
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 29 September 2023
Sec. Microbial Symbioses
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1147505
This article is part of the Research TopicEnvironmental Pollution and the Human Microbiota in Health and DiseaseView all 4 articles
 Shagun Shukla1,2
Shagun Shukla1,2 Ankita Srivastava3
Ankita Srivastava3 Digvijay Verma3
Digvijay Verma3 Siddhartha Gangopadhyay2,4
Siddhartha Gangopadhyay2,4 Anchal Chauhan2,4
Anchal Chauhan2,4 Vikas Srivastava2
Vikas Srivastava2 Savita Budhwar5
Savita Budhwar5 Dushyant Tyagi6
Dushyant Tyagi6 Deepak Chand Sharma1*
Deepak Chand Sharma1*Introduction: Approximately 200 million people worldwide are affected by arsenic toxicity emanating from the consumption of drinking water containing inorganic arsenic above the prescribed maximum contaminant level. The current investigation deals with the role of prenatal arsenic exposure in modulating the gut microbial community and functional pathways of the host.
Method: 16S rRNA-based next-generation sequencing was carried out to understand the effects of in utero 0.04 mg/kg (LD) and 0.4 mg/kg (HD) of arsenic exposure. This was carried out from gestational day 15 (GD-15) until the birth of pups to understand the alterations in bacterial diversity.
Results: The study focused on gestational exposure to arsenic and the altered gut microbial community at phyla and genus levels, along with diversity indices. A significant decrease in firmicutes was observed in the gut microbiome of mice treated with arsenic. Functional analysis revealed that a shift in genes involved in crucial pathways such as insulin signaling and non-alcoholic fatty liver disease pathways may lead to metabolic diseases in the host.
Discussion: The present investigation may hypothesize that in utero arsenic exposure can perturb the gut bacterial composition significantly as well as the functional pathways of the gestationally treated pups. This research paves the way to further investigate the probable mechanistic insights in the field of maternal exposure environments, which may play a key role in epigenetic modulations in developing various disease endpoints in the progeny.
Arsenic is a common toxic contaminant found in the environment and is categorized as a group I carcinogen (Bhattacharya et al., 2007; IARC Working Group on the Evaluation of Carcinogenic Risks to Humans, 2012). Many populations around the world are exposed to above-permissible levels of arsenic via drinking water [World Health Organization and the U.S. Environmental Protection Agency (EPA)]. The established guidelines for arsenic are <10 μg/L (Hughes et al., 2011). Various epidemiological and animal studies have linked chronic arsenic exposure with cancers of various organs such as the skin, bladder, and liver, along with non-cancer endpoints such as diabetes, obesity, hypertension, and cardiovascular diseases (van de Wiele et al., 2010; Hughes et al., 2011; Gribble et al., 2012; Naujokas et al., 2013; Grau-Perez et al., 2018). Recently, many studies have shown the correlation between arsenic exposure and metabolic disorders (Paul et al., 2011; Maull et al., 2012; Castriota et al., 2018). Trillions of microbes are present in the human gut and work in symbiosis with the host to maintain vital functions such as food digestion, metabolic processing, immune system development, epithelial homeostasis, and xenobiotic biotransformation (Ley et al., 2006a,b; Young et al., 2008; Qin et al., 2010; Nicholson et al., 2012; Jandhyala et al., 2015). In recent years, many reports have indicated a link between perturbed gut microbiota and the cause of many diseases such as cancer, diabetes, cardiovascular diseases, and obesity (Ley et al., 2005, 2006b; Wang et al., 2011; Qin et al., 2012; Roderburg and Luedde, 2014; Gentile and Weir, 2018; Guglielmi, 2018). Besides, the composition of gut microbiota is known to be affected by a variety of external factors such as antibiotics, food, stress, drugs, and environmental chemicals such as arsenic, mercury, and hydrocarbons (Liebert et al., 1997; van de Wiele et al., 2005, 2010; Pinyayev et al., 2011; Choi et al., 2013; Zhang et al., 2015a,b; An et al., 2019). Linking altogether, it can be hypothesized that arsenic has the potential to perturb the composition of the gut microbiota and thus alter the metabolic state of the organisms.
Some findings are able to establish the relationship between arsenic exposure, gut microbiota, and metabolic disorders in the adult population (Lu et al., 2014; Hur and Lee, 2015; Brabec et al., 2020; Yang et al., 2021). The gut microbiota of the infant is primarily established during birth and early life and is transferred vertically from the mother (Yao et al., 2021). Alteration of the gut microbiota and metabolic disorders due to prenatal arsenic exposure has not yet been well-studied. Therefore, the present investigation planned to study the effect of prenatal arsenic exposure on the gut microbiota and metabolic profile of resident microbes.
Female Balb/C mice (6–8 weeks old) weighing between 25 and 28 g were acquired from the animal housing and rearing facility of CSIR-IITR, Lucknow, India. Animals were kept in polypropylene cages at 25°C with 12 h light/12 h dark cycles and had ad libitum access to purified water and a standard chow diet. The female mice were divided into three groups (n = 10 in each group), viz., (i) control (without arsenic exposure), (ii) 0.04 mg/kg as exposure group (low dose LD), and (iii) 0.4 mg/kg as arsenic group (high dose HD). A working dose of arsenic, i.e., 0.04 and 0.4 mg/kg, was freshly prepared by dissolving sodium (meta) arsenite (NaAsO2) in water and was given daily via oral gavage from GD-15 (gestational day minus 15; i.e., 15 days prior to mating) until delivery (i.e., GD-21; Sharma et al., 2022). Water was administered orally to the control group. Animals were set for mating in a 2:1 (female:male) ratio after 15 days of dosing and on getting a positive plug; the males were separated. The pups were housed with their mothers for weaning until 21 days after birth. After weaning, the male and female offspring were segregated, and three male pups from each group were sacrificed at 6 weeks. For studying the bacterial diversity, gut microbiota samples were procured by collecting fecal samples from the respective groups. The fecal collection was carried out under aseptic conditions using a sterile spatula. All the experimental procedures performed in this study were approved by the Institutional Animal Ethical Committee (IAEC) of the Council of Scientific and Industrial Research—Indian Institute of Toxicology Research (CSIR-IITR), Lucknow, India.
Fecal samples of 6-week-old mouse offspring were collected via standard protocol, and their metagenomic DNA was extracted using a QIAamp Power Fecal Pro DNA Kit—QIAGEN (Germany) and stored at −20°C until use. The extracted metagenomes were processed for amplification of the bacterial-specific 16S rRNA V3-V4 hypervariable region. Amplicon library was prepared by Qubit quantification of 40 ng of extracted fecal metagenome for amplifying the V1 − 3F (5′AGAGTTTGATGMTGGCTCAG3′) and V (5′TTACCGCGGCMGCSGGCAC3′) hypervariable regions of the 16S rRNA encoding gene using the TAQ Master MIX (Biokart India Pvt. Ltd., India). Furthermore, next-generation sequencing (NGS) was performed using the Illumina Miseq platform with a 2 × 300 paired-end V3 sequencing kit at Biokart India Pvt. Ltd., Bangalore, Karnataka, India.
