- 1Institute of Animal Husbandry and Veterinary Medicine, Fujian Academy of Agricultural Sciences, Fuzhou, China
- 2Fujian Animal Diseases Control Technology Development Center, Fuzhou, China
This study aimed to characterize two novel mcr-1 variants, mcr-1.35 and mcr-1.36, which originated from Moraxella spp. that were isolated from diseased pigs in China. The Moraxella spp. carrying novel mcr-1 variants were subjected to whole-genome sequencing (WGS) and phylogenetic analysis based on the 16S rRNA gene. The mcr-1 variants mcr-1.35 and mcr-1.36 were characterized using phylogenetic analysis, a comparison of genetic environments, and protein structure prediction. The WGS indicated that two novel mcr-1 variants were located in the chromosomes of three Moraxella spp. with a genetic environment of mcr-1-pap2. In addition to the novel colistin resistance genes mcr-1.35 and mcr-1.36, the three Moraxella spp. contained other antimicrobial resistance genes, including aac(3)-IId, tet(O), sul2, floR, and blaROB-3. A functional cloning assay indicated that either the mcr-1.35 or mcr-1.36 gene could confer resistance to colistin in Escherichia coli DH5α and JM109. The nucleotide sequences of mcr-1.35 and mcr-1.36 presented 95.33 and 95.33% identities, respectively, to mcr-1.1. The phylogenetic analysis showed that mcr-1.35 and mcr-1.36 were derived from Moraxella spp. that belonged to subclades that were different from those of the mcr-1 variants (mcr-1.1 to mcr-1.34 except mcr-1.10) originating from Enterobacteriaceae. The deduced amino acid sequences of MCR-1.35 (MCR-1.36) showed 96.67% (96.49%), 82.59% (82.04%), 84.07% (83.52%), 55.52% (55.17%), 59.75% (59.57%), and 61.88% (61.69%) identity to MCR-1.10, MCR-2.2, MCR-6.1, MCR-LIN, MCR-OSL, and MCR-POR, respectively, that originated from Moraxella sp. Notably, protein structure alignment showed only a few changes in amino acid residues between MCR-1.1 and MCR-1.35, as well as between MCR-1.1 and MCR-1.36. In conclusion, this study identified Moraxella spp. carrying two novel mcr-1 variants, mcr-1.35 and mcr-1.36, conferring resistance to colistin, which were isolated from pig farms in China. In addition, mcr-like variants were observed to be located in the chromosomes of some species of Moraxella isolated from pig samples.
Introduction
The overuse and misuse of antibiotics have resulted in the emergence of multidrug-resistant bacteria that pose a serious threat to people worldwide (Boucher et al., 2009; Nordmann and Poirel, 2016; Pham Thanh et al., 2016). Colistin is one of the last-line treatment options against infection by multidrug-resistant and carbapenem-resistant Gram-negative bacteria (Liu et al., 2016). However, the emergence of mobile colistin-resistant mcr-1 genes threatens the clinical efficacy of colistin. Epidemiological studies have found the mcr-like genes in Enterobacteriaceae, Moraxellaceae, Vibrionaceae, and Pseudomonadaceae (Ling et al., 2017; Caselli et al., 2018; Ling et al., 2020). To date, variants from mcr-1.1 to mcr-1.34 and from mcr-2 to mcr-10 have been discovered in over 40 countries across five continents (Wang et al., 2018; Ling et al., 2020; Wang et al., 2020). With the exception of mcr-1.10, which was found in Moraxella spp., variants from mcr-1.1 to mcr-1.34 have been identified in various Enterobacteriaceae of different origins (AbuOun et al., 2017; Ling et al., 2020; Wang et al., 2020). Other mcr genes, including mcr-5, mcr-8, mcr-9, and mcr-10, have also been described in Enterobacteriaceae. Moreover, mcr-3 and mcr-7 were reported in Aeromonas spp. (Ling et al., 2020), and mcr-4 was found in Pseudomonas spp. (Ling et al., 2020). Additionally, a genetic and bioinformatics analysis suggested that Moraxella spp. could be considered a potential source of MCR-1/2-like determinants (Kieffer et al., 2017). Moreover, several variants of the mcr-like genes, including mcr-1.10, mcr-2.2, and mcr-6.1, were first found in Moraxella sp. (AbuOun et al., 2017).
In addition to colistin resistance genes, Moraxella spp. are also known to contain ampicillin, penicillin, quinolone, macrolide, and tetracycline resistance genes, among others (Flamm et al., 2012; AbuOun et al., 2017; Krol-Turminska et al., 2020; Raveendran et al., 2020; Zhang et al., 2022). Bacteria of the genus Moraxella are aerobic, rod-shaped, opportunistic Gram-negative pathogens (Vela et al., 2010; Embers et al., 2011). The genus contains 18 species, including pathogens that cause infection in humans and animals (Woodbury et al., 2009; Kubota et al., 2012). For instance, Moraxella lacunata can cause infections in the eye and the upper respiratory tract (Woodbury et al., 2009; Mehmeti et al., 2021), while Moraxella osloensis and Moraxella catarrhalis, which are usually isolated from the human respiratory tract, can cause meningitis and pneumonia (Murphy and Parameswaran, 2009; Lee et al., 2017). Overall, Moraxella spp. pose a serious threat to human and animal health.
In the present study, we characterized two novel mcr-1 variants, mcr-1.35 and mcr-1.36, which originated from Moraxella spp. isolated from pigs with respiratory tract disease in swine farms in China.
Materials and methods
Isolation, purification, and identification of Moraxella sp.
A total of 312 nasal swabs were collected from pigs with a respiratory disease from 15 swine farms in Fujian Province, China (Supplementary Table S1). Subsequently, the samples were transferred into tryptic soy broth (TSB) (OXOID Ltd., UK). These TSB samples were then vortexed and plated onto tryptic soy agar (TSA) (OXOID Ltd., UK) supplemented with 5% defibrillated sheep blood (Zhengzhou Jiulong Biotechnology Co., Ltd., China) and 5% bovine serum (Zhejiang Tianhang Biotechnology Co., Ltd., China). A single colony was purified, after which PCR was performed to identify the 16S rRNA gene homology of the isolates using the primers 27F/1492R (Supplementary Table S2). Subsequently, the isolates identified as Moraxella spp. were further studied.
PCR screening of mcr-1 in Moraxella spp. and antimicrobial susceptibility testing of mcr-1-positive Moraxella sp. isolates
PCR was performed to detect the presence of mcr-1 in Moraxella sp. isolates using the primer mcr-1-F/R (Supplementary Table S2). The minimum inhibitory concentrations (MICs) of mcr-1-positive Moraxella sp. against 12 antibiotics were determined using the microbroth dilution method according to the Clinical and Laboratory Standards Institute (Wayne, 2020). The Escherichia coli ATCC 25922 strain was used for quality control.
Phylogenetic analysis of amino acid sequences of the mcr-like variants and the 16S rRNA gene sequences from Moraxella spp.
The phylogenetic analysis of the deduced amino acid sequences of the mcr-like variants and the 16S rRNA gene from Moraxella spp. was performed using MEGA 11.0 software (Tamura et al., 2021). The mcr-like variants included mcr-1.35, mcr-1.36, mcr-1.10, mcr-2.2, and mcr-6.1 polymyxin resistance genes identified in Moraxella spp. and those identified in M. osloensis, Moraxella lincolnii, and Moraxella porci (mcrOSL, mcrLIN, and mcrPOR, respectively) (Kieffer et al., 2017). In addition to the 16S rRNA genes of mcr-1-positive Moraxella spp. isolated in this study, other 16S rRNA genes from different Moraxella species were obtained from the NCBI database.
Phylogenetic analysis of all mcr-1 gene variants
The phylogenetic analysis of the complete sequences of mcr-1.35 and mcr-1.36 found in Moraxella spp., as well as the complete sequences of mcr-1.1 to mcr-1.34 registered in GenBank, were performed using MEGA 11.0 software (Supplementary Table S3; Tamura et al., 2021).
