
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Microbiol., 22 May 2023
Sec. Infectious Agents and Disease
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1178538
This article is part of the Research TopicInsights in Infectious Agents and Disease: 2022View all 41 articles
The aquatic bacterium Vibrio cholerae is the etiological agent of the diarrheal disease cholera, which has plagued the world for centuries. This pathogen has been the subject of studies in a vast array of fields, from molecular biology to animal models for virulence activity to epidemiological disease transmission modeling. V. cholerae genetics and the activity of virulence genes determine the pathogenic potential of different strains, as well as provide a model for genomic evolution in the natural environment. While animal models for V. cholerae infection have been used for decades, recent advances in this area provide a well-rounded picture of nearly all aspects of V. cholerae interaction with both mammalian and non-mammalian hosts, encompassing colonization dynamics, pathogenesis, immunological responses, and transmission to naïve populations. Microbiome studies have become increasingly common as access and affordability of sequencing has improved, and these studies have revealed key factors in V. cholerae communication and competition with members of the gut microbiota. Despite a wealth of knowledge surrounding V. cholerae, the pathogen remains endemic in numerous countries and causes sporadic outbreaks elsewhere. Public health initiatives aim to prevent cholera outbreaks and provide prompt, effective relief in cases where prevention is not feasible. In this review, we describe recent advancements in cholera research in these areas to provide a more complete illustration of V. cholerae evolution as a microbe and significant global health threat, as well as how researchers are working to improve understanding and minimize impact of this pathogen on vulnerable populations.
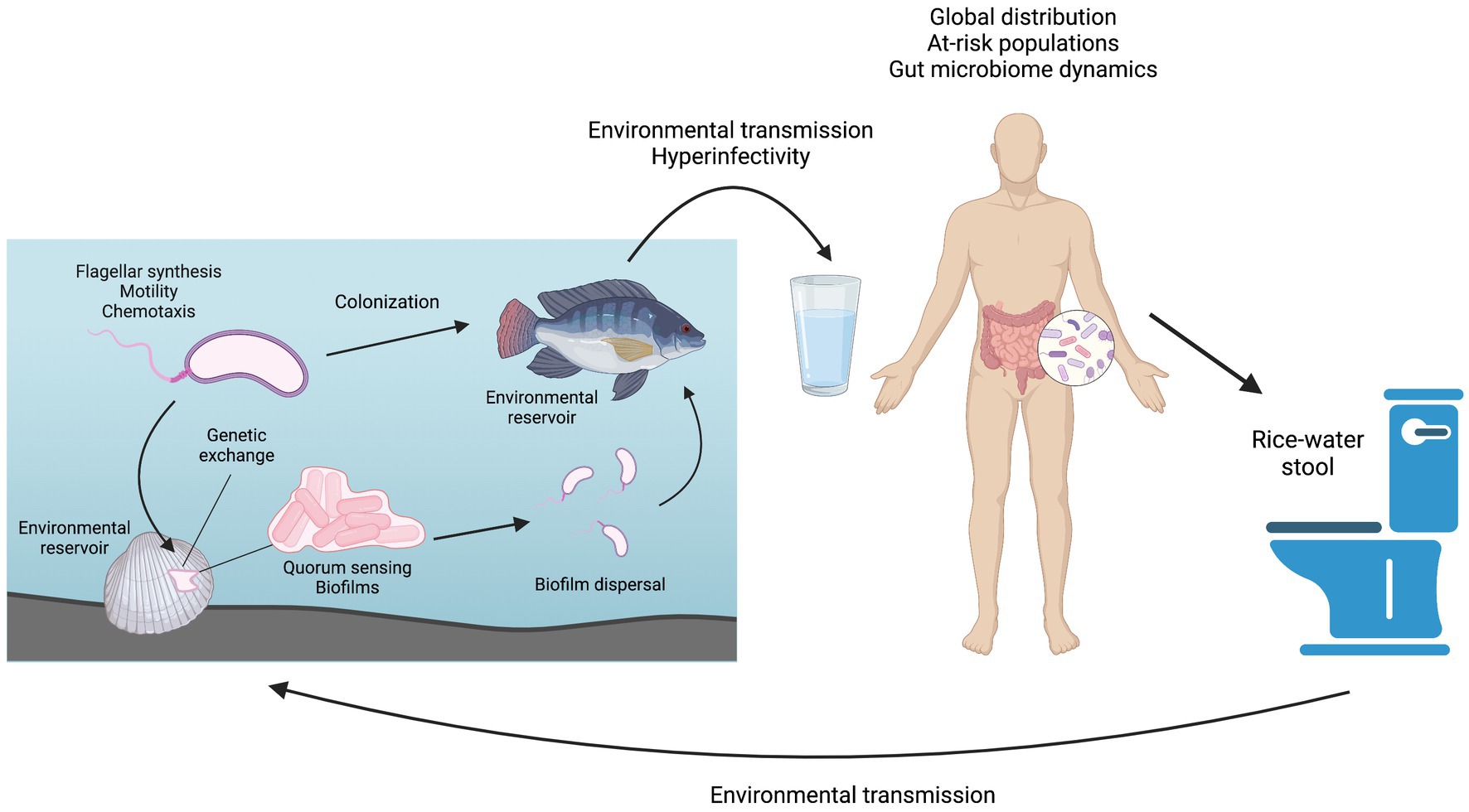
Graphical Abstract. Overview of the Vibrio cholerae life cycle and the topics covered in this review. Figure prepared using BioRender.
Cholera is an acute diarrheal disease with 2.9 million cases and 95,000 deaths estimated to occur each year in at least 47 countries across the world (Ali et al., 2015). The majority of severe cases occur in children under 5 years old. Cholera is often described as a disease of inequity, disproportionately affecting the poorest populations of a country or community (GTFCC, 2017). This longstanding disease has been thoroughly studied in a wide range of research fields from basic science to therapeutics. Mechanisms for prevention, intervention, and possible elimination of cholera have been clearly described and continue to be investigated but lack practical implementation in many vulnerable populations. The etiological agent of cholera is the gram-negative bacterium Vibrio cholerae, specifically those strains belonging to serogroups O1 and O139. Strains belonging to other serogroups may cause less severe non-cholera diarrhea or no disease symptoms at all and are collectively referred to as non-O1/non-O139 strains. V. cholerae is highly motile in aquatic environments, using a single, polar flagellum to propel itself (Echazarreta and Klose, 2019). This bacterium is also readily found in biofilms that form on hard surfaces, i.e., rocks or pipes, as well as in association with shellfish and vertebrate fish (Baine et al., 1974; Silva and Benitez, 2016). Humans can become infected with V. cholerae O1/O139 by consuming contaminated food or water, granting the bacteria access to the small intestine. Here, the bacteria aggregate using the toxin co-regulated pilus (TCP) and other colonization factors to colonize the intestine in a non-invasive manner (Almagro-Moreno et al., 2015; Silva and Benitez, 2016). Once established in the small intestine, the pathogen induces production of cholera toxin (CT), which results in an ion imbalance in the host intestine, leading to the rapid loss of fluids and electrolytes and potentially deadly dehydration via profuse, watery diarrhea (Thiagarajah and Verkman, 2005). Bacteria are disseminated from the intestine with the diarrhea, known as “rice water stool,” of an infected patient and exhibit hypervirulence for a limited time period (Butler et al., 2006). In many cases, cholera-containing fecal matter then contaminates a shared drinking water source, enabling infection with V. cholerae O1/O139 to spread rapidly through an entire community in the form of localized outbreaks (GTFCC, 2017). This massive fluid loss can lead to hypovolemia and can be up to 50% lethal if untreated (Sack et al., 2004). Fortunately, fatality rates drop significantly, to just 1%–2%, with standard treatment using an oral rehydration solution (ORS) (Desjeux et al., 1997). Prompt implementation of ORS therapy counteracts the fluid and electrolyte loss caused by V. cholerae infection and keeps the patient alive while the body naturally clears the infection over a period of several days.
Over the past 200 years, seven pandemics of cholera have been recorded, though instances of cholera-like illness have been described for millennia (Blake, 1994). The long-established nature of this disease has enabled evolutionary differentiation of the pathogen into thousands of strains ranging from environmental strains to those capable of causing endemic and pandemic cholera.
Cholera is estimated to cost $2 billion each year in global healthcare costs and loss of productivity (GTFCC, 2017). As global society becomes increasingly interconnected, cholera is perpetuated by human travel and transmission to naïve populations, lack of adequate infrastructure or disruption of existing infrastructure due to poverty, war, or natural disaster, and shifting weather patterns resulting from climate change. Fortunately, an abundance of research continues to emerge to better describe this pathogen and the disease it causes. This review aims to describe recent advancements in cornerstone cholera research related to genetic evolution of V. cholerae strains, molecular mechanisms of pathogenesis, improvements in animal models, pathogen interactions with the host microbiome and immune response, and disease epidemiology, presentation, and mitigation initiatives.
The Vibrio cholerae genome is approximately 4.0 MB in size and organized into two, distinct circular chromosomes (Trucksis et al., 1998; Heidelberg et al., 2000). Nearly 75% of the genome is contained on chromosome 1 with the remainder on chromosome 2, though a few isolates have been identified that contain a single, fused chromosome and are termed Natural Single Chromosome Vibrio (Chapman et al., 2015; Johnson et al., 2015; Yamamoto et al., 2018; Sozhamannan and Waldminghaus, 2020). While strain-specific differences exist, the entire V. cholerae genome generally contains between 3,600 and 3,900 coding sequences (Heidelberg et al., 2000; Thompson et al., 2011). Chromosome 1 serves as the core genome, encoding most of the essential housekeeping genes and conserved virulence genes. In contrast, chromosome 2 encodes many hypothetical proteins and open reading frames that appear to have been obtained from external sources through horizontal gene transfer events (Heidelberg et al., 2000). While many chromosome 2 genes serve redundant or unknown functions, at least a dozen essential genes have been identified on this chromosome and encode ribosomal proteins L35 and L20 as well as an NAD synthetase and ParA family protein involved in chromosome partitioning (Cameron et al., 2008; Hui et al., 2010; Chao et al., 2013; Kamp et al., 2013). Strains of V. cholerae are classified into serogroups according to the unique structure of the O-antigen associated with the strain’s outer membrane lipopolysaccharide (LPS) molecule. In a recent study, Murase et al. (2022) explored the genetic relatedness of all 210 reported serogroups and identified critical distinctions in structural biosynthesis gene clusters on both chromosomes. While only the O1 and O139 serogroups have been known to cause pandemic cholera, members of the remaining serogroups have had significant impacts as environmental strains. While not typically life-threatening, non-O1/non-O139 serogroups can cause sporadic cases of non-cholera diarrhea, sometimes closely resembling cholera, and some have been shown to act as evolutionary intermediaries in virulence gene acquisition via homologous recombination and horizontal gene transfer (Li et al., 2014, 2019a).
In the environment, V. cholerae is often associated with chitinous surfaces, such as those found on mollusks and other shellfish, and with phytoplankton (Baine et al., 1974; Tamplin et al., 1990; Hounmanou et al., 2019). Vertebrate fish have more recently been proposed as potential V. cholerae reservoirs, as both environmental and toxigenic strains have been isolated from numerous fish species, including tilapia (Oreochromis niloticus) (Senderovich et al., 2010; Halpern and Izhaki, 2017). Zebrafish (Danio rerio) have been shown to be naturally susceptible to infection and colonization by V. cholerae and have been developed as a V. cholerae model (Runft et al., 2014; Mitchell et al., 2017; Mitchell and Withey, 2018; Nag et al., 2018b, 2020). Aquatic reservoirs harboring both environmental and pandemic V. cholerae strains provide rich conditions for genetic evolution, and many non-O1/non-O139 have been found to contain partial pathogenicity islands and fully intact virulence genes, including the Vibrio seventh pandemic islands 1 and 2 (VSP-1, VSP-2), toxin co-regulated pilin (tcpA), hemolysin A (hlyA), and the Type 6 secretion system (T6SS) (Li et al., 2014, 2019a; Yan et al., 2022; Santoriello et al., 2023).
A mixed-transmission dynamic model of V. cholerae developed by Mavian et al. (2020) made use of spatiotemporal V. cholerae distribution following the single-source introduction in Haiti to model the establishment of aquatic reservoirs and the potential for evolutionary gene transfer events. These reservoirs are particularly relevant in cholera-endemic countries where seasonal cholera blooms result in multiple, periodic outbreaks with strains of varying virulence. Ongoing susceptibility to infection despite previous exposure to V. cholerae has been associated with serotype switching—a phenomenon in which V. cholerae O1 serogroup strains can express alternative surface antigens to present as either Ogawa, Inaba, or very rarely Hikojima serotypes—and is likely enabled by gene transfer under selective conditions in environmental reservoirs during off-peak cholera seasons (Baddam et al., 2020; Ramamurthy et al., 2020). Non-O1/non-O139 strains that primarily exist in the environment may use the same virulence genes in a different manner. For example, the recently identified T6SS gene cluster known as Aux3 has been shown to readily excise from the genome and recombine in a new location. However, this activity is typically only observed in environmental strains, while pandemic strains have integrated the feature into the chromosome (Santoriello et al., 2020).
From 1817 until the 1960s, pandemic cholera was caused by the V. cholerae O1 Classical biotype, characterized by the presence of a distinctive CT, TCP, and Vibrio pathogenicity island (VPI). Cholera in the current, ongoing seventh pandemic has been caused by a new O1 biotype, El Tor. El Tor biotype emerged as the primary causative agent of pandemic cholera beginning in 1961 and is defined by its resistance to polymyxin B, production of hemolysin A, and presence of two unique pathogenicity islands, VSP-1 and VSP-2, all of which Classical biotype lacks (Chart, 2012). Several diagnostic methods have been developed to distinguish between Classical and El Tor biotypes in recent years. PCR-based genotypic assays typically screen for specific sequence variations in virulence genes including tcpA, ctxA, ctxB, and toxR, while another genome-based method targets unique small RNA genes (Crumfield et al., 2018; Greig et al., 2018; Ahmed et al., 2019). Biotype can be distinguished phenotypically by evaluating antibiotic and phage susceptibility, capability for hemolysis and proteolysis, and variations in metabolism of citrate and glucose (Crumfield et al., 2018; Lee et al., 2020). One simple diagnostic measure for distinguishing between Classical and El Tor biotype strains in clinical settings has been the susceptibility of Classical strains to polymyxin B while El Tor strains have demonstrated resistance to this antibiotic, though this may not always be a reliable metric as El Tor strains continue to evolve (Crumfield et al., 2018).
When V. cholerae was first introduced to Haiti, it had devastating effects on the population which, initially, was largely attributed to the naivety of the previously unexposed region. While this certainly played a role in the rapid transmission and severe disease observed during this outbreak, Haitian variants of El Tor have been identified as more virulent than their southeastern Asian predecessors inducing elevated levels of inflammation and damage to the intestinal mucosa (Ghosh et al., 2019). These variants have also demonstrated greater production of CT, increased motility, and enhanced colonization dynamics in both human disease and animal host models (Satchell et al., 2016; Ghosh et al., 2019). Interestingly, recent epidemics in India and West Bengal have been caused by El Tor strains that more closely resemble Haitian variant strains in their genetic profile but also exhibit polymyxin B-sensitivity (Samanta et al., 2018; Shaw et al., 2022). These polymyxin B-sensitive strains have also exhibited hypervirulent traits compared to El Tor strains previously isolated in the same regions of southeast Asia (Samanta et al., 2018). These Classical-like features of more recently evolved El Tor isolates will likely require the development of new measures for phenotypic identification of biotype, particularly in resource-limited settings. Additionally, five clinical isolates from Kolkata, India revealed the inability to replicate the cholera toxin phage (CTXΦ) for the secretion of infectious particles by some El Tor variants, a feature that was commonly observed in El Tor strains between the 1970s and early 2010s (Ochi et al., 2021).
