- 1State Key Laboratory of Soil and Sustainable Agriculture, Institute of Soil Science, Chinese Academy of Sciences, Nanjing, China
- 2University of Chinese Academy of Sciences, Beijing, China
- 3Ecological Experimental Station of Red Soil, Chinese Academy of Sciences, Yingtan, China
- 4College of Land Resources and Environment, Jiangxi Agricultural University, Nanchang, China
The process of carbon (C) sequestration plays an important role in soil fertility and productivity, yet most studies have focused on the individual role of the bacterial community. However, an in-depth mechanistic understanding of how soil nematodes interact with the bacterial community to regulate soil C accumulation is still lacking. We conducted a 10-year field experiment to explore the nematode and bacterial communities and determine the influence of nematode-bacteria interactions on C mineralization, microbial metabolic quotient (qCO2), and carbon use efficiency (CUE) under the organic material amendments, including chemical fertilizers with straw (NS), chemical fertilizers with straw and pig manure (NSM), and chemical fertilizer with straw biochar (NB). Here, our results showed the abundance of bacterial and nematode communities was significantly higher under NS, NSM, and NB treatments than under chemical fertilizers (N) treatment, with the highest abundance under the NSM treatment. The enrichment index and functional dispersion index were significantly higher under NSM treatment than under N, NS, and NB treatments, while the channel index followed the opposite pattern. Structural equation modeling indicated that the potential predation pressure induced by nematodes may improve bacterial abundance, with positive cascading effects on C sequestration. Collectively, our study highlights the functional importance of nematode-microorganism interactions in mediating C dynamics under organic material amendments.
Introduction
Soil organic carbon (SOC) reservoir is the major component of the carbon (C) pool in terrestrial ecosystems and plays an important role in soil fertility and crop yield (Cook-Patton et al., 2020). Excessive use of chemical fertilizers degrades soil structure and reduces C content, while the application of organic materials, especially straw and manure, is considered an effective measure to improve soil quality and C sequestration (Wei et al., 2021). Straw itself is rich in mineral elements and nutrients and is often combined with chemical fertilizers to simultaneously improve C turnover and accumulation (Berhane et al., 2020). Pig manure efficiently increases easily decomposable particulate organic carbon due to its high nitrogen content and low carbon to nitrogen ratio (Gross and Glaser, 2021). A comprehensive understanding of the biological mechanisms of C turnover is paramount to quantifying C accumulation potential in agricultural ecosystems.
Microorganisms are the primary C decomposers in agricultural ecosystems (Zechmeister-Boltenstern et al., 2015). External carbon sources cause a series of changes in the abundance and composition of the soil microbial community, thereby affecting microbial metabolic quotient and C mineralization. Microbial biomass carbon is an essential component of the C pool (Fan et al., 2021). Microorganisms alter the amount and chemical composition of soil organic matter through decomposition, respiration, growth, and death (Min et al., 2021). While C measurements can represent changes in the magnitude of C storage, indicators related to microbial biomass and respiration can predict long-term trends in C sequestration (Zhou et al., 2017). The ratio of soil microbial respiration to growth (qCO2) indicates the energy required to maintain microbial biomass (Fließbach et al., 2000), with the higher qCO2 indicating faster nutrient turnover and lower carbon use efficiency (CUE) (Wardle and Ghani, 1995). It is generally accepted that increasing microbial CUE can enhance C sequestration (Sinsabaugh et al., 2013). However, the potential mechanism by which CUE varies with substrate quality and predation pressure needs to be further investigated (Dijkstra et al., 2015; Kane et al., 2022).
Traditional research has mainly focused on the individual role of the soil microbial community in C turnover and accumulation, but the understanding of how the interactions between soil animals and microorganisms affect C dynamics under the organic material amendments is still emerging (Van Den Hoogen et al., 2019). Nematodes are widely known as the most abundant soil invertebrates and contribute significantly to C decomposition and cycling through their interactions with the bacterial community (Jiang et al., 2013; Pausch et al., 2016; Zhang et al., 2021). Nematode predation alters the diversity and structure of the bacterial community and limits bacterial community size and physiology, thereby affecting C-related microbial functions (Grandy et al., 2016; Wang et al., 2021) A global-scale study further highlights the importance of nematode assemblages in soil food webs, nutrient cycling, and ecosystem functioning (Van Den Hoogen et al., 2019). Based on their board distribution and feeding traits, the ecological indices of nematode communities are well established as indicators of soil environmental conditions and food web status (Ferris et al., 2001). Although a rapidly growing literature has investigated the nematode-microbe interactions, there is still insufficient research on the direction and strength of nematode-microbe interactions on the regulation of C dynamics at different fertility levels (Grandy et al., 2016). Therefore, it is necessary to explore the role of nematodes on microorganisms to better understand the ecological role of nematode predation in mediating C dynamics.
Here, we aimed to examine how the influence of nematode predation affects C mineralization and sequestration by affecting the bacterial community in response to the organic material amendments. We used a field experiment under five fertilization treatments, and focused on answering the following two questions: (1) how the diversity, abundance, and composition of bacterial and nematode communities varied with the addition of different organic materials? and (2) whether and to what extent the nematode-bacteria interactions mediated C sequestration subject to organic material amendments? To address these questions, we examined the bacterial community structure by using Illumina sequencing, identified nematodes under a microscope, and determined qCO2 and CUE. We hypothesized that the nematode predation on bacterial community would improve C turnover and accumulation under organic material amendments.
Materials and methods
Description of the field experiment
The long-term experiment was established at the Yingtan National Agro-Ecosystem Observation and Research Station of the Chinese Academy of Sciences, Jiangxi Province (E116°55′30″, N28°15′20″). The climate of the region is characterized as warm and monsoon with a mean annual temperature of 17.6°C and a mean annual precipitation of 1795 mm. The soil is an acid loamy clay derived from Ferric Acrisols in the FAO classification system and Udic Ferralsols in the Chinese Soil Taxonomy.
The field experiment was initiated in 2011, and included five treatments with three replications in a randomized design: (1) no fertilizer (CK); (2) NPK chemical fertilizers (N); (3) NPK chemical fertilizers with straw (NS); (4) NPK chemical fertilizers with straw and pig manure (NSM); and (5) NPK chemical fertilizers with straw biochar (NB). Each of the 15 plots was applied to 20 m long and 5 m wide. The monoculture corn (cultivar Suyu 24) was annually planted in April and harvested in July. Except for CK, urea (N 150 kg hm−2, Shanxi Weihe Heavy Chemical Co., Ltd.), calcium–magnesium-phosphate fertilizer (P2O5 75 kg hm−2, Yifeng Fertilizer Co. Ltd.), and potassium fertilizer (K2O 60 kg hm−2, Mosaic Potash Esterhazy LP.) were once applied in each treatment. The amount of corn straw, pig manure, and straw biochar was applied as 1,000 kg C hm−2 year−1.
Soil sampling and chemical analysis
Soil samples were collected after the corn harvest at the end of July 2020. A total of 10 sampling points were taken from 0 to 20 cm topsoil samples using an S-shaped sampling method, and 1 kg soil samples were taken using the quartering method after evenly mixing. After that, fresh soil samples were placed in clean bags on ice and immediately transported to the laboratory. Soil samples in each treatment were separated into two subsamples for analysis of soil chemical properties, and the bacterial and nematode communities, respectively.
Soil pH was determined using a glass electrode at a water: soil ratio of 2.5:1 (v/w). Soil organic carbon (SOC) and total potassium (TK) were measured by the Walkley-Black wet digestion method and flame photometry, respectively (Jackson, 1958). Total phosphorus (TP) and total nitrogen (TN) were determined by molybdenum-blue colorimetry and Kjeldahl digestion, respectively (Zheng et al., 2009). Cation exchange capacity (CEC) was determined by the ammonium acetate saturation method (Kitsopoulos, 1999). Microbial biomass carbon (MBC) was measured by the chloroform fumigation incubation method (Vance et al., 1987).
Quantitative PCR and illumina sequencing
The DNA from the soil sample was extracted following standard procedures of the DNeasy PowerSoil Extraction Kit (MoBio Laboratories, Carlsbad, CA, USA). DNA quality and content were checked using a NanoDrop spectrophotometer. The copy number of the bacterial community was determined by quantitative PCR (qPCR) using universal primers 515F and 907R (Biddle et al., 2008). Each sample was amplified in a 30 μl reaction mix containing 15 μL 2 × qPCR Mix, 0.5 μl forward primer (10 μM), 0.5 μl reverse primer (10 μM), and 2 μL template DNA. The qPCR conditions were: initial denaturation at 95°C for 10 min, followed by 40 cycles of 95°C for 30 s, 60°C for 30 s, 72°C for 30 s, and a final extension at 72°C for 10 min. The qPCR amplification was performed in triplicate with efficiencies >95% and r2 values >0.99.
The amplicon fragments using the same primer pairs were subjected to high-throughput sequencing on the Illumina MiSeq. Raw sequencing data were processed using the Quantitative Insights into Microbial Ecology (QIIME) pipeline (version 1.9.1). Sequences with masses less than 20 and lengths less than 300 bp were removed, matched to different samples based on specific tags, and assigned to separate files for the bacterial 16S rRNA gene. The remaining sequences were chimera removed and clustered into operational taxonomic units (OTUs) at 97% similarity using the Usearch algorithm. The representative sequence for each OTU with the highest abundance was then chosen. Taxonomic assignments were performed against the Silva database (version 138) for the bacteria community. The command “alpha_diversity.py” of the QIIME software was used to calculate the Shannon index of bacterial alpha-diversity after the rarefaction of all samples to the same sequencing depth of 28,883 sequences.
