- 1Innovative Team of Antimicrobial Peptides and Alternatives to Antibiotics, Feed Research Institute, Chinese Academy of Agricultural Sciences, Beijing, China
- 2Gene Engineering Laboratory, Feed Research Institute, Chinese Academy of Agricultural Sciences, Beijing, China
- 3Key Laboratory of Feed Biotechnology, Ministry of Agriculture and Rural Affairs, Beijing, China
- 4The School of Medicine, Medical Sciences and Nutrition, University of Aberdeen, Aberdeen, United Kingdom
- 5S-Inova Biotech, Universidade Católica Dom Bosco, Campo Grande, MS, Brazil
- 6Centro de Análises Proteômicas e Bioquímicas Programa de Pós-Graduação em Ciências Genômicas e Biotecnologia, Universidade Católica de Brasília, Brasília, DF, Brazil
- 7Machine Biology Group, Departments of Psychiatry and Microbiology, Institute for Biomedical Informatics, Institute for Translational Medicine and Therapeutics, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, United States
- 8Departments of Bioengineering and Chemical and Biomolecular Engineering, School of Engineering and Applied Science, University of Pennsylvania, Philadelphia, PA, United States
- 9Penn Institute for Computational Science, University of Pennsylvania, Philadelphia, PA, United States
Editorial on the Research Topic
Community series in antimicrobial peptides: Molecular design, structure function relationship and biosynthesis optimization
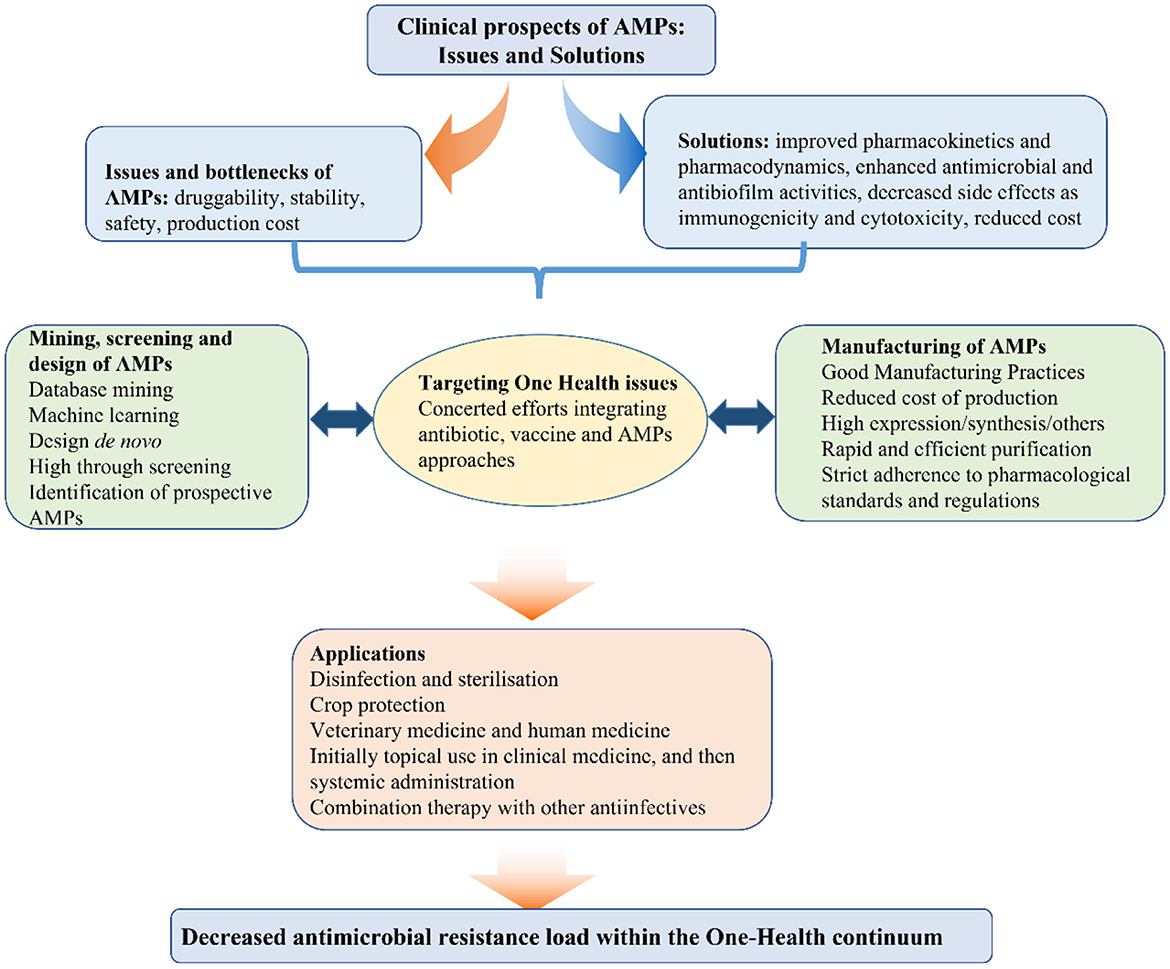
The continuous rise in antimicrobial resistance during the last decades has significantly contributed to the R&D of alternatives such as antimicrobial peptides (AMPs). In the first volume of this topic, we proposed a combinatorial approach involving AMPs, antimicrobials and vaccines, which would be instrumental for the prevention and treatment of human and animal diseases within the One Health framework (Figure 1) (Wang et al., 2018, 2019; Cardoso et al., 2019a; Yang et al., 2019; Ma et al., 2021; Wu et al., 2021; Zheng et al., 2021; Hao et al., 2022). Compared to conventional antimicrobials, AMPs possess certain advantages such as high penetration and internalization, in some cases decreased likelihood of resistance emergence among bacterial pathogens, and lower probability of accumulation in tissues (Wang et al., 2019, 2022; Aminov, 2022). Selective inhibition of bacterial pathogens without causing significant cytotoxic effects, is not only an essential requirement but also a critical challenge for the R&D of AMPs. The charge, special amino acid (aa) residues, hydrophobicity/hydrophilicity ratio and secondary structure directly affect the antibacterial activity, stability and cytotoxicity of AMPs. Thus, the discovery of new AMPs by natural screening, database mining and machine learning, in addition to the rational structural design of these agents could greatly contribute to translating them from lab to clinic. These exciting new developments are highlighted in 21 papers published in the second volume of the community series of Research Topics devoted to AMPs.
Discovery of natural AMPs
