- 1Department of Oral Mucosa, Shanghai Stomatological Hospital, Fudan University, Shanghai, China
- 2Shanghai Key Laboratory of Craniomaxillofacial Development and Diseases, Shanghai Stomatological Hospital, Fudan University, Shanghai, China
- 3Translational Medicine Center, Guangdong Women and Children Hospital, Guangzhou, China
Several studies have shown that the dysregulation of the oral microbiota plays a crucial role in human health conditions, such as dental caries, periodontal disease, oral cancer, other oral infectious diseases, cardiovascular diseases, diabetes, bacteremia, and low birth weight. The use of traditional detection methods in conjunction with rapidly advancing molecular techniques in the diagnosis of harmful oral microorganisms has expanded our understanding of the diversity, location, and function of the microbiota associated with health and disease. This review aimed to highlight the latest knowledge in this field, including microbial colonization; the most modern detection methods; and interactions in disease progression. The next decade may achieve the rapid diagnosis and precise treatment of harmful oral microorganisms.
1. Introduction
Microorganisms residing in the oral cavity are significant components in altering the balance between health and sickness, and include several hundred to thousands of diverse species. The mouth contains bacteria, archaea, protozoa, fungi, and viruses, although each has its own unique characteristic “fingerprint,” usually living in symbiotic harmony with the host (Palmer, 2014; Nobbs and Jenkinson, 2015; Nearing et al., 2020). In the last decade, Human oral microbiome database (HOMD) has been created to provide the scientific community with comprehensive curated information on the bacterial species inhabited in the human aerodigestive tract. Potentially, 774 oral bacterial species exist, approximately 58% of which are officially named, 16% are unnamed but can be cultivated, and the remaining 26% are known only as uncultivated phylotypes (Human Oral microbiome Database. Available at http://www.homd.org), which is currently identified with advances in microscopy and other approaches. Therefore, our understanding of microorganisms has become increasingly profound. An imbalance in a complex ecological community may affect oral and systemic diseases (Abusleme et al., 2013; Hong et al., 2015; Gomes-Filho et al., 2016; Slocum et al., 2016; Xiao et al., 2017; Duncan et al., 2019; Hu et al., 2019; Li et al., 2019; Yu et al., 2020; Corralo et al., 2021). Distinguishing regularly colonizing microorganisms from harmful ones is an important task in this field. Even detecting harmful microorganisms does not necessarily lead to disease. Linking the rapid detection of harmful oral microorganisms with disease diagnosis and precise treatment is the main focus of our efforts. In this review, we focused on the microbial variety in the mouth, presented the microbial community of diverse oral niches, and confirmed the relationship between dysbiosis and oral or systemic disorders.
2. Characteristics of oral microbiota
2.1. Composition
2.1.1. Bacteria
Bacteria comprise the majority of oral organisms. In 1881, Robert Koch developed a solidified culture medium and a reproducible technology for growing and isolating pure cultures of microbiota with gelatin and later agar, which advanced the identification, analysis, and classification of different microbes (Brock, 1961). In addition, these uncultivable phylotypes are widely observed through molecular taxonomy, particularly 16 s rRNA profiling, next-generation sequencing (NGS) technology, DNA microarrays, and metagenome sequencing (Willse et al., 2005; Claesson et al., 2010; Ghurye et al., 2016; Yang et al., 2016; Vincent et al., 2017; Miao et al., 2018; Gu et al., 2019). The HOMD includes 789 taxa in 19 phyla, including Bacteroidetes, Chlamydiae, Chloroflexi, Chlorobi, Actinobacteria, Firmicutes, Fusobacteria, Gracilibacteria (GN02), Saccharibacteria (TM7), Spirochaetes, WPS-2, Proteobacteria, Euryarchaeota, Synergistetes, SR1, Cyanobacteria, Ignavibacteriae, Lentisphaerae, and Tenericutes (HOMD).1
2.1.2. Fungi
In recent reviews, the oral presence of fungi is purportedly associated with pathologies, since its colonization is presented as a cause of disorders (Jenkinson, 2011; Wright et al., 2013; Chevalier et al., 2018; Anuta et al., 2022; Cannon, 2022). However, an alternative role for fungi in good oral health was reported by Ghannoum et al. (2010). They revealed the “basal” oral microbiota profile in 20 healthy individuals, and demonstrated that a total number of 101 fungal species are identified. It has also been reported that the number of species present in each participant ranged from 9 to 23. And Candida species were the most frequently obtained (isolated from 75% of subjects), followed by Aureobasidium, Cladosporium, Cryptococcus, Saccharomycetals, Aspergillus, Fusarium. Four of these predominant genera are pathogenic in humans (Ghannoum et al., 2010). In another study, at least 81 genera and 154 fungal species were observed in 30 adult individuals (15 with oral health and 15 with periodontitis), and Candida and Aspergillus were the most abundant genera, followed by Penicillium, Schizophyllum, Rhodotorula, and Gibberella. Aspergillus niger was the most frequently identified species in the samples (Peters et al., 2017).
2.1.3. Viruses
Most viruses present in the mouth are associated with various diseases. Human papilloma virus (HPV) infection can cause many oral lesions, the well-known benign clinical presentations are oral papilloma, condyloma, and focal epithelial hyperplasia (Jiang and Dong, 2017; Syrjanen, 2018; Moran-Torres et al., 2021; Santacroce et al., 2021). Persistent HPV infection is mandatory in HPV-induced malignancies. Herpes simplex virus contributes to herpetic gingivostomatitis, herpes labialis, herpes genitalis, herpetic paterecleris, and mucocutaneous orofacial disease (Dai and Zhou, 2018; Crimi et al., 2019; Atyeo et al., 2021). HIV-related lesions include oral hairy leukoplakia, oral candidiasis, linear gingival erythema, necrotizing ulcerative periodontitis, Kaposi sarcoma, and non-Hodgkin lymphoma (Moyes et al., 2016; Ottria et al., 2018; Polvora et al., 2018; Santella et al., 2019; Sharma Mahendra et al., 2020; Perez Rosero et al., 2021).
2.1.4. Protozoa
Compared with other microbial groups, protozoa constitute a small fraction. The first studied and the most parasite in the oral cavity is Entamoeba gingivalis, followed by Trichomona tenax (Abualqomsaan et al., 2010; Ghabanchi et al., 2010; Rashidi Maybodi et al., 2016; Yazar et al., 2016; Fadhil Ali Malaa et al., 2022). These protozoa are mainly non-pathogenic commensals under normal conditions. This is characteristic of patients with poor oral hygiene and partial or total edentulism. A Jordanian study designed to evaluate the prevalence of E. gingivalis and T. tenax in the oral cavity of 53 patients with gingivitis, 90 patients with periodontitis, and 94 healthy subjects found 88.9% periodontitis patients infected with E. gingivalis, 84.9% among patients with gingivitis, and 47.9% in healthy controls. For T. tenax, the percentage was 25.6% among periodontitis patients, 5.7% among gingivitis participants, and 3.2% in the control group. Colonization by E. gingivalis and T. tenax was significantly correlated with periodontal disease compared to the healthy cohort (Yaseen et al., 2021).
2.2. Colonization
Recent estimates reveal that the microbial community or biofilm is functionally and structurally organized and attached to surfaces such as the cheek, tongue, hard and soft palate, teeth, and gingival tissue (Zaura et al., 2009; Arweiler and Netuschil, 2016). The microbiota that reside and prevail on particular surfaces depends on the anatomic and biological features of each site (Mager et al., 2003, 2005). As they are easily sampled and accessible, microbiology can be the most studied.
2.2.1. Saliva
Saliva represents the “planktonic phase” of microorganisms. Each milliliter of saliva contained up to 109 microbiota, similar to the bacterial laboratory fluid cultures. Since saliva is swallowed continually, 5 g of bacteria probably flows into the stomach daily. As a result, saliva is not assumed to contain its own inhabitant microbiota. Although most populations share similar salivary microorganisms, differences in salivary composition exist between individuals. Moreover, the microbial composition did not exhibit geographical distribution features. Salivary organisms may be indicators of disease diagnosis. For instance, patients with dental caries, pulpitis, chronic periodontitis, and oral potentially malignant disorders have a distinctive salivary bacterial composition or distribution in healthy individuals (Mager et al., 2005; Belstrom et al., 2014, 2015; Gao et al., 2016; Wang X. et al., 2017; Khurshid et al., 2018).
2.2.2. Surfaces of hard tissues
Non-shedding surfaces of natural teeth or artificial material surfaces of dentures, oral implants, orthodontic appliances, and tooth fillings may be a stable location for the formation of dental plaque, which is a structurally and functionally organized microbial biofilm (Marsh, 2006; Costalonga and Herzberg, 2014). Biofilm formation is divided into supragingival and subgingival biofilms according to the location.
Supragingival plaque (above the gum line) contained a mixture of facultative and anaerobic species. In a Turkish study involving 26 caries-active patients and 26 caries-free controls, testing for dental supragingival plaque, the bacteria responsible for caries was defined as Anaeroglobus, Atopobium, Bifidobacterium, Centipeda, Cryptobacterium, Desulfobulbus, Filifactor, Howardella, Lactobacillus, Leptotrichiaceae (unclassified), Megasphaera, Mycoplasma, Olsenella, Phocaeicola, Propionibacterium, Pseudoramibacter, Scardovia, Schwartzia, Treponema, and Veillonellaceae (unclassified; Celik et al., 2021). Another study identified 13 genera that were highly abundant in supragingival plaque samples, including Corynebacterium, Capnocytophaga, Fusobacterium, Leptotrichia, Actinomyces, Streptococcus, Neisseria, Haemophilus/Aggr, Porphyromonas, Rothia, Lautropia, Veillonella, Prevotella (Mark Welch et al., 2016).
Subgingival plaque (below the gum line) contains more anaerobic genera owing to extreme circumstances, causing periodontitis. Griffen et al. (2012) identified 123 species at drastically higher levels in subjects with chronic periodontitis and 53 species in healthy individuals. Spirochaetes, Synergistetes, and Bacteroidetes were more abundant in the disease group, whereas Proteobacteria were significantly more abundant in healthy controls. Within the phylum, Firmicutes, Clostridia, Negativicutes, and Erysipelotrichia were correlated with disease, while the class Bacilli was health-associated (Griffen et al., 2012). Another study characterizing the subgingival microbiota of 22 patients with periodontal disease and 10 healthy individuals suggested that periodontitis communities had higher percentages of Spirochetes, Synergistetes, Firmicutes, and Chloroflexi, among other taxa, whereas the percentages of Actinobacteria, particularly Actinomyces, were higher in healthy individuals (Abusleme et al., 2013).
2.2.3. Surfaces of soft tissues
The colonization of microbiota on shedding surfaces, such as lips, cheeks, and palates, is limited owing to bacterial monolayers (He et al., 2015). In contrast, the papillary surface of the tongue has multiple layers of microbiota because of the safe environment provided by the papillae. In addition, tongue microbiota populations are closely related to oral malodor (Tanaka et al., 2004). A study designed by Kazor et al. (2003) to analyze the bacterial diversity on the surface of the tongue dorsum showed that the species predominant in healthy individuals were Streptococcus salivarius, Rothia mucilaginosa, and an uncharacterized species of Eubacterium (strain FTB41; Kazor et al., 2003). Although a large number of bacteria may reside on the tongue, epithelial desquamation restricts microbial colonization on mucosal surfaces. The epithelial shedding squamae was ingested with inhabitant microbes. Therefore, residents need to read the newly developed epithelium (Figures 1, 2).
2.3. Mutual translocation
While some species prefer specific habitats, others are found in multiple microniches within the oral cavity. According to physical and morphological criteria, the oral cavity can be divided into several major ecological communities, including the palate, dorsum of the tongue, floor of the mouth, buccal epithelium, supragingival plaque, and periodontal pocket. For example, P. gingivalis, P. intermedia, A. actinomycetemcomitans, Spirochetes are able to locate all of these areas (Marsh et al., 2009). A specific example revealed P. melaninogenica not only in pockets and supragingival plaques, but also in the saliva, tonsils, and other mucosal sites (Quirynen et al., 2001). The findings show that intraoral translocation of periodontopathogens inhibits guided tissue regeneration and locally applied antibiotics, thus jeopardizing the outcome of periodontal therapy (Quirynen et al., 2001).