Carbon use efficiency and microbial metabolic quotient
Soil microbial growth rate and respiration rate were determined by the 18O labeling method to determine carbon use efficiency (CUE) (Zheng et al., 2019). Briefly, 1 g of precultured soil sample was added into a 2 mL injection bottle, three replicates were set for each sample, and H218O (97.0%, ABX, Israel) was added to one of them to label the soil with 18O. The abundance was adjusted to 20%, and an equal volume of deionized water was added to another portion as a natural 18O natural abundance control. After the added water was thoroughly mixed with the soil, the sample was placed in a 50 mL headspace vial, and aluminum caps were crimped on to seal the headspace vials. The headspace vial gas was then immediately replaced with CO2-free air to limit the CO2 concentration in the vial. A blank headspace vial without soil was used as a blank control throughout the experiment. After all manipulations, the incubation vials were incubated at 15°C for 24 h. After the incubation, the gas in the headspace vial was collected using a syringe with a three-way valve, and the CO2 concentration was measured using a gas chromatograph (GC-7890B, Agilent, USA). After freeze-drying the soil samples in the injection vials, the DNA was extracted and transferred to a silver cup, and then dried at 45°C. The 18O abundance and oxygen content were determined by an elemental analyzer coupled to an isotope ratio mass spectrometer using the EA-IRMS system (Thermo Scientific, TX, USA). The newly produced DNA content (DNAproduced, μg) of the soil samples during the 24-h incubation period was calculated as follows:
In the formula, Ototal (μg) is the oxygen content in the dried sample DNA; at%excess is the difference between the abundance of 18O in the labeled and unlabeled samples; at%label is the abundance of 18O in the final soil solution.
The ratio of MBC to DNA content of each precultured sample was calculated as a conversion factor (fDNA), and fDNA was used to convert the newly generated DNA content in the sample during the culture period into MBC content, thereby calculating the microbial growth rate (Growth, ng C· g−1 h−1) formula is as follows:
The calculation formula of microbial respiration rate (Respiration, ng C g−1 h−1) is as follows:
where ∆CO2 (ppm) is the increase in CO2 concentration in the closed headspace vessel during the incubation period of the soil sample; M is the molar mass of carbon (12.01 g mol−1); V (l) is the total volume of gas; DW (g) is the soil dry weight; t (h) is incubation time.
The formula for calculating the metabolic entropy (qCO2, ng C μg MBC−1 h−1) is as follows:
Carbon mineralization
The determination of C mineralization adopts an indoor constant temperature culture, and C mineralization was determined by the alkali absorption technique: weigh 100 g of fresh soil in a 500 mL culture bottle and spread it on the bottom of the bottle. The water holding capacity is calculated when the maximum water holding capacity is 60%. Then, an absorption bottle containing 20 mL of NaOH solution at a concentration of 0.5 mol L−1 was placed in the culture bottle, the culture bottle was sealed with a cap, and placed in a constant temperature incubator at 25°C for 30 days under dark conditions. The solution in the absorption bottle was taken out, washed completely into the conical flask, and 20 mL of 1 mol L−1 BaCl2 solution and two drops of phenolphthalein indicator were added. The remaining NaOH was titrated and neutralized with 0.4 mol L−1 HCl solution until the red color disappeared, and the CO2 release by HCl consumption was calculated.
where cHCl is the concentration of hydrochloric acid (mol l−1), V0 is the volume of the blank titration, and V1 is the volume of hydrochloric acid consumed.
Nematode community and ecological indices
Baermann funnel method was used to extract nematodes (Cesarz et al., 2019). Briefly, 100 g of soil was spread evenly on the funnel device and then water was added to submerge the soil. The spring clip was opened to obtain an aqueous solution of nematodes after 72 h. The nematodes were killed at 60°C and stored in a 4% formalin solution. At least 150 nematodes per sample were collected and placed under a light microscope for nematode identification. Nematodes were classified into four trophic guilds based on morphology (Yeates, 1998), including bacterivores (Ba), fungivores (Fu), plant-parasites (Pp), and omnivores-predators (OP). Predation pressure of nematodes on bacteria was calculated as the ratio of the number of nematodes to bacterial copy number (Mathisen et al., 2016). Furthermore, we selected the nematode indices to indicate the food web status, including Shannon–Wiener index, channel index (CI), enrichment index (EI), structure index (SI), functional dispersion index (FDis), and free-living nematode maturity index (MI) (Ferris et al., 2001). The corresponding calculation formulas are as follows:
In these formulas, pi is the proportion of individuals of the ith taxon in the total number of nematodes; Ba, Fu, and PP represent the number of bacterivorous, fungivorous, and plant-parasitic nematodes, respectively; e, b, and s are enriched components (Ba1 and Fu2), basic components (Ba2 and Fu2), structural components (Ba3-Ba5, Fu3-Fu5, Om3-Om5, and Ca2-Ca5), respectively. The CI is between 0 and 100, greater than 50 indicates that the fungal degradation pathway is dominant, and less than 50 indicates that the bacterial degradation pathway is dominant. EI and SI vary from 0 to 100, with higher EI and SI indicating better nutritional status and better food web connectivity, respectively. Predation pressure refers to the predation pressure of bacterivorous nematodes on bacteria.
Statistical analyses
Significant difference analysis of the data was performed using one-way ANOVA with SPSS 24.0 (Kim, 2017). Spearman’s rank correlation test was used to measure the correlations between soil properties, bacterial and nematode communities, and C mineralization (Sedgwick, 2014). The varpart function in the “vegan” package was used to determine the contribution of different types of factors to bacterial diversity, community composition, and C mineralization. Non-metric multidimensional scaling (NMDS) analysis based on Bray-Curtis distance was conducted to characterize differences in the structure of bacterial and nematode communities (Cox and Ferry, 1993). Redundancy analysis was used to estimate the relationships between soil variables, bacterial community, and nematode assemblage (Capblancq et al., 2018). The envfit function in the “vegan” package was used to perform a permutation test to test for potentially significant effects of soil properties and nematode index on community structure (Oksanen, 2015). The random forest model was constructed using the package “randomForest” of the R software (Liaw and Wiener, 2002). The “rfUtilities” and “rfPermute” packages were used to test the p value of each variable and the model, and to check the effect of each factor on the C mineralization. Structural equation modeling (SEM) was constructed to assess the potential effects of soil factors, and the bacterial and nematode communities on C mineralization. The R-packaging “lavan” was used for SEM analysis, and the data used in the model were standardized (Mamet et al., 2019).
Results
Soil chemical properties and carbon mineralization
One-way ANOVA showed that soil chemical properties were significantly changed under different organic material amendments (Figure 1). Soil pH ranged from 4.4 to 4.8, and was significantly (p = 0.048) higher under NB treatment than under the other four treatments. Soil organic carbon (SOC) was significantly increased (p < 0.001) under NS, NSM, and NB treatments compared to CK and N treatments. Total phosphorus (TP) and total nitrogen (TN) were the highest under the NSM treatment and the lowest under CK treatment. There was no significant (p > 0.05) difference in cation-exchange capacity (CEC) and total potassium (TK) among all treatments. Carbon mineralization (CM) under NSM treatment was significantly (p < 0.05) higher than that under N, NS, and NB treatments (Supplementary Figure S1A). Carbon use efficiency (CUE) under N and NSM treatments was significantly (p < 0.05) improved compared to CK, NS, and NB treatments (Supplementary Figure S1B).
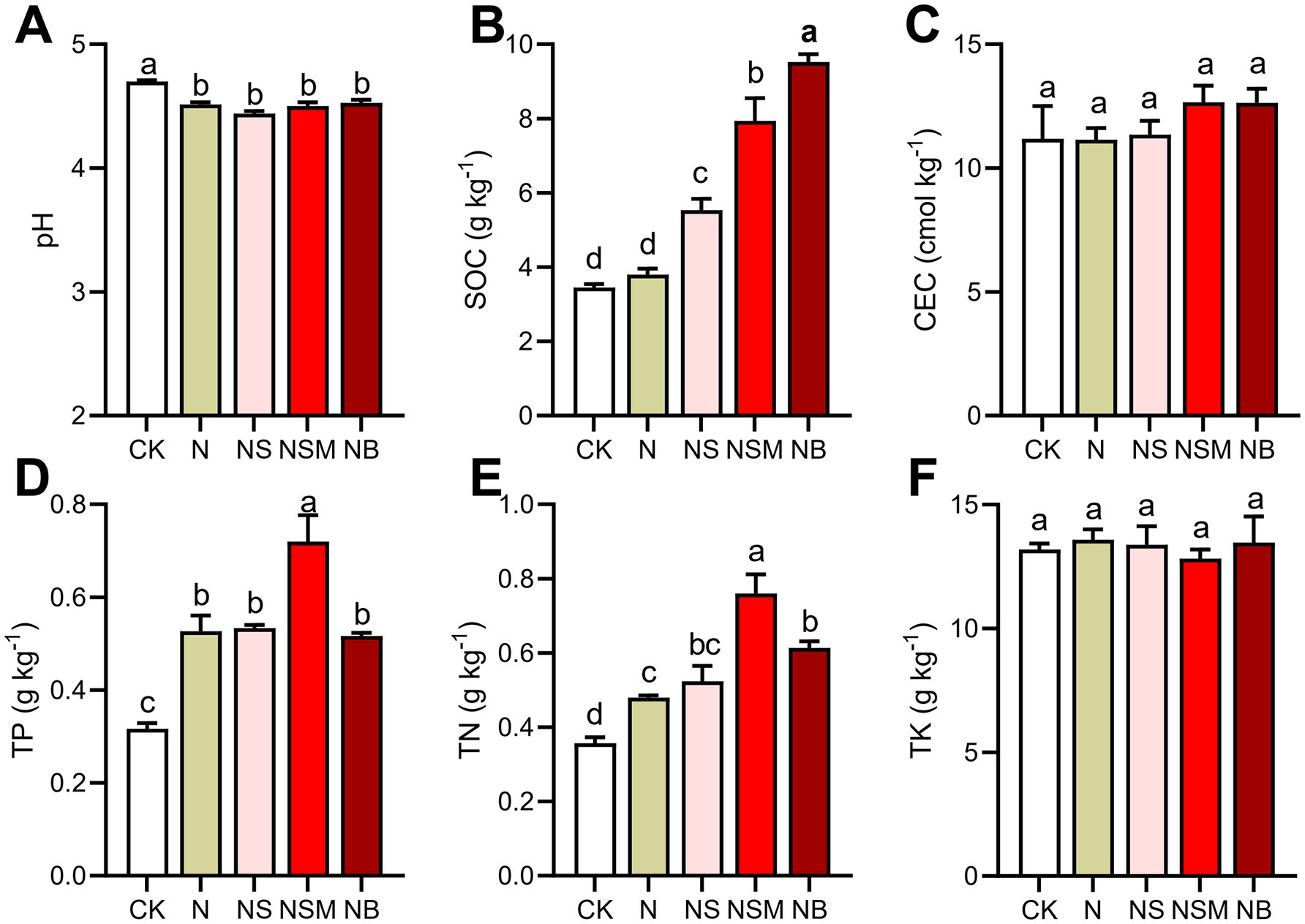
Figure 1. Soil chemical properties under the five fertilization treatments, including (A) pH, (B) soil organic carbon (SOC), (C) cation-exchange capacity (CEC), (D) total phosphorus (TP), (E) total nitrogen (TN), (F) total potassium (TK). Different lowercase letters indicate significant differences based on Tukey’s HSD test (p < 0.05). CK, no fertilizer; N, chemical fertilizers; NS, chemical fertilizers with straw; NSM, chemical fertilizers with straw and pig manure; NB, chemical fertilizers with biochar.
