al., 2011). The introduced enzyme showed an in vivo hydrogenase activity which connected to the light-dependent reactions of PETC under anoxic conditions. The heterologous hydrogenase supported limited growth in light using H2 as the sole source of reducing equivalents. Moreover, the importance of adequate coupling between the non-native hydrogenases and the native electron carriers was noted, where an expression of a non-native ferredoxin improved the H2 production. Comparably, the native [FeFe] hydrogenase operon from the bacterium Shewanella oneidensis MR-1 was expressed in heterocysts of the filamentous cyanobacterium Nostoc PCC 7120, leading to the assembly of an active enzyme (Gärtner et al., 2012). In a similar fashion, the native maturation machinery and of hydrogenase HydA of Clostridium acetobutylicum has been reported to yield an active [FeFe] hydrogenase in Nostoc PCC 7120 using the heterocyst specific nifH promoter. Furthermore, by co-expressing a GlbN cyanoglobin from Nostoc commune to decrease the oxygen levels, in vivo H2 production was observed concomitantly with oxygenic photosynthesis in the vegetative cells of the filaments (Avilan et al., 2018). Earlier reports of active [FeFe] hydrogenases in cyanobacteria without the introduction of the specific [FeFe] hydrogenase maturation machinery (Asada et al., 2000; Berto et al., 2011) requires further confirmations.
However, it is possible to circumvent the biological maturation of [FeFe] hydrogenase by using a semi-synthetic activation approach. This procedure consists on the heterologous expression of the inactive [FeFe] apo-hydrogenase in living cells, followed by subsequent incubation with a synthetic [2Fe]H subcluster mimics, leading to the formation of an active H-cluster and rendering a mature holo-[FeFe] hydrogenase. Semi-synthetic [FeFe] hydrogenase maturation was first demonstrated in E. coli (Khanna et al., 2017). The mechanism by which the synthetic cofactor enters the cell and activates the enzyme remains to be elucidated, but it has been found to occur on a minute-time scale (Mészáros et al., 2018). Later work expanded its utilization to cyanobacteria (Wegelius et al., 2018, 2021). The resulting non-native, semisynthetic enzyme unmistakably links to the native metabolism of the photosynthetic cell and retain its H2 production capacity for several days with an activity based on availability of electrons.
To date studies on [FeFe] hydrogenase expression in cyanobacteria has focused almost exclusively on representative examples of “prototypical” Group A [FeFe] hydrogenases. However, Wegelius et al. also reported on the variance in H2 production arising from employing [FeFe] hydrogenases from different phylogenetic groups (Wegelius et al., 2021), and showed that the Group D enzyme Thermoanaerobacter mathranii HydS displayed distinct differences in its hydrogen gas production profile relative to the Group A enzymes from C. reinhardtii and Solobacterium moorei.
Heterologous expression of [NiFe] hydrogenases provides the possibility to take advantage of the native cyanobacterial [NiFe] hydrogenase maturation machinery. Thus, it circumvents the challenge of efficiently co-expressing the hydrogenase, together with its maturation machinery (HydEFG) required to obtain active [FeFe] hydrogenases. As such, it provides a potentially more straightforward way to study the effect of introducing enzymes with improved properties or to engineer fusion proteins between the native cyanobacterial [NiFe] hydrogenase and different components of the PETC, which in turn can guide efforts related to [FeFe] hydrogenase.
Early research reported the expression of heterologous [NiFe] hydrogenases from Alteromonas macleodii and the bidirectional [NiFe] hydrogenase from Thiocapsa roseopersicina in a mutant Synechococcus elongatus PCC 7942 lacking the native bidirectional hydrogenase structural genes (ΔHoxYH). Although the proteins did not show any activity in vivo, in vitro H2 production confirmed the successful activation of the two hydrogenases, and the coupling of the catalytic subunits (HoxYH) from T. roseopersicina to the native diaphorase HoxEFU subunits. However, cloning of multiple hydrogenase accessory genes from native organisms was still required to achieve the assembly and activation of functional [NiFe] hydrogenases, highlighting the diversity of hydrogenase maturation mechanisms even within the realm of [NiFe] hydrogenases (Weyman et al., 2011).
In similar endeavors, O2-tolerant, and NAD(H)-dependent hydrogenase from Ralstonia eutropha (ReSH) was expressed in SynechocystisPCC 6803 featuring a knockout of the native hydrogenase (Lupacchini et al., 2021). In this study, the engineered strain was able to continuously produce H2 under illumination at a higher rate than the wild-type (WT) and for up to 20 h under illumination. This result indicates that the native maturation machinery was sufficient to mature the heterologously expressed [NiFe] hydrogenase without the requirement of its accessory hyp-genes. The sustained H2 production was attributed with the higher O2 tolerance of the ReSH. In addition, the ReSH expressing strain was able to use H2 as sole electron source, enabling autotrophic growth, even when water oxidation at PSII was blocked by DCMU (3-(3,4-dichlorophenyl)-1,1-dimethylurea) addition. This indicates clear coupling between the introduced hydrogenase and the native cyanobacterial redox metabolism. However, due to the NAD(H) dependent nature of ReSH, a high intracellular NADH/NAD+ ratio and efficient NADH supply are required for H2 production. This was achieved by glucose supplementation and blocking of any other metabolic electron sinks such as nitrate or carbonate/CO2 assimilation, where the cells were unable to produce H2 in the absence of both conditions. In the absence of glucose supplementation and presence of the native electron sinks for assimilation pathways no H2 production was observed. Overall, these results demonstrate the possibility of expressing heterologous O2-tolerant hydrogenases in cyanobacteria, and how the native cyanobacterial maturation machinery can activate heterologous [NiFe] hydrogenases. Nonetheless, it is also clear that further engineering of the electron channeling towards the hydrogenase is required for the development of an efficient H2 production system.
A complementary strategy is focused on enhancing the electron supply to the hydrogenase. PSI-hydrogenase fusion constructs have been considered an appealing solution. Critically this design could allow to intercept electrons from the PETC before they reach the Fd pool, thus avoiding the need for extensive engineering of hydrogenase coupling with the redox metabolism and competition with other native electron sinks. Following this principle, light-driven H2 production from the Desulfovibrio vulgaris [NiFe] hydrogenases fused to cyt-c3 and PsaE subunit of PSI was reported in vitro (Ihara et al., 2006). This study highlighted the suitability of stromal PSI subunits as targets for PSI-Hydrogenase fusion proteins, as well as the importance of considering electron transport pathways from PSI to the fused hydrogenase moiety. More interestingly, the native [NiFe] hydrogenase of Synechocystis PCC 6803 (SynH) was fused to PSI PsaD subunit and overexpressed in a Synechocystis PCC 6803 mutant lacking the native hydrogenase (Appel et al., 2020). To date, this study reported the highest headspace H2 concentration achieved using Synechocystis PCC 6803 (> 500 μM) and the only report of H2 production in vivo using a PSI-hydrogenase fusion construct. The study shows how in spite of the reversible nature of SynH, H2 uptake is virtually avoided in the engineered PSI-SynH system. This indicate that on the contrary to previous hydrogenase expression strategies, PSI-Hydrogenase fusion proteins can alter the enzyme’s native bias by impeding reversed electron flow from the hydrogenase moiety back to PSI. Additionally, the study addressed the relevance of the PsaD-SynH fusion protein design, aiming to place the exposed external [4Fe4S] cluster of Synechocystis PCC 6803 native hydrogenase closer to the FB and FA [4Fe4S] clusters of the PSI PsaC subunit. The expressed SynYH hydrogenase corresponds only to the catalytic subunits of the native [NiFe] hydrogenase, reducing its ability to interact with native electron donors such as ferredoxin, thus reducing the possibility of hydrogen oxidation. The observed results indicate that a direct electron transfer from the [4Fe4S] clusters of the PSI PsaC subunit towards the [4Fe4S] cluster of the hydrogenase is the main, if not the only, electron source for the catalytic activity. These findings open the possibility of further engineering PSI-Hydrogenase systems where hydrogenases with desirable features are fused to PSI, as well as developing new fusion constructs with improved electron transfer properties. The latter could be achieved by reducing the distance between the PsaC [4Fe4S] donor clusters and the acceptor [4Fe4S] cluster of the hydrogenase (Appel et al., 2020) as well as via the introduction of electron carriers within the fusion construct’s structure (Walters and Golbeck, 2020; Günzel et al., 2022). Similarly, coupling of the PSI PsaC subunit’s FB [4Fe4S] cluster to a Clostridium acetobutylicum HydA using a dithiol-based molecular wire has been reported (Lubner et al., 2011). The resulting biological/organic nanoconstruct exhibited a 2-fold increase in overall photosynthetic electron turnover in PSI and high light-driven H2 production using an in vitro assay. These results demonstrate the relevance of PSI-Hydrogenase fusions as a promising way to overcome diffusional limitations of soluble redox carriers as well as limiting competition with other competing pathways. Undoubtedly, recent discoveries about hydrogenases’ interactions with native electron carriers will help guiding more rational engineering designs (Rumpel et al., 2015).
It worth noting that these studies highlighted that their reported photobiological H2 production titers where achieved in controlled experimental conditions avoiding O2 inactivation. In this context, electrons are normally obtained from other sources of reducing equivalents (mainly glucose) and transferred to the PETC via fermentative pathways (Appel et al., 2020; Lupacchini et al., 2021). These findings accentuate the relevance of combining the described engineering strategies with further engineering in order to remove the limitations imposed by O2 production in PSII.
