
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 24 April 2023
Sec. Infectious Agents and Disease
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1160073
 Esra Ekinci1*†
Esra Ekinci1*† Laura Willen1†
Laura Willen1† Juan Pablo Rodriguez Ruiz2
Juan Pablo Rodriguez Ruiz2 Kirsten Maertens1
Kirsten Maertens1 Liesbet Van Heirstraeten2
Liesbet Van Heirstraeten2 Gabriela Serrano3
Gabriela Serrano3 Magali Wautier3
Magali Wautier3 Ariane Deplano3
Ariane Deplano3 Herman Goossens2
Herman Goossens2 Pierre Van Damme1
Pierre Van Damme1 Philippe Beutels4
Philippe Beutels4 Surbhi Malhotra-Kumar2
Surbhi Malhotra-Kumar2 Delphine Martiny3
Delphine Martiny3 Heidi Theeten1
Heidi Theeten1Background: Non-typeable Haemophilus influenzae has become increasingly important as a causative agent of invasive diseases following vaccination against H. influenzae type b. The emergence of antibiotic resistance underscores the necessity to investigate typeable non-b carriage and non-typeable H. influenzae (NTHi) in children.
Methods: Nasopharyngeal swab samples were taken over a three-year period (2016–2018) from 336 children (6–30 months of age) attending daycare centers (DCCs) in Belgium, and from 218 children with acute otitis media (AOM). Biotype, serotype, and antibiotic resistance of H. influenzae strains were determined phenotypically. Mutations in the ftsI gene were explored in 129 strains that were resistant or had reduced susceptibility to beta-lactam antibiotics. Results were compared with data obtained during overlapping time periods from 94 children experiencing invasive disease.
Results: Overall, NTHi was most frequently present in both carriage (DCC, AOM) and invasive group. This was followed by serotype “f” (2.2%) and “e” (1.4%) in carriage, and “b” (16.0%), “f” (11.7%), and “a” (4.3%) in invasive strains. Biotype II was most prevalent in all studied groups, followed by biotype III in carriage and I in invasive strains. Strains from both groups showed highest resistance to ampicillin (26.7% in carriage vs. 18.1% in invasive group). A higher frequency of ftsI mutations were found in the AOM group than the DCC group (21.6 vs. 14.9% – p = 0.056). Even more so, the proportion of biotype III strains that carried a ftsI mutation was higher in AOM compared to DCC (50.0 vs. 26.3% – p < 0.01) and invasive group.
Conclusion: In both groups, NTHi was most frequently circulating, while specific encapsulated serotypes for carriage and invasive group were found. Biotypes I, II and III were more frequently present in the carriage and invasive group. The carriage group had a higher resistance-frequency to the analyzed antibiotics than the invasive group. Interestingly, a higher degree of ftsI mutations was found in children with AOM compared to DCC and invasive group. This data helps understanding the H. influenzae carriage in Belgian children, as such information is scarce.
The nasopharynx is a major reservoir for bacterial pathogens that can lead to upper respiratory tract infections in children and nasopharyngeal carriage of pathogens is an important source of transmission to other susceptible groups, mainly elderly (Aniansson et al., 1992). Haemophilus influenzae is one of the most important bacterial colonizers of the nasopharynx and is a frequent cause of acute otitis media (AOM) and sinusitis (Hashida et al., 2008; Ortiz-Romero et al., 2017). H. influenzae can exist without a capsule (non-typeable H. influenzae) or can be capsulated by six different polysaccharide capsules (i.e., serotypes a to f; Pittman, 1931), with the most virulent one being H. influenzae serotype b (Hib). Up until the introduction of the H. influenzae vaccine — that included the Hib capsule — into the national childhood vaccination programs in 1993 (Jacquinet et al., 2021), Hib was the most common cause of invasive H. influenzae disease in young children, including meningitis (Pittman, 1931; Peltola et al., 1977; Peltola, 2000). The routine administration of Hib vaccines led to a significant decline in invasive Hib disease in all age groups, through direct and indirect (herd immunity) protection (Swingler et al., 2007); but this in turn made room for non-typeable H. influenzae (NTHi) cases as well as non-b-serotype cases to increase (Madore, 1996; Agrawal and Murphy, 2011). In Belgium, most (76.8%) of the invasive H. influenzae strains were reported as non-typeables during the period 2017–2021 (Grammens et al., 2017, 2018; Jacquinet et al., 2021). However, typeable serotypes, such as serotype “f” (9.7%), serotype “b” (5.8%), serotype “a” (2.7%), serotype “e” (1.1%) and serotype “d” (0.4%), were also found to be the cause of invasive infections in children and elderly during the same period (Grammens et al., 2017, 2018; Jacquinet et al., 2021). These observations demonstrate the importance of conducting further research into the circulating serotypes of H. influenzae.
H. influenzae strains can acquire antibiotic resistance by two different mechanisms; either by producing beta-lactamase enzymes which hydrolyze beta-lactam antibiotics (Medeiros et al., 1986; McElligott et al., 2020) or by acquiring mutations in the genes encoding for penicillin-binding protein (PBP) protein (Ubukata et al., 2001, 2002). Mutations in the ftsI gene, which encodes PBP3, lowers the binding affinity of beta-lactam antibiotics to PBP3 (Mendelman et al., 1987); hence, the diversity of mutations in the ftsI gene lead to several profiles that can affect differently the beta-lactam antibiotics, making macrolides and quinolones more important alternative treatment strategies (Tanaka et al., 2019). Beta-lactamase-negative ampicillin resistant H. influenzae or BLNAR is one of the problems for the treatment of H. influenzae with beta-lactam antibiotics. Specific amino acid substitutions have been identified by sequencing of the ftsI gene, producing a BLNAR phenotype (Yalçın et al., 2022). BLNARS are divided into groups I, II, III and III-like defined by the amino acid mutation patterns at specific sites (Hotomi et al., 2007). Group III and III-like (referred as high-BLNAR) have usually a higher ampicillin MIC (higher level of ampicillin resistant) compared to group I and II (Han et al., 2019). High BLNARs are changing worldwide, but more rapidly in Asia than in Europe and USA (Ubukata et al., 2001; Hasegawa et al., 2003; García-Cobos et al., 2007; Barbosa et al., 2011; Skaare et al., 2014; Honda et al., 2018).
