- 1Beijing Key Laboratory of Agricultural Genetic Resources and Biotechnology, Institute of Biotechnology, Beijing Academy of Agriculture and Forestry Sciences, Beijing, China
- 2Beijing Key Laboratory for Source Control Technology of Water Pollution, Engineering Research Center for Water Pollution Source Control and Eco-remediation, Beijing Forestry University, Beijing, China
Antibiotic resistance genes (ARGs) and antibiotic-resistant bacteria (ARB) in animal feces can be released into the atmosphere via aerosolization, posing a high health risk to farm workers. So far, little attention has been paid to the characterization of the aerosolization process. In this study, fecal and fine particulate matter (PM2.5) samples were collected from 20 animal farms involving swine, cattle, layers, and broilers, and the ARGs, ARB, and human pathogenic bacteria (HPB) were loaded in these two media. The results showed that approximately 70% of ARGs, 60% of ARBs, and 43% of HPBs were found to be preferential aerosolization. The bioaerosolization index (BI) of target 30 ARGs varied from 0.04 to 460.07, and the highest value was detected from tetW. The highest BI values of erythromycin- and tetracycline-resistant bacteria were for Kocuria (13119) and Staphylococcus (24746), respectively, and the distribution of BI in the two types of dominant ARB was similar. Regarding the bioaerosolization behavior of HPB, Clostridium saccharolyticum WM1 was the most easily aerosolized pathogen in swine and broiler farms, and Brucella abortus strain CNM 20040339 had the highest value in cattle and layer farms. Notably, the highest BI values for ARGs, ARB, and HPB were universally detected on chicken farms. Most ARGs, ARB, and HPB positively correlated with animal age, stocking density, and breeding area. Temperature and relative humidity have significant effects on the aerosolization behavior of targets, and the effects of these two parameters on the same target are usually opposite. The results of this study provide a basis for a better understanding of the contribution of animal feces to airborne ARGs and HPBs in farms, as well as for controlling the transport of the fecal microbiome to the environment through the aerosolization pathway.
1. Introduction
The expansion of centralized animal feeding operations (CAFOs) is one of the main causes of increased environmental antimicrobial resistance (AMR) (Zhu et al., 2013). Globally, 73% of all antimicrobials sold on Earth are used in animals raised for food (Van Boeckel et al., 2019), and this value accounts for approximately 80% in some developed countries such as Canada and the US (Tang et al., 2017; Xiong et al., 2018). The total annual consumption of antibiotics in animal husbandry in China is 97,000 tons (Song et al., 2020), ranking first worldwide. The abuse of antibiotics promotes the development of AMR and the emergence of novel resistance mechanisms in farmed animals (Van Boeckel et al., 2019). As an important AMR reservoir (Xie et al., 2019), animal gut flora are continuously released into the environment through excreta (mainly feces), and the animal-related AMR elements contained therein would pollute terrestrial and aquatic ecosystems (Zhao et al., 2021), including soil (Yang et al., 2022), groundwater (Zainab et al., 2020), and crops (He et al., 2016).
In recent years, the air has been proven to be an important pathway for AMR transmission from livestock to humans (de Rooij et al., 2019) through exposure to fecal ARGs via dust (Munk et al., 2018). Recent research has indicated that the structure of the antibiotic resistome in animal feces is correlated with that in farm dust (Luiken et al., 2020), and the average contribution rate of pig manure to airborne bacteria varies seasonally (Song et al., 2021; Bai et al., 2022). To date, studies on the connection between AMR in feces and air have focused on their communities (resistomes and bacterial microbiomes), and few studies have explored this relationship based on individual targets. This limits our understanding of the aerosolization process of specific AMR elements in feces, which is of great significance for certain worrying and high-risk ARGs, ARBs, and HPBs.
The aerosolization behavior was used to evaluate the microbial release ability of solids or liquids (Moletta et al., 2010). The earliest study on bacterial aerosolization was conducted using the ratio of the concentration of culturable bacteria in the air to that in natural waters (Parker et al., 1983). Although preferential aerosolization has been detected at the bacterial phylum level (Moletta et al., 2007), the aerosolization behavior of bacterial species is influenced by multiple factors (Parker et al., 1983). Recently, to evaluate the effectiveness of dissemination, an increasing number of researchers have begun to focus on the aerosolization behavior of microorganisms in biological contamination sources, including solid waste compost (He et al., 2019), animal manure compost (Wang R. et al., 2022), and sludge compost (Lu et al., 2021). However, there have been no studies on bacterial escape from animal feces, particularly regarding the aerosolization behavior of AMR elements and their associated factors.
In this study, animal feces were assumed to be the main source of airborne AMR in CAFOs. A systematic study was conducted on the aerosolization behavior of ARGs, ARBs, and HPBs and the corresponding potential factors from feces to PM2.5. The aim was to describe the overall characteristics of the animal-specific aerosolization behavior of target elements, determine the preferential aerosolization of targets based on their bioaerosolization indices, and explore the key parameters that affect aerosolization behavior.
2. Materials and methods
2.1. Sample collection
In this study, fecal and PM2.5 samples were collected in the Pinggu, Miyun, and Huairou districts of Beijing, China. In total, 40 samples (20 fecal and 20 air) were collected from four types of animal farms (swine, cattle, layer, and broiler) and five farms for each animal species. The animal breeding houses were semi-enclosed with natural ventilation. Information on animal breeding and environmental indices was recorded while collecting the samples, and the details of the sampling site were described in a previously published article (Xin et al., 2022).
The fecal samples were collected using a 5-point sampling method, with 10 g of feces at each point, and a total of 50 g of feces were collected from each farm and mixed well. PM2.5 samples (1.5 m above the ground) were collected using medium-volume (100 L/min) air samplers (2030A, Laoying, Qingdao, China). The particle separation device [with a 50% cut point diameter (Da50) = 2.5 ± 0.2 μm] allowed particulate matter with aerodynamic diameters of ≤2.5 to be collected on the filters. The sampling time was approximately 48 h. The blank filters used for sampling were 90 mm micro-quartz fiber filters (Ahlstrom Munktell, Falun, Sweden), and the filters were dried and sterilized in a muffle furnace at 400 °C for 5 h before sampling. The collected feces and filters were placed in an ice box and returned to the laboratory within 5 h and stored at −80 °C. A particle mass counter (MetOne, OR, USA) was used to measure the PM2.5 mass concentration, temperature, and relative humidity of air in the breeding house.
For culturable airborne ARBs, an Anderson six-stage sampler was used to collect aerosols 1.5 m above the ground inside the breeding houses. The device contains 400 pores at each stage, with pore size decreasing from > 7.0 μM at stage I to 4.7–7.0 μM at stage II, 3.3–4.7 μM at stage III, 2.1–3.3 μM at stage IV, 1.1–2.1 μM at stage V, and 0.65–1.1 μM at stage VI. Aerosols collected during stages IV–VI were considered to be fine particulate matter (Gao et al., 2015). Samples for the analysis of antibiotic-resistant bacteria were collected for 2 min, at 28.3 L/min. The sampling pump was calibrated using a flowmeter (Yuyao, Zhejiang, China) before sampling. Air samples were collected on two types of media (20 ml) simultaneously, including LB nutrient agar enriched with tetracycline or erythromycin. Tetracycline was dissolved using ultrapure water and filtered through a 0.22 μM PES filter, and erythromycin was dissolved using 95% ethanol and filtered through a 0.22 μM Nylon filter. The corresponding antibiotic solutions were added to the sterilized LB nutrient agar medium, and the working concentrations of tetracycline and erythromycin were 16 and 48 mg/L, respectively (Yang et al., 2016). Three parallel samples were collected for each medium type. Dishes were then sealed, transported immediately to the laboratory, and cultured for 48 h at 28 °C.
2.2. DNA extraction and droplet digital PCR
For PM2.5 samples, one-eighth of a micro-quartz fiber filter was added into 50 ml of sterile 1 × PBS buffer and centrifuged at a low speed of 200 g at 4 °C for 3 h. After vortex shaking, the resuspension was filtered through a 0.2 μM PES filter membrane, and DNA was extracted from the particulate matter on this membrane (Jiang et al., 2015). For the culturable total and antibiotic-resistant bacteria, all colonies from three replicates were scraped down, transferred into 15 ml sterile centrifuge tubes containing PBS buffer, and centrifuged at 12,000 rpm for 1 min. DNA was extracted from 300 μl of the resulting pellet. For fecal samples, 0.5 g of well-mixed fecal samples were considered for DNA extraction from the corresponding animal feces. DNA extraction for the above samples was performed using FastDNA® SPIN Kit for Soil (MP Biomedicals, CA, USA). The concentration and purity of the extracted DNA were determined by Qubit® dsDNA High Sensitivity Assay Kit (Thermo Fisher Scientific, USA).
