
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 30 March 2023
Sec. Microbiotechnology
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1138401
This article is part of the Research Topic Microbial Co-cultures: A New Era of Synthetic Biology and Metabolic Engineering View all 16 articles
The consumption of various enzymes in industrial applications around the world has increased immensely. Nowadays, industries are more focused on incorporating microbial enzymes in multiple processes to avoid the hazardous effects of chemicals. Among these commercially exploited enzymes, proteases are the most abundantly used enzymes in different industries. Numerous bacterial alkaline proteases have been studied widely and are commercially available; however, fungi exhibit a broader variety of proteases than bacteria. Additionally, since fungi are often recognized as generally regarded as safe (GRAS), using them as enzyme producers is safer than using bacteria. Fungal alkaline proteases are appealing models for industrial use because of their distinct spectrum of action and enormous diversity in terms of being active under alkaline range of pH. Unlike bacteria, fungi are less studied for alkaline protease production. Moreover, group of fungi growing at alkaline pH has remained unexplored for their capability for the production of commercially valuable products that are stable at alkaline pH. The current review focuses on the detailed classification of proteases, the production of alkaline proteases from different fungi by fermentation (submerged and solid–state), and their potential applications in detergent, leather, food, pharmaceutical industries along with their important role in silk degumming, waste management and silver recovery processes. Furthermore, the promising role of alkali–tolerant and alkaliphilic fungi in enzyme production has been discussed briefly. This will highlight the need for more research on fungi growing at alkaline pH and their biotechnological potential.
Enzymes are biocatalysts and are involved in nearly all biological reaction. Enzymes have been used in beer, wine, vinegar production, and cheese making since prehistoric time. The enzymes used in these processes were not pure and well–characterized. They were generally produced by micro–organisms that were spontaneously growing. Later, selected strains of micro–organisms were being used to produce enzymes on a large scale, followed by their purification. This development has made a remarkable contribution to the rectification of industrial processes. Further development of industrial enzymes has been revolutionized through directed mutation, protein engineering, and genetic engineering (Sharma, 2019). Currently, various enzymes are used in industries like lipases, proteases, amylases, cellulases, xylanases, etc. However, proteases remain the dominant type of enzyme as they are extensively valuable for multiple processes in detergents, dairy, food, paper, and pulp industries. Proteases hold about 60% shares of total enzymes sold commercially every year (Rao et al., 1998; Savitha et al., 2011; Figure 1). In 2019, the worldwide protease market was 2.76 billion USD, and expected to increase over the period of 2019–2024 with the annual growth rate of 6.1% (Choudhary et al., 2022). Among various proteases, alkaline proteases contribute the largest sector of the enzyme market, especially in the detergent industry (Abidi et al., 2011). Alkaline proteases are active at neutral to alkaline pH range and require either Asp-His-Ser triad (serine protease) or metal ions (metalloprotease) to act on the substrate (Choudhary et al., 2022). Alkaline proteases are preferred over the other types of proteases because of their ability to sustain under the alkaline pH without losing the action specificity. Owing to this fact, they have long been used in various industries, exclusively in the detergent industry. As an excellent and diverse enzyme reservoir, micro–organisms are extensively used for enzyme production to meet the current demand for proteases in various industries. Consequently, researchers keep searching for novel micro-organisms secreting alkaline proteases with desirable properties (Matkawala et al., 2021). In 1971, the first report was published regarding the production of alkaline protease by bacterium (Bacillus sp. strain 221) (Horikoshi, 1999). Since then, several bacterial and fungal genera have been studied for alkaline protease activity and exploited for their commercial production. In industrial processes, fungal proteases are the choice of enzymes over bacterial enzymes as they can be produced using low–cost substrate coupled with high and rapid productivity (Naeem et al., 2022). One more advantage of fungi over bacteria is biomass separation from production media; the mycelia can be easily removed from the broth, simplifying downstream processes (Souza et al., 2015; Arya et al., 2021). Therefore, the demand for fungal proteases for their extensive applications in various industries is increasing worldwide. The present review article deals with fungal alkaline proteases, their production, characterization, and applications in different fields.
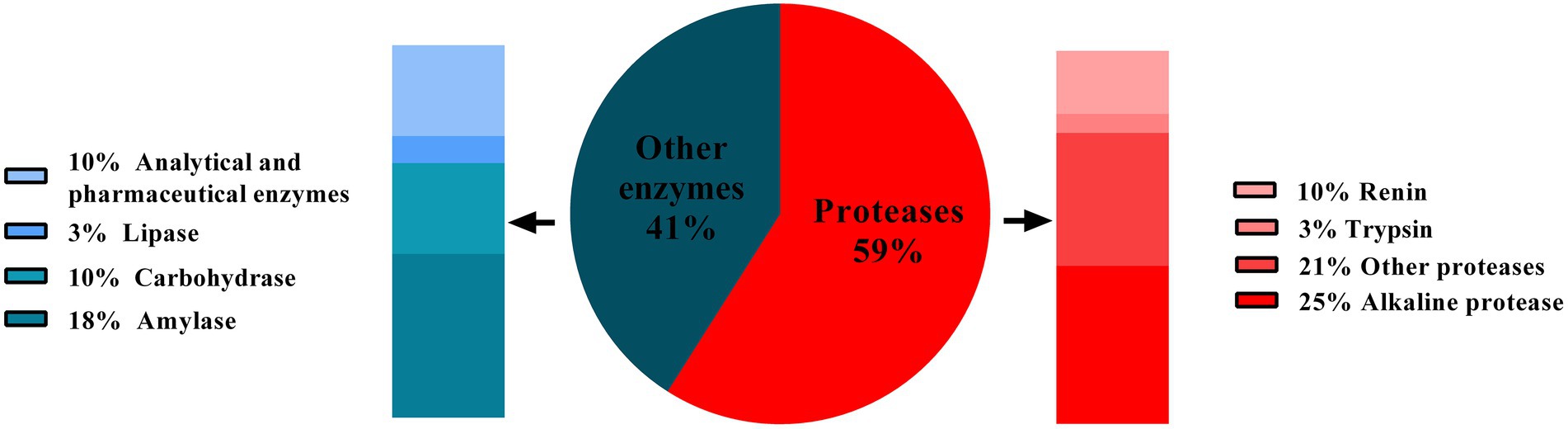
Figure 1. Contribution of types of proteases (red portion) in annual sales of the total enzyme. Source: adapted and modified from reference (Rao et al., 1998).
