
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 17 April 2023
Sec. Microbial Physiology and Metabolism
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1177123
This article is part of the Research Topic The Discovery of Bioactive Natural Products: The Isolation and Structural Elucidation, Genome Mining, OSMAC Strategy, and Biotransformation View all 15 articles
Lichen natural products are a tremendous source of new bioactive chemical entities for drug discovery. The ability to survive in harsh conditions can be directly correlated with the production of some unique lichen metabolites. Despite the potential applications, these unique metabolites have been underutilized by pharmaceutical and agrochemical industries due to their slow growth, low biomass availability, and technical challenges involved in their artificial cultivation. At the same time, DNA sequence data have revealed that the number of encoded biosynthetic gene clusters in a lichen is much higher than in natural products, and the majority of them are silent or poorly expressed. To meet these challenges, the one strain many compounds (OSMAC) strategy, as a comprehensive and powerful tool, has been developed to stimulate the activation of silent or cryptic biosynthetic gene clusters and exploit interesting lichen compounds for industrial applications. Furthermore, the development of molecular network techniques, modern bioinformatics, and genetic tools is opening up a new opportunity for the mining, modification, and production of lichen metabolites, rather than merely using traditional separation and purification techniques to obtain small amounts of chemical compounds. Heterologous expressed lichen-derived biosynthetic gene clusters in a cultivatable host offer a promising means for a sustainable supply of specialized metabolites. In this review, we summarized the known lichen bioactive metabolites and highlighted the application of OSMAC, molecular network, and genome mining-based strategies in lichen-forming fungi for the discovery of new cryptic lichen compounds.
Plant-derived natural products or their derivatives were a valuable source of therapeutic agents and played a key role in the treatment of various diseases, e.g., cancer chemotherapy, infectious diseases, cardiovascular diseases, and lipid metabolism disorders (Mann, 2002; Newman and Cragg, 2012; Atanasov et al., 2015; Waltenberger et al., 2016). However, natural plant product-based drug discovery has some difficulties because of technical barriers to screen natural products in high-throughput assays against molecular targets and synthetic compounds not meeting the expectations of an increasing number of new drugs reaching the market (Atanasov et al., 2015), thus scientists have had to turn their attention to other organisms. Microbes have proven to be a bountiful source of secondary metabolites that have been successfully developed as crucial drug leads. We have already known that the structures of over 80,000 natural products from microbes (Demain, 2014) and over 80% of the antibiotics are produced by microbes (de Lima Procópio et al., 2012) since the discovery of penicillin in 1928 (Demain, 2014). Due to the extensive use of antibiotics for common infections, pathogens are showing high resistance (Harikumar and Krishanan, 2022); therefore, there is an urgent need for finding novel drugs. Grube made a point of view that microbial symbioses have a high potential leading to a wide variety of unique structures and metabolic activities (Grube and Berg, 2009).
Several studies have shown that lichens are productive organisms for the synthesis of a broad range of secondary metabolites. Lichen is a stable community in the ecosystem of the Earth's biosphere, which is composed of a mutualistic relationship between fungi and algae or between fungi and cyanobacteria (Figure 1). However, the identity of lichens is considered based on the fungal partner, and to date, the predominant records of lichens that have been identified are ascomycetes in nature. Thus, the term “lichen-forming fungi (LFF)” refer to the fungi that live in lichen thallus throughout the entire life cycle by establishing a co-benefit symbiotic relationship without causing any adverse effect and are different from those endolichenic fungi (Muggia et al., 2009).
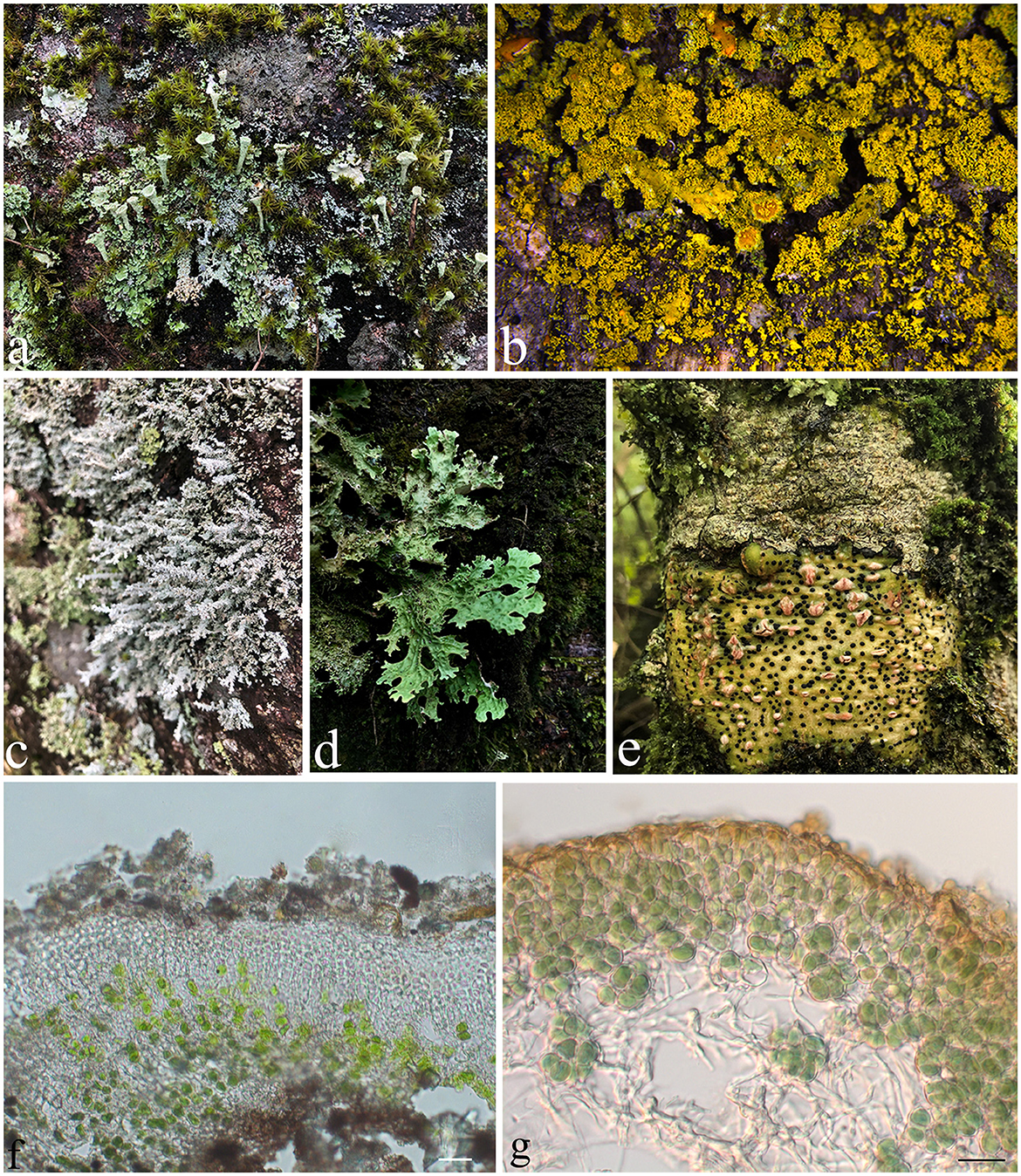
Figure 1. The diversity of lichens. (a) Cladonia sp.; (b) Candelaria sp.; (c) Stereocaulon sp.; (d) Sticta sp.; (e) Pyrenula sp.; (f) Thallus of Endocarpon pallidulum in vertical section, with green algae Diplosphaera chodatii as the photobiont; (g) Thallus of Peltula submarginata in vertical section, with cyanobacteria Chroococcales as the photobiont. Photos (b, f) were taken by Xue XD and Zhang TT, respectively, and photos (a, c–e) were taken by Yang QX. Scale bars: (f) = 20 μm, (g) = 10 μm.
Evidence from lichen fossils indicated that the interactions between fungi and algae have existed for at least 400 million years (Lücking and Nelsen, 2018), and studies have shown that lichen occurs over 10% of the terrestrial surface, especially extreme and aggressive environmental conditions that are not conducive to individual survival, such as extreme cold Arctic and Antarctic regions (Lee et al., 2014), hot and arid deserts (Kranner et al., 2008), alpine areas with strong UV irradiation, and on rocks or non-fertile soils (de Vera et al., 2003; Seymour et al., 2005; Boustie et al., 2010; Nguyen et al., 2013). This tendency of lichens to tolerate the extreme environment can be correlated with the production of both a unique and diverse range of metabolites known as lichen substances (Schweiger et al., 2022). Fungi and algae co-evolved unique biosynthetic pathways and metabolic mechanisms to synthesize these complex metabolites over a long period of time. Primary and secondary metabolisms are the two main groups of lichen compounds. Primary metabolism is the basic substance constituting the structure of lichen and includes proteins, amino acids, carotenoids, polysaccharides, and vitamins (Goga et al., 2020; Packiam and Perumal, 2022). The fungal filaments provide small, structurally complex, water-insoluble, and crystalline secondary metabolism, which can comprise up to 20% of the dry mass of thallus weight (Nguyen et al., 2013; González-Burgos et al., 2019; Zhao et al., 2021). Unlike primary metabolites, lichen secondary metabolism is not directly involved in growth but synthesized for algae or cyanobacteria protection (Muggia et al., 2009).
Lichens are known to produce small molecular compounds such as aliphatic and aromatic compounds, thus far, over 1,000 compounds have been identified (Shrestha and St. Clair, 2013). According to biosynthetic origins and chemical structures, lichen compounds were classified (Culberson and Elix, 1989), which were synthesized by acetyl-malonate, mevalonate, and shikimate pathways existing in all organisms as key routes for natural metabolism. The biosynthesis of lichen depsides, depsidones, dibenzofurans, chromones, xanthones, and anthraquinones occurs via the acetyl-malonate pathway, by which most bioactive compounds are synthesized, with coenzyme A as the precursor and polyketide synthase (PKS) as the responsible enzyme (Ibrahim et al., 2018). The most common lichen compounds synthesized by this pathway include evernic acid (Muggia et al., 2009), lecanoric acid (Lawrey, 1986), gyrophoric acid (Garima et al., 2022a), atranorin (Lawrey, 1986; Majchrzak-Celinska et al., 2022), thamnolic acid (Culberson et al., 1986; Jeong et al., 2021), umbilicaric acid (Posner et al., 1991; Yoshimura et al., 1994), protocetraric acid (Nishanth et al., 2015), fumarprotocetraric acid (Igoli et al., 2014; Ranković and Mišić, 2014), stictic acid (Bellio et al., 2017; Pejin et al., 2017), usnic acid (Moreira et al., 2015; Sepahvand et al., 2021), lepraric acid (Aberhart et al., 1969; Murugan et al., 2021), and thiophanic acid (Arshad et al., 1968; Dayan and Romagni, 2001). Usnic acid, one of the most common, isolated, and discussed lichen compounds, is well-known as an antibiotic with many pharmacological activities including antibacterial, antiprotozoal, anti-cytotoxic, anti-proliferative, antioxidant, and anti-inflammatory (Cocchietto et al., 2002). The mechanisms of bioactivity of usnic acid modify the structures of proteins causing irreversible changes and may even produce apoptosis. Lichen also produces an array of secondary metabolites derived from the mevalonate pathway, which play essential roles in the regulation of cell growth and development, and the products appear to be potentially interesting therapeutic targets for many areas of research such as oncology, autoimmune disorder, atherosclerosis, and Alzheimer disease (Buhaescu and Izzedine, 2007). The mevalonate pathway is mainly associated with the production of terpenes, steroids, and carotenoids (Goga et al., 2020). Until now, more than 20 different triterpene compounds from lichens have been reported. The shikimic acid pathway, ubiquitous in microorganisms and plants, provides precursors for the biosynthesis of primary metabolites such as aromatic amino acids and folic acid (Wilson et al., 1998). This pathway is mainly related to pulvinic acid and terphenylquinone pigments (Edwards et al., 2003), which help lichen adapt to UV stress by absorption and re-emitting the UV radiation as fluorescence or heat (Nguyen et al., 2013). The representative structures of lichen natural products are shown in Figure 2.