Raw reads obtained from the Illumina sequencing were further denoised using DADA2 in QIIME2 (2020.11) for quality filtering, removal of chimeric sequences, and generation of Amplicon Sequence Variants (ASVs; Bolyen et al., 2019). Further good-quality reads obtained after denoising were processed for taxonomy assignment in QIIME2 (2020.11) by training a feature classifier in the SILVA 16S database to obtain various taxa (phylum, class, order, family, and genus) level distributions for each sample with 99% sequence identity (Quast et al., 2013).
Various diversity indices (Chao1, Shannon, Simpson, Pielou evenness, observed features, and Faith_pd) were calculated to analyse the bacterial diversity of each sample. The data have been interpreted for different outputs such as GraphPad Software, v. 6.0; San Diego, CA; Microsoft Excel; NCSS V20.0.2; and STAMP V2.1.3 (Parks et al., 2014). The functional prediction of the 16S rDNA genes was performed using the Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt 2.0; version 2.4.1) algorithm (Douglas et al., 2020).
The generated ASVs were analyzed for the overall bacterial composition in the gut microbiota of control and prenatally arsenic-treated animals at various taxon levels. The statistically significant differences at the level of tier I, tier II, and tier III were performed using a two-sided G-test (w/Yates)+Fisher's test at a 95% confidence interval to analyse the PICRUSt2 output using STAMP (https://stamp-software.com/). Data were depicted as mean ± SE normalized to control values in the figures. Comparative analyses among the three groups were performed using a one-way ANOVA (p < 0.05) using GraphPad software (GraphPad Software, v. 6.0; San Diego, CA) and Microsoft Excel.
Amplicon libraries achieved from nine samples (three male samples from each group) revealed a total of 9,91,471 raw reads, which were converted to 5,12,543 high-quality non-chimeric reads using the DADA2 denoiser in the QIIME2 pipeline to generate amplicon sequence variants (ASVs) for taxonomy assignment (Supplementary Table 1).
On taxonomic assignment, the filtered reads were assigned into nine phyla, 13 classes, 25 orders, 35 families, 53 genera, and 27 species.
Overall data analysis revealed that Firmicutes (79.56%) were the most abundant phyla in the gut microbiota followed by Bacteroidota (10.57%), Actinobacteriota (5.58%), Verrucomicrobiota (2.14%), Cyanobacteria (1.29%), Patescibacteria (1.12%), and Desulfobacterota (1.01%). Other phyla were Deferribacterota (0.40%) and Proteobacteria (0.34%), which contributed <1%. Phyla Verrucomicrobiota, Cyanobacteria, Deferribacterota, and Proteobacteria did not appear in the control animals and were present only in either of the prenatally arsenic-treated groups (Figure 1A).
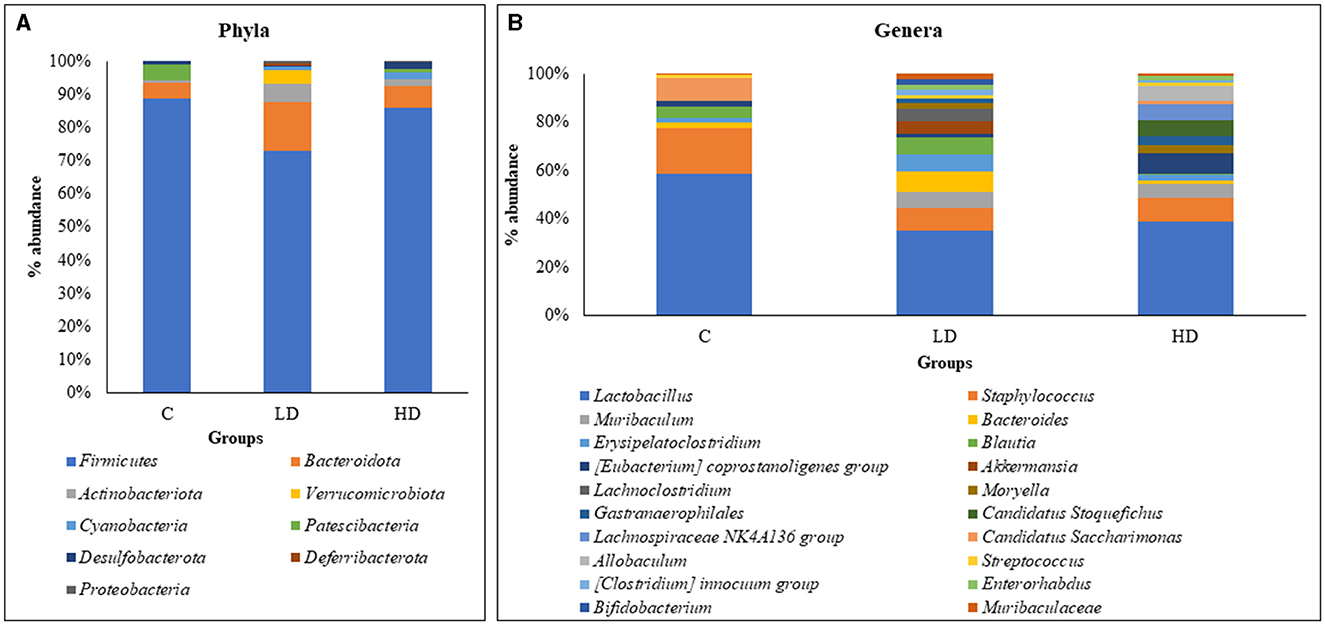
Figure 1. Relative percentage abundance at the level of phyla between C (control), LD (low dose), and HD (high dose) groups (A). Relative percentage abundance of top 20 genera among the three groups (C, LD, and HD) (B).
Of the 53 genera, Lactobacillus (33.87%) was the most abundant in the gut microbiota, followed by Staphylococcus (9.38%), Muribaculum (4.65%), Bacteroides (4.52%), Erysipelatoclostridium (4.4%), Blautia (4.28%), Coprostanoligenes [Eubacterium] (2.79%), Akkermansia (2.76%), Lachnoclostridium (2.36%), and Moryella (2.06%), which constituted the top 10 genera. Thirty-six genera were absent in the control group and observed in prenatally arsenic-treated groups only. Four genera (Roseburia, Odoribacter, Prevotella, and RF39) were detected only in the control group, while the remaining genera exhibited a portion below 2%, thus exhibiting low significance in modulating the physiology of the gut. The top 20 genus abundances have been shown in Figure 1B.
Out of the top nine phyla, Firmicutes (88.66%) and Patescibacteria (4.74%) showed relatively high dominance in the control group, i.e., without arsenic dose. Whereas in the LD group, Bacteroidota (14.78%), Actinobacteriota (5.27%), Verrucomicrobiota (4.02%), Deferribacterota (0.76%), and Proteobacteria (0.51%) were relatively dominant. In the HD group, Cyanobacteria (2.19%) and Desulfobacterota (2.28%) were relatively dominant.
