- 1Beijing Advanced Innovation Center for Food Nutrition and Human Health, Beijing Engineering and Technology Research Center of Food Additives, Beijing Technology and Business University, Beijing, China
- 2Department of Agriculture, University of Ioannina, Ioannina, Greece
- 3Department of Biotechnology, Faculty of Science and Technology, University of Central Punjab, Lahore, Pakistan
- 4Department of Pharmacology and Toxicology, College of Pharmacy, King Saud University, Riyadh, Saudi Arabia
The comparative genomic analysis of Lactiplantibacillus plantarum YW11 (L. plantarum YW11) isolated from Tibetan kefir involves comparison of the complete genome sequences of the isolated strain with other closely related L. plantarum strains. This type of analysis can be used to identify the genetic diversity among strains and to explore the genetic characteristics of the YW11 strain. The genome of L. plantarum YW11 was found to be composed of a circular single chromosome of 4,597,470 bp with a G + C content of 43.2%. A total of 4,278 open reading frames (ORFs) were identified in the genome and the coding density was found to be 87.8%. A comparative genomic analysis was conducted using two other L. plantarum strains, L. plantarum C11 and L. plantarum LMG21703. Genomic comparison revealed that L. plantarum YW11 shared 72.7 and 75.2% of gene content with L. plantarum C11 and L. plantarum LMG21703, respectively. Most of the genes shared between the three L. plantarum strains were involved in carbohydrate metabolism, energy production and conversion, amino acid metabolism, and transcription. In this analysis, 10 previously sequenced entire genomes of the species were compared using an in-silico technique to discover genomic divergence in genes linked with carbohydrate intake and their potential adaptations to distinct human intestinal environments. The subspecies pan-genome was open, which correlated with its extraordinary capacity to colonize several environments. Phylogenetic analysis revealed that the novel genomes were homogenously grouped among subspecies of l Lactiplantibacillus. L. plantarum was resistant to cefoxitin, erythromycin, and metronidazole, inhibited pathogens including Listeria monocytogenes, Clostridium difficile, Vibrio cholera, and others, and had excellent aerotolerance, which is useful for industrial operations. The comparative genomic analysis of L. plantarum YW11 isolated from Tibetan kefir can provide insights into the genetic characteristics of the strain, which can be used to further understand its role in the production of kefir.
1. Introduction
Lactic acid bacteria (LAB) are a group of microorganisms that are found everywhere in nature. Since they possess probiotic and antimicrobial properties, various species of LAB are added to a wide variety of foods to provide consumers with the opportunity to reap the associated health benefits (Sarwar et al., 2018; Rodrigo-Torres et al., 2019; Brandt et al., 2020; Tenea and Ortega, 2021). In addition, LAB species help to the safety of food by preventing the spread of microbes that cause unwanted spoiling or that are harmful. These organisms are also commonly employed in the fermentation procedures that are used to produce food. Barrangou et al. (2011), Fernández et al. (2013), Swain et al. (2014), Yépez et al. (2017), Adesulu-Dahunsi et al. (2018), and Goel et al. (2020). Additionally, LAB species are used in the medical and pharmaceutical industries, as well as in healthcare. LAB have demonstrated a variety of health promoting properties which can be used against intestinal illness, including inflammatory bowel diseases (IBD), as a result of their demonstrated antibacterial, immune modulating, and ability to control gut flora activities and these have been confirmed by different researchers (Yonekura et al., 2009; Hojsak et al., 2010; Kwon et al., 2010; Joo et al., 2011; Zhang et al., 2018). This is because these bacterial strains can regulate gut flora and control the bacteria that live there. Lactobacillus species are generally regarded as safe (GRAS) (Sarwar et al., 2018) for their usage in human bodies as well as their use in the food sector for an extended period as starter cultures. Among the lactobacillus species, lactiplantibacillus plantarum (previously known as lactobacillus plantarum) is the species that has received the most attention from researchers. It is possible to obtain lactiplantibacillus plantarum from a variety of sources, such as plant matter, fermented foods (yoghurt, pickles, cheese), meat products, fruit juices, the gastrointestinal tract of both humans and animals, and wine. In addition to that, this species is very useful for the fermentation of a wide range of foods (Stefanovic et al., 2017; Zhou et al., 2021; Wang et al., 2022).
The probiotic qualities of these strains are primarily responsible for all these activities, and because to the health promoting features that these strains possess, they have garnered the interest of researchers from all over the world (Jeong et al., 2022). More than 90 percent of the market for probiotics throughout the world was held by human products in 2021. According to the findings of experts, the worldwide market for probiotics might be worth $3.5 billion by the year 2026 (Wang et al., 2021). L. plantarum is a bacterium that dwells in the gastrointestinal tract (Kleerebezem et al., 2003; Aziz et al., 2022). It may be found in nearly any kind of environment. There are approximately 400 different bacterial species that make up the human stomach related framework. Some of these bacteria include L. acidophilus (Hatami et al., 2022), L. pentosus, L. brevis, L. lactis (Ashaolu and Reale, 2020), L. amylovorus, L. casei, L. bulgaricus (Albayrak and Duran, 2021), L. fermentum, L. plantarum and L. rhamnosus produces extracellular, exopolysaccharides, bacteriocins and lipoteichoic acids (Gupta et al., 2021).
The growing number of lactiplantibacillus strain genome sequences has shown their genetic potential for probiotic characteristics and adaptation to varied environmental conditions and stressors. Our Tibetan kefir strain Lactiplantibacillus plantarum YW11 regulates modulatory effects on gut dysbacteriosis, improves immunological response, and reduces inflammatory bowel illness, according to our newest findings (IBD) (Jian et al., 2020; Zhang et al., 2020). In addition to that it was also evident from another study that the L. plantarum YW11 has good tolerance to acid and bile stress (Jian et al., 2017). Correspondingly, we have also found that L. plantarum YW11 may be employed as a functional agent in the production of fermented dairy products with better textural stability and bioactivities such as cholesterol reducing, antioxidant, and antibiofilm properties (Zhang et al., 2020, 2022). Similarly, this strain L. plantarum YW11 has the competency of biotransformation of linoleic acid (LA) into conjugated linoleic acid (CLA) (Aziz et al., 2020). Most study has focused on viable probiotic strain effects and mechanisms. Scientists are growing interested in employing probiotics as immunologically active, microbiologically non-viable medications. It may be more effective, viable, and safer for therapeutic probiotic usage due to safety problems with the active metabolic form favoring bacterial translocation. The risk favored active metabolic form may explain these advantages. However, its genetic base for probiotic properties and adaptability is still mostly recognized (Moradi et al., 2020; Teame et al., 2020; De Jesus et al., 2022). Genomic-level studies can provide insights into the primary genetic factors and molecular mechanisms associated with the probiotic characteristics of these microorganisms, such as gastrointestinal tract survival, pathogen inhibition, and immunoregulation GIT survival, pathogen inhibition, and immunoregulation (Ventura et al., 2012; Salvetti and O’Toole, 2018; Castro-López et al., 2021).
