
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 12 May 2023
Sec. Extreme Microbiology
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1156033
This article is part of the Research Topic Microorganisms in Polar Regions: Understanding Their Survival Strategies for a Sustainable Future View all 12 articles
The McMurdo Dry Valleys of Antarctica experience a range of selective pressures, including extreme seasonal variation in temperature, water and nutrient availability, and UV radiation. Microbial mats in this ecosystem harbor dense concentrations of biomass in an otherwise desolate environment. Microbial inhabitants must mitigate these selective pressures via specialized enzymes, changes to the cellular envelope, and the production of secondary metabolites, such as pigments and osmoprotectants. Here, we describe the isolation and characterization of a Gram-negative, rod-shaped, motile, red-pigmented bacterium, strain DJPM01, from a microbial mat within the Don Juan Pond Basin of Wright Valley. Analysis of strain DJMP01’s genome indicates it can be classified as a member of the Massilia frigida species. The genome contains several genes associated with cold and salt tolerance, including multiple RNA helicases, protein chaperones, and cation/proton antiporters. In addition, we identified 17 putative secondary metabolite gene clusters, including a number of nonribosomal peptides and ribosomally synthesized and post-translationally modified peptides (RiPPs), among others, and the biosynthesis pathway for the antimicrobial pigment prodigiosin. When cultivated on complex agar, multiple prodiginines, including the antibiotic prodigiosin, 2-methyl-3-propyl-prodiginine, 2-methyl-3-butyl-prodiginine, 2-methyl-3-heptyl-prodiginine, and cycloprodigiosin, were detected by LC–MS. Genome analyses of sequenced members of the Massilia genus indicates prodigiosin production is unique to Antarctic strains. UV-A radiation, an ecological stressor in the Antarctic, was found to significantly decrease the abundance of prodiginines produced by strain DJPM01. Genomic and phenotypic evidence indicates strain DJPM01 can respond to the ecological conditions of the DJP microbial mat, with prodiginines produced under a range of conditions, including extreme UV radiation.
Upwards of 80% of Earth’s environments are permanently cold (<5°C), including polar, alpine, maritime, and atmospheric regions (De Maayer et al., 2014). Understanding how life survives at low temperatures is crucial to our understanding of life itself, as well as how life might exist on other planets. On the Antarctic continent, the McMurdo Dry Valleys (MDV) constitute the largest ice-free region (Levy, 2013). The MDV region is the coldest, driest desert on our planet, and experiences average annual air and absolute surficial soil temperatures ranging from −26.8 to −16.5°C and −59.8 to +31.1°C, respectively (Obryk et al., 2020), and receive a maximum of ~50 mm of precipitation in the form of snowfall annually (Fountain et al., 2009). Additionally, the MDV experiences long periods of sustained light and dark, due to its extreme southern latitude, and high levels of UV-A and UV-B radiation from thinning in the ozone layer (Frederick and Snell, 1988).
Antarctic microbial mats provide a unique system for studying the effects of UV radiation and other adaptations due to their high biomass compared to other microbial niches within the MDV (Davey and Clarke, 1992). The formation of these communities is reliant on photosynthetic cyanobacteria, including either Nostoc species or Phormidium and Oscillatoria species, depending on the geomorphology and hydrology of the niche (Alger et al., 1997). Other photosynthetic organisms, such as green algae and diatoms, can be found in Antarctic mats, as well as heterotrophic bacteria and microscopic invertebrates (Andriuzzi et al., 2018). Microbial mats that occupy perennial stream channels and shallow ponds are particularly exposed to extremes of seasonal variation and experience fluctuations in meltwater flow from glaciers due to changes in temperature and solar angle, which in turn can lead to changes in nutrient availability and community structure (Zoumplis et al., 2023). In the winter, communities are exposed to months of darkness, desiccation, and subzero temperatures. MDV mats in stream channels and surface ponds lie dormant for ~10 months of the year due to the lack of water and sunlight, followed by rapid reactivation in response to rehydration by glacial meltwater (McKnight et al., 2007). In addition to freezing and dehydration, MDV mats are exposed to intense ultraviolet (UV) radiation. This UV exposure causes DNA damage and the production of reactive oxygen species (ROS; Castenholz and Garcia-Pichel, 2012). UV radiation has been proposed as a major limiting factor on growth for communities within the MDV (Tosi et al., 2005).
Microbial mats are scattered throughout the MDV, largely concentrated in meltwater streams and lake margins (Power et al., 2020) that wax and wane in volume in response to climate (Mikucki et al., 2010). Within the South Fork of the Wright Valley is a near-saturated CaCl2 brine pond, known as Don Juan Pond (DJP). DJP is bounded to the west by a rock glacier, which provides a source of meltwater to the basin (Figure 1). While there are conflicting reports regarding the habitability of the brine (Toner et al., 2017), the DJP basin hosts a dense microbial mat that borders the rock glacier (Figure 1B). This mat was first described as approximately 500–600 m2 and 2–5 mm thick (Siegel et al., 1979). Microscopic investigation of the mat revealed an abundance of cyanobacteria phenotypically similar to Oscillatoria, in addition to the green algae Chlorella and Dunaliella, diatoms, invertebrates, and bacteria (Siegel et al., 1983). The extent to which the saturated brine of DJP might influence the mat is currently unknown, though it is possible that dried salts are deposited in the mat by aeolian processes (Diaz et al., 2018).
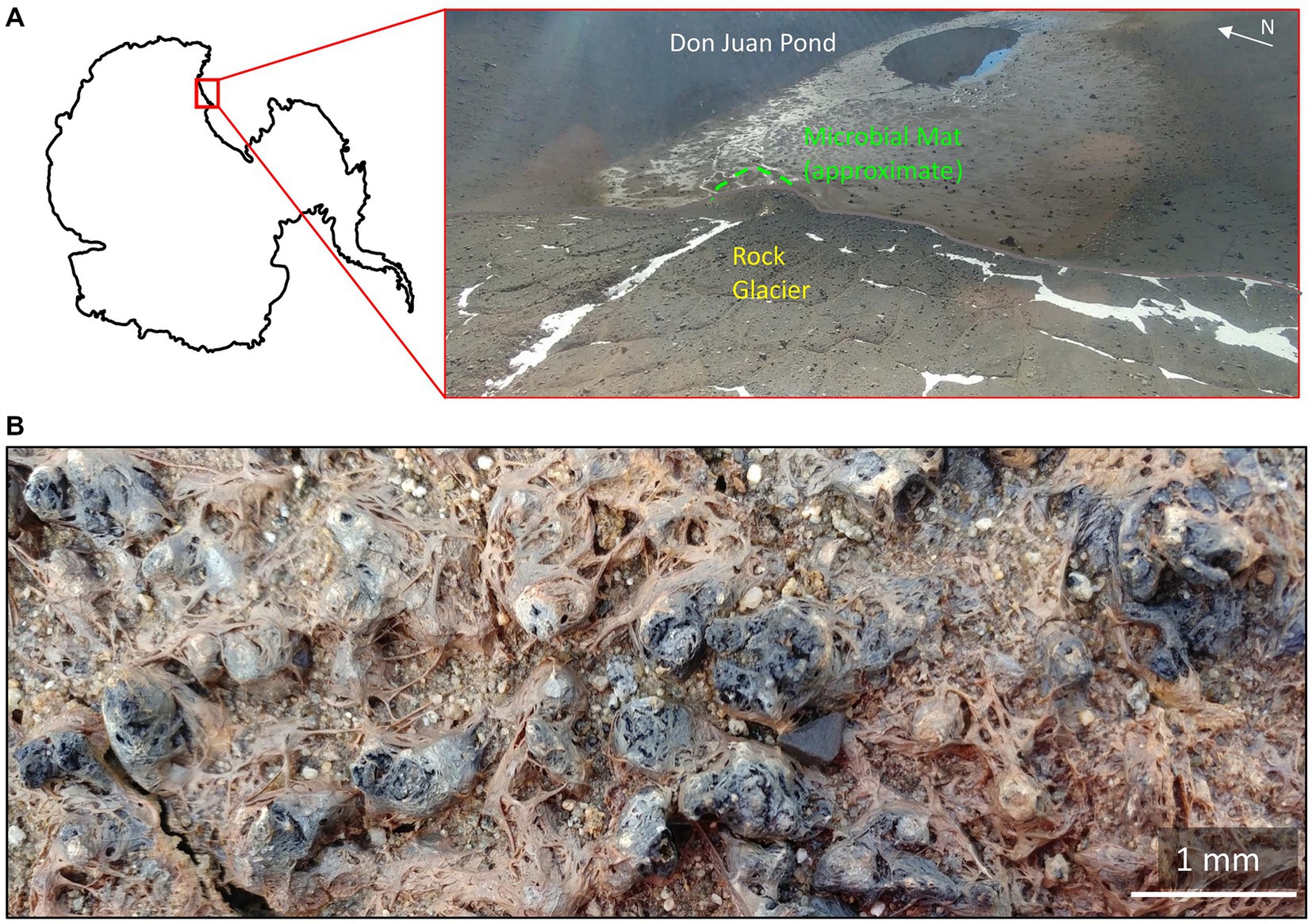
Figure 1. The Don Juan Pond (DJP) microbial mat (A) Location of the microbial mat within DJP basin. Approximate location of the basin in Antarctica is inset. (B) Close up of the DJP basin microbial mat collected for enrichment cultures that yielded strain DJPM01.
The genus Massilia was first described in 1998 in a report on the isolation of Massilia timonae from the blood sample of an immunocompromised patient (La Scola et al., 1998). The genus belongs to the Oxalobateraceae family and to date includes 61 validly published species (January 2023).1 Massilia species have been isolated from disparate environments across the globe and have been implicated in human disease (La Scola et al., 1998; Van Craenenbroeck et al., 2011; Kämpfer et al., 2012; Chiquet et al., 2015) and as a common member of rhizosphere communities of Arabidopsis (Tkacz et al., 2015) and wheat (Schlatter et al., 2020). Rhizosphere Massilia are of interest due to their possible role in plant development through the production of phytohormones and solubilization of phosphates, among other activities (Ofek et al., 2012; Schlatter et al., 2020). Several Massilia have also been isolated from the cryosphere, including from Arctic (Son et al., 2021; Dahal et al., 2021a), Antarctic (Holochová et al., 2020; Sedláček et al., 2022), and alpine regions (Shen et al., 2013, 2015; Guo et al., 2016; Gu et al., 2017; Wang et al., 2018).
While their ecological role in polar environments is not as well understood, Massilia species have been detected in diverse Antarctic niches. For example, Massilia ranged from 54 to 79% of culturable community members in both closed and open cryoconite holes in the Larsemann Hills, Dronning Maud Land, and Amery Ice Shelf regions (Sanyal et al., 2018). Sommers et al. (2019) identified Massilia as one of the ten most abundant sequences in cryoconite water collected from Taylor Glacier, a glacier within the MDV. These authors suggested that Massilia may play a role in cryoconite colonization. Massilia have also been isolated from microbial mats in both peninsular and continental Antarctica, including other ice-free regions such as the Schirmacher and Syowa Oasis (Peeters et al., 2012).
Here we characterize a red-pigmented Massilia species, strain DJPM01, isolated from the DJP microbial mat. We identified genomic evidence for environmental adaptations in strain DJPM01 and compared these findings with other Antarctic Massilia species as well as the taxonomic distribution of secondary metabolite biosynthetic gene clusters (BGCs) within the Massilia genus. We then analyzed cultures of strain DJPM01 for the presence and relative abundance of pigments when grown under ecologically relevant UV-A radiation exposure to further our understanding of microbial responses in extreme environments.
Samples of a microbial mat were collected in the South Fork of the Wright Valley at the terminus of a rock glacier in the Don Juan Pond (DJP) basin, Victoria Land, Antarctica (77.56293 S, 161.1751 E; Figure 1). Mat material was collected on 5 December 2018 using individually wrapped sterile scoops (SP Bel-Art; Wayne, NJ) and transferred into sterile Whirl-Pak bags (Whirl-Pak; Madison, WI). In situ temperature, pH, and salinity were measured at the time of collection using a vented Aqua TROLL 500 Multiparameter Sonde (in-situ), configured with pH/ORP, conductivity, rugged dissolved oxygen (RDO), and turbidity modules. Samples were stored and shipped cool (4°C) until later processing at the University of Tennes(see Knoxville, TN, United States).
Mat material was incubated in R2A broth (Difco; Becton, Dickinson and Company; Franklin, NJ) at ~10°C for 10 days, after which the broth was used to inoculate R2A agar plates (BD Difco; Franklin, NJ). Bright red-pigmented colonies appeared on the plate after 4 days of incubation at 10°C and were selected for further isolation through multiple restreaks on R2A agar. Gram staining and microscopy was performed on strain DJPM01 using the American Society of Microbiology Gram stain protocol (Smith and Hussey, 2005) and observed with an Axio Imager M2 microscope (Zeiss; Oberkochen, Germany). Motility assays were conducted by spot inoculation of strain DJPM01 onto 0.5% agar R2A plates. To assess resistance to UV-C light, R2A agar plates of strain DJPM01 were prepared and exposed UV light (254 nm) as described by Hirsch et al. (2004). Plates were exposed for 10, 20, 30, 60, 120, and 300 s and left to incubate in the dark at 10°C for 10 days. Strain DJPM01 was incubated in R2A broth under a nitrogen headspace to test for anaerobic growth. To test the effects of temperature on the growth rate of strain DJPM01, incubations were performed in 96-well plates. Briefly, 180 μL R2A broth was aliquoted into each well. Strain DJPM01 was grown to late log phase and inoculated into 48 wells of a 96-plate (10% inoculum). Plates were incubated at 4, 10, 15, 20, and 25°C, and full spectra were collected for each plate once per day for 14 days. Outer wells were removed from analysis to control for unequal levels of evaporation.
