- 1School of Life Science and Agricultural Engineering, Nanyang Normal University, Nanyang, China
- 2Research Center of Henan Provincial Agricultural Biomass Resource Engineering and Technology, Nanyang Normal University, Nanyang, China
- 3School of Water Resources and Environment Engineering, Nanyang Normal University, Nanyang, China
The genus Bannoa is a small group of ballistoconidium-forming yeasts in the family Erythrobasidiaceae (Cystobasidiomycetes). Prior to this study, seven species belonging to this genus have been described and published. In this study, phylogenetic analyzes of Bannoa based on the combined sequences of the small ribosomal subunit (SSU) rRNA gene, the internal transcribed spacer (ITS) regions, the D1/D2 domains of the large subunit rRNA gene (LSU) and the translation elongation factor 1-α gene (TEF1-α) were conducted. Three new species, namely B. ellipsoidea, B. foliicola, and B. pseudofoliicola, were delimited and proposed based on morphological and molecular evidence. B. ellipsoidea was found to be closely related to the type strains of B. guamensis, B. hahajimensis, and B. tropicalis, but with 0.7–0.9% divergence (4–5 substitutions) in the LSU D1/D2 domains and 3.7–4.1% divergence (19–23 substitutions and one−two gaps) in the ITS regions. B. foliicola was found to belong to the same clade as B. pseudofoliicola from which it differed by 0.4% divergence (two substitutions) in the LSU D1/D2 domains and 2.3% divergence (13 substitutions) in the ITS regions. The distinguishing morphological characteristics of the three new species, with respect to closely related taxa, are discussed. The identification of these new taxa significantly increases the number of Bannoa that have been described on the surface of plant leaves. Additionally, a key for the identification of Bannoa species is provided.
Introduction
The genus Bannoa, belonging to Erythrobasidiales, is a kind of ballistoconidium-forming Basidiomycota. It was established by Hamamoto et al. (2002) based on a single species, Bannoa hahajimensis, which was isolated from dead leaves of plants collected in the Ogasawara Islands, Japan. Additionally, the anamorphic species obtained in this study were described as three new species of Sporobolomyces, namely S. bischofiae, S. ogasawarensis, and S. syzygii. However, these three species formed a monophyletic cluster with Bannoa hahajimensis in the Erythobasidium clade based on sequence analyzes of the SSU rRNA gene (Hamamoto et al., 2002), the D1/D2 domains of the LSU rRNA gene (Hamamoto, 2011), and a combined gene dataset of SSU rRNA, ITS regions, LSU rRNA, and first and second codon of TEF1-α (Nagahama et al., 2006). This monophyly was confirmed by Wang et al. et al. based on a seven-gene dataset consisting of SSU rRNA, D1/D2 LSU rRNA domains, ITS regions, RPB1, RPB2, TEF1-α and CYTB (Wang et al., 2015a). Recently, the genus Bannoa was emended by Wang et al. based on the phylogenetic analysis of a seven-gene dataset (Wang et al., 2015b). In this seven-gene phylogeny, the genus represented a well-supported clade containing a teleomorphic species B. hahajimensis and three Sporobolomyces species, including S. bischofiae, S. ogasawarensis, and S. syzygii, which were formerly recognized as the Bannoa clade (Hamamoto et al., 2002; Nagahama et al., 2006; Hamamoto, 2011; Wang et al., 2015a). Recent incorporations into the genus are B. guamensis, B. rosea, and B. tropicalis isolated from the surfaces of diseased and healthy leaves of plants in the Euphorbiaceae, Asteraceae, and Poaceae plant families (Parra and Aime, 2019). The genus Bannoa was located in the family Erythrobasidiaceae but was phylogenetically distinguished from other recognized species in the genus Erythrobasidium (Wang et al., 2015b).
Yeasts in the genus Bannoa are best known for their orange to salmon-red appearance in culture (Hamamoto, 2011; Wang et al., 2015b). The sexual morph of Bannoa is characterized by the production of unicellular basidia on a clamp connection formed after mating, while the asexual morph is characterized by budding cells, some of which are ballistoconidia that can be ovoidal or ellipsoidal (Hamamoto et al., 2002; Hamamoto, 2011; Parra and Aime, 2019).
The worldwide species diversity of Bannoa has not been extensively studied. Until now, only seven species have been accepted in the genus Bannoa (Wang et al., 2015b; Parra and Aime, 2019). Among them, B. hahajimensis, B. ogasawarensis, and B. syzygii are reported to occur in China and Japan (Zang et al., 1998; Chiang et al., 2001; Hamamoto et al., 2002; Li et al., 2020), while B. bischofiae has only been identified in Japan (Hamamoto et al., 2002), and B. guamensis, B. rosea, and B. tropicalis have been isolated in South America (Parra and Aime, 2019). In addition, several studies suggest the existence of other potentially new species that could belong to this genus (Matheny et al., 2006; Vaïtilingom et al., 2012; Wang et al., 2015a,b; Parra and Aime, 2019; Li et al., 2020).
During investigations of basidiomycetous yeasts from the Henan Province, central China, seven ballistoconidium-forming yeasts were collected. Morphological characteristics and phylogenetic analyzes based on SSU, ITS, LSU D1/D2, and TEF1-α sequences indicated that these yeasts represented three undescribed species of the Bannoa genus. This study confirms the taxonomic affinities of these new species, explores the species diversity of Bannoa in central China, and infers evolutionary relationships between representative species of Bannoa.
Materials and methods
Sample collection and yeast isolation
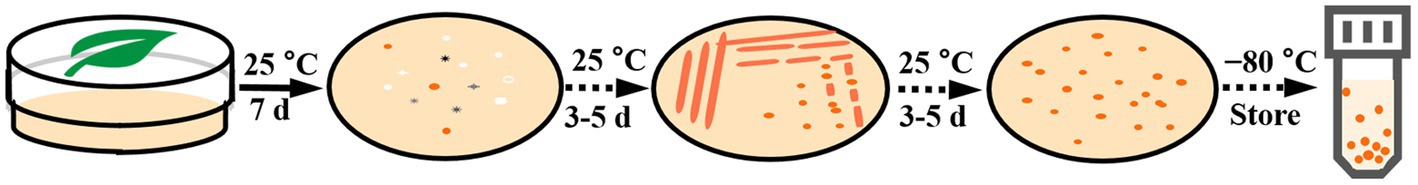
Plant leaves were collected from the Baotianman Nature Reserve (33°30′44″N, 111°55′47″E) in the Henan Province of central China. The protected area measures 4,285 ha and is classified as a world biosphere reserve by the United Nations Educational, Scientific and Cultural Organization (UNESCO). The local climate is a typical transitional climate spanning a northern subtropical zone to a warm temperate zone, with cold and dry winters and fresh and rainy summers, and with annual mean temperatures around 15°C (Hu et al., 2022). Plant leaf samples were stored in sterile flasks and transported under refrigerated conditions within 24 h of collection. Yeast strains were isolated from plant leaves by the ballistospore-fall method as previously described (Nakase and Takashima, 1993; Hu et al., 2022). The semi-withered leaf samples were stuck with vaseline to the inside of the lid of petri plates containing yeast malt (YM) agar (0.3% yeast extract, 0.3% malt extract, 0.5% peptone, 1% glucose, and 2% agar) and incubated at 25°C for 7 days, and the media was refreshed daily until colonies gradually formed. Different yeast morphotypes were picked and purified by streaking onto fresh YM agar. All purified yeast strains were suspended in YM broth supplemented with 20% (v/v) glycerol and stored at −80°C (Figure 1). Cultures of isolates obtained for this study were preserved in the Microbiology Lab, at Nanyang Normal University, Henan, China. All isolates used in this study and their origins are described in Table 1.
Morphological and physiological characterization
Morphological and physiological traits of the yeast strains were characterized according to methods established by Kurtzman et al. (2011). Colony traits were observed on YM agar after 2 weeks of incubation at 25°C. Mycelium formation was investigated by cultivation on corn meal (CM) agar (2% cornmeal infusion and 2% agar) in slide culture at 25°C for 2 weeks. The sexual processes of all strains were examined for individual strains and strain pairs on potato dextrose agar (PDA) (20% potato infusion, 2% glucose, and 1.5% agar), CM agar, and yeast carbon base plus 0.01% ammonium sulfate (YCBS) agar at 25°C for 2 months (Hamamoto et al., 2002; Parra and Aime, 2019). Glucose fermentation was carried out in a liquid medium using Durham fermentation tubes. Carbon and nitrogen source assimilation tests were conducted in liquid medium and starved inocula were used for the nitrogen assimilation tests (Kurtzman et al., 2011). Cycloheximide resistance tests were also performed in a liquid medium, while urea hydrolysis assays were conducted on agar slants. Acid production and reaction with diazonium blue B (DBB) were investigated on a solid medium in petri dishes. Growth at various temperatures (15, 20, 25, 30, 35, and 37°C) was determined by cultivation on YM agar. Cell morphology was examined using a Leica DM 2500 microscope (Leica Microsystems GmbH, Wetzlar, Germany) with a Leica DFC295 digital microscope color camera using bright field, phase contrast, and differential interference contrast (DIC) optics. Novel taxonomic descriptions and proposed names were deposited in MycoBank (http://www.mycobank.org; 8 January 2023).