Lichen cells contain many types of natural metabolites and other bioactive molecules, receiving increased attention due to their industrial, pharmaceutical, biotechnological, medical, and cosmetics applications (Elkhateeb and Daba, 2019). Many studies have supported the potentiality of different lichen species to produce unique natural compounds with different physicochemical and biological activities (Hamida et al., 2021). Their utilization in folklore as medicines in the treatment of diverse diseases, such as arthritis, alopecia, constipation, kidney diseases, leprosy, pharyngitis rabies, infection, worm, and infestation for several centuries, has been recorded in different pharmacopeias by native Americans, Haitian, Indians, Chinese, and Europeans (Romagni and Dayan, 2002; Elkhateeb and Daba, 2019). In Table 1, the known lichen natural products with different bioactivities were summarized, among which most were isolated from the natural lichen thallus.
Despite the great potential of lichen bioactivity, lichens have been long neglected by mycologists and overlooked by the pharmaceutical industry. One reason is their slow growth in nature and difficult culture in either fermenters or glasshouses, or even cultivated in the open air, and has scarcely been studied from a biochemical perspective; another reason is due to difficulties in obtaining them in sufficient quantities and purities for structural elucidation and pharmacological research. These circumstances prompted lichenologists to develop more suitable strategies to look for more lichen compounds in categories and activities.
To improve the production of a wider range of natural products from a microorganism, different culturing conditions are generally used. Bode et al. refer to the fact that a single strain is capable of producing a diverse array of structural compounds under specific growth conditions (Zahroh et al., 2022), however, never produces the entire compounds at the same time under one set of environmental conditions because it is not matching with a cost between energetic and metabolism (Zarins-Tutt et al., 2016). Very small changes in the cultivation parameters, such as culture medium composition, pH, temperature of growth, salinity, aeration, and even the shape of culture vessel used, can completely alter, induce, or optimize the physiology of a microbial strain and in turn significantly affect the biosynthesis of such metabolites (Bode et al., 2002). For example, bioactivity-guided isolation of the fungus Aspergillus versicolor KU258497 yielded two new and six known cryptic metabolites when co-cultivated with the bacterium Bacillus subtilis 168 trpC2 on solid rice medium, among which one new compound showed strong cytotoxic activity against the mouse lymphoma cell line L5178Y (Abdelwahab et al., 2018). A corresponding strategy named OSMAC (one strain of many compounds) opened up a new industrial production avenue for compounds needed. For lichens, the majority of the bioactive compounds are exclusively produced by the LFF; however, there are some instances where the symbiotic photobiont, particularly cyanobacteria also engaged in the production of some key secondary metabolites (Cox et al., 2005).
It has been indicated that there is a strong application basis of OSMAC in discovering lichen natural products. Especially, lichenologists from all over the world are becoming more and more interested in not only physiology but also metabolite production (Crittenden and Porter, 1991), thus, they hoped that mycelium from the lichen thallus may be free-grown on artificial medium and produced lichen compounds without the algal or cyanobacterial partners. The tissue culture method invented by Yoshimura et al. (1993), a technique for the separation of the LFF from lichen thallus and culturing it alone, greatly pushed the fulfillment of this process.
However, growth rate and metabolite yield in LFF are inverse relationships (Timsina et al., 2013) and are influenced by culturing conditions, such as the availability and type of carbon and nitrogen source (Calvo et al., 2002; Behera et al., 2006; Verma et al., 2012). Simple sugar such as glucose, sucrose, and polyethylene glycol as sole carbon sources supported high LFF growth and production of usnic acid in Usnea ghattensis culture, in contrast, nitrogen sources such as amino acids (glycine, asparagine, alanine, or vitamins), especially glycine, supported the LFF growth but did not well-support usnic acid production (Behera et al., 2006). A strain of LFF isolated from a thallus of Parmotrema reticulatum was cultured on different culture media, and all the colonies developed well; however, atranorin, the major cortical lichen depside, was only detected when the colonies were grown over 5 and 10 months on solid LB medium. By comparison, colonies grown on MEYE and MY10 with a gradually dry treatment did not synthesize any lichen secondary metabolite of polyketides but primary triacylglycerides and fatty acids as the major metabolites (Fazio et al., 2009).
Bu'Lock proposed that mycelial growth was slow under conditions of poor nutrition, but secondary metabolism could be induced (Bu'Lock et al., 1974), which is related to the carbon-nutrient balance hypothesis (Bryant et al., 1983). From LFF of Endocarpon pusillum cultured on the optimized PDB, nine secondary metabolites including two new isoindolin-1-ones were detected, while three known compounds and a new naphthoquinone were isolated from the rice culture (Liu R. D. et al., 2022). Temperature is another important factor influencing LFF cultivation and chemical diversity in secondary metabolism. It is due to higher or lower temperatures that will inhibit the enzyme secretion of LFF (Feller et al., 1994).
Therefore, it is greatly deserved to expect more lichen natural products will be discovered by OSMAC strategy after changing and improving a series of cultural conditions. However, there is an inevitable problem existing in this process, that is, what is the relationship between more lichen products and the valuable bioactivity of these products because lichen secondary metabolism mainly originates from the fungal partner, i.e., LFF, but is produced when the organisms are in symbiotic association (Moreira et al., 2015). Poor nutrition sources and slow growth rate are the natural factors in LFF decided by the characteristics of lichen symbiosis. Heterodea muelleri in the field produced diffractaic and barbatic acids, whereas the LFF cultures did not contain any detectable secondary metabolites (Hager et al., 2008). The study of temperature by Hamada in LFF and lichen thallus of Ramalina siliquosa examined changing of polyketides, that is, the quantity of depside produced by LFF of Ramalina siliquosa was the highest at optimal culture conditions (Hamada, 1989), on the contrary, depsidone was increased in R. siliquosa thallus (Hamada, 1981). Brunauer showed that the LFF of Xanthoria elegans produces secondary metabolism that is not present in the naturally collected lichen thallus by HPLC examination because the presence of gene clusters enables LFF to produce a potentially larger variety of polyketides than thallus (Brunauer et al., 2007). Another example, two new dibenzofurans, isostrepsilic acid and hypostrepsilic acid, are synthesized in large quantities by LFF culture of Umbilicaria orientalis on malt-yeast extract medium containing sugar alcohols, but they have not been produced by this lichen in the field (Kon et al., 1997). The production of lichen compounds is based on the resistance to extreme environments, and if the environment changes, the interactions between lichen and the environment will change, similarly, LFF is out of symbiosis when they grow on the culture medium and equal to the environment change; thus, the compounds of LFF are different from lichen symbiont, and correspondingly, some lichen bioactive compounds cannot be detected in the LFF cultures (Dayan and Romagni, 2001).
However, when quantitatively and variously LFF metabolites were obtained after more focusing on fermentation broth and mycelium, they were often found different from those contained in the natural lichen thallus (Miyagawa et al., 1993), while the reason why we are interested in the lichen natural products is due to the bioactive metabolites produced by the symbiotic lichen thallus. Whether the bioactivity of more and different lichen natural products produced in the LFF by OSMAC strategy is much better than those less in the lichen thallus is not fully understood. Anyway, there would be a long way to go on well balancing this current conflict. Well understanding and solving this problem still need to establish on the more and more discovering of lichen natural products through OSMAC under breaking the lichen symbiosis, which is also closely related to the requirement of increasing the growth rate of LFF. Here, we present Table 2 to show some optimized media and cultural conditions being reported.
In the search for secondary metabolisms, analytical methods must be determined to use for the detection of the compounds (Scherlach and Hertweck, 2009; Tarkka et al., 2009; Palazzotto and Weber, 2018; Manish and Yogesh, 2019). The methods for identification and determining lichen metabolites in the liquid or solid medium are traditionally chemical empirical processes, which include classic spot tests, micro-crystallography, TLC, and HPLC. Kim confirmed the metabolite of Cladonia rangiferina by using HPLC and reported that usnic acid could not be found in C. rangiferina despite the gene cluster producing usnic acid being observed in the genome (Kim et al., 2021). However, these cheap but not sensitive enough methods will fail when the quantities of metabolites are below the detection limit or when the similar retention time of other metabolites overlaps (Egbert et al., 2022).
Mass spectrometry (MS) is a fast, modern, and simple tool for the structure identification of chemical substances (Wambui et al., 2021), and many lichen compounds and functional groups have been identified using MS (Huneck, 1999). In recent years, several studies focusing on lichen chemistry highlighted the use of a range of hyphenated technology. Mass spectrometry (MS), due to its sensitivity and Nuclear Magnetic Resonance (NMR) spectroscopy coupled with chromatographic techniques, has been recognized as a key technology to study metabolomics (Krug and Muller, 2014). It has been well-demonstrated that liquid chromatography (LC)–MS/MS is considerably more sensitive for the analysis of usnic acid (Cansaran et al., 2006; Sveshnikova et al., 2019; Xu et al., 2022).
Mass spectrometry (MS)/MS-based molecular networking and extensive spectroscopic analyses involving GIAO (Gauge-Independent Atomic Orbital) NMR shift calculation led to the isolation and identification of novel quinoid lichen pigments (Lagarde et al., 2021). However, although MS is the most sensitive and powerful method which detects and elucidates extremely low-abundance metabolites occurring in natural product research, it does not provide any information concerning the spatial and temporal distribution of metabolites. Mass Spectrometry Imaging (MSI) visualizes the production, location, and distribution of metabolites, which is newly used in lichens to visualize the accumulation of various polyketides such as parietin, physodic acid, atranorin, and pinastric acid in different tissues of the lichen and localize the phenolic compounds (Gadea et al., 2020). Desorption electrospray ionization-imaging mass spectrometry (DESI-IMS) realized the spatial distribution of usnic acid in cross-sections of the lichen thallus (Xu et al., 2022).
Some other MS-based metabolomics tools, such as electron ionization-mass spectrometry (EI-MS) (Kai et al., 2017), high performance liquid chromatography-diode array detector-mass spectrometry (HPLC-DAD-MS) (Castro et al., 2017), electron spray ionization-mass spectrometry (HESI-MS/MS) fragmentation patterns (Castro et al., 2017), and liquid chromatography-diode array detector-tandem mass spectrometry (UPLC-PDA-MS/MS) (Kumar et al., 2018), also help to identify various novel lichen metabolites and increase the understanding on a complex biological system.
To facilitate the lichen chemistry, an open-access MS/MS-based library with 250 metabolites known as the Lichen DataBase (LDB) was published by Olivier–Jimenez team. To aid this area of research, the MetaboLights database was generated containing the MS spectra of metabolites. Complementing this, the GNPS platform (CCMSLIB00004751209 to CCMSLIB00004751517) contains the merged spectra of these metabolites within a metadata file. Such a fundamental database empowers research on lichen chemistry by prioritizing novel metabolites (Olivier-Jimenez et al., 2019).
