- 1Department of Pharmacy, Shanghai Tenth People's Hospital, Tongji University School of Medicine, Shanghai, China
- 2Department of Physiology and Pharmacology, School of Basic Medicine and Clinical Pharmacy, China Pharmaceutical University, Nanjing, China
- 3Department of Vascular Diseases, Shanghai TCM-Integrated Hospital, Shanghai University of Traditional Chinese Medicine, Shanghai, China
Candida parapsilosis is an emerging major human fungal pathogen. Echinocandins are first-line antifungal drugs for the treatment of invasive Candida infections. In clinical isolates, tolerance to echinocandins in Candida species is mostly due to point mutations of FKS genes, which encode the target protein of echinocandins. However, here, we found chromosome 5 trisomy was the major mechanism of adaptation to the echinocandin drug caspofungin, and FKS mutations were rare events. Chromosome 5 trisomy conferred tolerance to echinocandin drugs caspofungin and micafungin and cross-tolerance to 5-flucytosine, another class of antifungal drugs. The inherent instability of aneuploidy caused unstable drug tolerance. Tolerance to echinocandins might be due to increased copy number and expression of CHS7, which encodes chitin synthase. Although copy number of chitinase genes CHT3 and CHT4 was also increased to the trisomic level, the expression was buffered to the disomic level. Tolerance to 5-flucytosine might be due to the decreased expression of FUR1. Therefore, the pleiotropic effect of aneuploidy on antifungal tolerance was due to the simultaneous regulation of genes on the aneuploid chromosome and genes on euploid chromosomes. In summary, aneuploidy provides a rapid and reversible mechanism of drug tolerance and cross-tolerance in C. parapsilosis.
Introduction
In recent years, due to HIV infection, solid organ and stem cell transplantation, intensified chemotherapy, and immunosuppression for autoimmune diseases, the at-risk immunocompromised population is steadily increasing. Opportunistic fungal infections are emerging as an important public health concern and cost burden (Benedict et al., 2019). Candida, Aspergillus, and Mucor species are the most frequently isolated fungi in immunocompromised patients (Badiee and Hashemizadeh, 2014). Candida infections are most often caused by C. albicans (Pappas et al., 2018), but non-albicans Candida (NAC) species, such as C. glabrata, C. parapsilosis, C. tropicalis, and C. krusei, are also increasingly reported as a cause of superficial infections as well as disseminated and deep-tissue infections (Whaley et al., 2016). However, currently, there are only four classes of antifungal drugs: polyenes, azoles, 5-flucytosine, and echinocandins. Echinocandins are the first-line drugs for the treatment of bloodstream Candida infections (Pappas et al., 2016).
In yeasts, the target protein of echinocandins is encoded by FKS genes. Mutations of FKS genes, especially in the hot spot regions, are the major cause of resistance to echinocandins [reviewed in Perlin (2015)]. In C. parapsilosis, caspofungin (CSP)-resistant clinical isolates (Chen et al., 2010; Pfeiffer et al., 2010; Pfaller et al., 2011; Siopi et al., 2022) and mutants obtained during in vitro evolution (Chassot et al., 2016; Papp et al., 2018; Arastehfar et al., 2021) also usually bear mutations in hot spot regions of FKS genes. But in some isolates, resistance is due to mutations outside of the hot spot regions or there are no mutations in FKS genes (Berrio et al., 2018). Furthermore, mutations outside of the hot spot regions of FKS genes have been associated with increased tolerance, but not resistance to echinocandins (Daneshnia et al., 2022).
5-flucytosine (5FC) is a prodrug. It enters fungal cells via the cytosine permease. Then, it is converted into toxic 5-fluorouracil (5FU) by cytosine deaminase. 5FU is further processed by uracil phosphoribosyltransferase, and the product inhibits both DNA and protein synthesis (Vermes et al., 2000). In the C. parapsilosis genome, cytosine permease, cytosine deaminase, and uracil phosphoribosyltransferase are encoded by FCY2/CPAR2_806580, FCA1/CPAR2_602820, and FUR1/CPAR2_502030, respectively. 5FC is active against Candida species and Cryptococcus species, but monotherapy of 5FC usually results in the rapid development of resistance, and the resistance is usually due to loss-of-function mutations of genes involved in the uptake and intracellular metabolism of 5FC (Whelan, 1987; Hope et al., 2004; Papon et al., 2007; Billmyre et al., 2020; Chang et al., 2021). C. parapsilosis clinical isolates are generally susceptible to 5FC (Barchiesi et al., 2000; Cuenca-Estrella et al., 2001). The report of 5FC resistance in C. parapsilosis is very limited, possibly because the monotherapy of 5FC against C. parapsilosis infections is not recommended in the clinic.
In addition to genetic mutations, aneuploidy, which is a cellular state of unbalanced chromosome copy number, is considered a prevalent strategy of rapid adaptation to stresses including antifungal agents in fungal pathogens [reviewed in Tsai and Nelliat (2019)]. For example, in C. albicans, different stresses can select the same aneuploidy, thereby causing cross-adaptation (Yang et al., 2013, 2017, 2019, 2021b). Furthermore, in addition to the direct regulation of genes on the aneuploid chromosome (Kabir et al., 2005; Suwunnakorn et al., 2016; Yang et al., 2019, 2021b), aneuploidy also indirectly regulates genes on euploid chromosomes. For example, the copy number of Chr5 negatively regulates the expression of SOU1 on Chr4. SOU1 encodes a sorbose reductase required for L-sorbose utilization (Greenberg et al., 2005). Therefore, in the Chr5x1 strain, SOU1 is upregulated, thereby enabling C. albicans to utilize L-sorbose as the sole carbon source (Janbon et al., 1998). However, in C. parapsilosis, reports of aneuploidy formation are very limited. Previously we found aneuploidy was the predominant mechanism of adaptation to ER stress inducer tunicamycin and sphingolipid biosynthesis inhibitor aureobasidin A in C. parapsilosis. Furthermore, both stresses selected Chr6x3 adaptors, and Chr6x3 conferred cross-tolerance to tunicamycin and aureobasidin A (Yang et al., 2021c). The role of aneuploidy in the adaptation of C. parapsilosis to commonly used antifungal drugs is still largely unknown.
Antifungal resistance is usually defined as “the ability to grow at antifungal drug concentrations above a defined antifungal susceptibility breakpoint”. The extent of resistance can be measured by broth microdilution assay, which determines the minimal inhibitory concentration (MIC) of antifungals that inhibits fungal growth (Fisher et al., 2022). Disk diffusion assay (DDA) is an official method and is one of the most widely used methods in many clinical microbiology laboratories for routine antimicrobial sensitivity testing. According to the standards published by the Clinical and Laboratory Standards Institute (CLSI), in a DDA experiment, agar plates are inoculated with a standardized number of cells. A paper disk containing the test chemical is placed on the surface of the medium. After the incubation of the plates under suitable conditions, the diameters of the zone of inhibition (ZOI) are measured (CLSI, 2012). This method is fast, simple, and low cost, and the results are easy to interpret. However, growth inside the ZOI is not considered.
Recently, Judith Berman lab developed a new method to analyze and interpret the DDA results. They performed DDAs using disks containing fluconazole, a fungistatic drug against C. albicans. They defined the growth of subpopulations of C. albicans cells inside the ZOI as “antifungal tolerance” (Rosenberg et al., 2018). By definition, antifungal tolerance is “a characteristic of drug-susceptible genotypes to grow slowly at or above inhibitory drug concentrations” (Berman and Krysan, 2020; Fisher et al., 2022). A pipeline called diskImageR was developed to analyze DDA data (Gerstein et al., 2016; Rosenberg et al., 2018; Xu et al., 2021). diskImageR quantifies the radius (RAD) of the ZOI as a parameter that relates to the MIC, and the fraction of growth (FoG) within the ZOI as a parameter that measures tolerance. RAD20, the RAD value corresponding to the point where 20% growth reduction occurs, and FoG20, the area under the curve at the RAD threshold, divided by the maximum growth, were usually used as measurements of resistance and tolerance, respectively (Gerstein et al., 2016; Rosenberg et al., 2018; Xu et al., 2021). However, for fungicidal drugs, such as CSP, growth in the presence of the drug without change in RAD or FoG is also considered tolerance (Berman and Krysan, 2020).
In this study, we investigated how C. parapsilosis adapted to CSP and the impact of adaptation on tolerance to other antifungal drugs. We randomly analyzed 30 adaptors and we got mainly chromosome 5 trisomy (Chr5x3, n = 29) and occasionally chromosome 1 trisomy (Chr1x3, n = 1). Genetic mutation of FKS genes was not detected. Chr5x3 conferred tolerance, not resistance, to echinocandin drugs CSP and micafungin (MCF), as well as cross-tolerance to 5FC, but it also caused hypersensitivity to fluconazole (FLC). The Chr5x3 adaptor was unstable. It spontaneously reverted to chromosome 5 disomy (Chr5x2), and tolerance to echinocandins and 5FC was concomitantly lost. Aneuploidy simultaneously upregulated the expression of genes on the aneuploid chromosome and genes on other chromosomes, including genes associated with tolerance to echinocandins, 5FC and FLC. Further exposure of one Chr1x3 adaptor to CSP also selected mostly Chr5x3 adaptors, thereby causing cross-tolerance to echinocandins and 5FC. Therefore, we posit that Chr5x3 provides a rapid and reversible strategy of adaptation to CSP and cross-adaptation to 5FC in C. parapsilosis.