DNA extraction, PCR amplification, and sequencing
Genomic DNA was extracted from the yeasts using the Ezup Column Yeast Genomic DNA Purification Kit according to the manufacturer’s protocol (Sangon Biotech, China). Briefly, yeast cells were homogenized in snailase reaction buffe, followed by using with solution to subsequently remove protein. The resulting DNA was further purified by adsorption column and resuspended in 50 μL TE Buffer. A total of four nuclear loci were sequenced, including the small ribosomal subunit (SSU) rRNA gene, the internal transcribed spacer (ITS) regions, the D1/D2 domains of the large subunit (LSU) rRNA gene, and the translation elongation factor 1-α gene (TEF1-α). The primer pairs NS1/NS8 (White et al., 1990), ITS1/ITS4 (White et al., 1990), NL1/NL4 (Kurtzman and Robnett, 1998), and EF1-526F/EF1-1567R (Rehner and Buckley, 2005) were used for amplifying SSU, ITS, LSU, andTEF1-α, respectively.
PCR was conducted as described by Toome et al. (2013) for SSU, ITS, and LSU. For TEF1-α, a touchdown PCR protocol was used as described by the research group (Wang et al., 2014). PCR products were directly purified and sequenced by Sangon Biotech Inc. (Shanghai, China). We determined the identity and accuracy of the newly obtained sequences by comparing them to sequences in GenBank and assembled them using BioEdit (Hall, 1999). Newly obtained sequences were then submitted to GenBank (https://www.ncbi.nlm.nih.gov/genbank/; Table 2).
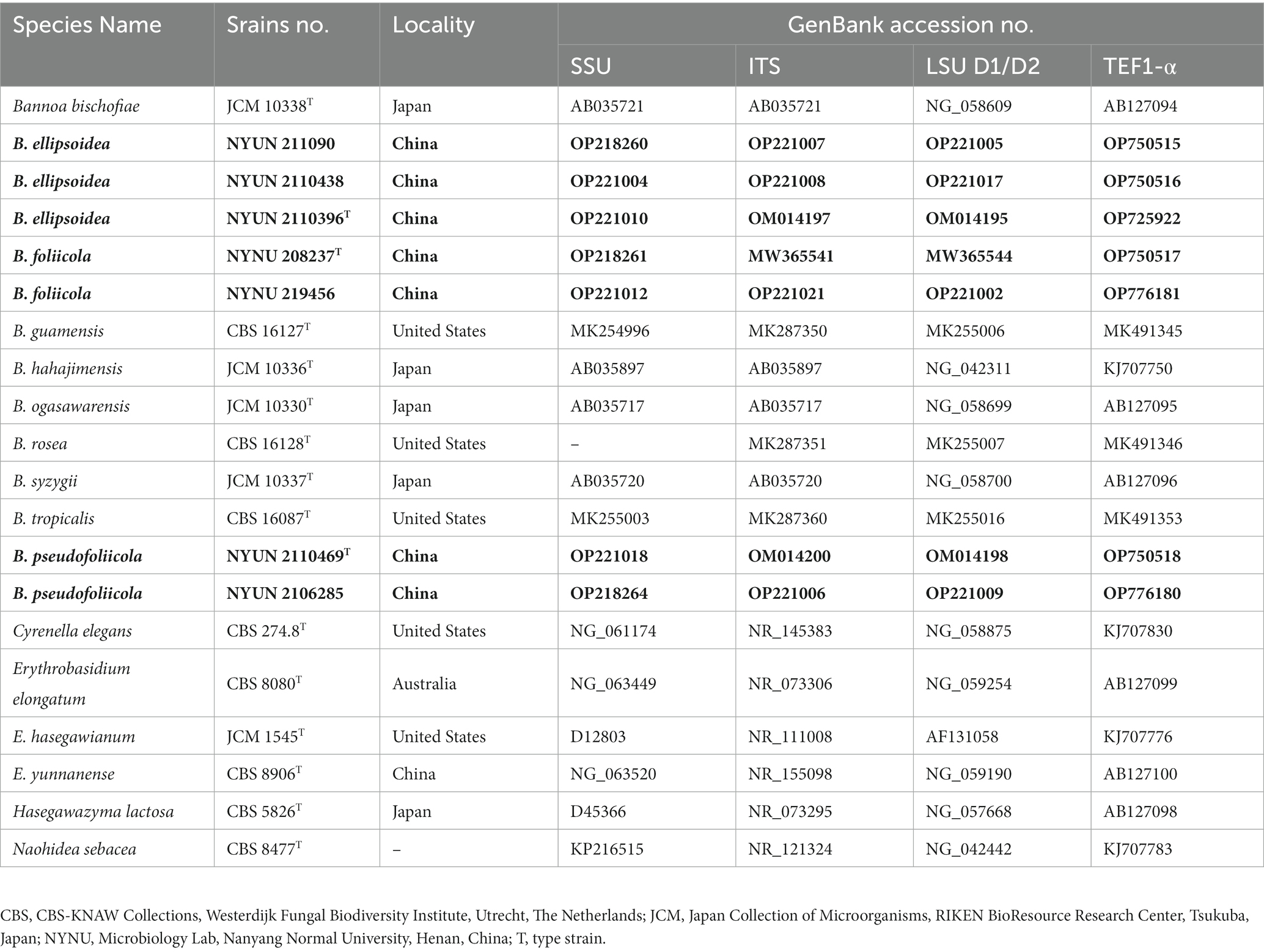
Table 2. Sequences used for the molecular phylogenetic analysis Entries in bold were newly generated in this study.
Phylogenetic analysis
The phylogenetic relationship of the new Bannoa species and related species was determined by analysis of concatenated sequence datasets of four loci (SSU, ITS, LSU D1/D2, and TEF1-α). Naohidea sebacea CBS 8477 was used as the outgroup, according to Wang et al. (2015a). For the dataset, each gene region was aligned using MAFFT v7.110 with default settings (Katoh and Standley, 2013), and then manually adjusted in BioEdit 7.1.3.0 (Hall, 1999). The“Concatenate Sequence” function in PhyloSuite v1.16 was used to combine the aligned sequences of the different loci (Zhang et al., 2020). Manual gap adjustments were done to improve the alignment and ambiguously aligned regions were also excluded. Multi-locus phylogenetic analyzes were performed with MEGA11 software (Tamura et al., 2021) using the neighbor-joining (NJ) and maximum likelihood (ML) methods. The Kimura-2 parameter distance correction (Saitou and Nei, 1987) and the general time reversible (GTR) models (Nei and Kumar, 2000) were, respectively, used for the NJ and ML analyzes. Confidence levels of the clades were estimated from bootstrap analysis (1,000 replicates) (Felsenstein, 1985).
Results
Molecular phylogenetic analysis
The combined SSU + ITS + LSU D1/D2 + TEF1-α dataset was analyzed to infer the taxonomic positions of the novel species described in this study. The dataset included sequences from 20 fungal samples representing 16 taxa including the outgroup, Naohidea sebacea CBS 8477T. After alignment and trimming, the combined dataset contained 3,075 characters including gaps with 1,396 characters for SSU, 428 characters for ITS, 504 characters for LSU, and 747 characters for TEF1-α alignment, of which 358 characters were parsimony-informative. The best model for the SSU + ITS + LSU D1/D2 + TEF1-α dataset estimated and applied in the ML analysis was GTR + I + G with equal frequency of nucleotides. ML and NJ analyzes produced almost identical topologies, thus only the ML generated tree is displayed with ML and NJ supported values above 50% at the nodes (Figure 2). Our phylogenetic analyzes indicated that Bannoa formed a monophyletic group with high support (100% ML; 100% NJ; Figure 2). Ten phylogenetic species of the genus Bannoa are delineated, three of which were new: B. ellipsoidea, B. foliicola, and B. pseudofoliicola.