With the rapid development and decreasing cost of genome sequencing and analysis in the early 2000s, extensive genomic comparison analyses have described the evolution of the El Tor biotype and its evolutionary successors in detail. Whole-genome studies have revealed the similarities and differences in key virulence factors, including the CTXΦ, and have used these data to characterize the evolution and spread of El Tor variants into three waves (Kim et al., 2014). Hu et al. (2016) detailed key genomic events between the early 1900s and the 1960s that enabled El Tor’s maturation from a relatively benign form of V. cholerae to the virulent pathogen credited with causing the ongoing 7th pandemic of cholera. These events included the acquisition of an El Tor-specific tcpA gene that enabled human colonization, pathogenicity islands VSP-1 and VSP-2, and an El Tor-specific CTXΦ. Since this initial characterization of the lineage leading to the 7th pandemic by El Tor, others have explored genetic variation within El Tor strains to assess virulence trait acquisition. Large genomic fragments carrying genes for antimicrobial resistance, including the integrative and conjugative element (ICE) known as the SXT element, have been acquired by some El Tor strains isolated from the natural environment, and afford resistance to tetracycline, streptomycin, and even chloramphenicol (Ahmed et al., 2005; Sarkar et al., 2019).
An analysis of over 300 V. cholerae O1 strains revealed El Tor strains typically contained more virulence-related genes than Classical strains, as well as the presence of many redundant genes across the El Tor genome (Li et al., 2019b). While V. cholerae readily takes up DNA from the environment, defense mechanisms have also been developed to prevent the acquisition of unwanted or detrimental features. Both small, multicopy- and large, low copy number plasmids can be degraded by defense mechanisms identified as DdmABC and DdmDE should they prove to be detrimental to overall fitness of the V. cholerae host cell (Jaskólska et al., 2022). Phage defense in El Tor strains has been attributed in part to activity of genes on the VSP-1 and VSP-2 pathogenicity islands, including an antiviral cytidine deaminase which disrupts normal availability of nucleotides to deprive infecting phage of necessary components to replicate (Hsueh et al., 2022; O’Hara et al., 2022).
The natural habitat of V. cholerae in biofilms is often composed of richly diverse communities of Vibrio species and other aquatic bacteria. Some of the species present in these biofilms are capable of natural competence and can release significant amounts of DNA (>100 μg/ml) into the environment. Biofilms readily form on chitinous surfaces, a biopolymer that has been shown to induce natural competence in some V. cholerae strains (Baur et al., 1996; Meibom et al., 2005). Additionally, V. cholerae killing of non-kin bacterial competitors mediated by the type six secretion system (T6SS) enables the uptake of large DNA fragments (>150 Kbp) following lysis of the target cell (Joshi et al., 2017; Matthey et al., 2019). Exposure to high concentrations of free DNA in these chitin-rich conditions enables the rapid genomic diversification of V. cholerae which is modeled in the evolution of El Tor strains.
In the aquatic environment, V. cholerae does not produce the human-specific virulence factors that are required to cause cholera. After ingestion by a human, the bacteria sense signals in the gut that initiate a complex cascade of transcription factors that ultimately induce production of the major virulence factors, CT and TCP, together with a collection of other accessory virulence factors (Matson et al., 2007). At the top of the cascade are transcription factors AphA and AphB (Kovacikova and Skorupski, 1999; Skorupski and Taylor, 1999; Kovacikova and Skorupski, 2001; Kovacikova et al., 2004). AphA is translated at low cell density, and its genomic targets were recently described using CHiP-Seq (Haycocks et al., 2019). AphB senses low oxygen and low pH and becomes active as V. cholerae passes through the stomach and into the upper small intestine (Kovacikova et al., 2010). AphA and AphB work together to activate transcription of the next level of the cascade, which is composed of the TcpPH and ToxRS pairs of integral membrane proteins. While ToxRS is thought to be constitutively produced, TcpPH production requires the activity of AphA and AphB. When produced, TcpPH senses the bile salt taurocholate (Yang et al., 2013; Xue et al., 2016), whereas ToxRS senses other bile salts in the intestinal lumen (Midgett et al., 2017; Bina et al., 2021). TcpP and ToxR then bind directly to the promoter of the master virulence regulator, ToxT, and activate its production (Krukonis et al., 2000; Krukonis and DiRita, 2003; Goss et al., 2010; Fan et al., 2014; Haas et al., 2015). ToxT binds directly to the promoters upstream of ctxAB and tcpA and activates their transcription. However, ToxT activity is repressed by unsaturated fatty acid components of bile to prevent virulence factor production in the lumen of the small intestine (Chatterjee et al., 2007; Koestler and Waters, 2014; Plecha and Withey, 2015). As motile V. cholerae enters the mucus layer, the large fatty acids cannot penetrate, and ToxT becomes activated by the presence of bicarbonate, which is secreted by epithelial cells (Abuaita and Withey, 2009). Unsaturated fatty acids and bicarbonate have opposing roles in affecting the affinity of ToxT for its DNA binding sites (Withey and DiRita, 2006; Thomson and Withey, 2014; Plecha and Withey, 2015; Thomson et al., 2015). Thus, CT and TCP production only occurs when V. cholerae has reached the ideal location for colonization, within the intestinal mucus layer and close to the epithelial surface in crypts, where CT can enter cells and play its toxic role, resulting in voluminous watery diarrhea (Millet et al., 2014).
Vibrio cholerae has two distinct phases in its lifecycle: the highly motile, free-swimming state, and the sessile, virulent state. Motility is important in an aqueous environment, while attachment and biofilm formation is necessary for colonization in the human small intestine or on the surfaces of fish, plankton, and other chitinous material (Tamplin et al., 1990; Donlan and Costerton, 2002; Senderovich et al., 2010; Hathroubi et al., 2017). Responding to environmental signaling is crucial for V. cholerae survival, and robust methods of inverse regulation over virulence factors and motility are required.
In an aqueous environment, V. cholerae is highly motile due to a single, sheathed, polar flagellum. The flagellum is powered by a protein complex in the membrane, called the motor, that uses the transmembrane sodium motive force to generate torque (Terashima et al., 2008; Halang et al., 2013). This causes rotational movements that can propel V. cholerae up to 60 cell-body lengths per second (McCarter, 2001). Other movements include twitching motility, which is dependent on pili, and gliding motility, which is independent of pili or flagella (Mattick, 2002; Nan and Zusman, 2011). This can be utilized by V. cholerae for moving in different media or for adherence (Butler and Camilli, 2005). The V. cholerae flagellum is being explored for its role in colonization and pathogenicity. Expression of major V. cholerae virulence factors have long been known to be inversely regulated with expression of flagellar genes, and V. cholerae that are actively colonizing intestinal epithelium typically do not have flagella.
The V. cholerae flagellum has recently been shown to secrete MakA, a motility-associated toxin. From the same gene cluster, proteins MakA, MakB, and MakE can form a tripartite cytolytic toxin in vitro, via membrane binding and assembly of a pore (Nadeem et al., 2021). The activation or presence of a flagellum can also influence biofilm development in an inverse manner. V. cholerae mutants that lacked a functional flagellum formed colonies with a morphological switch to a rugose colony, which is associated with expression of extracellular polysaccharides similar to biofilms (Echazarreta and Klose, 2019).
Movement of the V. cholerae flagellum is another area of study. The flagellum is perpetually rotating, and sodium concentration of the environment directly affects swimming speed (Halang et al., 2013; Grognot et al., 2021). V. cholerae is able to perform different swim patterns that result in either a more random dispersal or a more targeted movement near surfaces. This may be an advantage, perhaps in the event of chemotaxis, outcompeting other bacteria, or to find a viable surface for attachment (Utada et al., 2014; Grognot et al., 2021). V. cholerae has most chemotaxis genes organized into 3 operons, which allow motile V. cholerae to adjust its direction according to environmental signals (Heidelberg et al., 2000; Butler and Camilli, 2004) Similarly, non-chemotactic V. cholerae El Tor biotype mutants outcompeted wild type strains in the infant mouse small intestine, indicating that chemotaxis significantly inhibits colonization overall. However, colonization was localized aberrantly (Butler and Camilli, 2005). In particular, smooth swimming is crucial to competition in the small intestine of infant mice (Butler and Camilli, 2004). Chemotaxis is also relevant to differentiate among surfaces. Valiente et al. identified an accessory colonization gene in V. cholerae O1 El Tor, acfC, that encodes a methyl-accepting chemotaxis protein. In vitro, AcfC induced chemotaxis towards intestinal mucin but not chitin (Valiente et al., 2018). In the environment, V. cholerae uses the flagellum to swim to chitinous surfaces, and attaches irreversibly with the flagellum and mannose-sensitive hemagglutinin (MSHA) type IV pili (Utada et al., 2014). The second messenger cyclic di-GMP (c-di-GMP) is crucial to how V. cholerae responds to the environment, affecting the concentration and binding of transcriptional regulators (Conner et al., 2017; Homma and Kojima, 2022). CsrA is an RNA-binding protein that regulates c-di-GMP metabolism, which inversely regulates flagellar gene expression in V. cholerae, and directly regulates virulence gene expression (Tischler and Camilli, 2005; Jonas et al., 2008; Butz et al., 2021). Low levels of c-di-GMP promote FlrA, which is required for flagellar gene expression and motility (Yildiz and Visick, 2009; Conner et al., 2017). High levels of c-di-GMP repress motility and virulence, and activate biofilm matrix production, as CsrA controls polysaccharide production depending on the V. cholerae growth phase (Hunter et al., 2014; Conner et al., 2017; Echazarreta and Klose, 2019).
Once attached to a biotic or an abiotic surface, V. cholerae switches from a free-swimming planktonic bacterium to form aggregates. This transition initiates colonization. Depending on cell density, bacteria can communicate in a cell-to-cell manner to coordinate responses to the environment via a process called quorum sensing (Papenfort and Bassler, 2016). This relies on the secretion and detection of diffusible signaling molecules called autoinducers.
In V. cholerae, there are four histidine kinases that act as redundant quorum sensing receptors, LuxPQ, CqsS, CqsR and VpsS (Watve et al., 2020). Four synthases, LuxM, LuxS, Cqs, and Tdh, produce autoinducers AI-1, AI-2, CAI-1, and DPO (Mukherjee and Bassler, 2019). At a low cell density, autoinducer concentration is low, and virulence genes are expressed (Watve et al., 2020). A phosphorylation cascade activates production of AphA, a transcription factor at the top of the complex virulence regulatory cascade (Haycocks et al., 2019; Mukherjee and Bassler, 2019; Mashruwala and Bassler, 2020). At high cell density, the autoinducer concentration is high, which promotes cell-to-cell coordination (Mashruwala and Bassler, 2020). In V. cholerae, the quorum sensing regulator HapR is produced to repress AphA synthesis and reduce virulence gene expression, as has been demonstrated in Drosophila and other models (Rutherford et al., 2011). HapR also represses expression of Vibrio polysaccharide (VPS) matrix enzymes, as demonstrated in a Drosophila host (Zhu et al., 2002; Vance et al., 2003; Kamareddine et al., 2018). Judger et al. suggests that quorum sensing activates HapR to repress tryptophan uptake, which cascades to activate a commensal relationship from the Drosophila host innate immune system via serotonin production (Jugder et al., 2022).
The presence or absence of autoinducers can indicate environmental changes. CAI-1 is used for intra-genus communication, and AI-2 and DPO are used for inter-species communication. In anaerobic conditions, V. cholerae produces DPO but not CAI-1, while the opposite is true in aerobic conditions (Mashruwala and Bassler, 2020). This signaling is relevant to a free-swimming lifestyle in water, versus the anaerobic conditions of the human small intestine, for example.
The presence of mucin or chitin causes V. cholerae to grow outward from the founder cell, with the resulting mechanical pressure causing surface adhesion (Valiente et al., 2018; Yan and Bassler, 2019). High cell density induces high levels of c-di-GMP, which repress flagellar genes. VpsR and VpsT then activate to form a surface-associated aggregation called a biofilm (Hunter et al., 2014; Silva and Benitez, 2016; Conner et al., 2017). Biofilm cells are in a three-dimensional matrix made of polysaccharides, proteins, and extracellular DNA. This protective matrix allows for surface adhesion, enzyme proximity for metabolism, and potential horizontal gene transfer (Flemming and Wingender, 2010). V. cholerae in biofilm has also been shown to be hyper-infectious when compared to free-swimming V. cholerae (Tamayo et al., 2010). Human stool from confirmed cholera cases has both planktonic bacteria and clumped bacteria, indicating that aggregation or biofilm-like behavior may be occurring in the human gut (Faruque et al., 2006; Nelson et al., 2007).
A redundant set of four proteases control the timing of V. cholerae to either form a scaffold or use motility to aggregate into the scaffold (Jemielita et al., 2018). This further illuminates how V. cholerae persists in the water and how it colonizes the human small intestine. By culturing V. cholerae on different surfaces and comparing the subsequent biofilms to free-swimming cells when infecting infant mouse intestine, biofilm cells enhanced expression of virulence factors including the TCP (Gallego-Hernandez et al., 2020). The TCP is a major V. cholerae virulence factor required for human colonization (Kaper et al., 1995; Thelin and Taylor, 1996; Teschler et al., 2015).
Biofilm formation can also depend on extracellular components and certain proteases. In one study, extracellular signaling molecule accumulation was prevented via a continuously refreshed system. This identified a difference in biofilm gene regulation in V. cholerae, depending on dynamic versus static conditions (Seper et al., 2014). Extracellular nucleases were also found to regulate the amount of extracellular DNA involved in both developing and dispersing biofilm (Seper et al., 2011). These nucleases were also necessary for hyper-infectivity from biofilm-formed V. cholerae. Kitts et al. identified a calcium-dependent protease, LapG, which cleaves adhesins FrhA and CraA. A V. cholerae mutant lacking LapG increased the amount of biofilm formed, indicating the protease mediation in biofilm formation (Kitts et al., 2019).
Through mutagenesis, proteins from three classes of genes were identified to play a sequential role in biofilm dispersal: signal-transduction proteins, matrix-degradation enzymes, and motility factors (Bridges and Bassler, 2021). Dispersal is the last step following the sessile, biofilm phase in the V. cholerae life cycle. Mechanical changes in the environment such as flow rate can regulate V. cholerae biofilm production or dispersal. In nutrient medium with an increased flow rate, biofilm mass increased. If the flow rate was slowed or stopped, V. cholerae within biofilms dispersed from the outer surface (Singh et al., 2017).
Access to soluble nutrients also determines dispersal. RpoS protein, an alternate sigma factor, regulates a generalized stress response during starvation and is essential for V. cholerae to detach from mucus when it reaches a high cell density (Nielsen et al., 2006; Müller et al., 2007). During the stressor of low nutrient conditions, elevated RpoS was detected (Singh et al., 2017). Species-specific polyamines are also involved in biofilm development and repression (Lee et al., 2009). Periplasmic polyamine sensor MbaA, and polyamine reporter PotD1 regulate V. cholerae biofilm dispersal, with PotD1 being essential to activating dispersal (Bridges and Bassler, 2021). Excreted V. cholerae from humans, mice, or other animals has shown hyper-infectivity during the transmission to the next host. Detachment and dispersal from the biofilm into the environment are also linked to the hyper-infectivity of V. cholerae during transmission to the host from the environment (Alam et al., 2005; Butler et al., 2006; Tamayo et al., 2010).
