- 1Department of Veterinary Medicine and Animal Sciences, Università degli Studi di Milano, Lodi, Italy
- 2Laboratorio di Malattie Infettive degli Animali, Università degli Studi di Milano, Lodi, Italy
- 3Italian National Research Council, Institute of Agricultural Biology and Biotechnology, Lodi, Italy
- 4Quality Milk Production Services, Animal Health Diagnostic Center, Cornell University, Ithaca, NY, United States
Accurate and precise differentiation of staphylococci isolated from milk is of importance for udder health management. In particular, the rapid and specific identification of Staphylococcus aureus plays an essential role in the prevention and treatment programs for bovine mastitis. Plasma gelatinization in coagulase assays is routinely used to discriminate S. aureus from other species by detecting the presence of extracellular free staphylocoagulase. However, rarely occurring coagulase-deficient S. aureus strains can be responsible for clinical and subclinical mastitis cases. By investigating S. aureus isolates from a single herd over a 10-year period we identified the persistence of a phenotypically coagulase-negative S. aureus strain and pinpointed the possible cause to a single base pair deletion in the coa gene sequence. Our results support the need to integrate primary biochemical tests with molecular/sequence analysis approaches for correctly identifying and discriminating atypical S. aureus in bovine herds, as the coagulase test alone may fail to detect persistent mastitis-causing strains.
1. Introduction
The family Staphylococcaceae comprises the genus Staphylococcus, a diverse group of gram-positive bacteria globally recognized as commensal colonizers of humans and warm-blooded animals (Founou et al., 2018). In food-producing animals, their principal reservoirs are the skin and mucosa of pigs, chickens, sheep, goats, and cows (Huber et al., 2011). Among them, the majority of species shares the inability to clot rabbit plasma and are referred to as coagulase-negative staphylococci (CoNS). Though CoNS are frequently detected in dairy cattle (De Buck et al., 2021), they are considered “minor pathogens” (Hamel et al., 2020; Ryman et al., 2021). On the other hand, coagulase-positive staphylococci (CoPS) including Staphylococcus aureus, S. intermedius and S. hyicus, are ubiquitous and highly versatile microorganisms implicated in a large variety of infections, ranging from dermatitis to septicemia, with S. aureus being the best-known of these pathogens.
In dairy cows, S. aureus is predominantly classified as a contagious mastitis causative agent whose presence is more frequently associated with subclinical than clinical cases, although staphylococcal infections may also occur in clinical forms (Schroeder, 2012). This microorganism is characterized by lower recovery rates than the other staphylococci, despite the efforts in controlling its presence and spread in dairy herds (Exel et al., 2022). The accurate and precise differentiation of staphylococci isolated from milk samples has therefore a major impact on udder health management. In particular, the rapid and specific identification of S. aureus plays an essential role in bovine mastitis prevention and treatment programs, at the point that CoNS are currently referred as Non-aureus Staphylococci (NAS), stressing this dichotomy. As this species differs from other staphylococcal species for being generally β-hemolytic, the hemolysis detection can also represent a fast and cost-effective method for testing the presumptive S. aureus presence in primary cultures, despite the low sensitivity and specificity (Ryman et al., 2021). However, previous studies demonstrated that almost 20–25% of S. aureus isolates from bovine intramammary infections (IMIs) show no visible β-hemolysis on blood agar plates (Ryman et al., 2021). On the other hand, as several NAS species also show β-hemolysis, selective media like Baird-Parker + RPF (Rabbit Plasma Fibrinogen) Agar or Mannitol Salt Agar (MSA) can be alternatively used for the differential growth of staphylococci (Pumipuntu et al., 2017). Nevertheless, the former works through the same principle of coagulase activity and the latter yields positive reaction also with some NAS.
Therefore, in diagnostic laboratories using primary and secondary biochemical tests for bacterial species identification, the coagulase assay is most frequently used to differentiate S. aureus from NAS (Peetermans et al., 2015). The bound form or clumping factor can be detected by a slide test, while extracellular free S. aureus coagulase (staphylocoagulase) can be detected by plasma gelatinization in a standard coagulase test tube (CTT), normally prepared from rabbit or horse whole blood (Peetermans et al., 2015). This enzyme can prime the non-proteolytic activation of prothrombin and cleavage of fibrinogen to promote coagulation (McAdow et al., 2012). Nevertheless, some S. aureus strains may not express this major characteristic, producing false negative results (Akineden et al., 2011). These CTT-negative S. aureus could therefore be erroneously classified as NAS and managed as such, with a potential for a spread within the herd (Akineden et al., 2011; Sunagar et al., 2013).
