
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 28 April 2023
Sec. Infectious Agents and Disease
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1144026
This article is part of the Research TopicCoronaviruses from the One Health PerspectiveView all 5 articles
 Ye Li1
Ye Li1 K. T. Ash1,2
K. T. Ash1,2 Dominique C. Joyner1,2
Dominique C. Joyner1,2 Daniel E. Williams3
Daniel E. Williams3 I. Alamilla4
I. Alamilla4 P. J. McKay4
P. J. McKay4 C. Iler5
C. Iler5 B. M. Green6
B. M. Green6 F. Kara-Murdoch2,3
F. Kara-Murdoch2,3 C. M. Swift2,3
C. M. Swift2,3 Terry C. Hazen1,2,6,7,8,9*
Terry C. Hazen1,2,6,7,8,9*Introduction: Although severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) RNA has been frequently detected in sewage from many university dormitories to inform public health decisions during the COVID-19 pandemic, a clear understanding of SARS-CoV-2 RNA persistence in site-specific raw sewage is still lacking. To investigate the SARS-CoV-2 RNA persistence, a field trial was conducted in the University of Tennessee dormitories raw sewage, similar to municipal wastewater.
Methods: The decay of enveloped SARS-CoV-2 RNA and non-enveloped Pepper mild mottle virus (PMMoV) RNA was investigated by reverse transcription-quantitative polymerase chain reaction (RT-qPCR) in raw sewage at 4°C and 20°C.
Results: Temperature, followed by the concentration level of SARS-CoV-2 RNA, was the most significant factors that influenced the first-order decay rate constants (k) of SARS-CoV-2 RNA. The mean k values of SARS-CoV-2 RNA were 0.094 day−1 at 4°C and 0.261 day−1 at 20°C. At high-, medium-, and low-concentration levels of SARS-CoV-2 RNA, the mean k values were 0.367, 0.169, and 0.091 day−1, respectively. Furthermore, there was a statistical difference between the decay of enveloped SARS-CoV-2 and non-enveloped PMMoV RNA at different temperature conditions.
Discussion: The first decay rates for both temperatures were statistically comparable for SARS-CoV-2 RNA, which showed sensitivity to elevated temperatures but not for PMMoV RNA. This study provides evidence for the persistence of viral RNA in site-specific raw sewage at different temperature conditions and concentration levels.
Wastewater-based epidemiology (WBE) of severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) is monitored from a large scale of wastewater treatment plants (Sherchan et al., 2020; Ahmed et al., 2020a, 2021), hospital sewers (Zhang et al., 2020; Gonçalves et al., 2021), rivers (Haramoto et al., 2020; Rimoldi et al., 2020), natural lakes (Gerrity et al., 2021), to a small scale of residential buildings (Wong et al., 2021), and university dormitories (Betancourt et al., 2021; Scott et al., 2021; Jain et al., 2022; Mangwana et al., 2022), the latter of which is the focus of the current study. As the wastewater from university dormitories is mainly generated from washing and bathing and sometimes contains kitchen wastewater, while the contributor to influent sewage treatment wastewater is from residential (private residences, dormitories, hotels, and residential care facilities) and commercial facilities (including hospitals), which is much more complex than the wastewater from university dormitories. Apart from the wastewater composition between university dormitories and wastewater treatment plants, the bias also appears in evaluating the decay rate of SARS-CoV-2 RNA due to the duration between excretion in feces and wastewater sampling.
Recently, most research has focused on the persistence of SARS-CoV-2 RNA in wastewater influents at different temperatures. Ahmed et al. (2020b) found that the first-order decay rate constants (k) of SARS-CoV-2 RNA were 0.084 day−1 at 4°C, 0.114 day−1 at 15°C, 0.183 day−1 at 25°C, and 0.286 day−1 at 37°C in wastewater. It has been observed that the decay of SARS-CoV-2 RNA happened at 4°C over 28 days but not at −20°C or −75°C (Hokajärvi et al., 2021). Furthermore, some researchers investigated the infectivity of SARS-CoV-2, the decay rate of which was much faster than its RNA decay in the wastewater influents (De Oliveira et al., 2021; Hokajärvi et al., 2021). However, these studies were conducted with spiking exogenous SARS-CoV-2, which could not completely illustrate the natural decay of SARS-CoV-2 in wastewater. Wurtzer et al. (2021) pointed out that there was not only one form of SARS-CoV-2 RNA present in wastewater, and the intact structure of the spiking SARS-CoV-2 might lead to slower decay in wastewater. Only Weidhaas et al. (2021) and Yang et al. (2022) conducted studies with endogenous SARS-CoV-2 RNA. Yang et al. (2022) found that the k values of endogenous SARS-CoV-2 RNA were 0.134 day−1 at 4°C and 0.274 day−1 at 26°C. Interestingly, much higher k values of the endogenous SARS-CoV-2 RNA were observed in the study of Weidhaas et al. (2021), in which the k values ranged from 2.16 to 4.32 day−1 at 4°C to 35°C in wastewater. However, relatively little is known about the persistence of endogenous SARS-CoV-2 RNA in raw sewage from the specific site, which could infer if the decay of SARS-CoV-2 RNA happened in-sewer travel time at some temperature conditions.
