
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 02 February 2023
Sec. Food Microbiology
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1117644
This article is part of the Research Topic Targeting Gut Microbiota Modulation by Dietary Supplementation to Improve Metabolic Diseases View all 7 articles
 Xiao-Lei Zhang1†
Xiao-Lei Zhang1† Lei Chen2,3,4†
Lei Chen2,3,4† Jiang Yang1
Jiang Yang1 Shan-Shan Zhao2,3,4
Shan-Shan Zhao2,3,4 Shi Jin1
Shi Jin1 Na Ao1
Na Ao1 Jing Yang2,3,4
Jing Yang2,3,4 Hui-Xin Liu1,2,3,4*
Hui-Xin Liu1,2,3,4* Jian Du1*
Jian Du1*Background: Non-alcoholic fatty liver disease (NAFLD) represents a severe public health problem. Dysbiosis of gut microbiome has been identified as one of the key environmental factors contributing to NAFLD. As an essential nutrition, Vitamin D (VD) plays an important role in regulating gut microbiota based on its receptor (Vitamin D Receptor, VDR) which is highly expressed in the gastrointestinal tract.
Methods: Rats were fed with HFD (high-fat diet) for 12 weeks. And the rats were treated with VD two times a week by intraperitoneal injection for 12 weeks. H&E staining combined with plasma biochemical index was performed to characterize pathological changes and function of the liver. Fecal microbiota 16S rRNA gene sequencing and metabolomics were taken to reveal the altered gut microbiota and metabolites.
Result: The VD alleviates the HFD-induced lipid accumulation in the liver as well as decreases the levels of amlodipine besylate (ALT) and amlodipine aspartate (AST). VD supplement decreased the ratio of phylum Firmicutes/Bacteroidetes (F/B) but increased alpha diversity. In addition, the VD treatment improved the HFD-induced gut microbiota by increasing the Prevotella and Porphyromonadaceae and decreasing Mucispirillum, Acetatifactor, Desulfovibrio, and Oscillospira abundance. Furthermore, the capability of tyrosine metabolism, tryptophan metabolism, arginine biosynthesis, and sphingolipid metabolism was enhanced after VD treatment. Consistently, Prevotella positively correlated with tryptophan metabolism and sphingolipid metabolism. Importantly, the Prevotella abundance was positively associated with serotonin, melatonin, tryptamine, L-arginine, and 3-dehydrosphinganine which synthesize from tryptophan, tyrosine, arginosuccinate, and serine, respectively.
Conclusion: VD treatment inhibited HFD-induced NAFLD accompany by dysbiosis gut microbiota and metabolites, suggesting that VD supplement could be a potential intervention used for NAFLD treatment by targeting the specific microbiota.
The gut microbiome has been identified as an essential mediator in the occurrence and development of non-alcoholic fatty liver disease (NAFLD), the most prevalent chronic liver disease, by regulating host energy and metabolism (Brunt et al., 2015; Sookoian et al., 2020). Human cross-sectional studies show an increased ratio of phylum Firmicutes/Bacteroidetes (F/B) and a decreased butyrate-producing bacteria Ruminococcaceae involved in NAFLD progression (Mouzaki et al., 2013). The clinical study discovered that the Proteobacteria phylum was enriched in patients with NAFLD (Aron-Wisnewsky et al., 2020a). Gut microbiota may modulate NAFLD through carbohydrate absorption and nutrient metabolism as well as producing short-chain fatty acids (SCFAs), which link the gut microbiota and physiology, especially hepatic gluconeogenesis. For example, gut microbiota Enterococcus metabolize trimethyllysine (TML) to N,N,N-trimethyl-5-aminovaleric acid (TMAVA), which could aggravate fatty liver (Zhao et al., 2020). Also, the gut microbiota metabolite indole alleviates diet-induced NAFLD via resisting inflammatory responses (Ma et al., 2020). In addition, fecal microbiota transplantation (FMT) from high-fat diet-fed mice accelerates steatosis and impairs insulin (Bauer et al., 2022). Furthermore, FMT could decrease fat accumulation in the liver by improving gut microbiota dysbiosis, thus attenuating fatty liver disease (Aron-Wisnewsky et al., 2020b). A previous study revealed that Lactobacillus supplement-driven reprogramming of gut microbiome and metabolome ameliorates the progression of NAFLD (Yu et al., 2021). Thus, the gut microbiota might be the novel therapeutic concept for counteracting the development of NAFLD.
Vitamin D (VD) and its active form 1,25-dihydroxyvitamin D (1,25D) inhibit immune responses and indirectly attenuate immune responses by increasing IL-10 production in macrophages, dendritic cells, and T cells (Cantorna et al., 2019). Our previous study showed that VD supplement may attenuate diet-induced liver injury by inhibiting pyroptosis (Zhang et al., 2021c). Other mechanisms underlying VD therapy against NAFLD should be considered. Furthermore, the serum 1,25D level is correlated with the α-diversity of gut microbiota and butyrate-producing bacteria, leading to better gut microbial health (Thomas et al., 2020). Importantly, the vitamin D receptor (VDR) is highly expressed in the gastrointestinal tract where it regulates gene expression (Thomas et al., 2020). A significant shift in the microbiota and serum measurements of selected bile acids and fatty acids was discovered in Vdr knockout mice compared to control mice (Wang et al., 2016). Thus, a VD supplement might alleviate NAFLD by restoring the dysbiosis gut microbiota and metabolism.
Here, the high-fat diet (HFD)-induced NAFLD rat model was adopted to treat VD by intraperitoneal injection. VD supplement obviously reversed the serum transaminases, hepatic fat deposits, and hepatocyte membrane destruction induced by HFD. In addition, VD decreased plasma glucose profiles. Furthermore, VD treatments stimulated the growth of Porphyromonadaceae and Prevotella but decreased Mucispirillum, Acetatifactor, Desulfovibrio, and Oscillospira. The tyrosine metabolism, tryptophan metabolism, arginine biosynthesis, and sphingolipid metabolism were enhanced by VD treatments. Of note, sphingolipid metabolism was positively correlated with Prevotella. Taken together, targeting gut microbiota and the sphingolipid metabolism pathway may be a feasible preventive strategy for patients with NAFLD.
Five-week-old male SD rats were housed in a specific pathogen-free environment at 20–22°C with 12-h light–dark cycles. To establish the NAFLD model, rats were fed with an HFD (HFD group, protein, 20%; fat 60%, carbohydrates, 20%) after 1 week of adaptation for 12 weeks. Normal diet combined with VD supplement (ND + VD) and HFD with VD supplement (HFD + VD) groups’ rats were treated with 1,25(OH)2D3 (5 μg/kg, Cayman, USA) two times a week by intraperitoneal injection for 12 weeks. Rats in ND and HFD groups were intraperitoneally injected with an equivalent volume of vehicle. During the experiments, body weight and food intake were assayed weekly. All animal procedures used in this study were conducted according to the Guide for Care and Use of Laboratory Animals and approved by the ethics committee of China Medical University (No. 2018162). At the end of the experiment, animals were euthanized to harvest liver, blood, and feces for further analysis.
Plasma glucose, amlodipine besylate (ALT), amlodipine aspartate (AST), triglyceride (TG), and total cholesterol (TC) levels were quantified according to the manufacturer’s instructions (Nanjing Jiancheng, China).
Histological changes in liver tissues were detected using hematoxylin–eosin (H&E) staining. The fixed liver tissues were embedded in paraffin and cut into 5-μm thick slices.
The liver sections were analyzed by transmission electron microscopy.
Stool DNA was isolated, and the V3 + V4 hypervariable regions of the bacterial 16S rRNA gene were amplified using primers CCTACGGGNGGCWGCAG(F) and GACTACNVGGGTATCTAAT(R). After PCR products were purified, the positive amplicon sequencing was performed on the Illumina MiSeq PE300 platform. After cutting off barcodes, QIIME V.2.0. was used for analyzing raw sequencing reads (Caporaso et al., 2010; Yang et al., 2022). Uparse software (version 7.1) was used to cluster the same operational taxonomic units (OTUs) with ≥97% similarity sequences. The Greengenes 16S rRNA gene reference database was adopted to classify OTU taxonomically.
The OTU numbers of each sample were flattened, and the alpha diversity of fecal bacteria was calculated based on the normalized OTU table by R package Vegan as previous methods (Sheng et al., 2017; Wang et al., 2018; Zhang et al., 2022). Principal coordinate analysis (PCoA) and similarities (PERMANOVA) were used to reveal the difference in stool microbiome profile based on the OTU level, which was analyzed by R package Vegan (Liu et al., 2022; Zhao et al., 2022). In addition, the sequence of OTUs in samples was used to predict the function of the intestinal microbiome by PICRUST2 as previously described (Hu et al., 2015; Liu et al., 2016), and significantly changed KEGG pathways were tested by Kruskal–Wallis and post-hoc Dunn’s test.
