
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 20 February 2023
Sec. Biology of Archaea
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1126218
This article is part of the Research TopicMolecular Biology of Archaea - 2022View all 19 articles
The hyperthermophilic archaeon Thermococcus kodakarensis utilizes amino acids as a carbon and energy source. Multiple aminotransferases, along with glutamate dehydrogenase, are presumed to be involved in the catabolic conversion of amino acids. T. kodakarensis harbors seven Class I aminotransferase homologs on its genome. Here we examined the biochemical properties and physiological roles of two Class I aminotransferases. The TK0548 protein was produced in Escherichia coli and the TK2268 protein in T. kodakarensis. Purified TK0548 protein preferred Phe, Trp, Tyr, and His, and to a lower extent, Leu, Met and Glu. The TK2268 protein preferred Glu and Asp, with lower activities toward Cys, Leu, Ala, Met and Tyr. Both proteins recognized 2-oxoglutarate as the amino acceptor. The TK0548 protein exhibited the highest kcat/Km value toward Phe, followed by Trp, Tyr, and His. The TK2268 protein exhibited highest kcat/Km values for Glu and Asp. The TK0548 and TK2268 genes were individually disrupted, and both disruption strains displayed a retardation in growth on a minimal amino acid medium, suggesting their involvement in amino acid metabolism. Activities in the cell-free extracts of the disruption strains and the host strain were examined. The results suggested that the TK0548 protein contributes to the conversion of Trp, Tyr and His, and the TK2268 protein to that of Asp and His. Although other aminotransferases seem to contribute to the transamination of Phe, Trp, Tyr, Asp, and Glu, our results suggest that the TK0548 protein is responsible for the majority of aminotransferase activity toward His in T. kodakarensis. The genetic examination carried out in this study provides insight into the contributions of the two aminotransferases toward specific amino acids in vivo, an aspect which had not been thoroughly considered thus far.
Aminotransferases, or transaminases, catalyze the reversible transfer of an amino group from a donor to a keto group of an acceptor compound (Toyokawa et al., 2021; Koper et al., 2022). The reaction is dependent on pyridoxal 5′-phosphate (PLP). Many aminotransferases recognize the α-amino group of α-amino acids and the keto group of 2-oxoacids, but some recognize the amino group on side chains (Koszelewski et al., 2010; Malik et al., 2012; Zheng et al., 2018) or substrates other than α-amino acids (Gomm and O'Reilly, 2018; Kelly et al., 2020). The enzymes play a pivotal role in the carbon and nitrogen metabolism in a wide range of organisms. Aminotransferases are a focus of attention due to their involvement in a variety of diseases (Kunutsor et al., 2013; Montioli et al., 2021) and are also attractive enzymes for their use in biocatalysis (Malik et al., 2012; Gomm and O'Reilly, 2018; Kelly et al., 2020).
Aminotransferases can be divided into four classes (Class I to IV) based on their primary structure (Mehta et al., 1993). On the other hand, PLP-dependent enzymes can be classified by their folds (Fold type I to VII; Jansonius, 1998; Schneider et al., 2000; Koper et al., 2022). Aminotransferase Class I, II, and IV share a common ancestor with a Fold type I whereas Class III aminotransferases are members of Fold type IV PLP enzymes (Mehta et al., 1993). Class I enzymes constitute the majority of aminotransferases, including aromatic aminotransferases, aspartate aminotransferases, and alanine aminotransferases. Class II includes enzymes that can recognize amino groups other than the α-amino group of amino acids and utilize substrates such as γ-aminobutyric acid, or GABA, and ornithine. Class III enzymes with the distinct fold type IV utilize D-amino acids and branched-chain amino acids. Class IV enzymes utilize Ser, O-phosphoserine or Asp as the amino donor (Mehta et al., 1993; Mehta and Christen, 2000; Koper et al., 2022).
Thermococcales consists of three genera, Pyrococcus, Thermococcus, and Palaeococcus, and members readily utilize amino acids and peptides as a carbon and energy source for growth (Zillig et al., 1983; Fiala and Stetter, 1986; Takai et al., 2000). Different enzymes and pathways involved in amino acid catabolism and biosynthesis have been studied in these organisms. In particular, the enzymatic properties of a wide range of aminotransferases from Pyrococcus furiosus (Andreotti et al., 1995; Ward et al., 2000, 2002), Pyrococcus horikoshii (Matsui et al., 2000; Ura et al., 2001; Okada et al., 2012, 2014), Thermococcus litoralis (Andreotti et al., 1994; Sakuraba et al., 2004, 2008), Thermococcus sp. CKU-1 (Uchida et al., 2014) and Thermococcus kodakarensis (Kanai et al., 2015; Zheng et al., 2018) have been reported. Our group has been examining amino acid metabolism in T. kodakarensis (Shikata et al., 2007; Yokooji et al., 2013; Awano et al., 2014). In addition to the genome sequence (Fukui et al., 2005), a versatile genetic system (Sato et al., 2003, 2005; Matsumi et al., 2007; Santangelo et al., 2008, 2010) allows us to evaluate the physiological roles of individual genes in this organism.
A biochemical and genetic examination of enzymes/genes involved in Glu metabolism in T. kodakarensis confirmed the presence of glutamate dehydrogenase (TK1431), catalyzing an NADP-dependent interconversion between Glu and 2-oxoglutarate (Yokooji et al., 2013). The functions of 2-oxoglutarate:ferredoxin oxidoreductase (TK1123-TK1125, TK1131) and ADP-forming succinyl-CoA synthetase (TK1880, TK0943; Shikata et al., 2007) were also identified, constituting a route from Glu to succinate. The latter two reactions lead to the generation of reduced ferredoxin and ATP coupled to 2-oxoacid degradation, providing reducing equivalents and chemical energy (Yokooji et al., 2013). The presence of four 2-oxoacid:ferredoxin oxidoreductases and five ADP-forming acyl-CoA synthetases in T. kodakarensis suggest that other amino acids are catabolized through similar reactions. Based on the substrate specificities of the acyl-CoA synthetases (Mai and Adams, 1996; Glasemacher et al., 1997; Musfeldt et al., 1999; Shikata et al., 2007; Awano et al., 2014), it was suggested that the following amino acids are subject to catabolism; Ala, Val, Leu, Ile, Met, Phe, Tyr, Trp, Glu/Gln, Cys, and His (Awano et al., 2014). However, dehydrogenases acting on amino acids other than Glu such as aspartate dehydrogenase and alanine dehydrogenase have been genetically confirmed to be absent in T. kodakarensis (Yokooji et al., 2013). This implies that 2-oxoacid generation from amino acids relies on aminotransferases. In order to better understand which amino acids are subject to catabolic degradation and which aminotransferases are involved, here we carried out a biochemical and genetic examination on two aminotransferases in T. kodakarensis encoded by TK0548 and TK2268.
Figure 1 shows a phylogenetic tree of Class I aminotransferase homologs from four selected Thermococcales species, T. kodakarensis, P. furiosus, P. horikoshii, and T. litoralis, from which multiple aminotransferases have been experimentally studied. A clade including TK1990, a representative of a Class IV protein encoding cysteine desulfurase (Hidese et al., 2014), was used as the outgroup. There are eight groups (G1–G8) in which homologs occur in ten or more genomes of the 40 Thermococcales species examined. Among the eight groups, members of G1 (39 genomes among the 40 genomes examined harbor homologs; 39/40), G2 (40/40), G3 (40/40), G6 (40/40), and G8 (35/40) are present in most or all of the Thermococcales genomes used in our analysis. Members of G4 (12/40), G5 (10/40), and G7 (20/40) are less distributed. T. litoralis harbors a higher number of Class I aminotransferase homologs compared to the other three species. Homologs of OCC_03517 are not found on any of the other Thermococcales genomes, but homologs of OCC_10965 (3/40), OCC_02240 (10/40, G5), and OCC_11814 (5/40) can be identified in other species. A detailed phylogenetic tree based on sequences of all Class I aminotransferase homologs from Thermococcales species is shown in Supplementary Figure S1.
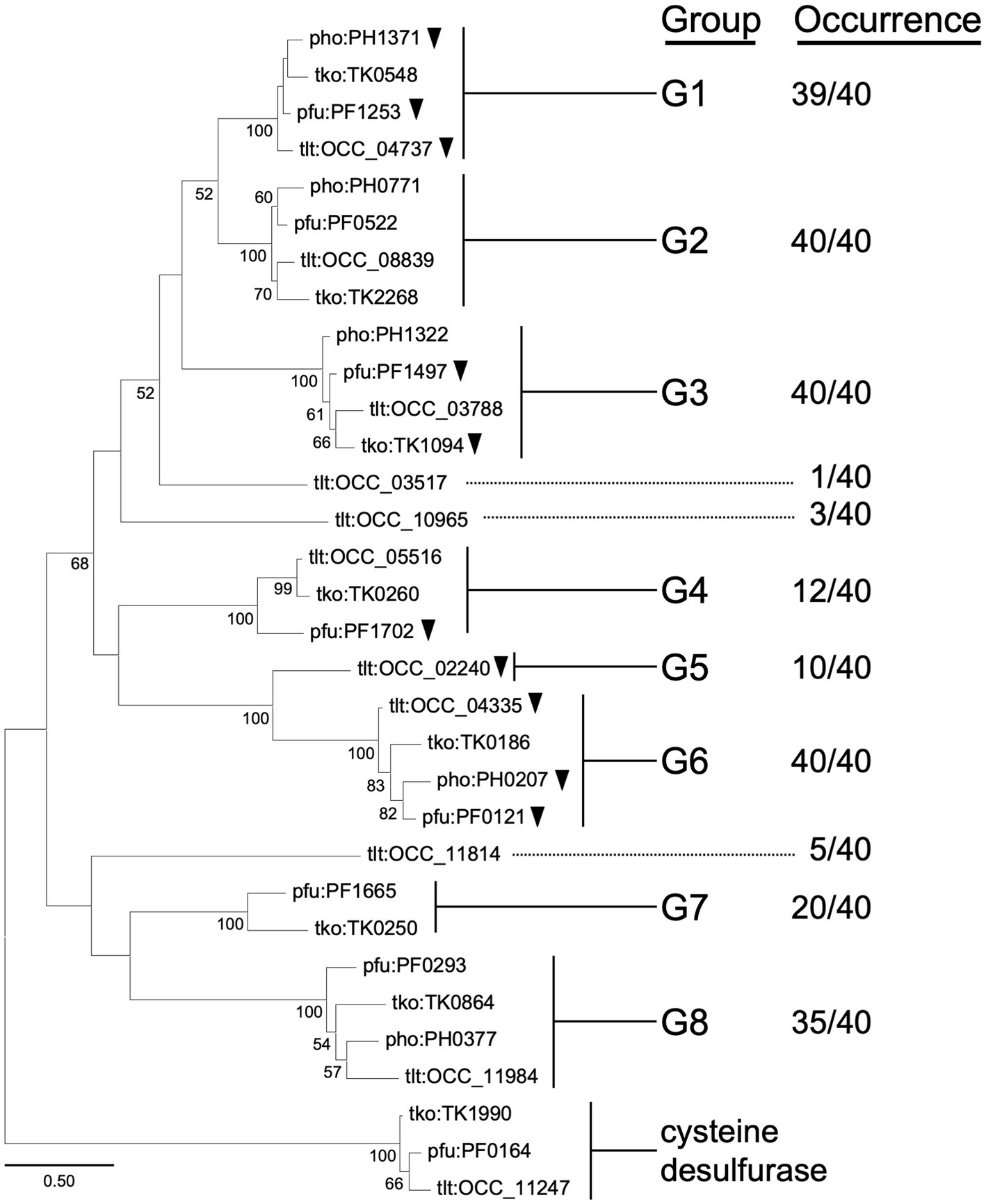
Figure 1. Phylogenetic analysis of Class I aminotransferases from selected species of Thermococcales. Amino acid sequences were collected from T. kodakarensis, T. litoralis, P. furiosus, and P. horikoshii. Homologs of cysteine desulfurase (TK1990) were added into the dataset as an outgroup. The sequences were aligned using MUSCLE algorithm (Edgar, 2004). The phylogenetic analysis was performed using the Maximum Likelihood method and JTT matrix-based model (Jones et al., 1992). The tree with the highest log likelihood (−9712.68) is shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Bootstrap values above 50 are shown. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the JTT model, then selecting the topology with superior log likelihood value. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. This analysis involved 32 amino acid sequences. All positions containing gaps and missing data were eliminated (complete deletion option). There were a total of 242 positions in the final dataset. Evolutionary analyses were conducted in MEGA11 (Tamura et al., 2021). Homologs that have been characterized genetically or biochemically are indicated with black arrowheads. Information on the groups or occurrence is indicated in the text.