Green algae harbor natively highly active [FeFe] hydrogenases and the associated HydEFG maturation machinery. Early studies of hydrogenase-driven H2 production in green algae traditionally focused on developing cultivation protocols that shifts cell metabolism towards anoxygenic conditions thereby inducing H2 production using nutrient-depleted media, as well as controlled pulse-illumination procedures (Melis et al., 2000; Kosourov et al., 2018). However, the growing availability of engineering tools developed during the last decade has led to several non-native hydrogenase expressions and other hydrogenase engineering strategies reported in green algae with promising results. Table 1 provides a summary of achievements on H2 production when expressing non-native hydrogenases in oxygenic photosynthetic microorganisms.
Chien et al. reported an up-to 10-fold increase in H2 production in engineered Chlorella DT strains. This was achieved by cloning and overexpression its principal native [FeFe] hydrogenase (Chien et al., 2012). Other engineering efforts focused on enhancing the redox coupling of the hydrogenase to PSI, aiming to reduce competition for PSI with Ferredoxin-NADP+ reductase (FNR). A synthetic ferredoxin (Fd) - [FeFe] hydrogenase protein was designed by fusing native Fdx1 and HydA from C. reinhardtii. Strains expressing the fusion constructs showed 4.5-fold increase in specific H2 production and surprisingly an enhanced O2 tolerance (Eilenberg et al., 2016). A later report studied in deeper detail the in vivo performance of the Fdx1-HydA1 synthetic protein and aimed to enhance H2 production by screening mutants overexpressing the fusion construct. Developed strains exhibited higher protein expression levels and longer-lasting sustained H2 production. However, this study also reported that up to 85% of the synthetic Fdx1-HydA1 fusion remained in the inactive apo-protein form, indicating inefficient in vivo hydrogenase maturation (Weiner et al., 2018). These findings highlight the importance of further synthetic protein engineering to optimize not only protein expression but also allow efficient hydrogenase maturation. More recently, a mutant ferredoxin with reduced electron transfer to FNR (Fdx1 D19A D58A) was fused to HydA1 using two different, 15 or 25 amino acids (aa), linkers. The obtained strains overexpressing the fusion constructs showed up to a 4.6-fold increase in H2 production when compared with a strain expressing only HydA1 at comparable levels (Rumpel et al., 2014).
Similarly, to studies in cyanobacteria, PSI-hydrogenase fusion constructs have been expressed in green algae, leading to enhanced in vivo H2 evolution. These efforts have primarily focused on C. reinhardtii which contains two native [FeFe] hydrogenases commonly denoted as HydA1 and HydA2. In vitro studies have suggested that HydA1 is primarily involved in H2 production, while HydA2 appears more biased towards H2 oxidation (Hambourger et al., 2008). In these studies, the stromal PsaC subunit of PSI was fused to C. reinhardtii HydA2. The engineered strain showed fermentative H2 production, reaching approximately 60% of WT activity in anoxic dark conditions, indicating that the HydA2 moiety was still able to accept electrons from Fd. Furthermore, the strain showed a 7-fold lower abundance of the PSI-HydA2 construct compared to PSI in wild-type cells. The reduced PSI pool lead to a downregulation of native PSII activity which alters PETC regulation, mainly constraining the LET and consequently reducing the O2 evolution. This lowered O2 production may reduce the hydrogenase inactivation thereby increasing observed H2 production. Overall, the expression of PSI-HydA2 fusion allowed a sustained H2 production upon illumination at rates comparable to the transient H2 production of the WT, arguably mainly due to the changes in PETC regulation. More recently, the potentially more efficient HydA1 was selected and a PSI-HydA1 fusion protein was designed and expressed in C. reinhardtii with the aim to increase H2 production (Engelbrecht et al., 2021). The obtained strain accumulated 5-times more PSI-HydA1 protein than the previous PSI-HydA2 strain and exhibited a comparable interaction with native Fd in dark anoxic conditions. However, the turnover frequency of the PSI-HydA1 chimera turned out to be 50% of the previous PSI-HydA2 construct, and overall H2 production by the fusion protein was 10-times lower than the previous PSI-HydA2 strain. These activities appear to conflict with the reported intrinsic biases of HydA1 of HydA2 (Engelbrecht et al., 2021). However, the results can largely be rationalized to higher abundance of PSI in the PSI-HydA1 fusion protein strains, releasing the constrains on PSII-driven O2 evolution. Furthermore, the report shows how the native HydEFG machinery was able to activate the apo-PsaC-HydA1 chimera expressed before the induction of anoxic conditions, reaching turnover rates above 90% of the fully active form. Additionally, it was also shown that upon O2 inactivation PSI-HydA1, the active site can be reinserted into the same PSI-HydA1 chimera by the maturases, inducing up to 60% of original hydrogenase activity recovery after 4 h of anaerobic adaptation (Kanygin et al., 2020, 2022).
It should be noted that in contrast to the [NiFe] hydrogenase-PSI fusion expressed in cyanobacteria (Appel et al., 2020), which was unable to produce H2 under dark fermentative conditions, the fusion constructs expressed in green algae (Kanygin et al., 2020, 2022) exhibited H2 production in darkness. Thus, it is unclear which is the main electron source of the hydrogenase. The reported dark fermentative H2 production of the PSI-HydA1/2 engineered strains indicate that the ferredoxin docking site is probably still free in the fusion protein, allowing the hydrogenase to accept electrons from ferredoxin. Consequently, the enhanced H2 production could be caused from the localization of the hydrogenase moiety in the proximal environment of PSI, thereby improving its access to reduced ferredoxin. Alternatively, it is also possible that the hydrogenase accepts electrons directly from the FeS cluster FB of the proximal PsaC subunit, or even a combination of both formerly described mechanisms. Overall, in the case of PSI-hydrogenase fusions, proximity to the FB [4Fe4S] cluster of the PSI PsaC subunit has been shown to be a critical requirement. The closeness to PSI ensures higher interaction with the reduced FeS clusters of the PsaC subunit or Fd and limits FNR coupling to PSI, broadening electron supply from Fd towards the hydrogenase (Moal and Lagoutte, 2012; Xiong et al., 2021).
Herein we describe the most relevant strategies and breakthroughs that has been reported to increase photobiological H2 production beyond the expression of hydrogenases. In spite of the diversity of the different strategies, they can be grouped into three main focus areas: Light harvesting engineering, electron transport rewiring, and intracellular O2 concentration reduction. Figure 1 provides a summary of discussed strategies and breakthroughs to increase photobiological production beyond the expression of hydrogenases.
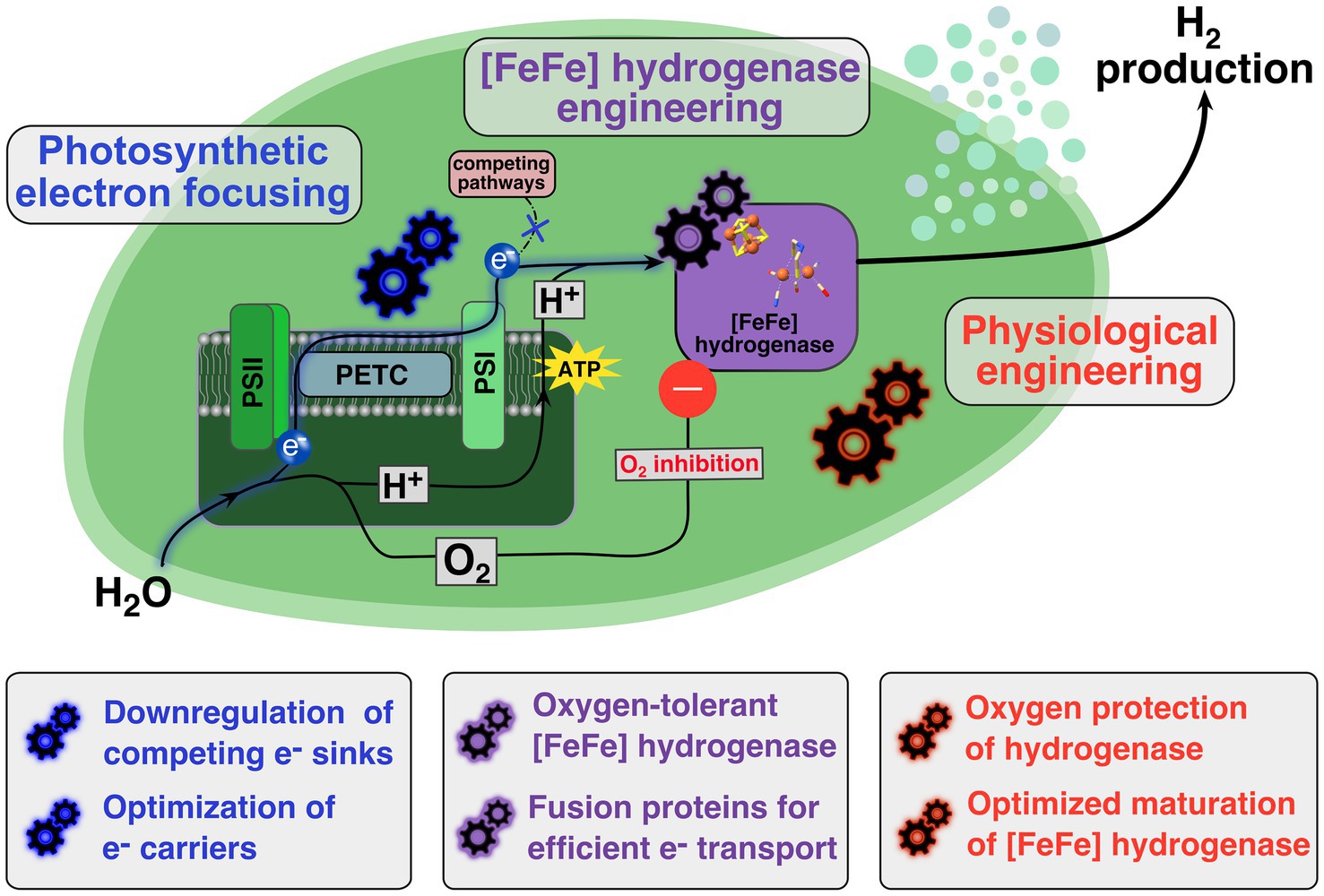
Figure 1. Schematic overview of engineering targets in photoautotrophic prokaryote for establishing efficient in vivo hydrogen production. Upon light-induced water splitting at photosystem II (PSII) oxygen is released and electrons are transferred via the photosynthetic electron transport chain (PETC) to photosystem I (PSI) where a cytoplasmatic electron carrier accepts these electrons. The photosynthetically-driven proton buildup in the thylakoids is enabling the ATP synthesis via ATP synthases at the thylakoid membrane. A heterologously expressed [FeFe] hydrogenase (purple box) uses the photosynthetically generated electrons and protons as substrates for a photobiological hydrogen production. To establish an efficient production system several features need to be targeted or introduced. Firstly, through photosynthetic electron focusing (blue gearwheels) the electrons transport from the photosynthetic machinery to the [FeFe] hydrogenase needs to be optimized. Secondly, the heterologously expressed [FeFe] hydrogenase has to be engineered (purple gearwheels) to work efficiently in the in vivo context. Especially, the oxygen-induced inhibition needs to be addressed by finding and introducing oxygen-insensitive or at least oxygen-sensitive tolerant enzyme variants into the in vivo system. Thirdly, physiological engineering efforts (red gearwheels) are necessary to assure that expressed [FeFe] hydrogenases can be activated and stay active for an extended period of time, as well as implementing further physiological modifications to protect the expressed hydrogenase from O2.
