- 1School of Basic Medical Sciences, Guizhou Medical University, Guiyang, China
- 2Department of Clinical Laboratory, The First People’s Hospital of Guiyang, Guiyang, Guizhou, China
- 3School of Public Health, The Key Laboratory of Environmental Pollution Monitoring and Disease Control, Ministry of Education, Guizhou Medical University, Guiyang, China
- 4Laboratory of Bacterial Infectious Disease of Experimental Center, Guizhou Provincial Centre for Disease Control and Prevention, Guiyang, Guizhou, China
Introduction: One of the main pathogens responsible for human hand, foot, and mouth disease (HFMD), coxsackievirus A16, has put young children’s health at danger, especially in countries in the Asia-Pacific region. Early quick identification is essential for the avoidance and control of the disorder since there are no vaccinations or antiviral medications available to prevent and manage CVA16 infection.
Methods: Here, we describe the creation of an easy, speedy, and accurate CVA16 infection detection approach using lateral flow biosensors (LFB) and reverse transcriptionmultiple cross displacement amplification (RT-MCDA). A group of 10 primers was developed for the RT-MCDA system in order to amplify the genes in an isothermal amplification device while targeting the highly conserved region of the CVA16 VP1 gene. Then, without requiring any extra tools, RT-MCDA amplification reaction products might well be detected by visual detection reagent (VDR) and LFB.
Results: The outcomes showed that 64°C within 40 min was the ideal reaction setting for the CVA16-MCDA test. Target sequences with <40 copies might be found using the CVA16-MCDA. There was no cross-reaction among CVA16 strains and other strains. The findings demonstrated that the CVA16-MCDA test could promptly and successfully identify all of the CVA16-positive (46/220) samples identified by the traditional real-time quantitative polymerase chain reaction (qRT-PCR) assays for 220 clinical anal swab samples. The whole process, such as the processing of the sample (15 min), the MCDA reaction (40 min), and the documenting of the results (2 min), could be finished in 1 h.
Conclusion: The CVA16-MCDA-LFB assay, which targeted the VP1 gene, was an efficient, simple, and highly specific examination that might be used extensively in rural regions’ basic healthcare institutions and point-of-care settings.
Introduction
Coxsackievirus A16 (CVA16) is a non-enveloped positive single-stranded RNA molecule of about 7,400 nucleotides (nt), which consists of a lengthy open reading frame (ORF) flanked by 5′ and 3′ untranslated regions (UTRs). It belongs to the genus Enterovirus and the family Picornaviridae (Sickles et al., 1955). In South Africa in 1955, CVA16 was first isolated. For the last several years, enterovirus 71 (EVA71), CVA6, CVA16, CVA10, and other enteroviruses have all been main pathogens responsible for HFMD in young children and infants (Xu et al., 2012; Zhang et al., 2019). Although CVA16 infections often produce minimal symptoms, in previous decades, outbreaks have been related to more serious diseases, including encephalitis, aseptic meningitis, and even fatal cases of CVA16 (Sun et al., 2014; Li et al., 2022). Additionally, CVA16 infection may result in a wide range of medical symptoms, including herpangina and upper and lower respiratory disorders, as well as serious consequences, including meningitis, myelitis, paralysis, encephalitis, and even death (Chen et al., 2019).
According to earlier research, CVA16 infection accounts for around 21% of serious HFMD patients with neurological consequences (Xu et al., 2012). Affected regions face a major public health risk from CVA16 viral infection. No efficient antiviral medications versus CVA16 have been produced to yet. In order to avoid severe cases of HFMD, lower death rates, and stop the spread of the illness in China, it is crucial to create a simple, quick, and reliable identification test for CVA16.
The two main principles of standard CVA16 detection techniques are viral isolation culture and serological testing (Xu et al., 2011). Furthermore, due to complicated processes, poor specificities, and sensitivities, which are either time-consuming or have a high false positive rate, viral isolation and serodiagnosis are not quick and accurate enough. The quantitative real-time fluorescent reverse transcription polymerase chain reaction (qRT-PCR), which is based on nucleic acid amplification, has a significant function in the identification of CVA16 and has fixed the issues with the other techniques stated above (Cui et al., 2013). A qRT-PCR technique, however, needs costly laboratory apparatus and knowledgeable operators, which are not obtainable in resource-limited settings. To resolve these issues, it’s crucial to develop a quick, cheap, and visible CVA16 detection technique.
Multiple cross displacement amplification (MCDA), a novel, inexpensive, quick, easy, and effective isothermal amplification approach identical to LAMP that has been widely employed in the identification of various pathogens, such as bacteria, viruses, and fungi, has been developed to address the limitations of traditional identification (Li et al., 2019; Wang et al., 2021; Chen et al., 2021b). To accomplish targeted amplification using the MCDA approach, 10 primers that bind to 10 different regions of the target sequence must be designed. A number of methods, like turbidimetry, agarose gel, and colorimetric markers (malachite green reagent), have been used to evaluate MCDA results in the past (Wang et al., 2015). However, the findings of these approaches may be easily misunderstood since it is difficult to discriminate between non-specific and specific amplification. The validation of nucleic acid labeled amplification products may be performed using a simple, sensitive, and highly focused lateral flow biosensor (LFB), which was developed and implemented to address these shortcomings (Jiang et al., 2022). Compared to these techniques, LFB is more precise, objective, inexpensive, and simple.
The current study’s objective was to establish a CVA16-MCDA-LFB approach that is quick, portable, sensitive, user-friendly, and can ensure accurate CVA16 identification. Additionally, we used the CVA16-MCDA-LFB assay to identify the rectal swabs in this case, confirming its appropriateness for point-of-care quick detection of HFMD.
Materials and methods
Reagents and instruments
Kits for extracting virus RNA nucleic acids were acquired from Tianlong Technology Co., Ltd. (Xi an tian long, ShanXi, China). From Beijing Baitaike Biotech Co., Ltd., a genomic DNA kit for nucleic acid extraction and purification was obtained (Beijing, China). HUIDEXIN Biotech Co., Ltd. (Tianjin, China) supplied the visual detection reagent (VDR), the polymer nanoparticle-based lateral flow biosensor (LFB), and isothermal amplification kits (RNA/DNA Universal). CVA16 qPCR diagnosis kits were acquired from Jin Hao Gene Co., Ltd. (Beijing, China). Nucleic acid concentration and purity were assessed using the Nano-Drop ND-2000 (Beijing, China) at A260/280.