We considered the physiological roles of these groups of aminotransferases by first taking into account their gene location and previous biochemical studies on aminotransferases from members of Thermococcales. In terms of gene location, we observed that TK0250 (G7) is included within the His biosynthesis gene cluster (TK0242-TK0251) in T. kodakarensis. We also observed a complete co-occurrence between the TK0250 homologs and the His biosynthesis operon in Thermococcales members (Supplementary Table S1). A similar situation was observed for TK0260 (G4). The gene is situated in a gene cluster (TK0259-TK0261) involved in the biosynthesis of Phe/Tyr in T. kodakarensis, and co-occurrence is observed between TK0260 homologs and the gene cluster in Thermococcales species (Supplementary Table S2). These observations suggest that the physiological roles of members of G4 and G7 are related to Phe/Tyr and His biosynthesis, respectively. The members of G8 from Thermococcales have not been characterized, but display 30% identity to Thr decarboxylase (encoded by MM2060) from Methanosarcina mazei (Tavares et al., 2018, 2019) and are clustered with genes related to cobalamin salvage. By contrast, members of G1, G2, G3, G5, and G6 did not show a tendency to be included in a particular biosynthesis operon or gene cluster.
Members of G1, G2, G3, and G6 are widely distributed among Thermococcales species, and a number of members have been biochemically characterized. In particular, members of G1 and G6 have been relatively well studied. In the case of G1, the structure of the PH1371 protein from P. horikoshii has been elucidated, and the amino donors most recognized were Tyr, Phe, Glu, His, and Trp (Matsui et al., 2000). The PF1253 protein from P. furiosus, referred to as AroAT II, displayed preference to Phe, Tyr and Trp, with highest kcat/Km toward Phe (Ward et al., 2002). The OCC_04737 protein from T. litoralis, referred to as ArAT II, also displayed similar properties, preferring Phe, Tyr, and Trp (Andreotti et al., 1994). In the case of G6, the PF0121 protein from P. furiosus, referred to as ArAT or AroAT I, displayed significant activity toward Phe, Trp, and Tyr (Andreotti et al., 1995). The OCC_04335 protein from T. litoralis, referred to as ArAT I, also recognized Phe, Tyr, and Trp (Andreotti et al., 1994). The PH0207 protein displays kynurenine aminotransferase activity which is involved in the degradation of Trp (Okada et al., 2012), and the crystal structure has been determined (Chon et al., 2005; Okada et al., 2014). Concerning other groups, the PF1497 protein from P. furiosus (G3), referred to as AlaAT, displays highest activity with Ala. The enzyme did not display activity toward Phe and Tyr (Ward et al., 2000). A genetic analysis on TK1094 from T. kodakarensis (G3) suggested a role in the conversion between pyruvate and Ala (Kanai et al., 2015). The PF1702 protein (G4), referred to as AspAT, displays highest activity toward Asp and Glu, with low activities to a broad range of amino acids (Ward et al., 2002).
We carried out further analyses on TK2268 and TK0548. The TK2268 protein is a member of G2, which does not have any member that has been experimentally characterized. Determining its biochemical properties would contribute to our understanding of amino acid metabolism in T. kodakarensis. We also examined the TK0548 protein, the most closely related protein to the TK2268 protein (45.4% identical). None of the G1 and G2 genes have been examined genetically.
In order to obtain recombinant proteins, the TK0548 and TK2268 genes were expressed in Escherichia coli. In the case of TK0548, soluble protein was obtained, and the recombinant TK0548 protein was purified through heat treatment, anion exchange chromatography and gel-filtration chromatography. In the case of TK2268, expression in E. coli resulted in the formation of inclusion bodies. The gene was thus expressed in the native host T. kodakarensis under the control of a strong constitutive promoter of the cell surface glycoprotein gene (csg, TK0895; Yokooji et al., 2009). Sequences to incorporate a His6-tag on the C-terminus of the TK2268 protein were introduced. The soluble TK2268 protein obtained in the cell extract of T. kodakarensis cells was purified using a nickel affinity column and gel filtration chromatography. Both proteins were subjected to SDS-PAGE, confirming their apparent homogeneities (Figure 2).
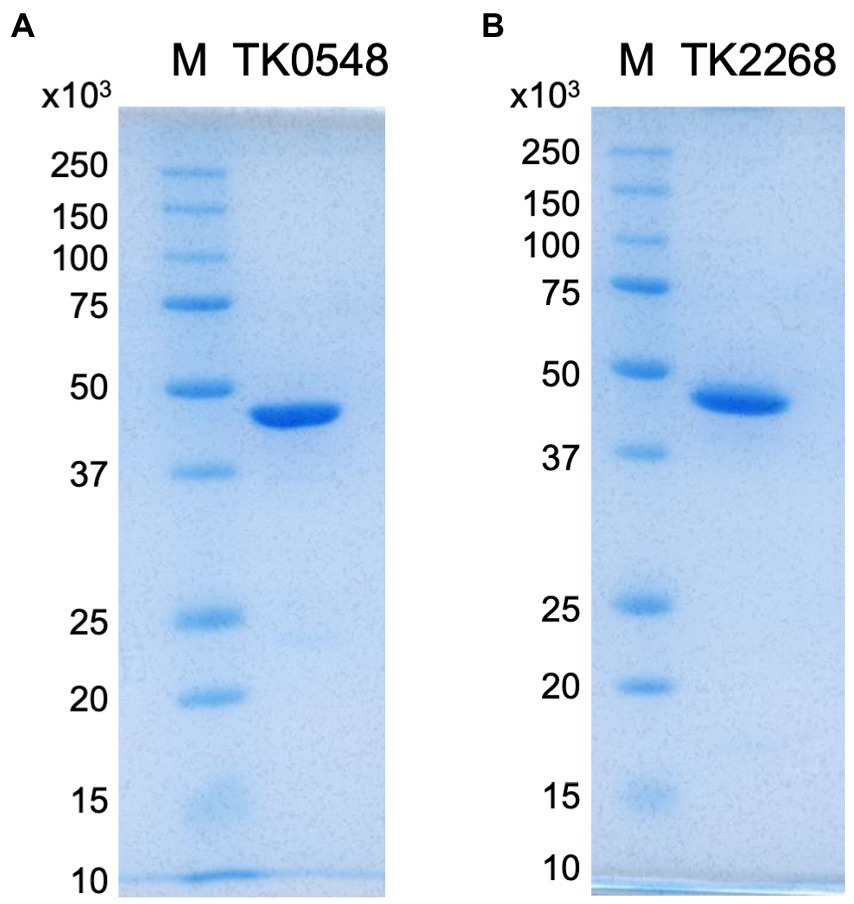
Figure 2. SDS-PAGE analyses of the purified TK0548 (A) and TK2268 (B) recombinant proteins. Two micrograms of purified TK0548 and TK2268 recombinant proteins were applied to each gel. Gels were stained with Coomassie Brilliant Blue. M indicates molecular weight markers.
The molecular masses of the purified, recombinant TK0548 and TK2268 proteins were examined with gel-filtration chromatography. The estimated molecular mass of the TK0548 protein was 85 kDa, and considering that the calculated molecular mass of the monomer was 43,608 Da, this suggested that the TK0548 protein was a dimer. In the case of the TK2268 protein, we consistently observed two peaks, corresponding to estimated molecular masses of 98 kDa and 327 kDa. As the calculated molecular mass of the monomer was 45,116 Da, the result suggested that the TK2268 protein forms a dimer unit, which may then further assemble to form an octamer.
The purified TK0548 and TK2268 proteins were examined for aminotransferase activity. We used Glu (10 mM) as the amino group donor and pyruvate (10 mM) as the amino acceptor. We observed aminotransferase activity in both proteins. When PLP was omitted from the reaction, we observed a partial decrease in activity in both cases (TK0548: 13% decrease, TK2268: 50% decrease) compared to those observed with the addition of PLP, suggesting that the TK0548 and TK2268 proteins are PLP-dependent aminotransferases. The only partial decrease in activity is most likely due to PLP bound to the proteins when they were produced in their respective host cells, E. coli (TK0548) and T. kodakarensis (TK2268). To further support the necessity of PLP for the reactions, we added hydroxylamine, a PLP inhibitor (Kito et al., 1978) to the enzymes in the absence of supplemental PLP. In this case, we observed a further 93% decrease in activity of the TK0548 protein and a 78% decrease in activity of the TK2268 protein with the addition of hydroxylamine (Supplementary Figure S2).
We first carried out an initial screening to identify the amino acids recognized by the TK0548 and TK2268 proteins. 2-Oxoglutarate or pyruvate was used as the amino acceptor, and the production of Glu or Ala, respectively, was examined after 15 min. The substrates were present in the reaction mixture at a concentration of 10 mM. As shown in Figure 3A, the TK0548 protein utilized Phe, Tyr, His, and Trp as an amino donor, as well as Met, Glu, and Leu to a lower extent. In the case of the TK2268 protein, Asp and Glu were utilized, along with Tyr, Leu, Ala, Met, and Cys (Figure 3B).