The existence of antibiotic resistance of H. influenzae underscores the necessity to investigate typeable non-b carriage and non-typeable H. influenzae in children. This study describes for the first time the carriage and antimicrobial susceptibility profile of H. influenzae strains in samples obtained from healthy Belgian children attending daycare centers (DCC) and children with acute otitis media (AOM), and compares it to Belgian children experiencing invasive disease caused by both non-typeable and encapsulated H. influenzae.
The study protocol, the informed consent forms, questionnaires and sample collection were approved by the ethics committee of University of Antwerp (UAntwerpen) and the Antwerp University Hospital (UZA; ID 15/45/471 and ID 18/31/355). Written informed consent was obtained from the infants’ parents or legal representatives.
Haemophilus influenzae carriage strains were collected from children sampled in the ongoing Belgian nasopharyngeal carriage study (Np carriage study) during the sample collection periods 2016–2018 (Wouters et al., 2018; Ekinci et al., 2022). Samples were either taken from healthy children attending randomly selected DCCs or from children presenting with AOM at a physician taking part in the study (convenience sample, yearly average of 17 centers geographically equally spread across Belgium). The sample collection period covered October to May/June for children with AOM and November to March for children in DCC (except for the baseline collection between March–June in 2016). Background characteristics (including demographics, antibiotic use, and clinical data on AOM if applicable) from the NP carriage study (Wouters et al., 2018; Ekinci et al., 2022). No data was present on Hib vaccination status or date for any of the included children.
A total number of 554 strains, collected between 2016 and 2018 for the NP carriage study, were analyzed. These strains include a stratified selection of strains from children attending DCC (n = 336) and all H. influenzae strains present from children suffering from AOM (n = 218), from the same collection period. To make an adequate comparison between samples taken from healthy children attending DCCs and AOM-children, samples were stratified on age, sex, vaccination status and AB treatment within the two groups were selected. The selected children aged between 6 to 30 months.
Microbiological results for all invasive strains (CSF, blood) isolated in Belgian children under five between 2015 and 2018 (referred to as the “invasive group”) were provided by the Belgian National Reference Centre (NRC) for Haemophilus influenzae including biotype, serotype, ftsI sequencing (when performed), AST to ampicillin, amoxicillin-clavulanic-acid, cefotaxime and ciprofloxacin.
Parents or legal representatives were asked to fill out a questionnaire before sampling. A single nasopharyngeal (NP) swab was taken with a flocked nylon swab and transported in 1 ml STGG (Skim milk – Tryptone – Glucose – Glycerol) at 2–8° C and cultured or stored at-80° C within 24 h.
Analyses were performed at the Belgian National Reference Center (NRC) for Haemophilus influenzae. Before testing, strains were subcultured twice onto chocolate agar (in-house preparation).
All isolates were identified by MALDI-TOF mass spectrometry using a Microflex LT spectrometer and the Biotyper 4.2.80 database (Bruker Daltonics, Bremen, Gerrmany). Biotypes and serotypes of H. influenzae strains were determined using biochemical tests based on the production of indole, urease and ornithine decarboxylase (Diatabs, Rosco Diagnostica, Albertslund, Denmark) and Difco Haemophilus antiserum agglutination kit (Becton Dickinson, Erembodegem, Belgium), respectively, and following manufacturer’s instructions.
Antimicrobial susceptibility testing were performed following EUCAST V9.0 guidelines and clinical breakpoints. A disk diffusion test with benzylpenicillin 1 unit disk and a beta-lactamase detection using BBL paper disks (Becton Dickinson) were performed in parallel with the susceptibility testing for ampicillin, amoxicillin-clavulanic acid and cefuroxime; ciprofloxacin was also tested. All minimum inhibitory concentrations (MIC) were determined by gradient strips antimicrobial susceptibility testing (E-test, bioMérieux, Marcy l’Etoile, France).
The transpeptidase domain of the ftsI gene was sequenced as described by Ubukata et al. (2001) for all strains showing lowered susceptibilities to beta-lactams (MIC ampicillin > = 1 μg/ml combined with a negative beta-lactamase test, MIC amoxicillin-clavulanic acid > = 2 μg/ml, resistance to cefuroxime). Briefly, a 621 bp fragment is amplified by PCR and sequenced. Sequences are compared to the “wild-type” reference isolate Rd. KW20 (ATCC ® 51907) sequence (NCBI L42023.1). The amino acids substitutions are detected and listed using the BioNumerics™ software 7.6.2 (bioMérieux). BLNAR strains were classified into three groups based on amino acid substitutions: in group I (substitution in His 517 to Arg-517), in group II (substitution Lys-526 to Asn-526), and group III (substitutions in Met-377 to Ile, Ser-385 to Thr, and Leu-389 to Phe; Ubukata et al., 2001).
Sample size was based on the number of H. influenzae strains obtained from children with AOM during the period 2016–2018 (n = 218). Similar numbers (n = 336) of H. influenzae strains obtained from children attending DCC during the period 2016–2018 were selected.
The Chi-Square (Chi2) Test was used to assess significant differences between different analyzed groups (α = 0.05). On serotype level Chi2 Test is performed on NTHi versus encapsulated H. influenzae. On biotype level Chi2 Test is performed on (in our study) frequently circulating biotypes (biotypes I, II, III) versus less frequently circulating biotypes (biotype IV, V, VI, VII).
The carriage group consisted of 554 samples divided in 336 samples obtained from healthy children attending DCC and 218 samples obtained from children suffering from AOM (a non-invasive disease). Children are defined as healthy if they either do not have a disease condition or they have a disease that is fully controlled which allows the child to attend the DCC. Results from the carriage groups are compared to the “invasive” group, consisting of 94 strains.
Non-typeable H. influenzae was most prevalent in both carriage (96.4%) and invasive strains (68.1%; Table 1). In the carriage group, also serotype “f” (2.2%) and serotype “e” (1.8%) were present, while in the invasive group, serotype “b” (16.0%), serotype “f” (11.7%), and serotype “a” (4.3%) were found (Table 1; Figure 1). The distribution of the different serotypes was similar between AOM and DCC children and was also similar across the different seasons (Supplementary Figure 1). The proportion of typeable serotypes was significantly higher in the invasive group compared to the carriage group (Table 1; Figure 1).