3. An overview of experimental approaches to study oral microbiota
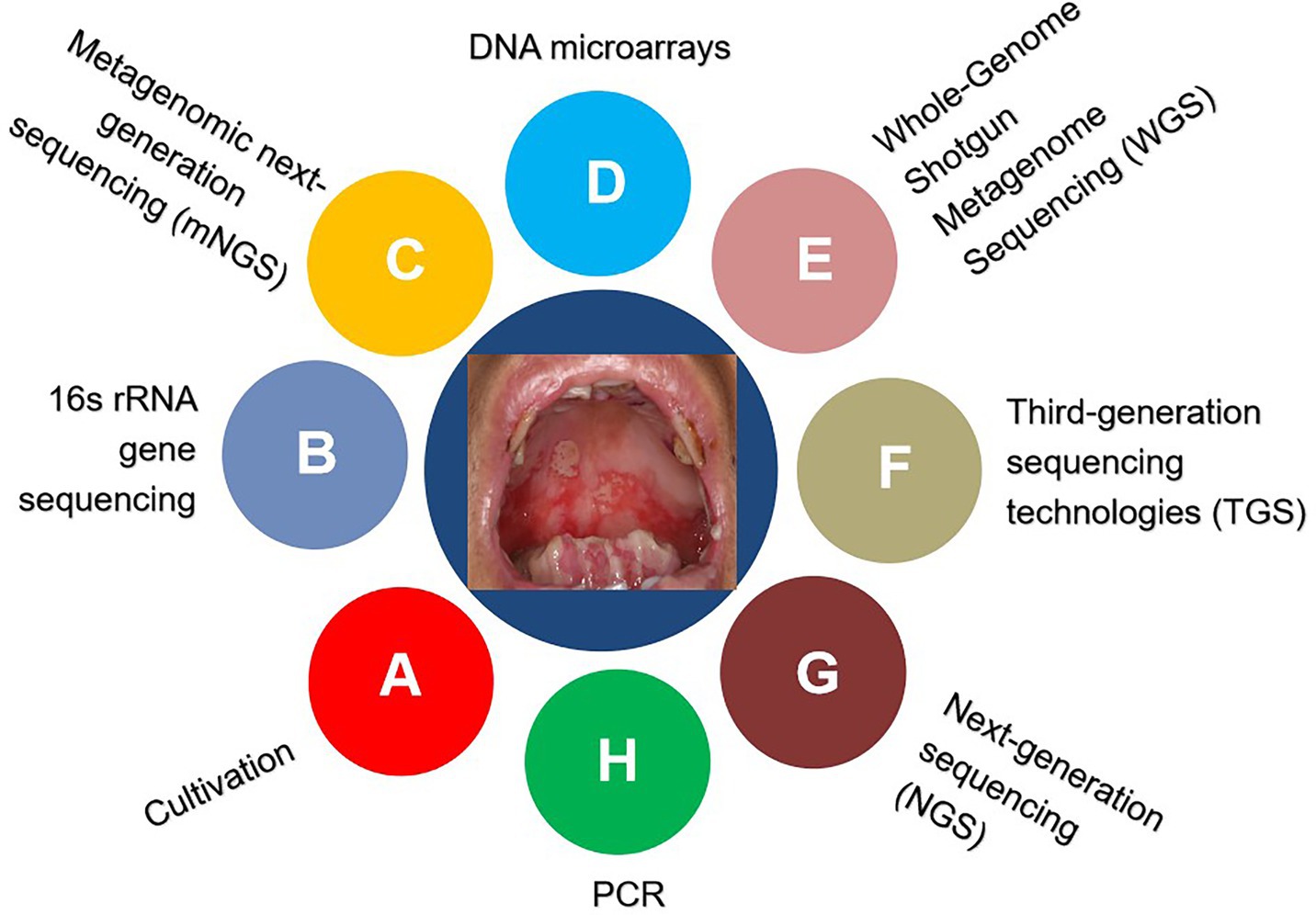
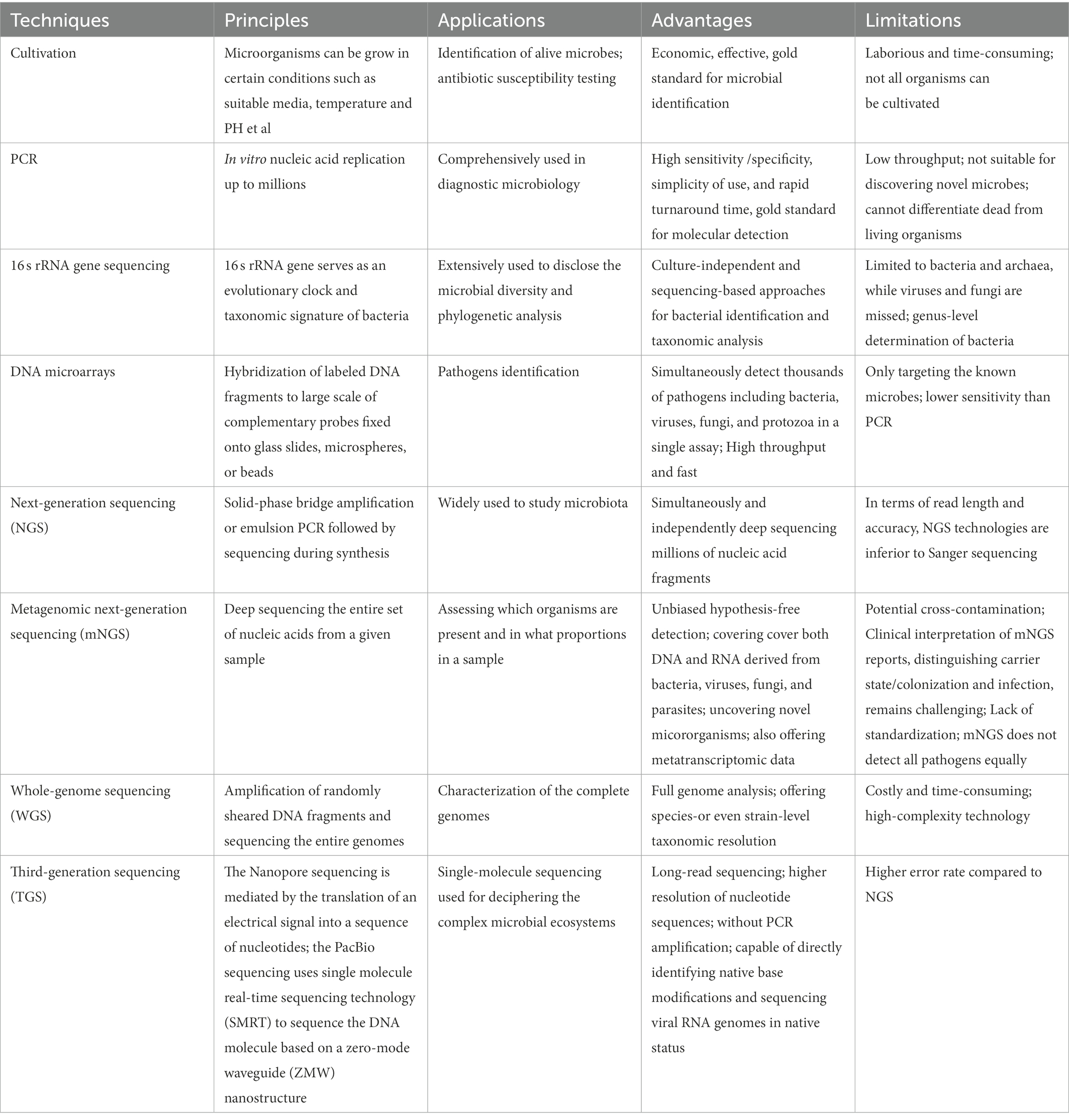
Historically, the cultivation of oral bacterial species requires distinctive conditions, such as biochemically defined media, an anaerobic environment, proper incubation temperature, and diverse PH (Brock, 1961; Kilian, 2007; Loeffelholz and Sanden, 2007; Marmonier, 2007; Vartoukian et al., 2010). However, not all bacteria can be cultivated in the laboratory (Black, 1886; Wade, 2002, 2013; Sizova et al., 2012; Leys et al., 2013; Siqueira and Rocas, 2013). The currently developed culture-independent approaches enable us to identify, classify, and characterize the uncultivable microbiological genera, as summarized in Table 1 and Figure 1 (Liu et al., 2012; Lagier et al., 2015; Krishnan et al., 2017; Benn et al., 2018; Verma et al., 2018).
3.1. Cultivation
A pure microbial culture is essential for research on its pathogenesis, antibiotic resistance, and invasive potential, to improve the knowledge and therapy of related infectious diseases. Overall, there are four primary elements involved in the growth of bacteria: nutrients, temperature, atmosphere, and incubation time. However, culture should be more pertinently accommodated for particular species in some cases to provide an optimal environment for their growth. Despite prominent advances in bacterial culture over the past decades, several unsatisfactory results remain. On the one hand, there are some limitations to displaying microbial diversity. However, a large number of organisms may not be cultivated by the conventional method because of their association with other microbiota. Instead of isolation, oral bacteria survive and develop in a complicated community called biofilms (Jenkinson, 2011; Soro et al., 2014). To date, almost 50% of oral microbial species detected through culture-independent gene sequencing techniques remain resistant to cultivation (Sizova et al., 2012; Siqueira and Rocas, 2013; Wade, 2013).
3.2. Polymerase chain reaction
Polymerase chain reaction is a revolutionary in vitro replication technique that enables the amplification of targeted genes up to millions in a 2–3 h timeframe. Owing to its high sensitivity/specificity, simplicity of use, and rapid turnaround time, PCR has been comprehensively used in diagnostic microbiology. In recent decades, multiple PCR variations have been developed for various purposes, including nested PCR, asymmetric PCR, qualitative PCR, and reverse transcription-PCR (Sakamoto et al., 2005). For example, Deng et al. (2012) established a novel strategy for rapid colorimetric analysis of Bacillus anthracis by combining asymmetric PCR with gold nanoparticles. To improve the positivity rate of skeletal tuberculosis diagnosis, which is usually performed by histopathology, researchers have investigated the clinical utility of qPCR-based diagnosis of skeletal tuberculosis using formalin-fixed paraffin-embedded tissues. Compared to that of traditional acid-fast bacillus staining, qPCR offers superior accuracy for mycobacterial detection in clinical samples (He et al., 2022). With the advent of the 2019 coronavirus disease (COVID-19) pandemic, RT-PCR is currently considered the gold standard for COVID-19 detection (El-Kafrawy et al., 2021). Importantly, real-time PCR/RT-PCR can determine experimental results without post-PCR procedures, such as gel electrophoresis, minimizing potential cross-contamination. Notably, molecular methods cannot be used to differentiate between dead and living organisms. In addition, PCR is not well suited for identifying novel microbes. In addition, it only detects a single or small number of gene regions/loci of interest at a time, as with multiplex PCR (Lochman et al., 2019). Therefore, high-throughput approaches are required.
3.3. 16S rRNA gene sequencing
As discussed above, culture-dependent methods and PCR-based techniques have limitations in the characterization of bacterial diversity. Owing to developments in sequencing-based approaches, such as 16 s rRNA gene sequencing, this weakness can now be overcome. The 16S rRNA gene is one of the most commonly analyzed marker genes because its functional consistency of 16S rRNA enables it to serve as an evolutionary clock and taxonomic signature in bacterial systematics (Woo et al., 2008). 16S rRNA gene sequencing has been extensively used to determine microbial diversity and for phylogenetic analyses (Janda and Abbott, 2007). PCR-amplified 16S fragments were cloned, sequenced, and clustered based on sequence similarity to infer likely taxonomy. Olson et al. (2011) identified different bacterial signatures in the subgingival plaque of subjects with low or high oral disease by broad-range PCR amplification of the 16S rRNA gene with universal bacterial primers followed by clone-by-clone sequencing. They found that high oral disease exhibited markedly increased bacterial diversity and included an elevated frequency of Clostridiales cluster bacteria, indicating that this atypical bacterial signature is associated with high oral disease (Olson et al., 2011). Another study revealed that anaerobes and oral bacteria are more frequently identified in patients with community-acquired pneumonia than previously thought. These findings strongly suggest that these bacteria may play important in pneumonia (Yamasaki et al., 2013).
Notably, the use of 16S rRNA gene sequencing for bacterial identification can usually be determined accurately at the genus level and not at the species level (Kilian et al., 2016). In addition, this technique is limited to bacteria and archaea, but cannot detect viruses and fungi. Nuclear ribosomal Internal Transcribed Spacer (ITS) regions, including ITS1 and ITS2, are relatively conserved among fungi. Therefore, DNA metabarcoding markers can be used to characterize the diversity and composition of fungal communities (Mbareche et al., 2020).
3.4. DNA microarray
In contrast to low-throughput molecular techniques, which generally discover a limited number of targets simultaneously, DNA microarrays offer the benefits of broad coverage, speed, and moderate cost. In recent decades, microarrays have become a viable alternative, enabling simultaneous detection and quantification of hundreds or thousands of targets in a single assay. As a three-dimensional array, suspension microarrays have faster hybridization kinetics and more flexibility in array construction than solid-phase arrays. Briefly, the procedure consists of broad-range PCR, probe design, probe immobilization to microspheres/beads, and molecular hybridization.
Wang et al. (2002) constructed a DNA microarray system, designated ViroChip, for virus identification. Later, GreeneChip and MDA microarrays were developed to detect several thousand pathogens, including viruses, bacteria, fungi, and protozoa (Palacios et al., 2007; Gardner et al., 2010). Common features of these platforms are the use of long (60-mer) oligonucleotide probes and random amplification of nucleic acids. Recently, Huang et al. (2013) developed a high-density microarray platform for large-scale vertebrate pathogen discovery based on similar strategies. This EOPM Chip can detect and distinguish all 2,554 known vertebrate virus species, 124 bacterial genera, 38 fungal genera, and 47 parasitic genera at the species or genus level, nearly all of which are known. Despite its lower sensitivity than that of PCR, this system provides sufficient sensitivity to identify the key pathogens responsible for the clinical symptoms. The authors successfully identified adenovirus causing an outbreak of flu-like infections, as well as cardiovirus in a hand-foot-and-mouth juvenile patient in clinical samples, demonstrating its potential for clinical applications. Theoretically, this cutting-edge platform can be applied for the molecular detection of oral microbes.
The Human Oral Microbe Identification Microarray (HOMIM), a 16S rRNA-based mid-density array containing more than 400 probes, was developed and utilized to analyze oral bacteria. To improve our in-depth understanding of the relationship between the oral microbiota and human health, oral diseases such as periodontitis were investigated using the HOMIM technology (Lourenco et al., 2014). HOMIM analysis of salivary microbiota revealed that a total of 16 bacterial species showed differences between pancreatic cancer samples and controls (Farrell et al., 2012). Advances in NGS have resulted in the upgrading of the HOMIM platform by HOMINGS,2 covering approximately 600 oral bacterial taxa to date.
