- 1California Institute of Technology, Jet Propulsion Laboratory, Pasadena, CA, United States
- 2Department of Plant Science, School of Life Sciences, University of Hyderabad, Hyderabad, India
- 3Department of Physiology and Biophysics, and the WorldQuant Initiative for Quantitative Prediction, Weill Cornell Medicine, New York, NY, United States
- 4NASA Ames, Moffet Field, CA, United States
A single strain from the family Paenibacillaceae was isolated from the wall behind the Waste Hygiene Compartment aboard the International Space Station (ISS) in April 2018, as part of the Microbial Tracking mission series. This strain was identified as a gram-positive, rod-shaped, oxidase-positive, catalase-negative motile bacterium in the genus Cohnella, designated as F6_2S_P_1T. The 16S sequence of the F6_2S_P_1T strain places it in a clade with C. rhizosphaerae and C. ginsengisoli, which were originally isolated from plant tissue or rhizosphere environments. The closest 16S and gyrB matches to strain F6_2S_P_1T are to C. rhizosphaerae with 98.84 and 93.99% sequence similarity, while a core single-copy gene phylogeny from all publicly available Cohnella genomes places it as more closely related to C. ginsengisoli. Average nucleotide identity (ANI) and digital DNA–DNA hybridization (dDDH) values to any described Cohnella species are <89 and <22%, respectively. The major fatty acids for strain F6_2S_P_1T are anteiso-C15:0 (51.7%), iso-C16:0 (23.1%), and iso-C15:0 (10.5%), and it is able to metabolize a wide range of carbon compounds. Given the results of the ANI and dDDH analyses, this ISS strain is a novel species within the genus Cohnella for which we propose the name Cohnella hashimotonis, with the type strain F6_2S_P_1T (=NRRL B-65657T and DSMZ 115098T). Because no closely related Cohnella genomes were available, this study generated the whole-genome sequences (WGSs) of the type strains for C. rhizosphaerae and C. ginsengisoli. Phylogenetic and pangenomic analysis reveals that F6_2S_P_1T, C. rhizosphaerae, and C. ginsengisoli, along with two uncharacterized Cohnella strains, possess a shared set of 332 gene clusters which are not shared with any other WGS of Cohnella species, and form a distinct clade branching off from C. nanjingensis. Functional traits were predicted for the genomes of strain F6_2S_P_1T and other members of this clade.
Introduction
The International Space Station (ISS) is an orbital space habitat which has been continuously inhabited by astronauts for 22 years (Robinson et al., 2006) and is therefore the only current model for how the microbiome of a space habitat evolves over time. Because the ISS is hermetically sealed against the vacuum of space, sources of its microbiome must necessarily be the human commensals from the astronauts, new equipment and supplies, or experiments conducted on board (Checinska Sielaff et al., 2019; Urbaniak et al., 2022). With a number of both governmental and commercial closed-habitat space facilities planned for both earth and lunar orbit in the near future, studying the microbiome of this unique habitat is vital to understanding both how astronaut health is affected by the presence of bacteria and fungi exposed to the space environment, and how those microbes can be utilized for biotechnological applications. Part of this effort should include the whole-genome sequencing (WGS) and taxonomic classification of microbes isolated from the ISS, to track their sources, spread, interactions, and potential applications. Until the mid-2010s, monitoring of bacteria and fungi aboard the ISS was mainly done via traditional culturing and identification using Sanger sequencing methods, to ensure crew health and food and water safety (La Duc et al., 2004; Checinska et al., 2015). More recently, a series of NASA Space Biology-funded missions known as Microbial Tracking 1 and 2 investigated the metagenomes of ISS surfaces and performed WGS on all isolated bacteria and fungi (Checinska Sielaff et al., 2016; Venkateswaran et al., 2017; Urbaniak et al., 2018; Bijlani et al., 2020; Daudu et al., 2020; Solomon et al., 2020; Blachowicz et al., 2021; Simpson et al., 2021a,b), in order to fully characterize the ISS microbiome and how it changes over time (Singh et al., 2018; Checinska Sielaff et al., 2019; Urbaniak et al., 2022). In the ongoing Microbial Tracking investigation of the ISS, spore-forming bacteria have been isolated multiple times from the genera Bacillus and Paenibacillus; however, this is the first instance that a bacterium from the genus Cohnella has been isolated from an ISS surface. As part of the Microbial Tracking 2 mission series, during April of 2018, astronauts sampled various surfaces aboard the ISS (Urbaniak et al., 2022) with pre-packaged, pre-sterilized wipes, including the panel behind the Waste Hygiene Compartment (WHC). Upon return to Earth, the strain F6_2S_P_1T was isolated from the WHC sample wipe, on blood agar incubated at 37°C. Preliminary 16S rRNA sequence analysis placed strain F6_2S_P_1T in the genus Cohnella.
The genus Cohnella (family Paenibacillaceae, order Bacillales, phylum Firmicutes/Bacillota) was first proposed by Kampfer et al. (2006) as distinct from the genus Paenibacillus, with the description of Cohnella thermotolerans (type species, isolated during hygiene checks at a starch production company in Sweden) and the reclassification of Paenibacillus hongkongensis as Cohnella hongkongensis (Teng et al., 2003). Cohnella species are differentiated from Paenibacillus due to high iso-C16:0 content in their fatty acid profiles (Kampfer et al., 2006). Members of Cohnella are rod-shaped, usually endospore-forming, and gram-positive.
Cohnella species have been first isolated from a wide variety of environmental samples, most usually in soil (Kampfer et al., 2014), but also in plant tissue (Garcia-Fraile et al., 2008; Flores-Felix et al., 2014) and compost heaps (He et al., 2013). Species from this genus are used in industrial applications including fermentation, biofuel production, and wastewater treatment bioreactors (Lin et al., 2020; Hero et al., 2021; Chen et al., 2022; Mohammadi et al., 2022). As with the genus Paenibacillus, many members of the genus Cohnella are strongly associated with the plant rhizosphere environment, particularly that of legume species (Niang et al., 2018). Multiple novel Cohnella species have been discovered in the root nodules of leguminous plants, although only two species are known to fix nitrogen (Garcia-Fraile et al., 2008; Wang et al., 2012, 2015; Flores-Felix et al., 2014; Takada et al., 2019). Cohnella species are also thought to play an important role in recycling plant biomass within soil, with multiple members of the genus possessing genes for degradation of hemicellulose, cellulose, and xylan (Khianngam et al., 2010; Hameed et al., 2013; Hero et al., 2021; Johnson et al., 2021). Overall, while Cohnella species have been occasionally detected in the blood and oral cavities of immune-compromised or otherwise unhealthy individuals (Teng et al., 2003; Abou Abdallah et al., 2019; Kim et al., 2021), they do not appear to be common human commensals.
In this study, we completed the characterization study of strain F6_2S_P_1T and describe it here with three main objectives. The first objective was to establish Cohnella sp. F6_2S_P_1T as a novel species, for which chemotaxonomic, phenotypic, phylogenetic (using taxonomic marker genes), and comparative genomic analyses were carried out. In addition, initial taxonomic analysis for strain F6_2S_P_1T showed that it was most closely related to four Cohnella species without publicly available genomes, two of which are officially recognized species (Kim et al., 2010; Kampfer et al., 2014). Thus, the strains Cohnella ginsengisoli DSM 18997T and Cohnella rhizosphaerae DSM 28161T were acquired from the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ) and whole-genome-sequenced, and assembled draft genomes were generated for these species.
The second objective was to discover the phylogenetic placement of strain F6_2S_P_1T within the genus Cohnella, for which we performed multiple phylogenetic analyses as well as a pangenome, and a genome-based tree containing all shared single-copy core genes from the genus Cohnella. The third objective was to explore the functional traits predicted by the genome of strain F6_2S_P_1T. Because strain F6_2S_P_1T is part of a branch of the genus Cohnella which is only distantly related to the rest of the genus, the functional traits predicted for the entire closely related clade of which strain F6_2S_P_1T is a member were characterized.
Materials and methods
Sample collection and isolation
Methods for preparing sampling kits and processing samples were similar to those used in the Microbial Tracking 1 mission (Singh et al., 2018; Checinska Sielaff et al., 2019). For each of five flight missions for Microbial Tracking 2, Sterile TexTra™ 10 TX3224 Dry Cleanroom Wipers were moistened with 15 ml of sterile phosphate-buffered saline (PBS), folded into quarters and placed in sterile Ziploc baggies, which were later used to assemble sampling kits at NASA Ames. Sampling kits were transported to the ISS, where astronauts collected surface particulates from a square meter area of eight surfaces: (1) the cupola, (2) the WHC, (3) the advanced resistive exercise device, (4) dining table, (5) Zero-Gravity Stowage Rack overhead 4, (6) permanent multipurpose module Port 1, (7) laboratory overhead 3, and (8) crew quarters bump-out exterior wall (see Singh et al., 2018 for detailed surface descriptions). For each surface, the astronaut tasked with sampling donned a new pair of sterile gloves, removed and unfolded a sterile wipe from its bag, and wiped a square meter area of the surface three times, folding the wipe in half for the second pass and into quarters for the third pass; each time, the surface was sampled in a different direction (horizontally, vertically, and diagonally) using the new previously unexposed surface of the wipe. Additionally, a wipe was removed from its bag and briefly exposed to the air, as an environmental control. A total of nine (eight locations and one control) samples were produced for each flight and stored at 4°C aboard the ISS and during transport to earth.
Upon delivery to Jet Propulsion Laboratory, ISS wipes and unopened control wipes were processed for the isolation of environmental microbes within a 12-h period. Each wipe was placed in a sterile 500-ml bottle with 200-ml sterile PBS and shaken vigorously for 2 min. PBS was then concentrated using an Innovaprep CP-150 (Innovaprep LLC, Drexel, Missouri, United States) with a 0.2 μm polysulfone hollow fiber concentrating pipette tip (Innovaprep LLC, SKU CC08022-10). Using additional sterile PBS, dilutions of 10−1 through 10−6 were prepared and plated onto Reasoner's 2 agar (R2A, BD Diagnostics Cat # 218263), potato dextrose agar (BD Diagnostics Cat #213400) with 0.05 g/L chloramphenicol, and blood agar (Hardy Diagnostics, #A10) media. Five colonies from each sample on each media type were selected and stored in one-tenth strength tryptic soy agar (TSA) stabs. Strain F6_2S_P_1T was isolated from blood agar inoculated with the PBS concentrate from Location 2 (wall behind WHC), collected during Flight 6 (third flight in the Microbial Tracking 2 mission series).
DNA extraction and whole-genome sequencing
The ZymoBIOMICS DNA MagBead kit was used to extract genomic DNA from strain F6_2S_P_1T according to the manufacturer's instructions. An Illumina Nextera DNA Flex library preparation kit was used to prepare the library for WGS as in Simpson et al. (2021b). The library was sequenced as part of a run on a NovaSeq 6000 S4 flow cell paired-end 2 × 150-bp platform. FastQC v0.11.7 (Bioinformatics, 2011) was used to quality filter and trim reads, and fastp v0.20 (Chen et al., 2018) was used to remove adapter sequences. A draft genome was assembled to the scaffold level using SPAdes v3.11.1 (Bankevich et al., 2012). Assembly quality, including number of contigs, genome size, and N50 value, was assessed using QUAST v5.0.2 (Gurevich et al., 2013). Default settings were used for all steps except for fastp, which included 512 adapters screening.
A secondary round of sequencing for strain F6_2S_P_1T, as well as C. ginsengisoli DSM 18997T and C. rhizosphaerae DSM 28161T, was conducted using Oxford Nanopore sequencing. C. ginsengisoli DSM 18997T and C. rhizosphaerae DSM 28161T were ordered from the DSMZ and cryostocks prepared for long term storage. To produce a more resolved genome for Cohnella sp. F6_2S_P_1T and to provide WGS, all strains were re-streaked from cryostock on TSA and genomic DNA was extracted using a Zymo QuikSpin DNA extraction kit. Nanopore sequencing libraries were prepared using an SQK-LSK110 Ligation Sequencing kit (Oxford Nanopore) and sequenced on an R9.4.1 flow cell, with base calling via MinKNOW/Guppy. Porechop (Wick et al., 2017a) and Nanofilt (De Coster et al., 2018) were used to remove adapter sequences and filter reads using the settings “-q 10 -l 200.” A hybrid genome assembly for Cohnella sp. F6_2S_P_1T was generated via Unicycler (Wick et al., 2017b) using Nanopore reads to polish the Illumina assembly. Flye (Kolmogorov et al., 2019) followed by polishing with Medaka (Oxford Nanopore Technologies) was used to assemble draft genomes for type strains of C. ginsengisoli and C. rhizosphaerae from Nanopore reads, and assembly quality was assessed using QUAST v5.0.2 (Gurevich et al., 2013).