As the ultimate driver of photobiological H2 production is light energy, enhancing the cells ability to harvest and utilize light more efficiently could enhance the electron availability for hydrogen production. The main strategies have focused on three directions: Broadening the light spectra utilization, reducing the light harvesting antennas for efficient light distribution and stronger illumination tolerance as well as optimizing the electron transport chain and reducing photoprotective energy dissipation.
The far-red longest-wavelength absorbing chlorophyll known to date is chlorophyll F (Chlf). A Chlf synthase gene (chlF) of Fischerella thermalis PCC 7521 was heterologously expressed in Synechoccocus elongatus PCC 7002, leading to the incorporation of Chlf into the native PSI, functionally connected to the other pigments in the complex. Mutant strains lacking PSII and grown in far-red light achieved up to 50% of the Chlf content found in native far-red acclimated PSI. Although, the heterologous Chlf molecules were not as red-shifted as in their native environment, the assembly of a reduced amount of Chlf allowed to extend the photosynthetically active radiation up to 750 nm (Tros et al., 2020).
Light-harvesting complexes have evolved to maximize solar energy capture. However, under strong illumination, the light-harvesting machinery absorb photons beyond the limits of photosystems capacity. Furthermore, large light-harvesting complexes also increase the shading effects in photobioreactors, causing to uneven light transfer.
It has been reported that truncation of light harvesting complexes in Synechocystis PCC 6803 leads to higher biomass accumulation and glycogen production while maintaining growth rates comparable to the WT. The effects were similar for deletion of phycocyanin (Δcpc) (Lea-Smith et al., 2014) or a phycobilisomes anchor protein (ΔapcE) (Joseph et al., 2014). Similar truncation strategies has also been reported to increase photosynthesis efficiency and biomass accumulation in green algae, extensively reviewed by Kumar et al. (2021). The potential influence of mutant cyanobacterial strains with unanchored phycobilisomes (ΔapcE), phycocyanin knockout (Δcpc), or whole phycobilisomes deletion (PAL mutant) have been studied. Overall antennae truncation lead to more resistant strains against light stress as well as reduced energy requirements for antenna synthesis and PSII repair. Additionally, mutants exhibited physiological adaptations upregulating linear electron transport (LET) and enhanced PSII expression and enhanced PSI turnover (Bernát et al., 2009). Nonetheless a higher number of PSII centers also leads to increased O2 evolution, an effect that should be considered when coupling LET to O2-sensitive hydrogenases.
A potential strategy towards enhancing light energy utilization is the expression of proteorhodopsin - retinal complexes (PRrC). PRrC acts as light-driven proton pumps with maximum absorption peaks between 490 and 520 nm, matching with the fraction of light normally reflected by photosynthetic chlorophylls (450–550 nm). To date, PRrC systems have been successfully expressed in SynechocystisPCC 6803 and obtained strains were able to stimulate photoheterotrophic growth in a PSI deficient strain. Nonetheless further growth improvements were halted by the kinetic limitations of the native NDH-1 complex for reversed NADP+ reduction. Surprisingly, although the provided additional PMF can increase the synthesis of ATP, thus enhancing LET via NADPH:ATP ratio balancing, during illumination with green light LET was reduced, mainly due to increased proton motive force (PMF) giving rise to cytochrome b6/f complex inhibition (Chen et al., 2017, 2019). This finding indicate that the integration of this systems should aim for a holistic comprehension of the overall bioenergetic context of the cells. In light of these findings, PRrC expression may expand the overall harvested light energy by filling the photosynthetic green gap with alternative light-driven proteins.
Solar energy is harvested by the photosynthetic apparatus and converted into reducing power. However, in contrast to the fixed carbon which will be converted into different metabolites and can be stored, reducing power in the form of electrons must be used upon generation. In order to keep generation and demand(s) balanced they are channeled to available electron sinks. In photosynthetic microorganisms the four main electron sinks are: Carbon fixation (the Calvin–Benson–Bassham cycle, CBB), nitrate reduction pathways, oxygen reduction pathways (respiration or photoprotective mechanisms) and hydrogen production (Kosourov et al., 2021). Furthermore, it has been demonstrated that preferential channeling of photosynthetic electrons towards native growth supporting pathways (mainly carbon fixation) hinders hydrogen production and promote hydrogen uptake even before the O2-driven hydrogenase deactivation takes place (Milrad et al., 2018). Therefore, rewiring the redox metabolism for a broader electron distribution towards hydrogen production is a crucial need for efficient photobiological H2 production. Multiple strategies exist to address these requirements.
Nitrogen, specifically nitrate assimilation reactions account 16–20% of the photosynthetic electrons (Grund et al., 2019). Thus, inhibition of nitrate and nitrite reduction reactions has shown to translate into increased H2 production (Gutthann et al., 2007). A Synechocystis PCC 6803 mutant with the nitrate and nitrite reductase genes knocked-out showed an up to 140-fold increase in H2 production from the native bidirectional [NiFe] hydrogenase when compared to the WT cells. Interestingly, when cells where grown in media lacking nitrate, the engineered cells showed a 10-fold increase in H2 production compared to WT cells. Since the transcript levels for the native hydrogenase where comparable between WT and the mutant strain, the increased H2 production can be attributed to an increased availability of electrons (Baebprasert et al., 2011).
Photoprotective mechanisms allows photosynthetic cells to cope with sudden changes in light irradiation and subsequent variations in reducing power generation. However, they are also responsible for significant energy dissipation. In fact, most of these enzymes catalyze O2 reduction at the expense of H+ and electron consumption which could be otherwise redirected to H2 production. Several studies have reported the relevance of flavodiiron proteins (FDPs) (Santana-Sanchez et al., 2019) and terminal oxidases (Lea-Smith et al., 2013) as crucial mechanisms enabling cell survival in natural environments. Mutant strains lacking FDPs or terminal oxidases did not show significant growth defects when grown in controlled illumination conditions, highlighting their main role as electron-release valves under periods of reducing power overproduction.
Early studies in cyanobacteria showed improvements in H2 production in a Synechocystis PCC 6803 mutant lacking a functional NAD(P)H dehydrogenase complex NDH-1. Mutant strains sustained significant H2 production in the light. Since NDH-1 has been shown to be one of the major contributors to PMF formation by cyclic electron transport (CET) in Synechocystis PCC 6803 (Miller et al., 2021), the mutants are unable to pump protons, causing to severe alteration of PETC regulation and eventually increasing H2 production due to the long-term accumulation a highly reduced NADPH pool during illumination, being sufficient to activate NADPH-dependent H2 production by the native reversible hydrogenases (Cournac et al., 2004). Similarly, H2 production in Synechocystis PCC 6803 knock-out strains for terminal cytochrome c oxidase (Δcox), terminal quinol oxidase (Δcyd), and alternative respiratory terminal oxidase (ΔARTO) all exhibited increased H2 production reaching up to 12-fold from WT levels for the triple mutant. Comparatively, the NDH-1 complex deficient mutant showed a 36-fold increase in total H2 evolution but the production rate was lower when compared to previous mutant strains (Gutthann et al., 2007). While the mechanism behind the enhanced H2 production in the terminal oxidase mutants seems to be mainly derived from enhanced electron supply to the hydrogenase, NDH-1 mutants may have other mechanisms. In the NDH1-mutants a lowered consumption of NADPH and reduced PMF derived from CET leads to an accumulation of NADPH which may be the main cause for enhanced production. Thus, it should be considered that altering the functioning of these respiratory photoprotective systems may not only increase H2 production, but also have a profound impact in overall redox balance and PETC regulation specially in more complex hosts as green algae (Elman et al., 2022). Other studies showed how deletion of FDPs in green algae lead to up more than 2-fold increase in H2 production under controlled pulse-illumination protocols. In addition, the lack of FDPs lead to a delayed activation of carbon fixation pathways (Jokel et al., 2019).
