
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 02 June 2023
Sec. Microbiological Chemistry and Geomicrobiology
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1152818
This article is part of the Research Topic Insights in Microbiological Chemistry and Geomicrobiology : 2022/2023 View all 11 articles
Diversity patterns and community assembly of soil microorganisms are essential for understanding soil biodiversity and ecosystem processes. Investigating the impacts of environmental factors on microbial community assembly is crucial for comprehending the functions of microbial biodiversity and ecosystem processes. However, these issues remain insufficiently investigated in related studies despite their fundamental significance. The present study aimed to assess the diversity and assembly of soil bacterial and fungal communities to altitude and soil depth variations in mountain ecosystems by using 16S and ITS rRNA gene sequence analyses. In addition, the major roles of environmental factors in determining soil microbial communities and assembly processes were further investigated. The results showed a U-shaped pattern of the soil bacterial diversity at 0–10 cm soil depth along altitudes, reaching a minimum value at 1800 m, while the fungal diversity exhibited a monotonically decreasing trend with increasing altitude. At 10–20 cm soil depth, the soil bacterial diversity showed no apparent changes along altitudinal gradients, while the fungal Chao1 and phylogenetic diversity (PD) indices exhibited hump-shaped patterns with increasing altitude, reaching a maximum value at 1200 m. Soil bacterial and fungal communities were distinctively distributed with altitude at the same depth of soil, and the spatial turnover rates in fungi was greater than in bacteria. Mantel tests suggested soil physiochemical and climate variables significantly correlated with the β diversity of microbial community at two soil depths, suggesting both soil and climate heterogeneity contributed to the variation of bacterial and fungal community. Correspondingly, a novel phylogenetic null model analysis demonstrated that the community assembly of soil bacterial and fungal communities were dominated by deterministic and stochastic processes, respectively. The assembly processes of bacterial community were significantly related to the soil DOC and C:N ratio, while the fungal community assembly processes were significantly related to the soil C:N ratio. Our results provide a new perspective to assess the responses of soil microbial communities to variations with altitude and soil depth.
The spatial distribution patterns of biome structure and diversity, as well as their underlying mechanisms that regulate spatiotemporal changes are the core theme of biogeography (Green and Bohannan, 2006; Chu et al., 2020). Numerous biogeographical studies have focused on the responses of ecosystems to large-scale environmental changes (Fukami and Wardle, 2005; Walker et al., 2010). Altitude gradients provided a verification platform for models to accurately predict biodiversity responses to global change (McCain and Colwell, 2011). Soil microorganisms play an essential role in biogeochemical cycles within terrestrial ecosystems and are sensitive to environmental changes. Therefore, it is crucial to reveal the fundamental mechanisms underlying the microbial community diversity and distribution patterns to assess their relationships with community stability and ecosystem functions, which are the main central goal in community ecology (Hanson et al., 2012; Meyer et al., 2018). In this context, spatial variations in the soil microbial community at large spatial scales have been continually investigated in different ecosystems, including farmland (Constancias et al., 2015), woodland (Singh et al., 2014; Schnecker et al., 2015), grassland (Baker et al., 2009), tundra (Bjork et al., 2008; Kim et al., 2014), and deserts (Andrew et al., 2012). Studies have highlighted the inconsistent conclusions on the altitudinal distribution patterns of microbial communities, such as monotonous (Bryant et al., 2008; Wang et al., 2011), unimodal (Singh et al., 2012), concave (Singh et al., 2014), and no altitude pattern (Shen et al., 2013). Although the influence of spatial distance on microbial distribution was confirmed (Whitaker et al., 2003; Papke and Ward, 2004), few studies have investigated the impacts of spatial distance on microbial evolution in mountain ecosystems considering the strong diffusion ability of soil microorganisms.
Natural selection, dispersal, drift, species formation, and extinction rates showed the strongest effects on microbial altitude diversity patterns (Kassen and Rainey, 2004; Hanson et al., 2012; Nemergut et al., 2013). Previous studies have showed that microbial communities are influenced by both biotic factors (e.g., terrestrial vegetation characteristics) and abiotic factors (e.g., climatic factors and soil physicochemical properties) (Bezemer et al., 2010; Orwin et al., 2010; Thoms et al., 2010; de Vries et al., 2012). Previous studies have revealed clearly spatial distribution patterns of soil microbial communities and diversity (Green et al., 2004; Fierer and Jackson, 2006; Fierer et al., 2009; Lauber et al., 2009; Nemergut et al., 2011). Soil pH, C:N ratio, and vegetation types were the key factors controlling the altitudinal distribution pattern of soil microbial diversity (Rousk et al., 2010; Shen et al., 2016; Bahram et al., 2018).
The Changbai Mountain area is one of the most biodiversity-rich temperate regions, making it a suitable area for studying biodiversity patterns (Yang and Xu, 2003; Shen et al., 2014; Zou et al., 2017). The main objective of this study is to investigate variations in soil microbial (bacterial and fungal) community structures and their assemblies at four altitudes (800, 1200, 1800, and 2300 m) and two soil depths (0–10 cm and 10–20 cm), as well as their responses to environmental factors on Changbai Mountain. The distance-decay relationships (DDRs) were used to explore changes in spatial turnover rates, while a null model was applied to measure microbial assemblies in different soil samples. The following hypotheses were examined in this study: (i) Altitudinal gradients and soil depths have a substantial impact on the diversities and communities of soil microorganism, as well as the spatial turnover rates of their communities. (ii) Altitudinal gradients and soil depths can distinctly influence the mechanisms of microbial community assembly, which are shaped by distinct environmental factors.
This study was conducted in autumn 2021 on Changbai Mountain (126°55′-129°00′E; 41°23′- 42°36′N), Jilin, northeast China. The study area has a temperate continental monsoon climate with altitudinal variability, whereas the winter is long and cold, and the summer is short and cool. The annual temperature and precipitation range from 2.9 to 4.8°C and 632 to 1154 mm, respectively. The main vegetation types, which exhibit an altitudinal distribution from low 800 m to high 2300 m, consist of mixed coniferous broad-leaved forest (MCB), coniferous spruce forest (CS), Erman’s birch forest (EB), and alpine tundra (AT; Figure 1). The climate and main vegetation characteristics of each habitat are reported in Supplementary Table S1. Three plots (25 m × 25 m each) were established for each vegetation type. To minimize the heterogeneity, we divided each plot into three subplots for soil sampling. The collected soil samples from the three subplots were mixed to obtain one composite soil sample from each plot. In total, 72 subplots were established in the field experiment (4 altitudes × 3 subplots × 3 replicates × 2 depths), from which 24 composite soil samples were collected (4 altitudes × 2 depths × 3 replicates). The collected composite soil samples were placed in plastic bags on ice and transported to the laboratory for further analysis. Soil samples were sieved through 2-mm sieve after the removal of visible debris, such as roots and litter and stored at 4°C for chemical analysis or at −80°C for DNA extraction.
Soil pH was measured using a PHS-3C pH meter (PHS-3C pH acidometer) after shaking the soil solution (soil: water = 1: 5) for 30 min. Soil TC and TN contents were determined using a Carlo Erba FLASHEA 1112 CHN-S analyzer according to the instruction manual. Soil total organic (TOC) contents were determined using the external heating-potassium dichromate titration method. Soil total phosphorus (TP) contents were determined using the nitric acid-perchloric acid digestion method combined with a continuous flow analytical system (San++ System, Skalar, Holland) according to the instruction manual. Soil dissolved organic carbon (DOC) contents were determined using a total organic carbon analyzer (TOC VCPH, Shimadzu) according to the instruction manual.
DNA was first extracted from 0.5 g soil samples using the FastDNA SPIN Kit for soil (MP Biomedicals, Santa Ana, CA, United States), then purified using the UltraClean Soil DNA Kit (MOBIO Laboratories, Carlsbad, CA, United States) following the manufacturer’s instructions. Bacterial 16S rRNA gen V3–V4 region was amplified using the 338F and 806R primers, while the ITS1 region of fungal ITS gene were amplified using the ITS1F and ITS2R primers (Adams et al., 2013). The PCR reactions were performed in triplicate. The PCR product was first extracted from 2% agarose gel, then purified using the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, United States) and quantified using an Quantus™ Fluorometer (Promega, United States). NEXTFLEX® Rapid DNA-Seq Kit was used to build the library. Purified amplicons were pooled in equimolar amounts and paired-end sequencing was performed on an Illumina MiSeq platform (Illumina, San Diego, United States) in the Majorbio Bio-Pharm Technology Co. Ltd. (Shanghai, China) according to its standard protocols.
Raw sequence data were processed and analyzed using the QIIME software package (Caporaso et al., 2010). The extraction of non-repeated sequences from the optimized sequence was convenient for reducing the redundant calculation in the intermediate process of analysis. Operational taxonomic unit (OTU) clustering was performed according to 97% similarity on non-repeated sequences (excluding single sequence). Chimeras were removed prior to the clustering process to obtain a representative OTU sequence. In order to classify species according to OTU sequences, the RDP classifier Bayesian algorithm was used to classify the representative OTU sequences at a 97% sequence similarity level.
The one-way analysis of variance (ANOVA), with the least significant difference (LSD) method, was performed using SPSS 19.0 to determine whether the differences between treatments were significant at the p < 0.05 level. The analysis of similarity (ANOSIM) was performed based on the Bray-Curtis distance using the vegan package in R (Version 3.3.1). The Mantel test was performed based on Bray-Curtis in QIIME software (Version 1.9.1). QIIME was used to calculate the β diversity distance matrix, while the vegan package was used for Nonmetric Multidimensional Scaling (NMDS). The Shannon and Chao1 indices were calculated in R to estimate α-diversity of the soil bacterial and fungal communities. The Phylogenetic diversity (PD) indices were analyzed on the online tool of Majorbio Cloud Platform.1 Distance-based redundancy analysis (db-RDA) was performed using the vegan package in R. The distance-decay relationships (DDRs) were investigated based on taxonomic dissimilarities (Sorensen distance) (Oksanen et al., 2020).
The slope of the ordinary least-squares regression between the log-transformed geographic distance and microbial community similarity was used to quantify the distance-decay rate of the microbial community using the vegan R package (Dixon, 2003). One-way ANOVA was performed to examine the significance of the slopes of the two curves using the lsmeans R package (Lenth, 2016). Null modeling-based approaches were performed to infer community assembly mechanisms (Chase and Myers, 2011; Stegen et al., 2013; Chase et al., 2019). First, representative OTU sequences were introduced into MEGA software (Version 10.2.4) to build a phylogenetic tree using the maximum likelihood method, then the ape R package was loaded to read the phylogenetic tree (.tre) and OTU-table (.csv) files. The mean nearest phylogenetic taxon distance (MNTD) and nearest taxon index (NTI) indices were calculated using the Picante package (Kembel et al., 2010). An average NTI value greater than 0 indicates sample aggregation and the next step calculation can be performed, while an average NTI value lower than 0 indicates sample dispersion requiring further re-grouping. In addition, the βMNTD and βNTI indices of different samples were calculated using the same calculation method. The last operation is the RCbray index (Stegen et al., 2013). The Picante package was used to further quantify the deterministic (heterogeneous and homogenous selections) and randomness processes (diffusion limits and co-diffusion). The null model, based on the βNTI and RCbray indices, was applied to quantify the contributions of the deterministic and randomness processes to soil microbial community assembly. βNTI values that were lower than −2 or higher than 2 indicated that the communities were driven by homogeneous or heterogenous processes. In addition, a |βNTI| value lower than 2 suggested a stochastic assembly process of microbial community, and the diffusion limit (RCbray >0.95), isotropic diffusion (RCbray < −0.95), and uncertain process (−0.95 < RCbray <0.95) were determined according to the size of RCbray index in the assembly processes of microbial communities (Stegen et al., 2015).
