
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 18 May 2023
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1158659
Extended-spectrum beta-lactamase-producing Gram-negative bacteria are common in the community and hospitals. To monitor ESBLs mediated by the CTX-M genotype, we collected clinical ESBL pathogenic strains from a hospital in central China and observed a strain of Escherichia coli, namely Ec15103 carrying blaCTX–M–14, blaCTX–M–64 and blaTEM–1, isolated from the blood of a 7-day-old infant in 2015. Strain Ec15103 contains two drug resistance plasmids: pEc15103A, an IncFI-type plasmid that cannot be conjugatively transferred and carries the drug resistance genes blaTEM–1, aacC2, aadA5, sul1, mph(A), sul2, strAB, and tetA(A); and pEc15103B, an IncK2/Z-type plasmid that carries the conjugation transfer gene and blaCTX–M–14. In addition, blaCTX–M–64 is located on the chromosome of Ec15103, and it is the first report of pathogen with blaCTX–M–64 located on its chromosome (the search terms used “blaCTX-M-64” and “chromosome”). blaCTX–M–14 and blaCTX–M–64 are carried by ISEcp1-mediated transposon Tn6503a and Tn6502, respectively. The conjugation transfer ability of pEc15103B was significantly inhibited by zidovudine (AZT) and linoleic acid (LA) and that expression of blaCTX–M–14, blaCTX–M–64 and blaTEM–1 at the mRNA level did not change based on the concentration of cefotaxime or ampicillin. Co-occurrence of blaCTX–M–14 and blaCTX–M–64 in a single isolate will enhance the drug resistance of bacteria, and the presence of blaCTX–M–64 in the chromosome may make the resistance more maintain. This fact will facilitate its dissemination and persistence under different antimicrobial selection pressures. It is essential to prevent these strains from further spreading in a hospital environment.
Refractory infections caused by multidrug-resistant (MDR) bacteria are rapidly increasing worldwide, especially for β-lactamase-producing Gram-negative bacteria (Hussain et al., 2021). These Gram-negative bacteria hydrolyze β-lactams, which renders them resistant to β-lactam antibiotics. According to Ambler’s classification, β-lactamases can be divided into four categories (Ambler et al., 1991): A (serine penicillinases), B (metallo-β-lactamases), C (cephalosporinases), and D (oxacillinases). Extended-spectrum beta-lactamases (ESBLs) are primarily plasmid-mediated, hydrolyzing penicillins, cephalosporins, and monocyclic beta-lactams aztreonam but not cephamycins and carbapenem enzymes. ESBLs can be inhibited by β-lactamase inhibitors, mainly of the class A and D enzymes. Previously, the TEM type and SHV type were the most common genotypes of ESBLs, but since the discovery of the CTX-M β-lactamase, this type has rapidly disseminated worldwide and become the most common ESBL genotype (Bonnet, 2004). It is denoted as CTX-M because it is resistant to cefotaxime but sensitive to ceftazidime (Bauernfeind et al., 1990); it is mainly encoded by plasmids that can be conjugated and transferred. The CTX-M genotype is found in 238 species, and subtypes1 are divided into 6 groups according to amino acid heterogeneity: Group 1, Group 2, Group 8, Group 9, Group 25, and KLUC Group (Rossolini et al., 2008).
ESBL-producing Gram-negative bacteria are common in the community and hospitals (Pitout et al., 2005). They cause a relatively high number of clinical infections, which are usually treated with beta-lactamase inhibitors or carbapenems. Therefore, ESBL-producing strains increase the use of carbapenem antibiotics. As this will further accelerate the evolution of bacterial resistance under antibiotic selection pressure (Bevan et al., 2017), monitoring ESBL strains is particularly important.
To monitor ESBLs mediated by the CTX-M genotype, we collected clinical ESBL-producing isolates from a hospital in central China. In this study, Escherichia coli Ec15103, which carried both blaCTX–M–14 and blaCTX–M–64, was characterized. Moreover, a detailed genetic characterization of pEc15103A and pEc15103B was revealed. blaCTX–M–14 was located on IncK2/Z-type plasmid pEc15103B, however, blaCTX–M–64 was harbored in the chromosome.
Escherichia coli Ec15103 was isolated from the blood of an infected 7-day-old infant at a Chinese teaching hospital in 2015. Bacterial species identification was performed using a Bruker MALDI Biotyper (Bruker Daltonics, Bremen, Germany). E. coli Ec600 (LacZ–, NalR, RifR) was used as the recipient bacterium for plasmid conjugation transfer in this study. Competent E. coli EPI300 was used as the recipient bacterium for plasmid chemical transformation. The strains used in this study can be found in Supplementary Table 1.
The ESBL confirmation test used the disc diffusion Antibiotic Sensitivity test (Drieux et al., 2008) and refers to the American Clinical and Laboratory Standards Institute 2020 (CLSI) standard (CLSI, 2020). The zone of inhibition is used to determine the susceptibility of bacteria to an antibiotic. Antimicrobial susceptibility disks (Kangtai Biotechnology, Wenzhou, China) containing ceftazidime (30 μg), cefotaxime (30 μg), ceftazidime/clavulanic acid (30 μg/10 μg), or cefotaxime/clavulanic acid (30 μg/10 μg) were placed on Mueller-Hinton plates (MH plates, Autobio, Zhengzhou, China) coated on the surface with a bacterial suspension (0.5 McFarland turbidity). The size of the inhibition zone was measured after 16–18 h of incubation at 35°C. Measured the inhibition zone around a disk of cephalosporin and around a disk of the same cephalosporin plus clavulanate. Depending on the disk type, a difference of ≥5 mm between the two diameters are considered as indicating ESBL production.
Plasmid conjugal transfer experiments were carried out with E. coli Ec600 as the recipient and strain Ec15103 as the donor. The recipient and donor bacteria were inoculated in Lysogeny Broth (LB, Solarbio Science & Technology, Beijing, China) and incubated at 37°C and 200 rpm overnight. A total of 50 μL of the overnight cultured bacterial suspension was inoculated in 5 mL of LB broth at 37°C and 200 rpm followed by growth to OD600≈1.0, and then 500 μL of donor and recipient bacteria were mixed. The mixture was spotted onto a 1-cm2 filter membrane placed on LB nutrient agar (Solarbio Science & Technology, Beijing, China). The plates were incubated at 37°C for 12∼18 h, then the cells were washed from the filter membrane to 1 mL of LB broth. After 1,000-fold dilution, 100 μL was spread on LB nutrient agar medium containing 100 μg/mL ampicillin (Solarbio Science & Technology, Beijing, China) and 750 μg/mL rifampicin (Solarbio Science & Technology, Beijing, China). The colonies after overnight culture at 37°C were transconjugants (Ec15103-Ec600), as verified by PCR.
Plasmid DNA of E. coli Ec15103 was extracted using a large plasmid extraction kit (Mei5 Biotechnology, Beijing, China), and plasmid transformation was carried out using competent EPI300 cells as the recipient bacteria. First, competent EPI300 cells were taken out from -80°C and quickly placed on ice. After 5 min, the bacteria had thawed, the plasmid DNA of E. coli Ec15103 was added, and the mixture was allowed to stand on ice for 30 min. Then, the samples were placed in a water bath at 42°C for 60 s, quickly placed on ice, and allowed to stand for 5 min. Next, 800 μL of LB broth was added to the EP tube and incubated at 37°C for 60 min at 200 rpm, followed by centrifugation at 6,000 rpm for 5 min to collect the bacteria. Approximately 100 μL of the supernatant was retained to resuspend the bacteria and spread the cells on LB medium containing 100 μg/mL ampicillin. The colonies after overnight culture at 37°C were transformants (Ec15103-EPI300-1, Ec15103-EPI300-2), as verified by PCR.