AMPs are short natural molecules, which are encountered in the majority of living organisms and serve as a first line of defense. According to the Antimicrobial Peptide Database (APD, https://aps.unmc.edu/), AMPs have been discovered from six life kingdoms: bacteria, archaea, protists, fungi, plants, and animals. They are mostly of animal origin, with 74% of known natural AMPs isolated from this source (Wang et al., 2016; Wang, 2022). AMPs in animals serve as host defense peptides to prevent pathogen invasion. Liu M. et al. established the coding sequences (CDS) and deduced the full-length amino acid sequences of novel hepcidin peptides from Antarctic Notothenioid Fish. The mature hepcidin peptides four disulphide bonds, differing from the typical defensins (α, β, and Θ) with three such bonds. This AMP was successfully expressed in Escherichia coli and displayed a broad-spectrum antibacterial activity. Microorganisms also produce AMPs to defend their ecological niches. Compared to animal AMPs, however, the biosynthetic pathways of microbial AMPs are divided into ribosomally produced and non-ribosomally produced peptides, yielding a great diversity of structural types of AMPs. Wu Y.-p. et al. isolated a cyclic non-ribosomal lipopeptide polymyxin A1 from Paenibacillus thiaminolyticus and determined the biosynthetic gene cluster for its synthesis, which included five open reading frames (ORFs). The lipopeptide structure confers stability and strong activity against Gram-negatove bacteria. This AMP, however, should be further tested for its efficacy and toxicity in vivo. Bacteriocins are ribosomally produced AMPs, which primary function is to inhibit competing strains present within the same ecological niche. Thus, they display a narrow activity range, essentially directed toward close relatives. This property could be advantageous, allowing precision therapy and infection control. Vogel et al. isolated a bacteriocin Angicin from Streptococcus anginosus, which is not subjected to posttranslational modifications (Vogel et al., 2021). Angicin displays no cytotoxicity toward eukaryotic cells since it precisely targets the bacterial mannose phosphotransferase system (Man-PTS), and there is no identified target in eukaryotic cells (Vogel et al.). Previous studies on AMPs have been mainly focused on their antimicrobial effect against bacteria, but some recent works have also involved fungi. Fungi are frequently plant pathogens, and there has been a considerable interest in using microorganisms or their compounds as a sustainable bio-control measure for the protection of plants against fungal diseases. Zhu H. et al. identified the antifungal tetrapeptide His-Ala-Phe-Lys (Hafk) from the bacterium Burkholderia arboris by using a Tn5 transposon mutation library. Inactivation or deletion of the cobA gene resulted in a reduced antifungal activity and significantly decreased the production of Hafk. Thus, the Hafk peptide has a significant potential as a biocontrol agent for crop fungal diseases. Presently, however, the natural reservoirs of AMPs such as the marine environment have not been sufficiently explored. The discovery and development of novel AMPs from under-explored ecological niches would certainly contribute to the health initiative within the One Health framework (Travis et al., 2018; Lazzaro et al., 2020; Hao et al., 2022).
