- Blis Technologies Ltd., South Dunedin, New Zealand
The human oral cavity contains a diversity of microbial habitats that have been adopted and adapted to as homeland by an amazingly heterogeneous population of microorganisms collectively referred to as the oral microbiota. These microbes generally co-habit in harmonious homeostasis. However, under conditions of imposed stress, as with changes to the host’s physiology or nutritional status, or as a response to foreign microbial or antimicrobial incursions, some components of the oral “microbiome” (viz. the in situ microbiota) may enter a dysbiotic state. This microbiome dysbiosis can manifest in a variety of guises including streptococcal sore throats, dental caries, oral thrush, halitosis and periodontal disease. Most of the strategies currently available for the management or treatment of microbial diseases of the oral cavity focus on the repetitive “broad sweep” and short-term culling of oral microbe populations, hopefully including the perceived principal pathogens. Both physical and chemical techniques are used. However, the application of more focused approaches to the harnessing or elimination of key oral cavity pathogens is now feasible through the use of probiotic strains that are naturally adapted for oral cavity colonization and also are equipped to produce anti-competitor molecules such as the bacteriocins and bacteriocin-like inhibitory substances (viz BLIS). Some of these probiotics are capable of suppressing the proliferation of a variety of recognized microbial pathogens of the human mouth, thereby assisting with the restoration of oral microbiome homeostasis. BLIS K12 and BLIS M18, the progenitors of the BLIS-producing oral probiotics, are members of the human oral cavity commensal species Streptococcus salivarius. More recently however, a number of other streptococcal and some non-streptococcal candidate oral probiotics have also been promoted. What is becoming increasingly apparent is that the future for oral probiotic applications will probably extend well beyond the attempted limitation of the direct pathological consequences of oral microbiome dysbiosis to also encompass a plethora of systemic diseases and disorders of the human host. The background to and the evolving prospects for the beneficial modulation of the oral microbiome via the application of BLIS-producing S. salivarius probiotics comprises the principal focus of the present review.
The human oral cavity as a microbial habitat
The indigenous microbes of the oral cavity are predominantly and distinctively site-specific in their habitat, their tropisms being determined by characteristic physical, physiological, and biological compatibilities that are the outcome of thousands of years of co-evolution with their human host. Interestingly the in vogue descriptor for the oral microbial community has progressively evolved over the years from “oral microflora” to “oral microbiota” to the presently favored “oral microbiome.” Every human harbors a personalized oral cavity microbiome and its role when present in finely balanced equilibrium is critical for health maintenance. Unfortunately, at times, our oral microbiome can become destabilized in the face of therapeutic, dietary or host physiological changes and this dysbiosis can become a source of harm to host tissues, both oral and systemic.
A core human microbiome seems likely to be common to all individuals (Turnbaugh et al., 2007). The human oral microbiome database (Gao et al., 2018) lists approximately 700 bacterial species, only half of which are at present officially named. Included are 43 Streptococcus species, 26 of which are named. As the largest bacterial group in the oral cavity the streptococci inevitably are a significant contributor to our oral health (Dewhirst et al., 2010). The core oral microbiome develops as a direct response to a unique combination of host lifestyle and genetic determinants and the essence of its composition is likely to be as specific to ourselves as is our fingerprint (McLean et al., 2022). The foundations of the oral microbiome are established in the perinatal period and comprise an impressive repertoire of microbes that are indigenous to this habitat (Kaan et al., 2021; Kageyama et al., 2022). At birth, the baby may harbor some microbes obtained from the mother during passage through the birth canal, but soon will also acquire from its various close contacts a rich menagerie of attached, planktonic and intracellular microbes. The pioneer colonizing microbes mirror those of the parents-especially the mother (Carlsson et al., 1970; Tagg et al., 1983). Fine-tuning of the microbiome however, will continue throughout life in response to physiological changes and natural aging processes and sometimes it will incur major (potentially detrimental) population shifts (dysbiosis) following host disease, major dietary shifts or antibiotic interventions.
The oral cavity is the body’s principal entry portal for novel microbes and each day a myriad will enter from the environment or from other animal contacts (mostly human) seeking a suitable niche for multiplication of their own kind. Some of these microbes upon entering the oral cavity will attach, some will invade, but most pass through unimpeded–and all (or their products) potentially contribute to the ecology of the oral microbiome. Planktonic bacterial cells may attach either directly or to saliva-coated oral cavity surfaces or indirectly via binding to other bacterial cells that have already colonized (Kolenbrander et al., 2002). Dental biofilms are multispecies ecosystems in which various oral species, tethered by salivary proteins and microbial exopolysaccharides, can interact cooperatively or competitively. Biofilms are able to resist some mechanical stress and their inhabitants can often shun antimicrobial treatments. Adhesion via coaggregation can be critical for the early retention of newly-introduced microbes on surfaces and may facilitate their subsequent colonization. Communication within the biofilm occurs via a number of mechanisms including metabolic synergies, genetic exchanges, inhibitor interactions, and quorum sensing, the later occurring in response to cell density changes and potentially having an influence both on virulence and bacteriocin production (Sedgley et al., 2008; Mignolet et al., 2018, 2019; Hols et al., 2019). Although biofilm-located microbes appear relatively refractory to the action of most chemotherapeutic agents there are indications that some bacteriocins may have efficacy for the treatment of biofilms, especially when these are used in combination with other agents known to function as stressors for the cell envelopes of planktonic cells (Mathur et al., 2018).
Increased knowledge of the principles of microbial ecology has heightened our awareness of the importance of the intimate and interdependent relationships between the human host and its oral microbiome. Subtle changes within the microbiome environment can directly influence gene expression, metabolic activity, inter-microbial competitiveness and ultimately the composition of the microbiome–and this in turn may have significant consequences for the physical health and perhaps even the emotional wellbeing of the host. Probiotic incursions within the resident oral cavity microbiome have historically largely been in the form of the fleeting passage of intestinal probiotics en route to the gastrointestinal tract. It has been only relatively recently that purposeful probiotic interventions have been developed using oral cavity sourced lineages of microorganisms such as the BLIS-producing S. salivarius probiotics that are already naturally pre-programmed for functional residence within the human oral cavity.
Some terminology updates
Microbiota and microbiome
The human microbiota is the collection of microorganisms, predominantly bacteria–but also including minority populations of archaea, fungi, viruses, mycoplasma, and protozoa that live in intimate association with the human body (Li X. et al., 2022). The term refers to the ecological community of commensal, symbiotic, and pathogenic microorganisms that are now recognized to be major determinants of both our health and disease (Lederberg and McCray, 2001). The distinction between the terms microbiome and microbiota has often been misunderstood. However, one contemporary view is that the microbiome encapsulates the microbiota (the community of microorganisms) together with their “theater of activity,” the latter comprising the microbial structural elements, metabolites/signal molecules and the surrounding environmental conditions (Berg et al., 2020). The microbiome provides various traits that humans have not been required to evolve for themselves. Indeed, the expression of the human metagenome (the genome of the entire human microbiome) provides a wide variety of important resources not associated with human cellular activity. Sleator has used the descriptor “human superorganism” to refer to the communal group of cells (only ca. 10% of which are human) that (largely) work in synchrony for the benefit of the collective entity—i.e., a fully-functional human being (Sleator, 2010). Other researchers favor the use of the term “human supraorganism” (Turnbaugh et al., 2007). Hill (2021) has promoted the concept that the human microbiome be considered to be an essential virtual organ, the integrity of which should be protected in the face of therapeutic (e.g., broad-spectrum antibiotic) interventions. This supports the value of adopting more selective (or targeted) microbial culling strategies using relatively narrow spectrum inhibitors such as the bacteriocins and BLIS to suppress pathogen numbers within the microbiome. By minimizing disruption to the existing microbiome composition, the proliferation of disease-initiating outgrowths of potentially pathogenic microbes (such as those present in a “carriage” state) is inhibited.
Dysbiosis
Dysbiosis can be viewed as a reduction in microbial diversity associated with a loss of beneficial microbes and an increase in pathobionts (viz. organisms that are typically indigenous to a microbiota, but sometimes becoming pathogenic and contributing to disease). Dysbiosis typically ensues either following or in association with disruption to microbial homeostasis due to an imbalance within the microbiome resulting either from underlying compositional or metabolic changes. Some factors triggering dysbiosis within the oral microbiome include antibiotic medications, dietary shifts (e.g., elevated sucrose or alcohol intake), a reduction in saliva flow and a proliferation of microbial pathogens. Many dental diseases and also an increasing number of systemic diseases of the human host (Peng et al., 2022; Xu et al., 2022) are now considered to be a consequence of dysbiotic shifts within the normally stable resident oral microbiome (Radaic and Kapila, 2021). The fostering of populations of health-enhancing species and functions via the judicious application of probiotics can help to mitigate against disease expression (Hoare et al., 2017; Rosier et al., 2017).
Probiotics
Although the formal definition of probiotic (viz. “pro-life”) has undergone a series of evolutionary changes over the years, the contemporary view is that probiotics are “live microorganisms that, when administered in adequate amounts, confer a health benefit to the host,” a term adopted by the World Health Organisation (WHO) (Reid, 2005; Teughels et al., 2008). The associated guidelines for use of the term “probiotic” stipulate the need for both a specific microbial strain designation and for at least one study demonstrating a health benefit to be performed in the target host species in order for the strain to be appropriately referred to as a probiotic. Additionally, more specific guidelines for the scientifically acceptable application of the term probiotic to a microbe have also been proposed (Hill et al., 2014). Unfortunately, these guidelines are not always adhered to and all too often candidate strains are prematurely promoted as probiotics prior to the demonstration that their administration actually confers a health benefit to the proposed target species (Reid et al., 2019). Use of the qualifier term “potential” or “putative” should always be adopted until proof of probiotic efficacy is obtained. As the public acceptance and adoption of probiotics continues to escalate, it remains of paramount importance to give stringent regulatory attention to the building of robust safety profiles for all candidate strains (van den Nieuwboer and Claassen, 2019).
It is now evident that the beneficial actions of probiotics can be attributed to a variety of factors including their metabolic characteristics, the molecules presented on the cell surface and their secreted products. Integral cell components such as the DNA and peptidoglycan may also contribute toward probiotic efficacy (Bermudez-Brito et al., 2012). Important attributes for any microbe proposed for use as a probiotic in humans include: (a) relatively targeted anti-pathogen activity (b) absence of cytotoxicity for human tissues (c) being of a species that is consistently prevalent within the human microbiota and (d) exhibiting a capability of integrating and persisting within the human microbiome.
Beneficial outcomes from the use of probiotics are principally derived from their:
(1) Modulation of the host immune system (Raheem et al., 2021).
(2) Molecular interactions with other microbes (commensals or pathogens) (Mignolet et al., 2018, 2019; Hols et al., 2019).
(3) Influences on microbial products (e.g., toxins), host cell products (e.g., bile salts) or food components (e.g., via enzyme activity) (Petrova et al., 2022).
Prebiotics, synbiotics, postbiotics, and oral probiotics
Prebiotics
Prebiotics were instigated by Gibson and Roberfroid (1995) as a dietary means of altering the human colonic microbiota toward a more favorable community structure. The International Scientific Association for Probiotics and Prebiotics (ISAPP) has published a consensus view on prebiotics refining the definition to “a substrate that is selectively utilized by host microorganisms, conferring a health benefit” (Gibson et al., 2017). This updated definition now includes provision for non-carbohydrate compounds, and the site of their action is not limited to the gastrointestinal tract, nor is its type limited to food. Researchers are increasingly exploring the potential roles of prebiotics in increasing the beneficial activities of oral probiotics and in modulating the composition and host interactivity of the oral microbiome (Devine and Marsh, 2009).
Synbiotics
Synbiotics were initially conceived of as a combination of probiotics and prebiotics and are now defined as “a mixture comprising live microorganisms and substrate(s) selectively utilized by host microorganisms that confers a health benefit on the host” (Swanson et al., 2020). A synergistic synbiotic is a synbiotic for which the substrate is designed to be selectively utilized by the co-administered microorganisms (Swanson et al., 2020). Simply put, the microbial component does not necessarily have to be a stand-alone probiotic and the non-digestible substrate does not necessarily have to be a stand-alone prebiotic, but, if together they provide a health benefit, then the mixture can be called a synbiotic.
Postbiotics
In 2019 an ISAPP expert panel proposed a consensus definition of postbiotics as “preparations of inanimate microorganisms and/or their components that confer a health benefit on the host” (Salminen et al., 2021b). Postbiotics contain inactivated microbial cells or cell components, with or without their metabolites, and these must be demonstrated to contribute to observed health benefits in the target host (species and subpopulation). The panel also noted that some confusion may have been created in the past through the use of a wide variety of other designations for these agents such as paraprobiotics, parapsychobiotics, ghost probiotics, zombie probiotics, metabiotics, tyndallized probiotics and bacterial lysates. Nevertheless, proponents of the term paraprobiotics have maintained that the definition of these as “non-viable microbial cells (either intact or broken), or crude cell extracts, which, when administered (orally or topically) in adequate amounts, confer a benefit on the human or animal consumer” is distinctive from that proposed for “postbiotics” (Taverniti and Guglielmetti, 2011; Siciliano et al., 2021). However, despite some strong support for retention of “paraprobiotic” as a distinctive term (Aguilar-Toalá et al., 2021) the ISAPP panel has more recently reaffirmed its recommendation for the unified application of the term postbiotic for all of these products (Salminen et al., 2021a; Vinderola et al., 2022).
Oral probiotics
Unlike most of the traditional intestinal tract-derived probiotics that are principally intended to provide gut-related health benefits, the oral probiotics are originally sourced from the oral microbiota. The beneficial outcomes of oral probiotics primarily (but not necessarily exclusively) result from their interaction with microbial and/or host cells within the oral cavity (Hale et al., 2016). This distinguishes bona fide oral probiotics from the more traditional “orally administered probiotics” (such as those present in yogurts or in products predominantly comprising intestinal lactobacilli and bifidobacteria) that in general do not persistently localize within the oral microbiota following their ingestion. Interestingly, many of the earlier endeavors to identify bacteria capable of fulfilling the functional role of oral probiotics had focused upon re-evaluating some of the already well-established intestinal probiotics, in spite of the inability of these strains to achieve any substantial persistent colonization within the oral microbiota.
Bacteriocins, BLIS, RiPPs, and AMPs
A remarkably heterogeneous array of chemical entities have been shown to be inhibitory to the growth of microorganisms (Figure 1). The peptidic nature of some introduces a degree of specificity to their antimicrobial activity.
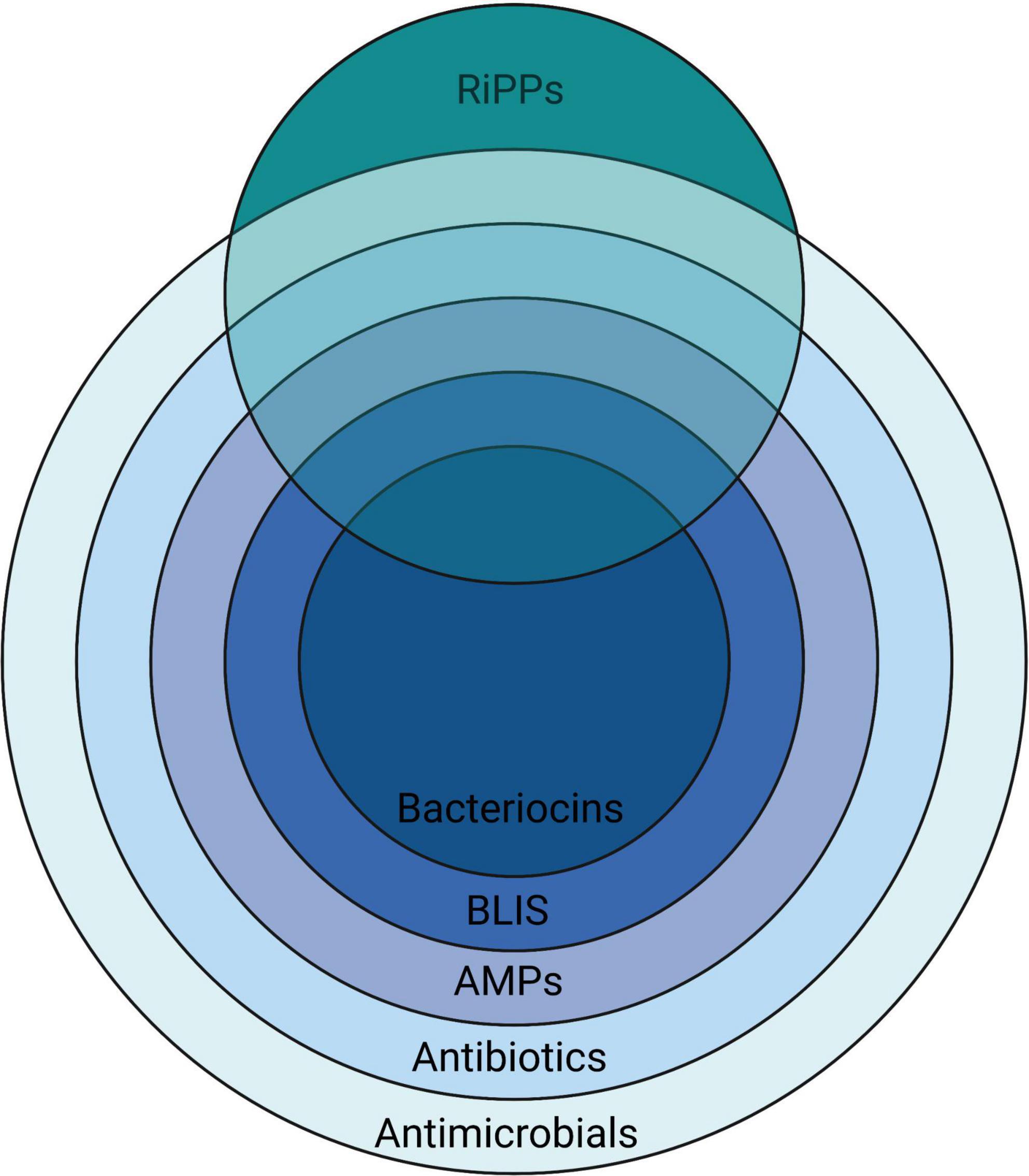
Figure 1. A nested overview of the contemporary usage of the terms used to describe various chemical agents having activity against microorganisms, illustrating how they relate to the more broadly applicable terms antibiotics (literally “opposing life”–used for any substance active against microbes, but in current usage referring to naturally-produced agents) and antimicrobials—a term that is also inclusive of synthetic substances. Created with BioRender.com.
