
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 22 March 2023
Sec. Evolutionary and Genomic Microbiology
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1166267
This article is part of the Research Topic Insights on Fungal Diversity of Ascomycetes and Basidiomycetes: Taxonomy and Interaction with Their Host View all 24 articles
Two new wood-inhabiting fungi from China, Steccherinum juniperi and S. incrustans, in the family Steccherinaceae are described and illustrated based on morphological and molecular analyses. The species S. juniperi was found growing on the rotten wood of Juniperus in Qinghai Province, China, while S. incrustans was collected on rotten angiosperm wood in Yunnan Province, China. The characteristics of S. juniperi include annual, resupinate basidiomata with a buff yellow fresh pore surface that becomes apricot orange when bruised, angular pores of 3–6 per mm, subicular generative hyphae sometimes covered with crystals, the presence of encrusted skeletocystidia in tube trama only, fusiform to slim clavate cystidioles, and ellipsoid basidiospores measuring as 3–4 × 2–3 μm. The characteristics of S. incrustans include annual, resupinate basidiomata with a buff yellow or pinkish buff to clay buff dried pore surface, angular pores (8–10 per mm), generative hyphae in trama frequently covered with crystals, the presence of encrusted skeletocystidia in tube trama and hymenium, and ellipsoid basidiospores (3.5–4.5 × 2.5–3.5 μm). Phylogenetic analysis based on a combined 2-locus dataset [ITS1-5.8S-ITS2 (ITS) + nuclear large subunit RNA (nLSU)] shows that the two species are members of Steccherinum, and they are compared with morphologically similar and related species of this genus, respectively. In addition, two new combinations from Junghuhnia, transferred to Steccherinum as S. austrosinense and S. nandinae, are proposed based on examination of their type materials and phylogenetic analysis.
Steccherinum Gray was established by Gray (1821), with S. ochraceum (Pers. ex J.F. Gmel.) Gray selected as its type. It is the largest genus in the Steccherinaceae (Polyporales) and has a worldwide distribution, with ~76 species accepted by Index Fungorum (http://www.indexfungorum.org/; accessed on 1 January 2023) and MycoBank (https://www.mycobank.org; accessed on 1 January 2023). Dai (2011) summarized corticioid and hydnoid fungi in China and 12 species of Steccherinum were mentioned. An identification key to 15 species of Steccherinum recorded from China was provided (Wan and Yuan, 2013).
Steccherinum is characterized by the resupinate to effuse-reflexed basidiomata with poroid or odontioid to hydnoid hymenophore, a monomitic or dimitic hyphal structure with thick-walled skeletal hyphae; most species have clamped generative hyphae, encrusted or smooth skeletocystidia, and smooth, thin-walled, ellipsoid basidiospores (Maas Geesteranus, 1974; Eriksson et al., 1984; Miettinen et al., 2012).
Miettinen et al. (2012) carried out a multigene phylogenetic analysis (ITS + nLSU + mtSSU + atp6 + tef1) for Steccherinaceae and proposed the monophyletic Steccherinum clade (Figure 4 in Miettinen et al., 2012). Liu and Dai (2021) thought that the limit of the genus Steccherinum in Miettinen et al. (2012) is reasonable, described S. fragile Z.B. Liu & Y.C. Dai, and proposed S. subcollabens (F. Wu et al.) Z.B. Liu & Y.C. Dai within the Steccherinum clade in their phylogenetic analysis of ITS + nLSU. Subsequently, Wu et al. (2021) described S. puerense Y.X. Wu et al. and S. rubigimaculatum Y.X. Wu et al. Dong et al. (2022) described S. hirsutum Y.X. Wu & C.L. Zhao and S. yunnanense Y.X. Wu & C.L. Zhao based on their phylogenetic analyses.
During investigations on the diversity of wood-rotting fungi from China, three resupinate polypore specimens were collected from Yunnan Province and Qinghai Province. Their morphology corresponded to the concept of Steccherinum. To confirm their affinity, phylogenetic analysis based on the ITS and nLSU rDNA sequences was carried out. Both morphological characteristics and molecular evidence demonstrated that these three specimens represent two new species of Steccherinum, which we describe in the present study. In addition, we studied the type specimens of Junghuhnia austrosinensis F. Wu et al. and J. nandinae F. Wu et al. They were transferred to Steccherinum based on morphological and phylogenetic analyses.
Macro-morphological descriptions were based on dry herbarium specimens and field notes. Microscopic measurements and drawings were prepared from slide preparations of dried tissues stained with Cotton Blue and Melzer's reagent as described by Dai (2010). Pores were measured by subjectively choosing as straight a line of pores as possible and measuring how many per mm. The following abbreviations are used in the description: CB = Cotton Blue; CB+ = cyanophilous in Cotton Blue; CB– = acyanophilous in Cotton Blue; IKI = Melzer's reagent; IKI– = neither amyloid nor dextrinoid in Melzer's reagent; KOH = 5% potassium hydroxide; n (a/b) = number of spores (a) measured from given number of specimens (b); L = spore length (arithmetic average of all the spores); W = spore width (arithmetic average of all the spores); and Q = variation in the L/W ratios between the specimens studied. When the variation in spore size is shown, 5% of the measurements were excluded from each end of the range, and these values are shown in parentheses. Special color terms follow Petersen (1996), and then, herbarium abbreviations follow Thiers (2018). Voucher specimens from the study were deposited in the herbarium of the Institute of Microbiology, Beijing Forestry University (BJFC).