Pan-probiosis, which compares the genomes of numerous probiotic bacterial strains, employs comparative genomics as an additional tool. This research aims to identify the best probiotic bacteria strains. Pan-genomic derivatives are a technique for discovering genes linked with probiotic properties that are either conserved across all strains of a certain bacteria or unique to a given genus or species (Rodenes et al., 2022). All known bacterial strains either have these genes, or all but one of them do not. Through the integration of phylogenomic research, studies can link genotypes and phenotypes to specific strains, enabling the use of those strains for specialized medical or biotechnological applications. This line of reasoning has been used by researchers to explain the probiotic profile of the L. plantarum YW11 strain. Some of the researchers used a particular technique, while others went in a completely different direction (Shin et al., 2022). Recent studies on them have given us more information about the probiotic potential of recently discovered species like Lactobacillus helveticus (Alessandri et al., 2022). It is difficult to assert that we have a firm grasp on the subject given the genetic pathways used to metabolize a wide variety of carbohydrates in the gut microbiota of newborns and adults, as well as the organism’s genomic plasticity. Even though we now have a better understanding of how L. plantarum YW11 adapts to the human GI tract, it would be premature to say that we currently have a firm grasp on the topic. The adaptability of the creature’s genome is responsible for this special quality. Therefore, it is crucial that this research includes genomes with distinctive traits. The genetic foundations of L. plantarum YW11, which survives in the various ecological niches that make up the human gut microbiome, are being investigated using comparative genomic analysis (Alessandri et al., 2022; Asarina et al., 2022; Chaudhary et al., 2022; Hebert and Meglécz, 2022; McPherson et al., 2022; Valdez-Baez et al., 2022; Wang et al., 2022; Xiang and Li, 2022). When analyzing these genetic roots, this context is very important. It also contains four additional strains that were isolated from young people in Chile and demonstrated a broad range of adaptability to the host using an in-silico method. Chile provided the first mention of the appearance of these novel strains (Alessandri et al., 2022; McPherson et al., 2022; Wang et al., 2022).
To this end, we attempted to characterize the functional genes of L. plantarum YW11 and other genomes with reported probiotic effects, in addition to other biological traits that may relate to the distinct host health advantages of this strain. We also checked for things like hydrophobic cell walls, antibiotic resistance, and antagonistic potential, and we examined cell growth. In that study, L. plantarum was shown to be resistant to the antibiotics cefoxitin, erythromycin, and metronidazole; to have a high inhibition rate against pathogens (including Listeria monocytogenes, Clostridium difficile, Vibrio cholera, and others); and to have a high aerotolerance, which is an advantageous property for industrial processes (Alessandri et al., 2022; Asarina et al., 2022; McPherson et al., 2022; Valdez-Baez et al., 2022; Wang et al., 2022). The genome project’s findings may shed light on some of these traits and mechanisms, paving the way for future research. The purpose of this research was to conduct a comparative genome analysis of this strain with 10 previously sequenced whole genomes of the species, and by searching for genes associated with favorable features.
2. Materials and methods
2.1. Analysis of Lactiplantibacillus plantarum YW11 comparative genome
The whole of the L. plantarum YW11 genome was submitted to GenBank and assigned the accession number.1 The nucleotide FASTA format was utilized in order to retrieve all 10 of the entire genome sequences of L. plantarum that can be found in the NCBI GenBank database (Hebert and Meglécz, 2022). All genomes were annotated using Prokka v1.14.5. The L. plantarum YW11 genome, along with the other fully sequenced genomes of the species, was used to conduct a synteny analysis. Several whole-genome sequence alignments were performed with the help of the implemented version of Mauve (v2.4) (Chaudhary et al., 2022).
2.2. Antibiotic resistance genes prediction
The NCBI-AMRFinderPlus, CARD, ARG-ANNOT, Resfinder, and MEGARES 2.0 databases were searched using the ABRIcate v1.0.1 software (Xiang and Li, 2022) in order to locate antibiotic resistance genes for the purpose of validating the accuracy of antibiotic resistance gene prediction (last update of databases: September 2022).
2.3. Taxonomy, phylogenomics, and evolutionary analysis
Calculations were made to determine the average levels of nucleotide similarity (ANI) between the 10 genomes of L. plantarum and the outgroup species (Alkalay-Oren et al., 2022). The phylogenetic tree was constructed by applying the Codon Tree Test method developed by the Pathosystems Resource Integration Center (PATRIC) (2viewed on 28 September 2022) to many genes, each of which only had a single copy of the gene (Spergser et al., 2022). This allowed for the phylogenomic tree to be accurate and reliable. The RaxML program utilized a total of 100 repetitions in order to calculate the support values (Batarseh et al., 2022).
2.4. Pangenome analysis
Data for 10 genomes retrieved from the NCBI RefSeq database were analyzed by panX to do the computation for the pangenome size (Ding et al., 2018). The analysis with the default settings and an identity cut-off of 99% was run. This was done while taking into consideration an abnormally high average GC content, which was equal to two times the standard deviation. The Cluster of Orthologous Genes (COG) designations were used to carry out the functional analysis and to explore the evolutionary relationships between gene clusters, and to identify potentially related gene clusters (Kamau et al., 2020). The analysis was used to illustrate the number of distinct genes possessed by each L. plantarum strain, and to analyze the biosynthetic pathways of gene clusters, and to identify potential new pathways.
2.5. Identifying genes related to probiotic features
The research that has been conducted on the genera L. plantarum and Lactobacillus has resulted in the discovery of genes that are involved in the mechanisms of adhesion, resistance to stress conditions (acid, bile salts, heat, and osmotic), the repair and protection of DNA and proteins, and the production of vitamins. These genes are also responsible to produce vitamins. Using a piece of software known as the Basic Local Alignment Search Tool (BLAST),3 we were able to match the protein sequences of these genes with the genome that we are now researching (Gaina et al., 2022). The alignment has to achieve at least 70% identity and a cutoff of 1E5 to be successful.
3. Results
3.1. Antibiotic resistance genes prediction
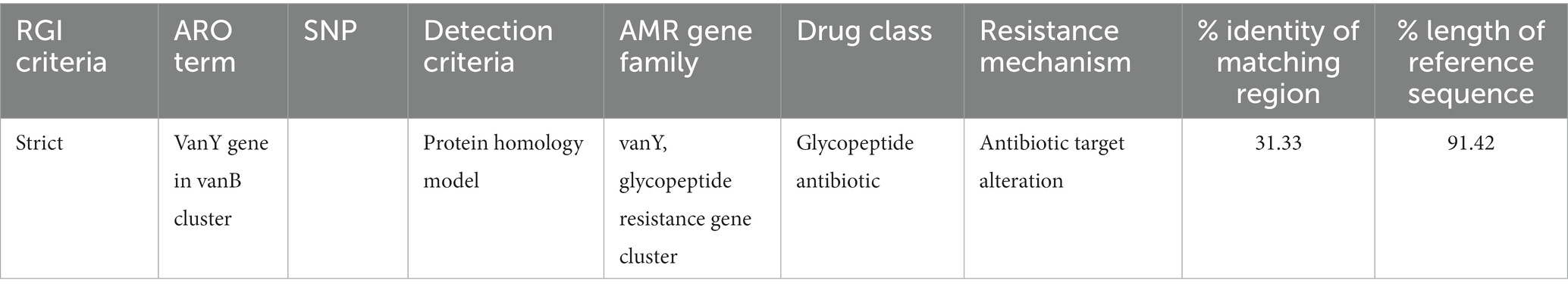
The discovery of genes that confer resistance to antibiotics led to the identification of two genes in total: vanY and vanB in Figure 1. The significance of the coverage percentage for each and every hit was more than 91.42 percent (Table 1).
