- 1Intracellular Pathogens Research Laboratory, Comparative Medicine Institute, College of Veterinary Medicine, North Carolina State University, Raleigh, NC, United States
- 2Department of Medical Sciences, School of Veterinary Medicine, University of Wisconsin, Madison, WI, United States
- 3Department of Clinical Sciences, College of Veterinary Medicine, North Carolina State University, Raleigh, NC, United States
Surveillance of the fleas and flea-borne pathogens infecting cats is important for both human and animal health. Multiple zoonotic Bartonella and Rickettsia species are known to infect the most common flea infesting cats and dogs worldwide: Ctenocephalides felis, the cat flea. The ability of other flea species to transmit pathogens is relatively unexplored. We aimed to determine cat host and flea factors independently associated with flea Bartonella and Rickettsia infection. We also assessed flea and cat infection by flea-host pair and location. To accomplish these aims, we performed qPCR for the detection of Bartonella, hemotropic Mycoplasma, Rickettsia, and Wolbachia DNA using paired cat and flea samples obtained from free-roaming cats presenting for spay or neuter across four locations in the United States. A logistic regression model was employed to identify the effect of cat (sex, body weight, geographic location, and Bartonella, hemotropic Mycoplasma, and Rickettsia spp., infection) and flea (clade and Rickettsia and Wolbachia infection) factors on C. felis Bartonella clarridgeiae infection. From 189 free roaming cats, we collected 84 fleas: Ctenocephalides felis (78/84), Cediopsylla simplex (4/84), Orchopeas howardi (1/84), and Nosopsyllus fasciatus (1/84). Ctenocephalides felis were phylogenetically assigned to Clades 1, 4, and 6 by cox1 gene amplification. Rickettsia asembonensis (52/84) and B. clarridgeiae (16/84) were the most common pathogenic bacteria detected in fleas. Our model identified host cat sex and weight as independently associated with B. clarridgeiae infection in fleas. Rickettsia asembonensis (52/84), Rickettsia felis (7/84) and Bartonella henselae (7/84) were detected in specific clades: R. felis was detected only in Clades 1 and 6 while B. henselae and R. asembonensis were detected only in Clade 4. Wolbachia spp., also displayed clade specificity with strains other than Wolbachia wCfeT only infecting fleas from Clade 6. There was poor flea and host agreement for Bartonella spp., infection; however, there was agreement in the Bartonella species detected in cats and fleas by geographic location. These findings reinforce the importance of considering reservoir host attributes and vector phylogenetic diversity in epidemiological studies of flea-borne pathogens. Widespread sampling is necessary to identify the factors driving flea-borne pathogen presence and transmission.
1. Introduction
Ctenocephalides felis, the cat flea, is the most common ectoparasite of the domestic dog and cat worldwide (Rust, 2017). Other flea species such as Cediopsylla simplex, Ctenocephalides canis, Echidnophaga gallinacea, Nosopsyllus fasciatus, and Pulex spp., are occasionally reported as ectoparasites of the domestic cat (Akucewich et al., 2002; Thomas et al., 2016; Abdullah et al., 2019). Little is known about the disease-causing capacity of these other flea species as a primary cause of skin irritation or a vector for pathogen transmission. The cat flea alone has the capacity to vector multiple zoonotic organisms (Bartonella spp., Dipylidium caninum, Rickettsia spp.; Rust, 2017). The factors influencing cat flea Bartonella and Rickettsia infection are currently unknown but have important implications for risk assessment and disease epidemiology in animals and humans.
The three Bartonella spp., associated with C. felis and their cat hosts are Bartonella henselae, Bartonella clarridgeiae, and Bartonella koehlerae (Chomel et al., 1996; Kordick et al., 1997; Avidor et al., 2004; Taber et al., 2022). Infection with these species is a cause of both animal and human disease with numerous manifestations including endocarditis, nervous system, rheumatological, and vascular diseases (Breitschwerdt, 2017; Álvarez-Fernández et al., 2018; Canneti et al., 2019). Studies analyzing the Bartonella spp., infection status of cats and their fleas generally report poor agreement between cat and flea pairs (Chomel et al., 1996; La Scola et al., 2002; Gutiérrez et al., 2015; Mifsud et al., 2020; Azrizal-Wahid et al., 2021). The role of host factors (e.g., body weight, sex) in association with flea Bartonella spp., infection were not investigated in these studies. One study investigating C. felis phylogenetic diversity and pathogen infection failed to identify associations with specific haplotypes (Azrizal-Wahid et al., 2021). Global sampling and cox1 gene sequencing by Lawrence et al. identified 8 distinct, bioclimatically limited C. felis clades (Lawrence et al., 2019). These include the temperate (Clades 1 and 2), tropical (Clades 3–6), and African (Clades 7 and 8) clades. A limited number of studies have utilized these established clades to shed light on flea Bartonella spp., infection: B. clarridgeiae infection has been reported in Clades 1, 3, 4, and 6, B. henselae infection has been reported in Clades 1, 4, and 6, and B. koehlerae infection has been reported in Clades 1 and 4 (Šlapeta and Šlapeta, 2016; Chandra et al., 2017; Manvell et al., 2022).
Rickettsia felis, Rickettsia asembonensis, and Candidatus ‘Rickettsia senegalensis’ comprise a group known as the Rickettsia felis-like organisms (RFLO) which are known to be vectored by C. felis (Reif and Macaluso, 2009; Legendre and Macaluso, 2017; Maina et al., 2019). Clinical signs of RFLO infection in humans are generally non-specific, and similar to those caused by other Rickettsia spp., such as acute headache, nausea, pyrexia, rash, and muscle, back, and joint pain (Richter et al., 2002; Richards et al., 2010). RFLO are occasionally detected in cats (Mullins et al., 2018; Phoosangwalthong et al., 2018; Maina et al., 2019) however previous studies suggested that the domestic dog or the flea could serve as a reservoir host (Barrs et al., 2010; Hii et al., 2011; Ng-Nguyen et al., 2020). Phylogenetic studies have identified R. asembonensis infection in Clade 4, and R. felis infection in Clades 1 and 6 (Manvell et al., 2022). Clade 3 C. felis were infected with an unconfirmed RFLO (Šlapeta and Šlapeta, 2016). Otherwise, there is limited information on the risk factors for flea infection with RFLO.
Hemotropic Mycoplasma species (hMyc) are occasionally, but not always detected in the cat flea and the cat flea has been proposed as a potential vector for hMyc (Lappin et al., 2006; Barrs et al., 2010; Persichetti et al., 2016; Abdullah et al., 2019). However, in laboratory experiments C. felis did not transmit hMyc efficiently (Woods et al., 2006) and other studies have suggested fighting as a mechanism of transmission among cats and potentially to humans (Barker, 2019; Hattori et al., 2020; Tasker, 2022). The manifestation of hMyc infection in animals and humans appears to primarily be pyrexia and hemolytic anemia with the ability to cause severe and potentially life-threatening disease in a subset of cases (Barker, 2019; Hattori et al., 2020; Tasker, 2022).
Another flea-associated genus of interest is Wolbachia, which infect a majority of arthropod and helminth species and manipulate insect reproduction, vector competence, and vector efficiency on a strain specific basis (Werren et al., 2008). Three Wolbachia strains have been isolated from C. felis: wCfeF, wCfeJ, and wCfeT (Driscoll et al., 2020; Khoo et al., 2020). Limited research has investigated the C. felis associated Wolbachia strains and their effect on vector and pathogen success, despite representing a potential opportunity for vector and/or pathogen control, as has been accomplished in mosquitos (Dorigatti et al., 2018). Investigating coinfection of Wolbachia and pathogenic Bartonella or Rickettsia spp., in C. felis may be important for flea-borne pathogen epidemiology. Microbiome analysis has revealed widespread coinfection of C. felis with Bartonella, Rickettsia, and Wolbachia, but this observation does not eliminate the possibility that a specific Wolbachia strain may impact vector pathogen acquisition, maintenance, and/or transmission in C. felis (Manvell et al., 2022).
Given the existing knowledge gaps surrounding flea-borne pathogen coinfection and associations with flea and reservoir host factors, we aimed to identify the cat (sex, body weight, geographic location, and Bartonella, hemotropic Mycoplasma, and Rickettsia spp., infection) and flea (phylogenetic clade and Bartonella, Rickettsia, and Wolbachia co-infection) factors influencing Bartonella and Rickettsia presence in fleas collected from free-roaming cats. We tested three specific hypotheses regarding the most common flea Bartonella spp., B. clarridgeiae: (1) location and flea clade are independently associated with flea B. clarridgeiae infection, (2) fleas collected from B. clarridgeiae infected cat hosts are more likely to be infected with B. clarridgeiae, and (3) flea coinfection with Rickettsia and Wolbachia spp., is associated with flea B. clarridgeiae infection.
2. Materials and methods
2.1. Sample collection
With the assistance of veterinarians and staff at partner Trap-Neuter-Release (TNR) programs, fleas and cat tissue were collected at four locations in North Carolina, Virginia, and Wisconsin under North Carolina State University IACUC protocol #19–003 and #21–468, and University of Wisconsin IACUC protocol #V006461. Samples collected from Washington, NC were obtained through collaboration with Paws and Love, Inc. in March 2019. Collections from Raleigh, NC were performed at the North Carolina State University Veterinary Hospital in April through June 2022. Collections in Orange, VA were performed at Paradocs Animal Hospital with the assistance of the Orange County Humane Society TNR program in May 2019. Collections from Madison, WI were performed in collaboration with the Madison Cat Project in March through August 2021. Cats presenting for routine spay and neuter through each local TNR program were selected for participation regardless of sex or apparent flea presence. Cats were excluded if eartip tissue was not obtained.
In order to collect fleas, volunteers were instructed to comb all cats regardless of apparent parasites or flea dirt with special attention paid to the tail base and ventral regions. Each cat was assigned their own flea comb and collection bag into which combings were placed. Fleas and combs were then frozen until the flea species was identified and DNA extracted.
Sample size calculations were based on pilot data on the proportion of cats with fleas able to be collected (estimated 40%) and estimating that approximately 50% of fleas would have PCR amplifiable Bartonella spp., DNA or Rickettsia spp., DNA. Logistically, using our sampling design with partner TNR organizations, it appeared feasible to collect samples from approximately 40 cats over a 1–6 month period. Therefore, with a total sample of 160 cats and 80 fleas (40 cats and 20 fleas per sampling location), we would have 80% power to detect an odds ratio of 4 or greater between the proportion of fleas with and without pathogen DNA and proposed binary explanatory variables (cat sex, cat infection with Bartonella spp., flea pathogen co-infection, flea Wolbachia co-infection). This was expected to be a large enough difference to be clinically and practically relevant for this exploratory study.
2.2. Fleas
Upon receiving fleas at the NCSU Intracellular Pathogens Research Laboratory, fleas were identified to the species level with the assistance of Dr. James Flowers, Clinical Professor of Parasitology (North Carolina State University, College of Veterinary Medicine). Fleas were individually washed and crushed according to previously reported protocol (Manvell et al., 2022). DNA was then extracted utilizing the Qiagen DNeasy Blood & Tissue Kit (Qiagen, Valencia, CA, United States) following the manufacturer’s tissue extraction protocol. DNA concentration and purity were determined spectrophotometrically (Thermo Fisher Scientific, Waltham, MA, United States). DNA from 18% (15/84) of fleas used for this study’s sample was also used for a previously published manuscript analyzing the C. felis microbiome (Manvell et al., 2022).