Animal models for cholera research can be divided into two categories: mammalian models and non-mammalian models. These models, their uses, and their advantages and disadvantages are summarized in Table 1. The most commonly used mammalian model for enteric diseases generally is the specific-pathogen free (SPF) adult mouse (Mus musculus), but these hosts can be resistant to intestinal colonization by some strains of orally inoculated V. cholerae, due in part to the presence of gut microbiota (Sit et al., 2019). The streptomycin-treated adult mouse is sometimes used as a model for V. cholerae colonization. However, in this model V. cholerae colonizes the colon but not the small intestine (SI), where V. cholerae colonizes during human infection. This model is TCP-independent, and there are no detectable disease symptoms (Nygren et al., 2009; Wang et al., 2018). Pretreatment with clindamycin or ketamine to disrupt intestinal microbiota and motility can permit oral V. cholerae infection in the adult mouse (Olivier et al., 2009; You et al., 2019). Clindamycin is used as an antibiotic, like streptomycin, to clear the gut microbiota, and ketamine is a dissociative anesthetic that can be used to slow down the bowel movement and give V. cholerae a better chance to colonize. The most widely used mammalian model for V. cholerae colonization is the 3- to 5-day-old infant mouse, where oral challenge of V. cholerae can initiate TCP-dependent colonization of the SI (not the colon) and CT-dependent fluid accumulation within 16 h (Angelichio et al., 1999; Ritchie and Waldor, 2009). Advanced microscopy of the infant mouse small intestinal tissue has revealed the localization patterns of V. cholerae in the intestine control virulence gene regulation during infection (Millet et al., 2014; Gallego-Hernandez et al., 2020). Different competition studies in the infant mouse model have recently emphasized the contributions of fatty acid and carbon metabolism, cell wall maintenance, and V. cholerae LPS modifications during intestinal colonization (Hayes et al., 2017; Wang et al., 2018; Fleurie et al., 2019; Yang et al., 2020; Sit et al., 2021).
Two adult rabbit surgical models have been widely used for decades to test the colonization and fluid secretion resulting from V. cholerae infection. The removable intestinal tie adult rabbit diarrhea (RITARD) model is used for studying the toxin-mediated diarrheal disease caused by V. cholerae (Spira et al., 1981; Sinha et al., 2015). The ileal loop model is a widely used surgical model to study the fluid accumulation following by V. cholerae colonization by surgically creating sealed loops in the small intestine of the adult rabbit (Burrows and Musteikis, 1966; Mondal et al., 2014; Withey et al., 2015). These adult rabbit surgical models have largely been replaced by the infant rabbit (Oryctolagus cuniculus) model of cholera, which exhibits rapid CT-dependent lethal diarrheal illness, along with TCP-dependent SI colonization (Ritchie et al., 2010). High numbers of V. cholerae can be readily collected from the infant rabbit diarrheal fluid, which is quite similar to human rice water stool in chemical composition (Ritchie et al., 2010). Infant rabbit studies have revealed the genetic landscape of colonization factors in V. cholerae through transposon-insertion sequencing (Tn-Seq) screens; however, these screens can be largely limited by bottleneck effects in infant mice (Fu et al., 2013; Kamp et al., 2013; Pritchard et al., 2014; Hubbard et al., 2018). V. cholerae RNA-seq, metabolomic, and proteomic datasets, as well as insights into V. cholerae population dynamics during infection, have been successfully studied in the infant rabbit model (Mandlik et al., 2011; Abel et al., 2015; Zoued et al., 2021). Recently, a transcriptomic study in infant rabbits revealed novel roles for CT in the shaping of the pathogen’s nutritional microenvironment (Rivera-Chavez and Mekalanos, 2019).
There are several non-mammalian species for studying V. cholerae as well, such as fruit flies (Drosophila melanogaster), nematodes (Caenorhabditis elegans), wax moths (Galleria mellonella), and zebrafish (Danio rerio). The intestinal anatomy and immune system of Drosophila is similar to mammals, enabling the use of flies to investigate pathogenicity, quorum sensing, and host response to V. cholerae infection (Hang et al., 2014; Vanhove et al., 2017; Kamareddine et al., 2018; Davoodi and Foley, 2019). The introduction of CT can induce death in flies although the infection and host-killing by V. cholerae in these models is neither CT-dependent nor TCP-dependent (Blow et al., 2005). Even with these limitations, this model is useful as it offers reproducibility, low cost, and easy genetic manipulation compared to mammalian hosts. Cholera research in C. elegans is important for studying the known and presumed accessory V. cholerae virulence factors and toxins (Vaitkevicius et al., 2006; Logan et al., 2018). The ability of V. cholerae to kill G. mellonella larvae and form biofilms in these hosts imitates the pathogenic potential with mammalian model organisms (Nuidate et al., 2016; Bokhari et al., 2017).
Vibrio cholerae persists in the aquatic environment between outbreaks where they fight with different predators and environmental stressors. Some models were developed to study the environmental survival of V. cholerae. The mannose-sensitive hemagglutinin (MSHA) type IV pilus is crucial for attachment and initiation of colonization of V. cholerae in the pharynx of the worm, C. elegans, which could be linked to a fitness advantage of V. cholerae upon contact with bacterium-grazing nematodes (List et al., 2018). The soft-shelled turtle has potential value as an animal model to study the colonization and the transmission of V. cholerae as V. cholerae can colonize both on the dorsal side surface and in the intestine of turtles. MSHA is necessary for body surface colonization whereas, toxin-coregulated pili (TCP) or N-acetylglucosamine-binding protein A (GbpA) play important roles for colonization in the intestine (Wang et al., 2017). The type VI secretion system (T6SS) of V. cholerae is capable of conferring virulence toward eukaryotic and prokaryotic hosts. VasX, a functional protein for T6SS confers the virulence of V. cholerae toward Dictyostelium discoideum, a species of soil-dwelling amoeba commonly known as slime mold (MacIntyre et al., 2010; Miyata et al., 2011). These models are also very important to demonstrate the lifestyle of aquatic organisms in the presence of the aquatic bacterium, V. cholerae.
Zebrafish (D. rerio) are an emerging non-mammalian model to investigate V. cholerae pathogenesis, including colonization, transmission, host response, and competition with intestinal microbiota. The zebrafish is a natural host for V. cholerae which means V. cholerae can colonize the zebrafish intestine without any modification to the host’s microbiota (Runft et al., 2014). V. cholerae infection can be initiated through a natural, oral route in zebrafish, which eventually leads to human cholera-like symptoms including liquid stool in excreted water (cloudy water) and secretion of mucin and protein in excreted water within 24 h of initial exposure (Runft et al., 2014; Mitchell et al., 2017; Nag et al., 2018b). Zebrafish also show promise to study innate and adaptive immune responses during V. cholerae infection (Farr et al., 2021, 2022). Studies in the zebrafish model have been informative regarding V. cholerae’s interaction with gut microbiota and the involvement of T6SS in this interaction (Logan et al., 2018; Breen et al., 2021b). However, as most non-mammalian studies remain to be validated in either human or small mammal cholera models, their applicability to our understanding of human cholera may be limited (Sit et al., 2022).
The human gut microbiome contains the majority of commensal bacteria in the body (Qin et al., 2010; Sender et al., 2016). In humans, the dominant phyla are Firmicutes and Bacteroidetes, though the composition of the gut microbiome can vary among individuals. Intrinsic factors, including host genetics, age, and sex, and extrinsic factors, such as diet and lifestyle, all affect the gut microbiome (Arumugam et al., 2011; Faith et al., 2011; Schmidt et al., 2018). The mucus lining of the gut is also an important component. The mucus layer acts as a natural barrier to protect the intestinal epithelium. It is primarily made of heavily-glycosylated proteins called mucins that are metabolized by the resident bacteria that reside in the intestinal mucus layer. Therefore, the mucus itself must be present to maintain a diverse microbiome (Kashyap et al., 2013; Li et al., 2015; Sicard et al., 2017).
In terms of microbiome models to study V. cholerae outside of human fecal samples, experimentation is limited. Mammalian animal models commonly used for V. cholerae infection are not ideal for observing microbiome dynamics after colonization. Infant and adult mice, or the adult rabbits used in ileal loop or RITARD models, lack a complex gut microbiome due to age, antibiotic use for colonization, or surgery, respectively (Sawasvirojwong et al., 2013; Matson, 2018). One option is a human or mouse fecal transplant into axenic or gnotobiotic mice to observe V. cholerae colonization in the presence of a “humanized” mouse microbiome (You et al., 2019; Alavi et al., 2020). Non-mammalian animal models that are colonized by V. cholerae present an advantage due to the fecundity and shorter development period. The Drosophila model possesses a simple gut microbiome of low diversity that can be easily manipulated (Wong et al., 2011). The zebrafish model has the advantage of being a natural V. cholerae host, and zebrafish are able to keep complex intestinal microbiomes largely intact throughout the entire period of V. cholerae exposure, infection, and clearance; the noninvasive method of infection is particularly relevant (Senderovich et al., 2010; Wong et al., 2011; Runft et al., 2014; Breen et al., 2021a). The affordability of high-throughput sequencing of the 16S ribosomal subunit has allowed for more in-depth microbiome studies within the past decade; therefore, the diversity and dynamics of gut microbiomes in relation to pathogens and general perturbation is of growing interest (Gibbons and Gilbert, 2015).
Depending on the biotype or strain, V. cholerae can utilize multiple defenses against the gut microbiome. All V. cholerae strains have the type six secretion system (T6SS) which, if functional, directly injects toxic effectors into eukaryotic cells and bacteria (Bachmann et al., 2015). The T6SS is one method V. cholerae uses to interact with the host and its resident microbiota. The bacterial dynamics can be complex. In the Drosophila model, V. cholerae with a functional T6SS inhibited host intestinal repair only when three common fly commensals were present altogether, rather than individually (Fast et al., 2020). In infant mice, V. cholerae expressing T6SS were able to compete against commensal Escherichia coli, with the E. coli demonstrating a 300-fold drop in CFU count per small intestine homogenate when compared to the group infected with V. cholerae expressing a defective T6SS (Zhao et al., 2018). This suggests that T6SS plays a significant role in colonization of the murine gut. Conversely, T6SS is not necessary for V. cholerae colonization of the conventionally-raised adult zebrafish gut. However, when comparing fish infected with T6SS-deleted V. cholerae versus wildtype, the amount of other Vibrio spp. increased. Perhaps V. cholerae uses T6SS as a form of competition to prevent commensal Vibrio species from proliferating (Unterweger et al., 2014; Breen et al., 2021b). In gnotobiotic zebrafish larvae, the T6SS of V. cholerae O1 El Tor biotype strain promoted an increase in intestinal movement to expel the inoculated commensal, Aeromonas veronii. This clearance then allowed for V. cholerae colonization (Logan et al., 2018).
Vibrio cholerae can interact with specific bacterial species of the microbiome to potentially improve its own colonization. In one study using predictive taxa from an algorithm analyzing fecal samples from Bangladeshi household contacts of cholera patients, certain bacterial species present were selected for in vitro study. V. cholerae was then exposed to these species and grown in nutrient-poor or nutrient-rich culture. V. cholerae growth with P. aminovorans in nutrient-rich culture was significantly increased when compared to growth with other species identified from the collected fecal samples (Midani et al., 2018). This data, albeit in vitro, could further support the idea that interactions with certain members of the microbiome are advantageous for V. cholerae.
Vibrio cholerae can also use gut components to compete against other bacteria. One survival study loaded V. cholerae with either a mucin or a gelatin control, and compared the recovered bacterial load from each condition. An O1 strain of V. cholerae, C6706, normally represses its T6SS in laboratory settings. In the presence of mucin, C6706 activated its T6SS to compete against other strains of V. cholerae that maintained an inactivated T6SS. These results did not occur in the gelatin control. This indicates a component of the mucin was necessary to activate a functional T6SS in C6706 (Bachmann et al., 2015).
Once V. cholerae reaches the upper small intestine in humans, it comes into contact with the relatively sparse duodenal microbiota. Fecal sample culture studies characterized the microbiome composition during and after diarrheal symptoms. The gut mucosa sloughs off, resulting in the characteristic rice-water stool. The immediate effect is a drastic decrease in diversity, primarily due to the physical efflux of the mucosa containing the resident gut microbes. Stool collected from Bangladeshi adults during the acute phase of diarrheal symptoms showed dominance of V. cholerae bacteria and a significant decrease in non-Vibrio bacteria (Hsiao et al., 2014; David et al., 2015). Hsiao et al. (2014) also detected 343 bacterial species that colonized the human gut during and after diarrheal symptoms, suggesting some bacteria are able to remain and recolonize following V. cholerae infection. Hours after oral rehydration therapy (ORS) was started, Streptococcus and Fusobacterium species dominated the microbiome. This overall composition was significantly different from the healthy adult microbiome controls. One caveat of human fecal studies is that V. cholerae colonizes the upper SI and not the colon, whereas most bacteria harvested in fecal samples are from the colon, so direct effects of V. cholerae on the SI microbiome are difficult to assess.
Effects of V. cholerae infection on the composition of the intestinal microbiome are also observed in the zebrafish model. Depending on the strain of V. cholerae used and whether a functional T6SS was present or absent, the microbiome profile of adult zebrafish was transiently changed following infection (Breen et al., 2021b). Using quantitative PCR (qPCR), some V. cholerae strains were also determined to cause an increase in overall bacterial load of the fish intestine.
The gut has a variety of defenses that can work against non-commensals. One important factor in the human intestine is bile, with bile acid being one major component used in digestion to solubilize lipids. Resident gut bacteria can metabolize these bile acids via bile salt hydrolases (Hung and Mekalanos, 2005; Song et al., 2019). This bile acid deconjugation can inhibit V. cholerae T6SS expression, perhaps due to a carboxylic acid group present in a bile salt, although the mechanism of action is not known (Bachmann et al., 2015). The resident gut microbiome can also produce components against non-commensal bacteria. Two human commensal Bifidobacterium species and a commensal B. subtile species were able to individually inhibit or decrease V. cholerae T6SS-mediated killing (Bachmann et al., 2015).
Another gut commensal, E. coli, can produce a genotoxin called colibactin, which is commonly associated with damaging DNA in host epithelial cells (Dougherty and Jobin, 2021). Through transposon mutagenesis, Chen et al. identified three E. coli mutants from mouse small intestine that were unable to compete against V. cholerae. These mutants all had a disruption of the polyketide synthase island, which encodes colobactin. This carried over to analyzing published shotgun metagenomics sequencing of fecal samples from households with a confirmed cholera case. When compared to asymptomatic or uninfected fecal samples, those collected from symptomatic individuals had significantly lower relative abundance reads of clb, a synthase involved in activating colibactin synthesis (Chen et al., 2022). Probiotic E. coli strains have also been found to be protective against V. cholerae infection using the zebrafish model (Nag et al., 2018a).
Blautia obeum, of the core human gut phyla Firmicutes, may also play a role in a V. cholerae infection. B. obeum produces the DPO autoinducer, which activates a regulatory cascade that may inhibit V. cholerae biofilm formation and toxin production (Papenfort et al., 2017). DPO can also inhibit AphA, a transcription factor that regulates virulence gene expression in V. cholerae (Herzog et al., 2019). In homogenized suckling mouse intestine colonized with a reporter V. cholerae and B. obeum, expression of TcpA, the primary structural subunit of the toxin co-regulated pilus (TCP) required for colonization, was significantly reduced. Conversely, in human fecal samples, a higher abundance of V. cholerae was associated with a lower amount of B. obeum (Alavi et al., 2020).