Total genomic DNA was extracted from dried specimens using a CTAB Rapid Plant Genome Extraction Kit (Aidlab Biotechnologies Company, Ltd., Beijing, China) according to the manufacturer's instructions with some modifications (Li et al., 2014). The ITS regions were amplified with primers ITS4 and ITS5 (White et al., 1990). The nLSU regions were amplified with primers LR0R and LR7 (Vilgalys and Hester, 1990).
The polymerase chain reaction (PCR) procedure for the ITS was as follows: initial denaturation at 95°C for 3 min, followed by 35 cycles at 94°C for 40 s, 54°C for 45 s, and 72°C for 1 min, and a final extension of 72°C for 10 min. The PCR procedure for the nLSU was as follows: initial denaturation at 94°C for 1 min, followed by 35 cycles at 94°C for 30 s, 48°C for 1 min, and 72°C for 1.5 min, and a final extension of 72°C for 10 min (Zhao et al., 2015). Aliquots of PCR products were examined on 2% agarose gels stained with GelStar Nucleic Acid Gel Stain (Lonza Rockland, Inc., Rockland, YN, USA) and examined under UV light. The sequencing of the PCR products was conducted by the Beijing Genomics Institute, Beijing, China, with the same primers used in the PCR reactions. Species were identified by sequence comparison with accessions in the NCBI databases using the BLAST program.
Phylogenetic trees were constructed using ITS + nLSU rDNA sequences, and phylogenetic analyses were performed with the maximum likelihood (ML), maximum parsimony (MP), and Bayesian inference (BI) methods. Sequences of the species and strains were primarily adopted from ITS-based and 28S-based tree topology as described by Liu and Dai (2021). New sequences generated in this study, along with reference sequences retrieved from GenBank (https://www.ncbi.nlm.nih.gov/genbank/; Table 1), were aligned by MAFFT 7 (Katoh et al., 2019; http://mafft.cbrc.jp/alignment/server/) using the “G-INS-i” strategy and manually adjusted in BioEdit 7.2.5 (Hall, 1999). Unreliably aligned sections were removed before the analyses, and efforts were made to manually inspect and improve the alignment. The data matrix was edited in Mesquite 3.70 (https://www.mesquiteproject.org/; Maddison and Maddison, 2021). The sequence alignment was deposited at TreeBase (Submission ID: 30018). According to Miettinen et al. (2012), Junghuhnia crustacea (Jungh.) Ryvarden also belongs to the family Steccherinaceae and is not close to the Steccherinum clade, thus sequences of Junghuhnia crustacea obtained from GenBank were used as out-groups to root the trees in the ITS + nLSU analysis.
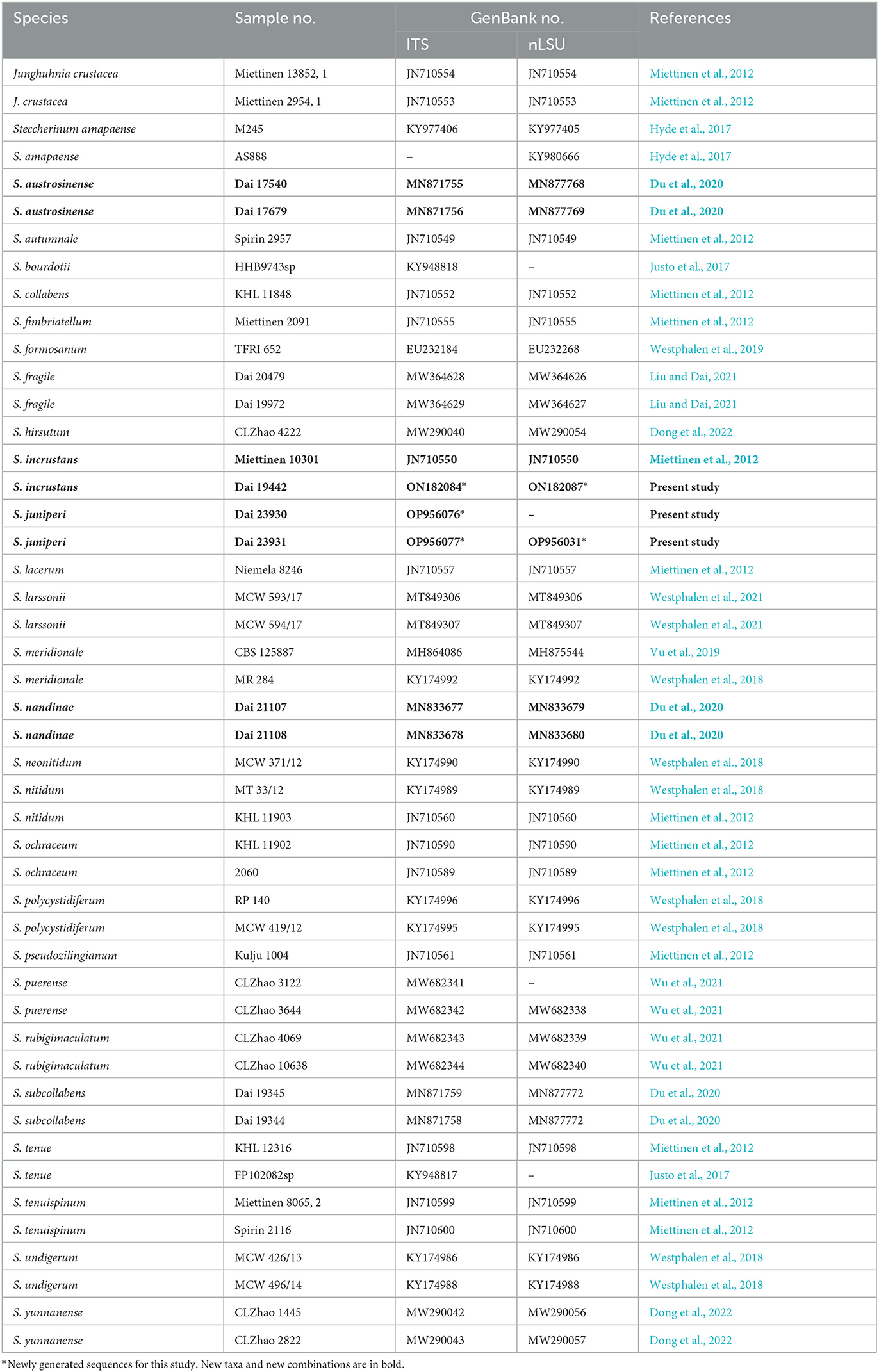
Table 1. List of species, specimens, and GenBank accession numbers of the sequences used in this study.
