
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Microbiol. , 07 June 2023
Sec. Evolutionary and Genomic Microbiology
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1168246
This article is part of the Research Topic Insights in Evolutionary & Genomic Microbiology: 2022 View all 11 articles
A variety of bacteria in the environment can utilize xenobiotic compounds as a source of carbon and energy. The bacterial strains degrading xenobiotics are suitable models to investigate the adaptation and evolutionary processes of bacteria because they appear to have emerged relatively soon after the release of these compounds into the natural environment. Analyses of bacterial genome sequences indicate that horizontal gene transfer (HGT) is the most important contributor to the bacterial evolution of genetic architecture. Further, host bacteria that can use energy effectively by controlling the expression of organized gene clusters involved in xenobiotic degradation will have a survival advantage in harsh xenobiotic-rich environments. In this review, we summarize the current understanding of evolutionary mechanisms operative in bacteria, with a focus on biphenyl/PCB-degrading bacteria. We then discuss metagenomic approaches that are useful for such investigation.
Xenobiotic compounds are man-made chemicals that are present at unnaturally high concentrations in the natural environment. A variety of bacteria in the environment can utilize various xenobiotic compounds as a source of carbon and energy (Van der Meer et al., 1992). Phylogenetically unrelated bacterial strains often share similar metabolic pathways and enzyme systems for the degradation of xenobiotic compounds. It is believed that bacteria have acquired the ability to degrade even xenobiotic compounds they have never encountered (Nagata et al., 2015; Jeffries et al., 2018; Miglani et al., 2022). The bacterial strains degrading xenobiotics are suitable models to investigate the adaptation and evolutionary processes of bacteria because they appear to have emerged relatively soon after the release of these compounds into the natural environment.
Polychlorinated biphenyls (PCBs) are xenobiotic compounds in which the aromatic biphenyl carbon skeleton contains between one and 10 chlorine atoms. The high chemical stability, superhydrophobicity, and toxicity of PCBs make them some of the most serious and persistent environmental pollutants (Abraham et al., 2002; Furukawa and Fujihara, 2008). It is therefore somewhat surprising that many microbes that are capable of degrading PCBs have been identified. A number of biphenyl-utilizing bacteria with the ability to degrade PCBs have been isolated and characterized (Pieper and Seeger, 2008). Lignin is a complex substance with a phenylpropane structure; it contains various biphenyl molecules and is widely distributed throughout the earth. Biphenyl-degrading bacteria are thought to be responsible for the final stage of lignin degradation (Iram et al., 2021). Since biphenyl dioxygenase, the first enzyme in the biphenyl catabolic pathway, hydroxylates plant-derived flavonoids (Zubrova et al., 2021) and its homologous enzyme oxidizes dehydroabietic acid (Witzig et al., 2007), a biphenyl-degrading pathway might be involved in the degradation of plant secondary metabolites other than lignin. Thus, the ancestry of biphenyl-utilizing bacteria and their catabolic genes is quite ancient, and the genes may be distributed across a wide range of bacteria. Further, they would have the potential to adapt to different aromatic compounds, including PCBs. Therefore, the biphenyl/PCB-degradation system in bacteria appear to be a suitable model for the study of microbial adaptive evolution.
Recently, many bacterial genomes and metagenomes derived from environments contaminated with xenobiotic compounds have been analyzed at an accelerated pace (Garrido-Sanz et al., 2018; Hirose et al., 2019; Miglani et al., 2022). Much evidence has been found to support the idea that different biphenyl/PCB-degrading bacteria have evolved in the environment through different processes. In this review manuscript, we summarize the diversity, recruitment, and expression of degradation genes for biphenyl/PCB, shedding light on the sophistication of degradation gene systems and the adaptive evolution of these host bacteria.
Mobilization of the catabolic genes in bacteria can be accomplished through a variety of mobile genetic elements (MGEs), including plasmid, transposon, and integrative and conjugative elements (ICEs). These genes are modified and rearranged in different ways in host bacterial cells. Degradation gene clusters for biphenyl/PCB (bph) are often located on MGEs, and can be transferred between bacterial cells, conferring degradation capacity to non-degrading bacteria (Van der Meer et al., 1992; Tsuda et al., 1999; Springael and Top, 2004; Satola et al., 2013).
Plasmids are mobile genetic elements that facilitate rapid adaptation and evolution by conjugative transfer between bacterial cells in the environment (Smillie et al., 2010; Aminov, 2011; Rodríguez-Beltrán et al., 2020). Catabolic plasmids contain the complete set of genes encoding the enzymes for the degradation of a xenobiotic compound. They are relatively large (more than 50 kb), due to the presence of numerous insertion sequences (ISs) and transposons. The important characteristic of catabolic plasmids is incompatibility. That is, plasmids are classified into incompatibility (Inc) groups based on their replication and partitioning systems; two plasmids of the same group cannot replicate in the same cell and are considered incompatible (Top and Springael, 2003; Popowska and Krawczyk-Balska, 2013; Shintani et al., 2015). Genes for xenobiotic degradation are often found on broad-host-range IncP-1 plasmids, such as pSS60 (Burlage et al., 1990), pBRC60 (Burlage et al., 1990) and pJP4 (Newby et al., 2000).
A variety of PCB-degrading phenotypes have also been attributed to catabolic plasmids (Furukawa and Fujihara, 2008). The bph genes of thermophilic Geobacillus sp. strain JF8 are located on a plasmid pBt40 (Mukerjee-dhar et al., 2005). Rhodococcus sp. RHA1 harbors large linear plasmids, including pRHL1 (1,100 kb), pRHL2 (450 kb), and pRHL3 (330 kb), and its bph genes are mainly located on the pRHL1 (Shimizu et al., 2001). Many other bph gene clusters have been identified on the linear mega-plasmids of different Rhodococcus species (Taguchi et al., 2004; Garrido-Sanz et al., 2020). Both the order and sequence of the bph genes have been shown to differ among rhodococci, and there is evidence of recombination around bph gene clusters, such as insertion of transpose (Taguchi et al., 2007). These findings suggest that these bph gene clusters evolved separately and were spread in rhodococci by horizontal transfer. Cupriavidus sp. SK-4, a PCB-degrading strain reported to utilize di-ortho-substituted biphenyl, was found to harbor a single plasmid: pSK4. Experimental results showed that pSK4 could be mobilized into Pseudomonas putida MB1335 and the PCB-degrading enzymes could be expressed in this strain (Ilori et al., 2015). Bacterial plasmids are important vehicles for horizontal gene transfer (Redondo-Salvo et al., 2020), and can therefore play a key role in the evolution of catabolic pathways and their hosts.
Transposons are defined segments of DNA that are able to move from one genetic location to another target location in the absence of any nucleotide sequence homology. Most bacterial transposable elements, including ISs and transposons, can be traditionally classified into three classes: class I, class II, and conjugative transposons (Tan, 1999; Tsuda et al., 1999). However, a revised classification system has been proposed wherein conjugative transposons, genomic islands, and integrative plasmids would be collectively called ICEs (Burrus and Waldor, 2004; Johnson and Grossman, 2015).
Bacterial class I elements include the simple ISs, which carry only the genetic determinants for transposition (usually transposase) and composite transposons, in which various genetic traits unrelated to transposition are flanked by two copies of very similar ISs in direct or inverted orientation (Ross et al., 2021). Bacterial class II transposons generally carry the genes for their transposition (transposase, and resolvase) and one or more phenotypic traits between their terminal inverted repeats (Ross et al., 2021). It has been reported that several bacterial class II transposons play a crucial role in the widespread distribution of various catabolic gene clusters, such as the cluster of genes encoding toluene, naphthalene, and carbazole degradation pathways (Tsuda et al., 1999; Sota et al., 2006; Nojiri, 2012). Only a few bacterial transposons carrying catabolic genes for biphenyl/PCB have been experimentally proven to be mobile. An example of a functional catabolic transposon is Tn5280, which was identified on plasmid pP51 of chlorobenzene-utilizing Pseudomonas sp. strain P51 (Tan, 1999; Top et al., 2002). Tn5280 transposes randomly at different chromosomal sites in P. putida KT2442 (van der Meer et al., 1991). This strongly suggests that some of the PCB degradation genes in strain P51 originated from a toluene or benzene degradation pathway, probably by HGT.
