
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 28 February 2023
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1133590
This article is part of the Research TopicHigh-level Antimicrobial Resistance or Hypervirulence in Emerging and Re-emerging “Super-Bug” Foodborne Pathogens: Detection, Mechanism, and Dissemination from Omics InsightsView all 16 articles
 Chunyang Wu1†
Chunyang Wu1† Ying Zhou2†
Ying Zhou2† Wenxiu Ai3
Wenxiu Ai3 Yinjuan Guo2
Yinjuan Guo2 Xiaocui Wu2
Xiaocui Wu2 Bingjie Wang2
Bingjie Wang2 Huilin Zhao2
Huilin Zhao2 Lulin Rao4
Lulin Rao4 Xinyi Wang2
Xinyi Wang2 Jiao Zhang2
Jiao Zhang2 Fangyou Yu2,4*
Fangyou Yu2,4* Liangxing Wang1*
Liangxing Wang1*The emergence of carbapenem-resistant Klebsiella pneumoniae (CRKP) strains and restricted therapeutic options pose a global threat to public health. Aminoglycosides are a wise choice, which can effectively reduce the mortality rate when combined with β-lactam drugs. However, in this study, we identified a ST15-KL112 CRKP FK3006 which not only exhibited resistance to carbapenems, but also exhibited high level resistance to aminoglycosides. In addition to the multidrug resistant phenotype, FK3006 also owned typical pathogenic characteristic, including hypermucoviscosity and hypervirulence phenotypes. According to the whole-genome sequencing, one pLVPK-like virulence plasmid, and three key resistant plasmids (blaOXA-232, blaCTX-M-15, and rmtF) were observed in FK3006. Compared to other typical ST15 CRKP, the presence of pLVPK-like virulence plasmid (p3006-2) endowed the FK3006 with high virulence features. High siderophore production, more cell invasive and more resistant to serum killing was observed in FK3006. The Galleria mellonella infection model also further confirmed the hypervirulent phenotype of FK3006 in vivo. Moreover, according to the conjugation assay, p3006-2 virulence plasmid also could be induced transfer with the help of conjugative IncFIIK p3006-11 plasmid (blaCTX-M-15). In addition to the transmissible plasmid, several insertion sequences and transposons were found around blaCTX-M-15, and rmtF to generate the mobile antimicrobial resistance island (ARI), which also make a significant contribution to the dissemination of resistant determinants. Overall, we reported the uncommon co-existence of blaOXA-232, rmtF-encoding plasmids, and pLVPK-like virulence plasmid in ST15-KL112 K. pneumoniae. The dissemination threatens of these high-risk elements in K. pneumoniae indicated that future studies are necessary to evaluate the prevalence of such isolates.
The emergence of carbapenem-resistant Klebsiella pneumoniae (CRKP) has become a major challenge facing clinical management and global public health, because of the extremely limited antibiotic therapy options (Ernst et al., 2020). Aminoglycosides are important options for treating infections caused by CRKP and are generally administered in combination with β-lactam agents and tigecycline (Daikos et al., 2014; Karaiskos et al., 2019). However, increasing rates of aminoglycoside resistance in CRKP have been reported in recent years, posing a new challenge for treatment (Galani et al., 2019). Hence, verifying the related mechanism and demonstrating the potential of the spread of these resistant phenotypes in clinical isolate are vital clues to solving antibiotic resistance.
It has been highlighted Klebsiella pneumoniae carbapenamase (KPC) is the most prevalent in China, it is noteworthy that OXA-48-like carbapenemases are common carbapenemases in Enterobacterales in certain regions of the world (Pitout et al., 2019). To date, several variants that differ from OXA-48 by only a few amino acids and display similar enzymatic profiles with OXA-48 have been identified. OXA-232 differs from OXA-48 by five amino acid substitutions, exhibiting a lower ability to hydrolyze carbapenems but greater hydrolytic activities against penicillin than OXA-48 (Potron et al., 2013; Miltgen et al., 2020). Since the first report of an OXA-232-producing K. pneumoniae strain in China in 2016, such isolates has became epidemic in China, and usually associated with a clonal dissemination of ST15 K. pneumoniae (Yin et al., 2017; Wang et al., 2022). Most aminoglycoside resistance mechanisms were associated with the aminoglycoside-modifying enzymes, among which only 16S rRNA methyltransferase (16S-RMTase)-encoding genes could mediate high-level resistance to aminoglycosides (Ramirez and Tolmasky, 2010). Among these genes, rmtB and armA present the most widespread 16S rRNA methylase genes, with rmtF is rarely reported (Nagasawa et al., 2014). Notably, these antibiotic resistances could be carried by various mobile genetic elements (MGEs), once these resistant elements co-existence in one host, the therapeutic options would be very limited.
Klebsiella pneumoniae has an exceptional ability to acquire exogenous resistance-encoding and hypervirulence-encoding genetic elements. For a long period, K. pneumoniae did not simultaneously encode the phenotypes of multidrug-resistance (MDR) and hypervirulence. However, carbapenem-resistant hypervirulent K. pneumoniae (hv-CRKP) isolates have been increasingly reported in recent years. The epidemic of hv-CRKP strains has emerged as a worldwide public health concern as they may cause untreatable, severe infections (Yang et al., 2021).
Although the hv-CRKP has increased rapidly, most were associated with the KPC carbapenemases and ST11 K. pneumoniae, the ST15-OXA-232 hv-CRKP was uncommon. In this study, we identified a hypermucovious multidrug-resistant ST15 K. pneumoniae (FK3006), exhibiting resistance to both aminoglycosides and ertapenem. Then, we applied whole-genome-sequencing (WGS) to explore the potential molecular mechanisms and observed four key plasmids. We also applied the conjugation assay to further determine the dissemination of these high-risk determinants and verified the relevant virulence phenotype of FK3006. In addition to the plasmids, we described other related MEGs through genetic comparisons as well. Overall, our goal was to report and describe a clinical hypervirulent carbapenem-resistant K. pneumoniae clearly, and emphasize the possible risk of these strains.
First, the 207 CRKP isolates from patients at 12 tertiary China teaching hospitals in eight Chinese provinces were collected From January 2015 to May 2021. And then the detached samples were cultured on blood agar plate and identified by MALDI-TOF MS. FK3006 was isolated from the sputum sample, for it is the only isolate harboring the co-existence of blaOXA-232, blaCTX-M-15, rmtF, and virulence plasmid analyzed by WGS. K. pneumoniae strain 3036 (FK3036) was used as control strain for the virulence-negative resistant strain (ST15, none Virulence factors) and NUTH-K2044(ST23) was used as a virulence-positive control (Supplementary Table S1). Plasmid conjugation was performed with Escherichia coli 600(EC600, rifampicin-resistant), which was used as the recipient strain.