Viral strains and clinical specimens
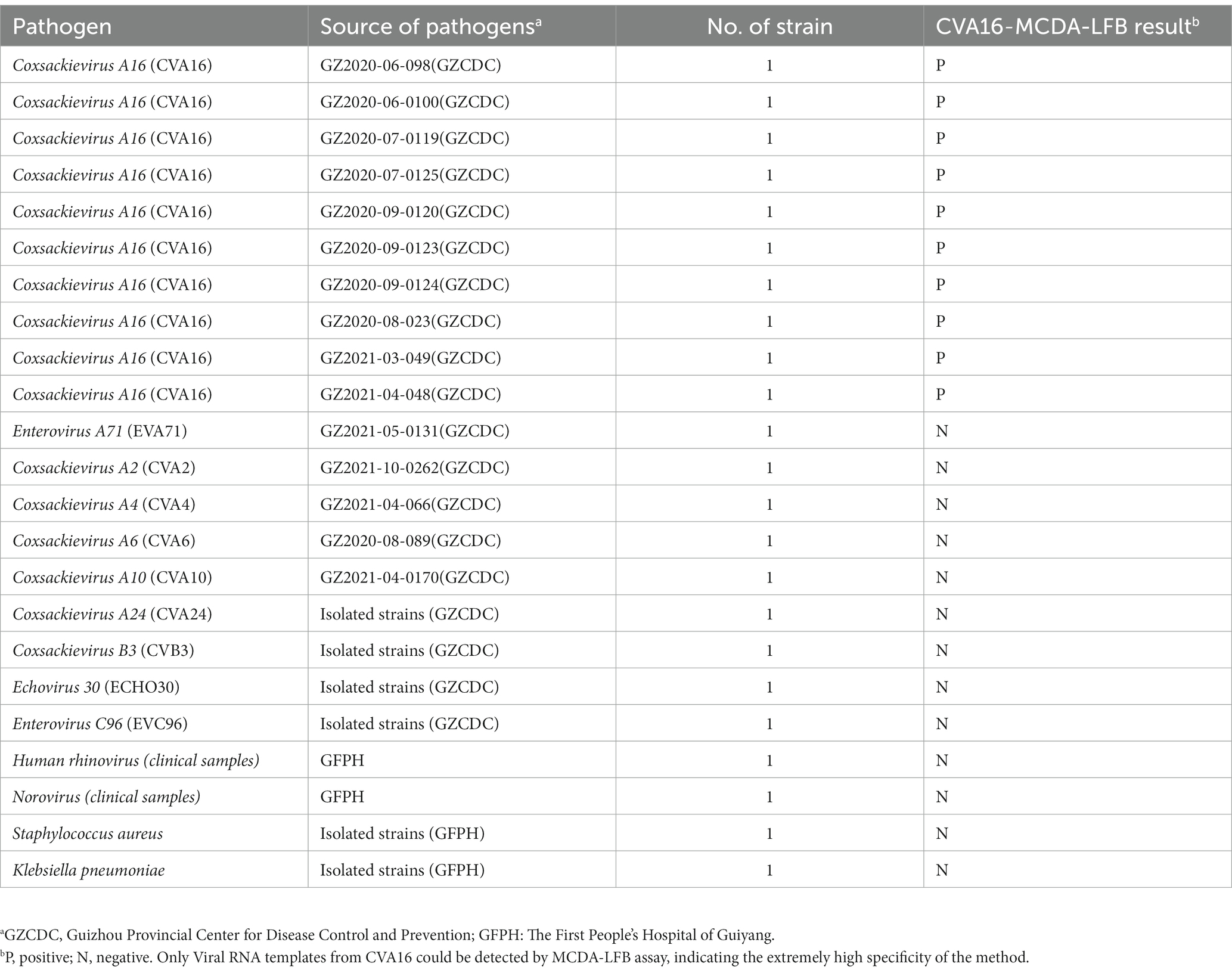
During the monitoring of HFMD in Guizhou Province from 2019 to 2021, enterovirus strains were obtained from individuals with HFMD there. To extract enteroviruses, human rhabdomyosarcoma (RD) cells were employed. Specimens that caused a cytopathic effect (CPE) following three passages were deemed positive and kept at −80°C until RNA purification. At 36°C, several bacterial strains were inoculated into the nutrient agar plate. All colonies were obtained independently after 2 days of pure culture. Utilizing a panel of common enteroviruses obtained from HFMD patients and validated by qRT-PCR, this assay’s specificity was assessed. In Table 1, every pathogen was illustrated. Additionally, 220 clinical specimens were taken from patients who had been hospitalized at the First People’s Hospital of Guiyang with suspected HFMD. All clinical samples were examined concurrently by commercial qRT-PCR and MCDA tests in order to assess CVA16-MCDA clinical performance. The First People’s Hospital of Guiyang’s Ethics Committee provided its authorization for this investigation (Ethical approval No.G2020-S001). According to the Helsinki Declaration, patients who supplied anal swab specimens provided written informed permission.
Nucleic acid extraction
Following the manufacturer’s recommendations, viral RNA was extracted from 200 μL of cell culture supernatant and anal swabs of suspected HFMD cases using an RNA extraction kit (Xi an tian long, ShanXi, China). Viral RNA was then eluted in 50 μL of nuclease-free water for use instantly or storage at −80°C. According to the manufacturer’s directions, DNA was isolated from each strain utilizing Baitaike DNA extraction kit (Beijing, China). Purity and concentration were then determined using a Nanodrop 2000 (Beijing, China) at A260/280. Before usage, the extracted DNA was kept at −20°C.
CVA16-MCDA primer design and standard plasmid construction
Primer Explorer 51 was used to create a set of 10 primers depending on the conserved area of the capsid protein VP1 gene (Genbank Accession: GQ279371.1) of CVA16 and the pathway of MCDA (Wang et al., 2015). These primers’ hybrids and hairpin structures were examined utilizing design tools for integrated DNA technologies. Using the BLAST analysis method, the CVA16-MCDA primers’ specificity was validated. Tsingke Biotechnology Co., Ltd. (Kunming, China) with HPLC purification grade was employed to create all primers used. Tsingke Biotechnology Co., Ltd. (Kunming, China) was employed to chemically synthesize and clone a 344-bp VP1 coding region containing the target sequence into the pUC57 plasmid (herein referred to as pUC57-CVA16-VP1), which contained the amplification target of the aforementioned MCDA primers, in order to enhance the MCDA assay and assess the limit of detection. Figure 1 and Table 2 illustrate the sequence details, locations, and alterations of the primers employed in this investigation.
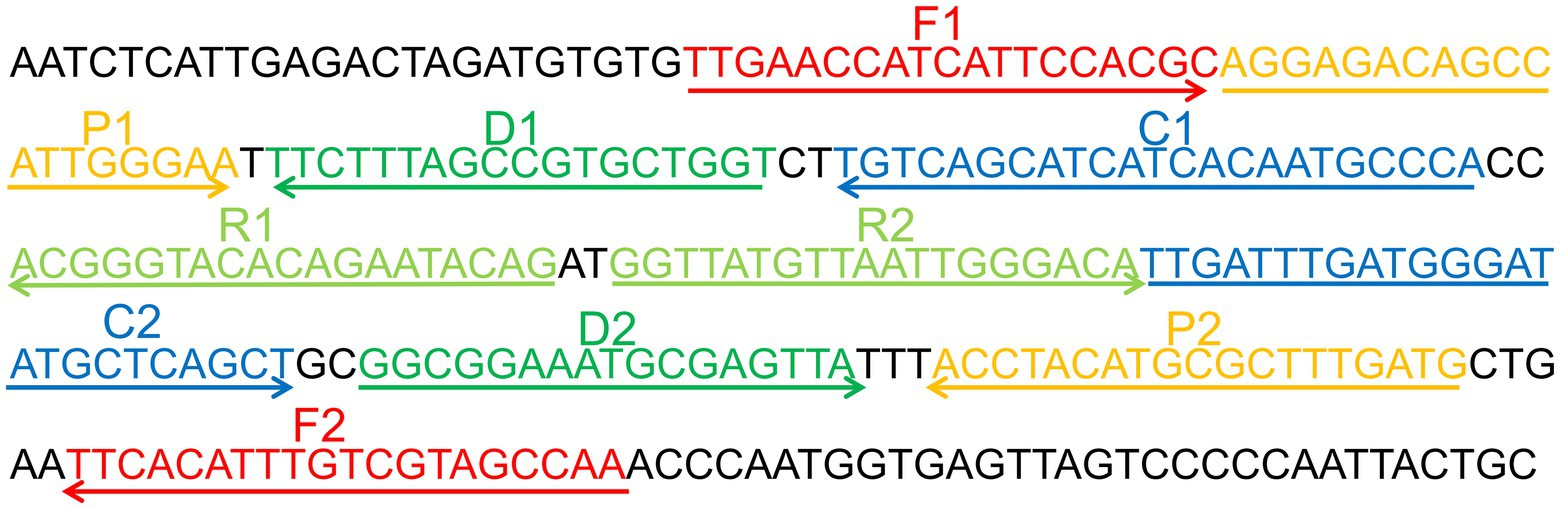
Figure 1. Appropriate primer design for CVA16-MCDA-LFB assay. The nucleotide sequences of the sense strand of VP1 are listed. Right arrows and left arrows indicate sense and complementary sequences that are used.