Structure and function of AMPs
Natural AMPs are the product of long-term evolution and they have evolved to perform certain functions such as providing protection to against infectious invasion or occupation of ecological niches. These functions are not always in-line with our needs, and here we can investigate the structure-and-function relationships of AMPs in order to improve their characteristics for our purposes. Mehamycin, a drosomycin-type antifungal peptide (DTAFP), belongs to the defensin-type family present in plants and ecdysozoans. By analyzing sequence and structural features of Mehamycin and other peptides in the DTAFP family, an 18-aa residue single Disulfide Bridge-linked Domain (sDBD) insert was identified (Gu et al.). Mutational analysis suggested a key role played by this insert in broadening the antimicrobial spectrum, accelerating pathogen eradication and thus conferring an evolutionary advantage. Identification of allosteric residues uncovered the structure-and-function trade-off. Besides the effect of peptide segments on structure and function of AMPs, single aa residues may also affect their biological function. This especially concerns aa with unique properties such as hydrophobic and basic aa. Sultana et al. investigated the role of basic aa residues, K58 and K59 and the N-terminal α-helix containing residues K7 and K30, in the antimicrobial activity of Angiogenin 4. Mutations in these positions resulted in reduced antimicrobial activity against Salmonella Typhimurium. Thus, the critical basic aa residues with different functionalities rather than overall electrostatic interactions play a key role in cell binding and disruption of the bacterial membrane integrity by Angiogenin 4. Optimized AMPs may be obtained by rational design by rearranging hydrophilic and hydrophobic residues, changing net charge or through conformational changes (de Moraes et al.; Wu R. et al.; Li et al.; Yuan et al.).
Computational mining of AMPs
At present, thousands of identified AMP sequences are deposited in public AMPs databases such as antimicrobial peptides database (APD, https://aps.unmc.edu/prediction), collection of anti-microbial peptides (CAMP, http://www.camp.bicnirrh.res.in/), database of antimicrobial activity and structure of peptides (DBAASP, https://www.dbaasp.org/home), and database of antimicrobial peptides (dbAMP, http://csb.cse.yzu.edu.tw/dbAMP/, all accessed on 14 December 2022). Their structure-and-function relationships, however, are not explored to a level that would allow their further improvement and optimization (Porto et al., 2018; Torres et al., 2021; Wan et al., 2022). In the post-genomic era, the growing number of sequences deposited in databases has become a new rich resource for discovery, modification and redesign of novel AMPs (Torres et al., 2022). Tools for such analyses include Multiple Descriptor Multiple Strategy (MultiDS) screening system and multi-task learning (MTL). They are based on physicochemical and structural parameters, strategies, and algorithms for the rapid search of new candidate AMPs from genome sequences, and these systems introduce the relationship between MIC values and other parameters, providing a new perspective for improving the antibacterial activity and other key properties of AMPs (Lee et al.; Liu L. et al.). AMPs identified by genome-based screening systems were homologous to annotated and unannotated natural AMPs, and the de novo design methods were implemented for optimal AMP structures. Therefore, a comprehensive screening system based on bioinformatic and artificial intelligence tools enable a high-throughput prediction of novel functional AMPs with a high potential and applicability for further wet lab work (Cardoso et al., 2020; Torres et al., 2021).
