- School of Life Sciences, Chongqing University, Chongqing, China
Cancer and microbial infections are significant worldwide health challenges. Numerous studies have demonstrated that bacteria may contribute to the emergence of cancer. In this review, we assemble bacterial species discovered in various cancers to describe their variety and specificity. The relationship between bacteria and macrophages in cancer is also highlighted, and we look for ample proof to establish a biological basis for bacterial-induced macrophage polarization. Finally, we quickly go over the potential roles of metabolites, cytokines, and microRNAs in the regulation of the tumor microenvironment by bacterially activated macrophages. The complexity of bacteria and macrophages in cancer will be revealed as we gain a better understanding of their pathogenic mechanisms, which will lead to new therapeutic approaches for both inflammatory illnesses and cancer.
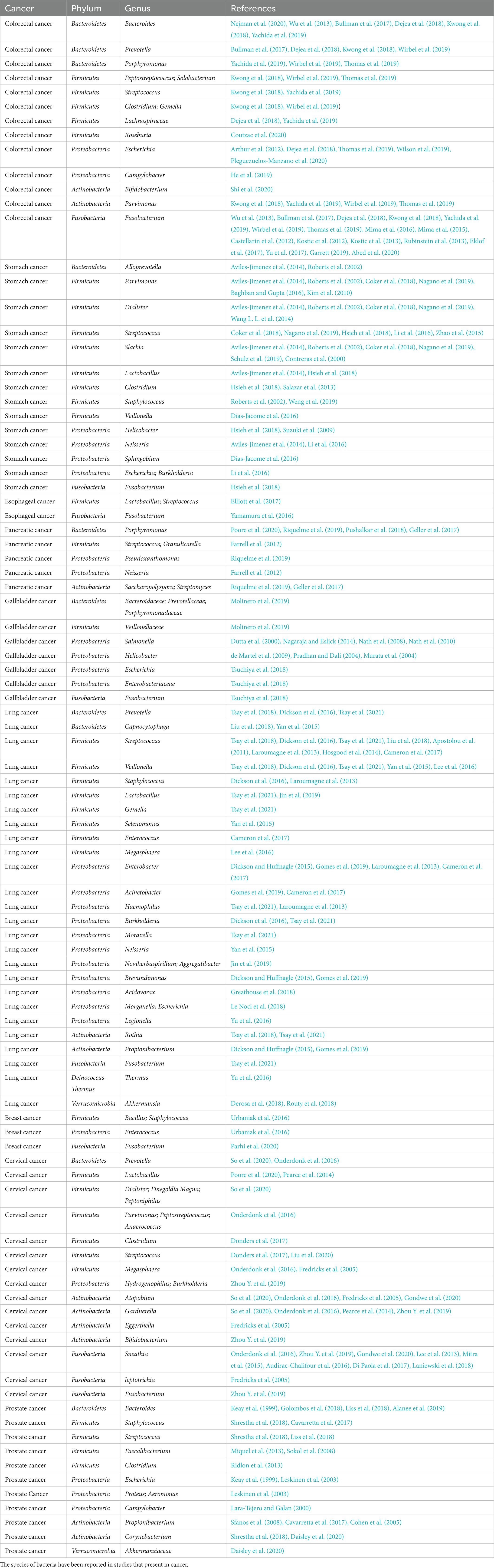
1. Bacterial diversity in different cancers
The immune system was traditionally thought to render tumors sterile. Thanks to technological advancements, numerous investigations in recent years have discovered that bacteria are prevalent in cancer. Many tumors’ early stages are challenging to recognize, and most malignancies have metastasized by the time of initial diagnosis. The classification of these bacteria may be able to provide us with some information. Testing of bacterial DNA has shown that each cancer type, including those not directly related to the external environment, has a different bacterial composition (Nejman et al., 2020). The goal of this section is to give a summary of the bacteria found in cancer tissue. To gather and compile the microorganisms, we specialize in nine common cancer types: colorectal, gastric, esophageal, pancreatic, gallbladder, lung, breast, cervical, and prostate (Table 1). Pathogenic bacteria and human commensal microorganisms frequently coexist in the enormous and complex microbial community that makes up the human gastrointestinal tract. The gastrointestinal microbiome has a significant impact on metabolic health and general health, and it is also the microbiome that has been studied the most in-depth and is used as a model to study host-microbiota interactions and disorders. In addition to being infrequently researched, the quantity of microbiota living in the other organs is substantially lower than that of the gut and stomach (Sepich-Poore et al., 2021). It’s uncertain how many and how diverse the bacteria are in cancer samples as compared to samples taken from healthy people. The quantity and diversity of bacteria are greater in breast tumor samples than in healthy, normal breast samples (Nejman et al., 2020). Breast cancer tissues of various grades and histological classifications and normal breast tissue differ greatly in terms of their bacterial composition (Nejman et al., 2020). Instead, the lung cancer tissue microbiome is less varied than the corresponding normal tissue microbiome (Peters et al., 2019). In reality, only one specific bacterial species, Helicobacter pylori, which is linked to gastric cancer designated by the World Health Organization as a class I carcinogen (Lunn et al., 2022), and research into other bacterial species’ potential roles as biomarkers in the majority of cancer types has not yet produced any conclusive findings (Scott et al., 2019). There is a large variety of microbial taxa with variable abundance but little overlap in studies of males with prostate cancer (Shrestha et al., 2018). Not only is there a microbiota within cancer, but crosstalk occurs in all organs of the body. The proliferation and composition of bacteria where not in direct contact with the outside world, to some extent, represent the bacteria that can be transferred from one organ to another. The microbiological makeup of the lungs is more similar to that of the oropharynx, and enteric organisms are the primary source of bacterial DNA in cancer patients’ pancreas tissue (Dickson and Huffnagle, 2015). Thanks to developments in polymerase chain reaction and metagenomics, we can now identify microbes more precisely. The researchers distinguish the microbiota of lung cancer tissue using different biological materials, such as bronchial or bronchoalveolar lavage fluid or sputum (Gomes et al., 2019), and the microbiota of prostate cancer using feces or urine microbiomes (Sfanos et al., 2008). But in every experiment, skin-associated germs could contaminate the reagents by transferring them from the personnel’s skin. Cancer-associated microbes in general are sometimes difficult to distinguish, and the study of particular bacteria in malignancies is still in its early stages.
2. Macrophages in the tumor microenvironment
Macrophages are multipurpose immune cells that perform a variety of tasks, such as regulating tissue homeostasis, protecting against infections, and accelerating wound healing (Wynn and Vannella, 2016). Macrophages are found in peripheral organs because these immunological sentinel cells are crucial in keeping an eye out for invasive infections in the surrounding tissue (Sfanos et al., 2008). When a host is infected by a pathogen, monocytes are drawn to the invasion sites and cytokines are released, which prompt additional immune responses from other immune cells. Indeed, bacteria and their metabolites have recently been shown to affect macrophages and tumor microenvironments. Numerous disorders, including cancer and infections for which there is yet no clear direct link, are affected by macrophage activation. Here, we discuss macrophages from three angles: their origin, their activation indicators, and the bacterial collection that causes them to become polarized. We just briefly touch on the preceding two aspects, because they have recently been discussed (Murray et al., 2014; Wynn and Vannella, 2016; Shapouri-Moghaddam et al., 2018; Christofides et al., 2022). Instead, we focus on the results of macrophage polarization caused by certain bacteria.
2.1. The source of macrophages
All tissues have macrophages, a kind of leukocyte that is divided into various subpopulations according to where it is found and how it functions. Macrophages come from two different origins. On the other hand, tissue-resident macrophages derived from erythro-myeloid progenitors in the yolk sac and fetal liver, or monocyte–macrophage DC progenitors in the bone marrow (Cassetta and Pollard, 2020). Peripheral blood monocytes, which are drawn to tissues by chemokines, can also develop into tissue-resident macrophages (Long et al., 2019). One of the numerous and varied cell groups that make up the tumor microenvironment and can affect tumor formation is the tumor-associated macrophages that populate the tumor tissue (Vitale et al., 2019). It is firmly established that TAMs influence tumor development, immunological control, tumor angiogenesis, and metastasis in the tumor microenvironment (Lin et al., 2019). Although the precise timing and process of this remain unknown, the bulk of TAMs are typically produced from blood monocytes, and tumor monocytes recruited via chemokines like CCL2 enter the tumor to develop into TAMs (Mantovani et al., 2008). Additionally, macrophages in metastatic tumors often referred to as metastasis-associated macrophages (MAMs), have different phenotypes and roles from those in primary tumors. TAMs states in patients have predictive relevance, according to some research, as their abundance correlates with various clinical outcomes (Pittet et al., 2022).
2.2. The polarization and markers of macrophages
Macrophage polarization is a biological process that eventually displays a certain phenotype after functionally responding to microenvironmental signals found in particular tissues. M1/M2 is acknowledged as the most straightforward word to describe macrophage phenotypes, based on the types of activation signals, such as immunological signals, tumor metabolism signals, and cell death signals. The M1 macrophage, a type of classical activation macrophage, plays a crucial role in anti-tumor immunity as well as mediating the host’s defense against a variety of bacteria, protozoa, and viruses. It is also involved in several chronic inflammatory and autoimmune illnesses (Murray and Wynn, 2011). Conversely, the M2 macrophage is an alternatively activated macrophage that devours fragmented and apoptotic cells and has anti-inflammatory and pro-angiogenic activity to control wound healing (Murray and Wynn, 2011). Depending on the activating stimuli received, M2 macrophages have been further divided into M2a, M2b, M2c, and M2d (Shapouri-Moghaddam et al., 2018). TAMs are a polarized novel subset of the M2 macrophage population, which was named M2d in recent studies (Mantovani et al., 2002; Duluc et al., 2007). M1-polarized cells produce ROS and NO more effectively, as well as pro-inflammatory cytokines like TNF-α, IL-1, and IL-6, as well as chemokines like CXCL8, CCL2, CXCL9, and CXCL10. M2-polarized cells express the mannose receptor, which triggers the production of chemokines including CCL17, CCL18, CCL22, and CCL24. They also produce anti-inflammatory cytokines like IL-10 and TGF-β. The expression of macrophage activation indicators, as well as cytokines, chemokines, and other secreted mediators, have all been thoroughly discussed in these papers (Mantovani et al., 2004; Murray et al., 2014; Shapouri-Moghaddam et al., 2018). TAM subpopulations are also first categorized as M1 and M2 macrophages based on the expression of certain markers, with functions assumed to be anti-tumor/anti-inflammation and pro-tumor/pro-inflammation development, respectively. It is usually determined that the majority of TAMs isolated from primary and metastatic cancers exhibit a suppressive M2-like phenotype (Murray and Wynn, 2011). However, because numerous subsets of TAMs display the indicators of both M1-and M2-polarization signatures, this simple nomenclature is unable to discriminate between the varied phenotypes of TAMs (Laviron and Boissonnas, 2019). TAMs are not precisely divided into the M1 and M2 phenotypes in vivo. How to select reliable biomarkers to classify TAMs in the marker database remains the main issue of research nowadays.
2.3. Macrophage polarization by bacteria
In response to cues from the immediate milieu, macrophages can change from one functional phenotype to another. Particular signaling sources like pathogenic sources might cause phenotypic flipping in macrophage populations and promote the development of tumors. Although most microorganisms are phagocytosed and killed by macrophages, some bacteria live in macrophages as opportunistic residents and utilize them for replication (Geller et al., 2017). M1 macrophages are mediated by microbial stimuli, including intracellular bacteria, to support cytotoxic activity and infection resistance. To thrive in the microenvironment, some bacteria can increase M2 polarization or interfere with M1 polarization. To avoid cytotoxic effects and circumvent the cellular immune response, microbes like Mycobacterium tuberculosis may mediate M2-polarized macrophages (Kaufmann, 2016). Table 2 lists the phenotypes of macrophages in response to bacterial pathogens in the context of oncology. Our understanding of functional markers may be too simplistic, while the use of complex markers may be confusing for researchers outside of immunology. All things considered, we continue to define pro-tumor/pro-inflammatory macrophages as M1 and anti-tumor/anti-inflammation macrophages as M2. However, it should not be forgotten that a particular live scene is unlikely to fall exactly into the combinations in Table 2, and a deeper study is needed to obtain further information and standardization (Figure 1).
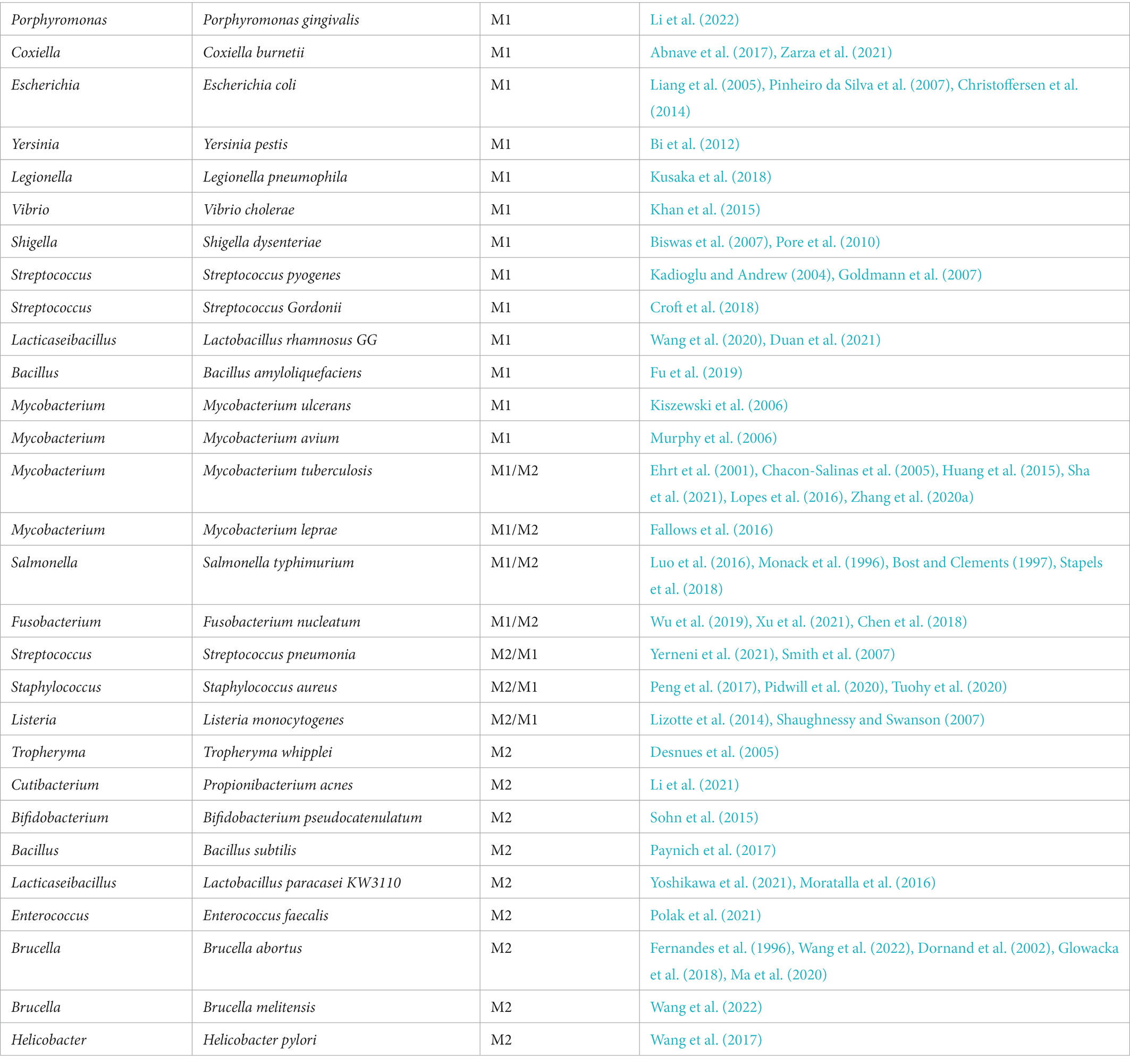
Table 2. An aggregate list of bacterial-driven macrophage polarization that has been studied is currently available.