More recently, a new phylogenetic DNA microarray (OralArray) containing 22 probe sets targeting bacteria was designed for the systematic analysis of bacterial communities in different microbial ecosystems, including the oral microbiota (Parolin et al., 2017). Unlike other DNA microarrays, whose discriminatory power is based on DNA hybridization, OralArray is based on Ligation Detection Reaction technology associated with Universal Arrays, which possesses excellent discriminative power and good performance in terms of sensitivity. Validation of OralArray on oral samples demonstrated its ability to efficiently detect the most representative bacterial populations colonizing the oral cavity under physiological and pathological conditions.
3.5. Next-generation sequencing
Continuous improvements in DNA sequencing technology over the past few decades have dramatically improved molecular diagnostics. In contrast to first-generation sequencing, NGS technologies are characterized by “sequencing during synthesis” and “massive parallel sequencing” with the ability to simultaneously and independently sequence millions of nucleic acid fragments. NGS enables direct and comprehensive investigation of microbial communities in a single test and provides deeper insights into the microorganisms involved in human diseases (Jo et al., 2016). In recent decades, NGS has gained wide usage in many scenarios owing to its high-throughput capacity, fast turnaround time, and high cost-effectiveness. For example, high-throughput sequencing of the V3-V4 hypervariable regions of the 16S rRNA gene on the Illumina Miseq PE300 platform revealed clear oral microbial dysbiosis among individuals with severe dental caries of primary Sjögren’s syndrome (pSS). Moreover, Veillonella has been proposed as a potential biomarker in patients with pSS (Zhou et al., 2018). These findings may facilitate our understanding of the link between microbial communities and oral homeostasis and human diseases. To study and clarify whether saliva represents a suitable sample for monitoring supragingival microbiota in healthy children, the 16S rRNA gene amplicon was subjected to NGS using the Illumina Miseq system. Sequence reads were clustered into operational taxonomic units (OTUs) after mapping to bacterial reference databases and representative OTU sequences were used for taxonomic identification and abundance estimation. Compared with supragingival plaque, saliva had a less even and less diverse community. Rothia and Streptococcus were found to discriminate between saliva and plaques. In contrast, supragingival microbiota also exhibited positive associations with salivary microbiota (Shi et al., 2018). It would be interesting to examine the impact of different delivery modes on oral microbiota in healthy infants. 16S rRNA gene-based high-throughput sequencing revealed that newborns delivered via cesarean section and individuals delivered vaginally had distinct oral bacteria. The dominant bacteria in the cesarean section group included Petrimonas, Bacteroides, Desulfovibrio, Pseudomonas, and Staphylococcus. The most abundant genera in the vaginal delivery group were Lactobacillus, Prevotella, and Gardnerella spp. Moreover, the results suggest that repeated sterilization of the vulva might influence infants’ oral microbiota, and attention should be paid to clinical practice (Li et al., 2018b). Similar studies using this technique have been reported previously (Ren et al., 2017; Richards et al., 2017).
Because of the exceptional increase in the number of sequence reads and the reasonable cost, NGS has been widely used to assess the diversity and composition of bacterial ecosystems. However, NGS-based 16S rRNA gene sequencing still focuses only on bacteria and does not cover fungi and viruses. NGS technologies are inferior to Sanger sequencing in terms of the read length and accuracy. In addition, chimera generation during PCR and the intrinsic error rate of sequencing are major concerns. Optimization of the algorithms and experimental procedures is required to solve these problems.
3.6. Metagenomic next-generation sequencing
Metagenomic NGS is defined as the process of profiling the entire set of nucleic acids from a given sample that may contain completely different kingdoms of microbes. It sequences and assigns millions of reads to corresponding reference genomes for taxonomic analysis. This powerful technique allows for an unbiased approach to assess the presence of microorganisms and their proportions in a sample. Unlike targeted methods (closed-ended analysis), unbiased hypothesis-free detection (open-ended analysis) is a major advantage of NGS. This technique is sometimes called shotgun sequencing; therefore, it can cover both DNA and RNA derived from bacteria, archaea, viruses, fungi, and parasites, making the diagnosis of mixed infections/co-infections possible. mNGS is akin to “searching for a needle in a haystack” screening technique and theoretically it can detect almost any pathogens in a single run that can never be achieved by traditional approaches (Simner et al., 2018; Gu et al., 2019). During the incipient stage of the COVID-19 pandemic, mNGS offers an accurate and rapid diagnosis compared to previous methods (Chen et al., 2020).
Because mNGS transforms our understanding of the microbial makeup of a mixed sample and the correlation between microbial inhabitants and human diseases, it has been extensively applied to various clinical scenarios (Palacios et al., 2008; Li et al., 2018a; Wang et al., 2019, 2020; Qian et al., 2020; Gu et al., 2021). In addition to depicting microbiota DNA sequences, mNGS can also offer metatranscriptomic data, providing higher-dimensional information. Thus, it can address several issues, such as the microbes present, their functions, and how they interact. Undoubtedly, the integration of the metagenome and metatranscriptome will allow researchers to systematically characterize the complex functions of human ecosystems and vastly expand our awareness of microbe-microbe and microbe-host interactions.
Relatively limited data is available regarding its application in oral microbiota analysis (Zhang Y. et al., 2021). Song et al. (2022) used mNGS to investigate potential pathogens in patients with complex oral mucosal infections and oral leukoplakia. mNGS of oral mucosal tissues identified Candida albicans, human gamma herpesvirus 4, and many other pathogens within 36 h (Song et al., 2022). These findings demonstrate that mNGS is an efficient and sensitive tool capable of simultaneously detecting almost all bacteria, viruses, and fungi potentially associated with disease progression. In addition, mNGS is less affected by antibiotic use and has a higher positive detection rate than conventional detection methods. Metagenomic analysis of dental swabs and plaques from patients with periodontal disease has revealed a core community of disease-associated microbes. Some functional genes and metabolic pathways involved in glycan biosynthesis and bacterial chemotaxis were over-represented in the periodontal disease microbiota. This work enriches the understanding of the oral microbial community structure and metabolic variation that is closely associated with periodontal health (Wang et al., 2013).
Although mNGS can produce in-depth, unbiased information and uncover novel organisms, some issues and obstacles need to be solved before high-complexity technology can become the mainstream and route approach in real settings (Han et al., 2019). First, given the excellent analytical sensitivity of mNGS, cross-contamination during specimen collection, library preparation, assay runs, and bioinformatics classification is a major concern. Quality controls are needed to check “splashome” and “kitome” contamination batch by batch, otherwise, its specificity remains the proverbial elephant in the room. Second, mNGS did not detect all pathogens equally. For example, mycobacteria may be more difficult to detect because routine lysis and extraction are difficult to release from mycobacteria. Consequently, even a few reads of mycobacteria are often considered positive, and other methods are required for confirmation. The clinical interpretation of mNGS reports, distinguishing carrier state/colonization from infection, remains challenging. In fact, mNGS lacks methodological standardization and consensus criteria for positive/negative results, making the interpretation of the findings somewhat subjective. Bioinformatics analysis is sophisticated, even for professionals. Sometimes, mNGS reports need to be reevaluated. It is essential to highlight that mNGS alone does not establish pathogenicity, and that close cooperation between clinicians and technicians/bioinformaticians is required to discuss the positive, inconclusive, and negative results of mNGS. Currently, no mNGS tests have been approved by the Food and Drug Administration. However, it may be worthwhile because some diagnoses are made solely by mNGS, whereas no other tests provide useful information. Taken together, mNGS has potential as a front-line diagnostic tool in the near future; however, more effort and research are needed to improve its clinical utility.
3.7. Whole-genome sequencing
Metagenomics involves WGS where the sequences of entire genomes are randomly sheared and determined by a “shotgun” strategy. WGS can potentially offer species-or even strain-level taxonomic resolution, whereas high-throughput sequencing of the 16S rRNA gene normally profiles the taxonomic composition at the genus or species level in microbiota analyses. Usually, this is preferred term when this technique is used for human whole-genomic analysis.
3.8. Third-generation sequencing technologies
Owing to technical restrictions, targeting sub-region sequencing for short fragments represents a historical compromise. For example, most of the 16 s rRNA gene sequencing based on NGS only amplifies a portion of the marker gene, providing relatively limited information for bacterial community characterization. The advent of innovative long-read sequencing technologies, including the PacBio and Oxford nanopore platforms, has changed this and opened new avenues for microbiota studies (Jiao et al., 2013; Li et al., 2016). In addition to maintaining the features of high-throughput and in-depth sequencing, the read lengths are up to 1.0–1.5 × 104 and 2–5 × 103 for PacBio and Oxford nanopore single-molecule sequencing platforms, respectively (Mosher et al., 2014; Laver et al., 2015; van Dijk et al., 2018). However, TGS is capable of routinely generating reads in excess of 1,500 bp and completely covering the full 16S rRNA gene (Franzen et al., 2015; Schloss et al., 2016; Wagner et al., 2016). Previous studies have shown that TGS of the full-length (V1-V9) 16S rRNA gene using the PacBio technique generated sufficient accuracy to resolve subtle nucleotide variations for taxonomic analysis at species or even strain levels (Earl et al., 2018; Johnson et al., 2019). Moreover, TGS has additional advantages over NGS platforms for deciphering complex microbial ecosystems. For example, nanopore sequencing is mediated by the translation of an electrical signal into a sequence of nucleotides and does not involve nucleic acid amplification. In addition, nanopore sequencing can directly identify native base modifications and sequence viral RNA genomes in native status (Quick et al., 2016; Rand et al., 2017; Simpson et al., 2017; Garalde et al., 2018; Liu et al., 2019; Freed et al., 2020). Consequently, this substantially eliminates the biases introduced by PCR or reverse transcription. Many critical human diseases are caused by emerging RNA viruses such as COVID-19, severe acute respiratory syndrome, and Ebola.
To assess the performance of different high-throughput sequencing platforms in analyzing oral microbial communities and their relation to health, saliva samples from five healthy preschool children were collected and subjected to sequencing on the MiSeq (targeting V3-V4) and PacBio RS II (targeting V1-V9) platforms. Although the PacBio system provided a lower amount of clean data, it had a longer read length and higher resolution of nucleotide sequences at the species or strain level than those of NGS (Wang Y. et al., 2017; Zhang et al., 2020). Long-read sequencing of two independent oral S. epidermidis isolates was conducted using the Oxford Nanopore MinION sequencing platform. TGS results revealed that both isolates harbored a novel structural organization. The identification of independent genetic evolution further illustrates the diversity of arginine catabolic mobile element elements in S. epidermidis, which may confer a selective advantage to oral S. epidermidis in dental plaque (McManus et al., 2019).
The main drawback of TGS is its high error rate, which has been significantly improved (Rang et al., 2018). The state-of-the-art methodologies open up more possibility of identifying microorganisms as a second wave of technical advances in the recent decade (Ciuffreda et al., 2021), it should have a wider application in medical practice.