The minimal bacterial Shannon, Chao1, and PD diversity indices at 0–10 cm depth were observed at 1800 m, showing a U-shaped altitudinal pattern, while the fungal α-diversity indices decreased with altitude (Figure 2 and Supplementary Figure S1). The bacterial diversity showed no obvious altitude pattern at 10–20 cm depth, while the fungal Chao1 and PD indices showed hump patterns with increasing altitude (Figure 2 and Supplementary Figure S2). The two most abundant bacterial phyla were Acidobacteria and Proteobacteria, accounting for 48.41–64.58% of the pyrosequences, followed in abundance by Actinobacteriota (12.31–27.48%), Chloroflexi (2.34–14.33%), Verrucomicrobiota (2.50–4.78%; Figure 3). The two most abundant fungal phyla were Basidiomycota and Ascomycota, accounting for 69.82–99.37% of the pyrosequences, followed by Mortierellomycota (0.52–21.14%), unclassified_k__Fungi (0.1–8.96%), and Rozellomycota (0.01–4.52%; Figure 3). The relative abundance of Proteobacteria at 0–10 cm depth was significantly higher than that at 10–20 cm soil depth at all altitudes (F = 13.77, p < 0.05, 800 m; F = 18.77, p < 0.05, 1800 m; F = 15.35, p < 0.05, 2300 m), except for 1200 m (Supplementary Figure S3). At 1800 m, the relative abundance of Basidiomycota at 0–10 cm soil depth was significantly lower (F = 9.17, p < 0.05) than that at 10–20 cm soil depth (Supplementary Figure S3). The NMDS plots showed different soil microbial compositions in the collected soil samples with respect to different altitudes (Figure 4). In addition, both soil depths showed significant variability along the altitude gradient (Figure 4 and Table 1).
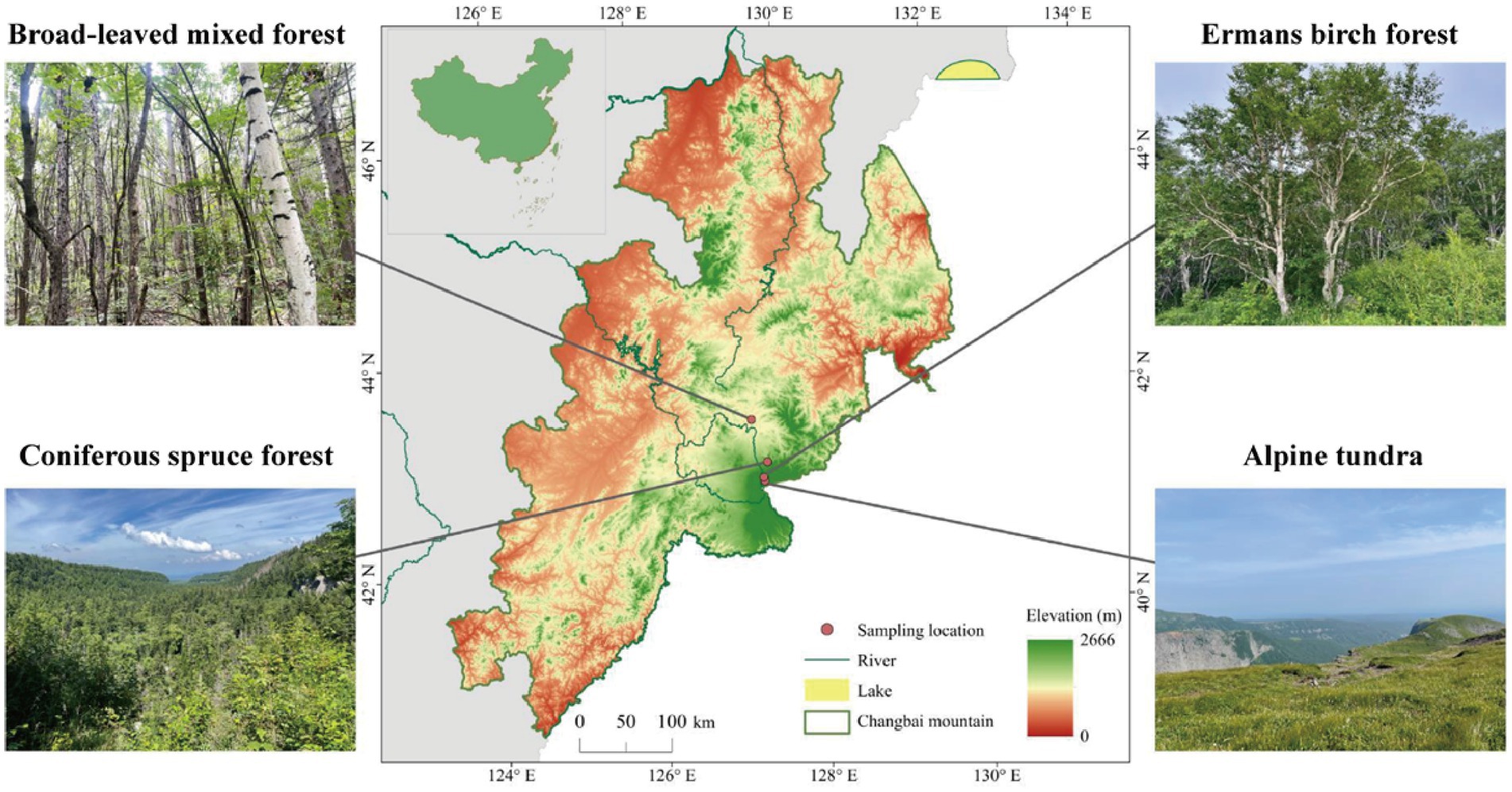
Figure 1. Vegetation types at the study sites on the northern slope of Changbai Mountain, northeast China.
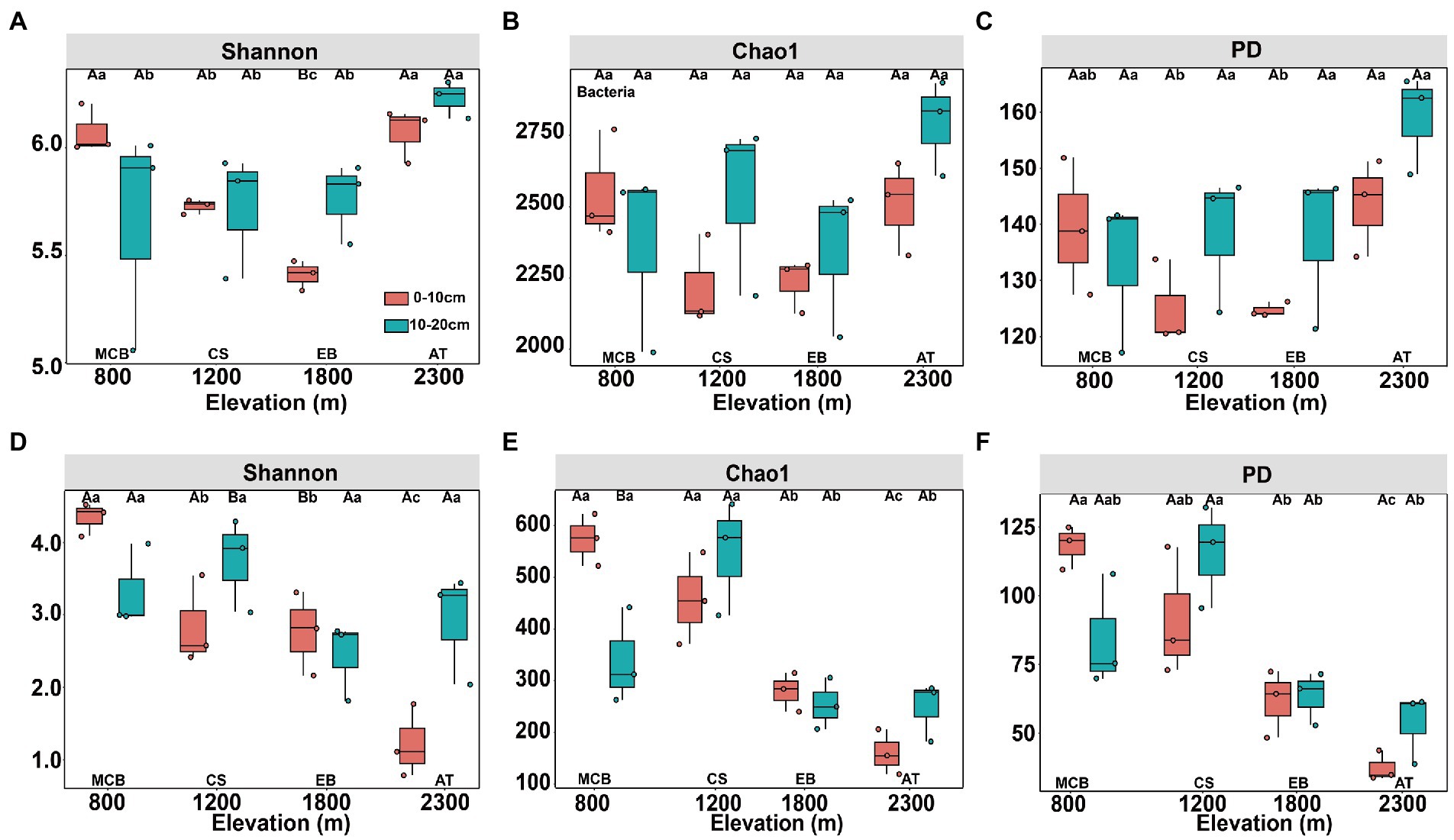
Figure 2. Changes in diversity of soil bacterial (A–C) and fungal (D–F) communities along altitudinal gradient and soil depth on Changbai Mountain. Capital letters indicate significant differences (p < 0.05) between soil depth treatments, lower case letters indicate significant differences (p < 0.05) between altitude treatments.
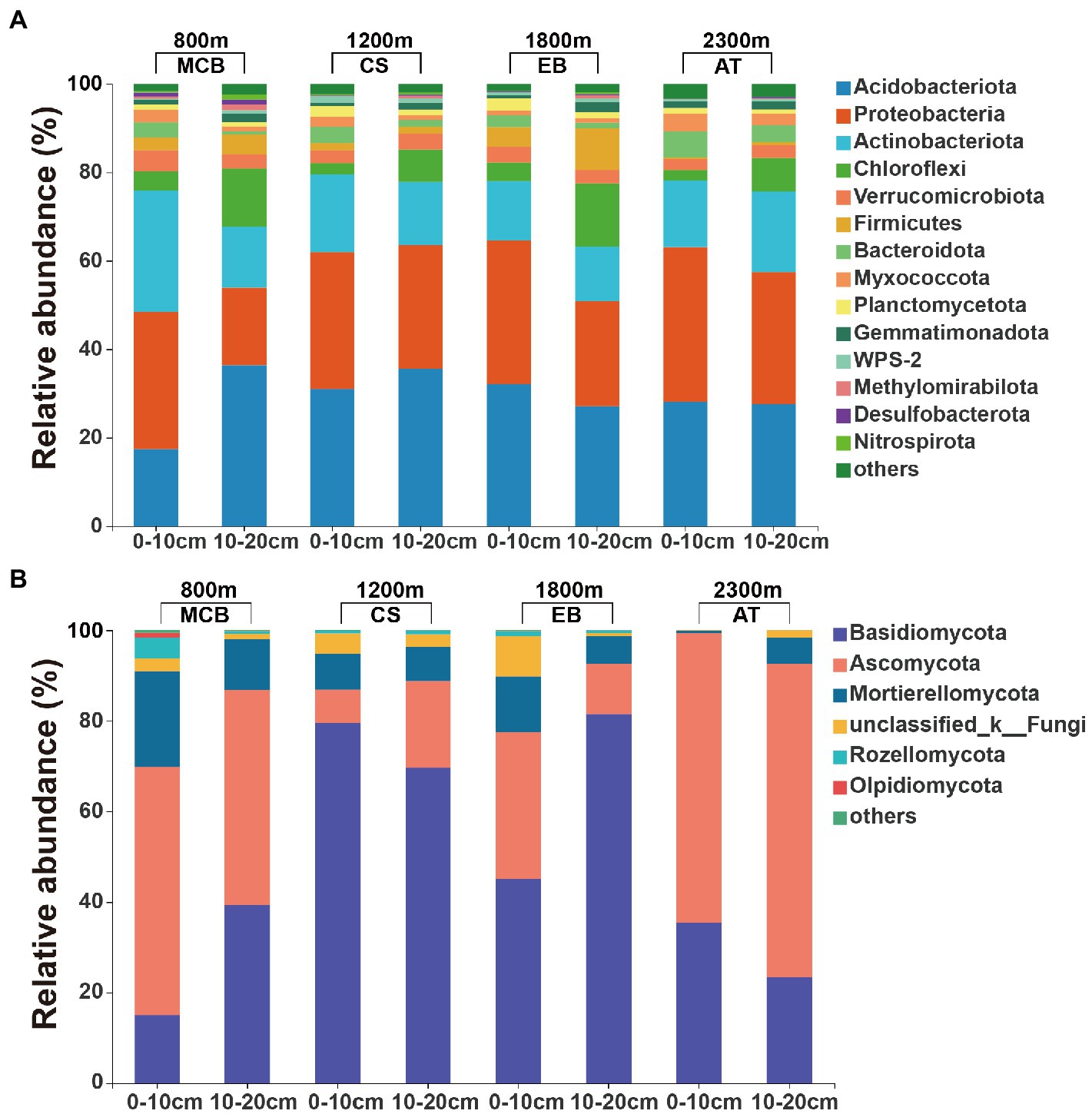
Figure 3. Relative abundances of the major soil (A) bacterial and (B) fungal communities at the phylum level.