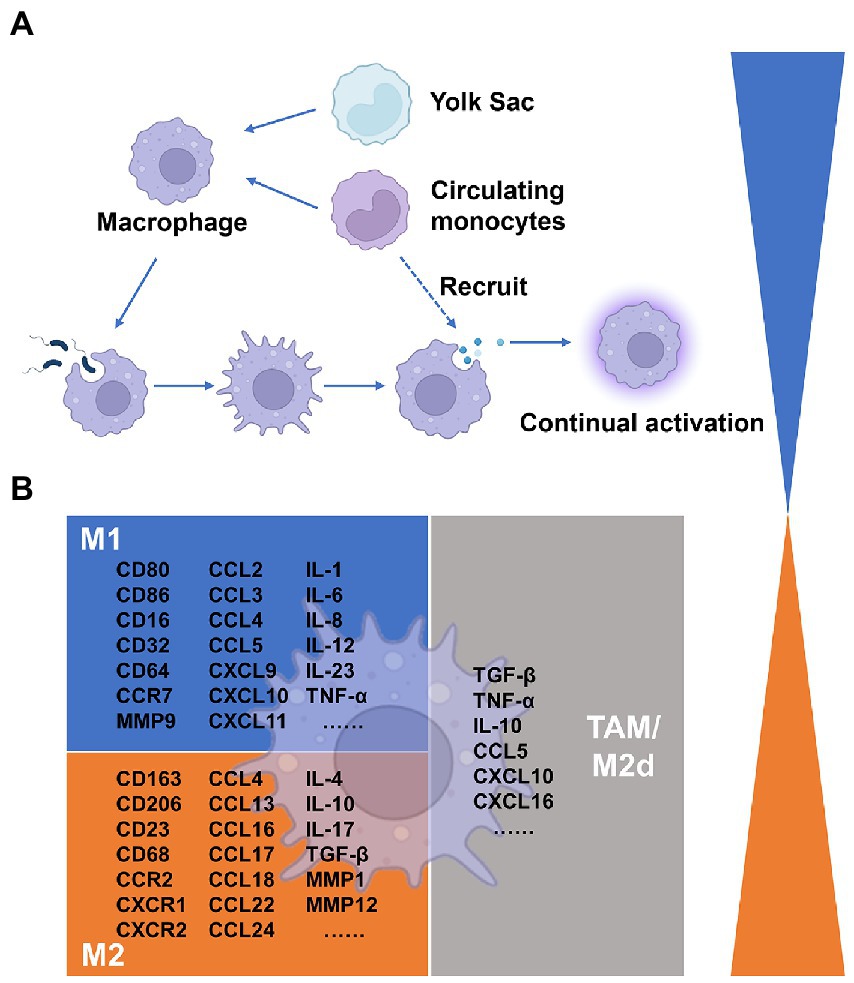
Figure 1. Summary of macrophages in the tumor microenvironment. (A) Origins of Macrophages. (B) Cytokines associated with macrophage differentiation to the classically and alternatively activated subsets.
3. Molecular mechanisms involved in bacterial-driven macrophage polarization
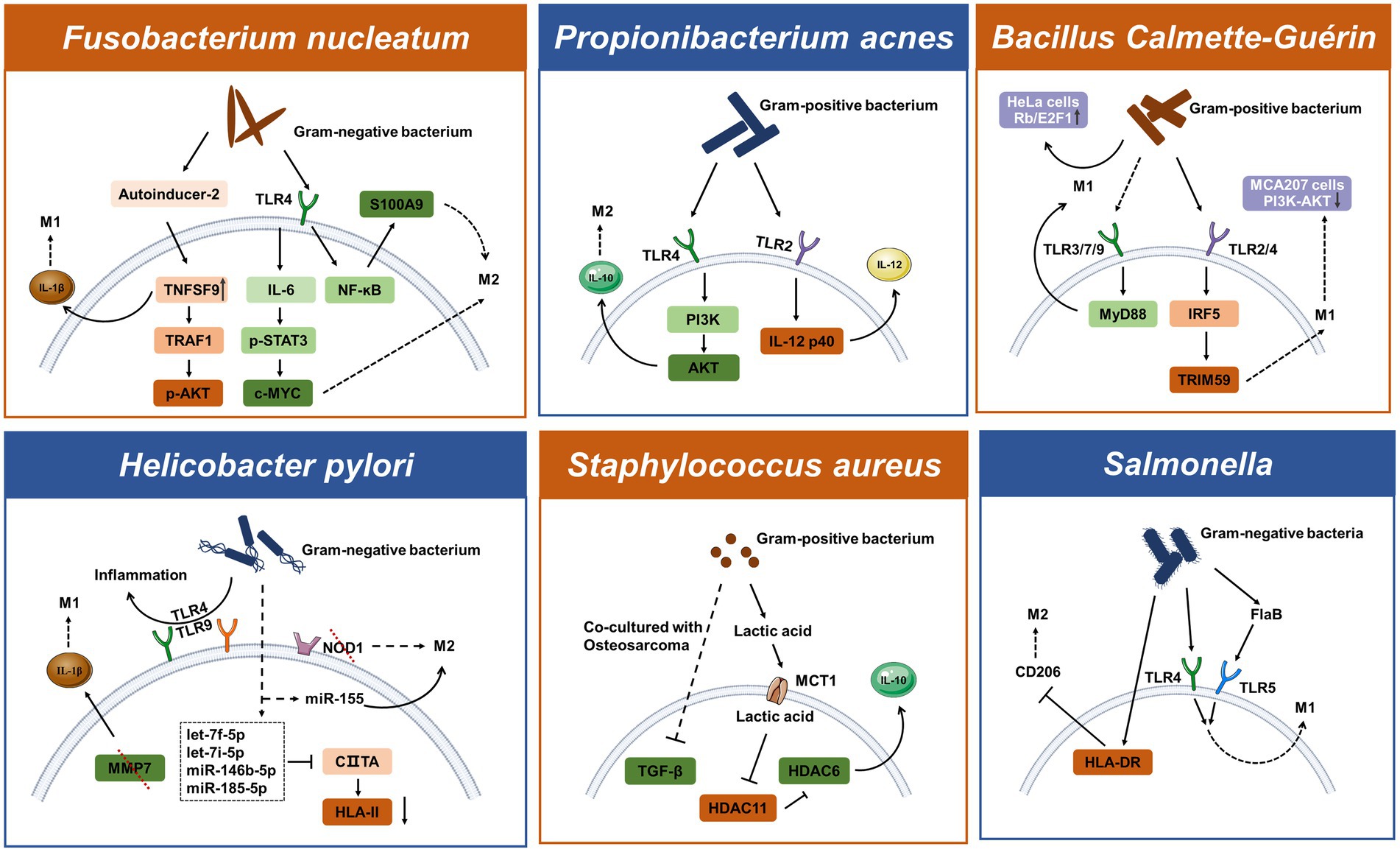
Since the microbiota has existed in the gut since human birth, the immune system and bacteria may have always been connected. Different bacterial species use both common and distinctive intrinsic mechanisms to either support or kill cancer. When it comes to host-pathogen interactions, living bacteria or bacterial components typically trigger innate immune cell reactions and cause immune cells, such as monocytes and macrophages, to migrate to tumors. Several bacteria are known to be connected to cancer, such as Helicobacter pylori and Salmonella typhi that have been shown to affect tumor growth (Lax and Thomas, 2002). However, the method by which bacteria in tumors interact with macrophages to select for the M1/M2 activation pathway is rarely discussed. In this part, we examine the molecular processes by which typical bacteria influence the polarization of macrophages in tumors, focusing on some of the well-known strains, such as Fusobacterium nucleatum, Helicobacter pylori, and Propionibacterium acnes.
3.1. Bacteria significantly linked to cancer
3.1.1. Fusobacterium nucleatum
A member of the bacterial genus that may cause cancer is called Fusobacterium nucleatum, a Gram-negative anaerobic bacterium. Fusobacterium nucleatum has been discovered as a periodontal pathogen and has been preferentially isolated from the oral cavity. Additionally, it has been extensively addressed how F. nucleatum and colorectal cancer are related (Hashemi Goradel et al., 2019). The Toll-like receptors recognize the molecular characteristics of pathogens, and each TLR elicits a different cellular response to the pathogen. In the microenvironment of colorectal tumors, F. nucleatum has been shown to enhance macrophage M2 polarization through a TLR4-dependent mechanism. Infection with F. nucleatum may also activate the IL-6/p-STAT3/c-MYC signaling pathway in macrophages in a TLR4-dependent manner (Chen et al., 2018). The study further illustrates that F. nucleatum promotes M2 macrophage polarization through activation of the TLR4/NF-κB/S100A9 cascade (Hu et al., 2021). The transcriptional stimulation of downstream NF-κB and STAT3, which can activate the survival pathway of tumor cells, is one of the main functions of TLR signaling. Fusobacterium nucleatum has been demonstrated to promote the growth of colorectal cancer via stimulating TLR4 signaling to MyD88, which then triggers the nuclear factor NF-κB and miR21 production (Yang et al., 2017). Fusobacterium nucleatum regulates miR-1,322/CCL20 through the NF-κB signaling pathway in colorectal cancer cells ultimately inducing macrophage M2 polarization (Xu et al., 2021). By releasing bioactive chemicals, bacteria can impact the host or nearby cells. A potential new marker for colorectal cancer is AI-2 in the gut microbiota (Li et al., 2019). Interestingly, AI-2 of F. nucleatum can promote macrophage M1 polarization via TNFSF9/IL-1β signaling (Wu et al., 2019).
3.1.2. Helicobacter pylori
Helicobacter pylori is a Gram-negative bacterium that when infected can cause chronic gastritis and subsequently increase the risk of developing gastric tumors in infected patients. M2-polarized macrophages identified by the CD163 molecule were substantially expressed in gastric cancer and lowly expressed in marginal tissues, implying that macrophage polarization is intimately related to gastric cancer (Zhu et al., 2020). Macrophages detect the presence of pathogen-associated molecular patterns from H. pylori using PRRs such as TLRs and NLRs. Inflammation caused by H. pylori infection is associated with the expression of TLR4 and TLR9. TLR9 is found in the intracellular compartment and can recognize nucleic acids from bacteria. TLR4 plays a major role in the inflammation of the superficial gastric lining, whereas TLR9 plays a major role in the inflammation of gastric cancer (Wang T. R. et al., 2014). Bacterial peptidoglycan particles are detectable by NOD1. NOD1 collaborates with TLRs to detect bacteria and mediate the production of inflammatory factors. Loss of NOD1 accelerates stomach carcinogenesis in a mouse model. The wild-type phenotype of macrophages rapidly changed from M2 to M1 after the H. pylori infection. While wild-type macrophages convert to a mixed M1-M2 phenotype after infection with H. pylori, NOD1-deficient macrophages exhibit a more pronounced M2 phenotype (Suarez et al., 2019). Another study found that the deletion of MMP7 boosted M1 macrophage polarization and raised the risk of gastric cancer brought on by H. pylori (Krakowiak et al., 2015). Certain miRNAs may play a role in the bacterial infection’s ability to persist. Helicobacter pylori can upregulate miRNAs targeting CIITA, thereby suppressing HLAII expression on macrophages which plays a key role in the presentation of antigens to T lymphocytes (Codolo et al., 2019; Coletta et al., 2021). After an H. pylori infection, macrophages regulate the release of proinflammatory cytokines via the increased expression of miR-155 (Yao et al., 2015).
3.2. Bacteria significantly linked to infection
3.2.1. Propionibacterium acnes
Propionibacterium acnes is a Gram-positive bacterium known as a cutaneous commensal. However, it can also manifest as an opportunistic pathogen, which gives the impression of intrusion. Through TLR4/PI3K/Akt signaling, P. acnes encourages M2 macrophage polarization in gastric cancer (Li et al., 2021). Regardless of the existence of malignancy, P. acnes infection can be found in the macrophages and epithelial cells of the prostate gland. However, persistent inflammation is linked to P. acnes-positive macrophage populations and is most likely a factor in the development of cancer. Both TLR4 and TLR2 are capable of identifying lipids and the LPS that Gram-negative bacteria generate. Interestingly, Kim et al. showed that P. acnes triggered an inflammatory response in macrophages by the activation of TLR2 while the TLR ligand may be the peptidoglycan (Kim et al., 2002). Due to the late discovery of the pathogenicity of P. acnes, little is known about this bacterium.
3.2.2. Staphylococcus aureus
Staphylococcus aureus is the leading causative agent in pneumonia and is initially cleared from the bloodstream by liver macrophages also called Kupffer cells. These infected cells will spread intracellular S. aureus throughout the body if they are unable to kill it, leading to disseminated infection. The virulence regulation of S. aureus is more sophisticated than that of many other bacterial pathogens. When combined with a strong Arg-1 induction, S. aureus biofilms can reduce iNOS expression and drive M2 macrophage polarization (Thurlow et al., 2011). In extramammary Paget S disease, S. aureus may be exacerbated by IL-17 and M2 macrophage polarization (Tuohy et al., 2020; Sakamoto et al., 2021). TGF-β levels were lower and inflammatory cytokines were more prominent in macrophages exposed to S. aureus in co-culture with osteosarcoma (Tuohy et al., 2020). The majority of the time, significant expression of conventional HDAC enzymes are linked to cancer, and it frequently indicates advanced disease and a poor prognosis for the patient. Interestingly, S. aureus-derived lactate inhibits the negative regulator HDAC11 to augment leukocyte IL-10 production in an HDAC6-dependent manner in the mouse prosthetic joint infection model (Heim et al., 2020). IL-10 expression correlated with the expression of HDAC6 and HDAC11 was also reported in M. tuberculosis infection (Wang et al., 2018).
3.3. Engineered bacteria
3.3.1. Bacillus Calmette-Guérin
Natural bacteria have been modified to acquire therapeutic functions as a result of the development of bioengineering technology. Bacillus Calmette-Guérin, a vaccine against tuberculosis, contains live-attenuated and non-toxic M. tuberculosis. The most advanced immunotherapy now available for non-muscle-invasive bladder cancer is BCG (Seow et al., 2010). To prevent the growth of malignancies, BCG instructs monocyte precursor cells to differentiate into functioning mature macrophages (Italiani and Boraschi, 2014). The pathogen BCG activates the MyD88 signaling pathway downstream of the cell surface TLRs, which in turn activates NF-κB and encourages cytokine transcription (de Queiroz et al., 2021). Through the TLR2/TLR4/IRF5 pathway, TRIM59 expression is elevated in BCG-activated macrophages (Jin et al., 2017). TRIM59 is a membrane protein expressed on macrophages that can increase the M1-polarized macrophages inside the tumor (Tian et al., 2019). In addition, BCG inhibits cervical carcinoma progression by promoting M1 macrophage polarization and inhibiting the pro-tumor activation of M2 macrophages via the Rb/E2F1 signaling pathway in Hela cells (Liu et al., 2021).