Maximum likelihood analysis was conducted using RAxML-HPC 8.2.3 (Stamatakis, 2014) and RAxML-HPC through the CIPRES Science Gateway 3.3 (Miller et al., 2010; http://www.phylo.org). Statistical support values were obtained using non-parametric bootstrapping with 1,000 replicates. The BI analysis was performed with MrBayes 3.2.7a (Ronquist et al., 2012). Four Markov chains were run for two runs from random starting trees for 1 million generations until the split deviation frequency value <0.01, and the trees were sampled at every 1,000 generations. The first 25% of the sampled trees were discarded as burn-in, and the remaining ones were used to reconstruct a majority rule consensus tree and calculate the Bayesian posterior probabilities (BPP) of the clades.
Maximum parsimony analysis was applied to the ITS + nLSU dataset sequences. The approaches to phylogenetic analysis were conducted as described by Liu et al. (2022), and the tree was constructed using PAUP* 4.0β10 (Swofford, 2002). All the characters were equally weighted, and gaps were treated as missing data. Trees were inferred using the heuristic search option with tree bisection and reconnection branch swapping, and 1,000 random sequence addition maxtrees were set to 5,000. Branches of zero length were collapsed, and all the parsimonious trees were saved. Clade robustness was assessed using a bootstrap analysis with 1,000 replicates (Felsenstein, 1985). Descriptive tree statistics, including the consistency index (CI), homoplasy index (HI), rescaled consistency index (RC), retention index (RI), and tree length (TL), were calculated for each maximum parsimonious tree generated.
A total of 24 models of evolution were scored using PAUP* 4.0β10 (Swofford, 2002). Optimal substitution models for the combined dataset were then determined using the Akaike information criterion implemented in MrModeltest 2.3 (Posada and Crandall, 1998; Nylander, 2004). The model GTR + I + G was selected in the ML and BI analyses.
Branches are labeled with ML bootstrap ≥ 70%, MP bootstrap ≥ 50%, and BPP ≥ 0.95, respectively. FigTree 1.4.4 (Rambaut, 2018) was used to visualize the resulting tree.
The combined ITS + nLSU dataset included sequences from 47 specimens representing 27 species (Table 1). The dataset had an aligned length of 2,044 characters, of which 1,544 were constant, 96 were variable but parsimony-uninformative, and 404 were parsimony-informative. MP analysis yielded an equally parsimonious tree (CI = 0.524, HI = 0.476, RC = 0.403, RI = 0.768, TL = 1,288). ML analysis resulted in the best tree, and Bayesian and MP analyses resulted in a similar topology to the ML analysis, with an average standard deviation of split frequencies of 0.007184 (BI). Hence, the ML tree is shown combined with the support values from the MP and BI analyses.
The phylogeny (Figure 1) inferred from the ITS and nLSU sequences demonstrated that the new species (Steccherinum juniperi and S. incrustans) and new combinations (S. austrosinense and S. nandinae) clustered in Steccherinum clade, and thus, they are described and proposed herein.
