- 1Institute of Medicinal Biotechnology, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China
- 2State Key Laboratory of Dao-di Herb, Beijing, China
- 3Yunnan Provincial Key Laboratory of Entomological Biopharmaceutical R&D, Dali University, Dali, China
Five Gram-stain-positive, aerobic, non-motile actinobacterial strains designated as CPCC 205763T, CPCC 203386T, CPCC 205716T, CPCC 203406T, and CPCC 203407 were obtained from different ecosystems associated with four kinds of Chinese traditional medicinal plants. The 16S rRNA gene sequences of these five strains showed closely related to members of the genus Herbiconiux of the family Microbacteriaceae, with the highest similarities of 97.4–99.7% to the four validly named species of Herbiconiux. In the phylogenetic trees based on 16S rRNA gene sequences and the core genome, these isolates clustered into the clade of the genus Herbiconiux within the lineage of the family Microbacteriaceae. The overall genome relatedness indexes (values of ANI and dDDH) and the phenotypic properties (morphological, physiological and chemotaxonomic characteristics) of these isolates, readily supported to affiliate them to the genus Herbiconiux, representing four novel species, with the isolates CPCC 203406T and CPCC 203407 being classified in the same species. For which the names Herbiconiux aconitum sp. nov. (type strain CPCC 205763T = I19A-01430T = CGMCC 1.60067T), Herbiconiux daphne sp. nov. (type strain CPCC 203386T = I10A-01569T = DSM 24546T = KCTC 19839T), Herbiconiux gentiana sp. nov. (type strain CPCC 205716T = I21A-01427T = CGMCC 1.60064T), and Herbiconiux oxytropis sp. nov. (type strain CPCC 203406T = I10A-02268T = DSM 24549T = KCTC 19840T) were proposed, respectively. In the genomes of these five strains, the putative encoding genes for amidase, endoglucanase, phosphatase, and superoxidative dismutase were retrieved, which were classified as biosynthetic genes/gene-clusters regarding plant growth-promotion (PGP) functions. The positive results from IAA-producing, cellulose-degrading and anti-oxidation experiments further approved their potential PGP bio-functions. Pangenome analysis of the genus Herbiconiux supported the polyphasic taxonomy results and confirmed their bio-function potential.
Highlights
• The members of the genus Herbiconiux were mostly found to inhabit the niches associated with plants.
• However, the microbe-plant interaction still has not been investigated exhaustively due to lack of enough Herbiconiux strains as materials. In this study, five strains designated as CPCC 205763T, CPCC 203386T, CPCC 205716T, CPCC 203406T, and CPCC 203407 were obtained from different ecosystems associated with four kinds of Chinese traditional medicinal plants.
• The 16S rRNA gene sequences information and the values of ANI and dDDH by comparing their genome sequences together with the phenotypic (morphological, physiological and chemotaxonomic) properties, readily supported to affiliate them to the genus Herbiconiux representing four novel species.
• In the genomes of these five strains, we detected the encoding genes for aldehyde dehydrogenase (EC 1.2.1.3) and amidase (EC 3.5.1.4) coding gene amiE involved in indole-3-acetic acid (IAA) biosynthesis pathway, furthermore, experiments confirmed that these strains produced IAA, an important hormone for plant growth.
• As a reward, these five strains exhibited anti-oxidation ability, which we proposed to be endowed by these anti-inflammatory herbs that shared the common ecosystems with these strains. Our study primarily accumulated valuable actinobacterial resources to exemplify microbe-plant interaction.
Introduction
Anti-inflammatory herbs have been widely used in Chinese typical ethnic drugs. Even nowadays, these ethnic drugs have still contributed significantly to the health of the people living in the marginal impoverished regions. We supposed that microorganisms associated with the anti-inflammatory herbs may gain some potential abilities to produce anti-inflammatory substances resulted from their co-evolution with these plants. During screening of the microorganisms with anti-inflammatory activities, five Herbiconiux spp. were collected. We carried out polyphasic taxonomic study on these strains. The detailed phenotypic and genotypic properties of these strains supported us to propose four novel species of the genus Herbiconiux.
Previously, Herbiconiux spp. were classified as members of the genus Leifsonia within the family Microbacteriaceae (Park et al., 1994). Later, combined the phylogenetic analysis based on 16S rRNA gene sequences and the chemotaxonomic characterization, Behrendt et al. (2011) reclassified Leifsonia ginsengi as Herbiconiux ginsengi. In 2012, two more species, Herbiconiux moechotypicola and Herbiconiux flava were identified, and accordingly, the description of the genus was emended (Kim et al., 2012). Currently, four species with valid scientific names have been accommodated in the genus Herbiconiux,1 sharing the taxonomic characteristics as follows. The peptidoglycan is of the B2γ type with D-AND L-2,4-diaminobutyric acid as the diagnostic diamino acids, and glycine, alanine and threo-3-hydroxyglutamic acid may present. The predominant menaquinone is MK-11; major amounts of MK-10 may also be present. Cellular fatty acids mainly comprise iso-and anteiso-branched fatty acids, with anteiso-C15:0 as a major component. Cyclohexyl-C17:0 may be present. The DNA G + C content is 69–71% (Hamada et al., 2012).
It is so interesting that except the type strain of H. moechotypicola, all of the type strains of other three species of the genus Herbiconiux were isolated from the ecosystems associated with plants. In this study, five more isolates representing four novel Herbiconiux species were obtained from the ecosystems involved in anti-inflammatory herbs.
The primary goal of the present research was to study the properties of Herbiconiux members inhabiting Chinese traditional medicinal plants relating niches, to explore the genotypic properties regarding plant growth-promotion functions. As a result, in the genomes of strains CPCC 205763T representing Herbiconiux aconitum sp. nov., CPCC 203386T representing Herbiconiux daphne sp. nov., CPCC 205716T representing Herbiconiux gentiana sp. nov., and CPCC 203406T and CPCC 203407 representing Herbiconiux oxytropis sp. nov., putative encoding genes for endoglucanase, phosphatase, superoxidative dismutase and amidase were retrieved, and the corresponding phenotypic experiments further validated the putative functions. On the one hand, the microbes harbor the abilities to synthesize plant growth hormone, indole-3-acetic acid (IAA), remove excessive harmful oxygen negative ions, transform the substrates that are hard to be assimilated directly by plants, to promote plant growth; on the other hand, these anti-inflammatory plants may horizontally transfer the abilities to produce anti-inflammatory substances to the microorganisms. Our study preliminarily exemplified microbe-plant interaction.
Materials and methods
Collection of samples, isolation and identification of Herbiconiux strains
The rhizosphere soil sample attached to the medicinal plant Aconitum carmichaelii was collected from Yili Valley (42°34′37′′ N, 81°13′19′′ E, 1,878 mH), Xinjiang Province, north-west China, labeled as IMB15132S; the rhizosphere soil sample of the plant Gentiana rigescens was collected from Leigong Mountain (26°23′38′′ N, 107°35′49′′ E, 2,071 mH), Kaili, Guizhou Province, south-west China (labeled as IMB20178S). The herb of Daphne aurantiaca labeled as IMB12384P was obtained from the Shangrila Alpine Botanical Garden (28°39′18′′ N, 100°18′19′′ E, 3,343 mH), Yunnan Province, south-west China. The leaf of a medicinal plant Oxytropis falcata (labeled as IMB11009P) was collected from Bujiangda county (29°55′47′′ N, 93°12′41′′ E, 3,992 mH), Tibet, west China.
The rhizosphere soil samples IMB15132S and IMB20178S were put into the sterilized envelopes and then taken to the laboratory within 1 week after collection. Isolation of microorganisms from these soils was carried out following the procedure described by Jiang et al. (2021). The medicinal plant samples IMB12384P and IMB11009P were sealed with sterile wax at the incisions before they were sent to the laboratory. The follow-up work was performed as described by Deng et al. (2022) to acquire the endophytic bacterial strains. Distinct colonies picked from the isolation plates were streaked into newly prepared PYG agar plates (g l−1; peptone 3, yeast extract 5, glycerol 10, betaine hydrochloride 1.25, sodium pyruvate 1.25, agar 15, pH 7.2), respectively. The purified isolates were maintained on PYG slants at 4°C and also as glycerol suspensions (20%, v/v) at –80°C.
The Herbiconiux spp. were primarily identified according to the 16S rRNA gene sequence comparison following next steps. Genomic DNA was extracted according to the manual of a commercial genomic DNA extraction kit (TianGen, China). PCR amplification of the strains’ 16S rRNA genes was carried out using the bacterial universal primers 27F and 1492R (Li et al., 2007). The obtained sequences were compared with available 16S rRNA gene sequences from GenBank using the BLAST program and the EzTaxon-e server2 to determine an approximate taxonomic affiliation (Yoon et al., 2017a). Multiple sequence alignment and analysis of the data were performed by using the molecular evolutionary genetics analysis (MEGA) software package version X (Kumar et al., 2018). The phylogenetic trees were reconstructed by the software package MEGA version X using the neighbor-joining (Kimura, 1979), and confirmed by maximum-likelihood (Felsenstein, 1981) and maximum-parsimony (Kluge and Farris, 1969) tree-making methods. Bootstrap analysis with 1,000 replicates was performed to obtain the confidence level of the branches (Felsenstein, 1985).
The reference strains of Herbiconiux ginsengi KCTC 19440T and Herbiconiux moechotypicola KCTC 19653T were obtained from KCTC (Korean Collection for Type Cultures), and Herbiconiux flava NBRC 16403T and Herbiconiux solani NBRC 106740T were acquired from NBRC (NITE Biological Resource Center), respectively, and they were included in some assays in parallel.
Polyphasic taxonomic study
Morphological and physiological characterization
Growth conditions of the strains were tested using ISP 2 (Shirling and Gottlieb, 1966), Tryptic soy agar (TSA, Difco), Reasoner’s 2A agar (R2A, Difco), nutrition agar (NA) and PYG agar. The growth of the strains was monitored at 4, 10, 15, 20, 28, 30, 32, 35, 37 and 40°C using TSB medium for cultivation for 2 weeks. The pH range (5.0–11.0, at intervals of 1 pH unit) for growth was observed in TSB using the buffer system described by Xu et al. (2005). Tolerance to NaCl was examined using TSB as the basal medium with different total NaCl concentrations [0–10% (w/v; at 1% intervals)]. Colony characteristics were recorded after 3-day incubation on TSA at 28°C. The Gram reaction was tested by the Gram-staining method as described by Magee et al. (1975), and the capsule-staining was performed according to the protocols described previously (Breakwell et al., 2009). The cells were observed using light microscopy (BH-2, Olympus). Motility of cells was examined on TSA semi-solid agar medium (0.3%, w/v) and then double checked using hanging drop method (Bernardet et al., 2002). The morphological characteristics of the exponentially-growing cells were observed using transmission electron microscopy (JEOL JEM-1010).
The assimilation of carbon compounds by the isolates was tested at 28°C using Biolog GEN III Microplates and observed in an Omnilog device (BIOLOG Inc., Hayward, CA, United States). Other metabolic properties were examined by API 50CH, API 20NE and API ZYM test kits (bioMerieux) according to the manufacturer’s instructions. Results were evaluated after incubation at 28°C for 72–144 h. Activities of catalase, oxidase and urease, hydrolysis of Tweens and starch were investigated according to the procedures as previously described (Zhou et al., 1998; Yuan et al., 2008). The cellulose degradation activity was examined using CMC-Na screening medium (Reinhold-Hurek et al., 1993). The ability of the strains to produce IAA (indole-3-acetic acid) was tested using colorimetric methods (Bric et al., 1991) and recorded as described by Jiang et al. (2022). The scavenging effect of these strains on DPPH (2,2-diphenyl-1-picrylhydrazyl) radical was studied following the method described earlier (Singh and Rajini, 2004) and slightly modified according to the manufacture’s instruction of DPPH scavenging kit (Yuanye Bio-Technology Co., Ltd., Shanghai, China) as follows. Cells were harvested and washed with 0.1 M PBS solution for twice, and then suspended in 0.1 M PBS solution to the final concentration of 105 cells ml−1. The cells were sonicated in ice bath at 200 W for 3 s, followed by 10 s break. Repeated 30 rounds. The broken cells were discarded by centrifugation at 10000 r.p.m. for 10 min to collect the supernatant. Then added 50 μl of the supernatant into 450 μl of DPPH solution (0.1 mM in 95% ethanol) and incubated the mixture in dark at room temperature for 30 min. The absorbance of the resulting solution was read at 517 nm against a blank. The scavenging activity of vitamin C standard solution (2, 4, 6, 8, 10 mg L−1) was taken as the abscissa and the absorbance value of the corresponding concentration at 517 nm as the ordinate to draw a scatter diagram, and a linear trend line was added to obtain the scavenging activity of vitamin C standard curve. The radical scavenging activity was measured as a decrease in the absorbance of DPPH and was calculated using the following equation:
Ab, Ac and As is the absorbance of the blank (PBS), control (ethanol) and the sample, respectively.
