- 1Department of Clinical Laboratory, The Second Affiliated Hospital of Soochow University, Suzhou, Jiangsu, China
- 2Department of Clinical Laboratory, The Affiliated Suzhou Hospital of Nanjing Medical University, Suzhou Municipal Hospital, Gusu School, Nanjing Medical University, Suzhou, Jiangsu, China
- 3Sichuan Academy of Medical Science and Sichuan Provincial People's Hospital, Chengdu, Sichuan, China
- 4Center of Medical Laboratory, The General Hospital of Ningxia Medical University, Yinchuan, China
- 5Department of Laboratory Medicine, The First Affiliated Hospital of Sun Yat-sen University, Guangzhou, Guangdong, China
- 6Laboratory Medicine Center, Aviation General Hospital, Beijing, China
- 7Department of Laboratory Medicine, The First Affiliated Hospital of Kunming Medical University, Kunming, Yunnan, China
- 8Department of Medical Affairs, Danaher Diagnostic Platform/Cepheid (People's Republic of China), New York, NY, United States
- 9Center for Discovery and Innovation, Hackensack-Meridian Health, Nutley, NJ, United States
- 10Hackensack Meridian School of Medicine, Seton Hall University, Nutley, NJ, United States
Carbapenem-resistant (CR) Klebsiella oxytoca complex can be associated with high mortality, emerging as a new threat to the public health. K. oxytoca complex is phylogenetically close to K. pneumoniae, one of most common species associated with multidrug resistance in Enterobacterale. The latest research showed that K. oxytoca is a complex of six species. Currently, the epidemiological and genomic characteristics of CR K. oxytoca complex in China are still unclear. Here, we conducted a multi-center study on 25 CR K. oxytoca complex collected from five representative regions in China. These isolates were, respectively, recovered from respiratory tract (12 cases, 48.0%), abdominal cavity (5 cases, 20.0%), blood (4 cases, 16.0%), urine tract (3 cases, 12.0%) and skin or soft tissue (1 cases, 4.0%). Among them, 32.0% (8/25) of patients infected with K. oxytoca complex had a poor prognosis. In this study, three K. oxytoca complex species were detected, namely K. michiganensis, K. oxytoca and K. pasteurii, among which K. michiganensis was the most common. Three carbapenemase genes were identified, including blaNDM-1 (10, 38.5%), blaKPC-2 (9, 34.6%) and blaIMP (6 blaIMP-4 and 1 blaIMP-8; 7, 26.9%). Subsequent multilocus sequence typing identified various sequence types (STs), among which ST43, ST92 and ST145 were relatively common. Different from the clonal dissemination of high-risk carbapenem-resistant K. pneumoniae strains, our research revealed a polyclonal dissemination characteristic of CR K. oxytoca complex in China. S1-nuclease PFGE and Southern blot experiment showed that carbapenemase genes were encoded in plasmids of different sizes. Two blaNDM-harboring plasmids were subsequently sequenced, and were characterized to be IncX3 and IncC incompatibility groups, respectively. This is the first multi-center study of CR K. oxytoca complex in China, which improved our understanding of the prevalence and antimicrobial resistance characteristics of CR K. oxytoca complex in China.
Introduction
Klebsiella oxytoca is an important member of the Klebsiella genus within the family Enterobacteriaceae. K. oxytoca is commonly found in the environment, including soil, water, and plants, but also can reside in the intestinal tract as a commensal bacterium (Podschun and Ullmann, 1998; Savino et al., 2009). In patients with a compromised immune system, especially those ICU patients and newborns, K. oxytoca can lead to diseases such as antibiotic-associated hemorrhagic colitis (AAHC), bloodstream infections, urinary tract infections, pneumonia, skin and soft tissue infections and intra-abdominal infections (Bouchillon et al., 2013; Hoban et al., 2014; Mineau et al., 2018; Ghasemian et al., 2019). Recently, new findings have significantly advanced our knowledge of this important pathogen. The latest research based on genomic taxonomy has shown that K. oxytoca is not a single species, but a complex composed of six species, namely K. oxytoca, K. grimontii, K. michiganensis, K. huaxiensis, K. pasteurii, and K. spallanzanii (Yang et al., 2022).
Klebsiella oxytoca complex is mainly reported in Asia-Western Pacific, Western Europe and North America, but rarely in Africa and South America (Yang et al., 2022), and the outbreaks usually involve strains with extended-spectrum beta-lactamases and carbapenamases (Decré et al., 2004; Hoenigl et al., 2012; Lowe et al., 2012). In recent years, carbapenem-resistant K. oxytoca (CRKO) complex strains were increasingly detected clinically associated with high mortality, emerging as a new threat to the public health (Hoenigl et al., 2012; Wang et al., 2017). In China, the isolation rate of CRKO complex appears to be relatively low. However, infections caused by CRKO complex frequently lead to serious clinical outcomes in hospitalized patients.
Although the clinical importance of CRKO complex is increasingly recognized, molecular epidemiological studies are still extremely limited in part due to the low detection rate. Up to now, large molecular epidemiological research of CRKO complex is still rare in China, despite that China is regarded as a CRE endemic region. In this study, we collected 25 clinical CRKO complex strains from five hospitals in Beijing, Chengdu, Guangzhou, Yinchuan and Suzhou in China, and conducted a multi-center genomic study, in order to understand the status of antimicrobial resistance and the molecular epidemiological characteristics of CRKO complex in China.
Materials and methods
Collection and identification of CRKO complex strains
Our multi-center study includes 5 representative cities in China, namely Beijing, Chengdu, Guangzhou, Suzhou and Yinchuan. All CRKO complex isolates from various clinical samples (sputum, blood and urine, etc.) in hospitalized patients with infections were included, and only the first eligible CRKO complex culture episode per patient was included. Preliminary bacterial species identification was carried out using automatic microbiology analyzer in participating hospitals, and CRKO complex was defined as resistant to at least of one carbapenem (imipenem or meropenem), based on the Clinical and Laboratory Standards Institute (CLSI) guideline. The minimal inhibitory concentrations (MICs) of CRKO complex against carbapenems were provided in the Supplementary Table 1. A total of 25 non-repetitive CRKO complex strains from July 2016 to March 2021 were collected. For this study, all isolates were also identified using matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS; Bruker Daltonics, Bremen, Germany) to verify their species.
Antibiotic susceptibility testing
MICs of isolates against commonly used antibiotics were determined by the Phoenix 100 Automated Microbiology System and the results were interpreted according to the Clinical and Laboratory Standards Institute (CLSI) guidelines (CLSI, 2020). A total of 16 antibiotics were tested, including carbapenems (imipenem and meropenem), cephalosporins (cefazoline, ceftriaxone, ceftazidime, cefuroxime, and cefotaxime), β-lactam inhibitor complex (ampicillin-sulbactam, amoxicillin-clavulanic acid and piperacillin-tazobactam), aminoglycosides (gentamicin and amikacin), monocyclic β-lactams (aztreonam), fluoroquinolones (ciprofloxacin and levofloxacin) and sulphamethoxazole/grimethoprin. For strains only producing KPC-2, novel β-lactam/β-lactamase inhibitor ceftazidime-avibactam susceptibility experiment was also carried out by disk diffusion method.
Detection of carbapenemase genes
Carbapenemase genes (blaKPC, blaNDM, blaIMP, blaVIM and blaOXA-48) were detected with polymerase chain reaction (PCR; Queenan and Bush, 2007; Nordmann et al., 2011) and other resistance genes were further verified by whole-genome sequencing.