Out of the 53 genera, Lactobacillus (52.77%), Staphylococcus (17.25%), Candidatus Saccharimonas (8.78%), Alistipes (3.41%), Desulfovibrio (0.82%), Roseburia (0.83%), Odoribacter (0.77%), Prevotella (0.43%), and RF39 (0.27%) showed relatively high dominance in the control group. In the LD group, 29 genera showed relative dominance, out of which 15 were more than 1%. Muribaculum (5.61%), Bacteroides (6.87%), Erysipelatoclostridium (6.08%), Blautia (6.29%), Akkermansia (4.74%), Lachnoclostridium (4.05%), Streptococcus (1.41%), [Clostridium] innocuum group (2.05%), Bifidobacterium (2.19%), Muribaculaceae (1.75%), Phascolarctobacterium (1.96%), [Ruminococcus] gnavus group (1.86%), Corynebacterium (1.22%), [Ruminococcus] torques group (1.09%), and Prevotellaceae UCG-001 (1.02%) were relatively dominant genera. Fourteen dominant genera had <1%. In the HD group, [Eubacterium] coprostanoligenes group (7.22%), Moryella (2.90%), Gastranaerophilales (2.97%), Candidatus Stoquefichus (5.63%), Lachnospiraceae NK4A136 group (5.60%), Allobaculum (5.20%), Enterorhabdus (1.93%), Erysipelotrichaceae UCG-003 (2.72%), Dubosiella (3.07%), Clostridia UCG-014 (1.60%), Enterococcus (1.45%), Lawsonia (2.44%), Lactococcus (1.27%), Anaerostipes (1.28%), and Clostridium sensu stricto 1 (0.43%) were relatively dominant genera.
Various diversity indices were analyzed for control and prenatally arsenic-exposed groups, i.e., LD and HD (Figure 2; Table 1). Prenatally arsenic-exposed groups LD and HD showed a higher richness of bacterial diversity than the control group. The highest average Chao1-index was observed in the LD group (49.33), followed by the HD (44.0.0) and the control groups (15). The highest average Shannon index was also observed in the LD group (5.01) as compared to the control (3.21) and the HD groups (2.81). Similarly, the average Simpson index was also maximum for the LD group (0.96) in comparison to the HD (0.86) and control groups (0.57). Average Pielou evenness was highest in the LD group (0.90), followed by the control (0.84) and the HD groups (0.50). Average observed features were also higher in the LD (48.67) and the HD (43.00) groups compared to the control group (15.00). Average Faith_pd was also measured, which was higher in the HD (8.37) and the LD (7.27) groups compared to the control (4.58) group.
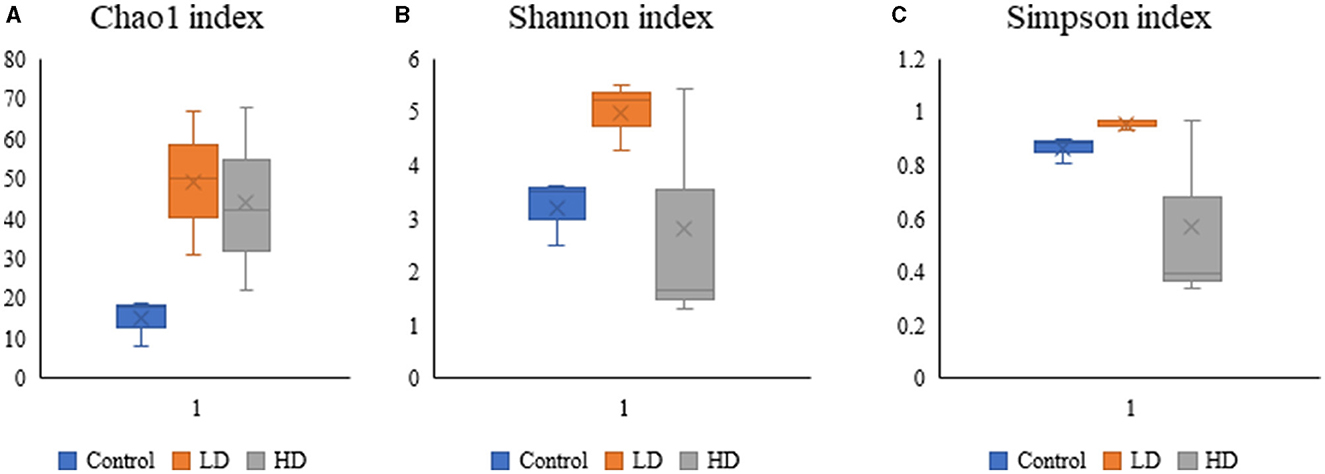
Figure 2. Box-plot analysis of the alpha diversity indices [Chao1 (A), Shannon (B), and Simpson (C)] among the three groups (C, LD, and HD).
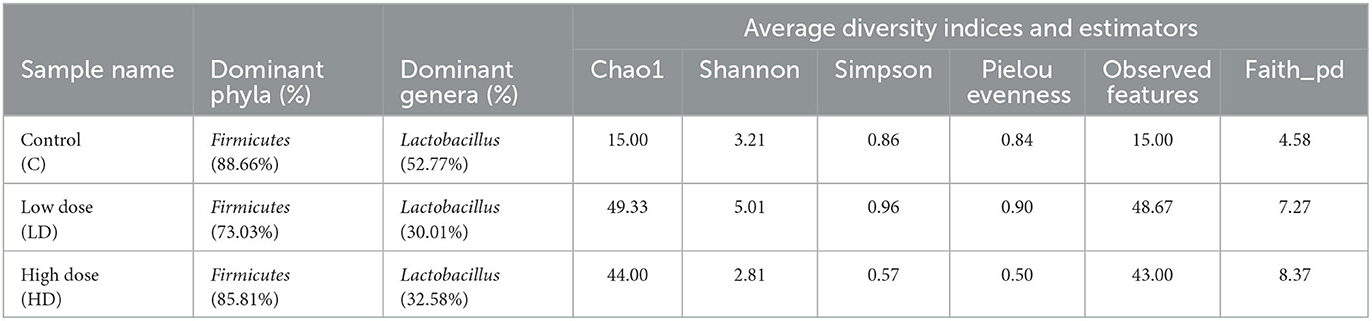
Table 1. Various diversity indices and estimators obtained among control (C) and arsenic treated groups (LD and HD).
A total of nine bacterial phyla were identified among the three groups. Of them, only four phyla (Firmicutes, Bacteroidota, Actinobacteriota, and Desulfobacterota) were present in the gut microbiota of both the control and arsenic-exposed groups (LD and HD). At the genus level, 53 genera were identified among the three groups where Lactobacillus, Staphylococcus, Bacteroides, Erysipelatoclostridium, Blautia, Coprostanoligenes group, Streptococcus, Muribaculaceae, and Desulfovibrio were observed in the gut microbiota of both the control and the treated groups.