The MICs of the FK3006, EC600, and transconjugants (FK3006JH-1, FK3006JH-2) were determined by standard broth microdilution method following the Clinical and Laboratory Standards Institute guidelines (CLSI, 2020), except for colistin and tigecycline, for which the European Committee on Antimicrobial Susceptibility Testing (EUCAST) breakpoints were used (EUCAST, 2020). Each AST was independently repeated three times in our study. Escherichia coli ATCC 25922 was used as the quality control organism in MIC determination.
The horizontal transferability of blaOXA-232, blaCTX-M-15, rmtF, and iucA were examined using the conjugation assay with E. coli 600. The FK3006 was used as donor strain, and E. coli 600 was used as the recipient strain. The recipients and donors were cultured in Luria-Bertani broth (37°C) until logarithmic phase (OD600 = 0.4–0.6), mixed in a ratio of 2:1 (200 μl,100uL) in 4 ml LB broth for 24 h. And then the serial dilutions were plated on selective media with appropriate antibiotics (gentamicin, 8 μg/ml[rmtF]; ampicillin, 100 μg/ml[blaCTX-M-15]; meropenem, 1 μg/ml[blaOXA-232]; dipotassium tellurite, 8 μg/ml[iucA]; rifampicin, 600 μg/ml). The frequency of plasmid transfer was calculated as the number of transconjugants per recipient. The transconjugants acquiring gene were confirmed by PCR and primers are listed in Supplementary Table S2.
After the plasmid was confirmed to be obtained, we applied growth curve assays to investigate fitness. Transconjugants (FK3006JH-1 and FK3006JH-2) and EC600 were cultured in LB medium overnight, then diluted to an OD600 of 0.01 and grown at 37°C for 24 h. Culture densities were determined by measuring the OD600 every 1 h for the first 12 h and then 24 h (Liu et al., 2016).
Bacterial genomic DNA of FK3006 was isolated using the Qiagen DNA extraction Kit (Qiagen, Germany) and the genome sequencing was then performed by the PacBio Sequel platform and the Illumina NovaSeq 6,000 platform. CANU (version 1.7.1) software was applied to assemble the data acquired by PacBio platform sequencing. The ORF prediction was measured in SnapGene (version 4.2.4). Resistant plasmid replicons were predicted using the PlasmidFinder tool1. To verify whether the plasmid was also a conjugative plasmid, VRprofile2 and OriT Finder website3 were performed to analysis of the four conjugal modules in the plasmid, including the relaxase gene, the origin of transfer site (oriT), the type IV secretion system gene cluster (T4SS), and the type IV coupling protein (T4CP) gene. The transposons (Tns) and insertion sequences (ISs) were also annotated and determined through the VRprofile and ISFinder4. We used BLAST Ring Image Generator (BRIG) to determine similar plasmids by comparing their identities and coverages. The circular representation was performed by Proksee5. The gene environments surrounding the antibiotic resistance gene was analyzed by Easyfig software. VFDB6 was performed to annotate the virulence factors.
Briefly, bacterial clones were diluted with saline to a concentration of approximately 108 CFU/ml, and then 1 μl bacterial clones were dropped on CAS and King’s B (2:1) plates. After incubation for 24 h at 37°C, siderophore production was determined by the presence of an orange halo around the bacterial colonies (Zhou et al., 2022).
Caterpillars of Galleria mellonella were stored at 4°C before use for ensuring their health and fitness. Caterpillars were selected weighing 200–250 mg each for the study. For the FK3006, EC600, FK3006JH-1 and FK3006JH-2 groups, the caterpillars were injected with 10 μl (~1 × 106 CFU) bacterial suspension at the left proleg, while the control groups were injected with PBS or empty syringe. A minimum of 10 caterpillars was used in each treatment group; they then were kept in culture at 37°C and inspected for 144 h. The survival rates of each group were recorded for each day (Li et al., 2020).
Briefly, serum was separated from blood samples and stored at-80°C. An inoculum of 25 μl (~1 × 106 CFU) bacteria prepared from the mid-log phase was incubated with 75 μl pooled human sera. The mixture was taken at 0, 1, 2, and 3 h, and then incubated on the MHA plate. The numbers of viable bacteria were determined at 24 h (Liu et al., 2019).
Approximately 1 × 105 A549 human lung epithelial cells (ATCC CCL-185) were grown in each well of 24-well plates in DMEM medium containing 10% fetal calf serum at 37°C with 5% CO2 for 12 h and then incubated for 21 h at 37°C with 2 × 107 CFU bacteria (Gottig et al., 2016). After centrifugation (3,000 rpm, 5 min, 4°C), the LDH in the supernatant was measured using the LDH Cytotoxicity Assay kit according to the instructions (Solarbio BC0685).
The S1-Nuclease Pulsed Field gel electrophoresis (S1-PFGE) was used to determine the existence of plasmids profiles in donor strains, and recipient strains. In short, the isolates were embedded in 1% Seakem Gold agarose and digested with S1 nuclease (Takara, Dalian, China). PFGE analysis was carried out using a CHEF-Mapper system (Bio-Rad) for 19 h with a switch time 4.0–40 s (parameters: 14°C, voltage 6 V/cm, and electric field angle 120°). The XbaI digested DNA of Salmonella serotype Braenderup H9812 was considered as a molecular size marker (Ai et al., 2021).
Data analyses were executed with the software GraphPad Prism 8.0.2. Results are shown as a two-tailed non-parametric Student’s test. For in vivo and vitro experiments survival data were analyzed by the Log Rank test (Mantel-Cox). P < 0.05 was considered to be statistically significant.
The complete nucleotide sequences of the chromosome and p3006-2, p3006-3, p3006-7, and p3006-11 plasmid were deposited as GenBank accession numbers JANCTJ010000000 consists of sequences JANCTJ010000001-JANCTJ010000014.
To characterize the antibiotic-resistant phenotype of FK3006, the 18 antibiotics susceptibility was tested in this strain. Results showed that FK3006 was a multi-drug resistant strain, which not only exhibited high-level resistance to both aminoglycoside antibiotics and a series of β-lactam antibiotics, but also was resistant to ciprofloxacin, sulfamethoxazole, minocycline, and piperacillin-tazobactam (Table 1). Although it was sensitive to imipenem, it still exhibited low-level resistance to other carbapenems including meropenem and ertapenem.
According to the subsequent WGS-based analysis, we further found that FK3006 belonged to ST15-KL112 isolates, a typical MDR clone. Moreover, 13 resistant elements, three resistant plasmids, and one virulence plasmid were detected in this isolate (Table 2). In addition, three key resistance genes played an important role in the acquisition of resistance to aminoglycoside antibiotics (rmtF), β-lactam antibiotics (blaCTX-M-15), and carbapenems (blaOXA-232). Moreover, rmpA2 as a critical factor activating the expression of capsular polysaccharides (CPS) genes which is responsible for CPS biosynthesis was detected in the IncFIB(K)/HI1B plasmid of FK3006. The iucABCD-iutA operon which encoding proteins necessary for aerobactin siderophore biosynthesis was also found in p3006-2.