Blood metabolite signatures were identified by LC-MS between HFD and HFD + VD groups samples. The processed data such as m/z, RT, and normalized peak area percentages were imported into SIMCA to identify metabolites. The HMDB database was adopted to map and identify the metabolites. Partial least squares discriminant analysis (PLS-DA) was used to reveal the metabolite changes in groups by R package ropls, and the abundance of significant metabolites with variable important in projection (VIP) ≥1 and p-value (Wilcoxon’s test) < 0.05 was selected for enrichment analysis. The enrichment pathway of differential plasma metabolite profile between the two groups was analyzed by MetaboAnalyst 5.0, respectively.1
The relationship of KEGG pathways of bacterial predicted function and remarkable changed bacterial, alpha diversity of microbiota and microbiome as well as blood metabolites and gut bacterial were analyzed by Spearman’s correlation and Mantel’s test based on R package dplyr and ggcor. Meanwhile, the co-occurrence network of metabolites of key pathways and changed bacteria was constructed and visualized by R package igraph and Cytoscape, respectively.
For statistics in multiple groups, we utilized Kruskal–Wallis and two-way ANOVA tests to evaluate the difference among groups, respectively. Post-hoc Dunn’s test, Tukey’s multiple-comparison test, and Wilcoxon’s test were performed to analyze the difference between the two groups. P-values < 0.05 were considered statistically significant. Error bars indicate mean ± standard error of mean (SEM).
As shown in Figures 1A–C, the HFD remarkably increased body weight (BW), liver/BW ratio, and fasting serum glucose more than ND. The VD treatment significantly decreased the aforementioned index relative to HFD-fed rats. In addition, progressive lipid droplet accumulation was observed in the livers of the HFD group via H&E staining (Figures 1D–E). Furthermore, the VD could inhibit hepatocyte swelling and membrane rupturing compared to the livers of the HFD group, as revealed by electron microscopy (Figure 1F). Consistent with the histological changes, serum ALT, AST, TG, and TC were also significantly decreased after VD intervention compared to those of the HFD-fed rats (Figures 1G–J).
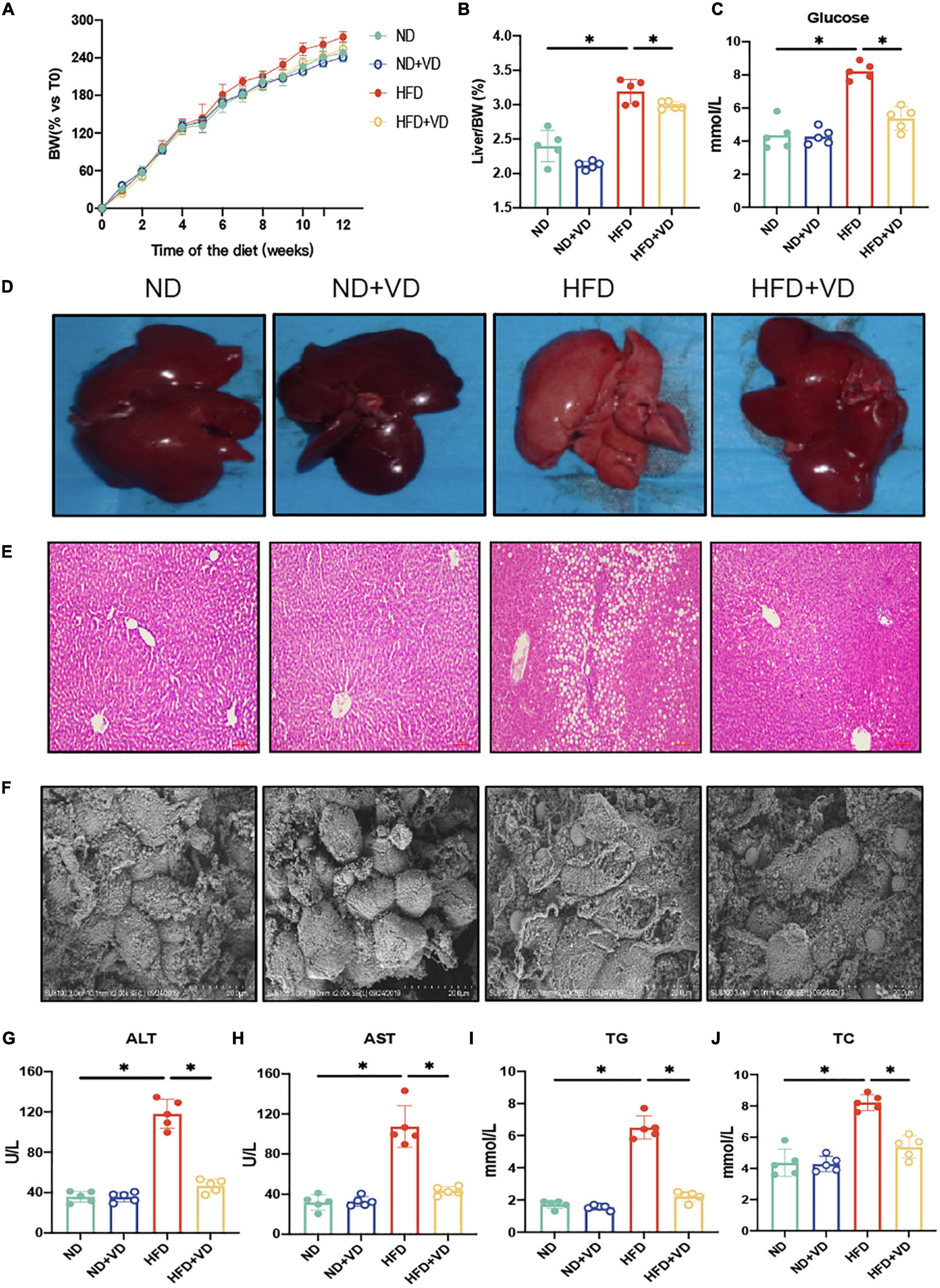
Figure 1. Vitamin D (VD) supplement alleviates high-fat diet (HFD)-induced non-alcoholic fatty liver. (A) The body weight (BW) of male rats fed indicated diet was measured weekly for 12 weeks and expressed as percentage vs. time 0. Data are mean values ± SEM (n = 5). (B,C) The liver organ index and fasting blood glucose in four groups. (D–F) Representative gross liver morphology, hematoxylin–eosin (H&E)-stained liver sections, and electron microscope picture of the liver. (G–J) The plasma amlodipine besylate (ALT), amlodipine aspartate (AST), total cholesterol (TC), and triglyceride (TG) levels in indicated groups. *P < 0.05. P-values are derived from ordinary two-way ANOVA followed by Tukey’s multiple-comparison test.
As shown in Figure 2A, the shared OTUs between HFD + VD and ND (2,787 = 1,721 + 155 + 749 + 162) were more than that between HFD + VD and HFD (2,212 = 1,721 + 155 + 241 + 95). Furthermore, the chao1 index in the HFD group significantly decreased compared to that in the ND group (Figure 2B), but the increased chao1 index was observed in the rats after VD treatment. We also found that VD intervention could significantly decrease the phylum Firmicutes/Bacteroides ratio which was induced by HFD (Figure 2C). In addition, the PCoA analysis revealed clear distinct discrimination in four groups (Figure 2D). Specifically, there was a closer distance between HFD + VD and ND rats than that between HFD + VD and HFD (Figure 2D), and the richness index was driven by the F/B ratio (Figure 2D). Moreover, Barnesiella and Porphyromonadaceae_unclassified genus were positively correlated with alpha diversity (Figure 2E). The negative association between alpha diversity and Clostridium, Odoribacter, and Oscillibacter genera was also revealed by Spearman’s correlation analysis (Figure 2E). It was noted that the VD treatment increased the proportion of phylum Bacteroidetes compared to HFD-fed rats (Figure 2F). The decrease in phylum Firmicutes was also observed in rats of HFD + VD relative to that in the HFD group (Figure 2F). At the genus level, the VD supplement reversed the decreased abundance of Porphyromonadaceae_unclassified and Prevotella genus, which only showed in the HFD group (Figure 2G). More importantly, the VD supplement could also boost the abundance of Lactobacillus genus even feeding with HFD (Figure 2G).
