- Department of Food Science and Technology, Seoul National University of Science and Technology, Seoul, Republic of Korea
Geobacillus stearothermophilus is a highly thermophilic, spore-forming Gram-positive bacterium that causes flat sour spoilage in low-acid canned foods. To address this problem, we isolated G. stearothermophilus-infecting phage GR1 from the soil and characterized its endolysin LysGR1. Phage GR1 belongs to the Siphoviridae family and possesses a genome of 79,387 DNA bps with 108 putative open reading frames. GR1 demonstrated a very low degree of homology to previously reported phages, indicating that it is novel. The endolysin of GR1 (LysGR1) contains an N-terminal amidase domain as an enzymatically active domain (EAD) and two C-terminal LysM domains as a cell wall binding domain (CBD). Although GR1 is specific to certain strains of G. stearothermophilus, LysGR1 showed a much broader lytic range, killing all the tested strains of G. stearothermophilus and several foodborne pathogens, such as Clostridium perfringens, Listeria monocytogenes, and Escherichia coli O157:H7. LysGR1_EAD, alone, also exhibits lytic activity against a wide range of bacteria, including Bacillus cereus, which is not terminated by a full-length endolysin. Both LysGR1 and its EAD effectively remove the G. stearothermophilus biofilms and are highly thermostable, retaining about 70% of their lytic activity after a 15-min incubation at 70°C. Considering the high thermal stability, broad lytic activity, and biofilm reduction efficacy of LysGR1 and its EAD, we hypothesize that these enzymes could act as promising biocontrol agents against G. stearothermophilus and as foodborne pathogens.
1. Introduction
Geobacillus stearothermophilus is a thermophilic, aerobic, spore-forming bacterium widely distributed throughout environments. A high initial load of G. stearothermophilus can cause flat sour spoilage in low-acid canned foods when its spores survive canning and ultra-high temperature processes, followed by germination and outgrowth (Wells-Bennik et al., 2019). In addition, it often forms biofilms in dairy manufacturing plants, which causes significant financial losses in the food industry (Andre et al., 2013; Batt and Tortorello, 2014). Although heat sterilization has been used to control G. stearothermophilus, its use is challenging because it can damage the textures, tastes, colors, and nutritional value of foods (Block, 2001). Thus, the development of novel antimicrobial agents against G. stearothermophilus has become of interest.
Bacteriophages and phage-derived proteins have gained growing attention as natural antimicrobial agents (Mahony et al., 2020). Phages are viruses found ubiquitously in nature that infect and replicate only in bacterial cells (Stone et al., 2019). At the end of the bacteriophage’s lytic cycle, phages produce peptidoglycan hydrolases, called endolysins, which cleave the cell wall in the host cell to release progeny virions (Fischetti, 2010). Endolysins from Gram-positive bacteria infecting phages generally harbor two distinct domains: a cell wall binding domain (CBD) and an enzymatically active domain (EAD) (Rahman et al., 2021). While EADs cleave the specific bonds constituting the bacterial cell wall, CBDs recognize and bind to the specific receptor of the bacterial cell wall (Loessner et al., 2002). Due to their potent lytic action and proteinaceous nature, endolysins have the potential to be used as food preservatives. Indeed, a number of studies have tested the use of phage endolysins to control diverse food-borne pathogens, including Staphylococcus aureus, Listeria monocytogenes, Clostridium perfringens (Chang et al., 2017; Van Tassell et al., 2017; Cho et al., 2021) in food matrices.
Despite of the importance of G. stearothermophilus in food industry, only a few G. stearothermophilus phages and their endolysins have been previously reported (Egbert and Mitchell, 1967; Rabussay et al., 1970; Skowron et al., 2018). Jin et al. (2013) isolated a bacteriophage GVE2 infecting Geobacillus sp. E263 from a deep-see hydrothermal vent and characterized the roles of its endolysin. Zebrowska et al. (2022) characterized the biochemical properties of a thermostable endolysin of phage TP-84, which infects G. stearothermophilus, and tested its potential as a disinfectant. To develop efficient endolysin-based antimicrobials, new G. stearothermophilus phages should be isolated and more in-depth studies of their endolysins are required. In this study, we isolated a novel G. stearothermophilus phage GR1 and characterized its endolysin LysGR1. Here, LysGR1 and its EAD exhibited much broader lytic activities compared to the high host specificity of GR1. Both recombinant proteins have high thermal stability in the temperature range of −20°C–70°C and effectively removed the G. stearothermophilus biofilms. Our findings may facilitate the development of promising biocontrol agents against G. stearothermophilus.
2. Materials and methods
2.1. Bacterial strains and plasmids
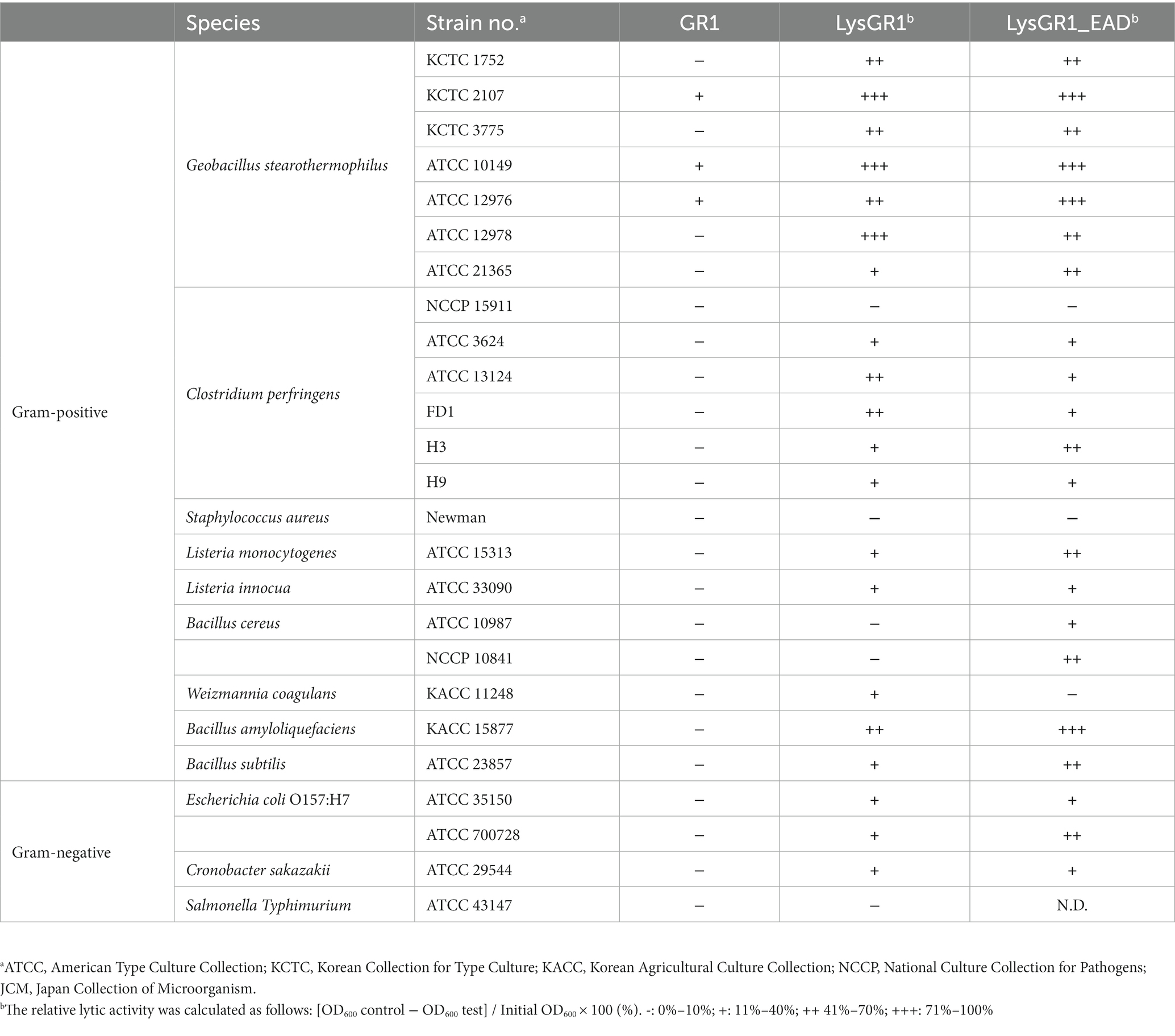
The bacterial strains used in this study are listed in Table 1. Geobacillus stearothermophilus ATCC 10149 was used for the isolation and propagation of GR1. All G. stearothermophilus strains were grown under agitation in Tryptic Soy Broth (TSB) at 50°C. Bacillus cereus, Salmonella, Escherichia coli, and Shigella flexneri strains were grown under agitation in Luria–Bertani (LB) broth at 37°C. Listeria and C. perfringens strains were grown in Brain Heart Infusion (BHI) broth at 37°C with shaking and anaerobic conditions, respectively. Bacillus strains, except B. cereus, Cronobacter sakazakii, Yersinia enterocolitica, Enterococcus faecalis, S. aureus, and Pseudomonas strains were grown in TSB at 37°C under agitation. The agar media were generated by supplementation of 1.5% agar with the broth medium. All media were purchased from Difco (Detroit, MI, USA). Bacterial stocks were stored at −80°C with 15% glycerol.
