- 1State Key Laboratory of Geobiology and Environmental Geology, China University of Geosciences, Wuhan, China
- 2School of Environmental Studies, China University of Geosciences, Wuhan, China
- 3Department of Microbiology, Ohio State University, Columbus, OH, United States
Karst caves are usually considered as natural laboratories to study pristine microbiomes in subsurface biosphere. However, effects of the increasingly detected nitrate in underground karst ecosystem due to the acid rain impact on microbiota and their functions in subsurface karst caves have remained largely unknown. In this study, samples of weathered rocks and sediments were collected from the Chang Cave, Hubei province and subjected to high-throughput sequencing of 16S rRNA genes. The results showed that nitrate significantly impacted bacterial compositions, interactions, and functions in different habitats. Bacterial communities clustered according to their habitats with distinguished indicator groups identified for each individual habitat. Nitrate shaped the overall bacterial communities across two habitats with a contribution of 27.2%, whereas the pH and TOC, respectively, structured bacterial communities in weathered rocks and sediments. Alpha and beta diversities of bacterial communities increased with nitrate concentration in both habitats, with nitrate directly affecting alpha diversity in sediments, but indirectly on weathered rocks by lowering pH. Nitrate impacted more on bacterial communities in weathered rocks at the genus level than in sediments because more genera significantly correlated with nitrate concentration in weathered rocks. Diverse keystone taxa involved in nitrogen cycling were identified in the co-occurrence networks such as nitrate reducers, ammonium-oxidizers, and N2-fixers. Tax4Fun2 analysis further confirmed the dominance of genes involved in nitrogen cycling. Genes of methane metabolism and carbon fixation were also dominant. The dominance of dissimilatory and assimilatory nitrate reduction in nitrogen cycling substantiated nitrate impact on bacterial functions. Our results for the first time revealed the impact of nitrate on subsurface karst ecosystem in terms of bacterial compositions, interactions, and functions, providing an important reference for further deciphering the disturbance of human activities on the subsurface biosphere.
1. Introduction
Karst caves in central China are likely impacted by nitrogen leaching, mainly in the form of nitrate, originating from non-point sources such as agricultural industry and atmospheric deposition (Liao et al., 2018). Due to the thin soil layer in karst areas and the hydraulic connection between the surface water and groundwater, nitrate can enter the subsurface karst system through karst pipes. Increasing nitrogen inputs from anthropogenic sources can reduce biodiversity and enhance greenhouse gas fluxes (e.g., nitrous oxide, methane, carbon dioxide), leading to ecosystem degradation (Bobbink et al., 1998; Liu and Greaver, 2009; Van den Heuvel et al., 2011; Soons et al., 2017). However, the effects of nitrate infiltration on the microbial community composition and functions in karst caves remain poorly understood to date.
Nitrogen infiltration may weaken the carbon sink effect in karst caves through abiotic and biotic outcomes. On the one hand, the presence of exogenous nitrate enhances the dissolution of carbonate rock, releasing CO2 to the atmosphere (Spence and Telmer, 2005; Liu et al., 2008). On the other hand, methane-oxidizing bacteria (MOB), particularly the Upland Soil Cluster (USC), have been shown to oxidize atmospheric methane (1.8–2.0 ppm) in caves and help maintain caves as atmospheric methane sinks (Zhao et al., 2018; Cheng et al., 2021a). The net methane consumption may, however, be subject to competitive inhibition of the methane monooxygenase (MMO) by elevated concentration of NH4+ or through the stimulation of methanogenic archaea (Dunfield and Knowles, 1995; King and Schnell, 1998; Bodelier and Laanbroek, 2004).
Nitrogen deposition in soils has increased with the development of industry and agriculture (Shi et al., 2016; Zhou et al., 2017; Hu et al., 2021). Inputs of excessive N are known to cause soil acidification (Raza et al., 2020) and alter the composition and activities of soil microbial communities (Shi et al., 2016; Zhou et al., 2017; Hu et al., 2021) and thus their functions in natural ecosystems. Increased nitrate input can affect other soil chemical and biological properties.
It has been reported that the abundance of Actinobacteria and Proteobacteria typically increases with high N availability, whereas oligotrophic taxa such as Acidobacteria exhibit an opposite pattern (Fierer et al., 2012b; Hester et al., 2018; Nie et al., 2018). Rhizobiales, a group of soil borne diazotrophs belonging to the Proteobacteria, have been reported to be negatively correlated with elevated concentration of soil NH4+ and NO3− (Che et al., 2018; Wang J. et al., 2021). Nitrogen fertilization decreased the bacterial diversity in soil perhaps by stimulating the growth of nitrophilous taxa and via competitive exclusion of other species (Campbell et al., 2010). Excessive nitrate input increases N2O emissions due to the decreasing pH, thus causing incomplete denitrification and making caves as greenhouse gas sources (Liu and Greaver, 2009; Van den Heuvel et al., 2011; Brenzinger et al., 2015). Nitrogen application has been shown to decrease the diversity of soil bacterial communities because of low pH levels (Yao et al., 2014), but opposite effects on bacterial diversity in natural ecosystems have also been reported in the literature. In general, microbiological changes upon nitrogen fertilization are difficult to deconstruct because of the complex interactions and associations in the soil–plant-rhizosphere-microbial community tangle.
In subsurface caves, environmental variables can significantly shape microbial communities. For example, microbial communities therein show a high habitat specialization and respond differently to environmental variables (Yang et al., 2021). TOC and pH have been reported to modulate bacterial communities in the Heshang Cave and the Xincuntun Cave across different habitats (Yun et al., 2016a; Cao et al., 2021). In the Luohandu Cave, for example, electrical conductivity and dissolved oxygen appear to influence microbial communities in dripping water and pool water, and those in sediments and weathered rocks are shaped by temperature, Ca/Mg, and sulfate (Yang et al., 2021), but these variables give little insight into understanding the underlying microbial biology. Our knowledge about how nitrate input impacts microbial compositions and functions in cave ecosystems is still far limited.
Cave microorganisms can be involved in nitrogen cycling via multiple ways. Dong et al. (2020) reported that in the Zhijin Cave bacteria participate in nitrogen fixation, denitrification, dissimilatory and assimilatory nitrate reduction, and comammox as indicated by PICRUSt (Dong et al., 2020). In addition, nitrification has also been reported in the Panlong Cave in the Guangxi Province (Zeng et al., 2022) and the Heshang Cave in the Hubei Province (Zhao et al., 2016). Several nitrate-reducing bacteria have been identified as keystone species in the co-occurrence network of cave ecosystems, such as Gaiella (Cheng et al., 2021a) and Salinarimonas (Dong et al., 2020). Autotrophic Nitrospira and Nitrospirae may play an essential role in sustaining the primary production through CO2 fixation coupled with nitrite oxidation in cave ecosystems (Ortiz et al., 2014; Lavoie et al., 2017; Kato et al., 2018; Ma et al., 2021). As a favorable electron acceptor, the input of nitrate can stimulate anaerobic substrate oxidations coupled with nitrate reduction and thus activate other geochemical processes (Xu et al., 2014).
Therefore, we hypothesize that nitrate input into the oligotrophic caves will significantly impact on subsurface microbial communities and stimulate the biogeochemical processes related to nitrogen as well as others. To test our hypothesis, we collected weathered rock and sediment samples in the Chang Cave, located in the western Hubei Province, which has a history of exposure to acid rain. Our results provide focus on variations in bacterial communities across different habitats and their responses to nitrate input, filling the gaps of anthropogenic impacts in the subsurface karst biosphere.
2. Materials and methods
2.1. Cave description
The Chang Cave (30°39′26.01″N, 109°58′27.49″E) is located in Jianshi County, Hubei Province, China, where has a typical subtropical monsoon climate (Supplementary Figure S1A). The average annual temperature is 13°C–18°C, and the average annual rainfall is 1,200–1,800 mm. The precipitation in late April to September accounts for about 70% of the total annual precipitation. The Chang Cave is a pristine cave with an average temperature of 15°C–18°C and close to saturated relative humidity. Multiple dripping points are observed here and there inside the cave, and the water is slightly alkaline with a pH of 7.5–8.5.
2.2. Sample collection
Samples of weathered rocks and loose sediments on the ground were collected with an interval of 50 m inward to the cave at 10 sampling sites in January 2020 (Supplementary Figure S1B). The weathered surface of the cave wall (0–1 cm depth) and loose sediments on the floor (to a depth of 2 cm) were collected. A five-point sampling strategy was exploited, and triplicate samples of weathered rocks and sediments were collected at each sampling site. All samples were transported under refrigeration within 24 h of collection to the Geomicrobiology Laboratory in China University of Geosciences (Wuhan). Samples were kept at −80°C upon arrival until analyzed.
2.3. Physicochemical analysis
All samples were freeze-dried (Alpha 12 LD freeze-dryer; Martin Christ, Osterode am Harz, Germany) and passed through a sterile 200-mesh sieve. Total organic carbon (TOC) content and C/N ratio were analyzed with a C-S analyzer (EA 4000, Analytik Jena AG, Jena, Germany) after acidification with 3 M HCl. One gram soil samples were mixed with 5 mL ultrapure water followed by 10 min vortex and subsequent centrifugation at 6,800 × g for 10 min (Yun et al., 2016b). Filtrates (0.22 μm membrane) were used for the pH measurement and dissolved ion analysis. The pH was measured with a multi-parameter water quality detector (HACH, Loveland, CO). The analysis of dissolved anions and cations was performed using anionic chromatography (ICS-600, Thermo Scientific, Waltham, MA) and ICP-OES (iCAP 7,600+, Thermo Scientific), respectively.
2.4. DNA extraction and 16S rRNA gene sequencing
Total nucleic acids were extracted from 0.5 g dry weight samples (freeze-dried and sieved through 200 mesh) using the MoBio PowerSoil DNA Kit (Qiagen, Redwood City, CA) following the manufacturer’s instructions. The concentration and quality of DNA were measured by a Nanodrop 2000 spectrophotometer (ND2000; Thermo Scientific) and visualized by 2% agarose gel electrophoresis. Bacterial diversity was examined via high throughput sequencing of the 16S rRNA genes with the primer set of 338F (5′-ACTCCTACGGGAGGCAGCA-3′) and 806R (5′-GGAC TACHVGGGTWTCTAAT-3′) on the Illumina Miseq platform (Shanghai Personal Biotechnology, Co., Ltd., Shanghai). The thermal cycling steps of the 16S rRNA genes were an initial denaturation at 98°C for 2 min, followed by 25 cycles of 98°C for 15 s, 52°C for 30 s, 72°C for 30 s and a final extension at 72°C for 5 min.
2.5. Sequence analysis
Raw sequence data were quality filtered and analyzed using QIIME 2 (Bolyen et al., 2019). Reads were processed by removing barcodes, primers, and low-quality sequences (with an average quality score < 30), in which chimera removed with DADA2 plugin. Subsequently ASVs (Amplicon Sequence Variants) representative sequences and feature tables were generated to annotate in the SILVA database release 134 (DeSantis et al., 2006). These samples were resampled to the same level of sequencing to avoid the impact of sequencing depth on microbial communities.
