- 1Department of Clinical Laboratory, The Second Affiliated Hospital of Soochow University, Suzhou, Jiangsu, China
- 2Department of Geriatrics, The Second Affiliated Hospital of Soochow University, Suzhou, Jiangsu, China
- 3Department of General Practice, The Second Affiliated Hospital of Soochow University, Suzhou, Jiangsu, China
- 4Hackensack Meridian Health Center for Discovery and Innovation, Nutley, NJ, United States
- 5Hackensack Meridian School of Medicine, Seton Hall University, Nutley, NJ, United States
Introduction: The raise of multi-drug resistant bacteria involving carbapenem, colistin, or tigecycline resistance constitutes a threat to public health, which partly results from the transmission of corresponding mobile resistance genes, such as blaKPC and blaNDM for carbapenem, mcr for colistin, and tmexCD-toprJ gene cluster for tigecycline. Herein, we described the emergence of an Aeromonas veronii strain HD6454 co-harboring blaKPC–2, mcr-3.17, and tmexC3.2-tmexD3.3-toprJ1b gene cluster from hospital sewage.
Methods: Whole genome sequencing (WGS) was used to determine the genome sequence of HD6454, and the detailed genomic analysis of genetic elements or regions carrying key antimicrobial resistance genes (ARGs) from HD6454 were performed. Cloning experiment was conducted to confirm the function of key ARGs in mediating antimicrobial resistance. Conjugation experiment was conducted to determine the mobility of the plasmid.
Results: The results showed that this strain belonged to a novel sequence type (ST) variant ST1016, and carried 18 important ARGs. Among them, the blaKPC–2 was carried by non-self-transmissible IncP-6 plasmid, while tmexC3.2-tmexD3.3-toprJ1b gene cluster and mcr-3.17 were carried by integrative and mobilizable element (IME) or IME-related region in chromosome. The mcr-3.17, mcr-3.6, and mcr-3-like3 genes were further inferred to originate from IMEs of Aeromonas species. Additionally, for the first time, the mcr-3.17 was confirmed to confer low-level resistance to colistin under inducible expression, while tmexC3.2-tmexD3.3-toprJ1b gene cluster was confirmed to confer low-level resistance to tigecycline.
Discussion: This is the first report of a strain co-harboring blaKPC–2, mcr-3.17, and tmexC3.2-tmexD3.3-toprJ1b gene cluster. Although the resistance and/or mobility of these ARGs are limited in this strain, the emergence of this multiple important ARGs-carrying strain deserves further attention.
Introduction
The rising bacterial resistance to carbapenems is a worldwide threat to public health. Carbapenem resistance is mainly caused by the expression of carbapenemase-encoding genes (blaKPC, blaNDM, blaVIM, etc.) (Nordmann and Poirel, 2019). Meanwhile, colistin and tigecycline are considered as the last-resort for the treatment of life-threatening infections caused by multi-drug resistant Gram-negative bacteria, especially the carbapenem-resistant strains (Doi, 2019; Gonzalez-Avila et al., 2021). However, global emergence of colistin/tigecycline-resistant pathogens has been increasingly reported (El-Sayed Ahmed et al., 2020; Yaghoubi et al., 2021), which partly results from the transmission of mobile resistance genes, including mcr for colistin (Anyanwu et al., 2020), and tet(X) (Fang et al., 2020) and resistance-nodulation-division (RND) efflux pump gene cluster tmexCD-toprJ (Lv et al., 2020; Wang et al., 2021b,c) for tigecycline. Co-carriers of carbapenem-resistant and colistin-resistance genes [such as blaKPC and mcr-3.3 (Tang et al., 2020)], or carbapenem-resistant and tigecycline-resistance genes [such as blaNDM–4, tet(X), and tmexCD3-toprJ3 (Hirabayashi et al., 2021)] in bacteria have been reported. These co-carriers of antibiotic resistance genes (ARGs) reduce the options of clinical antibiotic treatment, which raise a significant concern.
Aeromonas species are Gram-negative bacteria, and mainly infect aquatic organisms (Fernández-Bravo and Figueras, 2020). Aeromonas species can cause intestinal and extra-intestinal infections in human (Gowda et al., 2015). Although Aeromonas species widely distribute in diverse ecosystems, they are more commonly isolated from water, such as drinking water, seawater and hospital sewage (Fernández-Bravo and Figueras, 2020). Among them, hospital sewage plays an important role in the spread of ARGs in Aeromonas (Karkman et al., 2018; Zhang et al., 2020).
The potential association between Aeromonas species and carbapenem/tigecycline/colistin genes has been reported. Firstly, Nwaiwu and Aduba (2020) discovered that blaKPC–2 gene was the most carbapenemase-encoding gene carried by Aeromonas species plasmids within GenBank (9.52%, 10/105). Secondly, tmexCD-toprJ-carrying Aeromonas strains have also been gradually reported (Dong et al., 2022; Wu et al., 2023). Thirdly, since the first mobile colistin resistance (mcr) gene mcr-1 was characterized in Enterobacteriaceae in 2016 (Liu et al., 2016), additional nine mobile colistin resistance genes (mcr-2∼mcr-10) and various variants have been reported (El-Sayed Ahmed et al., 2020; Wang et al., 2020a). Among them, mcr-3 is considered most likely to originate from Aeromonas species (Yin et al., 2017; Shen et al., 2018). Further investigation could help clarify the mechanism of transmission of mcr-3 in Aeromonas species. Additionally, most mcr-3 variants have only been characterized by sequence analysis, their contributions to colistin resistance are not described.
In this study, we describe the emergence of an Aeromonas veronii strain co-harboring blaKPC–2, mcr-3.17, and tmexC3.2-tmexD3.3-toprJ1b gene cluster from hospital sewage for the first time. A detailed genomic dissection analysis was conducted to decipher the genomic characteristics of this strain.
Materials and methods
Bacterial strains
The A. veronii strain, HD6454, was isolated from the raw sewage in sewage conduit under washbasin in digestive department of the Second Affiliated Hospital of Soochow University (Suzhou, China) in July 2019. The detailed isolation methodology was described previously (Wen et al., 2022). Bacterial species identification was carried out using Matrix-Assisted Laser Desorption/Ionization Time of Flight Mass Spectrometry (MALDI-TOF-MS). The presence of carbapenemase genes (blaKPC, blaNDM, blaVIM, blaOXA–48–like, and blaIMP), mcr genes and tmexCD-toprJ was determined by PCR amplification as described previously (Lv et al., 2020; Tang et al., 2020).
Sequencing and sequence assembly
Bacterial genomic DNA was isolated using the Omega Bio-Tek Bacterial DNA Kit (Doraville, GA, USA), and sequenced from a sheared DNA library with average size of 10 kb on a Nanopore PromethION platform (Oxford Nanopore Technologies, OX, UK), as well as a paired-end library with an average insert size of 350 bp on a NovaSeq sequencer (Illumina, CA, USA). A hybrid assembly was then conducted by Unicycler 0.4.91 using both paired-end short Illumina reads and the long Nanopore reads.
Multi-locus sequence typing (MLST) analysis and phylogenetic analysis
The sequence type (ST) of the A. veronii strain HD6454 was identified according to the online multi-locus sequence typing (MLST) scheme.2 Amino acid sequences of MCR variants and MCR-3-like variants were aligned using ClustalW in MEGAX 10.1.8 (Kumar et al., 2018). Unrooted maximum-likelihood phylogenetic trees were further generated using MEGAX 10.1.8 with a bootstrap iteration of 1,000.
Sequence annotation and comparison
Open reading frames (ORFs) and pseudogenes were predicted using RAST 2.0 (Brettin et al., 2015) combined with BLASTP/BLASTN searches (Boratyn et al., 2013) against the UniProtKB/Swiss-Prot database (Boutet et al., 2016) and the RefSeq database (O’Leary et al., 2016). Annotation of resistance genes, mobile elements, and other features were carried out using the online databases including CARD (Jia et al., 2017), ResFinder 4.1 (Zankari et al., 2012), Danmel (Wang et al., 2022), ISfinder (Siguier et al., 2006), and Tn Number Registry (Tansirichaiya et al., 2019). Multiple and pairwise sequence comparisons were performed using BLASTN. Gene organization diagrams were drawn through scripts from Danmel (Wang et al., 2022), and displayed using Inkscape 1.0.3 Prediction of protein secondary structure was performed using ESPript 3.0 (Robert and Gouet, 2014) and displayed using Phyre 2.0 (Kelley et al., 2015).
Cloning experiments
The construct of recombinant plasmid was performed through seamless cloning by ClonExpress Ultra One Step Cloning Kit (Vazyme, China). The details were described in Supplementary text, and the primers used herein were shown in Supplementary Table 1. The resulting recombinant plasmids were transformed through heat shock into Escherichia coli DH5α, and 100 μg/ml ampicillin was used for the transformant selection. The transformants DH5α/pUC18, DH5α/pBAD24 and DH5α/pBAD24-mcr-1.1 were also constructed as controls. Successful transformants were confirmed by PCR, and Sanger sequencing on a 3730XL sequencer (ABI, Boston, MA, USA). Induction of the pBAD24 vector was performed using MH II broth (Cation-Adjusted) supplemented with 0.4% L-arabinose as previously described (Kieffer et al., 2019).
