
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol., 02 June 2023
Sec. Microbiological Chemistry and Geomicrobiology
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1163896
This article is part of the Research TopicMicrobial Ecological and Biogeochemical Processes in the Soil-Vadose Zone-Groundwater HabitatsView all 12 articles
Coastal wetlands (CW) are the junction of the terrestrial and marine ecosystems and have special ecological compositions and functions, which are important for maintaining biogeochemical cycles. Microorganisms inhabiting in sediments play key roles in the material cycle of CW. Due to the variable environment of CW and the fact that most CW are affected by human activities and climate change, CW are severely degraded. In-depth understanding of the community structure, function, and environmental potential of microorganisms in CW sediments is essential for wetland restoration and function enhancement. Therefore, this paper summarizes microbial community structure and its influencing factors, discusses the change patterns of microbial functional genes, reveals the potential environmental functions of microorganisms, and further proposes future prospects about CW studies. These results provide some important references for promoting the application of microorganisms in material cycling and pollution remediation of CW.
CW are the transitional regions between the terrestrial and marine ecosystems, mainly including shallow seas, estuaries, mangroves, salt marshes, deltas, etc. CW have the vegetated zones (mangroves, salt marshes, and seagrass beds) and non-vegetated zones (mudflats and sandy beaches), which are critical areas connecting land, freshwater habitats, and the ocean (Levin et al., 2001). CW can provide many facilities for human activities such as fishing and breeding (Zhang and Shao, 2013), and also protect coastal zones in flooding (Narayan et al., 2017). Moreover, CW richen in biodiversity, material cycling, energy flow, and species migration and evolution, with high primary productivity (Cui et al., 2016). CW are the most vulnerable ecosystems due to ocean dynamics, river disturbance, and human activities (Wang et al., 2022), and its degradation (such as biodiversity decline, ecosystem function loss, and coastal vegetation reduction) may lead to biological invasions, water quality deterioration, and reduced coastal protection from flooding and storm events (Barbier et al., 2011).
Microorganisms are an important component of wetland ecosystems and play key roles in biogeochemical cycles (DeLong et al., 2006). Microbial community structure has significant differences depending on different soil properties and vegetation types. Soil properties and plant types are the main determinants of microbial community structure (Yu et al., 2014). The interdependence between plants and microorganisms has a critical role in regulating ecosystem services such as nutrient cycling, productivity, and pollutants degradation (Abdu et al., 2017). Microorganisms can decompose soil organic matter, promote sulfate reduction, sulfide/sulfur oxidation, iron reduction, nitrification, pollutant degradation, and help improve soil structure and enhance ecosystem stability (Behera et al., 2017). CW exhibit strong nutrient and salinity gradients due to freshwater and seawater interactions, affecting soil microbial composition. Furthermore, these changes in microorganisms can lead to the variations of the function and structure of CW (Webster et al., 2015).
Despite the importance of microorganisms in CW (Figure 1), few studies have reviewed and summarized them. Therefore, the purpose of this study is: (1) to provide an overview of microorganisms in CW sediments; and (2) to identify future research directions and possible difficulties. In this paper, we summarize the community structure characteristics, functional genetic variation and potential environmental functions of microorganisms in CW.
The special soil characteristics and hydrological conditions of CW constitute a unique microbial community (Peralta et al., 2010). Linking microbial communities to physical, chemical, and biological factors can explore the drivers of microbial community formation (Fierer and Jackson, 2006), which is important for the restoration of environmental functions in wetland ecosystems.
Microbial species are abundant in CW, which are mainly divided into bacteria, archaea, and fungi (Table 1). Among them, bacterial communities have the highest richness, followed by archaeal and fungal communities (Cheung et al., 2018). Although the composition is the same at community level, there is some variation in microbial composition among different CW and different times of wetlands (Adame et al., 2012).
Proteobacteria is the most abundant bacterial phyla in CW sediments (Ling et al., 2015), and mainly includes α-, β-, γ-, δ-, and ε-Proteobacteria (Hu et al., 2014), and their composition varies somewhat in different wetland types. For example, γ-Proteobacteria dominates in coastal zone of Yellow River Delta, whereas γ- and δ-Proteobacteria dominate in brine-freshwater zone (Hu et al., 2016). ε-Proteobacteria is dominant Proteobacteria in Jiuduansha Wetlands of Yangtze Estuary, while γ- and β-Proteobacteria are abundant in Jiangyanan Shoal of the river (Fei Xi et al., 2014). On the contrary, Firmicutes is the dominant phylum near salt flats of the Yangtze River Delta (Zou et al., 2020). In addition to Proteobacteria, Actinobacteria, Chloroflexi, Bacteroidetes, Firmicutes, Acidobacteria, and Planctomycetes are also the main phyla in CW (Zhang et al., 2017; An et al., 2019). Although bacterial compositions are relatively similar, there are some differences in different wetlands types. Moreover, archaea are also an important component of microbial communities and play an important role in biogeochemical cycles of CW (Narrowe et al., 2017). The dominant phyla of archaea are mainly Euryarchaeota, Thaumarchaeota, Bathyarchaeota and Grenarchaeota (Zhao et al., 2020; Chi et al., 2021a,b,c,d). Fungi as an important component of microorganisms and its community are essential for maintaining soil versatility (Li H. et al., 2019). Ascomycota and Basidiomycota are the dominant taxa in CW (Mohapatra et al., 2021). Among them, Dothidomycetes and Sordariomycetes are the dominant classes, and the dominant orders include Pleosporales, Agaricales, and Capnodiales (Ye et al., 2022). Moreover, many fungi cannot be attributed to the known phyla (Cheung et al., 2018).
Microorganisms in CW sediments are influenced by soil physicochemical properties, including salinity, pH, and nutrients (Jackson and Vallaire, 2009). These properties can affect microbial growth and metabolism as well as microbial activity (Figure 2).
Salinity can directly affect abiotic and biotic processes, and is considered as a major driver of ecosystem structure and function (Brucet et al., 2012). Previous study showed that salinity affected microbial communities and the associated biogeochemical cycles (Chambers et al., 2011). In general, salinity elevation usually has a negative impact on microorganisms, and low salinity environments are suited for microbial growth (Hu et al., 2016). High salinity can affect CW ecosystems through inhibiting plant growth and heterotrophic metabolism, and reducing soil quality and heterotrophic bacterial diversity (Abed et al., 2007). Microbial community structure varies along salinity gradients (Yang et al., 2018). It was found that halophilic bacteria such as Fodinibius, Alkalilimnicola, Phycisphaera and Gp21 were abundant in high-salinity zone of the Yangtze River Delta, and the dominant genera in the transition zone were Rhodocyclus, Flavobacterium and Shin (Shinell; Li J. et al., 2019).
pH has a significant effect on microbial community (Rousk et al., 2010). The bacterial composition and diversity in various ecosystems respond strongly to soil pH (Shen et al., 2013). Nitrospirae was lower in saline wetlands with high pH than in freshwater wetlands with low pH, and Nitrospirae was significantly negatively correlated with pH (Chi et al., 2021a,b,c,d).