2.2. Isolation of bacteriophage GR1
To isolate Geobacillus-infecting phage, a soil sample was collected from the Gyeongchun Line railroad in Seoul, South Korea. A total of 25 g of the solid sample was mixed with 225 mL of Butterfield’s phosphate-buffered dilution water (0.25 M KH2PO4 adjusted to pH 7.2 with NaOH) in a sterile bag and homogenized using a Bag Mixer 400 CC (Interscience Laboratory Inc., St. Nom, France). Next, 10 mL of the diluted sample was mixed with 10 mL of 2× TSB with 5 mM of MgCl2 and CaCl2; then, the mixture was incubated after inoculation with 200 μL of the overnight cultured G. stearothermophilus ATCC 10149 and shaken for 24 h at 50°C. After incubation, 200 μL of chloroform was added to the overnight cultured mixture, and the mixture was incubated under agitation at 50°C for 10 min. After centrifugation (15,000 × g for 10 min), the supernatant was filtered using a 0.22 μm pore size filter (Sartorius Stedim Biotech, Göttingen, Germany). Then, 10-fold serial dilutions of the filtrate were spotted on G. stearothermophilus ATCC 10149 embedded in 0.4% soft TSB agar. The plate was incubated at 50°C for 24 h. To isolate and purify the bacteriophages, the overlay assay was performed as previously described (Kong and Ryu, 2015). The phage resuspension was overlaid on 0.4% soft TSB agar embedded with the host bacterium. This process was repeated at least three times to purify a single phage.
2.3. Phage propagation and purification
To propagate the isolated phage, the lysate of a single bacteriophage GR1 plaque was inoculated with the G. stearothermophilus ATCC 10149 culture (OD600 = 0.5) and incubated at 50°C for 4 h. After centrifugation (15,000 × g at 4°C for 5 min), the supernatant was filtered using a 0.22 μm pore size filter. The filtrate phage particles were concentrated as previously described (Kong and Ryu, 2015), with some modifications. Purified phages were dialyzed in 1 L of SM buffer in snakeskin dialysis tubing (molecular weight cut off (MWCO) 3.5 kDa, Thermo Fisher Scientific, Rockford, USA). The titers of the concentrated phage stocks were determined by the spotting assay and the phage stocks were stored at 4°C for use in further experiments.
2.4. Host range analysis
Host range analysis was performed as previously described (Na et al., 2016).
2.5. Morphological analysis by transmission electron microscopy
Morphological analysis was performed similarly to a previous study (Lee et al., 2019). Drops of purified phage GR1 stock (about 1.3 × 1010 PFU/mL) were placed on the prepared grids and incubated at room temperature for 1 min. After incubation, the excess phage suspension was removed using filter paper and the phage particles were negatively stained with the same volume of 2% uranyl acetate (pH 4.0) for 20 s; the excess solution was removed as described above. GR1 was examined by transmission electron microscopy (Libra 120 model, Carl Zeiss, Oberkochen, Germany) at 120 kV. GR1 was classified according to the International Committee on Taxonomy of Viruses (ICTV) classification.
2.6. Genome sequencing and in silico analysis
Phage DNA extraction was performed using a Phage DNA Isolation Kit (Norgen Biotek Corporation, Thorold, Ontario, Canada), according to the manufacturer’s instructions. The purified genomic DNA was sequenced using the Illumina Novaseq 6,000 platform at LabGenomics, South Korea, and assembled with a Unicycler assembler v0.4.8 at Sanigen, South Korea. The functions of the predicted open reading frames (ORFs) were determined using BLASTP and InterProScan databases (Altschul et al., 1990). The presence of tRNA-encoding genes was examined using the tRNAscan-SE database. A circular genome map was generated using the CGview server database (Stothard and Wishart, 2005). The amino acid sequence alignments of endolysins were conducted using ClustalX2 (Larkin et al., 2007).
2.7. Cloning, expression, and purification of LysGR1 and its EAD, EGFP-CBD fusion protein
The endolysin gene (LysGR1) and its enzymatically active domain (LysGR1_EAD), cell wall binding domain (LysGR1_CBD) were amplified from the genomic DNA of the GR1 phage by the polymerase chain reaction (PCR) (primers used in this study are indicated in Supplementary Table 1). The gene fragments encoding LysGR1 and LysGR1_EAD were treated with NcoI and SalI and cloned into pET28a (Novagen, Madison, WI, USA). The gene fragments encoding LysGR1_CBD were digested with BamHI and SalI and subcloned into pET28a::EGFP (Kong et al., 2015). To purify each protein, a hexahistidine (6 × -His) tag was added to the C-terminus of LysGR1 and its EAD, the N-terminus of the EGFP-CBD fusion protein, respectively. The plasmid with the correct insert was transformed into E. coli BL21 (DE3). The expression of the proteins was induced by 0.5 mM of isopropyl β-D-1- thiogalactopyranoside (IPTG) (Sigma-Aldrich, St. Louis, MO), and adjusted the OD600 from 0.6 to 0.8, followed by incubation for 20 h at 18°C with agitation. After centrifugation (4,000 × g at 4°C for 15 min), bacterial cells were resuspended in 5 mL of lysis buffer (50 mM Tris-Cl, pH 8.0, 200 mM NaCl). Cells were lysed by sonication (Sonics & Materials, Inc., CT, USA) for 6 min using intervals of 4 s on and 6 s off. To eliminate the insoluble materials, centrifugation was conducted at 21,000 × g and 4°C for 1 h, and the supernatant was collected and purified using 500 μL of nickel–nitrilotriacetic acid (Ni–NTA) agarose (Qiagen, Hilden, Germany). The concentration of proteins was examined by Bradford assay, while the mass, purity, and solubility were assessed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Purified LysGR1 and LysGR1_EAD were stored in lysis buffer at −80°C, while purified EGFP-CBD was stored in storage buffer (50 mM Tris-Cl (pH 8.0), 200 mM NaCl, 50% glycerol) at −20°C, respectively.
2.8. Turbidity reduction assay
Bacterial cells were grown to the exponential phase (approximately OD600 = 0.6) and resuspended in reaction buffer (20 mM Tris-Cl, pH 8.0). The purified endolysin (LysGR1) and its EAD were added to a final concentration of 1.6 μM, and the OD600 values were monitored over time at 45°C using a SpectraMax i3x plate reader (Molecular Devices, Sunnyvale, CA). The relative lytic activity was calculated when the OD600 of the endolysin-treated group (experimental OD600) reached the lowest value using the equation as follows:
The effect of pH on the enzymatic activity of LysGR1 and LysGR1_EAD was assessed, as previously described (Ha et al., 2018), yet with some modifications. An amount of 1.6 μM of endolysin was added to Bacillus amyloliquefaciens KACC 15877 cells suspended in Britton–Robinson universal buffer (0.04 M H3PO4, 0.04 M H3BO3, 0.04 M CH3COOH, and 0.2 M NaCl) with various pH buffers, which ranged between 6 and 10. To evaluate the effect of the temperature on LysGR1 and its EAD lytic activity, preincubated LysGR1 and its EAD at different temperatures (−20–80°C for 15 min) were used. The effect of NaCl on the enzymatic activity of LysGR1 and its EAD was tested with the reaction buffer using various NaCl concentrations (0–1,000 mM).
2.9. Time-killing assay
The time-killing assay was conducted as previously described with some modifications (Lee et al., 2021). The exponentially growing G. stearothermophilus ATCC 10149 cells (OD600 = 0.6) were washed once and resuspended to OD600 = 1.0 by PBS buffer and treated with 1.6 μM of LysGR1 and EAD, respectively. PBS buffer used as a control. After treatment, the mixture was incubated at 50°C for 1 h. The cell aliquots were harvested at the indicated time points (0, 10, 60 min) and diluted, plated in tryptic soy agar plates for enumeration. The number of viable cells was assessed by counting the colonies, and time-killing assay was performed in triplicate.
