
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 05 April 2023
Sec. Terrestrial Microbiology
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1134331
There are serious soil sickness in ginseng cultivation. Crop rotation is an effective agricultural management to improve soil sustainability and reduce soil sickness. To explore an appropriate ginseng rotation system, Medicago sativa (alfalfa) and Perilla frutescens (perilla) were planted on ginseng cultivation soil for 1 year to evaluate the improvement effect of both. Through chemical analysis and high-throughput sequencing technology, we found that after alfalfa and perilla cultivation for one-year, various nutrients and enzyme activities in ginseng cultivation soil were significantly improved. In addition, perilla significantly increased the diversity and richness of soil fungal communities. Cultivation of alfalfa and perilla significantly changed the composition of soil bacterial and fungal communities and significantly reduced the abundance of the potentially pathogenic fungi Ilyonectria. Further pot experiments also showed that the improved soil could significantly increase root activity of ginseng plant after two plants were planted. It should be noted that, unlike alfalfa, perilla decreased soil electrical conductivity, increased soil organic matter, soil urease, and may significantly improve the diversity and richness of soil fungal community. Moreover, in the pot experiment, the root fresh weight of ginseng cultured in perilla treated soil increased significantly. This study highlights that perilla may have better soil improvement effect than alfalfa and it has the potential to be used in the soil improvement of ginseng cultivation.
Ginseng (Panax ginseng Meyer) is one of the most famous Chinese herbal medicine in China with a history of thousands of years (Xiang et al., 2008; Yun, 2009; Yang et al., 2017). Ginseng is now widely cultivated in northeast China, Korea, and Japan (Baeg and So, 2013; Ru et al., 2015). Artificial cultivation of ginseng usually takes 4 to 6 years and requires extremely high soil quality (Proctor and Bailey, 1987). More importantly, due to allelopathy and soil fertility changes, ginseng is subject to substantial soil sickness, which seriously affects the yield of ginseng (Xiao et al., 2016). After ginseng has been planted, the soil is often called “old ginseng soil.” In this soil, it is difficult for ginseng to grow again and difficult for many other crops to grow normally, which affects the utilization rate of land. So far, the mechanism underlying soil sickness of ginseng is not completely clear. Scholars believe that the occurrence of soil sickness may be related to the effect of ginseng exudates on soil’s physical and chemical properties and microbial community (Zhang and Xue, 2010; Li Q. et al., 2021).
In fact, research has proven that farming systems relying on crop rotation to improve plant nutrition and control weeds, pests and plant pathogens are an ancient but attractive and environmentally friendly approach (Altieri, 2018). Implementing scientific crop rotation or effective soil improvement methods is the most effective measure to reduce or eliminate soil sickness (Li C. et al., 2021). The underlying mechanisms include improving soil physicochemical properties, increasing the diversity of soil microbiota, and associated beneficial microorganisms, and breaking the natural life cycle of soil-borne and airborne pathogens, as well as reducing the harmful effects of autotoxic compounds released by crops (Mäder et al., 2002; Peters et al., 2003; Gong et al., 2021; Woo et al., 2022). Therefore, it has become vital to find one or several crops that can generally grow after ginseng cultivation and positively impact the soil’s physicochemical properties and microbial community. Furthermore, the relevant research is of great significance to the development of the ginseng crop rotation system and the improvement of ginseng cultivation soil. Perilla (Perilla frutescens L.) is widely used as medicine and food in various Asian countries (Ahmed, 2019; Wei et al., 2022). It is also an adaptive and large biomass plant (Xiao et al., 2020). In previous studies, American ginseng (Panax quinquefolium L.)-perilla rotation could effectively resolve the continuous cropping problem of American ginseng (Zhao et al., 2005). Recent studies have found that perilla has a good potential in dealing with environmental pollution (Shu et al., 2021). The legume alfalfa (Medicago sativa L.) is a “microbial hotspot crop” (Baslam et al., 2012). Cultivation of alfalfa produces a large amount of root biomass, generating root exudates that recruit soil bacterial populations to the rhizosphere, so it is often used in crop rotations (Tesfaye et al., 2003; Samaddar et al., 2021). Previous studies have shown that alfalfa fixes atmosphere N2 to reduce or eliminate the need for synthetic N fertilizer and effectively increase crop yields in crop rotations (Fernandez et al., 2019). More importantly, perilla and alfalfa often grow in wild or semi-wild forms in ginseng fields. Therefore, these two plants may not be sensitive to the allelopathic effect produced by ginseng. At present, the effects of planting other crops on soil fertility, physicochemical properties, and microbial communities of soils after ginseng cultivation are not clear. In addition, there are no reports on the application of perilla or alfalfa to improve the soil after ginseng cultivation.
In this context, it is reasonable to speculate that the cultivation of perilla or alfalfa may improve the soil quality of ginseng cultivation. Therefore, in this study, perilla and alfalfa were used, respectively, to conduct one-year planting experiments on ginseng cultivation soil. Moreover, physical and chemical properties and microbial communities in perilla and alfalfa cultivated soils were analyzed, and compared with that in the untreated ginseng cultivation soil. Through this work, we will achieve the following objectives: (a) to compare the soil fertility and bacterial and fungal community composition among ginseng cultivation soil planted with perilla for 1 year, ginseng cultivation soil planted with alfalfa for 1 year, and untreated ginseng cultivation soil; (b) to analyze the improvement effect of two crops, perilla and alfalfa, on ginseng cultivation soil, and to provide data and theoretical basis for ginseng cultivation soil improvement and ginseng crop rotation strategy.
The field experiment was carried out on an experimental farm in Choushui Village, Fusong County, Jilin Province, China (42.40′ N and 127.09′ E). Fusong region is the production area of genuine ginseng medicinal materials and is known as the hometown of ginseng in China (Liu et al., 2011). In detail, the experimental site of this study has a temperate monsoon climate at an altitude of 410 m. The average annual temperature is 4°C, with an average temperature of −33°C in January and 30°C in July. The average annual frost-free period is 120 days, and the average annual precipitation is 800 mm.
The experiment field was cultivated for 4 years from 2017 to 2020, and the ginseng was taken out in 2020. The whole process of ginseng cultivation strictly follows the local standards of “ginseng safe production technical specification of pesticide application (DB22/T 1233–2019).”
The field where ginseng had been cultivated was then divided into 3 groups with 4 plots in each group: (1) no treatment (i.e., control check; n = 4); (2) perilla cultivation (n = 4); (3) alfalfa cultivation (n = 4). There are 12 plots in total, each with an area of 10 m2 and with distance of 0.5 m between adjacent plots. In addition, the same group of plots are not directly adjacent (detailed plots distribution is shown in Supplementary Figure S1). Perilla and alfalfa were cultivated for 1 year (from August 2020 to August 2021). The seeds of Perilla frutescens Britt. var. frutescens and Medicago sativa subsp. falcata were purchased from Changjingyuanlin Co. LTD (Jiangsu, China). Simply put, after the field was prepared, the seeds were sown. Considering production requirements, the growth density of perilla is about 5 plants per square meter, and that of alfalfa is about 150 plants per square meter. All crops were planted simultaneously and irrigated (it is usually watered every 5 days to keep the soil moist and drained when it rains more) to ensure their survival during the beginning of the growing period. Moreover, all experiment plots were not treated with fertilizer or other manual interventions except for regular weed cleaning.
Samples of soil were collected from each plot 1 year after planting on the same day, 5 August 2021. Sampling was carried out based on the above experimental design. In each plot, five subsamples were randomly collected at a depth of 0–20 cm, which were mixed into one soil sample. Therefore, we finally obtained four soil samples from each treatment. After all soil samples were passed through a 2 mm sieve, part of each sample was frozen at −80°C for DNA extraction. The other part was air-dried at room temperature to determine chemical properties and enzyme activities.
The soil pH of all samples was determined (soil-water ratio of 1:2.5) by using a pH meter. Moreover, soil electrical conductivity (EC) was determined by a conductivity meter (soil-water ratio of 1:5). The potassium dichromate oxidation-external heating method was used to measure soil organic matter (Fan et al., 2017). Soil available nitrogen was analyzed using the alkaline hydrolysis method (Gianello and Bremner, 1986). Soil available phosphorus was determined by the NaHCO3 extraction molybdenum antimony colorimetry method (Olsen, 1954). Soil available potassium was determined by the NH4OAc extraction fame photometric method (Yan et al., 2021b).
The soil urease, acid phosphatase, sucrase, catalase, and laccase activities were determined using the soil enzyme kit from Solarbio Science and Technology Co. (Beijing, China). Briefly, the soil urease assay was performed with method of indophenol blue colorimetry; the determination of acid phosphatase was based on the disodium phenyl phosphate colorimetry; sucrase activity was estimated by the 3,5-dinitrosalicylic acid colorimetry; the activity of catalase was determined by KMnO4 titration; the laccase activity was determined by ABTS colorimetry.
The bacterial and fungal DNA from 0.5 g of soil was extracted in triplicate using A.E.Z.N.A. Soil DNA Kit (Omega Bio-tek Inc., United States), followed by agarose gel electrophoresis to detect the quality of DNA (Pan et al., 2020). Then, the DNA concentration was determined by NanoDrop (2000) UV–vis spectrophotometer (Thermo Scientific, United States). To identify bacteria, the V3–V4 regions primers 338F (50-ACTCCTACGGGAGGCAGCAG-30) and 806R (50-GGACTACHVGGGTWTCTAAT-30) were selected to amplify the soil bacterial 16S rRNA gene (Zhang et al., 2020). For fungi, the Internal Transcribed Spacer (ITS) regions of the fungal gene were amplified with the primers ITS1F (5′-CTTGGTCATTTAGAGG AAGTAA-3′)—ITS2R (5′-GCTGCGTTCTTCATCGATGC-3′; Liang et al., 2021). The PCR amplification was performed as follows: 95°C for 3 min, 95°C for 30 s, 55°C for 30 s, and 72°C for 45 s (27 cycles for bacteria and 35 cycles for fungi), followed by 10 min at 72°C.