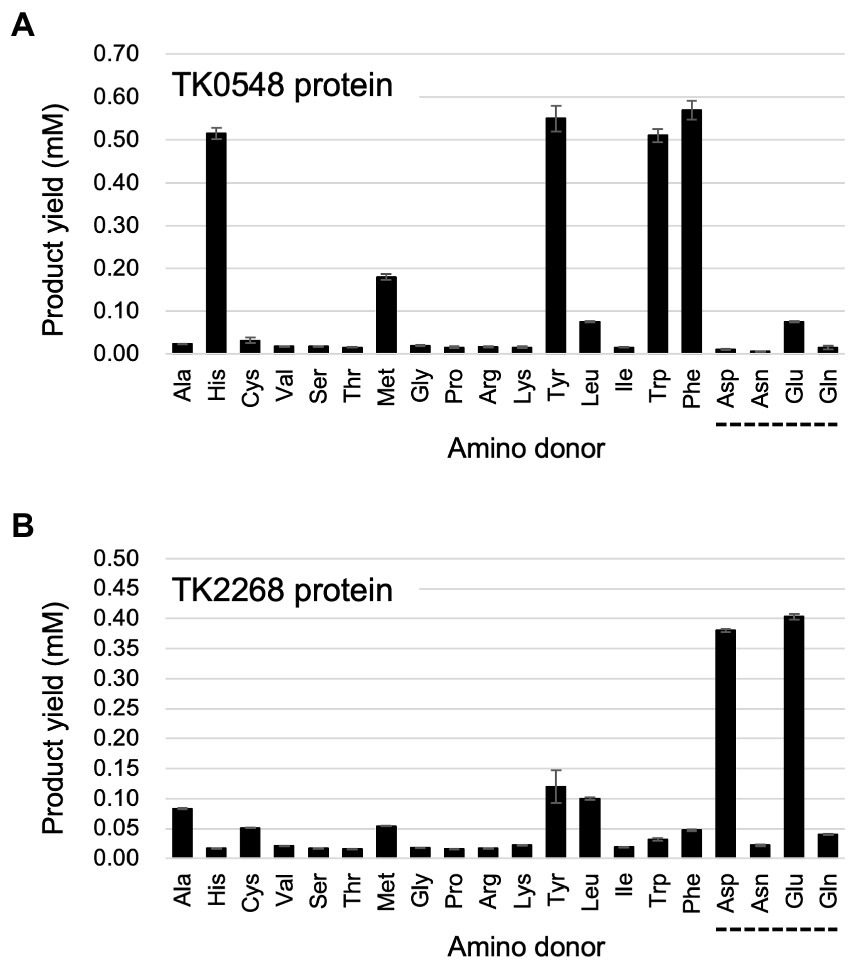
Figure 3. Examination of the amino donors recognized by the recombinant TK0548 (A) and TK2268 (B) proteins. Each protein was incubated for 15 min at 80°C with the indicated amino acids and 2-oxoglutarate and the generation of Glu was examined. The amino acids indicated with a dotted line below them were examined using pyruvate as the amino acceptor, and the generation of Ala was examined. Amino donors and acceptors were added at a concentration of 10 mM, with the exception of Tyr (6 mM). The results are the means of three independent assays and error bars indicate standard deviations.
Focusing on the amino acids that were recognized by the proteins, we examined enzyme activity. The amino acids and amino acceptors were constant at 10 mM. As shown in Figure 4A, the TK0548 protein exhibited highest activity toward Tyr, followed by Phe, Trp, and His. Activity toward Leu, Met, and Glu were much lower. On the other hand, the TK2268 protein displayed highest activity toward Glu, followed by Asp. Lower levels of activity were observed using Cys, Leu, Ala, Met, and Tyr (Figure 4B).
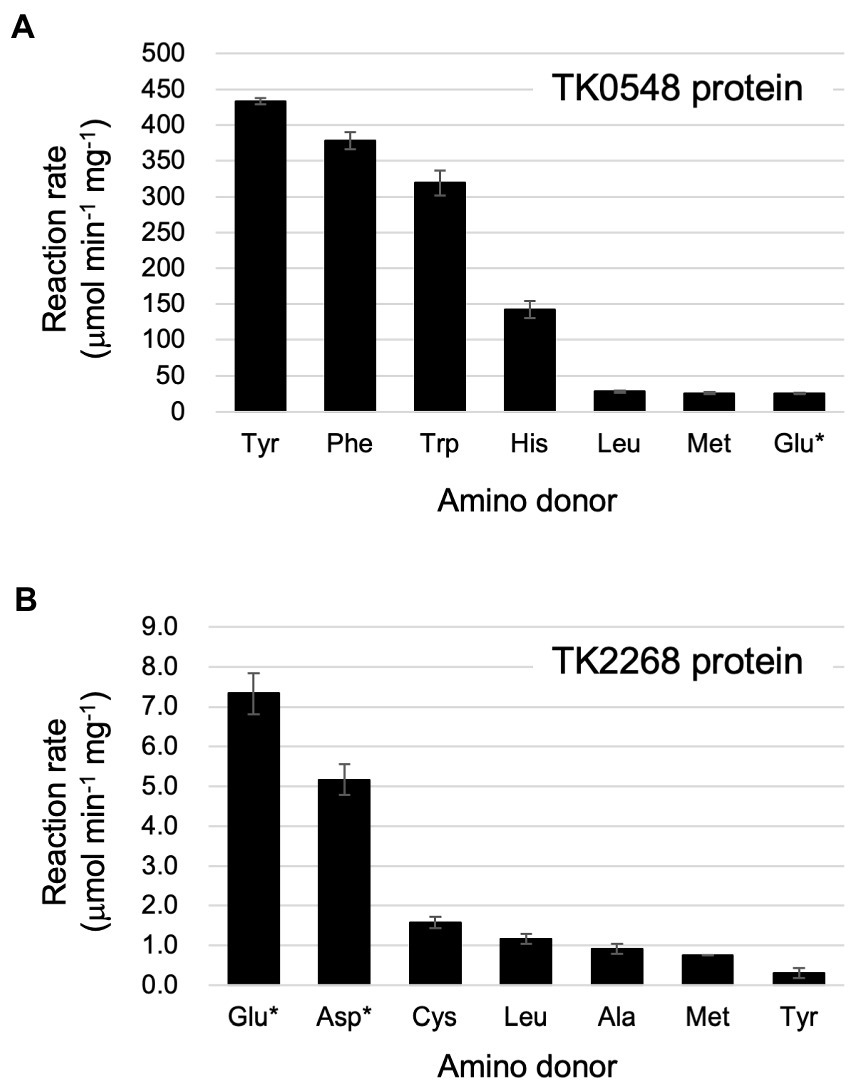
Figure 4. Reaction rates of the recombinant TK0548 (A) and TK2268 (B) proteins with various amino donors. Each protein was incubated for varying periods of time at 80°C with the indicated amino acids and 2-oxoglutarate or pyruvate (indicated with asterisks), and the reaction rates of Glu or Ala production, respectively, were calculated. Amino donors and acceptors were added at a concentration of 10 mM (Tyr: 6 mM). The results are the means of three independent assays and error bars indicate standard deviations.
Kinetic analyses were performed on the substrates that resulted in relatively high levels of activity. For the TK0548 protein, activities were first measured for varying concentrations of pyruvate or 2-oxoglutarate in the presence of 10 mM Phe. As shown in Table 1, the TK0548 protein exhibited a higher Vmax and lower Km toward 2-oxoglutarate when compared to pyruvate. The kcat/Km value toward 2-oxoglutarate was over 70-fold higher than that for pyruvate. We next examined activity with varying concentrations of Phe, Tyr, Trp, His, and Met in the presence of 10 mM 2-oxoglutarate (Table 1). The highest kcat/Km value was observed with Phe, with high values also observed for Tyr and Trp. A lower kcat/Km value was observed with His, and that for Met was extremely low. The results suggested that the TK0548 protein mainly utilizes the aromatic amino acids Phe, Tyr, and Trp as substrates, with His also a possible substrate.
Concerning the TK2268 protein, Leu (50 mM) was used as the amino donor to measure activity with varying concentrations of pyruvate and 2-oxoglutarate. The kcat/Km value toward 2-oxoglutarate was higher than that toward pyruvate (Table 2). This was mainly due to differences in the Km value, as their Vmax values were comparable. As the product of the aminotransferase reaction with 2-oxoglutarate would not be possible with Glu as the amino donor, 10 mM pyruvate was used as the amino acceptor to examine the activities with varying concentrations of Glu, Asp, and Leu. As a result, relatively high kcat/Km values were observed for both Asp and Glu compared to Leu and Tyr. The results suggest that the TK2268 protein is an aminotransferase with specificity toward the acidic amino acids Glu and Asp.
To understand the contribution of the TK0548 and TK2268 genes to amino acid catabolism in T. kodakarensis, we disrupted each gene using T. kodakarensis KU216 (ΔpyrF) as a host strain. Five transformants were chosen for each gene disruption and their loci were examined by PCR. Examples of transformants whose TK0548 gene (Figure 5A) or TK2268 gene (Figure 5B) was disrupted are shown. The respective loci were sequenced, confirming that gene disruption had occurred as intended.
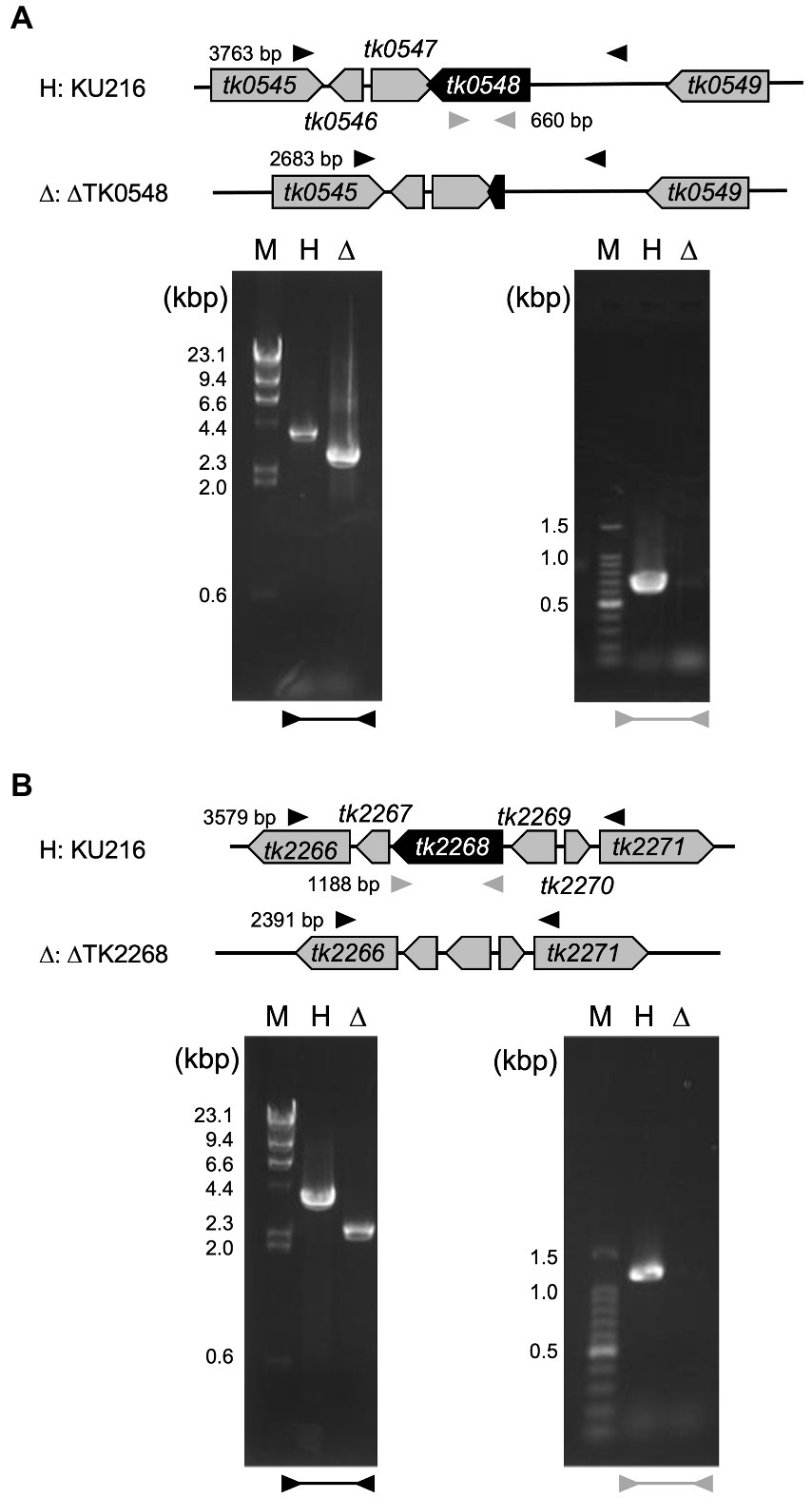
Figure 5. Confirmation of TK0548 (A) and TK2268 (B) gene disruption by PCR. Predicted gene loci before and after gene disruption are shown above the gels. Arrowheads indicate the position of primers outside the homologous regions used for recombination (black) and inside the coding regions (gray). Amplified fragments were subjected to agarose gel electrophoresis along with DNA markers (M). H, host strain KU216; Δ, gene disruption strains.