Inter-individual differences were observed at the phyla and genera levels among the control and arsenic-treated groups. At the phyla level, Cyanobacteria and Proteobacteria were seen in both the arsenic-treated groups, while their occurrence was missing in the control group. Moreover, Verrucomicrobiota and Deferribacterota were found only in the LD group. Whereas Patescibacteria was detected only in the control and HD groups. At the genera level, Muribaculum, Moryella, Gastranaerophilales, [Clostridium] innocuum group, Enterorhabdus, Dubosiella, Clostridia UCG-014, Enterococcus, Prevotellaceae UCG-003, Gordonibacter, and Clostridium sensu stricto 1 were present in both the arsenic-treated groups but were not present in the control group. Candidatus Stoquefichus, Lachnospiraceae NK4A136 group, Allobaculum, Lawsonia, and Anaerostipes were the genera that were only present in the HD group.
The PICRUSt 2 (version 2.4.1) analysis was performed in QIIME2 on a command basis for exploring the bacterial functional role in the treated groups, and the control groups showed significant variations in the functionality. In tier I, the majority of the genes were involved in the metabolic pathways (50.05%), followed by the genes of genetic information processing (18.08%), environmental information processing (15.46%), cellular processes (7.67%), human diseases (6.02%), and organismal systems (2.73%; Supplementary Figure 1). Tier I analysis revealed that in a dose-dependent gene expression in all the functional groups, the HD group showed the highest value, followed by the LD group, and then the control group. Tier II analysis showed a significant abundance for the genes that belong to carbohydrate metabolism (23.08%), translation (49.23%), membrane transport (65.83%), cellular community prokaryotes (54.23%), endocrine system (41.54%), and antimicrobial drug resistance (31.15%; Supplementary Figures 2A–C, 3A–C). Twelve relevant pathways were chosen for further investigation. A comparative analysis of these genes showed that most pathways (10) were not altered due to arsenic treatment, either at lower or higher doses of arsenic. Only genes involved in non-alcoholic fatty liver diseases and insulin signaling pathways showed a decrease in their relative counts as compared to the control groups. In addition, neither of the genes was significantly different among the groups (p > 0.05).
It was demonstrated that an environmentally relevant level of arsenic exposure during the gestational period can perturb the normal community composition and functional pathways in the mouse gut microbiome. It has been previously described in chronic arsenic exposure studies that patterns of energy metabolism genes were altered (Lu et al., 2014; Chi et al., 2017), and genes involved in LPS synthesis (Chi et al., 2017; Yang et al., 2021), oxidative stress responses, and DNA repair were broadly increased due to long-term exposure to arsenic (Lu et al., 2014; Chi et al., 2017, 2018). In addition, arsenic exposure also enriched genes that encode conjugative transposon proteins, components of the multidrug efflux system, and the synthesis of multiple vitamins. As the exposure to low, environmentally relevant doses of arsenic during the gestation period culminates in the disruption of glucose homeostasis and energy metabolism-related genes, the current study explored whether it also affects the establishment and maintenance of the natural gut microbiome or not. The results of the study provide a new understanding of the effects of arsenic on the gut microbiome, especially at environmentally relevant doses. High-throughput 16S rRNA gene sequencing was used for profiling to study the impact of arsenic exposure on the gut microbiome and its metabolic profiles. In a study, 16S RNA-based analysis of the gut microbiome unveiled that perturbation in the gut microbiome enhances the arsenic bioaccumulation of its toxicity (Coryell et al., 2018). Our observations clearly show that arsenic exposure induced a noteworthy change in the gut microbiome composition of prenatally treated male mice, indicating that arsenic exposure not only disturbs gut bacteria at the abundance level but also substantially alters the metabolomic profile of the host, resulting in the disturbance of host metabolite homeostasis after arsenic exposure. Chi et al. (2017) observed that the 100-ppb arsenic dose for 13 weeks was sufficient to alter the bacterial diversity of the gut microbiome of mice. Therefore, the current findings may provide mechanistic insights regarding perturbations of the gut microbiome as a new mechanism of environmental chemical-induced human diseases. It may be hypothesized that these alterations might be due to the vertical transfer of arsenic-exposed gut microbiota from the mother to F1-generation after gestational arsenic exposure. In the F1-generation, which has acquired arsenic-exposed microbiota from its mother, along with other adaptations and epigenetic modifications, an increase in the diversity of gut microbiota was observed. In the normal gut microbiota, the relative abundance of both Firmicutes and Bacteroidetes was recorded, but a shift in the relative abundance of Firmicutes and Bacteroidetes was observed in obese vs. lean mice, with a statistically significant reduction in Bacteroidetes and an increase in Firmicutes in obese mice. Overall analysis revealed that Firmicutes, Bacteroidota, and Actinobacteriota constitute the major phyla as reported in several investigations on arsenic toxicity (Thursby and Juge, 2017; Binda et al., 2018; Magne et al., 2020) in the gut microbiome. The members of these phyla are prominently present in the gut microbiota of humans as well as mice. Firmicutes and Bacteroidetes exhibit a plethora of enzymes involved in carbohydrate metabolism (Turnbaugh et al., 2006). Moreover, the prevalence of Lactobacillus, Staphylococcus, Muribaculum, and Bacteroides-like genera further corroborates the findings of several investigations (Bervoets et al., 2013; Harakeh et al., 2016; Crovesy et al., 2017, 2020; Halawa et al., 2019; Tokarek et al., 2021; Zheng et al., 2022). Guo et al. reported an increase in Firmicutes and Proteobacteria and a decrease in Bacteroidetes after arsenic exposure, which is consistent with changes observed here in the two lowest doses (Richardson et al., 2018). In the current study, the highest dose also showed an increase in Proteobacteria, but not in Firmicutes or Bacteroidetes populations (Richardson et al., 2018).