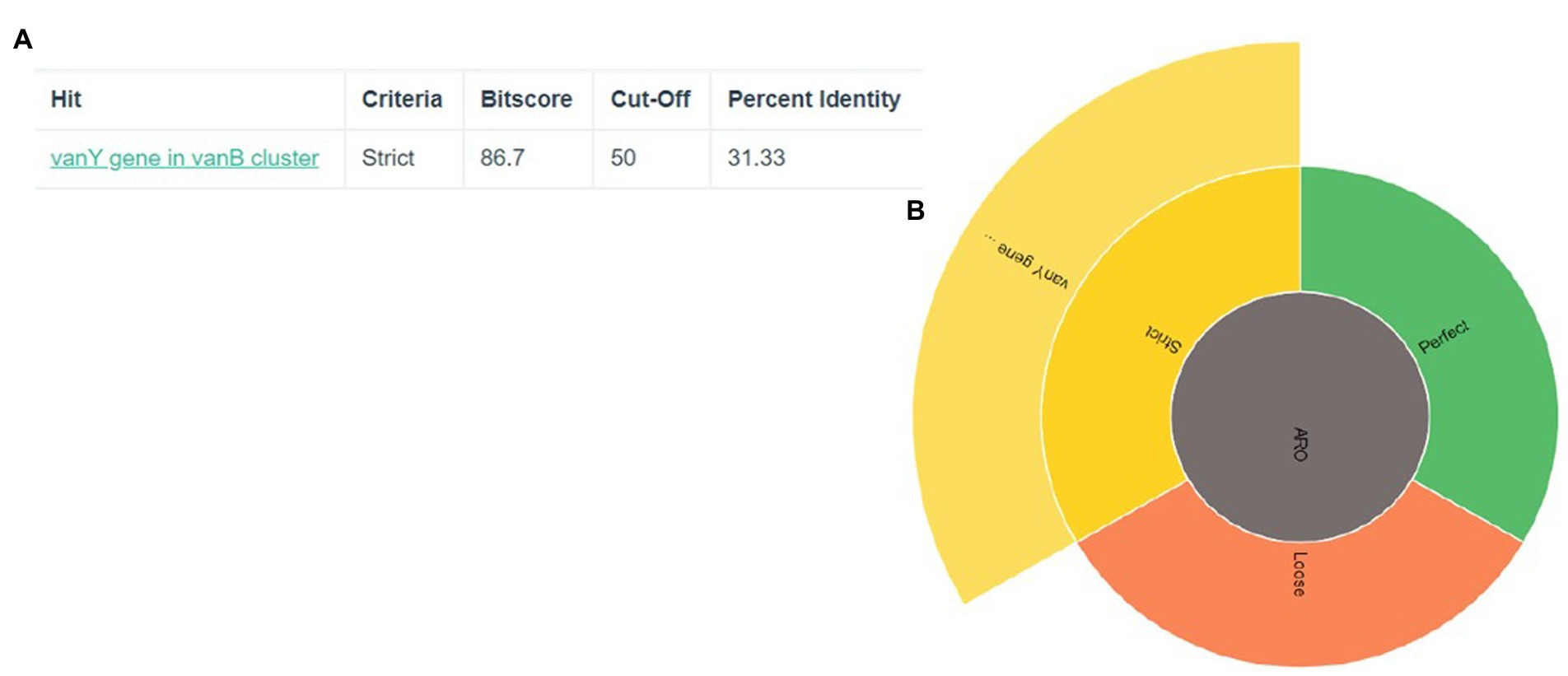
Figure 1. Prediction of antibiotic resistance genes (A) The criteria, cut-off and percent identity (B) the vanY gene show in strict area of ARO.
3.2. 3.2. Multiple whole genome sequence alignments
L. plantarum YW11 had a circular chromosome that was 2.99 Mbp in size and had 44.5% GC in its genome when it was completely sequenced. The genome assembly started off with a total of six contigs and a N50 value of 2,991,907. However, after the gaps in the sequence were filled in, it was able to retrieve the whole genome in a single contig. During the annotation procedure, a total of 2,832 genes, 68 transfer RNAs, 16 ribosomal RNAs, and 1986 CDS were found. The CDS represented 907 putative proteins. Concerning the origin of the data, most samples were collected from the feces of children, while just a few were taken from the feces of adults, vagina, the environment, and human breast milk. L. plantarum YW11 demonstrated collinearity of the gene blocks with most of the other genomes that were assessed while the conservation of the structure of the genome was being evaluated. In this regard, additional L. plantarum strains exhibited both a major and a small inversion in the genome’s core region, respectively.
3.3. Phylogenomic analysis
The phylogenetic tree organized the genomes into clusters according to their prior taxonomic structure. This was done to represent the divergence that occurred among the branches of the subspecies that all descended from the same ancestor as shows in Figure 2. We found, as was to be predicted, that most genomes were located in a manner that allowed for uniform segregation into subspecies plantarum taxonomic categories.