2.3. Tissues
When presenting for spay and neuter, free-roaming cats regularly have one eartip removed while under anesthesia to allow them to be visually identified as spayed/neutered following release. Instead of discarding the eartip, we collected and froze the tissue which was later dissected utilizing a scalpel and forceps disinfected with 94% ethanol between each sample. DNA extraction from tissues was performed utilizing the Qiagen DNeasy Blood & Tissue Kit (Qiagen, Valencia, CA, United States) following the manufacturer’s protocol. DNA concentration and purity were determined spectrophotometrically (Thermo Fisher Scientific, Waltham, MA, United States). DNA from eartip tissue from 39% (73/189) of the cats reported in this study, including 32% (17/53) of flea infested cats, were previously published in a manuscript comparing the presence of flea-borne pathogens in eartip and reproductive tissues (Manvell et al., 2021).
2.4. Polymerase chain reaction
Quantitative real-time PCR (qPCR) for Bartonella, Rickettsia, hMyc, and Wolbachia spp., was performed utilizing the primers listed below (Table 1). Following genus level qPCR, positive samples underwent Sanger sequencing. For these sequences, species and strain identity was determined by alignment with NCBI Basic Local Alignment Search Tool (BLAST). If 23S-5S Rickettsia qPCR returned a sequence that was not readable (e.g., overlapping peaks, inappropriate length) or had an inappropriate melting temperature, R. felis and R. asembonensis specific primers targeting the ompA gene were employed to confirm the infecting species with an expected length of 222 and 183 base pairs, respectively. Primer development was performed in AlignX utilizing R. asembonensis (GenBank MK923742.1) and R. felis (GenBank MG818714.1). Specificity was determined by in silico analysis with comparison to 22 Rickettsia spp., including Rickettsia rickettsii (DQ002504.1), Rickettsia parkeri (U43801.1), and Rickettsia rhipicephali (U43803.1). Primers were validated with C. felis samples of known infection status including eight R. asembonensis and eight R. felis infected C. felis, and numerous uninfected C. felis.
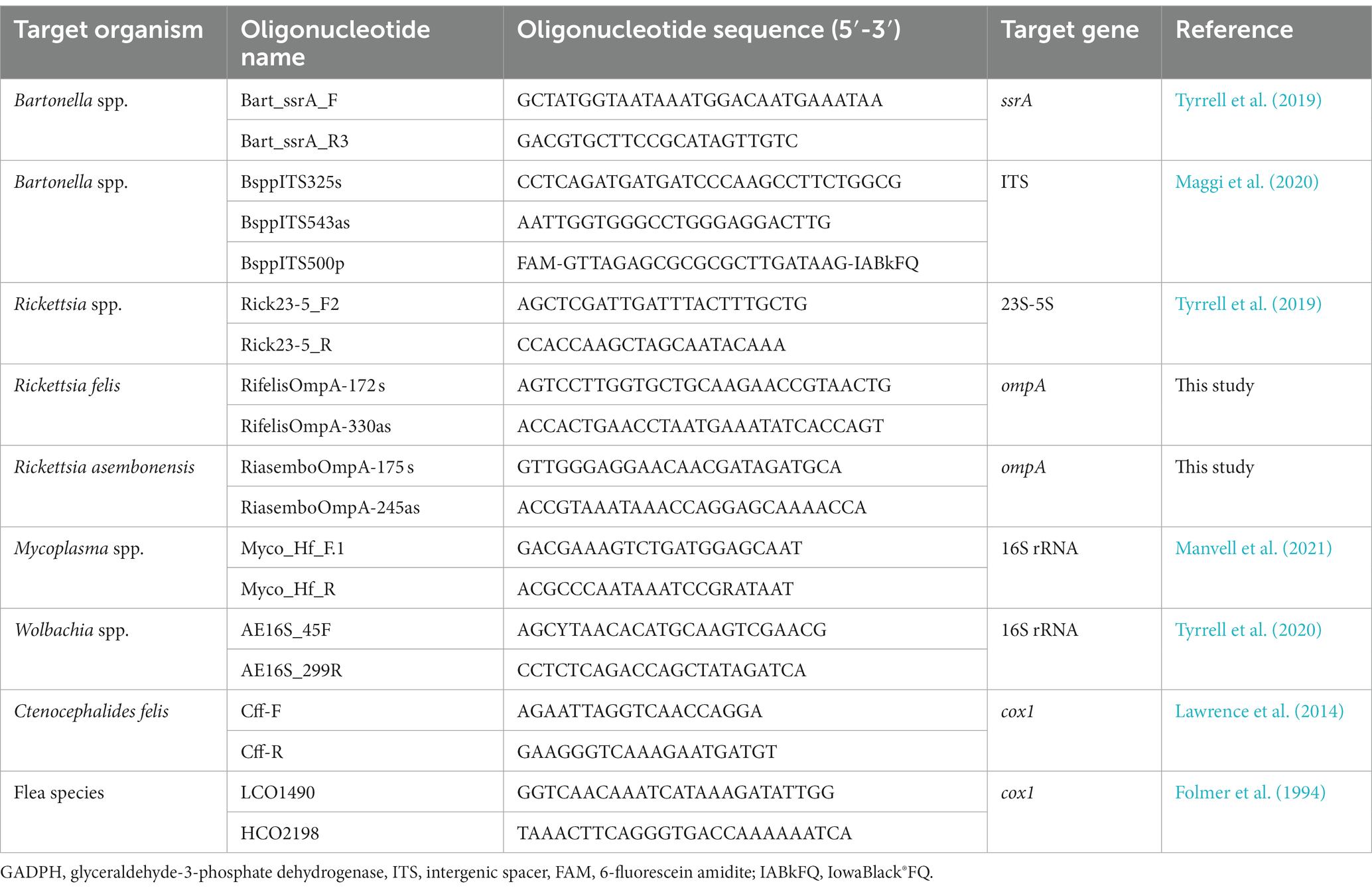
Table 1. Primers targeting housekeeping genes and pathogens including primer name, sequence, gene target, and reference to conditions.
Flea phylogeny was assessed by conventional PCR amplification of the cox1 gene. Ctenocephalides felis DNA was amplified using the Cff-F and Cff-R primers (Lawrence et al., 2014) while other flea species were amplified using the LCO1490 and HCO2198 primers (Folmer et al., 1994).
Each run included two negative controls: a No Template Control (NTC) consisting of nuclease free water and DNA from cat blood confirmed to be negative for the genus of interest. Each run also included a positive control plasmid. The Bartonella spp., intergenic spacer (ITS) qPCR utilized B. henselae culture as a positive control. Negative control DNA and positive control plasmids were obtained from the Vector Borne Disease Diagnostics Lab (VBDDL) at North Carolina State University.
2.5. Statistical methods
Data handling was performed in R version 4.1.0 (R Foundation for Statistical Computing, Vienna, Austria) using the here (Müller, 2020), janitor (Firke, 2021), reshape2, stats, stringr, and tidyverse (Wickham et al., 2019; Wickham, 2021) packages. Phylogenetic analysis was performed in R using the ape (Paradis and Schliep, 2019), bios2mds, Biostrings (Pagès et al., 2021), haplotypes, ips, irr (Gamer et al., 2019), msa (Bodenhofer et al., 2015), pegas (Paradis, 2010; Tsirogiannis and Sandel, 2016, 2017), PhyloMeasures, and seqinr packages. Visualization was performed in R using the cowplot (Wilke, 2020), ggsci (Xiao, 2018), ggplot2 (Wickham, 2016), and ggpubr packages. Fisher’s exact tests were employed to assess five different associations between categorical variables. Comparisons included: (1) Proportion of fleas with any Bartonella spp., by geographic location (4 locations); (2) proportion of fleas with any Rickettsia spp., by geographic location (4 locations); (3) proportion of fleas with any Wolbachia spp., by geographic location (4 locations); (4) proportion of fleas with R. asembonensis by presence of any Bartonella spp (any Bartonella spp., present/absent); (5) proportion of fleas with any Bartonella spp., by presence of any Wolbachia spp (any Wolbachia spp., present/absent). p values <0.05 were considered significant; due to the exploratory nature of this analysis and to prevent overly conservative p values, correction for multiple comparisons was not performed.
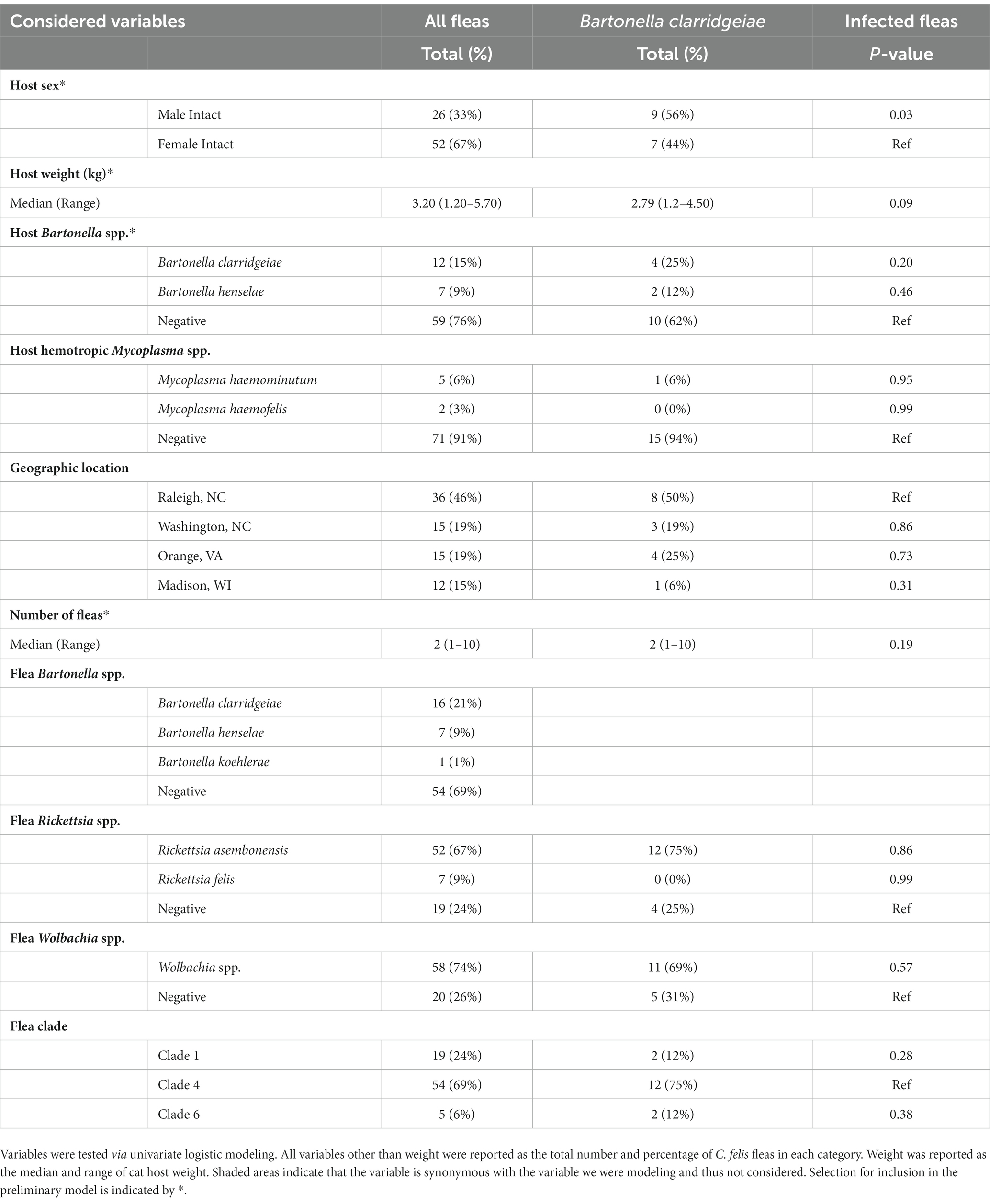
A multivariable logistic regression model was developed to identify the variables associated with B. clarridgeiae infection in Ctenocephalides felis fleas. Variables considered for inclusion are shown in Table 2, and included geographic location, cat hMyc, cat Bartonella, flea clade, and flea Rickettsia infection as categorical variables, cat sex and flea Wolbachia infection as binary variables, and cat weight as a continuous variable. Variables were first compared individually with the outcome of interest (flea B. clarridgeiae infection) via univariate logistic regression model and those with a p-value less than 0.25 were selected for inclusion in the preliminary model. The number of fleas collected from each flea’s host was included to control for the collection of multiple fleas from the same host. Additional models were created which systematically removed and replaced individual variables. These additional models were compared to the preliminary model on the basis of Akaike information criterion (AIC) and a Hosmer-Lemeshow Goodness of Fit test (GOF). When the removal of a variable resulted in a significantly different fit (GOF p < 0.05) or reduced AIC that variable was retained in the final model. The odds ratio (OR) and 95% confidence interval is reported for variables selected for the final model.