In accordance with the Nomenclature Committee of the International Union of Biochemistry and Molecular Biology (IUBMB), proteases belong to subgroup 4 of hydrolases (group 3) (Barrett, 1997). The proteases have been primarily categorized based on three essential criteria: (1) site of action; (2) chemical nature of the active site; (3) evolutionary relationship with respect to their structure. On the basis of site of action, proteases are classified as exo–peptidases and endo–peptidases. Proteases acting near the termini of the polypeptide chain are placed under the group of exo–peptidases. In contrast, endo–peptidases act on the peptide bond in the internal region of the polypeptide chain, as free amino acid or carboxyl group negatively affects the enzyme activity. Exo–peptidases are further clustered into carboxypeptidases (acting near C terminus) and aminopeptidases (acting near N terminus). Based on the chemical nature of the active site, proteases were previously grouped into four major groups: serine proteases, cysteine proteases, aspartic proteases and, metalloproteases (Rawlings and Barrett, 1993). After discovering three more types of proteolytic enzymes, (1) threonine protease, (2) glutamic protease and, (3) asparagine peptide lyase, the hierarchy of proteolytic enzyme was made in 1993 was redefined. As per the online database for peptidases and their inhibitors (MEROPS), seven types of proteolytic enzymes have been described (Rawlings et al., 2018). Serine proteases contain serine residue at their catalytic site used for their catalytic activity (Polgár, 2005). They show their activity at both neutral and alkaline pH with an optimum range of 7.0 to 11.0. Serine proteases that are effective at highly alkaline pH are clustered as serine alkaline proteases, which is a major subclass of serine proteases with optimum pH around 10. Subtilisin of bacterial origin is the second–largest subclass of serine proteases effective at alkaline pH (Rao et al., 1998; Ekici et al., 2008). Second group, Cysteine proteases, also known as thiol proteases contain catalytic triad of amino acids, Cys-His-Asn (Verma et al., 2016). They are optimally active at neutral pH (Barrett and Rawlings, 2001). The third group, Aspartic proteases, has a dyad of two highly conserved aspartic acid residues (Friedman and Caflisch, 2010). Being maximally active at low pH (3.0–4.0), they are usually known as acidic proteases. Fourth group, Metallo–proteases are known for a characteristic of their obligation for divalent metal ion (zinc, cobalt, copper, nickel, manganese, and iron) to activate water which acts as a nucleophile to catalyze the reaction (Rawlings and Barrett, 1995). In 1995, threonine protease was discovered as the fifth catalytic type of protease while resolving the proteasome structure from Thermoplasma acidophilum. Out of fourteen, three subunits of peptidase of proteasome had N–terminal threonine that participated in catalysis by acting as a nucleophile (Brannigan et al., 1995; Seemüller et al., 1995). The sixth type of protease was identified in 2004 as glutamic peptidases. The catalytic site of glutamic protease has a catalytic dyad of glutamic acid and glutamine (Fujinaga et al., 2004). In 2011, a new type of proteolytic enzyme termed asparagine peptide lyase was discovered, which uses asparagine residue of their active site for peptide bond cleavage. The active site of this enzyme contains Asp-Asn amino acids. Unlike all other proteolytic enzymes, this type of enzyme does not require hydrolysis (use of water molecule for lysis) to catalyze the reaction. Hence, they are not peptidase but peptide lyase (Rawlings et al., 2011). Detailed classification of these proteolytic enzymes according to the MEROPS database has been summarized in Table 1 (Fernandes et al., 2023).
Based on the evolutionary relationship, proteases are divided into different clans (also known as super families) that represent various protease families sharing a common ancestor in the evolution process (Argos, 1987). As per the recent data available on MEROPS database (Rawlings et al., 2018), proteolytic enzymes have been divided into 271 families, which are assembled into 56 clans. As of now, 53 families of serine peptidases, 96 families of cysteine peptidases, 16 families of aspartic peptidases, 76 families of metallopeptidases, 6 families of threonine peptidases, 2 families of glutamic peptidases, 10 families of asparagine lyases, 2 families of mixed catalytic type and 10 of unknown catalytic type have been identified (Rawlings and Bateman, 2019).
Alkaline proteases can be synthesized on a bulk scale using fungal isolates by submerged (Smf) and solid–state fermentation (SSF). Different fungi have been reported showing alkaline protease production potential (Table 2). Various cost–effective substrates like wheat bran, oil seed cakes, soybean bran, rice bran have been reported to give a considerable yield of the enzyme under SSF (Germano et al., 2003; Agrawal et al., 2004; Xiao et al., 2015; Novelli et al., 2016; Ram and Yepuru, 2018; Rukmi and Purwantisari, 2020). Although SSF offers volumetric productivity using cheaper substrates, superior monitoring and process control are still associated with Smf. Among fungi, Aspergillus sp. was found to be more prominent in the production of alkaline proteases. In a study, the regulation of alkaline protease production has been analyzed using both wild–type and mutant Aspergillus nidulans. This study describes an experiment to show the effect of carbon, nitrogen, and sulfur compounds on enzyme production regulation; when any of these compounds in the medium is withheld (Cohen, 1973). In another report, alkaline protease obtained from the cultures of Aspergillus niger strain Z1 in Czapek dox medium containing 1% casein can withstand the temperature up to 90°C and retained 48.4% of its original activity for 15 min. This alkaline protease has been reported to have its optimum activity at temperature 40°C and pH 9.0 (Coral et al., 2003). Aspergillus tamarii was tested for alkaline protease production, and its growth conditions were optimized in solid–state fermentation using wheat bran and submerged fermentation to maximize yield. The isolate was found to produce maximum enzyme at pH 9.0 and temperature 30°C in both fermentation types and required 65% moisture content in wheat bran (Anandan et al., 2007). Moreover, in the first report aimed to optimize production of alkaline protease by Aspergillus clavatus, it has been reported that the enzyme was highly active at pH 9.5 and 40°C temperature. The isolate showed the highest proteolytic activity (38 U/mL), when the medium was supplemented with 1% glucose and casein. Among tested nitrogen sources, NH4NO3 (0.2% w/v) was reported as the best nitrogen source and showed 27 U/mL enzyme activity (Tremacoldi and Carmona, 2005).
Further a report is available based on activity of bleach stable alkaline protease by the newly isolated Aspergillus clavatus ES1 (Hajji et al., 2007). The enzyme showed a 7.5–fold rise in specific activity when purified by acetone precipitation, gel filtration (Sephadex G–100), and ion–exchange chromatography (CM–Sepharose), with 29% recovery having optimum activity at 50°C and pH 8.5. In another study, alkaline protease obtained from Aspergillus terreus was studied for its use in detergent formulation. This monomeric enzyme with molecular weight 16 ± 1 kDa showed 148.9 U/mg enzyme activity. Casein was the best substrate over gelatin with 12.8 U/mL Vmax and 5.4 mg/mL Km. The enzyme was active from pH 8.0–12.0, showing optimum activity at pH 11.0 (Niyonzima and More, 2015). Additionally a literature is available (Ke et al., 2018) in which the successful expression of truncated alkaline protease in Pichia pastoris KM71 from Aspergillus sojae GIM3.33 using the RT–PCR technique has been described. This recombinant alkaline protease had 400.4 ± 40.5 U/mL enzyme activity up to three days after induction using methanol and was optimally active at pH 10.0 and temperature 45°C. A recent study was focused on the production of alkaline protease using Aspergillus sp. isolated from an Ethiopian food, Injera, and the use of response surface methodology (RSM) for process optimization. RSM optimization found that pH 8.24, 30.5°C, and 0.316% sucrose concentration gave the maximum enzyme activity (166.4221 U/mL) (Mustefa Beyan et al., 2021).