We have identified three key resistant plasmids, and one virulence plasmid in FK3006. As plasmids are often transmissible between bacteria, we made a detailed analysis of these plasmids, aiming to further clarify potential resistance and virulence dissemination threats of FK3006. In FK3006, we observed three resistant plasmids: p3006-3, p3006-7, and p3006-11. p3006-11 (blaCTX-M-15) was a typical IncFIIK-type MDR plasmid and shared high identity with the mobilizable plasmid pKP7450-3(identity 99.95%, blaCTX-M-15, IncFII(K), CP090471.1), as well as the mobilizable plasmid pMS3802-CTXM-vir (identity 99.59%, blaCTX-M-15, IncHI1B(pNDM-MAR), CP068016.1; Figure 1A). p3006-11 harbored four conjugation modules, holding the potential to self-transfer. Hence, we applied conjugation assay to imitate and evaluate the dissemination ability of p3006-11 plasmid. We found the p3006-11 was successfully transferred from FK3006 to E. coli EC600 (1.1 × 10−6-9.7 × 10−5). Moreover, the S1-PFGE pattern (Figure 2A) and MICs of FK3006JH-1(EC600 harboring blaCTX-M-15 plasmid) also confirmed that the resistance phenotype dissemination of FK3006 (Table 1). Notably, the obtain of p3006-11 did not affect the growth of E. coli 600, which ensuring the stable existence of the resistance plasmid (Supplementary Figure S1).
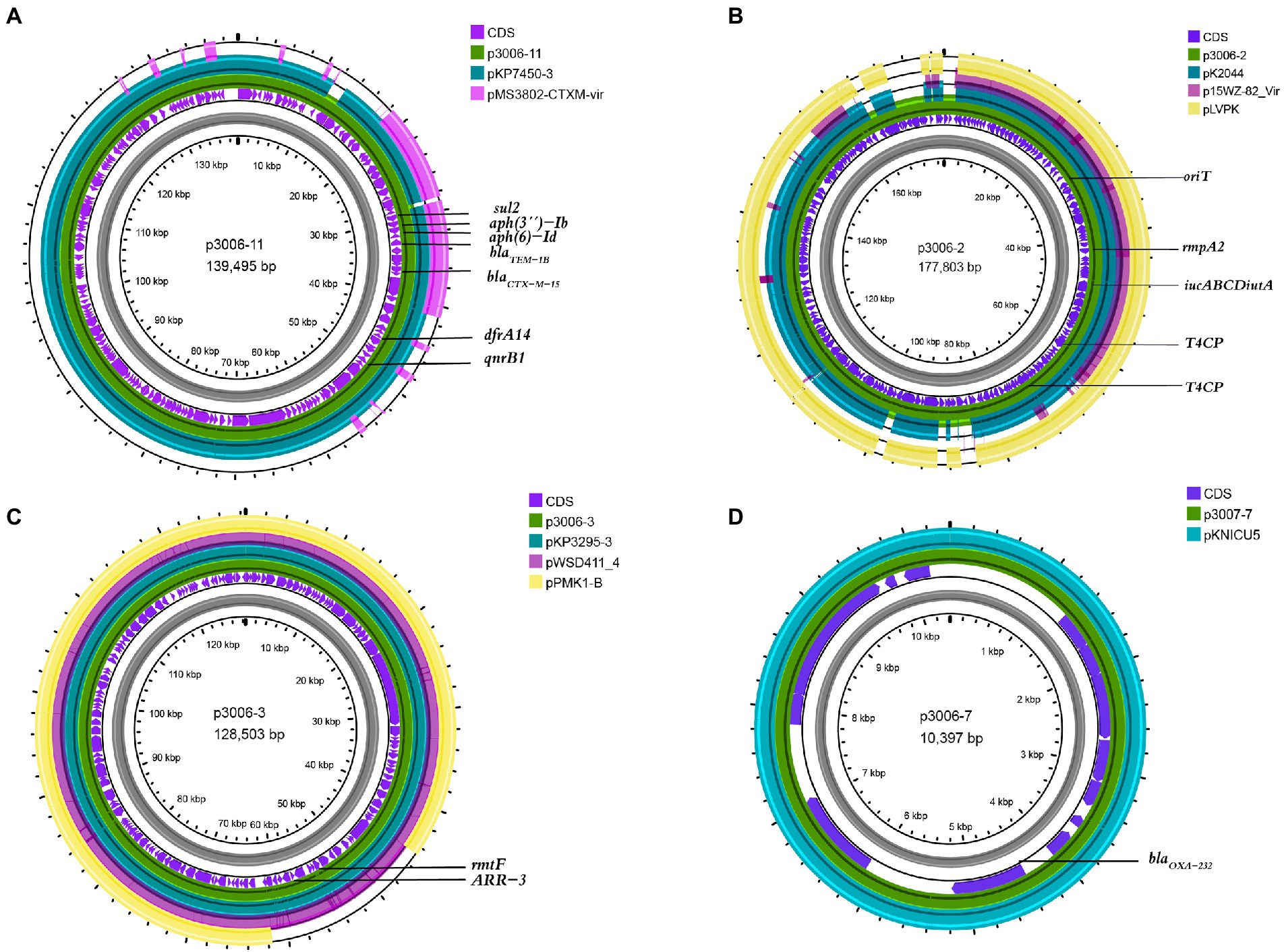
Figure 1. Comparative analysis of p3006-2, p3006-3, and p3006-7 plasmids with other reference plasmids. (A) Genome alignment was performed with p3006-11(JANCTJ010000011), pKP7450-3(99.95%, CP090471.1), and pMS3802-CTXM-vir(99.59%, CP068016.1). (B) Genome alignment was performed with p3006-2(JANCTJ010000002), virulent plasmid pK2044 (99.46%, CP026012.1), mobilisable virulence plasmid p15WZ-82-Vir(99.53%, CP032356.1), and pLVPK(99.51%, AY378100). (C) Genome alignment was performed with p3006-3(JANCTJ010000003), pKP3295-3(99.98%, CP079728.1), pWSD411_4(99.98%, CP045677.1), and mobilisable plasmid pPMK1-B(99.96%, CP008931.1). (D) Genome alignment was performed with p3006-7(JANCTJ010000007), and pKNICU5(KY454616.1).
