- 1Institute of Microbiology, School of Ecology and Nature Conservation, Beijing Forestry University, Beijing, China
- 2National Institute of Occupational Health and Poison Control, Chinese Center for Disease Control and Prevention, Beijing, China
In this study, Podoscypha was taxonomically and phylogenetically evaluated. In total, five specimens collected from the tropical areas of Yunnan Province in Southwest China were studied. In combination with morphological observations and phylogenetic analyses based on ITS and LSU loci, two new species and one new subspecies, Podoscypha subinvoluta, P. tropica, and P. petalodes subsp. cystidiata, respectively, were discovered. The illustrated descriptions of the new species and subspecies are provided. Moreover, the main morphological differences between related species are discussed.
1. Introduction
Podoscypha Pat. belongs to Podoscyphaceae D.A. Reid, Polyporales Gäum., and approximately 50 species have been accepted in the genus worldwide (Patouillard, 1900; Boidin, 1959, 1960; Reid, 1965; Dhingra, 1987; Ryvarden, 1997, 2015, 2020; Douanla-Meli and Langer, 2004; Drechsler-Santos et al., 2007; Bernicchia and Gorjón, 2010; Wu et al., 2019). The genus is characterized by flabelliform to infundibuliform, stipitate to shortly stipitate or sessile basidiocarps, hymenophore smooth to more or less rugose, a dimitic hyphal structure with clamped generative hyphae, thin-to thick-walled cystidia and hyaline, thin-walled, smooth, ellipsoid to cylindrical, acyanophilous, and negative in Melzer’s reagent basidiospores (Bernicchia and Gorjón, 2010; Wu et al., 2019).
Phylogenetic studies have revealed that Podoscypha belongs to Podoscyphaceae, Polyporales (Larsson, 2007; Sjökvist et al., 2012; Binder et al., 2013; Justo et al., 2017; Wu et al., 2019). The approximately 12 Podoscypha species examined showed that Podoscypha belonged to the residual polyporoid clade and appeared to group with Abortiporus biennis (Bull.) Singer (Binder et al., 2013; Justo et al., 2017; Wu et al., 2019). However, no sequences of the type species P. nitidula (Berk.) Pat. were examined. Further studies are needed for constructing a natural phylogeny of Podoscypha with other related genera.
By the end of 2022, only four Podoscypha species: Podoscypha brasiliensis D.A. Reid, P. elegans (G. Mey.) Pat., P. simulans (D.A. Reid) Sheng H. Wu, and P. yunnanensis C.L. Zhao had been discovered in China (Wu, 2003; Dai, 2011; Wu et al., 2019). During field investigations on macrofungi diversity in the tropical areas in Yunnan Province, China, several collections of Podoscypha were obtained. Morphological and phylogenetic analyses showed that these represented two new species and one new subspecies. In this study, the species are described with illustrated morphological description, phylogeny, and comparisons with morphologically similar or phylogenetically related species.
2. Materials and methods
2.1. Morphology
All the studied specimens were processed and deposited in the National Institute of Occupational Health and Poison Control, Chinese Center for Disease Control (NIOHP, China CDC). Macromorphological descriptions were based on the field notes and color photographs of basidiocarps. Color codes were matched based on Petersen (1996). The microscopic procedure was performed according to Reid (1965), Douanla-Meli and Langer (2004), and Wu et al. (2019). Sections were studied at a magnification of ×1,000 with a Nikon E 80i microscope and phase-contrast illumination. Line drawings were made with the aid of a drawing tube. When describing basidiospores, the abbreviation n/m/p means that “n” basidiospores from “m” basidiomata of “p” collections were measured. The dimensions for basidiospores are given using the following notation form (a−) b−c (−d). The range “b−c” contains a minimum of 90% of the measured values, and extreme values (a, d) are given in parentheses. L means the spore length (arithmetic average of all spores). W means the spore width (arithmetic average of all spores). Q is the “length/width ratio” of a spore in side view; Qm means the average Q of all basidiospores measured ± sample standard deviation. In the text, the following abbreviations were used: IKI = Melzer’s reagent, IKI− = both in amyloid and non-dextrinoid, KOH = 5% potassium hydroxide, CB = cotton blue, CB+ = cyanophilous, and CB− = acyanophilous.
2.2. DNA extraction, PCR amplification, and phylogenetic analysis
The Phire® Plant Direct PCR Kit (Finnzymes Oy, Finland) was used to obtain PCR products from dried specimens, according to the manufacturer’s instructions with some modifications (Li et al., 2015). ITS5/ITS4 were used to amplify the internal transcribed spacer (ITS) regions (White et al., 1990). LR0R/LR5 (Vilgalys and Hester, 1990) were used to amplify nuclear large subunit (LSU) rDNA. The PCR procedure for amplifying ITS and LSU sequences was as follows: initial denaturation at 98°C for 5 min; 35 cycles at 98°C for 5 s, 58°C and 52°C for 5 s, and 72°C for 5 s; and a final extension at 72°C for 10 min, respectively. The PCR products were qualified by electrophoresis and sequenced at Sangon Biotech, China, with the same primers. All newly generated sequences were first aligned and then deposited in GenBank. The results indicated that they were all Podoscypha species.
In addition to the sequences obtained from this study, the sequences of other taxa in Podoscypha were downloaded from GenBank, and Abortiporus biennis was selected as the outgroup for phylogenetic analysis, mainly based on the reports of Binder et al. (2013) and Wu et al. (2019). The detailed information is shown in Table 1. The sequences were aligned using ClustalX 1.83 (Chenna et al., 2003) and optimized manually in BioEdit 7.0.5.3 (Hall, 1999) prior to the phylogenetic analysis.
Maximum parsimony (MP) bootstrap analysis was performed using PAUP* 4.0b10 (Swofford, 2002); Bayesian inference (BI) analysis was performed using MrBayes 3.1.2 (Ronquist and Huelsenbeck, 2003); maximum likelihood (ML) analysis was performed using RAxML 7.2.6 (Stamatakis, 2006) for the phylogenetic analysis of the aligned datasets.
For analyzing BI, the best-fit models of nucleotide substitution were selected by the hierarchical likelihood ratio tests (hLRT; Huelsenbeck and Crandall, 1997; Posada and Crandall, 2001) using MrModelTest 2.2 (Posada and Crandall, 1998; Nylander, 2004). The best-fit models were GTR + G for ITS and LSU. Four Markov chains were set to run for 2 and 4 million generations for the ITS and LSU datasets, respectively, and then, automatically terminated using the stoprul and stopval commands when the standard deviation of the split frequencies fell below 0.01, with sampling for every 100th generation. The first 25% of trees were discarded (Ronquist and Huelsenbeck, 2003). Phylogenetic trees were visualized using TreeView (Page, 1996). Branches that received bootstrap supports for MP (≥75%), BPP (≥0.95), and ML (≥75%) were considered significantly supported.