2.6. Statistical analysis
Alpha diversities were conducted in QIIME2 (Bolyen et al., 2019). Beta diversity analyses were performed to explore the differences in bacterial composition across habitats and sampling sites, which were visualized by non-metric multidimensional scaling (NMDS) based on the Bray–Curtis dissimilarity matrix using the vegan package (Oksanen et al., 2018) in R (R Core Team, 2020). Plots of linear regression were used to reveal the bacterial diversity along the cave depth using the R package ggplot2 (Wickham, 2016). The LEFSe analysis1 was used to construct the linear discriminant analysis (LDA) model to find out significant differences in bacterial taxa between weathered rocks and sediments (Segata et al., 2011). The analyzed geochemical parameters were assessed for the variable inflation factor (VIF) using the Vegan package of R Project to make all the remaining environmental variables exhibited maximum VIF values of no higher than 10 (Yu et al., 2016). Redundancy analysis (RDA) was performed to determine the most significant physicochemical parameters that shaped composition and structure of bacterial communities using CANOCO 5.0. Mechanistic study of the effect of nitrate on the α-diversity and β-diversity was conducted using piecewise structural equation simulation (piecewiseSEM) method with the R package piecewiseSEM (Lefcheck and Freckleton, 2015). The genera present in more than 20% of samples with a relative abundance of >0.05% were retained, and those significantly associated with nitrate (r > |0.5|, p < 0.05) were then used to construct the nitrate interaction networks. Co-occurrence network analysis of significant taxon may help to explore the structure of complex microbial communities across spatial or temporal gradients. Only ASVs with a proportion above 0.05% across all samples and occurring in more than 20% of samples were retained in co-occurrence networks. The co-occurrence network was constructed based on the Spearman rank correlations with a coefficient > |0.7| and a p-value <0.01 (Liu et al., 2020). Visualization of the robust pairwise correlations of the ASVs was implemented by Gephi (version 0.9.2) software (Bastian et al., 2009). According to within-module connectivity (Zi) and among-module connectivity (Pi) thresholds, all nodes were classified into four groups: peripherals (Zi ≤ 2.5 and Pi ≤ 0.62), connectors (Zi ≤ 2.5 and Pi > 0.62), module hubs (Zi > 2.5 and Pi ≤ 0.62), and network hubs (Zi > 2.5 and Pi > 0.62; Olesen et al., 2007), where connectors, module hubs, and network hubs were considered as keystone taxa in the network (Zhou et al., 2010; Fan et al., 2018). The metabolic function profiles were predicted using the FAPROTAX (Louca et al., 2016) and the R package Tax4Fun2 (Wemheuer et al., 2020). Histogram of the KEGG level-3 functional pathways and nitrogen metabolism genes and heatmaps of functions predicted by FAPROTAX were generated in R (R Core Team, 2020). Statistical analysis of metagenomic profiles (STAMP) was used to analyze the differential metabolic profiles between the weathered rocks and sediments (Parks et al., 2014).
3. Results
3.1. Physicochemical properties of samples in the Chang Cave
The pH values of weathered rocks and sediments were significantly different (p < 0.05), but both were slightly alkaline (pH 7.69–9.21 in weathered rock versus pH 7.78–8.42 in sediments; Table 1). TOC concentrations in sediments were higher than those in weathered rocks from S1 to S5, while the opposite pattern was observed in S6–S10. The C/N ratios in sediments were highly variable and the average value (5.85 ± 2.18) were higher than that in weathered rocks (5.25 ± 0.89). The nitrate concentrations in weathered rock samples (mean 1152.95 mg·kg−1) were significantly higher than those in sediments (mean 348.78 mg·kg−1) as proved by Kruskal-Wallis test (p < 0.01; Table 1).
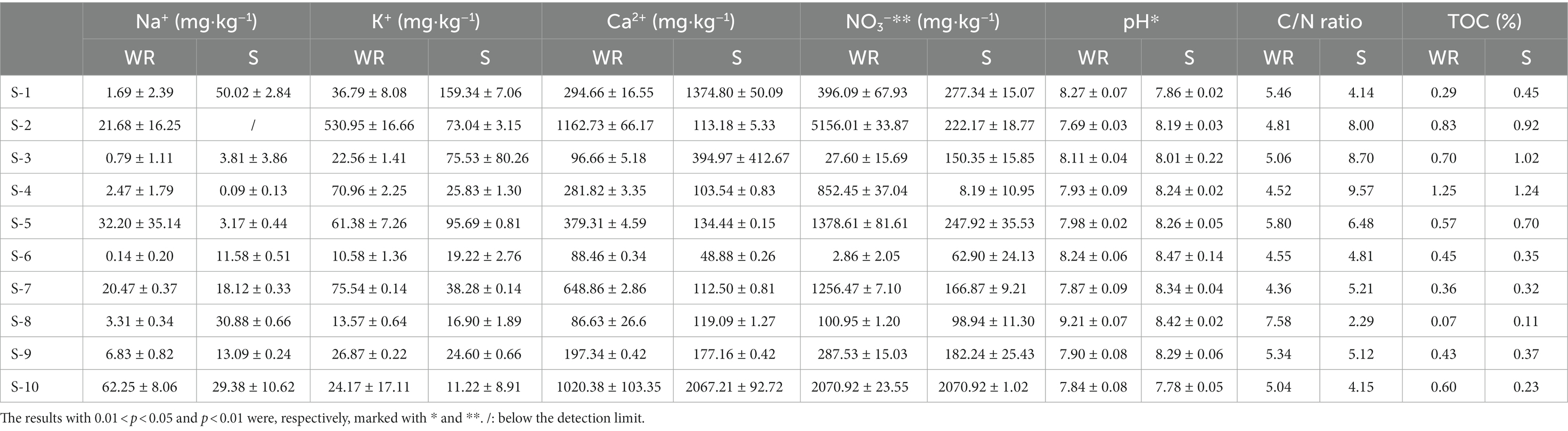
Table 1. Physicochemical properties of samples collected from weathered rocks and sediments in the Chang Cave, Hubei province, central China.
3.2. Habitat specialization of bacterial communities
At the class level, Actinobacteria was dominant in weathered rocks (Figure 1A), with relative abundance ranging from 30.96% to 77.77%, followed by Thermoleophilia with a relative abundance of 6.13% to 22.44%. In sediments, Gammaproteobacteria and Actinobacteria dominated bacterial communities with minor amounts of NC10 groups (0.38%–3.08%). The abundance of Rubrobacteria (2.26%–8.35%) in weathered rocks was significantly higher than that in sediments (0.05%–0.33%).
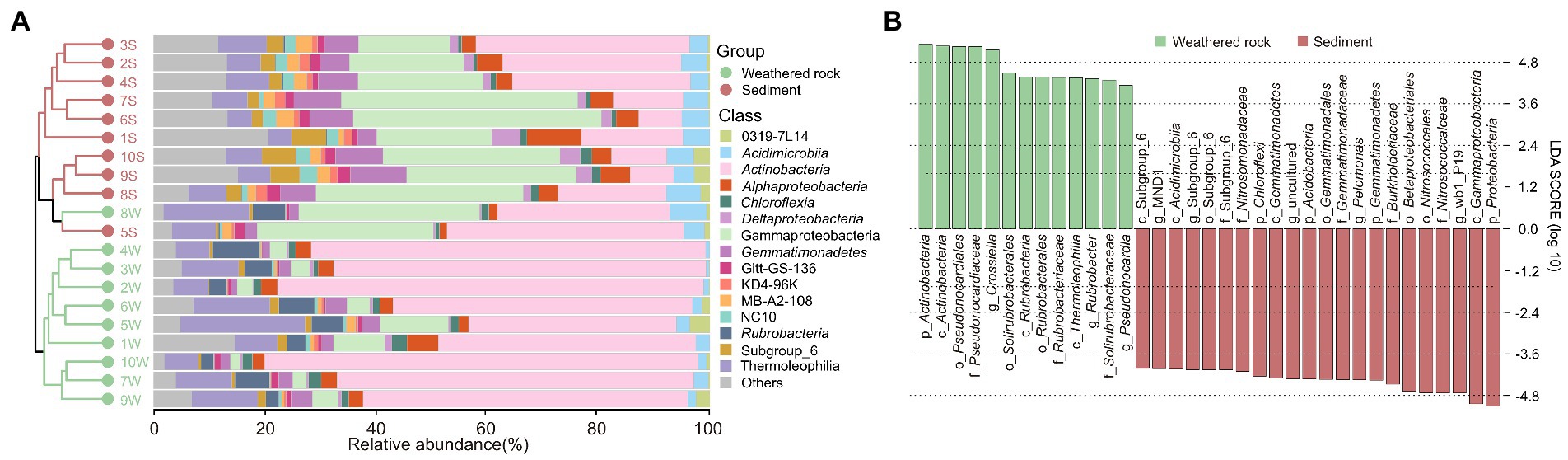
Figure 1. Compositions of bacterial communities at the class level (A) and microbial indicator groups in weathered rocks and sediments as indicated by least discriminant analysis with a LDA score > 4 (B) in the Chang Cave, Hubei province, China.
Least discriminant analysis effect size distinguished indicator taxa in weathered rocks and sediments from the phylum to genus levels (Figure 1B). At the phylum level, Actinobacteria was specific to weathered rocks, whereas Chloroflexi, Acidobacteria, Gemmatimonadetes, and Proteobacteria were associated with sediments. Pseudonocardiales, Solirubrobacterales, and Rubrobacterales were indicator orders in weathered rocks, and subgroup_6, Gemmatimonadates, Betaproteobacteriales, and Nitrosococcales were indicators in sediments. Crossiella, Rubrobacter, and Pseudonocardia were indicator genera in weathered rocks, whereas MND1, subgroup_6, Pelomonas, and wb1_R19 were in sediments.
Overall, the alpha diversity in sediments was significantly higher than that in weathered rocks as indicated by alpha diversity indexes (Supplementary Figure S2). The alpha diversity (Chao1 and Richness) indexes of weathered rocks were significantly negatively correlated with pH (Figure 2A), and those in sediments positively correlated with nitrate (Figure 2B). Bacterial communities in weathered rocks and sediments were clearly separated by the NMDS ordination plot (Figure 2C). NMDS1 distinguished bacterial communities in different habitats. Bacterial communities in sediments were located in the second and third quadrants and those in weathered rocks in the first and fourth quadrants. Bacterial communities within an individual habitats were separated along NMDS2 by different sampling sites (Figure 2C).