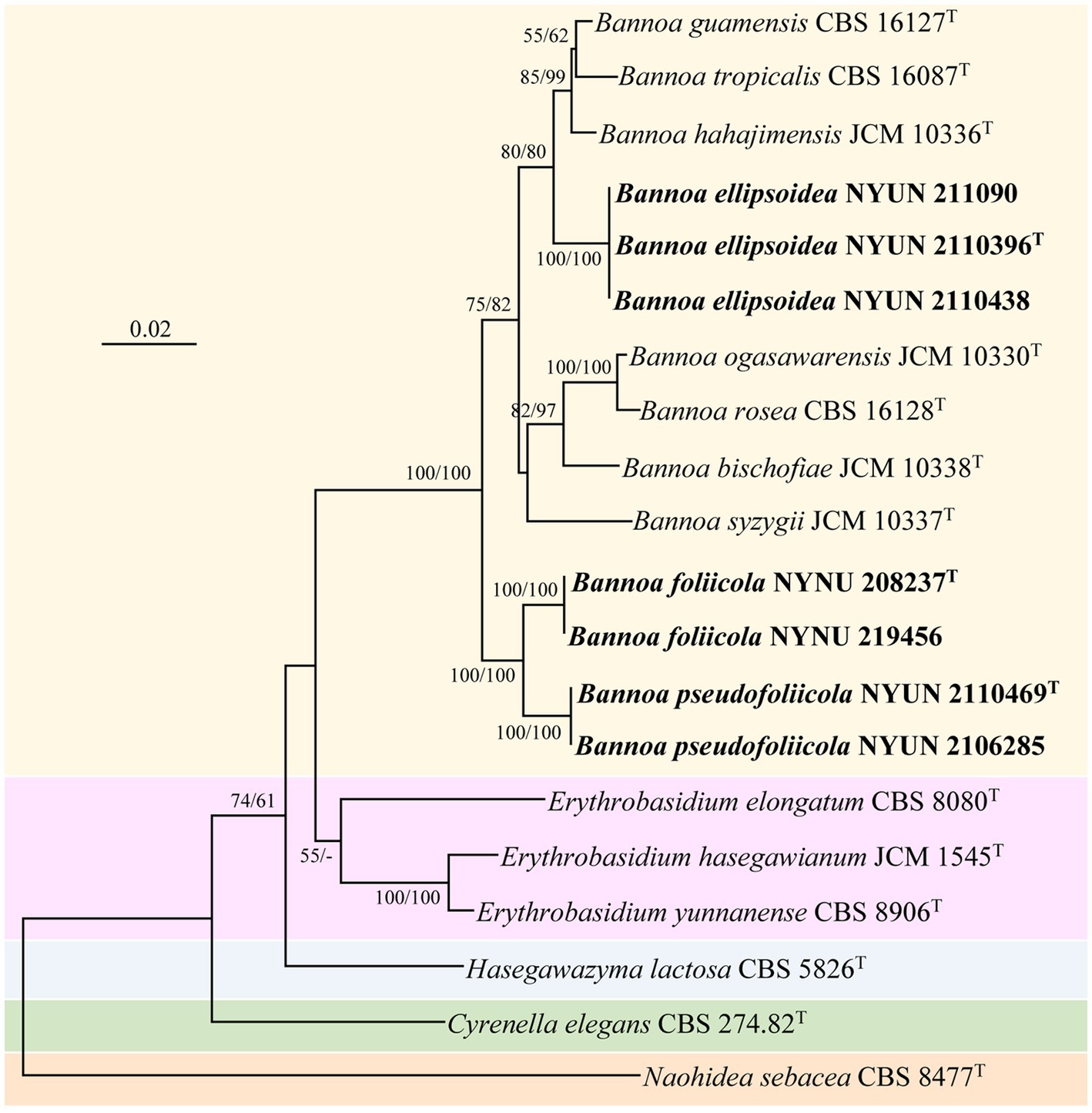
Figure 2. Maximum likelihood (ML) phylogram of Bannoa species based on the combined SSU + ITS + LSU D1/D2 + tef1 sequence dataset. Naohidea sebacea CBS 8477T was used as the outgroup. ML and neighbor-joining (NJ) bootstrap support values above 50% are shown at the nodes (ML/NJ). The novel strains described in this study are shown in bold.
Bannoa ellipsoidea was grouped with the three described species, B. guamensis, B. hahajimensis, and B. tropicalis (Hamamoto et al., 2002; Parra and Aime, 2019), with strong support (80% ML; 80% NJ; Figure 2). The LSU D1/D2 sequences of B. ellipsoidea differed by 0.7% divergence (4 substitutions) from the type strain of B. tropicalis, and by 0.9% divergence (5 substitutions) from the type strains of B. guamensis and B. hahajimensis. Surprisingly, significant sequence differences between B. ellipsoidea and its close relatives, B. guamensis, B. hahajimensis, and B. tropicalis, were observed in the ITS regions. In these regions, B. ellipsoidea differed from the type strain of B. tropicalis by 3.7% divergence (19 substitutions and one gaps). Similarly, B. ellipsoidea exhibited 3.7% divergence (21 substitutions and two gaps) from the type strain of B. guamensis and 4.1% divergence (23 substitutions and one gap) from the type strain of B. hahajimensis in the ITS regions. According to the basidiomycetous yeast species thresholds proposed by Fell et al. (2000), Scorzetti et al. (2002), and Vu et al. (2016), strains that differ by two or more nucleotide substitutions in the D1/D2 domains or 1–2% nucleotide differences in the ITS regions may represent different taxa. The differences in both the LSU D1/D2 and ITS sequences were significant enough for B. ellipsoidea to be considered a distinct Bannoa species.
Bannoa foliicola formed a subclade with B. pseudofoliicola described in this study with full support (100% ML; 100% NJ; Figure 2). B. foliicola differed from its nearest phylogenetic neighbor, B. pseudofoliicola CBS 6936T, by 0.4% divergence (2 substitutions) in the LSU D1/D2 domains and 2.3% divergence (13 substitutions) in the ITS regions. According to the criteria mentioned above, this data clearly supports the distinction between B. foliicola and B. pseudofoliicola at the species level.
Phenotypic characterization
All strains of the three new species formed orange-colored colonies (Figures 3A, 4A, 5A) and ovoidal, ellipsoidal, and cylindrical vegetative cells (Figures 3B, 4B, 5B) like other Bannoa species. Ballistoconidia were formed and were ovoidal and ellipsoidal in shape (Figures 3C, 4C, 5C). B. ellipsoidea produced more abundant ballistoconidia than strains of the other two new species. Sexual structures were not observed in the cultures of single or mixed strains on PDA, CM agar, or YCBS agar.
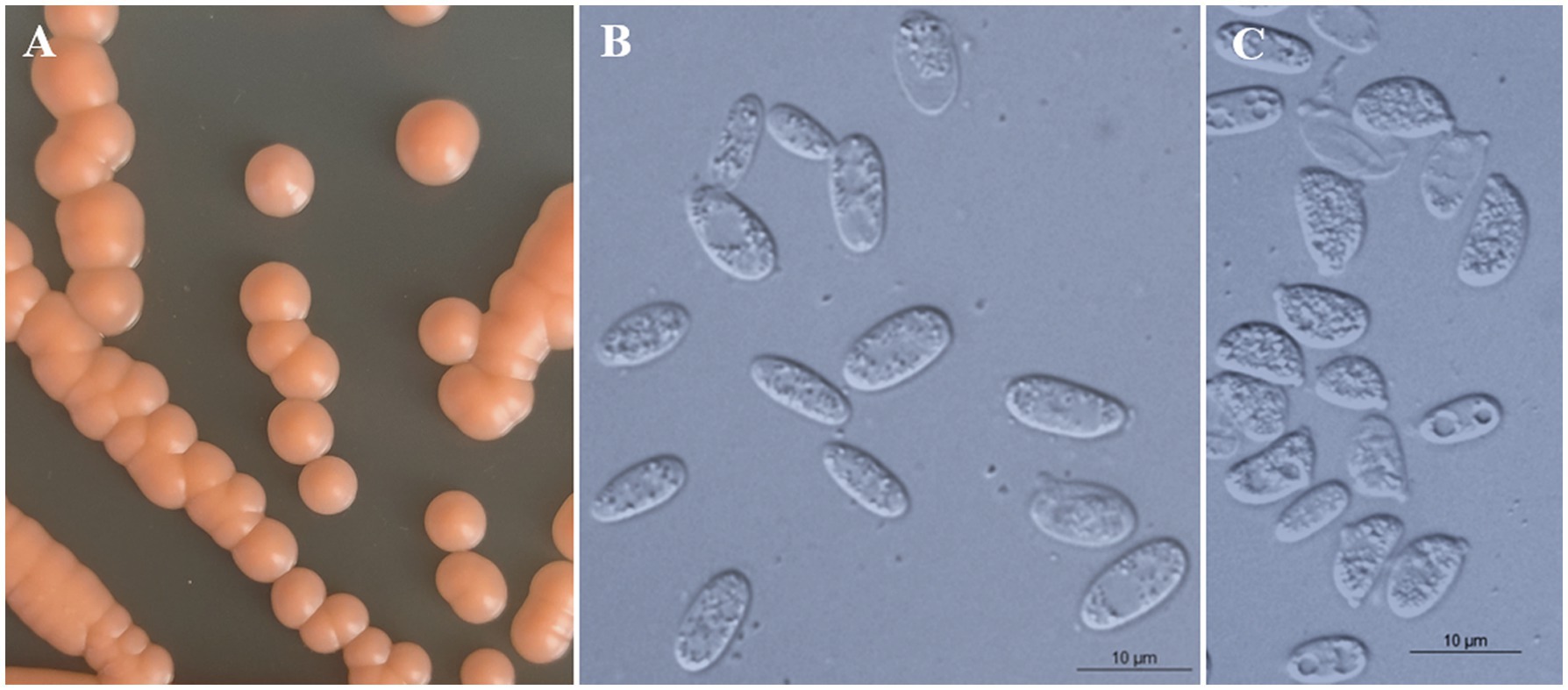
Figure 3. Morphological characteristics of Bannoa ellipsoidea sp. nov. (GDMCC 2.279, holotype). (A) Colony morphology on YM agar after growth for 7 d at 25°C. (B) Budding cells after growth for 7 d in YM broth at 25°C. (C) Ballistoconidia on CM agar after growth for 7 d at 25°C. Scale bars = 10 μm.