The PCR products were purified by an AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, United States) and quantified by the quantitative analyzer™ Fluorometer (Promega, United States). All purified PCR products were used for Illumina MiSeq high-throughput sequencing with Illumina’s MiSeq PE300 platform at the Majorbio Bio-pharm Technology Co., Ltd. (Shanghai, China). The raw sequencing data have been uploaded to the NCBI Sequence Read Archive database (Bacterial Accession Number: PRJNA820351; Fungal Accession Number: PRJNA820493).
Paired-end reads were assigned to samples based on their unique barcode and truncated by cutting off the barcode and primer sequence. Raw reads were filtered to obtain high-quality reads with the QIIME1 software (version 1.9.1, http://qiime.org/install/index; Caporaso et al., 2010). The high-quality reads with >97% sequence similarity were classified into different operational taxonomic units (OTUs) using UPARSE (version 7.1, http://www.drive5.com/uparse/; Edgar, 2013). The taxonomy of each bacterial and fungal gene sequence was analyzed using the Ribosomal Database Project (RDP) Classifier (version 2.11, http://rdp.cme.msu.edu/) against the Silva128 16S rRNA database1 and Unite 7.0 ITS fungi database,2 respectively, with a 70% confidence threshold (Cole et al., 2009; Pan et al., 2020).
Soil bacterial functions were predicted using the PICRUSt tool based on 16S rRNA sequencing data, and biological functions were annotated in the KEGG database (Kanehisa and Goto, 2000; Langille et al., 2013; Gong et al., 2021). In addition, the FUNGuild tool was used for the functional prediction and classification of soil fungi (Nguyen et al., 2016; Schmidt et al., 2019).
To further verify the improvement effect of alfalfa and perilla cultivation on ginseng cultivation soil, a pot experiment was conducted. After the one-year field experiment was finished, 20 kg of soil were collected treatment (no treatment, alfalfa, and perilla cultured treatments) for the pot experiment. To put it simply, a total of 18 polypropylene pots (diameter 24 cm, height 15 cm) were divided into 3 groups, with 6 pots in each group. Each group was loaded with corresponding soil collected in the field, 2,500 g of soil in each pot. Then, 2-year-old ginseng seedlings with similar root development and growth were selected for cleaning, and their roots were disinfected with sodium hypochlorite (Sun et al., 2018). Three ginseng seedlings were planted in each pot for 6 weeks. Watering and weeding were applied regularly.
After 6 weeks of growth, ginseng plant was collected and the aboveground part was removed from the stem base to obtain ginseng roots. The biomass (fresh weight and dry weight) of each group was measured. In addition, the root activity was determined by the 2,3,5-triphenyl tetrazolium chloride (TTC) method (Zhan et al., 2021). Root activity is expressed as the mass of triphenyl formazan produced per gram of fresh root per hour.
Based on OTUs, the alpha diversity of microbial communities was quantified, including richness (ACE and Chao1) and diversity (Shannon and Simpson) indexes. Beta-diversity was used for comparative analysis of microbial community composition in different groups. Principal co-ordinates analysis (PCoA) and redundancy analysis (RDA) was conducted using the R software (version 4.1.0, https://cran.r-project.org/; Dixon, 2003). LEfSe analysis (Linear Discriminant Analysis Effect Size) was used to elucidate the biomarkers in each group.
Data on soil physical and chemical properties, enzyme activities, and soil microbial alpha diversity were analyzed using the SPSS software (IBM Corporation, Armonk, NY, United States). The results were expressed as the arithmetic mean value ± standard deviation. The differences in the means were compared by the One-way ANOVA at p < 0.05.
Analysis of soil properties showed that pH of the perilla group was significantly higher than that of the control group, while the alfalfa group showed no significant change (Figure 1A). The EC values in the perilla group were significantly lower than those in the control group; in contrast, the EC values in the alfalfa group were significantly higher than those in the control group (Figure 1B). Soil organic matter was significantly higher in the perilla group than that in the control group (Figure 1C). Soil available nitrogen and available potassium were significantly higher in both the perilla and alfalfa groups than that in the control group (Figures 1D,F). And, available potassium was significantly higher in the alfalfa group than that in the perilla group (Figure 1F). Furthermore, there was no significant difference in available phosphorus among all the groups (Figure 1E).
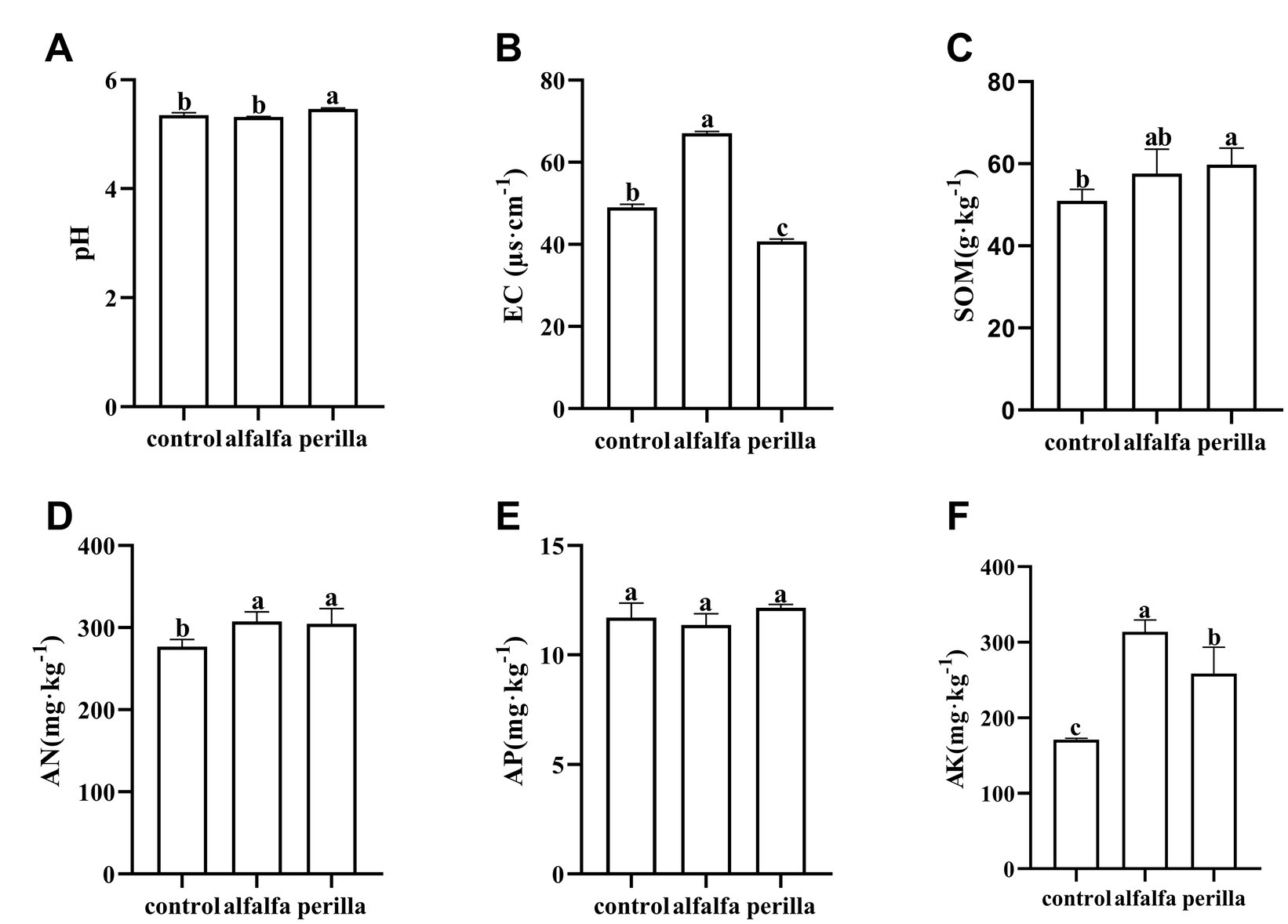
Figure 1. Physicochemical properties and nutrient content of the soil in different treatments. Control, no treatment; Alfalfa, alfalfa cultivation; Perilla, perilla cultivation. (A) pH; (B) EC, electric conductivity; (C) SOM, soil organic matter; (D) AN, available nitrogen; (E) AP, available phosphorus; (F) AK, available potassium. Error represents the standard deviation; different letters indicate a significant difference at the p < 0.05 level.
The soil urease activity in the perilla group was significantly higher than that in the control group (Figure 2A). In addition, the acid phosphatase and laccase activities in both the perilla and alfalfa groups were significantly higher than those in the control group, and the acid phosphatase activity in alfalfa was also significantly higher than that in the perilla group (Figures 2B,E). In contrast, the sucrase activity in the perilla group was significantly lower than that in the control group (Figure 2C). The catalase activity of the perilla group was significantly higher than that of the alfalfa group. In contrast, the catalase activity of these two groups was not significantly different from that of the control group (Figure 2D).