Integrative and conjugative elements (ICEs) are bacterial mobile genetic elements that are excised from the chromosome by site-specific recombination between the attachment ends (attL and attR) mediated by an ICE-specific integrase (Figure 1; Delavat et al., 2017). The excised ICE molecule undergoes single-strand cleavage at the origin of transfer (oriT). TraI (relaxase) binds to the oriT of the circular intermediate of the ICE, cleaves one strand and binds to the 5′ end, which is then recognized by the VirD4 complex and transported through the mating pair formation system (MPF) to a recipient cell. VirD4 and VirB4 are large (> 70 kDa) ATPase proteins, and VirB4 is a component of MPF (Christie et al., 2014). The double-stranded DNA is reconstituted and site-specifically recombines with the recipient’s attB attachment site on the chromosome to become re-integrated (Guglielmini et al., 2011). The integration sites in the genome of the recipient cell, such as the structural genes for tRNAs, have been reported for many conjugative transposons (Wozniak and Waldor, 2010; Delavat et al., 2017).
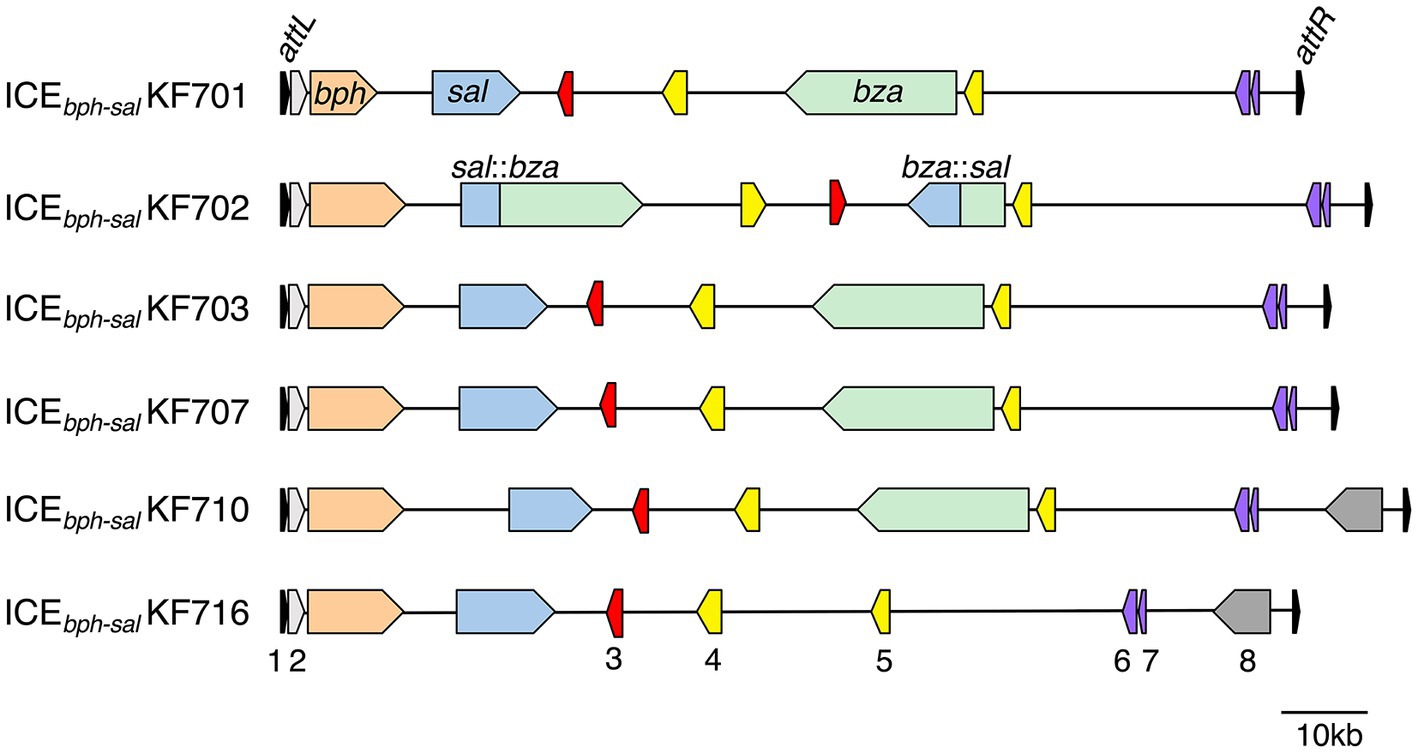
Figure 1. Organization of the ICEbph-sal in KF strains. ICEbph-salKF701, ICEbph-salKF702, ICEbph-salKF703, ICEbph-salKF707, and ICEbph-salKF710 carry the int gene, bph genes, sal genes, and bza genes. ICEbph-salKF716 carries the int gene, bph, and sal genes, but not the bza genes. The bza genes and sal genes in ICEbph-salKF702 are formed in a fusion gene. 1, tRNA-Gly (CCC) genes (partial) (black); 2, int genes (grey); 3, traI genes (red); 4, VirB4 components of the type IV secretory pathway (yellow); 5, a VirD4 component of the type IV secretory pathway (yellow); 6, parB genes (purple); 7, parA genes (purple); and 8, YbhFSR ABC-type transporter (dark grey). The attL (18 bp) site, including the 18 bp of the 3′ end of the tRNA-Gly gene, and attR (18 bp) site are indicated.
Although ICEs often carry cargo genes involved in pathogenicity, antibiotic resistance and heavy metal resistance to endow hosts with phenotypes beneficial for niche adaptation (Bellanger et al., 2014), they are also known to carry cargo genes for the catabolism of xenobiotic compounds (Hirose, 2023). ICEclc is a well-known ICE in xenobiotic biodegradation and carries cargo genes involved in the metabolism of chlorocatechol (clc genes) and aminophenol. The element was originally identified in the 3-chlorobenzoate-degrader Pseudomonas knackmussii B13 (Gaillard et al., 2006), and was almost identical to the ICEs inserted into the chromosome of Paraburkholderia xenovorans (formerly Burkholderia xenovorans) LB400 (ICEclc-LB400) (Chain et al., 2006) and P. aeruginosa JB2 (ICEclc-JB2) (Obi et al., 2018). P. xenovorans LB400 is also a well-characterized PCB-degrading bacteria, in which chlorobenzoate is formed during the degradation of PCB (Mondello, 1989). This bacterial strain provide insight into the roles of ICEs in the evolution of catabolic pathways for the biodegradation of chlorinated aromatic compounds. On degradative ICEs, catabolic gene products (e.g., bph, nah, and sal) that are related to the same substrate always share nearly 100% identity, whereas the sequences of the gene products of the transmission module exhibit variations (Mohapatra et al., 2022; Hirose, 2023). Although it has been suggested that there is a mechanism governing catabolic gene insertion and exchange on ICEs, there is little evidence to support this idea.