4. Microbial dysbiosis and oral pathology
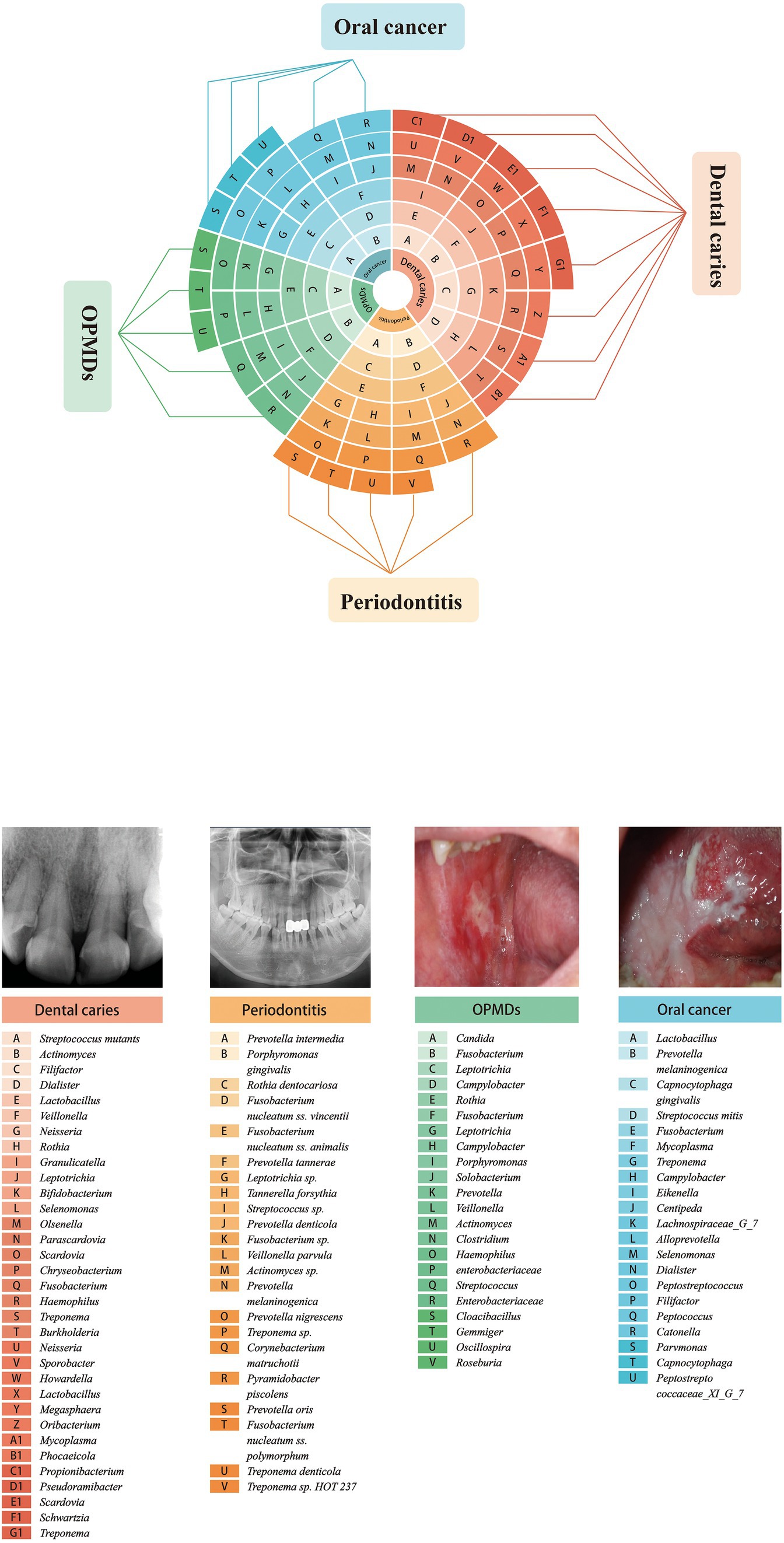
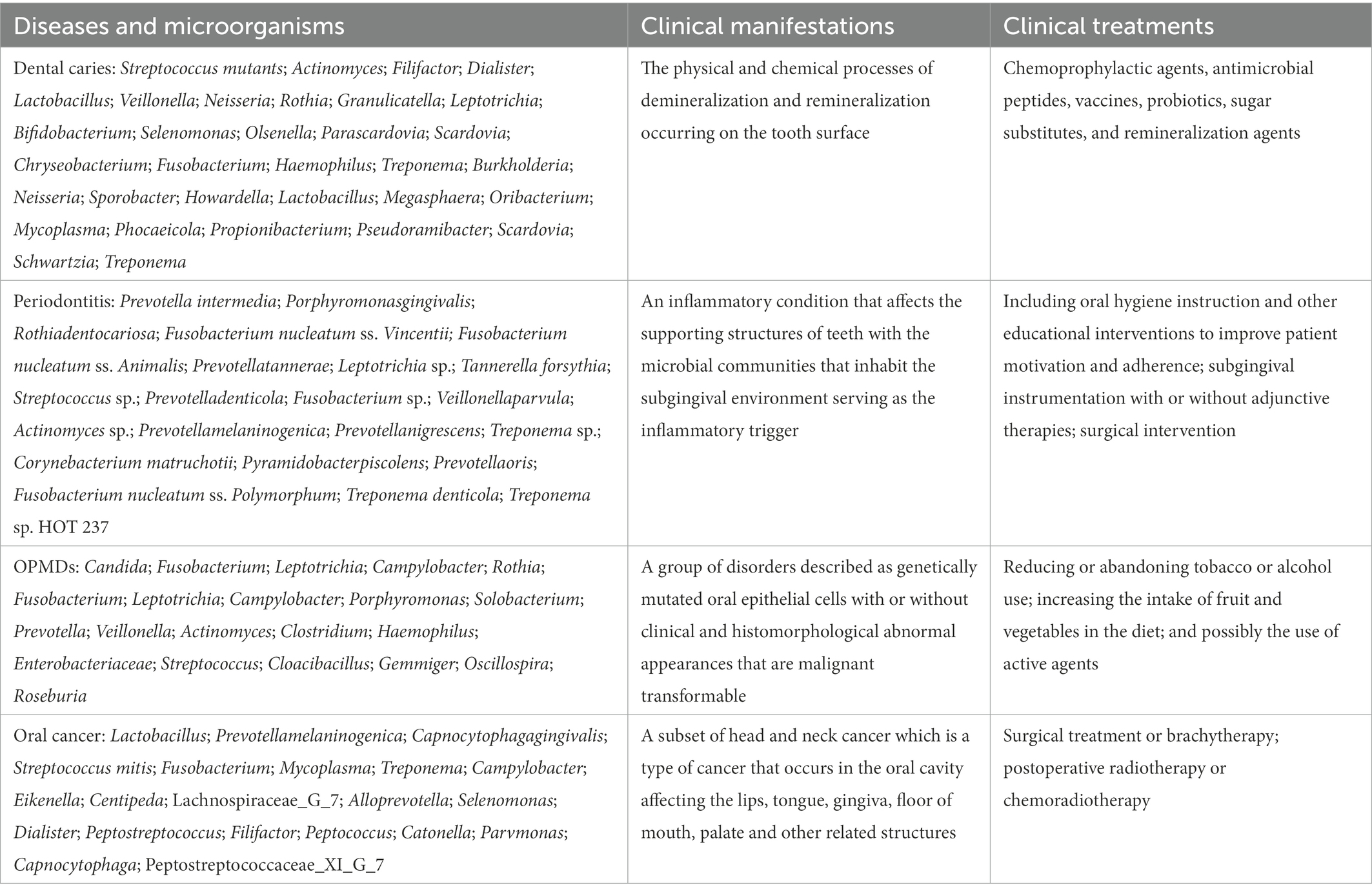
4.1. Dental caries
Dental caries, one of the most prevalent oral diseases, usually leads to tooth pain and pulp or periapical infections. This process involves three reversible stages. In the first stage, acidification is mild and infrequent, resulting from a microbial biofilm containing mainly non-mutans Streptococcus and Actinomyces (Takahashi and Nyvad, 2008). This is consistent with the balance of the demineralization/remineralization equilibrium or the conversion of the mineral balance toward net mineral gain. Once a continuous sugar supply is established, acidification changes moderately and frequently, which may improve the capability of non-mutans bacteria to produce acid and resist PH reduction. Therefore, non-mutans streptococci and other aciduric strains may selectively increase (Aas et al., 2005, 2008). These acidogenic and aciduric processes of the microorganism may shift the demineralization/remineralization equilibrium toward net mineral loss, resulting in the progression of dental caries (Becker et al., 2002; Martin et al., 2002; Munson et al., 2004; Mantzourani et al., 2009). Under continuous and prolonged acidic conditions, aciduric bacteria such as mutans streptococci, Lactobacillus, and Actinomyces may become dominant through acid-induced selection (Takahashi and Nyvad, 2011). Environmental acidification is a major factor in phenotypic and genotypic changes in microorganisms during the initiation and development of caries.
4.2. Periodontitis
Gingivitis is a reversible inflammation caused by the accumulation of bacterial plaque in the gingival tissue. The microbiota involved are commensal microbiota, such as Actinomyces species, F. nucleatum, Prevotella intermedia, Bacteroides, Capnocytophaga, and Eikenella (Huang et al., 2011; Igic et al., 2012).
Gingivitis may develop into periodontitis, resulting from loss of control. Periodontal disease is a chronic irreversible inflammation caused by the destruction of gum tissue, alveolar bone, and tooth loss. P. intermedia, F. nucleatum, Peptostreptococcus micros and Prevotella nigrescens, defined as microbiota of the “orange complex,” as well as the “red complex,” composed of Treponema denticola (T. denticola), Porphyromonas gingivalis (P. gingivalis), and Tannerella forsythia (T. forsythia) are the most commonly associated with periodontitis (Socransky et al., 1998). Although these microbial species are present in low numbers in healthy individuals, they are thought to be responsible for the occurrence and progression of the disease (Kumar et al., 2003). These complex microorganisms fall or disappear (below the detection limit) after efficient treatment.
Moreover, the presence of fungi, protozoa, viruses, and methanogenic archaea is highly correlated with the severity of chronic periodontitis (Horz et al., 2015; Zhu et al., 2015; Lauritano et al., 2016; Jabri et al., 2021).
4.3. Oral potentially malignant disorders
Oral potentially malignant disorders (OPMDs) are a series of lesions or conditions that may undergo malignant transformation to oral cancer. These diseases are most commonly represented as clinically white patches (oral lichen planus, leukoplakia), red patches (erythroplakia), or red and white patches (erythroleukoplakia). Although the etiopathogenesis of OPMDs is intricate and unclear, contemporary studies have suggested that microbial dysbiosis influences disease development through molecular mechanisms. Significant evidence supports that Candida colonization is regularly correlated with oral leukoplakia (OL), which is called as “Candida leukoplakia,” with infiltration of hyphae in the superficial epithelium (Abdulrahim et al., 2013; Gupta et al., 2019; Weerasekera et al., 2021). C. albicans is the most common Candida species associated with leukoplakia, and its genotype A strains are more frequently associated with OL transformation (Alnuaimi et al., 2015). Over the last decade, increased attention has been paid to microbial infections in the pathogenesis of oral lichen planus. Recent evidence indicates that bacteria are abundant throughout the epithelium and lamina propria of OLP tissues and are positively associated with the levels of infiltrated T cells and CD3+, CD4+, and CD8+ cells. Therefore, microorganisms may play a key role in OLP onset (Choi et al., 2016; Bombeccari et al., 2017; Baek and Choi, 2018; Yu et al., 2020). The microbiota varies among different OLP types, with higher levels of bacteria and lower levels of fungi. Higher enrichment of the fungi Candida and Aspergillus was identified in patients with reticular OLP than in healthy controls. In addition, Alternaria and Sclerotiniaceaeunidentified were significantly more abundant in patients with erosive OLP (Li et al., 2019).
4.4. Oral cancer
Oral squamous cell carcinoma (OSCC) is the most common malignant neoplasm of the oral cavity, accounting for 90% of head and neck cancers. The 5-year survival rate has remained at 50% within the last few decades due to asymptomatic presentation and poor prognosis. The driving factors vary significantly, including smoking, alcohol intake, and betel nut chewing. Recent studies have suggested that changes in the composition of the oral microbiota may play a role in the induction of oral cancer (Mager et al., 2005; Hooper et al., 2009; Schmidt et al., 2014; Srinivasprasad et al., 2015; Deo and Deshmukh, 2020; Stasiewicz and Karpinski, 2022). P. gingivalis, a critical pathogen in periodontal disease, is a significant trigger agent for oral cancer, and its colonization in tumor tissues is correlated with poor survival in patients with carcinoma (Karmakar et al., 2020; Nwizu et al., 2020; Kavarthapu and Gurumoorthy, 2021; Soder et al., 2021). A study has shown that the microbial communities in OSCC patients at stage 4 became more complicated compared to those in healthy controls. In addition, microbiological populations were significantly altered by the development of cancer from the early stage to the late stage. At the phylum level, the oral samples showed large changes in the abundances of Actinobacteria, Bacteroidetes, and Fusobacteria. The number of Fusobacteria increased significantly during cancer progression in the healthy controls. Although the abundance of dominant genera such as Fusobacteria increased with cancer development, the abundance of Haemophilus, Streptococcus, Actinomyces, and Porphyromonas decreased. Parvimona micra, Haemophilus influenzae, Fusobacterium periodonticum, Streptococcus constellatus, and Filifactor alocis progressively increased with oral cancer development (Yang et al., 2018). Another study revealed that the microbial composition of five genera, Enterococcus, Parvimonas, Peptostreptococcus, Bacillus, and Slackia, showed relatively significant differences between epithelial precursor lesions and cancer tissues. This shift in microbiota might serve as a potential biomarker for monitoring the occurrence and prognosis (Lee et al., 2017). The highly correlated organisms are summarized in Table 2 and Figure 2.
5. Oral microbiota and systemic condition
5.1. Atherosclerotic disease
Atherosclerotic disease, which is characterized by the accumulation of lipids and recruitment of T cells into the arterial wall, is considered a chronic inflammatory disorder. Although a number of traditional risk factors, such as diabetes, obesity, high blood pressure, and smoking are widely accepted, oral infection is increasingly manifested in atherosclerotic plaque samples (Fernandes et al., 2014; Aarabi et al., 2015; Slocum et al., 2016; Atarbashi-Moghadam et al., 2018; Herrera et al., 2020). Chhibber-Goel et al. (2016) demonstrated the presence of 23 oral commensal bacteria, either individually or in coexistence, within carotid endarterectomy biopsies from patients undergoing surgical procedures. Of these 23 bacteria, five were unique to coronary plaques, including P. gingivalis, P. endodontalis, P. intermedia, C. rectus, and P. nigrescens. The remaining 18 genera, otherwise confirmed in non-cardiac organs, are responsible for over 30 non-cardiac lesions (Chhibber-Goel et al., 2016).
5.2. Adverse pregnancy outcomes
Adverse pregnancy outcomes are a broad term containing preeclampsia, low birth weight, preterm premature rupture of membranes, early-onset neonatal sepsis, miscarriage, stillbirth, and fetal growth retardation (Figuero et al., 2020). The relationship between specific pathogenic bacteria and APO has been known for a long time (Komine-Aizawa et al., 2019; Bobetsis et al., 2020; Xu and Han, 2022). The prevalent oral species are Filifactor alocis, Fusobacterium nucleatum, Porphyromonas gingivalis, Campylobacter rectus, T. Denticola, and T. Forsythia, among others (Cobb et al., 2017). Stimulated fetal inflammatory and immune responses may ultimately increase the potential for APO owing to the virulent properties assigned to microbiota in the intrauterine environment.