2.10. Biofilm reduction assay on 24-well polystyrene surface
The biofilm removal assay was performed as previously described, although with some modifications (Cha et al., 2019). To determine the biofilm reduction capacity of LysGR1 and LysGR1_EAD, the G. stearothermophilus ATCC 12980 (= KCTC 1752) strain was incubated in TSB medium and sub-cultured into each well of a 24-well polystyrene microplate to generate biofilms. After incubation of the microplate for 48 h at 50°C, each well was washed at least three times with PBS, and the washed plate was incubated for 30 min at 50°C to fix the biofilms. After fixing the biofilms, serially diluted LysGR1 and LysGR1_EAD were added to each well, in PBS, to a final volume of 1 mL per well, while PBS was also used as the negative control. The microplate was incubated for 1 h at 50°C, and each well of the microplate was washed three times with PBS, then, stained with 0.1% crystal violet solution. The excess crystal violet solution was removed through additional washing with PBS, and the stained biofilms were solubilized with 33% acetic acid. The absorbance of bacteria in the biofilm was measured at 570 nm, and the remained biomass was presented as the A570 value. The relative A570 was calculated as follows: the A570 test (endolysin added or buffer only) divided by the A570 control (buffer only).
2.11. EGFP-fusion LysGR1_CBD binding assay
The binding capacity of the EGFP fusion LysGR1_CBD (EGFP-CBD) was assessed, as previously described (Loessner et al., 2002). Both an LSM 800 (Carl Zeiss, Germany) and an Eclipse Ti2-E fluorescent microscope (Nikon, Japan) were used in this assay.
2.12. Accession numbers
The GenBank accession numbers of GR1 and LysGR1 are OK896991 and UDY80750, respectively.
3. Results
3.1. Isolation and characterization of phage GR1
Geobacillus stearothermophilus phage GR1 was isolated from roadside soil, in Seoul, using G. stearothermophilus ATCC 10149 as a host. Transmission electron microscopy (TEM) showed that GR1 belongs to the Siphoviridae family, which has an icosahedral head with a diameter of 88 ± 9 nm (n = 10) and a non-contractile tail with a length of 187 ± 44 nm (n = 10) (Figure 1A). GR1 produced large, clear plaques against its host strain (Figure 1B) and infected three out of the seven strains of G. stearothermophilus (Table 1). GR1 did not infect other Gram-positive or Gram-negative bacteria, indicating that it has a high host specificity.
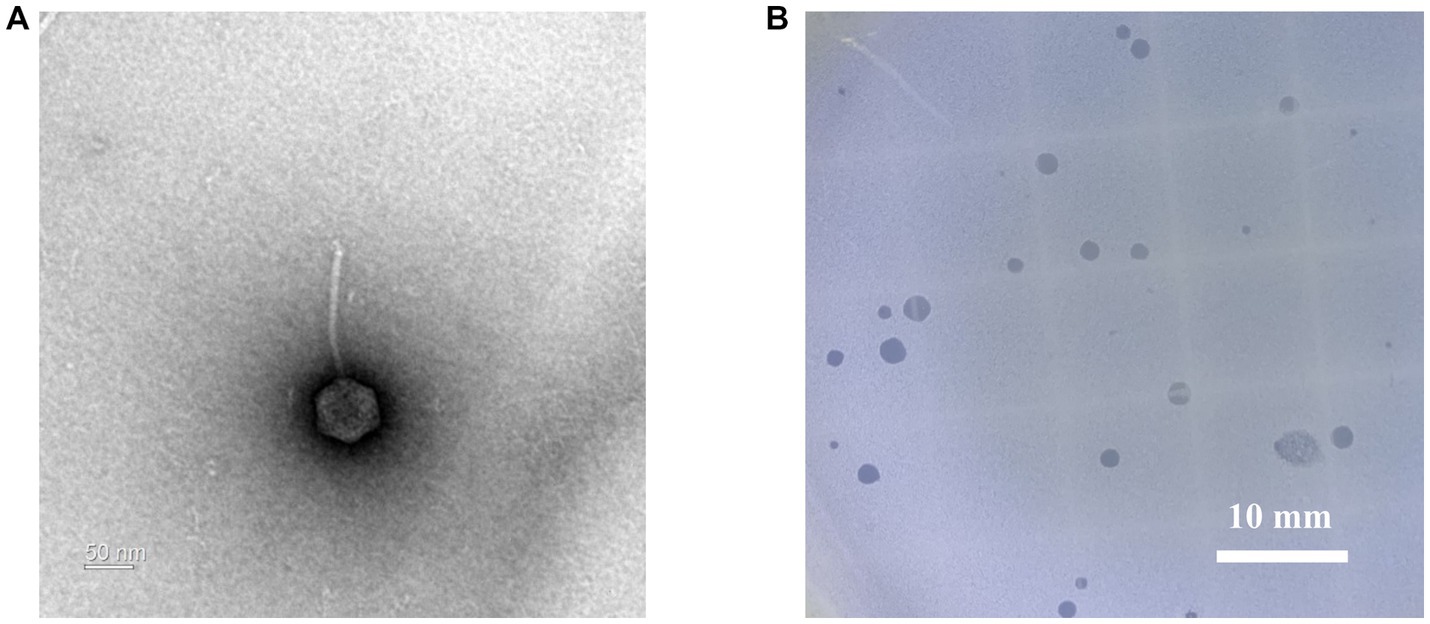
Figure 1. Morphological analysis of G. stearothermophilus infecting phage GR1. (A) A transmission electron microscopy image of GR1. (B) Plaque morphology of GR1.
The complete genome of phage GR1 comprises 79,387 bps of double-stranded DNA, with a 32.34% average G + C content, 108 putative open reading frames (ORFs), and 1 tRNA. The functional ORFs were categorized into six different groups according to their function: phage DNA packaging, phage structure, host lysis, nucleotide metabolism, additional function, and hypothetical proteins (Figure 2). The presence of an integrase and a tyrosine-type recombinase suggest that GR1 is a temperate phage. Other genes encoding the major capsid protein, putative adhesin, and terminase large subunit were also identified in the phage. BLASTN analysis revealed that there are no genetically similar phages, indicating that GR1 is a novel phage.
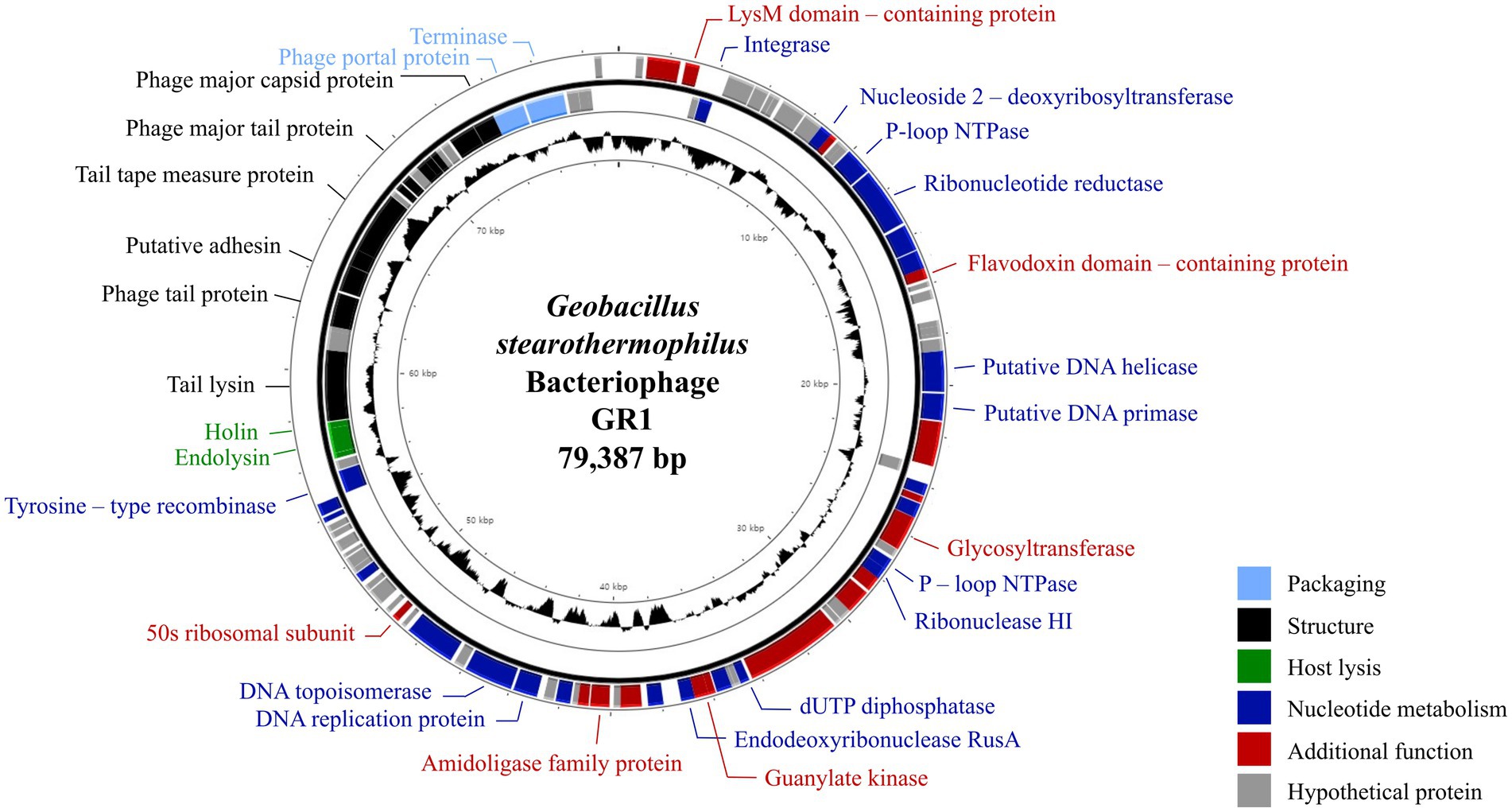
Figure 2. The genome map of GR1. Each ORF color indicates its function: deep blue: nucleotide metabolism; sky blue: packaging; green: host lysis; black: structural proteins; red: additional functions; grey: hypothetical proteins. The inner circle with the black graph shows the GC contents.