3. Results
3.1. Phylogenetic analyses
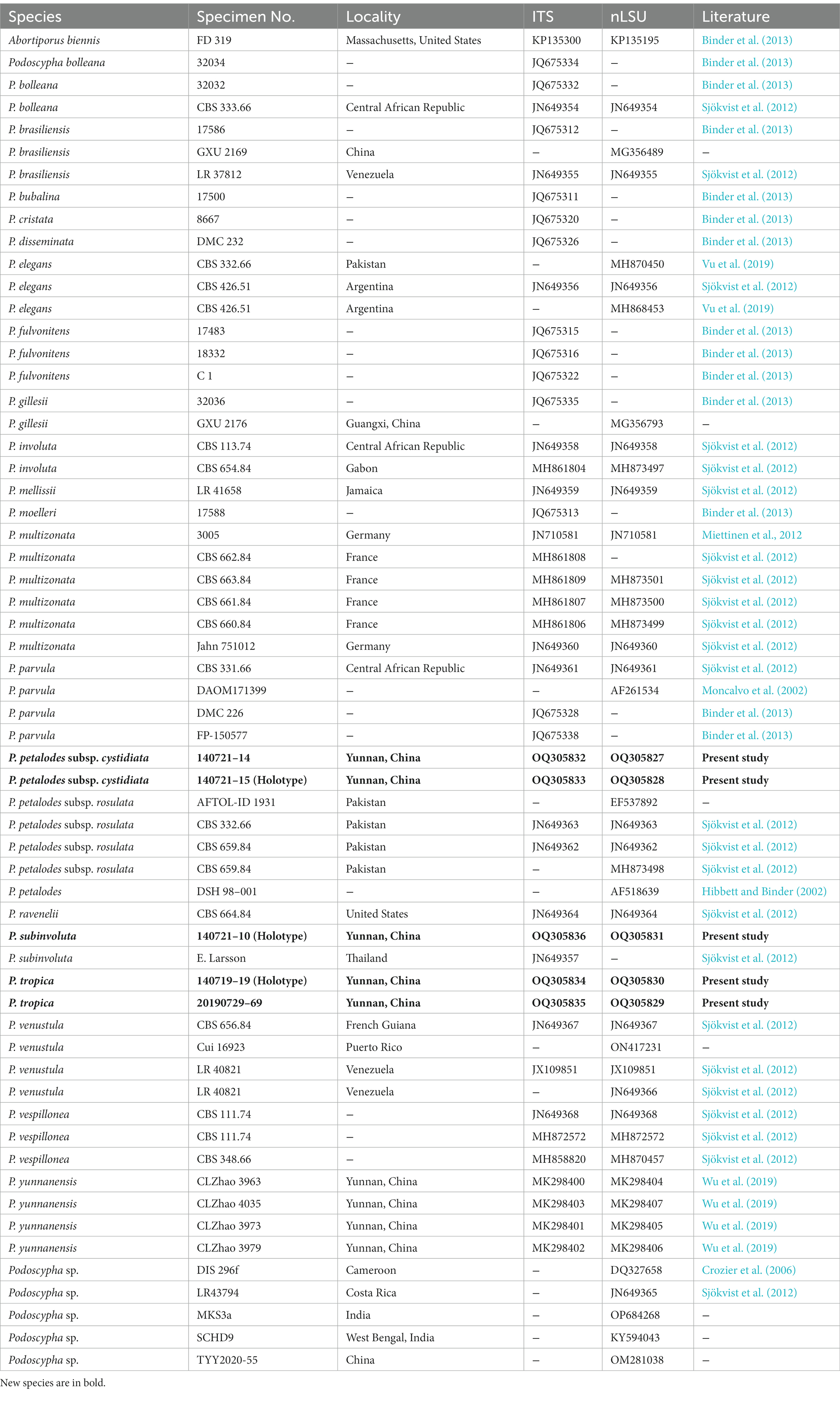
The ITS dataset consisted of 44 sequences representing 22 taxa, of which 21 were Podoscypha. The aligned length of the ITS sequence was 835 bp, which contained 447 parsimony informative sites. MP analysis revealed two equally parsimonious trees (tree length = 1,541, CI = 0.635, RI = 0.816, RC = 0.518, and HI = 0.316). BI and ML analyses showed almost identical tree topologies as MP analysis, with an average standard deviation of split frequencies of 0.007834 for BI analysis. Additionally, the MP tree is presented with the support values from MP, BI, and ML analyses (Figure 1).