Nutrients can also affect microbial growth. Total organic matter, total nitrogen and total phosphorus in the samples are usually tested as quantitative indicators of nutrient content when conducting experiments (Wang et al., 2012). The content of available nutrients affects microbial activity, and the addition of nutrients can effectively increase the abundance of bacterial strains (Meng et al., 2016). The unique structural composition of microbial communities in intertidal sediments of the Yellow River Delta is nutrient-related, and many saprophytic microorganisms are enriched (Zhang et al., 2017).
Vegetation has important influences on microbial community. Plants can create a unique environment for rhizosphere microorganisms (Grayston et al., 1998). Studies have shown that nutrient acquisition strategies of plant can drive the structural and functional formation of soil surface microbes and that changes in vegetation lead to changes in soil microbial diversity and function (Bahram et al., 2020). Root-mediated changes in soil can provide oxygen or other substrates for soil microbes (Noll et al., 2005) and also alter microbial community (Lipson et al., 2015). Plants alter the physicochemical conditions of sub-canopy soils (Menon et al., 2013), such as leaf litter can improve soil fertility and plant roots can release a variety of compounds into the surrounding soils (Garbeva et al., 2004). Roots of woody plants, such as mangroves, have different chemical (Perry and Mendelssohn, 2009) and physiological properties (Skelton and Allaway, 1996), and can transport different root secretions (Bertin et al., 2003). Differences of microbial community composition in mangrove- and swamp-dominated soils in Florida may be due to differences in root secretions or oxygen availability between vegetation types (Barreto et al., 2018). The photosynthesis of plants lead to the adsorption of cyanobacteria on plant rhizomes with more than 50% abundance in Yellow River Delta (Li et al., 2021).
Microbially mediated nitrogen cycle is one of the important components of biogeochemical cycles in CW (see Figure 3A). Among them, denitrification and dissimilatory nitrate reduction to ammonia (DNRA) processes are particularly important, and the end-products of these pathways have different effects on ecosystem nitrogen effectiveness (Morina and Franklin, 2022).
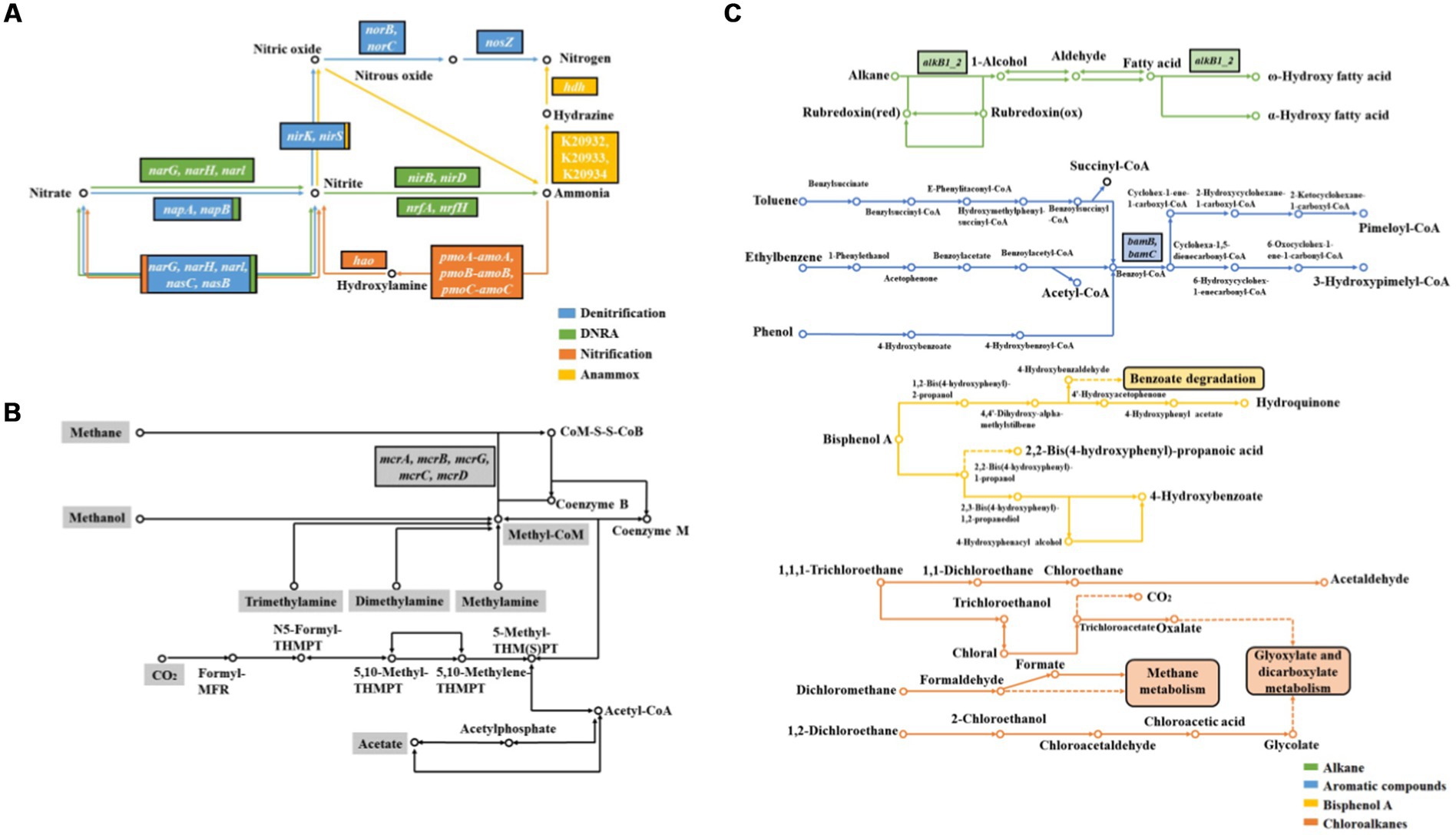
Figure 3. KEGG pathway of nitrogen cycle (A), methanogenesis (B) and organics degradation (C) in CW.