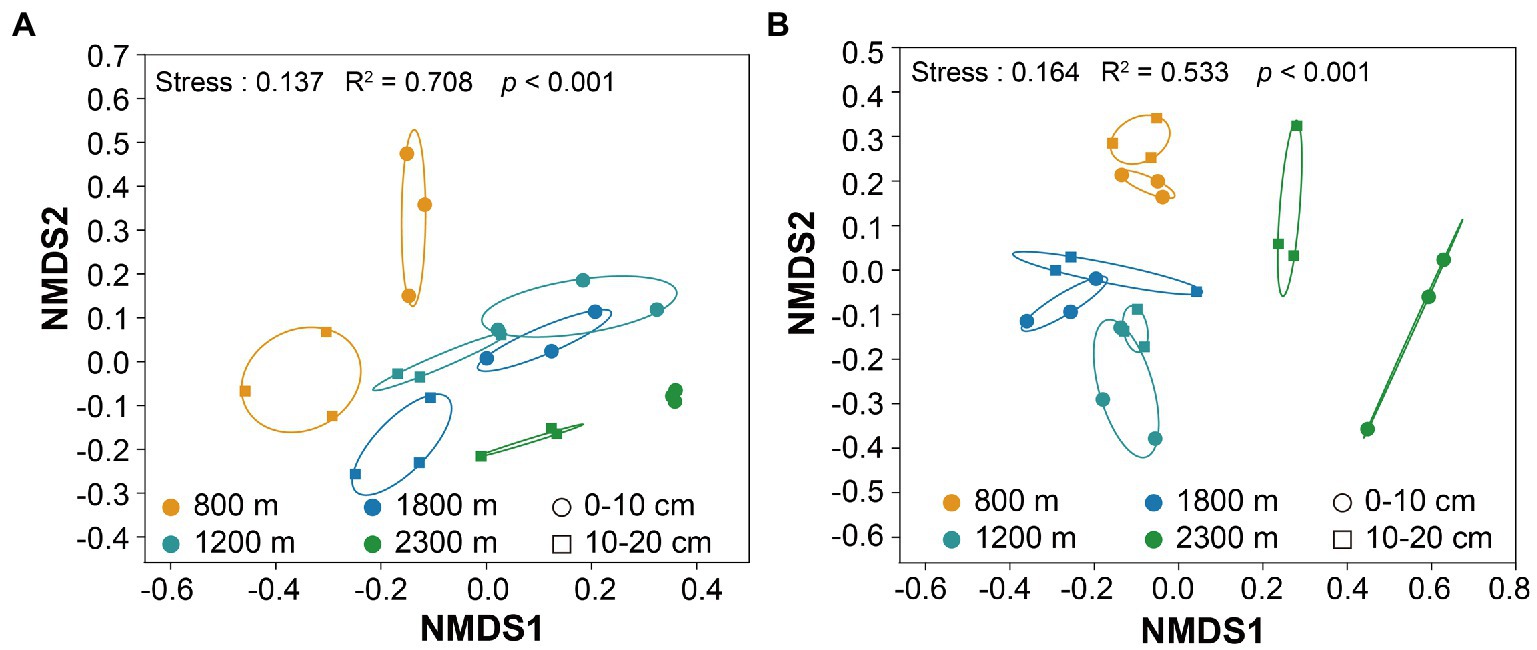
Figure 4. Non-metric multidimensional scaling (NMDS) of the bacterial (A) and fungal (B) communities along altitudes and soil depths.
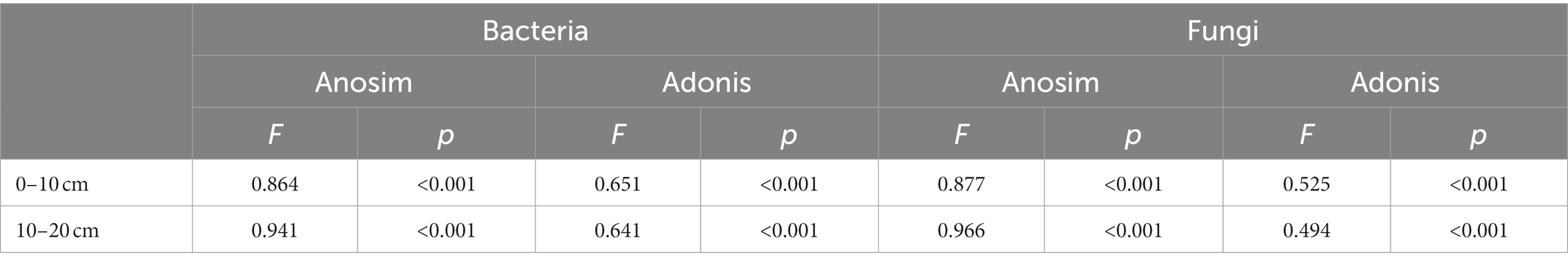
Table 1. Anosim and Adonis analysis of altitude and soil depth in shaping the bacterial and fungal communities structure.
The Mental test showed significant correlations between community composition and environmental variables (Table 2). The C:N ratio showed significant positive correlations with the soil bacterial community compositions at 0–10 cm (r = 0.757, p < 0.001) and 10–20 cm (r = 0.387, p < 0.05) soil depths (Table 2). Soil TP contents exhibited a significant positive correlation with the soil fungal community composition at 0–10 cm soil depth (r = 0.451, p < 0.001), while TOC was the most relevant soil variables to the soil fungal community at 10–20 cm soil depth (r = 0.341, p < 0.01; Table 2). Distance-based redundancy analysis (db-RDA) demonstrated the stronger effects of the soil pH and C:N ratio on the bacterial and fungal communities, respectively (Figure 5). Among the abiotic factors, C:N ratio and soil pH were the two major soil variables contributing to the bacterial α-diversity variation (Figure 6A), while C:N ratio and altitude were the two major soil variables contributing to the fungal α-diversity variations (Figure 6B).
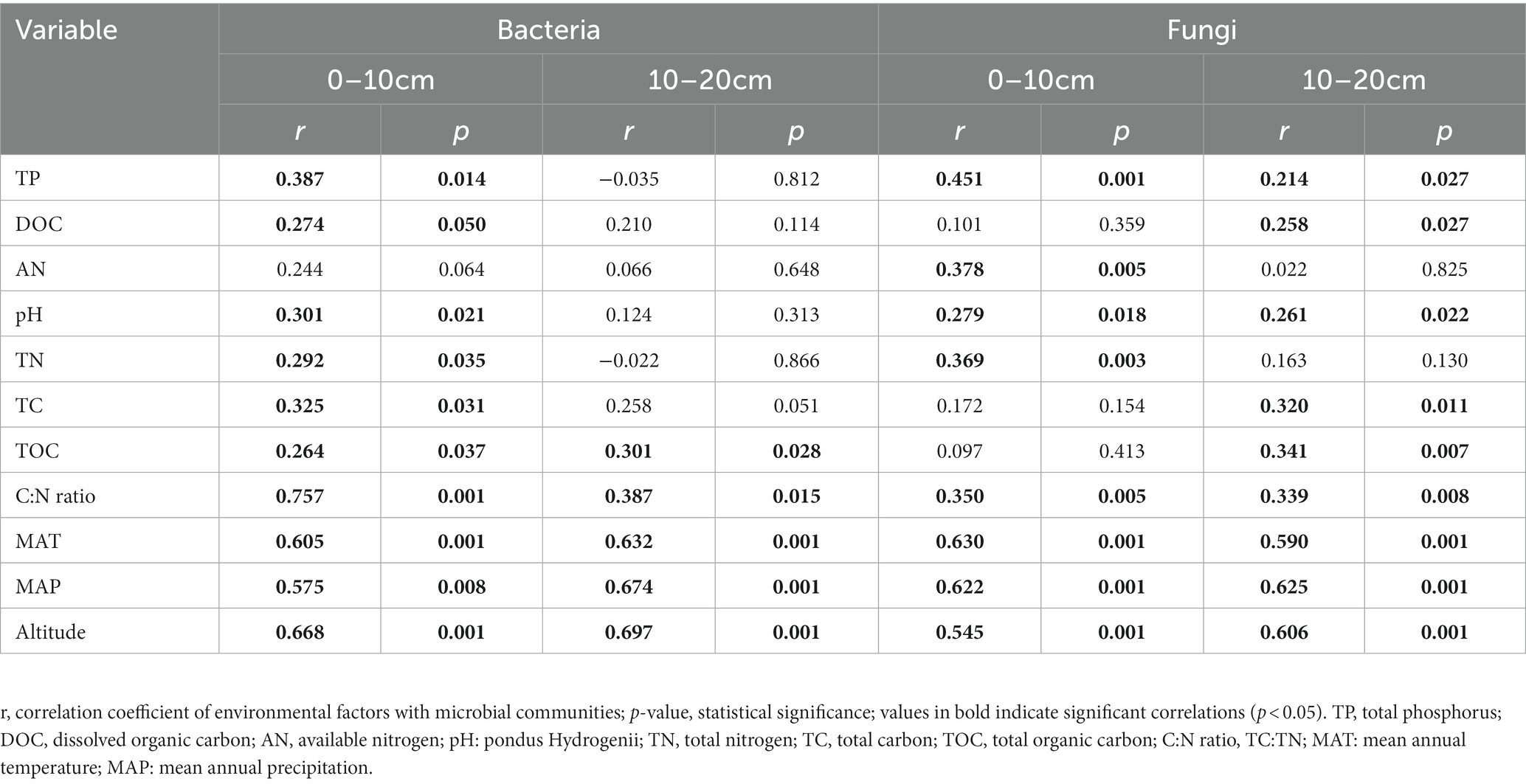
Table 2. Results of Mantel test on the correlation between community composition (Bray-Curtis) and environmental variables for bacteria and fungi.
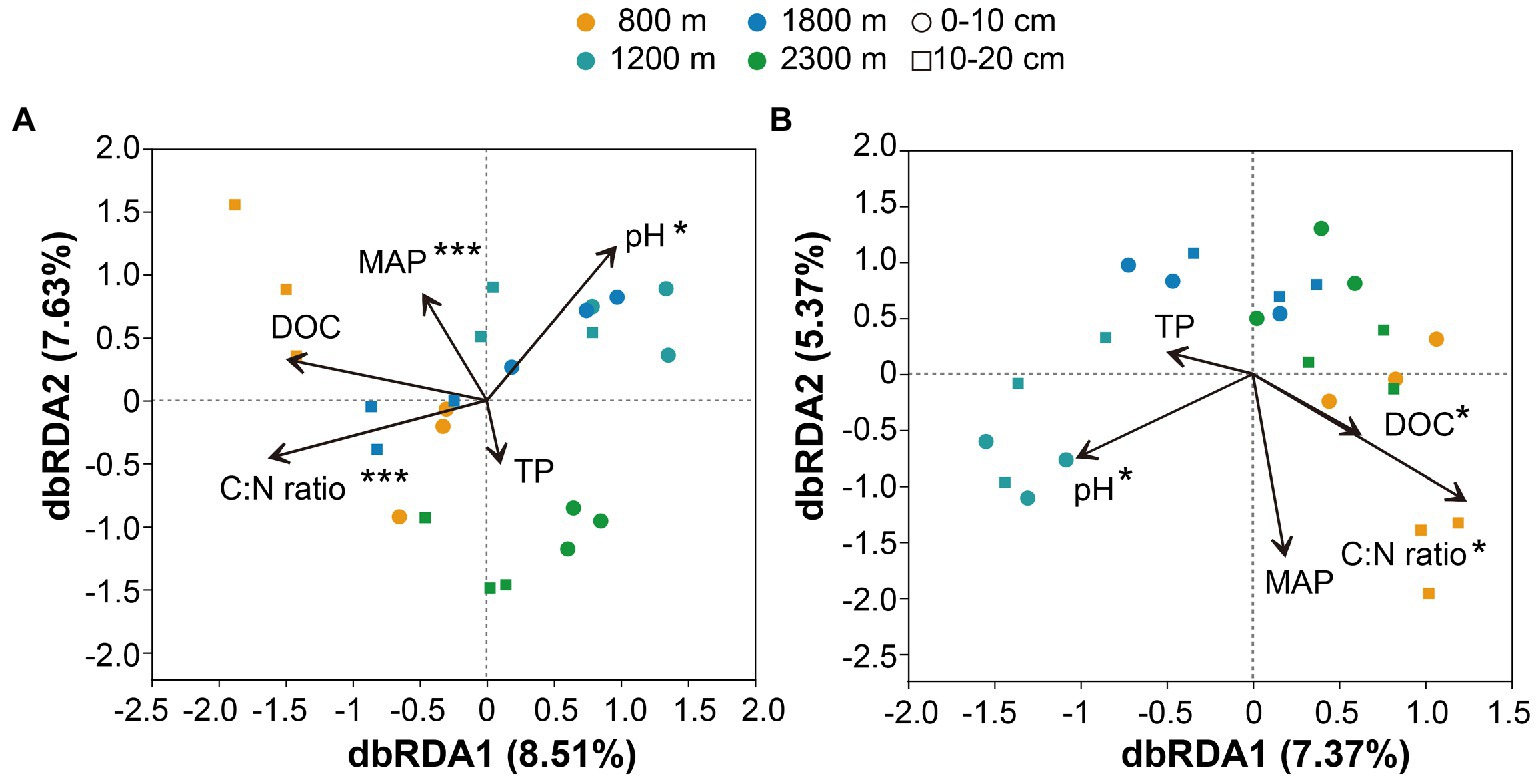
Figure 5. Distance-based redundancy analysis of soil bacterial (A) and fungal (B) communities along altitudinal gradient and soil depth as related to environmental variables. pH: pondus Hydrogenii; DOC, dissolved organic carbon; TP, total phosphorus; C:N ratio, TC/TN; MAP, mean annual precipitation. *p<0.05; **p<0.01; ***p<0.001.