Nematode community composition and ecological indices
Among all treatments, the dominant trophic guilds of the nematode community were bacterivores (Ba, 47.6%), followed by fungivores (Fu, 25.7%), omnivores-predators (OP, 13.2%) and plant parasites (PP, 13.3%) (Figure 2A). The dominant genus of bacterivorous nematodes was Cephalobus, followed by Eucephalobus and Protorhabditis (Figure 2A). Nonmetric multidimensional scale (NMDS) analysis showed that the fertilization treatments significantly changed the composition of the nematode community (Figure 2B). Nematode community compositions showed that nematode assemblages under organic treatments were significantly (p < 0.05) distinct from that under CK treatment (Figure 2B). The numbers of total nematodes and bacterivores were significantly (p < 0.05) increased under NSM treatment than other treatments (Supplementary Figure S2A). Metabolic footprints of bacterivores and omnivores-predators were significantly (p < 0.05) increased under NSM treatment than N, NS, and NB treatments, while there was no significant difference in metabolic footprints of fungivores among different treatments (Supplementary Figure S2B). We further found that the ecological indices of the nematode community significantly (p < 0.05) varied among fertilization treatments. Nematode diversity, as indicated by the Shannon index, was significantly (p < 0.05) higher under NSM treatments than under N treatment (Figure 3A). Functional dispersion index (FDis) was significantly (p < 0.05) improved by NS and NSM treatments compared to CK treatment, while channel index (CI) followed the opposite trend (Figures 3B,C). The maturity index (MI) of free-living nematodes under NS and NB treatments was significantly (p < 0.05) higher than that under CK and N treatments (Figure 3D). The enrichment index (EI) and structure index (SI) were significantly (p < 0.05) higher under NSM treatment than under CK and N treatments (Figures 3E,F).
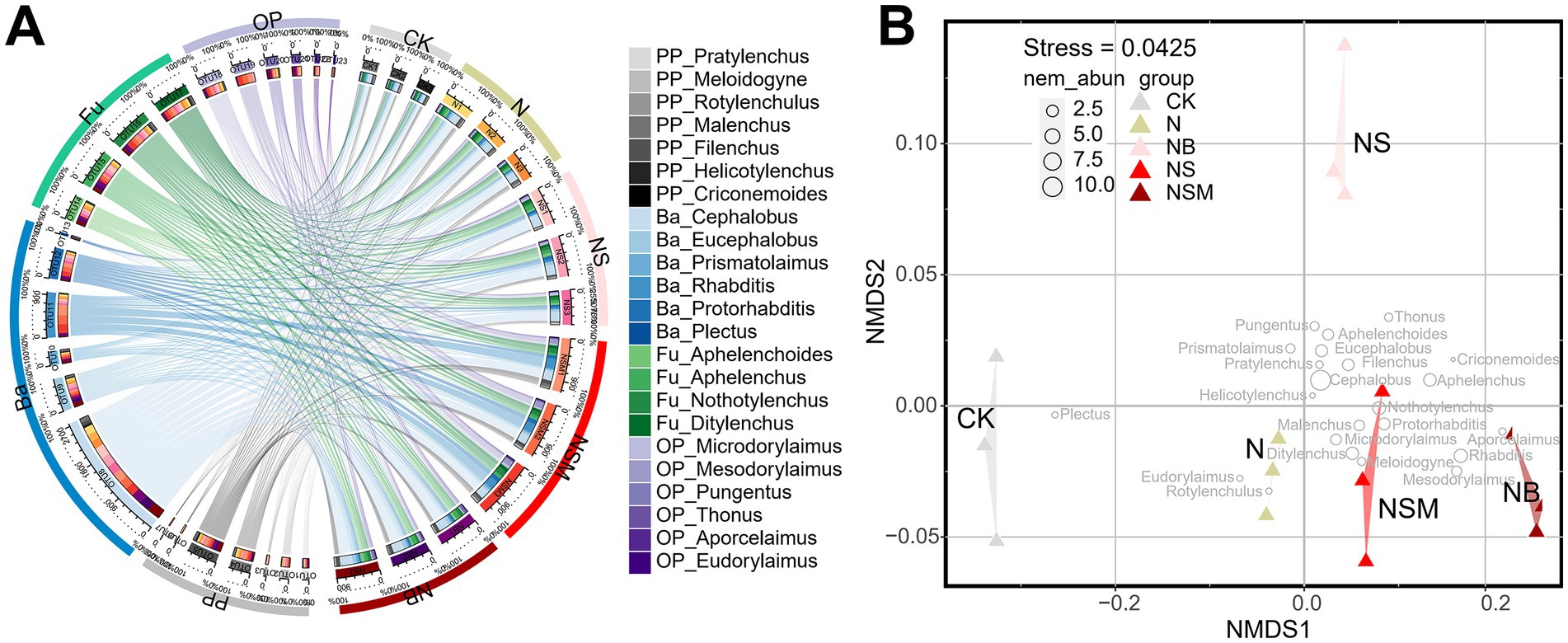
Figure 2. Chord diagram showing the relative abundances of nematode at the genus level under different fertilization treatments (A). The non-metric multi-dimensional scaling plot (NMDS) depicts the distance between the samples based on Bray-Curtis similarities (B). Different circle sizes represent the relative abundance of nematodes. CK, no fertilizer; N, chemical fertilizers; NS, chemical fertilizers with straw; NSM, chemical fertilizers with straw and pig manure; NB, chemical fertilizers with biochar.
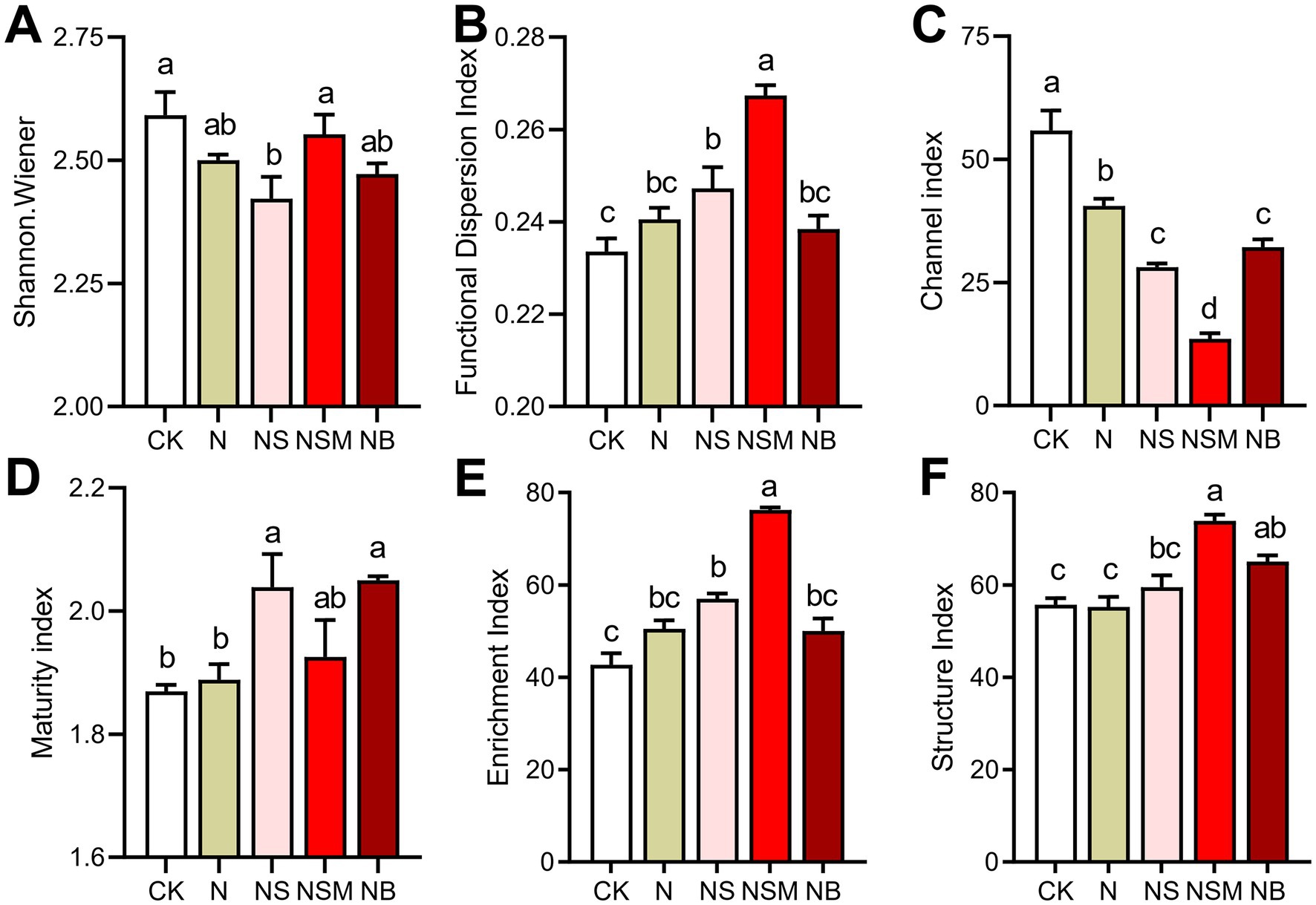
Figure 3. Nematode ecological index under the five fertilization treatments, including (A) Shannon–Wiener index, (B) functional dispersion index (FDis), (C) Channel index (CI), (D) maturity index (MI) of free-living nematodes, (E) Enrichment index (EI) and (F) Structure index (SI). Different lowercase letters indicate significant differences based on Tukey’s HSD test (p < 0.05). CK, no fertilizer; N, chemical fertilizers; NS, chemical fertilizers with straw; NSM, chemical fertilizers with straw and pig manure; NB, chemical fertilizers with biochar.
