- 1State Key Laboratory of Materials-Oriented Chemical Engineering, College of Food Science and Light Industry, Nanjing Tech University, Nanjing, China
- 2Engineering Laboratory for Industrial Microbiology Molecular Beeding of Anhui Province, College of Biologic and Food Engineering, Anhui Polytechnic University, Wuhu, China
Cofactors are crucial chemicals that maintain cellular redox balance and drive the cell to do synthetic and catabolic reactions. They are involved in practically all enzymatic activities that occur in live cells. It has been a hot research topic in recent years to manage their concentrations and forms in microbial cells by using appropriate techniques to obtain more high-quality target products. In this review, we first summarize the physiological functions of common cofactors, and give a brief overview of common cofactors acetyl coenzyme A, NAD(P)H/NAD(P)+, and ATP/ADP; then we provide a detailed introduction of intracellular cofactor regeneration pathways, review the regulation of cofactor forms and concentrations by molecular biological means, and review the existing regulatory strategies of microbial cellular cofactors and their application progress, to maximize and rapidly direct the metabolic flux to target metabolites. Finally, we speculate on the future of cofactor engineering applications in cell factories.
Introduction
Traditional petrochemical manufacturing has several disadvantages, the most evident of which is irreversible environmental contamination, which is a significant breach of the original aim of harmonious cohabitation between humans and nature. Therefore, bioconversion, which allows the production of chemical products from renewable carbon sources, has become an ideal technology (Bruschi et al., 2011). With the booming development of synthetic biology and metabolic engineering technologies in recent years, the application of microbial cell factories to manufacture diverse high-value-added compounds has become a research hotspot (Zhang et al., 2018; Liu et al., 2021b). Artemisinin (Paddon et al., 2013), vinblastine (Zhang J. et al., 2022), ginsenoside (Wang P. et al., 2019), hyaluronic acid (Wang Y. et al., 2020) polyketides(Wang W. et al., 2020), and squalene (Zhu et al., 2021) have all been full-synthesized or semi-synthesized by researchers.
Cofactors are frequently required for cell-based biotransformation processes (Duetz et al., 2001; Drepper et al., 2006), proteases have a chemical structure that comprises polypeptides and certain non-protein structures, and one refers to these non-protein structures such as metal ions, metal–organic/inorganic complexes, and organic small molecules collectively as cofactors. Cofactors are required for enzymes to fulfill their catalytic functions, and they combine with the original enzyme to generate the entire enzyme, which then performs the enzyme’s normal physiological responsibilities (McNaught and Wilkinson, 1997). Common cofactors include acetyl coenzyme A (acetyl-CoA), NAD(P)H/NAD(P)+, and ATP/ADP, of NAD(P)H/NAD(P)+ and ATP/ADP-dependent metabolic reactions involving up to 1,610 enzymes such as transferases, oxidoreductases, lyases, ligases, isomerase, and hydrolases. Acetyl-CoA, NAD(P)H/NAD(P)+, and ATP/ADP play direct roles in the metabolism and transport of sugars, proteins, and lipids (Liu et al., 2018), which in turn affect cellular physiological functions. When these cofactors are created and consumed by cellular metabolism, their redox state is disrupted, resulting in consequences such as sluggish cell growth and decreased biosynthesis. Adjusting the concentration and form of cofactors can alter the distribution of material metabolic flux, push metabolism toward maximum target products, accelerate glycolysis, and preserve the complex intracellular structure (Selles et al., 2018). Modification strategies such as modification of endogenous or exogenous cofactor metabolic pathways, conversion between cofactors, and fine-tuning of transcription factors (Wang et al., 2017) can effectively maintain intracellular redox balance, and cofactor engineering has emerged as a powerful tool for increasing production capacity (Li et al., 2018). In this article, we summarize the physiological functions of common cofactors as well as the existing regulatory strategies of microbial cellular cofactors, as well as the progress of their applications, to serve as a reference for the efficient synthesis of target metabolites and to look ahead to the future development of cofactor engineering applications in cell factories.
Cofactor physiological functions
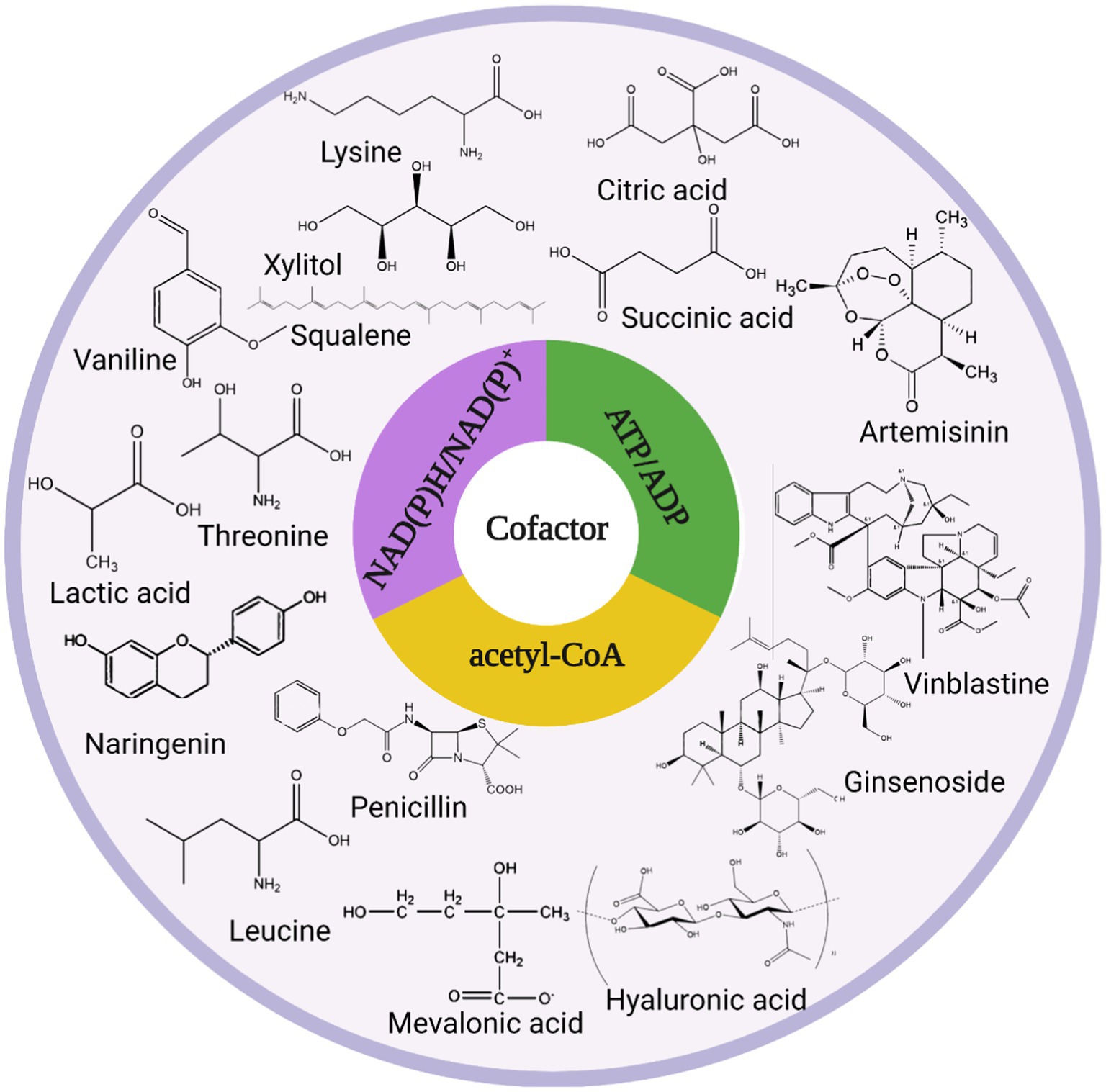
The intracellular redox state is closely related to cofactors, which can influence cellular metabolism, signal transduction, and material transport through cofactor regeneration, thereby affecting cellular physiological functions, and cofactors are also carriers of biological redox reactions and important factors in energy transfer (Wang et al., 2013). Cofactors can be divided into three broad categories based on their chemical structure and their role in enzyme-catalyzed reactions as described below: (i) catalytic cofactors, which are found in the active center of the enzyme and work together to catalyze the reaction. (ii) carrier cofactors, which are frequently used as carriers of electrons and atoms. (iii) substrate cofactors, which serve as raw materials for the synthesis of certain specific biological small molecular compounds and decrease as the reaction proceeds (Kulkarni and Deshpande, 2007). This article introduces three cofactors that play important roles in microbial cell metabolism: acetyl-CoA, NAD(P)H/NAD(P)+, and ATP/ADP, and their metabolic pathway in microbial cell metabolism is shown in Figure 1.
Acetyl coenzyme A
The acetyl-CoA synthetic pathway connects the various metabolic reactions occurring in the cytoplasm, nucleus, and mitochondria and can provide the cell with both a carbon source and energy (Wang J. et al., 2020). Second, acetyl-CoA serves as a precursor for the synthesis of isoprenoids, fatty acids and their derivatives, terpenoids, flavonoids, polyketides, etc. For example, acetyl-CoA generates isoprene and terpenoids through the MVA pathway (Liu H. et al., 2020). Acetyl-CoA can modify post-translational proteins and regulate cellular protein biological activity and stability (Su et al., 2016). Acetyl-CoA maintains the balance between cell proliferation and apoptosis by acting as both a metabolic intermediate and a second messenger (Wagner et al., 2017).
NAD(P)H/NAD(P)+
Intracellular NADH is mainly derived from glycolysis, fatty acid oxidation, and the tricarboxylic acid cycle, and pyruvate is converted to acetyl-CoA and NADH under aerobic conditions catalyzed by the pyruvate dehydrogenase complex. NAD(P)H/NAD(P)+ has a wide range of functions and can participate in approximately 1,500 enzymatic reactions in microbial metabolism, more than 100 of which have been linked to NADPH (Lee et al., 2013). NAD(P)H and NAD(P)+ play important roles in microbial cells as electron donors and acceptors, generating energy through electron transfer and participating in aerobic respiratory fermentation (Xu et al., 2020). Under aerobic conditions, NADH generates ATP via the electron transport chain (Curtabbi and Enriquez, 2022). Under anaerobic conditions, NADH is oxidized in the presence of acetaldehyde dehydrogenase and lactate dehydrogenase, when ATP is produced mainly through phosphorylation at the substrate level. NADPH homeostasis is regulated by a variety of signaling pathways and several metabolic enzymes that occur when cancer cells are altered and regulating NADPH in cancer cells is beneficial for cancer cell elimination (Pietrocola et al., 2015).