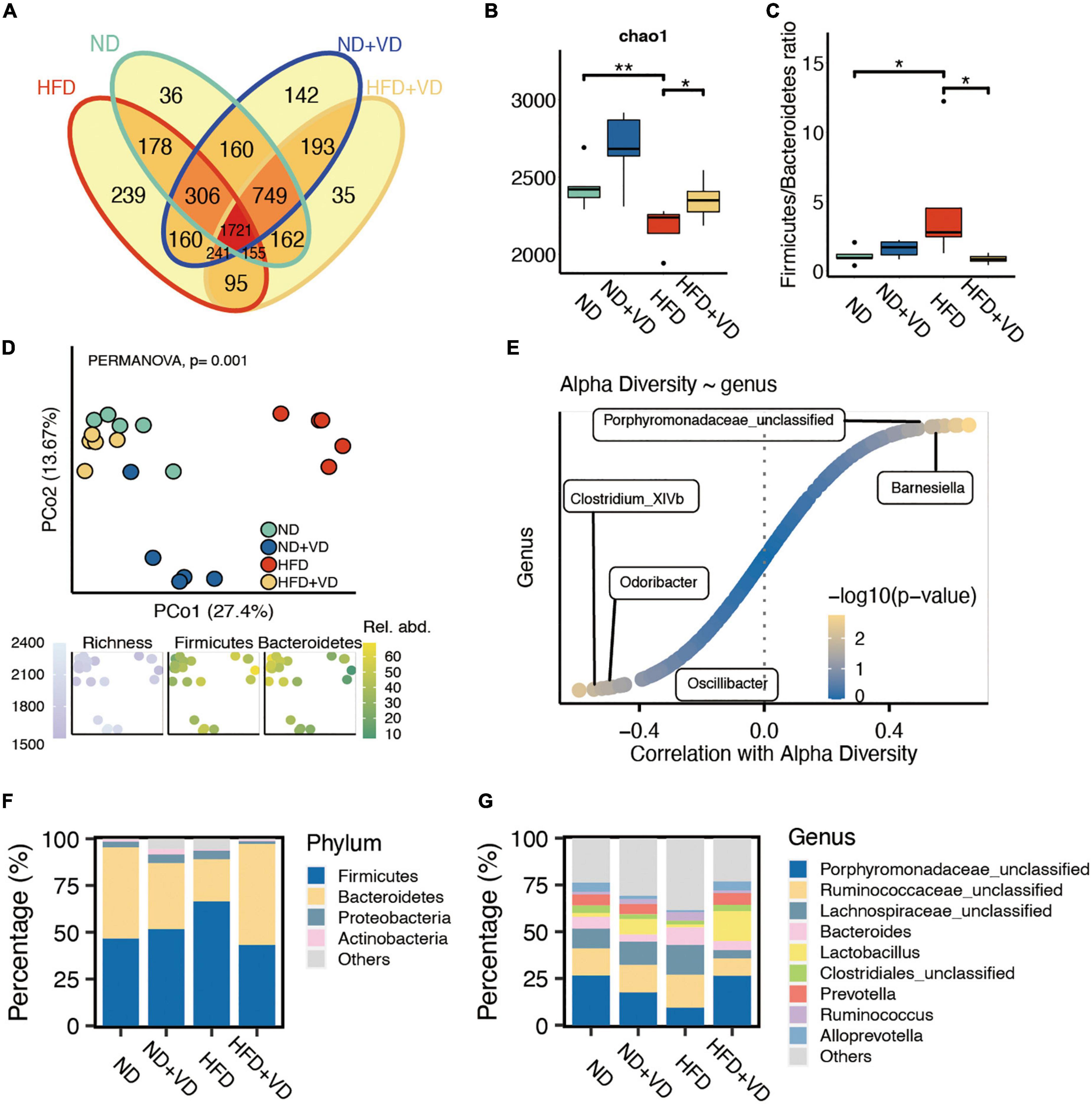
Figure 2. Altered fecal microbiota after vitamin D (VD) treatment. (A) The shared and unique observed operational taxonomic units (OTUs) are in four groups. (B) The alpha diversity difference among groups. (C) The comparison of phylum Firmicutes/Bacteroidetes ratio in four group samples. (D) Principal coordinate analysis (PCoA) reveals a clear distinct difference in gut microbiota in four groups. (E) The correlation between alpha diversity and alerted gut microbiome. (F,G) Stacked bar plots depicting the percentage of phylum (left) and genus (right) of intestinal bacteria in the screening results. *P < 0.05, **P < 0.01. Kruskal–Wallis test with post-hoc Dunn’s test.
Next, we compared the species level change after VD treatment. As shown in Figure 3A, HFD significantly increased the abundance of Mucispirillum, Acetatifactor, Desulfovibrio, Oscillospira, Clostridium_XlVb, Eisenbergiella, Odoribacter, and Robinsoniella species, but the aforementioned bacteria were all downregulated after VD treatment. However, the supplement of VD reversed the decreased proportion of Porphyromonadaceae_unclassified, Prevotella, Alloprevotella, and Barnesiella which were induced by HFD (Figure 3B).
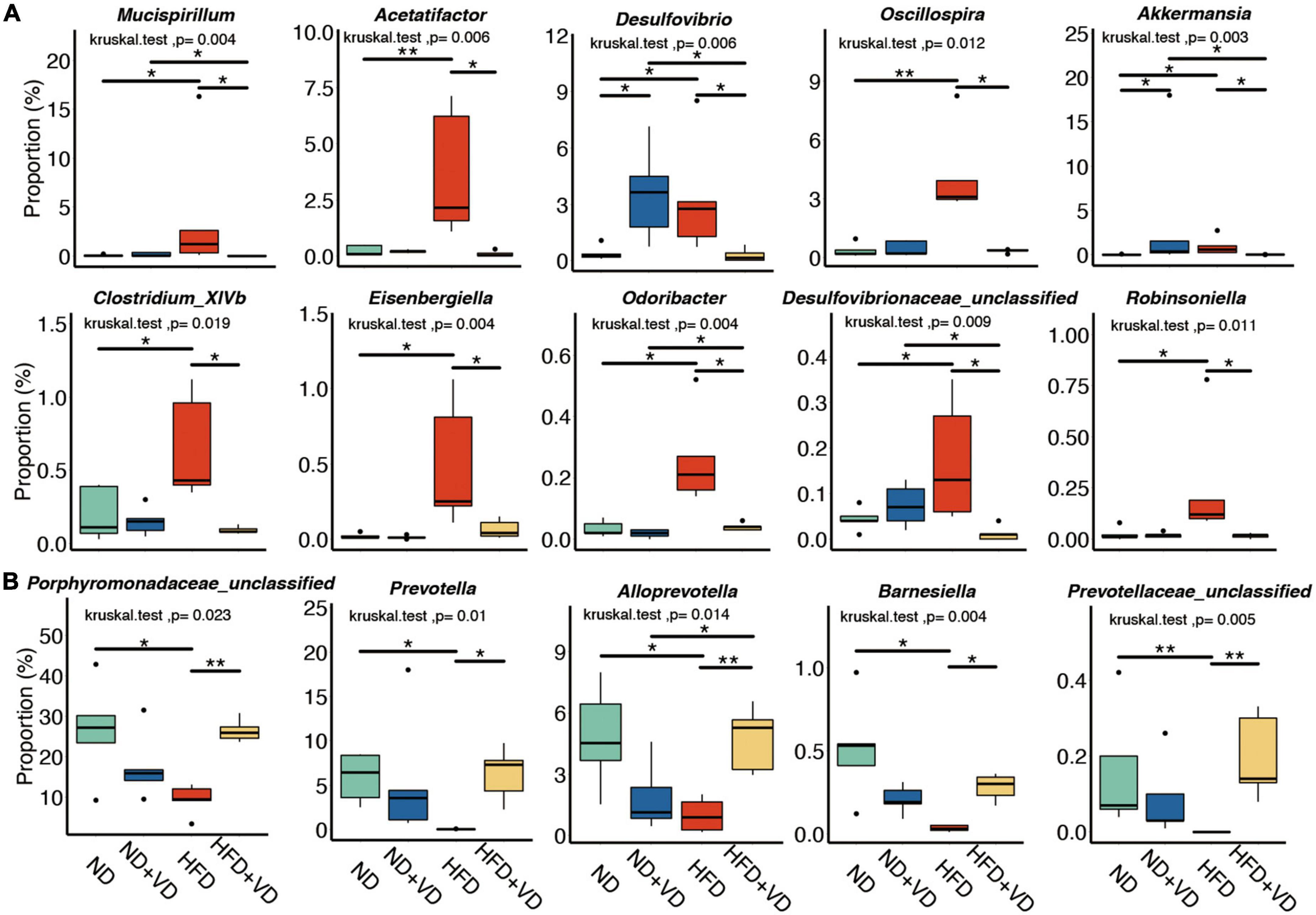
Figure 3. Identification of specific altered bacteria for vitamin D (VD) intervention. (A,B) Significantly changed gut microbiome in four groups. *P < 0.05, **P < 0.01. Kruskal–Wallis test with post-hoc Dunn’s test.
Furthermore, the predicted function of gut microbiota revealed that there was a clear distinct change in metabolism pathways. As shown in Figure 4A, apart from 25 pathways overlapping between HFD compared to ND and HFD + VD, the VD treatment uniquely changed the 56 pathways relative to HFD. Specifically, linoleic acid metabolism, ether lipid metabolism, and sphingolipid metabolism pathways were enriched in both two comparisons (Figure 4B), and the histidine metabolism and riboflavin metabolism were uniquely changed after VD and HFD supplement, respectively (Figure 4B). Importantly, arachidonic acid metabolism, linoleic acid metabolism, ether lipid metabolism, and sphingolipid metabolism which belong to lipid metabolism were increased after VD treatment (Figure 4C). Decreased amino acid metabolism, containing lysine degradation and phenylalanine metabolism, was downregulated upon VD supplement. Finally, the consisting higher levels of Porphyromonadaceae_unclassified, Prevotella, Alloprevotella, and Barnesiella after VD treatment were positively correlated with lipid metabolism, which negatively associated with Mucispirillum, Acetatifactor, and Desulfovibrio abundance (Figure 4C).
