
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 04 May 2023
Sec. Evolutionary and Genomic Microbiology
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1170790
 Peng Wu1
Peng Wu1 Wenqi Xiao2
Wenqi Xiao2 Yingyong Luo2
Yingyong Luo2 Zhuang Xiong2
Zhuang Xiong2 Xiaodie Chen2
Xiaodie Chen2 Jing He2
Jing He2 Ajia Sha2
Ajia Sha2 Mingying Gui1†*
Mingying Gui1†* Qiang Li2†*
Qiang Li2†*Introduction: Codon usage bias is a prevalent phenomenon observed across various species and genes. However, the specific attributes of codon usage in the mitochondrial genome of Ganoderma species remain unknown.
Methods: In this study, we investigated the codon bias of 12 mitochondrial core protein-coding genes (PCGs) in 9 Ganoderma species, including 13 Ganoderma strains.
Results: The codons of all Ganoderma strains showed a preference for ending in A/T. Additionally, correlations between codon base composition and the codon adaptation index (CAI), codon bias index (CBI) and frequency of optimal codons (FOP) were identified, demonstrating the impact of base composition on codon bias. Various base bias indicators were found to vary between or within Ganoderma strains, including GC3s, the CAI, the CBI, and the FOP. The results also revealed that the mitochondrial core PCGs of Ganoderma have an average effective number of codons (ENC) lower than 35, indicating strong bias toward certain codons. Evidence from neutrality plot and PR2-bias plot analysis indicates that natural selection is a major factor affecting codon bias in Ganoderma. Additionally, 11 to 22 optimal codons (ΔRSCU>0.08 and RSCU>1) were identified in 13 Ganoderma strains, with GCA, AUC, and UUC being the most widely used optimal codons in Ganoderma. By analyzing the combined mitochondrial sequences and relative synonymous codon usage (RSCU) values, the genetic relationships between or within Ganoderma strains were determined, indicating variations between them. Nevertheless, RSCU-based analysis illustrated the intra- and interspecies relationships of certain Ganoderma species.
Discussion: This study deepens our insight into the synonymous codon usage characteristics, genetics, and evolution of this important fungal group.
The genetic data present in the DNA of organic material is converted into a sequence of 20 amino acids through the mechanisms of transcription and translation (Iriarte et al., 2021; Parvathy et al., 2022). The 64 triplet codon arrangements found in DNA are responsible for encoding 20 standard amino acids, with 61 triplets dedicated to this purpose. Eighteen of the amino acids are represented by multiple codons, while tryptophan and methionine are typically encoded by a single codon. The degeneracy of the genetic code allows the same amino acid to be represented by synonymous codons or divergent codons (Krasovec and Filatov, 2019; Pepe and Keersmaecker, 2020). Synonymous codons are not usually used randomly or with equal probability; rather, some codons are more frequently utilized than others to encode amino acids. This phenomenon is known as codon usage bias (CUB) (Biro, 2008; Behura and Severson, 2013). Synonymous codons frequently appear with different frequencies in different genes, different organisms, or even the same gene from different species (Akashi, 1997; Choudhury et al., 2018; Camiolo et al., 2019). Substituting one codon for another synonymous codon will not result in any alteration of the primary sequence of amino acids or peptides, which is primarily caused by mutations in the gene coding region, particularly mutations in the second or third nucleotides of the codon (De Mandal et al., 2020; Bailey et al., 2021). Evolutionary processes can lead to variability in synonymous codons due to a synonymous mutation or ‘silent mutation’, which does not have any impact on functionality (Deng et al., 2020; De Oliveira et al., 2021). It can be concluded that codon usage bias is the consequence of mutation patterns that are unevenly distributed. As some codons are more prone to mutation than others, selection is able to maintain this bias (Rao et al., 2011; Trotta, 2013). GC heterogeneity and gBGC can lead to codon usage bias, which is a consequence of local recombination rate-based codon usage (Palidwor et al., 2010; Oldfield et al., 2020). Mutations, natural selection, and genetic drift are all factors that contribute to the development of synonymous codons and thus have a considerable effect on genome evolution (Li et al., 2018; Li B. et al., 2022). It has been proposed that interspecific differences in codon usage can be explained by a mutation mechanism in which codon bias is caused by the rate of repair of nucleotide bias or point mutations (LaBella et al., 2019; Lamolle et al., 2022; Li et al., 2022c, 2023b). Natural selection theory postulates that synonymous mutations that have an effect on an organism’s ability to adapt will be either favored or disfavored over the course of evolution, resulting in changes in codon usage in genomes or genes (Dhindsa et al., 2020; Cope and Shah, 2022).
Codon bias can have a significant impact on a variety of cellular processes and characteristics, such as transcription, translation efficiency and accuracy, mRNA stability, and protein expression, structure, function, and folding during cotranslation (Chaney and Clark, 2015; Fages-Lartaud et al., 2022). Codon bias has an influence on transcription due to its effect on chromatin structure and mRNA folding, which in turn affects translation efficiency by influencing the rate of translation elongation (Hia et al., 2019; Hia and Takeuchi, 2021). The genome has adapted to transcription and translation procedures through the development of codon bias. This increases the effectiveness of studies on the molecular evolution of genes by allowing selection of genes that do not alter amino acids (Yao et al., 2019; Yu et al., 2021). Furthermore, codon bias analysis can reveal related organisms (Li et al., 2023a), as they often use similar codons (Prabha et al., 2017; Rahman et al., 2022). By utilizing genetic engineering and recombinant DNA technology, the expression of heterologous genes can be enhanced by optimizing the codons of the genes that code for the most highly expressed proteins (López et al., 2020; Liu et al., 2021; Bao et al., 2023). The advancement of high-throughput sequencing technology has made codon bias analysis an indispensable tool for exploring species evolution, environmental adaptation, and genetics (Lal et al., 2016; Masłowska-Górnicz et al., 2022; Wu et al., 2023). However, the genetic features of codon bias in large, higher fungi are not yet fully understood (Jiang et al., 2017).
Eukaryotes rely on the mitochondrial genome, which is often referred to as the ‘second genome’, for their growth, development, and ability to adjust to the environment (Latorre-Pellicer et al., 2016). Fungi possess a set of 15 core protein-coding genes (PCGs), namely, atp6, atp8, atp9, cob, cox1, cox2, cox3, nad1, nad2, nad3, nad4, nad4L, nad5, nad6, and rps3 (Li et al., 2020a, 2022a,b). Variations in the mitochondrial genome are essential for the stability, resilience, and adaptability of eukaryotic cells, in addition to their development (Hayashi et al., 2016; Chen et al., 2020; Cai et al., 2021). Ganoderma is a genus of fungi that is part of the Ganodermataceae family, a group of fungi with a wide variety of species (Kwon et al., 2016; Sun et al., 2022a,c). It is believed that the genus Ganoderma encompasses more than 250 distinct species (Wang et al., 2012; Kües et al., 2015; Jargalmaa et al., 2017; Sun et al., 2022c). At present, the genomics and cultivation of Ganoderma genus are receiving increasing attention, promoting the development and utilization of Ganoderma resources (Si et al., 2021; Sun et al., 2022b). It has been observed that some species from the Ganoderma genus can be detrimental to woody plants, such as G. boninense (Dhillon et al., 2021), G. steyaertanum (Hidayati et al., 2014), G. mastoporum (Page et al., 2018), and G. philippii (Francis et al., 2014). Numerous Ganoderma species are renowned for their medicinal properties in traditional Asian medicine (Paterson, 2006; Bishop et al., 2015; Zhu et al., 2015). Some Ganoderma species, including G. lucidum, G. leucocontextum, G. sinense, and G. lingzhi, have been found to have a variety of beneficial ingredients that are of great economic and medical value (Chan et al., 2015; Chien et al., 2015; Gao et al., 2020; Zheng et al., 2020). The bioactive components of Ganoderma have drawn increasing attention from the Western medical and wellness industries, leading to a multibillion dollar market value in 2017 (Jargalmaa et al., 2017). To date, the codon usage, genetic properties, and evolutionary development of the mitochondrial core proteins in Ganoderma have not been explored.
This study investigated the utilization of synonymous codons of mitochondrial core PCGs between and within 13 Ganoderma strains, including G. applanatum, G. leucocontextum, G. tsugae, G. sinense, G. subamboinense s118, G. calidophilum, G. meredithae, G. lucidum KC763799, G. lucidum s26, G. lucidum s37, G. lingzhi s62, G. lingzhi s74, and G. lingzhi s8. Utilizing relative synonymous codon usage (RSCU) data, we deduced the phylogenetic relationship of different Ganoderma strains and compared it with the phylogenetic relationship established through mitochondrial genome sequence inference. This is the first study to investigate the synonymous codon usage patterns of this important group of edible and medicinal fungi, which will contribute to a better understanding of the evolution, genetics, and species differentiation of Ganoderma and other related species.
To date, 13 full Ganoderma mitochondrial genomes have been added to the NCBI database, 9 of which were reported in our prior research (Li et al., 2019b, 2022d). We first downloaded the G. applanatum, G. leucocontextum, G. tsugae, G. calidophilum, G. subamboinense, G. meredithae, G. sinense, G. lucidum, G. lucidum s26, G. lucidum s37, G. lingzhi s62, G. lingzhi s74, and G. lingzhi s8 mitochondrial genomes from the NCBI database under the accession numbers KR109212, MH252534, MH252533, MH252535, MW752412, KP410262, KF673550, KC763799, MH252532, MW752414, MW752415, MW752413, and MH252531, respectively (Li et al., 2013; Wang et al., 2016a,b; Utomo et al., 2019; Meng et al., 2022). We then obtained the core protein-coding sequence in the mitochondrial genomes of 13 Ganoderma strains. Core protein-coding genes with a sequence length of less than 300 bp were excluded from the subsequent analysis (Bu et al., 2021). Consequently, we identified 12 core protein-coding genes in each Ganoderma strain for further study, comprising atp6, cob, cox1, cox2, cox3, nad1, nad2, nad3, nad4, nad5, nad6, and rps3.
The GC3s metric is utilized to quantify the number of codons with guanine and cytosine at the third synonymous position, with the exception of Met, Trp, and stop codons (Huo et al., 2021; Xu et al., 2021). The codon adaptation index (CAI) is a numerical value used to gage the propensity for codons to be utilized in genes that have been expressed at high levels (Sharp and Li, 1987). The CAI is a measure that indicates the frequency of synonymous codon usage and is expressed as a numerical value from 0 to 1.0, with higher values indicating a higher frequency. The codon bias index (CBI) is a measure used to assess gene expression based on the relative abundance of optimal codons in a gene. The frequency of optimal codons (FOP) is calculated by dividing the number of optimal codons by the total number of synonymous codons in the gene. The effective number of codons (ENC) is a metric for evaluating the number of codons present in a gene, which can range from 20 to 61. A score of 20 implies that only one codon is allocated for each amino acid, whereas a result of 61 implies that each codon is employed with equal frequency. The ENC value is a measure of codon usage preference; a low value (below 35) signifies a strong preference, while a higher value (above 35) indicates a weaker preference. The relative synonymous codon usage (RSCU) is calculated by dividing the probability of the same codons encoding the same amino acid. Codon bias is deemed to be positive when the RSCU value is above 1 and negative when the value is below 1. The general average hydropathicity (GRAVY) value is calculated by adding up the hydropathy values of all the amino acids in the polymerase gene sequences and then multiplying the sum by the number of residues in the gene sequences. This value ranges from −2 to 2, with positive values indicating hydrophobic proteins and negative values indicating hydrophilic proteins. The aromaticity (AROMO) value is a metric of the frequency of aromatic amino acids (Phe, Tyr, and Trp). GRAVY and AROMO are also metrics of amino acid usage, and any changes in the amino acid composition will have an impact on the results of codon usage. All codon usage indicators can be derived through the use of CodonW1.4.2 (Peden, 1999) or the CAIcal server (Puigbo et al., 2008).