Recombinant AMPs expression
Although chemical synthesis is an important method for the preparation of short AMPs, the high manufacturing cost is a key limiting factor, particularly for peptides > 35 aa residues and with post-translational modifications (Deng et al., 2017; Cao et al., 2018; Wibowo and Zhao, 2019). Recombinant expression systems are widely used to produce various polypeptides and proteins. For example, Bacillus is an excellent host that can express heterologous proteins and also produce endogenous AMPs (Ren et al.). It is worth highlighting that there is currently no universal approach to express various AMPs, and the scope and applicability of each system is limited which it is based on special vector construction involving element reform and optimization, well-resistance selection for expression host suicide from AMPs, exact cleavage and secretion, and easy purification (Mao et al., 2014; Zhang et al., 2014; Teng et al., 2015; Li et al., 2017a,b, 2020; Wang et al., 2017; Cao et al., 2018; de Oliveira et al., 2020; Liu et al., 2021; Torres et al., 2021, 2022).
Effects of AMPs on bacteria at different growth stages
In multicellular organisms AMPs are part of innate immunity and thus serve as the first line of defense against pathogens. Compared to traditional antimicrobials, AMPs are characterized by more narrow mutant selection windows and lesser chances of emergence of bacterial resistance (Rodríguez-Rojas et al., 2014, 2018; Yu et al., 2018; Liu et al., 2021; Zheng et al., 2021; Wu et al., 2022). Activities of AMPs are usually evaluated in vitro with exponentially growing bacteria, but under natural conditions, bacterial growth rates are much slower (Savageau, 1983; Spaulding et al., 2017). Bacteria in stationary phase, for instance, are significantly less susceptible to antimicrobials compared to exponentially growing bacteria (Gutierrez et al., 2017; Mccall et al., 2019). Using five different AMPs and three antibiotics, Rodríguez-Rojas and Roll demonstrated that AMPs possess a better bactericidal effect on non-dividing bacteria compared to antibiotics. The authors reasoned that AMPs were selected as an antimicrobial defense strategy by metazoans precisely in part due to this desirable activity against non-dividing bacteria.
Conclusions
Pathogen resistance to antimicrobials, especially multi-drug resistance, poses a serious worldwide public health concern due to the higher morbidity and mortality rates caused by these infections. Alternatives to antimicrobials such as AMPs attract attention due to their multifactorial mechanism of action, low propensity to select for bacterial resistance, intracellular antibacterial activity, and special synergistic with conventional antimicrobials, among other advantages (Travis et al., 2018; Wang et al., 2018; Cardoso et al., 2019b; Lazzaro et al., 2020; Ageitos et al., 2022; Aminov, 2022; Hao et al., 2022; Zhu R. et al.). Thus, the discovery, modification, reformation and de novo design of AMPs represent an exciting approach for infection management and control. With the use of omics technologies, combined with synthetic biology approaches and gene editing and artificial intelligence tools, the increasing number of novel AMPs with high antimicrobial efficiency and low cytotoxicity can now be mined and identified for a potential use (Melo et al., 2021; Palmer et al., 2021). It must be not overlooked that AMPs, as a part of innate immunity, play a significant role in immune responses, which may occasionally be detrimental to the host. Thus, defining the antimicrobial and immune stimulation boundaries in order to limit the latter is a priority when designing new AMPs.
Currently, some AMPs are undergoing phase II-III clinical trials (Jiang et al., 2021). Most of them are used topically for wound and skin infections. The main reason for the topical use is to restrict systemic effects that could be detrimental for the host because of the impact of AMPs on the immune system. Compared to traditional antimicrobials, many AMPs derived from animals have immune functions besides their antibacterial effect (Ganz, 2003; Nesa et al., 2020). Thus, systemic application of AMPs may potentially display side effects resulting from their innate immunomodulatory properties. In order to be considered for systemic administration, AMPs should lack off-target effects, possess desirable bioavailability, stability, and half-life profiles, and optimal pharmacokinetic methods should be established (Zheng et al.). From a synthetic biology perspective, manufacturing of AMPs is not problematic since numerous toolkits are currently available (Cao et al., 2018; Hao et al., 2018). However, the choice of expression systems for AMPs should be determined based on desired properties such as the range of microorganisms targeted, the kind of application envisaged, possibility and feasibility of heterogenous expression of these peptides, and a reasonable and competitive cost of manufacturing once AMPs are ready for clinical applications (Zhang et al., 2011; Mao et al., 2014; Teng et al., 2015; Li et al., 2017b; Cao et al., 2018; de Oliveira et al., 2020; Hao et al., 2022).