Pepper mild mottle virus (PMMoV) and SARS-CoV-2 are single-strand RNA viruses, while PMMoV is a non-enveloped virus, and SARS-CoV-2 is an enveloped virus wrapped with the phospholipid layer. It has been reported that the reduction of the enveloped virus was more efficient than non-enveloped ones due to the protection of the genome from degradation by the capsid (Ye et al., 2016). In recent years, some studies have aimed to use models to normalize the concentration of SARS-CoV-2 RNA by the concentration of PMMoV RNA to find the relationship between the concentration of SARS-CoV-2 RNA in wastewater and COVID-19 incident cases. Some parameters the models needed could be directly obtained from the SARS-CoV-2 related experiments, like flow rate and the decay rate of SARS-CoV-2 RNA; others need to infer from PMMoV RNA concentration, like fecal loads and viral shedding rates. Hence, it is also essential to study the decay of PMMoV RNA to rectify the variation of RNA recovery between samples and to detect the decay of RNA during transport and storage. Many studies focused on finding the relationship between the RNA concentrations of these two viruses, but limited information on PMMoV RNA persistence in raw wastewater compared to SARS-CoV-2 RNA. Kitajima et al. (2018) have shown that there was slight decay of PMMoV RNA in wastewater with seasonal variation. Sala-Comorera et al. (2021) found that PMMoV RNA was stable and persisted longer in aquatic environments with no significant decay. Burnet et al. (2023) highlighted no decay signal of PMMoV RNA in wastewater influent at all temperature conditions (4°C, 12°C, and 20°C) over 1 month.
Critical gaps exist in our understanding of the persistence of enveloped SARS-CoV-2 RNA and non-enveloped PMMoV RNA in site-specific raw sewage. We investigated the k values of these two viral RNA in raw sewage from the university dormitories using reverse transcription-quantitative polymerase chain reaction (RT-qPCR) for at least 29 days, with attention to the effects of representative temperatures in cold and temperate regions (4°C and 20°C) and different concentration levels of SARS-CoV-2 RNA (102 to 104 copies/L). This study could enhance further understanding of the persistence of enveloped SARS-CoV-2 and non-enveloped PMMoV RNA in the real site-specific raw sewage system. The original surveillance study at this university (Ash et al., 2023) was used to identify select samples that would provide an array of initial concentrations used for these long-term decay studies.
During the summer of 2020, aiming to gradually end working remotely for both students and employees, the University of Tennessee administration decided to apply WBE for the early detection of COVID-19 cases in student dorms. Since each dorm has a known student number, these students could represent the health situation of the overall campus. Sewage samples were collected weekly to monitor 18 dorms, 15 Fraternities, and 14 Sororities, from 14 September 2020 to 21 September 2021, to track SARS-CoV-2 RNA in sewage during the academic semester. In order to sample from specific buildings with defined student populations, samples were collected from the downstream of dispense valves or sewer manholes just before being merged or mixed with other sewer lines. Grab samples (>50 mL) were collected from the manhole using a stainless-steel telescopic rod pole swivel dipper water swing sampler and submerged into the flowing sewage or using a sterile Nalgene bottle to collect sewage from the valve. Samples collecting started at 8:00 am. All samples were transported to the BSL-2 laboratory in a cooler containing ice within 3 h for immediate processing.
The initial concentrations of SARS-CoV-2 and PMMoV RNA were determined immediately by 50 ml of well-mixed raw sewage samples at time zero (t = 0) within 5 h of sample collection. Remain samples were incubated in the dark at 4°C and 20°C to evaluate the temperature effect on the decay of SARS-CoV-2 and PMMoV RNA in sewage. On specific days, sewage samples were retrieved from each temperature condition.
Sewage samples were pasteurized for 2 h at 60°C, followed by centrifuging at 5,000 × g for 10 min, and then filtration through a 0.45 and 0.22 μm nitrocellulose filter to remove large suspended particulate matter. The filtered samples were concentrated using ultrafiltration with an Amicon Ultra-15 filtration device (EMD Millipore, Burlington, MA). Centrifuge the Amicon Ultra at 4,000 × g for 30 min (Swing-arm rotor) or 5,000 × g for 20 min (Fixed-angle rotor) at room temperature. The concentrate was transferred to (~250 μL) 2 mL DNA LoBind tubes, and RNA exactions were carried out using a Qiagen viral RNA Mini Kit (Qiagen, Valencia, CA, United States; Ash et al., 2021). Briefly, a volume of 60 μL of RNA was extracted using the Qiagen viral RNA Mini Kit following the instructions of the manufacturer from a homogenized sample. DNase/RNase-free water was used as extraction negative control. All RNA samples were stored at −80°C and subjected to RT-qPCR analysis within 1 day of RNA extraction.
RT-qPCR was used to quantify the concentrations of SARS-CoV-2 and PMMoV RNA in each sample. CDC primer/probe assays SARS-CoV-2 N1was used, which was quantified using the TaqPath 1-Step RT-qPCR Master Mix, CG (Thermo Fisher Scientific) on an Applied Biosystems QuantStudios 7 Pro Real-Time PCR System instrument (Ash et al., 2023). Each 20 μL reaction contained 5 μL of 4X Master Mix (Thermo Fisher Scientific), 0.25 μL of 10 μmol/L probe, 1 μL each of 10 μmol/L forward and reverse primers, 7.75 μL of nuclease-free water, and 5 μL of nucleic acid extract. Reagents were pipetted into 96-well plates and vortexed for 10 s. Thermocycling conditions were as follows: uracil-DNA glycosylase incubation for 2 min at 25°C, reverse transcription for 15 min at 50°C, activation of the Taq enzyme for 2 min at 95°C, and two-step cycling for 3 s at 95°C and 30 s at 55°C for 45 cycles. A positive test result was defined as an exponential fluorescent curve that crossed the threshold within 40 cycles (cycle threshold [Ct] <40).