The standard MCDA-LFB assay
The CVA16-MCDA amplification was conducted in a one step reaction of 25 μL, including the final concentration of each replacement primers (F1 and F2) contains 0.4 μM and 0.8 μM each of amplification primers (C1∗, C2, R1, R2, D1∗, and D2) and 1.6 μM each of cross primers (CP1 and CP2), 12.5 μL of 2 × reaction mix, 1 μL (10 U) Bst 4.0 DNA polymerase, 1 μL (10 U) of AMV reverse transcriptase (only utilized for RNA temperate) and template (1 μL for each standard plasmid, 5 μL RNA for clinical samples), the total volume was made up to 25 μL with double distilled water (ddH2O). Also, 1 μL ddH2O was functioned as blank control, and virus RNA of EVA71 and CVA6 was employed as a negative control. To run program, the mixtures were incubated for 1 h at 64°C and then for 5 min at 80°C to stop the reaction.
CVA16-MCDA products detection
The CVA16-MCDA products were identified and validated utilizing three different MCDA detection techniques, including real-time turbidity LA-500 (Eiken Chemical Co., Ltd., Japan), LFB, and colorimetric indicator VDR. Turbidity >0.1 was regarded as a positive outcome for the real-time turbidity approach. The positive reaction solution changed noticeably from colorless to light green when VDR was used, whereas the negative and blank controls maintained colorless. While the CL and TL showed simultaneously during the LFB test, demonstrating positive findings, only the CL was seen during the negative amplification.
Optimal temperature of CVA16-MCDA assay
The amplification effectiveness of the MCDA reaction system was significantly influenced by temperature. The CVA16-MCDA assay’s optimal amplification temperature range was 61–68°C (at 1°C intervals). As negative controls (NC), amplification mixes containing 5 μL of EVA71 template were employed. As a blank control, 1 μL of double-distilled water (DW) was utilized. The Real-time Turbidimeter LA-500 was used to detect the CVA16-MCDA amplified products, and a threshold value of >0.1 within 1 h was considered a positive response.
Limit of detection and optimal isothermal amplification time of CVA16-MCDA assay
The initial pUC57-CVA16-VP1 concentration was 4 × 108 copies/μL in order to assess LoD of MCDA assay for CVA16. Then, 10-fold serial dilutions with TE buffer (4 × 106 to 4 × 100 copies) of pUC57-CVA16-VP1 DNA were employed to evaluate LoD of the CVA16-MCDA assays. In the amplification reaction system, 1 μL of the diluted plasmid was supplied as a template. Double distilled water was utilized as the negative control since the plasmid was employed as a template, and the technique for sensitivity testing did not include AMV reverse transcriptase. CVA16-MCDA reaction time was also optimized, and four different times (20–50 min, with 10 min intervals) were compared under identical reaction situations. All amplified products at each time point, including 20, 30, 40, and 50 min, were observed utilizing LFB, and each amplification time was evaluated three times minimum in the present investigation.
Specificity of CVA16-MCDA assay
Ten CVA16 strains and 13 non-CVA16 pathogens that were amplified under ideal circumstances were used to compare the analytical specificity of the CVA16-MCDA test. LFB was used to observe all MCDA products, and each test was performed at three times minimum.
Application of CVA16-MCDA-LFB detection in clinical samples
Typically 220 anal swab specimens from the First People’s Hospital in Guiyang were obtained to investigate the use of CVA16-MCDA detection in clinical samples. The samples were identified employing CVA16-MCDA-LFB and the qRT-PCR assay, and the outcomes were compared. From Jin hao Gene (Beijing, China), commercial qRT-PCR test kits for enteroviruses were acquired. The 7500 real-time PCR technology (Applied Biosystems, United States) was used to perform the qRT-PCR reactions. All processes were conducted following the manufacturer’s directions. The CVA16-MCDA-LFB determination was conducted, as previously mentioned.
Results
Confirmation of effectiveness of MCDA-LFB assay for the detection of CVA16
To determine if the MCDA primers (Table 2) for the CVA16 assay are valid. pUC57-CVA16-VP1 DNA was employed as a template for CVA16-MCDA mixes, which were processed for 60 min at a constant temperature of 64°C. Then, two alternative techniques, including VDR and LFB, were utilized to monitor the amplification products. The outcomes revealed that the nucleic acid from CVA16 provided positive results but not those from CVA6, EVA71, and the blank controls (Figures 2A,B). As a result, the CVA16-MCDA primers used in the present investigation to identify the VP1 gene were valid to create the CVA16-MCDA-LFB assay.
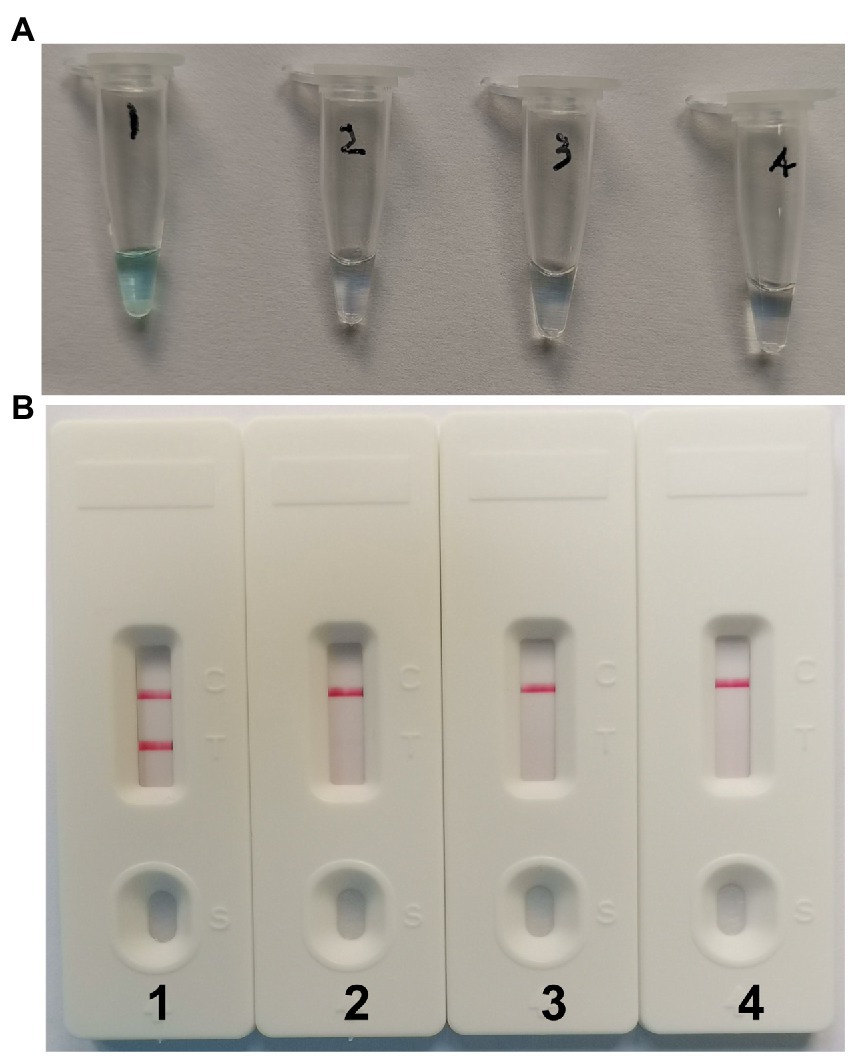
Figure 2. Identification and confirmation of CVA16-MCDA-LFB products. (A) The visible color changes of amplification products of CVA16-MCDA-LFB assay were analyzed by VDR. (B) The products of CVA16-MCDA-LFB were visually detected with Lateral flow biosensor. Tube 1/biosensor 1, positive amplification of pUC57-CVA16-VP1 DNA; tube 2/biosensor 2, negative control of virus RNA of EVA71; tube 3/biosensor 3, negative control of virus RNA of CVA6; tube 4/biosensor 4, blank control (double-distilled water, DW).
