- 1Foodborne and Waterborne Diseases Research Center, Research Institute for Gastroenterology and Liver Diseases, Shahid Beheshti University of Medical Sciences, Tehran, Iran
- 2Department of Life Science Engineering, Faculty of New Sciences and Technologies, University of Tehran, Tehran, Iran
- 3Biosensor Research Center, Endocrinology and Metabolism Molecular–Cellular Sciences Institute, Tehran University of Medical Sciences, Tehran, Iran
- 4Endocrinology and Metabolism Research Center, Endocrinology and Metabolism Research Institute, Tehran University of Medical Sciences, Tehran, Iran
Drought and limited sufficient water resources will be the main challenges for humankind during the coming years. The lack of water resources for washing, bathing, and drinking increases the use of contaminated water and the risk of waterborne diseases. A considerable number of waterborne outbreaks are due to protozoan parasites that may remain active/alive in harsh environmental conditions. Therefore, a regular monitoring program of water resources using sensitive techniques is needed to decrease the risk of waterborne outbreaks. Wellorganized point-of-care (POC) systems with enough sensitivity and specificity is the holy grail of research for monitoring platforms. In this review, we comprehensively gathered and discussed rapid, selective, and easy-to-use biosensor and nanobiosensor technologies, developed for the early detection of common waterborne protozoa.
1. Introduction
Healthy water resources are still one of the main challenges in most countries, particularly less developed regions in dry parts of the world (WHO/UNICEF, 2014; Andrade et al., 2018). The lack of sufficient and appropriate water resources increases the use of contaminated water sources and wastewater for drinking, washing, and irrigation. Accordingly, employing contaminated water not only directly increases the risk of contamination by waterborne pathogens but also enhances the risk of contamination of vegetables with waterborne pathogens, particularly in farmlands irrigated using wastewater (Karanis et al., 2006; Javanmard et al., 2019, 2020).
Waterborne diseases include a large group of illnesses with mild to severe symptoms, which are caused by a broad spectrum of pathogens including bacteria, fungi, parasites, and viruses (Kalyoussef and Feja, 2014). Parasites, both protozoa and helminths, are important group of foodborne and waterborne pathogens, which can be transmitted from wild and domestic animals and contaminated water resources to humans (Sharma and Mutharasan, 2013; Vasilescu and Marty, 2016; Pazoki et al., 2020). The transmission route of foodborne and waterborne protozoa (FWP) is mostly oral-fecal and infection occurs when cysts or oocysts of a parasite are unintentionally ingested by a host (Kalyoussef and Feja, 2014).
Waterborne protozoa are responsible for a considerable number of outbreaks in the world. Regarding a recently published systematic review, 86.7% of eastern African countries reported at least one waterborne parasite from 1953 to 2019 (Ngowi, 2020). However, there is no periodical monitoring strategy for the detection of waterborne parasites (particularly protozoa) in less-developed regions like African countries. In contrast, outbreak surveillance in Europe between 2000 and 2007 indicated the presence of 354 and 70 outbreaks, related to drinking water and bathing water, respectively, in which protozoa were responsible for 17 (4.7%) drinking water and 38 (54.3%) bathing water outbreaks (ENHIS, 2009). Moreover, it was estimated that ~7,150,000 (95% CrI 3,880,000–12,000,000; 21.3%) illnesses in the United States in 2014 were associated with waterborne agents, of which Giardia lamblia and Cryptosporidium spp., were responsible for 415,000 (95% CrI 140,000–816,000) and 322,000 (95% CrI 61,700–993,000) illnesses, respectively (Collier et al., 2021). In addition, more than 10% of the total waterborne outbreaks from 1991 to 2008 were attributed to parasitic agents, particularly protozoa (Karanis et al., 2006; Efstratiou et al., 2017). More recently, Cryptosporidium spp., G. lamblia, Cyclospora cayetanensis, Toxoplasma gondii, Blastocystis sp., Entamoeba histolytica, microsporidia, and Naegleria fowleri were reported as the main causative agents detected in 251 waterborne outbreaks from 2017 to 2020 (Ma et al., 2022). Interestingly, most waterborne outbreaks due to protozoa have been reported in developed countries, indicating the importance of periodical monitoring of water resources, as well as diagnostic capabilities (Ma et al., 2022).
Significant progress has been made in recent decades in developing portable, reusable, and effective miniaturized systems or point-of-care (POC) platforms. POC tests can be performed outside a clinical laboratory setting, at or near the site of patient care. In addition, along with climate change, particularly in recent years, the risk of transmission of infectious diseases and the number of areas, which were previously unaffected by specific infectious diseases, have increased (Mora et al., 2022). In contrast, global warming, as a coming challenge in the world, decreases access to healthy water resources and increases emerging water and food safety concerns (Duchenne-Moutien and Neetoo, 2021). In fact, drought due to climate changes increases seasonal water resources, which aggregates animals and human communities in a region, recycled water resources, groundwater, and even lagoon water, and therefore, the risk of transmission of potentially pathogenic microorganisms, particularly parasites, from animals to humans (Titcomb et al., 2021). Moreover, because of the presence of a resistant stage, cyst/oocyst/egg, in the life cycle of parasites, particularly protozoa, these microorganisms endure harsh conditions such as drought much more than other microorganisms. All of these reasons highlight the importance of protozoa infections and the development of POC techniques for the detection of these parasites in the future. Guidelines, commonly needed for establishing well-organized POC systems, are presented by the World Health Organization (WHO). These guidelines are identified as ASSURED, in which the abbreviation ASSURED stands for affordable, sensitive, specific, user-friendly, rapid, equipment-free, or minimal, and delivered to those who require them (Syedmoradi et al., 2017, 2021; Omidfar et al., 2020).
This study highlights the need for rapid, selective, and easy-to-use technology for the early detection of common waterborne parasitic pathogens. The purpose of the current review is to provide a comprehensive overview of conventional methods and emerging biosensors and nanobiosensors, with a focus on recent advances in smart-based devices.
2. A brief look at the significant waterborne parasites
Although a broad spectrum of parasites is reported from waterborne outbreaks, Cryptosporidium spp., G. lamblia, T. gondii, and E. histolytica are among the most frequently detected waterborne parasites in the world (Al-Shamiri et al., 2010; Robert-Gangneux and Dardé, 2012; Plutzer and Karanis, 2016; Sarkari et al., 2016; Ma et al., 2022). Nevertheless, microsporidia, Blastocystis sp., Dientamoeba fragilis, Balantidium coli, C. cayetanensis, and Isospora belli are the neglected waterborne parasites (Karanis et al., 2006; Plutzer and Karanis, 2016). Regarding the worldwide waterborne outbreaks reported by Baldursson and Karanis (2011), from 2004 to 2010, Cryptosporidium spp., and G. lamblia were the major causative agents in 60.3% (120) and 35.2% (70) of 199 outbreaks, respectively, while T. gondii, C. cayetanensis, Acanthamoeba spp., E. histolytica, and Blastocystis sp., were the other reported agents. Recently, an update on waterborne outbreaks due to parasites from 2017 to 2020 signified the high prevalence of Cryptosporidium spp., and G. lamblia as the major reported agents, followed by D. fragilis, T. gondii, C. cayetanensis, Blastocystis sp., E. histolytica, N. fowleri, and microsporidia (Ma et al., 2022).
2.1. Cryptosporidium spp.
Cryptosporidium spp. are apicomplexan protozoa, with several known species that infect humans and many other vertebrates, and are transmitted via the fecal-oral route through ingesting oocytes in contaminated food or water (Leitch and He, 2012; Gerace et al., 2019; Zahedi and Ryan, 2020). Cryptosporidium spp. can infect both immunocompetent and immunocompromised individuals, of which two species, Cryptosporidium parvum and Cryptosporidium hominis, are the most prevalent species in humans (Mmbaga and Houpt, 2017).
Cryptosporidium spp. are known as the main protozoa parasite reported from waterborne outbreaks (Zahedi and Ryan, 2020; Gururajan et al., 2021; Zahedi et al., 2021). A most recent study reported that, from 251 waterborne outbreaks with parasitic agents, Cryptosporidium was identified among 198 outbreaks (Ma et al., 2022). C. parvum is a zoonotic species, which is isolated from humans and a broad spectrum of animals; therefore, there is an increased risk of contamination of water resources not only by human feces but also by excreted oocysts from free-range animals (Fernández et al., 2021; Mohammad Rahimi et al., 2022). In addition, it was documented that routine wastewater treatment processes including sedimentation, activated sludge, chlorination, and filtrations are not enough to eliminate Cryptosporidium oocysts from water samples (Sroka et al., 2013; Ramo et al., 2017), which increases the concern for the contamination of downstream farmlands irrigated with treated wastewater (Javanmard et al., 2020).
2.2. Giardia lamblia
G. lamblia (also known as Giardia intestinalis and Giardia duodenalis) is an anaerobic-flagellated non-invasive protozoan, which can infect the small intestine and, in a few cases, the distal small bowel, cecum, stomach, and pancreas of humans and many other vertebrates (Einarsson et al., 2016; Bahramdoost et al., 2021). G. lamblia is more prevalent in children living in developing countries and is classified as FWP. This protozoan is transmitted via the fecal-oral route through ingestion of the cystic form in contaminated food or water (Bello et al., 2011).
G. lamblia is the second-most reported protozoa from waterborne outbreaks worldwide (Karanis et al., 2006; Efstratiou et al., 2017; Ma et al., 2022). Similar to Cryptosporidium spp., the main reason for the high prevalence of G. lamblia in waterborne outbreaks is the capability of this protozoan to remain viable during water treatment processes (Sroka et al., 2013; Ramo et al., 2017). Large waterborne outbreaks due to G. lamblia have been reported all over the world, particularly in developed countries. Nygård et al. (2006) documented a large outbreak of giardiasis among at least 1,300 cases in Norway that was linked to leakage of wastewater pipes and insufficient wastewater treatment. Recently, a large giardiasis outbreak related to tap water occurred in Bologna Province, Italy, and the presence of G. lamblia was documented in 228 stool samples (Resi et al., 2021). However, the number of reported outbreaks due to G. lamblia and Cryptosporidium spp., in developed countries is significantly higher than in less-developed regions, which could be due to the use of more sensitive detection technologies in developed countries (Ma et al., 2022).
2.3. Toxoplasma gondii
T. gondii is an obligate intracellular parasite, which infects most warm-blooded animals, and Felidae family members are its definitive hosts (Tenter et al., 2000; Mendez and Koshy, 2017). T. gondii is transmitted via several routes including vertical transmission (Kanková and Flegr, 2007; Robbins et al., 2012; Chaudhry et al., 2014), transfusion and needle stick (Foroutan-Rad et al., 2016), and fecal-oral route via eating or drinking oocyst-contaminated food and water (Hill and Dubey, 2002). However, T. gondii is considered a neglected waterborne protozoan (Baldursson and Karanis, 2011; Karanis et al., 2013; Plutzer and Karanis, 2016). Nevertheless, in an outbreak related to drinking water in the Champagne-Ardenne region, France, Villena et al. (2004) detected T. gondii DNA in 10/125 analyzed samples. Then, Aubert and Villena (2009) analyzed water samples, which were collected in 2001 in Champagne-Ardenne, France, and characterized T. gondii DNA in 37/482 environmental samples. These two studies suggested the high contamination of water samples with T. gondii in the studied region in France. The presence of T. gondii DNA in wastewater samples in Germany was as high as that reported in France. Accordingly, Gallas-Lindemann et al. (2013) scrutinized influent and effluent samples of wastewater in Germany and reported the presence of T. gondii DNA in 8/83 (9.6%) samples using loop-mediated isothermal amplification (LAMP). Microscopically, oocyst-like positive samples for T. gondii were also detected in environmental water samples collected in the Galápagos Islands, Ecuador, although molecular identification of the samples was not successful (Verant et al., 2014). T. gondii DNA was also identified using real-time PCR in 2/8 wastewater samples, together with G. lamblia, E. coli, Entamoeba dispar, Entamoeba hartmanni, Blastocystis sp., and Acanthamoeba spp. in Spain (Moreno-Mesonero et al., 2022). These studies, as well as a large outbreak due to the consumption of drinking water contaminated with T. gondii, which was reported in 2018 from Santa Maria, Brazil (Minuzzi et al., 2021), highlight the importance of waterborne toxoplasmosis and the neglected role of T. gondii in waterborne outbreaks due to insufficient detection techniques.
3. Available diagnostic techniques for the detection of waterborne parasites
Diagnosis plays a critical role in the discovery of new pathogens, monitoring and surveillance, the prediction of epidemics and pandemics due to emerging and re-emerging pathogens, and antibiotic resistance (Mohammad Rahimi et al., 2019). In recent decades, various methods have been developed for the diagnosis of intestinal parasites. However, conventional diagnostic methods are still employed for the detection of intestinal parasites, particularly in less-developed regions (Mohammad Rahimi et al., 2019). Accordingly, the detection of G. lamblia, Cryptosporidium spp. and E. histolytica is mainly based on the optical detection of cysts/oocysts and trophozoites of parasites using microscopy (Destura et al., 2015; Ricciardi and Ndao, 2015; Hooshyar et al., 2019), while the most common diagnostic method for T. gondii is immunoassay techniques (Elgun and Koltas, 2011; Rostami et al., 2018).
Despite the advantages of microscopic methods, there are some limitations such as the technique being time-consuming and optical skills of laboratory technicians (Laude et al., 2016; Sakamoto et al., 2018). In addition, due to the low number of FWP in a large volume of environmental samples like water, employing microscopic techniques is an important challenge. For example, the concentration of G. lamblia in water samples has been reported to be 0.01 to 100 cysts/L (WHO, 2014); thus, developing methods for the detection of 1 cyst/oocyst of G. lamblia and Cryptosporidium spp., in 10–100 L of water samples is desirable, particularly when we consider that Cryptosporidium spp., and G. lamblia are among the waterborne pathogens with a high priority and with infectivity dosage less than 10 oocysts/cysts (WHO, 2014). In contrast, the development of molecular techniques in recent decades has overcome the limitations of conventional methods and has provided more sensitivity and specificity for the detection of pathogens (Tavares et al., 2011).
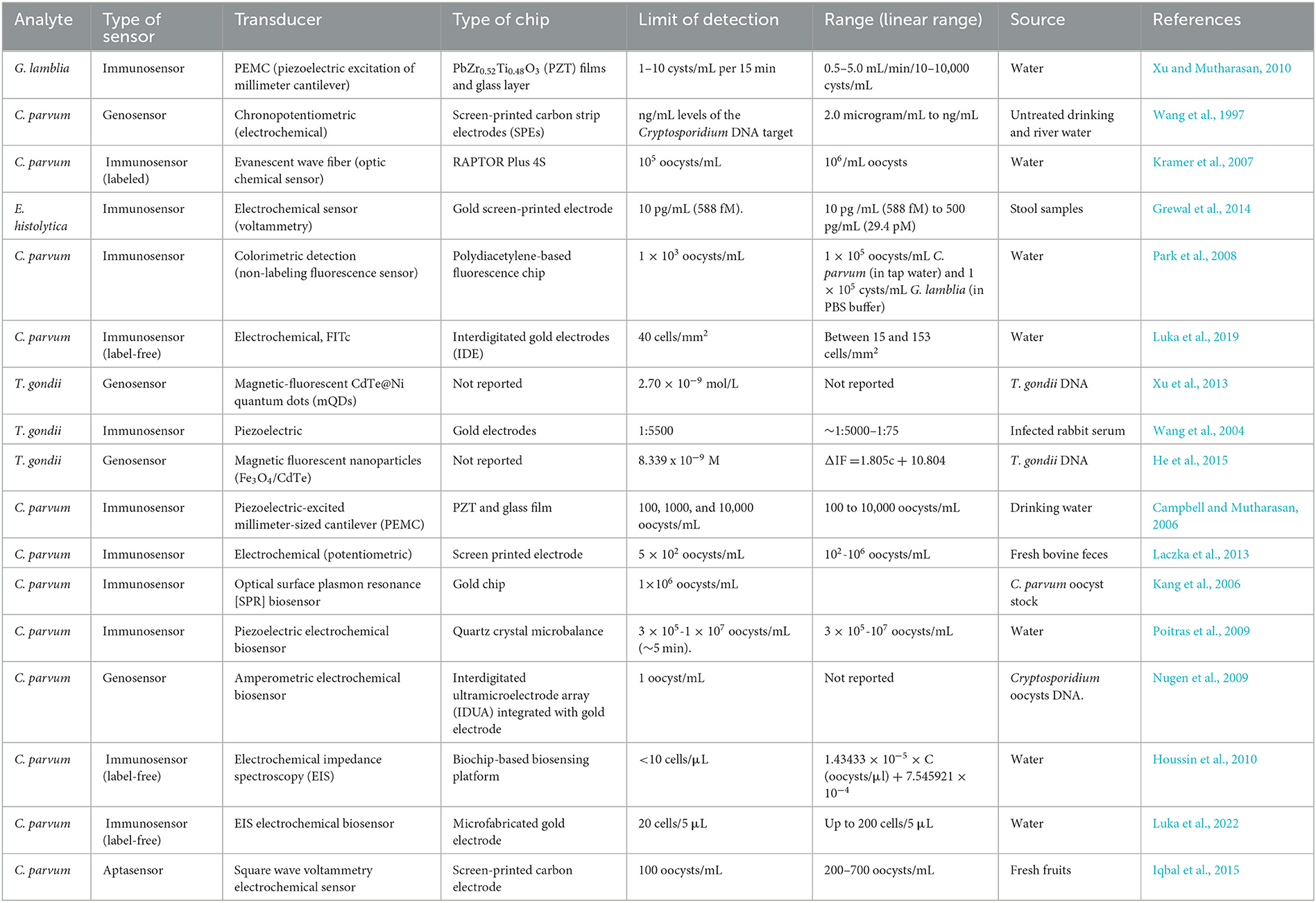
Although progress has been made in molecular techniques, there are still some disadvantages and limitations that restrict the application of molecular techniques. Multiple steps including DNA or RNA extraction, primer design challenges, false results due to undesirable primer interactions, and expensive equipment are the common challenges facing molecular methods (Garibyan and Avashia, 2013; Khurana and Chaudhary, 2018). Regarding the abovementioned limitations of conventional and advanced molecular methods, designing high-efficiency and field-adopted diagnostic devices with a simple user interface and a rapid protocol is the main priority (Luka et al., 2019) (Table 1).
In recent years, advanced devices have been developed to overcome the limitations of available techniques. For example, the sensitivity of microscopic techniques is not high enough, and a well-trained technician is needed to reduce the possibility of a false report. In addition, the staining procedure may take much time; these reasons are challenges at the time of outbreaks. In contrast to microscopic methods, serological and molecular techniques are not labor intensive, provide high sensitivity and specificity, and do not need a microscopist. Nevertheless, serological methods may provide cross reaction, and are not suitable for screening a population at the time of outbreaks or most intestinal FWP. In addition, due to expensive equipment and instruments for molecular methods, and the need for a well-equipped laboratory, most molecular-based approaches are not suitable for investigation of an outbreak (Mohammad Rahimi et al., 2019; Mahdavi Abhari et al., 2023). Therefore, focus has been dramatically increased on biosensors, such as POC tests, which can provide enough sensitivity and specificity, without the need for a well-trained technician or a specific facility.