PMMoV was also quantified by RT-qPCR using the TaqPath 1-Step RT-qPCR Master Mix, CG (Thermo Fisher Scientific) on a QuantStudios 7 Pro instrument. Each 20 μL reaction contained 5 μL of 4X Master Mix (Thermo Fisher Scientific), 0.5 μl of 10 μmol/L probe, 1.8 μL each of 10 μmol/L forward and reverse primers, 8.9 μL of nuclease-free water, and 2 μL of nucleic acid extract. Reagents were pipetted into 96-well plates and vortexed for 10 s. Thermocycling conditions were as follows: uracil-DNA glycosylase incubation for 2 min at 25°C, reverse transcription for 15 min at 50°C, activation of the Taq enzyme for 10 min at 95°C, and two-step cycling for 30 s at 95°C and 1 min at 60°C for 40 cycles.
A series of three positive and negative controls (Mastermix + DNase/RNase-free water) were included in each RT-qPCR run. All RT-qPCR reactions were performed in triplicate. The results were applied only if the positive control was positive and the negative control was negative. The sample was defined as positive only if all replicates were positive, each of which should be within the linear range of the standard curve. The efficiency of the N1 standard curve was 94.669% (R2 = 1). The final quantification of SARS-CoV-2 RNA was the mean of three replicates of virus copies. RT-qPCR outputs were converted to copies per liter. The detection limit of SARS-CoV-2 and PMMoV was 20 and 10 copies/L in this study.
The k and T90 were calculated for each temperature in Sigma Plot 14_5 by Eqs. (1, 2), respectively.
where k is the first-order decay rate. Ct and C0 are the concentration of the viral RNA at time t and time zero, respectively. T90 is the days needed to achieve a 90% reduction in initial concentration. The fit of the model is evaluated by r2.
In this study, the significant factors on k values were determined by a multiple linear regression model [Eq. (3)]. It could explain the impact of influential variables (temperature, concentration levels of SARS-CoV-2, decay rate constant of PMMoV RNA, and sampling sites) as independent data on the first decay rate constants of k, which also included the interaction effects between different influential variables.
where xi is the influential variable, a is the intercept, bi is the regression coefficient, and ε is the regression residual.
All linear fitting and statistical analyses were performed using SPSS (version 26). One-way ANOVA with the Kruskal–Wallis H test was used to evaluate the effect of different SARS-CoV-2 concentration levels and sampling sites, as well as the Mann–Whitney U test to compare differences between different temperatures. The paired t-test was used to compare the decay of SARS-CoV-2 and PMMoV RNA (α = 0.05 for both tests). All statistical differences were determined by p < 0.05.
The first-order decay rate model fits most decay curves of SARS-CoV-2 RNA (r2 = 0.512 to 0.996; Table 1), a bad fit for most decay curves of PMMoV RNA (r2 = 0 to 0.996; Table 1). The decay curves with smaller r2 had smaller k values, indicating limited decay with time. The k and T90 for SARS-CoV-2 and PMMoV RNA are shown in Table 1.
The multiple regression model revealed that the influential variables, e.g., temperature, PMMoV RNA, concentration levels of SARS-CoV-2 RNA, and the interaction between temperature and concentration levels of SARS-CoV-2 were significant for the first decay rate constants [F (4, 29) = 9.084, p-value < 0.0001]. The sampling site was removed from the model due to no significance. The modified model indicated that temperature, PMMoV RNA, and the concentration levels of SARS-CoV-2 RNA remained significant (t = 3.850, 2.185, −2.817, and the corresponding p-value was 0.001, 0.009, and 0.038, respectively) but not the interaction term, suggesting that the effect of temperature is rarely dependent on the concentration levels of SARS-CoV-2. Therefore, the regression model only included temperature, PMMoV RNA, and the concentration levels of SARS-CoV-2 as influential variables. The intercept (a) and coefficients (btemperature, blog10 kPMMoV, and bconcentration level of SARS-CoV-2) of the final regression model were − 0.714, 0.039, 0.035, and − 0.304, respectively. The F-ratio in the ANOVA table showed that the independent variables significantly predicted the dependent variable, F (3, 27) = 10.767, p-value < 0.0001. The positive regression coefficient indicated that the first decay rate constants increased with temperature and were higher at the high decay rate of PMMoV. The negative regression coefficient indicated that k was higher at the low-concentration level of SARS-CoV-2 than at the high level.
The k and T90 values of SARS-CoV-2 RNA are summarized in Table 1, and the decay curves of SARS-CoV-2 RNA are shown in Figure 1. The average initial concentrations (mean ± standard deviation) of SARS-CoV-2 RNA from high to low concentration levels ranged from 4.39 ± 4.19 to 2.65 ± 2.51 log10 copies/L. The mean k values were 0.367, 0.169, and 0.091 day−1 with the corresponding T90 values of 6.27, 13.62, and 25.30 days at high-, medium-, and low-concentration levels of SARS-CoV-2 RNA, respectively. The k values from high to low concentration levels of SARS-CoV-2 in sewage were significantly decreased (Kruskal–Wallis H = 7.48, p-value = 0.024). The temperature remained stable at 4.0°C ± 1°C and 20°C ± 2°C throughout the experiment. The mean k values were 0.094 day−1 at 4°C and 0.261 day−1 at 20°C. The decay characteristic of SARS-CoV-2 RNA at high temperature (20°C) was significantly faster compared to the low temperature (4°C; Mann–Whitney U = 59.00, p-value = 0.003). In addition, the mean T90 values of the SARS-CoV-2 RNA in raw sewage were 24.47 and 8.84 days, corresponding to 4°C and 20°C. When comparing decay rate constants among different concentration levels of SARS-CoV-2 at the same temperature, there were no significant differences for both viral RNA at 4 and 20°C (Table 1). The mean k values for SARS-Co-2 RNA at 4 and 20°C were 0.313 and 0.421 day−1 at the high-concentration level of SARS-CoV-2, 0.043 and 0.295 day−1 at the median concentration level, and 0.036 and 0.145 day−1 at the low-concentration level. The T90 values were 37.37, 118.75, and 120.87 days at 4°C and 9.13, 20.96, and 42.66 days at 20°C corresponding to the concentration levels of SARS-CoV-2 RNA at high, medium, and low, respectively. The decay of high-concentration levels of SARS-CoV-2 RNA was much faster than that with low-concentration levels of SARS-CoV-2 RNA at both low and high temperatures in raw sewage.