S1-nuclease PFGE and southern blot
In order to study the size of the plasmids harboring resistance genes (blaKPC, blaNDM, blaIMP), S1-nuclease PFGE and Southern blot experiment was performed on 12 selected strains. The genomic DNA was digested with the endonuclease S1 and then electrophoresed under the following conditions: voltage 6 V/cm, electrophoresis conditions of initial switching time of 2.2 s and final switching time of 63.8 s, running time 18 h, temperature 14°C. Plasmid DNA was then transferred from the gel to the nylon membrane by siphoning. The hybridization probes were prepared and labeled using PCR Dig Probe Synthesis kit (Roche), primer sequences were shown in Supplementary Table 2. DIG Wash and Block Buffer Set (Roche) was used for subsequent washing and blocking, and finally developed by chemiluminescence using CDP-Star. Salmonella strain H9812 was used as a size marker.
Whole genome sequencing (WGS) and bioinformatic analysis
WGS was performed on 25 CRKO complex strains. Genomic DNA was extracted using the Axygen DNA Mini Kit (Axygen, America) and sequenced by the Illumina Miseq system (Illumina, CA, USA) using a paired-end library with an average insert size of 350 bp. The quality filtered reads were assembled de novo utilizing SPAdes v3.11 (Bankevich et al., 2012). Average nucleotide identity (ANI) analysis was used to define species. To identify the acquired antimicrobial resistance (AMR) genes, all isolate genomes were genotyped for resistance by ResFinder, a Center for Genomics Epidemiology (CGE) bioinformatics tool (Zankari et al., 2012). The MLST of CRKO complex strains were analyzed by PubMLST1 (Jolley et al., 2018).
The heatmap of genotypic profile and phylogenetic tree of CRKO complex
The phylogenetic relationship of all CRKO complex isolates was conducted with MUMer 3.0, ClonalFrameML and MEGA7.0 as described previously (Liang et al., 2019). The heatmap of genotypic profile and phylogenetic tree were drawn and improved on iTOL (doi: 10.1093/nar/gkab301) and Inkscape v1.02 (Letunic and Bork, 2021). Annotation of resistance genes was carried out using the online database Resfinder (Zankari et al., 2012).
Plasmid sequencing and bioinformatic analysis
To further detail the characteristics of blaNDM-harboring plasmids, plasmid conjugation experiments were performed using the mixed broth culture method as previously described (Chen et al., 2014). Transconjugants were identified by detecting carbapenemase genes using PCR. Two representative plasmids isolated from the transconjugants were sequenced by next generation sequencing (NGS). Plasmid DNA was extracted from zygotes using Qiagen Plasmid Midi Kit (Qiagen, Valencia, CA, United States). The sequencing results were latter assembled de novo utilizing SPAdes v3.11 (Bankevich et al., 2012). Further assembly was obtained by comparing contigs to reference plasmid sequences (> 99% identical), with additional exanimating of overlapped paired ends and gap closure by PCR.
The plasmid replicon types were analyzed using the PlasmidFinder3 (Carattoli et al., 2014). Open reading frames (ORFs) and pseudogenes of assembled plasmid sequences were predicted using RAST 2.0 (Brettin et al., 2015) combined with BLASTP/BLASTN searches (Boratyn et al., 2013). The annotation of resistance genes, plasmid replicon types, mobile elements and other features were carried out using the online databases including CARD (Jia et al., 2017), ResFinder (Zankari et al., 2012), PlasmidFinder (Carattoli et al., 2014), ISfinder (Siguier et al., 2006), INTEGRALL (Moura et al., 2009), and Tn Number Registry (Tansirichaiya et al., 2019). Gene organization diagrams were drawn in Inkscape 0.48.1 software4 (Jolley and Maiden, 2010).
Nucleotide sequence accession numbers
The sequences of the 25 CRKO complex strains were submitted to GenBank under BioProject PRJNA925298. The sequences of two mapped plasmids were submitted to GenBank under accession number OQ338157-OQ338158.
Results
Characteristics of bacterial isolates
We collected a total of 25 non-repetitive CRKO complex strains from five hospitals in five China cities (Beijing, Chengdu, Guangzhou, Suzhou, and Yinchuan). The clinical and molecular characteristics were shown in Table 1. The sequencing analysis results showed that 16 strains were K. michiganensis, 5 strains were K. oxytoca, and 4 strains were K. pasteurii. These isolates were recovered from respiratory tract (12 cases, 48.0%), abdominal cavity (5 cases, 20.0%), blood (4 cases, 16.0%) and urine tract (3 cases, 12.0%), and one case was from skin or soft tissue. 32.0% (8/25) of patients had a poor prognosis.
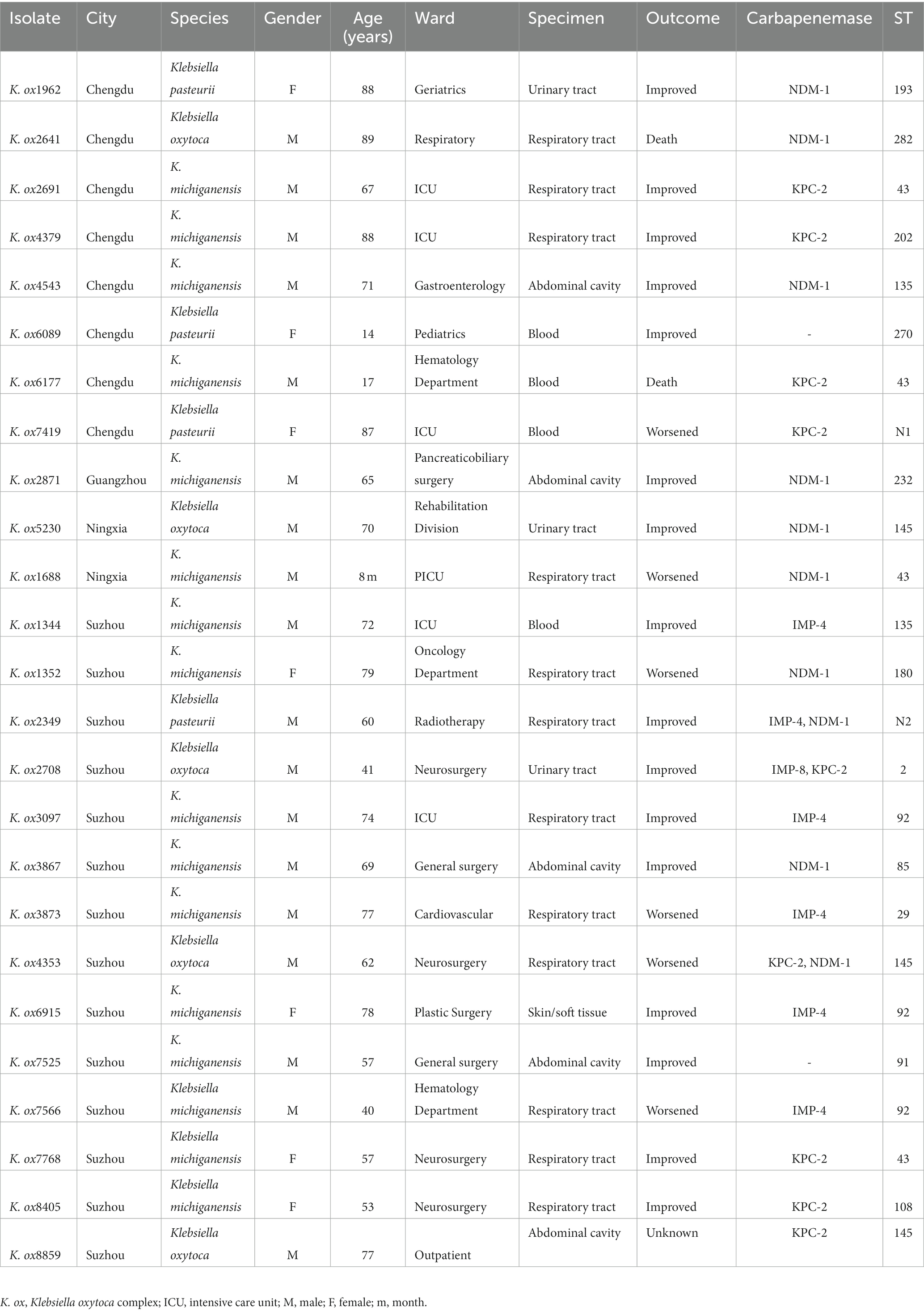
Table 1. Clinical and molecular characteristics of carbapenem-resistant (CR) Klebsiella oxytoca complex.