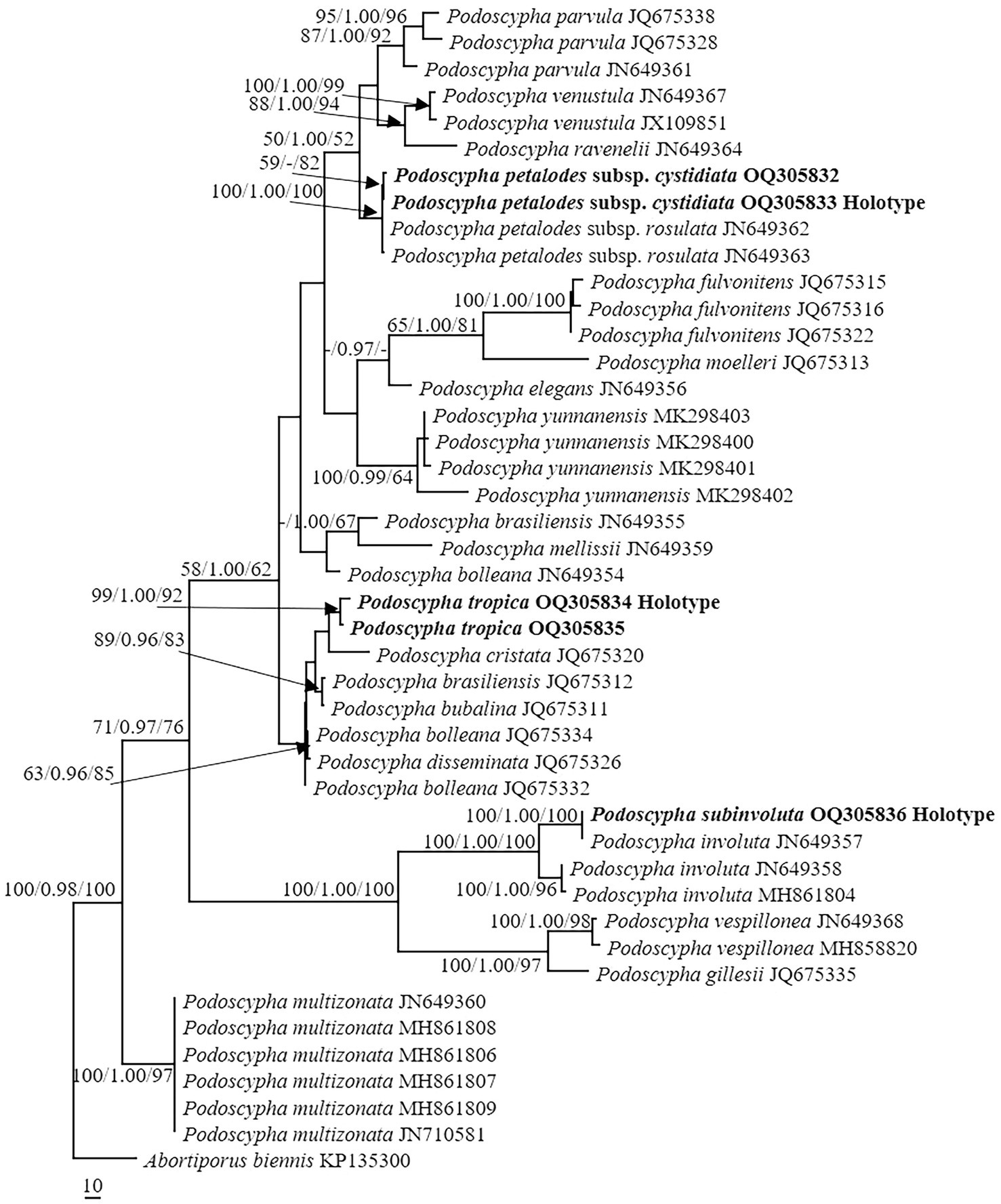
Figure 1. Phylogeny of Podoscypha by maximum parsimony (MP) analyses based on internal transcribed spacer (ITS) sequences. Branches are labeled with MP bootstrap >50%, Bayesian posterior probabilities >0.95, and maximum likelihood bootstrap >50%. New species and subspecies are indicated in bold.
The LSU dataset consisted of 44 sequences, representing approximately 18 taxa, of which 17 were Podoscypha. The aligned length of the LSU sequence was 926 bp, which contained 72 parsimony informative sites. MP analysis revealed 44 equally parsimonious trees (tree length = 161, CI = 0.745, RI = 0.917, RC = 0.684, HI = 0.255). BI and ML analyses resulted in almost identical tree topologies to MP analysis, with an average standard deviation of split frequencies of 0.006556 for BI analysis. Additionally, the MP tree is presented with the support values from MP, BI, and ML analyses (Figure 2).
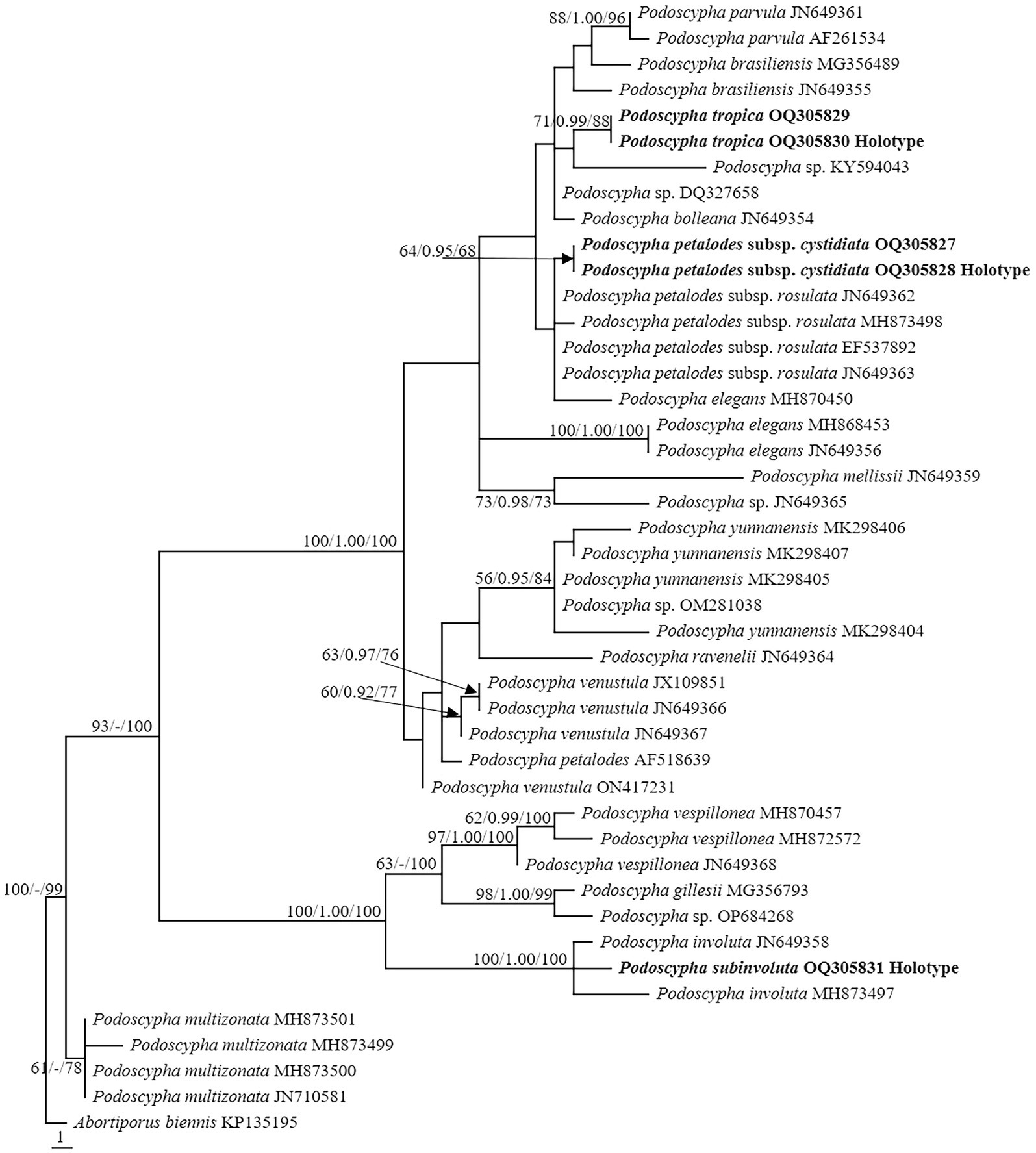
Figure 2. Phylogeny of Podoscypha by maximum parsimony (MP) analyses based on nuclear large subunit (LSU) sequences. Branches are labeled with MP bootstrap >50%, Bayesian posterior probabilities >0.95, and maximum likelihood bootstrap >50%. New species and subspecies are indicated in bold.