Bacteriocins
The term bacteriocin was first used by Jacob et al. (1953) to categorize the then newly-discovered group of colicins and colicin-like bactericidal antibiotics produced by certain members of the Enterobacteriaceae. The typically plasmid-encoded synthesis of these bacteriocins was shown sometimes to be lethal to the producing cell in a process of altruistic behavior referred to as “programmed cell death” and their adsorption to sensitive cells was dependant on the presence of specific receptors. The bacteriocins were differentiated from most of the “classical” chemotherapeutic antibiotics that were also being discovered at around that time because of their proteinaceous composition and also the relatively narrow targeting of their anti-bacterial activity which typically was focused upon susceptible bacteria of species either the same as or closely related to that of the bacteriocin producer (Jacob et al., 1953).
BLIS
This term was initially devised as an acronym for “Bacteriocin-Like Inhibitory Substances” and its intended application was as a preliminary descriptor for proteinaceous antibiotic activities produced by bacteria when tested in vitro against some closely related bacteria (Tagg, 1991, 1992). BLIS have been defined as “bacterial peptide or protein molecules, released extracellularly, that in low concentrations are able to kill certain other closely related bacteria by a mechanism against which the producer cell exhibits a degree of specific immunity”. This definition is not restrictive to substances having plasmid-borne genetic determinants. Nor does it imply a requirement for lethal biosynthesis, specific receptors or a particularly narrow activity spectrum, all of which were cornerstone elements of the original specifications for bacteriocins (Jacob et al., 1953).
RiPPs
The ribosomally synthesized and post-translationally modified peptides (RiPPs) are a diverse class of natural products of ribosomal origin. Consisting now of more than 20 sub-classes, RiPPs are produced by prokaryotes, eukaryotes, and archaea, and some possess a wide range of alternative biological functions, besides anti-microbial activity. Some RiPPs such as the lantibiotics also classify as bacteriocins because of their demonstrable targeted bactericidal activity (Arnison et al., 2013; Li and Rebuffat, 2020).
AMPs
The antimicrobial peptides (AMPs) are small, generally cationic, peptides from a wide variety of sources that can have relatively broad spectrum inhibitory activity against microorganisms (including bacteria, viruses, parasites, and fungi) by targeting membranes or specific intracellular components (Hale and Hancock, 2007; Omardien et al., 2016). The AMPs have been classified on the basis of their (1) source, (2) activity, (3) structural characteristics, and (4) amino acid-rich species (Huan et al., 2020).
An overview of the bacteriocins of gram-positive bacteria
The first documentation of antagonistic interactions between bacteria has been attributed to Louis Pasteur. In his Florey (1946), commented that Pasteur and Joubert (1877) had reported that the growth of the anthrax Bacillus in urine was inhibited by the concomitant presence of “common bacteria” (probably Escherichia coli) and that this perhaps justified “the highest hopes for therapeutics” (translated). Subsequently, numerous attempts were made to control infections such as diphtheria and anthrax by dosing patients or vulnerable contacts with a variety of non-pathogenic antagonistic microorganisms. The mechanisms of these putative “microbial interference” activities were never fully understood (Florey, 1946). With the discovery of penicillin by Fleming in 1929 the antibiotic era was launched and interest quickly waned in the development of microbial interference as a strategy for infection prevention. The documented investigation of bacteriocins actually dates from 1925, when Gratia reported that filtrates from cultures of Escherichia coli V inhibited the growth of some other E. coli strains (Gratia, 1925). The active principle was later named “colicine” by Gratia and Fredericq (1946). Bacteriocinogenicity (the production of bacteriocins) has subsequently been characterized as an apparently essential survival characteristic for both Gram-negative and Gram-positive bacteria (Klaenhammer, 1988; Riley and Wertz, 2002). Similar molecules, named the archaeocins (Price and Shand, 2000), are also produced by members of the Archaea. For bacteriocin-producing bacteria there is inevitably a trade-off between the benefits to be obtained from their increased competitiveness versus the fitness cost incurred by the heightened metabolic load associated with the production of the bacteriocin molecules and also the mandatory homologous bacteriocin immunity (viz. protective shield) in the host cell (Blanchard et al., 2016). An important consideration is the relative benefit to the cell of constitutive bacteriocin production compared to quorum sensing controlled production. Interestingly it appears that in most natural ecosystems the cost to the producer bacterium of cell density-mediated control over constitutive bacteriocin production and the associated host cell specific bacteriocin immunity is the more energetically favorable option (Blanchard et al., 2016).
The apparently ubiquitous production of bacteriocins by all members of naturally-occurring populations of bacteria infers and indeed validates their significance as indispensable survival attributes for bacteria in highly competitive natural ecosystems and highlights their contribution to the maintenance of long-term population stability within microbial communities via the suppression of over-exuberant proliferation by other family members competing for the same ecological niche (García-Curiel et al., 2021; Heilbronner et al., 2021). From the perspective of the producer cell, bacteriocin production must be tightly regulated since this is an energetically expensive activity and also potentially autotoxic should bacteriocin levels exceed the specific bacteriocin immunity capabilities of the producer cell. Understanding the conditions triggering induction in situ is important in order to regulate/optimize bacteriocin expression in the host tissues. More recently, a growing awareness of the mammalian host cell signaling (modulating) activities of molecules previously characterized as bacteriocins has raised important questions about the underlying raison d’être for these molecules (Lin et al., 2019; Małaczewska et al., 2019). Is the modulation (beneficial or otherwise) of the human host’s physiology/immunity/brain function actually the primary evolutionary selection for the expression of these molecules by our indigenous microbiota? Perhaps the anti-competitor (bacteriocin) activity of some of these molecules is an auxiliary function, aiding the retention of bacterial populations capable of expressing these molecules at levels that are most appropriate for their primary human cell signaling functions?
Several bacteriocin typing schemas were developed in the pre-genomic era for use as epidemiological tools to help track the movements of populations of coliform bacteria (Hamon, 1961) and Pseudomonas aeruginosa (Gillies and Govan, 1966; Tagg and Mushin, 1971), particularly within hospital settings. These had as their basis the in vitro demonstration of characteristic patterns of bacteriocin-mediated inhibitory activity produced by the test strains against specified sets of “indicator” bacteria. Nisin (named for Group N inhibitory substance) (Mattick et al., 1947) was isolated from Lactococcus lactis (Rogers and Whittier, 1928; Whitehead, 1933) and soon became established as the progenitor and indeed still remains the flagship molecule of the lantibiotic class of bacteriocins–molecules containing distinctive lanthionine or beta methyl lanthionine-mediated intra-molecular cross-linkages (Guder et al., 2000). The practical development of nisin as a food preservative followed rapidly (Delves-Broughton et al., 1996) and its successful commercial application actually pre-dates Florey’s development of penicillin (Florey, 1946). Subsequently, nisin and nisin-based probiotics have been widely used to obtain clinical benefits in the treatment of a variety of oral and systemic diseases (Nguyen et al., 2020).
It was soon recognized that other Gram-positive bacteria and especially the so-called lactic acid bacteria (LAB) are prolific producers of bacteriocin-like inhibitory substances (viz. BLIS) (Tagg et al., 1976; Jack et al., 1995; Heng et al., 2007c). The genus Streptococcus became an early focus for studies of the LAB bacteriocins, and an initial impetus was the search for a streptococcal bacteriocin targeting that classical human pathogen, Streptococcus pyogenes, as part of a strategy to limit the occurrence of rheumatic fever in school aged children (Tagg and Dierksen, 2003). Perhaps anomalously, the first of the streptococcal bacteriocins to be definitively characterized was actually produced by a strain of Streptococcus pyogenes. This bacteriocin, streptococcin A-FF22 (SA-FF22), was shown to be a lantibiotic, very closely resembling nisin (Tagg and Wannamaker, 1978). Subsequently, a streptococcal “BLIS fingerprinting” scheme was devised (Tagg and Bannister, 1979), modeled upon the principles that had been previously developed for the bacteriocin typing of Gram-negative bacteria. BLIS fingerprinting had as its basis the in vitro detection of both the production of (P-typing) and sensitivity to (S-typing) streptococcal BLIS activities. Use of this methodology soon established that the production of BLIS activity within the streptococci was both frequent and heterogeneous (Nes et al., 2007; Tagg, 2009) and that isolates of Streptococcus salivarius, Streptococcus uberis and also of the mutans streptococcus cluster were particularly prolific BLIS producers (Tagg, 1992).
The recommendations for a classification framework for inhibitory agents conforming to the overarching characteristics of the bacteriocins continue to evolve as the depth of knowledge of their structural and biological characteristics and their genetic basis has increased. Klaenhammer (1993) initially attempted to introduce some order into the classification of the known bacteriocins of the LAB by proposing four major classes. Class I—post-translationally modified bacteriocins, such as nisin; Class II—small (<10 kDa) heat-stable membrane-active bacteriocins; Class III—larger (>30 kDa) heat-labile bacteriocins and Class IV—complex bacteriocins composed of essential lipid or carbohydrate moieties in addition to protein. Class II was further subdivided into IIa (anti-listerial peptides having the amino acid motif YGNGV/L in the N-terminal part of the peptide), IIb (two-component peptides) and IIc (thiol-activated peptides requiring reduced cysteine residues for activity) (Figure 2).
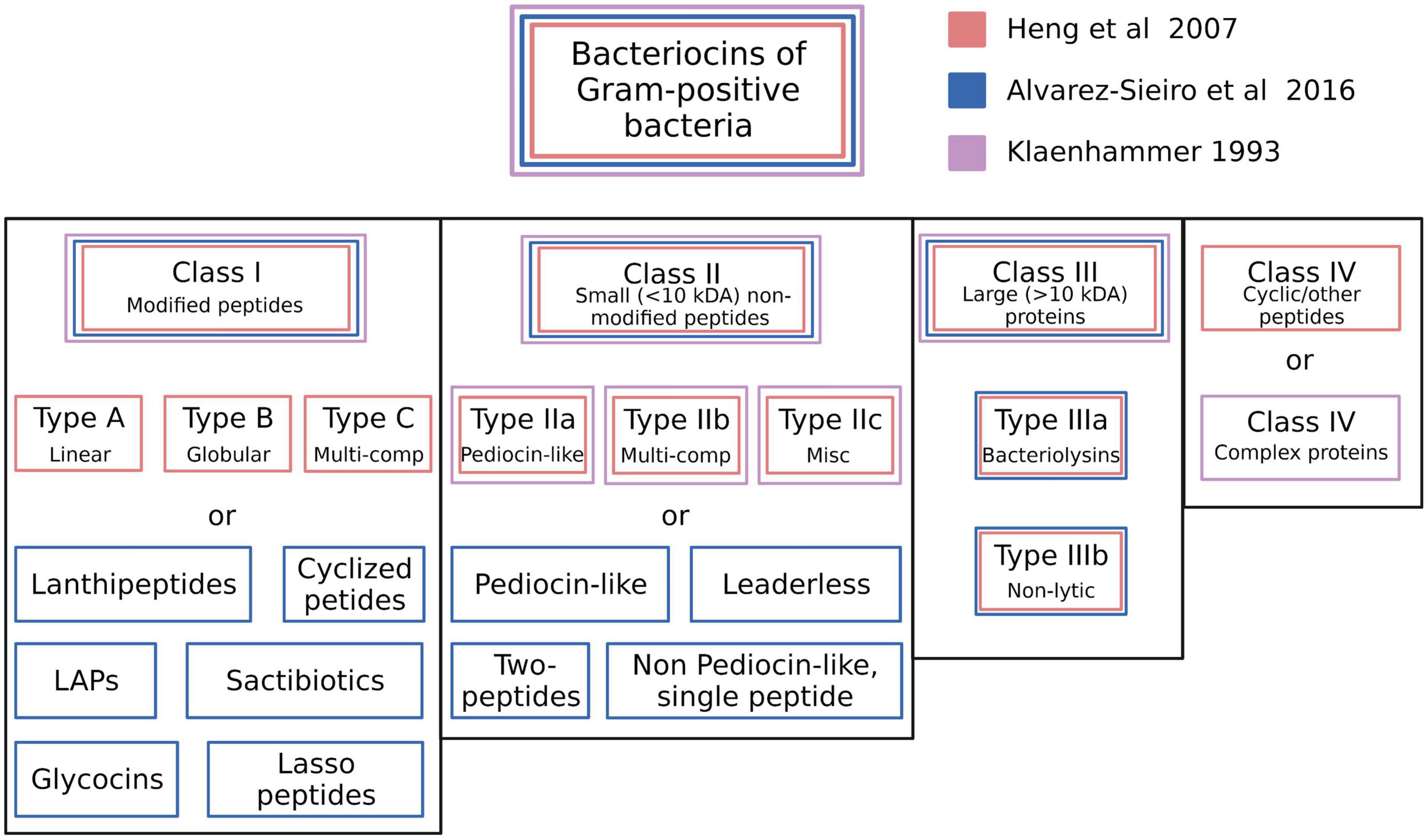
Figure 2. Schematic illustration of the bacteriocin typing schemes proposed by Klaenhammer (1993), Heng et al. (2007c), and Alvarez-Sieiro et al. (2016). The proposed classes for each scheme are color coded. Created with BioRender.com.
Subsequently, Heng et al. (2007c) proposed categorizing the then-known bacteriocins of Gram-positive bacteria according to a revised schema incorporating some of the cornerstone elements of the original Klaenhammer classification. Changes to the Klaenhammer scheme included: (1) elimination of class IV (the chemically complex bacteriocins) (2) Class III was subdivided into IIIa (bacteriolysins) and IIIb (non-lytic proteins) (3) the cyclic bacteriocins from Klaenhammer’s Class II now constituted a newly-defined Class IV (Figure 2). It was at around this time that the phenotypic BLIS fingerprinting of streptococcal isolates of diverse species and from a wide variety of sources was being used in the Tagg laboratory as a preliminary screening tool to facilitate the detection and differentiation of novel streptococcal bacteriocins. An outcome of this BLIS phenotype-driven strategy, the principles of which were essentially as more recently outlined by Twomey et al. (2021) was the de novo detection, purification and characterization of proteinaceous inhibitory molecules belonging to all four of the then-known classes of bacteriocins, as follows:
Class I (lantibiotics): SA-FF22 (Jack et al., 1994), streptin (Wescombe and Tagg, 2003), salivaricin A (Ross et al., 1993), salivaricin A1 (Johnson et al., 1979; Simpson et al., 1995; Tagg, 2004), salivaricins A2-A5 (Wescombe et al., 2006b), salivaricin B (Hyink et al., 2007), salivaricin G32 (Wescombe et al., 2012a), salivaricin 9 (Wescombe et al., 2011), salivaricin E (Walker et al., 2016), Smb (Hyink et al., 2005), BHT-A (Hyink et al., 2005), mutacin K8 (Robson et al., 2007) and nisin U (Wirawan et al., 2006). Subsequent studies have disclosed that transmissible megaplasmids present in some Streptococcus salivarius appear to be particularly adept at integrating (and expressing) lantibiotic loci (Wescombe et al., 2006a; Hyink et al., 2007; Barbour et al., 2020).
Class II (small (<10 kDa) non-modified peptides): mutacin N (Hale et al., 2004), mutacin IV and mutacin V (Hale et al., 2005), BHT-B (Hyink et al., 2005), streptocins STH1 and STH2 (Heng et al., 2007b), ubericin A (Heng et al., 2007a).
Class IIIa (large bacteriolytic proteins): zoocin A (Simmonds et al., 1997) and stellalysin (Heng et al., 2006b).
Class IIIb (large non-bacteriolytic proteins): dysgalacticin (Heng et al., 2006a,b), corynicin JK and enterococcin V583 (Swe et al., 2007) and streptococcin A-M57 (Heng et al., 2004).
Class IV (cyclic peptides): uberolysin (Wirawan et al., 2007).
The basic framework of the Heng bacteriocin classification scheme was subsequently adopted and adapted by several other research groups (Johnson et al., 2018; Ge et al., 2019). More recently however, the discovery of several structurally novel antimicrobial peptides produced by Gram-positive bacteria that also conform to the basic characteristics of the bacteriocins has prompted recommendations for further refinements to the classification of these molecules. For example, Alvarez-Sieiro et al. (2016) incorporated several additional subgroups within Klaenhammer’s Classes I and II, whilst also supporting the elimination of Klaenhammer’s Class IV (Figure 2). Their now-expanded Class I broadly comprised bacteriocin molecules that are ribosomally synthesized together with leader peptides which serve for enzyme recognition, transport, and for keeping the propeptide entities inactive throughout a variety of post-translational modifications that provide them with uncommon amino acids and structures (e.g., lanthionine, heterocycles, head-to-tail cyclization, glycosylations etc.). It should be noted that the Class I bacteriocins also cluster taxonomically within the rapidly expanding family of RiPPS (Arnison et al., 2013). Proposed subclasses within the Class I bacteriocin category were (i) lanthipeptides (Lagedroste et al., 2020), (ii) cyclized peptides (equivalent to Class IV of Heng et al. (2007c) (iii) LAPs, (iv) sactibiotics, (v) glycocins, and (vi) lasso peptides. Class II (the small unmodified bacteriocins) were subclassified as (i) pediocin-like (ii) two-peptide (iii) leaderless, and (iv) non-pediocin-like single peptides.
Subsequently, Acedo et al. (2018) and Zimina et al. (2020) have essentially concurred with the Alvarez-Sieiro et al. (2016) classification recommendations. In both cases retention of the >10 kDa Mr protein (i.e., Class III) category of bacteriocins was supported. On the other hand, Cotter et al. (2012) have argued for exclusion of the >10 kDa Mr proteins from classification as bacteriocins. It is our view however, that limiting the bacteriocins to molecules of mass <10 kDa would actually exclude most of the progenitor bacteriocins of Gram negative bacteria (viz the colicins) from classification as bacteriocins, as well as the growing collection of relatively large non-bacteriolytic proteinaceous bacteriocins, such as dysgalacticin (Heng et al., 2006a), streptococcin A-M57 (Heng et al., 2004) and the tailocins, now also shown to be produced by Gram positive bacteria (Acedo et al., 2018). The classical strategies employed for the screening, identification, purification, and characterization of bacteriocins and some recommendations for their refinement have been reviewed by Zou et al. (2018).
In summary, it is clear that microorganisms have evolved a broad repertoire of antimicrobial agents to support their survival, some of which conform to the cornerstone characteristics of the originally defined bacteriocin molecules viz. bacterial products having a bactericidal (“cin”) mode of action and with an underlying proteinaceous composition (i.e., they are primarily products of ribosomal synthesis). A number of apparent anomalies are evident, however. For example, the lantibiotic salivaricin A is bacteriostatic for most S. pyogenes and this is due to the widespread distribution of the Sal A immunity locus in S. pyogenes (Upton et al., 2001). Also, some of the so-called R23 bacteriocins produced by Lactiplantibacillus plantarum strain R23 have been found to have a bacteriostatic mode of action (Barbosa et al., 2021). In examples such as these there are undoubtedly strong host bacterium survival benefits linked to the modulated expression of the bacteriocins and/or bacteriocin immunity.