Whole-genome sequencing analysis
The DNA of three Moraxella spp. was extracted using a bacterial DNA extraction kit (OMEGA, USA). The whole genomes of these Moraxella sp. were sequenced using the Illumina Hiseq and Oxford Nanopore MinIon platforms, and complete sequences were obtained by hybrid assembly using Unicycler version 0.5.0 (Wick et al., 2017). Functional element prediction was conducted using Prokka (Seemann, 2014). The sequences were annotated by comparing the predicted gene sequences with functional databases such as RefSeq (O'Leary et al., 2016) and CARD (Alcock et al., 2020) with BLAST+. The genetic context of mcr-1 was analyzed using Easyfig (Sullivan et al., 2011).
Functional cloning of novel mcr-1 variants
A 1,623-bp DNA fragment of mcr-1.35 and mcr-1.36 was amplified from Moraxella spp. FZFQ2102, FZLJ2107, and FZLJ2109 using the primer mcr1L-F/R (Supplementary Table S2) and then cloned into pUC19 to obtain pUC19-mcr-1.35 and pUC19-mcr-1.36. The vector was transformed into E. coli DH5α and E. coli JM109. The MICs of polymyxin B against DH5α and JM109 harboring pUC19-mcr-1.35, pUC19-mcr-1.36, and pUC19 were determined by the microbroth dilution method.
Protein structure analysis of MCR-1 variants from Moraxella sp.
The structure of lipooligosaccharide phosphoethanolamine transferase A (EptA) (PDB accession number 5FGN) was obtained from the Protein Data Bank.1 The structures of MCR-1 variants were generated according to their amino acid sequences using the comparative protein-modeling SWISS-MODEL server (Waterhouse et al., 2018). These structures were then analyzed and visualized using PyMOL software.
Nucleotide sequence accession numbers
The complete nucleotide sequences of Moraxella spp. FZFQ2102, FZLJ2107, and FZLJ2109 were deposited in GenBank under the accession numbers CP099960, CP101111, and CP101112, respectively.
Results
Identification of mcr-1-positive Moraxella sp.
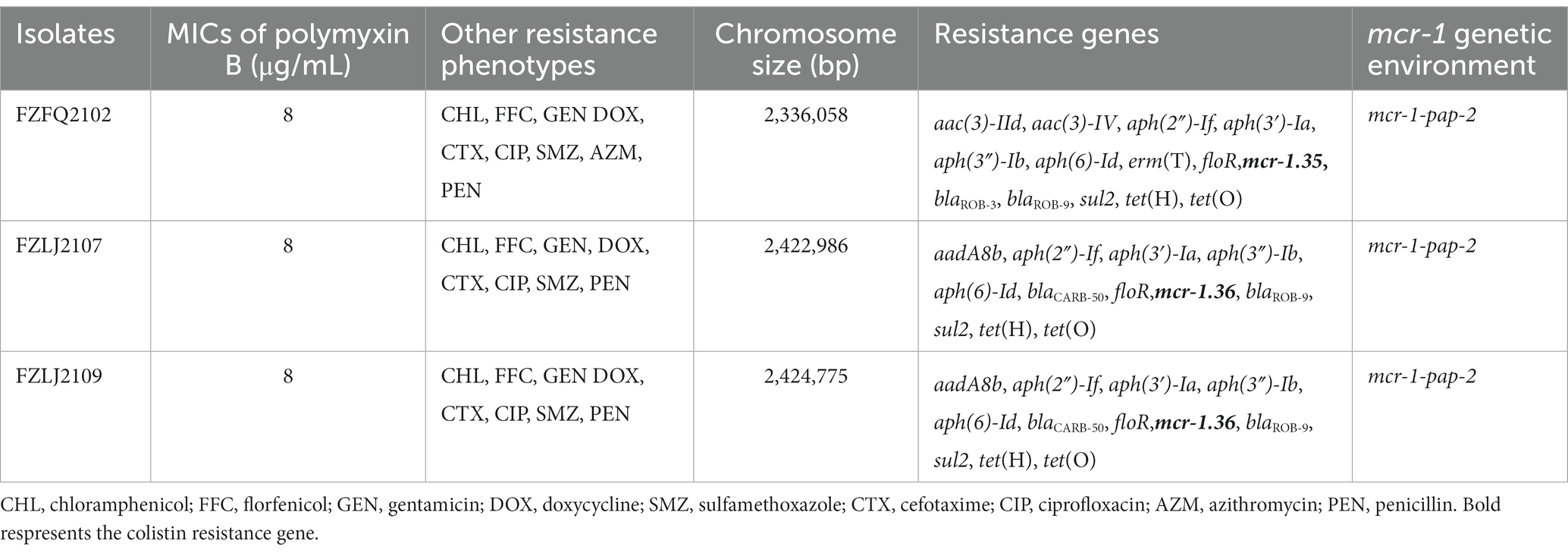
In total, 58 isolates that belonged to Moraxellae were isolated and identified from the 312 samples obtained from 15 swine farms in Fujian Province, China (Supplementary Table S1). Of these 58 isolates, three were identified as positive for mcr-1 and were named FZFQ2102, FZLJ2107, and FZLJ2109, respectively (Table 1).
Due to FZFQ2102, FZLJ2107, and FZLJ2109 being identified as Moraxella sp. by PCR, the phylogenetic analysis was performed using the 16S rRNA genes of the Moraxella sp. obtained in this study and those in the NCBI database. According to 16S rRNA sequence identity, the phylogenetic tree of Moraxella sp. was divided into four groups: M1, M2, M3, and M4. The Moraxella sp. FZFQ2102 shared a close phylogenetic relationship with Moraxella nonliquefaciens and Moraxella nasovis. Moreover, the Moraxella spp. FZLJ2107 and FZLJ2109 shared a close phylogenetic relationship with M. porci and Moraxella pluranimalium (Figure 1).