It is known that some biphenyl/PCB catabolic genes, bph, are horizontally transferred via ICEs. Several of the degradative ICEs carrying the bph gene belong to either of two groups, the Tn4371 family or ICEclc family, whose respective members share a common core region. The transfer module is required for the conjugal transfer from donor to recipient forming the type IV secretion system (T4SS) (Bellanger et al., 2014; Delavat et al., 2017). Many components required for conjugal transfer constitute a “core region” that is conserved among ICE family members. Tn4371 is the first ICE carrying bph genes and was found in the chromosome of Cupriavidus oxalacticus A5 (Springael et al., 1993). The chromosome of Acidovorax sp. strain KKS102 contains ICEKKS1024677 (Ohtsubo et al., 2012), which belongs to the Tn4371 family (Toussaint et al., 2003). ICEKKS1024677 is known to be transferred by conjugation to a wide range of bacteria across the genera via a circular intermediate. The bph genes of Cupriavidus basilensis KF708 and Commamonas testosteroni KF712 are also located on an ICE. The genes ICEbphKF708 and ICEbphKF712, which carry the bph genes of KF708 (Suenaga et al., 2015) and KF712 (Hirose et al., 2015a), are almost identical to ICEKKS1024677 and Tn4371, respectively. A new ICEclc family carrying bph genes and salicylic acid catabolic genes, sal, was found in the PCB-degrading strain Pseudomonas stutzeri KF716 (Hirose et al., 2015b). The ICEbph-salKF716 contains common core regions that show homology with those of ICEclc from P. knackmussii B13 (Gaillard et al., 2006) and ICEXTD from Azoarcus sp. CIB (Zamarro et al., 2016). A comparison of the genetic loci revealed that several putative ICEs from P. putida B6-2 (Tang et al., 2011), P. alcaliphila JAB1 (Ridl et al., 2018), P. stutzeri AN10 (Brunet-Galmés et al., 2012), and P. stutzeri 2A20 (Heinaru et al., 2016) had core regions highly conserved with those of ICEbph-salKF716, along with a variable region encoding the catabolic genes for phenol, naphthalene and biphenyl. ICEbph-salKF716 was reported to have been transferred from P. stutzeri KF716 to P. aeruginosa PAO1 via a circular extrachromosomal intermediate form (Hirose et al., 2021). These reports demonstrate that ICE subfamily members that share core regions highly conserved with those of ICEbph-salKF716 are widely distributed among aromatic-degrading bacteria.
In the past few years, several novel studies have provided insights into the diversity and evolution of biphenyl/PCB catabolic genes in the process of adaptation to environmental niches (Hirose et al., 2019, 2021). Furukawa et al. determined the whole genomes of 10 biphenyl/PCB-degrading bacterial strains (KF strains) isolated from a biphenyl-contaminated soil sample (Furukawa et al., 1989). Genome analyses revealed that all 10 strains had the bph genes, while seven strains also had sal genes (Figure 1). A series of ICEs named ICEbph-sals that were larger (more than 110 kb) than many other ICEs (Bellanger et al., 2014) contained highly conserved bph genes and sal genes (Hirose et al., 2019). Most of these ICEbph-sals possessed benzoate catabolic genes encoding the extradiol cleavage (bza) pathway. The fusion gene cluster of sal:bza and bza:sal was found in ICEbph-salKF702 (Fujihara et al., 2015), and was likely generated by homologous recombination between the sal and the bza genes.
ICEbph-sal of P. putida KF715 existed both in an extrachromosomal circular form (referred to as ICEbph-sal [circular] or pKF715A; hereinafter called pKF715A) and an integrated form in the chromosome (referred to as ICEbph-salKF715 [integrated]) in stationary phase culture (Suenaga et al., 2017). The ICEbph-sal KF715 was transferred at high frequency into P. putida AC30 and P. putida KT2440, and it was stably maintained in a circular form, pKF715A. The pKF715A in these transconjugant strains was further transferred into P. putida F39/D, and it existed in an integrated form in the chromosome. The structural features of bph and its flanking regions between KF701 and KF715 were almost identical, indicating that the bph-sal clusters were horizontally transferred to one another at this site between KF701 and KF715. Various mobile genes encoding transposases and a retron encoding retron-type reverse transcriptases have been shown to be inserted in the bph-sal clusters of ICEbph-sals. It has been hypothesized that these inserted sequences contribute to the exchange of the bph gene with the upper nah operon, the counterpart of the bph gene in the nah-sal cluster which is involved in naphthalene catabolism (Hirose et al., 2021; Mohapatra et al., 2022). These studies demonstrate that the bph gene clusters have integrative functions, are transferred among soil bacteria by various MGEs, including plasmids, transposons, and ICEs, and are diversified through modification.
The regulatory systems of aromatic compound-degradation genes are crucial evidence of the evolution of xenobiotic compound-degradation genes such as biphenyl, naphthalene, and phenol. In general, the expression of these degradation genes is controlled by one or more regulatory proteins (Diaz and Prieto, 2000). The regulatory protein binds to the effector, the substrate itself, or intermediates, thereby affecting the transcription of the operons. The many PCB catabolic operons in Gram-negative bacteria are regulated by regulatory proteins belonging to the GntR family (Merlin et al., 1997; Watanabe et al., 2000; Ohtsubo et al., 2001). In general, the regulators belonging to the GntR family acts as repressors (Tropel and van der Meer, 2004). The regulators bind to the operator region in the absence of substrates (biphenyl/PCBs) and inhibit the transcription of their operon (Figure 2). One such strain, KKS102, possesses a bph operon consisting of bphSEFGA1A2A3BCA4R. This operon is downregulated by a BphS belonging to the GntR family in the absence of HOPD (2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoic acid), an intermediate of biphenyl catabolism (Ohtsubo et al., 2001). The bph genes in Tn4371 isolated from C. oxalacticus A5 consisting of bphEFGA1A2A3BCD are also downregulated by BphS belonging to the GntR family (Merlin et al., 1997). These degrading microorganisms are thought to reduce energy consumption by suppressing the transcription of degradation genes in the absence of substrates.
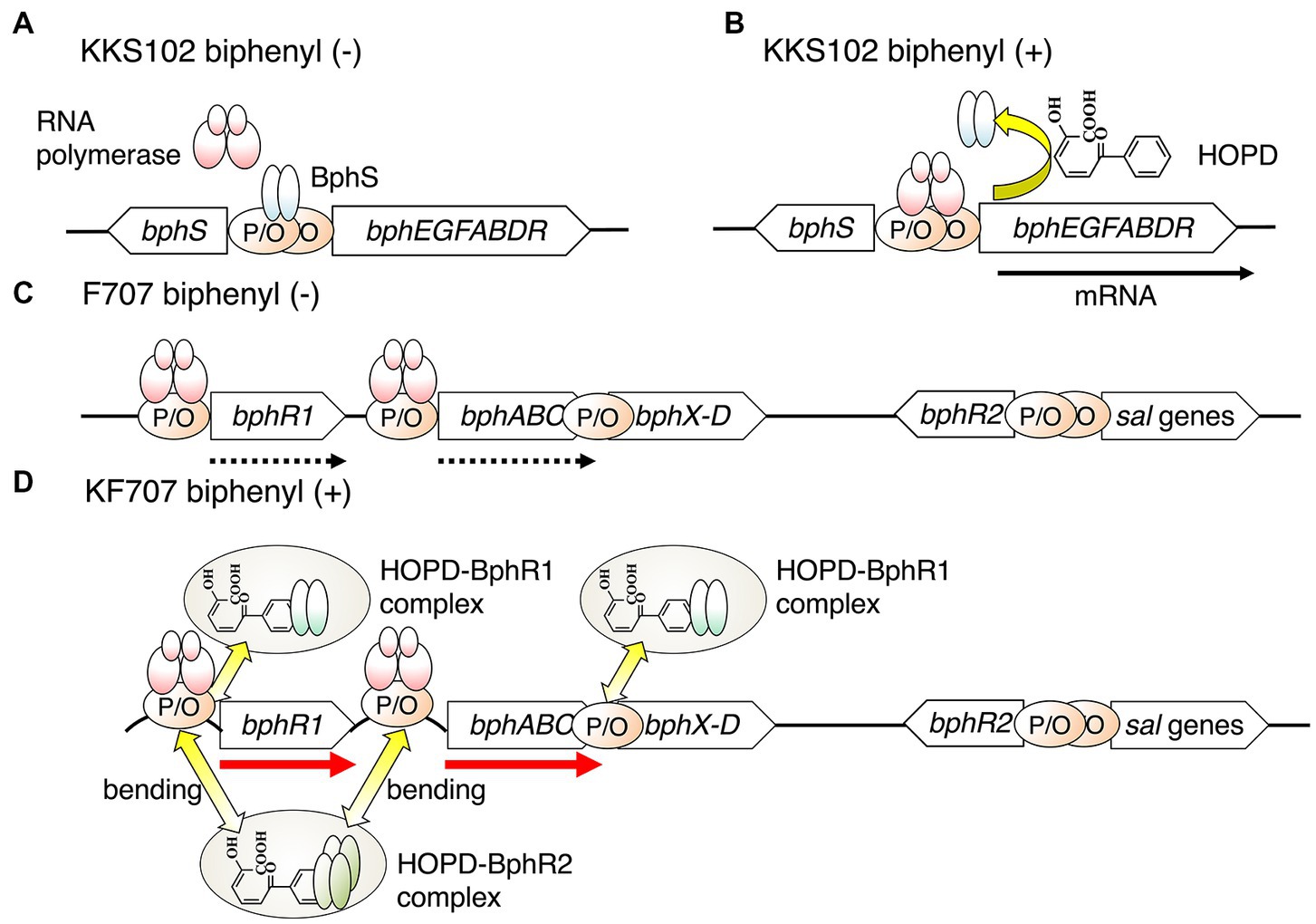
Figure 2. A transcriptional model of the bph gens in KKS102 in the absence (A) and in the presence (B) of biphenyl, and in KF707 in the absence (C) and in the presence of biphenyl (D). The black arrow shows induced transcription, the dotted arrow constitutive transcription, and the red arrow upregulated transcription. P, promoter; O, operator.