The bacteriocins, defined as such for our own convenience, of course represent only one component of an inevitably seamless continuum of bacterial antimicrobial activities. The abundance of whole genome sequence data expedites the detection and genetic analysis of putative bacteriocin clusters. With the advent and widespread application of genome mining technology, the knowledge base concerning the variety and distribution of bacteriocin-like determinants within the bacteria has flourished and a number of data base tools have been introduced to assist with the detection and cataloging of bacteriocin gene clusters (Sandiford, 2017; van Heel et al., 2018; Blin et al., 2021). Although the identification of putative new bacteriocins by database mining has been a promising innovation, the practical potential and the microbiome significance of these determinants is of course difficult to evaluate in the absence of suitable expression systems. The presence of bacteriocin, AMP and RiPP clusters can now be screened for using a variety of bioinformatic tools and databases, including antiSMASH 6.0 (Blin et al., 2021), BACTIBASE (Hammami et al., 2010), APD3–an antimicrobial peptide database (Wang et al., 2016, 2022), LABiocin–a database designed specifically for lactic acid bacterial bacteriocins (Kassaa et al., 2019) and BAGEL4—a web server available to mine RiPPs and bacteriocins (van Heel et al., 2018). InterProScan (Jones et al., 2014) can be used to assess the putative function of encoded proteins and identify protein domains and key sites (Javan et al., 2018). Fields et al. (2020) developed an in vitro pipeline for peptide design and testing using machine learning to predict 20-mer candidate peptides for synthesis followed by laboratory evaluation of their antimicrobial and cytotoxicity activities. Indeed, today, with the widespread availability of contemporary genome mining/annotation/cataloging tools, most putative bacteriocin loci are first detected not by classical phenotypic strain screening approaches, but in database formats prior to the subsequent confirmation of their phenotypic expression.
Some practical applications of bacteriocins in health management
In recent years, the recognition of the relatively narrow activity spectra, high molecular stability and low antigenicity of many of the bacteriocins has led to increased interest within the scientific community in the potential for their development as novel therapeutic agents for infection control in humans and animals (O’Connor et al., 2020). Indeed, cognizance of the central role of the microbiota in human and animal health (Hernández-González et al., 2021) and a growing awareness of the fact that bacteriocins cause much less collateral damage to the host microbiome than classical antibiotics makes them increasingly a highly desirable therapeutic option (Desriac et al., 2010; Svetoch and Stern, 2010; Cotter et al., 2012; Upton et al., 2012; Meade et al., 2020; Soltani et al., 2021; Fernandes and Jobby, 2022). State of the art innovations are now under development to optimize the delivery of bacteriocin preparations (Radaic et al., 2020). The development of novel antibiotics is not keeping pace with resistance development, and this has led to a revival of practical interest in bacteriocins or bacteriotherapy for infection control. Also, it is now recognized that the application of personalized cocktail combinations of probiotics for infection control has considerable potential as a tool to help limit the emergence of antibiotic resistance (Hols et al., 2019). Simons et al. (2020) have provided a comprehensive overview of the bacteriocins of Gram positive and Gram negative bacteria and a commentary about their potential importance in the search for agents having activity against multi-drug resistant bacteria. Apart from their potent antibacterial activity [with minimum inhibitory concentrations (MICs) often in the nanomolar range], some bacteriocins have also been shown to have direct antiviral activity (Tiwari et al., 2020; Todorov et al., 2021).
It is being increasingly recognized that a number of the molecules characterized as “bacteriocins” because of their documented ability to suppress the in vitro growth of prokaryote competitor bacteria may also be adept at physically and functionally interacting with eukaryotic cells of the host–potentially prompting their applications for the treatment of cancers (Kaur and Kaur, 2015; Rodrigues et al., 2019; Sadri et al., 2022) or for modulating neuronal cell activity (Bowland and Weyrich, 2022). Notably certain lantibiotics have been shown to exert beneficial therapeutic and immunomodulatory effects (van Staden et al., 2021) and recently, both salivaricin A2 and salivaricin B (the two key lantibiotic products of S. salivarius BLIS K12), have been shown to reduce disease severity in an animal model of rheumatoid arthritis (Li X. et al., 2022). Indeed, it is evident that some of these molecules may perform key functions for the human host relating more to their specific eukaryotic cell interactions than to their anti-prokaryotic cell activities. Understanding and purposefully manipulating these bacteriocin-mediated activities now provides an innovative contemporary strategy for medicine.
The influence of probiotic-mediated BLIS activity on the composition and functionality of the oral microbiome
In the past, bacteriocin production has largely been considered as a desirable rather than a key probiotic trait (Hegarty et al., 2016). The prospering and indeed the very survival of a bacterium within highly competitive microbial populations is significantly influenced by the controlled expression of its repertoire of bacteriocin determinants. It is evident however, that bacteriocins will only provide an advantage if (a) the benefit of their anti-competitor activity exceeds the metabolic cost of their production, (b) key mutualistic partner bacteria are not targeted and (c) any major competitor bacteria within that ecosystem cannot develop bacteriocin resistance (Heilbronner et al., 2021). It is recognized that bacteriocins are only one component of the biotoxic arsenal of microorganisms. Barbour et al. (2022) have recently reviewed current knowledge of the contribution of a wide variety of microbial metabolites to a microbe’s life and death within the oral microbiome and the implications of this (both good and bad) for the human host.
The expression of bacteriocins and of the homologous specific bacteriocin immunity by a bacterium is an energy demanding process requiring the coordinated action of multiple gene products. The high cost to the cell of bacteriocin activity infers that bacteriocinogenicity city must confer a significant survival benefit upon the host bacterium. It is evident that the proportion of bacteriocin-producing S. salivarius is very high and in most cases the genetic determinants for their sometimes multiple bacteriocins appear to be localized to (>100 kbp) transmissible megaplasmids (Wescombe et al., 2006a,2010). S. salivarius megaplasmids seem to serve as genetic receptacles for the acquisition and expression of a heterogeneous array of bacteriocin loci (Walker et al., 2016), putatively gathered from a variety of other oral streptococci.
The type-A lantibiotic salivaricin A produced by the S. salivarius probiotics BLIS K12 and BLIS M18 functions by forming pores in the cytoplasmic membranes of target (competitor?) bacteria (Geng et al., 2018). Pore formation, however, only occurs if there is a sufficiently high potential difference across the target cell membrane (Wescombe et al., 2009). On the other hand, salivaricin B (produced by BLIS K12, but not by BLIS M18) interferes with cell wall biosynthesis and specifically with septum formation. It does not appear to penetrate the cytoplasmic membrane (Barbour et al., 2016). These observations support the hypothesis that these lantibiotics may help modulate the oral microbiota composition by killing relatively rapidly multiplying competitor bacteria—such as, for example, pathogenic S. pyogenes during the acute stage of a streptococcal throat infection. Interestingly, with the notable exception of serotype M4 strains, it appears that most S. pyogenes harbor and express salivaricin A immunity determinants (viz SalY- a membrane-spanning region of a putative efflux pump) which may provide these S. pyogenes with cross protection against eukaryotic cationic peptides (Phelps and Neely, 2007). Another consequence of this is that salivaricin A is bacteriostatic for these S. pyogenes (Upton et al., 2001). Perhaps this represents a satisfactory evolutionary arrangement for both S. pyogenes and its human host? In an oral microbiome permeated by salivaricin A these S. pyogenes will be relatively constrained in their replication (as in a non-disease-causing carriage state)—but nevertheless some cells will survive! It is evident and indeed should be anticipated that pathogenic bacteria including the opportunistic beta hemolytic streptococci (Vogel and Spellerberg, 2021) are also equipped with their own bacteriocin armory and so the outcome in terms of clonal survival will inevitably also be influenced by a variety of additional factors including the relative cell numbers, adhesion avidity, nutritional requirements and host immune defenses etc.
Bacteriocin production is strongly influenced by both genetic and environmental factors and in order to conserve energy, bacteriocin expression can temporarily be turned off (Wescombe et al., 2006b). Alternatively, bacteriocin genetic loci may be either inactivated (via mutation) or completely lost (as happens in S. salivarius upon megaplasmid elimination) should that bacteriocin fail to confer a survival advantage to the host bacterium. Some bacteriocins inhibit at high concentrations, but have a signaling function when present in low concentrations (Fajardo et al., 2008). Bacteriocins produced by probiotic strains can act as quorum sensing molecules or auto-inducing peptides (Mignolet et al., 2018, 2019; Hols et al., 2019). The targeted killing capability of bacteriocins produced in situ by probiotic bacteria can potentially serve to confer a very substantial anti-infection benefit to the host.
Most probiotic bacteria currently under development are now routinely examined for their expression of bacteriocins targeting microbes that are potentially either pathogenic for the human host or that may contribute to the formation of dysbiotic populations within the host’s indigenous microbiota. The importance of bacteriocinogenicity as a keystone probiotic trait is now well-established (Hegarty et al., 2016). It should be noted however, that the exhibiting of bacteriocin-mediated antagonism in vitro by a probiotic candidate strain does not necessarily correlate with competitiveness in vivo as demonstrated in mouse model studies by Culp et al. (2022).
In summary, bacteriocin production may facilitate: (i) the de novo introduction of the bacteriocin producer bacterium within a pre-established microbiome (ii) resisting the invasion of a niche colonized by the bacteriocin producer by other competitor bacteria, including potential pathogens (iii) beneficial modulation of the host immune system (Dobson et al., 2012).
Desirable characteristics of an oral probiotic
Initially, many of the candidate probiotics examined for their potential to produce health benefits in the oral cavity were of intestinal origin. Pragmatically, in some cases, this may have been because these strains had already satisfied the prerequisite safety and regulatory hurdles. However, intuitively it seems that when seeking a probiotic for efficacious application in the oral cavity, a specialist oral cavity bacterium may be anticipated to be best equipped to do the job, since they have been evolutionarily conditioned for attachment and proliferation within an oral niche and their repertoire of anti-competitor molecules seems more likely to be focused against other microbes that are aggressive oral microbiome insurgents.
The established requirements for efficacious oral probiotics such as viability, adherence ability, provision of health benefits, safety, and delivery efficiency mirror the established requirements for intestinal probiotics (Papadimitriou et al., 2015; How and Yeo, 2021). Additional key attributes for an oral cavity probiotic for use in humans include:
(i) Anti-competitor (viz anti-potential oral cavity pathogen) activity (Figures 3, 4).
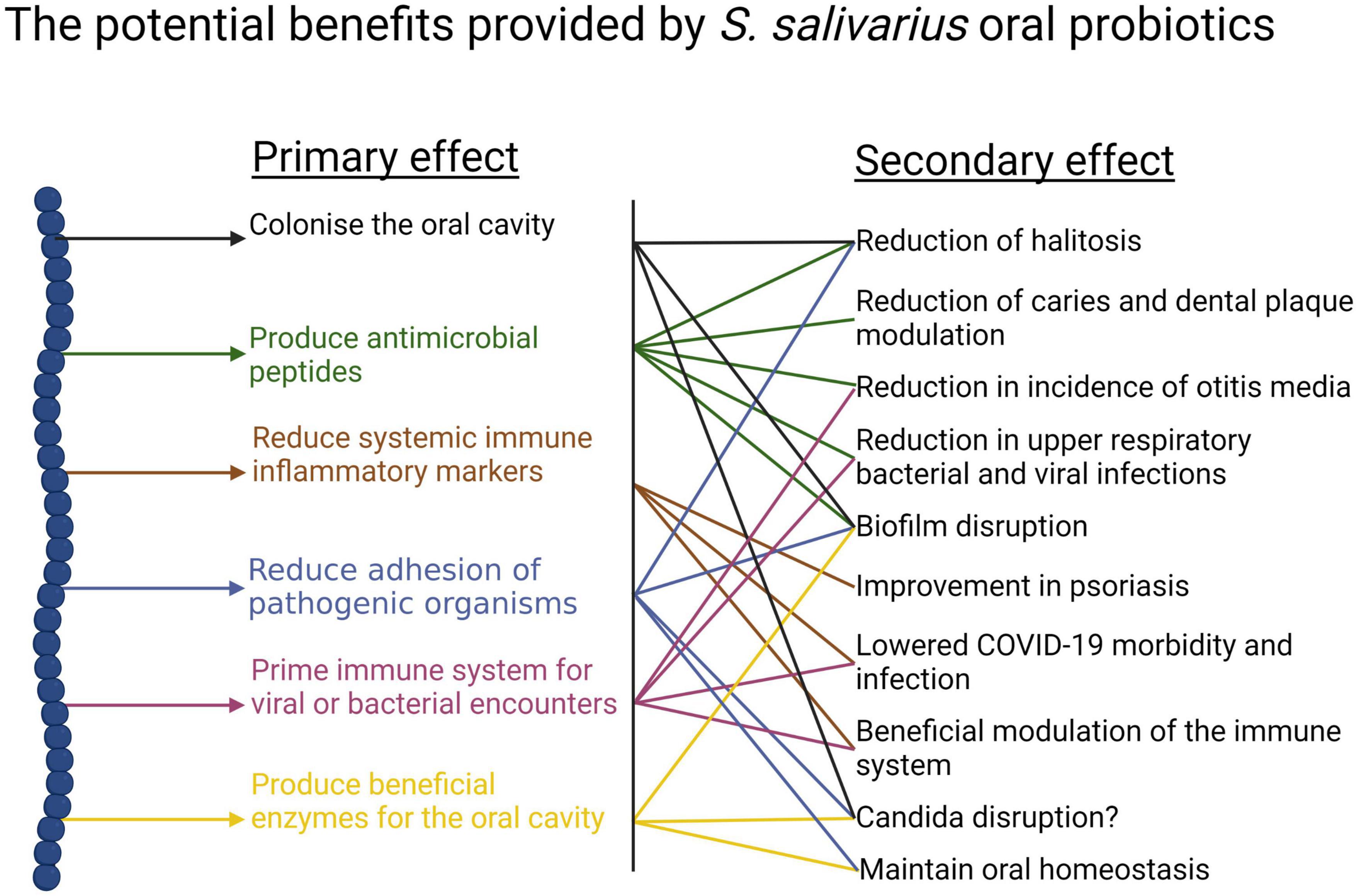
Figure 3. Beneficial activities (primary effects) and outcomes (secondary effects) within the human oral cavity and beyond that have been associated with the use of the S. salivarius probiotics BLIS K12 and BLIS M18. Created with BioRender.com.
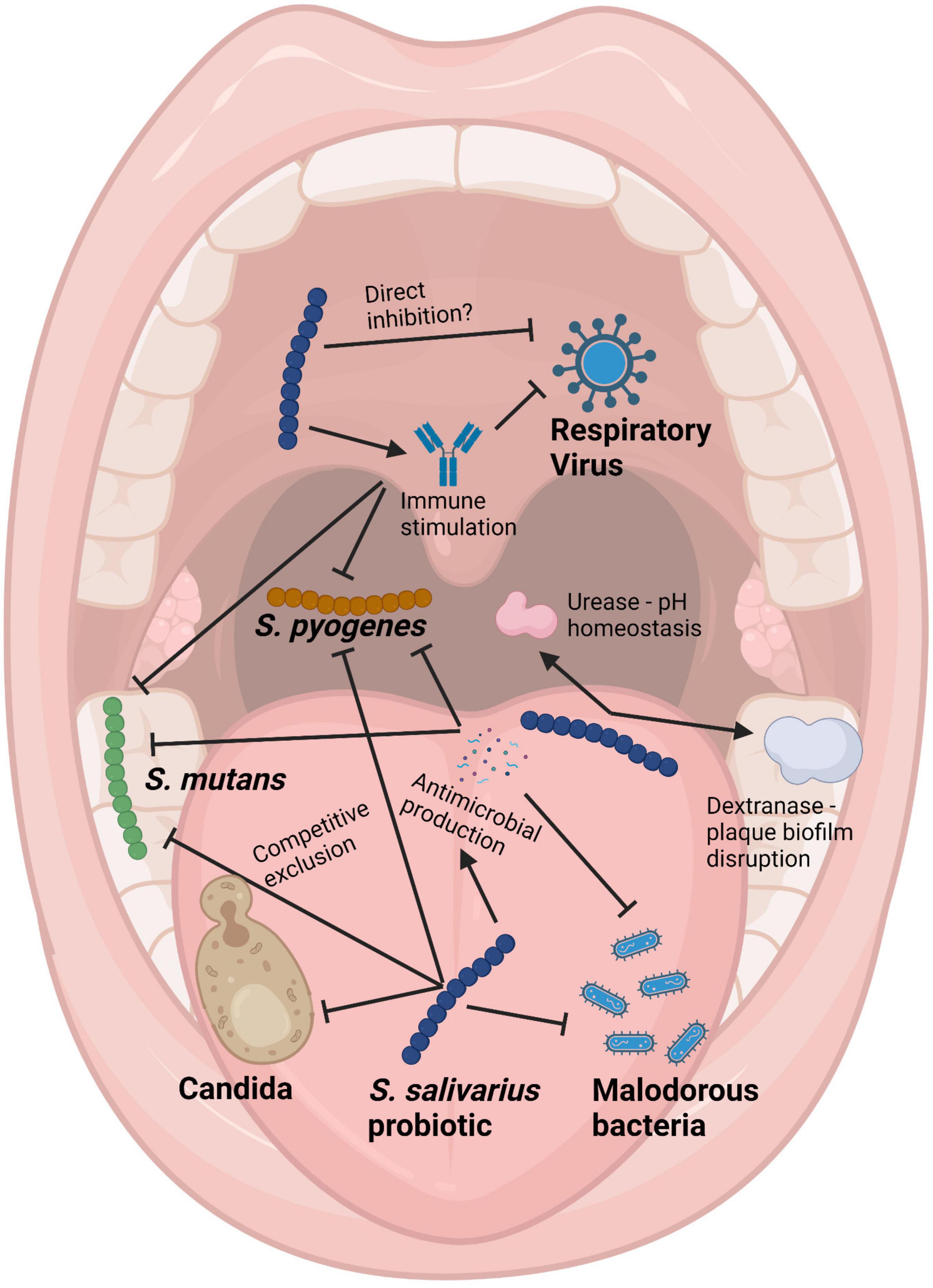
Figure 4. Schematic of the mouth showing some of the health benefits to the host provided by Streptococcus salivarius probiotics. Pointed arrows represent production of molecules, while blunt arrows signify inhibition. Created with BioRender.com.
(ii) High natural oral cavity prevalence of the probiotic species in healthy humans.
(iii) Long term persistence in the oral cavity.