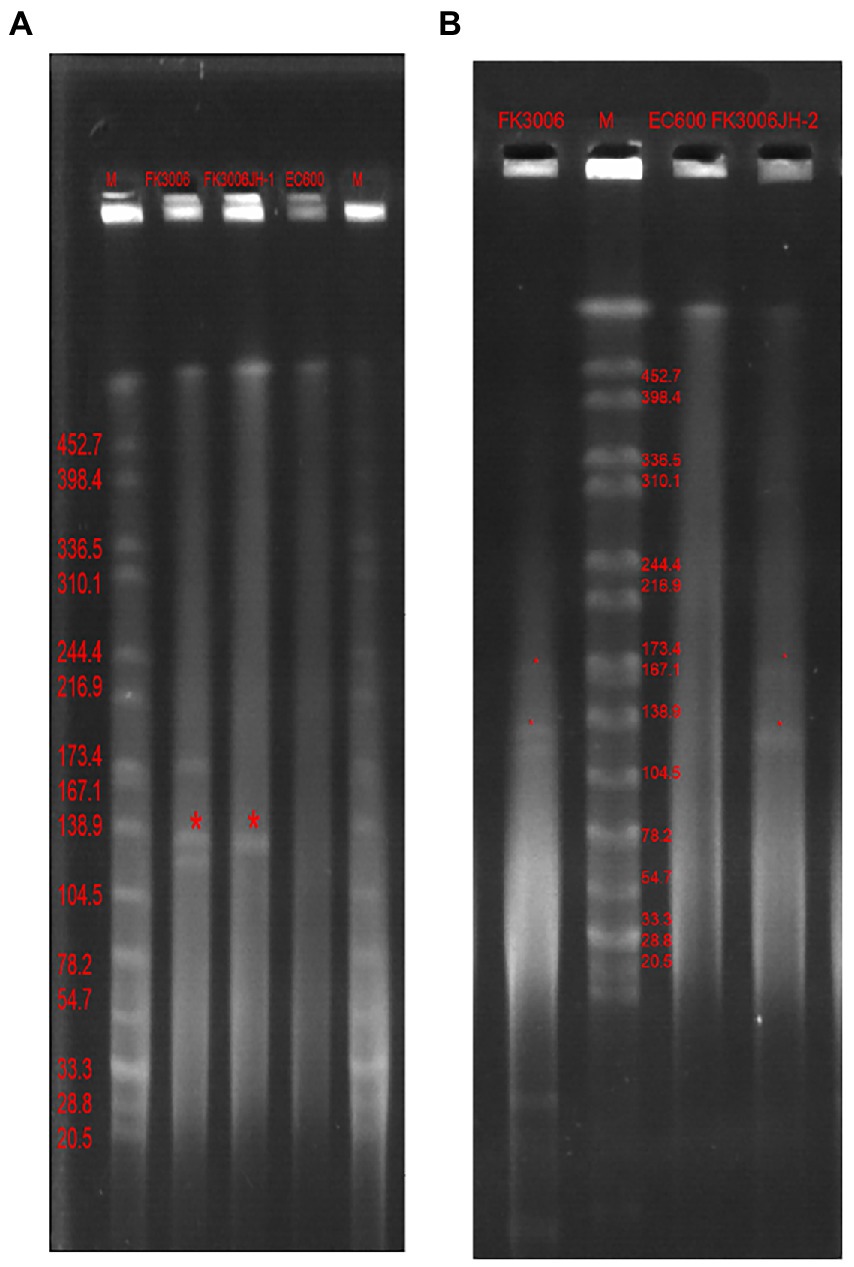
Figure 2. S1-PFGE profiles of FK3006, its transconjugants and E.coli 600 Lane marker was XbaI digested genomic DNA from Salmonella Braenderup H9812. (A) The transconjugant(FK3006JH-1, the serial dilutions were plated on selective media with ampicillin, 100 μg/ml) only had one plasmid, with the size of ~139 kb(p3006-11, blaCTX-M-15). (B) The transconjugant (FK3006JH-2, the serial dilutions were plated on selective media with dipotassium tellurite, 8 μg/ml) had two plasmid, with the size of ~139 kb(p3006-11, blaCTX-M-15) and ~ 178 kb(p3006-2, iucA).
p3006-2 was a typical IncFIB(K)/HI1B type virulence plasmid and shared ~99% identity with pK2044 (CP026012.1) and pLVPK (AY378100) plasmids (Figure 1B). Notably, p3006-2 also harbored the core oriT site and T4CP, which were identical to pK2044 and pLVPK. Previous studies have confirmed the pK2044 and pLVPK virulence plasmid could transferred from hypervirulent K. pneumoniae (hvKP) to ST11 CRKP and E. coli strains with the help of a self-transferable IncFIIK plasmid. In this study, we also observed that the p3006-2 virulence plasmid could be induced transfer with the help of p3006-11 plasmid (8.3 × 10−8-3.2 × 10−7; Figure 2B).
p3006-3 plasmid harbored rmtF gene and belonged to IncFIB type plasmid (Figure 1C), which was nearly identical (99.98%) to the human K. pneumoniae plasmid pKP3295-3 (rmtF, IncFIB(pKPHS1) CP079728.1) and pWSD411-4 (rmtF, IncFIB(pKPHS1), CP045677.1) previously reported in Hangzhou, China. Meanwhile, p3006-3 plasmid was similar to the pPMK1-B(coverage 86%, identity 99.96%, IncFIB(pKPHS1), CP008931.1) which also could be self-transferred to the recipient strain except for a multidrug resistance region of p3006-3 (Shi et al., 2020).
p3006-7 plasmid was a 10,397 bp circular molecule, harboring blaOXA-232 resistance element, and contained no necessary elements for transmission by bioinformatic analysis. The conjugation assay suggested that p3006-7 was not self-transferable. According to the genomic comparison, we found p3006-7 was almost identical to pKNICU5 plasmid (identity 99.93%, ColKP3, KY454616.1), isolated from the first OXA-232-producing K. pneumoniae strain in China (Figure 1D; Yin et al., 2017). As the self-transmissible modules were usually absent in such ColKP3 type plasmid, the blaOXA-232 genes may be mainly clonal dissemination.
The existence of pLVPK-like plasmid p3006-2 and the hypermucoviscosity indicated that the FK3006 may be a hypervirulent strain, here we applied several experiments to confirm the virulent phenotype. We found that FK3006 strain have a survival of about 74% after 60 min of incubation with the serum, which was significantly higher than that of the hypervirulence-negative resistant strain FK3036 (ST15 CRKP; Figure 3A). Moreover, there was a significant increase of siderophore production in FK3006 (d = 25 mm), compared with FK3036 (d = 10 mm), which did not have the iuc operon (Figure 3B). A549 human lung epithelial cells were infected with FK3006 (LDH, 1.70 μmol/l) led to cell LDH release of 76.9% compared with the positive control strain NUTH-K2044 group (LDH, 2.21 μmol/l). And, the negative control FK3036 group only release 0.85 μmol/l LDH. We also applied G. mellonella infection model to analyze the virulence of FK3006 in vivo. Testing showed that the survival rate of FK3006 group was 20% at 72 h, which was almost identical to that of a virulent strain of NUTH-K2044 (Figure 3C). A whole genome BLAST search was performed against VFDB to validate virulence factors harbored by the FK3006 (Supplementary Table S3). These results were in accordance with the fact that FK3006 populations were typical hypervirulent strains.