4. Biosensors: Development and types
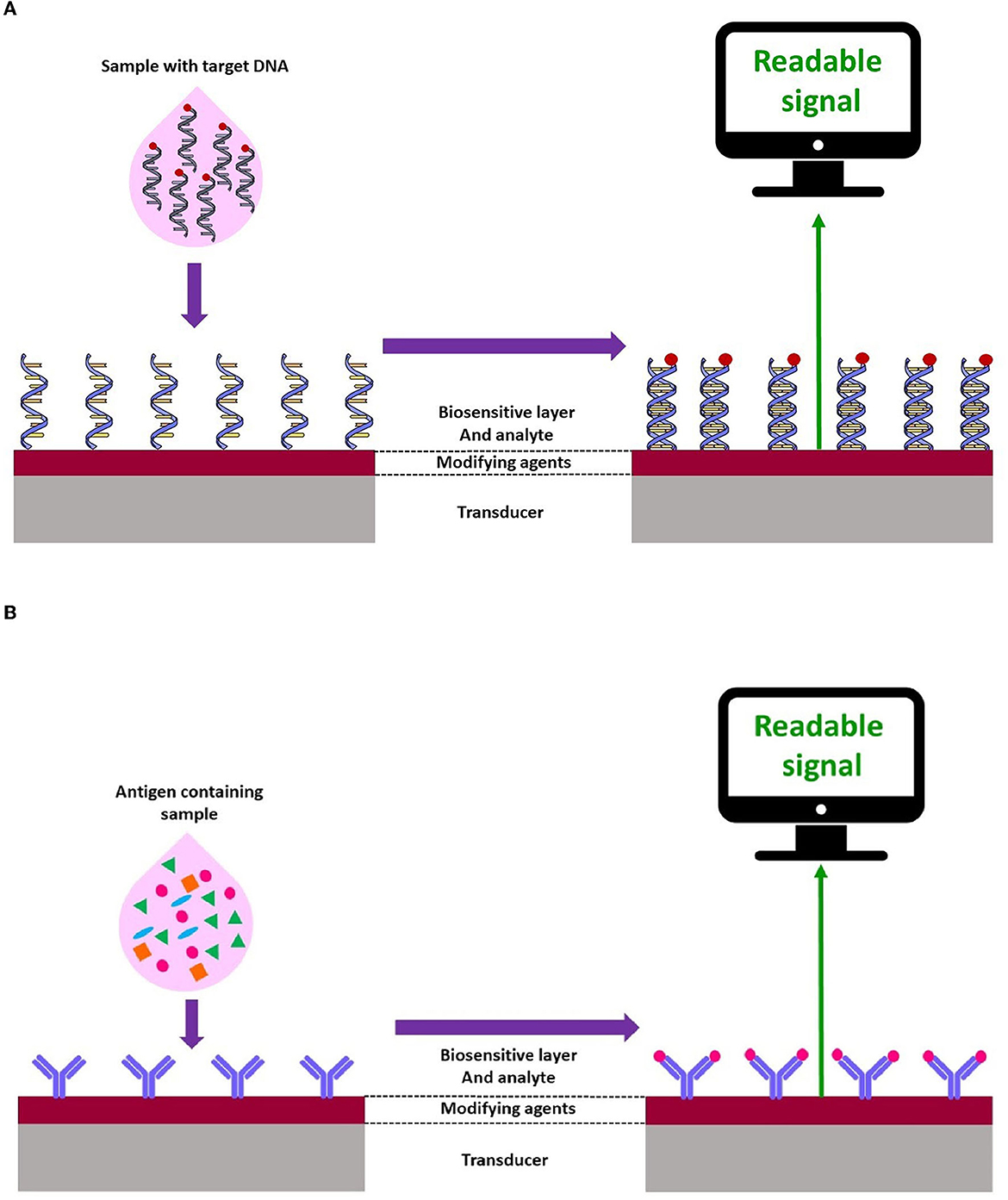
The importance of food and water safety in various industries has led to the mining and improvement of nanoscale analytical devices known as nanobiosensors. Due to the numerous advantages of these devices such as portability, low cost, rapid assay time, ease of use, and high selectivity and sensitivity (Terry et al., 2005; Ahmadi et al., 2022; Khoshfetrat et al., 2022; Saeidi et al., 2022), particularly for the detection of infectious agents and pollutants in the environment (Pejcic et al., 2006; Sin et al., 2014; Ahmadi et al., 2021c), focus on the fabrication of biosensors as a diagnostic technology for the detection of different analytes in food, water, and environmental samples has intensely increased (Terry et al., 2005; Pejcic et al., 2006; Metkar and Girigoswami, 2018; Salouti and Khadivi derakhshan, 2020) (Figure 1).
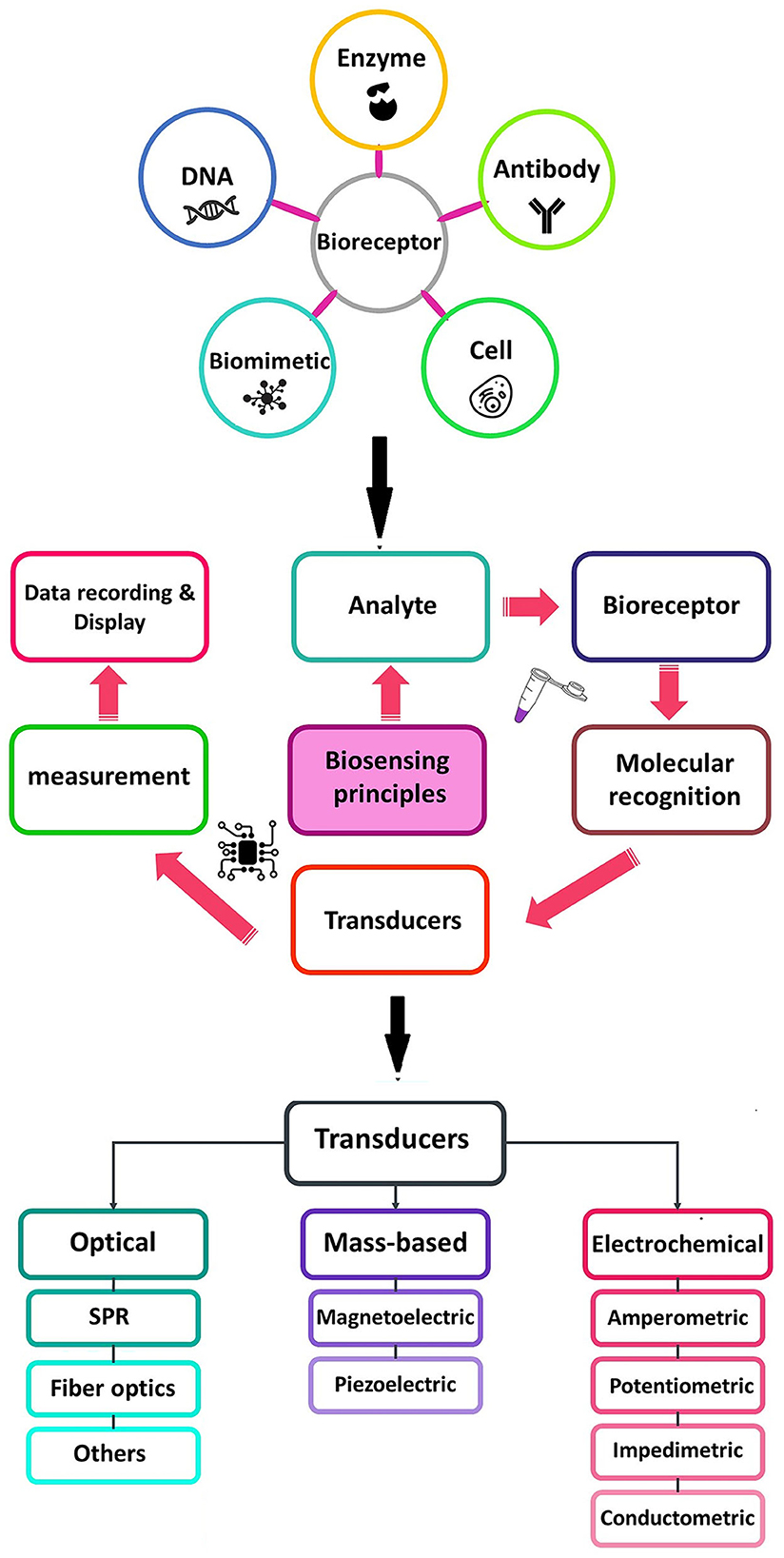
Figure 1. Schematic diagram of a typical biosensor consisting of various types of bioreceptors and transducers used in the biosensors.
A biosensor consists of at least two functional elements, namely, a molecular recognition element (receptor), which selectively interacts with its target analyte, and a physicochemical transducer. Biological elements are generally classified into enzymes, antibodies, and nucleic acids. A transducer is a component of biosensors which plays an important role in the signal detection process, and converts biological responses into a measurable signal with high quality (Khodaei et al., 2019; Ahmadi et al., 2021a,b).
Biosensors are generally classified into label-free and labeled. The latter biosensor employs labeled molecules for the detection of a target (Proll et al., 2007; Rhouati et al., 2016). Common labeling platforms are fluorescence or luminescence labeling, radiolabeling, isotope labeling, and enzymes (Deline and Nason, 2019; Ranjbar Bahadori et al., 2021). In these procedures, the final sensor signal represents the number of labels bound to target molecules. As a drawback, label-based technologies are labor- and cost-intensive and time-consuming (Cunningham and Laing, 2008). In addition, labeling of biomolecules can block active binding sites and alter the binding properties (Schöning and Poghossian, 2018).
In contrast, label-free biosensing technologies do not employ labels to facilitate measurements and instead incorporate the intrinsic physical properties of an analyte, such as the molecular weight, size, charge, electrical impedance, dielectric permittivity, and refractive index of a sample. In recent years, label-free biosensors have been developed due to their ability for rapid and inexpensive bio-detection in small reaction volumes (Schöning and Poghossian, 2018). Moreover, they can be integrated into lab-on-a-chip platforms, allowing monitoring of target analytes in real time. Label-free biosensors are usually designed based on optical, electrical or electrochemical, and acoustic parameters (Citartan et al., 2013).
Based on the biological elements, biosensors are categorized into genosensors, immunosensors, and aptasensors (Low et al., 2012; Kokkinos et al., 2016; Campuzano et al., 2017; Mohammed et al., 2017; Felix and Angnes, 2018).
In genosensors, oligonucleotide sequences (DNA or RNA) are usually employed as bio-receptors, which are immobilized onto the transducer surface and hybridized with the single-stranded target DNA. In fact, the oligonucleotide sequences, such as a probe, recognize the analyte (sample DNA or RNA) by matching with the complementarity sequences. Genosensor-based devices are widely used for the detection of a broad spectrum of pathogens (Drummond et al., 2003; Babkina and Budnikov, 2006; Gao et al., 2010; Mohammed et al., 2017) (Figure 2A).
Immunosensors play an important role in the evaluation of specific elements in biological fluids. Currently, such assays have been extensively utilized in food safety and environmental analysis (Felix and Angnes, 2018; Hosu et al., 2018) (Figure 2B).
A single-stranded functional nucleic acid or peptide with a strong receptor property is recognized as an aptamer. Aptamers are usually constructed from combinatorial single-stranded libraries by the systematic evolution of ligands using the exponential enrichment (SELEX) method, and are applied to detect multiple target analytes (Shamah et al., 2008; Liu et al., 2020). Aptamer-based techniques have been applied for the detection of numerous pathogens such as human immunodeficiency virus (HIV), hepatitis B virus (HBV), hepatitis C virus (HCV), Mycobacterium, Salmonella, Listeria, Staphylococcus, Clostridium, Bacillus, Escherichia, Aspergillus, Penicillium, SARS-CoV, influenza virus, respiratory syncytial virus (RSV), Trypanosome, Plasmodium, Cryptosporidium, and Leishmania (Cho et al., 2011; Nagarkatti et al., 2012; Martín et al., 2013; Iqbal et al., 2015; Babamiri et al., 2018; Lavania et al., 2018; Li et al., 2018, 2020; Suh et al., 2018; Wei et al., 2018; Xi et al., 2018; Cai et al., 2019; Singh et al., 2019; Zou et al., 2019). The number of aptasensor-based studies for the detection of FWP is low. In this regard, Iqbal et al. (2015) developed an electrochemical nanomaterial-based aptasensor using a gold nanoparticle (NP)-modified screen-printed carbon electrode (SPCE) to detect C. parvum oocysts in spiked fresh fruits. In this system, 14 aptamer colons were discovered and anti-aptamer and thiolated DNA primers were mixed to produce a hybrid compound that was assembled onto the SPCE. The fabricated aptasensor recognized C. parvum with a wide dynamic range from 150 to 800 oocysts and a detection limit of ~100 oocysts. This study suggested promising findings for the detection of C. parvum in food products (Iqbal et al., 2015) (Table 2; Figure 3).
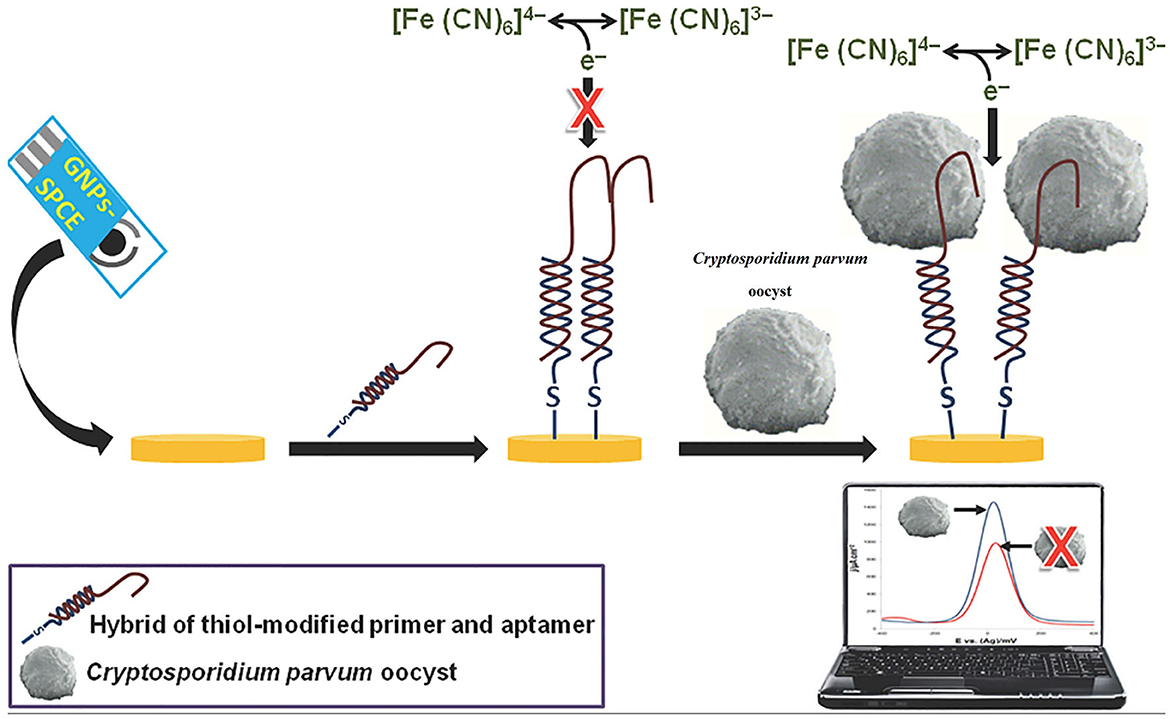
Figure 3. Illustration of a screen-printed aptamer-based electrochemical biosensing platform for the detection of C. parvum oocysts. Reproduced from Iqbal et al. (2015).
5. A brief look at nanomaterials incorporated into sensors
Improvements in nanotechnology science have provided the opportunity for researchers to work at nanoscale levels. Nanomaterials range from 1 to 100 nm and are classified into various groups, namely, nanoparticles (NPs), quantum dots (QDs), carbon nanotubes (CNTs), graphene, graphene oxide (GO), and nanochannels (Avant et al., 2019; Saleem and Zaidi, 2020; Pandey and Chusuei, 2021). Incorporating NPs with a biosensor system is performed for modifying and improving the sensor platforms and to overcome the limitations of conventional diagnosis tools (Luo et al., 2006). NPs are employed in the main types of biosensor systems including electrochemical, calorimetric, optical, and acoustic (Malik et al., 2013). NPs have critical functions such as reducing the time of reactions, catalysis, and immobilization of biomolecules (nucleic acid, antibody, and enzymes) into the electrochemical biosensors (Luo et al., 2006). In addition, due to their large specific surface, NPs are useful for improving electron transfer between biomolecules and the surface electrode (Cho et al., 2020). Common NPs include metal NPs (Au, Ag, and Pt), oxide NPs (SiO2, TiO2, ZrO2, and MnO2), and semiconductor NPs (CdS and PbS); metal NPs are more common due to their excellent catalytic properties in catalyzing electrochemical reactions (Tang et al., 2005). Moreover, silver and gold NPs have excellent conductivity properties, which enhance electron transfer between biomolecules and electrodes (Alaqad and Saleh, 2016). Moreover, gold NPs play an important role in increasing the sensitivity of electrochemical biosensors (Saha et al., 2012).
Magnetic NPs (MNPs) are used for designing magnetic biosensors, which have been broadly applied in medical areas such as diagnostic assays, DNA or RNA isolation, magnetic resonance imaging (MRI), and drug delivery (Khoo et al., 1997; Kudr et al., 2017; Wang et al., 2017; Ali et al., 2021). MNPs are a class of nanomaterials composed of metals such as cobalt, nickel, and iron, with paramagnetic, ferromagnetic, and superparamagnetic properties (Aboul-Enein et al., 1999; Akbarzadeh et al., 2012). Magnetic iron oxide NPs are employed for biomedical applications such as magnetic separation (Wu et al., 2015). For example, immunomagnetic separation (IMS) is now employed as a standard method for the detection and separation of Cryptosporidium spp., and G. lamblia oocysts/cysts from 10 to 150 L water samples (USEPA, 2012), although several studies have been conducted to increase the recovery rate of the method using either additional concentration methods or alternative elution (Hu et al., 2004; Fradette and Charette, 2022).
Oxide NPs have several chemical properties and possess a high surface energy (Stankic et al., 2016). For example, MnO2 NPs can directly react with biomolecules (Vukojević et al., 2018). In addition, oxide NPs, for instance, SiO2 NPs, can also be used as labels for biomolecules. SiO2 NPs, as an oligonucleotide label, have been used for electrochemical sensitive detection in genosensors and immunosensors (Ma et al., 2008; Wang et al., 2013).