Denitrification plays a key role in nitrogen removal (van Breemen et al., 2002), and there are some important metabolic enzymes, including nitrate reductase (Nar), nitrite reductase (Nir), nitric oxide reductase (Nor), and nitrous oxide reductase (Nos; Levy-Booth et al., 2014). Among these, Nir catalyzes the rate-limiting step in denitrification, encoding by the nirK and/or nirS genes. A previous study found that the enzyme genes associated with denitrification decreased with increasing distance from the river bank of Yellow River, and reached their highest levels at distances of 0–50 m (except 0 m; Li W. et al., 2019). Saline plants had no significant effect on the abundances of denitrification genes nirK, nirS, and nosZ in Suncheon Bay, South Korea (Chaudhary et al., 2018). Mesosaline soils affect negatively on nirS and nirK genes compared to freshwater soils in the east coast of the United States (Morina and Franklin, 2022). Moreover, the diversity of nirS genes in Chinese CW exhibited significant latitudinal heterogeneity, and it is speculated that temperature rather than salinity contributes significantly to the latitudinal distribution of nirS-based denitrifying bacteria (Gao et al., 2016).
DNRA can convert nitrate nitrogen to ammonia nitrogen, and is one of the potentially important nitrogen cycling processes in CW. The reduction of NO2− to NH4+ in DNRA is catalyzed by nitrite reductase, encoding by the nrfA gene. The abundance of nirS denitrifying bacteria is much greater than that of nrfA-DNRA microorganisms in the Chesapeake Bay watershed of United States, suggesting that denitrification is the primary nitrate reduction process (Franklin et al., 2017). The abundance of nrfA genes was low in tidal freshwater marshes in South Carolina of United States, suggesting weak DRNA process (Minick et al., 2019).
Nitrification and anammox are also important processes in the nitrogen cycle (Kuypers et al., 2018), but these two processes are weakly in CW ecosystems (Zhang X. et al., 2019). Functional genes associated with nitrification are mainly genes encoding ammonia monooxygenase (amoA/B/C) and hydroxylamine dehydrogenase (hao; Mohapatra et al., 2021), while those associated with anammox are mainly genes encoding hydrazine synthase (hszA; Harhangi et al., 2012). The abundance of functional genes associated with nitrification in intertidal wetlands disturbed by crabs is increased compared to the surrounding undisturbed sediments (An et al., 2021). Moreover, the copy number of nitrification genes is significantly higher in oily marshes (Bae et al., 2018).
Methane production is a major process in anaerobic carbon-cycle of CW, and methanogenic bacteria are the main microorganisms involved in this process, encoding by mcrA gene (see Figure 3B; Oremland et al., 1982). A previous study showed that the abundance of methanogenic genes in wetlands affected by runoff and tidal seawater increased with distance from the river bank, while gene abundance in tidal wetlands increased first and then decreased in Yellow River Delta (Chi et al., 2021a,b,c,d). The abundance of mcrA was significantly lower in oiled marshes compared to non-oiled marshes along the United States coast (Bae et al., 2018).
A large number of organic pollutants from human activities are released into wetlands with industrial development, adversely affecting the surrounding ecosystems (Qian et al., 2016). Petroleum hydrocarbons are the main pollutants that affect the material cycle and ecosystem function of wetlands (Yuan et al., 2014). Indigenous microorganisms in wetlands can degrade petroleum hydrocarbons, and lots of hydrocarbon-degrading bacteria isolated from petroleum-contaminated soils play key roles in petroleum hydrocarbons degradation (Tiralerdpanich et al., 2018). Therefore, the level of petroleum hydrocarbon contamination in different wetland soils can affect microbial community, which leads to changes in metabolic functions (see Figure 3C).
Both alkB and CYP 153A1 genes encoding alkane hydroxylases are enriched in tidal marshs from the Coacheco River in the United States under chronically contaminated petroleum hydrocarbons such as gasoline, n-hexane, and dodecane (Ní Chadhain et al., 2018). The gene alkB involving in aerobic alkanes degradation has high copy number in oil-bearing coastal salt marshes of the United States, whereas bamA related to anaerobic aromatics degradation has low copy number (Bae et al., 2018). Genes associated with the degradation of alkanes, cycloalkanes, aromatic carboxylic acids, chlorinated aromatics, polycyclic aromatic hydrocarbons, and other aromatic hydrocarbons are significantly reduced in salt marshes of Gulf Coast during oil concentration reduction (Beazley et al., 2012). The initial dioxygenase and open-loop dioxygenase associated with phenanthrene (PHE) degradation were expressed under PHE contamination in CW, indicating the presence of aerobic PHE degradation (Chi et al., 2021a,b,c,d).
Microorganisms contribute significantly to ecological functions (e.g., carbon and nitrogen cycle processes) in CW (Figure 4), which are critical in retaining chemical contaminants (e.g., organic pollutants) and excess nutrients (Horton et al., 2019).
Previous studies have shown that the species composition and spatio-temporal dynamics of soil microbial communities are related to habitat characteristics, plant types, and human interferences (Eddie et al., 2010). Microorganisms are highly sensitive to environmental changes and thus can be an ideal indicator for environment monitoring (Santos et al., 2010; Yang et al., 2012). The indicative effects of microbial communities are various (Table 2). Fungal community composition in different habitats varies and could be used as a bioindicator to assess the restoration process of mangrove ecosystems in Jiulongjiang estuary (Yu et al., 2014). Fungi as indicator species of P. australis soils is found in restoration area of the Yangtze River Delta (Ma et al., 2017). Differences in microbial community over a short period in Florida suggest that they can serve as early warning signals for sea-level rise (Chambers et al., 2016). The ratio of ammonia to nitrate nitrogen in CW of Pearl River Delta significantly affect bacterial community composition, and thus anaerobic ammonia-oxidizing bacteria is a bioindicator of terrestrial nitrogen input or pollution (Han and Gu, 2015).
Microorganisms have become popularly alternatives for pollutant bioremediation because they are environmentally friendly and cost-effective (Macaulay, 2015). Hydrocarbon-degrading microorganisms are ubiquitous in many environments (Head et al., 2006). Most studies on microbial hydrocarbon degradation have focused on environments highly exposed to hydrocarbons, such as areas surrounding oil deposits and hydrocarbon spills. Pseudomonas-type alkane-degrading bacteria are enriched in marshes nearby oil contamination, suggesting that oil degradation is important at this zone (Ní Chadhain et al., 2018). Bacterial community in sediments of Mexican coastal zone can degrade toluene, naphthalene, chloroalkanes, and chlorinated alkanes, but has low removals of aromatics, fluorobenzoates, and xylenes (Reyes-Sosa et al., 2018). Proteobacteria is responsible for the degradation of some phenolic compounds including bisphenol A (BPA) in mangrove of Shenzhen, and shows significant variation with BPA biodegradation (Tong et al., 2021). The correlation between fungal abundance and phenol oxidase activities in the Mai Po wetlands of Hong Kong suggests that fungi can contribute to soluble phenols reduction (Luo et al., 2018). The relative abundance of hydrocarbon-degrading bacteria (Proteobacteria, Actinobacteria, and Bacteroidetes) in hydrocarbon-contaminated sediments increases in salt marshes along the Gulf of Mexico (Beazley et al., 2012). PAHs-degrading bacteria (Proteobacteria, Bacteroidetes, Firmicutes, and Chloroflexi) in mangrove increase under polycyclic aromatic hydrocarbons contamination in the Jiulongjiang estuary (Su et al., 2016).