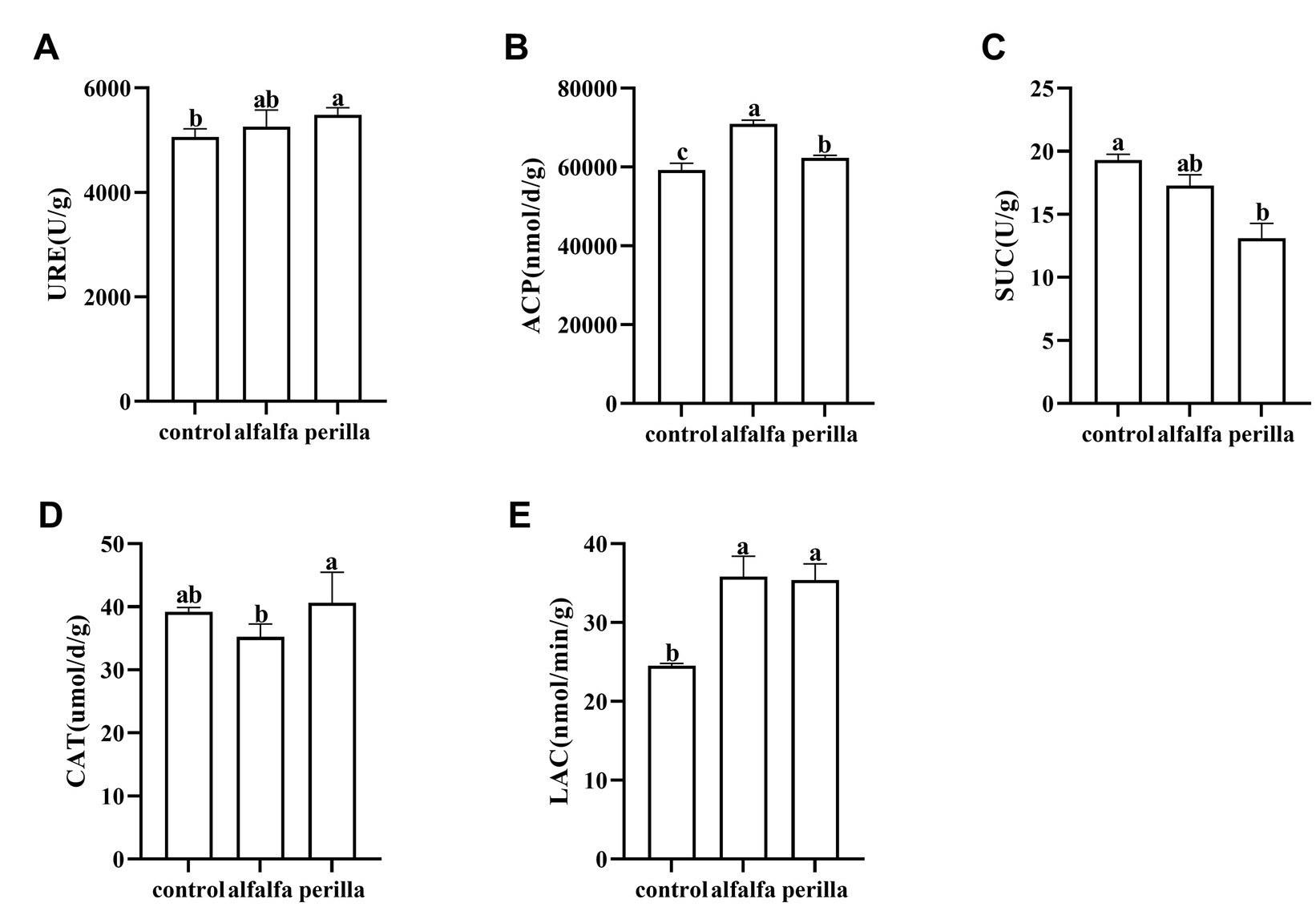
Figure 2. Enzyme activities of the soil in different treatments. Control, no treatment; Alfalfa, alfalfa cultivation; perilla, perilla cultivation. (A) URE, urease; (B) ACP, acid phosphatase; (C) SUC, sucrase; (D) CAT, catalase; (E) LAC, laccase. Error represents the standard deviation; different letters indicate a significant difference at the p < 0.05 level.
There were no significant differences in Shannon, Simpson, ACE, and Chao1 indexes of bacteria among different treatments. Furthermore, the coverage index of each group was more than 0.996 (Figure 3A). In terms of fungi, the Shannon index was significantly higher in the perilla group than that in the control group, while the Simpson index was not significantly different among the groups. Both ace and chao1 indexes were significantly higher in the perilla group than that in the control and alfalfa groups. In addition, the coverage indexes of the three groups were all greater than 0.994 (Figure 3B). Overall, alfalfa cultivation had no significant effect on the diversity and richness of microbial communities in ginseng cultivation soils. In contrast, the cultivation of perilla increased the diversity and richness of fungal communities in ginseng cultivation soils.
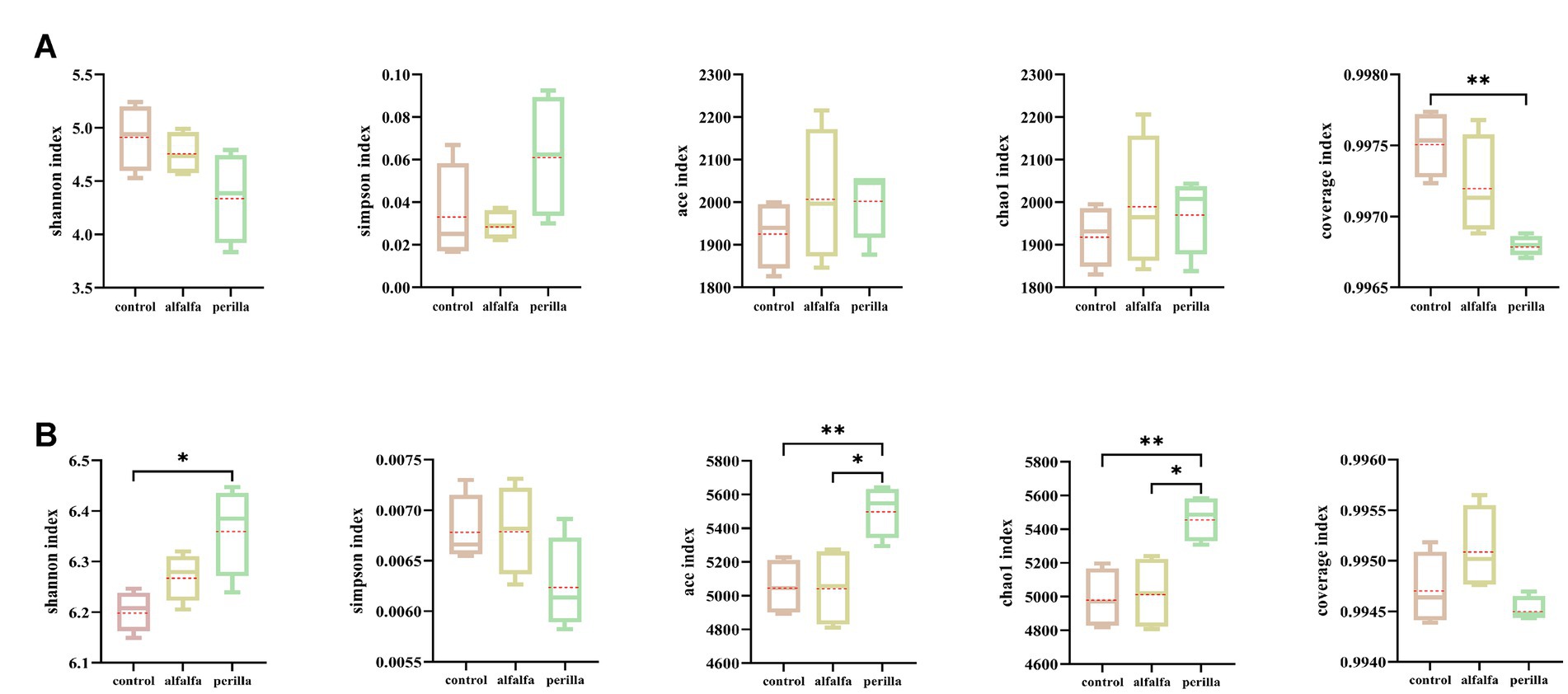
Figure 3. Soil (A) bacterial and (B) fungal alpha diversity in different treatments. Control, no treatment; Alfalfa, alfalfa cultivation; Perilla, perilla cultivation. “*“and “**” indicate a significant difference at the p < 0.05 and p < 0.01 level, respectively.
The relative abundance of bacterial and fungal communities at the phylum level and genus level were counted for each group. The relative abundance at the bacterial phylum level showed that Actinobacteria were the dominant phylum, followed by Proteobacteria and Acidobacteriota (Figure 4A). In addition, we found that Actinobacteria and Proteobacteria were significantly higher in the alfalfa group than that in the other two groups. On the contrary, the abundance of Acidobacteriota in the alfalfa group was significantly lower than that in the other two groups (Supplementary Figure S2). The most abundant bacterial at the genus level were Candidatus Udaeobacter, followed by norank-o-Gaiellales and Gaiella (Figure 4C). For fungi, the highest relative abundance at the phylum level was found in Ascomycota, followed by Basidiomycota and Mortierellomycota (Figure 4B). The Mortierellomycota abundance in the alfalfa and perilla groups was significantly lower than that in the control group (Supplementary Figure S3). The fungal genus with the highest relative abundance was Mortierella, followed by Trichocladium and Saitozyma (Figure 4D). We also found that the abundance of a common ginseng pathogen genus, Ilyonectria, was significantly lower in the alfalfa and perilla groups than that in the control group (Supplementary Figure S4).