Bacterial antimicrobial susceptibility test
The susceptibility of colistin and tigecycline was carried by the minimum inhibitory concentrations (MICs) method. Results were interpreted according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST).4 The susceptibility of other antimicrobial agents (Supplementary Table 2) was carried by using the Phoenix System-M50 automatic microbiology analyzer (BD, USA). Results were interpreted according to the 2020 Clinical and Laboratory Standards Institute (CLSI) guidelines. The susceptibility test through MIC method was repeated three times to ensure the accuracy of the result. The E. coli ATCC 25922 was used as the quality control.
Conjugal transfer
Conjugal transfer experiment was carried out with sodium azide-resistant E. coli J53 as the recipient, and the HD6454 isolate as the donor. In brief, overnight cultures of the HD6454 and J53 were mixed at a ratio of 1:1 in LB broth, and the mixture was then spotted on a hydrophilic nylon membrane filter with a 0.45 μm pore size (Millipore) that was placed on LB agar plate and then incubated for mating at 37 C for 18 h. Bacteria were washed from filter membrane and spotted on LB agar plate, and 200 μg/ml sodium azide (for J53) together with 2 μg/ml imipenem (for blaKPC) was used for selecting an E. coli transconjugant carrying blaKPC.
Nucleotide sequence accession numbers
The complete chromosome and plasmid sequences of HD6454 were submitted to GenBank under accession numbers CP079823-CP079826, respectively.
Results
Characteristics of HD6454
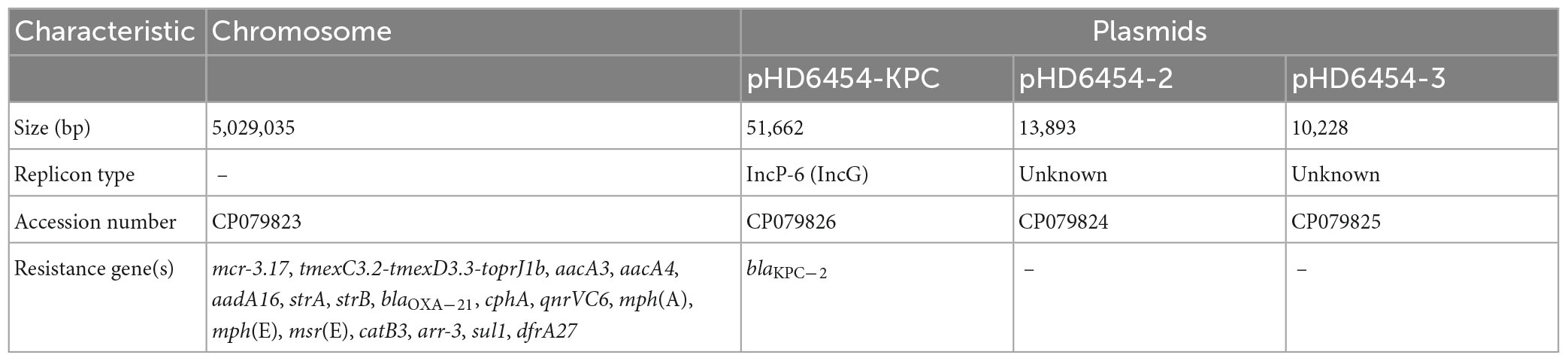
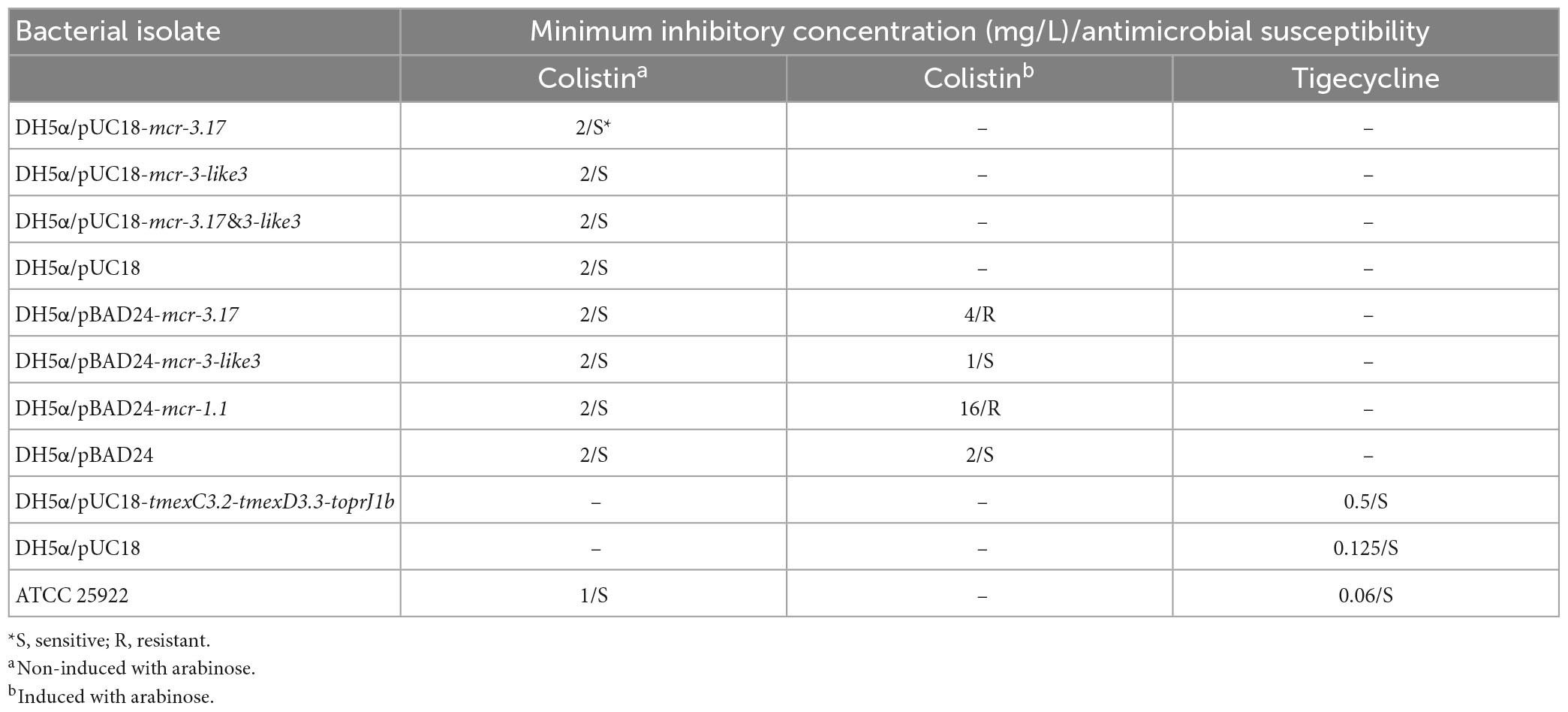
The A. veronii strain HD6454 was isolated from the raw sewage in sewage conduit under washbasin of digestive department, and belonged to a novel ST variant ST1016. The strain harbored a 5,029,035-bp chromosome genome and three plasmids: pHD6454-KPC, pHD6454-2, and pHD6454-3 (Table 1). Meanwhile, HD6454 carried 18 important ARGs, including blaKPC–2, mcr-3.17, and tmexC3.2-tmexD3.3-toprJ1b gene cluster. Additionally, a mcr-3-like gene was found downstream of the mcr-3.17 gene. This mcr-3-like gene was further designated as mcr-3-like3 as previously described (Shen et al., 2018). Susceptibility testing results showed that HD6454 was multidrug-resistant, including carbapenems (Supplementary Table 2). However, despite carrying mcr-3.17 and tmexC3.2-tmexD3.3-toprJ1b gene cluster, HD6454 was susceptible to colistin (2 mg/L) and tigecycline (0.125 mg/L) based on the EUCAST clinical breakpoints (see footnote 4).
Functional identification of mcr-3.17 and mcr-3-like3
The MCR-3.17 shared 89.24% amino acid sequence similarity to MCR-3.1 (accession number KY924928), while MCR-3-like3 shared 84.29% amino acid sequence similarity to MCR-3.1. The amino acid sequence of MCR-3.17 shared the highest similarity (93.15%) with MCR-3.6 (accession number MF598076) among all MCR-3 variants. The phylogenetic analysis also showed that, compared with other MCR-3 variants, the evolution relationship between MCR-3.6 and MCR-3.17 was the closest (Supplementary Figure 1). Nevertheless, MCR-3.17 was still different from MCR-3.6 in predicted protein secondary structure (Supplementary Figure 2). Previous studies showed that MCR proteins were predicted to have two conservative domains, the membrane-anchored domain (approximate residues range for MCR-3 was 1–172) and the soluble catalytic domain (approximate residues range for MCR-3 was 173 to the last) (Yin et al., 2017; Carroll et al., 2019). Compared with MCR-3.6, 33 mutations in amino acid sequences of the membrane-anchored domain of MCR-3.17 were identified, and further led the loss of two β-sheet areas in protein secondary structure, while MCR-3.17 also had four mutations in amino acid sequences of the soluble catalytic domain, and further led the increase of a 310-helix area in protein secondary structure (Supplementary Figure 2).