On the other hand, in Pseudomonas furukawaii KF707 (formerly P. pseudoalcaligenes KF707) (Kimura et al., 2018), a bph gene cluster consisting of bphR1A1A2A3A4BCX0X1X2X3D is upregulated by BphR1 (Watanabe et al., 2000). This regulator protein also belongs to the GntR family and acts as an activator having an effect opposite that of KKS102 (Ohtsubo et al., 2001). The details of this regulator protein will be described later. In addition, bph genes in P. xenovorans LB400 are also upregulated by ORF0, which belongs to the GntR family (Denef et al., 2004). This phenomenon is likely caused by the positional relationship of the promoter and operator of bph genes (Figure 2). In strain KKS102, the promoter and operator sequence of bphS exists between the bphS and bphEGFABDR genes, and thus the binding of BphS on the operator inhibits the binding of RNA polymerase on the promoter sequence of the bphEGFABDR genes (Ohtsubo et al., 2001). On the other hand, in strain KF707, the promoter and operator regions of the bphR1 and bphA genes do not overlap, and thus the binding of BphR1 on the bphR1 operator does not inhibit the binding of RNA polymerase. The binding of a complex of HOPD-BphR1 and HOPD-BphR2 on the operator could induce the bending of DNA molecules and enhance the binding of RNA polymerase on the promoter region of bph genes (Figure 2). Therefore, the expression of the bph operon is greatly enhanced in the presence of biphenyl in strain KF707 (Watanabe et al., 2000, 2003). Although the regulatory system for biphenyl/PCB degradation differs between strains KKS102 and KF707, in both cases the regulatory gene plays an important role in the efficient use of energy for growth and in the survival of the host in polluted environments.
The KF707 strain has another putative regulator, BphR2, that belongs to the LysR family and acts as an activator of the bph upper operon (bphR1 and bphABC) (Figure 2). The bphR2 gene is located far downstream (6.6 kb) of the bph genes and just upstream of the salicylate catabolic (sal) genes (Fujihara et al., 2006). BphR2 binds to the operator regions of bphR1 and bphA1 and activates the transcription of the bph upper operon. Interestingly, BphR2 also activates the transcription of sal genes in the presence of hydroxy muconate semi-aldehyde, the intermediate of salicylate. This regulatory behavior is the same as that of NahR, the regulator of the naphthalene catabolic (nah) operon. Actually, the bphR2 and sal genes of strain KF707 have high similarity to the sal genes of the nah gene cluster containing nahR on P. stutzeri AN10 (Bosch et al., 1999). This indicates that the bph-sal gene cluster of strain KF707 might have evolved through module exchange between the nah gene and bph gene in the nah-sal cluster.
Several investigations into the regulation of biphenyl/PCB-degrading Gram-positive bacteria have also been reported. In strain R. jostii, RHA1 possesses diverse biphenyl/PCB-degradation genes that encode multiple isozymes for each metabolism step and are distributed among multiple clusters (Takeda et al., 2004). The transcriptions of these genes are induced by dual regulatory systems. In the presence of biphenyl, a BphS1T1 two-component system induced five biphenyl-degrading gene clusters. And another two-component system, BphS2T2, also induced the biphenyl-degrading genes. However, in these cases the effector molecule was not biphenyl but other aromatic compounds such as ethylbenzene, benzene and so on (Takeda et al., 2010). And the transcriptions of bpd genes in another biphenyl/PCB-degrading gram-positive bacteria, Rhodococcus sp. strain M5, are regulated by a BpdST two-component system that is induced by the biphenyl (Labbé et al., 1997).
In order to survive under adverse conditions, bacteria transitorily induce the expression of particular genes to deal with environmental stresses. Tandem repeats (TRs) of short nucleotide sequences are often found in the intergenic regions in bacterial genomes (van Belkum et al., 1998). Several studies have reported that the rearrangement of TRs is involved in the gene expression of bacteria at either the transcriptional or translational level (Gemayel et al., 2010; Zhou et al., 2014).
Recently, a novel TR sequence, T(G/A)ACATG(A/C)T, and polymorphisms consisting of a number of repeats located in the region upstream of catechol 2,3-dioxygenase (C23O)-encoding genes were identified by metagenomic analysis (Suenaga et al., 2022). The level of protein expression of C23O dramatically increased as the number of TRs increased, reaching a maximum value with three and four repeats. Experimental results indicated that this nonanucleotide TR would affect the translational efficiency of the gene expression system. A metagenomic sample was collected from activated sludge used to treat industrial wastewater that contained mono- and polycyclic aromatic chemicals (Suenaga et al., 2009). C23O is a key enzyme in the degradation of aromatic compounds because catecholic compounds are the common intermediates in the degradation pathways (Figure 3; Hirose et al., 1994). In this harsh, aromatic-rich environment, the increase in C23O activity realized by adjusting the number of TRs is probably providing the host microbes a survival advantage. In fact, a metagenomic analysis demonstrated that, among the polymorphisms consisting of a number of repeats, tandem repeats of three and four, which indicate higher C23O enzyme activity, dominate in this environment (Suenaga et al., 2022).