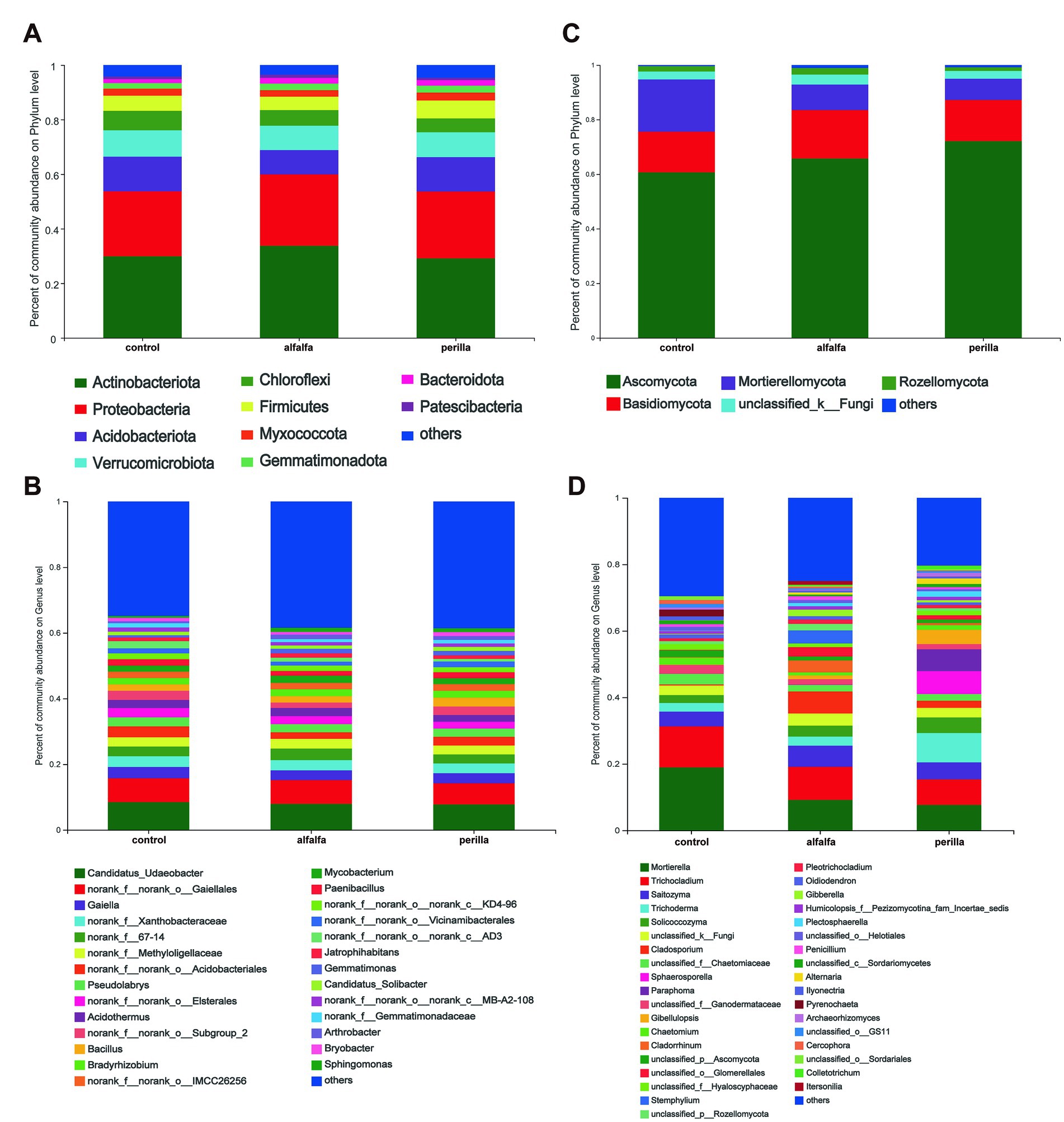
Figure 4. Relative abundances of (A) bacterial and (B) fungal phylum level and (C) bacterial and (D) fungal genus level in different treatments. Control, no treatment; Alfalfa, alfalfa cultivation; Perilla, perilla cultivation.
PCoA and ANOSIM at the OTU level clearly distinguished between soil bacterial (R = 0.773, p = 0.001) and fungal (R = 0.683, p = 0.001) communities in different groups (Figure 5). The first two axes (PC1 and PC2) explained 43.12% and 18.76% in the bacterial community, respectively (Figure 5A). The PCoA of the fungal community was variance explained by PC1 and PC2 for 33.46% and 21.73%, respectively (Figure 5B).

Figure 5. PCoA of (A) bacterial and (B) fungal communities in different treatments. Control, no treatment; Alfalfa, alfalfa cultivation; Perilla, perilla cultivation.
The Lefse tool analyzed indicator microbes in each microbial community group. As shown in Figure 6A, the indicator bacteria of the control group are the order Elsterales, the phylum Acidobacteriota (including the orders Subgroup 2 and Acidobacteriales), and the phylum Chloroflexi (including the class AD3). The indicator bacteria of the alfalfa group are the phylum Proteobacteria (including the orders Sphingomonadales and Burkholderiales), the phylum Bacteroidota and the phylum Actinobacteriota (including the orders Micrococcales and Corynebacteriales). Furthermore, the indicator bacterial taxa of the perilla group are the family Bacillaceae and the order Vicinamibacteria (including the order Vicinamibacterales; Figure 6A).

Figure 6. LEfSe analysis of (A) bacterial and (B) fungal communities in different treatments. Control, no treatment; Alfalfa, alfalfa cultivation; Perilla, perilla cultivation.
In the Lefse analysis of fungal, the indicator taxa of the control group are the phylum Mortierellomycota, the phylum Leotiomycetes (including the orders Helotiales and Chaetothyriales), the order Agaricales, and the families Cucurbitariaceae, Clavicipitaceae, and Bionectriaceae. The indicator fungi of the alfalfa group are the phylum Microbotryomycetes (including the orders Heterogastridiales and Cystofilobasidiales), the order Capnodiales (including the families Cladosporiaceae and Dissoconiaceae), and the families Didymellaceae, Pleosporaceae, and Nectriaceae. The indicator fungi of the perilla group are the classes Pezizomycetes, Archaeorhizomycetes, and the order Glomerellales (including the genera Gibellulopsis and Plectosphaerella; Figure 6B). The LDA results for each group of bacterial and fungal communities are shown in Supplementary Figures S5A,B, respectively.
The first two axes of the RDA model accounted for 50.92% and 37.59% of the total variance in the bacterial and fungal communities, respectively (Figures 7A,B). The degree of influence of environmental factors on the bacterial community was as follows: EC (R2 = 0.8976, p = 0.001) > pH (R2 = 0.8651, p = 0.001) > available phosphorus (R2 = 0.5346, p = 0.031) > available potassium (R2 = 0.5350, p = 0.051) > available nitrogen (R2 = 0.2823, p = 0.223) > soil organic matter (R2 = 0.2581, p = 0.262; Supplementary Table S1). On the other hand, pH (R2 = 0.6504, p = 0.002) and EC (R2 = 0.4233, p = 0.029) were the key chemical factors influencing soil fungal communities (Supplementary Table S2).

Figure 7. RDA analysis of (A) bacterial and (B) fungal and Pearson’s correlation analysis of (C) bacterial and (D) fungal with soil variables. “*,” “**,” and “***” indicate a significant difference at the p < 0.05, p < 0.01, and p < 0.001 level, respectively. Control, no treatment; Alfalfa, alfalfa cultivation; Perilla, perilla cultivation.
The Pearson correlation analysis between bacterial phylum levels and soil environmental factors showed that the relative abundance of the Actinobacteriota was significantly positively correlated with EC (p < 0.01) and acid phosphatase (p < 0.01) and negatively correlated with pH (p < 0.01). The relative abundance of Proteobacteria was significantly and positively correlated with EC (p < 0.01), available potassium (p < 0.01), and acid phosphatase (p < 0.001). In addition, Acidobacteriota was significantly negatively correlated with EC (p < 0.001), available potassium (p < 0.05) and acid phosphatase (p < 0.001; Figure 7C). In terms of fungi, the relative abundance of Ascomycota was significantly positively correlated with urease (p < 0.05), which in turn was significantly negatively correlated with sucrase (p < 0.05). Basidiomycota was significantly negatively correlated with available phosphorus (p < 0.05). The relative abundance of Mortierellomycota was significantly positively correlated with sucrase (p < 0.05), while it was significantly negatively correlated with both AK (p < 0.05), LAC (p < 0.001) and acid phosphatase (p < 0.05; Figure 7D).
There are six main types of primary functional layers, including Cellular Processes, Environmental Information Processing, Genetic Information Processing, Human Diseases, Metabolism, and Organismal Systems (Figure 8). Further, 41 secondary functional layers were found, and the top 20 with the highest abundance are shown in Supplementary Figure S6. Among them, the abundance of the alfalfa group in Carbohydrate Metabolism, Replication and Repair, and Energy Metabolism pathways was significantly lower than that of the control group (p < 0.05). Functional prediction analysis of each group of fungal communities was carried out by FUNGuild. The top 10 fungal functional groups are shown in Table 1. In this result, Endophyte/Litter Saprotroph/Soil Saprotroph/Undefined Saprotroph was significantly higher in the control group than in the other two groups (p < 0.05). On the contrary, Animal Pathogen/Endophyte/Lichen Parasite/Plant Pathogen/Wood Saprotroph in the control group was also significantly lower than that in the other two groups (p < 0.05). In addition, Fungal Parasite/Undefined Saprotroph was significantly higher in the alfalfa group than that in the control group (p < 0.05).
