- 1School of Ecology and Nature Conservation, Institute of Microbiology, Beijing Forestry University, Beijing, China
- 2International Exchange and Cooperation Department, Kunming University, Kunming, Yunnan, China
- 3Department of Horticulture and Food, Guangdong Eco-Engineering Polytechnic, Guangzhou, China
Polyporales is a diverse group of Agaricomycetes including more than 2,500 species belonging to 255 genera and 18 families. Recently, many studies focused on the classification of Polyporales, but the familial placements of some taxa remain uncertain. In this study, two new families, Climacocystaceae and Gloeoporellaceae of Polyporales, are proposed based on morphological characters and molecular data. Phylogenetic analyses of the two new families are inferred from the DNA sequences of the internal transcribed spacer regions (ITS), the large subunit of nuclear ribosomal RNA gene (nLSU), the largest subunit of RNA polymerase II gene (RPB1), the second largest subunit of RNA polymerase II gene (RPB2), and the translation elongation factor 1-α gene (TEF1). Furthermore, the divergence time of Polyporales was estimated as an additional taxonomic criterion based on the conserved regions of five DNA fragments (5.8S, nLSU, RPB1, RPB2, and TEF1). Bayesian evolutionary analysis revealed that the ancestor of Polyporales splits with a mean stem age of 136.53 Mya with a 95% highest posterior density (HPD) of 118.08–158.06 Mya. The mean stem ages of the families within Polyporales originated between 66.02 and 119.22 Mya, of which Climacocystaceae occurred in a mean stem age of 77.49 Mya with a 95% HPD of 61.45–93.16 Mya, and Gloeoporellaceae occurred in a mean stem age of 88.06 Mya with a 95% HPD of 67.15–107.76 Mya.
Introduction
Polyporales Gäum is one of the major orders of Basidiomycota (Kirk et al., 2008). Most species of the Polyporales are saprotrophic wood-decay fungi, which can cause white decay or brown decay of wood, and play a vital role in the degradation and reduction of forest ecosystems. Moreover, some species of Polyporales are edible fungi, medicinal fungi, or forest pathogens (Dai et al., 2007; Rajchenber and Robledo, 2013; Wu et al., 2019). Due to their important ecological functions and economic values, Polyporales had been extensively studied, and their members were increased rapidly. In Kirk et al. (2008), Polyporales contain about 1,800 species, 216 genera, and 13 families, while in He et al. (2019), about 2,500 species, 285 genera, and 18 families are included in Polyporales.
Previously, the establishment of families in Polyporales was basically based on morphological characteristics. Polyporaceae Fr. ex Corda is the oldest family in Polyporales, which was proposed by Fries (1838) to include all fungi with poroid hymenophores. Then, Irpicaceae Spirin and Zmitr., Meruliaceae Rea, Podoscyphaceae D.A. Reid, Sparassidaceae Herter, and Steccherinaceae Parmasto were proposed successively and still legitimately exist in the current concept of Polyporales (Herter, 1910; Rea, 1922; Reid, 1965; Parmasto, 1968; Spirin, 2003; Justo et al., 2017). Jülich (1981) proposed a considerable number of families of Basidiomycetes, some of which belong to Polyporales were rarely used and treated as synonyms.
Since the 21st century, DNA sequencing and phylogenetic techniques have been widely used in the systematic study of Polyporales (Binder et al., 2005; Larsson, 2007; Miettinen et al., 2012). Binder et al. (2013) presented a phylogenetic and phylogenomic overview of the Polyporales and listed 40 validly published and legitimate family names. Zhao et al. (2015) introduced a new family Fragiliporiaceae Y-CD, B-KC, and C. L. Zhao based on the combination of morphological characters and molecular data. Justo et al. (2017) provided a phylogenetic overview of Polyporales, 18 clades in the Polyporales were assigned at the family level; the climacocystis clade (Climacocystis Kotl. and Pouzar, Diplomitoporus Domański) and Tyromyces merulinus (Berk.) G. Cunn. cannot be assigned to a family within Polyporales. He et al. (2019) carried out an outline of all genera of Basidiomycota; in which 19 families were placed in Polyporales, including 18 families accepted by Justo et al. (2017) and Fragiliporiaceae. However, the genera Climacocystis and Diplomitoporus remained with an uncertain familial placement. Liu et al. (2022) presented a systematic classification and phylogenetic relationships of the brown-rot fungi within the Polyporales; the study showed that 29 clades are assigned a family name, including four new brown-rot fungal families, viz., Auriporiaceae B.K. Cui, Shun Liu & Y.C. Dai, Piptoporellaceae B.K. Cui, Shun Liu & Y.C. Dai, Postiaceae B.K. Cui, Shun Liu & Y.C. Dai and Taiwanofungaceae B.K. Cui, Shun Liu & Y.C. Dai. They focused on the phylogenetic relationships of the brown-rot fungi within the Polyporales, and the number and composition of white-rot fungi family were consistent with the study of He et al. (2019).
Fruiting body types and wood decay types are two key traits in the evolutionary origins and genetic bases of fungi (Nagy et al., 2017). The Polyporales is a diverse group of Agaricomycetes, not only in molecular sequences but also in morphological characteristics (fruiting body: Resupinate, effused-reflexed, pileate-sessile, pileate-stipitate, cauliflower-like, etc.; Hymenophores: Poroid, daedaleoid, hydnoid, lamellate, labyrinthine, odontoid, etc.). Moreover, Polyporales include two types of wood decay fungi, white-rot fungi and brown-rot fungi (Binder et al., 2013; Justo et al., 2017). The varied fruiting body types and wood decay types indicate that there are complex evolutionary relationships among the members of Polyporales. Recently, divergence time was used as important criteria for the classification and estimation of evolutionary time in Basidiomycota (Chen et al., 2015; Song et al., 2016; Zhao et al., 2016, 2017; Song and Cui, 2017; He et al., 2019; Zhu et al., 2019; Wu et al., 2020; Wang et al., 2021; Ji et al., 2022).
During the investigations of wood decay fungi, abundant samples of Climacocystis, Diplomitoporus, and Gloeoporellus Zmitr. were collected. To determine their phylogenetic positions within Polyporales, phylogenetic analyses were carried out based on the combined sequence datasets of ITS + nLSU + RPB1 and ITS + nLSU + RPB1 + RPB2 + TEF1. In addition, divergence time, as an additional criterion, was estimated by the molecular clock analyses with 5-gene loci (5.8S, nLSU, RPB1, RPB2, and TEF1).
Materials and methods
Morphological studies
The specimens used in this study are deposited at the herbarium of the Institute of Microbiology, Beijing Forestry University, China (BJFC). Morphological studies and abbreviations of this study followed the study of Sun et al. (2020) and Ji et al. (2022).
Molecular studies and phylogenetic analysis
The approaches for DNA extraction and polymerase chain reaction (PCR) used in this study with some modifications followed the study of Cui et al. (2019) and Liu et al. (2021). The primer pairs are ITS5 and ITS4 for ITS regions, LR0R and LR7 for nLSU regions, RPB1-Af and RPB1-Cr for the RPB1 gene, bRPB2-6F and bRPB2-7R for the RPB2 gene, and EF1-983F and EF1-1567R for TEF1 (White et al., 1990; Matheny et al., 2002; Matheny, 2005; Rehner and Buckley, 2005). The PCR products were purified and sequenced at the Beijing Genomics Institute (BGI), China. All newly generated sequences were deposited at GenBank (Table 1).
Table 1. A list of species, specimens, and GenBank accession number of sequences used for phylogenetic analyses in this study.
In the phylogenetic analyses, we selected exact and more gene fragments of representative species from previous studies. Other sequences were obtained from GenBank (Table 1). The sequences used in this study were aligned in MAFFT 7 (Katoh and Standley, 2013)1 and then manually adjusted in BioEdit (Hall, 1999). Each alignment sequence was spliced with Mesquite (Maddison and Maddison, 2017). The missing sequences and ambiguous nucleotides were coded as “N.”
The phylogenetic analysis methods used in this study followed Shen et al. (2019) and Sun et al. (2022). The sequences of Heterobasidion annosum (Fr.) Bref. and Stereum hirsutum (Willd.) Pers. were obtained as outgroups for the phylogenetic analyses following Binder et al. (2013) and Justo et al. (2017). The congruences of gene sequence datasets were evaluated with the incongruence length difference (ILD) test (Farris et al., 1994) with PAUP* 4.0b10 (Swofford, 2002), under 1,000 homogeneity replicates. Maximum parsimony analysis was applied to the combined gene dataset and the tree construction procedure was performed in PAUP* version 4.0b10. Clade robustness was assessed using a bootstrap (BT) analysis with 1,000 replicates (Felsenstein, 1985). Descriptive tree statistic tree length (TL), consistency index (CI), retention index (RI), rescaled consistency index (RC), and homoplasy index (HI) were calculated for each Most Parsimonious Tree (MPT) generated.