Materials and methods
Strains and growth conditions
The strains used in this study are listed in Supplementary Table 1. C. parapsilosis clinical isolate #12108 was used as the wild-type strain. The stock culture was preserved in 25% of glycerol and maintained at −80°C. Cells were routinely grown in the yeast extract–peptone–dextrose (YPD) media (1% [w/v] yeast extract, 2% [w/v] peptone, and 2% [w/v] D-glucose) at 37°C in a shaking incubator at 150–200 rpm. For solid medium, 2% [w/v] agar was added. SD agar plates (0.67% [wt/vol] yeast nitrogen base without amino acids, 2% [wt/vol] D-glucose, and 2% [wt/vol] agar) were used for testing tolerance to 5FC. Drugs were dissolved in dimethyl sulfoxide (DMSO) and stored at −20°C. The concentrations of echinocandins, 5FC, and FLC were 10, 0.5, and 40 mg/ml, respectively.
Obtaining caspofungin adaptors
Cells were suspended in distilled water. Cell density was determined by using a hemocytometer and was adjusted to 1 × 107 cells/ml. A total of 100 μl of cell suspension were spread on YPD plates supplemented with 100, 200, and 400 ng/ml of CSP. On day 5, only ~368 colonies (adaptors) appeared on the plate with 400 ng/ml of CSP, while on other plates, we saw lawn growth. A total of 30 adaptors (TJ60–TJ89) were randomly chosen from the plate with 400 ng/ml of CSP. Each adaptor was streaked from the drug plate onto the YPD plate. The plates were incubated at 37°C for 3 days. Four to six colonies with similar sizes were selected and frozen in 1 ml of 25% glycerol at −80°C.
Spot assay
Cells were suspended in distilled water and adjusted to 1 × 107 cells/ml. A total of 3 μl of 10-fold serial dilutions were spotted on YPD or SD plates with or without drugs (control) at 37°C and photographed after 3 days.
Growth curves
Cells were suspended in YPD broth. Cell densities were adjusted to 2.5 × 103 cells/ml in YPD broth with or without test drugs in a 96-well plate. The plate was incubated at 37°C. OD595 was monitored in a Tecan plate reader (Infinite F200 PRO, Tecan, Switzerland) at 15 min time intervals for 48 h. Data are represented as the mean ± SD of three biological replicates.
Colony instability assay
As described previously (Yang et al., 2021d), Chr1x3 and Chr5x3 adaptors were streaked from−80°C freezer to YPD agar and incubated at 37°C for 72 h. One small colony was randomly chosen and suspended in distilled water. Cells were diluted with distilled water and ~200 cells were spread on a YPD plate and incubated at 37°C for 72 h. One small (S) colony and one large (L) colony were randomly chosen for further studies.
Disk diffusion assays
The CLSI M44-A2 guidelines (CLSI, 2012) for the antifungal disk diffusion susceptibility testing were followed with slight modifications. Strains were grown on agar plates, and cell density was adjusted to 1 × 106 cells/ml as described earlier. A total of 100 μl of cell suspension was plated on plates. One paper disk (GE Healthcare, USA) was placed in the center of each plate. The plates were then incubated for 72 h and photographed.
DNA-seq
The test strains were grown on YPD plates at 37°C at a density of ~200 colonies per plate. Colonies were collected by centrifugation in a microfuge at 3,000 rpm for 1 min. Genomic DNA was extracted manually using the phenol–chloroform method (Selmecki et al., 2015). The genomic DNA library was prepared by BGI (Wuhan, China) according to their standard preparation protocol. Approximately 1 μg of genomic DNA was randomly fragmented with a Covaris LE220. Fragments (300–400 bp) were selected, end-repaired, and 3' adenylated using the Agencourt AMPure XP-Medium kit, then ligated to adaptors. The ligation products were amplified by PCR. After purification, the PCR products were heat denatured, then circularized with a splint oligo sequence. The single-strand circular DNA (ssCirDNA) was formatted as the final library, qualified by QC, and then sequenced by BGISEQ-500. ssCir DNA molecules formed a DNA nanoball (DNB) containing more than 300 copies through rolling-cycle replication. The DNBs were loaded into a patterned nano array by using high-density DNA nanochip technology and were sequenced on the BGISEQ-500 platform using BGISEQ-500 high-throughput sequencing kit (PE100). Finally, pair-end 100 bp reads were obtained by combinational probe-anchor synthesis (cPAS). Raw FASTQ files were uploaded to YMAP (version 1.0) (http://lovelace.cs.umn.edu/Ymap/) (Abbey et al., 2014). Read depth was plotted as a function of chromosome position using the CDC317 reference genome of C. parapsilosis (http://www.candidagenome.org/download/sequence/C_parapsilosis_CDC317/current/). Contig005504, contig005569, contig005806, contig005807, contig005809, contig006110, contig006139, and contig006372 were renamed to chromosome 1, chromosome 2, chromosome 3, chromosome 4, chromosome 5, chromosome 6, chromosome 7, and chromosome 8, respectively. Chromosome end bias and GC content bias were corrected by YMAP (Abbey et al., 2014).
RNA-seq
RNA-seq was performed as described previously (Yang et al., 2021b). Strains were streaked onto YPD plates from the −80°C freezer. After 72 h of incubation at 37°C, several colonies of similar sizes were chosen. Colonies were suspended in distilled water and adjusted to 1 × 104 cells/ml. A total of 100 μl of cell suspension were spread on YPD plates. The plates were incubated at 37°C for 72 h. Cells were collected by centrifugation, washed, and flash-frozen in liquid nitrogen. The total RNA was extracted for nine independent samples, corresponding to three conditions and three biological replicates. Total RNA extraction and purification, library construction, and sequencing were performed as described in Yang et al. (2013). Raw sequence files (.fastq files) underwent quality control analysis using the FastQC tool (http://www.bioinformatics.babraham.ac.uk/projects/fastqc). Reads were mapped to the C. parapsilosis CDC317 reference genome (http://www.candidagenome.org/download/sequence/C_parapsilosis_CDC317/current/). Differential gene expression profiling was carried out using DESeq2 (Love et al., 2014) with standard parameters. Genes with false discovery rate (FDR)-adjusted P-value (<0.05) and expression fold changes of more than 1.3 or <-1.3 were considered differentially expressed.
Reverse transcriptase PCR (RT-PCR)
Cells were grown under the same experimental conditions as in RNA-seq. RT-qPCR was performed in 96-well plates (Bio-Rad) on the CFX Touch 96-well Real-Time Systems (Bio-Rad). Primer sequences are listed in Supplementary Table 2. The reaction mix was performed using 5 μl of iTaq Universal SYBR Green Supermix (Bio-Rad), 2 μl of 2 μM primer mix, 2 μl of a diluted 1:10 cDNA, and water to make up the final volume to 10 μl. Cycling conditions were 95 °C for 3 min, 40 cycles of 95 °C for 5 s, and 60 °C for 30 min. Melt curve analysis conditions were 5 s at 95°C and then 5 s each at 0.5°C increments between 60°C and 95°C. ACT1 (CPAR2_201570) was the internal control. Fold change was calculated using the 2−ΔΔCt method (Livak and Schmittgen, 2001). All RT-PCR experiments were performed using three biological and three technical replicates.
Statistical analysis
The significance of differences between growth curves was performed using Tukey's honest significant difference (HSD) test.
Results
Aneuploidy enables adaptation to the lethal amount of caspofungin in C. parapsilosis
In order to obtain CSP adaptors, ~1 million cells of C. parapsilosis clinical isolate #12108 were spread on the YPD plate supplemented with 400 ng/ml of CSP. After 5 days of incubation at 37°C, ~368 colonies (adaptors) were obviously visible on the plate (Figure 1A). We randomly picked up 30 adaptors (TJ60–TJ89, Supplementary Figure 1) with different colony sizes on the drug plate. Whole genome sequencing indicated that all the adaptors were aneuploid: 29 had trisomy of chromosome 5 (Chr5x3), and 1 (TJ74) had trisomy of chromosome 1 (Chr1x3) (Figure 1B). Of note, there are three FKS genes in C. parapsilosis genome: GSC1/CPAR2_106400, GSL1/CPAR2_109680, and GSL2/CPAR2_804030. Sequences of these three FKS genes were visualized in Integrative Genomics Viewer (IGV), and we did not detect any mutation in the 30 adaptors. GSC1 and GSL1 are on Chr2, and GSL2 is on Chr4, but none of the adaptors had aneuploidy of Chr2 or Chr4. Therefore, Chr5x3 was the major mechanism of adaptation to CSP. Mutations of FKS genes and aneuploidy of chromosomes on which the FKS genes reside were not detected.