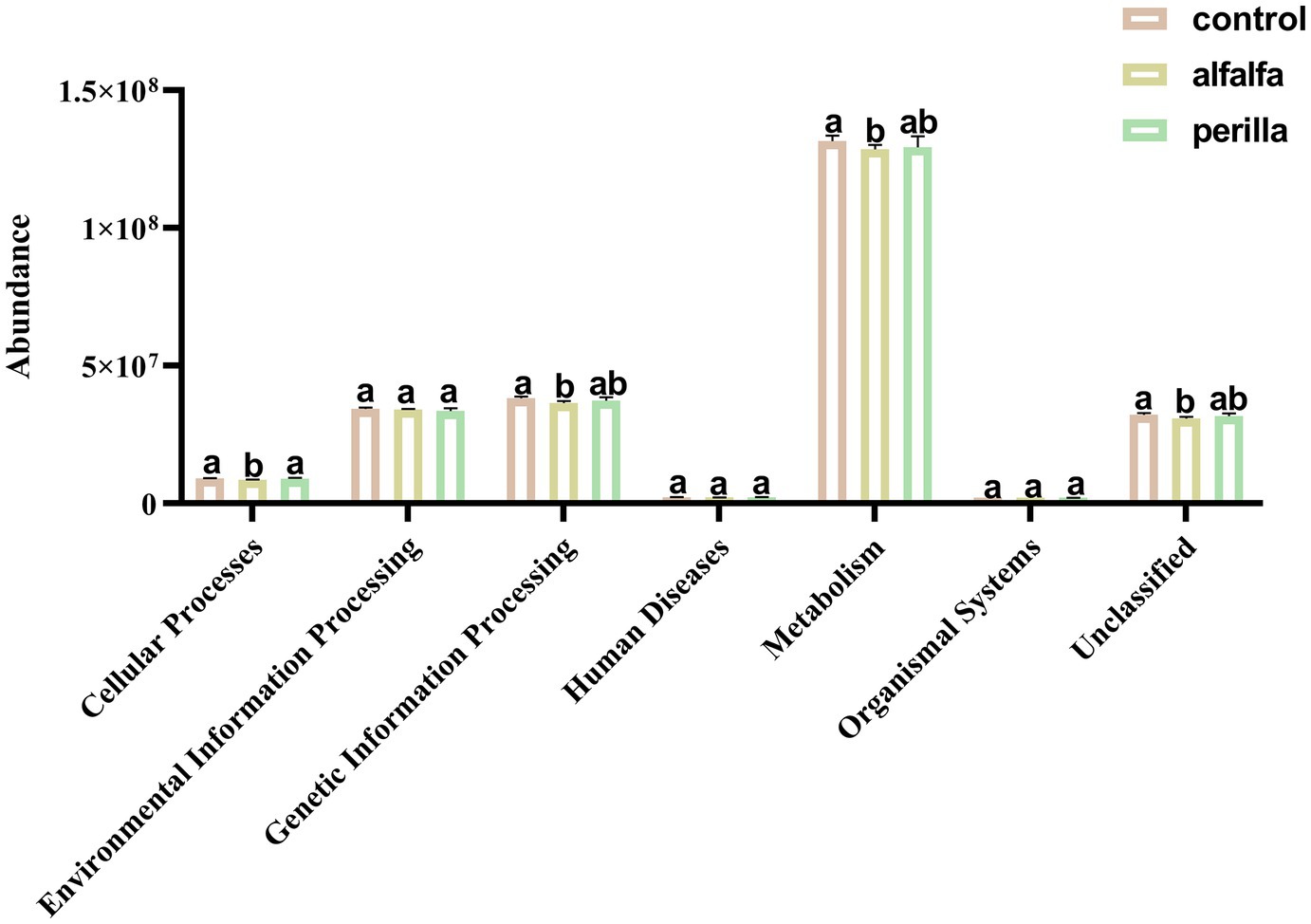
Figure 8. Soil bacterial function prediction in different treatments (Hierarchy level 1). Different letters indicate a significant difference at the p < 0.05 level. Control, no treatment; Alfalfa, alfalfa cultivation; Perilla, perilla cultivation.
We conducted a pot experiment to detect root biomass and root activity of ginseng cultivated in different treatment groups of soil. After sampling, the biomass of fresh and dry root was determined (Figure 9A). The fresh weight of ginseng root in perilla group was significantly higher than that in control group, but there was no significant difference between the other groups. The root activity of ginseng cultured in soil improved by 1 year-cultivation of alfalfa and perilla was significantly higher than that of untreated soil (control; p < 0.05; Figure 9B).
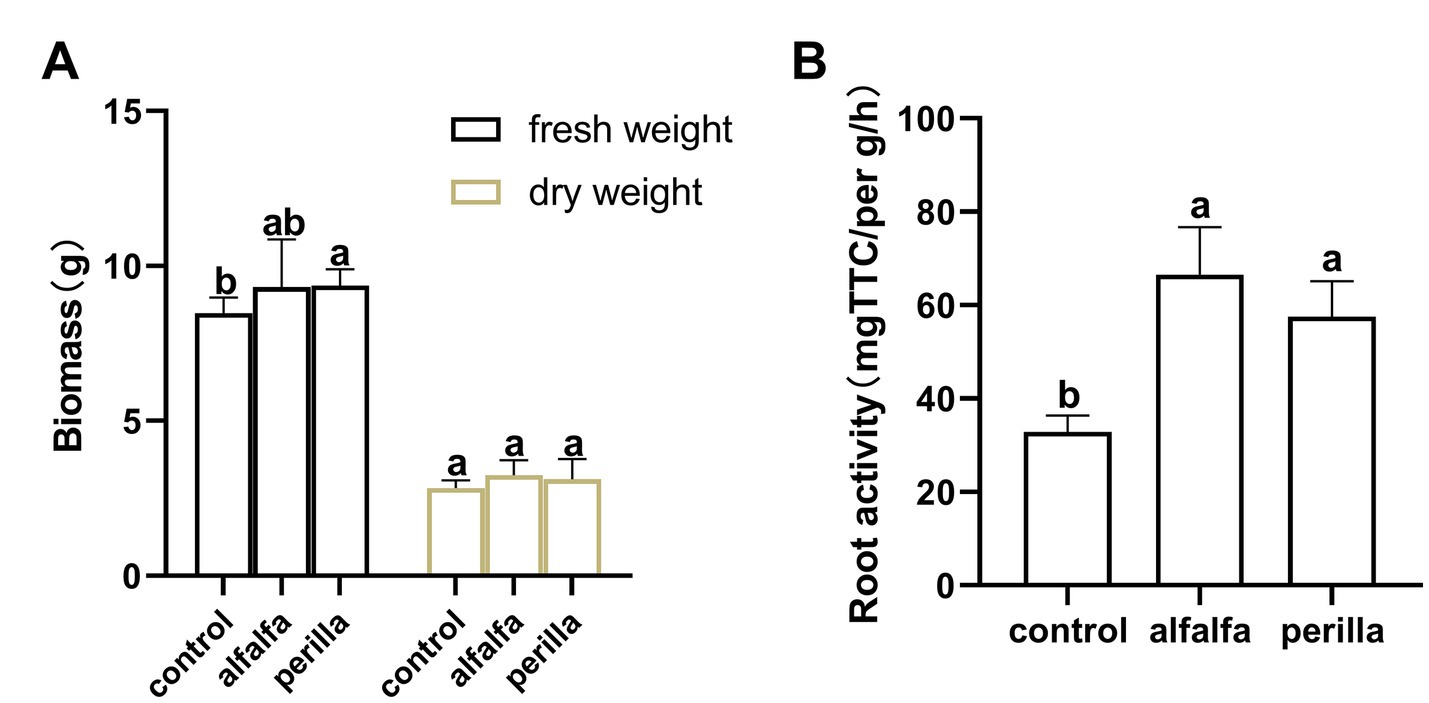
Figure 9. (A) Ginseng root biomass and (B) root activity in different treatments. Different letters indicate a significant difference at the p < 0.05 level. Control, no treatment; Alfalfa, alfalfa cultivation; Perilla, perilla cultivation.
The long term ginseng cultivation can lead to severe soil sickness, and crop rotation is an effective agricultural management to mitigate soil sickness. In the context of the ban on ginseng cultivation in deforestation and the limited land suitable for ginseng cultivation, finding a suitable crop rotation strategy has become an essential task for the sustainable development of the ginseng industry. In this study, we conducted one-year cultivation of alfalfa and perilla on ginseng cultivation soil in northeast China to analyze the effects of both cultivations on ginseng cultivation soil.
Environmental variables are important indicators for assessing soil quality (Schloter et al., 2003; Chen et al., 2020). Long-term ginseng cultivation can lead to soil acidification, while the heavy use of chemical fertilizers during cultivation can also cause the soil to be susceptible to salinization (Shi et al., 2009). Soil pH has a great influence on the growth and development of ginseng. Previous studies have shown that ginseng is generally suitable for growing in soil with a pH value of 5.0 to 6.5 (Li, 1995). In the soil with strong acidity, ginseng is more susceptible to external stress (You et al., 2015; Wang et al., 2019). In this study, we found that the pH of ginseng cultivation soil increased significantly, while soil salinity (indicated by EC) decreased significantly after 1 year of perilla cultivation. These results indicated that perilla cultivation could effectively improve soil acidification and salinization of ginseng cultivation. The alfalfa cultivation had no significant effect on soil acidity, but it may aggravate soil salinization. The critical difference between these two plants may be related to their differences in root exudates, root associated microbiome and enzyme activities. It is possible that more K ions were solubilized in alfalfa root zone. Although there are few reports on ginseng cultivation and soil salinization, the positive effects of perilla on the pH and EC of ginseng cultivation soils are of interest. Previous studies suggested that high nutrient concentration is the ideal growth condition for ginseng (Sun et al., 2017). This study focused on changes in available nutrients and soil organic matter. Alfalfa and perilla cultivation significantly increased the contents of available nitrogen and available potassium in the soil, and perilla cultivation also considerably increased soil organic matter (Figures 1C,D,F). Previous reports also suggested that alfalfa could improve nitrogen availability in the soil (Bhandari et al., 2020; Samaddar et al., 2021). These results indicated that both plants had positive effects on soil available nutrient pools of ginseng cultivation. It is worth emphasizing that alfalfa and perilla exerted opposite effects on soil EC in this study. Moreover, perilla had a more positive effect on soil EC and organic matter compared to alfalfa. These results will be useful for the future development of fertilization strategies for ginseng rotation systems.