Besides Aspergillus, very few other fungal genera have been reported for alkaline protease production. In a study, assessment of the protease production by Fusarium oxysporum f. sp. dianthi race 2 (Fod) using cell wall extracts of susceptible and resistant cultivars of carnation (Dianthus caryophyllus L.) has been described (Rodríguez et al., 2017). This study was aimed to reveal the protease–assisted pathogenicity in Fod–carnation interaction. In the presence of cell wall extract of susceptible carnation, Fod produced maximum protease enzymes. Serine protease (trypsin), one of the three isoenzymes secreted by the fungus, was further studied for its identification and biochemical characterization. The enzyme was highly active at pH 8.0 and 50°C temperature, which was then purified using gel filtration (Sephadex G–75), DEAE–cellulose ion exchange, and affinity chromatography (Benzamidine Sepharose 6B). The molecular weight of this protease was 54 kDa, showing Km of 0.31 mg/mL and Vmax of 24.7 μmol/min. Further an interesting study is available on alkaline protease produced by Clonostachys rosea (syn. Gliocladium roseum) in which the enzyme was studied for its role in infecting nematodes. Its biochemical characterization has been performed upon purification of enzyme using ammonium sulfate precipitation, HiLoad 16/10 Phenyl Sepharose FF column, and anion exchange column (SOURCE 15Q 4.6/100 PE column). The enzyme exhibited highest activity at 60°C temperature and pH 9.0–10.0. Nematicidal studies revealed that the crude enzyme was superior in nematode immobilization than the purified enzyme with 80 ± 5% nematicidal activity (Li et al., 2006). Further the fibrinolytic alkaline protease (M.W. 27 kD) produced from Fusarium sp. BLB isolated from plant leaf (Hibiscus) has been characterized (Ueda et al., 2007). Purification of the enzyme was carried out by (NH4)2SO4 precipitation and column chromatography (CM–Toyopearl 650 M and Superdex 75). The enzyme had its maximum fibrinolytic activity at pH 9.5 and temperature 50°C. In another study, from the culture of Penicillium chrysogenum X5 the thermostable serine alkaline protease was purified, and its biochemical characterization was done. The results revealed that the enzyme was active at 10.0 pH and 80°C temperature. Enzyme purification was done by following three steps which include heat treatment at 80°C/10 min, (NH4)2SO4 precipitation (30–50%) and dialysis, and anion exchange chromatography (UNO Q–12) using the FPLC system. To evaluate its compatibility with laundry detergents, the enzyme was tested for its wash performance by using it as an additive with detergents. Results revealed that, the enzyme improved the performance of laundry detergent to clean the egg and blood stains on the fabric (Benmrad et al., 2018). A new alkaline protease obtained from Penicillium nalgiovense showed its optimum activity at pH 8.0, 35°C temperature, and 0.25 M NaCl concentration. (NH4)2SO4 precipitation, dialysis, and ultrafiltration increased the enzyme activity by 12–fold. The molecular mass (45.2 kDa) was confirmed by ESI–MS analysis (Papagianni and Sergelidis, 2014). One more species of Penicillium has been studied for the production of alkaline protease using SSF (Xiao et al., 2015). From fermented fish sauce, 30 fungi were isolated based on morphology, among which only P. citrinum YL–1 showed enzyme activity. Under Plackett–Burman design, three significant variables like peptone concentration, initial pH, and moisture content were chosen for increasing enzyme yield. The influence of these variables on production of alkaline protease was studied using Box–Behnken design. Another study, focused on fungal isolate Conidiobolus coronatus from Entomophthorales order, showed the usage of alkaline protease for gelatin degradation and silver recovery from x–ray film. The obtained enzyme was active at 40°C and pH 9.0 with 1.35 U/mL specific activity (Shankar et al., 2010). Scopulariopsis spp. was used to produce alkaline protease with a molecular mass of 15 ± 1 kD showing 138.1 U/mg specific activity. The optimum pH of this enzyme was 9.0, while the enzyme was active and stable between the pH 8.0 to 12.0. The enzyme was optimally active at temperature 50°C. The enzyme had 4.3 mg/mL Km and 15.9 U/mL Vmax, using casein as a substrate. As the enzyme was found to be stable in the presence of surfactants, oxidizing and bleaching agents, active at high temperatures and alkaline pH, exhibiting wide range of substrate specificity and compatibility with detergents, its consumption in the detergent formulation has been highly recommended (Niyonzima and More, 2014).
Fungal alkaline proteases have a several applications, predominantly in the detergent and food industries. Fungal alkaline proteases are envisioned to have wide–ranging uses in other fields like bioremediation and leather treatment, etc. (Kumar and Takagi, 1999). Table 3 highlights some of the crucial uses of fungal alkaline proteases.
Proteases are essential and standard additives in detergents, as they can remove all kinds of proteinaceous materials (Nehra et al., 2002). The first enzymatic formulation named “Brunus” was prepared in 1913, containing crude pancreatic extract and sodium carbonate (Razzaq et al., 2019). A detergent containing a bacterial enzyme with the trade name “BIO–40” was marketed for the first time in 1956 (Kumar et al., 2008). Proteases act against a wide range of substrates, making it worthwhile to remove stains of foodstuff, blood, and body secretions. Characteristics such as stability at alkaline pH and high temperature, ability to withstand with surfactants, oxidizing and chelating agents will make alkaline proteases an indispensable candidate for their application in the detergent industry (Sharma, 2019).
Many researchers studied the compatibility of alkaline protease with detergents to make it sound as a detergent additive. In a report it is presented that the alkaline protease produced by Conidiobolus coronatus (NCL 86.8.20) retained its 90% of activity at a lower concentration (0.05 mg/mL–1) in commercial detergent solution after 1 h of incubation at 40°C; suggesting its possible use in detergents (Phadatare et al., 1993). Alkaline serine–protease of Aspergillus clavatus ES1 showed extreme stability (100%) in non–ionic surfactant [5% (v/v) tween 80 and triton X–100]; however, enzyme retained its 90% of activity when tested with 0.1% SDS (strong anionic surfactant). Also, the enzyme showed moderate stability towards 1% (w/v) and 2% (w/v) sodium perborate, retaining 71 and 53% activity, respectively (Hajji et al., 2007). In another study, similar results were obtained for the protease produced by Scopulariopsis spp., where an increase in protease activity with Tween–80, SDS, and Triton X–100 with retention of 50% enzyme activity in the commercial detergents (enzymatic and nonenzymatic) was obtained (Niyonzima and More, 2014). Another alkaline protease of Aspergillus species (A. niger and A. terreus) retained 50–80% of its initial activity in the presence of various commercially available detergents such as Tide, Hattic, Savo, Surf, Henko, Persil, Wheel, and Aerial (Ali, 2008; Devi et al., 2008). Further a team of researchers studied the compatibility of enzymes produced by two fungal isolates, Graphium putredinis, Trichoderma harzianum, and an intergenetically developed fusant. All three enzymes were stable towards SDS and sodium perborate with retention of 58.25–73.82% and 61.58–70.24% of residual activity, respectively, while enzyme from fusant was highly stable than parents with 76.74% of activity when tested for its compatibility with commercial detergent Rin Advanced (Savitha et al., 2011). Similar studies were focused on the compatibility and incorporation of alkaline protease produced by Aspergillus niger and Aspergillus sp. DHE7 as an additive in detergent formulation (Pundir et al., 2012; El-Ghonemy and Ali, 2021). Recent literature highlighted the introduction of third-generation cold-adapted alkaline proteases to the detergent industry which were found to have excellent activity and stability in surfactants and bleaches (Suresh and Dass, 2022).