Considering these findings, deletion of respiratory and/or photoprotective electron sinks are powerful strategies to enhance H2 production by the increasing electron availability. Nonetheless, the impact of these deletions on cell fitness and robustness should being considered. We note that potential fitness-rescue effects may be observed when deletions are combined with introduced engineered electron sink such as a hydrogenase.
Carbon fixation is essential for cell growth, where key reactions in the CBB cycle are energetically expensive consuming under ideal conditions 3 mol of ATP and 2 mol of NADPH per every mol of CO2 fixed. Consequently, it is not surprising that early studies on hydrogenase-catalyzed H2 production mainly focused on avoiding CBB activation via different physiological regulation mechanisms (Melis et al., 2000; Cournac et al., 2004). More recently, an RNA interference was used to downregulate expression of FNR in Chlamydomonas reinhardtii. Despite the small differences (12–18%) in autotrophic growth rates between WT and downregulated FNR mutants, a 60% reduction in Rubisco levels, 40% lower O2 evolution at PSII, and up to 140% higher starch degradation rates were observed which lead to an earlier onset of anaerobiosis and increased electron supply to the hydrogenases, translating into a 2.5-fold increase in overall H2 production (Sun et al., 2013). To our knowledge this is the only report of successful FNR regulation control in green algae via genetic engineering, leading to higher H2 production. In a similar fashion, a petH gene merodiploid Synechocystis PCC 6803 mutant, also exhibited up to 4-fold higher photobiological H2 production than the wild-type. In this study, the partial knock-out of the native FNR not only enhanced the H2 production but also lead to a longer duration of the transitory hydrogen production (Gutekunst et al., 2014). In this light, FNR downregulation is another promising strategy to increase photobiological hydrogen production.
As the ultimate aim of STH is using sunlight to source H2 from water, the process should mostly rely on LET where photosynthetic electrons are mostly sourced from water oxidation at PSII. However, increased PSII activity translates in higher O2 production, impairing H2 production due to the O2-sensitive nature of most hydrogenases (Vuorijoki et al., 2017; Kanygin et al., 2020, 2022). Albeit the relevance of dealing with O2 inactivation has been widely remarked, there are only a few studies reporting strategies tackling the O2-sensitivity issue by other means than expressing more O2-tolerant hydrogenases or anoxic cultivation conditions.
One possibility consists in expressing hydrogenases within an anoxic environment. In this regard, the heterocysts of filamentous nitrogen-fixing cyanobacteria could be used (Gärtner et al., 2012; Avilan et al., 2018). Although most described strategies showed limited H2 production, indicating that further engineering of the described systems is required. Similarly, expression of hydrogenases within anoxic bacterial microcompartments have already shown promising results in non-photosynthetic organisms. Particularly, carboxysomes are a very interesting scaffold for further engineering. Carboxysomes, present in all cyanobacteria, are polyhedral protein microcompartments that are selectively permeable to HCO3− and H+ but not to O2 or CO2. Natively carboxysomes are formed by the self-assembly of different protein subunits, incorporating bicarbonate transporters, carbonic anhydrase and Rubisco (ribulose-1,5-bisphosphate carboxylase/oxygenase) within the compartment as a way to provide an anoxic environment and enhance carbon fixation. Interestingly, carboxysomes can be engineered to also incorporate heterologous enzymes within them. Due to the additional protection against O2 they are emerging as promising scaffolds for improving hydrogenase activity. α-carboxysome shells have been engineered to co-encapsulate a protein fusion composed of the [FeFe] hydrogenase HydA1 from Chlamydomonas reinhardtii and its native Fd together with FNR from E. coli. In this system, NADPH is used as soluble electron donor and FNR enables NADPH dependent electron transfer to the hydrogenase. Interestingly, this system exhibited increased H2 production under aerobic conditions both in vivo and in vitro (Li et al., 2020). Similarly, the catalytic subunits of the NADPH-dependent [NiFe] hydrogenase HyaAB from E. coli where expressed inside α-carboxysome shells. The assembled systems showed increased thermal stability and O2 tolerance both in vivo and in vitro when compared with the free enzyme causing a significant increase in the measured H2 production (Jiang et al., 2023). Nonetheless, this improvements in O2 tolerance were more significant for the [NiFe] hydrogenase HyaAB than the [FeFe] hydrogenase HydA1. These results indicate that even when protected in a carboxysome shell, the hydrogenase intrinsic O2 sensitivity still a critical factor in the catalytic system performance. Additionally, it should be noted that the dissolved O2 levels in the aerobic cultures were significantly lower than atmospheric, due to the respiratory metabolism of E. coli. Because of this, although highly promising, whether these strategies can be transferred to photosynthetic microorganisms and efficiently coupled to native redox metabolism remains to be verified.
An artificial O2 consumption pathway was integrated in Chlamydomonas chloroplasts, expressing E. coli pyruvate oxidase and Synechococcus elongatus PCC 7942 catalase under the control of a strong heat shock inducing promoter. The mutant strain exhibited higher oxygen consumption rate without significant growth defects or disruptions in the overall photosynthetic rate. The reduced intracellular O2 concentration increased hydrogen production at moderate light intensities (30 μE) by 2-fold factor compared to WT. However, at higher light intensities (100 μE) O2 production at PSII overcame the effects of the implemented O2 consumption pathway, indicating the need of further refinement of the system (Xu et al., 2011). Recently, efficient O2 consuming devices have been designed and characterized in E. coli (Pacheco et al., 2018). However, to our knowledge there are no reports about their implementation in any photosynthetic microorganism. In addition, respiration in photosynthetic organisms plays a key role in regulating intracellular O2 (Shimakawa et al., 2020) and could potentially also be considered a potential engineering target. Eventually, the expression of oxygen-binding proteins has also been proven to reduce hydrogenase inhibition. The heterologous expression and assembly of the oxygen binding protein leghemoglobin from Glycine max, in chloroplasts of C. reinhardtii decreased the intracellular O2 content and increased H2 production rate up to 1.5-fold WT levels (Wu et al., 2010). Similar effects were observed for the expression of cyanoglobin in heterocysts of Nostoc PCC 7120 (Avilan et al., 2018).
Hydrogenases, their maturation machinery and electron carrier proteins all contain FeS clusters. Which is why it is expected that engineered strains overexpressing hydrogenases will feature a higher demand of FeS clusters. Therefore increasing native FeS cluster biosynthesis could potentially improve the system performance, as it has been already shown for nitrogenase enzymes (Li et al., 2016). In addition, as demonstrated by Akhtar and Jones (2008), deletion of iscR encoding the main FeS cluster biosynthesis pathway (ISC) repressor in E. coli increased hydrogenase activity and H2 accumulation. Besides the common heterotrophic pathways, photosynthetic organisms use the sulfur utilization factor (SUF) pathway as the main FeS cluster biosynthesis route and exhibit up to a 10-fold higher FeS cluster biosynthesis than E. coli (Gao, 2020). Similar studies in Synechocystis PCC 6803, showed that deletion of the SUF pathway repressor under sufficient iron conditions (SufR) lead to an increased FeS cluster assembly while maintaining overall cell fitness (Vuorijoki et al., 2017). In the light of these findings, deregulation of FeS cluster biosynthesis may be used as a promising strategy alleviate limitations in FeS cluster availability for expressed heterologous hydrogenases and the accessory proteins required for its functioning.
Future extensively engineered strains may exhibit limited growth, hindering the development of robust bioprocess design. This issue can be alleviated by implementing control systems for the expression of the introduced genetic elements. In this regard, the utilization of recently stationary phase promoters (Madsen et al., 2021) can drive the development of 2-stage cultivation strategies where initial growth is decoupled from further H2 production (Burg et al., 2016; Monshupanee et al., 2016). Additionally utilization of an O2-inducible promoter (Immethun et al., 2016) could be used for fine-control of O2 protective engineered systems.
Future endeavors towards cost-effective photobiological H2 production must rely on a holistic approach combining the strategies highlighted in this review. Three main engineering directions have been identified. Namely, the expression of non-native hydrogenase with improved features, extensive modifications of PETC for focusing electrons towards the heterologous hydrogenase, and further physiological engineering strategies to improve the overall performance of the producing strains and the expressed hydrogenase.
Firstly, H2 productivity is highly dependent on the hydrogenase activity, requiring enzymes with displaced catalytic bias towards H2 production combined with O2 insensitivity. In this light [FeFe] hydrogenases are the most promising biocatalysts, for which recent research reported novel heterologous hydrogenases with intrinsic O2 tolerance as well as engineered fusion hydrogenase proteins leading to improved H2 production. Regarding fusion hydrogenases, it has been shown that protein design is critical. Any hydrogenase fusion protein must ensure efficient maturation of the hydrogenase H-domain and integration within the host bioenergetic machinery, ideally establishing unidirectional electron transport towards H2 production.
Secondly, efficient focusing of photosynthetic electrons towards the hydrogenase can be achieved via the implementation of the formerly described strategies. Nonetheless, implementing multiple engineering strategies could significantly impact the overall photosynthesis physiology, leading to, e.g., alterations in PSI to PSII ratios and consequently the balance between PMF, LET and CET. These effects are especially relevant for PSI-hydrogenase fusion proteins and knock-out strains of respiratory terminal oxidases and FDPs. Due to potential synergistic effects this should be considered.