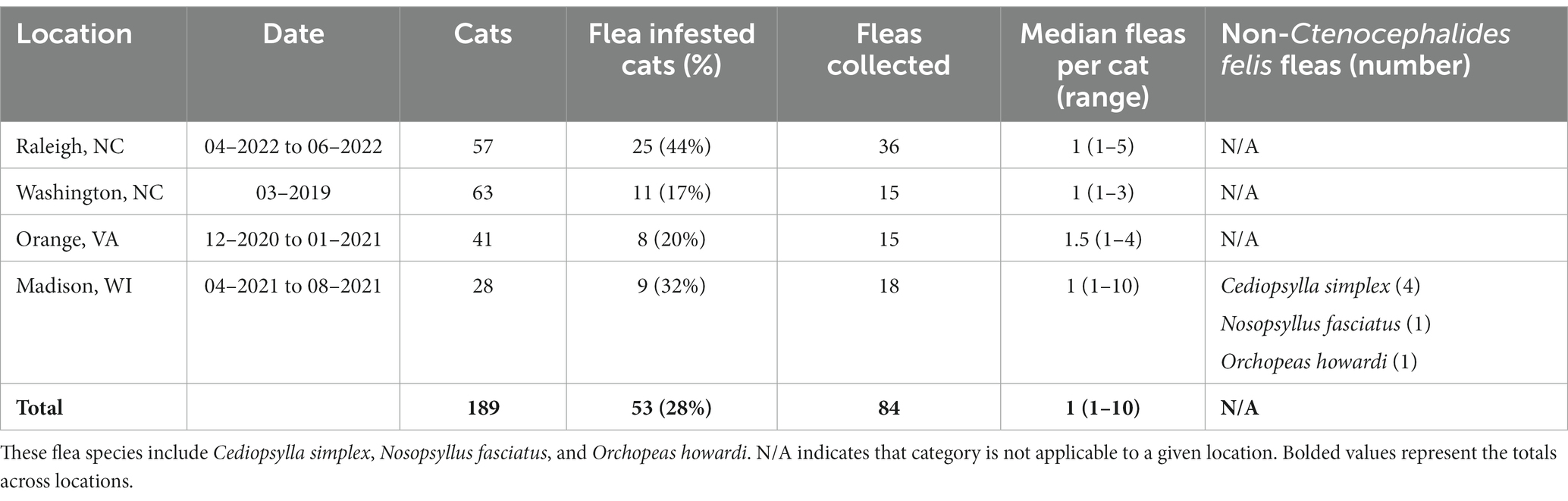
Table 2. Summary of sampling based on geographic location of origin including the dates of sampling, total number of cats, number of flea infested cats, total number of fleas, median number of fleas per infested cat (and range), and summary of non-Ctenocephalides felis sampled fleas.
Following selection of the final model, additional models were created to test three specific hypotheses: (1) location and flea clade are independently associated with flea B. clarridgeiae infection, (2) fleas collected from B. clarridgeiae infected cat hosts are more likely to be infected with B. clarridgeiae, and (3) flea coinfection with Rickettsia and Wolbachia spp., is associated with flea B. clarridgeiae infection. These models included the selected variables for the final model, the number of fleas per host, and the variables of interest in each hypothesis.
3. Results
In total we obtained a tissue sample from 189 cats, 53 of which had one or more fleas collected. The number of fleas and cats sampled at each location is shown in Table 2.
3.1. Flea characteristics and PCR results
By far the most common flea species collected was C. felis (93%, 78/84); however, other flea species were collected from cats in Madison, WI including Cediopsylla simplex (5%, 4/84), Orchopeas howardi (1/84), and Nosopsyllus fasciatus (1/84). No cat was infested with more than one flea species. The C. felis collected during this study were assigned to Clades 1, 4, and 6, as defined by Lawrence et al. (2019). The proportion of each C. felis clade by location are displayed in Figure 1. Cediopsylla simplex fleas were assigned to three haplotypes (Accession ID: OP713785, OP713787, OP713885) with no homologous submissions in GenBank (421/427, 98.5%, Accession ID: HM398833.1). The one N. fasciatus cox1 sequence (Accession ID: OP713901) was novel with only 86% (358/417) homology with N. fasciatus sequences from Belgium (Accession ID: LT158040). The one O. howardi flea (Accession ID: OP737457) displayed 99% (423/427) homology to Orchopeas caedens (Accession ID: HM398830.1) in the absence of available O. howardi sequences for comparison.
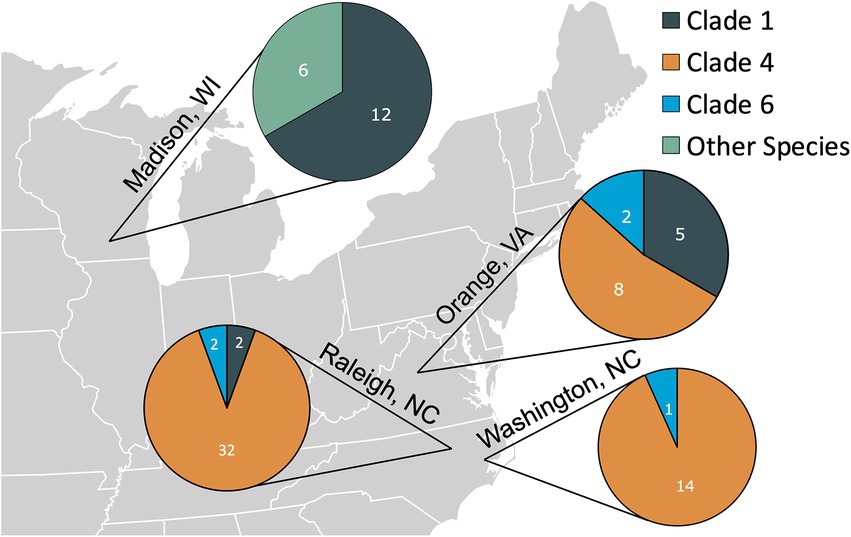
Figure 1. The number of fleas from each location assigned to the Ctenocephalides felis clades defined by Lawrence et al. (2019) or belonging to other flea species (Cediopsylla simplex, Orchopeas howardi, and Nosopsylus fasciatus).
The percentage of fleas infected with each bacterial genus is shown in Figure 2. A single C. felis contained a Wolbachia spp., that was distinct from previously described strains (Accession ID: OP731570). The O. howardi (n = 2) and N. fasciatus (n = 1) fleas were infected with Wolbachia pipientis, or a strain not able to be differentiated over the amplified region of the 16S gene. Hemotropic Mycoplasma spp., DNA was not amplified from fleas of any species.
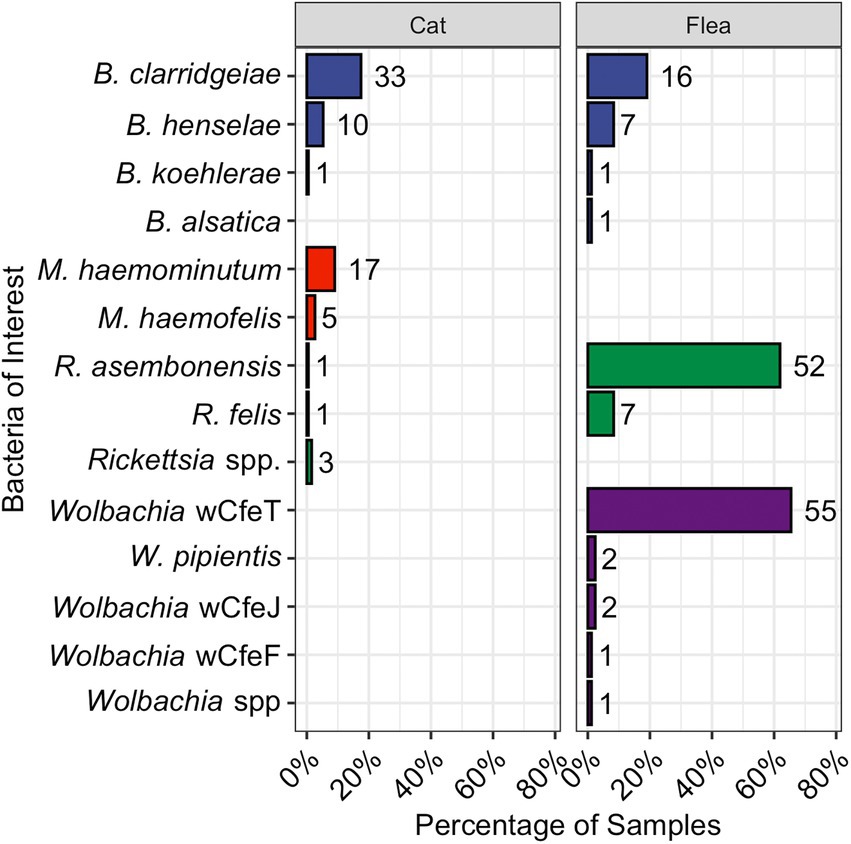
Figure 2. Bar chart displaying the proportion of cats (left) or fleas (right) infected with the various pathogens and bacteria as reported by qPCR. The total number is displayed to the right of each bar. Bartonella spp., are in blue, hemotropic Mycoplasma spp., in red, Rickettsia spp., in green, and Wolbachia spp., in purple.
3.2. Cat characteristics and PCR results
Female (51%, 97/189) and male (49%, 92/189) cats were sampled approximately equally. The percentage of cats infected with each bacterial genus is shown in Figure 2. Bartonella spp., were the most common flea-borne pathogen detected in the cats. Five cats (all from Raleigh, NC) were infected with Rickettsia spp. This includes two previously undescribed Rickettsia spp. amplicons with 100% homology over the 283 base pair sequence (Accession ID: OP744991). The closest available GenBank match was R. felis (Accession ID: KJ796446.1) with low query cover. Attempts to amplify a product using R. felis ompA specific primers were unsuccessful. The second unknown Rickettsia spp., was amplified from a single cat (Accession ID: OP744990) and most closely aligned with Rickettsia spp. isolates from Ixodes scapularis in New York (221/226, 98%, Accession ID: MN704870.1) and Amblyomma americanum in New York and North Carolina (221/226, 98%, Accession ID: KJ796407.1; Lee et al., 2014).