The complexity of the human gut microbiome is difficult to standardize, not only in composition but in computation. Variation in bioinformatics analysis can lead to different conclusions. The more common method of clustering 16S rRNA sequences based on 97% identity generates operational taxonomic units (OTUs). This analysis uses the Mothur pipeline as a clustering method. A newer denoising method, DADA2, generates amplicon sequence variants (ASVs) that provide higher sensitivity and variation specificity. These methods used on 16S rRNA gene amplicon datasets can lead to different results in taxonomic assignments, alpha diversity, and beta diversity than the OTU method on the same dataset (Prodan et al., 2020; Straub et al., 2020; Chiarello et al., 2022). Low abundance OTUs and ASVs can also skew the data, which may be relevant during the decreased gut microbiota following mucosal efflux (Prodan et al., 2020). Additionally, the widely used method of 16S rRNA gene sequencing is not reliably species-specific. This can pose a problem in gut microbiome identification, as this microbiome is composed of an estimated 1013 microbial cells with thousands of bacterial species (Luckey, 1972; Leviatan et al., 2022).
As previously mentioned, V. cholerae strains of the O1 serogroup have been subdivided into two biotypes: Classical and El Tor. Though rarely occurring now, Classical infections were characterized by robust action of the CT and severe diarrheal symptoms lasting about 3 days (Hu et al., 2016). Initial infections with the El Tor biotype were mild, often failing to result in any cholera symptoms, but these infections were persistent, with V. cholerae isolates found in the stool of infected individuals for 1–2 weeks. The binding subunit of CT encoded by early El Tor strains differs structurally by 2 amino acids compared to the Classical CT which is hypothesized to account for the disparity in disease severity exhibited by Classical and El Tor infections (Raychoudhuri et al., 2009; Baek et al., 2020). Acquisition of the Classical CTXΦ by El Tor in the early 2000s led to increased disease severity comparable to that caused by Classical strains and of prolonged duration as was characteristic of early El Tor strains (Na-Ubol et al., 2011; Hu et al., 2016). Previous exposure to V. cholerae causing symptomatic cholera provides protective immunity against subsequent exposure, though cross-protection is not generally achieved for other serotypes or in cases where initial exposure did not produce symptoms (Leung and Matrajt, 2021).
Vibrio cholerae O1 outbreaks largely occur in areas with poor sanitation, limited infrastructure, and minimal or no access to safe drinking water (GTFCC, 2017). Nearly all countries with high cholera burdens also suffer below-average access to basic water and sanitation services (GTFCC, 2017; WHO and UNICEF, 2017). Latin America and the Caribbean, Eastern, Southeastern, Western, Central, and Southern Asia, Northern Africa and Sub-Saharan Africa, and Oceania are all regions identified as lacking a basic drinking water service and/or sanitation services as of 2015 (WHO and UNICEF, 2017). Modelling estimates have revealed 69 countries with endemic cholera—defined as having predicted cholera cases in at least 3 years of a 5-year study period—including, but not limited to, Nepal, China, Indonesia, and several countries in Sub-Saharan Africa (Ali et al., 2015). Additionally, India, Ethiopia, Nigeria, Haiti, the Democratic Republic of the Congo, Tanzania, Kenya, and Bangladesh were identified as having >100,000 annual cases in the same study. In 2016, 80% of all reported cases were located in Haiti, the Democratic Republic of the Congo (DRC), Yemen, Somalia, and the United Republic of Tanzania (WHO, 2017). It is important to note that the vast majority of cholera cases are not reported for many reasons, including lack of laboratory diagnostic capabilities or reporting systems, fear of criticism due to a lack of proper infrastructure, and concern for disrupting trade partnerships or tourism initiatives. A review published in 2020 revealed estimated case numbers in the cholera-endemic countries of India, Pakistan, and the Philippines were 5.8-, 6.7-, and 20.8 times greater than the actual reported number of cases, respectively (Ganesan et al., 2020). The use of modeling to predict cholera outbreaks and regions at heightened risk is a truly noteworthy advancement for epidemiological studies and surveillance efforts (Allan et al., 2016; Ali et al., 2017; Camacho et al., 2018). Endemic cholera regions experience cholera “blooms,” or periods during which the number of V. cholerae infections increase dramatically in response to seasonal blooming of zooplankton and phytoplankton which feed copepods that support V. cholerae abundance in the environment (Epstein, 1993; Constantin de Magny and Colwell, 2009). This series of events often coincides with weather patterns such as monsoon season in the Bay of Bengal region (Lobitz et al., 2000). Several studies have emerged in the last two decades investigating the role of a changing global climate on V. cholerae environmental presence, exogenous gene acquisition, and vector-borne dissemination of this pathogen and the connection to global cholera outbreaks (Lipp et al., 2002; Jutla et al., 2010).
In addition to those living in under-resourced regions, displaced populations, and refugee settings have also experienced significant cholera outbreaks in recent years (Shannon et al., 2019). Cholera outbreaks in South Sudan, Yemen, Cameroon, Nigeria, Tanzania, Uganda, Haiti, and Iraq pose a significant threat to these humanitarian aid settings. Both refugee and impoverished populations lack ready access to rehydration therapy and services and therefore are at higher risk for severe disease and death (CDC, 2020). Children under 5 years of age face disproportionate incidence of infection with V. cholerae O1 and experience more severe disease (Deen et al., 2008). Healthcare workers or cholera response workers face an increased risk of exposure to V. cholerae O1 infection as well, along with leisure travelers who do not follow food or water safety guidelines or practice proper hygiene. Additional risk factors for more severe disease include individuals with blood type O, achlorhydria, or chronic medical conditions (CDC, 2020). Disaster relief operations also need to be aware of the potential for unintentional transmission of cholera to naïve populations and prepare accordingly. Genetic analysis suggests the introduction of O1/O139 V. cholerae strains to regions where it is nonendemic is almost exclusively due to human movement (Domman et al., 2017; Weill et al., 2017; Weil et al., 2019).
Cholera in Haiti over the past decade has clearly demonstrated how devastating the introduction of V. cholerae to a naïve population can be. V. cholerae was introduced to Haiti following an influx of international aid workers who responded to the catastrophic 7.0 magnitude earthquake in 2010 (Chin et al., 2010; Hendriksen et al., 2011; Frerichs et al., 2012; Katz et al., 2013; Orata et al., 2014). Already one of the poorest countries in the western hemisphere, Haiti also suffered from near total loss of infrastructure, access to clean water, and rampant crime in the wake of this natural disaster. Given this was the first introduction of V. cholerae to the Haitian people, the death toll rose rapidly to claim the lives of at least 100,000 people (Vega Ocasio et al., 2023). Cholera has since become endemic to this previously unexposed region. After three consecutive years of zero reported cases, however, the country was declared free of cholera by the Haitian Prime Minister, Dr. Ariel Henry in February 2022. Recent prolonged periods of violence and social unrest have disrupted water treatment infrastructure and restricted access of both citizens and aid workers to the resources needed for effective public health prevention measures against cholera. As a result, an outbreak in late September 2022 has already surged to over 20,000 suspected cases of cholera reported in Haiti as of January 3, 2023 (Vega Ocasio et al., 2023). Nearly 80% of patients were hospitalized, and a very high case-fatality rate of 3.0% has been observed. This rapid resurgence of the bacterial pathogen after a prolonged period of near silence suggests environmental reservoirs of V. cholerae have been well-established since its initial introduction, and eradication of the disease is unlikely if the underlying risk factors are not resolved.
Despite over 150 years of academic study, cholera persists as a significant health hazard in over 60 countries. Several prevention and intervention campaigns to eliminate cholera are currently used around the world. In endemic settings, the use of rapid, affordable, and accurate serotyping techniques is critical for identifying pandemic cholera in the field and implementing swift public health response measures. A lateral flow dipstick method, Cas12a-assisted rapid isothermal detection (CARID), was recently developed to identify O1 and O139 serogroups in complex samples using recombinase-aided amplification and CRISPR-Cas (Lu et al., 2022). Of note, the World Health Organization assembled the Global Task Force on Cholera Control (GTFCC) which has established a roadmap to ending cholera by 2030 and is referenced several times in this text. Disease prevention efforts include implementation of improved drinking water sources and sanitation services and instilling proper hygiene habits, often referred to as the WASH (water, sanitation, and hygiene) campaign (GTFCC, 2017). Educating vulnerable populations about daily practices that can be readily implemented to reduce risk for infection remains one of the most effective methods for disease prevention. The other major area of focus for disease prevention is the stockpiling and administration of cholera vaccines to at-risk populations.
Three oral cholera vaccines (OCVs) are currently approved by the WHO: ShancholTM, Euvichol-Plus®, and Dukoral® (WHO, 2019). The former two OCVs can be administered to adults and children over the age of 1 year, while the latter OCV is approved for individuals starting at age 2 years. Despite the heightened risk posed to very young children by cholera, no vaccine has been approved for infants under 1 year of age. A stockpile of Shanchol™ and Euvichol-Plus® OCVs has been established for use in mass vaccination campaigns, but it is insufficient to cover all those needing it. A major drawback to all current OCVs is the duration of protection. At most, these OCVs provide approximately 70% protection for 3 years, and in areas most affected by cholera, access to preventative medical care is limited or not financially feasible (WHO, 2019). Few advancements have been made in the realm of cholera therapeutics. ORS therapy has remained the standard of care for cholera cases for decades despite the proposal of other intervention methods including administration of unsaturated fatty acids, specifically linoleic acid, to prevent CT production by V. cholerae O1 or a commensal E. coli with glucose probiotic to inhibit colonization (Plecha and Withey, 2015; Withey et al., 2015; Nag et al., 2018a). Expanding beyond ORS has potential to reduce disease severity, shorten duration of the infection, or prevent infection altogether.
Though V. cholerae O1 infection continues to threaten the health and safety of impoverished or displaced populations, elimination of cholera remains an achievable goal for public health professionals. Recommendations for achieving this goal include implementation of educational programs for at-risk populations, installation or maintenance of safe drinking water sources and sanitation services, optimization of disease surveillance methods, expansion of cholera therapeutics, and increased accessibility to and development of new cholera vaccines, particularly designed for young children and to afford increased duration of protection.
From genomics to therapeutic development, V. cholerae research continues to make great strides towards a more complete understanding of how this pathogen survives in the aquatic environment, interacts with hosts, and causes disease. Analyses of evolutionary genetics have identified critical events that must occur for V. cholerae to transition from an environmental marine microbe to the pandemic strains that plague countries around the world today. Novel potential reservoirs in which V. cholerae continues to acquire new genetic material and gain fitness in the environment and the host are being identified in vertebrate fish in addition to the well-established reservoirs of copepods, phytoplankton, and shellfish. Studies focusing on motility, chemotaxis, and biofilm formation provide key insights to how V. cholerae survives in the environment and navigates colonization of the human host. Animal models in mice and rabbits offer means to study CT production and virulence gene expression in mammals while the zebrafish model can be used to gain a well-rounded perspective of natural infection including interactions with other members of the normal microbiome. Both the direct action of the T6SS and indirect activity of rapid fluid loss have been shown to disrupt the composition of the normal intestinal microbiota and open a new avenue of exploration regarding interspecies dynamics during V. cholerae infection. Increased understanding of microbiome composition that affords resistance against infection with V. cholerae offers potential for use of probiotics to reduce risk of infection and limit disease severity. Disease presentation has shifted with the changing genetic composition of El Tor variants, which exhibit various antimicrobial resistance patterns and a range of symptom severity depending on the exact etiological agent. Advances in cholera disease modeling have enabled better prediction of outbreak scenarios and, in turn, faster implementation of prevention initiatives and response efforts. Taken together, these advancements offer a holistic approach to the study of a persistent pathogen that has plagued the world for centuries. Continued efforts to explore V. cholerae dynamics as a model for genomic evolution, bacterial pathogenesis, and its role as a natural member of the aquatic ecosystem are needed, both to advance knowledge in the basic sciences and to develop relevant, effective public health measures to protect the most vulnerable populations.
MW led the writing of the manuscript. DN and IC contributed to writing the manuscript. JW contributed to the writing, edited the manuscript, and provided funding (grant numbers R01AI127390 and R21AI171072). All authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abel, S., Abel zur Wiesch, P., Chang, H. H., Davis, B. M., Lipsitch, M., and Waldor, M. K. (2015). Sequence tag-based analysis of microbial population dynamics. Nat. Methods 12, 223–226. doi: 10.1038/nmeth.3253
Abuaita, B. H., and Withey, J. H. (2009). Bicarbonate induces Vibrio cholerae virulence gene expression by enhancing ToxT activity. Infect. Immun. 77, 4111–4120. doi: 10.1128/IAI.00409-09
Ahmed, S. A., Raabe, C. A., Cheah, H. L., Hoe, C. H., Rozhdestvensky, T. S., and Tang, T. H. (2019). Utilization of small RNA genes to distinguish Vibrio cholerae biotypes via multiplex polymerase chain reaction. Am. J. Trop. Med. Hyg. 100, 1328–1334. doi: 10.4269/ajtmh.18-0525
Ahmed, A. M., Shinoda, S., and Shimamoto, T. (2005). A variant type of Vibrio cholerae SXT element in a multidrug-resistant strain of Vibrio fluvialis. FEMS Microbiol. Lett. 242, 241–247. doi: 10.1016/j.femsle.2004.11.012
Alam, A., Larocque, R. C., Harris, J. B., Vanderspurt, C., Ryan, E. T., Qadri, F., et al. (2005). Hyperinfectivity of human-passaged Vibrio cholerae can be modeled by growth in the infant mouse. Infect. Immun. 73, 6674–6679. doi: 10.1128/IAI.73.10.6674-6679.2005
Alavi, S., Mitchell, J. D., Cho, J. Y., Liu, R., Macbeth, J. C., and Hsiao, A. (2020). Interpersonal gut microbiome variation drives susceptibility and resistance to cholera infection. Cells 181, 1533–1546.e1513. doi: 10.1016/j.cell.2020.05.036
Ali, M., Nelson, A. R., Lopez, A. L., and Sack, D. A. (2015). Updated global burden of cholera in endemic countries. PLoS Negl. Trop. Dis. 9:e0003832. doi: 10.1371/journal.pntd.0003832
Ali, M., Sen Gupta, S., Arora, N., Khasnobis, P., Venkatesh, S., Sur, D., et al. (2017). Identification of burden hotspots and risk factors for cholera in India: an observational study. PLoS One 12:e0183100. doi: 10.1371/journal.pone.0183100
Allan, M., Grandesso, F., Pierre, R., Magloire, R., Coldiron, M., Martinez-Pino, I., et al. (2016). High-resolution spatial analysis of cholera patients reported in Artibonite department, Haiti in 2010–2011. Epidemics 14, 1–10. doi: 10.1016/j.epidem.2015.08.001
Almagro-Moreno, S., Pruss, K., and Taylor, R. K. (2015). Intestinal colonization dynamics of Vibrio cholerae. PLoS Pathog. 11:e1004787. doi: 10.1371/journal.ppat.1004787
Angelichio, M. J., Spector, J., Waldor, M. K., and Camilli, A. (1999). Vibrio cholerae intestinal population dynamics in the suckling mouse model of infection. Infect. Immun. 67, 3733–3739. doi: 10.1128/IAI.67.8.3733-3739.1999
Arumugam, M., Raes, J., Pelletier, E., Le Paslier, D., Yamada, T., Mende, D. R., et al. (2011). Enterotypes of the human gut microbiome. Nature 473, 174–180. doi: 10.1038/nature09944
Bachmann, V., Kostiuk, B., Unterweger, D., Diaz-Satizabal, L., Ogg, S., and Pukatzki, S. (2015). Bile salts modulate the mucin-activated type VI secretion system of pandemic Vibrio cholerae. PLoS Negl. Trop. Dis. 9:e0004031. doi: 10.1371/journal.pntd.0004031
Baddam, R., Sarker, N., Ahmed, D., Mazumder, R., Abdullah, A., Morshed, R., et al. (2020). Genome dynamics of Vibrio cholerae isolates linked to seasonal outbreaks of cholera in Dhaka, Bangladesh. mBio 11, e03339–e03319. doi: 10.1128/mBio.03339-19
Baek, Y., Lee, D., Lee, J., Yoon, Y., Nair, G. B., Kim, D. W., et al. (2020). Cholera toxin production in Vibrio cholerae O1 El tor biotype strains in single-phase culture. Front. Microbiol. 11:825. doi: 10.3389/fmicb.2020.00825
Baine, W., Mazzotti, M., Greco, D., Izzo, E., Zampieri, A., Angioni, G., et al. (1974). Epidemiology of cholera in Italy in 1973. Lancet 2, 1370–1374. doi: 10.1016/s0140-6736(74)92233-8
Baur, B., Hanselmann, K., Schlimme, W., and Jenni, B. (1996). Genetic transformation in freshwater: Escherichia coli is able to develop natural competence. Appl. Environ. Microbiol. 62, 3673–3678. doi: 10.1128/aem.62.10.3673-3678.1996
Bina, T. F., Kunkle, D. E., Bina, X. R., Mullett, S. J., Wendell, S. G., and Bina, J. E. (2021). Bile salts promote ToxR regulon activation during growth under virulence-inducing conditions. Infect. Immun. 89:e0044121. doi: 10.1128/IAI.00441-21
Blake, P. A. (1994). “Historical perspectives on pandemic cholera,” in Vibrio cholerae and cholera. 1st edition. eds. I. K. Wachsmuth, P. A. Blake, and Ø. Olsvik (ASM Press), 291–295.