In addition to MS, NMR spectroscopy has also been widely used to determine the structure of organic molecules, which is typically coupled with LC/GC for quantitative analysis of low molecular weight compounds. Metabolite profiles in crude extracts of Xanthoria elegans thalli during hydration and dehydration were assessed by using 31P- and 13C-NMR, and approximately 30 metabolites were identified and quantified (Aubert et al., 2007). The 1H-NMR spectra of Xanthoria parietina and Peltigera horizontalis crude extract displayed lichen-specific features with strong signals and confirmed untargeted analysis on a quantitative basis (Eisenreich et al., 2011). Moreover, NMR demonstrates which atoms are present in neighboring groups. Ultimately, NMR can provide information on how many atoms are present in each of these environments (Altemimi et al., 2017). Other imaging techniques such as Raman microscopic analysis can provide time-resolved information about the distribution of major compounds in lichens (Edwards et al., 1997; Liao et al., 2010; Gadea et al., 2020; Xu et al., 2022). FTIR imaging and Raman microscopy were used to localize the presence of usnic acid in Cladonia arbuscular, C. Uncialis, and C. sulphurina (Liao et al., 2010).
If it is the common fact that is not easily solved now about the conflict between more lichen natural products by OSMAC from LFF cultures and uncertain or not very well bioactivity compared with those isolated from lichen thallus, genome mining-based strategy will be a more explicit way to discover lichen natural products. With the development of bioinformatics and the applying next-generation sequencing data, there has indeed been more focus on natural product discovery based on genomics (Garima et al., 2022b; Luo et al., 2022). Genome mining has become a powerful tool to discover compounds, identify cryptic biosynthetic gene clusters, characterize the potential biosynthetic pathways, and predict the skeletal structure of the relative products (Liu Q. et al., 2022; Liu T. et al., 2022; Kalra et al., 2023). An increasing understanding of high-quality genome sequencing and genome mining techniques coupled with the introduction of powerful computational toolkits facilitates the process of connecting these gene clusters with key compounds (Li et al., 2016). Comparing the traditional method for the identification of biosynthetic gene clusters by using MS and NMR-based, in silico genome mining has become a crucial strategy for the discovery and characterization of gene clusters (Alam et al., 2022). Many web portals contain databases and tools to identify the metabolites by using BLAST, Diamond, and HMMer alignment tools. After uploading the genome to websites, the results of the detection and characterization of secondary metabolites are achieved soon. AntiSMASH (Medema et al., 2011), PRISM (Skinnider et al., 2017), and MIBiG (Kautsar et al., 2020) are representative in silico tools for predicting various types of gene clusters, and they were developed to automate biosynthetic gene clusters instead of much manual intervention in genome sequences (Kenshole et al., 2021). Among those three tools, antiSMASH is the largest database of biosynthetic gene cluster analysis, PKS, and non-ribosomal peptide synthase (NRPS) substrate specificity prediction, as well as known and unknown biosynthetic gene clusters comparison (Medema et al., 2011). In addition, antiSMASH was used to predict the molecular structure sequence database. In silico genome tool with antiSMASH and BLAST2GO programs investigated the type I-PKS module candidates in nine publicly available LFF genomes (Erken et al., 2021). In addition, rule-based tools such as antiSMASH, ClusterFinder, and RNNs, and machine learning tools have been developed to allow the identification of unknown biosynthetic gene clusters (Cimermancic et al., 2014; Hannigan et al., 2019). However, a much higher false-positive rate than the rule-based tools is the weakness of machine learning-based genome mining tools. Open sources such as Prodigal and automated annotation help reduce false-positive identification. All these softwares are powerful tools that help to make genome mining in silico of the interesting LFF.
The genome mining study of a few publicly available LFF genomes suggested the importance of genome mining at the strain level, as it increases the likelihood that researchers discover useful derivatives of known secondary metabolites. An integrated approach utilizing genomics and metabolomics is needed to study the lichen complex systems. Recently, genome mining and comparative genomics strategy were used to assess biosynthetic gene clusters and putative regulators of LFF Evernia prunastri and Pseudevernia furfuracea. The results showed that the NR-PKS from LFF Pseudevernia furfuracea produces depside lecanoric acid, which has never been detected from lichen thallus in nature (Calchera et al., 2019). Genome mining analysis based on a homology searching approach revealed that enzymes of grayanic acid, patulin, and betaenones A-C biosynthesis are encoded by Cladonia uncialis genome (Bertrand et al., 2018), and the result corresponds with Shishido et al. (2021). The uptake of advanced analytical techniques and next-generation computational tools brought a breakthrough in lichen chemistry and resulted in the identification of various novel compounds. Moreover, understanding the genetic components leading to the biosynthesis of these metabolites provides an opportunity to exploit their commercial utilization by employing synthetic biology approach.
In LFF, polyketides are the most common class of secondary metabolites. With the help of gene knockout studies, it has been observed that cryptic PKS gene codes for PKS required for the biosynthesis of the representative polyketide orsellinic acid. Polyketides synthesized by three types of multidomain polyketide synthases in the sequential acyl acetyl-polymalonyl pathway are major, structurally diverse classes of natural products (Lin and Qu, 2022; Liu Q. et al., 2022). In the case of bacteria and fungi, PKSs belong to types I and II, while type III is present in higher plants. Despite structural differences, almost all PKSs biosynthesize polyketides via sequential decarboxylative Claisen condensation of acyl-coenzyme A (CoA) precursors and use ketoacyl synthase to catalyze the C–C bond formed during carbon chain assembly, and this process is as similar as fatty acid synthases (Lin and Qu, 2022). From the ecological perspective, these polyketide-based secondary metabolites afford a large amount of cytotoxic or antibiotic compounds to adapt to the competitive living environment. Many of these compounds or their derivatives have emerged as clinically useful drugs or are promising drug candidates. Genetic regulation study of lichen or LFF secondary metabolism is at an early stage, and as time passes and technology advances, more and more research will be covered in this field (Valarmathi et al., 2009; Calchera et al., 2019; Singh et al., 2021). Recently, several LFF PKS genes have been cloned, such as type I NR-PKS gene XsmPKSI from Xanthoria substrigosa (Hametner and Stocker-Wörgötter, 2015); three new NR-PKS genes such as UlPKS2, UlPKS4, and UlPKS6 from Usnea longissima (Wang et al., 2014); and XsePKS1 from Xanthoria semiviridis (Chooi et al., 2008). In addition, some studies of polyketide synthase genes have also focused on phylogenomic analysis (Proctor et al., 2007; Wang et al., 2018; Kealey et al., 2021). The increasing number of phylogenomic analyses shows that a single fungal genome may contain more than one PKS gene, and each species of fungi can produce more than one polyketide or polyketide family (Stocker-Wörgötter, 2008). For example, 12 PKS genes have been identified in Cladonia grayi (Shukla et al., 2010). Armaleo et al. (2011) identified a likely orcinol decided PKS and other pathway genes in its metabolic cluster, and it was the first genetic evidence for a complete depside/depsidone biosynthetic pathway. Experimental data that seven complete non-reducing and nine highly-reducing PKS genes indicated Nppks7 was a new PKS that participated in usnic acid biosynthesis in LFF Nephromopsis pallescens (Wang et al., 2018).
The complex lichen biology as filamentous fungi, transcriptionally silent, and trace expression make an artificial synthesis of interesting secondary metabolites difficult (Harvey et al., 2018). Heterologous expression of biosynthetic gene clusters in a non-natural host or model system expedites natural product discovery, elucidation, and mass production. The apposite choice of host is one of the keys to successful heterologous expression. Due to many advantages, such as fast growth, high cell density, low cost, simple cultivation medium, fast transformation procedure, and ability to process and correctly splice introns, Saccharomyces cerevisiae (Kealey et al., 2021), Fusarium venenatum (Sinnemann et al., 2000), Aspergillus nidulans (Sinnemann et al., 2000), A. niger (Sinnemann et al., 2000), A. oryzae (Gagunashvili et al., 2009), and Neurospora crassa are the experimentally well-developed strains and are considered as the potential hosts for the expression of lichen DNA (Qiao et al., 2019). In other expression systems, to avoid the influence of the surrogate host's metabolism on heterologous biosynthesis, Ascochyta rabiei, chosen as the host, is a genetically tractable, wild-type plant-pathogenic fungus without the biosynthetic gene cluster of phytotoxic solanapyrones (Kim et al., 2021).
Using PKS genes as a heterologous expression of genes for filamentous fungal secondary metabolites has been widely reported (Gressler et al., 2011; Sakai et al., 2012; Qian et al., 2020). Although PKS genes of lichen and filamentous fungi showed the greatest homology, only a few PKS genes have been isolated and characterized functionally from lichen or LFF. As the first PKS gene from Solorina crocea LFF, PyrG encoding decarboxylase was functionally expressed under its own promoter in A. nidulans (Sinnemann et al., 2000). The result indicated that a heterologous expression system is a useful tool for the functional characterization of genes. Another example is that two pairs of degenerated primers have been used to locate and clone PKS genes containing a CMeT domain from Xanthoparmelia semiviridis (Chooi et al., 2008). Early functional research of lichen PKS genes mainly focused on symbiosis, physiology, and biochemistry because all the studies that attempted to express the PKS gene of unique lichen compounds failed (Chooi et al., 2008). Until recently, de novo biosynthetic PKS genes of atranorin and lecanoric acid have been successfully heterologously expressed (Kealey et al., 2021; Kim et al., 2021). Atranorin is one of the most concerned lichen compounds. The results from lichens such as Cladonia, Stereocaulon alpinum (Kim et al., 2021), and Bacidia rubella (Gerasimova et al., 2022) revealed that the PKS23 gene (atr1), a cytochrome P450 gene (art2) for oxidation, an O-methyltransferase (OMT) gene (atr3), and transporter gene (atr4) were involved in producing atranorin.
Lichen secondary metabolites are of major interest due to their applicability as therapeutic agents. However, a special way of symbiosis, extreme living environment, and slow growth of lichen limit the constant need for lichen compounds in industry and pharmacy. The analysis of genome sequence revealed that there exist silent biosynthetic gene clusters, which are usually not expressed until being activated, leading to the discovery of lichen compounds much inadequate. OSMAC strategy is a powerful and mature method for enhancing the chemodiversity of LFF natural compounds, under which new drugs could be obtained by manipulating nutritional or environmental factors of fermentation to activate silent gene clusters. However, sometimes, the results of the OSMAC strategy are not very satisfying because it has a limited capacity to mimic the complexities of natural environmental changes. MS-based molecular network strategy further facilitates lichen chemistry, especially linked to a series of databases such as LDB and MetaboLights. Genome- and bioinformatics-based genome mining strategy not only makes up for the difficulties and shortcomings of the OSMAC strategy but also strongly pushes the identification of biosynthetic gene clusters and increases the rate of discovery of new products. Genome mining strategy, covering several different usage cases in animal, plant, and microbe, shows diverse ways, in which genomic data can be used to uncover new secondary metabolites, improves our understanding of their biosynthesis, and uncovers long-term biosynthetic mysteries, but for lichen, it is just the beginning. Nowadays, various strategies for inducing the expression of silent biosynthetic gene clusters have been developed. However, each strategy has its own limitations, and no strategy could be universal for all strains. Furthermore, significant advances are needed in terms of the enrichment of the database for lichen metabolites together with the general standardization of different generations of data. A combination of OSMAC, molecular network, and genome mining-based strategies will be greatly helpful to predict the biosynthesis and accumulation of specific natural products, discover numerous novel secondary metabolites with a range of attractive bioactivities, and pursue the establishment and maintenance of the lichen symbiosis.
XW: conceptualization. MR: writing—original draft preparation. SJ, YW, XP, FP, and XW: writing—reviewing and editing. All authors read and approved the manuscript.
This study was supported by the National Natural Science Foundation of China (grant numbers 32070096, 32170082, and 32270007) and the Space Application System of China Manned Space Program (KJZ-YY-WSM05).
