
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 11 May 2023
Sec. Systems Microbiology
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1139810
Two novel Gram-positive bacteria, designated strains REN8T and REN14T, were isolated from baijiu pit mud in Sichuan Province, China. REN8T achieved the best growth at 37°C, a pH of 8.0, and a NaCl concentration of 2%, while REN14T displayed optimal growth at 37°C, a pH of 6.0, and a NaCl concentration of 1%. 16S rRNA and genomic phylogenetic analysis showed that REN8T and REN14T were clustered with the genus Planococcus. The genomic DNA G + C contents of REN8T and REN14T were 46.7 and 45.1 mol%, respectively. The dDDH and ANI values were 24.5 and 80.43% between REN8T and P. salinarum (the most closely related type strain) and 25.1 and 82.42% between REN14T and P. soli (the most closely related type strain). Genomic analysis showed that several carbohydrate-active enzymes and secondary metabolite gene clusters existed in REN8T and REN14T. Chemotaxonomic characteristics of REN8T and REN14T included major fatty acids, predominant menaquinones, and polar lipids, all of which were consistent with the genus Planococcus. Based on the polyphasic taxonomic method, these two strains represent two novel species of the genus Planococcus; the name Planococcus beigongshangi sp. nov. is proposed for the type strain REN8T (=JCM 33964T = GDMCC 1.2213T), and the name Planococcus beijingensis sp. nov. is proposed for the type strain REN14T (=JCM 34410T = GDMCC 1.2209T). The addition of REN8T and REN14T might improve the quality of huangjiu by considerably increasing the amino acid nitrogen content and acidity and decreasing the bioamine content, with no significant change in alcohol content.
The genus Planococcus was first described by Migula in 1894 (Approved Lists 1980) (Kocur et al., 1970). To date, the genus Planococcus encompasses 24 species with validly published and correct names [LPSN—List of Prokaryotic names with Standing in Nomenclature (dsmz.de)]. The genus Planococcus is widely distributed in different regions and environments, especially marine and high salt areas. All members of the genus are characterized as aerobic, shaped as cocci or short rods, and generally motile, Gram-positive to Gram-variable and nonendospore forming (Gupta and Patel, 2020).
Baijiu is a traditional fermented alcoholic beverage in China (Xu et al., 2021). Pit mud, as a place for anaerobic fermentation of baijiu, contains abundant microbial resources, which play crucial roles in the unique flavor formation of baijiu (Qian et al., 2021). Pit mud can provide a suitable environment for the survival of microorganisms, and the microbial community in pit mud undergoes complex energy metabolism, which is very important to the quality of baijiu (Wang et al., 2020). Pit mud can be continuously used for decades; therefore, recycled pit mud forms a unique and complex microbial community (Tao et al., 2017). Screening and identification of new microbial resources from pit mud are helpful for further understanding the unique flavor of baijiu. Moreover, numerous microbial resources have additional functions in other fields, such as medicine and biocontrol (Yu et al., 2022). Huangjiu is a kind of brewing wine made with wheat and water as raw materials through the joint metabolism, fermentation, and interaction of molds, yeast, and bacteria through a unique process (Yu et al., 2023). Different from baijiu, huangjiu belongs to fermented wine and baijiu belongs to distilled wine. Therefore, bacteria were isolated and cultured from pit mud to uncover new bacterial species and study the relationship between bacteria and huangjiu flavor components.
In this study, two new members of the genus Planococcus were isolated from baijiu pit mud (Supplementary Figure S1 shows the isolation site of the two strains and baijiu production process) in Sichuan Province, China. The two new strains named REN8T and REN14T were identified based on a polyphasic taxonomic approach. Finally, the names Planococcus beigongshangi for strain REN8T and Planococcus beijingensis for strain REN14T were proposed.
Samples of baijiu pit mud were collected in 2019 from Sichuan Province, China (30°05′N 102°54′E). Samples were treated and diluted as previously described (Sun et al., 2021). Then, the mud suspension was spread on trypticase soy agar (TSA) and R2A agar medium. A light yellow single colony named strain REN8T was isolated after incubating on TSA at 30°C for 2 days. Another light yellow single colony named strain REN14T was isolated after incubating on R2A at 37°C for 2 days. REN8T and REN14T were preserved in a 30% glycerol suspension at −80°C for further analysis.
Genomic DNA of REN8T and REN14T was extracted by a TIANGEN Bacterial DNA Kit (TIANGEN Biotech Beijing Co., Ltd., Beijing, China). Universal primer pairs of 27F and 1492R were used to amplify the 16S rRNA gene of REN8T and REN14T by PCR with the following program: 94°C for 5 min; 35 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 3 min; and a final extension at 72°C for 10 min. After electrophoresis detection, the PCR products were sequenced by Sangon Biotech Co., Ltd. (Shanghai, China). The NCBI BLAST algorithm and EzBioCloud server were used to analyze the 16S rRNA gene similarity of REN8T and REN14T (Yoon et al., 2017). 16S rRNA gene sequences of REN8T, REN14T and other closely related type strains were aligned with the CLUSTALX program (Thompson et al., 1997). Phylogenetic analysis of REN8T and REN14T was conducted by the neighbor-joining method (Saitou and Nei, 1987) using the Kimura two-parameter model with 1,000 replicate bootstrap values by MEGA 11 software (Kimura, 1980; Tamura et al., 2021).
Whole-genome sequencing of REN8T and REN14T was conducted at Shanghai Majorbio Bio-Pharm Technology Co., Ltd. (Shanghai, China). Then, SOAPdenovo was used to assemble the clean reads (Li et al., 2008, 2010). The average nucleotide identity (ANI) between REN8T and REN14T, REN8T and the closest species Planococcus salinarum ISL-16T (Yoon et al., 2010), and REN14T and the closest species Planococcus soli XN13T (Luo et al., 2014) were calculated using the ANI calculator (Yoon et al., 2017). The digital DNA–DNA hybridization (dDDH) values were also determined by Genome-to-Genome Distance Calculator 2.1 (Auch et al., 2010). The genomic DNA G + C content of REN8T and REN14T was calculated (Aziz et al., 2008). The genomic phylogenetic analysis among REN8T, REN14T and related species was also constructed using PhyloPhlAn software (Asnicar et al., 2020).
Genes from REN8T and REN14T were annotated to the NR, Swiss-Prot, Pfam, COG, GO, and KEGG databases (Ashburner et al., 2000; Bairoch and Apweiler, 2000; Kanehisa and Goto, 2000; Deng et al., 2006; Jensen et al., 2008; Finn et al., 2014). The genomic characteristics of REN8T and REN14T were analyzed by CGView software (Stothard and Wishart, 2005). The numbers of carbohydrate-active enzymes (CAZy) and secondary metabolite gene clusters were analyzed by the CAZy database and antiSMASH, respectively (Lombard et al., 2014; Blin et al., 2021).