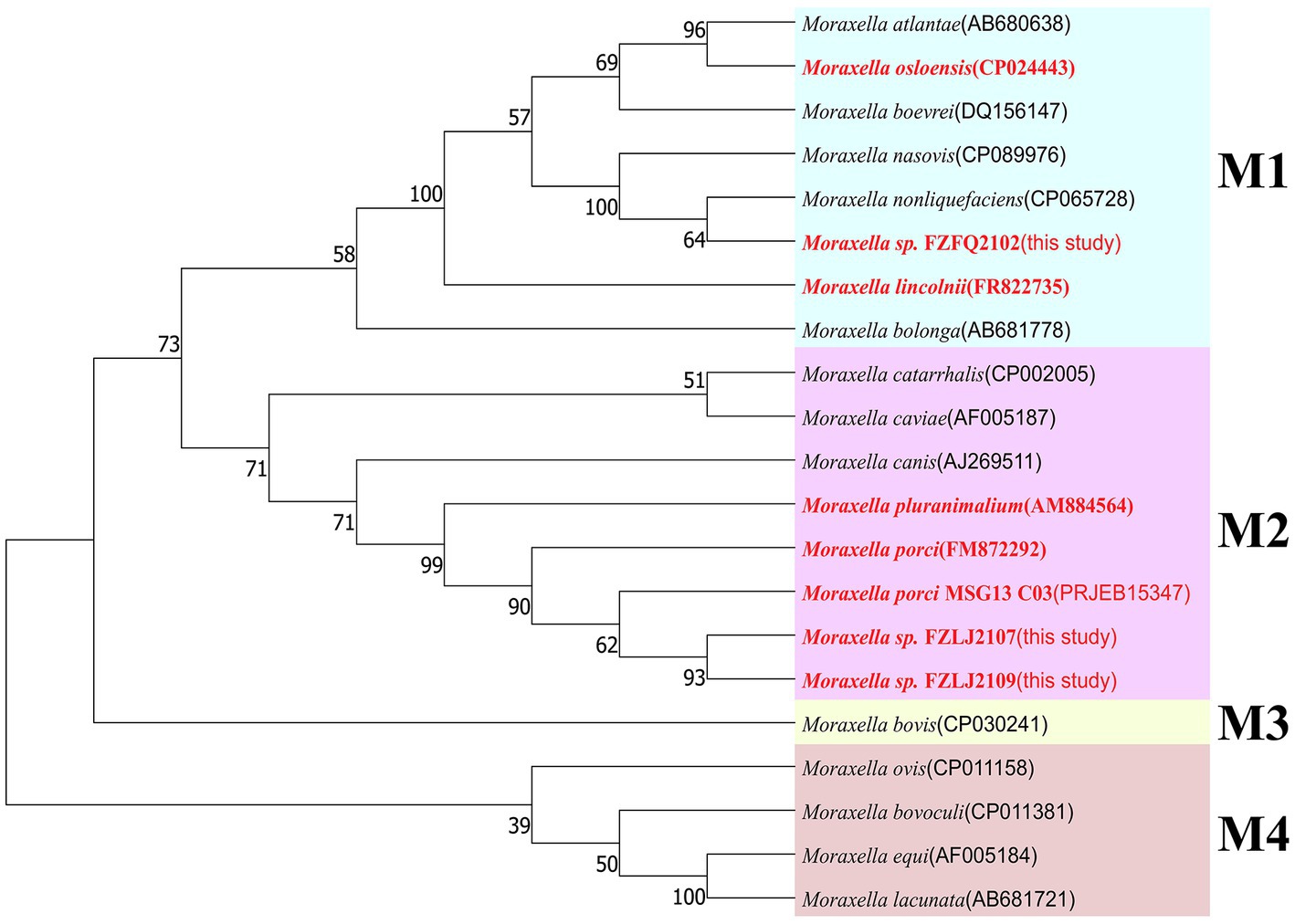
Figure 1. Phylogenetic tree of different Moraxella species based on 16S rRNA. The 16S rRNA genes of the three mcr-1-positive Moraxella spp. isolated in this study were compared with those of Moraxella spp. from the NCBI database. The neighbor-joining phylogeny was constructed using MEGA 11.0 with the 16S rRNA gene sequence of Moraxella spp. with the mcr gene (in red) and without the mcr gene (in black). Branch labels correspond to bootstrap support percentages out of 1,000 replicates. The tree is divided into four groups: M1 (light blue area), M2 (pink area), M3 (light yellow area), and M4 (light orange area).
Two novel colistin resistance gene mcr-1 variants in Moraxella sp.
Between 2015 and 2022, an increasing number of mcr-like variants were found in Moraxella sp. (AbuOun et al., 2017; Kieffer et al., 2017). The three mcr-1-positive Moraxella sp. isolated in this study demonstrated multidrug resistance, including resistance to polymyxin B (MIC = 8 μg/mL) (Table 1). Sequence analysis indicated that the three Moraxella sp. carried two mcr-1 variants. BLASTn results showed that the nucleotide sequence homology of the two mcr-1 variants with mcr-1 was 95.33% (Table 2). Furthermore, the phylogenetic analysis suggested that the nucleotide sequence identities of the two mcr-1 variants from Moraxella sp. FZFQ2102, FZLJ2107, and FZLJ2109 were distinct from those of mcr-1.1–mcr-1.34 (Figure 2). Therefore, the two novel mcr-1 variants were named mcr-1.35 and mcr-1.36.
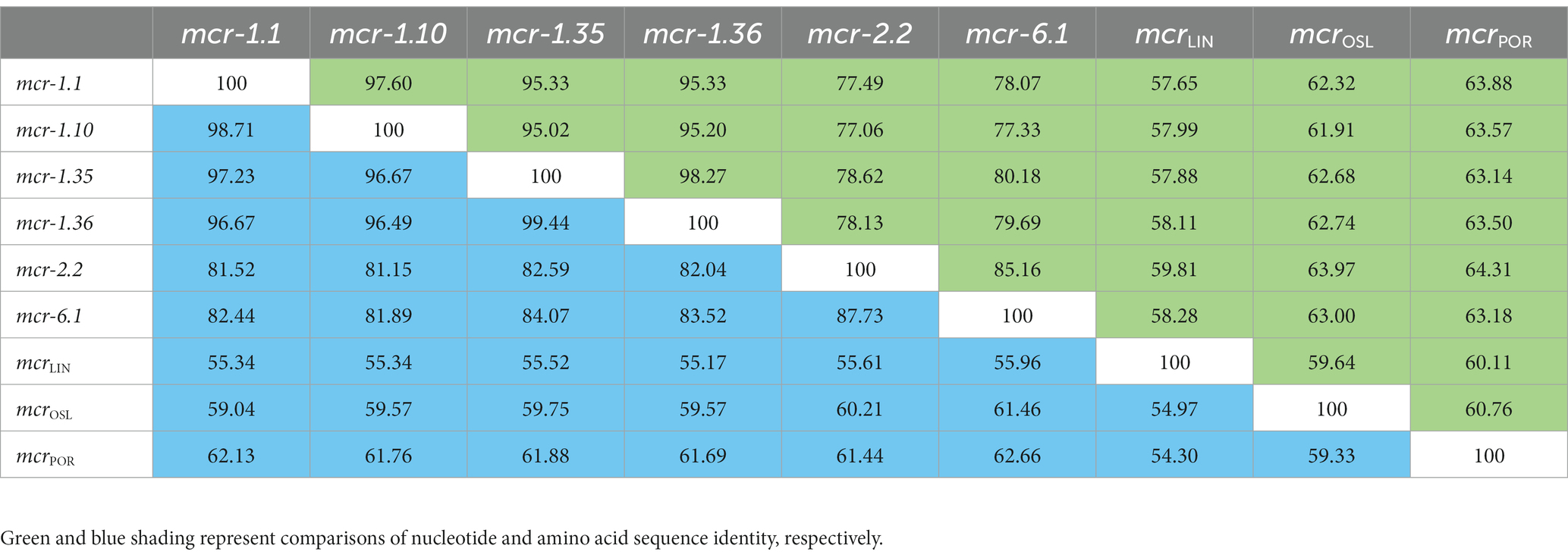
Table 2. Pairwise comparisons of mcr-1.1 and mcr-like variants in Moraxella sp. based on nucleotide and amino acid sequence identity (%).