XP and FP were employed by Jiangxi Xiankelai Biotechnology Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abdelwahab, M. F., Kurtán, T., Mándi, A., Müller, W. E. G., Fouad, M. A., Kamel, M. S., et al. (2018). Induced secondary metabolites from the endophytic fungus Aspergillus versicolor through bacterial co-culture and OSMAC approaches. Tetrahedron Lett. 59, 2647–2652. doi: 10.1016/j.tetlet.2018.05.067
Aberhart, D. J, Overton, K. H., and Huneck, S. (1969). Studies on lichen substances. Part LXII. Aromatic constituents of the lichen Roccella fuciformis DC. A revised structure for lepraric acid. J. Chem. Soc. 5, 704–707. doi: 10.1039/j39690000704
Alam, K., Hao, J., Zhong, L., Fan, G., Ouyang, Q., Islam, M. M., et al. (2022). Complete genome sequencing and in silico genome mining reveal the promising metabolic potential in Streptomyces strain CS-7. Front. Microbiol. 13, 939919. doi: 10.3389/fmicb.2022.939919
Altemimi, A., Lakhssassi, N., Baharlouei, A., Watson, D. G., and Lightfoot, D. A. (2017). Phytochemicals: extraction, isolation, and identification of bioactive compounds from plant extracts. Plants 6, 42. doi: 10.3390/plants6040042
Armaleo, D., Sun, X., and Culberson, C. (2011). Insights from the first putative biosynthetic gene cluster for a lichen depside and depsidone. Mycologia 103, 741–754. doi: 10.3852/10-335
Arshad, M., Devlin, J. P., Olli, W. D., and Wheele, R. E. (1968). The constitution of sordidone and its relation to thiophanic acid. Chem. Commun. 3, 154–155. doi: 10.1039/c19680000154
Atalay, F., Halici, M. B., Mavi, A., Cakir, A., Odabasoglu, F., Kazaz, C., et al. (2011). Antioxidant phenolics from Lobaria pulmonaria (L.) Hoffm. and Usnea ongissima Ach. lichen species. Turk. J. Chem. 35, 647–661. doi: 10.3906/kim-1008-847
Atanasov, A. G., Waltenberger, B., Pferschy-Wenzig, E. M., Linder, T., Wawrosch, C., Uhrin, P., et al. (2015). Discovery and resupply of pharmacologically active plant-derived natural products: a review. Biotechnol. Adv. 33, 1582–1614. doi: 10.1016/j.biotechadv.2015.08.001
Aubert, S., Juge, C., Boisson, A. M., Gout, E., and Bligny, R. (2007). Metabolic processes sustaining the reviviscence of lichen Xanthoria elegans (Link) in high mountain environments. Planta 226, 1287–1297. doi: 10.1007/s00425-007-0563-6
Bayir, Y., Odabasoglu, F., Cakir, A., Aslan, A., Suleyman, H., Halici, M., et al. (2006). The inhibition of gastric mucosal lesion, oxidative stress and neutrophil-infiltration in rats by the lichen constituent diffractaic acid. Phytomedicine 13, 584–590. doi: 10.1016/j.phymed.2005.07.002
Behera, B. C., Verma, N., Sonone, A., and Makhija, U. (2006). Experimental studies on the growth and usnic acid production in “lichen” Usnea ghattensis in vitro. Microbiol. Res. 161, 232–237. doi: 10.1016/j.micres.2005.08.006
Bellio, P., Di Pietro, L., Mancini, A., Piovano, M., Nicoletti, M., Brisdelli, F., et al. (2017). SOS response in bacteria: Inhibitory activity of lichen secondary metabolites against Escherichia coli RecA protein. Phytomedicine 29, 11–18. doi: 10.1016/j.phymed.2017.04.001
Bertrand, R. L., Abdel-Hameed, M., and Sorensen, J. L. (2018). Lichen biosynthetic gene clusters part II: homology mapping suggests a functional diversity. J. Nat. Prod. 81, 732–748. doi: 10.1021/acs.jnatprod.7b00770
Bessadottir, M., Eiriksson, F. F., Becker, S., Ogmundsdottir, M. H., Omarsdottir, S., Thorsteinsdottir, M., et al. (2015). Anti-proliferative and pro-apoptotic effects of lichen-derived compound protolichesterinic acid are not mediated by its lipoxygenase-inhibitory activity. Prostaglandins Leukot. Essent. Fatty Acids 98, 39–47. doi: 10.1016/j.plefa.2015.04.009
Bessadottir, M., Skuladottir, E. A., Gowan, S., Eccles, S., Ogmundsdottir, S., Ogmundsdottir, H. M., et al. (2014). Effects of anti-proliferative lichen metabolite, protolichesterinic acid on fatty acid synthase, cell signalling and drug response in breast cancer cells. Phytomedicine 21, 1717–1724. doi: 10.1016/j.phymed.2014.08.006
Bode, H., Bethe, B., Hofs, R., and Zeeck, A. (2002). Big effects from small changes: possible ways to explore nature's chemical diversity. Chem. BioChem. 3, 619–627. doi: 10.1002/1439-7633(20020703)3:7<619::AID-CBIC619>3.0.CO;2-9
Boustie, J., Tomasi, S., and Grube, M. (2010). Bioactive lichen metabolites: alpine habitats as an untapped source. Phytochem. Rev. 10, 287–307. doi: 10.1007/s11101-010-9201-1
Brunauer, G., Hager, A., Grube, M., Türk, R., and Stocker-Wörgötter, E. (2007). Alterations in secondary metabolism of aposymbiotically grown mycobionts of Xanthoria elegans and cultured resynthesis stages. Plant Physiol. Biochem. 45, 146–151. doi: 10.1016/j.plaphy.2007.01.004
Bryant, J. P., Stuart Chapin, F., and Klein, D. R. (1983). Carbon nutrient balance of boreal plants in relation to vertebrate herbivory. Oikos 40, 357–368. doi: 10.2307/3544308
Buhaescu, I., and Izzedine, H. (2007). Mevalonate pathway: a review of clinical and therapeutical implications. Clin. Biochem. 40, 575–584. doi: 10.1016/j.clinbiochem.2007.03.016
Bu'Lock, J. D., Detroy, R. W., Hoštálek, Z., and Munim-Al-Shakarchi, A. (1974). Regulation of secondary biosynthesis in Gibberella fujikuroi. Trans. Br. Mycol. Soc. 62, 377–389. doi: 10.1016/S0007-1536(74)80046-X
Burlando, B., Ranzato, E., Volante, A., Appendino, G., Pollastro, F., Verotta, L., et al. (2009). Antiproliferative effects on tumour cells and promotion of keratinocyte wound healing by different lichen compounds. Planta Med. 75, 607–613. doi: 10.1055/s-0029-1185329
Calchera, A., Dal Grande, F., Bode, H. B., and Schmitt, I. (2019). Biosynthetic gene content of the ‘perfume lichens' Evernia prunastri and Pseudevernia furfuracea. Molecules 24, 203. doi: 10.3390/molecules24010203
Calvo, A. M., Wilson, R. A., Bok, J. W., and Keller, N. P. (2002). Relationship between secondary metabolism and fungal development. Microbiol. Mol. Biol. Rev. 66, 447–459. doi: 10.1128/MMBR.66.3.447-459.2002
Cansaran, D., Kahya, D., Yurdakulola, E., and Atakol, O. (2006). Identification and quantitation of usnic acid from the lichen Usnea species of Anatolia and antimicrobial activity. Z. Naturforsch. C. J. Biosci. 61, 773–776. doi: 10.1515/znc-2006-11-1202
Castro, O. N., Benites, J., Rodilla, J., Santiago, J. C., Simirgiotis, M., Sepulveda, B., et al. (2017). Metabolomic analysis of the lichen Everniopsis trulla using ultra high performance liquid chromatography-quadrupole-orbitrap mass spectrometry (UHPLC-Q-OT-MS). Chromatographia 80, 967–973. doi: 10.1007/s10337-017-3304-4
Chon, S. H., Yang, E. J., Lee, T., and Song, K. S. (2016). SS-secretase (BACE1) inhibitory and neuroprotective effects of p-terphenyls from Polyzellus multiplex. Food Funct. 7, 3834–3842. doi: 10.1039/c6fo00538a
Chooi, Y. H., Stalker, D. M., Davis, M. A., Fujii, I., Elix, J. A., Louwhoff, S. H., et al. (2008). Cloning and sequence characterization of a non-reducing polyketide synthase gene from the lichen Xanthoparmelia semiviridis. Mycol. Res. 112, 147–161. doi: 10.1016/j.mycres.2007.08.022
Cimermancic, P., Medema, M. H., Claesen, J., Kurita, K., Wieland Brown, L. C., Mavrommatis, K., et al. (2014). Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters. Cell 158, 412–421. doi: 10.1016/j.cell.2014.06.034
Claudia, C., Xavier, B., Jabrane, A, Katy, V., Daniel, G., Patrick, L., et al. (2018). Lobaric acid and pseudodepsidones inhibit NF-κB signaling pathway by activation of PPAR-γ. Bioorg. Med. Chem. Lett. 26, 5845–5851. doi: 10.1016/j.bmc.2018.10.035
Çobanglu, G., Sesal, C., Gölmen, B., and Çakar, S. (2010). Evaluation of the antimicrobial properties of some lichens. South West J. Hortic. Biol. Environ. 1, 153–158.
Cocchietto, M., Skert, N., Nimis, P. L., and Sava, G. (2002). A review on usnic acid, an interesting natural compound. Naturwissenschaften 89, 137–146. doi: 10.1007/s00114-002-0305-3
Cox, P. A., Banack, S. A., Murch, S. J., Rasmussen, U., Tien, G., Bidigare, R. R., et al. (2005). Diverse taxa of cyanobacteria produce beta-N-methylamino-L-alanine, a neurotoxic amino acid. Proc. Natl. Acad. Sci. U. S. A. 102, 5074–5078. doi: 10.1073/pnas.0501526102
Crittenden, P. D., and Porter, N. (1991). Lichen-forming fungi: potential sources of novel metabolites. Trends Biotechnol. 9, 409–414. doi: 10.1016/0167-7799(91)90141-4
Culberson, C. F., Culberson, W. L., and Johnson, A. (1986). Two new lichen products, elatinic acid and methyl barbatate, from the genus Haematomma (Ascomycotina, Haematommataceae). Mycologia 78, 888–891. doi: 10.1080/00275514.1986.12025345
Culberson, C. F., and Elix, J. A. (1989). “Lichen substances,” in Methods in Plant Biochemistry, ed J. B. Harborne (London: Academic Press), 509–535.