In leather–processing industries, alkaline proteases have extensive applications in various processes like soaking, debating, and depilating of skin and hides. This enzymatic treatment removes unwanted pigments, increases the skin area, and produces a clean hide (Arora, 2003). Alkaline proteases accelerate the dehairing process, as the attack of protease on the hair follicle at higher pH makes the hair removal process easy. Conventionally, this has been carried out using chemicals like a saturated solution of lime and sodium sulfide to treat an animal hide; but this is a very expensive and produces intensely polluting effluent (Anwar and Saleemuddin, 1998). A very few studies have been reporting the application of fungal alkaline protease in the leather industry. A study was aimed on the potential use of alkaline protease produced by Aspergillus flavus in the tannery as a depilation agent (Malathi and Chakraborty, 1991). Also, from experimental data, it was confirmed that enzymatic treatment increased the tensile strength, stitch tear strength, bursting strength of leather as compared to control. A similar study has reported using alkaline protease as a dehairing agent in tannery (Pal et al., 1996). Further, the potential use of protease obtained from Conidiobolus coronatus has been studied in soaking, dehairing, and bating animal skin/ hide (Khandelwal, 2013).
The production of cheese is the primary application of proteases in the dairy industry. For good flavor and texture development, proteolysis plays a vital role (Fox, 1982). Protease hydrolyses kappa casein and stabilizes micelle formation, which prevents coagulation. Proteases obtained from fungi such as A. oryzae, Rhizomucor miechie, R. pusillus are extensively used as coagulants for cheese production in the dairy industry (Neelakantan et al., 1999). Besides the dairy industry, alkaline protease was used in meat tenderization because of its potential to hydrolyze connective tissue and muscle fiber (Ellaiah et al., 2002).
The vast diversity and specificity of fungal proteases are significant advantages in developing effective therapeutic agents. Protease obtained from Aspergillus oryzae has been applied to cure the digestive disorders like lytic enzyme deficiency syndromes by its oral administration (Rani et al., 2012). Alkaline protease produced by Aspergillus niger LCF 9 is used for therapeutic application because of its collagenolytic activity (Barthomeuf et al., 1992). Fibrinogen, fibrin, and blood clot were hydrolyzed efficiently by proteases obtained from Aspergillus strains (El-Shora and Metwally, 2008). Treatments for illnesses including Dupuytren’s disease, Peyronie’s disease, wound healing, burns, glaucoma, intervertebral disc herniation, debridement, keloid, vitrectomy, and cellulite involve alkaline protease with collagenase activity (Gurumallesh et al., 2019). In burns and wound treatment, immobilized subtilisins have been formulated as soft gel-based formulae, ointments, and bandage materials (Matkawala et al., 2021).
Keratin is the primary protein found in waste from the poultry and leather industry. Because of its compactly packed polypeptide, which is stabilized by strong disulfide bonds and some weak interaction, makes it difficult to degrade. Fungal alkaline proteases have been accessed by many researchers for the degradation of keratin. A study has been done on the degradation of keratin using proteases from five species of Aspergillus (A. flavus, A. niger, A. fumigatus, A. terreus, and A. nidulans). Among them, protease from A. niger degraded maximum keratin of chicken feathers with 28 μg/mL of cysteine release followed by A. flavus, A. fumigatus, A. nidulans, and A. terreus (Kim, 2003). Protease from A. oryzae has also been reported to degrade feather keratin with the release of various essential (threonine, isoleucine, methionine, leucine, tyrosine, valine, histidine, and lysine) and non–essential (glycine, serine, alanine, phenylalanine, glutamic acid, aspartic acid, and arginine) amino acids and ammonia. Released amino acids and ammonia from the waste can be further used as fertilizer or additives in feed (Ali et al., 2011). In another study, ten dermatophytes from the soil was isolated and studied to biodegrade human and animal hair. Among ten dermatophytes, Chrysosporium indicum degraded human hair with maximum weight loss (56.66%); whereas, Microsporum gypseum and Trichophyton verrucosum degraded animal hair with 49.34% weight loss (Sharma et al., 2011).
The proteases are significant candidates in the silk industry for silk degumming or sericin removal from silk. Rough texture of the raw silk fiber is due to the presence of sericin in the peripheral region of fiber. Degumming of silk before dyeing, helps to improve the sheen, texture, and color of cloth. Various fungal proteases have been used for silk degumming. The literature is available on the comparative study of silk degumming using Marseille’s soap and enzyme from Conidiobolus species (Gulrajani et al., 2000). Fungal protease showed 19.8% weight loss after incubation of 3 h at 37°C, while degumming with Marseille’s soap gave 20.5% weight loss on the incubation of 1.5 h at boiling temperature. Successful degumming at a lower temperature will make enzymatic degumming a cost–effective process compared to chemical degumming. One more study based on a similar approach reported comparing degumming by six microbial proteases (4 fungal and 2 actinomycetes) and conventional method using alkali and soap. Among tested proteases of microbial origin, Conidiobolus brefeldianus MTCC 5185 and Actinomycete–2 (BOA–3) showed best results with 21.10 ± 0.67 and 21.78 ± 0.99% weight loss, respectively, in a short time which is similar to the weight loss (21.40 ± 0.75%) by conventional method (More et al., 2013).
The photography industry uses a large quantity of silver in the preparation of light–sensitive emulsion. Used X–ray film has been found to contain around 1.5 to 2.0% silver in the gelatin layers. Silver recovery from X–ray films by conventional methods, mainly by combustion of X–ray films, causes pollution in environment. Hence, the hydrolysis of the gelatin present on the X–ray films using fungal alkaline proteases can be used as an alternative option for silver recovery (Sharma et al., 2019). In one of the studies, protease obtained from Conidiobolus coronatus showed 5% weight loss in x–ray film with the silver recovery of 3.87% (w/w) of sludge weight and 0.2% (w/w) of x–ray film weight, respectively (Shankar et al., 2010). Similarly from one more study, the recovery of 0.135 gm of silver from 40 gm of x–ray film has been reported with 0.335% yield using protease produced by Aspergillus versicolor PF/F/107, suggesting an eco–friendly way to the silver from used x–ray films (Choudhary, 2013).
Primarily most of the fungal species are known to grow at weakly acidic to neutral pH and only handful of researchers has reported on the fungi which can grow at alkaline pH as well. Consequently, these groups of alkaliphilic and alkali–tolerant fungi have remained unexplored for their capability for production of commercially valuable products that are stable at alkaline pH.
A research done in Japan on the distribution of alkalophilic and alkali–tolerant fungi from the limestone caves soil in Japan revealed that approximately one–third (30.8%) of the isolates had the optimum pH in the alkaline range. Fungal species belonging to genera Acremonium and Chrysosporium were found to be predominant at alkaline pH. (Nagai et al., 1998). A recent study for understanding the evolution alkaliphily in fungi has described the diversity of weak alkali–tolerant to alkaliphilic fungi from soils around the basin of soda lakes in Asia and Africa. It shows the wide of alkaliphilic trait throughout ascomycota division of fungi. Fungi belonging to Emericellopsis lineage (Emericellopsis alkalina) of order Hypocreales, along with fungi from Plectosphaerellaceae (Sodiomyces species, Acrostalagmus luteoalbus), Pleosporaceae (Alternaria sect. Soda), Chaetomiaceae (Thielavia sp.) families, were found to be strong alkalitolerants and effective alkaliphiles. Moderate alkali–tolerant fungi included members of genus Scopulariopsis, Fusarium, Cladosporium and Acremonium–like species. While, Penicillium sp., Purpureocillium lilacinum, and Alternaria alternata species showed weak alkali tolerance (Grum Grzhimaylo et al., 2016). In another such study from Thailand, 490 fungi were isolated from various mycological samples, including tree–holes, roots, leaf litter, wood, and soil at pH 11. A total of 324 (66%) isolates were screened for alkaline enzymes like Arabinanase, amylase, potato–galactanase, and protease. This screening revealed that most of these alkali–tolerant fungal isolates were able to produce at least one of the listed enzymes, and a few strains were positive for the activity of all four enzymes (Kladwang et al., 2003).