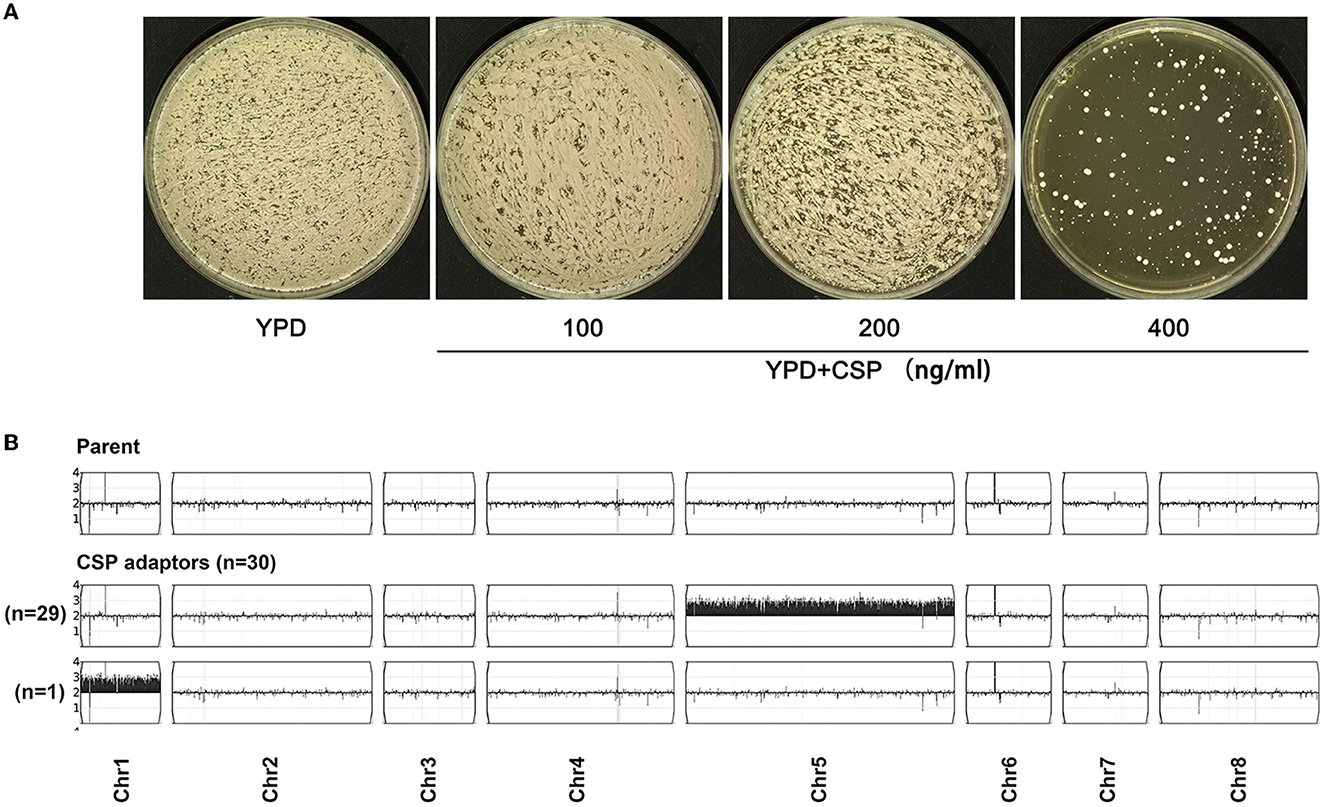
Figure 1. In C. parapsilosis, caspofungin adaptors are aneuploid. Approximately 1 million cells of C. parapsilosis strain #12108 were spread on YPD plates with or without caspofungin (CSP). The plates were incubated at 37°C for 5 days (A). On the plate with 400 ng/ml of CSP, randomly 30 adaptors were chosen and sequenced. The karyotypes were visualized using Ymap (B). Read depth (normalized to that of the diploid parent) is shown on the y-axis on a log2 scale converted to absolute copy numbers (1–4). The number of adaptors bearing the same aneuploidy was also shown in the figure.
Fitness loss and gain in caspofungin adaptors
It is known that in Saccharomyces cerevisiae and C. albicans, aneuploids usually have fitness loss in rich medium in the absence of stress (Pavelka et al., 2010; Yang et al., 2021d). In C. albicans, gain-of-function mutations of genes associated with resistance to azoles also have fitness costs in vitro and in vivo (Sasse et al., 2012; Hill et al., 2015; Popp et al., 2017). We asked if the aneuploid CSP adaptors also had fitness loss. Growth curves of the adaptors and the parent were measured in YPD broth (Figure 2A). The Chr5x3 adaptor TJ60 and the Chr1x3 adaptor TJ74 were significantly less fit than the parent (p < 0.0001 and p < 0.05, respectively. Tukey's HSD test).

Figure 2. Fitness loss and gain in caspofungin adaptors. The fitness cost of adaptation to caspofungin (CSP) was evaluated by growing strains in YPD broth without stress (A). Optical density at 595 nm (OD595) was measured every 1 h for 48 h at 37°C using a Tecan plate reader (Infinite F200 PRO, Tecan, Switzerland). Data are represented as the mean ± SD of three biological repeats. * and **** indicate p-values < 0.05, and p < 0.0001, respectively, as determined by Tukey's HSD test. In (B), disk diffusion assays were performed to compare Chr1x3 and Chr5x3 adaptors to parent for resistance to CSP and micafungin (MCF). The paper disks contained 50 μg of CSP or MCF as indicated in the figure. In (C), 3 μl of 10-fold serial dilutions were spotted on YPD plates. Drug concentrations are shown in the figure. In (B) and (C), the plates were incubated at 37°C for 72 h and then photographed.
We investigated whether the adaptors gained resistance or tolerance to CSP. DDAs with disks containing CSP were performed. In DDAs, the radius of the zone of inhibition (RAD) is reverse proportional to MIC (Milici et al., 2007). Here, we found that none of the adaptors had an obvious change in RAD as compared to the parent (Figure 2B). diskImageR analysis of DDA pictures indicated the parent, the Chr1x3 adaptor, and the Chr5x1 adaptor had the same RAD value of 9.5 ± 0.7. When tested with another echinocandin drug micafungin (MCF), the parent and the adaptors had the same RAD value of 12.0 ± 0.0. Therefore, the CSP adaptors did not develop resistance to CSP or MCF (Figure 2B). This is consistent with the finding that none of the adaptors had mutations of FKS genes, since FKS mutations in clinical isolates of Candida spp. usually cause increased resistance to CSP (Balashov et al., 2006; Garcia-Effron et al., 2010; Imtiaz et al., 2012; Beyda et al., 2014; Marti-Carrizosa et al., 2015; Naicker et al., 2016; Szymankiewicz et al., 2021).
To confirm the development of CSP resistance and FKS mutations were rare events, we repeated this experiment by testing more adaptors. Approximately 1 million cells of #12108 were spread on YPD plates supplemented with 400 ng/ml of CSP. Randomly 192 were tested with DDAs and none of them had reduced RAD (data not shown).
In C. albicans, strains tolerant to azole drugs fluconazole (FLC) (Rosenberg et al., 2018) or ketoconazole (Xu et al., 2021) have increased FoG20 when tested by DDA with disks containing azoles, as indicated by colonies growing inside of the zone of inhibition. However, here we found none of the CSP adaptors had colonies growing inside of the zone of inhibition (Figure 2B). Therefore, the CSP adaptors do not have reduced RAD20 or increased FoG20.
Previously we found spot assay was a sensitive method to detect improved ability to grow in the presence of CSP in C. albicans (Yang et al., 2019). Here, we investigated whether spot assay could detect CSP tolerance in the adaptors. Spot assay indicated that the Chr5x3 adaptor (TJ60) grew better than the parent in the presence of CSP and MCF. Furthermore, spot assay indicated that all 29 Chr5x3 adaptors grew better than the parent in the presence of CSP and MCF (Supplementary Figure 1). However, the Chr1x3 adaptor (TJ74) did not grow better than the parent (Figure 2C). Thus, all CSP adaptors were aneuploid and had fitness cost in rich medium in the absence of stress; however, Chr5x3 enabled better fitness in the presence of CSP and MCF.
Trisomy of chromosome 5 causes cross-tolerance to 5-flucytosine
Previously we found aneuploidy caused cross-tolerance to unrelated stresses in C. albicans (Yang et al., 2013, 2019, 2021b), C. parapsilosis (Yang et al., 2021c), and Cryptococcus neoformans (Yang et al., 2021a). Here, we investigated whether the aneuploid CSP adaptors caused cross-tolerance to other antifungal drugs. One Chr5x3 adaptor (TJ60) and one Chr1x3 adaptor (TJ74) were compared to the parent. DDAs with disks containing FLC or 5FC were performed.
When tested with FLC, the parent, Chr5x3 adaptor and Chr1x3 adaptor had a RAD20 of 17.0 ± 0.0, 23.5 ± 0.7, and 18.5 ± 0.7, respectively (Figure 3A). Therefore, Chr5x3 caused hypersensitivity to FLC, and Chr1x3 caused slightly increased sensitivity to FLC. When tested with 5FC, the parent, Chr5x3 adaptor, and Chr1x3 adaptor did not show an obvious change in RAD. However, inside the zone of inhibition, the Chr5x3 adaptor had lawn growth, while the Chr1x3 adaptor and the parent exhibited clear zones (Figure 3A). We investigated to what extent Chr5x3 can tolerate 5FC. Spot assay indicated that the growth of both parent and Chr1x3 was completely inhibited by 0.25 μg/ml of 5FC on the SD plate, but Chr5x3 could tolerate up to 128 μg/ml of 5FC (Figure 3B). Therefore, Chr5x3 caused hypersensitivity to FLC and tolerance to 5FC.