Maximum likelihood (ML) analysis was performed with RAxML-HPC v. 8.2.3 (Stamatakis, 2014) with 1,000 ML searches under the GTRGAMMA model, and only the maximum likelihood best tree from all searches was kept. Bayesian inference (BI) was performed using MrBayes v. 3.2 (Ronquist and Huelsenbeck, 2003) with four simultaneous independent chains for all datasets, performing five million generations until the split deviation frequency value of <0.01, and sampled every 100th generation. The first 25% of sampled trees were discarded as burn-in, while the remaining ones were used to calculate Bayesian posterior probabilities (BPPs) of the clades.
Phylogenetic trees were inferred from the combined sequences datasets of ITS + nLSU + RPB1 and ITS + nLSU + RPB1 + RPB2 + TEF1. Trees were viewed in FigTree v1.4.4.2 Branches that received bootstrap supports for maximum parsimony (MP), maximum likelihood (ML), and Bayesian posterior probabilities (BPP) greater than or equal to 75% (MP and ML) and 0.95 (BPP) were considered as significantly supported, respectively.
Divergence time estimation
Three fossil calibrations, Archaeomarasmius leggetti Hibbett, D. Grimaldi and Donoghue, Quatsinoporites cranhamii S. Y. Sm., Currah and Stockey, and Paleopyrenomycites devonicus Taylor, Hass, Kerp, M. Krings and Hanlin, were used in the divergence time estimating. Archaeomarasmius leggetti was used as the representative of the minimum age of Agaricales at 90–94 Mya (Hibbett et al., 1997); Q. cranhamii was the representative of the minimum age of Hymenochaetales at 113 Mya (Smith et al., 2004); P. devonicus was used as the representative of the minimum age between Basidiomycota and Ascomycota at 400 Mya (Taylor et al., 2005; Berbee and Taylor, 2010). Divergence time is estimated with the BEAST v1.8.0 software package (Drummond et al., 2012) with 5.8S, nLSU, RPB1, RPB2, and TEF1 sequences representing main lineages in Basidiomycota (Table 1).
All the DNA sequences of 5.8S, nLSU, RPB1, RPB2, and TEF1 were aligned in MAFFT 7 (Katoh and Standley, 2013) and manually adjusted in BioEdit (Hall, 1999). ModelTest 3.7 was used to estimate the rate of evolutionary changes at nucleic acids with the GTR substitution model (Posada and Crandall, 1998). BEATUti v2 was used to generate the BEAST XML input file. A log-normal distribution is employed for molecular clock analysis (Drummond and Rambaut, 2007). The clock model was set to an uncorrelated lognormal relaxed clock (Drummond et al., 2006; Lepage et al., 2007). A Yule speciation model is selected as the prior choice assuming a constant speciation rate per lineage. Gamma prior distribution was used for fossil node calibrations (shape = 1.0, scale = 50.0), and the offset was set at 90.0, 113.0, and 400.0 for Agaricales, Hymenochaetales, and Basidiomycota, respectively (Sánchez-Ramírez et al., 2014). All the ucld. mean parameters for different genes were set to gamma prior distribution, shape = 1.0, scale = 0.001, and offset = 0.0 (Sánchez-Ramírez et al., 2014). Overall, four independent Markov chain Monte Carlo (MCMC) chains of 100 million generations were conducted and saving trees every 5,000th generation. The log file is analyzed in Tracer v1.6 to confirm that the estimated effective sample size (ESS) is ≥2003. A maximum clade credibility (MCC) tree is summarized in TreeAnnotator, removing the first 10% of the sampled trees as burn-in and setting a posterior probability limit of 0.80, and viewed in FigTree v1.4.4.
Results
Phylogeny
The combined 3-gene (ITS + nLSU + RPB1) sequence dataset had an aligned length of 3,289 characters, including gaps (628 characters for ITS, 1,333 characters for nLSU, and 1,328 characters for RPB1), of which 1,415 characters were constant, 130 were variable and parsimony-uninformative, and 1,744 were parsimony-informative. MP analysis yielded 54 equally parsimonious trees (TL = 25,472, CI = 0.154, RI = 0.653, RC = 0.101, HI = 0.846). The best-fit evolutionary models applied in the Bayesian analyses were selected by MrModeltest2 v. 2.3 for each region of the three genes, and the model for ITS, nLSU, and RPB1 was GTR + I + G with an equal frequency of nucleotides. This model was applied in the Bayesian analyses for the combined dataset. ML analysis resulted in a similar topology as MP and Bayesian analyses, and only the ML topology is shown in Figure 1.
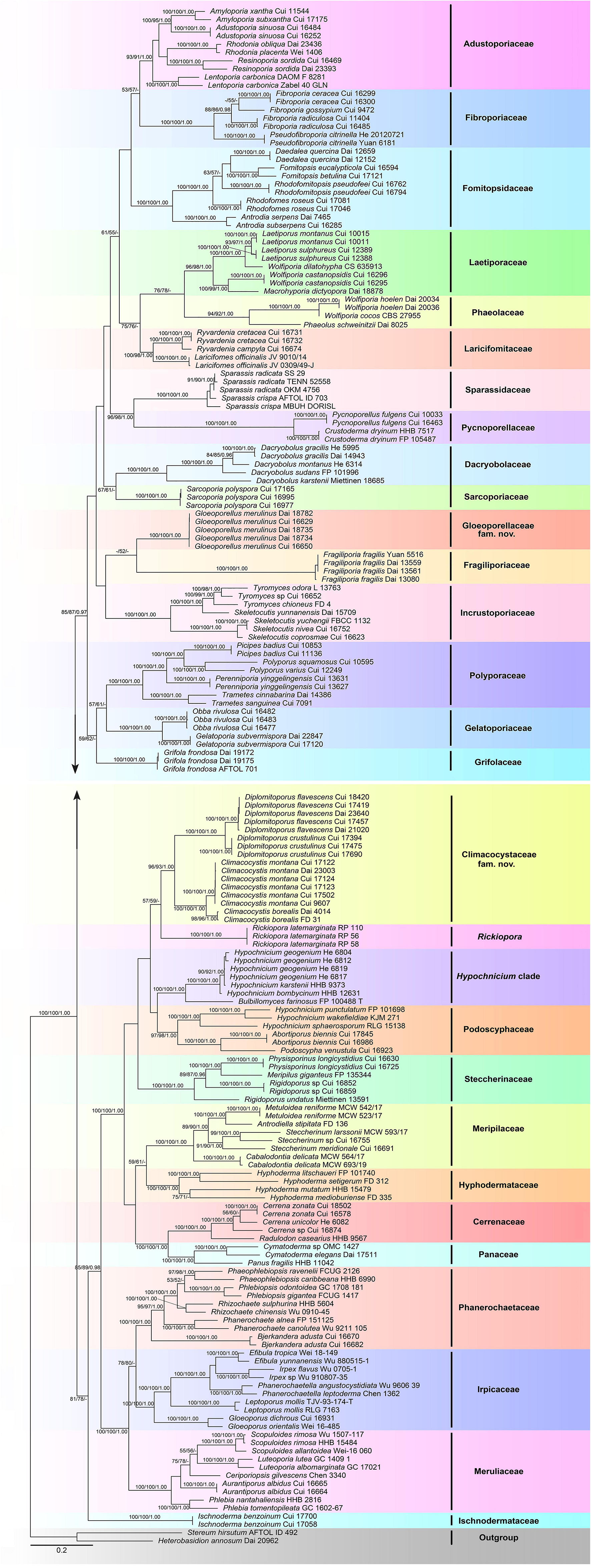
Figure 1. Maximum likelihood tree illustrating the phylogeny of the Polyporales based on the combined sequence dataset of ITS + nLSU + RPB1. Branches are labeled with parsimony bootstrap proportions higher than 50%, maximum likelihood bootstrap higher than 50% and Bayesian posterior probabilities more than 0.90, respectively.
The combined 5-gene (ITS + nLSU + RPB1 + RPB2 + TEF1) sequence dataset had an aligned length of 4,849 characters, including gaps (628 characters for ITS, 1,333 characters for nLSU, 1,328 characters for RPB1, 1,020 characters for RPB2, and 540 characters for TEF1), of which 1,980 characters were constant, 177 were variable and parsimony-uninformative, and 2,692 were parsimony-informative. MP analysis yielded 80 equally parsimonious trees (TL = 41,080, CI = 0.149, RI = 0.634, RC = 0.094, HI = 0.851). The best-fit evolutionary models applied in the Bayesian analyses were selected by MrModeltest2 v. 2.3 for each region of the five genes, and the model for ITS, LSU, RPB1, RPB2, and TEF1 was GTR + I + G with an equal frequency of nucleotides. This model was applied in the Bayesian analyses for the combined dataset. ML analysis resulted in a similar topology as MP and Bayesian analyses, and only the ML topology is shown in Figure 2.