A neutrality plot (GC12 vs. GC3) can be employed to evaluate the balance between mutation and selection when codon bias is created. GC12 is the mean GC content in the codon’s first and second positions (GC1 and GC2), while GC3 is the GC content in the third position. A strong association between GC12 and GC3 implies that the species is primarily under mutation pressure; in contrast, a lack of correlation suggests that the main driving force is natural selection.
The ENC-GC3s plot is a tool commonly used to determine whether the codon usage of a particular gene is affected by mutation alone or by other factors, such as natural selection. This plot consists of ENC values on the ordinate and GC3s values on the abscissa, with a predicted curve calculated by a specific formula. If the points are distributed around the expected curve, it can be concluded that mutation pressure is the primary factor in the formation of codon bias. However, if the points deviate significantly from the expected curve, it is likely that other factors, such as natural selection, are at play in the formation of codon bias.
The ENCRatio quantifies the divergence between the forecasted and actual ENCs.
Parity Rule 2 bias (PR2-bias) plot analysis can be used to identify the magnitude and direction of gene bias. This plot is based on A3/(A3+ U3) versus G3/(G3 + C3), and the center point of the plot indicates that the codon has no usage bias, with A = T and C = G.
Correspondence analysis (COA) is a widely accepted multivariate statistical analysis method used to recognize codon usage patterns. All genes were plotted in a 59-dimensional space, considering the 59 sense codons (Met and Trp excluded). This approach can detect the main trends in codon usage in the core CDSs of Ganoderma and arrange codons along the axis based on their RSCU value.
The genes were arranged in descending order of their ENC values, and 10% of the genes from the beginning and end of the list were chosen to form a dataset of high- and low-expression genes. The difference in RSCU (ΔRSCU) between the two datasets was then calculated, with a ΔRSCU value of more than 0.08 indicating a codon with high expression. Codons with RSCU values higher than 1 were regarded as high-frequency codons. A codon with both ΔRSCU>0.08 and RSCU>1 was identified as an optimal codon.
Codon usage-based and mitochondrial sequence-based methods were applied to compare the phylogenetic relationships of Ganoderma strains. Using the RSCU values of the 13 Ganoderma strains, SPSS v19.0 software was used to apply a hierarchical clustering method to depict the relationships between the various Ganoderma strains as a tree. Utilizing the procedure documented in our earlier studies, we also constructed phylogenetic trees of the 13 Ganoderma strains based on the combined mitochondrial gene datasets (Li et al., 2019a, 2020b). The alignment of individual mitochondrial genes was first conducted through MAFFT v7.037 (Katoh et al., 2017), and then the aligned sequences were merged into one set with the use of Sequence Matrix v1.7.8 (Vaidya et al., 2011). A partition homogeneity test was applied to detect any potential inconsistencies in the phylogenies of different mitochondrial genes. Partition Finder 2.1.1 (Lanfear et al., 2017) was employed to ascertain the best model of partitioning and evolution for the combined mitochondrial gene set. The phylogenetic tree was constructed using MrBayes v3.2.6 based on the Bayesian inference (BI) method (Ronquist et al., 2012). Two independent runs, each with four chains (three heated and one cold), were conducted for a total of 2 million generations. Samples were taken every 1,000 generations, and the first 25% of these samples were discarded as burn-in. The remaining trees were used to calculate Bayesian posterior probabilities (BPPs) in a 50% majority-rule consensus tree.
The analysis of the codon usage of 12 core mitochondrial PCGs from 13 Ganoderma strains revealed that the average length of these genes ranged from 363 bp to 1983 bp, with nad3 having the shortest average length and nad5 having the longest. Out of these 12 core PCGs, 9 genes had varying sequence lengths among the different Ganoderma strains, while the atp6, cox3 and nad2 genes had the same gene length across all 13 strains. The nad6 gene showed the greatest length variation, with a maximum difference of 180 bp. Furthermore, these 12 core PCGs was found to be rich in T bases, with an average content of 39.77%, followed by A bases, with an average content of 33.14%. The G and C base contents were relatively low, with averages of 14.14 and 12.96%, respectively. The average GC content of core PCGs ranged from 20.07 to 33.84%, with rps3 having the lowest GC content and cox1 having the highest. These results demonstrate that different Ganoderma strains display considerable differences in base composition and gene length, even within the same species, such as G. lucidum and G. lingzhi.
The GC1, GC2 and GC3 contents of the 12 core PCGs in the 13 Ganoderma strains were 33.70, 33.55 and 13.65%, respectively (Figure 1). The average GC3s value of these 12 PCGs was 11.97%, indicating that the mitochondrial core PCGs of Ganoderma tend to terminate with an A or a T base. Furthermore, the indices of A3s, T3s, G3s, and C3s of the 12 core PCGs of the Ganoderma strains showed that the codons were more likely to end with A, followed by T, C and G, with values of 60.48, 47.00, 9.33, and 3.60%, respectively. An analysis of the codon bias of 12 core PCGs in the 13 Ganoderma strains was conducted. The CAI values of the core PCGs ranged from 0.099 to 0.182, with nad2 having the lowest value and cox2 having the highest. G. lingzhi s74 had the lowest CAI value, while G. calidophilum had the highest, demonstrating that they had a strong codon bias. The CBI values of the 13 Ganoderma strains ranged from −0.204 to −0.181, with G. sinense having the lowest value and G. tsugae having the highest. The core PCGs of the Ganoderma strains showed a range of CBI values from −0.297 to −0.107, with nad2 having the lowest and atp6 having the highest. The FOP values of the 12 core PCGs ranged from 0.213 to 0.327, with nad2 having the lowest and rps3 having the highest. G. sinense had the lowest FOP value, while G. tsugae had the largest. The GRAVY values of the 12 core PCGs were mostly positive, indicating that they were likely hydrophobic proteins, with the exception of rps3, which was considered hydrophilic. The AROMO values of the PCGs ranged from 0.110 to 0.168, with cob having the highest and rps3 having the lowest. The GRAVY values of the 13 Ganoderma strains ranged from 0.774 to 0.831, with an average of 0.820. The three G. lucidum strains showed considerable variations in the ENC, GC3s, GC, and AROMO indicators, while the three G. lingzhi strains showed variations in CBI, FOP, ENC, GC3s, GC, GRAVY, and AROMO values, indicating that the frequency of synonymous codon usage also varied within Ganoderma species.
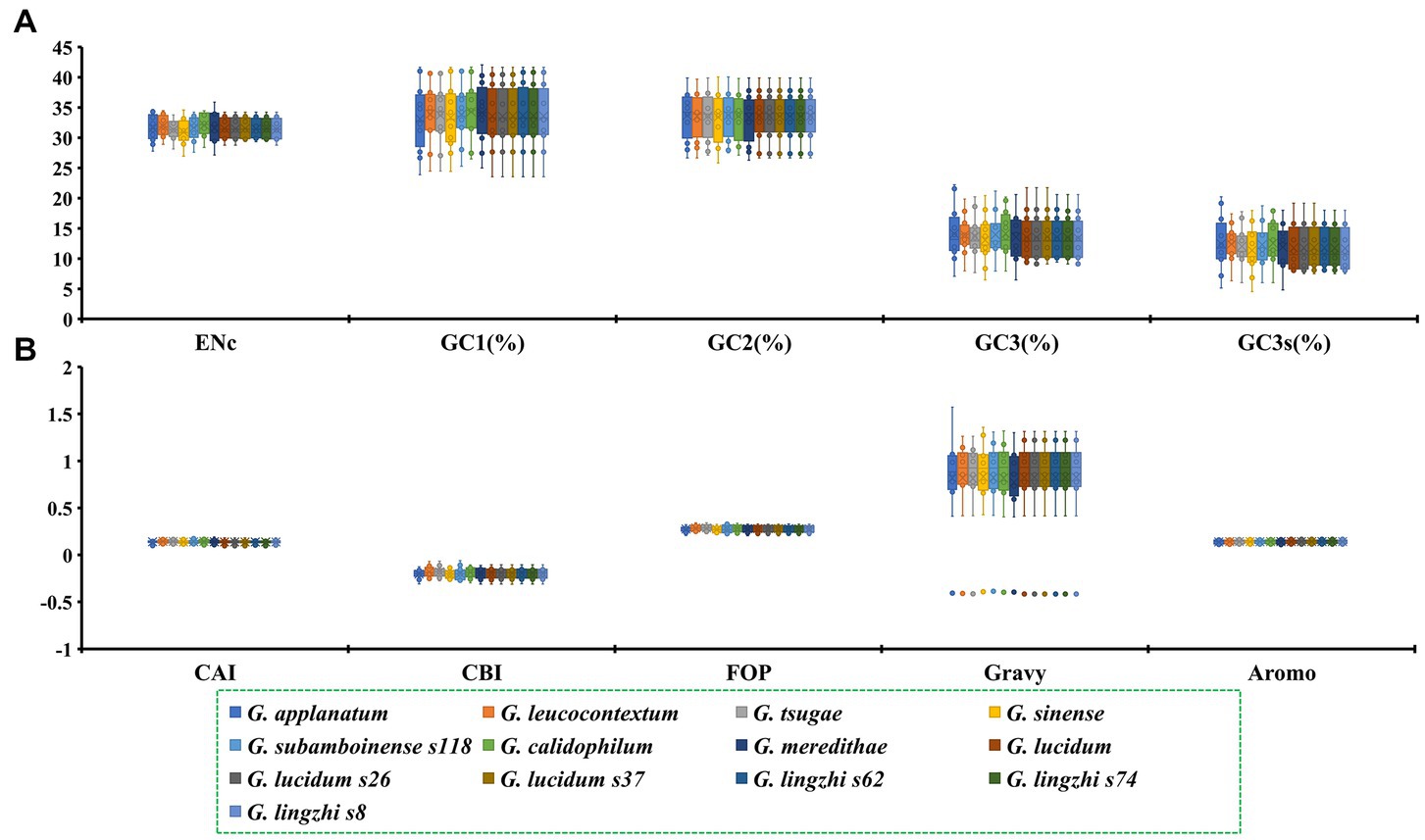
Figure 1. The codon usage indicators of twelve core protein-coding genes in 13 Ganoderma strains. (A) ENc, GC1, GC2, GC3, GC3s indicators. (B) CAI, CBI, FOP, Gravy, Aromo indicators.
A strong association was observed between the GC1 content of mitochondrial codons and GC3, GC3s, GC, and AROMO values in all 13 Ganoderma strains (p < 0.05; Figure 2). Additionally, a significant correlation was found between the GC2 content and GC content (p < 0.01). Moreover, the GC3 content was significantly correlated with GC3s and the GC content (p < 0.01), and it was also found to influence the ENC value in 11 out of the 13 Ganoderma strains, with the exceptions of G. lingzhi s62 and G. lingzhi s74. Additionally, the GC3s content was significantly associated with the ENC value in 7 Ganoderma strains (p < 0.05), with the exceptions of G. lucidum and G. lingzhi. Furthermore, the GC content was significantly correlated with the AROMO values in 12 out of the 13 Ganoderma strains (p < 0.05), with the exception of G. sinense. Moreover, the CAI of mitochondrial codons was significantly correlated with the CBI and FOP in all 13 Ganoderma strains (p < 0.05). A significant correlation was also observed between the CBI value and FOP value in all Ganoderma strains (p < 0.05).