3.4. Average nucleotide identity
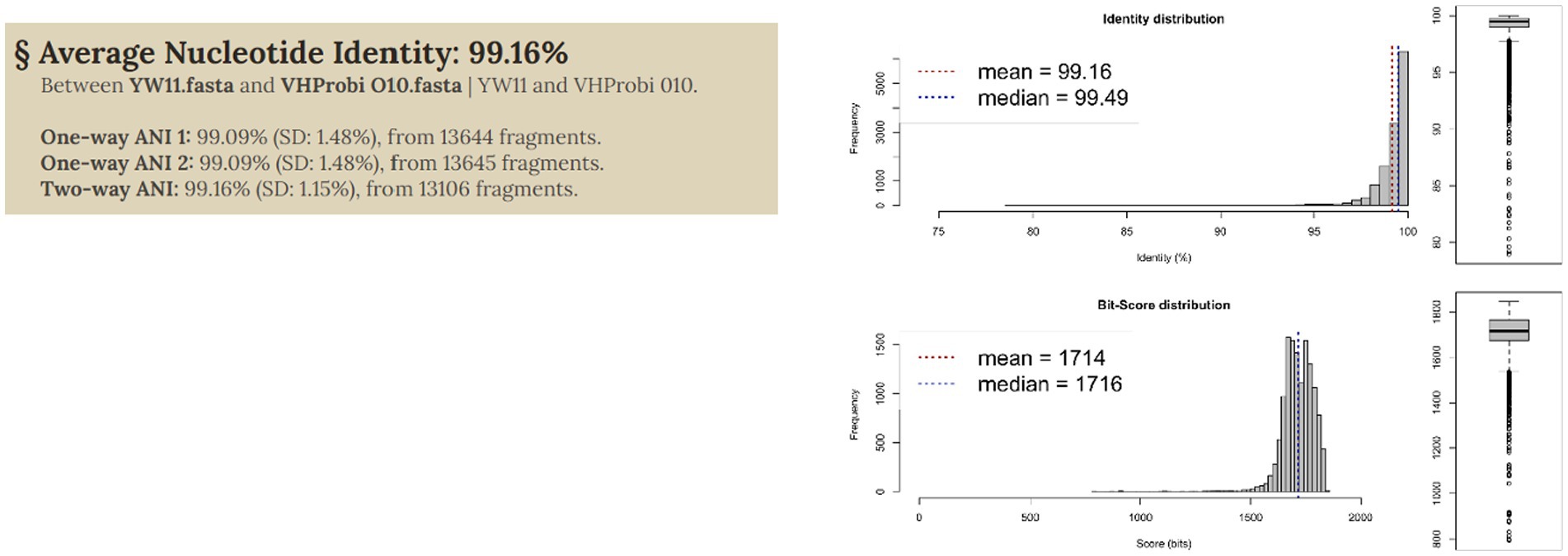
In order to assess the genomic link between the several L. plantarum genomes, an average nucleotide identity (ANI) analysis was carried out on each of the genomes that were selected for this research (both from public databases and the novel strains). This was done in order to define the genomic relationship among the L. plantarum genomes. The genomes were found to be substantially grouped into an ANI structure, with values reaching more than 0.991 as shows in Figure 3. It is interesting to note that some genomes came out with an ANI range lower than 0.994. Other strains of L. plantarum, for example, were isolated from calf feces and had the lowest ANI value. This elucidates the genetic difference that exists between strains that occupy the animal gut microbiome and those that inhabit the human gut microbiome. When it came to L. plantarum YW11, the ANI value was close to 0.995.
Figure 3 illustrates the taxonomic characterization achieved by doing similarity comparisons based on ANI values that were computed for each of the 10 strains of L. plantarum. In every single one of the comparisons that L. plantarum YW11 underwent with other strains of L. plantarum, the ANI values ranged between 0.94 and 0.96 when grouped with these strains, showing its high level of nucleotide similarity with this species.
3.5. Probiotic genes identification
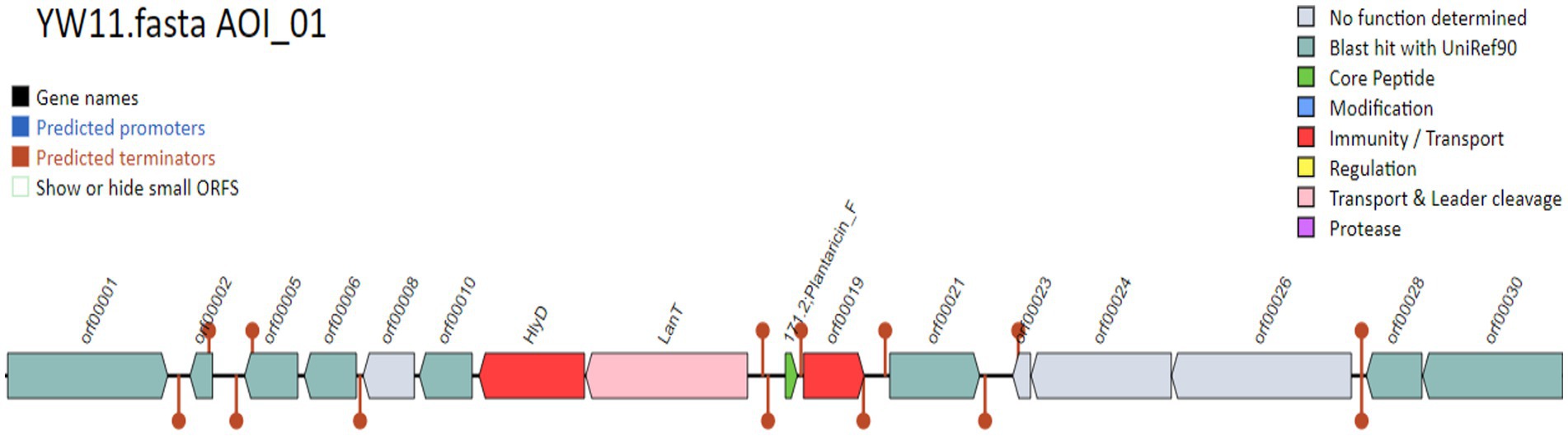
According to the display run summary and file information in Tables 2,3, the investigation uncovered a total of 16 condensed genes that were associated with adhesion. Among these genes were sequences that codified for sortases. The condensed genes include Immunity protein membrane-bound protease CAAX family, Glycerol uptake facilitator protein 3 OS = Lactobacillus plantarum, DNA helicase IV, and Alpha-glycerophosphate oxidase as display in Figure 4. Two sequences were codified for Accessory factor for ABC-transporter PlnH and Bacteriocin ABC-transporter, ATP-binding and permease protein PlnG shown in Table 4.
3.6. Binary pan genome
The calculation of the size of the pangenome found a total of 4,477 genes, based on how they were distributed throughout the 10 genomes. According to the ANI analysis, the fact that L. plantarum YW11 formed a well-supported clade with L. plantarum YW11 81 in the phylogenomic tree that was constructed using single-copy genes suggests that this strain is closely related as shows in Figure 5B. While performing experiment, L. plantarum strains were shown to be effective probiotics and revealed a connection to other clades as display in Figure 5A.