Likewise, it must be considered the physiological context of the producing strain. In this regard further host engineering should be implemented to ensure maximum hydrogenase activity. Therefore, establishing an efficient and orthogonal maturation systems is evidently required, especially in the case of [FeFe] hydrogenases expression in cyanobacteria. Moreover, O2 protection strategies should be implemented to avoid irreversible hydrogenase inactivation, as this is likely to remain an issue even in designed enzymes.
Eventually, it is crucial to distinguish between H2 production derived from the LET and other sources. Albeit many studies reported that reducing PSII activity correlated with higher H2 production due to reduced intracellular O2 concentrations this derives from overall lower water splitting at PSII and thus reduced LET, which ultimately limits the maximum theoretical STH H2 production. This long-sustained paradox between LET activity and H2 production may be overcome by the implementation of O2 consuming device, pathway or the assembly of an O2 protection mechanism in vivo thereby avoiding hydrogenase inactivation and efficiently coupling water splitting with H2 production.
Finally, future efforts to improve photobiological H2 production should use a combination of strategies highlighted in the study. These strategies include expressing and efficiently activating heterologous hydrogenases, avoiding its O2-inactivation and levering the limitations of PETC towards increasing electron channeling to the hydrogenase. The described strategies can significantly impact overall photosynthesis physiology. Namely alterations in PSI to PSII ratios and balance between PMF, LET and CET. Thus, the interaction between the different engineering strategies must consider its synergistic effects towards the development of cost-effective photobiological H2 production processes.
All authors contributed to conception and design of the review. CS and JF wrote the first draft of the manuscript. GB and PL contributed to read, revision and proof-reading of the manuscript drafts. CS contributed to the literature sources collection regarding hydrogenases. JF contributed to the literature sources collection of photosynthesis re-engineering and heterologous expression of hydrogenases. All authors contributed to the article and approved the submitted version.
This work was supported by the European Union Horizon Europe - the Framework Programme for Research and Innovation (2021–2027) under the grant agreement number 101070948 (project PhotoSynH2 to GB and PL) and The Swedish Energy Agency (project number 48574-1 to GB).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Akhtar, M. K., and Jones, P. R. (2008). Deletion of iscr stimulates recombinant clostridial Fe-Fe hydrogenase activity and H2-accumulation in Escherichia Coli Bl21(De3). Appl. Microbiol. Biotechnol. 78, 853–862. doi: 10.1007/s00253-008-1377-6
Appel, J., Hueren, V., Boehm, M., and Gutekunst, K. (2020). Cyanobacterial in vivo solar hydrogen production using a photosystem i–hydrogenase (Psad-Hoxyh) fusion complex. Nat. Energy 5, 458–467. doi: 10.1038/s41560-020-0609-6
Asada, Y., Koike, Y., Schnackenberg, J., Miyake, M., Uemura, I., and Miyake, J. (2000). Heterologous expression of clostridial hydrogenase in the cyanobacterium Synechococcus PCC7942. Biochim. Biophys. Acta - Gene Struct. Expression 1490, 269–278. doi: 10.1016/S0167-4781(00)00010-5
Avilan, L., Roumezi, B., Risoul, V., Bernard, C. S., Kpebe, A., Belhadjhassine, M., et al. (2018). phototrophic hydrogen production from a clostridial [FeFe] hydrogenase expressed in the heterocysts of the cyanobacterium Nostoc PCC 7120. Appl. Microbiol. Biotechnol. 102, 5775–5783. doi: 10.1007/s00253-018-8989-2
Baebprasert, W., Jantaro, S., Khetkorn, W., Lindblad, P., and Incharoensakdi, A. (2011). Increased H2 production in the cyanobacterium Synechocystis sp. strain PCC 6803 by redirecting the electron supply via genetic engineering of the nitrate assimilation pathway. Metab. Eng. 13, 610–616. doi: 10.1016/j.ymben.2011.07.004
Berggren, G., Glover, S. D., and Cheah, M. H. (2022). “15.02 - hydrogenases and model complexes in bioorganometallic chemistry” in Comprehensive Organometallic Chemistry Iv. eds. G. Parkin, K. Meyer, and D. O’hare (Oxford: Elsevier)
Bernát, G., Waschewski, N., and Rögner, M. (2009). Towards efficient hydrogen production: the impact of antenna size and external factors on electron transport dynamics in Synechocystis PCC 6803. Photosynth. Res. 99, 205–216. doi: 10.1007/s11120-008-9398-7
Berto, P., D’adamo, S., Bergantino, E., Vallese, F., Giacometti, G. M., and Costantini, P. (2011). The cyanobacterium Synechocystis sp. PCC 6803 is able to express an active [FeFe]-hydrogenase without additional maturation proteins. Biochem. Biophys. Res. Commun. 405, 678–683. doi: 10.1016/j.bbrc.2011.01.095
Birrell, J. A., Rodríguez-Maciá, P., Reijerse, E. J., Martini, M. A., and Lubitz, W. (2021). The catalytic cycle of [FeFe] hydrogenase: a tale of two sites. Coord. Chem. Rev. 449:214191. doi: 10.1016/j.ccr.2021.214191
Böck, A., King, P. W., Blokesch, M., and Posewitz, M. C. (2006). Maturation of hydrogenases. Adv. Microb. Physiol. 51, 1–71. doi: 10.1016/S0065-2911(06)51001-X
Buhrke, T., Lenz, O., Krauss, N., and Friedrich, B. (2005). Oxygen tolerance of the H2-sensing [NiFe] hydrogenase from Ralstonia eutropha H16 is based on limited access of oxygen to the active site. J. Biol. Chem. 280, 23791–23796. doi: 10.1074/jbc.M503260200
Burg, J. M., Cooper, C. B., Ye, Z., Reed, B. R., Moreb, E. A., and Lynch, M. D. (2016). Large-scale bioprocess competitiveness: the potential of dynamic metabolic control in two-stage fermentations. Curr. Opin. Chem. Eng. 14, 121–136. doi: 10.1016/j.coche.2016.09.008
Bussiness-Research-Company (2023). Nuclear Electricity Global Market Report 2023 [Online]. Www.Thebusinessresearchcompany.Com: The Bussiness Research Company. Available at: https://www.Thebusinessresearchcompany.Com/Report/Nuclear-Electricity-Global-Market-Report [Accessed February 27, 2023].
Calusinska, M., Happe, T., Joris, B., and Wilmotte, A. (2010). The surprising diversity of clostridial hydrogenases: a comparative genomic perspective. Microbiology 156, 1575–1588. doi: 10.1099/mic.0.032771-0
Carrieri, D., Wawrousek, K., Eckert, C., Yu, J., and Maness, P.-C. (2011). The role of the bidirectional hydrogenase in cyanobacteria. Bioresour. Technol. 102, 8368–8377. doi: 10.1016/j.biortech.2011.03.103
Chen, Q., Arents, J., Ganapathy, S., de Grip, W. J., and Hellingwerf, K. J. (2017). Functional Expression of Gloeobacter Rhodopsin in Synechocystis sp. PCC6803. Photochem. Photobiol. 93, 772–781. doi: 10.1111/php.12745
Chen, Q., Arents, J., Schuurmans, J. M., Ganapathy, S., de Grip, W. J., Cheregi, O., et al. (2019). Combining retinal-based and chlorophyll-based (oxygenic) photosynthesis: Proteorhodopsin expression increases growth rate and fitness of a ∆PSI strain of Synechocystis sp. PCC6803. Metabolic Engg. 52, 68–76. doi: 10.1016/j.ymben.2018.11.002
Chien, L.-F., Kuo, T.-T., Liu, B.-H., Lin, H.-D., Feng, T.-Y., and Huang, C.-C. (2012). Solar-To-Bioh2 production enhanced by homologous overexpression of hydrogenase in green alga Chlorella sp. DT. Int. J. Hydrog. Energy 37, 17738–17748. doi: 10.1016/j.ijhydene.2012.09.068
Cohen, J., Kim, K., Posewitz, M., Ghirardi, M. L., Schulten, K., Seibert, M., et al. (2005). Molecular dynamics and experimental investigation of H2 and O2 diffusion in [Fe]-hydrogenase. Biochem. Soc. Trans. 33, 80–82. doi: 10.1042/BST0330080
Corrigan, P. S., Tirsch, J. L., and Silakov, A. (2020). Investigation of the unusual ability of the [FeFe] hydrogenase from Clostridium beijerinckii to access an O2-protected state. J. Am. Chem. Soc. 142, 12409–12419. doi: 10.1021/jacs.0c04964
Cournac, L., Guedeney, G., Peltier, G., and Vignais, P. M. (2004). Sustained photoevolution of molecular hydrogen in a mutant of Synechocystis sp. strain PCC 6803 deficient in the type i Nadph-dehydrogenase complex. J. Bacteriol. 186, 1737–1746. doi: 10.1128/JB.186.6.1737-1746.2003
Cracknell, J. A., Vincent, K. A., Ludwig, M., Lenz, O., Friedrich, B., and Armstrong, F. A. (2008). Enzymatic oxidation of H2 In atmospheric O2: the electrochemistry of energy generation from trace H2 by aerobic microorganisms. J. Am. Chem. Soc. 130, 424–425. doi: 10.1021/ja078299+
Ducat, D. C., Sachdeva, G., and Silver, P. A. (2011). Rewiring hydrogenase-dependent redox circuits in cyanobacteria. Proc. Natl. Acad. Sci. 108, 3941–3946. doi: 10.1073/pnas.1016026108
Eilenberg, H., Weiner, I., Ben-Zvi, O., Pundak, C., Marmari, A., Liran, O., et al. (2016). The dual effect of a ferredoxin-hydrogenase fusion protein in vivo: successful divergence of the photosynthetic electron flux towards hydrogen production and elevated oxygen tolerance. Biotechnol. Biofuels 9:182. doi: 10.1186/s13068-016-0601-3
Elman, T., Hoai Ho, T. T., Milrad, Y., Hippler, M., and Yacoby, I. (2022). Enhanced chloroplast-mitochondria crosstalk promotes ambient algal-H2 production. Cell Rep. Phys. Sci. 3:100828. doi: 10.1016/j.xcrp.2022.100828
Engelbrecht, V., Liedtke, K., Rutz, A., Yadav, S., Günzel, A., and Happe, T. (2021). One isoform for one task? The second hydrogenase Of Chlamydomonas reinhardtii prefers hydrogen uptake. Int. J. Hydrog. Energy 46, 7165–7175. doi: 10.1016/j.ijhydene.2020.11.231
Exxon-Mobil (2022). Energy Demand: Three Drivers [Online]. Exxon Mobil. Available at: https://Corporate.Exxonmobil.Com/Energy-And-Innovation/Outlook-For-Energy/Energy-Demand#Industrial [Accessed February 27, 2023].