5.3. Diabetes mellitus
Diabetes mellitus is a metabolic syndrome characterized by hyperglycemia caused by a defect in insulin secretion, decreased insulin action or both (Harreiter and Roden, 2019). Multiple studies have verified a bidirectional relationship between diabetes and periodontitis. Diabetes leads to a shift in connective tissue metabolism and consequently reduces the capacity of inflammation resolution and remodeling, which results in aggravated periodontal impairment. However, the periodontal condition in the host may negatively affect glycemic control in diabetic subjects and exacerbate diabetes progression and its complications (Lalla and Papapanou, 2011; Preshaw et al., 2012; Bascones-Martinez et al., 2015; Nascimento et al., 2018; Sanz et al., 2018; Baeza et al., 2020; Zhang X. et al., 2021). Casarin et al. (2013) reported dramatic differences in the subgingival ecosystem between individuals with and without diabetes. Compared to the levels in non-diabetic individuals, diabetes patients exhibited a higher abundance of Neisseria, TM7, Gemella, Eikenella, Selenomonas, Actinomyces, Capnocytophaga, Fusobacterium, Aggregatibacter, Veillonella, and Streptococcus genera and a lower abundance of Filifactor, Synergistetes, Tannerella, Eubacterium, Porphyromonas, and Treponema genera (Casarin et al., 2013).
6. Conclusion
The use of the current molecular-based detection approaches has dramatically expanded our awareness of the diversity, architecture, and function of oral microorganisms in health and diseases. Disease conditions supervene when the balance of the microbiota community is disrupted. However, the underlying mechanisms remain unclear. Further research is necessary to accurately identify the pathogenic microbiota. Numerous strategies for defining oral microbes have been investigated and developed. However, future studies must explore the security and efficacy of these methods.
Author contributions
XX, SL, and HD wrote the manuscript. YS prepared the figures. LZ and ZS reviewed the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Natural Science Foundation of Shanghai (No. 22ZR1454200) and Shanghai Stomatological Hospital Talent Project (No. SSDC-2019-RC01) to SL.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
Aarabi, G., Eberhard, J., Reissmann, D. R., Heydecke, G., and Seedorf, U. (2015). Interaction between periodontal disease and atherosclerotic vascular disease--fact or fiction? Atherosclerosis 241, 555–560. doi: 10.1016/j.atherosclerosis.2015.04.819
Aas, J. A., Griffen, A. L., Dardis, S. R., Lee, A. M., Olsen, I., Dewhirst, F. E., et al. (2008). Bacteria of dental caries in primary and permanent teeth in children and young adults. J. Clin. Microbiol. 46, 1407–1417. doi: 10.1128/JCM.01410-07
Aas, J. A., Paster, B. J., Stokes, L. N., Olsen, I., and Dewhirst, F. E. (2005). Defining the normal bacterial flora of the oral cavity. J. Clin. Microbiol. 43, 5721–5732. doi: 10.1128/JCM.43.11.5721-5732.2005
Abdulrahim, M. H., McManus, B. A., Flint, S. R., and Coleman, D. C. (2013). Genotyping Candida albicans from Candida leukoplakia and non-Candida leukoplakia shows no enrichment of multilocus sequence typing clades but enrichment of ABC genotype C in Candida leukoplakia. PLoS One 8:e73738. doi: 10.1371/journal.pone.0073738
Abualqomsaan, M., Toz, S. O., Yolasigmaz, A., and Turgay, N. (2010). The investigation of Entamoeba gingivalis and Trichomonas tenax in a group of patients with periodontal disease. Turkiye Parazitol. Derg. 34, 91–94.
Abusleme, L., Dupuy, A. K., Dutzan, N., Silva, N., Burleson, J. A., Strausbaugh, L. D., et al. (2013). The subgingival microbiome in health and periodontitis and its relationship with community biomass and inflammation. ISME J. 7, 1016–1025. doi: 10.1038/ismej.2012.174
Alnuaimi, A. D., Wiesenfeld, D., O'Brien-Simpson, N. M., Reynolds, E. C., and McCullough, M. J. (2015). Oral Candida colonization in oral cancer patients and its relationship with traditional risk factors of oral cancer: a matched case-control study. Oral Oncol. 51, 139–145. doi: 10.1016/j.oraloncology.2014.11.008
Anuta, V., Talianu, M. T., Dinu-Pirvu, C. E., Ghica, M. V., Prisada, R. M., Albu Kaya, M. G., et al. (2022). Molecular mapping of antifungal mechanisms accessing biomaterials and new agents to target oral candidiasis. Int. J. Mol. Sci. 23:7520. doi: 10.3390/ijms23147520
Arweiler, N. B., and Netuschil, L. (2016). The oral microbiota. Adv. Exp. Med. Biol. 902, 45–60. doi: 10.1007/978-3-319-31248-4_4
Atarbashi-Moghadam, F., Havaei, S. R., Havaei, S. A., Hosseini, N. S., Behdadmehr, G., and Atarbashi-Moghadam, S. (2018). Periopathogens in atherosclerotic plaques of patients with both cardiovascular disease and chronic periodontitis. ARYA Atheroscler. 14, 53–57. doi: 10.22122/arya.v14i2.1504
Atyeo, N., Rodriguez, M. D., Papp, B., and Toth, Z. (2021). Clinical manifestations and epigenetic regulation of oral herpesvirus infections. Viruses 13:681. doi: 10.3390/v13040681
Baek, K., and Choi, Y. (2018). The microbiology of oral lichen planus: is microbial infection the cause of oral lichen planus? Mol Oral Microbiol 33, 22–28. doi: 10.1111/omi.12197
Baeza, M., Morales, A., Cisterna, C., Cavalla, F., Jara, G., Isamitt, Y., et al. (2020). Effect of periodontal treatment in patients with periodontitis and diabetes: systematic review and meta-analysis. J. Appl. Oral Sci. 28:e20190248. doi: 10.1590/1678-7757-2019-0248
Bascones-Martinez, A., Munoz-Corcuera, M., and Bascones-Ilundain, J. (2015). Diabetes and periodontitis: a bidirectional relationship. Med. Clin. 145, 31–35. doi: 10.1016/j.medcli.2014.07.019
Becker, M. R., Paster, B. J., Leys, E. J., Moeschberger, M. L., Kenyon, S. G., Galvin, J. L., et al. (2002). Molecular analysis of bacterial species associated with childhood caries. J. Clin. Microbiol. 40, 1001–1009. doi: 10.1128/JCM.40.3.1001-1009.2002
Belstrom, D., Fiehn, N. E., Nielsen, C. H., Kirkby, N., Twetman, S., Klepac-Ceraj, V., et al. (2014). Differences in bacterial saliva profile between periodontitis patients and a control cohort. J. Clin. Periodontol. 41, 104–112. doi: 10.1111/jcpe.12190
Belstrom, D., Fiehn, N. E., Nielsen, C. H., Klepac-Ceraj, V., Paster, B. J., Twetman, S., et al. (2015). Differentiation of salivary bacterial profiles of subjects with periodontitis and dental caries. J. Oral Microbiol. 7:27429. doi: 10.3402/jom.v7.27429
Benn, A., Heng, N., Broadbent, J. M., and Thomson, W. M. (2018). Studying the human oral microbiome: challenges and the evolution of solutions. Aust. Dent. J. 63, 14–24. doi: 10.1111/adj.12565
Bobetsis, Y. A., Graziani, F., Gursoy, M., and Madianos, P. N. (2020). Periodontal disease and adverse pregnancy outcomes. Periodontol. 83, 154–174. doi: 10.1111/prd.12294
Bombeccari, G. P., Gianni, A. B., and Spadari, F. (2017). Oral Candida colonization and oral lichen planus. Oral Dis. 23, 1009–1010. doi: 10.1111/odi.12681
Cannon, R. D. (2022). Oral fungal infections: past, present, and future. Front. Oral Health 3:838639. doi: 10.3389/froh.2022.838639
Casarin, R. C., Barbagallo, A., Meulman, T., Santos, V. R., Sallum, E. A., Nociti, F. H., et al. (2013). Subgingival biodiversity in subjects with uncontrolled type-2 diabetes and chronic periodontitis. J. Periodontal Res. 48, 30–36. doi: 10.1111/j.1600-0765.2012.01498.x
Celik, Z. C., Cakiris, A., Abaci, N., Yaniikoglu, F., Ilgin, C., Ekmekci, S. S., et al. (2021). The complex microbiome of caries-active and caries-free supragingival plaques in permanent dentition. Niger. J. Clin. Pract. 24, 1535–1540. doi: 10.4103/njcp.njcp_49_21
Chen, L., Liu, W., Zhang, Q., Xu, K., Ye, G., Wu, W., et al. (2020). RNA based mNGS approach identifies a novel human coronavirus from two individual pneumonia cases in 2019 Wuhan outbreak. Emerg. Microbes Infect. 9, 313–319. doi: 10.1080/22221751.2020.1725399
Chevalier, M., Ranque, S., and Precheur, I. (2018). Oral fungal-bacterial biofilm models in vitro: a review. Med. Mycol. 56, 653–667. doi: 10.1093/mmy/myx111
Chhibber-Goel, J., Singhal, V., Bhowmik, D., Vivek, R., Parakh, N., Bhargava, B., et al. (2016). Linkages between oral commensal bacteria and atherosclerotic plaques in coronary artery disease patients. NPJ Biofilms Microbiomes 2:7. doi: 10.1038/s41522-016-0009-7
Choi, Y. S., Kim, Y., Yoon, H. J., Baek, K. J., Alam, J., Park, H. K., et al. (2016). The presence of bacteria within tissue provides insights into the pathogenesis of oral lichen planus. Sci. Rep. 6:29186. doi: 10.1038/srep29186
Ciuffreda, L., Rodriguez-Perez, H., and Flores, C. (2021). Nanopore sequencing and its application to the study of microbial communities. Comput. Struct. Biotechnol. J. 19, 1497–1511. doi: 10.1016/j.csbj.2021.02.020
Claesson, M. J., Wang, Q., O'Sullivan, O., Greene-Diniz, R., Cole, J. R., Ross, R. P., et al. (2010). Comparison of two next-generation sequencing technologies for resolving highly complex microbiota composition using tandem variable 16S rRNA gene regions. Nucleic Acids Res. 38:e200. doi: 10.1093/nar/gkq873
Cobb, C. M., Kelly, P. J., Williams, K. B., Babbar, S., Angolkar, M., and Derman, R. J. (2017). The oral microbiome and adverse pregnancy outcomes. Int. J. Women's Health 9, 551–559. doi: 10.2147/IJWH.S142730
Corralo, D. J., Ev, L. D., Dame-Teixeira, N., Maltz, M., Arthur, R. A., Do, T., et al. (2021). Functionally active microbiome in Supragingival biofilms in health and caries. Caries Res. 55, 603–616. doi: 10.1159/000518963
Costalonga, M., and Herzberg, M. C. (2014). The oral microbiome and the immunobiology of periodontal disease and caries. Immunol. Lett. 162, 22–38. doi: 10.1016/j.imlet.2014.08.017
Crimi, S., Fiorillo, L., Bianchi, A., D'Amico, C., Amoroso, G., Gorassini, F., et al. (2019). Herpes virus, Oral clinical signs and QoL: Systematic review of recent data. Viruses 11:463. doi: 10.3390/v11050463
Dai, X., and Zhou, Z. H. (2018). Structure of the herpes simplex virus 1 capsid with associated tegument protein complexes. Science 360:7298. doi: 10.1126/science.aao7298
Deng, H., Xu, Y., Liu, Y., Che, Z., Guo, H., Shan, S., et al. (2012). Gold nanoparticles with asymmetric polymerase chain reaction for colorimetric detection of DNA sequence. Anal. Chem. 84, 1253–1258. doi: 10.1021/ac201713t
Deo, P. N., and Deshmukh, R. (2020). Oral microbiome and oral cancer–the probable nexus. J. Oral Maxillofac Pathol. 24, 361–367. doi: 10.4103/jomfp.JOMFP_20_20
Duncan, H. F., Galler, K. M., Tomson, P. L., Simon, S., El-Karim, I., Kundzina, R., et al. (2019). European society of endodontology position statement: management of deep caries and the exposed pulp. Int. Endod. J. 52, 923–934. doi: 10.1111/iej.13080
Earl, J. P., Adappa, N. D., Krol, J., Bhat, A. S., Balashov, S., Ehrlich, R. L., et al. (2018). Species-level bacterial community profiling of the healthy sinonasal microbiome using Pacific biosciences sequencing of full-length 16S rRNA genes. Microbiome 6:190. doi: 10.1186/s40168-018-0569-2
El-Kafrawy, S. A., El-Daly, M. M., Hassan, A. M., Kaki, R. M., Abuzenadah, A. M., Kamal, M. A., et al. (2021). A direct method for RT-PCR detection of SARS-CoV-2 in clinical samples. Healthcare 9:37. doi: 10.3390/healthcare9010037
Fadhil Ali Malaa, S. B., Jwad, A. A. A. A., and Al-Masoudi, H. K. (2022). Assessment of Entamoeba gingivalis and Trichomonas tenax in patients with chronic diseases and its correlation with some risk factors. Arch. Razi. Inst. 77, 87–93. doi: 10.22092/ARI.2021.356549.1868
Farrell, J. J., Zhang, L., Zhou, H., Chia, D., Elashoff, D., Akin, D., et al. (2012). Variations of oral microbiota are associated with pancreatic diseases including pancreatic cancer. Gut 61, 582–588. doi: 10.1136/gutjnl-2011-300784
Fernandes, C. P., Oliveira, F. A., Silva, P. G., Alves, A. P., Mota, M. R., Montenegro, R. C., et al. (2014). Molecular analysis of oral bacteria in dental biofilm and atherosclerotic plaques of patients with vascular disease. Int. J. Cardiol. 174, 710–712. doi: 10.1016/j.ijcard.2014.04.201
Figuero, E., Han, Y. W., and Furuichi, Y. (2020). Periodontal diseases and adverse pregnancy outcomes: mechanisms. Periodontol 2000 83, 175–188. doi: 10.1111/prd.12295
Franzen, O., Hu, J., Bao, X., Itzkowitz, S. H., Peter, I., and Bashir, A. (2015). Improved OTU-picking using long-read 16S rRNA gene amplicon sequencing and generic hierarchical clustering. Microbiome 3:43. doi: 10.1186/s40168-015-0105-6
Freed, N. E., Vlkova, M., Faisal, M. B., and Silander, O. K. (2020). Rapid and inexpensive whole-genome sequencing of SARS-CoV-2 using 1200 bp tiled amplicons and Oxford Nanopore rapid barcoding. Biol. Methods Protoc. 5:bpaa014. doi: 10.1093/biomethods/bpaa014
Gao, X., Jiang, S., Koh, D., and Hsu, C. Y. (2016). Salivary biomarkers for dental caries. Periodontol 2000 70, 128–141. doi: 10.1111/prd.12100
Garalde, D. R., Snell, E. A., Jachimowicz, D., Sipos, B., Lloyd, J. H., Bruce, M., et al. (2018). Highly parallel direct RNA sequencing on an array of nanopores. Nat. Methods 15, 201–206. doi: 10.1038/nmeth.4577
Gardner, S. N., Jaing, C. J., McLoughlin, K. S., and Slezak, T. R. (2010). A microbial detection array (MDA) for viral and bacterial detection. BMC Genomics 11:668. doi: 10.1186/1471-2164-11-668
Ghabanchi, J., Zibaei, M., Afkar, M. D., and Sarbazie, A. H. (2010). Prevalence of oral Entamoeba gingivalis and Trichomonas tenax in patients with periodontal disease and healthy population in shiraz, southern Iran. Indian J. Dent. Res. 21, 89–91. doi: 10.4103/0970-9290.62821
Ghannoum, M. A., Jurevic, R. J., Mukherjee, P. K., Cui, F., Sikaroodi, M., Naqvi, A., et al. (2010). Characterization of the oral fungal microbiome (mycobiome) in healthy individuals. PLoS Pathog. 6:e1000713. doi: 10.1371/journal.ppat.1000713
Ghurye, J. S., Cepeda-Espinoza, V., and Pop, M. (2016). Metagenomic assembly: overview, challenges and applications. Yale J. Biol. Med. 89, 353–362.