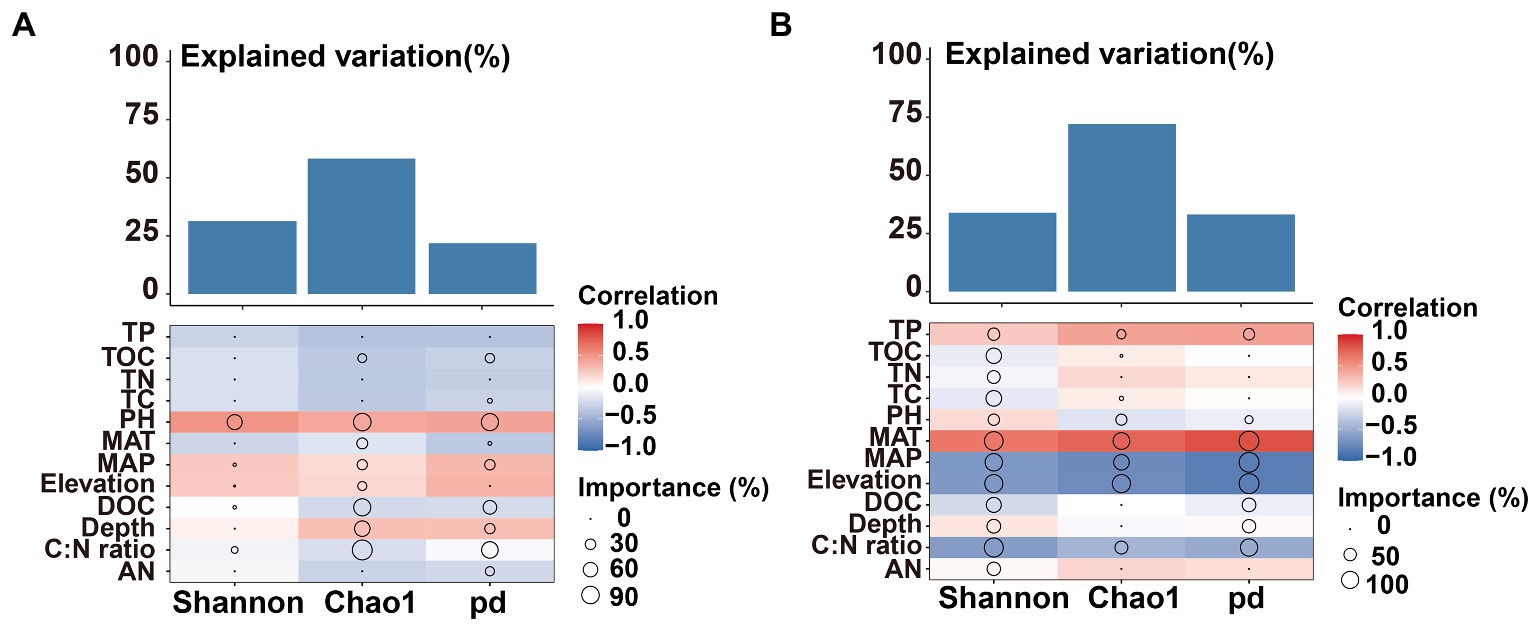
Figure 6. Contributions of environmental factors to bacterial bacterial (A) and fungal (B) α-diversity based on correlation and random forest model. Circle size represents the variable’s importance (i.e., percentage of increase of mean square error calculated via random forest model). Colors represent spearman’s correlations.
Community dissimilarity and geographic distance for each pairwise sample set showed a significant distance-decay relationship (DDR) of the microbial communities (Figures 7A,B). At the same soil depth, the spatial turnover rate of fungal community (β = 0.134–0.138) was higher than that of bacterial community (β = 0.056–0.065; Figures 7A,B). However, there was no significant difference in the turnover rates of soil bacterial and fungal communities between different soil depths (Figure 7C). Bacterial community assembly was determined by deterministic process (53.47%), while assembly processes of soil fungal communities was mainly determined by stochastic process (90.62%; Figure 8A). At 0–10 cm depth, the effect of the deterministic process (heterogeneous selection) on soil bacterial community assembly was dominated at the lowest altitude (800 m), while that of the stochastic process (undominated and dispersal limitation) exhibited a gradual dominance at the remaining altitudes (Figure 8B). The soil bacterial community assembly was mostly driven by the stochastic process at 10–20 cm soil depth (Figure 8B). In contrast, the fungal community assembly was dominated by the stochastic process among all selected altitudes and soil depths (Figure 8C).
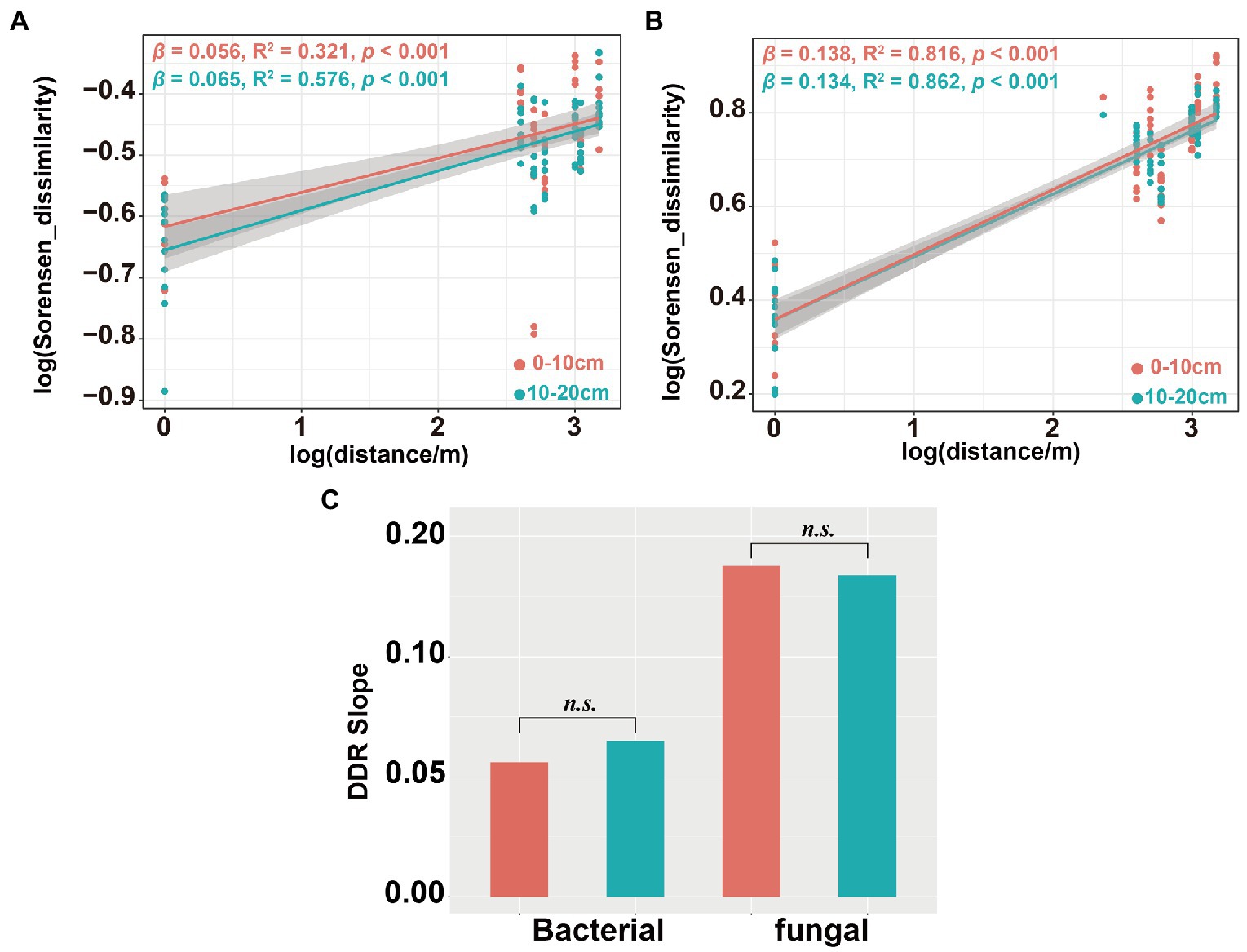
Figure 7. Distance-decay relationship (DDR) for the soil bacterial and fungal communities. DDR based on Sorensen dissimilarity along geographic distances for the bacterial community (A); DDR for the fungal community (B) and DDR slope significance test of soil bacterial and fungal communities (C). The slope β of DDR represent the spatial turnover rate of species.