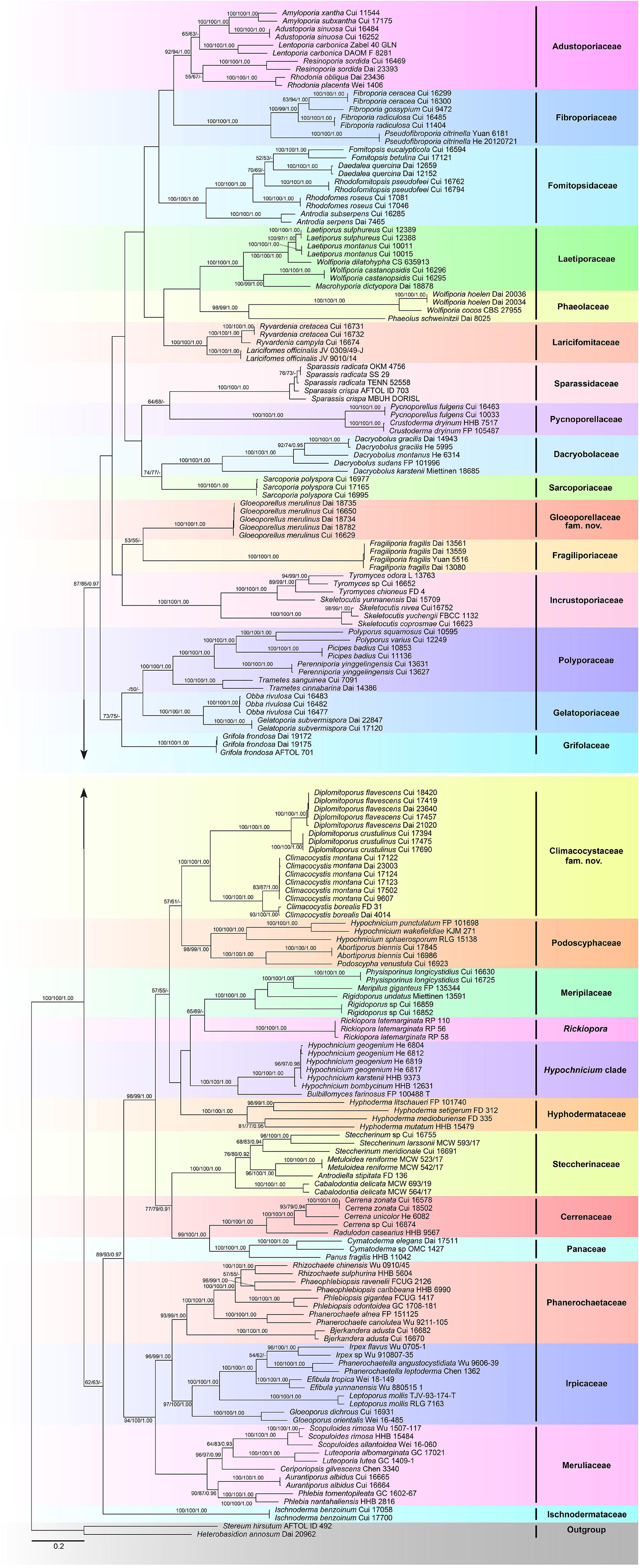
Figure 2. Maximum likelihood tree illustrating the phylogeny of the Polyporales based on the combined sequence dataset of ITS + nLSU + RPB1 + RPB2 + TEF1. Branches are labeled with parsimony bootstrap proportions higher than 50%, maximum likelihood bootstrap higher than 50%, and Bayesian posterior probabilities more than 0.90, respectively.
The phylogenetic trees inferred from ITS + nLSU + RPB1 and ITS + nLSU + RPB1 + RPB2 + TEF1 gene sequences were obtained from 185 fungal samples representing 113 taxa of Polyporales and two taxa of Russulales (Figures 1, 2). A total of 810 sequences derived from five gene loci (ITS, nLSU, RPB1, RPB2, and TEF1) were used to reconstruct the phylogenetic trees, of which 153 were newly generated. Phylogenetic analyses showed that 27 clades within Polyporales are assigned family names, including two new families, viz., Climacocystaceae fam. nov. and Gloeoporellaceae fam. nov., established for the climacocystis lineage (Climacocystis and Diplomitoporus) and Gloeoporellus merulinus (Berk.) Zmitr. (= Tyromyces merulinus) (Figures 1, 2).
Divergence time estimation
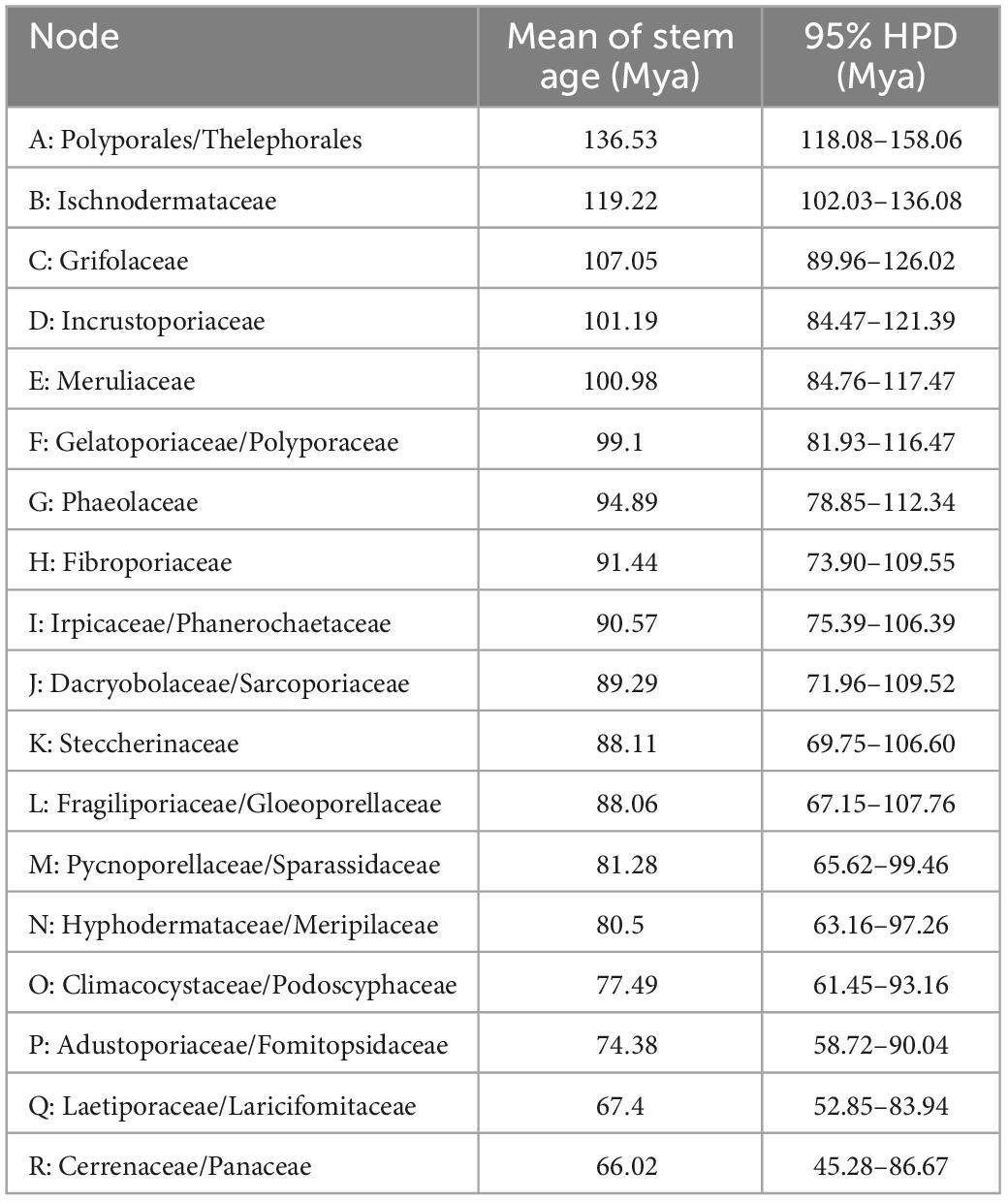
The combined dataset (5.8S, nLSU, RPB1, RPB2, and TEF1) for the molecular clock analysis includes 174 fungal samples representing 132 taxa, of which 123 fungal samples represent 87 taxa belonging to Polyporales. The MCMC tree (Figure 3) shows that the most recent ancestor of Polyporales evolved during the early Cretaceous, approximately 136.53 Mya with a 95% highest posterior density (HPD) of 118.08–158.06 Mya. The youngest families of Polyporales are Cerrenaceae Miettinen, Justo and Hibbett and Panacea Miettinen, Justo and Hibbett, occurring in a mean stem age of 66.02 Mya with a 95% HPD of 45.28–86.67 Mya, the oldest family of Polyporales is Ischnodermataceae Jülich, occurring in a mean stem age of 119.22 with a 95% HPD of 102.03–136.08 Mya, and the average divergence time of the families in Polyporales is 86.34 Mya. The estimated divergence times for families of Polyporales are summarized in Table 2.