Dayan, F. E., and Romagni, J. G. (2001). Lichens as a potential source of pesticides. Pestic. Outlook 12, 229–232. doi: 10.1039/b110543b
de Barros Alves, G. M., de Sousa Maia, M. B., de Souza Franco, E., Galvão, A. M., da Silva, T. G., Gomes, R. M., et al. (2014). Expectorant and antioxidant activities of purified fumarprotocetraric acid from Cladonia verticillaris lichen in mice. Pulm. Pharmacol. Ther. 27, 139–143. doi: 10.1016/j.pupt.2013.07.002
de Lima Procópio, R. E., da Silva, I. R., Martins, M. K., de Azevedo, J. L., and de Araújo, J. M. (2012). Antibiotics produced by Streptomyces. Braz. J. Infect. Dis. 16, 466–471. doi: 10.1016/j.bjid.2012.08.014
de Vera, J. P., Horneck, G., Rettberg, P., and Ott, S. (2003). The potential of the lichen symbiosis to cope with extreme conditions of outer space – I. Influence of UV radiation and space vacuum on the vitality of lichen symbiosis and germination capacity. Int. J. Astrobiol. 1, 285–293. doi: 10.1017/S1473550403001216
Demain, A. L. (2014). Importance of microbial natural products and the need to revitalize their discovery. J. Ind. Microbiol. Biotechnol. 41, 185–201. doi: 10.1007/s10295-013-1325-z
Dévéhat, F. L. L., Legouin, B., Couteau, C., Boustie, J., and Coiffard, L. (2013). Lichenic extracts and metabolites as UV filters. J. Photochem. Photobiol. B 120, 17–28. doi: 10.1016/j.jphotobiol.2013.01.009
Díaz, E. M., Zamora, J. C., Ruibal, C., Divakar, P. K., González-Benítez, N., Devehat, F. L., et al. (2020). Axenic culture and biosynthesis of secondary compounds in lichen symbiotic fungi, the Parmeliaceae. Symbiosis 82, 79–93. doi: 10.1007/s13199-020-0071903
Ebrahim, H. Y., Elsayed, H. E., Mohyeldin, M. M., Akl, M. R., Bhattacharjee, J., Egbert, S., et al. (2016). Norstictic acid inhibits breast cancer cell proliferation, migration, invasion, and in vivo invasive growth through targeting C-Met. Phytother. Res. 30, 557–566. doi: 10.1002/ptr.5551
Edwards, H. G. M., Newton, E. M., Wynn-Williams, D. D., and Coombes, S. R. (2003). Molecular spectroscopic studies of lichen substances 1: parietin and emodin. J. Mol. Struct. 648, 49–59. doi: 10.1016/S0022-2860(02)00384-8
Edwards, H. G. M., Russell, N. C. D., and Wynn-Williams, D. (1997). Fourier transform raman spectroscopic and scanning electron microscopic study of cryptoendolithic lichens from Antarctica. J. Raman Spectrosc. 28, 685–690. doi: 10.1002/(SICI)1097-4555(199709)28:9<685::AID-JRS160>3.0.CO;2-X
Egbert, S., Hoffman, J. R., McMullin, R. T., Lendemer, J. C., and Sorensen, J. L. (2022). Unraveling usnic acid: a comparison of biosynthetic gene clusters between two reindeer lichen (Cladonia rangiferina and C. uncialis). Fungal Biol. 126, 697–706. doi: 10.1016/j.funbio.2022.08.007
Eisenreich, W., Knispel, N., and Beck, A. (2011). Advanced methods for the study of the chemistry and the metabolism of lichens. Phytochem. Rev. 10, 445–456. doi: 10.1007/s11101-011-9215-3
Elkhateeb, W. A., and Daba, G. M. (2019). Lichens, an alternative drugs for modern diseases. Int. J. Res. Pharm. Biosci. 6, 5–9.
Emsen, B., Ozdemir, O., Engin, T., Togar, B., Cavusoglu, S., Turkez, H., et al. (2019). Inhibition of growth of U87MG human glioblastoma cells by Usnea longissima Ach. An. Acad. Bras. Cienc. 91, e20180994. doi: 10.1590/0001-3765201920180994
Erken, M. T., Cansaran-Duman, D., and Tanman, U. (2021). In silico prediction of type I PKS gene modules in nine lichenized fungi. Biotechnol. Biotechnol. Equip. 35, 358–365. doi: 10.1080/13102818.2021.1879679
Fazio, A. T., Bertoni, M. D., Adler, M. T., Ruiz, L. B., Rosso, M. L., Muggia, L., et al. (2009). Culture studies on the mycobiont isolated from Parmotrema reticulatum (Taylor) Choisy: metabolite production under different conditions. Mycol. Prog. 8, 359–365. doi: 10.1007/s11557-009-0609-1
Feller, G., Narinx, E., Arpigny, J. L., Zekhnini, Z., Swings, J., Gerday, C., et al. (1994). Temperature dependence of growth, enzyme secretion and activity of psychrophilic Antarctic bacteria. Appl. Microbiol. Biotechnol. 41, 477–479. doi: 10.1007/BF00939039
Furmanek, L., Czarnota, P., and Seaward, M. R. D. (2021). The effect of lichen secondary metabolites on Aspergillus fungi. Arch. Microbiol. 204, 100. doi: 10.1007/s00203-021-02649-0
Gadea, A., Fanuel, M., Le Lamer, A. C., Boustie, J., Rogniaux, H., Charrier, M., et al. (2020). Mass spectrometry imaging of sspecialized metabolites for predicting lichen fitness and snail foraging. Plants 9, 70. doi: 10.3390/plants9010070
Gagunashvili, A. N., Davidsson, S. P., Jonsson, Z. O., and Andresson, O. S. (2009). Cloning and heterologous transcription of a polyketide synthase gene from the lichen Solorina crocea. Mycol. Res. 113, 354–363. doi: 10.1016/j.mycres.2008.11.011
Gaikwad, S., Verma, N., Sharma, B. O., and Behera, B. C. (2014). Growth promoting effects of some lichen metabolites on probiotic bacteria. J. Food Sci. Technol. 51, 2624–2631. doi: 10.1007/s13197-012-0785-x
Garima, S., Anjuli, C., Dominik, M., Henrique, V., Jürgen, O., Imke, S., et al. (2022a). A candidate gene cluster for the bioactive natural product gyrophoric acid in lichen-forming fungi. Microbiol. Spectr. 10, e0010922. doi: 10.1128/spectrum.00109-22
Garima, S., Grande, F. D., and Schmitt, I. (2022b). Genome mining as a biotechnological tool for the discovery novel biosynthetic genes in lichens. Front. Fungal Biol. 3, 993171. doi: 10.1101/2022.05.04.490581
Gerasimova, J. V., Beck, A., Werth, S., and Resl, P. (2022). High diversity of type I polyketide genes in Bacidia rubella as revealed by the comparative analysis of 23 lichen genomes. J. Fungi 8, 449. doi: 10.3390/jof8050449
Gissurarson, S. R., Sigurdsson, S. B., Wagner, H., and Ingolfsdottir, K. (1997). Effect of lobaric acid on cysteinyl-leukotriene formation and contractile activity of guinea pig taenia coli. J. Pharmacol. Exp. Ther. 280, 770–773.
Goga, M., Elečko, J., Marcinčinová, M., Ručová, D., Bačkorová, M., and Bačkor, M. (2020). “Lichen metabolites: an overview of some secondary metabolites and their biological potential,” in Co-Evolution of Secondary Metabolites, eds J. M. Mérillon, and K. G. Ramawat (Switzerland: Springer Nature), 1–36.
Gollapudi, S. R., Telikepalli, H., Jampani, H. B., Mirhom, Y. W., Drake, S. D., Bhattiprolu, K. R., et al. (1994). Alectosarmentin, a new antimicrobial dibenzofuranoid lactol from the lichen, Alectoria sarmentosa. J. Nat. Prod. 57, 934–938.
González-Burgos, E., Fernández-Moriano, C., and Gómez-Serranillos, M. P. (2019). Current knowledge on Parmelia genus: ecological interest, phytochemistry, biological activities and therapeutic potential. Phytochemistry 165, 112051. doi: 10.1016/j.phytochem.2019.112051
Gressler, M., Zaehle, C., Scherlach, K., Hertweck, C., and Brock, M. (2011). Multifactorial induction of an orphan PKS-NRPS gene cluster in Aspergillus terreus. Chem. Biol. 18, 198–209. doi: 10.1016/j.chembiol.2010.12.011
Grube, M., and Berg, G. (2009). Microbial consortia of bacteria and fungi with focus on the lichen symbiosis. Fungal Biol. Rev. 23, 72–85. doi: 10.1016/j.fbr.2009.10.001
Hager, A., Brunauer, G., Turk, R., and Stocker-Worgotter, E. (2008). Production and bioactivity of common lichen metabolites as exemplified by Heterodea muelleri (Hampe) Nyl. J. Chem. Ecol. 34, 113–120. doi: 10.1007/s10886-007-9408-9
Hamada, N. (1981). The effect of temperature on the content of the medullary depsidone salazinic acid in Ramalina siliquosa (lichens). Can. J. Bot. 60, 383–385. doi: 10.1139/b82-053
Hamada, N. (1989). The effect of various culture conditions on depside production by an isolated lichen mycobiont. Bryologist 92, 310–313. doi: 10.2307/3243399
Hametner, C., and Stocker-Wörgötter, E. (2015). “Type I NR-PKS gene characterization of the cultured lichen mycobiont Xanthoparmelia substrigosa (Ascomycota),” in Recent Advances in Lichenology, eds D. K. Upreti, P. K., Divakar, V. Shukla, and R. Bajpai (Berlin: Springer), 95–110.
Hamida, R. S., Ali, M. A., Abdelmeguid, N. E., Al-Zaban, M. I., Baz, L., and Bin-Meferij, M. M. (2021). Lichens-a potential source for nanoparticles fabrication: a review on nanoparticles biosynthesis and their prospective applications. J. Fungi 7, 291. doi: 10.3390/jof7040291
Hannigan, G. D., Prihoda, D., Palicka, A., Soukup, J., Klempir, O., Rampula, L., et al. (2019). A deep learning genome-mining strategy for biosynthetic gene cluster prediction. Nucleic Acids Res. 47, 110. doi: 10.1093/nar/gkz654
Harikumar, G., and Krishanan, K. (2022). The growing menace of drug resistant pathogens and recent strategies to overcome drug resistance: a review. J. King Saud. Univ. Sci. 34, 101979. doi: 10.1016/j.jksus.2022.101979
Harvey, C. J. B., Tang, M., Schlecht, U., Horecka, J., Fischer, C. R., Lin, H. C., et al. (2018). HEx: a heterologous expression platform for the discovery of fungal natural products. Sci. Adv. 4, eaar5459. doi: 10.1126/sciadv.aar5459
Hong, J. M., Suh, S. S., Kim, T. K., Kim, J. E., Han, S. J., Youn, U. J., et al. (2018). Anti-cancer activity of lobaric acid and lobarstin extracted from the Antarctic lichen Stereocaulon alpnum. Molecules 23, 658. doi: 10.3390/molecules23030658
Huneck, S. (1999). The significance of lichens and their metabolites. Naturwissenschaften 86, 559–570. doi: 10.1007/s001140050676
Ibrahim, S. R. M., Mohamed, G., Al Haidari, R. A., El-Kholy, A. A., Zayed, M. F., and Khayat, M. T. (2018). Biologically active fungal depsidones: chemistry, biosynthesis, structural characterization, and bioactivities. Fitoterapia 129, 317–365. doi: 10.1016/j.fitote.2018.04.012
Igoli, J. O., Gray, A. I., Clements, C. J., Kantheti, P., and Singla, R. K. (2014). Antitrypanosomal activity & docking studies of isolated constituents from the lichen Cetraria islandica: possibly multifunctional scaffolds. Curr. Top. Med. Chem. 14, 1014–1021. doi: 10.2174/1568026614666140324122323
Ingolfsdottir, K., Gudmundsdottir, G. F., Ogmundsdottir, H. M., Paulus, K., Haraldsdottir, S., Kristinsson, H., et al. (2002). Effects of tenuiorin and methyl orsellinate from the lichen Peltigera leucophlebia on 5-/15-lipoxygenases and proliferation of malignant cell lines in vitro. Phytomedicine 9, 654–658. doi: 10.1078/094471102321616481
Ingolfsdottir, K., Wiedemann, B., Birgisdottir, M., Nenninger, A., Jonsdottir, S., and Wagner, H. (1997). Inhibitory effects of baeomycesic acid from the lichen Thamnolia subuliformis on 5-lipoxygenase in vitro. Phytomedicine 4, 125–128. doi: 10.1016/S0944-7113(97)80056-6
Ismed, F., Devehat, F. L., Rouaud, I., Ferron, S., Bakhtiar, A., and Boustie, J. (2017). NMR reassignment of stictic acid isolated from a Sumatran lichen Stereocaulon montagneanum (Stereocaulaceae) with superoxide anion scavenging activities. Z. Naturforsch. C. J. Biosci. 72, 55–62. doi: 10.1515/znc-2016-0148
Ivanova, V., Aleksieva, K., Kolarova, M., Chipeva, V., Schlegel, R., Schlegel, B., et al. (2002). Neuropogonines A, B and C, new depsidon-type metabolites from Neuropogon sp., an Antarctic lichen. Pharmazie 57, 73–74.