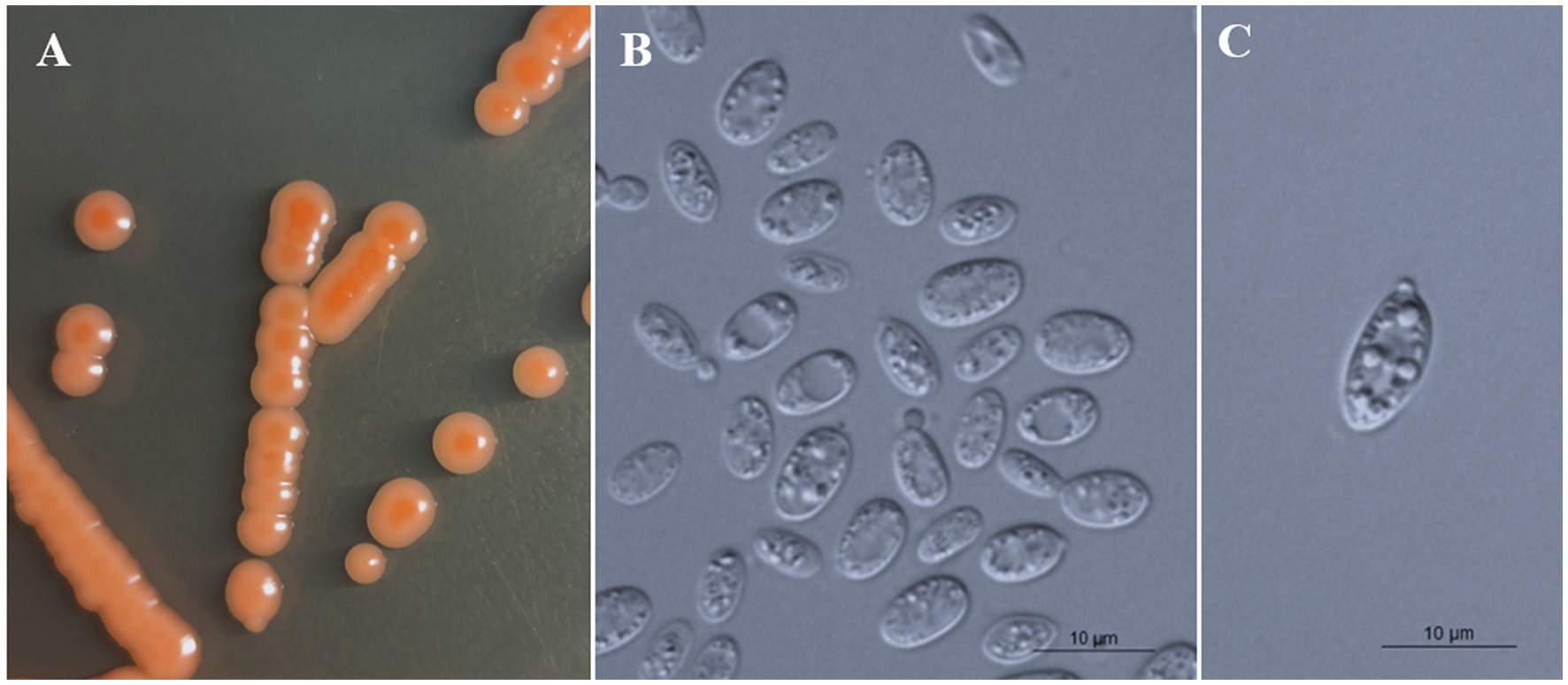
Figure 4. Morphological characteristics of Bannoa foliicola sp. nov. (CICC 33507, holotype). (A) Colony morphology on YM agar after growth for 7 d at 25°C. (B) Budding cells after growth for 7 d in YM broth at 25°C. (C) Ballistoconidia on CM agar after growth for 7 d at 25°C. Scale bars = 10 μm.
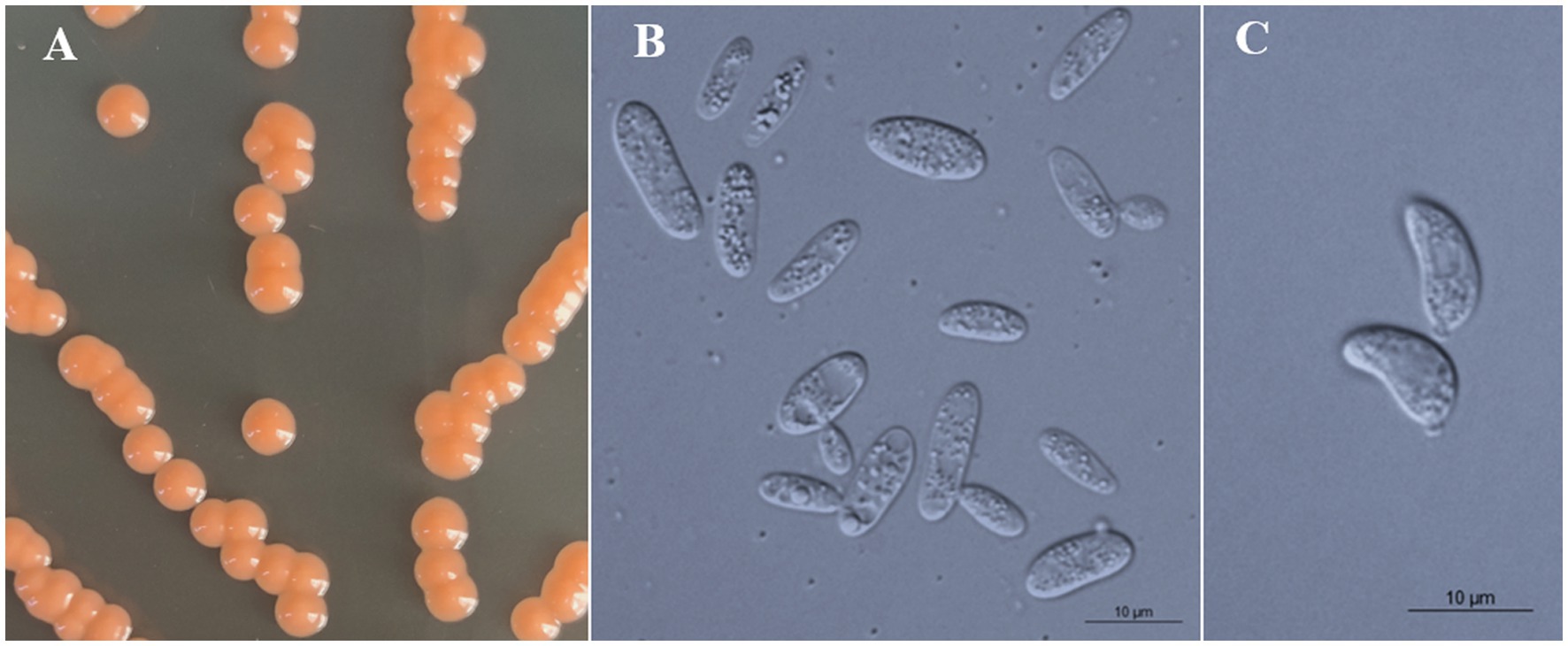
Figure 5. Morphological characteristics of Bannoa pseudofoliicola sp. nov. (GDMCC 2.270, holotype). (A) Colony morphology on YM agar after growth for 7 d at 25°C. (B) Budding cells after growth for 7 d in YM broth at 25°C. (C) Ballistoconidia on CM agar after growth for 7 d at 25°C. Scale bars = 10 μm.