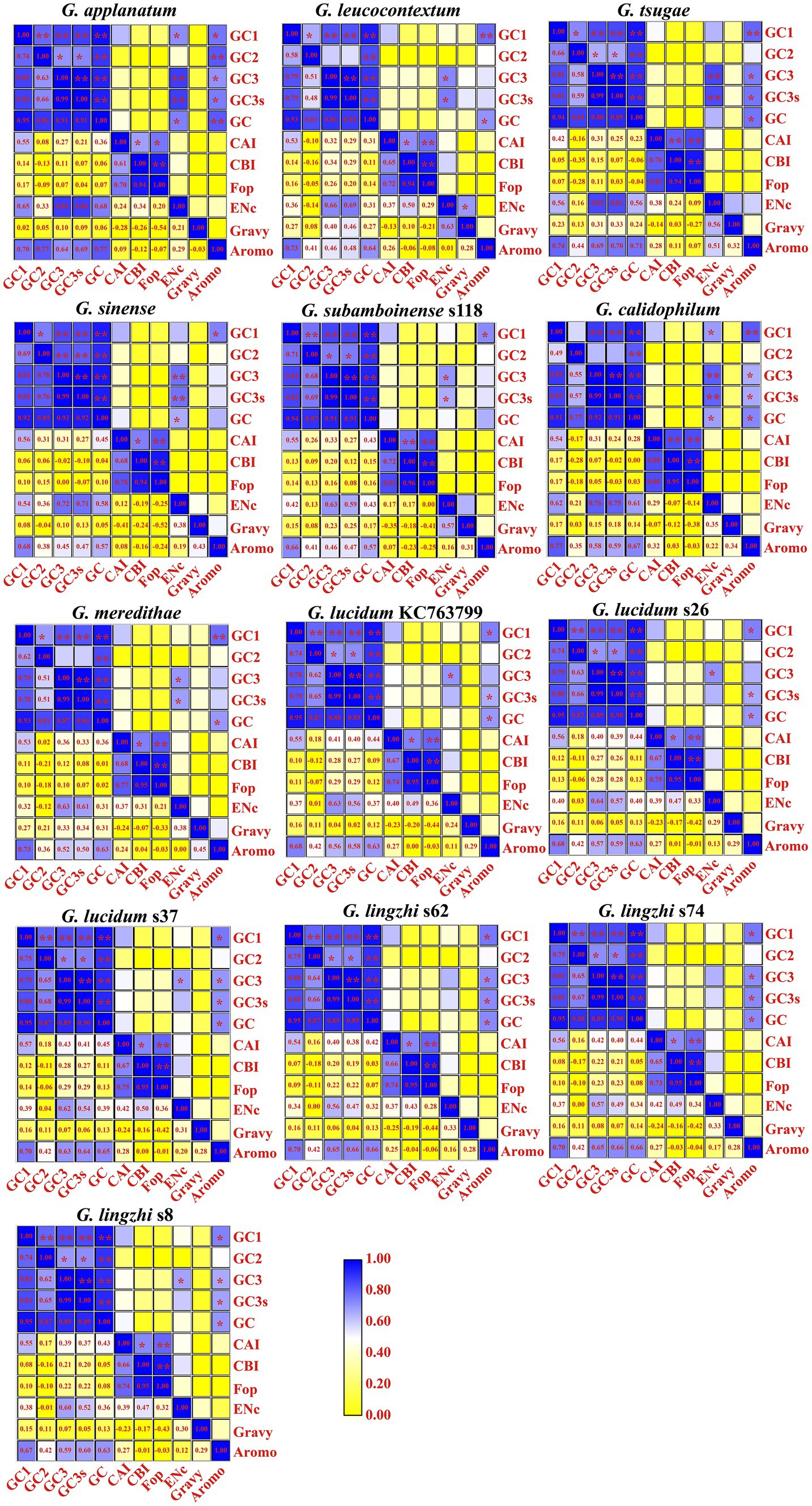
Figure 2. Pearson’s correlation analysis heatmap of codon usage indicators of 13 Ganoderma strains. The color of the blocks in the heatmap changes from yellow to blue, indicating an increase in the correlation coefficient. A single asterisk signifies a statistically significant correlation between the two (Continued)FIGURE 2 (Continued)indicators at the p < 0.05 level, while two asterisks symbolize a significant correlation between the two indicators at the p < 0.01 level. The 13 Ganoderma species are G. applanatum, G. leucocontextum, G. tsugae, G. sinense, G. subamboinense s118, G. calidophilum, G. meredithae, G. lucidum KC763799, G. lucidum s26, G. lucidum s37, G. lingzhi s62, G. lingzhi s74, and G. lingzhi s8, from left to right and from top to bottom.
Analysis of the neutrality plot between the GC12 and GC3 contents in Ganoderma mitochondrial codons revealed a weak positive correlation, with regression coefficients ranging from 0.79 to 0.97 and R2 values ranging from 0.5392 to 0.7496 (Figure 3). The GC12 content varied from 25.44 to 40.26%, and the GC3 content varied from 6.47 to 20.60%. Data points deviated from the trend line. Statistical analysis indicated that there was no significant correlation between the two values (p > 0.05), indicating that natural selection was the major factor influencing codon bias in Ganoderma.
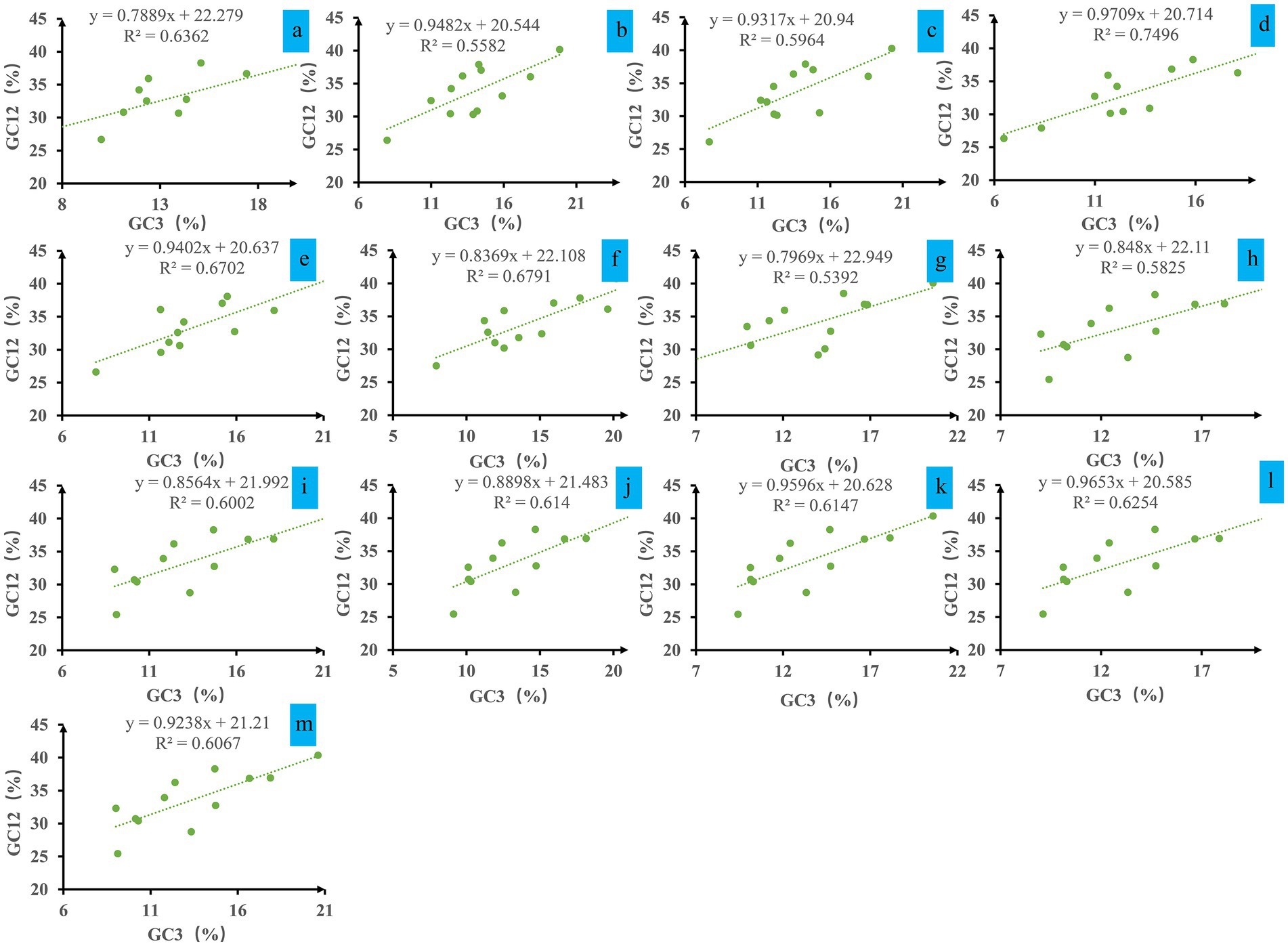
Figure 3. Neutrality plot analysis of GC12 and the third codon position (GC3) for the entire coding DNA sequence of 13 Ganoderma strains. (A) G. applanatum; (B) G. leucocontextum; (C) G. tsugae; (D) G. sinense; (E) G. subamboinense s118; (F) G. calidophilum; (G) G. meredithae; (H) G. lucidum KC763799; (I) G. lucidum s26; (J) G. lucidum s37; (K) G. lingzhi s62; (L) G. lingzhi s74; (M) G. lingzhi s8.
The average ENC value of the 12 core PCGs detected was 31.53, which is lower than 35, indicating a strong codon usage preference of proteins (Figure 1). The ENC values of the 13 Ganoderma strains ranged from 30.96 to 32.11, confirming the strong codon usage preference of Ganoderma species. The ENC plot showed that all Ganoderma genes were below the expected ENC-plot curve (Figure 4), suggesting that factors other than mutational pressure, such as natural selection, may be involved in codon bias formation.
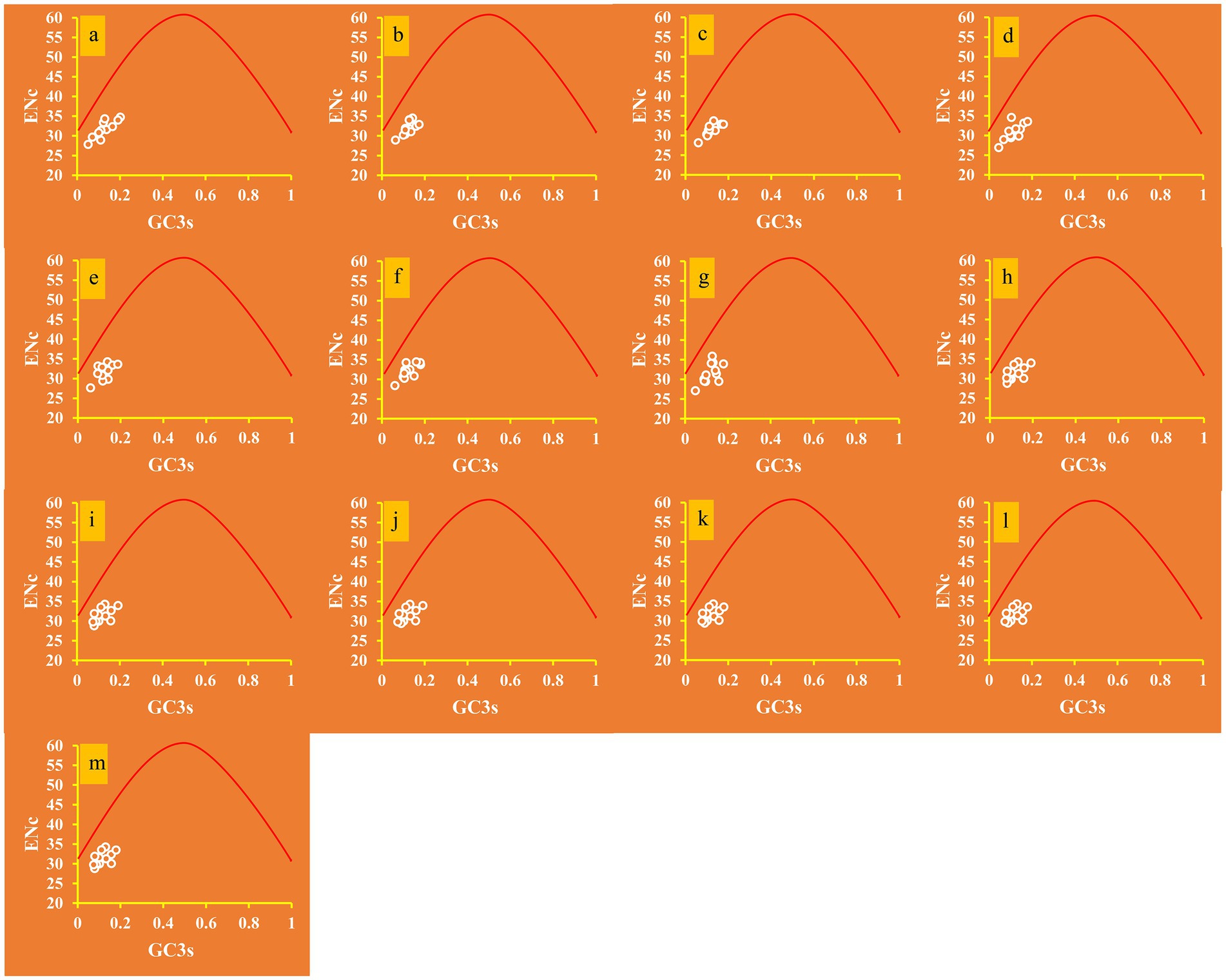
Figure 4. ENC-GC3 plot analysis of 12 core PCGs in 13 Ganoderma strains. The solid line represents the expected curve when codon usage bias is affected only by mutation pressure. (A) G. applanatum; (B) G. leucocontextum; (C) G. tsugae; (D) G. sinense; (E) G. subamboinense s118; (F) G. calidophilum; (G) G. meredithae; (H) G. lucidum KC763799; (I) G. lucidum s26; (J) G. lucidum s37; (K) G. lingzhi s62; (L) G. lingzhi s74; (M) G. lingzhi s8.