3.3.2. Salmonella
Salmonella species are facultative intracellular pathogenic bacteria that can invade and proliferate in macrophages and dendritic cells. In Salmonella-infected macrophages, the fatty acid regulator PPARδ is increased and may be linked to M2-polarized macrophages (Eisele et al., 2013) and the PI3K/Akt pathway in gallbladder cancer can facilitate migration and invasion due to CCL18 produced by M2 macrophages (Zhou Z. et al., 2019). While some studies suggest that engineered Salmonella bacteria help tumor-associated macrophage polarization (see Figure 2) to achieve enhanced antitumor immune response in vivo. The anti-tumor effect of engineered Salmonella also appears to induce infiltration of abundant immune cells through TLR4 signaling. Molecular mechanisms suggest that this may be due to the presence of LPS in the outer membrane of Gram-negative bacteria thereby activating the TLR4/MyD88 pathway that mediated CCL2 production (Akhter et al., 2018). Some of the attenuated Salmonella strains and their derivatives used as drug carriers have also been tested in early clinical trials. A newly engineered Salmonella typhimurium strain called YB1 was reported to induce enhanced HLA-DR expression and reduced CD206 expression, and to remodel macrophages from the M2-to M1-polarized (Yang et al., 2018). Additionally, heterologous flagellin from the bacterial pathogen Salmonella activates the TLR5 pathway and changes tumor-infiltrating macrophages into M1-polarized macrophages (Chen et al., 2021). Another engineered bacteria that secrete FlaB via dual pathways, TLR4 and TLR5, also leads to M1-polarized macrophages (Zheng J. H. et al., 2017).
4. Effect of activated macrophages on the tumor microenvironment
While studies targeting the direct relationship between cancer and microbiome are quite limited at present, there have been some interesting studies demonstrating that certain pathogenic processes, such as altered metabolic states and chronic inflammation, display commonality across cancers (Trinchieri, 2012; Andrejeva and Rathmell, 2017). Microbiota participates in shaping an immune-tolerant environment through the recruitment and activation of macrophages and is characterized by the accumulation of pro-inflammatory factors including metabolic intermediates and effectors. These pro-inflammatory factors aid in the development of cancer by promoting angiogenesis, chemoresistance, immune cell suppression, tumor invasion, and metastasis (Coussens and Werb, 2002).
4.1. Dynamic changes in macrophage metabolism
The components of pathogenic organisms, such as LPS are commonly used tools to activate macrophages. LPS stimulation and TLR activation induce a series of biochemical metabolic alterations in macrophages. Metabolomic analysis of LPS-activated macrophages shows downregulation of TCA cycle intermediates and upregulation of aerobic glycolysis, which correlates directly with the expression profiles of altered metabolites (Tannahill et al., 2013; Lauterbach et al., 2019). The TCA cycle is a fragmented process resulting in the secretion of large volumes of metabolites such as lactate and succinate. The production of lactate in LPS-activated macrophages is enhanced and it inhibits the motility of activated T cells in vitro (Haas et al., 2015) and the cytotoxic activity of CD8 + CTLs (Fischer et al., 2007). By activating HIF-1α and MAPK, abundant lactate causes macrophages to produce VEGF and ARG1. Both VEGF and ARG1 promote tumor progressions by inducing angiogenesis and arginase depletion. Succinate is a pro-inflammatory metabolite that inhibits prolyl hydroxylase activity and increases the production of ROS, which stabilizes HIF-1α (Liu et al., 2017). Several genes that promote tumor growth, including MMP9, are also activated by HIF (Zhang et al., 2015). Furthermore, considering that the HIF protein amount is under the control of iron-dependent prolyl hydroxylases (Bruick and McKnight, 2001) and Lcn-2 promotes downstream target gene activation (Bolignano et al., 2010), iron uptake is another mechanism behind the pro-tumorigenic activity of polarized macrophages. Iron is known to regulate the expression of several genes at the transcriptional level, most prominently via the generation of reactive oxygen species and their effects on the activity of NF-κB and other transcription factors (Templeton and Liu, 2003). Iron stimulates cell production of hydroxyl radicals through the overexpression of SOX9, which regulate tumor aggressiveness (Chanvorachote and Luanpitpong, 2016). Under infectious or inflammatory conditions, macrophages increase iron absorption while promoting inflammation (Jung et al., 2015). While macrophages infected with extracellular E. coli K88 reserve iron by elevating hepcidin transcription and increasing iron storage in cells, macrophages infected with intracellular S. typhimurium decrease free iron ions for intracellular bacterial proliferation and utilization (Gan et al., 2019). Reduced intracellular iron levels in macrophages prevent inflammatory cytokines like IL-6 and TNF-α from being translated (Wang et al., 2008). A key mechanism for pathogens to perturb the biochemical metabolism to promote their survival in macrophages is lipid metabolism. Lipid metabolism is a crucial way by which infections disrupt metabolic metabolism to aid in their survival in macrophages. Lipid droplets, which are now acknowledged as a well-established characteristic of many tumors, accumulate excessive amounts of lipids and cholesterol (Beloribi-Djefaflia et al., 2016). By changing the metabolism of host cells, M. tuberculosis encourages the production of macrophages with lipid bodies (Russell et al., 2009). Helicobacter pylori engagement of the intracellular NOD1 leads to the activation of NF-κB, which results in the up-regulation of COX-2 (Chang et al., 2004), and COX-2 plays a key role in the synthesis of lipid inflammatory mediators such as prostaglandins from arachidonic acid. In addition, microbial stimulation triggers the expression of SREBP-1a (Im et al., 2011) and the synthesis of phosphatidylcholine (Sanchez-Lopez et al., 2019), which is linked to the production of IL-1β and IL-18 (Oishi et al., 2017). Increases in dephosphorylation of SHP1 caused by higher levels of oxidative stress from fatty acid oxidation are correlated with tumor progression and involve a variety of immune cell types (Myers et al., 2020; Su et al., 2020).
4.2. Populations and expression of regulatory inflammatory factors in macrophage
In the context of immunity, activated macrophages undergo metabolic adjustments and modify the production of cytokines at the epigenetic, transcriptional, and post-translational levels in response to bacterial sensing. Furthermore, the release of increased concentrations of intermediates and effectors frequently controls the tumor immune microenvironment and aids in the development of tumors. TAMs attract naïve and Th2 lymphocytes and cause inefficient immunological reactions by secreting CCL17, CCL18, and CCL22 (Erreni et al., 2011). Additionally, through producing CCL18, which binds to PITPNM3 on the cancer cell membrane, TAMs in breast cancer increase the invasiveness of cancer cells (Chen et al., 2011). TAMs secrete PD-L1 to inhibit cytotoxic T cells and IL-10 to activate Treg (Zhu et al., 2016; Fang et al., 2021). Additionally, when PD-L1 is inhibited, TAMs may retain tumor immunosuppressive potential by boosting PD-L2 secretion (Umezu et al., 2019). TAMs can directly attract Treg cells to the site of the immunosuppressive milieu by generating CCL20 and CCL22, and they can also activate them by secreting IL-10 and TGF-β (Curiel et al., 2004; Biswas and Mantovani, 2010; Umezu et al., 2019). IL-6 and IL-10 are a group of cytokines, which are inducing tumor invasion and angiogenesis (Tamura et al., 2018). Recent studies showed that TAMs decrease E-cadherin by activating the TLR4/IL-10 signaling pathway promoting epithelial-to-mesenchymal transition in pancreatic cancer (Liu et al., 2013; Yao et al., 2018). Additionally, in transgenic mice models, IL-10 produced by TAMs is the key mediator in tumor resistance to paclitaxel and carboplatin (Ruffell et al., 2014). The IL-6 produced by TAMs promotes chemotherapy resistance in cancer cells by inhibiting the expression of miR-204-5p and activating the STAT3 pathway (Zhu et al., 2017).
4.3. MicroRNA in macrophage exosomes as critical regulators of the tumor microenvironment
Several proteins, including SHIP1, TAB2, and SOCS1, in the innate immune signaling pathways, are targeted by the miR-155 gene, which is increased in macrophages after LPS infection and alters affects the expression of inflammatory mediators (Androulidaki et al., 2009; Ceppi et al., 2009; Cremer et al., 2009). Through a post-transcriptional break, Salmonella can cause the let-7 family to suppress the expression of IL-6 and IL-10 (Schulte et al., 2011). In THP-1 cells infected with M. tuberculosis, miR-206 expression is noticeably elevated, and this raised miR-206 favorably regulates inflammatory cytokines and MMP9 via targeting TIMP3 (Fu et al., 2016). These are shown miRNA could regulate the modulation of innate immunity signaling pathways. The primary mechanism for extracellular miRNA synthesis uses exosomes with energy-dependent active secretion. Exosomes are extremely small extracellular vesicles that contain proteins, lipids and nucleic acids, among other active components (Tan et al., 2020). In pancreatic ductal adenocarcinoma, it has been discovered that TAM-EVs transport miR-501-3p to suppress TGFBR3 expression, activate the TGF-β pathway, and encourage tumor migration and invasion (Yin et al., 2019). The advancement of gastric cancer is aided by M2 macrophage-derived extracellular vesicles through a miR-130b-3p/MLL3/GRHL2 signaling cascade (Zhang et al., 2020b). Exosomal miRNAs may have an impact on the biology of different cell types in TME. The STAT3 pathway is inhibited and Treg/Th17 cell imbalance is produced by TAM-EVs enriched in miR-21-5p and miR-29a-3p (Zhou et al., 2018). MiR-29a-3p controlled the FOXO3/AKT/GSK3 axis to suppress the expression of PD-L1, and PD-L1 expression can impair CD8 + T cell activity, causing immunological escape (Lu et al., 2021). TAM-EVs also contribute significantly to the pathophysiology of tumor chemoresistance (Figure 3). MiR-21 from tumor-associated macrophages that is transferred exosomal provides cisplatin resistance on gastric cancer cells by enhanced activation of PI3K/AKT signaling pathway by down-regulation of PTEN (Zheng P. et al., 2017).
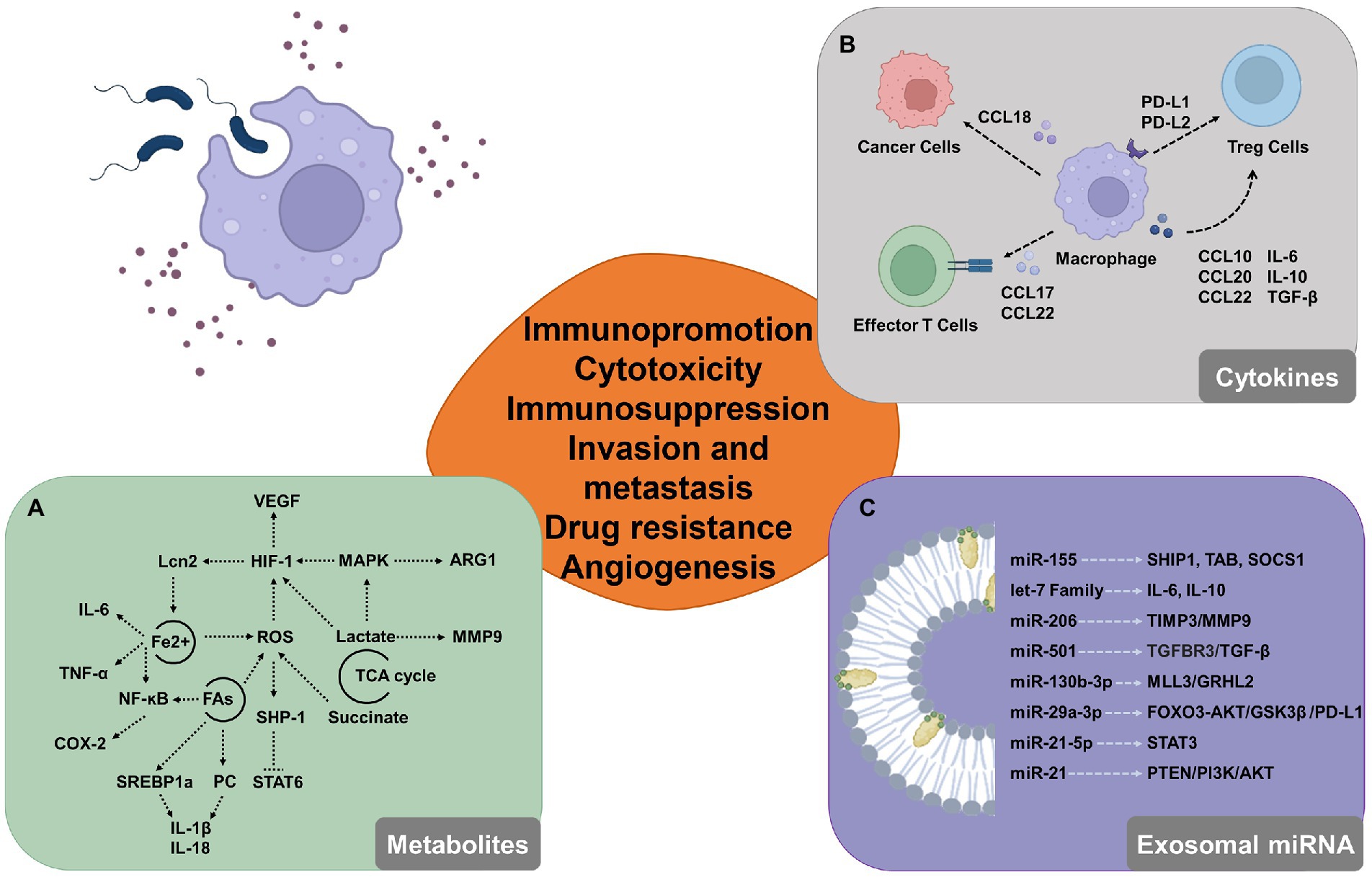
Figure 3. Potential involvement for macrophages that have been triggered by the microbiome in the tumor microenvironment. (A) By modifying the expression of intermediate metabolites, activated macrophages can modify glucose metabolism, iron cycling, and lipid metabolism and control the tumor microenvironment. (B) Activated macrophages release a variety of cytokines, chemokines, and growth factors that influence tumor formation by boosting cell proliferation and decreasing the activity of immune cells that can destroy tumors, like cytotoxic T cells. (C) Macrophage-derived exosomal miRNAs alter the immunological milieu by targeting proteins and activating molecules.
5. Discussion
Studies on the microbiome have progressed from focusing on the cultivation of oral and intestinal bacteria to mechanistically understanding the link between host and microbiome, and more recently, to microbial profiles of all ecological niches in the body. The use of specific microbial signatures of cancer types may improve early and minimally invasive diagnostic approaches, and in this review, we discuss the bacteria in different cancer. However, the part of cancer most clearly associated with bacterial species remains ill-defined. The speculation of the presence of microbiota within tumors was finally confirmed by sequencing-based diagnosis, but there are many challenges in discriminating tumor-specific bacteria. The most frequent issues are biased results brought on by the use of various tissue sample processing techniques and the confounding of chemicals or environmental pollutants. Exogenous bacteria can now be used as potential immunotherapeutic agents or as a neoadjuvant in the treatment of cancer (St Jean et al., 2008). We will be able to identify distinct pathways that can be exploited for diagnostic, preventative, and therapeutic purposes if we can more precisely identify a particular strain in the tumor.