Chemotaxonomic properties
Biomass for chemotaxonomic studies of the strains was obtained by cultivation in flasks on a rotary shaker (180 r.p.m.) using GYM broth at 28°C for 4 days except that cellular fatty acids extraction and analysis were conducted using the cultures harvested from Tryptic soy broth (TSB). The diagnostic isomers of diaminopimelic acid in the whole cell hydrolysates (4 N HCl, 100°C, 15 h) of these strains were subjected to thin-layer chromatography on cellulose plates using the solvent system of Schleifer and Kandler (1972). The sugar analysis of the whole cell hydrolysates followed procedures described by Staneck and Roberts (1974). Polar lipids were extracted and examined by two-dimensional TLC and identified using previously described procedures (Minnikin et al., 1984). Menaquinones were isolated using the method of Collins et al. (1977) and were analyzed by HPLC (Groth et al., 1997). Analysis of the cellular fatty acid pattern followed the described methods using the MIDI system (Microbial ID, Inc., Newark, Del; Kroppenstedt, 1985; Meier et al., 1993). MIDI Sherlock Version 6.0 and ACTIN1 database were employed for the analysis.
Genomic traits
Genome sequencing and assembly
The whole-genome sequencing was implemented using an Illumina HiSeq 4,000 system (Illumina, SanDiego, CA, United States) at the Beijing Genomics Institute (Beijing, China). Genomic DNA was sheared randomly to construct three read libraries with length of 300 bp by a Bioruptor ultrasonicator (Diagenode, Denville, NJ, United States) and physico-chemical methods. The paired-end fragment libraries were sequenced according to the Illumina HiSeq 4,000 system’s protocol. Raw reads of low quality from paired-end sequencing (those with consecutive bases covered by fewer than five reads) were discarded. The sequenced reads were assembled using SOAPdenovo v1.05 software. Digital DNA–DNA hybridization (dDDH) and average nucleotide identity (ANI) values between these strains and other related strains were calculated using the Genome-to-Genome Distance Calculator (GGDC, version 3.0; http://ggdc.dsmz.de/ggdc.php; Auch et al., 2010) and with the ezbiocloud platform (Yoon et al., 2017b), respectively.
Genome component prediction
The assembled genomic sequences of strains CPCC 205763T, CPCC 203386T, CPCC 205716T, CPCC 203406T, CPCC 203407 and H. moechotypicola KCTC 19653T were predicted by glimmer33 with Hidden Markov models and were functional annotated by the KEGG database (Kyoto Encyclopedia of Genes and Genomes). The assembled genomic sequences of other eight Herbiconiux strains (H. flava DSM 26474T, H. ginseng CGMCC 4.3491T, H. solani NBRC 106740T, Herbiconiux sp. L3-i23, Herbiconiux sp. SALV-R1, Herbiconiux sp. SYSU D00978, Herbiconiux sp. VKM Ac-1786 and Herbiconiux sp. VKM Ac-2,851) were downloaded from NCBI database and were also corrected as functionally annotated by the KEGG database. The Interpro database4 and the UniProt database5 were used for validation of putative functional genes. The tRNAscan-SE (Lowe and Eddy, 1997), RNAmmer, and the Rfam databases were employed for sorting of tRNA, rRNA and sRNAs, respectively.
Analysis of Pan-genome, functional genes and biosynthetic gene clusters
The protein sequences from each genome were followed by functional genes retrieval and pan-genome analysis. For pathway analysis, the predicted proteins sequences were uploaded to KEGG Automatic Annotation Server. Bacterial Pan-genome Analysis (BPGA) pipeline was applied for analysis of the genomic diversity of the Herbiconiux population. Pan-genome analysis was performed by BPGA 1.3 using default settings (Chaudhari et al., 2016). Predictions of gene clusters for natural products were performed using antiSMASH (antibiotic & Secondary Metabolite Analysis Shell, http://antismash.secondarymetabolites.org; Blin et al., 2021).
Results and analysis
Phenotypic characteristics
Strains designated as CPCC 205763T and CPCC 205716T were recovered from herb rhizospheric-niche samples IMB15132S and IMB20178S, respectively. Plant endophytic strains CPCC 203386T, CPCC 203406T, and CPCC 203407 were isolated from the samples IMB12384P (the stem of a medicinal plant Daphne aurantiaca) and IMB11009P (the leaves of Oxytropis falcata), respectively.
Good growth of these five isolates was observed on tested media ISP 2, TSA, R2A, and PYG, at 28–32°C and pH 7.0. NaCl was not required for growth. Light to bright yellow colonies formed on TSA with diameter of 0.8–1.2 mm in diameter. Capsulated cells were Gram-staining-positive, non-motile and rod-shaped. Detailed physiological and biochemical characteristics results from Biolog and API assay kits were listed in Table 1 and species description.
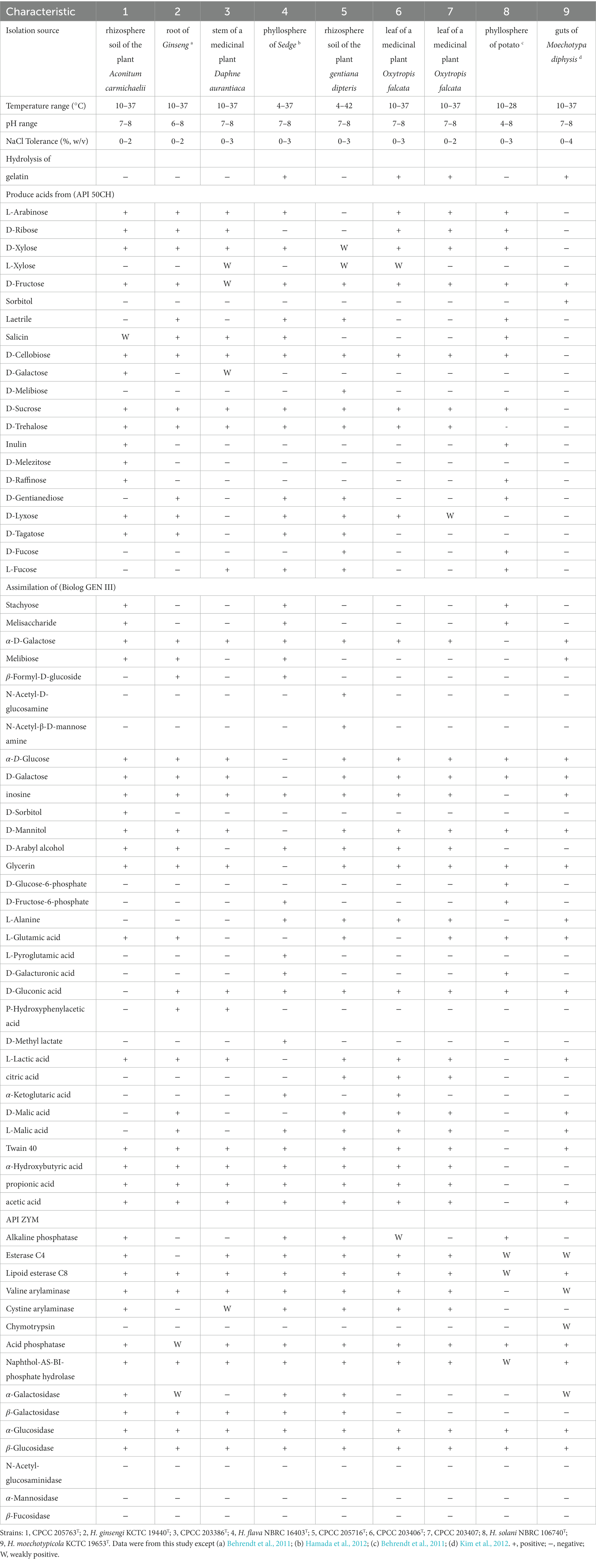
Table 1. Differential characteristics between strains CPCC 205763T, CPCC 203386T, CPCC 205716T, CPCC 203406T, CPCC 203407 and the closely related type strains of the genus Herbiconiux.
The clear transparent circle was observed around the strains on plate with CMC-Na as the sole carbon source, which demonstrated these strains were cellulose-degrading microbes. IAA was detected in the fermentation broth of strains CPCC 205763T, CPCC 203386T, CPCC 205716T, CPCC 203406T, and CPCC 203407. As shown in Supplementary Figure S1, the linear regression equation y = 0.0255x + 0.0411, r2 = 0.9992, gave a good fit. Accordingly, the IAA content produced by the test strains was calculated. The IAA content yielded from the fermentation broth of CPCC 205763T, CPCC 203386T, CPCC 205716T, CPCC 203406T, and CPCC 203407 was 5.89 ± 0.91 mg L−1, 0.94 ± 0.00 mg L−1, 4.61 ± 0.02 mg L−1, 2.17 ± 0.02 mg L−1 and 2.10 ± 0.05 mg L−1, respectively. Total DPPH scavenging potential of the test strains was measured and depicted in Supplementary Figure S2. The linear regression equation of the radical scavenging activity of vitamin C standard curve y = 11.65x + 17.724, r2 = 0.9999, had a good fit. Strains CPCC 205763T, CPCC 203386T, CPCC 205716T, CPCC 203406T and CPCC 203407 were capable of neutralizing the DPPH free radicals via hydrogen donating activity by 38.3 ± 0.9%, 78.6 ± 5.8%, 51.8 ± 3.3%, 65.4 ± 10.9%, and 33.8 ± 4.1% at the concentration of 105 cells mL−1, respectively. Accordingly, the Herbiconiux strains harbored a higher antioxidant activity than that of 4 mg L−1 vitamin C.
Chemotaxonomy
In the cell-wall peptidoglycan of these five strains, 2,4-diaminobutyric acid was detected as the diagnostic diamino acid. Diphosphatidylglycerol (DPG), phosphatidylglycerol (PG) and glycolipid (GL) were present in the polar lipids extract of the five strains (Supplementary Figure S3). The predominant respiratory quinone was detected as MK-11. In the acid profiles, anteiso-C15:0 was the major component (26–70%). In addition to anteiso-C15:0, strain CPCC 205763T contained C18: 1ɷ7c/C18: 1ɷ6c (28.1%), strain CPCC 203386T contained C17:1 ɷ9c (25.0%) and anteiso-C17:0 (11.1%), CPCC 205716T contained iso-C16:0 (20.6%), CPCC 203406T and CPCC 203407 contained anteiso-C17:0 and iso-C16:0 as the major fatty acids, respectively (Supplementary Table S1).
Phylogenetic analysis
The almost full-length 16S rRNA gene sequences (1,512 bp, 1,521 bp, 1,515 bp, 1,511 bp and 1,511 bp) of strains CPCC 205763T, CPCC 203386T, CPCC 205716T, CPCC 203406T, CPCC 203407 were obtained. The alignment results of the 16S rRNA gene sequences indicated that the five strains were member of the family Microbacteriaceae with closely related to the genus Herbiconiux. The 16S rRNA gene sequence of the strain CPCC 205763T showed the highest similarity with H. ginseng KCTC 19440T (99.2%), H. solani NBRC 106740T (98.3%), H. moechotypicola RB-62T (98.2%), H. flava NBRC 16403T (97.9%) and no more than 97.5% similarities with other type strains of the family Microbacteriaceae. The 16S rRNA gene sequences of the strains CPCC 203386T, CPCC 205716T, CPCC 203406T, CPCC 203407 showed highest similarities with H. flava NBRC 16403T (98.3–99.9%), H. ginseng KCTC 19440T (97.7–98.3%), H. moechotypicola RB-62T (97.7–98.1%) and H. solani NBRC 106740T (97.8–98.8%; Supplementary Table S2). In the phylogenetic tree based on the 16S rRNA gene sequences, these five strains fell in the genus Herbiconiux lineage. The strain CPCC 205763T formed a robust unique cluster with H. ginsengi KCTC 19440T, and the strains CPCC 203386T, CPCC 205716T, CPCC 203406T and CPCC 203407 formed a stable branch with the strains H. flava NBRC 16403T, H. solani NBRC 106740T and two unclassified strains of the genus Herbiconiux in the neighbor-joining tree (Figure 1), which showed almost the same case in the maximum-parsimony tree, maximum-likelihood tree and phylogenetic trees based on the concatenated core genes (Figure 2) and pan-matrix by pan-genome analysis (Supplementary Figure S4).
