- 1Institute of Medical Microbiology and Hospital Hygiene, Medical Faculty, Otto-von-Guericke University Magdeburg, Magdeburg, Germany
- 2Institute of Molecular and Clinical Immunology, Medical Faculty, Otto-von-Guericke University Magdeburg, Magdeburg, Germany
- 3Health Campus Immunology, Infectiology, and Inflammation (GCI3), Medical Center, Otto-von-Guericke University Magdeburg, Magdeburg, Germany
- 4CHaMP, Center for Health and Medical Prevention, Otto-von-Guericke-University Magdeburg, Magdeburg, Germany
- 5Department of Microbiology and Hospital Hygiene, Bundeswehr Central Hospital Koblenz, Koblenz, Germany
- 6Department of Microbiology and Hospital Hygiene, Bundeswehr Hospital Hamburg, Hamburg, Germany
- 7Institute for Medical Microbiology, Virology and Hospital Hygiene, University Medicine Rostock, Rostock, Germany
Extended spectrum beta-lactamases (ESBL) are frequently found in Enterobacterales isolates from Western Africa. However, information on the molecular epidemiology of regional ESBL-positive Enterobacterales strains is scarce. In order to provide epidemiological information, ESBL-positive Escherichia coli isolates from stool samples of European soldiers with diarrhea deployed to a field camp in Mali were subjected to whole-genome sequencing (Illumina MiSeq and Oxford Nanopore MinION) and antimicrobial susceptibility testing. With two exemptions, sequence-based analysis suggested an absence of transmission events between soldiers as indicated by a high genetic diversity of isolates and sequence types, confirming previous rep-PCR results. Third-generation cephalosporin resistance was associated with the presence of blaCTX-M-15 genes with (n = 14) and without (n = 5) co-occurring blaTEM-1b genes. Between 0 and 6 virulence and resistance plasmids per isolate were recorded. The detected resistance plasmids could be categorized into five types, which, in turn, share different sequence-identical segments, representing particular antimicrobial resistance gene-associated mobile genetic elements (MGEs). Phenotypic resistance rates within the 19 assessed isolates that showed distinguishable colony morphologies were 94.7% (18/19) against ampicillin-sulbactam and trimethoprim/sulfamethoxazole, 68.4% (13/19) against moxifloxacin, 31.6% (6/19) against ciprofloxacin, 42.1% (8/19) against gentamicin, 31.6% (6/19) against tobramycin, and 21.1% (4/19) against piperacillin-tazobactam and fosfomycin. Virulence-associated genes mediating infectious gastroenteritis were rarely detected. The gene aggR, which is characteristic for enteroaggregative E. coli, was only detected in one single isolate. In summary, we found a variety of different strains and clonal lineages of ESBL-carrying E. coli. Transmission either between soldiers or from common contaminated sources was demonstrated in two cases and played only a minor role in this military field camp, while there were indications that resistance gene bearing MGEs had been exchanged between antimicrobial resistance gene-(ARG-)carrying plasmids.
1. Introduction
Antimicrobial resistance (AMR) is quite common in Sub-Saharan Africa, particularly in Gram-negative rods, such as Enterobacterales (Tompkins et al., 2021). To counteract this challenge, the implementation of antibiotic stewardship programs in Africa has been suggested (Akpan et al., 2020). However, evidence-guided optimized prescription of antimicrobial therapy requires knowledge on regionally common susceptibility patterns. Therefore, AMR surveillance is crucial for this purpose (Akpan et al., 2020).
Antimicrobial resistance surveillance data from war- and crisis-haunted Sub-Saharan regions like Western African Mali are largely unavailable. Data on antimicrobial resistance and susceptibility mainly arise from arbitrarily distributed, small prevalence assessments. Next to information on extended spectrum beta-lactamase- (ESBL-)positive Enterobacterales (Sangare et al., 2015), resistance rates for other microorganisms such as Neisseria gonorrhoeae (Jary et al., 2021), pneumococci (Campbell et al., 2004), Haemophilus influenzae (Pécoul et al., 1991; Sow et al., 2005), and Staphylococcus aureus (Ruimy et al., 2008) are also scarce. In comparison to the latter, Malian studies on Enterobacterales with resistance against third-generation cephalosporins are generally more frequent (Sangare et al., 2015; Nkansa-Gyamfi et al., 2019). In a Malian orphanage, ESBL-positive Enterobacterales colonized the inhabitants’ and the workers’ skin to a proportion that they could be virtually considered as standard bacterial flora (Tandé et al., 2009). Consequently, in case of international adoption, nosocomial spread of such bacteria is a frequently observed phenomenon (Tandé et al., 2010). While such colonization was usually harmless in immunocompetent individuals (Tandé et al., 2009), however, immunocompromised hosts with tuberculosis and acquired immunosuppression suffered from severe superinfection due to ESBL-positive Klebsiella spp. (Meli et al., 2020). Among Enterobacterales isolated from Malian blood cultures, ESBL-carriage in about two out of three isolates has been repeatedly described (Sangare et al., 2016, 2017; Sangaré et al., 2017). Next to typical ESBL-harboring Enterobacterales like Escherichia spp. and Klebsiella spp., ESBL enzymes are also common in Salmonella enterica in Mali and in the Senegal (Boisramé-Gastrin et al., 2011; Harrois et al., 2014).
In a surveillance study conducted between December 2013 and August 2014 with European soldiers deployed to Mali on the European Union Training Mission (EUTM), ESBL-positive Enterobacterales were isolated from 13 out of 48 stool samples obtained from patients with diarrhea during the study interval (Frickmann et al., 2015). With the exemption of a single Klebsiella pneumoniae isolate, only ESBL-positive E. coli isolates were detected indicating multiple colonization events in superficial rep-PCR-based typing (Frickmann et al., 2015). The multiple transmission events were likely to be linked to environmental sources, since ESBL-positive Enterobacterales were detected in environmental samples and also in the food sources of the mission headquarters (Maaßen et al., 2017; Frickmann et al., 2018a). Genes from the blaCTX-M group 1 were reported to be responsible for the observed third-generation cephalosporin resistance (Hagen et al., 2015). Despite high enteric colonization rates of the European soldiers on deployment in Mali with ESBL-positive Enterobacterales (Frickmann et al., 2015), low detectable colonization of about 7% (3/43 samples) was recorded in German military returnees during screenings in the course of returnee assessments about 3 months after deployment to Mali (Frickmann et al., 2016; Pankok et al., 2022a). This finding is well in line with previous reports indicating that enteric colonization with ESBL-positive Enterobacterales is transient and that rates of detection considerably drop in the first 3 months after returning from international journeys (Ruppé et al., 2015; Arcilla et al., 2017; Frickmann et al., 2018b).
In our study, we performed whole-genome sequencing of ESBL-positive E. coli isolates cultured from the stool samples of the European soldiers deployed to Mali (Frickmann et al., 2015; Hagen et al., 2015) and analyzed the epidemiological relationship between isolates and the presence of molecular resistance and virulence determinants. In addition to virulence factors and antimicrobial resistance genes (ARGs) that are chromosomally encoded, the focus was on the analysis of antimicrobial resistance gene-(ARG-) bearing resistance plasmids. The potential abundance of highly virulent clonal lineages of E. coli was addressed (Denamur et al., 2021). Accordingly, our study provides important additional information on the local epidemiology in Mali.
2. Materials and methods
2.1. Study site and origin of the isolates
Escherichia coli isolates with resistance against third-generation cephalosporins had been collected in a previous surveillance study, which assessed stool samples of European soldiers with diarrhea during their deployment in Mali between December 2013 and August 2014, by isolation from Brilliance ESBL selective agar (Oxoid, Basingstoke, United Kingdom) as described before (Frickmann et al., 2015). Brilliance ESBL selective agar does not specifically select diarrheagenic E. coli and culture-based screening for them was not the purpose of the study approach. Forty-eight diarrhea stool samples were collected in the military Camp of Koulikoro about 60 km from the Malian capital Bamako, where the training site of the EUTM Mali mission was located. Koulikoro is a small community at the river Niger with about 50,000 inhabitants (latitude and longitude 12.52 N 7.34 W, Figure 1). During the sampling period, about 600 European soldiers were deployed on the EUTM Mali mission and changed about every 4–6 months.