Main resistance genes (blaCTX–M–1G, blaCTX–M–9G, and blaTEM–1) were screened by PCR (for the primers used, see Supplementary Table 2), and the PCR products were sent to Beijing Genomics Institute (BGI, Zhengzhou, China) for sequencing. The sequencing results were compared with the NCBI database to identify the main resistance genes.
The minimum inhibitory concentrations (MICs) values of 25 antibiotics (see Table 1) in this study were evaluated by the broth microdilution method and determined according to Clinical and Laboratory Standards Institute 2020 guidelines (CLSI, 2020).
Whole-genome DNA of E. coli Ec15103 was extracted with OMEGA D3350 Bacterial DNA Kit (Omega Bio-Tek, Norcross, GA, USA). The E. coli strain Ec15103 genome was sequenced using a PacBio Sequel II and DNBSEQ platform at Beijing Genomics Institute (BGI, Shenzhen, China). Four SMRT cells Zero-Mode Waveguide arrays of sequencing were used with the PacBio platform to generate the subread set. PacBio subreads (length < 1 kb) were removed. The program Canu was used for self-correction. Draft genomic unitigs, which are uncontested groups of fragments, were assembled using Canu, a high-quality corrected circular consensus sequence subread set. To improve the accuracy of the genome sequences, GATK2 was applied for single-base corrections. Genes were predicted with GeneMarkS™ and RAST and further annotated with BLASTP and BLASTN against the UniProt and NR databases. Annotation of mobile elements was based on the databases ISfinder, INTEGRALL, and Tn Number Registry. Gene organization diagrams were drawn with Inkscape version 0.48.
The genome sequence of strain Ec15103 has been submitted to GenBank: the accession number of the Ec15103 chromosome is CP104274; the accession numbers of pEc15103A and pEc15103B are ON324203 and ON324204, respectively.
Since zidovudine (AZT, Solarbio Science & Technology, Beijing, China) and linoleic acid (LA, Solarbio Science & Technology, Beijing, China) have different solubilities, we used two methods for inhibiting plasmid conjugation transfer. E. coli Ec600 was used as the recipient, and strain Ec15103 was used as the donor. (I) (AZT): The recipient and donor bacteria were inoculated in LB broth at 37°C and 200 rpm overnight. A total of 5 μL of the overnight bacteria was inoculated in 5 mL of LB broth and incubated at 37°C and 200 rpm to OD600≈1.0, and then 500 μL of donor and recipient bacteria were mixed. A total of 10 μL of the mixture was spotted on nitrocellulose membrane (4 cm2 size, 0.45 μm pore size) placed on LB nutrient agar medium, and the different groups of LB nutrient agar contained different concentrations of AZT. The plates were incubated at 37°C for 12∼18 h, then the cells were washed from the filter membrane to 1 mL of LB broth. After 1,000-fold dilution, 100 μL was spread onto LB nutrient agar medium containing 100 μg/mL ampicillin and 750 μg/mL rifampicin. The colony after 24–28 h of culture at 37°C was considered the transconjugant (Ec15103-Ec600). (II) (LA): Recipient and donor bacteria were inoculated in LB broth and incubated at 37°C and 200 rpm overnight. A total of 5 μL of the overnight bacterial culture was inoculated in 5 mL of LB broth and incubated at 37°C and 200 rpm to OD600≈1.0, and then 500 μL of donor and recipient bacteria were mixed. A 10 μL aliquot was added to 5 mL LB broth and cultured overnight at 37°C and 200 rpm. The different groups of LB broth contained different concentrations of LA, which was dissolved in 30% DMSO (Solarbio Science & Technology, Beijing, China). A total of 100 μl of the bacteria were cultured overnight and diluted 100-fold, and taken 100 μL to spread onto LB nutrient agar medium containing 100 μg/mL ampicillin and 750 μg/mL rifampicin. The colony after 24–28 h of culture at 37°C was considered the transconjugant (Ec15103-Ec600). All plasmid conjugation transfer inhibition assays were completed using a minimum of three independent experiments, with three biological replicates per experiment.
RNA was extracted from 2 mL of logarithmic phase culture grown in LB broth (OD600≈1.0) using a kit (Mei5 Biotechnology, Beijing, China). After reverse transcription real-time quantitative PCR with the SYBR Green method (Pfaffl, 2001), a reaction system with a total volume of 20 μL was established. E. coli 16S rRNA was used as a reference gene to observe relative transcription levels of the target genes blaCTX–M–64, blaCTX–M–14, and blaTEM–1 in strains Ec15103, Ec15103-Ec600, and Ec15103-EPI300-1 at different concentrations of cefotaxime (CTX, Solarbio Science & Technology, Beijing, China) or ampicillin (AMP, for the primers used, see Supplementary Table 2). No antibiotics was used as the control group. The 2–ΔΔCt method, corrected for different primer efficiencies and reference genes, was employed. All mRNA expression assays were completed with a minimum of three independent experiments, with at least three biological replicates per experiment.
Statistical analysis was performed using GraphPad Prism 8 and SPSS software. All data are presented as the mean standard deviation.
For strain Ec15103, the inhibition zone for cefotaxime was almost 6 mm, and that for cefotaxime/clavulanic acid was 26 mm. The inhibition zone for ceftazidime was 15 mm; that for ceftazidime/clavulanic acid was 23 mm (Figure 1A). The inhibition zone diameter was 5 mm larger than without clavulanic acid, indicating that strain Ec15103 produces ESBL.
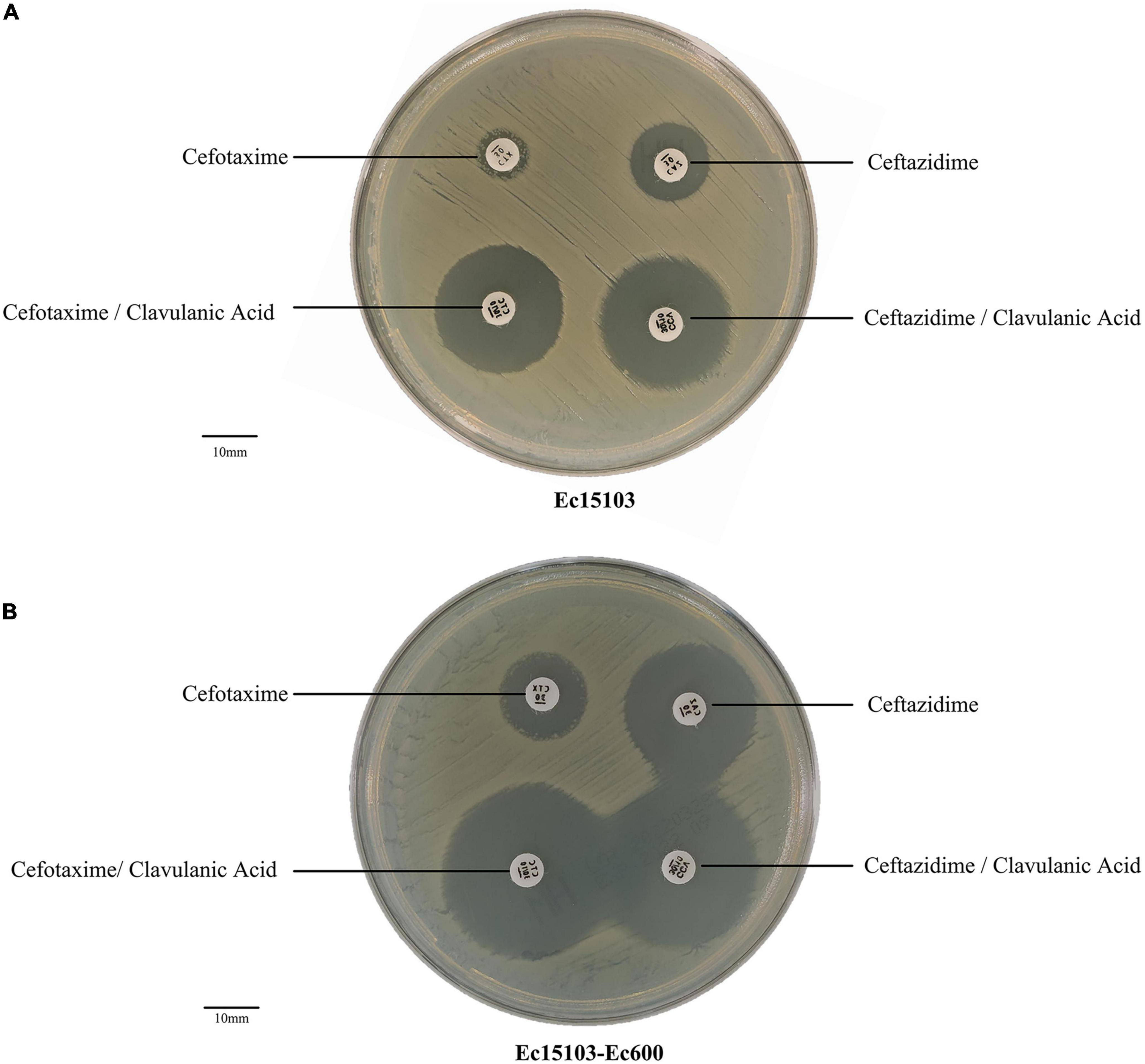
Figure 1. ESBL confirmatory test. (A) The ESBL confirmation test of Ec15103. (B) The ESBL confirmation test of Ec15103-Ec600. Each experiment was conducted with three parallel replicates, and only one representative figure is shown.