It is difficult for probiotic cells seeded into the human host in a planktonic state to integrate within a pre-established microbial community. Because of this, some key characteristics of an effective oral probiotic will include strong anti-competitor (e.g., bacteriocin or BLIS) repertoires as well as surface structures facilitating their specific adhesion within the oral cavity (Figures 3, 4). Other keystone attributes include an ability to beneficially modulate the functionality of human host cells, potentially protecting the host against inflammation/apoptosis induced by pathogens or in the promotion of homeostasis (Cosseau et al., 2008; Guglielmetti et al., 2010) and the release of enzymes beneficial for the host such as urease (countering acidogenesis) (Chen et al., 2000) and dextranase (Walker et al., 2016) (reducing plaque accumulation) (Figures 3, 4).
Scientific and public interest in the potential for further applications of probiotics as medical therapies is now steadily increasing. Progress in the past has been constrained by:
(i) Lack of standardization, making it difficult to compare and replicate results from different studies.
(ii) Limited understanding of mechanisms of action in providing health benefits, making it difficult to design targeted therapies and predict outcomes.
(iii) Inadequate clinical evidence due to small sample sizes, heterogeneous study populations and inconsistent outcomes.
(iv) Regulatory hurdles. Since probiotics are considered to be dietary supplements rather than drugs it is more difficult to ensure the quality and safety of probiotic products.
Pioneer applications of streptococci as oral probiotics
Many of the microbes consistently found to inhabit the human oral cavity have specifically evolved to operate optimally only within their human host and amongst the more than 700 species already identified, the streptococci are numerically prominent. Following birth, members of the species Streptococcus salivarius are amongst the first to inhabit the oral cavity and they play an important role in the underlying assembly of the oral microbiota (Abranches et al., 2018). The basis for selection of streptococcal strains for potential use as oral probiotics in so-called “bacteriotherapy” (Bellussi et al., 2019) or “replacement therapy” (Tagg and Dierksen, 2003) was not informed by in depth knowledge of the molecular basis for the in vitro observations of their anti-competitor activity against potentially pathogenic microbes. The underlying objective behind these oral microbiota modulation endeavors was to achieve implantation and persistence within the normal microbiota of relatively innocuous “effector” bacteria that could either competitively exclude or at least prevent the outgrowth of potentially disease-causing bacteria, without significantly disturbing the balance of the existing microbial ecosystem.
The principal target in the earliest applications of streptococcal probiotics was that classical human pathogen, S. pyogenes. Sanders et al. (1977) speculated that the relatively reduced occurrence of S. pyogenes pharyngitis episodes in adults by comparison to children may be attributable to adults having larger pharyngeal populations of non-hemolytic streptococci that were capable of producing bactericidal activity against S. pyogenes. Later, these researchers described a low molecular weight pantothenic acid antagonist that they named enocin as a S. salivarius product potentially contributing to anti-S. pyogenes activity (Sanders and Sanders, 1982). Meanwhile, Sprunt and Leidy (1988) demonstrated that a single inoculation with alpha hemolytic streptococcus strain 215 appeared to help beneficially restore the “protective balance” of the nasopharyngeal microbiota in infants. Subsequently, Grahn and Holm (1983) reported that the prevalence of oral alpha-hemolytic streptococci inhibitory in vitro to S. pyogenes was lower in children who became infected during a community outbreak of streptococcal tonsillitis. In follow-up studies these researchers showed that the use of an oral spray containing four strains of alpha hemolytic streptococci following a preliminary course of antibiotics was effective in reducing subsequent tonsillitis episodes in children (Roos et al., 1993, 1996; Falck et al., 1999). One of the inhibitory strains, the colicin V-encoding Streptococcus oralis 89a (Sidjabat et al., 2016), has subsequently also been co-administered with S. salivarius 24SMB in an on-going series of probiotic studies (see later). Prominent among the pioneering studies of use of S. salivarius to reduce tooth decay was the rough-colony forming (on sucrose-containing medium) strain TOVE-R. TOVE-R was found to competitively displace S. mutans and S. sobrinus from the teeth of rats and thereby inhibit tooth decay (Tanzer et al., 1985a,b). In none of these pioneering studies was the identity of the anti-streptococcal activity clearly defined.
BLIS-producing Streptococcus salivarius as oral probiotics for humans
Streptococcus salivarius is perhaps the most innocuous of all the bacterial species known to colonize the human oral and nasopharyngeal mucosae in large numbers. Infants typically acquire the mother’s predominant strain of S. salivarius within hours of birth (Carlsson et al., 1970; Tagg et al., 1983). S. salivarius is present at levels of up to 1 × 107 colony forming units (cfu) per milliliter of saliva and based on the daily estimated average consumption of saliva in adults (up to 1.5 liters), large numbers (estimates of 1 × 1010 cfu) are ingested daily. Within the oral microbiota, S. salivarius has been shown to be especially abundant on the dorsum of the tongue (Mark Welch et al., 2019). S. salivarius is also present in breast milk, an important source of the bacterium in the early months of life (Tagg et al., 1983) and it is also a significant and apparently influential member of the intestinal microbiota (Hols et al., 2019). A recent case control microbiome study has indicated that S. salivarius populations (a) are relatively over-represented in the nasopharyngeal microbiotas of healthy subjects and (b) can demonstrate in vitro inhibitory activity against respiratory pathobionts (Jörissen et al., 2021). Depletion of S. salivarius populations can lead to unbalanced “dysbiotic” overgrowth of the potentially harmful Candida spp. and various black-pigmented anaerobes, manifesting clinically as oral thrush (Liljemark and Gibbons, 1973) and halitosis (Kazor et al., 2003), respectively. It has been speculated that due to its strategic intra-oral cavity location, versatile and abundant anti-competitor weaponry and its substantial numbers, S. salivarius may have an important population surveillance and management (i.e., “sentinel”) role within the oral microbiota (Tagg and Dierksen, 2003). A recent microbiome analysis of children susceptible to chronic otitis media with effusion has also pointed to the important role of S. salivarius as a health-associated and prevalent inhabitant of the human nasopharynx (Jörissen et al., 2021).
S. pyogenes infections in school aged children and their major sequel rheumatic fever was the primary focus of our own pioneering search for an oral probiotic. Seeding of the oral microbiota with a probiotic having anti-S. pyogenes activity was reasoned to potentially provide a means of reducing the occurrence of streptococcal pharyngitis and also an alternative to the currently mandatory antibiotic prophylaxis regimens for the prevention of rheumatic fever recurrences. In the absence of an effective vaccine, S. pyogenes infections and their sequelae continue to cause widespread illness and mortality. Not all children appear equally susceptible to S. pyogenes infection. Our underlying hypothesis was that children experiencing fewer acquisitions of S. pyogenes were naturally protected due to their harboring of indigenous oral cavity populations of bacteria producing anti-S. pyogenes BLIS activity (Tagg, 2009). Our preliminary studies (Tagg and Russell, 1981; Dempster and Tagg, 1982) had indicated that S. pyogenes was particularly susceptible in vitro to an apparently wide variety of the BLIS activities produced by the human oral cavity commensal species S. salivarius. These observations encouraged us to conduct a 6-years longitudinal study of one hundred and three 5–6 years old Dunedin children, monitoring the occurrence of S. pyogenes pharyngitis episodes and their salivary content of BLIS-producing S. salivarius (Tagg et al., 1990). Throat cultures were plated on blood agar (to detect S. pyogenes) and saliva samples on Mitis Salivarius agar were screened for colonies of oral streptococci producing anti-S. pyogenes BLIS activity in simultaneous and deferred antagonism tests. These studies demonstrated that children experiencing relatively fewer S. pyogenes infections often harbored populations of S. salivarius displaying a strong (P-type 677) spectrum of inhibitory activity when tested in vitro using the Tagg and Bannister (1979) BLIS “fingerprinting” assay. The designation P-type 677 equates to inhibition of eight of the nine standard indicator bacteria used in the “fingerprinting” protocol.
The P-type 677 S. salivarius appear typically to be producers of the lantibiotic salivaricin A (Ross et al., 1993). Attention however, soon became focused upon one child in the study who for more than 12 months had retained a predominant oral S. salivarius population producing especially strong (P-type 777) (Figure 5) BLIS activity (i.e., inhibitory to all nine of the standard indicator bacteria), during which period they had experienced no new S. pyogenes infections. S. salivarius strain K12 (viz BLIS K12), selected as the prototype of these P-type 777 isolates, was shown to produce the lantibiotic salivaricin B in addition to salivaricin A. In a follow-up 10 months study of 780 school children 11% had predominant P-type 226 S. salivarius populations, 9% yielded P-type 677 isolates (i.e., putative salivaricin A producers) and around 20% of the children had S. salivarius of various other P-type designations. The group of children harboring P-type 677 S. salivarius populations experienced significantly fewer S. pyogenes infections than did the other children in the study (Dierksen and Tagg, 2000). Meanwhile, a survey of saliva samples from 180 young adults showed that 43% had BLIS-producing S. salivarius of 13 different P-type designations. Of these subjects, 19 (10.5%) yielded P-type 677 S. salivarius and no P-type 777 isolates were detected (Tagg et al., 1983). Examination of 14 independently-sourced S. salivarius producing especially strong (P-type 777) BLIS activity showed that they all harbored mega-plasmids, nine of which encoded various combinations of the lantibiotics salivaricin A, salivaricin B, and streptin (Wescombe et al., 2006a). Megaplasmids appear to be a major asset for S. salivarius and are clearly important for the assembly of their extensive and versatile BLIS portfolios.
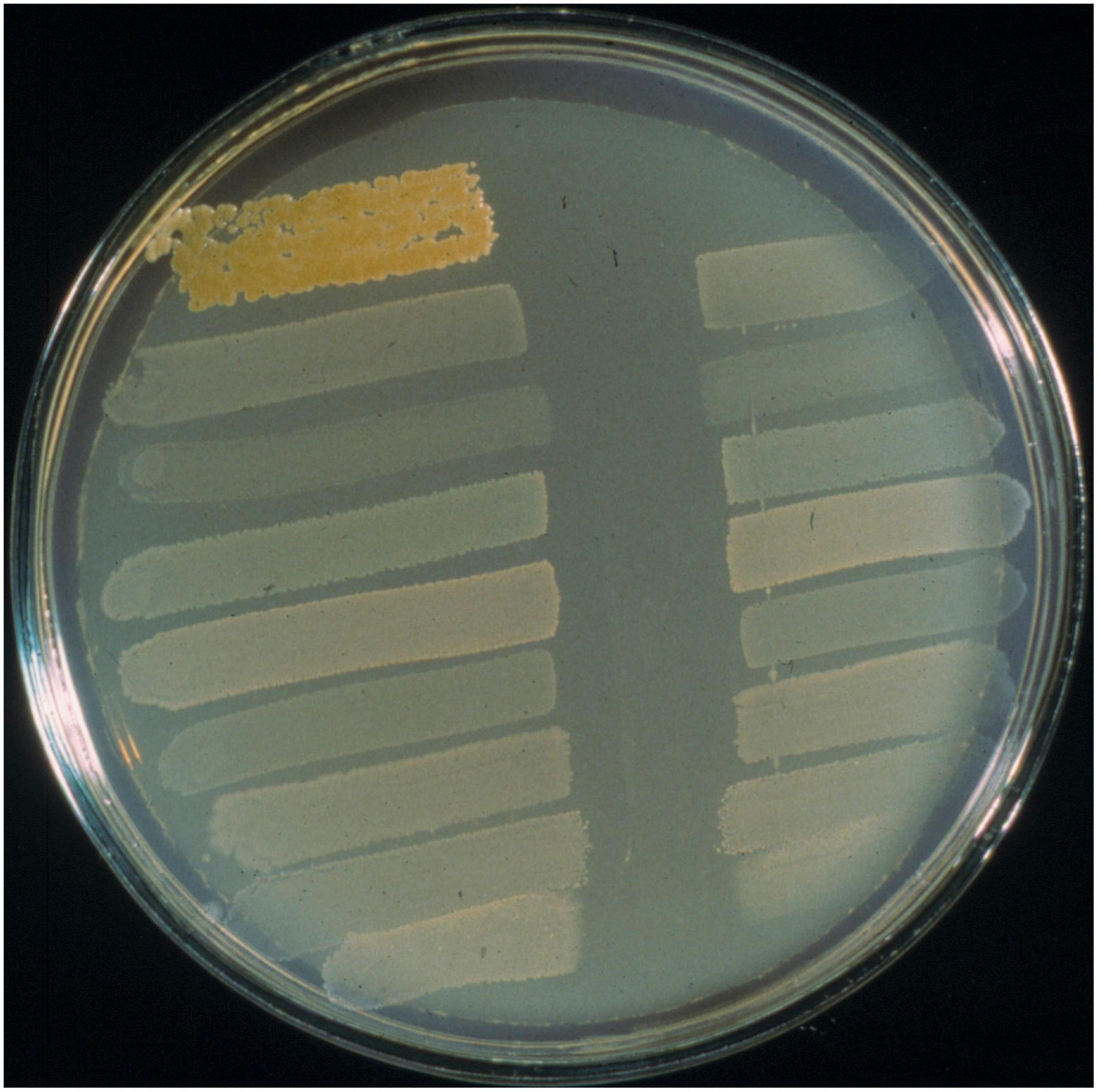
Figure 5. BLIS “fingerprinting” assay of S. salivarius BLIS K12 using the deferred antagonism test methodology as described by Tagg and Bannister (1979). Interference with the growth of all nine standard indicators (as shown here) is referred to as BLIS P-type 777 activity.
Streptococcus salivarius BLIS K12 was initially focused upon as the preferred candidate for development as an oral cavity probiotic because of its especially strong in vitro anti-S. pyogenes activity and its apparently safe and protective long-term persistence within the oral microbiota of its original source human subject (Burton et al., 2011). Extensive safety studies affirmed its “generally recognized as safe” (or “GRAS”) with no objection in the USA and in 2001 S. salivarius BLIS K12™ was distributed commercially by the New Zealand company BLIS Technologies Ltd (Dunedin, New Zealand)1 as the international prototype of the BLIS-producing oral probiotics. BLIS K12 also (a) binds with strong avidity to human epithelial cells in vitro and prevents attachment of S. pyogenes (Fiedler et al., 2013) (b) persists following ingestion in the human oral cavity (Horz et al., 2007) (c) reduces in vitro biofilm formation by S. mutans and several other oral pathogens (Stašková et al., 2021) and reduces the immune activation induced by periodontal disease pathogens (MacDonald et al., 2021). Subsequently BLIS K12 has been found to effect a broad spectrum of health benefits (Figures 3, 4) including the reduction of upper respiratory tract bacterial and viral infections in children (Di Pierro et al., 2014) and adults (Di Pierro et al., 2013), alleviation of halitosis (Burton et al., 2006), reduction of secretory otitis media (Di Pierro et al., 2015a) and Candida albicans infections (Ishijima et al., 2012), beneficial modulation of the immune system (Cosseau et al., 2008; Laws et al., 2021, 2022), stimulation of antiviral immune defenses (Bouwer et al., 2013) and the countering of a wide variety of pathologies that are linked to oral microbiota dysbiosis. The reader is referred to Supplementary Table A which documents the >60 publications focusing on probiotic applications of S. salivarius BLIS K12.
From the perspective of assisting with the maintenance of good oral health however, one apparent significant deficiency in the antimicrobial spectrum of BLIS K12 was the mutans streptococci, recognized as the principal etiological agents of dental caries. Our initial endeavors to identify a streptococcus having strong in vitro BLIS activity against mutans streptococci (James and Tagg, 1988) pinpointed Streptococcus zooepidemicus 4881, a producer of the potent Class IIIa bacteriocin zoocin A (Simmonds et al., 1997). This strain however, had low potential for oral probiotic development since the species has some disease associations and it cannot be considered to be an oral cavity commensal. A more ecologically compatible probiotic candidate strain given preliminary consideration was Streptococcus sanguinis K11 (Skilton and Tagg, 1992). Unfortunately however, the BLIS activity of strain K11 (streptococcin san-K11) appeared limited to the mutans streptococcus species Streptococcus rattus. At this time the strongest S. salivarius producer of in vitro anti-mutans BLIS activity was the P-type 777 strain Min5, but unfortunately its activity seemed limited to the mutans streptococcus species Streptococcus cricetus (James and Tagg, 1988).
Some S. salivarius isolates have been shown to release into human saliva quantities of the enzymes urease (Chen et al., 2000; Burton et al., 2013) and dextranase (Igarashi et al., 2001; Walker et al., 2016), which may potentially modulate dental plaque acidification and accumulation, respectively. In view of this it was decided to also incorporate evaluation of these enzyme activities as components the phenotypic screen of this laboratories BLIS-producing S. salivarius for an appropriate anti-mutans probiotic candidate. Some strains, such as S. salivarius JH (Walker et al., 2016), although exhibiting very potent anti-mutans BLIS activity had to be dismissed from further consideration due to their concomitant production of hemolytic activity—a characteristic of some S. salivarius (Tompkins and Tagg, 1989).
Ultimately, S. salivarius BLIS M18™ was focused upon for commercial development due to its unusually strong (for S. salivarius) activity against the mutans streptococcus species S. mutans and S. sobrinus, in addition to being a strong producer of both urease and dextranase enzyme activities. Four bacteriocin loci have been identified in the BLIS M18 genome: salivaricin A2, salivaricin 9, and salivaricin MPS (all megaplasmid-encoded) and the chromosomally-encoded lantibiotic salivaricin M which appears to be responsible for much of the observed anti-mutans streptococcal activity of this bacterium (Heng et al., 2011; Wescombe et al., 2012b). The range of specific probiotic applications of BLIS M18 have largely been prompted by idiosyncrasies of its BLIS spectrum (Supplementary Table B). Successful outcomes have been reported not only for its impact on Cariogram index (Di Pierro et al., 2015b; Tandelilin et al., 2018; Poorni et al., 2022), reduction of gingivitis/periodontitis (Scariya et al., 2015) and halitosis (Benic et al., 2019; Yoo et al., 2020) and modulation of black staining plaque (Gobbi et al., 2020), but also for its unanticipated postbiotic activity against colon cancer cells (Karaçam and Tunçer, 2022) and the important Gram-negative pathogens P. aeruginosa and Klebsiella pneumoniae (Tunçer and Karaçam, 2020).
It should be noted that in the case of both BLIS K12 and BLIS M18 the primary criterion in the process of strain selection was the in vitro detection of a S. salivarius producing novel BLIS activities against potentially pathogenic or dysbiosis-associated target species within the oral and nasopharyngeal microbiotas. Subsequently however, the spectrum of microbial dysbiosis and pathologies of the human host that have been beneficially modulated through the use of these BLIS-producing S. salivarius probiotics has expanded to include the complete life-span of the human host (Figure 6). Although a steadily increasing number of S. salivarius isolates are now being touted as potential probiotic candidates, very few have as yet progressed for evaluation in human trials (Supplementary Table C). Indeed, the only other S. salivarius to have as yet been widely applied as a probiotic is S. salivarius 24SMB, more recently in combination with S. oralis 89a in nasal spray formulations (Marchisio et al., 2015; Manti et al., 2020). Unlike strains BLIS K12 and BLIS M18, S. salivarius 24SMB is megaplasmid-negative and does not encode any lantibiotic activities (Vertillo Aluisio et al., 2022). It does however, appear to encode a novel blpU-K bacteriocin activity and two class-IIb bacteriocins. The companion probiotic, S. oralis 89a, on the other hand is a producer of colicin V, so called originally because of the association of the ColV plasmid with increased virulence in Escherichia coli strains (Smith and Huggins, 1976) and possibly accounting for the observed relatively strong activity of this strain against Gram negative bacteria (Sidjabat et al., 2016).