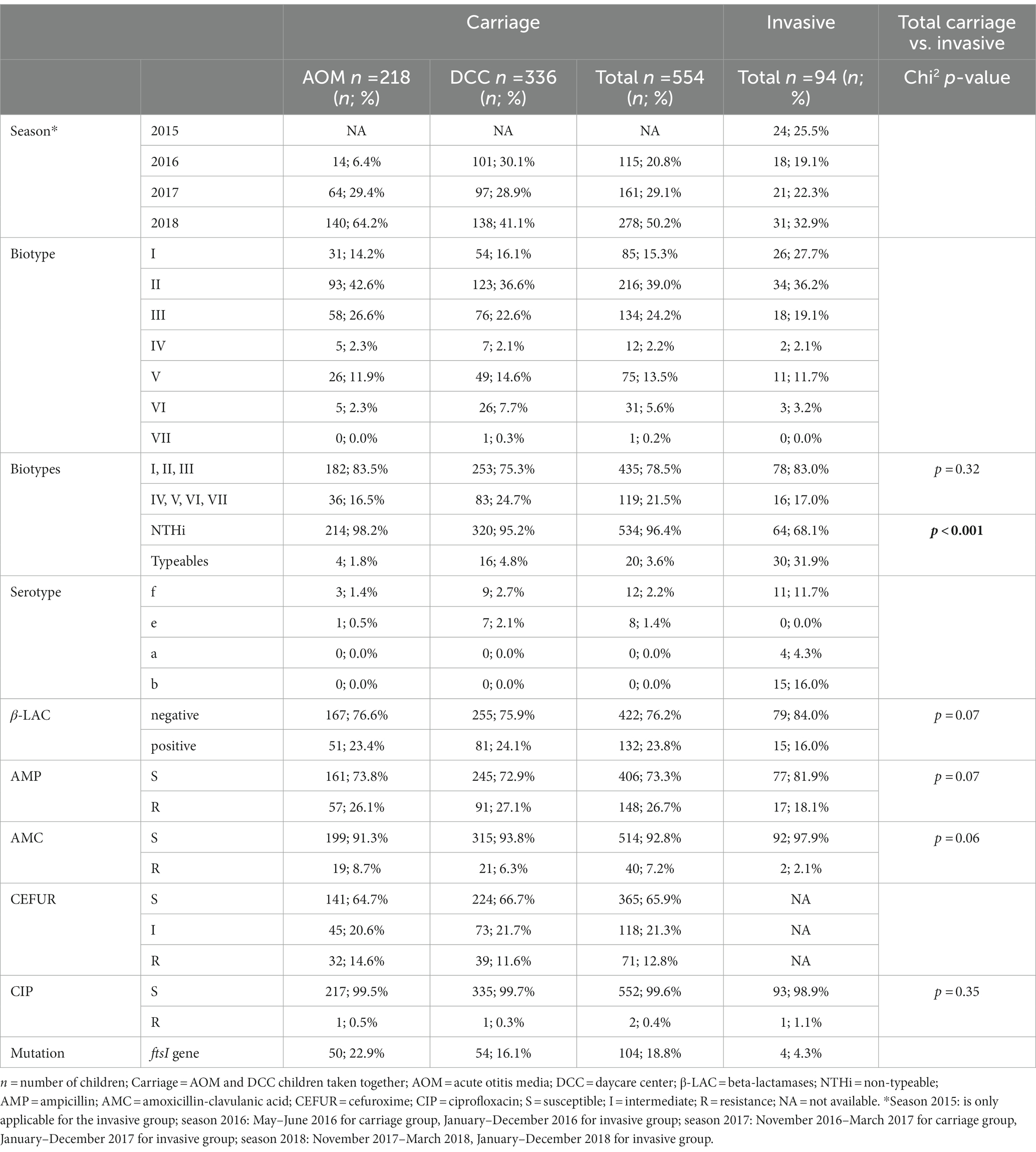
Table 1. Characteristics of Haemophilus influenzae strains in the carriage (AOM and DCC) and invasive group.
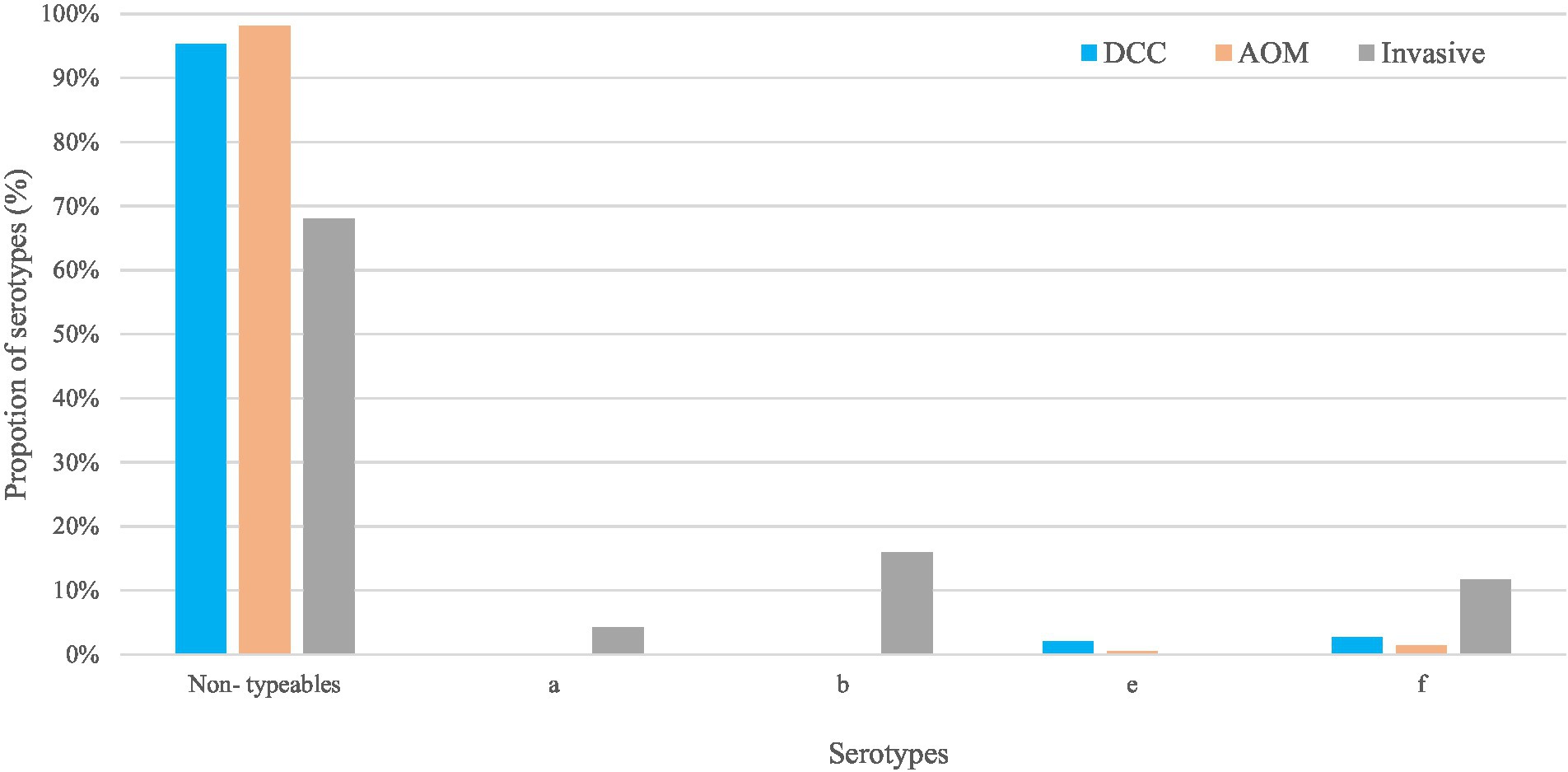
Figure 1. Non-typeable and typeable of Haemophilus influenzae in the carriage (AOM and DCC) and invasive group. Overall proportions of circulating serotypes in the carriage groups AOM (in orange) and DCC (in blue) as well as in the invasive group (in grey) detected in strains collected during the period 2016–2018 for the carriage group and 2015–2018 for the invasive group.