ATP/ADP
Substrate-level phosphorylation is an important pathway for ATP production, especially when oxidative phosphorylation is restricted. Two moles of ATP are produced per mole of glucose in the glycolytic pathway catalyzed by glycerol-3-acid phosphokinase and pyruvate kinase. The cofactor ATP/ADP, which is generated by substrate-level and oxidative phosphorylation, can enter the metabolic network of microorganisms in a variety of forms such as substrates, products, activators, and inhibitors, control the physiological functions of the cell and contribute to the formation of the cytoskeletal system (Zhou et al., 2009). ATP can power almost all cells, and sufficient ATP must be created to allow normal cell biosynthesis and cell maintenance (Yu et al., 2018). Aspergillus niger (A. niger) strain DS03043 is capable of producing glucoamylase, and 62% of the total ATP produced in its cells is used for biomass formation, while the remaining 27% may be used for non-growth maintenance (Lu et al., 2015). ATP can regulate the rate of cellular metabolism. The rate of glycolysis is determined by the demand for total cellular ATP rather than the expression of glycolysis-related enzymes (Hackett et al., 2016). The activity of essential enzymes in the tricarboxylic acid cycle is inhibited when ATP concentration is too high (Wang W. et al., 2020). ATP is a metabolic activator of lactate dehydrogenase (Willquist and van Niel, 2010).
Regulation of cofactor metabolic balance and its application
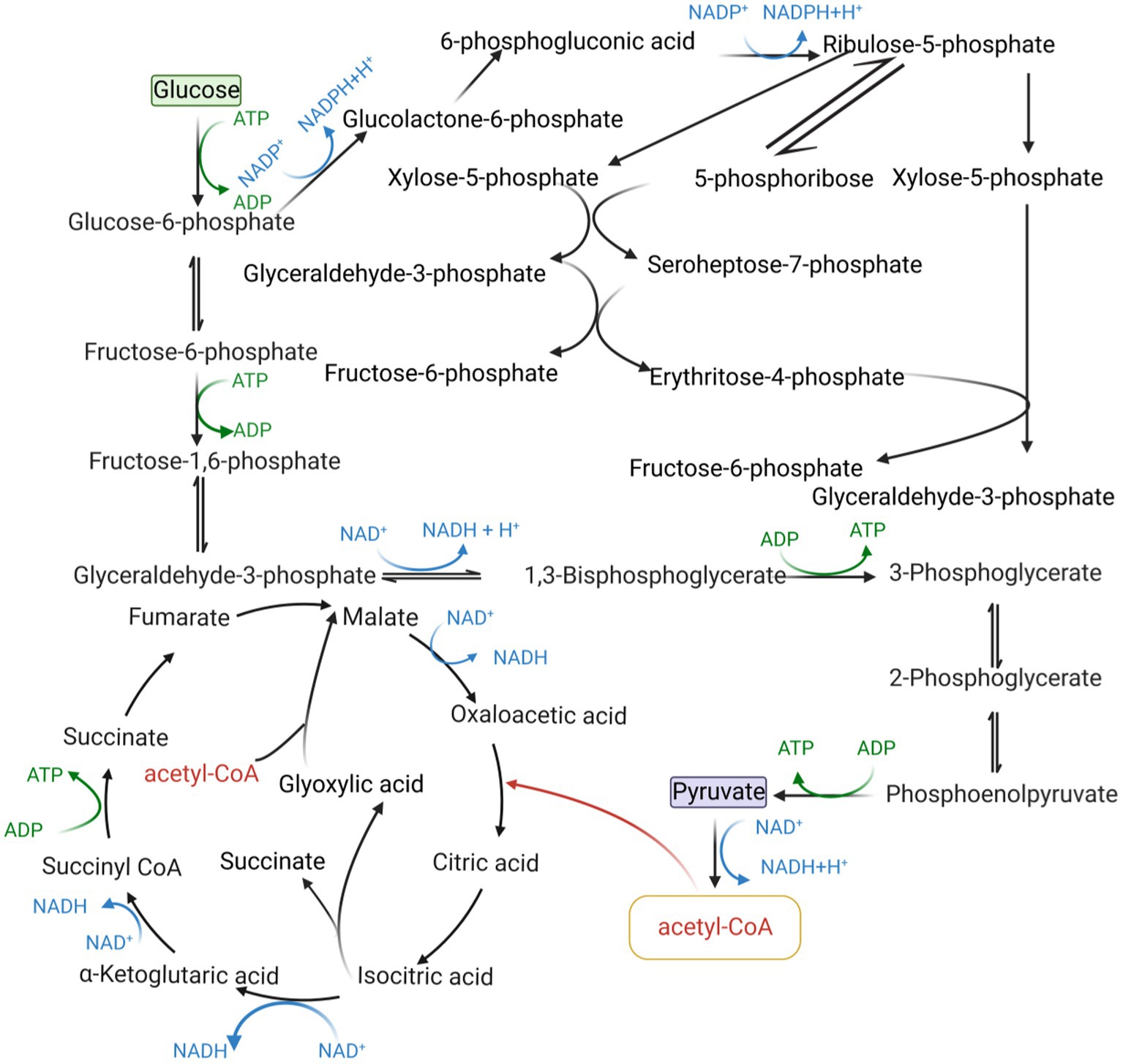
Metabolic engineering is based on intracellular biochemical reactions that regulate the concentration and form of the cellular metabolic network to maximize the production of the target product. The flow of product generation in microbial metabolism is shown in Figure 2.
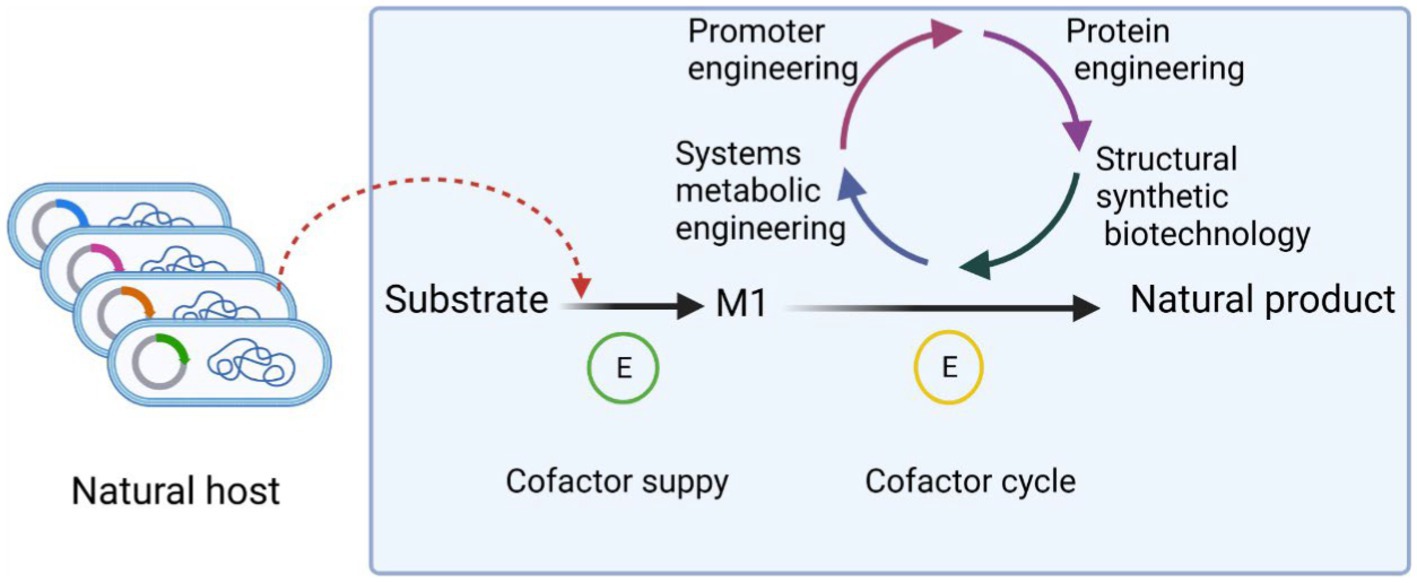
Figure 2. Process of product production in microbial metabolism. Cofactors play an important role in biosynthesis. First of all, the substrate was used to generate M1, which is necessary for the growth of microorganisms, through the combined action of cofactors, etc. The precursor M1 is then used as the precursor to synthesize substances with more complex chemical structure and no obvious physiological function for the microorganism. The cofactors of the whole synthesis process can be regulated by promoter engineering, protein engineering, structural synthetic biotechnology, system metabolic engineering and so on. These strategies make cofactor transition quickly and help to improve the cofactors balance.