As determined by PCR screening and sequencing of the major plasmid-borne ESBL and carbapenem resistance genes, strain Ec15103 harbored blaCTX–M–14, blaCTX–M–64, and blaTEM–1 but not any of the other genes tested (Figure 2). Plasmid conjugation transfer was carried out using Ec15103 as the donor strain and Ec600 (LacZ–, NalR, RifR) as the recipient strain. However, only one resistance marker, blaCTX–M–14, was transferred from strain Ec15103 to Ec600 through conjugation, generating the transconjugant Ec15103-Ec600 (Figure 2). Additionally, we performed an ESBL confirmation test for Ec15103-Ec600, obtaining an inhibition zone for cefotaxime of 13 mm, for cefotaxime/clavulanic acid of 28 mm, for ceftazidime of 23 mm, and for ceftazidime/clavulanic acid of 30 mm (Figure 1B). As the inhibition zone diameter was 5 mm larger with clavulanic acid than without it, strain Ec15103-Ec600 also shows ESBL enzymatic activity. Both Ec15103 and Ec15103-Ec600 were found to be resistant to ampicillin, ampicillin/sulbactam, cefazolin, cefuroxime, ceftriaxone, and cefotaxime but susceptible to cefoxitin, piperacillin/tazobactam, cefoperazone/sulbactam, meropenem, imipenem, amikacin, polymyxin B, nitrofurantoin, minocycline, and chloramphenicol. Ec15103 was resistant to ceftazidime, cefepime, gentamicin, ciprofloxacin, and cotrimoxazole, whereas Ec15103-Ec600 was not (Table 1).
Furthermore, we extracted plasmid from strain Ec15103 and transformed it into a recipient strain, competent E. coli EPI300 (λ-rpsL (StrR), trfA), via chemical transformation. Based on PCR screening, two different transformed strains were detected: Ec15103-EPI300-1 with only one resistance marker, blaTEM–1, and Ec15103-EPI300-2 with blaTEM–1 and blaCTX–M–14 (Figure 2). Their MICs differed from those mentioned above (Table 1). Strain Ec15103-EPI300-1 was found to be resistant to ampicillin, ampicillin/sulbactam, gentamicin and cotrimoxazole, and Ec15103-EPI300-2 to ampicillin, ampicillin/sulbactam, cefazolin, cefuroxime, ceftriaxone, cefotaxime, gentamicin and cotrimoxazole. Both strains were sensitive to ceftazidime, cefepime, and ciprofloxacin.
Whole-genome sequencing (WGS) of strain Ec15103 produced a 4991.97-kb circular chromosome and two plasmids: pEc15103A and pEc15103B. The chromosome has a mean G+C content of 50.6% and consists of 5,121 open reading frames (ORFs). Plasmid pEc15103A is 104.00 kb in length, with an average G+C content of 52.1%, and contains 119 ORFs (Figure 3A). The pEc15103B genome consists of an 87.62-kb circular DNA molecule with an average G+C content of 52.6% and a total of 96 ORFs (Figure 4A). Each plasmid can be divided into backbone and accessory modules. The pEc15103A backbone (Figure 3A) is composed of DNA regions for plasmid replication (repB and repE) and plasmid maintenance (parA, parB, yubM, ssb, etc.). pEc15103A contains five distinct accessory modules (Figure 3A): the MDR region, ΔIS1294-ΔIS2 region, ΔISKpn26-ΔIS1203 region, IS1A, and ΔIS640. The drug resistance genes are all located in the MDR region, including blaTEM–1, aacC2, aadA5, sul1, mph(A), sul2, strAB, and tetA(A). The pEc15103B backbone contains DNA regions for plasmid replication (repY, repZ), plasmid maintenance (yacA, yacB, yagA, etc.), conjugal transfer (trs, trb, pil, etc.) and transfer (imp, yfa to yfh, yga to ygg, etc.). pEc15103B harbors only two accessory modules, ΔTn6503a and IS26, and the drug resistance gene blaCTX–M–14 is located in the former (Figure 4A).