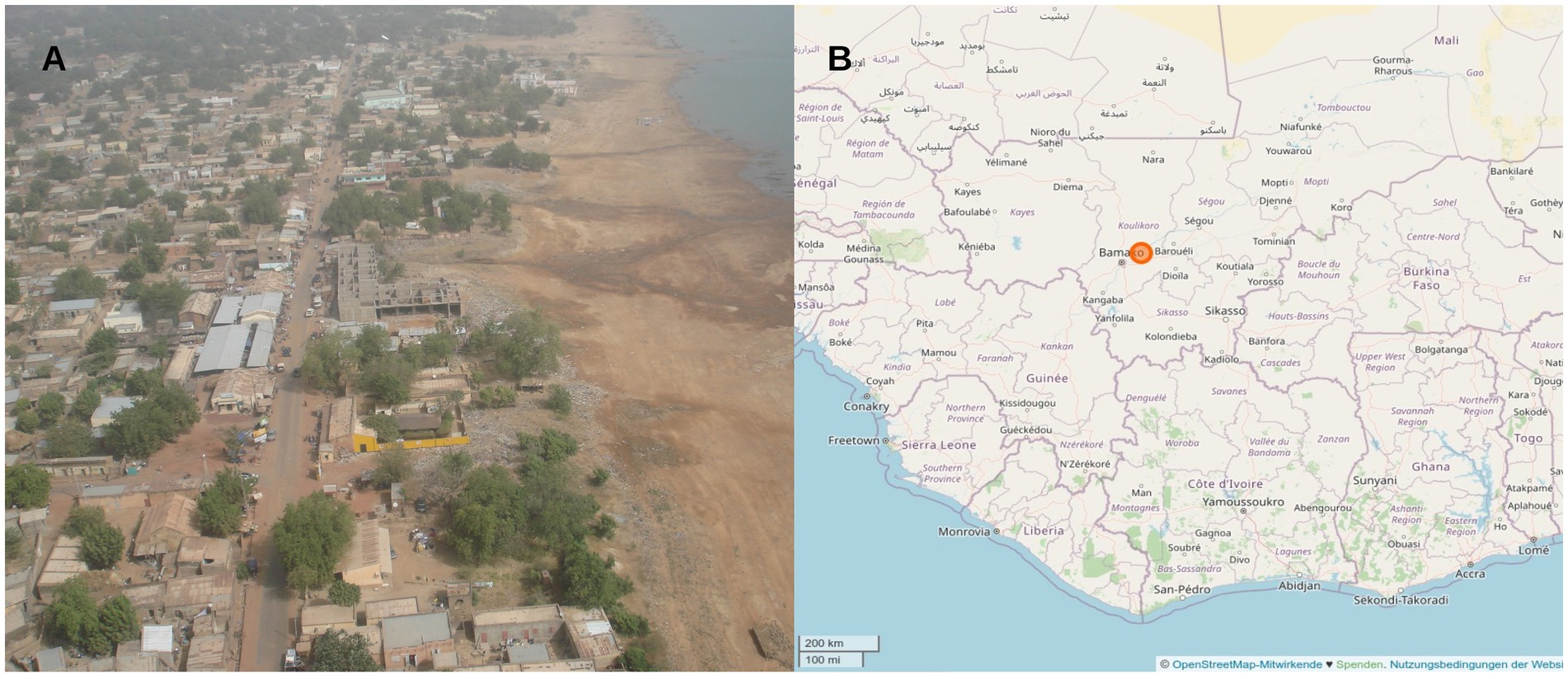
Figure 1. (A) Aerial view of Koulikoro, where the main training site of the EUTM mission was located. (B) Location of the study site ca. 60 km north-east from the Malian capital Bamako.
The strains had been isolated at the Department of Tropical Medicine at the Bernhard Nocht Institute, Bundeswehr Hospital Hamburg, Germany and stored as Microbank™-cryostocks (Pro-Lab Diagnostics, Round Rock, Texas, United States) at −80°C in the strain collection of the Department of Microbiology and Hospital Hygiene, Bundeswehr Central Hospital Koblenz, Germany, which is in charge for population-based microbiological assessments and deployment-associated microbiology in the German armed forces. Superficial molecular assessment of the isolates by rep-PCR typing had been initially performed after sampling (Frickmann et al., 2015; Hagen et al., 2015). Cultivation prior to DNA extraction was performed on Columbia agar enriched with 5% sheep blood at 37°C in a CO2-enriched atmosphere for 24–48 h.
2.2. Mass-spectrometry based species identification and phenotypic resistance testing
At the Medical Faculty of the Otto-von-Guericke University Magdeburg, all isolates were confirmed as E. coli by matrix-assisted laser-desorption-ionization time-of-flight mass-spectrometry (MALDI-TOF-MS) using a Vitek MS mass spectrometer with the corresponding VITEK MS Knowledge Base V3.2 (bioMérieux, Mary-l’Étoile, France). Phenotypic susceptibility testing was performed using the Micronaut Microdilution System (MERLIN Diagnostika GmbH, Bornheim-Hersel, Germany) employing MCN-S MH Hannover GN3 plates (E1-247-100) according to the manufacturer’s instructions. The results were interpreted according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) V12.0 (2022) standard.
2.3. DNA extraction, library preparation, and whole-genome sequencing
Isolation of bacterial DNA was performed via the cetyltrimethylammonium bromide (CTAB) method as described previously by Bechmann et al. (2022). Library preparation for Illumina paired-end sequencing was performed using the TruePrep DNA Library Prep Kit V2 for Illumina (1°ng; Vazyme Biotech Co. Ltd., Nanjing, China). Libraries were barcoded using the Nextera XT Index Kit (Illumina, San Diego, United States) and sequenced using the MiSeq Reagent Kit v2 (500-cycles, Illumina) as described by the manufacturer. Barcoded libraries for Nanopore long-read sequencing were prepared using the Rapid Barcoding Kit 96 (SQK-RBK110.96) according to the manufacturer’s instructions and sequenced on a R9.4.1 flow cell (FLO-MIN106) on the MinION platform (Oxford Nanopore technologies ltd., Oxford, United Kingdom).
2.4. Genome assembly
Illumina paired-end reads were preprocessed using fastp (https://github.com/OpenGene/fastp, v0.23.2), and filtlong (parameters: --min_length 1,000 --keep_percent 95, https://github.com/rrwick/Filtlong, v0.2.1) was used for long reads. Genomes were assembled using a long read consensus assembly approach presented in the trycycler software package (https://github.com/rrwick/Trycycler, v0.5.3), and cross-validated with results from the short read assembler SKESA (Shovill/SKESA, v2.4.0; Souvorov et al., 2018), as well as the hybrid assembler unicycler (https://github.com/rrwick/Unicycler, v0.4.8). In short, long reads were sampled into 20 unique subsets of reads with a 100x sequencing depth and each sets of four subsets were independently assembled using canu v1.8.1 (Koren et al., 2017), flye v2.9.1-b1780 (Kolmogorov et al., 2019), miniasm v0.3,1 minipolish v0.1.3 (Wick and Holt, 2019), necat v0.0.1,2 and raven v1.8.13 to generate 20 independent long-read assemblies. Subsequently, the trycycler functions cluster, reconcile, msa, partition, and consensus were applied on the set of assemblies for each sample to generated consensus assemblies. Finally, the consensus assemblies were polished using the long read polisher medaka4 and short read polisher polypolish v0.5.05 (Wick and Holt, 2022).
2.5. Annotations
The final assemblies were annotated using the NCBI Prokaryotic Genome Annotation Pipeline (Tatusova et al., 2016; Haft et al., 2018; Li et al., 2021) and further characterizations comprised plasmid identification and annotation (MOB-suite, PlasmidFinder, MobileElementFinder; Camacho et al., 2009; Carattoli et al., 2014; Robertson and Nash, 2018; Johansson et al., 2021), antimicrobial resistance (AMR) gene detection (AMRFinderPlus v3.11.2; Resfinder v4.1, PointFinder, ResFinder FG V1.0; Sommer et al., 2009; Zankari et al., 2012, 2017; Pehrsson et al., 2016; Feldgarden et al., 2021), and virulence gene detection (Virulence Finder; Joensen et al., 2014; Malberg Tetzschner et al., 2020).
2.6. Core genome multi locus sequence typing
Core genome multi-locus sequence typing (cgMLST) from assembled E. coli genomes was conducted using 2,520 cgMLST tragets provided by Ridom SeqSphere+ (v8.5.1,_2022–11). (I) MST for 16 samples was based on 2,520 columns, pairwise ignoring missing values (Jünemann et al., 2013; Weber et al., 2019) with a logarithmic scale. Distance was based on columns from E. coli cgMLST (2513) and E.coli MLST by Warwick (7). (II) MST for 55 samples (including reference datasets from Niger, Ghana, and Germany) was based on 2,274 columns, pairwise ignoring missing values. Distance was based on columns from E. coli cgMLST (2,274).
2.7. Ethics statement
Ethical clearance for the assessment of the surveillance data in an anonymized way without requirement of informed consent was provided by the ethics committee of the medical association of Hamburg (WF-037/14) on 20th January 2015 in line with National German laws. The study was conducted according to the guidelines of the Declaration of Helsinki.
3. Results
3.1. Phenotypic resistance patterns
From 12 of 48 soldiers (25%) with diarrhea on the EUTM mission, a total of 19 E. coli isolates were selected from ESBL screening agar based on the exhibited specific colony morphology. Thereby, the number of different morphotypes ranged from 1 to 4, comprising eight individuals with one morphotype, two individuals with two morphotypes, and one individuum each with three and with four morphotypes, respectively. All isolates were confirmed as E. coli by matrix-assisted laser-desorption-ionization time-of-flight mass-spectrometry (MALDI-TOF-MS). Automated antimicrobial susceptibility testing using VITEK-2 AST-N263 cards with interpretation according to the EUCAST 2022 standard confirmed ESBL phenotypes in all instances (positive ESBL screening test). Resistance in 100% (19/19) of the isolates was recorded against ampicillin, cefuroxime, cefotaxime, and ceftazidime; 94.7% (18/19) against ampicillin-sulbactam and trimethoprim/sulfamethoxazole (5.3% = 1/19 susceptible a high dosage), 68.4% (13/19) against moxifloxacin, 42.1% (8/19) against gentamicin, 31.6% (6/19) against ciprofloxacin (26.3% = 5/19 susceptible at high dosage) and tobramycin, and 21.1% (4/19) against piperacillin-tazobactam and fosfomycin. Thus, a high percentage of piperacillin-tazobactam susceptible and trimethoprim/sulfamethoxazole resistant ESBL-isolates was observed. All isolates (19/19) were susceptible toward imipenem, meropenem, ertapenem, tigecycline, and colistin. Details on the susceptibility patterns of the isolates are provided in Table 1.