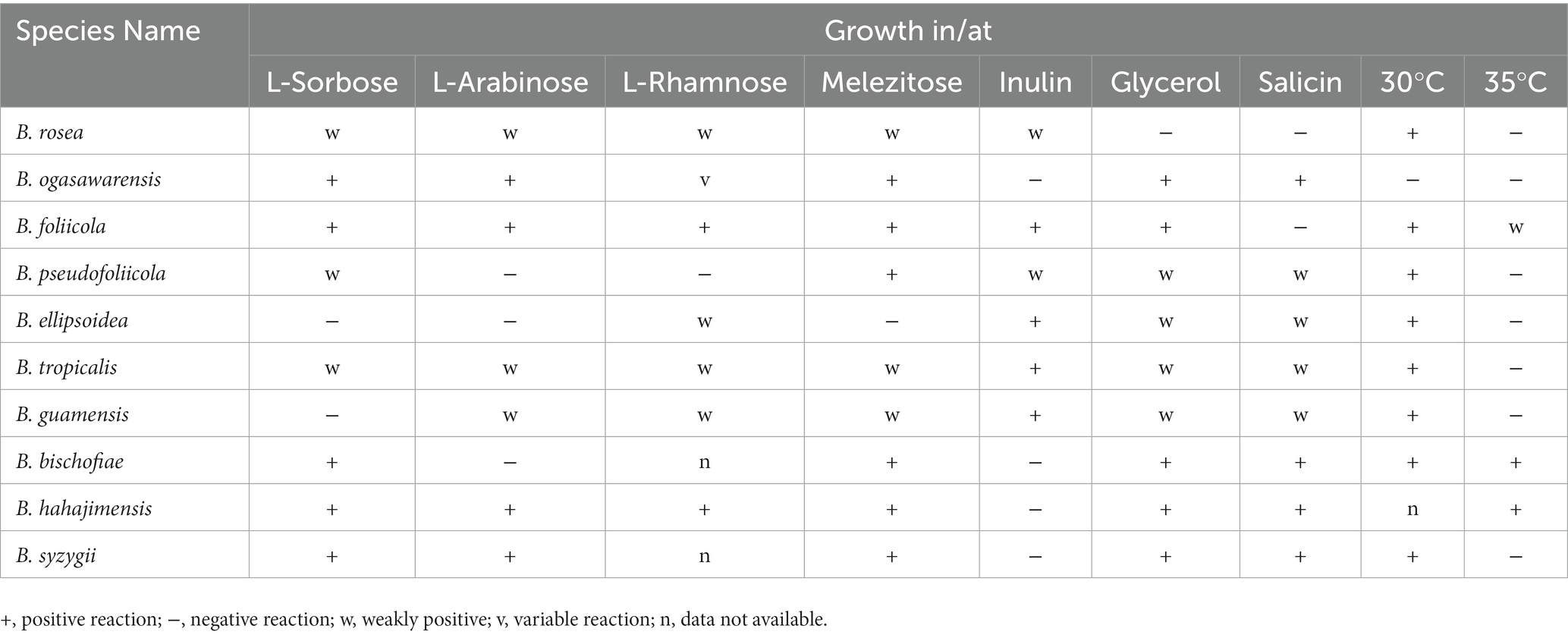
Phenotypic characteristics that differed between B. ellipsoidea, B. foliicola, and B. pseudofoliicola and their closely related species in the genus Bannoa are described in Table 3. B. ellipsoidea physiologically differed from B. guamensis, B. hahajimensis, and B. tropicalis in its inability to utilize L-arabinose, sucrose, maltose, melibiose, and D-mannitol (Table 3). Similarly, B. foliicola could be differentiated from B. pseudofoliicola by its ability to assimilate L-arabinose, D-arabinose, L-rhamnose, lactose, erythritol, galactitol, DL-lactate, succinate, and creatine and its inability to assimilate salicin. In addition, B. foliicola could grow at 35°C, while B. pseudofoliicola could not.
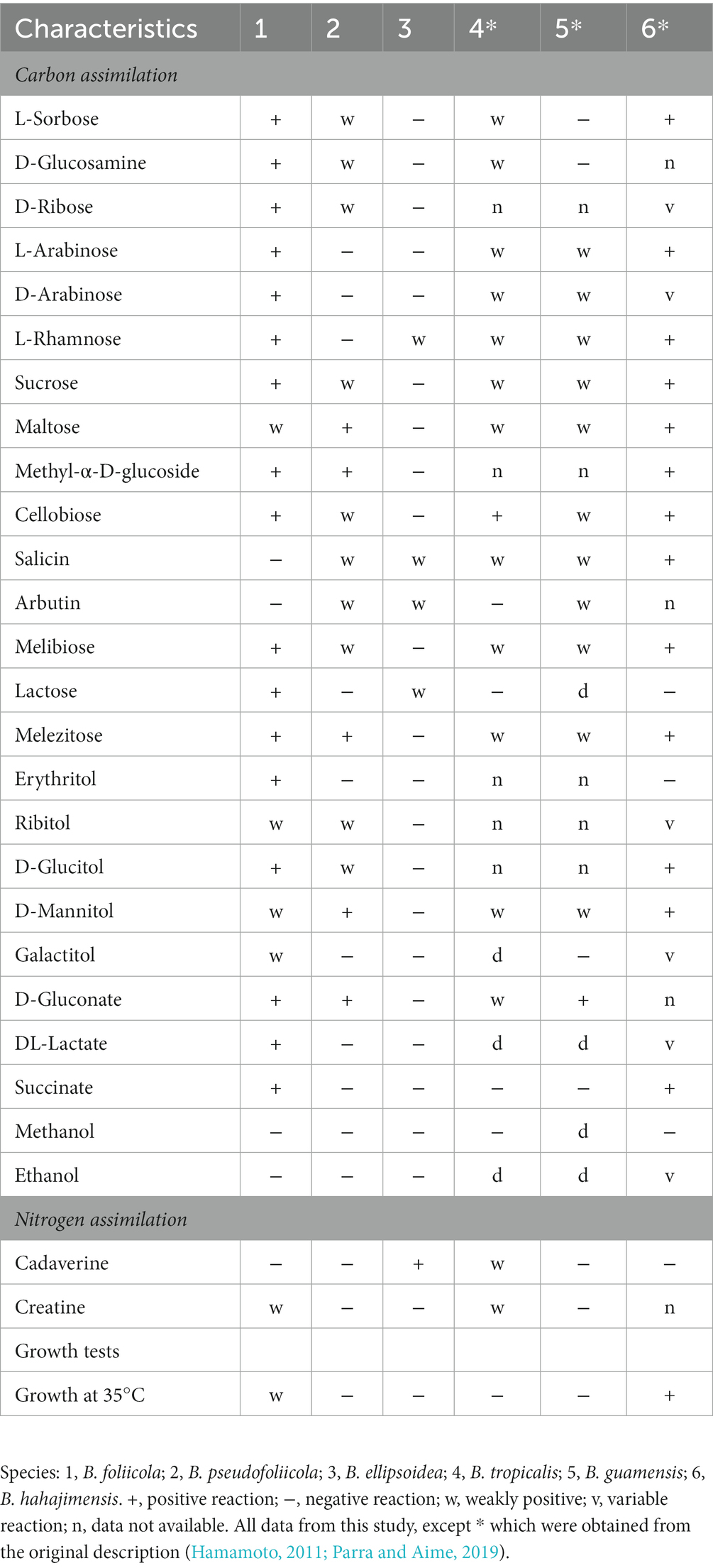
Table 3. Phenotypic characteristics that differed between the three novel species and their most closely related taxa.
Taxonomy
Bannoa ellipsoidea C.Y. Chai & F.L. Hui, sp. nov., Figure 3.
MycoBank: MB 847130.
Etymology: the specific epithet “ellipsoidea” refers to the ellipsoidal vegetative cells of the type strain.