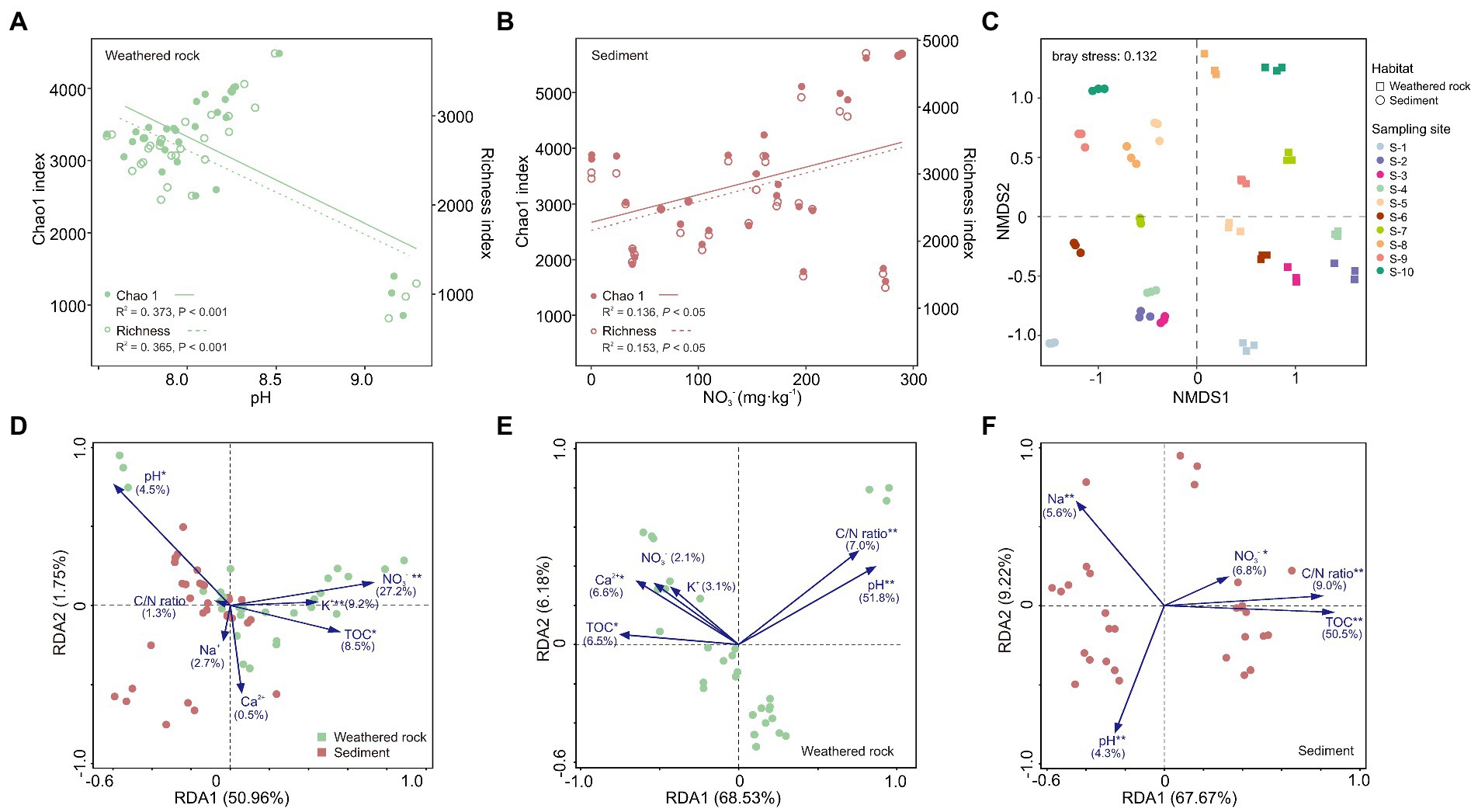
Figure 2. Bacterial diversities in weathered rock and sediment samples and redundancy analysis (RDA) of bacterial communities and environmental variables in the Chang Cave, Hubei province. (A) Linear regression analysis of alpha diversity indexes and pH in weathered rocks. Solid circles and solid lines: Chao1 index; hollow circles and dashed lines: Richness index. (B) Linear regression analysis of alpha diversity indexes and nitrate in sediments. (C) Non-metric multidimensional scaling (NMDS) ordination plot of bacterial communities based on Bray-Curtis dissimilarities. The squares represent weathered rock, the circles represent sediments, and the colors distinguish sampling locations. Redundancy analysis between environmental variables and bacterial communities at the class level in all samples (D), weathered rocks (E) and sediments (F). Significance level: p < 0.05, *; p < 0.01, **; p < 0.001, ***. Green: weathered rocks; red, sediments.
3.3. Environmental impacts on bacterial community structures
The pH and concentrations of TOC, nitrate, and K+ were demonstrated to significantly impact bacterial communities in caves with a contribution of 27.2% by NO3− (Figure 2D) across different habitats. As for each individual habitats, pH and TOC were, respectively, the most important environmental factors shaping the community structure in weathered rocks with an explanation of 51.8% (Figure 2E) and with an explanation of 50.5% in sediments (Figure 2F). In addition, NO3−, pH, C/N ratio and Na+ also significantly impacted bacterial communities in sediments.
Piecewise structural equation modeling (piecewise SEM) analysis revealed that the pH significantly and negatively shaped the alpha diversity (Chao1 and Richness indexes) of bacterial communities in weathered rocks and explained 37% of the variation (Figure 3A). Both NO3− and TOC negatively impacted bacterial communities indirectly via influencing the pH (Figure 3A). Nitrate was significantly and positively correlated with Chao1 and Richness indexes in sediments (Figure 3B). In addition, TOC and Chao1 indexes also showed a significant positive correlation. The improved piecewise SEM model explained 30% and 15% variation in Chao1 and Richness indexes in sediments, respectively (Figure 3B). Structural equation model showed that nitrate positively correlated with NMDS1 and explained 32% of its variation, which distinguished bacterial communities in different habitats. TOC was negatively related to NMDS2, and explained 21% of the variation, which distinguished bacterial communities at different sampling sites within each habitat (Figure 3C).
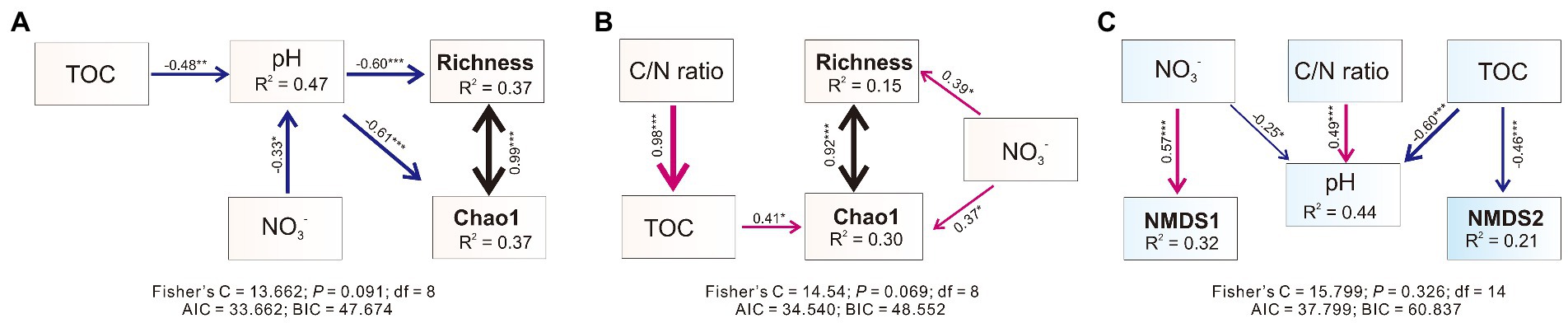
Figure 3. Piecewise structural equation modeling (piecewiseSEM) describing the effect of multiple physiochemical parameters on the microbial alpha [in weathered rocks (A), sediments (B)] and beta (C) diversities. Numbers adjacent to arrows are indicative of the effect size of the relationship. The width of the arrows is proportional to the strength of the relationship. Red arrows indicate a significant positive correlation, while blue arrows indicate a negative relationship. R2 denote the proportion of variance explained by the predictors. Significance level: p < 0.05, *; p < 0.01, **; p < 0.001, ***.
Networks were constructed between bacteria and NO3− to further explore the potential impact of NO3− on specific microbial groups. In total, 57 and 10 genera with significant Spearman’s correlation (|r| > 0.5, p < 0.05) with NO3− were included in the network of weathered rock (NW) and sediment (NS), respectively (Figures 4A,B). There were 20 (35.09%) and 6 (60%) genera showed positive correlations with NO3− in NW and NS, respectively. Taxonomically, the 57 genera mainly belonged to Proteobacteria (28), Actinobacteria (11), Acidobacteria (10), Gemmatimonadetes (3), and Chloroflexi (2) in NW, whereas the 10 genera in NS were mainly affiliated with Actinobacteria (4) and Proteobacteria (2). Novosphingobium (negatively), Quadrisphaera (positively), and uncultured Gemmatimonadaceae (negatively) were detected both in NW and NS and showed the same trend with NO3− concentration. Subgroup_6, Subgroup_7, Subgroup_12, Subgroup_22, RB41, JGI_0001001-H03, Bryobacter, Candidatus_Solibacter, and uncultured species belonged to Acidobacteria were negatively associated with NO3− in NW. Besides three genera of Thermoleophilia class in the weathered rock, there were eight and four genera of Actinobacteria positively correlated with NO3− in weathered rock and sediment, respectively. Nitrospira was only detected in NW with a negative correlation with NO3− (Figure 4A). Species in Gammaproteobacteria and Deltaproteobacteria were mainly negatively associated with NO3−, while Alphaproteobacteria showed more diverse response to NO3−.
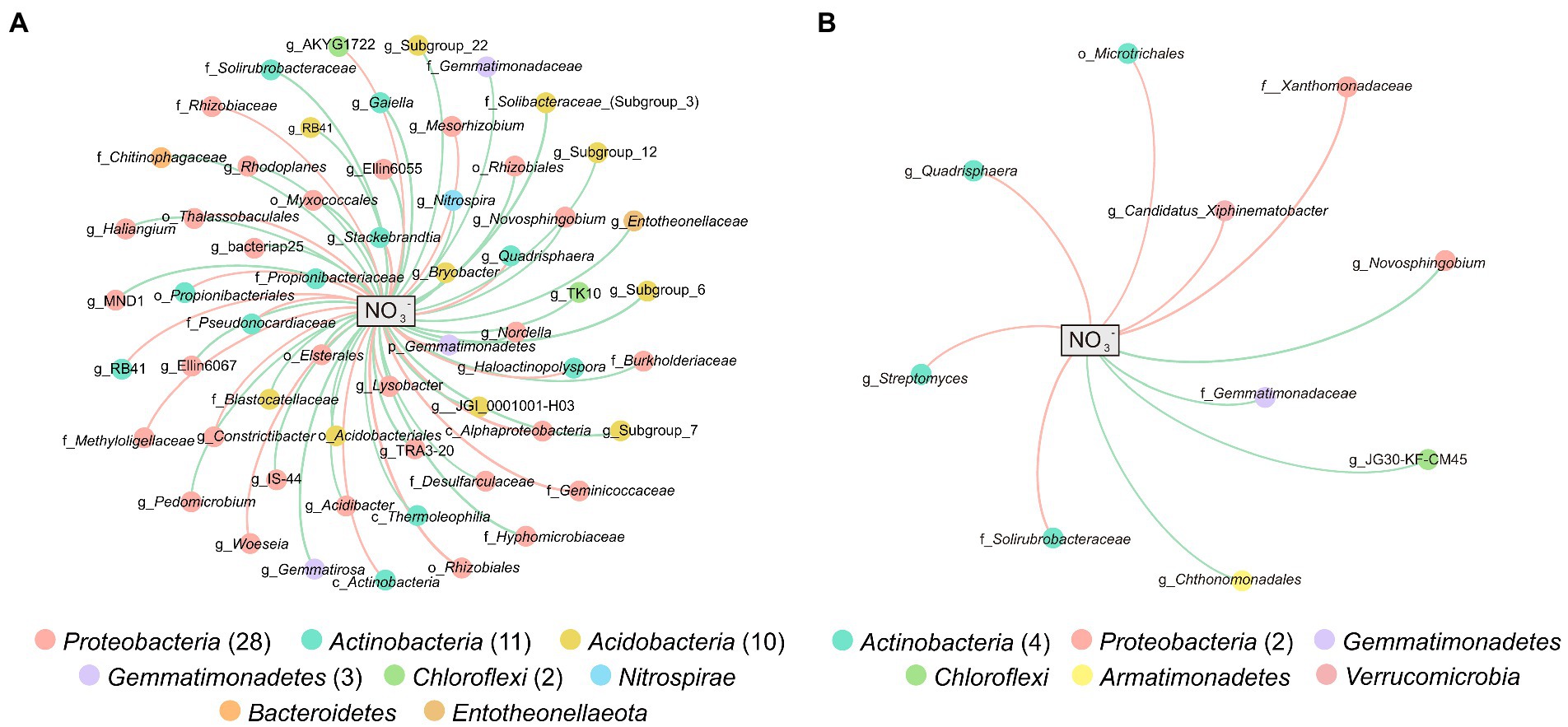
Figure 4. Networks between microbial genera and NO3− in weathered rocks (A) and sediments (B). Nodes are colored by microbial phyla, and the connection edge represents a significant correlation (|r| > 0.5, p < 0.05) with NO3− based on pairwise Spearman’s correlations. Positive correlations are in red and negative correlations are in green.