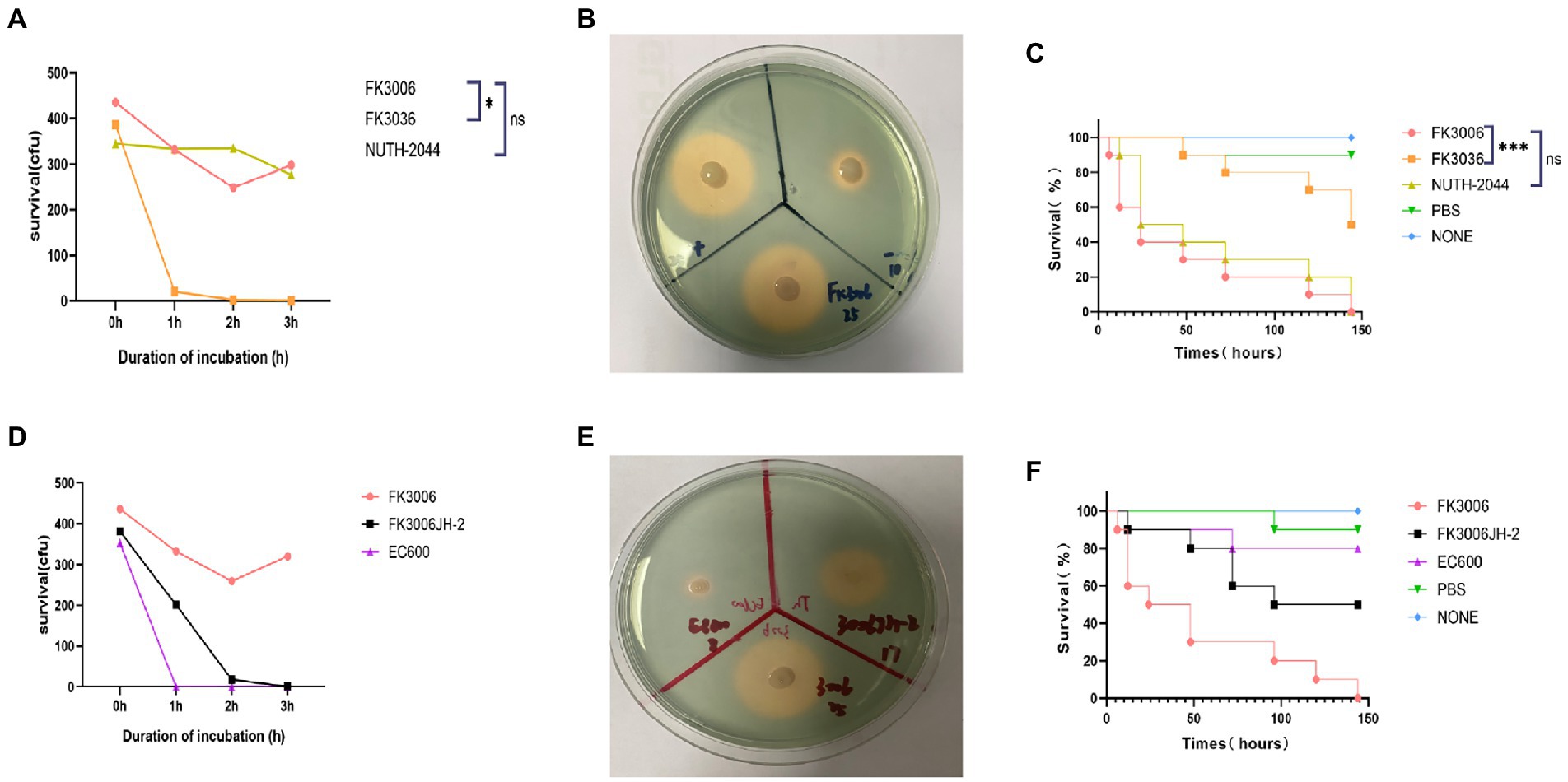
Figure 3. The virulence analysis of FK3006. (A) Serum killing assay of FK3006, FK3036 and NUTH-K2044 strains. Survival of each strain was evaluated by enumerating viable counts on MHA agar plate for 0, 1, 2, and 3 h of incubation in the pooled human serum at 37°C. There was a difference (p < 0.05) in the growth of the strain FK3006 and FK3036 *p < 0.05. (B) Siderophore production of FK3006, FK3036 and NTUH-K2044. FK3036 and NUTH-K2044 were used as negative and positive control, respectively. An orange halos zone indicated Siderophore production. (C) Survival rates of G. mellonella infected with FK3006, FK3036, NUTH-K2044, PBS, and none. Log-rank Mantel-638 Cox test was performed for analysis of the indicated curves. A difference (p < 0.001) was observed between FK3006 and FK3036 *** p < 0.001. (D) Serum killing assay of FK3006, FK3036JH-2 and EC600 strains. (E) Siderophore production of FK3006(25 mm), FK3006JH-2(17 mm) and EC600(8 mm). An orange halos zone indicated Siderophore production. (F) Survival rates of G. mellonella infected with FK3006, FK3006JH-2, EC600, PBS, and none.
Notably, according to the conjugation assay, we observed the virulence plasmid p3006-2 could successfully transferred to the recipient strain EC600, and obtained the transconjugant FK3006JH-2. We found that FK3006JH-2(iucA) strains have a survival of about 53% after 60 min of incubation with the serum, which was significantly higher than that of the recipient strain EC600 (Figure 3D). When the EC600 obtained the p3006-2 plasmid, siderophore production also increased with the diameter of the halo increased ~2-fold (Figure 3E). G. mellonella infection model showed that the survival rate of FK3006JH-2 group was significantly lower than that of EC600 group (Figure 3F). All these results indicated that the virulence features of the FK3006 could be transferred to the recipients through the key virulence plasmid p3006-2.