Acetyl coenzyme A
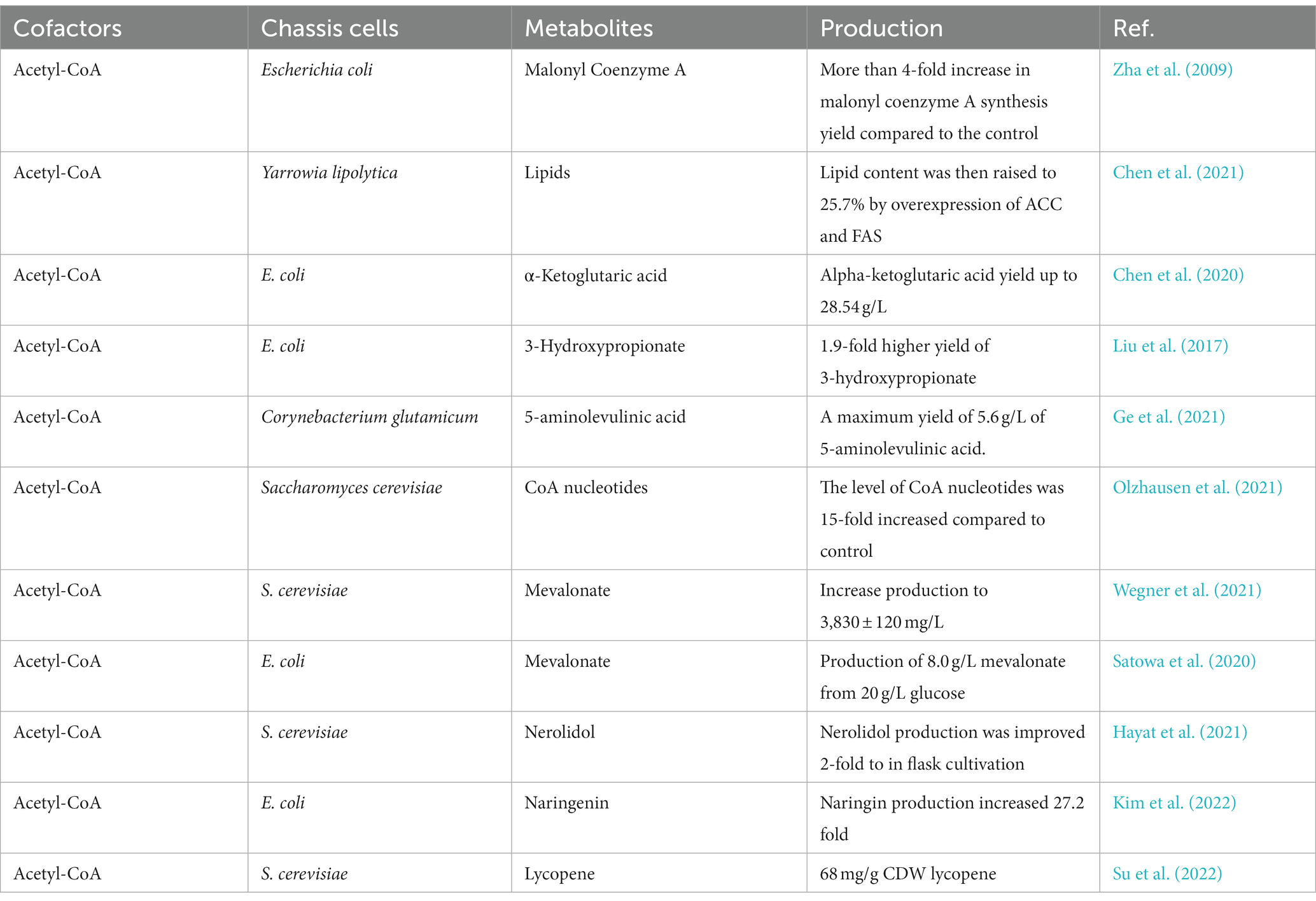
Acetyl-CoA, as an intermediary product of cellular metabolism, serves as a critical hub in microbial metabolism and is connected to the glycolytic, TCA cycle, amino acid, and fatty acid synthesis pathways (Farmer and Liao, 1997; Sagmeister et al., 2014). The most common current regulatory strategy regarding acetyl-CoA is to regulate it through the acetate pathway. This section focuses on acetyl-CoA’s metabolic regulatory strategy and its application in biosynthesis. Some of the regulatory strategies of acetyl-CoA and their applications are listed in Table 1.
Regulation of acetic acid metabolic pathway
Acetyl-CoA and acetic acid can be converted to each other, acetic acid is catalyzed by acetyl-CoA synthase (ACS) to generate acetyl-CoA, while acetyl-CoA is catalyzed by phosphoryl acetyltransferase and acetic acid kinase to produce acetic acid. By overexpressing ACS in E. coli, Zha et al. (2009) not only reduced acetic acid accumulation but also enhanced the metabolic flux of acetyl-CoA to the target product. Chen X. et al. (2020) overexpressed ACS to improve the availability of acetyl-CoA, resulting in α-keto glutaric acid production of up to 28.54 g/L. Chen et al. (2021) accumulated 9.2% of microbial lipids from acetate in shake flask fermentation by overexpressing ACS in Yarrowia lipolytica (Y. lipolytica). The lipid content was then raised to 25.7% by overexpression of acetyl-CoA carboxylase (ACC) and fatty acid synthase (FAS) in Y. lipolytica.
Regulation of glyoxalate metabolic pathway
Isocitrate lyase, isocitrate dehydrogenase, and malate synthase are key enzymes in the glyoxylate cycle. IclR can inhibit the expression of the aceBAK operon, which encodes these three enzymes. The iclR gene was knocked out, which activated the glyoxylate branch and enhanced the acetyl-CoA synthesis metabolic flux, resulting in a 1.9-fold increase in the yield of 3-hydroxypropionate with acetyl-CoA as a precursor (Liu et al., 2017). Ge et al. (2021) obtained 3.86 g/L of 5-aminolevulinic acid in Corynebacterium glutamicum (C. glutamicum) by alleviating the competition between glyoxylate and TCA cycle and knocking out the gene encoding isocitrate dehydrogenase, with a maximum yield of 5.6 g/L of 5-aminolevulinic acid after optimization of fermentation conditions.
Other metabolic regulatory pathways
Acetyl-CoA, as an important hub of biosynthesis, can be regulated in a variety of ways such as overexpression of related pathway genes, modification of related pathway enzymes, the introduction of exogenous metabolic pathways, and redistribution of metabolic fluxes in order to obtain more target products.
Zhang M. et al. (2022) expressed the phosphoketolase encoding gene (pK) and the pyruvate decarboxylase encoding gene (pdC) in a glucose-suppressed mutant and fermented it for 168 h in a 10 L bioreactor, generating 55.38 g/L of liamocin and 25.10 g/L of cell dry weight from 117.27 g/L of glucose. Olzhausen et al. (2021) increased acetyl-CoA biosynthesis in Saccharomyces cerevisiae (S. cerevisiae) by overexpressing a dysregulated pantothenic acid kinase gene and engineering the coenzyme A biosynthetic pathway. Wegner et al. (2021) increased coenzyme A biosynthesis by overexpression of pantothenic acid kinase and pantothenic acid supplementation, further increasing production to 3,830 ± 120 mg/L. Yu et al. (2021) enhanced γ-aminobutyric acid production in E. coli BL21 by modifying the enzyme of the coenzyme factor pyridoxal 5′-phosphate regeneration pathway.
Satowa et al. (2020) disrupted citrate synthase expression and reduced flux to the tricarboxylic acid cycle, resulting in intracellular acetyl-CoA levels 7-fold higher than in the wild-type strain. This strain produced 8.0 g/L mevalonate from 20 g/L glucose. Zhu et al. (2021) introduced the partial MVA pathway of mevalonate in acetyl-CoA mitochondria and increased mevalonate synthesis in the cytoplasm, which resulted in the squalene titer of 21.1 g/L with a specific squalene titer of 437.1 mg/g dcw. Hayat et al. (2021) introduced heterologous acetylating aldehyde dehydrogenase and phosphoketolase pathways for acetyl-CoA synthesis to enhance acetyl-CoA production to increase the yield of the nerolidol.
By fine-tuning the expression of pckA (encoding phosphoenolpyruvate carboxykinase) and redistributing flow between naringenin biosynthesis and cell growth at the isocitrate node, Kim et al. (2022) avoided massive loss of oxaloacetate. When compared to the unoptimized strain, the flux-optimized strain produced 97.02 mg/L naringenin with 21.02 mg naringenin/g acetate, a 27.2-fold increase in naringenin production (and a 38.3-fold increase in acetate production). Su et al. (2022) developed a novel strategy to balance acetyl-CoA metabolism and increase the amount of downstream products. First, the combination of acetaldehyde dehydrogenase and acetyl-CoA thiolase was optimized to redirect acetyl-CoA flux to the target pathway, resulting in a 21-fold increase in mevalonate production. Secondly, growth defects were mitigated by pathway engineering and evolutionary engineering to achieve a 10-fold increase in maximum productivity. ACC was then dynamically down-regulated as a complementary acetyl-CoA pathway, resulting in a more than 2-fold increase in yield. Finally combining the most efficient and complementary acetyl-CoA pathway, the final strain produced 68 mg/g CDW lycopene.
NAD(P)H/NAD(P)+
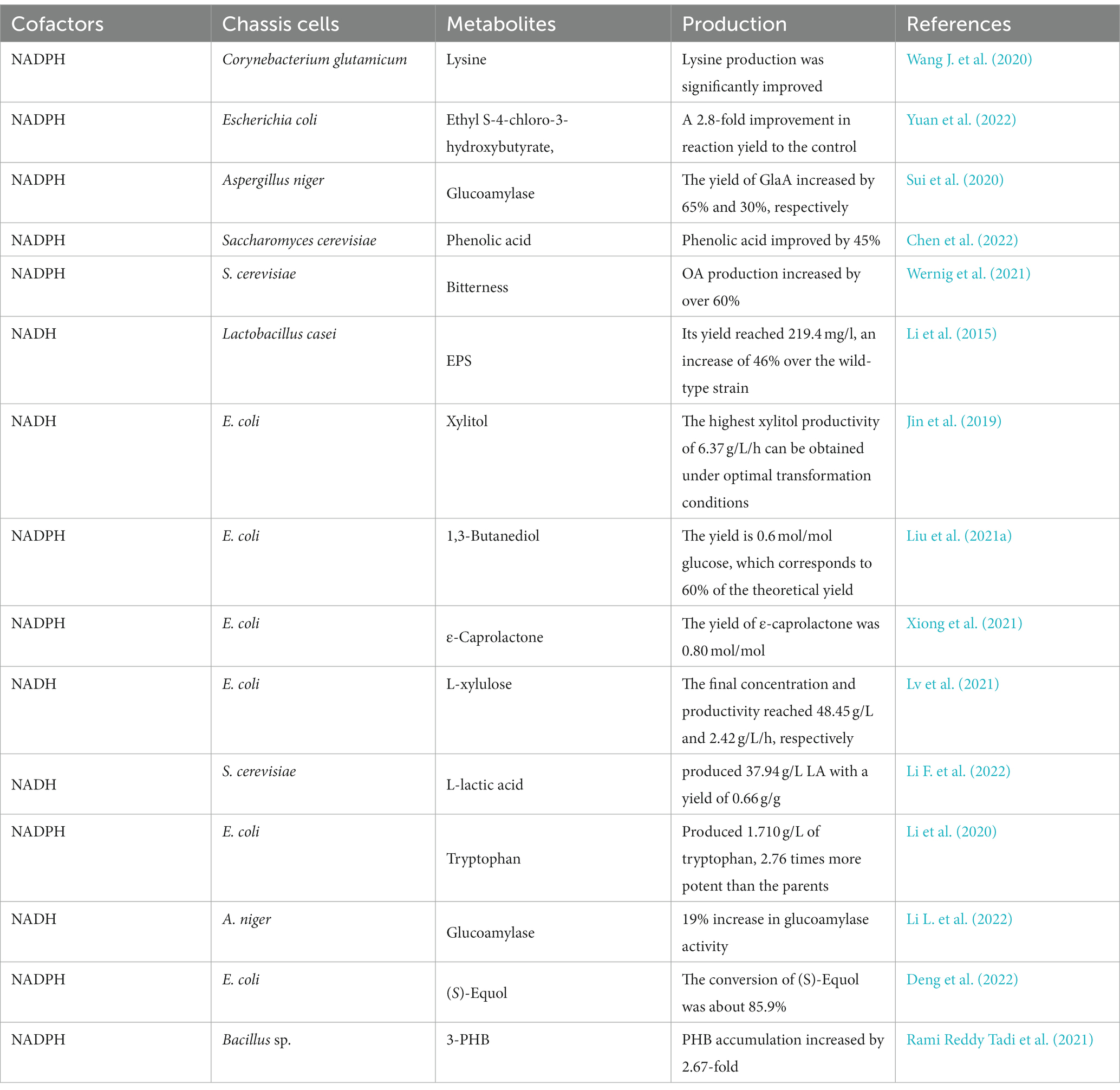
Many carbon metabolic pathways in microorganisms can synthesize NAD(P)H. Among them, the pentose phosphate pathway is the main metabolic pathway for the synthesis of NADPH, and glycolysis is the main metabolic pathway for the synthesis of NADH. Under aerobic conditions, pyruvate is changed to acetyl-CoA and NADH catalyzed by the pyruvate dehydrogenase complex, and the tricarboxylic acid cycle can generate NADH and NADPH. Non-metabolic coupled expression NADH oxidoreductase may lead to disruption of cellular metabolism, which in turn affects microbial growth and the synthesis of metabolites. Reconstructing the cofactor production pathway can effectively avoid the resulting disruption of cellular metabolism. This section focuses on the metabolic regulatory strategy of NAD(P)H/NAD(P)+ and its application in biosynthesis. Some of the NAD(P)H/NAD(P)+ regulatory strategies and their applications are listed in Table 2.