To characterize the profiles of ARGs in the fecal and air samples, the concentrations of 16S rRNA and 30 subtypes of ARGs encoding resistance to eight antimicrobial classes (Xin et al., 2022) were performed by the QX200 Droplet Digital™ PCR system. The annealing temperatures of ddPCR, sequences of the forward and reverse primers, and resistance mechanism of the 30 ARGs have been reported in previous studies (Gao et al., 2018; Ding et al., 2020). The ddPCR and data collection processes were based on previously published articles (Gao et al., 2018; Ding et al., 2020; Xin et al., 2022). The abundance of each ARG was normalized to “relative abundance” (copies per 16S rRNA gene) by dividing the copy number of each ARG by the number of 16S rRNA genes.
2.3. Illumina sequencing and data processing
Using Illumina sequencing, the structures of the unculturable and culturable bacterial communities in fecal and PM2.5 samples were determined. The V3–V4 hypervariable region of the bacterial 16S rRNA was amplified with the universal primers 338F (5′-ACTCCTACGGGAGGCAGA-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) using an ABI GeneAmp® 9700 PCR thermocycler (ABI, CA, United States), and the amplification reaction and conditions were conducted following previous studies (Chen et al., 2019; Xin et al., 2022). PCR products were purified and sequenced on an Illumina PE300 platform (Majorbio, Shanghai, China). Sequencing data were analyzed following previously described approaches (Gao et al., 2018; Xin et al., 2022). Raw sequencing data were uploaded to the NCBI Sequence Read Archive (SRA) database with registration numbers SRP079945 for culturable ARBs and SRP079949 for HPBs.
2.4. Identification of human pathogenic bacteria
Human pathogenic bacteria in fecal and air samples were determined based on a database of bacterial pathogens reported in an earlier study (Chen et al., 2016), in which the 16S rRNA gene sequences of bacterial pathogens were all available from NCBI (http://www.ncbi.nlm.nih.gov/). The 16S rRNA gene sequences of each sample were blasted against those of bacterial pathogens in the database, with an E-value of < 1 × 10−10. The results of BLAST hits were then filtered to identify the HPB with a sequence similarity threshold of > 99%.
2.5. Data analysis
The bioaerosolization index (BI) was used to quantify the aerosolization behavior of the microorganisms. The BI calculation is presented in Eq. (1), which is a concentration factor (Moletta et al., 2010).
where RAPM2.5 is the relative abundance of ARGs, ARBs, and HPBs in PM2.5, and RAfeces is the relative abundance of ARGs, ARBs, and HPBs in the fecal samples.
Heatmaps were conducted using R software (“vegan” and “pheatmap” packages). Line plots merged with scatter plots were drawn using the OriginPro 2022 software. Partial least squares path modeling (PLS-PM) was used to detect the causal relationships between the influencing factors and aerosolization behavior. All data were standardized before establishing a priori models using Z-scores in SPSS26.0. PLS-PM was conducted with the “plspm” package in the R software. PLS-PM analysis generally used goodness of fit (GOF) to judge the overall model fit: the larger the GOF value, the higher the model fit; GOF = 0.10, the model fit is low; GOF = 0.25, the model fit is medium; and GOF = 0.36, the model fit is high (Henseler and Sarstedt, 2013). For the correlation bubble plot, the package “vegan” was used to calculate “Spearman” correlations between environmental factors and BI values, and the package “ggplot2” was used to make the bubble plot.
3. Results and discussions
3.1. An abundance of antibiotic resistance genes, antibiotic-resistant bacteria, and human pathogenic bacteria
The bacterial biomass in animal feces ranged from 3.79 × 109 to 2.40 × 1010 copies/g as indicated by the concentration of 16S rRNA, and the average concentration of 30 ARGs was 2.88 ± 1.92 × 107 copies/g (Figure 1). Overall, ARGs encoding aminoglycoside resistance had the highest mean concentrations, with values of 6.46 ± 4.23 × 107 copies/g, and the lowest mean concentration (4.84 ± 2.13 × 106 copies/g) was observed for ARGs encoding resistance to multidrug-resistant classes. This distribution pattern is consistent with that of previous studies on swine feces (Zhang et al., 2019; Yue et al., 2021), poultry feces (McEachran et al., 2015), and cattle feces (Qian et al., 2018; Zhu et al., 2021; Yuan et al., 2022) because aminoglycosides are the most widely used antibiotics in the animal farming industry in China (Zhao et al., 2021). The concentrations of specific ARGs in the feces varied depending on the animal species; mean concentrations of ARGs in the feces of layer (2.66 ± 4.84 × 107 copies/g) and broilers (3.65 ± 5.88 × 107 copies/g) were significantly higher than those of swine and cattle (p < 0.05), which further confirmed the severity of AMR contamination in poultry farms (Cheng et al., 2013; Qian et al., 2018). In addition to the type and amount of antibiotics used in animal breeding processes, differences in the structure of the intestinal flora of animals may also play an important role in the different concentrations of fecal ARGs (Zhao et al., 2010).
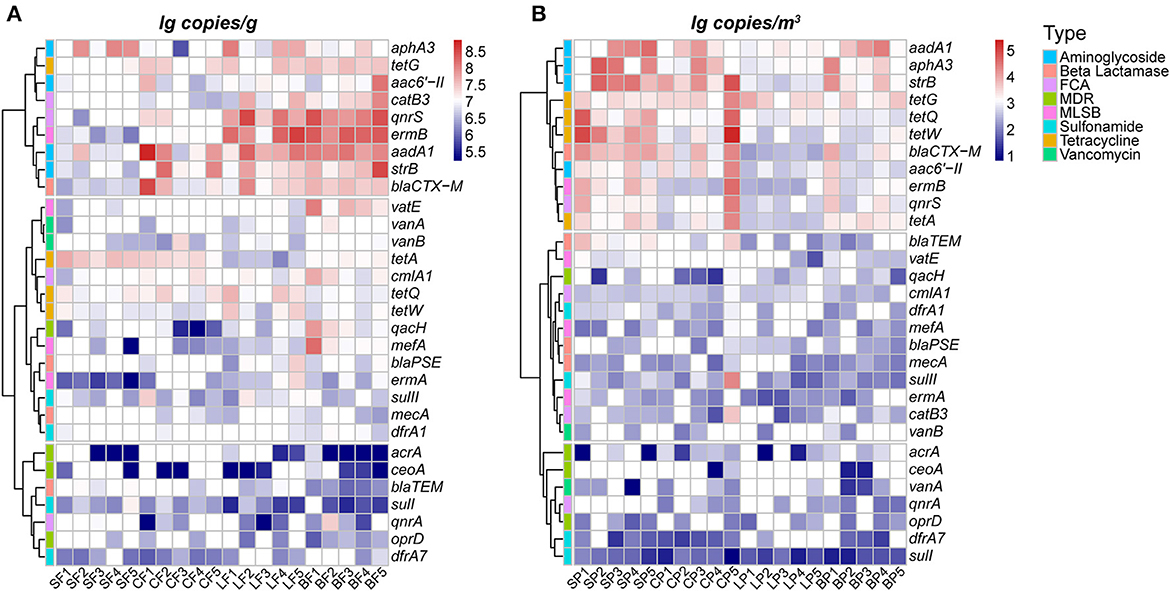
Figure 1. Abundance (log 10 transformed concentration) of 30 ARGs subtypes in animal fecal (A) and PM2.5 (B) samples by animal species. The first letter of the sample name represents the category of animals: S, swine; C, cattle; L, layer; B, broiler. The second letter represents the category of media: F, feces; P, PM2.5.
In terms of PM2.5 samples, the mean concentrations of 16S rRNA gene and ARGs were 3.40 ± 2.04 × 105 copies/m3 (ranged from 8.89 × 104 to 1.84 × 106 copies/m3) and 3.94 ± 1.73 × 103 copies/m3 (from 8.33 × 100 to 1.84 × 106 copies/m3), respectively (Figure 1B). Similar distributions were also detected in pigs (Song et al., 2021), chicken (Xu et al., 2022), and cattle farms (Bai et al., 2022), with the highest and lowest concentrations of airborne ARGs detected from those encoded aminoglycosides (1.20 ± 0.61 × 104 copies/m3) and multidrug resistance (2.5 ± 1.44 × 102 copies/m3), respectively. The animal-specific concentration of airborne ARGs was observed in PM2.5 such that the highest mean value was also determined in layer (2.56 ± 1.13 × 104 copies/m3) and broiler (5.64 ± 1.14 × 104 copies/m3) farms. According to the results in Figure 1, these synchronous concentrations of ARGs in PM2.5 and fecal samples can be used to account for the contribution of feces to airborne ARGs in animal farms.