Morphological characteristics of REN8T and REN14T were observed under light and transmission electron microscopes. An anaerobic system (Thermo Scientific, United States) filled with tri-mixture anaerobic gas (85:5:10 mixture of N2, CO2, and H2) was used to evaluate the anaerobic growth ability of REN8T and REN14T. The Gram reaction of REN8T and REN14T was performed according to the nonstaining method (Buck, 1982). The morphology and color of REN8T and REN14T on TSA medium were observed by the naked eye. Four media, LB (Luria-Bertani) agar, R2A agar, NA (nutrient agar) and TSA, were used to assess the growth ability of REN8T and REN14T. The growth ability of REN8T and REN14T at different temperatures (10, 20, 28, 30, 37, 42, 45, 50, and 55°C), different NaCl tolerances (0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 15, and 20% w/v), and different pH values (3–12, with the pH increments using the following biological buffer system: pH 3.0–5.0, 2 M sodium acetate; pH 6.0–7.0, 1 M KH2PO4, 1 M NaOH; pH 8.0, 1 M Tris–HCl; pH 9.0–10.0, 1.5 M NaHCO3, 1.5 M Na2CO3; and 11–12, 0.5 M NaOH) (Xu et al., 2005) were determined. The catalase activity and hydrolysis capacity of REN8T and REN14T were investigated as previously described (Sun et al., 2012). API ZYM and API 20NE kits (bioMérieux, France) were used to investigate the enzyme activities and biochemical characteristics of REN8T and REN14T. GENIII Micro Plates (Biolog) were used to detect the basic biochemical tests and carbon source oxidation tests of REN8T and REN14T.
The fatty acids of REN8T and REN14T were treated and detected by the Microbial Identification System (MIDI) (Sasser, 1990). The polar lipids of REN8T and REN14T were investigated by two-dimensional thin-layer chromatography (TLC) (Minnikin et al., 1984). Respiratory quinones of REN8T and REN14T were extracted, examined and analyzed by high-performance liquid chromatography (HPLC) (Hu et al., 2001).
Wheat qu is produced by naturally cultivating or purebred breeding microorganisms on broken raw wheat grains. Wheat was ground, and 20–30% water was added. Then, the mixtures were evenly stirred, treaded and stacked. Wheat qu was finally obtained after natural fermentation at 45–50°C.
One kilogram of millet and 3 kg of water were mixed and thoroughly cooked. After adding 0.15% saccharifying enzyme at 60°C for 30 min, REN8T and REN14T were inoculated and cultured for 48 h at 37°C. The fortified qu was prepared as follows: wheat was ground, and 20–30% water, 1% REN8T and REN14T culture solutions were added. Then, the mixtures were evenly stirred, treaded and stacked, and natural fermentation was performed at 45–50°C. Finally, qu fortified with REN8T was named QR8, qu fortified with REN14T was named QR14, and wheat qu was named Qu1.
Huangjiu was brewed using the following steps: millet cooking, saccharification, fermentation, clarification and sterilization.
Several indices of huangjiu, including the bioamine content, volatile substances, amino acid nitrogen content, nonsugar solid content, total sugars, alcohol, and acidity, were measured. The bioamine content was detected by HPLC according to the national standard (GB 5009.208–2016). The volatile substances in huangjiu were determined as previously described by Xing and Ren (2019). Amino acid nitrogen, nonsugar solid content, total sugars, alcohol and acidity were detected according to national standards (GB/T 13662–2018). The differences in bioamine content, volatile substances, amino acid nitrogen, nonsugar solid content, total sugars, alcohol and acidity between huangjiu brewed by wheat qu and huangjiu brewed by fortified qu were compared. All experiments were repeated three times, and a significance analysis with one-way ANOVA in SPSS Statistics was also conducted.
The 16S rRNA gene sequence of REN8T was 1,514 bp. The 16S rRNA gene sequence of REN14T was 1,407 bp. Based on the 16S rRNA phylogenetic analysis, REN8T and REN14T fell within the genus Planococcus (Figure 1). Genome-based phylogenetic analysis showed that REN8T and REN14T were clustered with the genus Planococcus (Figure 2).
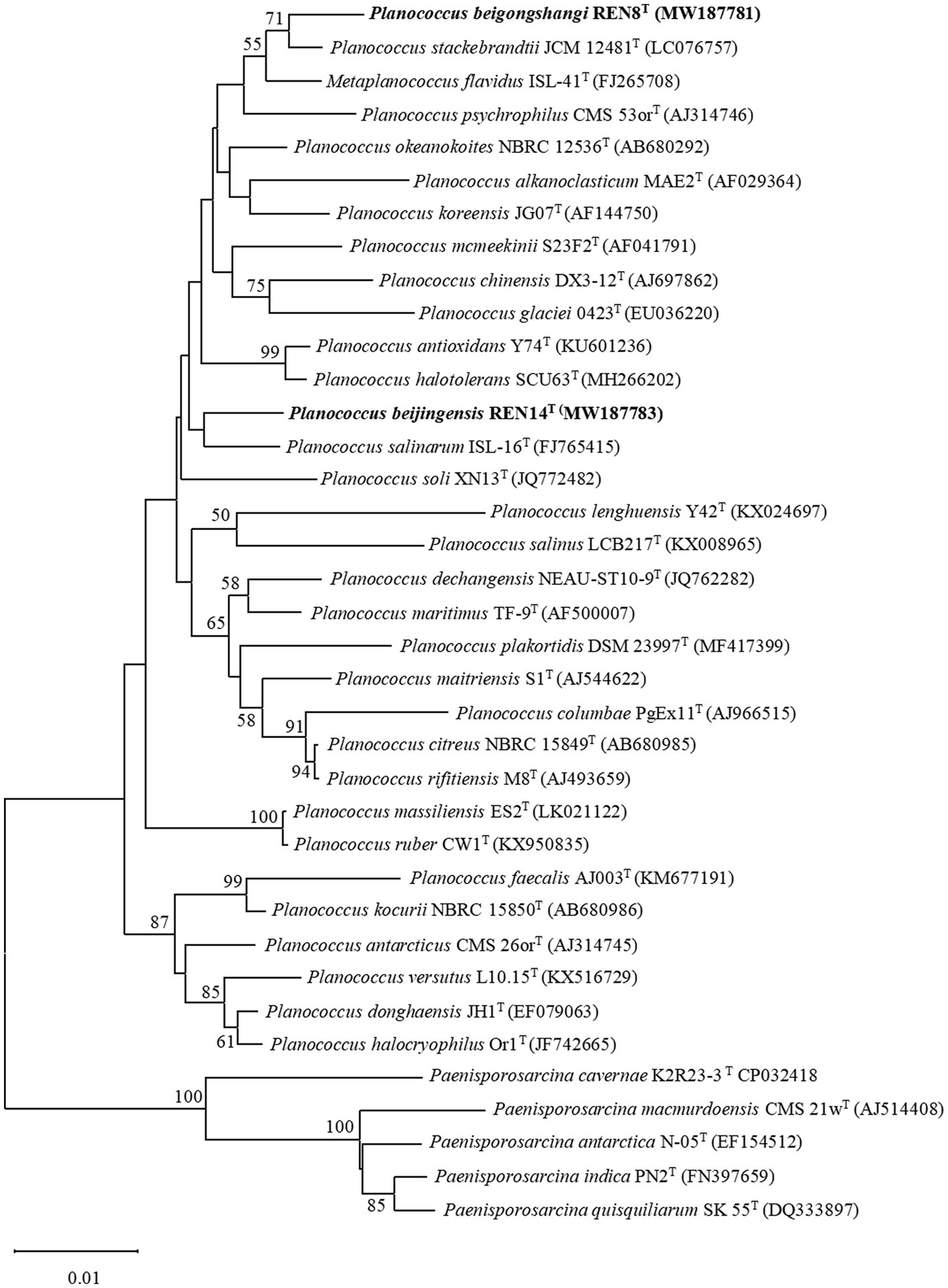
Figure 1. Phylogenetic analysis of the relationships of strains REN8T and REN14T and closely related species based on 16S rRNA gene sequences. The Kimura two-parameter model was used based on neighbor-joining analysis. Numbers at nodes are bootstrap values (%) based on 1,000 replications. Bar, 0.002 substitutions per nucleotide position.