3.2. Identification of endolysin LysGR1 and its EAD
The putative endolysin gene was identified in the genome of the bacteriophage GR1 and named LysGR1. Pfam and BLASTP analyses revealed that LysGR1 is a putative N-acetylmuramoyl-L-alanine amidase and contains an N-terminal amidase_3 domain (PF01520) as an enzymatically active domain (EAD), and C-terminal tandem repeats of LysM motifs (PF01476) as a putative cell wall binding domain (CBD) (Figure 3A). Amino acid sequence analysis revealed that LysGR1 only shares 27%, 32%, and 34% identities with lysins from the Bacillus megaterium phage G (GenBank protein ID: YP_009015333.1), Sporosarcina phage Lietuvens (GenBank protein ID: QIG62545.1), and Bacillus phage vB_BcM_Sam112 (GenBank protein ID: QGF21730.1), respectively (Figure 3B), which confirms its novelty. To test the lytic activity, we cloned the recombinant LysGR1 and its EAD (amino acids 1–194, denoted hereafter as LysGR1_EAD) to contain a C-terminal His-tag. Both proteins were expressed in their soluble form in E. coli and were purified by Ni-NTA affinity chromatography, which resulted in a homogeneous preparation (Supplementary Figure 1). The protein yield of LysGR1 was relatively lower than LysGR1_ EAD: 0.6 mg of LysGR1 vs. 5 mg of its EAD from 50 mL of E. coli cell lysates. A turbidity reduction assay showed that the lytic activity of LysGR1 against G. stearothermophilus ATCC 10149 cells is dose-dependent (Figure 3C), while it also exhibited a broad lytic spectrum against all the tested G. stearothermophilus strains (Table 1). In addition to G. stearothermophilus, LysGR1 killed various Bacillus spp. including, C. perfringens, L. monocytogenes, E. coli O157:H7, and C. sakazakii (Supplementary Figure 2).
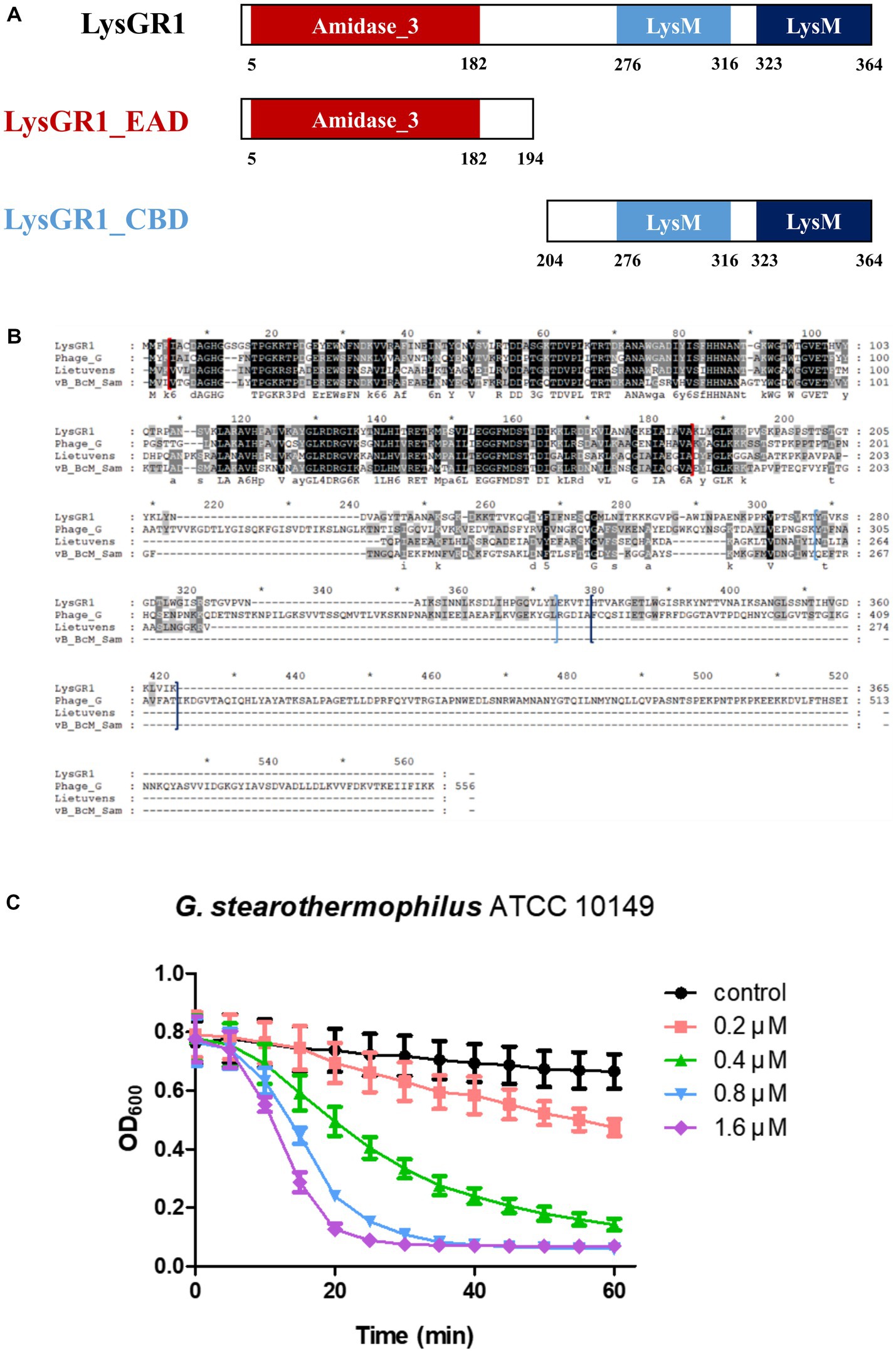
Figure 3. Modular structure of a novel endolysin LysGR1. (A) Schematic representation of LysGR1 and its EAD and CBD. LysGR1 has a conserved amidase domain as an EAD at the N-terminus and tandem repeats of LysM motifs as a putative CBD at the C-terminus. LysGR1_EAD contains only an amidase-3 domain, and the CBD construct contains two LysM domains. (B) Amino acid sequence alignment of LysGR1 with other endolysins from Bacillus and Sporosarcina infecting phages. Phage_G: N-acetylmuramoyl-L-alanine amidase from Bacillus megaterium phage G, Lietuvens: N-acetylmuramoyl-L-alanine amidase from Sporosarcina phage Lietuvens, vB_BcM_Sam: N-acetylmuramoyl-L-alanine amidase from Bacillus phage vB_BcM_Sam112. Each domain of LysGR1 indicated with colored square bracket: red: Amidase_3; light blue: first LysM; dark blue: second LysM. Conserved residues are shaded in black and gray (dark gray: > 70% conserved; light grey: > 40% conserved). (C) Dose-dependent lytic activity of recombinant LysGR1 against G. stearothermophilus ATCC 10149 cells.