The ΔTK0548 and ΔTK2268 disruption strains were first grown in the nutrient-rich ASW-YT-m1-S0 medium containing yeast extract and tryptone (Figure 6A). The growth of both disruption strains did not display significant differences to that of the host strain KU216. Therefore, in order to increase the dependency of growth on amino acid catabolism, the strains were grown in ASW-AA-m1-S0(+Ura) medium (Figure 6B). This medium is a synthetic medium with amino acids as the only major carbon and energy source. In this case, we observed a retardation in growth in both gene disruption strains. Cell yields of the cultures eventually reached similar levels, but the gene disruption strains took a 4-h longer period of time to reach maximum cell density. The results suggest that both proteins are involved in the utilization of amino acids for growth of T. kodakarensis.
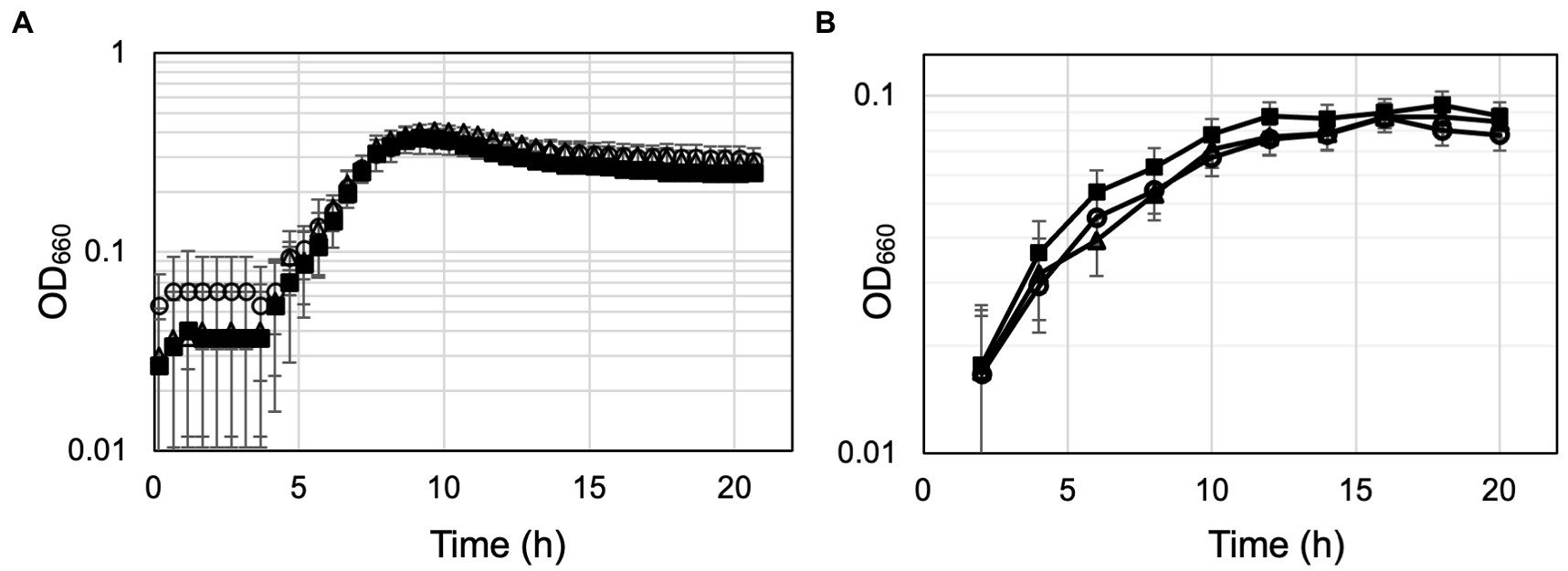
Figure 6. Growth properties of the host strain KU216 and the gene disruption strains. Growth of the host KU216 strain (closed squares) and the ΔTK0548 (open triangles) and ΔTK2248 (open circles) gene disruption strains were examined in a nutrient-rich ASW-YT-m1-S0 medium containing yeast extract and tryptone (A), and in a synthetic amino acid medium ASW-AA-m1-S0(+Ura) (B). Growth was measured at 85°C. Error bars indicate the standard deviations of three independent culture experiments. The vertical axis is represented in logarithmic scale.
The substrate specificities of multiple aminotransferases from members of Thermococcales have been determined in vitro. However, the contribution of each protein in the conversion of a particular amino acid in vivo cannot be determined by biochemical properties alone, as multiple aminotransferases, in some cases with overlapping substrate specificities or different expression levels, are present in the cell. We thus measured and compared aminotransferase activity in the cell extracts of T. kodakarensis host strain and gene disruption strains. As in vitro studies indicated that the TK0548 protein preferred Trp, Phe, Tyr, and His, while the TK2268 protein recognized Asp and Glu, activities in the cell extracts toward Trp, Phe, Tyr, His, Asp, and Glu were measured. We also used Ile as an amino donor. In vitro studies indicated that the recognition of both TK0548 and TK2268 proteins toward Ile was minimal (Figure 3), so the effects of TK0548 and TK2268 disruption on intracellular aminotransferase activity toward Ile would be expected to be low. As shown in Figure 7, aminotransferase activity for all seven amino acids was clearly observed in KU216 cell extracts. We observed that the levels of aminotransferase activity toward different amino acids greatly differed. Activities toward Phe or Glu were particularly high, whereas that toward Asp was notably low, two orders of magnitude lower than those observed for Phe or Glu. When we examined the effects of gene disruption, the disruption of TK0548 and TK2268 had no effect on intracellular aminotransferase activity toward Ile, consistent with our in vitro results that neither enzyme recognized Ile (Figure 3). We next examined differences observed upon gene disruption, focusing on those with p values below 0.01. We observed that the disruption of TK2268 resulted in a 35% decrease in aminotransferase activity toward Asp, one of the substrates preferred by the protein in in vitro experiments. Disruption of TK2268 did not affect the Glu aminotransferase activity in cell extracts, suggesting that the contribution of the protein to this activity is relatively small. This is not surprising though, as in vitro analysis indicated that the activity of the TK2268 protein toward Glu and Asp are comparable (Table 2). As the total Glu aminotransferase activity in T. kodakarensis cell extracts is over 100-fold higher than that toward Asp, a decrease in Glu aminotransferase activity comparable to the levels toward Asp would correspond to only a very small fraction of the total activity toward Glu. We also observed a decrease in His aminotransferase activity, but this could not be explained by the results of in vitro analysis. When TK0548 was disrupted, decreases in activity toward Trp (32%), Tyr (64%), and His (89%) were observed. The TK0548 protein seems to be a major contributor for Tyr and His aminotransferase activity in T. kodakarensis.
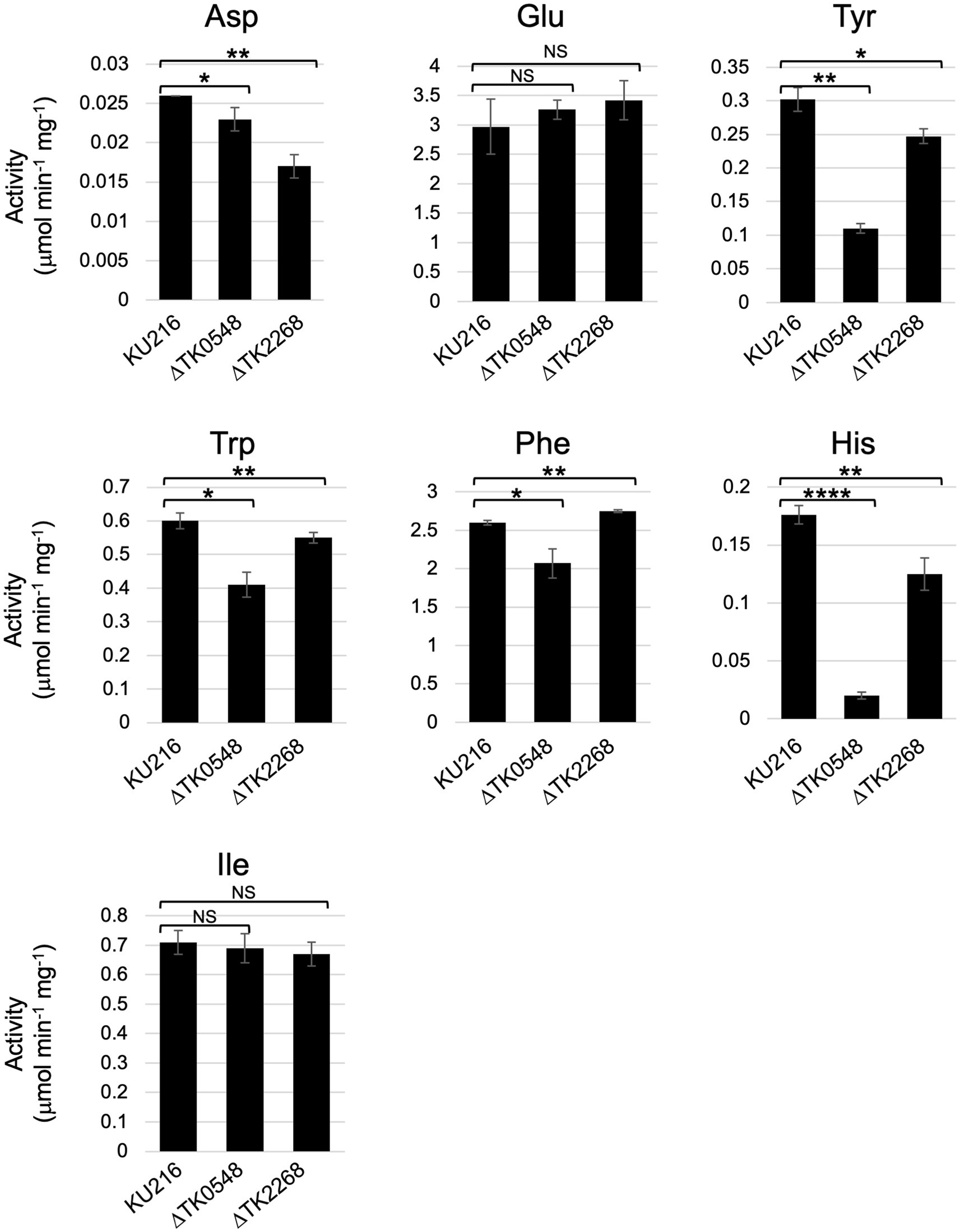
Figure 7. Aminotransferase activity in T. kodakarensis cell extracts. Cell extracts were incubated with different amino acids and 2-oxoglutarate, or pyruvate (for Glu and Asp), at 80°C for various periods of time. Activity was calculated based on the generation of Glu or Ala. Amino donors and acceptors were added at a concentration of 10 mM (Tyr: 6 mM). 0.01 < p < 0.05*; 0.001 < p < 0.01**; 0.0001 < p < 0.001***; p < 0.0001****; not significant: NS.