Fourmond, V., Baffert, C., Sybirna, K., Dementin, S., Abou-Hamdan, A., Meynial-Salles, I., et al. (2013). The mechanism of inhibition by H2 of H2-evolution by hydrogenases. Chem. Commun. 49, 6840–6842. doi: 10.1039/c3cc43297a
Gao, F. (2020). Iron–sulfur cluster biogenesis and iron homeostasis in cyanobacteria. Front. Microbiol. 11:165. doi: 10.3389/fmicb.2020.00165
Gärtner, K., Lechno-Yossef, S., Cornish, A. J., Wolk, C. P., and Hegg, E. L. (2012). Expression of Shewanella oneidensis Mr-1 [FeFe]-hydrogenase genes in Anabaena sp. strain PCC 7120. Appl. Environ. Microbiol. 78, 8579–8586. doi: 10.1128/AEM.01959-12
Gauquelin, C., Baffert, C., Richaud, P., Kamionka, E., Etienne, E., Guieysse, D., et al. (2018). Roles of the f-domain in [FeFe] hydrogenase. Biochim. Biophys. Acta - Bioenerg. 1859, 69–77. doi: 10.1016/j.bbabio.2017.08.010
Goldet, G., Brandmayr, C., Stripp, S. T., Happe, T., Cavazza, C., Fontecilla-Camps, J. C., et al. (2009). Electrochemical kinetic investigations of the reactions of [FeFe]-hydrogenases with carbon monoxide and oxygen: comparing the importance of gas tunnels and active-site electronic/redox effects. J. Am. Chem. Soc. 131, 14979–14989. doi: 10.1021/ja905388j
Greening, C., Biswas, A., Carere, C. R., Jackson, C. J., Taylor, M. C., Stott, M. B., et al. (2016). Genomic And metagenomic surveys of hydrogenase distribution indicate H2 is a widely utilised energy source for microbial growth and survival. ISME J. 10, 761–777. doi: 10.1038/ismej.2015.153
Grimm, A., De Jong, W. A., and Kramer, G. J. (2020). Renewable hydrogen production: a techno-economic comparison of photoelectrochemical cells and photovoltaic-electrolysis. Int. J. Hydrog. Energy 45, 22545–22555. doi: 10.1016/j.ijhydene.2020.06.092
Grinter, R., Kropp, A., Venugopal, H., Senger, M., Badley, J., Cabotaje, P., et al. (2023). Energy extraction from air: structural basis of atmospheric hydrogen oxidation. Nature 615, 541–547. doi: 10.1038/s41586-023-05781-7
Grund, M., Jakob, T., Wilhelm, C., Bühler, B., and Schmid, A. (2019). Electron balancing under different sink conditions reveals positive effects on photon efficiency and metabolic activity of Synechocystis sp. PCC 6803. Biotechnol. Biofuels 12:43. doi: 10.1186/s13068-019-1378-y
Günzel, A., Engelbrecht, V., and Happe, T. (2022). Changing the tracks: screening for electron transfer proteins to support hydrogen production. J. Biological Inorganic Chem. 27, 631–640. doi: 10.1007/s00775-022-01956-1
Gutekunst, K., Chen, X., Schreiber, K., Kaspar, U., Makam, S., and Appel, J. (2014). The bidirectional nife-hydrogenase in Synechocystis sp. PCC 6803 is reduced by flavodoxin and ferredoxin and is essential under mixotrophic, nitrate-limiting conditions. J. Biol. Chem. 289, 1930–1937. doi: 10.1074/jbc.M113.526376
Gutthann, F., Egert, M., Marques, A., and Appel, J. (2007). Inhibition of respiration and nitrate assimilation enhances photohydrogen evolution under low oxygen concentrations in Synechocystis sp. PCC 6803. Biochim. Biophys. Acta - Bioenerg. 1767, 161–169. doi: 10.1016/j.bbabio.2006.12.003
Hallenbeck, P. C., and Benemann, J. R. (2002). Biological hydrogen production; fundamentals and limiting processes. Int. J. Hydrog. Energy 27, 1185–1193. doi: 10.1016/S0360-3199(02)00131-3
Hambourger, M., Gervaldo, M., Svedruzic, D., King, P. W., Gust, D., Ghirardi, M., et al. (2008). [FeFe]-Hydrogenase-catalyzed H2 production in a photoelectrochemical biofuel cell. J. Am. Chem. Soc. 130, 2015–2022. doi: 10.1021/ja077691k
IEA (2021). Global Electricity Demand Is Growing Faster Than Renewables, Driving Strong Increase In Generation From Fossil Fuels [Online]. https://www.Iea.Org/: International Energy Agency. Available at: https://www.Iea.Org/News/Global-Electricity-Demand-Is-Growing-Faster-Than-Renewables-Driving-Strong-Increase-In-Generation-From-Fossil-Fuels [Accessed February 27, 2023].
Ihara, M., Nishihara, H., Yoon, K.-S., Lenz, O., Friedrich, B., Nakamoto, H., et al. (2006). Light-driven hydrogen production by a hybrid complex of a [NiFe]-hydrogenase and the cyanobacterial photosystem I. Photochem. Photobiol. 82, 676–682. doi: 10.1562/2006-01-16-RA-778
Immethun, C. M., Ng, K. M., Delorenzo, D. M., Waldron-Feinstein, B., Lee, Y.-C., and Moon, T. S. (2016). Oxygen-responsive genetic circuits constructed in Synechocystis sp. PCC 6803. Biotechnol. Bioeng. 113, 433–442. doi: 10.1002/bit.25722
Jiang, Q., Li, T., Yang, J., Aitchison, C. M., Huang, J., Chen, Y., et al. (2023). Synthetic engineering of a new biocatalyst encapsulating [NiFe]-hydrogenases for enhanced hydrogen production. J. Materials Chem. B 11, 2684–2692. doi: 10.1039/D2TB02781J
Jokel, M., Nagy, V., Tóth, S. Z., Kosourov, S., and Allahverdiyeva, Y. (2019). Elimination of the flavodiiron electron sink facilitates long-term H2 photoproduction in green algae. Biotechnol. Biofuels 12:280. doi: 10.1186/s13068-019-1618-1
Joseph, A., Aikawa, S., Sasaki, K., Matsuda, F., Hasunuma, T., and Kondo, A. (2014). Increased biomass production and glycogen accumulation in apce gene deleted Synechocystis sp. PCC 6803. AMB Express 4:17. doi: 10.1186/s13568-014-0017-z
Kanygin, A., Milrad, Y., Thummala, C., Reifschneider, K., Baker, P., Marco, P., et al. (2020). Rewiring photosynthesis: a photosystem i-hydrogenase chimera that makes H2 in vivo. Energy Environ. Sci. 13, 2903–2914. doi: 10.1039/C9EE03859K
Kanygin, A., Smith, A., Nagy, V., Tóth, S. Z., and Redding, K. E. (2022). Interplay between hydrogen production and photosynthesis in a green alga expressing an active photosystem i-hydrogenase chimera. Int. J. Hydrog. Energy 47, 21969–21983. doi: 10.1016/j.ijhydene.2022.03.096
Khanna, N., Esmieu, C., Mészáros, L. S., Lindblad, P., and Berggren, G. (2017). In vivo activation of an [FeFe] hydrogenase using synthetic cofactors. Energy Environm. Sci. 10, 1563–1567. doi: 10.1039/C7EE00135E
Khanna, N., and Lindblad, P. (2015). Cyanobacterial hydrogenases and hydrogen metabolism revisited: recent progress and future prospects. Int. J. Mol. Sci. 16, 10537–10561. doi: 10.3390/ijms160510537
King, P. W., Posewitz, M. C., Ghirardi, M. L., and Seibert, M. (2006). Functional studies of [FeFe] hydrogenase maturation in an Escherichia coli biosynthetic system. J. Bacteriol. 188, 2163–2172. doi: 10.1128/JB.188.6.2163-2172.2006
Koo, J., and Swartz, J. R. (2018). System analysis and improved [FeFe] hydrogenase O2 tolerance suggest feasibility for photosynthetic H2 production. Metab. Eng. 49, 21–27. doi: 10.1016/j.ymben.2018.04.024
Kosourov, S., Böhm, M., Senger, M., Berggren, G., Stensjö, K., Mamedov, F., et al. (2021). Photosynthetic hydrogen production: novel protocols, promising engineering approaches and application of semi-synthetic hydrogenases. Physiol. Plant. 173, 555–567. doi: 10.1111/ppl.13428