The relative abundances of the top 30 erythromycin- and tetracycline-resistant bacterial genera in fecal and PM2.5 samples, respectively, were analyzed using a heatmap (Figures 2A, B). Overall, similar dominant genera with comparable relative abundances were detected in the two ARB types, either in the air or fecal samples. However, the distribution of dominant ARBs in the air differed from that in the fecal samples. The 30 most dominant erythromycin-resistant bacteria belonged to four bacterial phyla (Figure 2). For erythromycin-resistant bacteria, Proteobacteria (53.1 ± 13.0%) was the predominant bacterial phylum in fecal samples, followed by Bacteroidetes (28.3 ± 17.7%) and Firmicutes (15.6 ± 15.1%). The three dominant phyla for fecal tetracycline-resistant bacteria were also Proteobacteria, Bacteroidetes, and Firmicutes with similar relative abundances of 41.3 ± 19.3%, 37.5 ± 31.0%, and 17.0 ± 9.82%, respectively. Firmicutes (36.4 ± 12.8%) was the predominant erythromycin-resistant bacterial phylum in PM2.5 samples, followed by Actinobacteria (27.0 ± 18.2%), Proteobacteria (24.6 ± 14.5%), and Bacteroidetes (12.0 ± 8.1%). For tetracycline-resistant bacteria, Firmicutes (50.8 ± 28.6%), Bacteroidetes (19.6 ± 13.5%), Proteobacteria (18.5 ± 11.7%), and Actinobacteria (10.7 ± 6.2%) were the most dominant in the air. According to previous studies on the air environment of poultry and pig farms (Wang et al., 2019; Bai et al., 2022; Jezak and Kozajda, 2022), Proteobacteria, Firmicutes, and Actinobacteria were also detected as the dominant bacteria in feces or air using molecular biological methods. Although similar dominant phyla were detected between the air and fecal ARBs, there were differences in their specific relative abundances. This phenomenon has also been previously reported for Firmicutes and Bacteroidetes on layer and broiler farms (Hong et al., 2012; Wery, 2014).
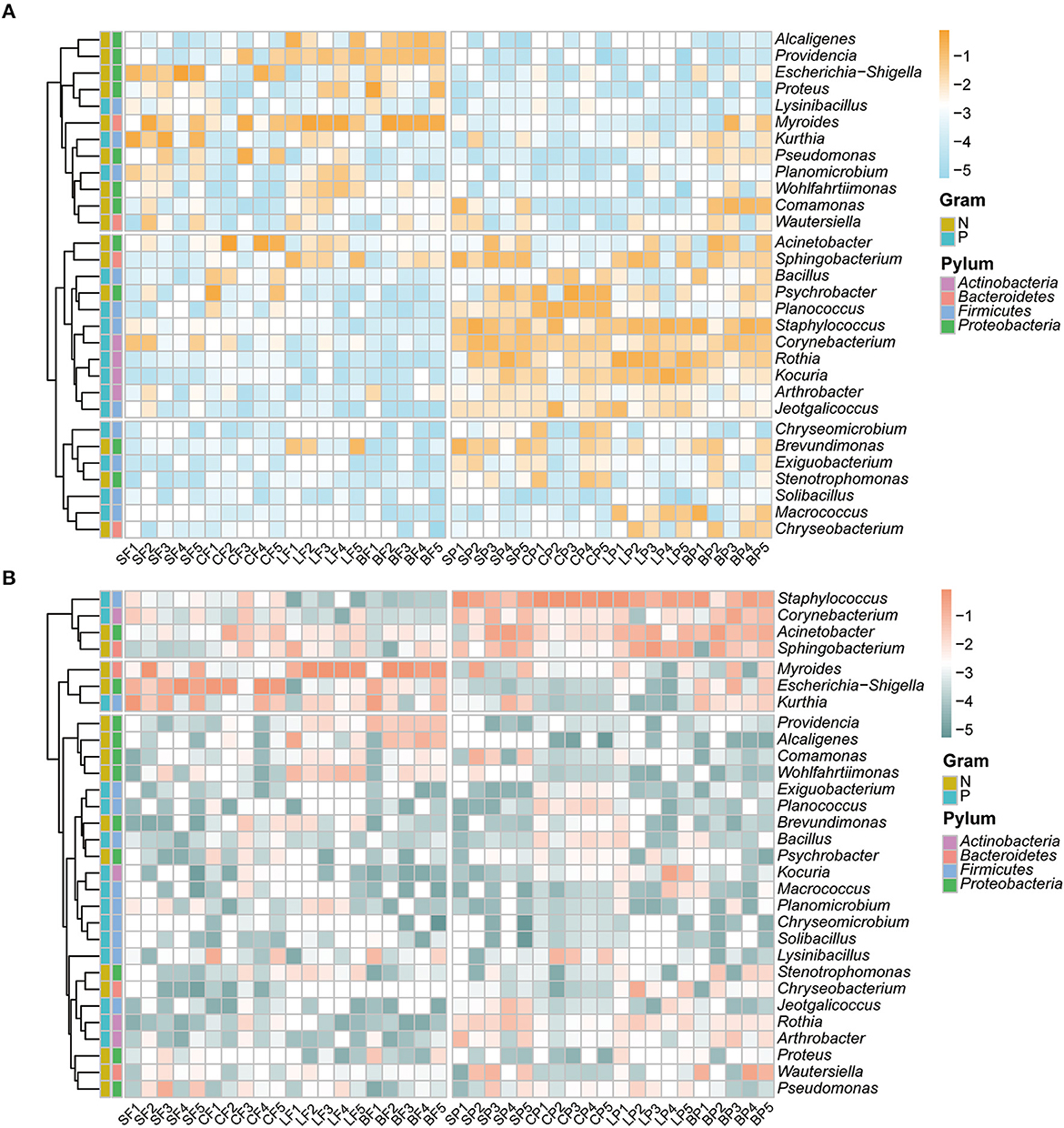
Figure 2. Relative abundance (log 10 transformed) of top 30 erythromycin- (A) and tetracycline-resistant bacterial genera (B) in feces and fine particles by animal species. The first letter of the sample name represents the category of animals: S, swine; C, cattle; L, layer; B, broiler. The second letter represents the category of media: F, feces; P, PM2.5.
As shown in Figure 2, Myroides and Escherichia–Shigella were the top two prevalent genera in fecal samples, and the third dominant genera among the erythromycin- and tetracycline-resistant bacteria was Acinetobacter (8.52 ± 2.09%) and Kurthia (10.4 ± 8.2%), respectively. However, distinctive dominant bacterial genera were found in PM2.5 samples that Staphylococcus was consistently detected as dominant ARBs in both erythromycin- and tetracycline-resistant bacteria with relative abundance of 15.6 ± 12.6% and 43.6 ± 28.5%, respectively. Previous studies on airborne ARBs have mostly focused on urban areas (Wang M. et al., 2022), which have also detected the prevalence of antibiotic-resistant Staphylococcus in the urban air of Beijing. As a bacterium that contains important potentially pathogenic species in humans and animals, the universal existence of antibiotic-resistant Staphylococcus in the air has given us a warning. The remaining prevalent genera for erythromycin-resistant bacteria were Rothia (11.5 ± 1.29%) and Psychrobacter (9.01 ± 1.47%). The tetracycline-resistant bacteria were Acinetobacter (13.4 ± 12.5%) and Sphingobacterium (14.8 ± 10.8%). Those genera, including Bacillus, Streptomyces, Kocuria, and Pseudomonas have also been reported as the dominant culturable bacterial genera in polluted air in Beijing during the heating season (Mao et al., 2019).
A total of 44 potential bacterial pathogenic species belonging to 22 genera were identified, and their total relative abundances in fecal and airborne bacteria were 39.7 ± 18.2% and 37.2 ± 13.4%, respectively. There were no significant differences in the distribution of human pathogenic bacteria in the feces and air of the animal farms. The top 30 species are shown in Figure 3. The prevalent pathogens were generally detected from Enterococcus faecalis, Acinetobacter sp., and Clostridium difficile in both air and fecal samples. Other dominant pathogenic species in fecal and PM2.5 samples were Streptococcus gallolyticus (8.4 ± 3.3%) and Clostridium saccharolyticum (6.1 ± 4.2%), respectively. Streptococcus gallolyticus was more abundant in swine feces. It is a causative agent in ruminants and birds and may cause digestive disorders such as lactic acidosis (Sitthicharoenchai et al., 2022). The relative abundance of Clostridium saccharolyticum was high in all air samples in this study, and it was also detected in high abundance in poultry feces in a previous study (Luo et al., 2013). Clostridium saccharolyticum can cause intestinal and soft tissue infections in animals and humans (Luo et al., 2013).