Biochemical studies have been carried out on wealth of aminotransferases from Thermococcales species. Proteins from T. kodakarensis, P. furiosus, P. horikoshii, and T. litoralis that are classified in the Class I to Class IV aminotransferases are listed in Table 3. Some proteins have been shown to catalyze reactions other than transamination, and include racemases, decarboxylases, desulfurases, hydroxymethyltransferases and others. Concerning the aminotransferases that have been biochemically examined, the amino donor and acceptor molecules are indicated. The TK0548 protein was shown here to prefer the aromatic amino acids Phe, Tyr and Trp, which is consistent with other members of G1. To a lower degree, the protein also recognized His. The PH1371 protein also displays activity toward His (Matsui et al., 2000). The activities of the PF1253 (Ward et al., 2002) and OCC_04737 (Andreotti et al., 1994) proteins toward His have not been examined. In the case of the TK2268 protein, it is the first characterized representative of G2, and displays specificity toward the acidic amino acids Asp and Glu. Although the range of substrates examined in separate studies differ, enzymes that belong to the same group have been shown to display similar substrate specificities, such as the four enzymes from G1 and the three enzymes from G6. Our results with the TK2268 protein thus raise the possibilities that members of G2 recognize acidic amino acids as amino donors.
In order to understand the contribution of each enzyme to the transamination of a specific amino acid in vivo, we examined and compared specific aminotransferase activities among cells of the host strain KU216 and ΔTK0548 and ΔTK2268 disruption strains. This takes into account not only the substrate specificity and activity levels of each enzyme, but also their expression levels in the cell. The results suggested that TK0548 contributed to the aminotransferase activity toward Phe and Trp, and to a higher degree Tyr. The considerable levels of activity still observed in the disruption strains most likely reflects the activity of the TK0186 protein, a member of G6 (Table 3). Interestingly, the TK0548 protein seems to be the predominant His aminotransferase in T. kodakarensis, accounting for approximately 90% of the activity in cell extracts.
Amino acid catabolism in members of the Thermococcales proceeds via amino acid, 2-oxoacid, acyl-CoA and acid (Figure 8). Concerning the conversion from amino acids to 2-oxoacids, our group has previously shown that Glu is the only amino acid that is converted by a dehydrogenase, a glutamate dehydrogenase encoded by TK1431. All other amino acids that are converted to 2-oxoacids would have to rely on aminotransferases (Yokooji et al., 2013). The 2-oxoacids are converted to acyl-CoAs via 2-oxoacid:ferredoxin oxidoreductases. There are seven potential sets of genes encoding 2-oxoacid:ferredoxin oxidoreductases in members of the Thermococcales, and four protein complexes from P. furiosus have been biochemically examined; pyruvate:ferredoxin oxidoreductase (POR), 2-ketoisovalerate:ferredoxin oxidoreductase (VOR), indolepyruvate:ferredoxin oxidoreductase (IOR), and 2-ketoglutarate:ferredoxin oxidoreductase (KGOR) (Blamey and Adams, 1993; Mai and Adams, 1994; Heider et al., 1996; Mai and Adams, 1996). The KGOR homolog in T. kodakarensis has been genetically examined, confirming its role in Glu metabolism, converting 2-oxoglutarate to succinyl-CoA (Yokooji et al., 2013). Finally, the acyl-CoAs are hydrolyzed by NDP-forming acyl-CoA synthetases. There are five NDP-forming acyl-CoA synthetases in Thermococcales; ACS I, ACS II, ACS III, succinyl-CoA synthetase (SCS), and 2-(imidazol-4-yl)acetyl-CoA synthetase (ICS). ACS I and ACS II from P. furiosus and ACS II, ACS III, SCS and ICS from T. kodakarensis have been biochemically examined (Mai and Adams, 1996; Glasemacher et al., 1997; Musfeldt et al., 1999; Shikata et al., 2007; Awano et al., 2014). The involvement of SCS in Glu metabolism has been genetically confirmed (Yokooji et al., 2013). As the NDP-forming acyl-CoA synthetases catalyze the reaction in which substrate-level phosphorylation occurs, it is possible to predict the amino acids that are subject to this catabolism based on the substrate specificities of the acyl-CoA synthetases. The substrates recognized by the five acyl-CoA synthetases combined suggest that the amino acids that are subject to this mode of catabolism are Ala, Val, Ile, Leu, Met, Phe, Tyr, Trp, Glu/Gln, Cys, and His (Awano et al., 2014). Aminotransferases that recognize Ala, Val, Ile, Leu, Met, Phe, Tyr, Trp, and Glu/Gln have been reported, as well as 2-oxoacid:ferredoxin oxidoreductases that act on the corresponding 2-oxoacids after transamination. However, aminotransferases and 2-oxoacid:ferredoxin oxidoreductases that are related to Cys and His degradation are still not known. In particular, ICS displays a remarkably high specificity toward 2-(imidazol-4-yl)-acetate, suggesting that it is involved solely in His metabolism (Awano et al., 2014). The results of this study strongly suggest that the TK0548 protein and ICS are metabolically linked in His catabolism (Figure 8). To a certain extent, the TK0548 protein also contributes in the catabolism of aromatic amino acids, particularly Tyr. Finally, the specificities of the five acyl-CoA synthetases combined raise the possibilities that Asp, Asn, Gly, Lys, Arg, Pro, Ser, and Thr may not be directed to the catabolic degradation involving 2-oxoacid:ferredoxin oxidoreductases and NDP-forming acyl-CoA synthetases, and this may be related to the notably low Asp aminotransferase activity observed in T. kodakarensis cell extracts and the TK2268 protein. As we have previously shown that T. kodakarensis does not harbor an Asp dehydrogenase, our present knowledge suggests that Asp is not catabolized through the pathway involving 2-oxoacid:ferredoxin oxidoreductases and NDP-forming acyl-CoA synthetases. We also could not find an aspartate ammonia lyase homolog on the genome, which would lead to the generation of fumarate. In addition, T. kodakarensis possesses a fumarase homolog, but lacks a number of homologs of the citric acid cycle including succinate dehydrogenase, malate dehydrogenase and citrate synthase. Other than the TK2268 protein, enzymes potentially related to oxaloacetate metabolism are a putative fumarase, malic enzyme (Fukuda et al., 2005), and phosphoenolpyruvate carboxykinase (Fukuda et al., 2004). T. kodakarensis may display a C4-compound metabolism distinct to those found in bacteria. Further understanding of oxaloacetate metabolism may provide valuable clues to elucidate the metabolism of Asp and the physiological function of TK2268. We would like to note, however, that we cannot rule out the possibility that the TK2268 protein recognizes a substrate completely different from those considered in this study.
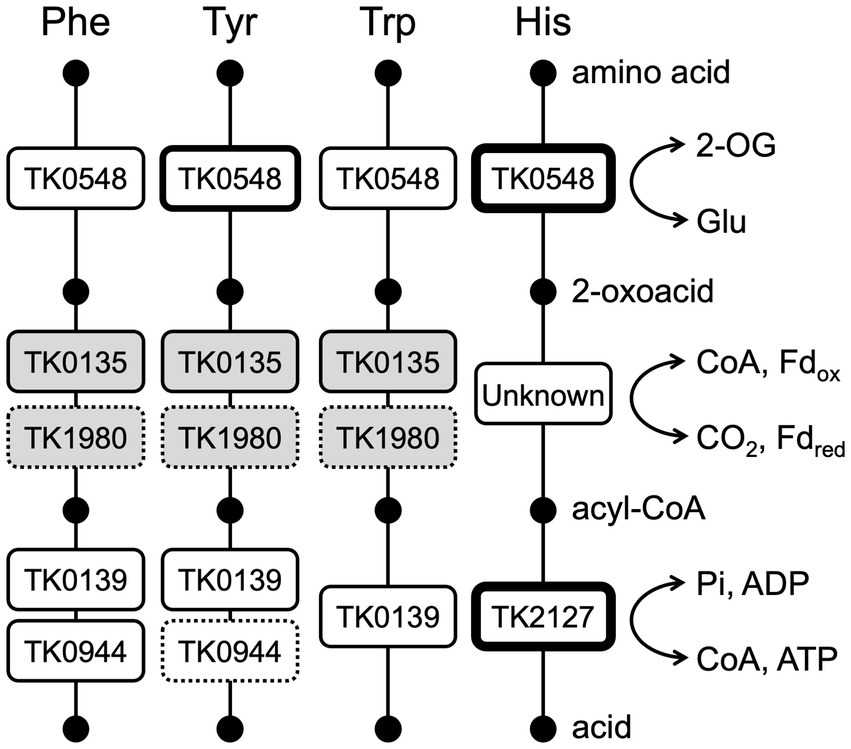
Figure 8. A diagram illustrating the involvement of aminotransferases, 2-oxoacid:ferredoxin oxidoreductases, and acyl-CoA synthetases in the catabolism of Phe, Tyr, Trp, and His in T. kodakarensis. Enzymes with greater roles in each step are indicated with boxes with thicker lines, and those with minor roles are indicated by dotted lines. Gene numbers of the acyl-CoA synthetases indicate those of the α subunit. TK0135 represents TK0135-0136, TK1980 represents TK1978-1981. The genes in white boxes are based on biochemical and genetic evidence and the shaded boxes are predicted based on similarity to the experimentally examined enzymes from P. furiosus. 2-OG, 2-oxoglutarate; CoA, coenzyme A; Fd, ferredoxin; Pi, phosphate.
Thermoccocus kodakarensis was isolated from Kodakara Island, Kagoshima, Japan (Morikawa et al., 1994; Atomi et al., 2004). T. kodakarensis KU216 (Sato et al., 2003, 2005) and derivative strains were cultivated under strictly anaerobic conditions at 85°C in nutrient-rich medium (ASW-YT-m1-S0 or ASW-YT-m1-pyruvate) or synthetic medium (ASW-AA-m1-S0). ASW-YT-m1-S0, ASW-YT-m1-pyruvate, and ASW-AA-m1-S0 are modified versions of ASW-YT-S0, ASW-YT-pyruvate, and ASW-AA-S0 media, respectively. ASW-YT-S0 was composed of 0.8 × artificial seawater (ASW) (Robb and Place, 1995), 5 g L−1 yeast extract, 5 g L−1 tryptone, and 2 g L−1 elemental sulfur. In ASW-YT-m1-S0, 20 μM KI, 20 μM H3BO3, 10 μM NiCl2, and 10 μM Na2WO4 were supplemented. In ASW-YT-m1-pyruvate medium, elemental sulfur was replaced with 5 g L−1 sodium pyruvate. ASW-AA-S0 was composed of 0.8 × ASW, a mixture of 20 amino acids, modified Wolfe’s trace minerals and a mixture of vitamins (Sato et al., 2003). In ASW-AA-m1-S0, 20 μM KI, 20 μM H3BO3, 10 μM NiCl2, and 10 μM Na2WO4 were supplemented, and the concentrations of L-arginine hydrochloride and L-valine were increased (from 125 mg L−1 to 250 mg L−1 and from 50 mg L−1 to 200 mg L−1, respectively). When cells without a pyrF gene were grown, 10 μg mL−1 uracil was added to make ASW-AA-m1-S0(+Ura). To remove oxygen in the medium, 5% (w/v) Na2S solution was added until the medium became colorless. Resazurine (0.5 mg L−1) was also added to all media as an oxygen indicator. For solid medium used to isolate transformants, 10 g L−1 gelrite, 7.5 g L−1 5-fluoroorotic acid (5-FOA), 10 μg mL−1 uracil, 4.5 mL of 1 M NaOH and 0.2% (v/v) polysulfide solution (10 g Na2S 9H2O and 3 g sulfur flowers in 15 mL H2O) rather than elemental sulfur was supplemented to ASW-AA-m1 medium. Escherichia coli DH5α (Takara Bio, Kusatsu, Japan) and BL21-Codonplus(DE3)-RIL strains (Agilent Technologies, Santa Clara, CA) were cultivated at 37°C in Lysogeny broth (LB) medium supplemented with ampicillin (100 mg L−1). E. coli DH5α was used for recombinant plasmid construction and E. coli BL21-Codonplus (DE3)-RIL was used for heterologous gene expression. Chemicals were purchased from Wako Pure Chemicals (Osaka, Japan) or Nacalai Tesque (Kyoto, Japan) unless mentioned otherwise.