Bacterial community and metabolic activities
Organic material amendments significantly (p < 0.05) differed the bacterial communities from those under CK and N treatments (Figure 4A). Bacterial community compositions showed that bacterial assemblages under four fertilization treatments were significantly (p < 0.05) distinct from that under CK treatment. At the phylum level, the bacterial community was dominated by Chloroflexi (30.5%), Proteobacteria (23.8%), Acidobacteria (15.1%), and Actinobacteria (14.0%) (Figure 4B). The relative abundance of Proteobacteria and Actinobacteria was significantly (p < 0.01) enriched under NS and NSM treatments compared to CK and NB treatments, while Chloroflexi and Acidobacteria were significantly higher under NB treatment (Figure 4B). The copy number and Shannon index of the bacterial community were significantly increased (p < 0.05) under NS, NSM, and NB treatments compared to CK and N treatments, while the microbial metabolic quotient (qCO2) followed the opposite trend (Figure 5).
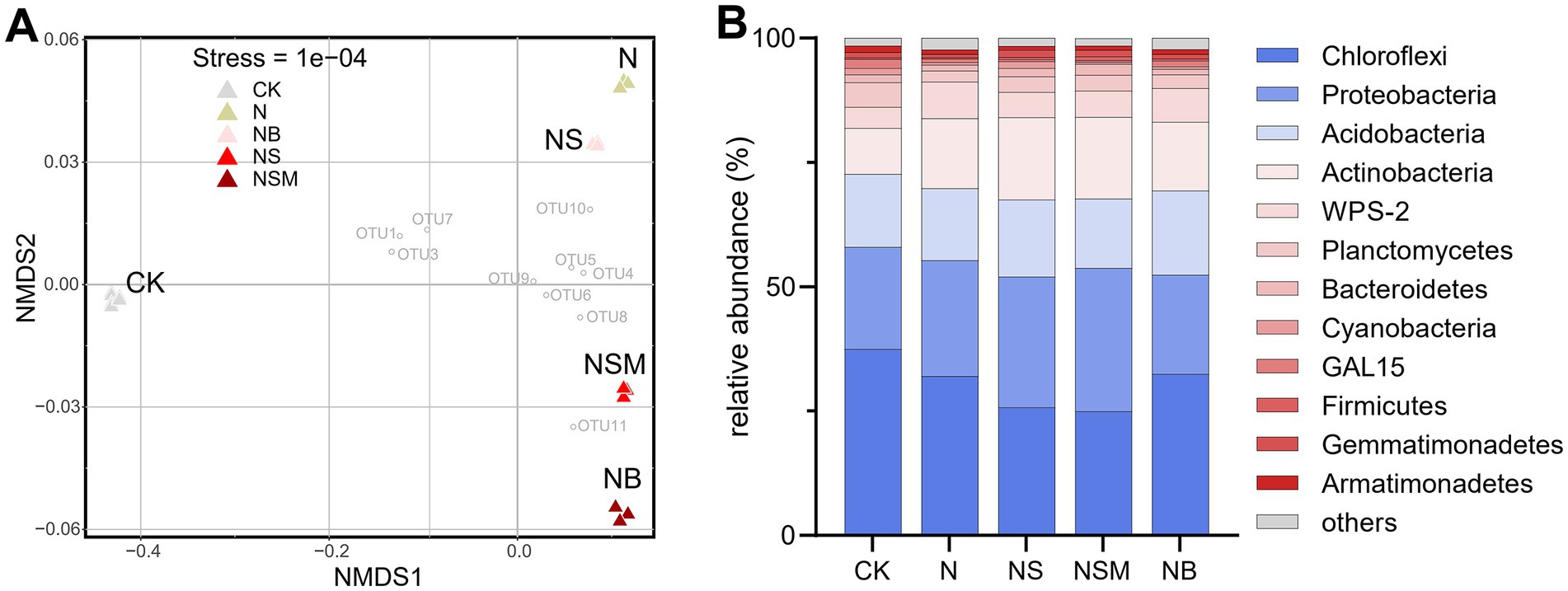
Figure 4. The non-metric multi-dimensional scaling plot (NMDS) of bacterial community structures of the five treatments (A). The mean relative abundance of bacterial phyla within each treatment (B). CK, no fertilizer; N, chemical fertilizers; NS, chemical fertilizers with straw; NSM, chemical fertilizers with straw and pig manure; NB, chemical fertilizers with biochar.
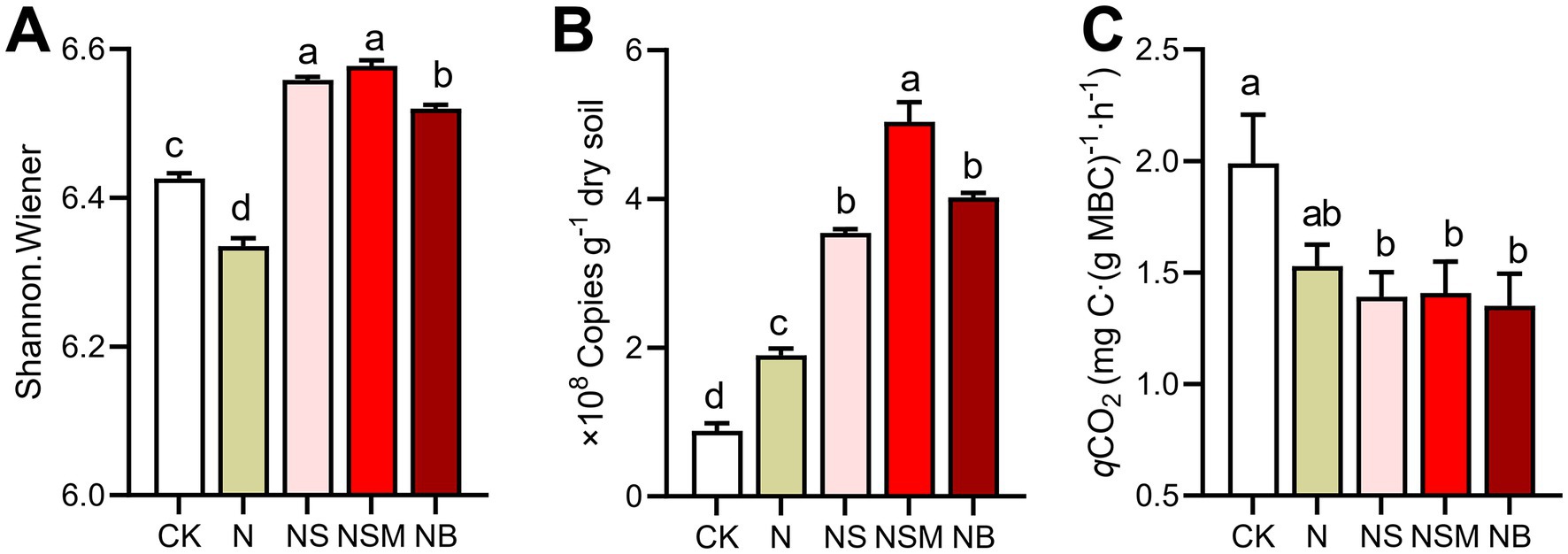
Figure 5. The diversity (A, Shannon index) and abundance (B, copy numbers of 16S RNA) of bacterial communities and microbial metabolic quotient (C, qCO2) under the five fertilization treatments. Different lowercase letters indicate significant differences based on Tukey’s HSD test (p < 0.05). CK, no fertilizer; N, chemical fertilizers; NS, chemical fertilizers with straw; NSM, chemical fertilizers with straw and pig manure; NB, chemical fertilizers with biochar.
Redundancy analysis showed that all parameters explained 77.8% of the variation in the bacterial communities (Figure 6A). The main grouping factors under NS and NSM treatments were Protorhabditis and OP, and Proteobacteria and Actinobacteria were clustered with a high relative abundance of bacterivores and Protorhabditis. The envfit test indicated that soil chemical factors (TP, pH, and TN) and nematode assemblage (the numbers of total nematodes, bacterivores, omnivores-predators, Protorhabditis and Cephalobus, CI, and predation pressure) had significant effects on the bacterial community composition (r2 = 0.61 to 0.87, p < 0.05) (Figure 6B). Variance partitioning analyses showed that the interactions between the environmental factors and nematode indices could explain 61% of the variation of bacterial communities, while the two independent explanations were only 6 and 8% (Figure 6C). Environmental factors and nematode indices can independently explain 15 and 17% of the variation of bacterial biomass, respectively, and interactions between the two major components were up to 25% (Figure 6D).