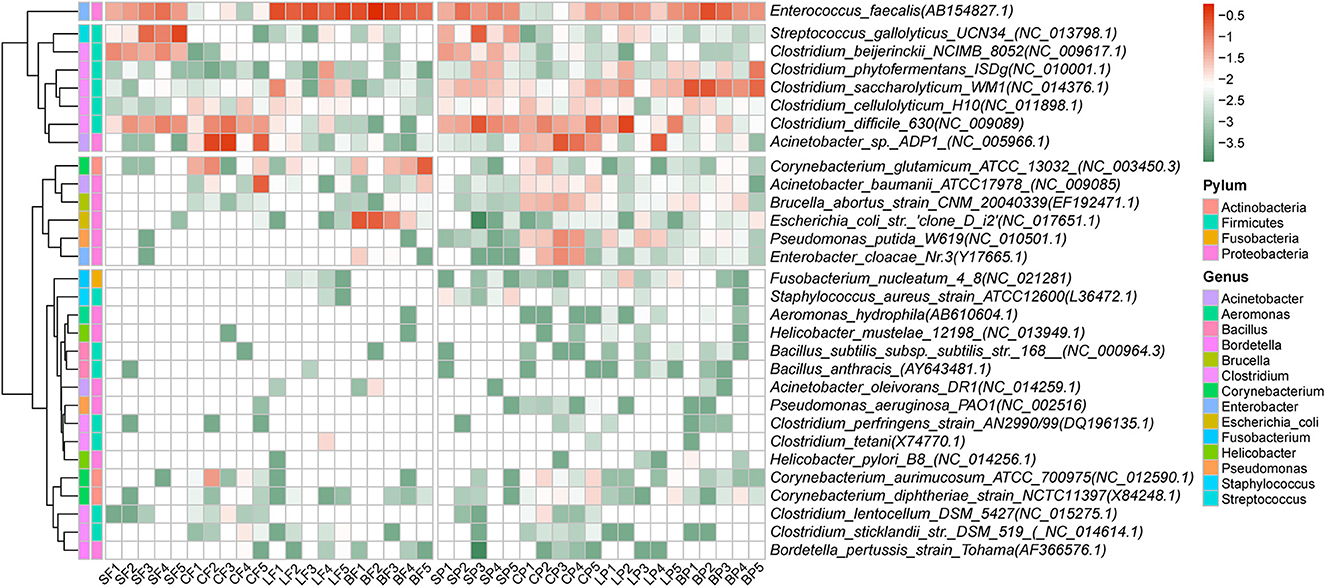
Figure 3. Relative abundance of dominant 30 human pathogenic bacteria in fecal and PM2.5 samples by animal species.
The average relative abundance of 44 bacterial pathogenic species ranged from 0.002% to 16.3% in fecal samples and from 0.001 to 7.2% in air samples. Although no significant differences in the overall relative abundance among the four animals were detected in either feces or air samples (p = 0.08), some specific pathogenic species in layer and broiler farms (both in feces and air) were significantly higher than those in swine feces, such as Enterococcus faecalis (p < 0.05). Acinetobacter sp. was the most dominant pathogenic bacterium in cattle feces but was almost non-existent in swine feces. In this study, Clostridium difficile was abundant in swine and cattle feces, which confirmed that C. difficile is mainly distributed in the feces of large animals (Baverud et al., 2003). Clostridium difficile is a zoonotic bacterium (Rodriguez et al., 2016). Consumption of C. difficile-contaminated foods by vulnerable people with gastrointestinal disorders may lead to infection or asymptomatic transmission in the community (Rodriguez et al., 2016).
3.2. Aerosolization behavior of antibiotic resistance genes, antibiotic resistance bacteria, and human pathogenic bacteria
In this study, the bioaerosolization index (BI) was used to indicate the aerosolization behavior of ARGs, ARBs, and HPBs in feces, and the distribution patterns of their corresponding BI values are shown in Figure 4. When the log BI values were >0, the targets were more likely to aerosolize from animal feces into the air.
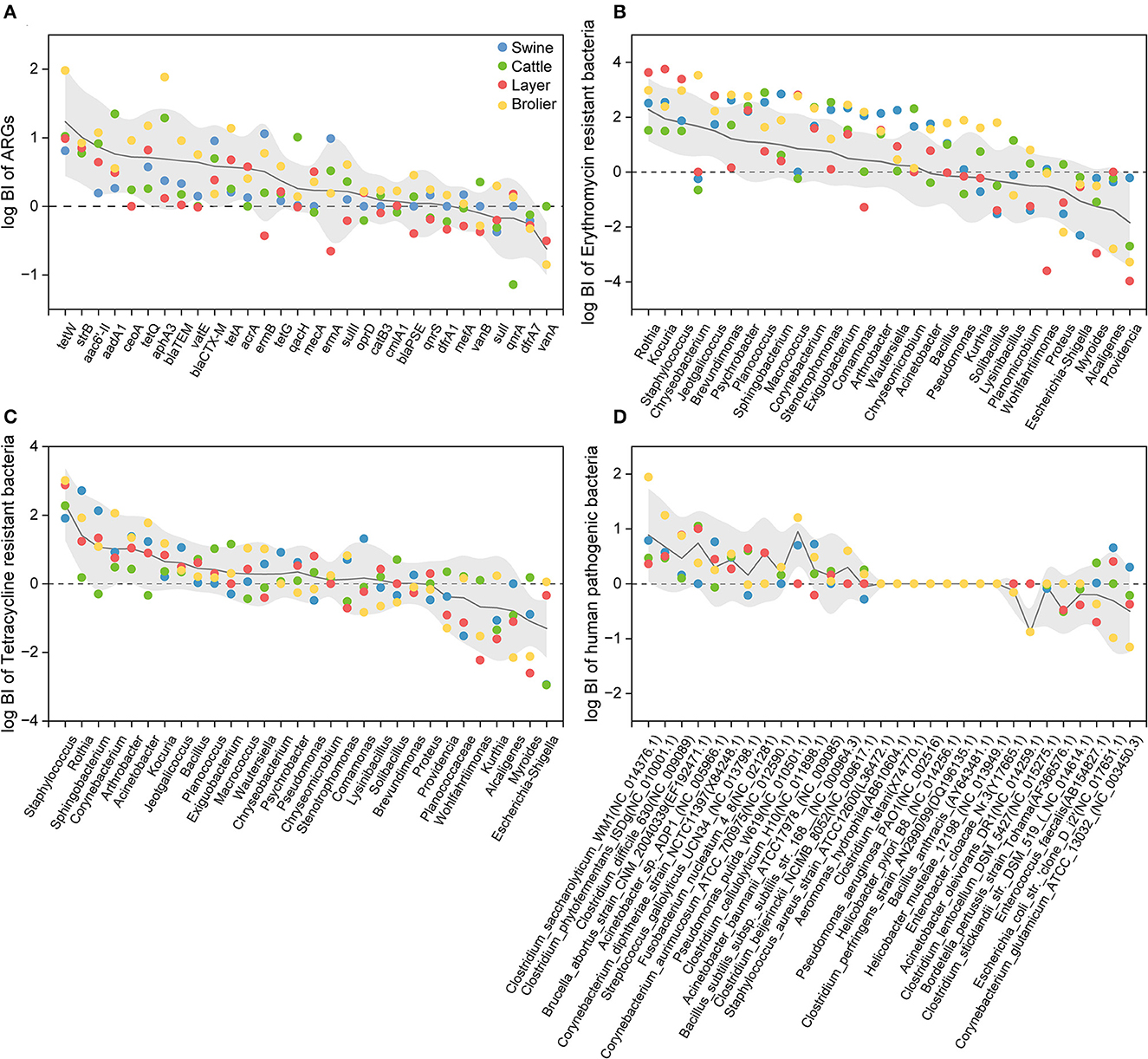
Figure 4. Bioaerosolization index (BI) of top 30 bacterial genera of ARGs (A), erythromycin- (B) and tetracycline-resistant bacterial genera (C), and top 30 human pathogenic bacteria (D). The BI value was log-transformed.
The aerosolization behavior of ARGs in the feces is shown in Figure 4A. Overall, the BI values of the ARGs ranged from 0.04 to 460, and the highest (tetW) and lowest (sulI) BI values were detected in broiler and swine farms. Among the 30 ARGs investigated, 73.3% of the ARGs could be preferentially aerosolized (log BI > 0), in which the genes that encoded the aminoglycoside and tetracycline resistance were more likely to be aerosolized with the log BI values greater than 0. A similarly high relative abundance of ARGs in the air has been previously reported in wastewater treatment plants (Yang et al., 2018), indicating that the preferential aerosolization of certain ARGs may be a ubiquitous phenomenon. However, although vanA and vanB, which encode vancomycin (known as “a last-resort drug”), were detected in both feces and air, these two genes were mostly difficult to aerosolize (except vanB on the cattle farm). The current results indicate that when evaluating the potential contribution of animal feces to the surrounding airborne AMR, it is necessary to comprehensively consider both the concentration of target pollutants in potential sources and their corresponding aerosolization capacity.
The number of preferentially aerosolized ARGs varied by animal species in the following order: broilers (27), cattle (21), layer chickens (19), and swine (18). The highest BI value observed at broiler, layer, swine, and cattle farms were tetW (log BI = 1.97 ± 0.68), tetW (log BI = 1.23 ± 0.79), ermB (log BI = 1.06 ± 0.86), and aadA1 (log BI = 1.35 ± 0.49), respectively. It is noteworthy that all the above genes are high-risk ARGs (Zhang et al., 2021), indicating their varying potential enhanced risk when transmitted through the air in different farms. We also found that the BI of the same ARG varied according to animal species. Macrolide-lincosamide-streptogramin B (MLSB) genes (ermB, ermA, and mefA) were difficult to aerosolize in layer farms (log BI < 0), whereas their log BI in other animal farms was > 0. The aerosolization behavior of FCA resistance genes also showed different trends in different animal farms, especially qnrS and qnrA. These two genes were classified as rank I high-risk ARGs and both showed passive aerosolization on cattle farms, whereas they were preferentially aerosolized on broiler farms. In contrast, the log BI values of vanB for vancomycin-resistance genes were greater than 0 only on cattle farms. This suggests that the aerosolization behavior of ARGs may be affected by the animal species, and the transmission risk caused by different types of animals is not completely consistent.
