- Research Group of Industrial Microbiology and Food Biotechnology (IMDO), Faculty of Sciences and Bioengineering Sciences, Vrije Universiteit Brussel, Brussels, Belgium
Industrial production of Gouda cheeses mostly relies on a rotated use of different mixed-strain lactic acid bacteria starter cultures to avoid phage infections. However, it is unknown how the application of these different starter culture mixtures affect the organoleptic properties of the final cheeses. Therefore, the present study assessed the impact of three different starter culture mixtures on the batch-to-batch variations among Gouda cheeses from 23 different batch productions in the same dairy company. Both the cores and rinds of all these cheeses were investigated after 36, 45, 75, and 100 weeks of ripening by metagenetics based on high-throughput full-length 16S rRNA gene sequencing accompanied with an amplicon sequence variant (ASV) approach as well as metabolite target analysis of non-volatile and volatile organic compounds. Up to 75 weeks of ripening, the acidifying Lactococcus cremoris and Lactococcus lactis were the most abundant bacterial species in the cheese cores. The relative abundance of Leuconostoc pseudomesenteroides was significantly different for each starter culture mixture. This impacted the concentrations of some key metabolites, such as acetoin produced from citrate, and the relative abundance of non-starter lactic acid bacteria (NSLAB). Cheeses with the least Leuc. pseudomesenteroides contained more NSLAB, such as Lacticaseibacillus paracasei that was taken over by Tetragenococcus halophilus and Loigolactobacillus rennini upon ripening time. Taken together, the results indicated a minor role of leuconostocs in aroma formation but a major impact on the growth of NSLAB. The relative abundance of T. halophilus (high) and Loil. rennini (low) increased with ripening time from rind to core. Two main ASV clusters of T. halophilus could be distinguished, which were differently correlated with some metabolites, both beneficial (regarding aroma formation) and undesirable ones (biogenic amines). A well-chosen T. halophilus strain could be a candidate adjunct culture for Gouda cheese production.
1. Introduction
Gouda cheese production is a complex interplay of a mixed-strain starter culture, non-starter microorganisms, and time. A typical Gouda cheese starter culture, used in lyophilized or frozen form, contains (undefined) strains of the lactic acid bacteria (LAB) Lactococcus lactis subsp. lactis (Lc. lactis), Lactococcus cremoris subsp. cremoris (Lc. cremoris), Lactococcus lactis subsp. lactis biovar diacetylactis (Lc. lactis biovar diacetylactis), and Leuconostoc mesenteroides and/or Leuconostoc lactis (Düsterhöft et al., 2017). The main role of this mesophilic starter culture is to lower the pH of the fermenting milk by converting lactose into lactate, which is almost completed after 10 h. Besides acidification by the homofermentative Lactococcus, citrate utilization by Lc. lactis biovar diacetylactis and/or leuconostocs occurs, which results into buttery flavor compounds, such as diacetyl, acetoin, and/or 2,3-butanediol (Starrenburg and Hugenholtz, 1991; Smit et al., 2005; Steele et al., 2013; Düsterhöft et al., 2017). In addition, all strains contribute to flavor development through proteolysis, peptidolysis, and amino acid conversions. The heterofermentative leuconostocs produce few eyes because of carbon dioxide formation through heterolactic fermentation and citrate conversion. These subprocesses of fermentation and ripening determine the complex and dynamic flavor profile of Gouda cheese, which is characterized by a sweet and mild taste.
The application of the same LAB starter cultures in a Gouda cheese production environment increases the risk of a bacteriophage infection, which may lead to failed fermentation processes and thus economical losses (Mahony and van Sinderen, 2014). Starter cultures consisting of a complex mixture of strains are in general less phage-sensitive, but a starter culture with only few strains guarantees a more consistent end-product quality (Erkus et al., 2013; Spus et al., 2015). Therefore, most cheese factories apply a starter culture rotation with three to five starter cultures with different phage sensitivity profiles (Høier et al., 2010; Parente et al., 2017; Frantzen et al., 2018). However, little is known about the impact of the use of rotating starter cultures on the final organoleptic properties of Gouda cheese throughout various batch productions.
Although mixed-strain LAB starter cultures are used to steer the Gouda fermentation and ripening process, mesophilic non-starter lactic acid bacteria (NSLAB) originate from the milk or the cheese production equipment or environment (e.g., brine bath and ripening room) and are able to grow to counts as high as 108 CFU/g during the ripening phase (Fitzsimons et al., 2001; Van Hoorde et al., 2008; Fox et al., 2017; Gobbetti et al., 2018). In general, Lacticaseibacillus paracasei and Lactiplantibacillus plantarum are the main NSLAB across various cheese types ripened for 4 months or longer, including Gouda cheese (Gobbetti et al., 2015). Positive contributions of NSLAB to cheese flavor are well known, as they hydrolyze peptides and convert the resulting free amino acids into flavor compounds, which however depends on the production batch (Montel et al., 2014; Gobbetti et al., 2015). Moreover, in some cases, they cause off-flavors or undesired gas production (Martley and Crow, 1993; Oberg et al., 2016). To take benefit from the NSLAB regarding flavor formation, but to avoid batch variability, selected NSLAB are added as adjunct cultures in some cheese productions (Antonsson et al., 2003; Van Hoorde et al., 2010b; O’Brien et al., 2017; Randazzo et al., 2021). Additionally, Enterococcus durans, Enterococcus faecalis, Enterococcus faecium, Lacticaseibacillus rhamnosus, Latilactobacillus curvatus, Lentilactobacillus parabuchneri, and Pediococcus pentosaceus may be part of the Gouda cheese NSLAB communities (Van Hoorde et al., 2008, 2010a). Also, culture-dependent methods detect a higher relative abundance of NSLAB in aged Gouda cheese compared to culture-independent approaches, because the starter culture strains occur in a viable but not culturable (VBNC) state during ripening that can only be detected through DNA sequencing (Ganesan et al., 2007; Ruggirello et al., 2014). Finally, given the reclassification of the Lactobacillus genus (Zheng et al., 2020), an update is needed to assess the full diversity of lactobacilli in Gouda cheese.
Whereas culture-dependent methods are more laborious and only isolate microorganisms that can grow on the agar media and under the incubation conditions applied, resulting into an inherent selection bias, culture-independent methods are more suited to investigate the taxonomical diversity of a microbial ecosystem (Ercolini, 2013; De Filippis et al., 2017; Levante et al., 2021). Most metagenetic high-throughput amplicon-based sequencing studies on cheese target only a part of the 16S rRNA gene, as has been performed, for instance, for Irish artisan cheeses (Quigley et al., 2012), soft Herve cheese (Delcenserie et al., 2014), Portuguese Pico cheese (Riquelme et al., 2015), traditional Brazilian cheeses (Kamimura et al., 2019), Italian mozzarella (Marino et al., 2019), Gouda cheeses (Park et al., 2019), and Grana Padano cheeses (Zago et al., 2021). Alternatively, the microbiota of some cheeses has been investigated by high-throughput sequencing of the whole 16S rRNA gene, using PacBio technology, which allows a more accurate species-level identification (Jin et al., 2018; Yang et al., 2020). The latter study has also attempted to link the most prevailing species with sensory profiles generated with an electric nose (Yang et al., 2020). However, since the metagenetic sequence reads were clustered into operational taxonomic units (OTUs) instead of amplicon sequence variants (ASVs), taxonomic assignment could not be performed below species level (Callahan et al., 2017). Indeed, ASVs allow a single-nucleotide resolution that can resolve taxonomical identification at species and sometimes even strain level (Callahan et al., 2019; Johnson et al., 2019). In that perspective, a meta-analysis of cheese microbiomes has indicated a significantly different volatile profile for strains of the same species, encompassing Brevibacterium linens, Lc. lactis, and Streptococcus thermophilus involved in artisan Irish cheese production (Walsh et al., 2020), which emphasizes the relevance of obtaining a reliable taxonomical resolution below species level.
Besides an accurate taxonomic assignment of the microbial dynamics during Gouda cheese ripening, a thorough metabolomic analysis as a function of the ripening time is needed, in particular to be able to make the link between the microbial taxa and flavor compounds produced unequivocally. For example, research on smear-ripened cheddar cheeses has correlated both Debaryomyces hansenii and Glutamicibacter arilaitensis with alcohols and carboxylic acids, and the latter species with ketones as well, whereas B. linens and Geotrichum candidum have been correlated with sulfur compounds (Bertuzzi et al., 2018). An in-depth study of Gouda cheese has investigated the sensory and chemical properties of 36 productions from five countries and indicated the age of the cheeses as most determining factor to explain differences in concentrations of flavor compounds formed during ripening (Jo et al., 2018). Lactic acid has been identified as the main organic acid in Gouda cheeses. Acetic acid, diacetyl, 2-and 3-methylbutanal, and ethyl butyrate are among the most important aroma-active compounds. This is in line with other studies that have indicated acetoin (buttery notes), lactones (fruity/sweet/buttery notes), methylketones (floral/nutty notes), and short-chain fatty acids (cheesy/rancid notes) as additional major constituents of Gouda cheeses (Dirinck and De Winne, 1999; Van Leuven et al., 2008; Van Hoorde et al., 2010b; Jung et al., 2013; Ruyssen et al., 2013). Sulfur compounds, such as dimethyl sulfides and methional, are present at relatively low concentrations, but do have an impact on the Gouda cheese flavor (Van Leuven et al., 2008; Jo et al., 2018). In addition to their contribution to the basic cheese taste (Toelstede and Hofmann, 2008), amino acids are precursors for volatile organic compounds that contribute to the cheese aroma, such as higher alcohols, higher aldehydes, and esters (Smit et al., 2005). However, they can also be the precursors of biogenic amines, which have a toxicological potential, especially histamine and tyramine (Komprda et al., 2007; Linares et al., 2012). All these compounds can be produced by a wide range of LAB species, which is considered to be a strain-dependent trait (Barbieri et al., 2019; Walsh et al., 2020). However, few studies have focused on potential differences in microbial and metabolite compositions of the core and rind of the cheeses investigated (Calasso et al., 2016; Dugat-Bony et al., 2016; Salazar et al., 2018).
Whereas several studies on Gouda cheese have focused on its microbial composition with limited focus on the metabolites produced, other studies have investigated a restricted part of the metabolome with limited attention for the microbial composition. Therefore, the present study aimed at a systematic assessment of the overall microbiome (below species level) and metabolome (including endogenous and microbial substrates and metabolites, both volatile and non-volatile ones) as well as their link with the application of rotated starter cultures and/or cheese cores and rinds of Gouda cheeses from 23 different production batches, throughout their whole ripening time and made in the same dairy factory.
2. Materials and methods
2.1. Cheese production, selection, and sampling
Gouda cheeses (23 production batches) were produced in a European dairy factory between mid-July and mid-August 2018 from standardized and pasteurized milk, according to a standard Gouda manufacturing process (Düsterhöft et al., 2017), using three different, commercial, frozen, mesophilic, undefined, mixed-strain LAB starter cultures (A, B, and C), composed of Lc. lactis, Lc. cremoris, Lc. lactis biovar diacetylactis, and Leuconostoc spp. As these starter culture mixtures were used in a rotation system, the corresponding Gouda cheeses are further referred to as Gouda A (8 production batches, numbered A1–A8), B (7, B1–B7), and C (8, C1–C8) cheeses (Figure 1). All starter culture mixtures were from the same starter culture manufacturer. Cheeses of each batch production were organoleptically assessed by the company’s tasting panel after 26 weeks of ripening and scored as less than average (L), good (G), or exceptionally good (E). For further analysis, for each starter culture used, three cheese wheels with sensory score L and three wheels with sensory score G of the different production batches were selected, resulting in 18 cheese wheels. Additionally, five cheese wheels with sensory score E were selected, irrespective of the starter culture used. This selection resulted in a total of 23 Gouda cheese production batches for metagenetic and metabolomic analysis, which was carried out as a function of the ripening time. The analyses were represented by cheeses of a ripening time ready for sale (36 and 45 weeks) and long-ripened cheeses (75 and 100 weeks). Batches with sensory score G were also analyzed after 26 (time point of sensory assessment) and 31 weeks of ripening.
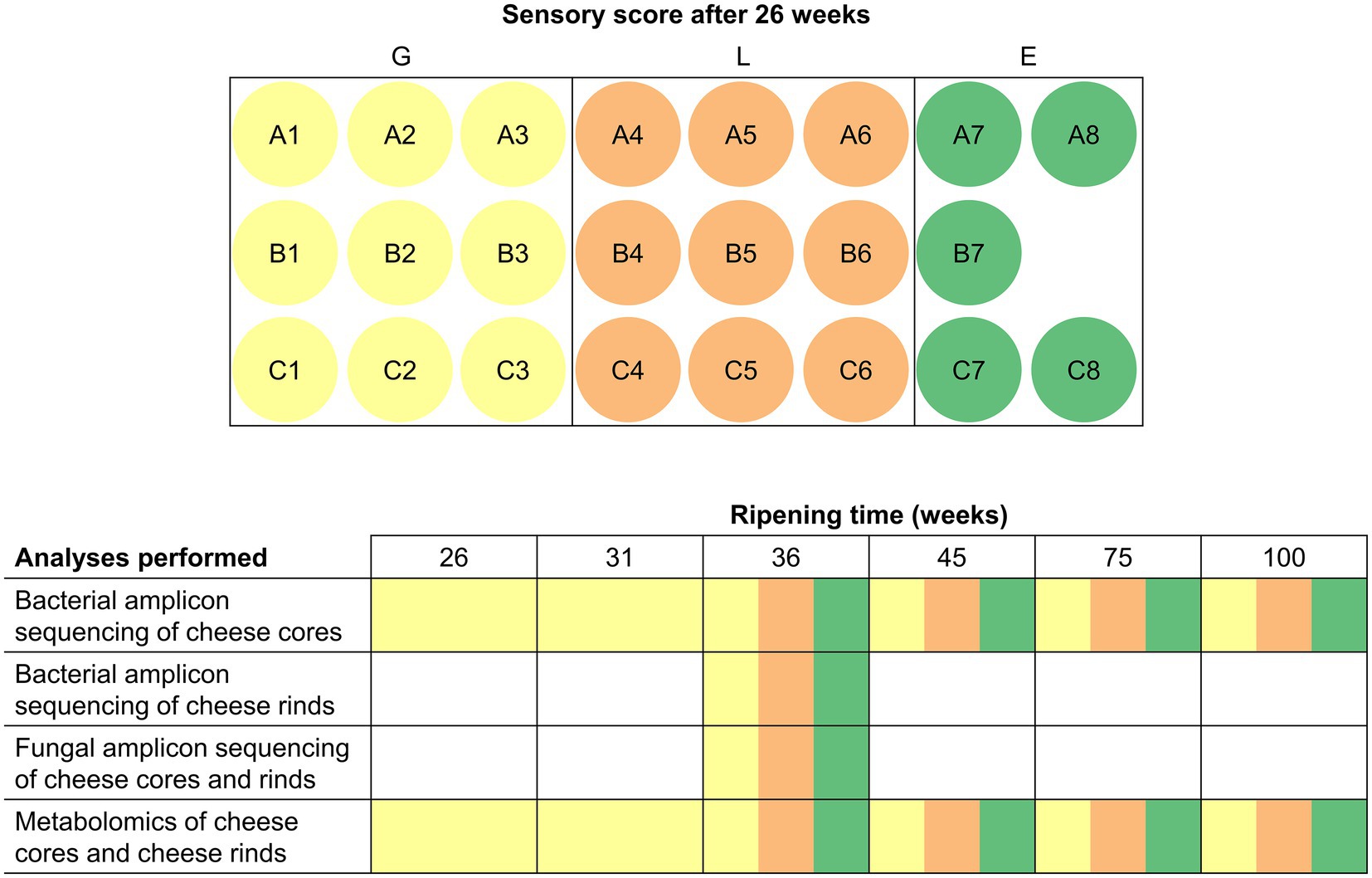
Figure 1. Overview of all cheese batch productions investigated during the present study and the multiphasic analyses performed. Each letter (A, B, and C) indicates the starter culture mixture applied that was used in a rotation system. The color indicates the sensory score given after 26 weeks of ripening: yellow, good (G); orange, less than average (L); green, exceptionally good (E). All cheese batch productions were investigated after 36, 45, 75, and 100 weeks of ripening. Wheels from batches with sensory score G were additionally investigated after 26 and 31 weeks of ripening.
All selected cheese wheels were vacuum-packed and stored at 4°C in the factory before their transport to and further sampling in the research group’s laboratory. Therefore, a cheese wheel was cut into two halves and a slice of 3 cm from the border of one halve was taken. Three pieces of approximately 40 g of the core were cut out aseptically from this slice, which were further cut into smaller pieces, and then pooled. For the rind samples, pieces of approximately 60 g of the top and the bottom of the remaining halve were pooled. Ten grams of pooled sample, either core or rind, were transferred to a stomacher bag (Seward, Worthing, West Sussex, United Kingdom) and mixed at medium speed with 90 mL of a sterile sodium citrate solution (2%, m/v; Sigma-Aldrich, St. Louis, MO, United States) in a stomacher (Laboratory Blender Stomacher 400, Seward) for 5 min. This suspension was centrifuged at 4,500 × g for 20 min and the cell pellets obtained were divided into four aliquots. The pellets were resuspended in 1 mL of sorbitol buffer [1.2 M sorbitol (VWR International, Darmstadt, Germany) and 50 mM Tris-base (Calbiochem, San Diego, CA, United States); pH 7.5], centrifuged at 6,000 × g for 10 min, and the resulting cell pellets were stored at −25°C before further DNA extraction.
2.2. Metagenetic analysis
2.2.1. DNA extraction
Whole-community DNA extraction of the cheese samples was performed as described previously (Vermote et al., 2018). Briefly, the cell pellets obtained as mentioned above were submitted to an enzymatic digestion [one incubation step with lyticase (Sigma-Aldrich) and Zymolyase (G-Biosciences, St. Louis, MO, United States) and another one with lysozyme (Merck, Darmstadt, Germany) and mutanolysin (Sigma-Aldrich)], a chemical/enzymatic treatment [with sodium dodecyl sulfate (Sigma-Aldrich) and proteinase K (Merck)], and a mechanical disruption by vortexing in the presence of acid-washed glass beads (Sigma-Aldrich), followed by protein removal using a mixture of chloroform, phenol and isoamyl alcohol (Sigma-Aldrich), a treatment with RNase (Roche, Basel, Switzerland), and DNA purification with a DNeasy Blood & Tissue Kit (Qiagen, Venlo, Netherlands). The DNA purity was assessed using a NanoDrop 2000 spectrophotometer and the DNA concentrations were measured with a Qubit 2.0 fluorometer using a Qubit dsDNA HS Assay Kit (all from Thermo Fisher Scientific, Carlsbad, CA, United States).