Gomes-Filho, I. S., Pereira, E. C., Cruz, S. S., Adan, L. F., Vianna, M. I., Passos-Soares, J. S., et al. (2016). Relationship among Mothers' glycemic level, periodontitis, and birth weight. J. Periodontol. 87, 238–247. doi: 10.1902/jop.2015.150423
Griffen, A. L., Beall, C. J., Campbell, J. H., Firestone, N. D., Kumar, P. S., Yang, Z. K., et al. (2012). Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME J. 6, 1176–1185. doi: 10.1038/ismej.2011.191
Gu, W., Deng, X., Lee, M., Sucu, Y. D., Arevalo, S., Stryke, D., et al. (2021). Rapid pathogen detection by metagenomic next-generation sequencing of infected body fluids. Nat. Med. 27, 115–124. doi: 10.1038/s41591-020-1105-z
Gu, W., Miller, S., and Chiu, C. Y. (2019). Clinical metagenomic next-generation sequencing for pathogen detection. Annu. Rev. Pathol. 14, 319–338. doi: 10.1146/annurev-pathmechdis-012418-012751
Gupta, V., Abhisheik, K., Balasundari, S., Devendra, N. K., Shadab, K., and Anupama, M. (2019). Identification of Candida albicans using different culture media and its association in leukoplakia and oral squamous cell carcinoma. J. Oral Maxillofac Pathol. 23, 28–35. doi: 10.4103/jomfp.JOMFP_157_18
Han, D., Li, Z., Li, R., Tan, P., Zhang, R., and Li, J. (2019). mNGS in clinical microbiology laboratories: on the road to maturity. Crit. Rev. Microbiol. 45, 668–685. doi: 10.1080/1040841X.2019.1681933
Harreiter, J., and Roden, M. (2019). Diabetes mellitus-definition, classification, diagnosis, screening and prevention (update 2019). Wien. Klin. Wochenschr. 131, 6–15. doi: 10.1007/s00508-019-1450-4
He, G., Chen, C. Y., Zhang, X., Ding, P. P., Hu, C. Z., Huang, X. F., et al. (2022). Clinical performance of quantitative PCR for the molecular identification of skeletal tuberculosis from formalin-fixed paraffin-embedded tissues. BMC Infect. Dis. 22:651. doi: 10.1186/s12879-022-07641-7
He, J., Li, Y., Cao, Y., Xue, J., and Zhou, X. (2015). The oral microbiome diversity and its relation to human diseases. Folia Microbiol. 60, 69–80. doi: 10.1007/s12223-014-0342-2
Herrera, D., Molina, A., Buhlin, K., and Klinge, B. (2020). Periodontal diseases and association with atherosclerotic disease. Periodontol 2000 83, 66–89. doi: 10.1111/prd.12302
Hong, B. Y., Furtado Araujo, M. V., Strausbaugh, L. D., Terzi, E., Ioannidou, E., and Diaz, P. I. (2015). Microbiome profiles in periodontitis in relation to host and disease characteristics. PLoS One 10:e0127077. doi: 10.1371/journal.pone.0127077
Hooper, S. J., Wilson, M. J., and Crean, S. J. (2009). Exploring the link between microorganisms and oral cancer: a systematic review of the literature. Head Neck 31, 1228–1239. doi: 10.1002/hed.21140
Horz, H. P., Robertz, N., Vianna, M. E., Henne, K., and Conrads, G. (2015). Relationship between methanogenic archaea and subgingival microbial complexes in human periodontitis. Anaerobe 35, 10–12. doi: 10.1016/j.anaerobe.2015.02.008
Hu, L., He, C., Zhao, C., Chen, X., Hua, H., and Yan, Z. (2019). Characterization of oral candidiasis and the Candida species profile in patients with oral mucosal diseases. Microb. Pathog. 134:103575. doi: 10.1016/j.micpath.2019.103575
Huang, S., Yang, F., Zeng, X., Chen, J., Li, R., Wen, T., et al. (2011). Preliminary characterization of the oral microbiota of Chinese adults with and without gingivitis. BMC Oral Health 11:33. doi: 10.1186/1472-6831-11-33
Huang, W., Yang, Y., Zhang, X., Zhao, C., Yin, A., Zhang, X., et al. (2013). An easy operating pathogen microarray (EOPM) platform for rapid screening of vertebrate pathogens. BMC Infect. Dis. 13:437. doi: 10.1186/1471-2334-13-437
Igic, M., Kesic, L., Lekovic, V., Apostolovic, M., Mihailovic, D., Kostadinovic, L., et al. (2012). Chronic gingivitis: the prevalence of periodontopathogens and therapy efficiency. Eur. J. Clin. Microbiol. Infect. Dis. 31, 1911–1915. doi: 10.1007/s10096-011-1520-7
Jabri, B., Iken, M., Achmit, M., Rida, S., and Ennibi, O. K. (2021). Occurrence of Candida albicans in periodontitis. Int. J. Dent. 2021:5589664. doi: 10.1155/2021/5589664
Janda, J. M., and Abbott, S. L. (2007). 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: pluses, perils, and pitfalls. J. Clin. Microbiol. 45, 2761–2764. doi: 10.1128/JCM.01228-07
Jenkinson, H. F. (2011). Beyond the oral microbiome. Environ. Microbiol. 13, 3077–3087. doi: 10.1111/j.1462-2920.2011.02573.x
Jiang, S., and Dong, Y. (2017). Human papillomavirus and oral squamous cell carcinoma: a review of HPV-positive oral squamous cell carcinoma and possible strategies for future. Curr. Probl. Cancer 41, 323–327. doi: 10.1016/j.currproblcancer.2017.02.006
Jiao, X., Zheng, X., Ma, L., Kutty, G., Gogineni, E., Sun, Q., et al. (2013). A benchmark study on error assessment and quality control of CCS reads derived from the PacBio RS. J. Data Mining Genomics Proteomics,: 136. doi: 10.4172/2153-0602.1000136
Jo, J. H., Kennedy, E. A., and Kong, H. H. (2016). Research techniques made simple: bacterial 16S ribosomal RNA gene sequencing in cutaneous research. J. Invest. Dermatol. 136, e23–e27. doi: 10.1016/j.jid.2016.01.005
Johnson, J. S., Spakowicz, D. J., Hong, B. Y., Petersen, L. M., Demkowicz, P., Chen, L., et al. (2019). Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat. Commun. 10:5029. doi: 10.1038/s41467-019-13036-1
Karmakar, S., Kar, A., Thakur, S., and Rao, V. U. S. (2020). Periodontitis and oral cancer-a striking link. Oral Oncol. 106:104630. doi: 10.1016/j.oraloncology.2020.104630
Kavarthapu, A., and Gurumoorthy, K. (2021). Linking chronic periodontitis and oral cancer: a review. Oral Oncol. 121:105375. doi: 10.1016/j.oraloncology.2021.105375
Kazor, C. E., Mitchell, P. M., Lee, A. M., Stokes, L. N., Loesche, W. J., Dewhirst, F. E., et al. (2003). Diversity of bacterial populations on the tongue dorsa of patients with halitosis and healthy patients. J. Clin. Microbiol. 41, 558–563. doi: 10.1128/JCM.41.2.558-563.2003
Khurshid, Z., Zafar, M. S., Khan, R. S., Najeeb, S., Slowey, P. D., and Rehman, I. U. (2018). Role of salivary biomarkers in Oral cancer detection. Adv. Clin. Chem. 86, 23–70. doi: 10.1016/bs.acc.2018.05.002
Kilian, M. (2007). “Haemophilus” in Manual of clinical microbiology. eds. P. R. Murray, E. J. Baron, J. H. Jorgensen, M. L. Landry, and M. A. Pfaller. 9th ed (Washington, DC: ASM Press), 636–648.
Kilian, M., Chapple, I. L., Hannig, M., Marsh, P. D., Meuric, V., Pedersen, A. M., et al. (2016). The oral microbiome–an update for oral healthcare professionals. Br. Dent. J. 221, 657–666. doi: 10.1038/sj.bdj.2016.865
Komine-Aizawa, S., Aizawa, S., and Hayakawa, S. (2019). Periodontal diseases and adverse pregnancy outcomes. J. Obstet. Gynaecol. Res. 45, 5–12. doi: 10.1111/jog.13782
Krishnan, K., Chen, T., and Paster, B. J. (2017). A practical guide to the oral microbiome and its relation to health and disease. Oral Dis. 23, 276–286. doi: 10.1111/odi.12509
Kumar, P. S., Griffen, A. L., Barton, J. A., Paster, B. J., Moeschberger, M. L., and Leys, E. J. (2003). New bacterial species associated with chronic periodontitis. J. Dent. Res. 82, 338–344. doi: 10.1177/154405910308200503
Lagier, J. C., Edouard, S., Pagnier, I., Mediannikov, O., Drancourt, M., and Raoult, D. (2015). Current and past strategies for bacterial culture in clinical microbiology. Clin. Microbiol. Rev. 28, 208–236. doi: 10.1128/CMR.00110-14
Lalla, E., and Papapanou, P. N. (2011). Diabetes mellitus and periodontitis: a tale of two common interrelated diseases. Nat. Rev. Endocrinol. 7, 738–748. doi: 10.1038/nrendo.2011.106
Lauritano, D., Lo Muzio, L., Gaudio, R. M., Lo Russo, L., Mucchi, D., Nardi, G. M., et al. (2016). The ecological catastrophe of oral diseases: a possible link between periodontitis and protozoa. J. Biol. Regul. Homeost. Agents 30, 143–147.
Laver, T., Harrison, J., O'Neill, P. A., Moore, K., Farbos, A., Paszkiewicz, K., et al. (2015). Assessing the performance of the Oxford Nanopore technologies MinION. Biomol. Detect Quant. 3, 1–8. doi: 10.1016/j.bdq.2015.02.001
Lee, W. H., Chen, H. M., Yang, S. F., Liang, C., Peng, C. Y., Lin, F. M., et al. (2017). Bacterial alterations in salivary microbiota and their association in oral cancer. Sci. Rep. 7:16540. doi: 10.1038/s41598-017-16418-x
Leys, E. J., Griffen, A. L., Beall, C., and Maiden, M. F. (2013). “Isolation, classification, and identification of oral microorganisms” in Oral microbiology and immunology. eds. R. J. Lamont, G. N. Hajishengallis, and H. F. Jenkinson (Washington: ASM Press), 77–96.