Author contributions
The first draft text of this editorial was written by NY as assistant of JW and his co-editors with their guide and direction. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (Grants No. 31872393, 2018–2022), National Key R&D Plan - High Expression of Thiopeptides and their Analogs (2022YFC2105000-03, 2022–2026), the National Agricultural Science and Technology Innovation Program (ASTIP) of Chinese Academy of Agricultural Sciences (CAAS-ASTIP-2017-FRI-02, 2013), and its key projects of Alternatives to Antibiotics for Animal (Grant No. CAAS-ZDRW202111, 2021–2023) and Feed (Grant No. CAAS-ZDXT2018008, 2018–2020) Usages. OF was supported by CAPES, CNPq, FAPDF, and FUNDECT. CF-N was supported by the AIChE Foundation, a BBRF Young Investigator Grant, the Dean's Innovation Fund of the Perelman School of Medicine at the University of Pennsylvania, and other funds from the National Institute of General Medical Sciences of the National Institutes of Health (R35GM138201) and the Defense Threat Reduction Agency (DTRA; HDTRA11810041 and HDTRA1-21-1-0014).
Acknowledgments
We would like to sincerely thank a total of 145 authors of 22 papers and over 70 peer editors and reviewers for their valuable professional contributions into the second volume of this Research Topic - Community Series in Antimicrobial Peptides: Molecular Design, Structure Function Relationship, and Biosynthesis Optimization, along with the staff of Frontiers in Microbiology, and also team supports of four Topic editors JW, RA, OF, and CF-N, including Dr. Ruoyu Mao's contribution into the first draft text of “about this Research Topic” under guide and direction of JW.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ageitos, L., Torres, M., and de la Fuente-Nunez, C. (2022). Biologically active peptides from venoms: applications in antibiotic resistance, cancer, and beyond. Int. J. Mol. Sci. 23, 15437. doi: 10.3390/ijms232315437
Aminov, R. (2022). Editorial: insights in antimicrobials, resistance, and chemotherapy: 2021. Front. Microbiol. 13:1037326. doi: 10.3389/978-2-83250-596-0
Cao, J., de la Fuente-Nunez, C., Ou, R. W., Torres, M., Pande, S. G., Sinskey, A. J., et al. (2018). Yeast-based synthetic biology platform for antimicrobial peptide production. ACS Synth. Biol. 7, 896–902. doi: 10.1021/acssynbio.7b00396
Cardoso, M. H., Meneguetti, B. T., Costa, B. O., Buccini, D. F., Oshiro, K. G. N., Preza, S. L. E., et al. (2019a). Non-lytic antibacterial peptides that translocate through bacterial membranes to act on intracellular targets. Int. J. Mol. Sci. 20, 4877. doi: 10.3390/ijms20194877
Cardoso, M. H., Orozco, R. Q., Rezende, S. B., Rodrigues, G., Oshiro, K. G. N., Cândido, E. S., et al. (2020). Computer-aided design of antimicrobial peptides: are we generating effective drug candidates? Front. Microbiol. 10, 3097.doi: 10.3389/fmicb.2019.03097
Cardoso, M. H., Santos, V., Costa, B. O., Buccini, D. F., Rezende, S. B., Porto, W. F., et al. (2019b). A short peptide with selective anti-biofilm activity against Pseudomonas aeruginosa and Klebsiella pneumoniae carbapenemase-producing bacteria. Microb. Pathog. 135, 103605–103605. doi: 10.1016/j.micpath.2019.103605
de Oliveira, K. B. S., Leite, M. L., Rodrigues, G. R., Duque, H. M., da Costa, R. A., Cunha, V. A., et al. (2020). Strategies for recombinant production of antimicrobial peptides with pharmacological potential. Exp. Rev. Clin. Pharmacol. 13, 367–390. doi: 10.1080/17512433.2020.1764347