Quantum dots are semiconductor nanocrystals made up of a reactive core, which contains semiconductor particles such as cadmium selenide (CdSe), cadmium telluride (CdTe), indium phosphide (InP), or zinc selenide (ZnSe). However, QDs, as ideal materials, have been widely used in the development of sensing technology due to their extraordinary chemical properties such as excellent optical aspects (Ding et al., 2022).
Carbon nanotubes are the most popular advanced sensing technology, and have recently attracted interest for their unique properties such as excellent electronic conductivity features and large surface-to-volume ratios (Zaporotskova et al., 2016). Nanotubes have cylindrical structures with several hexagonal graphite planes rolled in tubes, which are divided into single-walled NTs (SWNTs) and multi-walled NTs (MWNTs), based on the number of walls (Saxena and Srivastava, 2020).
Graphene and GO nanomaterials present unique chemical and electrical features, which have highlighted them as promising materials to improve signal responses in novel sensing technologies such as electrochemical biosensors, fluorescence resonance energy biosensors transfer (FRET), laser desorption/ionization mass spectrometry (LDI-MS), and surface-enhanced Raman spectroscopy (SERS) (Chauhan et al., 2017; Janegitz et al., 2017; Morales-Narváez et al., 2017). GO possesses a hydrophobic domain structure and hydrophilic oxygen-containing functional groups, which provide good biocompatibility and water dispersibility (Ghulam et al., 2022). However, their features, including high surface area and a high affinity for a variety of biomolecules (antibodies, enzymes, DNA, cells, and proteins), have made them ideal for next-generation biosensors (Lee et al., 2016).
The recent trends are the use of both single and array nanochannels for electrical biosensing applications. Graphene and its analogs are among the emerging materials used to obtain nanochannels (de la Escosura-Muñiz and Merkoçi, 2012). The applications of nanochannels are focused on the detection of DNA, protein, virus, toxin, and other analytes (Wang et al., 2015; Sun et al., 2016; Shiohara et al., 2022).
6. Applications of biosensors based on transducer types
The biosensor system employs a sensing technique and reacts with an analyte to produce a measurable electrochemical, electrical, mechanical, optical, or thermal signal (Mehrotra, 2016; Naresh and Lee, 2021). Biosensors can also be classified as electrochemical, optical or mechanical biosensors (Cammann, 1977; Thevenot et al., 1999; Ronkainen et al., 2010; Bermejo et al., 2011; Monosik et al., 2012; Ozdemir et al., 2013).
6.1. Electrochemical
In recent years, most studies on biosensors have focused on electrochemical systems (Ronkainen et al., 2010; Low et al., 2012; Kokkinos et al., 2016). The wide practical fabrication and usage of these biosensors are based on their advantages such as feasibility, portable, rapidness, low fabrication cost, simplicity of operation, and high selectivity of this system, which make these sensors quite desirable and attractive for the POC approach. Electrodes play an important role in the performance of electrochemical cells and biosensors. The electrode structure and properties influence the cost, sensitivity, selectivity, and limit of detection (LoD) of these biosensors (Faulkner and Bard, 2002; Cesewski and Johnson, 2020). In this regard, a label-free interdigitated-based capacitive biosensor was designed on interdigitated gold electrodes for the detection of Cryptosporidium oocysts in water samples (Luka et al., 2019). In this study, a capture probe, anti-Cryptosporidium monoclonal antibodies (IgG3) and bovine serum albumin (BSA), was employed to increase the specificity and to avoid non-specific interactions. The linear detection range for this technique was 15–153 oocysts/mm2 with a detection limit of 40 oocysts/mm2 (Luka et al., 2019).
Potentiometric biosensors have also been used for the detection of waterborne protozoa. Laczka et al. (2013) reported a novel electrochemical approach based on a potentiometric immunosensor for the rapid detection of C. parvum based on (HRP)-labeled secondary antibody, which was able to detect 5 × 102 Cryptosporidium oocysts/mL in 60 min. In comparison to available ELISA techniques, Laczka et al. (2013) improved the LoD from 100- to 1,000-fold for the detection of oocysts, without the need for any specific antibody. In a study performed by Wang et al. (1997), a new electrochemical hybridization biosensor based on screen-printed carbon strip electrodes (SPEs) by the chronopotentiometry approach, as an electrochemical technique, was fabricated to detect a short specific nucleotide sequence of Cryptosporidium in untreated drinking and river water using the chronopotentiometric (CP) transduction method. This approach was able to discriminate Cryptosporidium DNA with an extremely low LoD, 50 ng/mL, and a short hybridization time of the probe, 3 min (Wang et al., 1997). Chronopotentiometry is a galvanostatic method that is used to study the mechanism and kinetics of chemical reactions with a constant level of current for a given period of time (Lingane and Peters, 1971; Kinyua Muthuri et al., 2021).
Electrochemical impedance spectroscopy (EIS) has been designed as a highly effective method based on label-free methods for the detection of biomolecules. It is used to investigate binding events that occur at the electrode surface (Magar et al., 2021). In the field of parasitology, Grewal et al. (2014) developed a nano-yeast-single-chain Fv (scFv) affinity reagents on a low-cost commercial gold screen-printed electrode for the sensitive detection of E. histolytica cyst antigens in stool samples at concentrations down to 10 pg/mL in buffer, with an inter-assay reproducibility of (% RSD, n = 3) 4.1%. A number of studies have also utilized this method for the detection of Cryptosporidium spp. A non-labeled detection system using a polydiacetylene (PDA)-based fluorescence chip based on a colorimetric detection system was developed for the detection of C. parvum with an LoD of 1 × 103 oocysts/mL (Park et al., 2008). The main advantages of this study were real-time detection of Cryptosporidium spp. oocysts, rapidness, simplicity, and no need for any labeling or staining for analyses (Park et al., 2008). Houssin et al. (2010) fabricated a label-free EIS biochip-based biosensing platform for the detection of Cryptosporidium in water samples using EIS via an interdigitated microelectrode array with an LoD lower than 10 cells/μL. The authors suggested that this method could be proposed as an alternative technique to current staining procedures, which was able to differentiate live and dead oocysts based on electrical impedances between 10 kHz and 100 kHz (Houssin et al., 2010). More recently, Luka et al. (2022) reported a chip-based electrochemical biosensor for the sensitive and label-free detection of Cryptosporidium oocysts in water samples based on anti-Cryptosporidium monoclonal antibodies (IgG3). This novel platform was a fast, real-time, and inexpensive tool, which was utilized to measure C. parvum in the range of 0–300 oocysts, with an LoD of ~20 oocysts/5 μL (Figure 4).
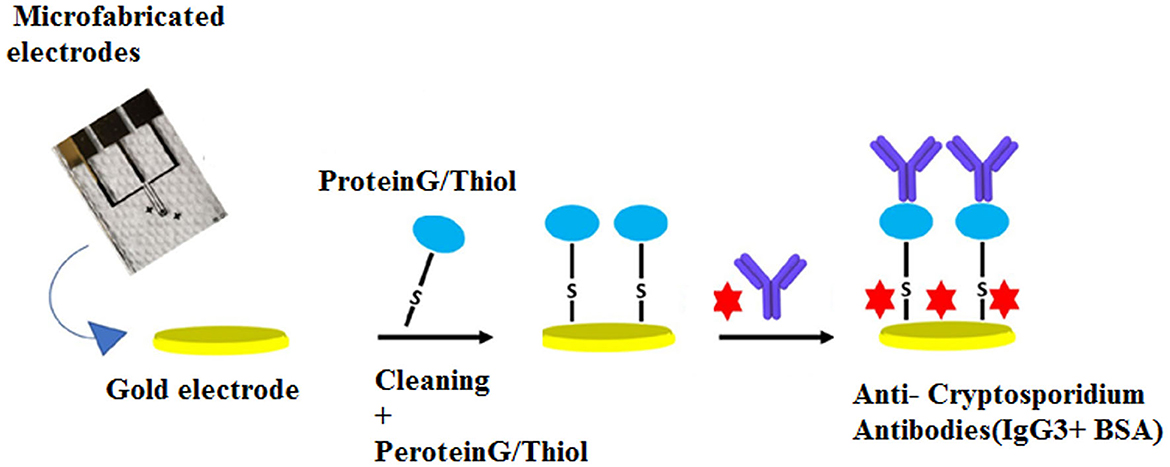
Figure 4. Chip-based device for label-free detection of Cryptosporidium oocysts in water. The figure shows the schematic process of the immobilization of anti-Cryptosporidium antibodies onto the Au electrode. Reproduced from Luka et al. (2022).
6.2. Optical (fluorescence, chemiluminescence-based biosensor, and surface plasmon resonance [SPR]) biosensors
The main components of an optical diagnostic device are a light source, optical transmission medium (fiber, waveguide, etc.), immobilizedm biological recognition element (enzymes, antibodies, or microbes), and optical detection system (Chen and Wang, 2020). An optical sensor converts light rays into electronic signals via measuring the physical quantity of light and translating it into a readable signal (Deshmukh et al., 2020). Optical sensing technologies are also divided into label-based techniques such as fluorescent labeling and label-free methods (Tang et al., 2010; Bermejo et al., 2011).
Fiber optic biosensors (FOBs), as fluorescence-based optical biosensors, are increasingly being employed for the detection of foodborne and waterborne pathogens. This technique utilizes antibodies or other molecules to capture the target pathogen from a sample (Narayanaswamy, 2006; Hayman, 2008). In this regard, Kramer et al. (2007) developed an optical sensor (rapid automated FOB assay) based on a sandwich immunoassay using anti-Cryptosporidium oocyst polyclonal and monoclonal antibodies to detect the parasite in potable water. In this study, the polyclonal antibody captured the target pathogen and marked it with a cyanine 5-labeled (Cy5) detector monoclonal antibody. The LoD was 105 oocysts/mL, while a 10-fold increase in sensitivity was achieved using the polyclonal antibody followed by boiling samples before the detection (Kramer et al., 2007); however, owing to the low infectivity dosage of Cryptosporidium spp., and the low concentration of oocysts in water samples, concentration and preparation steps before employing detection techniques are still required (WHO, 2014). To overcome the short lifetime of the excited state limitation of fluorescence (Berezin and Achilefu, 2010), a luminescence process, in which a photon may be released after any time, was developed (Gaft et al., 2015). Chemiluminescence-based biosensors are another type of optical sensing device (Aboul-Enein et al., 1999), which measure the rate of photon production and generate light through a chemical reaction (Kim et al., 2021). In this optical biosensor, the analyte interacts with the immobilized biomolecule, which is marked with chemiluminescence species. Some advantages of chemiluminescence tools are high sensitivity for the detection of pathogens, fast dynamic response, and a wide calibration range (Yan et al., 2021).
As a label-free-based biosensor, Luka et al. (2019) fabricated an interdigitated-based capacitive biosensor to detect Cryptosporidium oocysts in water samples. In this system, the number of Cryptosporidium oocysts captured on the surface of the electrode was identified by means of a fluorescein isothiocyanate (FITC) immunofluorescence assay. The result of this study indicated an LoD of 40 cells/mm2 and a linear range of detection between 15 and 153 cells/mm2 in environmental water samples. Briefly, anti-Cryptosporidium monoclonal antibodies (IgG3), as the capture biomolecules, were attached to the interdigitated gold electrodes (IDE) using the protein G/thiol. Finally, upon the formation of the Cryptosporidium-antibody complex, changes in the capacitive/dielectric properties were detected (Figure 5).
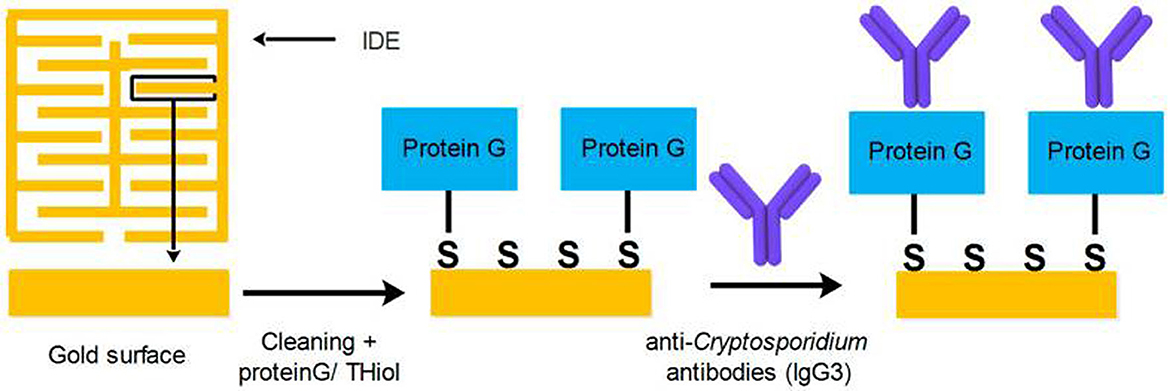
Figure 5. Procedure applied for covering the surface of IDE with SAM, and the attachment of anti-Cryptosporidium antibodies. Reproduced from Luka et al. (2019).
SPR biosensors have been developed based on refractive index to increase the sensitivity of optical biosensors (Zeng et al., 2021). This system is a label-free optical phenomenon without radioactivity and fluorescence, which is recently considered a very powerful tool to study the interactions between the analyte and biorecognition molecules. This type of biosensor has remarkable advantages such as high sensitivity and specificity, label-free measurement, real-time analysis, and high-throughput capacity (Olaru et al., 2015). In addition to the common analytical applications, SPR devices are suitable for food safety monitoring and environmental applications (Olaru et al., 2015). In this regard, Kang et al. (2006) developed a flow-type SPR biosensor for the rapid detection of Cryptosporidium oocysts. Accordingly, an SPR biosensor was designed based on mixed self-assembled monolayers (SAMs) using 3-mercaptopropanol (3-MPOH) and 11-mercaptoundecanoic acid (11-MUA). These groups enhance the accessibility of analytes to the sensor surface using biotin–streptavidin biomolecules. This system was able to identify C. parvum oocysts in real time with an LoD of 1 × 106 oocyst/mL, and the sensitivity was increased to ~1 × 102 oocyst/mL using biotin–streptavidin biomolecules (Figure 6).
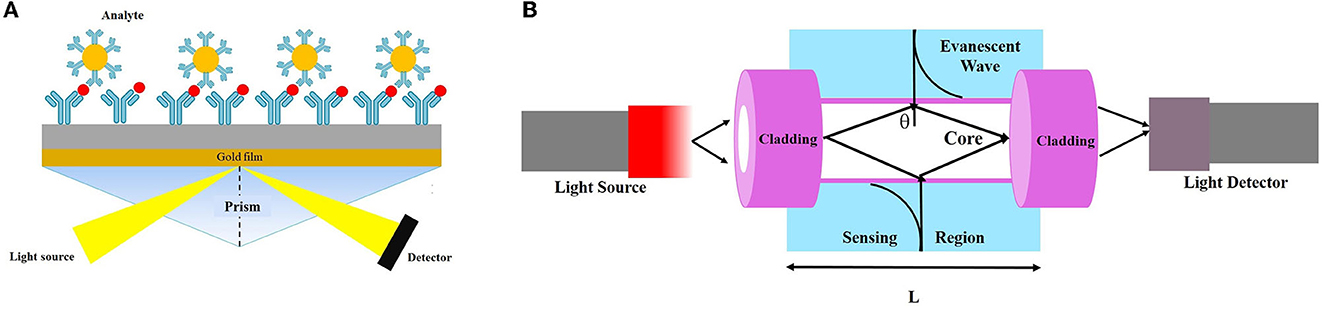
Figure 6. Schematic view of (A) evanescent wave fiber optic chemical sensor and (B) detection-based SPR technique.
6.3. Mechanical biosensors
Mechanical biosensors are sensitive to alterations in mechanical characteristics. These types of assays play a critical role in different bioanalytical settings (Arlett et al., 2011; Zhang and Hoshino, 2019). The functions of mechanical biosensors are mostly based on either the induced stress on the cantilever platform or the alteration in the resonant frequency of a mass-spring device (Xu et al., 2019; Chalklen et al., 2020).
The piezoelectric system is a class of mass-based biosensors, which measures changes in the oscillating crystal resonance frequency due to the interaction between bioreceptor and biological elements (antibodies, enzymes, and antigens) (Nicu et al., 2005). Various piezoelectric (like quartz crystal) (Lim et al., 2020; Wu et al., 2021) and biosensing materials have been used in piezoelectric biosensors (Skládal, 2016).
Piezoelectric quartz crystal (PQC) immunosensors, as mass-sensitive devices, have been fabricated to calculate the quantification of biomolecular interactions (Bunde et al., 1998; O'Sullivan and Guilbault, 2000). Wang et al. (2004) developed a new, simple, rapid, and highly sensitive technique that was a promising alternative approach to detect anti-T. gondii antibodies (TgAbs) in clinical samples. The authors demonstrated that the latex piezoelectric immunoassay (LPEIA) was improved by using gold NPs, as an alternative to latex particles.
Another type of piezoelectric biosensor is a piezoelectric-excited millimeter-sized cantilever (PEMC) sensor that consists of a piezoelectric and a borosilicate glass layer with a sensing area (Zuehlke, 2022). A PEMC sensor was fabricated to detect the waterborne parasite, G. lamblia, in aquatic samples (Xu and Mutharasan, 2010). The resonant frequency of the sensor was continuously monitored using monoclonal antibodies against G. lamblia cysts, which were immobilized on PEMC sensors. In this procedure, 1–10,000 G. lamblia cysts/mL samples in a flow interacted with the antibody-immobilized sensor, and, upon binding cysts to the antibody, and changes in the resonant frequency of the cantilever sensor were continuously recorded. This method detected 10 cysts/mL for 15 min. Similarly, a PEMC biosensor was designed using immobilized IgM to detect C. parvum oocyst in a flow configuration at 1 mL/min. The PEMC detected C. parvum at 100, 1,000, and 10,000 oocysts/mL in less than 1 min. The resonance frequency response of the sensor was logarithmically correlated with the concentration of C. parvum oocysts, and due to the high sensitivity and specificity, it was employed for monitoring drinking water (Campbell and Mutharasan, 2008).