3.3. Flea-cat PCR agreement
Nineteen percent (10/53) of flea infested cats were infected with Bartonella spp. Of these 10 Bartonella spp. infected cats with fleas, 6 yielded one or more flea that was infected with a Bartonella spp.; however, that flea often had a different Bartonella spp. based upon PCR amplification and sequencing than the host cat (3/6, Figures 3, 4B). A majority of Bartonella spp. infected fleas were collected from Bartonella spp., qPCR negative cats (68%, 17/25, Figure 3).
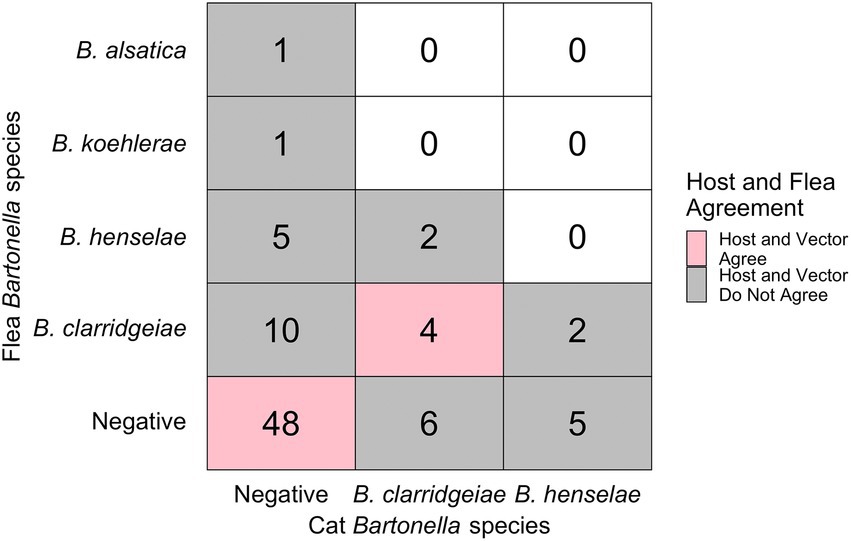
Figure 3. Chart displaying the number of fleas infected with each Bartonella spp. (y-axis) and their host’s infection status (x-axis). Pink indicates host-vector agreement while grey indicated host-vector disagreement regarding pathogen infection status. Number indicates the number of fleas (not cats) in each category.
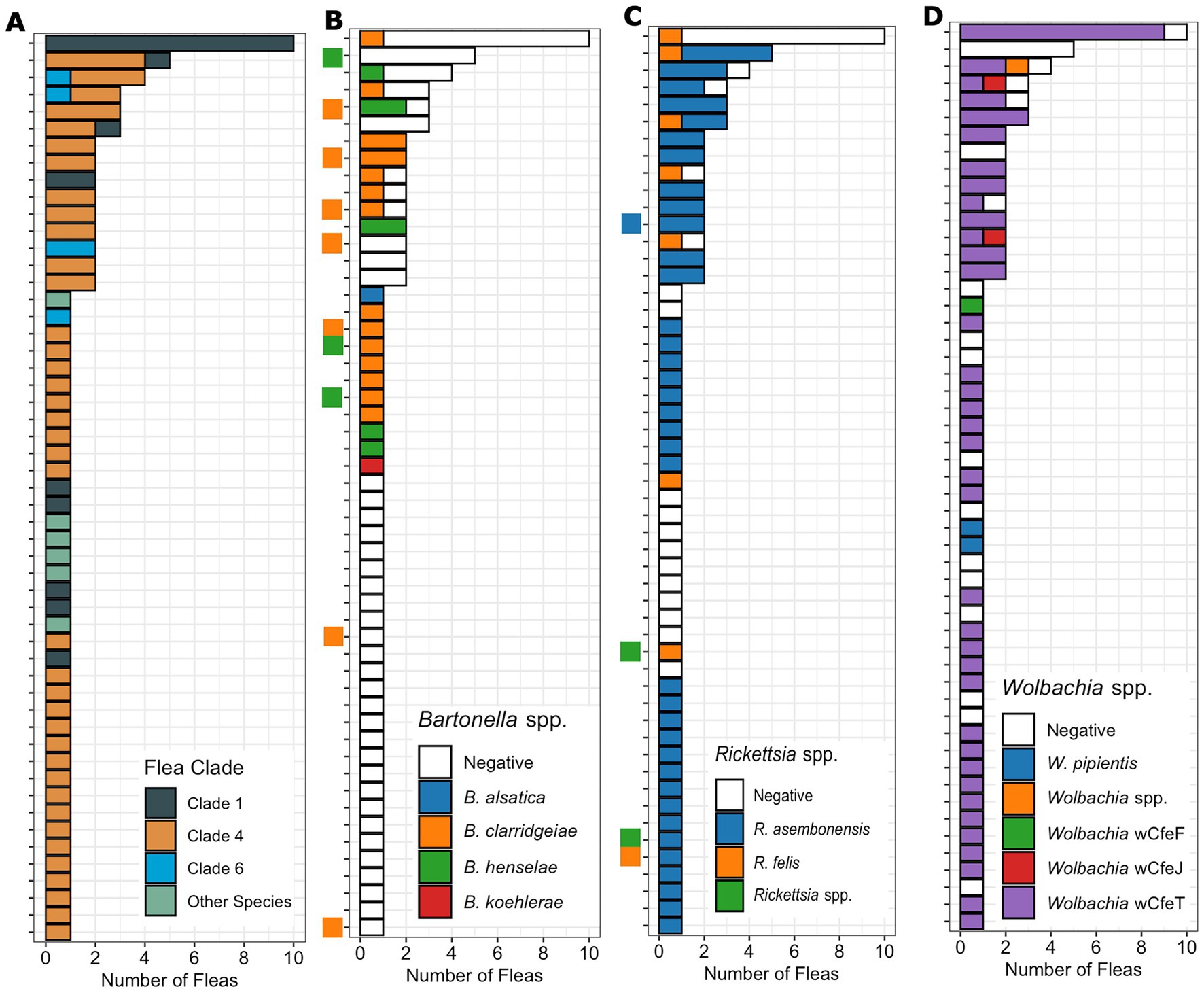
Figure 4. Bar chart displaying the number of fleas (x-axis) from each host (y-axis) by flea species or clade (A) as well as the Bartonella (B), Rickettsia (C), and Wolbachia (D) species or strains identified by qPCR. The cat host Bartonella (C) and Rickettsia (D) are indicated by a small square on the y-axis with uninfected cats left blank. The cat host is kept consistent across figure sections to allow comparison on a specific cat host.
Nine percent (5/53) of flea infested cats were infected with Rickettsia spp. A majority of Rickettsia spp. infected fleas were collected from Rickettsia spp. qPCR negative cats (68%, 54/80, Figure 4C). All five Rickettsia spp. infected cats yielded one or more Rickettsia spp. infected flea, but only one of these cats had the same Rickettsia spp. in tissue and flea based on PCR amplification and sequencing.
Hemotropic Mycoplasma DNA was amplified from cats, but not from fleas. Wolbachia spp. DNA was amplified only from fleas, but not from cats.
There were 15 cats with more than one flea collected; flea clade, Bartonella spp. Rickettsia spp. and Wolbachia spp. PCR results from each flea is shown in Figure 4. For cats with multiple fleas, infestation by C. felis fleas from multiple clades was common (27%, 4/15, Figure 4A). Infestation of a cat with fleas containing different Bartonella spp. was not detected (Figure 4B). In cats with more than one flea collected, often only a single flea was infected with Bartonella spp. (6/10). In contrast, for Rickettsia and Wolbachia spp., cats with multiple fleas frequently had multiple fleas positive (Figures 4C,D). Twelve cats (80%) had multiple fleas infected with Rickettsia spp., and twelve cats (80%) had multiple fleas infected with Wolbachia spp. Infestation with fleas infected with different Rickettsia spp. (2 cats) or Wolbachia (3 cats) strains on the same cat was also detected.
3.4. Associations between pathogens in fleas and explanatory factors
Infection with Bartonella in fleas, considering all three species together in the analysis, was not significantly associated with geographic location (p = 0.068, Figure 5A). When aggregated by geographic location, the same Bartonella spp. were found in cats and fleas, except for B. alsatica that was only found in one C. simplex (Supplementary Figure S1) and no cats in Madison WI. Furthermore, the relative proportion of Bartonella spp. agreed between sample types (flea and cat) at all locations except for Washington, NC, where B. henselae prevalence exceeded B. clarridgeiae in fleas, but not in cats (Supplementary Figure S1). In contrast, Rickettsia spp. infection in fleas, considering all species together, was significantly associated with geographic location (p < 0.0001, Figure 5B). Wolbachia infection in fleas was not associated with geographic location (p = 0.43, Figure 5C).
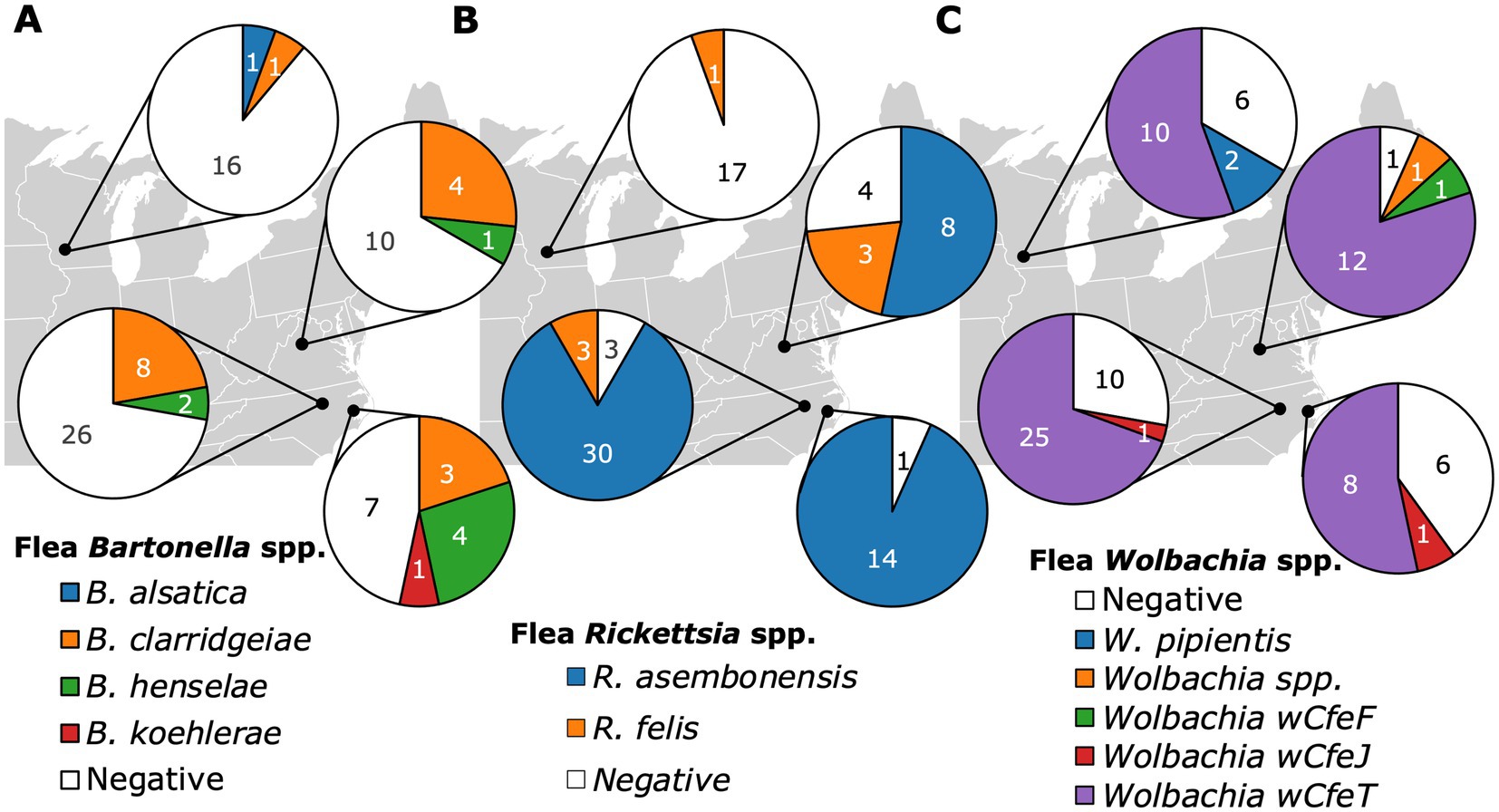
Figure 5. Pie charts displaying the Bartonella (A), Rickettsia (B), and Wolbachia (C) infection status of fleas based on their geographic location of origin including Raleigh NC, Washington NC, Orange VA, and Madison WI.