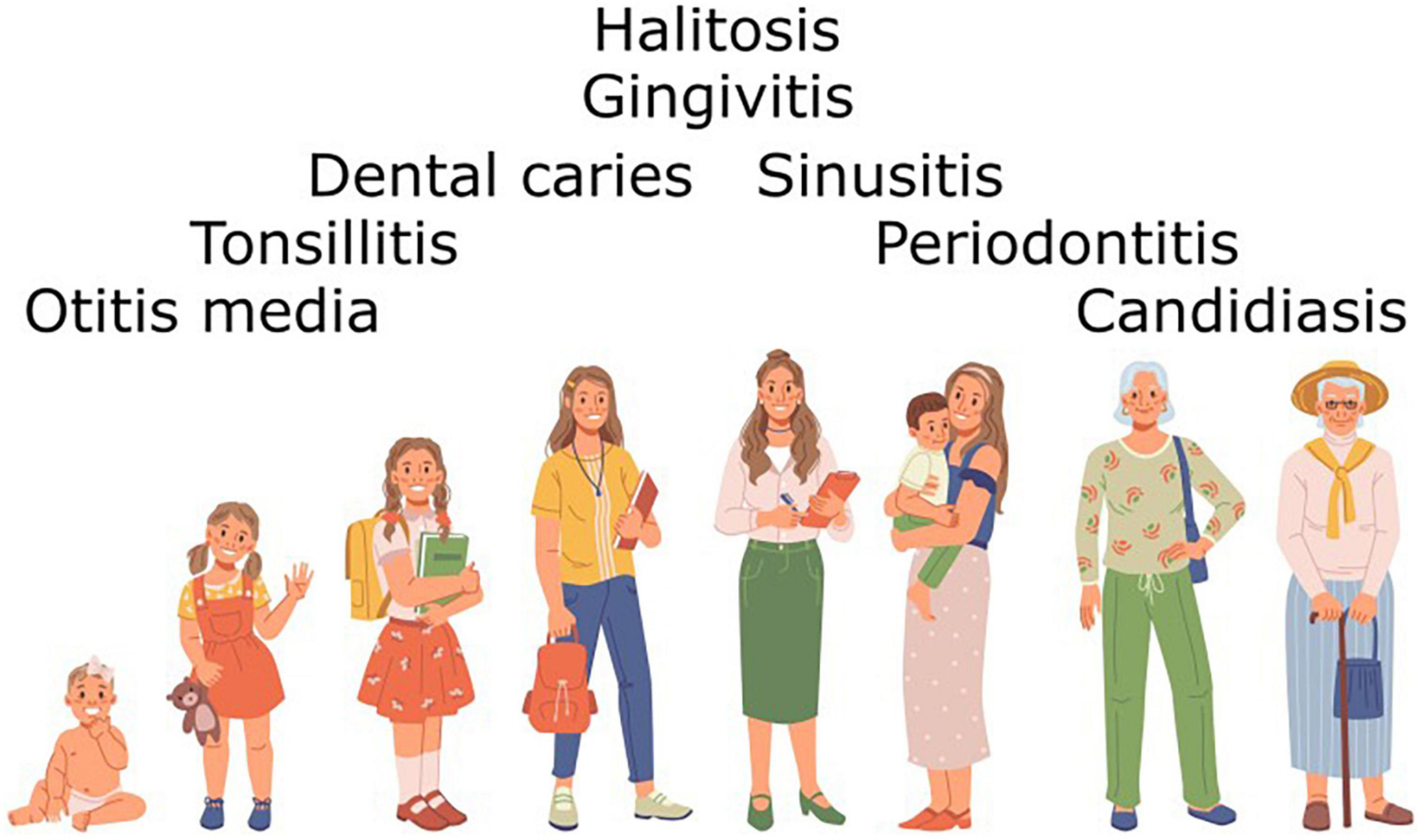
Figure 6. Diseases presenting at different stages of the life of the human host that have already been shown to be beneficially modulated by administration of BLIS-producing S. salivarius probiotics.
Several strains of streptococcal species other than S. salivarius have either been marketed as probiotics or have been promoted as potential probiotics (Supplementary Table D). Foremost amongst these is the commercial oral probiotic product Probiora 3, which comprises a mixture of S. rattus JH145, S. oralis KJ3sm, and Streptococcus uberis KJ2sm (Zahradnik et al., 2009). Other streptococcal species that have been promoted for potential probiotic development include S. cristatus, S. dentisani, S. sanguinis, S. parasanguinis, S. gordonii, S. mitis, S. oligofermentans, S. symci, Streptococcus strain C17T, and Streptococcus sp. A12. Indeed, a steadily-increasing assemblage of streptococcal isolates from the human oral microbiota and various other sources have either already been given consideration or are currently under development for use as oral probiotics (Hale et al., 2016).
Future prospects for oral probiotics
Much human illness is linked either directly (e.g., dental caries, periodontal disease, candidosis) or indirectly to the development of oral microbiota disequilibria (Figure 6). Not only can this contribute directly to the development of dental caries, periodontal disease and candidosis, but it may also facilitate initiation of the transition between carriage state and active infection by bacteria such as S. pyogenes as well as possibly influencing the onset of a wide variety of non-transmissible diseases. Indeed a surfeit of a number of periodontal pathogens has been identified as risk factors for an increasing repertoire of systemic diseases (Bourgeois et al., 2019). Consequently, research into the scientifically orchestrated beneficial applications of oral probiotics has received a substantial fillip as the evidence base underpinning the importance of establishing and maintaining a health-promoting oral microbiota continues to strengthen. Future research directions potentially facilitating the more widespread practical application of oral probiotics to the management and treatment of diseases of the human oral cavity and beyond include:
(i) Identification of optimal strains, dosage levels and delivery mechanisms.
(ii) Investigation of long-term safety.
(iii) Conducting of large-scale, well-controlled clinical trials.
(iv) Further development of regulatory guidelines.
With the advent of the antibiotic era, the further development of bacterial interference-based strategies for infection control was largely put “on hold.” More recently however, the efficacy of many therapeutic antibiotics has diminished due to the wide-spread propagation of antibiotic resistance. Combined with that there has been an associated resurgence of many infectious diseases and an increase in the numbers of immunologically compromised and aged human hosts. It is within this disturbing setting that probiotics are now increasingly being reconsidered either as an alternative or as a supplement to existing chemotherapies. With increasing interest now in probiotic interventions for the specific modulation of microbial dysbiosis in a wide variety of body tissues, attention is being focused upon the “mining” of the predominant microbes that are indigenous to these tissues as a prime source of potential probiotics that are optimally equipped to colonize and aid re-equilibration of the populations of microbes incumbent to these tissues.
Recently there has been renewed interest in understanding microbial behavior in natural habitats. Many chronic diseases can have complex etiologies (sometimes not conforming to classic Koch postulates) and there is mounting evidence for microbial interventions in a plethora of pathologies in the human host (Thomas et al., 2021), many of which are potentially either instigated or sustained by dysbiosis within the oral microbiome (Le Bars et al., 2017; Tiwari et al., 2020; Maitre et al., 2021b; Nguyen et al., 2021; Todorov et al., 2021; Gao et al., 2022; Rismayuddin et al., 2022; Sankarapandian et al., 2022). We all harbor personalized microbiomes based within our oral cavity biofilms, the balanced operations of which are a critical factor in our health status within the mouth -and beyond. The oral microbiome also has a “dark side” however, capable of eliciting disease—both oral and systemic. Ecological shifts within the microbiome may allow pathogenic microbes to elicit disease either due to their reinvigorated multiplication following emergence from a carriage status within the microbiome or due to their relatively unchallenged uptake following de novo entry into the oral cavity. As more detailed information becomes available concerning the population composition and dynamics of the oral microbiome and its genetic complement, this will lead to the informed development of more effective therapeutic and diagnostic approaches (Gao et al., 2018). Ultimately and inevitably, this should then contribute to the development of personalized health management programmes for the entire human/microbiome entity–or super organism. Already there is recognition of the potential for oral microbiome analysis in early life to identify possible microbial harbingers of future disease development in children followed by the administration of tailor made blends of specific oral probiotic “combos,” the composition of which can be adjusted in response to periodic monitoring of the composition of the host’s microbiome (Wescombe et al., 2009; Xiao et al., 2020).
The oral microbiome functions as a critical checkpoint for microbial visitors—harmless or harmful. Will refugee microbes gain acceptance and be assimilated within the oral microbiome or will they pass through for subsequent fitness checks in other parts of the body? Sophisticated profiling of the oral microbiome may indicate carriage of hibernating predators that are potentially capable of becoming a source of infection for the host or their close contacts. Alternatively, this profiling exercise may indicate levels of particular microbes that are potentially capable of adversely influencing host physiology. Indeed, both gingivitis and periodontitis have been linked to a two-way effect between oral and systemic disease (Bui et al., 2019). Furthermore, while the literature on the gut-brain axis is rapidly growing there is now also an undercurrent of scientific probing into the influence of oral microbes on brain function (Maitre et al., 2021a; Narengaowa et al., 2021). Recently, for example Ahrens et al. (2022) pointed to a potential association between an oral microbiome deficiency of the species Alloprevotella rava and suicidal ideation in young adults.
Streptococcus salivarius probiotics have already found successful application to the beneficial modulation of a wide spectrum of microbial dysbiosis impacting on human health. The principal oral bacterial infections currently identified and successfully targeted for probiotic intervention have been streptococcal pharyngitis, otitis media, halitosis and dental caries (Figure 6; Tagg, 2016; Hoare et al., 2017; Bustamante et al., 2020). Perhaps the time is now ripe for the accelerated development and practical application of BLIS-producing S. salivarius probiotics to the biotherapeutic control of other diseases and maladies of complex etiologies resulting from more subtle disturbances within the indigenous microbiota (Tagg, 2016). The targeted probiotic approach to microbiome management is cost effective, easily implemented and does not present many of the complications associated with immunization (hypersensitivity or antigenic cross-reactivity), chemotherapy (resistance development and direct toxicity) or other medications. The considerable scope for BLIS-producing S. salivarius to be beneficially utilized throughout the life of their natural human host perhaps encourages consideration of them as the prototype oral probiotics for all ages.
Author contributions
JT conceived the scope of the manuscript and wrote the first draft. LH designed and prepared the figures, and assisted with the compilation of references. RJ assisted with the text editing and structure. JH advised on text content and flow. All authors contributed to the article and approved the submitted version.
Conflict of interest
JT, LH, RJ, and JH are current employees of Blis Technologies Ltd., the company responsible for the development and commercial distribution of the original BLIS-producing oral probiotics.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1161155/full#supplementary-material
Footnotes
References
Abranches, J., Zeng, L., Kajfasz, J. K., Palmer, S. R., Chakraborty, B., Wen, Z. T., et al. (2018). Biology of oral streptococci. Microbiol. Spectr. 6. doi: 10.1128/MICROBIOLSPEC.GPP3-0042-2018
Acedo, J. Z., Chiorean, S., Vederas, J. C., and van Belkum, M. J. (2018). The expanding structural variety among bacteriocins from Gram-positive bacteria. FEMS Microbiol. Rev. 42, 805–828. doi: 10.1093/FEMSRE/FUY033
Aguilar-Toalá, J. E., Arioli, S., Behare, P., Belzer, C., Berni Canani, R., Chatel, J. M., et al. (2021). Postbiotics—when simplification fails to clarify. Nat. Rev. Gastroenterol. Hepatol. 18, 825–826. doi: 10.1038/S41575-021-00521-6
Ahrens, A. P., Sanchez-Padilla, D. E., Drew, J. C., Oli, M. W., Roesch, L. F. W., and Triplett, E. W. (2022). Saliva microbiome, dietary, and genetic markers are associated with suicidal ideation in university students. Sci. Rep. 12, 1–24. doi: 10.1038/s41598-022-18020-2
Alvarez-Sieiro, P., Montalbán-López, M., Mu, D., and Kuipers, O. P. (2016). Bacteriocins of lactic acid bacteria: Extending the family. Appl. Microbiol. Biotechnol. 100, 2939–2951. doi: 10.1007/S00253-016-7343-9
Arnison, P. G., Bibb, M. J., Bierbaum, G., Bowers, A. A., Bugni, T. S., Bulaj, G., et al. (2013). Ribosomally synthesized and post-translationally modified peptide natural products: Overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 30, 108–160. doi: 10.1039/C2NP20085F
Barbosa, J., Albano, H., Silva, B., Almeida, M. H., Nogueira, T., and Teixeira, P. (2021). Characterization of a Lactiplantibacillus plantarum r23 isolated from arugula by whole-genome sequencing and its bacteriocin production ability. Int. J. Environ. Res. Public Health 18:5515. doi: 10.3390/IJERPH18115515/S1
Barbour, A., Elebyary, O., Fine, N., Oveisi, M., and Glogauer, M. (2022). Metabolites of the oral microbiome: Important mediators of multi-kingdom interactions. FEMS Microbiol. Rev. 46:fuab039. doi: 10.1093/FEMSRE/FUAB039
Barbour, A., Tagg, J., Abou-Zied, O. K., and Philip, K. (2016). New insights into the mode of action of the lantibiotic salivaricin B. Sci. Rep. 6:31749. doi: 10.1038/SREP31749
Barbour, A., Wescombe, P., and Smith, L. (2020). Evolution of lantibiotic salivaricins: New weapons to fight infectious diseases. Trends Microbiol. 28, 578–593. doi: 10.1016/J.TIM.2020.03.001
Bellussi, L. M., Passali, F. M., Ralli, M., Vincentiis, M. D. E., Greco, A., and Passali, D. (2019). An overview on upper respiratory tract infections and bacteriotherapy as innovative therapeutic strategy. Eur. Rev. Med. Pharmacol. Sci. 23, 27–38. doi: 10.26355/EURREV_201903_17345
Benic, G. Z., Farella, M., Morgan, X. C., Viswam, J., Heng, N. C., Cannon, R. D., et al. (2019). Oral probiotics reduce halitosis in patients wearing orthodontic braces: A randomized, triple-blind, placebo-controlled trial. J. Breath Res. 13:36010. doi: 10.1088/1752-7163/AB1C81
Berg, G., Rybakova, D., Fischer, D., Cernava, T., Vergès, M.-C. C., Charles, T., et al. (2020). Microbiome definition re-visited: Old concepts and new challenges. Microbiome 8, 1–22. doi: 10.1186/S40168-020-00875-0
Bermudez-Brito, M., Plaza-Díaz, J., Muñoz-Quezada, S., Gómez-Llorente, C., and Gil, A. (2012). Probiotic mechanisms of action. Ann. Nutr. Metab. 61, 160–174. doi: 10.1159/000342079
Blanchard, A. E., Liao, C., and Lu, T. (2016). An ecological understanding of quorum sensing-controlled bacteriocin synthesis. Cell. Mol. Bioeng. 9, 443–454. doi: 10.1007/S12195-016-0447-6
Blin, K., Shaw, S., Kloosterman, A. M., Charlop-Powers, Z., van Wezel, G. P., Medema, M. H., et al. (2021). antiSMASH 6.0: Improving cluster detection and comparison capabilities. Nucleic Acids Res. 49, W29–W35. doi: 10.1093/NAR/GKAB335
Bourgeois, D., Inquimbert, C., Ottolenghi, L., and Carrouel, F. (2019). Periodontal pathogens as risk factors of cardiovascular diseases, diabetes, rheumatoid arthritis, cancer, and chronic obstructive pulmonary disease-is there cause for consideration? Microorganisms 7:424. doi: 10.3390/MICROORGANISMS7100424
Bouwer, A. L., Saunderson, S. C., Dunn, A. C., Lester, K. L., Crowley, L. R., Jack, R. W., et al. (2013). Rapid interferon-gamma release from natural killer cells induced by a streptococcal commensal. J. Interferon Cytokine Res. 33, 459–466. doi: 10.1089/JIR.2012.0116
Bowland, G. B., and Weyrich, L. S. (2022). The oral-microbiome-brain axis and neuropsychiatric disorders: An anthropological perspective. Front. Psychiatry 13:810008. doi: 10.3389/FPSYT.2022.810008
Bui, F. Q., Almeida-da-Silva, C. L. C., Huynh, B., Trinh, A., Liu, J., Woodward, J., et al. (2019). Association between periodontal pathogens and systemic disease. Biomed. J. 42, 27–35. doi: 10.1016/J.BJ.2018.12.001
Burton, J. P., Cowley, S., Simon, R. R., McKinney, J., Wescombe, P. A., and Tagg, J. R. (2011). Evaluation of safety and human tolerance of the oral probiotic Streptococcus salivarius K12: A randomized, placebo-controlled, double-blind study. Food Chem. Toxicol. 49, 2356–2364. doi: 10.1016/j.fct.2011.06.038
Burton, J. P., Drummond, B. K., Chilcott, C. N., Tagg, J. R., Thomson, W. M., Hale, J. D. F., et al. (2013). Influence of the probiotic Streptococcus salivarius strain M18 on indices of dental health in children: A randomized double-blind, placebo-controlled trial. J. Med. Microbiol. 62(Pt 6), 875–884. doi: 10.1099/jmm.0.056663-0
Burton, J., Chilcott, C., Moore, C., Speiser, G., and Tagg, J. (2006). A preliminary study of the effect of probiotic Streptococcus salivarius K12 on oral malodour parameters. J. Appl. Microbiol. 100, 754–764. doi: 10.1111/J.1365-2672.2006.02837.X
Bustamante, M., Oomah, B., Mosi-Roa, Y., Rubilar, M., and Burgos-Díaz, C. (2020). Probiotics as an adjunct therapy for the treatment of halitosis, dental caries and periodontitis. Probiotics Antimicrob. Proteins 12, 325–334. doi: 10.1007/S12602-019-9521-4
Carlsson, J., Grahnén, H., Jonsson, G., and Wikner, S. (1970). Early establishment of Streptococcus salivarius in the mouth of infants. J. Dent. Res. 49, 415–418. doi: 10.1177/00220345700490023601
Chen, Y. Y. M., Weaver, C. A., and Burne, R. A. (2000). Dual functions of Streptococcus salivarius urease. J. Bacteriol. 182:4667. doi: 10.1128/JB.182.16.4667-4669.2000
Cosseau, C., Devine, D. A., Dullaghan, E., Gardy, J. L., Chikatamarla, A., Gellatly, S., et al. (2008). The commensal Streptococcus salivarius K12 downregulates the innate immune responses of human epithelial cells and promotes host-microbe homeostasis. Infect. Immun. 76, 4163–4175. doi: 10.1128/IAI.00188-08
Cotter, P. D., Ross, R. P., and Hill, C. (2012). Bacteriocins—a viable alternative to antibiotics? Nat. Rev. Microbiol. 11, 95–105. doi: 10.1038/nrmicro2937
Culp, D. J., Hull, W., Schultz, A. C., Bryant, A. S., Lizarraga, C. A., Dupuis, M. R., et al. (2022). Testing of candidate probiotics to prevent dental caries induced by Streptococcus mutans in a mouse model. J. Appl. Microbiol. 132, 3853–3869. doi: 10.1111/JAM.15516
Delves-Broughton, J., Blackburn, P., Evans, R. J., and Hugenholtz, J. (1996). Applications of the bacteriocin, nisin. Antonie van Leeuwenhoek 69, 193–202. doi: 10.1007/BF00399424
Dempster, R. P., and Tagg, J. R. (1982). The production of bacteriocin-like substances by the oral bacterium Streptococcus salivarius. Arch. Oral Biol. 27, 151–157. doi: 10.1016/0003-9969(82)90136-4
Desriac, F., Defer, D., Bourgougnon, N., Brillet, B., Le Chevalier, P., and Fleury, Y. (2010). Bacteriocin as weapons in the marine animal-associated bacteria warfare: Inventory and potential applications as an aquaculture probiotic. Mar. Drugs 8, 1153–1177. doi: 10.3390/MD8041153
Devine, D. A., and Marsh, P. D. (2009). Prospects for the development of probiotics and prebiotics for oral applications. J. Oral Microbiol. 1, 3402–3413. doi: 10.3402/JOM.V1I0.1949
Dewhirst, F. E., Chen, T., Izard, J., Paster, B. J., Tanner, A. C. R., Yu, W. H., et al. (2010). The human oral microbiome. J. Bacteriol. 192:5002. doi: 10.1128/JB.00542-10
Di Pierro, F., Adami, T., Rapacioli, G., Giardini, N., and Streitberger, C. (2013). Clinical evaluation of the oral probiotic Streptococcus salivarius K12 in the prevention of recurrent pharyngitis and/or tonsillitis caused by Streptococcus pyogenes in adults. Expert Opin. Biol. Ther. 13, 339–343. doi: 10.1517/14712598.2013.758711
Di Pierro, F., Colombo, M., Zanvit, A., Risso, P., and Rottoli, A. S. (2014). Use of Streptococcus salivarius K12 in the prevention of streptococcal and viral pharyngotonsillitis in children. Drug Healthc. Patient Saf. 6, 15–20. doi: 10.2147/dhps.s59665
Di Pierro, F., Di Pasquale, D., and Di Cicco, M. (2015a). Oral use of Streptococcus salivarius K12 in children with secretory otitis media: Preliminary results of a pilot, uncontrolled study. Int. J. Gen. Med. 8, 303–308. doi: 10.2147/IJGM.S92488
Di Pierro, F., Zanvit, A., Nobili, P., Risso, P., and Fornaini, C. (2015b). Cariogram outcome after 90 days of oral treatment with Streptococcus salivarius M18 in children at high risk for dental caries: Results of a randomized, controlled study. Clin. Cosmet. Investig. Dent. 7, 107–113. doi: 10.2147/CCIDE.S93066
Dierksen, K., and Tagg, J. (2000). “The influence of indigenous bacteriocin-producing Streptococcus salivarius on the acquisition of Streptococcus pyogenes by primary school children in Dunedin, New Zealand,” in Streptococci and streptococcal diseases entering the new millenium, eds D. Martin and J. Tagg (Auckland: Securacopy), 81–85.