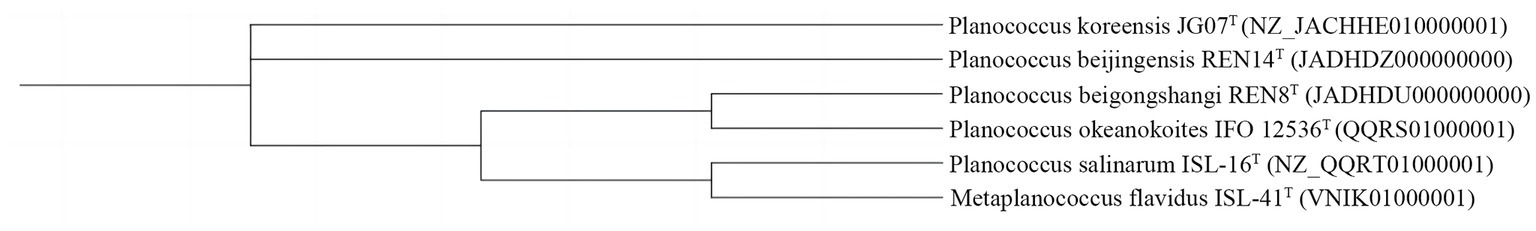
Figure 2. Phylogenetic analysis of the relationships of strains REN8T and REN14T and closely related species based on genomic analysis.
Genome information of REN8T and REN14T is shown in Table 1 (RAST Server—RAST Annotation Server).1 The genomic DNA G + C contents of REN8T and REN14T were 46.7 and 45.1 mol%, respectively, which fell within the genus Planococcus (34.8–51.0 mol%). The dDDH values between REN8T and P. salinarum and between REN8T and REN14T were 24.5 and 20.1%, respectively. The dDDH value between REN14T and P. soli was 25.1%. The ANI values between REN8T and P. salinarum and between REN8T and REN14T were 80.43 and 74.7%, respectively. The ANI value between REN14T and P. soli was 82.42%. The dDDH and ANI values of REN8T and REN14T and other strains of the same genus are shown in Table 2 [Type Strain Genome Server (dsmz.de)]. Based on the dDDH and ANI analysis, REN8T and REN14T were regarded as two novel species in the genus Planococcus.
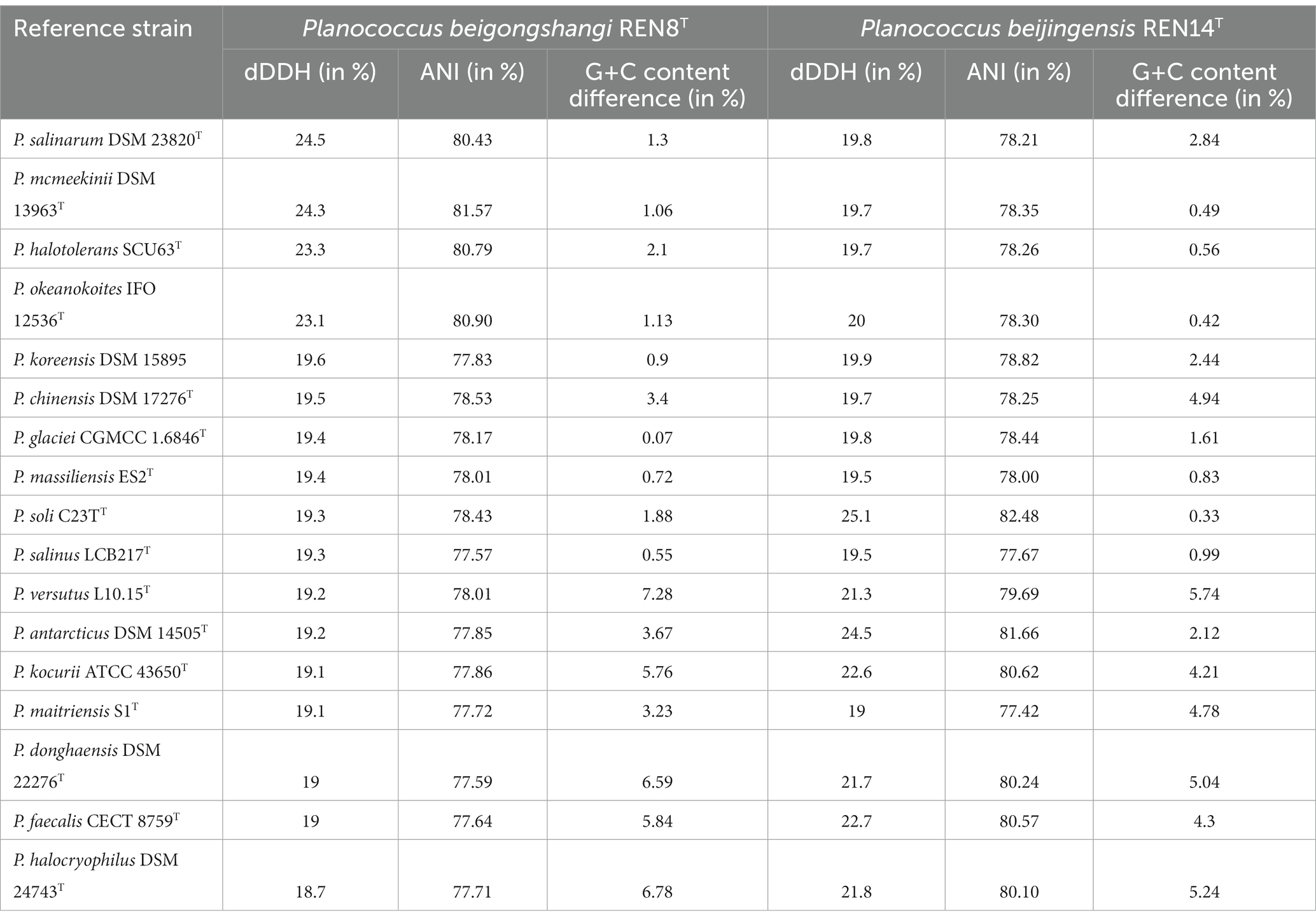
Table 2. Digital DNA–DNA hybridization (dDDH) and average nucleotide identity (ANI) values between REN8Tand REN14T and type strains of closely related species.
The genomic information of REN8T and REN14T is shown in Figures 3A,B. In total, 3,130 (NR), 2,454 (Swiss-Prot), 2,685 (Pfam), 2,806 (COG), 618 (GO), and 1,676 (KEGG) genes were annotated in REN8T. Twenty-three carbohydrate esterases, 23 glycosyl transferases, and 14 glycoside hydrolases were found in REN8T. Two terpenes and one T3PKS secondary metabolite cluster were found in REN8T. A total of 3,583 (NR), 2,922 (Swiss-Prot), 3,154 (Pfam), 3,275 (COG), 2,656 (GO), and 1,946 (KEGG) genes were annotated in REN14T. There were 28 glycosyl transferases, 26 carbohydrate esterases, and 25 glycoside hydrolases in REN14T. REN14T had one cluster of lanthipeptide-class-II secondary metabolites and two terpenes.