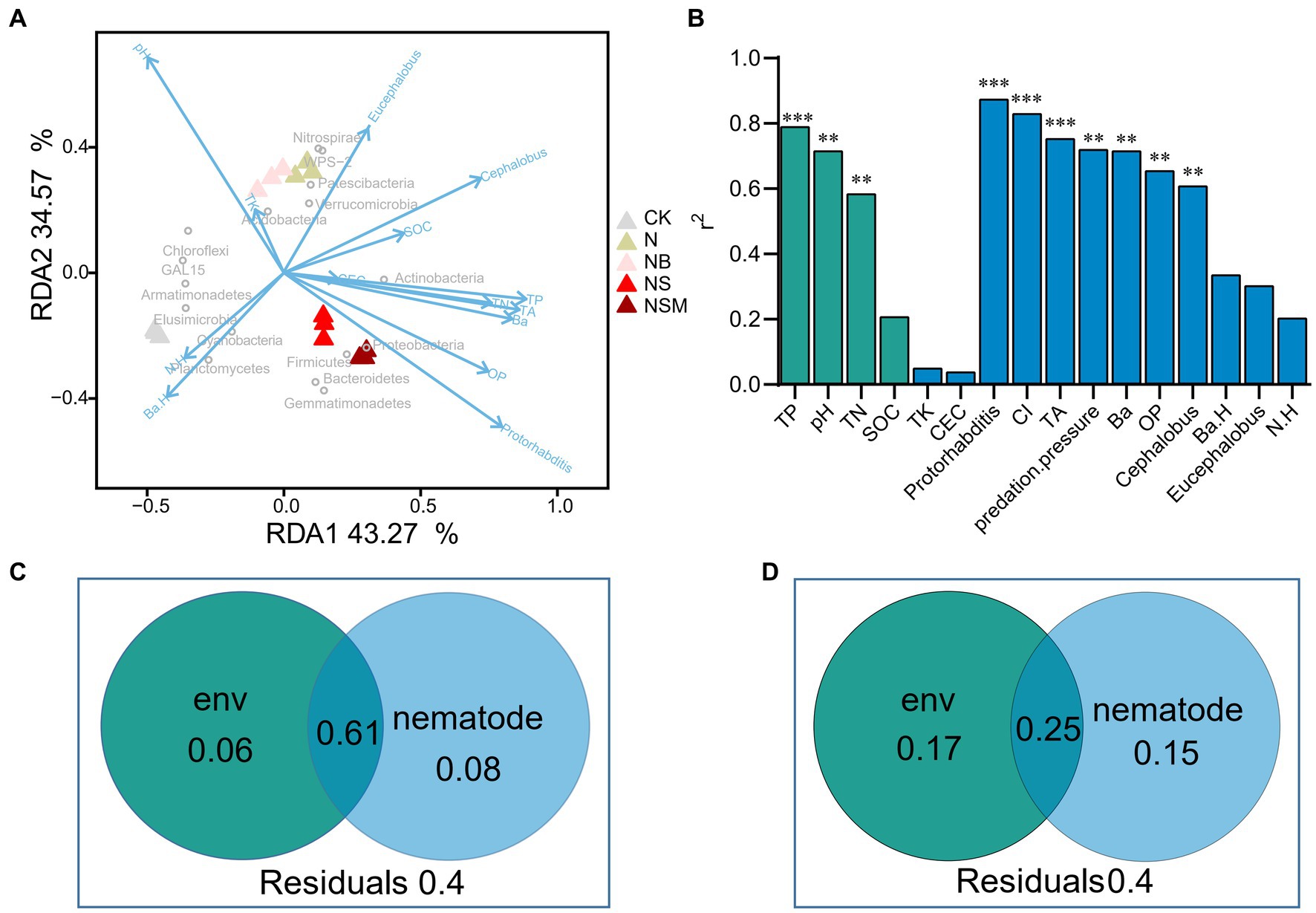
Figure 6. Redundancy analysis (RDA) indicates the relationships of soil properties and nematode indices with bacterial community (A). Contributions of each factor to community structure based on envfit test (Monte Carlo permutation test) (B). The proportion of soil chemical properties and nematodes explaining the composition (C, first principal coordinates, PCoA1) and abundance (D, copy number of 16S rRNA) of bacterial community. Soil chemical properties include soil pH, soil organic carbon (SOC), total nitrogen (TN), total phosphorus (TP), total potassium (TK), and cation-exchange capacity (CEC). The nematode community include the number of total nematodes (TA), bacterivores (Ba), and omnivores-predators (OP), and diversity (Shannon index, N.H). *p < 0.05, **p < 0.01, ***p < 0.001.
Potential nematode-bacteria interactions affected C sequestration
Correlation heatmap analysis showed that the bacterial diversity and abundance and CM were significantly positively correlated with the numbers of total, bacterivorous and omnivorous-predatory nematodes, predation pressure, EI, and FDis (r = 0.66 to 0.89, p < 0.05), but negatively correlated with CI (r = −0.84 to −0.87, p < 0.05) (Figure 7A). In contrast, the bacterial community composition (PCOA1) (r = −0.76 to −0.83, p < 0.05) and qCO2 (r = −0.50 to −0.64, p < 0.06) exhibited negative relationships with the abundance of total and bacterivorous nematodes, predation pressure, SI, and FDis. Variance partitioning analyses showed that soil chemical factors, nematode indices, and bacterial characteristics individually explained 36, 15, and 10% of the variances in CM, and interactively contributed to 15% of the observed variation (Figure 7B). Specifically, random forest analysis soil TN (6.5%), TP (7.1%), the abundance (4.7%) and composition (8.6%) of the bacterial community, and the numbers of total nematode (7.6%), bacterivores (7.5%), and omnivores-predators (6.7%) contributed significantly (p < 0.05) to C mineralization (Figure 7C). Structural equation modeling indicated that the nematode assemblage indirectly affected soil C sequestration by regulating bacterial community structure and biomass (Figure 7D). Specifically, the bacterial community was positively associated with the nematode assemblage (r = 0.42, p < 0.001), but negatively associated with qCO2 (r = −0.94, p < 0.001).
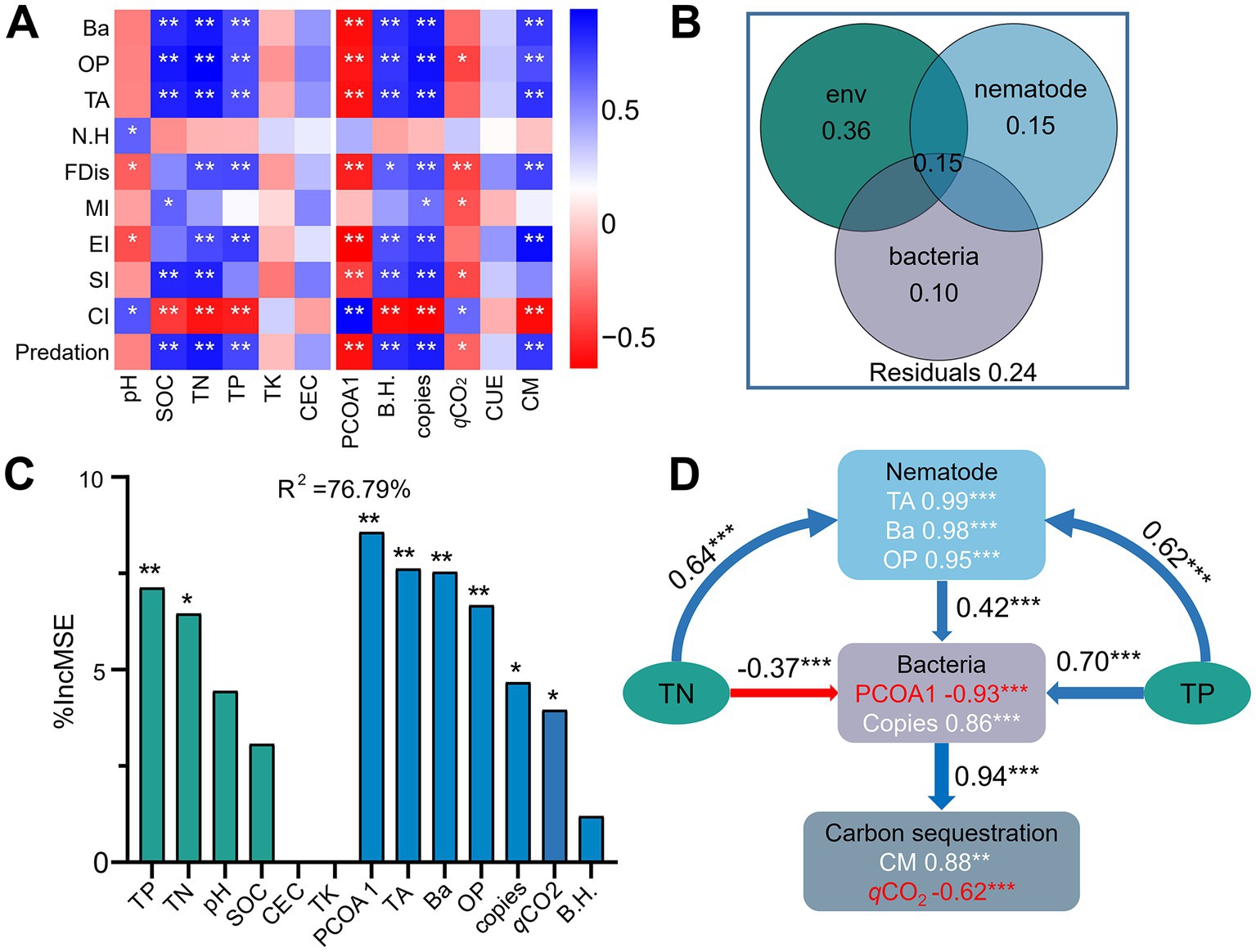
Figure 7. Spearman’s correlation coefficients between soil properties, the bacterial and nematode communities, and carbon mineralization (A). Blue and red denote positive and negative correlations, respectively. * p < 0.05, ** p < 0.01, *** p < 0.001. Contribution of soil chemical properties and the bacterial and nematode communities to carbon mineralization using variance partitioning analysis (B). Mean predictor importance (% of increased mean square error, MSE) of soil chemical properties and the bacterial and nematode communities on carbon mineralization using random forest modeling (C). Structural equation modeling shows the direct and indirect effects of soil properties and the bacterial and nematode communities on carbon sequestration (D). Soil chemical properties include soil pH, soil organic carbon (SOC), total nitrogen (TN), total phosphorus (TP), total potassium (TK), and cation-exchange capacity (CEC). The bacterial community include diversity (Shannon index, B.H), abundance (copy number of 16S rRNA) and composition (first principal coordinates, PCoA1). The nematode community include the number of total nematodes (TA), bacterivores (Ba), and omnivores-predators (OP), and diversity (Shannon index, N.H), Channel index (CI), Enrichment Index (EI) Structure Index (SI), functional dispersion index (FDis), and free-living nematode maturity index (MI) Carbon sequestration is indicated by carbon mineralization (CM) and microbial metabolic quotient (qCO2).