This dig at the literature highlights the gap in research to produce alkaline enzymes, especially industrially important alkaline proteases from alkaliphilic and alkali–tolerant fungi. This will lead to the production of more promising alkaline enzymes and can ease the wide range of industrially important reactions that are done at alkaline conditions.
Global consumption of various enzymes, especially microbial proteases, in industrial applications is increasing because of their wide range of applications in different industries. To meet the existing demand and rapid and easy production, the exploitation of other alternative microbial sources like fungi becomes essential. Fungal proteases are emerging as the best alternative and possess numerous commercial applications. The present comprehensive review describes the alkaline proteases obtained from fungi of different genera and their potential applications in various industries. Meager studies of fungal alkaline proteases leave the scope for research in alkaline proteases from fungi for their industrial applications, which will be helpful for future research worldwide. This review also highlights the need to study the alkaliphily trait spread among fungi, the diversity of alkali-tolerant and alkaliphilic fungi, and their use for the production of various alkaline enzymes. Moreover, due to their intrinsic adaptation to withstand the alkalinity, these enzymes can be used as a promising alternative for current alkaline enzymes produced by fungi growing at acidic to neutral pH.
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Authors thank Director, MACS’ Agharkar Research Institute, Pune for providing the necessary facilities to carry out the research work. KP also acknowledges the S. P. Pune University for providing the admission to Ph.D. degree and University Grant Commission (UGC), New Delhi for granting Senior Research Fellowship (SRF).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abidi, F., Chobert, J. M., Haertlé, T., and Marzouki, M. N. (2011). Purification and biochemical characterization of stable alkaline protease Prot–2 from Botrytis cinerea. Process Biochem. 46, 2301–2310. doi: 10.1016/j.procbio.2011.09.010
Abraham, L. D., and Breuil, C. (1996). Isolation and characterization of a subtilisin–like serine proteinase secreted by the sap–staining fungus Ophiostoma piceae. Enzym. Microb. Technol. 18, 133–140. doi: 10.1016/0141-0229(95)00098-4
Agrawal, D., Patidar, P., Banerjee, T., and Patil, S. (2004). Production of alkaline protease by Pddenicillium sp. under SSF conditions and its application to soy protein hydrolysis. Process Biochem. 39, 977–981. doi: 10.1016/S0032-9592(03)00212-7
Ali, U. F. (2008). Utilization of whey amended with some agro–industrial by–products for the improvement of protease production by aspergillus terreus and its compatibility with commercial detergents. Res. J. Agric. Biol. Sci. 4, 886–891.
Ali, T. H., Ali, N., and Mohamed, L. A. (2011). Production, purification and some properties of extracellular keratinase from feathers–degradation by aspergillus oryzae NRRL–447. J. Appl. Sci. Environ. Sanit. 6, 123–136.
Anandan, D., Marmer, W. N., and Dudley, R. L. (2007). Isolation, characterization and optimization of culture parameters for production of an alkaline protease isolated from aspergillus tamarii. J. Ind. Microbiol. Biotechnol. 34, 339–347. doi: 10.1007/s10295-006-0201-5
Anwar, A., and Saleemuddin, M. (1998). Alkaline proteases: a review. Bioresour. Technol. 64, 175–183. doi: 10.1016/S0960-8524(97)00182-X
Argos, P. (1987). A sensitive procedure to compare amino acid sequences. J. Mol. Biol. 193, 385–396. doi: 10.1016/0022-2836(87)90226-9
Arya, P. S., Yagnik, S. M., Rajput, K. N., Panchal, R. R., and Raval, V. H. (2021). Understanding the basis of occurrence, biosynthesis, and implications of thermostable alkaline proteases. Appl. Biochem. Biotechnol. 193, 4113–4150. doi: 10.1007/s12010-021-03701-x
Barrett, A. J. (1997). Nomenclature Committee of the International Union of biochemistry and molecular biology (NC-IUBMB). Enzyme nomenclature. Recommendations 1992. Supplement 4: corrections and additions. Eur. J. Biochem. 250, 1–6. doi: 10.1111/j.1432-1033.1997.001_1.x
Barrett, A. J., and Rawlings, N. D. (2001). Evolutionary lines of cysteine peptidases. Biol. Chem. 382, 727–734. doi: 10.1515/bchm.2001.382.5.727
Barthomeuf, C., Pourrat, H., and Pourrat, A. (1992). Collagenolytic activity of a new semi–alkaline protease from aspergillus Niger. J. Ferment. Bioeng. 73, 233–236. doi: 10.1016/0922-338X(92)90168-T
Benmrad, M. O., Moujehed, E., Elhoul, M. B., Mechri, S., Bejar, S., Zouari, R., et al. (2018). Production, purification, and biochemical characterization of serine alkaline protease from Penicillium chrysogenum strain X5 used as excellent bio–additive for textile processing. Int. J. Biol. Macromol. 119, 1002–1016. doi: 10.1016/j.ijbiomac.2018.07.194
Brannigan, J. A., Dodson, G., Duggleby, H. J., Moody, P. C., Smith, J. L., Tomchick, D. R., et al. (1995). A protein catalytic framework with an N-terminal nucleophile is capable of self-activation. Nature 378, 416–419. doi: 10.1038/378416a0
Cao, L., Tan, H., Liu, Y., Xue, X., and Zhou, S. (2008). Characterization of a new keratinolytic Trichoderma atroviride strain F6 that completely degrades native chicken feather. Lett. Appl. Microbiol. 46, 389–394. doi: 10.1111/j.1472-765X.2008.02327.x
Chakrabarti, S. K., Matsumura, N., and Ranu, R. S. (2000). Purification and characterization of an extracellular alkaline serine protease from aspergillus terreus (IJIRA 6.2). Curr. Microbiol. 40, 239–244. doi: 10.1007/s002849910048
Chandra Behera, B., Kumar Sethi, B., Mohapatra, S., Thatoi, H., and Ranjan, M. R. (2021). Bio-production of alkaline protease by Trichoderma longibrachiatum and Penicillium rubidurum using different agro-industrial products. Nov. Res. Microbiol. J. 5, 1241–1255. doi: 10.21608/nrmj.2021.178300
Chellappan, S., Jasmin, C., Basheer, S. M., Elyas, K. K., Bhat, S. G., and Chandrasekaran, M. (2006). Production, purification and partial characterization of a novel protease from marine Engyodontium album BTMFS10 under solid state fermentation. Process Biochem. 41, 956–961. doi: 10.1016/j.procbio.2005.10.017
Choudhary, V. (2013). Recovery of silver from used X–ray films by aspergillus versicolor protease. J. Acad. Ind. Res. 2, 39–41.