Phylogenetic analysis
Phylogenetic analysis using 16S rRNA gene sequences was carried out by extracting the 16S rDNA sequence from the WGS of strain F6_2S_P_1T and aligning it to the NCBI nucleotide database via blast search. The top 30 matching 16S rRNA sequences were retrieved. In addition, all publicly available genomes from the genus Cohnella were downloaded, and their 16S rDNA sequences were extracted using a blast wrapper script. 16S rRNA genes from the species Paenibacillus barengoltzii, Paenibacillus terrae, Thermoflavimicrobial dichotomicus, and Thermoactinomyces vulgaris were used to root the tree. Similarly, gyrB sequences from strain F6_2S_P_1T and from all publicly available Cohnella genomes, and the outgroups T. dichotomicus and T. vulgaris were extracted with the same wrapper script. For both gyrB and 16S trees, alignment was performed in R using the DECIPHER package (Wright, 2016). Trimming and tree-building using maximum likelihood and the substitution model GTR + G + I were performed using the phangorn package, including model selection using AIC values and 1,000 bootstrap replicates (Schliep, 2011). Phylogenetic trees based on 16S rRNA and gyrB genes were visualized using ggtree (Yu et al., 2017).
A genome-based phylogenetic tree was also generated using genes detected from the pangenomic analysis, which contained all publicly available Cohnella genomes, plus those of strain F6_2S_P_1T, C. ginsengisoli, and C. rhizosphaerae generated during this study (see below section for methods for pangenomic tree generation). Aligned, single-copy core genes which were shared by all species and had a geometric homogeneity of 1, but a functional homogeneity below 0.9, were extracted from the Cohnella genus pangenome into a concatenated FASTA file. A maximum likelihood genome-based tree was generated using raxml v8.2.12 (Stamatakis, 2014; Kozlov et al., 2019) with a GTR-GAMMA substitution model and the option “autoMRE” to automatically choose the optimal number of bootstrap replicates. The tree was visualized using the online platform Interactive Tree of Life (iTOL) (Letunic and Bork, 2007).
Additionally, average nucleotide identity (ANI) and digital DNA–DNA hybridization (dDDH) were calculated for strain F6_2S_P_1T against all sequenced and described Cohnella species using fastANI v1.33 (Jain et al., 2018) and DSMZ's online Genome-to-Genome Distance Calculator v3.0 (Meier-Kolthoff et al., 2022).
Light microscopy and SEM
A liquid culture of strain F6_2S_P_1T was incubated at 80°C for 10 min, then plated on R2A and grown at 30°C for 5 days to induce endospore formation. Endospore staining with malachite green and safranin was performed using the Schaeffer–Fulton method (Schaeffer and Fulton, 1933). Light microscopy and phase contrast images were taken on an Olympus BX53 microscope with an Olympus DP25 camera using Olympus cellSens software.
To prepare a colony of strain F6_2S_P_1T for SEM, the strain was streaked out on TSA and allowed to grow at 30°C for 48 h. A pure colony was then suspended in chilled 2.5% glutaraldehyde (Ted Pella Inc., Redding, CA, United States) in 0.1M sodium cacodylate (NaCaco) (Sigma Aldrich) solution for 1 h. The sample was then filtered through a 0.2 μm Isopore filter membrane (MilliporeSigma, Burlington, MA, United States) and moved into a 1.5 ml centrifuge tube. The sample was then incubated in 0.1 M NaCaco solution at 4°C for 10 min and then replaced with fresh 0.1 M NaCaco solution; this replacement was repeated three times. The sample was then dehydrated in a series of increasing IPA solutions mixed with water for 10 min each. The series was 50, 70, 80, 90, 95, and 100%, with the final 100% IPA rinsing occurring three times. The specimen was stored at 4°C in 100% IPA until it was critical point dried in a Tousimis Automegasamdri 915B critical point dryer (Rockville, MD, United States). Samples were adhered with carbon tape to SEM stubs (Ted Pella Inc.) and then underwent ~12 nm thick carbon coating with a Leica EM ACE600 Carbon Evaporator (Deerfield, IL, United States). SEM micrographs were captured on a FEI Quanta 200F microscope (Thermo Fisher, Waltham, MA, United States) located at the California Institute of Technologies Kavli Nanoscience Institute.
Pangenomic analysis and other genome characterization
Pangenomic analysis was carried out using anvi'o v7.1 (Eren et al., 2015) similar to methods described in Singh et al. (2021). Open reading frames were annotated with COG, KEGG, and KOfam database pathways/functions, and ANI between all genomes was additionally calculated. Two pangenomes were generated: one including all publicly available Cohnella genomes including unnamed species and one containing only named species. The function anvi-compute-functional-enrichment-in-pan was used to calculate functional enrichment in the C. ginsengisoli clade compared to the rest of the genus Cohnella.
The genome of the strain F6_2S_P_1T was also uploaded and annotated using the online Rapid Annotation using Subsystems Technology (RAST platform) (Aziz et al., 2008). The presence of biosynthetic gene clusters (BGCs) which might produce secondary metabolites was predicted in the strain F6_2S_P_1T genome using the antiSMASH pipeline (Blin et al., 2021). The presence of antimicrobial resistance or virulence genes was predicted by the ABRicate pipeline (Seemann, 2018).
To evaluate the diversity of carbohydrate-degrading and other carbohydrate-cycling enzymes (Carbohydrate-Active Enzymes, i.e., CAZymes) (Drula et al., 2022), the dbcan3 stand-alone tool (Zhang et al., 2018) was run against all publicly available genomes in the genera Cohnella and Paenibacillus as well as the genomes of strain F6_2S_P_1T, C. ginsengisoli, and C. rhizosphaerae. The results were parsed to generate a count matrix for the number of each CAZyme family/subfamily predicted in the genome of each species. All statistics and plots from analysis of this data were generated in R [v 4.2.2, “Innocent and Trusting,” R Core Team (2022)] utilizing the Tidyverse package (Wickham et al., 2019). An NMDS ordination (stress = 12.9, k = 3) was generated via vegan (Oksanen et al., 2019) using Bray–Curtis distance after standardization of each CAZyme category to between 0 and 1, and individual Kruskal–Wallis tests were performed for each CAZyme testing mean abundance between members of the C. ginsengisoli clade and other species of Cohnella and Paenibacillus with Bonferroni correction for multiple testing. Additionally, a heatmap of abundance for each category was generated using the pheatmap package (Kolde and Kolde, 2018) after standardization of the data using robust centered log ratios.
Phenotypic characterization
Growth temperature was determined by inoculating both solid (R2A plates) and liquid (trypticase soy broth (TSB), BD Diagnostics Cat # 257107) media in 15 ml loose-capped centrifuge tubes with strain F6_2S_P_1T and growing at 4, 15, 25, 30, 37, and 45°C. Plates and tubes were checked for growth daily for 7 days and incubation was halted if growth was observed. Samples grown at 4 and 15°C were incubated for additional time before final growth assessment: 2 weeks total for growth at 15°C and 4 weeks total at 4°C. Salt tolerance was determined by inoculating strain F6_2S_P_1T onto R2A with 0, 1, 2, 3, 4, and 5% added NaCl, as well as agar containing only peptone plus 0 or 1% NaCl, and examining growth after 7 days of incubation at 30°C. Oxidase activity was determined by testing solid culture with OxiDrops™ liquid oxidase reagent (Hardy Diagnostics). Catalase activity was determined by mixing colonies of strain F6_2S_P_1T with hydrogen peroxide on a sterile glass slide and monitoring for the presence of effervescence. pH tolerance (4–10) was tested by adjusting the pH of TSB broth with biological buffers as in Xu et al. (2005).
Mapping of metagenomic reads to genome
Metagenomic reads from ISS surfaces from Microbial Tracking 1 (Checinska Sielaff et al., 2019) and Microbial Tracking 2 (Urbaniak et al., 2022) were mapped to the genome of Cohnella sp. F6_2S_P_1T using Bowtie2 (Langmead and Salzberg, 2012), using the flags “—no-mixed” (no unpaired alignments) and “—no-discordant” (only matches where both forward and reverse reads map to the same area are considered). The number of matches where both a forward and reverse read mapped to the Cohnella sp. F6_2S_P_1T genome were reported as a percentage of total reads.
Chemotaxonomic analysis
Cells grown in TSB medium were harvested when growth of the cultures reached approximately 70% of the maximal optical density (exponential growth phase) and used for analysis of cellular fatty acids, polar lipids, and quinones, which was done as described previously (Ramaprasad et al., 2015). In brief, for cellular fatty acids analysis, 40 mg of bacterial cells were subjected to a series of four different reagents followed by saponification and methylation of fatty acids, thus enabling their cleavage from lipids. The fatty acid methyl esters thus obtained were analyzed by gas chromatography. The peaks obtained were then labeled and the equivalent chain length values were computed as published elsewhere (MIDI, 2012).
The polar lipid profile was analyzed by extracting polar lipids with methanol/chloroform/saline (2:1:0.8, by vol.) from 1 g freeze-dried bacterial cells. Separation of lipids was done by two-dimensional chromatography on a silica gel TLC plate (Kieselgel 60 F254; Merck) using chloroform–methanol–water (75:32:4 by vol.) in the first dimension and chloroform–methanol–acetic acid–water (86:16:15:4 by vol.) in the second dimension. Total polar lipid profiles were detected by spraying with 6% ethanolic molybdophosphoric acid. The respiratory isoprenoid quinone was extracted with chloroform/methanol mixture (2:1, v/v), evaporated under a vacuum, re-extracted with acetone, and analyzed using high-performance lipid chromatography according to previously reported methods (Ramaprasad et al., 2018). Polyamines were analyzed as described by Busse and Auling (1988) and Busse et al. (1997). The peptidoglycan of cells of strain F6_2S_P_1T was isolated following disruption of the cells by shaking with glass beads and subsequent trypsin digestion, according to the method of Schleifer (1985). Cell wall was hydrolyzed for amino acid analyses as described by Schleifer and Kandler (1972). Amino acids in cell-wall hydrolysates were analyzed by high performance liquid chromatography (HPLC) (Mckerrow et al., 2000; Ramaprasad et al., 2015). Carbon substrate utilization profile analysis was carried out as per the BioLog protocol provided by the manufacturer for Firmicutes using the GEN III MicroPlate test assay with a Biolog system (Hayward, CA, USA).
Results and discussion
Cohnella sp. F6_2S_P_1T isolated from the ISS was subjected to polyphasic taxonomic characterization to determine its phylogenetic affiliation. In addition, WGSs of C. ginsengisoli and C. rhizosphaerae were generated and compared with the genome of Cohnella sp. F6_2S_P_1T to identify the variable, conserved, and distinctive genomic traits.
Genome characteristics and relatedness indices
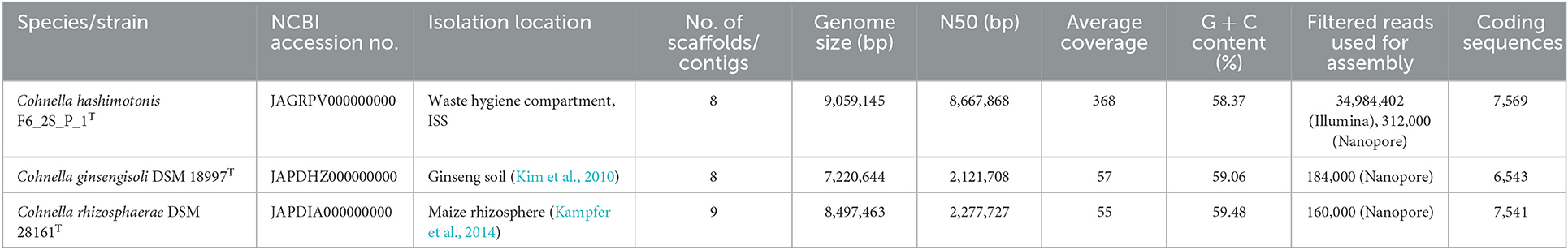
Assembly statistics for Cohnella sp. F6_2S_P_1, C. ginsengisoli, and C. rhizosphaerae are summarized in Table 1. Genome sizes are 9.06 Mb for strain F6_2S_P_1, 7.22 Mb for C. ginsengisoli, and 8.5 Mb for C. rhizosphaerae. GC content ranged from 58.37% for Cohnella sp. F6_2S_P_1T to 59.49% for C. rhizosphaerae. In general, among 39 sequenced Cohnella genomes, genome size varied from 4.0 to 9.4 Mb. This genome variation should be investigated for any functional genes acquired or shed over time.
ANI values between Cohnella sp. F6_2S_P_1T and C. ginsengisoli, and C. rhizosphaerae were 88.4 and 88.0%, respectively (Table 2). ANI between Cohnella sp. F6_2S_P_1T and all other named Cohnella species for which a genome was publicly available was <79.4% (Table 2). We also tested ANI between Cohnella sp. F6_2S_P_1T and available genomes for unnamed Cohnella species and found that Cohnella sp. OV330 and Cohnella sp. GbtcB17 were also closely related, with ANIs of 88.9 and 88.0%, respectively. This was also the trend for C. ginsengisoli and C. rhizosphaerae, which showed ANI to each other and to Cohnella sp. OV330 and Cohnella sp. GbtcB17 ranging from 88 to 92%, and which was otherwise most closely related to C. nanjingensis with an ANI value of ~79%. The dDDH results showed a similar pattern, as values were 35 and 34.5% between Cohnella sp. F6_2S_P_1T and C. ginsengisoli, and C. rhizosphaerae, respectively, and were otherwise below 22% (Table 2).