Blow, N. S., Salomon, R. N., Garrity, K., Reveillaud, I., Kopin, A., Jackson, F. R., et al. (2005). Vibrio cholerae infection of Drosophila melanogaster mimics the human disease cholera. PLoS Pathog. 1:e8. doi: 10.1371/journal.ppat.0010008
Bokhari, H., Ali, A., Noreen, Z., Thomson, N., and Wren, B. W. (2017). Galleria mellonella is low cost and suitable surrogate host for studying virulence of human pathogenic Vibrio cholerae. Gene 628, 1–7. doi: 10.1016/j.gene.2017.07.019
Breen, P., Winters, A. D., Theis, K. R., and Withey, J. H. (2021a). Vibrio cholerae infection induces strain specific modulation of the zebrafish intestinal microbiome. Infect. Immun. 89:e0015721. doi: 10.1128/iai.00157-21
Breen, P., Winters, A. D., Theis, K. R., and Withey, J. H. (2021b). The Vibrio cholerae type six secretion system is dispensable for colonization but affects pathogenesis and the structure of zebrafish intestinal microbiome. Infect. Immun. 89:e0015121. doi: 10.1128/IAI.00151-21
Bridges, A. A., and Bassler, B. L. (2021). Inverse regulation of Vibrio cholerae biofilm dispersal by polyamine signals. Elife 10:e65487. doi: 10.7554/eLife.65487
Burrows, W., and Musteikis, G. M. (1966). Cholera infection and toxin in the rabbit ileal loop. J. Infect. Dis. 116, 183–190. doi: 10.1093/infdis/116.2.183
Butler, S. M., and Camilli, A. (2004). Both chemotaxis and net motility greatly influence the infectivity of Vibrio cholerae. Proc. Natl. Acad. Sci. U. S. A. 101, 5018–5023. doi: 10.1073/pnas.0308052101
Butler, S. M., and Camilli, A. (2005). Going against the grain: chemotaxis and infection in Vibrio cholerae. Nat. Rev. Microbiol. 3, 611–620. doi: 10.1038/nrmicro1207
Butler, S. M., Nelson, E. J., Chowdhury, N., Faruque, S. M., Calderwood, S. B., and Camilli, A. (2006). Cholera stool bacteria repress chemotaxis to increase infectivity. Mol. Microbiol. 60, 417–426. doi: 10.1111/j.1365-2958.2006.05096.x
Butz, H. A., Mey, A. R., Ciosek, A. L., Crofts, A. A., Davies, B. W., and Payne, S. M. (2021). Regulatory effects of CsrA in Vibrio cholerae. MBio 12:e03380-20. doi: 10.1128/mBio.03380-20
Camacho, A., Bouhenia, M., Alyusfi, R., Alkohlani, A., Naji, M. A. M., de Radigues, X., et al. (2018). Cholera epidemic in Yemen, 2016-18: an analysis of surveillance data. Lancet Glob. Health 6, e680–e690. doi: 10.1016/S2214-109X(18)30230-4
Cameron, D. E., Urbach, J. M., and Mekalanos, J. J. (2008). A defined transposon mutant library and its use in identifying motility genes in Vibrio cholerae. Proc. Natl. Acad. Sci. U. S. A. 105, 8736–8741. doi: 10.1073/pnas.0803281105
CDC (2020). Sources of infection and risk factors [online]. Atlanta, GA: United States Centers for Disease Control and Prevention.
Chao, M. C., Pritchard, J. R., Zhang, Y. J., Rubin, E. J., Livny, J., Davis, B. M., et al. (2013). High-resolution definition of the Vibrio cholerae essential gene set with hidden Markov model-based analyses of transposon-insertion sequencing data. Nucleic Acids Res. 41, 9033–9048. doi: 10.1093/nar/gkt654
Chapman, C., Henry, M., Bishop-Lilly, K. A., Awosika, J., Briska, A., Ptashkin, R. N., et al. (2015). Scanning the landscape of genome architecture of non-O1 and non-O139 Vibrio cholerae by whole genome mapping reveals extensive population genetic diversity. PLoS One 10:e0120311. doi: 10.1371/journal.pone.0120311
Chart, H. (2012). “Vibrio, mobiluncus, gardnerella and spirillum: Cholera; vaginosis; rat bite fever” in Medical microbiology. eds. D. Greenwood, M. Barer, R. Slack, and W. Irving. 18th ed (Edinburgh: Churchill Livingstone), 314–323.
Chatterjee, A., Dutta, P. K., and Chowdhury, R. (2007). Effect of fatty acids and cholesterol present in bile on expression of virulence factors and motility of Vibrio cholerae. Infect. Immun. 75, 1946–1953. doi: 10.1128/IAI.01435-06
Chen, J., Byun, H., Liu, R., Jung, I. J., Pu, Q., Zhu, C. Y., et al. (2022). A commensal-encoded genotoxin drives restriction of Vibrio cholerae colonization and host gut microbiome remodeling. Proc. Natl. Acad. Sci. U. S. A. 119:e2121180119. doi: 10.1073/pnas.2121180119
Chiarello, M., McCauley, M., Villéger, S., and Jackson, C. R. (2022). Ranking the biases: the choice of OTUs vs. ASVs in 16S rRNA amplicon data analysis has stronger effects on diversity measures than rarefaction and OTU identity threshold. PLoS One 17:e0264443. doi: 10.1371/journal.pone.0264443
Chin, C.-S., Sorenson, J., Harris, J. B., Robins, W. P., Charles, R. C., Jean-Charles, R. R., et al. (2010). The origin of the Haitian cholera outbreak strain. N. Engl. J. Med. 364, 33–42. doi: 10.1056/NEJMoa1012928
Conner, J. G., Zamorano-Sánchez, D., Park, J. H., Sondermann, H., and Yildiz, F. H. (2017). The ins and outs of cyclic di-GMP signaling in Vibrio cholerae. Curr. Opin. Microbiol. 36, 20–29. doi: 10.1016/j.mib.2017.01.002
Constantin de Magny, G., and Colwell, R. R. (2009). Cholera and climate: a demonstrated relationship. Trans. Am. Clin. Climatol. Assoc. 120, 119–128.
Crumfield, K. D., Carignan, B. M., and Son, M. S. (2018). “Genotypic and phenotypic assays to distinguish Vibrio cholerae biotype” in Vibrio Cholerae: methods and protocols. ed. A. E. Sikora (New York, NY: Springer New York), 11–28.
David, L. A., Weil, A., Ryan, E. T., Calderwood, S. B., Harris, J. B., Chowdhury, F., et al. (2015). Gut microbial succession follows acute secretory diarrhea in humans. MBio 6, e00381–e00315. doi: 10.1128/mBio.00381-15
Davoodi, S., and Foley, E. (2019). Host-microbe-pathogen interactions: a review of Vibrio cholerae pathogenesis in drosophila. Front. Immunol. 10:3128. doi: 10.3389/fimmu.2019.03128
Deen, J. L., von Seidlein, L., Sur, D., Agtini, M., Lucas, M. E., Lopez, A. L., et al. (2008). The high burden of cholera in children: comparison of incidence from endemic areas in Asia and Africa. PLoS Negl. Trop. Dis. 2:e173. doi: 10.1371/journal.pntd.0000173
Desjeux, J. F., Briend, A., and Butzner, J. D. (1997). Oral rehydration solution in the year 2000: pathophysiology, efficacy and effectiveness. Baillieres Clin. Gastroenterol. 11, 509–527. doi: 10.1016/s0950-3528(97)90029-4
Domman, D., Quilici, M.-L., Dorman, M. J., Njamkepo, E., Mutreja, A., Mather, A. E., et al. (2017). Integrated view of Vibrio cholerae in the Americas. Science 358, 789–793. doi: 10.1126/science.aao2136
Donlan, R. M., and Costerton, J. W. (2002). Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15, 167–193. doi: 10.1128/CMR.15.2.167-193.2002
Dougherty, M. W., and Jobin, C. (2021). Shining a light on colibactin biology. Toxins (Basel) 13:346. doi: 10.3390/toxins13050346
Echazarreta, M. A., and Klose, K. E. (2019). Vibrio flagellar synthesis. Front. Cell. Infect. Microbiol. 9:131. doi: 10.3389/fcimb.2019.00131
Epstein, P. R. (1993). Algal blooms in the spread and persistence of cholera. Biosystems 31, 209–221. doi: 10.1016/0303-2647(93)90050-M
Faith, J. J., McNulty, N. P., Rey, F. E., and Gordon, J. I. (2011). Predicting a human gut microbiota’s response to diet in gnotobiotic mice. Science 333, 101–104. doi: 10.1126/science.1206025
Fan, F., Liu, Z., Jabeen, N., Birdwell, L. D., Zhu, J., and Kan, B. (2014). Enhanced interaction of Vibrio cholerae virulence regulators TcpP and ToxR under oxygen-limiting conditions. Infect. Immun. 82, 1676–1682. doi: 10.1128/IAI.01377-13
Farr, D., Nag, D., Chazin, W. J., Harrison, S., Thummel, R., Luo, X., et al. (2022). Neutrophil-associated responses to Vibrio cholerae infection in a natural host model. Infect. Immun. 90:e0046621. doi: 10.1128/iai.00466-21
Farr, D. A., Nag, D., and Withey, J. H. (2021). Characterization of the immune response to Vibrio cholerae infection in a natural host model. Front. Cell. Infect. Microbiol. 11:722520. doi: 10.3389/fcimb.2021.722520
Faruque, S. M., Biswas, K., Udden, S. M., Ahmad, Q. S., Sack, D. A., Nair, G. B., et al. (2006). Transmissibility of cholera: in vivo-formed biofilms and their relationship to infectivity and persistence in the environment. Proc. Natl. Acad. Sci. U. S. A. 103, 6350–6355. doi: 10.1073/pnas.0601277103
Fast, D., Petkau, K., Ferguson, M., Shin, M., Galenza, A., Kostiuk, B., et al. (2020). Vibrio cholerae-symbiont interactions inhibit intestinal repair in drosophila. Cell Rep. 30, 1088–1100.e1085. doi: 10.1016/j.celrep.2019.12.094
Flemming, H.-C., and Wingender, J. (2010). The biofilm matrix. Nat. Rev. Microbiol. 8, 623–633. doi: 10.1038/nrmicro2415
Fleurie, A., Zoued, A., Alvarez, L., Hines, K. M., Cava, F., Xu, L., et al. (2019). A Vibrio cholerae BolA-like protein is required for proper cell shape and cell envelope integrity. MBio 10:e00790-19. doi: 10.1128/mBio.00790-19
Frerichs, R. R., Keim, P. S., Barrais, R., and Piarroux, R. (2012). Nepalese origin of cholera epidemic in Haiti. Clin. Microbiol. Infect. 18, E158–E163. doi: 10.1111/j.1469-0691.2012.03841.x
Fu, Y., Waldor, M. K., and Mekalanos, J. J. (2013). Tn-Seq analysis of Vibrio cholerae intestinal colonization reveals a role for T6SS-mediated antibacterial activity in the host. Cell Host Microbe 14, 652–663. doi: 10.1016/j.chom.2013.11.001
Gallego-Hernandez, A. L., DePas, W. H., Park, J. H., Teschler, J. K., Hartmann, R., Jeckel, H., et al. (2020). Upregulation of virulence genes promotes Vibrio cholerae biofilm hyperinfectivity. Proc. Natl. Acad. Sci. U. S. A. 117, 11010–11017. doi: 10.1073/pnas.1916571117
Ganesan, D., Gupta, S. S., and Legros, D. (2020). Cholera surveillance and estimation of burden of cholera. Vaccine 38, A13–A17. doi: 10.1016/j.vaccine.2019.07.036
Ghosh, P., Sinha, R., Samanta, P., Saha, D. R., Koley, H., Dutta, S., et al. (2019). Haitian variant Vibrio cholerae O1 strains manifest higher virulence in animal models. Front. Microbiol. 10:111. doi: 10.3389/fmicb.2019.00111
Gibbons, S. M., and Gilbert, J. A. (2015). Microbial diversity--exploration of natural ecosystems and microbiomes. Curr. Opin. Genet. Dev. 35, 66–72. doi: 10.1016/j.gde.2015.10.003
Goss, T. J., Seaborn, C. P., Gray, M. D., and Krukonis, E. S. (2010). Identification of the TcpP-binding site in the toxT promoter of vibrio cholerae and the role of ToxR in TcpP-mediated activation. Infect. Immun. 78, 4122–4133. doi: 10.1128/IAI.00566-10
Greig, D. R., Hickey, T. J., Boxall, M. D., Begum, H., Gentle, A., Jenkins, C., et al. (2018). A real-time multiplex PCR for the identification and typing of Vibrio cholerae. Diagn. Microbiol. Infect. Dis. 90, 171–176. doi: 10.1016/j.diagmicrobio.2017.11.017
Grognot, M., Mittal, A., Mah’moud, M., and Taute, K. M. (2021). Vibrio cholerae motility in aquatic and mucus-mimicking environments. Appl. Environ. Microbiol. 87:e0129321. doi: 10.1128/AEM.01293-21
GTFCC (2017). “Ending cholera—a global roadmap to 2030 ”. (Geneva, Switzerland: World Health Organization).