Deng, T., Ge, H., He, H., Liu, Y., Zhai, C., and Feng, L. (2017). The heterologous expression strategies of antimicrobial peptides in microbial systems. Protein Expres. Purif. 140, 5259. doi: 10.1016/j.pep.2017.08.003
Ganz, T. (2003). Defensins: antimicrobial peptides of innate immunity. Nat. Rev. Immunol. 3, 710–720. doi: 10.1038/nri1180
Gutierrez, A., Jain, S., Bhargava, P., Hamblin, M., Lobritz, M. A., and Collins, J. J. (2017). Understanding and sensitizing density-dependent persistence to quinolone antibiotics. Mol. Cell 68, 1147–1154.e3. doi: 10.1016/j.molcel.2017.11.012
Hao, Y., Wang, J., de la Fuente-Nunez, C., and Franco, O. L. (2022). Editorial: antimicrobial peptides: molecular design, structure-function relationship, and biosynthesis optimization. Front. Microbiol. 13, 888540. doi: 10.3389/fmicb.2022.888540
Hao, Y., Yang, N., Teng, D., Wang, X. M., Mao, R. Y., and Wang, J. H. (2018). A review of the design and modification of lactoferricins and their derivatives. Biometals 31,331–341. doi: 10.1007/s10534-018-0086-6
Jiang, Y. J., Chen, Y. Y., Song, Z. Y., Tan, Z. Z., and Cheng, J. J. (2021). Recent advances in design of antimicrobial peptides and polypeptides toward clinical translation. Adv. Drug. Deliver. Rev. 170, 261–280. doi: 10.1016/j.addr.2020.12.016
Lazzaro, B. P., Zasloff, M., and Rolff, J. (2020). Antimicrobial peptides: application informed by evolution. Science 368, 487–492. doi: 10.1126/science.aau5480
Li, B., Yang, N., Wang, X., Hao, Y., Mao, R., Li, Z., et al. (2020). An enhanced variant designed from DLP4 cationic peptide against Staphylococcus aureus CVCC 546. Front. Microbiol. 11, 1057. doi: 10.3389/fmicb.2020.01057
Li, Z., Mao, R., Teng, D., Hao, Y., Chen, H., Wang, X., et al. (2017a). Antibacterial and immunomodulatory activities of insect defensins-DLP2 and DLP4 against multidrug-resistant Staphylococcus aureus. Sci. Rep. 7, 12124. doi: 10.1038/s41598-017-10839-4
Li, Z., Wang, X. M., Wang, X., Teng, D., Mao, R. Y., Hao, Y., et al. (2017b). Research advances on plectasin and its derivatives as new potential antimicrobial candidates. Process Biochem. 56,62–70. doi: 10.1016/j.procbio.2017.02.006
Liu, H., Yang, N., Teng, D., Mao, R., Hao, Y., Ma, X., et al. (2021). Design and pharmacodynamics of recombinant fungus defensin NZL with improved activity against Staphylococcus hyicus in vitro and in vivo. Int. J. Mol. Sci. 22, 5435. doi: 10.3390/ijms22115435
Ma, X., Yang, N., Mao, R., Hao, Y., Yan, X., Teng, D., et al. (2021). The pharmacodynamics study of insect defensin DLP4 against toxigenic Staphylococcus hyicus ACCC 61734 in vitro and vivo. Front. Cell. Infect. Microbiol. 11, 638598. doi: 10.3389/fcimb.2021.638598
Mao, R. Y., Teng, D., Wang, X. Y., Zhang, Y., Jiao, J., Cao, X. T., et al. (2014). Optimization of expression conditions for a novel NZ2114-derived antimicrobial peptide-MP1102 under the control of the GAP promoter in Pichia pastoris X-33. BMC Microbiol. 15, 57. doi: 10.1186/s12866-015-0389-5
Mccall, I. C., Shah, N., Govindan, A., Baquero, F., and Levin, B. R. (2019). Antibiotic killing of diversely generated populations of nonreplicating bacteria. Antimicrob. Agents Chemother. 63, e02360–e02318. doi: 10.1128/AAC.02360-18
Melo, M., Maasch, J., and de la Fuente-Nunez, C. (2021). Accelerating antibiotic discovery through artificial intelligence. Commun. Biol. 4, 1050. doi: 10.1038/s42003-021-02586-0