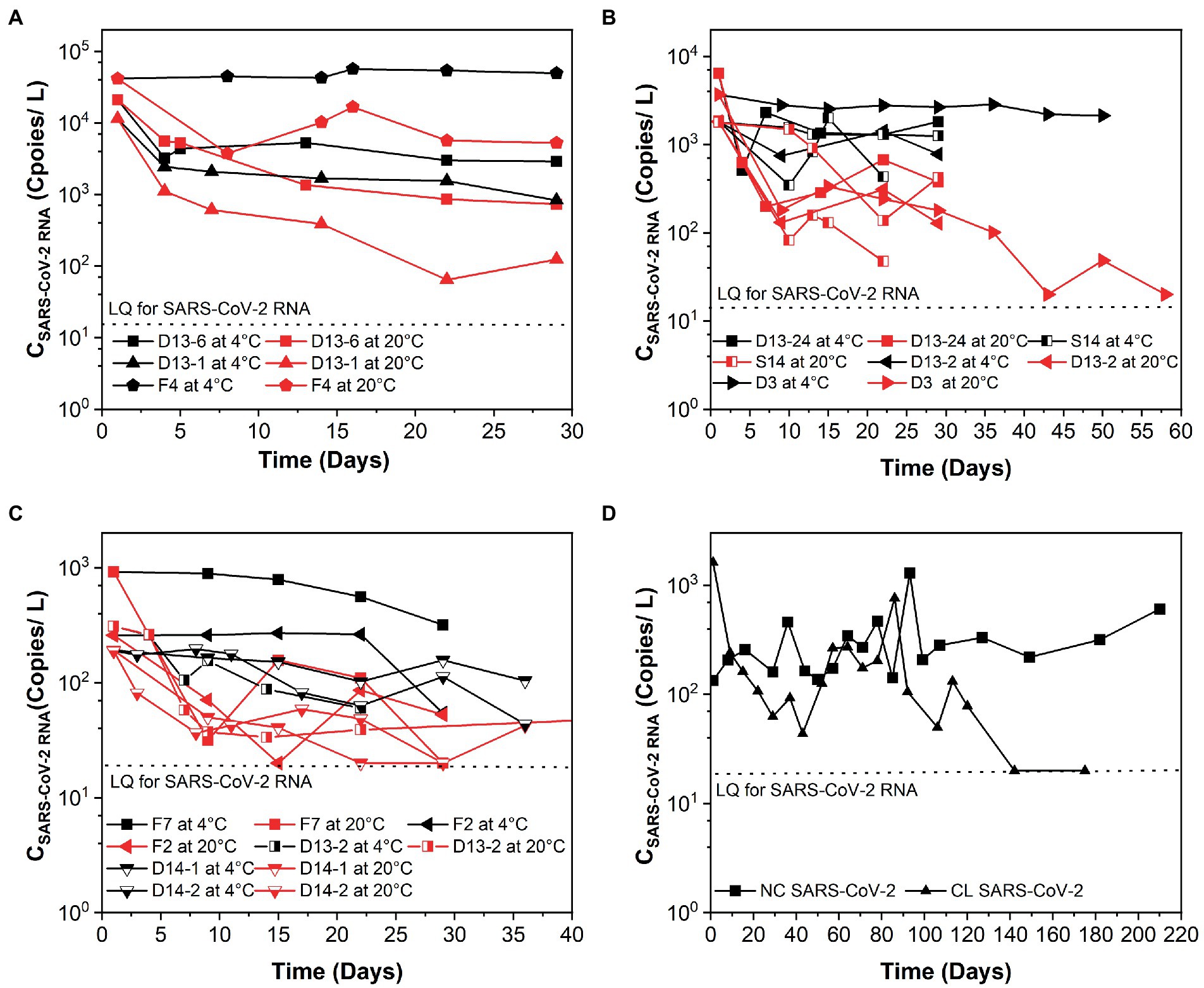
Figure 1. Persistence of SARS-CoV-2 over time (days) in raw sewage from university dormitories. (A) High-concentration level of SARS-CoV-2 (CSARS-CoV-2 RNA > 104 copies/L), (B) Medium-concentration level of SARS-CoV-2 (104 copies/L > CSARS-CoV-2 RNA > 103 copies/L), (C) Low-concentration level of SARS-CoV-2 (103 copies/L > CSARS-CoV-2 RNA > 102 copies/L), (D) Long time decay of SARS-CoV-2 at 4°C. LQ means the limit of quantification in RT-qPCR. F4, D13-6, D13-1, D13-24-1, D3, S14, D13-2-1, D13-24-2, S12-1, F7, S12-2, D13-2-2, F2, D14-1, D14-2, NC, and CL correspond to different campus samples.
Table 1 summarizes the k and T90 values of PMMoV RNA, and Figure 2 shows the persistence of PMMoV RNA in raw sewage. The mean initial concentrations (mean ± standard deviation) of PMMoV RNA ranged from 5.17 ± 4.80 to 5.60 ± 5.70 log10 copies/L at different concentration levels of SARS-CoV-2 RNA from high to low. The decay of PMMoV RNA was limited at all concentration levels of SARS-CoV-2 over at least 29 days. The mean k values ranged from 0.012 to 0.154 day−1 with T90 values from 14.95 to 191.88 days at different concentration levels of SARS-CoV-2 RNA, with no statistical difference. The mean k values were 0.05 day−1 at 4°C and 0.09 day−1 at 20°C, with the corresponding T90 values of 46.15 and 25.94 days. The decay characteristic of PMMoV RNA at different temperatures showed no statistical difference.