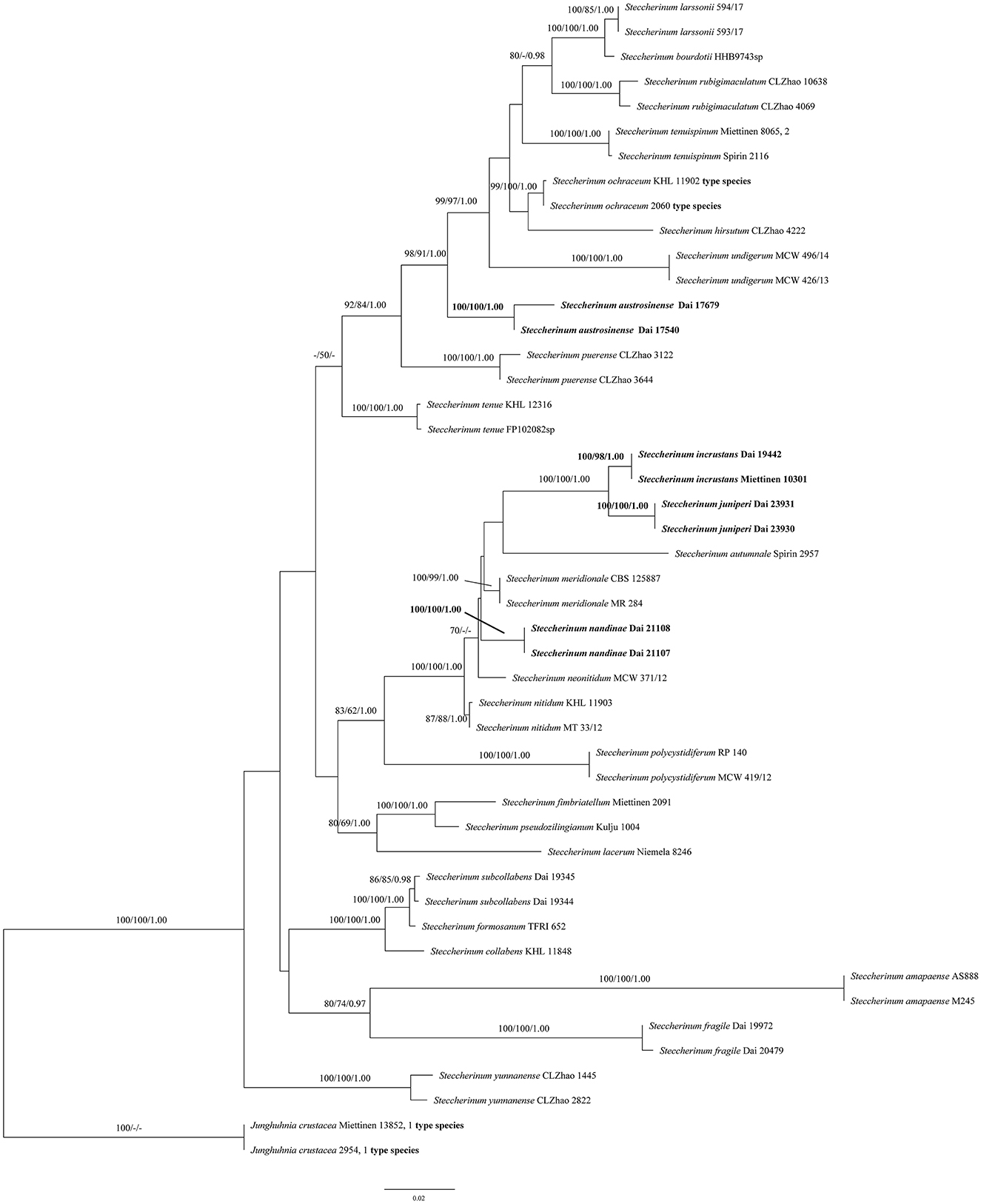
Figure 1. Phylogeny of Steccherinum generated by maximum likelihood (ML) analysis based on combined ITS and nLSU rDNA sequences. Branches are labeled with ML bootstrap >70%, maximum parsimony bootstrap >50%, and Bayesian posterior probabilities >0.95, respectively. New species and new combinations are in bold.
Steccherinum incrustans Z.B. Liu, Y.C. Dai & Jing Si, sp. nov.
MycoBank: MB847683.

Figure 2. Basidiomata of Steccherinum incrustans (Holotype, Dai 19442). (Scale bar = 1.0 cm). Photographed by Meng Zhou.
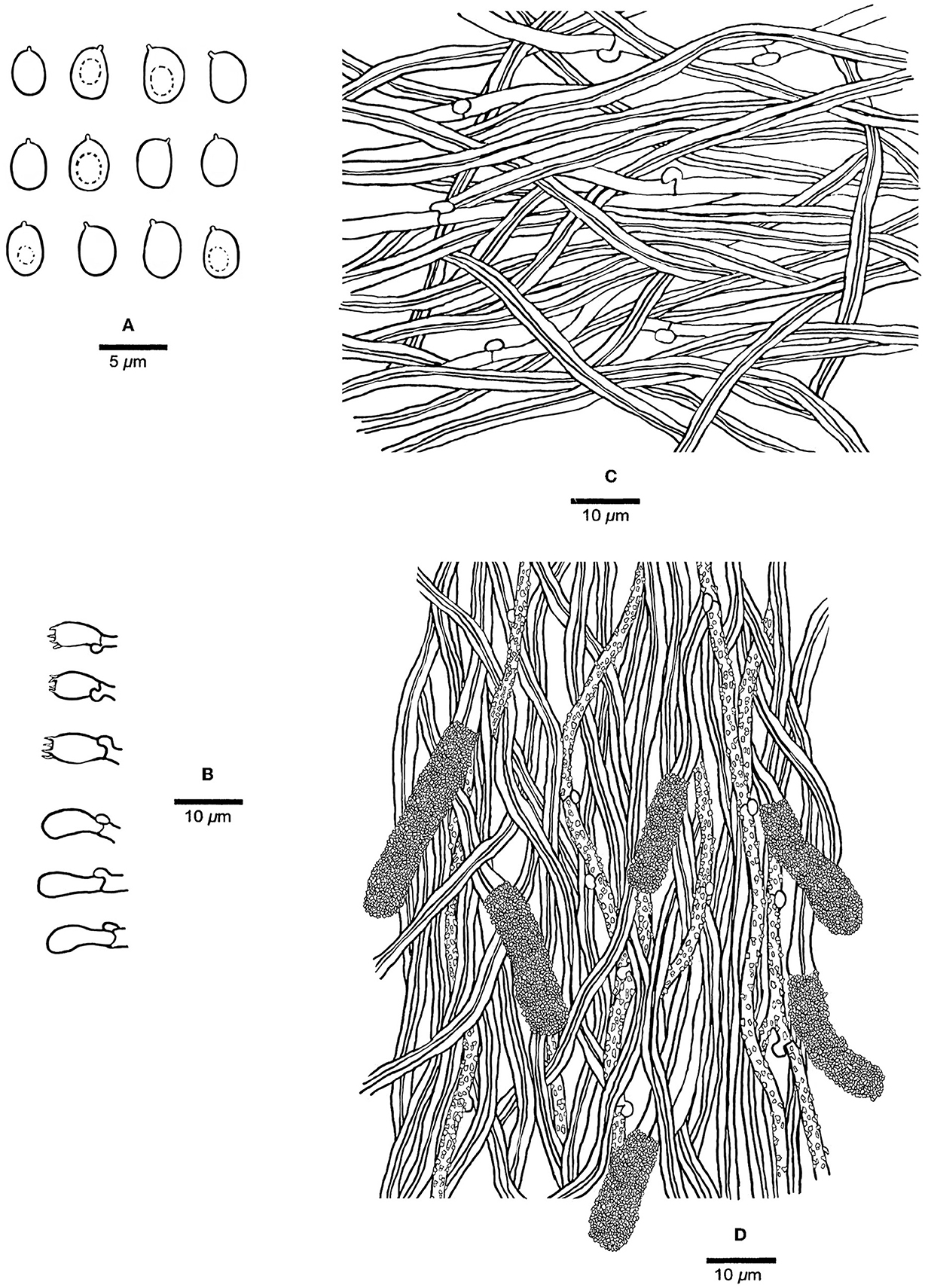
Figure 3. Microscopic structures of Steccherinum incrustans (Holotype, Dai 19442). (A) Basidiospores. (B) Basidia and basidioles. (C) Hyphae from subiculum. (D) Skeletocystidia and hyphae from trama. Drawn by Meng Zhou.
Holotype—China. Yunnan Province, Jinghong, Xishuangbanna Tropical Botanical Garden, on rotten angiosperm wood, 16.XII.2018, Dai 19442 (BJFC027910).
Etymology—Incrustans (Lat.): referring to the species having encrusted generative hyphae in trama.
Fruiting body—Basidiomata annual, resupinate, difficult to separate from the substrate, soft corky when fresh, hard corky when dry, up to 11 cm long, 2 cm wide, and ~1.5 mm thick at the center; pore surface buff yellow or pinkish buff to clay buff upon drying; sterile margin indistinct; pores angular, 8–10 per mm; dissepiments thin, entire; subiculum very thin to almost absent, paler than tubes, nearly 0.2 mm thick; tubes concolorous with poroid surface, up to 1.3 mm long.
Hyphal structure—Hyphal system dimitic; generative hyphae with clamp connections; skeletal hyphae dominant, CB+, IKI–; tissues unchanged in KOH.
Subiculum—Generative hyphae hyaline, thin-walled, unbranched, 2–3 μm in diam; skeletal hyphae dominant, hyaline, thick-walled with a narrow lumen, unbranched, flexuous, interwoven, 2.5–3.5 μm in diam.