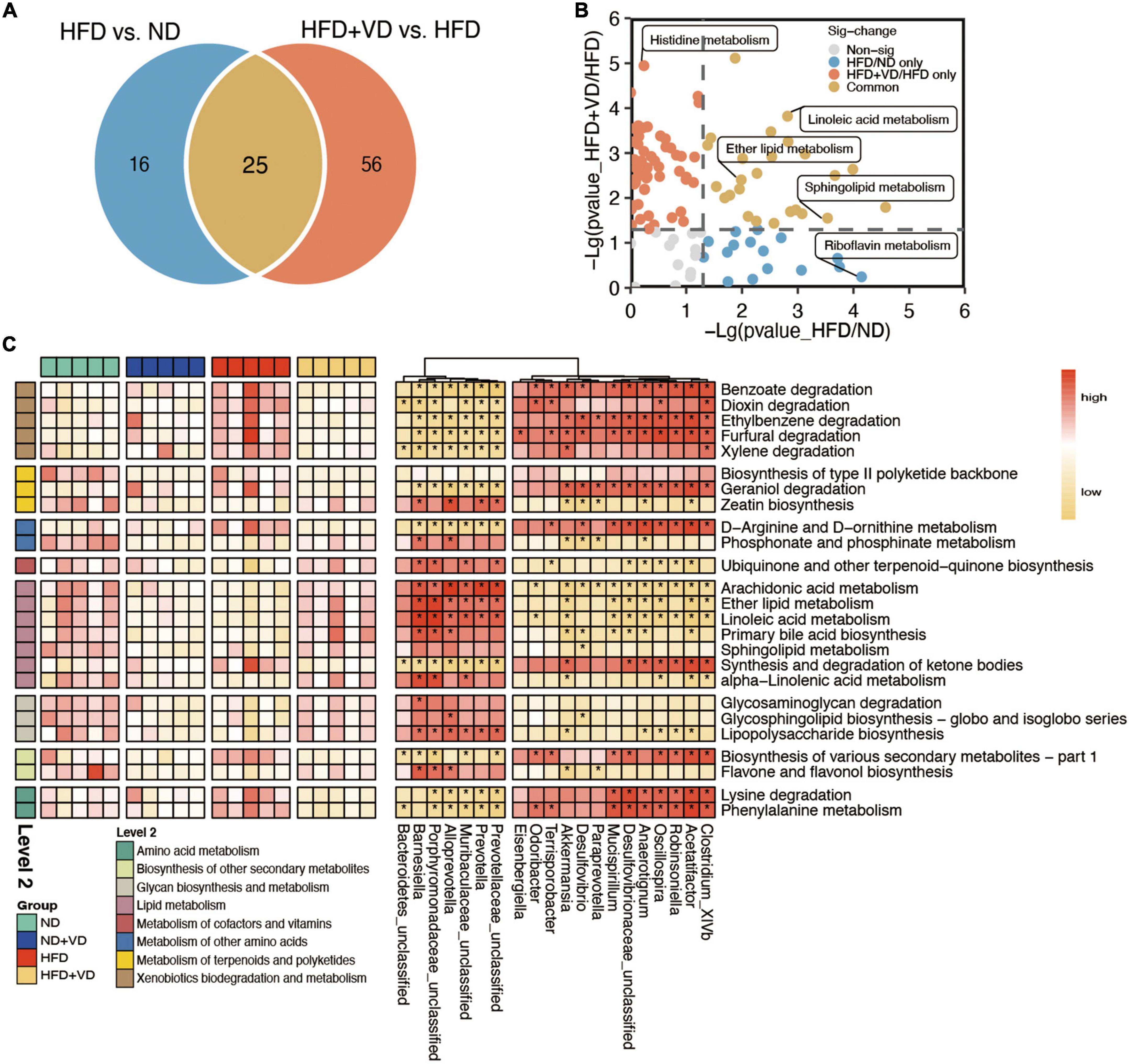
Figure 4. The characteristic function of gut microbiota. (A) Venn diagrams illustrating the number of predicted pathways significantly changed between high-fat diet (HFD) vs. ND and HFD + VD vs. HFD. (B) The clear distinct difference between HFD vs. ND and HFD + VD compared to HFD, respectively. (C) The predicted function of bacteria and the connection between the predicated function of intestinal microbiota and altered gut microbiota. Red denotes positive correlations. Yellow denotes negative correlations. *P < 0.01.
Plasma samples from the samples were analyzed by the global metabolite panel, which identified a significant change in 1,098 (up) and 1,474 (down) features after VD treatment (Figure 5A). As shown in Figure 5B, the L-valine, L-leucine, and L-isoleucine were significantly decreased after VD treatment. Also, the sphingosine, L-serine, and sphinganine were decreased in the HFD + VD group, but a higher level of 3-dehydrosphinganine, vitamin D3, L-arginine, and dopamine was observed after VD intervention. The metabolic set analysis revealed that the changed metabolites are mainly enriched in amino acids, peptides, benzoic acids, monosaccharides, amines, and indoles (Figure 5C). The changed metabolites are shown in Supplementary Table 1. Furthermore, the KEGG pathway enrichment indicated tyrosine metabolism, tryptophan metabolism, arginine biosynthesis, valine, leucine, and isoleucine biosynthesis, and sphingolipid metabolism pathways were remarkably altered after VVD treatment (Figure 5D).
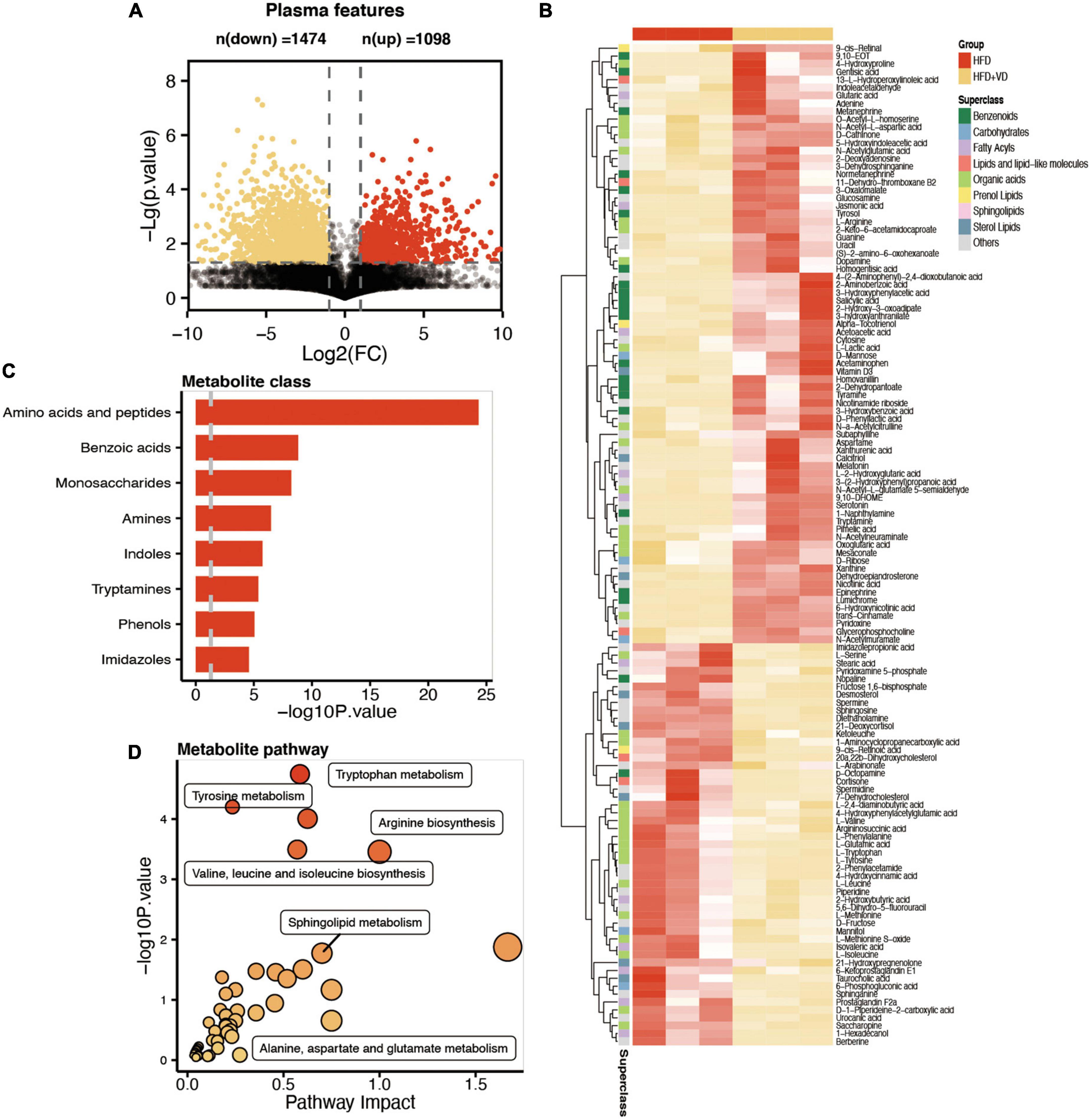
Figure 5. Comparative metabolomics analysis determines the change in plasma metabolites in high-fat diet (HFD) + VD from HFD. (A) The volcano plot shows the number of dysregulated metabolite features between the two groups. (B) The heatmap shows the annotated changed metabolites in the plasma profile. (C,D) The metabolic set and pathway enrichment based on the different metabolites.