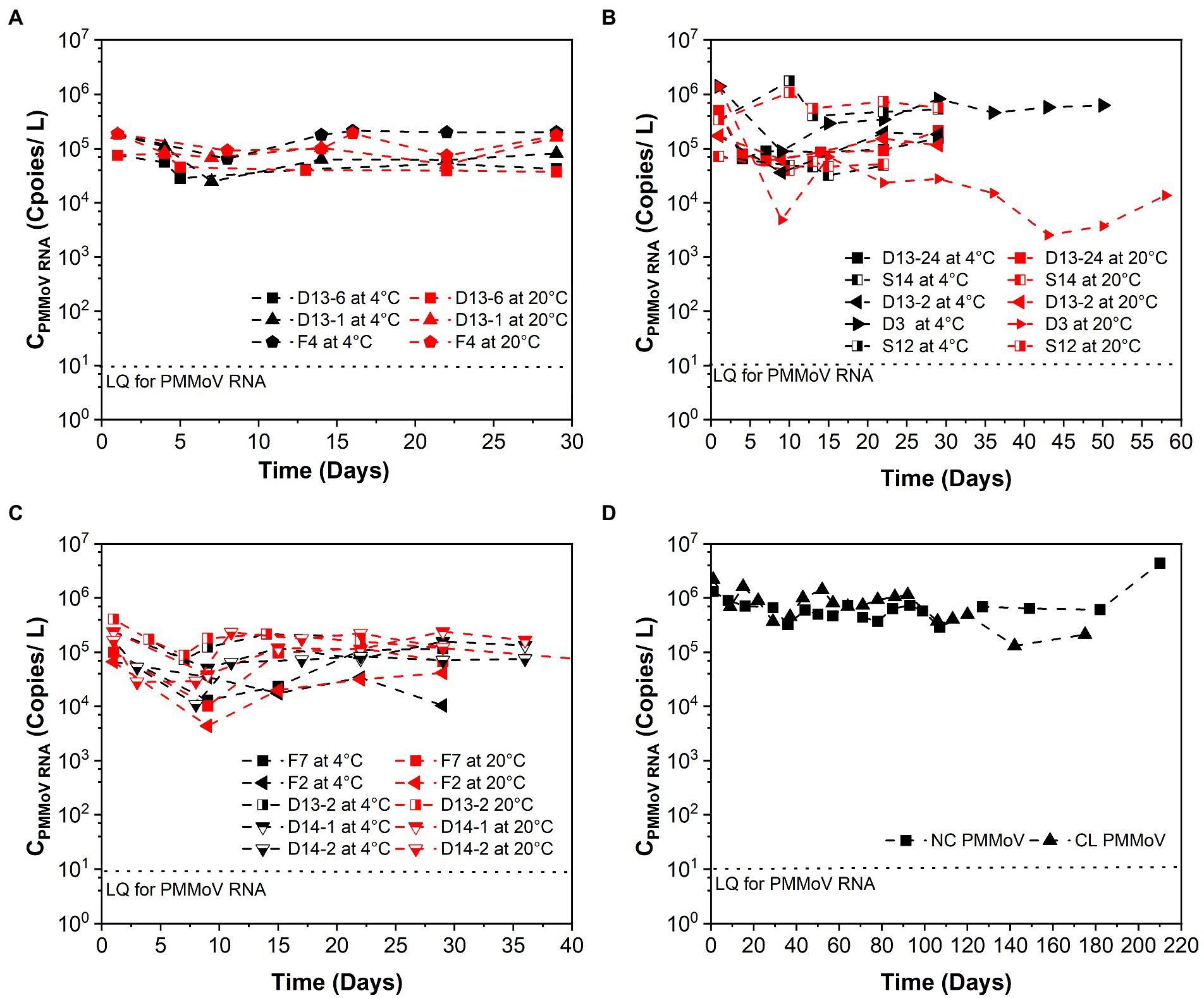
Figure 2. Persistence of PMMoV RNA over time (days) in raw sewage from university dormitories. (A) High-concentration level of SARS-CoV-2 (CSARS-CoV-2 RNA > 104 copies/L), (B) Medium-concentration level of SARS-CoV-2 (104 copies/L > CSARS-CoV-2 RNA > 103 copies/L), (C) Low-concentration level of SARS-CoV-2 (103 copies/L > CSARS-CoV-2 RNA > 102 copies/L), (D) Long time decay of SARS-CoV-2 at 4°C. LQ means the limit of quantification in RT-qPCR. F4, D13-6, D13-1, D13-24-1, D3, S14, D13-2-1, D13-24-2, S12-1, F7, S12-2, D13-2-2, F2, D14-1, D14-2, NC, and CL correspond to different campus samples.
In contrast to SARS-CoV-2 RNA, PMMoV RNA persisted for longer periods at both temperatures evaluated. The mean k value for PMMoV RNA at 4 and 20°C was 0.018 and 0.011 day−1 at the high-concentration level of SARS-CoV-2, 0.101 and 0.207 day−1 at the median concentration level, and 0.014 and 0.010 day−1 at the low-concentration level, respectively. T90 values of PMMoV RNA were 127.92, 22.80, and 164.47 days at 4°C and 209.33, 11.12, and 230.26 days at 20°C from the high to the low concentration level of SARS-CoV-2, respectively.