The topology of both ITS and LSU sequences was similar to the results of Wu et al. (2019). The phylogeny inferred from the ITS showed that Podoscypha petalodes subsp. Cystidiata formed a moderately supported lineage (59/−/82, Figure 1) and then clustered with P. petalodes subsp. rosulata with high support (100/1.00/100, Figure 1). LSU analysis revealed that two sequences of P. petalodes subsp. cystidiata grouped together (64/0.95/68, Figure 2) and then weakly grouped with P. petalodes subsp. rosulata. One sequence was labeled as P. elegans (MH870450). Podoscypha subinvoluta clustered with one sequence of P. involuta (JN649357) with high support (100/1.00/100, Figure 1), sister to the lineage of two other sequences of P. involuta in ITS analysis (Figure 1). In the LSU analysis, P. subinvoluta grouped with two sequences of P. involuta with high support (100/1.00/100, Figure 2). In total, two sequences of P. tropica clustered together with high support in both ITS and LSU analyses (ITS: 99/1.00/92, LSU: 71/0.99/88, Figures 1, 2) and then grouped with P. cristata, P. brasiliensis, P. bubalina, P. bolleana, P. disseminata, and Podoscypha sp. (KY594043, Figures 1, 2).
3.2. Taxonomy
Podoscypha petalodes subsp. cystidiata Jing Si & Hai J. Li subsp. nov. (Figures 3, 4).
Figure 4. Microscopic structures of Podoscypha petalodes subsp. cystidiata (140721–15, holotype). (A) Basidiospores, (B) hyphae from context, (C) cystidia, (D) basidia, basidioles, and gloeocystidia from the hymenium, (E) pilocystidia, and (F) caulocystidia. (Bars: (A) = 5 μm, (B–F) = 10 μm). Drawn by Hai-Jiao Li.
MycoBank no.: MB847438.
Diagnosis—Podoscypha petalodes subsp. cystidiata differs from P. petalodes subsp. floriformis, P. petalodes subsp. petalodes, and P. petalodes subsp. rosulata in having thin-to thick-walled cystidia in the hymenium.
Holotype—China. Yunnan Province, Xishuangbanna, Mengla County, Xishuangbanna Tropical Botanical Garden, on the base of a living angiosperm tree, 140721–15 (GenBank accession numbers: OQ305833-ITS, OQ305828-LSU).
Etymology—cystidiata referring to the presence of cystidia in the hymenium.
Habitat and distribution—Gregarious on angiosperm trees or fallen angiosperm branches, at present only discovered from its type locality, summer.
Known distribution—China (Yunnan).
Description—Basidiocarps annual, gregarious, stipitate, without odor or taste, corky when fresh and hard corky upon drying. Pilei spathulate to flabelliform, up to 4 cm high and 5 cm wide. Pileal surface glabrous, concentrically and radially zonate; cinnamon, yellowish brown, pinkish buff to clay-buff at the center and near the stipe when fresh, cinnamon, yellowish brown to orange-brown when dry, and gradually lightening toward the margin. The margin is cream to buff when fresh, pinkish buff to cinnamon-buff when dry and wavy. Hymenophore surface smooth, concentrically and radially zonate, cream, buff, to cinnamon-buff when fresh, flesh-pink to clay-buff when dry. Stipe surface glabrous, cinnamon, yellowish brown, pinkish buff to clay-buff near the cap, and dark brown to black toward the base when fresh; the color is similar to the pileal surface when dry, up to 3 cm long and 1.5–4 mm in diameter.
Hyphal structure—dimitic, generative hyphae with clamps, hyaline, thin-to thick-walled, branched; skeletal hyphae colorless, thick-walled with a wide to narrow lumen, unbranched to rarely branched; IKI−, CB−, tissues unchanged in KOH. Generative hyphae in the pileal context dominant, hyaline, thin-to thick-walled, branched, almost parallel along the pileal surface, 2–5 μm in diameter; skeletal hyphae in the pileal context hyaline, thick-walled, with a wide to narrow lumen, unbranched to rarely branched, 2–4 μm in diameter; generative hyphae in the stipe trama similar to that in the pileal context, 3–4 μm in diameter; skeletal hyphae in the stipe trama similar to that in the pileal context, 1.5–3.5 μm in diameter.
Microstructure—Basidia clavate to cylindrical, with four sterigmata and a basal clamp, 20–25 × 4–5 μm; basidioles dominant, in shape similar to basidia, but slightly smaller; Gloeocystidia abundant, thin-walled, cylindrical to subcylindrical, usually swollen near the base and with obtuse apex, traversing the entire width of the thickened hymenium, 105–155 × 10–13 μm; Cystidia hyaline, fusoid, thin-to thick-walled, 30–64 × 7–9 μm; Pilocystidia hyaline, clavate to cylindrical, thick-walled, with or without secondary septata, sometimes encrusted with irregular crystals, 45–101 × 9–13 μm; Caulocystidia more or less similar to pilocystidia, 58–110 × 5–13 μm; Basidiospores subglobose, broadly ellipsoid, ellipsoid to ovate, hyaline, thin-walled, smooth, often monoguttulate, IKI−, CB−, [60/2/2] (3.5−) 3.8–4.8 (−5) × 3–4 (−4.5) μm, L = 4.2 μm, W = 3.54 μm, Q = (1.05−) 1.08–1.34 (−1.4), and Qm = 1.19 ± 0.09.
Additional specimen examined (paratype)—China. Yunnan Province, Xishuangbanna, Mengla County, Xishuangbanna Tropical Botanical Garden, on fallen angiosperm branches, 140721-14 (GenBank accession numbers: OQ305832-ITS, OQ305827-LSU).
Podoscypha subinvoluta Jing Si & Hai J. Li sp. nov. (Figures 5, 6).