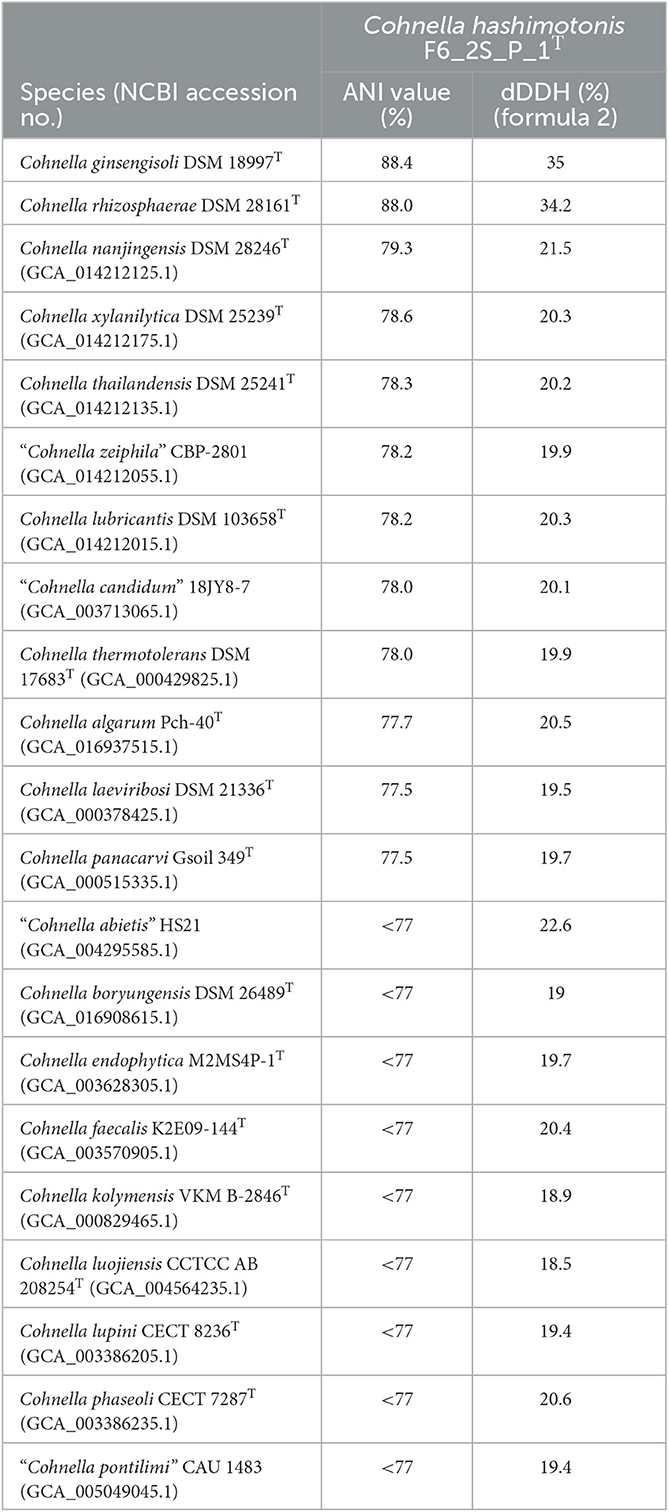
Table 2. Average nucleotide identity and digital DNA hybridization values between strain F6_2S_P_1T and other named members of the genus Cohnella.
Phylogeny of strain F6_2S_P_1T
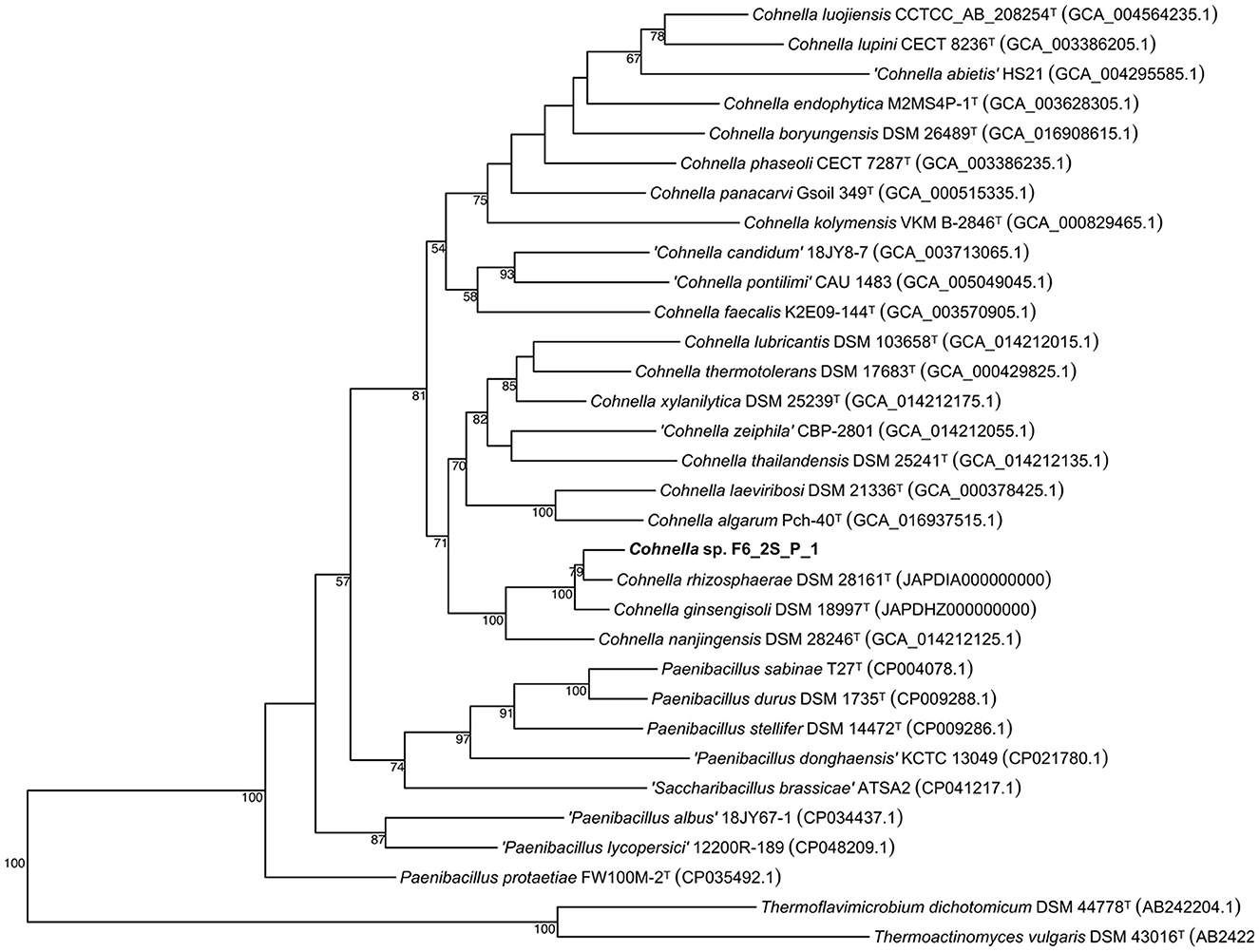
A 16S rDNA-based phylogenetic tree of all Cohnella species, plus the species P. barengoltzii, P. terrae, T. dichotomicus, and T. vulgaris as outgroups, shows that strain F6_2S_P_1T is placed within a clade containing the officially named species C. rhizosphaerae and C. ginsengisoli, as well as yet-to-be described species “C. plantaginis” and “C. capsici” (Figure 1). The 16S rDNA sequence similarities of strain F6_2S_P_1T with the members of this clade ranged from 98.84 to 97.37%. “C. plantaginis” and “C. capsici” form their own well-supported branch within this clade, while C. rhizosphaerae is basal to strain F6_2S_P_1T and C. ginsengisoli is the most basal species of the clade (Figure 1). However, branch support for the divergence of strain F6_2S_P_1T from C. rhizosphaerae was low (50) and not shown in Figure 1. A gyrB-based phylogeny was also generated using all publicly available Cohnella genomes plus the outgroups T. dichotomicus and T. vulgaris (Figure 2). The gyrB sequence similarities of strain F6_2S_P_1T with the members of this clade ranged from 93.99 to 93.55%. The gyrB tree supports the position of strain F6_2S_P_1T as non-basal to C. rhizosphaerae and C. ginsengisoli, with C. ginsengisoli as the most basal species in this branch of the genus Cohnella. Since WGSs of “C. plantaginis” and “C. capsici” are not available, they were not included in the gyrB phylogeny.
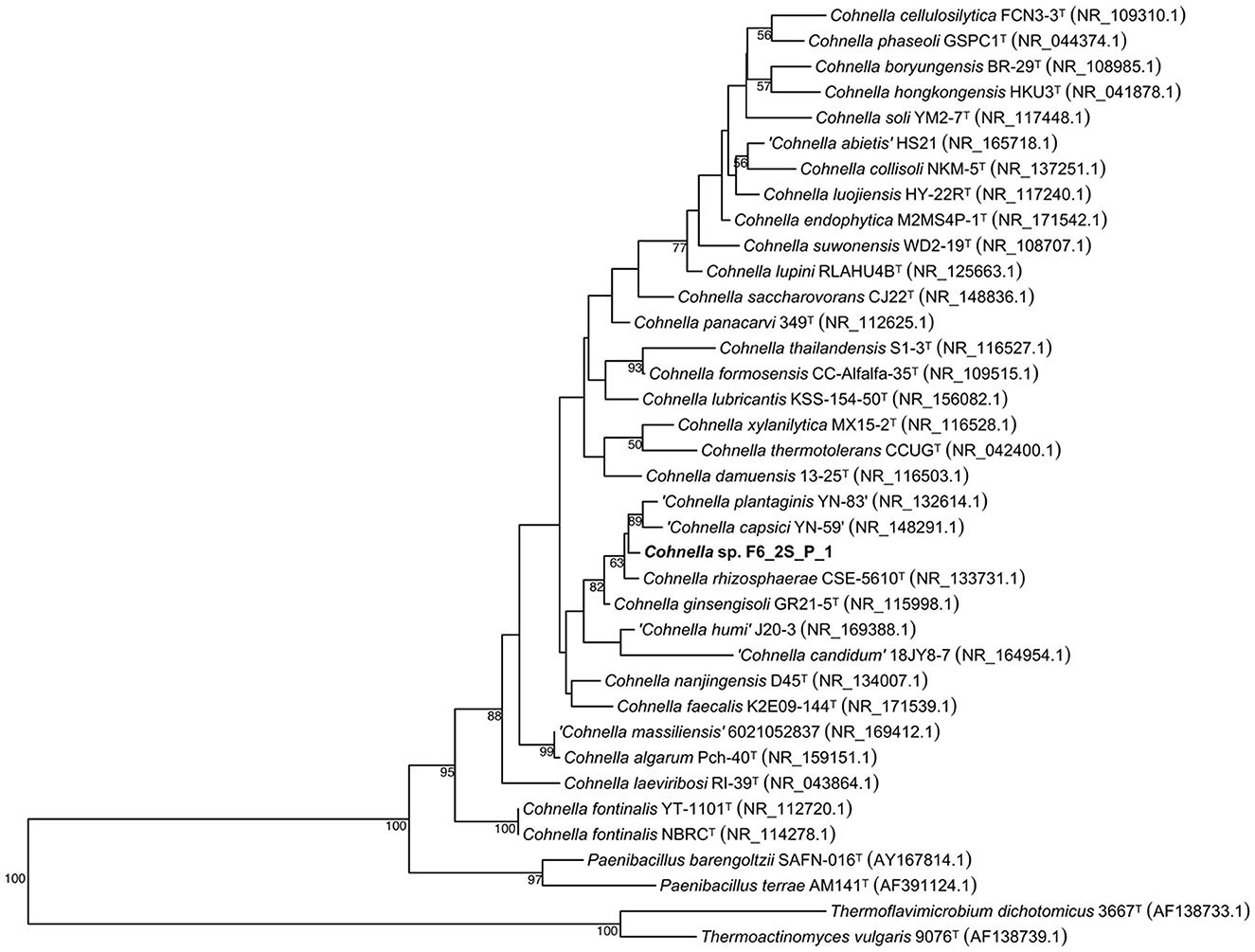
Figure 1. Maximum likelihood phylogenetic tree based on 16S rRNA gene sequences, showing the inclusion of Cohnella sp. F6_2S_P_1T in a branch of the genus including species isolated from plant tissue or rhizosphere soil.
The genome-based tree (Figure 3) for all sequenced and fully described Cohnella species was based on a concatenated alignment of 14 gene clusters containing a total of 294 single-copy core genes common to all species in the genus. This tree supports the position of C. rhizosphaerae as basal to C. ginsengisoli, in contrast to the 16S and gyrB trees. Nevertheless, the position of strain F6_2S_P_1T remains the same. A second genome-based tree was generated using a second pangenome analysis including genomes of all Cohnella species along with yet-to-be described species (Supplementary Figure S1), which showed that Cohnella sp. OV330 and Cohnella sp. GbtcB17 are also part of this clade, with Cohnella sp. GbtcB17 as the most basal species and Cohnella sp. OV330 as basal to C. ginsengisoli and strain F6_2S_P_1T.