Dobson, A., Cotter, P. D., Ross, R. P., and Hill, C. (2012). Bacteriocin production: A probiotic trait? Appl. Environ. Microbiol. 78, 1–6. doi: 10.1128/AEM.05576-11
Fajardo, A., Martínez-Martín, N., Mercadillo, M., Galán, J. C., Ghysels, B., Matthijs, S., et al. (2008). The neglected intrinsic resistome of bacterial pathogens. PLoS One 3:e1619. doi: 10.1371/JOURNAL.PONE.0001619
Falck, G., Grahn-Håkansson, E., Holm, S. E., Roos, K., and Lagergren, L. (1999). Tolerance and efficacy of interfering alpha-streptococci in recurrence of streptococcal pharyngotonsillitis: A placebo-controlled study. Acta Otolaryngol. 119, 944–948. doi: 10.1080/00016489950180333
Fernandes, A., and Jobby, R. (2022). Bacteriocins from lactic acid bacteria and their potential clinical applications. Appl. Biochem. Biotechnol. 194, 4377–4399. doi: 10.1007/S12010-022-03870-3
Fiedler, T., Riani, C., Koczan, D., Standar, K., Kreikemeyer, B., and Podbielski, A. (2013). Protective mechanisms of respiratory tract streptococci against Streptococcus pyogenes biofilm formation and epithelial cell infection. Appl. Environ. Microbiol. 79:1265. doi: 10.1128/AEM.03350-12
Fields, F. R., Freed, S. D., Carothers, K. E., Hamid, M. N., Hammers, D. E., Ross, J. N., et al. (2020). Novel antimicrobial peptide discovery using machine learning and biophysical selection of minimal bacteriocin domains. Drug Dev. Res. 81, 43–51. doi: 10.1002/DDR.21601
Florey, H. W. (1946). The use of micro-organisms for therapeutic purposes. Yale J. Biol. Med. 19:101.
Gao, L., Cheng, Z., Zhu, F., Bi, C., Shi, Q., and Chen, X. (2022). The oral microbiome and its role in systemic autoimmune diseases: A systematic review of big data analysis. Front. Big Data 5:927520. doi: 10.3389/FDATA.2022.927520
Gao, L., Xu, T., Huang, G., Jiang, S., Gu, Y., and Chen, F. (2018). Oral microbiomes: More and more importance in oral cavity and whole body. Protein Cell 9:488. doi: 10.1007/S13238-018-0548-1
García-Curiel, L., del Rocío López-Cuellar, M., Rodríguez-Hernández, A. I., and Chavarría-Hernández, N. (2021). Toward understanding the signals of bacteriocin production by Streptococcus spp. and their importance in current applications. World J. Microbiol. Biotechnol. 37:15. doi: 10.1007/S11274-020-02973-5
Ge, J., Kang, J., and Ping, W. (2019). Effect of acetic acid on bacteriocin production by Gram-positive bacteria. J. Microbiol. Biotechnol. 29, 1341–1348. doi: 10.4014/jmb.1905.05060
Geng, M., Austin, F., Shin, R., and Smith, L. (2018). Covalent structure and bioactivity of the type AII lantibiotic salivaricin A2. Appl. Environ. Microbiol. 84, e02528–17. doi: 10.1128/AEM.02528-17
Gibson, G. R., and Roberfroid, M. B. (1995). Dietary modulation of the human colonic microbiota: Introducing the concept of prebiotics. J. Nutr. 125, 1401–1412. doi: 10.1093/JN/125.6.1401
Gibson, G. R., Hutkins, R., Sanders, M.-E., Prescott, S. L., Reimer, R. A., Salminen, S. J., et al. (2017). Expert consensus document: The international scientific association for probiotics and prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat. Rev. Gastroenterol. Hepatol. 14, 491–502. doi: 10.1038/nrgastro.2017.75
Gillies, R. R., and Govan, J. R. (1966). Typing of Pseudomonas pyocyanea by pyocine production. J. Pathol. Bacteriol. 91, 339–345. doi: 10.1002/PATH.1700910207
Gobbi, E., de Francesco, M. A., Piccinelli, G., Caruso, A., Bardellini, E., and Majorana, A. (2020). In vitro inhibitory effect of two commercial probiotics on chromogenic actinomycetes. Eur. Arch. Paediatr. Dent. 21, 673–677. doi: 10.1007/s40368-020-00512-2
Grahn, E., and Holm, S. E. (1983). Bacterial interference in the throat flora during a streptococcal tonsillitis outbreak in an apartment house area. Zentralbl. Bakteriol. Mikrobiol. Hyg. A 256, 72–79. doi: 10.1016/S0174-3031(83)80054-7
Gratia, A. (1925). Sur un remarquable example d’antagonisme entre deux souches de colibacille. Compt. Rend. Soc. Biol. 93, 1040–1042.
Gratia, A., and Fredericq, P. (1946). Diversité des souches antibiotiques de B. Coli et étendue variable de leur champ d’action. C. R. Seances Soc. Biol. Fil. 140, 1032–1033.
Guder, A., Wiedemann, I., and Sahl, H.-G. (2000). Posttranslationally modified bacteriocins—the lantibiotics. Pept. Sci. 55, 62–73. doi: 10.1002/1097-0282200055:1<62::AID-BIP60<3.0.CO;2-Y
Guglielmetti, S., Taverniti, V., Minuzzo, M., Arioli, S., Stuknyte, M., Karp, M., et al. (2010). Oral bacteria as potential probiotics for the pharyngeal mucosa. Appl. Environ. Microbiol. 76, 3948–3958. doi: 10.1128/AEM.00109-10
Hale, J. D. F., and Hancock, R. E. W. (2007). Alternative mechanisms of action of cationic antimicrobial peptides on bacteria. Expert Rev. Anti Infect. Ther. 5, 951–959. doi: 10.1586/14787210.5.6.951
Hale, J. D. F., Balakrishnan, M., and Tagg, J. R. (2004). Genetic basis for mutacin N and of its relationship to mutacin I. Indian J. Med. Res. 119, 247–251.
Hale, J. D. F., Ting, Y. T., Jack, R. W., Tagg, J. R., and Heng, N. C. K. (2005). Bacteriocin (Mutacin) Production by Streptococcus mutans genome sequence reference strain UA159: Elucidation of the antimicrobial repertoire by genetic dissection. Appl. Environ. Microbiol. 71:7613. doi: 10.1128/AEM.71.11.7613-7617.2005
Hale, J. D. F., Wescombe, P. A., Tagg, J. R., and Heng, N. C. K. (2016). “Streptococcal bacteriocin-producing strains as oral probiotic agents,” in The bacteriocins: Current knowledge and future prospects, eds R. L. Dorit, S. M. Roy, and M. A. Riley (Poole: Caister Academic Press), 103–126. doi: 10.21775/9781910190371.06
Hammami, R., Zouhir, A., Le Lay, C., Ben Hamida, J., and Fliss, I. (2010). BACTIBASE second release: A database and tool platform for bacteriocin characterization. BMC Microbiol. 10:22. doi: 10.1186/1471-2180-10-22/FIGURES/2
Hegarty, J. W., Guinane, C. M., Ross, R. P., Hill, C., and Cotter, P. D. (2016). Bacteriocin production: A relatively unharnessed probiotic trait? F1000Res 5:2587. doi: 10.12688/f1000research.9615.1
Heilbronner, S., Krismer, B., Brötz-Oesterhelt, H., and Peschel, A. (2021). The microbiome-shaping roles of bacteriocins. Nat. Rev. Microbiol. 19, 726–739. doi: 10.1038/S41579-021-00569-W
Heng, N. C. K., Burtenshaw, G. A., Jack, R. W., and Tagg, J. R. (2004). Sequence analysis of pDN571, a plasmid encoding novel bacteriocin production in M-type 57 Streptococcus pyogenes. Plasmid 52, 225–229. doi: 10.1016/J.PLASMID.2004.08.002
Heng, N. C. K., Haji-Ishak, N. S., Kalyan, A., Wong, A. Y. C., Lovrić, M., Bridson, J. M., et al. (2011). Genome sequence of the bacteriocin-producing oral probiotic Streptococcus salivarius strain M18. J. Bacteriol. 193, 6402–6403. doi: 10.1128/JB.06001-11
Heng, N. C. K., Swe, P. M., Ting, Y. T., Dufour, M., Baird, H. J., Ragland, N. L., et al. (2006b). The large antimicrobial proteins (bacteriocins) of streptococci. Int. Congr. Ser. 1289, 351–354. doi: 10.1016/J.ICS.2005.11.020
Heng, N. C. K., Ragland, N. L., Swe, P. M., Baird, H. J., Inglis, M. A., Tagg, J. R., et al. (2006a). Dysgalacticin: A novel, plasmid-encoded antimicrobial protein (bacteriocin) produced by Streptococcus dysgalactiae subsp. equisimilis. Microbiology (Reading) 152, 1991–2001. doi: 10.1099/MIC.0.28823-0
Heng, N. C. K., Wescombe, P. A., Burton, J. P., Jack, R. W., and Tagg, J. R. (2007c). “The diversity of bacteriocins in Gram-positive bacteria,” in Bacteriocins, eds M. A. Riley and M. A. Chavan (Berlin: Springer), 45–92. doi: 10.1007/978-3-540-36604-1_4
Heng, N. C. K., Tagg, J. R., and Tompkins, G. R. (2007b). Competence-dependent bacteriocin production by Streptococcus gordonii DL1 (Challis). J. Bacteriol. 189:1468. doi: 10.1128/JB.01174-06
Heng, N. C. K., Burtenshaw, G. A., Jack, R. W., and Tagg, J. R. (2007a). Ubericin A, a class IIa bacteriocin produced by Streptococcus uberis. Appl. Environ. Microbiol. 73, 7763–7766. doi: 10.1128/AEM.01818-07
Hernández-González, J. C., Martínez-Tapia, A., Lazcano-Hernández, G., García-Pérez, B. E., and Castrejón-Jiménez, N. S. (2021). Bacteriocins from lactic acid bacteria. A powerful alternative as antimicrobials, probiotics, and immunomodulators in veterinary medicine. Animals (Basel) 11:979. doi: 10.3390/ANI11040979
Hill, C. (2021). Microbiome and infection: A case for “selective depletion”. Ann. Nutr. Metab. 77, 4–9. doi: 10.1159/000516399
Hill, C., Guarner, F., Reid, G., Gibson, G. R., Merenstein, D. J., Pot, B., et al. (2014). Expert consensus document: The international scientific association for probiotics and prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 11, 506–514. doi: 10.1038/nrgastro.2014.66
Hoare, A., Marsh, P. D., and Diaz, P. I. (2017). Ecological therapeutic opportunities for oral diseases. Microbiol. Spectr. 5, 1–24. doi: 10.1128/MICROBIOLSPEC.BAD-0006-2016
Hols, P., Ledesma-García, L., Gabant, P., and Mignolet, J. (2019). Mobilization of microbiota commensals and their bacteriocins for therapeutics. Trends Microbiol. 27, 690–702. doi: 10.1016/j.tim.2019.03.007
Horz, H. P., Meinelt, A., Houben, B., and Conrads, G. (2007). Distribution and persistence of probiotic Streptococcus salivarius K12 in the human oral cavity as determined by real-time quantitative polymerase chain reaction. Oral Microbiol. Immunol. 22, 126–130. doi: 10.1111/j.1399-302X.2007.00334.x
How, Y. H., and Yeo, S. K. (2021). Oral probiotic and its delivery carriers to improve oral health: A review. Microbiology (Reading) 167. doi: 10.1099/MIC.0.001076
Huan, Y., Kong, Q., Mou, H., and Yi, H. (2020). Antimicrobial peptides: Classification, design, application and research progress in multiple fields. Front. Microbiol. 11:2559. doi: 10.3389/FMICB.2020.582779/BIBTEX
Hyink, O., Balakrishnan, M., and Tagg, J. R. (2005). Streptococcus rattus strain BHT produces both a class I two-component lantibiotic and a class II bacteriocin. FEMS Microbiol. Lett. 252, 235–241. doi: 10.1016/J.FEMSLE.2005.09.003
Hyink, O., Wescombe, P. A., Upton, M., Ragland, N., Burton, J. P., and Tagg, J. R. (2007). Salivaricin A2 and the novel lantibiotic salivaricin B are encoded at adjacent loci on a 190-kilobase transmissible megaplasmid in the oral probiotic strain Streptococcus salivarius K12. Appl. Environ. Microbiol. 73, 1107–1113. doi: 10.1128/AEM.02265-06
Igarashi, T., Yano, Y., Yamamoto, A., Sasa, R., and Goto, N. (2001). Identification of Streptococcus salivarius by PCR and DNA probe. Lett. Appl. Microbiol. 32, 394–397. doi: 10.1046/J.1472-765X.2001.00928.X
Ishijima, S. A., Hayama, K., Burton, J. P., Reid, G., Okada, M., Matsushita, Y., et al. (2012). Effect of Streptococcus salivarius K12 on the In vitro growth of Candida albicans and its protective effect in an oral candidiasis model. Appl. Environ. Microbiol. 78, 2190–2199. doi: 10.1128/AEM.07055-11
Jack, R. W., Tagg, J. R., and Ray, B. (1995). Bacteriocins of gram-positive bacteria. Microbiol. Rev. 59, 171–200. doi: 10.1128/MR.59.2.171-200.1995
Jack, R., Benz, R., Tagg, J., Sahl, H.-G., and Jack, R. W. (1994). The mode of action of SA-FF22, a lantibiotic isolated from Streptococcus pyogenes strain FF22. Eur. J. Biochem. 219, 699–705. doi: 10.1111/j.1432-1033.1994.tb19986.x
Jacob, F., Lwoff, A., Siminovitch, A., and Wollman, E. (1953). [Definition of some terms relative to lysogeny]. Ann. Inst. Pasteur (Paris) 84, 222–224.