Antimicrobial susceptibility tests
The antimicrobial susceptibility of three K. oxytoca complex was similar but slightly different (Table 2). All three complex were highly resistant to cephalosporins, penicillin inhibitor complexes and carbapenems, and the susceptibility to the monocyclic β-lactam aztreonam was also low (only 25% of K. michiganensis strains were sensitive). Aminoglycosides in general have a high sensitivity, especially amikacin, but in K. oxytoca, both aminoglycosides and fluoroquinolones were less sensitive than those in other two complex isolates. For sulphamethoxazole/grimethoprin, only part of strains in K. michiganensis were susceptible. Overall, K. oxytoca appears to be resistant to more antibiotics than K. michiganensis and K. pasteurii. However, due to the limited number of K. pasteurii and K. oxytoca strains in our collection, the drug resistance profile should to be confirmed by further studies.
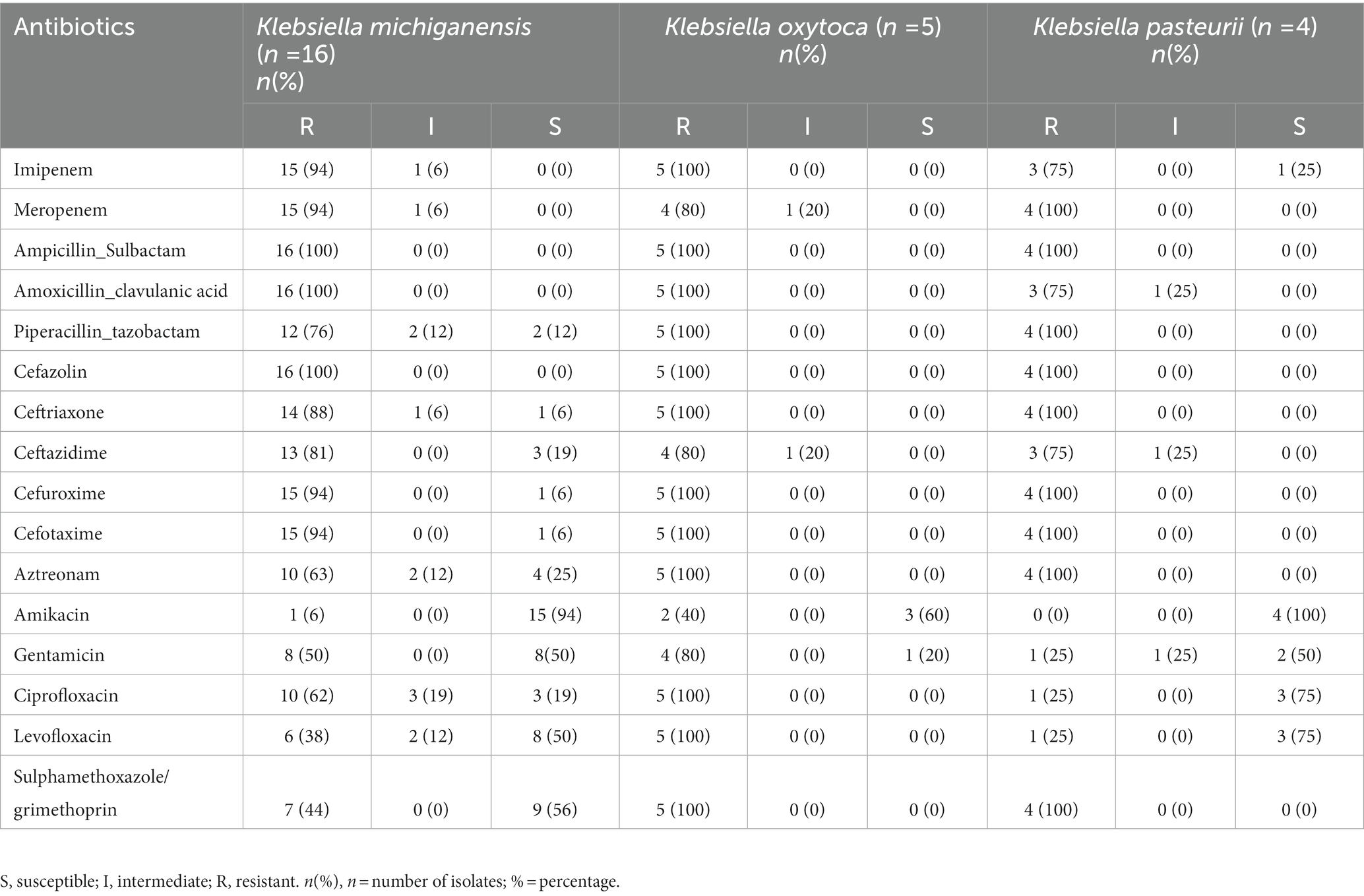
Table 2. Antimicrobial susceptibility of Klebsiella oxytoca complex against different antimicrobial agents.