Viral RNA from SARS-CoV-2 and PMMoV remained detectable for at least 29 days. SARS-CoV-2 RNA was relatively stable with faster decay rates at 20°C than at 4°C in our study. In contrast to SARS-CoV-2, PMMoV persisted for longer periods at both temperature conditions. The decay rates for SARS-CoV-2 and PMMoV were significantly different at both temperatures evaluated (paired t-test, t = 3.733, p-value < 0.002, at 4°C and paired t-test, t = 3.441, p-value < 0.004, at 20°C). In addition, we noted that regardless of other factors, there was a significant difference between the decay characteristics of SARS-CoV-2 and PMMoV RNA (paired t-test, t = 5.153, p-value < 0.001).
Several studies have reported higher persistence of SARS-CoV-2 RNA in wastewater at low temperatures compared to high temperatures, which was consistent with our research. It was observed that SARS-CoV-2 RNA could be detectable over 100 days at 4°C (Figure 1D). As Aboubakr et al. (2021) summarized that the structure of enveloped viruses like coronaviruses was fragile, and the way they infected their host cells made them susceptible to heat. As the morphology and chemical structure of the SARS-CoV-2 virus is similar to other coronaviruses, the possible explanation for this trend was reported as due to the denaturation of proteins and increased activity of extracellular enzymes under higher temperature, which facilitated the decay of SARS-CoV-2 (Aquino de Carvalho et al., 2017). Ahmed et al. (2020b) reported that the k values of spiked with gamma-irradiated SARS-CoV-2 were 0.084 day−1 at 4°C, 0.114 day−1 at 15°C, 0.183 day−1 at 25°C, and 0.286 day−1 at 37°C in wastewater. The decay rate constants of SARS-CoV-2 RNA in the study of Ahmed et al. (2020b) were slightly higher than our results. The reason for this could be due to the different sample preparation. Wurtzer et al. (2021) pointed out that there was not only one form of SARS-CoV-2 RNA present in sewage, including integrated viruses, genomic RNA protected within an infectious or non-infectious structure, and free total or partial genomic RNA. Yang et al. (2022) inferred that the integrity of the viral structure might be better than in raw wastewater, so the incomplete viral structure made viral RNA easier to degrade. Future study is necessary to investigate the influential elements that differentiate the decay between endogenous and exogenous SARS-CoV-2 RNA in sewage. In addition, Hokajärvi et al. (2021) conducted the study with exogenous SARS-CoV-2 RNA and showed that it decayed over 28 days at 4°C but not at −20°C or −75°C. Recently, Yang et al. (2022) observed that the k values of endogenous SARS-CoV-2 RNA were 0.134 and 0.274 day−1 at 4°C and 26°C, respectively. These values were slightly higher than our studies at similar temperature conditions. Interestingly, Weidhaas et al. (2021) obtained much higher k values of the endogenous SARS-CoV-2 RNA than our study, in which the k values ranged from 2.16 to 4.32 day−1 at 4 to 35°C in wastewater. This may be due to the experimental durations. From Figures 1A–C, we could also observe that at the very beginning, SARS-CoV-2 RNA decayed much faster than in the following days. In addition, Roldan-Hernandez et al. (2022) reported that the short length of the study could observe limited decay, which affected the k. The duration of our study was at least 29 days, but just 4 and 1 days for the studies of Hokajärvi et al. (2021) and Yang et al. (2022), respectively.
We had similar results with Bivins et al. (2020) regarding the decay rate of different SARS-CoV-2 concentrations, who revealed that the k of the high titer SARS-CoV-2 concentration was 0.67, which is around 7 times higher than the low titer decay rate in 20°C raw sewage samples over the experiment duration of 7 days, the reason for which is unknown.
The decay characteristic of PMMoV RNA at different temperatures showed no statistical difference. This was in agreement with the study of Burnet et al. (2023), who found no significant decay signal of PMMoV RNA in wastewater influent at all temperature conditions (4°C, 12°C, and 20°C) over 1 month. Kitajima et al. (2018) summarized that there was a slight seasonal variation of PMMoV RNA in wastewater. In addition, Sala-Comorera et al. (2021) found that PMMoV RNA was stable and persisted longer in aquatic environments, and no significant decline was observed. From these studies, PMMoV can be considered a conservative marker with respect to the virus reduction (Kitajima et al., 2018; Sala-Comorera et al., 2021). As PMMoV is a non-enveloped virus, it has been reported that the long persistence of the non-enveloped virus was due to lack of a structure named lipid bilayer outside the viral protein capsid that is sensitive to the detergents and organic solvents in the wastewater (Vidaver et al., 1973).
It has been reported that enveloped viruses were commonly thought to be less persistent than non-enveloped viruses due to the structure of a lipid layer, which is unstable at high temperatures and increases the decay of the virus RNA. Furthermore, the wastewater detergent and other chemical agents could be penetrated the lipid layer, which led to the degrading of the viral envelope (Wurtzer et al., 2021).
This study conducted a field trial on the persistence of enveloped SARS-CoV-2 and non-enveloped PMMoV RNA in raw sewage from university dormitories. In our study, SARS-CoV-2 RNA decayed significantly faster at 20°C than at 4°C. The decay rate constants of the high-concentration levels of SARS-CoV-2 RNA were significantly higher than the lower levels at both temperature conditions and could be detected at least 29 days at all concentration levels. Due to the prolonged persistence of SARS-CoV-2 RNA at 4°C, it suggested that sewage samples could be stored at 4°C without significant degradation for a considerable length of time. This is particularly important for laboratories where resources and throughput are limited, and sample storage is unavoidable. We also need to pay attention to the decay of SARS-CoV-2 RNA at higher concentration levels and higher temperatures for long in-sewer traveling time. In contrast to enveloped SARS-CoV-2 RNA, non-enveloped PMMoV RNA could be highly persistent in raw sewage at both temperature conditions. Future studies will need to identify why the decay of SARS-CoV-2 RNA at different concentration levels is significant.