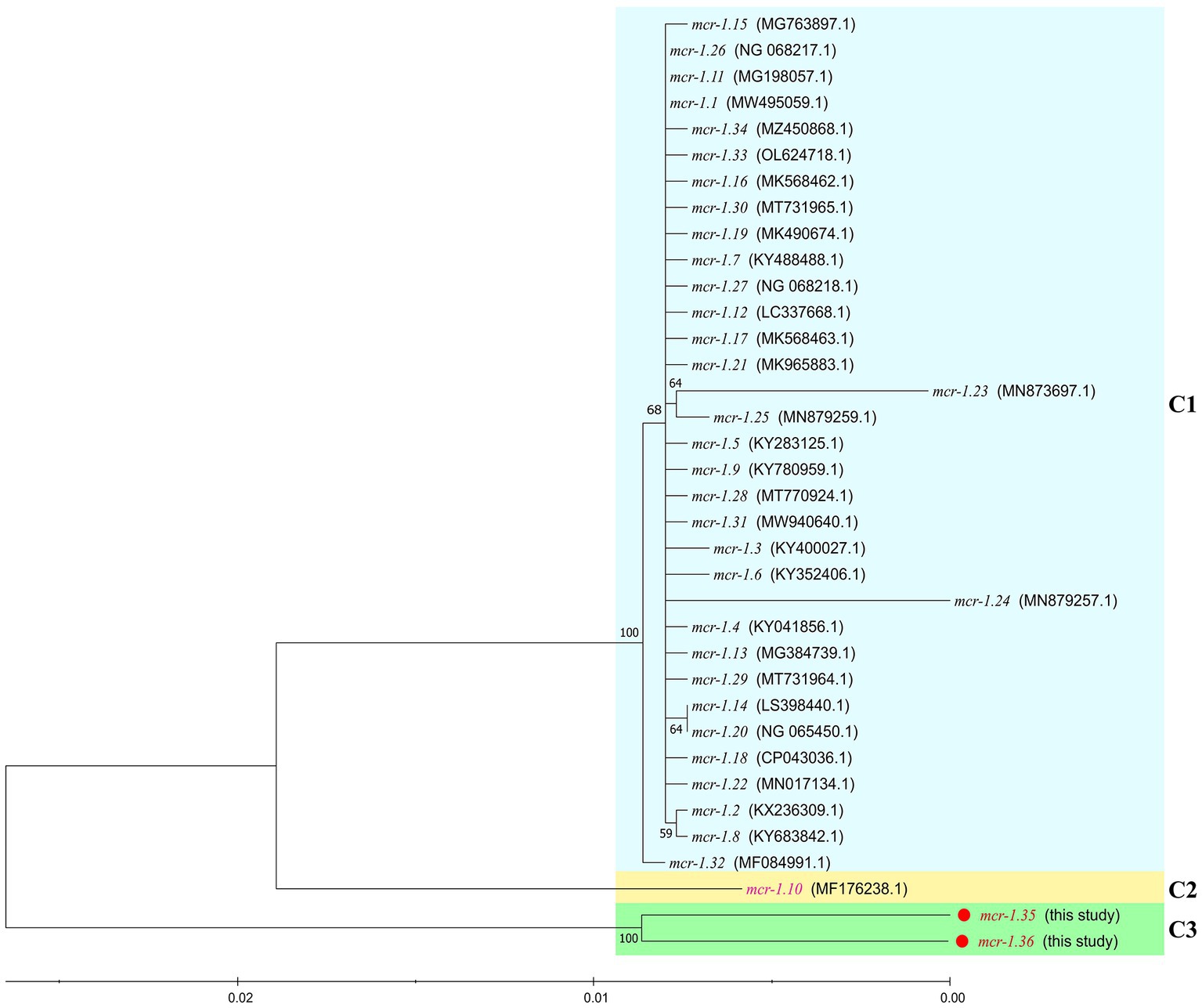
Figure 2. Comparison between mcr-1.35 and mcr-1.36 and all previously described mcr-1 subtypes, based on nucleotide sequences. The neighbor-joining phylogeny was constructed using MEGA 11.0 with the nucleotide sequences of the novel colistin-resistance genes mcr-1.35 and mcr-1.36 (in red) and all previously described the mcr-1 gene subtypes (mcr-1.1 to mcr-1.34 [in black and pink]). Branch labels correspond to bootstrap support percentages out of 1,000 replicates. The tree is divided into three groups: C1, which contains the mcr-1 gene subtypes from other bacterial species (light blue area), C2, which contains the mcr-1.10 gene from Moraxella spp. (light yellow area), and C3, which contains the mcr-1.35 and mcr-1.36 genes from Moraxella spp. (green area).
Characterization of the three mcr-1-positive Moraxella sp.
Whole-genome sequencing (WGS) suggested that the three Moraxella sp., FZFQ2102, FZLJ2107, and FZLJ2109, have 2,336,058-bp, 2,422,986-bp, and 2,424,775-bp chromosomes, respectively, and no plasmids (Table 1). WGS also showed that the three Moraxella sp. possessed multiple drug-resistance genes. The Moraxella sp. FZFQ2102 contained the colistin resistance gene mcr-1.35; the aminoglycoside resistance genes aac(3)-IId, aac(3)-IV, aph(2″)-If, aph(3′)-Ia, aph(3″)-Ib, and aph(6)-Id; the macrolide resistance gene ermT; the tetracycline resistance genes tet(H) and tet(O); the sulfonamide resistance gene sul2; the phenicol resistance gene floR; and the β-lactamase resistance genes blaROB-3 and blaROB-9. Moreover, the Moraxella sp. FZLJ2107 and FZLJ2109 contained the colistin resistance gene mcr-1.36; the aminoglycoside resistance genes aadA8b, aph(2″)-If, aph(3′)-Ia, aph(3″)-Ib, and aph(6)-Id; the tetracycline resistance genes tet(H) and tet(O); the sulfonamide resistance gene sul2; the phenicol resistance gene floR; and the β-lactamase resistance genes blaROB-9 and blaCARB-50 (Table 1).
The analysis of the context of the mcr-1 variants suggested that their flanking regions were highly similar among the Moraxella sp. The mcr genes in the Moraxella sp. were downstream of a site-specific recombinase gene (ssr) and upstream of the pap-2 gene. However, the genetic environment of mcr-1 variants in other mcr-1-carrying bacteria differed from that of the Moraxella sp. Interestingly, mcr-1 variants had a common genetic unit: mcr-1-pap2 (Figure 3).
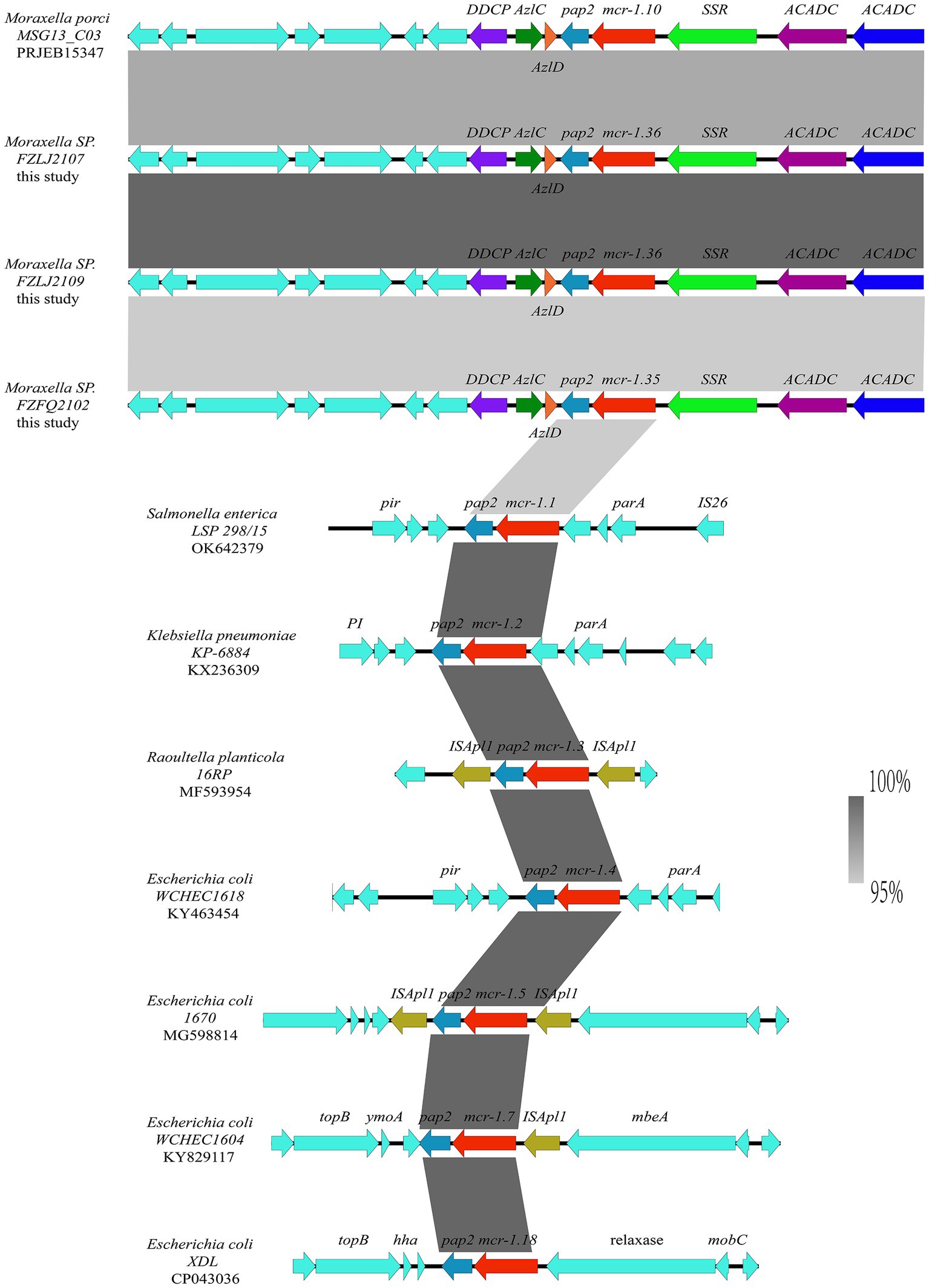
Figure 3. Comparison between the genetic context of the mcr-1 subtypes from Moraxella spp. and those from other bacterial species. The extents and directions of genes are shown by arrows labeled with gene names, with the mcr-1 subtypes represented by red arrows and the pap2 gene represented by a light blue arrow. Other flanking genes have been annotated. The shadow parallelograms between each sequence denote sequence identity.