Pentose phosphate pathway
The pentose phosphate pathway(PPP) is the main metabolic pathway for NADPH synthesis, and the key enzymes are glucose-6-phosphate dehydrogenase (G6PDH) and 6-phosphogluconate dehydrogenase (6PGDH), so overexpression of G6PDH and 6PGDH can effectively increase the flux of PPP and intracellular NADPH levels. The transcriptional regulator Cgl2680 is an important regulator of NADPH levels and L-lysine biosynthesis in C. glutamicum. By blocking the glycolytic pathway and overexpressing the PPP in lysine-producing strains and knocking out the gene Cgl2680, Cgl2680-deficient strains of C. glutamicum showed increased production of L-lysine and L-leucine as well as increased H2O2 tolerance (Wang L. et al., 2020). Insufficient intracellular NAD(P)H is a major issue limiting the reduced product synthesis by whole cell biocatalysis or microbial cell factory. Yuan et al. (2022) improved the PPP by overexpressing additional G6PDH encoding gene zwf and glucokinase encoding gene glk, thereby increasing the supply of NADPH, and intracellular NADPH content increased significantly from 150.3 to 681.8 mol/L, which was 4.5-fold higher than the control. It was applied to enhance the reduction process of 4-chloroacetoacetate to generate ethyl S-4-chloro-3-hydroxybutyrate, resulting in a 2.8-fold improvement in reaction yield. Sui et al. (2020) enhanced overexpression of the gndA gene (G6PDH) and maeA gene (NADP-dependent malic enzyme) in the PPP, which increased intracellular NADPH by 45% and 66%, respectively, while glucoamylase yields increased by 65% and 30%, respectively. The regeneration of NADPH was promoted by the non-oxidative step of PPP, which facilitated the synthesis of phenolic acid, and the NADPH regeneration strategy increased phenolic acid by 45% compared to the initial strain (Chen et al., 2022). Wernig et al. (2021) redirected glucose flux to the oxidative branch of the PPP and overexpressed heterologous phosphoketolase/phosphotransacetylase to improve the availability of NADPH and acetyl-CoA in the strain while eliminating eight-carbon fatty acid octanoic acid degradation, and these modifications resulted in more than 60% increase in octanoic acid production during glucose consumption compared to the parental strain.
New pathways for cofactor generation
Synthetic biology has provided technical principles for the optimization of key enzymes of metabolic pathways and improvement of cofactor regeneration pathways, and has increased the efficiency of carbon and energy utilization. It was shown that the expression of isozymes that do not produce additional cofactors such as NAD(P)H/NAD(P)+ can achieve a balance of cofactors while ensuring normal cell growth compared to endogenous enzymes, the introduction of metabolic synthetic pathways of other strains has become an effective means to address the low yield of target products (Chen R. et al., 2020).
Li et al. (2015) cloned a NADH oxidase gene from Streptococcus mutans and overexpressed it in Lactobacillus casei (L. casei) LC2W under the control of the constitutive promoter P23, and the NADH oxidase activity of the recombinant strain LC-nox was 0.854 U/mL, which was nearly 20-fold higher compared to the wild type. The overexpression of NADH oxidase resulted in a 22% reduction in lactate production in the recombinant strain. It was proposed that more carbon sources could be saved and used for extracellular polysaccharide (ESP) biosynthesis with a yield of 219.4 mg/L, a 46% increase over the wild-type strain. Babaei et al. (2019) further increased succinate flux by overexpressing genes in the glyoxylate pathway and the oxidative TCA branch, and expressing phosphoenolpyruvate carboxykinase from succinate-producing Actinobacillus succinogenes. Transient adaptation to glucose reduced the lag phase of the strain and increased its tolerance to high glucose concentrations. The resulting strain produced 7.8 ± 0.0 g/L succinate in shake flasks without pH control with a glucose yield of 0.105 g/g. Jin et al. (2019) could completely convert 150 g/L xylose to xylitol by co-expression of xylose reductase and glucose dehydrogenase in E. coli and could achieve a xylitol productivity of 21.2 g/L/h by in vitro biotransformation. The enzyme activity reached 1,533 U/L after optimization of induction conditions. It was possible to completely convert 200 g/L xylose to xylitol, and the highest xylitol productivity of 6.37 g/L/h was obtained under optimal conversion conditions. Tesfay et al. (2021) introduced NAD+ regenerating enzyme (NADH oxidase) from Streptococcus mutans ATCC 25175 together with xylitol-4-dehydrogenase from Pantoea ananatis into E. coli, co-expression resulted in an increase in L-xylulose concentration and productivity from xylitol as well as intracellular NAD+ concentration. In a 1 L bioconversion system, the final concentration and productivity of L-xylulose from 50 g/L xylitol reached 48.45 g/L and 2.42 g/L·h, respectively.
In addition, screening and modification of enzymes for application in the synthesis of compounds has become a means of enhancing the target product. Shen et al. (2021) identified 6 genes encoding NADPH-depleting enzymes and 19 ATP-depleting enzymes by CRISPRi screening. The deletion of yahK(encoding the NADPH-depleting enzyme), and fecE(encoding the ATP-depleting enzyme) increased the yield of 4-hydroxyphenylacetic acid from 6.32 to 7.76 g/L. Liu et al. (2021a) first performed screening for key pathway enzymes, then increased NADPH supply by cofactor engineering, followed by optimization of fermentation conditions, and the engineered E. coli strain could efficiently produce (R)-1,3-butanediol at a yield of 0.6 mol/mol glucose, equivalent to 60% of the theoretical yield. Lv et al. (2021) showed that the native pntAB encoding pyridine nucleotide transhydrogenase in E. coli was selected from five NADPH regeneration genes to supplement redox cofactor NADPH for the conversion of p-coumaric acid to caffeic acid during ferulic acid biosynthesis. Slivinskaya et al. (2022) modified the cofactor specificity of E. coli GAPDH by amino acid substitutions at positions 34, 188 and 189, several mutant enzymes with dual NAD+/NADP+ cofactor specificity were obtained and their kinetic parameters were determined. Overexpression of genes encoding mutant GAPDHs with dual cofactor specificity produce d in E. coli cells producing L-lysine, L-threonine and L-proline resulted in a significant increase in the accumulation of the corresponding amino acids in the culture medium. Xiong et al. (2021) combined alcohol dehydrogenase (ADH) with cyclohexanone monooxygenase (CHMO) in E. coli to obtain a self-sufficient NADPH cofactor regeneration system. In addition, by modifying the ribosome binding site, improved variants with better substrate tolerance and higher catalytic activity for ε-caprolactone were created. The best mutant strain produced 0.80 mol/mol of ε-caprolactone using 60 mM cyclohexanol as a substrate. In four sequential batches, the designed whole-cell biocatalyst produced 126 mM ε-caprolactone with a high molar yield of 0.78 mol/mol.
The introduction of new reactions/pathways, or the reconstruction of metabolic pathways followed by the modified key enzymes has also been shown by many researchers to be an effective means of enhancing the target product. Harth et al. (2020) generated the necessary redox cofactor by combining the reduction reaction of GalA with the oxidation reaction of the sugar alcohol sorbitol (which has a higher reduced state than glucose). Gu et al. (2019) reconstructed the metabolic pathway in GlcNAc-producing strains by introducing the pyruvate iron oxidoreductase PorAB, malate dehydrogenase BmqO, and glyceraldehyde-3-phosphate dehydrogenase GoR to achieve the catalytic processes of pyruvate to acetyl-CoA, malate to oxaloacetate and glyceraldehyde-3-phosphate to 3-phosphoglycerate, respectively, avoiding the excess of NADH and thus achieving intracellular redox homeostasis, ultimately increasing the fermentation yield of GlcNAc by 4.06-fold. The yield of Menaquinone-7 (MK-7) in Bacillus subtilis is low, and Ding et al. (2022) made MK-7 production of 39.01 mg/L by overexpressing key rate-limiting enzymes such as 1-deoxyxylulose-5-phosphate synthase, isopentenyl-diphosphate delta-isomerase, 1-deoxyxylulose-5-phosphate reductase. Next, after stoichiometric calculation and optimization of the cofactor regeneration pathway, two NADPH regeneration systems were constructed to enhance the endogenous cofactor regeneration pathway, and heterologous NADH kinase was introduced to increase the availability of NADPH in MK-7 biosynthesis. After three Design-Build-Test-Learn cycles, MK-7 reached a concentration of 53.07 mg/L after flask fermentation. In addition, the artificially constructed cofactor regeneration system resulted in a 9.15% reduction of NADH-dependent by-product lactate in the fermentation broth. This led to a reduction in energy loss and an increase in carbon conversion.