Tubes—Generative hyphae hyaline, thin-walled, rarely branched, frequently, and strongly encrusted with crystals, 1–2.5 μm in diam; skeletal hyphae dominant, hyaline, thick-walled with a narrow lumen, unbranched, flexuous, interwoven, 2–3 μm in diam. Skeletocystidia present in the hymenium and trama, abundant, clavate to cylindrical, thick-walled with a narrow lumen, originated from tramal skeletal hyphae, then projecting from hymenium, strongly encrusted in the obtuse apex, 15–35 × 5–10 μm (encrusted part); cystidioles absent; basidia barrel-shaped, hyaline, with a basal clamp connection and four sterigmata, 9–13 × 4–5.5 μm; basidioles dominant, clavate similar with basidia in length.
Spores—Basidiospores ellipsoid with an apiculus, hyaline, thin-walled, smooth, some with a medium guttule, IKI–, CB–, (3–)3.5–4.5(−4.7) × (2.2–)2.5–3.5(−3.8) μm, L = 3.98 μm, W = 2.90 μm, Q = 1.37 (n = 60/1).
Steccherinum juniperi Z.B. Liu, Y.C. Dai & Jing Si, sp. nov.
MycoBank: MB847674.

Figure 4. Basidiomata of Steccherinum juniperi (Paratype, Dai 23930) (Scale bar = 1.0 cm). Photographed by Yu-Cheng Dai.

Figure 5. Microscopic structures of Steccherinum juniperi (Holotype, Dai 23931). (A) Basidiospores. (B) Basidia. (C) Basidioles. (D) Cystidioles. (E) Hyphae from subiculum. (F) Hyphae from trama. Drawn by Meng Zhou.
Holotype—China. Qinghai Province, Yushu, Leba Valley, on rotten wood of Juniperus, 5.VIII.2022, Dai 23931 (BJFC039175).
Etymology—Juniperi (Lat.): referring to the species growing on Juniperus.
Fruiting body—Basidiomata annual, resupinate, difficult to separate from the substrate, soft corky when fresh, hard corky when dry, up to 10 cm long, 2 cm wide, and ~2.5 mm thick at the center; pore surface buff yellow when fresh, apricot orange when bruised, buff to honey yellow upon drying; sterile margin distinct, cream and nearly 1 mm width; pores angular, 3–6 per mm; dissepiments thin, entire; subiculum very thin to almost absent, paler than tubes, nearly 0.5 mm thick; tubes concolorous with poroid surface, up to 2 mm long.