Functional confirmation of mcr-1.35 and mcr-1.36
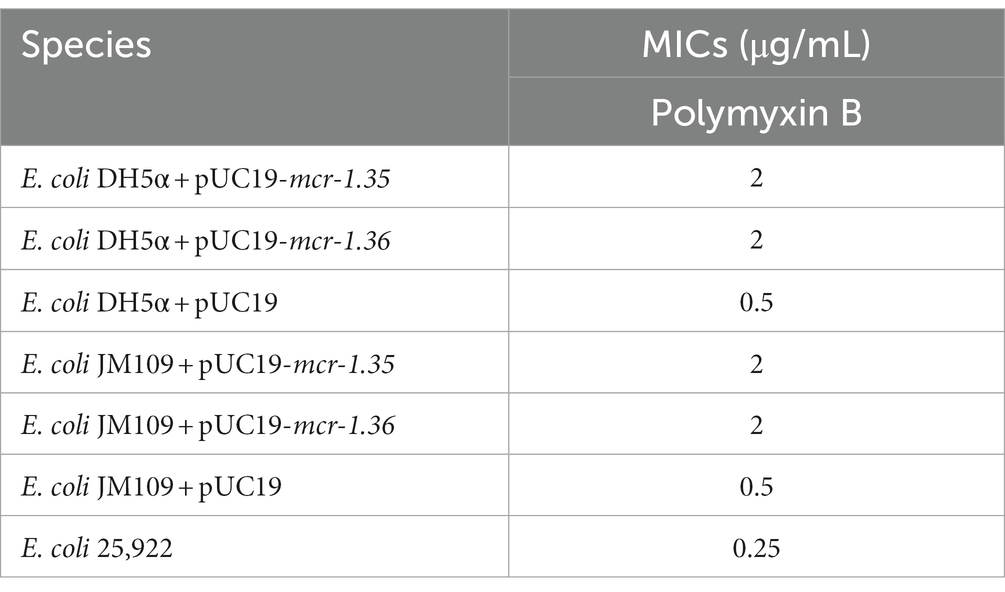
BLASTn analysis confirmed that two 1,623-bp open reading frames (ORFs) encoded the putative phosphoethanolamine transferase, which showed high nucleotide sequence identity to the original mcr-1.1 gene. The amino acid sequences of MCR-1.35 and MCR-1.36 showed 97.23 and 96.67% identity, respectively, to that of MCR-1.1 (Table 2). To confirm the function of the putative phosphoethanolamine transferase, the recombinant vectors pUC19-mcr-1.35 and pUC19-mcr-1.36 were constructed and subsequently transformed into E. coli DH5α and JM109. Compared with E. coli DH5α and JM109 carrying pUC19 alone, the expression of these ORFs in E. coli DH5α and JM109 led to 4-fold increases in the MIC of polymyxin B, which further identified the function of the mcr-1.35 and mcr-1.36 genes (Table 3).
Analysis of mcr-like variants from Moraxella sp.
BLASTn results showed that mcr-1.35 and mcr-1.36 exhibited more than 95% nucleotide sequence identity to mcr-1.10, indicating a close phylogenetic relationship. Mcr-1.35 and mcr-1.36 also exhibited 78.62 and 78.13% nucleotide sequence identity to mcr-2.2 and 80.18 and 79.69% nucleotide sequence identity to mcr-6.1, respectively. Furthermore, mcr-1.35 (mcr-1.36) showed 57.88% (58.11%), 62.68% (62.74%), and 63.14% (63.50%) nucleotide sequence identity to the mcrLIN, mcrOSL, and mcrPOR genes found in M. osloensis, M. porci, and M. lincolnii, respectively. Additionally, mcr-1.35 exhibited 98.27% nucleotide sequence identity to mcr-1.36 in this study (Table 2).
Compared with other phosphoethanolamine transferases in Moraxella sp., MCR-1.35, and MCR-1.36 aligned closely with an MCR-1.10 found in M. porci isolated from a pig in Great Britain (> 96%). MCR-1.35 and MCR-1.36 also exhibited 82.59 and 82.04% amino acid identity to MCR-2.2 and 84.07 and 83.52% amino acid identity to MCR-6.1, found in Moraxella pluranimalium isolated from pigs in Europe, respectively. In addition, MCR-1.35 (MCR-1.36) showed 55.52% (55.17%), 59.75% (59.57%), and 61.88% (61.69%) amino acid sequence identity to the MCR-OSL, MCR-POR, and MCR-LIN proteins found in M. osloensis, M. porci, and M. lincolnii, respectively (Table 2; Figure 4).
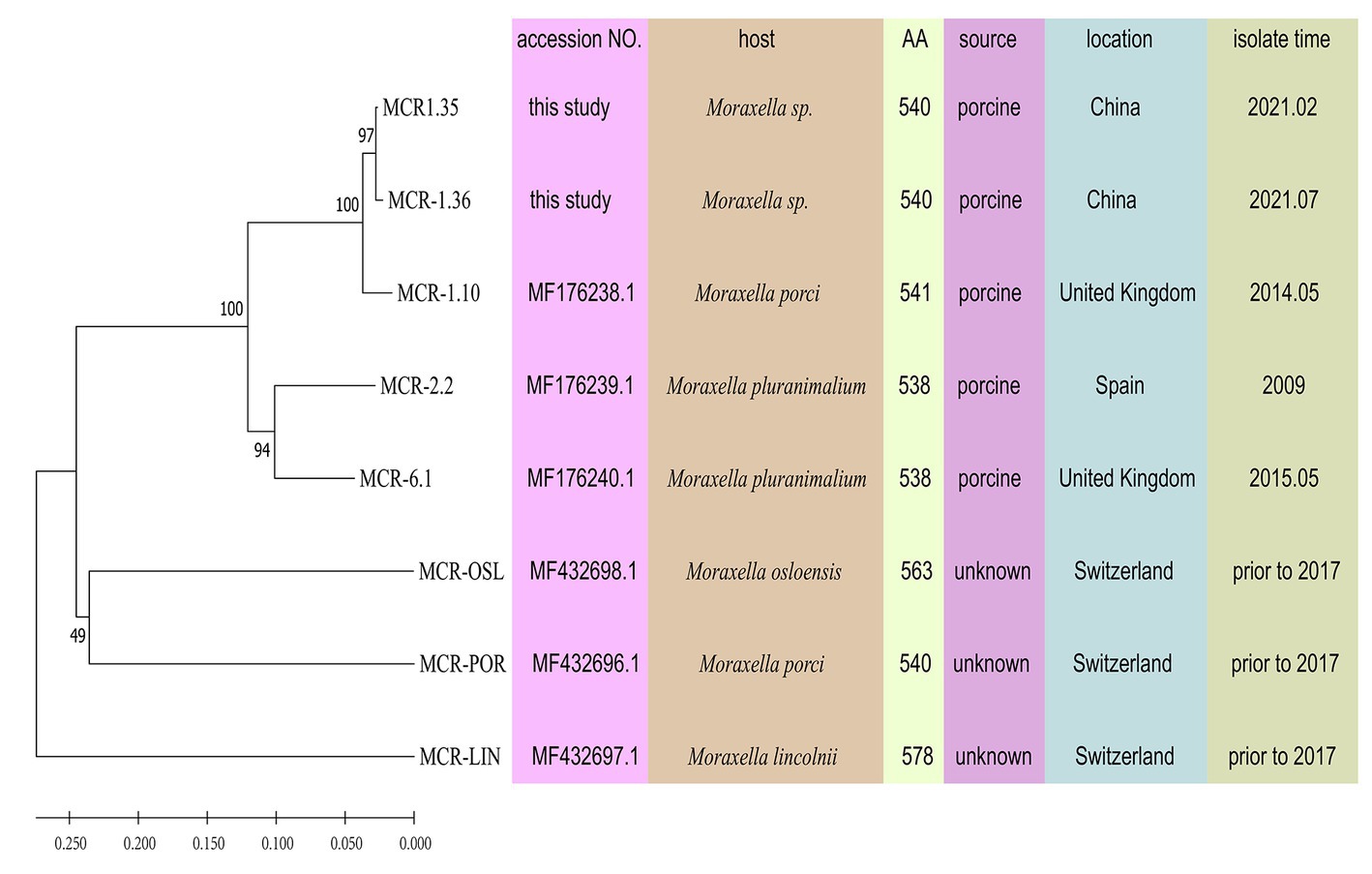
Figure 4. Phylogeny of MCR-like variants from Moraxella spp. determined based on amino acid sequences using MEGA 11.0. MCR-OSL, MCR-POR, and MCR-LIN are polymyxin resistance determinants that were identified in M. osloensis, M. porci and M. lincolnii, respectively. Some parameters relevant to the MCR proteins are presented, including bacterial host, the number of amino acids, isolate source, isolate location, and isolate time.