Soil enzymes are derived primarily from soil microorganisms and play an essential role in decomposing organic substrates and biogeochemical cycles (Zhao et al., 2018). Soil urease plays a vital role in the nitrogen cycle by catalyzing the hydrolysis of urea in the soil to produce carbon dioxide and ammonia (Mobley and Hausinger, 1989; Cai et al., 2015). Soil acid phosphatase activity directly affects the hydrolysis process of soil phosphorus compounds. Thus, the increase in soil pH and available nitrogen after perilla cultivation may be related to the increased activity of urease. Both plants are beneficial for phosphorus biotransformation in ginseng cultivation soils. Soil sucrase catalyzes the release of fructose and glucose from sucrose to provide carbon sources for plants and microorganisms (Antonious, 2003). Moreover, the cultivation of alfalfa and perilla significantly increased laccase activity, and laccase has an essential role in forming soil humus and organic matter (Theuerl and Buscot, 2010; Park et al., 2021). Soil catalase can drive the decomposition and transformation of peroxides in soil (Mobley and Hausinger, 1989; Zhu et al., 2021). However, the sucrase activity of the perilla group was lower than that of the untreated ginseng cultivation soil (Figure 2C). In short, alfalfa and perilla cultivation changed the physicochemical properties and enzyme activities of soil under ginseng cultivation to varying degrees, indicating that alfalfa and perilla cultivation may potentially improve soil properties by affecting material circulation and energy metabolism. Moreover, unlike alfalfa, perilla had a more positive effect on soil urease and catalase.
Plants greatly influence soil microbial communities, and different plant species recruit specific microbial groups (Berendsen et al., 2012). We found that the cultivation of alfalfa and perilla did not significantly change the alpha diversity indexes of soil bacterial communities in ginseng cultivation (Figure 3A). However, for soil fungal communities, perilla cultivation significantly increased the Shannon index, ace index, and chao1 index (Figure 3B). Therefore, these two plants could not affect the diversity and richness of bacterial microbial community in ginseng cultivation soil. Compared with alfalfa, perilla may cause strong fungal microbial selection, increase the diversity and richness of the fungal community in ginseng cultivation soil, and thus improve the ecological environment of ginseng cultivation soil.
The relative abundance of Actinobacteriota and Proteobacteria increased significantly after 1 year of cultivation of alfalfa, the three most abundant bacterial phyla in the soil, while the relative abundance of Acidobacteriota decreased. Interestingly, the cultivation of perilla did not change their abundance (Supplementary Figure S2). Proteobacteria was also one of the dominant bacteria in each group of soils. Proteobacteria exhibit extreme metabolic diversity, and their members are essential for soil nutrient cycling, mainly in organic phosphate solubilization and soil nitrogen fixation (Long et al., 2018; Ren et al., 2018). Actinobacteriota contains a variety of organic matter-degrading bacteria that can maintain high levels of carbon sources in the soil and are critical to the soil carbon cycle (Samaddar et al., 2019). This is why all groups had a higher proportion of Actinobacteriota in the soil. It is worth noting that after 1 year of perilla planting, the content of organic matter in soil was significantly higher than that in untreated soil, but the abundance of Actinobacteriota in the two groups was not significantly different. This result suggests that there are other potential reasons for the increase in organic matter in Perilla planting soil. In addition, Acidobacteriota can also degrade organic matter in the apoplastic matter and maintain an effective nutrient supply to the soil (Ward et al., 2009). Among the three phyla with the highest fungal community abundance, cultivation of alfalfa and perilla only significantly reduced the relative abundance of Mortierellomycota (Supplementary Figure S3). In previous studies, Mortierellomycota, as saprophytic fungi, played a leading role in the decomposition of soil organic matter (Cong et al., 2020). The decrease in Mortierellomycota abundance may be due to the effect of alfalfa and perilla cultivation on soil properties and the special rhizosphere exudates. The accumulation of pathogens in the soil is an essential factor causing soil sickness and continuous cropping disorder. The function of Ilyonectria taxa has yet to be fully understood and needs to be further verified. So far, the Ilyonectria isolates that have been reported to infect ginseng are divided into four species: I. robusta, I. mors-panacis, I. panacis, and I. crassa (Cabral et al., 2012; Farh et al., 2018). Ilyonectria may be associated with a variety of soil-borne diseases of ginseng, and its reduced abundance has significant implications for soil health (Liu et al., 2019; Guan et al., 2020; Bischoff Nunes and Goodwin, 2022). It is noteworthy that the cultivation of alfalfa and perilla can effectively reduce the relative abundance of the fungal genus Ilyonectria in soil (Supplementary Figure S4; Farh et al., 2018). The possible reason is that the planting of two plants can effectively interrupt the life cycle of this potential pathogen, which has important significance for the sickness of ginseng cultivation soil.
LEfSe tool was used to analyze bacterial and fungal community. The specialized communities that represent microbial communities with statistically significant differences are referred to as indicator microbes (Yan et al., 2021a). LEfSe identified microbial taxa with LDA scores of 3.5 or above. In our results, Actinobacteriota and Chloroflexi are indicators of control group bacteria. Proteobacteria, Bacteroidota, and Acidobacteriota are the indicator bacteria of the alfalfa group. In the perilla group, class Vicinamibacteria and family Bacillaceae are indicator microbial taxa (Figure 6A). These results also demonstrated that alfalfa and perilla cultivation changed the bacterial community in ginseng cultivation soil. In Lefse analysis of fungal communities, only Mortierellomycota was also screened as an indicator of the control group at the phylum level (Figure 6B). The PCoA and ANOSIM analysis results in this study provide ample evidence that alfalfa and perilla caused profound changes in the composition and structure of microbial communities in ginseng cultivation soil (Figure 5).
To sum up, the microbial communities of ginseng cultivation soil with 1 year of cultivation of alfalfa and perilla were significantly different from those of untreated soil. Cultivation of alfalfa and perilla may modify soil material cycling and energy flow by altering the abundance of some microbial taxa in the soil. In addition, the two plants had different effects on the microbial community, which was related to the different metabolic characteristics of the plant roots (Bi et al., 2021). These changes in microbial communities reflect the potential of both plants, especially perilla, to be applied in alleviating soil sickness in ginseng cultivation.
The close interaction between soil environmental factors and microbial communities has been well-documented by numerous studies (Guo et al., 2019; Neupane et al., 2021). The RDA results showed that soil properties contributed more than 50% and 37% to the alteration of bacterial and fungal microbial community composition, respectively (Figures 7A,B). This result suggests that these soil properties play a dominant role in constructing microbial community structures in ginseng cultivation soils. Pearson correlation analysis showed that soil variables such as pH, available nutrients, and soil enzyme activity had significant positive or negative correlations with the dominant microbial community (Figures 7C,D). Moreover, the ecological functions of these dominant microbial phyla in soils have been well-reported (Yan et al., 2021a).
Actinobacteriota was positively correlated with EC and acid phosphatase but negatively correlated with pH. Proteobacteria was positively correlated with EC, available potassium, and acid phosphatase. On the contrary, Acidobacteriota was negatively correlated with these three environmental factors (Figure 7C). These results indicate some critical environmental factors, especially EC and acid phosphatase, have essential effects on bacteria communities in ginseng cultivation soil. The environmental factors significantly correlated with the dominant phylum of fungi, especially Mortierellomycota, were mainly concentrated in soil enzyme activity (Figure 7D). These results indicate that different microorganisms are not equally sensitive to environmental factors. On the other hand, after 1 year of alfalfa and perilla cultivation, dominant bacteria and fungi showed different growth patterns in the soil and played an important role in improvement. The cultivation of two kinds of plants changed the soil environment of ginseng cultivation soil. It provided a different living environment for soil microorganisms, including the difference in pH and available nutrient content, which led to the formation of different microbial networks in the soil. Therefore, alfalfa and perilla may be beneficial for soil health and ecosystem sustainability of ginseng cultivation soils.
Finally, we performed a 6-week pot experiment in which ginseng was grown in different treated soils, and their root activity and biomass were analyzed. The growth and activity of roots directly affect the nutrient level and yield of plants. The root is also an essential medicinal part of ginseng. Compared with untreated ginseng cultivation soil, root activity of 2-year-old ginseng seedlings was significantly improved after 1 year of cultivation with alfalfa and perilla. Meanwhile, the fresh weight of ginseng root in the perilla group was significantly higher than that in untreated soil. There are important interactions among plant roots, soil microorganisms, and soil properties, which are of great significance to nutrient cycling, soil enzymes, and microbial community (Feng et al., 2019). This result further suggests the effect of alfalfa and perilla cultivation on soil properties, enzyme activities, and microorganisms. These factors may favor nutrient transformation and root growth.
The effects of the two plants on EC were the opposite. In addition, they all increased the available nitrogen, available potassium, acid phosphatase, and laccase of the soil, but only perilla significantly increased the pH, soil organic matter, and urease. Perilla cultivation significantly increased the richness and diversity of the fungal community but had no significant effect on the bacterial community. Cultivation of alfalfa and perilla significantly altered microbial community composition and reduced the abundance of ginseng pathogenic fungus Ilyonectria in soil. Generally, perilla has a better improvement effect on soil for ginseng cultivation than alfalfa. Finally, pot experiments showed that the improved soil could significantly improve root activity.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/, PRJNA820351 and https://www.ncbi.nlm.nih.gov/, PRJNA820493.
XB and XS: conceptualization. YZ, KZ, and XY: data curation. LZ: investigation and supervision. XB and QL: validation, and writing-review and editing. XB and XY: writing - original draft. All authors contributed to the article and approved the submitted version.
This research work was supported by the Project of Jilin Provincial Development and Reform Commission (2021C007), the Horizontal Research Project of Jilin Medical University (2021-HX002), the Science and Technology Research Project of Education Department of Jilin Province, China (JJKH20230549KJ), and Jilin Medical University Doctoral Research Startup Fund Project (JYBS2021031LK, JYBS2021026LK, and JYBS2021032LK).