The optimal temperature of CVA16-MCDA-LFB assay
With 4 × 106 copies/μL of pUC57-CVA16-VP1 DNA as the template, the CVA16-MCDA reaction was conducted from 61 to 68°C to validate the ideal temperature for isothermal amplification. Real-time turbidity LA-500 was employed to observe every reaction. Typically 64°C was determined to be the ideal reaction temperature for CVA16-MCDA amplification since this temperature allowed for the quickest achievement of the 0.1 absorbance threshold value that indicated positive amplification from the CVA16-MCDA reaction (Figure 3). In this investigation, the following CVA16-MCDA-LFB reaction was performed at 64°C.
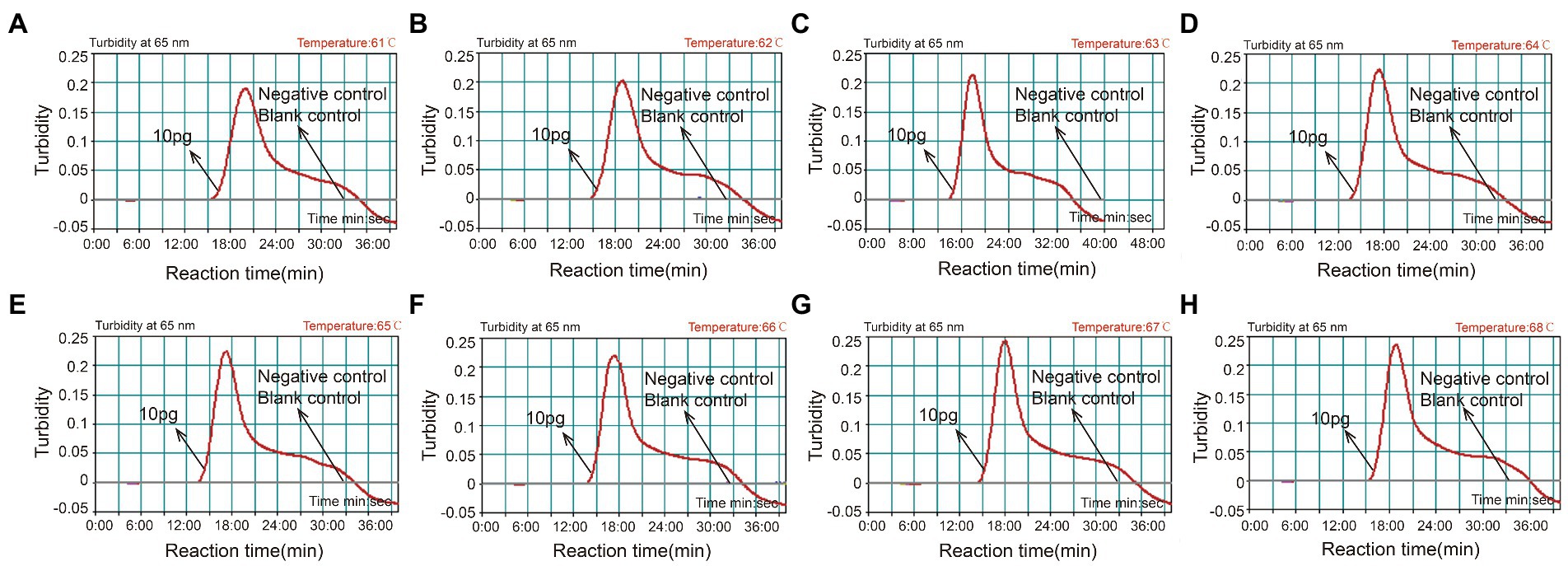
Figure 3. Optimal amplification temperature for CVA16-MCDA-LFB assay. By using a real-time measurement to monitor the turbidity of CVA16-MCDA-LFB reactions. The corresponding curves were displayed in the panels. The negative control was virus RNA of EVA71, and the blank control was sterile double-distilled water. Abscissa represents reaction time (min), ordinate represents turbidity. The threshold value was 0.1, and the turbidity >0.1 was considered as positive amplification. Eight kinetic curves (A-H) were generated from 61 to 68°C (1°C intervals), with 4 × 106 copies/μL of pUC57-CVA16-VP1 DNA per reaction.
Limit of detection and optimized time of MCDA for CVA16 detection
A serial dilution of the pUC57-CVA16-VP1 (4 × 106, 4 × 105, 4 × 104, 4 × 103, 4 × 102, 4 × 101 and 4 × 100 copies per microliter) was employed in MCDA assays to define CVA16-MCDA-LFB assay LoD. When the dilution passed 4 × 101 copies/μL, as illustrated in Figure 4, MCDA tubes displayed colorless and only control band in LFB. Bright blue MCDA tubes were shown for other dilutions, and two red bands—one representing the control line and the other representing the test line—were noticed in the LFB.
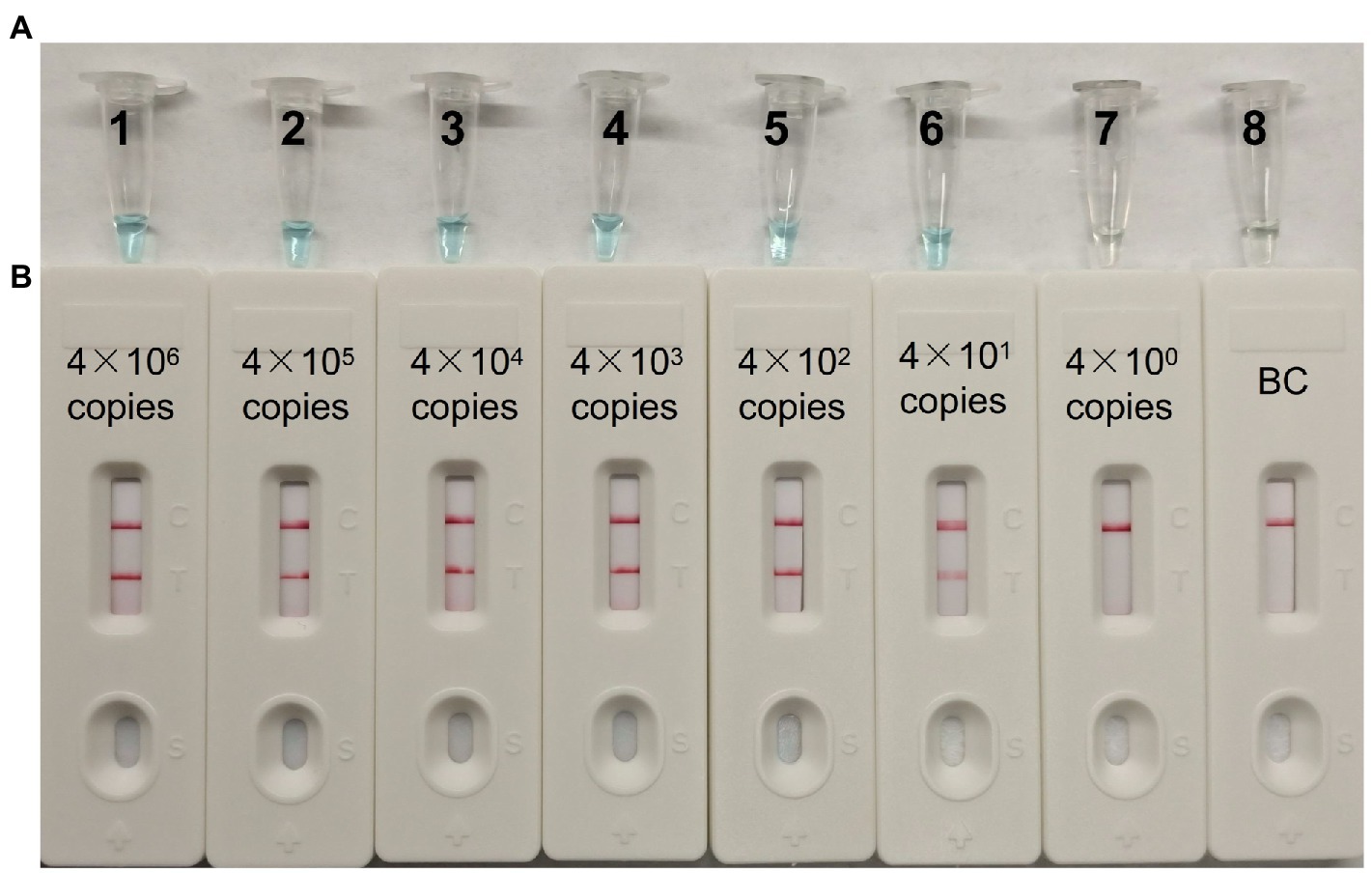
Figure 4. The detection limit of CVA16-MCDA-LFB assays. Two measurement techniques, including (A) a colorimetric indicator (VDR) and (B) lateral flow biosensor. A series of dilutions (4 × 106–4 × 100 copies) of pUC57-CVA16-VP1 DNA and a blank control (BC) double-distilled water were operated according to standard MCDA-LFB reactions.