In this study, pET21a(+) was used as an expression vector for TK0548 which was amplified from genomic DNA of T. kodakarensis KU216 using the primer set TK0548F/TK0548R (Table 4). The restriction enzyme sites NdeI and BamHI were incorporated into the 5′- and 3′-termini of the fragments, respectively, during PCR. The amplified product and pET21a(+) were digested with NdeI and BamHI, ligated using Ligation high Ver. 2, and introduced into E. coli DH5α cells. Positive colonies were selected by PCR analysis and confirmed by DNA sequencing. Plasmids were introduced into E. coli BL21-Codonplus (DE3)-RIL for gene expression.
The TK2268 gene was amplified from the genomic DNA of T. kodakarensis KU216 using the primer set TK2268F/R (Table 4). NdeI and SalI sites as well as a sequence to introduce a C-terminal His6-tag were incorporated during amplification. The amplified product was digested with NdeI and SalI and inserted into a T. kodakarensis-E. coli shuttle plasmid previously used for heterologous expression of TK2101 and TK1211 (Zheng et al., 2018, 2021). After confirming the absence of unintended mutations by DNA sequencing, the plasmid was introduced into T. kodakarensis KPD2 (ΔpyrF, ΔpdaD, ΔchiA) for gene expression. pdaD corresponds to TK0149, and disruption of this gene results in agmatine auxotrophy (Fukuda et al., 2008). For transformation, T. kodakarensis KPD2 was grown in ASW-YT-m1-S0 medium supplemented with agmatine (1.0 mM) at 85°C for 12 h. Cells were harvested by centrifugation (12,000 × g, 5 min, 4°C), and resuspended in 200 μL 0.8 × ASW-m1, followed by incubation on ice for 30 min. After mixing with 3.0 μg of the expression plasmid, the mixture was further incubated on ice for 1 h. Cells were inoculated into 20 mL ASW-YT-m1-S0 medium. After incubation at 85°C for 24 h, a 200-μL aliquot was further inoculated into 20 mL ASW-YT-m1-S0 medium. After incubation at 85°C 24 h, cells were spread onto solid ASW-YT-m1-S0 medium. After incubation at 85°C for 24 h, transformants displaying agmatine prototrophy were isolated and cultivated in ASW-YT-m1-S0. The presence of recombinant plasmids and absence of unintended mutation were confirmed by PCR and DNA sequencing, respectively.
Transformants with TK0548 expression plasmid were cultivated in LB medium (100 mg L−1 ampicillin and 30 mg L−1 chloramphenicol) at 37°C until the OD660 reached 0.6. Heterologous gene expression was induced by adding isopropyl-1-thio-β-D-galactopyranoside (IPTG) to a final concentration 0.1 mM followed by cultivation at 18°C for 20 h. Cells were harvested via centrifugation (12,000 × g, 15 min, 4°C), and resuspended in 50 mM HEPES buffer (containing 150 mM NaCl, pH 7.5). Sonication was used to lyse the cells and the insoluble cell debris was separated by centrifugation at 12,000 × g for 15 min at 4°C. The soluble cell extract was heat treated at 85°C for 15 min, and the thermolabile proteins derived from the host were removed by centrifugation (12,000 × g, 15 min, 4°C). The supernatant was loaded onto an anion exchange column (Resource Q) equilibrated with 50 mM HEPES buffer (pH 7.5). Protein was eluted with a linear gradient of 0 to 1.0 M NaCl. Fractions that contain TK0548 protein were collected and concentrated with an Amicon Ultra centrifugal filter unit (MWCO 10000). The resulting protein solution was applied to a Superdex 200 10/300 Gl gel-filtration column (GE Healthcare) with a mobile phase of 50 mM HEPES buffer (containing 150 mM NaCl, pH7.5) at a flow rate of 0.7 mL min−1.
The TK2268 gene expression strain was cultivated in ASW-YT-m1-pyruvate medium at 85°C for 20 h, and cells were collected by centrifugation (6,000 × g, 15 min, 4°C). After washing with 0.8 × ASW-m1, cells were resuspended in binding buffer (50 mM HEPES buffer, 20 mM imidazole, 500 mM KCl, 10% (v/v) glycerol, pH 7.5), then disrupted by sonication. Insoluble cell debris was removed by centrifugation (12,000 × g, 15 min, 4°C). The soluble cell extract was applied to a His GraviTrap column (GE Healthcare) which had been equilibrated with binding buffer. The TK2268 protein with a His6-tag at its C terminus was eluted by elution buffer (50 mM HEPES buffer, 500 mM imidazole, 500 mM KCl, 10% (v/v) glycerol, pH 7.5). After concentrating the eluate, it was applied to a Superdex 200 10/300 Gl gel-filtration column (GE Healthcare). The TK2268 protein was eluted with a mobile phase of 50 mM HEPES buffer (pH7.5) containing 500 mM KCl and 10% (v/v) glycerol, at a flow rate of 0.7 mL min−1.
For examining the molecular mass of proteins, Blue 2000 was used to examine the void volume of the column, and ribonuclease A (13.7 kDa), carbonic anhydrase (29 kDa), ovalbumin (44 kDa), conalbumin (75 kDa), aldolase (158 kDa), and ferritin (440 kDa; GE Healthcare) were used as standards. The mobile phase was 50 mM HEPES buffer (pH7.5) containing 500 mM KCl, 10% (v/v) glycerol, and the flow rate was 0.7 mL min−1. Protein concentrations were determined with a Protein Assay kit (Bio-Rad, Hercules, CA) using bovine serum albumin as standard.
Gene disruption strains were constructed using T. kodakarensis KU216 (ΔpyrF), which shows uracil auxotrophy, as a host strain. For disrupting the TK0548 gene, the region from the start codon to base number 1080 of TK0548 gene was deleted instead of the stop codon to avoid disturbing expression of the overlapping downstream gene. In the case of TK2268, the entire coding region, along with 9 bases of its 3′-flanking region, was deleted. The TK0548 and TK2268 genes along with their 5′- and 3′-flanking regions (~1.0 kbp) were amplified from the genome of T. kodakarensis KU216 using the primer sets dTK0548F1/R1 and dTK2268F1/R1 (Table 4). The amplified products were inserted in the HincII site of the plasmid pUD3 which contains the pyrF gene of T. kodakarensis inserted in the ApaI site of pUC118 (Yokooji et al., 2009). Inverse PCR was performed with the primer sets dTK0548F2/0548R2 and dTK2268F2/2268R2 (Table 4) to remove sequences from the recombinant plasmid. The sequences of relevant regions were confirmed by DNA sequencing.
Thermococcus kodakarensis KU216 was cultivated in ASW-YT-m1-S0 medium for 12 h. Cells were harvested and resuspended in 200 μL of 0.8 × ASW-m1, then kept on ice for 30 min. After addition of 3.0 μg of the disruption plasmid and further incubation on ice for 1 h, cells were cultivated in ASW-AA-m1-S0 medium without uracil for 48 h at 85°C. A 200 μL aliquot was inoculated into fresh ASW-AA-m1-S0 medium and further cultivated under the same conditions to enrich transformants displaying uracil prototrophy. The culture was spread onto ASW-YT-m1 solid medium supplemented with 7.5 g L−1 5-FOA and 60 mM NaOH. Only cells that have undergone a pop-out recombination can grow in the presence of 5-FOA. After cultivation at 85°C for 48 h, colonies were selected, and their genotypes were analyzed by PCR using primer sets dTK0548outF/0548outR and dTK2268outF/2268outR (Table 4). Transformants that led to amplification of DNA products with the expected size were chosen and cultivated in ASW-YT-m1-S0 medium. Gene disruption was also confirmed by DNA sequencing.
Initial examination of the aminotransferase activity of TK0548 and TK2268 proteins were carried out with Glu and pyruvate as amino donor and amino acceptor, respectively. Aminotransferase activity was measured at 80°C unless mentioned otherwise. The standard reaction mixture of TK0548 protein contained 20 μM PLP, 10 mM Glu, 10 mM pyruvate, 6.24 mM NaCl, and 0.4 μg mL−1 recombinant protein in 50 mM HEPES buffer (pH7.4). The standard reaction mixture of TK2268 protein contained 20 μM PLP, 10 mM Glu, 10 mM pyruvate, 8.8 mM KCl, 0.18% (v/v) glycerol and 4 μg mL−1 recombinant protein in 50 mM HEPES buffer (pH7.4). When the PLP-dependency of activity was measured, PLP was omitted and activity with or without 10 mM hydroxylamine was measured. When PLP was omitted without addition of hydroxylamine, the amino donor and acceptor were 10 mM Leu and 10 mM 2-oxoglutarate, respectively. In the presence of hydroxylamine, the TK0548 protein reaction was measured with Phe and 2-oxoglutarate, while the TK2268 protein reaction was measured with Asp and pyruvate. The reaction mixture without amino donor and amino acceptor was pre-incubated at 80°C for 2 min, and the amino donor and acceptor were added to start the reaction. After further incubation at 80°C for 5 or 15 min, the reaction was stopped through cooling the reaction mixture on ice for 10 min. Proteins were removed with an Amicon Ultra-0.5 centrifugal filter unit with an Ultracel-10 membrane (Millipore). The formation of Glu and Ala was detected and quantified by HPLC after derivatization. The derivatization mixture (100 μL) contained 10 μL of reaction mixture, 70 μL of solution B [borate sodium hydroxide buffer (0.4 M, pH 10.4)] and 20 μL of solution A (8 mg o-phthalaldehyde and 10 mg N-acetylcysteine were dissolved in 1 mL methanol). After derivatization for 5 min at room temperature, an aliquot (10 μL) of the solution was applied to a COSMOSIL 5C18-PAQ packed column (4.6ID × 250 mm) using a Nexera X2 liquid chromatography system with a fluorescence detector RF-20A XS (Shimadzu, Kyoto, Japan). Compounds were eluted with a solution of 20 mM sodium acetate (pH 5.6) and methanol at a flow rate of 0.7 mL min−1. The excitation and emission wavelength were 350 and 450 nm, respectively.
To screen the amino acids recognized by TK0548 and TK2268 proteins, 20 amino acids were used as amino donor, and 2-oxoglutarate or pyruvate was used as amino acceptor. To analyze the aminotransferase activity of TK0548 protein, Tyr, Phe, Trp, His, Leu, Met, and Glu were chosen as amino donor, and 2-oxoglutarate or pyruvate was used as amino acceptor. For the TK2268 protein, Glu, Asp, Cys, Leu, Ala, Met, and Tyr were chosen as amino donor, and 2-oxoglutarate or pyruvate was used as amino acceptor. The standard reaction mixture was incubated at 80°C for 3, 5, and 7 min (reaction mixture with His and Glu were incubated at 80°C for 1, 2, 3 min) to confirm that product formation was linear with time.