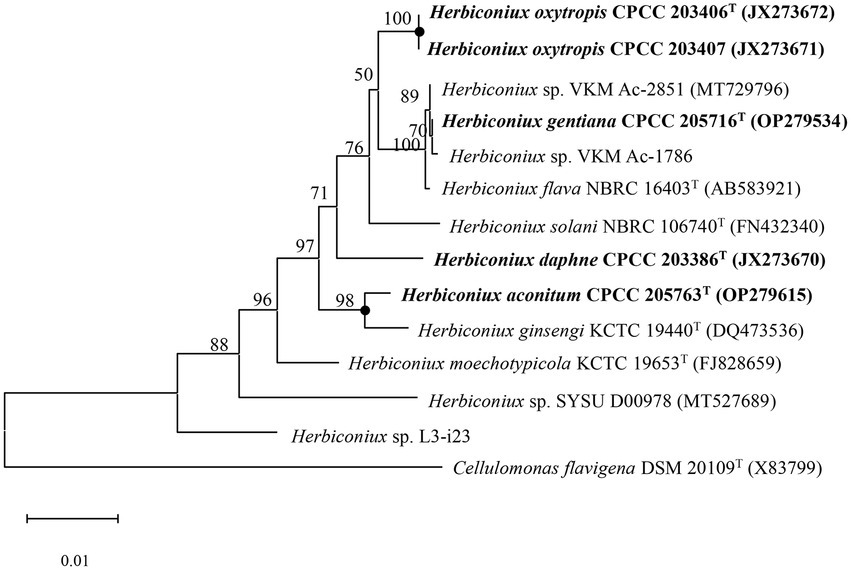
Figure 1. Neighbour-joining phylogenetic tree based on 16S rRNA gene sequences of strains CPCC 205763T, CPCC 203386T, CPCC 205716T, CPCC 203406T, CPCC 203407, and related species of the genus Herbiconiux. Filled circles indicate that the corresponding nodes were also recovered in the trees generated with the maximum-likelihood and maximum-parsimony methods. Bootstrap values (those above 50%) are shown as percentages of 1,000 replicates. Cellulomonas flavigena DSM 20109T (GenBank accession no. X83799) was used as an outgroup. Bar, 0.01 nt substitution per nt.
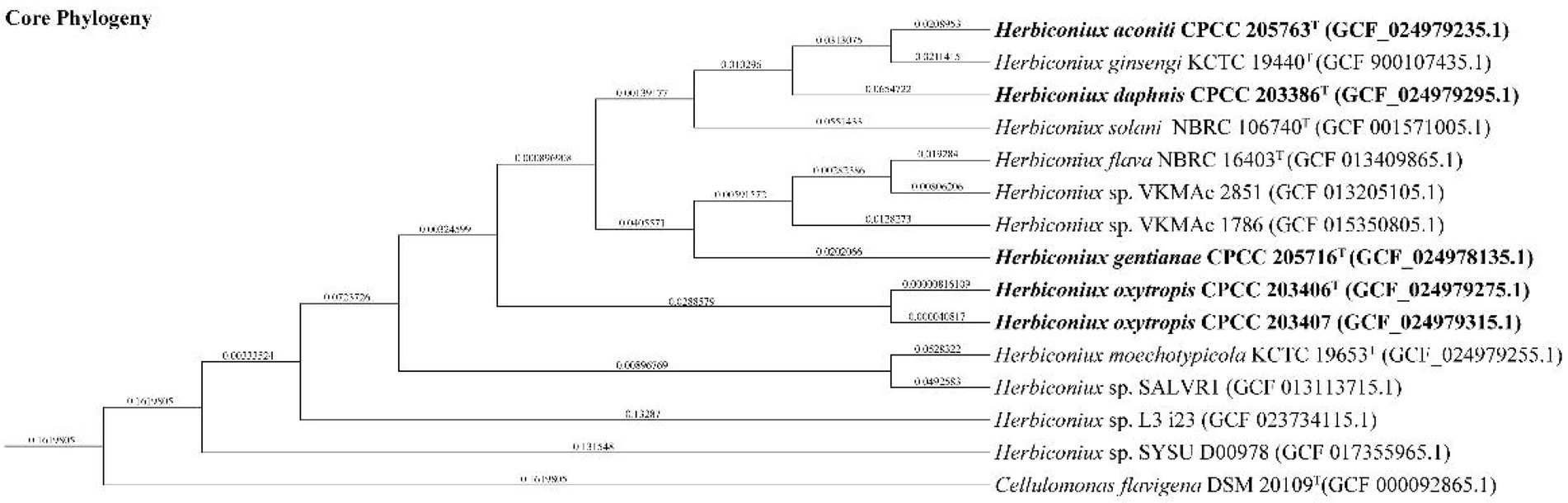
Figure 2. Phylogenetic tree constructed by BPGA showing the relationship of the newly proposed species with other species of the genus Herbiconiux based on concatenated core genes.
The genomic G + C content of these five strains ranged in 68.6–71.0%. ANI values calculated between strains CPCC 205763T, CPCC 203386T, CPCC 205716T, CPCC 203406T, and other type strains of Herbiconiux species were all less than 89.2%, and the corresponding dDDH values were below 68.1% (Supplementary Table S3). These values were lower than the thresholds used to delineate bacterial species (i.e., ANI < 95% and dDDH <70%; Kim et al., 2012). While strains CPCC 203407 and CPCC 203406T shared a high ANI value of 100% and a high dDDH value of 99.9% (Supplementary Table S3), which were consistent with the high level of 16S rRNA gene sequence similarity (100%) between the two strains, consistently indicating the assignment of these two strains to a same species of the genus Herbiconiux (Kim et al., 2012).
Filled circles indicate that the corresponding nodes were also recovered in the trees generated with the maximum-likelihood and maximum-parsimony methods. Bootstrap values (those above 50%) are shown as percentages of 1,000 replicates. Cellulomonas flavigena DSM 20109T (GenBank accession no. X83799) was used as an outgroup. Bar, 0.01 nt substitution per nt.
Genome features
The whole genomes of strains CPCC 205763T, CPCC 203386T, CPCC 205716T, CPCC 203406T, CPCC 203407, H. flava DSM 26474T, H. ginsengi CGMCC 4.3491T, H. moechotypicola KCTC 19653T, H. solani NBRC 106740T, Herbiconiux sp. L3-i23, Herbiconiux sp. SALV-R1, Herbiconiux sp. SYSU D00978, Herbiconiux sp. VKM Ac-1786 and Herbiconiux sp. VKM Ac-2,851 contained 3,952, 5,647, 3,674, 3,987, 3,945, 3,678, 4,594, 4,087, 3,688, 3,057, 4,305, 2,646, 3,742, and 4,143 genes, respectively. The detailed genomic characteristics of these 14 strains were summarized in Supplementary Table S4. In the 14 genomes of the genus Herbiconiux, the putative encoding genes for alkyl hydroperoxide reductase E (ahpE), catalase (katA, katC), manganese catalase (ydbD), redox-sensitive transcriptional activator SoxR (soxR) and superoxide dismutase (sodA) were retrieved. These enzymes might help these microorganisms to relieve the stress of excessive oxygen anion concentration that released from the medicinal plants into the environments. The carotenoid biosynthesis related genes (crtB and crtI), indole-3-acetic acid producing related genes (aldH, amiE, ddc maoI), iron-siderophore transport system permease protein associated encoding genes (entS, fecE, fecF, fepB, fepD, fepG, yclQ, yfhA, yfiY, yfiZ, and yusV) and phosphate-solubilizing encoding genes (phnB, phoA, phoB, phoH, phoP, phoR, phoU, ppk, pstA, pstB, pstC, and pstS) were also sorted (Figure 3).
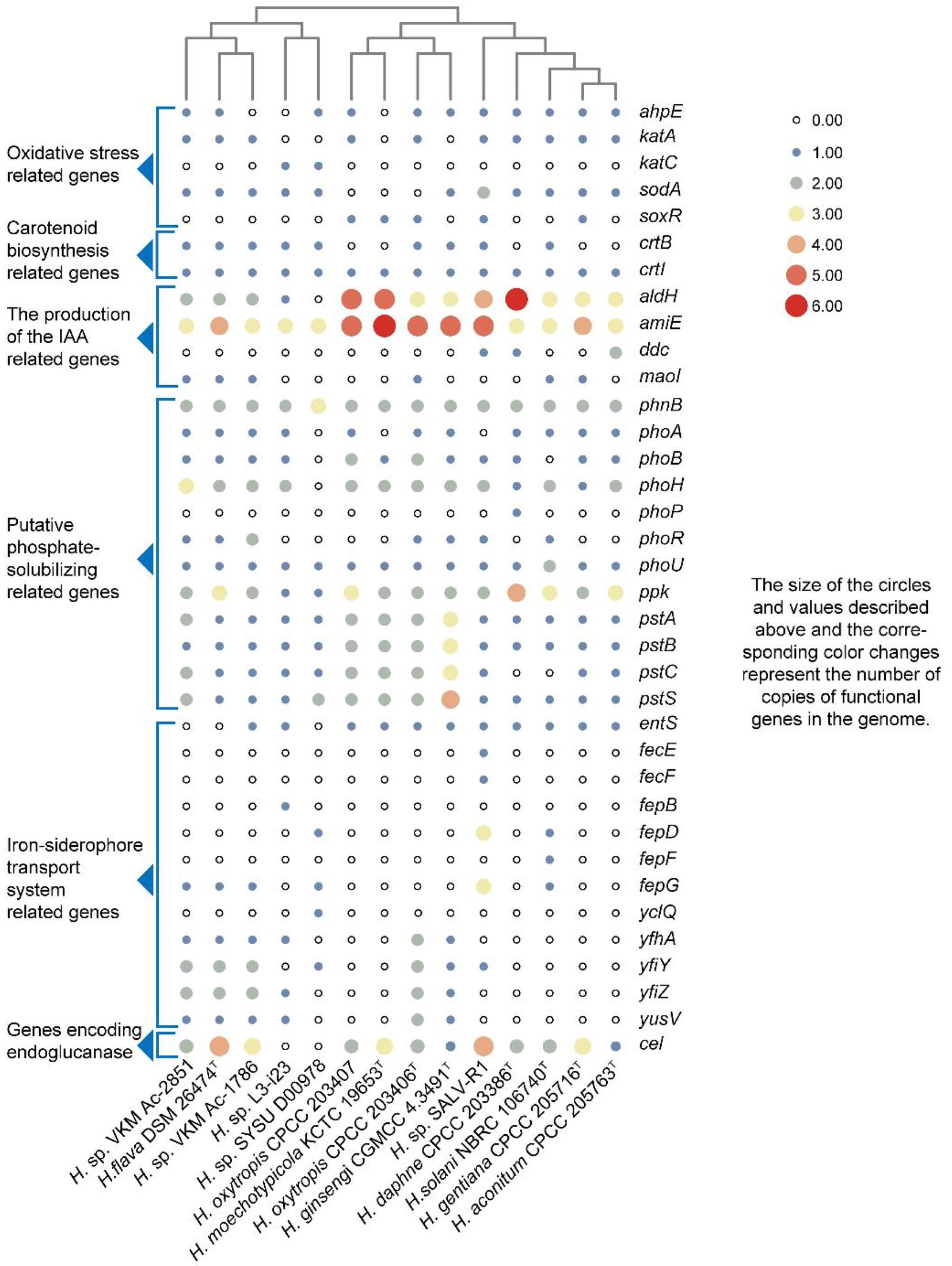
Figure 3. Heatmap of putative functional genes predicted from the 14 genomes of the genus Herbiconiux according to the copy number of the genes from the KEGG annotation.
Pangenome analysis of the genus Herbiconiux
A total of 51,271 protein-coding genes (Table 2) were sorted from the genomes of these 14 strains of the genus Herbiconiux, which were divided into 15,209 homologous families by cluster analysis. According to the presence or absence of a certain homologous gene in these 14 genomes, we defined the homologous gene families conservation values (HGFCV, be abbreviated to CV). Each homologous family was given a conserved value (CV) based on its frequency of occurrence in each of the 14 genomes. Histograms were constructed according to different CVs (Figure 4A). In the pan-genome profile of the genus Herbiconiux, there were a total of 753 core genes commonly shared by these 14 strains (CV = 14), accounting for about 5.0% of the total number of homologous gene families. The accessory genes (6,160 genes; CV = 2 ~ 13) accounted for about 40.5% of the homologous gene families in the genus Herbiconiux. The proportion of the unique genes (8,296 genes; CV = 1) was about 54.5%.