The ENCRatio values for all core PCGs were found to be greater than the expected values (Supplementary Figure S1), ranging from 18.22 to 19.50%. This suggests that GC3s play an essential role in the formation of codon bias. Consequently, it can be deduced that natural selection is a major factor in the formation of Ganoderma codon bias.
An analysis of Parity Rule 2 (PR2) plots was conducted to ascertain any biases in the mitochondrial genes of Ganoderma (Figure 5). The plot was centered on 0.5 and divided into four quadrants. The results indicated a strong preference for An over T and C over G at the third base of the mitochondrial codon of Ganoderma. Most of the dots were found in the fourth quadrant, followed by the third quadrant. With the exception of G. applanatum and G. subamboinense, the Ganoderma strains were not located in the first or second quadrant. This suggests a clear codon usage preference in Ganoderma species.
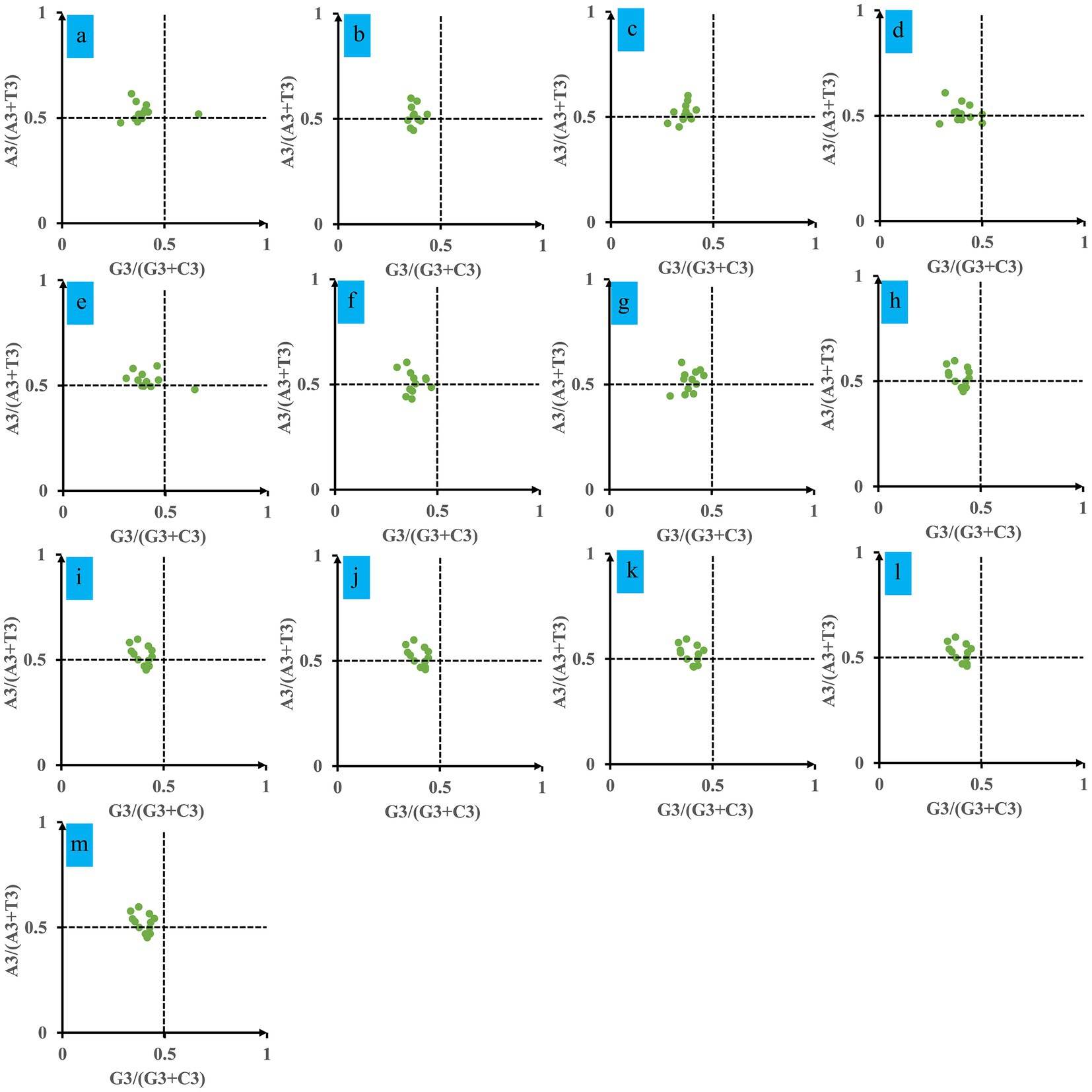
Figure 5. Parity Rule 2 (PR2) plot analysis of 12 core PCGs in 13 Ganoderma strains. (A) G. applanatum; (B) G. leucocontextum; (C) G. tsugae; (D) G. sinense; (E) G. subamboinense s118; (F) G. calidophilum; (G) G. meredithae; (H) G. lucidum KC763799; (I) G. lucidum s26; (J) G. lucidum s37; (K) G. lingzhi s62; (L) G. lingzhi s74; (M) G. lingzhi s8.
We conducted a correspondence analysis (COA) based on the RSCU values of mitochondrial genes from 13 Ganoderma strains to further analyze codon biases (Figure 6). Axis 1, Axis 2, Axis 3 and Axis 4 were the main contributors to variance, with average contribution rates of 29.15, 17.00, 10.82 and 9.11%, respectively. Axis 1 had the highest contribution to variance. Pearson correlation analysis revealed a significant correlation between Axis 1 and GC3s, CAI, CBI, and ENC values. Moreover, we observed considerable variation in the rps3 gene and other core PCGs, indicating the differentiation of synonymous codon usage of core PCGs.
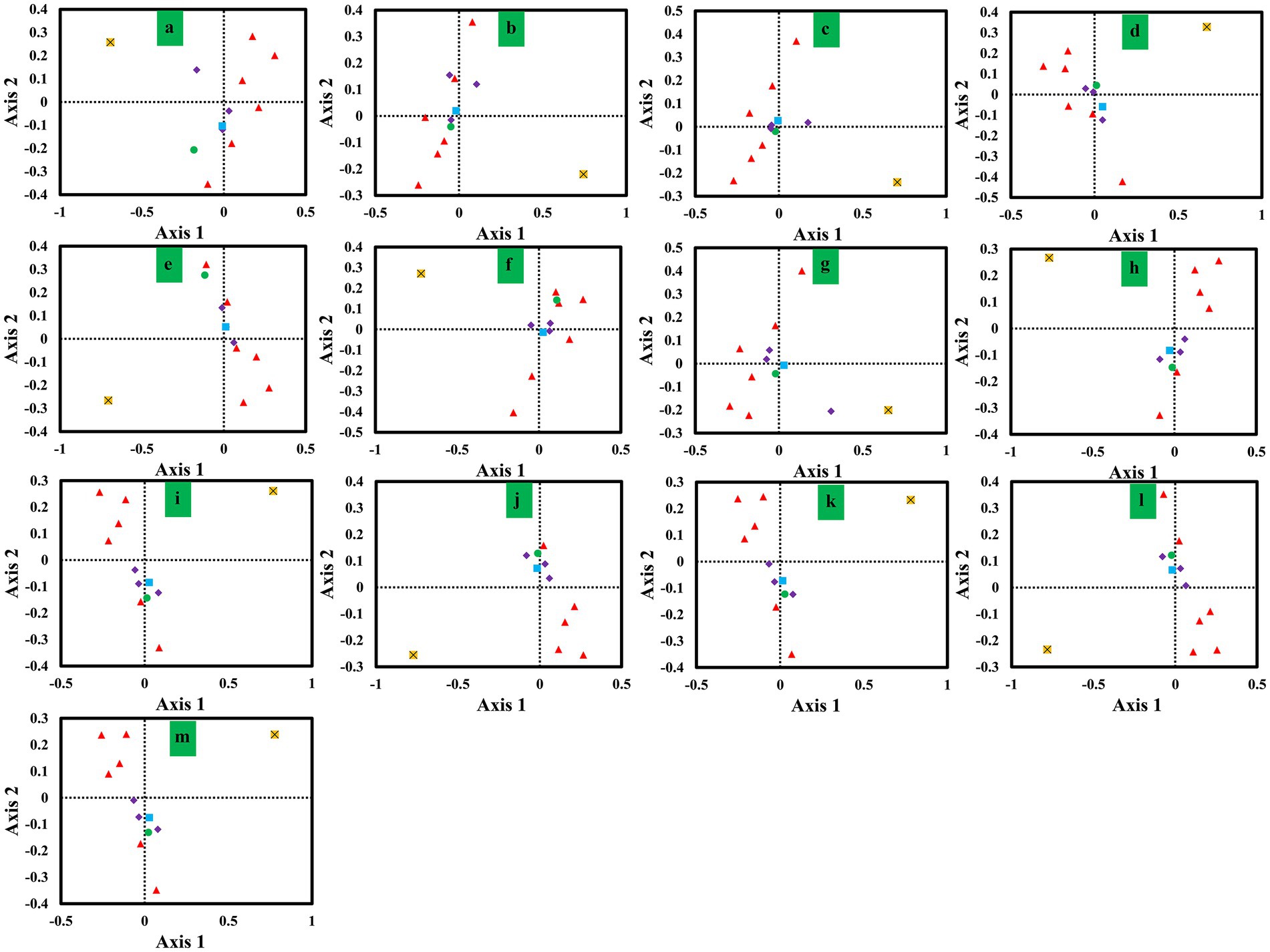
Figure 6. Correspondence analysis (COA) based on the relative synonymous codon usage (RSCU) values of 12 mitochondrial genes from 13 Ganoderma strains. Purple represents the cox gene, red represents the nad gene, green represents the atp6 gene, blue represents the cob gene, and yellow represents the rps3 gene. (A) G. applanatum; (B) G. leucocontextum; (C) G. tsugae; (D) G. sinense; (E) G. subamboinense s118; (F) G. calidophilum; (G) G. meredithae; (H) G. lucidum KC763799; (I) G. lucidum s26; (J) G. lucidum s37; (K) G. lingzhi s62; (L) G. lingzhi s74; (M) G. lingzhi s8.