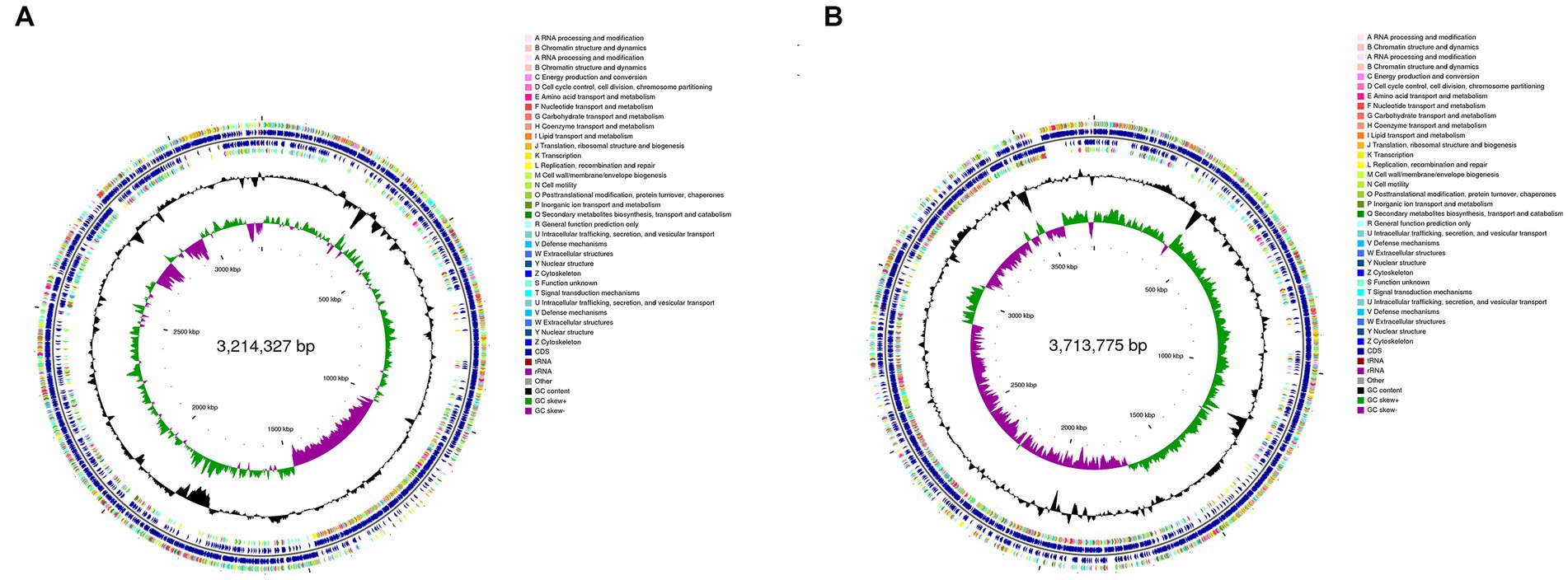
Figure 3. Circular plot from the genomes of REN8T and REN14T. Different loop levels represent relevant genomic information. (A) Genome of REN8T. (B) Genome of REN14T.
The basic characteristics of REN8T were as follows: Gram-positive, aerobic, and motile bacteria with short rods. The size of REN8T ranged from 0.4 to 0.6 m in width and 0.8 to 1.3 m in length (Supplementary Figure S2A). The diameter of REN14T was 1.0–2.0 μm (Supplementary Figure S2B), with the basic characteristics of Gram-positive, coccoid, aerobic and motile bacteria. After incubation for 2 days on TSA medium, colonies of REN8T and REN14T were circular, slightly convex, glistening, smooth and light yellow in color. REN8T and REN14T were able to grow on LB agar, R2A agar, TSA medium, and NA medium, and TSA medium was optimal. The growth temperature, pH and NaCl concentration range of REN8T was 28–37°C, 7.0–11.0 and 0–6% (w/v), respectively, with the optimal conditions being 37°C, a pH of 8.0 and 2% NaCl. Meanwhile, REN14T exhibited a growth range of 20–45°C, a pH of 5.0–12.0 and 0–15% (w/v) NaCl, with the optimal conditions being 37°C, a pH of 6.0 and 1% NaCl. The biochemical characteristics of REN8T, REN14T and other highly related species are shown in Table 3.
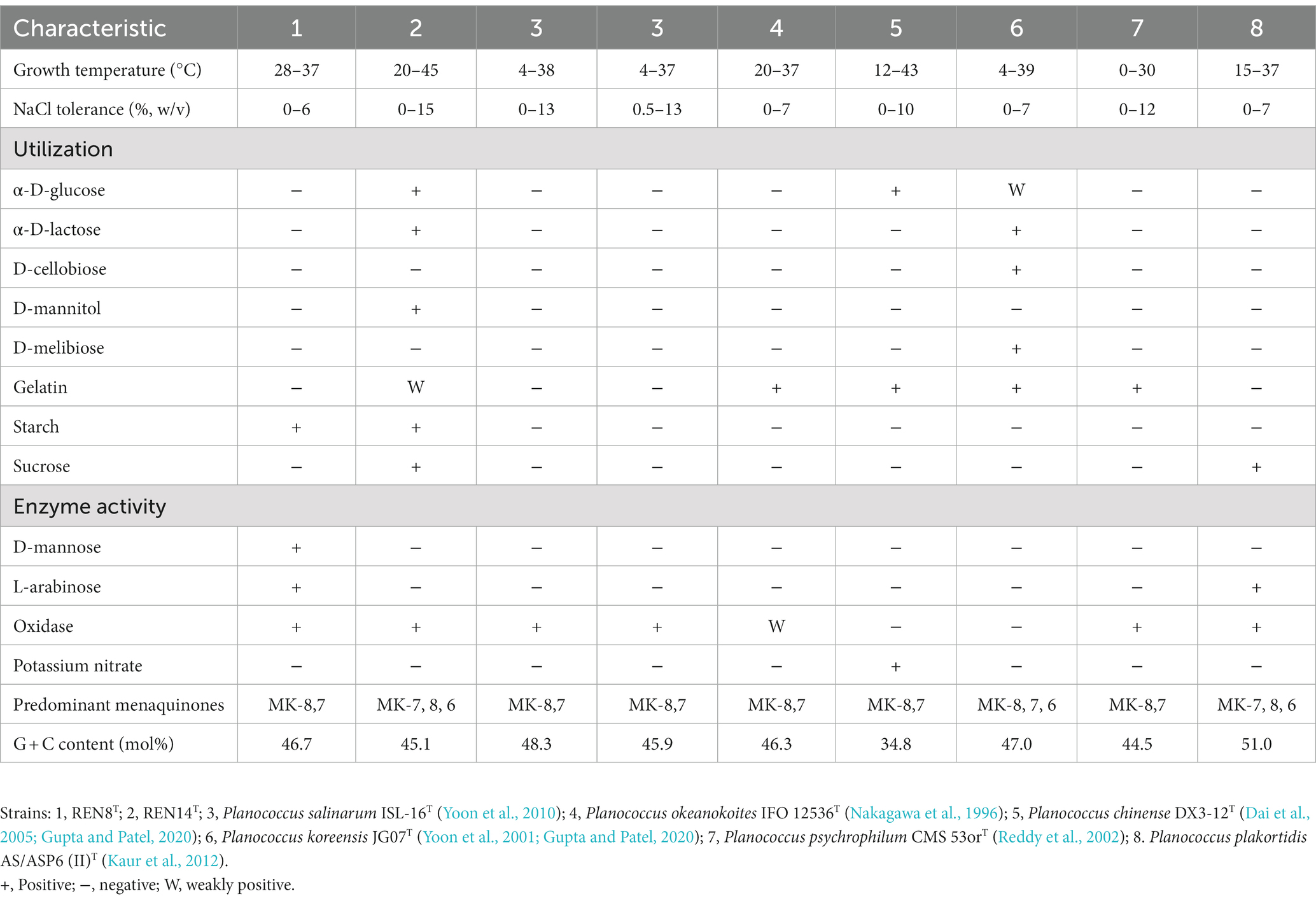
Table 3. Comparison of the phenotypic characteristics of strains REN8T and REN14T and type strains of closely related species.