As shown in Figures 4B, C, the aerosolization of the top 30 erythromycin- and tetracycline-resistant bacterial genera were similar; more than half of the ARBs showed preferential aerosolization, with an average BI of 17 and 18. According to previous research on the aerosolization behavior of bacteria, 11 and 10 genera with BI values higher than 0 were detected in vegetable composting (He et al., 2019) and sludge composting processes (Lu et al., 2021), respectively. One possible explanation for the high BI value of ARBs is that some bacteria have been reported to become more resistant to harsh environments after acquiring antibiotic resistance (Yang et al., 2016). The increased survivability of ARBs in the air was reflected in the higher BI values in this study.
The preferred aerosolized erythromycin-resistant bacteria were Rothia (log BI = 2.28 ± 1.14). Staphylococcus shows preferential aerosolization behavior in both erythromycin- and tetracycline-resistant bacteria with log BI values of 1.94 ± 1.11 and 2.52 ± 1.02, respectively. This may explain the prevalence of Staphylococcus (Figure 2). Although Myroides and Escherichia–Shigella were the dominant bacterial genera in two types of fecal ARBs (Figure 2), it seemed that they were difficult to aerosolize in the animal farm with BI values lower than 0. The same bacterial genera with resistance to different antibiotics show distinguished aerosolized behaviors that the log BI value of erythromycin- and tetracycline-resistant Acinetobacter were−0.17 ± 0.09 and 0.88 ± 0.63, respectively. Although the characteristics of the bacterium itself are considered the main determinants of its aerosolization behavior (Lu et al., 2021), the results of this study indicated that other factors might also play a non-negligible role.
In terms of human pathogenic bacteria, 14 of the top 30 bacteria were prone to preferential aerosolization (log BI > 0). Clostridium saccharolyticum WM1 was the most easily aerosolized pathogen in swine and broiler farms with log BI = 0.79 ± 0.50 and 1.94 ± 0.68, respectively. Brucella abortus strain CNM 20040339 had the highest value at cattle and layer farms with log BI = 1.05 ± 0.49 and log BI = 1.01 ± 0.28. Brucella abortus mainly infects animals, such as swine and cattle, and some studies have found that it may damage the reproductive system and joints of humans and animals (Aliyev et al., 2022). Brucella abortus mainly infected cattle and sheep breeders, slaughterers, and processors, and has been shown to be transmitted by aerosols (Kahl-McDonagh et al., 2007).
The aerosolization behavior of the pathogens was also animal-specific. Streptococcus gallolyticus was preferentially aerosolized in cattle and layer farms, with log BI = 0.60 ± 0.31 and 0.64 ± 0.11. Corynebacterium glutamicum ATCC 13032 gave priority to aerosols in swine farms but to passive aerosols in other animal farms. Among the 44 detected pathogenic bacterial species, some pathogens not only cause animal disease outbreaks and affect the production of meat or eggs but also pose potential risks to human health. For example, Clostridium difficile showed a high abundance in the air with preferential aerosolization behavior (mean log BI = 0.46 ± 0.32). As one of the most common pathogens in nosocomial infections, Clostridium difficile accounts for 15% of all pathogens associated with healthcare-related infections (Magill et al., 2018). The escape of Clostridium difficile from animal feces may pose a threat to the health of workers and residents around farms and even lead to community transmission. Preferentially aerosolized Streptococcus gallolyticus in cattle and layer farms also calls attention because it can cause meningitis, arthritis, and septicemia in pigs and meningitis and toxic shock syndrome in humans, which could lead to death in severe cases (Nguyen et al., 2021).
3.3. Potential factors affecting the aerosolization behavior of antibiotic resistance genes, antibiotic resistance bacteria, and human pathogenic bacteria
According to the distribution of BI involving ARGs, ARBs, and HPBs from the four types of animal farms (Figure 4), fluctuating values of the three types of targets were observed in different animal farms, even in the same target. In addition to the abovementioned animal types, it was assumed that other parameters might also affect the aerosolization process, including breeding-related parameters (age, animal number, breeding area, and stocking density) and environment-related parameters (temperature, relative humidity, and PM2.5). Details are described in our previous study (Xin et al., 2022).
Partial least squares path modeling (PLS-PM) was used to differentiate the effects of breeding, environmental, and animal types on aerosolization behavior. As shown in Figure 5, the GOF of the PLS-PM model was greater than 0.36, indicating that the parameters examined in this study significantly affected the variation in BI values (Henseler and Sarstedt, 2013). Overall, the aerosolization behavior of the targets was significantly influenced by the animal type (except for the abundance of tetracycline-resistant bacteria), which confirmed the results discussed above. In addition, the positive correlations between environmental factors and the aerosolization behaviors of ARGs and ARBs were all significant. The influence on HPB was also positive, with a path coefficient of 0.16. The results indicated that increasing the temperature, relative humidity, and PM2.5 mass concentration in the air might promote the escape of microorganisms in feces. High values of environmental factors within the ranges used in this study increase the survival ability of organisms (Kathiriya et al., 2021). The two types of ARBs showed the same negative correlation with breeding-related parameters. Notably, all the factors investigated here can accelerate the aerosol process of ARGs and HPBs.
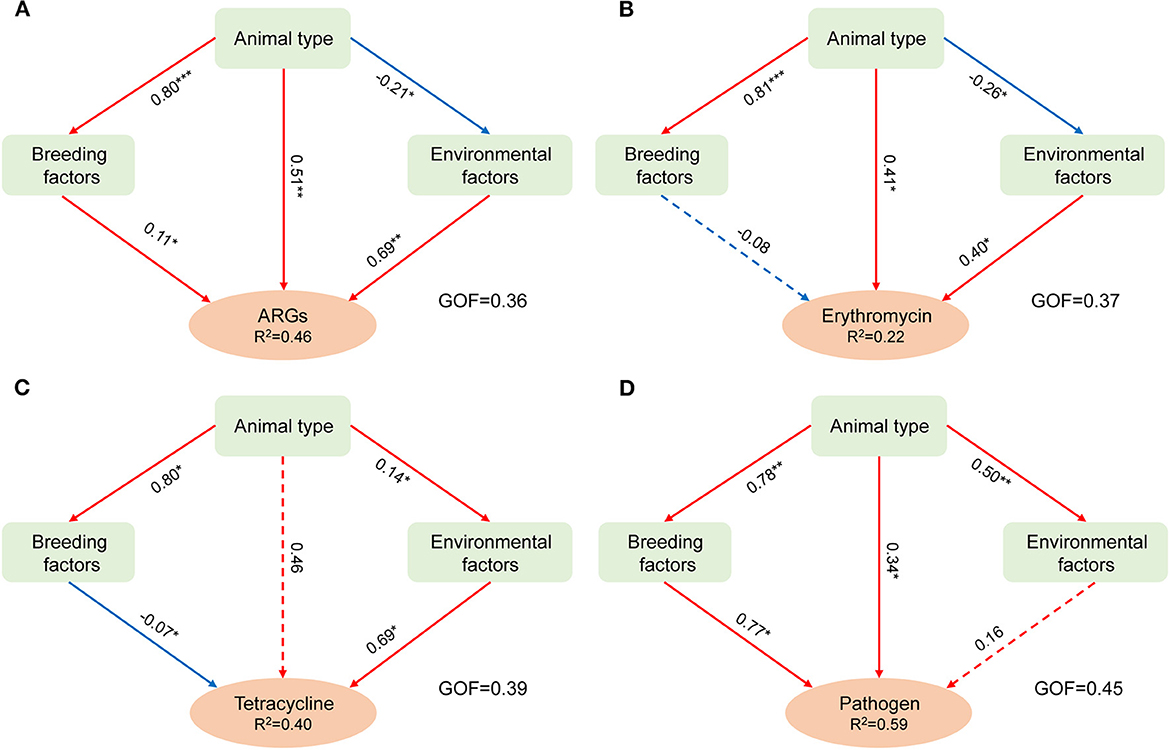
Figure 5. Main factors influencing the aerosolization behavior of antibiotic-resistance genes (A), erythromycin-resistant bacterial genera (B), tetracycline-resistant bacterial genera (C), and human pathogenic bacteria (D) using partial least square path modeling (PLS-PM). The red and blue arrows represented the positive and negative effects, respectively. The dotted and solid lines indicated significance (*p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001). GOF indicated the overall measure of model fit (Henseler and Sarstedt, 2013).
Previous studies have discussed the potential factors for bacteria or fungi escaping animal manure composting (Wang R. et al., 2022) and vegetable waste composting (He et al., 2019), mainly focusing on the physicochemical properties of the composting pile, such as the concentration of heavy metals and other influencing factors (such as C/N, DOC, and pH) on aerosolization behavior. The results of this study improved the role of influencing factors on the aerosol behavior of four types of targets based on a comprehensive discussion of the possible effects of environmental and breeding-related parameters, which provides an important reference basis for the targeted control of the escape of AMR elements and HPBs.