Analysis of the relative synonymous codon usage (RSCU) of the 13 Ganoderma strains revealed 29 high-frequency codons shared among the species (Figure 7). Of these, 12 ended in C, 7 in A, 6 in G, and only 4 in T, indicating a preference for high-frequency codons ending in C. Furthermore, 27, 22, 21, 18, 22, 21, 21, 22, 22, 21, 21, 21, and 22 highly expressed codons (ΔRSCU>0.08) were identified in the Ganoderma strains (Figure 8) G. applanatum, G. leucocontextum, G. tsugae, G. sinense, G. subamboinense s118, G. calidophilum, G. meredithae, G. lucidum KC763799, G. lucidum s26, G. lucidum s37, G. lingzhi s62, G. lingzhi s74, and G. lingzhi s8, respectively. G. lucidum and G. lingzhi exhibited differences in the number and types of highly expressed codons within the respective species. Comparative analysis revealed 20, 16, 11, 14, 18, 14, 16, 22, 22, 21, 21, 21, and 22 optimal codons (ΔRSCU>0.08 and RSCU>1) in G. applanatum, G. leucocontextum, G. tsugae, G. sinense, G. subamboinense s118, G. calidophilum, G. meredithae, G. lucidum KC763799, G. lucidum s26, G. lucidum s37, G. lingzhi s62, G. lingzhi s74, and G. lingzhi s8, respectively. GCA, AUC, and UUC were the most frequently used codons of all 13 strains, followed by GAU, which was used as the optimal codon for 12 strains. GAG and GUC were both used as the optimal codons for the two species. Significant differences in the use of optimal codons were observed within or between G. lucidum and G. lingzhi species. CUA and GUA were chosen as the optimal codons for G. lucidum KC763799, G. lucidum s26, and G. lingzhi s8, while AGU was the optimal codon for G. lucidum s37, G. lingzhi s62, and G. lingzhi s74.
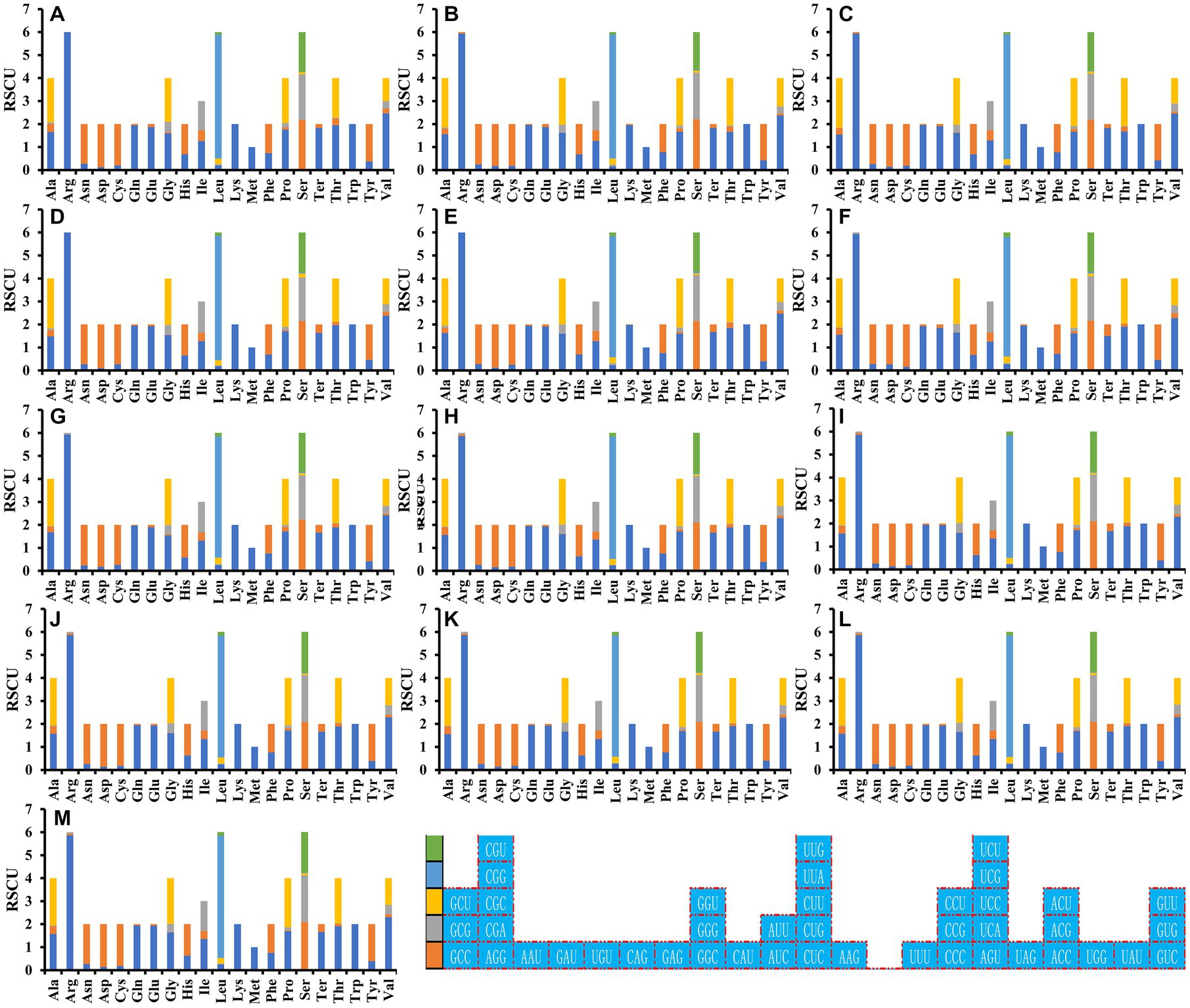
Figure 7. Relative synonymous codon usage (RSCU) analysis of 12 mitochondrial genes from 13 Ganoderma strains. (A) G. applanatum; (B) G. leucocontextum; (C) G. tsugae; (D) G. sinense; (E) G. subamboinense s118; (F) G. calidophilum; (G) G. meredithae; (H) G. lucidum KC763799; (I) G. lucidum s26; (J) G. lucidum s37; (K) G. lingzhi s62; (L) G. lingzhi s74; (M) G. lingzhi s8.

Figure 8. Optimal codon analysis of 13 Ganoderma strains (ΔRSCU>0.08 and RSCU>1) are marked in purple. Highly expressed codons (ΔRSCU>0.08) are marked in yellow, and high-frequency codons (RSCU>1) are marked in blue.
The Bayesian inference (BI) method was utilized to construct phylogenetic trees of 13 Ganoderma strains based on the combined mitochondrial gene set (Figure 9A). All the clades were well supported, with a BPP value of 1. The BI method identified G. leucocontextum as the sister species of G. tsugae and G. sinense as the sister species of G. subamboinense. Additionally, the six G. lucidum and G. lingzhi strains were grouped in the same evolutionary clade, demonstrating their close phylogenetic relationship. In contrast to the phylogenetic relationships inferred from sequences, those inferred from RSCU values had some discrepancies (Figure 9B), such as the phylogenetic status of G. calidophilum. Nevertheless, the RSCU-based tree also clearly revealed the close relationship between G. leucocontextum and G. tsugae, as well as between G. sinense and G. subamboinense. Furthermore, the relationships within G. lucidum and G. lingzhi were also clearly inferred using the RSCU method.
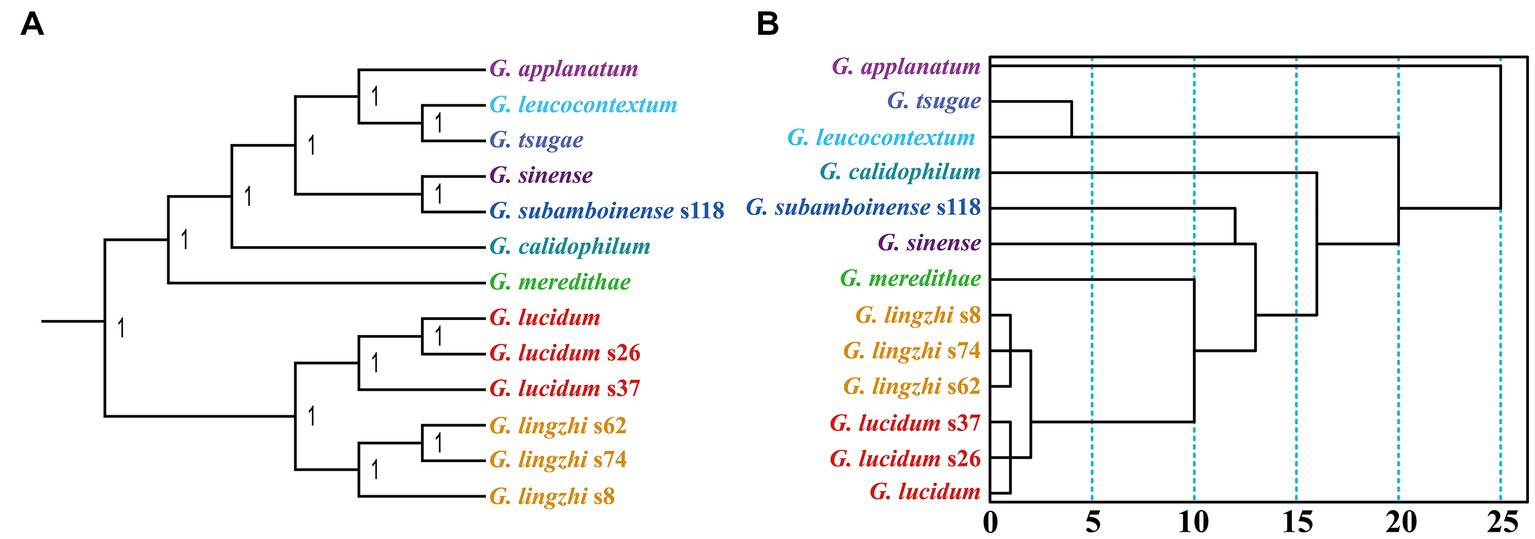
Figure 9. Relationship inference of 13 Ganoderma strains based on the Bayesian inference (BI) (A) method and relative synonymous codon usage (RSCU)-based hierarchical clustering (B).
Advances in sequencing technology have allowed scientists to obtain the genetic sequences of a range of species and genome types, such as the nuclear genome, chloroplast genome and mitochondrial genome (Xu et al., 2015; Barbhuiya et al., 2020; Chakraborty et al., 2020; Tu et al., 2022). Genetic information analysis has revealed that the usage of synonymous codons varies between different species, with some codons being utilized more often than others (Liu et al., 2020). Mutations and selection are the most influential factors in codon usage bias; however, other elements, such as gene base composition, gene length, gene expression level, tRNA abundance, amino acid hydrophobicity, and aromaticity, also have an effect (Arella et al., 2021; Begum and Chakraborty, 2022; Liu et al., 2022). Analyzing the codon bias of various species can provide an understanding of the genetic structure and evolutionary trend of the species (Xing et al., 2020; Xiao et al., 2022). The Ganoderma genus encompasses numerous species, some of which are employed in traditional Chinese medicine and others characterized as plant-infecting fungi (Kües et al., 2015; Zhang et al., 2019). In recent times, the cultivation of edible and medicinal Ganoderma fungi has significantly increased in both economic and medicinal value (Lin, 2019; Li Z. et al., 2019). The exogenous synthesis of the medicinal components of Ganoderma has also become a major research focus (Kladar et al., 2016; Gong et al., 2019; Angulo-Sanchez et al., 2022; Bao et al., 2022). Examining the codon bias of Ganoderma species could further our comprehension of the evolution, genetics, and biosynthesis of active ingredients of Ganoderma species. However, the codon usage of organelle genomes of higher fungi, especially Ganoderma species, has not been extensively researched.