For taxonomic identification, DNA was extracted from bacterial colonies using a DNeasy PowerSoil PowerLyzer Kit (Qiagen; Hilden, Germany) following the manufacturer’s protocol with the addition of a modified bead beating step as follows: 45 m sec−1 for 25 sec followed by 5 min rest at 10°C (FastPrep-24 Bead Lysis System; MP Biomedical; Santa Anna, CA). The 16S ribosomal RNA (rRNA) gene was amplified from extracted DNA using the following combination of primers: bacterial 27F (5’-AGAGTTTGATCCTGGCTCAG-3′; Weisburg et al., 1991) and 806R primers (5’-GGACTACNVGGGTWTCTAAT-3′; Apprill et al., 2015) and 515F (5’-GTGCCAGCMGCCGCGGTA-3′; Caporaso et al., 2011) and 1492R primers (5’-GGTTACCTTG TTACGACTT-3′; Weisburg et al., 1991). The resulting PCR products were purified using a Wizard SV Gel and PCR Clean-Up System Kit (Promega; Madison, WI) before Sanger sequencing, performed by Eurofins Genomics (Louisville, KY). Returned sequences were aligned using Geneious v6.1.8 (for a sequence length of 1,388 bp; Kearse et al., 2012) and compared to the NCBI nucleotide collection type material database (Federhen, 2012). Publicly available sequences for Massilia 16S rRNA genes were obtained from the RefSeq database (O'Leary et al., 2016), aligned, trimmed to the length of the shortest sequence (1,335 bp), and used to construct a maximum likelihood tree in MEGA v11.0.13 (Tamura et al., 2021). Additionally, strain DJPM01 was compared to all NCBI reference genomes within the Oxalobateraceae family using whole genome average nucleotide identity (ANI) through the enveomics collection (Rodriguez-R and Konstantinidis, 2016). Digital DNA–DNA hybridization (DDH) for all reference genomes available in the Oxalobacteraceae family was calculated using the Genome-to-Genome Distance Calculator (GGDC) v3.0 (Meier-Kolthoff et al., 2022). A final phylogenetic tree of RefSeq reference genomes was generated using OrthoFinder v2.5.4 (Emms and Kelly, 2019) with default settings. Estimation of abundance was performed using a 16S community amplicon sequence variant (ASV) library generated from the DJP mat (Schuler, 2022) using the Basic Local Alignment Search Toolkit (BLAST; Altschul et al., 1990). The final 16S rRNA gene sequence for strain DJPM01 was uploaded to GenBank under accession number OQ346149.
Total genomic DNA was extracted from microbial biomass using a cetrimonium bromide (CTAB) buffer/organic phase extraction protocol (as described by Campen, 2015) for whole genome sequencing. Quality and quantity of extracted DNA was determined by a Qubit 4 fluorometer (Invitrogen Carlsbad, CA) and NanoDrop 2000 spectrophotometer (Thermo Scientific; Waltham, MA, United States). High molecular weight genomic DNA (25 ng/μL) was sequenced by MR DNA (Shallowater, TX). Library preparation was performed using the Nextera DNA Flex Library Preparation Kit (Illumina Inc.; San Diego, CA) and quality was confirmed using the Agilent 2,100 Bioanalyzer (Agilent Technologies; Santa Clara, CA). The library was diluted to 0.6 nM before sequencing on the NovoSeq platform (Illumina; San Diego, CA; paired end, 500 cycles). Sequencing adapters and low-quality reads were trimmed from using Trimmomatic v0.36 (Bolger et al., 2014) on default parameters. Trimmed reads were assembled using Unicycler v0.4.8 (Wick et al., 2017) with a maximum contig size of 500 bp and assembly quality was assessed using QUAST v5.0.2 (Gurevich et al., 2013) and checkM v1.1.3 (Parks et al., 2015). The genome draft was annotated by Prokka v1.13.7 (Seemann, 2014) and the resulting annotation was analyzed using GhostKOALA v2.2 (Kanehisa et al., 2016). Putative CRISPR-Cas sequences were identified using CRISPRCasFinder v4.2.20 (Couvin et al., 2018) and putative prophage sequences were identified by VirSorter2 v2.2.3 (Guo et al., 2021), both using the Proksee interface.2 Putative secondary metabolite BGCs were identified via antiSMASH v6.0.0 (Blin et al., 2021) using default parameters. RefSeq Massilia genomes were annotated using Prokka v1.13.7, GhostKOALA v2.2, and antiSMASH v6.0.0 and all Massilia genomes in both NCBI and JGI databases were aligned against the pig operon of Serratia sp. strain ATCC 39006 to search for prodigiosin-specific biosynthetic genes (Harris et al., 2004). Putative prodigiosin operons were compared by pairwise alignment using SimpleSynteny v1.5 (Veltri et al., 2016). Statistical analyses of gene abundance were performed using R v4.1.2 (R Core Team, 2018). The final draft genome assembly was uploaded to GenBank under accession number JAQOZV000000000.
A lawn of strain DJPM01 was grown on R2A agar plates in quadruplicate and incubated under a UV-A light (320–400 nm, peak absorbance 395 nm; Supplementary Figure S1; SANSI Lighting; Shanghai, China). Nalgene petri dishes (Thermo Scientific; Waltham, MA) made of polymethylpentene were utilized to increase UV penetration compared to standard polystyrene petri dishes (Peng et al., 2011). Control plates were inoculated, wrapped in foil, and placed inside two black bags prior to incubation. Uninoculated media controls were also incubated under both light and dark treatments. All plates were incubated at 10°C for 11 days. Following incubation, agar was sterilely sliced into strips for storage at −80°C. Agar strips from each plate were extracted four times with 10 mL of high performance liquid chromatography (HPLC)-grade ethyl acetate (Fisher Scientific; Hampton, NH) and sonicated for 10 min. Organic phases were pooled and diluted to 50 mL with ethyl acetate. Authentic prodigiosin (1) standard (P0103; Sigma-Aldrich, St. Louis, MO) was diluted 100 μg mL−1 in liquid chromatography/mass spectrometry (LC/MS)-grade methanol (Fisher Scientific; Hampton, NH) and exposed to UV-A radiation overnight to investigate potential breakdown products. UV-A exposed and unexposed prodigiosin (1) standard solutions were diluted to 10 μg mL−1 prior to LC/MS analysis.
Extracts were analyzed on a Thermo Scientific (Waltham, MA) Q-exactive HF-X Hybrid Quadrupole-Orbitrap mass spectrometer interfaced with a Vanquish Horizon Ultra high performance liquid chromatography (UHPLC) system and VH-D10-A UV detector using a Waters (Milford, MA) HSS T3 C18 column (1.8 μm, 2.1 × 100 mm) with a VanGuard FIT precolumn. Using a flow rate of 0.5 mL min−1, samples (2 μL) were injected onto the column prewarmed to 40°C. UHPLC separation was achieved using the following parameters: 2% acetonitrile: 98% water with 0.1% formic acid for 1 min, 2–40% over 4 min, 40–98% over 3 min, 98–2% over 0.2 min, and 2% for 2 min. The UV–Vis data were collected at 534 nm. The following electrospray ionization (ESI) settings were used in probe position D: 40 sheath gas flow, 8 auxiliary gas flow, 1 sweep gas flow, 3.5 kV spray voltage, 380°C capillary temperature, 50 radiofrequency (RF) funnel level, and 350°C auxiliary gas temperature. Data were evaluated using MS-DIAL (Tsugawa et al., 2015) and MPACT (Samples et al., 2022). See supplementary information for data processing methods.
A Waters Synapt G2-Si Q-ToF mass spectrometer coupled to an Acquity H-Class ultra performance liquid chromatography (UPLC) system was employed for LC/MS analysis of select samples and prodigiosin (1) standard using the previously described column and gradient. Positive mode MSE acquisition was used, using 0.1 s MS1 survey scans from m/z 150–2000 in resolution mode, followed by 0.1 s high energy MS2 scan from m/z 50–2000. Data were acquired using the following parameters: 2 kV capillary voltage, 100°C source temperature, 20 V sampling cone, 800 L h−1 desolvation gas flow, 80 V source offset, and 30 V MS2 collision energy. Real-time mass correction used a lockspray of 400 pg μL−1 leucine enkephalin solution (1:1 methanol to water solution with 0.1% formic acid). A 10 μL injection volume was used in the closed loop configuration.
At time of collection, the water within the mat was 7.7°C; collection site pH was 7.2 and salinity was 0.4 parts per thousand (ppt). Following isolation, the purity of the resulting axenic culture was confirmed by 16S rRNA gene analyses and microscopy, which revealed a single rod-shaped morphology. The resulting red-pigmented isolate was designated strain DJPM01.
Growth of strain DJPM01 on R2A yielded tall, convex, scarlet colonies with entire margins that spread outwards over time (Figure 2). Colonies were resistant to removal from the surface of the agar and remained aggregated when transferred to broth culture, unless thoroughly agitated through vortexing. Strain DJPM01 was determined to be Gram-negative using the standard Gram stain protocol (Smith and Hussey, 2005). Cells were rod-shaped and 4.65 μm in length on average (Figure 2A). Cultures exhibited growth from 4 to 25°C, but not at 30°C (Supplemental Figure S2). The fastest growth occurred at 10°C with a rate (μ) of 0.25 ± 0.04 day−1. Colonies were pigmented under all culture conditions, with some variation in color intensity and hue at warmer temperatures (Supplemental Figure S2). Growth was not detected under anaerobic conditions. Strain DJPM01 grew on R2A agar adjusted to pH 10.5. Abundance of colony forming units decreased following 30 s of exposure to UV-C radiation, however colonies were present on plates irradiated for up to 120 s. Evidence for motility was observed when grown on low density R2A agar. When grown in R2A broth, strain DJPM01 formed a robust pellicle at the surface and produced extracellular polymeric substances (Figure 2D).
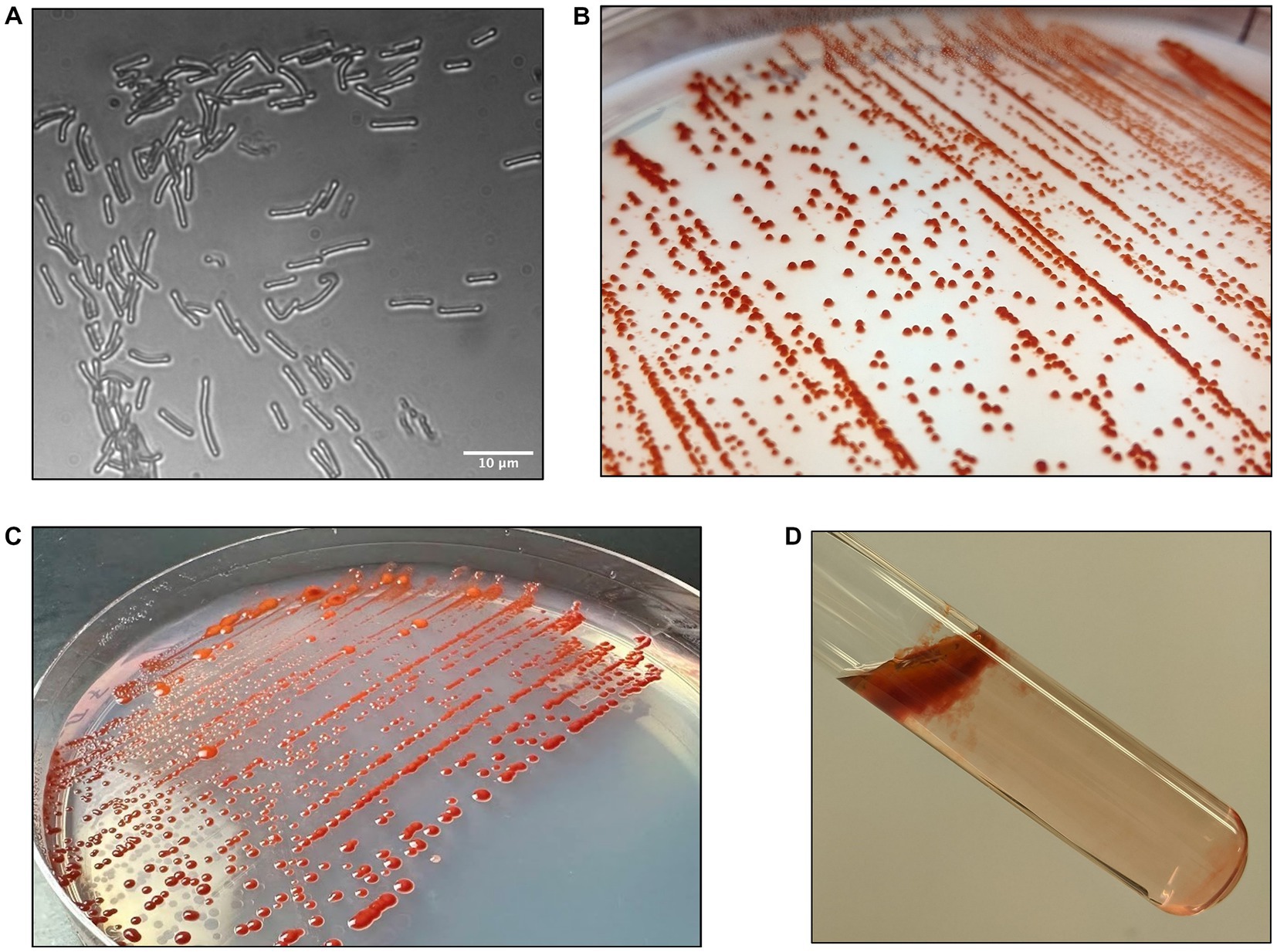
Figure 2. Morphology of strain DJPM01. (A) Light microscopy image of strain DJPM01 using phase shift. Cells were rods and 4.65 μm in length on average. (B,C) Strain DJPM01 grown on R2A agar at 10 (B) and 15 (C) °C. Colonies were robust and resisted removal from the agar. Bright red pigmentation was present at all temperatures. (D) Pellicle formation by strain DJPM01 in R2A broth, held at an angle.
Analysis of the trimmed 16S rRNA gene sequence (1,335 bp) placed strain DJPM01 within the Massilia genus (Supplementary Figure S3). Strain DJPM01 shared highest sequence identity with M. violaceinigra strain B2 (CP024608, 99.87%) originally isolated from glacial permafrost in the Xinjiang region of China (Wang et al., 2018). Strain DJPM01 also shared high sequence identity with several other cryosphere associated Massilia species, including M. aquatica strain CCM 8693 (MN612031, 99.87%), M. rubra strain CCM 8692 (MN611986, 99.80%), M. frigida strain CCM 8695 (MN612047, 99.80%), M. mucilaginosa strain 8,733 (MN612043, 99.67%), and M. atriviolacea strain SOD (NR_171529, 99.40%). Of these six isolates, four were isolated from James Ross Island on the Antarctic Peninsula (M. frigida, M. aquatica, M. rubra, and M. mucilaginosa; Holochová et al., 2020). M. atriviolacea, was first isolated from agricultural soils from Hefei, China (Yang et al., 2019). Comparison of the 16S rRNA sequence to the available DJP basin mat 16S ASV library suggests that strain DJPM01 is present in low abundance with 2,110 reads or ~0.03% relative abundance (Schuler, 2022).
Whole genome comparison placed strain DJPM01 within a distinct clade (≥87% ANI between clade members) including all currently identified Massillia with either a red or purple pigmented phenotype (Figure 3; Supplementary Table S1). This clade includes the six Massilia strains with highest 16S rRNA gene similarity to strain DJPM01, as well as M. antarctica strain P9640 (OM243919; Sedláček et al., 2022), which was also isolated from James Ross Island. All members of this clade have been shown to grow at or below 4°C, regardless of isolation locale. Strain DJPM01 shared highest ANI with M. frigida strain CCM8695 (97.36% identity). High sequence identity of strain DJPM01 to M. antarctica strain P9640 (91.92%) and M. violaceinigra strain B2 (91.38%) were also noted. Additionally, M. mucilaginosa strain CCM8733, M. atriviolacea strain SOD, M. rubra CCM 8692 (GCF 011682065), and M. aquatica CCM 8693 had relatively high sequence identity (87.94–88.41% similarity to strain DJPM01). Digital DNA–DNA hybridization (DDH) showed 75.6% similarity between strain DJPM01 and M. frigida strain CCM8695 (Supplementary Table S1).