Detection of Carbapenemase genes
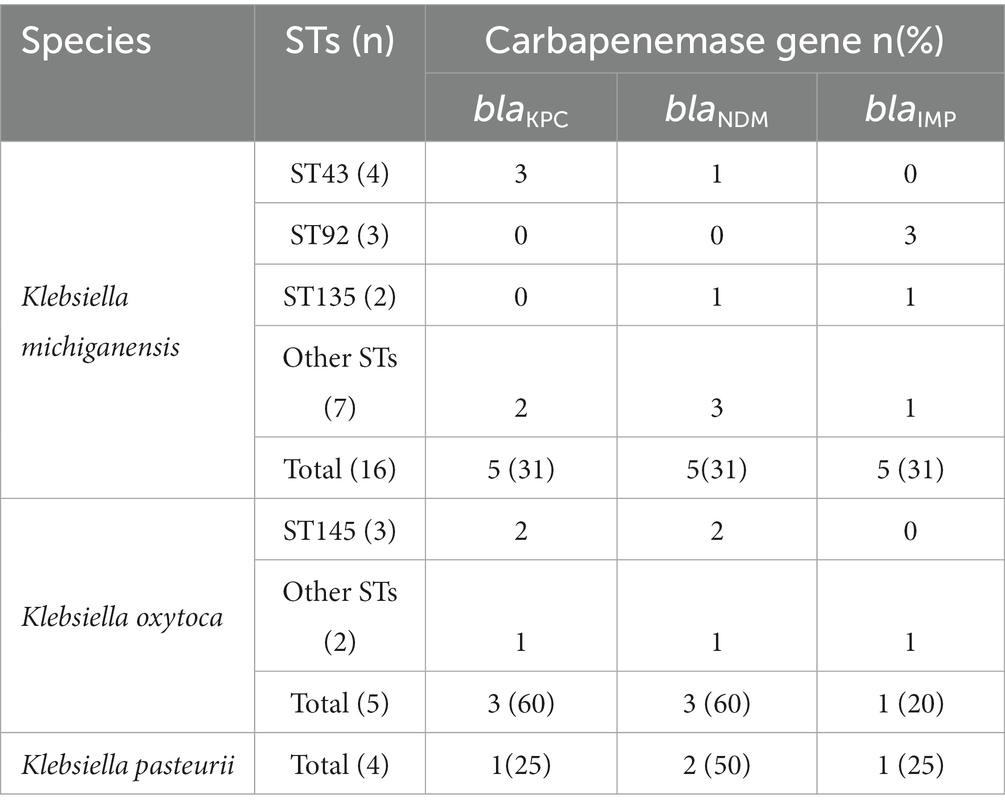
Carbapenemase genes were further tested in all isolates. A total of 23 out of the 25 (92.0%) strains were found to carry carbapenemase genes, including blaNDM-1 (10 strains, 38.5%), blaKPC-2 (9 strains, 34.6%) and blaIMP (6 blaIMP-4 and 1 blaIMP-8; 7 strains, 26.9%), among which three strains simultaneously contained two carbapenemase genes. Interestingly, strains harboring blaIMP were all isolated from Suzhou, accounting for 50% (7/14) of the total number of Suzhou isolates. blaVIM and blaOXA-48 were not detected in this study (Table 1). Notably, carbapenemase genes (blaKPC-2, blaNDM-1, and blaIMP) had significantly different distribution in the three K. oxytoca complex (Table 3). K. michiganensis had the highest detection rate of blaIMP-4, while carbapenemase genes in K. oxytoca were mainly blaKPC-2 and blaNDM-1, and only one strain carried blaIMP-8.
Other resistance genes of the isolates
As shown in Figure 1, In addition to the previously confirmed carbapenemase resistance genes by PCR, ESBL-encoding genes (blaTEM, blaSHV, and blaCTX − M) were also identified in K. oxytoca complex isolates. And the isolates were confirmed to harbor other resistance genes which mediate resistance to rifampicin (arr3), aminoglycoside (aac(3)-IIa, aac(3)-IId, aac(6′)-IIa, aac(6′)-IIc, aac(6′)-Ib-cr, aadA, etc), phenicols (catA, catB, cmlA1, floR), fosfomycin (fosA), trimethoprim (dfrA), macrolides (mph(A), mph(E), msr(E)), fluoroquinolones (qnrA1, qnrB4, qnrB6, qnrS1, qnrVC6), sulfonamides (sul1, sul2 and sul3), and tetracycline (tet(A) tet(B) and tet(D)). All strains contained efflux pump genes oqxA and oqxB, and K.ox7768 also carried colistin resistance gene mcr-9.
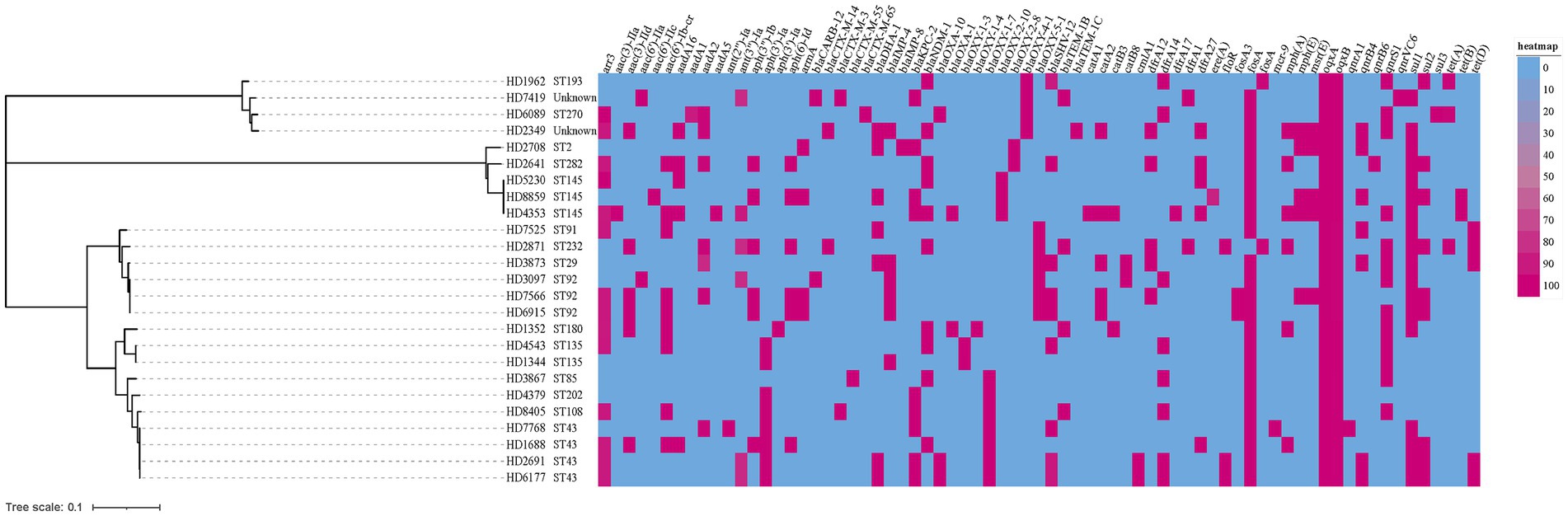
Figure 1. A heatmap of the genotypic profile and phylogenetic tree of all 25 carbapenem-resistant Klebsiella oxytoca (CRKO) complex. The cladogram on the left were phylogenetic tree indicating the cluster of isolates by STs. Bar corresponds to the scale of sequence divergence. The genotypic profile was represented as a gene present (red) or absent (blue). The resistance genes were indicated on the column header.
Multilocus sequence typing (MLST)
The results of multilocus sequence typing (MLST) analysis showed that 25 CRKO complex belonged to more than 15 different STs, of which 2 strains belonged to two new STs, named N1 and N2 herein (Figure 1). Except for ST43, ST92 and ST145, which were slightly common (n ≥ 3), other STs were mostly detected in a single strain. Results in Table 3 showed that ST43 and ST92 were the main STs of K. michiganensis, while ST145 was only detected in K. oxytoca.
Plasmid S1-PFGE and southern blot
Twelve carbapenemase-producing K. oxytoca complex isolates were selected for S1-PFGE and Southern blotting to characterize the plasmid sizes of carbapenemase genes. Results showed that plasmids harboring blaNDM ranged from 54.7 to 244.4 kb, with sizes of ~55 kb and ~ 150 kb more common. Plasmids harboring blaIMP were ~ 40 kb and ~ 173 kb, and plasmids carrying blaKPC were 78.2 ~ 104.5 kb (Supplementary Figures 1–3).