Haas, B. L., Matson, J. S., DiRita, V. J., and Biteen, J. S. (2015). Single-molecule tracking in live Vibrio cholerae reveals that ToxR recruits the membrane-bound virulence regulator TcpP to the toxT promoter. Mol. Microbiol. 96, 4–13. doi: 10.1111/mmi.12834
Halang, P., Leptihn, S., Meier, T., Vorburger, T., and Steuber, J. (2013). The function of the Na+−driven flagellum of Vibrio cholerae is determined by osmolality and pH. J. Bacteriol. 195, 4888–4899. doi: 10.1128/JB.00353-13
Halpern, M., and Izhaki, I. (2017). Fish as hosts of Vibrio cholerae. Front. Microbiol. 8:282. doi: 10.3389/fmicb.2017.00282
Hang, S., Purdy, A. E., Robins, W. P., Wang, Z., Mandal, M., Chang, S., et al. (2014). The acetate switch of an intestinal pathogen disrupts host insulin signaling and lipid metabolism. Cell Host Microbe 16, 592–604. doi: 10.1016/j.chom.2014.10.006
Hathroubi, S., Mekni, M. A., Domenico, P., Nguyen, D., and Jacques, M. (2017). Biofilms: microbial shelters against antibiotics. Microb. Drug Resist. 23, 147–156. doi: 10.1089/mdr.2016.0087
Haycocks, J. R. J., Warren, G. Z. L., Walker, L. M., Chlebek, J. L., Dalia, T. N., Dalia, A. B., et al. (2019). The quorum sensing transcription factor AphA directly regulates natural competence in Vibrio cholerae. PLoS Genet. 15:e1008362. doi: 10.1371/journal.pgen.1008362
Hayes, C. A., Dalia, T. N., and Dalia, A. B. (2017). Systematic genetic dissection of PTS in Vibrio cholerae uncovers a novel glucose transporter and a limited role for PTS during infection of a mammalian host. Mol. Microbiol. 104, 568–579. doi: 10.1111/mmi.13646
Heidelberg, J. F., Eisen, J. A., Nelson, W. C., Clayton, R. A., Gwinn, M. L., Dodson, R. J., et al. (2000). DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406, 477–483. doi: 10.1038/35020000
Hendriksen, R. S., Price, L. B., Schupp, J. M., Gillece, J. D., Kaas, R. S., Engelthaler, D. M., et al. (2011). population genetics of vibrio cholerae from nepal in 2010: evidence on the origin of the Haitian outbreak. mBio 2:e00157-11. doi: 10.1128/mBio.00157-11
Herzog, R., Peschek, N., Fröhlich, K. S., Schumacher, K., and Papenfort, K. (2019). Three autoinducer molecules act in concert to control virulence gene expression in Vibrio cholerae. Nucleic Acids Res. 47, 3171–3183. doi: 10.1093/nar/gky1320
Homma, M., and Kojima, S. (2022). Roles of the second messenger c-di-GMP in bacteria: focusing on the topics of flagellar regulation and vibrio spp. Genes Cells 27, 157–172. doi: 10.1111/gtc.12921
Hounmanou, Y. M. G., Mdegela, R. H., Dougnon, T. V., Madsen, H., Withey, J. H., Olsen, J. E., et al. (2019). Tilapia (Oreochromis niloticus) as a putative reservoir host for survival and transmission of Vibrio cholerae O1 biotype El tor in the aquatic environment. Front. Microbiol. 10:1215. doi: 10.3389/fmicb.2019.01215
Hsiao, A., Ahmed, A. M. S., Subramanian, S., Griffin, N. W., Drewry, L. L., Petri, W. A., et al. (2014). Members of the human gut microbiota involved in recovery from Vibrio cholerae infection. Nature 515, 423–426. doi: 10.1038/nature13738
Hsueh, B. Y., Severin, G. B., Elg, C. A., Waldron, E. J., Kant, A., Wessel, A. J., et al. (2022). Phage defence by deaminase-mediated depletion of deoxynucleotides in bacteria. Nat. Microbiol. 7, 1210–1220. doi: 10.1038/s41564-022-01162-4
Hu, D., Liu, B., Feng, L., Ding, P., Guo, X., Wang, M., et al. (2016). Origins of the current seventh cholera pandemic. Proc. Natl. Acad. Sci. U. S. A. 113, E7730–E7739. doi: 10.1073/pnas.1608732113
Hubbard, T. P., Billings, G., Dorr, T., Sit, B., Warr, A. R., Kuehl, C. J., et al. (2018). A live vaccine rapidly protects against cholera in an infant rabbit model. Sci. Transl. Med. 10:eaap8423. doi: 10.1126/scitranslmed.aap8423
Hui, M. P., Galkin, V. E., Yu, X., Stasiak, A. Z., Stasiak, A., Waldor, M. K., et al. (2010). ParA2, a Vibrio cholerae chromosome partitioning protein, forms left-handed helical filaments on DNA. Proc. Natl. Acad. Sci. U. S. A. 107, 4590–4595. doi: 10.1073/pnas.0913060107
Hung, D. T., and Mekalanos, J. J. (2005). Bile acids induce cholera toxin expression in Vibrio cholerae in a ToxT-independent manner. Proc. Natl. Acad. Sci. U. S. A. 102, 3028–3033. doi: 10.1073/pnas.0409559102
Hunter, J. L., Severin, G. B., Koestler, B. J., and Waters, C. M. (2014). The Vibrio cholerae diguanylate cyclase VCA0965 has an AGDEF active site and synthesizes cyclic di-GMP. BMC Microbiol. 14:22. doi: 10.1186/1471-2180-14-22
Jaskólska, M., Adams, D. W., and Blokesch, M. (2022). Two defence systems eliminate plasmids from seventh pandemic Vibrio cholerae. Nature 604, 323–329. doi: 10.1038/s41586-022-04546-y
Jemielita, M., Wingreen, N. S., and Bassler, B. L. (2018). Quorum sensing controls Vibrio cholerae multicellular aggregate formation. Elife 7:e42057. doi: 10.7554/eLife.42057
Johnson, S. L., Khiani, A., Bishop-Lilly, K. A., Chapman, C., Patel, M., Verratti, K., et al. (2015). Complete genome assemblies for two single-chromosome Vibrio cholerae isolates, strains 1154-74 (serogroup O49) and 10432-62 (serogroup O27). Genome Announc. 3:e00462-15. doi: 10.1128/genomeA.00462-15
Jonas, K., Edwards, A. N., Simm, R., Romeo, T., Römling, U., and Melefors, O. (2008). The RNA binding protein CsrA controls cyclic di-GMP metabolism by directly regulating the expression of GGDEF proteins. Mol. Microbiol. 70, 236–257. doi: 10.1111/j.1365-2958.2008.06411.x
Joshi, A., Kostiuk, B., Rogers, A., Teschler, J., Pukatzki, S., and Yildiz, F. H. (2017). Rules of engagement: the type VI secretion system in Vibrio cholerae. Trends Microbiol. 25, 267–279. doi: 10.1016/j.tim.2016.12.003
Jugder, B. E., Batista, J. H., Gibson, J. A., Cunningham, P. M., Asara, J. M., and Watnick, P. I. (2022). Vibrio cholerae high cell density quorum sensing activates the host intestinal innate immune response. Cell Rep. 40:111368. doi: 10.1016/j.celrep.2022.111368
Jutla, A. S., Akanda, A. S., and Islam, S. (2010). Tracking cholera in coastal regions using satellite observations. J. Am. Water Resour. Assoc. 46, 651–662. doi: 10.1111/j.1752-1688.2010.00448.x
Kamareddine, L., Wong, A. C. N., Vanhove, A. S., Hang, S., Purdy, A. E., Kierek-Pearson, K., et al. (2018). Activation of Vibrio cholerae quorum sensing promotes survival of an arthropod host. Nat. Microbiol. 3, 243–252. doi: 10.1038/s41564-017-0065-7
Kamp, H. D., Patimalla-Dipali, B., Lazinski, D. W., Wallace-Gadsden, F., and Camilli, A. (2013). Gene fitness landscapes of Vibrio cholerae at important stages of its life cycle. PLoS Pathog. 9:e1003800. doi: 10.1371/journal.ppat.1003800
Kaper, J. B., Morris, J. G. Jr., and Levine, M. M. (1995). Cholera. Clin. Microbiol. Rev. 8, 48–86. doi: 10.1128/CMR.8.1.48
Kashyap, P. C., Marcobal, A., Ursell, L. K., Smits, S. A., Sonnenburg, E. D., Costello, E. K., et al. (2013). Genetically dictated change in host mucus carbohydrate landscape exerts a diet-dependent effect on the gut microbiota. Proc. Natl. Acad. Sci. U. S. A. 110, 17059–17064. doi: 10.1073/pnas.1306070110
Katz, L. S., Petkau, A., Beaulaurier, J., Tyler, S., Antonova, E. S., Turnsek, M. A., et al. (2013). Evolutionary dynamics of Vibrio cholerae O1 following a single-source introduction to Haiti. mBio 4:e00398-13. doi: 10.1128/mBio.00398-13
Kim, E. J., Lee, D., Moon, S. H., Lee, C. H., Kim, S. J., Lee, J. H., et al. (2014). Molecular insights into the evolutionary pathway of Vibrio cholerae O1 atypical El tor variants. PLoS Pathog. 10:e1004384. doi: 10.1371/journal.ppat.1004384
Kitts, G., Giglio, K. M., Zamorano-Sanchez, D., Park, J. H., Townsley, L., Cooley, R. B., et al. (2019). A conserved regulatory circuit controls large adhesins in Vibrio cholerae. mBio 10. doi: 10.1128/mBio.02822-19
Koestler, B. J., and Waters, C. M. (2014). Intestinal GPS: bile and bicarbonate control cyclic di-GMP to provide Vibrio cholerae spatial cues within the small intestine. Gut Microbes 5, 775–780. doi: 10.4161/19490976.2014.985989
Kovacikova, G., Lin, W., and Skorupski, K. (2004). Vibrio cholerae AphA uses a novel mechanism for virulence gene activation that involves interaction with the LysR-type regulator AphB at the tcpPH promoter. Mol. Microbiol. 53, 129–142. doi: 10.1111/j.1365-2958.2004.04121.x
Kovacikova, G., Lin, W., and Skorupski, K. (2010). The LysR-type virulence activator AphB regulates the expression of genes in Vibrio cholerae in response to low pH and anaerobiosis. J. Bacteriol. 192, 4181–4191. doi: 10.1128/JB.00193-10
Kovacikova, G., and Skorupski, K. (1999). A Vibrio cholerae LysR homolog, AphB, cooperates with AphA at the tcpPH promoter to activate expression of the ToxR virulence cascade. J. Bacteriol. 181, 4250–4256. doi: 10.1128/JB.181.14.4250-4256.1999
Kovacikova, G., and Skorupski, K. (2001). Overlapping binding sites for the virulence gene regulators AphA, AphB and cAMP-CRP at the Vibrio cholerae tcpPH promoter. Mol. Microbiol. 41, 393–407. doi: 10.1046/j.1365-2958.2001.02518.x
Krukonis, E. S., and DiRita, V. J. (2003). DNA binding and ToxR responsiveness by the wing domain of TcpP, an activator of virulence gene expression in Vibrio cholerae. Mol. Cell 12, 157–165. doi: 10.1016/S1097-2765(03)00222-3
Krukonis, E. S., Yu, R. R., and Dirita, V. J. (2000). The Vibrio cholerae ToxR/TcpP/ToxT virulence cascade: distinct roles for two membrane-localized transcriptional activators on a single promoter. Mol. Microbiol. 38, 67–84. doi: 10.1046/j.1365-2958.2000.02111.x
Lee, D., Kim, E. J., Baek, Y., Lee, J., Yoon, Y., Nair, G. B., et al. (2020). Alterations in glucose metabolism in Vibrio cholerae serogroup O1 El tor biotype strains. Sci. Rep. 10:308. doi: 10.1038/s41598-019-57093-4
Lee, J., Sperandio, V., Frantz, D. E., Longgood, J., Camilli, A., Phillips, M. A., et al. (2009). An alternative polyamine biosynthetic pathway is widespread in bacteria and essential for biofilm formation in Vibrio cholerae. J. Biol. Chem. 284, 9899–9907. doi: 10.1074/jbc.M900110200
Leung, T., and Matrajt, L. (2021). Protection afforded by previous Vibrio cholerae infection against subsequent disease and infection: a review. PLoS Negl. Trop. Dis. 15:e0009383. doi: 10.1371/journal.pntd.0009383
Leviatan, S., Shoer, S., Rothschild, D., Gorodetski, M., and Segal, E. (2022). An expanded reference map of the human gut microbiome reveals hundreds of previously unknown species. Nat. Commun. 13:3863. doi: 10.1038/s41467-022-31502-1
Li, F., Du, P., Li, B., Ke, C., Chen, A., Chen, J., et al. (2014). Distribution of virulence-associated genes and genetic relationships in non-O1/O139 Vibrio cholerae aquatic isolates from China. Appl. Environ. Microbiol. 80, 4987–4992. doi: 10.1128/AEM.01021-14
Li, H., Limenitakis, J. P., Fuhrer, T., Geuking, M. B., Lawson, M. A., Wyss, M., et al. (2015). The outer mucus layer hosts a distinct intestinal microbial niche. Nat. Commun. 6:8292. doi: 10.1038/ncomms9292
Li, Z., Lu, X., Wang, D., Liang, W. L., Zhang, J., Li, J., et al. (2019a). Genomic comparison of serogroups O159 and O170 with other Vibrio cholerae serogroups. BMC Genomics 20:241. doi: 10.1186/s12864-019-5603-7
Li, Z., Pang, B., Wang, D., Li, J., Xu, J., Fang, Y., et al. (2019b). Expanding dynamics of the virulence-related gene variations in the toxigenic Vibrio cholerae serogroup O1. BMC Genomics 20:360. doi: 10.1186/s12864-019-5725-y
Lipp, E. K., Huq, A., and Colwell, R. R. (2002). Effects of global climate on infectious disease: the cholera model. Clin. Microbiol. Rev. 15, 757–770. doi: 10.1128/CMR.15.4.757-770.2002
List, C., Grutsch, A., Radler, C., Cakar, F., Zingl, F. G., Schild-Prufert, K., et al. (2018). Genes activated by Vibrio cholerae upon exposure to Caenorhabditis elegans reveal the mannose-sensitive hemagglutinin to be essential for colonization. mSphere 3:e00238-18. doi: 10.1128/mSphereDirect.00238-18
Lobitz, B., Beck, L., Huq, A., Wood, B., Fuchs, G., Faruque, A. S., et al. (2000). Climate and infectious disease: use of remote sensing for detection of Vibrio cholerae by indirect measurement. Proc. Natl. Acad. Sci. U. S. A. 97, 1438–1443. doi: 10.1073/pnas.97.4.1438
Logan, S. L., Thomas, J., Yan, J., Baker, R. P., Shields, D. S., Xavier, J. B., et al. (2018). The Vibrio cholerae type VI secretion system can modulate host intestinal mechanics to displace gut bacterial symbionts. Proc. Natl. Acad. Sci. U. S. A. 115, E3779–e3787. doi: 10.1073/pnas.1720133115
Lu, P., Chen, J., Li, Z., Li, Z., Zhang, J., Kan, B., et al. (2022). Visual identification and serotyping of toxigenic Vibrio cholerae serogroups O1 and O139 with CARID. Front. Cell. Infect. Microbiol. 12:863435. doi: 10.3389/fcimb.2022.863435
Luckey, T. D. (1972). Introduction to intestinal microecology. Am. J. Clin. Nutr. 25, 1292–1294. doi: 10.1093/ajcn/25.12.1292
MacIntyre, D. L., Miyata, S. T., Kitaoka, M., and Pukatzki, S. (2010). The Vibrio cholerae type VI secretion system displays antimicrobial properties. Proc. Natl. Acad. Sci. U. S. A. 107, 19520–19524. doi: 10.1073/pnas.1012931107
Mandlik, A., Livny, J., Robins, W. P., Ritchie, J. M., Mekalanos, J. J., and Waldor, M. K. (2011). RNA-Seq-based monitoring of infection-linked changes in Vibrio cholerae gene expression. Cell Host Microbe 10, 165–174. doi: 10.1016/j.chom.2011.07.007
Mashruwala, A. A., and Bassler, B. L. (2020). The Vibrio cholerae quorum-sensing protein VqmA integrates cell density, environmental, and host-derived cues into the control of virulence. mBio 11:e01572-20. doi: 10.1128/mBio.01572-20
Matson, J. S. (2018). “Infant mouse model of Vibrio cholerae infection and colonization” in Vibrio Cholerae: methods and protocols. ed. A. E. Sikora (New York, NY: Springer New York), 147–152.