Hyphal structure—Hyphal system dimitic; generative hyphae with clamp connections; skeletal hyphae dominant, IKI–, CB+; tissues unchanged in KOH.
Subiculum—Generative hyphae hyaline, thin- to slightly thick-walled, rarely branched, sometimes encrusted with crystals, 2–3.5 μm in diam; skeletal hyphae dominant, hyaline, thick-walled with a medium to narrow lumen, unbranched, flexuous, interwoven, 3–5 μm in diam.
Tubes—Generative hyphae hyaline, slightly thick-walled, occasionally branched, 2–3 μm in diam; skeletal hyphae dominant, hyaline, thick-walled with a medium lumen, unbranched, flexuous, interwoven, 2–4.5 μm in diam. Skeletocystidia present in trama only, abundant, clavate to cylindrical, thick-walled with a wide lumen, strongly encrusted in the obtuse apex, 20–120 × 7–10 μm (encrusted part); cystidioles present, fusiform to slim clavate, hyaline, thin-walled, 10–15 × 3–4 μm; basidia clavate, hyaline, with a basal clamp connection and four sterigmata, 12–13 × 4–6 μm; basidioles dominant, similar to basidia in shape, but slightly smaller.
Spores—Basidiospores ellipsoid with an apiculus, hyaline, thin-walled, smooth, some with a medium guttule, IKI–, CB–, 3–4(−4.8) × (1.8–)2–3(−3.2) μm, L = 3.57 μm, W = 2.46 μm, Q = 1.38–1.52 (n = 60/2).
Additional specimen (paratype) examined—China. Qinghai Province, Yushu, Leba Valley, on rotten wood of Juniperus, 5.VIII.2022, Dai 23930 (BJFC039174).
Steccherinum austrosinense (F. Wu, P. Du & X.M. Tian) Z.B. Liu, Y.C. Dai & Jing Si, comb. nov.
MycoBank: MB847672.
Basionym—Junghuhnia austrosinensis F. Wu, P. Du & X.M. Tian, MycoKeys 72: 5 (2020).
Materials studied—China. Yunnan Province, Jinghong, Virgin Forest Park, on fallen bamboo, 17.VI.2017, Dai 17540 (BJFC025072, holotype); Hainan Province, Wuzhishan County, Wuzhishan Forest Park, on fallen angiosperm branch, 9.IX.2019, Dai 17679 (BJFC025211, paratype).
Steccherinum nandinae (F. Wu, P. Du & X.M. Tian) Z.B. Liu, Y.C. Dai & Jing Si, comb. nov.
MycoBank: MB847673.
Basionym—Junghuhnia nandinae F. Wu, P. Du & X.M. Tian, MycoKeys 72: 8 (2020).
Materials studied—China. Chongqing, Nanchuan County, Jinfoshan Forest Park, on dead tree of Nandina domestica, 1.XI.2019, Dai 21107 (BJFC032766, holotype), Dai 21108 (BJFC032767, paratype).
In the present study, two new species (S. juniperi and S. incrustans) and two new combinations (S. austrosinense and S. nandinae) nested in the Steccherinum clade, based on the phylogenetic analysis of ITS + nLSU sequences data (Figure 1).
An ITS sequence JN710550 of the sample Miettinen 10301, named Junghuhnia cf. nitida from GenBank, is almost identical to Dai 19442 in the ITS regions, and the similarity between their sequences is up to 99.73%. Both samples were collected from Xishuangbanna, Yunnan Province, China. We believed that the sample Miettinen 10301 represented the same species as our specimen (Dai 19442), and they formed a lineage with strong supports (100% ML, 98% MP, and 1.00 BPP, Figure 1) in our phylogeny. Morphologically, S. incrustans can be distinguished from Junghuhnia nitida (Pers.) Ryvarden by having smaller pores (8–10 per mm vs. 5–7 per mm, Ryvarden and Johansen, 1980). In addition, S. incrustans differs from J. nitida by its tramal generative hyphae frequently covered with crystals, while they are smooth in J. nitida.
The phylogenetic analyses indicated that two specimens of S. juniperi formed a lineage with strong supports (100% ML, 100% MP, and 1.00 BPP) and grouped with S. incrustans with strong supports (100% ML, 100% MP, and 1.00 BPP) (Figure 1). Steccherinum juniperi differs from S. incrustans by its larger pores (3–6 per mm in S. juniperi vs. 8–10 per mm in S. incrustans). In addition, S. juniperi grows on gymnosperm in boreal forests, while S. incrustans grows on angiosperm in tropical forests.
Steccherinum juniperi, S. austrosinense, and S. neonitidum Westphalen & Tomšovský have poroid hymenophore and microscopically share cystidioles and similar sizes of basidiospores, but S. austrosinense and S. neonitidum have distinctly smaller pores (9–11 per mm in S. austrosinense, 8–10 per mm in S. neonitidum, vs. 3–6 per mm in S. juniperi). In addition, S. austrosinense and S. neonitidum grow on angiosperm, and their skeletocystidia are present in both tube trama and out of hymenium (Westphalen et al., 2018; Du et al., 2020), while S. juniperi grows on gymnosperm and only has skeletocystidia in tube trama.
Steccherinum collabens (Fr.) Vesterh. resembles S. juniperi in the field because they share similar pores when fresh and grow on gymnosperms; however, S. collabens has cylindrical to suballantoid basidiospores (3.2–3.6 × 1.4–1.7 μm, Niemelä, 2016).