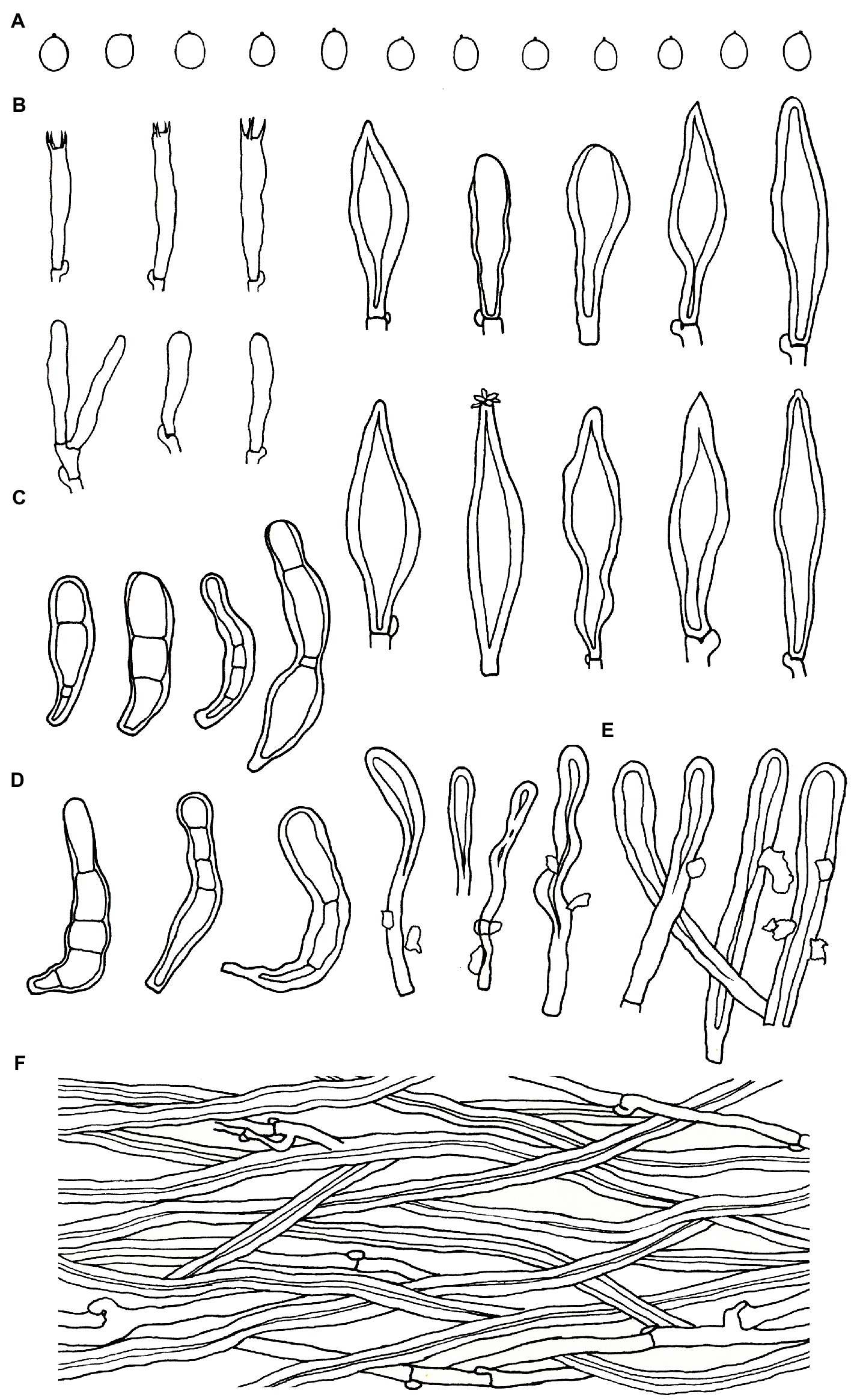
Figure 6. Microscopic structures of Podoscypha subinvoluta (140721-10, holotype). (A) Basidiospores, (B) basidia and basidioles, (C) cystidia, (D) pilocystidia, (E) caulocystidia, and (F) hyphae from context. (Bars: (A) = 5 μm, (B–F) = 10 μm). Drawn by Hai-Jiao Li.
MycoBank no.: MB847439.
Diagnosis—Podoscypha subinvolutais characterized by its partly tomentose, white to cream, buff, cinnamon, yellowish brown to clay-buff pileal surface, absence of gloeocystidia, thick-walled, mostly lanceolate cystidia, presence of pilocystidia and caulocystidia, small, thin-walled, ellipsoid to broadly ellipsoid basidiospores; discovered on angiosperms from tropical China.
Holotype—China. Yunnan Province, Xishuangbanna, Mengla County, Xishuangbanna Tropical Botanical Garden, on fallen angiosperm trunks, 140721-10 (GenBank accession numbers: OQ305836-ITS, OQ305831-LSU).
Etymology—subinvoluta referring to similarity with P. involuta.
Habitat and distribution—Gregarious on fallen angiosperm trunks, at present only discovered from its type locality, summer.
Known distribution—China (Yunnan).
Description—Basidiocarps annual, gregarious, stipitate, without odor or taste, corky when fresh and hard corky upon drying. Pilei flabelliform or reniform, up to 4.5 cm high and 6.5 cm wide. Pileal surface tomentose near the stipe and glabrous near the margin, concentrically and radially zonate; most parts white to cream, buff, cinnamon, yellowish brown to clay-buff near the stipe when fresh, flesh-pink to clay-pink to cinnamon when dry. Hymenophore surface smooth, cream, buff, to cinnamon when fresh, pinkish buff to clay-buff when dry. Stipe surface glabrous, cinnamon to yellowish brown when fresh, color similar to the pileal surface when dry, up to 1.2 cm long and 1.5–3 mm in diameter.
Hyphal structure—dimitic, generative hyphae with clamps, hyaline, thin-to thick-walled, branched; skeletal hyphae colorless, thick-walled with a wide to narrow lumen, unbranched to rarely branched; IKI−, CB−, tissues unchanged in KOH. Generative hyphae in the pileal context common, hyaline, thin-to thick-walled, branched, 2–5 μm in diameter; skeletal hyphae in the pileal context dominant, hyaline, thick-walled, with a wide to narrow lumen, unbranched to rarely branched, more or less parallel along the pileal surface, 3–6 μm in diameter; generative hyphae in the stipe trama similar to that in the pileal context, 3–4 μm in diameter; skeletal hyphae in the stipe trama similar to that in the pileal context, 4–7 μm in diameter, branched skeletal hyphae rare, thick-walled to subsoil, 2–3.5 μm in diameter, may be easily misinterpreted as binding hyphae.