The comparative analysis among the three groups (control, LD, and HD) showed the differences at the phyla level. Firmicutes were dominant in the control groups, which follows the routine pattern of the majority of the gut bacteriome (Chi et al., 2017). The depletion of Firmicutes in LD and HD groups indicates the effect of arsenic on the firmicute population and its alterations, which follows the findings of previous studies (Dheer et al., 2015; Wu et al., 2019). Lu et al. (2014) also reported a significant decrease in Firmicutes due to arsenic exposure, which plays a significant role in affecting energy harvesting pathways and short-chain fatty acid (SCFA) production. Several Firmicutes are known for butyrate production (Tremaroli and Bäckhed, 2012; Lu et al., 2014). SCFA such as propionate, butyrate, and acetate acts as the primary energy source for gut epithelial cells and promotes the first line of cellular defense (Vinolo et al., 2011). The abundance of Bacteroidota was prevalent in the LD group and exhibited a cluster of carbohydrate-utilizing enzymes such as Firmicutes. It may be due to the low dose of arsenic exposure to the LD group (0.04 mg/kg) as compared to the HD group (0.4 mg/kg). Dheer et al. (2015) observed a positive correlation between Bacteroidetes count and a higher dose of arsenic. In the HD group, a drastic depletion in Firmicutes, Bacteroidetes, and Actinobacteria was observed due to a higher dose of arsenic exposure. Studies examining alterations in the microbial composition after arsenic exposure have shown results that appear to conflict. However, Guo et al. found that providing mice with water containing arsenic increased the abundance of Firmicutes and decreased the abundance of Bacteroidetes (Richardson et al., 2018). Proteobacteria are often overrepresented in several intestinal and extraintestinal diseases, mostly with an inflammatory phenotype (Mukhopadhya et al., 2012; Rizzatti et al., 2017). At the late stages of life, the microbiota composition becomes less diverse and more dynamic, characterized by a higher Bacteroides to Firmicutes ratio and an increase in Proteobacteria (Biagi et al., 2010). Similar findings were observed at the genera level, where the control group harbored a higher count of the genera Firmicutes and Bacteroidetes in the control and LD groups. An increase in Lactobacillus in the control group was observed, whereas in the arsenic-exposed groups (LD and HD), the Lactobacillus count was significantly decreased. Lactobacillus spp. are known for their protective role in arsenic-induced health damage (Sanders et al., 2019; Du et al., 2020). Lactobacillus may acquire arsenic resistance by reverting the oxidative stress and the production of pro-inflammatory cytokines (de Matuoka et al., 2020). Du et al. (2020) observed the restoration of Lactobacillus spp. in the gut microbiome after 30 days of arsenic exposure in mice. Lactobacillus spp. exhibit antimicrobial and antioxidative probiotic activity to stabilize the gut microbiome (Sanders et al., 2019; Wu et al., 2019). The LD and HD groups showed several pathogenic bacteria, such as Streptococcus, Prevotellaceae, Corynebacterium, and Enterococcus, that altered bacterial diversity due to arsenic doses in mice. Besides, the overall diversity of the LD and HD groups showed variation compared to the control group. A long-term arsenic exposure of 6 months in adults reduced the count of Muribaculum; however, this genus was not observed in arsenic-treated groups only (Wang et al., 2020). Similarly, the count of Dubosiella significantly decreased after 30 days of arsenic exposure. Here, we observed the presence of Dubosiella in the treated group (Wang et al., 2020). The physiology of Dubosiella is not well-defined and needs to be further explored. Enterococcus, another unique genus of the treated group, belongs to the lactic acid bacteria (LAB) family, comprising both pathogenic and commensal microorganisms ubiquitous in the environment even as gut symbionts (Hanchi et al., 2018). Due to their tolerance to salts and acids, the strains of Enterococcus spp. are highly adapted to several food systems (Hanchi et al., 2018). Several arsenic-resistant strains of Enterococcus are known to assist in coping with high metal environments (Abrantes et al., 2011; Parsons et al., 2020).
In adult studies of metal exposures, a dose-dependent response in the alteration of the gut microbial community has been observed (Dheer et al., 2015; Gokulan et al., 2018). However, in the current investigation, a dose-dependent response in the alteration of gut microbiota was not detected. Whereas in metabolic pathways, there is an increase in pathways involved in carbohydrate metabolism, translation, membrane transport, cellular community prokaryotes, endocrine system, and antimicrobial drug resistance in a dose-dependent manner. This might be due to adaptation, epigenetic modifications, and transfer of altered microbial communities from directly arsenic-exposed mothers to the offspring during the gestation period, which needs to be further confirmed. The depletion of Roseburia, Ruminococcaceae, Odoribacter, and Prevotella-like genera in the control group identified them as arsenic-sensitive genera. Prevotella constitute the core bacteriome of the human gut (Nam et al., 2011; Bhute et al., 2016) and participate extensively in hydrolysing high-fiber-based polysaccharides. Prevotella spp. exhibit a repertoire of carbohydrate hydrolysing complexes (Yeoh et al., 2022). Roseburia is associated with the effects of colonic motility, immunity maintenance, and anti-inflammatory properties (Shao and Zhu, 2020). Roseburia spp. could also serve as biomarkers for symptomatic pathologies (e.g., gallstone formation) or as probiotics for the restoration of beneficial flora (Tamanai-Shacoori et al., 2017). The studies on Prevotella state that this genus was found to be enriched in the treated group. Whereas in the current investigation, it was present in the control group only (Wang et al., 2020).
In another study, Ashutosh et al. reported that Prevotella can suppress disease through the modulation of systemic immune responses (Mangalam et al., 2017). In addition, it was absent in the arsenic-treated group, which can play a role in metabolic deregulation. Whereas, Odoribacter showed a significant increase in the presence of cadmium and aluminum exposure (Zhai et al., 2017), which is contradictory to the current investigation, needs to be further examined, and may be attributed to the indirect effect of arsenic on prenatally treated mice.
The PICRUSt-based functional prediction showed the dominance of metabolic pathways (50.05%), which follows the pattern of several microbiome-based investigations. As it was a gut microbiome-based investigation, several genes of carbohydrate and fatty acid metabolism were assessed. It was observed that most pathways (10) were not altered due to arsenic treatment either at lower or higher doses of arsenic. Only genes involved in non-alcoholic fatty liver diseases and insulin-signaling pathways showed a decrease in their relative counts in the treated group as compared to the control groups. Frediani et al. (2018) reported a positive association between NAFLD and arsenic exposure. However, this investigation was not a microbiome-based study and therefore cannot be truly correlated with insulin-signaling pathways.
Overall, we can conclude that only gestational arsenic exposure is sufficient to alter the gut microbial community in the progeny. The effects of prenatal exposure are manifested through an increased gene expression profile in the gut microbiota with most of the genes clustered in metabolism and its pathways. The outcomes of the study illustrate that arsenic exposure perturbs the gut microbiome composition and associated metabolic profiles in mice, which represents an early and crucial step in understanding how arsenic exposure affects the gut microbiome and its functions.
The data presented in the study are deposited in the NCBI repository, SRA accession number PRJNA929678.
The animal study was reviewed and approved by the Ethical Committee, CSIR-Indian Institute of Toxicology Research, Lucknow.
SS and DCS conceived and planned the presented idea. SS, SG, AC, SB, and VS performed animal experiments and sequencing. SS, AS, DV, and DCS performed sequencing analysis and wrote the paper with input from all authors. DT supervised the statistical analysis. All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
We acknowledge CSIR-Indian Institute of Toxicology Research for funding this Research (OLP-19 project) to VS.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1147505/full#supplementary-material
Supplementary Figure 1. The PICRUSt analysis of tier I level shows the distribution of ASVs into various pathways according to the KEGG database.
Supplementary Figure 2. The PICRUSt analysis tier II shows the ASVs distribution in metabolic pathways (A), genetic information processing (B), and environmental information processing (C).
Supplementary Figure 3. The PICRUSt analysis tier II shows the ASVs distribution in cellular processes (A), human diseases (B), and organismal systems (C).