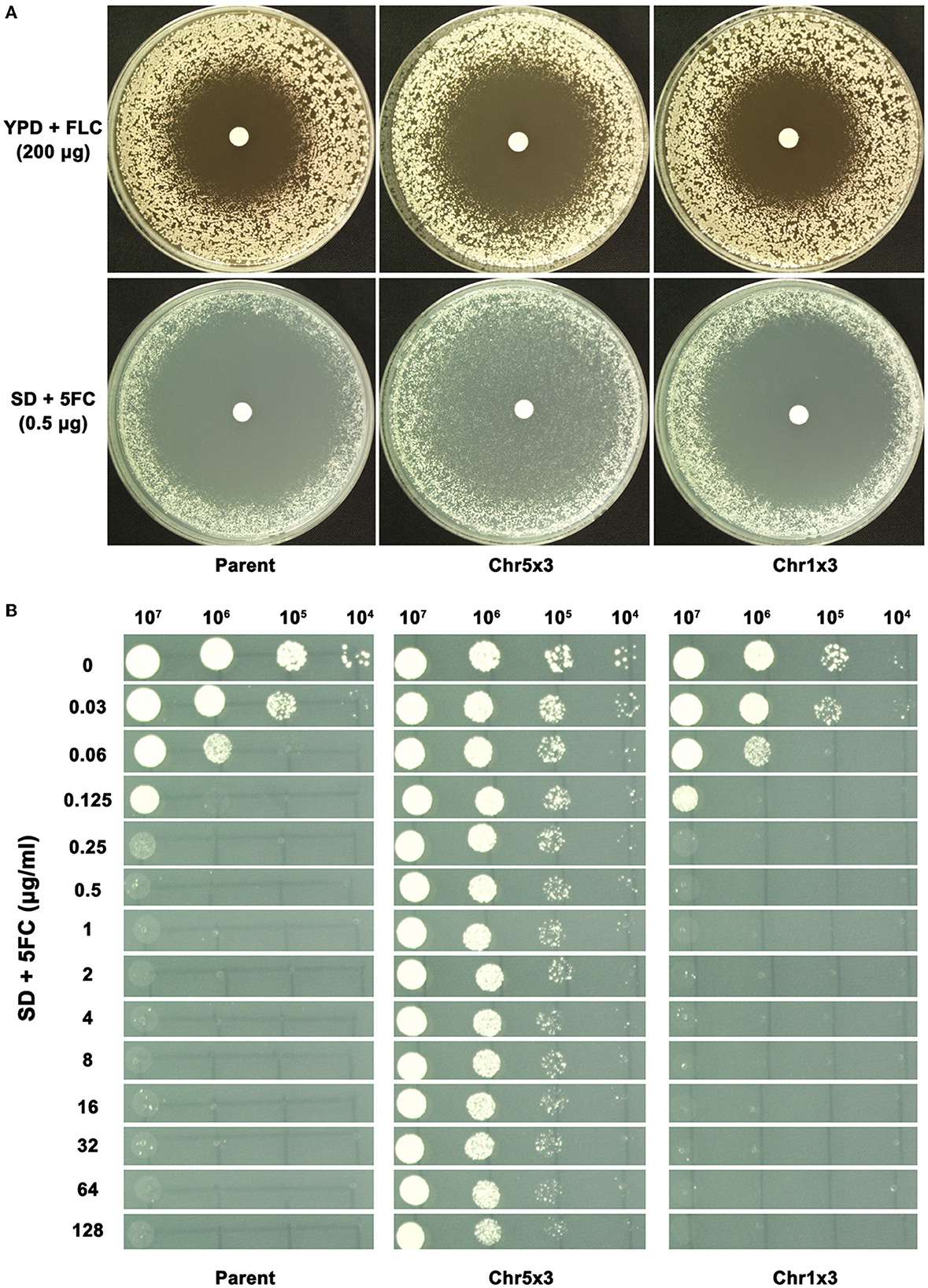
Figure 3. Chromosome 5 trisomy confers tolerance to 5-flucytosine. In (A), disk diffusion assays were performed to compare aneuploids and the parent for tolerance to fluconazole (FLC) and 5-flucytosine (5FC). YPD and SD plates were used for testing FLC and 5FC, respectively. In (B), spot assays were performed using a wide range of 5FC concentrations (0.03 μg/ml−128 μg/ml). A total of 3 μl of 10-fold serial dilutions were spotted on the plates. In (A) and (B), the plates were incubated at 37°C for 72 h and then photographed.
Cross-tolerance to caspofungin and 5-flucytosine is controlled by chromosome 5 copy number
We asked if aneuploids in C. parapsilosis were unstable. Approximately 200 cells of the Chr5x3 adaptor (TJ60) and Chr1x3 adaptor (TJ74) were spread on YPD plates. After incubation at 37°C for 48h, both adaptors exhibited colony instability: most colonies were small (indicated by cyan arrows in Figure 4A) and a few colonies were large (indicated by magenta arrows in Figure 4A). Whole-genome sequencing indicated that the large colonies were euploids (Figure 4B). Spot assay indicated that only the small colony of the Chr5x3 adaptor was tolerant to CSP, MCF, and 5FC. The large colony was not tolerant (Figure 4C). We investigated whether all 30 adaptors (TJ60–TJ89) were unstable. We found that all of them yielded small and large colonies on YPD plates (data not shown). Spot assay indicated that none of the large colonies was tolerant to CSP (Supplementary Figure 2). Therefore, in C. parapsilosis, aneuploids (Chr5x3 and Chr1x3) are unstable. Increased copy number of Chr5 causes tolerance to echinocandins and cross-tolerance to 5FC. Reversion of Chr5x3 to Chr5x2 is accompanied by the loss of tolerance and cross-tolerance to antifungal drugs.
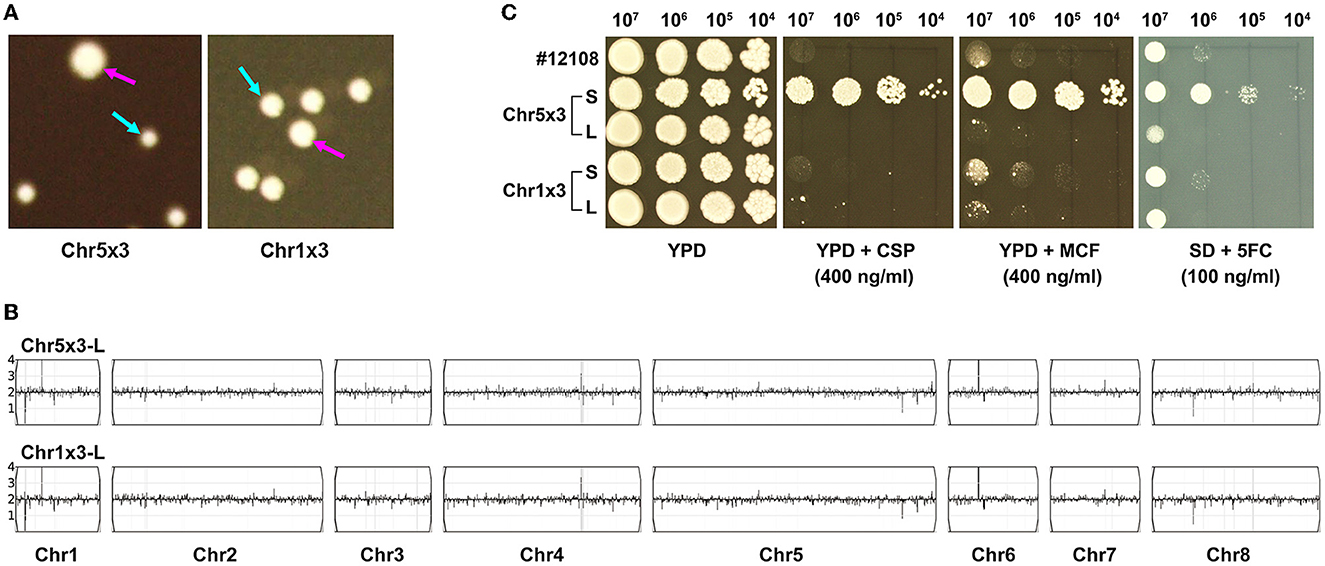
Figure 4. Genome and phenotype instability of aneuploids. Approximately 200 cells of aneuploid adaptors were spread on YPD plates. The plates were photographed after 72 h of incubation at 37°C. Cyan arrows indicate small (S) colonies and magenta arrows indicate large (L) colonies (A). The L colonies were sequenced, and the karyotypes were visualized using Ymap (B). For each adaptor, S and L colonies were compared for tolerance to caspofungin (CSP), micafungin (MCF), and 5-flucytosine (5FC) (C). Spot assays were performed using YPD plates supplemented with CSP or MCF, and the SD plate supplemented with 5FC. The plates were incubated at 37°C for 72 h and then photographed.
Aneuploidy causes the proportional change in transcriptomes
Since the CSP adaptors do not have FKS mutations, and FKS genes are not on the aneuploid chromosome, we investigated the mechanism of drug tolerance by performing RNA-seq and we compared the transcriptomes of CSP adaptors to the parent. Among the 1412 ORFs on Chr5, 744 were differentially expressed genes (q < 0.05) in the Chr5x3 adaptor as compared to the parent. Only 14 of the 744 ORFs were downregulated. All the remaining ORFs (98.1%) were upregulated. Among the 375 ORFs on Chr1, 253 were differentially expressed genes (q < 0.05) in the Chr1x3 adaptors as compared to the parent. All of them (100%) were upregulated. Therefore, both Chr1x3 and Chr5x1 caused proportional elevated transcription of the genes on the aneuploid chromosomes (Figure 5A).