3.4. Bacterial interactions in weathered rocks and sediments
Co-occurrence networks were conducted to reveal bacterial interactions. In total, there were 666 nodes (ASVs) and 8,217 edges in the co-occurrence network of weathered rocks (Co-NW; Figure 5A), whereas 744 nodes (ASVs) and 19,456 edges in the network for the sediment communities (Co-NS; Figure 5C). Most edges were positively linked (88.9% in Co-NW and 84.64% in Co-NS). Both networks showed good modularity with 6 main modules (Figures 5B,D). Nodes from the same or adjacent sampling sites tend to cluster in the same module. Topologically, Co-NW showed higher modularity (0.808), lower average clustering coefficient (0.549), higher average path length (3.511) than those in Co-NS (0.658, 0.553, 2.949; Table 2). Co-NW was more stable than Co-NS as estimated network stability by average degree.
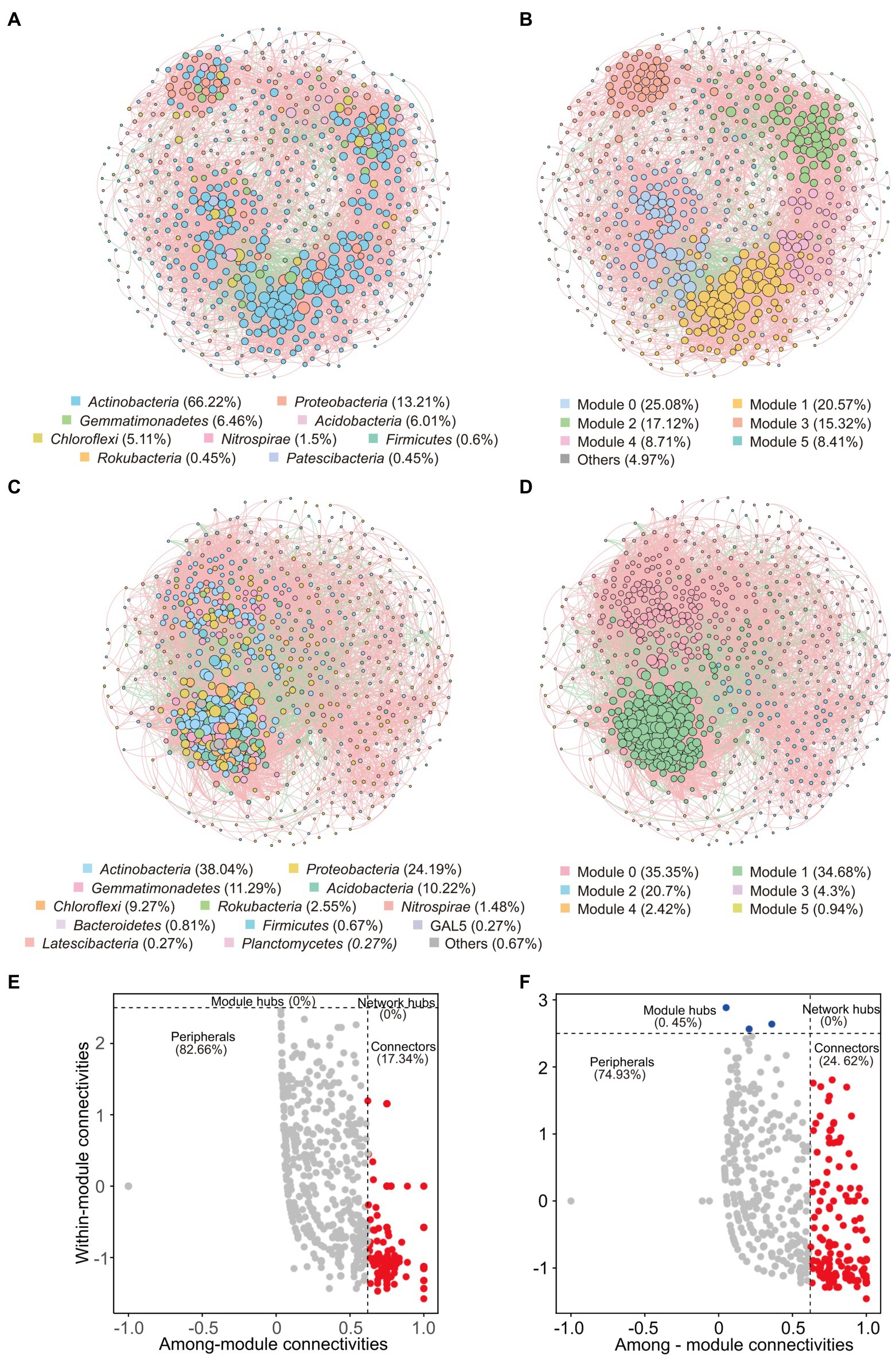
Figure 5. Co-occurrence networks of bacterial communities based on pairwise Spearman’s correlations between ASVs with a coefficient > |0.7| and a p-value < 0.01. The upper panel shows the network of weathered rocks with ASVs colored by taxonomy (A) and modularity (B). The lower panel shows the network of sediments with ASVs colored by taxonomy (C) and modularity (D). The size of each node is proportional to the number of connections. Red lines represent positive correlations and green lines represent negative correlations. Zi-Pi plots showing the distribution of ASVs with their topological roles in bacterial network of weathered rock (E) and sediment (F).
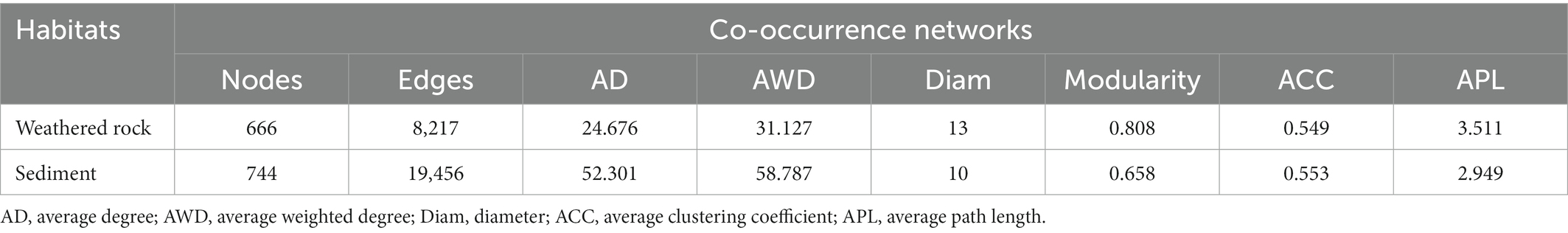
Table 2. Topological indices of co-occurrence networks in the Chang Cave, Hubei province, central China.
Among the nodes, 129/164 keystone taxa were identified as connectors in Co-NW and Co-NS, respectively (Figures 5E,F). The module hubs were only identified in sediments (3 keystone taxa, accounted for 0.45%; Figure 5F). Actinobacteria predominated in all keystone taxa, which accounted for 68.99% and 34.73% of all keystone taxa in Co-NW and Co-NS, respectively. Proteobacteria ranked the second and accounted for 1.88% and 8.41% in Co-NW and Co-NS, respectively. One and three nodes affiliated with NC10 group were identified as connectors in Co-NW and Co-NS, respectively.
3.5. Potential functions of bacterial communities
Bacterial functions were predicted with Tax4Fun based on 16S rRNA gene sequence data. Results showed that bacterial functions in weathered rocks and sediments were significantly different as indicated by the relative abundances of top 30 level-3 KEGG pathways and Wilcoxon rank-sum tests (Figure 6A). “Nitrogen metabolism” showed a high abundance in sediments (p < 0.05), whereas “methane metabolism” and “prokaryotic carbon fixation pathways” were more abundant in weathered rock samples (p < 0.05; Figure 6A). It is worth mentioning that “nitrogen metabolism” with a relative abundance of 1.98% was more abundant than “methane metabolism” (1.38%) and “prokaryotic carbon fixation pathways” (1.03%). This observation was also supported by the results from FARPROTAX functional prediction, which confirmed the relatively high abundance of functions related to nitrogen cycling (Supplementary Figure S4).
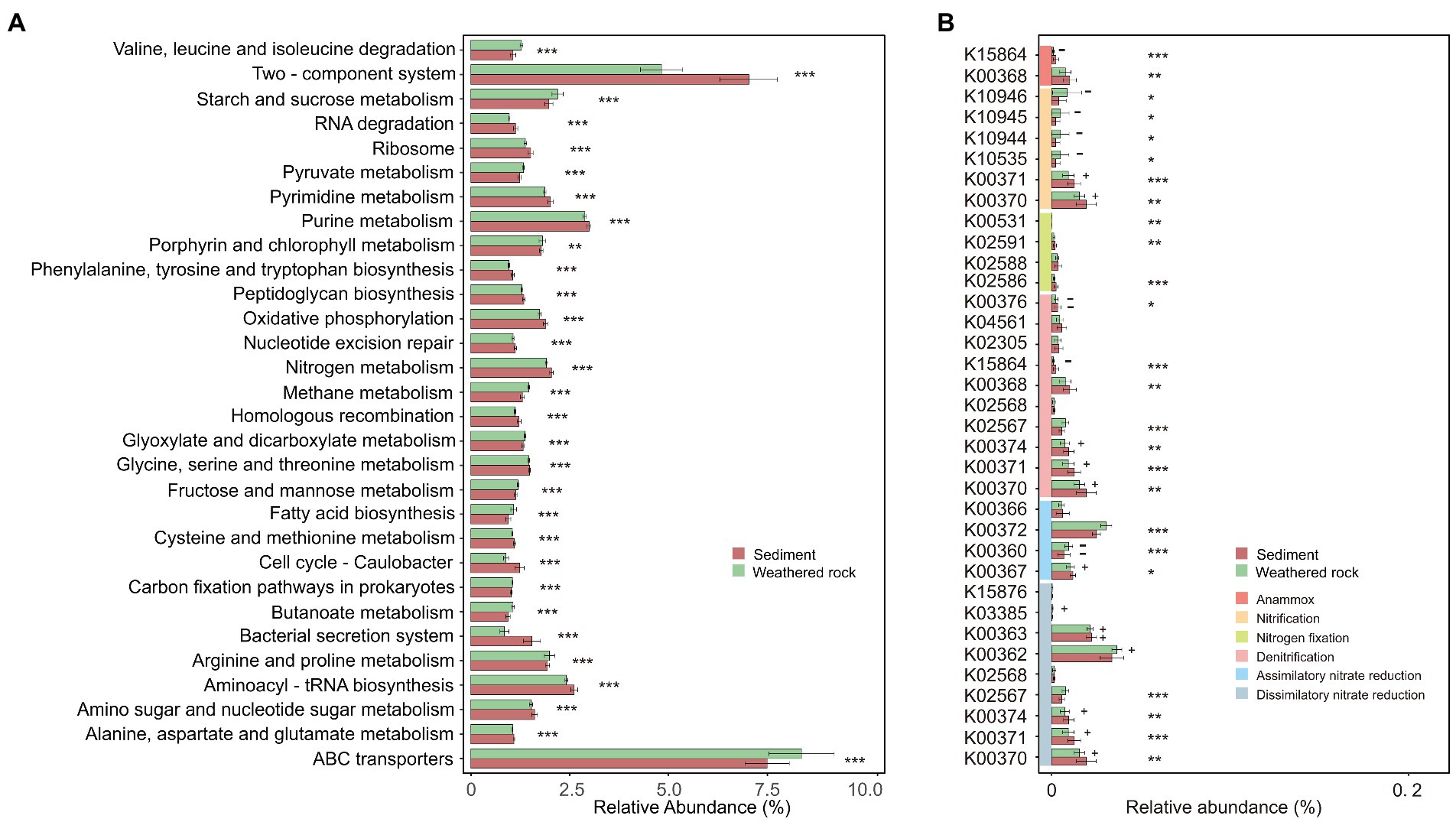
Figure 6. Relative abundance of the top 30 level-3 KEGG pathways (A) and nitrogen metabolism gene prediction (B) for bacterial communities in weathered rocks and sediments. Significance level: p < 0.05, *; p < 0.01, **; p < 0.001, ***. +/−: positively/negatively associated with nitrate.