The host’s metabolism and immunity can be altered by the microbiota and its secreted components, which in turn can affect antitumor immunity. Numerous pathways suggest that bacteria acting as foreign microorganisms may indirectly contribute to the beginning or development of cancer. Unexpectedly, bacteria may multiply in macrophages and control them via a variety of interference tactics (Rosenberger and Finlay, 2003). During tumorigenesis and regression, macrophage polarization appears to act as an intermediate process that is activated by certain signals. Macrophages exhibit different phenotypes after receiving multiple stimulations which act on different receptors and thus exert regulatory effects by acting on multiple signaling pathways. It is a complicated story about how bacteria and macrophages interact. It should be emphasized that host cells are frequently cultivated in vitro for the majority of research against pathogens, including bacteria. Macrophage polarization is a result, and the molecular pathways of bacteria-driven macrophage polarization ought to connect to a particular environment concurrently, as the tumor microenvironment is a special environment that has the potential to preferentially generate macrophage polarization. In this article, we went over the molecular mechanisms of bacterial-induced macrophage polarization as well as the impact of activated macrophages on the tumor microenvironment.
A causal link between microbial enrichment in cancer and cancer itself cannot be shown, but understanding the relationships between microbes, macrophages, and cancer cells requires in-depth functional analysis per microbial scale is necessary. On one hand, one way to increase the efficiency and safety of tumor-targeted medicines is to modify bacteria using research on such pathogen-macrophage interactions. On the other hand, molecular mechanisms of cell polarization provide information and guidance for switching macrophage polarity in cancer.
Author contributions
SX, YX, BF, DG, ZS, XL, and HW contributed to the study’s conception and design and commented on previous versions of the manuscript. Material preparation, data collection, and analysis were performed by BF, DG, and ZS. The first draft of the manuscript was written by SX and YX. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by National Natural Science Foundation of China (No. 81970008 and 82000020), Fundamental Research Funds for the Central Universities (2019CDYGZD009 and 2020CDJYGRH-1005), Natural Science Foundation of Chongqing, China (cstc2020jcyj-bshX0105 and cstc2020jcyj-msxmX0460) and Chongqing Talents: Exceptional Young Talents Project (No. cstc2021ycjh-bgzxm0099). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1115556/full#supplementary-material
References
Abed, J., Maalouf, N., Manson, A. L., Earl, A. M., Parhi, L., Emgard, J. E. M., et al. (2020). Colon cancer-associated fusobacterium nucleatum may originate from the Oral cavity and reach colon tumors via the circulatory system. Front. Cell. Infect. Microbiol. 10:400. doi: 10.3389/fcimb.2020.00400
Abnave, P., Muracciole, X., and Ghigo, E. (2017). Coxiella burnetii lipopolysaccharide: what do we know? Int. J. Mol. Sci. 18:2509. doi: 10.3390/ijms18122509
Akhter, N., Hasan, A., Shenouda, S., Wilson, A., Kochumon, S., Ali, S., et al. (2018). TLR4/MyD88 -mediated CCL2 production by lipopolysaccharide (endotoxin): implications for metabolic inflammation. J. Diabetes Metab. Disord. 17, 77–84. doi: 10.1007/s40200-018-0341-y
Alanee, S., El-Zawahry, A., Dynda, D., Dabaja, A., McVary, K., Karr, M., et al. (2019). A prospective study to examine the association of the urinary and fecal microbiota with prostate cancer diagnosis after transrectal biopsy of the prostate using 16sRNA gene analysis. Prostate 79, 81–87. doi: 10.1002/pros.23713
Andrejeva, G., and Rathmell, J. C. (2017). Similarities and distinctions of cancer and immune metabolism in inflammation and tumors. Cell Metab. 26, 49–70. doi: 10.1016/j.cmet.2017.06.004
Androulidaki, A., Iliopoulos, D., Arranz, A., Doxaki, C., Schworer, S., Zacharioudaki, V., et al. (2009). The kinase Akt1 controls macrophage response to lipopolysaccharide by regulating microRNAs. Immunity 31, 220–231. doi: 10.1016/j.immuni.2009.06.024
Apostolou, P., Tsantsaridou, A., Papasotiriou, I., Toloudi, M., Chatziioannou, M., and Giamouzis, G. (2011). Bacterial and fungal microflora in surgically removed lung cancer samples. J. Cardiothorac. Surg. 6:137. doi: 10.1186/1749-8090-6-137
Arthur, J. C., Perez-Chanona, E., Muhlbauer, M., Tomkovich, S., Uronis, J. M., Fan, T. J., et al. (2012). Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 338, 120–123. doi: 10.1126/science.1224820
Audirac-Chalifour, A., Torres-Poveda, K., Bahena-Roman, M., Tellez-Sosa, J., Martinez-Barnetche, J., Cortina-Ceballos, B., et al. (2016). Cervical microbiome and cytokine profile at various stages of cervical cancer: a pilot study. PLoS One 11:e0153274. doi: 10.1371/journal.pone.0153274
Aviles-Jimenez, F., Vazquez-Jimenez, F., Medrano-Guzman, R., Mantilla, A., and Torres, J. (2014). Stomach microbiota composition varies between patients with non-atrophic gastritis and patients with intestinal type of gastric cancer. Sci. Rep. 4:4202. doi: 10.1038/srep04202
Baghban, A., and Gupta, S. (2016). Parvimonas micra: a rare cause of native joint septic arthritis. Anaerobe 39, 26–27. doi: 10.1016/j.anaerobe.2016.02.004
Beloribi-Djefaflia, S., Vasseur, S., and Guillaumond, F. (2016). Lipid metabolic reprogramming in cancer cells. Oncogenesis. 5:e189. doi: 10.1038/oncsis.2015.49
Bi, Y., Wang, X., Han, Y., Guo, Z., and Yang, R. (2012). Yersinia pestis versus Yersinia pseudotuberculosis: effects on host macrophages. Scand. J. Immunol. 76, 541–551. doi: 10.1111/j.1365-3083.2012.02767.x
Biswas, A., Banerjee, P., Mukherjee, G., and Biswas, T. (2007). Porin of Shigella dysenteriae activates mouse peritoneal macrophage through toll-like receptors 2 and 6 to induce polarized type I response. Mol. Immunol. 44, 812–820. doi: 10.1016/j.molimm.2006.04.007
Biswas, S. K., and Mantovani, A. (2010). Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Nat. Immunol. 11, 889–896. doi: 10.1038/ni.1937
Bolignano, D., Donato, V., Lacquaniti, A., Fazio, M. R., Bono, C., Coppolino, G., et al. (2010). Neutrophil gelatinase-associated lipocalin (NGAL) in human neoplasias: a new protein enters the scene. Cancer Lett. 288, 10–16. doi: 10.1016/j.canlet.2009.05.027
Bost, K. L., and Clements, J. D. (1997). Intracellular salmonella Dublin induces substantial secretion of the 40-kilodalton subunit of interleukin-12 (IL-12) but minimal secretion of IL-12 as a 70-kilodalton protein in murine macrophages. Infect. Immun. 65, 3186–3192. doi: 10.1128/iai.65.8.3186-3192.1997
Bruick, R. K., and McKnight, S. L. (2001). A conserved family of prolyl-4-hydroxylases that modify HIF. Science 294, 1337–1340. doi: 10.1126/science.1066373
Bullman, S., Pedamallu, C. S., Sicinska, E., Clancy, T. E., Zhang, X., Cai, D., et al. (2017). Analysis of fusobacterium persistence and antibiotic response in colorectal cancer. Science 358, 1443–1448. doi: 10.1126/science.aal5240
Cameron, S. J. S., Lewis, K. E., Huws, S. A., Hegarty, M. J., Lewis, P. D., Pachebat, J. A., et al. (2017). A pilot study using metagenomic sequencing of the sputum microbiome suggests potential bacterial biomarkers for lung cancer. PLoS One 12:e0177062. doi: 10.1371/journal.pone.0177062
Cassetta, L., and Pollard, J. W. (2020). Tumor-associated macrophages. Curr. Biol. 30, R246–R248. doi: 10.1016/j.cub.2020.01.031
Castellarin, M., Warren, R. L., Freeman, J. D., Dreolini, L., Krzywinski, M., Strauss, J., et al. (2012). Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 22, 299–306. doi: 10.1101/gr.126516.111
Cavarretta, I., Ferrarese, R., Cazzaniga, W., Saita, D., Luciano, R., Ceresola, E. R., et al. (2017). The microbiome of the prostate tumor microenvironment. Eur. Urol. 72, 625–631. doi: 10.1016/j.eururo.2017.03.029
Ceppi, M., Pereira, P. M., Dunand-Sauthier, I., Barras, E., Reith, W., Santos, M. A., et al. (2009). MicroRNA-155 modulates the interleukin-1 signaling pathway in activated human monocyte-derived dendritic cells. Proc. Natl. Acad. Sci. U. S. A. 106, 2735–2740. doi: 10.1073/pnas.0811073106
Chacon-Salinas, R., Serafin-Lopez, J., Ramos-Payan, R., Mendez-Aragon, P., Hernandez-Pando, R., Van Soolingen, D., et al. (2005). Differential pattern of cytokine expression by macrophages infected in vitro with different mycobacterium tuberculosis genotypes. Clin. Exp. Immunol. 140, 443–449. doi: 10.1111/j.1365-2249.2005.02797.x
Chang, Y. J., Wu, M. S., Lin, J. T., Sheu, B. S., Muta, T., Inoue, H., et al. (2004). Induction of cyclooxygenase-2 overexpression in human gastric epithelial cells by helicobacter pylori involves TLR2/TLR9 and c-Src-dependent nuclear factor-kappaB activation. Mol. Pharmacol. 66, 1465–1477. doi: 10.1124/mol.104.005199
Chanvorachote, P., and Luanpitpong, S. (2016). Iron induces cancer stem cells and aggressive phenotypes in human lung cancer cells. Am. J. Physiol. Cell Physiol. 310, C728–C739. doi: 10.1152/ajpcell.00322.2015
Chen, T., Li, Q., Wu, J., Wu, Y., Peng, W., Li, H., et al. (2018). Fusobacterium nucleatum promotes M2 polarization of macrophages in the microenvironment of colorectal tumours via a TLR4-dependent mechanism. Cancer Immunol. Immunother. 67, 1635–1646. doi: 10.1007/s00262-018-2233-x
Chen, J., Qiao, Y., Chen, G., Chang, C., Dong, H., Tang, B., et al. (2021). Salmonella flagella confer anti-tumor immunological effect via activating Flagellin/TLR5 signalling within tumor microenvironment. Acta Pharm. Sin. B 11, 3165–3177. doi: 10.1016/j.apsb.2021.04.019
Chen, J., Yao, Y., Gong, C., Yu, F., Su, S., Chen, J., et al. (2011). CCL18 from tumor-associated macrophages promotes breast cancer metastasis via PITPNM3. Cancer Cell 19, 541–555. doi: 10.1016/j.ccr.2011.02.006
Christoffersen, T. E., Hult, L. T., Kuczkowska, K., Moe, K. M., Skeie, S., Lea, T., et al. (2014). In vitro comparison of the effects of probiotic, commensal and pathogenic strains on macrophage polarization. Probiotics Antimicrob Proteins. 6, 1–10. doi: 10.1007/s12602-013-9152-0
Christofides, A., Strauss, L., Yeo, A., Cao, C., Charest, A., and Boussiotis, V. A. (2022). The complex role of tumor-infiltrating macrophages. Nat. Immunol. 23, 1148–1156. doi: 10.1038/s41590-022-01267-2
Codolo, G., Toffoletto, M., Chemello, F., Coletta, S., Soler Teixidor, G., Battaggia, G., et al. (2019). Helicobacter pylori dampens HLA-II expression on macrophages via the up-regulation of miRNAs targeting CIITA. Front. Immunol. 10:2923. doi: 10.3389/fimmu.2019.02923
Cohen, R. J., Shannon, B. A., McNeal, J. E., Shannon, T., and Garrett, K. L. (2005). Propionibacterium acnes associated with inflammation in radical prostatectomy specimens: a possible link to cancer evolution? J. Urol. 173, 1969–1974. doi: 10.1097/01.ju.0000158161.15277.78
Coker, O. O., Dai, Z., Nie, Y., Zhao, G., Cao, L., Nakatsu, G., et al. (2018). Mucosal microbiome dysbiosis in gastric carcinogenesis. Gut 67, 1024–1032. doi: 10.1136/gutjnl-2017-314281
Coletta, S., Battaggia, G., Della Bella, C., Furlani, M., Hauke, M., Faass, L., et al. (2021). ADP-heptose enables helicobacter pylori to exploit macrophages as a survival niche by suppressing antigen-presenting HLA-II expression. FEBS Lett. 595, 2160–2168. doi: 10.1002/1873-3468.14156
Contreras, A., Doan, N., Chen, C., Rusitanonta, T., Flynn, M. J., and Slots, J. (2000). Importance of Dialister pneumosintes in human periodontitis. Oral Microbiol. Immunol. 15, 269–272. doi: 10.1034/j.1399-302x.2000.150410.x
Coussens, L. M., and Werb, Z. (2002). Inflammation and cancer. Nature 420, 860–867. doi: 10.1038/nature01322
Coutzac, C., Jouniaux, J. M., Paci, A., Schmidt, J., Mallardo, D., Seck, A., et al. (2020). Systemic short chain fatty acids limit antitumor effect of CTLA-4 blockade in hosts with cancer. Nat. Commun. 11:2168. doi: 10.1038/s41467-020-16079-x
Cremer, T. J., Ravneberg, D. H., Clay, C. D., Piper-Hunter, M. G., Marsh, C. B., Elton, T. S., et al. (2009). MiR-155 induction by F. novicida but not the virulent F. tularensis results in SHIP down-regulation and enhanced pro-inflammatory cytokine response. PLoS One 4:e8508. doi: 10.1371/journal.pone.0008508
Croft, A. J., Metcalfe, S., Honma, K., and Kay, J. G. (2018). Macrophage polarization alters Postphagocytosis survivability of the commensal Streptococcus gordonii. Infect. Immun. 86:e00858-17. doi: 10.1128/IAI.00858-17
Curiel, T. J., Coukos, G., Zou, L., Alvarez, X., Cheng, P., Mottram, P., et al. (2004). Specific recruitment of regulatory T cells in ovarian carcinoma fosters immune privilege and predicts reduced survival. Nat. Med. 10, 942–949. doi: 10.1038/nm1093
Daisley, B. A., Chanyi, R. M., Abdur-Rashid, K., Al, K. F., Gibbons, S., Chmiel, J. A., et al. (2020). Abiraterone acetate preferentially enriches for the gut commensal Akkermansia muciniphila in castrate-resistant prostate cancer patients. Nat. Commun. 11:4822. doi: 10.1038/s41467-020-18649-5
de Martel, C., Plummer, M., Parsonnet, J., van Doorn, L. J., and Franceschi, S. (2009). Helicobacter species in cancers of the gallbladder and extrahepatic biliary tract. Br. J. Cancer 100, 194–199. doi: 10.1038/sj.bjc.6604780
de Queiroz, N., Marinho, F. V., de Araujo, A., Fahel, J. S., and Oliveira, S. C. (2021). MyD88-dependent BCG immunotherapy reduces tumor and regulates tumor microenvironment in bladder cancer murine model. Sci. Rep. 11:15648. doi: 10.1038/s41598-021-95157-6
Dejea, C. M., Fathi, P., Craig, J. M., Boleij, A., Taddese, R., Geis, A. L., et al. (2018). Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science 359, 592–597. doi: 10.1126/science.aah3648