Jeong, M. H., Park, C. H., Kim, J. A., Choi, E. D., Kim, S., Hur, J. S., et al. (2021). Production and activity of cristazarin in the lichen-forming fungus Cladonia metacorallifera. J. Fungi 7, 601. doi: 10.3390/jof7080601
Joo, Y. A., Chung, H., Yoon, S., Park, J. I., Lee, J. E., Myung, C. H., et al. (2016). Skin barrier recovery by protease-activated receptor-2 antagonist lobaric acid. Biomol. Ther. 24, 529–535. doi: 10.4062/biomolther.2016.011
Kai, H., Kinoshita, K., Harada, H., Uesawa, Y., Maeda, A., Suzuki, R., et al. (2017). Establishment of a direct-injection electron ionization-mass spectrometry metabolomics method and Its spplication to lichen profiling. Anal. Chem. 89, 6408–6414. doi: 10.1021/acs.analchem.7b00077
Kalra, R., Conlan, X. A., and Goel, M. (2023). Recent advances in research for potential utilization of unexplored lichen metabolites. Biotechnol. Adv. 62, 108072. doi: 10.1016/j.biotechadv.2022.108072
Kautsar, S. A., Blin, K., Shaw, S., Navarro-Munoz, J. C., Terlouw, B. R., van der Hooft, J. J. J., et al. (2020). MIBiG 2.0: a repository for biosynthetic gene clusters of known function. Nucleic Acids Res. 48, 454–458. doi: 10.1093/nar/gkz882
Kealey, J. T., Craig, J. P., and Barr, P. J. (2021). Identification of a lichen depside polyketide synthase gene by heterologous expression in Saccharomyces cerevisiae. Metab. Eng. Commun. 13, e00172. doi: 10.1016/j.mec.2021.e00172
Kenshole, E., Herisse, M., Michael, M., and Pidot, S. J. (2021). Natural product discovery through microbial genome mining. Curr. Opin. Chem. Biol. 60, 47–54. doi: 10.1016/j.cbpa.2020.07.010
Kim, T. K. (2018). Total syntheses of lobaric acid and its derivatives from the Antarctic lichen Stereocaulon alpinum. J. Nat. Prod. 81, 1460–1467. doi: 10.1021/acs.jnatprod.8b00227
Kim, W., Liu, R., Woo, S., Kang, K. B., Park, H., Yu, Y. H., et al. (2021). Linking a gene cluster to atranorin, a major cortical substance of lichens, through genetic dereplication and heterologous expression. MBio 12, e0111121. doi: 10.1128/mBio.01111-21
Kon, Y., Adanil, H. K., Wardlaw, J. H., and Elix, J. A. (1997). Effects of culture conditions on dibenzofuran production by cultured mycobionts of lichens. Symbiosis 23, 97–106.
Kosanic, M., Manojlovic, N., Jankovic, S., Stanojkovic, T., and Rankovic, B. (2013). Evernia prunastri and Pseudoevernia furfuraceae lichens and their major metabolites as antioxidant, antimicrobial and anticancer agents. Food Chem. Toxicol. 53, 112–118. doi: 10.1016/j.fct.2012.11.034
Kowalski, M., Hausner, G., and Piercey-Normore, M. D. (2011). Bioactivity of secondary metabolites and thallus extracts from lichen fungi. Mycoscience 52, 413–418. doi: 10.1007/s10267-011-0118-3
Kranner, I., Beckett, R., Hochman, A. N., and Thomas, H. (2008). Desiccation-tolerance in lichens: a review. Bryologist 111, 576–593. doi: 10.1639/0007-2745-111.4.576
Krug, D., and Muller, R. (2014). Secondary metabolomics: the impact of mass spectrometry-based approaches on the discovery and characterization of microbial natural products. Nat. Prod. Rep. 31, 768–783. doi: 10.1039/c3np70127a
Kumar, K., Siva, B., Sarma, V. U. M., Mohabe, S., Reddy, A. M., Boustie, J., et al. (2018). UPLC-MS/MS quantitative analysis and structural fragmentation study of five Parmotrema lichens from the Eastern Ghats. J. Pharm. Biomed. Anal. 156, 45–57. doi: 10.1016/j.jpba.2018.04.017
Kwak, J. Y., Rhee, I. K., Lee, K. B., Hwang, J. S., Yoo, I. D., and Song, K. S. (1999). Thelephoric acid and kynapcin-9 in mushroom polyozellus multiflex Inhibit prolyl endopeptidase in vitro. J. Microbiol. Biotechnol. 9, 798–803.
Kwon, I. S., Yim, J. H., Lee, H. K., and Pyo, S. N. (2016). Lobaric acid inhibits VCAM-1 expression in TNF-α-stimulated vascular smooth muscle cells via modulation of NF-κB and MAPK signaling pathways. Biomol. Ther. 24, 25–32. doi: 10.4062/biomolther.2015.084
Lagarde, A., Mambu, L., Mai, P. Y., Champavier, Y., Stigliani, J. L., Beniddir, M. A., et al. (2021). Chlorinated bianthrones from the cyanolichen Nephroma laevigatum. Fitoterapia 149, 104811. doi: 10.1016/j.fitote.2020.104811
Lawrey, J. D. (1986). Biological role of lichen substances. Bryologist 89, 111–122. doi: 10.2307/3242751
Lee, H. W., Kim, J. W., Yim, J. H., Lee, H. K., and Pyo, S. N. (2019). Anti-Inflammatory activity of lobaric acid via suppressing NF-κB/MAPK pathways or NLRP3 inflammasome activation. Planta Med. 85, 302–311. doi: 10.1055/a-0777-2420
Lee, Y. M., Kim, E. H., Lee, H. K., and Hong, S. G. (2014). Biodiversity and physiological characteristics of Antarctic and Arctic lichens-associated bacteria. World J. Microbiol. Biotechnol. 30, 2711–2721. doi: 10.1007/s11274-014-1695-z
Li, Y. F., Tsai, K. J. S., Harvey, C. J. B., Li, J. J., Ary, B. E., Berlew, E. E., et al. (2016). Comprehensive curation and analysis of fungal biosynthetic gene clusters of published natural products. Fungal Genet. Biol. 89, 18–28. doi: 10.1016/j.fgb.2016.01.012
Liao, C., Piercey-Normore, M. D., Sorensen, J. L., and Gough, K. (2010). In situ imaging of usnic acid in selected Cladonia spp. by vibrational spectroscopy. Analyst 135, 3242–3248. doi: 10.1039/c0an00533a
Lin, Z., and Qu, X. D. (2022). Emerging diversity in polyketide synthase. Tetrahedron Lett. 110, 154183. doi: 10.1016/j.tetlet.2022.154183
Liu, Q., Zhang, D., Xu, Y., Gao, S., Gong, Y., Cai, X., et al. (2022). Cloning and functional characterization of the polyketide synthases based on genome mining of Preussia isomera XL-1326. Front. Microbiol. 13, 819086. doi: 10.3389/fmicb.2022.819086
Liu, R. D., Jiang, S. H., Chen, B. S., Paguirigan, J. A., Li, E. W., and Wei, J. C. (2022). Diversity of secondary metabolites from the lichen-forming fungus Endocarpon pusillum (Verrucariaceae, Ascomycota). Mycosystema 41, 1992–2003. doi: 10.13346/j.mycosystema.220118t
Liu, T., Ren, Z., Chunyu, W. X., Li, G. D., Chen, X., Zhang, Z. T. L., et al. (2022). Exploration of diverse secondary metabolites from Streptomyces sp. YINM00001, using genome mining and one strain many compounds approach. Front. Microbiol. 13, 831174. doi: 10.3389/fmicb.2022.831174
Lopes, T. I. B.C. R. G, Yoshida, N. C., and Honda N, K. (2008). Radical-scavenging activity of orsellinates. Chem. Pharm. Bull. 56, 1551–1554. doi: 10.1248/cpb.56.1551
Lücking, R., and Nelsen, M. P. (2018). “Ediacarans, protolichens, and lichen-derived Penicillium a critical reassessment of the evolution of lichenization in fungi,” in Transformative Paleobotany, eds M. Krings, C. J. Harper, N. R. Cúneo, and G. W. Rothwell (London: Academic Press), 551–590.
Luo, H., Li, C. T., Kim, J. C., Liu, Y. P., Jung, J. S., Koh, Y. J., et al. (2013). Biruloquinone, an acetylcholinesterase inhibitor produced by lichen-forming fungus Cladonia macilenta. J. Microbiol. Biotechnol. 23, 161–166. doi: 10.4014/jmb.1207.07016
Luo, H., Yamamoto, Y., Kim, J. A., Jung, J. S., Koh, Y. J., and Hur, J. S. (2009). Lecanoric acid, a secondary lichen substance with antioxidant properties from Umbilicaria antarctica in maritime Antarctica (King George Island). Polar Biol. 32, 1033–1040. doi: 10.1007/s00300-009-0602-9
Luo, M., Wang, M., Chang, S., He, N., Shan, G., Xie, Y., et al. (2022). Halogenase-targeted genome mining leads to the discovery of (+/-) pestalachlorides A1a, A2a, and their atropisomers. Antibiotics 11, 1304. doi: 10.3390/antibiotics11101304
Majchrzak-Celinska, A., Kleszcz, R., Studzinska-Sroka, E., Lukaszyk, A., Szoszkiewicz, A., Stelcer, E., et al. (2022). Lichen secondary metabolites inhibit the wnt/beta-catenin pathway in glioblastoma cells and improve the anticancer effects of temozolomide. Cells 11, 1084. doi: 10.3390/cells11071084
Manish, T., and Yogesh, J. (2019). “Future perspectives and challenges,” in Endolichenic Fungi: Present and Future Trends, eds T. Manishand J. Yogesh (Singapore: Springer), 171–180. doi: 10.1007/978-981-13-7268-1_8
Mann, J. (2002). Natural products in cancer chemotherapy:past, present and future. Cancer 2, 143–148. doi: 10.1038/nrc723
Manojlovic, N. T., Solujic, S., Sukdolak, S., and Krstic, L. J. (1998). Anthraquinones from the lichen Xanthoria parietina. J. Serb. Chem. Soc. 63, 7–11.