For kinetic analysis of the TK0548 protein reaction, reaction rates with various concentrations of Phe, Trp, Tyr, Met, and His were examined with 10 mM 2-oxoglutarate. Reaction rates with various concentrations of 2-oxoglutarate and pyruvate were examined with 10 mM Phe. For analysis of the TK2268 protein, reaction rates with various concentrations of Asp, Glu, Tyr, and Leu were examined with 10 mM pyruvate or 10 mM 2-oxoglutarate. Reaction rates with various concentrations of 2-oxoglutarate and pyruvate were examined with 10 mM Leu. Kinetic parameters were obtained by fitting the data to the Michaelis–Menten equation using IGORPRO, version 6.03 (Wave-Metrics, Lake, Oswego, OR).
Growth properties of the host strain KU216 and the ΔTK0548 and ΔTK2268 gene disruption strains were examined in ASW-YT-m1-S0 and ASW-AA-m1-S0(+Ura) media. Cells were precultured in the nutrient-rich medium ASW-YT-m1-S0 for 15 h until the stationary phase and inoculated into ASW-YT-m1-S0 medium or synthetic medium ASW-AA-m1-S0(+Ura). The OD660 of the culture was monitored.
Thermococcus kodakarensis KU216, ΔTK0548 and ΔTK2268 disruption strains were cultivated at ASW-YT-m1-pyruvate medium for 20 h and cells were collected by centrifugation (6,000 × g, 15 min, 4°C). Cells were disrupted by sonication and insoluble cell debris was removed (12,000 × g, 30 min, 4°C). After exchanging the buffer with 50 mM HEPES (containing 150 mM NaCl, pH7.4) using Amicon Ultra centrifugal filter unit (MWCO 10000), the aminotransferase activity in the cell-free extract was measured. For the aminotransferase activity toward His, Tyr, and Asp, the reaction mixture contained 20 μM PLP, 10 mM amino donor (the final concentration of Tyr was 6 mM), 10 mM amino acceptor (2-oxoglutarate or pyruvate), 5.76 mM NaCl and 0.384 mg mL−1 cell-free extracts in 50 mM HEPES buffer (pH7.4). For Trp, the reaction mixture contained 20 μM PLP, 10 mM amino donor, 10 mM amino acceptor (2-oxoglutarate), 2.88 mM NaCl and 0.182 mg mL−1 cell-free extracts in 50 mM HEPES buffer (pH7.4). In the case of Glu, Ile, and Phe, the reaction mixture contained 20 μM PLP, 10 mM amino donor, 10 mM amino acceptor (2-oxoglutarate or pyruvate), 0.576 mM NaCl and 0.0384 mg mL−1 cell-free extracts in 50 mM HEPES buffer (pH7.4). The reaction mixture without amino acceptor was pre-incubated at 80°C for 2 min, and the amino acceptor was added to start the reaction. After further incubation at 80°C for 1, 2, and 3 min (When Asp and Ile were used as amino donor, the reaction mixtures were incubated at 80°C for 4, 6, and 8 min and 2, 4, and 6 min, respectively). The reaction was stopped through cooling the reaction mixture on ice for 10 min. Proteins were removed with an Amicon Ultra-0.5 centrifugal filter unit with an Ultracel-10 membrane (Millipore). The formation of Glu and Ala was determined by HPLC after derivatization.
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
HA designed the experiments. YS carried out the biochemical and genetic experiments. YM carried out the bioinformatic analyses. All authors contributed in data analyses, writing the manuscript, and approved the submitted version.
This study was partially supported by JSPS KAKENHI grant numbers JP19H05679 (Post-Koch Ecology) and JP19H05684 to HA. This work was partially supported by JST SPRING, Grant Number JPMJSP2110 and JST, the establishment of university fellowships toward the creation of science technology innovation, Grant Number JPMJFS2123.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1126218/full#supplementary-material
Andreotti, G., Cubellis, M. V., Nitti, G., Sannia, G., Mai, X., Adams, M. W., et al. (1995). An extremely thermostable aromatic aminotransferase from the hyperthermophilic archaeon Pyrococcus furiosus. Biochim. Biophys. Acta 1247, 90–96. doi: 10.1016/0167-4838(94)00211-X
Andreotti, G., Cubellis, M. V., Nitti, G., Sannia, G., Mai, X., Marino, G., et al. (1994). Characterization of aromatic aminotransferases from the hyperthermophilic archaeon Thermococcus litoralis. Eur. J. Biochem. 220, 543–549. doi: 10.1111/j.1432-1033.1994.tb18654.x
Atomi, H., Fukui, T., Kanai, T., Morikawa, M., and Imanaka, T. (2004). Description of Thermococcus kodakaraensis sp. nov., a well studied hyperthermophilic archaeon previously reported as Pyrococcus sp. KOD1. Archaea 1, 263–267. doi: 10.1155/2004/204953
Awano, T., Wilming, A., Tomita, H., Yokooji, Y., Fukui, T., Imanaka, T., et al. (2014). Characterization of two members among the five ADP-forming acyl coenzyme a (acyl-CoA) synthetases reveals the presence of a 2-(Imidazol-4-yl)acetyl-CoA synthetase in Thermococcus kodakarensis. J. Bacteriol. 196, 140–147. doi: 10.1128/JB.00877-13
Blamey, J. M., and Adams, M. W. (1993). Purification and characterization of pyruvate ferredoxin oxidoreductase from the hyperthermophilic archaeon Pyrococcus furiosus. Biochim. Biophys. Acta 1161, 19–27. doi: 10.1016/0167-4838(93)90190-3
Chon, H., Matsumura, H., Koga, Y., Takano, K., and Kanaya, S. (2005). Crystal structure of a human kynurenine aminotransferase II homologue from Pyrococcus horikoshii OT3 at 2.20 a resolution. Proteins 61, 685–688. doi: 10.1002/prot.20614
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Fiala, G., and Stetter, K. O. (1986). Pyrococcus furiosus sp. nov. represents a novel genus of marine heterotrophic archaebacteria growing optimally at 100°C. Arch. Microbiol. 145, 56–61. doi: 10.1007/BF00413027
Fukuda, W., Fukui, T., Atomi, H., and Imanaka, T. (2004). First characterization of an archaeal GTP-dependent phosphoenolpyruvate carboxykinase from the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. J. Bacteriol. 186, 4620–4627. doi: 10.1128/JB.186.14.4620-4627.2004
Fukuda, W., Ismail, Y. S., Fukui, T., Atomi, H., and Imanaka, T. (2005). Characterization of an archaeal malic enzyme from the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. Archaea 1, 293–301. doi: 10.1155/2005/250757
Fukuda, W., Morimoto, N., Imanaka, T., and Fujiwara, S. (2008). Agmatine is essential for the cell growth of Thermococcus kodakaraensis. FEMS Microbiol. Lett. 287, 113–120. doi: 10.1111/j.1574-6968.2008.01303.x
Fukui, T., Atomi, H., Kanai, T., Matsumi, R., Fujiwara, S., and Imanaka, T. (2005). Complete genome sequence of the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1 and comparison with Pyrococcus genomes. Genome Res. 15, 352–363. doi: 10.1101/gr.3003105
Glasemacher, J., Bock, A. K., Schmid, R., and Schonheit, P. (1997). Purification and properties of acetyl-CoA synthetase (ADP-forming), an archaeal enzyme of acetate formation and ATP synthesis, from the hyperthermophile Pyrococcus furiosus. Eur. J. Biochem. 244, 561–567. doi: 10.1111/j.1432-1033.1997.00561.x
Gomm, A., and O'reilly, E. (2018). Transaminases for chiral amine synthesis. Curr. Opin. Chem. Biol. 43, 106–112. doi: 10.1016/j.cbpa.2017.12.007
Heider, J., Mai, X., and Adams, M. W. (1996). Characterization of 2-ketoisovalerate ferredoxin oxidoreductase, a new and reversible coenzyme A-dependent enzyme involved in peptide fermentation by hyperthermophilic archaea. J. Bacteriol. 178, 780–787. doi: 10.1128/jb.178.3.780-787.1996
Hidese, R., Inoue, T., Imanaka, T., and Fujiwara, S. (2014). Cysteine desulphurase plays an important role in environmental adaptation of the hyperthermophilic archaeon Thermococcus kodakarensis. Mol. Microbiol. 93, 331–345. doi: 10.1111/mmi.12662
Jansonius, J. N. (1998). Structure, evolution and action of vitamin B6-dependent enzymes. Curr. Opin. Struct. Biol. 8, 759–769. doi: 10.1016/S0959-440X(98)80096-1
Jones, D. T., Taylor, W. R., and Thornton, J. M. (1992). The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8, 275–282. doi: 10.1093/bioinformatics/8.3.275
Kanai, T., Simons, J. R., Tsukamoto, R., Nakajima, A., Omori, Y., Matsuoka, R., et al. (2015). Overproduction of the membrane-bound [NiFe]-hydrogenase in Thermococcus kodakarensis and its effect on hydrogen production. Front. Microbiol. 6:847. doi: 10.3389/fmicb.2015.00847
Kawakami, R., Kinoshita, C., Kawase, T., Sato, M., Hayashi, J., Sakuraba, H., et al. (2021). Characterization of a novel moderate-substrate specificity amino acid racemase from the hyperthermophilic archaeon Thermococcus litoralis. Biosci. Biotechnol. Biochem. 85, 1650–1657. doi: 10.1093/bbb/zbab078
Kawakami, R., Ohmori, T., Sakuraba, H., and Ohshima, T. (2015). Identification of a novel amino acid racemase from a hyperthermophilic archaeon Pyrococcus horikoshii OT-3 induced by D-amino acids. Amino. Acids. 47, 1579–1587. doi: 10.1007/s00726-015-2001-6
Kawakami, R., Ohshida, T., Sakuraba, H., and Ohshima, T. (2018). A novel PLP-dependent alanine/serine racemase from the hyperthermophilic archaeon Pyrococcus horikoshii OT-3. Front. Microbiol. 9:1481. doi: 10.3389/fmicb.2018.01481
Kawakami, R., Ohshida, T., Hayashi, J., Yoneda, K., Furumoto, T., Ohshima, T., et al. (2022). Crystal structure of a novel type of ornithine d-aminotransferase from the hyperthermophilic archaeon Pyrococcus horikoshii. Int. J. Biol. Macromol. 208, 731–740. doi: 10.1016/j.ijbiomac.2022.03.114
Kawakami, R., Sakuraba, H., Ohmori, T., and Ohshima, T. (2017). First characterization of an archaeal amino acid racemase with broad substrate specificity from the hyperthermophile Pyrococcus horikoshii OT-3. J. Biosci. Bioeng. 124, 23–27. doi: 10.1016/j.jbiosc.2017.02.004
Kelly, S. A., Mix, S., Moody, T. S., and Gilmore, B. F. (2020). Transaminases for industrial biocatalysis: novel enzyme discovery. Appl. Microbiol. Biotechnol. 104, 4781–4794. doi: 10.1007/s00253-020-10585-0
Kito, K., Sanada, Y., and Katunuma, N. (1978). Mode of inhibition of ornithine aminotransferase by L-canaline. J. Biochem. 83, 201–206. doi: 10.1093/oxfordjournals.jbchem.a131892
Koper, K., Han, S. W., Pastor, D. C., Yoshikuni, Y., and Maeda, H. A. (2022). Evolutionary origin and functional diversification of aminotransferases. J. Biol. Chem. 298:102122. doi: 10.1016/j.jbc.2022.102122