Although Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry (MALDI-TOF MS) technique is being increasingly applied in veterinary microbiology, many laboratories still carry out bacterial identification with primary biochemical tests as they are easy to set up and cost-effective. In these settings, coagulase deficiencies in S. aureus due to transcriptional or post-transcriptional alterations in the coa gene (McAdow et al., 2012) may therefore lead to an erroneous species identification, with adverse effects on udder health control programs.
In our laboratory work, we repeatedly isolated coagulase-deficient S. aureus from clinical and subclinical mastitis cases occurring in a single herd over a 10-year period. In this study, we present the detailed characterization of the coa gene of the coagulase-negative S. aureus strain isolated from this dairy farm with the aim of understanding the molecular basis for this phenotypic behavior and suggesting mitigation measures to possible identification bias.
2. Materials and methods
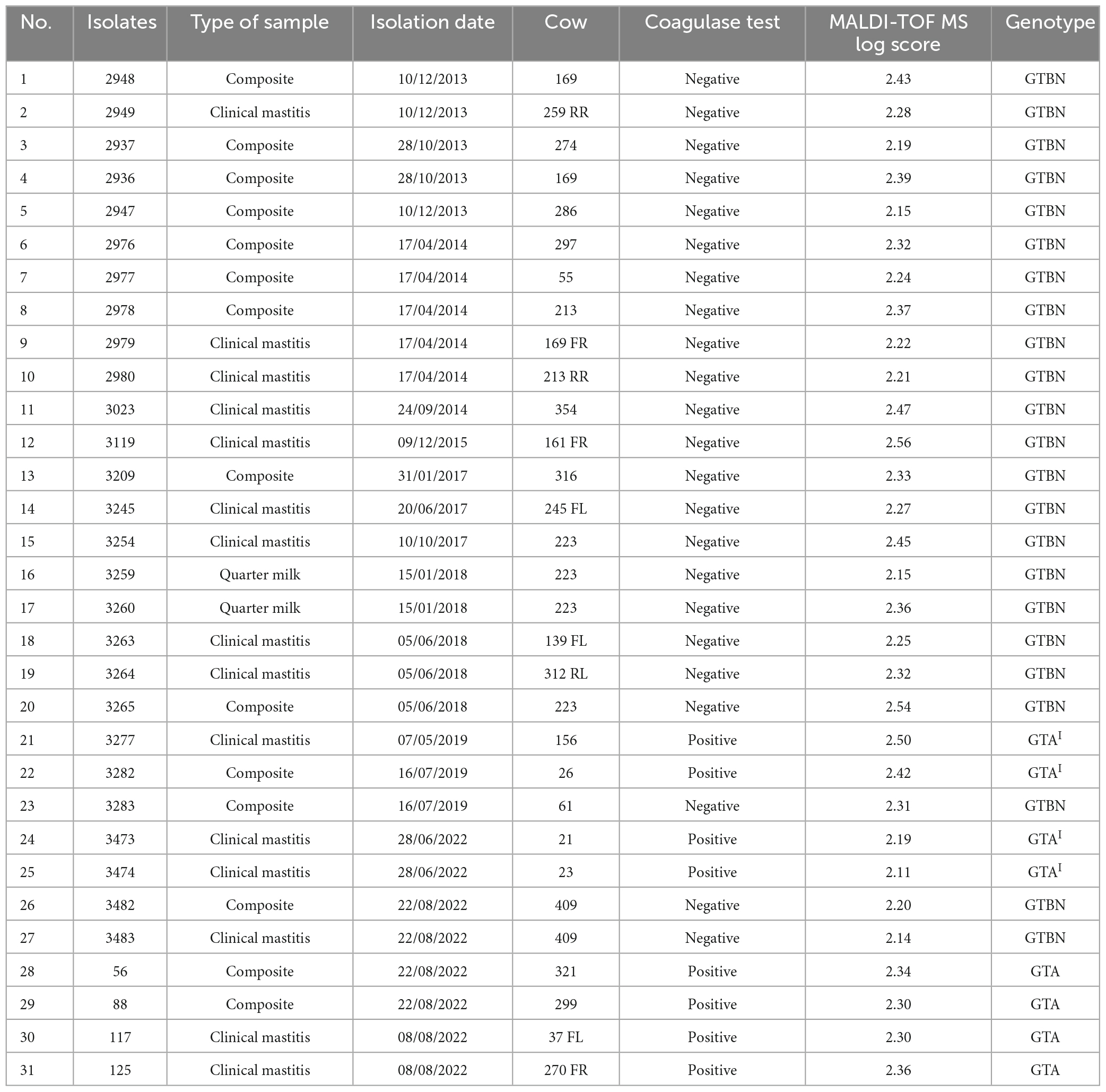
2.1. Sample collection and S. aureus identification
During the daily diagnostic activity at the Laboratorio di Malattie Infettive degli Animali (MiLab, Università degli Studi di Milano, Italy), from December 2013 to August 2022, a total of 31 staphylococcal isolates were obtained from as many milk samples from a single Italian dairy farm and presumptively identified as S. aureus. The samples (15 quarter clinical mastitis, 14 composite and 2 quarter milk samples with high somatic cell count, Table 1) were cultured and evaluated according to National Mastitis Council procedures (National Mastitis Council, 2017), including CTT (Pumipuntu et al., 2017). The typical colony morphology and the beta-hemolysis on blood agar suggested to subculture the isolates onto Mannitol Salt Agar (MSA, Oxoid, Basingstoke, United Kingdom) and Baird Parker with Rabbit Plasma (BP + RPF, Microbiol, Cagliari, Italy) for further characterization. All the isolates were frozen in Nutrient Broth (Microbiol, Cagliari, Italy) added with 15% glycerol (Carlo Erba Reagents, Milan, Italy) until further analyses.