For the CVA16-MCDA-LFB assay, four amplification times (20, 30, 40, and 50 min) were examined, respectively, at 64°C to determine the best amplification time. With LFB, the amplification products were observed. The outcomes showed that the pUC57-CVA16-VP1 LoD level (4 × 101 copies/μL) was evaluated while the amplification lasted 40 and 50 min (Figure 5). When the isothermal reaction was performed for 40 min at 64°C, the lowest template level (4 × 101 copies) showed two red bands (Test line and Control line). So, the ideal conditions for the remaining CVA16-MCDA-LFB tests in the present investigation were a reaction temperature of 64°C and an amplification period of 40 min. Thus, the whole diagnostic procedure of the CVA16-MCDA-LFB assay, including Sample collection (2 min), Virul RNA extraction (13 min), MCDA reaction (40 min) and result reporting (<2 min), can be completed within 60 min (Figure 6).
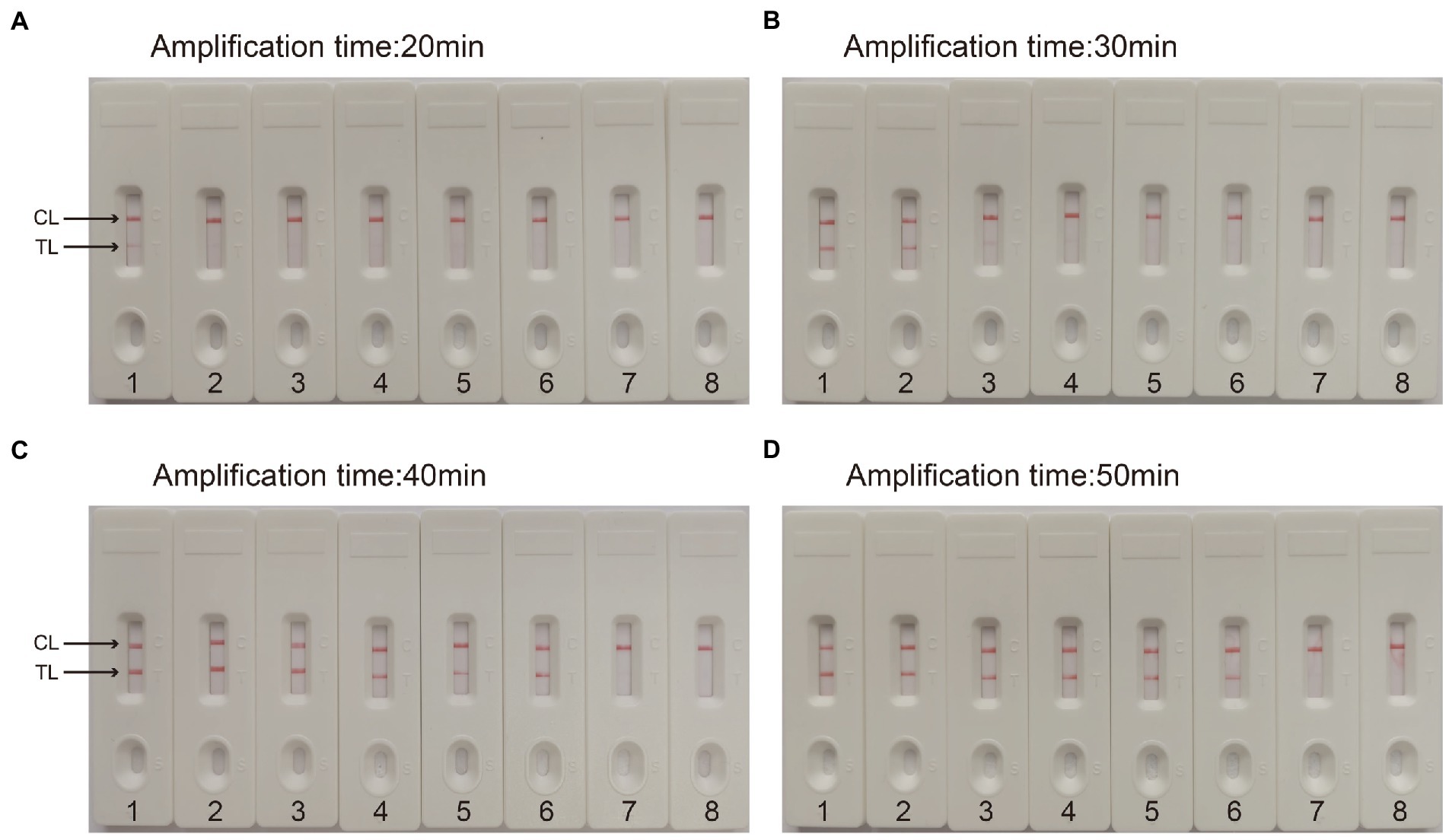
Figure 5. Optimal detection time required for CVA16-MCDA-LFB assay. Four different reaction times [(A) 20 min; (B) 30 min; (C) 40 min; (D) 50 min] were evaluated at 64°C. Biosensors 1–7 represent pUC57-CVA16-VP1 DNA levels of 4 × 106 to 4 × 100 copies per reaction, respectively; 8 represents a blank control double-distilled water.
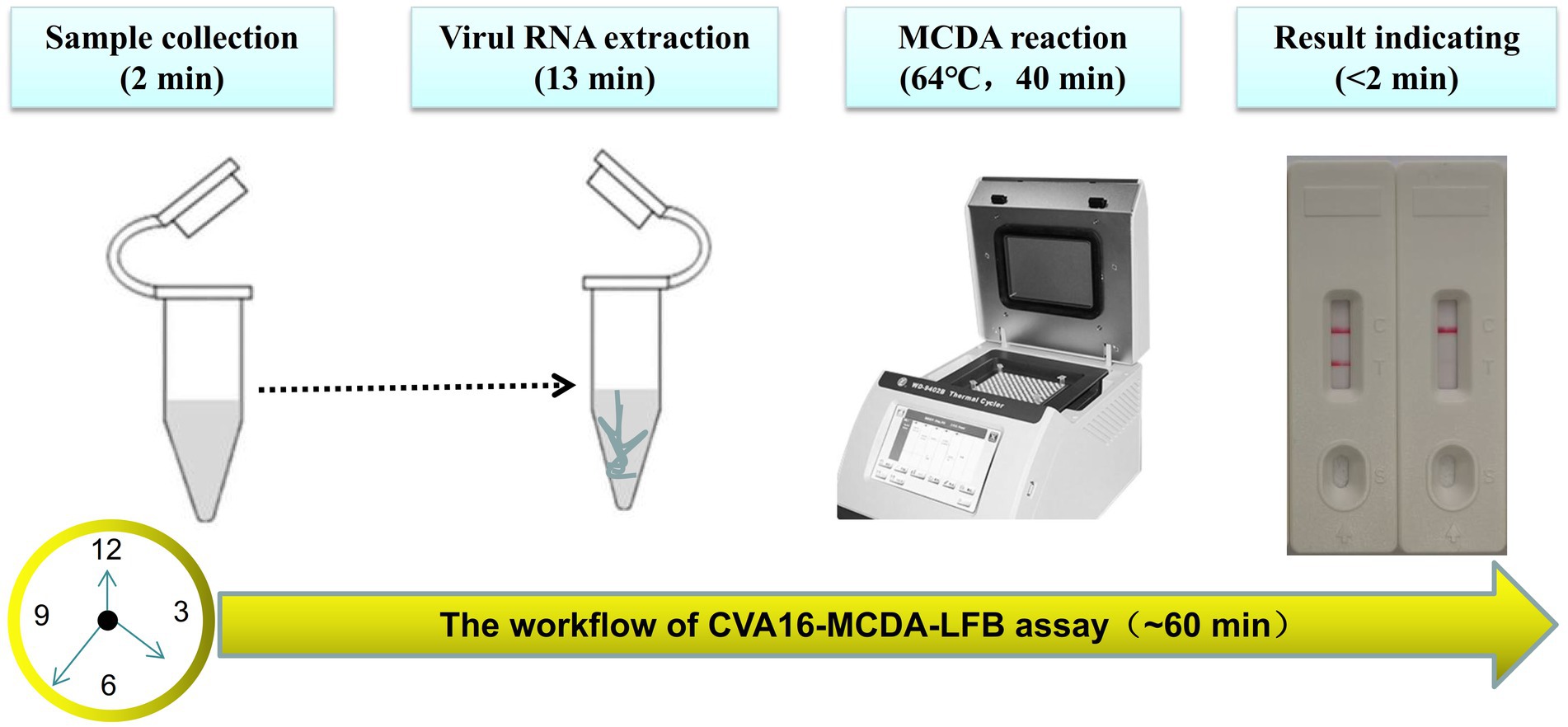
Figure 6. The workflow of CVA16-MCDA-LFB assay. Four steps, including sample collection (2 min), Virul RNA extraction (13 min), MCDA reaction (40 min) and result reporting (<2 min), are required for the CVA16-MCDA-LFB assay, and the total procedure can be completed within 60 min.