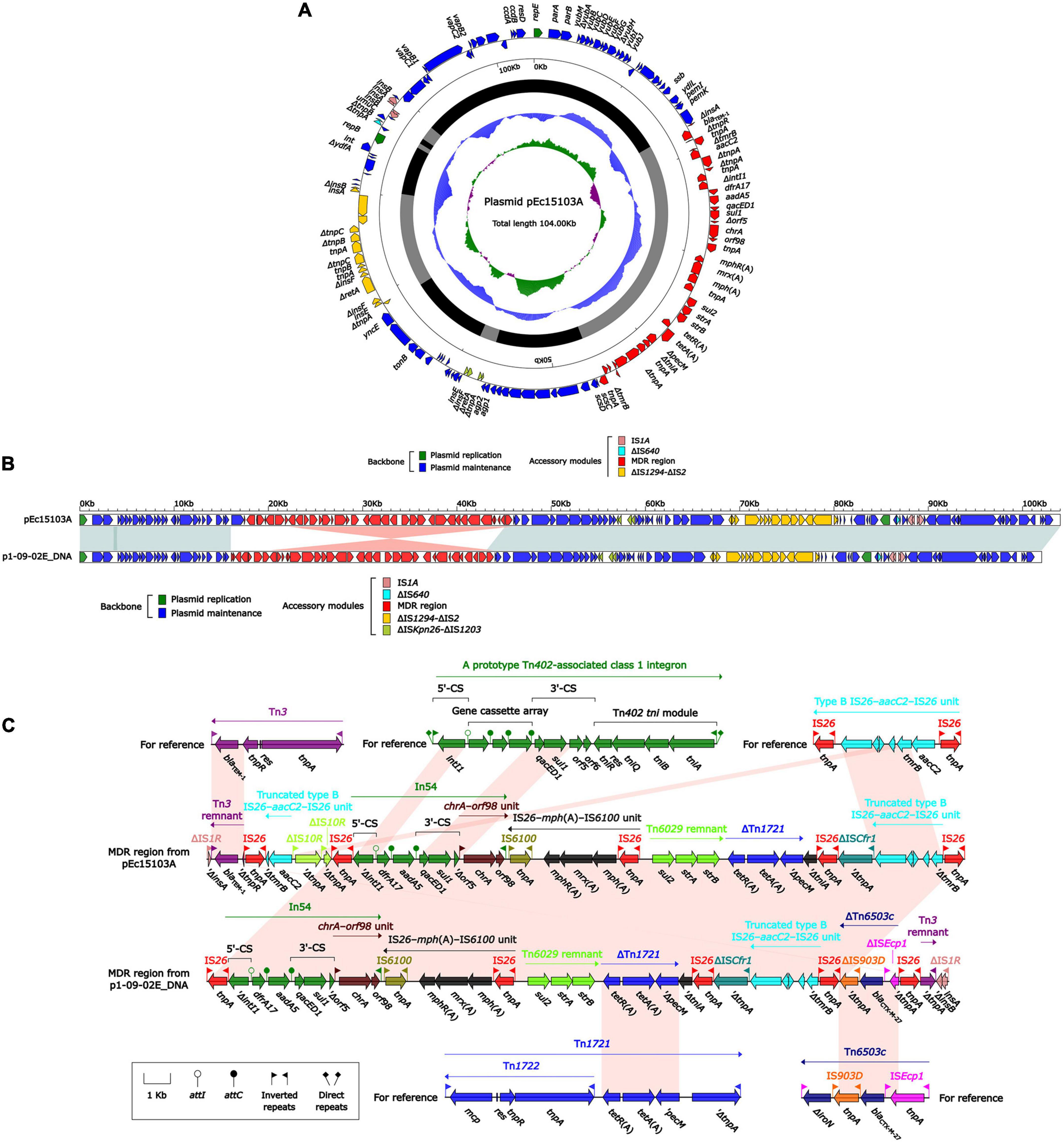
Figure 3. General Features of pEc15103A. (A) Schematic map of the sequence of plasmid pEc15103A. Genes are denoted by arrows and colored according to gene function classification. The innermost circle presents the GC skew [(G-C)/(G+C)] with a window size of 500 bp and a step size of 20 bp. The next-to-innermost circle presents the GC content. The backbone and accessory module regions are also shown. (B) Linear comparison of the sequenced plasmid pEc15103A. Genes are denoted by arrows colored according to gene function classification. Shaded regions denote regions of homology (>95% nucleotide identity). (C) Linear comparison of blaTEM–1 genetic surroundings. Genes are denoted by arrows and colored based on gene function classification.
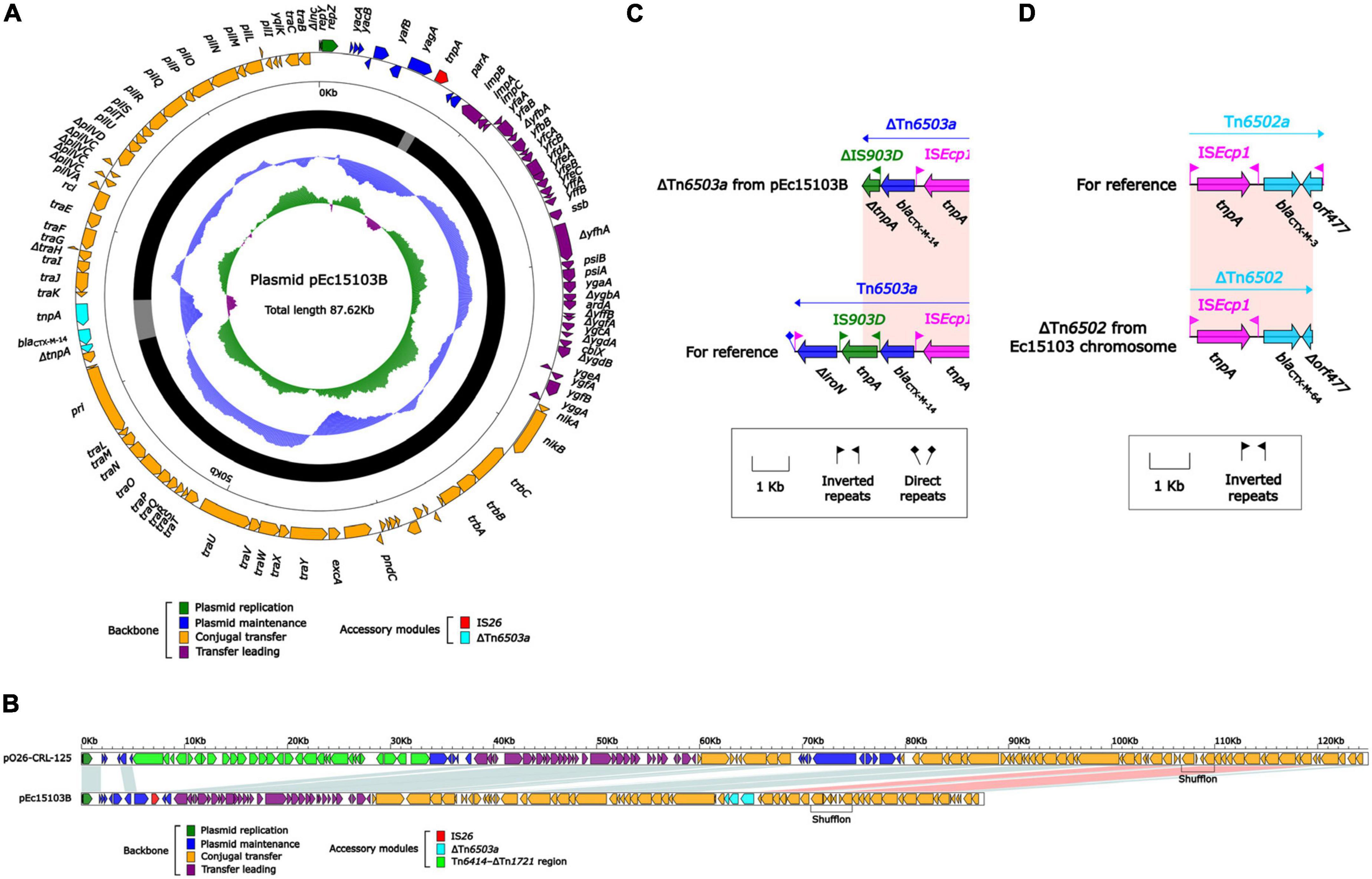
Figure 4. General features of pEc15103B and blaCTX–M–64. (A) Schematic map of the sequence of plasmid pEc15103B. Genes are denoted by arrows and colored according to gene function classification. The innermost circle presents the GC skew [(G-C)/(G+C)] with a window size of 500 bp and a step size of 20 bp. The next-to-innermost circle presents the GC content. The backbone and accessory module regions are also shown. (B) Linear comparison of the sequenced plasmid pEc15103B. Genes are denoted by arrows colored according to gene function classification. Shaded regions denote regions of homology (>95% nucleotide identity). (C) Linear comparison of blaCTX–M–14 genetic surroundings. Genes are denoted by arrows and colored based on gene function classification. (D) Linear comparison of blaCTX–M–64 genetic surroundings. Genes are denoted by arrows and colored based on gene function classification.
The overall structure of pEc15103A is most similar to that of p1-09-02E DNA (Mohsin et al., 2020) (GenBank accession number AP022651, 95% query coverage, and 99.99% maximum nucleotide identity). Therefore, p1-09-02E DNA was selected for comparative genomics analysis (Figure 3B). Both pEc15103A and p1-09-02E DNA are IncFI-type plasmids containing two replicons, RepFIA and RepFIB. Comparative genomic analysis showed that the backbone region of these two plasmids were essentially identical.