2.2.2. Metagenetics
To assess the bacterial members represented by the whole-community DNA isolated from the cheese samples, amplicon-based high-throughput sequencing was applied for the cheese core samples taken at all time points as well as for the cheese rind samples taken after 36 weeks of ripening. Therefore, the bacterial full-length 16S rRNA gene was amplified with the universal primer set 27F (AGRGTTYGATYMTGGCTCAG) and 1492R (RGYTACCTTGTTACGACTT) (Integrated DNA Technologies, Leuven, Belgium). Both primers were tagged with 5′ sample-specific barcodes to allow for multiplexed sequencing, according to the manufacturer’s instructions (PacBio, Menlo Park, CA, United States). PCR assays were performed using the KAPA HiFi DNA Polymerase (Hot Start and Ready Mix formulation; Roche), with 3 min of denaturation at 95°C, followed by 24 cycles of denaturation at 95°C for 30 s, annealing at 57°C for 30 s, and extension at 72°C for 60 s. Post-amplification quality control was performed by capillary electrophoresis, using a Bioanalyzer 2100 and a High Sensitivity DNA Kit (Agilent Technologies, Santa Clara, CA, United States). The DNA concentrations were measured with a Qubit 2.0 fluorometer. The amplicon sets were equimolarly pooled, a circular sequencing adaptor was ligated, and the resulting amplicon library was sequenced using a PacBio Sequel system in circular consensus mode (VIB Nucleomics Core Facility, Leuven, Belgium). Circular consensus sequences (CCS) were generated, using SMRT Link 4.0 (PacBio), with the minimum predicted accuracy set to 0.998. These long-read sequences were clustered into ASVs with the DADA2 R software package (version 1.14.1), updated for long amplicon reads (Callahan et al., 2019). The filtering parameter settings were minQ = 3, minLen = 1100, maxLen = 1600, maxN = 0, and maxEE = 2. In the dada function, the pool parameter was set to “pseudo” to perform the analysis twice. First, all samples were processed by the DADA2 algorithm independently and all ASVs obtained were then used as input for the second analysis, during which the algorithm considered all ASVs previously found as true ASVs. Chimeras were removed, using the removeBimeraDenovo function, with the parameter settings minFoldParentOverAbundance = 3.5 and method = “consensus.” Taxonomy was assigned with the SILVA database (version 138; Quast et al., 2013), using the assignTaxonomy function with the parameter minBoot = 50. Taken the recent elevation of Lc. lactis subsp. cremoris to the species level as Lactococcus cremoris (Li et al., 2021) into account, all Lactococcus ASVs were compared with the type strains, Lc. lactis ATCC 19435T and Lc. cremoris ATCC 19257T, and assigned to one of these species when the sequence identity was above 99.80%, or to Lactococcus lactis/cremoris otherwise.
To assess the fungal members represented by the whole-community DNA in the core and rind samples of the cheeses after 36 weeks of ripening, amplification of the internal transcribed spacer (ITS1) region of the fungal rRNA transcribed unit was performed with primer pair BITS1-B58S3 (Bokulich and Mills, 2013b). Further steps were performed using short-read sequencing (Illumina, San Diego, CA, United States) and sequence data analysis using DADA2, as described previously (De Bruyn et al., 2017). This allowed the identification at genus level. Only the forward reads were used, since the variable length of the ITS1 region did not allow a correct merging of the forward and reverse reads for all genera. The parameter settings maxN = 1, truncQ = 2, maxEE = 5, and minLen = 50 were applied. Taxonomy was assigned with the UNITE database (version 04.02.2020; Kõljalg et al., 2013).
All taxon data reported below are expressed as relative abundances of the total number of sequence reads.
2.2.3. Starter cultures
The original starter culture mixtures were used for whole-community DNA extraction and plating. Therefore, 200 g of frozen starter culture was resuspended in a stomacher bag with 1800 mL of a sterile saline solution (8.5 g/L of NaCl; Merck). This suspension was centrifuged to yield cell pellets, which were subjected to DNA extraction, followed by amplicon-based high-throughput sequencing of the full-length 16S rRNA gene (PacBio) and DADA2 processing, as described above.
Given the fact that very few sequence reads of leuconostocs were found, a plating of the starter culture mixture was carried out to obtain Leuconostoc isolates. Therefore, serial dilutions of the suspensions mentioned above were made with sterile saline, and 100 μL of each dilution was plated on MRSv agar [de Man-Rogosa-Sharpe (MRS) agar (Oxoid, Basingstoke, Hampshire, United Kingdom) supplemented with vancomycin (20 mg/L; Sigma Aldrich) and cycloheximide (200 mg/L; Merck)] to enhance the selection of leuconostocs and inhibit fungi, respectively. Plates were incubated at 30°C for 3 days. For identification, 24 colonies were randomly picked (eight per starter culture mixture) and transferred to liquid MRSv. After overnight incubation at 30°C, cell pellets were obtained by microcentrifugation at 6,000 × g for 10 min, which were washed in a Tris-ethylene diaminetetraacetic acid (EDTA)-sucrose buffer [50 mM Tris base (Calbiochem), 1 mM EDTA (Sigma-Aldrich), and 6.7% (m/v) sucrose (Merck); pH 8.0] and subjected to genomic DNA extraction with a Nucleospin 96 tissue kit (Macherey Nagel, Düren, Germany), according to the manufacturer’s instructions. For each isolate, amplicon-based high-throughput sequencing (PacBio) was applied as mentioned above to allow the detection of non-identical copies of the 16S rRNA gene within one isolate.
2.3. Meta-metabolomic analysis
2.3.1. Dry mass determinations
The dry mass of the cheese core and rind samples was determined in triplicate with the common oven-drying method. Therefore, approximately 3 g of cheese powder (see below) was dried in an oven (Heraeus, Hanau, Germany) at 105°C for 24 h. All quantifications of substrates and metabolites were expressed on a dry mass basis (m/m).
2.3.2. Cheese powder and extracts
To enable accurate chemical compound extractions, powders of the cheese samples were prepared. Therefore, cheese core and rind samples were frozen using liquid nitrogen (Air Liquide, Paris, France) and subsequently milled into a fine powder with a coffee grinder (De’Longhi KG49, Treviso, Italy). These cheese powders were subjected to two types of extractions. To assess organic acids, free amino acids, and biogenic amines, an aqueous extraction was performed, as described previously (Le Boucher et al., 2016; Zhang et al., 2019). Briefly, 1.0 g of cheese powder was mixed with 9.0 mL of ultrapure water (Milli-Q; Merck) on a rotating wheel at 30 rpm for 30 min at room temperature, followed by centrifugation at 1,000 × g for 5 min. Extracts were stored at −25°C until further analysis. To assess volatile organic compounds, ethyl acetate extracts were prepared by mixing 0.5 g of cheese powder with 9.5 mL of ethyl acetate (SupraSolv® grade; Merck) and supplemented with 100 μg/L of toluene-D8 (Sigma-Aldrich) as internal standard (IS). Ethyl acetate extracts were filtered with a Millex Syringe Driven Filter Unit (Millex; Merck) and immediately used for further analysis.
2.3.3. Free amino acid concentration determinations
The concentrations of all 20 proteinogenic amino acids as well as those of 4-aminobutyric acid, citrulline, and ornithine were quantified with external calibration, in triplicate, by ultra-performance liquid chromatography coupled to tandem mass spectrometry (UPLC-MS/MS), using an Acquity system equipped with an HSS T3 column (Waters, Milford, MA, United States), as described previously (Van der Veken et al., 2022). Minor modifications included a constant flow rate of 0.35 mL/min for the mobile phase and the following gradient: 0.0 to 1.0 min, 99% A and 1% B; 1.0 to 8.0 min, 30% A and 70% B; 8.1 min to 10.0 min, 100% B; and 10.1 to 25.0 min, 99% A and 1% B. Samples were prepared by the addition of 300 μL of acetonitrile (Merck) to 100 μL of aqueous extract, followed by microcentrifugation at 18,000 × g for 15 min. Then, 900 μL of ultrapure water with 0.10% formic acid and 8.0 mg/L of 2-aminobutyric acid (IS; Sigma-Aldrich) was added to 100 μL of supernatant, and the mixture was filtered with a 0.2-μm LG H-PTFE filter (Millex; Merck) before injection (10 μL) into the column.
2.3.4. Biogenic amine concentration determinations
The concentrations of agmatine, cadaverine, histamine, 2-phenylethylamine, putrescine, spermidine, spermine, tryptamine, and tyramine were quantified with external calibration, in triplicate, by UPLC-MS/MS, using an Acquity system equipped with an HSS T3 column (Waters), as described previously (Van der Veken et al., 2020). Samples were prepared by addition of 300 μL of acetonitrile (Merck) with 0.2% heptafluorobutyric acid (Sigma-Aldrich) to 300 μL of aqueous extracts, followed by microcentrifugation at 18,000 × g for 15 min, and filtering with a 0.2-μm LG H-PTFE filter (Millex; Merck) before injection (5 μL) into the column.
2.3.5. Organic acid concentration determinations
The concentrations of citric acid, fumaric acid, gluconic acid, glucuronic acid, hippuric acid, lactic acid, maleic acid, malic acid, malonic acid, orotic acid, oxalic acid, pyruvic acid, succinic acid, and uric acid were quantified with external calibration, in triplicate, by UPLC-MS/MS, using an Acquity system equipped with an HSS T3 column (Waters), as described previously (De Bruyn et al., 2017), except that eluent A contained 2% (v/v) methanol (Merck). Samples were prepared by addition of 700 μL of a 1:1 (v/v) mixture of ultrapure water and methanol (Merck), containing 20 mg/L of salicylic acid (Fluka, Buchs, Switzerland) as IS, to 100 μl of the aqueous extracts, followed by microcentrifugation at 18,000 × g for 15 min, and filtering with a 0.2-μm LG H-PTFE filter (Millex; Merck) before injection (2 μL) into the column.
The ratio between D-lactic acid and L-lactic acid was quantified, in duplicate, by UPLC-MS/MS, using an Acquity system equipped with an Astec Chirobiotic column (Supelco; Bellefonte, PA, United States). The mobile phase consisted of an isocratic flow of 15% ultrapure water with 33.3 mM ammonium acetate (VWR International) and 85% acetonitrile (Merck) at a constant flow rate of 0.6 mL/min. Samples were prepared by addition of 100 μL of aqueous extract to 900 μL of a solution containing 85% acetonitrile (Merck), 15% ultrapure water, 0.386 g/L of ammonium acetate (VWR International), and 4.0 mg/L of salicylic acid (IS; Fluka). The mixtures were vortexed for 5 min, followed by microcentrifugation at 18,000 × g for 15 min, and filtering with a 0.2-μm LG H-PTFE filter (Millex; Merck) before injection (10 μL) into the column.
2.3.6. Volatile organic compound concentration determinations
The concentrations of targeted volatile organic compounds present in ethyl acetate extracts were quantified with external standards by liquid injection gas chromatography with triple-quad tandem mass spectrometry (LI-GC-TQ-MS/MS), using a Trace 1,300 gas chromatograph equipped with a Dbwax-etr column (Thermo Fisher Scientific) and coupled to a TSQ 8000 EVO triple quadrupole mass spectrometer (Interscience, Breda, Netherlands), as described previously (Díaz-Muñoz et al., 2021).
2.4. Sensory analysis
Cheeses were organoleptically assessed by a company panel experienced in cheese tasting. This panel consisted of 10 people spread over sex and age. After 36, 45, 75, and 100 weeks of ripening, the panel members scored each Gouda cheese from each production batch individually on a scale from 0 to 20, whereafter Z-scores were calculated for all cheeses assessed by one person, using the formula Z-score = (x – μ)/σ, with μ the average and σ the standard deviation of all scores given by that person. Per cheese, the average Z-score of all panel members was then calculated and used as sensory quality criterium.
2.5. Statistical analysis
All statistical analyses were performed in R (version 4.1.0; R Core Team, 2021). One-way analysis of variance (ANOVA) was performed to compare differences in relative abundances of bacteria between starter culture rotations and ripening times, followed by post-hoc pairwise comparisons with Tukey’s test. A principal component analysis (PCA) was performed to compare species compositions of all cheese samples at all time points. A scaled PCA (Z-score transformation) was performed to compare Lactococcus ASVs at 36 weeks of ripening among batches and to compare all metabolites at 36, 45, 75, and 100 weeks of ripening, in each case between cores and rinds. For correlation analysis, only species with an average relative abundance of ≥0.2% across all cheese cores after 36, 45, 75, and 100 weeks of ripening were considered. Spearman correlation coefficients were calculated when data from different ripening times were used, whereas Pearson correlation coefficients were calculated for correlations with the sensory score at one ripening time. Spearman correlations between species and metabolites were visualized in a heatmap using the ComplexHeatmap package (version 2.0.0; Gu et al., 2016). Hierarchical clustering analysis was based on the Ward’s method (clustering method set to “Ward.D2”). For all statistical tests, results with a p-value < 0.05 were considered significantly different.
Microbial intra-sample diversity (alpha diversity) was assessed by calculating the inverse Simpson diversity index, applying the vegan package (version 2.5–7; Oksanen et al., 2020). Microbial inter-sample diversity (beta diversity) was assessed by conducting a permutational multivariate analysis of variance (PERMANOVA), based on Bray–Curtis dissimilarity scores, applying the RVAideMemoire package (version 0.9–80; Hervé, 2021).
3. Results
3.1. Metagenetics of cheese cores and rinds
Cores and rinds of Gouda cheeses from 23 cheese production batches, with sensory scores L, G or E, produced in the same dairy factory with starter culture mixture A, B or C, were investigated after 36, 45, 75, and 100 weeks of ripening through metagenetics (for the rinds only after 36 weeks of ripening) and meta-metabolomics. Nine cheese batches (score G) were also assessed after 26 and 31 weeks of ripening.
3.1.1. Bacterial species-level metagenetics
In general, the bacterial species diversity in the cheese cores was rather small for all LAB starter culture mixtures used throughout the ripening period (Figure 2). The most abundant bacterial species were Lc. lactis and Lc. cremoris. Leuconostoc pseudomesenteroides was the third most abundant bacterial species up to 45 weeks of ripening. The most abundant NSLAB species were Lacc. paracasei, which had its maximum relative abundance after 45 weeks of ripening, and Tetragenococcus halophilus, which was the most abundant NSLAB species after 100 weeks of ripening. Lactiplantibacillus plantarum, Loigolactobacillus rennini, and Staphylococcus equorum were always found at more than 15% of relative abundance in at least one cheese batch, whereas Lactococcus laudensis was found in almost all cheese batches, but always at a low relative abundance.
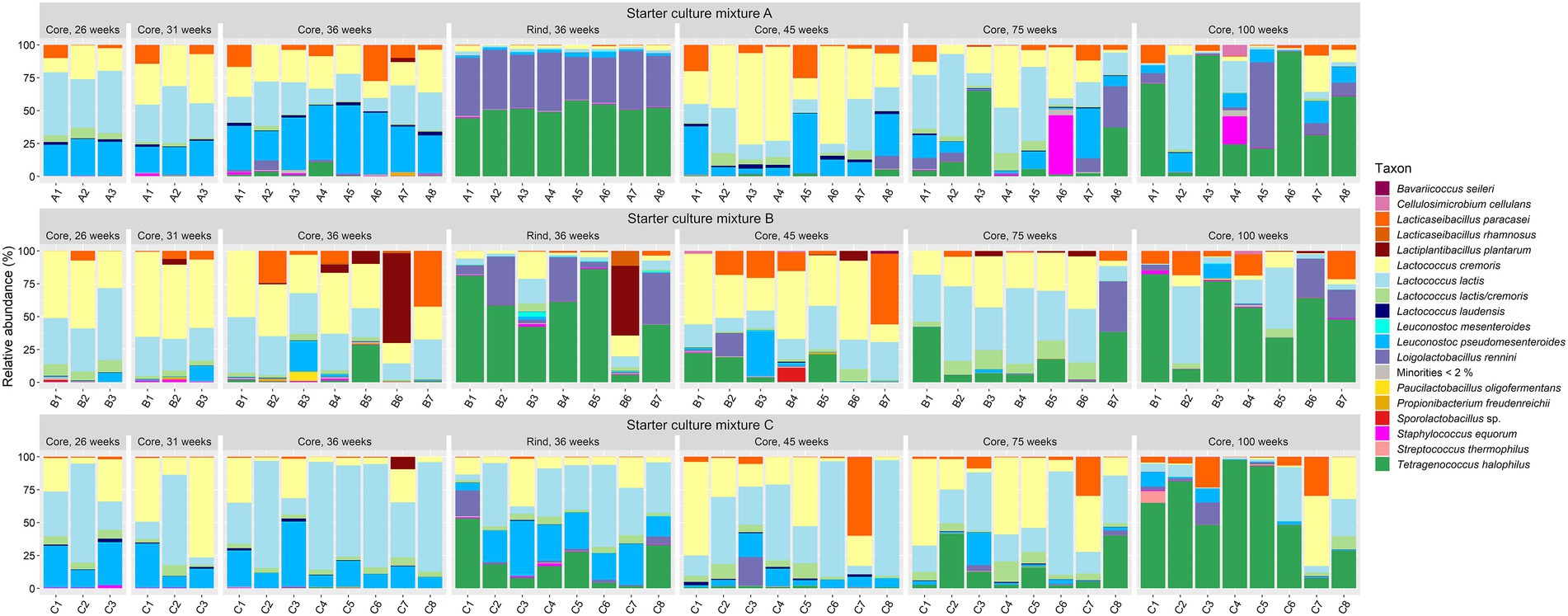
Figure 2. Bacterial species dynamics, expressed as relative abundances based on amplicon sequence variants of the full-length 16S rRNA gene, for cheese cores after 26, 31, 36, 45, 75, and 100 weeks of ripening and rinds after 36 weeks of ripening. The Gouda cheeses were made with three different mixed-strain starter cultures (A, B, and C). The numbers indicate the cheeses of the 23 different batch productions.
The cores of the Gouda C cheeses contained significantly more Lc. lactis compared to those of the other cheeses, whereas those of the Gouda B cheeses contained significantly more Lc. cremoris than those of the Gouda C cheeses after 36 weeks of ripening (Figure 2). After 26 and 36 weeks of ripening, the relative abundance of Leuc. pseudomesenteroides was significantly lower in the cores of the Gouda B cheeses than in those of the other ones. Only in one batch of the Gouda B cheeses (B3), Leuc. pseudomesenteroides was of high relative abundance throughout the ripening period. After 36 weeks of ripening, the Gouda A cheeses contained significantly more Leuc. pseudomesenteroides compared to the Gouda C cheeses, whereas the Gouda B cheeses contained significantly more NSLAB species than the Gouda C ones. Throughout the ripening period, Lc. laudensis was found in 24% of the cheese cores at a relative abundance of ≥1.0%, but after 36, 45, and 75 weeks of ripening, its relative abundance was significantly lower in the Gouda B cheeses compared to the Gouda A cheeses, and also significantly lower after 75 weeks of ripening compared to the Gouda C ones.