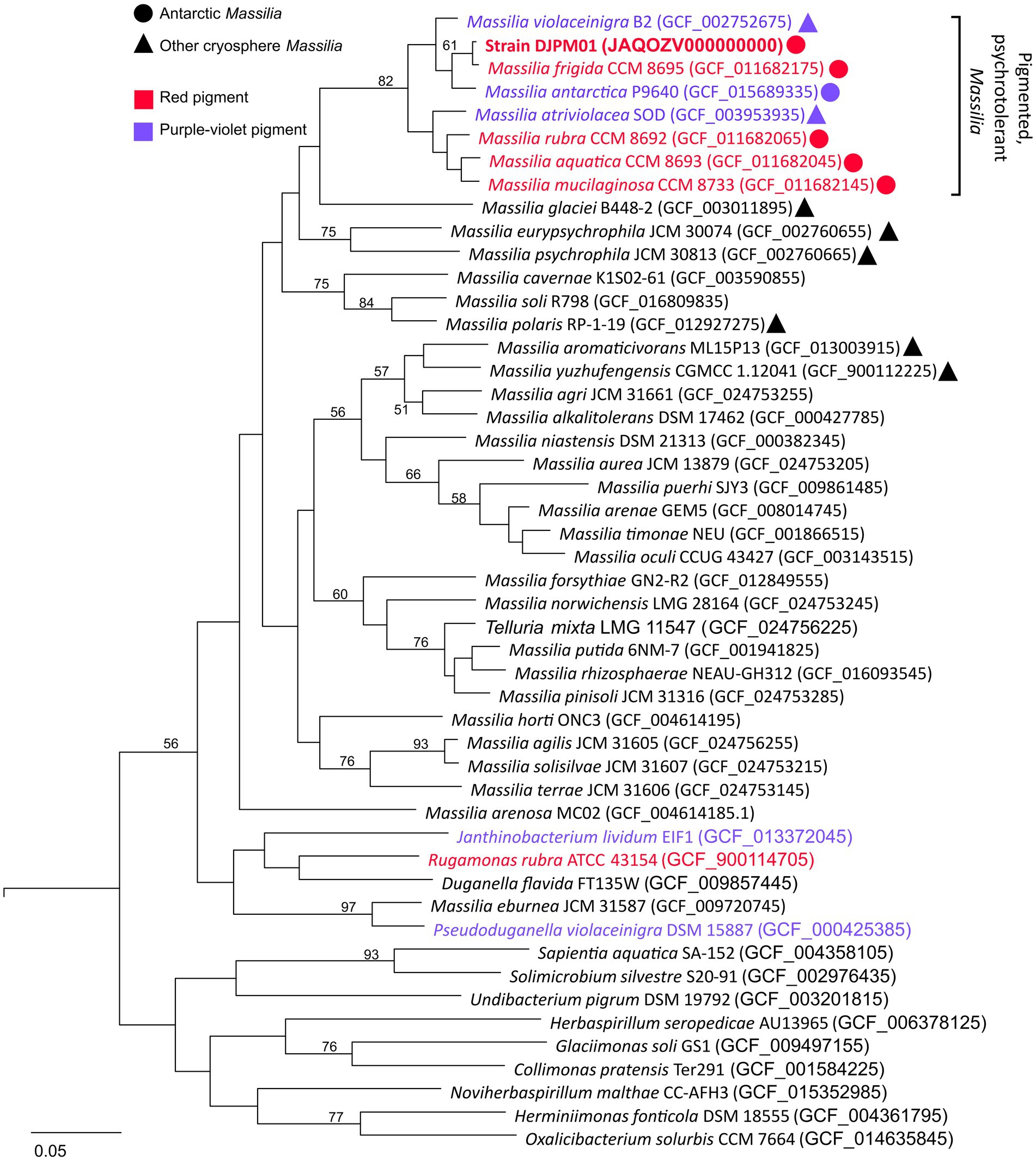
Figure 3. Multiple-gene tree of Oxalobacteraceae family members. Tree constructed using OrthoFinder v2.5.4 (Emms and Kelly, 2019). All Massilia with phenotypic and genotypic evidence for red (prodigiosin) or purple-violet (violacein) pigment production form a clade with >87% ANI between any given pair. All members of this clade were isolated cryosphere environments, including all five Antarctic Massilia. All Massilia genomes cluster into a single clade, with the exception of M. eburnea, originally identified as a species of Pseudoduganella (Lu et al., 2020). Node values represent bipartition support values >50. An Antarctic Shewanella species (strain BF02_Schw) was used as an outgroup (DQ677870.1).
Whole-genome sequencing resulted in 8,995,221 paired reads of which 8,693,443 (96.6%) pairs survived trimming with 0 N’s. Assembly yielded a genome 7.7 Gb in size across 236 contigs with 99.80% completeness (573 of 574 marker genes present) and 1.64% contamination (10 marker genes with two copies each; Figure 4). Following annotation, 6,626 predicted protein coding genes were identified, 3,037 of which had a non-hypothetical function (45.8% functional annotation). Of these genes, the largest functional categories within the KEGG database were protein families: signaling and cellular processes (361 genes), environmental information processing (339 genes), genetic information processing (339 genes), and carbohydrate metabolism (248 genes). The genome contains complete pathways for glycolysis and the TCA cycle, oxidative phosphorylation, flagellar motility, and denitrification. Additionally, we detected 9 putative CRISPR arrays and 4 Cas clusters (Figure 4). Both putative single- and double-stranded DNA phage sequences were found throughout the genome.
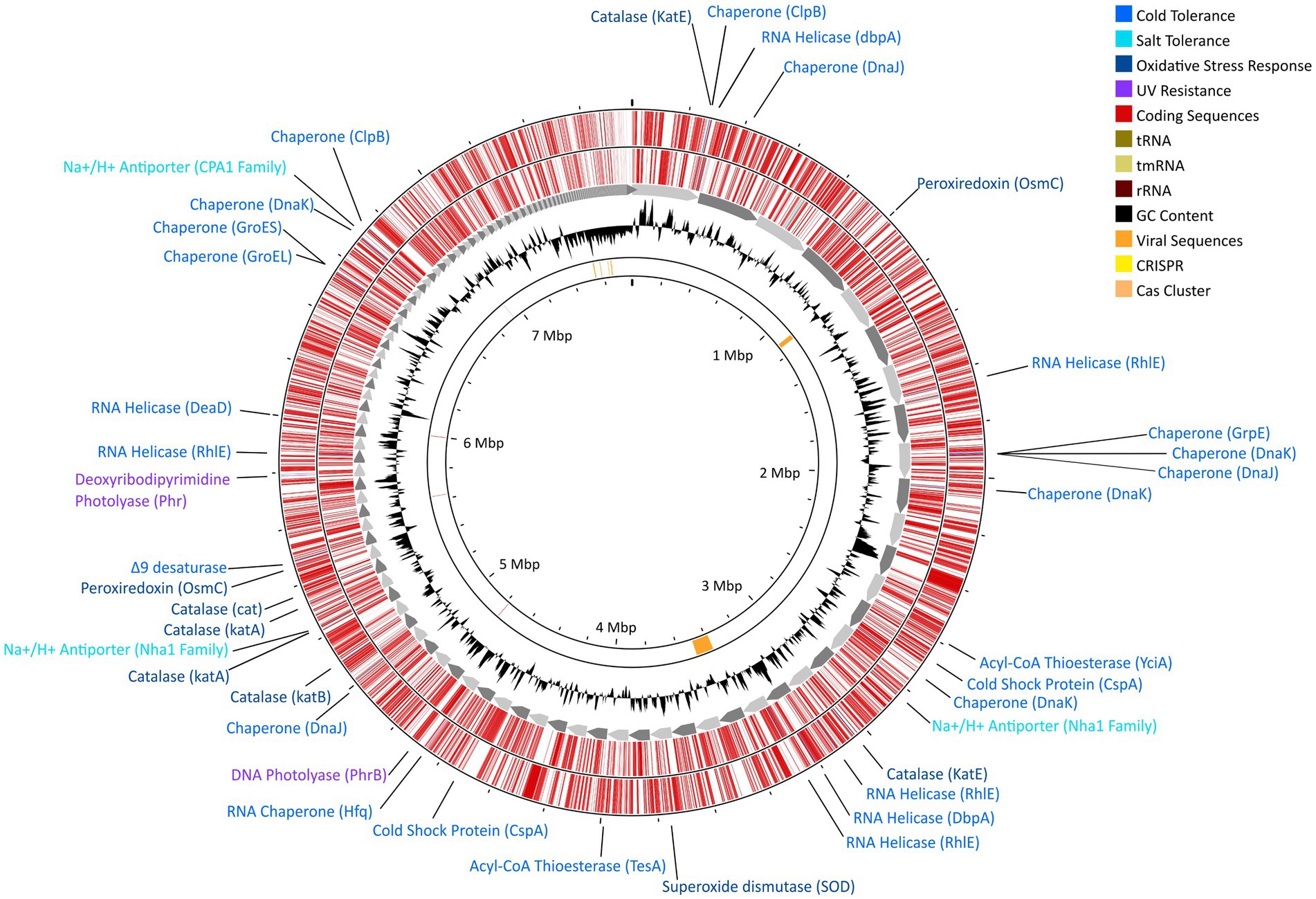
Figure 4. Gene map for strain DJPM01 highlighting proposed environmentally-relevant genes. Contigs are sorted by length and represented by grey arrows. Outer tracks depict coding sequences as annotated by Prokka v1.13.7 (Seemann, 2014), separated by strand. Genes related to environmentally-relevant stress adaptation and response are highlighted and labeled. Results of VirSorter2 v2.2.3 (Guo et al., 2021) and CRISPRCasFinder v4.2.20 (Couvin et al., 2018) are present on the inner ring.
Strain DJPM01 has genomic evidence for adaptation to cold environments (Figure 4; Supplementary Table S2). The genome contains genes homologous to those encoding protein chaperones, including two copies of ClpB (Mogk et al., 2003) and DnaJ (Qiu et al., 2006), three copies of DnaK (Thompson et al., 2012), and one copy each of GroEL (Lenz and Ron, 2014), GroES (Takei et al., 2012), and GrpE (Wu et al., 1996). There was also evidence for genes encoding cold shock proteins involved in repairing misfolded mRNA, such as ATP-dependent RNA helicases DpbA (Tsu et al., 2001), DeaD (Prud’homme-Généreux et al., 2004), and three copies of RhlE (Bizebard et al., 2004). Strain DJPM01 contained two copies of genes putatively encoding the cold shock protein CspA (La Teana et al., 1991). Three genes were also associated with the formation of unsaturated fatty acids, including a Δ9-desaturase (Li et al., 2009) and two acyl-CoA thioesterases, TesA (Cho and Cronan, 1993) and YciA (Postle and Good, 1985).
Several genes putatively involved in osmotic and oxidative stress were present as well (Figure 4; Supplementary Table S2). Strain DJPM01 has genomic evidence of several cation/proton antiporters in the NhaA (Vimont and Berche, 2000) and CPA1 families (Utsugi et al., 1998). Both the biosynthetic pathway and transporters for the compatible solute glutamate are present (Saum et al., 2006). Strain DJPM01 also contained genes for superoxide dismutase (Fee, 1991) and peroxiredoxin (Weber et al., 2006), which are enzymes known to combat oxidative stress, in addition to six copies of the catalase gene (von Ossowski et al., 1991). Several genes were associated with DNA repair (Figure 4; Supplementary Table S2). Strain DJPM01 encodes for several copies of genes for nucleotide excision, mismatch repair, and homologous and non-homologous recombination. The genome also encodes for two proteins associated with light-dependent DNA repair, deoxyribodipyrimidine photolyase (Sancar et al., 1984) and DNA photolyase (Zhang et al., 2013).
Annotation by antiSMASH (Blin et al., 2021) revealed a total of 17 putative biosynthetic gene clusters (BGCs), including high similarity matches to characterized gene clusters for aryl polyene APEEc (36%), violacein (100%), cephamycin C (36%), myxochelin (41%), iso-migrastatin (45%), and prodigiosin (42%; Supplementary Table S3). Of these genes, aryl polyenes have been proposed to mediate oxidative stress (Cimermancic et al., 2014), myxochelins are iron-chelating siderophores (Li et al., 2008), cephamycin C is a β-lactam antimicrobial (Alexander and Jensen, 1998), and iso-migrastatin is a glutaride-containing cytotoxic agent (Ju et al., 2005). The pigments violacein (blue-violet) and prodigiosin (1; red) have both been described to have antibiotic properties and UV-protective effects (Danevčič et al., 2016; Cauz et al., 2019). Closer analysis of the iso-migrastatin BGC using PRISM v4.4.5 (Skinnider et al., 2020) revealed this cluster encodes genes which are 58–72% identical (based on amino acid sequence) to those involved in the biosynthesis of the antifungal gladiofungin, however the cluster was on the end of a contig, missing roughly 15% of the third polyketide synthase gene (Supplemental Figure S4; Niehs et al., 2020).
To understand how the diversity of BGCs in DJPM01 compares to other Massilia genomes, putative secondary metabolite BGCs were identified in representative Massilia genomes from RefSeq using antiSMASH (Figure 5; O'Leary et al., 2016; Blin et al., 2021). Antarctic Massilia genomes contained an average of 14.5 secondary metabolite BGCs per genome, significantly higher than both mesophilic Massilia (9.2 BGCs per genome; Tukey, t = −4.95, p < 0.001) and Arctic and alpine Massilia (7.8 BGCs per genome; Tukey, t = −4.31, p < 0.001). This trend was still significant when normalized to genome size (ANOVA, F = 4.27, p = 0.023) and number of coding sequences per genome (ANOVA, F = 4.38, p = 0.021; Supplementary Table S4). Strain DJPM01 was the only Massilia to contain the putative gladiofungin polyketide gene cluster. The BGC for prodigiosin was only identified in Antarctic Massilia species (M. aquatica CCM 8693, M. frigida CCM 8695, M. mucilaginosa CCM 8733, and M. rubra CCM 8692).