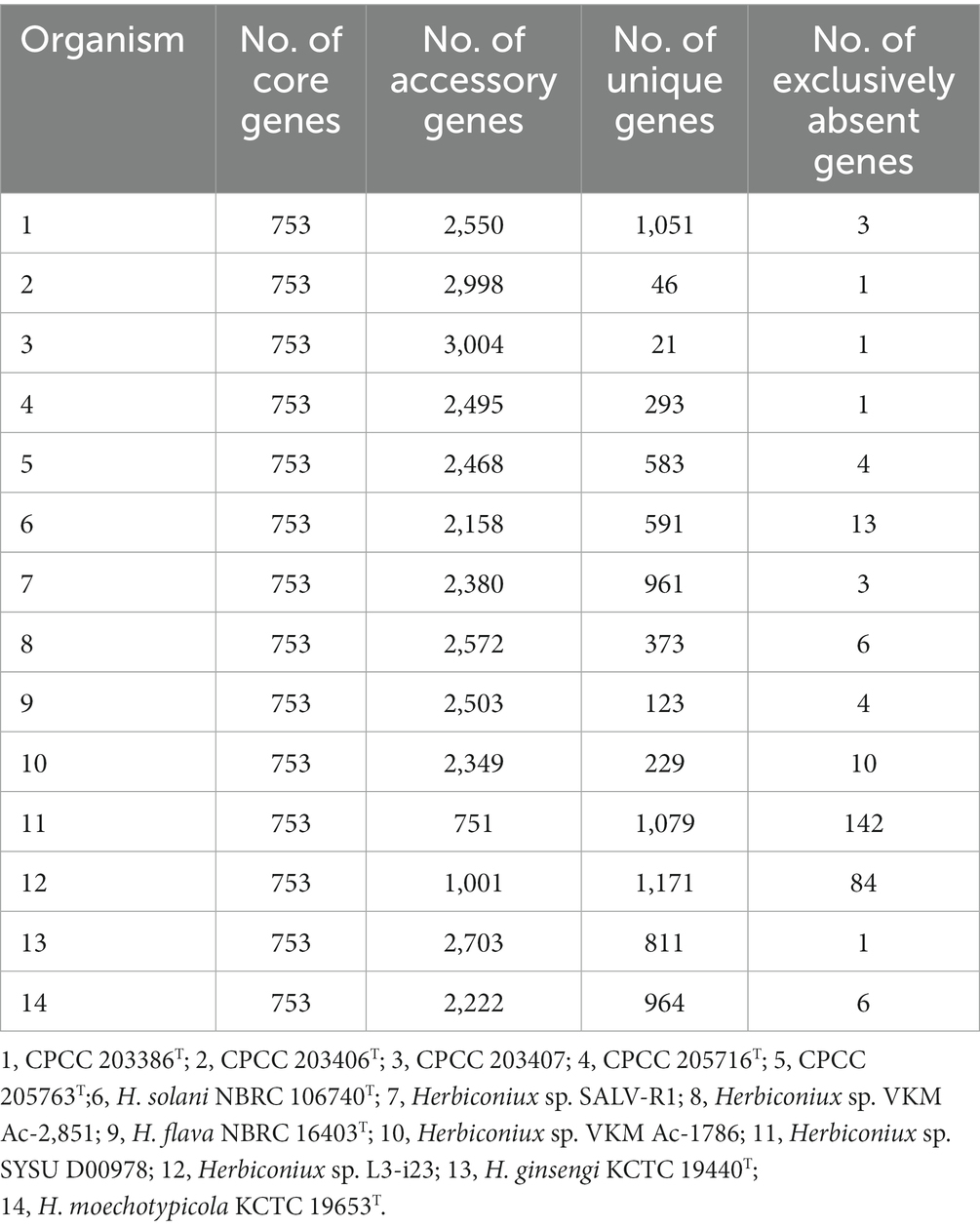
Table 2. Distribution of core, accessory, unique and exclusively absent genes among the genomes of 14 strains of the genus Herbiconiux using USEARCH clustering tool.
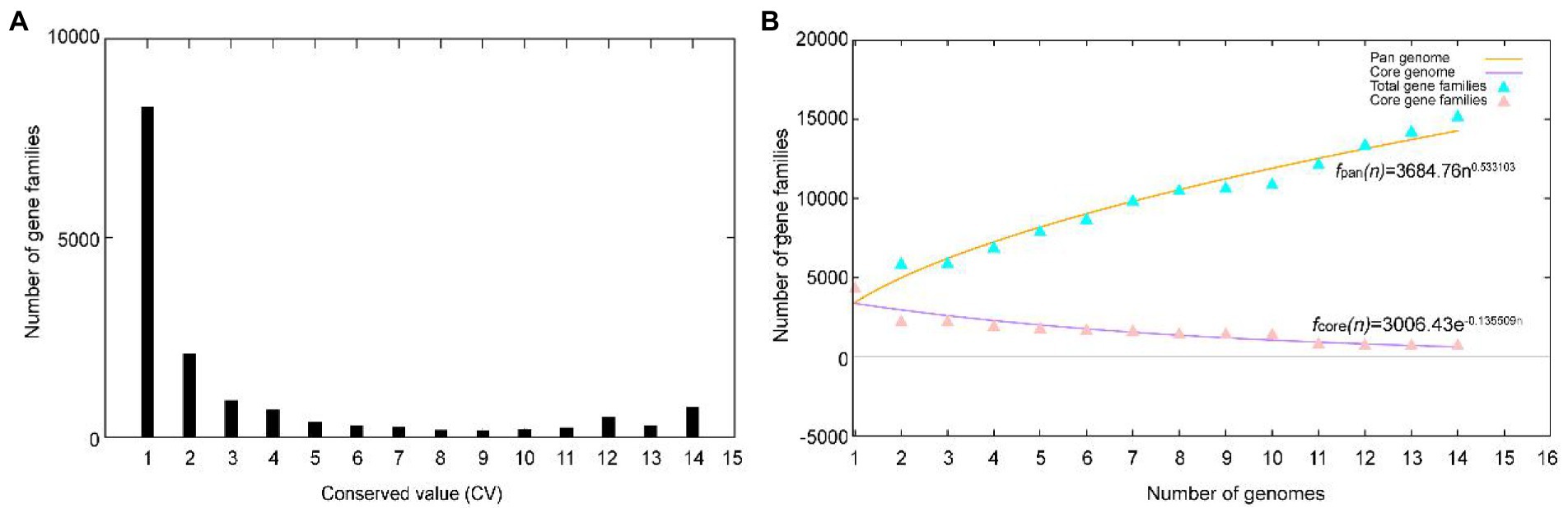
Figure 4. Overview of the pan-genomic results generated by BPGA using the 14 genomes of the genus Herbiconiux. (A) The gene family frequency spectrum. (B) The pan genome profile trends of the genus Herbiconiux obtained using clustering tools USEARCH.
The relationship between the pan-genome size and the number of genomes of the genus, and the relationship between the number of core genes and the number of genomes were deduced (Figure 4B) by using all the protein sequences extracted from these 14 strains of the genus Herbiconiux. The functional relationship between pan-genome size (f pan ) and the number of genomes (n) was obtained by fitting, as follows:
Out of 15,209 genes (clusters), BPGA could map 6,136 (40.3%) to KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways, i.e., core gene (822, 13.4%), accessory genes (2,735, 44.6%) and unique genes (2,579, 42.0%). After filtering some KEGG pathway related to eukaryotes, we obtained an overview on the metabolic pathway (>1%) corresponding to the gene(s) in the pan-genome of the genus Herbiconiux. A large number of core genes (787) were involved in carbohydrate metabolism (17.5%), some other elementary metabolism (biosynthesis of amino acids, 8.4%; carbon metabolism, 5.0%; 2-oxocarboxylic acid metabolism, 2.0%; fatty acid metabolism, 0.6% and degradation of aromatic compounds, 0.1%; 16.1%), amino acid metabolism (13.1%), translation (7.8%), energy metabolism (7.5%), nucleotide metabolism (6.5%), metabolism of cofactors and vitamins (4.8%), replication and repair (4.7%), membrane transport (3.4%), folding, sorting and degradation (2.7%), metabolism of other amino acids (2.4%), signal transduction (2.2%), lipid metabolism (2.2%), metabolism of terpenoids and polyketides (1.8%), biosynthesis of other secondary metabolites (1.7%), xenobiotics biodegradation and metabolism (1.5%), and glycan biosynthesis and metabolism (1.4%). Accessory and unique genes appeared to be enriched in carbohydrate metabolism, amino acid metabolism, membrane transport, as well as other elementary metabolism. Among the accessory genes (2,551), the major portion of genes seemed related to carbohydrate metabolism (19.0%), amino acid metabolism (14.0%), membrane transport (13.0%), some other elementary metabolism (carbon metabolism, 4.7%; biosynthesis of amino acids, 3.1%; fatty acid metabolism, 1.5%; degradation of aromatic compounds, 1.3% and 2-oxocarboxylic acid metabolism, 0.5%; 11.0%), xenobiotics biodegradation and metabolism (6.2%), lipid metabolism (6.2%), metabolism of cofactors and vitamins (5.1%), energy metabolism (5.0%), nucleotide metabolism (3.4%), signal transduction (3.1%), biosynthesis of other secondary metabolites (2.5%), metabolism of other amino acids (2.4%), replication and repair (2.4%), metabolism of terpenoids and polyketides (1.9%), folding, sorting and degradation (1.5%) and glycan biosynthesis and metabolism (1.1%). Unique genes (2,421) seemed to be mainly enriched in carbohydrate metabolism (20.3%), membrane transport (13.1%), amino acid metabolism (12.0%), some other elementary metabolism (carbon metabolism, 3.9%; biosynthesis of amino acids, 3.2%; fatty acid metabolism, 1.7%; degradation of aromatic compounds, 1.3% and 2-oxocarboxylic acid metabolism, 0.7%; 10.8%), xenobiotics biodegradation and metabolism (8.5%), lipid metabolism (7.7%), metabolism of cofactors and vitamins (4.8%), energy metabolism (3.2%), signal transduction (3.1%), biosynthesis of other secondary metabolites (3.1%), nucleotide metabolism (2.4%), metabolism of other amino acids (2.3%), glycan biosynthesis and metabolism (2.0%), metabolism of terpenoids and polyketides (1.7%), cell motility (1.4%) and replication and repair (1.3%; Figure 5).
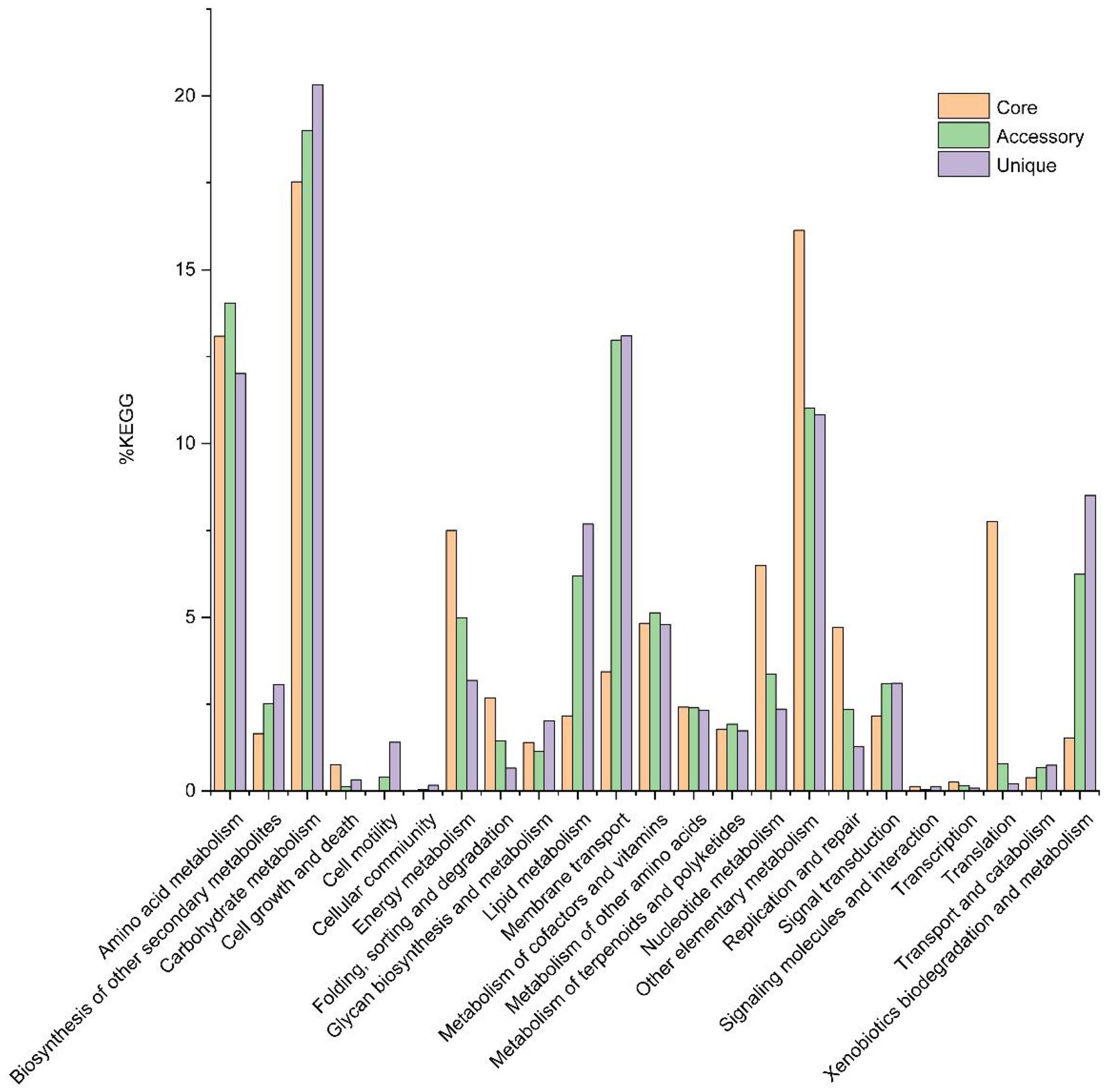
Figure 5. The assigned metabolic pathways associated with the core, accessory and unique genes among the genus Herbiconiux from the KEGG database (14 strains).