The cheese rinds (36 weeks of ripening) displayed a significantly lower relative abundance of Lc. lactis and Lc. cremoris compared to the cheese cores for all starter culture mixtures (Figure 2). Further, the bacterial diversity of the rinds of the Gouda A and B cheeses was different from those of the Gouda C cheeses. The Gouda A and B cheeses had a significantly higher relative abundance of Loil. rennini and T. halophilus in the rinds compared to the cores. Furthermore, the Gouda A cheeses contained less Lc. laudensis and Leuc. pseudomesenteroides in the rinds compared to the cores, whereas the Gouda B cheeses had more Leuc. mesenteroides in the rinds compared to the cores. Among the rinds, the Gouda C cheeses had more Lc. lactis and Leuc. pseudomesenteriodes and less T. halophilus compared to those of the other ones. The Gouda B rinds contained significantly more Leuc. mesenteroides compared to the other ones. All Gouda cheese rinds had significantly different levels of Loil. rennini, with the Gouda A rinds representing the highest and the Gouda C rinds the lowest relative abundances.
The alpha diversity of the bacterial species compositions displayed a bell shape as a function of the ripening time for the Gouda A and B cheeses, reaching its maximum at 36 and 45 weeks of ripening, respectively, whereas it was almost constant for the Gouda C cheeses (Supplementary Figure S1). This diversity index only differed significantly between cheeses made with different starter culture mixtures after 36 weeks of ripening in that the alpha diversity of the Gouda C cheeses was significantly lower in the cores and higher in the rinds compared to those of the other cheeses. The generally lower alpha diversity for the Gouda C cheeses compared to the other ones reflected a lower relative abundance of NSLAB species. The beta diversity of the bacterial species compositions among the Gouda cheeses roughly followed an upside down bell curve, with the highest similarities after 26 and 31 weeks of ripening, especially among the Gouda A and C cheeses, and after 75 and 100 weeks of ripening for all cheeses (Supplementary Figure S1). After 36 weeks of ripening, the Gouda A, B, and C cheeses had a significantly different bacterial species composition, for both the cores and the rinds when compared to each other. The bacterial species compositions were also significantly different between cores and rinds among the Gouda A and B cheeses.
A PCA indicated that Lc. lactis, Lc. cremoris, T. halophilus, Leuc. pseudomesenteroides, Lacc. paracasei, and Loil. rennini explained most of the species variance between the samples (Supplementary Figure S2). Most of the rinds of the 36-week Gouda A and B cheeses clustered with those of the cheeses of 100 weeks of ripening, based on the high relative abundances of T. halophilus and Loil. rennini. The cores of Gouda cheeses of 26, 31, 36, and 45 weeks of ripening were spread over three clusters, namely a cluster characterized by a high relative abundance of Lc. cremoris, a cluster of mainly Gouda C cheeses characterized by a high relative abundance of Lc. lactis, and a cluster of mainly cheeses ripened for 36 weeks and characterized by high relative abundances of Leuc. pseudomesenteroides, Lc. lactis, and Lc. cremoris.
Spearman correlation coefficients showed that Lc. lactis and Lc. cremoris were negatively correlated with T. halophilus and Loil. rennini (Supplementary Figure S3). The two latter species were positively correlated. Lactococcus laudensis was positively correlated with Lc. cremoris, Lc. lactis, and Leuc. pseudomesenteroides. Further, Lacp. plantarum, Lacc. rhamnosus, and Propionibacterium freudenreichii were positively correlated.
3.1.2. Bacterial ASV-level metagenetics
To allow insights below species level, the compositions of the ASVs obtained were assessed for all starter LAB and the most abundant NSLAB species.
3.1.2.1. Lactococcus
In the case of the genus Lactococcus, for all cheese cores and rinds, 21 ASVs belonged to Lc. cremoris, 6 ASVs to Lc. lactis, and 15 ASVs were considered as intermediate, as they displayed an identity varying between 99.39 and 99.73% with both type strains.
Concerning the starter culture mixtures, more than 80% of the ASVs belonged to Lc. cremoris, with the Lc. cremoris_01 ASV as the most abundant one, followed by the Lc. cremoris_02 ASV (Figure 3). The ASVs of Lc. lactis summed up to 5.7, 6.8, and 2.8% in starter culture mixture A, B, and C, respectively. The ASVs of Lc. laudensis were present in all starter culture mixtures at a relative abundance comparable to that of the leuconostocs (see below).
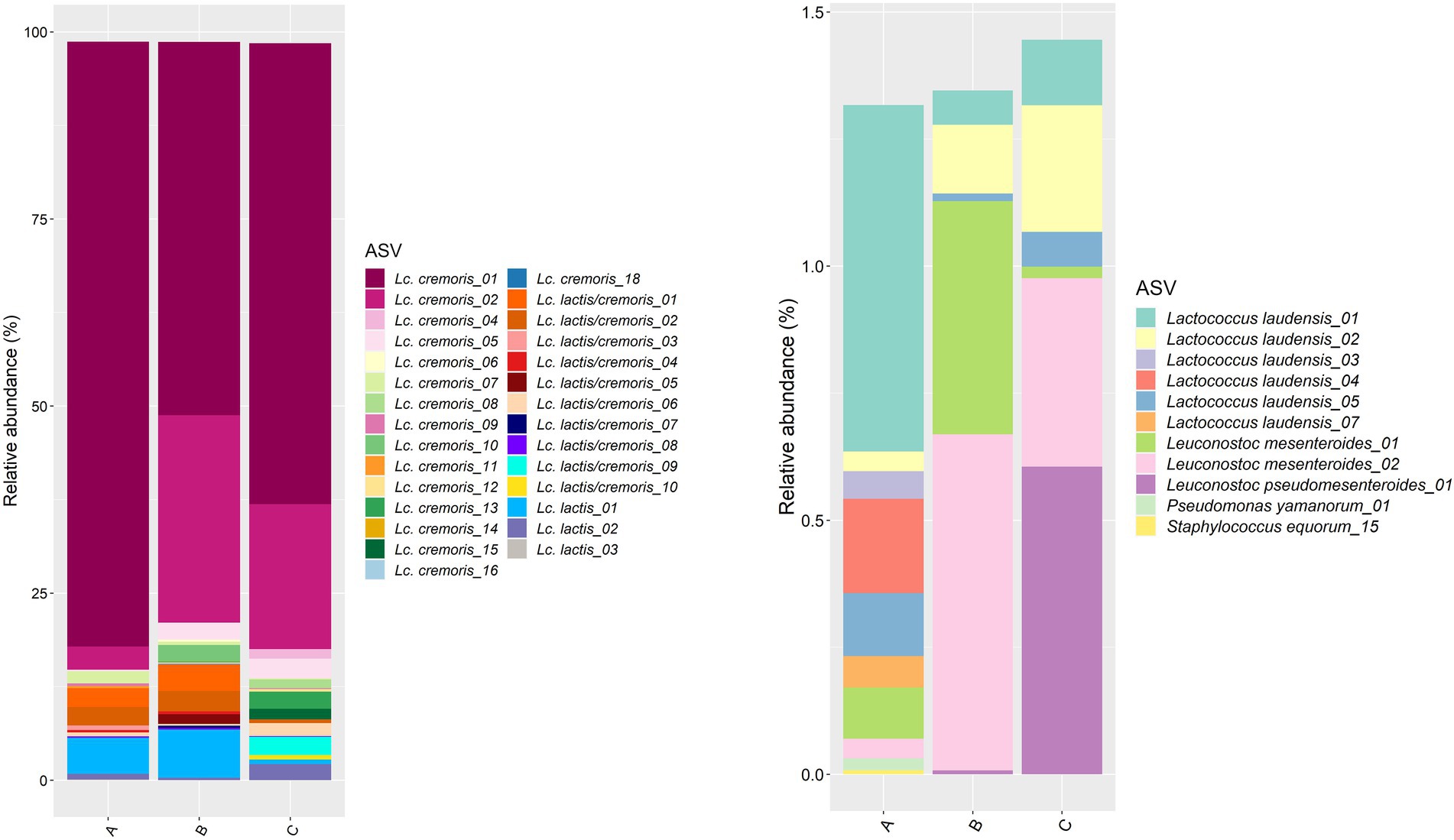
Figure 3. Taxonomic assessment on amplicon sequence variant (ASV) level of the bacterial full-length 16S rRNA gene for the three commercial starter culture mixtures (A, B, and C) investigated. Left, ASVs of Lactococcus cremoris and Lactococcus lactis (ASVs belonging to Lc. cremoris or Lc. lactis that could not be assigned to one of those species are indicated as Lc. lactis/cremoris). Right, ASVs of all other bacterial species identified in the starter culture mixtures.
A scaled PCA of all ASVs separated the 23 cheese production batches at 36 weeks of ripening based on starter culture mixture used; the ASVs corresponding with the cores and rinds showed no differences (Figure 4). The Gouda C cheeses had significantly less of the Lc. cremoris_01 ASV and more of the Lc. lactis_02 ASV after 36 weeks (both cores and rinds) and 45 weeks (cores) of ripening compared to the other cheeses (Supplementary Figure S4). Furthermore, the Gouda A cheeses had significantly less of the Lc. cremoris_02 ASV in the rinds than in those of the other cheeses after 36 weeks of ripening, whereas the Gouda C cheeses had then more of the Lc. cremoris_04 ASV in both cores and rinds than in those of the other cheeses. The Gouda B cheeses had more of the Lc. cremoris_06 ASV after 36 (both cores and rinds), 45 (cores), and 75 (cores) weeks of ripening than in those of the other cheeses. After 100 weeks of ripening, no significant differences between the Gouda cheeses was found. For the Gouda A cheeses, the relative abundance of the Lc. cremoris_01 ASV within the Lactococcus genus decreased as a function of the ripening time and was significantly lower after 75 weeks of ripening compared to that after 36 and 45 weeks of ripening and significantly lower after 100 weeks of ripening compared to 45 weeks. Among the Gouda B cheeses, the Lc. lactis_01 ASV increased as a function of the ripening time, which was significantly higher between 36 and 45 weeks of ripening compared to 100 weeks and after 45 weeks of ripening compared to 75 weeks. In contrast, the Lc. cremoris_01 ASV decreased as a function of the ripening time, and was significantly lower after 75 and 100 weeks of ripening compared to 36 and 45 weeks. In addition, the Lc. cremoris_02 ASV was significantly lower after 100 weeks of ripening compared to 45 weeks. Concerning the Gouda C cheeses, the relative abundances of the ASVs corresponding with Lc. cremoris and Lc. lactis were not significantly different during ripening.
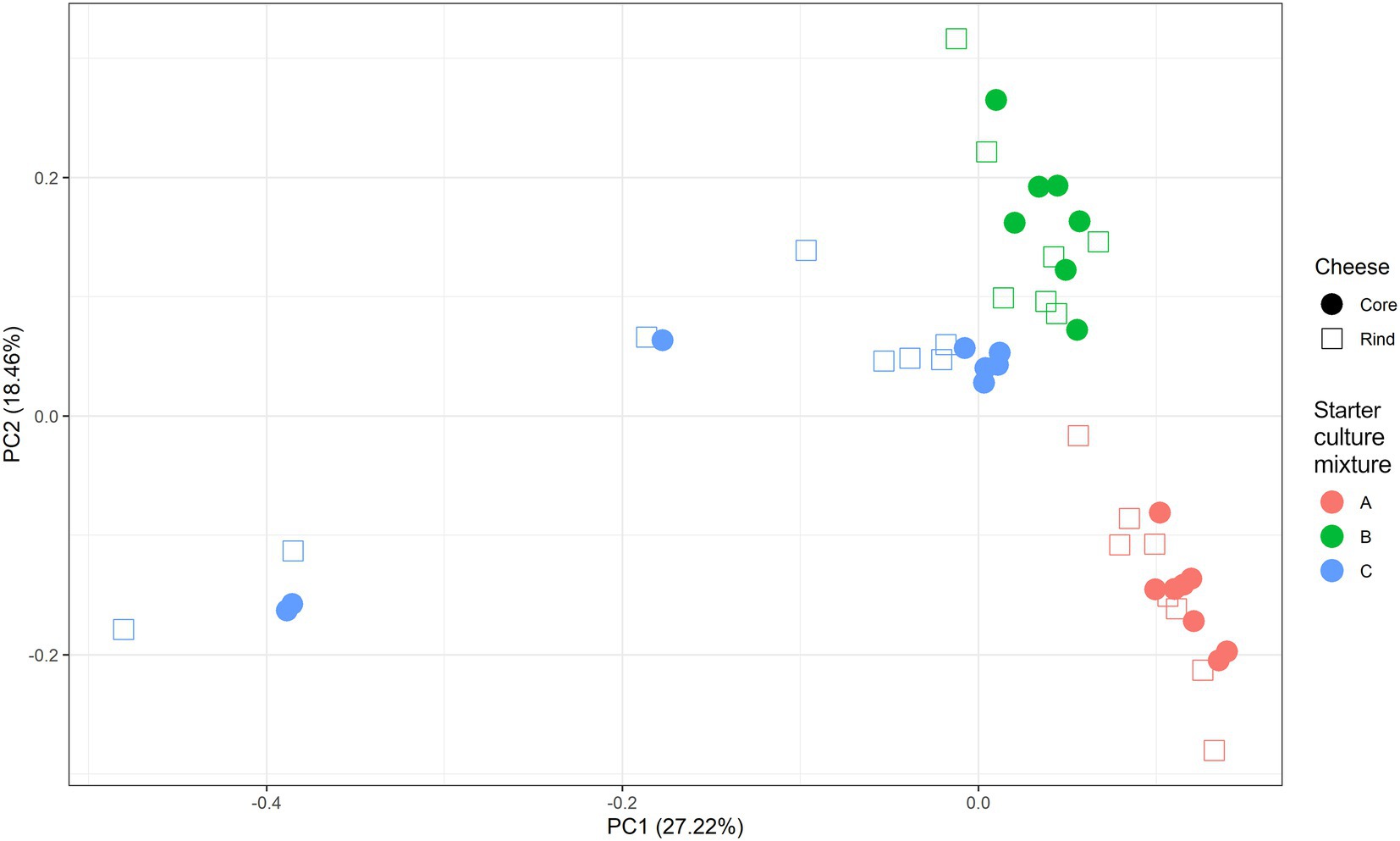
Figure 4. Scaled principal component analysis of all amplicon sequence variants based on the full-length 16S rRNA gene of Lactococcus cremoris and Lactococcus lactis identified in the cores (full symbols) and rinds (open symbols) from 23 different Gouda cheese batch productions made with three different mixed-strain starter cultures (A, red; B, green; C, blue) after 36 weeks of ripening.
3.1.2.2. Leuconostoc
The Gouda C cheeses mainly contained the Leuc. pseudomesenteroides_01 ASV (Supplementary Figure S4). This ASV was also the most abundant Leuconostoc ASV in the Gouda A and B cheeses. The Leuc. pseudomesenteroides_02 ASV was found in all Gouda A cheeses (but not at all ripening times), mostly in a ratio of 1:3 compared to the Leuc. pseudomesenteroides_01 ASV, whereas it appeared more randomly in all Gouda B cheeses (not at all ripening times) and in two Gouda C cheeses. The Leuc. pseudomesenteroides_03 ASV appeared at later time points of the ripening stage in most Gouda C cheeses, as well as in three Gouda A cheeses and at all ripening times in one Gouda B cheese.
All Leuc. mesenteroides ASVs were only found in the Gouda cheeses when the Leuconostoc genus was present at a low relative abundance (Supplementary Figure S4). This was also the case in the pure starter culture mixtures, displaying a relative abundance of 0.14, 1.13, and 1.00% of leuconostocs in starter culture mixture A, B and C, respectively. Moreover, the Leuc. pseudomesenteroides_02 and Leuc. pseudomesenteroides_03 ASVs were not found in the starter culture mixtures, despite their presence in the cheeses. Metagenetic analysis of the starter culture mixtures allowed to detect non-identical copies of the 16S rRNA gene within one isolate (50% of the isolates, Supplementary Figure S5). Indeed, 12 isolates from starter culture mixtures A and B contained both the Leuc. pseudomesenteroides_01 and Leuc. pseudomesenteroides_02 ASVs, in a ratio of 3:1, which was in accordance with a copy number of four for the 16S rRNA gene of Leuc. pseudomesenteroides (Stoddard et al., 2015). The other isolates each contained only one ASV, indicating that the four copies were identical. Hence, three different ASV clusters of Leuc. pseudomesenteroides were present. A first cluster (ASV cluster 1) contained only the Leuc. pseudomesenteroides_01 ASV, as found in the Gouda C cheeses. A second cluster (ASV cluster 2) contained the Leuc. pseudomesenteroides_01 and Leuc. pseudomesenteroides_02 ASVs in a 3:1 ratio, as found in the Gouda A cheeses. ASV cluster 3 represented the Leuc. pseudomesenteroides_03 ASV, which was spread among the Gouda A, B, and C cheeses.
Differences between ASVs present in the cores and rinds after 36 weeks of ripening were small for the Gouda C cheeses (Supplementary Figure S4). In the rinds of the Gouda A cheeses, the Leuc. mesenteroides_01 and Leuc. mesenteroides_02 ASVs appeared, whereas the rinds of the Gouda B cheeses had significantly more of the Leuc. mesenteroides_02 ASV and less of the Leuc. pseudomesenteroides_01 and Leuc. pseudomesenteroides_02 ASVs.
3.1.2.3. Lacticaseibacillus and Lactiplantibacillus
A total of 67 Lacc. paracasei, 22 Lacp. plantarum, eight Lacc. rhamnosus, and one Lacc. casei ASVs were found across all Gouda cheese cores and rinds, albeit that most were only found at low relative abundance (Supplementary Figure S6). The appearance of different Lacticaseibacillus and Lactiplantibacillus ASVs was not linked with the ripening time, the cheese cores or rinds, or the LAB starter culture mixtures applied, but these ASVs were production batch-dependent. In most cheese production batches, the same ASVs were found at different ripening times.
3.1.2.4. Tetragenococcus halophilus and Loigolactobacillus rennini
For T. halophilus, 108 different ASVs were found, which was the highest number of ASVs for any LAB species in the Gouda cheeses of the present study. The 10 most abundant T. halophilus ASVs could be grouped in two clusters of five highly correlated ASVs, found in equal ratios among the cheese production batches, namely ASV cluster 1 (ASVs T. halophilus_001–005) and ASV cluster 2 (ASVs T. halophilus_006–010) (Figure 5). Loigolactobacillus rennini had nine different ASVs, of which only three were highly abundant. They were found in a constant ratio of 3:1:1, suggesting one main ASV cluster.