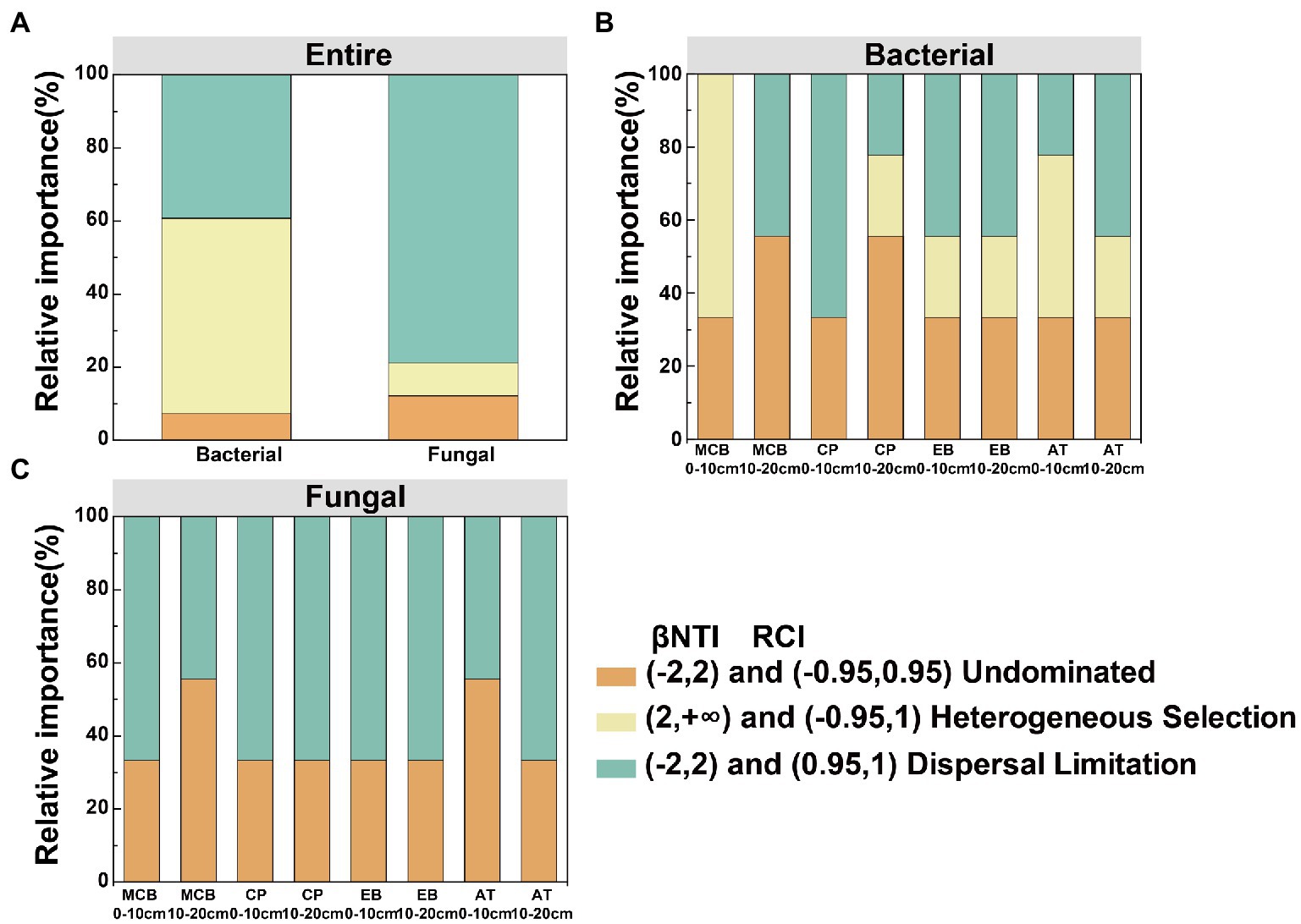
Figure 8. Deterministic and stochastic processes in the assembly of soil bacterial and fungal communities in the Changbai Mountain. These two processes were both detected using the β-Nearest Taxon Index (βNTI) and Raup-Crick Index (RCI). (A) Relative influences of deterministic and stochastic assembly processes in shaping a bacterial and fungal community. Relative influences of deterministic and stochastic assembly processes in shaping bacterial community (B) and fungal community (C) for different treatments.
The main explanatory soil variables were selected to determine the correlations between the assembly processes of microbial communities and environmental variables (based on Euclidean distance matrices; Table 3). For bacterial community, the significant positive correlations of βNTI with DOC (r2 = 0.206, p < 0.001) and C:N ratio (r2 = 0.141, p < 0.001; Figure 9). Whereas for fungal community, a significant positive correlation was observed between the soil C:N ratio and βNTI (r2 = 0.141, p < 0.001; Figure 9).
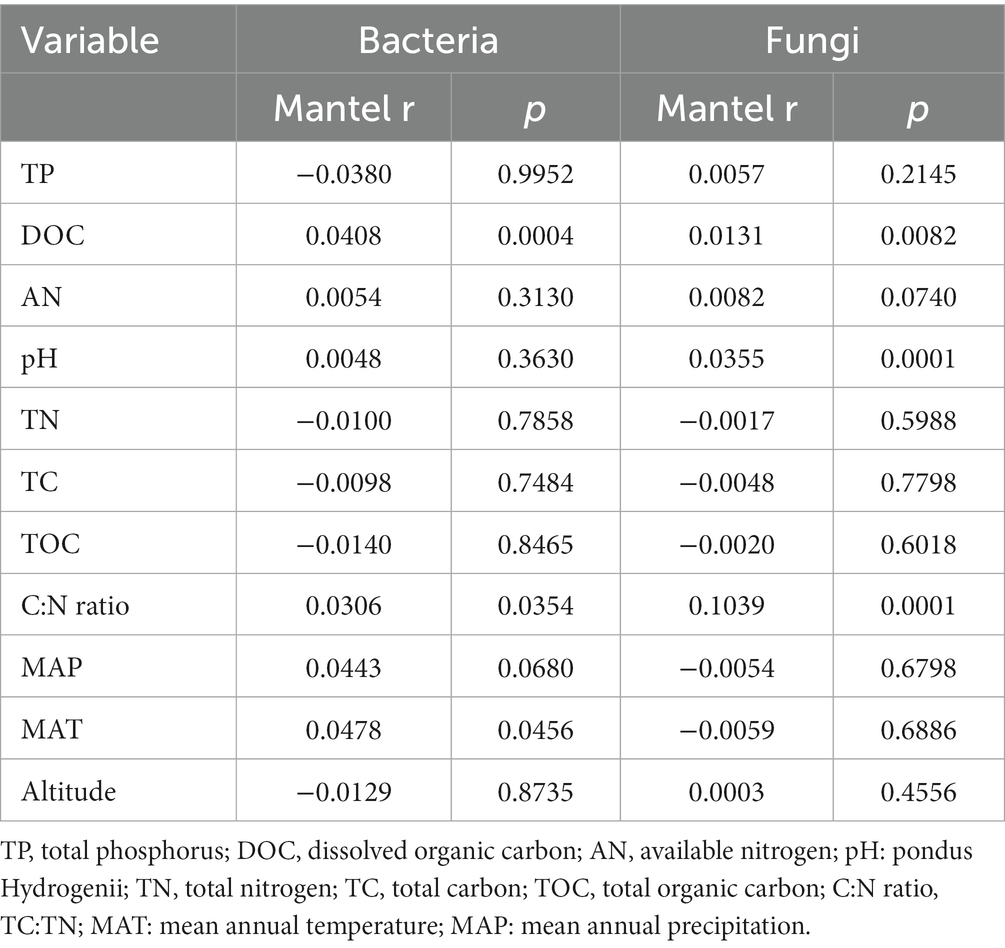
Table 3. Mantel tests of environmental variables against the phylogenetic turnover (β-nearest taxon index) of the soil bacterial and fungal communities.
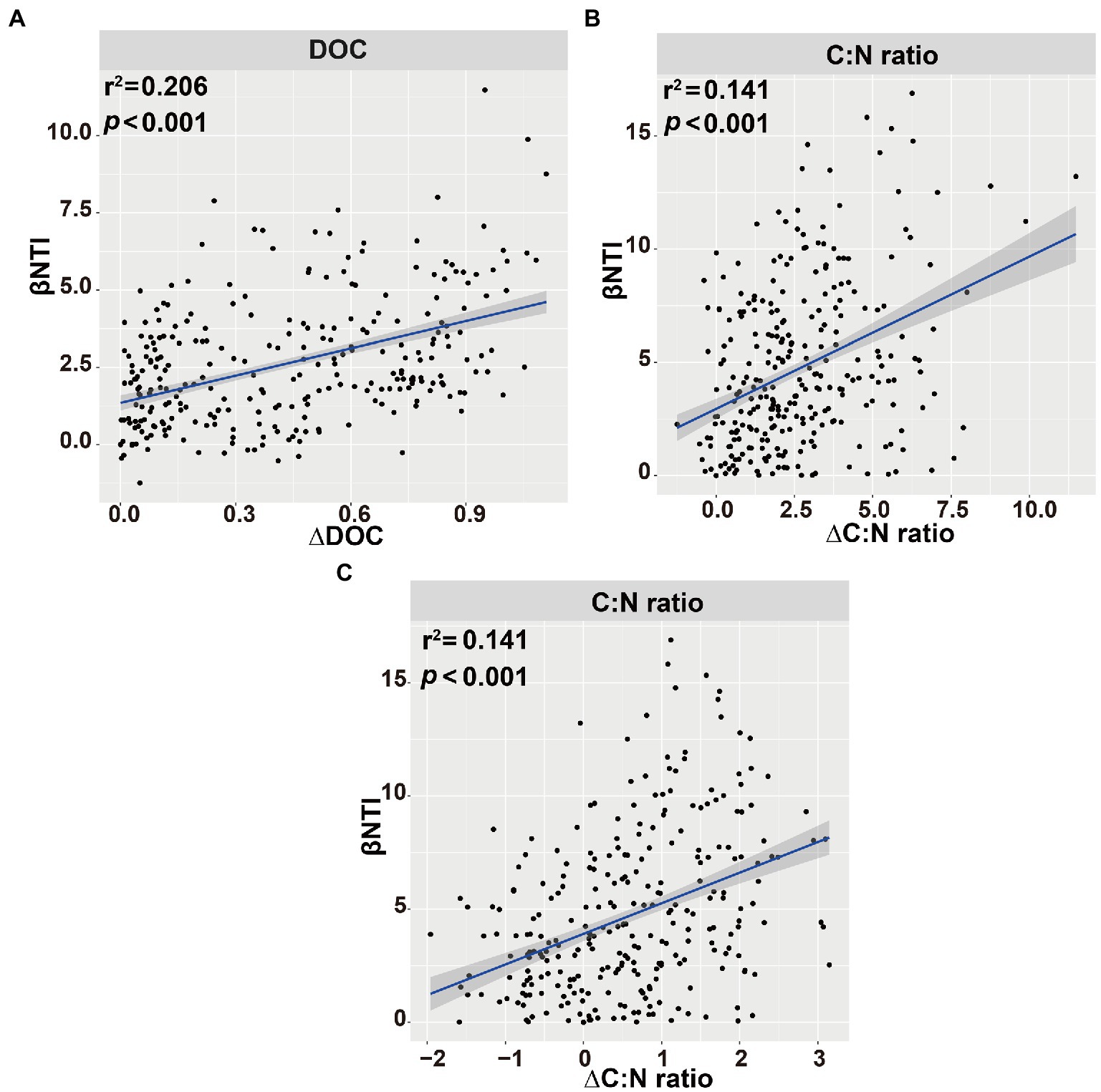
Figure 9. Linear regression analysis of the connections between β-nearest taxon index (βNTI) of soil bacterial (A,B) or fungal (C) communities and soil variables. Blue shaded area indicates 95% confidence interval (Spearman’s p < 0.05).
The multiple regression model (MRM) was used to further assess the relative contributions of each environmental factor to the soil bacterial and fungal communities (Table 4). According to the obtained results, the soil C:N ratio (R2 = 0.419, p < 0.001) and altitude (R2 = 0.534, p < 0.001) significantly explained the variations of soil bacterial community dissimilarities at the upper and deeper soil depths, respectively (Table 4). Whereas MAP significantly explained the variations in the soil fungal community dissimilarities at both soil depths (Table 4).
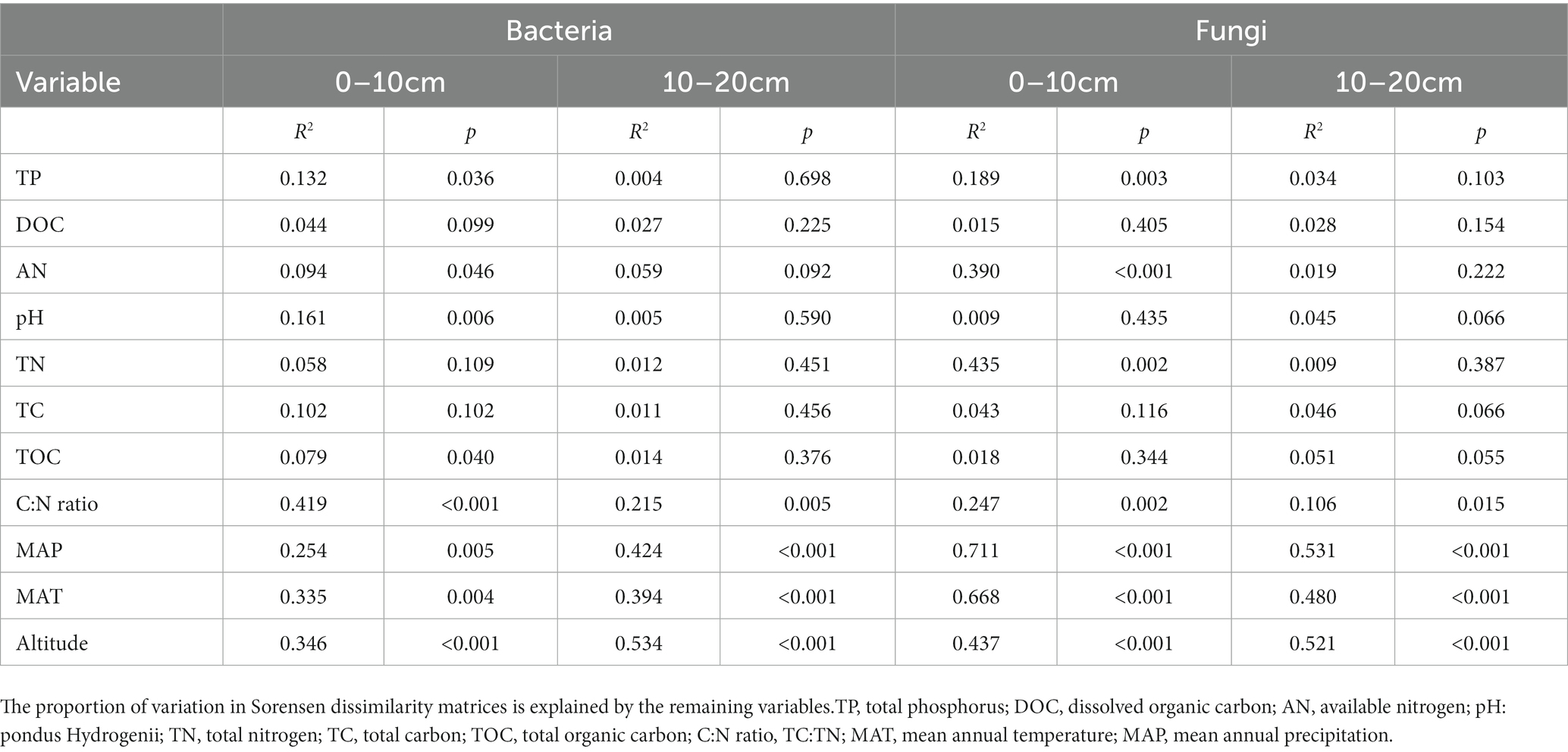
Table 4. Results of the multiple regression analysis on matrices analysis (MRM) for the microbial community composition.