Nesa, J., Sadat, A., Buccini, D. F., Kati, A., Mandal, A. K., and Franco, O. L. (2020). Antimicrobial peptides from Bombyx mori: a splendid immune defense response in silkworms. RSC Adv.10, 512–523. doi: 10.1039/C9RA06864C
Palmer, N., Maasch, J., Torres, M., and de la Fuente-Nunez, C. (2021). Molecular dynamics for antimicrobial peptide discovery. Infect. Immun. 89, 703–720. doi: 10.1128/IAI.00703-20
Porto, W. F., Irazazabal, L., Alves, E. S. F., Ribeiro, S. M., Matos, C. O., Pires, Á. S., et al. (2018). In silico optimization of a guava antimicrobial peptide enables combinatorial exploration for peptide design. Nat. Commun. 9,1490. doi: 10.1038/s41467-018-03746-3
Rodríguez-Rojas, A., Makarova, O., and Rolff, J. (2014). Antimicrobials, stress and mutagenesis. PLoS Pathog. 10, e1004445. doi: 10.1371/journal.ppat.1004445
Rodríguez-Rojas, A., Moreno-Morales, J., Mason, A. J., and Rolff, J. (2018). Cationic antimicrobial peptides do not change recombination frequency in Escherichia coli. Biol. Lett. 14, 20180006. doi: 10.1098/rsbl.2018.0006
Savageau, M. A. (1983). Escherichia coli habitats, cell types, and molecular mechanisms of gene control. The American Naturalist. 122, 732–744. doi: 10.1086/284168
Spaulding, C. N., Klein, R. D., Ruer, S., Kau, A. L., Schreiber, H. L., Cusumano, Z. T., et al. (2017). Selective depletion of uropathogenic E. coli from the gut by a FimH antagonist. Nature 546, 528–532. doi: 10.1038/nature22972
Teng, D., Xi, D., Zhang, J., Wang, X. M., Mao, R. Y., Zhang, Y., et al. (2015). Multiple copy number of target gene enhances plectasin secretion in Pichia pastoris X-33. Process Biochem. 50, 553–560. doi: 10.1016/j.procbio.2015.01.010
Torres, M., Cao, J., Franco, O. L., Lu, T. K., and de la Fuente-Nunez, C. (2021). Synthetic biology and computer-based frameworks for antimicrobial peptide discovery. ACS Nano. 15, 2143–2164. doi: 10.1021/acsnano.0c09509
Torres, M., Melo, M., Flowers, L., Crescenzi, O., Notomista, E., and de la Fuente-Nunez, C. (2022). Mining for encrypted peptide antibiotics in the human proteome. Nat. Biomed. Eng. 6, 67–75. doi: 10.1038/s41551-021-00801-1
Travis, A., Chernova, O., Chernov, V., and Aminov, R. (2018). Antimicrobial drug discovery: lessons of history and future strategies. Exp. Opin. Drug Disc. 13, 983–985. doi: 10.1080/17460441.2018.1515910
Vogel, V., Bauer, R., Mauerer, S., Schiffelholz, N., Haupt, C., Seibold, G. M., et al. (2021). Angicin, a novel bacteriocin of Streptococcus anginosus. Sci. Rep. 11, 24377. doi: 10.1038/s41598-021-03797-5
Wan, F., Kontogiorgos-Heintz, D., and de la Fuente-Nunez, C. (2022). Deep generative models for peptide design. Digit. Discov. 1, 195–208. doi: 10.1039/D1DD00024A
Wang, G., Li, X., and Wang, Z. (2016). APD3: the antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 44, 1087–1093. doi: 10.1093/nar/gkv1278
Wang, X., Teng, D., Wang, X. M., Hao, Y., Chen, H. X., Mao, R. Y., et al. (2019). Internalization, distribution, and activity of peptide H2 against the intracellular multidrug-resistant bovine mastitis-causing bacterium Staphylococcus aureus. Sci. Rep. 9, 7968. doi: 10.1038/s41598-019-44459-x
Wang, X., Wang, X. M., Teng, D., Hao, Y., and Jianhua, W. (2017). Research and development on lactoferrin and its derivatives in China from 2011–2015. Cell Biochem. Biol. 95, 162–170. doi: 10.1139/bcb-2016-0073