In order to determine the function of mcr-3.17 and mcr-3-like3 in mediating colistin resistance, the transformants DH5α/pUC18-mcr-3.17, DH5α/pUC18-mcr-3-like3 and DH5α/pUC18-mcr-3.17&mcr-3-like3 were obtained. However, MIC of these three transformants displayed no difference, compared with MIC of DH5α/pUC18 (Table 2). Considering probable variations in the promoter activities of mcr-3.17 and mcr-3-like3 in HD6545, the transformants DH5α/pBAD24-mcr-3.17 and DH5α/pBAD24-mcr-3-like3 were further constructed. In addition, transformants DH5α/pBAD24 and DH5α/pBAD24-mcr-1.1 were also constructed as controls. The antimicrobial susceptibility testing showed that DH5α/pBAD24-mcr-3.17 presented an MIC of 4 mg/L after induction of arabinose, while DH5α/pBAD24-mcr-1.1 had an MIC of 16 mg/L. On the contrary, DH5α/pBAD24-mcr-3-like3 presented a MIC of 1 mg/L after induction of arabinose (Table 2). This result suggested that mcr-3.17, but not mcr-3-like3, could confer low-level resistance to colistin.
Genetic characterization of mcr-3.17 and mcr-3-like3
The mcr-3.17 and mcr-3-like3 genes were located in a 46.5-kb mcr-3.17 region in the chromosome of HD6454 (Figure 1). The mcr-3-like3 gene was located immediately downstream of mcr-3.17, the nucleoside bases between them was 66 bp. Compared with the chromosome of a reference A. veronii strain JC529 (accession number CP058912), a 6.9-kb backbone region was replaced by the mcr-3.17 region in HD6454. This mcr-3.17 region contained a truncated Tn6378, a 25.3-kb region with multiple IS elements, and the “core mcr-3.17 region,” including truncated sulP gene, mcr-3.17 gene and mcr-3-like3–to–orf1128 region.
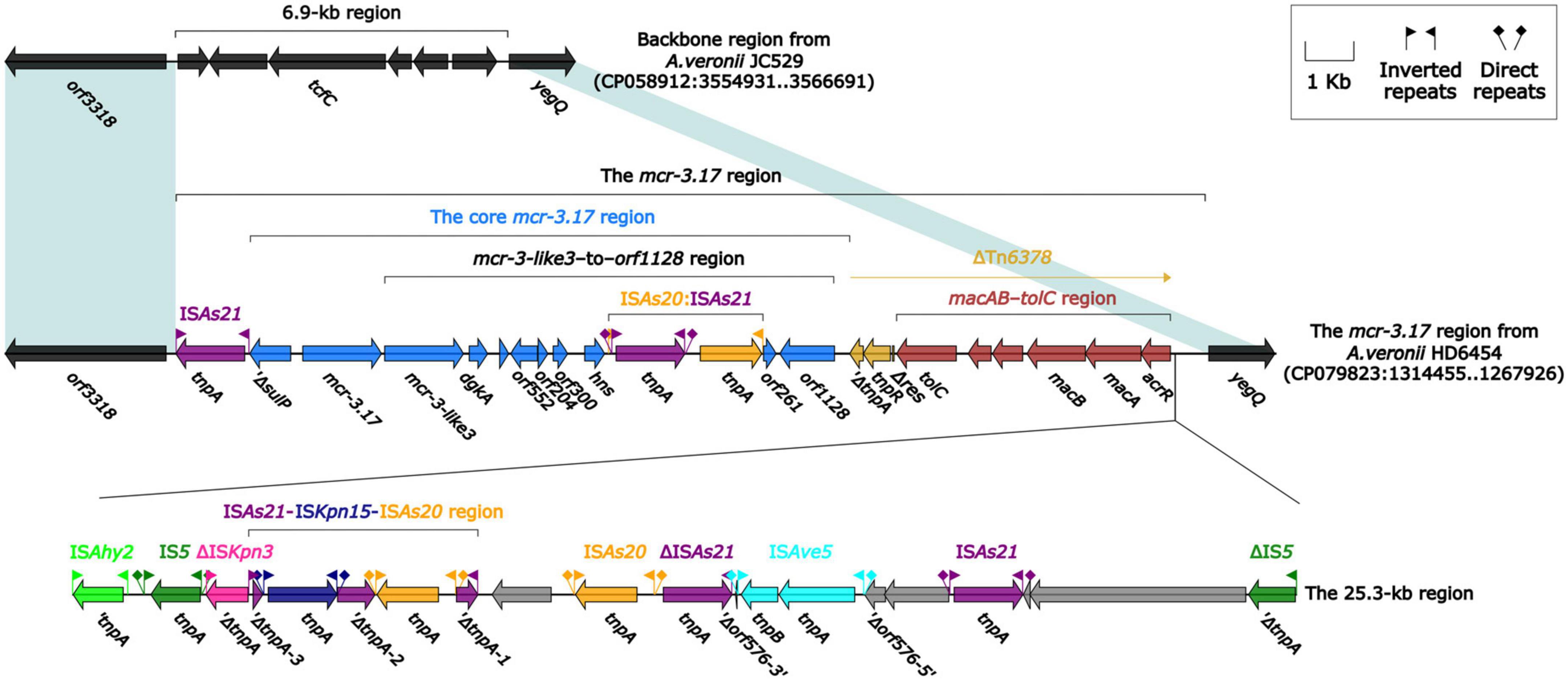
Figure 1. Organization of the mcr-3.17 region from HD6454, and comparison to related region. Genes are denoted by arrows. Genes, mobile genetic elements and other features are colored based on their functional classification. Shading denotes regions of homology (nucleotide identity ≥ 95%).
To further probe how this 46.5-kb mcr-3.17 region originated, a detailed genetic dissection analysis was applied to compare the genetic structure of HD6454 with additional 10 mcr-3.6 (n = 7) or mcr-3.17 (n = 3)-harboring genomes download form the GenBank (Supplementary Tables 3, 4). Similar core mcr-3.17 regions could be found in three mcr-3.17-carried contig fragments from Aeromonas allosaccharophila Z9-6 (Shen et al., 2018), and A. veronii CN17A0120 and ADV102 (Rangel et al., 2019; Figure 2). Truncated or complete usp–sulP region and truncated or complete mcr-3-like3–to–orf1128 regions could be found upstream and downstream of the mcr-3.17 gene from all mcr-3.17 regions in this study, respectively. However, due to the limited length of these contig assemblies, whether these core mcr-3.17 regions were carried by complete genetic elements or not could not be determined. Fortunately, an integrative and mobilizable element (IME) from A. sanarellii NS1 was found to share at least 96.71% identity with these core mcr-3.17 regions (Figure 2). This IME harbored mcr-3.6 and mcr-3-like3 genes, and was newly designated Tn7360. Similar usp–sulP region and truncated mcr-3-like3–to–orf1128 region could also be found in Tn7360. Therefore, the so-called core mcr-3.17 region was further named as Tn7360-related region. Moreover, except Tn7360, all mcr-3.6 and mcr-3-like3 genes were carried by IME Tn7361a/b (Shi et al., 2020) or related genetic elements and regions (Figure 3). The backbone sequence of Tn7361a/b was 95.60% identical to the backbone sequence of Tn7360 with 76% coverage. Tn7361a/b also harbored the complete mcr-3-like3–to–orf1128 region. Meanwhile, Tn7360 and Tn7361a/b were all integrated within the thyA gene with 6-bp direct repeats (DRs). In addition, truncated Tn7360 fragments were identified in other genetic elements, including composite transposon Tn6518 (Wang et al., 2020b) from A. veronii w55 and IME Tn6868 from A. hydrophila WP7-S18-ESBL-06.