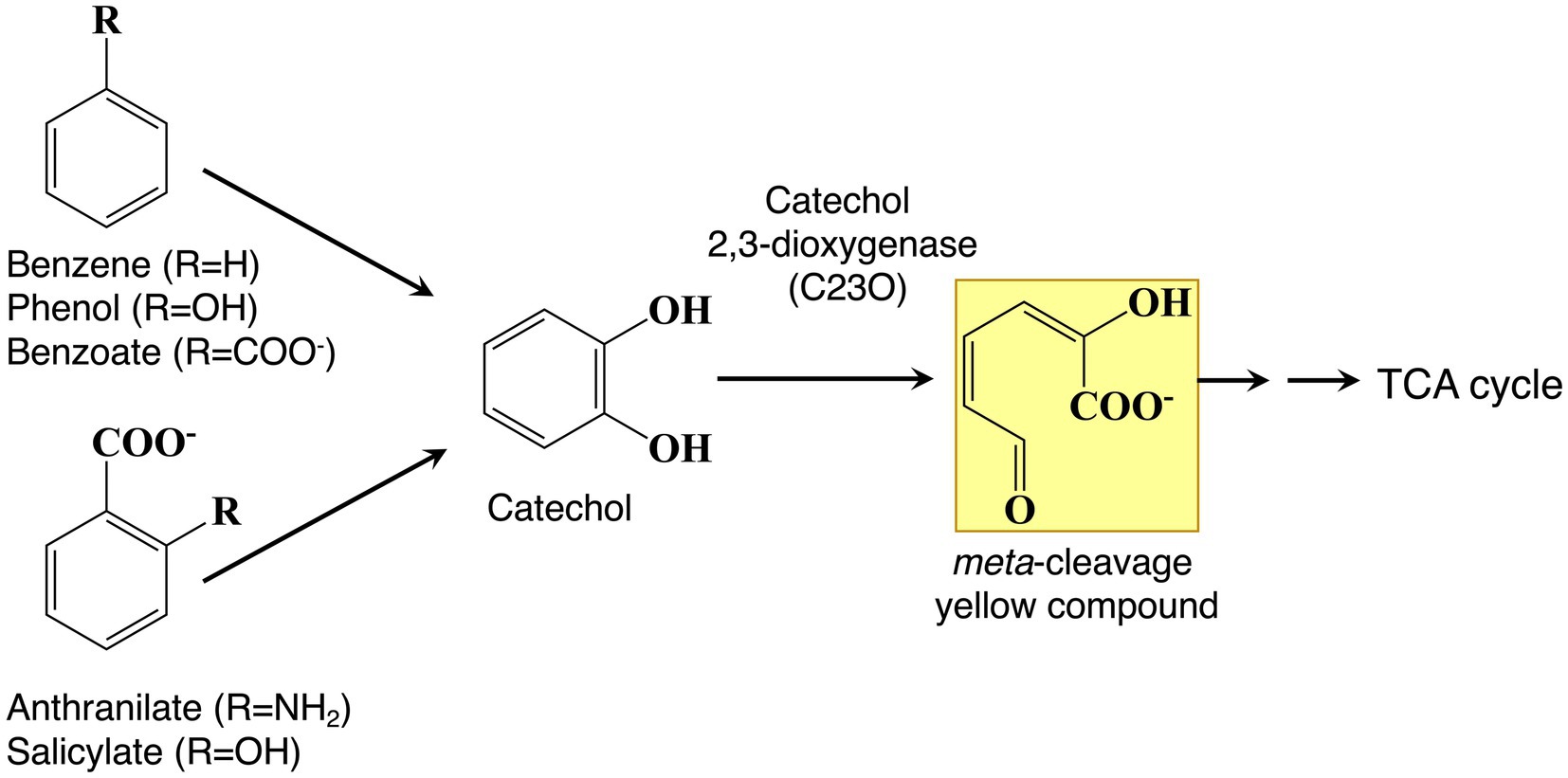
Figure 3. Bacterial degradation pathways of aromatic compounds. Aromatics are converted to catechol as a central intermediate. Catechol is cleaved by catechol 2,3-dioxygenase (C23O) to produce meta-cleavage yellow compounds, ultimately leading to TCA cycle intermediates.
In this review, based on the genome sequence of isolated xenobiotic compound- degrading bacteria, the current understanding of the evolutionary mechanisms occurring in degrading gene systems is presented. DNAs of particular interest, such as MGE, within the genomes of these bacterial strains have been a focus of recent studies and have been carefully analyzed. The selective collection of DNA molecules from the environment also seems to be efficient for the study of these evolutionary mechanisms. For instance, plasmids are a relatively undiscovered region of the genome and frequently contain genes that are essential for the survival of the host, such as genes involved in biodegradation and antibiotic resistance. Therefore, a metaplasmid or metamobilome study targeting total plasmid DNA in the environment is a more effective approach for understanding the content and composition of genes in microbial communities for key ecological processes (Suenaga, 2012; Alanin et al., 2021). The comprehensive acquisition of degradative genes by a metaplasmid approach at a single site will contribute to our understanding of their on-site adaptive evolution.
HS drafted the original manuscript. All authors critically reviewed and revised the manuscript draft and approved the final version for submission.
This work was performed as part of a project supported by JSPS KAKENHI and the Institution for Fermentation, Osaka (IFO).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abraham, W. R., Nogales, B., Golyshin, P. N., Pieper, D. H., and Timmis, K. N. (2002). Polychlorinated biphenyl-degrading microbial communities in soils and sediments. Curr. Opin. Microbiol. 5, 246–253. doi: 10.1016/S1369-5274(02)00323-5
Alanin, K. W. S., Jørgensen, T. S., Browne, P. D., Petersen, B., Riber, L., Kot, W., et al. (2021). An improved direct metamobilome approach increases the detection of larger-sized circular elements across kingdoms. Plasmid 115:102576. doi: 10.1016/j.plasmid.2021.102576
Aminov, R. I. (2011). Horizontal gene exchange in environmental microbiota. Front. Microbiol. 2:158. doi: 10.3389/fmicb.2011.00158
Bellanger, X., Payot, S., Leblond-Bourget, N., and Guédon, G. (2014). Conjugative and mobilizable genomic islands in bacteria: evolution and diversity. FEMS Microbiol. Rev. 38, 720–760. doi: 10.1111/1574-6976.12058
Bosch, R., García-Valdés, E., and Moore, E. R. (1999). Genetic characterization and evolutionary implications of a chromosomally encoded naphthalene-degradation upper pathway from Pseudomonas stutzeri AN10. Gene 236, 149–157. doi: 10.1016/S0378-1119(99)00241-3
Brunet-Galmés, I., Busquets, A., Peña, A., Gomila, M., Nogales, B., García-Valdés, E., et al. (2012). Complete genome sequence of the naphthalene-degrading bacterium Pseudomonas stutzeri AN10 (CCUG 29243). J. Bacteriol. 194, 6642–6643. doi: 10.1128/JB.01753-12
Burlage, R. S., Bemis, L. A., Layton, A. C., Sayler, G. S., and Larimer, F. (1990). Comparative genetic organization of incompatibility group P degradative plasmids. J. Bacteriol. 172, 6818–6825. doi: 10.1128/jb.172.12.6818-6825.1990
Burrus, V., and Waldor, M. K. (2004). Shaping bacterial genomes with integrative and conjugative elements. Res. Microbiol. 155, 376–386. doi: 10.1016/j.resmic.2004.01.012
Chain, P. S., Denef, V. J., Konstantinidis, K. T., Vergez, L. M., Agullo, L., Reyes, V. L., et al. (2006). Burkholderia xenovorans LB400 harbors a multi-replicon, 9.73-Mbp genome shaped for versatility. Proc. Natl. Acad. Sci. U. S. A. 103, 15280–15287. doi: 10.1073/pnas.0606924103
Christie, P. J., Whitaker, N., and González-Rivera, C. (2014). Mechanism and structure of the bacterial type IV secretion systems. Biochim. Biophys. Acta 1843, 1578–1591. doi: 10.1016/j.bbamcr.2013.12.019
Delavat, F., Miyazaki, R., Carraro, N., Pradervand, N., and van der Meer, J. R. (2017). The hidden life of integrative and conjugative elements. FEMS Microbiol. Rev. 41, 512–537. doi: 10.1093/femsre/fux0008
Denef, V. J., Park, J., Tsoi, T. V., Rouillard, J. M., Zhang, H., Wibbenmeyer, J. A., et al. (2004). Biphenyl and benzoate metabolism in a genomic context: outlining genome-wide metabolic networks in Burkholderia xenovorans LB400. Appl. Environ. Microbiol. 70, 4961–4970. doi: 10.1128/aem.70.8.4961-4970.2004
Diaz, E., and Prieto, M. A. A. (2000). Bacterial promoters triggering biodegradation of aromatic pollutants. Curr. Opin. Biotechnol. 11, 467–475. doi: 10.1016/S0958-1669(00)00126-9
Fujihara, H., Yamazoe, A., Hosoyama, A., Suenaga, H., Kimura, N., Hirose, J., et al. (2015). Draft genome sequence of Pseudomonas aeruginosa KF702 (NBRC 110665), a polychlorinated biphenyl-degrading bacterium isolated from biphenyl-contaminated soil. Genome Announc. 3, e00517–e00515. doi: 10.1128/genomeA.00517-15
Fujihara, H., Yoshida, H., Matsunaga, T., Goto, M., and Furukawa, K. (2006). Cross-regulation of biphenyl- and salicylate-catabolic genes by two regulatory systems in Pseudomonas pseudoalcaligenes KF707. J. Bacteriol. 188, 4690–4697. doi: 10.1128/JB.00329-06
Furukawa, K., and Fujihara, H. (2008). Microbial degradation of polychlorinated biphenyls: biochemical and molecular features. J. Biosci. Bioeng. 105, 433–449. doi: 10.1263/jbb.105.433
Furukawa, K., Hayase, N., Taira, K., and Tomizuka, N. (1989). Molecular relationship of chromosomal genes encoding biphenyl/poychlorinated biphenyl catabolism: some soil bacteria possess a highly conserved bph operon. J. Bacteriol. 171, 5467–5472. doi: 10.1128/jb.171.10.5467-5472.1989