For the carriage group, the most frequent biotypes were biotype II (39.0%), biotype III (24.2%), and biotype I (15.3%; Table 1; Figure 2). The distribution of the different biotypes was similar in the DCC and AOM population of the carriage group (Table 1) and did not differ across the different seasons (Supplementary Figure 2). For the invasive group, biotype II (36.2%), biotype I (27.7%), and biotype III (19.1%) were more frequently present (Table 1; Figure 2).
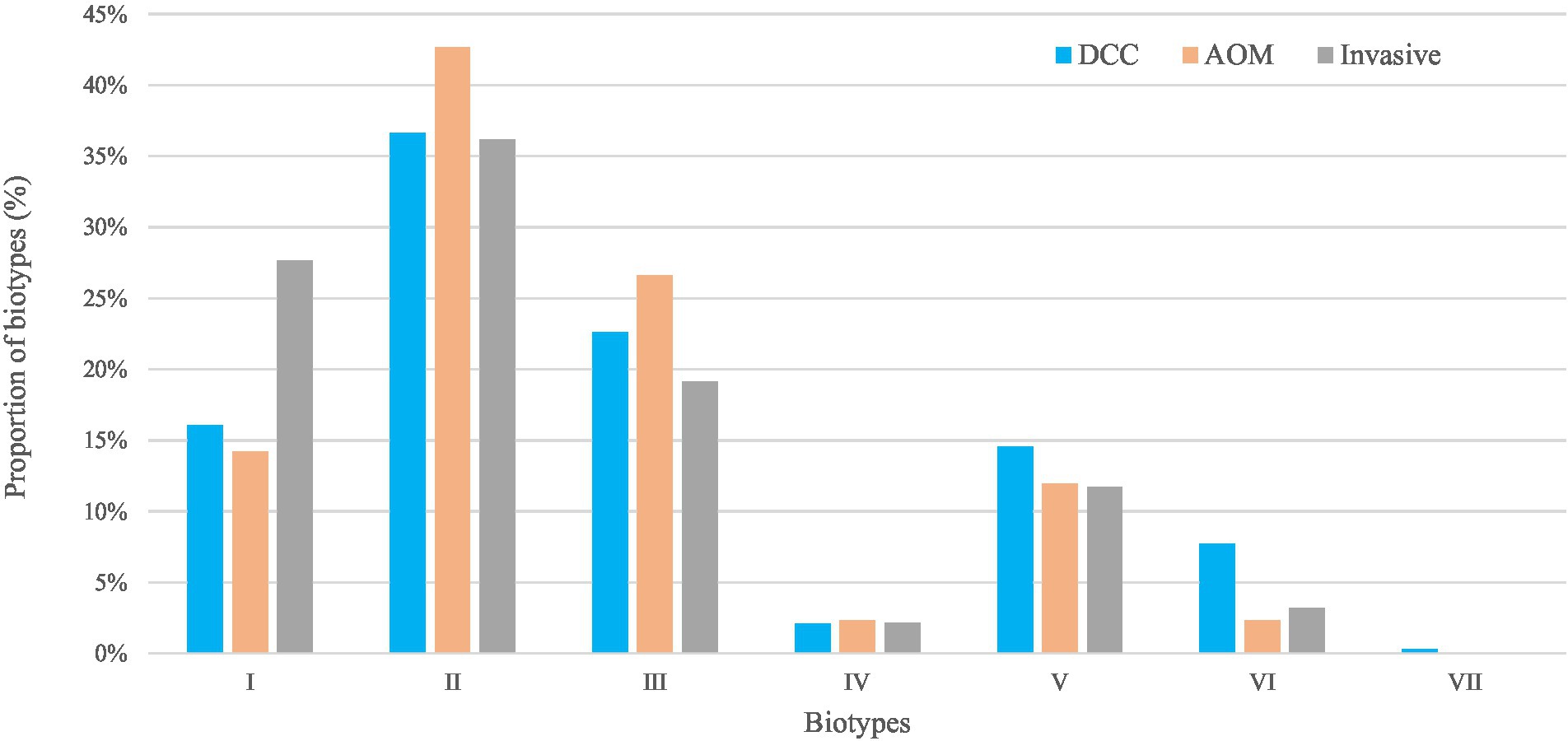
Figure 2. Biotypes of Haemophilus influenzae in the carriage (AOM and DCC) and invasive group. Overall proportions of circulating biotypes in the carriage groups AOM (in orange) and DCC (in blue) as well as in the invasive group (in grey) detected in strains collected during the period 2016–2018 for the carriage group and 2015–2018 for the invasive group.
H. influenzae strains from the carriage group showed resistance to ampicillin (26.7%), cefuroxime (12.8%), amoxicillin-clavulanic acid (7.2%), and ciprofloxacin (0.4%). No difference was observed in resistance of H. influenzae strains across the different seasons, nor between the AOM and DCC group. Also for the invasive group, resistance was found to ampicillin (18.1%), amoxicillin-clavulanic acid (2.1%), and ciprofloxacin (1.1%). No data was provided for cefuroxime in the invasive group (not systematically tested at that time by the NRC).