Bartonella clarridgeiae was the only Bartonella spp. detected in more than one C. felis clade as B. henselae and B. koehlerae were only detected in Clade 4 (Figure 6A), however infection with the genus Bartonella was not associated with C. felis clade (p = 0.28). Bartonella alsatica was detected only in a single C. simplex. Rickettsia spp. were strictly clade specific. Rickettsia asembonensis was detected in almost all fleas from Clade 4 (96%, 52/54) and R. felis was detected only in Clades 1 (32%, 6/19) and 6 (20%, 1/5, Figure 6B; p < 0.0001). Wolbachia spp. also displayed clade specificity with Wolbachia strain wCfeT being the only strain found in C. felis Clades 1 and 4 (Figure 6C). Despite being represented by the smallest sample size, Clade 6 C. felis had the greatest diversity of Wolbachia strains including strains wCfeF, wCfeJ, wCfeT, and a previously undescribed Wolbachia strain. The association of Rickettsia and Wolbachia with C. felis clade persisted even when fleas from diverse clades were infesting the same cat (Figure 4).
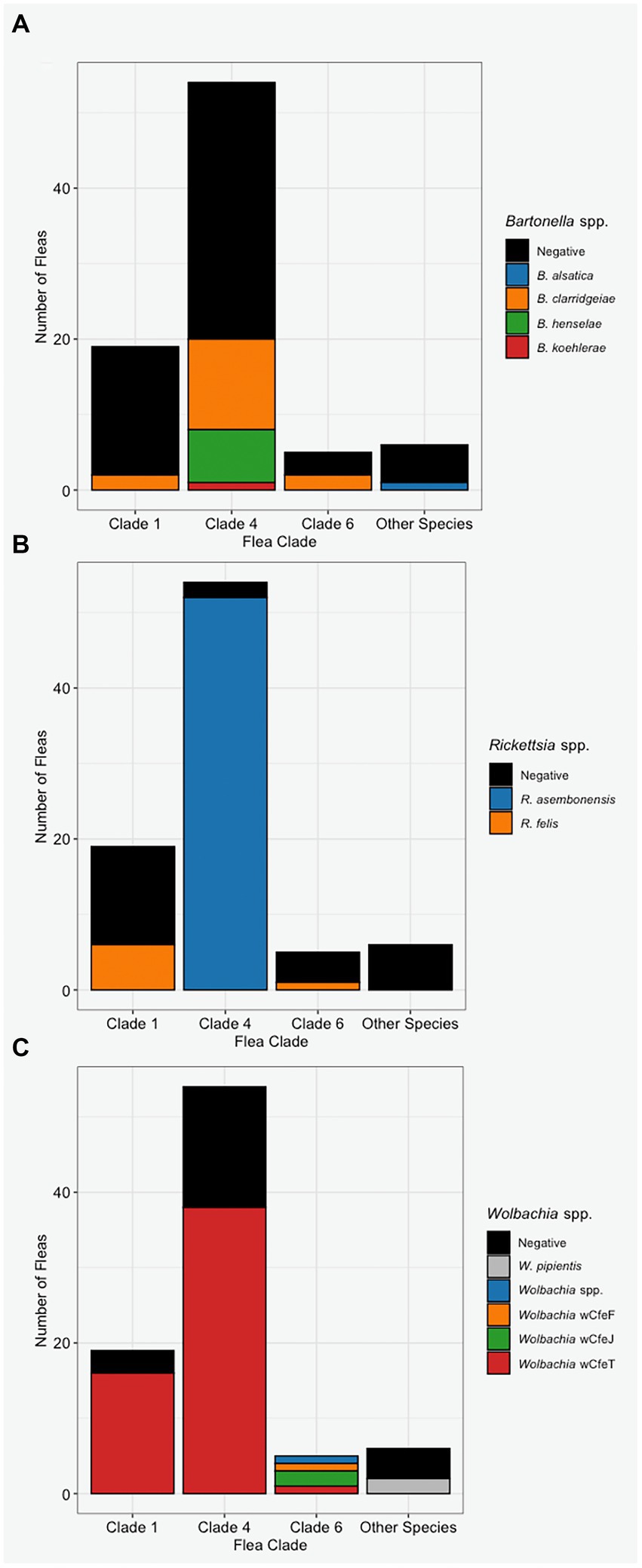
Figure 6. The Bartonella (A), Rickettsia (B), and Wolbachia (C) infection status of fleas based on their assigned clade or species (x-axis).
Bartonella spp. in fleas was associated with R. asembonensis coinfection (p = 0.030). Coinfected fleas included all B. henselae (n = 7) and B. koehlerae (n = 1), and a majority of B. clarridgeiae infected C. felis (75%, 12/16, Figure 7). No R. felis infected flea (n = 7) was coinfected with a Bartonella spp. A majority of Bartonella spp. infected fleas were also coinfected with Wolbachia spp. (68%, 17/25); however, this was not statistically significant compared to the proportion of all fleas infected with Wolbachia spp. (71%, 60/84; p = 0.79).
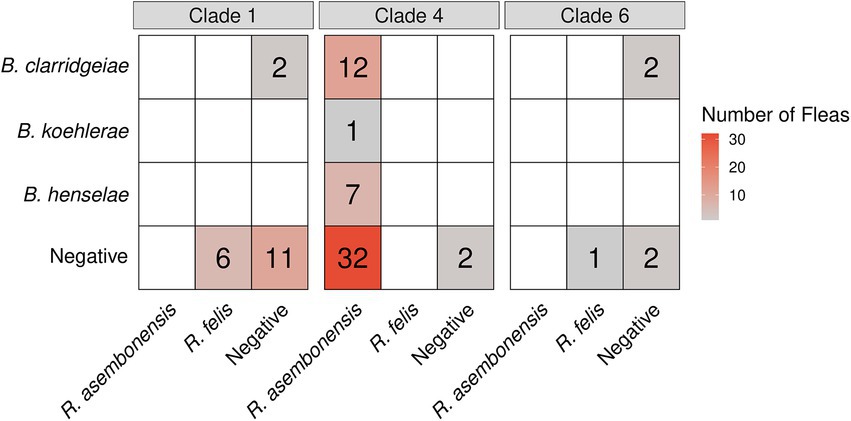
Figure 7. Heatmap displaying the Bartonella (y-axis) and Rickettsia (x-axis) coinfection status by Ctenocephalides felis clade.
A logistic regression model was developed to investigate potential independent associations between explanatory factors and flea B. clarridgeiae infection. Univariate associations with each variable of interest were calculated (Table 3), and the final multivariable model showed that cat sex and body weight were independently associated with C. felis B. clarridgeiae infection (Table 4). A higher proportion of male cats had fleas with B. clarridgeiae than female cats (OR 4.22, 95% CI 1.13–15.7), and for each 1 kg increase in cat host body weight, fleas had 0.41 lower odds of B. clarridgeiae infection (95% CI 0.17–0.98). When controlling for cat body weight, sex, and the number of fleas collected per cat, geographic location and flea clade were not associated with B. clarridgeiae infection of C. felis (Table 5). Similarly, when controlling for cat body weight, sex, and the number of fleas collected per cat, neither cat B. clarridgeiae infection nor flea infection status was associated with B. clarridgeiae infection of C. felis (Table 4).
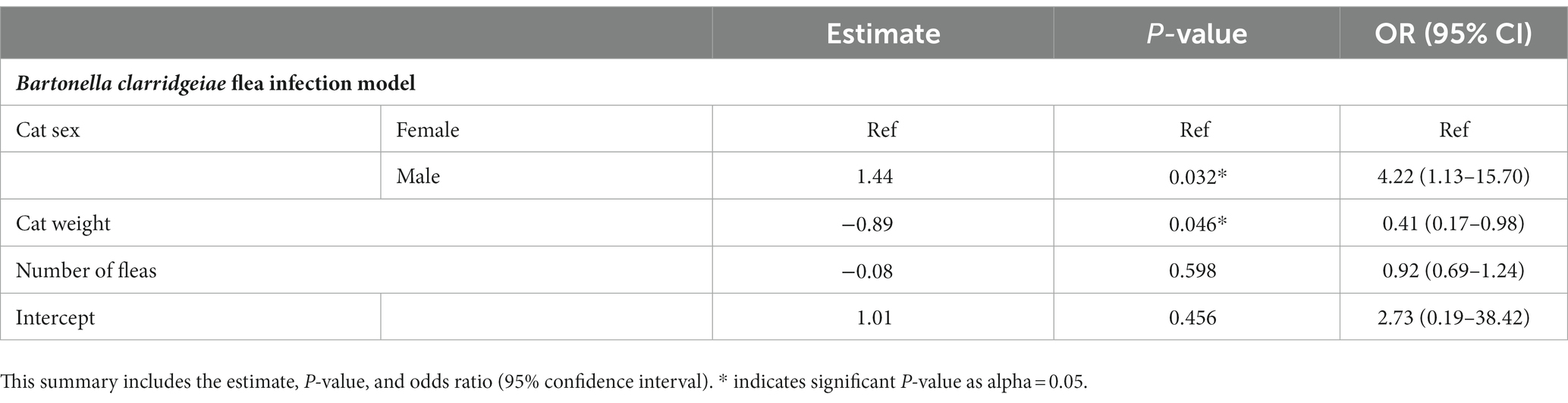
Table 4. Summary of the model selected to predict B. clarridgeiae infection in Ctenocephalides felis including only the variables significantly associated with host and flea characteristics and potentially confounding variable (number of fleas per host cat).