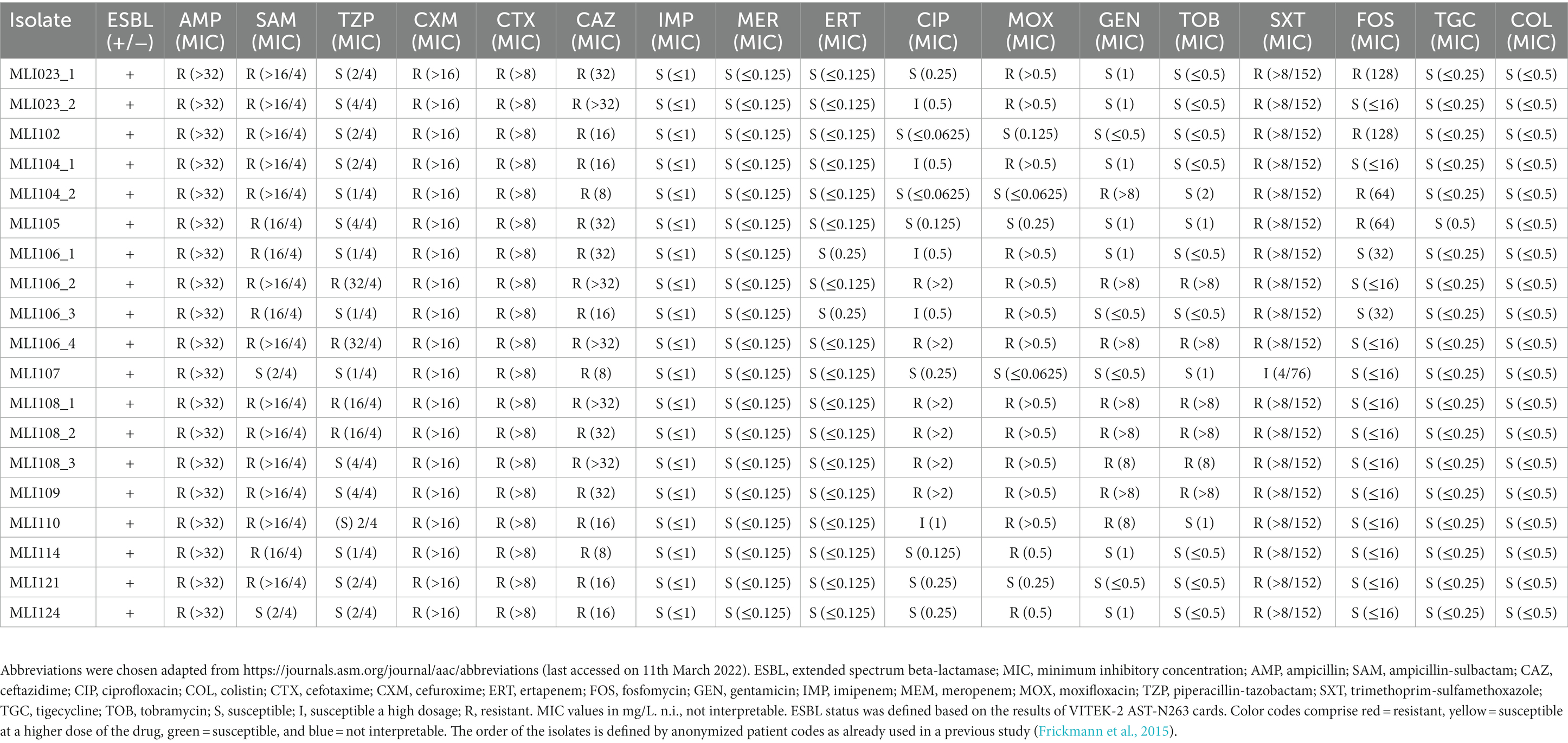
Table 1. Phenotypic resistance as measured for the assessed Escherichia coli isolates applying MICRONAUT microdilution broth plates (catalog number: E1-247-100) interpreted according to the EUCAST 2022 standard.
3.2. cgMLST and clonal lineages of the isolates
Assemblies of 16 isolates from the 12 ESBL positive soldiers were investigated via cgMLST-based analysis to exclude an outbreak. The recorded phylogenetic distance between the isolates ranged from 2 to 2,277 alleles as indicated by the cgMLST-based minimum spanning tree. Three pairs of isolates turned out to have the same cgMLST-based sequence type as well as on closer analysis the exact same genome sequence and were therefore considered identical, i.e., MLI23-1 and -2 (in spite of varying phenotypic susceptibility testing results, Table 1), MLI106-2 and -4, and MLI108-1 and -2 are copy strains; therefore, only one isolate was considered in the phylogenetic analysis (Figure 2). Genetically identical isolates, for which only colony-morphology had been different, were associated with the same soldier in all three instances. The isolates MLI104-1 and -2 originating from a single individual are very closely related, which is indicated by a difference of only two alleles. Close phylogenetic relationship between isolates from two different individuals indicative of direct transmission or common sources was detected in two cases. Thus, the isolates MLI114 and MLI121 differed by only 14 alleles and the isolates MLI102 and MLI109 differed by only three alleles within the 2,520 considered columns of the cgMLST scheme. Of note, the isolates MLI102 and MLI109 had been collected from samples taken within a 10-day-interval. For MLI114 and 121, details on a timely association cannot be provided due to incomplete data acquisition during the epidemiological assessment (Frickmann et al., 2015). The remaining isolates showed high genetic diversity, indicating that they originated from different sources and were not transmitted from soldier to soldier. This high diversity was confirmed by the high number of detected sequence types as shown in the Supplementary Table 1.
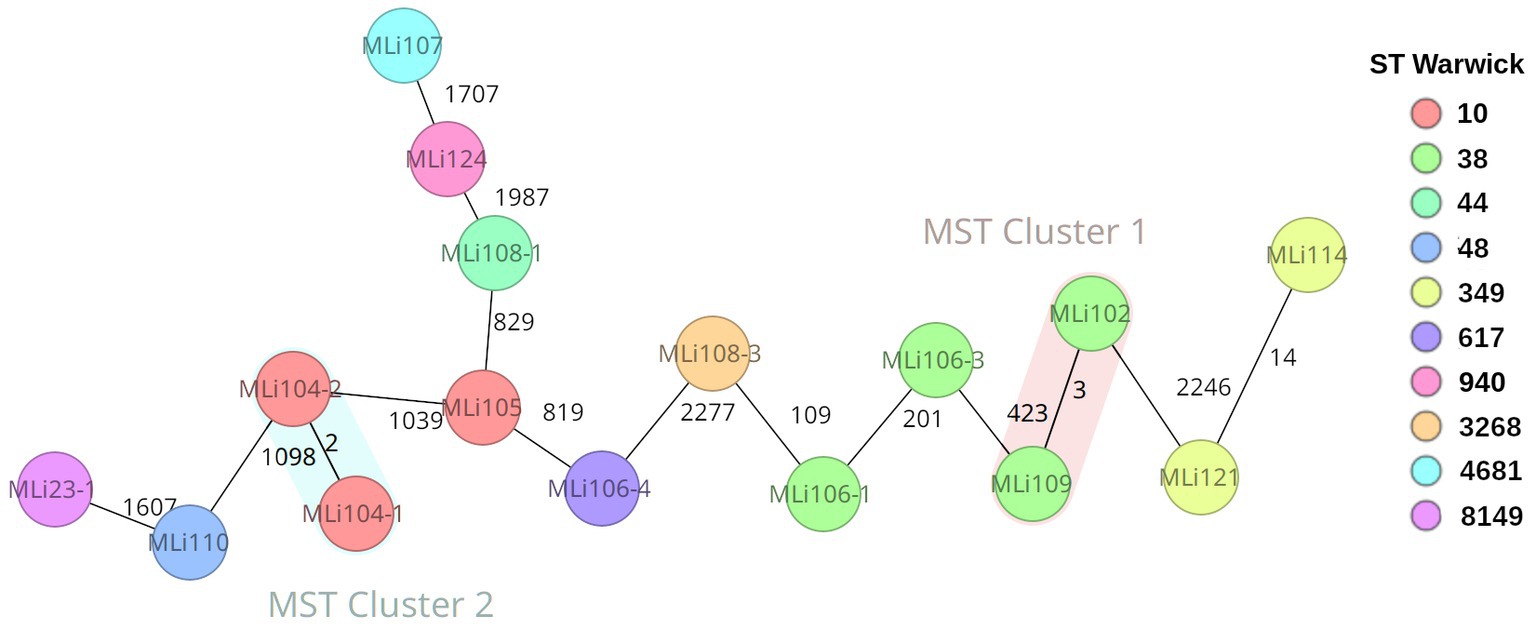
Figure 2. Ridom SeqSphere+ generated cgMLST-based minimum spanning tree showing the genetic distance of the Escherichia coli isolates indicated by numbers of allele differences in a total of 2,520 assessed columns (E. coli cgMLST: 2513; pairwise ignoring missing values, logarithmic scale, E. coli MLST Warwick: 7; cluster distance threshold 10). The nodes are colored by ST Warwick (see legend).
In order to gain at least an orienting impression of whether the isolates we analyzed originated in West Africa/Mali or were imported from Europe by the soldiers, we performed a further cgMLST analysis. Since there were no E. coli sequence data from Mali in Enterobase and NCBI GenBank so far, and no such sequence data were available from most neighboring countries either, we included a dozen E. coli sequences each from Niger and Ghana and 24 E. coli sequences from two different studies from Germany in a second cgMLST analysis (Supplementary Table 3; Supplementary Figure 1). In this analysis, only four clusters showed up at a cluster distance threshold of 10. These clusters include only two isolates each. These are the two clusters from our study mentioned above as well as one cluster formed by two isolates from Ghana and one cluster formed by two isolates from Germany. This means there was actually no evidence of importation of the E. coli isolates we considered in the study from Europe. Taking a higher perspective and looking at the studied isolates by origin in our cgMLST-based dendrogram, we recognized three superclusters. Thus, we found a supercluster on the right side of the tree of predominantly German isolates in which there were still three isolates from Ghana. The distance to the next isolate from Mali is 2,135 alleles. On the left side of the dendrogram is another cluster of predominantly German isolates together with isolate MLI124. The distance of this supercluster to the next isolate from Africa/Ghana is 1,556 alleles. In between is a supercluster containing 18 of 19 isolates from Mali and, in addition, all isolates/sequence types from Niger and eight of 11 sequence types from Ghana. Only two isolates from the German datasets are in this supercluster belonging to the sequence types ST 10 (17–04317) and ST 216 (17–041045), which are both globally distributed lineages. In summary, according to the limitations of the method, it can be assumed that the majority of the isolates really originated from Mali and only for isolate MLI124 there is a suspicion that it was imported from Europe.