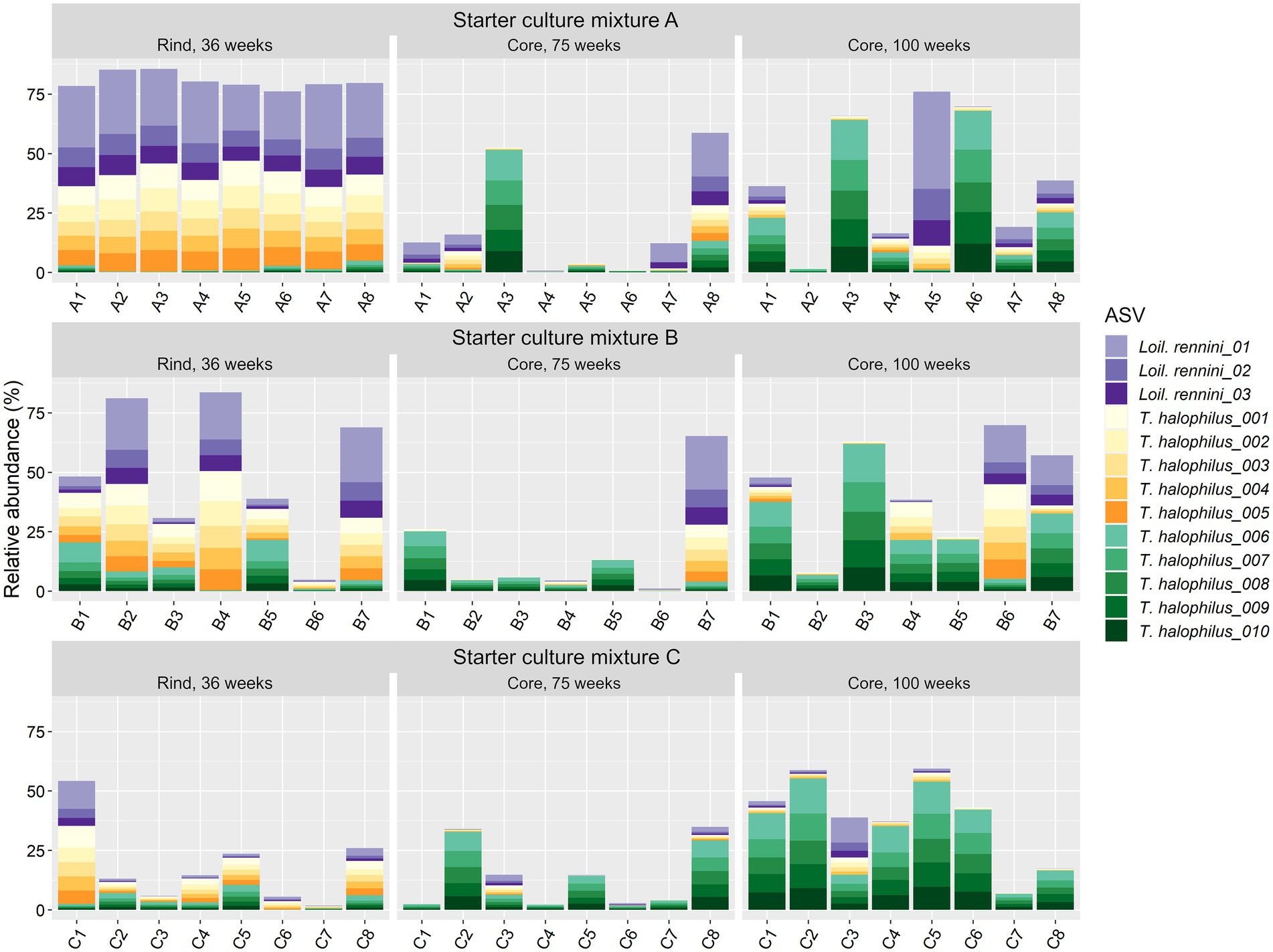
Figure 5. Relative abundance of the amplicon sequence variants (ASVs) based on the full-length 16S rRNA gene of the non-starter lactic acid bacteria (NSLAB) species Loigolactobacillus rennini (three most abundant ones) and Tetragenococcus halophilus (10 most abundant ones) identified in the Gouda cheeses from 23 different batch productions made with three different mixed-strain starter cultures (A, B, and C) after 36 weeks of ripening (rinds) and 75 and 100 weeks of ripening (cores). The numbers indicate the cheeses of the 23 different batch productions. The ASVs T. halophilus_001–005 form ASV cluster 1, whereas the ASVs T. halophilus_006–010 form ASV cluster 2.
3.1.3. Fungal genus-level metagenetics
As fungi were not expected to be abundant in the Gouda cheeses and since 36 weeks of ripening was considered as the most interesting time point regarding the bacterial species diversity, the fungal species diversity was only assessed at this ripening time. The main genera present were Debaryomyces and Saccharomyces (Figure 6). Both genera accounted for >75.0% of the relative abundances in 75% of all cheese production batches. Five other genera were present at more than 5.0% of relative abundance in at least one cheese batch, namely Alternaria, Candida, Hanseniaspora, Kazachstania, and Malassezia.
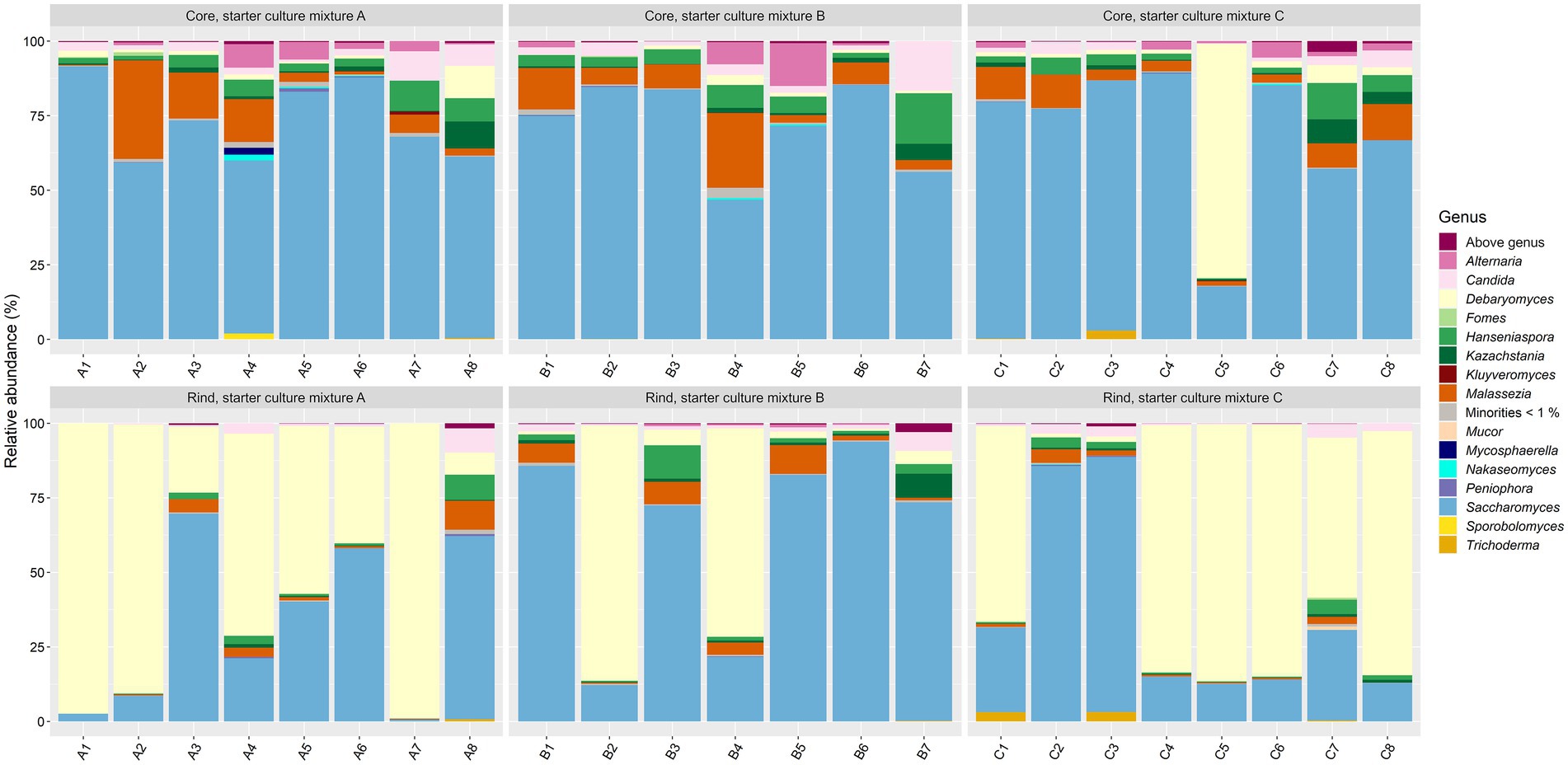
Figure 6. Taxonomic assessment on fungal genus level, expressed as relative abundances based on amplicon sequence variants (ASVs) of the internal transcribed spacer (ITS) region ITS1 of the rRNA transcribed unit, for cheese cores and rinds after 36 weeks of ripening. The Gouda cheeses were made with three different mixed-strain starter cultures (A, B, and C). The numbers indicate the cheeses of the 23 different batch productions.
The fungal alpha diversity was not significantly different between the Gouda A, B, and C cheeses for both cores and rinds, and was not significantly different between the cores and rinds among the Gouda cheeses made with the same starter culture mixture. The beta diversity of the fungal genera compositions between the cores and rinds was only significantly different for the Gouda A cheeses. For all cheese production batches, the relative abundance of the genus Saccharomyces was significantly higher in the cores compared to the rinds (average relative abundances of 71.3 and 43.0%, respectively), whereas the relative abundance of the genus Debaryomyces was significantly lower in the cores compared to the rinds (average relative abundances of 5.3 and 48.1%, respectively).
3.2. Meta-metabolomics of cheese cores and rinds
3.2.1. Free amino acids
Amino acids were the most abundant metabolites found in the Gouda cheese cores and rinds, with concentrations of total free amino acids ranging from 15.1 to 120.2 g/kg (Figure 7). Glutamic acid, lysine, leucine, proline, and valine were, in decreasing concentrations, the most abundant free amino acids among all cheese production batches and ripening times.
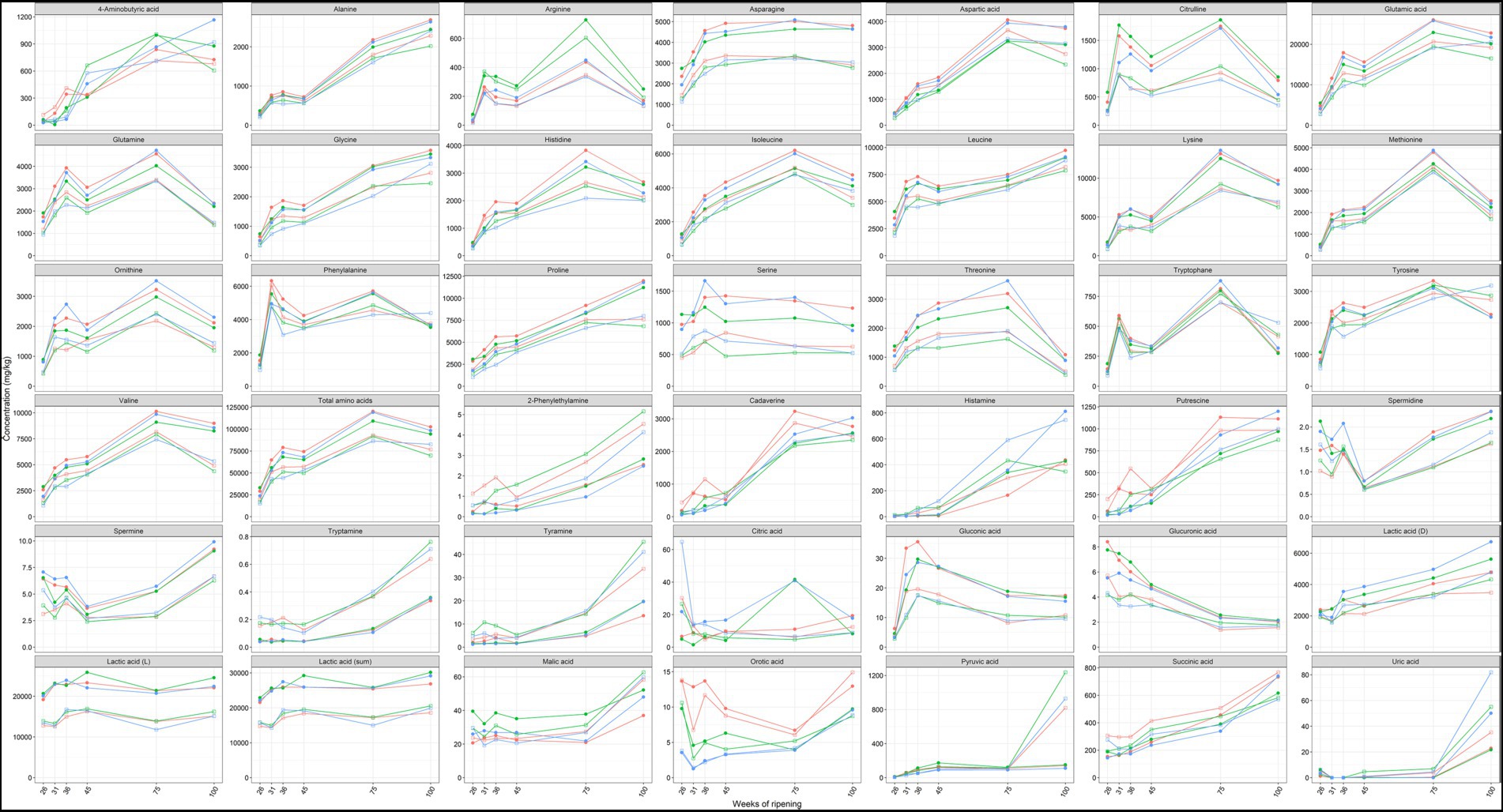
Figure 7. Average concentrations, expressed in mg/kg dry mass, of amino acids, biogenic amines, and organic acids in the cores (full symbols) and rinds (open symbols) of Gouda cheeses made with three different mixed-strain starter cultures (A, red; B, green; C, blue) as a function of the ripening time. The concentrations of cysteine, maleic acid, malonic acid, hippuric acid, and fumaric acid were below the limit of determination, whereas agmatine was not found.
The concentrations of all amino acids mentioned above displayed an increasing trend from 26 up to 75 weeks of ripening, both in the cheese cores and rinds, with a stagnation or small decrease between 36 and 45 weeks of ripening, followed by a major increase between 45 and 75 weeks. The concentrations were in general higher after 100 weeks of ripening than after 45 weeks but lower than after 75 weeks. The concentrations of most amino acids were significantly higher in the cheese cores compared to the rinds at all ripening times.
The concentrations of arginine were among all samples 88% higher in the Gouda B cheeses compared to the other ones, which was significant for the cheese rinds after 36 weeks of ripening and for all cheese cores and rinds after 45, 75, and 100 weeks of ripening. The concentrations of most other amino acids were lower in the Gouda B cheeses compared to the other ones.
3.2.2. Biogenic amines
The concentrations of cadaverine, histamine, 2-phenylethylamine, putrescine, tryptamine, and tyramine increased significantly in the Gouda cheese cores and rinds from 45 to 100 weeks of ripening, with the main increase between 45 and 75 weeks (Figure 7). In the case of cadaverine, histamine, and putrescine, concentrations around 1 g/kg were reached after 75 and 100 weeks of ripening. The concentrations of 2-phenylethylamine, tryptamine, and tyramine were significantly higher in the cheese rinds compared to the cores at all ripening times. A significantly higher concentration in the rind was also the case for histamine after 31, 36, 45, and 75 weeks of ripening and for putrescine and cadaverine after 36 and 45 weeks of ripening. The low concentrations of spermine and spermidine were significantly higher in the cheese cores at all ripening times, except for spermidine after 36 and 45 weeks of ripening.
The concentrations of cadaverine and putrescine were significantly higher in the Gouda A cheeses compared to the other ones, after 31 weeks of ripening in the cores and after 36 weeks in the cores and rinds.
3.2.3. Organic acids
Lactic acid was the main organic acid present in the Gouda cheeses, with an average concentration of 25.8 g/kg in the cores and 17.2 g/kg in the rinds (Figure 7). The concentrations of D-lactic acid and succinic acid increased between 45, 75, and 100 weeks of ripening in both the cores and rinds. The low concentrations of citric acid were more than twice as high in the cores of the Gouda C cheeses compared to those of the other ones after 26 and 36 weeks of ripening. Concerning the cheese rinds, the concentrations of pyruvic acid were significantly lower for the Gouda C cheeses compared to the other ones after 36 weeks of ripening. However, pyruvic acid only significantly increased in the rinds of all Gouda cheeses, with a ninefold increase between 75 and 100 weeks of ripening. Malic acid had higher concentrations in the cores of the Gouda B cheeses, which was significant after 26, 36, and 45 weeks of ripening. The Gouda A cheeses had higher concentrations of orotic acid, which was significant in the cores after 31 and 36 weeks of ripening and in the rinds after 36 and 45 weeks.
3.2.4. Volatile organic compounds
Acetic acid, acetoin, and 2,3-butanediol were the main volatile organic compounds found in the Gouda cheeses examined (Figure 8). The concentrations of these compounds were significantly higher in the cores than in the rinds at all ripening times. In contrast, the concentrations of benzaldehyde, δ-decalactone, ethyl dodecanoate, hexanal, octanoic acid, 2-pentanone, 2-phenylethanol, and trimethylpyrazine were significantly higher in the rinds for at least three ripening times. The concentrations of benzaldehyde, dimethylsulfone, δ-dodecalactone, phenyl acetaldehyde, 2-phenylethanol, tetramethylpyrazine, and all short-chain fatty acids increased as a function of the ripening time, mainly between 45 and 75 weeks.