Microstructure—Basidia clavate to cylindrical, with 4 sterigmata and a basal clamp, 18–25 × 4–5 μm; basidioles dominant, shape similar to basidia, but slightly smaller; Gloeocystidia not observed; Cystidia abundant, hyaline, thick-walled, mostly lanceolate, sometimes obpyriform or subcylindrical, rarely with crystals on the apex, which dissolve rapidly in KOH, may arise at any level in the thickened hymenium or buried under successive layers of basidia, 30–50 × 7–12 μm; Pilocystidia hyaline, clavate to cylindrical, thick-walled, with secondary septata, 20–64 × 6–9 μm; Caulocystidia form a paliform cutis, hyaline, clavate to cylindrical, thick-walled, encrusted with irregular crystals and strongly glutinized, 36–60 × 4–8 μm; Basidiospores ellipsoid to broadly ellipsoid, rarely subglobose, hyaline, thin-walled, smooth, IKI−, CB−, [40/1/1] 2.5–3.7 (−3.8) × 2–2.7 (−2.8) μm, L = 3.02 μm, W = 2.31 μm, Q = (1.07−) 1.14–1.52 (−1.73), and Qm = 1.31 ± 0.14.
Podoscypha tropica Jing Si & Hai J. Li sp. nov. (Figures 5, 6).
MycoBank no.: MB847440.
Diagnosis—Podoscypha tropica is characterized by a glabrous, yellowish-brown, pinkish-buff to brownish-orange pileal surface with cream, buff to pinkish buff margin, presence of gloeocystidia, pilocystidia, and caulocystidia without secondary septata, small, thin-walled, ellipsoid to broadly ellipsoid basidiospores; discovered on angiosperms from tropical China (Figures 7, 8).
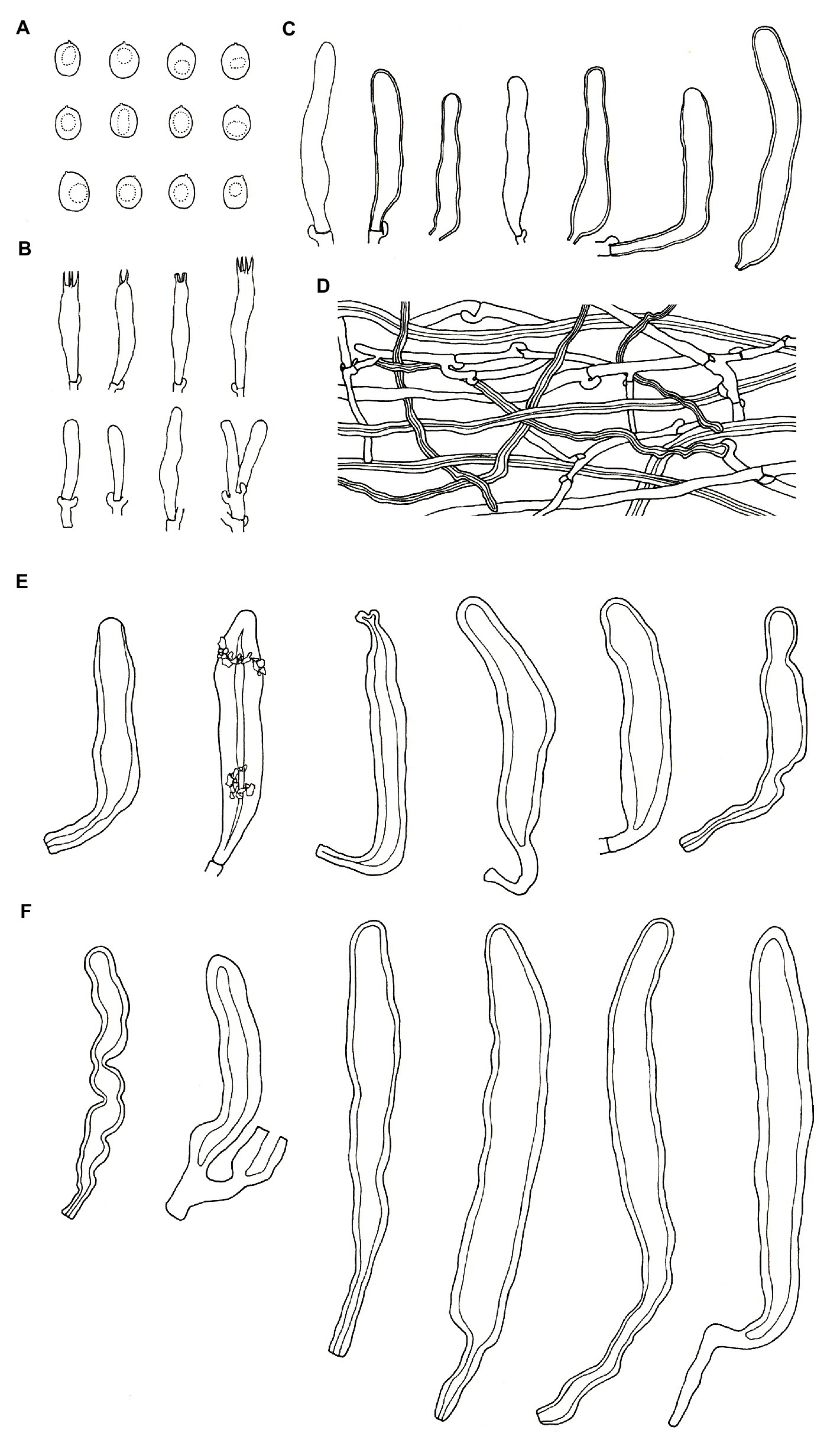
Figure 8. Microscopic structures of Podoscypha tropica (140719-19, holotype). (A) Basidiospores, (B) basidia and basidioles, (C) gloeocystidia, (D) hyphae from context, (E) pilocystidia, and (F) caulocystidia. (Bars: (A) = 5 μm, (B–F) = 10 μm). Drawn by Hai-Jiao Li.
Holotype—China. Yunnan Province, Xishuangbanna, Mengla County, Wangtianshu Park, N: 21°37′27.81″E: 101°35′13.12″, on fallen angiosperm trunks, 140719-19 (GenBank accession numbers: OQ305834-ITS, OQ305830-LSU).
Etymology—tropica referring to the species being distributed in the tropics.
Habitat and distribution—Gregarious on fallen angiosperm trunks or fallen angiosperm branches, at present only discovered from its type locality, summer.
Known distribution—China (Yunnan).
Description—Basidiocarps annual, gregarious, stipitate, without odor or taste, corky when fresh, and hard corky upon drying. Pilei spathulate, reniform, flabellate to pseudoinfundibuliform, up to 5 cm high and 6 cm wide. Pileal surface glabrous, yellowish brown, pinkish buff to brownish orange at the center and near the stipe when fresh, clay-buff to orange-brown when dry. The margin cream, buff to pinkish buff when fresh, clay-buff to orange-brown when dry, and strongly dentate. Hymenophore surface smooth, cream to buff when fresh, cinnamon to clay-buff when dry. Stipe surface glabrous, buff to straw-yellow on the upper part, and dark brown to fuscous toward the base when fresh, and the color slightly paler when dry, up to 5 cm long and 2–4 mm in diameter.