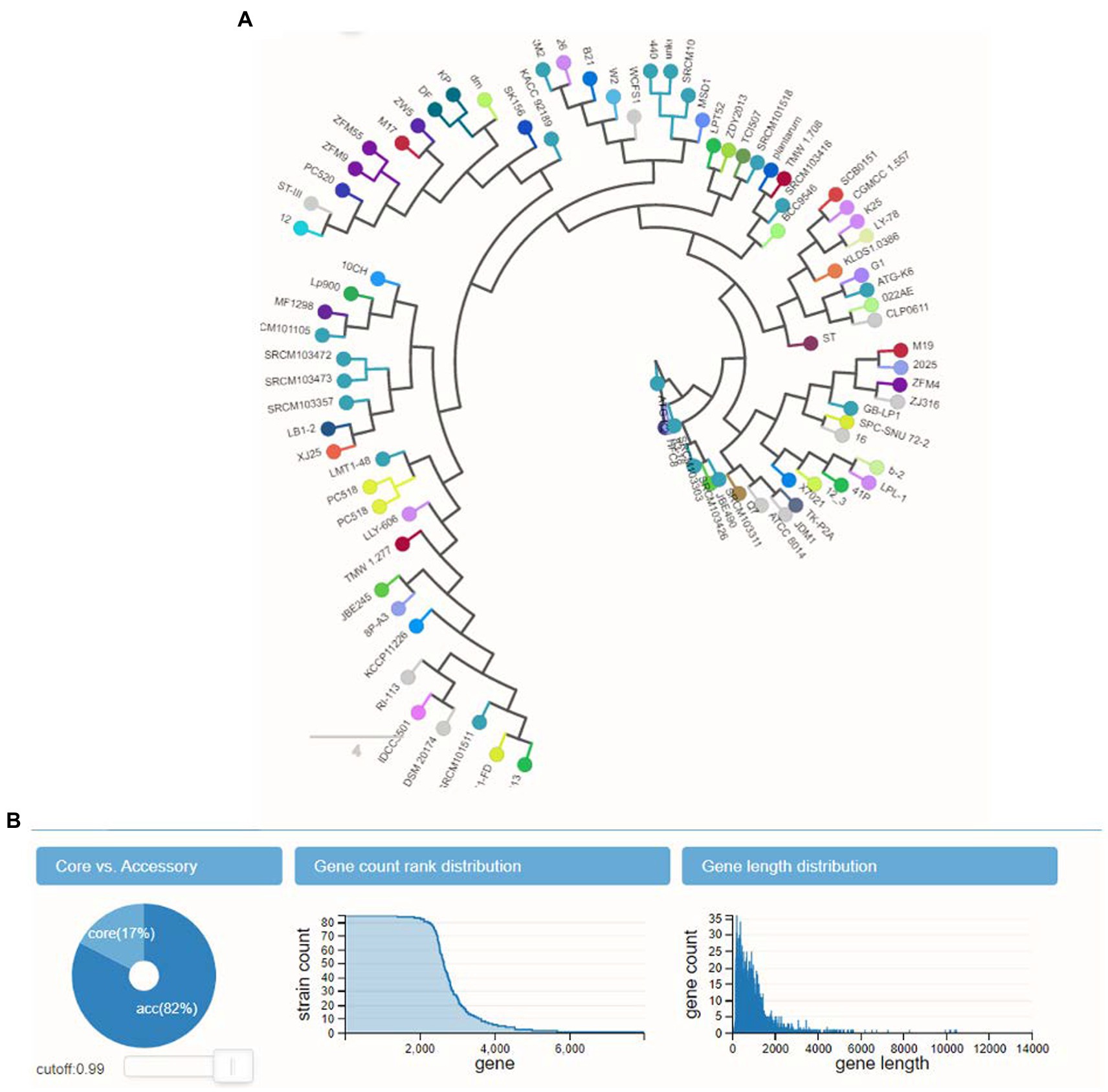
Figure 5. The panX outcomes for the pangenome analysis and exploration. (A) L. plantarum strains were shown to be effective probiotics and revealed a connection to other clades (B) the gene counts distribution and length distribution analysis.
4. Discussion
Comparative genomics studies on various strains of L. plantarum may provide information on how different taxonomic groups adapt to their habitat and which of their traits are required for such adaptations. These modifications might be related to the host or to the geological and geographical environment in which they dwell. The taxa of L. plantarum exhibit greater genomic variety than previously believed, according to earlier findings from pangenome research (Kamau et al., 2020; Gaina et al., 2022; Li et al., 2022). A closed pangenome is regarded as a finalized pangenome in which the number of genomes does not change even if new genomes are added to it, as opposed to an open pangenome, that expands every time a new genome is added. It has been proposed that whether the pangenome is open or closed is closely tied to the mode of life of the bacterial species being studied (Surve et al., 2022). Given this perspective, animals with an open pangenome are which live in various habitats and have a variety of genetic exchange pathways. Salmonellae, Escherichia coli, Helicobacter pylori, Streptococci, and Meningococci pangenomes are a few examples. As a result, they have a restricted selection of genes available to them. Examples of closed pangenomes are Mycobacterium TB, Bacillus anthracis, and Chlamydia trachomatis (Gaina et al., 2022; Li et al., 2022; Liu et al., 2022; Surve et al., 2022; Syrokou et al., 2022).
The gastrointestinal tract includes the oral cavity, large intestine, stomach, and small intestine of the human are some of the locations where L. plantarum YW11 may be found. It stands out among gut microbes because it is a major component of the gut microbiota in newborn humans and is frequently found in the gut microbiota of adults (Teame et al., 2020). Humans are the only species with this characteristic (Kim et al., 2022). Previous research indicates that by examining the core genome, which is a genetically conserved section, we may be able to identify subspecies-specific adaptations. The COGs discovered in each strain isolated from Chileans in this study were found to be among the higher percentages allocated to the functional category “carbohydrate transport and metabolism (G)” in the shell gene set (Li et al., 2022). The metabolism of carbohydrates falls under this category. To acquire nutrients and subsequently carve out an ecological niche for themselves, these functions are crucial in controlling the contact with the host and the environment (Carpi et al., 2022; Yin et al., 2022; Aziz et al., 2023).
It’s remarkable how different conclusions can be drawn from the phylogenetic analysis of L. plantarum YW11. The L. plantarum YW11 strain was classified as a subspecies of lactobacillus in both the phylogenetic tree we built and the original annotation. L. plantarum YW11, which was isolated from an infant’s gut microbiota, thrived in neutral HMOs such as LNT and LNnT. Previous studies suggest L. plantarum YW11 may be a niche adaptation rather than a horizontal gene transfer (Syrokou et al., 2022). L. plantarum YW11 also grouped further away from Chilean isolated strains with an ANI value <0.98. Its genome is closer to other L. plantarum strain, which was isolated from a calf’s stomach microbiota. Albert et al. study’s grouped the L. plantarum YW11 genome like the infantis subspecies. Although most L. plantarum genomes belong to the subspecies lactobacillus, some strains, such as YW11, may have had a unique genomic architecture to adapt to their ecological niches (Spergser et al., 2022).
5. Conclusion
In our recently published study we demonstrated that the L. plantarum YW11 genome we found exopolysaccharides including terpenes, T3PKS and RiPP like regions. On further investigations of this genome with other species, e.g, enterococcus, bacillus cereus and halo bacillus we noticed that L. plantarum YW11 genome has two bacteriocins Streptin and Ruminococcin-A, were further analyzed for their probiotic role via docking with virulent proteins of pathogenic bacterial species which confirmed that both bacteriocins are potent inhibitors of the target bacterial pathogens and help the human host elicit a strong immune response against pathogenic bacteria. In this study we found out that a carbohydrate enzyme in the L. plantarum YW11 genome. Similar enzymes were discovered in L. plantarum strains during previous studies. The previous research demonstrated that particular strains of L. plantarum may selectively constrain the development of the baby’s gut microbiota’s carbohydrate-mediated symbiosis. The conclusions that were reached from the study reflected these findings. L. plantarum’s metabolic abilities are critical for trophic interactions with other commensal bacterial populations, promoting a mutualistic environment in their host, allowing cross-feeding connections between microorganisms, and maintaining appropriate gut microbiome growth. Cross-feeding interactions are those in which one microbe consumes the nutrients from another microbe. This genome L. plantarum YW11 is of very great interest and can be helpful for food safety, food fermentation and food starter cultures. Moreover, its probiotic capabilities cannot be ignored and several in-vitro and in-vivo activities can be performed on it.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Author contributions
TA, MN, KJ, MS, and YZ: conceptualization. AA, MN, KJ, MS, and YZ: methodology and investigation. MA: software. AA: validation. TA: formal analysis and data curation. YZ, MA, and AA: resources. TA and MN: writing—original draft preparation and writing—review and editing. KJ, AS, AA, and MS: visualization. YZ: supervision and funding acquisition. AA and MA: project administration. All authors contributed to the article and approved the submitted version.