3.3. Bacteriolytic and bactericidal activity of LysGR1 and EAD
To determine the lytic and antimicrobial activity of LysGR1 and its EAD, the turbidity reduction assay and time-killing test were conducted, respectively. Compared to LysGR1, LysGR1_EAD also possesses a similar lytic range to LysGR1, but interestingly, it generally lyses bacterial cells more rapidly than its parental endolysin (Figure 4A). Moreover, LysGR1_EAD can lyse several B. cereus strains, which LysGR1 could not kill (Table 1). We also measured the number of viable bacterial cells to evaluate the bactericidal ability of LysGR1 and its EAD in a time-kinetic manner (Figure 4B). LysGR1 and EAD caused 4.4 and 5.4 log reduction, respectively, of G. stearothermophilus cells after the addition of 1.6 μM of each enzyme for 10 min at 50°C. At 1 h, no viable cells were detected after the treatment of EAD, while LysGR1-treated group showed 7.2 log reduction, indicating the antimicrobial potential of both LysGR1 and EAD.
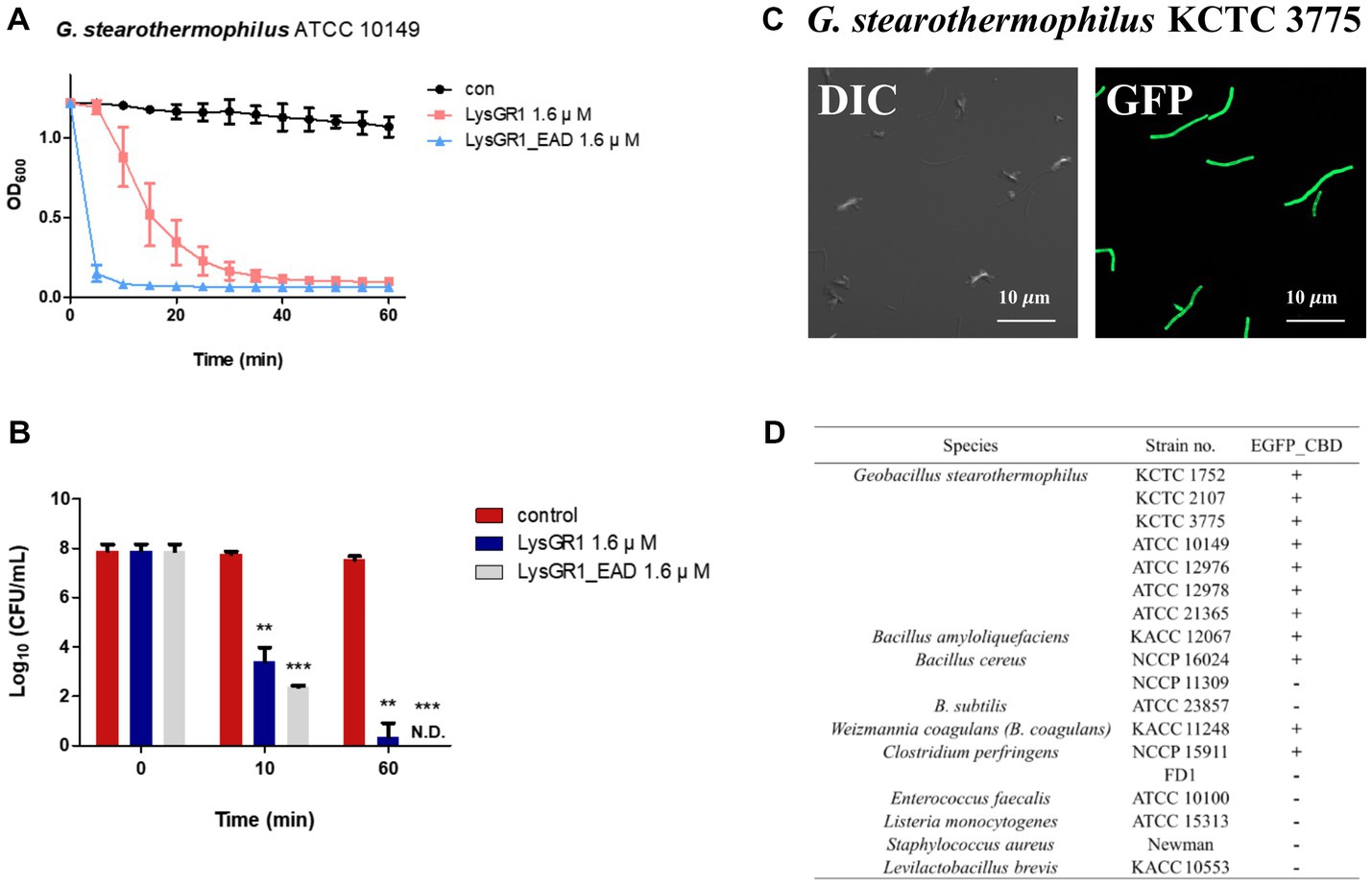
Figure 4. Antimicrobial and binding activity of LysGR1 and its domains. (A) Comparisons of the lytic activity of LysGR1 and its EAD against G. stearothermophilus ATCC 10149 cells. (B) Time-killing activity of LysGR1 and EAD against G. stearothermophilus ATCC 10149 cells was determined in PBS buffer. The error bars indicate the standard deviation and data were replicated for three independent times and analyzed based on the PBS results by t-test, *p < 0.05, **p < 0.01, ***p < 0.005. (C) Binding capacity of LysGR1_CBD toward G. stearothermophilus KCTC 3775 cells. (D) Binding spectrum of LysGR1_CBD.
3.4. Binding activity of LysGR1_CBD
To examine the binding ability of LysGR1, we fused EGFP to the C-terminal tandem repeats of LysM domains of LysGR1 (amino acids 204–364, hereafter called LysGR1_CBD). The fluorescence binding assay revealed that LysGR1_CBD could bind to all G. stearothermophilus strains tested (Figures 4C,D), proving the presence of CBD within LysGR1. LysGR1_CBD also binds to some strains of B. amyloliquefaciens, B. cereus, W. coagulans, and C. perfringens cells, suggesting that LysGR1_CBD targets broadly conserved cell wall structure of these bacteria.
3.5. Biochemical properties of LysGR1 and its EAD
Next, we evaluated the effects of temperature, NaCl, and pH on the lytic activity of LysGR1 and its EAD. The thermal stability was analyzed following the pre-incubation of the enzymes at each temperature for 15 min. Both LysGR1 and its EAD were not significantly affected by temperatures up to 60°C and retain more than 60% lytic activity after incubating at 70°C (Figures 5A,D), indicating their high thermal stabilities. Both enzymes also showed the highest lytic activity at 100–200 mM, while LysGR1_EAD was more sensitive to increasing NaCl concentrations than LysGR1 (Figures 5B,E). Furthermore, LysGR1 and its EAD were relatively stable under a wide range of pHs (pH: 6.0–10.0) (Figures 5C,F).