Specificity of MCDA-LFB for CVA16 detection
CVA16-MCDA-LFB test specificity was determined using 13 non-CVA16 pathogens and 10 isolated CVA16 strains (Table 1). The MCDA-LFB detection was conducted in the ideal situations indicated above. Employing LFB, two red lines were visible at the locations of Test line and Control line for the CVA16 strain, but only one line was observed at the position of C line for all non-CVA16 pathogens and the blank control (Figure 7), indicating that the CVA16-LAMP-LFB assay had 100% specificity.
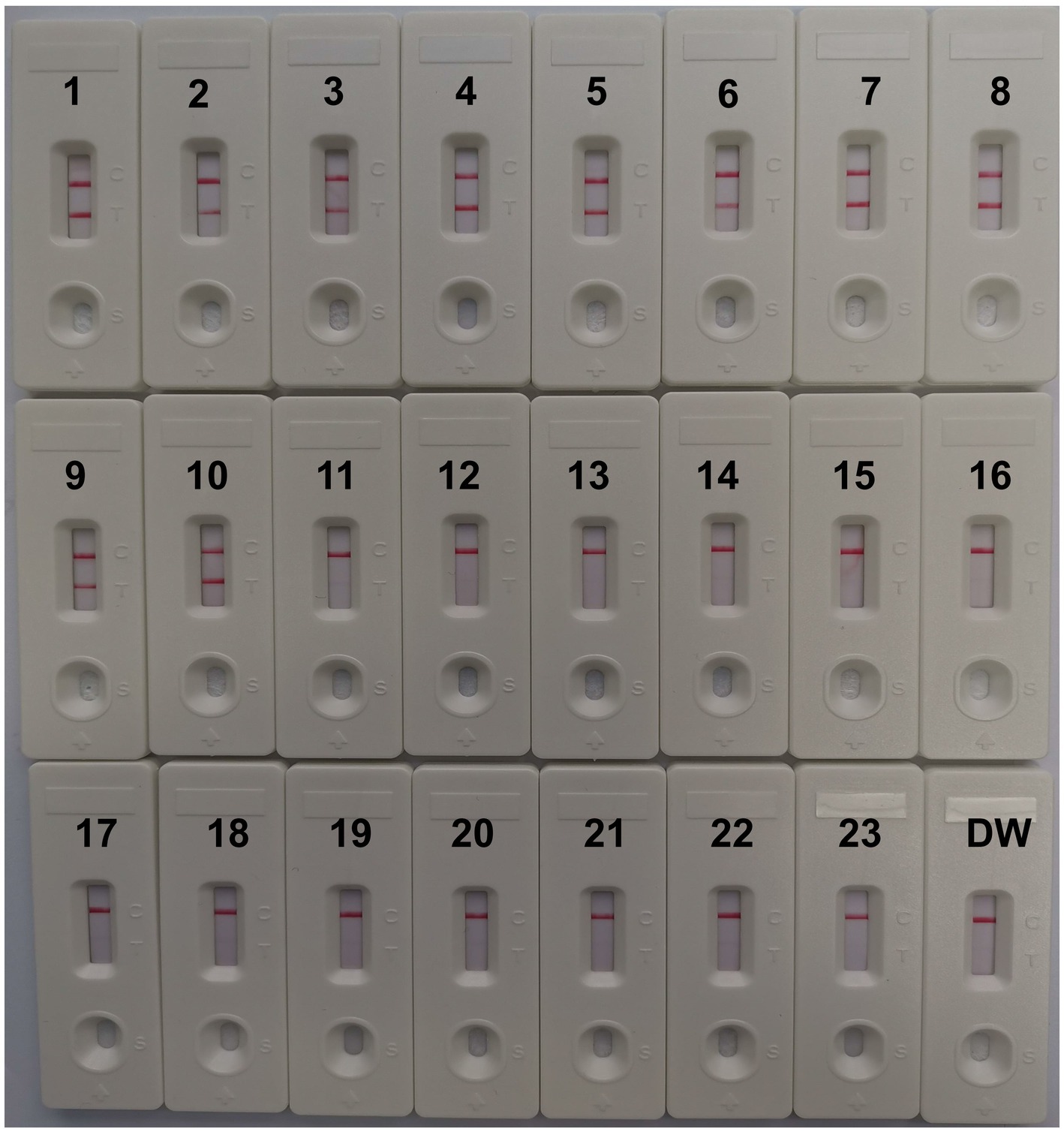
Figure 7. Specificity of LFB assays detecting CVA16-MCDA-LFB products. The CVA16-MCDA-LFB assay was evaluated with different genomic DNA as templates. Both the control line and the test line were visible in LFB for all CVA16, and only the control line was appeared in non-CVA16. 1–10, CVA16 isolated strains; 11, EVA71 isolated strains; 12, CVA2 isolated strains; 13, CVA4 isolated strains; 14, CVA6 isolated strains; 15, CVA10 isolated strains; 16, CVA24 isolated strains; 17, CVB3 isolated strains; 18, ECHO30 isolated strains; 19, EVC96 isolated strains; 20, Human rhinovirus clinical samples; 21, Norovirus clinical samples; 22, Staphylococcus aureus isolated strains; 23, Klebsiella pneumoniae isolated strains; 24, a blank control double-distilled water (DW).
Validation with clinical samples
Two hundred twenty suspected HFMD infection anal swab specimens were concurrently identified using CVA16-MCDA-LFB and real-time PCR (LoD: 500 copies) to further validate the use of CVA16-MCDA-LFB as a useful approach for CVA16 identification in clinics. Typically 46 of the 220 anal swab samples were positive for CVA16. Outcomes from the CVA16-MCDA-LFB assay perfectly matched those from the real-time PCR assay (Table 3). These results showed that the CVA16-MCDA-LFB assay provides a quick, accurate, and sensitive detection of CVA16 and can be used as a potential screening tool for CVA16 in a clinical and basic laboratory. It is particularly suitable for screening HFMD patients at the early stages of infection.