Derosa, L., Hellmann, M. D., Spaziano, M., Halpenny, D., Fidelle, M., Rizvi, H., et al. (2018). Negative association of antibiotics on clinical activity of immune checkpoint inhibitors in patients with advanced renal cell and non-small-cell lung cancer. Ann. Oncol. 29, 1437–1444. doi: 10.1093/annonc/mdy103
Desnues, B., Raoult, D., and Mege, J. L. (2005). IL-16 is critical for Tropheryma whipplei replication in Whipple's disease. J. Immunol. 175, 4575–4582. doi: 10.4049/jimmunol.175.7.4575
Di Paola, M., Sani, C., Clemente, A. M., Iossa, A., Perissi, E., Castronovo, G., et al. (2017). Characterization of cervico-vaginal microbiota in women developing persistent high-risk human papillomavirus infection. Sci. Rep. 7:10200. doi: 10.1038/s41598-017-09842-6
Dias-Jacome, E., Libanio, D., Borges-Canha, M., Galaghar, A., and Pimentel-Nunes, P. (2016). Gastric microbiota and carcinogenesis: the role of non-helicobacter pylori bacteria – a systematic review. Rev. Esp. Enferm. Dig. 108, 530–540. doi: 10.17235/reed.2016.4261/2016
Dickson, R. P., Erb-Downward, J. R., Martinez, F. J., and Huffnagle, G. B. (2016). The microbiome and the respiratory tract. Annu. Rev. Physiol. 78, 481–504. doi: 10.1146/annurev-physiol-021115-105238
Dickson, R. P., and Huffnagle, G. B. (2015). The lung microbiome: new principles for respiratory bacteriology in health and disease. PLoS Pathog. 11:e1004923. doi: 10.1371/journal.ppat.1004923
Donders, G. G. G., Bellen, G., Grinceviciene, S., Ruban, K., and Vieira-Baptista, P. (2017). Aerobic vaginitis: no longer a stranger. Res. Microbiol. 168, 845–858. doi: 10.1016/j.resmic.2017.04.004
Dornand, J., Gross, A., Lafont, V., Liautard, J., Oliaro, J., and Liautard, J. P. (2002). The innate immune response against Brucella in humans. Vet. Microbiol. 90, 383–394. doi: 10.1016/S0378-1135(02)00223-7
Duan, B., Shao, L., Liu, R., Msuthwana, P., Hu, J., and Wang, C. (2021). Lactobacillus rhamnosus GG defense against salmonella enterica serovar typhimurium infection through modulation of M1 macrophage polarization. Microb. Pathog. 156:104939. doi: 10.1016/j.micpath.2021.104939
Duluc, D., Delneste, Y., Tan, F., Moles, M. P., Grimaud, L., Lenoir, J., et al. (2007). Tumor-associated leukemia inhibitory factor and IL-6 skew monocyte differentiation into tumor-associated macrophage-like cells. Blood 110, 4319–4330. doi: 10.1182/blood-2007-02-072587
Dutta, U., Garg, P. K., Kumar, R., and Tandon, R. K. (2000). Typhoid carriers among patients with gallstones are at increased risk for carcinoma of the gallbladder. Am. J. Gastroenterol. 95, 784–787. doi: 10.1111/j.1572-0241.2000.01860.x
Ehrt, S., Schnappinger, D., Bekiranov, S., Drenkow, J., Shi, S., Gingeras, T. R., et al. (2001). Reprogramming of the macrophage transcriptome in response to interferon-gamma and mycobacterium tuberculosis: signaling roles of nitric oxide synthase-2 and phagocyte oxidase. J. Exp. Med. 194, 1123–1140. doi: 10.1084/jem.194.8.1123
Eisele, N. A., Ruby, T., Jacobson, A., Manzanillo, P. S., Cox, J. S., Lam, L., et al. (2013). Salmonella require the fatty acid regulator PPARdelta for the establishment of a metabolic environment essential for long-term persistence. Cell Host Microbe 14, 171–182. doi: 10.1016/j.chom.2013.07.010
Eklof, V., Lofgren-Burstrom, A., Zingmark, C., Edin, S., Larsson, P., Karling, P., et al. (2017). Cancer-associated fecal microbial markers in colorectal cancer detection. Int. J. Cancer 141, 2528–2536. doi: 10.1002/ijc.31011
Elliott, D. R. F., Walker, A. W., O'Donovan, M., Parkhill, J., and Fitzgerald, R. C. (2017). A non-endoscopic device to sample the oesophageal microbiota: a case-control study. Lancet Gastroenterol. Hepatol. 2, 32–42. doi: 10.1016/S2468-1253(16)30086-3
Erreni, M., Mantovani, A., and Allavena, P. (2011). Tumor-associated macrophages (TAM) and inflammation in colorectal cancer. Cancer Microenviron. 4, 141–154. doi: 10.1007/s12307-010-0052-5
Fallows, D., Peixoto, B., Kaplan, G., and Manca, C. (2016). Mycobacterium leprae alters classical activation of human monocytes in vitro. J. Inflamm (Lond). 13:8. doi: 10.1186/s12950-016-0117-4
Fang, W., Zhou, T., Shi, H., Yao, M., Zhang, D., Qian, H., et al. (2021). Progranulin induces immune escape in breast cancer via up-regulating PD-L1 expression on tumor-associated macrophages (TAMs) and promoting CD8(+) T cell exclusion. J. Exp. Clin. Cancer Res. 40:4. doi: 10.1186/s13046-020-01786-6
Farrell, J. J., Zhang, L., Zhou, H., Chia, D., Elashoff, D., Akin, D., et al. (2012). Variations of oral microbiota are associated with pancreatic diseases including pancreatic cancer. Gut 61, 582–588. doi: 10.1136/gutjnl-2011-300784
Fernandes, D. M., Jiang, X., Jung, J. H., and Baldwin, C. L. (1996). Comparison of T cell cytokines in resistant and susceptible mice infected with virulent Brucella abortus strain 2308. FEMS Immunol. Med. Microbiol. 16, 193–203. doi: 10.1111/j.1574-695X.1996.tb00136.x
Fischer, K., Hoffmann, P., Voelkl, S., Meidenbauer, N., Ammer, J., Edinger, M., et al. (2007). Inhibitory effect of tumor cell-derived lactic acid on human T cells. Blood 109, 3812–3819. doi: 10.1182/blood-2006-07-035972
Fredricks, D. N., Fiedler, T. L., and Marrazzo, J. M. (2005). Molecular identification of bacteria associated with bacterial vaginosis. N. Engl. J. Med. 353, 1899–1911. doi: 10.1056/NEJMoa043802
Fu, A., Mo, Q., Wu, Y., Wang, B., Liu, R., Tang, L., et al. (2019). Protective effect of bacillus amyloliquefaciens against salmonella via polarizing macrophages to M1 phenotype directly and to M2 depended on microbiota. Food Funct. 10, 7653–7666. doi: 10.1039/C9FO01651A
Fu, X., Zeng, L., Liu, Z., Ke, X., Lei, L., and Li, G. (2016). MicroRNA-206 regulates the secretion of inflammatory cytokines and MMP9 expression by targeting TIMP3 in mycobacterium tuberculosis-infected THP-1 human macrophages. Biochem. Biophys. Res. Commun. 477, 167–173. doi: 10.1016/j.bbrc.2016.06.038
Gan, Z., Tang, X., Wang, Z., Li, J., Wang, Z., and Du, H. (2019). Regulation of macrophage iron homeostasis is associated with the localization of bacteria. Metallomics 11, 454–461. doi: 10.1039/C8MT00301G
Garrett, W. S. (2019). The gut microbiota and colon cancer. Science 364, 1133–1135. doi: 10.1126/science.aaw2367
Geller, L. T., Barzily-Rokni, M., Danino, T., Jonas, O. H., Shental, N., Nejman, D., et al. (2017). Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science 357, 1156–1160. doi: 10.1126/science.aah5043
Glowacka, P., Zakowska, D., Naylor, K., Niemcewicz, M., and Bielawska-Drozd, A. (2018). Brucella – virulence factors, pathogenesis and treatment. Pol. J. Microbiol. 67, 151–161. doi: 10.21307/pjm-2018-029
Goldmann, O., von Kockritz-Blickwede, M., Holtje, C., Chhatwal, G. S., Geffers, R., and Medina, E. (2007). Transcriptome analysis of murine macrophages in response to infection with streptococcus pyogenes reveals an unusual activation program. Infect. Immun. 75, 4148–4157. doi: 10.1128/IAI.00181-07
Golombos, D. M., Ayangbesan, A., O'Malley, P., Lewicki, P., Barlow, L., Barbieri, C. E., et al. (2018). The role of gut microbiome in the pathogenesis of prostate cancer: a prospective. Pilot Study. Urology. 111, 122–128. doi: 10.1016/j.urology.2017.08.039
Gomes, S., Cavadas, B., Ferreira, J. C., Marques, P. I., Monteiro, C., Sucena, M., et al. (2019). Profiling of lung microbiota discloses differences in adenocarcinoma and squamous cell carcinoma. Sci. Rep. 9:12838. doi: 10.1038/s41598-019-49195-w
Gondwe, T., Ness, R., Totten, P. A., Astete, S., Tang, G., Gold, M. A., et al. (2020). Novel bacterial vaginosis-associated organisms mediate the relationship between vaginal douching and pelvic inflammatory disease. Sex. Transm. Infect. 96, 439–444. doi: 10.1136/sextrans-2019-054191
Greathouse, K. L., White, J. R., Vargas, A. J., Bliskovsky, V. V., Beck, J. A., von Muhlinen, N., et al. (2018). Interaction between the microbiome and TP53 in human lung cancer. Genome Biol. 19:123. doi: 10.1186/s13059-018-1501-6
Haas, R., Smith, J., Rocher-Ros, V., Nadkarni, S., Montero-Melendez, T., D'Acquisto, F., et al. (2015). Lactate regulates metabolic and pro-inflammatory circuits in control of T cell migration and effector functions. PLoS Biol. 13:e1002202. doi: 10.1371/journal.pbio.1002202
Hashemi Goradel, N., Heidarzadeh, S., Jahangiri, S., Farhood, B., Mortezaee, K., Khanlarkhani, N., et al. (2019). Fusobacterium nucleatum and colorectal cancer: a mechanistic overview. J. Cell. Physiol. 234, 2337–2344. doi: 10.1002/jcp.27250
He, Z., Gharaibeh, R. Z., Newsome, R. C., Pope, J. L., Dougherty, M. W., Tomkovich, S., et al. (2019). Campylobacter jejuni promotes colorectal tumorigenesis through the action of the cytolethal distending toxin. Gut 68, 289–300. doi: 10.1136/gutjnl-2018-317200
Heim, C. E., Bosch, M. E., Yamada, K. J., Aldrich, A. L., Chaudhari, S. S., Klinkebiel, D., et al. (2020). Lactate production by Staphylococcus aureus biofilm inhibits HDAC11 to reprogramme the host immune response during persistent infection. Nat. Microbiol. 5, 1271–1284. doi: 10.1038/s41564-020-0756-3
Hosgood, H. D. 3rd, Sapkota, A. R., Rothman, N., Rohan, T., Hu, W., Xu, J., et al. (2014). The potential role of lung microbiota in lung cancer attributed to household coal burning exposures. Environ. Mol. Mutagen. 55, 643–651. doi: 10.1002/em.21878
Hsieh, Y. Y., Tung, S. Y., Pan, H. Y., Yen, C. W., Xu, H. W., Lin, Y. J., et al. (2018). Increased abundance of clostridium and fusobacterium in gastric microbiota of patients with gastric cancer in Taiwan. Sci. Rep. 8:158. doi: 10.1038/s41598-017-18596-0
Hu, L., Liu, Y., Kong, X., Wu, R., Peng, Q., Zhang, Y., et al. (2021). Fusobacterium nucleatum facilitates M2 macrophage polarization and colorectal carcinoma progression by activating TLR4/NF-kappaB/S100A9 Cascade. Front. Immunol. 12:658681. doi: 10.3389/fimmu.2021.658681
Huang, Z., Luo, Q., Guo, Y., Chen, J., Xiong, G., Peng, Y., et al. (2015). Mycobacterium tuberculosis-induced polarization of human macrophage orchestrates the formation and development of tuberculous granulomas in vitro. PLoS One 10:e0129744. doi: 10.1371/journal.pone.0129744
Im, S. S., Yousef, L., Blaschitz, C., Liu, J. Z., Edwards, R. A., Young, S. G., et al. (2011). Linking lipid metabolism to the innate immune response in macrophages through sterol regulatory element binding protein-1a. Cell Metab. 13, 540–549. doi: 10.1016/j.cmet.2011.04.001
Italiani, P., and Boraschi, D. (2014). From monocytes to M1/M2 macrophages: phenotypical vs functional differentiation. Front. Immunol. 5:514. doi: 10.3389/fimmu.2014.00514
Jin, C., Lagoudas, G. K., Zhao, C., Bullman, S., Bhutkar, A., Hu, B., et al. (2019). Commensal microbiota promote lung cancer development via gammadelta T cells. Cells 176, 998–1013.e16. doi: 10.1016/j.cell.2018.12.040
Jin, Z., Tian, Y., Yan, D., Li, D., and Zhu, X. (2017). BCG increased membrane expression of TRIM59 through the TLR2/TLR4/IRF5 pathway in RAW264.7 macrophages. Protein Pept. Lett. 24, 765–770. doi: 10.2174/0929866524666170818155524
Jung, M., Mertens, C., and Brune, B. (2015). Macrophage iron homeostasis and polarization in the context of cancer. Immunobiology 220, 295–304. doi: 10.1016/j.imbio.2014.09.011
Kadioglu, A., and Andrew, P. W. (2004). The innate immune response to pneumococcal lung infection: the untold story. Trends Immunol. 25, 143–149. doi: 10.1016/j.it.2003.12.006
Kaufmann, S. H. (2016). Immunopathology of mycobacterial diseases. Semin. Immunopathol. 38, 135–138. doi: 10.1007/s00281-015-0547-8
Keay, S., Zhang, C. O., Baldwin, B. R., and Alexander, R. B. (1999). Polymerase chain reaction amplification of bacterial 16s rRNA genes in prostate biopsies from men without chronic prostatitis. Urology 53, 487–491. doi: 10.1016/S0090-4295(98)00553-6
Khan, J., Sharma, P. K., and Mukhopadhaya, A. (2015). Vibrio cholerae porin OmpU mediates M1-polarization of macrophages/monocytes via TLR1/TLR2 activation. Immunobiology 220, 1199–1209. doi: 10.1016/j.imbio.2015.06.009
Kim, J., Ochoa, M. T., Krutzik, S. R., Takeuchi, O., Uematsu, S., Legaspi, A. J., et al. (2002). Activation of toll-like receptor 2 in acne triggers inflammatory cytokine responses. J. Immunol. 169, 1535–1541. doi: 10.4049/jimmunol.169.3.1535
Kim, K. S., Rowlinson, M. C., Bennion, R., Liu, C., Talan, D., Summanen, P., et al. (2010). Characterization of Slackia exigua isolated from human wound infections, including abscesses of intestinal origin. J. Clin. Microbiol. 48, 1070–1075. doi: 10.1128/JCM.01576-09
Kiszewski, A. E., Becerril, E., Aguilar, L. D., Kader, I. T., Myers, W., Portaels, F., et al. (2006). The local immune response in ulcerative lesions of Buruli disease. Clin. Exp. Immunol. 143, 445–451. doi: 10.1111/j.1365-2249.2006.03020.x
Kostic, A. D., Chun, E., Robertson, L., Glickman, J. N., Gallini, C. A., Michaud, M., et al. (2013). Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 14, 207–215. doi: 10.1016/j.chom.2013.07.007
Kostic, A. D., Gevers, D., Pedamallu, C. S., Michaud, M., Duke, F., Earl, A. M., et al. (2012). Genomic analysis identifies association of fusobacterium with colorectal carcinoma. Genome Res. 22, 292–298. doi: 10.1101/gr.126573.111
Krakowiak, M. S., Noto, J. M., Piazuelo, M. B., Hardbower, D. M., Romero-Gallo, J., Delgado, A., et al. (2015). Matrix metalloproteinase 7 restrains helicobacter pylori-induced gastric inflammation and premalignant lesions in the stomach by altering macrophage polarization. Oncogene 34, 1865–1871. doi: 10.1038/onc.2014.135
Kusaka, Y., Kajiwara, C., Shimada, S., Ishii, Y., Miyazaki, Y., Inase, N., et al. (2018). Potential role of gr-1+ CD8+ T lymphocytes as a source of interferon-gamma and M1/M2 polarization during the acute phase of murine legionella pneumophila pneumonia. J. Innate Immun. 10, 328–338. doi: 10.1159/000490585