Functionally diverse microbial communities in CW contribute to the biogeochemical transformation of elements such as carbon (C) and nitrogen (N; Yuan et al., 2014; Yang et al., 2022), and these biological processes mainly include carbon formation and degradation, carbon fixation and nitrogen metabolism, methane metabolism, and exogenous biodegradation and metabolism (Mohapatra et al., 2021).
Halobacteria and Thaumarchaeota are found in Yellow River Delta which can fix CO2 (Hu et al., 2016). Compared to mangrove, invasive Spartina alterniflora significantly can increase CH4 emissions and decrease CO2 emissions (Han and Gu, 2015). CH4 production was high in soils with saline plants in Suncheon Bay, Korea (Chaudhary et al., 2018). Nutrient transformation is related to highly active and adaptive bacterial metabolic channels in Chinese coastal zone (Zhang et al., 2022). When exposed to unstable substrates, microbial respiration is much higher and can produce more CO2 in mangrove and marsh soils along the east coast of Florida, United States (Barreto et al., 2018). In Liaohe River estuary of China, there is a positive correlation between soil respiration rates and Clostridia abundance, suggesting that anaerobic carbon decomposition is important in brackish wetland soils (Yang et al., 2018).
Changes in biomass and community structure may enhance soil N sequestration due to the abilities of special heterotrophic metabolism and refractory organics degradation of soil microorganisms (Tang et al., 2011). Denitrification is the main mechanism for nitrogen removal in CW of Bohai Sea (Zhang Y. et al., 2019), and ammonia-oxidizing archaea and bacteria are also important in global nitrogen cycle (Bai et al., 2019). Nitrate reduction rates are associated with denitrifying bacterial community in the protected area in Spain, suggesting that microbial communities are closely associated with N2O emissions (Bañeras and Ruiz-Rueda, 2012). Nitrogen and phosphorus additions can increase microbial denitrification in the absence of salinity (Chi et al., 2021a,b,c,d). Nitrification and denitrification rates are more higher in intertidal areas disturbed by crabs, and this process greatly contribute to N2O emissions (An et al., 2021).
Microorganisms in CW sediments are highly biodiverse and spatial heterogeneous, and play a key role in maintaining biogeochemical cycles. With the rapid development of molecular biology technologies, our understanding of microbial community and their potential functions has grown substantially. However, several major challenges remain in CW studies.
(1) Accurate identification of key factors affecting microbial community structure. Microorganisms in coastal wetlands are important for maintaining the normal biogeochemical cycle, so it is important to explore the factors affecting the community structure. The most studies about influencing factors focused on single factor, ignoring a combination of multiple factors. Therefore, it is important to elucidate the dominant factors affecting microbial community, which will help to maintain the stability of coastal ecosystems, prevent CW destruction, and restore degraded CW.
(2) In-depth reveal the potential functions of microorganisms, and decipher the relationship between functional stability and microbial biodiversity. Microorganisms are important for environmental management due to the community and functional diversities. The stable microbial community not only ensure the normal biogeochemical cycle, but also decompose complex pollutants into harmless substances through metabolic activities. Furthermpre, clarifying the coupling relationship between microbial diversity and functional stability and parsing the function of biological elements in habitat function ascension will help to maintain and improve the stability of CW function.
Microorganisms in CW have become important players involving in biogeochemical cycles and potential solutions for the treatment of difficult-to-degrade pollutants. Microbial community structure usually rapidly changes in response to environmental changes. Therefore, they can be used as indicators to detect changes in CW. Our studies discuss the changes in microbial composition of CW, summarize the effects of different factors on microbial community structure and the important functional genes, and further reveal the potential environmental functions of coastal microbes. Microbial communities involving in organic pollutant degradation and material cycling of CW have been well developed, but their functions and the relationship between functional stability and microbial biodiversity need to be further explored.
SL: data curation and analysis and writing - original draft. HL: conceptualization, methodology, resources, supervision, writing - review and editing, project administration, and funding acquisition. HW, BY, and AS: conceptualization, methodology, data curation, and writing - review and editing. All authors contributed to the article and approved the submitted version.
This work was financially supported by the National Key R&D Program of China (no. 2022YFF1300901), National Natural Science Foundation of China (no. 42077353), and Natural Science Foundation of Jilin Province (no. 20230101100JC).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abdu, N., Abdullahi, A. A., and Abdulkadir, A. (2017). Heavy metals and soil microbes. Environ. Chem. Lett. 15, 65–84. doi: 10.1007/s10311-016-0587-x
Abed, R. M. M., Kohls, K., and de Beer, D. (2007). Effect of salinity changes on the bacterial diversity, photosynthesis and oxygen consumption of cyanobacterial mats from an intertidal flat of the Arabian gulf. Environ. Microbiol. 9, 1384–1392. doi: 10.1111/j.1462-2920.2007.01254.x
Adame, M. F., Reef, R., Herrera-Silveira, J. A., and Lovelock, C. E. (2012). Sensitivity of dissolved organic carbon exchange and sediment bacteria to water quality in mangrove forests. Hydrobiologia 691, 239–253. doi: 10.1007/s10750-012-1071-7