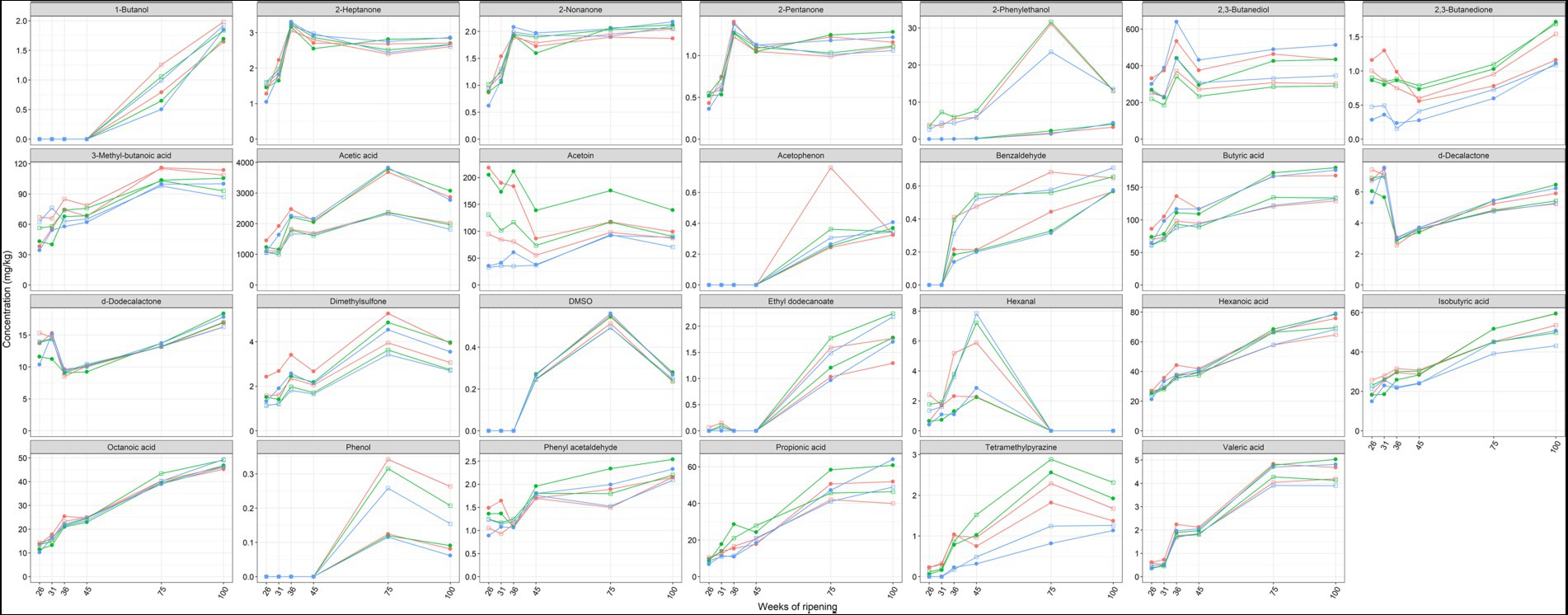
Figure 8. Average concentrations, expressed in mg/kg dry mass, of volatile organic compounds in the cores (full symbols) and rinds (open symbols) of Gouda cheeses made with three different mixed-strain starter cultures (A, red; B, green; C, blue) as a function of the ripening time.
The concentrations of acetoin were lower in the Gouda C cheeses, in both cores and rinds, compared to the other ones at all ripening times. This was threefold lower in the cores after 26 and 31 weeks of ripening and in both the cores and rinds after 36 weeks. In general, the Gouda B cheeses had the highest acetoin concentrations. A similar trend was found for the concentrations of 2,3-butanedione. In contrast, the concentrations of 2,3-butanediol were higher in the Gouda C cheeses compared to the other ones, which was significant in the cores and rinds after 36 weeks of ripening. The concentrations of dimethyl sulfone were significantly higher in the cores of the Gouda A cheeses compared to those of the other ones after 26, 31, and 36 weeks of ripening.
3.3. Impact of the starter culture mixture and species
After 36 weeks of ripening, a PCA on the scaled values of all metabolites clustered the cheese production batches mainly according to the starter culture mixture used (Figure 9). The effect of the starter culture mixture decreased during ripening. At all ripening times, a clear distinction was found between cheese cores and rinds.
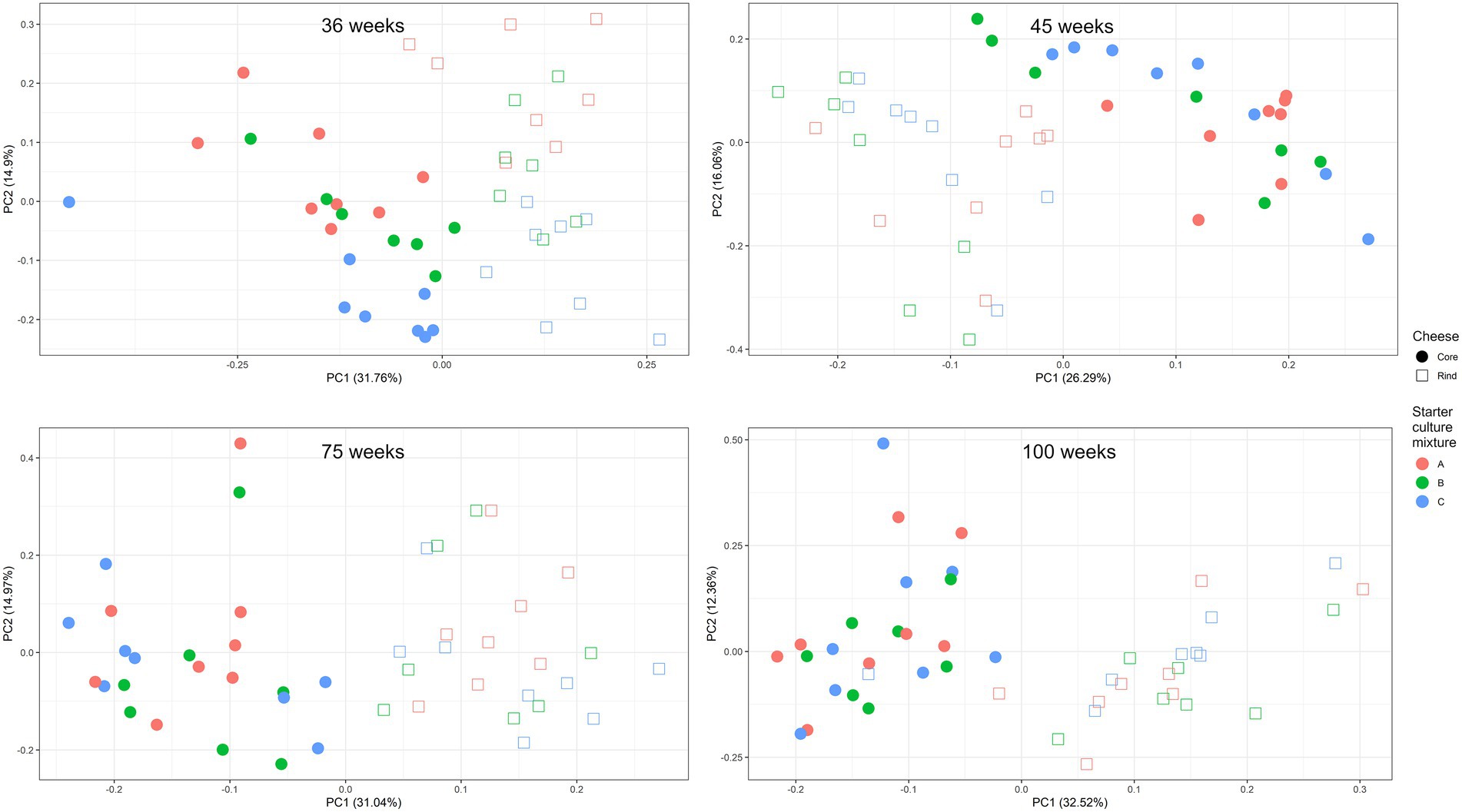
Figure 9. Scaled principal component analysis of the concentrations of the non-volatile and volatile organic compounds of Figures 6, 7, measured in the cores (full symbols) and rinds (open symbols) of Gouda cheeses made with three different mixed-strain starter cultures (A, red; B, green; C, blue), according to the ripening time.
According to the clustered heatmap of Spearman correlations, two main clusters could be distinguished that correlated the metabolites and main species throughout ripening (Figure 10). One cluster contained all Lactococcus and Leuconostoc species as well as S. thermophilus, whereas the other cluster contained P. freudenreichii, St. equorum, and the most abundant NSLAB. All species of the former cluster were positively correlated with L-lactic acid, whereas the species Lacc. paracasei, T. halophilus, and Loil. rennini were positively correlated with D-lactic acid. Most amino acids were positively correlated with Lacc. paracasei and T. halophilus. Serine, threonine, and citrulline were positively correlated with Lactococcus. All biogenic amines, except for spermine and spermidine, were positively correlated with Loil. rennini and T. halophilus. These LAB species were also positively correlated with dry mass, reflecting their osmotolerance. Tetragenococcus halophilus was positively correlated with short-chain fatty acids and, in general, with most metabolites that increased during ripening.
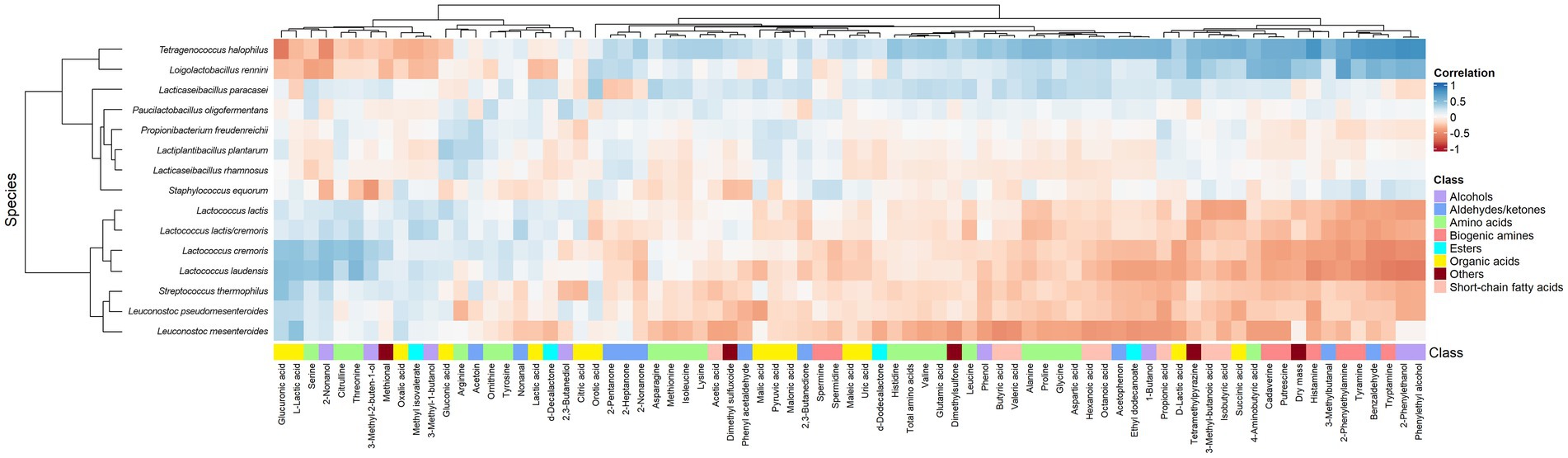
Figure 10. Spearman correlations between the relative abundances of the major bacterial species and the metabolite concentrations found in the Gouda cheeses from 23 different batch productions at all ripening times. Metabolites without any significant correlation are not shown.
Intra-species differences were also found. Acetoin and its related metabolites, 2,3-butanediol and 2,3-butanedione, and citric acid had different correlations among ASVs of Lc. lactis, Lc. cremoris and Leuc. pseudomesenteroides (Table 1), whereas the two main T. halophilus ASV clusters had different correlations with specific amino acids, biogenic amines, and other aroma compounds after 75 weeks of ripening (Table 2). Since it was hypothesized that the ASV clusters of Leuc. pseudomesenteroides did not straightforwardly correspond with the three ASVs identified, correlations with the Leuc. pseudomesenteroides_01 ASV differed from those with ASV cluster 1. The differences were most pronounced when only the cheese cores of 36 weeks of ripening were considered. ASV cluster 1 and ASV cluster 3 were negatively correlated with the total amino acid concentration and acetoin, but positively with lactic acid, whereas the opposite was the case for ASV cluster 2, suggesting that the latter was more related to flavor formation, whereas the former clusters participated more in acidification (Table 3). This was also found for the higher concentrations of most amino acids at 36 weeks of ripening for the Gouda A cheeses containing ASV cluster 2.
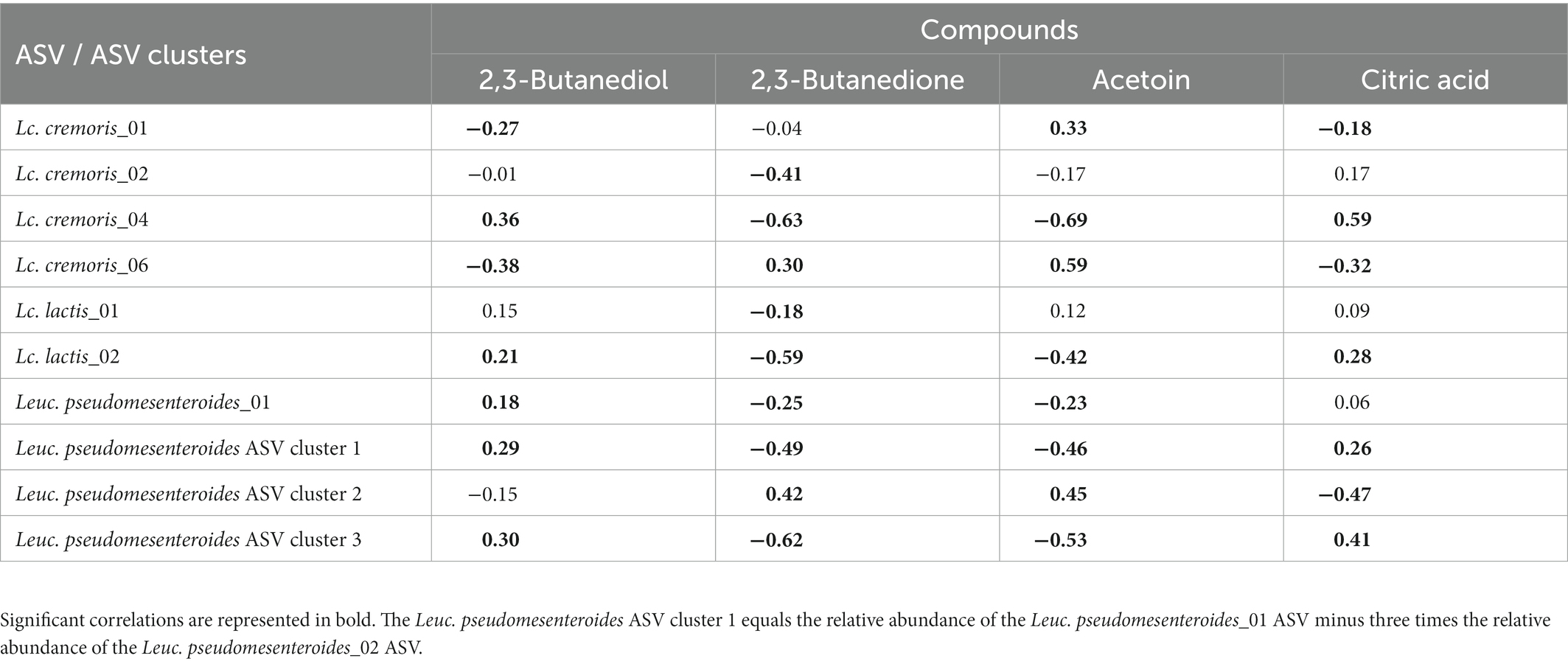
Table 1. Spearman correlations between acetoin concentrations and other pyruvate metabolites and a selection of significantly different amplicon sequence variants (ASVs) of the starter culture mixtures applied.
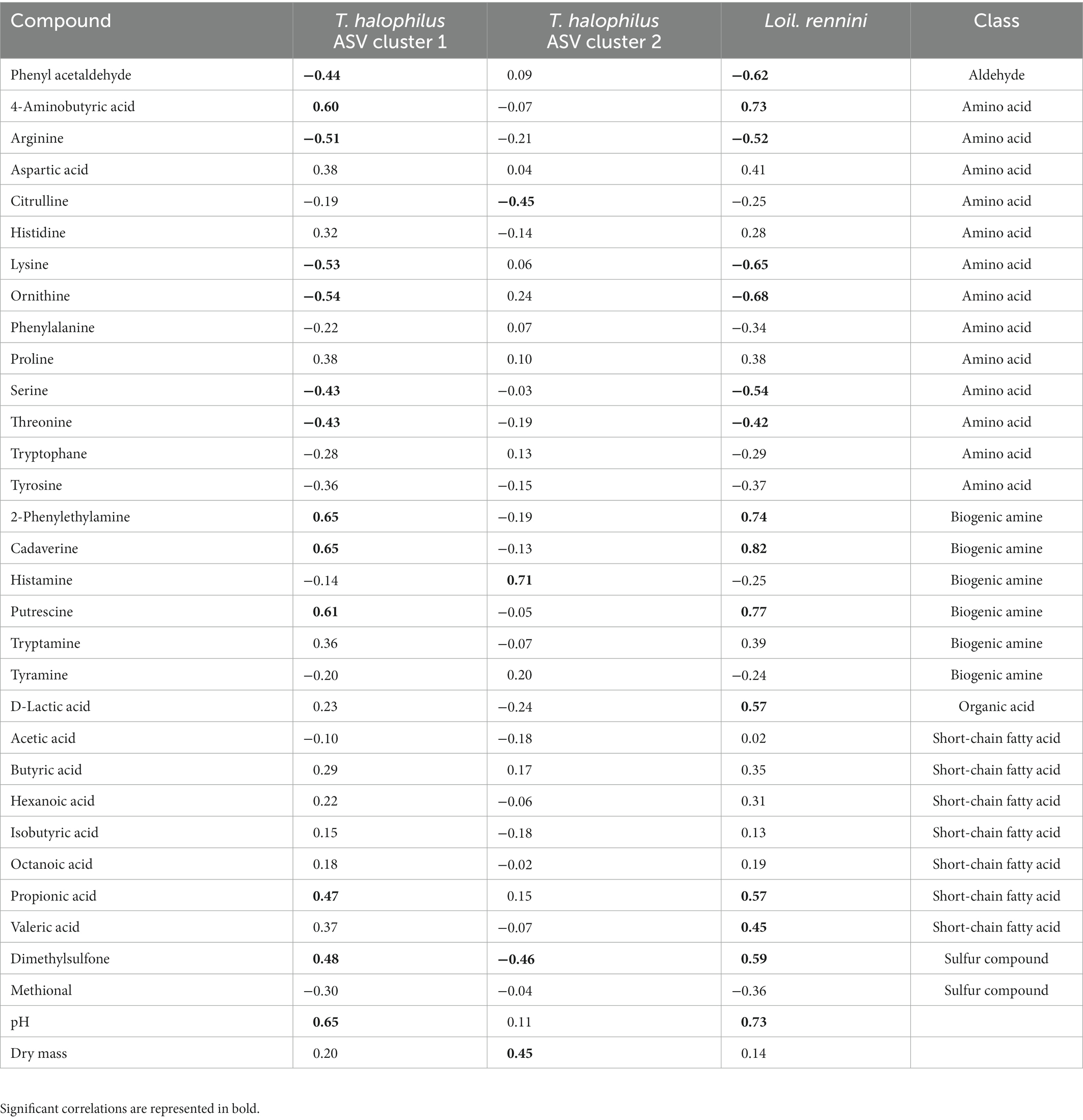
Table 2. Spearman correlations between compounds and the two main Tetragenococcus halophilus ASV clusters and Loigolactobacillus rennini after 75 weeks of ripening.