Kwong, T. N. Y., Wang, X., Nakatsu, G., Chow, T. C., Tipoe, T., Dai, R. Z. W., et al. (2018). Association between bacteremia from specific microbes and subsequent diagnosis of colorectal cancer. Gastroenterology 155, 383–390.e8. doi: 10.1053/j.gastro.2018.04.028
Laniewski, P., Barnes, D., Goulder, A., Cui, H., Roe, D. J., Chase, D. M., et al. (2018). Linking cervicovaginal immune signatures, HPV and microbiota composition in cervical carcinogenesis in non-Hispanic and Hispanic women. Sci. Rep. 8:7593. doi: 10.1038/s41598-018-25879-7
Lara-Tejero, M., and Galan, J. E. (2000). A bacterial toxin that controls cell cycle progression as a deoxyribonuclease I-like protein. Science 290, 354–357. doi: 10.1126/science.290.5490.354
Laroumagne, S., Lepage, B., Hermant, C., Plat, G., Phelippeau, M., Bigay-Game, L., et al. (2013). Bronchial colonisation in patients with lung cancer: a prospective study. Eur. Respir. J. 42, 220–229. doi: 10.1183/09031936.00062212
Lauterbach, M. A., Hanke, J. E., Serefidou, M., Mangan, M. S. J., Kolbe, C. C., Hess, T., et al. (2019). Toll-like receptor signaling rewires macrophage metabolism and promotes histone acetylation via ATP-citrate Lyase. Immunity 51, 997–1011.e7. doi: 10.1016/j.immuni.2019.11.009
Laviron, M., and Boissonnas, A. (2019). Ontogeny of tumor-associated macrophages. Front Immunol. 10:1799. doi: 10.3389/fimmu.2019.01799
Lax, A. J., and Thomas, W. (2002). How bacteria could cause cancer: one step at a time. Trends Microbiol. 10, 293–299. doi: 10.1016/S0966-842X(02)02360-0
Le Noci, V., Guglielmetti, S., Arioli, S., Camisaschi, C., Bianchi, F., Sommariva, M., et al. (2018). Modulation of pulmonary microbiota by antibiotic or probiotic aerosol therapy: a strategy to promote immunosurveillance against lung metastases. Cell Rep. 24, 3528–3538. doi: 10.1016/j.celrep.2018.08.090
Lee, J. E., Lee, S., Lee, H., Song, Y. M., Lee, K., Han, M. J., et al. (2013). Association of the vaginal microbiota with human papillomavirus infection in a Korean twin cohort. PLoS One 8:e63514. doi: 10.1371/journal.pone.0063514
Lee, S. H., Sung, J. Y., Yong, D., Chun, J., Kim, S. Y., Song, J. H., et al. (2016). Characterization of microbiome in bronchoalveolar lavage fluid of patients with lung cancer comparing with benign mass like lesions. Lung Cancer 102, 89–95. doi: 10.1016/j.lungcan.2016.10.016
Leskinen, M. J., Rantakokko-Jalava, K., Manninen, R., Leppilahti, M., Marttila, T., Kylmala, T., et al. (2003). Negative bacterial polymerase chain reaction (PCR) findings in prostate tissue from patients with symptoms of chronic pelvic pain syndrome (CPPS) and localized prostate cancer. Prostate 55, 105–110. doi: 10.1002/pros.10218
Li, J. J., Liu, Y., Song, L. T., Li, C. Y., and Jiang, S. Y. (2022). Regulation of microRNA-126 on the polarization of human macrophages stimulated by Porphyromonas gingivalis lipopolysaccharide. Zhonghua Kou Qiang Yi Xue Za Zhi 57, 390–396. doi: 10.3760/cma.j.cn112144-20210701-00310
Li, Q., Peng, W., Wu, J., Wang, X., Ren, Y., Li, H., et al. (2019). Autoinducer-2 of gut microbiota, a potential novel marker for human colorectal cancer, is associated with the activation of TNFSF9 signaling in macrophages. Onco. Targets. Ther. 8:e1626192. doi: 10.1080/2162402X.2019.1626192
Li, S. J., Sun, S. J., Gao, J., and Sun, F. B. (2016). Wogonin induces Beclin-1/PI3K and reactive oxygen species-mediated autophagy in human pancreatic cancer cells. Oncol. Lett. 12, 5059–5067. doi: 10.3892/ol.2016.5367
Li, Q., Wu, W., Gong, D., Shang, R., Wang, J., and Yu, H. (2021). Propionibacterium acnes overabundance in gastric cancer promote M2 polarization of macrophages via a TLR4/PI3K/Akt signaling. Gastric Cancer 24, 1242–1253. doi: 10.1007/s10120-021-01202-8
Liang, M. D., Bagchi, A., Warren, H. S., Tehan, M. M., Trigilio, J. A., Beasley-Topliffe, L. K., et al. (2005). Bacterial peptidoglycan-associated lipoprotein: a naturally occurring toll-like receptor 2 agonist that is shed into serum and has synergy with lipopolysaccharide. J. Infect. Dis. 191, 939–948. doi: 10.1086/427815
Lin, Y., Xu, J., and Lan, H. (2019). Tumor-associated macrophages in tumor metastasis: biological roles and clinical therapeutic applications. J. Hematol. Oncol. 12:76. doi: 10.1186/s13045-019-0760-3
Liss, M. A., White, J. R., Goros, M., Gelfond, J., Leach, R., Johnson-Pais, T., et al. (2018). Metabolic biosynthesis pathways identified from fecal microbiome associated with prostate cancer. Eur. Urol. 74, 575–582. doi: 10.1016/j.eururo.2018.06.033
Liu, J., Luo, M., Zhang, Y., Cao, G., and Wang, S. (2020). Association of high-risk human papillomavirus infection duration and cervical lesions with vaginal microbiota composition. Ann. Transl. Med. 8:1161. doi: 10.21037/atm-20-5832
Liu, L., Shi, W., Xiao, X., Wu, X., Hu, H., Yuan, S., et al. (2021). BCG immunotherapy inhibits cancer progression by promoting the M1 macrophage differentiation of THP1 cells via the Rb/E2F1 pathway in cervical carcinoma. Oncol. Rep. 46:245. doi: 10.3892/or.2021.8196
Liu, H. X., Tao, L. L., Zhang, J., Zhu, Y. G., Zheng, Y., Liu, D., et al. (2018). Difference of lower airway microbiome in bilateral protected specimen brush between lung cancer patients with unilateral lobar masses and control subjects. Int. J. Cancer 142, 769–778. doi: 10.1002/ijc.31098
Liu, P. S., Wang, H., Li, X., Chao, T., Teav, T., Christen, S., et al. (2017). Alpha-ketoglutarate orchestrates macrophage activation through metabolic and epigenetic reprogramming. Nat. Immunol. 18, 985–994. doi: 10.1038/ni.3796
Liu, C. Y., Xu, J. Y., Shi, X. Y., Huang, W., Ruan, T. Y., Xie, P., et al. (2013). M2-polarized tumor-associated macrophages promoted epithelial-mesenchymal transition in pancreatic cancer cells, partially through TLR4/IL-10 signaling pathway. Lab. Investig. 93, 844–854. doi: 10.1038/labinvest.2013.69
Lizotte, P. H., Baird, J. R., Stevens, C. A., Lauer, P., Green, W. R., Brockstedt, D. G., et al. (2014). Attenuated listeria monocytogenes reprograms M2-polarized tumor-associated macrophages in ovarian cancer leading to iNOS-mediated tumor cell lysis. Onco. Targets. Ther. 3:e28926. doi: 10.4161/onci.28926
Long, K. B., Collier, A. I., and Beatty, G. L. (2019). Macrophages: key orchestrators of a tumor microenvironment defined by therapeutic resistance. Mol. Immunol. 110, 3–12. doi: 10.1016/j.molimm.2017.12.003
Lopes, R. L., Borges, T. J., Zanin, R. F., and Bonorino, C. (2016). IL-10 is required for polarization of macrophages to M2-like phenotype by mycobacterial DnaK (heat shock protein 70). Cytokine 85, 123–129. doi: 10.1016/j.cyto.2016.06.018
Lu, L., Ling, W., and Ruan, Z. (2021). TAM-derived extracellular vesicles containing microRNA-29a-3p explain the deterioration of ovarian cancer. Mol. Ther. Nucleic. Acids 25, 468–482. doi: 10.1016/j.omtn.2021.05.011
Lunn, R. M., Mehta, S. S., Jahnke, G. D., Wang, A., Wolfe, M. S., and Berridge, B. R. (2022). Cancer Hazard evaluations for contemporary needs: highlights from new National Toxicology Program Evaluations and methodological advancements. J. Natl. Cancer Inst. 114, 1441–1448. doi: 10.1093/jnci/djac164
Luo, F., Sun, X., Qu, Z., and Zhang, X. (2016). Salmonella typhimurium-induced M1 macrophage polarization is dependent on the bacterial O antigen. World J. Microbiol. Biotechnol. 32:22. doi: 10.1007/s11274-015-1978-z
Ma, Z., Li, R., Hu, R., Deng, X., Xu, Y., Zheng, W., et al. (2020). Brucella abortus BspJ is a Nucleomodulin that inhibits macrophage apoptosis and promotes intracellular survival of Brucella. Front. Microbiol. 11:599205. doi: 10.3389/fmicb.2020.599205
Mantovani, A., Allavena, P., Sica, A., and Balkwill, F. (2008). Cancer-related inflammation. Nature 454, 436–444. doi: 10.1038/nature07205
Mantovani, A., Sica, A., Sozzani, S., Allavena, P., Vecchi, A., and Locati, M. (2004). The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 25, 677–686. doi: 10.1016/j.it.2004.09.015
Mantovani, A., Sozzani, S., Locati, M., Allavena, P., and Sica, A. (2002). Macrophage polarization: tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol. 23, 549–555. doi: 10.1016/S1471-4906(02)02302-5
Mima, K., Nishihara, R., Qian, Z. R., Cao, Y., Sukawa, Y., Nowak, J. A., et al. (2016). Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut 65, 1973–1980. doi: 10.1136/gutjnl-2015-310101
Mima, K., Sukawa, Y., Nishihara, R., Qian, Z. R., Yamauchi, M., Inamura, K., et al. (2015). Fusobacterium nucleatum and T cells in colorectal carcinoma. JAMA Oncol. 1, 653–661. doi: 10.1001/jamaoncol.2015.1377
Miquel, S., Martin, R., Rossi, O., Bermudez-Humaran, L. G., Chatel, J. M., Sokol, H., et al. (2013). Faecalibacterium prausnitzii and human intestinal health. Curr. Opin. Microbiol. 16, 255–261. doi: 10.1016/j.mib.2013.06.003
Mitra, A., MacIntyre, D. A., Lee, Y. S., Smith, A., Marchesi, J. R., Lehne, B., et al. (2015). Cervical intraepithelial neoplasia disease progression is associated with increased vaginal microbiome diversity. Sci. Rep. 5:16865. doi: 10.1038/srep16865
Molinero, N., Ruiz, L., Milani, C., Gutierrez-Diaz, I., Sanchez, B., Mangifesta, M., et al. (2019). The human gallbladder microbiome is related to the physiological state and the biliary metabolic profile. Microbiome 7:100. doi: 10.1186/s40168-019-0712-8
Monack, D. M., Raupach, B., Hromockyj, A. E., and Falkow, S. (1996). Salmonella typhimurium invasion induces apoptosis in infected macrophages. Proc. Natl. Acad. Sci. U. S. A. 93, 9833–9838. doi: 10.1073/pnas.93.18.9833
Moratalla, A., Caparros, E., Juanola, O., Portune, K., Puig-Kroger, A., Estrada-Capetillo, L., et al. (2016). Bifidobacterium pseudocatenulatum CECT7765 induces an M2 anti-inflammatory transition in macrophages from patients with cirrhosis. J. Hepatol. 64, 135–145. doi: 10.1016/j.jhep.2015.08.020
Murata, H., Tsuji, S., Tsujii, M., Fu, H. Y., Tanimura, H., Tsujimoto, M., et al. (2004). Helicobacter bilis infection in biliary tract cancer. Aliment. Pharmacol. Ther. 20, 90–94. doi: 10.1111/j.1365-2036.2004.01972.x
Murphy, J. T., Sommer, S., Kabara, E. A., Verman, N., Kuelbs, M. A., Saama, P., et al. (2006). Gene expression profiling of monocyte-derived macrophages following infection with Mycobacterium avium subspecies avium and Mycobacterium avium subspecies paratuberculosis. Physiol. Genomics 28, 67–75. doi: 10.1152/physiolgenomics.00098.2006
Murray, P. J., Allen, J. E., Biswas, S. K., Fisher, E. A., Gilroy, D. W., Goerdt, S., et al. (2014). Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity 41, 14–20. doi: 10.1016/j.immuni.2014.06.008
Murray, P. J., and Wynn, T. A. (2011). Protective and pathogenic functions of macrophage subsets. Nat. Rev. Immunol. 11, 723–737. doi: 10.1038/nri3073
Myers, D. R., Abram, C. L., Wildes, D., Belwafa, A., Welsh, A. M. N., Schulze, C. J., et al. (2020). Shp1 loss enhances macrophage effector function and promotes anti-tumor immunity. Front. Immunol. 11:576310. doi: 10.3389/fimmu.2020.576310
Nagano, T., Otoshi, T., Hazama, D., Kiriu, T., Umezawa, K., Katsurada, N., et al. (2019). Novel cancer therapy targeting microbiome. Onco. Targets. Ther. 12, 3619–3624. doi: 10.2147/OTT.S207546
Nagaraja, V., and Eslick, G. D. (2014). Systematic review with meta-analysis: the relationship between chronic salmonella typhi carrier status and gall-bladder cancer. Aliment. Pharmacol. Ther. 39, 745–750. doi: 10.1111/apt.12655
Nath, G., Singh, Y. K., Kumar, K., Gulati, A. K., Shukla, V. K., Khanna, A. K., et al. (2008). Association of carcinoma of the gallbladder with typhoid carriage in a typhoid endemic area using nested PCR. J. Infect. Dev. Ctries. 2, 302–307. doi: 10.3855/jidc.226
Nath, G., Singh, Y. K., Maurya, P., Gulati, A. K., Srivastava, R. C., and Tripathi, S. K. (2010). Does salmonella Typhi primarily reside in the liver of chronic typhoid carriers? J. Infect. Dev. Ctries. 4, 259–261. doi: 10.3855/jidc.820
Nejman, D., Livyatan, I., Fuks, G., Gavert, N., Zwang, Y., Geller, L. T., et al. (2020). The human tumor microbiome is composed of tumor-type-specific intracellular bacteria. Science 368, 973–980. doi: 10.1126/science.aay9189
Oishi, Y., Spann, N. J., Link, V. M., Muse, E. D., Strid, T., Edillor, C., et al. (2017). SREBP1 contributes to resolution of pro-inflammatory TLR4 signaling by reprogramming fatty acid metabolism. Cell Metab. 25, 412–427. doi: 10.1016/j.cmet.2016.11.009
Onderdonk, A. B., Delaney, M. L., and Fichorova, R. N. (2016). The human microbiome during bacterial vaginosis. Clin. Microbiol. Rev. 29, 223–238. doi: 10.1128/CMR.00075-15
Parhi, L., Alon-Maimon, T., Sol, A., Nejman, D., Shhadeh, A., Fainsod-Levi, T., et al. (2020). Breast cancer colonization by fusobacterium nucleatum accelerates tumor growth and metastatic progression. Nat. Commun. 11:3259. doi: 10.1038/s41467-020-16967-2
Paynich, M. L., Jones-Burrage, S. E., and Knight, K. L. (2017). Exopolysaccharide from Bacillus subtilis induces anti-inflammatory M2 macrophages that prevent T cell-mediated disease. J. Immunol. 198, 2689–2698. doi: 10.4049/jimmunol.1601641
Pearce, M. M., Hilt, E. E., Rosenfeld, A. B., Zilliox, M. J., Thomas-White, K., Fok, C., et al. (2014). The female urinary microbiome: a comparison of women with and without urgency urinary incontinence. MBio 5, e01283–e01214. doi: 10.1128/mBio.01283-14
Peng, K. T., Hsieh, C. C., Huang, T. Y., Chen, P. C., Shih, H. N., Lee, M. S., et al. (2017). Staphylococcus aureus biofilm elicits the expansion, activation and polarization of myeloid-derived suppressor cells in vivo and in vitro. PLoS One 12:e0183271. doi: 10.1371/journal.pone.0183271
Peters, B. A., Hayes, R. B., Goparaju, C., Reid, C., Pass, H. I., and Ahn, J. (2019). The microbiome in lung cancer tissue and recurrence-free survival. Cancer Epidemiol. Biomark. Prev. 28, 731–740. doi: 10.1158/1055-9965.EPI-18-0966