Discussion
Bacterial community varied with organic material amendments
We revealed that organic material amendments could significantly promote soil fertility by improving SOC, TN, and TP. Corn straw and pig manure are rich in soil organic matter and nutrients, and biochar is a carbon-rich material. The addition of these organic materials greatly increases SOC content (Cooper et al., 2011; Sandhu et al., 2017). Notably, SOC, TP, and TN were significantly increased under NSM treatment compared to N treatment, but not significantly increased under NS treatment. The simultaneous utilization of straw and manure reduces the C/N ratio of straw, resulting in faster decomposition and turnover rates of soil nutrients (Zheng et al., 2022a). Compared with corn straw, manure treatments can substantially enhance soil phosphorus content due to the high proportion of phosphorus. The NSM treatment significantly increased the contents of TN and TP by improving the organic N mineralization of straw compared with NS and NB treatments. In particular, the more reasonable ratio of corn straw and pig manure leads to a higher efficiency of the positive priming effect on straw decomposition, and a higher level of soil fertility (Abdalla et al., 2022). Soils treated with straw and pig manure enrich the food resources for microbial growth, and stimulate bacterial biomass and diversity (Hussain et al., 2019). Biochar with a good porous structure may provide habitats for microorganisms that are protected from predators, thereby preserving bacterial biomass within the biochar (Pietikäinen et al., 2000). Compared to NB treatment, we found that lower SOC under NSM treatment was not consistent with the trend of bacterial abundance, which may be partially explained by the alleviation of nutrient limitation (Wei et al., 2020). Organic material amendments promote soil structural complexity and heterogeneity, and better soil quality contributes to biodiversity by increasing spatial niches (Bebber and Richards, 2022). Soils with more abundant resources and higher nutrient availability can support higher bacterial diversity (Clarke and Gaston, 2006).
Nematode community affected by organic material amendments
We observed that the numbers of total nematodes, bacterivores, and omnivores-predators were considerably increased under NS, NSM, and NB treatments due to the improved food sources and better habitats. Organic material amendments not only create better habitats for nematodes and facilitate their movement through soil pore water, but also improve the ability of nematodes to capture abundant food resources (bacteria) due to the increased soil organic matter (Liu et al., 2016; Jiang et al., 2018b). In addition, our results determined that fertilization practices increased the metabolic footprint of four nematode groups, suggesting that more carbon flowed into the soil nematode community for nematode reproduction (Zhang et al., 2016). Straw and manure amendments increased the functional dispersion index (FDis), indicating weaker species competition within a community and more complete utilization of available resources (Prada-Salcedo et al., 2021). Organic material amendments result in a more bacterially dominated fast cycle decomposition pathway, as indicated by the high number of bacterial-feeding nematodes and low CI value (Zhong et al., 2017). The interaction between EI and SI has been widely used to indicate resource enrichment and food web status (Ferris et al., 2001). The higher values of EI and SI under NSM treatment represent better nutrient status and a more complex soil food web. The higher MI value supported that the food web was more mature under the NS and NB treatments, whereas the lower MI value under NSM treatment was mainly due to the increased presence of opportunistic bacterivorous nematodes. For example, Rhabditis and Protorhabditis are capable of rapidly reproducing and responding to bacteria when food resources are abundant at high nutrient levels (Hu and Qi, 2010).
Nematode-bacteria interactions enhanced C sequestration
Microbial metabolic quotient (qCO2) and carbon use efficiency (CUE) are commonly indicated as the rate of C storage in diverse ecosystems, with lower qCO2 reflecting better soil biophysical conditions and higher CUE reflecting higher metabolic efficiency (Chavarria et al., 2018; Feng et al., 2021). We suggested that the lower qCO2 value probably indicates better soil biophysical conditions caused by the organic material amendments (NS, NSM, and NB treatments). The variation of qCO2 may also be related to microbial community composition and biomass, and the values of lower qCO2 and higher CUE indicate a higher C sequestration potential (Ros et al., 2006; Liu et al., 2020). Phosphorus (P) limitation may have a greater impact on CUE in red soils (Feng et al., 2021). Since biochar is a reluctant compound with extremely limited P content, bacteria need to secrete more extracellular enzymes to acquire nutrients, resulting in a lower CUE value (Allison, 2005; Spohn, 2016). Biochar amendment can induce competition with potential keystone taxa to promote the abundance and diversity of the bacterial community, thereby reducing carbohydrate catabolism and qCO2 (Chen et al., 2019).
Soil bacterial community plays a crucial role in organic matter decomposition and nutrient availability (Steenwerth et al., 2002). Our results indicated that the nematode predation may increase the abundance of the bacterial community, with positive cascading effects on C sequestration. Numerous studies have pointed out that the bacterial community composition and biomass can mediate qCO2 and CUE, leading to variations in C mineralization processes (Spohn and Chodak, 2015; Fanin and Bertrand, 2016). Although the higher bacterial abundance indicates that more microorganisms are involved in the C decomposition, qCO2 was generally found to decrease with increasing microbial biomass. Nematodes are thought to be functionally critical in microbial food webs. Bacterivorous nematodes are primarily involved in regulating rates of C mineralization and altering pathways of energy flow and nutrient cycling by affecting the abundance and composition of the bacterial community (Hungate et al., 2021; Tian et al., 2022). We found that the bacterial biomass significantly increased with the increased predation pressure, with the highest biomass under NSM treatment. Trophic dynamics suggest that the prey biomass at a given trophic level is maximal at a moderate level of predation (Carpenter et al., 1985). Bacterivores can positively influence the diversity and abundance of the bacterial community by providing new niches for colonization and by creating physical and behavioral refuges for prey (Cressman and Garay, 2009; Zheng et al., 2022b). Selective predation by bacterivorous nematodes alters the bacterial community, specifically promoting the relative abundance of Proteobacteria to alter C mineralization (Neidig et al., 2011). Protorhabditis shows a strong effect on Gram-negative bacteria (e.g., Proteobacteria), which may be related to their thinner cell wall that makes them more easily digestible by predators (Wilson, 1993). In this case, nematode-bacteria interactions significantly increase the metabolic capacities of six substrate groups of carbon sources, including carbohydrates, carboxylic acids, amino acids, polymers, phenolic acids, and amines (Jiang et al., 2018b). In addition, bacterivore predation further shows a positive relationship with the size and turnover rate of C pools and contributes positively to C sequestration (Zhang et al., 2013; Jiang et al., 2018a). The C sequestration efficiency was improved under NSM treatment, which could be further explained by the bacterial characteristics and nematode indices. The higher level of SOC and TP implied sufficient nutrient resources, which was supported by the higher EI value under NSM treatment than under CK treatment. However, the lower CI indicates the superiority in ensuring less carbon flux to higher trophic levels and the degradation of more labile carbon in the bacterial decomposition channel (Tabarant et al., 2011).
Conclusion
Our findings indicated that organic material amendments enhanced the abundance of bacterial and nematode communities, and significantly altered the structure of bacterial and nematode communities. The higher values of EI, SI, and MI indicated a more mature and stable food web structure, while the low value of CI indicated dominant bacterial decomposition pathways under organic material amendments. We found that nematode predation could increase bacterial abundance and enhance C sequestration, thereby facilitating C accumulation and storage. Taken together, our research integrates the ecology of nematode assemblages with empirical evidence to provide insights into nematode predation driving C dynamics under organic material amendments. As such, a comprehensive understanding of the biological mechanisms of C sequestration may have important implications for the development of sustainable agroecosystems.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
YJ and BS designed all the experiments. GS, LL, and YJ wrote the manuscript. GS, GZ, ZZ, JZ, and YS were responsible for performing the field and lab experiments. All authors analyzed all data, discussed the results, critically reviewed the manuscript, and approved its publication. All authors read and approved the final manuscript.