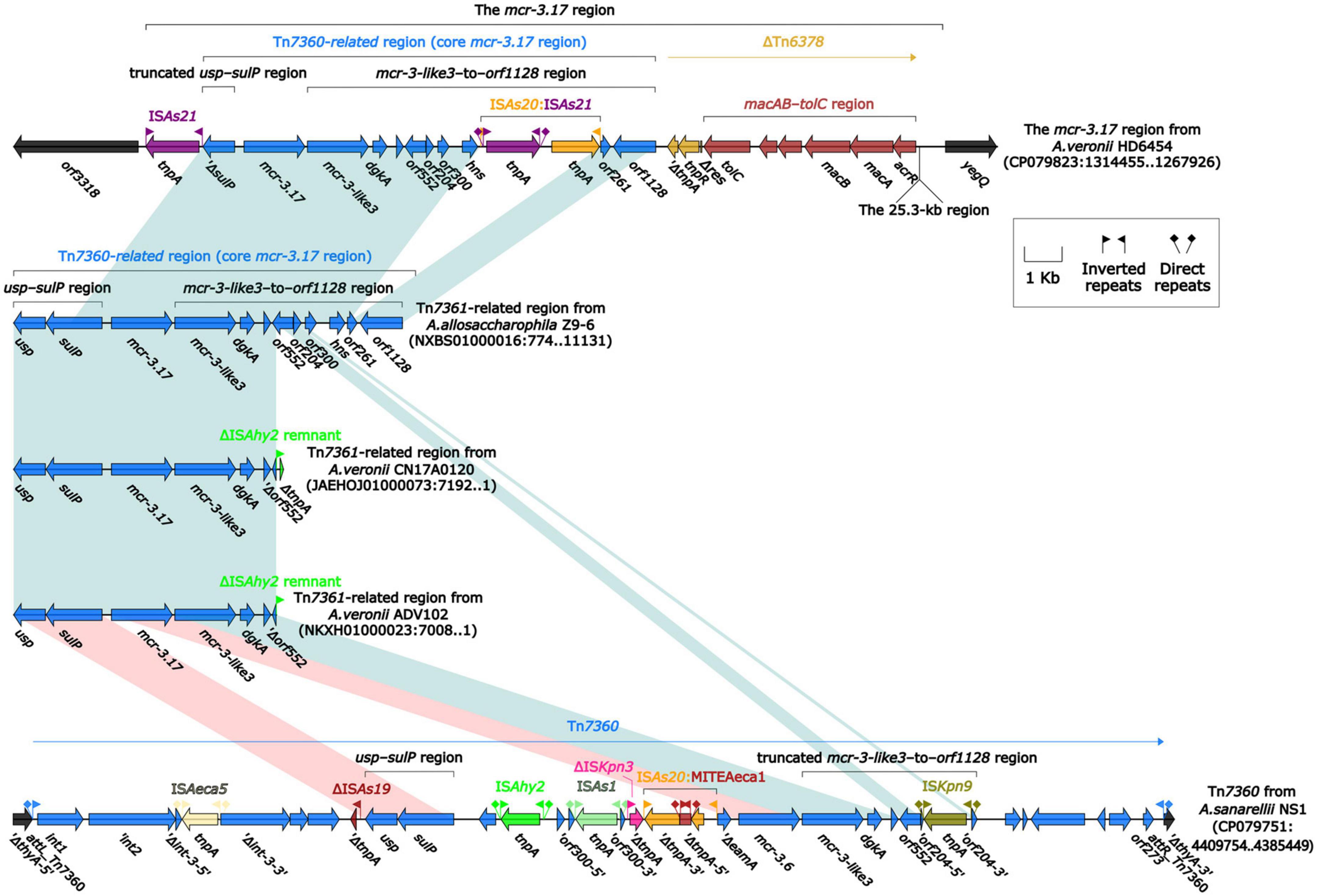
Figure 2. Organization of Tn7360-related regions, and comparison to Tn7360. The detailed information of strains is described in Supplementary Tables 3, 4. Genes are denoted by arrows. Genes, mobile genetic elements and other features are colored based on their functional classification. Shading denotes regions of homology (light blue: ≥95% nucleotide identity; light red: 88–95% nucleotide identity).
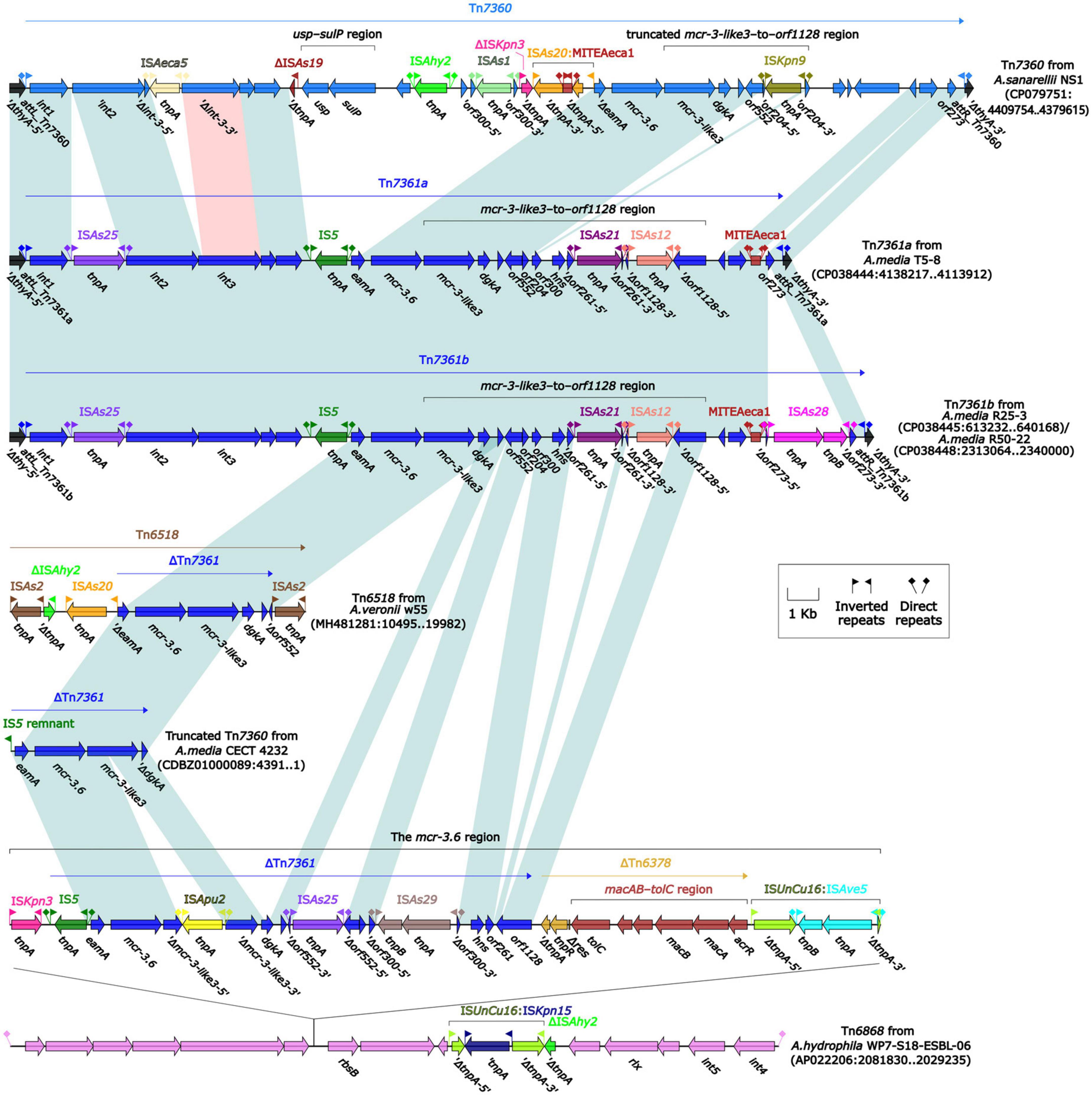
Figure 3. Organization of mcr-3.6-carrying genetic elements or related regions. The detailed information of strains is described in Supplementary Tables 3, 4. Genes are denoted by arrows. Genes, mobile genetic elements and other features are colored based on their functional classification. Shading denotes regions of homology (light blue: ≥95% nucleotide identity; light red: 88–95% nucleotide identity).
In summary, all mcr-3.6, mcr-3.17 and mcr-3-like3 characterized in this study were located within IME Tn7360/Tn7361 or related genetic elements and regions from Aeromonas species.
Functional identification of tmexC3.2-tmexD3.3-toprJ1b gene cluster
A tnfxB3-tmexCD3-toprJ1-like gene cluster was found in the chromosome of strain HD6454 initially. The sequence of this cluster was of 100% identical to the one carried by chromosome of A. caviae WCW1-2 (accession number CP039832) (Wang et al., 2021b). Furthermore, according to the naming scheme previously described (Wang et al., 2021a): (1) compared with the tnfxB3 gene from Proteus cibarius strain SDQ8C180-2T (accession number CP073356), one mutation was identified in the tnfxB3-like gene from HD6454 (predicted to encode Thr46Ala), and this tnfxB3-like gene was thus named tnfxB3.2; (2) compared with the tmexD3 gene from strain SDQ8C180-2T, one mutation was identified in the tmexD3-like gene from HD6454 (predicted to encode Val56Glu). This tmexD3-like gene was also different from the tmexD3.2 gene in strain P. mirabilis SDY9C89-2 (accession number MZ004963), so it was named tmexD3.3; (3) the tmexC3-like gene and toprJ1-like gene from HD6454 were identical to the tmexC3.2 gene from Pseudomonas aeruginosa strain AHM8C91AI (accession number JAGSOC000000000) and the toprJ1b gene from strain SDQ8C180-2T, respectively. In general, this tnfxB3-tmexCD3-toprJ1-like gene cluster from HD6454 was identified as tnfxB3.2-tmexC3.2-tmexD3.3-toprJ1b finally.
In order to determine the function of tmexC3.2-tmexD3.3-toprJ1b in mediating tigecycline resistance, the transformants DH5α/pUC18-tmexC3.2-tmexD3.3-toprJ1b was obtained. The antimicrobial susceptibility testing showed that DH5α/pUC18-tmexC3.2-tmexD3.3-toprJ1b presented an MIC of 0.5 mg/L to tigecycline, while DH5α/pUC18 had an MIC of 0.125 mg/L (Table 2). This result suggested that tmexC3.2-tmexD3.3-toprJ1b could confer low-level resistance to tigecycline.
Genetic characterization of tmexC3.2-tmexD3.3-toprJ1b gene cluster
The tmexCD1-toprJ1 gene cluster was firstly identified in the structure “int1-int2-hp1-hp2-tnfxB1-tmexCD1-toprJ1,” and this structure was further carried by Tn5393 (Lv et al., 2020). Subsequently, this structure along with its attachment site at the left/right end (attL/R) was defined as relaxosome-missing IME Tn6855 (Yu et al., 2021b). Further genetic dissection analysis showed that the tnfxB3.2-tmexC3.2-tmexD3.3-toprJ1b gene cluster from HD6454 was located within an IME Tn6855 variant, which was integrated in the umuC gene of the novel IME Tn7379 with 6-bp DRs (Figure 4). This Tn6855 variant differed from Tn6855 by the substitution of tnfxB1-tmexCD1-toprJ1 gene cluster to tnfxB3.2-tmexC3.2-tmexD3.3-toprJ1b. Additionally, some other tnfxB3-tmexCD3-toprJ1b gene clusters were also located within similar Tn6855 variants (Figure 4), and integrated within the umuC genes as previously reported (Wang et al., 2021a,b).