The MDR region of pEc15103A contains the following resistance gene modules: a Tn3 remnant, a truncated type B IS26–aacC2–IS26 unit, In54, IS26–mph(A)–IS6100 unit, a Tn6029 remnant, and ΔTn1721 (Figure 3C). Tn3 is a unit transposon; for pEc15103A, Tn3 is truncated by IS26 to form the Tn3 remnant. The Tn3 remnant includes a 38-bp IRR, the blaTEM–1 gene encoding β-lactam resistance, and a truncated tnpR (Partridge and Hall, 2005). The IS26–aacC2–IS26 unit, firstly observed in plasmid pSTMDT12_L DNA (GenBank accession number AP011958), is a structure containing the aminoglycoside resistance gene aacC2, and the core structure of the resistance unit is retained in pEc15103A. In54 is a class 1 integron containing a 5′-conserved segment (5′-CS), variable region, and 3′-conserved segment (3′-CS). The variable region of In54 contains two gene cassettes carrying dfrA17, which encodes quaternary ammonium resistance, and aadA5, which encodes aminoglycoside resistance (Changkaew et al., 2014). IS26–mph(A)–IS6100 unit is a macrolide resistance vector formed by two insertion sequences, IS26 and IS6100, that combined the macrolide resistance genes mph(A), mrx, and mphR(A) (Partridge, 2011). In pEc15103A, IS26–mph(A)–mrx–mphR(A)–IS6100 is located downstream of In54, and this maybe occurred through IS6100-mediated recombination (Feng et al., 2017). Tn6029 is a composite transposon carrying the β-lactamase gene blaTEM–1, the sulfonamide resistance gene sul2, and the streptomycin resistance gene strAB; the structure is IS26–ΔTn2–IS26–repA–repC–sul2–strAB–IS26, where ΔTn2 is the residue at the 3’ end of Tn2, and the structure is tnpR–blaTEM–1–IRR (Reid et al., 2015). The structure of the Tn6029 remnant in pEc15103A is sul2–strAB. Tn1721 is a Tn3 family transposon containing the class A tetracycline resistance module tetA(A)–tetR(A) (Schmitt et al., 1979), and its linear structure is IRL–mcp–res–tnpR–tnpA–IRR-1–tetR(A)–tetA(A)–pecM–ΔtnpA–IRR-2. In pEc15103A, the genes associated with Tn1721 transposition were missing, and the remaining structure of Tn1721 is tetR(A)–tetA(A)–ΔpecM.
The MDR region of p1-09-02E DNA and pEc15103A are similar, and there is only one modular difference: the 4.7-kb 5′-terminal structure of the MDR region of pEc15103A is lost in p1-09-02E DNA, meanwhile, a unique structure (ΔTn6503c–IS26–Tn3 remnant–ΔIS1R) is added in the 3′-end of MDR region in p1-09-02E DNA. Tn6503c is an ISEcp1-mediated transposon and displays an ISEcp1-blaCTX–M–27-IS903D-ΔiroN structure (Cheng et al., 2019). For p1-09-02E DNA, Tn6503c is truncated by two copies of IS26, and the line structure of ΔTn6503c is ΔISEcp1- blaCTX–M–27-ΔIS903D.
pEc15103B is an IncK2/Z plasmid. In this study, the IncK2/Z reference plasmid pO26-CRL-125 (Venturini et al., 2013) (GenBank accession number KC340960) was included in the comparative genomic analysis (Figure 4B). The results showed that pEc15103B contains the conserved IncK2/Z-type plasmid backbone region. There were two accessory regions in pEc15103B, that was IS26 and ΔTn6503a. The blaCTX–M–14 is located in ΔTn6503a (Figure 4C). Tn6503a is almost identical to Tn6503c except that blaCTX–M–27 mutates into blaCTX–M–14. The 3′ end of Tn6503a is truncated in pEc15103B, mainly manifesting as deletion of ΔiroN and truncation of IS903D.
In addition, the chromosome of strain Ec15103 contains the drug resistance gene blaCTX–M–64, which is located in ΔTn6502 (Figure 4D). The structure of ΔTn6502 is ISEcp1-blaCTX–M–3-orf477; in the Ec15103 chromosome, orf477 in this transposon is truncated, and blaCTX–M–3 has undergone mutation of individual bases, forming the structure ISEcp1-blaCTX–M–64-Δorf477.
Currently, plasmid-mediated intraspecific and interspecific horizontal gene transfer accounts for the majority of the prevalence and spread of antibiotic resistance genes (Alekshun and Levy, 2007; Carattoli, 2013). However, it has been found that some drugs can inhibit plasmid conjugation transfer (Liu et al., 2020). Using strain Ec15103 as the donor strain and Ec600 as the recipient strain, we performed inhibition of plasmid conjugation transfer experiments as described by Buckner et al. (2020). Compared with the LB broth control group, the number of monoclonal clones of the conjugative transfer strain in the AZT drug group decreased in a concentration-dependent manner, even when the concentration was as low as 0.008 μg/mL, the number of monoclonal clones of the conjugative transfer strain was significantly reduced (P < 0.01; Figure 5A). When the concentration was 0.064 μg/mL, the number of monoclonal clones of the conjugative transfer strain was close to 0, which was very significant (P < 0.0001; Figure 5A). Because LA is insoluble in water, we used 30% DMSO as the solvent, and the results between the 30% DMSO group and the LB broth group were not significantly different (P > 0.05; Figure 5B). When the concentration of LA was 1 mM, the number of monoclonal clones of the conjugative transfer strain was significantly reduced (P < 0.001; Figure 5B), and the number of monoclonal clones of the conjugative transfer strain was further reduced when the concentration of LA was 2 mM, 3 mM, and 6 mM (P < 0.0001; Figure 5B), with a significant difference compared with the 1 mM group (P < 0.01; Figure 5B).
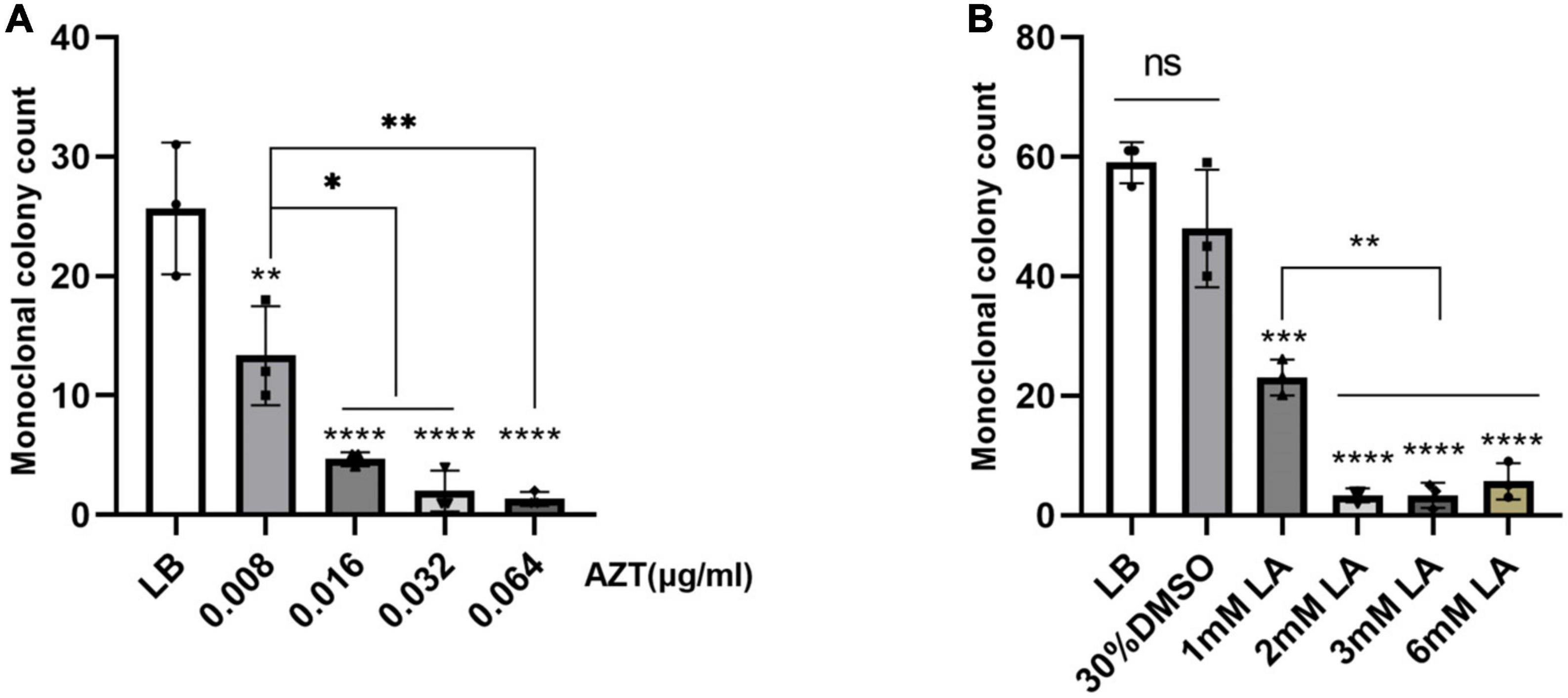
Figure 5. Inhibition of plasmid conjugation transfer experiments. The effect of drugs on plasmid conjugation transfer was evaluated by comparing the monoclonal colony count of conjugants under different drug concentrations. The LB group was the blank control group, and LA was dissolved in 30% DMSO. (A) The effect of zidovudine (AZT) on plasmid conjugation transfer; AZT was dissolved in water. (B) Effects of linoleic acid (LA) on the conjugative transfer of plasmids; LA was dissolved in 30% DMSO. Data show the mean standard deviation from a minimum of three independent experiments, each with a minimum of three biological replicates. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001, statistical method using one-way ANOVA.