Secondary metabolite biosynthesis gene clusters analysis
The results from antiSMASH database showed that in these 14 strains of the genus Herbiconiux, three to eight secondary metabolite gene clusters with moderate similarities to previously described secondary metabolite biosynthetic gene clusters were retrieved. These gene clusters exhibited 1–100% similarities to previously reported secondary metabolite biosynthetic gene clusters, such as carotenoid, pyoverdin, microansamycin, tiancimycin, rustmicin, herboxidiene, divergolide A/ divergolide B/ divergolide C/ divergolide D, cypemycin, alkylresorcinol gene clusters and other unidentified secondary metabolite clusters attributable to NAPAA, redox-cofactor, RiPP-like, RRE-containing, NRPS-like, bottromycin, and T3PKS types, respectively (Supplementary Table S5).
Discussion
Obviously, the 16S rRNA gene sequences alignment and the phylogenetic analysis indicated these five isolates affiliated to the genus Herbiconiux, which was supported by the chemotaxonomic traits. The genome relatedness indexes, i.e., values of ANI and dDDH between strains CPCC 205763T, CPCC 203386T, CPCC 205716T, CPCC 203406T, and all the type strains of the validly named species in the genus Herbiconiux well differentiated them from each other. The ANI and dDDH values between strain CPCC 203406T and CPCC 203407 suggested to classify these two strains into a same species. Integrated the morphological properties, physiologic characteristics, chemotaxonomic profiles and phylogenetic analysis of these five strains, it is reasonable to propose four novel species of the genus Herbiconiux, i.e., Herbiconiux aconitum sp. nov. with CPCC 205763T as the type strain, Herbiconiux daphne sp. nov. with CPCC 203386T as the type strain, Herbiconiux gentiana sp. nov. with CPCC 205716T as the type strain, and Herbiconiux oxytropis sp. nov. with CPCC 203406T as the type strain.
Rhizospheric and endophytic microorganisms live together with the plants for a time, and co-evolve with the host plant, where microbes and plants shared the common special mico-ecosystem, so that they form a mutually beneficial symbiotic relationship during the evolution process. On the one hand, plants provide photosynthetic products and minerals for the growth of these microbes. In turn, once a kind of beneficial bacteria colonizes in a niche associated with a plant, it can influence the physiologic traits of the plant through various mechanisms. In this study, strains CPCC 205763T, CPCC 203386T, CPCC 205716T, CPCC 203406T, and CPCC 203407 were isolated from different ecosystems associated with four kinds of Chinese traditional medicinal plants, Aconitum carmichaelii, Daphne aurantiaca, Gentiana rigescens and Oxytropis falcate, respectively. As well, the strains Herbiconiux flava DSM 26474T, Herbiconiux ginsengi CGMCC 4.3491T, Herbiconiux solani NBRC 106740T, Herbiconiux sp. SALV-R1, Herbiconiux sp. VKM Ac-1786 and Herbiconiux sp. VKM Ac-2,851 were reported to inhabit the niches associated with plants. Based on the genomic information, we summarized genetic characteristics of these Herbiconiux spp. The detailed phenotypic properties illustrated the abilities of these strains to make contribution to their associated medicinal plants. For instance, indole-3-acetic acid (IAA), a kind of plant hormone, plays an important role in plant-microbial interactions, especially, in promoting the growth of plants. The phenotypic assays confirmed that strains CPCC 205763T, CPCC 203386T, CPCC 205716T, CPCC 203406T, and CPCC 203407 could produce IAA. In the genomic category of these five strains, the related encoding genes involved in the IAA-producing pathway were retrieved. For instance, the encoding genes for aldehyde dehydrogenase (EC 1.2.1.3; aldH), amidase (Unwin et al., 2004), aromatic-L-amino-acid/L-tryptophan decarboxylase (ddc) and monoamine oxidase (MAO, aofH). What’s more, in the core genome of the genus Herbiconiux, the amidase coding gene (Unwin et al., 2004) was retrieved. In addition, according to the distribution of IAA-producing genes in the five strains, the pathway of IAA production by the annotation of the KEGG database could be predicted (Figure 6).
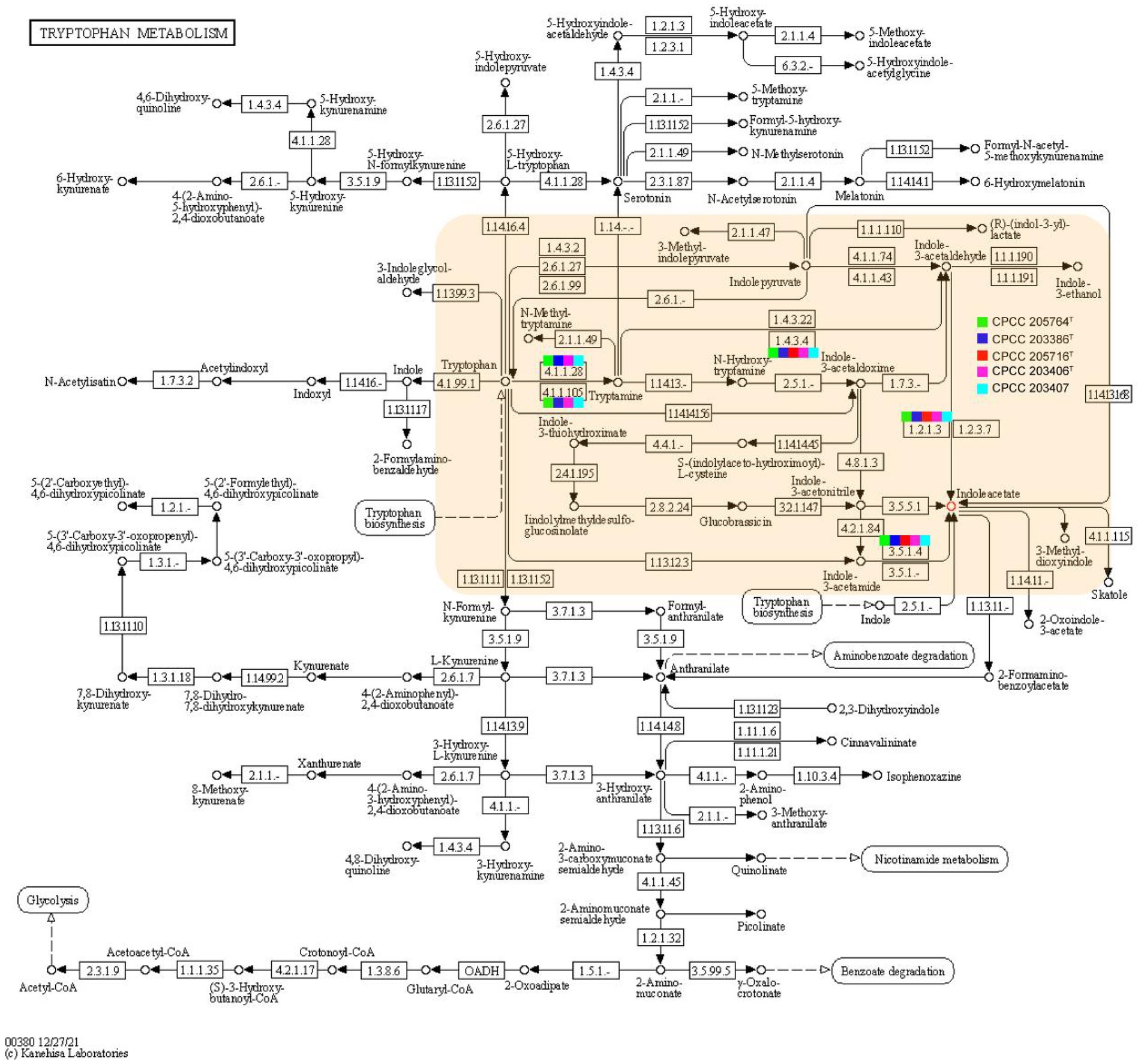
Figure 6. Putative overview of indole-3-acetic acid-producing pathway in the tryptophan metabolism pathways of the strains CPCC 205763T, CPCC 203386T, CPCC 205716T, CPCC 203406T, and CPCC 203407. The squares colored in green, blue, red, pink and light blue represents the strain CPCC 205763T, CPCC 203386T, CPCC 205716T, CPCC 203406T, and CPCC 203407, respectively.
The metabolites produced by rhizospheric microbes or endophytes, such as IAA, can stimulate the growth and development of the plants that shared the common ecosystems with them, accordingly, improve the resistance of the plants to biotic or abiotic stresses. Therefore, some metabolites from these microorganisms are of significance for plant growth and development. In order to explore the useful metabolite-producing candidates, microbiologists are increasingly employing genome sequencing of a wide variety of such microbes. Here, we identified biosynthetic gene clusters (BGCs) by antiSMASH. From the antiSMASH results of these strains, we found that 100% (14/14) of the Herbiconiux genomes contain the carotenoid gene clusters (Table 3), with the 50 –66% similarities. The carotenoids belong to the isoprenoids and contain eight isoprene units, namely, tetraterpenoids. For one thing, carotenoids have the ability to absorb and transfer electrons, and play an important role in scavenging superoxide anion free radicals produced during photosynthesis (Bartley and Scolnik, 1995). In addition, carotenoids, such as astaxanthin (Chang and Xiong, 2020), fucoxanthin and zeaxanthin (Firdous et al., 2015) were reported to have anti-inflammatory functions. In this research, the four kinds of Chinese traditional medicinal plants, Aconitum carmichaelii (Zhao et al., 2018), Daphne aurantiaca (Nie et al., 2021), Gentiana rigescens (Zhao et al., 2015) and Oxytropis falcate (Chen et al., 2011), were reported to have anti-inflammatory activities. Could we suppose that these microbes isolated from these ecological niches might be endowed with the anti-inflammatory traits by their corresponding medicinal plants?