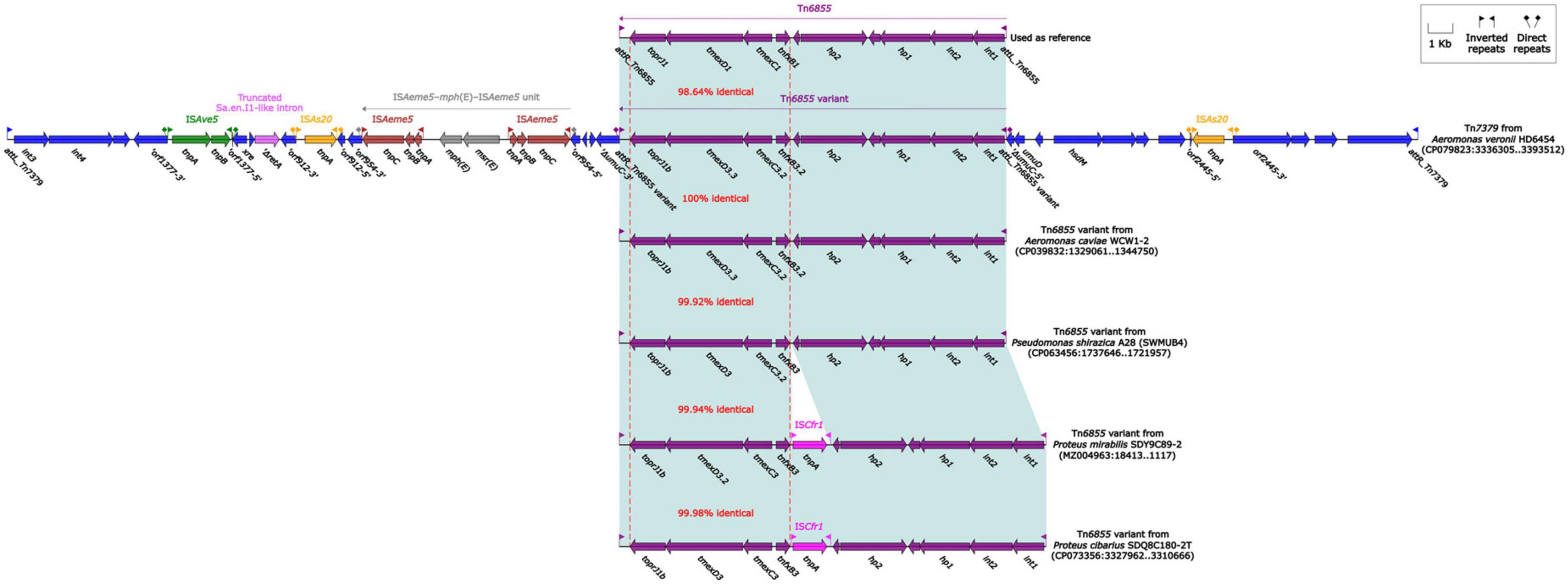
Figure 4. Organization of Tn7379 from HD6454, and comparison to related Tn6855 and its variants. Genes are denoted by arrows. Genes, mobile genetic elements and other features are colored based on their functional classification. Shading denotes regions of homology (light blue: ≥98% nucleotide identity). The accession number of Tn6855 (Yu et al., 2021b) used as reference is MK347425.
The blaKPC–2-carrying IncP-6 plasmid pHD6454-KPC
The blaKPC–2 was carried by a 51.66-kb IncP-6 plasmid, which was assigned the name pHD6454-KPC. The modular structure of pHD6454-KPC was divided into the backbone and three accessory modules (ISPa19, Tn5563b and the blaKPC–2 region) which were resulted from exogenous DNA regions insertion at different sites of the backbone (Supplementary Figure 3). The plasmid pHD6454-KPC shared 99.99% nucleotide identity to the IncP-6 reference plasmid p10265-KPC from Pseudomonas aeruginosa 10,265 with 76% coverage (accession number KU578314) (Dai et al., 2016), while the backbone of them were 99.98% nucleotide identical, along with 99% coverage. This showed that these two plasmids were mainly different in accessory modules (Figure 5). The unit transposon Tn5563b in pHD6454-KPC differed from Tn5563a in p10265-KPC by the insertion of a merT-harboring region (inserted regionmerT, 12.90 kb) (Figure 6A). The inserted regionmerT was flanked by 9-bp DRs, and may originated from the transposition and homologous recombination of an unknown-type plasmid. Additionally, the blaKPC–2 region in pHD6454-KPC was composed of a truncated blaKPC–2-carrying unit transposon Tn6296 (Wang et al., 2017) and a truncated unit transposon Tn6376b, and was almost identical to the one in p10265-KPC (Figure 6B).
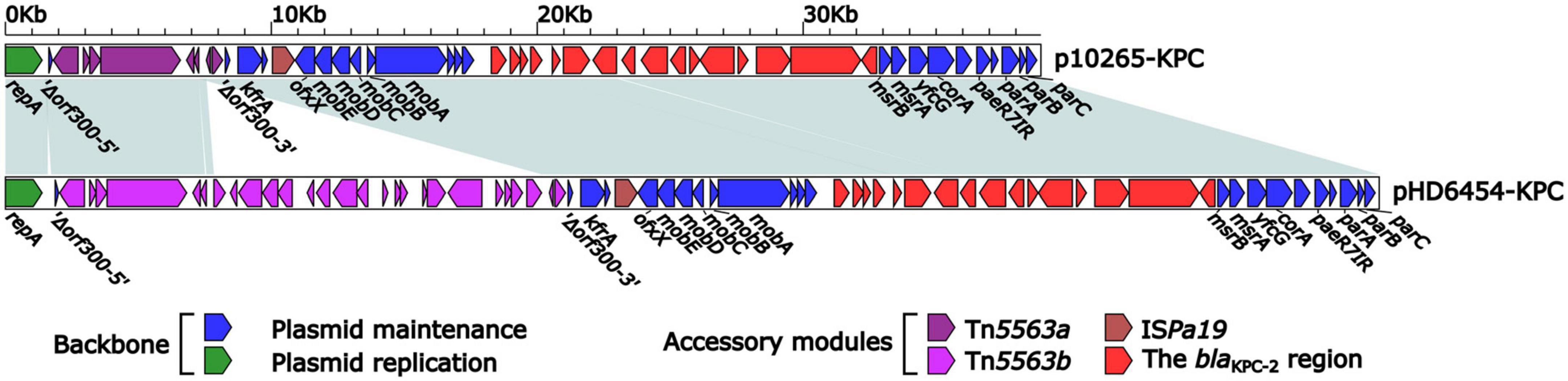
Figure 5. Linear comparison of IncP-6 plasmids pHD6454-KPC and p10265-KPC. Genes are denoted by arrows. Genes, mobile genetic elements and other features are colored based on function classification. Shading regions denote homology of two plasmids (light blue: ≥99% nucleotide identity).
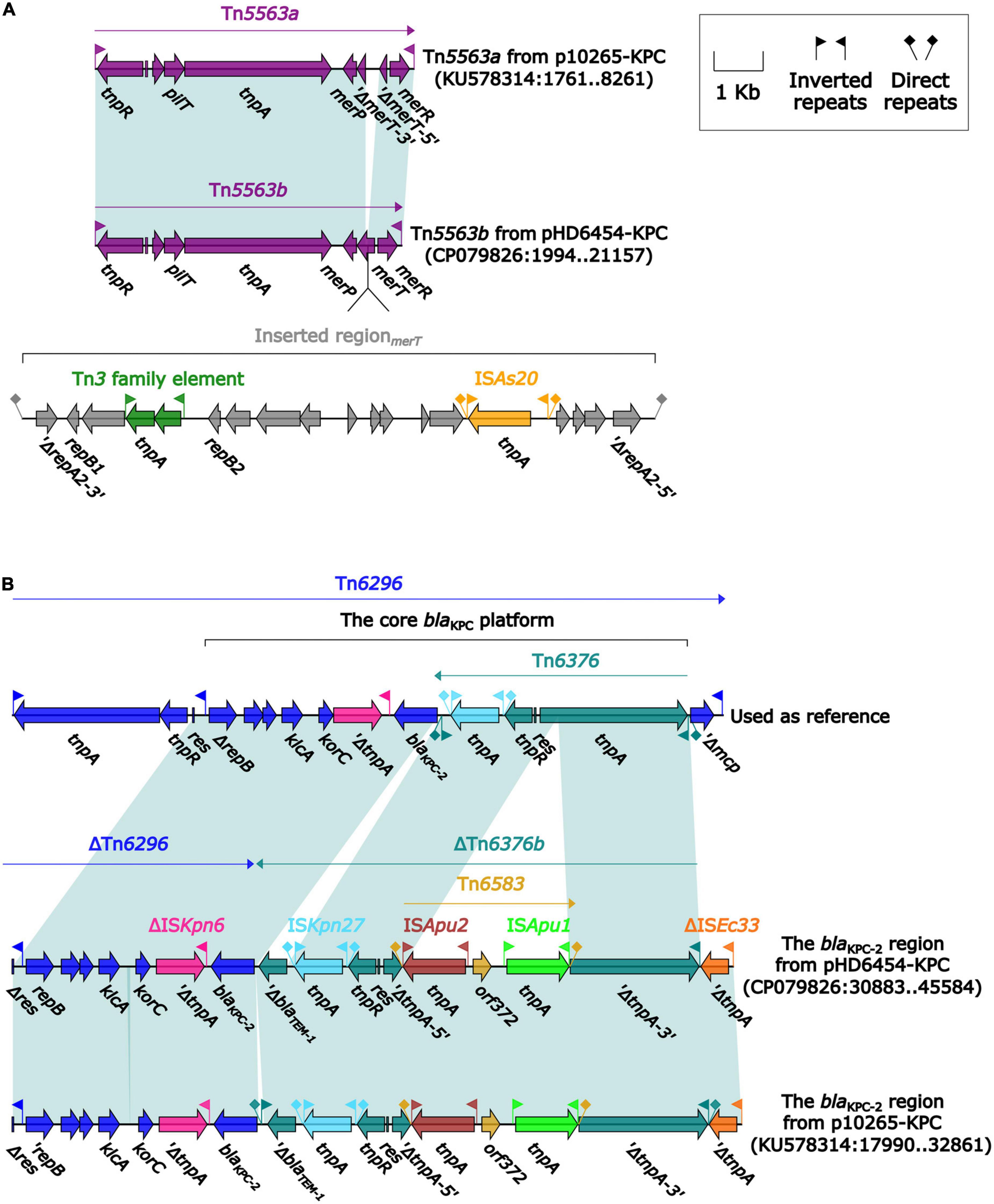
Figure 6. Organization of (A) the blaKPC–2 region and (B) Tn5563b from pHD6454-KPC, and comparison to related genetic elements. Genes are denoted by arrows. Genes, mobile genetic elements and other features are colored based on their functional classification. Shading denotes regions of homology (light blue: ≥ 99% nucleotide identity). The accession number of Tn6296 (Wang et al., 2017) used as reference are FJ628167.