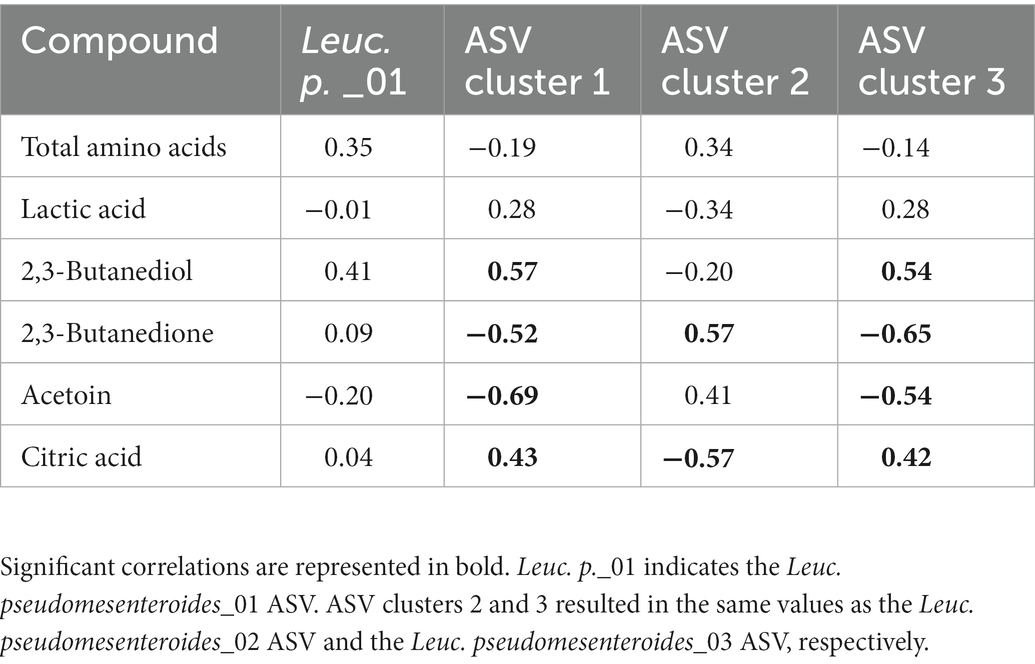
Table 3. Spearman correlations between some metabolite concentrations and Leuconostoc pseudomesenteroides amplicon sequence variants (ASVs) and ASV clusters, for Gouda cheese cores after 36 weeks of ripening.
3.4. Sensory analysis
The initial classification of the Gouda cheeses of 23 production batches after 26 weeks of ripening was mainly in line with the sensory scores obtained after 36 and 45 weeks of ripening (Supplementary Table S1). However, after 75 and 100 weeks of ripening, the Gouda cheeses with sensory score E had a lower average Z-score compared to those with sensory scores G and L. Moreover, 1, 6, 27, and 17 metabolites were significantly correlated with the sensory scores of Gouda cheeses of 36, 45, 75, and 100 weeks of ripening, respectively (Supplementary Table S2). These correlations often changed with the ripening time, as illustrated with the amino acids arginine and lysine that negatively correlated with the sensory scores after 45 weeks of ripening and positively after 75 and 100 weeks of ripening. The biogenic amines cadaverine, 2-phenylethylamine and putrescine, as well as 4-aminobutyric acid, had the strongest negative correlation with the sensory scores of cheeses of 75 and 100 weeks of ripening, whereas those compounds had no significant correlation with sensory scores after 36 and 45 weeks of ripening. At early ripening times, their concentrations were much lower and hence below their threshold values. Acetic acid, citrulline, gluconic acid, lactic acid, methional, phenylacetaldehyde, serine, and threonine were among the metabolites with a positive correlation with the sensory scores of the cheeses.
On species level, Lc. cremoris and Loil. rennini had a positive and negative correlation with the sensory scores of the cheeses after 75 weeks of ripening, respectively (Supplementary Table S3). At that ripening time, both LAB species were negatively correlated. Loigolactobacillus rennini was positively correlated with some biogenic amines and 4-aminobutyric acid; those compounds were negatively correlated with the sensory scores of the Gouda cheeses.
On ASV level, more significant correlations were found. In the case of Lc. cremoris, Lacc. paracasei and T. halophilus, opposite correlations were found within those species at different ripening times (Supplementary Table S4). The T. halophilus ASV cluster 1 had a strong negative correlation with the sensory scores of the 75-week cheeses and was positively correlated with Loil. rennini, whereas the T. halophilus ASV cluster 2 had no significant correlation with the sensory scores of the cheeses neither with the LAB species Loil. rennini (Figure 5).
4. Discussion
The present study deals with the investigation of batch-to-batch variations of a Gouda-type cheese produced on large scale in a European dairy factory, with special attention for the rotated starter culture mixtures applied, the ripening time, and the microbial and metabolite compositions of the cores and rinds of the cheese wheels. Although the commercial supplier of the mixed-strain starter cultures guaranteed that the starter culture mixtures A, B, and C used in the present study lead to similar end-products, significant differences were found between the Gouda A, B, and C cheeses produced, not only regarding their microbiome but also their metabolome. The main acidifiers, Lc. lactis and Lc. cremoris, did occur in all cheeses, albeit not in the same relative abundances, but species of the heterofermenter Leuconostoc did not, at least not to the same extent or according to the starter culture mixture used. The data of the present study further indicated that the dairy factory involved harbored a house microbiota that was mainly composed of Lacc. paracasei, T. halophilus, and Loil. rennini.
For the bacterial metagenetic analysis, a high-throughput, full-length 16S rRNA gene sequencing and ASV analysis was applied, which allowed accurate identifications at both species and strain level (Callahan et al., 2019). The big advantage of such a metagenetics approach lays in the single-nucleotide resolution, which even resulted in the detection of highly correlated, non-identical ASVs, probably belonging to one strain or cluster of strains. However, this deeper resolution also led to new challenges. For example, clusters with shared ASVs made further analysis less straightforward. Additionally, regardless of the precision, the use of only one gene might give a biased view on the bacterial diversity regarding subspecies level.
Most differences in microbiome and metabolome among the Gouda cheeses produced with the three different mixed-strain starter cultures applied were found after 36 weeks of ripening, as illustrated by the bacterial beta diversity and the scaled PCA of metabolites that grouped these cheese production batches according to the starter culture mixture used. At later ripening times, differences between the cheese production batches made with the same starter culture mixture increased and became more important than differences between the three groups of Gouda cheeses produced. This corroborated with a higher relative abundance of NSLAB that had a low diversity on species level across batches, but a striking diversity on ASV level. A similar situation has been described for a cheddar cheese factory, where cheese batches produced in different vats during the same day, or in the same vat during different days, have less than half of the NSLAB strains encountered as identical (Williams et al., 2002). The existence of a house microbiota and its role in the propagation of NSLAB in the factory and cheeses produced has been described frequently (Mounier et al., 2006; Bokulich and Mills, 2013a; Delcenserie et al., 2014; Calasso et al., 2016). Indeed, cheeses produced later during the day tend to have a higher bacterial diversity, suggesting a gradual accumulation of NSLAB in the production facility (O’Sullivan et al., 2015). This house microbiota was probably a main source of NSLAB contamination and associated variability for the 23 Gouda cheeses investigated.
The acidifying lactococci, Lc. cremoris and Lc. lactis, were the most abundant bacterial species in the Gouda cheeses produced, the former LAB species being present in higher relative abundances in the starter culture mixtures applied. The typical differences regarding salt tolerance and arginine conversion between these homofermentative LAB species were not always confirmed. For example, all Gouda cheese production batches had the same ASV profiles in the cores and rinds after 36 weeks of ripening, although it could be expected that the more salty rind would contain more ASVs of the more salt-tolerant Lc. lactis. Additionally, high arginine concentrations were not correlated with Lc. cremoris, although this species can not metabolize arginine (Schleifer et al., 1985). Yet, the higher arginine concentrations of the Gouda B cheeses could be linked with their slightly higher relative abundances of Lc. cremoris. Alternatively, in most cheese production batches, the relative abundance of Lc. cremoris decreased in favor of Lc. lactis, in particular between 45 and 100 weeks of ripening, which has to be ascribed to the faster speed of lysis of Lc. cremoris than Lc. lactis (Erkus et al., 2013; Fox et al., 2017).
Two ASVs of Lc. cremoris, Lc. cremoris_01 and Lc. cremoris_06, showed a high correlation with acetoin. However, acetoin is normally produced from citrate by Lc. lactis biovar diacetylactis, likely suggesting that the acetoin-producing strains lysed before 26 weeks of ripening. Alternatively, citrate-metabolizing and non-metabolizing Lc. lactis strains could share the same ASVs, which made it impossible to find meaningful correlations. Possibly, a substantial fraction of the Lactococcus reads was from cells in a VBNC state, as serine was one of the few amino acids positively correlated with Lactococcus and serine and methionine are particularly produced when Lactococcus enters a VBNC state (Ganesan et al., 2007).
In general, the different Lactococcus ASVs identified in the present study allowed to link each cheese production batch with its starter culture mixture applied, which can be of use for food authentication, although the differences in ASVs could not explain metabolic differences between the Gouda cheese production batches. However, it is well known that the Lactococcus genotype is often not in line with its phenotype (Tailliez et al., 1998; Fernández et al., 2011; Cavanagh et al., 2015), which has been reflected in the different reclassifications of Lactococcus species over time (Orla-Jensen, 1919; Garvie and Farrow, 1982; Schleifer et al., 1985; Li et al., 2021).
Considering the non-lactococci in the starter cultures used, a striking difference was the very low relative abundance of leuconostocs in the Gouda B cheeses, although they were present in the original starter culture mixtures applied. The Gouda A cheeses had a higher and approximately the same level of leuconostocs as the Gouda C cheeses. Yet, the presence of the Leuc. pseudomesenteroides_01 and Leuc. pseudomesenteroides_02 ASVs in the starter culture mixtures was lower than 0.008% (based on the total number of sequence reads). In general, undefined Gouda cheese starter cultures contain a low share of leuconostocs (Smid et al., 2014; Porcellato and Skeie, 2016; Frantzen et al., 2017). Overall, Leuc. mesenteroides was more abundant in the starter culture mixtures used, but it was marginally present in the Gouda cheeses produced compared to Leuc. pseudomesenteroides. In older literature and product information sheets, Leuc. mesenteroides subsp. cremoris and Leuconostoc lactis are considered as the main (sub)species in Gouda cheeses and mesophilic Gouda cheese starter cultures, but during the last 15 years, isolation of Leuc. mesenteroides subsp. mesenteroides, Leuc. mesenteroides subsp. dextranicum, and Leuc. pseudomesenteroides is more common (Frantzen et al., 2017). Of course, some Leuc. pseudomesenteroides isolates have also been wrongly identified as Leuc. mesenteroides, due to the use of solely partial 16S rRNA gene sequencing and/or phenotypical tests (Frantzen et al., 2017). Apart from a different effect on cheese metabolites, Leuc. pseudomesenteroides grows better and has a larger genome size compared to Leuc. mesenteroides subsp. cremoris, suggesting a higher survival potential during cheese ripening (Pedersen et al., 2016; Frantzen et al., 2017).
The three different ASV clusters of Leuc. pseudomesenteroides found in the Gouda cheeses of the present study correlated with either acidification and the lack of an efficient citrate metabolism (ASV clusters 1 and 3) or amino acid expulsion and flavor formation (ASV cluster 2). However, the lactococci associated with each Leuconostoc ASV cluster could be responsible for these differences as well. Indeed, leuconostocs are not believed to directly contribute to lactose fermentation (Ardö and Varming, 2010). Also, they lack cell envelope proteinases, making them dependent on peptide release from lactococci, and although they have only limited aminopeptidolytic activity, they can increase the concentrations of some amino acids extracellularly (Pedersen et al., 2016). Further, whereas the concentrations of most amino acids were lower in the Gouda B cheeses compared to the other ones, reflecting the lower relative abundance of leuconostocs, these cheeses contained the highest acetoin and 2,3-butanedione concentrations. This could be ascribed to the citrate-metabolizing Lc. lactis strains abundant or present in those cheeses, questioning the need of Leuconostoc for the production of these aroma compounds. Indeed, Lc. lactis biovar diacetylactis can even produce more acetoin compared to Leuconostoc (Starrenburg and Hugenholtz, 1991; Hugenholtz, 1993). However, the Gouda B cheeses with a low relative abundance of Leuconostoc contained more NSLAB, suggesting that inhibition of the growth of undesired bacteria seems to be an interesting contribution of Leuconostoc, an effect that has been described before (Martley and Crow, 1993). This effect was stronger for strains of the ASV cluster 1 in the Gouda C cheeses, compared to those of ASV cluster 2 in the Gouda A cheeses.
The increase of the relative abundances of the NSLAB species from 5 to 20% between 26 and 45 weeks of Gouda cheese ripening was in line with other culture-independent Gouda cheese studies (Porcellato and Skeie, 2016; Park et al., 2019). Lacticaseibacillus paracasei was the most abundant NSLAB species during the first 45 weeks of ripening, confirming previous reports (Van Hoorde et al., 2008; Kołakowski et al., 2012). It was represented by 10 of ASVs, differing between the Gouda cheese production batches and ripening times, but often occurring in a similar ratio, suggesting the presence of non-identical copies of the 16S rRNA gene within one strain. Possibly, the cheese manufacturing environment contained a high diversity of Lacc. paracasei and each production batch got inoculated with some strains that prevailed during ripening and contributed to the release of amino acids and the production of volatile organic compounds. In support of these beneficial properties, the use of Lacc. paracasei as adjunct culture for cheese production is well-known (Antonsson et al., 2003; Van Hoorde et al., 2010b; Stefanovic et al., 2018; Asahina et al., 2020; Bancalari et al., 2020; Randazzo et al., 2021).
Tetragenococcus halophilus was the most abundant NSLAB species in multiple Gouda cheeses after 75 and 100 weeks of ripening. This LAB species has been found culture-independently in Polish Oscypek cheese (Alegría et al., 2012), Spanish Cabrales and Manchego-type cheeses (Diaz et al., 2016; Botello-Morte et al., 2022), the rind of Gouda cheese (Salazar et al., 2018), and Irish hard cheese (only genus level; Quigley et al., 2012), but always at low relative abundances. Culture-dependently, it has been isolated from Mexican Cotija cheese (Morales et al., 2011), Brie de Meaux cheese (Unno et al., 2020), and Spanish blue-veined cheeses (Rodríguez et al., 2022). In these culture-dependent studies, T. halophilus has only been picked from media enriched with 2.5, 5.0, or 7.0% (m/v) NaCl. Besides a halophilic character, this LAB species displays an alkaliphilic behavior, as growth is better at pH 8.0 than at pH 7.0, whereas no growth occurs at pH 5.0 (Justé et al., 2012; Unno et al., 2020). Except for its salt dependency in agar media, its rare isolation could be ascribed to its prevalence in cheese rinds or long-ripened cheeses, which give lower counts with a culture-dependent analysis and fewer cells for proper DNA extraction with a culture-independent analysis.
The presence of T. halophilus in cheeses probably originates from the brine bath, as its occurrence in the latter was shown both culture-dependently and culture-independently (data not shown). Indeed, it has been found as a prevalent species in Belgian, Italian, and Danish brines (Marino et al., 2017; Haastrup et al., 2018; Vermote et al., 2018). One Italian brine shows a relative abundance of more than 50% (on sequence read basis) of Tetragenococcus and has a pH of 4.6, whereas the pH of the other brines varies between 4.8 and 5.7, suggesting that either growth is still possible at a pH lower than 5.0 or that no growth but survival occurs thanks to its salt tolerance. Since T. halophilus was already abundant in the Gouda cheese rinds of the present study after 36 weeks of ripening, it could likely proliferate and migrate from the brine to the cheese rind and finally to the core upon further cheese ripening. Furthermore, strong correlations were found between T. halophilus and the concentrations of free amino acids and short-chain fatty acids, which was in line with the proteolytic and lipolytic properties of isolates from Mexican cheeses (Morales et al., 2011). These properties could make T. halophilus interesting as an adjunct starter culture for (Gouda) cheese production. This LAB species is already used as primary starter culture for the production of oriental fermented salted food products, such as soy sauce, fish sauce, and shrimp paste (Justé et al., 2014). However, some T. halophilus strains harbor histidine and/or tyrosine decarboxylase genes, allowing histamine and/or tyramine production, respectively (Diaz et al., 2016; Chun et al., 2019). Alternatively, well-chosen strains would even lower biogenic amine formation, probably indirectly through the repression of other tetragenococci that produce biogenic amines (Udomsil et al., 2011; Kim et al., 2019), which supports the need to perform a genome sequencing and functional analysis approach of potential adjunct culture strains before their application.
Although T. halophilus was represented by more than 100 different ASVs in the Gouda cheeses examined, only two main ASV clusters occurred after 75 weeks of ripening, which confirmed its strain-dependent properties. Whereas the T. halophilus ASV cluster 1 was mainly found in cheeses with higher pH (which may have been caused by biogenic amine formation) and higher concentrations of short-chain fatty acids, the T. halophilus ASV cluster 2 was mainly found in cheeses with higher dry mass, reflecting the acid-sensitive and lipolytic character of strains of the former cluster and the osmotolerance of those of the latter one. In addition, the T. halophilus ASV cluster 1 strongly correlated with Loil. rennini and high and low concentrations of several biogenic amines and their precursor amino acids, respectively, after 75 weeks of ripening. The T. halophilus ASV cluster 2 was negatively correlated with histidine and positively correlated with histamine, suggesting the presence of a histidine decarboxylase gene that was expressed after 75 and 100 weeks of ripening. Indeed, histidine decarboxylase genes can be present in T. halophilus genomes, whereas other decarboxylase genes have not been found in the pangenome composed of 15 T. halophilus strains (Satomi et al., 2011; Chun et al., 2019).
Putrescine and cadaverine negatively impacted the sensory scores of the Gouda cheeses examined, whereas histamine did not. However, consumption of foods with a high histamine concentration can result in histamine poisoning (Özogul and Özogul, 2019). In the Gouda cheeses of the present study, the histamine concentrations were low up to 45 weeks of ripening, but they were around the Codex Alimentarius limit of 200 mg/kg (wet mass) established for fish and fish products after 75 and 100 weeks of ripening. Hence, because of a correlation of both ASV clusters 1 and 2 with the presence of several biogenic amines, a careful selection should be made when strains of these clusters are considered to be used as adjunct cultures. Indeed, in the case of the T. halophilus ASV cluster 1, the presence of biogenic amines could be related to the presence of Loil. rennini, and in the case of the T. halophilus ASV cluster 2, histamine production was most likely related to T. halophilus. However, the use of T. halophilus as adjunct culture could be made possible after curing of the plasmid encoding histamine production (Satomi et al., 2011; Botello-Morte et al., 2022).