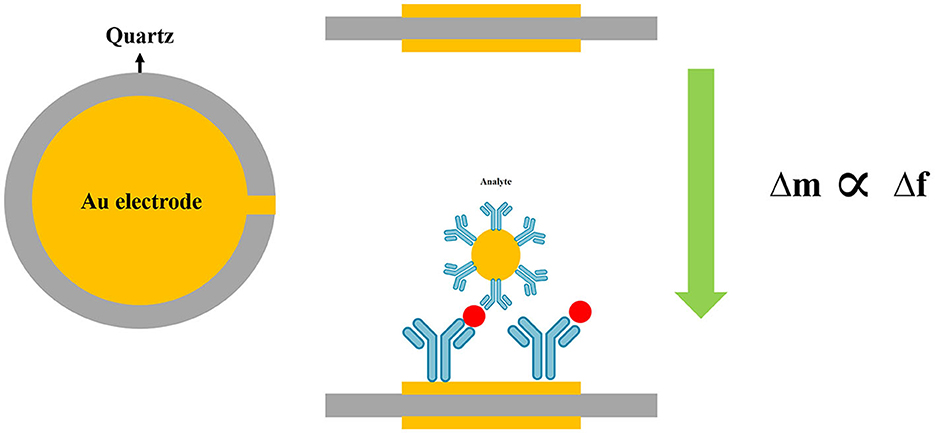
A quartz crystal microbalance (QCM) with dissipation monitoring (QCM-D) was employed to detect Cryptosporidium oocysts in water samples (Poitras et al., 2009). Water samples are usually contaminated by a wide range of microorganisms, including bacteria, viruses, and parasites, that may cause interference during the detection of target pathogens in the biosensing system. To overcome this limitation, the QCM-D was used as a platform for the specific binding of C. parvum to an antibody-covered gold-coated crystal surface to increase the specificity of the method. This technique was able to detect oocyst concentrations from 3 × 105 to 1 × 107 per mL of water with a rapid operation (~5 min) (Poitras et al., 2009) (Figure 7).
MNP-based approaches are able to rapidly detect FWP with high sensitivity and selectivity (Akbarzadeh et al., 2012). MNPs have been intensively studied due to their capability to be employed in many areas such as magnetic storage devices, optical magnetic materials, magnetic separation, and DNA-targeted diagnosis (Duguet et al., 2006; Reddy et al., 2012). In the field of parasitology, in a study developed by Xu et al. (2013), magnetic-fluorescent CdTe@Ni quantum dots (mQDs) were utilized to design a sensitive nanobiosensor based on fluorescence resonance energy transfer (FRET) in order to detect T. gondii DNA. In this study, mQDs and commercial BHQ2 were the energy donors and acceptors, respectively. To produce a sensing probe, sCdTe@Ni mQDs and BHQ2 were used to label a stem-loop T. gondii DNA oligonucleotide at the 5′ and 3′ ends, respectively. This system was able to detect the target DNA of T. gondii with a LoD of ~2.70 × 10−9 mol/L. In addition, a study developed by He et al. (2015) detected T. gondii using the quenching of magnetic fluorescence NPs (Fe3O4/CdTe) based on CdTe QDs, which were synthesized using 3-mercaptopropionic (MPA) capping for T. gondii DNA detection, with a LoD of 8.339 × 10−9 M of DNA. In this study, similar to Xu et al. (2013), a stem-loop T. gondii DNA oligonucleotide was employed, which was conjugated to Fe3O4/CdTe at the 5′ end as the energy donor and BHQ2 at the 3′ end as the acceptor.
7. Microfluidic devices
Microfluidic systems are geometrically small scale (typically sub-millimeter) and can be incorporated with biosensor systems. Theoretically, microfluidic devices are comprised of thin grooves or small wells, channels, micro-channels, and chambers. These devices are rapid and accurate, and are increasingly employed for the detection of waterborne pathogens (Woolley and Mathies, 1994; Stone et al., 2004; Fiorini and Chiu, 2005; Chin et al., 2012).
In a biosensor-independent manner, there is a commercial microfluidic device for the detection of Cryptosporidium oocysts in water samples with on-chip integrated sample preparation features, named CryptoDetect CARD. This technology involves integrated immunomagnetic separation (IMS); however, the technology needs more development to specify the LoD and sensitivity, and the need for sample preparation, filtration, and concentration still limits its use (Rheonix, 2011).
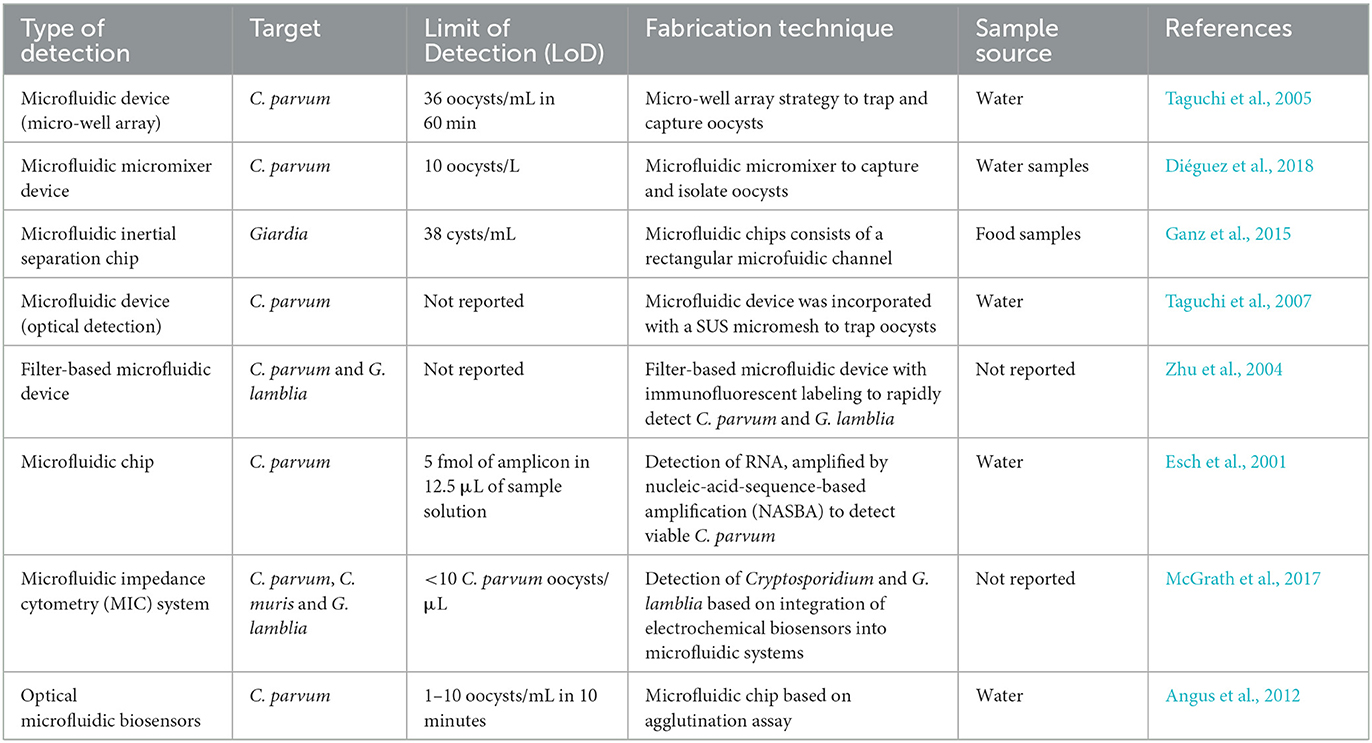
As a first strategy, hydrodynamic trapping together with immunofluorescence detection was utilized (Zhu et al., 2004; Taguchi et al., 2005, 2007; Lay et al., 2008; Mudanyali et al., 2010). In this regard, Taguchi et al. (2005) developed a micro-well array strategy to capture oocysts. For trapping oocysts in wells, micro-wells with a 10 μm or 30 μm diameter and a 10 μm depth were developed to capture oocysts of Cryptosporidium. This technology was able to detect the oocysts in very small sample volumes and could therefore be used instead of visual inspection of microscope slides. The micro-wells were coated with streptavidin and anti-C. parvum antibodies, and the samples containing C. parvum oocysts (107 oocysts/mL) suspended in PBS were simply deposited onto the array. This approach was able to detect Cryptosporidium oocysts for 60 min at a maximum flow rate of 350 μL/min (5 mL could be processed in under 15 min), with a LoD of 36 oocysts/mL (Taguchi et al., 2005). Diéguez et al. (2018) purposed a disposable microfluidic micromixer, which was able to specifically capture, isolate, and concentrate Cryptosporidium from water samples. This designed device was able to analyze the quantification of captured oocysts using immunofluorescence microscopy, as well as an imaging flow cytometer. In addition, the microfluidic micromixer device provided a rapid and efficient detection method, with a capture efficiency of 96% compared to other available laboratory-scale technologies for the detection of Cryptosporidium oocysts (Figure 8A).
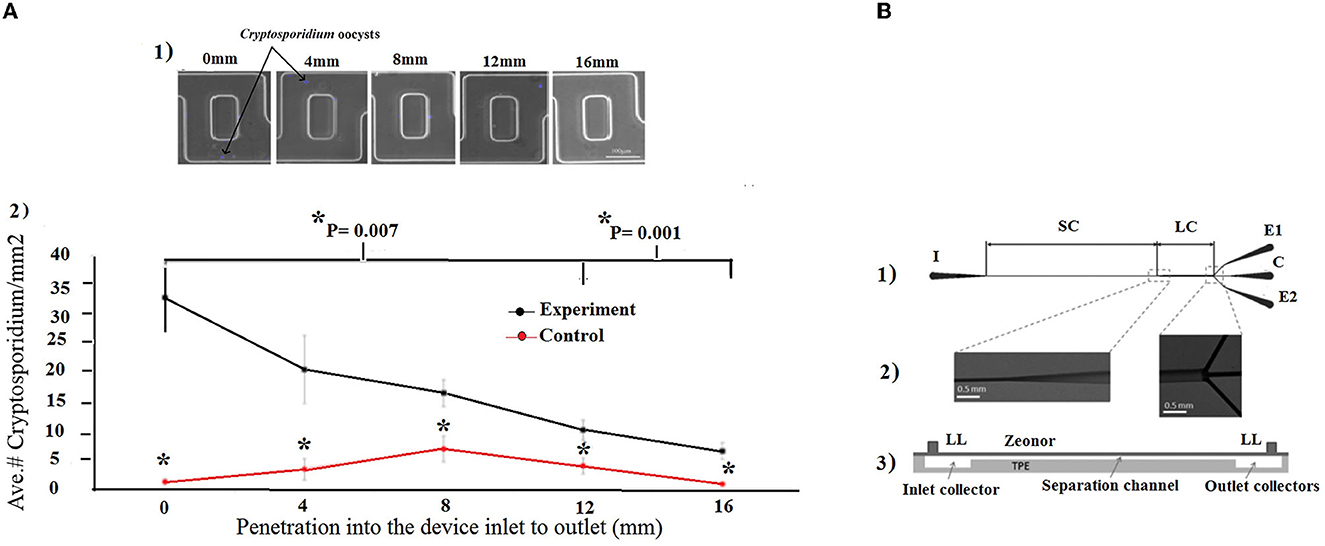
Figure 8. (A) Microfluidic micromixer device. (1) The captured oocysts at different lengths in the device were systematically counted randomly in 10 areas. (2) The number of oocysts per area was averaged at each length. *indicates statistical significancy. Reprinted from Diéguez et al. (2018). (B) Microfluidic chip. (1) The inlet (I) port allows entry into the separation channel (SC), which then widens into a large channel (LC) that splits into three outlet collectors (C, E1, and E2). (2) Scanning electron microscopy images, and (3) schematic view of a vertical section, the Zeonor-TPE chip assembly with the Luer lock (LL) ports, separation channel, and input and output collectors. Reproduced from Ganz et al. (2015) with permission from the American Society of Microbiology (ASM) publication.
In addition, a microfluidic inertial separation chip was designed and fabricated for the separation of Giardia cysts from food samples (Ganz et al., 2015). The microfluidic chips consisted of an inlet, a main separation channel with a rectangular microfluidic channel, and a large channel, which was divided into three smaller channels connected to three output channels. The method was very efficient and specific for G. lamblia, and recovered an average of 68.4% of cysts, with a LoD of 38 cysts from a 25 g lettuce sample (Ganz et al., 2015) (Figure 8B).
Hydrodynamic trapping of Cryptosporidium oocysts, either in wells or filters, through pre-filter structures or a raindrop filter, was also developed. This microfluidic device was incorporated into a SUS micromesh to capture C. parvum oocysts. Trapped C. parvum oocysts were visualized by fluorescent staining. The concentration of added C. parvum oocysts and oocysts detected by the SUS micromesh was linearly correlated within the range of 18–200 oocysts/mL. The results of this technique were in agreement with the direct immunofluorescence assay coupled with the immunomagnetic separation (DFA-IMS) method, while the recovery of SUS micromesh (93%) was higher than DFA-IMS (90%), suggesting that the SUS micromesh is a promising procedure for counting C. parvum oocysts (Taguchi et al., 2007) (Figure 9).
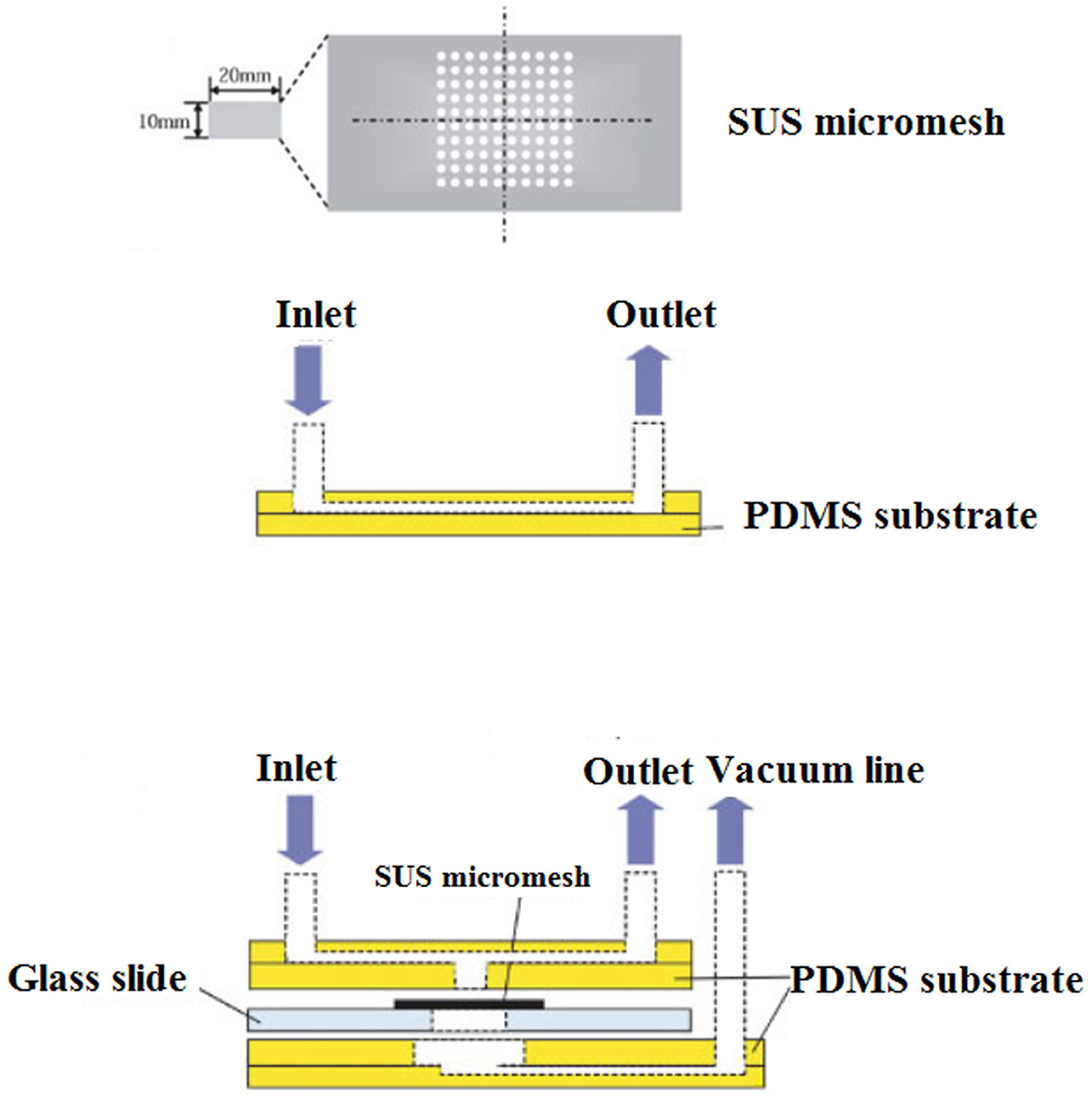
Figure 9. Schematic overview of the experimental setup of a micromesh microfluidic. (1) View of the SUS micromesh. (2) Side view of a PDMS microfluidic device equipped with the SUS micromesh, and (3) side view of a PDMS microfluidic device with microchannel, sample inlet, and outlet. Reproduced from Taguchi et al. (2007) with permission from Wiley & Sons.
As a second strategy, it was illustrated that trapping of Cryptosporidium oocysts using sieves or filters may increase the sensitivity of microfluidic systems (Zhu et al., 2004; Lay et al., 2008). A fully automated system consisting of a filtration unit and pumping system (1,000 L within 24 h), complemented by a microfluidic chip, Crypto-Tect bio-slide, was developed by the Shaw Water Ltd. Company, which stained and counted Cryptosporidium oocysts with a LoD slightly higher than 10 oocysts (Shaw, 2009), while 1,000 L drinking water was concentrated to a 1.5 mL sample, which was suitable for introducing to the microfluidic system. In addition, a microfluidic device based on the detection of C. parvum and G. lamblia oocysts/cysts using positive pressure was developed that identified C. parvum and G. lamblia fluorescent labels, while the staining solution was 10 to 100 times more diluted than the recommended concentration in the conventional glass method (Zhu et al., 2004).
Microfluidic trapping devices can also be integrated with on-chip molecular methods for further applications (Mahdavi Abhari et al., 2023). Molecular sensing techniques include pre-amplification of the microorganism genomic material, either via fluorescence or electrochemical tools. These reliable and rapid detection techniques still require genomic materials. The detection of C. parvum in water resources still requires the parasite to be collected and concentrated from a large water sample volume. In fact, during the analysis of water samples, the numbers of recovered parasites are usually low and cannot be detected without DNA amplification. To overcome this limitation, Esch et al. (2001), developed a microfluidic chip, which was amplified by nucleic-acid-sequence-based amplification (NASBA), using DNA-modified liposomes to detect RNA in viable C. parvum. A NASBA-generated amplicon was placed between the capture and reporter probes in a microfluidic channel. To generate fluorescence, reporter probes were tagged with carboxyfluorescein-filled liposomes, which increased the sensitivity of detection, even in very low concentrations of targets. The LoD of the microfluidic chip was reported to be 5 fmol of amplicon in 12.5 μL of sample solution (Figure 10).