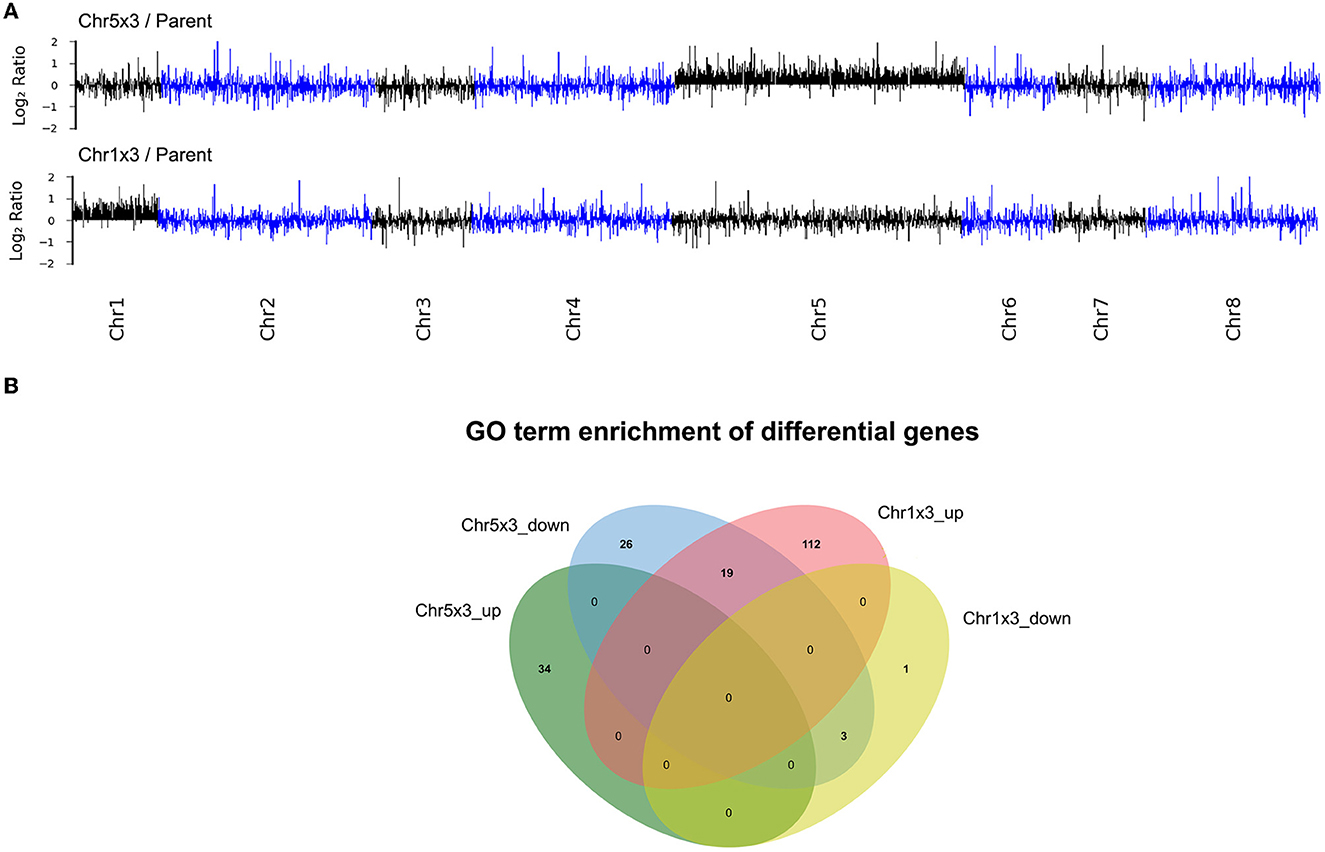
Figure 5. Transcriptional consequences of aneuploidy. Transcriptomes of Chr5x3 and Chr1x3 adaptors were compared to the diploid parent. Log2 ratios were plotted as a function of gene position on the chromosomes (A). Differential genes were analyzed for gene ontology (GO) enrichments. Lists of up and downregulated genes were analyzed separately. Venn diagram was used to illustrate the difference and commonality of the GO enrichments in genes upregulated in Chr1x3 (Chr1x3_up) and Chr5x3 (Chr5x3_up), as well as genes downregulated in Chr1x3 (Chr1x3_down) and Chr5x3 (Chr5x3_down) as compared to parent (B).
Among genes upregulated in the Chr5x3 adaptor, processes associated with peroxisome, DNA repair, cell cycle, and catabolism were significantly enriched. In contrast, among genes upregulated in the Chr1x3 adaptor, processes associated with biosynthesis including organic substances, cellular nitrogen compounds, and macromolecules were significantly enriched (Supplementary Table 3). Several processes were commonly downregulated in Chr5x3 and Chr1x3 adaptors, including protein folding (GOID: 0006457), mitochondrion organization (GODI: 0007005), and protein import into mitochondrial intermembrane space (GOID: 0045041). Some GO terms were significantly enriched in Chr5x3 downregulated genes and enriched in Chr1x3 upregulated genes, including ribosome biogenesis (GOID: 0042254), ribosome localization (GOID: 0033750), ribosomal subunit export from the nucleus (GOID: 0000054), peptide biosynthetic process (GOID:0043043), and translation (GOID: 0006412) (Figure 5B).
We investigated whether genes associated with β-1,3-glucan synthesis and chitin synthesis and degradation were differentially regulated in the Chr5x3 adaptor TJ60. None of the FKS genes (GSC1, GSL1, and GSL2) was upregulated in the Chr5x3 adaptor. Among the genes encoding chitin synthase (CHS1/CPAR2_800050, CHS2/CPAR2_701490, CHS3/CPAR2_801800, CHS4/CPAR2_807030, CHS5/CPAR2_210990, CHS7/CPAR2_212710, and CHS8/CPAR2_502940), only one gene, CHS7 was significantly upregulated in the Chr5x3 adaptor. CHS7 is on Chr5. Among the genes encoding chitinases (CHT2/CPAR2_502140, CHT3/CPAR2_200660, and CHT4/CPAR2_211950), two genes, CHT3 and CHT4 are on Chr5 but their transcription was compensated to the disomic level (Supplementary Table 4). Therefore, we posit that Chr5x3 enables tolerance to CSP via increasing copy number and transcription of CHS7 and reducing the transcription of CHT3 and CHT4, thereby increasing chitin content in the cell wall.
We also investigated whether genes associated with 5FC tolerance were downregulated in the Chr5x3 adaptor. FUR1 was significantly downregulated in the Chr5x3 adaptor. FCA1 and FCY2 did not exhibit obvious change (Supplementary Table 4). FUR1 is on Chr7. We posit that Chr5x3 causes 5FC tolerance via indirectly downregulating the transcription of the FUR1 gene on the euploid chromosome.
Compared to parent, several ERG genes were downregulated in Chr5x3 adaptor, but not in Chr1x3 adaptor, including ERG3/CPAR2_105550, ERG5/CPAR2_703970, ERG8/CPAR2_400710, and ERG11/CPAR2_303740. ERG11 was upregulated in the Chr1x3 adaptor. ERG1/CPAR2_210480, ERG4/CPAR2_502980, and ERG13/CPAR2_701400 were upregulated in the Chr1x3 adaptor, but not in the Chr5x3 adaptor. ERG12/CPAR2_803530 was downregulated in both Chr1x3 and Chr5x3 adaptors. UPC2/CPAR2_207280 encodes a transcription factor that positively regulates ergosterol biosynthetic genes. In Chr5x3 but not in the Chr1x3 adaptor, UPC2 was upregulated. In addition, CDR1/CPAR2_405290, which encodes the drug efflux pump, was downregulated in the Chr5x3 adaptor, but not in the Chr1x3 adaptor, as compared to the parent (Supplementary Table 4). Taken together, we posit that Chr5x3 causes hypersensitivity to FLC via decreasing the expression of ERG genes and CDR1.
We visualized the genome sequencing data of the Chr5x3 adaptor and the parent in IGV, and we did not see mutations of genes ERG3, ERG11, or CDR1
The expression profile of genes in RNA-seq results was validated by reverse transcriptase PCR (RT-PCR). Six genes were tested, including CHS7, CHT3, CHT4, FUR1, ERG11, and CDR1 (Supplementary Figure 4).
Chr1x3 strain adapts to caspofungin mainly via amplification of Chr5
We investigated how a Chr1x3 strain would adapt to CSP. Approximately one million cells of adaptor TJ74 were spread on the YPD plate supplemented with 400 ng/ml of CSP. Randomly 30 adaptors (TJ2267–TJ2296) were chosen. Spot assay indicated that 22 of the 30 adaptors grew better than TJ74 and #12108 in the presence of CSP (Supplementary Figure 3). We sequenced all 22 tolerant adaptors. Based on the karyotypes, the tolerant adaptors were categorized into four classes: Class 1 adaptors (n = 17) had Chr5x3 alone. Class 2 adaptor (n = 1) had Chr5x3, segmental trisomy of Chr3 (SegChr3x3, from 0.43 Mb till the right telomere), and segmental trisomy of Chr8 (SegChr8x3, from left telomere till 0.87 Mb). Class 3 adaptor (n = 1) had Chr5x3, segmental trisomy of Chr1 (from 0.40 Mb till the right telomere), and segmental trisomy of Chr2 (SegChr2x3, from left telomere to 0.25 Mb). Class 4 adaptors (n = 3) had Chr1x3, SegChr3x3, and SegChr8x3 (Figure 6A). Of note, none of the 22 tolerant adaptors had FKS mutations. Although GSC1 and GSL1 are on Chr2, in the Class 3 adaptor, which had SegChr2x3, the amplified region of Chr2 did not encompass GSC1 and GSL1. Therefore, the Chr1x3 strain adapted to CSP mainly via losing Chr1x3 and gaining Chr5x3 (19 out of 22) or maintaining Chr1x3 but gaining SegChr3x3 and SegChr8x3. Genetic mutation or elevated copy number of FKS genes did not happen.