Next, as illustrated in Figure 6A, the pathway analysis revealed that VD treatment increased the metabolizing ability of tryptophan, which may lead to the production of more tryptamine, serotonin, and melatonin, and the enhanced capability of tyrosine metabolism was also observed in the HFD + VD group (Figure 6B). Furthermore, VD intervention induced the N-acetylglutamate and arginine biosynthesis from glutamate and L-argininosuccinate, respectively (Figure 6C).
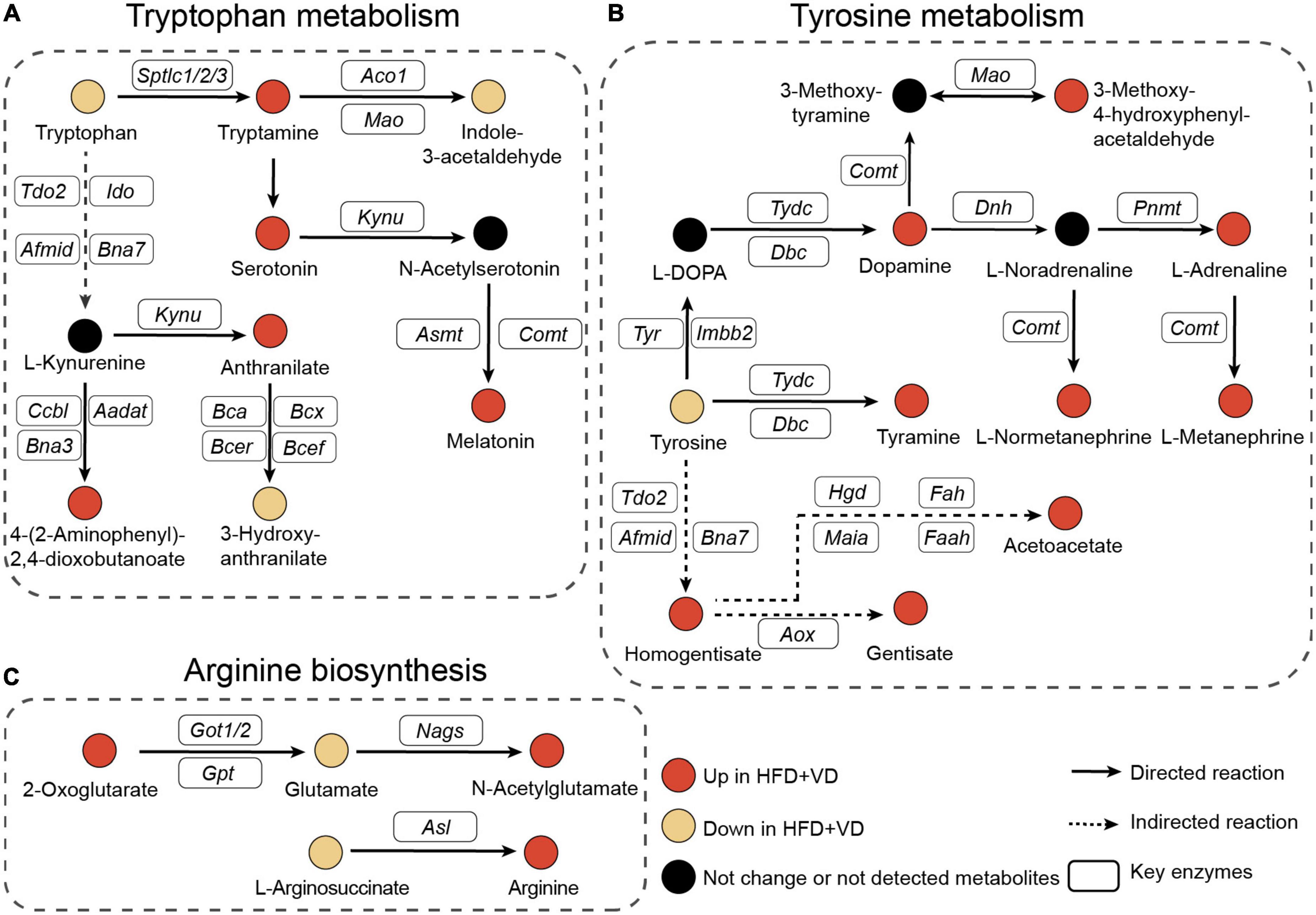
Figure 6. Pathway’s analysis. (A–C) Illustrative examples of metabolite changes in tryptophan metabolism, tyrosine metabolism, and arginine biosynthesis pathways.
To further explore the potential influence of VD in both gut microbiota and plasma metabolites, the Spearman analysis was performed to connect the changed metabolites and bacteria. As shown in Figure 7A, L-tryptophan, L-tyrosine, L-glutamic acid, L-phenylalanine, L-isoleucine, L-leucine, L-valine, and sphinganine, which were downregulated after VD treatment, were positively associated with Clostridium_XlVb, Paraprevotella, and Mucispirillum, which were lower in the HFD + VD group. Prevotella, Prevotellaceae_unclassified, and Alloprevotella were correlated with dopamine, L-arginine, indoleacetaldehyde, and 3-dehydrosphinganine. In addition, Mucispirillum, Desulfovibrio, and Akkermansia maintained a negative association with 4-hydroxyproline and tyramine. The enriched Oscillospira, Robinsoniella, and Acetatifactor in the HFD group were negatively correlated with upregulated 2-aminobenzoic acid, epinephrine, tryptamine, serotonin, and melatonin after VD treatment. It was noted that the integrated analysis of significantly changed pathways revealed that sphingolipid metabolism overlaps with microbiome and metabolite (Figure 7B). Furthermore, Mantel’s test and Spearman’s analysis were taken to associate the metabolites which belong to sphingolipid metabolism and altered gut bacterial (Figures 7C, D). Specifically, HFD + VD groups’ rats enriched in Prevotella and Alloprevotella were negatively correlated with Acetatifactor, Oscillospira, and Desulfovibrio, while maintaining a strong positive association with 3-dehydrosphinganine. Importantly, even though VD treatment induced the 3-dehydrosphinganine directly synthesis from L-serine and palmitoyl-CoA, the following metabolism to produce sphinganine and sphingosine was downregulated in the HFD + VD group (Figure 7E).