Sequencing of blaNDM-harboring plasmids
S1-PFGE and Southern blotting experiments showed that ~55 and ~ 150 kb were two plasmid sizes with relatively high frequency. We successfully obtained the transconjugants by conjugation experiments and sent two blaNDM-harboring plasmids (pHD1962-NDM, ~55 kb and pHD1688-NDM, ~150 kb) for further sequencing. Results showed that pHD1962-NDM was encoded by an IncX3 plasmid of 53.87 kb. Except for blaNDM-1, it also carried another β-lactamase gene blaSHV-12 (Figure 2A). The pHD1962-NDM carries an AMR region, which comprised of IS26–blaSHV-12–IS26 unit, truncated Tn125 variant, IS3000 and truncated Tn3. The blaNDM-1 is located on a truncated Tn125 variant (Figure 3). Compared with the reference Tn125 (Genbank accession number JN872328), the ISAba125 downstream ISCR21 was lost in this truncated Tn125 variant, while an IS5 was inserted in the ISAba125 upstream of blaNDM-1. pHD1688-NDM was encoded by an IncC plasmid of 146.25 kb, in which blaNDM-1 and several other resistance genes were co-located in the 27.3 kb MDR region (Figure 2B). Additionally, blaNDM-1 is also located on a truncated Tn125, which is captured by the integron In1201 (Figure 3). Both two plasmids had conjugal transfer regions (Figure 2), and conjugation experiments also confirmed that these plasmids were capable of horizontal transfer between strains.
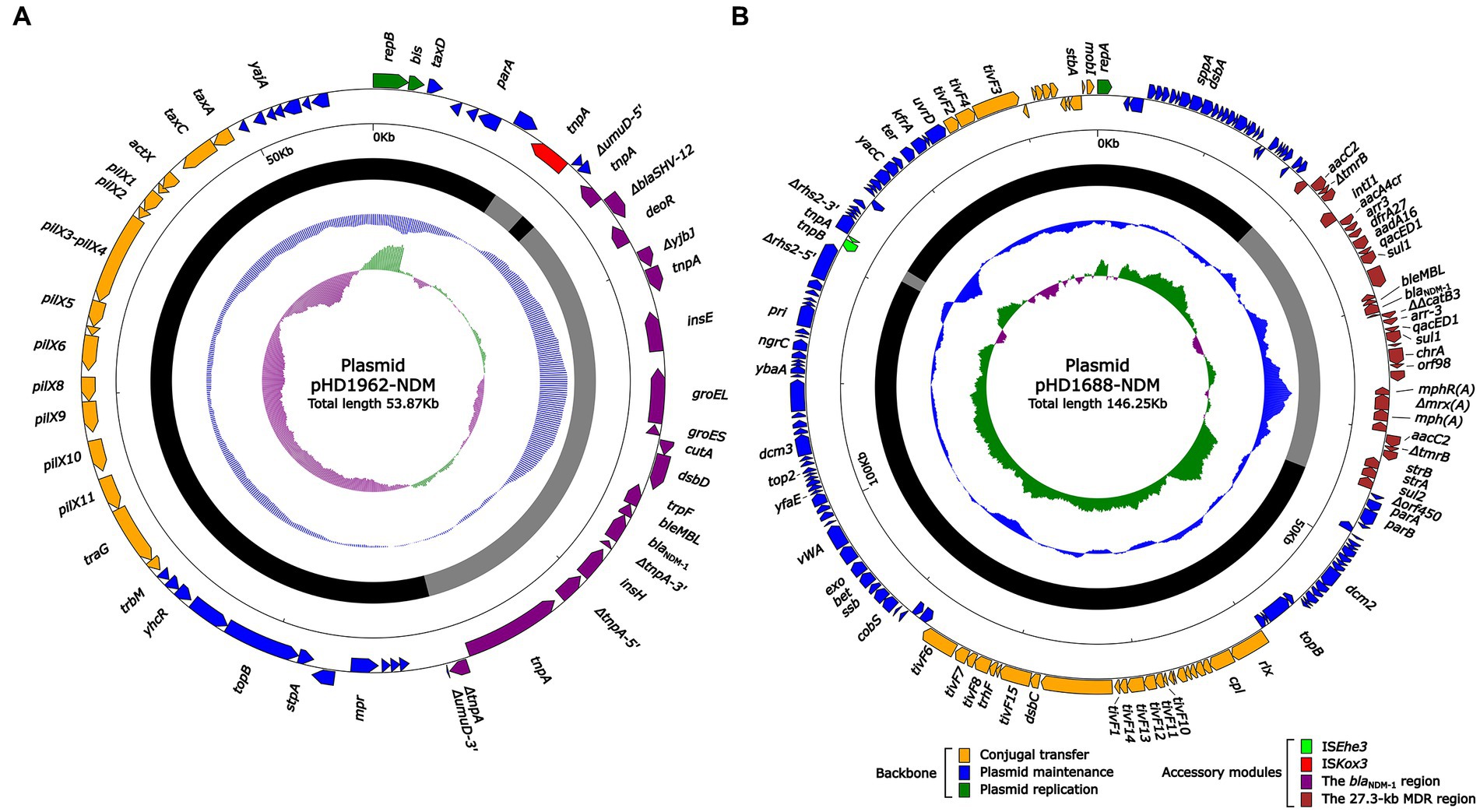
Figure 2. Schematic diagrams of plasmid pHD1962-NDM (A) and pHD1688-NDM (B). Genes of different functions are denoted by arrows and presented in various colors. The circles show (from outside to inside): predicted coding sequences, scale in 10 kb, backbone (black) and accessory module (gray) regions, GC content and GC skew [(G–C)/(G + C)].