KEGG functional genes related to “nitrogen metabolism” fell into six categories, which included dissimilatory/assimilatory nitrate reductions, denitrification, nitrogen fixation, nitrification, and anammox (Figure 6B). The nitrite reductase (NADH) [EC:1.7.1.15] had the highest relative abundance, followed by nitrate reductase [EC:1.7.5.1] and nitrite oxidoreductase [1.7.99.-]. The key genes responsible for nitrification and dissimilatory nitrate reduction were negatively and positively correlated with nitrate, respectively, in weathered rocks. Based on the results from FARPROTAX, weathered rocks had a higher abundance of nitrate reduction compared to those in sediments (3.90% and 0.68% in average, respectively). Sediments showed a higher abundance of aerobic ammonia oxidation and nitrification (7.66% and 8.90% in average, respectively) than those in weathered rocks (0.69% and 0.97% in average, respectively; Supplementary Figure S5).
4. Discussion
Following Europe and North America, China has the third largest acid rain region with acid rain mainly in the south of the Yangtze River (Shu et al., 2019). Despite the dominance of sulfate in acid rain, the ratio of SO42−/NO3− decreased by 81.9% in the period of 1998–2018, indicating the increase of NO3− (Xuan et al., 2021). The annual average of wet nitrogen deposition increased from 11.11 to 13.87 kg ha−1 year−1 in China (Jia et al., 2014; Zhu et al., 2015). Dry nitrogen deposition mainly occurred in the North, East and Central China, and the average annual increase of dry deposition in the last decade was 1–2 kg N ha−1 year−1 (Jia et al., 2016). The Chang Cave is located in the acid rain area with high deposition of dry/wet nitrogen, receiving large amounts of exotic nitrate (Supplementary Figure S3; Jia et al., 2016; Yun et al., 2016a). The forest litter in overlying soil above the Chang Cave may also be a nitrate source (Hill, 1981), which can be transferred in to the cave by water along with organic-rich ammonia or ammonium to the dry passages (Pace, 1971; Hill, 1981; Hill et al., 1983). Previous studies have shown that rapid percolation of nitrate-rich water leads to elevated nitrate concentrations in the cavernous limestone (Knox and Moody, 1991; Boyer and Pasquarell, 1996). Other cave nitrate sources include bacterial nitrogen fixation (Faust, 1949, 1968; Lewis, 1992), bat guano (Hill, 1987), ammonium-urea from amberat (cave rat feces and urine; Moore and Sullivan, 1978), fertilizers, and sewage (Hess, 1900; Hill, 1981). Understanding of the impact of nitrate input on the microbial composition and functions in subsurface caves will help elucidate disturbance by human activities of ecological functions in the subsurface biosphere.
4.1. Nitrate impact differently on bacterial communities in weathered rocks and sediments
The application of nitrogen fertilizers is well-known to influence the structure of soil bacterial communities (Fierer et al., 2012a; Zhou et al., 2015). However, the effect of nitrate on microbial communities has often been neglected in previous studies related to karst caves. Nitrate input with acid rain or fertilization in overlying soils may result in the alteration of microbial diversity, microbial composition, and microbial functions in karst caves, which subsequently may have profound feedback on global climate change.
Nitrate contributed most to the distinguished bacterial communities in different habitats (Figures 2D–F) and resulted in unique indicator groups within each habitat. Actinomyces, Crossiella, and Rubrobacteria were indicator groups in weathered rocks, with Betaproteobacteriales, Acidobacteria, Nitrosococcales, Nitrosococcaceae, and Nitrosomonadaceae in sediments. Most indicator groups were related to nitrogen cycling. For example, the indicator phyla/order in weathered rock Actinobacteria is reported to be a major component of nitrate reducers in low-temperature peat and permafrost (Steven et al., 2008; Palmer et al., 2012; Palmer and Horn, 2012). Crossiella, an aerobic, non-motile actinomycete and a nitrate reducer, has already been described in subsurface environments (Portillo et al., 2009). In sediments Betaproteobacteriales are capable of denitrification, ammonia oxidation, nitrite oxidation, and nitrogen assimilation (Kalyuzhnaya et al., 2006). The subgroup_6 of Acidobacteria responds sensitively to high nitrogen availability in wetland soil (Hester et al., 2018). Nitrosococcales, Nitrosococcaceae, and Nitrosomonadaceae are ammonia-oxidizers (Ward et al., 2021). In addition, there were significant changes in the abundance of other bacteria due to the high input of exotic nitrate.
Besides the differences in indicator groups, bacterial communities also significantly differed in the alpha diversity and beta diversity between sediments and weathered rocks as indicated by alpha diversity indexes (Supplementary Figure S2) and NMDS analysis (Figure 2C). The alpha diversity of bacterial communities was higher in sediments than in the weathered rocks in the Chang Cave, consistent with observations in other caves (Ma et al., 2021; Ai et al., 2022). Moreover, our observation of the difference in the beta diversity between weathered rocks and sediments was also reported in other caves in Guilin city (Cao et al., 2021; Yang et al., 2021). These differences in bacterial diversities are likely results from the nitrate impact.
Nitrate may impact on bacterial alpha diversities via indirect or direct pathways in different habitats. In weathered rocks nitrate directly altered pH and thus indirectly impacted on bacterial diversity index (Figure 3A). In contrast, nitrate directly impacted bacterial diversity index in sediments (Figure 3B). Nitrate serves as a nutrient and electron acceptor in oligotrophic ecosystems (Spilde et al., 2005; Barton et al., 2010; Kimble et al., 2018). High nitrogen availability may reduce the abundance of nitrogen-fixing microbes (Berthrong et al., 2014) and stimulate nitrophilous taxa. Nitrifiers and denitrifiers which use inorganic N as energy sources or electron acceptors may competitively exclude other bacterial taxa (Campbell et al., 2010). Karst ecosystems based on chemoautotrophy are limited by the availability of inorganic energy sources (Fe, S, and N especially; Engel, 2007). Input of exogenous NO3− increases the available nutrient content and may promote microbial development including but not limited to those involved in the nitrogen cycling. This presumption was supported by the significant positive correlation between bacterial alpha diversity and nitrate in the sediments (Figures 2B, 3B), a result from the direct impact of nitrate input. Nitrate input may decrease the pH, which primarily alters the composition of soil microbial communities (Zhang et al., 2014; Tian and Niu, 2015). Long-term N deposition at supersaturation decreases the microbial diversity due to soil acidification (Chung et al., 2007; Wang et al., 2018a,b), an indirect impact of nitrate on the bacterial diversity due to lowered pH.
Different from the soil systems, carbonates in karst caves showed strong buffer effect to nitrate input, which result in a slight drop in pH, thus favoring for cave microorganisms exposed to long-term alkaline stress. Bacterial diversity is the highest in neutral soils and decreases as soils become more alkaline (Fierer and Jackson, 2006). Alkaline conditions in karst caves may pose prolonged alkaline stress to microorganisms, and slight decrease in pH may benefit cave microbiota and lead to increasing bacterial diversity. The structural equation model analysis supported the decrease in the pH due to nitrate input, revealing a significant negative correlation between nitrate and pH in weathered rocks (Figures 2A, 3A). The negative correlation between the pH and alpha diversity confirmed that nitrate input increased the bacterial alpha diversity by lowering the pH (Figure 3A).
More genera significantly correlated with nitrate in the weathered rocks compared to sediments with the same criteria (Figure 4), suggesting stronger impact of nitrate on weathered rock. The input of nitrate had different effects on bacteria performing different functions or even a similar function. Except for Lysobacter, Mesorhizobium, and Woeseia, the bacteria in the N-cycle were negatively associated with nitrate, such as Haliangium, MND1, Gaiella, IS-44, Ellin6067, Nitrospira, Nordella, and Novosphingobium. Woeseia, IS-44, Ellin6067, and Nitrospira are involved in ammonia oxidation (Zhao et al., 2020; McCormick et al., 2021; Wang L. et al., 2021), and Haliangium is capable of nitrification–denitrification (Li et al., 2018). Gaiella and MND1 can reduce nitrate to nitrite (Albuquerque et al., 2011; McCormick et al., 2021). Lysobacter, Mesorhizobium, Nordella, and Novosphingobium are diazotrophic (Kaneko et al., 2000; Addison et al., 2007; Iwata et al., 2010; Cao et al., 2022). There are negative correlations between nitrate and bacteria involved in the Fe-Mn cycle, such as the iron-reducing bacteria Acidibacter and TRA3-20 and the manganese-oxidizing bacterium Pedomicrobium (Ridge et al., 2007; Nicomrat et al., 2008; Falagan and Johnson, 2014). Constrictibacter and Stackebrandtia that are responsible for cellulose decomposition and chitobiose degradation, respectively (Kang et al., 2012; Wang et al., 2019; Carrasco and Preston, 2020), showed significantly positive correlations with nitrate possibly resulting from alleviation of nitrogen limitation. Negative correlations between nitrate and Acidobacteriia, Deltaproteobacteria, Gammaproteobacteria, Gemmatimonadetes, and Nitrospira were also confirmed by Spearman’s tests, and Actinobacteria and Chloroflexia were positively associated with nitrate (Supplementary Table S1). Due to the positive and negative responses of microbial groups to nitrate concentration, nitrate input may subsequently alter their relative abundances as well as the bacterial composition and functions. Bacteria in weathered rocks were more impacted by nitrate due to higher contents of nitrate as opposed to those in sediments.
4.2. Bacterial interactions and functions
Consistent with previous studies (Cao et al., 2021; Ma et al., 2021; Yang et al., 2021), nodes in the co-occurrence network of weathered rocks and sediments were mostly positively linked (88.9% and 84.64%, respectively; Figures 5A–D), which suggested a prevalence of collaboration rather than competition to overcome nutrient limitation in oligotrophic caves. The higher modularity of Co-NW indicated that bacterial populations form closer collective structures, in which they mainly cooperated and established mutualistic relationships to maintain the stability of the ecosystem. More connectors were detected in Co-NW than in Co-NS, but module hubs were only detected in the sediments, suggesting a less hub-based and more connected structure in the weathered rocks.