2.2. DNA extraction and molecular identification
DNA was extracted using the DNA isolation system kit (Clonit, Medical System, Genova, Italy) according to the protocol described by Cremonesi et al. (2006), starting from step 2. DNA quality and quantity were measured using a NanoDrop ND-1000 spectrophotometer (Nano-Drop Technologies, Wilmington, DE); the samples were stored at −20°C until further use.
To check the specificity of the isolates, the DNA extracted was amplified with nuc (thermonuclease coding gene) and coa (coagulase coding gene) primers, as previously described (Cremonesi et al., 2005). As positive controls, S. aureus reference strains (ATCC 19040, ATCC 19041, ATCC 19048, ATCC 700699) were used in each PCR assay. All amplified PCR fragments were separated by 2% agarose gel electrophoresis (GellyPhor, Euroclone, Milan, Italy), stained with ethidium bromide (0.05 mg/mL; Sigma-Aldrich), and visualized under UV transilluminator (BioView Ltd., Nes Ziona, Israel). A 100-bp DNA ladder (Finnzymes, Espoo, Finland) was included in each gel.
2.3. Confirmation through MALDI-TOF MS
MALDI-TOF MS (Bruker Daltonik GmbH, Bremen, Germany) has been introduced in routine mastitis diagnostics at MiLab since January 2021. All the isolates were analyzed following the protocol described in Monistero et al. (2021) and Rosa et al. (2022).
2.4. RS-PCR typing
All the 31 isolates were also genotyped by RS-PCR as previously described with a detailed working protocol (Fournier et al., 2008; Graber, 2016). The method is based on the amplification of the 16S–23S rRNA intergenic spacer region. The PCR products were analyzed using the miniaturized electrophoresis system DNA 7500 LabChip (Agilent Technologies, Santa Clara, CA). Genotypes were inferred from the electrophoresis profile using the Mahal software, which is freely available online (Graber, 2016).1
2.5. Library preparation, sequencing, bioinformatics analysis
The DNA was amplified for the entire coagulase gene by using the following primers designed by the Primer3 programme2 : FORWARD = GCCGCTTTAATACCAGCAAC; REVERSE = CTTCCGATTGTTCGATGCTT (amplicon size = 2268 bp). The PCR amplifications were performed in 25 μl volumes per sample. A total of 12.5 μl of GoTaq® Long PCR Master Mix, 2X (Promega Corporation, Madison, USA) and 0.2 μl of each primer (100 μM) were added to 2 μl of genomic DNA (5 ng/μl). A first amplification step was performed in an Applied Biosystem 2700 thermal cycler (ThermoFisher Scientific). Samples were denatured at 95°C for 2 min, followed by 30 cycles with a denaturing step at 94°C for 30 s, annealing at 55°C for 30 s and extension at 68°C for 2 min, with a final extension at 72°C for 10 min. The amplicons were then cleaned with Agencourt AMPure XP (Beckman, Coulter Brea, CA, USA) and libraries were prepared following the Nextera XT DNA library Prep Kit Protocol (Illumina, San Diego, CA, USA), using a tagmentation of 15 min with PdmI enzyme. The libraries obtained were quantified by Real Time PCR with KAPA Library Quantification Kits (Kapa Biosystems, Inc., MA, USA), pooled in equimolar proportion and sequenced in one MiSeq (Illumina) run with 2 × 300-base paired-end reads. FASTQ files were mapped with BWA-MEM2 against staphylocoagulase GenBank locus sequence LOCUS JN861807 (strain MSSA_129) on the Galaxy Platform (Afgan et al., 2018) and reads coverage was visualized using IGV (Robinson et al., 2011).