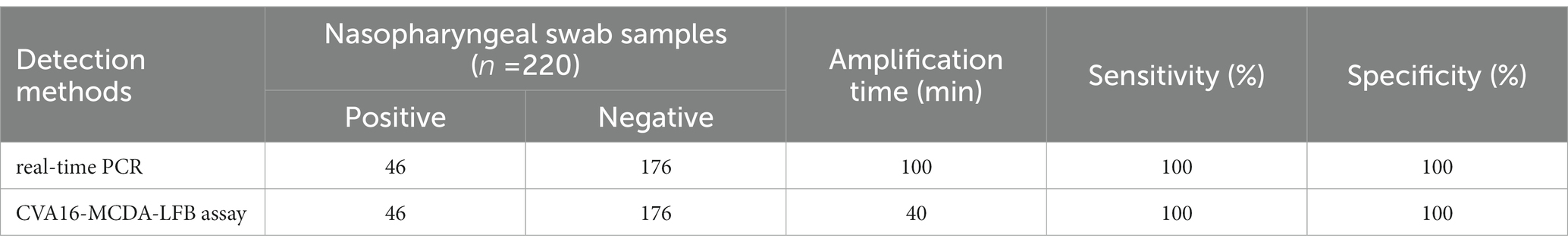
Table 3. Comparison of real-time PCR and CVA16-MCDA-LFB methods to identify CVA16 in clinical samples.
Discussion
One of the most prevalent diseases in the world is HFMD. According to studies by Zhuang et al. (2015) and Esposito and Principi (2018), it disproportionately affects young children under the age of five. The Chinese Ministry of Health closely monitors it and has classified it as a category III notifiable infectious illness since 2008. (Nishimura and Shimizu, 2012). The two main pathogens of HFMD are enterovirus 71 (EVA71) and CVA16. Since the EVA71 vaccination has been available since late 2016, the proportion of HFMD cases related to EVA71 has significantly dropped (Takahashi et al., 2016; Han et al., 2020). Nonetheless, the CVA16 epidemic persists, and more than 22% of all HFMD cases are recorded annually in China (Yi et al., 2021). As a result, CVA16 is a significant public health issue, and there is no available medication or vaccination to prevent it. Therefore, it’s crucial to differentiate CVA16 quickly, precisely, and sensitively in order to stop and manage HFMD epidemics. Traditional detection techniques, such as viral isolation culture and serological testing, along with PCR-based technologies, often cannot meet the necessities for rapid identification in terms of time and sensitivity. Therefore, it is necessary to create a new technique to easily, quickly, sensitively, and precisely detect and diagnose CVA16.
Here, we describe the use of LFB and MCDA to identify CVA16. The MCDA is an isothermal amplification technique that does not need specialized tools or expert knowledge, only a basic water bath or heater (Jiao et al., 2019). A total of 10 specific primers were created using the MCDA-LFB approach to identify 10 distinct regions of the target sequence. We have chosen the highly conserved area of VP1 gene, which is very crucial for the immunogenicity of CVA16 vaccine since the primary neutralizing epitopes are situated in this region (Shi et al., 2013). Amplification products could be detected by the LFB labeled with FAM and biotin; it took about 2 min to read the LFB’s findings. Compared to MCDA methods used in earlier studies, the CVA16-MCDA method identified outcomes in LFB, which excludes the demands of specific reagents (e.g., pH indicators), difficult processes (e.g., electrophoresis), and expensive devices (e.g., real-time turbidity), was quick, sensitive, simple for using, and not prone to error (Wang et al., 2018; Zhao et al., 2019; Chen et al., 2021a).
In the CVA16-MCDA-LFB test, we altered the reaction temperature and reaction time to find the perfect situations. At 64°C, CVA16-MCDA-LFB has a greater amplification efficiency than it has at other temperatures. Additionally, because of CVA16-MCDA-great LFB’s sensitivity, the findings demonstrated that it was possible to detect the LoD level of 4 × 101 copies/μL of pUC57-CVA16-VP1 during continuous 40-min amplification. This suggests that the MCDA-LFB technique is a quick, accurate, and simple way to find small quantities of CVA16. The CVA16-MCDA-LFB test has a high specificity in addition to its excellent sensitivity. The positive findings for the CVA16-MCDA-LFB assay specificity test were obviously obtained from CVA16 strains but not from non-CVA16 strains. More crucially, we were able to identify Nasopharyngeal swab specimens from clinics using the CVA16-MCDA-LFB test. Compared with real-time PCR method, CVA16-MCDA-LFB technique is more time-saving, with 100% positive and 100% negative rates, real-time PCR needs 3–4 h during the whole process (Guney et al., 2003; Cui et al., 2013). However, even with sample preparation, the CVA16-MCDA-LFB test procedure’s overall detection time was <1 h. It was shown that our established approach for detecting CVA16 is quicker than methods based on culture and real-time PCR. Antibiotic exposure may be decreased when doctors are able to provide targeted medications to patients more rapidly since the CVA16-MCDA-LFB test can reveal findings in only 1 h. This makes this technique potentially valuable for detecting target pathogens.
The CVA16-MCDA-LFB technique in the present investigation also has significant restrictions. First, the recently discovered CVA16-MCDA-LFB detection is a qualitative evaluation of the pathogen and is unable to measure the pathogen quantity in specimens. The MCDA-LFB assay will be used in a more accurate investigation to assess CVA16 in clinical samples. Second, for LFB detection, CVA16-MCDA amplifications must be eliminated from the reaction tube since carry-over contamination is a major issue with this approach. However, spraying 70% ethanol and sodium hypochlorite solution is also useful in avoiding DNA contamination after identification has been finished. Additionally, MCDA is an isothermal amplification method that needs many primer pairs; contamination may easily happen and provide false findings. We will thus address the aforementioned issues in subsequent investigations.
Conclusion
A visual, quick, easy, and sensitive CVA16-MCDA-LFB assay based on the VP1 gene was successfully created in the current investigation to detect CVA16. When compared to real-time PCR molecular diagnostic tests, the MCDA-LFB assay avoids complex procedures and does not demand costly equipment or experienced technical staff. The LoD of the new test for detecting CVA16 in an isolated sample was as low as 40 copies, indicating that the MCDA-LFB assay was very sensitive. More significantly, the new test may decrease detection time and support clinicians in providing targeted treatments for patients with HFMD more rapidly, particularly in resource-poor regions.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding authors.
Ethics statement
The study was approved by the Human Ethics Committee of the First People’s Hospital of Guiyang (approval no. G2020-S001) and acts following the Declaration of Helsinki. The monitoring stations deleted all identifying information from the people suspected of having HFMD infection prior to obtaining the samples/isolates and conducting the study. The First People’s Hospital in Guiyang’s Human Ethics Committee waived the patient’s informed consent.