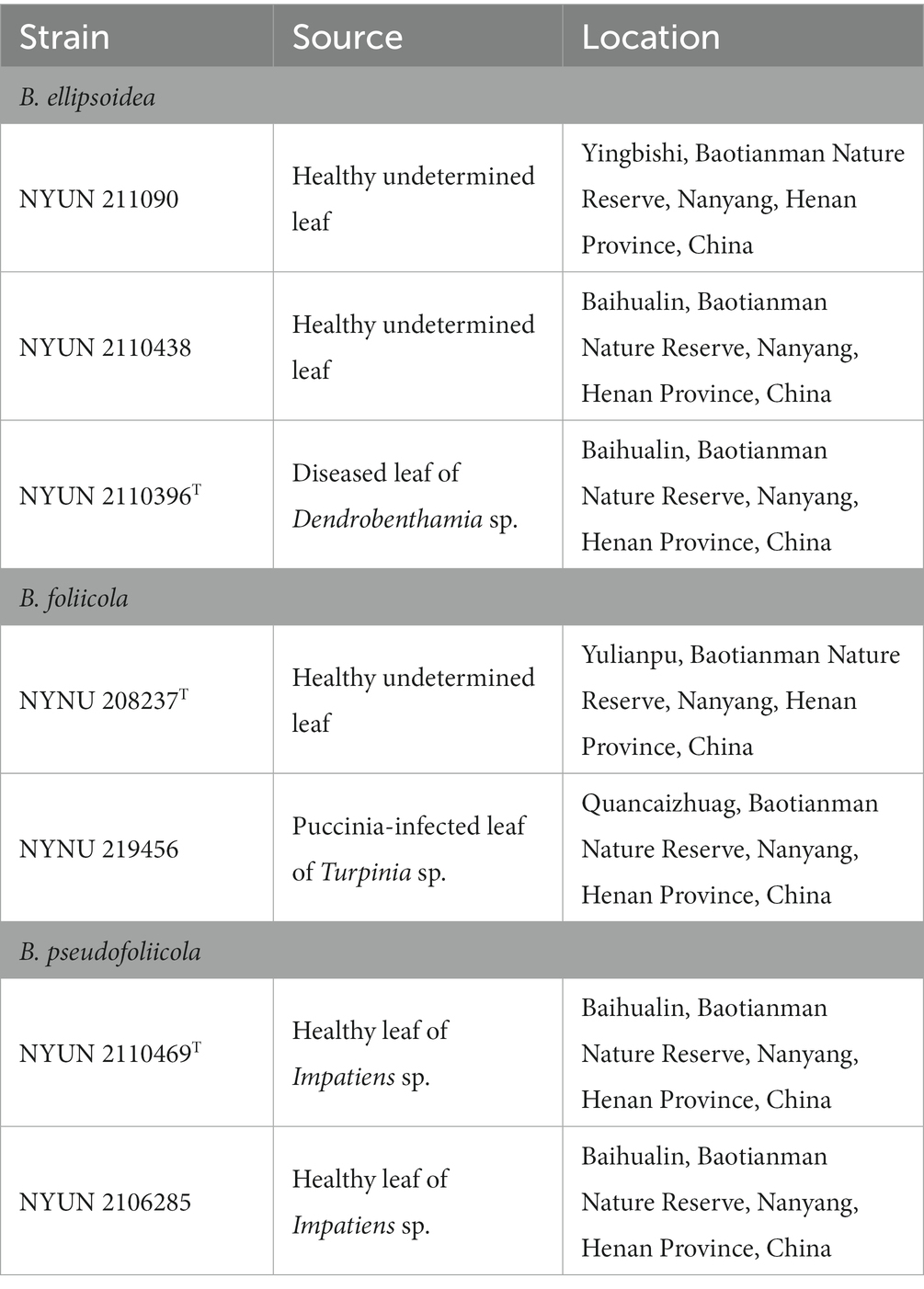
Typus: China, Henan Province, Neixiang County, Baotianman Nature Reserve, in phylloplane from diseased leaf of Dendrobenthamia sp., October 2021, L. Zhang and H. zhang, NYUN 2110396 (holotype GDMCC 2.279T preserved as a metabolically inactive state, culture ex-type JCM 35734).
Description: On YM agar, after 7 d at 25°C, colonies are orange, smooth, glistening and butyrous in texture (Figure 3A). The margin is entire. In YM broth, after 7 d at 25°C, cells are ellipsoidal and cylindrical, 3.2–4.1 × 8.3–9.5 μm and single, budding is polar (Figure 3B). After 1 mo at 25°C, a ring and sediment are present. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on PDA, CM agar and YCBS agar. Ballistoconidia are ovoid and ellipsoidal, 5.0–6.3 × 7.4–13.3 μm (Figure 3C). Glucose fermentation is absent. Glucose, inulin, raffinose (weak), galactose (weak), lactose (weak), trehalose (weak), salicin (weak), L-rhamnose (weak), D-xylose (weak) and glycerol (weak) are assimilated as sole carbon. Sucrose, melibiose, maltose, melezitose, methyl-α-D-glucoside, cellobiose, L-sorbose, L-arabinose, D-arabinose, D-ribose, methanol, ethanol, erythritol, ribitol, galactitol, D-mannitol, D-glucitol, myo-inositol, DL-lactate, succinate, citrate, D-gluconate, D-glucosamine, D-glucono-1,5-lactone, 2-keto-D-gluconate, 5-keto-D-gluconate and D-glucuronate are not assimilated. Cadaverine is assimilated as sole nitrogen sources. Nitrate, nitrite, ethylamine, L-lysine, creatine, creatinine, glucosamine, imidazole and D-tryptophan are not assimilated. Maximum growth temperature is 30°C. Starch-like substances are not produced. Urease activity is positive. Diazonium Blue B reaction is positive.
Additional strain examined: China, Henan Province, Neixiang County, Baotianman Nature Reserve, in phylloplane from healthy undetermined leaf, October 2021, L. Zhang and H. zhang, NYUN 211090 and NYUN 2110438.
GenBank accession numbers: holotype GDMCC 2.279T (SSU: OP221010, ITS: OM014197, D1/D2: OM014195, TEF1-α: OP725922); additional strains NYUN 211090 (SSU: OP218260, ITS: OP221007, D1/D2: OP221005, TEF1-α: OP750515) and NYUN 2110438 (SSU: OP221004, ITS: OP221008, D1/D2: OP221017, TEF1-α: OP750516).
Bannoa foliicola C.Y. Chai & F.L. Hui, sp. nov., Figure 4.
MycoBank: MB 847131.
Etymology: the specific epithet “foliicola” refers to the substrate origin of the type strain, leaves.
Typus: China, Henan Province, Neixiang County, Baotianman Nature Reserve, in phylloplane from healthy undetermined leaf, July 2016, L. Zhang and H. zhang, NYNU 208237 (holotype CICC 33507T preserved as a metabolically inactive state, culture ex-type CBS 16656).
Description: On YM agar, after 7 d at 25°C, colonies are orange to salmon-red, smooth, glistening and butyrous in texture (Figure 4A). The margin is entire. In YM broth, after 7 d at 25°C, cells are ovoid and ellipsoidal, 2.1–4.4 × 6.4–8.0 μm and single, budding is polar (Figure 4B). After 1 mo at 25°C, a ring and sediment are present. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on PDA, CM agar and YCBS agar. Ballistoconidia are ovoid and ellipsoidal, 5.2–6.9 × 10.8–14.3 μm (Figure 4C). Glucose fermentation is absent. Glucose, inulin, sucrose, raffinose, melibiose, galactose (weak), lactose, trehalose, maltose (weak), melezitose, methyl-α-D-glucoside, cellobiose, L-sorbose, L-rhamnose, D-xylose, L-arabinose, D-arabinose, D-ribose, glycerol, erythritol, ribitol (weak), galactitol (weak), D-mannitol (weak), D-glucitol, DL-lactate, succinate, D-gluconate, D-glucosamine, 2-keto-D-gluconate and D-glucuronate are assimilated as sole carbon sources. Salicin, methanol, ethanol, myo-inositol, citrate, D-glucono-1,5-lactone and 5-Keto-D-gluconate are not assimilated. L-Lysine, creatine (weak), glucosamine and D-tryptophan are assimilated as sole nitrogen sources. Nitrate, nitrite, ethylamine, cadaverine, creatinine and imidazole are not assimilated. Maximum growth temperature is 35°C. Growth in vitamin-free medium is positive. Starch-like substances are not produced. Urease activity is positive. Diazonium Blue B reaction is positive.
Additional strain examined: China, Henan Province, Neixiang County, Baotianman Nature Reserve, in phylloplane from puccinia-infected leaf of Turpinia sp., October 2021, L. Zhang and H. zhang, NYNU 219456.
GenBank accession numbers: holotype CICC 33507T (SSU: OP218261, ITS: MW365541, D1/D2: MW365544, TEF1-α: OP750517); additional strain NYNU 219456 (SSU: OP221012, ITS: OP221021, D1/D2: OP221002, TEF1-α: OP776181).
Bannoa pseudofoliicola C.Y. Chai & F.L. Hui, sp. nov., Figure 5.
MycoBank: MB 847132.
Etymology: the specific epithet “pseudofoliicola” refers to the similar colony morphology to that of Bannoa foliicola.
Typus: China, Henan Province, Neixiang County, Baotianman Nature Reserve, in phylloplane from healthy undetermined leaf, October 2021, L. Zhang and H. zhang, NYNU 2110469 (holotype GDMCC 2.270T preserved as a metabolically inactive state, culture ex-type JCM 35726).
Description: On YM agar, after 7 d at 25°C, colonies are orange, smooth, glistening and butyrous in texture (Figure 5A). The margin is entire. In YM broth, after 7 d at 25°C, cells are ellipsoidal and cylindrical, 3.2–3.7 × 7.7–12.6 μm and single, budding is polar (Figure 5B). After 1 mo at 25°C, a pellicle and sediment are present. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on PDA, CM agar and YCBS agar. Ballistoconidia are ovoid and ellipsoidal, 4.7–7.0 × 9.0–11.4 μm (Figure 5C). Glucose fermentation is absent. Glucose, inulin (weak), sucrose (weak), raffinose (weak), melibiose (weak), galactose (weak), trehalose, maltose, melezitose, methyl-α-D-glucoside, cellobiose (weak), salicin (weak), L-sorbose (weak), D-xylose (weak), D-ribose (weak), glycerol (weak), ribitol (weak), D-mannitol, D-glucitol (weak), D-gluconate, D-glucosamine (weak), 2-keto-D-gluconate and D-glucuronate are assimilated as sole carbon sources. Lactose, L-rhamnose, L-arabinose, D-arabinose, methanol, ethanol, erythritol, galactitol, myo-inositol, DL-lactate, succinate, citrate, D-glucono-1,5-lactone and 5-keto-D-gluconate are not assimilated. Creatinine (weak), glucosamine (weak) and D-tryptophan (weak) are assimilated as sole nitrogen sources. Nitrate, nitrite, ethylamine, L-lysine, cadaverine, creatine and imidazole are not assimilated. Maximum growth temperature is 30°C. Growth in vitamin-free medium is positive. Starch-like substances are not produced. Urease activity is positive. Diazonium Blue B reaction is positive.