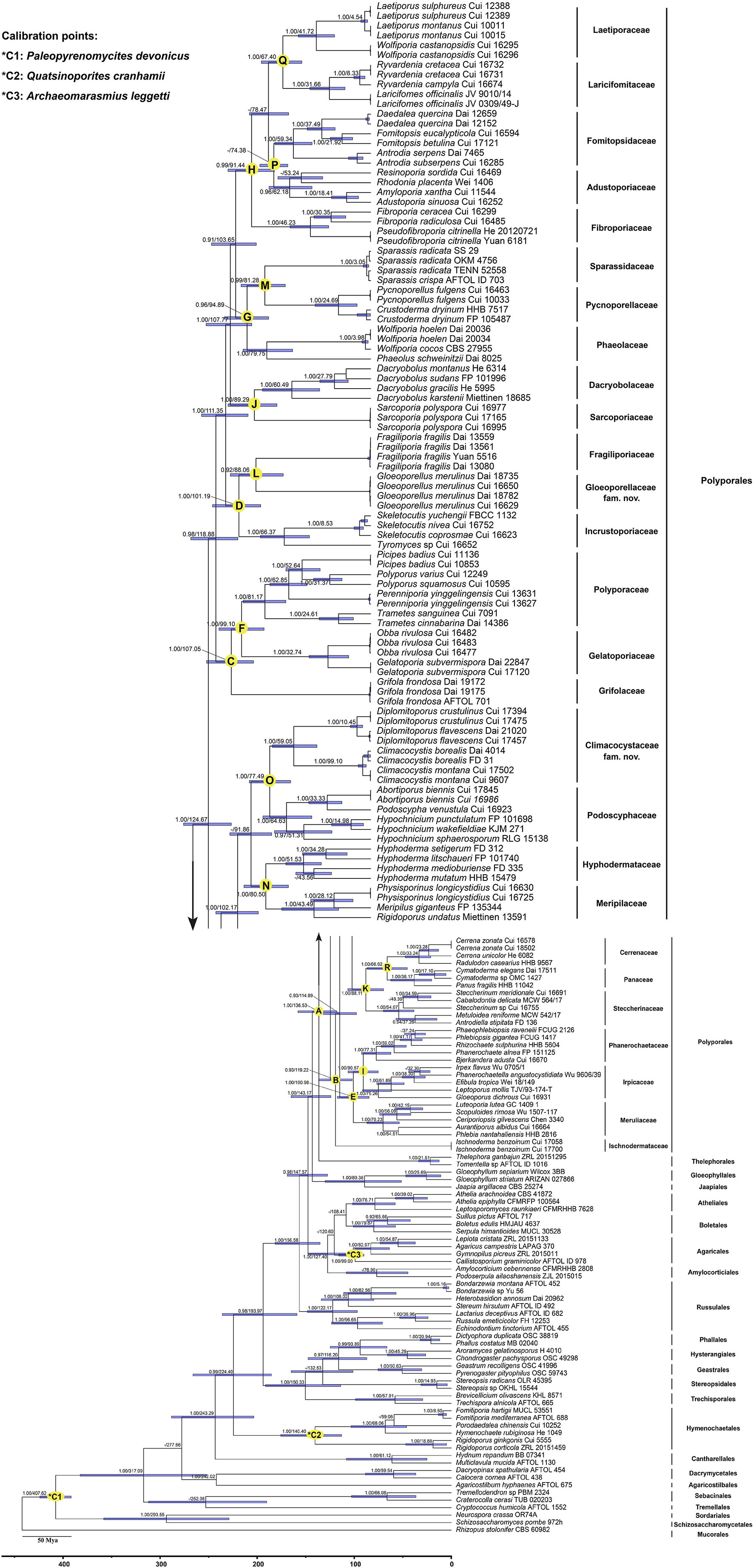
Figure 3. Divergence time estimation of families within Polyporales from Bayesian evolutionary analysis sampling tree based on the combined sequence dataset of 5.8S + nLSU + RPB1 + RPB2 + TEF1. Posterior probabilities not less than 0.80 and the mean ages of each node are annotated. The 90% highest posterior densities of divergence time estimation are marked by horizontal bars.
Taxonomy
Climacocystaceae B.K. Cui, Shun Liu & Y.C. Dai, fam. nov.
MycoBank: MB 840367
Type genus: Climacocystis.
Diagnosis: Basidiomata annual, pileate, resupinate to effused-reflexed, soft corky and watery when fresh, brittle, corky to hard corky when dry. Hymenophores poroid. Hyphal system monomitic, dimitic to trimitic; generative hyphae with clamp connections, skeletal hyphae IKI–, CB–. Cystidia present or absent, cystidioles occasionally present. Basidiospores broadly ellipsoid to globose, colorless, thin- to slightly thick-walled, smooth, IKI–, CB–. Causing a white rot.
Genera: Climacocystis, Diplomitoporus.
Climacocystis Kotl. and Pouzar, Ceská Mykologie 12 (2): 95, 1958.
MycoBank: MB 17325
Type species: Climacocystis borealis (Fr.) Kotl. and Pouzar.
Diagnosis: Basidiomata annual, pileate, sessile to laterally substipitate, usually imbricate, soft and watery when fresh, corky to hard corky and light in weight when dry. Pileus applanate, fan-shaped to dimidiate. Pileal surface white to cream, tomentose to hirsute when fresh, becoming cream, yellowish-brown to orange-brown, glabrous or tufted with short stiff hairs when dry, often radially furrowed, azonate; margin acute. Pore surface white to cream when fresh, becoming cream, clay-buff to orange-brown when dry; pores angular or irregular; dissepiments thin, entire to lacerate. Context white to clay-buff, corky to hard corky. Tubes white, clay-buff to orange-brown, corky to hard corky. Hyphal system monomitic; generative hyphae with clamp connections, IKI–, CB–; tissues unchanged in KOH. Cystidia present, ventricose, colorless, thin- to thick-walled, smooth or apically encrusted. Basidia clavate, colorless, thin-walled. Basidiospores ellipsoid to subcylindrical, colorless, thin-walled, smooth, IKI–, CB–. Causing a white rot.
Notes: Climacocystis was established by Kotlába and Pouzar (1958) and typified by C. borealis, which is widely distributed in the northern hemisphere (Gilbertson and Ryvarden, 1986; Núñez and Ryvarden, 2001; Dai, 2012). Song et al. (2014) carried out taxonomic and phylogenetic studies on Climacocystis in China, and C. montana B-KC and JS described high elevations in southwestern China based on morphological and molecular characteristics. Currently, two species are accepted in Climacocystis, including C. borealis and C. montana. Basidiomata of C. borealis and C. montana are shown in Figure 4.

Figure 4. Basidiomata of Climacocystis and Diplomitoporus species: C. borealis (A,B: Dai 13028); C. montana (C,D: Cui 17502); D. crustulinus (E: Cui 17394; F: Cui 17475); D. flavescens (G: Dai 22798; H: Dai 23640). Scale bars: A = 2 cm; B,C,D = 3 cm; E,F,G,H = 1.5 cm.
Specimens examined: Climacocystis borealis. CHINA. Heilongjiang Province, Yichun, Wuying, Fenglin Nature Reserve, on stump of Pinus sp., 8 September 2002, Dai 3703 (BJFC 000443). FINLAND. Helsinki, Vantaa, Tamisto Nature Reserve, on fallen trunk of Picea sp., 22 September 2010, Dai 11798 (BJFC 008905); on stump of Picea sp., 15 November 2011, Dai 12681 (BJFC 012265). SWITZERLAND. Geneva, on living tree of Picea sp., 25 November 2012, Dai 13208 (BJFC 014072). Climacocystis montana. CHINA. Xizang Autonomous Region (Tibet), Leiwuqi County, on fallen trunk of Picea sp., 22 September 2010, Cui 9603 (BJFC 008541, holotype), Cui 9610 (BJFC 008548), Cui 9612 (BJFC 008550), Cui 9607 (BJFC 008545). Sichuan Province, Jiuzhaigou County, Jiuzhaigou Nature Reserve, on fallen trunk of Picea sp., 11 October 2012, Cui 10603 (BJFC 013528). Yunnan Province, Shangri-La County, Pudacuo National Park, on root of Picea sp., 17 September 2018, Cui 17122 (BJFC 030422), Cui 17123 (BJFC 030423), Cui 17124 (BJFC 030424), 13 August 2019, Cui 17502 (BJFC 034361); on root of Picea sp., 7 September 2021, Dai 23003 (BJFC 037576).
Diplomitoporus Domanski, Acta Societatis Botanicorum Poloniae 39: 191, 1970.
MycoBank: MB 17515
Type species: Diplomitoporus flavescens (Bres.) Domański.
Diagnosis: Basidiomata annual, resupinate to effuse-reflexed, fibrous, soft corky when fresh, brittle to hard corky when dry. Pore surface white, cream to straw-yellow when fresh, becoming cream, ochraceous to dark ochraceous when dry; pores round to angular. Context cream to ochraceous, brittle to corky. Tubes concolorous with the pore surface, brittle to hard corky. Hyphal system dimitic to trimitic; generative hyphae with clamp connections; skeletal hyphae IKI–, CB–. Cystidia absent; cystidioles occasionally present, subclavate to fusoid, colorless, thin-walled, smooth. Basidia clavate, subclavate to subglobose, colorless, thin-walled. Basidiospores allantoid to ellipsoid or globose, colorless, thin- to slightly thick-walled, smooth, IKI–, CB–. Causing a white rot.