The altitudinal diversity pattern is one of the most fundamental patterns in animal and plant biogeography (Denef et al., 2009). Here we observed that soil bacterial diversity appeared a U-shaped pattern with altitude at 0–10 cm soil depth, which is in line with the previous results (Singh et al., 2014). The α-diversity of soil bacterial communities did not show an obvious distribution pattern with altitudes at 10–20 cm soil depth, which was consistent with the results revealed in previous studies (Fierer et al., 2011). The peak pattern (Miyamoto et al., 2014; Wang et al., 2015; Ogwu et al., 2019) or U-shaped pattern (Liu et al., 2018) of soil fungal community were observed in previous studies, showing the highest and lowest diversity observed at mid-altitude. The α-diversity of fungal community at 0–10 cm soil depth exhibited a monotonically decreasing trend with altitude (Figure 2 and Supplementary Figure S2), while the Chao1 and PD indices of the fungal communities at 10–20 cm soil depth showed a hump-shaped pattern (Figure 2 and Supplementary Figure S3). In addition, the significant differences of ectomycorrhizal fungi distribution between soil depths might resulted in different substrate utilization patterns (Dickie et al., 2002).
Both bacterial and fungal communities were significantly different among the selected altitudes and soil depth (Figure 4 and Table 1). We found bacterial and fungal communities have significant correlation with altitude (Table 2). Previous research found that bacterial community composition varied with elevation (Fierer et al., 2011). The fungal community composition had significant differences along the altitude gradient (Kivlin et al., 2014), which might be due to the different selection of soil habitats by different fungal groups, with obvious ecological niche differentiation (Cook and Quinn, 1995). It had been reported that the spatial distribution pattern of fungal community was closely related to environmental heterogeneity (pH, soil nutrition and altitude) (Cox et al., 2010; Polme et al., 2013). Acidobacteria and Proteobacteria were the major soil microorganisms on Changbai Mountain (Figure 3), which was consistent with the findings reported in previous studies (Shen et al., 2013). This might be due to the crucial role of Acidobacteria in organic matter decomposition and nutrient cycling (Eichorst et al., 2018). Proteobacteria have high morphological and metabolic diversity, which enables them to easily utilize soil organic carbon and nitrogen (Bryant and Frigaard, 2006; Mukhopadhya et al., 2012). Basidiomycetes and Ascomycetes were the dominant soil fungal groups due to the presence of a thicker litter layer in the upper soil layer than in the deeper soil layer. Indeed, Basidiomycetes can decompose complex lignocellulosic components (Lundell et al., 2010). Ascomycetes can use more resources to better tolerate environmental pressure (Yang et al., 2017; Egidi et al., 2019).
Soil nitrogen (N) and phosphorus (P) are important nutrients for bacterial growth and activity, influencing the spatial distribution of soil microorganisms (Wang et al., 2018; Zhang et al., 2019). Soil fungal diversity in forests was related to plant diversity, temperature, and altitude (Geml et al., 2014; Wang et al., 2015; Wan and He, 2020). Previous studies have pointed out that soil pH was a main factor affected the distribution pattern of soil microbial communities along altitudinal gradients (Baath and Anderson, 2003; Lauber et al., 2009; Delgado-Baquerizo et al., 2017). Climatic factors (MAT and MAP) were dominant variables affected the diversity and community composition of bacteria (Singh et al., 2014; Xu et al., 2014; Nottingham et al., 2018; Frindte et al., 2019). The results of this study showed significant positive correlations of TP, DOC, pH, TN, TC, TOC, C:N ratio, MAT, MAP and altitudes with the bacterial community composition at upper soil depth, while TOC, C:N ratio, MAT, MAP and altitudes exhibited significant positive correlations with the bacterial community at deeper soil depth (Table 2). It is noteworthy that the soil fungal community composition exhibited significant positive correlations with soil TP and TOC contents at the upper and deeper soil depths, respectively (Table 2). Soil pH leads to niche differentiation of soil microbial and controls their patterns along altitude gradients (Shen et al., 2013; Bahram et al., 2018). Whereas soil C:N ratio and phosphorus contents could significantly affect the soil fungal diversity in forests and farmland (Lauber et al., 2013; Shen et al., 2017; Li et al., 2018; Delgado-Baquerizo and Eldridge, 2019). The importance of MAT had been widely reported at both horizontal and vertical scales (Yang et al., 2014; Delgado-Baquerizo et al., 2016; Zhou et al., 2016). Among the abiotic factors considered in this study, soil pH and C:N ratio were the major factors contributing to the variation in the soil bacterial α-diversity (Figure 6A), which is consistent with the results observed in Southwest Tibet (Peay et al., 2017; Shen et al., 2019), indicating that soil pH was the main factor controlling soil microbial diversity. Besides soil pH, soil C:N ratio and altitude showed correlations with microbial diversity (Singh et al., 2014; Shen et al., 2017; Li et al., 2018; Delgado-Baquerizo and Eldridge, 2019). This study demonstrated the contributions of the soil C:N ratio and altitudes to the variations in the fungal α-diversity (Figure 6B).
Several studies have shown that soil microbial community have obvious spatial scale patterns in DDRs (Soininen et al., 2007; Morlon et al., 2008; Deng et al., 2016). We observed steeper turnover slopes in the fungal community (β = 0.134–0.138) than that in the bacterial community (β = 0.056–0.065). This indicates greater dissimilarity of the soil fungal community over geographic distance than that of the soil bacterial community (Figures 7A,B). In addition, we also observed significant DDR for both bacterial (0–10 cm: R2 = 0.321, p < 0.001;10–20 cm: R2 = 0.576, p < 0.001) and fungal (0–10 cm: R2 = 0.816, p < 0.001;10–20 cm: R2 = 0.862, p < 0.001) communities, but soil depth had no significant effects on spatial turnover rate of either bacterial or fungal communities (Figure 7C). These spatial turnover patterns might be due to vegetation characteristics, geographic distance, and environmental conditions (Noss, 1990; Whittaker et al., 2001). Several studies have highlighted two primary factors that alter the spatial turnover rates of microorganisms (Deng et al., 2016). The first factor was niche selection which affects soil microorganism species through environmental heterogeneity, leading to variations in the community composition (Bell, 2010). The second factor was the restriction of spatial microbial dispersion (Cho and Tiedje, 2000; Peay et al., 2010), resulting in disparate soil microbial community (Zhou and Ning, 2017).
Quantifying the relative contributions of the microbial community to its construction provides an understanding of the spatiotemporal distribution patterns of the microbial community (Stegen et al., 2012). Our observation that deterministic assembly in the soil bacterial community was the dominant process (53.47%) and random assembly contributed substantially to the soil fungal community (90.62%; Figure 8), which was in line with the findings of previous studies (Zhang et al., 2018; Liu et al., 2020). This pointed out that soil bacterial community assembly was mainly driven by a deterministic process. Soil fungi require external forces to reproduce, such as wind, underground mycelial proliferation, plant transplantation, and animal activities (Liu et al., 2020).
It is essential to reveal the factors that lead to the dominance of different microbial community assembly processes. Previous studies have pointed out that assembly processes were related to soil chemical properties, such as pH, and NH4+-N (Jiang et al., 2019; Wan et al., 2021). In this study, the βNTI of bacteria was associated with the soil DOC contents and C:N ratio, while that of fungi was associated with the soil C:N ratio, suggesting that the assembly processes of soil bacterial and fungal communities were affected by soil parameters (Figure 9). Soil organic carbon and nitrogen contents could be readily absorbed by Proteobacteria (Bryant and Frigaard, 2006; Mukhopadhya et al., 2012). The relative abundance of Bacteroidetes was positively correlated with resource availability, which could preferentially consume soil active organic carbon (Fierer et al., 2007).
Our study revealed changes in the soil microbial community along the altitudinal gradient and soil depth on Changbai Mountain, as well as the main mechanisms controlling the soil microbial community assembly. The obtained results revealed significant differences in the diversity, community composition, and structure of soil microorganisms between altitudes and soil depths. The soil bacterial and fungal communities exhibited clear spatial patterns without showing differences in their spatial turnover rates between soil depths. The community assembly of soil bacteria and fungi were dominated by deterministic and stochastic processes, respectively. The assembly processes of soil bacterial and fungal communities differed along the altitude gradient and soil depth, which were due to the spatial variability in soil factors that control the aggregation of bacterial and fungal communities.
The data presented in the study are deposited in the National Center for Biotechnology Information (NCBI) repository, accession number PRJNA936073.
YK and HW designed the study, conducted the statistical analysis, and wrote the manuscript. YK, YZ, QW, QG, KL, and YL performed the field investigation and collected the data. All authors contributed to the article and approved the submitted version.
This work was supported by the National Natural Science Foundation of China (U20A2083), the National Key R&D Program of China (2022YFF1300900), and Jilin Scientific and Technological Development Program (20210509037RQ).