Funding
This study was supported by the National Key Research and Development Program (2022YFD1900603), National Natural Science Foundation of China (41922048, 42177298, and 42107336), Jiangxi Provincial Natural Science Foundation (20224BAB215054), the Youth Innovation Promotion Association of CAS (Y2021084), Double Thousand Plan of Jiangxi Province, and the Science and Technology Project of Jiangsu Province (BE2022393).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1155088/full#supplementary-material
References
Abdalla, K., Sun, Y., Zarebanadkouki, M., Gaiser, T., Seidel, S., and Pausch, J. (2022). Long-term continuous farmyard manure application increases soil carbon when combined with mineral fertilizers due to lower priming effects. Geoderma 428:116216. doi: 10.1016/j.geoderma.2022.116216
Allison, S. D. (2005). Cheaters, diffusion and nutrients constrain decomposition by microbial enzymes in spatially structured environments. Ecol. Lett. 8, 626–635. doi: 10.1111/j.1461-0248.2005.00756.x
Bebber, D. P., and Richards, V. R. (2022). A meta-analysis of the effect of organic and mineral fertilizers on soil microbial diversity. Appl. Soil Ecol. 175:104450. doi: 10.1016/j.apsoil.2022.104450
Berhane, M., Xu, M., Liang, Z., Shi, J., Wei, G., and Tian, X. (2020). Effects of long-term straw return on soil organic carbon storage and sequestration rate in North China upland crops: a meta-analysis. Glob. Chang. Biol. 26, 2686–2701. doi: 10.1111/gcb.15018
Biddle, J. F., Fitz-Gibbon, S., Schuster, S. C., Brenchley, J. E., and House, C. H. (2008). Metagenomic signatures of the Peru margin subseafloor biosphere show a genetically distinct environment. Proc. Natl. Acad. Sci. U. S. A. 105, 10583–10588. doi: 10.1073/pnas.0709942105
Capblancq, T., Luu, K., Blum, M. G., and Bazin, E. (2018). Evaluation of redundancy analysis to identify signatures of local adaptation. Mol. Ecol. Resour. 18, 1223–1233. doi: 10.1111/1755-0998.12906
Carpenter, S. R., Kitchell, J. F., and Hodgson, J. R. (1985). Cascading trophic interactions and lake productivity. Bioscience 35, 634–639. doi: 10.2307/1309989
Cesarz, S., Schulz, A. E., Beugnon, R., and Eisenhauer, N. (2019). Testing soil nematode extraction efficiency using different variations of the Baermann-funnel method. Soil Org. 91, 61–72. doi: 10.25674/so91201
Chavarria, D. N., Pérez-Brandan, C., Serri, D. L., Meriles, J. M., Restovich, S. B., Andriulo, A. E., et al. (2018). Response of soil microbial communities to agroecological versus conventional systems of extensive agriculture. Agric. Ecosyst. Environ. 264, 1–8. doi: 10.1016/j.agee.2018.05.008
Chen, L., Jiang, Y., Liang, C., Luo, Y., Xu, Q., Han, C., et al. (2019). Competitive interaction with keystone taxa induced negative priming under biochar amendments. Microbiome 7:77. doi: 10.1186/s40168-019-0693-7
Clarke, A., and Gaston, K. J. (2006). Climate, energy and diversity. P. Roy. Soc. B-Biol. Sci. 273, 2257–2266. doi: 10.1098/rspb.2006.3545
Cook-Patton, S. C., Leavitt, S. M., Gibbs, D., Harris, N. L., Lister, K., Anderson-Teixeira, K. J., et al. (2020). Mapping carbon accumulation potential from global natural forest regrowth. Nature 585, 545–550. doi: 10.1038/s41586-020-2686-x
Cooper, J. M., Burton, D., Daniell, T. J., Griffiths, B. S., and Zebarth, B. J. (2011). Carbon mineralization kinetics and soil biological characteristics as influenced by manure addition in soil incubated at a range of temperatures. Eur. J. Soil Biol. 47, 392–399. doi: 10.1016/j.ejsobi.2011.07.010
Cox, T. F., and Ferry, G. (1993). Discriminant analysis using non-metric multidimensional scaling. Pattern Recogn. 26, 145–153. doi: 10.1016/0031-3203(93)90096-F
Cressman, R., and Garay, J. (2009). A predator–prey refuge system: evolutionary stability in ecological systems. Theor. Popul. Biol. 76, 248–257. doi: 10.1016/j.tpb.2009.08.005
Dijkstra, P., Salpas, E., Fairbanks, D., Miller, E. B., Hagerty, S. B., van Groenigen, K. J., et al. (2015). High carbon use efficiency in soil microbial communities is related to balanced growth, not storage compound synthesis. Soil Biol. Biochem. 89, 35–43. doi: 10.1016/j.soilbio.2015.06.021
Fanin, N., and Bertrand, I. (2016). Aboveground litter quality is a better predictor than belowground microbial communities when estimating carbon mineralization along a land-use gradient. Soil Biol. Biochem. 94, 48–60. doi: 10.1016/j.soilbio.2015.11.007
Fan, X., Gao, D., Zhao, C., Wang, C., Qu, Y., Zhang, J., et al. (2021). Improved model simulation of soil carbon cycling by representing the microbially derived organic carbon pool. ISME J. 15, 2248–2263. doi: 10.1038/s41396-021-00914-0
Feng, J., Zeng, X. M., Zhang, Q., Zhou, X. Q., Liu, Y. R., and Huang, Q. (2021). Soil microbial trait-based strategies drive metabolic efficiency along an altitude gradient. ISME Commun. 1:71. doi: 10.1038/s43705-021-00076-2
Ferris, H., Bongers, T., and de Goede, R. G. (2001). A framework for soil food web diagnostics: extension of the nematode faunal analysis concept. Appl. Soil Ecol. 18, 13–29. doi: 10.1016/S0929-1393(01)00152-4
Fließbach, A., Mäder, P., and Niggli, U. (2000). Mineralization and microbial assimilation of 14C-labeled straw in soils of organic and conventional agricultural systems. Soil Biol. Biochem. 32, 1131–1139. doi: 10.1016/S0038-0717(00)00028-6
Grandy, A. S., Wieder, W. R., Wickings, K., and Kyker-Snowman, E. (2016). Beyond microbes: are fauna the next frontier in soil biogeochemical models? Soil Biol. Biochem. 102, 40–44. doi: 10.1016/j.soilbio.2016.08.008
Gross, A., and Glaser, B. (2021). Meta-analysis on how manure application changes soil organic carbon storage. Sci. Rep. 11:5516. doi: 10.1038/s41598-021-82739-7
Hu, C., and Qi, Y. (2010). Effect of compost and chemical fertilizer on soil nematode community in a Chinese maize field. Eur. J. Soil Biol. 46, 230–236. doi: 10.1016/j.ejsobi.2010.04.002
Hungate, B. A., Marks, J. C., Power, M. E., Schwartz, E., van Groenigen, K. J., Blazewicz, S. J., et al. (2021). The functional significance of bacterial predators. MBio 12, e00466–e00421. doi: 10.1128/mBio.00466-21
Hussain, A., Zahir, Z. A., Ditta, A., Tahir, M. U., Ahmad, M., Mumtaz, M. Z., et al. (2019). Production and implication of bio-activated organic fertilizer enriched with zinc-solubilizing bacteria to boost up maize (Zea mays L.) production and biofortification under two cropping seasons. Agronomy 10:39. doi: 10.3390/agronomy10010039
Jiang, Y., Qian, H., Wang, X., Chen, L., Liu, M., Li, H., et al. (2018a). Nematodes and microbial community affect the sizes and turnover rates of organic carbon pools in soil aggregates. Soil Biol. Biochem. 119, 22–31. doi: 10.1016/j.soilbio.2018.01.001
Jiang, Y., Zhou, H., Chen, L., Yuan, Y., Fang, H., Luan, L., et al. (2018b). Nematodes and microorganisms interactively stimulate soil organic carbon turnover in the macroaggregates. Front. Microbiol. 9:2803. doi: 10.3389/fmicb.2018.02803
Jiang, Y., Sun, B., Jin, C., and Wang, F. (2013). Soil aggregate stratification of nematodes and microbial communities affects the metabolic quotient in an acid soil. Soil Biol. Biochem. 60, 1–9. doi: 10.1016/j.soilbio.2013.01.006
Kane, J. L., Kotcon, J. B., Freedman, Z. B., and Morrissey, E. M. (2022). Fungivorous nematodes drive microbial diversity and carbon cycling in soil. Ecology 104:e3844. doi: 10.1002/ecy.3844
Kim, T. K. (2017). Understanding one-way ANOVA using conceptual figures. Korean J. Anesthesiol. 70, 22–26. doi: 10.4097/kjae.2017.70.1.22
Kitsopoulos, K. P. (1999). Cation-exchange capacity (CEC) of zeolitic volcaniclastic materials: applicability of the ammonium acetate saturation (AMAS) method. Clay Clay Miner. 47, 688–696. doi: 10.1346/CCMN.1999.0470602
Liaw, A., and Wiener, M. (2002). Classification and regression by randomForest. R News 2, 18–22. doi: 10.1023/A:1010933404324
Liu, T., Chen, X., Hu, F., Ran, W., Shen, Q., Li, H., et al. (2016). Carbon-rich organic fertilizers to increase soil biodiversity: evidence from a meta-analysis of nematode communities. Agric. Ecosyst. Environ. 232, 199–207. doi: 10.1016/j.agee.2016.07.015
Liu, Z., Wu, X., Liu, W., Bian, R., Ge, T., Zhang, W., et al. (2020). Greater microbial carbon use efficiency and carbon sequestration in soils: amendment of biochar versus crop straws. GCB Bioenergy 12, 1092–1103. doi: 10.1111/gcbb.12763
Mamet, S. D., Redlick, E., Brabant, M., Lamb, E. G., Helgason, B. L., Stanley, K., et al. (2019). Structural equation modeling of a winnowed soil microbiome identifies how invasive plants re-structure microbial networks. ISME J. 13, 1988–1996. doi: 10.1038/s41396-019-0407-y