We would like to thank Majorbio Bio-pharm Technology Co., Ltd. (Shanghai, China) for help with the data analysis of high-throughput sequencing.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1134331/full#supplementary-material
Ahmed, H. M. (2019). Ethnomedicinal, phytochemical and pharmacological investigations of Perilla frutescens (L.). Britt. Molecules 24:102. doi: 10.3390/molecules24010102
Altieri, M. A. (2018). Agroecology: The science of sustainable agriculture. Boca Raton, FL: CRC Press.
Antonious, G. F. (2003). Impact of soil management and two botanical insecticides on urease and Invertase activity. J. Environ. Sci. Health B 38, 479–488. doi: 10.1081/PFC-120021667
Baeg, I. H., and So, S. H. (2013). The world ginseng market and the ginseng (Korea). J. Ginseng Res. 37, 1–7. doi: 10.5142/jgr.2013.37.1
Baslam, M., Erice, G., and Goicoechea, N. (2012). Impact of arbuscular mycorrhizal fungi (AMF) and atmospheric CO2 concentration on the biomass production and partitioning in the forage legume alfalfa. Symbiosis 58, 171–181. doi: 10.1007/s13199-012-0199-6
Berendsen, R. L., Pieterse, C. M. J., and Bakker, P. A. H. M. (2012). The rhizosphere microbiome and plant health. Trends Plant Sci. 17, 478–486. doi: 10.1016/j.tplants.2012.04.001
Bhandari, K. B., West, C. P., and Acosta-Martinez, V. (2020). Assessing the role of interseeding alfalfa into grass on improving pasture soil health in semi-arid Texas High Plains. Appl. Soil Ecol. 147:103399. doi: 10.1016/j.apsoil.2019.103399
Bi, B., Wang, K., Zhang, H., Wang, Y., Fei, H., Pan, R., et al. (2021). Plants use rhizosphere metabolites to regulate soil microbial diversity. Land Degrad. Dev. 32, 5267–5280. doi: 10.1002/ldr.4107
Bischoff Nunes, I., and Goodwin, P. H. (2022). Interaction of ginseng with Ilyonectria root rot pathogens. Plan. Theory 11:2152. doi: 10.3390/plants11162152
Cabral, A., Groenewald, J. Z., Rego, C., Oliveira, H., and Crous, P. W. (2012). Cylindrocarpon root rot: multi-gene analysis reveals novel species within the Ilyonectria radicicola species complex. Mycol. Prog. 11, 655–688. doi: 10.1007/s11557-011-0777-7
Cai, Z., Li, S., Zhang, W., Ma, J., Wang, J., Cai, J., et al. (2015). Effects of the novel pyrimidynyloxybenzoic herbicide ZJ0273 on enzyme activities, microorganisms and its degradation in Chinese soils. Environ. Sci. Pollut. Res. 22, 4425–4433. doi: 10.1007/s11356-014-3674-1
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Chen, D., Duan, Y., Jin, Y., Yang, Y., and Yuan, L. (2020). Soil quality and microbiology in response to fertilizations in a paddy-upland rotation with multiple crops and frequent tillage. Exp. Agric. 56, 227–238. doi: 10.1017/S0014479719000322
Cole, J. R., Wang, Q., Cardenas, E., Fish, J., Chai, B., Farris, R. J., et al. (2009). The ribosomal database project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37, D141–D145. doi: 10.1093/nar/gkn879
Cong, P., Wang, J., Li, Y., Liu, N., Dong, J., Pang, H., et al. (2020). Changes in soil organic carbon and microbial community under varying straw incorporation strategies. Soil Tillage Res. 204:104735. doi: 10.1016/j.still.2020.104735
Dixon, P. (2003). VEGAN, a package of R functions for community ecology. J. Veg. Sci. 14, 927–930. doi: 10.1111/j.1654-1103.2003.tb02228.x
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Fan, D., Zhang, Y., Qin, S., and Wu, B. (2017). Relationships between Artemisia ordosica communities and environmental factors following sand-dune stabilization in the mu us desert, Northwest China. J. For. Res. 28, 115–124. doi: 10.1007/s11676-016-0289-z
Farh, M. E.-A., Kim, Y.-J., Kim, Y.-J., and Yang, D.-C. (2018). Cylindrocarpon destructans/Ilyonectria radicicola-species complex: causative agent of ginseng root-rot disease and rusty symptoms. J. Ginseng Res. 42, 9–15. doi: 10.1016/j.jgr.2017.01.004
Feng, Y., Hu, Y., Wu, J., Chen, J., Yrjälä, K., and Yu, W. (2019). Change in microbial communities, soil enzyme and metabolic activity in a Torreya grandis plantation in response to root rot disease. For. Ecol. Manage. 432, 932–941. doi: 10.1016/j.foreco.2018.10.028
Fernandez, A. L., Fabrizzi, K. P., Tautges, N. E., Lamb, J. A., and Sheaffer, C. C. (2019). Cutting management and alfalfa stand age effects on organically grown corn grain yield and soil N availability. Renewable Agricult Food Syst 34, 144–154. doi: 10.1017/S1742170517000394
Gianello, C., and Bremner, J. (1986). Comparison of chemical methods of assessing potentially available organic nitrogen in soil. Commun. Soil Sci. Plant Anal. 17, 215–236. doi: 10.1080/00103628609367709
Gong, X., Shi, J., Zhou, X., Yuan, T., Gao, D., and Wu, F. (2021). Crop rotation with cress increases cucumber yields by regulating the composition of the Rhizosphere soil microbial community. Front. Microbiol. 12:631882. doi: 10.3389/fmicb.2021.631882
Guan, Y., Chen, M., Ma, Y., Du, Z., Yuan, N., Li, Y., et al. (2020). Whole-genome and time-course dual RNA-Seq analyses reveal chronic pathogenicity-related gene dynamics in the ginseng rusty root rot pathogen Ilyonectria robusta. Sci. Rep. 10:1586. doi: 10.1038/s41598-020-58342-7
Guo, J., Wang, G., Wu, Y., Shi, Y., Feng, Y., and Cao, F. (2019). Ginkgo agroforestry practices alter the fungal community structures at different soil depths in eastern China. Environ. Sci. Pollut. Res. 26, 21253–21263. doi: 10.1007/s11356-019-05293-w
Kanehisa, M., and Goto, S. (2000). KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. doi: 10.1093/nar/28.1.27
Langille, M. G., Zaneveld, J., Caporaso, J. G., McDonald, D., Knights, D., Reyes, J. A., et al. (2013). Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 31, 814–821. doi: 10.1038/nbt.2676
Li, T. S. (1995). Asian and American ginseng—a review. HortTechnology 5, 27–34. doi: 10.21273/HORTTECH.5.1.27
Li, C., Chen, G., Zhang, J., Zhu, P., Bai, X., Hou, Y., et al. (2021). The comprehensive changes in soil properties are continuous cropping obstacles associated with American ginseng (Panax quinquefolius) cultivation. Sci. Rep. 11, 1–14. doi: 10.1038/s41598-021-84436-x
Li, Q., Zhan, Y., Xie, H., Yan, N., Wang, E., and Chen, C. (2021). Potential causes and recovery of soil sickness from the Panax ginseng cultivation. Allelopath. J. 54, 105–120. doi: 10.26651/allelo.j/2021-54-1-1352
Liang, J., Yu, X., Cao, Y., Zhang, J., Yan, N., Xue, L., et al. (2021). Effect of quicklime on microbial Community in Strong Acidic Soil. J. Soil Sci. Plant Nutr. 21, 1771–1781. doi: 10.1007/s42729-021-00478-0
Liu, S.-Q., Ren, Y.-Y., and Xu, H.-B. (2011). Effect of thinning flower buds on fruit and seed traits and root yield of Panax ginseng C.a. Meyer. Ind. Crop Prod. 33, 559–565. doi: 10.1016/j.indcrop.2010.12.024
Liu, D., Sun, H., and Ma, H. (2019). Deciphering microbiome related to rusty roots of Panax ginseng and evaluation of antagonists against pathogenic Ilyonectria. Front. Microbiol. 10:1350. doi: 10.3389/fmicb.2019.01350
Long, X.-E., Yao, H., Huang, Y., Wei, W., and Zhu, Y.-G. (2018). Phosphate levels influence the utilisation of rice rhizodeposition carbon and the phosphate-solubilising microbial community in a paddy soil. Soil Biol. Biochem. 118, 103–114. doi: 10.1016/j.soilbio.2017.12.014
Mäder, P., Fliessbach, A., Dubois, D., Gunst, L., Fried, P., and Niggli, U. (2002). Soil fertility and biodiversity in organic farming. Science 296, 1694–1697. doi: 10.1126/science.1071148
Mobley, H., and Hausinger, R. (1989). Microbial ureases: significance, regulation, and molecular characterization. Microbiol. Rev. 53, 85–108. doi: 10.1128/mr.53.1.85-108.1989
Neupane, A., Bulbul, I., Wang, Z., Lehman, R. M., Nafziger, E., and Marzano, S.-Y. L. (2021). Long term crop rotation effect on subsequent soybean yield explained by soil and root-associated microbiomes and soil health indicators. Sci. Rep. 11:9200. doi: 10.1038/s41598-021-88784-6
Nguyen, N. H., Song, Z., Bates, S. T., Branco, S., Tedersoo, L., Menke, J., et al. (2016). FUNGuild: an open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 20, 241–248. doi: 10.1016/j.funeco.2015.06.006
Olsen, S. R. (1954). Estimation of available phosphorus in soils by extraction with sodium bicarbonate. Washington, DC: US Department of Agriculture.