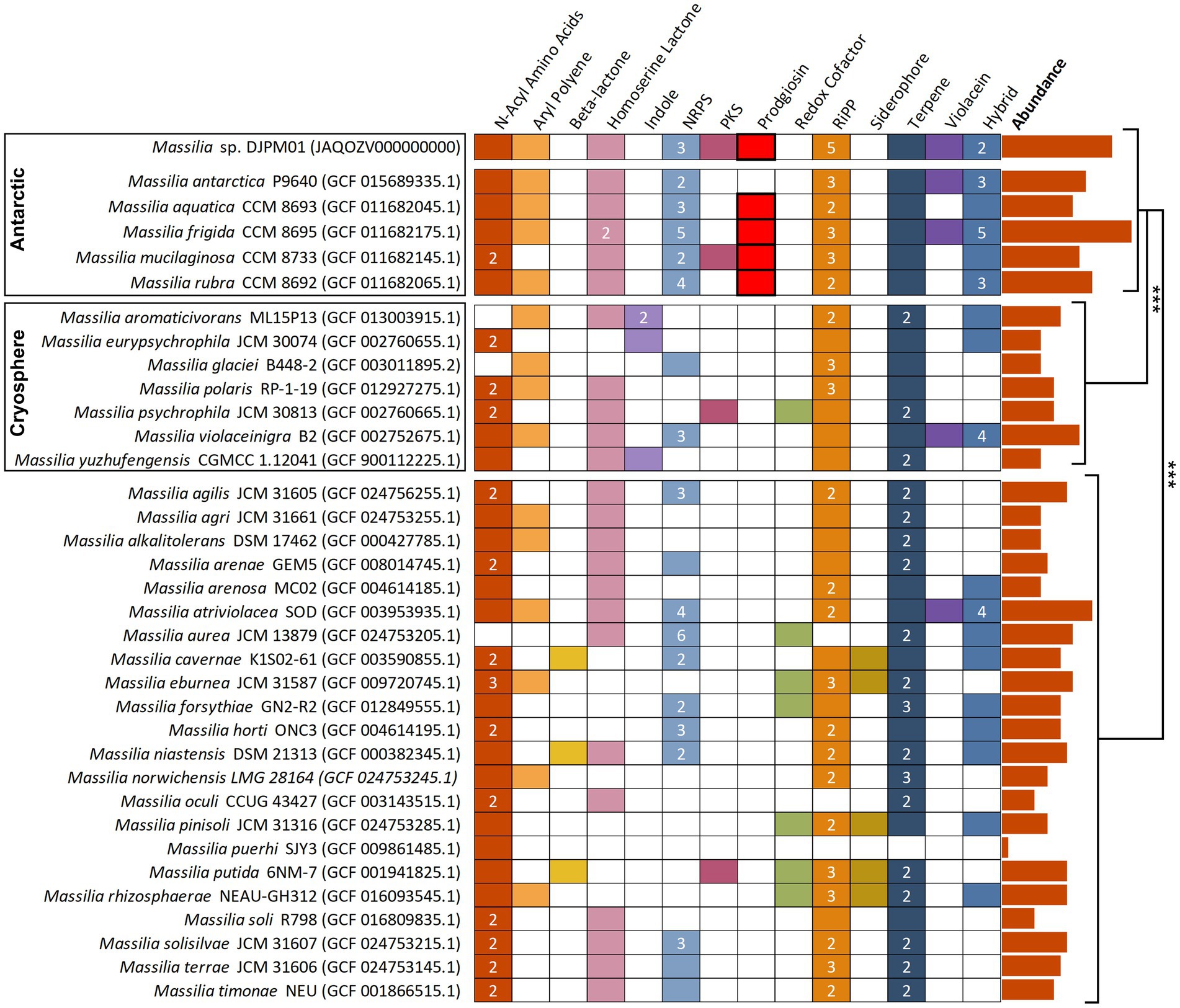
Figure 5. Annotated secondary metabolite BGS in Massilia reference genomes, sorted by isolation environment. Clusters have been sorted into broad categories, with hybrid clusters containing motifs of multiple categories (i.e., PKS-NRPS). Numbers represent the total abundance of clusters when multiple are present within the genome. Prodigiosin and violacein BGCs were confirmed via BLAST. Absolute abundance of clusters are plotted to the right of the figure and are significantly different between isolation environments (Tukey). NRPS, nonribosomal peptide synthetase; PKS, polyketide synthase; RiPP, ribosomally synthesized and post-translationally modified peptide.
Genes for Serratia sp. strain ATCC 39006 prodigiosin BGC were aligned to all Massilia genomes available on NCBI (Harris et al., 2004). Only five genomes were identified to contain BGCs with high sequence identity to the prodigiosin BGC, with the highest bit scores for pigC, pigD, and pigE (Supplementary Figure S5A). Of these five organisms, four were Massilia species originally isolated from James Ross Island. The fifth genome was reported as M. violaceinigra strain sipir (GCF 022811105.1) in NCBI; the metadata associated with this entry indicated it was isolated from freshwater. Our analyses indicated this genome had >97% ANI to the Antarctic M. aquatica CCM 8693, which also has genotypic and phenotypic evidence for prodigiosin production. Given the uncertainty about the phylogeny of this strain we excluded it from further analyses. The four Antarctic Massilia prodigiosin BGCs had high sequence identity (93.59–100.00% identity) at the hypothetical gene level to strain DJPM01 (Supplementary Figure S5B).
Gene annotations were compared between strain DJPM01 and M. frigida stain CCM 8695. Both strains shared 1990 genes with non-hypothetical functions (97.9%), with 41 genes unique to strain DJPM01 and 51 genes unique to M. frigida. Genes unique to strain DJPM01 included two genes associated with lipopolysaccharide biosynthesis (hddA and hddC), one copy of fatty acid elongation gene fabI, and three genes associated with toxin-antitoxin systems (fitA, fitB, and yefM). Both genomes contain genes associated with type IV pilin production, multifunctional fibers produced on cell surfaces, though M. frigida does not have a copy of mshC (Toma et al., 2002). An additional 91 genes had a higher copy number in strain DJPM01 and 102 genes had lower copy number in strain DJPM01. Strain DJPM01 had 2 additional copies of genes for catalase, and four fewer copies of the fatty acid biosynthesis gene fabB. When compared to non-Antarctic species, Antarctic Massilia tended to, on average, have slightly more genes associated with fatty acid biosynthesis (especially fab genes; 4 more genes), protein chaperones (1.4 more genes), and catalase (1.4 more genes).
LC/MS and UV–Vis spectroscopic analyses of extracted DJPM01 agar plates resulted in the detection of prodigiosin (1), which was confirmed with an authentic standard (Figures 6A,B). Prodigiosin (1, m/z 324.2072; calculated for C20H26N3O [M + H]+, 324.2076) was detected with a characteristic UV–Vis spectrum (λmax = 540 nm; Figures 6A,B; Supplementary Figure S6; Andreyeva and Ogorodnikova, 2015). MS/MS spectra of the peak identified as prodigiosin (1) were congruent with the fragmentation pattern of the authentic standard. Furthermore, a number of distinct peaks with similar UV–Vis absorbances were detected in chromatograms with masses consistent with prodiginines based on mass defect values and even [M + H]+ masses. Metabolomic analyses indicated the presence of numerous features consistent with those of 2-methyl-3-propyl prodiginine (2), 2-methyl-3-butyl prodiginine (3), 2-methyl-3-heptylprodiginine (4), cycloprodigiosin (5), and other uncharacterized prodiginines (Figure 6D; Supplementary Table S5).
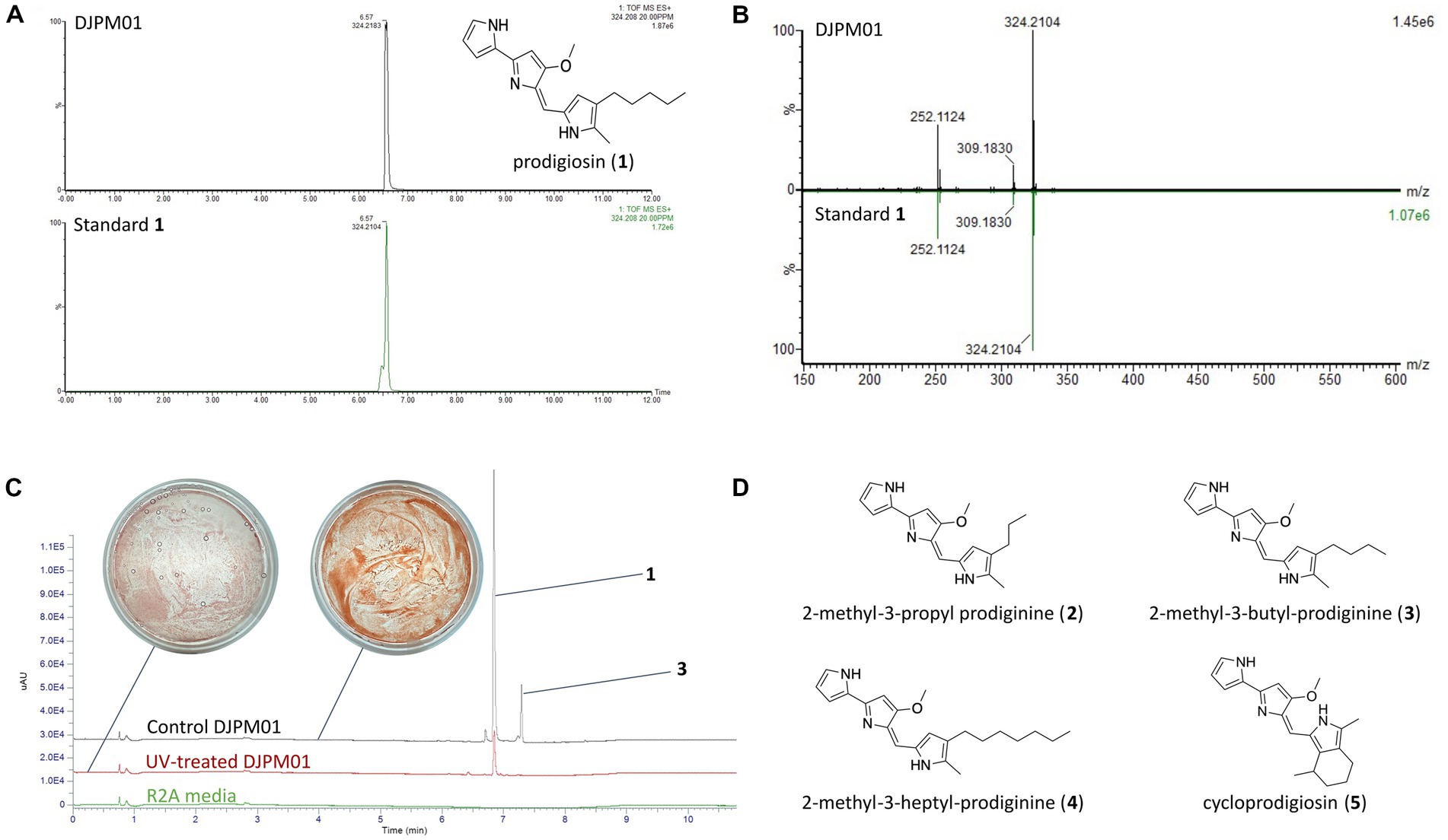
Figure 6. Detection of prodigiosin and derivatives in strain DJPM01 extracts. (A) Extracted ion chromatograms (m/z 324.208) of prodigiosin (1) in DJPM01 R2A agar extracts (black) compared to a prodigiosin standard (green). (B) Mirrored MS/MS mass spectra of prodigiosin (1) in DJPM01 agar extracts and prodigiosin standard. UV–Vis spectra of prodigiosin (1) in DJPM01 agar extracts (black) and prodigiosin standard (green) showing identical spectra. (C) Stacked UV trace diagram for plate extracts from control and UV-treated cultures. Dominant peaks corresponding to prodigiosin (1) and 2-methyl-3-butyl-prodiginine (3). Images of UV-exposed (left) and control (right) plates following 11 days of incubation. (D) Molecular structures of detected prodigiosin derivatives.
Incubation under UV light yielded a less-saturated, darker-hued pigmentation compared to control plates grown in the dark, with no UV exposure (Figure 6C). LC/MS analyses of UV-treated strain DJPM01 indicated statistically significant differences between UV-treated, control, and media blank samples (Supplementary Figure S7). A statistically significant decrease was observed in prodigiosin (1; q < 0.001) and other metabolites (Figures 6C, 7). Mass spectral analyses suggested that these uncharacterized prodiginines may be cyclized, reduced, oxygenated, and otherwise modified variations of prodigiosin. LC/MS analysis of a UV-treated prodigiosin (1) standard indicated a conversion of the compound to a non-pigmented degradation product with an m/z of 354.2169 [M + H]+ and probable molecular formula of C21H28N3O2 (Supplementary Figure S8). Additionally, several other features with mass shifts consistent with addition of CH2O to major prodiginines were observed. The only compound unique to the UV-treated strain DJPM01 was glycerophosphoserine (m/z 522.2831; calculated [M + H]+ for C24H45NO9P, 522.2832;). Masses congruent to those of arylpolyenes and migrastatin (m/z 512.2617; calculated [M + H]+ for C24H45NO9P, 522.2832) were also detected. No LC/MS or UV–Vis spectroscopic evidence of other pigments was found.