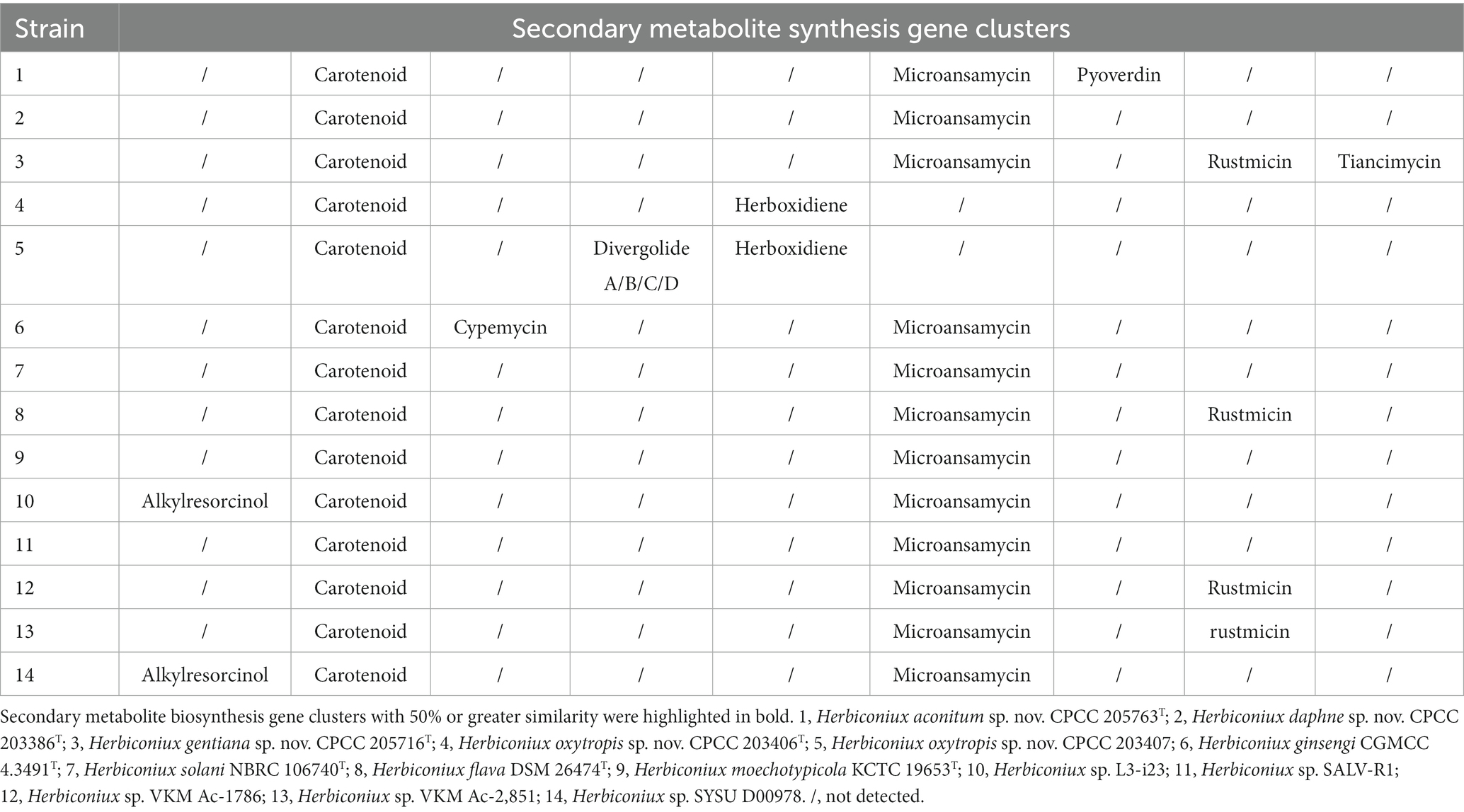
Table 3. Comparative results of secondary metabolite biosynthesis gene clusters predicted from the 14 strains’ genomes of the genus Herbiconiux.
From the antiSMASH results of the strain CPCC 203406T and CPCC 203407, the herboxidiene gene clusters were retrieved. Herboxidiene (GEX1A) is a potent phytotoxic polyketone compound. On the one hand, as a potent splicing inhibitor in plants, GEX1A could trigger abiotic stress responses and ABA (abscisic acid) signaling in plants. Splicing stress signaling generated by GEX1A treatment is differentially regulated to ensure plant adaptation to stress conditions (AlShareef et al., 2017). On the other hand, as a novel polyketide which selectively and effectively controls several annual weed species, GEX1A could improve the biological competitiveness of the medicinal plant. In the antiSMASH results of the strain Herbiconiux sp. L3-i23 and Herbiconiux sp. SYSU D00978, the BGCs of alkylresorcinol were identified. Alkylresorcinols are phenolic lipids widely distributed in plants and bacteria. Different degrees of substitution yield various compounds that have been reported to possess antioxidant (Gliwa et al., 2011), anti-inflammatory (Wei et al., 2022), antimicrobial and antitumor activities. Given a variety of alkylresorcinols compounds have antimicrobial activity, it is possible to conclude that these compounds act as defensive agents in plants. Moreover, phenolic compounds (such as 5-n-alkylresorcinols, phlorizin, resveratrol, ferulic acid, et al.) are part of the plant’s protection system against various pests and are therefore considered natural alternatives to protective agents (Patzke and Schieber, 2018). In the annotation results of the KEGG, the spermidine/putrescine transport related genes (potA, potB, potC, and potD) were retrieved in the genomes of the new species. In plants, it has been shown that increasing polyamine levels minimizes harmful effects caused by biotic and abiotic stresses (Yoda et al., 2003).
Reactive oxygen species (ROS), generated by the plant, might be neutralized by the production of enzymes such as superoxide dismutases, catalases and alkyl hydroperoxide reductases in microorganisms. In the genomes of the strains CPCC 205763T, CPCC 203386T, CPCC 205716T, CPCC 203406T, CPCC 203407, and other members of the genus Herbiconiux included in our study, various antioxidant encoding genes were retrieved, such as genes coding for superoxide dismutase (sodA), putative manganese catalase (ydbD), catalase coding gene (katA), alkyl hydroperoxide reductase E (ahpE) and redox-sensitive transcriptional activator SoxR (soxR). The carotenoids and phytohormones mentioned above also play an important role in antioxidant activities. Therefore, the strains of the genus Herbiconiux may play an important role in their associated plants responding to oxidative stress.
Microorganisms promote the growth of their associated plants by obtaining nutrients from the environments. Such as the acquisition of organic phosphorus and inorganic phosphorus. At the genomic level, phosphate starvation-inducible protein coding gene (phoH), alkaline phosphatase coding genes (phoA, phoB), phosphate transport-related genes (phnB, phoP, phoR, phoU, pstA, pstB, pstC, and pstS) and polyphosphate kinase coding gene (ppk) were retrieved from all these 14 genomes. We also found some genes (entS) related to the production and export of the siderophore enterobactin and iron-siderophore transport system permease protein related genes (fecE, fecF, fepB, fepD, fepF, and fepG.) in the genomes of the genus Herbiconiux (Figure 3). Accordingly, we speculate that the new species of genus Herbiconiux may have beneficial effects on the growth of their associated medicinal plants, in addition to production of IAA, partially through the production of phytohormones and siderophore, solubilization of phosphorus, or by production of SOD to partly mitigate the oxidative pressure.
Conclusion
Our research investigated the taxonomic characteristics of Herbiconiux, and discovered the genetic basis of Herbiconiux producing secondary metabolites, based on pan-genome analysis and experimental validation, specifically, the PGP function of the Herbiconiux spp. associated with plants. Results from this study indicated diversity of novel Herbiconiux members are abundant in the ecosystems associated with plants, and are a group of plants-friendly microbes. Based on these results, we expect to accumulation of sufficient Herbiconiux cultures from diverse ecosystems to compare the components of secondary metabolite gene clusters in different Herbiconiux species, and to reveal new gene elements associated with secondary metabolism and explore important secondary metabolites from Herbiconiux.
Description of Herbiconiux aconitum sp. nov.
Herbiconiux aconitum (a.co.ni’ti. N.L. gen. n. aconiti, of Aconitum, a plant genus name, referring to the site related to a plant of Aconitum carmichaelii, from which the type strain was isolated).
Cells are aerobic, Gram-staining-positive, rod-shaped. Colonies are 0.8–1.0 mm in diameter, convex with an entire margin, glistening, viscous and bright yellow on TSA medium. Growth temperature and pH range for growth are 10–37°C and pH 7.0–8.0, with optimum growth at 28–30°C and pH 7.0. Cannot tolerate >2% (w/v) NaCl. Positive for oxidase, catalase, acid phosphatase, alkaline phosphatase, cystine arylamidase, esterase (C4), esterase lipase (C8), leucine arylamidase, naphthol-AS-BI-phosphohydrolase, valine arylamidase, α-galactosidase, α-glucosidase, β-galactosidase, and β-glucosidase, negative for lipase (C14), N-acetyl-β-glucosamimidase, α-chymotrypsin, α-fucosidase, α-mannosidase, β-glucuronidase and gelatin hydrolyzation. Utilizes acetic acid, acetoacetic acid, D-arabitol, D-cellobiose, dextrin, D-fructose, D-galactose, D-maltose, D-mannitol, D-mannose, D-melibiose, D-raffinose, D-salicin, D-sorbitol, D-trehalose, D-turanose, gentiobiose, glycerol, glycyl-L-proline, inosine, L-fucose, L-glutamic acid, L-lactic acid, L-rhamnose, N-acetyl-D-galactosamine, pectin, propionic acid, stachyose, sucrose, tween 40, α-D-glucose, α-D-lactose, α-hydroxy-butyric acid, and α-keto-butyric acid as the sole carbon source; acid is produced from arbutin, D-cellobiose, D-fructose, D-galactose, D-glucose, D-lactose, D-lyxose, D-maltose, D-mannitol, D-mannose, D-melezitose, D-raffinose, D-ribose, D-saccharose, D-tagatose, D-trehalose, D-turanose, D-xylose, esculin ferric citrate, glycerol, inulin, L-arabinose, L-rhamnose, but not from amidon, amygdalin, D-arabinose, D-arabitol, D-ardonitol, D-fucose, D-melibiose, D-sorbitol, dulcitol, erythritol, gentiobiose, glycogen, inositol, L-arabitol, L-fucose, L-xylose, mehyl-α-D-glucopyranoside, mehyl-α-D-mannopyranoside, mehyl-β-D-xylopyranoside, N-acetylglucosamine, potassium 2-ketogluconate, potassium 5-ketogluconate, potassium, gluconate, or xylitol. In the API 20NE test system, positive for aesculin hydrolysis, β-galactosidase, and assimilation of adipic acid, DL-malic acid, L-arabinose and potassium gluconate, but negative for arginine dihydrolase, D-mannitol, glucose fermentation, indole production, nitrate reduction, protease, urease, and assimilation of capric acid and phenylacetic acid. Diphosphatidylglycerol and phosphatidylglycerol are detected in the polar lipids extraction. The predominant quinone is MK-11. The fatty acid profile consists of the predominant components (> 10%) anteiso-C15:0 and summed feature 8 (C18:1 ɷ7c and/or C18:1 ɷ6c). Glucose as cell-wall sugars. The diagnostic diamino acids in the cell-wall peptidoglycan are alanine, glutamic acid and glycine. The type strain is CPCC 205763T (= I19A-01430T = CGMCC 1.60067T), isolated from a rhizosphere soil sample of the plant Aconitum carmichaelii collected from Xinjiang Province, north-west China. The genomic G + C content of the type strain is 68.2%.
Description of Herbiconiux daphne sp. nov.
Herbiconiux daphne (daph’nis. N.L. gen. n. daphnis, of Daphne, a plant genus name, referring to the isolation of the type strain from a plant of Daphne aurantiaca).