To clarify the specific factors affecting the aerosolization behavior of dominant targets, the relationships between each parameter and the BI value of ARGs, ARB, and HPB were further analyzed using a correlation heatmap and Spearman's correlation coefficient as shown in Figure 6. According to the relationship between the seven factors and the BI of 30 ARGs in Figure 6A, animal number (24) and stocking density (26) were positively correlated with the aerosolization of the majority of genes, which were usually the subtypes with high concentrations in animal feces (Figure 1). These results were observed mainly in poultry farms. Chicken farms with large depositories or high breeding densities usually have a high cleaning frequency, which may promote the escape of ARGs (Liu et al., 2018). Animal age mostly negatively affected the BI of ARGs (Figure 6A) and two types of ARBs (Figures 6B, C), which may be related to the decreasing of fecal bacterial AMR as animal breeding time (Berge et al., 2005; Gaire et al., 2022).
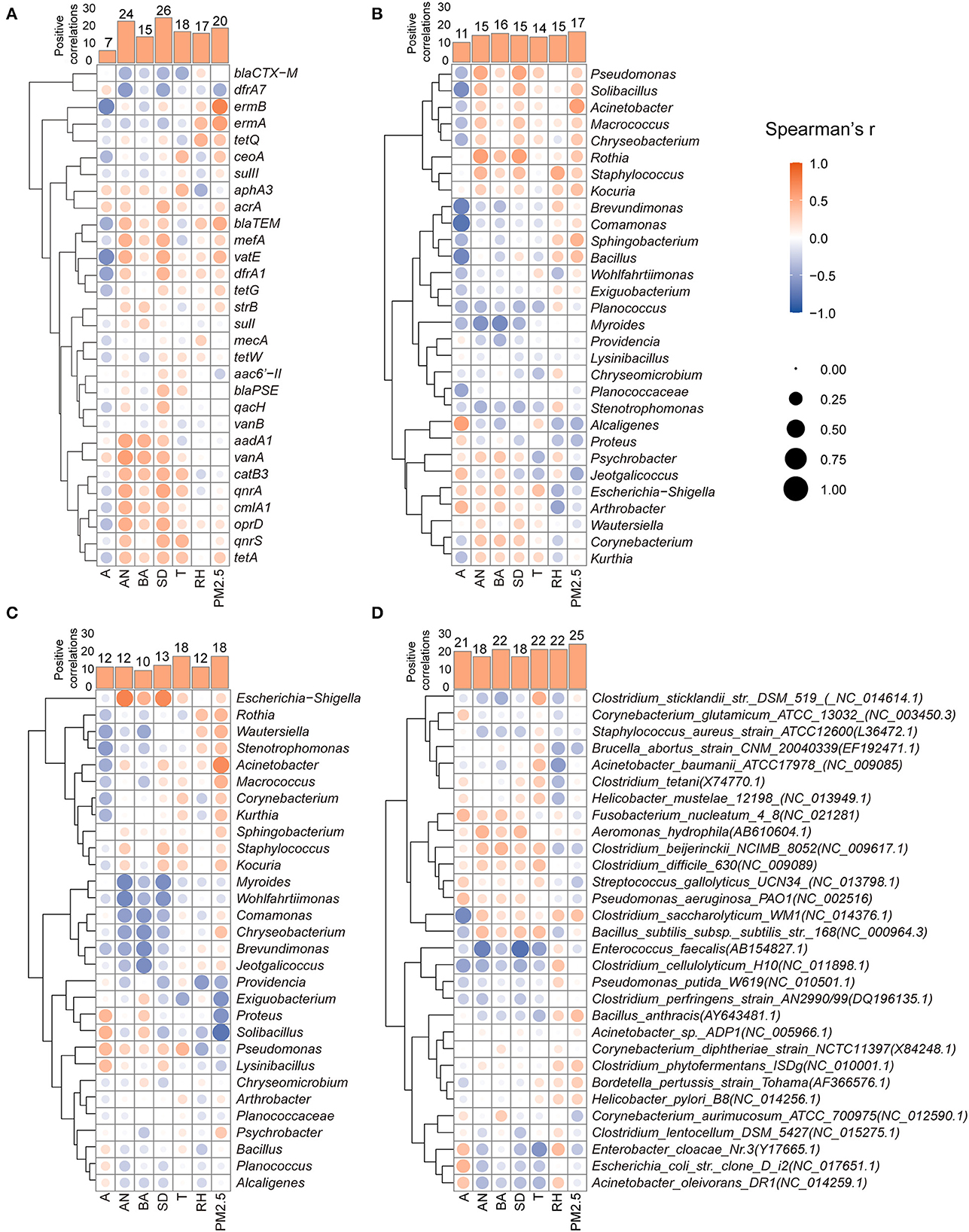
Figure 6. Factors associated with the aerosolization behavior of specific ARGs (A), erythromycin-resistant bacterial genera (B), tetracycline-resistant bacterial genera (C), and human pathogenic bacteria (D). (A, Age; AN, animal number; BA, breeding area; SD, stocking density; T, temperature; RH, relative humidity; PM2.5, PM2.5 mass concentration).
As shown in Figures 6B, C, the potential factors had a relatively neutral impact on the BI value, and no obvious promoting or inhibiting trends were found in this study. The number of positive correlations between the BI and temperature (18) was comparable to that between the BI and humidity (17). However, the effects of temperature and relative humidity on the BI values of the same ARGs showed opposite trends. This may be due to the interactive effects of temperature and relative humidity on bioaerosol activity (Dungan, 2010), which may inactivate microbial proteins under their synergistic influence. Although the effects of temperature and relative humidity on bacterial aerosolization require further investigation, the results of the present study suggest that farms can regulate air temperature and relative humidity to control the aerosolization behavior of airborne bacteria. In addition, some breeding-related parameters, such as the animal number and stocking density, had similar effects on certain types of ARBs. For example, erythromycin- and tetracycline-resistant Escherichia–Shigella, Staphylococcus, Kurthia, and Pseudomonas had positive correlations with the animal number, breeding area, and stocking density.
For HPBs, the results of the correlation heatmap (Figure 6D) showed that most parameters were positively correlated with the aerosolization behavior of HPBs; in particular, the PM2.5 mass concentration was positively correlated with the BI of 25 HPBs. Overall, PM2.5 mainly promotes the biological aerosolization process. Previous studies have shown that the abundance and diversity of airborne ARGs in the environment increase correspondingly with PM2.5 mass concentrations (Sun et al., 2020). The air environment on animal farms is filled with various pollutants, including both inert and active substances, which may provide protection and nutrition to airborne microorganisms, resulting in increased survivability in the air (Sun et al., 2020). Previous studies have shown that the proportion of pathogenic bacteria (Streptococcus pneumoniae) increases with increasing PM levels (Cao et al., 2014), and the increased survival of microorganisms in the air is reflected in the increased BI value. For preferentially aerosolized HPBs (BI > 0), the aerosolization behaviors of Staphylococcus aureus and Escherichia coli were mainly positively correlated with RH and PM2.5, and Pseudomonas aeruginosa and Acinetobacter baumannii were mainly positively correlated with breeding factors.
4. Conclusion
Based on the analysis of the aerosolization behavior and influencing factors of ARGs, ARBs, and HPBs in animal feces from four types of animal farms, the results of this study showed that most of the targets in animal feces were aerosolized, which included human pathogenic bacteria and high-risk ARGs. The average bioaerosolization index of ARGs, ARBs, and HPBs in broiler farms was generally the highest, whereas that in cattle farms was the lowest. Breeding-related parameters had a much more significant effect on aerosolization. The results will help comprehensively assess the potential health risks of fecal microorganisms transported by air paths and their contribution to the surrounding environment. At the same time, it provides technical parameters for the targeted control of the escape of pathogenic bacteria and high-risk ARGs.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Author contributions
HX: writing—original draft preparation, investigation, and formal analysis. TQ and YG: investigation and data curation. HG: investigation and software. LZ: writing—review and editing and conceptualization. MG: writing—review and editing, project administration, and funding acquisition. All the authors contributed to the manuscript and approved the submitted version.