James, S. M., and Tagg, J. R. (1988). A search within the Genera Streptococcus, Enterococcus and Lactobacillus for organisms inhibitory to mutans streptococci. Microb. Ecol. Health Dis. 1, 153–162. doi: 10.3109/08910608809141531
Javan, R. R., van Tonder, A. J., King, J. P., Harrold, C. L., and Brueggemann, A. B. (2018). Genome sequencing reveals a large and diverse repertoire of antimicrobial peptides. Front. Microbiol. 9:2012. doi: 10.3389/FMICB.2018.02012
Johnson, D. W., Tagg, J. R., and Wannamaker, L. W. (1979). Production of a bacteriocine-like substance by group-A streptococci of M-type 4 and T-pattern 4. J. Med. Microbiol. 12, 413–427. doi: 10.1099/00222615-12-4-413
Johnson, E. M., Jung, D. Y. G., Jin, D. Y. Y., Jayabalan, D. R., Yang, D. S. H., and Suh, J. W. (2018). Bacteriocins as food preservatives: Challenges and emerging horizons. Crit. Rev. Food Sci. Nutr. 58, 2743–2767. doi: 10.1080/10408398.2017.1340870
Jones, P., Binns, D., Chang, H. Y., Fraser, M., Li, W., McAnulla, C., et al. (2014). InterProScan 5: Genome-scale protein function classification. Bioinformatics 30, 1236–1240. doi: 10.1093/BIOINFORMATICS/BTU031
Jörissen, J., van den Broek, M. F. L., de Boeck, I., van Beeck, W., Wittouck, S., Boudewyns, A., et al. (2021). Case-control microbiome study of chronic otitis media with effusion in children points at Streptococcus salivarius as a pathobiont-inhibiting species. mSystems 6, e00056–21. doi: 10.1128/MSYSTEMS.00056-21
Kaan, A. M., Kahharova, D., and Zaura, E. (2021). Acquisition and establishment of the oral microbiota. Periodontol 2000 86, 123–141. doi: 10.1111/PRD.12366
Kageyama, S., Furuta, M., Takeshita, T., Ma, J., Asakawa, M., and Yamashita, Y. (2022). High-level acquisition of maternal oral bacteria in formula-fed infant oral microbiota. mBio 13:e0345221. doi: 10.1128/mbio.03452-21
Karaçam, S., and Tunçer, S. (2022). Exploiting the acidic extracellular pH: Evaluation of Streptococcus salivarius M18 postbiotics to target cancer cells. Probiotics Antimicrob. Proteins 14:e0345221. doi: 10.1007/S12602-021-09806-3
Kassaa, I., Rafei, R., Moukhtar, M., Zaylaa, M., Gharsallaoui, A., Asehraou, A., et al. (2019). LABiocin database: A new database designed specifically for lactic acid bacteria bacteriocins. Int. J. Antimicrob. Agents 54, 771–779. doi: 10.1016/J.IJANTIMICAG.2019.07.012
Kaur, S., and Kaur, S. (2015). Bacteriocins as potential anticancer agents. Front. Pharmacol. 6:272. doi: 10.3389/FPHAR.2015.00272
Kazor, C. E., Mitchell, P. M., Lee, A. M., Stokes, L. N., Loesche, W. J., Dewhirst, F. E., et al. (2003). Diversity of bacterial populations on the tongue dorsa of patients with halitosis and healthy patients. J. Clin. Microbiol. 41, 558–563. doi: 10.1128/JCM.41.2.558-563.2003
Klaenhammer, T. R. (1988). Bacteriocins of lactic acid bacteria. Biochimie 70, 337–349. doi: 10.1016/0300-9084(88)90206-4
Klaenhammer, T. R. (1993). Genetics of bacteriocins produced by lactic acid bacteria. FEMS Microbiol. Rev. 12, 39–85. doi: 10.1111/J.1574-6976.1993.TB00012.X
Kolenbrander, P. E., Andersen, R. N., Blehert, D. S., Egland, P. G., Foster, J. S., and Palmer, R. J. Jr. (2002). Communication among oral bacteria. Microbiol. Mol. Biol. Rev. 66:486. doi: 10.1128/MMBR.66.3.486-505.2002
Lagedroste, M., Reiners, J., Knospe, C. V., Smits, S. H. J., and Schmitt, L. (2020). A structural view on the maturation of lanthipeptides. Front. Microbiol. 11:1183. doi: 10.3389/FMICB.2020.01183
Laws, G. A., Harold, L. K., Tagg, J. R., and Hale, J. D. F. (2022). Interferon gamma response in human saliva following exposure to the oral probiotic Streptococcus salivarius BLIS K12. Probiotics Antimicrob. Proteins. doi: 10.1007/S12602-022-10010-0 [Epub ahead of print].
Laws, G. L., Hale, J. D. F., and Kemp, R. A. (2021). Human systemic immune response to ingestion of the oral probiotic Streptococcus salivarius BLIS K12. Probiotics Antimicrob. Proteins 13, 1521–1529. doi: 10.1007/s12602-021-09822-3
Le Bars, P., Matamoros, S., Montassier, E., Le Vacon, F., Potel, G., Soueidan, A., et al. (2017). The oral cavity microbiota: Between health, oral disease, and cancers of the aerodigestive tract. Can. J. Microbiol. 63, 475–492. doi: 10.1139/CJM-2016-0603
Lederberg, J., and McCray, A. T. (2001). ’Ome sweet ’omics—a genealogical treasury of words. Scientist 15:8.
Li, J., Jin, J., Li, S., Zhong, Y., Jin, Y., Zhang, X., et al. (2022). Tonsillar microbiome-derived lantibiotics induce structural changes of IL-6 and IL-21 receptors and modulate host immunity. Adv. Sci. 9:2202706. doi: 10.1002/ADVS.202202706
Li, X., Liu, Y., Yang, X., Li, C., and Song, Z. (2022). The oral microbiota: Community composition, influencing factors, pathogenesis, and interventions. Front. Microbiol. 13:895537. doi: 10.3389/FMICB.2022.895537
Li, Y., and Rebuffat, S. (2020). The manifold roles of microbial ribosomal peptide–based natural products in physiology and ecology. J. Biol. Chem. 295:34. doi: 10.1074/JBC.REV119.006545
Liljemark, W. F., and Gibbons, R. J. (1973). Suppression of Candida albicans by human oral streptococci in gnotobiotic mice. Infect. Immun. 8, 846–849. doi: 10.1128/IAI.8.5.846-849.1973
Lin, D., Hutchison, K. E., Portillo, S., Vegara, V., Ellingson, J. M., Liu, J., et al. (2019). Association between the oral microbiome and brain resting state connectivity in smokers. Neuroimage 200, 121–131. doi: 10.1016/J.NEUROIMAGE.2019.06.023
MacDonald, K. W., Chanyi, R. M., Macklaim, J. M., Cadieux, P. A., Reid, G., and Burton, J. P. (2021). Streptococcus salivarius inhibits immune activation by periodontal disease pathogens. BMC Oral Health 21:245. doi: 10.1186/s12903-021-01606-z
Maitre, Y., Mahalli, R., Micheneau, P., Delpierre, A., Guerin, M., Amador, G., et al. (2021b). Pre and probiotics involved in the modulation of oral bacterial species: New therapeutic leads in mental disorders? Microorganisms 9:1450. doi: 10.3390/MICROORGANISMS9071450
Maitre, Y., Mahalli, R., Micheneau, P., Delpierre, A., Amador, G., and Denis, F. (2021a). Evidence and therapeutic perspectives in the relationship between the oral microbiome and Alzheimer’s disease: A systematic review. Int. J. Environ. Res. Public Health 18:11157. doi: 10.3390/IJERPH182111157
Małaczewska, J., Kaczorek-Łukowska, E., Wójcik, R., Rękawek, W., and Siwicki, A. K. (2019). In vitro immunomodulatory effect of nisin on porcine leucocytes. J. Anim. Physiol. Anim. Nutr. 103, 882–893. doi: 10.1111/JPN.13085
Manti, S., Parisi, G. F., Papale, M., Licari, A., Salpietro, C., Miraglia del Giudice, M., et al. (2020). Bacteriotherapy with Streptococcus salivarius 24SMB and Streptococcus oralis 89a nasal spray for treatment of upper respiratory tract infections in children: A pilot study on short-term efficacy. Ital. J. Pediatr. 46:42. doi: 10.1186/S13052-020-0798-4
Marchisio, P., Santagati, M., Scillato, M., Baggi, E., Fattizzo, M., Rosazza, C., et al. (2015). Streptococcus salivarius 24SMB administered by nasal spray for the prevention of acute otitis media in otitis-prone children. Eur. J. Clin. Microbiol. Infect. Dis. 34, 2377–2383. doi: 10.1007/S10096-015-2491-X
Mark Welch, J. L., Dewhirst, F. E., and Borisy, G. G. (2019). Biogeography of the oral microbiome: The site-specialist hypothesis. Annu. Rev. Microbiol. 73, 335–358. doi: 10.1146/ANNUREV-MICRO-090817-062503
Mathur, H., Field, D., Rea, M. C., Cotter, P. D., Hill, C., and Ross, R. P. (2018). Fighting biofilms with lantibiotics and other groups of bacteriocins. NPJ Biofilms Microbiomes 4:9. doi: 10.1038/s41522-018-0053-6
Mattick, A. T. R., Hirsch, A., and Berridge, N. J. (1947). Further observations on an inhibitory substance (nisin) from lactic streptococci. Lancet 2, 5–8. doi: 10.1016/S0140-6736(47)90004-4
McLean, A. R., Torres-Morales, J., Dewhirst, F. E., Borisy, G. G., and Mark Welch, J. L. (2022). Site-tropism of streptococci in the oral microbiome. Mol. Oral Microbiol. 37, 229–243. doi: 10.1111/OMI.12387
Meade, E., Slattery, M. A., and Garvey, M. (2020). Bacteriocins, potent antimicrobial peptides and the ‘ht against multi drug resistant species: Resistance is futile? Antibiotics 9:32. doi: 10.3390/ANTIBIOTICS9010032
Mignolet, J., Cerckei, G., Damoczi, J., Ledesma-Garcia, L., Sass, A., Coenye, T., et al. (2019). Subtle selectivity in a pheromone sensor triumvirate desynchronizes competence and predation in a human gut commensal. eLife 8:e47139. doi: 10.7554/ELIFE.47139
Mignolet, J., Fontaine, L., Sass, A., Nannan, C., Mahillon, J., Coenye, T., et al. (2018). Circuitry rewiring directly couples competence to predation in the gut dweller Streptococcus salivarius. Cell Rep. 22, 1627–1638. doi: 10.1016/j.celrep.2018.01.055
Narengaowa, Kong, W., Lan, F., Awan, U. F., Qing, H., and Ni, J. (2021). The oral-gut-brain AXIS: The influence of microbes in Alzheimer’s disease. Front. Cell. Neurosci. 15:113. doi: 10.3389/FNCEL.2021.633735/BIBTEX
Nes, I. F., Diep, D. B., and Holo, H. (2007). Bacteriocin diversity in Streptococcus and Enterococcus. J. Bacteriol. 189:1189. doi: 10.1128/JB.01254-06
Nguyen, T., Brody, H., Lin, G. H., Rangé, H., Kuraji, R., Ye, C., et al. (2020). Probiotics, including nisin-based probiotics, improve clinical and microbial outcomes relevant to oral and systemic diseases. Periodontol 2000 82, 173–185. doi: 10.1111/PRD.12324
Nguyen, T., Brody, H., Radaic, A., and Kapila, Y. (2021). Probiotics for periodontal health—current molecular findings. Periodontol 2000 87, 254–267. doi: 10.1111/prd.12382
O’Connor, P. M., Kuniyoshi, T. M., Oliveira, R. P., Hill, C., Ross, R. P., and Cotter, P. D. (2020). Antimicrobials for food and feed; a bacteriocin perspective. Curr. Opin. Biotechnol. 61, 160–167. doi: 10.1016/J.COPBIO.2019.12.023
Omardien, S., Brul, S., and Zaat, S. A. J. (2016). Antimicrobial activity of cationic antimicrobial peptides against gram-positives: Current progress made in understanding the mode of action and the response of bacteria. Front. Cell Dev. Biol. 4:111. doi: 10.3389/FCELL.2016.00111
Papadimitriou, K., Zoumpopoulou, G., Foligné, B., Alexandraki, V., Kazou, M., Pot, B., et al. (2015). Discovering probiotic microorganisms: In vitro, in vivo, genetic and OMICS approaches. Front. Microbiol. 6:58. doi: 10.3389/FMICB.2015.00058
Peng, X., Cheng, L., You, Y., Tang, C., Ren, B., Li, Y., et al. (2022). Oral microbiota in human systematic diseases. Int. J. Oral Sci. 14, 1–11. doi: 10.1038/s41368-022-00163-7
Petrova, P., Arsov, A., Tsvetanova, F., Parvanova-Mancheva, T., Vasileva, E., Tsigoriyna, L., et al. (2022). The complex role of lactic acid bacteria in food detoxification. Nutrients 14:2038. doi: 10.3390/NU14102038
Phelps, H. A., and Neely, M. N. (2007). SalY of the Streptococcus pyogenes lantibiotic locus is required for full virulence and intracellular survival in macrophages. Infect. Immun. 75:4541. doi: 10.1128/IAI.00518-07
Poorni, S., Nivedhitha, M., Srinivasan, M., and Balasubramaniam, A. (2022). Effect of probiotic Streptococcus salivarius K12 and M18 lozenges on the cariogram parameters of patients with high caries risk: A randomised control trial. Cureus 14:e23282. doi: 10.7759/cureus.23282
Price, L. B., and Shand, R. F. (2000). Halocin S8: A 36-amino-acid microhalocin from the haloarchaeal strain S8a. J. Bacteriol. 182, 4951–4958. doi: 10.1128/JB.182.17.4951-4958.2000
Radaic, A., and Kapila, Y. L. (2021). The oralome and its dysbiosis: New insights into oral microbiome-host interactions. Comput. Struct. Biotechnol. J. 19, 1335–1360. doi: 10.1016/J.CSBJ.2021.02.010
Radaic, A., de Jesus, M. B., and Kapila, Y. L. (2020). Bacterial anti-microbial peptides and nano-sized drug delivery systems: The state of the art toward improved bacteriocins. J. Control. Release 321, 100–118. doi: 10.1016/J.JCONREL.2020.02.001
Raheem, A., Liang, L., Zhang, G., and Cui, S. (2021). Modulatory effects of probiotics during pathogenic infections with emphasis on immune regulation. Front. Immunol. 12:616713. doi: 10.3389/FIMMU.2021.616713
Reid, G. (2005). The importance of guidelines in the development and application of probiotics. Curr. Pharm. Des. 11, 11–16. doi: 10.2174/1381612053382395
Reid, G., Gadir, A. A., and Dhir, R. (2019). Probiotics: Reiterating what they are and what they are not. Front. Microbiol. 10:424. doi: 10.3389/FMICB.2019.00424
Riley, M. A., and Wertz, J. E. (2002). Bacteriocins: Evolution, ecology, and application. Annu. Rev. Microbiol. 56, 117–137. doi: 10.1146/ANNUREV.MICRO.56.012302.161024
Rismayuddin, N. A. R., Mohd Badri, P. E. A., Ismail, A. F., Othman, N., Bandara, H. M. H. N., and Arzmi, M. H. (2022). Synbiotic Musa acuminata skin extract and Streptococcus salivarius K12 inhibit candida species biofilm formation. Biofouling 38, 614–627. doi: 10.1080/08927014.2022.2105142
Robson, C. L., Wescombe, P. A., Klesse, N. A., and Tagg, J. R. (2007). Isolation and partial characterization of the Streptococcus mutans type AII lantibiotic mutacin K8. Microbiology (Reading) 153, 1631–1641. doi: 10.1099/MIC.0.2006/003756-0
Rodrigues, G., Silva, G. G. O., Buccini, D. F., Duque, H. M., Dias, S. C., and Franco, O. L. (2019). Bacterial proteinaceous compounds with multiple activities toward cancers and microbial infection. Front. Microbiol. 10:1690. doi: 10.3389/FMICB.2019.01690/BIBTEX
Rogers, L. A., and Whittier, E. O. (1928). Limiting factors in the lactic fermentation. J. Bacteriol. 16, 211–229. doi: 10.1128/JB.16.4.211-229.1928
Roos, K., Holm, S. E., Grahn, E., and Lind, L. (1993). Alpha-streptococci as supplementary treatment of recurrent streptococcal tonsillitis: A randomized placebo-controlled study. Scand. J. Infect. Dis. 25, 31–35. doi: 10.1080/00365549309169666
Roos, K., Holm, S. E., Grahn-Håkansson, E., and Lagergren, L. (1996). Recolonization with selected alpha-streptococci for prophylaxis of recurrent streptococcal pharyngotonsillitis–a randomized placebo-controlled multicentre study. Scand. J. Infect. Dis. 28, 459–462. doi: 10.3109/00365549609037940
Rosier, B. T., Marsh, P. D., and Mira, A. (2017). Resilience of the oral microbiota in health: Mechanisms that prevent dysbiosis. J. Dent. Res. 97, 371–380. doi: 10.1177/0022034517742139
Ross, K. F., Ronson, C. W., and Tagg, J. R. (1993). Isolation and characterization of the lantibiotic salivaricin A and its structural gene salA from Streptococcus salivarius 20P3. Appl. Environ. Microbiol. 59, 2014–2021. doi: 10.1128/AEM.59.7.2014-2021.1993
Sadri, H., Aghaei, M., and Akbari, V. (2022). Nisin induces apoptosis in cervical cancer cells via reactive oxygen species generation and mitochondrial membrane potential changes. Biochem. Cell Biol. 100, 136–141. doi: 10.1139/BCB-2021-0225
Salminen, S., Collado, M. C., Endo, A., Hill, C., Lebeer, S., Quigley, E. M. M., et al. (2021b). The international scientific association of probiotics and prebiotics (ISAPP) consensus statement on the definition and scope of postbiotics. Nat. Rev. Gastroenterol. Hepatol. 18, 649–667. doi: 10.1038/s41575-021-00440-6
Salminen, S., Collado, M. C., Endo, A., Hill, C., Lebeer, S., Quigley, E. M. M., et al. (2021a). Reply to: Postbiotics—when simplification fails to clarify. Nat. Rev. Gastroenterol. Hepatol. 18, 827–828. doi: 10.1038/S41575-021-00522-5
Sanders, C. C., and Sanders, W. E. (1982). Enocin: An antibiotic produced by Streptococcus salivarius that may contribute to protection against infections due to group A streptococci. J. Infect. Dis. 146, 683–690. doi: 10.1093/INFDIS/146.5.683
Sanders, C. C., Nelson, G. E., and Sanders, W. E. (1977). Bacterial interference. IV. Epidemiological determinants of the antagonistic activity of the normal throat flora against group A streptococci. Infect. Immun. 16, 599–603. doi: 10.1128/IAI.16.2.599-603.1977
Sandiford, S. K. (2017). Genome database mining for the discovery of novel lantibiotics. Expert Opin. Drug Discov. 12, 489–495. doi: 10.1080/17460441.2017.1303475
Sankarapandian, V., Maran, B. A. V., Rajendran, R. L., Jogalekar, M. P., Gurunagarajan, S., Krishnamoorthy, R., et al. (2022). An update on the effectiveness of probiotics in the prevention and treatment of cancer. Life 12:59. doi: 10.3390/LIFE12010059
Scariya, L., Nagarathna, D. V., and Varghese, M. (2015). Probiotics in periodontal therapy. Int. J. Pharma Bio Sci. 6, 242–250. doi: 10.5455/musbed.20141106034910
Sedgley, C. M., Lee, E. H., Martin, M. J., and Flannagan, S. E. (2008). Antibiotic resistance gene transfer between Streptococcus gordonii and Enterococcus faecalis in root canals of teeth ex vivo. J. Endod. 34, 570–574. doi: 10.1016/j.joen.2008.02.014
Siciliano, R. A., Reale, A., Mazzeo, M. F., Morandi, S., Silvetti, T., and Brasca, M. (2021). Paraprobiotics: A new perspective for functional foods and nutraceuticals. Nutrients 13:1225. doi: 10.3390/nu13041225
Sidjabat, H. E., Håkansson, E. G., and Cervin, A. (2016). Draft genome sequence of the oral commensal Streptococcus oralis 89a with interference activity against respiratory pathogens. Genome Announc. 4, e01546–15. doi: 10.1128/GENOMEA.01546-15
Simmonds, R. S., Simpson, W. J., and Tagg, J. R. (1997). Cloning and sequence analysis of zooA, a Streptococcus zooepidemicus gene encoding a bacteriocin-like inhibitory substance having a domain structure similar to that of lysostaphin. Gene 189, 255–261. doi: 10.1016/S0378-1119(96)00859-1
Simons, A., Alhanout, K., and Duval, R. E. (2020). Bacteriocins, antimicrobial peptides from bacterial origin: Overview of their biology and their impact against multidrug-resistant bacteria. Microorganisms 8:639. doi: 10.3390/microorganisms8050639
Simpson, W. J., Ragland, N. L., Ronson, C. W., and Tagg, J. R. (1995). A lantibiotic gene family widely distributed in Streptococcus salivarius and Streptococcus pyogenes. Dev. Biol. Stand. 85, 639–643.