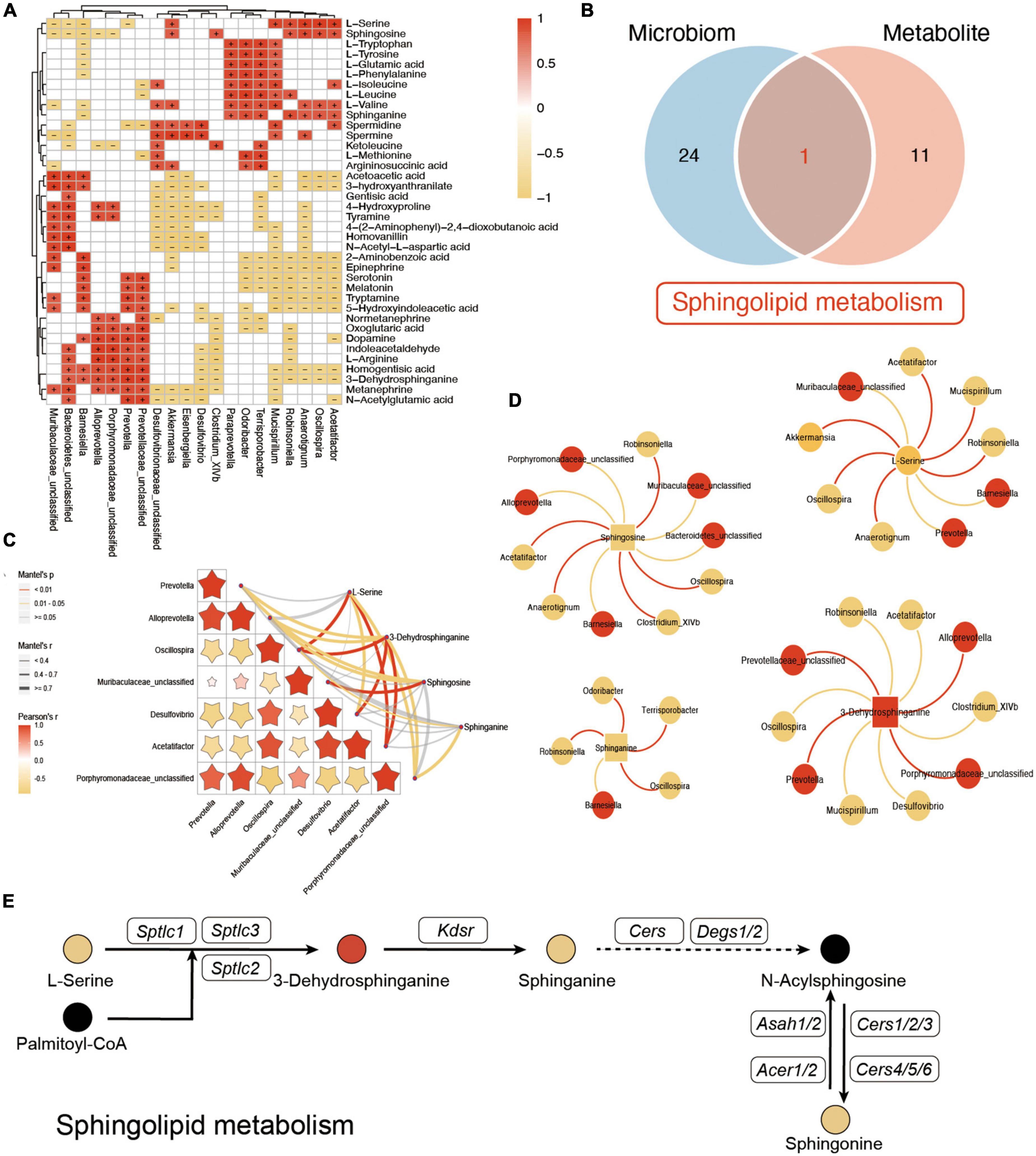
Figure 7. Integrated analysis between microbiome and metabolites. (A) The Spearman correlation analysis between the significantly changed microbiome and metabolites. + and – indicate the significant positive and negative association, respectively. (B) Venn diagram of significant differential KEGG pathways in functional analysis of gut microbiota and the metabolic pathway based on the metabolites. (C,D) Mantel’s analysis and interaction network plots of enriched metabolites in the shared pathways with microbes that participate in its metabolism. Red colors indicate significantly upregulated metabolites or microbes, while yellow colors indicate significantly downregulated metabolites or microbes. Red and yellow lines indicate the positive correlation and negative correlation, respectively. (E) Illustrative example of metabolite changes in the sphingolipid metabolism pathway.
It has been well known that the high prevalence of NAFLD is becoming a key health concern globally (Draijer et al., 2019). More importantly, emerging evidence has demonstrated that gut microbiota is involved in NAFLD progression (Leung et al., 2022). VD, as an essential nutrient, benefits the bone growth. Additionally, low serum VD levels are at increased risk of multiple adverse health outcomes including obesity, incident diabetes, and autoimmune diseases (Barbachano et al., 2017). However, limited studies have been focused on the correlation between VD and NAFLD so far. Our previous study focuses on the effect of VD on hepatic injury, lipid accumulation, activation of the NLRP3 inflammasome, and pyroptosis (Zhang et al., 2021c). The dysbiosis of gut microbiota indicated the potential therapy against NAFLD. In this study, we further investigated the beneficial effects of VD on NAFLD by regulating the gut microbiota and metabolism. Our results indicate that the supplement of VD ameliorated the HFD-induced liver function and reversed the hepatic steatosis by modulating the gut microbiota.
The VD treatment significantly decreased the F/B ratio, the indicator of calorie absorption capability (Komaroff, 2017). The increased Chao1 index of gut microbiota in the HFD + VD group indicated that the VD treatment could restore the decreased species diversity induced by HFD. The Bacteroidetes phylum, usually decreased in patients with NAFLD, (Del Chierico et al., 2017) was reduced in the HFD group but could be reversed by VD supplement. At the genus level, the Lactobacillus, as documented to ameliorate the progression of NAFLD through the modulation of the gut microbiome in mice, (Jang et al., 2019; Lee et al., 2020) could also be induced after VD treatment. Similar results in rats showed that VD could restore HFD-induced gut microbiota dysbiosis by increasing the relative abundance of Lactobacillus and decreasing the relative abundance of Acetatifactor, Oscillibacter, and Flavonifractor (Zhang et al., 2021c). Furthermore, Porphyromonadaceae and Ruminococcaceae, which have been identified as butyrate-generating bacteria with health benefit effects, (Zhang et al., 2021a) were increased by VD supplement in HFD rats. Also, the Prevotella, negatively correlated with NAFLD severity, (Schwimmer et al., 2019) was increased after VD intervention. In addition, Mucispirillum, Desulfovibrio, and Desulfovibrionaceae, enriched in the high-fat/high-cholesterol-induced NAFLD mice, (Zhang et al., 2021b) were decreased after VD treatment. The increased level of Acetobacter, which could induce gut dysfunction, however, was decreased in the HFD + VD group. Further function analysis of gut microbiota indicates that the VD treatment significantly increased the linoleic acid metabolism capability of gut microbiota. Consistent with our findings, it has been reported that HFD could inhibit linoleic acid metabolism by altering the gut microbiota (Miyamoto et al., 2019). Taken together, our findings indicate that the VD could alleviate the NAFLD based on gut microbiota restoration.
The VD treatment reversed the metabolic dysbiosis as shown by metabolism analysis. Patients with metabolic syndrome could decrease the capacity of the microbiota to metabolize tryptophan into derivatives (Natividad et al., 2018; Wrzosek et al., 2021). However, we found that tryptophan metabolism was enhanced after VD treatment. Higher levels of tryptophan derivatives, such as tryptamine, serotonin, anthranilate, and melatonin, were observed in HFD + VD rats. Other reports have identified tryptamine as a metabolite that depends on the microbiota and is depleted under an HFD (Krishnan et al., 2018). Recently, research also revealed that serotonin and melatonin could regulate appetite and safeguard against fatty liver, respectively (Li et al., 2019). In addition, a previous study also revealed that tyrosine levels were positively associated with the presence of NAFLD (Gobeil et al., 2022). The lower level of tyrosine and a higher level of its derivatives imply that the VD treatment promotes the capability of tyrosine metabolism. Another study found that the bacteria in patients who received the weight-loss intervention had a higher capacity to produce tyramine from tyrosine (Li et al., 2021). The gut microbiota-derived dopamine could also regulate food seeking (Fernandes et al., 2020). Furthermore, an increase in adrenaline, normetanephrine, and changes in cecal microbiota, can induce the energy metabolism of fat tissue (Larabee et al., 2020). Other authors have proposed that Akkermansia muciniphila benefits metabolic syndrome by specific modulation of acetoacetate (Depommier et al., 2021). It has been documented that a higher level of glutamate increases the risk for obesity and metabolic syndrome and promotes hepatic gluconeogenesis (Andres-Hernando et al., 2021; Zhuang et al., 2021). However, the VD treatment not only inhibits the glutamate synthesis from 2-oxoglutarate but also promotes its metabolism to produce N-acetylglutamate. Dietary arginine could prevent intestinal inflammation in mice (Alexander et al., 2022). The arginine metabolism, as a potential immunomodulatory pathway, is mediated by Bifidobacterium longum and A. muciniphila (Strazar et al., 2021). However, the VD treatment promotes arginine synthesis from L-argininosuccinate. Thus, these results suggest that VD treatment may not only reverse the gut microbiota disorder but also restore metabolic dysbiosis.
Importantly, the integrated analysis of gut microbiota and metabolites also revealed a positive correlation between tryptophan, tyrosine, glutamic acid, phenylalanine, BCAAs, sphinganine, spermidine, and Mucispirillum, suggesting that the elevated level of these metabolites in HFD rats may be due to the enrichment of Mucispirillum. Increasing evidence has demonstrated that gut microbiota-derived sphingolipids could modulate hepatic metabolism (Le et al., 2022). Sphingolipids were reported to be generated by Bacteroidetes, the dominant phylum of the gut microbiome (Johnson et al., 2020). Our results showed that 3-dehydrosphinganine was positively correlated with Prevotella under the Bacteroidetes phylum. It has been reported that the pathway for bacterial sphingolipid synthesis is common in Prevotella (Heaver et al., 2022). Our results indicated that the enhancement of Prevotella abundance after VD treatment might promote the 3-dehydrosphinganine synthesis from serine. Sphingosine could regulate intestinal immune cells to protect the gut from infection; (Sun et al., 2022) however, sphingosine was also enriched in patients with NAFLD (Oh et al., 2020). In the present study, the positive correlation between sphingosine and Oscillospira suggests that the decline of Oscillospira after VD treatment may promote sphingosine synthesis. However, whether Prevotella and Oscillospira affect 3-dehydrosphinganine and sphingosine synthesis needs further study. This study has a few limitations. First, the role of vitamin D in obesity was different between men and women, and our study only uncovered the effects of VD on gut microbiota and metabolism in NAFLD without gender-dependent manners. A second limitation of this study is linked to the number of detected metabolites, which could be further improved by using high-resolution mass spectrometry, which accurately distinguishes isomer metabolites, especially lipids and their derivatives. Even though the aim of this study was to reveal the previously unknown VD supplement characteristics based on gut microbiota and metabolic profile to provide the clue for NAFLD prevention and treatment, it is a rather descriptive, and follow-up study on one of the hypotheses indicated by the omics analysis, such as fecal microbiota transplantation from vitamin D-supplemented rats to NAFLD rats or germ-free animal model validation, needs to be further studied.