The genomic dissection analysis showed that pHD6454-KPC lost the tra module, which was consistent with other IncP-6 plasmids as described previously (Dai et al., 2016; Hu et al., 2019). Repeated conjugation attempts failed to transfer pHD6454-KPC from strain HD6454 into J53, which matched the sequence analysis result.
Discussion
The mcr-3 genes have spread widely into diverse environmental niches by horizontal and vertical transfer (Anyanwu et al., 2020). Since the mcr-3.1 gene was initially identified in China in 2017, at least 40 non-redundant mcr-3 variants have been reported in Asia, North America, Africa, and Europe (El-Sayed Ahmed et al., 2020; Snyman et al., 2021; Stosic et al., 2021; Yu et al., 2021a). Among them, mcr-3.17 was only reported once in 2018, which was identified in A. allosaccharophila isolated from chicken meat in China (Shen et al., 2018). However, neither the detailed genomic structure of mcr-3.17 nor the resistant phenotype of mcr-3.17 to colistin have been confirmed in the previous study. In this study, mcr-3.17 was found in the hospital environment, and was confirmed to confer low-level resistance to colistin only under inducible expression. The mcr-3.6 gene has been proved to confer high-level resistance to colistin (Wang et al., 2020b). Although mcr-3.6 gene shares the highest similarity with mcr-3.17 among all mcr-3 variants, they still show several differences in both amino acid sequence and protein secondary structure, which may explain the low-level resistance to colistin of mcr-3.17. However, the key mutations affecting the level of resistance of mcr-3 variants to colistin remain unknown and need to be further investigated. As for mcr-3-like gene, a total of four mcr-3-like variants have been reported (Ling et al., 2017; Shen et al., 2018), which are all downstream of mcr-3 variants. Moreover, the nucleoside bases between these mcr-3-like variants and the corresponding mcr-3 variants upstream of them are always 66 bp. Previous studies have preliminarily proved that mcr-3-like1 and mcr-3-like3 could not mediate the MIC changes in recipient strains (Ling et al., 2017; Wang et al., 2020b). In this study, whether mcr-3-like3 gene was cloned into the cloning vector pUC18 together with its upstream promoter-proximal region or induced in the expression vector pBAD24, it could also not mediate the resistance to colistin. This result further proves that the MCR-3-like3 has no resistance activity to colistin.
Several tmexCD3-toprJ1b variants have been identified in various species, including Aeromonas spp. (Hirabayashi et al., 2021; Wang et al., 2020b,2021a,c; Wu et al., 2023). Although the identical sequence has been reported, the tmexC3.2-tmexD3.3-toprJ1b gene cluster was named systematically and was confirmed to confer low-level resistance to tigecycline for the first time in this study.
It was noteworthy that, despite carrying mcr-3.17 and tmexC3.2-tmexD3.3-toprJ1b gene cluster, HD6454 was susceptible to colistin and tigecycline. Since mcr-3.17 could confer low-level resistance to colistin only under inducible expression, namely, the high expression level of mcr-3.17, the phenomenon that HD6454 was susceptible to colistin might result from the low expression level of mcr-3.17 in HD6454. Meanwhile, tnfxB3 gene has been confirmed to have the transcriptional repression function to downstream tmexCD3-toprJ1b (Wang et al., 2021a). Hence, the transcriptional repression function of tnfxB3.2 may explain the phenomenon that HD6454 was susceptible to tigecycline, which deserves further study.
Integrative and mobilizable elements are not self-transmissible, and their intercellular mobility is achieved with utilization of the conjugation machinery of unrelated co-resident conjugative element (Guédon et al., 2017). In this study, both tnfxB3.2-tmexC3.2-tmexD3.3-toprJ1b gene cluster and mcr-3.17 were identified to be associated with IMEs. Among them, tnfxB3.2-tmexC3.2-tmexD3.3-toprJ1b gene cluster was carried by Tn6855 variant. Such Tn6855 variants could further integrate into the umuC gene of various genetical elements, for example, SXT/R391 family integrative and conjugative elements (Wang et al., 2021a,c), a IncC-IncX3 hybrid plasmid pNUITM-VK5_mdr (Hirabayashi et al., 2021), and another IME Tn7379 identified herein. Meanwhile, all mcr-3.6, mcr-3.17 and mcr-3-like3 characterized in this study were located within IME Tn7360/Tn7361 or related genetic elements [such as composite transposon Tn6518 (Wang et al., 2020b) and IME Tn6868] and regions. This shows that IME serves as an important carrier and mediator in the transmission of tnfxB3-tmexCD3-toprJ1b gene cluster and partial mcr-3 variants.
Due to the lack of a tra module encoding primary pilus, IncP-6 plasmid was previously considered to be not self-transmissible (Dai et al., 2016; Hu et al., 2019). However, IncP-6 plasmid harbors conserved par-rep regions for partition-replication and mob gene module for mobilization, which allows it to transfer if the right self-transmissible plasmids are co-resident (Dai et al., 2016). This explains why the same blaKPC–2 encoding IncP-6 plasmid could be found in non-clonal different isolates within various species from clinical or environmental sources (Yao et al., 2017). Similarly, the backbone sequence of the blaKPC–2-carrying pHD6454-KPC found in this study was almost identical to the IncP-6-type plasmid p10265-KPC firstly reported in China (Dai et al., 2016). Moreover, the accessory modules of pHD6454-KPC also seemed to evolved from the ones in p10265-KPC (Dai et al., 2016) undergoing the events of gene acquisition and deletion. Therefore, the potential transmission of these blaKPC–2-carrying IncP-6 plasmids in China should be closely monitored.
To the best of our knowledge, this is the first report of a strain co-harboring blaKPC–2, mcr-3.17, and tmexC3.2-tmexD3.3-toprJ1b gene cluster. Several factors may contribute to the emergence of this A. veronii strain HD6454. Firstly, mcr-3 is considered that most likely originated from Aeromonas species (Yin et al., 2017; Shen et al., 2018). All the 11 strains carrying mcr-3.6 or mcr-3.17 characterized in this study are Aeromonas species. Secondly, acquisition of exogenous DNA is a general property of Aeromonas environmental isolates (Huddleston et al., 2013), while the potential transmission of mcr-3-carrying or tmexCD3-toprJ1b-carrying IMEs (Guédon et al., 2017), and blaKPC–2-carrying IncP-6 plasmid increases the possibility of obtaining exogenous DNA by Aeromonas isolates. Thirdly, these ARGs and related genetic elements seem to be mainly distributed in China. Among them, blaKPC–2-bearing IncP-6 plasmid has been reported to be the most detected in China (Hu et al., 2019). Eight of the 11 strains carrying mcr-3.6 or mcr-3.17 characterized in this study are isolated from clinical or environmental sources in China. Furthermore, the current reports of tmexCD3-toprJ1b gene cluster are mainly based on strains from China (Wang et al., 2020b,2021a,c). These factors also show that Aeromonas species may act as important vectors for the dissemination of ARGs in China.