An, Z., Gao, D., Chen, F., Wu, L., Zhou, J., Zhang, Z., et al. (2021). Crab bioturbation alters nitrogen cycling and promotes nitrous oxide emission in intertidal wetlands: influence and microbial mechanism. Sci. Total Environ. 797:149176. doi: 10.1016/j.scitotenv.2021.149176
An, J., Liu, C., Wang, Q., Yao, M., Rui, J., Zhang, S., et al. (2019). Soil bacterial community structure in Chinese wetlands. Geoderma 337, 290–299. doi: 10.1016/j.geoderma.2018.09.035
Bae, H.-S., Huang, L., White, J. R., Wang, J., DeLaune, R. D., and Ogram, A. (2018). Response of microbial populations regulating nutrient biogeochemical cycles to oiling of coastal saltmarshes from the Deepwater horizon oil spill. Environ. Pollut. 241, 136–147. doi: 10.1016/j.envpol.2018.05.033
Bahram, M., Netherway, T., Hildebrand, F., Pritsch, K., Drenkhan, R., Loit, K., et al. (2020). Plant nutrient-acquisition strategies drive topsoil microbiome structure and function. New Phytol. 227, 1189–1199. doi: 10.1111/nph.16598
Bai, Y., Mei, L., Zuo, W., Zhang, Y., Gu, C., Shan, Y., et al. (2019). Response of bacterial communities in coastal mudflat saline soil to sewage sludge amendment. Appl. Soil Ecol. 144, 107–111. doi: 10.1016/j.apsoil.2019.07.007
Bañeras, L., and Ruiz-Rueda, O. (2012). The role of plant type and salinity in the selection for the denitrifying community structure in the rhizosphere of wetland vegetation. Int. Microbiol. 15, 89–99. doi: 10.2436/20.1501.01.162
Barbier, E. B., Hacker, S. D., Kennedy, C., Koch, E. W., Stier, A. C., and Silliman, B. R. (2011). The value of estuarine and coastal ecosystem services. Ecol. Monogr. 81, 169–193. doi: 10.1890/10-1510.1
Barreto, C. R., Morrissey, E. M., Wykoff, D. D., and Chapman, S. K. (2018). Co-occurring mangroves and salt marshes differ in microbial community composition. Wetlands 38, 497–508. doi: 10.1007/s13157-018-0994-9
Beazley, M. J., Martinez, R. J., Rajan, S., Powell, J., Piceno, Y. M., Tom, L. M., et al. (2012). Microbial community analysis of a coastal salt Marsh affected by the Deepwater horizon oil spill. PLoS One 7:e41305. doi: 10.1371/journal.pone.0041305
Behera, P., Mahapatra, S., Mohapatra, M., Kim, J. Y., Adhya, T. K., Raina, V., et al. (2017). Salinity and macrophyte drive the biogeography of the sedimentary bacterial communities in a brackish water tropical coastal lagoon. Sci. Total Environ. 595, 472–485. doi: 10.1016/j.scitotenv.2017.03.271
Bertin, C., Yang, X., and Weston, L. A. (2003). The role of root exudates and allelochemicals in the rhizosphere. Plant Soil 256, 67–83. doi: 10.1023/A:1026290508166
Brucet, S., Boix, D., Nathansen, L. W., Quintana, X. D., Jensen, E., Balayla, D., et al. (2012). Effects of temperature, salinity and fish in structuring the macroinvertebrate Community in Shallow Lakes: implications for effects of climate change. PLoS One 7:e30877. doi: 10.1371/journal.pone.0030877
Chambers, L. G., Guevara, R., Boyer, J. N., Troxler, T. G., and Davis, S. E. (2016). Effects of salinity and inundation on microbial community structure and function in a mangrove peat soil. Wetlands 36, 361–371. doi: 10.1007/s13157-016-0745-8
Chambers, L. G., Reddy, K. R., and Osborne, T. Z. (2011). Short-term response of carbon cycling to salinity pulses in a freshwater wetland. Soil Sci. Soc. Am. J. 75, 2000–2007. doi: 10.2136/sssaj2011.0026
Chaudhary, D. R., Kim, J., and Kang, H. (2018). Influences of different halophyte vegetation on soil microbial Community at Temperate Salt Marsh. Microb. Ecol. 75, 729–738. doi: 10.1007/s00248-017-1083-y
Cheung, M. K., Wong, C. K., Chu, K. H., and Kwan, H. S. (2018). Community structure, dynamics and interactions of Bacteria, Archaea and Fungi in subtropical coastal wetland sediments. Sci. Rep. 8:14397. doi: 10.1038/s41598-018-32529-5
Chi, Z., Hou, L., Li, H., Li, J., Wu, H., and Yan, B. (2021b). Elucidating the archaeal community and functional potential in two typical coastal wetlands with different stress patterns. J. Clean. Prod. 285:124894. doi: 10.1016/j.jclepro.2020.124894
Chi, Z., Hou, L., Li, H., Wu, H., and Yan, B. (2021c). Indigenous bacterial community and function in phenanthrene-polluted coastal wetlands: potential for phenanthrene degradation and relation with soil properties. Environ. Res. 199:111357. doi: 10.1016/j.envres.2021.111357
Chi, Z., Ju, S., Li, H., Li, J., Wu, H., and Yan, B. (2021a). Deciphering edaphic bacterial community and function potential in a Chinese delta under exogenous nutrient input and salinity stress. Catena 201:105212. doi: 10.1016/j.catena.2021.105212
Chi, Z., Wang, W., Li, H., Wu, H., and Yan, B. (2021d). Soil organic matter and salinity as critical factors affecting the bacterial community and function of Phragmites australis dominated riparian and coastal wetlands. Sci. Total Environ. 762:143156. doi: 10.1016/j.scitotenv.2020.143156
Cui, B., He, Q., Gu, B., Bai, J., and Liu, X. (2016). China’s coastal wetlands: understanding environmental changes and human impacts for management and conservation. Wetlands 36, 1–9. doi: 10.1007/s13157-016-0737-8
DeLong, E. F., Preston, C. M., Mincer, T., Rich, V., Hallam, S. J., Frigaard, N. U., et al. (2006). Community genomics among stratified microbial assemblages in the Ocean’s interior. Science 311, 496–503. doi: 10.1126/science.1120250
Eddie, B., Juhl, A., Krembs, C., Baysinger, C., and Neuer, S. (2010). Effect of environmental variables on eukaryotic microbial community structure of land-fast Arctic Sea ice. Environ. Microbiol. 12, 797–809. doi: 10.1111/j.1462-2920.2009.02126.x
Fei Xi, X., Wang, L., Jun Hu, J., Shu Tang, Y., Hu, Y., Hua Fu, X., et al. (2014). Salinity influence on soil microbial respiration rate of wetland in the Yangtze River estuary through changing microbial community. J. Environ. Sci. 26, 2562–2570. doi: 10.1016/j.jes.2014.07.016