Koszelewski, D., Tauber, K., Faber, K., and Kroutil, W. (2010). ω-Transaminases for the synthesis of non-racemic alpha-chiral primary amines. Trends Biotechnol. 28, 324–332. doi: 10.1016/j.tibtech.2010.03.003
Kunutsor, S. K., Apekey, T. A., and Walley, J. (2013). Liver aminotransferases and risk of incident type 2 diabetes: a systematic review and meta-analysis. Am. J. Epidemiol. 178, 159–171. doi: 10.1093/aje/kws469
Mai, X., and Adams, M. W. (1994). Indolepyruvate ferredoxin oxidoreductase from the hyperthermophilic archaeon Pyrococcus furiosus. J. Biol. Chem. 269, 16726–16732. doi: 10.1016/S0021-9258(19)89451-6
Mai, X., and Adams, M. W. (1996). Purification and characterization of two reversible and ADP-dependent acetyl coenzyme a synthetases from the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol. 178, 5897–5903. doi: 10.1128/jb.178.20.5897-5903.1996
Malik, M. S., Park, E. S., and Shin, J. S. (2012). Features and technical applications of ω-transaminases. Appl. Microbiol. Biotechnol. 94, 1163–1171. doi: 10.1007/s00253-012-4103-3
Matsui, I., Matsui, E., Sakai, Y., Kikuchi, H., Kawarabayasi, Y., Ura, H., et al. (2000). The molecular structure of hyperthermostable aromatic aminotransferase with novel substrate specificity from Pyrococcus horikoshii. J. Biol. Chem. 275, 4871–4879. doi: 10.1074/jbc.275.7.4871
Matsumi, R., Manabe, K., Fukui, T., Atomi, H., and Imanaka, T. (2007). Disruption of a sugar transporter gene cluster in a hyperthermophilic archaeon using a host-marker system based on antibiotic resistance. J. Bacteriol. 189, 2683–2691. doi: 10.1128/JB.01692-06
Mehta, P. K., and Christen, P. (2000). The molecular evolution of pyridoxal-5′-phosphate-dependent enzymes. Adv. Enzymol. Relat. Areas Mol. Biol. 74, 129–184. doi: 10.1002/9780470123201.ch4
Mehta, P. K., Hale, T. I., and Christen, P. (1993). Aminotransferases: demonstration of homology and division into evolutionary subgroups. Eur. J. Biochem. 214, 549–561. doi: 10.1111/j.1432-1033.1993.tb17953.x
Montioli, R., Bellezza, I., Desbats, M. A., Borri Voltattorni, C., Salviati, L., and Cellini, B. (2021). Deficit of human ornithine aminotransferase in gyrate atrophy: molecular, cellular, and clinical aspects. Biochim. Biophys. Acta Proteins Proteom. 1869:140555. doi: 10.1016/j.bbapap.2020.140555
Morikawa, M., Izawa, Y., Rashid, N., Hoaki, T., and Imanaka, T. (1994). Purification and characterization of a thermostable thiol protease from a newly isolated hyperthermophilic Pyrococcus sp. Appl. Environ. Microbiol. 60, 4559–4566. doi: 10.1128/aem.60.12.4559-4566.1994
Musfeldt, M., Selig, M., and Schonheit, P. (1999). Acetyl coenzyme a synthetase (ADP forming) from the hyperthermophilic archaeon Pyrococcus furiosus: identification, cloning, separate expression of the encoding genes, acdAI and acdBI, in Escherichia coli, and in vitro reconstitution of the active heterotetrameric enzyme from its recombinant subunits. J. Bacteriol. 181, 5885–5888. doi: 10.1128/JB.181.18.5885-5888.1999
Okada, K., Angkawidjaja, C., Koga, Y., and Kanaya, S. (2014). Structural and mechanistic insights into the kynurenine aminotransferase-mediated excretion of kynurenic acid. J. Struct. Biol. 185, 257–266. doi: 10.1016/j.jsb.2014.01.009
Okada, K., Angkawidjaja, C., Koga, Y., Takano, K., and Kanaya, S. (2012). Characteristic features of kynurenine aminotransferase allosterically regulated by (alpha)-ketoglutarate in cooperation with kynurenine. PLoS One 7:e40307. doi: 10.1371/journal.pone.0040307
Robb, F., and Place, A. (1995). Media for Thermophiles. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
Sakuraba, H., Kawakami, R., Takahashi, H., and Ohshima, T. (2004). Novel archaeal alanine:glyoxylate aminotransferase from Thermococcus litoralis. J. Bacteriol. 186, 5513–5518. doi: 10.1128/JB.186.16.5513-5518.2004
Sakuraba, H., Yoneda, K., Takeuchi, K., Tsuge, H., Katunuma, N., and Ohshima, T. (2008). Structure of an archaeal alanine: glyoxylate aminotransferase. Acta Crystallogr. D Biol. Crystallogr. 64, 696–699. doi: 10.1107/S0907444908006732
Santangelo, T. J., Cubonova, L., and Reeve, J. N. (2008). Shuttle vector expression in Thermococcus kodakaraensis: contributions of cis elements to protein synthesis in a hyperthermophilic archaeon. Appl. Environ. Microbiol. 74, 3099–3104. doi: 10.1128/AEM.00305-08
Santangelo, T. J., Cubonova, L., and Reeve, J. N. (2010). Thermococcus kodakarensis genetics: TK1827-encoded β-glycosidase, new positive-selection protocol, and targeted and repetitive deletion technology. Appl. Environ. Microbiol. 76, 1044–1052. doi: 10.1128/AEM.02497-09
Sato, T., Fukui, T., Atomi, H., and Imanaka, T. (2003). Targeted gene disruption by homologous recombination in the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. J. Bacteriol. 185, 210–220. doi: 10.1128/JB.185.1.210-220.2003
Sato, T., Fukui, T., Atomi, H., and Imanaka, T. (2005). Improved and versatile transformation system allowing multiple genetic manipulations of the hyperthermophilic archaeon Thermococcus kodakaraensis. Appl. Environ. Microbiol. 71, 3889–3899. doi: 10.1128/AEM.71.7.3889-3899.2005
Schneider, G., Kack, H., and Lindqvist, Y. (2000). The manifold of vitamin B6 dependent enzymes. Structure 8, R1–R6. doi: 10.1016/S0969-2126(00)00085-X
Shikata, K., Fukui, T., Atomi, H., and Imanaka, T. (2007). A novel ADP-forming succinyl-CoA synthetase in Thermococcus kodakaraensis structurally related to the archaeal nucleoside diphosphate-forming acetyl-CoA synthetases. J. Biol. Chem. 282, 26963–26970. doi: 10.1074/jbc.M702694200
Takai, K., Sugai, A., Itoh, T., and Horikoshi, K. (2000). Palaeococcus ferrophilus gen. Nov., sp. nov., a barophilic, hyperthermophilic archaeon from a deep-sea hydrothermal vent chimney. Int. J. Syst. Evol. Microbiol. 50 Pt 2, 489–500. doi: 10.1099/00207713-50-2-489
Tamura, K., Stecher, G., and Kumar, S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Tavares, N. K., Stracey, N., Brunold, T. C., and Escalante-Semerena, J. C. (2019). The L-Thr kinase/L-Thr-phosphate decarboxylase (CobD) enzyme from Methanosarcina mazei Go1 contains metallocenters needed for optimal activity. Biochemistry 58, 3260–3279. doi: 10.1021/acs.biochem.9b00268
Tavares, N. K., Zayas, C. L., and Escalante-Semerena, J. C. (2018). The Methanosarcina mazei MM2060 gene encodes a bifunctional kinase/decarboxylase enzyme involved in Cobamide biosynthesis. Biochemistry 57, 4478–4495. doi: 10.1021/acs.biochem.8b00546
Toyokawa, Y., Koonthongkaew, J., and Takagi, H. (2021). An overview of branched-chain amino acid aminotransferases: functional differences between mitochondrial and cytosolic isozymes in yeast and human. Appl. Microbiol. Biotechnol. 105, 8059–8072. doi: 10.1007/s00253-021-11612-4
Uchida, Y., Hayashi, H., Washio, T., Yamasaki, R., Kato, S., and Oikawa, T. (2014). Cloning and characterization of a novel fold-type I branched-chain amino acid aminotransferase from the hyperthermophilic archaeon Thermococcus sp. CKU-1. Extremophiles 18, 589–602. doi: 10.1007/s00792-014-0642-0
Ura, H., Harata, K., Matsui, I., and Kuramitsu, S. (2001). Temperature dependence of the enzyme-substrate recognition mechanism. J. Biochem. 129, 173–178. doi: 10.1093/oxfordjournals.jbchem.a002829
Ward, D. E., De Vos, W. M., and Van Der Oost, J. (2002). Molecular analysis of the role of two aromatic aminotransferases and a broad-specificity aspartate aminotransferase in the aromatic amino acid metabolism of Pyrococcus furiosus. Archaea 1, 133–141. doi: 10.1155/2002/959031
Ward, D. E., Kengen, S. W., Van Der Oost, J., and De Vos, W. M. (2000). Purification and characterization of the alanine aminotransferase from the hyperthermophilic archaeon Pyrococcus furiosus and its role in alanine production. J. Bacteriol. 182, 2559–2566. doi: 10.1128/JB.182.9.2559-2566.2000
Yokooji, Y., Sato, T., Fujiwara, S., Imanaka, T., and Atomi, H. (2013). Genetic examination of initial amino acid oxidation and glutamate catabolism in the hyperthermophilic archaeon Thermococcus kodakarensis. J. Bacteriol. 195, 1940–1948. doi: 10.1128/JB.01979-12
Yokooji, Y., Tomita, H., Atomi, H., and Imanaka, T. (2009). Pantoate kinase and phosphopantothenate synthetase, two novel enzymes necessary for CoA biosynthesis in the archaea. J. Biol. Chem. 284, 28137–28145. doi: 10.1074/jbc.M109.009696
Yoshida, A., Tomita, T., Atomi, H., Kuzuyama, T., and Nishiyama, M. (2016). Lysine biosynthesis of Thermococcus kodakarensis with the capacity to function as an ornithine biosynthetic system. J. Biol. Chem. 291, 21630–21643. doi: 10.1074/jbc.M116.743021
Zheng, R. C., Hachisuka, S. I., Tomita, H., Imanaka, T., Zheng, Y. G., Nishiyama, M., et al. (2018). An ornithine ω-aminotransferase required for growth in the absence of exogenous proline in the archaeon Thermococcus kodakarensis. J. Biol. Chem. 293, 3625–3636. doi: 10.1074/jbc.RA117.001222
Zheng, R. C., Lu, X. F., Tomita, H., Hachisuka, S. I., Zheng, Y. G., and Atomi, H. (2021). TK1211 encodes an amino acid racemase towards leucine and methionine in the hyperthermophilic archaeon Thermococcus kodakarensis. J. Bacteriol. 203, e00617–e00620. doi: 10.1128/JB.00617-20
Keywords: Archaea, aminotransferase, metabolism, enzyme, genetics
Citation: Su Y, Michimori Y and Atomi H (2023) Biochemical and genetic examination of two aminotransferases from the hyperthermophilic archaeon Thermococcus kodakarensis. Front. Microbiol. 14:1126218. doi: 10.3389/fmicb.2023.1126218
Received: 17 December 2022; Accepted: 31 January 2023;
Published: 20 February 2023.
Edited by:
Marleen van Wolferen, University of Freiburg, GermanyReviewed by:
Ivan A. Berg, University of Münster, GermanyCopyright © 2023 Su, Michimori and Atomi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Haruyuki Atomi, ✉ YXRvbWlAc2JjaGVtLmt5b3RvLXUuYWMuanA=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.