McGillick, B. E., Kumaran, D., Vieni, C., and Swaminathan, S. (2016). β-Hydroxyacyl-acyl carrier protein dehydratase (FabZ) from Francisella tularensis and Yersinia pestis: structure determination, enzymatic characterization, and cross-Inhibition studies. Biochemistry 55, 1091–1099. doi: 10.1021/acs.biochem.5b00832
Medema, M. H., Blin, K., Cimermancic, P., de Jager, V., Zakrzewski, P., Fischbach, M. A., et al. (2011). AntiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 39, 339–346. doi: 10.1093/nar/gkr466
Mishra, T., Shukla, S., Meena, S., Singh, R., Pal, M., Upreti, D. K., et al. (2017). Isolation and identification of cytotoxic compounds from a fruticose lichen Roccella montagnei, and it's in silico docking study against CDK-10. Braz. J. Pharmacogn. 27, 724–728. doi: 10.1016/j.bjp.2017.07.006
Miyagawa, H., Hamada, N., and Tamio Ueno, M. S. (1993). Hypostrepsilic acid, a new dibenzofuran from the culturei lichen mycobiont of Evernia esorediosa. Phytochem. Rev. 34, 589–591. doi: 10.1016/0031-9422(93)80057-Y
Moreira, A. S., Braz-Filho, R., Mussi-Dias, V., and Vieira, I. J. (2015). Chemistry and biological activity of Ramalina lichenized fungi. Molecules 20, 8952–8987. doi: 10.3390/molecules20058952
Muggia, L., Schmitt, I., and Grube, M. (2009). Lichens as treasure chests of natural products. SIM News 59, 85–97.
Murugan, M., Muthu, S., Rajendran, K., and Ponnusamy, P. (2021). Review: UV protection and anticancer properties of lichen secondary metabolites. Sci. Acad. 2, 1–29.
Neeraj, V., Behera, B., Parizadeh, H., and Sharma, B. O. (2011). Bactericidal activity of some lichen secondary compounds of Cladonia ochrocholra, Parmotrema nilgherrensis & Parmotrema sancti-angelii. Intl. J. Drug Dev. Res. 3, 222–232.
Newman, D. J., and Cragg, G. M. (2012). Natural products as sources of new drugs over the 30 years from 1981 to 2010. J. Nat. Prod. 75, 311–335. doi: 10.1021/np200906s
Nguyen, K. H., Chollet-Krugler, M., Gouault, N., and Tomasi, S. (2013). UV-protectant metabolites from lichens and their symbiotic partners. Nat. Prod. Rep. 30, 1490–1508. doi: 10.1039/c3np70064j
Nishanth, K. S., Sreerag, R. S., Deepa, I., Mohandas, C., and Nambisan, B. (2015). Protocetraric acid: an excellent broad spectrum compound from the lichen Usnea albopunctata against medically important microbes. Nat. Prod. Res. 29, 574–577. doi: 10.1080/14786419.2014.953500
Oettl, S. K., Gerstmeier, J., Khan, S. Y., Wiechmann, K., Bauer, J., Atanasov, A. G., et al. (2013). Imbricaric acid and perlatolic acid: multi-targeting anti-inflammatory depsides from Cetrelia monachorum. PLoS ONE 8, e76929. doi: 10.1371/journal.pone.0076929
Olaleye, D. O., Odaibo, G. N., Abonyi, D. O., Ibezim, E. C., Adikwu, M. U., Ofokansi, K. C., et al. (2007). In vitro evaluation of the antiviral activity of extracts from the lichen Parmelia perlata (L.) Ach. against three RNA viruses. J. Infect. Dev. Ctries. 1, 315–320. doi: 10.3855/jidc.370
Olivier-Jimenez, D., Chollet-Krugler, M., Rondeau, D., Beniddir, M. A., Ferron, S., Delhaye, T., et al. (2019). A database of high-resolution MS/MS spectra for lichen metabolites. Sci. Data 6, 294. doi: 10.1038/s41597-019-0305-1
Packiam, M., and Perumal, M. S. (2022). “Culture-independent and culture-dependent approaches in symbiont analysis: in Proteobacteria,” in Microbial Symbionts: Functions and Molecular Interactions on Host, ed D. Dharumadurai (San Diego, CA: Academic Press), 743–763.
Palazzotto, E., and Weber, T. (2018). Omics and multi-omics approaches to study the biosynthesis of secondary metabolites in microorganisms. Curr. Opin. Microbiol. 45, 109–116. doi: 10.1016/j.mib.2018.03.004
Paluszczak, J., Kleszcz, R., Studzinska-Sroka, E., and Krajka-Kuzniak, V. (2018). Lichen-derived caperatic acid and physodic acid inhibit Wnt signaling in colorectal cancer cells. Mol. Cell. Biochem. 441, 109–124. doi: 10.1007/s11010-017-3178-7
Papadopoulou, P., Tzakou, O., Vagias, C., Kefalas, P., and Roussis, V. (2007). Beta-orcinol metabolites from the lichen Hypotrachyna revoluta. Molecules 12, 997–1005. doi: 10.3390/12050997
Parrot, D., Peresse, T., Hitti, E., Carrie, D., Grube, M., and Tomasi, S. (2015). Qualitative and spatial metabolite profiling of lichens by a LC-MS approach combined with optimised extraction. Phytochem. Anal. 26, 23–33. doi: 10.1002/pca.2532
Pejin, B., Iodice, C., Bogdanović, G., Kojić, V., and Tešević, V. (2017). Stictic acid inhibits cell growth of human colon adenocarcinoma HT-29 cells. Arab. J. Chem. 10, 1240–1242. doi: 10.1016/j.arabjc.2013.03.003
Pichler, G., Candotto Carniel, F., Muggia, L., Holzinger, A., Tretiach, M., and Kranner, I. (2021). Enhanced culturing techniques for the mycobiont isolated from the lichen Xanthoria parietina. Mycol. Prog. 20, 797–808. doi: 10.1007/s11557-021-01707-7
Pierangelo, B., Bernardetta, S., Alisia, M., Pietro, D., Bottoni, L., Sabatini, C., et al. (2015). Interaction between lichen secondary metabolites and antibiotics against clinical isolates methicillin-resistant Staphylococcus aureus strains. Phytomedicine 22, 223–230. doi: 10.1016/j.phymed.2014.12.005
Posner, B., Feige, G. B., and Huneck, S. (1991). Studies on the chemistry of the lichen genus Umbilicaria Hoffm. Z. Nat. 47, 1–9. doi: 10.1515/znc-1992-1-202
Proctor, R. H., Butchko, R. A. E., Brown, D. W., and Moretti, A. (2007). Functional characterization, sequence comparisons and distribution of a polyketide synthase gene required for perithecial pigmentation in some Food Addit. Contam. 24, 1076–1087. doi: 10.1080/02652030701546495
Qian, Z., Bruhn, T., D'Agostino, P. M., Herrmann, A., Haslbeck, M., Antal, N., et al. (2020). Discovery of the streptoketides by direct cloning and rapid heterologous expression of a cryptic PKS II gene cluster from Streptomyces sp. Tu 6314. J. Org. Chem. 85, 664–673. doi: 10.1021/acs.joc.9b02741
Qiao, Y. M., Yu, R. L, and Zhu, P. (2019). Advances in targeting and heterologous expression of genes involved in the synthesis of fungal secondary metabolites. RSC Adv. 9, 35124–35134. doi: 10.1039/C9RA06908A
Rankovic, B., Kosanic, M., Stanojkovic, T., Vasiljevic, P., and Manojlovic, N. (2012). Biological activities of Toninia candida and Usnea barbata together with their norstictic acid and usnic acid constituents. Int. J. Mol. Sci. 13, 14707–14722. doi: 10.3390/ijms131114707
Ranković, B., and Mišić, M. (2014). The antimicrobial activity of the lichen substances of the lichens Cladonia furcata, Ochrolechia androgyna, Parmelia caperata and Parmelia conspresa. Biotechnol. Biotechnol. Equip. 22, 1013–1016. doi: 10.1080/13102818.2008.10817601
Rao, P. S., Sarma, K. G., and Seshadri, T. R. (1965). Chemical components of the Lobaria lichens from the weatern Himalayas. Curr. Sci. 34, 9–11.
Reddy, S. D., Silva, B., Kumar, K., Babu, V. S. P., Sravanthi, V., Boustie, J., et al. (2019). Comprehensive analysis of secondary metabolites in Usnea longissima (Lichenized Ascomycetes, Parmeliaceae) using UPLC-ESI-QTOF-MS/MS and pro-apoptotic activity of brbatic acid. Molecules 24, 2270. doi: 10.3390/molecules24122270
Rezanka, T., and Sigler, K. (2007). Hirtusneanoside, an unsymmetrical dimeric tetrahydroxanthone from the lichen Usnea hirta. J. Nat. Prod. 70, 1487–1491. doi: 10.1021/np070079m
Romagni, J. G., and Dayan, F. E. (2002). “Structural diversity of lichen metabolites and their potential use,” in Advances in Microbial Toxin Research and Its Biotechnological Exploitation, ed R. K. Upadhyay (New York, NY: Kluwer Academic/Plenum Publishers), 151–169.
Russo, A., Piovano, M., Lombardo, L., Garbarino, J., and Cardile, V. (2008). Lichen metabolites prevent UV light and nitric oxide-mediated plasmid DNA damage and induce apoptosis in human melanoma cells. Life Sci. 83, 468–474. doi: 10.1016/j.lfs.2008.07.012
Russo, A., Piovano, M., Lombardo, L., Vanella, L., Cardile, V., and Garbarino, J. (2006). Pannarin inhibits cell growth and induces cell death in human prostate carcinoma DU-145 cells. Anticancer Drugs 17, 1163–1169. doi: 10.1097/01.cad.0000236310.66080.ed
Sakai, K., Kinoshita, H., and Nihira, T. (2012). Heterologous expression system in Aspergillus oryzae for fungal biosynthetic gene clusters of secondary metabolites. Appl. Microbiol. Biotechnol. 93, 2011–2022. doi: 10.1007/s00253-011-3657-9
Scherlach, K., and Hertweck, C. (2009). Triggering cryptic natural product biosynthesis in microorganisms. Org. Biomol. Chem. 7, 1753–1760. doi: 10.1039/b821578b
Schmeda-Hirschmann, G., Tapia, A., Lima, B., Pertino, M., Sortino, M., Zacchino, S., et al. (2008). A new antifungal and antiprotozoal depside from the Andean lichen Protousnea poeppigii. Phytother. Res. 22, 349–355. doi: 10.1002/ptr.2321
Schweiger, A. H., Ullmann, G. M., Nurk, N. M., Triebel, D., Schobert, R., and Rambold, G. (2022). Chemical properties of key metabolites determine the global distribution of lichens. Ecol. Lett. 25, 416–426. doi: 10.1111/ele.13930
Seo, C., Sohn, J. H., Ahn, J. S., Yim, J. H., Lee, H. K., and Oh, H. (2009). Protein tyrosine phosphatase 1B inhibitory effects of depsidone and pseudodepsidone metabolites from the Antarctic lichen Stereocaulon alpinum. Bioorg. Med. Chem. Lett. 19, 2801–2803. doi: 10.1016/j.bmcl.2009.03.108
Sepahvand, A., Studzinska-Sroka, E., Ramak, P., and Karimian, V. (2021). Usnea sp.: antimicrobial potential, bioactive compounds, ethnopharmacological uses and other pharmacological properties; a review article. J. Ethnopharmacol. 268, 113656. doi: 10.1016/j.jep.2020.113656
Seymour, F. A., Crittenden, P. D., Dickinson, M. J., Paoletti, M., Montiel, D., Cho, L., et al. (2005). Breeding systems in the lichen-forming fungal genus Cladonia. Fungal Genet. Biol. 42, 554–563. doi: 10.1016/j.fgb.2005.03.006
Shishido, T. K., Wahlsten, M., Laine, P., Rikkinen, J., Lundell, T., and Auvinen, P. (2021). Microbial communities of Cladonia lichens and their biosynthetic gene clusters potentially encoding natural products. Microorganisms 9, 1347. doi: 10.3390/microorganisms9071347
Shrestha, G., and St. Clair, L. L. (2013). Lichens: a promising source of antibiotic and anticancer drugs. Phytochem. Rev. 12, 229–244. doi: 10.1007/s11101-013-9283-7
Shukla, V., Joshi, G. P., and Rawat, M. S. M. (2010). Lichens as a potential natural source of bioactive compounds: a review. Phytochem. Rev. 9, 303–314. doi: 10.1007/s11101-010-9189-6
Singh, G., Armaleo, D., Dal Grande, F., and Schmitt, I. (2021). Depside and depsidone synthesis in lichenized fungi comes into focus through a genome-wide comparison of the olivetoric acid and physodic acid chemotypes of Pseudevernia furfuracea. Biomolecules 11, 1445. doi: 10.3390/biom11101445
Sinnemann, S. J., Andresson, O. S., Brown, D. W., and Miao, V. P. W. (2000). Cloning and heterologous expression of Solorina crocea pyrG. Curr. Genet. 37, 333–338. doi: 10.1007/s002940050536
Skinnider, M. A., Merwin, N. J., Johnston, C. W., and Magarvey, N. A. (2017). PRISM 3: expanded prediction of natural product chemical structures from microbial genomes. Nucleic Acids Res. 45, W49–W54. doi: 10.1093/nar/gkx320
Stocker-Wörgötter, E. (2008). Metabolic diversity of lichen-forming ascomycetous fungi: culturing, polyketide and shikimate metabolite production, and PKS genes. Nat. Prod. Rep. 25, 188–200. doi: 10.1039/B606983P
Stocker-Wörgötter, E., and Hager, A. (2008). “Appendix: culture methods for lichens and lichen symbionts,” in Lichen Biology, ed T. H. Nash III (Cambridge: Cambridge University Press).