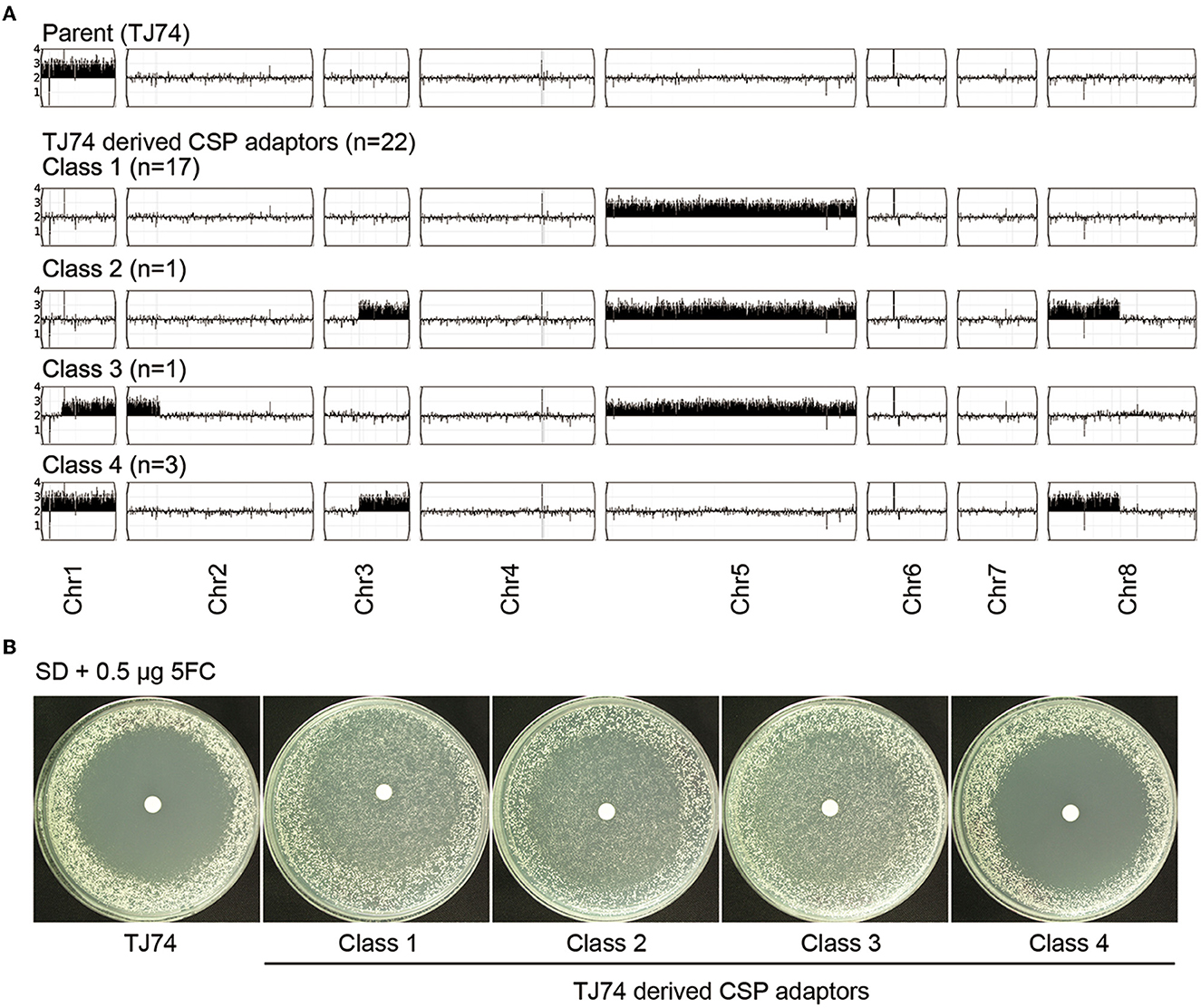
Figure 6. Adaptation of chromosome 1 trisomy strain to caspofungin. One chromosome 1 trisomy strain, TJ74, was spread on the YPD plate supplemented with 400 ng/ml caspofungin (CSP). Twenty-two out of 30 adaptors randomly chosen gained tolerance to CSP. All 22 tolerant adaptors were sequenced. Based on the pattern of aneuploidy, the adaptors were divided into four classes (A). Each class was tested for tolerance to 5-flucytosine (5FC) by disk diffusion assays. Only adaptors with Chr5x3 were tolerant to 5FC (B).
We enquired whether Chr1x3-derived CSP adaptors were cross-tolerant to 5FC. DDAs on SD plates with paper disks containing 5FC indicated that all Class 1, Class 2, and Class 3 adaptors were tolerant to 5FC and Class 4 adaptors were not (Figure 6B). Therefore, adaptors with Chr5x3 were cross-tolerant to CSP and 5FC. Adaptors without Chr5x3 were only tolerant to CSP but not tolerant to 5FC.
Discussion
In this study, we found that aneuploidy, Chr5x3 in particular, enabled rapid adaptation of both euploid and aneuploid C. parapsilosis strains to echinocandins and caused cross-tolerance to 5FC. Chr5x3 simultaneously upregulated the expression of genes on the aneuploid Chr5 and genes on euploid chromosomes. The large-scale effect of aneuploidy on whole-genome expression resulted in the pleiotropic effect on antifungal tolerance, including the development of cross-tolerance.
Of note, FKS mutations were not detected in the CSP adaptors, and FKS genes were not differentially expressed in the CSP adaptors as compared to the wild type. In C. albicans, increased expression of genes encoding chitin synthases or decreased expression of genes encoding chitinases causes tolerance to CSP (Walker et al., 2008; Suwunnakorn et al., 2016). Here we found that, among the genes encoding chitin synthases, CHS7 is on Chr5 and was significantly upregulated in the Chr5x3 adaptor compared to the parent. The increased expression of CHS7 might be due to increased copy number, since in yeast cells, aneuploidy usually caused the proportional change at the transcript level (Pavelka et al., 2010). Among the genes encoding chitinases, CHT3 and CHT4 are on Chr5, but the expressions of CHT3 and CHT4 in the Chr5x3 adaptor and in the parent were similar. Therefore, we posit that Chr5 confers CSP tolerance by upregulating the copy number and expression of CHS7 to the trisomic level and buffering the expression of CHT3 and CHT4 to the disomic level. It remains unclear how Chr5x3 selectively buffers the expression of CHT3 and CHT4, but not CHS7. Whether dosage compensation at the transcript level exists in yeast cells is still under debate (Hose et al., 2015; Torres et al., 2016).
Tolerance to 5FC in clinical isolates of Candida and Cryptococcus species is usually due to loss-of-function mutations of genes involved in the cellular uptake and intracellular metabolism of 5FC (Whelan, 1987; Hope et al., 2004; Papon et al., 2007; Billmyre et al., 2020; Chang et al., 2021). In C. albicans, the FUR1 gene is on Chr5. Previously we found decreased copy number of FUR1 via Chr5x1 conferred tolerance to 5FC (Yang et al., 2013). In C. parapsilosis, FUR1 is on Chr7. Here we found in C. parapsilosis that Chr5x3 downregulated the expression of FUR1. Thus, we posit on Chr5, there is a negative regulator of FUR1 on Chr7 in the C. parapsilosis genome, and Chr5x3 confers 5FC tolerance via downregulating the expression of FUR1.
The Chr5x3 adaptor was more susceptible to FLC, indicating that aneuploidy per se does not cause tolerance to antifungal agents. Altered ergosterol synthesis and drug efflux cause FLC tolerance in Candida spp. (Berkow and Lockhart, 2017). Here we found that several ERG genes were downregulated in the Chr5x3 adaptor, including ERG3, ERG5, ERG8, ERG11, and ERG12. In addition, CDR1, which encodes the drug efflux pump, was also downregulated.
Taken together, Chr5x3 enables cross-tolerance to CSP and 5FC and hypersensitivity to FLC via simultaneously regulating genes associated with antifungal tolerance on aneuploid chromosomes and on the remaining euploid chromosomes.
In fungi, aneuploids are inherently unstable. In C. albicans, C. auris, and Cryptococcus neoformans, drug-tolerant aneuploids spontaneously revert to euploids in the absence of a drug, and tolerance to drugs is concomitantly lost (Sionov et al., 2010; Yang et al., 2019; Bing et al., 2020). Here we found that aneuploid C. parapsilosis adaptors were also unstable. Single-time growth on the YPD plate was sufficient to induce the reversion of Chr1x3 and Chr5x3 adaptors to euploids. Cross-tolerance to CSP and 5FC in the Chr5x3 adaptor was concomitantly lost after it reverted to euploid. Therefore, in C. parapsilosis, aneuploidy is a reversible strategy of adaptation to stresses including antifungal drugs.
Conclusion
In summary, this study indicates that Chr5x3 is the major mechanism of rapid and reversible adaptation to CSP and cross-adaptation to 5FC in C. parapsilosis. The pleiotropic effect of aneuploidy on antifungal tolerance is via directly regulating genes on the aneuploidy chromosome and indirectly regulating genes on the euploid chromosomes. It will be interesting to investigate whether the formation of Chr5x3 happens in infected patients treated with CSP.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ebi.ac.uk/arrayexpress/, E-MTAB-12335; https://www.ebi.ac.uk/arrayexpress/, E-MTAB-12321; https://www.ebi.ac.uk/arrayexpress/, E-MTAB -12322.
Author contributions
FY and Y-yJ analyzed the data. FY wrote the manuscript. L-lS and HL carried out the research. T-hY and Y-bC helped to develop the experimental idea and design. Y-yJ and Y-bC funded the experiments. All authors approved the submitted version.
Funding
This manuscript was supported by the National Key Research and Development Program of China (2022YFC2303000), the National Natural Science Foundation of China (No. 82020108032), the Innovation Program of Shanghai Municipal Education Commission (No. 202101070007E00094) to Y-yJ, the National Natural Science Foundation of China (No. 81872910), and the Shanghai Key Basic Research Project (No. 19JC1414900) to Y-bC.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1137083/full#supplementary-material
References
Abbey, D. A., Funt, J., Lurie-Weinberger, M. N., Thompson, D. A., Regev, A., Myers, C. L., et al. (2014). YMAP: a pipeline for visualization of copy number variation and loss of heterozygosity in eukaryotic pathogens. Genome Med. 6, 100. doi: 10.1186/s13073-014-0100-8
Arastehfar, A., Daneshnia, F., Hilmioglu-Polat, S., Ilkit, M., Yasar, M., Polat, F., et al. (2021). Genetically related micafungin-resistant Candida parapsilosis blood isolates harbouring novel mutation R658G in hotspot 1 of Fks1p: a new challenge? J. Antimicrob. Chemother. 76, 418–422. doi: 10.1093/jac/dkaa419
Badiee, P., and Hashemizadeh, Z. (2014). Opportunistic invasive fungal infections: diagnosis and clinical management. Indian J. Med. Res. 139, 195–204.