The plasmid not only contains genes that promote the survival of the host but also carries other MGEs, such as Tn and IS, which also make a significant contribution to the dissemination of resistance genes. To thoroughly analyze the dissemination potential of rmtF and blaCTX-M-15 in FK3006, we also analyze each of the MGEs surrounding them. In p3006-11, the blaCTX-M-15, together with other antibiotic resistance genes (sul2-aph(3″)-Ib-aph(6)-Id-blaTEM-1B-dfrA14- qnrB1), was part of a large AMR of 37,526 bp that was bracketed by two opposite orientation copies of the IS26 (Figure 4A) and it was closely associated with the presence of class 1 integrons. ISEcp1, a member of the IS1380 family frequently upstream of the blaCTX-M-15, seems to be a crucial element in the mobilization and dissemination of blaCTX-M-15, as it not only located surrounding blaCTX-M-15 in the p3006-11 plasmid, but also surrounding the blaCTX-M-15 in the pE16K0288-1(CP052263.1, IncFIB(K), IncFII(K), Korea), pDA33141-217(CP029588.1, IncFIB(K), IncFII(K), Sweden) and pMS3802-CTX-M-Vir(CP068016.1, IncHI1B(pNDM-MAR), repB, Spain) (Upadhyay et al., 2015; Fu et al., 2020; Hernandez et al., 2021). Similar to pCRKP-1,215_1 (CP024839.1, IncFII(pKPX1), Korea) and pARLG-3,135-1(CP033947.1, IncFIB(pQil), IncFII(K), United States), the typical IS6100-rmtF-ARR-3 ARI was identified in p3006-3, containing class 1 integrons (Figure 4B). IS6100, like IS26, is a member of the IS6 family, and has previously been described as the most common IS element adjacent to rmtF (Mataseje et al., 2014). However, most of IS6100 appears in the vicinity of drug resistance genes alone unlike IS26 (Partridge et al., 2018).

Figure 4. Linear comparison of the blaCTX-M-15 and rmtF region. (A) The blaCTX-M-15 region was compared with the regions extracted from pE16K0288-1(CP052263.1), pDA33141-217(CP029588.1), and pMS3802-CTX-M-Vir(NZ_CP068016.1). (B) The rmtF region was compared with the regions extracted from MS6671-UAE(LN824138.1), pARLG-3,135-1(CP033947.1), and pCRKP-1,215-1(NZ_CP024839.1).
Globally, CRE including CRKP pose a major public health threat (Logan and Weinstein, 2017; Ernst et al., 2020). The notorious nosocomial pathogen hvKP exhibits enhanced virulence features and causes metastatic, and invasive infections (Russo and Marr, 2019). The phenotypes of MDR and hypervirulence in K. pneumoniae did not overlap for a long time as MDR phenotypes are often exhibited by classical K. pneumoniae (cKP) strains while the carriage of MDR genes in hvKP isolates was rare(Lee et al., 2017). However, more and more isolates with MDR and hypervirulence have been detected in the face of antibiotic selection pressure, this poses a wide array of problems for the treatment (Hennequin and Robin, 2016; Tang et al., 2020). According to epidemiological researches, most hv-CRKP were associated with blaKPC-2 and ST11 K. pneumoniae. However, in this study, we report an un-common co-existence of blaOXA-232, rmtF, and pLVPK-like virulence plasmid in a ST15 K. pneumoniae.
FK3006, which owned the typical hypermucoviscosity feature, was isolated from the sputum sample of a young patient admitted to the ICU direct postoperatively. In FK3006, we got three resistant plasmids: p3006-3 (rmtF, ARR-3), p3006-7(blaOXA-232) and p3006-11 (blaCTX-M-15), as well as a pLVPK-like virulence plasmid. The p3006-11 plasmid was conjugative and could be successfully self-transferred to EC600. Except for p3006-11 plasmid, other resistant plasmids were typed as non-mobile plasmid, as the core oriT site was absent in p3006-3 and p3006-7. Notably, although the p3006-2 virulence plasmid was typed as non-conjugative plasmid like pK2044 and pLVPK plasmid, we observed it could be induced mobilized with the help of p3006-11 plasmid as previously studied (Xu et al., 2021; Tian et al., 2022). The pLVPK-like virulence plasmids of K. pneumoniae are generally regarded as nonconjugative, these results indicated that the co-existence of IncFIIK resistant plasmid and pLVPK-like virulence plasmid would increase the risk for the virulence dissemination. Although previous studies have reported the co-existence of blaOXA-232 and virulence-like plasmid in a ST15 K. pneumoniae, such isolate did not exhibit the hypervirulent phenotype (Shu et al., 2019). However, in our study, the existence of pLVPK-like virulence plasmid endowed the typical hypervirulent characteristics to the FK3006, and such difference may be attributed to the expression variance or other unrecognized virulence determinants in FK3006.
The dissemination of resistance genes is not only via plasmids but also via other mobile structures. ISEcp1 was located in upstream of the blaCTX-M-15 gene, which was common in other reported blaCTX-M-15 elements. In addition, the promoter sequence-35(TTGAAA) and-10(TACAAT) regions in ISEcp1 provide a potential promoter for blaCTX-M-15 gene, inducing high expression of it (Ben Slama et al., 2011; Kieslich et al., 2016). Further, IS26 surrounds blaCTX-M-15 gene and plays a key role in the dissemination (Seo and Lee, 2021). IS26 can be located at the upstream of the CTX-M-15 gene, either alone or in combination with ISEcp1. It was reported that ISEcp1 was often truncated when it co-existed with IS26, and the truncation position was not fixed. Notably, the promoter sequence of ISEcp1 was preserved (Diestra et al., 2009). In our study, ISEcp1 was not truncated by IS26. All in all, the surrounding environment of blaCTX-M-15 contains a variety of transposons and integrons. Meanwhile, coexisting with other drug-resistant genes in the same plasmid makes it easier for them to survive in the environment.
Enterobacterales isolates producing rmtF used to be extremely rare in China, but in recent years relevant reports have emerged and are always accompanied by coproduction of OXA-232. Plasmids with rmtF gene often acquired through multiple mobile elements. In this study, the rmtF plasmid p3006-3 could not self-transferred to other isolates, but the MGEs surrounding the rmtF generated a mobile ARI. The genetic background of the rmtF is associated with insertion sequence IS6100. Previous studies showed Tn6229 was related to the Tn3 family and carried a class 1 integron harboring the rmtF and IS6100 (Mataseje et al., 2014). This ARI, together with several MGEs (IS6100 and Tn3), can form a highly active transmission among the strains, which is extremely harmful.
In this study, we report the uncommon co-existence of blaOXA-232, rmtF, and a movable pLVPK-like virulence plasmid a ST15 K. pneumoniae. The association of antibiotic resistance genes with mobile genetic elements in FK3006 could promote rapid emergence of hv-CRKP strains. Notably, we found the co-exsistence of resistance and virulence plasmid not only generated the high-risk hypervirulent multidrug-resistant phenotype, but also increased the transmission threaten of non-conjugative virulence plasmid. In the future, when plasmid analysis becomes a routine detection method for such high-risk bacteria in medical institutions, necessary interventions could be carried out as early as possible, and the mortality rate might be reduced.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
CW: conceptualization, data curation, formal analysis, methodology, and writing–original draft. YZ: data curation, methodology, and writing–original draft. WA and HZ: methodology. YG, BW, and XnW: software. XaW: formal analysis. LR and JZ: writing–original draft. FY: conceptualization, project administration, and writing–review and editing. LW: conceptualization and writing–review and editing. All authors contributed to the article and approved the submitted version.