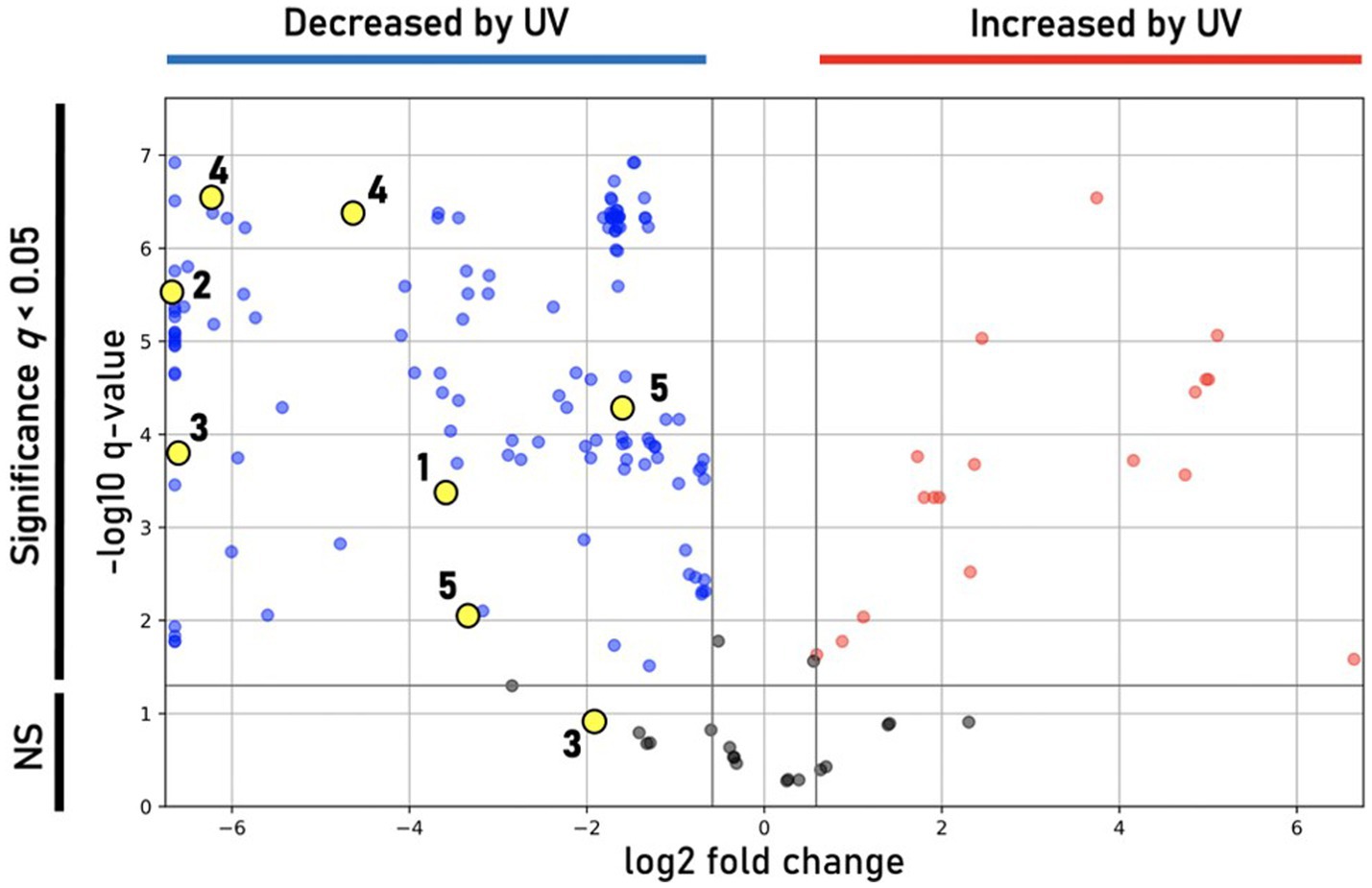
Figure 7. Differential features observed under UV treatment. Volcano plot showing statistically significant (q < 0.05) features in DJPM01 grown on R2A agar that are either increased (red) or decreased (blue) relative to control samples that were not irradiated with UV-A light. Numbered and unnumbered yellow dots correspond to known features and unknown, prodiginine-related features, respectively.
Massilia species have been identified in diverse ecological niches across Antarctica, though little is known about their function within cold systems. We isolated strain DJPM01 from a microbial mat which sits adjacent to one of the saltiest aquatic features on Earth, the hypersaline Don Juan Pond in the McMurdo Dry Valleys (Toner et al., 2017). We identified this aerobic, Gram negative bacterium as a member of the M. frigida species. This strain was isolated on R2A agar, which is commonly utilized to cultivate bacteria from Antarctic environments, though growth on this medium is not an indicator of abundance or importance to a given system (Pulschen et al., 2017). Strain DJPM01 is estimated to be present at low abundance (~0.03%) in the established microbial mat community; however, abundance and ecological importance are not mutually exclusive, as highlighted with the rare biosphere concept (Lynch and Neufeld, 2015). Members of the Massilia genus have been of interest for their agricultural and pharmaceutical applications (Lewin et al., 2021; Dahal et al., 2021b). Thus, we examined both potential adaptations to ecologically relevant conditions and production of microbial secondary metabolites, including the pigment prodigiosin, in strain DJPM01.
Modern prokaryotic species concepts strive to incorporate both the advanced sequencing tools available to interrogate microbial genomes as well as phenotypic assays (Rosselló-Móra and Amann, 2015). While specifics may be debated, it is generally agreed that species exhibit a large degree of genomic and phenotypic coherence (Rosselló-Móra and Amann, 2015). Conservative estimates indicate that a DDH of >70% (Tindall et al., 2010), ANI threshold of 95–96% (e.g., Konstantinidis and Tiedje, 2005; Konstantinidis et al., 2006), and 16S rRNA gene sequence identity of >98% (Kim et al., 2014) is appropriate for delineating prokaryotic species for taxonomic purposes. While strain DJPM01 has >99% sequence identity with several Massilia type strains, highest ANI (97.36%) and DDH (75.6%) were found with M. frigida strain CCM8695. Thus, we conclude that strain DJPM01 is a strain of this species group.
Strain DJPM01 shares phenotypic similarities with M. frigida strain CCM 8695 and other psychrotolerant strains, including colony morphology, motility, inability to grow at temperatures at or above 30°C, and sensitivity to UV-C irradiation (Holochová et al., 2020). Strain DJPM01 was more tolerant of alkaline growth conditions compared to reported observations of both psychrotolerant and mesophilic Massilia, though several Massilia species are also able to grow on media above pH 10, including M. alkalitolerans, which can survive on media adjusted to pH 12 (Xu et al., 2005). Based on 16S rRNA gene and whole-genome comparisons, strain DJPM01 forms a clade with seven named psychrotolerant Massilia, including the five described Antarctic species (Wang et al., 2018; Yang et al., 2019; Holochová et al., 2020; Sedláček et al., 2022). Of the Antarctic Massilia with published genome sequences, four have been described as producing a pink-red pigment when grown on R2A agar (Holochová et al., 2020) and contain the complete pathway for prodigiosin biosynthesis. The fifth, M. antarctica strain P9640, produces a blue-purple pigment, likely violacein (Sedláček et al., 2022). However, Massilia isolates beyond this clade (<82% ANI) have only been described as either unpigmented or as producing a yellow pigment, which could be carotenoids based on the putative BGCs present in their genomes (Gallego et al., 2006; Zhang et al., 2006).
The genome of strain DJPM01 contains several adaptations to life in an Antarctic microbial mat. Low temperatures can negatively impact transcription and translation, both through decreases in reaction rate and through changes in the secondary structure of DNA, RNA, and proteins (De Maayer et al., 2014). Strain DJPM01 encodes multiple sets of genes associated with either preventing the formation of transcription-inhibiting RNA secondary structures (cold shock proteins) or unraveling the structures that do form (RNA helicases; Siddiqui et al., 2013). Similarly, strain DJPM01 has genes encoding for several protein chaperones to ensure correct folding of polypeptide chains. Other genes, such as fab genes and a putative Δ9-desaturase, are involved in the biosynthesis of unsaturated fatty acids to increase membrane fluidity at low temperature (De Maayer et al., 2014). Cold-adapted organisms synthesize compatible solutes to prevent the formation of ice crystals within the cell and combat changes in salinity associated with freezing temperatures (Chin et al., 2010). Genes involved in the biosynthesis and transport of the compatible solute glutamate are present within the genome, as well as several cation/proton antiporters, all of which are utilized to counteract changes in osmotic pressure (Kuroda et al., 2005; Saum and Müller, 2008).
UV irradiation can lead to both DNA damage and the generation of ROS (Castenholz and Garcia-Pichel, 2012). Strain DJPM01 contained several genes for reducing ROS (e.g., catalase and peroxiredoxin) and for DNA repair, including two photolyases, which may enable photo-repair of DNA during long Antarctic days in the austral summer. UV stress can also be mediated through the production of radiation-absorbing pigments. Strain DJPM01 contains BGCs for prodigiosin and violacein, pigments associated with UV survival in other organisms, such as Serratia, Streptomyces, and Vibrio species (Borić et al., 2011; Stankovic et al., 2012; Cediel Becerra et al., 2022), and increases the UV-screening ability of commercial sunscreen (Suryawanshi et al., 2015). Violacein has been described in a number of Massilia species, including M. antarctica from James Ross Island and M. violaceinigra from Tianshan glacier, China (Agematu et al., 2011; Myeong et al., 2016; Wang et al., 2018; Yang et al., 2019; Sedláček et al., 2022). To date, prodigiosin has only been described in Antarctic Massilia species (Holochová et al., 2020). The prevalence of pigment biosynthesis genes in Antarctic Massilia suggests that both violacein and prodigiosin could play a role in adaptation to the environment. However, it remains unclear why only the Antarctic Massilia appear to possess genes for prodigiosin. Regardless, climatic change in the MDV likely involves dynamic changes in solar radiation (Obryk et al., 2018), thus the production of UV-screening pigments may not provide a universal advantage to the organisms in question.
In addition to pigments, several other putative secondary metabolite BGCs were identified in the genome, including a homoserine lactone BGC potentially involved in quorum sensing, and a putative siderophore with similarity to myxochelin, shown to play a role in iron acquisition (Kunze et al., 1989; Montgomery et al., 2013). Several RiPP clusters are also present, which produce diverse molecules with a wide variety of functions, including antibacterial or enzyme inhibiting activities (Arnison et al., 2013). Notably, strain DJPM01 contained a putative BGC involved in the biosynthesis of the antifungal polyketide gladiofungin (Niehs et al., 2020). Further analysis of PKS modules in this gene cluster supported its identification as encoding for gladiofungin biosynthetic machinery (Supplementary Figure S4). This is the first report of a Massilia with the genetic potential to produce this antifungal. Gladiofungin has been shown to inhibit the fungus Purpureocillium lilacinum (Niehs et al., 2020), which has been isolated from the Antarctic peninsula (Gonçalves et al., 2015). The gladiofungin BGC in strain DJPM01 may play a role in inhibition of fungal competitors.
Strain DJPM01 has a similar abundance of genes associated with cold adaptation when compared to other psychrotolerant Massilia species, including genes for protein chaperones, RNA helicases, and fatty acid biosynthesis. Minor differences exist between strain DJPM01 and the closely related M. frigida, including the presence of specific toxin-antitoxin genes, which are believed to play a role in stress response (Fucich and Chen, 2020). Compared to non-Antarctic Massilia, the six Massilia isolated from Antarctic regions contained a significantly greater abundance of secondary metabolite BGCs (Supplementary Table S4). The genetic potential to synthesize a wide arsenal of molecules in response to specific stressors under poly-extreme conditions would impart an ecological advantage (Giddings and Newman, 2015b; Sayed et al., 2020). Of the putatively identified BGCs, the myxochelin cluster is present across all six genomes, suggesting a strong selection pressure for iron acquisition (Kunze et al., 1989). Other gene clusters that are only present in one or two genomes, such as cephamycin and the gladiofungin we identified here, may play more specialized roles in competitive inhibition of other community members in situ. Secondary metabolism of Antarctic microbes remains poorly characterized and the abundance of putative BGCs that encode for yet uncharacterized molecules may be of interest in the search for novel bioactive compounds (Correa and Abreu, 2020). Extreme environments likely offer an untapped wealth of secondary metabolites that may prove useful in a number of industrial or biomedical applications (Giddings and Newman, 2015a; Sayed et al., 2020).
Our work provides the first molecular characterization of prodiginines produced by a member of the Massilia genus. Holochová et al. (2020) noted the pigmentation and BGC for prodigiosin in four Antarctic Massilia species, and Woodhams et al. (2018) examined a mesophilic Massilia (now Pseudoduganella; Lu et al., 2022) for the production of prodigiosins, however none were detected. In our study, mass defect analysis (Figure 8) identified several metabolites congruent with prodiginines detected within similar retention times (5–8 min), including analogs formed from either the addition of oxygen, reduction, or cyclization. Some of these metabolites are known compounds; however, most are unknown and warrant further investigation (He et al., 2022). Due to the high sequence similarity between putative prodigiosin BGCs in M. frigida, M. rubra, M. mucilaginosa, and M. aquatica, it is likely that all five red-pigmented Antarctic Massilia are genetically capable of producing related pigments.
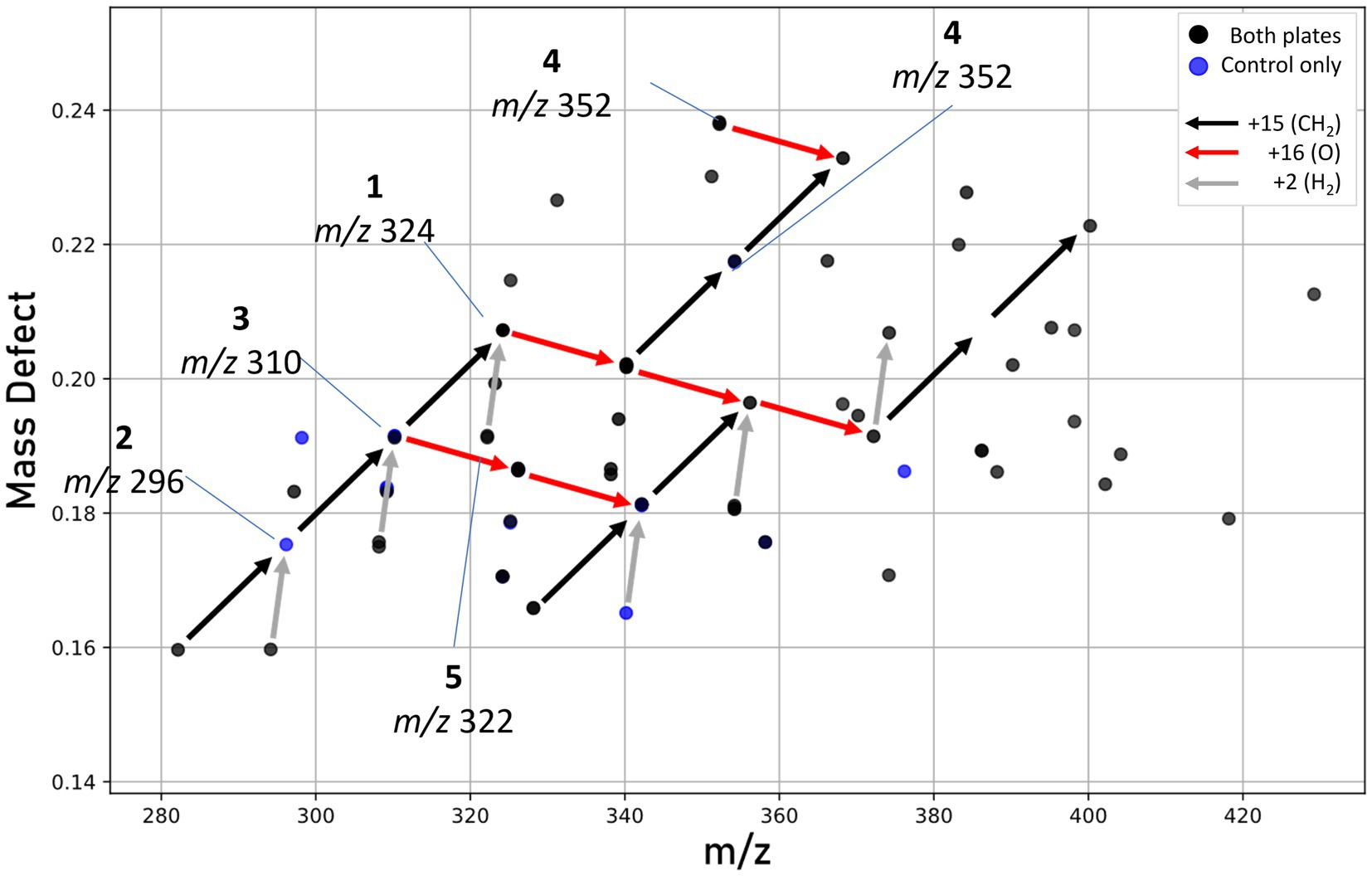
Figure 8. Mass defect plot showing features hypothesized to be prodiginines. Features seen in either both extracts (black dots) or control plate extracts only (blue dots). No unique prodiginine features were seen in UV-treated plates. Arrows represent mass defect vectors, with red denoting the addition of oxygen (+16), black the addition of a methyl group (+15), and gray a loss of hydrogen possibly indicating cyclization.