3.3. Identified antimicrobial resistance genes
In all 16 isolates (excluding the copy strains), third-generation cephalosporin resistance could be attributed either to blaCTX-M-15 alone (n = 4/16) or in combination with blaTEM-1b (n = 12/16). Details are provided in Figure 3, Supplementary Figure 2, and Supplementary Table 1, next to information on other identified antimicrobial resistance genes either encoded on the bacterial chromosomes or on plasmids. As shown there, the resistance-mediating genes blaEC and blaCTX-M-15 were the only ones occurring in all isolates. A fourth beta-lactamase gene blaOXA-1 could be detected in four isolates.
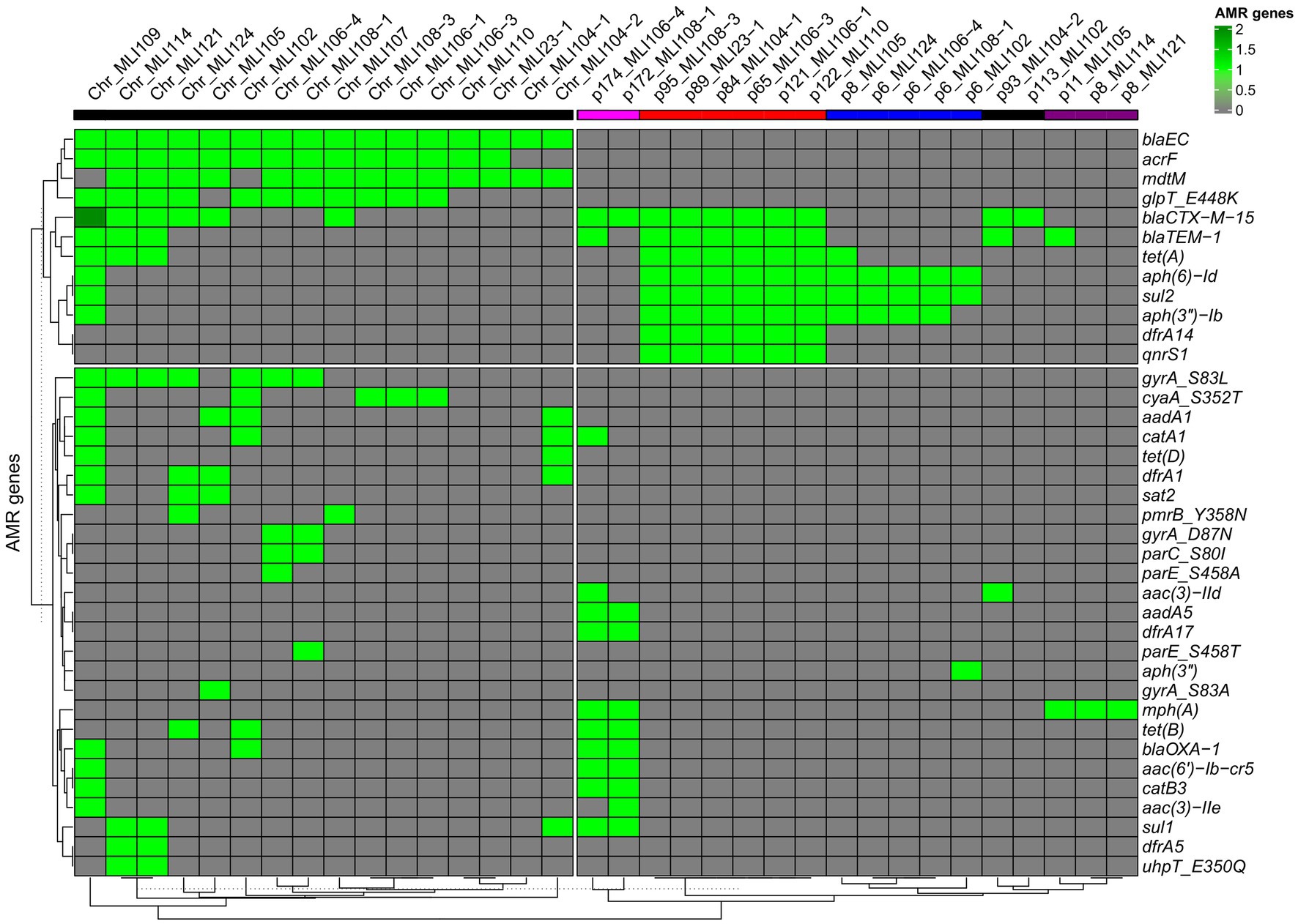
Figure 3. Heatmap of the associations of bacterial chromosomes (left side) as well as plasmids (right side) with encoded ARGs. The heatmap with the ARG/chromosome/plasmid sequence-based dendrograms were generated using the R package ComplexHeatmap [hierarchical clustering (hclust) with Euclidean distance]. A light green rectangle indicates the presence of a specific ARG, a dark green rectangle indicates two copies of a specific ARG. A gray rectangle indicates the absence of a particular ARG. The five different plasmid types, based on gene overlaps among each other, are indicated by different colored bars in the header of the heat map: type 1—pink, type 2—red, type 3—blue, type 4—black, and type 5—purple.
Of the isolates tested, 11/16 were found to be moxifloxacin resistant. Of the quinolone resistance genes detected, the vast majority were found in these 11 moxifloxacin resistant isolates: qnrS1 (7/11), gyrA S83L (7/11), gyrA D87N (4/11), parE S458T (2/11), parE S458A (1/11), and parC S80I (2/11). None of the moxifloxacin resistant isolates did not have at least one of these quinolone resistance determinants. Two of the moxifloxacin resistant isolates (MLI114 and MLI 124) were ciprofloxacin susceptible. Both isolates were positive for gyrA S83L only. Nevertheless, among the five quinolone susceptible isolates, there was one isolate (MLI105) that was gyrA S83A positive and one (MLI121) that was gyrA S83L positive.
Resistance to trimethoprim-sulfamethoxazole was attributable to the combined occurrence of the sulfonamide resistance genes sul1 (n = 5) and sul2 (n = 12) as well as the trimethoprim resistance genes dfrA1 (n = 4), dfrA5 (n = 2), dfrA14 (n = 6), and dfrA17 (n = 2). The only isolate that was trimethoprim-sulfamethoxazole susceptible at increased dosage, E. coli MLI107, did not exhibit any of these six ARGs.
Of the six gentamicin resistant isolates, five were positive for aph6-Id and aph3-Ib. In one isolate, aac3-IId, aadA5, and aac6-Ib-cr5 were found additionally. In a second isolate, aac3-IIe, aadA5, and aac6-Ib-cr5; and in a third aac3-IIe and aac6-Ib-cr5 were additional aminoglycoside-specific ARGs. In a single gentamicin-resistant isolate, aac3-IId was the only aminoglycoside-specific ARG.
Antimicrobial resistance genes conferring tetracycline resistance, which was not phenotypically tested, were tetA (n = 10), tetB (n = 4), and tetD (n = 2).
The pmrB gene, associated with polymyxin B resistance, was detected chromosomally encoded in two isolates. However, we only tested susceptibility to colistin (polymyxin E) and did not find phenotypically detectable resistance.
3.4. Resistance plasmids
As indicated in Figure 3, in the isolates investigated five different types of resistance plasmids were found according to the ARGs that were encoded on them. These numerical plasmid types assigned by us correlate only partially with the incompatibility types listed for each plasmid in Supplementary Table 1. The plasmids of the first plasmid type had a molecular weight of more than 170 kbp, belonged to the incompatibility types IncFIA, IncFIB, as well as IncFII and encoded the type-common resistance genes blaCTX-M-15, aadA5, dfrA17, mphA, tetB, blaOXA-1, aac(6′)-Ib-cr5, catB3, and sul1. The plasmids of the second plasmid type had a molecular weight of 65–122 kbp, belonged to the incompatibility types IncFIB or IncY and encoded the type-common ARGs blaCTX-M-15, blaTEM-1, tetA, aph(6)-Id, sul2, aph(3″)-(Ib), dfrA14, and qnrS1. The plasmids of the third plasmid type were much smaller, belonged to none of the incompatibility types, and had a molecular weight of 6–8 kbp and encoded the type-common ARGs aph(6)-Id, sul2, and aph(3″)-(Ib). The plasmids of the fourth plasmid type belonged to the incompatibility type IncI1 or to none incompatibility type, and had blaCTX-M-15 as the only common ARG at a molecular weight of 93–113 kbp, and the plasmids of the fifth plasmid type were again much smaller, belonged to none of the incompatibility types, had a molecular weight of 8–11 kpb and encoded mphA.