Beta-lactamases were present in 23.8 and 16.0% of the H. influenzae strains in the carriage and invasive group, respectively (Table 1). In the carriage (n = 132) as well as the invasive group (n = 15), all of the beta-lactamase positive strains showed resistance to ampicillin, while 3.0% (4/132) and 6.7% (1/15) of the strains were resistant to amoxicillin-clavulanic acid in the carriage and invasive group, respectively. Resistance to cefuroxime was found in 10.6% (14/132) of the beta-lactamase positive strains in the carriage group.
From the 554 samples from the carriage group, 129 resistant or “less” susceptible strains were reanalyzed and were checked on mutations in the ftsI gene. In 81.0% (104/129) of the strains, a mutation in the ftsI gene was found. No difference was observed in the incidence of the ftsI mutation over the different time periods. Of all ftsI mutated strains, 93.7% corresponded to the Ubukata-group 2 and a small proportion (3.1%) to the Ubukata-group 3. A higher frequency of ftsI mutations was found in the strains of children with AOM compared to children attending DCCs (21.6 vs. 14.9% – p = 0.056). Moreover, a higher proportion of biotype III strains carried a mutated ftsI genein the strains of children with AOM compared to children attending DCCs (50.0 vs. 26.3% – p < 0.01), while no difference in mutation frequency in other biotypes was seen. All strains carrying a mutated ftsI gene were non-typeable.
The highest resistance of the mutated strains in the carriage group was seen to cefuroxime (56.7%; 59/104), followed by amoxicillin-clavulanic acid (35.6%; 37/104) and ampicillin (27.8%; 29/10).
In the invasive group, 4.3% (4/94) of the strains showed a mutation in the ftsI gene, all corresponding to the Ubukata-group 2. Biotype II was most frequently (75%; 3/4) present in these strains followed by biotype III (25%; 1/4). As in the carriage group, all mutated strains were not typeable. The mutated strains in the invasive group were resistant to ampicillin (50.0%; 2/4) and amoxicillin-clavulanic acid (50.0%; 2/4).
This three-year study describes the carriage status and antimicrobial susceptibility profile of H. influenzae strains in samples obtained from Belgian children attending DCCs and children with AOM and compares it to the susceptibility profile of H. influenzae strains responsible for invasive disease in children.
As expected, non-typeable Haemophilus influenzae strains were predominantly present in the different subgroups of our study. Also, the NRC reports Belgium showed that mostly non-typeables were present in invasive disease (81/103–78.6% in 2017; 96/129–74.4% in 2018; 128/162–79.0% in 2019; 62/84–73.8% in 2020; 68/87–78.0% in 2021). It is already known that invasive H. influenzae serotype b (Hib) strains have been replaced by non-typeable H. influenzae in many countries since the introduction of the Hib conjugate vaccines (Van Eldere et al., 2014; Zarei et al., 2016; Slack, 2017; Slack et al., 2021). Also serotypes “a,” “e,” and “f” have been reported in different countries as disease causing serotypes, and are also serotypes found in our study (Ladhani et al., 2012; Palaniappan et al., 2020; Slack et al., 2021; Reilly et al., 2022). In North America, an increase in proportion of serotype “a” has been observed among invasive disease cases (Tsang and Ulanova, 2017; Slack et al., 2021; Soeters et al., 2021; Ulanova, 2021). Serotype “a” is also found as possible disease causing serotype in our study as this serotype is only found in the invasive group but not in the carriage group. In Europe, on the other hand, serotype “f” became the most frequent encapsulated serotype causing invasive disease after implementing Hib conjugate vaccine, but the numbers remain relatively low (Ladhani et al., 2012; Reilly et al., 2022), whereas there is also an increasing trend in infections caused by serotype “e” (Ladhani et al., 2012; Slack et al., 2021). In both groups of the current study, serotype “f” was quite frequently present, supporting the findings in Europe, while serotype “e” is (rarely) present only in the carriage group. Also according the reports of the NRC in Belgium, serotype “f” is the most prevalent serotype (12/103–11.7% in 2017; 14/129–10.9% in 2018, 15/162–9.3% in 2019; 9/84–10.7% in 2020 and 5/87–5.8% in 2021), while serotype “e” was rarely present (3/103–2.9% in 2017 and 3/129–2.3% in 2018; not present in 2019–2021; Grammens et al., 2017, 2018; Jacquinet et al., 2021).
The most frequently found biotype in this study is biotype II, accounting for more than 35% in each analyzed group (DCC, AOM as well as the invasive group). This was followed by biotype III, biotype I and biotype V which is in line with observations from other studies (Albritton et al., 1978; Righter and Luchsinger, 1988; Watson et al., 1988). Different studies have found that biotype I is often associated with pathogenicity in children, such as severe meningitis, regardless whether the presence of biotype I occurred simultaneously with the presence of serotype “b” (Kilian, 1976; Long et al., 1983). These findings are supported by the results that we found in this study – a higher proportion of biotype I is found in the invasive group (27.7 vs. 15.3%). Biotype II and biotype III are commensal to the upper respiratory tract and colonization does usually not progress to invasive disease (Kilian, 1976; Long et al., 1983). However, biotype II and biotype III may be involved in the pathogenesis of mucosal diseases such as sinusitis and otitis media as well as in the pathogenesis of invasive disease (Kilian, 1976; Long et al., 1983). Also in our study, biotype II and III were more frequently found in the AOM (42.6 and 26.6%) group compared to the DCC (36.6 and 22.6%) and invasive group (36.2 and 19.1%). In addition, it is more commonly seen that biotypes I, II, III, and serotype “b” strains show resistance to ampicillin compared to other biotypes and non-serotype “b” strains (Jain et al., 2006). Interestingly, we found a higher mutation frequency in the ftsI gene in the strains obtained from the AOM group, especially higher proportion of biotype III in this mutated strains compared to the strains obtained from the DCC group. It is also seen that strains carrying PBP3 alterations are more invasive, suggesting that these alterations may be involved in adhesion and invasion mechanisms (Okabe et al., 2010; Atkins et al., 2014; Singh et al., 2016).