Cofactor balance regulation
The balance of cofactor concentrations is an important condition for achieving enzyme catalytic efficiency; simply increasing cofactor concentrations alone does not necessarily improve metabolism, but may instead cause an imbalance in the intracellular environment. When the concentration of NADH is too high, the oxygen consumption of cellular respiration increases, substrate consumption is accelerated, and economic efficiency is reduced. The anabolic metabolism of N-acetylglucosamine leads to an excess of NADH, which is utilized for the synthesis of substances such as the NADH-dependent by-product 2,3-butanediol. Gu et al. (2019) first overexpressed NADH oxidase intracellularly to achieve intracellular redox balance, and when NADH oxidase was expressed using the strong constitutive promoter P43, intracellular redox balance could not be achieved due to overexpression of oxidase, and the fermentation yield of N-acetylglucosamine was only 0.86 g/L. Therefore, further regulation of NADH oxidase using promoter engineering resulted in a final fermentation yield of 12.46 g/L of N-acetylglucosamine. Li F. et al. (2022) regulated the redox genes to consume excessive NADH, and lactate production reached 0.04 g/g to 0.37 g/g in YNB medium. Subsequently, strain PK27 produced 37.94 g/L lactate in YPD medium with a yield of 0.66 g/g. Li et al. (2020) expressed mutated serA and thrA to increase the precursor supply of serine, resulting in the accumulation of 1.380 g/L tryptophan. Finally, to maintain cofactor balance, the genes sthA and pntAB encoding transhydrogenases were overexpressed. With sufficient amounts of precursors and balanced cofactors, the engineered strain produced 1.710 g/L tryptophan after 48 h of shake flask fermentation, which was 2.76-fold higher than the yield of the parental strain.
Other metabolic regulatory pathways
Insufficient supply of cofactors can limit the production of metabolites, and enhancing the cofactor synthesis pathway can address this limitation well. Li F. et al. (2022) led to a further 19% increase in glucoamylase activity by overexpressing mitochondrial NADH kinase (AN17) and malic enzyme (MaeA). Deng et al. (2022) optimized a cascade system by regulating the intensity of NADPH gene expression to produce a higher (S)-equol titer (3418.5 mg/L) with a conversion rate of approximately 85.9%. van Aalst et al. (2022) functional expression of the gene in anaerobic, slow-growing chemostat cultivation on glucose-sorbitol mixtures resulted in a 12-fold higher co-consumption rate of sorbitol than that observed in sorbitol-consuming reference strains. Rami Reddy Tadi et al. (2021) metabolic engineering of Bacillus megaterium through co-expression of the precursor (phbRBC) and NADPH cofactor regeneration (zwf) genes resulted in a 2.67-fold increase in PHB accumulation.
ATP/ADP
Currently, there is a better understanding of both material and energy metabolism, and ATP regulation strategies combined with synthetic biology are increasingly being applied in microbial cells to meet the need to enhance target products, etc. (Chen et al., 2017; Cai et al., 2018; Zhang et al., 2019). This section focuses on the metabolic regulatory strategies of ATP/ADP and their applications in biosynthesis.
Regulation of oxidative phosphorylation levels
Under aerobic conditions, ATP is synthesized mainly through the oxidative phosphorylation pathway, and therefore the concentration of ATP can be regulated by regulating oxidative phosphorylation (Liu et al., 2006; Hara and Kondo, 2015). Under anaerobic conditions, NADH is oxidized by actions such as acetaldehyde dehydrogenase or lactate dehydrogenase (Zhu and Shimizu, 2004; Zhang et al., 2006), and substrate level phosphorylation is the main source of ATP (Bai et al., 2020), which can be regulated by regulating NADH levels and thus ATP levels (Xu et al., 2017). Dank et al. (2022) discovered that when lactate was consumed under microaerobic conditions, pyruvate, propionate, and acetic acid were produced first, and when lactate was depleted, pyruvate and propionate were oxidized with 6-fold increase in vitamin B12 concentrations compared to anaerobic conditions, demonstrating the potential of propionate and pyruvate as carbon sources for vitamin B12 production. Thus, under microaerobic conditions, a fed-batch reactor with anaerobically precultured lactate-grown cells was fed propionate, resulting in enhanced biomass and vitamin B12 synthesis. Skorokhodova et al. (2021) investigated the anaerobic generation of pyruvate from glucose in recombinant E. coli strains with impaired fermentation ability during respiration. Nitrate functions as an external terminal electron acceptor, and enforced ATP hydrolysis causes the recombinant to consume significantly more glucose while maintaining substrate level conversion of the target product. Pyruvate is generated from glucose in yields of 1.77–1.78 mol/mol under anaerobic nitrate respiration and enforced ATP hydrolysis, and the recombinant is almost depleted of substrate with no or minimal by-product formation. Wang D. et al. (2019) knocked out the gene por1 encoding mitochondrial pore protein in Candida utilis, which increased intracellular NADH and ATP concentrations. The co-production of S-adenosylmethionine (SAM) and glutathione in the recombinant strain increased by 34.9% and 25.1%, respectively. Chen and Tan (2018) employed an ATP regulation strategy in yeast to boost SAM production by regulating NADH availability and oxygen delivery, and the SAM titer reached a high of 55 mg/L after 28 h of cultivation. Moreover, respiratory complex I plays a crucial role in regulating the cell growth and secondary metabolism of Monascus via by altering intracellular ROS and ATP levels (Cai et al., 2022).
Furthermore, ATP regulation in microorganisms can be performed by introducing related substrates or by overexpressing/knocking out genes encoding related substrates. Ebrahimzadeh et al. (2022) employed a simultaneous pulsed-feeding method of citrate and glutamate to boost poly-γ-glutamate synthesis in Bacillus licheniformis, obtaining approximately 88 ± 4 g/L of γ-glutamate under optimal conditions. Kang et al. (2013) added sodium citrate to Lactobacillus panis PM1 and increased pyruvate production, which in turn led to an increase in acetate and lactate production as well as Zhou et al. (2010) discovered that increased ATP supply improved Candida glabrata tolerance in acidic conditions. Xue et al. (2019) improved pullulan production and enhanced the capacity to bind CO and oxygen in Aureobasidium melanogenum P16 by overexpression of the optimized Vitreoscilla hemoglobin (VHb) gene and the native flavohemoglobin (FHb) gene. During a 10-liter fermentation, the engineered strains were 42.1%, 15.6%, and 40.0% higher than the parental strain in terms of pullulan, yield and productivity, respectively.
Metabolic regulation of ATP-related enzymes
ATP synthesis and regeneration are also closely related to ATP synthase and ATP/ADP-dependent enzymes in key pathways. Knockout of ethanol dehydrogenase in S. cerevisiae and overexpression of acetaldehyde dehydrogenase increased glycerol production (Cordier et al., 2007). Overexpression of the oxidase gene aox in A. niger resulted in increased citric acid production (Hou et al., 2018). Integration of the Arabidopsis-derived mitochondrial ATP6 gene into Candida utilis could increase intracellular glutathione and SAM production by 46.6% and 28.7%, respectively (Xu et al., 2018). Overexpression of phosphoenolpyruvate carboxykinase and triosephosphate isomerase enhances the pyruvate metabolic pathway in shake flask cultures, increasing phloroglucinol concentrations by 44% and 92%, respectively (Liu W. et al., 2020). Heterologous expression of PCK in Enterobacter aerogenes and knockout of glucose phosphotransferase increased succinate production (Tajima et al., 2015). The exogenous non-ATP-dependent pathway replaces the native ATP-dependent pathway in S. cerevisiae, resulting in a reconfiguration of the metabolic pathway from ethanol to platform compounds such as acetyl-CoA (Kozak et al., 2016). Yamanaka et al. (2010) controlled acidic pH, which stimulated ATP-dependent ε-poly-l-lysine synthase activity and largely enhanced the synthesis of the target compound ε-poly-l-lysine.
Summary and outlook
NADH, which interacts with related enzymes to affect ATP production under both aerobic and anaerobic conditions, is closely related to ATP synthesis (Hara and Kondo, 2015; Bai et al., 2020). Under aerobic conditions, NADH is oxidized via the electron transport chain to produce ATP, with oxygen serving as the terminal electron acceptor, while under anaerobic conditions, NADH is oxidized via the fermentation pathway by the action of acetaldehyde dehydrogenase or lactate dehydrogenase, where substrate-level phosphorylation is the main source of ATP. NADH-based strategies to regulate intracellular ATP are easier to manipulate and more efficient, and are suitable for upregulating or downregulating intracellular ATP levels. However, changes in NADH affect the intracellular redox state as well as cell growth metabolism and product synthesis. Therefore, alteration of NADH levels is not a direct method for ATP regulation, and a comprehensive consideration of whether the redox state of the cell is conducive to the synthesis of target metabolites is required.
With the rapid development of cofactor engineering, it has become an important research direction in the field of metabolic engineering and synthetic biology. Currently, the most common cofactors are acetyl-CoA, NAD(P)H/NAD(P)+, and ATP, and a variety of corresponding regulatory strategies have been applied to enhance the yield of target products. For example, single or combined metabolic regulatory strategies have been used to regulate cofactors or to specifically modify key enzymes of the cofactor production or consumption pathways. Although the construction of cofactor recycling systems, regulation of cofactor regeneration pathways, and modification of cofactor synthesis pathways have also achieved relatively significant results, these approaches also have various problems and drawbacks, such as the precise dynamic regulation of cofactors, the maintenance of a stable redox state in microbial cells, and the lack of understanding of the complex regulatory mechanisms of essential metabolism, etc., which hinder their application in practical production. In the future, it is expected to continue research on the regulation of cofactors to maximize and rapidly direct the metabolic flow to the target metabolites.