From the heat map in Figure 3, it can be already assumed that most ARGs of plasmid types 2, 3, and 4 are located on segments of different sizes with higher sequence identity. All six plasmids of type 2 share a plasmid segment that encodes the ARGs dfrA14, sul2, aph(3″) − Ib, aph(6) − Id, tetA/tetR, blaTEM − 1, blaCTX − M − 15, and qnrS1 (Figure 4). A subsegment containing the ARGs sul2, aph(3″) − (Ib), and aph(6) − Id was found in the five type 3 plasmids. Another subsegment encoding blaCTX-M-15 existed in the two type 4 plasmids. A subsegment encoding blaTEM-1 was found on the type 4 plasmid p93_MLI104-2, as well as on the type 2 plasmids. These (sub-) segments were most likely transferred together at one point of plasmid evolution on different mobile genetic elements, so that in particular the type 2 plasmids can be conceived as plasmid chimeras. As an example, we will take a closer look at the type 2 plasmid p122_MLI110. At position 15.208–63.996 there is a composite transposon, cn_48788_IS5075 (family IS110), encoding the ARGs sul2, aph(3″) − Ib, aph(6) − Id, tetA/tetR, blaTEM − 1, blaCTX − M − 15, and qnrS1. A second composite transposon, cn_27847_IS26 (family IS6), at position 12.611–40.458, covers the same ARGs. ARG dfrA14 is on a separate MGE, cn_4021_IS26 (family IS6) at position 9.410–13.431. In contrast, there was only one MGE on the type 4 plasmid p93_MLI104-2. While the composite transposon, cn_3146_IS26, at position 87.935–91.081, carried blaCTX − M − 15, the two ARGs blaTEM − 1 and aac(3)-IId were encoded on the remaining plasmid regions. The type 3 plasmid p6_MLI124, encoding sul2, aph(3″) − Ib, and aph(6) − Id, carried no MGE. Mobile genetic elements of all chromosomes and plasmids associated with antimicrobial resistance genes are listed in the Supplementary Table 1.
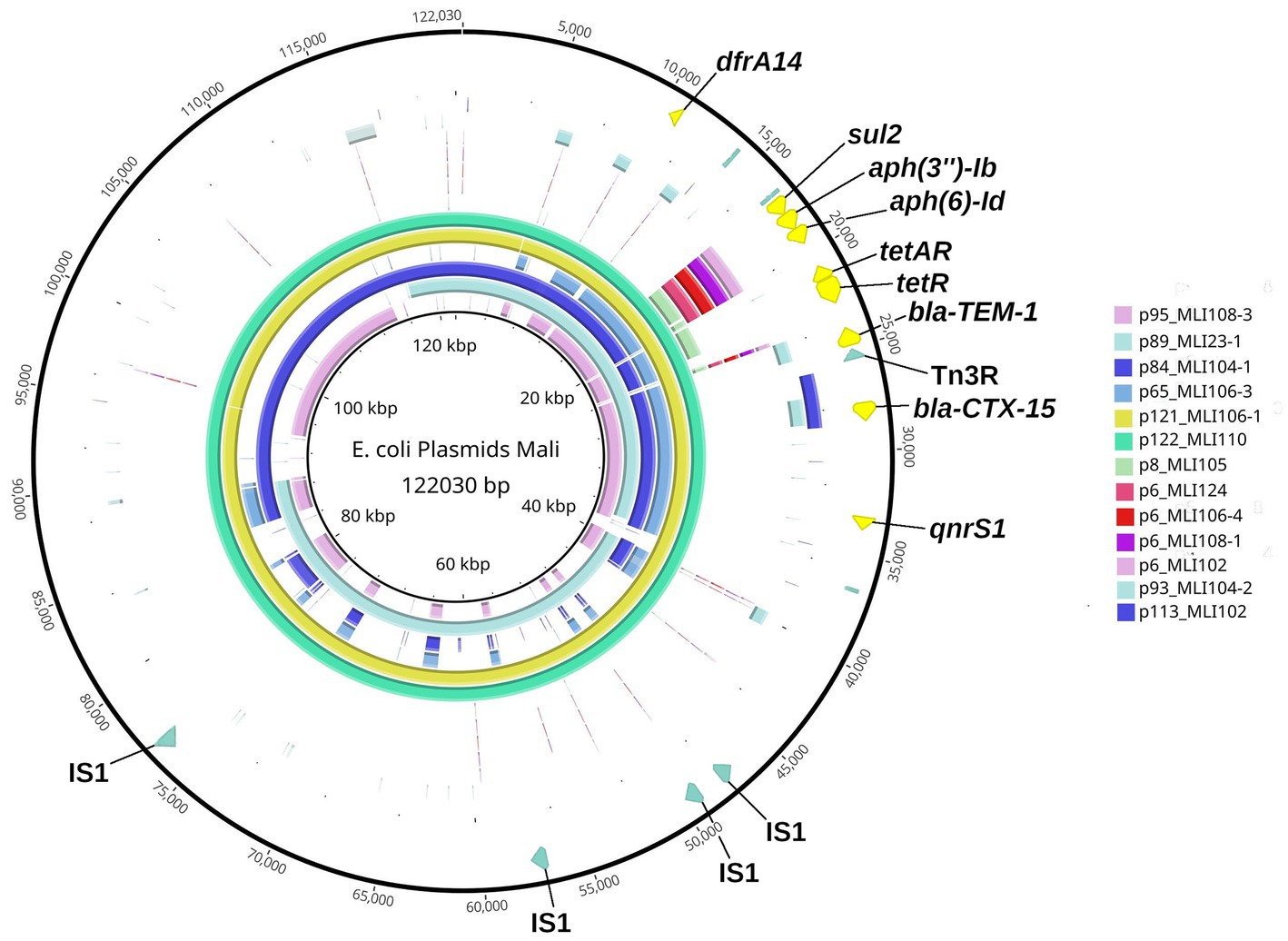
Figure 4. Blast Ring Image Generator-(BRIG-)generated graph depicting homologous plasmid segments of the plasmid types 2–4 detected in the Escherichia coli isolates from soldiers probed in Mali. The parameters used to generate the BRIG-graph were: BLAST type: blastn, upper identity threshold 70%, lower identity threshold 30%, ring size 30, and plasmid origin at 12 o’clock position. The largest plasmid of these three plasmid types, type 2 plasmid p122_MLI110, was chosen as reference (inner black circle) and is additionally shown in the sixth outer circle (mint-green). The individual plasmids are color coded as in the legend and the ARGs are annotated. Outermost Ring: plasmid-map of p122_MLI110 indicating the particular ARGs in yellow and specific MGE recognition sequences in olive green.
3.5. Identified virulence determinants
Recorded virulence factors mostly comprised genes affecting the bacterial iron metabolism, while less frequently observed genes influenced mechanisms like capsule formation and the excretion of substances from the cell. Details are provided in the Supplementary Table 2. Only in a single instance (MLI109) and thus well in line with a previous observation (Frickmann et al., 2015), the presence of the aggR gene confirmed the isolate as enteroaggregative Escherichia coli and thus as an isolate with enteropathogenic potential. The E. coli isolate MLI 109 carries one virulence plasmid on which the aggR gene was localized, which showed in a BLAST search a sequence identity >99% to pAA plasmids of other E. coli isolates deposited at NCBI GenBank. Consequently, the E. coli isolate MLI 109 is a typical representative of the EAEC (Enteroaggregative Escherichia coli) pathovar.
4. Discussion
In this study, we characterized 19 isolates (morphotypes) of third-generation cephalosporin resistant Escherichia coli from diarrhea stool samples of 12 European soldiers during the EUTM (European Training Mission) Mali mission between December 2013 and August 2014 (Frickmann et al., 2015; Hagen et al., 2015). Based on the results, the following conclusions appear justified.
First, a high molecular diversity of the isolates from the different individuals was confirmed, similar as suggested by rep-typing in a previous analysis (Frickmann et al., 2015). Only in two cases, a transmission from soldier to soldier or from common sources could be demonstrated. These results suggest that direct transmission between the different soldiers or transmission from a common source were just exceptions. The high genetic diversity of the isolates suggests multiple transmission events, most likely reflecting a high local prevalence of different ESBL-positive E. coli strains. This is underscored by the fact that genetically heterogenic third-generation cephalosporin-resistant E. coli isolates were found in stool samples from a single individual. Antibiotic exposure seemed to have played—if any—only a minor role in the acquisition of ESBL-positive E. coli by the assessed European soldiers suffering from diarrhea on tropical deployment in contrast to previous suggestions (Kantele et al., 2015). Therefore, no respective details have been presented. In short and as recorded for the previously described epidemiological assessment (Frickmann et al., 2015), only the patient from whom the isolate MLI-107 was isolated received treatment of traveler’s diarrhea with rifaximin, while malaria prophylaxis with 100 mg once daily doxycycline was taken by the patients providing the isolates MLI-104_1 and MLI-102_2 as well as MLI-108_1, MLI-108_2, and MLI-108_3. It may be speculated that the low-dose doxycycline application might have triggered the selection of MLI-104_1 and MLI-108_3 via the co-existing tet(A)-gene and the selection of MLI_104_2 via the co-existing tet(D)-gene, however, the extent of this effect remains speculative.
Second, next-generation sequencing analyses allowed a deeper insight into the molecular background of the recorded third-generation cephalosporin resistance than a previous multiplex PCR- and Sanger sequencing-based approach (Hagen et al., 2015). The ESBL-mediating gene blaCTX-M-15 [with (n = 14/19) or without (n = 5/19) blaTEM-1b] was detected in all isolates without exemption. This finding is expected, since blaCTX-M-15 is known to be highly prevalent in multiple countries of Sub-Saharan Africa (Mshana et al., 2013; Onduru et al., 2021). Interestingly, both chromosomal and plasmid-encoded blaCTX-M-15 genes were detected in the isolates, whereas a study in neighboring Ghana found blaCTX-M-15 encoded exclusively on plasmids in the E. coli isolates examined (Pankok et al., 2022b).
Third, the assessment of the sequence types did not indicate known pathogenic E. coli clonal lineages (Denamur et al., 2021) and in line with this, few virulence factors mediating enteropathogenicity were documented. Only one isolate was confirmed as enteroaggregative E. coli as shown in a previous publication (Frickmann et al., 2015), however, the etiological relevance of E. coli harboring this virulence determinant has been challenged recently (Tisdale et al., 2022).