The biosynthesis of prodigiosin may provide numerous advantages to the cold-adapted Massilia, which may serve as pioneer species during microbial mat establishment (Sommers et al., 2019). Given prodigiosin’s role as a UV-protectant in other microbial species (Borić et al., 2011; Stankovic et al., 2012; Cediel Becerra et al., 2022), we sought to determine the effect of intense UV-A radiation on pigment production by strain DJPM01. Following irradiation, we saw a decrease in the abundance of most metabolites, including prodigiosin (1), with the exception of an increase in an oxygenated prodiginine (m/z 340.2020), which may represent a UV photolysis product of 2-methyl-3-butyl-prodiginine (3). We did not detect the pigment violacein under any growth condition tested, despite strong genomic evidence for a violacein BGC. Thus, it is likely that this molecule may be produced under yet-unknown conditions, such as in response to predation by metazoans (Choi et al., 2015). The color change (Figure 6C) of UV-treated strain DJPM01 was likely related to prodiginine degradation, as the irradiated prodigiosin standard was observed to lose color intensity following 24-h of UV-A exposure (Supplementary Figure S8). As pigment degradation is likely the result of UV-A exposure, and the rate of photolysis is independent from molecular synthesis, transcriptional or translational response in prodigiosin biosynthesis upon UV exposure should be further investigated. Alternatively, Massilia within the natural mat environment may also benefit from other photoprotective mechanisms. Photoprotective compounds can provide a shielding effect to the entire community within microbial mats, so that not only the producers benefit from the UV protection (Gareia-Pichel and Castenholz, 1994).
In addition to UV-protection, prodigiosin has been linked to numerous bioactivities. For example, prodigiosin has been shown to function as an antimicrobial agent against both Gram positive and Gram negative bacteria (Danevčič et al., 2016), and thus may serve as a mediator of interspecies competition within the microbial mat environment. Prodigiosin has also been reported as a nematicidal agent (Habash et al., 2020). Nematodes in the MDV function as grazers within microbial mats, and have been identified as present within the DJP mat (Siegel et al., 1979; Andriuzzi et al., 2018), thus prodigiosin may serve as a defense against overgrazing. Additionally, psychrophilic organisms increase intracellular ATP concentrations to support metabolic activity at low temperatures (Amato and Christner, 2009). In high-density cultures of S. marcescens, accumulation of intracellular ATP occurred at a higher rate in prodigiosin pigmented cells, compared to non-pigmented cells (Haddix and Shanks, 2018). Thus, prodigiosin may also play an undescribed role in energetics and adaptation to low temperatures. More directed experiments are necessary to elucidate the ecological function of prodiginines in strain DJPM01 in situ.
In the coming decades, changes in solar radiation will impact MDV communities through both direct and indirect processes. The low albedo of MDV soils leads to increased levels of energy absorption compared to the surrounding ice (Dana et al., 1998) resulting in higher summer in situ temperatures (Fountain et al., 2014), which could lead to possible hydrological and geochemical changes, such as enhanced permafrost melting and salt mobilization (Ball and Virginia, 2012). Additionally, both anthropogenic and natural (i.e., volcanoes and wildfire) emissions drive variations in solar flux in the MDV, potentially resulting in a decrease UV radiation (Obryk et al., 2018). Thus, microbial community response to UV irradiation will likely vary with local climatic trends. Investigating the responses of individual community members to simulated stressors (i.e., UV radiation) can inform how natural microbial communities may respond to environmental variation associated with anthropogenic climate change.
Our characterization of Massilia frigida strain DJPM01 provides insight into the complex microbial interactions occurring in an Antarctic microbial mat and suggests multiple avenues for future research. In Antarctic Massilia, both pigment production and the high number of BGCs suggests Massilia may play a role in mediating microbial interactions in Antarctic ecosystems and highlights the importance of investigating Antarctic microbial isolates for their production of bioactive compounds. Prodigiosin has been implicated in bactericidal, nematocidal, and UV-screening activities, though the role it plays in situ in complex, multi-species Antarctic microbial mats warrants further investigation. Our data demonstrate that UV radiation impacts the accumulation of this pigment in Massilia, thus the abundance of this molecule may be influenced by increased variation in solar radiation. The changing climate in the MDV system is complex with effects on microbial communities uncertain; understanding the role of secondary metabolites in the MDV communities, especially under ecologically relevant conditions can inform how these communities will respond to environmental change.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/genbank/, JAQOZV000000000. https://www.ncbi.nlm.nih.gov/genbank/, OQ346149.
JAM and L-AG conceived of the project and acquired funding and resources. JAM collected and processed the samples. JS, L-AG, RS, and JAM contributed to the design of the experiments. JS performed the culture work, DNA extraction, genomic analyses, and wrote the first draft of the manuscript. JS and L-AG extracted samples for metabolomics. RS analyzed the metabolite samples. L-AG and RS interpreted the metabolomics data. All authors contributed to the article and approved the submitted version.
This work was funded by the National Science Foundation OPP #2148730 and OPP #1643687 (to JAM) and OPP #2148731 (to L-AG) with additional support from the University of Tennessee Office of Research, Innovation and Economic Development (to JAM).
We are grateful to the Antarctic Support Contractor and ANTAEM Team members for logistical support during sample collection. In addition, we thank Teresa Mccarrell for initiating enrichments and Thomas Hyde for his assistance with culture work.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1156033/full#supplementary-material
Agematu, H., Suzuki, K., and Tsuya, H. (2011). Massilia sp. BS-1, a novel violacein-producing bacterium isolated from soil. Biosci. Biotechnol. Biochem. 75, 2008–2010. doi: 10.1271/bbb.100729
Alexander, D. C., and Jensen, S. E. (1998). Investigation of the Streptomyces clavuligerus cephamycin C gene cluster and its regulation by the CcaR proteinbize. J. Bacteriol. 180, 4068–4079. doi: 10.1128/JB.180.16.4068-4079.1998
Alger, A. S., McKnight, D. M., Spaulding, S. A., Tate, C. M., Shupe, G. H., Welch, K. A., et al. (1997). Ecological Processes in a Cold Desert Ecosystem: The Abundance and Species Distribution of Algal Mats in Glacial Meltwater Streams in Taylor Valley. Boulder, CO: University of Colorado: Institute of Arctic and Alpine Research, Occasional Paper 51.
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Amato, P., and Christner, B. C. (2009). Energy metabolism response to low-temperature and frozen conditions in Psychrobacter cryohalolentis. Appl. Environ. Microbiol. 75, 711–718. doi: 10.1128/AEM.02193-08
Andreyeva, I. N., and Ogorodnikova, T. I. (2015). Pigmentation of Serratia marcescens and spectral properties of prodigiosin. Microbiology 84, 28–33. doi: 10.1134/S0026261715010026
Andriuzzi, W. S., Adams, B. J., Barrett, J. E., Virginia, R. A., and Wall, D. H. (2018). Observed trends of soil fauna in the Antarctic dry valleys: early signs of shifts predicted under climate change. Ecology 99, 312–321. doi: 10.1002/ecy.2090
Apprill, A., McNally, S., Parsons, R., and Weber, L. (2015). Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat. Microb. Ecol. 75, 129–137. doi: 10.3354/ame01753
Arnison, P. G., Bibb, M. J., Bierbaum, G., Bowers, A. A., Bugni, T. S., Bulaj, G., et al. (2013). Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 30, 108–160. doi: 10.1039/c2np20085f
Ball, B. A., and Virginia, R. A. (2012). Meltwater seep patches increase heterogeneity of soil geochemistry and therefore habitat suitability. Geoderma 189–190, 652–660. doi: 10.1016/j.geoderma.2012.06.028
Bizebard, T., Ferlenghi, I., Iost, I., and Dreyfus, M. (2004). Studies on three E. coli DEAD-box helicases point to an unwinding mechanism different from that of model DNA helicases. Biochemistry 43, 7857–7866. doi: 10.1021/bi049852s
Blin, K., Shaw, S., Kloosterman, A. M., Charlop-Powers, Z., van Wezel, G. P., Medema, M. H., et al. (2021). antiSMASH 6.0: improving cluster detection and comparison capabilities. Nucleic Acids Res. 49, W29–W35. doi: 10.1093/nar/gkab335
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Borić, M., Danevčič, T., and Stopar, D. (2011). Prodigiosin from Vibrio sp. DSM 14379; a new UV-protective pigment. Microb. Ecol. 62, 528–536. doi: 10.1007/s00248-011-9857-0
Campen, R. L. (2015). Investigating the Mode of Action of Tuberculosis Drugs Using Hypersensitive Mutants of Mycobacterium smegmatis. Dissertation. Wellington, NZ: Victoria University of Wellington.
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Lozupone, C. A., Turnbaugh, P. J., et al. (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. 108, 4516–4522. doi: 10.1073/pnas.1000080107
Castenholz, R. W., and Garcia-Pichel, F. (2012). “Cyanobacterial responses to UV radiation” in Ecology of Cyanobacteria II. ed. B. A. Whitton (Dordrecht: Springer Netherlands), 481–499.
Cauz, A. C. G., Carretero, G. P. B., Saraiva, G. K. V., Park, P., Mortara, L., Cuccovia, I. M., et al. (2019). Violacein targets the cytoplasmic membrane of Bacteria. ACS Infect. Dis. 5, 539–549. doi: 10.1021/acsinfecdis.8b00245
Cediel Becerra, J. D. D., Suescún Sepúlveda, J. A., and Fuentes, J. L. (2022). Prodigiosin production and Photoprotective/Antigenotoxic properties in Serratia marcescens indigenous strains from eastern cordillera of Colombia. Photochem. Photobiol. 98, 254–261. doi: 10.1111/php.13507
Chin, J. P., Megaw, J., Magill, C. L., Nowotarski, K., Williams, J. P., Bhaganna, P., et al. (2010). Solutes determine the temperature windows for microbial survival and growth. Proc. Natl. Acad. Sci. U. S. A. 107, 7835–7840. doi: 10.1073/pnas.1000557107
Chiquet, C., Boisset, S., Pechinot, A., Creuzot-Garcher, C., Aptel, F., and Bron, A. M. (2015). Massilia timonae as cause of chronic endophthalmitis following cataract surgery. J Cataract Refract Surg 41, 1778–1780. doi: 10.1016/j.jcrs.2015.07.016
Cho, H., and Cronan, J. E. (1993). Escherichia coli thioesterase I, molecular cloning and sequencing of the structural gene and identification as a periplasmic enzyme. J. Biol. Chem. 268, 9238–9245. doi: 10.1016/S0021-9258(18)98341-9
Choi, S. Y., Yoon, K., Lee, J. I., and Mitchell, R. J. (2015). Violacein: properties and production of a versatile bacterial pigment. Biomed. Res. Int. 2015, 1–8. doi: 10.1155/2015/465056
Cimermancic, P., Medema, M. H., Claesen, J., Kurita, K., Wieland Brown, L. C., Mavrommatis, K., et al. (2014). Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters. Cells 158, 412–421. doi: 10.1016/j.cell.2014.06.034
Correa, T., and Abreu, F. (2020). “Antarctic microorganisms as sources of Q21 biotechnological products” in Physiological and Biotechnological Aspects of Extremophiles. eds. R. Salwan and V. Sharma (Cambridge, MA: Academic Press), 269–284.
Couvin, D., Bernheim, A., Toffano-Nioche, C., Touchon, M., Michalik, J., Néron, B., et al. (2018). CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins. Nucleic Acids Res. 46, W246–W251. doi: 10.1093/nar/gky425
Dahal, R. H., Chaudhary, D. K., and Kim, J. (2021b). Genome insight and description of antibiotic producing Massilia antibiotica sp. nov., isolated from oil-contaminated soil. Sci. Rep. 11:6695. doi: 10.1038/s41598-021-86232-z
Dahal, R. H., Chaudhary, D. K., Kim, D.-U., and Kim, J. (2021a). Cold-shock gene cspC in the genome of Massilia polaris sp. nov. revealed cold-adaptation. Antonie Van Leeuwenhoek 114, 1275–1284. doi: 10.1007/s10482-021-01600-z
Dana, G. L., Wharton, R. A. Jr., and Dubayah, R. A. (1998). “Solar radiation in the Mcmurdo dry valleys, Antarctica” in Ecosystem Dynamics in a Polar Desert: The Mcmurdo Dry Valleys (Washington DC: American Geophysical Union), 39–64.
Danevčič, T., Vezjak, M. B., Zorec, M., and Stopar, D. (2016). Prodigiosin – a multifaceted Escherichia coli antimicrobial agent. PLoS One 11:e0162412. doi: 10.1371/journal.pone.0162412
Davey, M. C., and Clarke, K. J. (1992). Fine structure of a terrestrial cyanobacterial mat from Antarctica1. J. Phycol. 28, 199–202. doi: 10.1111/j.0022-3646.1992.00199.x
De Maayer, P., Anderson, D., Cary, C., and Cowan, D. A. (2014). Some like it cold: understanding the survival strategies of psychrophiles. EMBO Rep. 15, 508–517. doi: 10.1002/embr.201338170
Diaz, M. A., Adams, B. J., Welch, K. A., Welch, S. A., Opiyo, S. O., Khan, A. L., et al. (2018). Aeolian material transport and its role in landscape connectivity in the McMurdo dry valleys, Antarctica. J. Geophys. Res. Earth 123, 3323–3337. doi: 10.1029/2017JF004589
Emms, D. M., and Kelly, S. (2019). OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20:238. doi: 10.1186/s13059-019-1832-y
Federhen, S. (2012). The NCBI taxonomy database. Nucleic Acids Res. 40, D136–D143. doi: 10.1093/nar/gkr1178
Fee, J. A. (1991). Regulation of sod genes in Escherichia coli: relevance to superoxide dismutase function. Mol. Microbiol. 5, 2599–2610. doi: 10.1111/j.1365-2958.1991.tb01968.x
Fountain, A. G., Levy, J. S., Gooseff, M. N., and Van Horn, D. (2014). The McMurdo Dry Valleys: A landscape on the threshold of change. Geomorphology 225, 25–35. doi: 10.1016/j.geomorph.2014.03.044
Fountain, A. G., Nylen, T. H., Monaghan, A., Basagic, H. J., and Bromwich, D. (2009). Snow in the McMurdo dry valleys, Antarctica. Int. J. Climatol. doi: 10.1002/joc.1933
Frederick, J. E., and Snell, H. E. (1988). Ultraviolet radiation levels during the Antarctic spring. Science 241, 438–440. doi: 10.1126/science.241.4864.438
Fucich, D., and Chen, F. (2020). Presence of toxin-antitoxin systems in picocyanobacteria and their ecological implications. ISME J. 14, 2843–2850. doi: 10.1038/s41396-020-00746-4
Gallego, V., Sánchez-Porro, C., García, M. T., and Ventosa, A. (2006). Massilia aurea sp. nov., isolated from drinking water. Int. J. Syst. Evol. Microbiol. 56, 2449–2453. doi: 10.1099/ijs.0.64389-0
Gareia-Pichel, F., and Castenholz, R. W. (1994). “On the significance of solar ultraviolet radiation for the ecology of microbial mats” in Microbial Mats NATO ASI Series. eds. L. J. Stal and P. Caumette (Berlin, Heidelberg: Springer), 77–84.