VD supplements alleviated the hepatic lipid accumulation induced by HFD. The present analysis of rats’ fecal bacteria and metabolites revealed the function of VD supplements in restoring the gut microbiota and metabolism dysbiosis. VD treatment induced the abundance of Prevotella which positively correlated with serotonin, melatonin, tryptamine, L-arginine, and 3-dehydrosphinganine. The reduction of Mucispirillum after VD supplement kept a positive correlation with plasma tryptophan, tyrosine, glutamic acid, phenylalanine, BCAAs, sphinganine, and spermidine, indicating that the VD could promote the tryptophan metabolism, tyrosine metabolism, and arginine biosynthesis via inhibiting the proliferation of Mucispirillum. In addition, VD treatment promotes sphingolipid metabolism in both gut microbiota function and metabolite pathway. This comprehensive integrated microbiota and metabolomic analysis provide insights into the relationship between the VD, fecal microbiome, and the deregulation of metabolism in process of HFD-induced NAFLD, suggesting that VD supplement could be a potential intervention used for NAFLD treatment by targeting the specific microbiota.
The data presented in the study are deposited in the NCBI repository, accession number PRJNA928563.
This animal study was reviewed and approved by the Ethics Committee of China Medical University (No. 2018162).
X-LZ, JiaY, LC, S-SZ, SJ, and NA designed and performed the experiments. LC conducted the analyses and wrote the manuscript. X-LZ measured the fecal bacteria and metabolites. H-XL and JD conceived and supervised the study. All authors read and approved the final version of the manuscript.
This research received financial support from the General Project of Liaoning Provincial Department of Education under Grant No. LJKZ0758.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1117644/full#supplementary-material
Alexander, M., Ang, Q., Nayak, R., Bustion, A., Sandy, M., Zhang, B., et al. (2022). Human gut bacterial metabolism drives Th17 activation and colitis. Cell Host Microbe 30, 17–30e19. doi: 10.1016/j.chom.2021.11.001
Andres-Hernando, A., Cicerchi, C., Kuwabara, M., Orlicky, D., Sanchez-Lozada, L., Nakagawa, T., et al. (2021). Umami-induced obesity and metabolic syndrome is mediated by nucleotide degradation and uric acid generation. Nat. Metab. 3, 1189–1201. doi: 10.1038/s42255-021-00454-z
Aron-Wisnewsky, J., Vigliotti, C., Witjes, J., Le, P., Holleboom, A., Verheij, J., et al. (2020a). Gut microbiota and human NAFLD: Disentangling microbial signatures from metabolic disorders. Nat. Rev. Gastroenterol. Hepatol. 17, 279–297. doi: 10.1038/s41575-020-0269-9
Aron-Wisnewsky, J., Warmbrunn, M., Nieuwdorp, M., and Clement, K. (2020b). Nonalcoholic fatty liver disease: Modulating gut microbiota to improve severity? Gastroenterology 158, 1881–1898. doi: 10.1053/j.gastro.2020.01.049
Barbachano, A., Fernandez-Barral, A., Ferrer-Mayorga, G., Costales-Carrera, A., Larriba, M., and Munoz, A. (2017). The endocrine vitamin D system in the gut. Mol. Cell Endocrinol. 453, 79–87. doi: 10.1016/j.mce.2016.11.028
Bauer, K., Littlejohn, P., Ayala, V., Creus-Cuadros, A., and Finlay, B. (2022). Nonalcoholic fatty liver disease and the gut-liver axis: Exploring an undernutrition perspective. Gastroenterology 162, 1858–1875e1852. doi: 10.1053/j.gastro.2022.01.058
Brunt, E., Wong, V., Nobili, V., Day, C., Sookoian, S., Maher, J., et al. (2015). Nonalcoholic fatty liver disease. Nat. Rev. Dis. Primers 1:15080. doi: 10.1038/nrdp.2015.80
Cantorna, M., Rogers, C., and Arora, J. (2019). Aligning the paradoxical role of vitamin D in gastrointestinal immunity. Trends Endocrinol. Metab. 30, 459–466. doi: 10.1016/j.tem.2019.04.005
Caporaso, J., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F., Costello, E., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Del Chierico, F., Nobili, V., Vernocchi, P., Russo, A., De Stefanis, C., Gnani, D., et al. (2017). Gut microbiota profiling of pediatric nonalcoholic fatty liver disease and obese patients unveiled by an integrated meta-omics-based approach. Hepatology 65, 451–464. doi: 10.1002/hep.28572
Depommier, C., Everard, A., Druart, C., Maiter, D., Thissen, J., Loumaye, A., et al. (2021). Serum metabolite profiling yields insights into health promoting effect of A. muciniphila in human volunteers with a metabolic syndrome. Gut Microbes 13:1994270. doi: 10.1080/19490976.2021.1994270
Draijer, L., Benninga, M., and Koot, B. (2019). Pediatric NAFLD: An overview and recent developments in diagnostics and treatment. Expert Rev. Gastroenterol. Hepatol. 13, 447–461. doi: 10.1080/17474124.2019.1595589
Fernandes, A., Alves da Silva, J., Almeida, J., Cui, G., Gerfen, C., Costa, R., et al. (2020). Postingestive modulation of food seeking depends on vagus-mediated dopamine neuron activity. Neuron 106, 778–788e776. doi: 10.1016/j.neuron.2020.03.009
Gobeil, E., Maltais-Payette, I., Taba, N., Briere, F., Ghodsian, N., Abner, E., et al. (2022). Mendelian randomization analysis identifies blood tyrosine levels as a biomarker of non-alcoholic fatty liver disease. Metabolites 12:440. doi: 10.3390/metabo12050440
Heaver, S., Le, H., Tang, P., Basle, A., Mirretta Barone, C., Vu, D., et al. (2022). Characterization of inositol lipid metabolism in gut-associated Bacteroidetes. Nat. Microbiol. 7, 986–1000. doi: 10.1038/s41564-022-01152-6
Hu, Y., Chau, T., Liu, H., Liao, D., Keane, R., Nie, Y., et al. (2015). Bile acids regulate nuclear receptor (Nur77) expression and intracellular location to control proliferation and apoptosis. Mol. Cancer Res. 13, 281–292. doi: 10.1158/1541-7786.MCR-14-0230
Jang, H., Park, H., Kang, D., Chung, H., Nam, M., Lee, Y., et al. (2019). A protective mechanism of probiotic Lactobacillus against hepatic steatosis via reducing host intestinal fatty acid absorption. Exp. Mol. Med. 51, 1–14. doi: 10.1038/s12276-019-0352-x
Johnson, E., Heaver, S., Waters, J., Kim, B., Bretin, A., Goodman, A., et al. (2020). Sphingolipids produced by gut bacteria enter host metabolic pathways impacting ceramide levels. Nat. Commun. 11:2471. doi: 10.1038/s41467-020-16274-w
Komaroff, A. (2017). The microbiome and risk for obesity and diabetes. JAMA 317, 355–356. doi: 10.1001/jama.2016.20099
Krishnan, S., Ding, Y., Saedi, N., Choi, M., Sridharan, G., Sherr, D., et al. (2018). Gut microbiota-derived tryptophan metabolites modulate inflammatory response in hepatocytes and macrophages. Cell Rep. 23, 1099–1111. doi: 10.1016/j.celrep.2018.03.109
Larabee, C., Neely, O., and Domingos, A. (2020). Obesity: A neuroimmunometabolic perspective. Nat. Rev. Endocrinol. 16, 30–43. doi: 10.1038/s41574-019-0283-6
Le, H., Lee, M., Besler, K., and Johnson, E. (2022). Host hepatic metabolism is modulated by gut microbiota-derived sphingolipids. Cell Host Microbe 30, 798–808e797. doi: 10.1016/j.chom.2022.05.002
Lee, N., Yoon, S., Han, D., Gupta, H., Youn, G., Shin, M., et al. (2020). Lactobacillus and Pediococcus ameliorate progression of non-alcoholic fatty liver disease through modulation of the gut microbiome. Gut Microbes 11, 882–899. doi: 10.1080/19490976.2020.1712984
Leung, H., Long, X., Ni, Y., Qian, L., Nychas, E., Siliceo, S., et al. (2022). Risk assessment with gut microbiome and metabolite markers in NAFLD development. Sci. Transl. Med. 14:eabk0855. doi: 10.1126/scitranslmed.abk0855