Notes: Diplomitoporus was described by Domanski (1970) with D. flavescens as type species. Some species of Diplomitoporus have been transferred to other genera based on morphological or molecular evidence (Ghobad-Nejhad and Dai, 2010; Miettinen, 2012). Baltazar et al. (2014) reviewed the species of Diplomitoporus from Brazil and reported seven species of this genus in Brazil. In recent years, Ryvarden and co-authors described several Diplomitoporus species based on their morphological characteristics (Ryvarden, 2018, 2019, 2020; Decock and Ryvarden, 2020). Although 34 Diplomitoporus species are recorded in Index Fungorum,4 only the molecular sequences of D. crustulinus and D. flavescens are available in GenBank. Basidiomata of D. crustulinus and D. flavescens are shown in Figure 4.
Specimens examined: Diplomitoporus crustulinus. CHINA. Sichuan Province, Daocheng County, Yading Nature Reserve, on fallen branch of Picea sp., 11 August 2019, Cui 17394 (BJFC 034253); Jiulong County, on fallen branch of Abies sp., 12 September 2019, Cui 17690 (BJFC 034549). Yunnan Province, Shangri-La County, Pudacuo National Park, on fallen branch of Picea sp., 13 August 2019, Cui 17475 (BJFC 034334). Diplomitoporus flavescens. BELARUS. Brestskaya Voblasts, Belavezhskaya Pushcha National Park, on fallen trunk of Pinus sp., 18 October 2019, Dai 21020 (BJFC 032679). CHINA. Hebei Province, Zhuolu County, Xiaowutai Nature Reserve, on dead tree of Pinus sp., 9 September 2017, Dai 18096 (BJFC 025626), Dai 18097 (BJFC 025627). Jilin Province, Antu County, Changbaishan Nature Reserve, on fallen branch of Pinus sp., 20 September 2019, Dai 20846 (BJFC 032515). Sichuan Province, Xiangcheng County, on fallen branch of Pinus sp., 12 August 2019, Cui 17419 (BJFC 034278), Cui 17457 (BJFC 034316), Cui 17459 (BJFC 034318). Xizang Autonomous Region (Tibet), Linzhi, Bomi County, Bulang, on fallen branch of Pinus yunnanensis, 21 October 2021, Dai 23640 (BJFC 038212); on fallen trunk of Pinus yunnanensis, 21 October 2021, Dai 23650 (BJFC 038222); Chayu County, Cibagou Nature Reserve, on fallen branch of Pinus densata, 10 September 2020, Cui 18420 (BJFC 035281), Cui 18428 (BJFC 035289), Cui 18444 (BJFC 035305); Mangkang County, Jueba Mountain, on fallen branch of Pinus densata, 9 September 2020, Cui 18392 (BJFC 035253). Yunnan Province, Lanping County, Tongdian, Luoguqing, on fallen branch of Pinus yunnanensis, 3 September 2021, Dai 22798 (BJFC 037371).
Gloeoporellaceae B.K. Cui, Shun Liu & Y.C. Dai, fam. nov.
MycoBank: MB 840368
Type genus: Gloeoporellus.
Diagnosis: Basidiomata annual, resupinate to effused-reflexed, soft corky to corky when fresh, corky to fragile when dry. Hymenophores poroid. Hyphal system dimitic; generative hyphae with clamp connections, binding hyphae IKI–, CB+. Cystidia absent; cystidioles present. Basidiospores allantoid, colorless, thin-walled, smooth, IKI–, CB–. Causing a white rot.
Genus: Gloeoporellus.
Gloeoporellus Zmitr., Folia Cryptogamica Petropolitana 6: 85, 2018.
MycoBank: MB 827569
Type species: Gloeoporellus merulinus.
Diagnosis: Basidiomata annual, resupinate to effuse-reflexed, soft corky to corky when fresh, corky to fragile when dry. Pore surface buff-yellow, yellowish brown to apricot-orange when fresh, yellowish buff to orange-yellow when dry; pores round to angular. Context buff-yellow to orange-yellow, corky to fragile. Tubes concolorous with the pore surface, corky to fragile. Hyphal system dimitic; generative hyphae with clamp connections; binding hyphae IKI–, CB+. Cystidia absent; cystidioles present, tubular to fusoid, colorless, thin-walled, smooth. Basidia subclavate, colorless, thin-walled. Basidiospores allantoid to cylindrical, colorless, thin- to slightly thick-walled, smooth, IKI–, CB–. Causing a white rot.
Notes: Tyromyces merulinus was proposed by Cunningham (1965) as a new combination. This species distributes in Argentina, Australia, and New Zealand in the southern hemisphere, and the type locality is Tasmania, Australia (Cunningham, 1965). In Justo et al. (2017), Tyromyces merulinus cannot be placed with certainty in any of the recognized families. Zmitrovich (2018) presented the system of Polyporaceae and carried out the overview of the order Polyporales; Gloeoporellus was proposed as a new genus, with Tyromyces merulinus as type species. Zmitrovich (2018) and He et al. (2019) placed Gloeoporellus into Incrustoporiaceae Jülich, but without the support of phylogenetic analysis. Only one species, Gloeoporellus merulinus, is accepted in this genus now. Basidiomata of G. merulinus are shown in Figure 5.
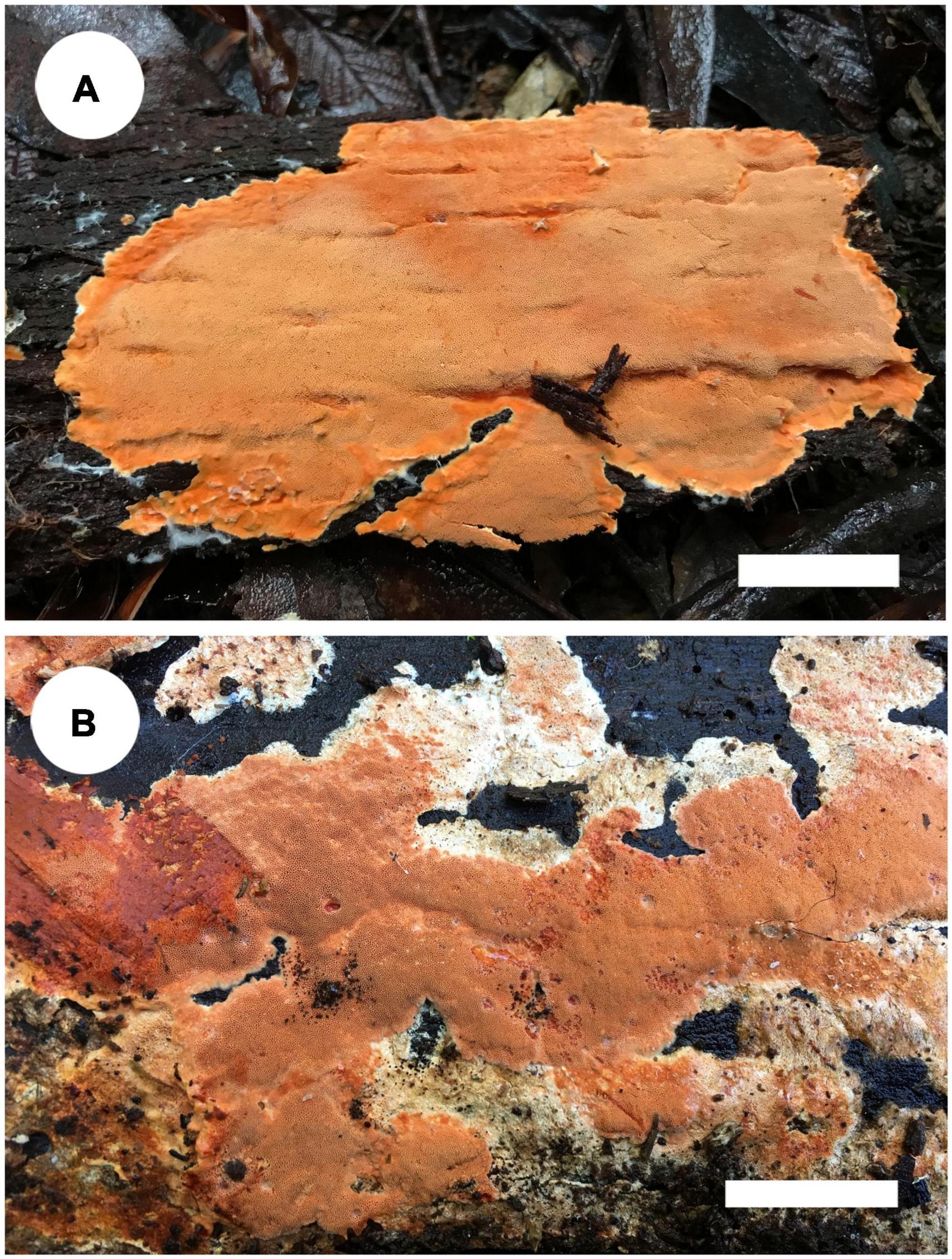
Figure 5. Basidiomata of Gloeoporellus species: G. merulinus (A: Cui 16724; B: Dai 18735). Scale bars: A,B = 2 cm.