Structure prediction and amino acid sequence analysis of MCR-1 variants from Moraxella sp.
MCR-1 variants from Moraxella sp. were aligned using BioEdit (Figure 5A). When aligned with MCR-1.1, MCR-1.10 presented six amino acid mutations at residues R11C, A23S, M155V, M234T, A354T, and A443T. Four residues, S23, V155, T354, and T443, were located in the α-helix of the secondary structure, while residues C11 and T234 were located in the random coil. However, compared with those of MCR-1.1, the amino acid sequences of MCR-1.35 exhibited changes at residues Q277, T354, T381, A491, S498, N499, N500, S501, S502, 503 deletion, F503, T506, S508, and A511. Three residues, T354, T506, and S508, were located in the α-helix on the secondary structure, while all other residues were located in the random coil. In addition, amino acid sequences of MCR-1.36 presented changes at residues S23, Q277, T354, T381, M451, A491, S498, N499, N500, S501, S502, 503 deletion, F503, T506, S508, and A511. In addition, the four residues S23, T354, T506, and S508 were located in the α-helix on the secondary structure, with all other residues located in the random coil (Figures 5C, D). Notably, protein structure alignment revealed only a few differences between MCR-1.1 and MCR-1.10, MCR-1.1 and MCR-1.35, and MCR-1.1 and MCR-1.36 (Figure 5D).
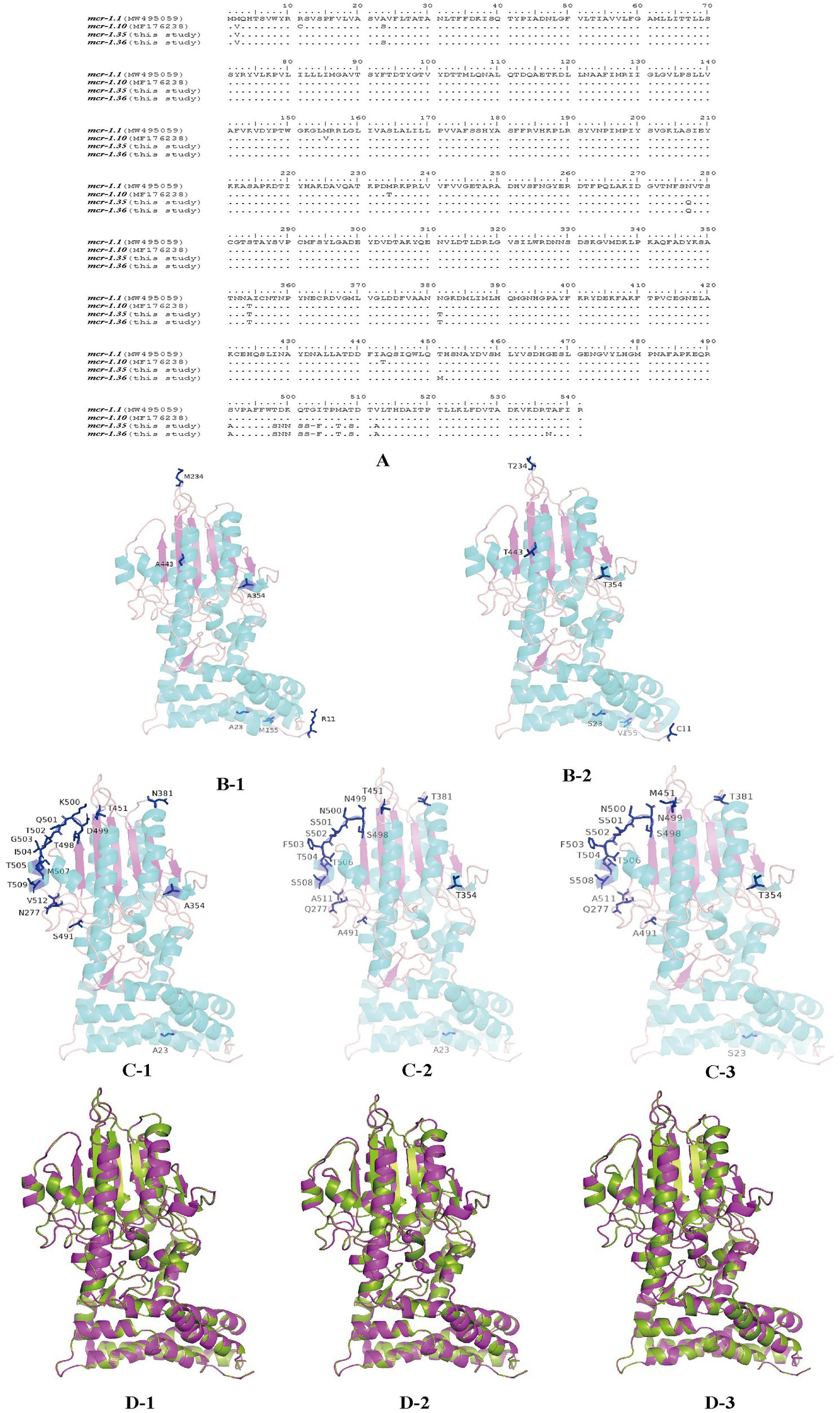
Figure 5. Structures of MCR-1 variants (MCR-1.1, MCR-1.10, MCR-1.35, and MCR-1.36) (A) Amino acid sequences of MCR-1.10, MCR-1.35, and MCR-1.36 compared with MCR-1.1. Amino acid residues are depicted in black, and the same amino acids are represented by dots in the alignment. (B) Structural models comparison of the MCR-1.1 protein (B-1) and MCR-1.10 (B-2) from Moraxella spp. based on lipooligosaccharide phosphoethanolamine transferase EptA. (C) Structural models comparison of the MCR-1.1 protein (C-1), and MCR-1.35 (C-2), MCR-1.36 (C-3) from Moraxella spp. based on lipooligosaccharide phosphoethanolamine transferase EptA. Models were constructed using the SWISS-MODEL server, and structures were viewed and edited using PyMOL2.5.4. Mutated amino acids are shown in the structural model. (D) Comparison between the structure of MCR-1.1 (in green) and that of MCR-1.10 (D-1), MCR-1.35 (D-2) and MCR-1.36 (D-3) (in pink).