The vegetation in Northwest China is relatively simple compared with the other parts of China, and a few limited new taxa of wood-habiting fungi were described from the area (Dai et al., 2007a,b, 2021), especially only a few species were recorded on Juniperus in China (Dai, 2012; Cui et al., 2019; Wu et al., 2022). Steccherinum juniperi is described as Juniperus in a dry environment of Northwest China, thus indicating that some special species adapted to the special host in the arid area.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, ON182084, ON182087, OP956031, OP956076, and OP956077.
Z-BL: investigation, software, data curation, visualization, and writing—original draft. Q-YZ and MZ: data curation and visualization. JS: visualization, supervision, writing—reviewing and editing, project administration, and funding acquisition. All authors contributed to the manuscript and approved the submitted version.
The research was supported by the National Natural Science Foundation of China (Nos. 32270016 and 32070016).
The authors would like to express their deep appreciation to Prof. Yu-Cheng Dai (Beijing Forestry University, China) for allowing us to study his specimens.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Cui, B. K., Li, H. J., Ji, X., Zhou, J. L., Song, J., Si, J., et al. (2019). Species diversity, taxonomy and phylogeny of Polyporaceae (Basidiomycota) in China. Fungal Divers. 97, 137–392. doi: 10.1007/s13225-019-00427-4
Dai, Y. C. (2010). Hymenochaetaceae (Basidiomycota) in China. Fungal Divers. 45, 131–343. doi: 10.1007/s13225-010-0066-9
Dai, Y. C. (2011). A revised checklist of corticioid and hydnoid fungi in China for 2010. Mycoscience 52, 69–79. doi: 10.1007/S10267-010-0068-1
Dai, Y. C. (2012). Polypore diversity in China with an annotated checklist of Chinese polypores. Mycoscience 53, 49–80. doi: 10.1007/s10267-011-0134-3
Dai, Y. C., Cui, B. K., and Yuan, H. S. (2007a). Notes on polypores from Gansu and Qinghai Province, northwest China. Cryptogamie Mycol. 28, 177–187.
Dai, Y. C., Wei, Y. L., Yuan, H. S., Huang, M. Y., and Penzina, T. (2007b). Polypores from Altay and Tian Mts. in Xinjiang, northwest China. Cryptogamie Mycol. 28, 269–279.
Dai, Y. C., Yang, Z. L., Cui, B. K., Wu, G., Yuan, H. S., Zhou, L. W., et al. (2021). Diversity and systematics of the important macrofungi in Chinese forests. Mycosystema 40, 770–805. doi: 10.13346/j.mycosystema.210036
Dong, J. H., Wu, Y. X., and Zhao, C. L. (2022). Two new species of Steccherinum (Polyporales, Basidiomycota) from southern China based on morphology and DNA sequence data. Mycoscience 63, 65–72. doi: 10.47371/mycosci.2022.02.002
Du, P., Wu, F., and Tian, X. M. (2020). Three new species of Junghuhnia (Polyporales, Basidiomycota) from China. MycoKeys 72, 1–16. doi: 10.3897/mycokeys.72.51872
Eriksson, J., Hjortstam, K., and Ryvarden, L. (1984). The Corticiaceae of North Europe 7. Schizopora-Suillosporium. Oslo: Fungiflora, 1–169.
Felsenstein, J. (1985). Confidence intervals on phylogenetics: an approach using bootstrap. Evolution 39, 783–791. doi: 10.2307/2408678
Gray, S. F. (1821). Natural Arrangement of British Plants. Vol. 1. London: Baldwin, Craddock and Joy, 1–824. doi: 10.5962/bhl.title.43804
Hall, T. A. (1999). Bioedit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98.
Hyde, K. D., Norphanphoun, C., Abreu, V. P., Bazzicalupo, A., Thilini Chethana, K. W., Clericuzio, M., et al. (2017). Fungal diversity notes 603–708: taxonomic and phylogenetic notes on genera and species. Fungal Divers. 87, 1–235. doi: 10.1007/s13225-017-0391-3
Justo, A., Miettinen, O., Floudas, D., Ortiz-Santana, B., Sjökvist, E., Lindner, D., et al. (2017). A revised family-level classification of the Polyporales (Basidiomycota). Fungal Biol. 121, 798–824. doi: 10.1016/j.funbio.2017.05.010
Katoh, K., Rozewicki, J., and Yamada, K. D. (2019). MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 20, 1160–1166. doi: 10.1093/bib/bbx108
Li, H. J., Cui, B. K., and Dai, Y. C. (2014). Taxonomy and multi-gene phylogeny of Datronia (Polyporales, Basidiomycota). Persoonia 32, 170–182. doi: 10.3767/003158514X681828
Liu, Z. B., and Dai, Y. C. (2021). Steccherinum fragile sp. nov. and S. subcollabens comb. nov. (Steccherinaceae, Polyporales), evidenced by morphological characters and phylogenetic analysis. Phytotaxa 483, 106–116. doi: 10.11646/phytotaxa.483.2.3
Liu, Z. B., Wu, Y. D., Zhao, H., Lian, Y. P., Wang, Y. R., Wang, C. G., et al. (2022). Outline, divergence times, and phylogenetic analyses of Trechisporales (Agaricomycetes, Basidiomycota). Front. Microbiol. 13, 818358. doi: 10.3389/fmicb.2022.818358
Maddison, W. P., and Maddison, D. R. (2021). Mesquite: A Modular System for Evolutionary Analysis, version 3.70. Available online at: http://www.mesquiteproject.org (accessed January 1, 2023).