The salt-tolerant Loil. rennini, which was prevalent in the Gouda cheese rinds after 36 weeks of ripening and in the cores of some cheese production batches after 75 and 100 weeks of ripening, was another prevalent NSLAB species. It was first isolated from rennet and held responsible for cracks and off-flavor defects in Dutch cheeses (Chenoll et al., 2006). It has also been found in Greek Kopanisti and Mana cheeses and in cheese brine (Asteri et al., 2009; Vermote et al., 2018). This LAB species, represented by a limited number of ASVs in the Gouda cheeses of the present study, correlated positively with different biogenic amines. Indeed, our investigation of the genome of a strain isolated from a 2-year Greek Kopanisti cheese (Kazou et al., 2017) revealed the presence of different decarboxylase genes, such as those coding for glutamate decarboxylase and ornithine decarboxylase, which yield 4-aminobutyric acid and putrescine, respectively. Moreover, a high correlation was found between Loil. rennini and a high pH after 75 weeks of ripening, likely to be ascribed to these decarboxylations.
The fungi found in the Gouda cheeses of the present study were in accordance with other reports on Debaryomyces as the most prevailing yeast in Gouda cheeses (Welthagen and Viljoen, 1998; Banjara et al., 2015). Whereas the latter studies did not differentiate between cheese cores and rinds, the present study showed that Debaryomyces was the most abundant yeast in the rind and Saccharomyces in the core of the Gouda cheeses produced. Debaryomyces has been found as a prevalent yeast in cheese brines and cheese production environments before (Mounier et al., 2006; Bokulich and Mills, 2013a; Vermote et al., 2018). Its higher salt tolerance and oxygen dependency than that of Saccharomyces (Sánchez et al., 2006) was reflected by the higher relative abundance of Debaryomyces in the Gouda cheese rinds and of Saccharomyces in the anaerobic cheese cores. The occurrence of yeasts in the Gouda cheeses of the present study may be linked with the production of different types of cheeses in the same dairy factory and their presence in cheese brines (Vermote et al., 2018). However, whereas some studies have mentioned yeast counts in Gouda cheeses up to 105 CFU/g (Welthagen and Viljoen, 1998; Kołakowski et al., 2012), plating of some cheese samples of the present study on yeast-peptone-dextrose agar did not result in growth (data not shown), indicating that yeasts were present in very low numbers.
5. Conclusion
Batch-to-batch variation in mature Gouda cheeses was significant on microbial and metabolite level. Even when the rotated starter culture mixtures (A, B, and C) were considered to be similar, differences in microbial compositions and metabolite concentrations were found. A metagenetic approach based on high-throughput full-length 16S rRNA gene sequencing, accompanied with an ASV analysis, allowed differentiation of the bacterial communities on intra-species level. Starter culture mixture B did not result in prevailing leuconostocs in the corresponding cheeses, which led to a higher relative abundance of NSLAB. Hence, the presence of leuconostocs seemed to inhibit the growth of NSLAB, an effect that especially occurred in the Gouda C cheeses. Batch-to-batch differences generally increased with ripening time and became then less related to the starter culture mixture applied. Yet, metabolite differences between cores and rinds of the Gouda cheeses were found at all ripening times. The bacterial composition of the rinds at 36 weeks of ripening resembled those of the cores at 75 and 100 weeks of ripening. Among the NSLAB, which originate from the house microbiota that might vary, but will most probably be independent of the season given the constant temperature under which the factory is operating, Lacc. paracasei was most abundant until 45 weeks of ripening. This was taken over by T. halophilus and to a lesser extent Loil. rennini, which both could be responsible for biogenic amine formation and a bad flavor. Strong correlations of T. halophilus with desirable metabolites regarding aroma formation may make it a candidate adjunct culture. Finally, to increase Gouda cheese quality standardization, the use of only one starter culture mixture is proposed, together with a potential adjunct culture. Starter culture mixture rotation could still be applied, which each time should lead to another cheese type; for instance, starter culture mixture A should only be used for young Gouda cheeses, B for matured Gouda cheeses, and C for extra-matured Gouda cheeses. Also, the metagenetic ASV approach applied in the present study will allow differentiation of mixed-strain starter cultures across Gouda cheese producers in particular and contribute to food authentication in general.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ebi.ac.uk/ena, PRJEB58546.
Author contributions
HD carried out the research and drafted the manuscript. SW and LDV supervised the research and revised and edited the manuscript. LDV was responsible for funding. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Research Council of the Vrije Universiteit Brussel (SRP7 and IOF3017 projects) and the Agency Flanders Innovation and Entrepreneurship (VLAIO).
Acknowledgments
The authors thank Maria de la Cruz Loayza for her contribution to the cheese sampling and metabolite target analysis as well as ing. Wim Borremans for his technical advice.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1128394/full#supplementary-material
References
Alegría, Á., Szczesny, P., Mayo, B., Bardowski, J., and Kowalczyk, M. (2012). Biodiversity in Oscypek, a traditional Polish cheese, determined by culture-dependent and-independent approaches. Appl. Environ. Microbiol. 78, 1890–1898. doi: 10.1128/AEM.06081-11
Antonsson, M., Molin, G., and Ardö, Y. (2003). Lactobacillus strains isolated from Danbo cheese as adjunct cultures in a cheese model system. Int. J. Food Microbiol. 85, 159–169. doi: 10.1016/S0168-1605(02)00536-6
Ardö, Y., and Varming, C. (2010). Bacterial influence on characteristic flavour of cheeses made with mesophilic DL-starter. Aust. J. Dairy Technol. 65:153.
Asahina, Y., Hagi, T., Kobayashi, M., Narita, T., Sasaki, K., Tajima, A., et al. (2020). Expression profiles of milk proteolysis-related genes in Lactobacillus paracasei EG9, a non-starter lactic acid bacterial strain, during Gouda-type cheese ripening. Int. Dairy J. 110:104812. doi: 10.1016/j.idairyj.2020.104812
Asteri, I. A., Robertson, N., Kagkli, D. M., Andrewes, P., Nychas, G., Coolbear, T., et al. (2009). Technological and flavour potential of cultures isolated from traditional Greek cheeses: a pool of novel species and starters. Int. Dairy J. 19, 595–604. doi: 10.1016/j.idairyj.2009.04.006
Bancalari, E., Montanari, C., Levante, A., Alinovi, M., Neviani, E., Gardini, F., et al. (2020). Lactobacillus paracasei 4341 as adjunct culture to enhance flavor in short ripened Caciotta-type cheese. Food Res. Int. 135:109284. doi: 10.1016/j.foodres.2020.109284
Banjara, N., Suhr, M. J., and Hallen-Adams, H. E. (2015). Diversity of yeast and mold species from a variety of cheese types. Curr. Microbiol. 70, 792–800. doi: 10.1007/s00284-015-0790-1
Barbieri, F., Montanari, C., Gardini, F., and Tabanelli, G. (2019). Biogenic amine production by lactic acid bacteria: a review. Foods 8, 1–27. doi: 10.3390/foods8010017
Bertuzzi, A. S., Walsh, A. M., Sheehan, J. J., Cotter, P. D., Crispie, F., McSweeney, P. L., et al. (2018). Omics-based insights into flavor development and microbial succession within surface-ripened cheese. mSystems 3:e00211-17. doi: 10.1128/mSystems.00211-17
Bokulich, N. A., and Mills, D. A. (2013a). Facility-specific “house” microbiome drives microbial landscapes of artisan cheesemaking plants. Appl. Environ. Microbiol. 79, 5214–5223. doi: 10.1128/AEM.00934-13
Bokulich, N. A., and Mills, D. A. (2013b). Improved selection of internal transcribed spacer-specific primers enables quantitative, ultra-high-throughput profiling of fungal communities. Appl. Environ. Microbiol. 79, 2519–2526. doi: 10.1128/AEM.03870-12
Botello-Morte, L., Moniente, M., Gil-Ramírez, Y., Virto, R., García-Gonzalo, D., and Pagán, R. (2022). Identification by means of molecular tools of the microbiota responsible for the formation of histamine accumulated in commercial cheeses in Spain. Food Control 133:108595. doi: 10.1016/j.foodcont.2021.108595
Calasso, M., Ercolini, D., Mancini, L., Stellato, G., Minervini, F., Di Cagno, R., et al. (2016). Relationships among house, rind and core microbiotas during manufacture of traditional Italian cheeses at the same dairy plant. Food Microbiol. 54, 115–126. doi: 10.1016/j.fm.2015.10.008
Callahan, B. J., McMurdie, P. J., and Holmes, S. P. (2017). Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J. 11, 2639–2643. doi: 10.1038/ismej.2017.119
Callahan, B. J., Wong, J., Heiner, C., Oh, S., Theriot, C. M., Gulati, A. S., et al. (2019). High-throughput amplicon sequencing of the full-length 16S rRNA gene with single-nucleotide resolution. Nucleic Acids Res. 47:e103. doi: 10.1093/nar/gkz569
Cavanagh, D., Casey, A., Altermann, E., Cotter, P. D., Fitzgerald, G. F., and McAuliffe, O. (2015). Evaluation of Lactococcus lactis isolates from nondairy sources with potential dairy applications reveals extensive phenotype-genotype disparity and implications for a revised species. Appl. Environ. Microbiol. 81, 3961–3972. doi: 10.1128/AEM.04092-14
Chenoll, E., Macián, C., and Aznar, R. (2006). Lactobacillus rennini sp. nov., isolated from rennin and associated with cheese spoilage. Int. J. Syst. Bacteriol. 56, 449–452. doi: 10.1099/ijs.0.64021-0
Chun, B. H., Han, D. M., Kim, K. H., Jeong, S. E., Park, D., and Jeon, C. O. (2019). Genomic and metabolic features of Tetragenococcus halophilus as revealed by pan-genome and transcriptome analyses. Food Microbiol. 83, 36–47. doi: 10.1016/j.fm.2019.04.009
De Bruyn, F., Zhang, S. J., Pothakos, V., Torres, J., Lambot, C., Moroni, A. V., et al. (2017). Exploring the impacts of postharvest processing on the microbiota and metabolite profiles during green coffee bean production. Appl. Environ. Microbiol. 83, 1–16. doi: 10.1128/AEM.02398-16
De Filippis, F., Parente, E., and Ercolini, D. (2017). Metagenomics insights into food fermentations. Microb. Biotechnol. 10, 91–102. doi: 10.1111/1751-7915.12421
Delcenserie, V., Taminiau, B., Delhalle, L., Nezer, C., Doyen, P., Crevecoeur, S., et al. (2014). Microbiota characterization of a Belgian protected designation of origin cheese, Herve cheese, using metagenomic analysis. J. Dairy Sci. 97, 6046–6056. doi: 10.3168/jds.2014-8225
Diaz, M., Ladero, V., Redruello, B., Sanchez-Llana, E., del Rio, B., Fernandez, M., et al. (2016). A PCR-DGGE method for the identification of histamine-producing bacteria in cheese. Food Control 63, 216–223. doi: 10.1016/j.foodcont.2015.11.035
Díaz-Muñoz, C., Van de Voorde, D., Comasio, A., Verce, M., Hernandez, C. E., Weckx, S., et al. (2021). Curing of cocoa beans: fine-scale monitoring of the starter cultures applied and metabolomics of the fermentation and drying steps. Front. Microbiol. 11:616875. doi: 10.3389/fmicb.2020.616875
Dirinck, P., and De Winne, A. (1999). Flavour characterisation and classification of cheeses by gas chromatographic-mass spectrometric profiling. J. Chromatogr. A 847, 203–208. doi: 10.1016/S0021-9673(99)00193-4
Dugat-Bony, E., Garnier, L., Denonfoux, J., Ferreira, S., Sarthou, A. S., Bonnarme, P., et al. (2016). Highlighting the microbial diversity of 12 French cheese varieties. Int. J. Food Microbiol. 238, 265–273. doi: 10.1016/j.ijfoodmicro.2016.09.026
Düsterhöft, E. M., Engels, W., and Huppertz, T. (2017). “Gouda and related cheeses” in Cheese: Chemistry, Physics and Microbiology. eds. P. L. H. McSweeney, P. F. Fox, P. Cotter, and D. Everett, vol. 2. 4th ed (San Diego, CA: Elsevier), 865–888.
Ercolini, D. (2013). High-throughput sequencing and metagenomics: moving forward in the culture-independent analysis of food microbial ecology. Appl. Environ. Microbiol. 79, 3148–3155. doi: 10.1128/AEM.00256-13
Erkus, O., De Jager, V. C. L., Spus, M., Van Alen-Boerrigter, I. J., Van Rijswijck, I. M. H., Hazelwood, L., et al. (2013). Multifactorial diversity sustains microbial community stability. ISME J. 7, 2126–2136. doi: 10.1038/ismej.2013.108
Fernández, E., Alegría, Á., Delgado, S., Martín, M. C., and Mayo, B. (2011). Comparative phenotypic and molecular genetic profiling of wild Lactococcus lactis subsp. lactis strains of the L. lactis subsp. lactis and L. lactis subsp. cremoris genotypes, isolated from starter-free cheeses made of raw milk. Appl. Environ. Microbiol. 77, 5324–5335. doi: 10.1128/AEM.02991-10
Fitzsimons, N. A., Cogan, T. M., Condon, S., and Beresford, T. (2001). Spatial and temporal distribution of non-starter lactic acid bacteria in Cheddar cheese. J. Appl. Microbiol. 90, 600–608. doi: 10.1046/j.1365-2672.2001.01285.x
Fox, P. F., Guinee, T. P., Cogan, T. M., and McSweeney, P. L. H. (2017). Fundamentals of Cheese Science. 2nd Edn., Springer, New York.
Frantzen, C. A., Kleppen, H. P., and Holo, H. (2018). Lactococcus lactis diversity in undefined mixed dairy starter cultures as revealed by comparative genome analyses and targeted amplicon sequencing of epsD. Appl. Environ. Microbiol. 84, 1–15. doi: 10.1128/AEM.02199-17
Frantzen, C. A., Kot, W., Pedersen, T. B., Ardö, Y. M., Broadbent, J. R., Neve, H., et al. (2017). Genomic characterization of dairy associated Leuconostoc species and diversity of leuconostocs in undefined mixed mesophilic starter cultures. Front. Microbiol. 8:132. doi: 10.3389/fmicb.2017.00132
Ganesan, B., Stuart, M. R., and Weimer, B. C. (2007). Carbohydrate starvation causes a metabolically active but nonculturable state in Lactococcus lactis. Appl. Environ. Microbiol. 73, 2498–2512. doi: 10.1128/AEM.01832-06
Garvie, E. I., and Farrow, J. A. (1982). Streptococcus lactis subsp. cremoris (Orla-Jensen) comb. nov., and Streptococcus lactis subsp. diacetilactis (Matuszewski et al.) nom. rev., comb. nov. Int. J. Syst. Bacteriol. 32, 453–455. doi: 10.1099/00207713-32-4-453
Gobbetti, M., De Angelis, M., Di Cagno, R., Mancini, L., and Fox, P. F. (2015). Pros and cons for using non-starter lactic acid bacteria (NSLAB) as secondary/adjunct starters for cheese ripening. Trends Food Sci. Technol. 45, 167–178. doi: 10.1016/j.tifs.2015.07.016
Gobbetti, M., Di Cagno, R., Calasso, M., Neviani, E., Fox, P. F., and De Angelis, M. (2018). Drivers that establish and assembly the lactic acid bacteria biota in cheeses. Trends Food Sci. Technol. 78, 244–254. doi: 10.1016/j.tifs.2018.06.010
Gu, Z., Eils, R., and Schlesner, M. (2016). Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847–2849. doi: 10.1093/bioinformatics/btw313
Haastrup, M. K., Johansen, P., Malskær, A. H., Castro-Mejía, J. L., Kot, W., Krych, L., et al. (2018). Cheese brines from Danish dairies reveal a complex microbiota comprising several halotolerant bacteria and yeasts. Int. J. Food Microbiol. 285, 173–187. doi: 10.1016/j.ijfoodmicro.2018.08.015
Hervé, M. (2021). RVAideMemoire: Testing and Plotting Procedures for Biostatistics. R Package Version 0.9–80. Available at: https://CRAN.R-project.org/package=RVAideMemoire (accessed September 2021).
Høier, E., Janzen, T., Rattray, F., Sørensen, K., Børsting, M. W., Brockmann, E., et al. (2010). “The production, application and action of lactic cheese starter cultures” in Technology of Cheesemaking. eds. B. A. Law and A. Y. Tamime. 2nd ed (Chichester: Wiley-Blackwell), 166–192.
Hugenholtz, J. (1993). Citrate metabolism in lactic acid bacteria. FEMS Microbiol. Rev. 12, 165–178. doi: 10.1111/j.1574-6976.1993.tb00017.x
Jin, H., Mo, L., Pan, L., Hou, Q., Li, C., Darima, I., et al. (2018). Using PacBio sequencing to investigate the bacterial microbiota of traditional Buryatian cottage cheese and comparison with Italian and Kazakhstan artisanal cheeses. J. Dairy Sci. 101, 6885–6896. doi: 10.3168/jds.2018-14403
Jo, Y., Benoist, D. M., Ameerally, A., and Drake, M. A. (2018). Sensory and chemical properties of Gouda cheese. J. Dairy Sci. 101, 1967–1989. doi: 10.3168/jds.2017-13637
Johnson, J. S., Spakowicz, D. J., Hong, B. Y., Petersen, L. M., Demkowicz, P., Chen, L., et al. (2019). Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat. Commun. 10, 1–11. doi: 10.1038/s41467-019-13036-1
Jung, H. J., Ganesan, P., Lee, S. J., and Kwak, H. S. (2013). Comparative study of flavor in cholesterol-removed Gouda cheese and Gouda cheese during ripening. J. Dairy Sci. 96, 1972–1983. doi: 10.3168/jds.2012-5644
Justé, A., Lievens, B., Rediers, H., and Willems, K. A. (2014). “The genus Tetragenococcus” in Lactic Acid Bacteria: Biodiversity and Taxonomy. eds. W. H. Holzapfel and B. J. B. Wood (Chichester: John Wiley & Sons), 213–227.