Kosourov, S., Jokel, M., Aro, E.-M., and Allahverdiyeva, Y. (2018). A new approach for sustained and efficient H2 photoproduction by Chlamydomonas reinhardtii. Energy Environ. Sci. 11, 1431–1436. doi: 10.1039/C8EE00054A
Kruse, O., Rupprecht, J., Mussgnug, J. H., Dismukes, G. C., and Hankamer, B. (2005). Photosynthesis: a blueprint for solar energy capture and biohydrogen production technologies. Photochem. Photobiol. Sci. 4, 957–970. doi: 10.1039/b506923h
Kubas, A., Orain, C., De Sancho, D., Saujet, L., Sensi, M., Gauquelin, C., et al. (2017). Mechanism of O2 diffusion and reduction in FeFe hydrogenases. Nat. Chem. 9, 88–95. doi: 10.1038/nchem.2592
Kumar, V., Sharma, N., Jaiswal, K. K., Vlaskin, M. S., Nanda, M., Tripathi, M. K., et al. (2021). Microalgae with a truncated light-harvesting antenna to maximize photosynthetic efficiency and biomass productivity: recent advances and current challenges. Process Biochem. 104, 83–91. doi: 10.1016/j.procbio.2021.03.006
Land, H., Senger, M., Berggren, G., and Stripp, S. T. (2020). Current state of [FeFe]-hydrogenase research: biodiversity and spectroscopic investigations. ACS Catal. 10, 7069–7086. doi: 10.1021/acscatal.0c01614
Lautier, T., Ezanno, P., Baffert, C., Fourmond, V., Cournac, L., Fontecilla-Camps, J. C., et al. (2011). The quest for a functional substrate access tunnel in Fefe hydrogenase. Faraday Discuss. 148, 385–407. doi: 10.1039/C004099C
Lea-Smith, D. J., Bombelli, P., Dennis, J. S., Scott, S. A., Smith, A. G., and Howe, C. J. (2014). Phycobilisome-deficient strains of Synechocystis sp. PCC 6803 have reduced size and require carbon-limiting conditions to exhibit enhanced productivity. Plant Physiol. 165, 705–714. doi: 10.1104/pp.114.237206
Lea-Smith, D. J., Ross, N., Zori, M., Bendall, D. S., Dennis, J. S., Scott, S. A., et al. (2013). Thylakoid terminal oxidases are essential for the cyanobacterium Synechocystis sp. PCC 6803 to survive rapidly changing light intensities. Plant Physiol. 162, 484–495. doi: 10.1104/pp.112.210260
Leroux, F., Dementin, S., Burlat, B., Cournac, L., Volbeda, A., Champ, S., et al. (2008). Experimental approaches to kinetics of gas diffusion in hydrogenase. Proc. Natl. Acad. Sci. 105, 11188–11193. doi: 10.1073/pnas.0803689105
Li, T., Jiang, Q., Huang, J., Aitchison, C. M., Huang, F., Yang, M., et al. (2020). Reprogramming bacterial protein organelles as a nanoreactor for hydrogen production. Nat. Commun. 11. doi: 10.1038/s41467-020-19280-0
Li, S., Li, F., Zhu, X., Liao, Q., Chang, J.-S., and Ho, S.-H. (2022). Biohydrogen production from microalgae for environmental sustainability. Chemosphere 291:132717. doi: 10.1016/j.chemosphere.2021.132717
Li, X.-X., Liu, Q., Liu, X.-M., Shi, H.-W., and Chen, S.-F. (2016). Using synthetic biology to increase nitrogenase activity. Microb. Cell Factories 15:43. doi: 10.1186/s12934-016-0442-6
Liebgott, P.-P., Leroux, F., Burlat, B., Dementin, S., Baffert, C., Lautier, T., et al. (2010). Relating diffusion along the substrate tunnel and oxygen sensitivity in hydrogenase. Nat. Chem. Biol. 6, 63–70. doi: 10.1038/nchembio.276
Lubitz, W., Ogata, H., Rüdiger, O., and Reijerse, E. (2014). Hydrogenases. Chem. Rev. 114, 4081–4148. doi: 10.1021/cr4005814
Lubner, C. E., Applegate, A. M., Knörzer, P., Ganago, A., Bryant, D. A., Happe, T., et al. (2011). Solar hydrogen-producing bionanodevice outperforms natural photosynthesis. Proc. Natl. Acad. Sci. 108, 20988–20991. doi: 10.1073/pnas.1114660108
Lui, J., Chen, W.-H., Tsang, D. C., and You, S. (2020). A critical review on the principles, applications, and challenges of waste-to-hydrogen technologies. Renew. Sust. Energ. Rev. 134:110365. doi: 10.1016/j.rser.2020.110365
Lukey, M. J., Roessler, M. M., Parkin, A., Evans, R. M., Davies, R. A., Lenz, O., et al. (2011). Oxygen-tolerant [NiFe]-hydrogenases: the individual and collective importance of supernumerary cysteines at the proximal Fe-S cluster. J. Am. Chem. Soc. 133, 16881–16892. doi: 10.1021/ja205393w
Lupacchini, S., Appel, J., Stauder, R., Bolay, P., Klähn, S., Lettau, E., et al. (2021). Rewiring cyanobacterial photosynthesis by the implementation of an oxygen-tolerant hydrogenase. Metab. Eng. 68, 199–209. doi: 10.1016/j.ymben.2021.10.006
Madsen, M. A., Hamilton, G., Herzyk, P., and Amtmann, A. (2021). Environmental regulation of Pndba600, an auto-inducible promoter for two-stage industrial biotechnology in cyanobacteria. Front. Bioeng. Biotechnol. 8:619055. doi: 10.3389/fbioe.2020.619055
Martini, M. A., Bikbaev, K., Pang, Y., Lorent, C., Wiemann, C., Breuer, N., et al. (2023). Binding of exogenous cyanide reveals new active-site states in [FeFe] hydrogenases. Chem. Sci. 14, 2826–2838. doi: 10.1039/D2SC06098A
Melis, A., Zhang, L., Forestier, M., Ghirardi, M. L., and Seibert, M. (2000). Sustained photobiological hydrogen gas production upon reversible inactivation of oxygen evolution in the green alga Chlamydomonas reinhardtii. Plant Physiol. 122, 127–136. doi: 10.1104/pp.122.1.127
Mészáros, L. S., Nemeth, B., Esmieu, C., Ceccaldi, P., and Berggren, G. (2018). In vivo Epr characterization of semi-synthetic [FeFe] hydrogenases. Angew. Chem. Int. Ed. 57, 2596–2599. doi: 10.1002/anie.201710740
Meyer, J. (2007). [FeFe] hydrogenases and their evolution: a genomic perspective. Cell. Molecul. Life Sci. 64, 1063–1084. doi: 10.1007/s00018-007-6477-4
Miller, N. T., Vaughn, M. D., and Burnap, R. L. (2021). Electron flow through Ndh-1 complexes is the major driver of cyclic electron flow-dependent proton pumping in cyanobacteria. Biochim. Biophys. Acta - Bioenerg. 1862:148354. doi: 10.1016/j.bbabio.2020.148354
Milrad, Y., Schweitzer, S., Feldman, Y., and Yacoby, I. (2018). Green algal hydrogenase activity is outcompeted by carbon fixation before inactivation by oxygen takes place. Plant Physiol. 177, 918–926. doi: 10.1104/pp.18.00229
Moal, G., and Lagoutte, B. (2012). Photo-induced electron transfer from photosystem i to NaDP+: characterization and tentative simulation of the in vivo environment. Biochim. Biophys. Acta - Bioenerg. 1817, 1635–1645. doi: 10.1016/j.bbabio.2012.05.015
Monshupanee, T., Nimdach, P., and Incharoensakdi, A. (2016). Two-stage (photoautotrophy and heterotrophy) cultivation enables efficient production of bioplastic poly-3-hydroxybutyrate in auto-sedimenting cyanobacterium. Sci. Rep. 6:37121. doi: 10.1038/srep37121
Morra, S. (2022). Fantastic [FeFe]-hydrogenases and where to find them. Front. Microbiol. 13:853626. doi: 10.3389/fmicb.2022.853626
Morra, S., Arizzi, M., Valetti, F., and Gilardi, G. (2016). Oxygen stability in the new [FeFe]-hydrogenase from Clostridium beijerinckii Sm10 (Cba5h). Biochemistry 55, 5897–5900. doi: 10.1021/acs.biochem.6b00780
Novus-Light-Technologies (2023). Renewable Energy Market To Grow 8.4% Cagr Through 2030 [Online]. Www.Novuslight.Com: Novus Light Technologies Today. Available at: https://www.Novuslight.Com/Renewable-Energy-Market-To-Grow-8-4-Cagr-Through-2030_N12240.Html [Accessed February 27, 2023].