Skilton, C. J., and Tagg, J. R. (1992). Production by Streptococcus sanguis of bacteriocin-like inhibitory substances (BLIS) with activity against Streptococcus rattus. Microb. Ecol. Health Dis. 5, 219–226. doi: 10.3109/08910609209141589
Sleator, R. (2010). The human superorganism—of microbes and men. Med. Hypotheses 74, 214–215. doi: 10.1016/J.MEHY.2009.08.047
Smith, H. W., and Huggins, M. B. (1976). Further observations on the association of the colicine V plasmid of Escherichia coli with pathogenicity and with survival in the alimentary tract. J. Gen. Microbiol. 92, 335–350. doi: 10.1099/00221287-92-2-335
Soltani, S., Hammami, R., Cotter, P., Rebuffat, S., Said, L., Gaudreau, H., et al. (2021). Bacteriocins as a new generation of antimicrobials: Toxicity aspects and regulations. FEMS Microbiol. Rev. 45:fuaa039. doi: 10.1093/FEMSRE/FUAA039
Sprunt, K., and Leidy, G. (1988). The use of bacterial interference to prevent infection. Can. J. Microbiol. 34, 332–338. doi: 10.1139/M88-061
Stašková, A., Sondorová, M., Nemcová, R., Kačírová, J., Mad’ar, M., and Karygianni, L. (2021). Antimicrobial and antibiofilm activity of the probiotic strain Streptococcus salivarius K12 against oral potential pathogens. Antibiotics 10:793. doi: 10.3390/antibiotics10070793
Svetoch, E. A., and Stern, N. J. (2010). Bacteriocins to control Campylobacter spp. In poultry–A review. Poult. Sci. 89, 1763–1768. doi: 10.3382/PS.2010-00659
Swanson, K. S., Gibson, G. R., Hutkins, R., Reimer, R. A., Reid, G., Verbeke, K., et al. (2020). The international scientific association for probiotics and prebiotics (ISAPP) consensus statement on the definition and scope of synbiotics. Nat. Rev. Gastroenterol. Hepatol. 17, 687–701. doi: 10.1038/s41575-020-0344-2
Swe, P. M., Heng, N. C. K., Ting, Y. T., Baird, H. J., Carne, A., Tauch, A., et al. (2007). ef1097 and ypkK encode enterococcin V583 and corynicin JK, members of a new family of antimicrobial proteins (bacteriocins) with modular structure from Gram-positive bacteria. Microbiology (Reading) 153, 3218–3227. doi: 10.1099/MIC.0.2007/010777-0
Tagg, J. (2004). Prevention of streptococcal pharyngitis by anti-Streptococcus pyogenes bacteriocin-like inhibitory substances (BLIS) produced by Streptococcus salivarius. Indian J. Microbiol. 119:13.
Tagg, J. R. (1992). “Bacteriocins of gram-positive bacteria: An opinion regarding their nature, nomenclature and numbers,” in Bacteriocins, microcins and lantibiotics, eds R. James, C. Lazdunski, and F. Pattus (Berlin: Springer), 33–35. doi: 10.1007/978-3-642-76974-0_5
Tagg, J. R. (2009). Streptococcal bacteriocin-llke inhibitory substances: Some personal insights into the bacteriocin-llke activities produced by streptococci good and bad. Probiotics Antimicrob. Proteins 1, 60–66. doi: 10.1007/s12602-008-9002-7
Tagg, J. R. (2016). Developing a bacteriotherapy-based approach to control streptococcal infections. Nutrafoods 15, 11–20.
Tagg, J. R., and Bannister, L. V. (1979). “Fingerprinting” β-haemolytic streptococci by their production of and sensitivity to bacteriocine-like inhibitors. J. Med. Microbiol. 12, 397–411. doi: 10.1099/00222615-12-4-397
Tagg, J. R., and Mushin, R. (1971). Epidemiology of Pseudomonas aeruginosa infection in hospitals. 1. Pyocine typing of Ps. aeruginosa. Med. J. Aust. 1, 847–852. doi: 10.5694/J.1326-5377.1971.TB87891.X
Tagg, J. R., and Russell, C. (1981). Bacteriocin production by Streptococcus salivarius strain P. Can. J. Microbiol. 27, 918–923. doi: 10.1139/M81-144
Tagg, J. R., and Wannamaker, L. W. (1978). Streptococcin A-FF22: Nisin-like antibiotic substance produced by a group A Streptococcus. Antimicrob. Agents Chemother. 14, 31–39. doi: 10.1128/AAC.14.1.31
Tagg, J. R., Pybus, V., Phillips, L. V., and Fiddes, T. M. (1983). Application of inhibitor typing in a study of the transmission and retention in the human mouth of the bacterium Streptococcus salivarius. Arch. Oral Biol. 28, 911–915. doi: 10.1016/0003-9969(83)90086-9
Tagg, J. R., Ragland, N. L., and Dickson, N. P. (1990). A longitudinal study of lancefield group a Streptococcus acquisitions by a group of young Dunedin schoolchildren. N. Z. Med. J. 103, 429–431.
Tagg, J., and Dierksen, K. (2003). Bacterial replacement therapy: Adapting “germ warfare” to infection prevention. Trends Biotechnol. 21, 217–223. doi: 10.1016/S0167-7799(03)00085-4
Tagg, J., Dajani, A., and Wannamaker, L. (1976). Bacteriocins of gram-positive bacteria. Bacteriol. Rev. 40, 722–756. doi: 10.1128/BR.40.3.722-756.1976
Tandelilin, R., Widita, E., Agustina, D., and Saini, R. (2018). The effect of oral probiotic consumption on the caries risk factors among high-risk caries population. J. Int. Oral Health 10:132. doi: 10.4103/jioh.jioh_82_18
Tanzer, J. M., Kurasz, A. B., and Clive, J. (1985a). Competitive displacement of mutans streptococci and inhibition of tooth decay by Streptococcus salivarius TOVE-R. Infect. Immun. 48, 44–50. doi: 10.1128/iai.48.1.44-50.1985
Tanzer, J. M., Kurasz, A. B., and Clive, J. (1985b). Inhibition of ecological emergence of mutans streptococci naturally transmitted between rats and consequent caries inhibition by Streptococcus salivarius TOVE-R infection. Infect. Immun. 49, 76–83. doi: 10.1128/IAI.49.1.76-83.1985
Taverniti, V., and Guglielmetti, S. (2011). The immunomodulatory properties of probiotic microorganisms beyond their viability (ghost probiotics: Proposal of paraprobiotic concept). Genes Nutr. 6:261. doi: 10.1007/S12263-011-0218-X
Teughels, W., van Essche, M., Sliepen, I., and Quirynen, M. (2008). Probiotics and oral healthcare. Periodontol 2000 48, 111–147. doi: 10.1111/J.1600-0757.2008.00254.X
Thomas, C., Minty, M., Vinel, A., Canceill, T., Loubières, P., Burcelin, R., et al. (2021). Oral microbiota: A major player in the diagnosis of systemic diseases. Diagnostics 11:1376. doi: 10.3390/DIAGNOSTICS11081376
Tiwari, S. K., Dicks, L. M. T., Popov, I. V., Karaseva, A., Ermakov, A., Suvorov, A., et al. (2020). Probiotics at war against viruses: What is missing from the picture? Front. Microbiol. 11:1877. doi: 10.3389/FMICB.2020.01877
Todorov, S. D., Tagg, J. R., and Ivanova, I. V. (2021). Could probiotics and postbiotics function as “silver bullet” in the post-COVID-19 era? Probiotics Antimicrob. Proteins 13, 1499–1507. doi: 10.1007/S12602-021-09833-0
Tompkins, G. R., and Tagg, J. R. (1989). The ecology of bacteriocin-producing strains of Streptococcus salivarius. Microb. Ecol. Health Dis. 2, 19–28. doi: 10.3109/08910608909140197
Tunçer, S., and Karaçam, S. (2020). Cell-free supernatant of Streptococcus salivarius M18 impairs the pathogenic properties of Pseudomonas aeruginosa and Klebsiella pneumonia. Arch. Microbiol. 202, 2825–2840. doi: 10.1007/S00203-020-02005-8
Turnbaugh, P. J., Ley, R. E., Hamady, M., Fraser-Liggett, C. M., Knight, R., and Gordon, J. I. (2007). The human microbiome project. Nature 449, 804–810. doi: 10.1038/NATURE06244
Twomey, E., Hill, C., Field, D., and Begley, M. (2021). Recipe for success: Suggestions and recommendations for the isolation and characterisation of bacteriocins. Int. J. Microbiol. 2021:9990635. doi: 10.1155/2021/9990635
Upton, M., Cotter, P., and Tagg, J. R. (2012). Antimicrobial peptides as therapeutic agents. In. J. Microbiol. 2012:326503. doi: 10.1155/2012/326503
Upton, M., Tagg, J. R., Wescombe, P., and Jenkinson, H. F. (2001). Intra- and interspecies signaling between Streptococcus salivarius and Streptococcus pyogenes mediated by SalA and SalA1 lantibiotic peptides. J. Bacteriol. 183, 3931–3938. doi: 10.1128/JB.183.13.3931-3938.2001
van den Nieuwboer, M., and Claassen, E. (2019). Dealing with the remaining controversies of probiotic safety. Benef. Microbes 10, 605–616. doi: 10.3920/BM2018.0159
van Heel, A. J., de Jong, A., Song, C., Viel, J. H., Kok, J., and Kuipers, O. P. (2018). BAGEL4: A user-friendly web server to thoroughly mine RiPPs and bacteriocins. Nucleic Acids Res. 46, W278–W281. doi: 10.1093/NAR/GKY383
van Staden, A. D. P., van Zyl, W. F., Trindade, M., Dicks, L. M. T., and Smith, C. (2021). Therapeutic application of lantibiotics and other lanthipeptides: Old and new findings. Appl. Environ. Microbiol. 87:e0018621. doi: 10.1128/AEM.00186-21
Vertillo Aluisio, G., Spitale, A., Bonifacio, L., Privitera, G. F., Stivala, A., Stefani, S., et al. (2022). Streptococcus salivarius 24SMBc genome analysis reveals new biosynthetic gene clusters involved in antimicrobial effects on Streptococcus pneumoniae and Streptococcus pyogenes. Microorganisms 10:2042. doi: 10.3390/microorganisms10102042
Vinderola, G., Sanders, M. E., and Salminen, S. (2022). The concept of postbiotics. Foods 11:1077. doi: 10.3390/FOODS11081077
Vogel, V., and Spellerberg, B. (2021). Bacteriocin production by beta-hemolytic streptococci. Pathogens 10:867. doi: 10.3390/PATHOGENS10070867
Walker, G. V., Heng, N. C. K., Carne, A., Tagg, J. R., and Wescombe, P. A. (2016). Salivaricin E and abundant dextranase activity may contribute to the anti-cariogenic potential of the probiotic candidate Streptococcus salivarius JH. Microbiology (Reading) 162, 476–486. doi: 10.1099/MIC.0.000237
Wang, G., Li, X., and Wang, Z. (2016). APD3: The antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 44, D1087–D1093. doi: 10.1093/NAR/GKV1278
Wang, G., Zietz, C. M., Mudgapalli, A., Wang, S., and Wang, Z. (2022). The evolution of the antimicrobial peptide database over 18 years: Milestones and new features. Protein Sci. 31, 92–106. doi: 10.1002/PRO.4185
Wescombe, P. A., and Tagg, J. R. (2003). Purification and characterization of streptin, a type A1 lantibiotic produced by Streptococcus pyogenes. Appl. Environ. Microbiol. 69, 2737–2747. doi: 10.1128/AEM.69.5.2737-2747.2003
Wescombe, P. A., Upton, M., Dierksen, K. P., Ragland, N. L., Sivabalan, S., Wirawan, R. E., et al. (2006b). Production of the lantibiotic salivaricin A and its variants by oral streptococci and use of a specific induction assay to detect their presence in human saliva. Appl. Environ. Microbiol. 72, 1459–1466. doi: 10.1128/AEM.72.2.1459-1466.2006
Wescombe, P. A., Burton, J. P., Cadieux, P. A., Klesse, N. A., Hyink, O., Heng, N. C. K., et al. (2006a). Megaplasmids encode differing combinations of lantibiotics in Streptococcus salivarius. Antonie van Leeuwenhoek 90, 269–280. doi: 10.1007/s10482-006-9081-y
Wescombe, P. A., Dyet, K. H., Dierksen, K. P., Power, D. A., Jack, R. W., Burton, J. P., et al. (2012a). Salivaricin G32, a homolog of the prototype Streptococcus pyogenes Nisin-Like lantibiotic SA-FF22, produced by the commensal species Streptococcus salivarius. Int. J. Microbiol. 2012:738503. doi: 10.1155/2012/738503
Wescombe, P. A., Hale, J. D., Heng, N. C., and Tagg, J. R. (2012b). Developing oral probiotics from Streptococcus salivarius. Future Microbiol. 7, 1355–1371. doi: 10.2217/fmb.12.113
Wescombe, P. A., Heng, N. C. K., Burton, J. P., and Tagg, J. R. (2010). Something old and something new: An update on the amazing repertoire of bacteriocins produced by Streptococcus salivarius. Probiotics Antimicrob. Proteins 2, 37–45. doi: 10.1007/s12602-009-9026-7
Wescombe, P. A., Heng, N. C. K., Burton, J. P., Chilcott, C. N., and Tagg, J. R. (2009). Streptococcal bacteriocins and the case for Streptococcus salivarius as model oral probiotics. Future Microbiol. 4, 819–835. doi: 10.2217/fmb.09.61
Wescombe, P. A., Upton, M., Renault, P., Wirawan, R. E., Power, D., Burton, J. P., et al. (2011). Salivaricin 9, a new lantibiotic produced by Streptococcus salivarius. Microbiology 157, 1290–1299. doi: 10.1099/mic.0.044719-0
Whitehead, H. R. (1933). A substance inhibiting bacterial growth, produced by certain strains of lactic streptococci. Biochem. J. 27, 1793–1800. doi: 10.1042/BJ0271793
Wirawan, R. E., Klesse, N. A., Jack, R. W., and Tagg, J. R. (2006). Molecular and genetic characterization of a novel nisin variant produced by Streptococcus uberis. Appl. Environ. Microbiol. 72, 1148–1156. doi: 10.1128/AEM.72.2.1148-1156.2006
Wirawan, R. E., Swanson, K. M., Kleffmann, T., Jack, R. W., and Tagg, J. R. (2007). Uberolysin: A novel cyclic bacteriocin produced by Streptococcus uberis. Microbiology (Reading) 153, 1619–1630. doi: 10.1099/MIC.0.2006/005967-0
Xiao, J., Fiscella, K. A., and Gill, S. R. (2020). Oral microbiome: Possible harbinger for children’s health. Int. J. Oral Sci. 12:12. doi: 10.1038/S41368-020-0082-X
Xu, X., Xiao, J., and Niu, Y. (2022). Editorial: The pivotal role of oral microbiota dysbiosis and microbiota-host interactions in diseases. Front. Cell. Infect. Microbiol. 12:947638. doi: 10.3389/FCIMB.2022.947638
Yoo, H. J., Jwa, S. K., Kim, D. H., and Ji, Y. J. (2020). Inhibitory effect of Streptococcus salivarius K12 and M18 on halitosis in vitro. Clin. Exp. Dent. Res. 6, 207–214. doi: 10.1002/cre2.269
Zahradnik, R. T., Magnusson, I., Walker, C., McDonell, E., Hillman, C. H., and Hillman, J. D. (2009). Preliminary assessment of safety and effectiveness in humans of ProBiora3TM, a probiotic mouthwash. J. Appl. Microbiol. 107, 682–690. doi: 10.1111/J.1365-2672.2009.04243.X
Zimina, M., Babich, O., Prosekov, A., Sukhikh, S., Ivanova, S., Shevchenko, M., et al. (2020). Overview of global trends in classification, methods of preparation and application of bacteriocins. Antibiotics (Basel) 9, 1–21. doi: 10.3390/ANTIBIOTICS9090553
Keywords: probiotics, BLIS, bacteriocins, oral cavity, Streptococcus salivarius, oral health, dental caries, halitosis
Citation: Tagg JR, Harold LK, Jain R and Hale JDF (2023) Beneficial modulation of human health in the oral cavity and beyond using bacteriocin-like inhibitory substance-producing streptococcal probiotics. Front. Microbiol. 14:1161155. doi: 10.3389/fmicb.2023.1161155
Received: 08 February 2023; Accepted: 03 March 2023;
Published: 28 March 2023.
Edited by:
Harsh Mathur, Teagasc Food Research Centre, IrelandReviewed by:
Koshy Philip, University of Malaya, MalaysiaGabriele Bierbaum, University Hospital Bonn, Germany
Santosh Kumar Tiwari, Maharshi Dayanand University, India
Copyright © 2023 Tagg, Harold, Jain and Hale. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: John R. Tagg, am9obi50YWdnQGJsaXMuY28ubno=
†ORCID: John R. Tagg, orcid.org/0000-0001-9477-6579; Liam K. Harold, orcid.org/0000-0002-7570-7348; Rohit Jain, orcid.org/0000-0002-7621-588X; John D. F. Hale, orcid.org/0000-0003-1187-6000
 John R. Tagg
John R. Tagg Liam K. Harold
Liam K. Harold Rohit Jain
Rohit Jain John D. F. Hale
John D. F. Hale