Choudhary, K., Mankar, M. K., and Sahay, S. (2022). “Extremophilic enzymes: catalytic features and industrial applications” in Extremophilic fungi: Ecology, Physiology and Applications (Singapore: Springer Nature Singapore), 273–314.
Cohen, B. L. (1973). Regulation of intracellular and extracellular neutral and alkaline proteases in aspergillus nidulans. Microbiology 79, 311–320. doi: 10.1099/00221287-79-2-311
Coral, G., Arikan, B. U., Unaldi, M. N., and Guvenmez, H. A. (2003). Thermostable alkaline protease produced by an aspergillus Niger strain. Ann. Microbiol. 53, 491–498.
Devi, M. K., Banu, A. R., Gnanaprabhal, G. R., Pradeep, B. V., and Palaniswamy, M. (2008). Purification, characterization of alkaline protease enzyme from native isolate aspergillus Niger and its compatibility with commercial detergents. Indian J. Sci. Technol. 1, 1–6. doi: 10.17485/ijst/2008/v1i7.8
Dozie, I. N., Okeke, C. N., and Unaeze, N. C. (1994). A thermostable, alkaline-active, keratinolytic proteinase from Chrysosporium keratinophilum. World J. Microbiol. Biotechnol. 10, 563–567. doi: 10.1007/BF00367668
Ekici, Ö. D., Paetzel, M., and Dalbey, R. E. (2008). Unconventional serine proteases: variations on the catalytic Ser/his/asp triad configuration. Protein Sci. 17, 2023–2037. doi: 10.1110/ps.035436.108
El-Ghonemy, D. H., and Ali, T. H. (2021). Effective bioconversion of feather–waste keratin by Thermo–surfactant stable alkaline keratinase produced from aspergillus sp. DHE7 with promising biotechnological application in detergent formulations. Biocatal. Agric. Biotechnol. 35:102052. doi: 10.1016/j.bcab.2021.102052
El-Khonezy, M. I., Elgammal, E. W., Ahmed, E. F., and Abd-Elaziz, A. M. (2021). Detergent stable thiol-dependant alkaline protease produced from the endophytic fungus aspergillus ochraceus BT21: purification and kinetics. Biocatal. Agric. Biotechnol. 35:102046. doi: 10.1016/j.bcab.2021.102046
Ellaiah, P., Srinivasulu, B., and Adinarayana, K. (2002). A review on microbial alkaline proteases. J. Sci. Ind. Res. 61, 690–704.
El-Shora, H. M., and Metwally, M. A. (2008). Production, purification and characterization of proteases from whey by some fungi. Ann. Microbiol. 58, 495–502. doi: 10.1007/BF03175548
Fernandes, V. D., Deepika, M., Ajith, S., and Pramod, T. (2023). “Versatile action, properties, application and mechanism of eco-friendly microbial enzyme-proteases” in Enzymes-Mechanisms and Action (Delhi India: Jaya Publishing House), 155.
Fox, P. F. (1982). Proteolysis in milk and dairy products. Biochem. Soc. Trans. 10, 282–284. doi: 10.1042/bst0100282
Friedman, R., and Caflisch, A. (2010). On the orientation of the catalytic dyad in aspartic proteases. Proteins 78, NA–1582. doi: 10.1002/prot.22674
Fujinaga, M., Cherney, M. M., Oyama, H., Oda, K., and James, M. N. (2004). The molecular structure and catalytic mechanism of a novel carboxyl peptidase from Scytalidium lignicolum. Proc. Natl. Acad. Sci. U. S. A. 101, 3364–3369. doi: 10.1073/pnas.0400246101
Germano, S., Pandey, A., Osaku, C. A., Rocha, S. N., and Soccol, C. R. (2003). Characterization and stability of proteases from Penicillium sp. produced by solid–state fermentation. Enzym. Microb. Technol. 32, 246–251. doi: 10.1016/S0141-0229(02)00283-1
Grum Grzhimaylo, A. A., Georgieva, M. L., Bondarenko, S. A., Debets, A. J., and Bilanenko, E. N. (2016). On the diversity of fungi from soda soils. Fungal Divers. 76, 27–74. doi: 10.1007/s13225-015-0320-2
Gulrajani, M. L., Agarwal, R., and Chand, S. (2000). Degumming of silk with a fungal protease. Indian J. Fibre Text Res. 25, 138–142.
Gurumallesh, P., Alagu, K., Ramakrishnan, B., and Muthusamy, S. (2019). A systematic reconsideration on proteases. Int. J. Biol. Macromol. 128, 254–267. doi: 10.1016/j.ijbiomac.2019.01.081
Hajji, M., Kanoun, S., Nasri, M., and Gharsallah, N. (2007). Purification and characterization of an alkaline serine–protease produced by a new isolated aspergillus clavatus ES1. Process Biochem. 42, 791–797. doi: 10.1016/j.procbio.2007.01.011
Horikoshi, K. (1999). Alkaliphiles: some applications of their products for biotechnology. Microbiol. Mol. Biol. Rev. 63, 735–750. doi: 10.1128/MMBR.63.4.735-750.1999
Ke, Y., Yuan, X., Li, J., Zhou, W., Huang, X., and Wang, T. (2018). High–level expression, purification, and enzymatic characterization of a recombinant aspergillus sojae alkaline protease in Pichia pastoris. Protein Expr. Purif. 148, 24–29. doi: 10.1016/j.pep.2018.03.009
Khandelwal, H. B. (2013). Production, purification and characterization of fungal alkaline protease and its applications. Ph.D. Thesis. Pune: University of Pune.
Khandelwal, H. B., More, S. V., Kalal, K. M., and Laxman, R. S. (2015). Eco–friendly enzymatic dehairing of skins and hides by C. brefeldianus protease. Clean Technol. Environ. Policy 17, 393–405. doi: 10.1007/s10098-014-0791-y
Kim, J. D. (2003). Keratinolytic activity of five aspergillus species isolated from poultry farming soil in Korea. Mycobiology 31, 157–161. doi: 10.4489/MYCO.2003.31.3.157
Kladwang, W., Bhumirattana, A., and Hywel Jones, N. (2003). Alkaline–tolerant fungi from Thailand. Fungal Divers. 13, 69–83.