Wang, X., Wang, X. M., Teng, D., Mao, R. Y., Hao, Y., Yang, et al. (2018). Increased intracellular activity of MP1102 and NZ2114 against Staphylococcus aureus in vitro and in vivo. Sci. Rep. 8, 4204. doi: 10.1038/s41598-018-22245-5
Wang, Z., Yang, N., Teng, D., Hao, Y., Li, T., Han, H., et al. (2022). Resistance response to arenicin derivatives in Escherichia coli. Appl. Microbiol. Biotechnol. 106, 211–226. doi: 10.1007/s00253-021-11708-x
Wibowo, D., and Zhao, C. X. (2019). Recent achievements and perspectives for large-scale recombinant production of antimicrobial peptides. Appl. Microbiol. Biotechn. 103, 659671. doi: 10.1007/s00253-018-9524-1
Wu, C. L., Peng, K. L., Yip, B. S., Chih, Y. H., and Cheng, J. W. (2021). Boosting synergistic effects of short antimicrobial peptides with conventional antibiotics against resistant bacteria. Front. Microbiol. 12, 747760. doi: 10.3389/fmicb.2021.747760
Wu, Y., Yang, N., Mao, R. Y., Hao, Y., Teng, D., and Wang, J. H. (2022). In vitro pharmacodynamics and bactericidal mechanism of fungal defensinderived peptides NZX and P2 against Streptococcus agalactiae. Microorganisms 10, 881. doi: 10.3390/microorganisms10050881
Yang, N., Teng, D., Mao, R. Y., Hao, Y., Wang, X., Wang, Z. L., et al. (2019). A recombinant fungal defensin-like peptide-P2 combats multidrug-resistant Staphylococcus aureus and biofilms. Appl. Microbiol. Biotechnol. 103, 5193–5213. doi: 10.1007/s00253-019-09785-0
Yu, G., Baeder, D. Y., Regoes, R. R., and Rolff, J. (2018). Predicting drug resistance evolution: insights from antimicrobial peptides and antibiotics. Proc. R. Soc. B Biol. Sci. 285, 5–7. doi: 10.1098/rspb.2017.2687
Zhang, J., Yang, Y., Teng, D., Tian, Z., Wang, S., and Wang, J. (2011). Expression of plectasin in Pichia pastoris and its characterization as a new antimicrobial peptide against Staphyloccocus and Streptococcus. Protein Expr. Purif. 78, 189–196. doi: 10.1016/j.pep.2011.04.014
Zhang, Y., Teng, D., Mao, R., Wang, X., Xi, D., Hu, X., et al. (2014). High expression of a plectasin-derived peptide NZ2114 in Pichia pastoris and its pharmacodynamics, postantibiotic and synergy against Staphylococcus aureus. Appl. Microbiol. Biotechnol. 98, 681–694. doi: 10.1007/s00253-013-4881-2
Keywords: antimicrobial peptides, mining and learning, structure function relationship, heterologous expression, druggability
Citation: Yang N, Aminov R, Franco OL, de la Fuente-Nunez C and Wang J (2023) Editorial: Community series in antimicrobial peptides: Molecular design, structure function relationship and biosynthesis optimization. Front. Microbiol. 14:1125426. doi: 10.3389/fmicb.2023.1125426
Received: 16 December 2022; Accepted: 03 January 2023;
Published: 16 January 2023.
Edited and reviewed by: Guangshun Wang, University of Nebraska Medical Center, United States
Copyright © 2023 Yang, Aminov, Franco, de la Fuente-Nunez and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianhua Wang,  d2FuZ2ppYW5odWFAY2Fhcy5jbg==;
d2FuZ2ppYW5odWFAY2Fhcy5jbg==;  d2FuZ2ppYW5odWEucGVraW5nQHFxLmNvbQ==; Rustam Aminov,
d2FuZ2ppYW5odWEucGVraW5nQHFxLmNvbQ==; Rustam Aminov,  cnVzdGFtLmFtaW5vdkBnbWFpbC5jb20=; Octavio Luiz Franco,
cnVzdGFtLmFtaW5vdkBnbWFpbC5jb20=; Octavio Luiz Franco,  b2NmcmFuY29AZ21haWwuY29t; Cesar de la Fuente-Nunez,
b2NmcmFuY29AZ21haWwuY29t; Cesar de la Fuente-Nunez,  Y2Z1ZW50ZUB1cGVubi5lZHU=
Y2Z1ZW50ZUB1cGVubi5lZHU=
 Na Yang1,2,3
Na Yang1,2,3 Rustam Aminov
Rustam Aminov Octavio Luiz Franco
Octavio Luiz Franco Cesar de la Fuente-Nunez
Cesar de la Fuente-Nunez Jianhua Wang
Jianhua Wang