Giddings, L.-A., and Newman, D. J. (2015a). “Bioactive compounds from extremophiles” in SpringerBriefs in Microbiology: Extremophilic Bacteria. eds. S. M. Tiquia-Arashiro and M. Mormile (Cham, Germany: Springer).
Giddings, L.-A., and Newman, D. J. (2015b). “Bioactive compounds from terrestrial extremophiles” in Springer Briefs in Microbiology: Extremophilic Bacteria. eds. S. M. Tiquia-Arashiro and M. Mormile (Cham, Germany: Springer).
Gonçalves, V. N., Carvalho, C. R., Johann, S., Mendes, G., Alves, T. M. A., Zani, C. L., et al. (2015). Antibacterial, antifungal and antiprotozoal activities of fungal communities present in different substrates from Antarctica. Polar Biol. 38, 1143–1152. doi: 10.1007/s00300-015-1672-5
Gu, Z., Liu, Y., Xu, B., Wang, N., Jiao, N., Shen, L., et al. (2017). Massilia glaciei sp. nov., isolated from the Muztagh glacier. Int. J. Syst. Evol. Microbiol. 67, 4075–4079. doi: 10.1099/ijsem.0.002252
Guo, J., Bolduc, B., Zayed, A. A., Varsani, A., Dominguez-Huerta, G., Delmont, T. O., et al. (2021). VirSorter2: a multi-classifier, expert-guided approach to detect diverse DNA and RNA viruses. Microbiome 9:37. doi: 10.1186/s40168-020-00990-y
Guo, B., Liu, Y., Gu, Z., Shen, L., Liu, K., Wang, N., et al. (2016). Massilia psychrophila sp. nov., isolated from an ice core. Int. J. Syst. Evol. Microbiol. 66, 4088–4093. doi: 10.1099/ijsem.0.001315
Gurevich, A., Saveliev, V., Vyahhi, N., and Tesler, G. (2013). QUAST: quality assessment tool for genome assemblies. Bioinformatics 29, 1072–1075. doi: 10.1093/bioinformatics/btt086
Habash, S. S., Brass, H. U. C., Klein, A. S., Klebl, D. P., Weber, T. M., Classen, T., et al. (2020). Novel Prodiginine derivatives demonstrate bioactivities on plants, nematodes, and Fungi. Front. Plant Sci. 11:579807. doi: 10.3389/fpls.2020.579807
Haddix, P. L., and Shanks, R. M. Q. (2018). Prodigiosin pigment of Serratia marcescens is associated with increased biomass production. Arch. Microbiol. 200, 989–999. doi: 10.1007/s00203-018-1508-0
Harris, A. K. P., Williamson, N. R., Slater, H., Cox, A., Abbasi, S., Foulds, I., et al. (2004). The Serratia gene cluster encoding biosynthesis of the red antibiotic, prodigiosin, shows species- and strain-dependent genome context variation. Microbiology 150, 3547–3560. doi: 10.1099/mic.0.27222-0
He, S., Li, P., Wang, J., Zhang, Y., Lu, H., Shi, L., et al. (2022). Discovery of new secondary metabolites from marine bacteria Hahella based on an omics strategy. Mar. Drugs 20:269. doi: 10.3390/md20040269
Hirsch, P., Gallikowski, C. A., Siebert, J., Peissl, K., Kroppenstedt, R., Schumann, P., et al. (2004). Deinococcus frigens sp. nov., Deinococcus saxicola sp. nov., and Deinococcus marmoris sp. nov., low temperature and draught-tolerating, UV-resistant bacteria from continental Antarctica. Syst. Appl. Microbiol. 27, 636–645. doi: 10.1078/0723202042370008
Holochová, P., Mašlaňová, I., Sedláček, I., Švec, P., Králová, S., Kovařovic, V., et al. (2020). Description of Massilia rubra sp. nov., Massilia aquatica sp. nov., Massilia mucilaginosa sp. nov., Massilia frigida sp. nov., and one Massilia genomospecies isolated from Antarctic streams, lakes and regoliths. Syst. Appl. Microbiol. 43:126112. doi: 10.1016/j.syapm.2020.126112
Ju, J., Lim, S.-K., Jiang, H., and Shen, B. (2005). Migrastatin and Dorrigocins are shunt metabolites of iso-Migrastatin. J. Am. Chem. Soc. 127, 1622–1623. doi: 10.1021/ja043808i
Kämpfer, P., Lodders, N., Martin, K., and Falsen, E. (2012). Massilia oculi sp. nov., isolated from a human clinical specimen. Int. J. Syst. Evol. Microbiol. 62, 364–369. doi: 10.1099/ijs.0.032441-0
Kanehisa, M., Sato, Y., and Morishima, K. (2016). BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J. Mol. Biol. 428, 726–731. doi: 10.1016/j.jmb.2015.11.006
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Kim, M., Oh, H.-S., Park, S.-C., and Chun, J. (2014). Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int. J. Syst. Evol. Microbiol. 64, 346–351. doi: 10.1099/ijs.0.059774-0
Konstantinidis, K. T., Ramette, A., and Tiedje, J. M. (2006). The bacterial species definition in the genomic era. Philos Trans R Soc B Biol Sci 361, 1929–1940. doi: 10.1098/rstb.2006.1920
Konstantinidis, K. T., and Tiedje, J. M. (2005). Genomic insights that advance the species definition for prokaryotes. Proc. Natl. Acad. Sci. 102, 2567–2572. doi: 10.1073/pnas.0409727102
Kunze, B., Bedorf, N., Kohl, W., Höfle, G., and Reichenbach, H. (1989). Myxochelin a, a new iron-chelating compound from Angiococcus disciformis (myxobacterales). Production, isolation, physico-chemical and biological properties. J. Antibiot. 42, 14–17. doi: 10.7164/antibiotics.42.14
Kuroda, T., Mizushima, T., and Tsuchiya, T. (2005). Physiological roles of three Na+/H+ Antiporters in the Halophilic bacterium Vibrio parahaemolyticus. Microbiol. Immunol. 49, 711–719. doi: 10.1111/j.1348-0421.2005.tb03662.x
la Scola, B., Birtles, R. J., Mallet, M.-N., and Raoult, D. (1998). Massilia timonae gen. Nov., sp. nov., isolated from blood of an immunocompromised patient with cerebellar lesions. J. Clin. Microbiol. 36, 2847–2852. doi: 10.1128/JCM.36.10.2847-2852.1998
La Teana, A., Brandi, A., Falconi, M., Spurio, R., Pon, C. L., and Gualerzi, C. O. (1991). Identification of a cold shock transcriptional enhancer of the Escherichia coli gene encoding nucleoid protein H-NS. Proc. Natl. Acad. Sci. 88, 10907–10911. doi: 10.1073/pnas.88.23.10907
Lenz, G., and Ron, E. Z. (2014). Novel interaction between the major bacterial heat shock chaperone (GroESL) and an RNA chaperone (CspC). J. Mol. Biol. 426, 460–466. doi: 10.1016/j.jmb.2013.10.018
Levy, J. (2013). How big are the McMurdo dry valleys? Estimating ice-free area using Landsat image data. Antarct. Sci. 25, 119–120. doi: 10.1017/S0954102012000727
Lewin, S., Francioli, D., Ulrich, A., and Kolb, S. (2021). Crop host signatures reflected by co-association patterns of keystone Bacteria in the rhizosphere microbiota. Environ Microbiome 16:18. doi: 10.1186/s40793-021-00387-w
Li, Y., Weissman, K. J., and Müller, R. (2008). Myxochelin biosynthesis: direct evidence for two- and four-Electron reduction of a carrier protein-bound Thioester. J. Am. Chem. Soc. 130, 7554–7555. doi: 10.1021/ja8025278
Li, Y., Xu, X., Dietrich, M., Urlacher, V. B., Schmid, R. D., Ouyang, P., et al. (2009). Identification and functional expression of a Δ9 fatty acid desaturase from the marine bacterium Pseudoalteromonas sp. MLY15. J. Mol. Catal. B Enzym. 56, 96–101. doi: 10.1016/j.molcatb.2008.07.012
Lu, H.-B., Cai, Z.-P., Yang, Y.-G., and Xu, M.-Y. (2020). Duganella rivus sp. nov., Duganella fentianensis sp. nov., Duganella qianjiadongensis sp. nov. and Massilia guangdongensis sp. nov., isolated from subtropical streams in China and reclassification of all species within genus Pseudoduganella. Antonie Van Leeuwenhoek 113, 1155–1165. doi: 10.1007/s10482-020-01422-5
Lu, H., Song, D., Deng, T., Mei, C., and Xu, M. (2022). Duganella vulcania sp. nov., Rugamonas fusca sp. nov., Rugamonas brunnea sp. nov. and Rugamonas apoptosis sp. nov., isolated from subtropical streams, and phylogenomic analyses of the genera Janthinobacterium, Duganella, Rugamonas, Pseudoduganella and Massilia. Int. J. Syst. Evol. Microbiol. 72:005407. doi: 10.1099/ijsem.0.005407
Lynch, M. D. J., and Neufeld, J. D. (2015). Ecology and exploration of the rare biosphere. Nat. Rev. Microbiol. 13, 217–229. doi: 10.1038/nrmicro3400
McKnight, D. M., Tate, C. M., Andrews, E. D., Niyogi, D. K., Cozzetto, K., Welch, K., et al. (2007). Reactivation of a cryptobiotic stream ecosystem in the McMurdo dry valleys, Antarctica: a long-term geomorphological experiment. Geomorphology 89, 186–204. doi: 10.1016/j.geomorph.2006.07.025
Meier-Kolthoff, J. P., Carbasse, J. S., Peinado-Olarte, R. L., and Göker, M. (2022). TYGS and LPSN: a database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes. Nucleic Acids Res. 50, D801–D807. doi: 10.1093/nar/gkab902
Mikucki, J., Lyons, W. B., Hawes, I., Lanoil, B. D., and Doran, P. T. (2010). “Saline lakes and ponds in the McMurdo dry valleys: ecological analogs to martian paleolake environments” in Life in Antarctic Deserts and Other Cold Dry Environments: Astrobiological Analogs. eds. P. T. Doran, W. B. Lyons, and D. M. McKnight (Cambridge, Massachusetts: Cambridge University Press), 160–194.
Mogk, A., Schlieker, C., Strub, C., Rist, W., Weibezahn, J., and Bukau, B. (2003). Roles of individual domains and conserved motifs of the AAA+ chaperone ClpB in Oligomerization, ATP hydrolysis, and chaperone activity*. J. Biol. Chem. 278, 17615–17624. doi: 10.1074/jbc.M209686200
Montgomery, K., Charlesworth, J. C., LeBard, R., Visscher, P. T., and Burns, B. P. (2013). Quorum sensing in extreme environments. Life 3, 131–148. doi: 10.3390/life3010131
Myeong, N. R., Seong, H. J., Kim, H.-J., and Sul, W. J. (2016). Complete genome sequence of antibiotic and anticancer agent violacein producing Massilia sp. strain NR 4-1. J. Biotechnol. 223, 36–37. doi: 10.1016/j.jbiotec.2016.02.027
Niehs, S. P., Kumpfmüller, J., Dose, B., Little, R. F., Ishida, K., Flórez, L. V., et al. (2020). Insect-associated Bacteria assemble the antifungal Butenolide Gladiofungin by non-canonical polyketide chain termination. Angew. Chem. Int. Ed. 59, 23122–23126. doi: 10.1002/anie.202005711
Obryk, M. K., Doran, P. T., Fountain, A. G., Myers, M., and McKay, C. P. (2020). Climate from the McMurdo dry valleys, Antarctica, 1986–2017: surface air temperature trends and redefined summer season. J. Geophys. Res. Atmos. 125:e2019JD032180. doi: 10.1029/2019JD032180
Obryk, M. K., Fountain, A. G., Doran, P. T., Lyons, W. B., and Eastman, R. (2018). Drivers of solar radiation variability in the McMurdo dry valleys, Antarctica. Sci Rep 8:5002. doi: 10.1038/s41598-018-23390-7
Ofek, M., Hadar, Y., and Minz, D. (2012). Ecology of root colonizing Massilia (Oxalobacteraceae). PLoS One 7:e40117. doi: 10.1371/journal.pone.0040117
O'Leary, N. A., Wright, M. W., Brister, J. R., Ciufo, S., Haddad, D., McVeigh, R., et al. (2016). Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 44, D733–D745. doi: 10.1093/nar/gkv1189
Parks, D. H., Imelfort, M., Skennerton, C. T., Hugenholtz, P., and Tyson, G. W. (2015). CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25, 1043–1055. doi: 10.1101/gr.186072.114
Peeters, K., Verleyen, E., Hodgson, D. A., Convey, P., Ertz, D., Vyverman, W., et al. (2012). Heterotrophic bacterial diversity in aquatic microbial mat communities from Antarctica. Polar Biol. 35, 543–554. doi: 10.1007/s00300-011-1100-4
Peng, J., Chou, K., Li, C., and Lee, S. (2011). Generation kinetics of color centers in irradiated poly(4-methyl-1-pentene). J. Appl. Phys. 110:063529. doi: 10.1063/1.3642957
Postle, K., and Good, R. F. (1985). A bidirectional rho-independent transcription terminator between the E. coli tonB gene and an opposing gene. Cells 41, 577–585. doi: 10.1016/s0092-8674(85)80030-1
Power, S. N., Salvatore, M. R., Sokol, E. R., Stanish, L. F., and Barrett, J. E. (2020). Estimating microbial mat biomass in the McMurdo dry valleys, Antarctica using satellite imagery and ground surveys. Polar Biol. 43, 1753–1767. doi: 10.1007/s00300-020-02742-y
Prud’homme-Généreux, A., Beran, R. K., Iost, I., Ramey, C. S., Mackie, G. A., and Simons, R. W. (2004). Physical and functional interactions among RNase E, polynucleotide phosphorylase and the cold-shock protein, CsdA: evidence for a ‘cold shock degradosome’. Mol. Microbiol. 54, 1409–1421. doi: 10.1111/j.1365-2958.2004.04360.x
Pulschen, A. A., Bendia, A. G., Fricker, A. D., Pellizari, V. H., Galante, D., and Rodrigues, F. (2017). Isolation of uncultured Bacteria from Antarctica using long incubation periods and low nutritional media. Front. Microbiol. 8:1346. doi: 10.3389/fmicb.2017.01346
Qiu, X.-B., Shao, Y.-M., Miao, S., and Wang, L. (2006). The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell. Mol. Life Sci. 63, 2560–2570. doi: 10.1007/s00018-006-6192-6
R Core Team (2018). R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. Available at: https://www.R-project.org/ (Accessed January 28, 2023).