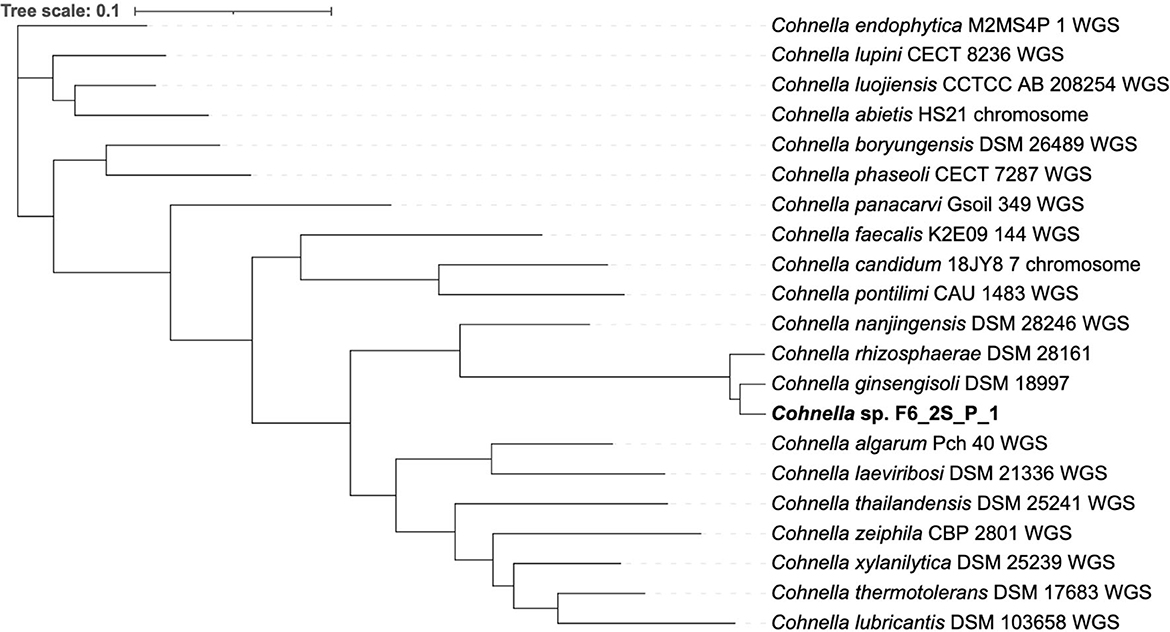
Figure 3. Phylogenetic tree based on 294 single-copy core genes shared by all available Cohnella species genomes.
Phenotypic and chemotaxonomic characterization
Light microscopy and staining with safranin and malachite green show that cells are rod-shaped and vary extensively in length and that strain F6_2S_P_1T forms ellipsoidal/oval endospores which quickly convert back to vegetative cells (Figure 4). SEM imaging reveals the presence of peritrichous flagella attached to F6_2S_P_1T cells (Figure 5A) and confirms the presence of elongated cells (Figure 5B).
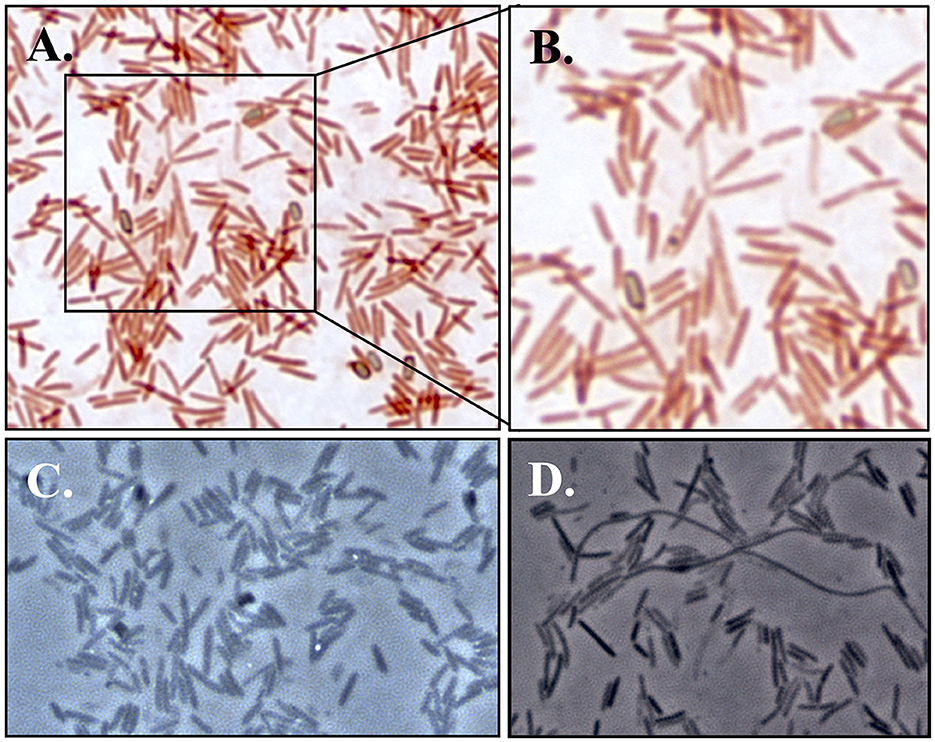
Figure 4. Light microscopy images of strain F6_2S_P_1T. (A) Bright field images at 1000X magnification with staining with safranin and malachite green for endospore visualization; (B) close-up of A featuring multiple endospores which are beginning to transition back to vegetative cells, as well as an endospore within a vegetative cell; (C) phase-contrast microscopy (1000X) of strain F6_2S_P_1T showing the presence of endospores; (D) phase-contrast microscopy (1000X) of strain F6_2S_P_1T showing the presence of elongated Spaghetti-like cells.
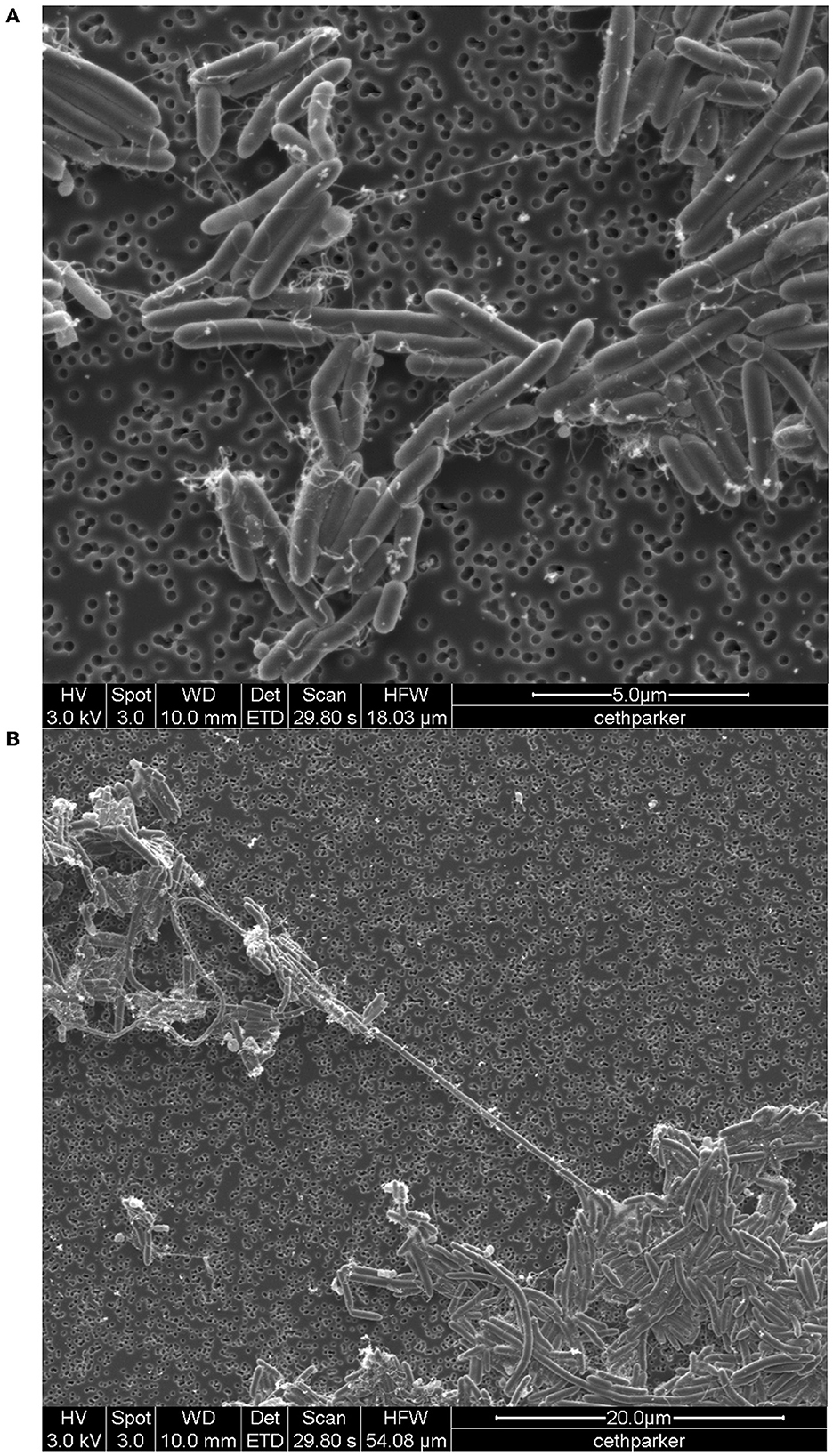
Figure 5. SEM image of Cohnella sp. F6_2S_P_1T, (A) with the visible presence of flagella and (B) showing long “spaghetti-like” cells.
Differentiating biochemical characteristics of Cohnella sp. F6_2S_P_1T with other Cohnella species are depicted in Table 3. Cohnella sp. F6_2S_P_1T grew at temperatures between 15 and 37°C, between pHs 6 and 8, and from 0 to 3% NaCl. Colonies were weakly oxidase-positive and catalase-negative. Catalase positive in C. ginsengisoli, C. plantaginis, and C. capsici; absence of growth at 3% NaCl and production of oxidase in C. ginsengisoli; absence of endospore formation in C. rhizosphaerae; nitrate reduction not seen in C. plantaginis and C. capsica were some of the phenotypic characteristics that could differentiate Cohnella sp. F6_2S_P_1T from other Cohnella species.
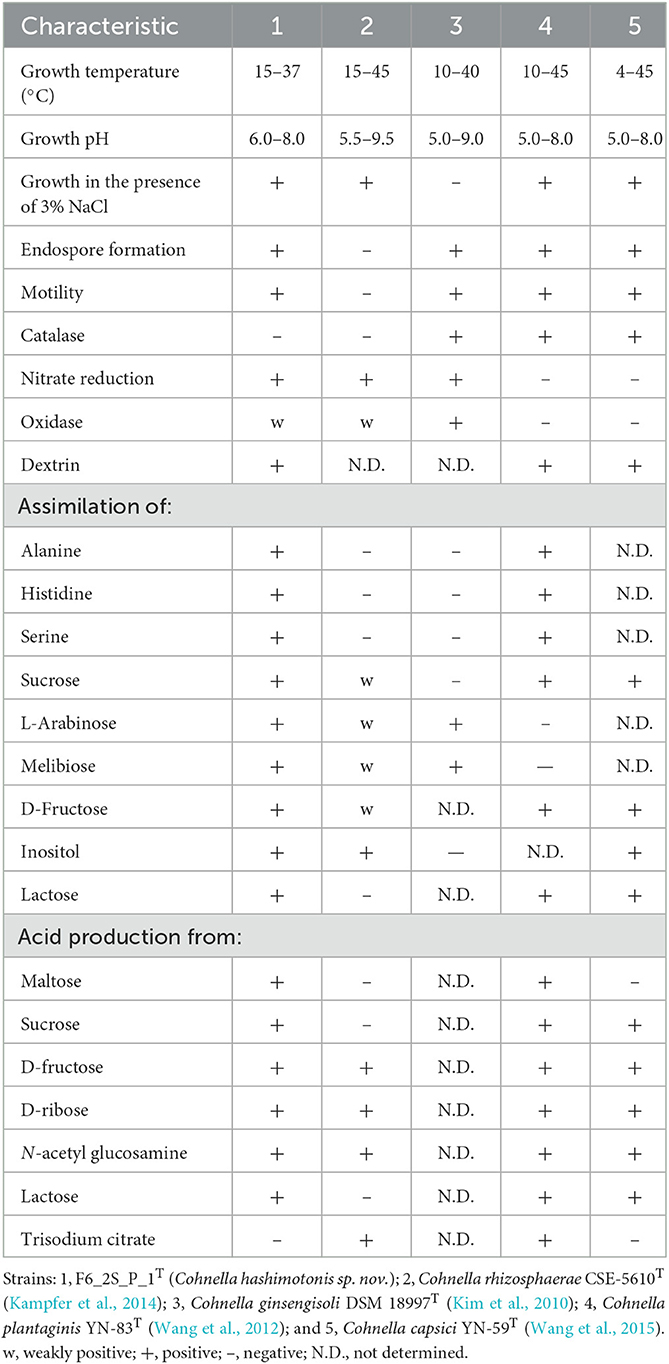
Table 3. Phenotypic characteristics of strain F6_2S_P_1T and closely related species of genus Cohnella.
Major fatty acids (more than 5% of the total) are C16:0, iso-C15:0, iso-C16:0, and anteiso-C15:0 (Table 4). Differences in fatty acid profile of strain F6_2S_P_1T with regard to C. rhizosphaerae CSE-5610T, Cohnella ginsengisoli DSM 18997T, C. plantaginis YN-83T, and C. capsici YN-59T are shown in Table 4. C. rhizosphaerae showed >25% of C16:0 saturated fatty acid, whereas other Cohnella species exhibited <10% production. Absence of branched fatty acid such as iso-C14:0 in Cohnella sp. F6_2S_P_1T and in C rhizosphaerae could be used to differentiate the novel species described in this study. However, production of iso-C16:0 and anteiso-C15:0 was very high in Cohnella sp. F6_2S_P_1T when compared to other described Cohnella species.