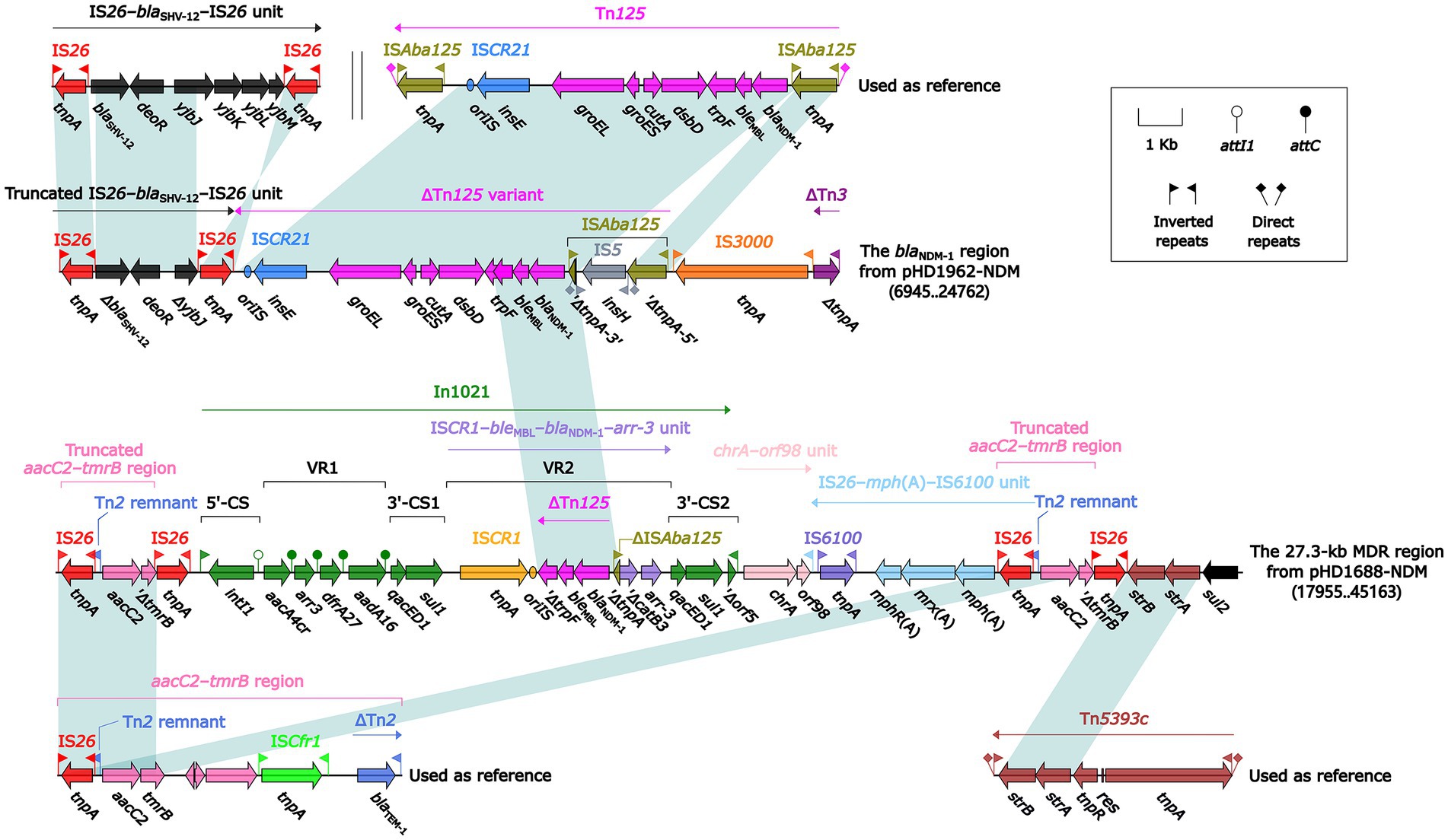
Figure 3. Linear comparison of the blaNDM-1 region, the 27.3-kb MDR region, and related mobile elements. Genes are denoted by arrows. Genes, mobile elements and other features are colored based on their functional classification. Shading denotes regions of homology (nucleotide identity ≥ 95%). Numbers in brackets indicate nucleotide positions within the plasmid pHD1962-NDM and pHD1688-NDM, respectively. The accession number of IS26–blaSHV-12–IS26 unit, Tn125, aacC2–tmrB region and Tn5393c used as reference are CP024828, JN872328, JX101693 and AF262622, respectively.
Discussion
Carbapenem-resistant K. oxytoca complex, harboring various carbapenemase genes, is spreading globally (Hoenigl et al., 2012; Nazik et al., 2014; Wang et al., 2017), but molecular epidemiological studies are still limited. We conducted a multi-center study on clinical CRKO complex collected from five representative regions of China to probe its molecular epidemiological characteristics. In our study, K. michiganensis was the most common species, accounting for 64.0% of the total isolates, followed by K. oxytoca (20.0%, 5/25) and K. pasteurii (16.0%, 4/25). K. grimontii, K. huaxiensis and K. spallanzanii were not detected. K. michiganensis is phylogenetically closest to K. oxytoca (Saha et al., 2013). Previous studies have confirmed K. michiganensis as the most common species through analysis of 162 K. oxytoca-related whole-genome sequences (Chen et al., 2020), and suggested that some conditions attributed to K. oxytoca may actually be caused by K. michiganensis. Although current research on K. michiganensis is limited, it is considered an emerging pathogen closely related to human infections (Chen et al., 2020).
In this study, carbapenemase genes were detected in 92.0% (23/25) isolates, indicating that carbapenemase was also the most important carbapenem resistance mechanism in K. oxytoca complex. Unlike K. pneumoniae, of which KPC was the most prevalent carbapenemase in China (Zhang et al., 2017), Class B MBLs were more commonly detected in K. oxytoca complex in our study. Class B MBLs include NDM, IMP and VIM, with NDM being the most widespread MBLs in Enterobacteriaceae. In addition to the K. oxytoca complex, NDM was also frequently identified in K. pneumoniae, E. coli, E. aerogenes, Serratia marcescens, Citrobacter freundii and E. cloacae (Zhang et al., 2017). Among MBLs, the isolation rate of IMP and VIM in Enterobacteriaceae was usually lower than NDM. However, compared with Enterobacteriaceae of the same period, we found that the isolation rate of blaIMP (26.9%) in K. oxytoca complex appears to be significantly higher. A nationwide surveillance of CRE strains in China also showed the similar result with the isolation rate of blaIMP in K. oxytoca complex was 25%, much higher than other CRE strains during the same period (Zhang et al., 2017). However, all the blaIMP-harboring strains in this study were isolated in Suzhou, accounting for 50.0% (7/14) of the total strains from this site, among which 3 strains belonged to ST92, and the rest were of independent STs. Moreover, due to the long separation interval of the three ST92 strains (>6 months) and the fact that the patients were from different wards, the possibility of clonal transmission was low. Ceftazidime-avibactam, a novel β-lactam/β-lactamase inhibitor, is highly active against KPC-producing CRE, and has been the first-line therapy for CRE infections in many hospitals. Ceftazidime-avibactam susceptibility experiment was carried out on 7 strains only producing KPC-2, and the results showed that all strains were sensitive, indicating that ceftazidime-avibactam still maintained high efficacy to KPC-producing K. oxytoca complex in vitro. In this study, three strains co-harboring both class A and class B carbapenemase genes were also detected, which may bring more challenges to the clinical treatment as these strains may confer to high level carbapenem resistance and resistance to novel β-lactam/β-lactamase inhibitors. K. ox6089 and K. ox7525 were two non-carbapenemase-producing strains. Although K. ox6089 was found to harbor ESBLs CTX-M-65, we suspect additional mechanisms, such as mutations of outer membrane proteins or overexpression of efflux pumps, may contribute to carbapenem resistance in these strains.
Although K. oxytoca complex and K. pneumonia both belong to the genus of Klebsiella, the epidemic characteristics of the two are significantly different. The global spread of carbapenem resistant K. pneumoniae is mainly driven by a few high-risk clone group (CG) strains (Wyres and Holt, 2016). In China, the clonal spread of ST11 is the key cause of carbapenem resistance in K. pneumoniae (Qi et al., 2010; Zhang et al., 2017). On the contrary, our research revealed a polyclonal dissemination characteristic of K. oxytoca complex in China. No major STs were detected in K. oxytoca complex and the three species all showed the characteristics of polyclonal transmission. However, the distribution of STs is related to the species of K. oxytoca complex. Among them, ST43 and ST92 were mainly identified in K. michiganensis, while ST145 was a slightly common ST in K. oxytoca. Our study also showed that carbapenemase types may be related to the STs of K. oxytoca complex. As ST43 mainly produced KPC and ST92 mainly produced MBLs, especially IMP.