In addition to the research on the regulation of cofactors, some progress has been made in the development of noncanonical cofactor systems in microorganisms. Black et al. (2020) demonstrated the development of an irregular redox cofactor system based on nicotinamide mononucleotide (NMN+). The key enzyme in this system is a calculated glucose dehydrogenase, which can be used to support a variety of redox chemistry in vitro. This work proves the effective use of noncanonical cofactors in biocatalysis and metabolic pathway design. Recent findings reignite the interest in engineering enzymes using noncanonical cofactors, which exhibit excellent industrial properties in vitro and can achieve specific electron transport in vivo (King et al., 2020). Wang et al. (2021) reprogrammed the substrate binding pockets of nicotinic acid mononucleotide (NaMN) adenylate transferase to combine to create NCD synthase (NcdS) so that cytidine triphosphate and nicotinamide mononucleotides exceed their regular substrates ATP and NaMN, respectively. Only the overexpression of NcdS in the model host E. coli promoted the production of intracellular NCD and reached a higher NCD level as high as 5.0 mM through further regulation. Finally, L-malic acid was converted into D-lactic acid by metabolic circuit mediated NCD connection, which confirmed the self-sufficiency of non-natural cofactors. NcdS and NCD-linked enzymes provide unique tools and opportunities for interesting research in chemical biology and synthetic biology.
Author contributions
YS and XL conceptualized the review, analyzed the data, and helped to write the manuscript. LJ, TZ, and BL helped to write the manuscript and prepared the figures. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Key Research and Development Program of China (2021YFC2102700), the National Natural, Science Foundation of China (31922070 and U2106228), the Jiangsu Synergetic Innovation Center for Advanced Bio-Manufacture (XTC2205), the Natural Science Foundation of the Jiangsu Higher Education Institutions of China (22KJB550008), and Scientific Research Start-up Fund for Introduced Talents of Anhui Polytechnic University (2022YOO068).
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Babaei, M., Rueksomtawin Kildegaard, K., Niaei, A., Hosseini, M., Ebrahimi, S., Sudarsan, S., et al. (2019). Engineering oleaginous yeast as the host for fermentative succinic acid production from glucose. Front. Bioeng. Biotechnol. 7, 361–374. doi: 10.3389/fbioe.2019.00361
Bai, Y., Li, X., Zhang, D., Hou, C., Zheng, X., Chen, L., et al. (2020). Effects of different atp contents on phosphorylation level of glycogen phosphorylase and its activity in lamb during incubation at 4 °C in vitro. Int. J. Food Sci. Technol. 55, 3000–3007. doi: 10.1111/ijfs.14565
Black, W. B., Zhang, L., Mak, W. S., Maxel, S., Cui, Y., King, E., et al. (2020). Engineering a nicotinamide mononucleotide redox cofactor system for biocatalysis. Nat. Chem. Biol. 16, 87–94. doi: 10.1038/s41589-019-0402-7
Bruschi, F., Dundar, M., Gahan, P. B., Gartland, K., Szente, M., Viola-Magni, M. P., et al. (2011). Biotechnology worldwide and the 'european biotechnology thematic network' association (ebtna). Curr. Opin. Biotechnol. 22, S7–S14. doi: 10.1016/j.copbio.2011.05.506
Cai, D., Chen, Y., He, P., Wang, S., Mo, F., Li, X., et al. (2018). Enhanced production of poly-γ-glutamic acid by improving atp supply in metabolically engineered bacillus licheniformis. Biotechnol. Bioeng. 115, 2541–2553. doi: 10.1002/bit.26774
Cai, X., Zhang, S., Lin, J., Wang, Y., Ye, F., Zhou, B., et al. (2022). Role of the gene ndufs8 located in respiratory complex i from monascus purpureus in the cell growth and secondary metabolites biosynthesis. J. Fungi. 8, 655–674. doi: 10.3390/jof8070655
Chen, X., Dong, X., Liu, J., Luo, Q., and Liu, L. (2020). Pathway engineering of escherichia coli for alpha-ketoglutaric acid production. Biotechnol. Bioeng. 117, 2791–2801. doi: 10.1002/bit.27456
Chen, R., Gao, J., Yu, W., Chen, X., Zhai, X., Chen, Y., et al. (2022). Engineering cofactor supply and recycling to drive phenolic acid biosynthesis in yeast. Nat. Chem. Biol. 18, 520–529. doi: 10.1038/s41589-022-01014-6
Chen, Y., and Tan, T. (2018). Enhanced s-adenosylmethionine production by increasing atp levels in baker's yeast (saccharomyces cerevisiae). J. Agric. Food Chem. 66, 5200–5209. doi: 10.1021/acs.jafc.8b00819
Chen, L., Yan, W., Qian, X., Chen, M., Zhang, X., Xin, F., et al. (2021). Increased lipid production inyarrowia lipolytica from acetate through metabolic engineering and cosubstrate fermentation. ACS Synth. Biol. 10, 3129–3138. doi: 10.1021/acssynbio.1c00405
Chen, R., Yang, S., Zhang, L., and Zhou, Y. J. (2020). Advanced strategies for production of natural products in yeast. Iscience. 23:100879. doi: 10.1016/j.isci.2020.100879
Chen, Y., Zhou, H., Wang, M., and Tan, T. (2017). Control of atp concentration in escherichia coli using an atp-sensing riboswitch for enhanced s-adenosylmethionine production. RSC Adv. 7, 22409–22414. doi: 10.1039/c7ra02538f
Cordier, H., Mendes, F., Vasconcelos, I., and François, J. M. (2007). A metabolic and genomic study of engineered saccharomyces cerevisiae strains for high glycerol production. Metab. Eng. 9, 364–378. doi: 10.1016/j.ymben.2007.03.002
Curtabbi, A., and Enriquez, J. A. (2022). The ins and outs of the flavin mononucleotide cofactor of respiratory complex i. IUBMB Life 74, 629–644. doi: 10.1002/iub.2600
Dank, A., Biel, G., Abee, T., and Smid, E. J. (2022). Microaerobic metabolism of lactate and propionate enhances vitamin b12 production in propionibacterium freudenreichii. Microb. Cell Fact. 21:225. doi: 10.1186/s12934-022-01945-8
Deng, H., Gao, S., Zhang, W., Zhang, T., Li, N., and Zhou, J. (2022). High titer of (s)-equol synthesis from daidzein in Escherichia coli. ACS Synth. Biol. 11, 4043–4053. doi: 10.1021/acssynbio.2c00378
Ding, X., Zheng, Z., Zhao, G., Wang, L., Wang, H., Yang, Q., et al. (2022). Bottom-up synthetic biology approach for improving the efficiency of menaquinone-7 synthesis in bacillus subtilis. Microb. Cell Fact. 21:101. doi: 10.1186/s12934-022-01823-3
Drepper, T., Eggert, T., Hummel, W., Leggewie, C., Pohl, M., Rosenau, F., et al. (2006). Neue biokatalysatoren für die weiße biotechnologie. Chem. Ing. Tech. 78, 239–248. doi: 10.1002/cite.200500197
Duetz, W. A., van Beilen, J. B., and Witholt, B. (2001). Using proteins in their natural environment: potential and limitations of microbial whole-cell hydroxylations in applied biocatalysis. Curr. Opin. Biotechnol. 12, 419–425. doi: 10.1016/s0958-1669(00)00237-8
Ebrahimzadeh, K. M., Bahrami, A., and Babaeipour, V. (2022). Enhanced production of poly-gamma-glutamic acid by bacillus licheniformis atcc 9945a using simultaneous pulse-feedings of citrate and glutamate. Prep. Biochem. Biotechnol. 52, 961–968. doi: 10.1080/10826068.2021.2021232
Farmer, W. R., and Liao, J. C. (1997). Reduction of aerobic acetate production by escherichia coli. Appl. Environ. Microbiol. 63, 3205–3210. doi: 10.1128/aem.63.8.3205-3210.1997
Ge, F., Li, X., Ge, Q., Zhu, D., Li, W., Shi, F., et al. (2021). Modular control of multiple pathways of corynebacterium glutamicum for 5-aminolevulinic acid production. AMB Express 11:179. doi: 10.1186/s13568-021-01335-0
Gu, Y., Lv, X., Liu, Y., Li, J., Du, G., Chen, J., et al. (2019). Synthetic redesign of central carbon and redox metabolism for high yield production of n-acetylglucosamine in bacillus subtilis. Metab. Eng. 51, 59–69. doi: 10.1016/j.ymben.2018.10.002
Hackett, S. R., Zanotelli, V. R., Xu, W., Goya, J., Park, J. O., Perlman, D. H., et al. (2016). Systems-level analysis of mechanisms regulating yeast metabolic flux. Science 354:aaf2786. doi: 10.1126/science.aaf2786
Hara, K. Y., and Kondo, A. (2015). Atp regulation in bioproduction. Microb. Cell Fact. 14:198. doi: 10.1186/s12934-015-0390-6
Harth, S., Wagner, J., Sens, T., Choe, J., Benz, J. P., Weuster-Botz, D., et al. (2020). Engineering cofactor supply and nadh-dependent d-galacturonic acid reductases for redox-balanced production of l-galactonate in saccharomyces cerevisiae. Sci. Rep. 10:19021. doi: 10.1038/s41598-020-75926-5
Hayat, I. F., Plan, M., Ebert, B. E., Dumsday, G., Vickers, C. E., and Peng, B. (2021). Auxin-mediated induction of gal promoters by conditional degradation of mig1p improves sesquiterpene production in saccharomyces cerevisiae with engineered acetyl-coa synthesis. J. Microbial. Biotechnol. 14, 2627–2642. doi: 10.1111/1751-7915.13880
Hou, L., Liu, L., Zhang, H., Zhang, L., Zhang, L., Zhang, J., et al. (2018). Functional analysis of the mitochondrial alternative oxidase gene (aox1) from aspergillus Niger cgmcc 10142 and its effects on citric acid production. Appl. Microbiol. Biotechnol. 102, 7981–7995. doi: 10.1007/s00253-018-9197-9