In the initial Np carriage study, frequent use of antibiotics (>3 treatments in 3 months) was an exclusion criterium in AOM children but not in DCC children. Children included from DCC turned out to have had more AB treatments than children included with AOM. However, AOM children may have used a broad-spectrum antibiotic. Measuring trends in the antimicrobial susceptibility of H. influenzae strains is gaining importance because of the increased use of existing antibiotics as well as development of novel antibiotics. To treat H. influenzae infections, aminopenicillins and cephalosporins are used as first choice, but there are mechanisms of resistance against these aminopenicillins (Schotte et al., 2019). As the number of ampicillin-resistant strains has increased, it was important to verify the effectiveness of cephalosporins (such as cefuroxime) to treat ampicillin-resistant H. influenzae infections (Deghmane et al., 2019; Wang et al., 2019; Nürnberg et al., 2021). This makes the development of novel antibiotic treatments important.
In conclusion, this study found that biotypes I, II and III are more frequently present in the carriage group as well as in the invasive group, while specific serotypes for carriage (serotype “e”) and invasive group (serotype “a” and “b”) were found. In addition, a higher antimicrobial resistance level for the analyzed antibiotics was observed in the carriage (DCC, AOM) group compared to the invasive group. In the strains of children suffering from AOM, a higher degree of mutations in the ftsI gene was found compared to the strains obtained from DCC children. The current study helps to lay the ground to understand the dynamics of H. influenzae carriage in Belgian children, as available data on remaining H. influenzae dynamics in a vaccinated child population (in a carriage as well as an invasive group) is scarce. This study provides an important first insight into the characteristics of circulating H. influenzae strains, especially in light of emerging antibiotic resistance to beta-lactams as they may have implications for the treatment of H. influenzae infections. It is important to monitor changes in the microbiology and epidemiology of H. influenzae as it can lead to clinical failure caused by antimicrobial resistance.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
The studies involving human participants were reviewed and approved by Ethics committee of University of Antwerp (UAntwerpen) and the Antwerp University Hospital (UZA; ID 15/45/471 and ID 18/31/355). Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin.
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
The study is supported by a research grant from Research Foundation Flanders (FWO Research Grant 1150017N, Antigoon ID 33341) and an investigator-initiated research grant from Pfizer.
We would like to thank all members of the expert advisory board (HG, R. Cohen, A. Finn, K. Van Herck, and D. Tuerlinckx) for their contribution to the study protocol and interpretation of the results; Research Link–ECSOR operating as the CRO; the cooperating nurses, physicians, ONE and Kind & Gezin for assistance in the recruitment and sampling; the children and their parents for their participation.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1160073/full#supplementary-material
Agrawal, A., and Murphy, T. F. (2011). Haemophilus influenzae infections in the H. influenzae type b conjugate vaccine era. J. Clin. Microbiol. 49, 3728–3732. doi: 10.1128/JCM.05476-11
Albritton, W. L., Penner, S., Slaney, L., and Brunton, J. (1978). Biochemical characteristics of Haemophilus influenzae in relationship to source of isolation and antibiotic resistance. J. Clin. Microbiol. 7, 519–523. doi: 10.1128/jcm.7.6.519-523.1978
Aniansson, G., Aim, B., Andersson, B., Larsson, P., Nylen, O., Peterson, H., et al. (1992). Nasopharyngeal colonization during the first year of life. J. Infect. Dis. 165, S38–S42. doi: 10.1093/infdis/165-Supplement_1-S38
Atkins, N. A., Kunde, D. A., Zosky, G., and Tristram, S. G. (2014). Genotypically defined β-lactamase-negative ampicillin-resistant isolates of non-typable Haemophilus influenzae are associated with increased invasion of bronchial epithelial cells in vitro. J. Med. Microbiol. 63, 1400–1403. doi: 10.1099/jmm.0.077966-0
Barbosa, A. R., Giufre, M., Cerquetti, M., and Bajanca-Lavado, M. P. (2011). Polymorphism in ftsI gene and {beta}-lactam susceptibility in Portuguese Haemophilus influenzae strains: clonal dissemination of beta-lactamase-positive isolates with decreased susceptibility to amoxicillin/clavulanic acid. J. Antimicrob. Chemother. 66, 788–796. doi: 10.1093/jac/dkq533
Deghmane, A. E., Hong, E., Chehboub, S., Terrade, A., Falguières, M., Sort, M., et al. (2019). High diversity of invasive Haemophilus influenzae isolates in France and the emergence of resistance to third generation cephalosporins by alteration of ftsI gene. J. Infect. 79, 7–14. doi: 10.1016/j.jinf.2019.05.007
Ekinci, E., van Heirstraeten, L., Willen, L., Desmet, S., Wouters, I., Vermeulen, H., et al. (2022). Serotype 19A and 6C account for one third of pneumococcal carriage among Belgian day-care children four years after a shift to a lower-valent PCV. J. Pediatric. Infect. Dis. Soc. 12, 36–42. doi: 10.1093/jpids/piac117
García-Cobos, S., Campos, J., Lázaro, E., Román, F., Cercenado, E., García-Rey, C., et al. (2007). Ampicillin-resistant non-beta-lactamase-producing Haemophilus influenzae in Spain: recent emergence of clonal isolates with increased resistance to cefotaxime and cefixime. Antimicrob. Agents Chemother. 51, 2564–2573. doi: 10.1128/AAC.00354-07
Grammens, D. M., Moens, C., and Wyndham-Thomas, C., (2017). Epidemiologische surveillance van invasieve infecties met Haemophilus influenzae – 2017. Sciensano: Belgium.
Grammens, D. M., Moens, C., and Wyndham-Thomas, C. (2018). Epidemiologische surveillance van invasieve infecties met Haemophilus influenzae – 2018. Sciensano: Belgium.