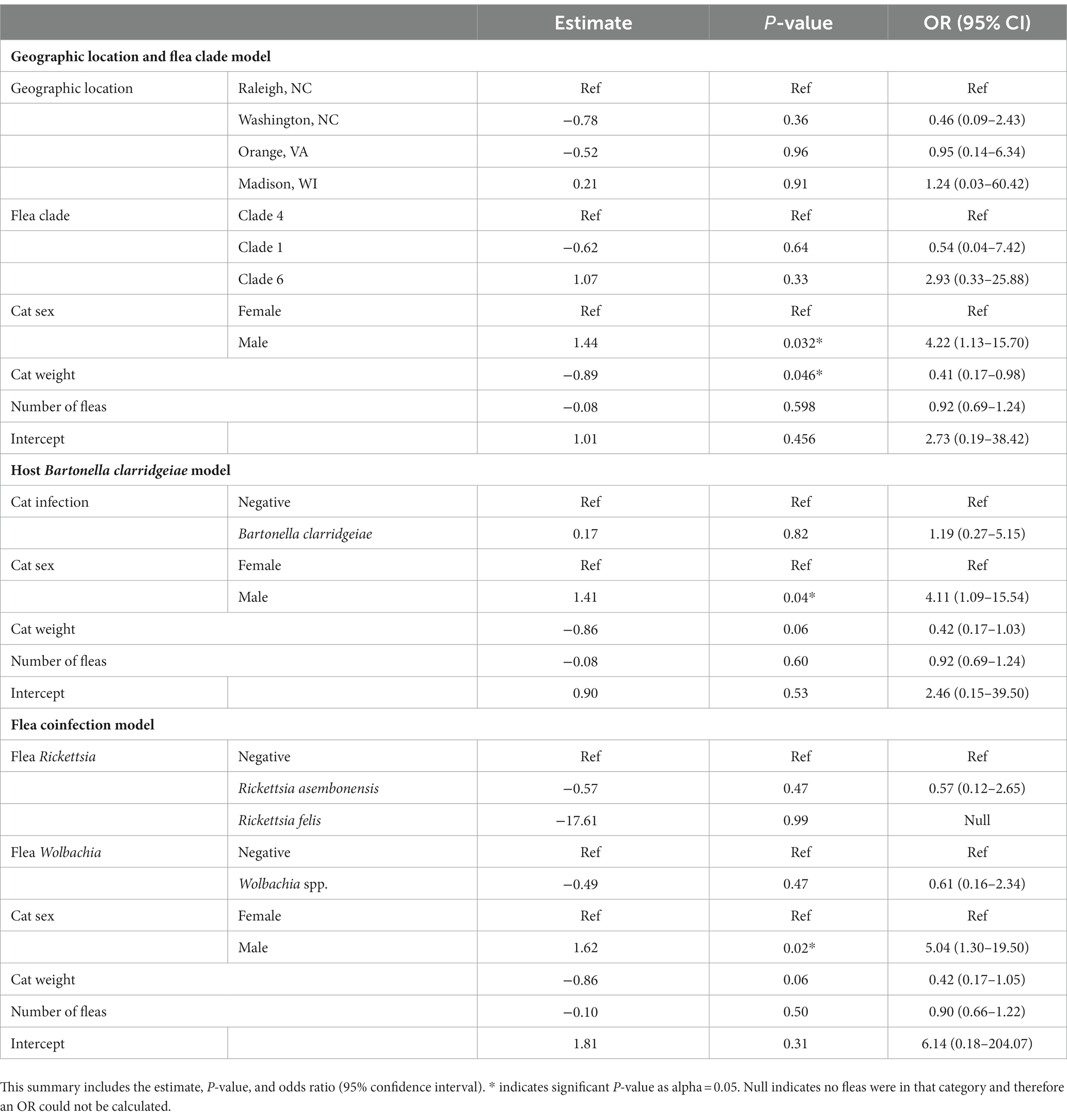
Table 5. Summary of the models created to assess the role of geographic location and flea clade, host B. clarridgeiae infection, and flea coinfection in Ctenocephalides felis B. clarridgeiae infection when cat sex, weight, and number of fleas are controlled for.
4. Discussion
This study found that Rickettsia asembonensis (62%) and Bartonella clarridgeiae (19%) were the most common pathogenic bacteria detected in fleas. R. asembonensis was detected exclusively in Clade 4 C. felis regardless of geographic location yet was only detected in one single cat. Bartonella clarridgeiae infection was detected in a similar proportion of C. felis (21%) and cat hosts (17%), but C. felis infection was only independently associated with cat sex (higher in male cats) and weight (higher in lighter cats), and not with cat infection status, geographic location, or flea clade.
Bartonella clarridgeiae was the most common Bartonella spp. infecting fleas and cats in this study. Interestingly, only cat host demographic factors (cat sex and body weight), and not flea factors such as flea clade or co-infection, nor cat Bartonella infection status were significantly independently associated with flea B. clarridgeiae infection. Unlike B. clarridgeiae, B. henselae and B. koehlerae were only detected in Clade 4 C. felis. Based upon previous studies detecting B. henselae in Clades 1, 3, and 6, and B. koehlerae in Clades 1 and 4, it is likely that these Bartonella spp., are not strictly clade specific (Šlapeta and Šlapeta, 2016; Manvell et al., 2022). Our failure to detect B. henselae in the Clade 1 and 6 fleas collected for this study may be due to relative rarity of infection in these clades combined with small sample size of Clade 1 (n = 17) and Clade 6 (n = 7) fleas or restriction of sampling sites to the eastern and midwestern United States. Further sampling of wild-caught fleas, as well as laboratory studies assessing the efficiency of Bartonella spp. acquisition by C. felis from diverse genetic backgrounds and coinfection status are warranted.
Despite our finding that cat sex and body weight were independently associated with flea B. clarridgeiae infection, there was poor agreement between individual cat and flea Bartonella spp., infection status. Only three of 10 Bartonella infected cats hosted fleas infected with the same Bartonella spp. In agreement with previous publications, we concluded that fleas collected from a specific cat do not provide insight into the Bartonella infection status of that cat (Chomel et al., 1996; La Scola et al., 2002; Gutiérrez et al., 2015; Mifsud et al., 2020). Furthermore, a higher percentage of Bartonella spp. uninfected (qPCR negative) cats (30%, 43/145) had fleas collected than Bartonella spp. infected cats (23%, 10/44). This finding reinforces that it is not necessary for cats to present with flea infestation to consider the possibility of Bartonella spp. infection. The inability for study personnel to collect fleas from Bartonella spp. infected cats may be indicative of highly efficient grooming by the cat host or the long duration of Bartonella infection in the cat, an important characteristic of their role as a reservoir host (Kordick and Breitschwerdt, 1988; Taber et al., 2022).
Flea Rickettsia spp. infection was significantly associated with flea phylogenetic clade. Rickettsia felis DNA was amplified only in Clades 1 (32%, 6/19) and 6 (20%, 1/5) while R. asembonensis was amplified from almost all C. felis from Clade 4 (96%, 52/54) but no C. felis from Clades 1 or 6. This clade specificity was previously reported in a larger sampling of C. felis from California, Louisiana, and North Carolina which included a portion of the fleas reported in this manuscript (Manvell et al., 2022). The detection of Rickettsia spp. in the tissue of 9% (5/56) of cats from Raleigh, NC was surprising considering that we did not detect Rickettsia spp. in cats from any other location (n = 133 cats). Some studies have failed to detect Rickettsia spp. in cats when performing PCR on blood (Hawley et al., 2007; Barrs et al., 2010); however, other studies report comparable infection rates to those found in Raleigh, NC (Mullins et al., 2018; Phoosangwalthong et al., 2018). The diversity of these Rickettsia spp. is of interest as we detected DNA sequences of two C. felis associated Rickettsia spp. (R. asembonensis and R. felis) and an unnamed Rickettsia spp. genetically most similar to Rickettsia spp. reported in ticks (Lee et al., 2014). Investigation of the relative clinical importance of these Rickettsia spp. for cats, humans, and other animals is warranted.
Our data supports the use of fleas as sentinels for population level cat Bartonella spp. presence and relative abundance. Within individual geographic locations, we reported complete agreement between fleas and cats for the presence of B. clarridgeiae, B. henselae, and B. koehlerae. Bartonella clarridgeiae was the most common Bartonella spp. in both cats and fleas from all locations except Washington, NC. Washington, NC displayed the highest proportion of B. henselae infection in both cats and fleas and was the only location where B. henselae was more common than B. clarridgeiae in fleas. Washington, NC was also the only location with documented B. koehlerae, which we found in both cats and fleas. Our data supports that flea sampling can provide insight into the Bartonella species diversity and relative abundance in a specific geographic area. This information is critical for regional diagnostic considerations and for the prevention of Bartonella spp. transmission to aberrant hosts (human, dog, etc) in which clinical manifestations are typically more severe and cryptic, with lower bacteremia hindering diagnosis.
The detection of C. simplex, N. fasciatus, and O. howardi infesting cats indicates the need for further investigation of the regional diversity of fleas infesting cats, the ability of these fleas to transmit pathogens, and the efficacy of flea control products for the control of these flea species. Each of these non-C. felis fleas was collected from a different cat, confirming that infestation of non-C. felis fleas on cats has occurred multiple times in this community. The capacity of these fleas to serve as a vector for disease transmission is essentially unknown. In this small sample size one C. simplex was infected with B. alsatica (20%, 1/5). This finding is of potential relevance to human medicine, as B. alsatica, a rabbit reservoir adapted species, has been reported in cases of endocarditis and host versus graft rejection (Raoult et al., 2006; Jeanclaude et al., 2009; Puges et al., 2019). Bartonella, hemotropic Mycoplasma, and Rickettsia spp. DNA was not amplified from N. fasciatus (n = 1) or O. howardi (n = 1). This may be due to a lack of vector competence, the presence of these flea species on atypical hosts that are not as efficient in maintaining and transmitting associated pathogens, or the small number of fleas tested in this study.
The most prevalent Wolbachia strain (wCfeT) did not display C. felis clade specificity, with detection in all clades sampled (1, 4, and 6), while other Wolbachia strains (wCfeF, wCfeJ, and an uncharacterized strain) were only detected in Clade 6. We propose that this finding may be related to increased diversification and evolution of Wolbachia within certain C. felis clades. Wolbachia species are a known endosymbiont of most insect species with certain strains causing profound effects in the species they infect (Werren et al., 2008). Genomic comparison of two C. felis associated Wolbachia (wCfeT and wCfeJ) strains indicated differential biotin synthesis, as well as cytoplasmic incompatibility-like genes with unknown implications for fleas or flea-borne pathogens (Driscoll et al., 2020). Flatau et al. (2018) reported that increased Wolbachia loads lowered reproductive success in Synosternus cleopatrae, a flea species infecting gerbils. This effect was observed only in laboratory fleas and not wild-caught fleas indicating the specificity of these associations which may be due to flea genetic diversity or variations in the flea associated microbiome. Further exploration of C. felis-Wolbachia relationships is necessary to define the evolutionary advantages or disadvantages conveyed by specific Wolbachia strains, as well as the ability of diverse C. felis clades to acquire and maintain various Wolbachia strains and the implications for flea-borne pathogen transmission.
We did not amplify hemotropic Mycoplasma spp. from the fleas in this study despite flea collection from hemotropic Mycoplasma infected cats, a finding also reported by other studies (Persichetti et al., 2016; Abdullah et al., 2019). It is unknown if this is due to the geographic location sampled, lack of flea pooling, or other methodological differences.