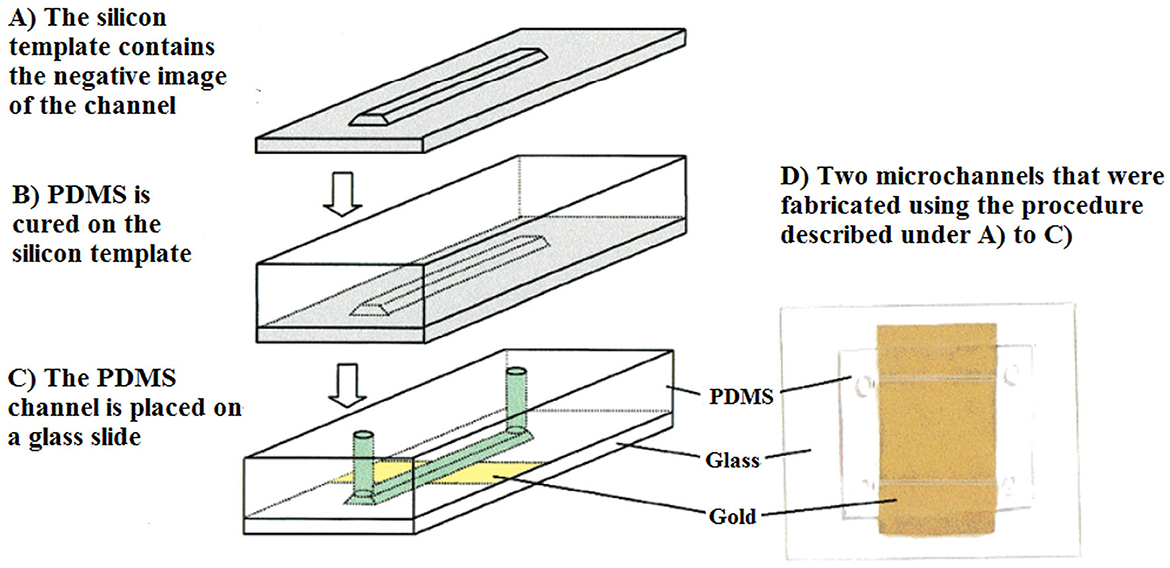
Figure 10. View of a PDMS microchannel. The channel was fabricated in silicon and placed on a glass slide, where gold was deposited. Reprinted with permission from Esch et al. (2001) copyright from the American Chemistry Society.
Recent developments have been presented in novel engineering systems for the detection of Cryptosporidium and G. lamblia based on the integration of electrochemical biosensors into microfluidic systems. A microfluidic impedance cytometry (MIC) system based on the detection of viable parasites was proposed and designed by McGrath et al. (2017), which was able to rapidly discriminate live and inactive C. parvum oocysts with over 90% certainty, and to identify the viability of Cryptosporidium and Giardia at the single (oo)cyst level (Figure 11; Table 3).
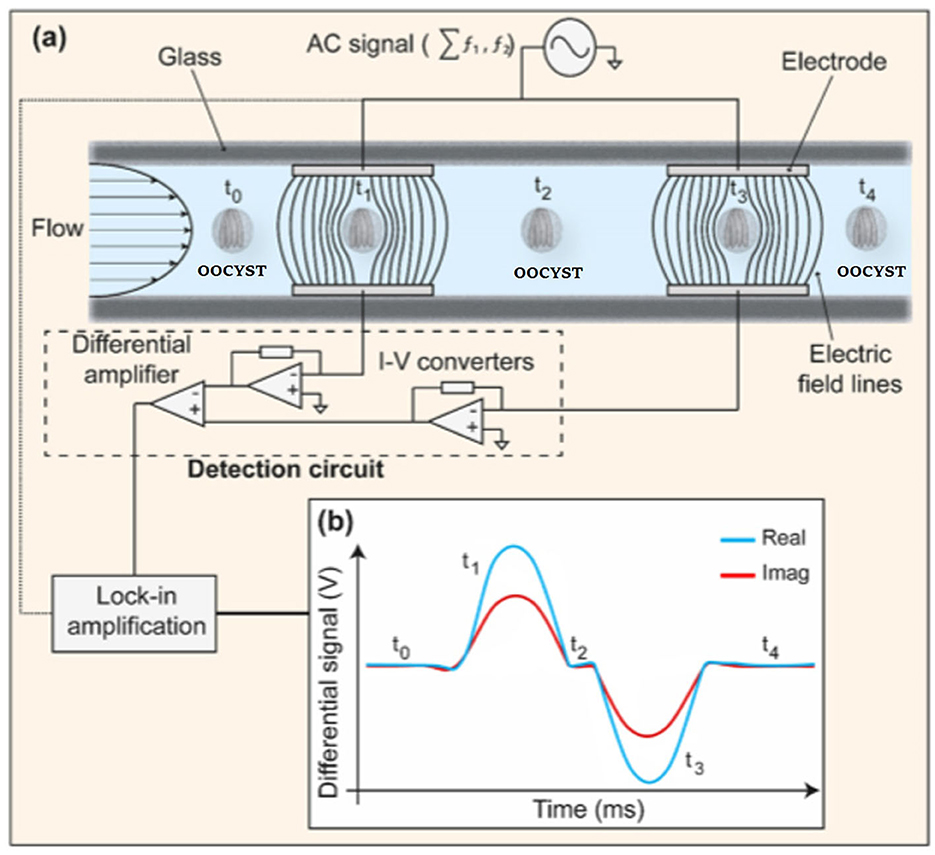
Figure 11. Schematic picture of a microfluidic impedance cytometer. (a) Two parallel facing electrodes. The electrodes were fabricated within a microfluidic channel. (b) The current flowing through the bottom electrodes was measured using a custom detection circuit. The circuit consists of trans-impedance amplifiers, which convert current (I) into voltage (V), and a differential amplifier. Reproduced from McGrath et al. (2017).
8. Smartphone microscopic method
In recent years, smartphone microscopic methods have been described and used as an alternative platform for the detection of targeted pathogens, incorporated with traditional optical microscopic methods. The strengths of these techniques are the low cost, small size, being portable, and easy transportation to rural and remote settings. In fact, portable devices that can transmit relevant data to remote experts have a large impact on the quantity and quality of care. In addition, cell phone cameras are the most ubiquitous optical sensors in the world (Breslauer et al., 2009; Rajchgot et al., 2017).
In the field of waterborne protozoa, Shrestha et al. (2020) developed a smartphone-based microscopic assay for the simultaneous detection of oocysts/cysts of Cryptosporidium and G. lamblia in vegetable and water samples. The device consisted of a ball lens 1 mm in diameter, an aluminum mounting plate to transform a smartphone, and a white LED as an illumination source. After concentration of oocysts and staining with Lugol's iodine, oocysts were counted using the smartphone microscope. In comparison to commercial bright field and fluorescence microscopes, the smartphone-based microscopic assay was a low-cost alternative for screening oocysts/cysts of Cryptosporidium spp., and G. lamblia. The LoD of Giardia ranged from 24 cysts/100 g for cucumber to 73 cysts/100 g for cabbage. The LoD for Cryptosporidium ranged from 11 oocysts/100 g for radish to 25 oocysts/100 g for cabbage, while the LoD of Cryptosporidium was lower than that of Giardia (Shrestha et al., 2020) (Figure 12).
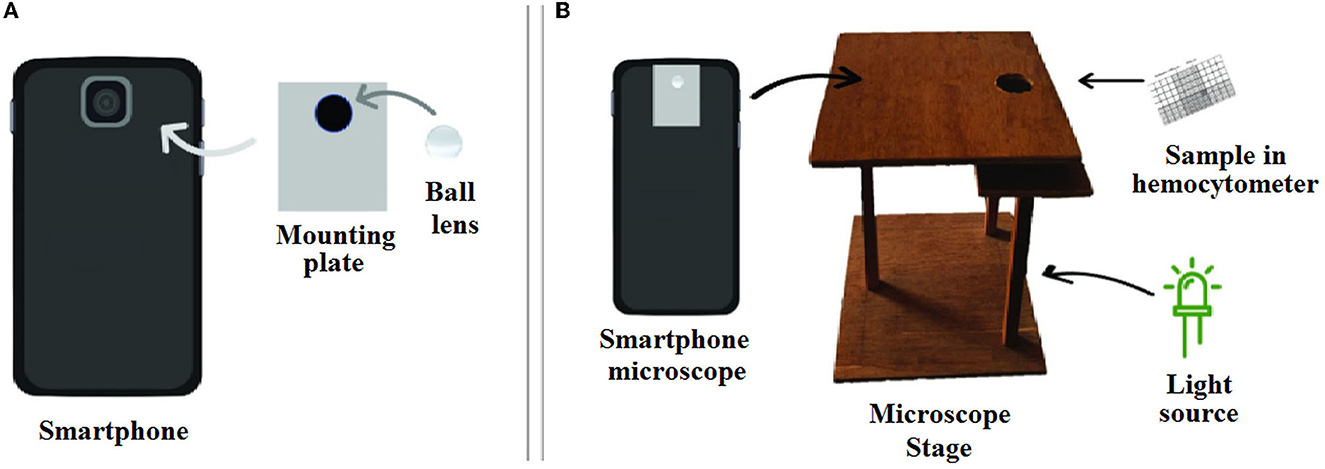
Figure 12. Schematic overview of a smartphone microscope setup. (A) Fabrication of ball lens onto mounting plate and the smartphone camera. (B) The set up of measurement system. Reproduced from Shrestha et al. (2020).
Recently, Luka et al. (2021) fabricated a 3D portable and smartphone-integrated on-chip colorimetric biosensor, which was invisible to the naked eye. In this regard, oligonucleotide-modified gold NPs (AuNPs) were used for the detection of Cryptosporidium RNA using UV–Vis spectroscopy. The color change of the AuNPs from red to blue after 5 min was an indicator for Cryptosporidium RNA. The advantages of these methods were the low sample volume (15 μL), short analysis time (~30 min), and high detection limit (5 μM) (Figure 13).
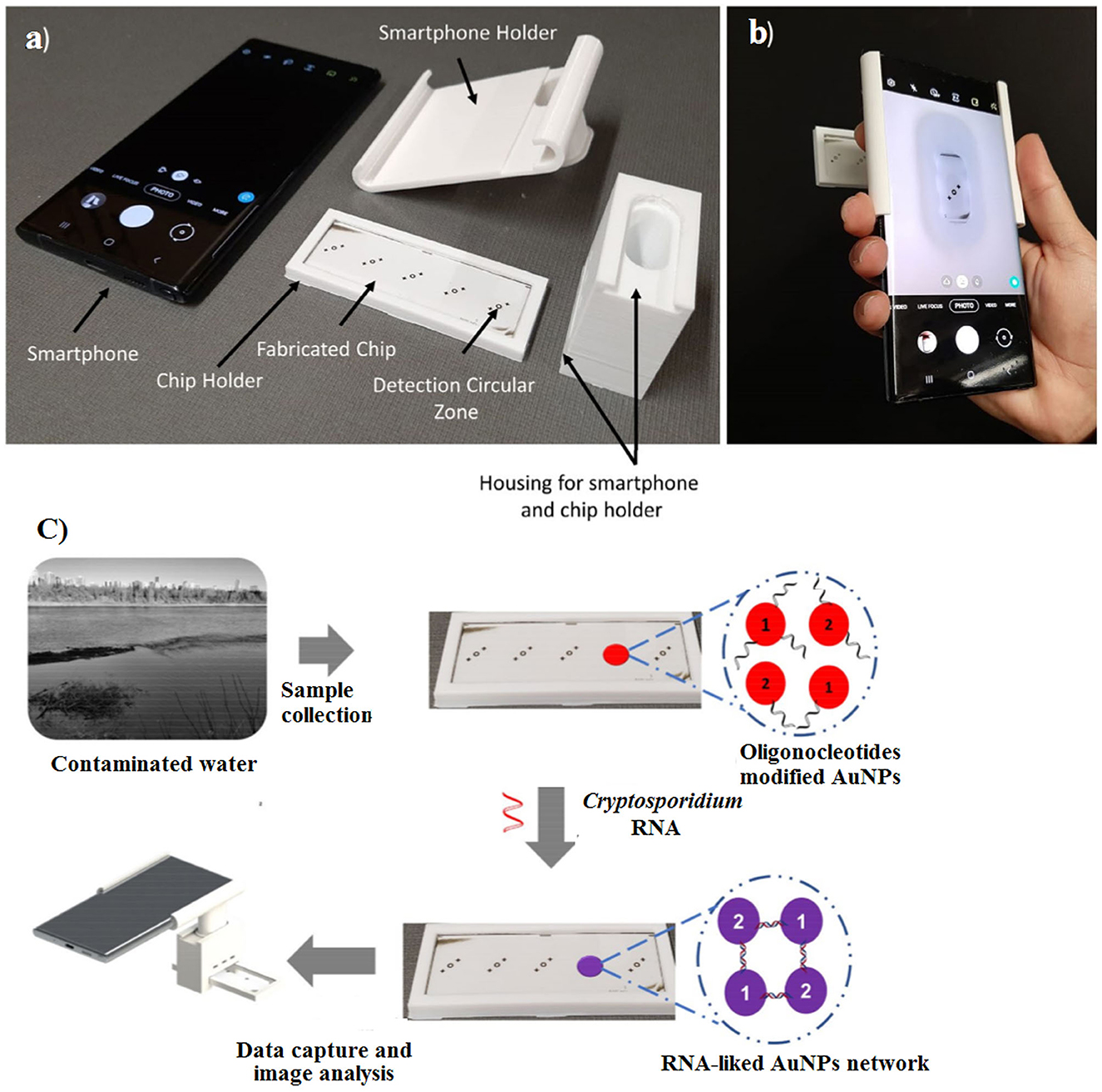
Figure 13. Fabricated chip and 3D portable holder assembly integrated with a smartphone. (a) The major components of the detection system, (b) the assembled detection system, and (c) schematic overview of sensing platform. Reproduced from Luka et al. (2021).
8.1. Future outlook
In recent years, advanced diagnostic procedures have been presented to overcome the limitations of available common techniques. Molecular biology assays, as gold standard methods, have been routinely utilized for rapid detection, identification, and differentiation of FWP. In addition, in recent decades, various studies have been conducted based on electrochemical and optical biosensors and nanobiosensors for the early detection of common waterborne pathogens. Although these methods frequently profit from good accuracy, reliability, and multiple sample processing, most of them suffer from the need for specialized expensive equipment, centralized services, infrastructure/or professional staff, and a lack of point-of-use (PoU) employment capabilities. The ASSURED criteria are an important guideline provided by the WHO for developing efficient POC devices to distinguish major human diseases (Syedmoradi et al., 2017; Ahmadi et al., 2020). Advances in digital health include mobile health, health information technology (IT), and wearable devices, and the acronym REASSURED (real-time connectivity, ease of specimen collection, affordable, sensitive, specific, user-friendly, rapid and robust, equipment-free or simple environmentally friendly, deliverable to end-users) has been offered for the development of diagnostic methods to address vital priorities such as global health emergencies (Land et al., 2019; Mahmoudi et al., 2022). Factors associated with the effective implementation of ASSURED diagnostic systems that should be considered in addressing POC diagnostic tests are real-time connectivity and ease of specimen collection (Land et al., 2019). Finally, for the successful diagnosis and management of infectious diseases, the necessity to fabricate smart biosensing systems is vital. We believe that smart biosensing platforms play an extremely significant role in diagnosing, as well as in predicting and controlling, future trends in infectious diseases, either epidemics or pandemics.
Author contributions
HM and KO: conceived, designed, reviewing, and editing the manuscript. SN: data gathering, literature review, and writing the manuscript. FS: illustrations and graphics. All authors read and approved the final version of the manuscript.
Funding
This study was financially supported by the Research Institute for Gastroenterology and Liver Diseases, Shahid Beheshti University of Medical Sciences (Grant Number: RIGLD-1126).
Acknowledgments
The authors thank all members of the Foodborne and Waterborne Diseases Research Center for their collaborations.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aboul-Enein, H., Stefan, R.-I., and Van Staden, J. (1999). Chemiluminescence-based (bio)sensors — an overview critical reviews in analytical chemistry. Crit. Rev. Anal. Chem. 29, 323–331. doi: 10.1080/10408349891199338
Ahmadi, A., Kabiri, S., and Omidfar, K. (2020). Advances in HbA1c Biosensor development based on field effect transistors: a review. IEEE Sens. J. 20, 8912–8921. doi: 10.1109/JSEN.2020.2987836
Ahmadi, A., Khoshfetrat, S. M., Kabiri, S., Dorraji, P. S., Larijani, B., and Omidfar, K. (2021a). Electrochemiluminescence paper-based screen-printed electrode for HbA1c detection using two-dimensional zirconium metal-organic framework/Fe(3)O(4) nanosheet composites decorated with Au nanoclusters. Mikrochim Acta 188, 296. doi: 10.1007/s00604-021-04959-y
Ahmadi, A., Khoshfetrat, S. M., Kabiri, S., Fotouhi, L., Dorraji, P. S., and Omidfar, K. (2021b). Impedimetric paper-based enzymatic biosensor using electrospun cellulose acetate nanofiber and reduced graphene oxide for detection of glucose from whole blood. IEEE Sens. J. 21, 9210–9217. doi: 10.1109/JSEN.2021.3053033
Ahmadi, A., Khoshfetrat, S. M., Mirzaeizadeh, Z., Kabiri, S., Rezaie, J., and Omidfar, K. (2022). Electrochemical immunosensor for determination of cardiac troponin I using two-dimensional metal-organic framework/Fe(3)O(4)-COOH nanosheet composites loaded with thionine and pCTAB/DES modified electrode. Talanta 237, 122911. doi: 10.1016/j.talanta.2021.122911
Ahmadi, A., Mirzaeizadeh, Z., and Omidfar, K. (2021c). Simultaneous detection of SARS-CoV-2 IgG/IgM antibodies, using gold nanoparticles-based lateral flow immunoassay. Monoclon Antibies Immunodiagn. Immunother. 40, 210–218. doi: 10.1089/mab.2021.0027
Akbarzadeh, A., Samiei, M., and Davaran, S. (2012). Magnetic nanoparticles: preparation, physical properties, and applications in biomedicine. Nanoscale Res. Lett. 7, 144. doi: 10.1186/1556-276X-7-144
Alaqad, K., and Saleh, T. A. (2016). Gold and silver nanoparticles: synthesis methods, characterization routes and applications towards drugs. J. Environ. Analyt. Toxicol. 2016, 4. doi: 10.4172/2161-0525.1000384
Ali, A., Shah, T., Ullah, R., Zhou, P., Guo, M., Ovais, M., et al. (2021). Review on recent progress in magnetic nanoparticles: synthesis, characterization, and diverse applications. Front. Chem. 9, 629054. doi: 10.3389/fchem.2021.629054
Al-Shamiri, A., Al-Zubairy, A., and Al-Mamari, R. (2010). The prevalence of Cryptosporidium spp. in children, Taiz district, Yemen. Iran J. Parasitol. 5, 26–32.