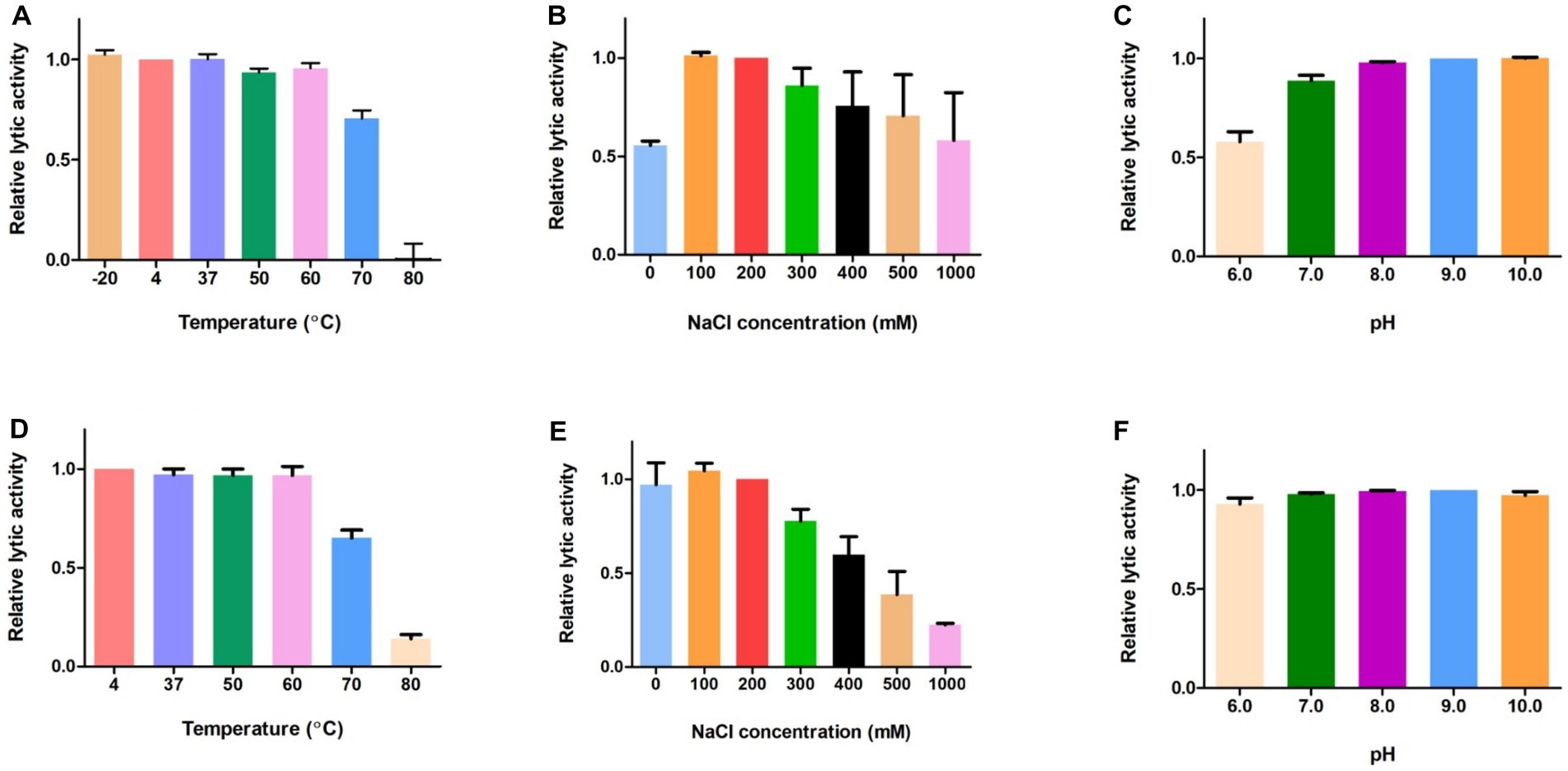
Figure 5. Biochemical properties of LysGR1 and its EAD. The effects of temperature (A), NaCl (B), and pH (C) on the lytic activity of LysGR1 and the effects of temperature (D), NaCl (E), and pH (F) on the lytic activity of LysGR1_EAD against B. amyloliquefaciens KACC 15877 cells. Each column represents the mean of triplicate experiments, and the error bars indicate the standard deviation.
3.6. Biofilm reduction activity of LysGR1 on 24-well polystyrene surface
The anti-biofilm activities of LysGR1 and LysGR1_EAD were tested against the G. stearothermophilus ATCC 12980 biofilms, which had been formed in 24-well polystyrene microplates. Both LysGR1 and its EAD showed dose-dependent biofilm reduction efficacies (Figures 6A,B). A total of 86.86 and 74.27% of the biofilms were removed following treatment with 1.6 μM LysGR1 and its EAD, respectively. This result suggests that LysGR1 and LysGR1_EAD could be used as natural anti-biofilm agents against G. stearothermophilus.
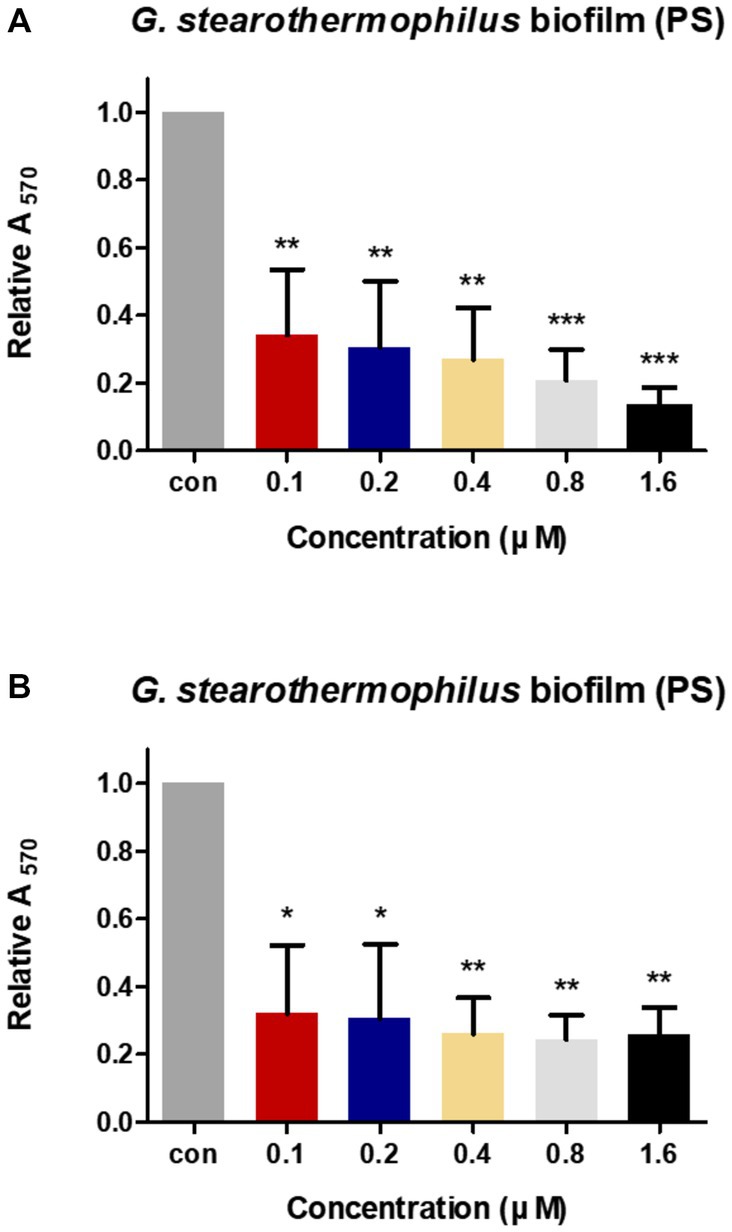
Figure 6. Biofilm reduction activity of LysGR1 (A) and its EAD (B) in 24-well polystyrene microplates. G. stearothermophilus ATCC 12980 biofilms on the surface of polystyrene were treated with various concentrations of LysGR1 and its EAD for 1 h. Each column represents the mean of triplicate experiments, and the error bars indicate the standard deviation. Data were analyzed based on the buffer-treated negative control by t-test, *p < 0.05, **p < 0.01, ***p < 0.005.
4. Discussion
Geobacillus stearothermophilus is a thermophilic, aerobic, spore-forming bacterium with a growth temperature range of 37°C–75°C (Nazina et al., 2001). Usually, G. stearothermophilus is responsible for flat-sour spoilage in low-acid canned foods because it produces heat-stable proteases, lipases, and short-chain fatty acids that sour the product without producing gases (Batt and Tortorello, 2014). Geobacillus stearothermophilus is also especially problematic in powdered milk processing because general thermal treatments are inefficient in the removal of highly heat-resistant spores (Burgess et al., 2010). During evaporation, the milk is exposed to temperatures between 45°C and 75°C, which are suitable for the germination and growth of residual G. stearothermophilus spores in manufacturing plants (Ruckert et al., 2004; Kumar et al., 2021). Furthermore, G. stearothermophilus is able to form biofilms resistant to cleaning chemicals and sanitizers, thus, becoming a continuous source of food contamination (Flint et al., 2001). Therefore, there is increasing demand to develop antimicrobial agents against G. stearothermophilus.