One major limitation of this study was that fleas were not collected from every cat that a tissue sample was obtained from. While the flea collection protocol was standardized, since the aim of the study was not to determine the prevalence of flea infestation in this cat population it is likely that fleas from cats with lower flea burdens or cats with fleas that were difficult to catch were underrepresented in this sample. The length of time cats were housed in traps prior to anesthesia may have also impacted whether fleas were able to be collected, since fleas may not have been found on cats with longer waiting times or more efficient groomers. Because of this uncertainty, we did not attempt to investigate flea presence or flea burden on individual cats as explanatory factors for flea-borne pathogens. Additionally, our small sample of non-C. felis fleas and C. felis from Clades 1 and 6 prevented a thorough investigation of the pathogen occurrence and prevalence of pathogen coinfection in these fleas. Samples were collected at different times from different locations, a potentially confounding factor for geographic location-based conclusions. Furthermore, our small sample of R. felis and B. henselae infected fleas prevented the application of a modeling approach for analysis of potential host and flea factors associated with infection. Our means of pathogen detection (qPCR) has imperfect sensitivity for detection of stealth pathogens (such as Bartonella spp.), likely resulting in under reporting infection in cat tissues and potentially fleas (Maggi et al., 2020; Lashnits et al., 2021). The collection of tissue and not blood samples may restrict comparison to other publications as tissue displays higher Bartonella spp. sensitivity by qPCR (Lashnits et al., 2021). Genus-specific PCR, such as the assays employed herein, are unable to efficiently detect coinfection with more than one species within the genus due to preferential amplification, so we were unable to evaluate coinfection in a single sample (flea or cat), a phenomenon known to occur with flea Wolbachia spp. (Driscoll et al., 2020).
In conclusion, our study documented a significant independent association of flea B. clarridgeiae infection with cat sex and body weight. The cause of this findings is currently unknown and warrants investigation. The lack of agreement between specific cat host and flea pairs was expected on the basis of previous literature; however, geographical correlations of Bartonella spp. presence supports the use of fleas as sentinels to detect the relative proportion of Bartonella spp. in circulation within specific geographic areas. The detection of fleas not traditionally associated with cats (C. simplex, O. howardi, and N. fasciatus) raises questions regarding the importance of these flea species as a cause of allergy or as vectors for pathogen transmission. The association of Rickettsia spp. with specific flea clades should encourage future research regarding flea phylogenetic and coinfection associations with flea-borne pathogen prevalence. Finally, the association of Bartonella spp. with R. asembonensis infection and C. felis genetic diversity should inspire future investigation of the effect of coinfection and vector diversity in pathogen acquisition and maintenance by this ubiquitous vector.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number (s) can be found at: https://doi.org/10.5061/dryad.0k6djhb43.
Ethics statement
The animal study was reviewed and approved by North Carolina State University IACUC protocol #19–003 and #21–468, and University of Wisconsin IACUC protocol #V006461.
Author contributions
CM, EB, and EL conceived and designed experiments, and acquired funding. CM and EL performed data curation, formal analysis, software development, data visualization, and original draft writing. CM, LK, YL, KF, RM, and EL performed sample collection and processing. KF, EB, RM, and EL performed project supervision and administration. All authors reviewed and approved the final manuscript.
Funding
This research was supported by the State of North Carolina and donations to the North Carolina State University College of Veterinary Medicine Bartonella Vector Borne Diseases Research Fund. A portion of this project was completed while CM was supported by the North Carolina State University’s Comparative Medicine Institute Summer Interdisciplinary Research Initiative and the NC State Molecular Biotechnology Training Program of the National Institutes of Health under award number 1T32GM133366. Portions of this project were also completed while EL research was supported by the Comparative Medicine and Translational Research Program of the National Institutes of Health under award number T32OD011130 and the University of Wisconsin School of Veterinary Medicine Companion Animal Fund.
Acknowledgments
This work would not have been possible without the veterinarians, veterinary technicians, and volunteers who assisted in the collection of flea and cat samples while attempting to control the impact of free-roaming cat populations. This includes Shauna Riggs and Joanne Prasse from Paradocs Animal Hospital and Adam Bauknecht from the Madison Cat Project. We would like to thank James Flowers for his assistance in flea identification.
Conflict of interest
In conjunction with S. Sontakke and North Carolina State University, EB holds US Patent No. 7,115,384 Media and Methods for Cultivation of Microorganisms, which was issued on October 3rd, 2006, and also co-founder, shareholder, and Chief Scientific Officer for Galaxy Diagnostics, a company that provides advanced diagnostic testing for the detection of Bartonella spp. infections.
The handling editor MA declared a past co-authorship with the EB and RM.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1137059/full#supplementary-material
References
Abdullah, S., Helps, C., Tasker, S., Newbury, H., and Wall, R. (2019). Pathogens in fleas collected from cats and dogs: distribution and prevalence in the United Kingdom. Parasit. Vectors 12:10. doi: 10.1186/s13071-019-3326-x
Akucewich, L. H., Philman, K., Clark, A., Gillespie, J., Kunkle, G., Nicklin, C. F., et al. (2002). Prevalence of ectoparasites in a population of feral cats from north Central Florida during the summer. Vet. Parasitol. 109, 129–139. doi: 10.1016/S0304-4017(02)00205-4
Álvarez-Fernández, A., Breitschwerdt, E. B., and Solano-Gallego, L. (2018). Bartonella infections in cats and dogs including zoonotic aspects. Parasit. Vectors 11:624. doi: 10.1186/s13071-018-3152-6
Avidor, B., Graidy, M., Efrat, G., Leibowitz, C., Shapira, G., Schattner, A., et al. (2004). Bartonella koehlerae, a new cat-associated agent of culture-negative human endocarditis. J. Clin. Microbiol. 42, 3462–3468. doi: 10.1128/JCM.42.8.3462
Azrizal-Wahid, N., Sofian-Azirun, M., and Low, V. L. (2021). Flea-borne pathogens in the cat flea Ctenocephalides felis and their association with mtDNA diversity of the flea host. Comp. Immunol. Microbiol. Infect. Dis. 75:101621. doi: 10.1016/j.cimid.2021.101621
Barker, E. N. (2019). Update on feline hemoplasmosis. Vet. Clin. North Am. Small Snim. Pract. 49, 733–743. doi: 10.1016/j.cvsm.2019.02.009
Barrs, V. R., Beatty, J. A., Wilson, B. J., Evans, N., Gowan, R., Baral, R. M., et al. (2010). Prevalence of Bartonella species, rickettsia felis, haemoplasmas and the Ehrlichia group in the blood of cats and fleas in eastern Australia. Aust. Vet. J. 88, 160–165. doi: 10.1111/j.1751-0813.2010.00569.x
Bodenhofer, U., Bonatesta, E., Horejš-Kainrath, C., and Hochreiter, S. (2015). Msa: an R package for multiple sequence alignment. Bioinformatics 31, btv494–btv499. doi: 10.1093/bioinformatics/btv494
Breitschwerdt, E. B. (2017). Bartonellosis, one health and all creatures great and small. Vet. Dermatol. 28, 96–e21. doi: 10.1111/vde.12413
Canneti, B., García, J. C. G., Cores, F. J., and Trigo, M. (2019). Neurological presentations of Bartonella henselae infection. Neurol. Sci. 40, 261–268. doi: 10.1007/s10072-018-3618-5
Chandra, S., Forsyth, M., Lawrence, A. L., Emery, D., and Slapeta, J. (2017). Cat fleas (Ctenocephalides felis) from cats and dogs in New Zealand: molecular characterization, presence of rickettsia felis and Bartonella clarridgeiae and comparison with Australia. Vet. Parasitol. 234, 25–30. doi: 10.1016/j.vetpar.2016.12.017
Chomel, B. B., Kasten, R. W., Floyd-Hawkins, K., Chi, B., Yamamoto, K., Roberts-Wilson, J., et al. (1996). Experimental transmission of Bartonella henselae by the cat flea. J. Clin. Microbiol. 34, 1952–1956. doi: 10.1128/jcm.34.8.1952-1956.1996
Dorigatti, I., McCormack, C., Nedjati-Gilani, G., and Ferguson, N. M. (2018). Using Wolbachia for dengue control: insights from modelling. Trends Parasitol. 34, 102–113. doi: 10.1016/j.pt.2017.11.002
Driscoll, T. P., Verhoeve, V. I., Brockway, C., Shrewsberry, D. L., Plumer, M., Sevdalis, S. E., et al. (2020). Evolution of Wolbachia mutualism and reproductive parasitism: insight from two novel strains that co-infect cat fleas. PeerJ 8:e10646. doi: 10.7717/peerj.10646
Flatau, R., Segoli, M., Khokhlova, I., and Hawlena, H. (2018). Wolbachia’s role in mediating its flea’s reproductive success differs according to flea origin. FEMS Microbiol. Ecol. 94:157. doi: 10.1093/femsec/fiy157
Folmer, O., Black, M., Hoeh, W., Lutz, R., and Vrijenhoek, R. (1994). DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 14, 275–140. doi: 10.1071/ZO9660275
Gamer, M., Lemon, J., Fellows, I., and Singh, P. (2019). Irr: Various coefficients of interrater reliability and agreement.