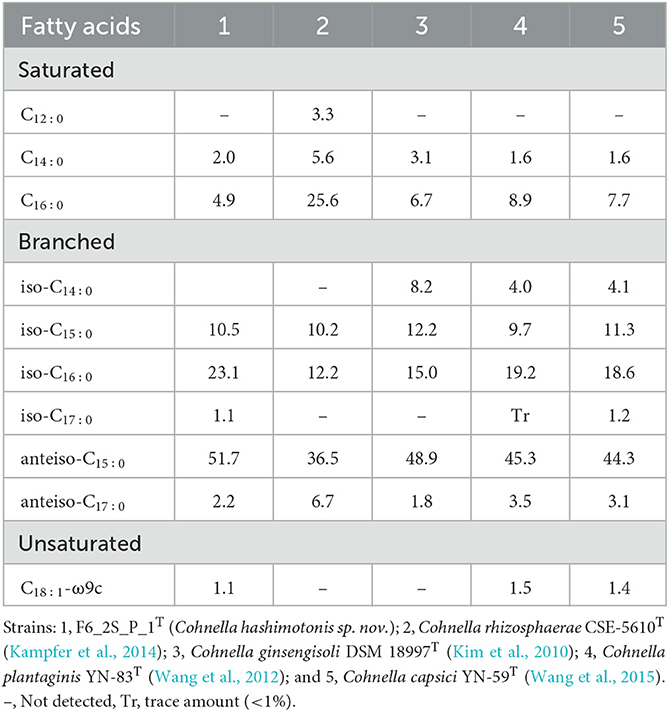
Table 4. Percentage of total cellular fatty acids for strain F6_2S_P_1T and closely related species of genus Cohnella.
The polar lipid profile was composed of diphosphatidylglycerol, phosphatidylglycerol, phosphatidylethanolamine, three aminophospholipids, a phospholipid, and minor amounts of unidentified polar lipid (Supplementary Figure S2). This profile is in agreement with other described members of the C. ginsengisoli clade (Kim et al., 2010; Kampfer et al., 2014; Wang et al., 2015).
The quinone system contained menaquinones MK-7 (98%) and MK-8 (2%). The cell-wall peptidoglycan of strain F6_2S_P_1T contained meso-diaminopimelic acid as the diamino acid, which is the case for other described members of the C. ginsengisoli clade where that data are reported. All other described members of the C. ginsengisoli clade also contain MK-7 as the dominant menaquinone (Kim et al., 2010; Kampfer et al., 2014; Wang et al., 2015). Descriptions of C. capsici and C. plantaginis do not report low abundance menaquinones, but the quinone system of F6_2S_P_1T is similar to that of C. rhizosphaerae (99.8% MK-7, 0.2% MK-8), while C. ginsengisoli has 93% MK-7 and 7% MK-6.
Comparative genomic analysis of C. hashimotonis and the C. ginsengisoli clade
The genome of strain F6_2S_P_1T contained 309 genes not aligned with other species of Cohnella, named or undescribed (Figure 6). Of these genes, 121 of them received an annotation from at least one predictive database. Annotated genes specific to strain F6_2S_P_1T were dominated by the COG pathways of carbohydrate and/or amino acid transport and metabolism (27 genes), transcription (10 genes), cell wall/membrane/envelope biogenesis (11 genes), and signal transduction mechanisms (+ transcription) (18 genes). The large number of carbon-cycling metabolic genes found in the genome of strain F6_2S_P_1T within Cohnella is unsurprising, given the diverse nature of the carbon substrates which strain F6_2S_P_1T, C. capsici, and C. plantaginis can metabolize compared to the fairly limited substrates utilized by C. rhizosphaerae and C. ginsengisoli (Table 3).
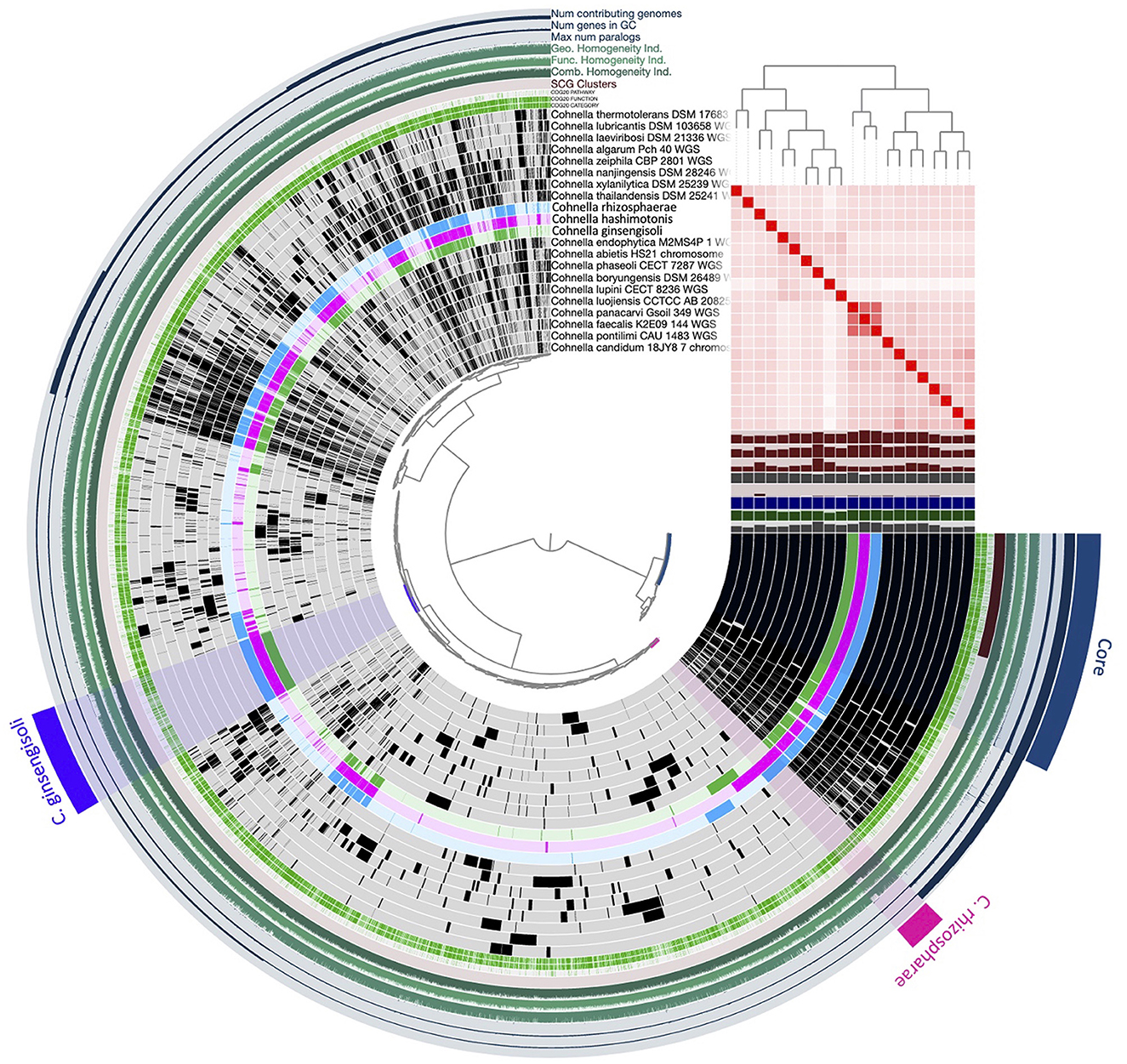
Figure 6. Pangenomic analysis of the genus Cohnella, including C. hashimotonis sp. nov. (pink) as well as its previously unsequenced closest relatives C. rhizosphaerae (blue) and C. ginsengisoli (green).
Because C. capsici and C. plantaginis are both characterized as fixing N2, the annotations for the genome of strain F6_2S_P_1T were searched extensively for genes having to do with nitrogen fixation, as well as blastn and blastx searches for key genes involved in N2 fixation. However, despite the presence of the gene nifU in most species of the genus Cohnella, there was no indication that strain F6_2S_P_1T encodes for N2 fixation.
Other genes of interest predicted by annotation on the RAST platform include multiple genes for endospore formation and for the plant growth hormone auxin, genes which are likely shared by many members of the genus Cohnella, which belongs to the order Bacillales (including many known spore formers) and which are common plant endophytes. BGCs which might produce secondary metabolites were predicted in the genome of strain F6_2S_P_1T via the antiSMASH pipeline, including two lassopeptides, two non-ribosomal peptide synthetase clusters, one agrD-like cyclic lactone autoinducer peptide, one protein cluster, and one heterocyst glycolipid synthase-like polyketide synthase. All are uncharacterized except the two lassopeptides; these are moderate matches to the lasso peptide paeninodin which was characterized from the species Paenibacillus dendritiformis (Zhu et al., 2016). Paeninodin and BGCs showing similarity to it have been predicted in multiple Bacillus and Paenibacillus endophyte species and are suspected to have biocontrol properties that combat plant pathogens (Muñoz et al., 2022; Semenzato et al., 2022). The genomes of C. ginsengisoli, C. rhizosphaerae, and strain F6_2S_P_1T all contained 2–3 predicted BGCs for paeninodin, with C. ginsengisoli containing one that is a 100% match, while C. nanjingensis (the most closely related species outside the C. ginsengisoli clade) contained a single paeninodin match. The only other match in any of these species to a known BGC is a predicted cluster in the C. rhizosphaerae which is a moderate match to ectoine. Other differences include multiple cyclic lactone autoinducer peptide BGCs predicted in the genome of C. nanjingensis (vs. a single one in all other tested genomes), and a siderophore predicted in the genome of C. rhizosphaerae. Strain F6_2S_P_1T was the only genome in the genus Cohnella which a heterocyst glycolipid synthase-like polyketide synthase was detected; these BGCs are used by cyanobacteria to compartmentalize N2 fixation and are activated in low-nitrogen conditions (Campbell et al., 1997), but as strain F6_2S_P_1T does not contain a full suite of genes for N2 fixation and the cluster is not a strong match, it is likely a producer of some kind of potentially uncharacterized lipid compound. No genes for antimicrobial resistance or virulence were predicted by the ABRicate pipeline.
The entire closely related branch of which strain F6_2S_P_1T is a member, which we term the C. ginsengisoli clade (due to its basal position and status as the first officially described species within the clade), contained 322 gene clusters not found in any other publicly available Cohnella genome. All members of this clade in addition to strain F6_2S_P_1T were originally isolated from plant tissue or the rhizosphere environment, including C. ginsengisoli (isolated from soil in which ginseng was grown, Kim et al., 2010), C. rhizosphaerae (isolated from healthy maize seed tissue, Kampfer et al., 2014), C. capsici (isolated from Capsicum rhizosphere, Wang et al., 2015), C. plantaginis (isolated from plantain rhizosphere, Wang et al., 2012), Cohnella sp. OV330 (isolated from poplar rhizosphere, JGI, 2015), and Cohnella sp. GbtcB17 (isolated from Ginko biloba root tissue, Zou et al., 2021). It is speculated that strain F6_2S_P_1T might have originated from the plants grown in ISS. Since shotgun metagenomics of plants associated samples (tissue, water, and soil) were not available, the source of this novel strain isolated from the WHC location could not be confirmed. Cohnella was one of the top 30 genera recently detected via 16S amplicon sequencing of plant tissue of chili peppers grown in the Advanced Plant Habitat from sterilized seeds aboard the ISS, though it was not a top genus detected in artificial soil medium (Khodadad et al., 2022). Cohnella was also not a dominant member of the microbiome of Outredgeous lettuce grown aboard the ISS (Khodadad et al., 2020). However, since in these ISS studies, a 250-bp 16S rRNA gene amplicon was used for the microbial diversity analyses, Cohnella species might have not been identified appropriately.
Functional enrichment analysis for the C. ginsengisoli clade compared to other Cohnella species using the COG20 and KOfam databases revealed hundreds of statistically significantly enriched predicted functions (p < 0.05 with FDR adjustment) annotating one or more gene clusters, including 11 functions which were detected in all C. ginsengisoli clade members (Supplementary Table S1) but in none of the rest of the genus. Most notably, C. ginsengisoli clade species were enriched in genes for heme oxygenase and for a bacteriophytochrome (annotated on NCBI as diguanylate cyclase), which were detected using both COG20 and KOfam database annotation. Consensus protein sequences for genes related to these functions, when aligned to the NCBI database, most closely match to the genome of Paenibacillus chitinolyticus (after matches to members of the C. ginsengisoli clade); heme oxygenase and diguanylate cyclase are located one after the other in the assembled chromosome of this species (NZ_CP026520.1). Heme oxygenase (HO-1) breaks down heme and releases iron and is a regulator of intracellular iron availability in plants and animals (Singh and Bhatla, 2022). Enhanced HO-1 expression in plants and animals is also associated with stress response as the resulting biliverdin and bilirubin scavenge ROS species (Singh and Bhatla, 2022); the bacterial species Paenibacillus polymyxa applied as a probiotic induces increased intestinal HO-1 expression as part of its anti-oxidant effects on gut health in animals (Wu et al., 2019). In bacteria, expression and production of heme oxygenase are associated with iron scavenging within a host, and heme oxygenase expression can have either beneficial anti-oxidant or enhanced pathogenic effects on plant or animal hosts (Richard et al., 2019). In non-photosynthetic bacteria, heme oxygenase also produces biliverdin which is used as the chromatophore in bacteriophytochromes (Takemoto et al., 2019). It is also possible that this HO-1/diguanylate cyclase complex acts as an oxygen sensor (Martínková et al., 2013). Other functions significantly enriched in this clade include stress response genes such as heat shock protein DnaJ. KEGG pathway analysis predicted two functional pathways in all C. ginsengisoli clade species which were not present in any other Cohnella species: a pathway for carbon fixation (incomplete reductive citrate cycle) and a pathway for plant terpenoid biosynthesis (specifically beta-carotene biosynthesis).