Matson, J. S., Withey, J. H., and DiRita, V. J. (2007). Regulatory networks controlling Vibrio cholerae virulence gene expression. Infect. Immun. 75, 5542–5549. doi: 10.1128/IAI.01094-07
Matthey, N., Stutzmann, S., Stoudmann, C., Guex, N., Iseli, C., and Blokesch, M. (2019). Neighbor predation linked to natural competence fosters the transfer of large genomic regions in Vibrio cholerae. Elife 8:e48212. doi: 10.7554/eLife.48212
Mattick, J. S. (2002). Type IV pili and twitching motility. Annu. Rev. Microbiol. 56, 289–314. doi: 10.1146/annurev.micro.56.012302.160938
Mavian, C., Paisie, T. K., Alam, M. T., Browne, C., Beau De Rochars, V. M., Nembrini, S., et al. (2020). Toxigenic Vibrio cholerae evolution and establishment of reservoirs in aquatic ecosystems. Proc. Natl. Acad. Sci. U. S. A. 117, 7897–7904. doi: 10.1073/pnas.1918763117
McCarter, L. L. (2001). Polar flagellar motility of the Vibrionaceae. Microbiol. Mol. Biol. Rev. 65, 445–462. doi: 10.1128/MMBR.65.3.445-462.2001
Meibom, K. L., Blokesch, M., Dolganov, N. A., Wu, C.-Y., and Schoolnik, G. K. (2005). Chitin induces natural competence in Vibrio cholerae. Science 310, 1824–1827. doi: 10.1126/science.1120096
Midani, F. S., Weil, A. A., Chowdhury, F., Begum, Y. A., Khan, A. I., Debela, M. D., et al. (2018). Human gut microbiota predicts susceptibility to Vibrio cholerae infection. J. Infect. Dis. 218, 645–653. doi: 10.1093/infdis/jiy192
Midgett, C. R., Almagro-Moreno, S., Pellegrini, M., Taylor, R. K., Skorupski, K., and Kull, F. J. (2017). Bile salts and alkaline pH reciprocally modulate the interaction between the periplasmic domains of Vibrio cholerae ToxR and ToxS. Mol. Microbiol. 105, 258–272. doi: 10.1111/mmi.13699
Millet, Y. A., Alvarez, D., Ringgaard, S., von Andrian, U. H., Davis, B. M., and Waldor, M. K. (2014). Insights into Vibrio cholerae intestinal colonization from monitoring fluorescently labeled bacteria. PLoS Pathog. 10:e1004405. doi: 10.1371/journal.ppat.1004405
Mitchell, K. C., Breen, P., Britton, S., Neely, M. N., and Withey, J. H. (2017). Quantifying Vibrio cholerae enterotoxicity in a zebrafish infection model. Appl. Environ. Microbiol. 83:e00783-17. doi: 10.1128/AEM.00783-17
Mitchell, K. C., and Withey, J. H. (2018). “Danio rerio as a native host model for understanding pathophysiology of Vibrio cholerae” in Vibrio Cholerae: methods and protocols. ed. A. E. Sikora (Totowa: Humana Press Inc), 97–102.
Miyata, S. T., Kitaoka, M., Brooks, T. M., McAuley, S. B., and Pukatzki, S. (2011). Vibrio cholerae requires the type VI secretion system virulence factor VasX to kill Dictyostelium discoideum. Infect. Immun. 79, 2941–2949. doi: 10.1128/IAI.01266-10
Mondal, M., Nag, D., Koley, H., Saha, D. R., and Chatterjee, N. S. (2014). The Vibrio cholerae extracellular chitinase ChiA2 is important for survival and pathogenesis in the host intestine. PLoS One 9:e103119. doi: 10.1371/journal.pone.0103119
Mukherjee, S., and Bassler, B. L. (2019). Bacterial quorum sensing in complex and dynamically changing environments. Nat. Rev. Microbiol. 17, 371–382. doi: 10.1038/s41579-019-0186-5
Müller, J., Miller, M. C., Nielsen, A. T., Schoolnik, G. K., and Spormann, A. M. (2007). vpsA- and luxO-independent biofilms of Vibrio cholerae. FEMS Microbiol. Lett. 275, 199–206. doi: 10.1111/j.1574-6968.2007.00884.x
Murase, K., Arakawa, E., Izumiya, H., Iguchi, A., Takemura, T., Kikuchi, T., et al. (2022). Genomic dissection of the Vibrio cholerae O-serogroup global reference strains: reassessing our view of diversity and plasticity between two chromosomes. Microb. Genom. 8:mgen000860. doi: 10.1099/mgen.0.000860
Nadeem, A., Nagampalli, R., Toh, E., Alam, A., Myint, S. L., Heidler, T. V., et al. (2021). A tripartite cytolytic toxin formed by Vibrio cholerae proteins with flagellum-facilitated secretion. Proc. Natl. Acad. Sci. U. S. A. 118:e2111418118. doi: 10.1073/pnas.2111418118
Nag, D., Breen, P., Raychaudhuri, S., and Withey, J. H. (2018a). Glucose metabolism by Escherichia coli inhibits Vibrio cholerae intestinal colonization of zebrafish. Infect. Immun. 86:e00486-18. doi: 10.1128/IAI.00486-18
Nag, D., Farr, D. A., Walton, M. G., and Withey, J. H. (2020). Zebrafish models for pathogenic Vibrios. J. Bacteriol. 202:e00165-20. doi: 10.1128/JB.00165-20
Nag, D., Mitchell, K., Breen, P., and Withey, J. H. (2018b). Quantifying Vibrio cholerae colonization and Diarrhea in the adult zebrafish model. J. Vis. Exp. 137:57767. doi: 10.3791/57767
Nan, B., and Zusman, D. R. (2011). Uncovering the mystery of gliding motility in the myxobacteria. Annu. Rev. Genet. 45, 21–39. doi: 10.1146/annurev-genet-110410-132547
Na-Ubol, M., Srimanote, P., Chongsa-Nguan, M., Indrawattana, N., Sookrung, N., Tapchaisri, P., et al. (2011). Hybrid & El tor variant biotypes of Vibrio cholerae O1 in Thailand. Indian J. Med. Res. 133, 387–394.
Nelson, E. J., Chowdhury, A., Harris, J. B., Begum, Y. A., Chowdhury, F., Khan, A. I., et al. (2007). Complexity of rice-water stool from patients with Vibrio cholerae plays a role in the transmission of infectious diarrhea. Proc. Natl. Acad. Sci. U. S. A. 104, 19091–19096. doi: 10.1073/pnas.0706352104
Nielsen, A. T., Dolganov, N. A., Otto, G., Miller, M. C., Wu, C. Y., and Schoolnik, G. K. (2006). RpoS controls the Vibrio cholerae mucosal escape response. PLoS Pathog. 2:e109. doi: 10.1371/journal.ppat.0020109
Nuidate, T., Tansila, N., Saengkerdsub, S., Kongreung, J., Bakkiyaraj, D., and Vuddhakul, V. (2016). Role of indole production on virulence of Vibrio cholerae using galleria mellonella larvae model. Indian J. Microbiol. 56, 368–374. doi: 10.1007/s12088-016-0592-6
Nygren, E., Li, B. L., Holmgren, J., and Attridge, S. R. (2009). Establishment of an adult mouse model for direct evaluation of the efficacy of vaccines against Vibrio cholerae. Infect. Immun. 77, 3475–3484. doi: 10.1128/IAI.01197-08
O’Hara, B. J., Alam, M., and Ng, W.-L. (2022). The Vibrio cholerae seventh Pandemic Islands act in tandem to defend against a circulating phage. PLoS Genet. 18:e1010250. doi: 10.1371/journal.pgen.1010250
Ochi, K., Mizuno, T., Samanta, P., Mukhopadhyay, A. K., Miyoshi, S.-I., and Imamura, D. (2021). Recent Vibrio cholerae O1 epidemic strains are unable to replicate CTXΦ prophage genome. mSphere 6, e00337–e00321. doi: 10.1128/mSphere.00337-21
Olivier, V., Queen, J., and Satchell, K. J. (2009). Successful small intestine colonization of adult mice by Vibrio cholerae requires ketamine anesthesia and accessory toxins. PLoS One 4:e7352. doi: 10.1371/journal.pone.0007352
Orata, F. D., Keim, P. S., and Boucher, Y. (2014). The 2010 cholera outbreak in Haiti: how science solved a controversy. PLoS Pathog. 10:e1003967. doi: 10.1371/journal.ppat.1003967
Papenfort, K., and Bassler, B. L. (2016). Quorum sensing signal-response systems in gram-negative bacteria. Nat. Rev. Microbiol. 14, 576–588. doi: 10.1038/nrmicro.2016.89
Papenfort, K., Silpe, J. E., Schramma, K. R., Cong, J. P., Seyedsayamdost, M. R., and Bassler, B. L. (2017). A Vibrio cholerae autoinducer-receptor pair that controls biofilm formation. Nat. Chem. Biol. 13, 551–557. doi: 10.1038/nchembio.2336
Plecha, S. C., and Withey, J. H. (2015). Mechanism for inhibition of Vibrio cholerae ToxT activity by the unsaturated fatty acid components of bile. J. Bacteriol. 197, 1716–1725. doi: 10.1128/JB.02409-14
Pritchard, J. R., Chao, M. C., Abel, S., Davis, B. M., Baranowski, C., Zhang, Y. J., et al. (2014). ARTIST: high-resolution genome-wide assessment of fitness using transposon-insertion sequencing. PLoS Genet. 10:e1004782. doi: 10.1371/journal.pgen.1004782
Prodan, A., Tremaroli, V., Brolin, H., Zwinderman, A. H., Nieuwdorp, M., and Levin, E. (2020). Comparing bioinformatic pipelines for microbial 16S rRNA amplicon sequencing. PLoS One 15:e0227434. doi: 10.1371/journal.pone.0227434
Qin, J., Li, R., Raes, J., Arumugam, M., Burgdorf, K. S., Manichanh, C., et al. (2010). A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65. doi: 10.1038/nature08821
Ramamurthy, T., Nandy, R. K., Mukhopadhyay, A. K., Dutta, S., Mutreja, A., Okamoto, K., et al. (2020). Virulence regulation and innate host response in the pathogenicity of Vibrio cholerae. Front. Cell. Infect. Microbiol. 10:572096. doi: 10.3389/fcimb.2020.572096
Raychoudhuri, A., Patra, T., Ghosh, K., Ramamurthy, T., Nandy, R. K., Takeda, Y., et al. (2009). Classical ctxB in Vibrio cholerae O1, Kolkata, India. Emerg. Infect. Dis. 15, 131–132. doi: 10.3201/eid1501.080543
Ritchie, J. M., Rui, H., Bronson, R. T., and Waldor, M. K. (2010). Back to the future: studying cholera pathogenesis using infant rabbits. mBio 1:e00047-10. doi: 10.1128/mBio.00047-10
Ritchie, J. M., and Waldor, M. K. (2009). Vibrio cholerae interactions with the gastrointestinal tract: lessons from animal studies. Curr. Top. Microbiol. Immunol. 337, 37–59. doi: 10.1007/978-3-642-01846-6_2
Rivera-Chavez, F., and Mekalanos, J. J. (2019). Cholera toxin promotes pathogen acquisition of host-derived nutrients. Nature 572, 244–248. doi: 10.1038/s41586-019-1453-3
Runft, D. L., Mitchell, K. C., Abuaita, B. H., Allen, J. P., Bajer, S., Ginsburg, K., et al. (2014). Zebrafish as a natural host model for Vibrio cholerae colonization and transmission. Appl. Environ. Microbiol. 80, 1710–1717. doi: 10.1128/AEM.03580-13
Rutherford, S. T., van Kessel, J. C., Shao, Y., and Bassler, B. L. (2011). AphA and LuxR/HapR reciprocally control quorum sensing in vibrios. Genes Dev. 25, 397–408. doi: 10.1101/gad.2015011
Sack, D. A., Sack, R. B., Nair, G. B., and Siddique, A. K. (2004). Cholera. Lancet 363, 223–233. doi: 10.1016/S0140-6736(03)15328-7
Samanta, P., Saha, R. N., Chowdhury, G., Naha, A., Sarkar, S., Dutta, S., et al. (2018). Dissemination of newly emerged polymyxin B sensitive Vibrio cholerae O1 containing Haitian-like genetic traits in different parts of India. J. Med. Microbiol. 67, 1326–1333. doi: 10.1099/jmm.0.000783
Santoriello, F. J., Kirchberger, P. C., Boucher, Y., and Pukatzki, S. (2023). Pandemic Vibrio cholerae acquired competitive traits from an environmental vibrio species. Life Sci. Alliance 6:e202201437. doi: 10.26508/lsa.202201437
Santoriello, F. J., Michel, L., Unterweger, D., and Pukatzki, S. (2020). Pandemic Vibrio cholerae shuts down site-specific recombination to retain an interbacterial defence mechanism. Nat. Commun. 11:6246. doi: 10.1038/s41467-020-20012-7
Sarkar, A., Morita, D., Ghosh, A., Chowdhury, G., Mukhopadhyay, A. K., Okamoto, K., et al. (2019). Altered integrative and conjugative elements (ICEs) in recent Vibrio cholerae O1 isolated from cholera cases, Kolkata, India. Front. Microbiol. 10:2072. doi: 10.3389/fmicb.2019.02072
Satchell, K. J. F., Jones, C. J., Wong, J., Queen, J., Agarwal, S., and Yildiz, F. H. (2016). Phenotypic analysis reveals that the 2010 Haiti cholera epidemic is linked to a hypervirulent strain. Infect. Immun. 84, 2473–2481. doi: 10.1128/IAI.00189-16
Sawasvirojwong, S., Srimanote, P., Chatsudthipong, V., and Muanprasat, C. (2013). An adult mouse model of Vibrio cholerae-induced Diarrhea for studying pathogenesis and potential therapy of cholera. PLoS Negl. Trop. Dis. 7:e2293. doi: 10.1371/journal.pntd.0002293
Schmidt, T. S. B., Raes, J., and Bork, P. (2018). The human gut microbiome: from association to modulation. Cells 172, 1198–1215. doi: 10.1016/j.cell.2018.02.044
Sender, R., Fuchs, S., and Milo, R. (2016). Are we really vastly outnumbered? Revisiting the ratio of bacterial to host cells in humans. Cells 164, 337–340. doi: 10.1016/j.cell.2016.01.013
Senderovich, Y., Izhaki, I., and Halpern, M. (2010). Fish as reservoirs and vectors of Vibrio cholerae. PLoS One 5:e8607. doi: 10.1371/journal.pone.0008607
Seper, A., Fengler, V. H. I., Roier, S., Wolinski, H., Kohlwein, S. D., Bishop, A. L., et al. (2011). Extracellular nucleases and extracellular DNA play important roles in Vibrio cholerae biofilm formation. Mol. Microbiol. 82, 1015–1037. doi: 10.1111/j.1365-2958.2011.07867.x
Seper, A., Pressler, K., Kariisa, A., Haid, A. G., Roier, S., Leitner, D. R., et al. (2014). Identification of genes induced in Vibrio cholerae in a dynamic biofilm system. Int. J. Med. Microbiol. 304, 749–763. doi: 10.1016/j.ijmm.2014.05.011
Shannon, K., Hast, M., Azman, A. S., Legros, D., McKay, H., and Lessler, J. (2019). Cholera prevention and control in refugee settings: successes and continued challenges. PLoS Negl. Trop. Dis. 13:e0007347. doi: 10.1371/journal.pntd.0007347