In addition, the analysis provided a deeper insight into locally abundant resistance gene-carrying plasmids as well as resistance genes mediating resistance to antimicrobial drugs other than third-generation cephalosporines. Thus, we were able to categorize the observed plasmids into five types, which, in turn, share different sequence-identical segments carrying specific ARGs. A broad variety of plasmid-encoded and chromosomal resistance genes as well as antimicrobial resistance gene-associated mobile genetic elements was detected, explaining the observed resistance against other none-beta-lactam drugs like fluoroquinolones, aminoglycosides, epoxide antibiotics (fosfomycin), and trimethoprim-sulfamethoxazole commonly used for the therapy of infections with Enterobacterales as well. This information contributes to the scarcely available data on AMR in Gram-negative bacteria in Mali.
Of note, occurrence of variable in vitro susceptibility toward piperacillin-tazobactam rather than general in-vitro resistance in spite of blaCTX-M-15-carriage is the rule and not the exemption as repeatedly shown in international literature (Bradford, 2001; Titelman et al., 2011; Rosenkilde et al., 2019). This is also true for the observed only partial correlation of cotrimoxazole resistance with the observed resistance mediating genes. The dfrA gene group encodes trimethoprim-insensitive homologs of the bacterial dihydrofolate reductase with varying levels of residual trimethoprim susceptibility (Lombardo et al., 2016; Ambrose and Hall, 2021). The common dfrA1 gene, for example, has been associated with 1,000-fold reduced susceptibility toward trimethoprim (Lombardo et al., 2016). The also observed genes sul1 and sul2 encode variants of the dihydropteroate synthase in the folic acid pathway mediating insensitivity toward sulfonamides (Sköld, 2000). Of note, however, sul genes have been reported to show a tremendous variance of expression in E. coli as a fitness cost compensation mechanism, a phenomenon, which is likely to be associated with the observed variance of cotrimoxazole resistance in this study (Zhou et al., 2021).
The study has a number of limitations. First, we analyzed a small number of isolates in depth, which limits representativeness. Second, the samples were collected several years ago and may not reflect strains currently occurring in Mali. Nevertheless, the sequences of the studied isolates may be useful as reference in AMR surveillance. Third, since there was no initial screening for ESBL carriage, we cannot exclude completely that some isolates were imported by the deployed soldiers. However, frequent personnel turnover is typical for a military camp. There are several factors speaking in favor of the hypothesis that at least a considerable proportion of ESBL-positive E. coli had been acquired on deployment in Mali. The additional cgMLST-based relationship analysis we performed showed, with the exception of only one isolate (MLI124), an arrangement of the isolates from Mali with isolates from Niger and Ghana in a supercluster that supported their West African origin as probable. Within the time period when the assessed isolates had been collected, the civilian study group of Lübbert and colleagues reported enteric colonization with ESBL-positive Enterobacterales in about one out of three travel returnees from Western Africa, while the average prevalence in the European standard population was considerably lower at this time (Lübbert et al., 2015). The prevalence within the screened soldier population matched this ESBL prevalence of the returnee’s in Lübbert’s study nearly exactly, suggesting a comparable underlying epidemiological background. While we are well aware that the details of individual spatial–temporal acquisition of each isolate cannot be defined with certainty and even comparisons with sequences deposited in available databases might be misleading, because the isolation site of a bacterium is not necessarily identical with the initial transmission site, we nevertheless feel justified by the abovementioned epidemiological considerations to assume a Malian origin of at least a major proportion of the assessed isolates. The practical relevance of such estimations remains nevertheless debatable, because transmission events can occur in both directions and so, mutual transmission events both from Malian individuals to European soldiers and vice versa have to be assumed and the survival of the transmitted strains in the respective populations necessarily depended on the individual strain’s evolutionary fitness.
5. Conclusion
With this study, we extended the knowledge on the molecular epidemiology of AMR in Western African Mali and detected a high local prevalence of blaCTX-M-15. This underscores the need for interventions that tackle the worldwide problem of AMR.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/genbank/, CP116913; https://www.ncbi.nlm.nih.gov/genbank/, CP116911; https://www.ncbi.nlm.nih.gov/genbank/, CP116912; https://www.ncbi.nlm.nih.gov/genbank/, CP116916; https://www.ncbi.nlm.nih.gov/genbank/, CP116914; https://www.ncbi.nlm.nih.gov/genbank/, CP116915; https://www.ncbi.nlm.nih.gov/genbank/, CP116919; https://www.ncbi.nlm.nih.gov/genbank/, CP116917; https://www.ncbi.nlm.nih.gov/genbank/, CP116918; https://www.ncbi.nlm.nih.gov/genbank/, CP116923; https://www.ncbi.nlm.nih.gov/genbank/, CP116920; https://www.ncbi.nlm.nih.gov/genbank/, CP116921; https://www.ncbi.nlm.nih.gov/genbank/, CP116922; https://www.ncbi.nlm.nih.gov/genbank/, CP116925; https://www.ncbi.nlm.nih.gov/genbank/, CP116924; https://www.ncbi.nlm.nih.gov/genbank/, CP116929; https://www.ncbi.nlm.nih.gov/genbank/, CP116926; https://www.ncbi.nlm.nih.gov/genbank/, CP116927; https://www.ncbi.nlm.nih.gov/genbank/, CP116928; https://www.ncbi.nlm.nih.gov/genbank/, CP116931; https://www.ncbi.nlm.nih.gov/genbank/, CP116930; https://www.ncbi.nlm.nih.gov/genbank/, CP117049; https://www.ncbi.nlm.nih.gov/genbank/, CP117046; https://www.ncbi.nlm.nih.gov/genbank/, CP117047; https://www.ncbi.nlm.nih.gov/genbank/, CP117048; https://www.ncbi.nlm.nih.gov/genbank/, CP117020; https://www.ncbi.nlm.nih.gov/genbank/, CP117017; https://www.ncbi.nlm.nih.gov/genbank/, CP117018; https://www.ncbi.nlm.nih.gov/genbank/, CP117019; https://www.ncbi.nlm.nih.gov/genbank/, CP117053; https://www.ncbi.nlm.nih.gov/genbank/, CP117050; https://www.ncbi.nlm.nih.gov/genbank/, CP117051; https://www.ncbi.nlm.nih.gov/genbank/, CP117052; https://www.ncbi.nlm.nih.gov/genbank/, CP116987; https://www.ncbi.nlm.nih.gov/genbank/, CP116994; https://www.ncbi.nlm.nih.gov/genbank/, CP116988; https://www.ncbi.nlm.nih.gov/genbank/, CP116989; https://www.ncbi.nlm.nih.gov/genbank/, CP116990; https://www.ncbi.nlm.nih.gov/genbank/, CP116991; https://www.ncbi.nlm.nih.gov/genbank/, CP116992; https://www.ncbi.nlm.nih.gov/genbank/, CP116993; https://www.ncbi.nlm.nih.gov/genbank/, CP117001; https://www.ncbi.nlm.nih.gov/genbank/, CP116995; https://www.ncbi.nlm.nih.gov/genbank/, CP116996; https://www.ncbi.nlm.nih.gov/genbank/, CP116997; https://www.ncbi.nlm.nih.gov/genbank/, CP116998; https://www.ncbi.nlm.nih.gov/genbank/, CP116999; https://www.ncbi.nlm.nih.gov/genbank/, CP117000; https://www.ncbi.nlm.nih.gov/genbank/, CP117006; https://www.ncbi.nlm.nih.gov/genbank/, CP117002; https://www.ncbi.nlm.nih.gov/genbank/, CP117003; https://www.ncbi.nlm.nih.gov/genbank/, CP117004; https://www.ncbi.nlm.nih.gov/genbank/, CP117005; https://www.ncbi.nlm.nih.gov/genbank/, CP117008; https://www.ncbi.nlm.nih.gov/genbank/, CP117007; https://www.ncbi.nlm.nih.gov/genbank/, CP117010; https://www.ncbi.nlm.nih.gov/genbank/, CP117009; https://www.ncbi.nlm.nih.gov/genbank/, CP117013; https://www.ncbi.nlm.nih.gov/genbank/, CP117011; https://www.ncbi.nlm.nih.gov/genbank/, CP117012; https://www.ncbi.nlm.nih.gov/genbank/, CP117016; https://www.ncbi.nlm.nih.gov/genbank/, CP117014; https://www.ncbi.nlm.nih.gov/genbank/, CP117015; https://www.ncbi.nlm.nih.gov/genbank/, JANLNI000000000; https://www.ncbi.nlm.nih.gov/genbank/, JANLNI010000027.1; https://www.ncbi.nlm.nih.gov/genbank/, JANLNI010000050.1; and https://www.ncbi.nlm.nih.gov/genbank/, JANLNI010000064.1.
Author contributions
HF, RH, and AZ: conceptualization. AT, PM, MR, and SK: methodology. MR: software. MR, KH, SK, and AZ: validation. MR, KH, and AZ: formal analysis. KH, RH, and AZ: investigation. HF, AZ, and AK: resources. MR, KH, AZ, and HF: data curation. HF and AZ: writing—original draft preparation, supervision, and project administration. KH, MR, PM, AT, SK, AK, HF, RH, and AZ: writing—review and editing. MR and KH: visualization. AK: funding acquisition. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by the Deutsche Forschungsgemeinschaft (DFG; grant number ZA 697/6–1).