In this study, we isolated the Geobacillus-infecting bacteriophage GR1 and characterized its endolysin LysGR1 and its EAD. GR1 has 79,387 bps of DNA, which represents the second largest genome size among Geobacillus phages, after the Geobacillus virus E3 (141,298 bp). BlastN analysis revealed that GR1 has a very low sequence similarity to previously reported Geobacillus phages indicating its novelty. LysGR1 shares some amino acid sequence similarities with other bacterial autolysins, although the overall identity is around 50% due to the low sequence homology of the CBD region. Interestingly, phage GR1 can only infect a few strains of G. stearothermophilus, whereas, LysGR1 and its EAD showed a much broader lytic range, including B. amyloliquefaciens, Bacillus subtilis, L. monocytogenes, C. perfringens, and are even capable of killing some Gram-negative bacteria without any pretreatment with outer membrane permeabilizers. In the case of Gram-negative bacteria-targeting endolysins, cationic peptides, such as poly-L-Arg and ε-poly-L-lysine, have been fused to endolysins to destabilize the negatively charged outer membranes of Gram-negative bacteria, which increases the chance of the lysins accessing the peptidoglycan layer (Gutierrez and Briers, 2021). Considering that LysGR1 has a net positive charge (pI = 9.75), owing to the high proportion of lysine residues (41/365 aa), we hypothesize that this basic property might partially work as an outer membrane permeabilizer. LysGR1 and its EAD also showed a similar lytic range and effectively removed the G. stearothermophilus biofilms on the polystyrene surface (over 75% of the biofilms). Compared to the EAD, LysGR1 tends to be more effective at lysing G. stearothermophilus cells (ATCC 12978, KCTC 3775) that are strongly bound with LysGR1_CBD. On the other hand, bacterial cells that have weak or no binding toward LysGR1_CBD are generally more susceptible to EAD than the full-length endolysin. These CBD-independent, high lytic activity of the EAD might arise from a positive net charge (pI = 9.54) and a small size that efficiently enable the penetration of EAD to peptidoglycan mesh (Demchick and Koch, 1996; Low et al., 2011). In this case, CBD rather hinders the hydrolytic activity of EAD when the CBD has no or less cognate target by enlarging the size of entire enzyme which is undesirable feature. It is also possible that EAD has its own cell wall binding ability as shown in a previous study (Mayer et al., 2011), which undoubtedly needs further experiments.
Although endolysins are considered promising eco-friendly antimicrobials due to their rapid action, specificity, and low risk of bacterial resistance, their low stabilities often pose a problem in their applications to the food industry (Lee et al., 2022). Indeed, endolysins from mesophilic bacteria infecting phages generally lose their hydrolytic function above 45°C–60°C (Heselpoth et al., 2015a; Oliveira et al., 2016; Ni et al., 2021). To address this, one research group tried to fuse the catalytic domains of thermophile lytic enzymes to the cell wall binding domains of C. perfringens endolysins to create an anti-clostridium agent (Swift et al., 2015, 2019). Alternative approaches, such as structure-based point mutations (Heselpoth et al., 2015b; Love et al., 2021) the in silico design of chimeric endolysins (Haddad Kashani et al., 2017) or stabilizer addition (Heselpoth et al., 2015a) have also been reported to improve the thermostability of the endolysins. Biochemical analysis revealed that LysGR1 and its EAD are highly thermostable, maintain their full lytic activity from 4°C to 60°C, and even retain over 60% of their lytic activity after incubation at 70°C for 15 min. In addition, LysGR1_EAD has a high protein yield (100 mg/L) through the E. coli T7-lac expression system, potentiating its possible use as a fusion partner for generating heat-stable chimeric antimicrobials. Overall, by possessing broad lytic activities and high stabilities, both LysGR1 and EAD could be promising biocontrol agents against both food-borne pathogens and food spoilage bacteria.
Data availability statement
The data presented in the study has been deposited in the GenBank database, under accession number OK896991.
Author contributions
DC and MK conceived and designed the experiments and wrote the manuscript. DC performed the experiments and analyzed the data. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT) (No., no. 2020R1C1C1008127).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1178748/full#supplementary-material
References
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Andre, S., Zuber, F., and Remize, F. (2013). Thermophilic spore-forming bacteria isolated from spoiled canned food and their heat resistance. Results of a French ten-year survey. Int. J. Food Microbiol. 165, 134–143. doi: 10.1016/j.ijfoodmicro.2013.04.019
Batt, C. A., and Tortorello, M. L. (2014). Encyclopedia of food microbiology, Amsterdam, Academic Press.
Block, S. S. (2001). Disinfection, sterilization, and preservation, Philadelphia, PA: Lippincott Williams & Wilkins.
Burgess, S. A., Lindsay, D., and Flint, S. H. (2010). Thermophilic bacilli and their importance in dairy processing. Int. J. Food Microbiol. 144, 215–225. doi: 10.1016/j.ijfoodmicro.2010.09.027
Cha, Y., Son, B., and Ryu, S. (2019). Effective removal of staphylococcal biofilms on various food contact surfaces by Staphylococcus aureus phage endolysin LysCSA13. Food Microbiol. 84:103245. doi: 10.1016/j.fm.2019.103245
Chang, Y., Yoon, H., Kang, D. H., Chang, P. S., and Ryu, S. (2017). Endolysin LysSA97 is synergistic with carvacrol in controlling Staphylococcus aureus in foods. Int. J. Food Microbiol. 244, 19–26. doi: 10.1016/j.ijfoodmicro.2016.12.007
Cho, J. H., Kwon, J. G., O'sullivan, D. J., Ryu, S., and Lee, J. H. (2021). Development of an endolysin enzyme and its cell wall-binding domain protein and their applications for biocontrol and rapid detection of Clostridium perfringens in food. Food Chem. 345:128562. doi: 10.1016/j.foodchem.2020.128562
Demchick, P., and Koch, A. L. (1996). The permeability of the wall fabric of Escherichia coli and Bacillus subtilis. J. Bacteriol. 178, 768–773. doi: 10.1128/jb.178.3.768-773.1996
Egbert, L. N., and Mitchell, H. K. (1967). Characteristics of Tphi3, a bacteriophage for Bacillus stearothermophilus. J. Virol. 1, 610–616. doi: 10.1128/jvi.1.3.610-616.1967
Fischetti, V. A. (2010). Bacteriophage endolysins: a novel anti-infective to control Gram-positive pathogens. Int. J. Med. Microbiol. 300, 357–362. doi: 10.1016/j.ijmm.2010.04.002
Flint, S., Palmer, J., Bloemen, K., Brooks, J., and Crawford, R. (2001). The growth of Bacillus stearothermophilus on stainless steel. J. Appl. Microbiol. 90, 151–157. doi: 10.1046/j.1365-2672.2001.01215.x
Gutierrez, D., and Briers, Y. (2021). Lysins breaking down the walls of Gram-negative bacteria, no longer a no-go. Curr. Opin. Biotechnol. 68, 15–22. doi: 10.1016/j.copbio.2020.08.014
Ha, E., Son, B., and Ryu, S. (2018). Clostridium perfringens Virulent Bacteriophage CPS2 and Its Thermostable Endolysin LysCPS2. Viruses 10:251. doi: 10.3390/v10050251
Haddad Kashani, H., Fahimi, H., Dasteh Goli, Y., and Moniri, R. (2017). A novel chimeric endolysin with antibacterial activity against methicillin-resistant Staphylococcus aureus. Front. Cell. Infect. Microbiol. 7:290. doi: 10.3389/fcimb.2017.00290
Heselpoth, R. D., Owens, J. M., and Nelson, D. C. (2015a). Quantitative analysis of the thermal stability of the gamma phage endolysin PlyG: a biophysical and kinetic approach to assaying therapeutic potential. Virology 477, 125–132. doi: 10.1016/j.virol.2014.11.003