To investigate the effect of antibiotics on expression of resistance genes at the mRNA level, different concentrations of antibiotics were used, and expression of resistance genes was determined by RT-qPCR. Analysis of blaCTX–M–14 and blaCTX–M–64 mRNA levels of strain Ec15103 at different concentrations of CTX showed that expression of blaCTX–M–14 and blaCTX–M–64 did not differ significantly in the presence of CTX (Figures 6A, B). Similarly, expression of blaCTX–M–14 did not change substantially in strain Ec15103-Ec600 in the presence of CTX (Figure 6C), nor did blaTEM–1 mRNA levels in strain Ec15103-EPI300-1 at different concentrations of AMP (Figure 6D).
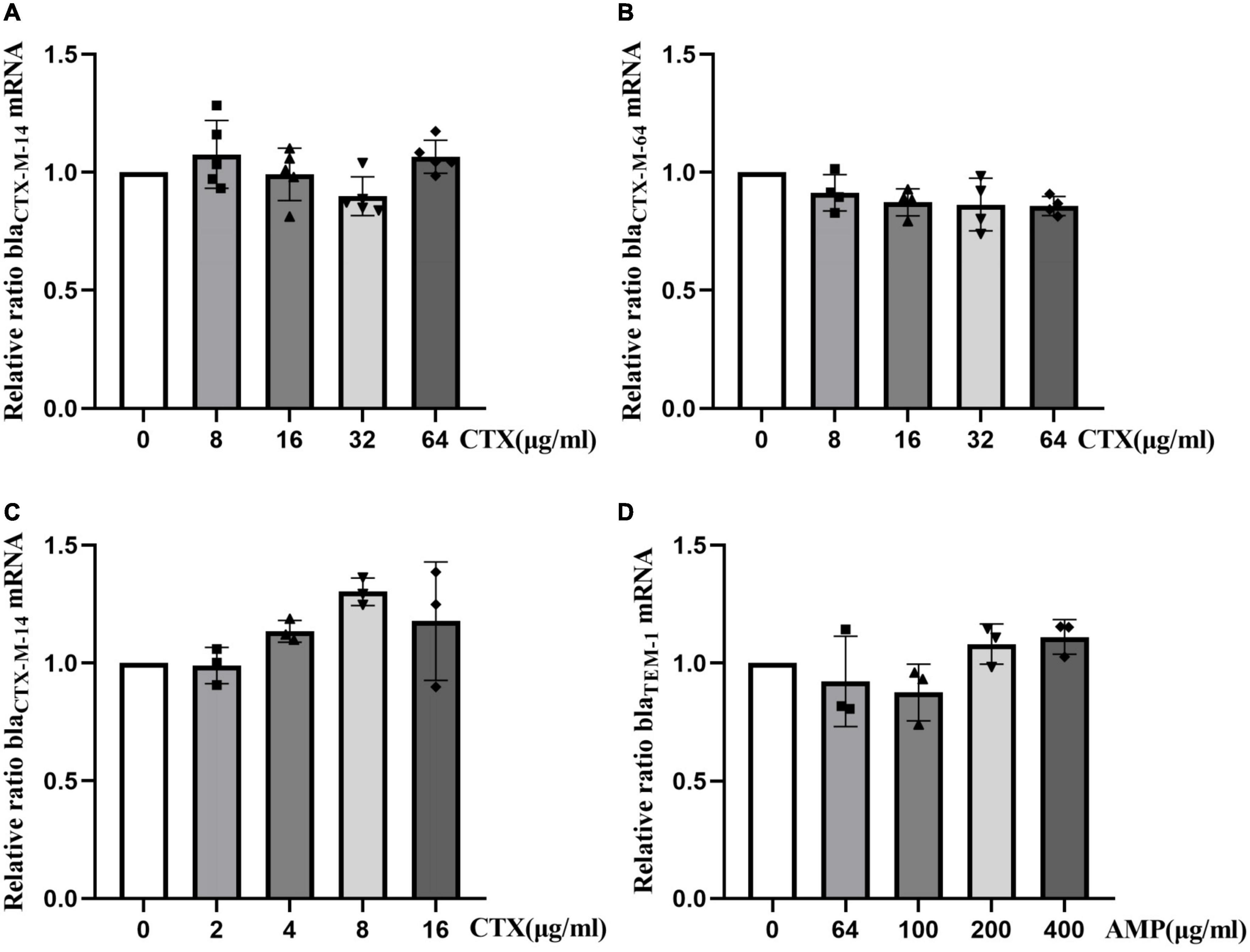
Figure 6. Relative changes in resistance gene mRNA levels in strains Ec15103, Ec15103-Ec600, and Ec15103-EPI300-1 under the influence of antibiotics. (A,B) Relative changes in blaCTX–M–14 and blaCTX–M–64 mRNA levels in strain Ec15103 under different concentrations of cefotaxime (CTX); blaCTX–M–14 is present on the Ec15103 plasmid pEc15103B and blaCTX–M–64 on the Ec15103 chromosome. (C) Relative changes in blaCTX–M–14 mRNA levels in strain Ec15103-Ec600 at different concentrations of CTX. blaCTX–M–14 is located on the Ec15103-Ec600 conjugate plasmid pEc15103B. (D) Relative changes in blaTEM–1 mRNA levels in strain Ec15103-EPI300-1 at different concentrations of ampicillin (AMP). blaTEM–1 is located on the Ec15103-EPI300-1 transconjugant plasmid pEc15103A. For at least two independent experiments, each with at least three biological replicates, the data shown represent the mean standard deviation using one-way ANOVA.
In this study, E. coli Ec15103 was isolated from the blood of a 7-day-old infant in 2015, and Ec15103 harbored two CTX-M genes: blaCTX–M–14 and blaCTX–M–64. Genome sequencing denoted two drug resistance plasmids: pEc15103A is an IncFI-type plasmid that cannot be conjugatively transferred and contains the drug resistance genes blaTEM–1, aacC2, aadA5, sul1, mph(A), sul2, strAB, and tetA(A); pEc15103B contains the conjugation transfer gene, is an IncK2/Z-type plasmid and contains blaCTX–M–14. In pEc15103B, blaCTX–M–14 is located in ΔTn6503a with a structure of ISEcp1-blaCTX–M–14-ΔIS903D. blaCTX–M–64 was located on the chromosome of strain Ec15103, and it is the first report of pathogen with blaCTX–M–64 located on its chromosome (the search terms used “blaCTX-M-64” and “chromosome”).