Specimens examined: Gloeoporellus merulinus. AUSTRALIA. Tasmania, Mount Field Forest, close to Mount National Park, on rotten wood of Nothofagus cunninghamii, 14 May 2018, Dai 18734 (BJFC 027203); on rotten wood of Nothofagus sp., 14 May 2018, Dai 18735 (BJFC 027204); on living tree of Eucalyptus sp., 14 May 2018, Cui 16676 (BJFC 029975); Hobart, Mount Wellington, on rotten wood of Acacia sp., 13 May 2018, Cui 16629 (BJFC 029928); Timbs Track, on fallen trunk of Nothofagus sp., 14 May 2018, Cui 16650 (BJFC 029949), Cui 16668 (BJFC 029967); Arve River, Streamside Nature Reserve, on dead tree of Eucalyptus sp., 15 May 2018, Dai 18782 (BJFC 027250); on fallen trunk of Eucalyptus sp., 15 May 2018, Cui 16724 (BJFC 030023).
Discussion
The Polyporales are a diverse group of Agaricomycetes, which have received extensive attention and studies. Some mycologists have attempted to adopt ribosomal RNA genes to study the phylogeny of Polyporales, but the results suggested that it is difficult to resolve the taxonomic structure of Polyporales (Larsson et al., 2004; Binder et al., 2005). Subsequently, protein-coding genes, including RPB1, RPB2, and TEF1, were applied to the phylogenetic study of Polyporales (Matheny et al., 2007; Binder et al., 2013; Zhao et al., 2015; Justo et al., 2017). To better verify the independent status of the two new families and provide more molecular data for future research, the phylogenetic analyses of Polyporales are inferred from the combined datasets of ITS + nLSU + RPB1 sequences (Figure 1) and ITS + nLSU + RPB1 + RPB2 + TEF1 sequences (Figure 2). The topological structures obtained from the phylogenetic analyses of the two sequence datasets are slightly different, probably due to the different gene fragments used in the phylogenetic analyses. The results showed that 27 lineages of Polyporales are recognized at the family level (Figures 1, 2), viz., Adustoporiaceae Audet, Cerrenaceae, Dacryobolaceae Jülich, Fibroporiaceae Audet, Fomitopsidaceae Jülich, Fragiliporiaceae, Gelatoporiaceae Miettinen, Justo and Hibbett, Grifolaceae Jülich, Hyphodermataceae Jülich, Incrustoporiaceae, Irpicaceae, Ischnodermataceae, Laetiporaceae Jülich, Laricifomitaceae Jülich, Meripilaceae Jülich, Meruliaceae, Panaceae, Phaeolaceae Jülich, Phanerochaetaceae Jülich, Podoscyphaceae, Polyporaceae, Pycnoporellaceae Audet, Sarcoporiaceae Audet, Sparassidaceae, Steccherinaceae, the climacocystis lineage (Climacocystis and Diplomitoporus) and Gloeoporellus merulinus (Tyromyces merulinus) could not be recognized in any existing families and they are proposed as two new families.
Binder et al. (2005) showed that Climacocystis nested inside the antrodia clade, but other analyses showed that Climacocystis grouped in the residual polyporoid clade (Miettinen et al., 2012; Binder et al., 2013; Justo et al., 2017). Justo et al. (2017) indicated that Climacocystis and Diplomitoporus grouped together with high support within the residual clade and could not be assigned to any recognized family of the Polyporales. Perhaps, previous studies lacked sufficient morphological features and molecular data to determine the family level of Climacocystis and Diplomitoporus, so their classifications at the family level were treated as incertae sedis (He et al., 2019). In the present study, Climacocystis and Diplomitoporus grouped together with high support within the residual clade (100% MP, 100% ML, 1.00 BPP; Figures 1, 2). This lineage has unique morphological characters and forms a well-supported clade. Phylogenetically, Climacocystaceae was closely related to the white-rot fungal families Hyphodermataceae, Meripilaceae, Podoscyphaceae, and Steccherinaceae (Figures 1, 2). Morphologically, Hyphodermataceae differs by having corticioid basidiomata, monomitic hyphal system, and thin-walled basidiospores; Meripilaceae differs in having monomitic or dimitic hyphal system without clamped generative hyphae; Podoscyphaceae differs by possessing mostly pileate basidiomata with smooth, ridged, or poroid hymenophore, dimitic or trimitic hyphal system, and thin-walled basidiospores; Steccherinaceae differs in possessing poroid or hydnoid hymenophore, dimitic hyphae system, mostly thin-walled, and rather small basidiospores (Reid, 1965; Parmasto, 1968; Jülich, 1981; Justo et al., 2017). Thus, a new family, Climacocystaceae, is proposed based on phylogenetic analyses and morphological characters. Rickiopora Westph., Tomšovskı, and Rajchenb. Were established by Westphalen et al. (2016) and related to Climacocystis. But in the current study, Rickiopora cannot be grouped with Climacocystaceae with high support (Figures 1, 2). Hypochnicium J. Erikss. and Bulbillomyces Jülich were grouped together and given an informal name as the hypochnicium clade, and the hypochnicium clade was closely related to the climacocystis clade without high support (Justo et al., 2017). In our current study, the hypochnicium clade is not closely related to the climacocystis clade (Figures 1, 2). Regarding the phylogenetic relationship between the climacocystis clade and related taxa, we found some differences between the current study and previous studies. This may be due to the difference in the number and composition of samples, gene fragments, and analysis methods used in the phylogenetic analysis. Nevertheless, all phylogenetic analyses support that the climacocystis clade cannot be placed in any recognized family and should be established as a new family.
Justo et al. (2017) revealed that Tyromyces merulinus is sister to Incrustoporiaceae, and this species cannot be assigned to a family within Polyporales. Subsequently, Gloeoporellus was proposed to accommodate Tyromyces merulinus (Zmitrovich, 2018) and was placed in the Incrustoporiaceae (Zmitrovich, 2018; He et al., 2019) without the verification of the phylogenetic analysis. In the present study, specimens of Gloeoporellus merulinus are grouped together with high support (100% MP, 100% ML, 1.00 BPP; Figures 1, 2), and are closely related to Fragiliporiaceae and Incrustoporiaceae without statistical support. Morphologically, Fragiliporiaceae resembles Gloeoporellaceae by having an annual growth habit, resupinate basidiomata, clamped generative hyphae, and thin-walled basidiospores. However, Fragiliporiaceae differs by its brittle basidiomata, grayish-buff to lavender pore surface when fresh, vinaceous gray to grayish brown when dry, larger pores, monomitic hyphal system, larger, and allantoid basidiospores (Zhao et al., 2015). Gloeoporellaceae and Incrustoporiaceae share poroid hymenophores, clamped generative hyphae and thin-walled basidiospores, but Incrustoporiaceae differs by having pileate, resupinate to effused-reflexed basidiomata, monomitic, dimitic to trimitic hyphal system, allantoid to ellipsoid or subglobose basidiospores, and tips of generative hyphae at tube mouths commonly with rose-thorn encrustations (Justo et al., 2017; Korhonen et al., 2018; Yuan et al., 2020). Thus, a new family, Gloeoporellaceae, is proposed to accommodate Gloeoporellus merulinus.
In the current study, the molecular clock analysis is executed to verify the taxonomic system for Polyporales with the estimated divergence time. The MCC tree (Figure 3) shows that the ancestor of Agaricales, Amylocorticiales, Cantharellales, Hymenochaetales, Hysterangiales, Phallales, Polyporales, Russulales, and Thelephorales split at about 120.6, 127.4, 243.29, 224.4, 93.89, 93.89, 136.53, 156.58, and 136.53 Mya, respectively; these data generally agree with previous studies (Chen et al., 2015; He et al., 2019; Wang et al., 2021; Ji et al., 2022). He et al. (2019) showed the mean of stem age of families within Agaricomycotina in a range of 27–259 Mya, among which the mean of stem age of six families in Polyporales ranged from 62 to 106 Mya. The current molecular clock analysis shows that the families in Polyporales diverged between 66.02 and 119.22 Mya (Figure 3 and Table 2), of which the Climacocystaceae occur in a mean stem age of 77.49 Mya and Gloeoporellaceae occur in a mean stem age of 88.06 Mya. According to the evolutionary divergence times, Climacocystaceae and Gloeoporellaceae could be recognized as independent families within Polyporales.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Author contributions
B-KC designed the experiment and conceived and supervised the work. SL, J-LZ, JS, Y-FS, Y-CD, and B-KC prepared the samples. SL and B-KC made the morphological examinations. SL performed the phylogenetic analyses. SL and J-LZ performed the molecular clock analysis. SL and JS performed the molecular sequencing. SL, J-LZ, JS, and B-KC wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the National Natural Science Foundation of China (nos. 32270010, U2003211, and 31870008) and Beijing Forestry University Outstanding Young Talent Cultivation Project (no. 2019JQ03016).