Studzinska-Sroka, E., Majchrzak-Celinska, A., Bandurska, M., Rosiak, N., Szwajgier, D., Baranowska-Wojcik, et al. (2022). Is caperatic acid the only compound responsible for activity of lichen Platismatia glauca within the nervous system? Antioxidants 11, 2069. doi: 10.3390/antiox11102069
Sun, Y. H., Li, J., Zhang, Y., Tu, Y. T., Huang, C. Z., Tao, J., et al. (2018). The polysaccharide extracted from Umbilicaria esculenta inhibits proliferation of melanoma cells through ROS-activated mitochondrial apoptosis pathway. Biol. Pharm. Bull. 41, 57–64. doi: 10.1248/bpb.b17-00562
Sveshnikova, N., Yuan, T., Warren, J. M., and Piercey-Normore, M. D. (2019). Development and validation of a reliable LC-MS/MS method for quantitative analysis of usnic acid in Cladonia uncialis. BMC Res. Notes 12, 550. doi: 10.1186/s13104-019-4580-x
Takenaka, Y., Morimoto, N., Hamada, N., and Tanahashi, T. (2011). Phenolic compounds from the cultured mycobionts of Graphis proserpens. Phytochemistry 72, 1431–1435. doi: 10.1016/j.phytochem.2011.04.017
Tanahashi, T., Takenaka, Y., Mizushina, Y., and Hamada, N. (2017). A cytotoxic pyranonaphthoquinone from cultured lichen mycobionts of Haematomma sp. Heterocycles 94, 1728. doi: 10.3987/COM-17-13704
Tarkka, M. T., Sarniguet, A., and Frey-Klett, P. (2009). Inter-kingdom encounters: recent advances in molecular bacterium-fungus interactions. Curr. Genet. 55, 233–243. doi: 10.1007/s00294-009-0241-2
Tatipamula, V. B., Vedula, G. S., and Sastry, A. V. S. (2019). Chemical and pharmacological evaluation of manglicolous lichen Roccella montagnei Bel em. D. D. Awasthi. Future. J. Pharm. Sci. 5, 8. doi: 10.1186/s43094-019-0009-6
Tay, T., Turk, A. O., Yilmaz, M., Turk, H., and Kivanc, M. (2004). Evaluation of the antimicrobial activity of the acetone extract of the lichen Ramalina farinacea and its (+)-usnic acid, norstictic acid, and protocetraric acid constituents. Z. Naturforsch. C. J. Biosci. 59, 384–388. doi: 10.1515/znc-2004-5-617
Tekiela, A., Furmanek, L., Andrusiewicz, M., Bara, G., Seaward, M. R. D., Kapusta, I., et al. (2021). Can lichen secondary compounds impact upon the pathogenic soil fungi Fusarium oxysporum and F. avenaceum? Folia Cryptog. Estonica. Fasc. 58, 165–181. doi: 10.12697/fce.2021.58.18
Thadhani, V. M., Choudhary, M. I., Ali, S., Omar, I., Siddique, H., and Karunaratne, V. (2011). Antioxidant activity of some lichen metabolites. Nat. Prod. Res. 25, 1827–1837. doi: 10.1080/14786419.2010.529546
Timsina, B. A., Sorensen, J. L., Weihrauch, D., and Piercey-Normore, M. D. (2013). Effect of aposymbiotic conditions on colony growth and secondary metabolite production in the lichen-forming fungus Ramalina dilacerata. Fungal Biol. 117, 731–743. doi: 10.1016/j.funbio.2013.09.003
Türk, H., Yilmaz, M., Tay, T., Türk, A. Ö., and Kivanç, M. (2006). Antimicrobial activity of extracts of chemical races of the lichen Pseudevernia furfuracea and their physodic acid, chloroatranorin, atranorin, and olivetoric acid. Zeitschrift Naturforschung C6, 499–507. doi: 10.1515/znc-2006-7-806
Umezawa, H., Shibamoto, N., Naganawa, H., Ayukawa, S., Matsuzaki, M., and Takeuchi, T. (1974). Isolation of lecanoric acid, an inhibitor of histidine decarboxylase from a fungus. J. Antibiot. 27, 587–596. doi: 10.7164/antibiotics.27.587
Valarmathi, R., Hariharan, G. N., Venkataraman, G., and Parida, A. (2009). Characterization of a non-reducing polyketide synthase gene from lichen Dirinaria applanata. Phytochemistry 70, 721–729. doi: 10.1016/j.phytochem.2009.04.007
Verma, N., Behera, B. C., and Joshi, A. (2012). Studies on nutritional requirement for the culture of lichen Ramalina nervulosa and Ramalina pacifica to enhance the production of antioxidant metabolites. Folia Microbiol. 57, 107–114. doi: 10.1007/s12223-012-0100-2
Verma, N., Behera, B. C., and Makhija, U. V. (2011). Studies on cytochromes of lichenized fungi under optimized culture conditions. Mycoscience 52, 65–68. doi: 10.47371/S10267-010-0070-7
Waltenberger, B., Mocan, A., Smejkal, K., Heiss, E. H., and Atanasov, A. G. (2016). Natural products to counteract the epidemic of cardiovascular and metabolic disorders. Molecules 21, 807. doi: 10.3390/molecules21060807
Wambui, J., Stevens, M. J. A., Sieber, S., Cernela, N., Perreten, V., and Stephan, R. (2021). Targeted genome mining reveals the psychrophilic clostridium estertheticum complex as a potential source for novel bacteriocins, including cesin A and estercticin A. Front. Microbiol. 12, 801467. doi: 10.3389/fmicb.2021.801467
Wang, Y., Geng, C. A., Yuan, X. L., Hua, M., Tian, F. H., and Li, C. T. (2018). Identification of a putative polyketide synthase gene involved in usnic acid biosynthesis in the lichen Nephromopsis pallescens. PLoS ONE 13, e0199110. doi: 10.1371/journal.pone.0199110
Wang, Y., Kim, J. A., Cheong, Y. H., Joshi, Y., Koh, Y. J., and Hur, J. S. (2011). Isolation and characterization of a reducing polyketide synthase gene from the lichen-forming fungus Usnea longissima. J. Microbiol. 49, 473–480. doi: 10.1007/s12275-011-0362-4
Wang, Y., Wang, J., Cheong, Y. H., and Hur, J. S. (2014). Three new non-reducing polyketide synthase genes from the lichen-forming fungus Usnea longissima. Mycobiology 42, 34–40. doi: 10.5941/MYCO.2014.42.1.34
Wilson, D. J., Patton, S., Florova, G., Hale, V., and Reynolds, K. A. (1998). The shikimic acid pathway and polyketide biosynthesis. J. Ind. Microbiol. Biotechnol. 20, 299–303. doi: 10.1038/sj.jim.2900527
Xu, M., Oppong-Danquah, E., Wang, X., Oddsson, S., Abdelrahman, A., Pedersen, S. V., et al. (2022). Novel methods to characterise spatial distribution and enantiomeric composition of usnic acids in four Icelandic lichens. Phytochemistry 200, 113210. doi: 10.1016/j.phytochem.2022.113210
Yoshimura, I., Kinoshita, Y., Yamamoto, Y., Huneck, S., and Yamada, Y. (1994). Analysis of secondary metabolites from lichen by high performance liquid chromatography with a photodiode array detector. Phytochem. Anal. 5, 197–205. doi: 10.1002/pca.2800050405
Yoshimura, I., Kurokawa, T., Yamamoto, Y., and Kinoshita, Y. (1993). Development of lichen thalli in vitro. Bryologist 96, 412–421. doi: 10.2307/3243871
Yousuf, S., Choudhary, M. I., and Rahman, A. U. (2014). Lichens: chemistry and biological activities. Nat. Prod. Chem. 43, 223–259. doi: 10.1016/B978-0-444-63430-6.00007-2
Zacharski, D. M., Esch, S., Konig, S., Mormann, M., Brandt, S., Ulrich-Merzenich, G., et al. (2018). beta-1,3/1,4-Glucan Lichenan from Cetraria islandica (L.) ACH induces cellular differentiation of human keratinocytes. Fitoterapia 129, 226–236. doi: 10.1016/j.fitote.2018.07.010
Zahroh, E. W., Ningsih, F., and Sjamsuridzal, W. (2022). Detection of antimicrobial compounds from thermophilic actinomycetesusing one strain many compounds(OSMAC) approach. J. Biol. Lingkungan 9, 76–94. doi: 10.31289/biolink.v9i1.6438
Zarins-Tutt, J. S., Barberi, T. T., Gao, H., Mearns-Spragg, A., Zhang, L. X., Newman, D. J., et al. (2016). Prospecting for new bacterial metabolites_ a glossary of approaches for inducing, activating and upregulating the biosynthesis of bacterial cryptic or silent Natural Products. Nat.Prod. Rop. 33, 54–72. doi: 10.1039/C5NP00111K
Zhang, T., and Wei, J. (2011). Survival analyses of symbionts isolated from Endocarpon pusillum Hedwig to desiccation and starvation stress. Sci. China Life Sci. 54, 480–489. doi: 10.1007/s11427-011-4164-z
Zhao, Y. S., Wang, M. F., and Xu, B. J. (2021). A comprehensive review on secondary metabolites and health-promoting effects of edible lichen. J. Funct. Foods 80, 104283. doi: 10.1016/j.jff.2020.104283
Keywords: lichen, natural products, bioactivity, PKS, OSMAC strategy, genome mining
Citation: Ren M, Jiang S, Wang Y, Pan X, Pan F and Wei X (2023) Discovery and excavation of lichen bioactive natural products. Front. Microbiol. 14:1177123. doi: 10.3389/fmicb.2023.1177123
Received: 01 March 2023; Accepted: 24 March 2023;
Published: 17 April 2023.
Edited by:
Dewu Zhang, Chinese Academy of Medical Sciences, ChinaCopyright © 2023 Ren, Jiang, Wang, Pan, Pan and Wei. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xinli Wei, d2VpeGxAaW0uYWMuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.