Different from K. pneumoniae, class B metallo-β lactamases (MBLs) were the most common carbapenemase in K. oxytoca complex, especially NDM-producing strains which have frequently been reported in hospitalized patients or hospital environment (Wang et al., 2017; Zhang et al., 2017; Pérez-Vazquez et al., 2019; Yang et al., 2022). In this study, two blaNDM-harboring plasmids were sequenced and characterized to be IncX3 and IncC incompatibility groups, respectively. In China, IncX3 is the most common type of blaNDM-harboring plasmid replicon in carbapenem-resistant Enterobacteriaceae (An et al., 2016; Wang et al., 2017). Conjugation experiments show that the IncX3 plasmid can be efficiently transferred to a variety of Enterobacteriaceae (Wang et al., 2018), which also makes the IncX3 plasmid an important vector for the global spread of blaNDM. In addition to the IncX3 plasmid, IncC plasmid carrying blaNDM was detected in this study. IncC is another common type of blaNDM-carrying plasmid. It has a broad host range. Except for Enterobacteriaceae, it is also found in the Morganellaceae and Vibrionaceae, contributing to the global spread of blaNDM (Wu et al., 2019). The IncC-type plasmid has been previously reported in K. oxytoca complex in Spain (Pérez-Vazquez et al., 2019). But in China, this was the first report about blaNDM-carrying IncC plasmid in K. oxytoca complex. In this study, conjugation experiments confirmed these plasmids had the ability of horizontal transfer between strains, which should be vigilant against the outbreaks of carbapenem-resistant bacteria caused by horizontal transmission of plasmids between strains.
The molecular epidemiological characteristics of CRKO complex in China appears to be significantly different from those in other countries. A study in Spain showed that ST2 dominated in K. oxytoca complex, while in China, ST43, ST92 and ST145 are relatively common, and the rest STs are distributed sporadically. Other studies also suggested limited international transmission of the same clone (Long et al., 2022), but close surveillance is needed to monitor the further spread of these multidrug resistant strains. In addition, the emergence of CP K. oxytoca in Spain was mainly due to the spread of isolates that produced VIM and OXA-48 (Pérez-Vazquez et al., 2019), while in China it was mainly NDM, KPC and IMP, and no isolates of VIM and OXA-48 were detected, revealing the complexity of the molecular resistance characteristics of K. oxytoca complex.
In summary, our study detected different carbapenemase types in various K. oxytoca complex STs from 5 regions in China, highlighting the extensive genetic diversity among K. oxytoca complex strains. Our study served as the first step towards detailing the genomic and clinical characterization of K. oxytoca complex strains in China. Given the close evolutionary relationship between K. oxytoca complex and K. pneumoniae, one of the most common MDR species, further comparison of the genomic and clinical characteristics between the two groups will likely help to unravel novel features associated with the epidemics of MDR K. pneumoniae and provide strategies to control the further spread of drug resistance.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
HY, MW, WJ, BH, FQ, and BS were responsible for the specimen collection and information collation. WW and XY were responsible for the experiment and article writing. Y-WT, LC and HD contributed to the revision of the article. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the Science Foundation of Jiangsu Province Health Department [ZDB2020014]; the Science Foundation of Suzhou Health Department [LCZX202106] and the Discipline Construction Program of the Second Affiliated Hospital of Soochow University [XKTJ-TD202001]; the Science and Technology Program of Suzhou (SLJ2022003, 2022SS41).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1153781/full#supplementary-material
Footnotes
References
An, J., Guo, L., Zhou, L., Ma, Y., Luo, Y., Tao, C., et al. (2016). NDM-producing Enterobacteriaceae in a Chinese hospital, 2014-2015: identification of NDM-producing Citrobacterwerkmanii and acquisition of blaNDM-1-carrying plasmid in vivo in a clinical Escherichia coli isolate. J. Med. Microbiol. 65, 1253–1259. doi: 10.1099/jmm.0.000357
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Boratyn, G. M., Camacho, C., Cooper, P. S., Coulouris, G., Fong, A., Ma, N., et al. (2013). BLAST: a more efficient report with usability improvements. Nucleic Acids Res. 41, W29–W33. doi: 10.1093/nar/gkt282
Bouchillon, S., Badal, R. E., Hoban, D. J., and Hawser, S. P. (2013). Antimicrobial susceptibility of inpatient urinary tract isolates of gram-negative bacilli in the United States: results from the study for monitoring antimicrobial resistance trends (SMART) program: 2009-2011. Clin. Ther. 35, 872–877. doi: 10.1016/j.clinthera.2013.03.022
Brettin, T., Davis, J. J., Disz, T., Edwards, R. A., Gerdes, S., Olsen, G. J., et al. (2015). RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 5:8365. doi: 10.1038/srep08365
Carattoli, A., Zankari, E., García-Fernández, A., Voldby, L. M., Lund, O., Villa, L., et al. (2014). In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903. doi: 10.1128/AAC.02412-14
Chen, Y., Brook, T. C., Soe, C. Z., O'Neill, I., Alcon-Giner, C., Leelastwattanagul, O., et al. (2020). Preterm infants harbour diverse Klebsiella populations, including atypical species that encode and produce an array of antimicrobial resistance- and virulence-associated factors. Microb. Genom. 6:e000377. doi: 10.1099/mgen.0.000377
Chen, L., Hu, H., Chavda, K. D., Zhao, S., Liu, R., Liang, H., et al. (2014). Complete sequence of a KPC-producing IncN multidrug-resistant plasmid from an epidemic Escherichia coli ST131 strain in China. Antimicrob. Agents Chemother. 58, 2422–2425. doi: 10.1128/AAC.02587-13
CLSI. (2020). Performance Standards for Antimicrobial Susceptibility Testing M100-30 th ed. Wayne, PA: CLSI.