Additional strain examined: China, Henan Province, Neixiang County, Baotianman Nature Reserve, in phylloplane from healthy undetermined leaf, October 2021, L. Zhang and H. zhang, NYUN 2106285.
GenBank accession numbers: holotype GDMCC 2.270T (SSU: OP221018, ITS: OM014200, D1/D2: OM014198, TEF1-α: OP750518); additional strain NYUN 2106285 (SSU: OP218264, ITS: OP221006, D1/D2: OP221009, TEF1-α: OP776180).
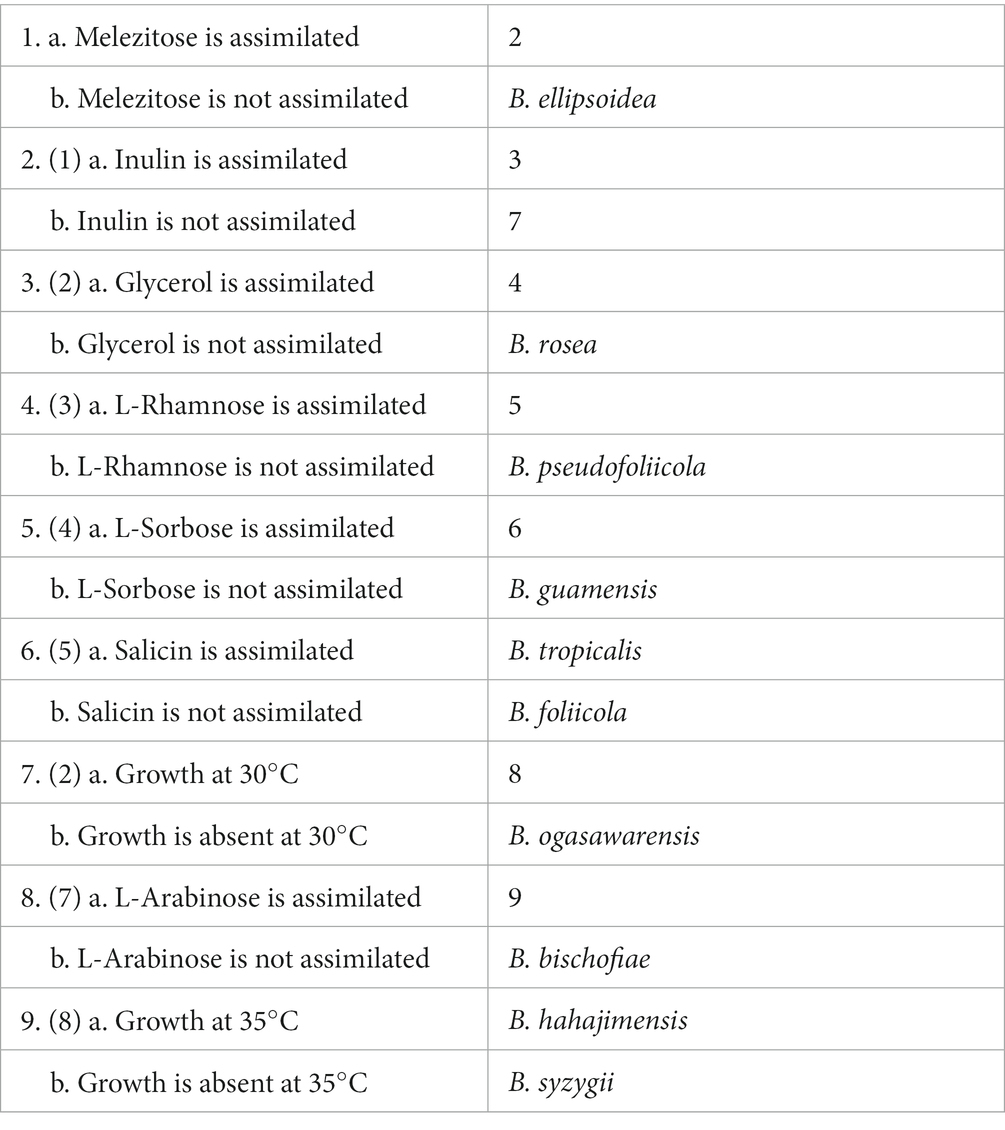
Key to species of Bannoa
The main trait comparisons and key identification of Bannoa are summarized below for the reader’s convenience (Table 4).
Discussion
In this study, three species of Bannoa from China, B. ellipsoidea, B. foliicola, and B. pseudofoliicola, are described as new species based on molecular analyzes and morphological features. Phylogenetic analyzes of these three new species of Bannoa confirmed that Bannoa is a monophyletic genus in a strongly supported clade that was similar to those described by Wang et al. (2015a,b) and Parra and Aime (2019). These three new species were clearly separated from other Bannoa species and from each other (Figure 2). Pairwise sequence comparisons of the LSU D1/D2 domains and the ITS regions between these three species and related species showed that they had lower similarity values than the common threshold for species demarcation in basidiomycetous yeast (Fell et al., 2000; Scorzetti et al., 2002; Vu et al., 2016). Additionally, they were highly similar in colony morphology and color, as well as in cell shape, but differed markedly from the closest related species in terms of their physiological and biochemical characteristics (Table 3). Phylogenetic analyzes coupled with morphological studies confirm the existence of these species in China.
Most reported Bannoa isolates were isolated from dead asymptomatic or infected leaves. B. bischofiae, B. hahajimensis, B. ogasawarensis, and B. syzygii were isolated from dead leaves of various plant species in the Ogasawara, Iriomote-Jima, and Yakushima islands of subtropical southwestern Japan (Hamamoto et al., 2002), or from the surface of leaves of Miscanthus and other unidentified plants in China (Zang et al., 1998; Chiang et al., 2001; Li et al., 2020). B. guamensis, B. rosea, and B. tropicalis were isolated from surfaces of diseased and healthy leaves of plants in the Euphorbiaceae, Asteraceae, and Poaceae plant families from the South Pacific Island of Guam and Guyana in South America (Parra and Aime, 2019). In addition, two strains of Bannoa, isolated from teliospores of Cintractia axicola in Panama (Matheny et al., 2006) and from cloud water samplings in France (Vaïtilingom et al., 2012), have been identified but not yet been named or extensively described (Parra and Aime, 2019). In the current study, we describe three new species of the Bannoa genus, B. ellipsoidea, B. foliicola and B. pseudofoliicola, isolated from healthy or diseased leaves collected in subtropical central China. Based on the evidence provided above, it is suggested that the phylloplane of plant leaves is the main habitat of yeasts in the genus Bannoa.
Members of Bannoa have not been sufficiently studied, and the diversity of Bannoa is poorly understood. All known Bannoa species, including B. ellipsoidea, B. foliicola and B. pseudofoliicola described in the present study, have a relatively narrow geographic distribution range, found primarily in subtropical Asia and tropics South America. However, some strains of Bannoa have been isolated in different parts of the world; for example, Bannoa sp. MP3490 (DQ631900) has been obtained from Panama, Bannoa aff. Ogasawarensis MCA7670 (MK990652) and Bannoa aff. Ogasawarensis MCA 7643 (MK990651) from Vanuatu, Bannoa aff. Guamensis MCA7799 (MK990655) from Cameroon, and Bannoa sp. BRIP 28272 (OK001795) from Australia. In addition, several environmental sequences of Bannoa have also been reported from Mexico (James et al., 2016), from Austria, and from Australia (Raghavendra et al., 2017). This suggests this genus could be broadly distributed and further large-scale studies are needed to explore the diversity and distribution of Bannoa species worldwide. Ultimately, these findings will greatly improve our understanding of the diversity, distribution and ecology of Bannoa.
In conclusion, three new Bannoa species isolated from the surface of plant leaves in China are identified based on morphology and phylogeny; viz. B. ellipsoidea, B. foliicola and B. pseudofoliicola. This study enriches the species diversity of the genus, which will also promote its taxonomy and phylogeny.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
C-YC and X-YC isolated the yeast strains and performed the taxonomic characterization. TL performed the phylogenetic analysis. C-YC drafted the manuscript and prepared the tables and figures. F-LH designed the study and revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by the National Natural Science Foundation of China (Project no. 31570021), Key scientific research projects of colleges and universities in Henan Province China (Project no. 23A210018) and the State Key Laboratory of Motor Vehicle Biofuel Technology, Henan Tianguan Enterprise Group Co., Ltd., China (no. 2018001).