We are very grateful to Xiuxue Chen for making the sampling point map on this paper. We also thank Dandan Liu, Weiqiang Guo, and Zhiyu Zhang assistance in with field soil sampling.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1152818/full#supplementary-material
Adams, R. I., Miletto, M., Taylor, J. W., and Bruns, T. D. (2013). Dispersal in microbes: fungi in indoor air are dominated by outdoor air and show dispersal limitation at short distances. ISME J. 7, 1262–1273. doi: 10.1038/ismej.2013.28
Andrew, D. R., Fitak, R. R., Munguia-Vega, A., Racolta, A., Martinson, V. G., and Dontsova, K. (2012). Abiotic factors shape microbial diversity in Sonoran Desert soils. Appl. Environ. Microbiol. 78, 7527–7537. doi: 10.1128/AEM.01459-12
Baath, E., and Anderson, T. H. (2003). Comparison of soil fungal/bacterial ratios in a pH gradient using physiological and PLFA-based techniques. Soil Biol. Biochem. 35, 955–963. doi: 10.1016/S0038-0717(03)00154-8
Bahram, M., Hildebrand, F., Forslund, S. K., Anderson, J. L., Soudzilovskaia, N. A., Bodegom, P. M., et al. (2018). Structure and function of the global topsoil microbiome. Nature 560, 233–237. doi: 10.1038/s41586-018-0386-6
Baker, K. L., Langenheder, S., Nicol, G. W., Ricketts, D., Killham, K., Campbell, C. D., et al. (2009). Environmental and spatial characterisation of bacterial community composition in soil to inform sampling strategies. Soil Biol. Biochem. 41, 2292–2298. doi: 10.1016/j.soilbio.2009.08.010
Bell, T. (2010). Experimental tests of the bacterial distance-decay relationship. ISME J. 4, 1357–1365. doi: 10.1038/ismej.2010.77
Bezemer, T. M., Fountain, M. T., Barea, J. M., Christensen, S., Dekker, S. C., Duyts, H., et al. (2010). Divergent composition but similar function of soil food webs of individual plants: plant species and community effects. Ecology 91, 3027–3036. doi: 10.1890/09-2198.1
Bjork, R. G., Bjorkman, M. P., Andersson, M. X., and Klemedtsson, L. (2008). Temporal variation in soil microbial communities in Alpine tundra. Soil Biol. Biochem. 40, 266–268. doi: 10.1016/j.soilbio.2007.07.017
Bryant, D. A., and Frigaard, N. U. (2006). Prokaryotic photosynthesis and phototrophy illuminated. Trends Microbiol. 14, 488–496. doi: 10.1016/j.tim.2006.09.001
Bryant, J. A., Lamanna, C., Morlon, H., Kerkhoff, A. J., Enquist, B. J., and Green, J. L. (2008). Microbes on mountainsides: contrasting elevational patterns of bacterial and plant diversity. Proc. Natl. Acad. Sci. U. S. A. 105, 11505–11511. doi: 10.1073/pnas.0801920105
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Chase, J. M., Kraft, N., Smith, K. G., Vellend, M., and Inouye, B. D. (2019). Using null models to disentangle variation in community dissimilarity from variation in a-diversity. Ecosphere 2, 1–11. doi: 10.1890/es10-00117.1
Chase, J. M., and Myers, J. A. (2011). Disentangling the importance of ecological niches from stochastic processes across scales. Philos. Trans. Roy. Soc. B. Biol. Sci. 366, 2351–2363. doi: 10.1098/rstb.2011.0063
Cho, J. C., and Tiedje, J. M. (2000). Biogeography and degree of endemicity of fluorescent Pseudomonas strains in soil. Appl. Environ. Microbiol. 66, 5448–5456. doi: 10.1128/AEM.66.12.5448-5456.2000
Chu, H., Gao, G. F., Ma, Y., Fan, K., and Delgado-Baquerizo, M. (2020). Soil microbial biogeography in a changing world: recent advances and future perspectives. mSystems 5:e00803-19. doi: 10.1128/mSystems.00803-19
Constancias, F., Saby, N. P., Terrat, S., Dequiedt, S., Horrigue, W., Nowak, V., et al. (2015). Contrasting spatial patterns and ecological attributes of soil bacterial and archaeal taxa across a landscape. Microbiology 4, 518–531. doi: 10.1002/mbo3.256
Cook, R. R., and Quinn, J. F. (1995). The influence of colonization in nested species subsets. Oecologia 102, 413–424. doi: 10.1007/BF00341353
Cox, F., Barsoum, N., Lilleskov, E. A., and Bidartondo, M. I. (2010). Nitrogen availability is a primary determinant of conifer mycorrhizas across complex environmental gradients. Ecol. Lett. 13, 1103–1113. doi: 10.1111/j.1461-0248.2010.01494.x
de Vries, F. T., Manning, P., Tallowin, J. R. B., Mortimer, S. R., Pilgrim, E. S., Harrison, K. A., et al. (2012). Abiotic drivers and plant traits explain landscape-scale patterns in soil microbial communities. Ecol. Lett. 15, 1230–1239. doi: 10.1111/j.1461-0248.2012.01844.x
Delgado-Baquerizo, M., and Eldridge, D. J. (2019). Cross-biome drivers of soil bacterial alpha diversity on a worldwide scale. Ecosystems 22, 1220–1231. doi: 10.1007/s10021-018-0333-2
Delgado-Baquerizo, M., Maestre, F. T., Reich, P. B., Jeffries, T. C., Gaitan, J. J., Encinar, D., et al. (2016). Microbial diversity drives multifunctionality in terrestrial ecosystems. Nat. Commun. 7:10541. doi: 10.1038/ncomms10541
Delgado-Baquerizo, M., Reich, P. B., Khachane, A. N., Campbell, C. D., Thomas, N., Freitag, T. E., et al. (2017). It is elemental: soil nutrient stoichiometry drives bacterial diversity. Environ. Microbiol. 19, 1176–1188. doi: 10.1111/1462-2920.13642
Denef, K., Roobroeck, D., Wadu, M., Lootens, P., and Boeckx, P. (2009). Microbial community composition and rhizodeposit-carbon assimilation in differently managed temperate grassland soils. Soil Biol. Biochem. 41, 144–153. doi: 10.1016/j.soilbio.2008.10.008
Deng, Q., Cheng, X. L., Hui, D. F., Zhang, Q., Li, M., and Zhang, Q. F. (2016). Soil microbial community and its interaction with soil carbon and nitrogen dynamics following afforestation in Central China. Sci. Total Environ. 541, 230–237. doi: 10.1016/j.scitotenv.2015.09.080
Dickie, I. A., Xu, B., and Koide, R. T. (2002). Vertical niche differentiation of ectomycorrhizal hyphae in soil as shown by T-RFLP analysis. New Phytol. 156, 527–535. doi: 10.1046/j.1469-8137.2002.00535.x
Dixon, P. (2003). VEGAN, a package of R functions for community ecology. J. Veg. Sci. 14, 927–930. doi: 10.1111/j.1654-1103.2003.tb02228.x
Egidi, E., Delgado-Baquerizo, M., Plett, J. M., Wang, J. T., Eldridge, D. J., Bardgett, R. D., et al. (2019). A few Ascomycota taxa dominate soil fungal communities worldwide. Nat. Commun. 10:2369. doi: 10.1038/s41467-019-10373-z
Eichorst, S. A., Trojan, D., Roux, S., Herbold, C., Rattei, T., and Woebken, D. (2018). Genomic insights into the Acidobacteria reveal strategies for their success in terrestrial environments. Environ. Microbiol. 20, 1041–1063. doi: 10.1111/1462-2920.14043
Fierer, N., Bradford, M. A., and Jackson, R. B. (2007). Toward an ecological classification of soil bacteria. Ecology 88, 1354–1364. doi: 10.1890/05-1839
Fierer, N., and Jackson, R. B. (2006). The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. U. S. A. 103, 626–631. doi: 10.1073/pnas.0507535103
Fierer, M. C., McCain, C. M., Meir, P., Zimmermann, M., Rapp, J. M., Silman, M. R., et al. (2011). Microbes do not follow the elevational diversity patterns of plants and animals. Ecology 92, 797–804. doi: 10.1890/10-1170.1
Fierer, N., Strickland, M. S., Liptzin, D., Bradford, M. A., and Cleveland, C. C. (2009). Global patterns in belowground communities. Ecol. Lett. 12, 1238–1249. doi: 10.1111/j.1461-0248.2009.01360.x
Frindte, K., Pape, R., Werner, K., Loffler, J., and Knief, C. (2019). Temperature and soil moisture control microbial community composition in an arctic-alpine ecosystem along elevational and micro-topographic gradients. ISME J. 13, 2031–2043. doi: 10.1038/s41396-019-0409-9
Fukami, T., and Wardle, D. A. (2005). Long-term ecological dynamics: reciprocal insights from natural and anthropogenic gradients. Proc. Roy. Soc. B. Biol. Sci. 272, 2105–2115. doi: 10.1098/rspb.2005.3277
Geml, J., Pastor, N., Fernandez, L., Pacheco, S., Semenova, T. A., Becerra, A. G., et al. (2014). Large-scale fungal diversity assessment in the Andean Yungas forests reveals strong community turnover among forest types along an altitudinal gradient. Mol. Ecol. 23, 2452–2472. doi: 10.1111/mec.12765
Green, J., and Bohannan, B. J. M. (2006). Spatial scaling of microbial biodiversity. Trends Ecol. Evol. 21, 501–507. doi: 10.1016/j.tree.2006.06.012
Green, J. L., Holmes, A. J., Westoby, M., Oliver, I., Briscoe, D., Dangerfield, M., et al. (2004). Spatial scaling of microbial eukaryote diversity. Nature 432, 747–750. doi: 10.1038/nature03034
Hanson, C. A., Fuhrman, J. A., Horner-Devine, M. C., and Martiny, J. B. H. (2012). Beyond biogeographic patterns: processes shaping the microbial landscape. Nat. Rev. Microbiol. 10, 497–506. doi: 10.1038/nrmicro2795
Jiang, Y., Song, H., Lei, Y., Korpelainen, H., and Li, C. (2019). Distinct co-occurrence patterns and driving forces of rare and abundant bacterial subcommunities following a glacial retreat in the eastern Tibetan Plateau. Biol. Fertil. Soils 55, 351–364. doi: 10.1007/s00374-019-01355-w
Kassen, R., and Rainey, P. B. (2004). The ecology and genetics of microbial diversity. Annu. Rev. Microbiol. 58, 207–231. doi: 10.1146/annurev.micro.58.030603.123654
Kembel, S. W., Cowan, P. D., Helmus, M. R., Cornwell, W. K., Morlon, H., Ackerly, D. D., et al. (2010). Picante: R tools for integrating phylogenies and ecology. Bioinformatics 26, 1463–1464. doi: 10.1093/bioinformatics/btq166
Kim, H. M., Jung, J. Y., Yergeau, E., Hwang, C. Y., Hinzman, L., Nam, S., et al. (2014). Bacterial community structure and soil properties of a subarctic tundra soil in council, Alaska. FEMS Microbiol. Ecol. 89, 465–475. doi: 10.1111/1574-6941.12362
Kivlin, S. N., Winston, G. C., Goulden, M. L., and Treseder, K. K. (2014). Environmental filtering affects soil fungal community composition more than dispersal limitation at regional scales. Fungal Ecol. 12, 14–25. doi: 10.1016/j.funeco.2014.04.004
Lauber, C. L., Hamady, M., Knight, R., and Fierer, N. (2009). Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microbiol. 75, 5111–5120. doi: 10.1128/AEM.00335-09
Lauber, C. L., Ramirez, K. S., Aanderud, Z., Lennon, J., and Fierer, N. (2013). Temporal variability in soil microbial communities across land-use types. ISME J. 7, 1641–1650. doi: 10.1038/ismej.2013.50
Lenth, R. V. (2016). Least-squares means: the R package lsmeans. J. Stat. Softw. 69, 1–33. doi: 10.18637/jss.v069.i01
Li, J. B., Shen, Z. H., Li, C. N., Kou, Y. P., Wang, Y. S., Tu, B., et al. (2018). Stair-step pattern of soil bacterial diversity mainly driven by pH and vegetation types along the elevational gradients of Gongga Mountain, China. Front. Virol. 9:569. doi: 10.3389/fmicb.2018.00569
Liu, Y. R., Delgado-Baquerizo, M., Wang, J. T., Hu, H. W., Yang, Z. M., and He, J. Z. (2018). New insights into the role of microbial community composition in driving soil respiration rates. Soil Biol. Biochem. 118, 35–41. doi: 10.1016/j.soilbio.2017.12.003
Liu, K. S., Liu, Y. Q., Hu, A. Y., Wang, F., Chen, Y. Y., Gu, Z. Q., et al. (2020). Different community assembly mechanisms underlie similar biogeography of bacteria and microeukaryotes in Tibetan lakes. FEMS Microbiol. Ecol. 96:fiaa071. doi: 10.1093/femsec/fiaa071
Lundell, T. K., Makela, M. R., and Hilden, K. (2010). Lignin-modifying enzymes in filamentous basidiomycetes - ecological, functional and phylogenetic review. J. Basic Microbiol. 50, 5–20. doi: 10.1002/jobm.200900338