Funding
This research work was financially supported by National Natural Science Foundation of China (Grant no. 31871823) and National Key Research and Development Program of China (2017YFE0131800).
Acknowledgments
The authors greatly acknowledge and express their gratitude to the Researchers Supporting Project number (RSP2023R462), King Saud University, Riyadh, Saudi Arabia.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
Adesulu-Dahunsi, A., Jeyaram, K., and Sanni, A. (2018). Probiotic and technological properties of exopolysaccharide producing lactic acid bacteria isolated from cereal-based nigerian fermented food products. Food Control 92, 225–231. doi: 10.1016/j.foodcont.2018.04.062
Albayrak, Ç. B., and Duran, M. (2021). Isolation and characterization of aroma producing lactic acid bacteria from artisanal white cheese for multifunctional properties. LWT Food Sci. Technol. 150:112053. doi: 10.1016/j.lwt.2021.112053
Alessandri, G., Lugli, G. A., Tarracchini, C., Rizzo, S. M., Argentini, C., Viappiani, A., et al. (2022). Disclosing the genomic diversity among members of the bifidobacterium genus of canine and feline origin with respect to those from human. Appl. Environ. Microbiol. 88, e02038–e02021. doi: 10.1128/aem.02038-21
Alkalay-Oren, S., Yerushalmy, O., Adler, K., Khalifa, L., Gelman, D., Coppenhagen-Glazer, S., et al. (2022). Complete genome sequence of pseudomonas aeruginosa bacteriophage PASA16, used in multiple phage therapy treatments globally. Microbiol. Resour. Announc. 11, e00092–e00022. doi: 10.1128/mra.00092-22
Asarina, S., Sariasih, S., and Kulsum, Y. (2022). In silico prediction of bacteriocin gene within the genus of Lactobacillus (Prediksi in silico gen bacteriocin pada genus lactobacillus). Jurnal Biologi Indonesia 18, 103–110. doi: 10.47349/jbi/18012022/103
Ashaolu, T. J., and Reale, A. (2020). A holistic review on Euro-Asian lactic acid bacteria fermented cereals and vegetables. Microorganisms 8:1176. doi: 10.3390/microorganisms8081176
Aziz, T., Naveed, M., Makhdoom, S. I., Ali, U., Mughal, M. S., Sarwar, A., et al. (2023). Genome investigation and functional annotation of Lactiplantibacillus plantarum YW11 revealing streptin and Ruminococcin-A as potent nutritive bacteriocins against gut symbiotic pathogens. Molecules 28:491. doi: 10.3390/molecules28020491
Aziz, T., Naveed, M., Sarwar, A., Makhdoom, S. I., Mughal, M. S., Ali, U., et al. (2022). Functional annotation of Lactiplantibacillus plantarum 13-3 as a Potential starter probiotic involved in the food safety of fermented products. Molecules 27:5399. doi: 10.3390/molecules27175399
Aziz, T., Sarwar, A., Fahim, M., Al Dalali, S., Din, Z. U., Ud Din, J., et al. (2020). In silico characterization of linoleic acid biotransformation to rumenic acid in food derived Lactobacillus plantarum YW11. Acta Biochim. Pol. 67, 99–109. doi: 10.18388/abp.2020_5095
Barrangou, R., Lahtinen, S., Ibrahim, F., and Ouwehand, A. (2011). “Genus Lactobacillus” in Lactic Acid Bacteria: Microbiological and Functional Aspects. eds. S. Lahtinen and A. Von Wrigh. 4th ed (Boca Raton, FL, USA: CRC Press).
Batarseh, T. N., Morales-Cruz, A., Ingel, B., Roper, M. C., and Gaut, B. S. (2022). Using genomes and evolutionary analyses to screen for host-specificity and positive selection in the plant pathogen Xylella fastidiosa. Appl. Environ. Microbiol. 88, e01220–e01222. doi: 10.1128/aem.01220-22
Brandt, K., Nethery, M. A., O’Flaherty, S., and Barrangou, R. (2020). Genomic characterization of Lactobacillus fermentum DSM 20052. BMC Genomics 21, 1–13. doi: 10.1186/s12864-020-6740-8
Carpi, F. M., Coman, M. M., Silvi, S., Picciolini, M., Verdenelli, M. C., and Napolioni, V. (2022). Comprehensive pan-genome analysis of Lactiplantibacillus plantarum complete genomes. J. Appl. Microbiol. 132, 592–604. doi: 10.1111/jam.15199
Castro-López, C., García, H. S., Guadalupe Martínez-Ávila, G. C., GonzálezCórdova, A. F., Vallejo-Cordoba, B., and Hernández-Mendoza, A. (2021). Genomics-based approaches to identify and predict the health-promoting and safety activities of promising probiotic strains - a probiogenomics review. Trends Food Sci. Technol. 108, 148–163. doi: 10.1016/j.tifs.2020.12.017
Chaudhary, N., Maurya, R. K., Singh, D., Mohan, B., and Taneja, N. (2022). Genome analysis and antibiofilm activity of phage 590b against multidrug-resistant and extensively drug-resistant uropathogenic Escherichia coli isolates, India. Pathogens 11:1448. doi: 10.3390/pathogens11121448
De Jesus, L. C. L., Aburjaile, F. F., Sousa, T. D. J., Felice, A. G., and Soares, S. D. C. (2022). Alcantara LCJ and Azevedo VADC genomic characterization of lactobacillus delbrueckii strains with probiotics properties. Front. Bioinform. 2:912795. doi: 10.3389/fbinf.2022.912795
Ding, W., Baumdicker, F., and Neher, R. A. (2018). panX: pan-genome analysis and exploration. Nucleic Acids Res. 9:e5. doi: 10.1093/nar/gkx977
Fernández, L., Langa, S., Martín, V., Maldonado, A., Jiménez, E., Martín, R., et al. (2013). The human milk microbiota: origin and potential roles in health and disease. Pharmacol. Res. 69, 1–10. doi: 10.1016/j.phrs.2012.09.001
Gaina, C. D., Sanam, M. U., Nalley, W. M., and Benu, I.. Molecular Identification of Bone Morphogenetic Protein-15 (BMP-15) gene of Sumba Ongole cattle in International Conference on Improving Tropical Animal Production for Food Security (ITAPS 2021) (2022). doi: 10.2991/absr.k.220309.070