We thank the authority of NTUH-K2044 by Jin-Town Wang from National Taiwan University Hospital.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1133590/full#supplementary-material
1. ^https://cge.cbs.dtu.dk/services/PlasmidFinder/
2. ^https://tool2-mml.sjtu.edu.cn/VRprofile/
3. ^https://tool-mml.sjtu.edu.cn/oriTfinder/oriTfinder.html
4. ^https://www-is.biotoul.fr/
5. ^https://proksee.ca/projects/10a9d9fa-c055-4dfe-b615-a55049475a52
Ai, W., Zhou, Y., Wang, B., Zhan, Q., Hu, L., Xu, Y., et al. (2021). First report of coexistence of blaSFO–1 and blaNDM–1 β-lactamase genes as well as colistin resistance gene mcr-9 in a transferrable plasmid of a clinical isolate of Enterobacter hormaechei. Front. Microbiol. 12:676113. doi: 10.3389/fmicb.2021.676113
Ben Slama, K., Ben Sallem, R., Jouini, A., Rachid, S., Moussa, L., Saenz, Y., et al. (2011). Diversity of genetic lineages among CTX-M-15 and CTX-M-14 producing Escherichia coli strains in a Tunisian hospital. Curr. Microbiol. 62, 1794–1801. doi: 10.1007/s00284-011-9930-4
CLSI. (2020). Performance Standards for Antimicrobial Susceptibility Testing. CLSI Supplement M100. 30th. Wayne: Clinical and Laboratory Standards Institute.
Daikos, G. L., Tsaousi, S., Tzouvelekis, L. S., Anyfantis, I., Psichogiou, M., Argyropoulou, A., et al. (2014). Carbapenemase-producing Klebsiella pneumoniae bloodstream infections: lowering mortality by antibiotic combination schemes and the role of carbapenems. Antimicrob. Agents Chemother. 58, 2322–2328. doi: 10.1128/AAC.02166-13
Diestra, K., Juan, C., Curiao, T., Moya, B., Miro, E., Oteo, J., et al. (2009). Characterization of plasmids encoding blaESBL and surrounding genes in Spanish clinical isolates of Escherichia coli and Klebsiella pneumoniae. J. Antimicrob. Chemother. 63, 60–66. doi: 10.1093/jac/dkn453
Ernst, C. M., Braxton, J. R., Rodriguez-Osorio, C. A., Zagieboylo, A. P., Li, L., Pironti, A., et al. (2020). Adaptive evolution of virulence and persistence in carbapenem-resistant Klebsiella pneumoniae. Nat. Med. 26, 705–711. doi: 10.1038/s41591-020-0825-4
EUCAST. (2020). Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 10.0. Available at: http://www.eucast.org (Accessed January 20, 2020).
Fu, Y., Xu, X., Zhang, L., Xiong, Z., Ma, Y., Wei, Y., et al. (2020). Fourth generation cephalosporin resistance among Salmonella enterica Serovar Enteritidis isolates in Shanghai, China conferred by Bla CTX-M-55 harboring plasmids. Front. Microbiol. 11:910. doi: 10.3389/fmicb.2020.00910
Galani, I., Nafplioti, K., Adamou, P., Karaiskos, I., Giamarellou, H., Souli, M., et al. (2019). Nationwide epidemiology of carbapenem resistant Klebsiella pneumoniae isolates from Greek hospitals, with regards to plazomicin and aminoglycoside resistance. BMC Infect. Dis. 19:167:167. doi: 10.1186/s12879-019-3801-1
Gottig, S., Riedel-Christ, S., Saleh, A., Kempf, V. A., and Hamprecht, A. (2016). Impact of blaNDM-1 on fitness and pathogenicity of Escherichia coli and Klebsiella pneumoniae. Int. J. Antimicrob. Agents 47, 430–435. doi: 10.1016/j.ijantimicag.2016.02.019
Hennequin, C., and Robin, F. (2016). Correlation between antimicrobial resistance and virulence in Klebsiella pneumoniae. Eur. J. Clin. Microbiol. Infect. Dis. 35, 333–341. doi: 10.1007/s10096-015-2559-7
Hernandez, M., Lopez-Urrutia, L., Abad, D., De Frutos Serna, M., Ocampo-Sosa, A. A., and Eiros, J. M. (2021). First report of an extensively drug-resistant ST23 Klebsiella pneumoniae of capsular serotype K1 co-producing CTX-M-15, OXA-48 and ArmA in Spain. Antibiotics (Basel) 10:157. doi: 10.3390/antibiotics10020157
Karaiskos, I., Lagou, S., Pontikis, K., Rapti, V., and Poulakou, G. (2019). The “old” and the “new” antibiotics for MDR gram-negative pathogens: for whom, when, and how. Front. Public Health 7:151. doi: 10.3389/fpubh.2019.00151
Kieslich, K., Littlejohns, P., and Weale, A. (2016). Drug appraisal issues must be resolved at policy level. BMJ 354:i4519. doi: 10.1136/bmj.i4519
Lee, C. R., Lee, J. H., Park, K. S., Jeon, J. H., Kim, Y. B., Cha, C. J., et al. (2017). Antimicrobial resistance of Hypervirulent Klebsiella pneumoniae: epidemiology, Hypervirulence-associated determinants, and resistance mechanisms. Front. Cell. Infect. Microbiol. 7:483. doi: 10.3389/fcimb.2017.00483
Li, G., Shi, J., Zhao, Y., Xie, Y., Tang, Y., Jiang, X., et al. (2020). Identification of hypervirulent Klebsiella pneumoniae isolates using the string test in combination with Galleria mellonella infectivity. Eur. J. Clin. Microbiol. Infect. Dis. 39, 1673–1679. doi: 10.1007/s10096-020-03890-z
Liu, D., Liu, Z. S., Hu, P., Cai, L., Fu, B. Q., Li, Y. S., et al. (2016). Characterization of surface antigen protein 1 (SurA1) from Acinetobacter baumannii and its role in virulence and fitness. Vet. Microbiol. 186, 126–138. doi: 10.1016/j.vetmic.2016.02.018
Liu, Y., Long, D., Xiang, T. X., Du, F. L., Wei, D. D., Wan, L. G., et al. (2019). Whole genome assembly and functional portrait of hypervirulent extensively drug-resistant NDM-1 and KPC-2 co-producing Klebsiella pneumoniae of capsular serotype K2 and ST86. J. Antimicrob. Chemother. 74, 1233–1240. doi: 10.1093/jac/dkz023