Fierer, N., and Jackson, R. B. (2006). The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. 103, 626–631. doi: 10.1073/pnas.0507535103
Franklin, R. B., Morrissey, E. M., and Morina, J. C. (2017). Changes in abundance and community structure of nitrate-reducing bacteria along a salinity gradient in tidal wetlands. Pedobiologia 60, 21–26. doi: 10.1016/j.pedobi.2016.12.002
Gao, J., Hou, L., Zheng, Y., Liu, M., Yin, G., Li, X., et al. (2016). nirS-encoding denitrifier community composition, distribution, and abundance along the coastal wetlands of China. Appl. Microbiol. Biotechnol. 100, 8573–8582. doi: 10.1007/s00253-016-7659-5
Garbeva, P., van Veen, J. A., and van Elsas, J. D. (2004). MICROBIAL DIVERSITY IN SOIL: selection of microbial populations by plant and soil type and implications for disease Suppressiveness. Annu. Rev. Phytopathol. 42, 243–270. doi: 10.1146/annurev.phyto.42.012604.135455
Grayston, S. J., Wang, S., Campbell, C. D., and Edwards, A. C. (1998). Selective influence of plant species on microbial diversity in the rhizosphere. Soil Biol. Biochem. 30, 369–378. doi: 10.1016/S0038-0717(97)00124-7
Han, P., and Gu, J.-D. (2015). Further analysis of Anammox bacterial community structures along an anthropogenic nitrogen-input gradient from the riparian sediments of the Pearl River Delta to the Deep-Ocean sediments of the South China Sea. Geomicrobiol J. 32, 789–798. doi: 10.1080/01490451.2014.1001502
Harhangi, H. R., le Roy, M., van Alen, T., Hu, B. L., Groen, J., Kartal, B., et al. (2012). Hydrazine synthase, a unique Phylomarker with which to study the presence and biodiversity of Anammox Bacteria. Appl. Environ. Microbiol. 78, 752–758. doi: 10.1128/AEM.07113-11
Head, I. M., Jones, D. M., and Röling, W. F. M. (2006). Marine microorganisms make a meal of oil. Nat. Rev. Microbiol. 4, 173–182. doi: 10.1038/nrmicro1348
Horton, D. J., Theis, K. R., Uzarski, D. G., and Learman, D. R. (2019). Microbial community structure and microbial networks correspond to nutrient gradients within coastal wetlands of the Laurentian Great Lakes. FEMS Microbiol. Ecol. 95:fiz033. doi: 10.1093/femsec/fiz033
Hu, Y., Wang, L., Tang, Y., Li, Y., Chen, J., Xi, X., et al. (2014). Variability in soil microbial community and activity between coastal and riparian wetlands in the Yangtze River estuary – potential impacts on carbon sequestration. Soil Biol. Biochem. 70, 221–228. doi: 10.1016/j.soilbio.2013.12.025
Hu, Y., Wang, L., Xi, X., Hu, J., Hou, Y., le, Y., et al. (2016). Effects of salinity on soil bacterial and archaeal community in estuarine wetlands and its implications for carbon sequestration: verification in the Yellow River Delta. Chem. Ecol. 32, 669–683. doi: 10.1080/02757540.2016.1177519
Jackson, C. R., and Vallaire, S. C. (2009). Effects of salinity and nutrients on microbial assemblages in Louisiana wetland sediments. Wetlands 29, 277–287. doi: 10.1672/08-86.1
Kuypers, M. M. M., Marchant, H. K., and Kartal, B. (2018). The microbial nitrogen-cycling network. Nat. Rev. Microbiol. 16, 263–276. doi: 10.1038/nrmicro.2018.9
Levin, L. A., Boesch, D. F., Covich, A., Dahm, C., Erséus, C., Ewel, K. C., et al. (2001). The function of marine critical transition zones and the importance of sediment biodiversity. Ecosystems 4, 430–451. doi: 10.1007/s10021-001-0021-4
Levy-Booth, D. J., Prescott, C. E., and Grayston, S. J. (2014). Microbial functional genes involved in nitrogen fixation, nitrification and denitrification in forest ecosystems. Soil Biol. Biochem. 75, 11–25. doi: 10.1016/j.soilbio.2014.03.021
Li, J., Chen, Q., Li, Q., Zhao, C., and Feng, Y. (2021). Influence of plants and environmental variables on the diversity of soil microbial communities in the Yellow River Delta wetland, China. Chemosphere 274:129967. doi: 10.1016/j.chemosphere.2021.129967
Li, H., Chi, Z., Li, J., Wu, H., and Yan, B. (2019). Bacterial community structure and function in soils from tidal freshwater wetlands in a Chinese delta: potential impacts of salinity and nutrient. Sci. Total Environ. 696:134029. doi: 10.1016/j.scitotenv.2019.134029
Li, J., Delgado-Baquerizo, M., Wang, J. T., Hu, H. W., Cai, Z. J., Zhu, Y. N., et al. (2019). Fungal richness contributes to multifunctionality in boreal forest soil. Soil Biol. Biochem. 136:107526. doi: 10.1016/j.soilbio.2019.107526
Li, W., Lv, X., Ruan, J., Yu, M., Song, Y. B., Yu, J., et al. (2019). Variations in soil bacterial composition and diversity in newly formed coastal wetlands. Front. Microbiol. 9:3256. doi: 10.3389/fmicb.2018.03256
Li, Y., Wang, Y., Xu, S., Hu, B., and Wang, Z. L. (2016). Effects of Mariculture and solar-salt production on sediment microbial community structure in a coastal wetland. J. Coast. Res. 33:573. doi: 10.2112/JCOASTRES-D-16-00093.1
Lin, G., He, Y., Lu, J., Chen, H., and Feng, J. (2021). Seasonal variations in soil physicochemical properties and microbial community structure influenced by Spartina alterniflora invasion and Kandelia obovata restoration. Sci. Total Environ. 797:149213. doi: 10.1016/j.scitotenv.2021.149213
Ling, Y.-C., Bush, R., Grice, K., Tulipani, S., Berwick, L., and Moreau, J. W. (2015). Distribution of iron- and sulfate-reducing bacteria across a coastal acid sulfate soil (CASS) environment: implications for passive bioremediation by tidal inundation. Front. Virol. 6:624. doi: 10.3389/fmicb.2015.00624
Lipson, D. A., Raab, T. K., Parker, M., Kelley, S. T., Brislawn, C. J., and Jansson, J. (2015). Changes in microbial communities along redox gradients in polygonized Arctic wet tundra soils: soil microbial communities along a redox gradient. Environ. Microbiol. Rep. 7, 649–657. doi: 10.1111/1758-2229.12301
Lu, G., Xie, B., Cagle, G. A., Wang, X., Han, G., Wang, X., et al. (2021). Effects of simulated nitrogen deposition on soil microbial community diversity in coastal wetland of the Yellow River Delta. Sci. Total Environ. 757:143825. doi: 10.1016/j.scitotenv.2020.143825