Shaw, S., Samanta, P., Chowdhury, G., Ghosh, D., Dey, T. K., Deb, A. K., et al. (2022). Altered molecular attributes and antimicrobial resistance patterns of Vibrio cholerae O1 El tor strains isolated from the cholera endemic regions of India. J. Appl. Microbiol. 133, 3605–3616. doi: 10.1111/jam.15794
Sicard, J. F., Le Bihan, G., Vogeleer, P., Jacques, M., and Harel, J. (2017). Interactions of intestinal bacteria with components of the intestinal mucus. Front. Cell. Infect. Microbiol. 7:387. doi: 10.3389/fcimb.2017.00387
Silva, A. J., and Benitez, J. A. (2016). Vibrio cholerae biofilms and cholera pathogenesis. PLoS Negl. Trop. Dis. 10:e0004330. doi: 10.1371/journal.pntd.0004330
Singh, P. K., Bartalomej, S., Hartmann, R., Jeckel, H., Vidakovic, L., Nadell, C. D., et al. (2017). Vibrio cholerae combines individual and collective sensing to trigger biofilm dispersal. Curr. Biol. 27, 3359–3366.e3357. doi: 10.1016/j.cub.2017.09.041
Sinha, R., Koley, H., Nag, D., Mitra, S., Mukhopadhyay, A. K., and Chattopadhyay, B. (2015). Pentavalent outer membrane vesicles of Vibrio cholerae induce adaptive immune response and protective efficacy in both adult and passive suckling mice models. Microbes Infect. 17, 215–227. doi: 10.1016/j.micinf.2014.10.011
Sit, B., Fakoya, B., and Waldor, M. K. (2022). Animal models for dissecting Vibrio cholerae intestinal pathogenesis and immunity. Curr. Opin. Microbiol. 65, 1–7. doi: 10.1016/j.mib.2021.09.007
Sit, B., Fakoya, B., Zhang, T., Billings, G., and Waldor, M. K. (2021). Dissecting serotype-specific contributions to live oral cholera vaccine efficacy. Proc. Natl. Acad. Sci. U. S. A. 118:e2018032118. doi: 10.1073/pnas.2018032118
Sit, B., Zhang, T., Fakoya, B., Akter, A., Biswas, R., Ryan, E. T., et al. (2019). Oral immunization with a probiotic cholera vaccine induces broad protective immunity against Vibrio cholerae colonization and disease in mice. PLoS Negl. Trop. Dis. 13:e0007417. doi: 10.1371/journal.pntd.0007417
Skorupski, K., and Taylor, R. K. (1999). A new level in the Vibrio cholerae ToxR virulence cascade: AphA is required for transcriptional activation of the tcpPH operon. Mol. Microbiol. 31, 763–771. doi: 10.1046/j.1365-2958.1999.01215.x
Song, Z., Cai, Y., Lao, X., Wang, X., Lin, X., Cui, Y., et al. (2019). Taxonomic profiling and populational patterns of bacterial bile salt hydrolase (BSH) genes based on worldwide human gut microbiome. Microbiome 7:9. doi: 10.1186/s40168-019-0628-3
Sozhamannan, S., and Waldminghaus, T. (2020). Exception to the exception rule: synthetic and naturally occurring single chromosome Vibrio cholerae. Environ. Microbiol. 22, 4123–4132. doi: 10.1111/1462-2920.15002
Spira, W. M., Sack, R. B., and Froehlich, J. L. (1981). Simple adult rabbit model for vibrio cholerae and enterotoxigenic Escherichia coli diarrhea. Infect. Immun. 32, 739–747. doi: 10.1128/iai.32.2.739-747.1981
Straub, D., Blackwell, N., Langarica-Fuentes, A., Peltzer, A., Nahnsen, S., and Kleindienst, S. (2020). Interpretations of environmental microbial community studies are biased by the selected 16S rRNA (gene) amplicon sequencing pipeline. Front. Microbiol. 11:550420. doi: 10.3389/fmicb.2020.550420
Tamayo, R., Patimalla, B., and Camilli, A. (2010). Growth in a biofilm induces a hyperinfectious phenotype in Vibrio cholerae. Infect. Immun. 78, 3560–3569. doi: 10.1128/IAI.00048-10
Tamplin, M. L., Gauzens, A. L., Huq, A., Sack, D. A., and Colwell, R. R. (1990). Attachment of Vibrio cholerae serogroup O1 to zooplankton and phytoplankton of Bangladesh waters. Appl. Environ. Microbiol. 56, 1977–1980. doi: 10.1128/aem.56.6.1977-1980.1990
Terashima, H., Kojima, S., and Homma, M. (2008). Flagellar motility in bacteria structure and function of flagellar motor. Int. Rev. Cell Mol. Biol. 270, 39–85. doi: 10.1016/s1937-6448(08)01402-0
Teschler, J. K., Zamorano-Sánchez, D., Utada, A. S., Warner, C. J., Wong, G. C., Linington, R. G., et al. (2015). Living in the matrix: assembly and control of Vibrio cholerae biofilms. Nat. Rev. Microbiol. 13, 255–268. doi: 10.1038/nrmicro3433
Thelin, K. H., and Taylor, R. K. (1996). Toxin-coregulated pilus, but not mannose-sensitive hemagglutinin, is required for colonization by Vibrio cholerae O1 El tor biotype and O139 strains. Infect. Immun. 64, 2853–2856. doi: 10.1128/iai.64.7.2853-2856.1996
Thiagarajah, J. R., and Verkman, A. S. (2005). New drug targets for cholera therapy. Trends Pharmacol. Sci. 26, 172–175. doi: 10.1016/j.tips.2005.02.003
Thompson, C. C., Marin, M. A., Dias, G. M., Dutilh, B. E., Edwards, R. A., Iida, T., et al. (2011). Genome sequence of the human pathogen Vibrio cholerae Amazonia. J. Bacteriol. 193, 5877–5878. doi: 10.1128/JB.05643-11
Thomson, J. J., Plecha, S. C., and Withey, J. H. (2015). A small unstructured region in Vibrio cholerae ToxT mediates the response to positive and negative effectors and ToxT proteolysis. J. Bacteriol. 197, 654–668. doi: 10.1128/JB.02068-14
Thomson, J. J., and Withey, J. H. (2014). Bicarbonate increases binding affinity of Vibrio cholerae ToxT to virulence gene promoters. J. Bacteriol. 196, 3872–3880. doi: 10.1128/JB.01824-14
Tischler, A. D., and Camilli, A. (2005). Cyclic diguanylate regulates Vibrio cholerae virulence gene expression. Infect. Immun. 73, 5873–5882. doi: 10.1128/IAI.73.9.5873-5882.2005
Trucksis, M., Michalski, J., Deng, Y. K., and Kaper, J. B. (1998). The Vibrio cholerae genome contains two unique circular chromosomes. Proc. Natl. Acad. Sci. U. S. A. 95, 14464–14469. doi: 10.1073/pnas.95.24.14464
Unterweger, D., Miyata, S. T., Bachmann, V., Brooks, T. M., Mullins, T., Kostiuk, B., et al. (2014). The Vibrio cholerae type VI secretion system employs diverse effector modules for intraspecific competition. Nat. Commun. 5:3549. doi: 10.1038/ncomms4549
Utada, A. S., Bennett, R. R., Fong, J. C. N., Gibiansky, M. L., Yildiz, F. H., Golestanian, R., et al. (2014). Vibrio cholerae use pili and flagella synergistically to effect motility switching and conditional surface attachment. Nat. Commun. 5:4913. doi: 10.1038/ncomms5913
Vaitkevicius, K., Lindmark, B., Ou, G., Song, T., Toma, C., Iwanaga, M., et al. (2006). A Vibrio cholerae protease needed for killing of Caenorhabditis elegans has a role in protection from natural predator grazing. Proc. Natl. Acad. Sci. U. S. A. 103, 9280–9285. doi: 10.1073/pnas.0601754103
Valiente, E., Davies, C., Mills, D. C., Getino, M., Ritchie, J. M., and Wren, B. W. (2018). Vibrio cholerae accessory colonisation factor AcfC: a chemotactic protein with a role in hyperinfectivity. Sci. Rep. 8:8390. doi: 10.1038/s41598-018-26570-7
Vance, R. E., Zhu, J., and Mekalanos, J. J. (2003). A constitutively active variant of the quorum-sensing regulator LuxO affects protease production and biofilm formation in Vibrio cholerae. Infect. Immun. 71, 2571–2576. doi: 10.1128/IAI.71.5.2571-2576.2003
Vanhove, A. S., Hang, S., Vijayakumar, V., Wong, A. C., Asara, J. M., and Watnick, P. I. (2017). Vibrio cholerae ensures function of host proteins required for virulence through consumption of luminal methionine sulfoxide. PLoS Pathog. 13:e1006428. doi: 10.1371/journal.ppat.1006428
Vega Ocasio, D., Juin, S., Berendes, D., Heitzinger, K., Prentice-Mott, G., Desormeaux, A. M., et al. (2023). Cholera outbreak - Haiti, September 2022–January 2023. MMWR Morb. Mortal. Wkly Rep. 72, 21–25. doi: 10.15585/mmwr.mm7202a1
Wang, H., Xing, X., Wang, J., Pang, B., Liu, M., Larios-Valencia, J., et al. (2018). Hypermutation-induced in vivo oxidative stress resistance enhances Vibrio cholerae host adaptation. PLoS Pathog. 14:e1007413. doi: 10.1371/journal.ppat.1007413
Wang, J., Yan, M., Gao, H., Lu, X., and Kan, B. (2017). Vibrio cholerae colonization of soft-shelled turtles. Appl. Environ. Microbiol. 83:e00713-17. doi: 10.1128/AEM.00713-17
Watve, S., Barrasso, K., Jung, S. A., Davis, K. J., Hawver, L. A., Khataokar, A., et al. (2020). Parallel quorum-sensing system in Vibrio cholerae prevents signal interference inside the host. PLoS Pathog. 16:e1008313. doi: 10.1371/journal.ppat.1008313
Weil, A. A., Becker, R. L., and Harris, J. B. (2019). Vibrio cholerae at the intersection of immunity and the microbiome. mSphere 4:e00597-19. doi: 10.1128/mSphere.00597-19
Weill, F.-X., Domman, D., Njamkepo, E., Tarr, C., Rauzier, J., Fawal, N., et al. (2017). Genomic history of the seventh pandemic of cholera in Africa. Science 358, 785–789. doi: 10.1126/science.aad5901
WHO and UNICEF (2017). “Progress on drinking water, sanitation, and hygiene: 2017 update and SDG baselines ”. (Geneva, Switzerland: World Health Organization and the United Nations Children’s Fund).
Withey, J. H., and DiRita, V. J. (2006). The toxbox: specific DNA sequence requirements for activation of Vibrio cholerae virulence genes by ToxT. Mol. Microbiol. 59, 1779–1789. doi: 10.1111/j.1365-2958.2006.05053.x
Withey, J. H., Nag, D., Plecha, S. C., Sinha, R., and Koley, H. (2015). Conjugated linoleic acid reduces cholera toxin production in vitro and in vivo by inhibiting Vibrio cholerae ToxT activity. Antimicrob. Agents Chemother. 59, 7471–7476. doi: 10.1128/AAC.01029-15
Wong, C. N., Ng, P., and Douglas, A. E. (2011). Low-diversity bacterial community in the gut of the fruitfly Drosophila melanogaster. Environ. Microbiol. 13, 1889–1900. doi: 10.1111/j.1462-2920.2011.02511.x
Xue, Y., Tu, F., Shi, M., Wu, C. Q., Ren, G., Wang, X., et al. (2016). Redox pathway sensing bile salts activates virulence gene expression in Vibrio cholerae. Mol. Microbiol. 102, 909–924. doi: 10.1111/mmi.13497
Yamamoto, S., Lee, K. I., Morita, M., Arakawa, E., Izumiya, H., and Ohnishi, M. (2018). Single circular chromosome identified from the genome sequence of the Vibrio cholerae O1 bv. El tor Ogawa strain V060002. Genome Announc. 6:e00564-18. doi: 10.1128/genomeA.00564-18
Yan, J., and Bassler, B. L. (2019). Surviving as a community: antibiotic tolerance and persistence in bacterial biofilms. Cell Host Microbe 26, 15–21. doi: 10.1016/j.chom.2019.06.002
Yan, L., Jin, Y., Zhang, B., Xu, Y., Peng, X., Qin, S., et al. (2022). Diverse aquatic animal matrices play a key role in survival and potential virulence of non-O1/O139 Vibrio cholerae isolates. Front. Microbiol. 13:896767. doi: 10.3389/fmicb.2022.896767
Yang, M., Liu, Z., Hughes, C., Stern, A. M., Wang, H., Zhong, Z., et al. (2013). Bile salt-induced intermolecular disulfide bond formation activates Vibrio cholerae virulence. Proc. Natl. Acad. Sci. U. S. A. 110, 2348–2353. doi: 10.1073/pnas.1218039110
Yang, S., Xi, D., Wang, X., Li, Y., Li, Y., Yan, J., et al. (2020). Vibrio cholerae VC1741 (PsrA) enhances the colonization of the pathogen in infant mice intestines in the presence of the long-chain fatty acid, oleic acid. Microb. Pathog. 147:104443. doi: 10.1016/j.micpath.2020.104443
Yildiz, F. H., and Visick, K. L. (2009). Vibrio biofilms: so much the same yet so different. Trends Microbiol. 17, 109–118. doi: 10.1016/j.tim.2008.12.004
You, J. S., Yong, J. H., Kim, G. H., Moon, S., Nam, K. T., Ryu, J. H., et al. (2019). Commensal-derived metabolites govern Vibrio cholerae pathogenesis in host intestine. Microbiome 7:132. doi: 10.1186/s40168-019-0746-y
Zhao, W., Caro, F., Robins, W., and Mekalanos, J. J. (2018). Antagonism toward the intestinal microbiota and its effect on Vibrio cholerae virulence. Science 359, 210–213. doi: 10.1126/science.aap8775
Zhu, J., Miller, M. B., Vance, R. E., Dziejman, M., Bassler, B. L., and Mekalanos, J. J. (2002). Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. Proc. Natl. Acad. Sci. U. S. A. 99, 3129–3134. doi: 10.1073/pnas.052694299
Keywords: cholera, Vibrio cholerae, animal models, bacterial pathogenesis, aquatic bacteria
Citation: Walton MG, Cubillejo I, Nag D and Withey JH (2023) Advances in cholera research: from molecular biology to public health initiatives. Front. Microbiol. 14:1178538. doi: 10.3389/fmicb.2023.1178538
Received: 03 March 2023; Accepted: 14 April 2023;
Published: 22 May 2023.
Edited by:
Axel Cloeckaert, Institut National de recherche pour l’agriculture, l’alimentation et l’environnement (INRAE), FranceReviewed by:
James E. Bina, University of Pittsburgh, United StatesCopyright © 2023 Walton, Cubillejo, Nag and Withey. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jeffrey H. Withey, andpdGhleUBtZWQud2F5bmUuZWR1
†Present Address: Dhrubajyoti Nag, Biology Department, City College of New York, New York, NY, United States
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.