Pidwill, G. R., Gibson, J. F., Cole, J., Renshaw, S. A., and Foster, S. J. (2020). The role of macrophages in Staphylococcus aureus infection. Front. Immunol. 11:620339. doi: 10.3389/fimmu.2020.620339
Pinheiro da Silva, F., Aloulou, M., Skurnik, D., Benhamou, M., Andremont, A., Velasco, I. T., et al. (2007). CD16 promotes Escherichia coli sepsis through an FcR gamma inhibitory pathway that prevents phagocytosis and facilitates inflammation. Nat. Med. 13, 1368–1374. doi: 10.1038/nm1665
Pittet, M. J., Michielin, O., and Migliorini, D. (2022). Clinical relevance of tumour-associated macrophages. Nat. Rev. Clin. Oncol. 19, 402–421. doi: 10.1038/s41571-022-00620-6
Pleguezuelos-Manzano, C., Puschhof, J., Rosendahl Huber, A., van Hoeck, A., Wood, H. M., Nomburg, J., et al. (2020). Mutational signature in colorectal cancer caused by genotoxic pks(+) E. coli. Nature 580, 269–273. doi: 10.1038/s41586-020-2080-8
Polak, D., Yaya, A., Levy, D. H., Metzger, Z., and Abramovitz, I. (2021). Enterococcus faecalis sustained infection induces macrophage pro-resolution polarization. Int. Endod. J. 54, 1840–1849. doi: 10.1111/iej.13574
Poore, G. D., Kopylova, E., Zhu, Q., Carpenter, C., Fraraccio, S., Wandro, S., et al. (2020). Microbiome analyses of blood and tissues suggest cancer diagnostic approach. Nature 579, 567–574. doi: 10.1038/s41586-020-2095-1
Pore, D., Mahata, N., Pal, A., and Chakrabarti, M. K. (2010). 34 kDa MOMP of Shigella flexneri promotes TLR2 mediated macrophage activation with the engagement of NF-kappaB and p38 MAP kinase signaling. Mol. Immunol. 47, 1739–1746. doi: 10.1016/j.molimm.2010.03.001
Pradhan, S. B., and Dali, S. (2004). Relation between gallbladder neoplasm and helicobacter hepaticus infection. Kathmandu Univ Med J (KUMJ). 2, 331–335
Pushalkar, S., Hundeyin, M., Daley, D., Zambirinis, C. P., Kurz, E., Mishra, A., et al. (2018). The pancreatic cancer microbiome promotes oncogenesis by induction of innate and adaptive immune suppression. Cancer Discov. 8, 403–416. doi: 10.1158/2159-8290.CD-17-1134
Ridlon, J. M., Ikegawa, S., Alves, J. M., Zhou, B., Kobayashi, A., Iida, T., et al. (2013). Clostridium scindens: a human gut microbe with a high potential to convert glucocorticoids into androgens. J. Lipid Res. 54, 2437–2449. doi: 10.1194/jlr.M038869
Riquelme, E., Zhang, Y., Zhang, L., Montiel, M., Zoltan, M., Dong, W., et al. (2019). Tumor microbiome diversity and composition influence pancreatic cancer outcomes. Cells 178, 795–806.e12. doi: 10.1016/j.cell.2019.07.008
Roberts, P. J., Dickinson, R. J., Whitehead, A., Laughton, C. R., and Foweraker, J. E. (2002). The culture of lactobacilli species in gastric carcinoma. J. Clin. Pathol. 55:477. doi: 10.1136/jcp.55.6.477
Rosenberger, C. M., and Finlay, B. B. (2003). Phagocyte sabotage: disruption of macrophage signalling by bacterial pathogens. Nat. Rev. Mol. Cell Biol. 4, 385–396. doi: 10.1038/nrm1104
Routy, B., Le Chatelier, E., Derosa, L., Duong, C. P. M., Alou, M. T., Daillere, R., et al. (2018). Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 359, 91–97. doi: 10.1126/science.aan3706
Rubinstein, M. R., Wang, X., Liu, W., Hao, Y., Cai, G., and Han, Y. W. (2013). Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/beta-catenin signaling via its FadA adhesin. Cell Host Microbe 14, 195–206. doi: 10.1016/j.chom.2013.07.012
Ruffell, B., Chang-Strachan, D., Chan, V., Rosenbusch, A., Ho, C. M., Pryer, N., et al. (2014). Macrophage IL-10 blocks CD8+ T cell-dependent responses to chemotherapy by suppressing IL-12 expression in intratumoral dendritic cells. Cancer Cell 26, 623–637. doi: 10.1016/j.ccell.2014.09.006
Russell, D. G., Cardona, P. J., Kim, M. J., Allain, S., and Altare, F. (2009). Foamy macrophages and the progression of the human tuberculosis granuloma. Nat. Immunol. 10, 943–948. doi: 10.1038/ni.1781
Sakamoto, R., Kajihara, I., Mijiddorj, T., Otsuka-Maeda, S., Sawamura, S., Nishimura, Y., et al. (2021). Existence of Staphylococcus aureus correlates with the progression of extramammary Paget's disease: potential involvement of interleukin-17 and M2-like macrophage polarization. Eur. J. Dermatol. 31, 48–54. doi: 10.1684/ejd.2021.3972
Salazar, C. R., Sun, J., Li, Y., Francois, F., Corby, P., Perez-Perez, G., et al. (2013). Association between selected oral pathogens and gastric precancerous lesions. PLoS One 8:e51604. doi: 10.1371/journal.pone.0051604
Sanchez-Lopez, E., Zhong, Z., Stubelius, A., Sweeney, S. R., Booshehri, L. M., Antonucci, L., et al. (2019). Choline uptake and metabolism modulate macrophage IL-1beta and IL-18 production. Cell Metab. 29, 1350–1362.e7. doi: 10.1016/j.cmet.2019.03.011
Schulte, L. N., Eulalio, A., Mollenkopf, H. J., Reinhardt, R., and Vogel, J. (2011). Analysis of the host microRNA response to salmonella uncovers the control of major cytokines by the let-7 family. EMBO J. 30, 1977–1989. doi: 10.1038/emboj.2011.94
Schulz, C., Schutte, K., Mayerle, J., and Malfertheiner, P. (2019). The role of the gastric bacterial microbiome in gastric cancer: helicobacter pylori and beyond. Ther. Adv. Gastroenterol. 12:1756284819894062. doi: 10.1177/1756284819894062
Scott, A. J., Alexander, J. L., Merrifield, C. A., Cunningham, D., Jobin, C., Brown, R., et al. (2019). International cancer microbiome consortium consensus statement on the role of the human microbiome in carcinogenesis. Gut 68, 1624–1632. doi: 10.1136/gutjnl-2019-318556
Seow, S. W., Cai, S., Rahmat, J. N., Bay, B. H., Lee, Y. K., Chan, Y. H., et al. (2010). Lactobacillus rhamnosus GG induces tumor regression in mice bearing orthotopic bladder tumors. Cancer Sci. 101, 751–758. doi: 10.1111/j.1349-7006.2009.01426.x
Sepich-Poore, G. D., Zitvogel, L., Straussman, R., Hasty, J., Wargo, J. A., and Knight, R. (2021). The microbiome and human cancer. Science 371:6536. doi: 10.1126/science.abc4552
Sfanos, K. S., Sauvageot, J., Fedor, H. L., Dick, J. D., De Marzo, A. M., and Isaacs, W. B. (2008). A molecular analysis of prokaryotic and viral DNA sequences in prostate tissue from patients with prostate cancer indicates the presence of multiple and diverse microorganisms. Prostate 68, 306–320. doi: 10.1002/pros.20680
Sha, S., Shi, Y., Tang, Y., Jia, L., Han, X., Liu, Y., et al. (2021). Mycobacterium tuberculosis Rv1987 protein induces M2 polarization of macrophages through activating the PI3K/Akt1/mTOR signaling pathway. Immunol. Cell Biol. 99, 570–585. doi: 10.1111/imcb.12436
Shapouri-Moghaddam, A., Mohammadian, S., Vazini, H., Taghadosi, M., Esmaeili, S. A., Mardani, F., et al. (2018). Macrophage plasticity, polarization, and function in health and disease. J. Cell. Physiol. 233, 6425–6440. doi: 10.1002/jcp.26429
Shaughnessy, L. M., and Swanson, J. A. (2007). The role of the activated macrophage in clearing listeria monocytogenes infection. Front. Biosci. 12, 2683–2692. doi: 10.2741/2364
Shi, Y., Zheng, W., Yang, K., Harris, K. G., Ni, K., Xue, L., et al. (2020). Intratumoral accumulation of gut microbiota facilitates CD47-based immunotherapy via STING signaling. J. Exp. Med. 217:e20192282. doi: 10.1084/jem.20192282
Shrestha, E., White, J. R., Yu, S. H., Kulac, I., Ertunc, O., De Marzo, A. M., et al. (2018). Profiling the urinary microbiome in men with positive versus negative biopsies for prostate cancer. J. Urol. 199, 161–171. doi: 10.1016/j.juro.2017.08.001
Smith, M. W., Schmidt, J. E., Rehg, J. E., Orihuela, C. J., and McCullers, J. A. (2007). Induction of pro- and anti-inflammatory molecules in a mouse model of pneumococcal pneumonia after influenza. Comp. Med. 57, 82–89.
So, K. A., Yang, E. J., Kim, N. R., Hong, S. R., Lee, J. H., Hwang, C. S., et al. (2020). Changes of vaginal microbiota during cervical carcinogenesis in women with human papillomavirus infection. PLoS One 15:e0238705. doi: 10.1371/journal.pone.0238705
Sohn, W., Jun, D. W., Lee, K. N., Lee, H. L., Lee, O. Y., Choi, H. S., et al. (2015). Lactobacillus paracasei induces M2-dominant Kupffer cell polarization in a mouse model of nonalcoholic steatohepatitis. Dig. Dis. Sci. 60, 3340–3350. doi: 10.1007/s10620-015-3770-1
Sokol, H., Pigneur, B., Watterlot, L., Lakhdari, O., Bermudez-Humaran, L. G., Gratadoux, J. J., et al. (2008). Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl. Acad. Sci. U. S. A. 105, 16731–16736. doi: 10.1073/pnas.0804812105
St Jean, A. T., Zhang, M., and Forbes, N. S. (2008). Bacterial therapies: completing the cancer treatment toolbox. Curr. Opin. Biotechnol. 19, 511–517. doi: 10.1016/j.copbio.2008.08.004
Stapels, D. A. C., Hill, P. W. S., Westermann, A. J., Fisher, R. A., Thurston, T. L., Saliba, A. E., et al. (2018). Salmonella persisters undermine host immune defenses during antibiotic treatment. Science 362, 1156–1160. doi: 10.1126/science.aat7148
Su, P., Wang, Q., Bi, E., Ma, X., Liu, L., Yang, M., et al. (2020). Enhanced lipid accumulation and metabolism are required for the differentiation and activation of tumor-associated macrophages. Cancer Res. 80, 1438–1450. doi: 10.1158/0008-5472.CAN-19-2994
Suarez, G., Romero-Gallo, J., Piazuelo, M. B., Sierra, J. C., Delgado, A. G., Washington, M. K., et al. (2019). Nod1 imprints inflammatory and carcinogenic responses toward the gastric pathogen helicobacter pylori. Cancer Res. 79, 1600–1611. doi: 10.1158/0008-5472.CAN-18-2651
Suzuki, M., Mimuro, H., Kiga, K., Fukumatsu, M., Ishijima, N., Morikawa, H., et al. (2009). Helicobacter pylori CagA phosphorylation-independent function in epithelial proliferation and inflammation. Cell Host Microbe 5, 23–34. doi: 10.1016/j.chom.2008.11.010
Tamura, R., Tanaka, T., Yamamoto, Y., Akasaki, Y., and Sasaki, H. (2018). Dual role of macrophage in tumor immunity. Immunotherapy 10, 899–909. doi: 10.2217/imt-2018-0006
Tan, S., Xia, L., Yi, P., Han, Y., Tang, L., Pan, Q., et al. (2020). Exosomal miRNAs in tumor microenvironment. J. Exp. Clin. Cancer Res. 39:67. doi: 10.1186/s13046-020-01570-6
Tannahill, G. M., Curtis, A. M., Adamik, J., Palsson-McDermott, E. M., McGettrick, A. F., Goel, G., et al. (2013). Succinate is an inflammatory signal that induces IL-1beta through HIF-1alpha. Nature 496, 238–242. doi: 10.1038/nature11986
Templeton, D. M., and Liu, Y. (2003). Genetic regulation of cell function in response to iron overload or chelation. Biochim. Biophys. Acta 1619, 113–124. doi: 10.1016/S0304-4165(02)00497-X
Thomas, A. M., Manghi, P., Asnicar, F., Pasolli, E., Armanini, F., Zolfo, M., et al. (2019). Metagenomic analysis of colorectal cancer datasets identifies cross-cohort microbial diagnostic signatures and a link with choline degradation. Nat. Med. 25, 667–678. doi: 10.1038/s41591-019-0405-7
Thurlow, L. R., Hanke, M. L., Fritz, T., Angle, A., Aldrich, A., Williams, S. H., et al. (2011). Staphylococcus aureus biofilms prevent macrophage phagocytosis and attenuate inflammation in vivo. J. Immunol. 186, 6585–6596. doi: 10.4049/jimmunol.1002794
Tian, Y., Jin, Z., Zhu, P., Liu, S., Zhang, D., Tang, M., et al. (2019). TRIM59: a membrane protein expressed on bacillus Calmette-Guerin-activated macrophages that induces apoptosis of fibrosarcoma cells by direct contact. Exp. Cell Res. 384:111590. doi: 10.1016/j.yexcr.2019.111590
Trinchieri, G. (2012). Cancer and inflammation: an old intuition with rapidly evolving new concepts. Annu. Rev. Immunol. 30, 677–706. doi: 10.1146/annurev-immunol-020711-075008
Tsay, J. J., Wu, B. G., Badri, M. H., Clemente, J. C., Shen, N., Meyn, P., et al. (2018). Airway microbiota is associated with upregulation of the PI3K pathway in lung cancer. Am. J. Respir. Crit. Care Med. 198, 1188–1198. doi: 10.1164/rccm.201710-2118OC
Tsay, J. J., Wu, B. G., Sulaiman, I., Gershner, K., Schluger, R., Li, Y., et al. (2021). Lower airway Dysbiosis affects lung cancer progression. Cancer Discov. 11, 293–307. doi: 10.1158/2159-8290.CD-20-0263
Tsuchiya, Y., Loza, E., Villa-Gomez, G., Trujillo, C. C., Baez, S., Asai, T., et al. (2018). Metagenomics of microbial communities in gallbladder bile from patients with gallbladder cancer or Cholelithiasis. Asian Pac. J. Cancer Prev. 19, 961–967. doi: 10.22034/APJCP.2018.19.4.961
Tuohy, J. L., Somarelli, J. A., Borst, L. B., Eward, W. C., Lascelles, B. D. X., and Fogle, J. E. (2020). Immune dysregulation and osteosarcoma: Staphylococcus aureus downregulates TGF-beta and heightens the inflammatory signature in human and canine macrophages suppressed by osteosarcoma. Vet. Comp. Oncol. 18, 64–75. doi: 10.1111/vco.12529
Umezu, D., Okada, N., Sakoda, Y., Adachi, K., Ojima, T., Yamaue, H., et al. (2019). Inhibitory functions of PD-L1 and PD-L2 in the regulation of anti-tumor immunity in murine tumor microenvironment. Cancer Immunol. Immunother. 68, 201–211. doi: 10.1007/s00262-018-2263-4
Urbaniak, C., Gloor, G. B., Brackstone, M., Scott, L., Tangney, M., and Reid, G. (2016). The microbiota of breast tissue and its association with breast cancer. Appl. Environ. Microbiol. 82, 5039–5048. doi: 10.1128/AEM.01235-16
Vitale, I., Manic, G., Coussens, L. M., Kroemer, G., and Galluzzi, L. (2019). Macrophages and metabolism in the tumor microenvironment. Cell Metab. 30, 36–50. doi: 10.1016/j.cmet.2019.06.001
Wang, L., Johnson, E. E., Shi, H. N., Walker, W. A., Wessling-Resnick, M., and Cherayil, B. J. (2008). Attenuated inflammatory responses in hemochromatosis reveal a role for iron in the regulation of macrophage cytokine translation. J. Immunol. 181, 2723–2731. doi: 10.4049/jimmunol.181.4.2723
Wang, Y., Li, Y., Li, H., Song, H., Zhai, N., Lou, L., et al. (2017). Brucella dysregulates monocytes and inhibits macrophage polarization through LC3-dependent autophagy. Front. Immunol. 8:691. doi: 10.3389/fimmu.2017.00691
Wang, T. R., Peng, J. C., Qiao, Y. Q., Zhu, M. M., Zhao, D., Shen, J., et al. (2014). Helicobacter pylori regulates TLR4 and TLR9 during gastric carcinogenesis. Int. J. Clin. Exp. Pathol. 7, 6950–6955.