Abrantes, M. C., de Fátima Lopes, M., and Kok, J. (2011). Impact of manganese, copper and zinc ions on the transcriptome of the nosocomial pathogen Enterococcus faecalis V583. PLoS ONE 6, 26519. doi: 10.1371/journal.pone.0026519
An, X., Bao, Q., Di, S., Zhao, Y., Zhao, S., Zhang, H., et al. (2019). The interaction between the gut Microbiota and herbal medicines. Biomed. Pharmacother. 118, 109252. doi: 10.1016/j.biopha.2019.109252
Bervoets, L., van Hoorenbeeck, K., Kortleven, I., van Noten, C., Hens, N., Vael, C., et al. (2013). Differences in gut microbiota composition between obese and lean children: a cross-sectional study. Gut Pathog. 5, 10. doi: 10.1186/1757-4749-5-10
Bhattacharya, P., Welch, A. H., Stollenwerk, K. G., McLaughlin, M. J., Bundschuh, J., and Panaullah, G. (2007). Arsenic in the environment: biology and chemistry. Sci. Tot. Environ. 379, 109–120. doi: 10.1016/j.scitotenv.2007.02.037
Bhute, S., Pande, P., Shetty, S. A., Shelar, R., Mane, S., Kumbhare, S. V., et al. (2016). Molecular characterization and meta-analysis of gut microbial communities illustrate enrichment of Prevotella and Megasphaera in Indian subjects. Front. Microbiol. 7, 660. doi: 10.3389/fmicb.2016.00660
Biagi, E., Nylund, L., Candela, M., Ostan, R., Bucci, L., Pini, E., et al. (2010). Through ageing, and beyond: gut microbiota and inflammatory status in seniors and centenarians. PLoS ONE 5, e7ec0607604d. doi: 10.1371/annotation/df45912f-d15c-44ab-8312-e7ec0607604d
Binda, C., Lopetuso, L. R., Rizzatti, G., Gibiino, G., Cennamo, V., and Gasbarrini, A. (2018). Actinobacteria: a relevant minority for the maintenance of gut homeostasis. Dig. Liver Dis. 50, 421–428. doi: 10.1016/j.dld.2018.02.012
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Brabec, J. L., Wright, J., Ly, T., Wong, H. T., McClimans, C. J., Tokarev, V., et al. (2020). Arsenic disturbs the gut microbiome of individuals in a disadvantaged community in Nepal. Heliyon 6, e03313. doi: 10.1016/j.heliyon.2020.e03313
Castriota, F., Acevedo, J., Ferreccio, C., Smith, A. H., Liaw, J., Smith, M. T., et al. (2018). Obesity and increased susceptibility to arsenic-related type 2 diabetes in Northern Chile. Environ. Res. 167, 248–254. doi: 10.1016/j.envres.2018.07.022
Chi, L., Bian, X., Gao, B., Tu, P., Ru, H., and Lu, K. (2017). The effects of an environmentally relevant level of arsenic on the gut microbiome and its functional metagenome. Toxicol. Sci. 160, 193. doi: 10.1093/toxsci/kfx174
Chi, L., Gao, B., Tu, P., Liu, C. W., Xue, J., Lai, Y., et al. (2018). Individual susceptibility to arsenic-induced diseases: the role of host genetics, nutritional status, and the gut microbiome. Mamm. Genome 29, 63–79. doi: 10.1007/s00335-018-9736-9
Choi, J. J., Eum, S. Y., Rampersaud, E., Daunert, S., Abreu, M. T., and Toborek, M. (2013). Exercise attenuates PCB-induced changes in the mouse gut microbiome. Environ. Health Perspect. 121, 725–730. doi: 10.1289/ehp.1306534
Coryell, M., McAlpine, M., Pinkham, N. V., McDermott, T. R., and Walk, S. T. (2018). The gut microbiome is required for full protection against acute arsenic toxicity in mouse models. Nat. Commun. 9, 5424. doi: 10.1038/s41467-018-07803-9
Crovesy, L., Masterson, D., and Rosado, E. L. (2020). Profile of the gut microbiota of adults with obesity: a systematic review. Eur. J. Clin. Nutr. 74, 1251–1262. doi: 10.1038/s41430-020-0607-6
Crovesy, L., Ostrowski, M., Ferreira, D. M. T. P., Rosado, E. L., and Soares-Mota, M. (2017). Effect of Lactobacillus on body weight and body fat in overweight subjects: a systematic review of randomized controlled clinical trials. Int. J. Obes. 41, 1607–1614. doi: 10.1038/ijo.2017.161
de Matuoka, E., Chiocchetti, G., Monedero, V., Zúñiga, M., Vélez, D., and Devesa, V. (2020). In-vitro evaluation of the protective role of Lactobacillus strains against inorganic arsenic toxicity. Probiot. Antimicrob. Proteins 6, 1484–1491. doi: 10.1007/s12602-020-09639-6
Dheer, R., Patterson, J., Dudash, M., Stachler, E. N., Bibby, K. J., Stolz, D. B., et al. (2015). Arsenic induces structural and compositional colonic microbiome change and promotes host nitrogen and amino acid metabolism. Toxicol. Appl. Pharmacol. 289, 397–408. doi: 10.1016/j.taap.2015.10.020
Douglas, G. M., Maffei, V. J., Zaneveld, J. R., Yurgel, S. N., Brown, J. R., Taylor, C. M., et al. (2020). PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 38, 685–688. doi: 10.1038/s41587-020-0548-6
Du, X., Zhang, J., Schramm, K.-W., Huang, Q., Tian, M., Zhang, X., et al. (2020). Persistence and reversibility of arsenic-induced gut microbiome and metabolome shifts in rats after 30-days recovery duration. ResearchSquare 2020, 25. doi: 10.21203/rs.3.rs-29645/v1
Frediani, J. K., Naioti, E. A., Vos, M. B., Figueroa, J., Marsit, C. J., and Welsh, J. A. (2018). Arsenic exposure and risk of nonalcoholic fatty liver disease (NAFLD) among U.S. adolescents and adults: an association modified by race/ethnicity, NHANES 2005-2014. Environ. Health 1, 17. doi: 10.1186/s12940-017-0350-1
Gentile, C. L., and Weir, T. L. (2018). The gut microbiota at the intersection of diet and human health. Science 362, 776–780. doi: 10.1126/science.aau5812
Gokulan, K., Arnold, M. G., Jensen, J., Vanlandingham, M., Twaddle, N. C., Doerge, D. R., et al. (2018). Exposure to arsenite in CD-1 mice during juvenile and adult stages: effects on intestinal microbiota and gut-associated immune status. mBio 9, 18. doi: 10.1128/mBio.01418-18
Grau-Perez, M., Navas-Acien, A., Galan-Chilet, I., Briongos-Figuero, L. S., Morchon-Simon, D., Bermudez, J. D., et al. (2018). Arsenic exposure, diabetes-related genes and diabetes prevalence in a general population from Spain. Environ. Pollut. 235, 948–955. doi: 10.1016/j.envpol.2018.01.008
Gribble, M. O., Howard, B., Umans, J. G., Shara, N. M., Francesconi, K. A., Goessler, W., et al. (2012). Arsenic exposure, diabetes prevalence, and diabetes control in the Strong Heart Study. Am. J. Epidemiol. 176, 865–874. doi: 10.1093/aje/kws153