Miettinen, O., Larsson, E., Sjökvist, E., and Larsson, K. H. (2012). Comprehensive taxon sampling reveals unaccounted diversity and morphological plasticity in a group of dimitic polypores (Basidiomycota, Polyporales). Cladistics 28, 251–270. doi: 10.1111/j.1096-0031.2011.00380.x
Miller, M. A., Pfeiffer, W., and Schwartz, T. (2010). “Creating the CIPRES Science Gateway for inference of large phylogenetic trees” in Proceedings of the Gateway Computing Environments Workshop (GCE) (New Orleans), 1–8. doi: 10.1109/GCE.2010.5676129
Nylander, J. A. A. (2004). MrModeltest v2. Uppsala: Program distributed by the author. Evolutionary Biology Centre, Uppsala University.
Petersen, J. H. (1996). The Danish Mycological Society's Colour-Chart. Greve: Foreningen til Svampekundskabens Fremme.
Posada, D., and Crandall, K. A. (1998). Modeltest: testing the model of DNA substitution. Bioinformatics 14, 817–818. doi: 10.1093/bioinformatics/14.9.817
Rambaut, A. (2018). Molecular Evolution, Phylogenetics and Epidemiology. FigTree ver. 1.4.4 Software. Available online at: http://tree.bio.ed.ac.uk/software/figtree/ (accessed January 1, 2023).
Ronquist, F., Teslenko, M., Mark, P., Avres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice, across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Ryvarden, L., and Johansen, I. (1980). A Preliminary Polypore Flora of East Africa. Oslo: Fungiflora, 1–636.
Stamatakis, A. (2014). RAxML Version 8: a tool for phylogenetic analyses and post analyses of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Swofford, D. L. (2002). PAUP*: phylogenetic analysis using parsimony (* and other methods), version 4.0 beta 10. Sunderland, MA: Sinauer Associates. doi: 10.1002/0471650129.dob0522
Thiers, B. (2018). Index Herbariorum: A Global Directory of Public Herbaria and Associated Staff. New York: New York Botanical Garden's Virtual Herbarium. Available online at: http://sweetgum.nybg.org/science/ih/ (accessed January 1, 2023).
Vilgalys, R., and Hester, M. (1990). Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 172, 4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990
Vu, D., Groenewald, M., De Vries, M., Gehrmann, T., Stielow, B., Eberhardt, U., et al. (2019). Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation. Stud. Mycol. 92, 135–154. doi: 10.1016/j.simyco.2018.05.001
Wan, X. Z., and Yuan, H. S. (2013). Hydnaceous fungi of China 5. Steccherinum (Basidiomycota, Meruliaceae) in China. Mycosystema 32, 1086–1096. doi: 10.13346/j.mycosystema.2013.06.017
Westphalen, M. C., Motato-Vásquez, V., Tomšovsk, M., and Gugliotta, A. M. (2021). Additions to the knowledge of hydnoid steccherinaceae: Cabalodontia, Etheirodon, Metuloidea, and Steccherinum. Mycologia 113, 791–806. doi: 10.1080/00275514.2021.1894536
Westphalen, M. C., Rajchenberg, M., Tomšovský, M., and Gugliotta, A. M. (2018). A re-evaluation of neotropical Junghuhnia s.lat. (Polyporales, Basidiomycota) based on morphological and multigene analyses. Persoonia 41, 130–141. doi: 10.3767/persoonia.2018.41.07
Westphalen, M. C., Tomšovský, M., Gugliotta, A. M., and Rajchenberg, M. (2019). An overview of Antrodiella and related genera of Polyporales from the Neotropics. Mycologia 111, 813–831. doi: 10.1080/00275514.2019.1633895
White, T. J., Bruns, T., Lee, S., and Taylor, J. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in PCR Protocols: A Guide to Methods and Applications, eds. M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (New York, Academic Press), 315–322. doi: 10.1016/B978-0-12-372180-8.50042-1
Wu, F., Zhou, L. W., Vlasák, J., and Dai, Y. C. (2022). Global diversity and systematics of Hymenochaetaceae with poroid hymenophore. Fungal Divers. 113, 1–192. doi: 10.1007/s13225-021-00496-4
Wu, Y. X., Dong, J. H., and Zhao, C. L. (2021). Steccherinum puerense and S. rubigimaculatum spp. nov. (Steccherinaceae, Polyporales), two new species from southern China. Nova Hedwig. 113, 243–258. doi: 10.1127/nova_hedwigia/2021/0636
Keywords: diversity, macrofungi, phylogenetic analysis, wood-rotting fungi, fungal resources
Citation: Liu Z-B, Zhou M, Zhang Q-Y and Si J (2023) A contribution to the genus Steccherinum (Steccherinaceae, Polyporales): Introducing two new species and two new combinations of the genus. Front. Microbiol. 14:1166267. doi: 10.3389/fmicb.2023.1166267
Received: 15 February 2023; Accepted: 27 February 2023;
Published: 22 March 2023.
Edited by:
Yong-Zhong Lu, Guizhou Institute of Technology, ChinaReviewed by:
Ming Zhang, Guangdong Academy of Science, ChinaCopyright © 2023 Liu, Zhou, Zhang and Si. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jing Si, amluZ3NpMTc4OEAxMjYuY29t
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.