Acknowledgments
We express our gratitude to Ms. Xing Ji (China), Chang-Ge Song (China), and Yan Wang (China) for their help during field collections and molecular studies. Drs. Genevieve Gates (Australia), Jun-Zhi Qiu (China), Li-Wei Zhou (China), Xiao-Lan He (China), Hai-Xia Ma (China), Yuan-Yuan Chen (China), Shi-Liang Liu (China), and Long-Fei Fan (China) are thanked for their companionship during field collections.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
AIC, Akaike information criterion; BI, Bayesian inference; BJFC, Herbarium of the Institute of Microbiology, Beijing Forestry University, China; BGI, Beijing Genomics Institute; BPP, Bayesian posterior probabilities; BT, bootstrap; CB, cotton blue; CB+, Cyanophilous; CB–, Acyanophilous; CI, consistency index; CTAB, cetyl trimethylammonium bromide; GTR + I + G, general time reversible + proportion invariant + gamma; HI, Homoplasy Index; hLRTs, hierarchical likelihood ratio tests; HPD, highest posterior density; IKI, Melzer’s reagent; IKI–, neither amyloid nor dextrinoid; ILD, incongruence length difference test; ITS, internal transcribed spacer; KOH, 5% potassium hydroxide; MCC, maximum clade credibility; MCMC, Markov Chain Monte Carlo; ML, maximum likelihood; MP, maximum parsimony; MPT, most parsimonious tree; nLSU, nuclear large subunit rDNA; PCR, polymerase chain reaction; RC, rescaled gonsistency index; RI, retention index; RPB1, DNA-directed RNA polymerase II subunit 1; RPB2, DNA-directed RNA polymerase II subunit 2; TEF1, translation elongation factor 1- α ; TL, tree length.
Footnotes
- ^ http://mafft.cbrc.jp/alignment/server/
- ^ http://tree.bio.ed.ac.uk/software/figtree/
- ^ http://tree.bio.ed.ac.uk/software/tracer/
- ^ http://www.indexfungorum.org
References
Baltazar, J. M., Ryvarden, L., and Gibertoni, T. B. (2014). Diplomitoporus (Polyporales. Basidiomycota) in Brazil revisited. Mycol. Prog. 13, 313–319. doi: 10.1007/s11557-013-0916-4
Berbee, M. L., and Taylor, J. W. (2010). Dating the molecular clock in fungi – how close are we? Fungal Biol. Rev. 24, 1–16. doi: 10.1016/j.fbr.2010.03.001
Binder, M., Hibbett, D. S., Larsson, K. H., Larsson, E., and Langer, E. (2005). The phylogenetic distribution of resupinate forms across the major clades of mushroom-forming fungi (Homobasidiomycetes). Syst. Biodivers. 3, 113–157. doi: 10.1017/S1477200005001623
Binder, M., Justo, A., Riley, R., Salamov, A., Lopez-Giraldez, F., Sjökvist, E., et al. (2013). Phylogenetic and phylogenomic overview of the Polyporales. Mycologia 105, 1350–1373. doi: 10.3852/13-003
Chen, J. J., Cui, B. K., Zhou, L. W., Korhonen, K., and Dai, Y. C. (2015). Phylogeny, divergence time estimation, and biogeography of the genus Heterobasidion (Basidiomycota, Russulales). Fungal Divers. 71, 185–200. doi: 10.1007/s13225-014-0317-2
Cui, B. K., Li, H. J., Ji, X., Zhou, J. L., Song, J., Si, J., et al. (2019). Species diversity, taxonomy and phylogeny of Polyporaceae (Basidiomycota) in China. Fungal Divers. 97, 137–392. doi: 10.1007/s13225-019-00427-4
Dai, Y. C. (2012). Polypore diversity in China with an annotated checklist of Chinese polypores. Mycoscience 53, 49–80. doi: 10.1007/s10267-011-0134-3
Dai, Y. C., Cui, B. K., Yuan, H. S., and Li, B. D. (2007). Pathogenic wood-decaying fungi in China. Forest Pathol. 37, 105–120. doi: 10.1111/j.1439-0329.2007.00485.x
Decock, C., and Ryvarden, L. (2020). Aphyllophorales of Africa 41 Some polypores from Gabon. Synop. Fungorum 42, 5–15.
Domanski, S. (1970). Wood-inhabiting fungi of Bialowieza virgin forests in Poland. XIII. Two species of Diplomitoporus Doman., gen. nov. Acta Soc. Bot. Pol. 39, 191–207. doi: 10.5586/asbp.1970.016
Drummond, A. J., and Rambaut, A. (2007). BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7:214. doi: 10.1186/1471-2148-7-214
Drummond, A. J., Ho, S. Y. W., Phillips, M. J., and Rambaut, A. (2006). Relaxed phylogenetics and dating with confidence. PLoS Biol. 4:e88. doi: 10.1371/journal.pbio.0040088
Drummond, A. J., Suchard, M. A., Xie, D., and Rambaut, A. (2012). Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29, 1969–1973. doi: 10.1093/molbev/mss075
Farris, J. S., Källersjö, M., Kluge, A. G., and Bult, C. (1994). Testing significance of incongruence. Cladistics 10, 315–319. doi: 10.1111/j.1096-0031.1994.tb00181.x
Felsenstein, J. (1985). Confidence intervals on phylogenies: An approach using the bootstrap. Evolution 39, 783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x
Ghobad-Nejhad, M., and Dai, Y. C. (2010). Diplomitoporus rimosus is found in Asia and belongs to the Hymenochaetales. Mycologia 102, 1510–1517. doi: 10.3852/10-025
Hall, T. A. (1999). Bioedit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98. doi: 10.1021/bk-1999-0734.ch008
He, M. Q., Zhao, R. L., Hyde, K. D., Begerow, D., Kemler, M., Yurkov, A., et al. (2019). Notes, outline and divergence times of Basidiomycota. Fungal Divers. 99, 105–367. doi: 10.1007/s13225-019-00435-4
Hibbett, D. S., Grimaldi, D., and Donoghue, M. J. (1997). Fossil mushrooms from Miocene and Cretaceous ambers and the evolution of Homobasidiomycetes. Am. J. Bot. 84, 981–991. doi: 10.2307/2446289
Ji, X., Zhou, J. L., Song, C. G., Xu, T. M., Wu, D. M., and Cui, B. K. (2022). Taxonomy, phylogeny and divergence times of Polyporus (Basidiomycota) and related genera. Mycosphere 13, 1–52. doi: 10.5943/mycosphere/13/1/1
Justo, A., Miettinen, O., Floudas, D., Ortiz-Santana, B., Sjökvist, E., Lindner, D., et al. (2017). A revised family-level classification of the Polyporales (Basidiomycota). Fungal Biol. 121, 798–824. doi: 10.1016/j.funbio.2017.05.010
Katoh, K., and Standley, D. M. (2013). MAFFT Multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kirk, P. M., Cannon, P. F., Minter, D. W., and Stalpers, J. A. (2008). Dictionary of the Fungi, 10th Edn. Oxon: CAB International.
Korhonen, A., Seelan, J. S. S., and Miettinen, O. (2018). Cryptic species diversity in polypores: The Skeletocutis nivea species complex. MycoKeys 36, 45–82. doi: 10.3897/mycokeys.36.27002
Kotlába, F., and Pouzar, Z. (1958). Polypori novi vel minus cogniti Cechoslovakiae III. Ceská Mykol. 12, 95–104.
Larsson, K. H. (2007). Re-thinking the classification of corticioid fungi. Mycol. Res. 111, 1040–1063. doi: 10.1016/j.mycres.2007.08.001
Larsson, K. H., Larsson, E., and Kõljalg, U. (2004). High phylogenetic diversity among corticioid homobasidiomycetes. Mycol. Res. 108, 983–1002. doi: 10.1017/S0953756204000851
Lepage, T., Bryant, D., Philippe, H., and Lartillot, N. A. (2007). A general comparison of relaxed molecular clock models. Mol. Biol. Evol. 24, 2669–2680. doi: 10.1093/molbev/msm193
Liu, S., Chen, Y. Y., Sun, Y. F., He, X. L., Song, C. G., Si, J., et al. (2022). Systematic classification and phylogenetic relationships of the brown-rot fungi within the Polyporales. Fungal Divers. doi: 10.1007/s13225-022-00511-2 [Online ahead of print].
Liu, S., Shen, L., Wang, Y., Xu, T., Gates, G., and Cui, B. (2021). Species diversity and molecular phylogeny of Cyanosporus (Polyporales. Basidiomycota). Front. Microbiol. 12:631166. doi: 10.3389/fmicb.2021.631166
Maddison, W. P., and Maddison, D. R. (2017). Mesquite: A modular system for evolutionary analysis, Version 3.2.