In plasmid conjugation transfer inhibition experiments, the conjugation transfer ability of plasmid pEc15103B was significantly inhibited by AZT and LA, which is consistent with the results of Buckner et al. (2020). In general, conjugative transfer of plasmids contributes significantly to the spread of drug resistance genes (Grohmann et al., 2003; Harrison and Brockhurst, 2012). In the post-antibiotic era, it seems reasonable to curb the spread of drug resistance genes and slow the pace of superbug production by inhibiting plasmid conjugation transfer, as the human intestine is a natural library for drug resistance gene spread (Huddleston, 2014). AZT was the first antiretroviral agent to be licensed for treating human immunodeficiency virus (HIV) infection (Rachlis, 1990), and 0.008 μg/mL is much lower than the dose of the drug in AIDS treatment. However, it can significantly inhibit the conjugative transfer of the plasmid. LA, which is an essential nutrient (Whelan and Fritsche, 2013), is very effective in inhibiting the conjugation and transfer of plasmids, and is an excellent choice. Nevertheless, these drugs may not be effective for the treatment of infection and will increase the patient’s treatment burden, and there is no animal study on their inhibitory effect.
In addition, we sought to determine whether the expression of these resistance genes increases with an increase in antibiotic concentration. Therefore, we conducted qPCR experiments to verify the relationship between expression of drug-resistance genes and the concentration of antibiotics at the mRNA level. Under the conditions of different concentrations of cefotaxime, mRNA expression levels of the genes blaCTX–M–14 and blaCTX–M–64 in strains Ec15103 and Ec15103-Ec600 did not change significantly. Similarly, under the effect of ampicillin, expression at the mRNA level of the blaTEM–1 gene of strain Ec15103-EPI300-1 was also unaffected. This is different from the description by Kjeldsen et al. (2015), probably because of differences between strains, and some studies had noted that relative expression at the mRNA level and the protein level of the blaCTX–M gene differ (Geyer et al., 2016).
Due to the presence of blaCTX–M–64 on the chromosome, strain Ec15103 exhibited resistant to ceftazidime; conversely, other conjugated and transformed strains were sensitive. blaCTX–M–64 was first reported in 2009 and found on a Shigella drug resistance plasmid in Japanese patients with diarrhea after travel to China (Nagano et al., 2009). It is considered to be a hybrid chimera of blaCTX–M–15 (CTX-M-1G) and blaCTX–M–14 (CTX-M-9G) and shows strong ceftazidime hydrolysis activity. There were relatively few subsequent reports of pathogenic bacteria carrying the blaCTX–M–64 resistance gene, and in most cases, it was found on plasmids. At present, it is generally believed that the CTX-M gene derive from the chromosome of Kluyvera spp (Poirel et al., 2002) and that it became incorporated on a plasmid through the action of mobile elements such as transposons and integrons. Therefore, it is generally believed that the CTX-M resistance genes in other bacteria are mainly located on plasmids (D’Andrea et al., 2013). However, there are an increasing number of reports of chromosomally located CTX-M genes (most of which are blaCTX–M–15 and blaCTX–M–14) recently (Irrgang et al., 2017; Zheng et al., 2021). Therefore, as the CTX-M drug resistance gene derives from the chromosome, it may again be inserted into the bacterial chromosome. In addition to E. coli, Klebsiella pneumoniae (Coelho et al., 2010), Acinetobacter baumannii (Potron et al., 2011), Proteus mirabilis (Song et al., 2011), and Salmonella (Zhang et al., 2019) all harbor CTX-M gene on the chromosome. This also demonstrates that insertion of blaCTX–M into the chromosome by a plasmid under the action of a transposon is not a random event. Indeed, bacterial resistance may be evolving in this direction. Because the presence of plasmids is not without cost to bacteria, those containing plasmids often have a relatively long replication cycle (Harrison and Brockhurst, 2012), and the plasmids may be lost during passage and reproduction. Once the resistance gene is inserted into the chromosome, there is no longer a cost, and the resistance gene will continue to replicate with each division cycle, presenting another challenge for antibiotic treatment.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, CP104274; https://www.ncbi.nlm.nih.gov/genbank/, ON324203; https://www.ncbi.nlm.nih.gov/genbank/, ON324204.
Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
LW and JF conceived the study. MY and JH performed the experiments. LW and MY analyzed the data. XL, CX, DL, and YM contributed reagents, materials, and analysis tools. LW, JF, and MY wrote the manuscript. All authors listed have made a substantial, direct, and intellectual contribution to the work, and approved it for publication.
This work was supported by the Science and Technology Department of Henan Province (212102310739) and the National Natural Science Foundation of China (82002207).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1158659/full#supplementary-material
Alekshun, M. N., and Levy, S. B. (2007). Molecular mechanisms of antibacterial multidrug resistance. Cell 128, 1037–1050. doi: 10.1016/j.cell.2007.03.004
Ambler, R. P., Coulson, A. F., Frère, J. M., Ghuysen, J. M., Joris, B., Forsman, M., et al. (1991). A standard numbering scheme for the class a beta-lactamases. Biochem. J. 276(Pt 1), 269–270.
Bauernfeind, A., Grimm, H., and Schweighart, S. (1990). A new plasmidic cefotaximase in a clinical isolate of Escherichia coli. Infection 18, 294–298. doi: 10.1007/BF01647010
Bevan, E. R., Jones, A. M., and Hawkey, P. M. (2017). Global epidemiology of CTX-M β-lactamases: Temporal and geographical shifts in genotype. J. Antimicrob. Chemother. 72, 2145–2155. doi: 10.1093/jac/dkx146
Bonnet, R. (2004). Growing group of extended-spectrum beta-lactamases: The CTX-M enzymes. Antimicrob. Agents Chemother. 48, 1–14. doi: 10.1128/AAC.48.1.1-14.2004
Buckner, M. M. C., Ciusa, M. L., Meek, R. W., Moorey, A. R., Mccallum, G. E., Prentice, E. L., et al. (2020). HIV drugs inhibit transfer of plasmids carrying extended-spectrum β-lactamase and carbapenemase genes. mBio 11:e03355-19.
Changkaew, K., Utrarachkij, F., Siripanichgon, K., Nakajima, C., Suthienkul, O., and Suzuki, Y. (2014). Characterization of antibiotic resistance in Escherichia coli isolated from shrimps and their environment. J. Food Protect. 77, 1394–1401. doi: 10.4315/0362-028X.JFP-13-510
Cheng, Q., Jiang, X., Xu, Y., Hu, L., Luo, W., Yin, Z., et al. (2019). Type 1, 2, and 1/2-hybrid IncC plasmids from China. Front. Microbiol. 10:2508. doi: 10.3389/fmicb.2019.02508
CLSI (2020). Performance Standards for Antimicrobial Susceptibility Testing: CLSI Supplement M100, 30th Edn. Wayne, PA: Clinical and Laboratory Standards Institute.
Coelho, A., González-López, J. J., MIRó, E., Alonso-TARRéS, C., Mirelis, B., Larrosa, M. N., et al. (2010). Characterisation of the CTX-M-15-encoding gene in Klebsiella pneumoniae strains from the Barcelona metropolitan area: Plasmid diversity and chromosomal integration. Int. J. Antimicrob. Agents 36, 73–78. doi: 10.1016/j.ijantimicag.2010.03.005
D’Andrea, M. M., Arena, F., Pallecchi, L., and Rossolini, G. M. (2013). CTX-M-type β-lactamases: A successful story of antibiotic resistance. Int. J. Med. Microbiol. 303, 305–317.