Gaillard, M., Vallaeys, T., Vorhölter, F. J., Minoia, M., Werlen, C., Sentchilo, V., et al. (2006). The CLC element of Pseudomonas sp. strain B13, a genomic island with various catabolic properties. J. Bacteriol. 188, 1999–2013. doi: 10.1128/jb.188.5.1999-2013.2006
Garrido-Sanz, D., Manzano, J., Martín, M., Redondo-Nieto, M., and Rivilla, R. (2018). Metagenomic analysis of a biphenyl-degrading soil bacterial consortium reveals the metabolic roles of specific populations. Front. Microbiol. 9:232. doi: 10.3389/fmicb.2018.00232
Garrido-Sanz, D., Sansegundo-Lobato, P., Redondo-Nieto, M., Suman, J., Cajthaml, T., Blanco-Romero, E., et al. (2020). Analysis of the biodegradative and adaptive potential of the novel polychlorinated biphenyl degrader Rhodococcus sp. WAY2 revealed by its complete genome sequence. Microbial Gemomics 6:363. doi: 10.1099/mgen.0.000363
Gemayel, R., Vinces, M. D., Legendre, M., and Verstrepen, K. J. (2010). Variable tandem repeats accelerate evolution of coding and regulatory sequences. Annu. Rev. Genet. 44, 445–477. doi: 10.1146/annurev-genet-072610-155046
Guglielmini, J., Quintais, L., Garcillán-Barcia, M. P., de la Cruz, F., and Rocha, E. P. C. (2011). The repertoire of ICE in prokaryotes underscores the Unity, diversity, and ubiquity of conjugation. PLoS Genet. 7:e1002222. doi: 10.1371/journal.pgen.1002222
Heinaru, E., Naanuri, E., Grünbach, M., Jõesaar, M., and Heinaru, A. (2016). Functional redundancy in phenol and toluene degradation in Pseudomonas stutzeri strains isolated from the Baltic Sea. Gene 589, 90–98. doi: 10.1016/j.gene.2016.05.022
Hirose, J. (2023). Diversity and evolution of integrative and conjugative elements involved in bacterial aromatic compound degradation and their utility in environmental remediation. Microorganisms 11:438. doi: 10.3390/microorganisms11020438
Hirose, J., Fujihara, H., Watanabe, T., Kimura, N., Suenaga, H., Futagami, T., et al. (2019). Biphenyl/PCB degrading bph genes of ten bacterial strains isolated from biphenyl-contaminated soil in Kitakyushu, Japan: comparative and dynamic features as integrative conjugative elements (ICEs). Genes (Basel) 10:404. doi: 10.3390/genes10050404
Hirose, J., Kimura, N., Suyama, A., Kobayashi, A., Hayashida, S., and Furukawa, K. (1994). Functional and structural relationship of various extradiol aromatic ring-cleavage dioxygenases of Pseudomonas origin. FEMS Microbiol. Lett. 118, 273–277. doi: 10.1111/j.1574-6968.1994.tb06840.x
Hirose, J., Watanabe, T., Futagami, T., Fujihara, H., Kimura, N., Suenaga, H., et al. (2021). A new ICEclc subfamily integrative and conjugative element responsible for horizontal transfer of biphenyl and salicylic acid catabolic pathway in the PCB-degrading strain Pseudomonas stutzeri KF716. Microorganisms 9:2462. doi: 10.3390/microorganisms9122462
Hirose, J., Yamazoe, A., Hosoyama, A., Kimura, N., Suenaga, H., Watanabe, T., et al. (2015a). Draft genome sequence of the polychlorinated biphenyl-degrading bacterium Comamonas testosteroni KF712 (NBRC 110673). Genome Announc. 3, e01214–e01215. doi: 10.1128/genomeA.01214-15
Hirose, J., Yamazoe, A., Hosoyama, A., Kimura, N., Suenaga, H., Watanabe, T., et al. (2015b). Draft genome sequence of the polychlorinated biphenyl-degrading bacterium Pseudomonas stutzeri KF716 (NBRC 110668). Genome Announc. 3:e01215. doi: 10.1128/genomeA.01215-15
Ilori, M. O., Picardal, F. W., Aramayo, R., Adebusoye, S. A., Obayori, O. S., and Benedik, M. J. (2015). Catabolic plasmid specifying polychlorinated biphenyl degradation in Cupriavidus sp. strain SK-4: mobilization and expression in a pseudomonad. J. Basic Microbiol. 55, 338–345. doi: 10.1002/jobm.201200807
Iram, A., Berenjian, A., and Demirci, A. (2021). A review on the utilization of lignin as a fermentation substrate to produce lignin-modifying enzymes and other value-added products. Molecules 26:2960. doi: 10.3390/molecules26102960
Jeffries, T. C., Rayu, S., Nielsen, U. N., Lai, K., Ijaz, A., Nazaries, L., et al. (2018). Metagenomic functional potential predicts degradation rates of a model organophosphorus xenobiotic in pesticide contaminated soils. Front. Microbiol. 9:147. doi: 10.3389/fmicb.2018.00147
Johnson, C. M., and Grossman, A. D. (2015). Integrative and conjugative elements (ICEs): what they do and how they work. Annu. Rev. Genet. 49, 577–601. doi: 10.1146/annurev-genet-112414-055018
Kimura, N., Watanabe, T., Suenaga, H., Fujihara, H., Futagami, T., Goto, M., et al. (2018). Pseudomonas furukawaii sp. nov., a polychlorinated biphenyl-degrading bacterium isolated from biphenyl-contaminated soil in Japan. Int. J. Syst. Evol. Microbiol. 68, 1429–1435. doi: 10.1099/ijsem.0.002670
Labbé, D., Garnon, J., and Lau, P. C. (1997). Characterization of the genes encoding a receptor-like histidine kinase and a cognate response regulator from a biphenyl/polychlorobiphenyl-degrading bacterium, Rhodococcus sp. strain M5. J. Bacteriol. 179, 2772–2776. doi: 10.1128/jb.179.8.2772-2776.1997
Merlin, J. W. L. C., Springael, D., Mergeay, M., and Toussaint, A. (1997). Organisation of the bph gene cluster of transposon Tn4371, encoding enzymes for the degradation of biphenyl and 4-chlorobiphenyl compounds. Mol. Gen. Genet. MGG 253, 499–506. doi: 10.1007/s004380050349
Miglani, R., Parveen, N., Kumar, A., Ansari, M. A., Khanna, S., Rawat, G., et al. (2022). Degradation of xenobiotic pollutants: an environmentally sustainable approach. Meta 12:818. doi: 10.3390/metabo12090818
Mohapatra, B., Malhotra, H., and Phale, P. S. (2022). Life within a contaminated niche: comparative genomic analyses of an integrative conjugative element ICEnahCSV86 and two genomic islands from Pseudomonas bharatica CSV86T suggest probable role in colonization and adaptation. Front. Microbiol. 13:928848. doi: 10.3389/fmicb.2022.928848