Luo, L., Wu, R., Gu, J. D., Zhang, J., Deng, S., Zhang, Y., et al. (2018). Influence of mangrove roots on microbial abundance and ecoenzyme activity in sediments of a subtropical coastal mangrove ecosystem. Int. Biodeterior. Biodegradation 132, 10–17. doi: 10.1016/j.ibiod.2018.05.002
Ma, Z., Zhang, M., Xiao, R., Cui, Y., and Yu, F. (2017). Changes in soil microbial biomass and community composition in coastal wetlands affected by restoration projects in a Chinese delta. Geoderma 289, 124–134. doi: 10.1016/j.geoderma.2016.11.037
Macaulay (2015). Understanding the behaviour of oil-degrading MICRO-organisms to enhance the microbial remediation of spilled petroleum. Appl. Ecol. Environ. Res. 13, 247–261. doi: 10.15666/aeer/1301_247262
Meng, L., Liu, H., Bao, M., and Sun, P. (2016). Microbial community structure shifts are associated with temperature, dispersants and nutrients in crude oil-contaminated seawaters. Mar. Pollut. Bull. 111, 203–212. doi: 10.1016/j.marpolbul.2016.07.010
Menon, R., Jackson, C. R., and Holland, M. M. (2013). The influence of vegetation on microbial enzyme activity and bacterial community structure in freshwater constructed wetland sediments. Wetlands 33, 365–378. doi: 10.1007/s13157-013-0394-0
Minick, K. J., Mitra, B., Noormets, A., and King, J. S. (2019). Saltwater reduces potential CO2 and CH4 production in peat soils from a coastal freshwater forested wetland. Biogeosciences 16, 4671–4686. doi: 10.5194/bg-16-4671-2019
Mohapatra, M., Yadav, R., Rajput, V., Dharne, M. S., and Rastogi, G. (2021). Metagenomic analysis reveals genetic insights on biogeochemical cycling, xenobiotic degradation, and stress resistance in mudflat microbiome. J. Environ. Manag. 292:112738. doi: 10.1016/j.jenvman.2021.112738
Morina, J. C., and Franklin, R. B. (2022). Intensity and duration of exposure determine prokaryotic community response to salinization in freshwater wetland soils. Geoderma 428:116138. doi: 10.1016/j.geoderma.2022.116138
Narayan, S., Beck, M. W., Wilson, P., Thomas, C. J., Guerrero, A., Shepard, C. C., et al. (2017). The value of coastal wetlands for flood damage reduction in the northeastern USA. Sci. Rep. 7:9463. doi: 10.1038/s41598-017-09269-z
Narrowe, A. B., Angle, J. C., Daly, R. A., Stefanik, K. C., Wrighton, K. C., and Miller, C. S. (2017). High-resolution sequencing reveals unexplored archaeal diversity in freshwater wetland soils: freshwater wetland archaeal diversity. Environ. Microbiol. 19, 2192–2209. doi: 10.1111/1462-2920.13703
Ní Chadhain, S. M., Miller, J. L., Dustin, J. P., Trethewey, J. P., Jones, S. H., and Launen, L. A. (2018). An assessment of the microbial community in an urban fringing tidal marsh with an emphasis on petroleum hydrocarbon degradative genes. Mar. Pollut. Bull. 136, 351–364. doi: 10.1016/j.marpolbul.2018.09.002
Noll, M., Matthies, D., Frenzel, P., Derakshani, M., and Liesack, W. (2005). Succession of bacterial community structure and diversity in a paddy soil oxygen gradient. Environ. Microbiol. 7, 382–395. doi: 10.1111/j.1462-2920.2005.00700.x
Oremland, R. S., Marsh, L. M., and Polcin, S. (1982). Methane production and simultaneous sulphate reduction in anoxic, salt marsh sediments. Nature 296, 143–145. doi: 10.1038/296143a0
Peralta, A. L., Matthews, J. W., and Kent, A. D. (2010). Microbial community structure and Denitrification in a wetland mitigation Bank. Appl. Environ. Microbiol. 76, 4207–4215. doi: 10.1128/AEM.02977-09
Perry, C. L., and Mendelssohn, I. A. (2009). Ecosystem effects of expanding populations of Avicennia germinans in a Louisiana salt marsh. Wetlands 29, 396–406. doi: 10.1672/08-100.1
Qian, X., Liang, B., Fu, W., Liu, X., and Cui, B. (2016). Polycyclic aromatic hydrocarbons (PAHs) in surface sediments from the intertidal zone of Bohai Bay, Northeast China: spatial distribution, composition, sources and ecological risk assessment. Mar. Pollut. Bull. 112, 349–358. doi: 10.1016/j.marpolbul.2016.07.040
Reyes-Sosa, M. B., Apodaca-Hernández, J. E., and Arena-Ortiz, M. L. (2018). Bioprospecting for microbes with potential hydrocarbon remediation activity on the northwest coast of the Yucatan peninsula, Mexico, using DNA sequencing. Sci. Total Environ. 642, 1060–1074. doi: 10.1016/j.scitotenv.2018.06.097
Rousk, J., Bååth, E., Brookes, P. C., Lauber, C. L., Lozupone, C., Caporaso, J. G., et al. (2010). Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J. 4, 1340–1351. doi: 10.1038/ismej.2010.58
Santos, H. F., Cury, J. C., Carmo, F. L., Rosado, A. S., and Peixoto, R. S. (2010). 18S rDNA sequences from microeukaryotes reveal oil indicators in mangrove sediment. PLoS One 5:e12437. doi: 10.1371/journal.pone.0012437
Shen, C., Xiong, J., Zhang, H., Feng, Y., Lin, X., Li, X., et al. (2013). Soil pH drives the spatial distribution of bacterial communities along elevation on Changbai Mountain. Soil Biol. Biochem. 57, 204–211. doi: 10.1016/j.soilbio.2012.07.013
Skelton, N. J., and Allaway, W. G. (1996). Oxygen and pressure changes measured in situ during flooding in roots of the Grey mangrove Avicennia marina (Forssk.) Vierh. Aquat. Bot. 54, 165–175. doi: 10.1016/0304-3770(96)01043-1
Su, J., Ouyang, W., Hong, Y., Liao, D., Khan, S., and Li, H. (2016). Responses of endophytic and rhizospheric bacterial communities of salt marsh plant (Spartina alterniflora) to polycyclic aromatic hydrocarbons contamination. J. Soils Sediments 16, 707–715. doi: 10.1007/s11368-015-1217-0
Tang, Y., Wang, L., Jia, J. W., Fu, X. H., le, Y. Q., Chen, X. Z., et al. (2011). Response of soil microbial community in Jiuduansha wetland to different successional stages and its implications for soil microbial respiration and carbon turnover. Soil Biol. Biochem. 43, 638–646. doi: 10.1016/j.soilbio.2010.11.035
Tiralerdpanich, P., Sonthiphand, P., Luepromchai, E., Pinyakong, O., and Pokethitiyook, P. (2018). Potential microbial consortium involved in the biodegradation of diesel, hexadecane and phenanthrene in mangrove sediment explored by metagenomics analysis. Mar. Pollut. Bull. 133, 595–605. doi: 10.1016/j.marpolbul.2018.06.015