Heselpoth, R. D., Yin, Y., Moult, J., and Nelson, D. C. (2015b). Increasing the stability of the bacteriophage endolysin PlyC using rationale-based FoldX computational modeling. Protein Eng. Des. Sel. 28, 85–92. doi: 10.1093/protein/gzv004
Jin, M., Ye, T., and Zhang, X. (2013). Roles of bacteriophage GVE2 endolysin in host lysis at high temperatures. Microbiology (Reading) 159, 1597–1605. doi: 10.1099/mic.0.067611-0
Kong, M., and Ryu, S. (2015). Bacteriophage PBC1 and its endolysin as an antimicrobial agent against Bacillus cereus. Appl. Environ. Microbiol. 81, 2274–2283. doi: 10.1128/AEM.03485-14
Kong, M., Sim, J., Kang, T., Nguyen, H. H., Park, H. K., Chung, B. H., et al. (2015). A novel and highly specific phage endolysin cell wall binding domain for detection of Bacillus cereus. Eur. Biophys. J. 44, 437–446. doi: 10.1007/s00249-015-1044-7
Kumar, M., Flint, S., Palmer, J., Chanapha, S., and Hall, C. (2021). Influence of the incubation temperature and total dissolved solids concentration on the biofilm and spore formation of dairy isolates of Geobacillus stearothermophilus. Appl. Environ. Microbiol. 87:e02311-20. doi: 10.1128/AEM.02311-20
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., Mcgettigan, P. A., Mcwilliam, H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Lee, D., Im, J., Na, H., Ryu, S., Yun, C. H., and Han, S. H. (2019). The novel enterococcus phage vB_EfaS_HEf13 has broad lytic activity against clinical isolates of Enterococcus faecalis. Front. Microbiol. 10:2877. doi: 10.3389/fmicb.2019.02877
Lee, C., Kim, H., and Ryu, S. (2022). Bacteriophage and endolysin engineering for biocontrol of food pathogens/pathogens in the food: recent advances and future trends. Crit. Rev. Food Sci. Nutr., 1–20. doi: 10.1080/10408398.2022.2059442
Lee, C., Kim, J., Son, B., and Ryu, S. (2021). Development of advanced chimeric endolysin to control multidrug-resistant Staphylococcus aureus through Domain Shuffling. ACS Infect. Dis. 7, 2081–2092. doi: 10.1021/acsinfecdis.0c00812
Loessner, M. J., Kramer, K., Ebel, F., and Scherer, S. (2002). C-terminal domains of Listeria monocytogenes bacteriophage murein hydrolases determine specific recognition and high-affinity binding to bacterial cell wall carbohydrates. Mol. Microbiol. 44, 335–349. doi: 10.1046/j.1365-2958.2002.02889.x
Love, M. J., Coombes, D., Manners, S. H., Abeysekera, G. S., Billington, C., and Dobson, R. C. J. (2021). The molecular basis for Escherichia coli O157:H7 phage FAHEc1 endolysin function and protein engineering to increase thermal stability. Viruses 13:1101. doi: 10.3390/v13061101
Low, L. Y., Yang, C., Perego, M., Osterman, A., and Liddington, R. (2011). Role of net charge on catalytic domain and influence of cell wall binding domain on bactericidal activity, specificity, and host range of phage lysins. J. Biol. Chem. 286, 34391–34403. doi: 10.1074/jbc.M111.244160
Mahony, J., Casey, E., and Van Sinderen, D. (2020). The Impact and Applications of Phages in the Food Industry and Agriculture. Viruses 12:210. doi: 10.3390/v12020210
Mayer, M. J., Garefalaki, V., Spoerl, R., Narbad, A., and Meijers, R. (2011). Structure-based modification of a Clostridium difficile-targeting endolysin affects activity and host range. J. Bacteriol. 193, 5477–5486. doi: 10.1128/JB.00439-11
Na, H., Kong, M., and Ryu, S. (2016). Characterization of LysPBC4, a novel Bacillus cereus-specific endolysin of bacteriophage PBC4. FEMS Microbiol. Lett. 363:fnw092. doi: 10.1093/femsle/fnw092
Nazina, T. N., Tourova, T. P., Poltaraus, A. B., Novikova, E. V., Grigoryan, A. A., Ivanova, A. E., et al. (2001). Taxonomic study of aerobic thermophilic bacilli: descriptions of Geobacillus subterraneus gen. nov., sp. nov. and Geobacillus uzenensis sp. nov. from petroleum reservoirs and transfer of Bacillus stearothermophilus, Bacillus thermocatenulatus, Bacillus thermoleovorans, Bacillus kaustophilus, Bacillus thermodenitrificans to Geobacillus as the new combinations G. stearothermophilus, G. th. Int. J. Syst. Evol. Microbiol. 51, 433–446. doi: 10.1099/00207713-51-2-433
Ni, P., Wang, L., Deng, B., Jiu, S., Ma, C., Zhang, C., et al. (2021). Characterization of a Lytic Bacteriophage against Pseudomonas syringae pv. actinidiae and Its Endolysin. Viruses 13:631. doi: 10.3390/v13040631
Oliveira, H., Vilas Boas, D., Mesnage, S., Kluskens, L. D., Lavigne, R., Sillankorva, S., et al. (2016). Structural and enzymatic characterization of ABgp46, a novel phage endolysin with broad anti-gram-negative bacterial activity. Front. Microbiol. 7:208. doi: 10.3389/fmicb.2016.00208
Rabussay, D., Zillig, W., and Herrlich, P. (1970). Characterization of the Bacillus stearothermophilus phage phimu-4 and its DNA. Virology 41, 91–100. doi: 10.1016/0042-6822(70)90057-7
Rahman, M. U., Wang, W., Sun, Q., Shah, J. A., Li, C., Sun, Y., et al. (2021). Endolysin, a promising solution against antimicrobial resistance. Antibiotics (Basel) 10:1277. doi: 10.3390/antibiotics10111277
Ruckert, A., Ronimus, R. S., and Morgan, H. W. (2004). A RAPD-based survey of thermophilic bacilli in milk powders from different countries. Int. J. Food Microbiol. 96, 263–272. doi: 10.1016/j.ijfoodmicro.2004.03.020
Skowron, P. M., Kropinski, A. M., Zebrowska, J., Janus, L., Szemiako, K., Czajkowska, E., et al. (2018). Sequence, genome organization, annotation and proteomics of the thermophilic, 47.7-kb Geobacillus stearothermophilus bacteriophage TP-84 and its classification in the new Tp84virus genus. PLoS One 13:e0195449. doi: 10.1371/journal.pone.0196798
Stone, E., Campbell, K., Grant, I., and Mcauliffe, O. (2019). Understanding and Exploiting Phage-Host Interactions. Viruses 11:567. doi: 10.3390/v11060567
Stothard, P., and Wishart, D. S. (2005). Circular genome visualization and exploration using CGView. Bioinformatics 21, 537–539. doi: 10.1093/bioinformatics/bti054
Swift, S. M., Reid, K. P., Donovan, D. M., and Ramsay, T. G. (2019). Thermophile lytic enzyme fusion proteins that target Clostridium perfringens. Antibiotics (Basel) 8:214. doi: 10.3390/antibiotics8040214
Swift, S. M., Seal, B. S., Garrish, J. K., Oakley, B. B., Hiett, K., Yeh, H. Y., et al. (2015). A thermophilic phage endolysin fusion to a Clostridium perfringens-specific cell wall binding domain creates an anti-clostridium antimicrobial with improved thermostability. Viruses 7, 3019–3034. doi: 10.3390/v7062758
Van Tassell, M. L., Ibarra-Sanchez, L. A., Hoepker, G. P., and Miller, M. J. (2017). Hot topic: antilisterial activity by endolysin PlyP100 in fresh cheese. J. Dairy Sci. 100, 2482–2487. doi: 10.3168/jds.2016-11990
Wells-Bennik, M. H. J., Janssen, P. W. M., Klaus, V., Yang, C., Zwietering, M. H., and Den Besten, H. M. W. (2019). Heat resistance of spores of 18 strains of Geobacillus stearothermophilus and impact of culturing conditions. Int. J. Food Microbiol. 291, 161–172. doi: 10.1016/j.ijfoodmicro.2018.11.005
Zebrowska, J., Zolnierkiewicz, O., Ponikowska, M., Puchalski, M., Krawczun, N., Makowska, J., et al. (2022). Cloning and characterization of a thermostable endolysin of bacteriophage TP-84 as a potential disinfectant and biofilm-removing biological agent. Int. J. Mol. Sci. 23:7612. doi: 10.3390/ijms23147612
Keywords: Geobacillus stearothermophilus, food spoilage, bacteriophage, endolysin, enzymatically active domain
Citation: Choi D and Kong M (2023) LysGR1, a novel thermostable endolysin from Geobacillus stearothermophilus bacteriophage GR1. Front. Microbiol. 14:1178748. doi: 10.3389/fmicb.2023.1178748
Edited by:
Ok Kyung Koo, Chungnam National University, Republic of KoreaReviewed by:
Jaewoo Bai, Seoul Women's University, Republic of KoreaFrank Oechslin, Laval University, Canada
Copyright © 2023 Choi and Kong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Minsuk Kong, a29uZ21pbjFAc2VvdWx0ZWNoLmFjLmty
 Dahee Choi
Dahee Choi Minsuk Kong
Minsuk Kong