Mitochondria are commonly known as the auxiliary genome of eukaryotes (Li et al., 2020c, 2023c). This study found considerable variation in the length and base composition of the mitochondrial core PCGs of various Ganoderma strains, even within the same species, suggesting the differentiation of Ganoderma mitochondrial genes. The majority of the differences in codon usage were observed at the third codon. It was also noted that the core PCGs of Ganoderma species typically terminate with A/T, which is consistent with the mitochondrial codon usage pattern observed in some other eukaryotes (Mazumder et al., 2021; Montana-Lozano et al., 2022). Additionally, there was a disparity in base usage between different species and genes. G. lucidum and G. lingzhi exhibited changes in various base bias indicators, such as CAI, CBI, FOP, ENC, and GC3s values, suggesting that the frequency of base synonymous codon usage varied within Ganoderma species. The results also showed a correlation between codon base composition and GC3s, CAI, CBI, and FOP values, suggesting that base composition has an impact on codon bias. A low ENC value (below 35) is indicative of a strong codon preference (Prabha et al., 2017; Pepe and Keersmaecker, 2020). The mitochondrial core PCGs of Ganoderma had an average ENC value of 31.53, which is lower than 35 and thus demonstrates a strong codon preference. The disparity between the expected and actual ENC values was considerable, with a range of 18.22 to 19.50%. The neutrality plot and PR2-bias plot analyzes also provided evidence that natural selection plays a role in the codon bias of Ganoderma, which is consistent with the results from the mitochondrial genomes of other species (Barbhuiya et al., 2021a,b). The findings of this study revealed that despite some discrepancies in codon usage indicators between or within Ganoderma species, all of the fungi experienced strong natural selection on their mitochondrial PCGs.
It is speculated that the ancestors of eukaryotes obtained mitochondria from bacteria (Lang et al., 1999), and most mitochondrial genes have been shifted to the nucleus (Adams and Palmer, 2003). Many eukaryotes still possess some core PCGs, tRNA genes and rRNA genes for energy production (Bullerwell et al., 2000; Costa et al., 2012), which can be employed as molecular indicators for phylogenetic analysis. The mitochondrial genome is seen as a beneficial asset for establishing the phylogenetic relationships between species (Li et al., 2015; Johri et al., 2019). A combined mitochondrial gene set was employed to analyze the genetic relationships between various Ganoderma species and strains, and the results demonstrated a high level of support for each evolutionary clade. The relative synonymous codon usage (RSCU) of a range of Ganoderma species and strains was evaluated to identify relationships between them, which diverged from those established from sequence-based data. The RSCU value also accurately reflected the genetic connections between or within some Ganoderma strains or species, which was consistent with the findings of previous studies (Crane et al., 2021; Gupta et al., 2021; Li et al., 2022d). In general, this research has improved our understanding of codon usage characteristics and genetic evolution of this higher fungal group and other related fungal species. Furthermore, it has been demonstrated that phylogenetic results based on RSCU values can be a useful supplement to those based on sequences.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number (s) can be found in the article/Supplementary material.
QL, PW, and MG: conceived and designed experiments. YL, AS, WX, ZX, JH, and XC: analyzed the data. PW and QL: wrote and reviewed the manuscript. MG: project management. All authors contributed to the article and approved the submitted version.
This study is supported by the Natural Science Foundation of Sichuan Province (no. 2023NSFSC1229) and Key R&D Project of Sichuan Provincial Department of Science and Technology (2023YFN0062).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1170790/full#supplementary-material
Adams, K. L., and Palmer, J. D. (2003). Evolution of mitochondrial gene content: gene loss and transfer to the nucleus. Mol. Phylogenet. Evol. 29, 380–395. doi: 10.1016/S1055-7903(03)00194-5
Akashi, H. (1997). Codon bias evolution in Drosophila. Population genetics of mutation-selection drift. Gene 205, 269–278. doi: 10.1016/S0378-1119(97)00400-9
Angulo-Sanchez, L. T., López-Peña, D., Torres-Moreno, H., Gutiérrez, A., Gaitán-Hernández, R., and Esqueda, M. (2022). Biosynthesis, gene expression, and pharmacological properties of triterpenoids of Ganoderma species (Agaricomycetes): a review. Int. J. Med. Mushrooms 24, 1–17. doi: 10.1615/IntJMedMushrooms.2022044016
Arella, D., Dilucca, M., and Giansanti, A. (2021). Codon usage bias and environmental adaptation in microbial organisms. Mol. Gen. Genomics. 296, 751–762. doi: 10.1007/s00438-021-01771-4
Bailey, S. F., Alonso Morales, L. A., and Kassen, R. (2021). Effects of synonymous mutations beyond codon Bias: the evidence for adaptive synonymous substitutions from microbial evolution experiments. Genome Biol. Evol. 13:evab141. doi: 10.1093/gbe/evab141
Bao, Z. J., Feng, H. Y., Tu, W. Y., Li, L. J., and Li, Q. (2022). Method and mechanism of chromium removal from soil: a systematic review. Environ. Sci. Pollut. Res. 29, 35501–35517. doi: 10.1007/s11356-022-19452-z
Bao, Z. J., Wang, X. M., Wang, Q. F., Zou, L., Peng, L. X., Li, L. J., et al. (2023). A novel method of domestication combined with ARTP to improve the reduction ability of Bacillus velezensis to Cr(VI). J. Environ. Chem. Eng. 11:109091. doi: 10.1016/j.jece.2022.109091
Barbhuiya, P. A., Uddin, A., and Chakraborty, S. (2020). Analysis of compositional properties and codon usage bias of mitochondrial CYB gene in anura, urodela and gymnophiona. Gene 751:144762. doi: 10.1016/j.gene.2020.144762
Barbhuiya, P. A., Uddin, A., and Chakraborty, S. (2021a). Codon usage pattern and evolutionary forces of mitochondrial ND genes among orders of class Amphibia. J. Cell. Physiol. 236, 2850–2868. doi: 10.1002/jcp.30050
Barbhuiya, P. A., Uddin, A., and Chakraborty, S. (2021b). Understanding the codon usage patterns of mitochondrial CO genes among amphibians. Gene 777:145462. doi: 10.1016/j.gene.2021.145462
Begum, N. S., and Chakraborty, S. (2022). Influencing elements of codon usage bias in Birnaviridae and its evolutionary analysis. Virus Res. 310:198672. doi: 10.1016/j.virusres.2021.198672
Behura, S. K., and Severson, D. W. (2013). Codon usage bias: causative factors, quantification methods and genome-wide patterns: with emphasis on insect genomes. Biol. Rev. Camb. Philos. Soc. 88, 49–61. doi: 10.1111/j.1469-185X.2012.00242.x
Biro, J. C. (2008). Does codon bias have an evolutionary origin? Theor. Biol. Med. Model. 5:16. doi: 10.1186/1742-4682-5-16
Bishop, K. S., Kao, C. H., Xu, Y., Glucina, M. P., Paterson, R. R., and Ferguson, L. R. (2015). From 2000years of Ganoderma lucidum to recent developments in nutraceuticals. Phytochemistry 114, 56–65. doi: 10.1016/j.phytochem.2015.02.015
Bu, Y., Wu, X., Sun, N., Man, Y., and Jing, Y. (2021). Codon usage bias predicts the functional MYB10 gene in Populus. J. Plant Physiol. 265:153491. doi: 10.1016/j.jplph.2021.153491
Bullerwell, C. E., Burger, G., and Lang, B. F. (2000). A novel motif for identifying rps3 homologs in fungal mitochondrial genomes. Trends Biochem. Sci. 25, 363–365. doi: 10.1016/S0968-0004(00)01612-1
Cai, N., Gomez-Duran, A., Yonova-Doing, E., Kundu, K., Burgess, A. I., Golder, Z. J., et al. (2021). Mitochondrial DNA variants modulate N-formylmethionine, proteostasis and risk of late-onset human diseases. Nat. Med. 27, 1564–1575. doi: 10.1038/s41591-021-01441-3
Camiolo, S., Toome-Heller, M., Aime, M. C., Haridas, S., Grigoriev, I. V., Porceddu, A., et al. (2019). An analysis of codon bias in six red yeast species. Yeast 36, 53–64. doi: 10.1002/yea.3359
Chakraborty, S., Yengkhom, S., and Uddin, A. (2020). Analysis of codon usage bias of chloroplast genes in Oryza species: codon usage of chloroplast genes in Oryza species. Planta 252:67. doi: 10.1007/s00425-020-03470-7
Chan, J. S., Asatiani, M. D., Sharvit, L. E., Trabelcy, B., Barseghyan, G. S., and Wasser, S. P. (2015). Chemical composition and medicinal value of the new Ganoderma tsugae var. jannieae CBS-120304 medicinal higher basidiomycete mushroom. Int. J. Med. Mushrooms 17, 735–747. doi: 10.1615/IntJMedMushrooms.v17.i8.40
Chaney, J. L., and Clark, P. L. (2015). Roles for synonymous codon usage in protein biogenesis. Annu. Rev. Biophys. 44, 143–166. doi: 10.1146/annurev-biophys-060414-034333
Chen, K., Lu, P., Beeraka, N. M., Sukocheva, O. A., Madhunapantula, S. V., Liu, J., et al. (2020). Mitochondrial mutations and mitoepigenetics: focus on regulation of oxidative stress-induced responses in breast cancers. Semin. Cancer Biol. 83, 556–569. doi: 10.1016/j.semcancer.2020.09.012
Chien, R. C., Tsai, S. Y., Lai, E. Y., and Mau, J. L. (2015). Antiproliferative activities of hot water extracts from culinary-medicinal mushrooms, Ganoderma tsugae and Agrocybe cylindracea (higher Basidiomycetes) on Cancer cells. Int. J. Med. Mushrooms 17, 453–462. doi: 10.1615/IntJMedMushrooms.v17.i5.50
Choudhury, M. N., Uddin, A., and Chakraborty, S. (2018). Nucleotide composition and codon usage bias of SRY gene. Andrologia 50:e12787. doi: 10.1111/and.12787
Cope, A. L., and Shah, P. (2022). Intragenomic variation in non-adaptive nucleotide biases causes underestimation of selection on synonymous codon usage. PLoS Genet. 18:e1010256. doi: 10.1371/journal.pgen.1010256
Costa, G. G., Cabrera, O. G., Tiburcio, R. A., Medrano, F. J., Carazzolle, M. F., Thomazella, D. P., et al. (2012). The mitochondrial genome of Moniliophthora roreri, the frosty pod rot pathogen of cacao. Fungal Biol. 116, 551–562. doi: 10.1016/j.funbio.2012.01.008
Crane, A., Versoza, C. J., Hua, T., Kapoor, R., Lloyd, L., Mehta, R., et al. (2021). Phylogenetic relationships and codon usage bias amongst cluster K mycobacteriophages. G3 (Bethesda) 11:jkab291. doi: 10.1093/g3journal/jkab291