Predictive analysis of the presence of CAZymes (carbohydrate-active enzymes, Drula et al., 2022) was followed by ordination (Figure 7; Supplementary Figure S3) and heatmap generation (Supplementary Figure S4). Both heatmaps and ordinations show that strain F6_2S_P1T and its closest relatives cluster tightly together, along with C. fermenti (isolated from a fermented vegetable, Lin et al., 2020), C. phaseoli (isolated from the root nodule of a legume species, Garcia-Fraile et al., 2008), C. zeiphila (isolated from corn tissue, Johnson et al., 2021), and several undescribed strains; members of the C. ginsengisoli clade, therefore, appear to be both taxonomically similar and have similar profiles of carbohydrate-cycling genes. PERMANOVA analysis confirmed that the centroid of the C. ginsengisoli clade was significantly different from that of other Cohnella and Paenibacillus species (p < 0.001). One axis out of three in the NMDS ordination clearly differentiates the C. ginsengisoli clade and the other Cohnella species mentioned above from the majority of Cohnella and Paenibacillus species (NMDS2, shown in Figure 7; Supplementary Figure S3); therefore, scores correlating CAZyme ordination vectors to this axis using the envfit function were assessed for significance, plotted, and ranked against the axis in question.
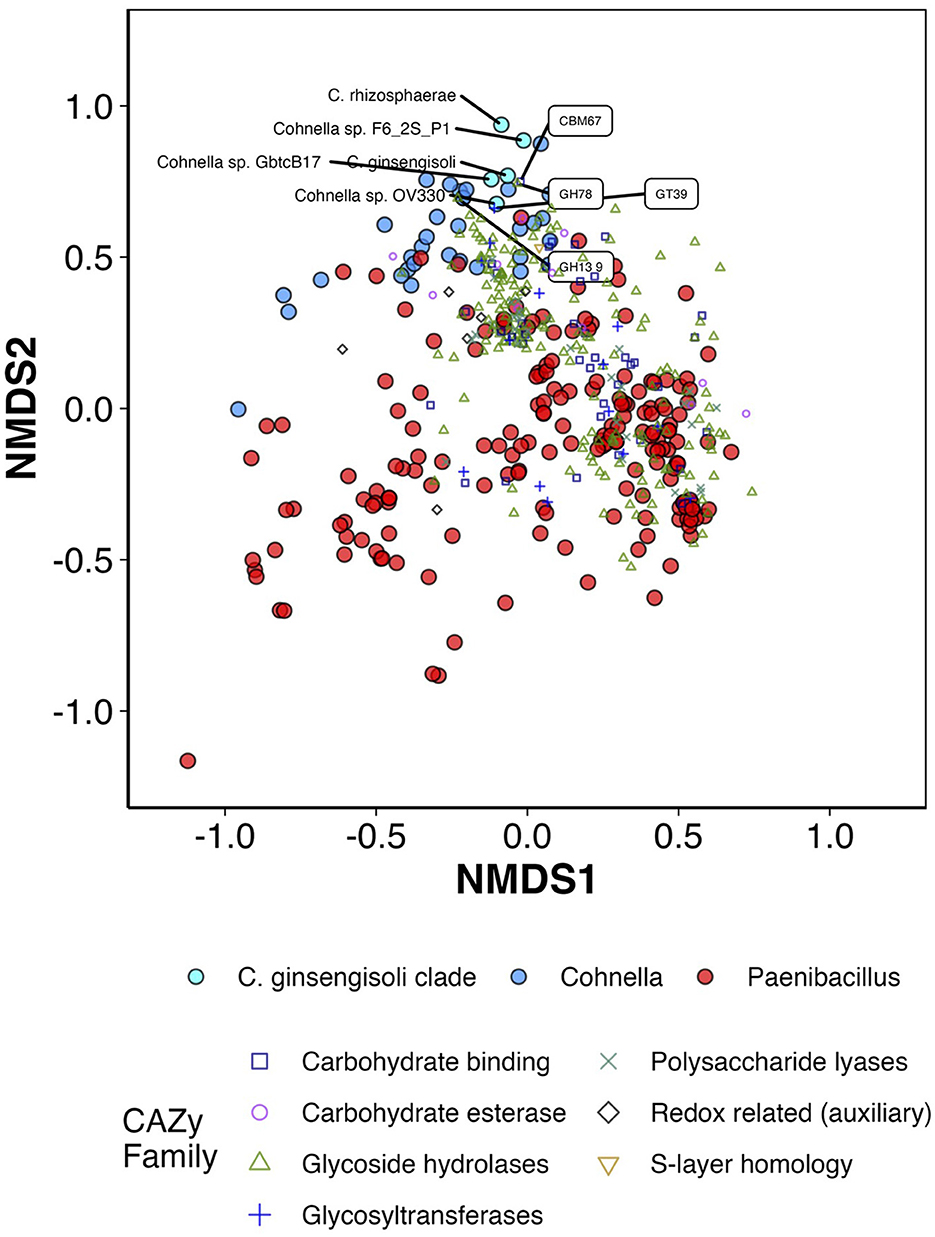
Figure 7. First two axes of NMDS ordination (standardization = “range,” distance = “Bray-Curtis,” maxit = 500, k = 3, stress = 0.129, R2 = 0.98) for counts of different predicted CAZymes in the genomes of Cohnella and Paenibacillus species. Also present in the figure are CAZyme vector correlations with axes 1 and 2 (not original coordinates) via envfit/Pearson's correlation with fdr correction + p < 0.01. Species belonging to the C. ginsengisoli clade and the top 4 CAZyme families/subfamilies positively correlated with axis 2 are labeled.
Most strongly associated with this axis and the C. ginsengisoli clade are the CAZyme families CBM67 and GH78, which were both identified as part of the enzyme α-L-rhamnosidase which alters flavonoids and other rhamnose-containing glycosides such as pectins in plant cell walls (Yadav et al., 2010). Flavonoids are phenolic compounds produced by plants that often have medicinal or flavorful properties. α-L-rhamnosidase cleaves terminal α-rhamnose residues and in doing so helps concentrate and improve the action of flavonoids; α-L-rhamnosidase from different bacterial species is used for many industrial applications such as improving wine aroma, sweetening fruit juice, and increasing concentration and efficacy of antibiotic and anti-cancer compounds (Xie et al., 2022). The C. ginsengisoli clade and other Cohnella species enriched in α-L-rhamnosidase enzyme modules are all confirmed or likely plant symbionts or feed on plant tissue; however, since the majority of Cohnella and Paenibacillus species appear to have at least some association with plant tissue or rhizosphere, there may be additional promising applications for these particular species in processing plant medicinal or other plant phenolic compounds.
CAZymes negatively associated with the C. ginsengisoli clade (and associated mainly with Paenibacillus species) include chitosanases (GH46 and CBM12), endo-β-N-acetylglucosaminidase (GH85), and β-1,4-mannosyl-glycoprotein β-1,4-N-acetylglucosaminyltransferase (GT17). Four CAZyme families associated with the C. ginsengisoli clade compared to other Cohnella and Paenibacillus species had an effect size of 0.5 or greater (Kruskal–Wallis); these were maltooligosaccharide-producing α-amylases (CG13_5), chondroitin AC lyase (PL8_3), and alginate lyases (PL34 and PL14_3).
Presence of Cohnella sp. F6_2S_P_1T in ISS metagenomes
Metagenomic reads from ISS surfaces, crew capsule, and ISS HEPA filter particulates (Venkateswaran et al., 2014; Checinska Sielaff et al., 2019; Urbaniak et al., 2022) were mapped to the genome of strain F6_2S_P_1T. Most metagenomes had <0.05% of reads that mapped to strain F6_2S_P_1T, with a maximum of 0.22% of reads mapping to strain F6_2S_P_1T for a low-read-number sample collected from the cupola of the ISS during Flight 4 of the Microbial Tracking 2 mission series. But upon examination of regions to which the metagenomic reads mapped to strain F6_2S_P_1T, 98+% of reads mapped to regions of 16S or 23S rRNA genes and were only considered significant by bowtie2 because of the conserved nature of the 16S and 23S rRNA genes. We therefore concluded that strain F6_2S_P_1T is not dominant in the ISS environmental surfaces or air particulates collected during the Microbial Tracking 1 and 2 mission series. However, molecular microbial diversity-based examination of plant and its associated materials using culturing and metagenomics would allow NASA to trace the source of this strain, since Cohnella species were reported to produce hormones that might promote growth of plants. For example, as mentioned above, Cohnella was an abundant genus in the microbiome of healthy plant tissue of chili plants grown aboard the ISS (Khodadad et al., 2022).
Conclusion
We used multiple means of characterization, including phenotypic, chemotaxonomic, phylogenetic, and functional genome-prediction methods to characterize Cohnella sp. F6_2S_P_1T and place it within the established taxonomy of the genus Cohnella. Based on these data, we propose that this novel species be named C. hashimotonis. We also characterized the branch of Cohnella within which C. hashimotonis is placed and predicted functional characteristics of C. hashimotonis and the C. ginsengisoli clade as a whole. All species of Cohnella closely related to C. hashimotonis are from the plant/rhizosphere environment and likely have mutualistic associations with plant roots or other tissue. It is likely, therefore, that C. hashimotonis was introduced to the ISS not as a human commensal but as a plant commensal during one of the many plant growth experiments conducted on board. Nevertheless, C. hashimotonis can clearly survive in the extreme conditions of the ISS built environment, including radiation exposure, desiccation, low nutrient availability, and microgravity, and is closely related to a number of rhizosphere bacteria; it is therefore a promising candidate for testing as a member of plant-growth-promoting consortia for future plant experiments in outer space.
Description of Cohnella hashimotonis sp. nov.
Cohnella hashimotonis (ha.shi.mo.to.nis. masc. n. hashimotonis named in honor of Professor Hideo Hashimoto, a food microbiologist from Hiroshima Univ., Japan). Cells are endospore-forming, aerobic, and gram-positive rods, 0.5–0.6 μm in width and 0.8–100+ μm in length. In addition to normal cells, elongated, spaghetti-like cells (Figures 4D, 5B) are observed in light microscopy and SEM images. Colonies are translucent cream-colored, acquiring a pinkish tinge after multiple days of growth, and grow well on R2A but poorly on TSA. Growth occurs at between 15 and 37°C, between pHs 6 and 8, and from 0 to 3% NaCl. Colonies are weakly oxidase-positive and catalase-negative. This species reduces nitrate to nitrite, produces dextrin from starch, and utilizes the following carbon substrates: alanine, histidine, serine, sucrose, L-arabinose, melibiose, D-fructose, inositol, and lactose. The following substances are utilized for acid production: maltose, sucrose, D-fructose, D-ribose, N-acetyl, glucosamine, lactose, and trisodium citrate. Major fatty acids composing C. hashimotonis are anteiso-C15:0 (51.7%), iso-C16:0 (23.1%), and iso-C15:0 (10.5%). The polar lipids are diphosphatidylglycerol, phosphatidylglycerol, phosphatidylethanolamine, three aminophospholipids, and a phospholipid. The major quinone system is menaquinones MK-7. The cell-wall peptidoglycan of strain contained meso-diaminopimelic acid as the diamino acid.
C. hashimotonis is closely related to C. rhizosphaerae and C. ginsengisoli based on 16 rRNA gene sequence similarity (<99%). However, the phylogenetic position of the strain F6_2S_P_1T was resolved as a novel species due to gyrB sequence similarity (<94%), ANI (<89%), and dDDH (<35%) analyses with any other sequenced Cohnella species. GC content is 58.37%. The type strain F6_2S_P_1T was isolated from the wall behind the Waste Hygiene Compartment, i.e., the space toilet, aboard the ISS, in 2018; its genome is available on NCBI, accession number JAGRPV000000000. The type strain F6_2S_P_1T is submitted to two culture collections (=NRRL B-65657T and DSMZ 115098T).
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
NS and KV managed the ISS strain collection and the ISS strain genome sequencing project. AS, VE, and KV conceived and designed the study. VE and AS performed the microbiological experiments and carried out the phenotypic assays. VE performed BioLog based biochemical characterization and chemotaxonomy. FK managed to coordinate in sample collection from ISS and developed sample collection protocols. CM and ND generated shotgun metagenome sequences using ONT platform. NS performed the original taxonomic analysis and de novo assembly. AS performed genomic, pangenomic, and metagenomic characterization, long-read assemblies, and light microscopy assays and wrote the manuscript. KV edited the manuscript. CP performed SEM on F6_2S_P_1T. All authors read and approved the final manuscript.