Hyphal structure—dimitic, generative hyphae with clamps, hyaline, thin-walled, branched; skeletal hyphae colorless, thick-walled with a wide to narrow lumen, unbranched to rarely branched; IKI−, CB−, tissues unchanged in KOH. Generative hyphae in the pileal context common, hyaline, thin-walled, branched, interwoven, 2–4 μm in diameter; skeletal hyphae in the pileal context dominant, hyaline, thick-walled, with a wide to the narrow lumen, unbranched to rarely branched, more or less parallel along the pileal surface, 2–5 μm in diameter; generative hyphae in the stipe trama similar to that in the pileal context, 2–4.5 μm in diameter; skeletal hyphae in the stipe trama similar to that in the pileal context, 2–4 μm in diameter.
Microstructure—Basidia clavate to cylindrical, with 4 sterigmata and a basal clamp, 25–33 × 4–5.5 μm; basidioles dominant, in shape similar to basidia, but slightly smaller; Gloeocystidia common, thin-to thick-walled, cylindrical to subcylindrical, usually swollen near the base and with obtuse apex, 45–75 × 7–10 μm; Cystidia absent; Pilocystidia hyaline, clavate to cylindrical, thick-walled, without secondary septata, sometimes encrusted with irregular crystals, 50–150 × 10–18 μm; Caulocystidia more or less similar to pilocystidia, 71–177 × 6–21 μm; Basidiospores ellipsoid to broadly ellipsoid, hyaline, thin-walled, smooth, often monoguttulate, IKI−, CB−, [60/2/2] 4.2–6.1 (−6.7) × (3−) 3.3–4.6 (−5) μm, L = 5.28 μm, W = 3.97 μm, Q = (1.09−) 1.14–1.53 (−1.83), and Qm = 1.34 ± 0.15.
Additional specimen examined (paratype)—China. Yunnan Province, Dehong, Ruili, Mengxiu Town, Nanjingli Village, Alt.: 1,247 m; N: 24°7′27″E: 97°49′28″, on fallen angiosperm branches, 20190729-69 (GenBank accession numbers: OQ305835-ITS, OQ305829-LSU).
4. Discussion
In total, three subspecies of Podoscypha petalodes have been described: P. petalodes subsp. floriformis, P. petalodes subsp. petalodes, and P. petalodes subsp. rosulate. All three have gloeocystidia, but without cystidia in the hymenium (Duss, 1904; Reid, 1965). The new subspecies, P. petalodes subsp. cystidiata, can be easily distinguished from the others by its fusoid, thin-to thick-walled cystidia.
Podoscypha subinvolutais similar to P. involuta by its partly tomentose concentrically and radially zonate pileal surface, mostly lanceolate cystidia, pilocystidia clavate to cylindrical, thick-walled, and small basidiospores; however, P. subinvoluta lacks gloeocystidia but has caulocystidia and larger basidiospores (2.5–3.7 × 2–2.7 μm vs. 2–3 × 1.75–2 μm, Reid, 1965). The partial ITS1 sequence of P. subinvoluta was obtained. Among the 279 bp aligned sequences, it clustered with one sequence of P. involuta (JN649357) from Thailand (only 2 bp sites were different), whereas it differed from the other two P. involuta sequences (JN649358 and MH861804) by 23 bp sites. Thus, we speculate that the specimen of P. involuta (JN649357) may be P. subinvoluta. The LSU sequence from the type specimen of P. subinvoluta grouped with two P. involuta sequences, and only 5 bp sites were different in the 915 bp aligned sequences.
In the phylogenetic analyses, P. tropica clustered with P. cristata, P. brasiliensis, P. bubalina, P. bolleana, P. disseminate, etc. Compared to the new species, P. cristata is lacking in pilocystidia and caulocystidia and has distinctly smaller basidiospores (4–4.75 × 1.75–2 μm, Reid, 1965). Podoscypha brasiliensis differs from the new species in having a minute tomentose pileal surface, the absence of pilocystidia and caulocystidia, and slightly longer and narrower basidiospores (5–7 × 3.5–4 μm, Reid, 1965). Podoscypha bubalina differs from P. tropica by having truly infundibuliform basidiocarps, the absence of pilocystidia, and smaller basidiospores (3.75–4.75 × 2.5–3.2 μm, Reid, 1965). Podoscypha bolleana is similar to P. tropica in having gloeocystidia, pilocystidia, and caulocystidia, but can be easily differentiated by its smaller basidiospores (4–5 × 2.75–3.2 μm, Reid, 1965). Similar to P. tropica, P. disseminata also has gloeocystidia, pilocystidia, and caulocystidia; however, P. disseminata has pilocystidia and caulocystidia with secondary septata, chlamydospores in the context, and slightly narrower basidiospores (4.5–6 × 3–4 μm, Douanla-Meli and Langer, 2004).
Podoscypha yunnanensis was a new species described in Yunnan Province, China (Wu et al., 2019). It has only caulocystidia without gloeocystidia and pilocystidia, which can be easily distinguished from the two new species and one new subspecies discovered in this study. The diversity of Podoscypha in China is poorly understood, especially in the subtropical and tropical regions. Further studies are required to reveal the species diversity in China.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/genbank/, OQ305832–OQ305836; https://www.ncbi.nlm.nih.gov/genbank/, OQ305827–OQ305831.
Author contributions
H-JL designed the research. H-JL, JS, Y-ZZ, and J-QL prepared the samples, conducted molecular experiments, and analyzed the data. H-JL and JS discussed the results and drafted and edited the manuscript. JS carried out the project administration and funding acquisition. All authors contributed to the manuscript and approved the submitted version.
Funding
The research was financed by the National Natural Science Foundation of China (32270016 and 32070016).
Acknowledgments
We express our gratitude to Yu-Cheng Dai (Beijing Forestry University, China) for improving the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Bernicchia, A., and Gorjón, S. P. (2010). Fungi Europaei 12: Corticiaceaes l. Edizioni Candusso, Lomazzo.
Binder, M., Justo, A., Riley, R., Salamov, A., López-Girálde, F., Sjökvist, E., et al. (2013). Phylogenetic and phylogenomic overview of the Polyporales. Mycologia 105, 1350–1373. doi: 10.3852/13-003
Boidin, J. (1959). Hétérobasidiomycèstes saprophytes et Homobasidiomycètesrésupinés: VI. Essai sur le genre Stereum sensulato. Rev. Mycol. 24, 197–225.