Keystone taxa in networks are considered to play critical ecological roles to sustain the health of the bacterial ecosystems. Most keystone taxa in this study were involved in nitrogen cycling, indicating the fundamental role of nitrogen in subsurface karst ecosystems (Supplementary Table S2). For example, the NC10 group as connectors in this work and previous studies (Cheng et al., 2021b; Yang et al., 2021) has been demonstrated to oxidize CH4 anaerobically coupled with nitrite reduction (Ettwig et al., 2008). This is a clear example the coupling of carbon and nitrogen cycling in the subsurface biosphere. Moreover, this also suggested a potential contribution by NC10 to anaerobic methane oxidation besides aerobic consumption of methane by the Upland Soil Cluster (Cheng et al., 2021a). Wb1-P19 is a genus named after an uncultured clone in cave water, which phylogenetically clusters with sulfur- or nitrite-oxidizing autotrophs (Holmes et al., 2001). Species in Conexibacter, Sphingomonas, Kribbella, Nocardioides, Amycolatopsis, Streptomyces, Actinophytocola, Aeromicrobium, Stackebrandtia, and Rubrobacter can reduce nitrate into nitrite (Pranamuda et al., 1997; Monciardini et al., 2003; Keulen et al., 2005; Labeda and Kroppenstedt, 2005; Prakash and Lal, 2006; Ramasamy et al., 2012; Seki et al., 2012; Suzuki, 2015; Bouznada et al., 2016; Guerrero-Cruz et al., 2019; Curtis et al., 2020). Nitrosospira is a ubiquitous ammonium-oxidizing bacterium (AOB) found in various environments (Norton et al., 2008). The comammox Nitrospira, capable of oxidizing ammonia completely to nitrate (Daims et al., 2015; van Kessel et al., 2015) was recently reported in the Heshang Cave (Ma et al., 2021) and served as connector in Co-NW in the Chang Cave. This expands our knowledge about the distribution of comammox in natural environments. Species in Gemmatimonas were found to reduce N2O (Chee-Sanford et al., 2019). Denitrification and dissimilatory nitrate reduction activities have been observed in Bacillus species (Heylen and Keltjens, 2012). Azotobacter paspali lacks the nitrate reductase enzyme and is able to fix nitrogen in the presence of nitrate (Stephan et al., 1997). The clade RB41 is a group of rhizospheric bacteria, some possibly assigned to Acidobacteria, that promote host plant uptake of nitrogen (Fu et al., 2021). Subgroup_2, Subgroup_6, Subgroup_7, Subgroup_13, and Solirubrobacter are thought to be related to nitrogen transformations (Liu et al., 2017; Gao et al., 2022). Genomic information indicated that Dyella has the potential to perform nitrate reduction (Too et al., 2018), and Ga0077536 is involved in nitrogen fixation (Gonzalez-Pimentel et al., 2021). 67–14 in the order of Solirubrobacterales is reported to be associated with autotrophic CO2 fixation and ferrous iron redox reactions (Rodriguez et al., 2022), commonly found in environments with limited concentrations of TOC (Filippini et al., 2019).
Consistent with the functions of keystone taxa in the co-occurrence networks, high relative gene abundances of nitrogen metabolism were indicated by Tax4Fun2, with the dominance of dissimilatory and assimilatory nitrate reductions (Figures 6A,B). Heatmap analysis also showed the significant enrichment for processes involved in the nitrogen cycle (Supplementary Figure S4). Nitrification is an important pathway in various ecosystems, but the input of exogenous nitrate in weathered rocks may have inhibited nitrification (Figure 6B), reducing the production of N2O as a by-product (Hirsch and Mauchline, 2015). Dissimilatory nitrate reduction to ammonium (DNRA) converts NO3− to NH4+, thus providing additional NH4+ to primary producers (Rütting et al., 2011; Minick et al., 2016; Pandey et al., 2020). DNRA can be coupled with the oxidation of various electron donors such as organic matter, methane, sulfur compounds, H2 or iron (Zumft, 1997; Cardoso et al., 2006; Weber et al., 2006; Haroon et al., 2013; Ettwig et al., 2016). The highest relative abundance of DNRA was found in weathered rocks in our study (Figure 6B). The assimilatory nitrate reduction process reduces NO3− to NH4+ by the nitrate reductase and assimilatory nitrite reductase with NADH as the reducing power. Nitrate reduction was more commonly observed in weathered rocks, whereas aerobic ammonia oxidation and nitrification were prevalent in the sediments (Supplementary Figure S5).
Besides the genes related to N-metabolism, high abundance of genes involved in methane metabolism and carbon fixation were also detected, indicating ecological roles in C1-cycling of cave microorganisms. Methane oxidizing bacteria, particularly the Upland Soil Cluster, responsible for the oxidation of trace levels of methane in the air, have been reported to be widely distributed in caves (Zhao et al., 2018; Cheng et al., 2021a), thus coinciding with our results of function prediction. In addition to USC, ASVs affiliated NC10 capable of coupling anaerobic oxidation of methane with nitrite reduction may also contribute to C1 and N cycles in the cave. This provides another clue to investigate the ecological function of cave as methane sink, and merits further studies. Genes involved in methane oxidation were most likely inhibited by exogenous nitrate input in the cave, especially in the weathered rock (Supplementary Table S4). Furthermore, negative correlations between nitrate concentration and the abundance of Gammaproteobacteria (including low-affinity methanotrophs) and NC10 were found (Mills et al., 2013; Webster et al., 2022; Supplementary Table S4). High-affinity methanotrophs, known as the Upland Soil Cluster (USC) α and γ clades, may be responsible for atmospheric-CH4 consumption in caves (Zhao et al., 2018; Cheng et al., 2021a), and their response to nitrate input is not known. Further study is needed to determine the specific effect of nitrate input on cave methane fluxes. Carbon fixation pathways were dominated by the reductive acetyl-CoA pathway (Wood-Ljungdahl pathway) and 3-hydroxypropionate bicycle (Supplementary Figure S6). The reductive acetyl-CoA pathway conserves energy from CO2 or C1 compounds, and is considered to be the primary route of CO2 fixation in Lava caves (Selensky et al., 2021). In karst caves, the high concentrations of CO2 and HCO3− and limited amount of organic matter derived from photosynthesis favor C1 fixation via the Calvin-Benson cycle. The 3-hydroxypropionate bicycle has been found in Chloroflexus (Hugler et al., 2002; Lavoie et al., 2017). Key enzymes were identified involving in reductive pentose phosphate cycle (Calvin cycle), reductive tricarboxylic acid (TCA) cycle, reductive acetyl-CoA pathway (Wood-Ljungdahl pathway), 3-hydroxypropionate bicycle, 3-hydroxypropionate/4-hydroxybutylate cycle, and dicarboxylate/4-hydroxybutyrate cycle (Supplementary Table S3). The absence of propionyl-CoA synthase in 3-hydroxypropionate bicycle serves as additional evidence to support autotrophic bacteria as the main primary producers in karst caves (Cañveras et al., 2001; Parker et al., 2013; Jones and Macalady, 2016; Dong et al., 2020). Nitrate was positively correlated with genes responsible for the Calvin-Benson cycle (Supplementary Table S3), employed by numerous groups of autotrophic bacteria such as H2-, Fe2+-, S-, and NH4+-oxidizers (Raven, 2009; Claassens et al., 2018). In contrast, nitrate was negatively correlated with the TCA cycle (Supplementary Table S3). The 3-hydroxypropionate bicycle genes were positively associated with nitrate in carbon fixation pathways in the Chang Cave, consistent with Chloroflexia abundance being enhanced by nitrate input (Supplementary Table S1). In contrast, genes of the reductive citrate cycle (Arnon-Buchanan cycle) were negatively associated with nitrate in the Chang cave (Supplementary Table S4). To be note, partially sequenced 16S rRNA genes fail to distinguish taxa beyond the genus level (Heidrich and Beule, 2022), and functional prediction accuracy depends on the size and research area of the reference gene database (Mongad et al., 2021). Uncertainty dose exist in functional predictions based on the partially sequenced 16S rRNA gene sequence data (Mongad et al., 2021; Heidrich and Beule, 2022), which can be overcome via metagenome sequencing in near future.
5. Conclusion
This study revealed highly diverse bacterial communities in the Chang Cave with strong habitat specialization. For the first time, we demonstrated the impact of nitrate on the composition, diversity, interaction and function of bacterial communities in karst caves. Nitrate shaped bacterial communities across different habitats with an explanation of 27.2%. It increased the bacterial alpha diversity directly in the sediments and indirectly in weathered rocks via lowering the pH. Nitrate also directly increased the beta diversity of bacterial communities, resulting in high habitat specialization. More genera were significantly correlated with nitrate in the weathered rocks than in the sediment samples, suggesting a strong impact of nitrate on weathered rocks. Keystone taxa involved in nitrogen cycling were detected such as Rokubacteriales belonging to NC10, capable of nitrite reduction coupling with anaerobic methane oxidation, and comammox Nitrospira, completely oxidizing ammonia to nitrate, and various nitrate-reducers. Results of function prediction also confirmed the dominance of the genes related to nitrogen metabolism followed by those in methane metabolism and carbon fixation. Elevated nitrate concentration may also have shifted the pathway of carbon fixation in caves, enhancing the Calvin-Benson cycle and 3-hydroxypropionate bicycle, but inhibiting the reductive tricarboxylic acid and reductive citrate cycles. The results enhance our understanding on the N-cycling in karst caves and offer a new window to study the impact of anthropogenic activities on subsurface biosphere and ecological functions of caves in terms of nitrogen and carbon cycling under the context of global change.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/, SRR22513012–SRR22513071.
Author contributions
XLiu, HW, and WW contributed to conception and design of the study. XLiu organized the database and performed the statistical analysis. WW, XC, and YW helped data mining. XLiu wrote the first draft of the manuscript. LL, QL, LM, and XLu wrote sections of the manuscript. HW and OT finalized the manuscript. All authors contributed to the manuscript revision, read, and approved the submitted version.
Funding
This work was supported by National Natural Science Foundation of China (no. 91951208).
Acknowledgments
We are grateful to Junhua Huang, Hongbin Zhang, and Shuyu Xue from China University of Geosciences (Wuhan) for providing the climatic description of the Chang Cave.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The handling editor YS declared a shared affiliation with the authors XLiu, HW, WW, XC, YW, QL, LL, LM, and XLu at the time of review.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1115449/full#supplementary-material
Footnotes
References
Addison, S. L., Foote, S. M., Reid, N. M., and Lloyd-Jones, G. (2007). Novosphingobium nitrogenifigens sp. nov., a polyhydroxyalkanoate-accumulating diazotroph isolated from a New Zealand pulp and paper wastewater. Int. J. Syst. Evol. Microbiol. 57, 2467–2471. doi: 10.1099/ijs.0.64627-0
Ai, J., Guo, J., Li, Y., Zhong, X., Lv, Y., Li, J., et al. (2022). The diversity of microbes and prediction of their functions in karst caves under the influence of human tourism activities-a case study of Zhijin cave in Southwest China. Environ. Sci. Pollut. Res. Int. 29, 25858–25868. doi: 10.1007/s11356-021-17783-x
Albuquerque, L., Franca, L., Rainey, F. A., Schumann, P., Nobre, M. F., and da Costa, M. S. (2011). Gaiella occulta gen. Nov., sp. nov., a novel representative of a deep branching phylogenetic lineage within the class Actinobacteria and proposal of Gaiellaceae fam. Nov. and Gaiellales Ord. Nov. Syst. Appl. Microbiol. 34, 595–599. doi: 10.1016/j.syapm.2011.07.001
Barton, H. A., Taylor, M. R., and Pace, N. R. (2010). Molecular phylogenetic analysis of a bacterial community in an oligotrophic cave environment. Geomicrobiol J. 21, 11–20. doi: 10.1080/01490450490253428
Bastian, M., Heymann, S., and Jacomy, M. (2009). "Gephi: an open source software for exploring and manipulating networks," in: Proceedings of the international AAAI conference on web and social media. Vol. 3. 361–362.