De Mandal, S., Mazumder, T. H., Panda, A. K., Kumar, N. S., and Jin, F. (2020). Analysis of synonymous codon usage patterns of HPRT1 gene across twelve mammalian species. Genomics 112, 304–311. doi: 10.1016/j.ygeno.2019.02.010
De Oliveira, J. L., Morales, A. C., Hurst, L. D., Urrutia, A. O., Thompson, C. R. L., and Wolf, J. B. (2021). Inferring adaptive codon preference to understand sources of selection shaping codon usage Bias. Mol. Biol. Evol. 38, 3247–3266. doi: 10.1093/molbev/msab099
Deng, Y., De Lima Hedayioglu, F., Kalfon, J., Chu, D., and Von Der Haar, T. (2020). Hidden patterns of codon usage bias across kingdoms. J. R. Soc. Interface 17:20190819. doi: 10.1098/rsif.2019.0819
Dhillon, B., Hamelin, R. C., and Rollins, J. A. (2021). Transcriptional profile of oil palm pathogen, Ganoderma boninense, reveals activation of lignin degradation machinery and possible evasion of host immune response. BMC Genomics 22:326. doi: 10.1186/s12864-021-07644-9
Dhindsa, R. S., Copeland, B. R., Mustoe, A. M., and Goldstein, D. B. (2020). Natural selection shapes codon usage in the human genome. Am. J. Hum. Genet. 107, 83–95. doi: 10.1016/j.ajhg.2020.05.011
Fages-Lartaud, M., Hundvin, K., and Hohmann-Marriott, M. F. (2022). Mechanisms governing codon usage bias and the implications for protein expression in the chloroplast of Chlamydomonas reinhardtii. Plant J. 112, 919–945. doi: 10.1111/tpj.15970
Francis, A., Beadle, C., Puspitasari, D., Irianto, R., Agustini, L., Rimbawanto, A., et al. (2014). Disease progression in plantations of Acacia mangium affected by red root rot (Ganoderma philippii). For. Pathol. 44, 447–459. doi: 10.1111/efp.12141
Gao, X., Qi, J. Y., Ho, C. T., Li, B., Mu, J. J., Zhang, Y. T., et al. (2020). Structural characterization and immunomodulatory activity of a water-soluble polysaccharide from Ganoderma leucocontextum fruiting bodies. Carbohydr. Polym. 249:116874. doi: 10.1016/j.carbpol.2020.116874
Gong, T., Yan, R., Kang, J., and Chen, R. (2019). Chemical components of Ganoderma. Adv. Exp. Med. Biol. 1181, 59–106. doi: 10.1007/978-981-13-9867-4_3
Gupta, S., Paul, K., and Roy, A. (2021). Codon usage signatures in the genus Cryptococcus: a complex interplay of gene expression, translational selection and compositional bias. Genomics 113, 821–830. doi: 10.1016/j.ygeno.2020.10.013
Hayashi, J. I., Hashizume, O., Ishikawa, K., and Shimizu, A. (2016). Mutations in mitochondrial DNA regulate mitochondrial diseases and metastasis but do not regulate aging. Curr. Opin. Genet. Dev. 38, 63–67. doi: 10.1016/j.gde.2016.03.004
Hia, F., and Takeuchi, O. (2021). The effects of codon bias and optimality on mRNA and protein regulation. Cell. Mol. Life Sci. 78, 1909–1928. doi: 10.1007/s00018-020-03685-7
Hia, F., Yang, S. F., Shichino, Y., Yoshinaga, M., Murakawa, Y., Vandenbon, A., et al. (2019). Codon bias confers stability to human mRNAs. EMBO Rep. 20:e48220. doi: 10.15252/embr.201948220
Hidayati, N., Glen, M., Nurrohmah, S. H., Rimbawanto, A., and Mohammed, C. L. (2014). Ganoderma steyaertanum as a root-rot pathogen of forest trees. For. Pathol. 44, 460–471. doi: 10.1111/efp.12142
Huo, X., Liu, S., Li, Y., Wei, H., Gao, J., Yan, Y., et al. (2021). Analysis of synonymous codon usage of transcriptome database in Rheum palmatum. PeerJ 9:e10450. doi: 10.7717/peerj.10450
Iriarte, A., Lamolle, G., and Musto, H. (2021). Codon usage Bias: an endless tale. J. Mol. Evol. 89, 589–593. doi: 10.1007/s00239-021-10027-z
Jargalmaa, S., Eimes, J. A., Park, M. S., Park, J. Y., Oh, S. Y., and Lim, Y. W. (2017). Taxonomic evaluation of selected Ganoderma species and database sequence validation. PeerJ 5:e3596. doi: 10.7717/peerj.3596
Jiang, W., Lv, B., Wu, X., Wang, J., Wu, G., Shi, C., et al. (2017). Analysis of synonymous codon usage patterns in the edible fungus Volvariella volvacea. Biotechnol. Appl. Biochem. 64, 218–224. doi: 10.1002/bab.1538
Johri, P., Marinov, G. K., Doak, T. G., and Lynch, M. (2019). Population genetics of Paramecium mitochondrial genomes: recombination, mutation spectrum, and efficacy of selection. Genome Biol. Evol. 11, 1398–1416. doi: 10.1093/gbe/evz081
Katoh, K., Rozewicki, J., and Yamada, K. D. (2017). MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 20, 1160–1166. doi: 10.1093/bib/bbx108
Kladar, N. V., Gavarić, N. S., and Božin, B. N. (2016). Ganoderma: insights into anticancer effects. Eur. J. Cancer Prev. 25, 462–471. doi: 10.1097/CEJ.0000000000000204
Krasovec, M., and Filatov, D. A. (2019). Evolution of codon usage Bias in diatoms. Genes (Basel) 10:894. doi: 10.3390/genes10110894
Kües, U., Nelson, D. R., Liu, C., Yu, G. J., Zhang, J., Li, J., et al. (2015). Genome analysis of medicinal Ganoderma spp. with plant-pathogenic and saprotrophic life-styles. Phytochemistry 114, 18–37. doi: 10.1016/j.phytochem.2014.11.019
Kwon, O. C., Park, Y. J., Kim, H. I., Kong, W. S., Cho, J. H., and Lee, C. S. (2016). Taxonomic position and species identity of the cultivated Yeongji “Ganoderma lucidum” in Korea. Mycobiology 44, 1–6. doi: 10.5941/MYCO.2016.44.1.1
Labella, A. L., Opulente, D. A., Steenwyk, J. L., Hittinger, C. T., and Rokas, A. (2019). Variation and selection on codon usage bias across an entire subphylum. PLoS Genet. 15:e1008304. doi: 10.1371/journal.pgen.1008304
Lal, D., Verma, M., Behura, S. K., and Lal, R. (2016). Codon usage bias in phylum Actinobacteria: relevance to environmental adaptation and host pathogenicity. Res. Microbiol. 167, 669–677. doi: 10.1016/j.resmic.2016.06.003
Lamolle, G., Iriarte, A., and Musto, H. (2022). Codon usage in the flatworm Schistosoma mansoni is shaped by the mutational bias towards a+T and translational selection, which increases GC-ending codons in highly expressed genes. Mol. Biochem. Parasitol. 247:111445. doi: 10.1016/j.molbiopara.2021.111445
Lanfear, R., Frandsen, P. B., Wright, A. M., Senfeld, T., and Calcott, B. (2017). PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 34, 772–773. doi: 10.1093/molbev/msw260
Lang, B. F., Gray, M. W., and Burger, G. (1999). Mitochondrial genome evolution and the origin of eukaryotes. Annu. Rev. Genet. 33, 351–397. doi: 10.1146/annurev.genet.33.1.351
Latorre-Pellicer, A., Moreno-Loshuertos, R., Lechuga-Vieco, A. V., Sanchez-Cabo, F., Torroja, C., Acin-Perez, R., et al. (2016). Mitochondrial and nuclear DNA matching shapes metabolism and healthy ageing. Nature 535, 561–565. doi: 10.1038/nature18618
Li, Q., Bao, Z., Tang, K., Feng, H., Tu, W., Li, L., et al. (2022a). First two mitochondrial genomes for the order Filobasidiales reveal novel gene rearrangements and intron dynamics of Tremellomycetes. IMA Fungus 13:7. doi: 10.1186/s43008-022-00094-2
Li, Q., He, X., Ren, Y., Xiong, C., Jin, X., Peng, L., et al. (2020a). Comparative Mitogenome analysis reveals mitochondrial genome differentiation in ectomycorrhizal and asymbiotic Amanita species. Front. Microbiol. 11:1382. doi: 10.3389/fmicb.2020.01382
Li, Z., Hu, W., Cao, X., Liu, P., Shang, Y., and Zhou, J. (2018). Synonymous codon usages as an evolutionary dynamic for Chlamydiaceae. Int. J. Mol. Sci. 19:4010. doi: 10.3390/ijms19124010
Li, Q., Li, L., Zhang, T., Xiang, P., Wu, Q., Tu, W., et al. (2022b). The first two mitochondrial genomes for the genus Ramaria reveal mitochondrial genome evolution of Ramaria and phylogeny of Basidiomycota. IMA Fungus 13:16. doi: 10.1186/s43008-022-00100-7
Li, Q., Luo, Y., Sha, A., Xiao, W., Xiong, Z., Chen, X., et al. (2023a). Analysis of synonymous codon usage patterns in mitochondrial genomes of nine Amanita species. Front. Microbiol. 14:1134228. doi: 10.3389/fmicb.2023.1134228
Li, Q., Ren, Y., Shi, X., Peng, L., Zhao, J., Song, Y., et al. (2019a). Comparative mitochondrial genome analysis of two ectomycorrhizal Fungi (Rhizopogon) reveals dynamic changes of intron and phylogenetic relationships of the subphylum Agaricomycotina. Int. J. Mol. Sci. 20:5167. doi: 10.3390/ijms20205167
Li, Q., Ren, Y., Xiang, D., Shi, X., Zhao, J., Peng, L., et al. (2020b). Comparative mitogenome analysis of two ectomycorrhizal fungi (Paxillus) reveals gene rearrangement, intron dynamics, and phylogeny of basidiomycetes. IMA Fungus 11:12. doi: 10.1186/s43008-020-00038-8
Li, H., Shao, R., Song, N., Song, F., Jiang, P., Li, Z., et al. (2015). Higher-level phylogeny of paraneopteran insects inferred from mitochondrial genome sequences. Sci. Rep. 5:8527. doi: 10.1038/srep08527
Li, B., Wu, H., Miao, Z., Hu, L., Zhou, L., and Lu, Y. (2022). Codon usage of hepatitis E viruses: a comprehensive analysis. Front. Microbiol. 13:938651. doi: 10.3389/fmicb.2022.938651
Li, Q., Xiang, P., Li, L., Zhang, T., Wu, Q., Bao, Z., et al. (2023b). Phosphorus mining activities alter endophytic bacterial communities and metabolic functions of surrounding vegetables and crops. Plant Soil 2023:5961. doi: 10.1007/s11104-023-05961-4
Li, Q., Xiang, D., Wan, Y., Wu, Q., Wu, X., Ma, C., et al. (2019b). The complete mitochondrial genomes of five important medicinal Ganoderma species: features, evolution, and phylogeny. Int. J. Biol. Macromol. 139, 397–408. doi: 10.1016/j.ijbiomac.2019.08.003
Li, Q., Xiang, P., Zhang, T., Wu, Q., Bao, Z. J., Tu, W. Y., et al. (2022c). The effect of phosphate mining activities on rhizosphere bacterial communities of surrounding vegetables and crops. Sci. Total Environ. 821:153479. doi: 10.1016/j.scitotenv.2022.153479
Li, Q., Xiao, W., Wu, P., Zhang, T., Xiang, P., Wu, Q., et al. (2023c). The first two mitochondrial genomes from Apiotrichum reveal mitochondrial evolution and different taxonomic assignment of Trichosporonales. IMA Fungus 14:7. doi: 10.1186/s43008-023-00112-x
Li, Q., Yang, L., Xiang, D., Wan, Y., Wu, Q., Huang, W., et al. (2020c). The complete mitochondrial genomes of two model ectomycorrhizal fungi (Laccaria): features, intron dynamics and phylogenetic implications. Int. J. Biol. Macromol. 145, 974–984. doi: 10.1016/j.ijbiomac.2019.09.188
Li, J., Zhang, J., Chen, H., Chen, X., Lan, J., and Liu, C. (2013). Complete mitochondrial genome of the medicinal mushroom Ganoderma lucidum. PLoS One 8:e72038. doi: 10.1371/journal.pone.0072038