Andrade, L., O'Dwyer, J., O'Neill, E., and Hynds, P. (2018). Surface water flooding, groundwater contamination, and enteric disease in developed countries: a scoping review of connections and consequences. Environ. Pollut. 236, 540–549. doi: 10.1016/j.envpol.2018.01.104
Angus, S., Kwon, H.-J., and Yoon, J.-Y. (2012). Low-level detection of Cryptosporidium parvum in field water using optical microfluidic biosensors. Prog. Biomed. Opt. Imaging Proc. SPIE 8229, 13. doi: 10.1117/12.909618
Arlett, J. L., Myers, E. B., and Roukes, M. L. (2011). Comparative advantages of mechanical biosensors. Nat. Nanotechnol. 6, 203–215. doi: 10.1038/nnano.2011.44
Aubert, D., and Villena, I. (2009). Detection of Toxoplasma gondii oocysts in water: proposition of a strategy and evaluation in Champagne-Ardenne region, France. Mem. Inst. Oswaldo Cruz. 104, 290–295. doi: 10.1590/S0074-02762009000200023
Avant, B., Bouchard, D., Chang, X., Hsieh, H. S., Acrey, B., Han, Y., et al. (2019). Environmental fate of multiwalled carbon nanotubes and graphene oxide across different aquatic ecosystems NanoImpact 13, 1–12. doi: 10.1016/j.impact.2018.11.001
Babamiri, B., Salimi, A., and Hallaj, R. (2018). A molecularly imprinted electrochemiluminescence sensor for ultrasensitive HIV-1 gene detection using EuS nanocrystals as luminophore. Biosens. Bioelectron. 117, 332–339. doi: 10.1016/j.bios.2018.06.003
Babkina, S. S., and Budnikov, G. K. (2006). Electrochemical biosensors based on nucleic acids and their use in bioaffinity assays for determining DNA and and its effectors. J Anal Chem 61, 728–739. doi: 10.1134/S1061934806080028
Bahramdoost, Z., Mirjalali, H., Yavari, P., and Haghighi, A. (2021). Development of HRM real-time PCR for assemblage characterization of Giardia lamblia. Acta. Trop. 224, 106109. doi: 10.1016/j.actatropica.2021.106109
Baldursson, S., and Karanis, P. (2011). Waterborne transmission of protozoan parasites: review of worldwide outbreaks - an update 2004-2010. Water Res. 45, 6603–6614. doi: 10.1016/j.watres.2011.10.013
Bello, J., Núñez, F. A., González, O. M., Fernández, R., Almirall, P., and Escobedo, A. A. (2011). Risk factors for Giardia infection among hospitalized children in Cuba. Ann. Trop. Med. Parasitol. 105, 57–64. doi: 10.1179/136485911X12899838413385
Berezin, M. Y., and Achilefu, S. (2010). Fluorescence lifetime measurements and biological imaging. Chem. Rev. 110, 2641–2684. doi: 10.1021/cr900343z
Bermejo, C., Haerizadeh, F., Takanaga, H., Chermak, D., and Frommer, W. B. (2011). Optical sensors for measuring dynamic changes of cytosolic metabolite levels in yeast. Nat protoc 6, 1806–1817. doi: 10.1038/nprot.2011.391
Breslauer, D. N., Maamari, R. N., Switz, N. A., Lam, W. A., and Fletcher, D. A. (2009). Mobile phone based clinical microscopy for global health applications. PLoS One 4, e6320. doi: 10.1371/journal.pone.0006320
Bunde, R. L., Jarvi, E. J., and Rosentreter, J. J. (1998). Piezoelectric quartz crystal biosensors. Talanta 46, 1223–1236. doi: 10.1016/S0039-9140(97)00392-5
Cai, R., Yin, F., Zhang, Z., Tian, Y., and Zhou, N. (2019). Functional chimera aptamer and molecular beacon based fluorescent detection of Staphylococcus aureus with strand displacement-target recycling amplification. Anal. Chim. Acta. 1075, 128–136. doi: 10.1016/j.aca.2019.05.014
Cammann, K. (1977). Bio-sensors based on ion-selective electrodes. Fresenius' Z.Anal Chem. 287, 1–9. doi: 10.1007/BF00539519
Campbell, G., and Mutharasan, R. (2006). Piezoelectric-excited millimeter-sized cantilever (PEMC) sensors detect Bacillus anthracis at 300 spores/mL. Biosens. Bioelectron. 21, 1684–1692. doi: 10.1016/j.bios.2005.08.001
Campbell, G. A., and Mutharasan, R. (2008). Near real-time detection of Cryptosporidium parvum oocyst by IgM-functionalized piezoelectric-excited millimeter-sized cantilever biosensor. Biosens. Bioelectron. 23, 1039–1045. doi: 10.1016/j.bios.2007.10.017
Campuzano, S., Yáñez-Sedeño, P., and Pingarrón, J. M. (2017). Electrochemical genosensing of circulating biomarkers. Sensors 17, 866. doi: 10.3390/s17040866
Cesewski, E., and Johnson, B. N. (2020). Electrochemical biosensors for pathogen detection. Biosens. Bioelectron. 159, 112214–112214. doi: 10.1016/j.bios.2020.112214
Chalklen, T., Jing, Q., and Kar-Narayan, S. (2020). Biosensors based on mechanical and electrical detection techniques. Sensors 20, 605. doi: 10.3390/s20195605
Chaudhry, S. A., Gad, N., and Koren, G. (2014). Toxoplasmosis and pregnancy. Can. Fam. Physician. 60, 334–336.
Chauhan, N., Maekawa, T., and Kumar, D. N. S. (2017). Graphene based biosensors—accelerating medical diagnostics to new-dimensions. J. Material Res. 32, 2860–2882. doi: 10.1557/jmr.2017.91
Chen, C., and Wang, J. (2020). Optical biosensors: an exhaustive and comprehensive review. Analyst 145, 1605–1628. doi: 10.1039/C9AN01998G
Chin, C. D., Linder, V., and Sia, S. K. (2012). Commercialization of microfluidic point-of-care diagnostic devices. Lab Chip 12, 2118–2134. doi: 10.1039/c2lc21204h
Cho, I.-H., Kim, D. H., and Park, S. (2020). Electrochemical biosensors: perspective on functional nanomaterials for on-site analysis. Biomaterial Res. 24, 6. doi: 10.1186/s40824-019-0181-y
Cho, S. J., Woo, H. M., Kim, K. S., Oh, J. W., and Jeong, Y. J. (2011). Novel system for detecting SARS coronavirus nucleocapsid protein using an ssDNA aptamer. J. Biosci. Bioeng. 112, 535–540. doi: 10.1016/j.jbiosc.2011.08.014
Citartan, M., Gopinath, S. C. B., Tominaga, J., and Tang, T.-H. (2013). Label-free methods of reporting biomolecular interactions by optical biosensors. Analyst 138, 3576–3592. doi: 10.1039/c3an36828a
Collier, S. A., Deng, L., Adam, E. A., Benedict, K. M., Beshearse, E. M., Blackstock, A. J., et al. (2021). Estimate of burden and direct healthcare cost of infectious waterborne disease in the United States. Emerg. Infect. Dis. 27, 140–149. doi: 10.3201/eid2701.190676
Cunningham, B., and Laing, L. (2008). Advantages and application of label-free detection assays in drug screening. Expert. Opin. Drug. Discov. 3, 891–901. doi: 10.1517/17460441.3.8.891
de la Escosura-Muñiz, A., and Merkoçi, A. (2012). Nanochannels preparation and application in biosensing. ACS Nano. 6, 7556–7583. doi: 10.1021/nn301368z
Deline, A., and Nason, J. (2019). Evaluation of labeling methods used for investigating the environmental behavior and toxicity of metal oxide nanoparticles. Environ. Sci. Nano 6, 19. doi: 10.1039/C8EN01187G
Deshmukh, K., Goel, N., and Patel, B. C. (2020). Optical sensors: overview, characteristics and applications. Adv. Modern Sens. 3, 27. doi: 10.1088/978-0-7503-2707-7ch3
Destura, R. V., Cena, R. B., Galarion, M. J. H., Pangilinan, C. M., Arevalo, G. M., Alba, R. O. C., et al. (2015). Advancing Cryptosporidium diagnostics from bench to bedside. Curr. Trop. Med. Rep. 2, 150–160. doi: 10.1007/s40475-015-0055-x
Diéguez, L., Winter, M., Molan, S., Monis, P., King, B., and Thierry, B. (2018). Disposable microfluidic micromixers for effective capture of Cryptosporidium parvum oocysts from water samples. J. Biol. Eng. 12, 4–4. doi: 10.1186/s13036-018-0095-6
Ding, R., Chen, Y., Wang, Q., Wu, Z., Zhang, X., Li, B., et al. (2022). Recent advances in quantum dots-based biosensors for antibiotics detection. J. Pharm. Anal. 12, 355–364. doi: 10.1016/j.jpha.2021.08.002
Drummond, T. G., Hill, M. G., and Barton, J. K. (2003). Electrochemical DNA sensors. Nat. Biotechnol. 21, 1192–1199. doi: 10.1038/nbt873
Duchenne-Moutien, R. A., and Neetoo, H. (2021). Climate change and emerging food safety issues: a review. J. Food Prot. 84, 1884–1897. doi: 10.4315/JFP-21-141
Duguet, E., Vasseur, S., Mornet, S., and Devoisselle, J.-M. (2006). Magnetic nanoparticles and their applications in medicine. Nanomed. 1, 157–168. doi: 10.2217/17435889.1.2.157
Efstratiou, A., Ongerth, J. E., and Karanis, P. (2017). Waterborne transmission of protozoan parasites: Review of worldwide outbreaks—an update 2011–2016. Water Res. 114, 14–22. doi: 10.1016/j.watres.2017.01.036
Einarsson, E., Ma'ayeh, S., and Svärd, S. G. (2016). An up-date on Giardia and giardiasis. Curr. Opin. Microbiol. 34, 47–52. doi: 10.1016/j.mib.2016.07.019
Elgun, G., and Koltas, I. S. (2011). Investigation of Cryptosporidium spp. antigen by ELISA method in stool specimens obtained from patients with diarrhea. Parasitol. Res. 108, 395–397. doi: 10.1007/s00436-010-2079-4
Esch, M. B., Locascio, L. E., Tarlov, M. J., and Durst, R. A. (2001). Detection of Viable Cryptosporidium parvum using dna-modified liposomes in a microfluidic chip. Anal. Chem. 73, 2952–2958. doi: 10.1021/ac001508n
Faulkner, L. R., and Bard, A. J. (2002). Electrochemical Methods: Fundamentals and Applications. London: John Wiley and Sons.
Felix, F. S., and Angnes, L. (2018). Electrochemical immunosensors—a powerful tool for analytical applications. Biosens. Bioelectron. 102, 470–478. doi: 10.1016/j.bios.2017.11.029
Fernández, P., Navarro, E., Remesar, S., García Dios, D., Martinez, N., Prieto, A., et al. (2021). The age-related Cryptosporidium species distribution in asymptomatic cattle from north-western Spain. Animals 11, 256. doi: 10.3390/ani11020256
Fiorini, G. S., and Chiu, D. T. (2005). Disposable microfluidic devices: fabrication, function, and application. BioTech 38, 429–446. doi: 10.2144/05383RV02
Foroutan-Rad, M., Majidiani, H., Dalvand, S., Daryani, A., Kooti, W., Saki, J., et al. (2016). Toxoplasmosis in blood donors: a systematic review and meta-analysis. Transfus. Med. Rev. 30, 116–122. doi: 10.1016/j.tmrv.2016.03.002
Fradette, M. S., and Charette, S. J. (2022). Working toward improved monitoring of Cryptosporidium and Giardia (oo)cysts in water samples: testing alternatives to elution and immunomagnetic separation from USEPA Method 16231. BMC Res. Notes. 15, 254. doi: 10.1186/s13104-022-06118-9
Gaft, M., Reisfeld, R., and Panczer, G. (2015). Modern Luminescence Spectroscopy of Minerals and Materials. New York, NY: Springer.
Gallas-Lindemann, C., Sotiriadou, I., Mahmoodi, M. R., and Karanis, P. (2013). Detection of Toxoplasma gondii oocysts in different water resources by loop mediated isothermal amplification (LAMP). Acta. Trop. 125, 231–236. doi: 10.1016/j.actatropica.2012.10.007
Ganz, K. R., Clime, L., Farber, J. M., Corneau, N., Veres, T., and Dixon, B. R. (2015). Enhancing the detection of Giardia duodenalis cysts in foods by inertial microfluidic separation. Appl. Environ. Microbiol. 81, 3925–3933. doi: 10.1128/AEM.03868-14
Gao, H. W., Qin, P., Lin, C., Shang, Z. M., and Sun, W. (2010). Electrochemical DNA biosensor for the detection of Listeria Monocytogenes using toluidine blue as a hybridization indicator. J. Iran Chem. Soc. 7, 119–127. doi: 10.1007/BF03245868
Garibyan, L., and Avashia, N. (2013). Polymerase chain reaction. J. Invest. Dermatol. 133, 1–4. doi: 10.1038/jid.2013.1
Gerace, E., Lo Presti, V. D. M., and Biondo, C. (2019). Cryptosporidium infection: epidemiology, pathogenesis, and differential diagnosis. Eur. J. Microbiol. Immunol. (Bp) 9, 119–123. doi: 10.1556/1886.2019.00019
Ghulam, A. N., Dos Santos, O. A. L., Hazeem, L., Pizzorno Backx, B., Bououdina, M., and Bellucci, S. (2022). Graphene Oxide (GO) materials-applications and toxicity on living organisms and environment. J. Funct. Biomater. 13, 2. doi: 10.3390/jfb13020077
Grewal, Y. S., Shiddiky, M. J., Spadafora, L. J., Cangelosi, G. A., and Trau, M. (2014). Nano-yeast-scFv probes on screen-printed gold electrodes for detection of Entamoeba histolytica antigens in a biological matrix. Biosens. Bioelectron. 55, 417–422. doi: 10.1016/j.bios.2013.12.043
Gururajan, A., Rajkumari, N., Devi, U., and Borah, P. (2021). Cryptosporidium and waterborne outbreaks—a mini review. Trop. Parasitol. 11, 11–15. doi: 10.4103/tp.TP_68_20
Hayman, R. B. (2008). “Fiber optic biosensors for bacterial detection,” in Principles of Bacterial Detection: Biosensors, Recognition Receptors and Microsystems, eds Zourob, M., Elwary, S., Turner, A. (New York, NY: Springer), 125–137.
He, L., Ni, L., Zhang, X., Zhang, C., Li, R., and Xu, S. (2015). Fluorescent detection of specific DNA sequences related to Toxoplasma gondii based on magnetic fluorescent nanoparticles Fe3O4/CdTe Biosensor. Int. J. Biochem. Res. Rev. 6, 130–139. doi: 10.9734/IJBCRR/2015/15254
Hill, D., and Dubey, J. (2002). Toxoplasma gondii: transmission, diagnosis and prevention. Clin. Microbiol. Infect. 8, 634–640. doi: 10.1046/j.1469-0691.2002.00485.x
Hooshyar, H., Rostamkhani, P., Arbabi, M., and Delavari, M. (2019). Giardia lamblia infection: review of current diagnostic strategies. Gastroenterol Hepatol Bed Bench 12, 3–12.