Mondello, F. J. (1989). Cloning and expression in Escherichia coli of Pseudomonas strain LB400 genes encoding polychlorinated biphenyl degradation. J. Bacteriol. 171, 1725–1732. doi: 10.1128/jb.171.3.1725-1732.1989
Mukerjee-dhar, G., Shimura, M., Miyazawa, D., Kimbara, K., and Hatta, T. (2005). Bph genes of the thermophilic PCB degrader, Bacillus sp. JF8: characterization of the divergent ring-hydroxylating dioxygenase and hydrolase genes upstream of the Mn-dependent BphC. Microbiology 151, 4139–4151. doi: 10.1099/mic.0.28437-0
Nagata, Y., Ohtsubo, Y., and Tsuda, M. (2015). Properties and biotechnological applications of natural and engineered haloalkane dehalogenases. Appl. Microbiol. Biotechnol. 99, 9865–9881. doi: 10.1007/s00253-015-6954-x
Newby, D. T., Josephson, K. L., and Pepper, I. L. (2000). Detection and characterization of plasmid pJP4 transfer to indigenous soil bacteria. Appl. Environ. Microbiol. 66, 290–296. doi: 10.1128/aem.66.1.290-296.2000
Nojiri, H. (2012). Structural and molecular genetic analyses of the bacterial carbazole degradation system. Biosci. Biotechnol. Biochem. 76, 1–18. doi: 10.1271/bbb.110620
Obi, C. C., Vayla, S., de Gannes, V., Berres, M. E., Walker, J., Pavelec, D., et al. (2018). The integrative conjugative element clc (ICEclc) of Pseudomonas aeruginosa JB2. Front. Microbiol. 9:1532. doi: 10.3389/fmicb.2018.01532
Ohtsubo, Y., Delawary, M., Kimbara, K., Takagi, M., Ohta, A., and Nagata, Y. (2001). BphS, a key transcriptional regulator of bph genes involved in polychlorinated biphenyl/biphenyl degradation in Pseudomonas sp. KKS102. J. Biol. Chem. 276, 36146–36154. doi: 10.1074/jbc.M100302200
Ohtsubo, Y., Ishibashi, Y., Naganawa, H., Hirokawa, S., Atobe, S., Nagata, Y., et al. (2012). Conjugal transfer of polychlorinated biphenyl/biphenyl degradation genes in acidovorax sp. strain KKS102, which are located on an integrative and conjugative element. J. Bacteriol. 194, 4237–4248. doi: 10.1128/JB.00352-12
Pieper, D. H., and Seeger, M. (2008). Bacterial metabolism of polychlorinated biphenyls. J. Mol. Microbiol. Biotechnol. 15, 121–138. doi: 10.1159/000121325
Popowska, M., and Krawczyk-Balska, A. (2013). Broad-host-range IncP-1 plasmids and their resistance potential. Front. Microbiol. 4:44. doi: 10.3389/fmicb.2013.00044
Redondo-Salvo, S., Fernández-López, R., Ruiz, R., Vielva, L., de Toro, M., Rocha, E. P. C., et al. (2020). Pathways for horizontal gene transfer in bacteria revealed by a global map of their plasmids. Nat. Commun. 11:3602. doi: 10.1038/s41467-020-17278-2
Ridl, J., Suman, J., Fraraccio, S., Hradilova, M., Strejcek, M., Cajthaml, T., et al. (2018). Complete genome sequence of Pseudomonas alcaliphila JAB1 (=DSM 26533), a versatile degrader of organic pollutants. Stand. Genomic Sci. 13:3. doi: 10.1186/s40793-017-0306-7
Rodríguez-Beltrán, J., Sørum, V., Toll-Riera, M., de la Vega, C., Peña-Miller, R., and San Millán, Á. (2020). Genetic dominance governs the evolution and spread of mobile genetic elements in bacteria. Proc. Natl. Acad. Sci. U. S. A. 117, 15755–15762. doi: 10.1073/pnas.2001240117
Ross, K., Varani, A. M., Snesrud, E., Huang, H., Alvarenga, D. O., Zhang, J., et al. (2021). TnCentral: a prokaryotic transposable element database and web portal for transposon analysis. mBio 12, e02060–e02021. doi: 10.1128/mBio.02060-21
Satola, B., Wübbeler, J. H., and Steinbüchel, A. (2013). Metabolic characteristics of the species Variovorax paradoxus. Appl. Microbiol. Biotechnol. 97, 541–560. doi: 10.1007/s00253-012-4585-z
Shimizu, S., Kobayashi, H., Masai, E., and Fukuda, M. (2001). Characterization of the 450-kb linear plasmid in a polychlorinated biphenyl degrader, Rhodococcus sp. strain RHA1. Appl. Environ. Microbiol. 67, 2021–2028. doi: 10.1128/AEM.67.5.2021-2028.2001
Shintani, M., Sanchez, Z. K., and Kimbara, K. (2015). Genomics of microbial plasmids: classification and identification based on replication and transfer systems and host taxonomy. Front. Microbiol. 6, 1–16. doi: 10.3389/fmicb.2015.00242
Smillie, C., Garcillán-Barcia, M. P., Francia, M. V., Rocha, E. P., and de la Cruz, F. (2010). Mobility of plasmids. Microbiol. Mol. Biol. Rev. 74, 434–452. doi: 10.1128/mmbr.00020-10
Sota, M., Yano, H., Ono, A., Miyazaki, R., Ishii, H., Genka, H., et al. (2006). Genomic and functional analysis of the IncP-9 naphthalene-catabolic plasmid NAH7 and its transposon Tn4655 suggests catabolic gene spread by a tyrosine recombinase. J. Bacteriol. 188, 4057–4067. doi: 10.1128/JB.00185-06
Springael, D., Kreps, S., and Mergeay, M. (1993). Identification of a catabolic transposon, Tn4371, carrying biphenyl and 4-chlorobiphenyl degradation genes in Alcaligenes eutrophus A5. J. Bacteriol. 175, 1674–1681. doi: 10.1128/jb.175.6.1674-1681
Springael, D., and Top, E. M. (2004). Horizontal gene transfer and microbial adaptation to xenobiotics: new types of mobile genetic elements and lessons from ecological studies. Trends Microbiol. 12, 53–58. doi: 10.1016/j.tim.2003.12.010
Suenaga, H. (2012). Targeted metagenomics: a high-resolution metagenomics approach for specific gene clusters in complex microbial communities. Environ. Microbiol. 14, 13–22. doi: 10.1111/j.1462-2920.2011.02438.x
Suenaga, H., Fujihara, H., Kimura, N., Hirose, J., Watanabe, T., Futagami, T., et al. (2017). Insights into the genomic plasticity of Pseudomonas putida KF715, a strain with unique biphenyl-utilizing activity and genome instability properties. Environ. Microbiol. Rep. 9, 589–598. doi: 10.1111/1758-2229.12561
Suenaga, H., Koyama, Y., Miyakoshi, M., Miyazaki, R., Yano, H., Sota, M., et al. (2009). Novel organization of aromatic degradation pathway genes in a microbial community as revealed by metagenomic analysis. ISME J. 3, 1335–1348. doi: 10.1038/ismej.2009.76