Berthrong, S. T., Yeager, C. M., Gallegos-Graves, L., Steven, B., Eichorst, S. A., Jackson, R. B., et al. (2014). Nitrogen fertilization has a stronger effect on soil nitrogen-fixing bacterial communities than elevated atmospheric CO2. Appl. Environ. Microbiol. 80, 3103–3112. doi: 10.1128/AEM.04034-13
Bobbink, R., Hornung, M., and Roelofs, J. M. (1998). The effects of air-borne nitrogen pollutants on species diversity in natural and semi-natural European vegetation. J. Ecol. 86, 717–738. doi: 10.1046/j.1365-2745.1998.8650717.x
Bodelier, P. L. E., and Laanbroek, H. J. (2004). Nitrogen as a regulatory factor of methane oxidation in soils and sediments. FEMS Microbiol. Ecol. 47, 265–277. doi: 10.1016/s0168-6496(03)00304-0
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Bouznada, K., Bouras, N., Schumann, P., Sproer, C., Sabaou, N., and Klenk, H. P. (2016). Actinophytocola algeriensis sp. nov., an actinobacterium isolated from Saharan soil. Int. J. Syst. Evol. Microbiol. 66, 2760–2765. doi: 10.1099/ijsem.0.001136
Boyer, D. G., and Pasquarell, G. C. (1996). Agricultural land use effects on nitrate concentrations in a mature karst aquifer. JAWRA J. Am. Water Resources Association 32, 565–573. doi: 10.1111/j.1752-1688.1996.tb04054.x
Brenzinger, K., Dorsch, P., and Braker, G. (2015). pH-driven shifts in overall and transcriptionally active denitrifiers control gaseous product stoichiometry in growth experiments with extracted bacteria from soil. Front. Microbiol. 6:961. doi: 10.3389/fmicb.2015.00961
Campbell, B. J., Polson, S. W., Hanson, T. E., Mack, M. C., and Schuur, E. A. (2010). The effect of nutrient deposition on bacterial communities in Arctic tundra soil. Environ. Microbiol. 12, 1842–1854. doi: 10.1111/j.1462-2920.2010.02189.x
Cañveras, C., Sanchez-Moral, S. V., Sloer, C., and Saiz-Jimenez, J. (2001). Microorganisms and microbially induced fabrics in cave walls. Geomicrobiol J. 18, 223–240. doi: 10.1080/01490450152467769
Cao, J., Cheng, X., Zeng, Z., Yang, Z., Liu, X., Wang, H., et al. (2021). Habitat specificity and co-occurrence network of bacterial communities in the Xincuntun cave, Guilin, Guangxi. Chin. Sci. Bull. 66, 4003–4016. doi: 10.1360/tb-2021-0021
Cao, P., Wei, X., Wang, G., Chen, X., Han, J., and Li, Y. (2022). Microbial inoculants and garbage fermentation liquid reduced root-knot nematode disease and as uptake in Panax quinquefolium cultivation by modulating rhizosphere microbiota community. Chin. Herb. Med. 14, 58–69. doi: 10.1016/j.chmed.2021.11.001
Cardoso, R. B., Sierra-Alvarez, R., Rowlette, P., Flores, E. R., Gomez, J., and Field, J. A. (2006). Sulfide oxidation under chemolithoautotrophic denitrifying conditions. Biotechnol. Bioeng. 95, 1148–1157. doi: 10.1002/bit.21084
Carrasco, J., and Preston, G. M. (2020). Growing edible mushrooms: a conversation between bacteria and fungi. Environ. Microbiol. 22, 858–872. doi: 10.1111/1462-2920.14765
Che, R., Deng, Y., Wang, W., Rui, Y., Zhang, J., Tahmasbian, I., et al. (2018). Long-term warming rather than grazing significantly changed total and active soil procaryotic community structures. Geoderma 316, 1–10. doi: 10.1016/j.geoderma.2017.12.005
Chee-Sanford, J., Tian, D., and Sanford, R. (2019). Consumption of N2O and other N-cycle intermediates by Gemmatimonas aurantiaca strain T-27. Microbiology 165, 1345–1354. doi: 10.1099/mic.0.000847
Cheng, X., Liu, X., Wang, H., Su, C., Zhao, R., Bodelier, P. L. E., et al. (2021a). USCgamma dominated community composition and cooccurrence network of methanotrophs and bacteria in subterranean karst caves. Microbiol. Spectr. 9:e0082021. doi: 10.1128/Spectrum.00820-21
Cheng, X., Yun, Y., Wang, H., Ma, L., Tian, W., Man, B., et al. (2021b). Contrasting bacterial communities and their assembly processes in karst soils under different land use. Sci. Total Environ. 751:142263. doi: 10.1016/j.scitotenv.2020.142263
Chung, H., Zak, D. R., Reich, P. B., and Ellsworth, D. S. (2007). Plant species richness, elevated CO2, and atmospheric nitrogen deposition alter soil microbial community composition and function. Glogal Change Biol. 13, 980–989. doi: 10.1111/j.1365-2486.2007.01313.x
Claassens, N. J., Sanchez-Andrea, I., Sousa, D. Z., and Bar-Even, A. (2018). Towards sustainable feedstocks: a guide to electron donors for microbial carbon fixation. Curr. Opin. Biotechnol. 50, 195–205. doi: 10.1016/j.copbio.2018.01.019
Curtis, S. M., Norton, I., Everest, G. J., Pelser, J. G., de Kock, M. C., and Meyers, P. R. (2020). Development of a Kribbella-specific isolation medium and description of Kribbella capetownensis sp. nov. and Kribbella speibonae sp. nov., isolated from soil. Antonie Van Leeuwenhoek 113, 617–628. doi: 10.1007/s10482-019-01365-6
Daims, H., Lebedeva, E. V., Pjevac, P., Han, P., Herbold, C., Albertsen, M., et al. (2015). Complete nitrification by Nitrospira bacteria. Nature 528, 504–509. doi: 10.1038/nature16461
DeSantis, T. Z., Hugenholtz, P., Larsen, N., Rojas, M., Brodie, E. L., Keller, K., et al. (2006). Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72, 5069–5072. doi: 10.1128/AEM.03006-05
Dong, Y., Gao, J., Wu, Q., Ai, Y., Huang, Y., Wei, W., et al. (2020). Co-occurrence pattern and function prediction of bacterial community in karst cave. BMC Microbiol. 20:137. doi: 10.1186/s12866-020-01806-7
Dunfield, P., and Knowles, R. (1995). Kinetics of inhibition of methane oxidation by nitrate, nitrite, and ammonium in a humisol. Appl. Environ. Microbiol. 61, 3129–3135. doi: 10.1128/AEM.61.8.3129-3135.1995
Engel, A. S. (2007). Observations on the biodiversity of sulfidic karst habitats. J. Cave Karst Studies 9, 187–206.
Ettwig, K. F., Shima, S., van de Pas-Schoonen, K. T., Kahnt, J., Medema, M. H., Op Den Camp, H. J. M., et al. (2008). Denitrifying bacteria anaerobically oxidize methane in the absence of Archaea. Environ. Microbiol. 10, 3164–3173. doi: 10.1111/j.1462-2920.2008.01724.x
Ettwig, K. F., Zhu, B., Speth, D., Keltjens, J. T., Jetten, M. S. M., and Kartal, B. (2016). Archaea catalyze iron-dependent anaerobic oxidation of methane. Proc. Natl. Acad. Sci. U. S. A. 113, 12792–12796. doi: 10.1073/pnas.1609534113
Falagan, C., and Johnson, D. B. (2014). Acidibacter ferrireducens gen. Nov., sp. nov.: an acidophilic ferric iron-reducing gammaproteobacterium. Extremophiles 18, 1067–1073. doi: 10.1007/s00792-014-0684-3
Fan, K., Weisenhorn, P., Gilbert, J. A., and Chu, H. (2018). Wheat rhizosphere harbors a less complex and more stable microbial co-occurrence pattern than bulk soil. Soil Biol. Biochem. 125, 251–260. doi: 10.1016/j.soilbio.2018.07.022
Fierer, N., and Jackson, R. B. (2006). The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. U. S. A. 103, 626–631. doi: 10.1073/pnas.0507535103
Fierer, N., Lauber, C. L., Ramirez, K. S., Zaneveld, J., Bradford, M. A., and Knight, R. (2012a). Comparative metagenomic, phylogenetic and physiological analyses of soil microbial communities across nitrogen gradients. ISME J. 6, 1007–1017. doi: 10.1038/ismej.2011.159
Fierer, N., Leff, J. W., Adams, B. J., Nielsen, U. N., Bates, S. T., Lauber, C. L., et al. (2012b). Cross-biome metagenomic analyses of soil microbial communities and their functional attributes. Proc. Natl. Acad. Sci. U. S. A. 109, 21390–21395. doi: 10.1073/pnas.1215210110
Filippini, G., Bugnot, A. B., Johnston, E. L., Ruszczyk, J., Potts, J., Scanes, P., et al. (2019). Sediment bacterial communities associated with environmental factors in intermittently closed and Open Lakes and lagoons (ICOLLs). Sci. Total Environ. 693:133462. doi: 10.1016/j.scitotenv.2019.07.268
Fu, Y., Kumar, A., Chen, L., Jiang, Y., Ling, N., Wang, R., et al. (2021). Rhizosphere microbiome modulated effects of biochar on ryegrass 15N uptake and rhizodeposited 13C allocation in soil. Plant Soil 463, 359–377. doi: 10.1007/s11104-021-04845-9
Gao, Y., Yuan, L., Du, J., Wang, H., Yang, X., Duan, L., et al. (2022). Bacterial community profile of the crude oil-contaminated saline soil in the Yellow River Delta natural reserve, China. Chemosphere 289:133207. doi: 10.1016/j.chemosphere.2021.133207
Gonzalez-Pimentel, J. L., Martin-Pozas, T., Jurado, V., Miller, A. Z., Caldeira, A. T., Fernandez-Lorenzo, O., et al. (2021). Prokaryotic communities from a lava tube cave in La Palma Island (Spain) are involved in the biogeochemical cycle of major elements. PeerJ 9:e11386. doi: 10.7717/peerj.11386
Guerrero-Cruz, S., Stultiens, K., van Kessel, M. A. H. J., Versantvoort, W., Jetten, M. S. M., Op den Camp, H. J. M., et al. (2019). Key physiology of a nitrite-dependent methane-oxidizing enrichment culture. Environ. Microbiol. 85:e00124-19. doi: 10.1128/AEM.00124-19
Haroon, M. F., Hu, S., Shi, Y., Imelfort, M., Keller, J., Hugenholtz, P., et al. (2013). Anaerobic oxidation of methane coupled to nitrate reduction in a novel archaeal lineage. Nature 500, 567–570. doi: 10.1038/nature12375
Heidrich, V., and Beule, L. (2022). Are short-read amplicons suitable for the prediction of microbiome functional potential? A critical perspective. iMeta 1:e38. doi: 10.1002/imt2.38
Hess, W. H. (1900). The origin of nitrates in cavern earths. J. Geol. 8, 129–134. doi: 10.1086/620781
Hester, E. R., Harpenslager, S. F., Diggelen, J. M. H. V., Lamers, L. L., Jetten, M. S. M., Lüke, C., et al. (2018). Linking nitrogen load to the structure and function of wetland soil and Rhizosphere microbial communities. mSystems 3, e00214–e00217. doi: 10.1128/mSystems.00214-17
Heylen, K., and Keltjens, J. (2012). Redundancy and modularity in membrane-associated dissimilatory nitrate reduction in bacillus. Front. Microbiol. 3:371. doi: 10.3389/fmicb.2012.00371
Hill, C. A. (1987). Geology of Carlsbad cavern and other caves in the Guadalupe Mountains, New Mexico and Texas New Mexico Bur Mines Miner Resour Bull 117. Socorro, NM: New Mexico Bureau of Mines and Mineral Resources. 150.