Wang, X., Wu, Y., Jiao, J., and Huang, Q. (2018). Mycobacterium tuberculosis infection induces IL-10 gene expression by disturbing histone deacetylase 6 and histonedeacetylase 11 equilibrium in macrophages. Tuberculosis (Edinb.) 108, 118–123. doi: 10.1016/j.tube.2017.11.008
Wang, B., Wu, Y., Liu, R., Xu, H., Mei, X., Shang, Q., et al. (2020). Lactobacillus rhamnosus GG promotes M1 polarization in murine bone marrow-derived macrophages by activating TLR2/MyD88/MAPK signaling pathway. Anim. Sci. J. 91:e13439. doi: 10.1111/asj.13439
Wang, Y., Xi, J., Yi, J., Meng, C., Zhao, X., Sun, Z., et al. (2022). Brucella induces M1 to M2 polarization of macrophages through STAT6 signaling pathway to promote bacterial intracellular survival. Res. Vet. Sci. 145, 91–101. doi: 10.1016/j.rvsc.2022.02.006
Wang, L. L., Yu, X. J., Zhan, S. H., Jia, S. J., Tian, Z. B., and Dong, Q. J. (2014). Participation of microbiota in the development of gastric cancer. World J. Gastroenterol. 20, 4948–4952. doi: 10.3748/wjg.v20.i17.4948
Weng, M. T., Chiu, Y. T., Wei, P. Y., Chiang, C. W., Fang, H. L., and Wei, S. C. (2019). Microbiota and gastrointestinal cancer. J. Formos. Med. Assoc. 118, S32–S41. doi: 10.1016/j.jfma.2019.01.002
Wilson, M. R., Jiang, Y., Villalta, P. W., Stornetta, A., Boudreau, P. D., Carra, A., et al. (2019). The human gut bacterial genotoxin colibactin alkylates DNA. Science 363:eaar7785. doi: 10.1126/science.aar7785
Wirbel, J., Pyl, P. T., Kartal, E., Zych, K., Kashani, A., Milanese, A., et al. (2019). Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nat. Med. 25, 679–689. doi: 10.1038/s41591-019-0406-6
Wu, J., Li, K., Peng, W., Li, H., Li, Q., Wang, X., et al. (2019). Autoinducer-2 of fusobacterium nucleatum promotes macrophage M1 polarization via TNFSF9/IL-1beta signaling. Int. Immunopharmacol. 74:105724. doi: 10.1016/j.intimp.2019.105724
Wu, N., Yang, X., Zhang, R., Li, J., Xiao, X., Hu, Y., et al. (2013). Dysbiosis signature of fecal microbiota in colorectal cancer patients. Microb. Ecol. 66, 462–470. doi: 10.1007/s00248-013-0245-9
Wynn, T. A., and Vannella, K. M. (2016). Macrophages in tissue repair, regeneration, and fibrosis. Immunity 44, 450–462. doi: 10.1016/j.immuni.2016.02.015
Xu, C., Fan, L., Lin, Y., Shen, W., Qi, Y., Zhang, Y., et al. (2021). Fusobacterium nucleatum promotes colorectal cancer metastasis through miR-1322/CCL20 axis and M2 polarization. Gut Microbes 13:1980347. doi: 10.1080/19490976.2021.1980347
Yachida, S., Mizutani, S., Shiroma, H., Shiba, S., Nakajima, T., Sakamoto, T., et al. (2019). Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nat. Med. 25, 968–976. doi: 10.1038/s41591-019-0458-7
Yamamura, K., Baba, Y., Nakagawa, S., Mima, K., Miyake, K., Nakamura, K., et al. (2016). Human microbiome fusobacterium Nucleatum in esophageal cancer tissue is associated with prognosis. Clin. Cancer Res. 22, 5574–5581. doi: 10.1158/1078-0432.CCR-16-1786
Yan, X., Yang, M., Liu, J., Gao, R., Hu, J., Li, J., et al. (2015). Discovery and validation of potential bacterial biomarkers for lung cancer. Am. J. Cancer Res. 5, 3111–3122.
Yang, Y., Weng, W., Peng, J., Hong, L., Yang, L., Toiyama, Y., et al. (2017). Fusobacterium nucleatum increases proliferation of colorectal cancer cells and tumor development in mice by activating toll-like receptor 4 signaling to nuclear factor-kappaB, and up-regulating expression of MicroRNA-21. Gastroenterology 152, 851–866.e24. doi: 10.1053/j.gastro.2016.11.018
Yang, M., Xu, J., Wang, Q., Zhang, A. Q., and Wang, K. (2018). An obligatory anaerobic salmonella typhimurium strain redirects M2 macrophages to the M1 phenotype. Oncol. Lett. 15, 3918–3922. doi: 10.3892/ol.2018.7742
Yao, Y., Li, G., Wu, J., Zhang, X., and Wang, J. (2015). Inflammatory response of macrophages cultured with helicobacter pylori strains was regulated by miR-155. Int. J. Clin. Exp. Pathol. 8, 4545–4554.
Yao, R. R., Li, J. H., Zhang, R., Chen, R. X., and Wang, Y. H. (2018). M2-polarized tumor-associated macrophages facilitated migration and epithelial-mesenchymal transition of HCC cells via the TLR4/STAT3 signaling pathway. World J. Surg. Oncol. 16:9. doi: 10.1186/s12957-018-1312-y
Yerneni, S. S., Werner, S., Azambuja, J. H., Ludwig, N., Eutsey, R., Aggarwal, S. D., et al. (2021). Pneumococcal extracellular vesicles modulate host immunity. MBio 12:e0165721. doi: 10.1128/mBio.01657-21
Yin, Z., Ma, T., Huang, B., Lin, L., Zhou, Y., Yan, J., et al. (2019). Macrophage-derived exosomal microRNA-501-3p promotes progression of pancreatic ductal adenocarcinoma through the TGFBR3-mediated TGF-beta signaling pathway. J. Exp. Clin. Cancer Res. 38:310. doi: 10.1186/s13046-019-1313-x
Yoshikawa, M., Yamada, S., Sugamata, M., Kanauchi, O., and Morita, Y. (2021). Dectin-2 mediates phagocytosis of lactobacillus paracasei KW3110 and IL-10 production by macrophages. Sci. Rep. 11:17737. doi: 10.1038/s41598-021-97087-9
Yu, G., Gail, M. H., Consonni, D., Carugno, M., Humphrys, M., Pesatori, A. C., et al. (2016). Characterizing human lung tissue microbiota and its relationship to epidemiological and clinical features. Genome Biol. 17:163. doi: 10.1186/s13059-016-1021-1
Yu, T., Guo, F., Yu, Y., Sun, T., Ma, D., Han, J., et al. (2017). Fusobacterium nucleatum promotes Chemoresistance to colorectal cancer by modulating autophagy. Cells 170, 548–563.e16. doi: 10.1016/j.cell.2017.07.008
Zarza, S. M., Mezouar, S., and Mege, J. L. (2021). From Coxiella burnetii infection to pregnancy complications: key role of the immune response of placental cells. Pathogens 10:627. doi: 10.3390/pathogens10050627
Zhang, Z., Amorosa, L. F., Coyle, S. M., Macor, M. A., Lubitz, S. E., Carson, J. L., et al. (2015). Proteolytic cleavage of AMPKalpha and intracellular MMP9 expression are both required for TLR4-mediated mTORC1 activation and HIF-1alpha expression in leukocytes. J. Immunol. 195, 2452–2460. doi: 10.4049/jimmunol.1500944
Zhang, Y., Li, S., Liu, Q., Long, R., Feng, J., Qin, H., et al. (2020a). Mycobacterium tuberculosis heat-shock protein 16.3 induces macrophage M2 polarization through CCRL2/CX3CR1. Inflammation 43, 487–506. doi: 10.1007/s10753-019-01132-9
Zhang, Y., Meng, W., Yue, P., and Li, X. (2020b). M2 macrophage-derived extracellular vesicles promote gastric cancer progression via a microRNA-130b-3p/MLL3/GRHL2 signaling cascade. J. Exp. Clin. Cancer Res. 39:134. doi: 10.1186/s13046-020-01626-7
Zhao, Q., Hu, H., Wang, W., Peng, H., and Zhang, X. (2015). Genome sequence of Sphingobium yanoikuyae B1, a polycyclic aromatic hydrocarbon-degrading strain. Genome Announc. 3:e01522-14. doi: 10.1128/genomeA.01522-14
Zheng, P., Chen, L., Yuan, X., Luo, Q., Liu, Y., Xie, G., et al. (2017). Exosomal transfer of tumor-associated macrophage-derived miR-21 confers cisplatin resistance in gastric cancer cells. J. Exp. Clin. Cancer Res. 36:53. doi: 10.1186/s13046-017-0528-y
Zheng, J. H., Nguyen, V. H., Jiang, S. N., Park, S. H., Tan, W., Hong, S. H., et al. (2017). Two-step enhanced cancer immunotherapy with engineered Salmonella typhimurium secreting heterologous flagellin. Sci. Transl. Med. 9:eaak9537. doi: 10.1126/scitranslmed.aak9537
Zhou, J., Li, X., Wu, X., Zhang, T., Zhu, Q., Wang, X., et al. (2018). Exosomes released from tumor-associated macrophages transfer miRNAs that induce a Treg/Th17 cell imbalance in epithelial ovarian cancer. Cancer Immunol. Res. 6, 1578–1592. doi: 10.1158/2326-6066.CIR-17-0479
Zhou, Z., Peng, Y., Wu, X., Meng, S., Yu, W., Zhao, J., et al. (2019). CCL18 secreted from M2 macrophages promotes migration and invasion via the PI3K/Akt pathway in gallbladder cancer. Cell. Oncol. (Dordr.) 42, 81–92. doi: 10.1007/s13402-018-0410-8
Zhou, Y., Wang, L., Pei, F., Ji, M., Zhang, F., Sun, Y., et al. (2019). Patients with LR-HPV infection have a distinct vaginal microbiota in comparison with healthy controls. Front. Cell. Infect. Microbiol. 9:294. doi: 10.3389/fcimb.2019.00294
Zhu, X., Shen, H., Yin, X., Long, L., Chen, X., Feng, F., et al. (2017). IL-6R/STAT3/miR-204 feedback loop contributes to cisplatin resistance of epithelial ovarian cancer cells. Oncotarget 8, 39154–39166. doi: 10.18632/oncotarget.16610
Zhu, Q., Wu, X., Tang, M., and Wu, L. (2020). Observation of tumor-associated macrophages expression in gastric cancer and its clinical pathological relationship. Medicine (Baltimore) 99:e19839. doi: 10.1097/MD.0000000000019839
Zhu, Q., Wu, X., Wu, Y., and Wang, X. (2016). Interaction between Treg cells and tumor-associated macrophages in the tumor microenvironment of epithelial ovarian cancer. Oncol. Rep. 36, 3472–3478. doi: 10.3892/or.2016.5136
Glossary
Keywords: bacteria, cancer, tumor-associated macrophages, M1/M2 macrophage polarization, tumor microenvironment
Citation: Xu S, Xiong Y, Fu B, Guo D, Sha Z, Lin X and Wu H (2023) Bacteria and macrophages in the tumor microenvironment. Front. Microbiol. 14:1115556. doi: 10.3389/fmicb.2023.1115556
Edited by:
Yuan Xiao, Shanghai Jiao Tong University, ChinaReviewed by:
Armando Rojas, Catholic University of the Maule, ChileSunil Banskar, University of Arizona, United States
Copyright © 2023 Xu, Xiong, Fu, Guo, Sha, Lin and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoyuan Lin, ✉ bHh5MjAxOUBjcXUuZWR1LmNu; Haibo Wu, ✉ aGJ3dTAyM0BjcXUuZWR1LmNu
†These authors have contributed equally to this work and share first authorship
 Shiyao Xu†
Shiyao Xu† Haibo Wu
Haibo Wu