Li, D., Tong, J., Li, Y., Meng, H., Ji, Q., Zhang, G., et al. (2019). Melatonin safeguards against fatty liver by antagonizing TRAFs-mediated ASK1 deubiquitination and stabilization in a beta-arrestin-1 dependent manner. J. Pineal Res. 67:e12611. doi: 10.1111/jpi.12611
Li, J., Ashrafian, H., Sarafian, M., Homola, D., Rushton, L., Barker, G., et al. (2021). Roux-en-Y gastric bypass-induced bacterial perturbation contributes to altered host-bacterial co-metabolic phenotype. Microbiome 9:139. doi: 10.1186/s40168-021-01086-x
Liu, H., Rocha, C., Dandekar, S., and Wan, Y. (2016). Functional analysis of the relationship between intestinal microbiota and the expression of hepatic genes and pathways during the course of liver regeneration. J. Hepatol. 64, 641–650. doi: 10.1016/j.jhep.2015.09.022
Liu, Y., Chen, L., Liu, L., Zhao, S., You, J., Zhao, X., et al. (2022). Interplay between dietary intake, gut microbiota, and metabolic profile in obese adolescents: Sex-dependent differential patterns. Clin. Nutr. 41, 2706–2719. doi: 10.1016/j.clnu.2022.10.009
Ma, L., Li, H., Hu, J., Zheng, J., Zhou, J., Botchlett, R., et al. (2020). Indole alleviates diet-induced hepatic steatosis and inflammation in a manner involving myeloid cell 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3. Hepatology 72, 1191–1203. doi: 10.1002/hep.31115
Miyamoto, J., Igarashi, M., Watanabe, K., Karaki, S., Mukouyama, H., Kishino, S., et al. (2019). Gut microbiota confers host resistance to obesity by metabolizing dietary polyunsaturated fatty acids. Nat. Commun. 10:4007. doi: 10.1038/s41467-019-11978-0
Mouzaki, M., Comelli, E., Arendt, B., Bonengel, J., Fung, S., Fischer, S., et al. (2013). Intestinal microbiota in patients with nonalcoholic fatty liver disease. Hepatology 58, 120–127. doi: 10.1002/hep.26319
Natividad, J., Agus, A., Planchais, J., Lamas, B., Jarry, A., Martin, R., et al. (2018). Impaired aryl hydrocarbon receptor ligand production by the gut microbiota is a key factor in metabolic syndrome. Cell Metab. 28, 737–749e734. doi: 10.1016/j.cmet.2018.07.001
Oh, T., Kim, S., Caussy, C., Fu, T., Guo, J., Bassirian, S., et al. (2020). A universal gut-microbiome-derived signature predicts cirrhosis. Cell Metab. 32, 878–888e876. doi: 10.1016/j.cmet.2020.10.015
Schwimmer, J., Johnson, J., Angeles, J., Behling, C., Belt, P., Borecki, I., et al. (2019). Microbiome signatures associated with steatohepatitis and moderate to severe fibrosis in children with nonalcoholic fatty liver disease. Gastroenterology 157, 1109–1122. doi: 10.1053/j.gastro.2019.06.028
Sheng, L., Jena, P., Hu, Y., Liu, H., Nagar, N., Kalanetra, K., et al. (2017). Hepatic inflammation caused by dysregulated bile acid synthesis is reversible by butyrate supplementation. J. Pathol. 243, 431–441. doi: 10.1002/path.4983
Sookoian, S., Salatino, A., Castano, G., Landa, M., Fijalkowky, C., Garaycoechea, M., et al. (2020). Intrahepatic bacterial metataxonomic signature in non-alcoholic fatty liver disease. Gut 69, 1483–1491. doi: 10.1136/gutjnl-2019-318811
Strazar, M., Temba, G., Vlamakis, H., Kullaya, V., Lyamuya, F., Mmbaga, B., et al. (2021). Gut microbiome-mediated metabolism effects on immunity in rural and urban African populations. Nat. Commun. 12:4845. doi: 10.1038/s41467-021-25472-z
Sun, R., Gu, X., Lei, C., Chen, L., Chu, S., Xu, G., et al. (2022). Neutral ceramidase-dependent regulation of macrophage metabolism directs intestinal immune homeostasis and controls enteric infection. Cell Rep. 38:110560. doi: 10.1016/j.celrep.2022.110560
Thomas, R., Jiang, L., Adams, J., Xu, Z., Shen, J., Janssen, S., et al. (2020). Vitamin D metabolites and the gut microbiome in older men. Nat. Commun. 11:5997. doi: 10.1038/s41467-020-19793-8
Wang, J., Thingholm, L., Skieceviciene, J., Rausch, P., Kummen, M., Hov, J., et al. (2016). Genome-wide association analysis identifies variation in vitamin D receptor and other host factors influencing the gut microbiota. Nat. Genet. 48, 1396–1406. doi: 10.1038/ng.3695
Wang, J., Zheng, J., Shi, W., Du, N., Xu, X., Zhang, Y., et al. (2018). Dysbiosis of maternal and neonatal microbiota associated with gestational diabetes mellitus. Gut 67, 1614–1625. doi: 10.1136/gutjnl-2018-315988
Wrzosek, L., Ciocan, D., Hugot, C., Spatz, M., Dupeux, M., Houron, C., et al. (2021). Microbiota tryptophan metabolism induces aryl hydrocarbon receptor activation and improves alcohol-induced liver injury. Gut 70, 1299–1308. doi: 10.1136/gutjnl-2020-321565
Yang, J., Chen, L., Shang, X., Chen, Y., Zhao, S., Jin, S., et al. (2022). Roux-en-Y gastric bypass-induced perturbative changes in microbial communities and metabolic pathways in rats. Front. Microbiol. 13:1034839. doi: 10.3389/fmicb.2022.1034839
Yu, J., Youn, G., Choi, J., Kim, C., Kim, B., Yang, S., et al. (2021). Lactobacillus lactis and Pediococcus pentosaceus-driven reprogramming of gut microbiome and metabolome ameliorates the progression of non-alcoholic fatty liver disease. Clin. Transl. Med. 11:e634. doi: 10.1002/ctm2.634
Zhang, Q., Wu, Z., Zhao, S., Yang, J., Chen, L., Wang, X., et al. (2022). Identification and spatial visualization of dysregulated bile acid metabolism in high-fat diet-fed mice by mass spectral imaging. Front. Nutr. 9:858603. doi: 10.3389/fnut.2022.858603
Zhang, X., Shang, X., Jin, S., Ma, Z., Wang, H., Ao, N., et al. (2021c). Vitamin D ameliorates high-fat-diet-induced hepatic injury via inhibiting pyroptosis and alters gut microbiota in rats. Arch. Biochem. Biophys. 705:108894. doi: 10.1016/j.abb.2021.108894
Zhang, S., Wu, P., Tian, Y., Liu, B., Huang, L., Liu, Z., et al. (2021a). Gut microbiota serves a predictable outcome of short-term low-carbohydrate diet (LCD) intervention for patients with obesity. Microbiol. Spectr. 9:e0022321. doi: 10.1128/Spectrum.00223-21
Zhang, X., Coker, O., Chu, E., Fu, K., Lau, H., Wang, Y., et al. (2021b). Dietary cholesterol drives fatty liver-associated liver cancer by modulating gut microbiota and metabolites. Gut 70, 761–774. doi: 10.1136/gutjnl-2019-319664
Zhao, M., Zhao, L., Xiong, X., He, Y., Huang, W., Liu, Z., et al. (2020). TMAVA, a metabolite of intestinal microbes, is increased in plasma from patients with liver steatosis, inhibits gamma-butyrobetaine hydroxylase, and exacerbates fatty liver in mice. Gastroenterology 158, 2266–2281e2227. doi: 10.1053/j.gastro.2020.02.033
Zhao, S., Chen, L., Yang, J., Wu, Z., Wang, X., Zhang, Q., et al. (2022). Altered gut microbial profile accompanied by abnormal fatty acid metabolism activity exacerbates endometrial cancer progression. Microbiol. Spectr. 10:e0261222. doi: 10.1128/spectrum.02612-22
Keywords: gut microbiota, NAFLD, vitamin D, sphingolipid metabolism, tyrosine metabolism
Citation: Zhang X-L, Chen L, Yang J, Zhao S-S, Jin S, Ao N, Yang J, Liu H-X and Du J (2023) Vitamin D alleviates non-alcoholic fatty liver disease via restoring gut microbiota and metabolism. Front. Microbiol. 14:1117644. doi: 10.3389/fmicb.2023.1117644
Received: 06 December 2022; Accepted: 10 January 2023;
Published: 02 February 2023.
Edited by:
Xiaojiao Zheng, Shanghai Jiao Tong University Affiliated Sixth People’s Hospital, ChinaReviewed by:
Yukiko Miyamoto, University of California, San Diego, United StatesCopyright © 2023 Zhang, Chen, Yang, Zhao, Jin, Ao, Yang, Liu and Du. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hui-Xin Liu,  bGl1aHhAY211LmVkdS5jbg==; Jian Du,
bGl1aHhAY211LmVkdS5jbg==; Jian Du,  ZHVqaWFuX2NtdTRoQDE2My5jb20=
ZHVqaWFuX2NtdTRoQDE2My5jb20=
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.