McCain, C. M., and Colwell, R. K. (2011). Assessing the threat to montane biodiversity from discordant shifts in temperature and precipitation in a changing climate. Ecol. Lett. 14, 1236–1245. doi: 10.1111/j.1461-0248.2011.01695.x
Meyer, K. M., Memiaghe, H., Korte, L., Kenfack, D., Alonso, A., and Bohannan, B. J. M. (2018). Why do microbes exhibit weak biogeographic patterns? ISME J. 12, 1404–1413. doi: 10.1038/s41396-018-0103-3
Miyamoto, Y., Nakano, T., Hattori, M., and Nara, K. (2014). The mid-domain effect in ectomycorrhizal fungi: range overlap along an elevation gradient on Mount Fuji, Japan. ISME J. 8, 1739–1746. doi: 10.1038/ismej.2014.34
Morlon, H., Chuyong, G., Condit, R., Hubbell, S., Kenfack, D., Thomas, D., et al. (2008). A general framework for the distance-decay of similarity in ecological communities. Ecol. Lett. 11, 904–917. doi: 10.1111/j.1461-0248.2008.01202.x
Mukhopadhya, I., Hansen, R., El-Omar, E. M., and Hold, G. L. (2012). IBD-what role do Proteobacteria play? Nat. Rev. Gastroenterol. Hepatol. 9, 219–230. doi: 10.1038/nrgastro.2012.14
Nemergut, D. R., Costello, E. K., Hamady, M., Lozupone, C., Jiang, L., Schmidt, S. K., et al. (2011). Global patterns in the biogeography of bacterial taxa. Environ. Microbiol. 13, 135–144. doi: 10.1111/j.1462-2920.2010.02315.x
Nemergut, D. R., Schmidt, S. K., Fukami, T., O'Neill, S. P., Bilinski, T. M., Stanish, L. F., et al. (2013). Patterns and processes of microbial community assembly. Microbiol. Mol. Biol. Rev. 77, 342–356. doi: 10.1128/MMBR.00051-12
Noss, R. F. (1990). Indicators for monitoring biodiversity: a hierarchical approach. Conserv. Biol. 4, 355–364. doi: 10.1111/j.1523-1739.1990.tb00309.x
Nottingham, A. T., Fierer, N., Turner, B. L., Whitaker, J., Ostle, N. J., McNamara, N. P., et al. (2018). Microbes follow Humboldt: temperature drives plant and soil microbial diversity patterns from the Amazon to the Andes. Ecology 99, 2455–2466. doi: 10.1002/ecy.2482
Ogwu, M. C., Takahashi, K., Dong, K., Song, H. K., Moroenyane, I., Waldman, B., et al. (2019). Fungal elevational Rapoport pattern from a High Mountain in Japan. Sci. Rep. 9:6570. doi: 10.1038/s41598-019-43025-9
Oksanen, J., Blanchet, F.G., Friendly, M., Kindt, R., and Wagner, H.H. (2020). Vegan: community ecology package. R Package Version 2.5-7. Available at: https://CRAN.R-project.org/package=vegan
Orwin, K. H., Bertram, J. E., Clough, T. J., Condron, L. M., Sherlock, R. R., O'Callaghan, M., et al. (2010). Impact of bovine urine deposition on soil microbial activity, biomass, and community structure. Appl. Soil Ecol. 44, 89–100. doi: 10.1016/j.apsoil.2009.10.004
Papke, R. T., and Ward, D. M. (2004). The importance of physical isolation to microbial diversification. FEMS Microbiol. Ecol. 48, 293–303. doi: 10.1016/j.femsec.2004.03.013
Peay, K. G., Garbelotto, M., and Bruns, T. D. (2010). Evidence of dispersal limitation in soil microorganisms: isolation reduces species richness on mycorrhizal tree islands. Ecology 91, 3631–3640. doi: 10.1890/09-2237.1
Peay, K. G., von Sperber, C., Cardarelli, E., Toju, H., Francis, C. A., Chadwick, O. A., et al. (2017). Convergence and contrast in the community structure of Bacteria, Fungi and Archaea along a tropical elevation-climate gradient. FEMS Microbiol. Ecol. 93:fix045. doi: 10.1093/femsec/fix045
Polme, S., Bahram, M., Yamanaka, T., Nara, K., Dai, Y. C., Grebenc, T., et al. (2013). Biogeography of ectomycorrhizal fungi associated with alders (Alnus spp.) in relation to biotic and abiotic variables at the global scale. New Phytol. 198, 1239–1249. doi: 10.1111/nph.12170
Rousk, J., Baath, E., Brookes, P. C., Lauber, C. L., Lozupone, C., Caporaso, J. G., et al. (2010). Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J. 4, 1340–1351. doi: 10.1038/ismej.2010.58
Schnecker, J., Wild, B., Takriti, M., Alves, R. J. E., Gentsch, N., Gittel, A., et al. (2015). Microbial community composition shapes enzyme patterns in topsoil and subsoil horizons along a latitudinal transect in Western Siberia. Soil Biol. Biochem. 83, 106–115. doi: 10.1016/j.soilbio.2015.01.016
Shen, C. C., Ge, Y., Yang, T., and Chu, H. Y. (2017). Verrucomicrobial elevational distribution was strongly influenced by soil pH and carbon/nitrogen ratio. J. Soils Sediments 17, 2449–2456. doi: 10.1007/s11368-017-1680-x
Shen, C. C., Liang, W. J., Shi, Y., Lin, X. G., Zhang, H. Y., Wu, X., et al. (2014). Contrasting elevational diversity patterns between eukaryotic soil microbes and plants. Ecology 95, 3190–3202. doi: 10.1890/14-0310.1
Shen, C. C., Shi, Y., Fan, K. K., He, J. S., Adams, J. M., Ge, Y., et al. (2019). Soil pH dominates elevational diversity pattern for bacteria in high elevation alkaline soils on the Tibetan Plateau. FEMS Microbiol. Ecol. 95:fiz003. doi: 10.1093/femsec/fiz003
Shen, C. C., Shi, Y., Ni, Y. Y., Deng, Y., Van Nostrand, J. D., He, Z. L., et al. (2016). Dramatic increases of soil microbial functional gene diversity at the treeline ecotone of Changbai Mountain. Front. Microbiol. 7:1184. doi: 10.3389/fmicb.2016.01184
Shen, C. C., Xiong, J. B., Zhang, H. Y., Feng, Y. Z., Lin, X. G., Li, X. Y., et al. (2013). Soil pH drives the spatial distribution of bacterial communities along elevation on Changbai Mountain. Soil Biol. Biochem. 57, 204–211. doi: 10.1016/j.soilbio.2012.07.013
Singh, D., Lee-Cruz, L., Kim, W.-S., Kerfahi, D., Chun, J.-H., and Adams, J. M. (2014). Strong elevational trends in soil bacterial community composition on Mt. Halla, South Korea. Soil Biol. Biochem. 68, 140–149. doi: 10.1016/j.soilbio.2013.09.027
Singh, D., Takahashi, K., and Adams, J. M. (2012). Elevational patterns in archaeal diversity on Mt. Fuji. PLoS One 7:e44494. doi: 10.1371/journal.pone.0044494
Soininen, J., McDonald, R., and Hillebrand, H. (2007). The distance decay of similarity in ecological communities. Ecography 30, 3–12. doi: 10.1111/j.0906-7590.2007.04817.x
Stegen, J. C., Lin, X. J., Fredrickson, J. K., Chen, X. Y., Kennedy, D. W., Murray, C. J., et al. (2013). Quantifying community assembly processes and identifying features that impose them. ISME J. 7, 2069–2079. doi: 10.1038/ismej.2013.93
Stegen, J. C., Lin, X. J., Fredrickson, J. K., and Konopka, A. E. (2015). Estimating and mapping ecological processes influencing microbial community assembly. Front. Microbiol. 6:370. doi: 10.3389/fmicb.2015.00370
Stegen, J. C., Lin, X. J., Konopka, A. E., and Fredrickson, J. K. (2012). Stochastic and deterministic assembly processes in subsurface microbial communities. ISME J. 6, 1653–1664. doi: 10.1038/ismej.2012.22
Thoms, C., Gattinger, A., Jacob, M., Thomas, F. M., and Gleixner, G. (2010). Direct and indirect effects of tree diversity drive soil microbial diversity in temperate deciduous forest. Soil Biol. Biochem. 42, 1558–1565. doi: 10.1016/j.soilbio.2010.05.030
Walker, L. R., Wardle, D. A., Bardgett, R. D., and Clarkson, B. D. (2010). The use of chronosequences in studies of ecological succession and soil development. J. Ecol. 98, 725–736. doi: 10.1111/j.1365-2745.2010.01664.x
Wan, W., Gadd, G. M., Yang, Y., Yuan, W., Gu, J., Ye, L., et al. (2021). Environmental adaptation is stronger for abundant rather than rare microorganisms in wetland soils from the Qinghai-Tibet Plateau. Mol. Ecol. 30, 2390–2403. doi: 10.1111/mec.15882
Wan, P., and He, R. R. (2020). Soil microbial community characteristics under different vegetation types at the national nature reserve of Xiaolongshan Mountains, Northwest China. Ecol Inform 55:101020. doi: 10.1016/j.ecoinf.2019.101020
Wang, J. T., Cao, P., Hu, H. W., Li, J., Han, L. L., Zhang, L. M., et al. (2015). Altitudinal distribution patterns of soil bacterial and archaeal communities along Mt. Shegyla on the Tibetan Plateau. Microb. Ecol. 69, 135–145. doi: 10.1007/s00248-014-0465-7
Wang, J. J., Soininen, J., Zhang, Y., Wang, B. X., Yang, X. D., and Shen, J. (2011). Contrasting patterns in elevational diversity between microorganisms and macroorganisms. J. Biogeogr. 38, 595–603. doi: 10.1111/j.1365-2699.2010.02423.x
Wang, Q., Wang, C., Yu, W. W., Turak, A., Chen, D. W., Huang, Y., et al. (2018). Effects of nitrogen and phosphorus inputs on soil bacterial abundance, diversity, and community composition in Chinese fir plantations. Front. Microbiol. 9:1543. doi: 10.3389/fmicb.2018.01543
Whitaker, R. J., Grogan, D. W., and Taylor, J. W. (2003). Geographic barriers isolate endemic populations of hyperthermophilic archaea. Science 301, 976–978. doi: 10.1126/science.1086909
Whittaker, R. J., Willis, K. J., and Field, R. (2001). Scale and species richness: towards a general, hierarchical theory of species diversity. J. Biogeogr. 28, 453–470. doi: 10.1046/j.1365-2699.2001.00563.x
Xu, M., Li, X. L., Cai, X. B., Gai, J. P., Li, X. L., Christie, P., et al. (2014). Soil microbial community structure and activity along a montane elevational gradient on the Tibetan Plateau. Eur. J. Soil Biol. 64, 6–14. doi: 10.1016/j.ejsobi.2014.06.002
Yang, T., Adams, J. M., Shi, Y., He, J. S., Jing, X., Chen, L. T., et al. (2017). Soil fungal diversity in natural grasslands of the Tibetan Plateau: associations with plant diversity and productivity. New Phytol. 215, 756–765. doi: 10.1111/nph.14606
Yang, Y., Gao, Y., Wang, S., Xu, D., Yu, H., Wu, L., et al. (2014). The microbial gene diversity along an elevation gradient of the Tibetan grassland. ISME J. 8, 430–440. doi: 10.1038/ismej.2013.146
Yang, X., and Xu, M. (2003). Biodiversity conservation in Changbai Mountain biosphere reserve, northeastern China: status, problem, and strategy. Biodivers. Conserv. 12, 883–903. doi: 10.1023/A:1022841107685
Zhang, Q., Goberna, M., Liu, Y. G., Cui, M., Yang, H. S., Sun, Q. X., et al. (2018). Competition and habitat filtering jointly explain phylogenetic structure of soil bacterial communities across elevational gradients. Environ. Microbiol. 20, 2386–2396. doi: 10.1111/1462-2920.14247
Zhang, B., Xue, K., Zhou, S. T., Che, R. X., Du, J. Q., Tang, L., et al. (2019). Phosphorus mediates soil prokaryote distribution pattern along a small-scale elevation gradient in Noijin Kangsang Peak, Tibetan Plateau. FEMS Microbiol. Ecol. 95:fiz076. doi: 10.1093/femsec/fiz076
Zhou, J. Z., Deng, Y., Shen, L. N., Wen, C. Q., Yan, Q. Y., Ning, D. L., et al. (2016). Temperature mediates continental-scale diversity of microbes in forest soils. Nat. Commun. 7:12083. doi: 10.1038/ncomms12083
Zhou, J. Z., and Ning, D. L. (2017). Stochastic community assembly: does it matter in microbial ecology? Microbiol. Mol. Biol. Rev. 81:e00002-17. doi: 10.1128/mmbr.00002-17
Keywords: altitudinal gradient, spatial scaling, assembly processes, soil depth, soil microbiota
Citation: Kang Y, Wu H, Zhang Y, Wu Q, Guan Q, Lu K and Lin Y (2023) Differential distribution patterns and assembly processes of soil microbial communities under contrasting vegetation types at distinctive altitudes in the Changbai Mountain. Front. Microbiol. 14:1152818. doi: 10.3389/fmicb.2023.1152818
Received: 28 January 2023; Accepted: 29 March 2023;
Published: 02 June 2023.
Edited by:
Ruiyong Zhang, Institute of Oceanology, Chinese Academy of Sciences (CAS), ChinaReviewed by:
Jian Liu, Shandong University, ChinaCopyright © 2023 Kang, Wu, Zhang, Wu, Guan, Lu and Lin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Haitao Wu, d3VoYWl0YW9AaWdhLmFjLmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.