Results similar to those of other members of the genus Planococcus were detected in REN8T and REN14T. The major fatty acids of REN8T were iso-C15:0 (33.15%), antesio-C15:0 (23.82%), iso-C17:0 (13.36%), and antesio-C17:0 (9.97%), and those of REN14T were iso-C15:0 (45.62%), antesio-C15:0 (26.56%), iso-C17:0 (9.38%), and antesio-C17:0 (6.47%). Although the fatty acid compositions of REN8T and REN14T were similar to those of the reference strains, their proportions varied (Table 4). The cellular polar lipids of REN14T were DPG, PE and PG, and REN8T had polar lipids of DPG, PE, PG and AL (Supplementary Figure S3). The predominant menaquinones of REN8T were MK-8:MK-7 = 28:72, with REN14T having menaquinones of MK-8:MK-7:MK-6 = 44:45:11.
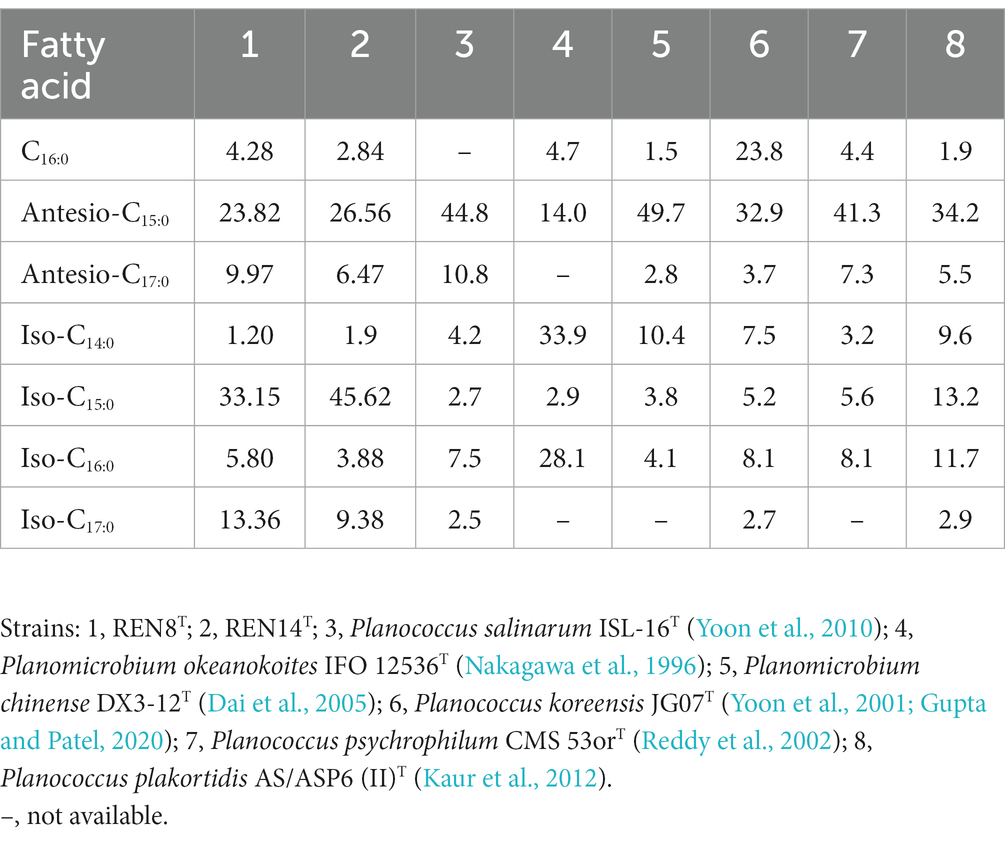
Table 4. Comparison of the fatty acid (%) profiles of strains REN8T and REN14T and type strains of closely related species.
Compared with wheat qu, fortified qu significantly reduced the content of bioamines. QR8 and QR14 reduced bioamines by 18.7 and 20.7%, respectively. Among these bioamines, histamine, spermine and spermidine were markedly reduced (Table 5).
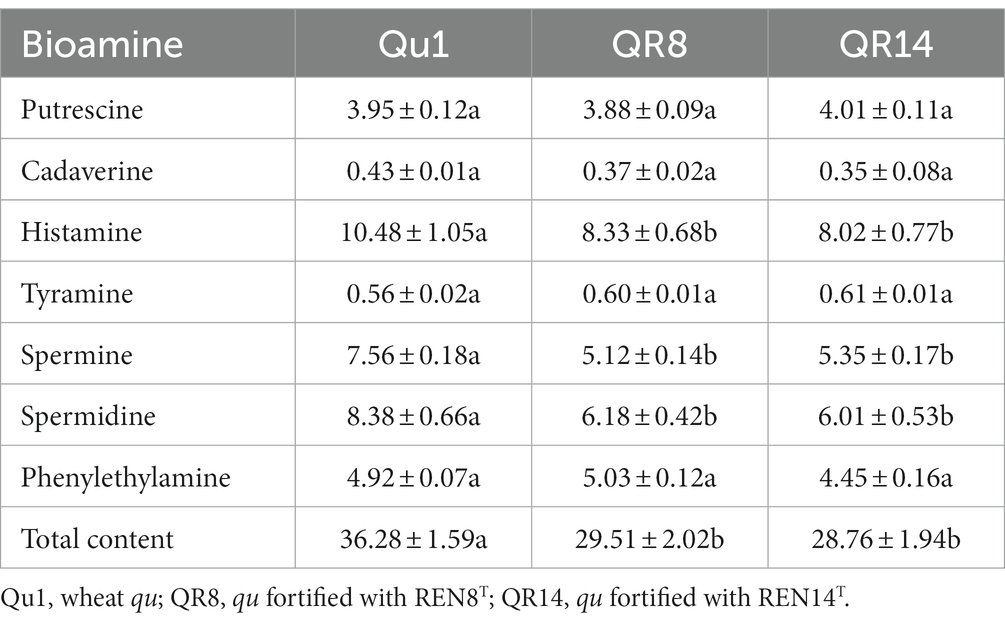
Table 5. Comparison of the bioamine content of wheat qu (Qu1) and wheat qu inoculated with REN8T (QR8) and REN14T (QR14).