Hosu, O., Selvolini, G., and Marrazza, G. (2018). Recent advances of immunosensors for detecting food allergens. Curr. Opin. Electrochem. 10, 149–156. doi: 10.1016/j.coelec.2018.05.022
Houssin, T., Follet, J., Follet, A., Dei-Cas, E., and Senez, V. (2010). Label-free analysis of water-polluting parasite by electrochemical impedance spectroscopy. Biosens. Bioelectron. 25, 1122–1129. doi: 10.1016/j.bios.2009.09.039
Hu, J., Feng, Y., Ong, S. L., Ng, W. J., Song, L., Tan, X., and Chu, X. (2004). Improvement of recoveries for the determination of protozoa Cryptosporidium and Giardia in water using method. J. Microbiol. Methods. 58, 321–325. doi: 10.1016/j.mimet.2004.04.013
Iqbal, A., Labib, M., Muharemagic, D., Sattar, S., Dixon, B., and Berezovski, M. (2015). Detection of Cryptosporidium parvum oocysts on fresh produce using dna aptamers. PLoS ONE 10, e0137455. doi: 10.1371/journal.pone.0137455
Janegitz, B. C., Silva, T. A., Wong, A., Ribovski, L., Vicentini, F. C., Taboada Sotomayor, M. D. P., et al. (2017). The application of graphene for in vitro and in vivo electrochemical biosensing. Biosens. Bioelectron. 89, 224–233. doi: 10.1016/j.bios.2016.03.026
Javanmard, E., Mirsamadi, E. S., Olfatifar, M., Ghasemi, E., Saki, F., Mirjalali, H., et al. (2020). Prevalence of Cryptosporidium and Giardia in vegetables in Iran: a nineteen-years meta-analysis review. J. Environ. Health Sci. Eng. 18, 1629–1641. doi: 10.1007/s40201-020-00493-w
Javanmard, E., Rahimi, H. M., Niyyati, M., Aghdaei, H. A., Sharifdini, M., Mirjalali, H., et al. (2019). Molecular analysis of Blastocystis sp. and its subtypes from treated wastewater routinely used for irrigation of vegetable farmlands in Iran. J. Water Health 17, 837–844. doi: 10.2166/wh.2019.045
Kalyoussef, S., and Feja, K. N. (2014). Foodborne illnesses. Adv. Pediatr. 61, 287–312. doi: 10.1016/j.yapd.2014.04.003
Kang, C., Lee, S., Park, T., and Sim, S. (2006). Performance enhancement of real-time detection of protozoan parasite, Cryptosporidium oocyst by a modified surface plasmon resonance (SPR) biosensor. Enzyme Microb. Technol. 39, 387–390. doi: 10.1016/j.enzmictec.2005.11.039
Kanková, S., and Flegr, J. (2007). Longer pregnancy and slower fetal development in women with latent “asymptomatic” toxoplasmosis. BMC Infect Dis. 7, 114. doi: 10.1186/1471-2334-7-114
Karanis, P., Aldeyarbi, H. M., Mirhashemi, M. E., and Khalil, K. M. (2013). The impact of the waterborne transmission of Toxoplasma gondii and analysis efforts for water detection: an overview and update. Environ. Sci. Pollut. Res. Int. 20, 86–99. doi: 10.1007/s11356-012-1177-5
Karanis, P., Kourenti, C., and Smith, H. (2006). Waterborne transmission of protozoan parasites: a worldwide review of outbreaks and lessons learnt. J. Water Health 5, 1–38. doi: 10.2166/wh.2006.002
Khodaei, R., Ahmady, A., Khoshfetrat, S. M., Kashanian, S., Tavangar, S. M., and Omidfar, K. (2019). Voltammetric immunosensor for E-cadherin promoter DNA methylation using a Fe(3)O(4)-citric acid nanocomposite and a screen-printed carbon electrode modified with poly(vinyl alcohol) and reduced graphene oxide. Mikrochim Acta. 186, 170. doi: 10.1007/s00604-019-3234-y
Khoo, V. S., Dearnaley, D. P., Finnigan, D. J., Padhani, A., Tanner, S. F., and Leach, M. O. (1997). Magnetic resonance imaging (MRI): considerations and applications in radiotherapy treatment planning. Radiother. Oncol. 42, 1–15. doi: 10.1016/S0167-8140(96)01866-X
Khoshfetrat, S. M., Seyed Dorraji, P., Shayan, M., Khatami, F., and Omidfar, K. (2022). Smartphone-based electrochemiluminescence for visual simultaneous detection of rassf1a and slc5a8 tumor suppressor gene methylation in thyroid cancer patient plasma. Analyt Chem. 94, 8005–8013. doi: 10.1021/acs.analchem.2c01132
Khurana, S., and Chaudhary, P. (2018). Laboratory diagnosis of cryptosporidiosis. Trop. Parasitol. 8, 2–7. doi: 10.4103/tp.TP_34_17
Kim, H. T., Jin, E., and Lee, M.-H. (2021). Portable chemiluminescence-based lateral flow assay platform for the detection of cortisol in human serum. Biosensors. 11, 21. doi: 10.3390/bios11060191
Kinyua Muthuri, L., Nagy, L., and Nagy, G. (2021). Chronopotentiometric method for assessing antioxidant activity: a reagentless measuring technique. Electrochem. Commun. 122, 106907. doi: 10.1016/j.elecom.2020.106907
Kokkinos, C., Economou, A., and Prodromidis, M. I. (2016). Electrochemical immunosensors: critical survey of different architectures and transduction strategies. TrAC Trends Anal. Chem. 79, 88–105. doi: 10.1016/j.trac.2015.11.020
Kramer, M. F., Vesey, G., Look, N. L., Herbert, B. R., Simpson-Stroot, J. M., and Lim, D. V. (2007). Development of a Cryptosporidium oocyst assay using an automated fiber optic-based biosensor. J. Biol. Eng. 1, 3–3. doi: 10.1186/1754-1611-1-3
Kudr, J., Haddad, Y., Richtera, L., Heger, Z., Cernak, M., Adam, V., et al. (2017). Magnetic nanoparticles: from design and synthesis to real world applications. Nanomaterials (Basel). 7, 243. doi: 10.3390/nano7090243
Laczka, O., Skillman, L., Ditcham, W. G., Hamdorf, B., Wong, D. K., Bergquist, P., et al. (2013). Application of an ELISA-type screen printed electrode-based potentiometric assay to the detection of Cryptosporidium parvum oocysts. J. Microbiol. Methods. 95, 182–185. doi: 10.1016/j.mimet.2013.08.014
Land, K. J., Boeras, D. I., Chen, X.-S., Ramsay, A. R., and Peeling, R. W. (2019). REASSURED diagnostics to inform disease control strategies, strengthen health systems and improve patient outcomes. Nat. Microb. 4, 46–54. doi: 10.1038/s41564-018-0295-3
Laude, A., Valot, S., Desoubeaux, G., Argy, N., Nourrisson, C., Pomares, C., et al. (2016). Is real-time PCR-based diagnosis similar in performance to routine parasitological examination for the identification of Giardia intestinalis, Cryptosporidium parvum/Cryptosporidium hominis and Entamoeba histolytica from stool samples? Evaluation of a new commercial multiplex PCR assay and literature review. Clin. Microbiol. Infect. 22, 190. doi: 10.1016/j.cmi.2015.10.019
Lavania, S., Das, R., Dhiman, A., Myneedu, V. P., Verma, A., Singh, N., et al. (2018). Aptamer-based tb antigen tests for the rapid diagnosis of pulmonary tuberculosis: potential utility in screening for tuberculosis. ACS Infect Dis. 4, 1718–1726. doi: 10.1021/acsinfecdis.8b00201
Lay, C., Teo, C. Y., Zhu, L., Peh, X. L., Ji, H. M., Chew, B. R., et al. (2008). Enhanced microfiltration devices configured with hydrodynamic trapping and a rain drop bypass filtering architecture for microbial cells detection. Lab Chip. 8, 830–833. doi: 10.1039/b800015h
Lee, J., Kim, J., Kim, S., and Min, D. H. (2016). Biosensors based on graphene oxide and its biomedical application. Adv Drug Deliv Rev. 105, 275–287. doi: 10.1016/j.addr.2016.06.001
Leitch, G. J., and He, Q. (2012). Cryptosporidiosis-an overview. J. Biomed. Res. 25, 1–16. doi: 10.1016/S1674-8301(11)60001-8
Li, H. Y., Jia, W. N., Li, X. Y., Zhang, L., Liu, C., and Wu, J. (2020). Advances in detection of infectious agents by aptamer-based technologies. Emerg. Microbes. Infect. 9, 1671–1681. doi: 10.1080/22221751.2020.1792352
Li, L., Li, Q., Liao, Z., Sun, Y., Cheng, Q., Song, Y., et al. (2018). Magnetism-resolved separation and fluorescence quantification for near-simultaneous detection of multiple pathogens. Anal. Chem. 90, 9621–9628. doi: 10.1021/acs.analchem.8b02572
Lim, H. J., Saha, T., Tey, B. T., Tan, W. S., and Ooi, C. W. (2020). Quartz crystal microbalance-based biosensors as rapid diagnostic devices for infectious diseases. Biosens. Bioelectron. 168, 112513–112513. doi: 10.1016/j.bios.2020.112513
Lingane, P. J., and Peters, D. G. (1971). Chronopotentiometry. C R C Crit. Rev. Anal. Chem. 1, 587–634. doi: 10.1080/10408347108085644
Liu, Q., Zhang, W., Chen, S., Zhuang, Z., Zhang, Y., Jiang, L., et al. (2020). SELEX tool: a novel and convenient gel-based diffusion method for monitoring of aptamer-target binding. J. Biol. Eng. 14, 1. doi: 10.1186/s13036-019-0223-y
Low, K. F., Chuenrangsikul, K., Rijiravanich, P., Surareungchai, W., and Chan, Y. Y. (2012). Electrochemical genosensor for specific detection of the food-borne pathogen, Vibrio cholerae. World J. Microbiol. Biotechnol. 28, 1699–1706. doi: 10.1007/s11274-011-0978-x
Luka, G., Samiei, E., Dehghani, S., Johnson, T., Najjaran, H., and Hoorfar, M. (2019). Label-free capacitive biosensor for detection of Cryptosporidium. Sensors (Basel) 19, 21. doi: 10.3390/s19020258
Luka, G. S., Najjaran, H., and Hoorfar, M. (2022). On-chip-based electrochemical biosensor for the sensitive and label-free detection of Cryptosporidium. Sci. Rep. 12, 6957. doi: 10.1038/s41598-022-10765-0
Luka, G. S., Nowak, E., Toyata, Q. R., Tasnim, N., Najjaran, H., and Hoorfar, M. (2021). Portable on-chip colorimetric biosensing platform integrated with a smartphone for label/PCR-free detection of Cryptosporidium RNA. Sci. Rep. 11, 23192. doi: 10.1038/s41598-021-02580-w
Luo, X., Morrin, A., Killard, A. J., and Smyth, M. R. (2006). Application of nanoparticles in electrochemical sensors and biosensors. Electroanal. Int. J. Devot. Fundam. Pract. Aspect Electroanalys. 18, 319–326. doi: 10.1002/elan.200503415
Ma, J.-Y., Li, M.-Y., Qi, Z.-Z., Fu, M., Sun, T.-F., Elsheikha, H. M., et al. (2022). Waterborne protozoan outbreaks: An update on the global, regional, and national prevalence from 2017 to 2020 and sources of contamination. Sci. Total Environ. 806, 150562. doi: 10.1016/j.scitotenv.2021.150562
Ma, Y., Jiao, K., Yang, T., and Sun, D. (2008). Sensitive PAT gene sequence detection by nano-SiO2/p-aminothiophenol self-assembled films DNA electrochemical biosensor based on impedance measurement. Sens. Actuator B: Chem. 131, 565–571. doi: 10.1016/j.snb.2007.12.046
Magar, H. S., Hassan, R. Y. A., and Mulchandani, A. (2021). Electrochemical impedance spectroscopy (eis): principles, construction, and biosensing applications. Sensors (Basel). 21, 12. doi: 10.3390/s21196578
Mahdavi Abhari, F., Niyyati, M., Assadzadeh Aghdaei, H., and Mirjalali, H. (2023). Loop mediated isothermal amplification for detection of foodborne parasites: a journey from lab to lab-on-a-chip. Food Control. 143, 109251. doi: 10.1016/j.foodcont.2022.109251
Mahmoudi, T., Naghdi, T., Morales-Narváez, E., and Golmohammadi, H. (2022). Toward smart diagnosis of pandemic infectious diseases using wastewater-based epidemiology. TrAC Trend. Anal. Chem. 153, 116635. doi: 10.1016/j.trac.2022.116635
Malik, P., Katyal, V., Malik, V., Asatkar, A., Inwati, G., and Mukherjee, T. K. (2013). Nanobiosensors: concepts and variations. ISRN Nanomater. 2013, 327435. doi: 10.1155/2013/327435
Martín, M. E., García-Hernández, M., García-Recio, E. M., Gómez-Chacón, G. F., Sánchez-López, M., and González, V. M. (2013). DNA aptamers selectively target Leishmania infantum h2a protein. PLOS ONE. 8, e78886. doi: 10.1371/journal.pone.0078886
McGrath, J. S., Honrado, C., Spencer, D., Horton, B., Bridle, H. L., and Morgan, H. (2017). Analysis of parasitic protozoa at the single-cell level using microfluidic impedance cytometry. Sci. Rep. 7, 2601. doi: 10.1038/s41598-017-02715-y
Mehrotra, P. (2016). Biosensors and their applications—a review. J. Oral. Biol. Craniofac. Res. 6, 153–159. doi: 10.1016/j.jobcr.2015.12.002
Mendez, O. A., and Koshy, A. A. (2017). Toxoplasma gondii: Entry, association, and physiological influence on the central nervous system. PLoS pathog. 13, e1006351. doi: 10.1371/journal.ppat.1006351
Metkar, S., and Girigoswami, K. (2018). Diagnostic biosensors in medicine- a review. Biocatal. Agric. Biotechnol. 17, 32. doi: 10.1016/j.bcab.2018.11.029
Minuzzi, C. E., Fernandes, F. D., Portella, L. P., Bräunig, P., Sturza, D. A. F., Giacomini, L., et al. (2021). Contaminated water confirmed as source of infection by bioassay in an outbreak of toxoplasmosis in South Brazil. Transbound. Emerg. Dis. 68, 767–772. doi: 10.1111/tbed.13741
Mmbaga, B. T., and Houpt, E. R. (2017). Cryptosporidium and Giardia infections in children: a review. Pediatr. Clin. North Am. 64, 837–850. doi: 10.1016/j.pcl.2017.03.014
Mohammad Rahimi, H., Pourhosseingholi, M. A., Yadegar, A., Mirjalali, H., and Zali, M. R. (2019). High-resolution melt curve analysis: A real-time based multipurpose approach for diagnosis and epidemiological investigations of parasitic infections. Comp. Immunol. Microbiol. Infect. Dis. 67, 101364. doi: 10.1016/j.cimid.2019.101364
Mohammad Rahimi, H., Soleimani Jevinani, S., Nemati, S., Sharifdini, M., Mirjalali, H., and Zali, M. R. (2022). Molecular characterization of Cryptosporidium skunk genotype in raccoons (Procyon lotor) in Iran: concern for zoonotic transmission. Parasitol. Res. 121, 483–489. doi: 10.1007/s00436-021-07367-6
Mohammed, A. M., Ibraheem, I. J., Obaid, A. S., and Bououdina, M. (2017). Nanostructured ZnO-based biosensor: DNA immobilization and hybridization. Sens. Bio-Sens. Res. 15, 46–52. doi: 10.1016/j.sbsr.2017.07.003
Monosik, R., Stredansky, M., Tkac, J., and Sturdik, E. (2012). Application of enzyme biosensors in analysis of food and beverages. Food Anal. Methods. 5, 40–53. doi: 10.1007/s12161-011-9222-4
Mora, C., McKenzie, T., Gaw, I. M., Dean, J. M., von Hammerstein, H., Knudson, T. A., et al. (2022). Over half of known human pathogenic diseases can be aggravated by climate change. Nat. Clim. Chang. 12, 869–875. doi: 10.1038/s41558-022-01426-1
Morales-Narváez, E., Baptista-Pires, L., Zamora-Gálvez, A., and Merkoçi, A. (2017). Graphene-based biosensors: going simple. Adv Mater. 29, 7. doi: 10.1002/adma.201604905
Moreno-Mesonero, L., Amorós, I., Moreno, Y., and Alonso, J. L. (2022). Simultaneous detection of less frequent waterborne parasitic protozoa in reused wastewater using amplicon sequencing and qPCR techniques. J. Environ. Manage 314, 115029. doi: 10.1016/j.jenvman.2022.115029
Mudanyali, O., Oztoprak, C., Tseng, D., Erlinger, A., and Ozcan, A. (2010). Detection of waterborne parasites using field-portable and cost-effective lensfree microscopy. Lab. Chip. 10, 2419–2423. doi: 10.1039/c004829a
Nagarkatti, R., Bist, V., Sun, S., Fortes de Araujo, F., Nakhasi, H. L., and Debrabant, A. (2012). Development of an aptamer-based concentration method for the detection of Trypanosoma cruzi in blood. PLoS One. 7, e43533. doi: 10.1371/journal.pone.0043533
Narayanaswamy, R. (2006). Optical Chemical Sensors and Biosensors for Food Safety and Security Applications.
Naresh, V., and Lee, N. (2021). A Review on biosensors and recent development of nanostructured materials-enabled biosensors. Sensors. 7, 21. doi: 10.3390/s21041109
Ngowi, H. A. (2020). Prevalence and pattern of waterborne parasitic infections in eastern Africa: a systematic scoping review. Food Waterborne Parasitol. 20, e00089. doi: 10.1016/j.fawpar.2020.e00089
Nicu, L., Guirardel, M., Chambosse, F., Rougerie, P., Hinh, S., Trévisiol, E., et al. (2005). Resonating piezoelectric membranes for microelectromechanically based bioassay: detection of streptavidin–gold nanoparticles interaction with biotinylated DNA. Sens. Actuat. B: Chem. 110, 125–136. doi: 10.1016/j.snb.2005.01.021
Nugen, S. R., Asiello, P. J., Connelly, J. T., and Baeumner, A. J. (2009). PMMA biosensor for nucleic acids with integrated mixer and electrochemical detection. Biosens. Bioelectron. 24, 2428–2433. doi: 10.1016/j.bios.2008.12.025
Nygård, K., Schimmer, B., Søbstad, Ø., Walde, A., Tveit, I., Langeland, N., et al. (2006). A large community outbreak of waterborne giardiasis-delayed detection in a non-endemic urban area. BMC Public Health 6, 141. doi: 10.1186/1471-2458-6-141
Olaru, A., Bala, C., Jaffrezic-Renault, N., and Aboul-Enein, H. Y. (2015). Surface plasmon resonance (SPR) biosensors in pharmaceutical analysis. Crit. Rev. Anal. Chem. 45, 97–105. doi: 10.1080/10408347.2014.881250
Omidfar, K., Ahmadi, A., Syedmoradi, L., Khoshfetrat, S. M., and Larijani, B. (2020). Point-of-care biosensors in medicine: a brief overview of our achievements in this field based on the conducted research in EMRI (endocrinology and metabolism research Institute of Tehran University of medical sciences) over the past fourteen years. J. Diabet. Metab. Disord. 2020, 1-5. doi: 10.1007/s40200-020-00668-0
O'Sullivan, C. K., and Guilbault, G. G. (2000). “Piezoelectric immunosensors: theory and applications,” in Biosensors and Their Applications, eds Yang, V. C., Ngo, T. T. (Boston, MA: Springer), pp. 159–173.