Pan, X., Zhang, S., Zhong, Q., Gong, G., Wang, G., Guo, X., et al. (2020). Effects of soil chemical properties and fractions of Pb, cd, and Zn on bacterial and fungal communities. Sci. Total Environ. 715:136904. doi: 10.1016/j.scitotenv.2020.136904
Park, H. J., Lee, Y. M., Do, H., Lee, J. H., Kim, E., Lee, H., et al. (2021). Involvement of laccase-like enzymes in humic substance degradation by diverse polar soil bacteria. Folia Microbiol. 66, 331–340. doi: 10.1007/s12223-020-00847-9
Peters, R. D., Sturz, A. V., Carter, M. R., and Sanderson, J. B. (2003). Developing disease-suppressive soils through crop rotation and tillage management practices. Soil Tillage Res. 72, 181–192. doi: 10.1016/S0167-1987(03)00087-4
Proctor, J. T. A., and Bailey, W. G. (1987). Ginseng: industry, botany, and culture. Hortic. Rev. 8, 187–236. doi: 10.1002/9781118060827.ch6
Ren, C., Wang, T., Xu, Y., Deng, J., Zhao, F., Yang, G., et al. (2018). Differential soil microbial community responses to the linkage of soil organic carbon fractions with respiration across land-use changes. For. Ecol. Manage. 409, 170–178. doi: 10.1016/j.foreco.2017.11.011
Ru, W., Wang, D., Xu, Y., He, X., Sun, Y. E., Qian, L., et al. (2015). Chemical constituents and bioactivities of Panax ginseng (C. A. Mey.). Drug Discov Ther 9, 23–32. doi: 10.5582/ddt.2015.01004
Samaddar, S., Schmidt, R., Tautges, N. E., and Scow, K. (2021). Adding alfalfa to an annual crop rotation shifts the composition and functional responses of tomato rhizosphere microbial communities. Appl. Soil Ecol. 167:104102. doi: 10.1016/j.apsoil.2021.104102
Samaddar, S., Truu, J., Chatterjee, P., Truu, M., Kim, K., Kim, S., et al. (2019). Long-term silicate fertilization increases the abundance of Actinobacterial population in paddy soils. Biol. Fertil. Soils 55, 109–120. doi: 10.1007/s00374-018-01335-6
Schloter, M., Dilly, O., and Munch, J. C. (2003). Indicators for evaluating soil quality. Agric. Ecosyst. Environ. 98, 255–262. doi: 10.1016/S0167-8809(03)00085-9
Schmidt, R., Mitchell, J., and Scow, K. (2019). Cover cropping and no-till increase diversity and symbiotroph:saprotroph ratios of soil fungal communities. Soil Biol. Biochem. 129, 99–109. doi: 10.1016/j.soilbio.2018.11.010
Shi, W.-M., Yao, J., and Yan, F. (2009). Vegetable cultivation under greenhouse conditions leads to rapid accumulation of nutrients, acidification and salinity of soils and groundwater contamination in south-eastern China. Nutr. Cycl. Agroecosyst. 83, 73–84. doi: 10.1007/s10705-008-9201-3
Shu, J., He, Q., Zheng, H., Liao, M. A., Kuang, N., and Huang, Z. (2021). Abscisic acid improves the ability of accumulator plant Perilla frutescens to perform phytoremediation in cadmium-contaminated soils. Int. J. Environ. Anal. Chem. 18, 1–10. doi: 10.1080/03067319.2021.1890058
Sun, H., Wang, Q.-X., Liu, N., Li, L., Zhang, C.-G., Liu, Z.-B., et al. (2017). Effects of different leaf litters on the physicochemical properties and bacterial communities in Panax ginseng-growing soil. Appl. Soil Ecol. 111, 17–24. doi: 10.1016/j.apsoil.2016.11.008
Sun, Z., Yang, L., Zhang, L., and Han, M. (2018). An investigation of Panax ginseng Meyer growth promotion and the biocontrol potential of antagonistic bacteria against ginseng black spot. J. Ginseng Res. 42, 304–311. doi: 10.1016/j.jgr.2017.03.012
Tesfaye, M., Dufault, N. S., Dornbusch, M. R., Allan, D. L., Vance, C. P., and Samac, D. A. (2003). Influence of enhanced malate dehydrogenase expression by alfalfa on diversity of rhizobacteria and soil nutrient availability. Soil Biol. Biochem. 35, 1103–1113. doi: 10.1016/S0038-0717(03)00162-7
Theuerl, S., and Buscot, F. (2010). Laccases: toward disentangling their diversity and functions in relation to soil organic matter cycling. Biol. Fertil. Soils 46, 215–225. doi: 10.1007/s00374-010-0440-5
Wang, Q., Sun, H., Xu, C., Ma, L., Li, M., Shao, C., et al. (2019). Analysis of rhizosphere bacterial and fungal communities associated with rusty root disease of Panax ginseng. Appl. Soil Ecol. 138, 245–252. doi: 10.1016/j.apsoil.2019.03.012
Ward, N. L., Challacombe, J. F., Janssen, P. H., Henrissat, B., Coutinho, P. M., Wu, M., et al. (2009). Three genomes from the phylum Acidobacteria provide insight into the lifestyles of these microorganisms in soils. Appl. Environ. Microbiol. 75, 2046–2056. doi: 10.1128/AEM.02294-08
Wei, M.-C., Wang, C.-S., Liou, R.-M., and Yang, Y.-C. (2022). Development and validation of a green and sustainable procedure for the preparation of Perilla frutescens extracts. Food Chem. 369:130929. doi: 10.1016/j.foodchem.2021.130929
Woo, S. L., De Filippis, F., Zotti, M., Vandenberg, A., Hucl, P., and Bonanomi, G. (2022). Pea-wheat rotation affects soil microbiota diversity, community structure, and Soilborne pathogens. Microorganisms 10:370. doi: 10.3390/microorganisms10020370
Xiang, Y.-Z., Shang, H.-C., Gao, X.-M., and Zhang, B.-L. (2008). A comparison of the ancient use of ginseng in traditional Chinese medicine with modern pharmacological experiments and clinical trials. Phytother. Res. 22, 851–858. doi: 10.1002/ptr.2384
Xiao, Q., Wang, Y., Lü, Q., Wen, H., Han, B., Chen, S., et al. (2020). Responses of glutathione and phytochelatins biosysthesis in a cadmium accumulator of Perilla frutescens (L.) Britt. Under cadmium contaminated conditions. Ecotoxicol. Environ. Saf. 201:110805. doi: 10.1016/j.ecoenv.2020.110805
Xiao, C., Yang, L., Zhang, L., Liu, C., and Han, M. (2016). Effects of cultivation ages and modes on microbial diversity in the rhizosphere soil of Panax ginseng. J. Ginseng Res. 40, 28–37. doi: 10.1016/j.jgr.2015.04.004
Yan, T., Xue, J., Zhou, Z., and Wu, Y. (2021a). Biochar-based fertilizer amendments improve the soil microbial community structure in a karst mountainous area. Sci. Total Environ. 794:148757. doi: 10.1016/j.scitotenv.2021.148757
Yan, T., Xue, J., Zhou, Z., and Wu, Y. (2021b). Effects of biochar-based fertilizer on soil bacterial network structure in a karst mountainous area. Catena 206:105535. doi: 10.1016/j.catena.2021.105535
Yang, Y., Ren, C., Zhang, Y., and Wu, X. (2017). Ginseng: an nonnegligible natural remedy for healthy aging. Aging Dis. 8, 708–720. doi: 10.14336/AD.2017.0707
You, J., Liu, X., Zhang, B., Xie, Z., Hou, Z., and Yang, Z. (2015). Seasonal changes in soil acidity and related properties in ginseng artificial bed soils under a plastic shade. J. Ginseng Res. 39, 81–88. doi: 10.1016/j.jgr.2014.08.002
Yun, T. K. (2009). Brief introduction of Panax ginseng C.a. Meyer. J. Korean Med. Sci. 16, S3–S5. doi: 10.3346/jkms.2001.16.S.S3
Zhan, Y., Yan, N., Miao, X., Li, Q., and Chen, C. (2021). Different responses of soil environmental factors, soil bacterial community, and root performance to reductive soil disinfestation and soil fumigant chloropicrin. Front. Microbiol. 12:796191. doi: 10.3389/fmicb.2021.796191
Zhang, W., Jia, X., Chen, S., Wang, J., Ji, R., and Zhao, L. (2020). Response of soil microbial communities to engineered nanomaterials in presence of maize (Zea mays L.) plants. Environ. Pollut. 267:115608. doi: 10.1016/j.envpol.2020.115608
Zhang, H., and Xue, Q. (2010). Research progress in control of continuous cropping obstacle in ginseng. Acta Agricult Jiangxi 22, 68–71.
Zhao, Q., Tang, J., Li, Z., Yang, W., and Duan, Y. (2018). The influence of soil Physico-chemical properties and enzyme activities on soil quality of saline-alkali Agroecosystems in Western Jilin Province, China. Sustainability 10:1529. doi: 10.3390/su10051529
Zhao, Y., Wang, Y., and Yang, J. (2005). Study on the rotation of crops among Panax quinquefolium, Perilla frutescens and Coix lacryma-jobi. China J Chin Materia Med 30, 12–15.
Keywords: ginseng cultivation soil, soil sickness, soil microbial community, alfalfa, perilla
Citation: Bian X, Yang X, Zhang K, Zhai Y, Li Q, Zhang L and Sun X (2023) Potential of Medicago sativa and Perilla frutescens for overcoming the soil sickness caused by ginseng cultivation. Front. Microbiol. 14:1134331. doi: 10.3389/fmicb.2023.1134331
Received: 30 December 2022; Accepted: 17 March 2023;
Published: 05 April 2023.
Edited by:
Entao Wang, National Polytechnic Institute,MexicoReviewed by:
Xiaojing Hu, Northeast Institute of Geography and Agroecology (CAS), ChinaCopyright © 2023 Bian, Yang, Zhang, Zhai, Li, Zhang and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xin Sun, c3VueGluYmhAMTI2LmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.