Koutb, M. M., Hassan, E. A., Morsy, F. M., and Bagy, M. M. (2022). Optimization of keratinase production by keratinolytic fungus Chrysosporium tropicum and its potentiality in bidegradation of chicken feathers. J. Umm Al-Qura Univ. Appl. Sci. 20, 1–7. doi: 10.1007/s43994-022-00020-7
Kumar, D., Savitri, T. N., Verma, R., and Bhalla, T. C. (2008). Microbial proteases and application as laundry detergent additive. Res. J. Microbiol. 3, 661–672. doi: 10.3923/jm.2008.661.672
Kumar, C. G., and Takagi, H. (1999). Microbial alkaline proteases: from a bioindustrial viewpoint. Biotechnol. Adv. 17, 561–594. doi: 10.1016/S0734-9750(99)00027-0
Lanka, S., Anjali, C. H., and Pydipalli, M. (2017). Enhanced production of alkaline protease by Aspergillus niger DEF 1 isolated from dairy form effluent and determination of its fibrinolytic ability. Afr. J. Microbiol. Res. 11, 440–449. doi: 10.5897/AJMR2016-8379
Laxman, R. S., Sonawane, A. P., More, S. V., Rao, B. S., Rele, M. V., Jogdand, V. V., et al. (2005). Optimization and scale up of production of alkaline protease from Conidiobolus coronatus. Process Biochem. 40, 3152–3158. doi: 10.1016/j.procbio.2005.04.005
Li, J., Yang, J., Huang, X., and Zhang, K. Q. (2006). Purification and characterization of an extracellular serine protease from Clonostachys rosea and its potential as a pathogenic factor. Process Biochem. 41, 925–929. doi: 10.1016/j.procbio.2005.10.006
Malathi, S., and Chakraborty, R. (1991). Production of alkaline protease by a new aspergillus flavus isolate under solid–substrate fermentation conditions for use as a depilation agent. Appl. Environ. Microbiol. 57, 712–716. doi: 10.1128/aem.57.3.712-716.1991
Matkawala, F., Nighojkar, S., Kumar, A., and Nighojkar, A. (2021). Microbial alkaline serine proteases: production, properties and applications. World J. Microbiol. Biotechnol. 37, 63–62. doi: 10.1007/s11274-021-03036-z
Mignon, B., Swinnen, M., Bouchara, J. P., Hofinger, M., Nikkels, A., Pierard, G., et al. (1998). Purification and characterization of a 315 kDa keratinolytic subtilisin-like serine protease from Microsporum canis and evidence of its secretion in naturally infected cats. Med. Mycol. 36, 395–404. doi: 10.1080/02681219880000631
More, S. V., Khandelwal, H. B., Joseph, M. A., and Laxman, R. S. (2013). Enzymatic degumming of silk with microbial proteases. J. Nat. Fibers 10, 98–111. doi: 10.1080/15440478.2012.761114
More, S. S., Lakshmi Sridhar, D., Prakash, S. N., Vishwakarma, J., and Umashankar, S. (2013). Purification and properties of a novel fungal alkaline keratinase from Cunninghamella echinulata. Turk J. Biochem. 38, 68–74. doi: 10.5505/tjb.2013.37928
Mustefa Beyan, S., Venkatesa Prabhu, S., Mumecha, T. K., and Gemeda, M. T. (2021). Production of alkaline proteases using aspergillus sp. isolated from Injera: RSM–GA based process optimization and enzyme kinetics aspect. Curr. Microbiol. 78, 1823–1834. doi: 10.1007/s00284-021-02446-4
Naeem, M., Manzoor, S., Abid, M. U. H., Tareen, M. B. K., Asad, M., Mushtaq, S., et al. (2022). Fungal proteases as emerging biocatalysts to meet the current challenges and recent developments in biomedical therapies: an updated review. J. Fungi 8:109. doi: 10.3390/jof8020109
Nagai, K., Suzuki, K., and Okada, G. (1998). Studies on the distribution of alkalophilic and alkali–tolerant soil fungi II: fungal flora in two limestone caves in Japan. Mycoscience 39, 293–298. doi: 10.1007/BF02464011
Neelakantan, S., Mohanty, A. K., and Kaushik, J. K. (1999). Production and use of microbial enzymes for dairy processing. Curr. Sci. 77, 143–148.
Nehra, K. S., Dhillon, S., Chaudhary, K., and Singh, R. (2002). Production of alkaline protease by aspergillus sp. under submerged and solid substrate fermentation. Indian J. Microbiol. 42, 43–47.
Niyonzima, F. N., and More, S. (2014). Purification and properties of detergent–compatible extracellular alkaline protease from Scopulariopsis spp. Prep. Biochem. Biotechnol. 44, 738–759. doi: 10.1080/10826068.2013.854254
Niyonzima, F. N., and More, S. S. (2015). Purification and characterization of detergent–compatible protease from aspergillus terreus gr. 3. Biotech 5, 61–70. doi: 10.1007/s13205-014-0200-6
Novelli, P. K., Barros, M. M., and Fleuri, L. F. (2016). Novel inexpensive fungi proteases: production by solid state fermentation and characterization. Food Chem. 198, 119–124. doi: 10.1016/j.foodchem.2015.11.089
Omrane Benmrad, M., Moujehed, E., Ben Elhoul, M., Zaraî Jaouadi, N., Mechri, S., Rekik, H., et al. (2016). A novel organic solvent–and detergent–stable serine alkaline protease from Trametes cingulata strain CTM10101. Int. J. Biol. Macromol. 91, 961–972. doi: 10.1016/j.ijbiomac.2016.06.025
Pal, S., Banerjee, R., Bhattacharyya, B. C., and Chakraborty, R. (1996). Application of a proteolytic enzyme in tanneries as a depilating agent. J. Am. Leather Chem. Assoc. 91, 59–63.
Palanivel, P., Ashokkumar, L., and Balagurunathan, R. (2013). Production, purification and fibrinolytic characterization of alkaline protease from extremophilic soil fungi. Int J Pharm. Bio. Sci 4, 101–110.
Papagianni, M., and Sergelidis, D. (2014). Purification and biochemical characterization of a novel alkaline protease produced by Penicillium nalgiovense. Appl. Biochem. Biotechnol. 172, 3926–3938. doi: 10.1007/s12010-014-0824-3
Phadatare, S. U., Deshpande, V. V., and Srinivasan, M. C. (1993). High activity alkaline protease from Conidiobolus coronatus (NCL 86.8.20): enzyme production and compatibility with commercial detergents. Enzym. Microb. Technol. 15, 72–76. doi: 10.1016/0141-0229(93)90119-M
Polgár, L. (2005). The catalytic triad of serine peptidases. Cell. Mol. Life Sci. 62, 2161–2172. doi: 10.1007/s00018-005-5160-x
Prolo, T., Izidoro, S. C., de Lima, V. A., Maia, G. A., and Knob, A. (2020). Adding value to a recalcitrant and problematic waste: the use of dog hair for fungal keratinolytic protease production. Biocatal. Biotransformation 38, 343–356. doi: 10.1080/10242422.2020.1746770
Pundir, R. K., Rana, S., and Tyagi, H. (2012). Studies on compatibility of fungal alkaline protease with commercially available detergents. Int. J. Modern Biochem. 1, 41–56.
Ram, R. M., and Yepuru, S. K. (2018). Production of alkaline pro tease from aspergillus oryzae isolated from seashore of bay of Bengal. J. Appl. Nat. Sci. 10, 1210–1215. doi: 10.31018/jans.v10i4.1905
Rani, M. R., Prasad, N. N., and Sambasivarao, K. R. (2012). Optimization of cultural conditions for the production of alkaline protease from a mutant aspergillus Flavus AS2. Asian J. Exp. Biol. Sci. 3, 565–576.