Balashov, S. V., Park, S., and Perlin, D. S. (2006). Assessing resistance to the echinocandin antifungal drug caspofungin in Candida albicans by profiling mutations in FKS1. Antimicrob Agents Chemother. 50, 2058–2063. doi: 10.1128/AAC.01653-05
Barchiesi, F., Arzeni, D., Caselli, F., and Scalise, G. (2000). Primary resistance to flucytosine among clinical isolates of Candida spp. J. Antimicrob. Chemother. 45, 408–409. doi: 10.1093/jac/45.3.408
Benedict, K., Jackson, B. R., Chiller, T., and Beer, K. D. (2019). Estimation of direct healthcare costs of fungal diseases in the United States. Clin. Infect. Dis. 68, 1791–1797. doi: 10.1093/cid/ciy776
Berkow, E. L., and Lockhart, S. R. (2017). Fluconazole resistance in Candida species: a current perspective. Infect. Drug Resist. 10, 237–245. doi: 10.2147/IDR.S118892
Berman, J., and Krysan, D. J. (2020). Drug resistance and tolerance in fungi. Nat. Rev. Microbiol. 18, 319–331. doi: 10.1038/s41579-019-0322-2
Berrio, I., Maldonado, N., De Bedout, C., Arango, K., Cano, L. E., Valencia, Y., et al. (2018). Comparative study of Candida spp. isolates: identification and echinocandin susceptibility in isolates obtained from blood cultures in 15 hospitals in Medellín, Colombia. J. Glob. Antimicrob. Resist. 13, 254–260. doi: 10.1016/j.jgar.2017.11.010
Beyda, N. D., John, J., Kilic, A., Alam, M. J., Lasco, T. M., Garey, K. W., et al. (2014). FKS mutant Candida glabrata: risk factors and outcomes in patients with candidemia. Clin. Infect Dis. 59, 819–825. doi: 10.1093/cid/ciu407
Billmyre, R. B., Applen Clancey, S., Li, L. X., Doering, T. L., and Heitman, J. (2020). 5.-fluorocytosine resistance is associated with hypermutation and alterations in capsule biosynthesis in Cryptococcus. Nat. Commun. 11, 127. doi: 10.1038/s41467-019-13890-z
Bing, J., Hu, T., Zheng, Q., Muñoz, J. F., Cuomo, C. A., Huang, G., et al. (2020). Experimental evolution identifies adaptive aneuploidy as a mechanism of fluconazole resistance in Candida auris. Antimicrob Agents Chemother. 65, e01466-20. doi: 10.1128/AAC.01466-20
Chang, Y. C., Lamichhane, A. K., Cai, H., Walter, P. J., Bennett, J. E., Kwon-Chung, K. J., et al. (2021). Moderate levels of 5-fluorocytosine cause the emergence of high frequency resistance in cryptococci. Nat. Commun. 12, 3418. doi: 10.1038/s41467-021-23745-1
Chassot, F., Venturini, T. P., Piasentin, F. B., Rossato, L., Fiorini, A., Svidzinski, T. I., et al. (2016). Exploring the in vitro resistance of Candida parapsilosis to echinocandins. Mycopathologia 181, 663–670. doi: 10.1007/s11046-016-0028-1
Chen, Y. C., Lin, Y. H., Chen, K. W., Lii, J., Teng, H. J., Li, S. Y., et al. (2010). Molecular epidemiology and antifungal susceptibility of Candida parapsilosis sensu stricto, Candida orthopsilosis, and Candida metapsilosis in Taiwan. Diagn. Microbiol. Infect. Dis. 68, 284–292. doi: 10.1016/j.diagmicrobio.2010.07.004
CLSI (2012). “Performance Standards for Antimicrobial Disk Susceptibility Tests, Approved Standard, 7thed., CLSI document M02-A11”. (Clinical and Laboratory Standards Institute, 950 West Valley Road, Suite 2500, Wayne, Pennsylvania 19087, USA).
Cuenca-Estrella, M., Díaz-Guerra, T. M., Mellado, E., and Rodríguez-Tudela, J. L. (2001). Flucytosine primary resistance in Candida species and Cryptococcus neoformans. Eur. J. Clin. Microbiol. Infect. Dis. 20, 276–279. doi: 10.1007/PL00011265
Daneshnia, F., de Almeida Júnior, J. N., Arastehfar, A., Lombardi, L., Shor, E., Moreno, L., et al. (2022). Determinants of fluconazole resistance and echinocandin tolerance in C. parapsilosis isolates causing a large clonal candidemia outbreak among COVID-19 patients in a Brazilian ICU. Emerg. Microbes Infect. 11, 2264–2274. doi: 10.1080/22221751.2022.2117093
Fisher, M. C., Alastruey-Izquierdo, A., Berman, J., Bicanic, T., Bignell, E. M., Bowyer, P., et al. (2022). Tackling the emerging threat of antifungal resistance to human health. Nat. Rev. Microbiol. 20, 557–571. doi: 10.1038/s41579-022-00720-1
Garcia-Effron, G., Chua, D. J., Tomada, J. R., DiPersio, J., Perlin, D. S., Ghannoum, M., et al. (2010). Novel FKS mutations associated with echinocandin resistance in Candida species. Antimicrob Agents Chemother 54, 2225–2227. doi: 10.1128/AAC.00998-09
Gerstein, A. C., Rosenberg, A., Hecht, I., and Berman, J. (2016). diskImageR: quantification of resistance and tolerance to antimicrobial drugs using disk diffusion assays. Microbiology 162, 1059–1068. doi: 10.1099/mic.0.000295
Greenberg, J. R., Price, N. P., Oliver, R. P., Sherman, F., and Rustchenko, E. (2005). Candida albicans SOU1 encodes a sorbose reductase required for L-sorbose utilization. Yeast 22, 957–969. doi: 10.1002/yea.1282
Hill, J. A., O'Meara, T. R., and Cowen, L. E. (2015). Fitness trade-offs associated with the evolution of resistance to antifungal drug combinations. Cell Rep. 10, 809–819. doi: 10.1016/j.celrep.2015.01.009
Hope, W. W., Tabernero, L., Denning, D. W., and Anderson, M. J. (2004). Molecular mechanisms of primary resistance to flucytosine in Candida albicans. Antimicrob. Agents Chemother. 48, 4377–4386. doi: 10.1128/AAC.48.11.4377-4386.2004
Hose, J., Yong, C. M., Sardi, M., Wang, Z., Newton, M. A., Gasch, A. P., et al. (2015). Dosage compensation can buffer copy-number variation in wild yeast. Elife 4, e05462. doi: 10.7554/eLife.05462.023
Imtiaz, T., Lee, K. K., Munro, C. A., MacCallum, D. M., Shankland, G. S., Johnson, E. M., et al. (2012). Echinocandin resistance due to simultaneous FKS mutation and increased cell wall chitin in a Candida albicans bloodstream isolate following brief exposure to caspofungin. J. Med. Microbiol. 61, 1330–1334. doi: 10.1099/jmm.0.045047-0
Janbon, G., Sherman, F., and Rustchenko, E. (1998). Monosomy of a specific chromosome determines L-sorbose utilization: a novel regulatory mechanism in Candida albicans. Proc. Natl. Acad. Sci. USA. 95, 5150–5155. doi: 10.1073/pnas.95.9.5150
Kabir, M. A., Ahmad, A., Greenberg, J. R., Wang, Y. K., and Rustchenko, E. (2005). Loss and gain of chromosome 5 controls growth of Candida albicans on sorbose due to dispersed redundant negative regulators. Proc. Natl. Acad. Sci. USA. 102, 12147–12152. doi: 10.1073/pnas.0505625102
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. doi: 10.1186/s13059-014-0550-8
Marti-Carrizosa, M., Sanchez-Reus, F., March, F., Canton, E., and Coll, P. (2015). Implication of Candida parapsilosis FKS1 and FKS2 mutations in reduced echinocandin susceptibility. Antimicrob. Agents Chemother. 59, 3570–3573. doi: 10.1128/AAC.04922-14
Milici, M. E., Maida, C. M., Spreghini, E., Ravazzolo, B., Oliveri, S., Scalise, G., et al. (2007). Comparison between disk diffusion and microdilution methods for determining susceptibility of clinical fungal isolates to caspofungin. J. Clin. Microbiol. 45, 3529–3533. doi: 10.1128/JCM.00826-07
Naicker, S. D., Magobo, R. E., Zulu, T. G., Maphanga, T. G., Luthuli, N., Lowman, W., et al. (2016). Two echinocandin-resistant Candida glabrata FKS mutants from South Africa. Med. Mycol. Case Rep. 11, 24–26. doi: 10.1016/j.mmcr.2016.03.004
Papon, N., Noël, T., Florent, M., Gibot-Leclerc, S., Jean, D., Chastin, C., et al. (2007). Molecular mechanism of flucytosine resistance in Candida lusitaniae: contribution of the FCY2, FCY1, and FUR1 genes to 5-fluorouracil and fluconazole cross-resistance. Antimicrob Agents Chemother. 51, 369–371. doi: 10.1128/AAC.00824-06
Papp, C., Kocsis, K., Tóth, R., Bodai, L., Willis, J. R., Ksiezopolska, E., et al. (2018). Echinocandin-induced microevolution of candida parapsilosis influences virulence and abiotic stress tolerance. mSphere 3, e00547–18. doi: 10.1128/mSphere.00547-18