For volatile substances, 104 volatile substances were detected in Qu1, while 106 and 109 kinds of volatile substances were detected in QR8 and QR14, respectively. Except for acid substances, other volatile substances and total content were not significantly altered among different types of huangjiu, which indicated that the addition of REN8T and REN14T did not influence the flavor quality of huangjiu fermentation (Tables 6, 7).
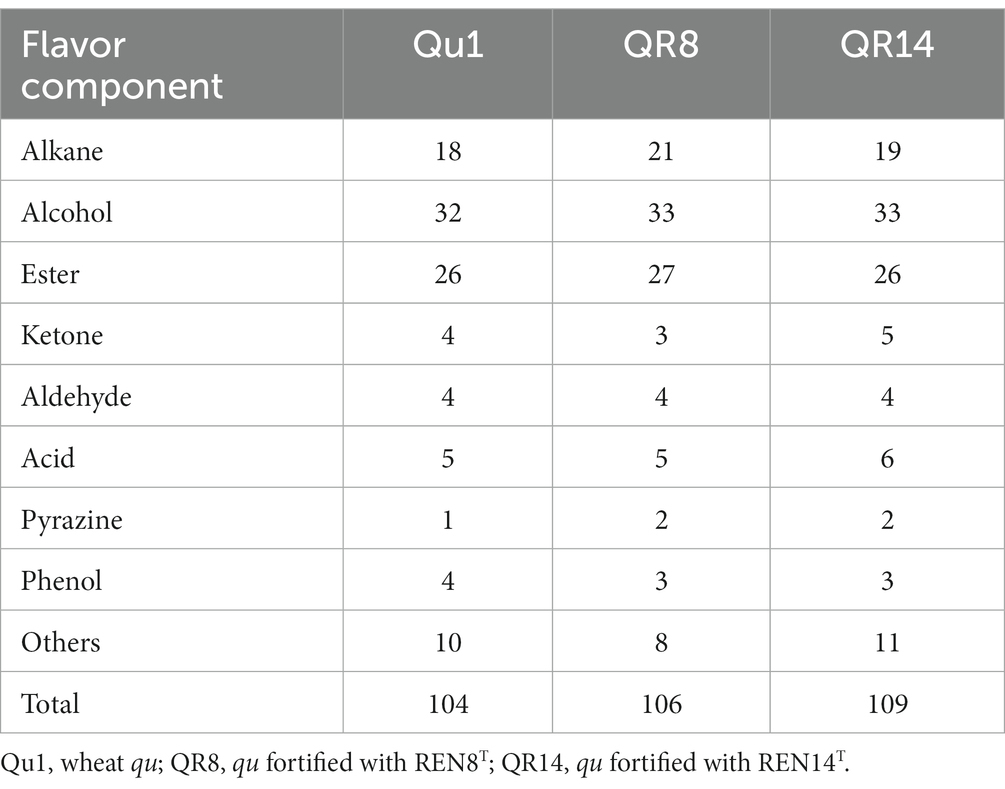
Table 6. Comparison of the varieties of flavor components in wheat qu (Qu1) and wheat qu inoculated with REN8T (QR8) and REN14T (QR14).
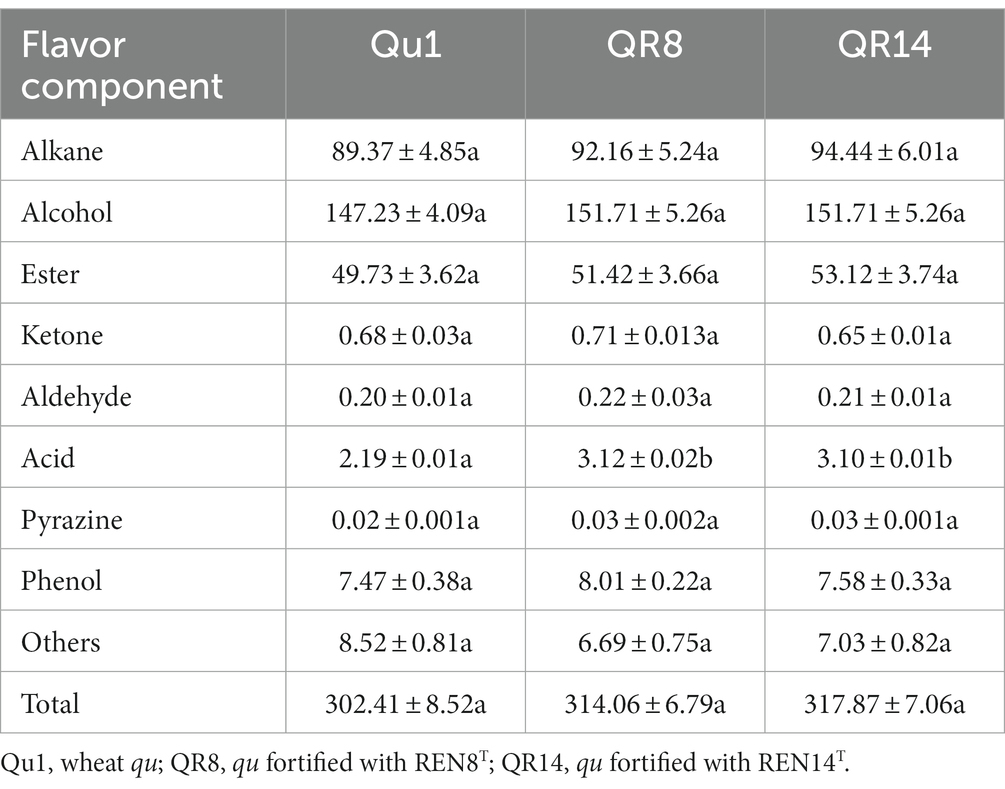
Table 7. Comparison of flavor component content of wheat qu (Qu1) and wheat qu inoculated with REN8T (QR8) and REN14T (QR14).
For other indices, the contents of alcohol, nonsugar solids and total sugars were not significantly altered after using QR8 and QR14, while amino acid nitrogen and acidity were dramatically improved (Table 8).
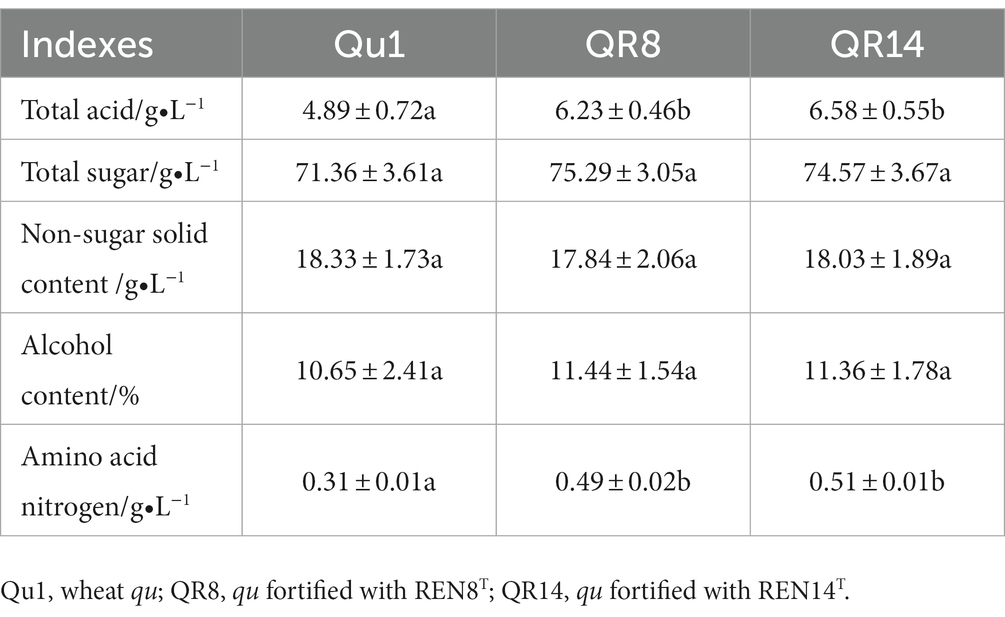
Table 8. Quality comparison of wheat qu (Qu1) and wheat qu inoculated with REN8T (QR8) and REN14T (QR14).