Rao, M. B., Tanksale, A. M., Ghatge, M. S., and Deshpande, V. V. (1998). Molecular and biotechnological aspects of microbial proteases. Microbiol. Mol. Biol. Rev. 62, 597–635. doi: 10.1128/MMBR.62.3.597-635.1998
Rawlings, N. D., and Barrett, A. J. (1993). Evolutionary families of peptidases. Biochem. J. 290, 205–218. doi: 10.1042/bj2900205
Rawlings, N. D., and Barrett, A. J. (1995). Evolutionary families of metallopeptidases. Methods Enzymol. 248, 183–228. doi: 10.1016/0076-6879(95)48015-3
Rawlings, N. D., Barrett, A. J., and Bateman, A. (2011). Asparagine peptide Lyases: a seventh catalytic type of proteolytic enzymes. J. Biol. Chem. 286, 38321–38328. doi: 10.1074/jbc.M111.260026
Rawlings, N. D., Barrett, A. J., Thomas, P. D., Huang, X., Bateman, A., and Finn, R. D. (2018). The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Res. 46, D624–D632. doi: 10.1093/nar/gkx1134
Rawlings, N. D., and Bateman, A. (2019). Origins of peptidases. Biochimie 166, 4–18. doi: 10.1016/j.biochi.2019.07.026
Razzaq, A., Shamsi, S., Ali, A., Ali, Q., Sajjad, M., Malik, A., et al. (2019). Microbial proteases applications. Front. Bioeng. Biotechnol. 7, 1–20. doi: 10.3389/fbioe.2019.00110
Rodríguez, K. L., Higuera, B. L., and Martínez, S. T. (2017). Induction of proteases secreted by fusarium oxysporum f. sp. Dianthi in the presence of carnation root cell walls. Biochemical characterization of a serine protease. J. Plant Pathol. 99, 609–617.
Rukmi, I., and Purwantisari, S. (2020). The production of alkaline protease from aspergillus flavus DUCC K225 on rice bran containing medium. J. Phys. Conf. Ser. 1524:012058. doi: 10.1088/1742-6596/1524/1/012058
Savitha, S., Sadhasivam, S., Swaminathan, K., and Lin, F. H. (2011). Fungal protease: production, purification and compatibility with laundry detergents and their wash performance. J. Taiwan Inst. Chem. Eng. 42, 298–304. doi: 10.1016/j.jtice.2010.05.012
Seemüller, E., Lupas, A., Stock, D., Löwe, J., Huber, R., and Baumeister, W. (1995). Proteasome from Thermoplasma acidophilum: a threonine protease. Science 268, 579–582. doi: 10.1126/science.7725107
Shankar, S., More, S. V., and Laxman, R. S. (2010). Recovery of silver from waste X–ray film by alkaline protease from Conidiobolus coronatus. J. Sci. Eng. Technol. 6, 60–69. doi: 10.3126/kuset.v6i1.3311
Shankar, S., Rao, M., and Laxman, R. S. (2011). Purification and characterization of an alkaline protease by a new strain of Beauveria sp. Process Biochem. 46, 579–585. doi: 10.1016/j.procbio.2010.10.013
Sharma, N. (2019). A review on fungal alkaline protease. J. Emerg. Technol. Innov. Res. 6, 261–273. doi: 10.1729/Journal.22354
Sharma, M., Gat, Y., Arya, S., Kumar, V., Panghal, A., and Kumar, A. (2019). A review on microbial alkaline protease: an essential tool for various industrial approaches. Ind. Biotechnol. 15, 69–78. doi: 10.1089/ind.2018.0032
Sharma, M., Sharma, M., and Rao, V. M. (2011). In vitro biodegradation of keratin by dermatophytes and some soil keratinophiles. Afr. J. Biochem. Res. 5, 1–6.
Sindhu, R., Suprabha, G. N., and Shashidhar, S. (2009). Optimization of process parameters for the production of alkaline protease from Penicillium godlewskii SBSS 25 and its application in detergent industry. Afr. J. Microbiol. Res. 3, 515–522.
Souza, P. M., Bittencourt, M. L., Caprara, C. C., Freitas, M. D., Almeida, R. P., Silveira, D., et al. (2015). A biotechnology perspective of fungal proteases. Braz. J. Microbiol. 46, 337–346. doi: 10.1590/S1517-838246220140359
Suresh, A. J., and Dass, R. S. (2022). “Cold-adapted fungi: evaluation and comparison of their habitats, molecular adaptations and industrial applications” in Survival Strategies in Cold-Adapted Microorganisms (Singapore: Springer), 31–61.
Suryawanshi, H. K., and Pandya, N. D. (2017). Screening, identification of alkaline proteases producing fungi from soil of different habitats of Amalner Tahsil [Maharashtra] and their applications. Int. J. Appl. Sci. Biotechnol. 5, 397–402. doi: 10.3126/ijasbt.v5i3.18304
Sutar, I. I., Srinivasan, M. C., and Vartak, H. G. (1992). Production of an alkaline proteinase from Conidiobolus coronatus and its use to resolve DL–phenylalanine and DL–phenylglycine. World J. Microbiol. Biotechnol. 8, 254–258. doi: 10.1007/BF01201873
Tremacoldi, C. R., and Carmona, E. C. (2005). Production of extracellular alkaline proteases by aspergillus clavatus. World J. Microbiol. Biotechnol. 21, 169–172. doi: 10.1007/s11274-004-2724-0
Tunga, R., Shrivastava, B., and Banerjee, R. (2003). Purification and characterization of a protease from solid state cultures of Aspergillus parasiticus. Process Biochem. 38, 1553–1558. doi: 10.1016/S0032-9592(03)00048-7
Ueda, M., Kubo, T., Miyatake, K., and Nakamura, T. (2007). Purification and characterization of fibrinolytic alkaline protease from fusarium sp BLB. Appl. Microbiol. Biotechnol. 74, 331–338. doi: 10.1007/s00253-006-0621-1
Verma, S., Dixit, R., and Pandey, K. C. (2016). Cysteine proteases: modes of activation and future prospects as pharmacological targets. Front. Pharmacol. 7, 1–12. doi: 10.3389/fphar.2016.00107
Xiao, Y. Z., Wu, D. K., Zhao, S. Y., Lin, W. M., and Gao, X. Y. (2015). Statistical optimization of alkaline protease production from Penicillium citrinum YL–1 under solid–state fermentation. Prep. Biochem. Biotechnol. 45, 447–462. doi: 10.1080/10826068.2014.923450
Zanphorlin, L. M., Facchini, F. D., Vasconcelos, F., Bonugli-Santos, R. C., Rodrigues, A., and Sette, L. D. (2010). Production, partial characterization, and immobilization in alginate beads of an alkaline protease from a new thermophilic fungus Myceliophthora sp. J. Microbiol. 48, 331–336. doi: 10.1007/s12275-010-9269-8
Keywords: alkaline proteases, classification, fermentation, alkaliphilic fungi, pH
Citation: Pawar KS, Singh PN and Singh SK (2023) Fungal alkaline proteases and their potential applications in different industries. Front. Microbiol. 14:1138401. doi: 10.3389/fmicb.2023.1138401
Received: 05 January 2023; Accepted: 09 March 2023;
Published: 30 March 2023.
Edited by:
Tarun Belwal, Zhejiang University, ChinaReviewed by:
Debdulal Banerjee, Vidyasagar University, IndiaCopyright © 2023 Pawar, Singh and Singh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sanjay Kumar Singh, c2tzaW5naEBhcmlwdW5lLm9yZw==; Paras Nath Singh, cG5zaW5naEBhcmlwdW5lLm9yZw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.