Author contributions
JC, YW, XT, and JL conceived and designed the experiments. JC, YZ, YW, and JL conducted the trials. JC, YW, and XT analyzed the data. YW, JC, YZ, and XT wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by a grant from the National Natural Science Foundation of China (grant no. 81860594), Zhu Ke He Tong [2021]-43-25 and Zhu Ke He Tong [2020]-10-6 from Science and Technology Department of Guiyang city of Guizhou Province, Qian Ke He [2018]1094, Qian Ke He Zhi Cheng [2021] yi ban 440 from Science and Technology Department of Guizhou Province, and The Project for the growth of Young Scientific and technological talents in ordinary Colleges and Universities in Guizhou Province (Qian jia he KY zi [2022] No. 231).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
Chen, X., Huang, L., Zhou, Q., Tan, Y., Tan, X., and Dong, S. (2021a). A nanoparticle-based biosensor combined with multiple cross displacement amplification for the rapid and visual diagnosis of Neisseria gonorrhoeae in clinical application. Front. Microbiol. 12:747140. doi: 10.3389/fmicb.2021.747140
Chen, L., Yao, X. J., Xu, S. J., Yang, H., Wu, C. L., Lu, J., et al. (2019). Molecular surveillance of coxsackievirus A16 reveals the emergence of a new clade in mainland China. Arch. Virol. 164, 867–874. doi: 10.1007/s00705-018-4112-3
Chen, X., Zhou, Q., Dong, S., Wang, S., Liu, R., Wu, X., et al. (2021b). Multiple cross displacement amplification linked with nanoparticles-based lateral flow biosensor in screening of hepatitis B virus in clinical application. Infect. Drug Resist. 14, 1219–1229. doi: 10.2147/idr.s297645
Cui, A., Xu, C., Tan, X., Zhang, Y., Zhu, Z., Mao, N., et al. (2013). The development and application of the two real-time RT-PCR assays to detect the pathogen of HFMD. PLoS One 8:e61451. doi: 10.1371/journal.pone.0061451
Esposito, S., and Principi, N. (2018). Hand, foot and mouth disease: current knowledge on clinical manifestations, epidemiology, aetiology and prevention. Eur. J. Clin. Microbiol. Infect. Dis. 37, 391–398. doi: 10.1007/s10096-018-3206-x
Guney, C., Ozkaya, E., Yapar, M., Gumus, I., Kubar, A., and Doganci, L. (2003). Laboratory diagnosis of enteroviral infections of the central nervous system by using a nested RT-polymerase chain reaction (PCR) assay. Diagn. Microbiol. Infect. Dis. 47, 557–562. doi: 10.1016/s0732-8893(03)00148-2
Han, Z., Song, Y., Xiao, J., Jiang, L., Huang, W., Wei, H., et al. (2020). Genomic epidemiology of coxsackievirus A16 in mainland of China, 2000-18. Virus Evol. 6:veaa084. doi: 10.1093/ve/veaa084
Jiang, L., Gu, R., Li, X., Song, M., Huang, X., and Mu, D. (2022). Multiple cross displacement amplification coupled with lateral flow biosensor (MCDA-LFB) for rapid detection of legionella pneumophila. BMC Microbiol. 22:20. doi: 10.1186/s12866-021-02363-3
Jiao, W. W., Wang, Y., Wang, G. R., Wang, Y. C., Xiao, J., Sun, L., et al. (2019). Development and clinical validation of multiple cross displacement amplification combined with nanoparticles-based biosensor for detection of mycobacterium tuberculosis: preliminary results. Front. Microbiol. 10:2135. doi: 10.3389/fmicb.2019.02135
Li, S., Liu, C., Liu, Y., Ma, Q., Wang, Y., and Wang, Y. (2019). Development of a multiple cross displacement amplification combined with nanoparticles-based biosensor assay to detect Neisseria meningitidis. IDR 12, 2077–2087. doi: 10.2147/idr.s210735
Li, J. F., Zhang, C. J., Li, Y. W., Li, C., Zhang, S. C., Wang, S. S., et al. (2022). Coxsackievirus A6 was the most common enterovirus serotype causing hand, foot, and mouth disease in Shiyan City, Central China. World J. Clin. Cases 10, 11358–11370. doi: 10.12998/wjcc.v10.i31.11358
Nishimura, Y., and Shimizu, H. (2012). Cellular receptors for human enterovirus species a. Front. Microbiol. 3:105. doi: 10.3389/fmicb.2012.00105
Shi, J., Huang, X., Liu, Q., and Huang, Z. (2013). Identification of conserved neutralizing linear epitopes within the VP1 protein of coxsackievirus A16. Vaccine 31, 2130–2136. doi: 10.1016/j.vaccine.2013.02.051
Sickles, G. M., Mutterer, M., Feorino, P., and Plager, H. (1955). Recently classified types of Coxsackie virus, group a; behavior in tissue culture. Proc. Soc. Exp. Biol. Med. 90, 529–531. doi: 10.3181/00379727-90-22088
Sun, T., Liu, Y., Zhang, Y., and Zhou, L. (2014). Molecular phylogeny of coxsackievirus A16. J. Clin. Microbiol. 52, 3829–3830. doi: 10.1128/jcm.01330-14
Takahashi, S., Liao, Q., Van Boeckel, T. P., Xing, W., Sun, J., Hsiao, V. Y., et al. (2016). Hand, foot, and mouth disease in China: Modeling epidemic dynamics of Enterovirus serotypes and implications for vaccination. PLoS Med. 13:e1001958. doi: 10.1371/journal.pmed.1001958
Wang, Y., Wang, Y., Ma, A. J., Li, D. X., Luo, L. J., Liu, D. X., et al. (2015). Rapid and sensitive isothermal detection of nucleic-acid sequence by multiple cross displacement amplification. Sci. Rep. 5:11902. doi: 10.1038/srep11902
Wang, Y., Yan, W., Fu, S., Hu, S., Wang, Y., Xu, J., et al. (2018). Multiple cross displacement amplification coupled with nanoparticles-based lateral flow biosensor for detection of Staphylococcus aureus and identification of methicillin-Resistant S. aureus. Front. Microbiol. 9:907. doi: 10.3389/fmicb.2018.00907
Wang, Y., Zhao, X., Cheng, J., Tang, X., Chen, X., Yu, H., et al. (2021). Development and application of a multiple cross displacement amplification combined with nanoparticle-based lateral flow biosensor assay to detect Candida tropicalis. Front. Microbiol. 12:681488. doi: 10.3389/fmicb.2021.681488
Xu, F., He, D., He, S., Wu, B., Guan, L., Niu, J., et al. (2011). Development of an IgM-capture ELISA for Coxsackievirus A16 infection. J. Virol. Methods 171, 107–110. doi: 10.1016/j.jviromet.2010.10.009
Xu, W., Liu, C. F., Yan, L., Li, J. J., Wang, L. J., Qi, Y., et al. (2012). Distribution of enteroviruses in hospitalized children with hand, foot and mouth disease and relationship between pathogens and nervous system complications. Virol. J. 9:8. doi: 10.1186/1743-422x-9-8
Yi, L., Zeng, H., Zheng, H., Peng, J., Guo, X., Liu, L., et al. (2021). Molecular surveillance of coxsackievirus A16 in southern China, 2008-2019. Arch. Virol. 166, 1653–1659. doi: 10.1007/s00705-021-05052-8
Zhang, M., Zhao, Y., Zhang, H., Lin, K., Liu, H., Zhang, J., et al. (2019). Molecular characterization of Coxsackievirus A16 strains isolated from children with severe hand, foot, and mouth disease in Yunnan, Southwest China, during 2009-2015. J. Med. Virol. 91, 155–160. doi: 10.1002/jmv.25297
Zhao, F., Niu, L., Yan, L., Nong, J., Wang, C., Wang, J., et al. (2019). Establishment and application of multiple cross displacement amplification coupled with lateral flow biosensor (MCDA-LFB) for visual and rapid detection of Candida albicans in clinical samples. Front. Cell. Infect. Microbiol. 9:102. doi: 10.3389/fcimb.2019.00102
Keywords: coxsackievirus A16, reverse transcription-multiple cross displacement amplification, hand, foot, and mouth disease, lateral flow biosensor, MCDA-LFB, limit of detection
Citation: Cheng J, Wang Y, Zhou Y, Lu J and Tang X (2023) Highly sensitive and rapid identification of coxsackievirus A16 based on reverse transcription multiple cross displacement amplification combined with nanoparticle-based lateral flow biosensor assay. Front. Microbiol. 14:1121930. doi: 10.3389/fmicb.2023.1121930
Edited by:
Jade L. L. Teng, The University of Hong Kong, Hong Kong SAR, ChinaReviewed by:
Xianfeng Zhou, Jiangxi University of Traditional Chinese Medicine, ChinaZhiqin Xie, Guangxi Veterinary Research Institute, China
Copyright © 2023 Cheng, Wang, Zhou, Lu and Tang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yu Wang, d2FuZ3pob25neXV3eUAxNjMuY29t; Xiaomin Tang, NTcwODE5MTA3QHFxLmNvbQ==
†These authors have contributed equally to this work
 Jinzhi Cheng1†
Jinzhi Cheng1† Yu Wang
Yu Wang Yuhong Zhou
Yuhong Zhou Jingrun Lu
Jingrun Lu Xiaomin Tang
Xiaomin Tang