2.6. Molecular phylogenetic analysis by the neighbor-joining method
CDS DNA sequences used for phylogenetic analysis from different S. aureus strains and subspecies were obtained from the NCBI database.3 Phylogenetic relationships were estimated in MEGAX (Kumar et al., 2018). DNA sequences (Supplementary material) were aligned by MUSCLE with default settings. Where a frameshift mutation was present, the whole sequence was used to allow for a full length alignment. Evolutionary relationships among coagulase sequences were inferred by using the Neighbor-Joining method and evolutionary distances were computed using the Maximum Composite Likelihood method and are in the units of the number of base substitutions per site. The rate variation among sites was modeled with a gamma distribution. The reliability of the phylogenetic tree was estimated by setting 1000 bootstrap replicates.
2.7. Sanger sequencing
All the samples were also analyzed by conventional sanger sequencing. The DNA was amplified for the coagulase gene by using the following primers designed in conserved regions: FORWARD = ATGGGATAACAAAGCAGATG; REVERSE = GGTTCTTCAACTTTCTTCTC (amplicon size = 900 bp). The PCR amplifications were performed in 25 μl volumes per sample. A total of 12.5 μl of PCR Master Mix, 2X (Thermofisher Scientific) and 0.2 μl of each primer (100 μM) were added to 2 μl of genomic DNA (5 ng/μl). The amplification step was performed in an Applied Biosystem 2700 thermal cycler (ThermoFisher Scientific). Samples were denatured at 95°C for 2 min, followed by 30 cycles with a denaturing step at 95°C for 1 min, annealing at 54°C for 1 min and extension at 72°C for 1 min, with a final extension at 72°C for 10 min. The amplicons were then cleaned with Wizard® SV Gel and PCR Clean-Up System (Promega Italia, Milan, Italy) and the cleaned products were sequenced by Eurofins Genomics (Ebersberg, Germany), following the instructions of the manufacturer.
3. Results and discussion
3.1. Bacteriological analysis and coagulase test
As previously described (Akineden et al., 2011), the discrimination between CoPS and CoNS catalase-positive cocci represents one of the most important routine procedures for identifying the etiological agents of contagious mastitis within a dairy herd. Coagulase is considered a virulence factor and staphylococci that are not able to produce this protein are reported to be less pathogenic. This is why the CTT for S. aureus coagulase activity remains the reference and the most used method in the laboratory routine. Although rare, some atypical S. aureus strains may occur (Akineden et al., 2011; Sunagar et al., 2013) and coagulase test specificity should be considered to avoid misclassification. From bacteriological analysis, 23 isolates (10 quarter clinical mastitis, 11 composite and 2 quarter high somatic cell samples, Table 1) resulted unexpectedly negative at the coagulase test after both 4 and 24 h of incubation at 37°C. The 23 CTT-negative isolates found in this study demonstrated the importance of applying appropriate screening testing to avoid S. aureus misclassification and to prevent staphylococcal infection spread within farms. The remaining 8 S. aureus analyzed were CTT-positive isolates producing a typical coagulase halo around the colonies on BP-RPF demonstrating lecithinase activity, whereas this was weak to absent for the 23 CTT-negative isolates. Aside from CTT results, all 31 S. aureus isolates evaluated in this study showed a positive reaction on MSA, proving the higher sensitivity of this selective medium compared to BP-RPF. As an initial screening method and compared to CTT, using MSA represents a low cost good choice for a more sensitive and still acceptably rapid S. aureus identification. However, due to the limited specificity, S. aureus identified through MSA positive results should be confirmed by further molecular methods (Pumipuntu et al., 2017).
3.2. Molecular characterization: amplification of nuc and coa genes and RS-PCR analysis
The identification as S. aureus was also confirmed by MALDI-TOF results, with scores >2.00 for all the isolates (Table 1). A misidentified S. aureus due to a negative CTT could drive to severe consequences as the other virulence factors activity (i.e., enterotoxins) could be conserved. The MALDI-TOF identification was further supported by PCR on the thermonuclease (nuc) and coagulase (coa) genes for all the 31 isolates evaluated in this study. Moreover, the RS-PCR carried out for verification of the genotypes circulating in the herd in the time frame evaluated in this study revealed that all the 23 coagulase-negative S. aureus belonged to genotype GTBN (Table 1), while the 8 coagulase-positive S. aureus belonged to genotypes GTA (N = 6 in 2022) and GTAI (n = 2 in 2019). Previous studies (Cremonesi et al., 2015; Cosandey et al., 2016) using the RS-PCR technique, however, demonstrated that S. aureus isolated from bovine IMIs are genetically heterogeneous, with some S. aureus genotypes showing a limited tendency to spread in the herd, as for GTBN, GTA and GTAI. The knowledge of the characteristics of S. aureus circulating in the herd might help to formulate strategies for focused treatment and control of disease (Monistero et al., 2018).