Colonies on TSA medium are light yellow, convex with an entire margin, glistening and viscous, with 0.6–0.8 mm in diameter after 5 days at 28°C (pH 7.0). Cells are aerobic, Gram-staining-positive, rod-shaped. Growth occurs at 10–37°C and pH 7.0–8.0, with optimum at 28–32°C and pH 7.0, respectively. NaCl is not required for growth, but NaCl tolerance is up to 3.0% (w/v). Positive for oxidase and catalase reaction, negative for gelatin hydrolyzation. In the API 20NE test system, positive for aesculin hydrolysis, β-galactosidase, and assimilation of L-arabinose, but negative for arginine dihydrolase, D-glucose, D-mannose, D-mannitol, indole production, glucose fermentation, maltose nitrate reduction, protease, urease, and assimilation of adipic acid, capric acid, DL-malic acid, phenylacetic acid, potassium gluconate and trisodium citrate. In the API 50 CHB test system, acid is produced only from arbutin, D-cellobiose, D-galactose, D-glucose, D-maltose, D-mannitol, D-mannose, D-ribose, D-saccharose, D-trehalose, D-turanose, D-xylose, esculin ferric citrate, L-arabinose, L-fucose, L-rhamnose and salicin. Positive for acid phosphatase, esterase (C4), esterase Lipase (C8), leucine arylamidase, naphthol-AS-BI-phosphohydrolase, valine arylamidase, α-glucosidase, β-galactosidase, β-glucosidase and β-glucuronidase. Can utilize acetic acid, acetoacetic acid, D-cellobiose, D-fructose, D-galactose, D-gluconic acid, D-glucuronic acid, D-maltose, D-mannitol, D-mannose, D-salicin, D-trehalose, D-turanose, gentiobiose, glycerol, glycyl-L-proline, inosine, L-fucose, L-lactic acid, L-rhamnose, N-acetyl-D-galactosamine, pectin, p-hydroxy-phenylacetic acid, propionic acid, sucrose, tween 40, α-D-glucose, α-D-lactose, α-hydroxy-butyric acid and α-keto-butyric acid as the sole carbon source, but cannot utilize 3-methyl glucose, bromo-succinic acid, citric acid, D-arabitol, D-aspartic acid, D-fructose-6-PO4, D-fucose, D-galacturonic acid, D-glucose-6-PO4, D-lactic acid methyl ester, D-malic acid, D-melibiose, D-raffinose, D-saccharic acid, D-serine, D-sorbitol, formic acid, gelatin, glucuronamide, L-alanine, L-arginine, L-aspartic acid, L-galactonic acid, lactone, L-glutamic acid, L-histidine, L-malic acid, L-pyroglutamic acid, L-serine, methyl pyruvate, mucic acid, myo-inositol, N-acetyl neuraminic acid, N-acetyl-D-glucosamine, N-acetyl-β-D-mannosamine, quinic acid, α-Keto-glutaric acid, β-Hydroxy-D,L-butyric acid, β-Methyl-D-Glucoside or γ-amino-butryric acid. Diphosphatidylglycerol and phosphatidylethanolamine are detected in the polar lipids extraction. The predominant quinone is MK-11. The major cellular fatty acids are anteiso-C15:0, C17:1 ɷ9c, anteiso-C17:0 and summed feature 8 (C18:1 ɷ7c and/or C18:1 ɷ6c). Glucose as cell-wall sugars. The diagnostic diamino acids in the cell-wall peptidoglycan are alanine, glutamic acid and glycine. The type strain is CPCC 203386T (=I10A-01569T = DSM 24546T = KCTC 19839T), isolated from the stem of a medicinal plant Daphne aurantiaca collected from Yunnan Province, south-west China. The genomic G + C content of the type strain is 65.3%.
Description of Herbiconiux gentiana sp. nov.
Herbiconiux gentiana (gen.ti.a’nae. N.L. gen. n. gentianae, of Gentiana, a plant genus name, referring to the site related to a plant of Gentiana rigescens, from which the type strain was isolated).
Cells are aerobic, Gram-staining-positive, rod-shaped. Colonies are yellowish, smooth, convex and circular with diameter of 0.4–0.8 mm after 5 days on TSA medium at 28°C. Growth occurs at 4–42°C and pH 7.0–8.0, optimally at 28°C and pH 7.0. Cells are able to tolerate up to 3% NaCl (w/v) on TSA medium and grow optimally without additional NaCl. Positive for oxidase and catalase reaction, negative for gelatin hydrolyzation. In API 50CH test strips, acid is produced by D-arabitol, D-cellobiose, D-fructose, D-galactose, D-glucose, D-lyxose, D-maltose, D-mannitol, D-mannose, D-ribose, D-saccharose, D-trehalose, D-turanose, D-xylose, esculin ferric citrate, glycerol, L-arabinose and L-rhamnose, but not from amidon, arbutin, D-arabinose, D-ardonitol, D-lactose, D-melezitose, D-raffinose, D-ribose, D-sorbitol, dulcitol, erythritol, glycogen, inositol, inulin, L-arabinose, L-sorbose, mehyl-α-D-glucopyranoside, mehyl-α-D-mannopyranoside, mehyl-β-D-xylopyranoside, N-acetylglucosamine, potassium 2-ketogluconate, salicin, or xylitol. According to the results from the API ZYM strips, alkaline phosphatase, cystine arylamidase, esterase (C4), esterase lipase (C8), leucine arylamidase, valine arylamidase, α-glucosidase and β-glucosidase are positive; negative for lipase (C14), N-acetyl-β-glucosamimidase, trypsin, α-chymotrypsin, α-mannosidase and β-glucuronidase. In the Biolog Gen III MicroPlate system, the following carbon sources are oxidized: acetic acid, acetoacetic acid, bromo-succinic acid, citric acid, D-arabitol, D-cellobiose, dextrin, D-fructose, D-galactose, D-gluconic acid, D-malic acid, D-maltose, D-mannitol, D-mannose, D-trehalose, D-turanose, gelatin, gentiobiose, glycerol, glycyl-L-proline, inosine, L-alanine, L-aspartic acid, L-fucose, L-glutamic acid, L-lactic acid, L-malic acid, L-rhamnose, pectin, propionic acid, sucrose, tween 40, α-D-glucose, α-hydroxy-butyric acid and β-hydroxy-D,L-butyric acid. In the API 20NE test system, positive for aesculin hydrolysis, β-galactosidase, and assimilation of D-glucose, potassium gluconate and adipic acid, but negative for arginine dihydrolase, D-mannose, maltose, adipic acid, glucose fermentation, indole production, nitrate reduction, protease, urease, and assimilation of capric acid, DL-malic acid and phenylacetic acid. Diphosphatidylglycerol and phosphatidylethanolamine are detected in the polar lipids extraction. Cells contain anteiso-C15:0 and iso-C16:0 as the predominant cellular fatty acids and glucose as cell-wall sugar. The diamino acid in the cell-wall peptidoglycan are lanine, glutamic acid and glycine. The predominant quinone is MK-11. The type strain is CPCC 205716T (= I21A-01427T = CGMCC 1.60064T), isolated from a rhizosphere soil sample of the plant Gentiana rigescens collected from Guizhou Province, south-west China. The genomic G + C content of the type strain is 70.8%.
Description of Herbiconiux oxytropis sp. nov.
Herbiconiux oxytropis (o.xy.tro’pis. N.L. gen. n. oxytropis, of Oxytropis, a plant genus name, referring to the isolation of the type strain from a plant of Oxytropis falcata).
Cells are aerobic, Gram-staining-positive and rod-shaped, form yellow-coloured colonies about 0.9–1.2 mm in diameter after growing 24 h at 28°C on TSA medium. Growth occurs at 10–37°C [optimum, 28°C and at pH 7.0–8.0 (pH 7.0)]. The range of NaCl for growth is 0–3.0% (w/v); optimum growth occurs without NaCl. Positive for oxidase, catalase reaction and gelatin hydrolyzation. In API ZYM strip test, activities of acid phosphatase, cystine arylamidase, esterase (C4), esterase lipase (C8), leucine arylamidase, naphthol-AS-BI-phosphohydrolase, valine arylamidase, α-glucosidase, and β-glucosidase are positive, but N-acetyl-β-glucosamimidase, α-chymotrypsin, α-fucosidase, α-galactosidase, α-mannosidase, β-galactosidase and β-glucuronidase are negative. In carbon source oxidation tests, 3-methyl glucose, acetic acid, acetoacetic acid, bromo-succinic acid, citric acid, D-arabitol, D-cellobiose, dextrin, D-fructose, D-galactose, D-gluconic acid, D-malic acid, D-maltose, D-mannitol, D-mannose, D-salicin, D-trehalose, D-turanose, gentiobiose, glycerol, glycyl-L-proline, inosine, L-alanine, L-aspartic acid, L-lactic acid, L-malic acid, L-rhamnose, methyl pyruvate, pectin, propionic acid, sucrose, tween 40, α-D-glucose, α-D-lactose, α-hydroxy-butyric acid, α-keto-butyric acid and α-keto-glutaric acid are oxidized. In API 50CH tests, acid is produced from D-arabitol, D-cellobiose, D-fructose, D-galactose, D-glucose, D-lyxose, D-maltose, D-mannitol, D-mannose, D-ribose, D-saccharose, D-trehalose, D-turanose, D-xylose, esculin ferric citrate, glycerol, L-arabinose and L-rhamnose, but not from amidon, amygdalin, amygdalin, arbutin, D-arabinose, D-ardonitol, D-fucose, D-lactose, D-melezitose, D-melibiose, D-raffinose, D-sorbitol, D-sorbitol, D-tagatose, dulcitol, erythritol, gentiobiose, glycogen, inositol, inulin, L-arabitol, L-fucose, L-sorbose, mehyl-α-D-glucopyranoside, mehyl-α-D-glucopyranoside, mehyl-α-D-mannopyranoside, mehyl-α-D-mannopyranoside, mehyl-β-D-xylopyranoside, N-acetylglucosamine, N-acetylglucosamine, potassium 2-ketogluconate, potassium 5-ketogluconate, potassium gluconate, salicin, or xylitol. In the API 20NE test system, positive for aesculin hydrolysis, β-galactosidase, and assimilation of D-glucose, D-mannitol, maltose, potassium gluconate and trisodium citrate, but negative for arginine dihydrolase, glucose fermentation, indole production, nitrate reduction, protease, urease, and assimilation of adipic acid, capric acid, D-mannose, DL-malic acid, L-arabinose and phenylacetic acid. Diphosphatidylglycerol and phosphatidylglycerol are detected in the polar lipids extraction. The predominant quinone is MK-11. Predominant cellular fatty acids are anteiso-C15:0, anteiso-C17:0 and iso-C16:0. Glucose, rhamnose and ribose as cell-wall sugars. The diagnostic diamino acids in the cell-wall peptidoglycan are alanine, glutamic acid and glycine. The type strain is CPCC CPCC 203406T (=I10A-02268T = DSM 24549T = KCTC 19840T), isolated from the leaf of a medicinal plant Oxytropis falcata collected from Tibet, west China. The genomic G + C content of the type strain is 70.1%.
Author’s note
The 16S rRNA gene sequences and the whole genome shotgun projects of the strains have been deposited at DDBJ/ENA/GenBank under the accession numbers as follows: CPCC 205763T (OP279615; JANLCM000000000), CPCC 203386T (JX273670; JANLCJ000000000), CPCC 205716T (OP279534; JANTEZ000000000), CPCC 203406T (JX273672; JANLCL000000000), and CPCC 203407 (JX273671; JANLCK000000000).
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
YD, Z-MJ, X-FH, and JS carried out the experiments. YD, Z-MJ, X-FH, and Y-QZ conceived the research, analyzed the data, and prepared the manuscript. L-YY and W-HL collected the samples. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by CAMS Innovation Fund for Medical Sciences (CIFMS, 2021-I2M-1-055), National Natural Science Foundation of China (32170021 and 81960712), Beijing Natural Science Foundation (5212018), Key project at central government level-the ability establishment of sustainable use for valuable Chinese medicine resources (2060302), and the National Infrastructure of Microbial Resources (NIMR-2021-3).
Acknowledgments
We sincerely thank Institute of Microbiology, Chinese Academy of Sciences, for the kind assistance in the cells’ morphology observation using transmission electron microscopy. In this published article, any mistakes in nomenclature were not related to Professors Oren and Schink but rather were the result of the authors’ oversight during the paper’s revision process. The authors had indeed received invaluable assistance from Professors Oren and Schink concerning nomenclature. Consequently, the authors express their sincere apologies to both professors for neglecting to integrate their feedback on the nomenclature.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1119226/full#supplementary-material
Abbreviations
ANI, average nucleotide identity; dDDH, digital DNA–DNA hybridization; IAA, indole-3-acetic acid; DPPH, 2,2-diphenyl-1-picrylhydrazyl.