Jin, L., Xu, W., Yang, B., Liu, Z., and Zheng, Y. (2019). Efficient biosynthesis of xylitol from xylose by coexpression of xylose reductase and glucose dehydrogenase in escherichia coli. Appl. Biochem. Biotechnol. 187, 1143–1157. doi: 10.1007/s12010-018-2878-0
Kang, T. S., Korber, D. R., and Tanaka, T. (2013). Contributions of citrate in redox potential maintenance and atp production: metabolic pathways and their regulation in lactobacillus panis pm1. Appl. Microbiol. Biotechnol. 97, 8693–8703. doi: 10.1007/s00253-013-5108-2
Kim, D. H., Hwang, H. G., and Jung, G. Y. (2022). Optimum flux rerouting for efficient production of naringenin from acetate in engineered escherichia coli. Biotechnol Biofuels Bioprod. 15:90. doi: 10.1186/s13068-022-02188-w
King, E., Maxel, S., and Li, H. (2020). Engineering natural and noncanonical nicotinamide cofactor-dependent enzymes: design principles and technology development. Curr. Opin. Biotechnol. 66, 217–226. doi: 10.1016/j.copbio.2020.08.005
Kozak, B. U., van Rossum, H. M., Niemeijer, M. S., van Dijk, M., Benjamin, K., Wu, L., et al. (2016). Replacement of the initial steps of ethanol metabolism in saccharomyces cerevisiae by atp-independent acetylating acetaldehyde dehydrogenase. FEMS Yeast Res. 16:w6. doi: 10.1093/femsyr/fow006
Kulkarni, N. S., and Deshpande, M. S. (2007). General enzymology ; Himalaya Pub. House: Mumbai, India
Lee, W., Kim, J., Park, E., Han, N. S., Kim, M., and Seo, J. (2013). Effects of nadh kinase on nadph-dependent biotransformation processes in escherichia coli. Appl. Microbiol. Biotechnol. 97, 1561–1569. doi: 10.1007/s00253-012-4431-3
Li, Z., Ding, D., Wang, H., Liu, L., Fang, H., Chen, T., et al. (2020). Engineering escherichia coli to improve tryptophan production via genetic manipulation of precursor and cofactor pathways. Synth. Syst. Biotechnol. 5, 200–205. doi: 10.1016/j.synbio.2020.06.009
Li, F. L., Shi, Y., Zhang, J. X., Gao, J., and Zhang, Y. W. (2018). Cloning, expression, characterization and homology modeling of a novel water-forming nadh oxidase from streptococcus mutans atcc 25175. Int. J. Biol. Macromol. 113, 1073–1079. doi: 10.1016/j.ijbiomac.2018.03.016
Li, N., Wang, Y., Zhu, P., Liu, Z., Guo, B., and Ren, J. (2015). Improvement of exopolysaccharide production in lactobacillus casei lc2w by overexpression of nadh oxidase gene. Microbiol. Res. 171, 73–77. doi: 10.1016/j.micres.2014.12.006
Li, F., Wei, X., Sun, Q., Guo, Y., and Liu, J. (2022). Production of l-lactic acid in saccharomyces cerevisiae through metabolic engineering and rational cofactor engineering. Sugar Tech. 24, 1272–1283. doi: 10.1007/s12355-022-01142-2
Li, L., Yu, L., Wang, B., and Pan, L. (2022). Impact of overexpressing nadh kinase on glucoamylase production inaspergillus Niger. J. Ind. Microbiol. Biotechnol. 49, kuac015–kuac025. doi: 10.1093/jimb/kuac015
Liu, Y., Cen, X., Liu, D., and Chen, Z. (2021a). Metabolic engineering of escherichia coli for high-yield production of (r)-1,3-butanediol. ACS Synth. Biol. 10, 1946–1955. doi: 10.1021/acssynbio.1c00144
Liu, Y., Cruz-Morales, P., Zargar, A., Belcher, M. S., Pang, B., Englund, E., et al. (2021b). Biofuels for a sustainable future. Cell 184, 1636–1647. doi: 10.1016/j.cell.2021.01.052
Liu, M., Ding, Y., Chen, H., Zhao, Z., Liu, H., Xian, M., et al. (2017). Improving the production of acetyl-coa-derived chemicals in escherichia coli bl21(de3) through iclr and arca deletion. BMC Microbiol. 17:10. doi: 10.1186/s12866-016-0913-2
Liu, L. M., Li, Y., Du, G. C., and Chen, J. (2006). Increasing glycolytic flux in torulopsis glabrata by redirecting atp production from oxidative phosphorylation to substrate-level phosphorylation. J. Appl. Microbiol. 100, 1043–1053. doi: 10.1111/j.1365-2672.2006.02871.x
Liu, J., Li, H., Zhao, G., Caiyin, Q., and Qiao, J. (2018). Redox cofactor engineering in industrial microorganisms: strategies, recent applications and future directions. J. Ind. Microbiol. Biotechnol. 45, 313–327. doi: 10.1007/s10295-018-2031-7
Liu, H., Wang, F., Deng, L., and Xu, P. (2020). Genetic and bioprocess engineering to improve squalene production in yarrowia lipolytica. Bioresour. Technol. 317:123991. doi: 10.1016/j.biortech.2020.123991
Liu, W., Zhang, R., Wei, M., Cao, Y., and Xian, M. (2020). Increasing the pyruvate pool by overexpressing phosphoenolpyruvate carboxykinase or triosephosphate isomerase enhances phloroglucinol production in escherichia coli. Biotechnol. Lett. 42, 633–640. doi: 10.1007/s10529-020-02812-5
Lu, H., Liu, X., Huang, M., Xia, J., Chu, J., Zhuang, Y., et al. (2015). Integrated isotope-assisted metabolomics and (13)c metabolic flux analysis reveals metabolic flux redistribution for high glucoamylase production by aspergillus Niger. Microb. Cell Fact. 14:147. doi: 10.1186/s12934-015-0329-y
Lv, H., Zhang, Y., Shao, J., Liu, H., and Wang, Y. (2021). Ferulic acid production by metabolically engineered escherichia coli. Bioresour. Bioprocess. 8:70. doi: 10.1186/s40643-021-00423-0
McNaught, A., and Wilkinson, A. (1997). Iupac compendium of chemical terminology, 2nd Edn.; Blackwell scientific publications: Oxford
Olzhausen, J., Grigat, M., Seifert, L., Ulbricht, T., and Schuller, H. J. (2021). Increased biosynthesis of acetyl-coa in the yeast saccharomyces cerevisiae by overexpression of a deregulated pantothenate kinase gene and engineering of the coenzyme a biosynthetic pathway. Appl. Microbiol. Biotechnol. 105, 7321–7337. doi: 10.1007/s00253-021-11523-4
Paddon, C. J., Westfall, P. J., Pitera, D. J., Benjamin, K., Fisher, K., McPhee, D., et al. (2013). High-level semi-synthetic production of the potent antimalarial artemisinin. Nature 496, 528–532. doi: 10.1038/nature12051
Pietrocola, F., Galluzzi, L., Bravo-San, P. J., Madeo, F., and Kroemer, G. (2015). Acetyl coenzyme a: a central metabolite and second messenger. Cell Metab. 21, 805–821. doi: 10.1016/j.cmet.2015.05.014
Rami Reddy Tadi, S., Dutt Ravindran, S., Balakrishnan, R., and Sivaprakasam, S. (2021). Recombinant production of poly-(3-hydroxybutyrate) by bacillus megaterium utilizing millet bran and rapeseed meal hydrolysates. Bioresour. Technol. 326:124800. doi: 10.1016/j.biortech.2021.124800
Sagmeister, P., Schimek, C., Meitz, A., Herwig, C., and Spadiut, O. (2014). Tunable recombinant protein expression with e. coli in a mixed-feed environment. Appl. Microbiol. Biotechnol. 98, 2937–2945. doi: 10.1007/s00253-013-5445-1
Satowa, D., Fujiwara, R., Uchio, S., Nakano, M., Otomo, C., Hirata, Y., et al. (2020). Metabolic engineering of e. coli for improving mevalonate production to promote nadph regeneration and enhance acetyl-coa supply. Biotechnol. Bioeng. 117, 2153–2164. doi: 10.1002/bit.27350
Selles, V. L., Kelly, C. L., Mordaka, P. M., and Heap, J. T. (2018). Review of nad(p)h-dependent oxidoreductases: properties, engineering and application. BBA-Proteins Proteomics. 1866, 327–347. doi: 10.1016/j.bbapap.2017.11.005
Shen, Y. P., Liao, Y. L., Lu, Q., He, X., Yan, Z. B., and Liu, J. Z. (2021). Atp and nadph engineering of escherichia coli to improve the production of 4-hydroxyphenylacetic acid using crispri. Biotechnol. Biofuels 14:100. doi: 10.1186/s13068-021-01954-6
Skorokhodova, A. Y., Gulevich, A. Y., and Debabov, V. G. (2021). Optimization of the anaerobic production of pyruvic acid from glucose by recombinant escherichia coli strains with impaired fermentation ability via enforced atp hydrolysis. Appl. Biochem. Microbiol. 57, 434–442. doi: 10.1134/S0003683821040153
Slivinskaya, E. A., Plekhanova, N. S., Altman, I. B., and Yampolskaya, T. A. (2022). Engineering of escherichia coli glyceraldehyde-3-phosphate dehydrogenase with dual nad(+)/nadp(+) cofactor specificity for improving amino acid production. Microorganisms. 10, 976–990. doi: 10.3390/microorganisms10050976
Su, B., Lai, P., Yang, F., Li, A., Deng, M. R., and Zhu, H. (2022). Engineering a balanced acetyl coenzyme a metabolism in saccharomyces cerevisiae for lycopene production through rational and evolutionary engineering. J. Agric. Food Chem. 70, 4019–4029. doi: 10.1021/acs.jafc.2c00531
Su, X., Wellen, K. E., and Rabinowitz, J. D. (2016). Metabolic control of methylation and acetylation. Curr. Opin. Chem. Biol. 30, 52–60. doi: 10.1016/j.cbpa.2015.10.030
Sui, Y. F., Schutze, T., Ouyang, L. M., Lu, H., Liu, P., Xiao, X., et al. (2020). Engineering cofactor metabolism for improved protein and glucoamylase production in aspergillus Niger. Microb. Cell Fact. 19:198. doi: 10.1186/s12934-020-01450-w
Tajima, Y., Yamamoto, Y., Fukui, K., Nishio, Y., Hashiguchi, K., Usuda, Y., et al. (2015). Impact of an energy-conserving strategy on succinate production under weak acidic and anaerobic conditions in enterobacter aerogenes. Microb. Cell Fact. 14:80. doi: 10.1186/s12934-015-0269-6