Justé, A., Van Trappen, S., Verreth, C., Cleenwerck, I., De Vos, P., Lievens, B., et al. (2012). Characterization of Tetragenococcus strains from sugar thick juice reveals a novel species, Tetragenococcus osmophilus sp. nov., and divides Tetragenococcus halophilus into two subspecies, T. halophilus subsp. halophilus subsp. nov. and T. halophilus subsp. flandriensis subsp. nov. Int. J. Syst. Evol. Microbiol. 62, 129–137. doi: 10.1099/ijs.0.029157-0
Kamimura, B. A., de Filippis, F., Sant’Ana, A. S., and Ercolini, D. (2019). Large-scale mapping of microbial diversity in artisanal Brazilian cheeses. Food Microbiol. 80, 40–49. doi: 10.1016/j.fm.2018.12.014
Kazou, M., Alexandraki, V., Pot, B., Tsakalidou, E., and Papadimitriou, K. (2017). Whole-genome sequence of the cheese isolate Lactobacillus rennini ACA-DC 565. Genome Announc. 5, 16–17. doi: 10.1128/genomeA.01579-16
Kim, K. H., Lee, S. H., Chun, B. H., Jeong, S. E., and Jeon, C. O. (2019). Tetragenococcus halophilus MJ4 as a starter culture for repressing biogenic amine (cadaverine) formation during saeu-jeot (salted shrimp) fermentation. Food Microbiol. 82, 465–473. doi: 10.1016/j.fm.2019.02.017
Kołakowski, P., Podolak, R., and Kowalska, M. (2012). Microbial profile of Gouda cheese during ripening in two independent chambers - a short report. Pol. J. Food Nutr. Sci. 62, 179–184. doi: 10.2478/v10222-012-0051-y
Kõljalg, U., Nilsson, R. H., Abarenkov, K., Tedersoo, L., Taylor, A. F. S., Bahram, M., et al. (2013). Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol. 22, 5271–5277. doi: 10.1111/mec.12481
Komprda, T., Smělá, D., Novická, K., Kalhotka, L., Šustová, K., and Pechová, P. (2007). Content and distribution of biogenic amines in Dutch-type hard cheese. Food Chem. 102, 129–137. doi: 10.1016/j.foodchem.2006.04.041
Le Boucher, C., Gagnaire, V., Briard-Bion, V., Jardin, J., Maillard, M. B., Dervilly-Pinel, G., et al. (2016). Spatial distribution of Lactococcus lactis colonies modulates the production of major metabolites during the ripening of a model cheese. Appl. Environ. Microbiol. 82, 202–210. doi: 10.1128/AEM.02621-15
Levante, A., Bertani, G., Bottari, B., Bernini, V., Lazzi, C., Gatti, M., et al. (2021). How new molecular approaches have contributed to shedding light on microbial dynamics in Parmigiano Reggiano cheese. Curr. Opin. Food Sci. 38, 131–140. doi: 10.1016/j.cofs.2020.11.005
Li, T. T., Tian, W. L., and Gu, C. T. (2021). Elevation of Lactococcus lactis subsp. cremoris to the species level as Lactococcus cremoris sp. nov. and transfer of Lactococcus lactis subsp. tructae to Lactococcus cremoris as Lactococcus cremoris subsp. tructae comb. nov. Int. J. Syst. Evol. 71:004727. doi: 10.1099/ijsem.0.004727
Linares, D. M., Del Río, B., Ladero, V., Martínez, N., Fernández, M., Martín, M. C., et al. (2012). Factors influencing biogenic amines accumulation in dairy products. Front. Microbiol. 3:180. doi: 10.3389/fmicb.2012.00180
Mahony, J., and van Sinderen, D. (2014). Current taxonomy of phages infecting lactic acid bacteria. Front. Microbiol. 5:7. doi: 10.3389/fmicb.2014.00007
Marino, M., Dubsky de Wittenau, G., Saccà, E., Cattonaro, F., Spadotto, A., Innocente, N., et al. (2019). Metagenomic profiles of different types of Italian high-moisture Mozzarella cheese. Food Microbiol. 79, 123–131. doi: 10.1016/j.fm.2018.12.007
Marino, M., Innocente, N., Maifreni, M., Mounier, J., Cobo-Díaz, J. F., Coton, E., et al. (2017). Diversity within Italian cheesemaking brine-associated bacterial communities evidenced by massive parallel 16S rRNA gene tag sequencing. Front. Microbiol. 8:2119. doi: 10.3389/fmicb.2017.02119
Martley, F. G., and Crow, V. L. (1993). Interactions between non-starter microorganisms during cheese manufacture and ripening. Int. Dairy J. 3, 461–483. doi: 10.1016/0958-6946(93)90027-W
Montel, M. C., Buchin, S., Mallet, A., Delbes-Paus, C., Vuitton, D. A., Desmasures, N., et al. (2014). Traditional cheeses: rich and diverse microbiota with associated benefits. Int. J. Food Microbiol. 177, 136–154. doi: 10.1016/j.ijfoodmicro.2014.02.019
Morales, F., Morales, J. I., Hernández, C. H., and Hernández-Sánchez, H. (2011). Isolation and partial characterization of halotolerant lactic acid bacteria from two Mexican cheeses. Appl. Biochem. Biotechnol. 164, 889–905. doi: 10.1007/s12010-011-9182-6
Mounier, J., Goerges, S., Gelsomino, R., Vancanneyt, M., Vandemeulebroecke, K., Hoste, B., et al. (2006). Sources of the adventitious microflora of a smear-ripened cheese. J. Appl. Microbiol. 101, 668–681. doi: 10.1111/j.1365-2672.2006.02922.x
O’Brien, E., Mills, S., Dobson, A., Serrano, L. M., Hannon, J., Ryan, S. P., et al. (2017). Contribution of the novel sulfur-producing adjunct Lactobacillus nodensis to flavor development in Gouda cheese. J. Dairy Sci. 100, 4322–4334. doi: 10.3168/jds.2016-11726
O’Sullivan, D. J., Cotter, P. D., O’Sullivan, O., Giblin, L., McSweeney, P. L. H., and Sheehan, J. J. (2015). Temporal and spatial differences in microbial composition during the manufacture of a continental-type cheese. Appl. Environ. Microbiol. 81, 2525–2533. doi: 10.1128/AEM.04054-14
Oberg, C. J., Oberg, T. S., Culumber, M. D., Ortakci, F., Broadbent, J. R., and McMahon, D. J. (2016). Lactobacillus wasatchensis sp. nov., a non-starter lactic acid bacteria isolated from aged cheddar cheese. Int. J. Syst. Evol. 66, 158–164. doi: 10.1099/ijsem.0.000689
Oksanen, J., Blanchet, F. G., Friendly, M., Kindt, R., Legendre, P., McGlinn, D., et al. (2020). vegan: Community Ecology Package. R Package Version 2.5–7. Available at: https://CRAN.R-project.org/package=vegan (accessed March 2022).
Özogul, Y. Ö., and Özogul, F. (2019). “Biogenic amines formation, toxicity, regulations in food” in Biogenic Amines in Food: Analysis, Occurrence and Toxicity. eds. B. Saad and R. Tofalo (London: Royal Society of Chemistry), 1–17.
Parente, E., Cogan, T. M., and Powell, I. B. (2017). “Starter cultures: general aspects” in Cheese. eds. P. L. H. McSweeney, P. F. Fox, P. Cotter, and D. Everett, vol. 1. 4th ed (San Diego, CA: Elsevier), 201–226.
Park, W., Yoo, J., Oh, S., Ham, J., Jeong, S., and Kim, Y. (2019). Microbiological characteristics of Gouda cheese manufactured with pasteurized and raw milk during ripening using next generation sequencing. Food Sci. Anim. Resour. 39, 585–600. doi: 10.5851/kosfa.2019.e49
Pedersen, T. B., Vogensen, F. K., and Ardö, Y. (2016). Effect of heterofermentative lactic acid bacteria of DL-starters in initial ripening of semi-hard cheese. Int. Dairy J. 57, 72–79. doi: 10.1016/j.idairyj.2016.02.041
Porcellato, D., and Skeie, S. B. (2016). Bacterial dynamics and functional analysis of microbial metagenomes during ripening of Dutch-type cheese. Int. Dairy J. 61, 182–188. doi: 10.1016/j.idairyj.2016.05.005
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucl. Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Quigley, L., O’Sullivan, O., Beresford, T. P., Ross, R. P., Fitzgerald, G. F., and Cotter, P. D. (2012). High-throughput sequencing for detection of subpopulations of bacteria not previously associated with artisanal cheeses. Appl. Environ. Microbiol. 78, 5717–5723. doi: 10.1128/AEM.00918-12
R Core Team (2021). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Randazzo, C. L., Liotta, L., De Angelis, M., Celano, G., Russo, N., Van Hoorde, K., et al. (2021). Adjunct culture of non-starter lactic acid bacteria for the production of Provola dei nebrodi PDO cheese: in vitro screening and pilot-scale cheese-making. Microorganisms 9, 1–18. doi: 10.3390/microorganisms9010179
Riquelme, C., Câmara, S., de Enes Dapkevicius, M. L. N., Vinuesa, P., da Silva, C. C. G., Malcata, F. X., et al. (2015). Characterization of the bacterial biodiversity in Pico cheese (an artisanal Azorean food). Int. J. Food Microbiol. 192, 86–94. doi: 10.1016/j.ijfoodmicro.2014.09.031
Rodríguez, J., González-Guerra, A., Vázquez, L., Fernández-López, R., Flórez, A. B., de la Cruz, F., et al. (2022). Isolation and phenotypic and genomic characterization of Tetragenococcus spp. from two Spanish traditional blue-veined cheeses made of raw milk. Int. J. Food Microbiol. 371:109670. doi: 10.1016/j.ijfoodmicro.2022.109670
Ruggirello, M., Dolci, P., and Cocolin, L. (2014). Detection and viability of Lactococcus lactis throughout cheese ripening. PLoS One 9:e0114280. doi: 10.1371/journal.pone.0114280
Ruyssen, T., Janssens, M., Van Gasse, B., Van Laere, D., Van der Eecken, N., De Meerleer, M. L., et al. (2013). Characterisation of Gouda cheeses based on sensory, analytical and high-field 1H nuclear magnetic resonance spectroscopy determinations: effect of adjunct cultures and brine composition on sodium-reduced Gouda cheese. Int. Dairy J. 33, 142–152. doi: 10.1016/j.idairyj.2013.04.009
Salazar, J. K., Carstens, C. K., Ramachandran, P., Shazer, A. G., Narula, S. S., Reed, E., et al. (2018). Metagenomics of pasteurized and unpasteurized Gouda cheese using targeted 16S rDNA sequencing. BMC Microbiol. 18:189. doi: 10.1186/s12866-018-1323-4
Sánchez, N. S., Calahorra, M., González-Hernández, J. C., and Peña, A. (2006). Glycolytic sequence and respiration of Debaryomyces hansenii as compared to Saccharomyces cerevisiae. Yeast 23, 361–374. doi: 10.1002/yea.1360
Satomi, M., Furushita, M., Oikawa, H., and Yano, Y. (2011). Diversity of plasmids encoding histidine decarboxylase gene in Tetragenococcus spp. isolated from Japanese fish sauce. Int. J. Food Microbiol. 148, 60–65. doi: 10.1016/j.ijfoodmicro.2011.04.025
Schleifer, K. H., Kraus, J., Dvorak, C., Kilpper-Bälz, R., Collins, M. D., and Fischer, W. (1985). Transfer of Streptococcus lactis and related streptococci to the genus Lactococcus gen. nov. Syst. Appl. Microbiol. 6, 183–195. doi: 10.1016/S0723-2020(85)80052-7
Smid, E. J., Erkus, O., Spus, M., Wolkers-Rooijackers, J. C. M., Alexeeva, S., and Kleerebezem, M. (2014). Functional implications of the microbial community structure of undefined mesophilic starter cultures. Microb. Cell Factories 13, S2–S9. doi: 10.1186/1475-2859-13-S1-S2
Smit, G., Smit, B. A., and Engels, W. J. M. (2005). Flavour formation by lactic acid bacteria and biochemical flavour profiling of cheese products. FEMS Microbiol. Rev. 29, 591–610. doi: 10.1016/j.fmrre.2005.04.002
Spus, M., Li, M., Alexeeva, S., Wolkers-Rooijackers, J. C. M., Zwietering, M. H., Abee, T., et al. (2015). Strain diversity and phage resistance in complex dairy starter cultures. J. Dairy Sci. 98, 5173–5182. doi: 10.3168/jds.2015-9535
Starrenburg, M., and Hugenholtz, J. (1991). Citrate fermentation by Lactococcus and Leuconostoc spp. Metab. Clin. Exp. 57, 3535–3540. doi: 10.1128/aem.57.12.3535-3540.1991
Steele, J., Broadbent, J., and Kok, J. (2013). Perspectives on the contribution of lactic acid bacteria to cheese flavor development. Curr. Opin. Biotechnol. 24, 135–141. doi: 10.1016/j.copbio.2012.12.001
Stefanovic, E., Kilcawley, K. N., Roces, C., Rea, M. C., O’Sullivan, M., Sheehan, J. J., et al. (2018). Evaluation of the potential of Lactobacillus paracasei adjuncts for flavor compounds development and diversification in short-aged Cheddar cheese. Front. Microbiol. 9:1506. doi: 10.3389/fmicb.2018.01506
Stoddard, S. F., Smith, B. J., Hein, R., Roller, B. R. K., and Schmidt, T. M. (2015). rrnDB: improved tools for interpreting rRNA gene abundance in bacteria and archaea and a new foundation for future development. Nucl. Acids Res. 43, D593–D598. doi: 10.1093/nar/gku1201
Tailliez, P., Tremblay, J., Ehrlich, S. D., and Chopin, A. (1998). Molecular diversity and relationship within Lactococcus lactis, as revealed by randomly amplified polymorphic DNA (RAPD). Syst. Appl. Microbiol. 21, 530–538. doi: 10.1016/S0723-2020(98)80065-9
Toelstede, S., and Hofmann, T. (2008). Quantitative studies and taste re-engineering experiments toward the decoding of the nonvolatile sensometabolome of Gouda cheese. J. Agric. Food Chem. 56, 5299–5307. doi: 10.1021/jf800552n
Udomsil, N., Rodtong, S., Choi, Y. J., Hua, Y., and Yongsawatdigul, J. (2011). Use of Tetragenococcus halophilus as a starter culture for flavor improvement in fish sauce fermentation. J. Agric. Food Chem. 59, 8401–8408. doi: 10.1021/jf201953v
Unno, R., Matsutani, M., Suzuki, T., Kodama, K., Matsushita, H., Yamasato, K., et al. (2020). Lactic acid bacterial diversity in Brie cheese focusing on salt concentration and pH of isolation medium and characterisation of halophilic and alkaliphilic lactic acid bacterial isolates. Int. Dairy J. 109:104757. doi: 10.1016/j.idairyj.2020.104757
Van der Veken, D., Benhachemi, R., Charmpi, C., Ockerman, L., Poortmans, M., Van Reckem, E., et al. (2020). Exploring the ambiguous status of coagulase-negative staphylococci in the biosafety of fermented meats: the case of antibacterial activity versus biogenic amine formation. Microorganisms 8:167. doi: 10.3390/microorganisms8020167
Van der Veken, D., Hollanders, C., Verce, M., Michiels, C., Ballet, S., Weckx, S., et al. (2022). Genome-based characterization of a plasmid-associated micrococcin P1 biosynthetic gene cluster and virulence factors in Mammaliicoccus sciuri IMDO-S72. Appl. Environ. Microbiol. 88:e02088-21. doi: 10.1128/aem.02088-21
Van Hoorde, K., Heyndrickx, M., Vandamme, P., and Huys, G. (2010a). Influence of pasteurization, brining conditions and production environment on the microbiota of artisan Gouda-type cheeses. Food Microbiol. 27, 425–433. doi: 10.1016/j.fm.2009.12.001
Van Hoorde, K., Van Leuven, I., Dirinck, P., Heyndrickx, M., Coudijzer, K., Vandamme, P., et al. (2010b). Selection, application and monitoring of Lactobacillus paracasei strains as adjunct cultures in the production of Gouda-type cheeses. Int. J. Food Microbiol. 144, 226–235. doi: 10.1016/j.ijfoodmicro.2010.05.007
Van Hoorde, K., Verstraete, T., Vandamme, P., and Huys, G. (2008). Diversity of lactic acid bacteria in two Flemish artisan raw milk Gouda-type cheeses. Food Microbiol. 25, 929–935. doi: 10.1016/j.fm.2008.06.006
Van Leuven, I., Van Caelenberg, T., and Dirinck, P. (2008). Aroma characterisation of Gouda-type cheeses. Int. Dairy J. 18, 790–800. doi: 10.1016/j.idairyj.2008.01.001
Vermote, L., Verce, M., De Vuyst, L., and Weckx, S. (2018). Amplicon and shotgun metagenomic sequencing indicates that microbial ecosystems present in cheese brines reflect environmental inoculation during the cheese production process. Int. Dairy J. 87, 44–53. doi: 10.1016/j.idairyj.2018.07.010
Walsh, A. M., Macori, G., Kilcawley, K. N., and Cotter, P. D. (2020). Meta-analysis of cheese microbiomes highlights contributions to multiple aspects of quality. Nat. Food 1, 500–510. doi: 10.1038/s43016-020-0129-3
Welthagen, J. J., and Viljoen, B. C. (1998). Yeast profile in Gouda cheese during processing and ripening. Int. J. Food Microbiol. 41, 185–194. doi: 10.1016/S0168-1605(98)00042-7
Williams, A. G., Choi, S. C., and Banks, J. M. (2002). Variability of the species and strain phenotype composition of the non-starter lactic acid bacterial population of cheddar cheese manufactured in a commercial creamery. Food Res. Int. 35, 483–493. doi: 10.1016/S0963-9969(01)00147-8
Yang, C., Zhao, F., Hou, Q., Wang, J., Li, M., and Sun, Z. (2020). PacBio sequencing reveals bacterial community diversity in cheeses collected from different regions. J. Dairy Sci. 103, 1238–1249. doi: 10.3168/jds.2019-17496
Zago, M., Bardelli, T., Rossetti, L., Nazzicari, N., Carminati, D., Galli, A., et al. (2021). Evaluation of bacterial communities of Grana Padano cheese by DNA metabarcoding and DNA fingerprinting analysis. Food Microbiol. 93:103613. doi: 10.1016/j.fm.2020.103613
Zhang, S. J., De Bruyn, F., Pothakos, V., Torres, J., Falconi, C., Moccand, C., et al. (2019). Following coffee production from cherries to cup: microbiological and metabolomic analysis of wet processing of Coffea arabica. Appl. Environ. Microbiol. 85:e02635-18. doi: 10.1128/AEM.02635-18
Zheng, J., Wittouck, S., Salvetti, E., Franz, C. M., Harris, H. M., Mattarelli, P., et al. (2020). A taxonomic note on the genus Lactobacillus: description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. 70, 2782–2858. doi: 10.1099/ijsem.0.004107
Keywords: starter culture rotation, metabolomics, cheese ripening, Gouda cheese core, Gouda cheese rind, high-throughput full-length 16S rRNA gene sequencing
Citation: Decadt H, Weckx S and De Vuyst L (2023) The rotation of primary starter culture mixtures results in batch-to-batch variations during Gouda cheese production. Front. Microbiol. 14:1128394. doi: 10.3389/fmicb.2023.1128394
Edited by:
Javier Carballo, University of Vigo, SpainReviewed by:
Erasmo Neviani, University of Parma, ItalyFranca Rossi, Experimental Zooprophylactic Institute of Abruzzo and Molise G. Caporale, Italy
Copyright © 2023 Decadt, Weckx and De Vuyst. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Luc De Vuyst, ✉ bHVjLmRlLnZ1eXN0QHZ1Yi5iZQ==
 Hannes Decadt
Hannes Decadt Stefan Weckx
Stefan Weckx Luc De Vuyst
Luc De Vuyst