3.3. Sequencing of the coa gene
Out of the 23 isolates with the same GTBN genotype, 4 (isolates number 1, 4, 5, 12) were randomly chosen and analyzed in one Miseq run in order to determine the nucleotide sequence of the coa gene. As previously described (Watanabe et al., 2005, 2009; Johler et al., 2012), this gene is known to be divided into six regions: the signal sequence, the D1 and D2 regions enabling contact with prothrombin, the central region, a repeat region, and the C-terminal sequence.
In order to choose a suitable reference for mapping, preliminary BLAST searches using individual reads from the FASTQ sequencing files were carried out against S. aureus from the GenBank database. Since all the reads used aligned to strain MSSA_129 (sequence JN861807, Johler et al., 2012), this strain was used as reference. The coa gene from the methicillin-susceptible S. aureus MSSA_129 strain was previously reported to harbor a single base deletion at position 653 within the D2 region of the gene, leading to a frameshift and premature termination of the resulting protein (Johler et al., 2012). The four GTBN S. aureus strains revealed the same sequence (Figure 1A), and complete identity to MSSA_129 around position 653 in the D2 region of the gene (Figure 1B and Supplementary material 2A). This was further confirmed in all the 23 GTBN isolates by Sanger sequencing, by using primers chosen in conserved regions between the sequences from GTBN of interest (isolates 1, 4, 5, 12) and sequences from non-GTBN genotypes (NCTC7485, ATCC6538, SA1428). Conversely, the deletion at position 653 was not observed in the GTA and GTAI coagulase sequences. As the MSSA_129 strain (Johler et al., 2012), also all our GTBN samples would therefore be characterized by a premature stop codon resulting in a predicted 223 aa peptide that is most likely not functional. By inspecting the rest of the sequence, no differences to the reference used were found in the central portion of the gene, not even at around 1,165–1,190 bp where fewer reads were available to ensure coverage (Figure 1C). Finally, sequence comparison revealed that the four GTBN strains could be genetically differentiated from the reference used (MSSA_129) by the presence of a SNP, (C to A) at position 1,835 (Figure 1D).

Figure 1. Molecular analysis of sequence variants observed in the GTBN coagulase genes. (A) Sequencing reads obtained from isolates GTBN#1, 4, 5, and 12 were mapped using MSSA_129 as reference sequence. Gray signals correspond to sequence identity with MSSA_129. The boxes show in more detail (B) the position of the single A deletion at position 653 bp in both the reference and the four samples (red arrowhead), (C) an area with drop in read coverage at around 1,165–1,190 and (D) a SNP (C to A) at position 1,835 common to all sequenced samples.
To gain insights into the variability of coa genes from different S. aureus strains and subspecies, the sequence from the GTBN isolates and that of MSSA_129 were aligned to 21 coa CDS obtained from GenBank (listed in Supplementary material), and their relationships were inferred by using the Neighbor-Joining method (Figure 2). This analysis showed that the sequences were separated in different clusters, and that those of GTBN isolates and MSSA_129 clustered closely, among others, with the two sequences Stp58 and Stp25 (Watanabe et al., 2009). These two were CTT-positive MSSA isolated in Japan (Watanabe et al., 2009). The Stp58 coa sequence was identical to the Stp25 coa sequence; their alignment revealed the lack of the single base deletion at position 653 bp observed in the 4 GTBN samples from this study (Supplementary material 2B), resulting in a predicted full length peptide in these two strains (Supplementary material 2C). Since Stp58 and Stp25 were reported as CTT-positive, this is further evidence that the base deletion is responsible for the lack of coagulase activity in GTBN 1, 4, 5, and 12.