Footnotes
1. ^https://lpsn.dsmz.de/genus/herbiconiux
2. ^http://eztaxon-e.ezbiocloud.net
3. ^http://www.cbcb.umd.edu/software/glimmer/
References
Alshareef, S., Ling, Y., Butt, H., Mariappan, K. G., Benhamed, M., and Mahfouz, M. M. (2017). Herboxidiene triggers splicing repression and abiotic stress responses in plants. BMC Genomics 18:260. doi: 10.1186/s12864-017-3656-z
Auch, A. F., Von Jan, M., Klenk, H. P., and Göker, M. (2010). Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand. Genomic Sci. 2, 117–134. doi: 10.4056/sigs.531120
Bartley, G. E., and Scolnik, P. A. (1995). Plant carotenoids: pigments for photoprotection, visual attraction, and human health. Plant Cell 7, 1027–1038. doi: 10.1105/tpc.7.7.1027
Behrendt, U., Schumann, P., Hamada, M., Suzuki, K. I., Spröer, C., Ulrich, A., et al. (2011). Reclassification of Leifsonia ginsengi (Qiu et al. 2007) as Herbiconiux ginsengi gen. nov., comb. nov. and description of Herbiconiux solani sp. nov., an actinobacterium associated with the phyllosphere of Solanum tuberosum L. Int. J. Syst. Evol. Microbiol. 61, 1039–1047. doi: 10.1099/ijs.0.021352-0
Bernardet, J.-F., Nakagawa, Y., and Holmes, B., Subcommittee on the taxonomy of Flavobacterium and Cytophaga-like bacteria of the International Committee on Systematics of Prokaryotes (2002). Proposed minimal standards for describing new taxa of the family Flavobacteriaceae and emended description of the family. Int. J. Syst. Evol. Microbiol. 52, 1049–1070. doi: 10.1099/00207713-52-3-1049
Blin, K., Shaw, S., Kloosterman, A. M., Charlop-Powers, Z., Van Wezel, G. P., Medema, M. H., et al. (2021). antiSMASH 6.0: improving cluster detection and comparison capabilities. Nucleic Acids Res. 49, W29–W35. doi: 10.1093/nar/gkab335
Breakwell, D. P., Moyes, R. B., and Reynolds, J. (2009). Differential staining of bacteria: capsule stain. Curr. Protoc. Microbiol. 15:A.3I.1-A.3I.4. doi: 10.1002/9780471729259.mca03is15
Bric, J. M., Bostock, R. M., and Silverstone, S. E. (1991). Rapid in situ assay for indoleacetic acid production by bacteria immobilized on a nitrocellulose membrane. Appl. Environ. Microbiol. 57, 535–538. doi: 10.1128/aem.57.2.535-538.1991
Chang, M. X., and Xiong, F. (2020). Astaxanthin and its effects in inflammatory responses and inflammation-associated diseases: recent advances and future directions. Molecules 25:5342. doi: 10.3390/molecules25225342
Chaudhari, N. M., Gupta, V. K., and Dutta, C. (2016). BPGA-an ultra-fast pan-genome analysis pipeline. Sci. Rep. 6:24373. doi: 10.1038/srep24373
Chen, Z. P., Qu, M. M., Chen, H. X., Liu, D., Xiao, Y. Y., Chen, J., et al. (2011). The studies of anti-inflammatory and analgesic activities and pharmacokinetics of Oxytropis falcate Bunge extraction after transdermal administration in rats. Fitoterapia 82, 426–433. doi: 10.1016/j.fitote.2010.11.026
Collins, M. D., Pirouz, T., Goodfellow, M., and Minnikin, D. E. (1977). Distribution of menaquinones in actinomycetes and corynebacteria. J. Gen. Microbiol. 100, 221–230. doi: 10.1099/00221287-100-2-221
Deng, Y., Han, X. F., Jiang, Z. M., Yu, L. Y., Li, Y., and Zhang, Y. Q. (2022). Characterization of three Stenotrophomonas strains and proposal of Stenotrophomonas mori sp. nov., and Stenotrophomonas lacuserhaii sp. nov. Front. Microbiol. 13:1056762. doi: 10.3389/fmicb.2022.1056762
Felsenstein, J. (1981). Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17, 368–376. doi: 10.1007/bf01734359
Felsenstein, J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791. doi: 10.2307/2408678
Firdous, A. P., Kuttan, G., and Kuttan, R. (2015). Anti-inflammatory potential of carotenoid meso-zeaxanthin and its mode of action. Pharm. Biol. 53, 961–967. doi: 10.3109/13880209.2014.950673
Gliwa, J., Gunenc, A., Ames, N., Willmore, W. G., and Hosseinian, F. S. (2011). Antioxidant activity of alkylresorcinols from rye bran and their protective effects on cell viability of PC-12 AC cells. J. Agric. Food Chem. 59, 11473–11482. doi: 10.1021/jf2023353
Groth, I., Schumann, P., Rainey, F. A., Martin, K., Schuetze, B., and Augsten, K. (1997). Demetria terragena gen. nov., sp. nov., a new genus of actinomycetes isolated from compost soil. Int. J. Syst. Bacteriol. 47, 1129–1133. doi: 10.1099/00207713-47-4-1129
Hamada, M., Komukai, C., Tamura, T., Evtushenko, L. I., Vinokurova, N. G., and Suzuki, K. I. (2012). Description of Herbiconiux flava sp. nov. and emended description of the genus Herbiconiux. Int. J. Syst. Evol. Microbiol. 62, 795–799. doi: 10.1099/ijs.0.031260-0
Jiang, Z. M., Deng, Y., Han, X. F., Su, J., Wang, H., Yu, L. Y., et al. (2022). Geminicoccus flavidas sp. nov. and Geminicoccus harenae sp. nov., two IAA-producing novel rare bacterial species inhabiting desert biological soil crusts. Front. Microbiol. 13:1034816. doi: 10.3389/fmicb.2022.1034816
Jiang, Z. M., Zhang, B. H., Sun, H. M., Zhang, T., Yu, L. Y., and Zhang, Y. Q. (2021). Properties of Modestobacter deserti sp. nov., a kind of novel phosphate-solubilizing actinobacteria inhabited in the desert biological soil crusts. Front. Microbiol. 12:742798. doi: 10.3389/fmicb.2021.742798
Kim, B. C., Park, D. S., Kim, H., Oh, H. W., Lee, K. H., Shin, K. S., et al. (2012). Herbiconiux moechotypicola sp. nov., a xylanolytic bacterium isolated from the gut of hairy long-horned toad beetles, Moechotypa diphysis (Pascoe). Int. J. Syst. Evol. Microbiol. 62, 90–95. doi: 10.1099/ijs.0.028357-0
Kimura, M. (1979). The neutral theory of molecular evolution. Sci. Am. 241, 98–126. doi: 10.1038/scientificamerican1179-98
Kluge, A. G., and Farris, J. S. (1969). Quantitative phyletics and the evolution of anurans. Syst. Biol. 18, 1–32. doi: 10.1093/sysbio/18.1.1
Kroppenstedt, R. M. (1985). Fatty acid and menaquinone analysis of actinomycetes and related organisms. Soc. Appl. Bacteriol. Tech. Ser. 20, 173–199.
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Li, W. J., Xu, P., Schumann, P., Zhang, Y. Q., Pukall, R., Xu, L. H., et al. (2007). Georgenia ruanii sp. nov., a novel actinobacterium isolated from forest soil in Yunnan (China), and emended description of the genus Georgenia. Int. J. Syst. Evol. Microbiol. 57, 1424–1428. doi: 10.1099/ijs.0.64749-0
Lowe, T. M., and Eddy, S. R. (1997). tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25, 955–964. doi: 10.1093/nar/25.5.955
Magee, C. M., Rodeheaver, G., Edgerton, M. T., and Edlich, R. F. (1975). A more reliable gram staining technic for diagnosis of surgical infections. Am. J. Surg. 130, 341–346. doi: 10.1016/0002-9610(75)90398-0
Meier, A., Kirschner, P., Schröder, K. H., Wolters, J., Kroppenstedt, R. M., and Böttger, E. C. (1993). Mycobacterium intermedium sp. nov. Int. J. Syst. Bacteriol. 43, 204–209. doi: 10.1099/00207713-43-2-204
Minnikin, D. E., O'Donnell, A. G., Goodfellow, M., Alderson, G., Athalye, M., Schaal, A., et al. (1984). An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J. Microbiol. Methods 2, 233–241. doi: 10.1016/0167-7012(84)90018-6
Nie, Y. W., Li, Y., Luo, L., Zhang, C. Y., Fan, W., Gu, W. Y., et al. (2021). Phytochemistry and pharmacological activities of the diterpenoids from the genus Daphne. Molecules 26:6598. doi: 10.3390/molecules26216598
Park, Y. H., Suzuki, K., Yim, D. G., Lee, K. C., Kim, E., Yoon, J., et al. (1994). Suprageneric classification of peptidoglycan group B actinomycetes by nucleotide sequencing of 5S ribosomal RNA. Antonie Van Leeuwenhoek 64, 307–313. doi: 10.1007/bf00873089
Patzke, H., and Schieber, A. (2018). Growth-inhibitory activity of phenolic compounds applied in an emulsifiable concentrate-ferulic acid as a natural pesticide against Botrytis cinerea. Food Res. Int. 113, 18–23. doi: 10.1016/j.foodres.2018.06.062
Reinhold-Hurek, B., Hurek, T., Claeyssens, M., and Van Montagu, M. (1993). Cloning, expression in Escherichia coli, and characterization of cellulolytic enzymes of Azoarcus sp., a root-invading diazotroph. J. Bacteriol. 175, 7056–7065. doi: 10.1128/jb.175.21.7056-7065.1993
Schleifer, K. H., and Kandler, O. (1972). Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol. Rev. 36, 407–477. doi: 10.1128/br.36.4.407-477.1972
Shirling, E. T., and Gottlieb, D. (1966). Methods for characterization of Streptomyces species. Int. J. Syst. Bacteriol. 16, 313–340. doi: 10.1099/00207713-16-3-313
Singh, N., and Rajini, P. (2004). Free radical scavenging activity of an aqueous extract of potato peel. Food Chem. 85, 611–616. doi: 10.1016/j.foodchem.2003.07.003
Staneck, J. L., and Roberts, G. D. (1974). Simplified approach to identification of aerobic actinomycetes by thin-layer chromatography. Appl. Microbiol. 28, 226–231. doi: 10.1128/am.28.2.226-231.1974
Unwin, J., Standage, S., Alexander, D., Hosted, T., Horan, A. C., and Wellington, E. M. (2004). Gene cluster in Micromonospora echinospora ATCC15835 for the biosynthesis of the gentamicin C complex. J. Antibiot. 57, 436–445. doi: 10.7164/antibiotics.57.436
Wei, Y., Yu, N., Wang, Z., Hao, Y., Wang, Z., Yang, Z., et al. (2022). Analysis of the multi-physiological and functional mechanism of wheat alkylresorcinols based on reverse molecular docking and network pharmacology. Food Funct. 13, 9091–9107. doi: 10.1039/d2fo01438f
Xu, P., Li, W. J., Tang, S. K., Zhang, Y. Q., Chen, G. Z., Chen, H. H., et al. (2005). Naxibacter alkalitolerans gen. nov., sp. nov., a novel member of the family 'Oxalobacteraceae' isolated from China. Int. J. Syst. Evol. Microbiol. 55, 1149–1153. doi: 10.1099/ijs.0.63407-0
Yoda, H., Yamaguchi, Y., and Sano, H. (2003). Induction of hypersensitive cell death by hydrogen peroxide produced through polyamine degradation in tobacco plants. Plant Physiol. 132, 1973–1981. doi: 10.1104/pp.103.024737
Yoon, S. H., Ha, S. M., Kwon, S., Lim, J., Kim, Y., Seo, H., et al. (2017a). Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 67, 1613–1617. doi: 10.1099/ijsem.0.001755
Yoon, S. H., Ha, S. M., Lim, J., Kwon, S., and Chun, J. (2017b). A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 110, 1281–1286. doi: 10.1007/s10482-017-0844-4
Yuan, L. J., Zhang, Y. Q., Guan, Y., Wei, Y. Z., Li, Q. P., Yu, L. Y., et al. (2008). Saccharopolyspora antimicrobica sp. nov., an actinomycete from soil. Int. J. Syst. Evol. Microbiol. 58, 1180–1185. doi: 10.1099/ijs.0.65532-0
Zhao, D., Shi, Y., Zhu, X., Liu, L., Ji, P., Long, C., et al. (2018). Identification of potential biomarkers from Aconitum carmichaelii, a traditional chinese medicine, using a Metabolomic approach. Planta Med. 84, 434–441. doi: 10.1055/s-0043-121708
Zhao, Y., Zhang, J., Jin, H., Zhang, J., Shen, T., and Wang, Y. (2015). Discrimination of Gentiana rigescens from different origins by Fourier transform infrared spectroscopy combined with chemometric methods. J. AOAC Int. 98, 22–26. doi: 10.5740/jaoacint.13-395
Keywords: Herbiconiux, pangenome, polyphasic taxonomy, medicinal plants, plant growth-promotion
Citation: Deng Y, Jiang Z-M, Han X-F, Su J, Yu L-Y, Liu W-H and Zhang Y-Q (2023) Pangenome analysis of the genus Herbiconiux and proposal of four new species associated with Chinese medicinal plants. Front. Microbiol. 14:1119226. doi: 10.3389/fmicb.2023.1119226
Edited by:
Pratiksha Singh, Guangxi University for Nationalities, ChinaReviewed by:
Hilal Ay, Ondokuz Mayıs University, TürkiyeMohini Prabha Singh, Punjab Agricultural University, India
Copyright © 2023 Deng, Jiang, Han, Su, Yu, Liu and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yu-Qin Zhang, ✉ eXpoYW5nQGltYi5wdW1jLmVkdS5jbg==
†These authors share first authorship
 Yang Deng1,2†
Yang Deng1,2† Yu-Qin Zhang
Yu-Qin Zhang