Li, C., Chng, K. R., Boey, E. J., Ng, A. H., Wilm, A., and Nagarajan, N. (2016). INC-Seq: accurate single molecule reads using nanopore sequencing. Gigascience 5:34. doi: 10.1186/s13742-016-0140-7
Li, H., Gao, H., Meng, H., Wang, Q., Li, S., Chen, H., et al. (2018a). Detection of pulmonary infectious pathogens from lung biopsy tissues by metagenomic next-generation sequencing. Front. Cell. Infect. Microbiol. 8:205. doi: 10.3389/fcimb.2018.00205
Li, H., Wang, J., Wu, L., Luo, J., Liang, X., Xiao, B., et al. (2018b). The impacts of delivery mode on infant's oral microflora. Sci. Rep. 8:11938. doi: 10.1038/s41598-018-30397-7
Li, Y., Wang, K., Zhang, B., Tu, Q., Yao, Y., Cui, B., et al. (2019). Salivary mycobiome dysbiosis and its potential impact on bacteriome shifts and host immunity in oral lichen planus. Int. J. Oral Sci. 11:13. doi: 10.1038/s41368-019-0045-2
Liu, H., Begik, O., Lucas, M. C., Ramirez, J. M., Mason, C. E., Wiener, D., et al. (2019). Accurate detection of m(6) a RNA modifications in native RNA sequences. Nat. Commun. 10:4079. doi: 10.1038/s41467-019-11713-9
Liu, L., Li, Y., Li, S., Hu, N., He, Y., Pong, R., et al. (2012). Comparison of next-generation sequencing systems. J. Biomed. Biotechnol. 2012:251364. doi: 10.1155/2012/251364
Lochman, J., Zapletalova, M., Poskerova, H., Izakovicova Holla, L., and Borilova Linhartova, P. (2019). Rapid multiplex real-time PCR method for the detection and quantification of selected cariogenic and periodontal bacteria. Diagnostics 10:8. doi: 10.3390/diagnostics10010008
Loeffelholz, M. J., and Sanden, G. N. (2007). “Bordetella” in Manual of clinical microbiology. eds. P. R. Murray, E. J. Baron, J. H. Jorgensen, M. L. Landry, and M. A. Pfaller. 9th ed (Washington, DC: ASM Press), 803–814.
Lourenco, T. G., Heller, D., Silva-Boghossian, C. M., Cotton, S. L., Paster, B. J., and Colombo, A. P. (2014). Microbial signature profiles of periodontally healthy and diseased patients. J. Clin. Periodontol. 41, 1027–1036. doi: 10.1111/jcpe.12302
Mager, D. L., Haffajee, A. D., Devlin, P. M., Norris, C. M., Posner, M. R., and Goodson, J. M. (2005). The salivary microbiota as a diagnostic indicator of oral cancer: a descriptive, non-randomized study of cancer-free and oral squamous cell carcinoma subjects. J. Transl. Med. 3:27. doi: 10.1186/1479-5876-3-27
Mager, D. L., Ximenez-Fyvie, L. A., Haffajee, A. D., and Socransky, S. S. (2003). Distribution of selected bacterial species on intraoral surfaces. J. Clin. Periodontol. 30, 644–654. doi: 10.1034/j.1600-051x.2003.00376.x
Mantzourani, M., Gilbert, S. C., Sulong, H. N., Sheehy, E. C., Tank, S., Fenlon, M., et al. (2009). The isolation of bifidobacteria from occlusal carious lesions in children and adults. Caries Res. 43, 308–313. doi: 10.1159/000222659
Mark Welch, J. L., Rossetti, B. J., Rieken, C. W., Dewhirst, F. E., and Borisy, G. G. (2016). Biogeography of a human oral microbiome at the micron scale. Proc. Natl. Acad. Sci. U. S. A. 113, E791–E800. doi: 10.1073/pnas.1522149113
Marmonier, A. (2007). “Diagnostic phénotypique” in Précis de bactériologie clinique. eds. J. Freney, F. Renaud, R. Leclercq, and P. Riegel (Paris, France: ESKA), 49–60.
Marsh, P. D. (2006). Dental plaque as a biofilm and a microbial community–implications for health and disease. BMC Oral Health 6:S14. doi: 10.1186/1472-6831-6-S1-S14
Marsh, P. D., Martin, M. V., Lewis, M. A. O., and Williams, D. W. (2009). Oral microbiology, 5th. Churchill Livingstone Elsevier, Edinburgh.
Martin, F. E., Nadkarni, M. A., Jacques, N. A., and Hunter, N. (2002). Quantitative microbiological study of human carious dentine by culture and real-time PCR: association of anaerobes with histopathological changes in chronic pulpitis. J. Clin. Microbiol. 40, 1698–1704. doi: 10.1128/JCM.40.5.1698-1704.2002
Mbareche, H., Veillette, M., Bilodeau, G., and Duchaine, C. (2020). Comparison of the performance of ITS1 and ITS2 as barcodes in amplicon-based sequencing of bioaerosols. PeerJ 8:e8523. doi: 10.7717/peerj.8523
McManus, B. A., O'Connor, A. M., Egan, S. A., Flanagan, P. R., and Coleman, D. C. (2019). First description of arginine catabolic mobile element (ACME) type VI harboring the kdp operon only in Staphylococcus epidermidis using short and long read whole genome sequencing: further evidence of ACME diversity. Infect. Genet. Evol. 71, 51–53. doi: 10.1016/j.meegid.2019.03.008
Miao, Q., Ma, Y., Wang, Q., Pan, J., Zhang, Y., Jin, W., et al. (2018). Microbiological diagnostic performance of metagenomic next-generation sequencing when applied to clinical practice. Clin. Infect. Dis. 67, S231–S240. doi: 10.1093/cid/ciy693
Moran-Torres, A., Pazos-Salazar, N. G., Tellez-Lorenzo, S., Jimenez-Lima, R., Lizano, M., Reyes-Hernandez, D. O., et al. (2021). HPV oral and oropharynx infection dynamics in young population. Braz. J. Microbiol. 52, 1991–2000. doi: 10.1007/s42770-021-00602-3
Mosher, J. J., Bowman, B., Bernberg, E. L., Shevchenko, O., Kan, J., Korlach, J., et al. (2014). Improved performance of the PacBio SMRT technology for 16S rDNA sequencing. J. Microbiol. Methods 104, 59–60. doi: 10.1016/j.mimet.2014.06.012
Moyes, D. L., Islam, A., Kohli, A., and Naglik, J. R. (2016). Oral epithelial cells and their interactions with HIV-1. Oral Dis. 22, 66–72. doi: 10.1111/odi.12410
Munson, M. A., Banerjee, A., Watson, T. F., and Wade, W. G. (2004). Molecular analysis of the microflora associated with dental caries. J. Clin. Microbiol. 42, 3023–3029. doi: 10.1128/JCM.42.7.3023-3029.2004
Nascimento, G. G., Leite, F. R. M., Vestergaard, P., Scheutz, F., and Lopez, R. (2018). Does diabetes increase the risk of periodontitis? A systematic review and meta-regression analysis of longitudinal prospective studies. Acta Diabetol. 55, 653–667. doi: 10.1007/s00592-018-1120-4
Nearing, J. T., DeClercq, V., Van Limbergen, J., and Langille, M. G. I. (2020). Assessing the variation within the Oral microbiome of healthy adults. mSphere 5. doi: 10.1128/mSphere.00451-20
Nobbs, A. H., and Jenkinson, H. F. (2015). Interkingdom networking within the oral microbiome. Microbes Infect. 17, 484–492. doi: 10.1016/j.micinf.2015.03.008
Nwizu, N., Wactawski-Wende, J., and Genco, R. J. (2020). Periodontal disease and cancer: epidemiologic studies and possible mechanisms. Periodontol 2000 83, 213–233. doi: 10.1111/prd.12329
Olson, J. C., Cuff, C. F., Lukomski, S., Lukomska, E., Canizales, Y., Wu, B., et al. (2011). Use of 16S ribosomal RNA gene analyses to characterize the bacterial signature associated with poor oral health in West Virginia. BMC Oral Health 11:7. doi: 10.1186/1472-6831-11-7
Ottria, L., Lauritano, D., Oberti, L., Candotto, V., Cura, F., Tagliabue, A., et al. (2018). Prevalence of HIV-related oral manifestations and their association with HAART and CD4+ T cell count: a review. J. Biol. Regul. Homeost. Agents 32, 51–59.
Palacios, G., Druce, J., Du, L., Tran, T., Birch, C., Briese, T., et al. (2008). A new arenavirus in a cluster of fatal transplant-associated diseases. N. Engl. J. Med. 358, 991–998. doi: 10.1056/NEJMoa073785
Palacios, G., Quan, P. L., Jabado, O. J., Conlan, S., Hirschberg, D. L., Liu, Y., et al. (2007). Panmicrobial oligonucleotide array for diagnosis of infectious diseases. Emerg. Infect. Dis. 13, 73–81. doi: 10.3201/eid1301.060837
Palmer, R. J. Jr. (2014). Composition and development of oral bacterial communities. Periodontol 2000 64, 20–39. doi: 10.1111/j.1600-0757.2012.00453.x
Parolin, C., Giordani, B., Nahui Palomino, R. A., Biagi, E., Severgnini, M., Consolandi, C., et al. (2017). Design and validation of a DNA-microarray for phylogenetic analysis of bacterial communities in different oral samples and dental implants. Sci. Rep. 7:6280. doi: 10.1038/s41598-017-06743-6
Perez Rosero, E., Heron, S., Jovel, J., O'Neil, C. R., Turvey, S. L., Parashar, P., et al. (2021). Differential signature of the microbiome and neutrophils in the oral cavity of HIV-infected individuals. Front. Immunol. 12:780910. doi: 10.3389/fimmu.2021.780910
Peters, B. A., Wu, J., Hayes, R. B., and Ahn, J. (2017). The oral fungal mycobiome: characteristics and relation to periodontitis in a pilot study. BMC Microbiol. 17:157. doi: 10.1186/s12866-017-1064-9
Polvora, T. L. S., Nobre, A. V. V., Tirapelli, C., Taba, M. Jr., Macedo, L. D., Santana, R. C., et al. (2018). Relationship between human immunodeficiency virus (HIV-1) infection and chronic periodontitis. Expert. Rev. Clin. Immunol. 14, 315–327. doi: 10.1080/1744666X.2018.1459571
Preshaw, P. M., Alba, A. L., Herrera, D., Jepsen, S., Konstantinidis, A., Makrilakis, K., et al. (2012). Periodontitis and diabetes: a two-way relationship. Diabetologia 55, 21–31. doi: 10.1007/s00125-011-2342-y
Qian, Y. Y., Wang, H. Y., Zhou, Y., Zhang, H. C., Zhu, Y. M., Zhou, X., et al. (2020). Improving pulmonary infection diagnosis with metagenomic next generation sequencing. Front. Cell. Infect. Microbiol. 10:567615. doi: 10.3389/fcimb.2020.567615
Quick, J., Loman, N. J., Duraffour, S., Simpson, J. T., Severi, E., Cowley, L., et al. (2016). Real-time, portable genome sequencing for Ebola surveillance. Nature 530, 228–232. doi: 10.1038/nature16996
Quirynen, M., De Soete, M., Dierickx, K., and van Steenberghe, D. (2001). The intra-oral translocation of periodontopathogens jeopardises the outcome of periodontal therapy. A review of the literature. J. Clin. Periodontol. 28, 499–507. doi: 10.1034/j.1600-051x.2001.028006499.x
Rand, A. C., Jain, M., Eizenga, J. M., Musselman-Brown, A., Olsen, H. E., Akeson, M., et al. (2017). Mapping DNA methylation with high-throughput nanopore sequencing. Nat. Methods 14, 411–413. doi: 10.1038/nmeth.4189
Rang, F. J., Kloosterman, W. P., and de Ridder, J. (2018). From squiggle to basepair: computational approaches for improving nanopore sequencing read accuracy. Genome Biol. 19:90. doi: 10.1186/s13059-018-1462-9
Rashidi Maybodi, F., Haerian Ardakani, A., Fattahi Bafghi, A., Haerian Ardakani, A., and Zafarbakhsh, A. (2016). The effect of nonsurgical periodontal therapy on Trichomonas tenax and Entamoeba gingivalis in patients with chronic periodontitis. J. Dent. 17, 171–176.
Ren, W., Zhang, Q., Liu, X., Zheng, S., Ma, L., Chen, F., et al. (2017). Exploring the oral microflora of preschool children. J. Microbiol. 55, 531–537. doi: 10.1007/s12275-017-6474-8
Richards, V. P., Alvarez, A. J., Luce, A. R., Bedenbaugh, M., Mitchell, M. L., Burne, R. A., et al. (2017). Microbiomes of site-specific dental plaques from children with different caries status. Infect. Immun. 85. doi: 10.1128/IAI.00106-17
Sakamoto, M., Umeda, M., and Benno, Y. (2005). Molecular analysis of human oral microbiota. J. Periodontal Res. 40, 277–285. doi: 10.1111/j.1600-0765.2005.00793.x
Santacroce, L., Di Cosola, M., Bottalico, L., Topi, S., Charitos, I. A., Ballini, A., et al. (2021). Focus on HPV infection and the molecular mechanisms of Oral carcinogenesis. Viruses 13:559. doi: 10.3390/v13040559
Santella, A. J., Matthews, A., Casa-Levine, C., Pizzitola, L., Doonachar, A., and Page, G. O. (2019). Oral rapid HIV testing: implementation experiences of dental hygiene faculty and students. J. Dent. Hyg. 93, 23–32.