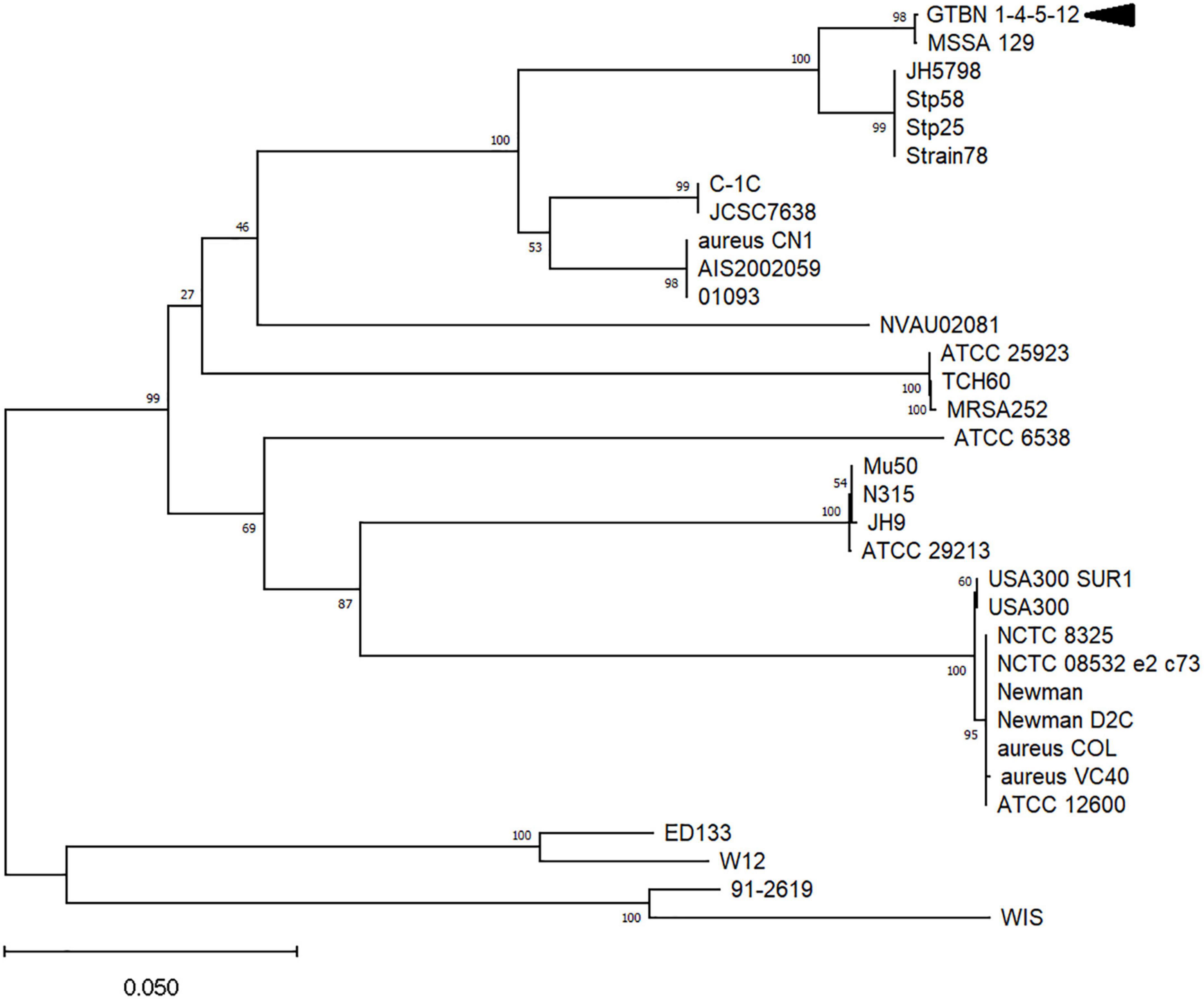
Figure 2. Evolutionary relationships among coagulase genes in different Staphylococcus aureus strains and subspecies. Only bootstrap values >50 are shown. The arrowhead indicates the sequence corresponding to the four isolates (GTBN#1, 4, 5, and 12).
The collection of samples over a 10-years period suggested the persistence of the low diffusive GTBN in the same herd; all these coagulase negative strains were characterized by an indel-mutation in coa gene. This study has potential limitations as analysis by using whole genome sequencing (WGS), cheaper today than few years ago, would provide deeper information on the atypical S. aureus circulating in bovine herds and allow to inspect the sequences of genes other than coagulase. However, the use of gene sequence analysis is nowadays still far from being a routine method, and in this specific case study this approach was enough to provide an answer to the issue encountered. As no single phenotypic test can guarantee reliable results in S. aureus detection, our work emphasizes the usefulness of MALDI-TOF or molecular methods in routine analyses to identify and discriminate atypical staphylococci from bovine mastitis cases. Where such advanced identification techniques are not yet available, combining the results of more than one test is crucial to avoid misidentification due to atypical strains. The knowledge about the epidemiology of coagulase–negative S. aureus genotypes might support control measures directed to reduce the spread of a contagious pathogen within dairy herds.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI SRA – SAMN32298094.
Author contributions
CL and PC designed and performed the experiments. SG helped with the data analysis. BC, PM, and MA gave advices to the researchers. VM, CL, SG, MA, and PC wrote the manuscript. All authors critically reviewed the manuscript and approved the final version.