Ozdemir, M. S., Marczak, M., Bohets, H., Bonroy, K., Roymans, D., Stuyver, L., et al. (2013). A label-free potentiometric sensor principle for the detection of antibody–antigen interactions. Anal. Chem. 85, 4770–4776. doi: 10.1021/ac400514u
Pandey, R. R., and Chusuei, C. C. (2021). Carbon Nanotubes, Graphene, and carbon dots as electrochemical biosensing composites. Molecules 26, 21. doi: 10.3390/molecules26216674
Park, C. K., Kang, C. D., and Sim, S. J. (2008). Non-labeled detection of waterborne pathogen Cryptosporidium parvum using a polydiacetylene-based fluorescence chip. Biotechnol. J. 3, 687–693. doi: 10.1002/biot.200700246
Pazoki, H., Niyyati, M., Javanmard, E., Lasjerdi, Z., Spotin, A., Mirjalali, H., et al. (2020). Isolation of N. philippinensis and N. americana strains from irrigation waters of farmland soils in Iran. Environ. Sci. Pollut. Res. Int. 27, 24568–24573. doi: 10.1007/s11356-020-08992-x
Pejcic, B., Marco, R. D., and Parkinson, G. (2006). The role of biosensors in the detection of emerging infectious diseases. Analyst. 131, 1079–1090. doi: 10.1039/b603402k
Plutzer, J., and Karanis, P. (2016). Neglected waterborne parasitic protozoa and their detection in water. Water. Res. 101, 318–332. doi: 10.1016/j.watres.2016.05.085
Poitras, C., Fatisson, J., and Tufenkji, N. (2009). Real-time microgravimetric quantification of Cryptosporidium parvum in the presence of potential interferents. Water Res. 43, 2631–2638. doi: 10.1016/j.watres.2009.03.021
Proll, G., Steinle, L., Pröll, F., Kumpf, M., Moehrle, B., Mehlmann, M., et al. (2007). Potential of label-free detection in high-content-screening applications. J. Chromatogr. A. 1161, 2–8. doi: 10.1016/j.chroma.2007.06.022
Rajchgot, J., Coulibaly, J. T., Keiser, J., Utzinger, J., Lo, N. C., Mondry, M. K., et al. (2017). Mobile-phone and handheld microscopy for neglected tropical diseases. PLoS Negl. Trop. Dis. 11, e0005550. doi: 10.1371/journal.pntd.0005550
Ramo, A., Del Cacho, E., Sánchez-Acedo, C., and Quílez, J. (2017). Occurrence and genetic diversity of Cryptosporidium and Giardia in urban wastewater treatment plants in north-eastern Spain. Sci. Total Environ. 598, 628–638. doi: 10.1016/j.scitotenv.2017.04.097
Ranjbar Bahadori, S., Mulgaonkar, A., Hart, R., Wu, C.-Y., Zhang, D., Pillai, A., et al. (2021). Radiolabeling strategies and pharmacokinetic studies for metal based nanotheranostics. WIREs Nanomed. Nanobiotechnol. 13, e1671. doi: 10.1002/wnan.1671
Reddy, L. H., Arias, J. L., Nicolas, J., and Couvreur, P. (2012). Magnetic nanoparticles: design and characterization, toxicity and biocompatibility, pharmaceutical and biomedical applications. Chem. Rev. 112, 5818–5878. doi: 10.1021/cr300068p
Resi, D., Varani, S., Sannella, A. R., De Pascali, A. M., Ortalli, M., Liguori, G., et al. (2021). A large outbreak of giardiasis in a municipality of the Bologna province, north-eastern Italy, November 2018 to April 2019. Euro. Surveill. 21, 26. doi: 10.2807/1560-7917.ES.2021.26.35.2001331
Rhouati, A., Catanante, G., Nunes, G., Hayat, A., and Marty, J. (2016). Label-free aptasensors for the detection of mycotoxins. Sensors. 16, 2178. doi: 10.3390/s16122178
Ricciardi, A., and Ndao, M. (2015). Diagnosis of parasitic infections:what's going on? J. Biomol. Screen. 20, 6–21. doi: 10.1177/1087057114548065
Robbins, J. R., Zeldovich, V. B., Poukchanski, A., Boothroyd, J. C., and Bakardjiev, A. I. (2012). Tissue barriers of the human placenta to infection with Toxoplasma gondii. Infect. Immun. 80, 418–428. doi: 10.1128/IAI.05899-11
Robert-Gangneux, F., and Dardé, M. L. (2012). Epidemiology of and diagnostic strategies for toxoplasmosis. Clin. Microbiol. Rev. 25, 264–296. doi: 10.1128/CMR.05013-11
Ronkainen, N. J., Halsall, H. B., and Heineman, W. R. (2010). Electrochemical biosensors. Chem. Soc. Rev. 39, 1747–1763. doi: 10.1039/b714449k
Rostami, A., Karanis, P., and Fallahi, S. (2018). Advances in serological, imaging techniques and molecular diagnosis of Toxoplasma gondii infection. Infection. 46, 303–315. doi: 10.1007/s15010-017-1111-3
Saeidi, M., Amidian, M. A., Sheybanikashani, S., Mahdavi, H., Alimohammadi, H., Syedmoradi, L., et al. (2022). Multilayered mesoporous composite nanostructures for highly sensitive label-free quantification of cardiac troponin-I. Biosensors (Basel) 12, 21. doi: 10.3390/bios12050337
Saha, K., Agasti, S. S., Kim, C., Li, X., and Rotello, V. M. (2012). Gold nanoparticles in chemical and biological sensing. Chem. Rev. 112, 2739–2779. doi: 10.1021/cr2001178
Sakamoto, S., Putalun, W., Vimolmangkang, S., Phoolcharoen, W., Shoyama, Y., Tanaka, H., et al. (2018). Enzyme-linked immunosorbent assay for the quantitative/qualitative analysis of plant secondary metabolites. J. Nat. Med. 72, 32–42. doi: 10.1007/s11418-017-1144-z
Saleem, H., and Zaidi, S. J. (2020). Developments in the application of nanomaterials for water treatment and their impact on the environment. Nanomaterials (Basel). 10, 9. doi: 10.3390/nano10091764
Salouti, M., and Khadivi derakhshan, F. (2020). Biosensors and Nanobiosensors in Environmental Applications, pp. 515–591.
Sarkari, B., Hosseini, G., Motazedian, M. H., Fararouei, M., and Moshfe, A. (2016). Prevalence and risk factors of intestinal protozoan infections: a population-based study in rural areas of Boyer-Ahmad district, Southwestern Iran. BMC Infect. Dis. 16, 703. doi: 10.1186/s12879-016-2047-4
Saxena, S., and Srivastava, A. K. (2020). Nano-Optics. Thomas, S., Grohens, Y., Vignaud, G., Kalarikkal, N. and James, J. (eds), (London: Elsevier), pp. 265-291.
Schöning, M., and Poghossian, A. (2018). “Advanced materials, devices and applications,” in Label-free Biosensing, eds Schöning, M., Poghossian, A. (Cham: Springer).
Shamah, S. M., Healy, J. M., and Cload, S. T. (2008). Complex target SELEX. Acc. Chem. Res. 41, 130–138. doi: 10.1021/ar700142z
Sharma, H., and Mutharasan, R. (2013). Review of biosensors for foodborne pathogens and toxins. Sens. Actuat. B: Chem. 183, 535–549. doi: 10.1016/j.snb.2013.03.137
Shiohara, A., Wojnilowicz, M., Lyu, Q., Pei, Y., Easton, C. D., Chen, Y., et al. (2022). SARS-CoV-2 virus detection via a polymeric nanochannel-based electrochemical biosensor. Small. 22, e2205281. doi: 10.1002/smll.202205281
Shrestha, R., Duwal, R., Wagle, S., Pokhrel, S., Giri, B., and Neupane, B. B. (2020). A smartphone microscopic method for simultaneous detection of (oo)cysts of Cryptosporidium and Giardia. PLoS Negl. Trop. Dis. 14, e0008560–e0008560. doi: 10.1371/journal.pntd.0008560
Sin, M. L. Y., Mach, K. E., Wong, P. K., and Liao, J. C. (2014). Advances and challenges in biosensor-based diagnosis of infectious diseases. Expert. Rev. Mol. Diagn. 14, 225–244. doi: 10.1586/14737159.2014.888313
Singh, N. K., Jain, P., Das, S., and Goswami, P. (2019). Dye coupled aptamer-captured enzyme catalyzed reaction for detection of pan malaria and P. falciparum species in laboratory settings and instrument-free paper-based platform. Anal Chem. 91, 4213–4221. doi: 10.1021/acs.analchem.9b00670
Skládal, P. (2016). Piezoelectric biosensors. TrAC Trends Analyt Chem. 79, 127–133. doi: 10.1016/j.trac.2015.12.009
Sroka, J., Stojecki, K., Zdybel, J., Karamon, J., Cencek, T., and Dutkiewicz, J. (2013). Occurrence of Cryptosporidium oocysts and Giardia cysts in effluent from sewage treatment plant from eastern Poland. Ann. Agric. Environ. Med. Spec. 1, 57–62.
Stankic, S., Suman, S., Haque, F., and Vidic, J. (2016). Pure and multi metal oxide nanoparticles: synthesis, antibacterial and cytotoxic properties. J. 14, 73. doi: 10.1186/s12951-016-0225-6
Stone, H. A., Stroock, A. D., and Ajdari, A. (2004). Engineering flows in small devices: microfluidics toward a lab-on-a-chip. Annu. Rev. Fluid Mech. 36, 381–411. doi: 10.1146/annurev.fluid.36.050802.122124
Suh, S. H., Choi, S. J., Dwivedi, H. P., Moore, M. D., Escudero-Abarca, B. I., and Jaykus, L. A. (2018). Use of DNA aptamer for sandwich type detection of Listeria monocytogenes. Anal. Biochem. 557, 27–33. doi: 10.1016/j.ab.2018.04.009
Sun, X., Yasui, T., Yanagida, T., Kaji, N., Rahong, S., Kanai, M., et al. (2016). Identifying DNA methylation in a nanochannel. Sci. Technol. Adv. Mater. 17, 644–649. doi: 10.1080/14686996.2016.1223516
Syedmoradi, L., Daneshpour, M., Alvandipour, M., Gomez, F. A., Hajghassem, H., and Omidfar, K. (2017). Point of care testing: the impact of nanotechnology. Biosens. Bioelectron. 87, 373–387. doi: 10.1016/j.bios.2016.08.084
Syedmoradi, L., Norton, M. L., and Omidfar, K. (2021). Point-of-care cancer diagnostic devices: from academic research to clinical translation. Talanta. 225, 122002. doi: 10.1016/j.talanta.2020.122002
Taguchi, T., Arakaki, A., Takeyama, H., Haraguchi, S., Yoshino, M., Kaneko, M., et al. (2007). Detection of Cryptosporidium parvum oocysts using a microfluidic device equipped with the SUS micromesh and FITC-labeled antibody. Biotechnol. Bioeng. 96, 272–280. doi: 10.1002/bit.21104
Taguchi, T., Takeyama, H., and Matsunaga, T. (2005). Immuno-capture of Cryptosporidium parvum using micro-well array. Biosens. Bioelectron. 20, 2276–2282. doi: 10.1016/j.bios.2004.10.017
Tang, D., Yuan, R., Chai, Y., and Fu, Y. (2005). Study on electrochemical behavior of a diphtheria immunosensor based on silica/silver/gold nanoparticles and polyvinyl butyral as matrices. Electrochem. Commun. 7, 177–182. doi: 10.1016/j.elecom.2004.12.004
Tang, Y., Zeng, X., and Liang, J. (2010). Surface plasmon resonance: an introduction to a surface spectroscopy technique. J. Chem. Educ. 87, 742–746. doi: 10.1021/ed100186y
Tavares, R., Staggemeier, R., Borges, A., Rodrigues, M., Castelan, L., Vasconcelos, J., et al. (2011). Molecular techniques for the study and diagnosis of parasite infection. J. Venomous Anim. Toxin Trop. Dis. 17, 239–248. doi: 10.1590/S1678-91992011000300003
Tenter, A. M., Heckeroth, A. R., and Weiss, L. M. (2000). Toxoplasma gondii: from animals to humans. Int. J. Parasitol. 30, 1217–1258. doi: 10.1016/S0020-7519(00)00124-7
Terry, L. A., White, S. F., and Tigwell, L. J. (2005). The application of biosensors to fresh produce and the wider food industry. J. Agric. Food Chem. 53, 1309–1316. doi: 10.1021/jf040319t
Thevenot, D. R., Toth, K., Durst, R. A., and Wilson, G. S. (1999). Electrochemical biosensors: recommended definitions and classification. Pure Appl. Chem. 71, 2333–2348. doi: 10.1351/pac199971122333
Titcomb, G., Mantas, J. N., Hulke, J., Rodriguez, I., Branch, D., and Young, H. (2021). Water sources aggregate parasites with increasing effects in more arid conditions. Nat. Commun. 12, 7066. doi: 10.1038/s41467-021-27352-y
Vasilescu, A., and Marty, J.-L. (2016). Electrochemical aptasensors for the assessment of food quality and safety. TrAC Trends Anal. Chem. 79, 60–70. doi: 10.1016/j.trac.2015.11.024
Verant, M. L., d'Ozouville, N., Parker, P. G., Shapiro, K., VanWormer, E., and Deem, S. L. (2014). Attempted detection of Toxoplasma gondii oocysts in environmental waters using a simple approach to evaluate the potential for waterborne transmission in the Galápagos Islands, Ecuador. Ecohealth. 11, 207–214. doi: 10.1007/s10393-013-0888-5
Villena, I., Aubert, D., Gomis, P., Ferté, H., Inglard, J. C., Denis-Bisiaux, H., et al. (2004). Evaluation of a strategy for Toxoplasma gondii oocyst detection in water. Appl. Environ. Microbiol. 70, 4035–4039. doi: 10.1128/AEM.70.7.4035-4039.2004
Vukojević, V., Djurdjić, S., Ognjanović, M., Fabián, M., Samphao, A., Kalcher, K., et al. (2018). Enzymatic glucose biosensor based on manganese dioxide nanoparticles decorated on graphene nanoribbons. J. Electroanal. Chem. 823, 610–616. doi: 10.1016/j.jelechem.2018.07.013
Wang, H., Zhang, Y., Yu, H., Wu, D., Ma, H., Li, H., et al. (2013). Label-free electrochemical immunosensor for prostate-specific antigen based on silver hybridized mesoporous silica nanoparticles. Anal. Biochem. 434, 123–127. doi: 10.1016/j.ab.2012.11.012
Wang, J., Ali, Z., Si, J., Wang, N., He, N., and Li, Z. (2017). Simultaneous extraction of DNA and RNA from hepatocellular carcinoma (Hep G2) based on silica-coated magnetic nanoparticles. J. Nanosci. Nanotechnol. 17, 802–806. doi: 10.1166/jnn.2017.12442
Wang, J., Rivas, G., Parrado, C., Cai, X., and Flair, M. N. (1997). Electrochemical biosensor for detecting DNA sequences from the pathogenic protozoan Cryptosporidium parvum. Talanta. 44, 2003–2010. doi: 10.1016/S0039-9140(96)02191-1
Wang, W., Lei, C., Li, J., Wu, Z., Shen, G., and Yu, R. (2004). A piezoelectric immunoagglutination assay for Toxoplasma gondii antibodies using gold nanoparticles. Biosens. Bioelectron. 19, 701–709. doi: 10.1016/S0956-5663(03)00265-3
Wang, Y., Montana, V., Grubišić, V., Stout, R. F. Jr, Parpura, V., and Gu, L. Q. (2015). Nanopore sensing of botulinum toxin type B by discriminating an enzymatically cleaved peptide from a synaptic protein synaptobrevin 2 derivative. ACS Appl. Mater. Interfaces. 7, 184–192. doi: 10.1021/am5056596
Wei, X., Zhou, W., Sanjay, S. T., Zhang, J., Jin, Q., Xu, F., et al. (2018). Multiplexed instrument-free bar-chart spinchip integrated with nanoparticle-mediated magnetic aptasensors for visual quantitative detection of multiple pathogens. Anal. Chem. 90, 9888–9896. doi: 10.1021/acs.analchem.8b02055
WHO/UNICEF (2014). Progress on Drinking Water and Sanitation: Update. (Geneva: World Health Organization), pp. 1–78.
Woolley, A. T., and Mathies, R. A. (1994). Ultra-high-speed DNA fragment separations using microfabricated capillary array electrophoresis chips. Proc. Natl. Acad. Sci. 91, 11348. doi: 10.1073/pnas.91.24.11348
Wu, W., Wu, Z., Yu, T., Jiang, C., and Kim, W. S. (2015). Recent progress on magnetic iron oxide nanoparticles: synthesis, surface functional strategies and biomedical applications. Sci. Technol. Adv. Mater. 16, 023501doi: 10.1088/1468-6996/16/2/023501
Wu, Y., Ma, Y., Zheng, H., and Ramakrishna, S. (2021). Piezoelectric materials for flexible and wearable electronics: a review. Mater. Des. 211, 110164. doi: 10.1016/j.matdes.2021.110164
Xi, Z., Gong, Q., Wang, C., and Zheng, B. (2018). Highly sensitive chemiluminescent aptasensor for detecting HBV infection based on rapid magnetic separation and double-functionalized gold nanoparticles. Sci. Rep. 8, 9444. doi: 10.1038/s41598-018-27792-5
Xu, J., Bertke, M., Wasisto, H. S., and Peiner, E. (2019). Piezoresistive microcantilevers for humidity sensing. J. Micromech. Microeng. 29, 053003. doi: 10.1088/1361-6439/ab0cf5
Xu, S., and Mutharasan, R. (2010). Rapid and Sensitive Detection of Giardia lamblia using a piezoelectric cantilever biosensor in finished and source waters. Environ. Sci. Technol. 44, 1736–1741. doi: 10.1021/es9033843
Xu, S., Zhang, C., He, L., Wang, T., Ni, L., Sun, M., et al. (2013). DNA Detection of Toxoplasma gondii with a magnetic molecular beacon probe via CdTe@Ni quantum dots as energy donor. J. Nanomater. 2013, 473703. doi: 10.1155/2013/473703
Yan, X., Shu, Q., Zhao, K., Xiao, Y., Ai, F., and Zheng, X. (2021). Chemiluminescence “signal-on-off” dual signals ratio biosensor based on single-stranded DNA functions as guy wires to detect EcoR V. Talanta. 235, 122749. doi: 10.1016/j.talanta.2021.122749
Zahedi, A., Monis, P., Deere, D., and Ryan, U. (2021). Wastewater-based epidemiology—surveillance and early detection of waterborne pathogens with a focus on SARS-CoV-2, Cryptosporidium and Giardia. Parasitol. Res. 120, 4167–4188. doi: 10.1007/s00436-020-07023-5
Zahedi, A., and Ryan, U. (2020). Cryptosporidium—an update with an emphasis on foodborne and waterborne transmission. Res. Vet. Sci. 132, 500–512. doi: 10.1016/j.rvsc.2020.08.002
Zaporotskova, I. V., Boroznina, N. P., Parkhomenko, Y. N., and Kozhitov, L. V. (2016). Carbon nanotubes: sensor properties. a review. Modern Electron. Mater. 2, 95–105. doi: 10.1016/j.moem.2017.02.002
Zeng, Y., Zhou, J., Sang, W., Kong, W., Qu, J., Ho, H.-P., et al. (2021). High-sensitive surface plasmon resonance imaging biosensor based on dual-wavelength differential method. Front. Chem. 9, 21. doi: 10.3389/fchem.2021.801355
Zhang, X., and Hoshino, K. (2019). Mechanical transducers: Cantilevers, acoustic wave sensors, and thermal sensors, pp. 311–412.
Zhu, L., Zhang, Q., Feng, H., Ang, S., Chau, F. S., and Liu, W. T. (2004). Filter-based microfluidic device as a platform for immunofluorescent assay of microbial cells. Lab. Chip. 4, 337–341. doi: 10.1039/b401834f
Zou, X., Wu, J., Gu, J., Shen, L., and Mao, L. (2019). Application of aptamers in virus detection and antiviral therapy. Front. Microbiol. 9, 10. doi: 10.3389/fmicb.2019.01462
Keywords: waterborne protozoa, point-of-care, biosensors, microfluidic, smartphone
Citation: Nemati S, Shalileh F, Mirjalali H and Omidfar K (2023) Toward waterborne protozoa detection using sensing technologies. Front. Microbiol. 14:1118164. doi: 10.3389/fmicb.2023.1118164
Received: 07 December 2022; Accepted: 30 January 2023;
Published: 24 February 2023.
Edited by:
Ons Bouchami, Universidade Nova de Lisboa, PortugalReviewed by:
Jess Vergis, Kerala Veterinary and Animal Sciences University, IndiaMatthew D. Moore, University of Massachusetts Amherst, United States
Mohamed Fethi Diouani, Institut Pasteur de Tunis, Tunisia
Laura Cerqueira, University of Porto, Portugal
Copyright © 2023 Nemati, Shalileh, Mirjalali and Omidfar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hamed Mirjalali, 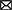 aGFtZWRtaXJqYWxhbGlAc2JtdS5hYy5pcg==; Kobra Omidfar,
aGFtZWRtaXJqYWxhbGlAc2JtdS5hYy5pcg==; Kobra Omidfar, 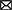 b21pZGZhckB0dW1zLmFjLmly
b21pZGZhckB0dW1zLmFjLmly
†These authors have contributed equally to this work
 Sara Nemati
Sara Nemati Farzaneh Shalileh2
Farzaneh Shalileh2 Hamed Mirjalali
Hamed Mirjalali Kobra Omidfar
Kobra Omidfar