Sanz, M., Ceriello, A., Buysschaert, M., Chapple, I., Demmer, R. T., Graziani, F., et al. (2018). Scientific evidence on the links between periodontal diseases and diabetes: consensus report and guidelines of the joint workshop on periodontal diseases and diabetes by the international diabetes federation and the European Federation of Periodontology. J. Clin. Periodontol. 45, 138–149. doi: 10.1111/jcpe.12808
Schloss, P. D., Jenior, M. L., Koumpouras, C. C., Westcott, S. L., and Highlander, S. K. (2016). Sequencing 16S rRNA gene fragments using the PacBio SMRT DNA sequencing system. PeerJ 4:e1869. doi: 10.7717/peerj.1869
Schmidt, B. L., Kuczynski, J., Bhattacharya, A., Huey, B., Corby, P. M., Queiroz, E. L., et al. (2014). Changes in abundance of oral microbiota associated with oral cancer. PLoS One 9:e98741. doi: 10.1371/journal.pone.0098741
Sharma Mahendra, V., Ranauta, A., Yuvraj, A., Santella, A. J., Taslim, A., and Doughty, J. (2020). The role of patient and public involvement in oral health and HIV/AIDS research, practice and policy. Oral Dis. 26, 117–122. doi: 10.1111/odi.13584
Shi, W., Tian, J., Xu, H., Zhou, Q., and Qin, M. (2018). Distinctions and associations between the microbiota of saliva and supragingival plaque of permanent and deciduous teeth. PLoS One 13:e0200337. doi: 10.1371/journal.pone.0200337
Simner, P. J., Miller, S., and Carroll, K. C. (2018). Understanding the promises and hurdles of metagenomic next-generation sequencing as a diagnostic tool for infectious diseases. Clin. Infect. Dis. 66, 778–788. doi: 10.1093/cid/cix881
Simpson, J. T., Workman, R. E., Zuzarte, P. C., David, M., Dursi, L. J., and Timp, W. (2017). Detecting DNA cytosine methylation using nanopore sequencing. Nat. Methods 14, 407–410. doi: 10.1038/nmeth.4184
Siqueira, J. F. Jr., and Rocas, I. N. (2013). As-yet-uncultivated oral bacteria: breadth and association with oral and extra-oral diseases. J. Oral Microbiol. 5. doi: 10.3402/jom.v5i0.21077
Sizova, M. V., Hohmann, T., Hazen, A., Paster, B. J., Halem, S. R., Murphy, C. M., et al. (2012). New approaches for isolation of previously uncultivated oral bacteria. Appl. Environ. Microbiol. 78, 194–203. doi: 10.1128/AEM.06813-11
Slocum, C., Kramer, C., and Genco, C. A. (2016). Immune dysregulation mediated by the oral microbiome: potential link to chronic inflammation and atherosclerosis. J. Intern. Med. 280, 114–128. doi: 10.1111/joim.12476
Socransky, S. S., Haffajee, A. D., Cugini, M. A., Smith, C., and Kent, R. L. Jr. (1998). Microbial complexes in subgingival plaque. J. Clin. Periodontol. 25, 134–144. doi: 10.1111/j.1600-051x.1998.tb02419.x
Soder, B., Kallmen, H., Yucel-Lindberg, T., and Meurman, J. H. (2021). Periodontal microorganisms and diagnosis of malignancy: a cross-sectional study. Tumour Biol. 43, 1–9. doi: 10.3233/TUB-200066
Song, Y., Song, F., Liu, S., Chen, S., and Song, Z. (2022). Rapid diagnosis of a complex oral mucosal infection using metagenomic next-generation sequencing: a case report. J. Int. Med. Res. 50:6679. doi: 10.1177/03000605221136679
Soro, V., Dutton, L. C., Sprague, S. V., Nobbs, A. H., Ireland, A. J., Sandy, J. R., et al. (2014). Axenic culture of a candidate division TM7 bacterium from the human oral cavity and biofilm interactions with other oral bacteria. Appl. Environ. Microbiol. 80, 6480–6489. doi: 10.1128/AEM.01827-14
Srinivasprasad, V., Dineshshankar, J., Sathiyajeeva, J., Karthikeyan, M., Sunitha, J., and Ragunathan, R. (2015). Liaison between micro-organisms and oral cancer. J. Pharm. Bioallied Sci. 7, 354–360. doi: 10.4103/0975-7406.163451
Stasiewicz, M., and Karpinski, T. M. (2022). The oral microbiota and its role in carcinogenesis. Semin. Cancer Biol. 86, 633–642. doi: 10.1016/j.semcancer.2021.11.002
Syrjanen, S. (2018). Oral manifestations of human papillomavirus infections. Eur. J. Oral Sci. 126, 49–66. doi: 10.1111/eos.12538
Takahashi, N., and Nyvad, B. (2008). Caries ecology revisited: microbial dynamics and the caries process. Caries Res. 42, 409–418. doi: 10.1159/000159604
Takahashi, N., and Nyvad, B. (2011). The role of bacteria in the caries process: ecological perspectives. J. Dent. Res. 90, 294–303. doi: 10.1177/0022034510379602
Tanaka, M., Yamamoto, Y., Kuboniwa, M., Nonaka, A., Nishida, N., Maeda, K., et al. (2004). Contribution of periodontal pathogens on tongue dorsa analyzed with real-time PCR to oral malodor. Microbes Infect. 6, 1078–1083. doi: 10.1016/j.micinf.2004.05.021
Van Dijk, E. L., Jaszczyszyn, Y., Naquin, D., and Thermes, C. (2018). The third revolution in sequencing technology. Trends Genet. 34, 666–681. doi: 10.1016/j.tig.2018.05.008
Vartoukian, S. R., Palmer, R. M., and Wade, W. G. (2010). Cultivation of a Synergistetes strain representing a previously uncultivated lineage. Environ. Microbiol. 12, 916–928. doi: 10.1111/j.1462-2920.2009.02135.x
Verma, D., Garg, P. K., and Dubey, A. K. (2018). Insights into the human oral microbiome. Arch. Microbiol. 200, 525–540. doi: 10.1007/s00203-018-1505-3
Vincent, A. T., Derome, N., Boyle, B., Culley, A. I., and Charette, S. J. (2017). Next-generation sequencing (NGS) in the microbiological world: how to make the most of your money. J. Microbiol. Methods 138, 60–71. doi: 10.1016/j.mimet.2016.02.016
Wade, W. (2002). Unculturable bacteria--the uncharacterized organisms that cause oral infections. J. R. Soc. Med. 95, 81–83. doi: 10.1177/014107680209500207
Wade, W. G. (2013). The oral microbiome in health and disease. Pharmacol. Res. 69, 137–143. doi: 10.1016/j.phrs.2012.11.006
Wagner, J., Coupland, P., Browne, H. P., Lawley, T. D., Francis, S. C., and Parkhill, J. (2016). Evaluation of PacBio sequencing for full-length bacterial 16S rRNA gene classification. BMC Microbiol. 16:274. doi: 10.1186/s12866-016-0891-4
Wang, S., Chen, Y., Wang, D., Wu, Y., Zhao, D., Zhang, J., et al. (2019). The feasibility of metagenomic next-generation sequencing to identify pathogens causing tuberculous meningitis in cerebrospinal fluid. Front. Microbiol. 10:1993. doi: 10.3389/fmicb.2019.01993
Wang, D., Coscoy, L., Zylberberg, M., Avila, P. C., Boushey, H. A., Ganem, D., et al. (2002). Microarray-based detection and genotyping of viral pathogens. Proc. Natl. Acad. Sci. U. S. A. 99, 15687–15692. doi: 10.1073/pnas.242579699
Wang, C., Huang, Z., Li, W., Fang, X., and Zhang, W. (2020). Can metagenomic next-generation sequencing identify the pathogens responsible for culture-negative prosthetic joint infection? BMC Infect. Dis. 20:253. doi: 10.1186/s12879-020-04955-2
Wang, X., Kaczor-Urbanowicz, K. E., and Wong, D. T. (2017). Salivary biomarkers in cancer detection. Med. Oncol. 34:7. doi: 10.1007/s12032-016-0863-4
Wang, J., Qi, J., Zhao, H., He, S., Zhang, Y., Wei, S., et al. (2013). Metagenomic sequencing reveals microbiota and its functional potential associated with periodontal disease. Sci. Rep. 3:1843. doi: 10.1038/srep01843
Wang, Y., Zhang, J., Chen, X., Jiang, W., Wang, S., Xu, L., et al. (2017). Profiling of oral microbiota in early childhood caries using single-molecule real-time sequencing. Front. Microbiol. 8:2244. doi: 10.3389/fmicb.2017.02244
Weerasekera, M. M., Wijesinghe, G. K., Sampath, A., Dilhari, A., Madhumal, T., Dilrukshi, R., et al. (2021). The genotypes and virulence attributes of C. albicans isolates from oral leukoplakia. Med. Oral Patol. Oral Cir. Bucal. 26, e786–e794. doi: 10.4317/medoral.24748
Willse, A., Chandler, D. P., White, A., Protic, M., Daly, D. S., and Wunschel, S. (2005). Comparing bacterial DNA microarray fingerprints. Stat. Appl. Genet. Mol. Biol. 4:1162. doi: 10.2202/1544-6115.1162
Woo, P. C., Lau, S. K., Teng, J. L., Tse, H., and Yuen, K. Y. (2008). Then and now: use of 16S rDNA gene sequencing for bacterial identification and discovery of novel bacteria in clinical microbiology laboratories. Clin. Microbiol. Infect. 14, 908–934. doi: 10.1111/j.1469-0691.2008.02070.x
Wright, C. J., Burns, L. H., Jack, A. A., Back, C. R., Dutton, L. C., Nobbs, A. H., et al. (2013). Microbial interactions in building of communities. Mol Oral Microbiol 28, 83–101. doi: 10.1111/omi.12012
Xiao, E., Mattos, M., Vieira, G. H. A., Chen, S., Correa, J. D., Wu, Y., et al. (2017). Diabetes enhances IL-17 expression and alters the Oral microbiome to increase its pathogenicity. Cell Host Microbe 22:e4, 120–128.e4. doi: 10.1016/j.chom.2017.06.014
Xu, B., and Han, Y. W. (2022). Oral bacteria, oral health, and adverse pregnancy outcomes. Periodontol 2000 89, 181–189. doi: 10.1111/prd.12436
Yamasaki, K., Kawanami, T., Yatera, K., Fukuda, K., Noguchi, S., Nagata, S., et al. (2013). Significance of anaerobes and oral bacteria in community-acquired pneumonia. PLoS One 8:e63103. doi: 10.1371/journal.pone.0063103
Yang, B., Wang, Y., and Qian, P. Y. (2016). Sensitivity and correlation of hypervariable regions in 16S rRNA genes in phylogenetic analysis. BMC Bioinformatics 17:135. doi: 10.1186/s12859-016-0992-y
Yang, C. Y., Yeh, Y. M., Yu, H. Y., Chin, C. Y., Hsu, C. W., Liu, H., et al. (2018). Oral microbiota community dynamics associated with Oral squamous cell carcinoma staging. Front. Microbiol. 9:862. doi: 10.3389/fmicb.2018.00862
Yaseen, A., Mahafzah, A., Dababseh, D., Taim, D., Hamdan, A. A., Al-Fraihat, E., et al. (2021). Oral colonization by Entamoeba gingivalis and Trichomonas tenax: a PCR-based study in health, gingivitis, and periodontitis. Front. Cell. Infect. Microbiol. 11:782805. doi: 10.3389/fcimb.2021.782805
Yazar, S., Cetinkaya, U., Hamamci, B., Alkan, A., Sisman, Y., Esen, C., et al. (2016). Investigation of Entamoeba gingivalis and Trichomonas tenax in periodontitis or gingivitis patients in Kayseri. Turkiye Parazitol. Derg. 40, 17–21. doi: 10.5152/tpd.2016.4351
Yu, F. Y., Wang, Q. Q., Li, M., Cheng, Y. H., Cheng, Y. L., Zhou, Y., et al. (2020). Dysbiosis of saliva microbiome in patients with oral lichen planus. BMC Microbiol. 20:75. doi: 10.1186/s12866-020-01733-7
Zaura, E., Keijser, B. J., Huse, S. M., and Crielaard, W. (2009). Defining the healthy "core microbiome" of oral microbial communities. BMC Microbiol. 9:259. doi: 10.1186/1471-2180-9-259
Zhang, Y., Qi, Y., Lo, E. C. M., McGrath, C., Mei, M. L., and Dai, R. (2021). Using next-generation sequencing to detect oral microbiome change following periodontal interventions: a systematic review. Oral Dis. 27, 1073–1089. doi: 10.1111/odi.13405
Zhang, J., Su, L., Wang, Y., and Deng, S. (2020). Improved high-throughput sequencing of the human oral microbiome: from Illumina to PacBio. Can. J. Infect. Dis. Med. Microbiol. 2020:6678872. doi: 10.1155/2020/6678872
Zhang, X., Wang, M., Wang, X., Qu, H., Zhang, R., Gu, J., et al. (2021). Relationship between periodontitis and microangiopathy in type 2 diabetes mellitus: a meta-analysis. J. Periodontal Res. 56, 1019–1027. doi: 10.1111/jre.12916
Zhou, Z., Ling, G., Ding, N., Xun, Z., Zhu, C., Hua, H., et al. (2018). Molecular analysis of oral microflora in patients with primary Sjogren's syndrome by using high-throughput sequencing. PeerJ 6:e5649. doi: 10.7717/peerj.5649
Keywords: oral cavity, microbiota, infectious diseases, rapid diagnosis, precise treatment
Citation: Xiao X, Liu S, Deng H, Song Y, Zhang L and Song Z (2023) Advances in the oral microbiota and rapid detection of oral infectious diseases. Front. Microbiol. 14:1121737. doi: 10.3389/fmicb.2023.1121737
Edited by:
George Grant, University of Aberdeen, United KingdomReviewed by:
Gislane Lelis Vilela de Oliveira, São Paulo State University, BrazilZhangyong Song, Southwest Medical University, China
Copyright © 2023 Xiao, Liu, Deng, Song, Zhang and Song. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhifeng Song, c3pmOTExMDYyN0AxNjMuY29t; Liang Zhang, emhhbmdsaWFuZzE5OTlAdHNpbmdodWEub3JnLmNu
†These authors have contributed equally to this work
 Xuan Xiao
Xuan Xiao Shangfeng Liu2†
Shangfeng Liu2†