Guglielmi, G. (2018). How gut microbes are joining the fight against cancer. Nature 557, 482–484. doi: 10.1038/d41586-018-05208-8
Halawa, M. R., El-Salam, M. A., Mostafa, B. M., and Sallout, S. S. (2019). The gut microbiome, Lactobacillus acidophilus; relation with type 2 diabetes mellitus. Curr. Diabetes Rev. 15, 480–485. doi: 10.2174/1573399815666190206162143
Hanchi, H., Mottawea, W., Sebei, K., and Hammami, R. (2018). The genus enterococcus: between probiotic potential and safety concerns-an update. Front. Microbiol. 9, 1791. doi: 10.3389/fmicb.2018.01791
Harakeh, S. M., Khan, I., Kumosani, T., Barbour, E., Almasaudi, S. B., Bahijri, S. M., et al. (2016). Gut microbiota: a contributing factor to obesity. Front. Cell Infect. Microbiol. 6, 95. doi: 10.3389/fcimb.2016.00095
Hughes, M. F., Beck, B. D., Chen, Y., Lewis, A. S., and Thomas, D. J. (2011). Arsenic exposure and toxicology: a historical perspective. Toxicol. Sci. 123, kfr184. doi: 10.1093/toxsci/kfr184
Hur, K. Y., and Lee, M. S. (2015). Gut microbiota and metabolic disorders. Diabetes Metab. J. 39, 198–203. doi: 10.4093/dmj.2015.39.3.198
IARC Working Group on the Evaluation of Carcinogenic Risks to Humans (2012). Arsenic, metals, fibres, and dusts. IARC Monogr. Eval. Carcinog. Risks Hum. 100, 11–465. Available online at: https://publications.iarc.fr/Book-And-Report-Series/Iarc-Monographs-On-The-Identification-Of-Carcinogenic-Hazards-To-Humans/Arsenic-Metals-Fibres-And-Dusts-2012
Jandhyala, S. M., Talukdar, R., Subramanyam, C., Vuyyuru, H., Sasikala, M., and Nageshwar Reddy, D. (2015). Role of the normal gut microbiota. World J. Gastroenterol. 21, 8787–8803. doi: 10.3748/wjg.v21.i29.8787
Ley, R. E., Bäckhed, F., Turnbaugh, P., Lozupone, C. A., Knight, R. D., and Gordon, J. I. (2005). Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. U. S. A. 102, 11070–11075. doi: 10.1073/pnas.0504978102
Ley, R. E., Peterson, D. A., and Gordon, J. I. (2006a). Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 124, 837–848. doi: 10.1016/j.cell.2006.02.017
Ley, R. E., Turnbaugh, P. J., Klein, S., and Gordon, J. I. (2006b). Microbial ecology: human gut microbes associated with obesity. Nature 444, 1022–1023. doi: 10.1038/4441022a
Liebert, C. A., Wireman, J., Smith, T., and Summers, A. O. (1997). Phylogeny of mercury resistance (mer) operons of gram-negative bacteria isolated from the fecal flora of primates. Appl. Environ. Microbiol. 63, 1066–1076. doi: 10.1128/aem.63.3.1066-1076.1997
Lu, K., Abo, R. P., Schlieper, K. A., Graffam, M. E., Levine, S., Wishnok, J. S., et al. (2014). Arsenic exposure perturbs the gut microbiome and its metabolic profile in mice: an integrated metagenomics and metabolomics analysis. Environ. Health Perspect. 122, 284–291. doi: 10.1289/ehp.1307429
Magne, F., Gotteland, M., Gauthier, L., Zazueta, A., Pesoa, S., Navarrete, P., et al. (2020). The firmicutes/bacteroidetes ratio: a relevant marker of gut dysbiosis in obese patients? Nutrients 12, 51474. doi: 10.3390/nu12051474
Mangalam, A., Shahi, S. K., Luckey, D., Karau, M., Marietta, E., Luo, N., et al. (2017). Human gut-derived commensal bacteria suppress CNS inflammatory and demyelinating disease. Cell. Rep. 20, 1269–1277. doi: 10.1016/j.celrep.2017.07.031
Maull, E. A., Ahsan, H., Edwards, J., Longnecker, M. P., Navas-Acien, A., Pi, J., et al. (2012). Evaluation of the association between arsenic and diabetes: a National Toxicology Program workshop review. Environ. Health Perspect. 120, 1104579. doi: 10.1289/ehp.1104579
Mukhopadhya, I., Hansen, R., El-Omar, E. M., and Hold, G. L. (2012). IBD-what role do Proteobacteria play? Nat. Rev. Gastroenterol. Hepatol. 9, 219–230. doi: 10.1038/nrgastro.2012.14
Nam, Y. D., Jung, M. J., Roh, S. W., Kim, M. S., and Bae, J. W. (2011). Comparative analysis of Korean human gut microbiota by barcoded pyrosequencing. PLoS ONE 6, e22109. doi: 10.1371/journal.pone.0022109
Naujokas, M. F., Anderson, B., Ahsan, H., Aposhian, H. V., Graziano, J. H., Thompson, C., et al. (2013). The broad scope of health effects from chronic arsenic exposure: update on a worldwide public health problem. Environ. Health Perspect. 121, 1205875. doi: 10.1289/ehp.1205875
Nicholson, J. K., Holmes, E., Kinross, J., Burcelin, R., Gibson, G., Jia, W., et al. (2012). Host-gut microbiota metabolic interactions. Science 336, 1262–1267. doi: 10.1126/science.1223813
Parks, D. H., Tyson, G. W., Hugenholtz, P., and Beiko, R. G. (2014). STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics 30, 3123–3124. doi: 10.1093/bioinformatics/btu494
Parsons, C., Lee, S., and Kathariou, S. (2020). Dissemination and conservation of cadmium and arsenic resistance determinants in Listeria and other Gram-positive bacteria. Mol. Microbiol. 113, 560–569. doi: 10.1111/mmi.14470
Paul, D. S., Walton, F. S., Saunders, R. J., and Stýblo, M. (2011). Characterization of the impaired glucose homeostasis produced in C57BL/6 mice by chronic exposure to arsenic and high-fat diet. Environ. Health Perspect. 119, 1003324. doi: 10.1289/ehp.1003324
Pinyayev, T. S., Kohan, M. J., Herbin-Davis, K., Creed, J. T., and Thomas, D. J. (2011). Preabsorptive metabolism of sodium arsenate by anaerobic microbiota of mouse cecum forms a variety of methylated and thiolated arsenicals. Chem. Res. Toxicol. 24, 475–477. doi: 10.1021/tx200040w
Qin, J., Li, R., Raes, J., Arumugam, M., Burgdorf, K. S., Manichanh, C., et al. (2010). A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65. doi: 10.1038/nature08821
Qin, J., Li, Y., Cai, Z., Li, S., Zhu, J., Zhang, F., et al. (2012). A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490, 55–60. doi: 10.1038/nature11450
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucl. Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Richardson, J. B., Dancy, B. C. R., Horton, C. L., Lee, Y. S., Madejczyk, M. S., Xu, Z. Z., et al. (2018). Exposure to toxic metals triggers unique responses from the rat gut microbiota. Sci. Rep. 8, 24931. doi: 10.1038/s41598-018-24931-w