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
YL and TH: writing. DW, IA, PM, DJ, and CI: collection. YL, KA, DW, DJ, IA, PM, CI, and BG: lab analysis. YL, KA, and TH: data Analysis. TH and DJ: project management. All authors contributed to the article and approved the submitted version.
The author would like to thank the Office of Research and Engagement of the University of Tennessee, Knoxville, for providing funding for the project.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Aboubakr, H. A., Sharafeldin, T. A., and Goyal, S. M. (2021). Stability of SARS-CoV-2 and other coronaviruses in the environment and on common touch surfaces and the influence of climatic conditions: a review. Transbound. Emerg. Dis. 68, 296–312. doi: 10.1111/tbed.13707
Ahmed, W., Angel, N., Edson, J., Bibby, K., Bivins, A., O’Brien, J. W., et al. (2020a). First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 728:138764. doi: 10.1016/j.scitotenv.2020.138764
Ahmed, W., Bertsch, P. M., Bibby, K., Haramoto, E., Hewitt, J., Huygens, F., et al. (2020b). Decay of SARS-CoV-2 and surrogate murine hepatitis virus RNA in untreated wastewater to inform application in wastewater-based epidemiology. Environ. Res. 191:110092. doi: 10.1016/j.envres.2020.110092
Ahmed, W., Tscharke, B., Bertsch, P. M., Bibby, K., Bivins, A., Choi, P., et al. (2021). SARS-CoV-2 RNA monitoring in wastewater as a potential early warning system for COVID-19 transmission in the community: a temporal case study. Sci. Total Environ. 761:144216. doi: 10.1016/j.scitotenv.2020.144216
Aquino De Carvalho, N., Stachler, E. N., Cimabue, N., and Bibby, K. (2017). Evaluation of Phi6 persistence and suitability as an enveloped virus surrogate. Environ. Sci. Technol. 51, 8692–8700. doi: 10.1021/acs.est.7b01296
Ash, K., Alamilla, I., Li, Y., Joyner, D., Williams, D., Mckay, P., et al. (2021). Coding-complete genome sequence of a SARS-CoV-2 variant obtained from raw sewage at the University of Tennessee—Knoxville Campus. Microbiol Res Announce 10, e01049–e01021. doi: 10.1128/MRA.01049-21
Ash, K. T., Li, Y., Alamilla, I., Joyner, D., Williams, D. E., Mckay, P., et al. (2023). SARS-CoV-2 raw wastewater surveillance from student residences on an urban university campus. Front. Microbiol. 14:1101205. doi: 10.3389/fmicb.2023.1101205
Betancourt, W. Q., Schmitz, B. W., Innes, G. K., Prasek, S. M., Brown, K. M. P., Stark, E. R., et al. (2021). COVID-19 containment on a college campus via wastewater-based epidemiology, targeted clinical testing and an intervention. Sci. Total Environ. 779:146408. doi: 10.1016/j.scitotenv.2021.146408
Bivins, A., Greaves, J., Fischer, R., Yinda, K. C., Ahmed, W., Kitajima, M., et al. (2020). Persistence of SARS-CoV-2 in water and wastewater. Environ. Sci. Technol. Lett. 7, 937–942. doi: 10.1021/acs.estlett.0c00730
Burnet, J.-B., Cauchie, H.-M., Walczak, C., Goeders, N., and Ogorzaly, L. (2023). Persistence of endogenous RNA biomarkers of SARS-CoV-2 and PMMoV in raw wastewater: impact of temperature and implications for wastewater-based epidemiology application. Sci. Total Environ. 857:159401. doi: 10.1016/j.scitotenv.2022.159401
De Oliveira, L. C., Torres-Franco, A. F., Lopes, B. C., Da Silva Santos, B. S. Á., Costa, E. A., Costa, M. S., et al. (2021). Viability of SARS-CoV-2 in river water and wastewater at different temperatures and solids content. Water Res. 195:117002. doi: 10.1016/j.watres.2021.117002
Gerrity, D., Papp, K., Stoker, M., Sims, A., and Frehner, W. (2021). Early-pandemic wastewater surveillance of SARS-CoV-2 in southern Nevada: methodology, occurrence, and incidence/prevalence considerations. Water Res X 10:100086. doi: 10.1016/j.wroa.2020.100086
Gonçalves, J., Koritnik, T., Mioč, V., Trkov, M., Bolješič, M., Berginc, N., et al. (2021). Detection of SARS-CoV-2 RNA in hospital wastewater from a low COVID-19 disease prevalence area. Sci. Total Environ. 755:143226. doi: 10.1016/j.scitotenv.2020.143226
Haramoto, E., Malla, B., Thakali, O., and Kitajima, M. (2020). First environmental surveillance for the presence of SARS-CoV-2 RNA in wastewater and river water in Japan. Sci. Total Environ. 737:140405. doi: 10.1016/j.scitotenv.2020.140405
Hokajärvi, A.-M., Rytkönen, A., Tiwari, A., Kauppinen, A., Oikarinen, S., Lehto, K.-M., et al. (2021). The detection and stability of the SARS-CoV-2 RNA biomarkers in wastewater influent in Helsinki, Finland. Sci. Total Environ. 770:145274. doi: 10.1016/j.scitotenv.2021.145274