Rodriguez-R, L. M., and Konstantinidis, K. T. (2016). The enveomics collection: a toolbox for specialized analyses of microbial genomes and metagenomes. PeerJ 4:e1900v1. doi: 10.7287/peerj.preprints.1900v1
Rosselló-Móra, R., and Amann, R. (2015). Past and future species definitions for bacteria and archaea. Syst. Appl. Microbiol. 38, 209–216. doi: 10.1016/j.syapm.2015.02.001
Samples, R., Puckett, S., and Balunas, M. (2022). MPACT: An advanced informatics tool for metabolomics and data visualization of specialized metabolites from complex microbial samples. ChemRxiv. [Preprint]. doi: 10.26434/chemrxiv-2022-r0xbx
Sancar, G. B., Smith, F. W., Lorence, M. C., Rupert, C. S., and Sancar, A. (1984). Sequences of the Escherichia coli photolyase gene and protein. J. Biol. Chem. 259, 6033–6038. doi: 10.1016/S0021-9258(18)91118-X
Sanyal, A., Antony, R., Samui, G., and Thamban, M. (2018). Microbial communities and their potential for degradation of dissolved organic carbon in cryoconite hole environments of Himalaya and Antarctica. Microbiol. Res. 208, 32–42. doi: 10.1016/j.micres.2018.01.004
Saum, S. H., and Müller, V. (2008). Regulation of osmoadaptation in the moderate halophile Halobacillus halophilus: chloride, glutamate and switching osmolyte strategies. Saline Systems 4:4. doi: 10.1186/1746-1448-4-4
Saum, S. H., Sydow, J. F., Palm, P., Pfeiffer, F., Oesterhelt, D., and Müller, V. (2006). Biochemical and molecular characterization of the biosynthesis of glutamine and glutamate, two major compatible solutes in the moderately Halophilic bacterium Halobacillus halophilus. J. Bacteriol. 188, 6808–6815. doi: 10.1128/JB.00781-06
Sayed, A. M., Hassan, M. H. A., Alhadrami, H. A., Hassan, H. M., Goodfellow, M., and Rateb, M. E. (2020). Extreme environments: microbiology leading to specialized metabolites. J. Appl. Microbiol. 128, 630–657. doi: 10.1111/jam.14386
Schlatter, D. C., Yin, C., Hulbert, S., and Paulitz, T. C. (2020). Core Rhizosphere microbiomes of Dryland wheat are influenced by location and land use history. Appl. Environ. Microbiol. 86:e02135-19. doi: 10.1128/AEM.02135-19
Schuler, C. (2022). Astrobiological Analogs: Exploring the Microbial Ecology of Cryptic Cryosphere Habitats. Dissertation. Knoxville, TN: University of Tennessee.
Sedláček, I., Holochová, P., Busse, H.-J., Koublová, V., Králová, S., Švec, P., et al. (2022). Characterisation of waterborne psychrophilic Massilia isolates with Violacein production and description of Massilia antarctica sp. nov. Microorganisms 10:704. doi: 10.3390/microorganisms10040704
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Shen, L., Liu, Y., Gu, Z., Xu, B., Wang, N., Jiao, N., et al. (2015). Massilia eurypsychrophila sp. nov. a facultatively psychrophilic bacteria isolated from ice core. Int. J. Syst. Evol. Microbiol. 65, 2124–2129. doi: 10.1099/ijs.0.000229
Shen, L., Liu, Y., Wang, N., Yao, T., Jiao, N., Liu, H., et al. (2013). Massilia yuzhufengensis sp. nov., isolated from an ice core. Int. J. Syst. Evol. Microbiol. 63, 1285–1290. doi: 10.1099/ijs.0.042101-0
Siddiqui, K. S., Williams, T. J., Wilkins, D., Yau, S., Allen, M. A., Brown, M. V., et al. (2013). Psychrophiles. Annu. Rev. Earth Planet. Sci. 41, 87–115. doi: 10.1146/annurev-earth-040610-133514
Siegel, B. Z., Mcmurty, G., Siegel, S. M., Chen, J., and Larock, P. (1979). Life in the calcium chloride environment of Don Juan pond, Antarctica. Nature 280, 828–829. doi: 10.1038/280828a0
Siegel, B. Z., Siegel, S. M., Chen, J., and LaRock, P. (1983). An extraterrestrial habitat on earth: the algal mat of Don Jaun pond. Adv. Space Res. 3, 39–42. doi: 10.1016/0273-1177(83)90171-0
Skinnider, M. A., Johnston, C. W., Gunabalasingam, M., Merwin, N. J., Kieliszek, A. M., MacLellan, R. J., et al. (2020). Comprehensive prediction of secondary metabolite structure and biological activity from microbial genome sequences. Nat. Commun. 11:6058. doi: 10.1038/s41467-020-19986-1
Smith, A. C., and Hussey, M. A. (2005). Gram Stain Protocols. American Society for Microbiology, 1:14.
Sommers, P., Darcy, J. L., Porazinska, D. L., Gendron, E. M. S., Fountain, A. G., Zamora, F., et al. (2019). Comparison of microbial communities in the sediments and water columns of frozen Cryoconite holes in the McMurdo dry valleys, Antarctica. Front Microbiol 10:65. doi: 10.3389/fmicb.2019.00065
Son, J., Lee, H., Kim, M., Kim, D.-U., and Ka, J.-O. (2021). Massilia aromaticivorans sp. nov., a BTEX degrading bacterium isolated from Arctic soil. Curr. Microbiol. 78, 2143–2150. doi: 10.1007/s00284-021-02379-y
Stankovic, N., Radulovic, V., Petkovic, M., Vuckovic, I., Jadranin, M., Vasiljevic, B., et al. (2012). Streptomyces sp. JS520 produces exceptionally high quantities of undecylprodigiosin with antibacterial, antioxidative, and UV-protective properties. Appl. Microbiol. Biotechnol. 96, 1217–1231. doi: 10.1007/s00253-012-4237-3
Suryawanshi, R. K., Patil, C. D., Borase, H. P., Narkhede, C. P., Stevenson, A., Hallsworth, J. E., et al. (2015). Towards an understanding of bacterial metabolites prodigiosin and violacein and their potential for use in commercial sunscreens. Int. J. Cosmet. Sci. 37, 98–107. doi: 10.1111/ics.12175
Takei, Y., Iizuka, R., Ueno, T., and Funatsu, T. (2012). Single-molecule observation of protein folding in symmetric GroEL-(GroES)2 complexes*. J. Biol. Chem. 287, 41118–41125. doi: 10.1074/jbc.M112.398628
Tamura, K., Stecher, G., and Kumar, S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Thompson, A. D., Bernard, S. M., Skiniotis, G., and Gestwicki, J. E. (2012). Visualization and functional analysis of the oligomeric states of Escherichia coli heat shock protein 70 (Hsp70/DnaK). Cell Stress Chaperones 17, 313–327. doi: 10.1007/s12192-011-0307-1
Tindall, B. J., Rosselló-Móra, R., Busse, H.-J., Ludwig, W., and Kämpfer, P. (2010). Notes on the characterization of prokaryote strains for taxonomic purposes. Int. J. Syst. Evol. Microbiol. 60, 249–266. doi: 10.1099/ijs.0.016949-0
Tkacz, A., Cheema, J., Chandra, G., Grant, A., and Poole, P. S. (2015). Stability and succession of the rhizosphere microbiota depends upon plant type and soil composition. ISME J. 9, 2349–2359. doi: 10.1038/ismej.2015.41
Toma, C., Kuroki, H., Nakasone, N., Ehara, M., and Iwanaga, M. (2002). Minor pilin subunits are conserved in Vibrio cholerae type IV pili. FEMS Immunol Med Microbiol 33, 35–40. doi: 10.1111/j.1574-695X.2002.tb00569.x
Toner, J. D., Catling, D. C., and Sletten, R. S. (2017). The geochemistry of Don Juan pond: evidence for a deep groundwater flow system in Wright Valley, Antarctica. Earth Planet. Sci. Lett. 474, 190–197. doi: 10.1016/j.epsl.2017.06.039
Tosi, S., Onofri, S., Brusoni, M., Zucconi, L., and Vishniac, H. (2005). Response of Antarctic soil fungal assemblages to experimental warming and reduction of UV radiation. Polar Biol. 28, 470–482. doi: 10.1007/s00300-004-0698-x
Tsu, C. A., Kossen, K., and Uhlenbeck, O. C. (2001). The Escherichia coli DEAD protein DbpA recognizes a small RNA hairpin in 23S rRNA. RNA 7, 702–709. doi: 10.1017/S1355838201010135
Tsugawa, H., Cajka, T., Kind, T., Ma, Y., Higgins, B., Ikeda, K., et al. (2015). MS-DIAL: data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat. Methods 12, 523–526. doi: 10.1038/nmeth.3393
Utsugi, J., Inaba, K., Kuroda, T., Tsuda, M., and Tsuchiya, T. (1998). Cloning and sequencing of a novel Na+/H+ antiporter gene from Pseudomonas aeruginosa. Biochim. Biophys. Acta 1398, 330–334. doi: 10.1016/s0167-4781(98)00058-x
Van Craenenbroeck, A. H., Camps, K., Zachée, P., and Wu, K. L. (2011). Massilia timonae infection presenting as generalized lymphadenopathy in a man returning to Belgium from Nigeria▿. J. Clin. Microbiol. 49, 2763–2765. doi: 10.1128/JCM.00160-11
Veltri, D., Wight, M. M., and Crouch, J. A. (2016). SimpleSynteny: a web-based tool for visualization of microsynteny across multiple species. Nucleic Acids Res. 44, W41–W45. doi: 10.1093/nar/gkw330
Vimont, S., and Berche, P. (2000). NhaA, an Na(+)/H(+) antiporter involved in environmental survival of Vibrio cholerae. J. Bacteriol. 182, 2937–2944. doi: 10.1128/JB.182.10.2937-2944.2000
von Ossowski, I., Mulvey, M. R., Leco, P. A., Borys, A., and Loewen, P. C. (1991). Nucleotide sequence of Escherichia coli katE, which encodes catalase HPII. J. Bacteriol. 173, 514–520. doi: 10.1128/jb.173.2.514-520.1991
Wang, H., Zhang, X., Wang, S., Zhao, B., Lou, K., and Xing, X.-H. (2018). Massilia violaceinigra sp. nov., a novel purple-pigmented bacterium isolated from glacier permafrost. Int. J. Syst. Evol. Microbiol. 68, 2271–2278. doi: 10.1099/ijsem.0.002826
Weber, A., Kögl, S. A., and Jung, K. (2006). Time-dependent proteome alterations under osmotic stress during aerobic and anaerobic growth in Escherichia coli. J. Bacteriol. 188, 7165–7175. doi: 10.1128/JB.00508-06
Weisburg, W. G., Barns, S. M., Pelletier, D. A., and Lane, D. J. (1991). 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173, 697–703. doi: 10.1128/jb.173.2.697-703.1991
Wick, R. R., Judd, L. M., Gorrie, C. L., and Holt, K. E. (2017). Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13:e1005595. doi: 10.1371/journal.pcbi.1005595
Woodhams, D. C., LaBumbard, B. C., Barnhart, K. L., Becker, M. H., Bletz, M. C., Escobar, L. A., et al. (2018). Prodigiosin, Violacein, and volatile organic compounds produced by widespread cutaneous Bacteria of amphibians can inhibit two Batrachochytrium fungal pathogens. Microb. Ecol. 75, 1049–1062. doi: 10.1007/s00248-017-1095-7
Wu, B., Wawrzynow, A., Zylicz, M., and Georgopoulos, C. (1996). Structure-function analysis of the Escherichia coli GrpE heat shock protein. EMBO J. 15, 4806–4816. doi: 10.1002/j.1460-2075.1996.tb00861.x
Xu, P., Li, W.-J., Tang, S.-K., Zhang, Y.-Q., Chen, G.-Z., Chen, H.-H., et al. (2005). Naxibacter alkalitolerans gen. Nov., sp. nov., a novel member of the family ‘Oxalobacteraceae’ isolated from China. Int. J. Syst. Evol. Microbiol. 55, 1149–1153. doi: 10.1099/ijs.0.63407-0
Yang, E., Zhao, M., Li, S., Wang, Y., Sun, L., Liu, J., et al. (2019). Massilia atriviolacea sp. nov., a dark purple-pigmented bacterium isolated from soil. Int. J. Syst. Evol. Microbiol. 69, 2135–2141. doi: 10.1099/ijsem.0.003449
Zhang, Y.-Q., Li, W.-J., Zhang, K.-Y., Tian, X.-P., Jiang, Y., Xu, L.-H., et al. (2006). Massilia dura sp. nov., Massilia albidiflava sp. nov., Massilia plicata sp. nov. and Massilia lutea sp. nov., isolated from soils in China. Int. J. Syst. Evol. Microbiol. 56, 459–463. doi: 10.1099/ijs.0.64083-0
Zhang, F., Scheerer, P., Oberpichler, I., Lamparter, T., and Krauß, N. (2013). Crystal structure of a prokaryotic (6-4) photolyase with an Fe-S cluster and a 6,7-dimethyl-8-ribityllumazine antenna chromophore. Proc. Natl. Acad. Sci. U. S. A. 110, 7217–7222. doi: 10.1073/pnas.1302377110
Keywords: prodigiosin biosynthesis, Antarctic Massilia, psychrophilic bacteria, whole-genome sequencing, Don Juan Pond Basin
Citation: Shaffer JMC, Giddings L-A, Samples RM and Mikucki JA (2023) Genomic and phenotypic characterization of a red-pigmented strain of Massilia frigida isolated from an Antarctic microbial mat. Front. Microbiol. 14:1156033. doi: 10.3389/fmicb.2023.1156033
Received: 01 February 2023; Accepted: 20 April 2023;
Published: 12 May 2023.
Edited by:
Trista J. Vick-Majors, Michigan Technological University, United StatesReviewed by:
Wei Li, Lawrence Livermore National Laboratory (DOE), United StatesCopyright © 2023 Shaffer, Giddings, Samples and Mikucki. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jill A. Mikucki, am1pa3Vja2lAdXRrLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.