Suenaga, H., Matsuzawa, T., and Sahara, T. (2022). Discovery by metagenomics of a functional tandem repeat sequence that controls gene expression in bacteria. FEMS Microbiol. Ecol. 98:37. doi: 10.1093/femsec/fiac037
Suenaga, H., Yamazoe, A., Hosoyama, A., Kimura, N., Hirose, J., Watanabe, T., et al. (2015). Draft genome sequence of the polychlorinated biphenyl-degrading bacterium Cupriavidus basilensis KF708 (NBRC 110671) isolated from biphenyl-contaminated soil. Genome Announc. 3, e00143–e00115. doi: 10.1128/genomeA.00143-15
Taguchi, K., Motoyama, M., Iida, T., and Kudo, T. (2007). Polychlorinated biphenyl/biphenyl degrading gene clusters in Rhodococcus sp. K37, HA99, and TA431 are different from well-known bph gene clusters of rhodococci. Biosci. Biotechnol. Biochem. 71, 1136–1144. doi: 10.1271/bbb.60551
Taguchi, K., Motoyama, M., and Kudo, T. (2004). Multiplicity of 2,3-dihydroxybiphenyl dioxygenase genes in the gram-positive polychlorinated biphenyl degrading bacterium Rhodococcus rhodochrous K37. Biosci. Biotechnol. Biochem. 68, 787–795. doi: 10.1271/bbb.68.787
Takeda, H., Shimodaira, J., Yukawa, K., Hara, N., Kasai, D., Miyauchi, K., et al. (2010). Dual two-component regulatory systems are involved in aromatic compound degradation in a polychlorinated-biphenyl degrader, Rhodococcus jostii RHA1. J. Bacteriol. 192, 4741–4751. doi: 10.1128/JB.00429-10
Takeda, H., Yamada, A., Miyauchi, K., Masai, E., and Fukuda, M. (2004). Characterization of transcriptional regulatory genes for biphenyl degradation in Rhodococcus sp. strain RHA1. J. Bacteriol. 186, 2134–2146. doi: 10.1128/JB.186.7.2134-2146.2004
Tan, H. M. (1999). Bacterial catabolic transposons. Appl. Microbiol. Biotechnol. 51, 1–12. doi: 10.1007/s002530051356
Tang, H., Yu, H., Li, Q., Wang, X., Gai, Z., Yin, G., et al. (2011). Genome sequence of Pseudomonas putida strain B6-2, a Superdegrader of polycyclic aromatic hydrocarbons and dioxin-like compounds. J. Bacteriol. 193, 6789–6790. doi: 10.1128/JB.06201-11
Top, E. M., and Springael, D. (2003). The role of mobile genetic elements in bacterial adaptation to xenobiotic organic compounds. Curr. Opin. Biotechnol. 14, 262–269. doi: 10.1016/s0958-1669(03)00066-1
Top, E. M., Springael, D., and Boon, N. (2002). Catabolic mobile genetic elements and their potential use in bioaugmentation of polluted soils and waters. FEMS Microbiol. Ecol. 42, 199–208. doi: 10.1111/j.1574-6941.2002.tb01009.x
Toussaint, A., Merlin, C., Benotmane, M. A., Monchy, S., Benotmane, M. A., Leplae, R., et al. (2003). The biphenyl- and 4-chlorobiphenyl-catabolic transposon Tn4371, a member of a new family of genomic islands related to IncP and Ti plasmids. Appl. Environ. Microbiol. 69, 4837–4845. doi: 10.1128/AEM.69.8.4837-4845.2003
Tropel, D., and van der Meer, J. R. (2004). Bacterial transcriptional regulators for degradation pathways of aromatic compounds. Microbiol. Mol. Biol. Rev. 68, 474–500. doi: 10.1128/MMBR.68.3.474-500.2004
Tsuda, M., Tan, H. M., Nishi, A., and Furukawa, K. (1999). Mobile catabolic genes in bacteria. J. Biosci. Bioeng. 87, 401–410. doi: 10.1016/S1389-1723(99)80086-3
van Belkum, A., Scherer, S., van Alphen, L., and Verbrugh, H. (1998). Short-sequence DNA repeats in prokaryotic genomes. Microbiol. Mol. Biol. Rev. 62, 275–293. doi: 10.1128/MMBR.62.2.275-293.1998
Van der Meer, J. R., Alexander, J. B., Zehnder, A. J. B., and De Vos, W. M. (1991). Identification of a novel composite transposable element, Tn5280, carrying chlorobenzene dioxygenase genes of Pseudomonas sp. strain P51. J. Bacterol. 173, 7077–7083. doi: 10.1128/jb.173.22.7077-7083.1991
Van der Meer, J. R., De Vos, W. M., Harayama, S., and Zehnder, A. J. B. (1992). Molecular mechanisms of genetic adaptation to xenobiotic compounds. Microbiol. Rev. 56, 677–694. doi: 10.1128/mmbr.56.4.677-694.1992
Watanabe, T., Fujihara, H., and Furukawa, K. (2003). Characterization of the second LysR-type regulator in the biphenyl-catabolic gene cluster of Pseudomonas pseudoalcaligenes KF707. J. Bacteriol. 185, 3575–3582. doi: 10.1128/JB.185.12.3575-3582.2003
Watanabe, T., Inoue, R., Kimura, N., and Furukawa, K. (2000). Versatile transcription of biphenyl catabolic bphOperon in Pseudomonas pseudoalcaligenes KF707*. J. Biol. Chem. 275, 31016–31023. doi: 10.1074/jbc.M003023200
Witzig, R., Aly, H. A., Strompl, C., Wray, V., Junca, H., and Pieper, D. H. (2007). Molecular detection and diversity of novel diterpenoid dioxygenase DitA1 genes from proteobacterial strains and soil samples. Environ. Microbiol. 9, 1202–1218. doi: 10.1111/j.1462-2920.2007.01242.x
Wozniak, R. A. F., and Waldor, M. K. (2010). Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat. Rev. Microbiol. 8, 552–563. doi: 10.1038/nrmicro2382
Zamarro, M. T., Martín-Moldes, Z., and Díaz, E. (2016). The ICEXTD of Azoarcus sp. CIB, an integrative and conjugative element with aerobic and anaerobic catabolic properties. Environ. Microbiol. 18, 5018–5031. doi: 10.1111/1462-2920.13465
Zhou, K., Aertsen, A., and Michiels, C. W. (2014). The role of variable DNA tandem repeats in bacterial adaptation. FEMS Microbiol. Rev. 38, 119–141. doi: 10.1111/1574-6976.12036
Keywords: xenobiotic compounds, degrading bacteria, mobile genetic elements, gene regulation and expression, adaptive evolution
Citation: Fujihara H, Hirose J and Suenaga H (2023) Evolution of genetic architecture and gene regulation in biphenyl/PCB-degrading bacteria. Front. Microbiol. 14:1168246. doi: 10.3389/fmicb.2023.1168246
Received: 17 February 2023; Accepted: 16 May 2023;
Published: 07 June 2023.
Edited by:
Feng Gao, Tianjin University, ChinaReviewed by:
Masataka Tsuda, Tohoku University, JapanCopyright © 2023 Fujihara, Hirose and Suenaga. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hikaru Suenaga, c3VlbmFnYS1oaWthcnVAYWlzdC5nby5qcA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.