Tesfay, M. A., Win, X., Lin, H., Liu, Y., Li, C., Lin, J., et al. (2021). Efficient l-xylulose production using whole-cell biocatalyst with nad+ regeneration system through co-expression of xylitol dehydrogenase and nadh oxidase in escherichia coli. Biochem. Eng. J. 175:108137. doi: 10.1016/j.bej.2021.108137
van Aalst, A. C. A., Mans, R., and Pronk, J. T. (2022). An engineered non-oxidative glycolytic bypass based on Calvin-cycle enzymes enables anaerobic co-fermentation of glucose and sorbitol by saccharomyces cerevisiae. Biotechnol Biofuels Bioproducts. 15:112. doi: 10.1186/s13068-022-02200-3
Wagner, G. R., Bhatt, D. P., O'Connell, T. M., Thompson, J. W., Dubois, L. G., Backos, D. S., et al. (2017). A class of reactive acyl-coa species reveals the non-enzymatic origins of protein acylation. Cell Metab. 25, 823–837.e8. doi: 10.1016/j.cmet.2017.03.006
Wang, M., Chen, B., Fang, Y., and Tan, T. (2017). Cofactor engineering for more efficient production of chemicals and biofuels. Biotechnol. Adv. 35, 1032–1039. doi: 10.1016/j.biotechadv.2017.09.008
Wang, X., Feng, Y., Guo, X., Wang, Q., Ning, S., Li, Q., et al. (2021). Creating enzymes and self-sufficient cells for biosynthesis of the non-natural cofactor nicotinamide cytosine dinucleotide. Nat. Commun. 12:2116. doi: 10.1038/s41467-021-22357-z
Wang, Y., Hu, L., Huang, H., Wang, H., Zhang, T., Chen, J., et al. (2020). Eliminating the capsule-like layer to promote glucose uptake for hyaluronan production by engineered corynebacterium glutamicum. Nat. Commun. 11:3120. doi: 10.1038/s41467-020-16962-7
Wang, J., Ledesma-Amaro, R., Wei, Y., Ji, B., and Ji, X. J. (2020). Metabolic engineering for increased lipid accumulation in yarrowia lipolytica—a review. Bioresour. Technol. 313:123707. doi: 10.1016/j.biortech.2020.123707
Wang, W., Li, S., Li, Z., Zhang, J., Fan, K., Tan, G., et al. (2020). Harnessing the intracellular triacylglycerols for titer improvement of polyketides in streptomyces. Nat. Biotechnol. 38, 76–83. doi: 10.1038/s41587-019-0335-4
Wang, D., Li, D., Zhang, G., Wang, C., and Wei, G. (2019). Disruption of por1 gene in candida utilis improves co-production of s-adenosylmethionine and glutathione. J. Biotechnol. 290, 16–23. doi: 10.1016/j.jbiotec.2018.12.005
Wang, Y., San, K. Y., and Bennett, G. N. (2013). Cofactor engineering for advancing chemical biotechnology. Curr. Opin. Biotechnol. 24, 994–999. doi: 10.1016/j.copbio.2013.03.022
Wang, P., Wei, W., Ye, W., Li, X., Zhao, W., Yang, C., et al. (2019). Synthesizing ginsenoside rh2 in saccharomyces cerevisiae cell factory at high-efficiency. Cell Discov. 5:5. doi: 10.1038/s41421-018-0075-5
Wang, L., Yu, H., Xu, J., Ruan, H., and Zhang, W. (2020). Deciphering the crucial roles of arac-type transcriptional regulator cgl2680 on nadph metabolism and l-lysine production in corynebacterium glutamicum. World J. Microbiol. Biotechnol. 36:82. doi: 10.1007/s11274-020-02861-y
Wegner, S. A., Chen, J., Ip, S. S., Zhang, Y., Dugar, D., and Avalos, J. L. (2021). Engineering acetyl-coa supply and erg9 repression to enhance mevalonate production in saccharomyces cerevisiae. J. Ind. Microbiol. Biotechnol. 48:b50. doi: 10.1093/jimb/kuab050
Wernig, F., Baumann, L., Boles, E., and Oreb, M. (2021). Production of octanoic acid in saccharomyces cerevisiae: investigation of new precursor supply engineering strategies and intrinsic limitations. Biotechnol. Bioeng. 118, 3046–3057. doi: 10.1002/bit.27814
Willquist, K., and van Niel, E. W. (2010). Lactate formation in caldicellulosiruptor saccharolyticus is regulated by the energy carriers pyrophosphate and atp. Metab. Eng. 12, 282–290. doi: 10.1016/j.ymben.2010.01.001
Xiong, J., Chen, H., Liu, R., Yu, H., Zhuo, M., Zhou, T., et al. (2021). Tuning a bi-enzymatic cascade reaction in escherichia coli to facilitate nadph regeneration for ε-caprolactone production. Bioresour. Bioprocess. 8:32. doi: 10.1186/s40643-021-00370-w
Xu, Y., Niu, X., Chen, H., Zhao, S., and Chen, X. (2017). Switch-on fluorescence sensor for ascorbic acid detection based on mos2 quantum dots-mno2 nanosheets system and its application in fruit samples. Chin. Chem. Lett. 28, 338–344. doi: 10.1016/j.cclet.2016.10.003
Xu, R., Wang, D., Wang, C., Zhang, G., and Wei, G. (2018). Improved s-adenosylmethionine and glutathione biosynthesis by heterologous expression of an atp6 gene in candida utilis. J. Basic Microbiol. 58, 875–882. doi: 10.1002/jobm.201800151
Xu, M., Wu, H., Shen, P., Jiang, X., Chen, X., Lin, J., et al. (2020). Enhancement of nadph availability for coproduction of coenzyme q(10) and farnesol from rhodobacter sphaeroides. J. Ind. Microbiol. Biotechnol. 47, 263–274. doi: 10.1007/s10295-020-02261-z
Xue, S., Jiang, H., Chen, L., Ge, N., Liu, G., Hu, Z., et al. (2019). Over-expression of vitreoscilla hemoglobin (vhb) and flavohemoglobin (fhb) genes greatly enhances pullulan production. Int. J. Biol. Macromol. 132, 701–709. doi: 10.1016/j.ijbiomac.2019.04.007
Yamanaka, K., Kito, N., Imokawa, Y., Maruyama, C., Utagawa, T., and Hamano, Y. (2010). Mechanism of -poly-l-lysine production and accumulation revealed by identification and analysis of an -poly-l-lysine-degrading enzyme. Appl. Environ. Microbiol. 76, 5669–5675. doi: 10.1128/AEM.00853-10
Yu, P., Ma, J., Zhu, P., Chen, Q., and Zhang, Q. (2021). Enhancing the production of γ-aminobutyric acid in escherichia coli bl21 by engineering the enzymes of the regeneration pathway of the coenzyme factor pyridoxal 5′-phosphate. World J. Microbiol. Biotechnol. 37:130. doi: 10.1007/s11274-021-03103-5
Yu, T., Zhou, Y. J., Huang, M., Liu, Q., Pereira, R., David, F., et al. (2018). Reprogramming yeast metabolism from alcoholic fermentation to lipogenesis. Cell 174, 1549–1558.e14. doi: 10.1016/j.cell.2018.07.013
Yuan, L., Qin, Y., Zou, Z., Appiah, B., Huang, H., Yang, Z., et al. (2022). Enhancing intracellular nadph bioavailability through improving pentose phosphate pathway flux and its application in biocatalysis asymmetric reduction reaction. J. Biosci. Bioeng. 134, 528–533. doi: 10.1016/j.jbiosc.2022.08.010
Zha, W., Rubin-Pitel, S. B., Shao, Z., and Zhao, H. (2009). Improving cellular malonyl-coa level in escherichia coli via metabolic engineering. Metab. Eng. 11, 192–198. doi: 10.1016/j.ymben.2009.01.005
Zhang, J., Hansen, L. G., Gudich, O., Viehrig, K., Lassen, L., Schrubbers, L., et al. (2022). A microbial supply chain for production of the anti-cancer drug vinblastine. Nature 609, 341–347. doi: 10.1038/s41586-022-05157-3
Zhang, H., Kang, X., Xiao, N., Gao, M., Zhao, Y., Zhang, B., et al. (2019). Intracellular expression of vitreoscilla haemoglobin improves lipid production in yarrowia lipolytica. Lett. Appl. Microbiol. 68, 248–257. doi: 10.1111/lam.13111
Zhang, Y., Li, Y., Du, C., Liu, M., and Cao, Z. A. (2006). Inactivation of aldehyde dehydrogenase: a key factor for engineering 1,3-propanediol production by klebsiella pneumoniae. Metab. Eng. 8, 578–586. doi: 10.1016/j.ymben.2006.05.008
Zhang, R., Li, C., Wang, J., Yang, Y., and Yan, Y. (2018). Microbial production of small medicinal molecules and biologics: from nature to synthetic pathways. Biotechnol. Adv. 36, 2219–2231. doi: 10.1016/j.biotechadv.2018.10.009
Zhang, M., Wang, Z., Chi, Z., Liu, G. L., and Chi, Z. M. (2022). Metabolic engineering of aureobasidium melanogenum 9-1 for overproduction of liamocins by enhancing supply of acetyl-coa and atp. Microbiol. Res. 265:127172. doi: 10.1016/j.micres.2022.127172
Zhou, J., Liu, L., and Chen, J. (2010). Improved atp supply enhances acid tolerance of candida glabrata during pyruvic acid production. J. Appl. Microbiol. 110, 44–53. doi: 10.1111/j.1365-2672.2010.04865.x
Zhou, J., Liu, L., Shi, Z., Du, G., and Chen, J. (2009). Atp in current biotechnology: regulation, applications and perspectives. Biotechnol. Adv. 27, 94–101. doi: 10.1016/j.biotechadv.2008.10.005
Zhu, Z. T., Du, M. M., Gao, B., Tao, X. Y., Zhao, M., Ren, Y. H., et al. (2021). Metabolic compartmentalization in yeast mitochondria: burden and solution for squalene overproduction. Metab. Eng. 68, 232–245. doi: 10.1016/j.ymben.2021.10.011
Keywords: cofactors, metabolic engineering, regulatory strategies, microbial cell factory, biological manufacturing
Citation: Sun Y, Zhang T, Lu B, Li X and Jiang L (2023) Application of cofactors in the regulation of microbial metabolism: A state of the art review. Front. Microbiol. 14:1145784. doi: 10.3389/fmicb.2023.1145784
Edited by:
Yongkun Lv, Zhengzhou University, ChinaReviewed by:
Chao Ye, Nanjing Normal University, ChinaDongbo Cai, Hubei University, China
Chenyi Li, College of Chemistry (UC), Berkeley, United States
Copyright © 2023 Sun, Zhang, Lu, Li and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiangfei Li, eGlhbmdmZWlsaUBtYWlsLmFocHUuZWR1LmNu
 Yang Sun
Yang Sun Ting Zhang1
Ting Zhang1 Ling Jiang
Ling Jiang