Drieux, L., Brossier, F., Sougakoff, W., and Jarlier, V. (2008). Phenotypic detection of extended-spectrum beta-lactamase production in Enterobacteriaceae: Review and bench guide. Clin. Microbiol. Infect. 14(Suppl. 1) 90–103. doi: 10.1111/j.1469-0691.2007.01846.x
Feng, J., Yin, Z., Zhao, Q., Zhao, Y., Zhang, D., Jiang, X., et al. (2017). Genomic characterization of novel IncFII-type multidrug resistant plasmids p0716-KPC and p12181-KPC from Klebsiella pneumoniae. Sci. Rep. 7:5830. doi: 10.1038/s41598-017-06283-z
Geyer, C. N., Fowler, R. C., Johnson, J. R., Johnston, B., Weissman, S. J., Hawkey, P., et al. (2016). Evaluation of CTX-M steady-state mRNA, mRNA half-life and protein production in various STs of Escherichia coli. J. Antimicrob. Chemother. 71, 607–616. doi: 10.1093/jac/dkv388
Grohmann, E., Muth, G., and Espinosa, M. (2003). Conjugative plasmid transfer in gram-positive bacteria. Microbiol. Mol. Biol. Rev. 67, 277–301.
Harrison, E., and Brockhurst, M. A. (2012). Plasmid-mediated horizontal gene transfer is a coevolutionary process. Trends Microbiol. 20, 262–267. doi: 10.1016/j.tim.2012.04.003
Huddleston, J. R. (2014). Horizontal gene transfer in the human gastrointestinal tract: Potential spread of antibiotic resistance genes. Infect. Drug Resist. 7, 167–176.
Hussain, H. I., Aqib, A. I., Seleem, M. N., Shabbir, M. A., Hao, H., Iqbal, Z., et al. (2021). Genetic basis of molecular mechanisms in β-lactam resistant gram-negative bacteria. Microb. Pathog. 158:105040.
Irrgang, A., Falgenhauer, L., Fischer, J., Ghosh, H., Guiral, E., Guerra, B., et al. (2017). CTX-M-15-producing E. coli isolates from food products in germany are mainly associated with an IncF-type plasmid and belong to two predominant clonal E. coli lineages. Front. Microbiol. 8:2318. doi: 10.3389/fmicb.2017.02318
Kjeldsen, T. S., Overgaard, M., Nielsen, S. S., Bortolaia, V., Jelsbak, L., Sommer, M., et al. (2015). CTX-M-1 β-lactamase expression in Escherichia coli is dependent on cefotaxime concentration, growth phase and gene location. J. Antimicrob. Chemother. 70, 62–70.
Liu, Y., Tong, Z., Shi, J., Jia, Y., Yang, K., and Wang, Z. (2020). Correlation between exogenous compounds and the horizontal transfer of plasmid-borne antibiotic resistance genes. Microorganisms 8:1211. doi: 10.3390/microorganisms8081211
Mohsin, M., Tanaka, K., Kawahara, R., Kondo, S., Noguchi, H., Motooka, D., et al. (2020). Whole-genome sequencing and comparative analysis of the genomes of Bacteroides thetaiotaomicron and Escherichia coli isolated from a healthy resident in Vietnam. J. Glob. Antimicrob. Resist. 21, 65–67. doi: 10.1016/j.jgar.2020.02.034
Nagano, Y., Nagano, N., Wachino, J., Ishikawa, K., and Arakawa, Y. (2009). Novel chimeric beta-lactamase CTX-M-64, a hybrid of CTX-M-15-like and CTX-M-14 beta-lactamases, found in a Shigella sonnei strain resistant to various oxyimino-cephalosporins, including ceftazidime. Antimicrob. Agents Chemother. 53, 69–74. doi: 10.1128/AAC.00227-08
Partridge, S. R. (2011). Analysis of antibiotic resistance regions in Gram-negative bacteria. FEMS Microbiol. Rev. 35, 820–855.
Partridge, S. R., and Hall, R. M. (2005). Evolution of transposons containing blaTEM genes. Antimicrob. Agents Chemother. 49, 1267–1268.
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45.
Pitout, J. D., Nordmann, P., Laupland, K. B., and Poirel, L. (2005). Emergence of Enterobacteriaceae producing extended-spectrum beta-lactamases (ESBLs) in the community. J. Antimicrob. Chemother. 56, 52–59. doi: 10.1093/jac/dki166
Poirel, L., KäMPFER, P., and Nordmann, P. (2002). Chromosome-encoded Ambler class A beta-lactamase of Kluyvera georgiana, a probable progenitor of a subgroup of CTX-M extended-spectrum beta-lactamases. Antimicrob. Agents Chemother. 46, 4038–4040. doi: 10.1128/AAC.46.12.4038-4040.2002
Potron, A., Munoz-Price, L. S., Nordmann, P., Cleary, T., and Poirel, L. (2011). Genetic features of CTX-M-15-producing Acinetobacter baumannii from Haiti. Antimicrob. Agents Chemother. 55, 5946–5948. doi: 10.1128/AAC.05124-11
Reid, C. J., Chowdhury, P. R., and Djordjevic, S. P. (2015). Tn6026 and Tn6029 are found in complex resistance regions mobilised by diverse plasmids and chromosomal islands in multiple antibiotic resistant Enterobacteriaceae. Plasmid 80, 127–137. doi: 10.1016/j.plasmid.2015.04.005
Rossolini, G. M., D’Andrea, M. M., and Mugnaioli, C. (2008). The spread of CTX-M-type extended-spectrum beta-lactamases. Clin. Microbiol. Infect. 14(Suppl. 1) 33–41.
Schmitt, R., Bernhard, E., and Mattes, R. (1979). Characterisation of Tn1721, a new transposon containing tetracycline resistance genes capable of amplification. Mol. General Genet. 172, 53–65. doi: 10.1007/BF00276215
Song, W., Kim, J., Bae, I. K., Jeong, S. H., Seo, Y. H., Shin, J. H., et al. (2011). Chromosome-encoded AmpC and CTX-M extended-spectrum β-lactamases in clinical isolates of Proteus mirabilis from Korea. Antimicrob Agents Chemother. 55, 1414–1419.
Venturini, C., Hassan, K. A., Roy, C., Paulsen, I. T., Walker, M. J., and Djordjevic, S. P. (2013). Sequences of two related multiple antibiotic resistance virulence plasmids sharing a unique IS26-related molecular signature isolated from different Escherichia coli pathotypes from different hosts. PLoS One 8:e78862. doi: 10.1371/journal.pone.0078862
Zhang, C. Z., Ding, X. M., Lin, X. L., Sun, R. Y., Lu, Y. W., Cai, R. M., et al. (2019). The emergence of chromosomally located bla (CTX-M-55) in Salmonella from foodborne animals in China. Front. Microbiol. 10:1268. doi: 10.3389/fmicb.2019.01268
Keywords: Escherichia coli, antimicrobial resistance, ESBLs, CTX-M-14, CTX-M-64
Citation: Yang M, Liu D, Li X, Xiao C, Mao Y, He J, Feng J and Wang L (2023) Characterizations of blaCTX–M–14 and blaCTX–M–64 in a clinical isolate of Escherichia coli from China. Front. Microbiol. 14:1158659. doi: 10.3389/fmicb.2023.1158659
Received: 04 February 2023; Accepted: 02 May 2023;
Published: 18 May 2023.
Edited by:
Asad U. Khan, Aligarh Muslim University, IndiaReviewed by:
Shazi Shakil, King Abdulaziz University, Saudi ArabiaCopyright © 2023 Yang, Liu, Li, Xiao, Mao, He, Feng and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Li Wang, d2FuZ2xpODUxMjE3QDE2My5jb20=; Jiao Feng, ZmVuZ2ppYW8tMTIxOUAxNjMuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.