In this study, two novel species were isolated and identified. Based on the polyphasic taxonomic method described above, including phylogenetic analysis, genomic analysis, phenotypic characteristics and chemotaxonomic characteristics, strains REN8T and REN14T are two novel species within the genus Planococcus. Therefore, two novel species named Planococcus beigongshangi sp. nov. and Planococcus beijingensis sp. nov. are proposed.
Excessive bioamine consumption is harmful to health. In huangjiu brewing, controlling the content of bioamines is very important. Screening of specific strains that could effectively decompose biogenic amines is crucial to controlling the content of bioamines in huangjiu brewing. In this research, the addition of REN8T and REN14T could improve the quality of huangjiu, significantly increasing amino acid nitrogen content and acidity and decreasing the bioamines content, with flavoring substances and the alcohol content showing no obvious alteration.
In terms of reducing bioamines, the addition of REN8T and REN14T could significantly reduce the content of bioamines. This could be because REN8T and REN14T effectively inhibited the growth of some microorganisms that produce bioamines, resulting in a decrease in bioamine content in huangjiu. The addition of REN8T and REN14T may influence the microecology of the brewing process, causing some infectious microorganisms to be inhibited and the content of amino acid nitrogen and acidity to be improved.
Two novel Gram-positive bacteria, designated strains REN8T and REN14T, were isolated from baijiu pit mud in Sichuan Province, China. 16S rRNA and genomic phylogenomic analysis showed that REN8T and REN14T belonged to the genus Planococcus. The dDDH values between REN8T and P. salinarum and between REN14T and P. soli were 24.5 and 25.1%, respectively. The ANI values between REN8T and P. salinarum and between REN14T and P. soli were 80.43 and 82.42%, respectively. Combined with morphological, physiological, biochemical properties, and chemotaxonomic characteristics including major fatty acids, polar lipids and predominant menaquinones, strain REN8T represents a novel species, and the name Planococcus beigongshangi sp. nov. was proposed for the type strain REN8T (=JCM 33964T = GDMCC 1.2213T) (Table 9). Strain REN14T also represents a novel species, and the name Planococcus beijingensis sp. nov. is proposed for the type strain REN14T (=JCM 34410T = GDMCC 1.2209T) (Table 10). The addition of REN8T and REN14T to huangjiu could improve its quality by increasing the amino acid nitrogen content and acidity while decreasing the bioamine content, and the flavoring substances and alcohol content were not obviously altered.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
SH, JW, and LL: method. SH, and MH: data curation. SH: writing—original draft preparation. QR: writing—review and editing, supervision, and investigation. All authors have read and agreed to the published version of the manuscript.
This work was supported by the National Key Research and Development Program of China (2018YFC1603606 and 2018YFC1603800).
We are grateful to Zou Xiaohui, associate researcher at the National Institute for Viral Disease Control and Prevention, China and CDC, for preparing the transmission electron microscope.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1139810/full#supplementary-material
Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., et al. (2000). Gene ontology: tool for the unification of biology. Nat. Genet. 25, 25–29. doi: 10.1038/75556
Asnicar, F., Thomas, A. M., Beghini, F., Mengoni, C., Manara, S., Manghi, P., et al. (2020). Precise phylogenetic analysis of microbial isolates and genomes from metagenomes using PhyloPhlAn 3.0. Nat. Commun. 11:2500. doi: 10.1038/s41467-020-16366-7
Auch, A. F., Klenk, H. P., and Goker, M. (2010). Standard operating procedure for calculating genome-to-genome distances based on high-scoring segment pairs. Stand. Genomic Sci. 2, 142–148. doi: 10.4056/sigs.541628
Aziz, R. K., Bartels, D., Best, A. A., DeJongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75
Bairoch, A., and Apweiler, R. (2000). The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 28, 45–48. doi: 10.1093/nar/28.1.45
Blin, K., Shaw, S., Kloosterman, A. M., Charlop-Powers, Z., van Wezel, G. P., Medema, M. H., et al. (2021). antiSMASH 6.0: improving cluster detection and comparison capabilities. Nucleic Acids Res. 49, W29–W35. doi: 10.1093/nar/gkab335
Buck, J. D. (1982). Nonstaining (KOH) method for determination of gram reactions of marine bacteria. Appl. Environ. Microbiol. 44, 992–993. doi: 10.1128/aem.44.4.992-993.1982
Dai, X., Wang, Y. N., Wang, B. J., Liu, S. J., and Zhou, Y. G. (2005). Planomicrobium chinense sp. nov., isolated from coastal sediment, and transfer of Planococcus psychrophilus and Planococcus alkanoclasticus to Planomicrobium as Planomicrobium psychrophilum comb. nov. and Planomicrobium alkanoclasticum comb. nov. Int. J. Syst. Evol. Microbiol. 55, 699–702. doi: 10.1099/ijs.0.63340-0
Deng, Y. Y., Li, J. Q., Wu, S. F., Zhu, Y. P., Chen, Y. W., and He, F. C. (2006). Integrated nr database in protein annotation system and its localization. Comput. Eng. 32, 71–74. doi: 10.3969/j.issn.1000-3428.2006.05.026
Finn, R. D., Bateman, A., Clements, J., Coggill, P., Eberhardt, R. Y., Eddy, S. R., et al. (2014). Pfam: the protein families database. Nucleic Acids Res. 42, D222–D230. doi: 10.1093/nar/gkt1223
Gupta, R. S., and Patel, S. (2020). Robust demarcation of the family Caryophanaceae (Planococcaceae) and its different genera including three novel genera based on phylogenomics and highly specific molecular signatures. Front. Microbiol. 10:2821. doi: 10.3389/fmicb.2019.02821
Hu, H. Y., Lim, B. R., Goto, N., and Fujie, K. (2001). Analytical precision and repeatability of respiratory quinones for quantitative study of microbial community structure in environmental samples. J. Microbiol. Methods 47, 17–24. doi: 10.1016/S0167-7012(01)00286-X
Jensen, L. J., Julien, P., Kuhn, M., von Mering, C., Muller, J., Doerks, T., et al. (2008). eggNOG: automated construction and annotation of orthologous groups of genes. Nucleic Acids Res. 36, D250–D254. doi: 10.1093/nar/gkm796
Kanehisa, M., and Goto, S. (2000). KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. doi: 10.1093/nar/28.1.27
Kaur, I., and Das, A. P. (2012). Planococcus plakortidis sp. nov., isolated from the marine sponge Plakortis simplex (Schulze). Int. J. Syst. Evol. Microbiol 62, 883–889. doi: 10.1099/ijs.0.029967-0