Conclusion
In conclusion, this study identified the A. veronii strain HD6454 co-harboring blaKPC–2, mcr-3.17, and tmexC3.2-tmexD3.3-toprJ1b gene cluster for the first time. The blaKPC–2 was carried by IncP-6 plasmid, while tmexC3.2-tmexD3.3-toprJ1b gene cluster and mcr-3.17 were carried by IME or IME-related region in chromosome. Although the resistance and/or mobility of these ARGs are limited in HD6454, the emergence of this multiple important ARGs-carrying strain deserves further attention.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
HD, LC, and ZZ conceived and designed the experiments. ZZ, JZ, and TW performed the experiments. ZZ, YW, and CY analyzed the genome sequence. ZZ and SW wrote the manuscript. HZ, SW, JL, YW, LC, and HD contributed other analysis or discussion. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the Science Foundation of Jiangsu Province Health Department (ZDB2020014), the Science Foundation of Suzhou Health Department (LCZX202106), the Science and Technology Program of Suzhou (SLJ2022003 and 2022SS41), and the Discipline Construction Program of the Second Affiliated Hospital of Soochow University (XKTJ-TD202001).
Acknowledgments
We thank Professor Dongsheng Zhou from State Key Laboratory of Pathogen and Biosecurity, Beijing Institute of Microbiology and Epidemiology for the assistance in the drawing of figures.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1115740/full#supplementary-material
Supplementary Figure 1 | Evolution relationships of the 54 MCR variants and MCR-3-like variants. Degree of support (percentage) for each cluster of associated taxa, as determined by bootstrap analysis, is shown next to each branch. Bar corresponds to scale of sequence divergence.
Supplementary Figure 2 | Comparison of amino acid sequences and protein secondary structures of MCR-3.6 and MCR-3.17. Within the amino acid sequence alignment, a strict identity was denoted by a red box with a white character. A yellow box around an amino acid residue denoted similarity across groups, while a residue in boldface denoted similarity within a group. Letter α, β, and η represented α-helix, β-sheet and 310-helix, respectively. Green digits below the alignment denote cysteine residues forming a disulfide bridge.
Supplementary Figure 3 | Schematic diagram of the plasmid pHD6454-KPC. Genes of different functions are denoted by arrows and presented in various colors. The circles show (from outside to inside): predicted coding sequences, scale in 10 kb, backbone (black) and accessory module (gray) regions, GC content and GC skew [(G–C)/(G + C)].
Footnotes
- ^ https://github.com/rrwick/Unicycler
- ^ https://pubmlst.org/
- ^ https://inkscape.org/en/
- ^ https://www.eucast.org/
References
Anyanwu, M. U., Jaja, I. F., and Nwobi, O. C. (2020). Occurrence and Characteristics of Mobile Colistin Resistance (mcr) Gene-Containing Isolates from the Environment: A Review. Int. J. Environ. Res. Public Health 17:1028. doi: 10.3390/ijerph17031028
Boratyn, G. M., Camacho, C., Cooper, P. S., Coulouris, G., Fong, A., Ma, N., et al. (2013). BLAST: a more efficient report with usability improvements. Nucleic Acids Res. 41, W29–W33. doi: 10.1093/nar/gkt282
Boutet, E., Lieberherr, D., Tognolli, M., Schneider, M., Bansal, P., Bridge, A. J., et al. (2016). UniProtKB/Swiss-Prot, the Manually Annotated Section of the UniProt KnowledgeBase: How to Use the Entry View. Methods Mol. Biol. 1374, 23–54.
Brettin, T., Davis, J. J., Disz, T., Edwards, R. A., Gerdes, S., Olsen, G. J., et al. (2015). RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 5:8365. doi: 10.1038/srep08365
Carroll, L. M., Gaballa, A., Guldimann, C., Sullivan, G., Henderson, L. O., and Wiedmann, M. (2019). Identification of Novel Mobilized Colistin Resistance Gene mcr-9 in a Multidrug-Resistant, Colistin-Susceptible Salmonella enterica Serotype Typhimurium Isolate. mBio 10:e00853-19. doi: 10.1128/mBio.00853-19
Dai, X., Zhou, D., Xiong, W., Feng, J., Luo, W., Luo, G., et al. (2016). The IncP-6 Plasmid p10265-KPC from Pseudomonas aeruginosa Carries a Novel ΔISEc33-Associated blaKPC–2 Gene Cluster. Front. Microbiol. 7:310. doi: 10.3389/fmicb.2016.00310
Doi, Y. (2019). Treatment Options for Carbapenem-resistant Gram-negative Bacterial Infections. Clin. Infect Dis. 69, S565–S575.
Dong, N., Zeng, Y., Wang, Y., Liu, C., Lu, J., Cai, C., et al. (2022). Distribution and spread of the mobilised RND efflux pump gene cluster tmexCD-toprJ in clinical Gram-negative bacteria: a molecular epidemiological study. Lancet Microbe 3, e846–e856. doi: 10.1016/S2666-5247(22)00221-X
El-Sayed Ahmed, M. A. E., Zhong, L. L., Shen, C., Yang, Y., Doi, Y., and Tian, G. B. (2020). Colistin and its role in the Era of antibiotic resistance: an extended review (2000-2019). Emerg. Microbes Infect. 9, 868–885. doi: 10.1080/22221751.2020.1754133
Fang, L. X., Chen, C., Cui, C. Y., Li, X. P., Zhang, Y., Liao, X. P., et al. (2020). Emerging High-Level Tigecycline Resistance: Novel Tetracycline Destructases Spread via the Mobile Tet(X). Bioessays 42:e2000014. doi: 10.1002/bies.202000014
Fernández-Bravo, A., and Figueras, M. J. (2020). An Update on the Genus Aeromonas: Taxonomy, Epidemiology, and Pathogenicity. Microorganisms 8:129. doi: 10.3390/microorganisms8010129
Gonzalez-Avila, L. U., Loyola-Cruz, M. A., Hernandez-Cortez, C., Bello-Lopez, J. M., and Castro-Escarpulli, G. (2021). Colistin Resistance in Aeromonas spp. Int. J. Mol. Sci. 22:5974.
Gowda, T. K., Reddy, V. R., Devleesschauwer, B., Zade, N. N., Chaudhari, S. P., Khan, W. A., et al. (2015). Isolation and Seroprevalence of Aeromonas spp. Among Common Food Animals Slaughtered in Nagpur, Central India. Foodborne Pathog. Dis. 12, 626–630. doi: 10.1089/fpd.2014.1922
Guédon, G., Libante, V., Coluzzi, C., Payot, S., and Leblond-Bourget, N. (2017). The Obscure World of Integrative and Mobilizable Elements, Highly Widespread Elements that Pirate Bacterial Conjugative Systems. Genes 8:337. doi: 10.3390/genes8110337
Hirabayashi, A., Dao, T. D., Takemura, T., Hasebe, F., Trang, L. T., Thanh, N. H., et al. (2021). A Transferable IncC-IncX3 Hybrid Plasmid Cocarrying blaNDM–4, tet(X), and tmexCD3-toprJ3 Confers Resistance to Carbapenem and Tigecycline. mSphere 6:e0059221. doi: 10.1128/mSphere.00592-21
Hu, X., Yu, X., Shang, Y., Xu, H., Guo, L., Liang, Y., et al. (2019). Emergence and Characterization of a Novel IncP-6 Plasmid Harboring blaKPC–2 and qnrS2 Genes in Aeromonas taiwanensis Isolates. Front. Microbiol. 10:2132. doi: 10.3389/fmicb.2019.02132
Huddleston, J. R., Brokaw, J. M., Zak, J. C., and Jeter, R. M. (2013). Natural transformation as a mechanism of horizontal gene transfer among environmental Aeromonas species. Syst. Appl. Microbiol. 36, 224–234. doi: 10.1016/j.syapm.2013.01.004
Jia, B., Raphenya, A. R., Alcock, B., Waglechner, N., Guo, P., Tsang, K. K., et al. (2017). CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 45, D566–D573. doi: 10.1093/nar/gkw1004
Karkman, A., Do, T. T., Walsh, F., and Virta, M. P. J. (2018). Antibiotic-Resistance Genes in Waste Water. Trends Microbiol. 26, 220–228.
Kelley, L. A., Mezulis, S., Yates, C. M., Wass, M. N., and Sternberg, M. J. (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858. doi: 10.1038/nprot.2015.053
Kieffer, N., Royer, G., Decousser, J. W., Bourrel, A. S., Palmieri, M., and Ortiz, et al. (2019). mcr-9, an Inducible Gene Encoding an Acquired Phosphoethanolamine Transferase in Escherichia coli, and Its Origin. Antimicrob. Agents Chemother. 63:e00965-19. doi: 10.1128/AAC.00965-19
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Ling, Z., Yin, W., Li, H., Zhang, Q., Wang, X., Wang, Z., et al. (2017). Chromosome-Mediated mcr-3 Variants in Aeromonas veronii from Chicken Meat. Antimicrob. Agents Chemother. 61:e001272-17. doi: 10.1128/AAC.01272-17
Liu, Y. Y., Wang, Y., Walsh, T. R., Yi, L. X., Zhang, R., Spencer, J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168. doi: 10.1016/S1473-3099(15)00424-7
Lv, L., Wan, M., Wang, C., Gao, X., Yang, Q., Partridge, S. R., et al. (2020). Emergence of a Plasmid-Encoded Resistance-Nodulation-Division Efflux Pump Conferring Resistance to Multiple Drugs, Including Tigecycline, in Klebsiella pneumoniae. mBio 11, e002930–19. doi: 10.1128/mBio.02930-19
Nordmann, P., and Poirel, L. (2019). Epidemiology and Diagnostics of Carbapenem Resistance in Gram-negative Bacteria. Clin. Infect. Dis. 69, S521–S528.