Han, M. S., Jung, H. J., Lee, H. J., and Choi, E. H. (2019). Increasing prevalence of group III penicillin-binding protein 3 mutations conferring high-level resistance to Beta-lactams among nontypeable Haemophilus influenzae isolates from children in Korea. Microb. Drug Resist. 25, 567–576. doi: 10.1089/mdr.2018.0342
Hasegawa, K., Yamamoto, K., Chiba, N., Kobayashi, R., Nagai, K., Jacobs, M. R., et al. (2003). Diversity of ampicillin-resistance genes in Haemophilus influenzae in Japan and the United States. Microb. Drug Resist. 9, 39–46. doi: 10.1089/107662903764736337
Hashida, K., Shiomori, T., Hohchi, N., Muratani, T., Mori, T., Udaka, T., et al. (2008). Nasopharyngeal Haemophilus influenzae carriage in Japanese children attending day-care centers. J. Clin. Microbiol. 46, 876–881. doi: 10.1128/JCM.01726-07
Honda, H., Sato, T., Shinagawa, M., Fukushima, Y., Nakajima, C., Suzuki, Y., et al. (2018). Multiclonal expansion and high prevalence of β-lactamase-negative Haemophilus influenzae with high-level ampicillin resistance in Japan and susceptibility to quinolones. Antimicrob. Agents Chemother. 62, e00851–e00818. doi: 10.1128/AAC.00851-18
Hotomi, M., Fujihara, K., Billal, D. S., Suzuki, K., Nishimura, T., Baba, S., et al. (2007). Genetic characteristics and clonal dissemination of beta-lactamase-negative ampicillin-resistant Haemophilus influenzae strains isolated from the upper respiratory tract of patients in Japan. Antimicrob. Agents Chemother. 51, 3969–3976. doi: 10.1128/AAC.00422-07
Jacquinet, D. M., Prevost, B., and Grammens, T. (2021). Epidemiologische surveillance van invasieve infecties met Haemophilus influenzae – 2019 to 2021. Sciensano: Belgium.
Jain, A., Kumar, P., and Awasthi, S. (2006). High ampicillin resistance in different biotypes and serotypes of Haemophilus influenzae colonizing the nasopharynx of healthy school-going Indian children. J. Med. Microbiol. 55, 133–137. doi: 10.1099/jmm.0.46249-0
Kilian, M. (1976). A taxonomic study of the genus Haemophilus, with the proposal of a new species. J. Gen. Microbiol. 93, 9–62. doi: 10.1099/00221287-93-1-9
Ladhani, S. N., Collins, S., Vickers, A., Litt, D. J., Crawford, C., Ramsay, M. E., et al. (2012). Invasive Haemophilus influenzae serotype e and f disease, England and Wales. Emerg. Infect. Dis. 18, 725–732. doi: 10.3201/eid1805.111738
Long, S. S., Teter, M. J., and Gilligan, P. H. (1983). Biotype of Haemophilus influenzae: correlation with virulence and ampicillin resistance. J. Infect. Dis. 147, 800–806. doi: 10.1093/infdis/147.5.800
Madore, D. V. (1996). Impact of immunization on Haemophilus influenzae type b disease. Infect. Agents Dis. 5, 8–20.
McElligott, M., Meyler, K., Bennett, D., Mulhall, R., Drew, R. J., and Cunney, R. (2020). Epidemiology of Haemophilus influenzae in the Republic of Ireland, 2010-2018. Eur. J. Clin. Microbiol. Infect. Dis. 39, 2335–2344. doi: 10.1007/s10096-020-03971-z
Medeiros, A. A., Levesque, R., and Jacoby, G. A. (1986). An animal source for the ROB-1 beta-lactamase of Haemophilus influenzae type b. Antimicrob. Agents Chemother. 29, 212–215. doi: 10.1128/AAC.29.2.212
Mendelman, P. M., Chaffin, D. O., Musser, J. M., de Groot, R., Serfass, D. A., and Selander, R. K. (1987). Genetic and phenotypic diversity among ampicillin-resistant, non-beta-lactamase-producing, nontypeable Haemophilus influenzae isolates. Infect. Immun. 55, 2585–2589. doi: 10.1128/iai.55.11.2585-2589.1987
Nürnberg, S., Claus, H., Krone, M., Vogel, U., and Lâm, T. T. (2021). Cefotaxime resistance in invasive Haemophilus influenzae isolates in Germany 2016-19: prevalence, epidemiology and relevance of PBP3 substitutions. J. Antimicrob. Chemother. 76, 920–929. doi: 10.1093/jac/dkaa557
Okabe, T., Yamazaki, Y., Shiotani, M., Suzuki, T., Shiohara, M., Kasuga, E., et al. (2010). An amino acid substitution in PBP-3 in Haemophilus influenzae associate with the invasion to bronchial epithelial cells. Microbiol. Res. 165, 11–20. doi: 10.1016/j.micres.2008.03.003
Ortiz-Romero, M. D. M., Espejo-García, M. P., Alfayate-Miguelez, S., Ruiz-López, F. J., Zapata-Hernandez, D., Gonzalez-Pacanowska, A. J., et al. (2017). Epidemiology of nasopharyngeal carriage by Haemophilus influenzae in healthy children: a study in the Mediterranean coast region. Pediatr. Infect. Dis. J. 36, 919–923. doi: 10.1097/INF.0000000000001625
Palaniappan, P. A., Mohamed Sukur, S., Liow, Y. L., Maniam, S., Sherina, F., and Ahmad, N. (2020). Carriage of Haemophilus influenzae among children attending childcare centres in Kuala Lumpur, Malaysia in the post vaccination era: a cross-sectional study. Vaccine 38, 8232–8237. doi: 10.1016/j.vaccine.2020.09.066
Peltola, H. (2000). Worldwide Haemophilus influenzae type b disease at the beginning of the 21st century: global analysis of the disease burden 25 years after the use of the polysaccharide vaccine and a decade after the advent of conjugates. Clin. Microbiol. Rev. 13, 302–317. doi: 10.1128/CMR.13.2.302
Peltola, H., Käythy, H., Sivonen, A., and Mäkelä, P. H. (1977). Haemophilus influenzae type b capsular polysaccharide vaccine in children: a double-blind field study of 100,000 vaccinees 3 months to 5 years of age in Finland. Pediatrics 60, 730–737. doi: 10.1542/peds.60.5.730
Pittman, M. (1931). Variation and type specificity in the bacterial species Hemophilus influenzae. J. Exp. Med. 53, 471–492. doi: 10.1084/jem.53.4.471
Reilly, A. S., McElligott, M., Mac Dermott Casement, C., and Drew, R. J. (2022). Haemophilus influenzae type f in the post-Haemophilus influenzae type b vaccination era: a systematic review. J. Med. Microbiol. 71. doi: 10.1099/jmm.0.001606