Funding
This study was supported by the National Natural Science Foundation of China (Grant Numbers 41961134033 and 51878053) and the Beijing Agriculture Innovation Consortium (Grant Number BJJQ-G08).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aliyev, J., Alakbarova, M., Garayusifova, A., Omarov, A., Aliyeva, S., and Fretin, D., et al. (2022). Identification and molecular characterization of Brucella abortus and Brucella melitensis isolated from milk in cattle in azerbaijan. BMC Vet. Res. 18, 71. doi: 10.1186/s12917-022-03155-1
Bai, H., He, L., Wu, D., Gao, F., Zhang, M., and Zou, H., et al. (2022). Spread of airborne antibiotic resistance from animal farms to the environment: dispersal pattern and exposure risk. Environ. Int. 158, 106927. doi: 10.1016/j.envint.2021.106927
Baverud, V., Gustafsson, A., Franklin, A., Aspan, A., and Gunnarsson, A. (2003). Clostridium difficile: prevalence in horses and environment, and antimicrobial susceptibility. Equine Vet. J. 35, 465–471. doi: 10.2746/042516403775600505
Berge, A. C., Atwill, E. R., and Sischo, W. M. (2005). Animal and farm influences on the dynamics of antibiotic resistance in faecal Escherichia coli in young dairy calves. Prev. Vet. Med. 69, 25–38. doi: 10.1016/j.prevetmed.2005.01.013
Cao, C., Jiang, W., Wang, B., Fang, J., Lang, J., and Tian, G., et al. (2014). Inhalable microorganisms in beijing's PM2.5 and PM10 pollutants during a severe smog event. Environ. Sci. Technol. 48, 1499–1507. doi: 10.1021/es4048472
Chen, M., Qiu, T., Sun, Y., Song, Y., Wang, X., and Gao, M. (2019). Diversity of tetracycline- and erythromycin-resistant bacteria in aerosols and manures from four types of animal farms in China. Environ. Sci. Pollut. R. 26, 24213–24222. doi: 10.1007/s11356-019-05672-3
Chen, Q., An, X., Li, H., Su, J., Ma, Y., and Zhu, Y. (2016). Long-term field application of sewage sludge increases the abundance of antibiotic resistance genes in soil. Environ. Int. 92-93, 1–10. doi: 10.1016/j.envint.2016.03.026
Cheng, W., Chen, H., Su, C., and Yan, S. (2013). Abundance and persistence of antibiotic resistance genes in livestock farms: a comprehensive investigation in eastern China. Environ. Int. 61, 1–7. doi: 10.1016/j.envint.2013.08.023
de Rooij, M. M. T., Hoek, G., Schmitt, H., Janse, I., Swart, A., and Maassen, C. B. M., et al. (2019). Insights into livestock-related microbial concentrations in air at residential level in a livestock dense area. Environ. Sci. Technol. 53, 7746–7758. doi: 10.1021/acs.est.8b07029
Ding, L. J., Zhou, X. Y., and Zhu, Y. G. (2020). Microbiome and antibiotic resistome in household dust from Beijing, China. Environ. Int. 139, 105702. doi: 10.1016/j.envint.2020.105702
Dungan, R. S. (2010). Board-invited review: fate and transport of bioaerosols associated with livestock operations and manures. J. Anim. Sci. 88, 3693–3706. doi: 10.2527/jas.2010-3094
Gaire, T. N., Noyes, N. R., Scott, H. M., Ericsson, A. C., Dunmire, K., and Tokach, M. D., et al. (2022). A longitudinal investigation of the effects of age, dietary fiber type and level, and injectable antimicrobials on the fecal microbiome and antimicrobial resistance of finisher pigs. J. Anim. Sci. 100, skac217. doi: 10.1093/jas/skac217
Gao, M., Qiu, T., Jia, R., Han, M., Song, Y., and Wang, X. (2015). Concentration and size distribution of viable bioaerosols during non-haze and haze days in Beijing. Environ. Sci. Pollut. R. 22, 4359–4368. doi: 10.1007/s11356-014-3675-0
Gao, M., Qiu, T., Sun, Y., and Wang, X. (2018). The abundance and diversity of antibiotic resistance genes in the atmospheric environment of composting plants. Environ. Int. 116, 229–238. doi: 10.1016/j.envint.2018.04.028
He, L., Ying, G., Liu, Y., Su, H., Chen, J., and Liu, S., et al. (2016). Discharge of swine wastes risks water quality and food safety: antibiotics and antibiotic resistance genes from swine sources to the receiving environments. Environ. Int. 92-93, 210–219. doi: 10.1016/j.envint.2016.03.023
He, P., Wei, S., Shao, L., and Lü, F. (2019). Aerosolization behavior of prokaryotes and fungi during composting of vegetable waste. Waste Manage. 89, 103–113. doi: 10.1016/j.wasman.2019.04.008
Henseler, J., and Sarstedt, M. (2013). Goodness-of-fit indices for partial least squares path modeling. Computation. Stat. 28, 565–580. doi: 10.1007/s00180-012-0317-1
Hong, P. Y., Li, X., Yang, X., Shinkai, T., Zhang, Y., and Wang, X., et al. (2012). Monitoring airborne biotic contaminants in the indoor environment of pig and poultry confinement buildings. Environ. Microbiol. 14, 1420–1431. doi: 10.1111/j.1462-2920.2012.02726.x
Jezak, K., and Kozajda, A. (2022). Occurrence and spread of antibiotic-resistant bacteria on animal farms and in their vicinity in Poland and Ukraine-review. Environ. Sci. Pollut. Res. Int. 29, 9533–9559. doi: 10.1007/s11356-021-17773-z
Jiang, W., Liang, P., Wang, B., Fang, J., Lang, J., and Tian, G., et al. (2015). Optimized DNA extraction and metagenomic sequencing of airborne microbial communities. Nat. Protoc. 10, 768–779. doi: 10.1038/nprot.2015.046
Kahl-McDonagh, M. M., Arenas-Gamboa, A. M., and Ficht, T. A. (2007). Aerosol infection of balb/c mice with Brucella melitensis and Brucella abortus and protective efficacy against aerosol challenge. Infect. Immun. 75, 4923–4932. doi: 10.1128/IAI.00451-07
Kathiriya, T., Gupta, A., and Singh, N. K. (2021). An opinion review on sampling strategies, enumeration techniques, and critical environmental factors for bioaerosols: an emerging sustainability indicator for society and cities. Environ. Technol. Innov. 21, 101287. doi: 10.1016/j.eti.2020.101287
Liu, H., Zhang, X., Zhang, H., Yao, X., Zhou, M., and Wang, J., et al. (2018). Effect of air pollution on the total bacteria and pathogenic bacteria in different sizes of particulate matter. Environ. Pollut. 233, 483–493. doi: 10.1016/j.envpol.2017.10.070
Lu, F., Hu, T., Wei, S., Shao, L., and He, P. (2021). Bioaerosolization behavior along sewage sludge biostabilization. Front. Env. Sci. Eng. 15. doi: 10.1007/s11783-020-1339-5
Luiken, R. E. C., Van Gompel, L., Bossers, A., Munk, P., Joosten, P., Hansen, R. B., et al. (2020). Farm dust resistomes and bacterial microbiomes in European poultry and pig farms. Environ. Int. 143, 105971. doi: 10.1016/j.envint.2020.105971
Luo, Y. H., Peng, H. W., Wright, A. D., Bai, S. P., Ding, X. M., and Zeng, Q. F., et al. (2013). Broilers fed dietary vitamins harbor higher diversity of cecal bacteria and higher ratio of Clostridium, faecalibacterium, and Lactobacillus than broilers with no dietary vitamins revealed by 16s rRNA gene clone libraries. Poult. Sci. 92, 2358–2366. doi: 10.3382/ps.2012-02935
Magill, S. S., O'Leary, E., Janelle, S. J., Thompson, D. L., Dumyati, G., and Nadle, J., et al. (2018). Changes in prevalence of health care-associated infections in U.S. Hospitals. N Engl J Med. 379, 1732–1744. doi: 10.1056/NEJMoa1801550
Mao, Y., Ding, P., Wang, Y., Ding, C., Wu, L., and Zheng, P., et al. (2019). Comparison of culturable antibiotic-resistant bacteria in polluted and non-polluted air in Beijing, China. Environ. Int. 131, 104936. doi: 10.1016/j.envint.2019.104936
McEachran, A. D., Blackwell, B. R., Hanson, J. D., Wooten, K. J., Mayer, G. D., and Cox, S. B., et al. (2015). Antibiotics, bacteria, and antibiotic resistance genes: aerial transport from cattle feed yards via particulate matter. Environ. Health Perspect. 123, 337–343. doi: 10.1289/ehp.1408555
Moletta, M., Bru-Adan, V., Delgenes, J., Hamelin, J., Wéry, N., and Godon, J. (2010). Selective microbial aerosolization in biogas demonstrated by quantitative PCR. Bioresource Technol. 101, 7252–7257. doi: 10.1016/j.biortech.2010.04.035
Moletta, M., Delgenes, J., and Godon, J. (2007). Differences in the aerosolization behavior of microorganisms as revealed through their transport by biogas. Sci. Total Environ. 379, 75–88. doi: 10.1016/j.scitotenv.2007.02.019
Munk, P., Knudsen, B. E., Lukjancenko, O., Duarte, A. S. R., Van Gompel, L., and Luiken, R. E. C., et al. (2018). Abundance and diversity of the faecal resistome in slaughter pigs and broilers in nine European countries. Nat. Microbiol. 3, 898–908. doi: 10.1038/s41564-018-0192-9
Nguyen, N., Luu, Y., Hoang, T. D., Nguyen, H. X., Dao, T. D., and Bui, V. N., et al. (2021). An epidemiological study of Streptococcus suis prevalence among swine at industrial swine farms in northern Vietnam. One Health. 13, 100254. doi: 10.1016/j.onehlt.2021.100254
Parker, B. C., Ford, M. A., Gruft, H., and Falkinham, J. R. (1983). Epidemiology of infection by nontuberculous mycobacteria. Iv. Preferential aerosolization of Mycobacterium intracellulare from natural waters. Am Rev Respir Dis. 128, 652–656.