Goel, A., Halami, P. M., and Tamang, J. P. (2020). Genome analysis of Lactobacillus plantarum isolated from some Indian fermented foods for bacteriocin production and probiotic marker genes. Front. Microbiol. 11:40. doi: 10.3389/fmicb.2020.00040
Gupta, S., Mohanty, U., and Majumdar, R. K. (2021). Isolation and characterization of lactic acid bacteria from traditional fermented fish product shidal of India with reference to their probiotic potential. LWT Food Sci. Technol. 146:111641. doi: 10.1016/j.lwt.2021.111641
Hatami, S., Yavarmanesh, M., and Sankian, M. (2022). Seyed Ali Issazadeh comparison of probiotic lactobacillus strains isolated from dairy and Iranian traditional food products with those from human source on intestinal microbiota using BALB/C mice model. Braz. J. Microbiol. 7, 390–404. doi: 10.1007/s42770-022-00790-6
Hebert, R., and Meglécz, E. (2022). NSDPY: a python package to download DNA sequences from NCBI. SoftwareX 18:101038. doi: 10.1016/j.softx.2022.101038
Hojsak, I., Snovak, N., Abdovic, S., Szajewska, H., Misak, Z., and Kolacek, S. (2010). Lactobacillus GG in the prevention of gastrointestinal and respiratory tract infections in children who attend day care centers: A randomized, double-blind, placebo-controlled trial. Clin. Nutr. 29, 312–316. doi: 10.1016/j.clnu.2009.09.008
Jeong, J. J., Park, H. J., Cha, M. G., Park, E., Won, S. M., Ganesan, R., et al. (2022). The Lactobacillus as a probiotic: focusing on liver diseases. Microorganisms 10:288. doi: 10.3390/microorganisms10020288
Jian, Z., Dongyan, C., Ming, Y., Yijiang, H., Yuanhua, Z., Zexuan, C., et al. (2020). Screening of folate-producing lactic acid bacteria and modulatory effects of folate-biofortified yogurt on gut dysbacteriosis of folate-deficient rats. Food Funct. 11, 6308–6318. doi: 10.1039/d0fo00480d
Jian, Z., Wenshen, Z., Xialei, G., Tingting, G., Zheng, Y., Yuetong, W., et al. (2017). Survival and Effect of Exopolysaccharide-Producing Lactobacillus plantarum YW11 on the Physicochemical Properties of Ice Cream. Pol. J. Food Nutr. Sci 67, 191–200. doi: 10.1515/pjfns-2017-0002
Joo, H. M., Hyun, Y. J., Myoung, K. S., Ahn, Y. T., Lee, J. H., Huh, C. S., et al. (2011). Lactobacillus johnsonii HY7042 ameliorates Gardnerella vaginalis-induced vaginosis by killing Gardnerella vaginalis and inhibiting NF-κB activation. Int. Immunopharmacol. 11, 1758–1765. doi: 10.1016/j.intimp.2011.07.002
Kamau, A., Kulmanov, M., Arold, S. T., Pain, A., Gojobori, T., and Duarte, C. M. (2020). Functional pangenome analysis suggests inhibition of the protein E as a readily available therapy for COVID-2019. BioRxiv. doi: 10.1101/2020.02.17.952895
Kim, E., Yang, S. M., Kim, D., and Kim, H. Y. (2022). Complete genome sequencing and comparative genomics of three potential probiotic strains, Lacticaseibacillus casei FBL6, Lacticaseibacillus chiayiensis FBL7, and Lacticaseibacillus zeae FBL8. Front. Microbiol. 12:4135. doi: 10.3389/fmicb.2021.794315
Kleerebezem, M., Boekhorst, J., Van Kranenburg, R., Molenaar, D., Juipers, O. P., et al. (2003). Complete genome sequence of Lactobacillus plantarum WCFS1. Proc. Natl. Acad. Sci. U. S. A. 100, 1990–1995. doi: 10.1073/pnas.0337704100
Kwon, H. K., Lee, C. G., So, J. S., Chae, C. S., Hwang, J. S., Sahoo, A., et al. (2010). Generation of regulatory dendritic cells and CD4+Foxp3+ T cells by probiotics administration suppresses immune disorders. Proc. Natl. Acad. Sci. U. S. A. 107, 2159–2164. doi: 10.1073/pnas.0904055107
Li, K., Wang, S., Liu, W., Kwok, L. Y., Bilige, M., and Zhang, W. (2022). Comparative genomic analysis of 455 Lactiplantibacillus plantarum isolates: Habitat-specific genomes shaped by frequent recombination. Food Microbiol. 104:103989. doi: 10.1016/j.fm.2022.103989
Liu, G., Liu, Y., Ro, K. S., Du, L., Tang, Y. J., Zhao, L., et al. (2022). Genomic characteristics of a novel strain Lactiplantibacillus plantarum X7021 isolated from the brine of stinky tofu for the application in food fermentation. LWT 15:113054. doi: 10.1016/j.lwt.2021.113054
McPherson, J., Hu, C., Begum, K., Wang, W., Lancaster, C., Gonzales-Luna, A. J., et al. (2022). Functional and metagenomic evaluation of ibezapolstat for early evaluation of anti-recurrence effects in Clostridioides difficile infection. Antimicrob. Agents Chemother. 66, e02244–e02221. doi: 10.1128/aac.02244-21
Moradi, M., Kousheh, S. A., Almasi, H., Alizadeh, A., Guimarães, J. T., Yılmaz, N., et al. (2020). Postbiotics produced by lactic acid bacteria: the next frontier in food safety. Compr. Rev. Food Sci. Food Saf. 19, 3390–3415. doi: 10.1111/1541-4337.12613
Rodenes, A., Ramo, B. C., Perez, B. A., Blanch, J. F., Vijayakumar, V., Superti, G., et al. (2022). Reclassification of probiotic Lactobacillus acidophilus NCIMB 30184 as Lactobacillus helveticus and Lactobacillus casei NCIMB 30185 as Lacticaseibacillus paracasei. bioRxiv. doi: 10.1101/2022.10.17.512536
Rodrigo-Torres, L., Yépez, A., Aznar, R., and Arahal, D. R. (2019). Genomic insights into five strains of Lactobacillus plantarum with biotechnological potential isolated from chicha, a traditional maize-based fermented beverage from Northwestern Argentina. Front. Microbiol. 10:2232. doi: 10.3389/fmicb.2019.02232
Salvetti, E., and O’Toole, P. W. (2018). The genomic basis of lactobacilli as health-promoting organisms. Microbiol. Spectr. 5, 49–71. doi: 10.1128/microbiolspec.BAD-0011-2016
Sarwar, A., Aziz, T., Din, J., Khalid, A., Rahman, T., and Daudzai, Z. (2018). Pros of lactic acid bacteria in microbiology: a review. Biomed. Lett. 4, 59–66.