Rizzatti, G., Lopetuso, L. R., Gibiino, G., Binda, C., and Gasbarrini, A. (2017). Proteobacteria: a common factor in human diseases. Biomed. Res. Int. 2017, 9351507. doi: 10.1155/2017/9351507
Roderburg, C., and Luedde, T. (2014). The role of the gut microbiome in the development and progression of liver cirrhosis and hepatocellular carcinoma. Gut. Microbes 5, 441–445. doi: 10.4161/gmic.29599
Sanders, M. E., Merenstein, D. J., Reid, G., Gibson, G. R., and Rastall, R. A. (2019). Probiotics and prebiotics in intestinal health and disease: from biology to the clinic. Nat. Rev. Gastroenterol. Hepatol. 16, 605–616. doi: 10.1038/s41575-019-0173-3
Shao, M., and Zhu, Y. (2020). Long-term metal exposure changes gut microbiota of residents surrounding a mining and smelting area. Sci. Rep. 10, 7. doi: 10.1038/s41598-020-61143-7
Sharma, V., Gangopadhyay, S., Shukla, S., Chauhan, A., Singh, S., Singh, R. D., et al. (2022). Prenatal exposure to arsenic promotes sterile inflammation through the Polycomb repressive element EZH2 and accelerates skin tumorigenesis in mouse. Toxicol. Appl. Pharmacol. 443, 116004. doi: 10.1016/j.taap.2022.116004
Tamanai-Shacoori, Z., Smida, I., Bousarghin, L., Loreal, O., Meuric, V., Fong, S. B., et al. (2017). Roseburia spp.: a marker of health? Fut. Microbiol. 12, 157–170. doi: 10.2217/fmb-2016-0130
Thursby, E., and Juge, N. (2017). Introduction to the human gut microbiota. Biochem. J. 474, 1823–1836. doi: 10.1042/BCJ20160510
Tokarek, J., Gadzinowska, J., Młynarska, E., Franczyk, B., and Rysz, J. (2021). What is the role of gut microbiota in obesity prevalence? A few words about gut microbiota and its association with obesity and related diseases. Microorganisms 10, 10052. doi: 10.3390/microorganisms10010052
Tremaroli, V., and Bäckhed, F. (2012). Functional interactions between the gut microbiota and host metabolism. Nature 489, 242–249. doi: 10.1038/nature11552
Turnbaugh, P. J., Ley, R. E., Mahowald, M. A., Magrini, V., Mardis, E. R., and Gordon, J. I. (2006). An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444, 1027–1031. doi: 10.1038/nature05414
van de Wiele, T., Gallawa, C. M., Kubachka, K. M., Creed, J. T., Basta, N., Dayton, E. A., et al. (2010). Arsenic metabolism by human gut microbiota upon in vitro digestion of contaminated soils. Environ. Health Perspect. 118, 1004–1009. doi: 10.1289/ehp.0901794
van de Wiele, T., Vanhaecke, L., Boeckaert, C., Peru, K., Headley, J., Verstraete, W., et al. (2005). Human colon microbiota transform polycyclic aromatic hydrocarbons to estrogenic metabolites. Environ. Health Perspect. 113, 6–10. doi: 10.1289/ehp.7259
Vinolo, M. A. R., Rodrigues, H. G., Nachbar, R. T., and Curi, R. (2011). Regulation of inflammation by short chain fatty acids. Nutrients 3, 858–876. doi: 10.3390/nu3100858
Wang, J., Hu, W., Yang, H., Chen, F., Shu, Y., Zhang, G., et al. (2020). Arsenic concentrations, diversity and co-occurrence patterns of bacterial and fungal communities in the feces of mice under sub-chronic arsenic exposure through food. Environ. Int. 138, 105600. doi: 10.1016/j.envint.2020.105600
Wang, Z., Klipfell, E., Bennett, B. J., Koeth, R., Levison, B. S., Dugar, B., et al. (2011). Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 472, 57–63. doi: 10.1038/nature09922
Wu, F., Yang, L., Islam, M. T., Jasmine, F., Kibriya, M. G., Nahar, J., et al. (2019). The role of gut microbiome and its interaction with arsenic exposure in carotid intima-media thickness in a Bangladesh population. Environ. Int. 123, 104–113. doi: 10.1016/j.envint.2018.11.049
Yang, Y., Chi, L., Lai, Y., Hsiao, Y. C., Ru, H., and Lu, K. (2021). The gut microbiome and arsenic-induced disease-iAs metabolism in mice. Curr. Environ. Health Rep. 8, 89–97. doi: 10.1007/s40572-021-00305-9
Yao, Y., Cai, X., Ye, Y., Wang, F., Chen, F., and Zheng, C. (2021). The role of microbiota in infant health: from early life to adulthood. Front. Immunol. 12, 708472. doi: 10.3389/fimmu.2021.708472
Yeoh, Y. K., Sun, Y., Ip, L. Y. T., Wang, L., Chan, F. K. L., Miao, Y., et al. (2022). Prevotella species in the human gut is primarily comprised of Prevotella copri, Prevotella stercorea and related lineages. Sci. Rep. 12, 9055. doi: 10.1038/s41598-022-12721-4
Young, V. B., Britton, R. A., and Schmidt, T. M. (2008). The human microbiome and infectious diseases: beyond koch. Interdiscip. Perspect. Infect. Dis. 2008, 296873. doi: 10.1155/2008/296873
Zhai, Q., Li, T., Yu, L., Xiao, Y., Feng, S., Wu, J., et al. (2017). Effects of subchronic oral toxic metal exposure on the intestinal microbiota of mice. Sci. Bull. 62, 831–840. doi: 10.1016/j.scib.2017.01.031
Zhang, J., Guo, Z., Xue, Z., Sun, Z., Zhang, M., Wang, L., et al. (2015a). A phylo-functional core of gut microbiota in healthy young Chinese cohorts across lifestyles, geography and ethnicities. ISME J. 9, 1979–1990. doi: 10.1038/ismej.2015.11
Zhang, S., Jin, Y., Zeng, Z., Liu, Z., and Fu, Z. (2015b). Subchronic exposure of mice to cadmium perturbs their hepatic energy metabolism and gut microbiome. Chem. Res. Toxicol. 28, 2000–2009. doi: 10.1021/acs.chemrestox.5b00237
Keywords: in-utero arsenic exposure, bacteriome, arsenic, 16S rRNA gene, insulin signaling
Citation: Shukla S, Srivastava A, Verma D, Gangopadhyay S, Chauhan A, Srivastava V, Budhwar S, Tyagi D and Sharma DC (2023) Analysis of gut bacteriome of in utero arsenic-exposed mice using 16S rRNA-based metagenomic approach. Front. Microbiol. 14:1147505. doi: 10.3389/fmicb.2023.1147505
Received: 23 January 2023; Accepted: 31 July 2023;
Published: 29 September 2023.
Edited by:
Jose Manuel Miranda, University of Santiago de Compostela, SpainReviewed by:
Alinne Castro, Dom Bosco Catholic University, BrazilCopyright © 2023 Shukla, Srivastava, Verma, Gangopadhyay, Chauhan, Srivastava, Budhwar, Tyagi and Sharma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Deepak Chand Sharma, ZGNzaGFybWFAZHNtbnJ1LmFjLmlu; ZGNzaGFybWFAZ21haWwuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.