Logan, L. K., and Weinstein, R. A. (2017). The epidemiology of Carbapenem-resistant Enterobacteriaceae: the impact and evolution of a global menace. J. Infect. Dis. 215, S28–S36. doi: 10.1093/infdis/jiw282
Mataseje, L. F., Boyd, D. A., Lefebvre, B., Bryce, E., Embree, J., Gravel, D., et al. (2014). Complete sequences of a novel blaNDM-1-harbouring plasmid from Providencia rettgeri and an FII-type plasmid from Klebsiella pneumoniae identified in Canada. J. Antimicrob. Chemother. 69, 637–642. doi: 10.1093/jac/dkt445
Miltgen, G., Cholley, P., Martak, D., Thouverez, M., Seraphin, P., Leclaire, A., et al. (2020). Carbapenemase-producing Enterobacteriaceae circulating in the Reunion Island, a French territory in the Southwest Indian Ocean. Antimicrob. Resist. Infect. Control 9:36. doi: 10.1186/s13756-020-0703-3
Nagasawa, M., Kaku, M., Kamachi, K., Shibayama, K., Arakawa, Y., Yamaguchi, K., et al. (2014). Loop-mediated isothermal amplification assay for 16S rRNA methylase genes in gram-negative bacteria. J. Infect. Chemother. 20, 635–638. doi: 10.1016/j.jiac.2014.08.013
Partridge, S. R., Kwong, S. M., Firth, N., and Jensen, S. O. (2018). Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 31:e00088. doi: 10.1128/CMR.00088-17
Pitout, J. D. D., Peirano, G., Kock, M. M., Strydom, K. A., and Matsumura, Y. (2019). The global ascendency of OXA-48-type carbapenemases. Clin. Microbiol. Rev. 33:e00102. doi: 10.1128/CMR.00102-19
Potron, A., Rondinaud, E., Poirel, L., Belmonte, O., Boyer, S., Camiade, S., et al. (2013). Genetic and biochemical characterisation of OXA-232, a carbapenem-hydrolysing class D beta-lactamase from Enterobacteriaceae. Int. J. Antimicrob. Agents 41, 325–329. doi: 10.1016/j.ijantimicag.2012.11.007
Ramirez, M. S., and Tolmasky, M. E. (2010). Aminoglycoside modifying enzymes. Drug Resist. Updat. 13, 151–171. doi: 10.1016/j.drup.2010.08.003
Russo, T. A., and Marr, C. M. (2019). Hypervirulent Klebsiella pneumoniae. Clin. Microbiol. Rev. 32:e00001. doi: 10.1128/CMR.00001-19
Seo, K. W., and Lee, Y. J. (2021). The occurrence of CTX-M-producing E. coli in the broiler parent stock in Korea. Poult. Sci. 100, 1008–1015. doi: 10.1016/j.psj.2020.09.005
Shi, Q., Han, R., Guo, Y., Zheng, Y., Yang, Y., Yin, D., et al. (2020). Emergence of ST15 Klebsiella pneumoniae clinical isolates producing plasmids-mediated RmtF and OXA-232 in China. Infect. Drug Resist. 13, 3125–3129. doi: 10.2147/IDR.S257298
Shu, L., Dong, N., Lu, J., Zheng, Z., Hu, J., Zeng, W., et al. (2019). Emergence of OXA-232 carbapenemase-producing Klebsiella pneumoniae that carries a pLVPK-like virulence plasmid among elderly patients in China. Antimicrob. Agents Chemother. 63:e02246. doi: 10.1128/AAC.02246-18
Tang, M., Kong, X., Hao, J., and Liu, J. (2020). Epidemiological characteristics and formation mechanisms of multidrug-resistant Hypervirulent Klebsiella pneumoniae. Front. Microbiol. 11:581543. doi: 10.3389/fmicb.2020.581543
Tian, D., Liu, X., Chen, W., Zhou, Y., Hu, D., Wang, W., et al. (2022). Prevalence of hypervirulent and carbapenem-resistant Klebsiella pneumoniae under divergent evolutionary patterns. Emerg Microbes Infect 11, 1936–1949. doi: 10.1080/22221751.2022.2103454
Upadhyay, S., Hussain, A., Mishra, S., Maurya, A. P., Bhattacharjee, A., and Joshi, S. R. (2015). Genetic environment of plasmid mediated CTX-M-15 extended Spectrum Beta-lactamases from clinical and food borne bacteria in north-eastern India. PLoS One 10:e0138056. doi: 10.1371/journal.pone.0138056
Wang, M., Guo, H., He, F., and Xu, J. (2022). Genomic and phylogenetic analysis of a multidrug-resistant Klebsiella pneumoniae ST15 strain co-carrying blaOXA-232 and blaCTX-M-15 recovered from a gallbladder infection in China. J. Glob. Antimicrob. Resist. 30, 228–230. doi: 10.1016/j.jgar.2022.06.023
Xu, Y., Zhang, J., Wang, M., Liu, M., Liu, G., Qu, H., et al. (2021). Mobilization of the nonconjugative virulence plasmid from hypervirulent Klebsiella pneumoniae. Genome Med. 13:119. doi: 10.1186/s13073-021-00936-5
Yang, X., Dong, N., Chan, E. W., Zhang, R., and Chen, S. (2021). Carbapenem resistance-encoding and virulence-encoding conjugative plasmids in Klebsiella pneumoniae. Trends Microbiol. 29, 65–83. doi: 10.1016/j.tim.2020.04.012
Yin, D., Dong, D., Li, K., Zhang, L., Liang, J., Yang, Y., et al. (2017). Clonal dissemination of OXA-232 Carbapenemase-producing Klebsiella pneumoniae in neonates. Antimicrob. Agents Chemother. 61:e00385. doi: 10.1128/AAC.00385-17
Keywords: blaOXA-232, rmtF, Klebsiella pneumoniae, carbapenemase, mobile element, plasmid
Citation: Wu C, Zhou Y, Ai W, Guo Y, Wu X, Wang B, Zhao H, Rao L, Wang X, Zhang J, Yu F and Wang L (2023) Co-occurrence of OXA-232, RmtF-encoding plasmids, and pLVPK-like virulence plasmid contributed to the generation of ST15-KL112 hypervirulent multidrug-resistant Klebsiella pneumoniae. Front. Microbiol. 14:1133590. doi: 10.3389/fmicb.2023.1133590
Received: 29 December 2022; Accepted: 09 February 2023;
Published: 28 February 2023.
Edited by:
Wei Wang, China National Center for Food Safety Risk Assessment, ChinaCopyright © 2023 Wu, Zhou, Ai, Guo, Wu, Wang, Zhao, Rao, Wang, Zhang, Yu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fangyou Yu, ✉ d3pqeHlmeUAxNjMuY29t; Liangxing Wang, ✉ d2FuZ2xpYW5neGluZ0B3emhvc3BpdGFsLmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.