Li, Q., Zhang, T., Li, L., Bao, Z., Tu, W., Xiang, P., et al. (2022d). Comparative Mitogenomic analysis reveals intraspecific, interspecific variations and genetic diversity of medical fungus Ganoderma. J. Fungi (Basel) 8:781. doi: 10.3390/jof8080781
Li, Z., Zhou, J., and Lin, Z. (2019). Development and innovation of Ganoderma industry and products in China. Adv. Exp. Med. Biol. 1181, 187–204. doi: 10.1007/978-981-13-9867-4_7
Lin, Z. (2019). Ganoderma (lingzhi) in traditional Chinese medicine and Chinese culture. Adv. Exp. Med. Biol. 1181, 1–13. doi: 10.1007/978-981-13-9867-4_1
Liu, H., Lu, Y., Lan, B., and Xu, J. (2020). Codon usage by chloroplast gene is bias in Hemiptelea davidii. J. Genet. 99:8. doi: 10.1007/s12041-019-1167-1
Liu, K., Ouyang, Y., Lin, R., Ge, C., and Zhou, M. (2022). Strong negative correlation between codon usage bias and protein structural disorder impedes protein expression after codon optimization. J. Biotechnol. 343, 15–24. doi: 10.1016/j.jbiotec.2021.11.001
Liu, Y., Yang, Q., and Zhao, F. (2021). Synonymous but not silent: the codon usage code for gene expression and protein folding. Annu. Rev. Biochem. 90, 375–401. doi: 10.1146/annurev-biochem-071320-112701
López, J. L., Lozano, M. J., Fabre, M. L., and Lagares, A. (2020). Codon usage optimization in the prokaryotic tree of life: how synonymous codons are differentially selected in sequence domains with different expression levels and degrees of conservation. MBio 11:e00766. doi: 10.1128/mBio.00766-20
Masłowska-Górnicz, A., Van Den Bosch, M. R. M., Saccenti, E., and Suarez-Diez, M. (2022). A large-scale analysis of codon usage bias in 4868 bacterial genomes shows association of codon adaptation index with GC content, protein functional domains and bacterial phenotypes. Biochim. Biophys. Acta Gene Regul. Mech. 1865:194826. doi: 10.1016/j.bbagrm.2022.194826
Mazumder, G. A., Uddin, A., and Chakraborty, S. (2021). Analysis of codon usage bias in mitochondrial CO gene among platyhelminthes. Mol. Biochem. Parasitol. 245:111410. doi: 10.1016/j.molbiopara.2021.111410
Meng, G., Ye, L., Ma, S., Wu, L., Deng, Y., and Wu, X. (2022). Intraspecific comparison of mitochondrial genomes reveals the evolution in medicinal fungus Ganoderma lingzhi. J. Biosci. Bioeng. 134, 374–383. doi: 10.1016/j.jbiosc.2022.07.003
Montana-Lozano, P., Balaguera-Reina, S. A., and Prada-Quiroga, C. F. (2022). Comparative analysis of codon usage of mitochondrial genomes provides evolutionary insights into reptiles. Gene 851:146999. doi: 10.1016/j.gene.2022.146999
Oldfield, C. J., Peng, Z., Uversky, V. N., and Kurgan, L. (2020). Codon selection reduces GC content bias in nucleic acids encoding for intrinsically disordered proteins. Cell. Mol. Life Sci. 77, 149–160. doi: 10.1007/s00018-019-03166-6
Page, D. E., Glen, M., Puspitasari, D., Rimbawanto, A., Ratkowsky, D., and Mohammed, C. (2018). Sexuality and mating types of Ganoderma philippii, Ganoderma mastoporum and Ganoderma australe, three basidiomycete fungi with contrasting ecological roles in south-east Asian pulpwood plantations. Australas. Plant Pathol. 47, 83–94. doi: 10.1007/s13313-017-0531-y
Palidwor, G. A., Perkins, T. J., and Xia, X. (2010). A general model of codon bias due to GC mutational bias. PLoS One 5:e13431. doi: 10.1371/journal.pone.0013431
Parvathy, S. T., Udayasuriyan, V., and Bhadana, V. (2022). Codon usage bias. Mol. Biol. Rep. 49, 539–565. doi: 10.1007/s11033-021-06749-4
Paterson, R. R. (2006). Ganoderma–a therapeutic fungal biofactory. Phytochemistry 67, 1985–2001. doi: 10.1016/j.phytochem.2006.07.004
Pepe, D., and Keersmaecker, K. D. E. (2020). Codon bias analyses on thyroid carcinoma genes. Minerva Endocrinol. 45, 295–305. doi: 10.23736/S0391-1977.20.03252-6
Prabha, R., Singh, D. P., Sinha, S., Ahmad, K., and Rai, A. (2017). Genome-wide comparative analysis of codon usage bias and codon context patterns among cyanobacterial genomes. Mar. Genomics 32, 31–39. doi: 10.1016/j.margen.2016.10.001
Puigbo, P., Bravo, I. G., and Garcia-Vallve, S. (2008). CAIcal: a combined set of tools to assess codon usage adaptation. Biol. Direct 3:38. doi: 10.1186/1745-6150-3-38
Rahman, S. U., Abdullah, M., Khan, A. W., Haq, M. I. U., Haq, N. U., Aziz, A., et al. (2022). A detailed comparative analysis of codon usage bias in Alongshan virus. Virus Res. 308:198646. doi: 10.1016/j.virusres.2021.198646
Rao, Y., Wu, G., Wang, Z., Chai, X., Nie, Q., and Zhang, X. (2011). Mutation bias is the driving force of codon usage in the Gallus gallus genome. DNA Res. 18, 499–512. doi: 10.1093/dnares/dsr035
Ronquist, F., Teslenko, M., Van Der Mark, P., Ayres, D. L., Darling, A., Hohna, S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Sharp, P. M., and Li, W. H. (1987). The codon adaptation index--a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 15, 1281–1295. doi: 10.1093/nar/15.3.1281
Si, J., Wu, Y., Ma, H. F., Cao, Y. J., Sun, Y. F., and Cui, B. K. (2021). Selection of a pH- and temperature-stable laccase from Ganoderma australe and its application for bioremediation of textile dyes. J. Environ. Manag. 299:113619. doi: 10.1016/j.jenvman.2021.113619
Sun, Y. F., Fang, Y. X., and Cui, B. K. (2022a). Taxonomy and phylogeny of Sanguinoderma rugosum complex with descriptions of a new species and a new combination. Front. Microbiol. 13:1087212. doi: 10.3389/fmicb.2022.1087212
Sun, Y. F., Lebreton, A., Xing, J. H., Fang, Y. X., Si, J., Morin, E., et al. (2022b). Phylogenomics and comparative genomics highlight specific genetic features in Ganoderma species. J. Fungi (Basel) 8:311. doi: 10.3390/jof8030311
Sun, Y. F., Xing, J. H., He, X. L., Wu, D. M., Song, C. G., Liu, S., et al. (2022c). Species diversity, systematic revision and molecular phylogeny of Ganodermataceae (Polyporales, Basidiomycota) with an emphasis on Chinese collections. Stud. Mycol. 101, 287–415. doi: 10.3114/sim.2022.101.05
Trotta, E. (2013). Selection on codon bias in yeast: a transcriptional hypothesis. Nucleic Acids Res. 41, 9382–9395. doi: 10.1093/nar/gkt740
Tu, W. Y., Cao, X. N., Cheng, J., Li, L. J., Zhang, T., Wu, Q., et al. (2022). Chinese baijiu: the perfect works of microorganisms. Front. Microbiol. 13:919044. doi: 10.3389/fmicb.2022.919044
Utomo, C., Tanjung, Z. A., Aditama, R., Buana, R. F. N., Pratomo, A. D. M., Tryono, R., et al. (2019). Complete mitochondrial genome sequence of the Phytopathogenic basidiomycete Ganoderma boninense strain G3. Microbiol. Resour. Announc. 8:e00968. doi: 10.1128/MRA.00968-18
Vaidya, G., Lohman, D. L., and Meier, R. (2011). SequenceMatrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 27, 171–180. doi: 10.1111/j.1096-0031.2010.00329.x
Wang, X. C., Shao, J., and Liu, C. (2016a). The complete mitochondrial genome of the medicinal fungus Ganoderma applanatum (Polyporales, Basidiomycota). Mitochondrial DNA A DNA Mapp. Seq. Anal. 27, 2813–2814. doi: 10.3109/19401736.2015.1053080
Wang, X. C., Wu, K., Chen, H., Shao, J., Zhang, N., Chen, X., et al. (2016b). The complete mitochondrial genome of the white-rot fungus Ganoderma meredithiae (Polyporales, Basidiomycota). Mitochondrial DNA A DNA Mapp. Seq. Anal. 27, 4197–4198. doi: 10.3109/19401736.2015.1022737
Wang, X. C., Xi, R. J., Li, Y., Wang, D. M., and Yao, Y. J. (2012). The species identity of the widely cultivated Ganoderma, 'G. lucidum' (Ling-zhi), in China. PLoS One 7:e40857. doi: 10.1371/journal.pone.0040857
Wu, Q., Li, L., Xiang, P., Zhang, T., Peng, L., Zou, L., et al. (2023). Phages in fermented foods: interactions and applications. Fermentation 9:201. doi: 10.3390/fermentation9030201
Xiao, G., Zhou, J., Huo, Z., Wu, T., Li, Y., Li, Y., et al. (2022). The shift in synonymous codon usage reveals similar genomic variation during domestication of Asian and African Rice. Int. J. Mol. Sci. 23:12860. doi: 10.3390/ijms232112860
Xing, Y., Gong, R., Xu, Y., Liu, K., and Zhou, M. (2020). Codon usage bias affects α-amylase mRNA level by altering RNA stability and cytosine methylation patterns in Escherichia coli. Can. J. Microbiol. 66, 521–528. doi: 10.1139/cjm-2019-0624
Xu, Q., Chen, H., Sun, W., Zhu, D., Zhang, Y., Chen, J. L., et al. (2021). Genome-wide analysis of the synonymous codon usage pattern of Streptococcus suis. Microb. Pathog. 150:104732. doi: 10.1016/j.micpath.2021.104732
Xu, W., Xing, T., Zhao, M., Yin, X., Xia, G., and Wang, M. (2015). Synonymous codon usage bias in plant mitochondrial genes is associated with intron number and mirrors species evolution. PLoS One 10:e0131508. doi: 10.1371/journal.pone.0131508
Yao, H., Chen, M., and Tang, Z. (2019). Analysis of synonymous codon usage Bias in Flaviviridae virus. Biomed. Res. Int. 2019:5857285. doi: 10.1155/2019/5857285
Yu, X., Liu, J., Li, H., Liu, B., Zhao, B., and Ning, Z. (2021). Comprehensive analysis of synonymous codon usage Bias for complete genomes and E2 gene of atypical porcine Pestivirus. Biochem. Genet. 59, 799–812. doi: 10.1007/s10528-021-10037-y
Zhang, J., Liu, Y., Tang, Q., Zhou, S., Feng, J., and Chen, H. (2019). Polysaccharide of Ganoderma and its bioactivities. Adv. Exp. Med. Biol. 1181, 107–134. doi: 10.1007/978-981-13-9867-4_4
Zheng, S., Zhu, N., Shi, C., and Zheng, H. (2020). Genomic data mining approaches for the discovery of anticancer peptides from Ganoderma sinense. Phytochemistry 179:112466. doi: 10.1016/j.phytochem.2020.112466
Keywords: codon usage, mitochondrial genome, genetics, natural selection, fungi
Citation: Wu P, Xiao W, Luo Y, Xiong Z, Chen X, He J, Sha A, Gui M and Li Q (2023) Comprehensive analysis of codon bias in 13 Ganoderma mitochondrial genomes. Front. Microbiol. 14:1170790. doi: 10.3389/fmicb.2023.1170790
Received: 21 February 2023; Accepted: 12 April 2023;
Published: 04 May 2023.
Edited by:
Baokai Cui, Beijing Forestry University, ChinaReviewed by:
Yifei Sun, Beijing Forestry University, ChinaCopyright © 2023 Wu, Xiao, Luo, Xiong, Chen, He, Sha, Gui and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mingying Gui, R3VpbWluZ3lpbmdAMTI2LmNvbQ==; Qiang Li, bGVlcTExMEAxMjYuY29t
Present addresses: Mingying Gui, School of Food and Biological Engineering, Chengdu University, Chengdu, Sichuan, ChinaQiang Li, School of Food and Biological Engineering, Chengdu University, Chengdu, Sichuan, China
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.