Decré, D., Burghoffer, B., Gautier, V., Petit, J. C., and Arlet, G. (2004). Outbreak of multi-resistant Klebsiella oxytoca involving strains with extended-spectrum beta-lactamases and strains with extended-spectrum activity of the chromosomal beta-lactamase. J. Antimicrob. Chemother. 54, 881–888. doi: 10.1093/jac/dkh440
Ghasemian, A., Mohabati Mobarez, A., Najar Peerayeh, S., Talebi Bezmin Abadi, A., Khodaparast, S., and Mahmood, S. S. (2019). Expression of adhesin genes and biofilm formation among Klebsiella oxytoca clinical isolates from patients with antibiotic-associated haemorrhagic colitis. J. Med. Microbiol. 68, 978–985. doi: 10.1099/jmm.0.000965
Hoban, D., Badal, R., Bouchillon, S., Hackel, M., Kazmierczak, K., Lascols, C., et al. (2014). In vitro susceptibility and distribution of β-lactamases in Enterobacteriaceae causing intra-abdominal infections in North America 2010–2011. Diagn. Microbiol. Infect. Dis. 79, 367–372. doi: 10.1016/j.diagmicrobio.2014.03.026
Hoenigl, M., Valentin, T., Zarfel, G., Wuerstl, B., Leitner, E., Salzer, H. J., et al. (2012). Nosocomial outbreak of Klebsiella pneumoniae carbapenemase-producing Klebsiella oxytoca in Austria. Antimicrob. Agents Chemother. 56, 2158–2161. doi: 10.1128/AAC.05440-11
Jia, B., Raphenya, A. R., Alcock, B., Waglechner, N., Guo, P., Tsang, K. K., et al. (2017). CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 45, D566–D573. doi: 10.1093/nar/gkw1004
Jolley, K. A., Bray, J. E., and Maiden, M. C. J. (2018). Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 3:124. doi: 10.12688/wellcomeopenres.14826.1
Jolley, K. A., and Maiden, M. C. (2010). BIGSdb: Scalable analysis of bacterial genome variation at the population leve. BMC Bioinformatics. 11:595. doi: 10.1186/1471-2105-11-595
Letunic, I., and Bork, P. (2021). Interactive tree of life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296. doi: 10.1093/nar/gkab301
Liang, Q., Jiang, X., Hu, L., Yin, Z., Gao, B., Zhao, Y., et al. (2019). Sequencing and genomic diversity analysis of IncHI5 plasmids. Front. Microbiol. 9:3318. doi: 10.3389/fmicb.2018.03318
Long, H., Hu, Y., Feng, Y., and Zong, Z. (2022). Genome analysis of Klebsiella oxytoca complex for antimicrobial resistance and virulence genes. Antimicrob. Agents Chemother. 66:e0218321. doi: 10.1128/aac.02183-21
Lowe, C., Willey, B., O'Shaughnessy, A., Lee, W., Lum, M., Pike, K., et al. (2012). Outbreak of extended-spectrum β-lactamase-producing Klebsiella oxytoca infections associated with contaminated handwashing sinks (1). Emerg. Infect. Dis. 18, 1242–1247. doi: 10.3201/eid1808.111268
Mineau, S., Kozak, R., Kissoon, M., Paterson, A., Oppedisano, A., Douri, F., et al. (2018). Emerging antimicrobial resistance among Escherichia coli strains in bloodstream infections in Toronto, 2006-2016: a retrospective cohort study. CMAJ Open 6, E580–E586. doi: 10.9778/cmajo.20180039
Moura, A., Soares, M., Pereira, C., Leitão, N., Henriques, I., and Correia, A. (2009). INTEGRALL: a database and search engine for integrons, integrases and gene cassettes. Bioinformatics 25, 1096–1098. doi: 10.1093/bioinformatics/btp105
Nazik, H., Aydin, S., and Albayrak, R. (2014). Detection and spread of oxa-48-producing Klebsiella oxytoca isolates in Istanbul, Turkey. Southeast Asian J Trop Med Public Health 45, 123–129.
Nordmann, P., Poirel, L., Carrër, A., Toleman, M. A., and Walsh, T. R. (2011). How to detect NDM-1 producers. J. Clin. Microbiol. 49, 718–721. doi: 10.1128/JCM.01773-10
Pérez-Vazquez, M., Oteo-Iglesias, J., Sola-Campoy, P. J., Carrizo-Manzoni, H., Bautista, V., Lara, N., et al. (2019). Characterization of Carbapenemase-producing Klebsiella oxytoca in Spain, 2016-2017. Antimicrob. Agents Chemother. 63, e02529–e02518. doi: 10.1128/AAC.02529-18
Podschun, R., and Ullmann, U. (1998). Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin. Microbiol. Rev. 11, 589–603. doi: 10.1128/CMR.11.4.589
Qi, Y., Wei, Z., Ji, S., Du, X., Shen, P., and Yu, Y. (2010). ST11, the dominant clone of KPC-producing Klebsiella pneumoniae in China. J. Antimicrob. Chemother. 66, 307–312. doi: 10.1093/jac/dkq431
Queenan, A. M., and Bush, K. (2007). Carbapenemases: the versatile beta-lactamases. Clin. Microbiol. Rev. 20, 440–458. doi: 10.1128/CMR.00001-07
Saha, R., Farrance, C. E., Verghese, B., Hong, S., and Donofrio, R. S. (2013). Klebsiella michiganensis sp. nov., a new bacterium isolated from a tooth brush holder. Curr. Microbiol. 66, 72–78. doi: 10.1007/s00284-012-0245-x
Savino, F., Cordisco, L., Tarasco, V., Calabrese, R., Palumeri, E., and Matteuzzi, D. (2009). Molecular identification of coliform bacteria from colicky breastfed infants. Acta Paediatr. 98, 1582–1588. doi: 10.1111/j.1651-2227.2009.01419.x
Siguier, P., Perochon, J., Lestrade, L., Mahillon, J., and Chandler, M. (2006). ISfinder: the reference Centre for bacterial insertion sequences. Nucleic Acids Res. 34, D32–D36. doi: 10.1093/nar/gkj014
Tansirichaiya, S., Rahman, M. A., and Roberts, A. (2019). P. the transposon registry. Mob DNA 10:40. doi: 10.1186/s13100-019-0182-3
Wang, Y., Tong, M. K., Chow, K. H., Cheng, V. C., Tse, C. W., Wu, A. K., et al. (2018). Occurrence of highly conjugative IncX3 epidemic plasmid carrying blaNDM in Enterobacteriaceae isolates in geographically widespread areas. Front. Microbiol. 9:2272. doi: 10.3389/fmicb.2018.02272
Wang, J., Yuan, M., Chen, H., Chen, X., Jia, Y., Zhu, X., et al. (2017). First report of Klebsiella oxytoca strain simultaneously producing NDM-1, IMP-4, and KPC-2 Carbapenemases. Antimicrob. Agents Chemother. 61, e00877–e00817. doi: 10.1128/AAC.00877-17
Wu, W., Feng, Y., Tang, G., Qiao, F., McNally, A., and Zong, Z. (2019). NDM metallo-β-lactamases and their bacterial producers in health care settings. Clin. Microbiol. Rev. 32, e00115–e00118. doi: 10.1128/CMR.00115-18
Wyres, K. L., and Holt, K. E. (2016). Klebsiella pneumoniae population genomics and antimicrobial-resistant cones. Trends Microbiol. 24, 944–956. doi: 10.1016/j.tim.2016.09.007
Yang, J., Long, H., Hu, Y., Feng, Y., McNally, A., and Zong, Z. (2022). Klebsiella oxytoca complex: update on taxonomy, antimicrobial resistance, and virulence. Clin. Microbiol. Rev. 35:e0000621. doi: 10.1128/CMR.00006-21
Zankari, E., Hasman, H., Cosentino, S., Vestergaard, M., Rasmussen, S., Lund, O., et al. (2012). Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67, 2640–2644. doi: 10.1093/jac/dks261
Keywords: Klebsiella oxytoca complex, resistance, carbapenemase gene, plasmid, MLST
Citation: Wan W, Yang X, Yu H, Wang M, Jia W, Huang B, Qu F, Shan B, Tang Y-W, Chen L and Du H (2023) Genomic characterization of carbapenem-resistant Klebsiella oxytoca complex in China: a multi-center study. Front. Microbiol. 14:1153781. doi: 10.3389/fmicb.2023.1153781
Edited by:
Haijian Zhou, China CDC, ChinaReviewed by:
Bing Gu, Guangdong Provincial People's Hospital, ChinaQing Yang, Zhejiang University, China
Copyright © 2023 Wan, Yang, Yu, Wang, Jia, Huang, Qu, Shan, Tang, Chen and Du. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hong Du, aG9uZ19kdUAxMjYuY29t
†These authors have contributed equally to this work
 Weimin Wan
Weimin Wan Xiaochun Yang2†
Xiaochun Yang2† Min Wang
Min Wang Bin Huang
Bin Huang Fen Qu
Fen Qu Yi-Wei Tang
Yi-Wei Tang Hong Du
Hong Du