Gutiérrez, R., Nachum-Biala, Y., and Harrus, S. (2015). Relationship between the presence of Bartonella species and bacterial loads in cats and cat fleas (Ctenocephalides felis) under natural conditions. Appl. Environ. Microbiol. 81, 5613–5621. doi: 10.1128/AEM.01370-15
Hattori, N., Kuroda, M., Katano, H., Takuma, T., Ito, T., Arai, N., et al. (2020). Candidatus mycoplasma haemohominis in human, Japan. Emerg. Infect. Dis. 26, 11–19. doi: 10.3201/eid2601.190983
Hawley, J. R., Shaw, S. E., and Lappin, M. R. (2007). Prevalence of rickettsia felis DNA in the blood of cats and their fleas in the United States. J. Feline Med. Surg. 9, 258–262. doi: 10.1016/j.jfms.2006.12.005
Hii, S. F., Kopp, S. R., Abdad, M. Y., Thompson, M. F., O’Leary, C. A., Rees, R. L., et al. (2011). Molecular evidence supports the role of dogs as potential reservoirs for rickettsia felis. Vect. Borne Zoonotic Dis. 11, 1007–1012. doi: 10.1089/vbz.2010.0270
Jeanclaude, D., Godmer, P., Leveiller, D., Pouedras, P., Fournier, P. E., Raoult, D., et al. (2009). Bartonella alsatica endocarditis in a French patient in close contact with rabbits. Clin. Microbiol. Infect. 15, 110–111. doi: 10.1111/j.1469-0691.2008.02187.x
Khoo, J. J., Kurtti, T. J., Husin, N. A., Beliavskaia, A., Lim, F. S., Zulkifli, M. M. S., et al. (2020). Isolation and propagation of laboratory strains and a novel flea-derived field strain of Wolbachia in tick cell lines. Microorganisms 8:19. doi: 10.3390/microorganisms8070988
Kordick, D. L., and Breitschwerdt, E. B. (1988). Persistent infection of pets within a household with three Bartonella species. Emerg. Infect. Dis. 4, 325–328. doi: 10.3201/eid0402.980225
Kordick, D. L., Hilyard, E. J., Hadfield, T. L., Wilson, K. H., Steigerwalt, A. G., Brenner, D. O. N. J., et al. (1997). Bartonella clarridgeiae, a newly recognized zoonotic pathogen causing inoculation papules, fever, and lymphadenopathy (cat scratch disease). J. Clin. Microbiol. 35, 1813–1818. doi: 10.1128/jcm.35.7.1813-1818.1997
La Scola, B., Davoust, B., Boni, M., and Raoult, D. (2002). Lack of correlation between Bartonella DNA detection within fleas, serological results, and results of blood culture in a Bartonella-infected stray cat population. Clin. Microbiol. Infect. 8, 345–351. doi: 10.1046/j.1469-0691.2002.00434.x
Lappin, M. R., Griffin, B., Brunt, J., Riley, A., Burney, D., Hawley, J., et al. (2006). Prevalence of Bartonella species, haemoplasma species, Ehrlichia species, Anaplasma phagocytophilum, and Neorickettsia risticii DNA in the blood of cats and their fleas in the United States. J. Feline Med. Surg. 8, 85–90. doi: 10.1016/j.jfms.2005.08.003
Lashnits, E., Neupane, P., Bradley, J. M., Richardson, T., Maggi, R. G., and Breitschwerdt, E. B. (2021). Comparison of serological and molecular assays for Bartonella species in dogs with hemangiosarcoma. Pathogens 10, 1–18. doi: 10.3390/pathogens10070794
Lawrence, A. L., Brown, G. K., Peters, B., Spielman, D. S., Morin-Adeline, V., and Šlapeta, J. (2014). High phylogenetic diversity of the cat flea (Ctenocephalides felis) at two mitochondrial DNA markers. Med. Vet. Entomol. 28, 330–336. doi: 10.1111/mve.12051
Lawrence, A. L., Webb, C. E., Clark, N. J., Halajian, A., Mihalca, A. D., Miret, J., et al. (2019). Out-of-Africa, human-mediated dispersal of the common cat flea, Ctenocephalides felis: the hitchhiker’s guide to world domination. Int. J. Parasitol. 49, 321–336. doi: 10.1016/j.ijpara.2019.01.001
Lee, S., Kakumanu, M. L., Ponnusamy, L., Vaughn, M., Funkhouser, S., Thornton, H., et al. (2014). Prevalence of Rickettsiales in ticks removed from the skin of outdoor workers in North Carolina. Parasit. Vectors 7:607. doi: 10.1186/s13071-014-0607-2
Legendre, K. P., and Macaluso, K. R. (2017). Rickettsia felis: a review of transmission mechanisms of an emerging pathogen. Trop. Med. Infect. Dis. 2:8. doi: 10.3390/tropicalmed2040064
Maggi, R. G., Richardson, T., Breitschwerdt, E. B., and Miller, J. C. (2020). Development and validation of a droplet digital PCR assay for the detection and quantification of Bartonella species within human clinical samples. J. Microbiol. Methods 176:106022. doi: 10.1016/j.mimet.2020.106022
Maina, A. N., Jiang, J., Luce-Fedrow, A. S., John, H. K., Farris, C. M., and Richards, A. L. (2019). Worldwide presence and features of flea-borne rickettsia asembonensis. Front. Vet. Sci. 5:8. doi: 10.3389/fvets.2018.00334
Manvell, C., Berman, H., Callahan, B., Breitschwerdt, E., Swain, W., Ferris, K., et al. (2022). Identification of microbial taxa present in Ctenocephalides felis (cat flea) reveals widespread co-infection and associations with vector phylogeny. Parasit. Vectors 15:398. doi: 10.1186/s13071-022-05487-1
Manvell, C., Ferris, K., Maggi, R., Breitschwerdt, E. B., and Lashnits, E. (2021). Prevalence of vector-borne pathogens in reproductive and non-reproductive tissue samples from free-roaming domestic cats in the South Atlantic United States. Pathogens 10:17. doi: 10.3390/pathogens10091221
Mifsud, M., Takacs, N., Gyurkovszky, M., Solymosi, N., and Farkas, R. (2020). Detection of flea-borne pathogens from cats and fleas in a Maltese shelter. Vect. Borne Zoonotic Dis. 20, 529–534. doi: 10.1089/vbz.2019.2553
Mullins, K. E., Maina, A. N., Krueger, L., Jiang, J., Cummings, R., Drusys, A., et al. (2018). Rickettsial infections among cats and cat fleas in Riverside County, California. Am. J. Trop. Med. 99, 291–296. doi: 10.4269/ajtmh.17-0706
Ng-Nguyen, D., Hii, S. F., Hoang, M. T. T., Nguyen, V. A. T., Rees, R., Stenos, J., et al. (2020). Domestic dogs are mammalian reservoirs for the emerging zoonosis flea-borne spotted fever, caused by Rickettsia felis. Sci. Rep. 10:4151. doi: 10.1038/s41598-020-61122-y
Pagès, H., Aboyoun, P., Gentleman, R., and DebRoy, S. (2021). Efficient manipulation of biological strings Biostrings.
Paradis, E. (2010). Pegas: an R package for population genetics with an integrated-modular approach. Bioinformatics 26, 419–420. doi: 10.1093/bioinformatics/btp696
Paradis, E., and Schliep, K. (2019). Ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 35, 526–528. doi: 10.1093/bioinformatics/bty633
Persichetti, M. F., Solano-Gallego, L., Serrano, L., Altet, L., Reale, S., Masucci, M., et al. (2016). Detection of vector-borne pathogens in cats and their ectoparasites in southern Italy. Parasit. Vectors 9:247. doi: 10.1186/s13071-016-1534-1
Phoosangwalthong, P., Hii, S. F., Kamyingkird, K., Kengradomkij, C., Pinyopanuwat, N., Chimnoi, W., et al. (2018). Cats as potential mammalian reservoirs for rickettsia sp. genotype RF2125 in Bangkok Thailand. Vet. Parasitol. Reg. Stud. Rep. 13, 188–192. doi: 10.1016/j.vprsr.2018.07.001
Puges, M., Ménard, A., Berard, X., Geneviève, M., Pinaquy, J. B., Edouard, S., et al. (2019). An unexpected case of Bartonella alsatica prosthetic vascular graft infection. Infect. Drug Res. 12, 2453–2456. doi: 10.2147/IDR.S206805
Raoult, D., Roblot, F., Rolain, J. M., Besnier, J. M., Loulergue, J., Bastides, F., et al. (2006). First isolation of Bartonella alsatica from a valve of a patient with endocarditis. J. Clin. Microbiol. 44, 278–279. doi: 10.1128/JCM.44.1.278-279.2006
Reif, K. E., and Macaluso, K. R. (2009). Ecology of Rickettsia felis: a review. J. Med. Entomol. 46, 723–736. doi: 10.1603/033.046.0402
Richards, A. L., Jiang, J., Omulo, S., Dare, R., Abdirahman, K., Ali, A., et al. (2010). Human infection with Rickettsia felis, Kenya. Emerg. Infect. Dis. 16, 1081–1086. doi: 10.3201/eid1607.091885
Richter, J., Fournier, P., Petridou, J., and Häussinger, D. (2002). Rickettsia felis infection acquired in Europe and documented by polymerase chain reaction. Emerg. Infect. Dis. 8, 207–208. doi: 10.3201/eid0802.010293
Rust, M. K. (2017). The biology and ecology of cat fleas and advancements in their pest management: a review. Insects 8:51. doi: 10.3390/insects8040118
Šlapeta, Š., and Šlapeta, J. (2016). Molecular identity of cat fleas (Ctenocephalides felis) from cats in Georgia, United States carrying Bartonella clarridgeiae, Bartonella henselae, and Rickettsia sp. RF2125. Vet. Parasitol. Reg. Stud. Rep. 3, 36–40. doi: 10.1016/j.vprsr.2016.06.005
Taber, R., Pankowski, A., Ludwig, A. L., Jensen, M., Magsamen, V., and Lashnits, E. (2022). Bartonellosis in dogs and cats, an update. Vet. Clin. North Am. Small Anim. Pract. 52, 1163–1192. doi: 10.1016/j.cvsm.2022.06.006
Tasker, S. (2022). Hemotropic mycoplasma. Vet. Clin. North Am. Small Anim. Pract. 52, 1319–1340. doi: 10.1016/j.cvsm.2022.06.010
Thomas, J. E., Staubus, L., Goolsby, J. L., and Reichard, M. (2016). Ectoparasites of free-roaming domestic cats in the Central United States. Vet. Parasitol. 228, 17–22. doi: 10.1016/j.vetpar.2016.07.034
Tsirogiannis, C., and Sandel, B. (2016). PhyloMeasures: a package for computing phylogenetic biodiversity measures and their statistical moments. Ecography 39, 709–714. doi: 10.1111/ecog.01814
Tsirogiannis, C., and Sandel, B. (2017). PhyloMeasures: Fast and exact algorithms for computing phylogenetic biodiversity measures.
Tyrrell, J. D., Qurollo, B. A., Mowat, F. M., and Kennedy-Stoskopf, S. (2020). Molecular prevalence of selected vector-borne organisms in captive red wolves (Canis rufus). J. Zoo Wildl. Med. 51, 663–667. doi: 10.1638/2019-0162
Tyrrell, J. D., Qurollo, B. A., Tornquist, S. J., Schlaich, K. G., Kelsey, J., Chandrashekar, R., et al. (2019). Molecular identification of vector-borne organisms in Ehrlichia seropositive Nicaraguan horses and first report of Rickettsia felis infection in the horse. Acta Trop. 200:105170. doi: 10.1016/j.actatropica.2019.105170
Werren, J. H., Baldo, L., and Clark, M. E. (2008). Wolbachia: master manipulators of invertebrate biology. Nat. Rev. Microbiol. 6, 741–751. doi: 10.1038/nrmicro1969
Wickham, H., Averick, M., Bryan, J., Chang, W., McGowan, L. D., François, R., et al. (2019). Welcome to the tidyverse. J. Open Source Softw. 4:1686. doi: 10.21105/joss.01686
Woods, J. E., Wisnewki, N., and Lappin, M. R. (2006). Attempted transmission of Candidatus Mycoplasma haemominutum and Mycoplasma haemofelis by feeding cats infected Ctenocephalides felis. Am. J. Vet. Res. 67, 494–497. doi: 10.2460/ajvr.67.3.494
Keywords: Bartonella, flea, Wolbachia, vector phylogenetics, flea associated Rickettsia, host-vector agreement, cat flea
Citation: Moore C, Breitschwerdt EB, Kim L, Li Y, Ferris K, Maggi R and Lashnits E (2023) The association of host and vector characteristics with Ctenocephalides felis pathogen and endosymbiont infection. Front. Microbiol. 14:1137059. doi: 10.3389/fmicb.2023.1137059
Edited by:
Marcos Rogério André, São Paulo State University, BrazilReviewed by:
Algimantas Paulauskas, Vytautas Magnus University, LithuaniaFrederic Beugnet, Boehringer Ingelheim, France
Monika Danchenko, University of South Alabama, United States
Copyright © 2023 Moore, Breitschwerdt, Kim, Li, Ferris, Maggi and Lashnits. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Erin Lashnits, bGFzaG5pdHNAd2lzYy5lZHU=
 Charlotte Moore
Charlotte Moore Edward B. Breitschwerdt
Edward B. Breitschwerdt Lisa Kim2
Lisa Kim2 Ricardo Maggi
Ricardo Maggi