Funding
Part of the research described in this publication was carried out at the Jet Propulsion Laboratory, California Institute of Technology, under a contract with National Aeronautics and Space Administration. This research was funded by a 2012 Space Biology NNH12ZTT001N grant no. 19-12829-26 under Task Order NNN13D111T award to KV, which also funded the post-doctoral fellowship for CP. The funders had no role in study design, data collection and interpretation, the writing of the manuscript, or the decision to submit the study for publication.
Acknowledgments
The authors would like to thank Camilla Urbaniak for isolating the strain, JAXA astronaut Kanai Norishige for collecting the relevant sample aboard the ISS, the Implementation Team at NASA Ames Research Center for coordinating this effort, and Zymo Research Corp. for extracting DNA. The authors acknowledge the Jet Propulsion Laboratory supercomputing facility staff, notably Narendra J. Patel (Jimmy) and Edward Villanueva, for their continuous support in providing the best possible infrastructure for BIGDATA analysis. CM thanks the Scientific Computing Unit (SCU) at WCM, the WorldQuant Foundation, NASA (NNX14AH50G, NNX17AB26G, NNH18ZTT001N-FG2, 80NSSC22K0254, and NNX16AO69A), the National Institutes of Health (R01MH117406), and the LLS (MCL7001-18, LLS 9238-16)© 2023 California Institute of Technology, Government sponsorship acknowledged.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1166013/full#supplementary-material
References
Abou Abdallah, R., Bou Khalil, J., Andrieu, C., Tomei, E., Armstrong, N., Fournier, P. E., et al. (2019). Draft genome and description of Cohnella massiliensis sp. nov., a new bacterial species isolated from the blood culture of a hemodialysis patient. Arch. Microbiol. 201, 305–312. doi: 10.1007/s00203-018-1612-1
Aziz, R. K., Bartels, D., Best, A. A., Dejongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST Server: rapid annotations using subsystems technology. BMC Genom. 9, 1–15. doi: 10.1186/1471-2164-9-75
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comp. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Bijlani, S., Singh, N. K., Mason, C. E., Wang, C. C., and Venkateswaran, K. (2020). Draft genome sequences of Sphingomonas species associated with the International Space Station. Microbiol. Resour. Announc. 9, e00578–e00520. doi: 10.1128/MRA.00578-20
Bioinformatics, B. (2011). FastQC: A Quality Control Tool for High Throughput Sequence Data. Cambridge: Babraham Institute.
Blachowicz, A., Singh, N. K., Wood, J. M., Debieu, M., O'hara, N. B., Mason, C. E., et al. (2021). Draft genome sequences of Aspergillus and Penicillium species isolated from the international space station and crew resupply vehicle capsule. Microbiol. Resour. Announc. 10, e01398–e01320. doi: 10.1128/MRA.01398-20
Blin, K., Shaw, S., Kloosterman, A. M., Charlop-Powers, Z., Van Wezel, G. P., Medema, M. H., et al. (2021). antiSMASH 6.0: improving cluster detection and comparison capabilities. Nucleic Acids Res. 49, W29–W35. doi: 10.1093/nar/gkab335
Busse, H.-J., Bunka, S., Hensel, A., and Lubitz, W. (1997). Discrimination of members of the family Pasteurellaceae based on polyamine patterns. Int. J. Syst. Evol. Microbiol. 47, 698–708. doi: 10.1099/00207713-47-3-698
Busse, J., and Auling, G. (1988). Polyamine pattern as a chemotaxonomic marker within the Proteobacteria. Syst. Appl. Microbiol. 11, 1–8. doi: 10.1016/S0723-2020(88)80040-7
Campbell, E. L., Cohen, M. F., and Meeks, J. C. (1997). A polyketide-synthase-like gene is involved in the synthesis of heterocyst glycolipids in Nostoc punctiforme strain ATCC 29133. Arch. Microbiol. 167, 251–258. doi: 10.1007/s002030050440
Checinska Sielaff, A., Singh, N. K., Allen, J. E., Thissen, J., Jaing, C., and Venkateswaran, K. (2016). Draft genome sequences of biosafety level 2 opportunistic pathogens isolated from the environmental surfaces of the International Space Station. Genome Announc. 4, 1–21. doi: 10.1128/genomeA.01263-16
Checinska Sielaff, A., Urbaniak, C., Mohan, G. B. M., Stepanov, V. G., Tran, Q., Wood, J. M., et al. (2019). Characterization of the total and viable bacterial and fungal communities associated with the International Space Station surfaces. Microbiome 7, 50. doi: 10.1186/s40168-019-0666-x
Checinska, A., Probst, A. J., Vaishampayan, P., White, J. R., Kumar, D., Stepanov, V. G., et al. (2015). Microbiomes of the dust particles collected from the International Space Station and Spacecraft Assembly Facilities. Microbiome 3, 50. doi: 10.1186/s40168-015-0116-3
Chen, L., Li, W., Zhao, Y., Zhou, Y., Zhang, S., and Meng, L. (2022). Isolation and application of a mixotrophic sulfide-oxidizing Cohnella thermotolerans LYH-2 strain to sewage sludge composting for hydrogen sulfide odor control. Bioresour. Technol. 345, 126557. doi: 10.1016/j.biortech.2021.126557
Chen, S., Zhou, Y., Chen, Y., and Gu, J. (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Daudu, R., Parker, C. W., Singh, N. K., Wood, J. M., Debieu, M., O'hara, N. B., et al. (2020). Draft genome sequences of Rhodotorula mucilaginosa Strains isolated from the International Space Station. Microbiol. Resour. Announc. 9, 00570-20. doi: 10.1128/MRA.00570-20
De Coster, W., D'hert, S., Schultz, D. T., Cruts, M., and Van Broeckhoven, C. (2018). NanoPack: visualizing and processing long-read sequencing data. Bioinformatics 34, 2666–2669. doi: 10.1093/bioinformatics/bty149
Drula, E., Garron, M.-L., Dogan, S., Lombard, V., Henrissat, B., and Terrapon, N. (2022). The carbohydrate-active enzyme database: functions and literature. Nucleic Acids Res. 50, D571–D577. doi: 10.1093/nar/gkab1045
Eren, A. M., Esen, Ö. C., Quince, C., Vineis, J. H., Morrison, H. G., Sogin, M. L., et al. (2015). Anvi'o: an advanced analysis and visualization platform for ‘omics data. PeerJ 3, e1319. doi: 10.7717/peerj.1319
Flores-Felix, J. D., Carro, L., Ramirez-Bahena, M. H., Tejedor, C., Igual, J. M., Peix, A., et al. (2014). Cohnella lupini sp. nov., an endophytic bacterium isolated from root nodules of Lupinus albus. Int. J. Syst. Evol. Microbiol. 64, 83–87. doi: 10.1099/ijs.0.050849-0
Garcia-Fraile, P., Velazquez, E., Mateos, P. F., Martinez-Molina, E., and Rivas, R. (2008). Cohnella phaseoli sp. nov., isolated from root nodules of Phaseolus coccineus in Spain, and emended description of the genus Cohnella. Int. J. Syst. Evol. Microbiol. 58, 1855–1859. doi: 10.1099/ijs.0.65468-0
Gurevich, A., Saveliev, V., Vyahhi, N., and Tesler, G. (2013). QUAST: quality assessment tool for genome assemblies. Bioinformatics 29, 1072–1075. doi: 10.1093/bioinformatics/btt086
Hameed, A., Hung, M.-H., Lin, S.-Y., Hsu, Y.-H., Liu, Y.-C., Shahina, M., et al. (2013). Cohnellaformosensis sp. nov., a xylanolytic bacterium isolated from the rhizosphere of Medicago sativa L. Int. J. Syst. Evol. Microbiol. 63, 2806–2812. doi: 10.1099/ijs.0.045831-0
He, Y., Xie, K., Xu, P., Huang, X., Gu, W., Zhang, F., et al. (2013). Evolution of microbial community diversity and enzymatic activity during composting. Res. Microbiol. 164, 189–198. doi: 10.1016/j.resmic.2012.11.001
Hero, J. S., Pisa, J. H., Romero, C. M., Nordberg Karlsson, E., Linares-Pasten, J. A., and Martinez, M. A. (2021). Endo-xylanases from Cohnella sp. AR92 aimed at xylan and arabinoxylan conversion into value-added products. Appl. Microbiol. Biotechnol. 105, 6759–6778. doi: 10.1007/s00253-021-11495-5
Jain, C., Rodriguez, R. L., Phillippy, A. M., Konstantinidis, K. T., and Aluru, S. (2018). High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 9, 5114. doi: 10.1038/s41467-018-07641-9
JGI (2015). Study: Populus Root and Rhizosphere Microbial Communities From Tennessee, USA, Organism: Cohnella sp. OV330. Available online at: https://gold.jgi.doe.gov/projects?id=Gp0110579 (accessed January 15, 2023).
Johnson, E. T., Dowd, P. F., Skory, C. D., and Dunlap, C. A. (2021). Description of Cohnella zeiphila sp. nov., a bacterium isolated from maize callus cultures. Antonie Van Leeuwenhoek 114, 37–44. doi: 10.1007/s10482-020-01495-2
Kampfer, P., Glaeser, S. P., Mcinroy, J. A., and Busse, H. J. (2014). Cohnella rhizosphaerae sp. nov., isolated from the rhizosphere environment of Zea mays. Int J Syst Evol Microbiol 64, 1811–1816. doi: 10.1099/ijs.0.060814-0
Kampfer, P., Rossello-Mora, R., Falsen, E., Busse, H. J., and Tindall, B. J. (2006). Cohnella thermotolerans gen. nov., sp. nov., and classification of ‘Paenibacillus hongkongensis' as Cohnella hongkongensis sp. nov. Int. J. Syst. Evol. Microbiol. 56, 781–786. doi: 10.1099/ijs.0.63985-0
Khianngam, S., Tanasupawat, S., Akaracharanya, A., Kim, K. K., Lee, K. C., and Lee, J. S. (2010). Cohnella xylanilytica sp. nov. and Cohnella terrae sp. nov., xylanolytic bacteria from soil. Int. J. Syst. Evol. Microbiol. 60, 2913–2917. doi: 10.1099/ijs.0.017855-0
Khodadad, C., Dixit, A. R., Hummerick, M., Spencer, L. E., Richards, J. T., Spern, C. J., et al. (2022). Microbial Analysis of Chile Peppers grown in NASA's Advanced Plant Habitat (APH) on the International Space Station. Houston, TX: ASGSR.
Khodadad, C. L., Hummerick, M. E., Spencer, L. E., Dixit, A. R., Richards, J. T., Romeyn, M. W., et al. (2020). Microbiological and nutritional analysis of lettuce crops grown on the international space station. Front. Plant Sci. 199, 199. doi: 10.3389/fpls.2020.00199
Kim, J. K., Choi, C.-H., Kim, D.-W., Kim, S. Y., Hwang, K. J., Lee, W.-K., et al. (2021). Cohnella cholangitidis sp. nov., a novel species of the genus Cohnella isolated from a clinical specimen in Korea. Archi. Microbiol. 203, 6053–6060. doi: 10.1007/s00203-021-02565-3
Kim, S. J., Weon, H. Y., Kim, Y. S., Anandham, R., Jeon, Y. A., Hong, S. B., et al. (2010). Cohnella yongneupensis sp. nov. and Cohnella ginsengisoli sp. nov., isolated from two different soils. Int. J. Syst. Evol. Microbiol. 60, 526–530. doi: 10.1099/ijs.0.013581-0
Kolde, R., and Kolde, M. R. (2018). Package ‘pheatmap'. R Package 1. Comprehensive R Archive Network.
Kolmogorov, M., Yuan, J., Lin, Y., and Pevzner, P. A. (2019). Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 37, 540–546. doi: 10.1038/s41587-019-0072-8
Kozlov, A. M., Darriba, D., Flouri, T., Morel, B., and Stamatakis, A. (2019). RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 35, 4453–4455. doi: 10.1093/bioinformatics/btz305
La Duc, M. T., Sumner, R., Pierson, D., Venkat, P., and Venkateswaran, K. (2004). Evidence of pathogenic microbes in the International Space Station drinking water: reason for concern? Habitation 10, 39–48. doi: 10.3727/154296604774808883
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Letunic, I., and Bork, P. (2007). Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23, 127–128. doi: 10.1093/bioinformatics/btl529
Lin, S. Y., Chen, W. M., Hameed, A., Huang, G. H., Hung, M. H., and Young, C. C. (2020). Cohnella fermenti sp. nov., isolated from a fermentation process. Int. J. Syst. Evol. Microbiol. 70, 2602–2610. doi: 10.1099/ijsem.0.004080
Martínková, M., Kitanishi, K., and Shimizu, T. (2013). Heme-based globin-coupled oxygen sensors: linking oxygen binding to functional regulation of diguanylate cyclase, histidine kinase, and methyl-accepting chemotaxis. J. Biol. Chem. 288, 27702–27711. doi: 10.1074/jbc.R113.473249
Mckerrow, J., Vagg, S., Mckinney, T., Seviour, E., Maszenan, A., Brooks, P., et al. (2000). A simple HPLC method for analysing diaminopimelic acid diastereomers in cell walls of Gram-positive bacteria. Lett. Appl. Microbiol. 30, 178–182. doi: 10.1046/j.1472-765x.2000.00675.x
Meier-Kolthoff, J. P., Carbasse, J. S., Peinado-Olarte, R. L., and Göker, M. (2022). TYGS and LPSN: a database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes. Nucleic Acids Res. 50, D801–D807. doi: 10.1093/nar/gkab902
MIDI (2012). Sherlock Microbial Identification System MIS Operating Manual. Available online at: http://midi-inc.com/pdf/Sherlock_MIS_Operating_Manual.pdf (accessed January 15, 2023).