Jain, N., Hamilton, D., Mital, S., Ilias, A., Brinkmann, M., and Mcphedran, K. (2022). Long-term passive wastewater surveillance of SARS-CoV-2 for seven university dormitories in comparison to municipal surveillance. Sci. Total Environ. 852:158421. doi: 10.1016/j.scitotenv.2022.158421
Kitajima, M., Sassi, H. P., and Torrey, J. R. (2018). Pepper mild mottle virus as a water quality indicator. NPJ Clean Water 1, 1–9. doi: 10.1038/s41545-018-0019-5
Mangwana, N., Archer, E., Muller, C. J., Preiser, W., Wolfaardt, G., Kasprzyk-Hordern, B., et al. (2022). Sewage surveillance of SARS-CoV-2 at student campus residences in the Western cape, South Africa. Sci. Total Environ. 851:158028. doi: 10.1016/j.scitotenv.2022.158028
Rimoldi, S. G., Stefani, F., Gigantiello, A., Polesello, S., Comandatore, F., Mileto, D., et al. (2020). Presence and infectivity of SARS-CoV-2 virus in wastewaters and rivers. Sci. Total Environ. 744:140911. doi: 10.1016/j.scitotenv.2020.140911
Roldan-Hernandez, L., Graham, K. E., Duong, D., and Boehm, A. B. (2022). Persistence of endogenous SARS-CoV-2 and pepper mild mottle virus RNA in wastewater-settled solids. ACS ES T Water 2, 1944–1952. doi: 10.1021/acsestwater.2c00003
Sala-Comorera, L., Reynolds, L. J., Martin, N. A., O’sullivan, J. J., Meijer, W. G., and Fletcher, N. F. (2021). Decay of infectious SARS-CoV-2 and surrogates in aquatic environments. Water Res. 201:117090. doi: 10.1016/j.watres.2021.117090
Scott, L. C., Aubee, A., Babahaji, L., Vigil, K., Tims, S., and Aw, T. G. (2021). Targeted wastewater surveillance of SARS-CoV-2 on a university campus for COVID-19 outbreak detection and mitigation. Environ. Res. 200:111374. doi: 10.1016/j.envres.2021.111374
Sherchan, S. P., Shahin, S., Ward, L. M., Tandukar, S., Aw, T. G., Schmitz, B., et al. (2020). First detection of SARS-CoV-2 RNA in wastewater in North America: a study in Louisiana, USA. Sci. Total Environ. 743:140621. doi: 10.1016/j.scitotenv.2020.140621
Vidaver, A. K., Koski, R., and Van Etten, J. L. (1973). Bacteriophage φ6: a lipid-containing virus of pseudomonas phaseolicola. J. Virol. 11, 799–805. doi: 10.1128/jvi.11.5.799-805.1973
Weidhaas, J., Aanderud, Z. T., Roper, D. K., Vanderslice, J., Gaddis, E. B., Ostermiller, J., et al. (2021). Correlation of SARS-CoV-2 RNA in wastewater with COVID-19 disease burden in sewersheds. Sci. Total Environ. 775:145790. doi: 10.1016/j.scitotenv.2021.145790
Wong, J. C. C., Tan, J., Lim, Y. X., Arivalan, S., Hapuarachchi, H. C., Mailepessov, D., et al. (2021). Non-intrusive wastewater surveillance for monitoring of a residential building for COVID-19 cases. Sci. Total Environ. 786:147419. doi: 10.1016/j.scitotenv.2021.147419
Wurtzer, S., Waldman, P., Ferrier-Rembert, A., Frenois-Veyrat, G., Mouchel, J.-M., Boni, M., et al. (2021). Several forms of SARS-CoV-2 RNA can be detected in wastewaters: implication for wastewater-based epidemiology and risk assessment. Water Res. 198:117183. doi: 10.1016/j.watres.2021.117183
Yang, S., Dong, Q., Li, S., Cheng, Z., Kang, X., Ren, D., et al. (2022). Persistence of SARS-CoV-2 RNA in wastewater after the end of the COVID-19 epidemics. J. Hazard. Mater. 429:128358. doi: 10.1016/j.jhazmat.2022.128358
Ye, Y., Ellenberg, R. M., Graham, K. E., and Wigginton, K. R. (2016). Survivability, partitioning, and recovery of enveloped viruses in untreated municipal wastewater. Environ. Sci. Technol. 50, 5077–5085. doi: 10.1021/acs.est.6b00876
Zhang, D., Ling, H., Huang, X., Li, J., Li, W., Yi, C., et al. (2020). Potential spreading risks and disinfection challenges of medical wastewater by the presence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) viral RNA in septic tanks of Fangcang hospital. Sci. Total Environ. 741:140445. doi: 10.1016/j.scitotenv.2020.140445
Keywords: SARS-CoV-2, PMMoV RNA, SARS-CoV-2 RNA, raw sewage, temperature, viral density
Citation: Li Y, Ash KT, Joyner DC, Williams DE, Alamilla I, McKay PJ, Iler C, Green BM, Kara-Murdoch F, Swift CM and Hazen TC (2023) Decay of enveloped SARS-CoV-2 and non-enveloped PMMoV RNA in raw sewage from university dormitories. Front. Microbiol. 14:1144026. doi: 10.3389/fmicb.2023.1144026
Received: 16 January 2023; Accepted: 03 April 2023;
Published: 28 April 2023.
Edited by:
Yashpal S. Malik, Indian Veterinary Research Institute (IVRI), IndiaReviewed by:
Krzysztof Treder, Plant Breeding and Acclimatization Institute, PolandCopyright © 2023 Li, Ash, Joyner, Williams, Alamilla, McKay, Iler, Green, Kara-Murdoch, Swift and Hazen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Terry C. Hazen, dGNoYXplbkB1dGsuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.