Acknowledgments
We are grateful to Nadja Schlüter, Francis Meier, and Sabine Jürgenfeld for excellent technical assistance.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1169829/full#supplementary-material
Footnotes
1. ^https://github.com/lh3/miniasm
2. ^https://github.com/xiaochuanle/NECAT
3. ^https://github.com/lbcb-sci/raven
References
Akpan, M. R., Isemin, N. U., Udoh, A. E., and Ashiru-Oredope, D. (2020). Implementation of antimicrobial stewardship programmes in African countries: a systematic literature review. J. Glob. Antimicrob. Resist. 22, 317–324. doi: 10.1016/j.jgar.2020.03.009
Ambrose, S. J., and Hall, R. M. (2021). dfrA trimethoprim resistance genes found in gram-negative bacteria: compilation and unambiguous numbering. J. Antimicrob. Chemother. 76, 2748–2756. doi: 10.1093/jac/dkab212
Arcilla, M. S., van Hattem, J. M., Haverkate, M. R., Bootsma, M. C. J., van Genderen, P. J. J., Goorhuis, A., et al. (2017). Import and spread of extended-spectrum β-lactamase-producing Enterobacteriaceae by international travellers (COMBAT study): a prospective, multicentre cohort study. Lancet Infect. Dis. 17, 78–85. doi: 10.1016/S1473-3099(16)30319-X
Bechmann, L., Böttger, R., Baier, C., Tersteegen, A., Bauer, K., Kaasch, A. J., et al. (2022). Serratia marcescens outbreak in a neonatal intensive care unit associated with contaminated donor milk. Infect. Control Hosp. Epidemiol. 5, 1–7. doi: 10.1017/ice.2022.187
Boisramé-Gastrin, S., Tandé, D., Münck, M. R., Gouriou, S., Nordmann, P., and Naas, T. (2011). Salmonella carriage in adopted children from Mali_ 2001-08. J. Antimicrob. Chemother. 66, 2271–2276. doi: 10.1093/jac/dkr307
Bradford, P. A. (2001). Extended-spectrum beta-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin. Microbiol. Rev. 14, 933–951. doi: 10.1128/CMR.14.4.933-951.2001
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421
Campbell, J. D., Kotloff, K. L., Sow, S. O., Tapia, M., Keita, M. M., Keita, T., et al. (2004). Invasive pneumococcal infections among hospitalized children in Bamako. Mali. Pediatr. Infect. Dis J. 23, 642–649. doi: 10.1097/01.inf.0000130951.85974.79
Carattoli, A., Zankari, E., García-Fernández, A., Voldby Larsen, M., Lund, O., Villa, L., et al. (2014). In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903. doi: 10.1128/AAC.02412-14
Denamur, E., Clermont, O., Bonacorsi, S., and Gordon, D. (2021). The population genetics of pathogenic Escherichia coli. Nat. Rev. Microbiol. 19, 37–54. doi: 10.1038/s41579-020-0416-x
Feldgarden, M., Brover, V., Gonzalez-Escalona, N., Frye, J. G., Haendiges, J., Haft, D. H., et al. (2021). AMRFinderPlus and the reference gene catalog facilitate examination of the genomic links among antimicrobial resistance, stress response, and virulence. Sci. Rep. 11:12728. doi: 10.1038/s41598-021-91456-0
Frickmann, H., Hagen, R. M., Geiselbrechtinger, F., and Hoysal, N. (2018a). Infectious diseases during the European Union training mission Mali (EUTM MLI)—a four-year experience. Mil. Med. Res. 5:19. doi: 10.1186/s40779-018-0166-5
Frickmann, H., Podbielski, A., and Kreikemeyer, B. (2018b). Resistant gram-negative Bacteria and diagnostic point-of-care options for the field setting during military operations. Biomed. Res. Int. 2018, 1–9. doi: 10.1155/2018/9395420
Frickmann, H., Warnke, P., Frey, C., Schmidt, S., Janke, C., Erkens, K., et al. (2015). Surveillance of food- and smear-transmitted pathogens in European soldiers with diarrhea on deployment in the tropics: experience from the European Union training Mission (EUTM) Mali. Biomed. Res. Int. 2015:573904. doi: 10.1155/2015/573904
Frickmann, H., Wiemer, D., Frey, C., Hagen, R. M., Hinz, R., Podbielski, A., et al. (2016). Low enteric colonization with multidrug-resistant pathogens in soldiers returning from deployments-experience from the years 2007-2015. PLoS One 11:e0162129. doi: 10.1371/journal.pone.0162129
Haft, D. H., DiCuccio, M., Badretdin, A., Brover, V., Chetvernin, V., O'Neill, K., et al. (2018). RefSeq: an update on prokaryotic genome annotation and curation. Nucleic Acids Res. 46, D851–D860. doi: 10.1093/nar/gkx1068
Hagen, R. M., Hinz, R., and Frickmann, H. (2015). β-Lactamases encoded by blaCTX-M Group I genes as determinants of resistance of ESBL-positive Enterobacteriaceae in European soldiers in tropical Mali. Eur. J. Microbiol. Immunol. 5, 281–284. doi: 10.1556/1886.2015.00037
Harrois, D., Breurec, S., Seck, A., Delauné, A., Le Hello, S., Pardos de la Gándara, M., et al. (2014). Prevalence and characterization of extended-spectrum β-lactamase-producing clinical Salmonella enterica isolates in Dakar, Senegal, from 1999 to 2009. Clin. Microbiol. Infect. 20, O109–O116. doi: 10.1111/1469-0691.12339
Jary, A., Teguete, I., Sidibé, Y., Kodio, A., Dolo, O., Burrel, S., et al. (2021). Prevalence of cervical HPV infection, sexually transmitted infections and associated antimicrobial resistance in women attending cervical cancer screening in Mali. Int. J. Infect. Dis. 108, 610–616. doi: 10.1016/j.ijid.2021.06.024
Joensen, K. G., Scheutz, F., Lund, O., Hasman, H., Kaas, R. S., Nielsen, E. M., et al. (2014). Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J. Clin. Microbiol. 52, 1501–1510. doi: 10.1128/JCM.03617-13
Johansson, M. H. K., Bortolaia, V., Tansirichaiya, S., Aarestrup, F. M., Roberts, A. P., and Petersen, T. N. (2021). Detection of mobile genetic elements associated with antibiotic resistance in Salmonella enterica using a newly developed web tool: MobileElementFinder. J. Antimicrob. Chemother. 76, 101–109. doi: 10.1093/jac/dkaa390
Jünemann, S., Sedlazeck, F. J., Prior, K., Albersmeier, A., John, U., Kalinowski, J., et al. (2013). Updating benchtop sequencing performance comparison. Nat. Biotechnol. 31, 294–296. doi: 10.1038/nbt.2522
Kantele, A., Lääveri, T., Mero, S., Vilkman, K., Pakkanen, S. H., Ollgren, J., et al. (2015). Antimicrobials increase travelers' risk of colonization by extended-spectrum betalactamase-producing Enterobacteriaceae. Clin. Infect. Dis. 60, 837–846. doi: 10.1093/cid/ciu957
Kolmogorov, M., Yuan, J., Lin, Y., and Pevzner, P. A. (2019). Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 37, 540–546. doi: 10.1038/s41587-019-0072-8
Koren, S., Walenz, B. P., Berlin, K., Miller, J. R., Bergman, N. H., and Phillippy, A. M. (2017). Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27, 722–736. doi: 10.1101/gr.215087.116
Li, W., O'Neill, K. R., Haft, D. H., DiCuccio, M., Chetvernin, V., Badretdin, A., et al. (2021). RefSeq: expanding the prokaryotic genome annotation pipeline reach with protein family model curation. Nucleic Acids Res. 49, D1020–D1028. doi: 10.1093/nar/gkaa1105
Lombardo, M. N., Dayanandan, N., Wright, D. L., and Anderson, A. C. (2016). Crystal structures of trimethoprim-resistant DfrA1 rationalize potent inhibition by Propargyl-linked Antifolates. ACS Infect. Dis. 2, 149–156. doi: 10.1021/acsinfecdis.5b00129
Lübbert, C., Straube, L., Stein, C., Makarewicz, O., Schubert, S., Mössner, J., et al. (2015). Colonization with extended-spectrum beta-lactamase-producing and carbapenemase-producing Enterobacteriaceae in international travelers returning to Germany. Int. J. Med. Microbiol. 305, 148–156. doi: 10.1016/j.ijmm.2014.12.001
Maaßen, W., Frey, C., Frickmann, H., and Erkens, K. (2017). Tropenmediziner der Bundeswehr: Einsatzerfahrungen in Mali. Flugmed. Tropenmed. Reisemed. 24, 81–88. doi: 10.1055/s-0043-105251
Malberg Tetzschner, A. M., Johnson, J. R., Johnston, B. D., Lund, O., and Scheutz, F. (2020). In silico genotyping of Escherichia coli isolates for Extraintestinal virulence genes by use of whole-genome sequencing data. J. Clin. Microbiol. 58, e01269–e01220. doi: 10.1128/JCM.01269-20
Meli, H., Cissoko, Y., Konaté, I., Soumaré, M., Fofana, A., Dembélé, J. P., et al. (2020). Tuberculosis and HIV coinfection complicated by nosocomial infection caused by Klebsiella pneumoniae: about 4 cases in a Department of Infectious diseases in Mali. Pan Afr. Med. J. 37:141. doi: 10.11604/pamj.2020.37.141.22716
Mshana, S. E., Hain, T., Domann, E., Lyamuya, E. F., Chakraborty, T., and Imirzalioglu, C. (2013). Predominance of Klebsiella pneumoniae ST14 carrying CTX-M-15 causing neonatal sepsis in Tanzania. BMC Infect. Dis. 13:466. doi: 10.1186/1471-2334-13-466
Nkansa-Gyamfi, N. A., Kazibwe, J., Traore, D. A. K., and Nji, E. (2019). Prevalence of multidrug-, extensive drug-, and pandrug-resistant commensal Escherichia coli isolated from healthy humans in community settings in low- and middle-income countries: a systematic review and meta-analysis. Glob. Health Action 12:1815272. doi: 10.1080/16549716.2020.1815272