Pappas, P. G., Kauffman, C. A., Andes, D. R., Clancy, C. J., Marr, K. A., Ostrosky-Zeichner, L., et al. (2016). Clinical practice guideline for the management of candidiasis: 2016 update by the infectious diseases society of America. Clin. Infect. Dis. 62, e1–50. doi: 10.1093/cid/civ933
Pappas, P. G., Lionakis, M. S., Arendrup, M. C., Ostrosky-Zeichner, L., and Kullberg, B. J. (2018). Invasive candidiasis. Nat. Rev. Dis. Primers 4, 18026. doi: 10.1038/nrdp.2018.26
Pavelka, N., Rancati, G., Zhu, J., Bradford, W. D., Saraf, A., Florens, L., et al. (2010). Aneuploidy confers quantitative proteome changes and phenotypic variation in budding yeast. Nature 468, 321–325. doi: 10.1038/nature09529
Perlin, D. S. (2015). Mechanisms of echinocandin antifungal drug resistance. Ann. NY Acad. Sci. 1354, 1–11. doi: 10.1111/nyas.12831
Pfaller, M. A., Diekema, D. J., Andes, D., Arendrup, M. C., Brown, S. D., Lockhart, S. R., et al. (2011). Clinical breakpoints for the echinocandins and Candida revisited: integration of molecular, clinical, and microbiological data to arrive at species-specific interpretive criteria. Drug Resist. Updat. 14, 164–176. doi: 10.1016/j.drup.2011.01.004
Pfeiffer, C. D., Garcia-Effron, G., Zaas, A. K., Perfect, J. R., Perlin, D. S., Alexander, B. D., et al. (2010). Breakthrough invasive candidiasis in patients on micafungin. J. Clin. Microbiol. 48, 2373–2380. doi: 10.1128/JCM.02390-09
Popp, C., Hampe, I. A. I., Hertlein, T., Ohlsen, K., Rogers, P. D., Morschhauser, J., et al. (2017). Competitive fitness of fluconazole-resistant clinical Candida albicans strains. Antimicrob Agents Chemother. 61, e00584-17. doi: 10.1128/AAC.00584-17
Rosenberg, A., Ene, I. V., Bibi, M., Zakin, S., Segal, E. S., Ziv, N., et al. (2018). Antifungal tolerance is a subpopulation effect distinct from resistance and is associated with persistent candidemia. Nat. Commun. 9, 2470. doi: 10.1038/s41467-018-04926-x
Sasse, C., Dunkel, N., Schafer, T., Schneider, S., Dierolf, F., Ohlsen, K., et al. (2012). The stepwise acquisition of fluconazole resistance mutations causes a gradual loss of fitness in Candida albicans. Mol. Microbiol. 86, 539–556. doi: 10.1111/j.1365-2958.2012.08210.x
Selmecki, A. M., Maruvka, Y. E., Richmond, P. A., Guillet, M., Shoresh, N., Sorenson, A. L., et al. (2015). Polyploidy can drive rapid adaptation in yeast. Nature 519, 349–352. doi: 10.1038/nature14187
Sionov, E., Lee, H., Chang, Y. C., and Kwon-Chung, K. J. (2010). Cryptococcus neoformans overcomes stress of azole drugs by formation of disomy in specific multiple chromosomes. PLoS Pathog. 6, e1000848. doi: 10.1371/journal.ppat.1000848
Siopi, M., Papadopoulos, A., Spiliopoulou, A., Paliogianni, F., Abou-Chakra, N., Arendrup, M. C., et al. (2022). Pan-echinocandin resistant C. parapsilosis Harboring an F652S Fks1 Alteration in a patient with prolonged echinocandin therapy. J. Fungi. 8, 931. doi: 10.3390/jof8090931
Suwunnakorn, S., Wakabayashi, H., and Rustchenko, E. (2016). Chromosome 5 of human pathogen Candida albicans carries multiple genes for negative control of Caspofungin and Anidulafungin susceptibility. Antimicrob Agents Chemother. 60, 7457–7467. doi: 10.1128/AAC.01888-16
Szymankiewicz, M., Kamecki, K., Jarzynka, S., Koryszewska-Baginska, A., Oledzka, G., Nowikiewicz, T., et al. (2021). Case report: echinocandin-resistance Candida glabrata FKS mutants from patient following radical cystoprostatectomy due to muscle-invasive bladder cancer. Front. Oncol. 11, 794235. doi: 10.3389/fonc.2021.794235
Torres, E. M., Springer, M., and Amon, A. (2016). No current evidence for widespread dosage compensation in S. cerevisiae. Elife 5, e10996. doi: 10.7554/eLife.10996
Tsai, H. J., and Nelliat, A. (2019). A double-edged sword: aneuploidy is a prevalent strategy in fungal adaptation. Genes 10, 787. doi: 10.3390/genes10100787
Vermes, A., Guchelaar, H. J., and Dankert, J. (2000). Flucytosine: a review of its pharmacology, clinical indications, pharmacokinetics, toxicity and drug interactions. J. Antimicrob. Chemother. 46, 171–179. doi: 10.1093/jac/46.2.171
Walker, L. A., Munro, C. A., Bruijn, d. e., Lenardon, I., McKinnon, M. D. A., and Gow, N.A. (2008). Stimulation of chitin synthesis rescues Candida albicans from echinocandins. PLoS Pathog. 4, e1000040. doi: 10.1371/journal.ppat.1000040
Whaley, S. G., Berkow, E. L., Rybak, J. M., Nishimoto, A. T., Barker, K. S., Rogers, P. D., et al. (2016). Azole Antifungal resistance in Candida albicans and emerging non-albicans Candida Species. Front. Microbiol. 7, 2173. doi: 10.3389/fmicb.2016.02173
Whelan, W. L. (1987). The genetic basis of resistance to 5-fluorocytosine in Candida species and Cryptococcus neoformans. Crit. Rev. Microbiol. 15, 45–56. doi: 10.3109/10408418709104447
Xu, Y., Lu, H., Zhu, S., Li, W. Q., Jiang, Y. Y., Berman, J., et al. (2021). Multifactorial mechanisms of tolerance to ketoconazole in Candida albicans. Microbiol Spectr. 9, e0032121. doi: 10.1128/Spectrum.00321-21
Yang, F., Gritsenko, V., Lu, H., Zhen, C., Gao, L., Berman, J., et al. (2021a). Adaptation to fluconazole via aneuploidy enables cross-adaptation to amphotericin b and flucytosine in cryptococcus neoformans. Microbiol Spectr. 9, e0072321. doi: 10.1128/Spectrum.00723-21
Yang, F., Gritsenko, V., Slor Futterman, Y., Gao, L., Zhen, C., Lu, H., et al. (2021b). Tunicamycin potentiates antifungal drug tolerance via aneuploidy in Candida albicans. MBio 12, e0227221. doi: 10.1128/mBio.02272-21
Yang, F., Kravets, A., Bethlendy, G., Welle, S., and Rustchenko, E. (2013). Chromosome 5 monosomy of Candida albicans controls susceptibility to various toxic agents, including major antifungals. Antimicrob Agents Chemother. 57, 5026–5036. doi: 10.1128/AAC.00516-13
Yang, F., Lu, H., Wu, H., Fang, T., Berman, J., Jiang, Y. Y., et al. (2021c). Aneuploidy underlies tolerance and cross-tolerance to drugs in Candida parapsilosis. Microbiol Spectr. 9, e0050821. doi: 10.1128/Spectrum.00508-21
Yang, F., Teoh, F., Tan, A. S. M., Cao, Y., Pavelka, N., Berman, J., et al. (2019). Aneuploidy enables cross-adaptation to unrelated drugs. Mol. Biol. Evol. 36, 1768–1782. doi: 10.1093/molbev/msz104
Yang, F., Todd, R. T., Selmecki, A., Jiang, Y. Y., Cao, Y. B., Berman, J., et al. (2021d). The fitness costs and benefits of trisomy of each Candida albicans chromosome. Genetics 218, iyab056. doi: 10.1093/genetics/iyab056
Yang, F., Zhang, L., Wakabayashi, H., Myers, J., Jiang, Y., Cao, Y., et al. (2017). Tolerance to caspofungin in Candida albicans is associated with at least three distinctive mechanisms that govern expression of FKS genes and cell wall remodeling. Antimicrob. Agents Chemother. 61, e00071-17. doi: 10.1128/AAC.00071-17
Keywords: Candida parapsilosis, caspofungin, aneuploidy, cross tolerance, 5-flucytosine
Citation: Sun L-l, Li H, Yan T-h, Cao Y-b, Jiang Y-y and Yang F (2023) Aneuploidy enables cross-tolerance to unrelated antifungal drugs in Candida parapsilosis. Front. Microbiol. 14:1137083. doi: 10.3389/fmicb.2023.1137083
Received: 04 January 2023; Accepted: 17 March 2023;
Published: 11 April 2023.
Edited by:
Sameh S. M. Soliman, University of Sharjah, United Arab EmiratesReviewed by:
Lisa Lombardi, University College Dublin, IrelandKin-Ming Tsui, University of British Columbia, Canada
Copyright © 2023 Sun, Li, Yan, Cao, Jiang and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Feng Yang, ZmVuZy55YW5nMDQwNUBnbWFpbC5jb20=; Yuan-ying Jiang, amlhbmd5eUB0b25namkuZWR1LmNu
†These authors have contributed equally to this work
 Liu-liu Sun1,2†
Liu-liu Sun1,2† Hao Li
Hao Li Tian-hua Yan
Tian-hua Yan Yong-bing Cao
Yong-bing Cao Yuan-ying Jiang
Yuan-ying Jiang Feng Yang
Feng Yang