Funding
This research was supported by the EU funding within the NextGenerationEU-MUR PNRR Extended Partnership initiative on Emerging Infectious Diseases (project no. PE00000007, INF-ACT).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1120305/full#supplementary-material
Footnotes
References
Afgan, E., Baker, D., Batut, B., van den Beek, M., Bouvier, D., Cech, M., et al. (2018). The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucl. Acids Res. 46, W537–W544. doi: 10.1093/nar/gky379
Akineden, O., Hassan, A. A., Schneider, E., and Usleber, E. (2011). A coagulase-negative variant of Staphylococcus aureus from bovine mastitis milk. J. Dairy Res. 78, 38–42. doi: 10.1017/S0022029910000774
Cosandey, A., Boss, R., Luini, M., Artursson, K., Bardiau, M., Breitenwieser, F., et al. (2016). Staphylococcus aureus genotype B and other genotypes isolated from cow milk in European countries. J. Dairy Sci. 99, 529–540. doi: 10.3168/jds.2015-9587
Cremonesi, P., Castiglioni, B., Malferrari, G., Biunno, I., Vimercati, C., Moroni, P., et al. (2006). Technical note: Improved method for rapid DNA extraction of mastitis pathogens directly from milk. J. Dairy Sci. 89, 163–169. doi: 10.3168/jds.S0022-0302(06)72080-X
Cremonesi, P., Luzzana, M., Brasca, M., Morandi, S., Lodi, R., Vimercati, C., et al. (2005). Development of a multiplex PCR assay for the identification of Staphylococcus aureus enterotoxigenic strains isolated from milk and dairy products. Mol. Cell Probes 19, 299–305. doi: 10.1016/j.mcp.2005.03.002
Cremonesi, P., Pozzi, F., Raschetti, M., Bignoli, G., Capra, E., Graber, H. U., et al. (2015). Genomic characteristics of Staphylococcus aureus strains associated with high within-herd prevalence of intramammary infections in dairy cows. J. Dairy Sci. 98, 6828–6838. doi: 10.3168/jds.2014-9074
De Buck, J., Ha, V., Naushad, S., Nobrega, D. B., Luby, C., Middleton, J. R., et al. (2021). Non-aureus Staphylococci and bovine health: Current understanding and knowledge gaps. Front. Vet. Sci. 8:658031. doi: 10.3389/fvets.2021.658031
Exel, C. E., Halasa, T., Koop, G., Steeneveld, W., Lam, T. J. G. M., Benedictus, L., et al. (2022). A stochastic modelling approach to determine the effect of diverse Staphylococcus aureus strains on the economic and epidemiological outcomes of mastitis intervention strategies in dairy cattle. Prev. Vet. Med. 199:105566. doi: 10.1016/j.prevetmed.2021.105566
Founou, L. L., Founou, R. C., Essack, S. Y., and Djoko, C. F. (2018). Mannitol-fermenting methicillin-resistant staphylococci (MRS) in pig abattoirs in Cameroon and South Africa: A serious food safety threat. Int. J. Food Microbiol. 285, 50–60. doi: 10.1016/j.ijfoodmicro.2018.07.006
Fournier, C., Kuhnert, P., Frey, J., Miserez, R., Kirchhofer, M., Kaufmann, T., et al. (2008). Bovine Staphylococcus aureus: Association of virulence genes, genotypes and clinical outcome. Res. Vet. Sci. 85, 439–448. doi: 10.1016/j.rvsc.2008.01.010
Graber, H. U. (2016). Genotyping of Staphylococcus aureus by ribosomal spacer PCR (RS-PCR). J. Vis. Exp. 117:54623. doi: 10.3791/54623
Hamel, J., Zhang, Y., Wente, N., and Krömker, V. (2020). Non-S. aureus staphylococci (NAS) in milk samples: Infection or contamination? Vet. Microbiol. 242:108594. doi: 10.1016/j.vetmic.2020.108594
Huber, H., Ziegler, D., Pflüger, V., Vogel, G., Zweifel, C., and Stephan, R. (2011). Prevalence and characteristics of methicillin-resistant coagulase-negative staphylococci from livestock, chicken carcasses, bulk tank milk, minced meat, and contact persons. BMC Vet. Res. 7:6. doi: 10.1186/1746-6148-7-6
Johler, S., Moser, M., Engl, C., Tasara, T., Corti, S., Chen, J., et al. (2012). A coagulase- and α-glucosidase-negative variant of Staphylococcus aureus: A challenge for routine microbiological diagnostics. J. Clin. Microbiol. 50, 1827–1828. doi: 10.1128/JCM.06345-11
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
McAdow, M., Missiakas, D. M., and Schneewind, O. (2012). Staphylococcus aureus secretes coagulase and von Willebrand factor binding protein to modify the coagulation cascade and establish host infections. J. Innate Immun. 4, 141–148. doi: 10.1159/000333447
Monistero, V., Barberio, A., Cremonesi, P., Castiglioni, B., Morandi, S., Lassen, D. C. K., et al. (2021). Genotyping and antimicrobial susceptibility profiling of Streptococcus uberis isolated from a clinical bovine mastitis outbreak in a dairy farm. Antibiotics (Basel) 10:644. doi: 10.3390/antibiotics10060644
Monistero, V., Graber, H. U., Pollera, C., Cremonesi, P., Castiglioni, B., Bottini, E., et al. (2018). Staphylococcus aureus isolates from bovine mastitis in eight countries: Genotypes, detection of genes encoding different toxins and other virulence genes. Toxins (Basel) 10:247. doi: 10.3390/toxins10060247
National Mastitis Council (2017). Laboratory handbook on bovine mastitis, 3rd Edn. Verona, WI: National Mastitis Council.