Onduru, O. G., Mkakosya, R. S., Aboud, S., and Rumisha, S. F. (2021). Genetic determinants of resistance among ESBL-producing Enterobacteriaceae in community and hospital settings in east, central, and southern Africa: A systematic review and Meta-analysis of prevalence. Can. J. Infect. Dis. Med. Microbiol. 2021, 1–9. doi: 10.1155/2021/5153237
Pankok, F., Fuchs, F., Loderstädt, U., Kaase, M., Balczun, C., Scheithauer, S., et al. (2022a). Molecular epidemiology of Escherichia coli with resistance against third-generation Cephalosporines isolated from deployed German soldiers-A retrospective assessment after deployments to the African Sahel region and other sites between 2007 and 2016. Microorganisms 10:2448. doi: 10.3390/microorganisms10122448
Pankok, F., Taudien, S., Dekker, D., Thye, T., Oppong, K., Wiafe Akenten, C., et al. (2022b). Epidemiology of plasmids in Escherichia coli and Klebsiella pneumoniae with acquired extended Spectrum Beta-lactamase genes isolated from chronic wounds in Ghana. Antibiotics 11:689. doi: 10.3390/antibiotics11050689
Pécoul, B., Varaine, F., Keita, M., Soga, G., Djibo, A., Soula, G., et al. (1991). Long-acting chloramphenicol versus intravenous ampicillin for treatment of bacterial meningitis. Lancet 338, 862–866. doi: 10.1016/0140-6736(91)91511-R
Pehrsson, E. C., Tsukayama, P., Patel, S., Mejía-Bautista, M., Sosa-Soto, G., Navarrete, K. M., et al. (2016). Interconnected microbiomes and resistomes in low-income human habitats. Nature 533, 212–216. doi: 10.1038/nature17672
Robertson, J., and Nash, J. H. E. (2018). MOB-suite: software tools for clustering, reconstruction and typing of plasmids from draft assemblies. Microb. Genom. 4:e000206. doi: 10.1099/mgen.0.000206
Rosenkilde, C. E. H., Munck, C., Porse, A., Linkevicius, M., Andersson, D. I., and Sommer, M. O. A. (2019). Collateral sensitivity constrains resistance evolution of the CTX-M-15 β-lactamase. Nat. Commun. 10:618. doi: 10.1038/s41467-019-08529-y
Ruimy, R., Maiga, A., Armand-Lefevre, L., Maiga, I., Diallo, A., Koumaré, A. K., et al. (2008). The carriage population of Staphylococcus aureus from Mali is composed of a combination of pandemic clones and the divergent Panton-valentine leukocidin-positive genotype ST152. J. Bacteriol. 190, 3962–3968. doi: 10.1128/JB.01947-07
Ruppé, E., Armand-Lefèvre, L., Estellat, C., Consigny, P. H., El Mniai, A., Boussadia, Y., et al. (2015). High rate of acquisition but short duration of carriage of multidrug-resistant Enterobacteriaceae after travel to the tropics. Clin. Infect. Dis. 61, 593–600. doi: 10.1093/cid/civ333
Sangare, S. A., Maiga, A. I., Guindo, I., Maiga, A., Camara, N., Dicko, O. A., et al. (2016). Prevalence of ESBL-producing Enterobacteriaceae isolated from blood cultures in Mali. J. Infect. Dev. Ctries. 10, 1059–1064. doi: 10.3855/jidc.7536
Sangare, S. A., Maiga, A. I., Guindo, I., Maiga, A., Camara, N., Savadogo, S., et al. (2015). Prevalence of extended-spectrum beta-lactamase-producing Enterobacteriaceae isolated from blood cultures in Africa. Med. Mal. Infect. 45, 374–382. doi: 10.1016/j.medmal.2015.08.003
Sangaré, S. A., Maïga, A. I., Maïga, A., Diallo, S., Camara, N., Savadogo, S., et al. (2017). Prevalence of extended-spectrum beta-lactamase phenotypes in enterobacteria isolated from blood cultures of patients at admission to the University Hospital of Bamako. Med. Sante Trop. 27, 170–175. doi: 10.1684/mst.2017.0681
Sangare, S. A., Rondinaud, E., Maataoui, N., Maiga, A. I., Guindo, I., Maiga, A., et al. (2017). Very high prevalence of extended-spectrum beta-lactamase-producing Enterobacteriaceae in bacteriemic patients hospitalized in teaching hospitals in Bamako. PLoS One 12:e0172652. doi: 10.1371/journal.pone.0172652
Sköld, O. (2000). Sulfonamide resistance: mechanisms and trends. Drug Resist. Updat. 3, 155–160. doi: 10.1054/drup.2000.0146
Sommer, M. O. A., Dantas, G., and Church, G. M. (2009). Functional characterization of the antibiotic resistance reservoir in the human microflora. Science 325, 1128–1131. doi: 10.1126/science.1176950
Souvorov, A., Agarwala, R., and Lipman, D. J. (2018). SKESA: strategic k-mer extension for scrupulous assemblies. Genome Biol. 19:153. doi: 10.1186/s13059-018-1540-z
Sow, S. O., Diallo, S., Campbell, J. D., Tapia, M. D., Keita, T., Keita, M. M., et al. (2005). Burden of invasive disease caused by Haemophilus influenzae type b in Bamako, Mali_ impetus for routine infant immunization with conjugate vaccine. Pediatr. Infect. Dis. J. 24, 533–537. doi: 10.1097/01.inf.0000164768.28135.0d
Tandé, D., Boisramé-Gastrin, S., Münck, M. R., Héry-Arnaud, G., Gouriou, S., Jallot, N., et al. (2010). Intrafamilial transmission of extended-spectrum-beta-lactamase-producing Escherichia coli and Salmonella enterica Babelsberg among the families of internationally adopted children. J. Antimicrob. Chemother. 65, 859–865. doi: 10.1093/jac/dkq068
Tandé, D., Jallot, N., Bougoudogo, F., Montagnon, T., Gouriou, S., and Sizun, J. (2009). Extended-spectrum beta-lactamase-producing Enterobacteriaceae in a Malian orphanage. Emerg. Infect. Dis. 15, 472–474. doi: 10.3201/eid1503.071637
Tatusova, T., DiCuccio, M., Badretdin, A., Chetvernin, V., Nawrocki, E. P., Zaslavsky, L., et al. (2016). NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 44, 6614–6624. doi: 10.1093/nar/gkw569
Tisdale, M. D., Tribble, D. R., Mitra, I., Telu, K., Kuo, H. C., Fraser, J. A., et al. (2022). TaqMan Array card testing of participant-collected stool smears to determine the pathogen-specific epidemiology of travellers' diarrhoea. J. Travel Med. 29:taab138. doi: 10.1093/jtm/taab138
Titelman, E., Iversen, A., Kahlmeter, G., and Giske, C. G. (2011). Antimicrobial susceptibility to parenteral and oral agents in a largely polyclonal collection of CTX-M-14 and CTX-M-15-producing Escherichia coli and Klebsiella pneumoniae. APMIS 119, 853–863. doi: 10.1111/j.1600-0463.2011.02766.x
Tompkins, K., Juliano, J. J., and van Duin, D. (2021). Antimicrobial resistance in Enterobacterales and its contribution to Sepsis in sub-saharan Africa. Front. Med. 8:615649. doi: 10.3389/fmed.2021.615649
Weber, R. E., Pietsch, M., Frühauf, A., Pfeifer, Y., Martin, M., Luft, D., et al. (2019). IS26-mediated transfer of blaNDM-1 as the Main route of resistance transmission during a polyclonal, multispecies outbreak in a German hospital. Front. Microbiol. 10:2817. doi: 10.3389/fmicb.2019.02817
Wick, R. R., and Holt, K. E. (2019). Benchmarking of long-read assemblers for prokaryote whole genome sequencing. F1000Res 8:2138. doi: 10.12688/f1000research.21782.4
Wick, R. R., and Holt, K. E. (2022). Polypolish: short-read polishing of long-read bacterial genome assemblies. PLoS Comput. Biol. 18:e1009802. doi: 10.1371/journal.pcbi.1009802
Zankari, E., Allesøe, R., Joensen, K. G., Cavaco, L. M., Lund, O., and Aarestrup, F. M. (2017). PointFinder: a novel web tool for WGS-based detection of antimicrobial resistance associated with chromosomal point mutations in bacterial pathogens. J. Antimicrob. Chemother. 72, 2764–2768. doi: 10.1093/jac/dkx217
Zankari, E., Hasman, H., Cosentino, S., Vestergaard, M., Rasmussen, S., Lund, O., et al. (2012). Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67, 2640–2644. doi: 10.1093/jac/dks261
Keywords: extended spectrum beta lactamase, ESBL, Enterobacterales, antibiotic resistance, surveillance, Sub-Saharan Africa, Mali, military personnel
Citation: Hoffmann K, Riediger M, Tersteegen A, Marquardt P, Kahlfuß S, Kaasch AJ, Hagen RM, Frickmann H and Zautner AE (2023) Molecular epidemiology of enterically colonizing Escherichia coli with resistance against third-generation cephalosporins isolated from stool samples of European soldiers with concomitant diarrhea on deployment in Western African Mali. Front. Microbiol. 14:1169829. doi: 10.3389/fmicb.2023.1169829
Edited by:
Shuchao Wang, Chinese Academy of Agricultural Sciences, ChinaReviewed by:
Torahiko Okubo, Hokkaido University, JapanInga Eichhorn, Robert Koch Institute (RKI), Germany
David Tribble, Uniformed Services University of the Health Sciences, United States
Copyright © 2023 Hoffmann, Riediger, Tersteegen, Marquardt, Kahlfuß, Kaasch, Hagen, Frickmann and Zautner. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Andreas E. Zautner, YXphdXRuZUBnd2RnLmRl
†These authors have contributed equally to this work
 Katharina Hoffmann1†
Katharina Hoffmann1† Matthias Riediger
Matthias Riediger Aljoscha Tersteegen
Aljoscha Tersteegen Pauline Marquardt
Pauline Marquardt Sascha Kahlfuß
Sascha Kahlfuß Hagen Frickmann
Hagen Frickmann Andreas E. Zautner
Andreas E. Zautner