Righter, J., and Luchsinger, I. (1988). Haemophilus influenzae from four laboratories in one Canadian city. J. Antimicrob. Chemother. 22, 333–339. doi: 10.1093/jac/22.3.333
Schotte, L., Wautier, M., Martiny, D., Piérard, D., and Depypere, M. (2019). Detection of beta-lactamase-negative ampicillin resistance in Haemophilus influenzae in Belgium. Diagn. Microbiol. Infect. Dis. 93, 243–249. doi: 10.1016/j.diagmicrobio.2018.10.009
Singh, N. K., Kunde, D. A., and Tristram, S. G. (2016). Effect of epithelial cell type on in vitro invasion of non-typeable Haemophilus influenzae. J. Microbiol. Methods 129, 66–69. doi: 10.1016/j.mimet.2016.07.021
Skaare, D., Anthonisen, I., Caugant, D. A., Jenkins, A., Steinbakk, M., Strand, L., et al. (2014). Multilocus sequence typing and ftsI sequencing: a powerful tool for surveillance of penicillin-binding protein 3-mediated beta-lactam resistance in nontypeable Haemophilus influenzae. BMC Microbiol. 14:131. doi: 10.1186/1471-2180-14-131
Slack, M. P. E. (2017). The evidence for non-typeable Haemophilus influenzae as a causative agent of childhood pneumonia. Pneumonia 9:9. doi: 10.1186/s41479-017-0033-2
Slack, M. P. E., Cripps, A. W., Grimwood, K., Mackenzie, G. A., and Ulanova, M. (2021). Invasive Haemophilus influenzae infections after 3 decades of Hib protein conjugate vaccine use. Clin. Microbiol. Rev. 34:e0002821. doi: 10.1128/CMR.00028-21
Soeters, H. M., Oliver, S. E., Plumb, I. D., Blain, A. E., Zulz, T., Simons, B. C., et al. (2021). Epidemiology of invasive Haemophilus influenzae serotype a disease-United States, 2008-2017. Clin. Infect. Dis. 73, e371–e379. doi: 10.1093/cid/ciaa875
Swingler, G., Fransman, D., and Hussey, G. (2007). Conjugate vaccines for preventing Haemophilus influenzae type B infections. Cochrane Database Syst. Rev. 2:Cd001729. doi: 10.1002/14651858.CD001729.pub2
Tanaka, E., Hara, N., Wajima, T., Ochiai, S., Seyama, S., Shirai, A., et al. (2019). Emergence of Haemophilus influenzae with low susceptibility to quinolones and persistence in tosufloxacin treatment. J. Glob. Antimicrob. Resist. 18, 104–108. doi: 10.1016/j.jgar.2019.01.017
Tsang, R. S. W., and Ulanova, M. (2017). The changing epidemiology of invasive Haemophilus influenzae disease: emergence and global presence of serotype a strains that may require a new vaccine for control. Vaccine 35, 4270–4275. doi: 10.1016/j.vaccine.2017.06.001
Ubukata, K., Shibasaki, Y., Yamamoto, K., Chiba, N., Hasegawa, K., Takeuchi, Y., et al. (2001). Association of amino acid substitutions in penicillin-binding protein 3 with beta-lactam resistance in beta-lactamase-negative ampicillin-resistant Haemophilus influenzae. Antimicrob. Agents Chemother. 45, 1693–1699. doi: 10.1128/AAC.45.6.1693-1699.2001
Ubukata, K., Sunakawa, K., Nonoyama, M., Iwata, S., Konno, M., Ubukata, K., et al. (2002). Differentiation of beta-lactamase-negative ampicillin-resistant Haemophilus influenzae from other H. influenzae strains by a disc method. J. Infect. Chemother. 8, 50–58. doi: 10.1007/s101560200006
Ulanova, M. (2021). Invasive Haemophilus influenzae serotype a disease in the H. influenzae serotype b conjugate vaccine era: where are we going? Clin. Infect. Dis. 73, e380–e382. doi: 10.1093/cid/ciaa868
Van Eldere, J., Slack, M. P. E., Ladhani, S., and Cripps, A. W. (2014). Non-typeable Haemophilus influenzae, an under-recognised pathogen. Lancet Infect. Dis. 14, 1281–1292. doi: 10.1016/S1473-3099(14)70734-0
Wang, H. J., Wang, C.-Q., Hua, C.-Z., Yu, H., Zhang, T., Zhang, H., et al. (2019). Antibiotic resistance profiles of Haemophilus influenzae isolates from children in 2016: a multicenter study in China. Can. J. Infect. Dis. Med. Microbiol. 2019:6456321. doi: 10.1155/2019/6456321
Watson, K. C., Kerr, E. J., and Baillie, M. (1988). Temporal changes in biotypes of Haemophilus influenzae isolated from patients with cystic fibrosis. J. Med. Microbiol. 26, 129–132. doi: 10.1099/00222615-26-2-129
Wouters, I., van Heirstraeten, L., Desmet, S., Blaizot, S., Verhaegen, J., Goossens, H., et al. (2018). Nasopharyngeal s. pneumoniae carriage and density in Belgian infants after 9 years of pneumococcal conjugate vaccine programme. Vaccine 36, 15–22. doi: 10.1016/j.vaccine.2017.11.052
Yalçın, M., Tristram, S., and Bozdoğan, B. (2022). Use of trans-complementation method to determine the effects of various ftsI mutations on β-lactamase-negative ampicillin-resistant (BLNAR) Haemophilus influenzae strains. Arch. Microbiol. 205:27. doi: 10.1007/s00203-022-03371-1
Keywords: Haemophilus influenzae, children, biotypes, serotypes, mutations, antibiotic susceptibility testing
Citation: Ekinci E, Willen L, Rodriguez Ruiz JP, Maertens K, Van Heirstraeten L, Serrano G, Wautier M, Deplano A, Goossens H, Van Damme P, Beutels P, Malhotra-Kumar S, Martiny D and Theeten H (2023) Haemophilus influenzae carriage and antibiotic resistance profile in Belgian infants over a three-year period (2016–2018). Front. Microbiol. 14:1160073. doi: 10.3389/fmicb.2023.1160073
Received: 06 February 2023; Accepted: 03 April 2023;
Published: 24 April 2023.
Edited by:
Jozsef Soki, University of Szeged, HungaryReviewed by:
Nina M. Van Sorge, Amsterdam University Medical Center, NetherlandsCopyright © 2023 Ekinci, Willen, Rodriguez Ruiz, Maertens, Van Heirstraeten, Serrano, Wautier, Deplano, Goossens, Van Damme, Beutels, Malhotra-Kumar, Martiny and Theeten. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Esra Ekinci, ZXNyYS5la2luY2lAdWFudHdlcnBlbi5iZQ==
†These authors share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.