Matheny, P. B. (2005). Improving phylogenetic inference of mushrooms with RPB1 and RPB2 nucleotide sequences (Inocybe, Agaricales). Mol. Phylogenet. Evol. 35, 1–20. doi: 10.1016/j.ympev.2004.11.014
Matheny, P. B., Liu, Y. J., Ammirati, J. F., and Hall, B. D. (2002). Using RPB1 sequences to improve phylogenetic inference among mushrooms (Inocybe, Agaricales). Am. J. Bot. 89, 688–698. doi: 10.2307/4131413
Matheny, P. B., Wang, Z., Binder, M., Curtis, J. M., Lim, Y. W., Nilsson, R. H., et al. (2007). Contributions of rpb2 and tef1 to the phylogeny of mushrooms and allies (Basidiomycota, Fungi). Mol. Phylogenet. Evol. 43, 430–451. doi: 10.1016/j.ympev.2006.08.024
Miettinen, O. (2012). Diplomitoporus dilutabilis belongs to Cinereomyces (Polyporales, Basidiomycota). Mycotaxon 121, 345–348. doi: 10.105248/121.345
Miettinen, O., Larsson, E., Sjökvist, E., and Larsson, K. H. (2012). Comprehensive taxon sampling reveals unaccounted diversity and morphological plasticity in a group of dimitic polypores (Polyporales, Basidiomycota). Cladistics 28, 251–270. doi: 10.1111/j.1096-0031.2011.00380.x
Nagy, László G, Tóth, Renáta, Kiss, Enikö, Slot, Jason, Gácser, Attila, and Kovács, Gábor M (2017). Six key traits of fungi: Their evolutionary origins and genetic bases. Microbiol Spectr. 5:FUNK–0036–2016. doi: 10.1128/microbiolspec.FUNK-0036-2016
Posada, D., and Crandall, K. A. (1998). Modeltest: Testing the model of DNA substitution. Bioinformatics 14, 817–818. doi: 10.1093/bioinformatics/14.9.817
Rajchenber, M., and Robledo, G. (2013). Pathogenic polypores in Argentina. For. Pathol. 43, 171–184. doi: 10.1111/efp.12032
Rea, C. (1922). British Basidiomycetae: A handbook to the larger British fungi. Cambridge: Cambridge University Press, 1–799.
Rehner, S. A., and Buckley, E. (2005). A Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: Evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 97, 84–98. doi: 10.1080/15572536.2006.11832842
Ronquist, F., and Huelsenbeck, J. P. (2003). MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574. doi: 10.1093/bioinformatics/btg180
Ryvarden, L. (2018). Studies in African Aphyllophorales 23 Some new species in Ceriporiopsis and Diplomitoporus. Synop. Fungorum 38, 12–19.
Ryvarden, L. (2019). Studies in Aphyllophorales of Africa 30 Some poroid species from Uganda. Synop. Fungorum 39, 41–45.
Sánchez-Ramírez, S., Tulloss, R. E., Amalfi, M., and Moncalvo, J. M. (2014). Palaeotropical origins, boreotropical distribution and increased rates of diversification in a clade of edible ectomycorrhizal mushrooms (Amanita section Caesareae). J. Biogeogr. 42, 351–363. doi: 10.1111/jbi.12402
Shen, L. L., Wang, M., Zhou, J. L., Xing, J. H., Cui, B. K., and Dai, Y. C. (2019). Taxonomy and phylogeny of Postia. Multi-gene phylogeny and taxonomy of the brown-rot fungi: Postia (Polyporales, Basidiomycota) and related genera. Persoonia 42, 101–126. doi: 10.3767/persoonia.2019.42.05
Smith, S. Y., Currah, R. S., and Stockey, R. A. (2004). Cretaceous and Eocene poroid hymenophores from Vancouver Island, British Columbia. Mycologia 96, 180–186. doi: 10.2307/3762001
Song, J., and Cui, B. K. (2017). Phylogeny, divergence time and historical biogeography of Laetiporus (Basidiomycota, Polyporales). BMC Evol. Biol. 17:102. doi: 10.1186/s12862-017-0948-5
Song, J., Chen, J. J., Wang, M., Chen, Y. Y., and Cui, B. K. (2016). Phylogeny and biogeography of the remarkable genus Bondarzewia (Basidiomycota, Russulales). Sci. Rep. 6:34568. doi: 10.1038/srep34568
Song, J., Chen, Y. Y., and Cui, B. K. (2014). Phylogeny and taxonomy of Climacocystis (Polyporales) in China. Cryptogamie Mycol. 35, 221–232. doi: 10.7872/crym.v35.iss3.2014.221
Stamatakis, A. (2014). RAxML Version 8: A tool for phylogenetic analyses and post analyses of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Sun, Y., Costa-Rezende, D. H., Xing, J., Zhou, J., Zhang, B., Gibertoni, T. B., et al. (2020). Multi-gene phylogeny and taxonomy of Amauroderma s. lat. (Ganodermataceae). Persoonia 44, 206–239. doi: 10.3767/persoonia.2020.44.08
Sun, Y., Xing, J., He, X., Wu, D., Song, C., Liu, S., et al. (2022). Species diversity, systematic revision and molecular phylogeny of Ganodermataceae (Polyporales, Basidiomycota) with an emphasis on Chinese collections. Stud. Mycol. 101, 287–415. doi: 10.3767/10.3114/sim.2022.101.05
Swofford, D. L. (2002). PAUP *: Phylogenetic analysis using parsimony (* and other methods), version 4.0b10. Sunderland: Sinauer Associates, doi: 10.1111/j.0014-3820.2002.tb00191.x
Taylor, T. N., Hass, H., Kerp, H., Krings, M., and Hanlin, R. T. (2005). Perithecial ascomycetes from the 400 million year old Rhynie chert: Anexample of ancestral polymorphism. Mycologia 97, 269–285. doi: 10.1080/15572536.2006.11832862
Wang, X. W., Tom, W. M., Liu, X. L., and Zhou, L. W. (2021). Towards a natural classification of Hyphodontia sensu lato and the trait evolution of basidiocarps within Hymenochaetales (Basidiomycota). J. Fungi 7:478. doi: 10.3390/jof7060478
Westphalen, M. C., Rajchenberg, M., Tomšovský, M., and Gugliotta, A. M. (2016). Extensive characterization of the new genus Rickiopora (Polyporales). Fungal Biol. 120, 1002–1009. doi: 10.1016/j.funbio.2016.05.001
White, T. J., Bruns, T., Lee, S., and Taylor, J. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in PCR Protocols: A Guide to Methods and Applications, eds M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (San Diego: Academic Press), 315–322. doi: 10.1016/B978-0-12-372180-8.50042-1
Wu, F., Yuan, Y., Chen, J. J., Cui, B. K., and Dai, Y. C. (2020). Terrestriporiaceae fam. nov., a new family of Russulales (Basidiomycota). Mycosphere 11, 2755–2766. doi: 10.5943/mycosphere/11/1/21
Wu, F., Zhou, L. W., Yang, Z. L., Bau, T., Li, T. H., Dai, Y. C., et al. (2019). Resource diversity of Chinese macrofungi: Edible, medicinal and poisonous species. Fungal Divers. 98, 1–76. doi: 10.1007/s13225-019-00432-7
Yuan, H. S., Lu, X., Dai, Y. C., Hyde, K. D., Kan, Y. H., Kušan, I., et al. (2020). Fungal diversity notes 1277–1386: taxonomi and phylogenetic contributions to fungal taxa. Fungal Divers. 104, 1–266. doi: 10.1007/s13225-020-00461-7
Zhao, C. L., Cui, B. K., Song, J., and Dai, Y. C. (2015). Fragiliporiaceae, a new family of Polyporales (Basidiomycota). Fungal Divers. 70, 115–126.
Zhao, R. L., Li, G. J., Sánchez-Ramírez, S., Stata, M., Yang, Z. L., Wu, G., et al. (2017). A six-gene phylogenetic overview of Basidiomycota and allied phyla with estimated divergence times of higher taxa and a phyloproteomics perspective. Fungal Divers. 84, 43–74. doi: 10.1007/s13225-017-0381-5
Zhao, R. L., Zhou, J. L., Chen, J., Margaritescu, S., Sánchez-Ramírez, S., Ramírez, S., et al. (2016). Towards standardizing taxonomic ranks using divergence times – a case study for reconstruction of the Agaricus taxonomic system. Fungal Divers. 78, 239–292. doi: 10.1007/s13225-016-0357-x
Zhu, L., Song, J., Zhou, J. L., Si, J., and Cui, B. K. (2019). Species diversity, phylogeny, divergence time and biogeography of the genus Sanghuangporus (Basidiomycota). Front. Microbiol. 10:812. doi: 10.3389/fmicb.2019.00812
Keywords: molecular clock, multi-gene phylogeny, new family, taxonomy, white-rot fungi
Citation: Liu S, Zhou J-L, Song J, Sun Y-F, Dai Y-C and Cui B-K (2023) Climacocystaceae fam. nov. and Gloeoporellaceae fam. nov., two new families of Polyporales (Basidiomycota). Front. Microbiol. 14:1115761. doi: 10.3389/fmicb.2023.1115761
Received: 04 December 2022; Accepted: 11 January 2023;
Published: 03 February 2023.
Edited by:
Yong-Zhong Lu, Guizhou Institute of Technology, ChinaReviewed by:
Gang Wu, Kunming Institute of Botany (CAS), ChinaXin-Zhan Liu, Institute of Microbiology (CAS), China
Taihui Li, Guangdong Academy of Science, China
Copyright © 2023 Liu, Zhou, Song, Sun, Dai and Cui. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bao-Kai Cui,  cuibaokai@bjfu.edu.cn
cuibaokai@bjfu.edu.cn
 Shun Liu
Shun Liu