Kimura, M. (1980). A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120. doi: 10.1007/BF01731581
Kocur, M., Zdena, P., Hodgkiss, W., and Martinec, T. (1970). The taxonomic status of the genus Planococcus Migula 1894. Int. J. Syst. Bacteriol. 20, 241–248. doi: 10.1099/00207713-20-3-241
Li, R., Li, Y., Kristiansen, K., and Wang, J. (2008). SOAP: short oligonucleotide alignment program. Bioinformatics 24, 713–714. doi: 10.1093/bioinformatics/btn025
Li, R., Zhu, H., Ruan, J., Qian, W., Fang, X., Shi, Z., et al. (2010). De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 20, 265–272. doi: 10.1101/gr.097261.109
Lombard, V., Ramulu, H. G., Drula, E., Coutinho, P. M., and Henrissat, B. (2014). The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42, D490–D495. doi: 10.1093/nar/gkt1178
Luo, X., Zhang, J., Li, D., Xin, Y., Xin, D., and Fan, L. (2014). Planomicrobium soli sp. nov., isolated from soil. Int. J. Syst. Evol. Microbiol. 64, 2700–2705. doi: 10.1099/ijs.0.055426-0
Minnikin, D. E., O’Donnell, A. G., Goodfellow, M., Alderson, G., Athalye, M., Schaal, A., et al. (1984). An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J. Microbiol. Methods 2, 233–241. doi: 10.1016/0167-7012(84)90018-6
Nakagawa, Y., Sakane, T., and Yokota, A. (1996). Emendation of the genus Planococcus and transfer of Flavobacterium okeanokoites Zobell and Upham 1944 to the genus Planococcus as Planococcus okeanokoites comb. nov. Int. J. Syst. Bacteriol. 46, 866–870. doi: 10.1099/00207713-46-4-866
Qian, W., Lu, Z. M., Chai, L. J., Zhang, X. J., Li, Q., Wang, S. T., et al. (2021). Cooperation within the microbial consortia of fermented grains and pit mud drives organic acid synthesis in strong-flavor Baijiu production. Food Res. Int. 147:110449. doi: 10.1016/j.foodres.2021.110449
Reddy, G. S. N., Prakash, J. S. S., Vairamani, M., Prabhakar, S., Matsumoto, G. I., and Shivaji, S. (2002). Planococcus antarcticus and Planococcus psychrophilus spp. nov. isolated from cyanobacterial mat samples collected from ponds in Antarctica. Extremophiles 6, 253–261. doi: 10.1007/s00792-001-0250-7
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425.
Sasser, M. (1990) Identification of bacteria by gas chromatog- raphy of cellular fatty acids. MIDI technical note 101. MIDI Inc., Newark, DE.
Stothard, P., and Wishart, D. S. (2005). Circular genome visualization and exploration using CGView Oxford University Press.
Sun, Z. B., Dai, F., Yan, Y., Guo, L. Y., Gu, H. Y., Xu, J. L., et al. (2021). Pseudoxanthomonas beigongshangi sp. nov., a novel bacteria with predicted nitrite and nitrate reduce ability isolated from pit mud of Baijiu. Antonie Van Leeuwenhoek 114, 1307–1314. doi: 10.1007/s10482-021-01603-w
Sun, Z. B., Zhang, H., Yuan, X. F., Wang, Y. X., Feng, D. M., Wang, Y. H., et al. (2012). Luteimonas cucumeris sp. nov., isolated a from cucumber leaf. Int. J. Syst. Evol. Microbiol. 62, 2916–2920. doi: 10.1099/ijs.0.037549-0
Tamura, K., Stecher, G., and Kumar, S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Tao, Y., Wang, X., Li, X., Wei, N., Jin, H., Xu, Z., et al. (2017). The functional potential and active populations of the pit mud microbiome for the production of Chinese strong-flavour liquor. Microb. Biotechnol. 10, 1603–1615. doi: 10.1111/1751-7915.12729
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F., and Higgins, D. G. (1997). The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. doi: 10.1093/nar/25.24.4876
Wang, X. J., Zhu, H. M., Ren, Z. Q., Huang, Z. G., Wei, C. H., and Deng, J. (2020). Characterization of microbial diversity and community structure in fermentation pit mud of different ages for production of strong-aroma Baijiu. Pol. J. Microbiol. 69, 1–14. doi: 10.33073/pjm-2020-018
Xing, X., and Ren, Q. (2019). Identification of a bacteriocin producing strain from huangjiuqu and study on its biogenic aminereduction efficiency. J. Food Sci. Technol. 37, 56–62. doi: 10.3969/j.issn.2095-6002.2019.02.009
Xu, P., Li, W. J., Tang, S. K., Zhang, Y. Q., Chen, G. Z., Chen, H. H., et al. (2005). Naxibacter alkalitolerans gen. Nov., sp. nov., a novel member of the family ‘Oxalobacteraceae’ isolated from China. Int. J. Syst. Evol. Microbiol. 55, 1149–1153. doi: 10.1099/ijs.0.63407-0
Xu, Y. Q., Wang, X. C., Liu, X., Li, X. T., Zhang, C. N., Li, W. W., et al. (2021). Discovery and development of a novel short-chain fatty acid ester synthetic biocatalyst under aqueous phase from Monascus purpureus isolated from Baijiu. Food Chem. 338:128025. doi: 10.1016/j.foodchem.2020.128025
Yoon, S., Ha, S., Kwon, S., Lim, J., Kim, Y., Seo, H., et al. (2017). Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 67, 1613–1617. doi: 10.1099/ijsem.0.001755
Yoon, J. H., Kang, S. S., Lee, K. C., Lee, E. S., Kho, Y. H., Kang, K. H., et al. (2001). Planomicrobium koreense gen. Nov., sp. nov., a bacterium isolated from the Korean traditional fermented seafood jeotgal, and transfer of Planococcus okeanokoites (Nakagawa et al. 1996) and Planococcus mcmeekinii (Junge et al. 1998) to the genus Planomicrobium. Int. J. Syst. Evol. Microbiol. 51, 1511–1520. doi: 10.1099/00207713-51-4-1511
Yoon, J. H., Kang, S. J., Lee, S. Y., Oh, K. H., and Oh, T. K. (2010). Planococcus salinarum sp. nov., isolated from a marine solar saltern, and emended description of the genus Planococcus. Int. J. Syst. Evol. Microbiol. 60, 754–758. doi: 10.1099/ijs.0.013136-0
Yu, H., Li, Q., Guo, W., Chen, C., Ai, L., and Tian, H. (2023). Dynamic analysis of volatile metabolites and microbial community and their correlations during the fermentation process of traditional Huangjiu (Chinese rice wine) produced around winter solstice. Food Chem X 18:100620. doi: 10.1016/j.fochx.2023.100620
Keywords: Planococcus beigongshangi , Planococcus beijingensis , baijiu , huangjiu , application
Citation: Hao S, Ren Q, Wang J, Li L and Huang M (2023) Two novel Planococcus species isolated from baijiu pit mud with potential application in brewing. Front. Microbiol. 14:1139810. doi: 10.3389/fmicb.2023.1139810
Received: 07 January 2023; Accepted: 21 April 2023;
Published: 11 May 2023.
Edited by:
Spyridon Ntougias, Democritus University of Thrace, GreeceReviewed by:
Hilal Ay, Yıldız Technical University, TürkiyeCopyright © 2023 Hao, Ren, Wang, Li and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qing Ren, cmVucWluZ0B0aC5idGJ1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.