Discussion
Moraxella spp. have been identified as potential sources of MCR-like polymyxin resistance determinants (Kieffer et al., 2017; Poirel et al., 2017). One study identified the mcr-like genes, including mcr-1.10 (M. porci), mcr-2.2 (M. pluranimalium), and mcr-6.1 (M. pluranimalium), in Moraxella spp. isolated from the cecal contents of healthy pigs on farms in Great Britain (AbuOun et al., 2017). Furthermore, the proteins MCR-POR (M. porci), MCR-OSL (M. osloensis), and MCR-LIN (M. lincolnii) in Moraxella spp. shared high amino acid identities with MCR-1/2-like (Kieffer et al., 2017). In the present study, the novel mcr-1 variants mcr-1.35 and mcr-1.36 were identified in Moraxella spp. recovered from the nasal swabs of pigs with respiratory diseases in farms in China. Notably, the emergence of different mcr-like variants in various Moraxella spp. indicates that the mcr-like variants were stored in the chromosomes of some Moraxella species.
Three Moraxella spp. containing novel mcr-1 variants showed resistance to polymyxin (Table 1). Additionally, the MIC of colistin for Moraxella spp. containing mcr-1.10, mcr-2.2, and mcr-6.1 ranged from 1 to 2 μg/mL (AbuOun et al., 2017), while the MIC of colistin for EptA-containing Moraxella spp. ranged from 2 to 64 μg/mL (AbuOun et al., 2017; Kieffer et al., 2017). Moreover, mcr-1.35, mcr-1.36, mcr-2.2, mcrosl, mcrlin, and mcrpor conferred resistance to polymyxin in E. coli (Kieffer et al., 2017; Poirel et al., 2017). In addition to colistin resistance genes (mcr-1.10, mcr-1.35, mcr-1.36, mcr-2.2, and mcr-6.1), Moraxella spp. also carried genes for β-lactam resistance (blaBRO-1 and blaBRO-2) aminoglycoside resistance (aac(3)-IId, aac(3)-IV, aph(2″)-If, aph(3′)-Ia, aph(3″)-Ib, and aph(6)-Id); tetracycline resistance [tet(B), tet(D), tet(L), and tet(O)]; chloramphenicol resistance (floR), sulfonamide resistance (sul2), and macrolide resistance (ermT) (Flamm et al., 2012; AbuOun et al., 2017; Krol-Turminska et al., 2020; Raveendran et al., 2020; Zhang et al., 2022). In recent studies, Moraxella spp. from human samples have proven to be mainly resistant to β-lactam, tetracycline, and macrolide (Flamm et al., 2012; Krol-Turminska et al., 2020; Raveendran et al., 2020; Zhang et al., 2022). The three Moraxella spp. from animal samples collected in this study showed multidrug resistance, indicating that Moraxella spp. pose potential threats to human and animal health.
The phylogenetic analysis showed that the mcr-1 variants (mcr-1.10, mcr-1.35, and mcr-1.36) that originated from Moraxella spp. belonged to subclades (C2 and C3 group) distinct from that of the mcr-1 variants originating from Enterobacteriaceae (C1 group) (Figure 2). The phylogenetic relationships between mcr-1.35, mcr-1.36, and mcr-1.1 differed, but the products of these genes are known to be responsible for polymyxin resistance (Liu et al., 2016). The comparisons of the nucleotide sequences showed that the context of the mcr-1 variants was similar among Moraxella spp. but differed from the genetic environment of the mcr-1 variants in other bacteria. These results indicate that mcr-1-pap2 acts as the genetic unit of the mcr-1 variants in Moraxella spp. (AbuOun et al., 2017; Poirel et al., 2017).
Compared with that of MCR-1.1, the amino acid sequences of MCR-1.35 and MCR-1.36 exhibited mutations located at the α-helix and random coil on the secondary structure. Previous research has suggested that the eight active sites (E246, T285, K333, H395, D465, H466, E468, and H478) located on the β-sheet of the protein’s secondary structure are essential for the activity of MCR-1 (Hu et al., 2016; Ma et al., 2016; Stojanoski et al., 2016; Hinchliffe et al., 2017). Notably, these β-sheet mutations were highly conserved in MCR-1.35 and MCR-1.36 ORFs (Carroll et al., 2019; Wang et al., 2020), indicating that the mutations of MCR-1.35 and MCR-1.36 have no influence on their polymyxin resistance activity. Furthermore, protein structure alignment indicated that the structures of MCR-1.35, MCR-1.36, and MCR-1.1 did not differ significantly.
In conclusion, this study identified the Moraxella spp. carrying two novel mcr-1 variants, mcr-1.35 and mcr-1.36, conferring resistance to colistin, which were isolated from pig farms in China. Moraxella spp. were considered potential sources of MCR-like determinants, and the mcr-like variants were observed to be located in the chromosomes of some Moraxella species isolated from pig samples. Therefore, further research on the mcr-like genes in Moraxella spp. may help us understand the evolution and spread of the mcr-like genes.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
LZ and YC designed the experiments. YC, RW, and HL performed the experiments. LW and XW prepared the tables and figures. YC, RW, QC, and RC prepared the manuscript. All authors read and approved the final version of the manuscript.
Funding
This study was supported by the National Nature Science Foundation of China (No. 31872502), basic scientific research projects of provincial public welfare scientific research institutions (Fujian Province), China (No. 2022R10260013), and the key project of the Fujian “5511” Collaborative Innovation Project, China (No. XTCXGC2021008).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1153740/full#supplementary-material
Footnotes
References
AbuOun, M., Stubberfield, E. J., Duggett, N. A., Kirchner, M., Dormer, L., Nunez-Garcia, J., et al. (2017). Mcr-1 and mcr-2 variant genes identified in Moraxella species isolated from pigs in Great Britain from 2014 to 2015. J. Antimicrob. Chemother. 72, 2745–2749. doi: 10.1093/jac/dkx286
Alcock, B. P., Raphenya, A. R., Lau, T. T. Y., Tsang, K. K., Bouchard, M., Edalamand, A., et al. (2020). CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 48, D517–D525. doi: 10.1093/nar/gkz935
Boucher, H. W., Talbot, G. H., Bradley, J. S., Edwards, J. E., Gilbert, D., Rice, L. B., et al. (2009). Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin. Infect. Dis. 48, 1–12. doi: 10.1086/595011
Carroll, L. M., Gaballa, A., Guldimann, C., Sullivan, G., Henderson, L. O., and Wiedmann, M. (2019). Identification of novel mobilized Colistin resistance gene mcr-9 in a multidrug-resistant, Colistin-susceptible Salmonella enterica serotype Typhimurium isolate. MBio 10:e00853–19. doi: 10.1128/mBio.00853-19
Caselli, E., D'Accolti, M., Soffritti, I., Piffanelli, M., and Mazzacane, S. (2018). Spread of mcr-1-driven Colistin resistance on hospital surfaces, Italy. Emerg. Infect. Dis. 24, 1752–1753. doi: 10.3201/eid2409.171386
Embers, M. E., Doyle, L. A., Whitehouse, C. A., Selby, E. B., Chappell, M., and Philipp, M. T. (2011). Characterization of a Moraxella species that causes epistaxis in macaques. Vet. Microbiol. 147, 367–375. doi: 10.1016/j.vetmic.2010.06.029
Flamm, R. K., Sader, H. S., Farrell, D. J., and Jones, R. N. (2012). Macrolide and tetracycline resistance among Moraxella catarrhalis isolates from 2009 to 2011. Diagn. Microbiol. Infect. Dis. 74, 198–200. doi: 10.1016/j.diagmicrobio.2012.06.007
Hinchliffe, P., Yang, Q. E., Portal, E., Young, T., Li, H., Tooke, C. L., et al. (2017). Insights into the mechanistic basis of plasmid-mediated Colistin resistance from crystal structures of the catalytic domain of MCR-1. Sci. Rep. 7:39392. doi: 10.1038/srep39392
Hu, M., Guo, J., Cheng, Q., Yang, Z., Chan, E. W. C., Chen, S., et al. (2016). Crystal structure of Escherichia coli originated MCR-1, a phosphoethanolamine transferase for Colistin resistance. Sci. Rep. 6:38793. doi: 10.1038/srep38793
Kieffer, N., Nordmann, P., and Poirel, L. (2017). Moraxella species as potential sources of MCR-like Polymyxin resistance determinants. Antimicrob. Agents Chemother. 61:e00129–17. doi: 10.1128/AAC.00129-17
Krol-Turminska, K., Olender, A., and Bogut, A. (2020). Tetracycline resistance in Moraxella catarrhalis clinical strains isolated in Poland. New Microbiol. 43, 103–106.