Boidin, J. (1960). Le genre Stereum Pers. S.L. au Congo belge. Bulletin du Jardin botanique de l'État a Bruxelles 30, 283–355. doi: 10.2307/3667306
Chenna, R., Sugawara, H., Koike, T., Lopez, R., Gibson, T. J., Higgins, D. G., et al. (2003). Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31, 3497–3500. doi: 10.1093/nar/gkg500
Crozier, J., Thomas, S. E., Aime, M. C., Evans, H. C., and Holmes, K. A. (2006). Molecular characterization of fungal endophytic morphospecies isolated from stems and pods of Theobroma cacao. Plant Pathol. 55, 783–791. doi: 10.1111/j.1365-3059.2006.01446.x
Dai, Y. C. (2011). A revised checklist of corticioid and hydnoid fungi in China for 2010. Mycoscience 52, 69–79. doi: 10.1007/s10267-010-0068-1
Douanla-Meli, C., and Langer, E. (2004). A taxonomic study of the family Podoscyphaceae (Basidiomycetes), new species and new records in Cameroon. Mycotaxon 90, 323–335.
Drechsler-Santos, E. R., Gibertoni, T. B., and de Queiroz Cavalcanti, M. A. (2007). Podoscypha aculeata, a new record for the neotropics. Mycotaxon 101, 69–72.
Hall, T. A. (1999). Bio edit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98.
Hibbett, D. S., and Binder, M. (2002). Evolution of complex fruiting-body morphologies in homobasidiomycetes. Proc. R. Soc. Lond. B Biol. Sci. 269, 1963–1969. doi: 10.1098/rspb.2002.2123
Huelsenbeck, J. P., and Crandall, K. A. (1997). Phylogeny estimation and hypothesis testing using maximum likelihood. Annu. Rev. Ecol. Syst. 28, 437–466. doi: 10.1146/annurev.ecolsys.28.1.437
Justo, A., Miettinen, O., Floudas, D., Ortiz-Santana, B., Sjökvist, E., Lindner, D., et al. (2017). A revised family-level classification of the Polyporales (Basidiomycota). Fungal Biol. 121, 798–824. doi: 10.1016/j.funbio.2017.05.010
Larsson, K. H. (2007). Re-thinking the classification of corticioid fungi. Mycol. Res. 111, 1040–1063. doi: 10.1016/j.mycres.2007.08.001
Li, H. J., Xie, J. W., Zhang, S., Zhou, Y. J., Ma, P. B., Zhou, J., et al. (2015). Amanita subpallidorosea, a new lethal fungus from China. Mycol. Prog. 14:43. doi: 10.1007/s11557-015-1055-x
Miettinen, O., Larsson, E., Sjökvist, E., and Larsson, K. H. (2012). Comprehensive taxon sampling reveals unaccounted diversity and morphological plasticity in a group of dimitic polypores (Polyporales, Basidiomycota). Cladistics 28, 251–270. doi: 10.1111/j.1096-0031.2011.00380.x
Moncalvo, J. M., Vilgalys, R., Redhead, S. A., Johnson, J. E., James, T. Y., Aime, M. C., et al. (2002). One hundred and seventeen clades of euagarics. Mol. Phylogenet. Evol. 23, 357–400. doi: 10.1016/S1055-7903(02)00027-1
Nylander, J. A. A. (2004). MrModeltest V2. Program Distributed by the Author. Uppsala: Evolutionary Biology Centre, Uppsala University.
Page, R. D. (1996). Tree view: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12, 357–358. doi: 10.1093/bioinformatics/12.4.357
Patouillard, N. T. (1900). Essaitaxonomique sur les Familles et les Genres des Hyménomycètes. Lucien Declume Press, Lons-Le-Saunier.
Petersen, J. H. (1996). The Danish Mycological Society's Colour-Chart. Foreningentil Svampekundskabens Fremme, Greve, Denmark.
Posada, D., and Crandall, K. A. (1998). MODELTEST: testing the model of DNA substitution. Bioinformatics 14, 817–818. doi: 10.1093/bioinformatics/14.9.817
Posada, D., and Crandall, K. A. (2001). Selecting the best-fit model of nucleotide substitution. Syst. Biol. 50, 580–601. doi: 10.1080/106351501750435121
Ronquist, F., and Huelsenbeck, J. P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574. doi: 10.1093/bioinformatics/btg180
Ryvarden, L. (2015). Type studies in Stereums. Lato 5. Species described by M.J. Berkeley. Synopsis Fungorum 33, 13–19.
Sjökvist, E., Larsson, E., Eberhardt, U., Ryvarden, L., and Larsson, K. H. (2012). Stipitate stereoid basidiocarps have evolved multiple times. Mycologia 104, 1046–1055. doi: 10.3852/11-174
Stamatakis, A. (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690. doi: 10.1093/bioinformatics/btl446
Swofford, D. L. (2002). PAUP: Phylogenetic Analysis Using Parsimony, Version 4.0 Beta 10. Sinauer Associates, Sunderland.
Vilgalys, R., and Hester, M. (1990). Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 172, 4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990
Vu, D., Groenewald, M., de Vries, M., Gehrmann, T., Stielow, B., Eberhardt, U., et al. (2019). Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation. Stud. Mycol. 92, 135–154. doi: 10.1016/j.simyco.2018.05.001
White, T. J., Bruns, T., Lee, S., and Taylor, J. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics” in PCR Protocols: A Guide to Methods and Applications. eds. M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (New York: Academic Press)
Keywords: macrofungi, species diversity, Podoscyphaceae, taxonomy, phylogeny, fungal resources
Citation: Si J, Zhang Y-Z, Liang J-Q and Li H-J (2023) Morphology and phylogeny identify two new species and one new subspecies of Podoscypha from Yunnan Province, Southwest China. Front. Microbiol. 14:1151365. doi: 10.3389/fmicb.2023.1151365
Edited by:
Ji-Chuan Kang, Guizhou University, ChinaReviewed by:
Yuanyuan Chen, Henan Agricultural University, ChinaChanglin Zhao, Southwest Forestry University, China
Copyright © 2023 Si, Zhang, Liang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jing Si, ✉ amluZ3NpMTc4OEAxMjYuY29t; Hai-Jiao Li, ✉ bGloYWlqaWFvNzE1QDEyNi5jb20=
 Jing Si
Jing Si Yi-Zhe Zhang
Yi-Zhe Zhang Jia-Qi Liang
Jia-Qi Liang Hai-Jiao Li
Hai-Jiao Li