Mohammadi, S., Tarrahimofrad, H., Arjmand, S., Zamani, J., Haghbeen, K., and Aminzadeh, S. (2022). Expression, characterization, and activity optimization of a novel cellulase from the thermophilic bacteria Cohnella sp. A01. Sci. Rep. 12, 10301. doi: 10.1038/s41598-022-14651-7
Muñoz, C. Y., Zhou, L., Yi, Y., and Kuipers, O. P. (2022). Biocontrol properties from phyllospheric bacteria isolated from Solanum lycopersicum and Lactuca sativa and genome mining of antimicrobial gene clusters. BMC Genom. 23, 1–12. doi: 10.1186/s12864-022-08392-0
Niang, N., Sandrine, D., Ibrahima, N., and Elisabeth, N. (2018). Genetic diversity of rhizobia and plant growth promoting rhizobacteria of soil under the influence of Piliostigma reticulatum (DC.) Hochst and their impact on shrub growth. Afr. J. Agri. Res. 13, 2668–2679. doi: 10.5897/AJAR2018.13494
Oksanen, J., Blanchet, F. G., Friendly, M., Kindt, R., Legendre, P., Mcglinn, D., et al. (2019). “vegan: Community Ecology Package”. R package version 2.5-6. Comprehensive R Archive Network.
R Core Team (2022). “R: a language and environment for statistical computing,” in R.F.F.S. Computing (Vienna).
Ramaprasad, E., Mahidhara, G., Sasikala, C., and Ramana, C. V. (2018). Rhodococcus electrodiphilus sp. nov., a marine electro active actinobacterium isolated from coral reef. Int. J. Syst. Evol. Microbiol. 68, 2644–2649. doi: 10.1099/ijsem.0.002895
Ramaprasad, E., Sasikala, C., and Ramana, C. V. (2015). Flectobacillus rhizosphaerae sp. nov., isolated from the rhizosphere soil of Oryza sativa (L.), and emended description of the genus Flectobacillus. Int. J. Syst. Evol. Microbiol. 65, 3451–3456. doi: 10.1099/ijsem.0.000432
Richard, K. L., Kelley, B. R., and Johnson, J. G. (2019). Heme uptake and utilization by gram-negative bacterial pathogens. Front. Cell. Infect. Microbiol. 9, 81. doi: 10.3389/fcimb.2019.00081
Robinson, J. A., Rhatigan, J. L., Baumann, D. K., Tate, J., and Thumm, T. (2006). International Space Station Research Summary Through Expedition 10. NASA/TP 213146. NASA.
Schaeffer, A. B., and Fulton, M. D. (1933). A simplified method of staining endospores. Science 77, 194–194. doi: 10.1126/science.77.1990.194
Schleifer, K. H. (1985). “Analysis of the chemical composition and primary structure of murein,” in Methods in Microbiology, ed T. Bergen (Elsevier), 1–376.
Schleifer, K. H., and Kandler, O. (1972). Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol. Rev. 36, 407–477. doi: 10.1128/br.36.4.407-477.1972
Schliep, K. P. (2011). phangorn: phylogenetic analysis in R. Bioinformatics 27, 592–593. doi: 10.1093/bioinformatics/btq706
Seemann, T. (2018). ABRicate: Mass Screening of Contigs for Antimicrobial and Virulence Genes. Melbourne, VIC: Department of Microbiology and Immunology, The University of Melbourne. Available online at: https://github.com/tseemann/abricate (accessed February 28, 2019).
Semenzato, G., Alonso-Vásquez, T., Del Duca, S., Vassallo, A., Riccardi, C., Zaccaroni, M., et al. (2022). Genomic analysis of endophytic bacillus-related strains isolated from the medicinal plant Origanum vulgare L. Revealed the presence of metabolic pathways involved in the biosynthesis of bioactive compounds. Microorganisms 10, 919. doi: 10.3390/microorganisms10050919
Simpson, A. C., Urbaniak, C., Bateh, J. R., Singh, N. K., Wood, J. M., Debieu, M., et al. (2021a). Draft genome sequences of fungi isolated from the international Space Station during the microbial tracking-2 experiment. Microbiol. Resour. Announc. 10, e00751–e00721. doi: 10.1128/MRA.00751-21
Simpson, A. C., Urbaniak, C., Singh, N. K., Wood, J. M., Debieu, M., O'hara, N. B., et al. (2021b). Draft genome sequences of various bacterial phyla isolated from the International Space Station. Microbiol. Resour. Announc. 10, e00214–00221. doi: 10.1128/MRA.00214-21
Singh, N., and Bhatla, S. C. (2022). Heme oxygenase-nitric oxide crosstalk-mediated iron homeostasis in plants under oxidative stress. Free Radic. Biol. Med. 182, 192–205. doi: 10.1016/j.freeradbiomed.2022.02.034
Singh, N. K., Lavire, C., Nesme, J., Vial, L., Nesme, X., Mason, C. E., et al. (2021). Comparative genomics of novel agrobacterium G3 strains isolated from the International Space Station and description of Agrobacterium tomkonis sp. nov. Front. Microbiol. 12, 765943. doi: 10.3389/fmicb.2021.765943
Singh, N. K., Wood, J. M., Karouia, F., and Venkateswaran, K. (2018). Succession and persistence of microbial communities and antimicrobial resistance genes associated with International Space Station environmental surfaces. Microbiome 6, 204. doi: 10.1186/s40168-018-0585-2
Solomon, S. A., Bharadwaj, A. R., Singh, N. K., Wood, J. M., Debieu, M., O'hara, N. B., et al. (2020). Draft genome sequences of Klebsiella species isolated from the International Space Station. Microbiol. Resour. Announc. 9, 00923-20. doi: 10.1128/MRA.00923-20
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Takada, K., Tanaka, N., Kikuno, H., Babil, P., Onjo, M., Park, B.-J., et al. (2019). Isolation of nitrogen-fixing bacteria from water yam (Dioscorea alata L.). Trop. Agric. Dev. 63, 198–203. doi: 10.11248/jsta.63.198
Takemoto, J. Y., Chang, C. W. T., Chen, D., and Hinton, G. (2019). Heme-derived bilins. Isr. J. Chem. 59, 378–386. doi: 10.1002/ijch.201800167
Teng, J., Woo, P., Leung, K., Lau, S., Wong, M., and Yuen, K. (2003). Pseudobacteraemia in a patient with neutropenic fever caused by a novel paenibacillus species: Paenibacillus hongkongensis sp. nov. Mol. Pathol. 56, 29. doi: 10.1136/mp.56.1.29
Urbaniak, C., Massa, G., Hummerick, M., Khodadad, C., Schuerger, A., and Venkateswaran, K. (2018). Draft genome sequences of two Fusarium oxysporum isolates cultured from infected Zinnia hybrida plants grown on the International Space Station. Genome Announc. 6, e00326-18. doi: 10.1128/genomeA.00326-18
Urbaniak, C., Morrison, M. D., Thissen, J. B., Karouia, F., Smith, D. J., Mehta, S., et al. (2022). Microbial tracking-2, a metagenomics analysis of bacteria and fungi onboard the International Space Station. Microbiome 10, 1–19. doi: 10.1186/s40168-022-01293-0
Venkateswaran, K., Checinska Sielaff, A., Ratnayake, S., Pope, R. K., Blank, T. E., Stepanov, V. G., et al. (2017). Draft genome sequences from a novel clade of Bacillus cereus sensu lato strains, isolated from the International Space Station. Genome Announc. 5, e00680–e00617. doi: 10.1128/genomeA.00680-17
Venkateswaran, K., Vaishampayan, P., Cisneros, J., Pierson, D. L., Rogers, S. O., and Perry, J. (2014). International Space Station environmental microbiome - microbial inventories of ISS filter debris. Appl. Microbiol. Biotechnol. 98, 6453–6466. doi: 10.1007/s00253-014-5650-6
Wang, L. Y., Chen, S. F., Wang, L., Zhou, Y. G., and Liu, H. C. (2012). Cohnella plantaginis sp. nov., a novel nitrogen-fixing species isolated from plantain rhizosphere soil. Antonie Van Leeuwenhoek 102, 83–89. doi: 10.1007/s10482-012-9715-1
Wang, L. Y., Wang, T. S., and Chen, S. F. (2015). Cohnella capsici sp. nov., a novel nitrogen-fixing species isolated from Capsicum annuum rhizosphere soil, and emended description of Cohnella plantaginis. Antonie Van Leeuwenhoek 107, 133–139. doi: 10.1007/s10482-014-0310-5
Wick, R. R., Judd, L. M., Gorrie, C. L., and Holt, K. E. (2017a). Completing bacterial genome assemblies with multiplex MinION sequencing. Microbial Genom. 3, e1005595. doi: 10.1099/mgen.0.000132
Wick, R. R., Judd, L. M., Gorrie, C. L., and Holt, K. E. (2017b). Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13, e1005595. doi: 10.1371/journal.pcbi.1005595
Wickham, H., Averick, M., Bryan, J., Chang, W., Mcgowan, L. D. A., François, R., et al. (2019). Welcome to the Tidyverse. J. Open Source Softw. 4, 1686. doi: 10.21105/joss.01686
Wright, E. (2016). Using DECIPHER v2.0 to analyze big biological sequence data in R. R J. 8, 352–359. doi: 10.32614/RJ-2016-025
Wu, Y., Wang, B., Zeng, Z., Liu, R., Tang, L., Gong, L., et al. (2019). Effects of probiotics Lactobacillus plantarum 16 and Paenibacillus polymyxa 10 on intestinal barrier function, antioxidative capacity, apoptosis, immune response, and biochemical parameters in broilers. Poult. Sci. 98, 5028–5039. doi: 10.3382/ps/pez226
Xie, J., Zhao, J., Zhang, N., Xu, H., Yang, J., Ye, J., et al. (2022). Efficient production of isoquercitin, icariin and icariside II by a novel thermostable α-l-rhamnosidase PodoRha from Paenibacillus odorifer with high α-1, 6-/α-1, 2-glycoside specificity. Enzyme Microb. Technol. 158, 110039. doi: 10.1016/j.enzmictec.2022.110039
Xu, P., Li, W.-J., Tang, S.-K., Zhang, Y.-Q., Chen, G.-Z., Chen, H.-H., et al. (2005). Naxibacter alkalitolerans gen. nov., sp. nov., a novel member of the family ‘Oxalobacteraceae' isolated from China. Int. J. Syst. Evol. Microbiol. 55, 1149–1153. doi: 10.1099/ijs.0.63407-0
Yadav, V., Yadav, P. K., Yadav, S., and Yadav, K. (2010). α-L-Rhamnosidase: a review. Process Biochem. 45, 1226–1235. doi: 10.1016/j.procbio.2010.05.025
Yu, G., Smith, D. K., Zhu, H., Guan, Y., and Lam, T. T. Y. (2017). ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 8, 28–36. doi: 10.1111/2041-210X.12628
Zhang, H., Yohe, T., Huang, L., Entwistle, S., Wu, P., Yang, Z., et al. (2018). dbCAN2: a meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 46, W95–W101. doi: 10.1093/nar/gky418
Zhu, S., Hegemann, J. D., Fage, C. D., Zimmermann, M., Xie, X., Linne, U., et al. (2016). Insights into the unique phosphorylation of the lasso peptide paeninodin. J. Biol. Chem. 291, 13662–13678. doi: 10.1074/jbc.M116.722108
Keywords: International Space Station (ISS), novel bacteria, Cohnella, space biology, pangenome analysis
Citation: Simpson AC, Eedara VVR, Singh NK, Damle N, Parker CW, Karouia F, Mason CE and Venkateswaran K (2023) Comparative genomic analysis of Cohnella hashimotonis sp. nov. isolated from the International Space Station. Front. Microbiol. 14:1166013. doi: 10.3389/fmicb.2023.1166013
Received: 14 February 2023; Accepted: 24 May 2023;
Published: 15 June 2023.
Edited by:
Javier Pascual, Darwin Bioprospecting Excellence, SpainReviewed by:
Rita Abou-Abdallah, IHU Mediterranee Infection, FranceSaeed Aminzadeh, National Institute for Genetic Engineering and Biotechnology, Iran
Copyright © 2023 Simpson, Eedara, Singh, Damle, Parker, Karouia, Mason and Venkateswaran. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kasthuri Venkateswaran, a2p2ZW5rYXRAanBsLm5hc2EuZ292; Christopher E. Mason, Y2htMjA0MkBtZWQuY29ybmVsbC5lZHU=
†ORCID: Kasthuri Venkateswaran orcid.org/0000-0002-8724-8177
 Anna C. Simpson1
Anna C. Simpson1 V. V. Ramprasad Eedara
V. V. Ramprasad Eedara Nitin K. Singh
Nitin K. Singh Ceth W. Parker
Ceth W. Parker Christopher E. Mason
Christopher E. Mason Kasthuri Venkateswaran
Kasthuri Venkateswaran