Kubota, H., Mitani, A., Niwano, Y., Takeuchi, K., Tanaka, A., Yamaguchi, N., et al. (2012). Moraxella species are primarily responsible for generating malodor in laundry. Appl. Environ. Microbiol. 78, 3317–3324. doi: 10.1128/AEM.07816-11
Lee, W. S., Hsueh, P. R., Yu, F. L., Chen, F. L., Hsieh, T. C., and Ou, T. Y. (2017). Moraxella osloensis bacteremia complicating with severe pneumonia in a patient with lung cancer. J. Microbiol. Immunol. Infect. 50, 395–396. doi: 10.1016/j.jmii.2015.03.005
Ling, Z., Yin, W., Li, H., Zhang, Q., Wang, X., Wang, Z., et al. (2017). Chromosome-mediated mcr-3 variants in Aeromonas veronii from chicken meat. Antimicrob. Agents Chemother. 61:e01272–17. doi: 10.1128/AAC.01272-17
Ling, Z., Yin, W., Shen, Z., Wang, Y., Shen, J., and Walsh, T. R. (2020). Epidemiology of mobile colistin resistance genes mcr-1 to mcr-9. J. Antimicrob. Chemother. 75, 3087–3095. doi: 10.1093/jac/dkaa205
Liu, Y. Y., Wang, Y., Walsh, T. R., Yi, L. X., Zhang, R., Spencer, J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168. doi: 10.1016/S1473-3099(15)00424-7
Ma, G., Zhu, Y., Yu, Z., Ahmad, A., and Zhang, H. (2016). High resolution crystal structure of the catalytic domain of MCR-1. Sci. Rep. 6:39540. doi: 10.1038/srep39540
Mehmeti, F., Cochard, B., Cuerel, C., De Coulon, G., Dayer, R., Steiger, C., et al. (2021). Moraxella lacunata subacute osteomyelitis in a child. J. Pediatric. Infect. Dis. Soc. 10, 161–163. doi: 10.1093/jpids/piz089
Murphy, T. F., and Parameswaran, G. I. (2009). Moraxella catarrhalis, a human respiratory tract pathogen. Clin. Infect. Dis. 49, 124–131. doi: 10.1086/599375
Nordmann, P., and Poirel, L. (2016). Plasmid-mediated colistin resistance: an additional antibiotic resistance menace. Clin. Microbiol. Infect. 22, 398–400. doi: 10.1016/j.cmi.2016.03.009
O'Leary, N. A., Wright, M. W., Brister, J. R., Ciufo, S., Haddad, D., McVeigh, R., et al. (2016). Reference sequence (RefSeq) database at NCBI: current status, taxonnomic expansion, and functional annotation. Nucleic Acids Res. 44, D733–D745. doi: 10.1093/nar/gkv1189
Pham Thanh, D., Thanh Tuyen, H., Nguyen Thi Nguyen, T., Chung The, H., Wick, R. R., Thwaites, G. E., et al. (2016). Inducible colistin resistance via a disrupted plasmid-borne mcr-1 gene in a 2008 Vietnamese Shigella sonnei isolate. J. Antimicrob. Chemother. 71, 2314–2317. doi: 10.1093/jac/dkw173
Poirel, L., Kieffer, N., Fernandez-Garayzabal, J. F., Vela, A. I., Larpin, Y., and Nordmann, P. (2017). MCR-2-mediated plasmid-borne polymyxin resistance most likely originates from Moraxella pluranimalium. J. Antimicrob. Chemother. 72, 2947–2949. doi: 10.1093/jac/dkx225
Raveendran, S., Kumar, G., Sivanandan, R. N., and Dias, M. (2020). Moraxella catarrhalis: a cause of concern with emerging resistance and presence of BRO Beta-lactamase gene-report from a tertiary care hospital in South India. Int. J. Microbiol. 2020:7316257. doi: 10.1155/2020/7316257
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Stojanoski, V., Sankaran, B., Prasad, B. V., Poirel, L., Nordmann, P., and Palzkill, T. (2016). Structure of the catalytic domain of the colistin resistance enzyme MCR-1. BMC Biol. 14:81. doi: 10.1186/s12915-016-0303-0
Sullivan, M. J., Petty, N. K., and Beatson, S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27, 1009–1010. doi: 10.1093/bioinformatics/btr039
Tamura, K., Stecher, G., and Kumar, S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Vela, A. I., Sanchez-Porro, C., Aragon, V., Olvera, A., Dominguez, L., Ventosa, A., et al. (2010). Moraxella porci sp. nov., isolated from pigs. Int. J. Syst. Evol. Microbiol. 60, 2446–2450. doi: 10.1099/ijs.0.016626-0
Wang, C., Feng, Y., Liu, L., Wei, L., Kang, M., and Zong, Z. (2020). Identification of novel mobile colistin resistance gene mcr-10. Emerg. Microbes. Infect. 9, 508–516. doi: 10.1080/22221751.2020.1732231
Wang, X., Wang, Y., Zhou, Y., Li, J., Yin, W., Wang, S., et al. (2018). Emergence of a novel mobile colistin resistance gene, mcr-8, in NDM-producing Klebsiella pneumoniae. Emerg. Microbes Infec. 7:122. doi: 10.1038/s41426-018-0124-z
Waterhouse, A., Bertoni, M., Bienert, S., Studer, G., Tauriello, G., Gumienny, R., et al. (2018). SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303. doi: 10.1093/nar/gky427
P. A. Wayne (2020) in Performance standards for antimicrobial disk and dilution susceptibility tests for Bacteria isolated from animals. ed. Clinical and laboratory standards institute (CLSI). 5th ed (Pittsburgh: CLSI Supplement VET01S; Clinical and Laboratory Standards Institute)
Wick, R. R., Judd, L. M., Gorrie, C. L., and Holt, K. E. (2017). Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13:e1005595. doi: 10.1371/journal.pcbi.1005595
Woodbury, A., Jorgensen, J., Owens, A., and Henao-Martinez, A. (2009). Moraxella lacunata septic arthritis in a patient with lupus nephritis. J. Clin. Microbiol. 47, 3787–3788. doi: 10.1128/JCM.01403-09
Keywords: pig, Moraxella , mcr-1 variant, colistin, resistance, mcr-1.35 , mcr-1.36
Citation: Che Y, Wu R, Li H, Wang L, Wu X, Chen Q, Chen R and Zhou L (2023) Characterization of two novel colistin resistance gene mcr-1 variants originated from Moraxella spp. Front. Microbiol. 14:1153740. doi: 10.3389/fmicb.2023.1153740
Edited by:
Liang-xing Fang, South China Agricultural University, ChinaReviewed by:
Jiyun Li, Hunan Agricultural University, ChinaTao He, Jiangsu Academy of Agricultural Sciences, China
Jing Wang, Yangzhou University, China
Copyright © 2023 Che, Wu, Li, Wang, Wu, Chen, Chen and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lunjiang Zhou, bHVuamlhbmdAMTYzLmNvbQ==
†These authors share first authorship
 Yongliang Che
Yongliang Che Renjie Wu1,2†
Renjie Wu1,2†