Qian, X., Gu, J., Sun, W., Wang, X. J., Su, J. Q., and Stedfeld, R. (2018). Diversity, abundance, and persistence of antibiotic resistance genes in various types of animal manure following industrial composting. J. Hazard. Mater. 344, 716–722. doi: 10.1016/j.jhazmat.2017.11.020
Rodriguez, C., Taminiau, B., Van Broeck, J., Delmee, M., and Daube, G. (2016). Clostridium difficile in food and animals: a comprehensive review. Adv. Exp. Med. Biol. 932, 65–92. doi: 10.1007/5584_2016_27
Sitthicharoenchai, P., Burrough, E. R., Arruda, B. L., Sahin, O., Dos, S. J., and Magstadt, D. R., et al. (2022). Streptococcus gallolyticus and bacterial endocarditis in swine, United States, 2015-2020. Emerging Infect. Dis. 28, 192–195. doi: 10.3201/eid2801.210998
Song, L., Wang, C., Jiang, G., Ma, J., Li, Y., and Chen, H., et al. (2021). Bioaerosol is an important transmission route of antibiotic resistance genes in pig farms. Environ. Int. 154, 106559. doi: 10.1016/j.envint.2021.106559
Song, Y., Han, Z., Song, K., and Zhen, T. (2020). Antibiotic consumption trends in China: evidence from six-year surveillance sales records in Shandong province. Front. Pharmacol. 11, 491. doi: 10.3389/fphar.2020.00491
Sun, X., Li, D., Li, B., Sun, S., Yabo, S. D., and Geng, J., et al. (2020). Exploring the disparity of inhalable bacterial communities and antibiotic resistance genes between hazy days and non-hazy days in a cold megacity in northeast China. J. Hazard. Mater. 398, 122984. doi: 10.1016/j.jhazmat.2020.122984
Tang, K. L., Caffrey, N. P., Nobrega, D. B., Cork, S. C., Ronksley, P. E., Barkema, H. W., et al. (2017). Restricting the use of antibiotics in food-producing animals and its associations with antibiotic resistance in food-producing animals and human beings: a systematic review and meta-analysis. Lancet Planet Health 1, e316–e327. doi: 10.1016/S2542-5196(17)30141-9
Van Boeckel, T. P., Pires, J., Silvester, R., Zhao, C., Song, J., and Criscuolo, N. G., et al. (2019). Global trends in antimicrobial resistance in animals in low- and middle-income countries. Science 365, aaw1944 doi: 10.1126/science.aaw1944
Wang, L., Wang, J., Wang, J., Zhu, L., Yang, L., and Yang, R. (2019). Distribution characteristics of antibiotic resistant bacteria and genes in fresh and composted manures of livestock farms. Sci. Total Environ. 695, 133781. doi: 10.1016/j.scitotenv.2019.133781
Wang, M., Yao, M., and Zhu, Y. (2022). Antibiotic resistance genes and antibiotic sensitivity in bacterial aerosols and their comparisons with known respiratory pathogens. J. Aerosol Sci. 161, 105931. doi: 10.1016/j.jaerosci.2021.105931
Wang, R., Yu, A., Qiu, T., Guo, Y., Gao, H., and Sun, X., et al. (2022). Aerosolization behaviour of fungi and its potential health effects during the composting of animal manure. Int. J. Env. Res. Pub. He. 19, 5644. doi: 10.3390/ijerph19095644
Wery, N. (2014). Bioaerosols from composting facilities–a review. Front. Cell. Infect. Microbiol. 4, 42. doi: 10.3389/fcimb.2014.00042
Xie, J., Jin, L., He, T., Chen, B., Luo, X., and Feng, B., et al. (2019). Bacteria and antibiotic resistance genes (ARGs) in pm2.5 from China: implications for human exposure. Environ. Sci. Technol. 53, 963–972. doi: 10.1021/acs.est.8b04630
Xin, H., Gao, M., Wang, X., Qiu, T., Guo, Y., and Zhang, L. (2022). Animal farms are hot spots for airborne antimicrobial resistance. Sci. Total Environ. 851, 158050. doi: 10.1016/j.scitotenv.2022.158050
Xiong, W., Sun, Y., and Zeng, Z. (2018). Antimicrobial use and antimicrobial resistance in food animals. Environ. Sci. Pollut. Res. Int. 25, 18377–18384. doi: 10.1007/s11356-018-1852-2
Xu, X., Zhou, W., Xie, C., Zhu, Y., Tang, W., and Zhou, X., et al. (2022). Airborne bacterial communities in the poultry farm and their relevance with environmental factors and antibiotic resistance genes. Sci. Total Environ. 846, 157420. doi: 10.1016/j.scitotenv.2022.157420
Yang, L., Zhou, S., Lin, C., Huang, X., Neilson, R., and Yang, X. (2022). Effects of biofertilizer on soil microbial diversity and antibiotic resistance genes. Sci. Total Environ. 820, 153170. doi: 10.1016/j.scitotenv.2022.153170
Yang, Q., Wang, R., Ren, S., Szoboszlay, M., and Moe, L. A. (2016). Practical survey on antibiotic-resistant bacterial communities in livestock manure and manure-amended soil. J. Environ. Sci. Health B. 51, 14–23. doi: 10.1080/03601234.2015.1080481
Yang, Y., Zhou, R., Chen, B., Zhang, T., Hu, L., and Zou, S. (2018). Characterization of airborne antibiotic resistance genes from typical bioaerosol emission sources in the urban environment using metagenomic approach. Chemosphere. 213, 463–471. doi: 10.1016/j.chemosphere.2018.09.066
Yuan, W., Zeng, X., Cao, Y., Yang, Q., Riaz, L., and Wang, Q. (2022). Distribution of antibiotic resistance genes from human and animal origins to their receiving environments: a regional scale survey of urban settings. Environ. Pollut. 293, 118512. doi: 10.1016/j.envpol.2021.118512
Yue, Z., Zhang, J., Zhou, Z., Ding, C., Wan, L., and Liu, J., et al. (2021). Pollution characteristics of livestock faeces and the key driver of the spread of antibiotic resistance genes. J. Hazard. Mater. 409, 124957. doi: 10.1016/j.jhazmat.2020.124957
Zainab, S. M., Junaid, M., Xu, N., and Malik, R. N. (2020). Antibiotics and antibiotic resistant genes (ARGs) in groundwater: a global review on dissemination, sources, interactions, environmental and human health risks. Water Res. 187, 116455. doi: 10.1016/j.watres.2020.116455
Zhang, A., Gaston, J. M., Dai, C. L., Zhao, S., Poyet, M., and Groussin, M., et al. (2021). An omics-based framework for assessing the health risk of antimicrobial resistance genes. Nat. Commun. 12, 4765. doi: 10.1038/s41467-021-25096-3
Zhang, J., Lu, T., Chai, Y., Sui, Q., Shen, P., and Wei, Y. (2019). Which animal type contributes the most to the emission of antibiotic resistance genes in large-scale swine farms in China? Sci. Total Environ. 658, 152–159. doi: 10.1016/j.scitotenv.2018.12.175
Zhao, L., Dong, Y. H., and Wang, H. (2010). Residues of veterinary antibiotics in manures from feedlot livestock in eight provinces of China. Sci. Total Environ. 408, 1069–1075. doi: 10.1016/j.scitotenv.2009.11.014
Zhao, Y., Yang, Q. E., Zhou, X., Wang, F., Muurinen, J., and Virta, M. P., et al. (2021). Antibiotic resistome in the livestock and aquaculture industries: status and solutions. Crit. Rev. Env. Sci. Tec. 51, 2159–2196. doi: 10.1080/10643389.2020.1777815
Zhu, T., Chen, T., Cao, Z., Zhong, S., Wen, X., and Mi, J., et al. (2021). Antibiotic resistance genes in layer farms and their correlation with environmental samples. Poult. Sci. 100, 101485. doi: 10.1016/j.psj.2021.101485
Keywords: animal farms, fine particulate matter, antibiotic resistance, human pathogenic bacteria, aerosolization behavior
Citation: Xin H, Qiu T, Guo Y, Gao H, Zhang L and Gao M (2023) Aerosolization behavior of antimicrobial resistance in animal farms: a field study from feces to fine particulate matter. Front. Microbiol. 14:1175265. doi: 10.3389/fmicb.2023.1175265
Received: 27 February 2023; Accepted: 28 March 2023;
Published: 20 April 2023.
Edited by:
Xu Yan, Henan Normal University, ChinaReviewed by:
Jianwei Liu, Beijing University of Civil Engineering and Architecture, ChinaYanjie Wang, Zhengzhou University, China
Copyright © 2023 Xin, Qiu, Guo, Gao, Zhang and Gao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liqiu Zhang, emhhbmdsaXFpdUBiamZ1LmVkdS5jbg==; Min Gao, Z2FvbWluQGJhYnJjLmFjLmNu
 Huibo Xin
Huibo Xin Tianlei Qiu1
Tianlei Qiu1