Ogata, H., Lubitz, W., and Higuchi, Y. (2016). Structure and function of [NiFe] hydrogenases. J. Biochem. 160, 251–258. doi: 10.1093/jb/mvw048
Pacheco, C. C., Büttel, Z., Pinto, F., Rodrigo, G., Carrera, J., Jaramillo, A., et al. (2018). Modulation of Intracellular O2 Concentration in Escherichia coli Strains Using Oxygen Consuming Devices. ACS Synthetic Bio. 7, 1742–1752. doi: 10.1021/acssynbio.7b00428
Pagnier, A., Balci, B., Shepard, E. M., Yang, H., Warui, D. M., Impano, S., et al. (2022). [FeFe]-hydrogenase: defined lysate-free maturation reveals a key role for lipoyl-H-protein in dtma ligand biosynthesis. Angew. Chem. Int. Ed. 61:E202203413. doi: 10.1002/anie.202203413
Pandey, K., Islam, S. T. A., Happe, T., and Armstrong, F. A. (2017). Frequency and potential dependence of reversible electrocatalytic hydrogen interconversion by [FeFe]-hydrogenases. Proc. Natl. Acad. Sci. 114, 3843–3848. doi: 10.1073/pnas.1619961114
Redding, K. E., Appel, J., Boehm, M., Schuhmann, W., Nowaczyk, M. M., Yacoby, I., et al. (2022). Advances and challenges in photosynthetic hydrogen production. Trends Biotechnol. 40, 1313–1325. doi: 10.1016/j.tibtech.2022.04.007
Rodríguez-Maciá, P., Reijerse, E. J., Van Gastel, M., Debeer, S., Lubitz, W., Rüdiger, O., et al. (2018). Sulfide protects [FeFe] hydrogenases from O2. J. Am. Chem. Soc. 140, 9346–9350. doi: 10.1021/jacs.8b04339
Roseboom, W., De Lacey, A. L., Fernandez, V. M., Hatchikian, E. C., and Albracht, S. P. J. (2006). The active site of the [FeFe]-hydrogenase from Desulfovibrio desulfuricans. Ii. redox properties, light sensitivity and co-ligand exchange as observed by infrared spectroscopy. J. Biol. Inorg. Chem. 11, 102–118. doi: 10.1007/s00775-005-0040-2
Rumpel, S., Siebel, J.F., Diallo, M., Farès, C., Reijerse, E. J., and Lubitz, W. (2015). Structural Insight into the Complex of Ferredoxin and [FeFe] Hydrogenase from Chlamydomonas reinhardtii. Chem. Bio. Chem. 16, 1663–1669. doi: 10.1002/cbic.201500130
Rumpel, S., Siebel, J. F., Farès, C., Duan, J., Reijerse, E., Happe, T., et al. (2014). Enhancing hydrogen production of microalgae by redirecting electrons from photosystem i to hydrogenase. Energy Environ. Sci. 7, 3296–3301. doi: 10.1039/C4EE01444H
Rutz, A., Das, C. K., Fasano, A., Jaenecke, J., Yadav, S., Apfel, U.-P., et al. (2023). Increasing the O2 resistance of the [FeFe]-hydrogenase Cba5h through enhanced protein flexibility. ACS Catal. 13, 856–865. doi: 10.1021/acscatal.2c04031
Santana-Sanchez, A., Solymosi, D., Mustila, H., Bersanini, L., Aro, E.-M., and Allahverdiyeva, Y. (2019). Flavodiiron proteins 1–to-4 function in versatile combinations in o2 photoreduction in cyanobacteria. elife 8:E45766. doi: 10.7554/eLife.45766
Schäfer, C., Friedrich, B., and Lenz, O. (2013). Novel, oxygen-insensitive group 5 [NiFe]-hydrogenase in Ralstonia Eutropha. Appl. Environ. Microbiol. 79, 5137–5145. doi: 10.1128/AEM.01576-13
Shimakawa, G., Kohara, A., and Miyake, C. (2020). Characterization of light-enhanced respiration in cyanobacteria. Int. J. Mol. Sci. 22:342. doi: 10.3390/ijms22010342
Sun, Y., Chen, M., Yang, H., Zhang, J., Kuang, T., and Huang, F. (2013). Enhanced H2 Photoproduction by down-regulation of ferredoxin-Nadp+ reductase (Fnr) in the green alga Chlamydomonas Reinhardtii. Int. J. Hydrog. Energy 38, 16029–16037. doi: 10.1016/j.ijhydene.2013.10.011
Touloupakis, E., Faraloni, C., Silva Benavides, A. M., and Torzillo, G. (2021). Recent achievements in microalgal photobiological hydrogen production. Energies 14:7170. doi: 10.3390/en14217170
Tros, M., Bersanini, L., Shen, G., Ho, M.-Y., Van Stokkum, I. H. M., Bryant, D. A., et al. (2020). Harvesting far-red light: functional integration of chlorophyll f into photosystem i complexes of Synechococcus sp. PCC 7002. Biochim. Biophys. Acta - Bioenerg. 1861:148206. doi: 10.1016/j.bbabio.2020.148206
Vignais, P. M., and Billoud, B. (2007). Occurrence, classification, and biological function of hydrogenases: an overview. Chem. Rev. 107, 4206–4272. doi: 10.1021/cr050196r
Vogt, S., Lyon, E. J., Shima, S., and Thauer, R. K. (2008). The exchange activities of [Fe] Hydrogenase (iron–sulfur-cluster-free hydrogenase) from methanogenic archaea in comparison with the exchange activities of [FeFe] and [NiFe] hydrogenases. J. Biol. Inorg. Chem. 13, 97–106. doi: 10.1007/s00775-007-0302-2
Volbeda, A., Charon, M.-H., Piras, C., Hatchikian, E. C., Frey, M., and Fontecilla-Camps, J. C. (1995). Crystal structure of the nickel–iron hydrogenase from Desulfovibrio gigas. Nature 373, 580–587. doi: 10.1038/373580a0
Volbeda, A., Garcin, E., Piras, C., De Lacey, A. L., Fernandez, V. M., Hatchikian, E. C., et al. (1996). Structure of the [NiFe] Hydrogenase Active Site: Evidence for Biologically Uncommon Fe Ligands. J. Am. Chem.l Soc. 118, 12989–12996. doi: 10.1021/ja962270g
Vuorijoki, L., Tiwari, A., Kallio, P., and Aro, E.-M. (2017). Inactivation of iron-sulfur cluster biogenesis regulator sufr in Synechocystis sp. PCC 6803 induces unique iron-dependent protein-level responses. Biochim. Biophys. Acta Gen. Subj. 1861, 1085–1098. doi: 10.1016/j.bbagen.2017.02.020
Walters, K. A., and Golbeck, J. H. (2020). Designing a modified clostridial 2[4Fe–4S] ferredoxin as a redox coupler to directly link photosystem I with a Pt nanoparticle. Photosynthesis Res. 143, 165–181. doi: 10.1007/s11120-019-00679-w
Wegelius, A., Khanna, N., Esmieu, C., Barone, G. D., Pinto, F., Tamagnini, P., et al. (2018). Generation of a functional, semisynthetic [FeFe]-hydrogenase in a photosynthetic microorganism. Energy Environ. Sci. 11, 3163–3167. doi: 10.1039/C8EE01975D
Wegelius, A., Land, H., Berggren, G., and Lindblad, P. (2021). Semisynthetic [FeFe]-hydrogenase with stable expression and H2 production capacity in a photosynthetic microbe. Cell Rep. Phys. Sci. 2:100376. doi: 10.1016/j.xcrp.2021.100376
Weiner, I., Shahar, N., Feldman, Y., Landman, S., Milrad, Y., Ben-Zvi, O., et al. (2018). Overcoming the expression barrier of the ferredoxin-hydrogenase chimera In Chlamydomonas reinhardtii supports a linear increment in photosynthetic hydrogen output. Algal Res. 33, 310–315. doi: 10.1016/j.algal.2018.06.011
Weyman, P. D., Vargas, W. A., Tong, Y., Yu, J., Maness, P. C., Smith, H. O., et al. (2011). Heterologous expression of alteromonas macleodii and Thiocapsa roseopersicina [NiFe] hydrogenases in Synechococcus elongatus. PLoS One 6:E20126. doi: 10.1371/journal.pone.0020126
Winkler, M., Duan, J., Rutz, A., Felbek, C., Scholtysek, L., Lampret, O., et al. (2021). A Safety cap protects hydrogenase from oxygen attack. Nat. Commun. 12:756. doi: 10.1038/s41467-020-20861-2
Wu, S., Yan, G., Xu, L., Wang, Q., and Liu, X. (2010). Improvement of hydrogen production with expression Of Lba gene in chloroplast of Chlamydomonas reinhardtii. Int. J. Hydrog. Energy 35, 13419–13426. doi: 10.1016/j.ijhydene.2009.11.118
Xiong, D., Happe, T., Hankamer, B., and Ross, I. L. (2021). Inducible high level expression of a variant Δd19a,D58a-ferredoxin-hydrogenase fusion increases photohydrogen production efficiency in the green alga Chlamydomonas reinhardtii. Algal Res. 55:102275. doi: 10.1016/j.algal.2021.102275
Keywords: photobiological hydrogen, [FeFe] hydrogenases, [NiFe] hydrogenases, cyanobacteria, green algae, heterologous expression, genetic engineering, photosynthesis
Citation: Schumann C, Fernández Méndez J, Berggren G and Lindblad P (2023) Novel concepts and engineering strategies for heterologous expression of efficient hydrogenases in photosynthetic microorganisms. Front. Microbiol. 14:1179607. doi: 10.3389/fmicb.2023.1179607
Received: 04 March 2023; Accepted: 09 June 2023;
Published: 12 July 2023.
Edited by:
Stefan Frielingsdorf, Technical University of Berlin, GermanyReviewed by:
Francesca Valetti, University of Turin, ItalyCopyright © 2023 Schumann, Fernández Méndez, Berggren and Lindblad. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Peter Lindblad, cGV0ZXIubGluZGJsYWRAa2VtaS51dS5zZQ==; Gustav Berggren, Z3VzdGF2LmJlcmdncmVuQGtlbWkudXUuc2U=
†These authors share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.