Nwaiwu, O., and Aduba, C. C. (2020). An in silico analysis of acquired antimicrobial resistance genes in Aeromonas plasmids. AIMS Microbiol. 6, 75–91. doi: 10.3934/microbiol.2020005
O’Leary, N. A., Wright, M. W., Brister, J. R., Ciufo, S., Haddad, D., Mcveigh, R., et al. (2016). Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 44, D733–D745. doi: 10.1093/nar/gkv1189
Rangel, L. T., Marden, J., Colston, S., Setubal, J. C., Graf, J., and Gogarten, J. P. (2019). Identification and characterization of putative Aeromonas spp. T3SS effectors. PLoS One 14, e0214035. doi: 10.1371/journal.pone.0214035
Robert, X., and Gouet, P. (2014). Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324. doi: 10.1093/nar/gku316
Shen, Y., Xu, C., Sun, Q., Schwarz, S., Ou, Y., Yang, L., et al. (2018). Prevalence and Genetic Analysis of mcr-3-Positive Aeromonas Species from Humans, Retail Meat, and Environmental Water Samples. Antimicrob. Agents Chemother. 62, e404–e418. doi: 10.1128/AAC.00404-18
Shi, Y., Tian, Z., Gillings, M. R., Zhang, Y., Zhang, H., Huyan, J., et al. (2020). Novel Transposon Tn6433 Variants Accelerate the Dissemination of tet(E) in Aeromonas in an Aerobic Biofilm Reactor under Oxytetracycline Stresses. Environ. Sci. Technol. 54, 6781–6791. doi: 10.1021/acs.est.0c01272
Siguier, P., Perochon, J., Lestrade, L., Mahillon, J., and Chandler, M. (2006). ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34, D32–D36.
Snyman, Y., Whitelaw, A. C., Barnes, J. M., Maloba, M. R. B., and Newton-Foot, M. (2021). Characterisation of mobile colistin resistance genes (mcr-3 and mcr-5) in river and storm water in regions of the Western Cape of South Africa. Antimicrob. Resist. Infect. Control 10, 96. doi: 10.1186/s13756-021-00963-2
Stosic, M. S., Leangapichart, T., Lunha, K., Jiwakanon, J., Angkititrakul, S., Järhult, J. D., et al. (2021). Novel mcr-3.40 variant co-located with mcr-2.3 and blaCTX–M–63 on an IncHI1B/IncFIB plasmid found in Klebsiella pneumoniae from a healthy carrier in Thailand. J. Antimicrob. Chemother. 76, 2218–2220. doi: 10.1093/jac/dkab147
Tang, L., Huang, J., She, J., Zhao, K., and Zhou, Y. (2020). Co-Occurrence of the blaKPC–2 and mcr-3.3 Gene in Aeromonas caviae SCAc2001 Isolated from Patients with Diarrheal Disease. Infect. Drug Resist. 13, 1527–1536. doi: 10.2147/IDR.S245553
Tansirichaiya, S., Rahman, M. A., and Roberts, A. P. (2019). The transposon registry. Mob. DNA 10:40.
Wang, C., Feng, Y., Liu, L., Wei, L., Kang, M., and Zong, Z. (2020a). Identification of novel mobile colistin resistance gene mcr-10. Emerg. Microbes Infect. 9, 508–516. doi: 10.1080/22221751.2020.1732231
Wang, X., Zhai, W., Wang, S., Shen, Z., Wang, Y., and Zhang, Q. (2020b). A Novel Transposon, Tn6518, Mediated Transfer of mcr-3 Variant in ESBL-Producing Aeromonas veronii. Infect Drug Resist 13, 893–899. doi: 10.2147/IDR.S239865
Wang, C. Z., Gao, X., Yang, Q. W., Lv, L. C., Wan, M., Yang, J., et al. (2021b). A Novel Transferable Resistance-Nodulation-Division Pump Gene Cluster, tmexCD2-toprJ2, Confers Tigecycline Resistance in Raoultella ornithinolytica. Antimicrob. Agents Chemother. 65, e2229–e2220. doi: 10.1128/AAC.02229-20
Wang, Q., Peng, K., Liu, Y., Xiao, X., Wang, Z., and Li, R. (2021c). Characterization of TMexCD3-TOprJ3, an RND-Type Efflux System Conferring Resistance to Tigecycline in Proteus mirabilis, and Its Associated Integrative Conjugative Element. Antimicrob. Agents Chemother. 65, e0271220. doi: 10.1128/AAC.02712-20
Wang, C. Z., Gao, X., Lv, L. C., Cai, Z. P., Yang, J., and Liu, J. H. (2021a). Novel tigecycline resistance gene cluster tnfxB3-tmexCD3-toprJ1b in Proteus spp. and Pseudomonas aeruginosa, co-existing with tet(X6) on an SXT/R391 integrative and conjugative element. J. Antimicrob. Chemother. 76, 3159–3167. doi: 10.1093/jac/dkab325
Wang, D., Zhu, J., Zhou, K., Chen, J., Yin, Z., Feng, J., et al. (2017). Genetic characterization of novel class 1 Integrons In0, In1069 and In1287 to In1290, and the inference of In1069-associated integron evolution in Enterobacteriaceae. Antimicrob. Resist. Infect. Control 6:84. doi: 10.1186/s13756-017-0241-9
Wang, P., Jiang, X., Mu, K., Jing, Y., Yin, Z., Cui, Y., et al. (2022). DANMEL: A manually curated reference database for analyzing mobile genetic elements associated with bacterial drug resistance. mLife 1, 460–464.
Wen, Y., Xie, X., Xu, P., Yang, C., Zhu, Z., Zhu, J., et al. (2022). NDM-1 and OXA-48-Like Carbapenemases (OXA-48, OXA-181 and OXA-252) Co-Producing Shewanella xiamenensis from Hospital Wastewater, China. Infect. Drug Resist. 15, 6927–6938.
Wu, Y., Dong, N., Cai, C., Zeng, Y., Lu, J., Liu, C., et al. (2023). Aeromonas spp. from hospital sewage act as a reservoir of genes resistant to last-line antibiotics. Drug Resist. Updat. 67:100925. doi: 10.1016/j.drup.2023.100925
Yaghoubi, S., Zekiy, A. O., Krutova, M., Gholami, M., Kouhsari, E., Sholeh, M., et al. (2021). Tigecycline antibacterial activity, clinical effectiveness, and mechanisms and epidemiology of resistance: narrative review. Eur. J. Clin. Microbiol. Infect. Dis. 41, 1003–1022. doi: 10.1007/s10096-020-04121-1
Yao, Y., Lazaro-Perona, F., Falgenhauer, L., Valverde, A., Imirzalioglu, C., Dominguez, L., et al. (2017). Insights into a Novel blaKPC–2-Encoding IncP-6 Plasmid Reveal Carbapenem-Resistance Circulation in Several Enterobacteriaceae Species from Wastewater and a Hospital Source in Spain. Front. Microbiol. 8:1143. doi: 10.3389/fmicb.2017.01143
Yin, W., Li, H., Shen, Y., Liu, Z., Wang, S., Shen, Z., et al. (2017). Novel Plasmid-Mediated Colistin Resistance Gene mcr-3 in Escherichia coli. mBio 8, e543–e517. doi: 10.1128/mBio.00543-17
Yu, T., Yang, H., Li, J., Chen, F., Hu, L., Jing, Y., et al. (2021b). Novel Chromosome-Borne Accessory Genetic Elements Carrying Multiple Antibiotic Resistance Genes in Pseudomonas aeruginosa. Front. Cell Infect. Microbiol. 11:638087. doi: 10.3389/fcimb.2021.638087
Yu, L., Kitagawa, H., Kayama, S., Hisatsune, J., Ohge, H., and Sugai, M. (2021a). Complete Genome Sequence of Aeromonas caviae Strain MS6064, a mcr-3-Carrying Clinical Isolate from Japan. Microbiol. Resour. Announc. 10, e1037–e1020. doi: 10.1128/MRA.01037-20
Zankari, E., Hasman, H., Cosentino, S., Vestergaard, M., Rasmussen, S., Lund, O., et al. (2012). Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67, 2640–2644.
Keywords: Aeromonas veronii, blaKPC–2, mcr-3.17, tmexCD3-toprJ1b, hospital sewage
Citation: Zhu Z, Wu S, Zhu J, Wang T, Wen Y, Yang C, Lv J, Zhang H, Chen L and Du H (2023) Emergence of Aeromonas veronii strain co-harboring blaKPC–2, mcr-3.17, and tmexC3.2-tmexD3.3-toprJ1b cluster from hospital sewage in China. Front. Microbiol. 14:1115740. doi: 10.3389/fmicb.2023.1115740
Received: 04 December 2022; Accepted: 02 May 2023;
Published: 17 May 2023.
Edited by:
Marta Tacão, University of Aveiro, PortugalReviewed by:
Pedro Teixeira, Chemistry and Technology Network (REQUIMTE), PortugalJian-Hua Liu, South China Agricultural University, China
Anna Luiza Bauer Canellas, Federal University of Rio de Janeiro, Brazil
Copyright © 2023 Zhu, Wu, Zhu, Wang, Wen, Yang, Lv, Zhang, Chen and Du. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hong Du, aG9uZ19kdUAxMjYuY29t
†These authors have contributed equally to this work
 Zhichen Zhu1†
Zhichen Zhu1† Jinnan Lv
Jinnan Lv Haifang Zhang
Haifang Zhang Hong Du
Hong Du