Peetermans, M., Verhamme, P., and Vanassche, T. (2015). Coagulase activity by Staphylococcus aureus: A potential target for therapy? Semin. Thromb. Hemost. 41, 433–444. doi: 10.1055/s-0035-1549849
Pumipuntu, N., Kulpeanprasit, S., Santajit, S., Tunyong, W., Kong-Ngoen, T., Hinthong, W., et al. (2017). Screening method for Staphylococcus aureus identification in subclinical bovine mastitis from dairy farms. Vet. World 10, 721–726. doi: 10.14202/vetworld.2017.721-726
Robinson, J. T., Thorvaldsdóttir, H., Winckler, W., Guttman, M., Lander, E. S., Getz, G., et al. (2011). Integrative genomics viewer. Nat. Biotechnol. 29, 24–26. doi: 10.1038/nbt.1754
Rosa, N. M., Penati, M., Fusar-Poli, S., Addis, M. F., and Tola, S. (2022). Species identification by MALDI-TOF MS and gap PCR-RFLP of non-aureus Staphylococcus, Mammaliicoccus, and Streptococcus spp. associated with sheep and goat mastitis. Vet. Res. 53:84. doi: 10.1186/s13567-022-01102-4
Ryman, V. E., Kautz, F. M., and Nickerson, S. C. (2021). Case study: Misdiagnosis of nonhemolytic Staphylococcus aureus isolates from cases of bovine mastitis as coagulase-negative staphylococci. Animals (Basel) 11:252. doi: 10.3390/ani11020252
Schroeder, J. W. (2012). Mastitis control programs: Bovine mastitis and milking management. AS1129 (Revised). Fargo, ND: NDSU Extension Circular, North Dakota State University, 1–15.
Sunagar, R., Deore, S. N., Deshpande, P. V., Rizwan, A., Sannejal, A. D., Sundareshan, S., et al. (2013). Differentiation of Staphylococcus aureus and Staphylococcus epidermidis by PCR for the fibrinogen binding protein gene. J. Dairy Sci. 96, 2857–2865. doi: 10.3168/jds.2012-5862
Watanabe, S., Ito, T., Sasaki, T., Li, S., Uchiyama, I., Kishii, K., et al. (2009). Genetic diversity of staphylocoagulase genes (coa): Insight into the evolution of variable chromosomal virulence factors in Staphylococcus aureus. PLoS One 4:e5714. doi: 10.1371/journal.pone.0005714
Keywords: coagulase gene, dairy cow, genotyping, sequencing, Staphylococcus aureus
Citation: Locatelli C, Gattolin S, Monistero V, Castiglioni B, Moroni P, Addis MF and Cremonesi P (2023) Staphylococcus aureus coa gene sequence analysis can prevent misidentification of coagulase-negative strains and contribute to their control in dairy cow herds. Front. Microbiol. 14:1120305. doi: 10.3389/fmicb.2023.1120305
Received: 11 December 2022; Accepted: 24 April 2023;
Published: 11 May 2023.
Edited by:
Qing Pan, Qingdao Agricultural University, ChinaReviewed by:
Marthie Magdaleen Ehlers, University of Pretoria, South AfricaJán Matiašovic, Veterinary Research Institute (VRI), Czechia
Copyright © 2023 Locatelli, Gattolin, Monistero, Castiglioni, Moroni, Addis and Cremonesi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Paola Cremonesi, cGFvbGEuY3JlbW9uZXNpQGliYmEuY25yLml0
†These authors have contributed equally to this work
 Clara Locatelli
Clara Locatelli Stefano Gattolin
Stefano Gattolin Valentina Monistero
Valentina Monistero Bianca Castiglioni
Bianca Castiglioni Paolo Moroni
Paolo Moroni Maria Filippa Addis
Maria Filippa Addis Paola Cremonesi
Paola Cremonesi