Tong, T., Li, R., Chen, J., Ke, Y., and Xie, S. (2021). Bisphenol a biodegradation differs between mudflat and mangrove forest sediments. Chemosphere 270:128664. doi: 10.1016/j.chemosphere.2020.128664
Torres, G. G., Figueroa-Galvis, I., Muñoz-García, A., Polanía, J., and Vanegas, J. (2019). Potential bacterial bioindicators of urban pollution in mangroves. Environ. Pollut. 255:113293. doi: 10.1016/j.envpol.2019.113293
van Breemen, N., Boyer, E. W., Goodale, C. L., Jaworski, N. A., Paustian, K., Seitzinger, S. P., et al. (2002). Where did all the nitrogen go? Fate of nitrogen inputs to large watersheds in the northeastern U.S.A. Biogeochemistry 57, 267–293. doi: 10.1023/A:1015775225913
Wang, Y., Sheng, H. F., He, Y., Wu, J. Y., Jiang, Y. X., Tam, N. F. Y., et al. (2012). Comparison of the levels of bacterial diversity in freshwater, intertidal wetland, and marine sediments by using millions of Illumina tags. Appl. Environ. Microbiol. 78, 8264–8271. doi: 10.1128/AEM.01821-12
Wang, X.-Q., Wang, Y. H., Song, Y. B., and Dong, M. (2022). Formation and functions of arbuscular mycorrhizae in coastal wetland ecosystems: a review. Ecosystem Health Sustainab 8:2144465. doi: 10.1080/20964129.2022.2144465
Ward, N. D., Morrison, E. S., Liu, Y., Rivas-Ubach, A., Osborne, T. Z., Ogram, A. V., et al. (2019). Marine microbial community responses related to wetland carbon mobilization in the coastal zone. Limnol. Oceanogr. Lett. 4, 25–33. doi: 10.1002/lol2.10101
Webster, G., O'Sullivan, L. A., Meng, Y., Williams, A. S., Sass, A. M., Watkins, A. J., et al. (2015). Archaeal community diversity and abundance changes along a natural salinity gradient in estuarine sediments. FEMS Microbiol. Ecol. 91, 1–18. doi: 10.1093/femsec/fiu025
Yang, J.-W., Brandon, M., Landais, A., Duchamp-Alphonse, S., Blunier, T., Prié, F., et al. (2022). Global biosphere primary productivity changes during the past eight glacial cycles. Science 375, 1145–1151. doi: 10.1126/science.abj8826
Yang, J., Yu, X., Liu, L., Zhang, W., and Guo, P. (2012). Algae community and trophic state of subtropical reservoirs in Southeast Fujian, China. Environ. Sci. Pollut. Res. 19, 1432–1442. doi: 10.1007/s11356-011-0683-1
Yang, J., Zhan, C., Li, Y., Zhou, D., Yu, Y., and Yu, J. (2018). Effect of salinity on soil respiration in relation to dissolved organic carbon and microbial characteristics of a wetland in the Liaohe River estuary, Northeast China. Sci. Total Environ. 642, 946–953. doi: 10.1016/j.scitotenv.2018.06.121
Ye, G., Chen, J., Yang, P., Hu, H. W., He, Z. Y., Wang, D., et al. (2022). Non-native plant species invasion increases the importance of deterministic processes in fungal community assembly in a coastal wetland. Microbial Ecol., 1–12. doi: 10.1007/s00248-022-02144-z
Yu, Y., Wang, H., Liu, J., Wang, Q., Shen, T., Guo, W., et al. (2012). Shifts in microbial community function and structure along the successional gradient of coastal wetlands in Yellow River estuary. Eur. J. Soil Biol. 49, 12–21. doi: 10.1016/j.ejsobi.2011.08.006
Yu, Z., Yang, J., Yu, X., Liu, L., and Tian, Y. (2014). Aboveground vegetation influences belowground microeukaryotic Community in a Mangrove Nature Reserve. Wetlands 34, 393–401. doi: 10.1007/s13157-013-0506-x
Yuan, H., Li, T., Ding, X., Zhao, G., and Ye, S. (2014). Distribution, sources and potential toxicological significance of polycyclic aromatic hydrocarbons (PAHs) in surface soils of the Yellow River Delta, China. Mar. Pollut. Bull. 83, 258–264. doi: 10.1016/j.marpolbul.2014.03.043
Zhang, G., Bai, J., Tebbe, C. C., Zhao, Q., Jia, J., Wang, W., et al. (2021). Salinity controls soil microbial community structure and function in coastal estuarine wetlands. Environ. Microbiol. 23, 1020–1037. doi: 10.1111/1462-2920.15281
Zhang, Z., Han, P., Zheng, Y., Jiao, S., Dong, H., Liang, X., et al. (2022). Spatiotemporal dynamics of bacterial taxonomic and functional profiles in estuarine intertidal soils of China coastal zone. Microb. Ecol. 85, 383–399. doi: 10.1007/s00248-022-01996-9
Zhang, Y., Ji, G., Wang, C., Zhang, X., and Xu, M. (2019). Importance of denitrification driven by the relative abundances of microbial communities in coastal wetlands. Environ. Pollut. 244, 47–54. doi: 10.1016/j.envpol.2018.10.016
Zhang, L., and Shao, H. (2013). Direct plant–plant facilitation in coastal wetlands: a review. Estuar. Coast. Shelf Sci. 119, 1–6. doi: 10.1016/j.ecss.2013.01.002
Zhang, X., Zhang, L., Zhang, L., Ji, Z., Shao, Y., Zhou, H., et al. (2019). Comparison of rhizosphere bacterial communities of reed and Suaeda in Shuangtaizi River estuary, Northeast China. Mar. Pollut. Bull. 140, 171–178. doi: 10.1016/j.marpolbul.2019.01.041
Zhang, H., Zheng, S., Ding, J., Wang, O., and Liu, F. (2017). Spatial variation in bacterial community in natural wetland-river-sea ecosystems. J. Basic Microbiol. 57, 536–546. doi: 10.1002/jobm.201700041
Zhao, Q., Zhao, H., Gao, Y., Zheng, L., Wang, J., and Bai, J. (2020). Alterations of bacterial and archaeal communities by freshwater input in coastal wetlands of the Yellow River Delta, China. Appl. Soil Ecol. 153:103581. doi: 10.1016/j.apsoil.2020.103581
Keywords: coastal wetlands, microorganisms, community structure, functional gene, environmental potential
Citation: Liang S, Li H, Wu H, Yan B and Song A (2023) Microorganisms in coastal wetland sediments: a review on microbial community structure, functional gene, and environmental potential. Front. Microbiol. 14:1163896. doi: 10.3389/fmicb.2023.1163896
Received: 11 February 2023; Accepted: 16 May 2023;
Published: 02 June 2023.
Edited by:
Ruiyong Zhang, Institute of Oceanology, Chinese Academy of Sciences (CAS), ChinaCopyright © 2023 Liang, Li, Wu, Yan and Song. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huai Li, bGlodWFpQGlnYS5hYy5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.