Shin, J. H., Chung, W. H., Park, Y. S., and Nam, Y. D. (2022). Detection and identification of Lactobacillus acidophilus species and its commercial probiotic strains using CRISPR loci-based amplicon analysis. LWT 171:114166. doi: 10.1016/j.lwt.2022.114166
Spergser, J., DeSoye, P., Ruppitsch, W., Rosel, A. C., Dinhopl, N., Szostak, M. P., et al. (2022). Mycoplasma tauri sp. nov.isolated from the bovine genital tract. Syst. Appl. Microbiol. 45:126292. doi: 10.1016/j.syapm.2021.126292
Stefanovic, E., Fitzgerald, G., and McAuliffe, O. (2017). Advances in the genomics and metabolomics of dairy lactobacilli: a review. Food Microbiol. 61, 33–49. doi: 10.1016/j.fm.2016.08.009
Surve, S., Shinde, D. B., and Kulkarni, R. (2022). Isolation, characterization and comparative genomics of potentially probiotic Lactiplantibacillus plantarum strains from Indian foods. Sci. Rep. 12, 1–6. doi: 10.1038/s41598-022-05850-3
Swain, M. R., Anandharaj, M., Ray, R. C., and Rani, R. P. (2014). Fermented fruits and vegetables of Asia: a potential source of probiotics. Biotechnol. Res. Int. 2014:250424. doi: 10.1155/2014/250424
Syrokou, M. K., Paramithiotis, S., Drosinos, E. H., Bosnea, L., and Mataragas, M. (2022). A comparative genomic and safety assessment of six lactiplantibacillus plantarum subsp. argentoratensis strains isolated from spontaneously fermented greek wheat sourdoughs for potential biotechnological application. Int. J. Mol. Sci. 23:2487. doi: 10.3390/ijms23052487
Teame, T., Wang, A., Xie, M., Zhang, Z., Yang, Y., Ding, Q., et al. (2020). Paraprobiotics and postbiotics of Probiotic Lactobacilli, their positive effects on the host and action mechanisms: a review. Front. Nutr. 7:570344. doi: 10.3389/fnut.2020.570344
Tenea, G. N., and Ortega, C. (2021). Genome Characterization of Lactiplantibacillus plantarum Strain UTNGt2 Originated from Theobroma grandiflorum (White Cacao) of Ecuadorian Amazon: antimicrobial peptides from safety to potential applications. Antibiotics 10:383. doi: 10.3390/antibiotics10040383
Valdez-Baez, J. L., De Jesus, L. C., Marques, P. H., da Silva Prado, L. C., Felice, A. G., Rodrigues, T. C., et al. (2022). Comparative genomics in probiotic bacteria lactic acid bacteria. Food Biotechnol. 103, 15611–15616. doi: 10.1073/pnas.0607117103
Ventura, M., Turroni, F., and van Sinderen, D. (2012). Probiogenomics as a tool to obtain genetic insights into adaptation of probiotic bacteria to the human gut. Bioeng. Bugs 3, 73–79. doi: 10.4161/bbug.18540
Wang, S. Y., Chen, Y. P., Huang, R. F., Wu, Y. L., Ho, S. T., Li, K. Y., et al. (2022). Subspecies classification and comparative genomic analysis of lactobacillus kefiranofaciens HL1 and M1 for Potential niche-specific genes and pathways. Microorganisms 10:1637. doi: 10.3390/microorganisms10081637
Wang, Y., Liang, Q., Lu, B., Shen, H., Liu, S., Shi, Y., et al. (2021). Whole-genome analysis of probiotic product isolates reveals the presence of genes related to antimicrobial resistance, virulence factors, and toxic metabolites, posing potential health risks. BMC Genomics 22:210. doi: 10.1186/s12864-021-08189-7
Wang, L., Zhao, Z., Zhao, L., Zhao, Y., Yang, G., Wang, C., et al. (2022). Lactobacillus plantarum DP189 Reduces _-SYN aggravation in MPTP-induced Parkinson’s disease mice via regulating oxidative damage, inflammation, and Gut microbiota disorder. J. Agric. Food Chem. 2, 1163–1173. doi: 10.1186/s12864-021-07539-9
Xiang, R., and Li, M. (2022). Detection of a Novel Metallo-β-Lactamase, CAM-2, in a metagenome-assembled genome from China. Microbiol. Spectr. 31, e00261–e00222.
Yépez, A., Luz, C., Meca, G., Vignolo, G., Mañes, J., and Aznar, R. (2017). Biopreservation potential of lactic acid bacteria from Andean fermented food of vegetal origin. Food Control 78, 393–400. doi: 10.1016/j.foodcont.2017.03.009
Yin, Z., Liu, X., Qian, C., Sun, L., Pang, S., Liu, J., et al. (2022). Pan-genome analysis of Delftia tsuruhatensis reveals important traits concerning the genetic diversity, pathogenicity, and biotechnological properties of the species. Microbiol. Spectr. 10, e02072–e02021. doi: 10.1128/spectrum.02072-21
Yonekura, S., Okamoto, Y., Okawa, T., Hisamitsu, M., Chazono, H., Kobayashi, K., et al. (2009). Effects of daily intake of Lactobacillus paracasei strain KW3110 on Japanese cedar pollinosis. Allergy Asthma Proc. 30, 397–405. doi: 10.2500/aap.2009.30.3256
Zhang, Z., Lv, J., Pan, L., and Zhang, Y. (2018). Roles, and applications of probiotic Lactobacillus strains. Appl. Microbiol. Biotechnol. 102, 8135–8143. doi: 10.1007/s00253-018-9217-9
Zhang, M., Xiaona, H., Aziz, T., Jian, Z., and Yang, Z. (2020). Exopolysaccharides from Lactobacillus plantarum YW11 improve immune response and ameliorate inflammatory bowel disease symptoms. Acta Biochim. Pol. 67, 485–493. doi: 10.18388/abp.2020_5371
Zhang, M., Yao, M., Lai, T., Zhao, H., Wang, Y., and Yang, Z. (2022). Response of Lactiplantibacillus plantarum NMGL2 to combinational cold and acid stresses during storage of fermented milk as analyzed by data-independent acquisition proteomics. Foods 10:1514. doi: 10.3390/foods10071514
Keywords: kefir, comparative genomics, cefoxitin, carbohydrate metabolism, genetics and genomics, pathogenic
Citation: Aziz T, Naveed M, Jabeen K, Shabbir MA, Sarwar A, Zhennai Y, Alharbi M, Alshammari A and Alasmari AF (2023) Integrated genome based evaluation of safety and probiotic characteristics of Lactiplantibacillus plantarum YW11 isolated from Tibetan kefir. Front. Microbiol. 14:1157615. doi: 10.3389/fmicb.2023.1157615
Edited by:
Waseem Safdar, National University of Medical Sciences (NUMS), PakistanReviewed by:
Ashwag Y. Shami, Princess Nourah Bint Abdulrahman University, Saudi ArabiaRewaa S. Jalal, University of Jeddah, Saudi Arabia
Nageen Hussain, University of the Punjab, Pakistan
Falak Zeb, University of Sharjah, United Arab Emirates
Copyright © 2023 Aziz, Naveed, Jabeen, Shabbir, Sarwar, Zhennai, Alharbi, Alshammari and Alasmari. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yang Zhennai, eWFuZ3poZW5uYWlAMTYzLmNvbQ==
 Tariq Aziz
Tariq Aziz Muhammad Naveed3
Muhammad Naveed3