Mathisen, P., Thelaus, J., de Luna, S. S., and Andersson, A. (2016). Rapid adaptation of predation resistance in bacteria isolated from a seawater microcosm. Aquat. Microb. Ecol. 78, 81–92. doi: 10.3354/ame01802
Min, K., Slessarev, E., Kan, M., McFarlane, K., Oerter, E., Pett-Ridge, J., et al. (2021). Active microbial biomass decreases, but microbial growth potential remains similar across soil depth profiles under deeply-vs. shallow-rooted plants. Soil Biol. Biochem. 162:108401. doi: 10.1016/j.soilbio.2021.108401
Neidig, N., Paul, R. J., Scheu, S., and Jousset, A. (2011). Secondary metabolites of Pseudomonas fluorescens CHA0 drive complex non-trophic interactions with bacterivorous nematodes. Microb. Ecol. 61, 853–859. doi: 10.1007/s00248-011-9821-z
Oksanen, J. (2015). Vegan: an introduction to ordination. Available at: https://cran.r-project.org/web/packages/vegan/vignettes/intro-vegan.pdf
Pausch, J., Hofmann, S., Scharroba, A., Kuzyakov, Y., and Ruess, L. (2016). Fluxes of root-derived carbon into the nematode micro-food web of an arable soil. Food Webs 9, 32–38. doi: 10.1016/j.fooweb.2016.05.001
Pietikäinen, J., Kiikkilä, O., and Fritze, H. (2000). Charcoal as a habitat for microbes and its effect on the microbial community of the underlying humus. Oikos 89, 231–242. doi: 10.1034/j.1600-0706.2000.890203.x
Prada-Salcedo, L. D., Wambsganss, J., Bauhus, J., Buscot, F., and Goldmann, K. (2021). Low root functional dispersion enhances functionality of plant growth by influencing bacterial activities in European forest soils. Environ. Microbiol. 23, 1889–1906. doi: 10.1111/1462-2920.15244
Ros, M., Klammer, S., Knapp, B., Aichberger, K., and Insam, H. (2006). Long-term effects of compost amendment of soil on functional and structural diversity and microbial activity. Soil Use Manag. 22, 209–218. doi: 10.1111/j.1475-2743.2006.00027.x
Sandhu, S. S., Ussiri, D. A., Kumar, S., Chintala, R., Papiernik, S. K., Malo, D. D., et al. (2017). Analyzing the impacts of three types of biochar on soil carbon fractions and physiochemical properties in a corn-soybean rotation. Chemosphere 184, 473–481. doi: 10.1016/j.chemosphere.2017.05.165
Sinsabaugh, R. L., Manzoni, S., Moorhead, D. L., and Richter, A. (2013). Carbon use efficiency of microbial communities: stoichiometry, methodology and modelling. Ecol. Lett. 16, 930–939. doi: 10.1111/ele.12113
Spohn, M., and Chodak, M. (2015). Microbial respiration per unit biomass increases with carbon-to-nutrient ratios in forest soils. Soil Biol. Biochem. 81, 128–133. doi: 10.1016/j.soilbio.2014.11.008
Spohn, M. (2016). Element cycling as driven by stoichiometric homeostasis of soil microorganisms. Basic Appl Ecol. 17, 471–478. doi: 10.1016/j.baae.2016.05.003
Steenwerth, K. L., Jackson, L. E., Calderón, F. J., Stromberg, M. R., and Scow, K. M. (2002). Soil microbial community composition and land use history in cultivated and grassland ecosystems of coastal California. Soil Biol. Biochem. 34, 1599–1611. doi: 10.1016/S0038-0717(02)00144-X
Tabarant, P., Villenave, C., Risede, J. M., Roger-Estrade, J., Thuries, L., and Dorel, M. (2011). Effects of four organic amendments on banana parasitic nematodes and soil nematode communities. Appl. Soil Ecol. 49, 59–67. doi: 10.1016/j.apsoil.2011.07.001
Tian, S., Zhu, B., Yin, R., Wang, M., Jiang, Y., Zhang, C., et al. (2022). Organic fertilization promotes crop productivity through changes in soil aggregation. Soil Biol. Biochem. 165:108533. doi: 10.1016/j.soilbio.2021.108533
Van Den Hoogen, J., Geisen, S., Routh, D., Ferris, H., Traunspurger, W., Wardle, D. A., et al. (2019). Soil nematode abundance and functional group composition at a global scale. Nature 572, 194–198. doi: 10.1038/s41586-019-1418-6
Vance, E. D., Brookes, P. C., and Jenkinson, D. S. (1987). Microbial biomass measurements in forest soils: the use of the chloroform fumigation-incubation method in strongly acid soils. Soil Biol. Biochem. 19, 697–702. doi: 10.1016/0038-0717(87)90051-4
Wang, S., Chen, X., Li, D., and Wu, J. (2021). Effects of soil organism interactions and temperature on carbon use efficiency in three different forest soils. Soil Ecol. Lett. 3, 156–166. doi: 10.1007/s42832-020-0067-x
Wardle, D. A., and Ghani, A. A. (1995). A critique of the microbial metabolic quotient (qCO2) as a bioindicator of disturbance and ecosystem development. Soil Biol. Biochem. 27, 1601–1610. doi: 10.1016/0038-0717(95)00093-T
Wei, W., Huangfu, C., Jia, Z., and Liu, S. (2021). Long-term organic amendments improved soil carbon sequestration to support crop production. J. Plant Nutr. Soil Sci. 184, 678–687. doi: 10.1002/jpln.202100128
Wei, X., Zhu, Z., Liu, Y., Luo, Y., Deng, Y., Xu, X., et al. (2020). C:N:P stoichiometry regulates soil organic carbon mineralization and concomitant shifts in microbial community composition in paddy soil. Biol. Fertil. Soils 56, 1093–1107. doi: 10.1007/s00374-020-01468-7
Wilson, J. R. (1993). “Organization of forage plant tissues” in Forage cell wall structure and digestibility. ed. H. G. Jung (Madison, WI: American Society of Agronomy Inc), 1–32.
Yeates, G. W. (1998). “Feeding in free-living soil nematodes: a functional approach” in The physiology and biochemistry of free-living and plant-parasitic nematodes (Wallingford, UK: CAB international), 245–269.
Zechmeister-Boltenstern, S., Keiblinger, K. M., Mooshammer, M., Peñuelas, J., Richter, A., Sardans, J., et al. (2015). The application of ecological stoichiometry to plant–microbial–soil organic matter transformations. Ecol. Monogr. 85, 133–155. doi: 10.1890/14-0777
Zhang, S., Li, Q., Lü, Y., Zhang, X., and Liang, W. (2013). Contributions of soil biota to C sequestration varied with aggregate fractions under different tillage systems. Soil Biol. Biochem. 62, 147–156. doi: 10.1016/j.soilbio.2013.03.023
Zhang, Z., Wang, H., Wang, Y., Zhang, X., Zhao, T., and Mahamood, M. (2021). Organic input practice alleviates the negative impacts of elevated ozone on soil microfood-web. J. Clean. Prod. 290:125773. doi: 10.1016/j.jclepro.2020.125773
Zhang, Z., Zhang, X., Mahamood, M., Zhang, S., Huang, S., and Liang, W. (2016). Effect of long-term combined application of organic and inorganic fertilizers on soil nematode communities within aggregates. Sci. Rep. 6:31118. doi: 10.1038/srep31118
Zheng, J., Tao, L., Dini-Andreote, F., Luan, L., Kong, P., Xue, J., et al. (2022a). Dynamic responses of ammonia-oxidizing archaea and bacteria populations to organic material amendments affect soil nitrification and nitrogen use efficiency. Front. Microbiol. 13:911799. doi: 10.3389/fmicb.2022.911799
Zheng, J., Dini-Andreote, F., Luan, L., Geisen, S., Xue, J., Li, H., et al. (2022b). Nematode predation and competitive interactions affect microbial-mediated phosphorus dynamics. MBio 13:e0329321. doi: 10.1128/mbio.03293-21
Zheng, Q., Hu, Y., Zhang, S., Noll, L., Böckle, T., Richter, A., et al. (2019). Growth explains microbial carbon use efficiency across soils differing in land use and geology. Soil Biol. Biochem. 128, 45–55. doi: 10.1016/j.soilbio.2018.10.006
Zheng, S., Hu, J., Chen, K., Yao, J., Yu, Z., and Lin, X. (2009). Soil microbial activity measured by microcalorimetry in response to long-term fertilization regimes and available phosphorous on heat evolution. Soil Biol. Biochem. 41, 2094–2099. doi: 10.1016/j.soilbio.2009.07.019
Zhong, S., Zeng, H. C., and Jin, Z. Q. (2017). Influences of different tillage and residue management systems on soil nematode community composition and diversity in the tropics. Soil Biol. Biochem. 107, 234–243. doi: 10.1016/j.soilbio.2017.01.007
Keywords: bacterial community, nematode assemblage, nematode indices, carbon mineralization, microbial metabolic quotient, organic material amendments
Citation: Shi G, Luan L, Zhu G, Zeng Z, Zheng J, Shi Y, Sun B and Jiang Y (2023) Interaction between nematodes and bacteria enhances soil carbon sequestration under organic material amendments. Front. Microbiol. 14:1155088. doi: 10.3389/fmicb.2023.1155088
Edited by:
Jesús Navas-Castillo, IHSM La Mayora, CSIC, SpainReviewed by:
Jia Liu, Jiangxi Academy of Agricultural Sciences (CAAS), ChinaJianping Wu, Yunnan University, China
Copyright © 2023 Shi, Luan, Zhu, Zeng, Zheng, Shi, Sun and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuji Jiang, eWpqaWFuZ0Bpc3Nhcy5hYy5jbg==
†These authors have contributed equally to this work
 Guangping Shi1,2†
Guangping Shi1,2† Lu Luan
Lu Luan Bo Sun
Bo Sun Yuji Jiang
Yuji Jiang