Acknowledgments
The authors are incredibly grateful to their colleagues at the School of Life Science and Agricultural Engineering, Nanyang Normal University, including Lin Zhang and Hao zhang for providing specimens; and to Ran-Ran Jia and Wen-Ting Hu for their assistance with morphological observations.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Chiang, Y., Chou, C., Lee, P., and Chiang, T. (2001). Detection of leaf-associated fungi based on PCR and nucleotide sequence of the ribosomal internal transcribed spacer (ITS) in Miscanthus. Bot. Bulletin-Academia Sinica Taipei 42, 39–44. doi: 10.4049/jimmunol.175.12.8242
Fell, J. W., Boekhout, T., Fonseca, A., Scorzetti, G., and Statzell Tallman, A. (2000). Biodiversity and systematics of basidiomycetous yeasts as determined by large-subunit rDNA D1/D2 domain sequence analysis. Int. J. Syst. Evol. Microbiol. 50, 1351–1371. doi: 10.1099/00207713-50-3-1351
Felsenstein, J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x
Hall, T. A. (1999). Bioedit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98.
Hamamoto, M. (2011). “Bannoa Hamamoto (2002)” in The yeasts, a taxonomic study. eds. C. P. Kurtzman, J. W. Fell, and T. Boekhout. 5th ed (Amsterdam: Elsevier), 1383–11386. doi: 10.1016/B978-0-444-52149-1.00103-8
Hamamoto, M., Thanh, V. N., and Nakase, T. (2002). Bannoa hahajimensis gen. nov., sp. nov., and three related anamorphs, Sporobolomyces bischofiae sp. nov., Sporobolomyces ogasawarensis sp. nov. and Sporobolomyces syzygii sp. nov yeasts isolated from plants in Japan. Int. J. Syst. Evol. Microbiol. 52, 1023–1032. doi: 10.1099/00207713-52-3-1023
Hu, W. T., Chu, S. B., Li, Y., and Hui, F. L. (2022). Hyphopichia xiaguanensis f.a., sp. nov., an ascomycetous yeast species isolated from plant leaves. Int. J. Syst. Evol. Microbiol. 72:5398. doi: 10.1099/ijsem.0.005398
James, T. Y., Marino, J. A., Perfecto, I., and Vandermeer, J. (2016). Identification of putative coffee rust mycoparasites using single molecule DNA sequencing of infected pustules. Appl. Environ. Microbiol. 82, 631–639. doi: 10.1128/AEM.02639-15
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kurtzman, C. P., Fell, J. W., and Boekhout, T. (2011). “Methods for isolation, phenotypic characterization and maintenance of yeasts” in The yeasts, a taxonomic study. eds. C. P. Kurtzman, J. W. Fell, and T. Boekhout. 5th ed (Amsterdam: Elsevier), 87–110. doi: 10.1016/B978-0-444-52149-1.00007-0
Kurtzman, C. P., and Robnett, C. J. (1998). Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie Van Leeuwenhoek 73, 331–371. doi: 10.1023/a:1001761008817
Li, A. H., Yuan, F. X., Groenewald, M., Bensch, K., Yurkov, A. M., and Li, K. (2020). Diversity and phylogeny of basidiomycetous yeasts from plant leaves and soil: proposal of two new orders, three new families, eight new genera and one hundred and seven new species. Stud. Mycol. 96, 17–140. doi: 10.1016/j.simyco.2020.01.002
Matheny, P. B., Gossmann, J. A., Zalar, P., Kumar, T. K. A., and Hibbett, D. S. (2006). Resolving the phylogenetic position of the Wallemiomycetes: an enigmatic major lineage of Basidiomycota. Can. J. Bot. 84, 1794–1805. doi: 10.1139/b06-128
Nagahama, T., Hamamoto, M., Nakase, T., Shimamura, S., and Horikoshi, K. (2006). Phylogenetic relationship within the Erythrobasidium clade: molecular phylogenies, secondary structure, and intron positions inferred from partial sequences of ribosomal RNA and elongation factor-1a genes. J. Gen. Appl. Microbiol. 52, 37–45. doi: 10.2323/jgam.52.37
Nakase, T., and Takashima, M. (1993). A simple procedure for the high frequency isolation of new taxa of ballistosporous yeasts living on the surfaces of plants. RIKEN Rev. 3, 33–34. doi: 10.4049/jimmunol.175.12.8242
Nei, M., and Kumar, S. (2000). Molecular Evolution and Phylogenetics. Oxford University Press. England.
Parra, P. P., and Aime, M. C. (2019). New species of Bannoa described from the tropics and the first report of the genus in South America. Mycologia 111, 953–964. doi: 10.1080/00275514.2019
Raghavendra, A. K., Bissett, A. B., Thrall, P. H., Morin, L., Steinrucken, T. V., Galea, V. J., et al. (2017). Characterisation of above-ground endophytic and soil fungal communities associated with dieback-affected and healthy plants in five exotic invasive species. Fungal Ecol. 26, 114–124. doi: 10.1016/j.funeco.2017.01.003
Rehner, S. A., and Buckley, E. A. (2005). Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 97, 84–98. doi: 10.3852/mycologia.97.1.84
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. doi: 10.1093/oxfordjournals.molbev.a040454
Scorzetti, G., Fell, J. W., Fonseca, A., and Statzell Tallman, A. (2002). Systematics of basidiomycetous yeasts: a comparison of large subunit D1/D2 and internal transcribed spacer rDNA regions. FEMS Yeast Res. 2, 495–517. doi: 10.1111/j.1567-1364.2002.tb00117.x
Tamura, K., Stecher, G., and Kumar, S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Toome, M., Roberson, R. W., and Aime, M. C. (2013). Meredithblackwellia eburneangen. et sp. nov., Kriegeriaceae fam. nov. and Kriegeriales Ord. nov. toward resolving higher-level classification in Microbotryomycetes. Mycologia 105, 486–495. doi: 10.3852/12-251
Vaïtilingom, M., Attard, E., Gaiani, N., Sancelme, M., Deguillaume, L., Flossmann, A. I., et al. (2012). Long-term features of cloud microbiology at the puy de Dôme (France). Atmos. Environ. 56, 88–100. doi: 10.1016/j.atmosenv.2012.03.072
Vu, D., Groenewald, M., Szöke, S., Cardinali, G., Eberhardt, U., Stielow, B., et al. (2016). DNA barcoding analysis of more than 9 000 yeast isolates contributes to quantitative thresholds for yeast species and genera delimitation. Stud. Mycol. 85, 91–105. doi: 10.1016/j.simyco.2016.11.007
Wang, Q. M., Groenewald, M., Takashima, M., Theelen, B., Han, P. J., Liu, X. Z., et al. (2015a). Phylogeny of yeasts and related taxa within Pucciniomycotina determined from multigene sequence analyses. Stud. Mycol. 81, 27–53. doi: 10.1016/j.simyco.2015.08.002
Wang, Q. M., Groenewald, M., Takashima, M., Theelen, B., Han, P. J., Liu, X. Z., et al. (2015b). Phylogenetic classification of yeasts and related taxa within Pucciniomycotina. Stud. Mycol. 81, 149–189. doi: 10.1016/j.simyco.2015.12.002
Wang, Q. M., Theelen, B., Groenewald, M., Bai, F. Y., and Boekhout, T. (2014). Moniliellomycetes and Malasseziomycetes, two new classes in Ustilaginomycotina. Persoonia 33, 41–47. doi: 10.3767/003158514X682313
White, T. J., Bruns, T., Lee, S., and Taylor, J. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics” in PCR Protocols: A Guide to Methods and Applications. eds. M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (San Diego, CA: Academic Press), 315–322.
Zang, W., Liu, C. Y., Xie, G. F., Shen, C., Liu, X. Z., Zou, H. J., et al. (1998). Resources and species diversity of the phyllosphere yeasts from the Yandang Mountains in Zhejiang Province. Acta Ecol. Sin. 38, 3920–3930. doi: 10.5846/stxb201705040827
Keywords: Basidiomycota, morphology, multigene analyzes, plant leaves, three new species
Citation: Chai C-Y, Lei T, Chu X-Y and Hui F-L (2023) Multi-gene phylogeny and taxonomy of the genus Bannoa with the addition of three new species from central China. Front. Microbiol. 14:1143156. doi: 10.3389/fmicb.2023.1143156
Edited by:
Rajesh Jeewon, University of Mauritius, MauritiusReviewed by:
Luca Roscini, University of Perugia, ItalyRegina Sharmila Dass, Pondicherry University, India
Copyright © 2023 Chai, Lei, Chu and Hui. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Feng-Li Hui, ZmVuZ2xpaHVpQHllYWgubmV0
 Chun-Yue Chai
Chun-Yue Chai Ting Lei1
Ting Lei1 Feng-Li Hui
Feng-Li Hui