Hill, C., Eller, P., Fliermans, C., and Hauer, P. (1983). Saltpeter conversion and the origin of cave nitrates. Nat. Geogr. Soc. Res. Rep. 15, 295–309.
Hirsch, P. R., and Mauchline, T. H. (2015). The importance of the microbial N cycle in soil for crop plant nutrition. Adv. Appl. Microbiol. 93, 45–71. doi: 10.1016/bs.aambs.2015.09.001
Holmes, A. J., Tujula, N. A., Holley, M., Contos, A., James, J. M., Rogers, P., et al. (2001). Phylogenetic structure of unusual aquatic microbial formations in Nullarbor caves, Australia. Environ. Microbiol. 3, 256–264. doi: 10.1046/j.1462-2920.2001.00187.x
Hu, Y., Chen, M., Yang, Z., Cong, M., Zhu, X., and Jia, H. (2021). Soil microbial community response to nitrogen application on a swamp meadow in the arid region of Central Asia. Front. Microbiol. 12:797306. doi: 10.3389/fmicb.2021.797306
Hugler, M., Menendez, C., Schagger, H., and Fuchs, G. (2002). Malonyl-coenzyme a reductase from Chloroflexus aurantiacus, a key enzyme of the 3-hydroxypropionate cycle for autotrophic CO2 fixation. J. Bacteriol. 184, 2404–2410. doi: 10.1128/JB.184.9.2404-2410.2002
Iwata, K., Azlan, A., Yamakawa, H., and Omori, T. (2010). Ammonia accumulation in culture broth by the novel nitrogen-fixing bacterium, Lysobacter sp. E4. J. Biosci. Bioeng. 110, 415–418. doi: 10.1016/j.jbiosc.2010.05.006
Jia, Y., Yu, G., Gao, Y., He, N., Wang, Q., Jiao, C., et al. (2016). Global inorganic nitrogen dry deposition inferred from ground- and space-based measurements. Sci. Rep. 6:19810. doi: 10.1038/srep19810
Jia, Y., Yu, G., He, N., Zhan, X., Fang, H., Sheng, W., et al. (2014). Spatial and decadal variations in inorganic nitrogen wet deposition in China induced by human activity. Sci. Rep. 4:3763. doi: 10.1038/srep03763
Jones, D. S., and Macalady, J. L. (2016). “The snotty and the stringy: energy for subsurface life in caves” in Their world: A diversity of microbial environments. ed. C. J. Hurst (Cham: Springer International Publishing), 203–224.
Kalyuzhnaya, M. G., Bowerman, S., Lara, J. C., Lidstrom, M. E., and Chistoserdova, L. (2006). Methylotenera mobilis gen. Nov., sp. nov., an obligately methylamine-utilizing bacterium within the family Methylophilaceae. Int. J. Syst. Evol. Microbiol. 56, 2819–2823. doi: 10.1099/ijs.0.64191-0
Kaneko, T., Nakamura, Y., Sato, S., Asamizu, E., Kato, T., Sasamoto, S., et al. (2000). Complete genome structure of the nitrogen-fixing symbiotic bacterium Mesorhizobium loti. DNA Res. 7, 331–338. doi: 10.1093/dnares/7.6.331
Kang, S. M., Khan, A. L., Hamayun, M., Hussain, J., Joo, G. J., You, Y. H., et al. (2012). Gibberellin-producing Promicromonospora sp. SE188 improves Solanum lycopersicum plant growth and influences endogenous plant hormones. J. Microbiol. 50, 902–909. doi: 10.1007/s12275-012-2273-4
Kato, S., Shibuya, T., Takaki, Y., Hirai, M., Nunoura, T., and Suzuki, K. (2018). Genome-enabled metabolic reconstruction of dominant chemosynthetic colonizers in deep-sea massive sulfide deposits. Environ. Microbiol. 20, 862–877. doi: 10.1111/1462-2920.14032
Keulen, G. V., Alderson, J., White, J., and Sawers, R. G. (2005). Nitrate respiration in the actinomycete Streptomyces coelicolor. Biochem. Soci Trans. 33, 210–212. doi: 10.1042/BST0330210
Kimble, J. C., Winter, A. S., Spilde, M. N., Sinsabaugh, R. L., and Northup, D. E. (2018). A potential central role of Thaumarchaeota in N-cycling in a semi-arid environment, Fort Stanton cave, Snowy River passage, New Mexico, USA. FEMS Microbiol. Ecol. 94:fiy173. doi: 10.1093/femsec/fiy173
King, G. M., and Schnell, S. (1998). Effects of ammonium and non-ammonium salt additions on methane oxidation by Methylosinus trichosporium OB3b and Maine forest soils. Appl. Environ. Microbiol. 64, 253–257. doi: 10.1128/AEM.64.1.253-257.1998
Knox, E., and Moody, D. (1991). “Influence of hydrology, soil properties, and agricultural land use on nitrogen in groundwater,” in Managing Nitrogen for Groundwater Quality and Farm Profitability. eds. R. F. Follett, D. R. Keeney, and R. M. Cruse (Madison, Wis, USA: Soil Science Society of America), 19–57.
Labeda, D. P., and Kroppenstedt, R. M. (2005). Stackebrandtia nassauensis gen. Nov., sp. nov. and emended description of the family Glycomycetaceae. Int. J. Syst. Evol. Microbiol. 55, 1687–1691. doi: 10.1099/ijs.0.63496-0
Lavoie, K. H., Winter, A. S., Read, K. J., Hughes, E. M., Spilde, M. N., and Northup, D. E. (2017). Comparison of bacterial communities from lava cave microbial mats to overlying surface soils from lava beds National Monument, USA. PLoS One 12:e0169339. doi: 10.1371/journal.pone.0169339
Lefcheck, J. S., and Freckleton, R. (2015). PIECEWISESEM: piecewise structural equation modelling in R for ecology, evolution, and systematics. Methods Ecol. Evol. 7, 573–579. doi: 10.1111/2041-210x.12512
Lewis, W. C. (1992). On the origin of cave saltpeter: a second opinion. Natl. Speleological Soc. Bull. 54, 28–30.
Li, L., Dong, Y., Qian, G., Hu, X., and Ye, L. (2018). Performance and microbial community analysis of bio-electrocoagulation on simultaneous nitrification and denitrification in submerged membrane bioreactor at limited dissolved oxygen. Bioresour. Technol. 258, 168–176. doi: 10.1016/j.biortech.2018.02.121
Liao, J., Hu, C., Wang, M., Li, X., Ruan, J., Zhu, Y., et al. (2018). Assessing acid rain and climate effects on the temporal variation of dissolved organic matter in the unsaturated zone of a karstic system from southern China. J. Hydrol. 556, 475–487. doi: 10.1016/j.jhydrol.2017.11.043
Liu, C., Dong, Y., Hou, L., Deng, N., and Jiao, R. (2017). Acidobacteria communityresponses to nitrogen dose and form in Chinese fir plantations in southern China. Curr. Microbiol. 74, 396–403. doi: 10.1007/s00284-016-1192-8
Liu, L., and Greaver, T. L. (2009). A review of nitrogen enrichment effects on three biogenic GHGs: the CO2 sink may be largely offset by stimulated N2O and CH4 emission. Ecol. Lett. 12, 1103–1117. doi: 10.1111/j.1461-0248.2009.01351.x
Liu, C., Jiang, Y., Tao, F., Lang, Y., and Li, S. (2008). Chemical weathering of carbonate rocks by sulfuric acid and the carbon cycling in Southwest China. Geochimica 4, 404–414. doi: 10.19700/j.0379-1726.2008.04.012
Liu, J., Zhu, S., Liu, X., Yao, P., Ge, T., and Zhang, X. H. (2020). Spatiotemporal dynamics of the archaeal community in coastal sediments: assembly process and co-occurrence relationship. ISME J. 14, 1463–1478. doi: 10.1038/s41396-020-0621-7
Louca, S., Parfrey, L. W., and Doebeli, M. (2016). Decoupling function and taxonomy in the global ocean microbiome. Science 353, 1272–1277. doi: 10.1126/science.aaf4507
Ma, L., Huang, X., Wang, H., Cheng, X., Liu, D., Lu, X., et al. (2021). Microbial interactions drive distinct taxonomic and potential metabolic responses to habitats in karst cave ecosystem. Microbiol. Spectr. 9, e01152–e01121. doi: 10.1128/Spectrum.01152-21
McCormick, N. E., Earle, M., Ha, C., Hakes, L., Evans, A., Anderson, L., et al. (2021). Biological and physico-chemical mechanisms accelerating the acclimation of Mn-removing biofilters. Water Res. 207:117793. doi: 10.1016/j.watres.2021.117793
Mills, C. T., Slater, G. F., Dias, R. F., Carr, S. A., Reddy, C. M., Schmidt, R., et al. (2013). The relative contribution of methanotrophs to microbial communities and carbon cycling in soil overlying a coal-bed methane seep. FEMS Microbiol. Ecol. 84, 474–494. doi: 10.1111/1574-6941.12079
Minick, K. J., Pandey, C. B., Fox, T. R., and Subedi, S. (2016). Dissimilatory nitrate reduction to ammonium and N2O flux: effect of soil redox potential and N fertilization in loblolly pine forests. Biol. Fertil. Soils 52, 601–614. doi: 10.1007/s00374-016-1098-4
Monciardini, P., Cavaletti, L., Schumann, P., Rohde, M., and Donadio, S. (2003). Conexibacter woesei gen. Nov., sp. nov., a novel representative of a deep evolutionary line of descent within the class Actinobacteria. Int. J. Syst. Evol. Microbiol. 53, 569–576. doi: 10.1099/ijs.0.02400-0
Mongad, D. S., Chavan, N. S., Narwade, N. P., Dixit, K., Shouche, Y. S., and Dhotre, D. P. (2021). MicFunPred: a conserved approach to predict functional profiles from 16S rRNA gene sequence data. Genomics 113, 3635–3643. doi: 10.1016/j.ygeno.2021.08.016
Moore, G., and Sullivan, G. (1978). "Speleology: the study of caves: Rev 2nd ed. St. Louis, MO". Cave Books, Inc.
Nicomrat, D., Dick, W. A., Dopson, M., and Tuovinen, O. H. (2008). Bacterial phylogenetic diversity in a constructed wetland system treating acid coal mine drainage. Soil Biol. Biochem. 40, 312–321. doi: 10.1016/j.soilbio.2007.08.009
Nie, Y., Wang, M., Zhang, W., Ni, Z., Hashidoko, Y., and Shen, W. (2018). Ammonium nitrogen content is a dominant predictor of bacterial community composition in an acidic forest soil with exogenous nitrogen enrichment. Sci. Total Environ. 624, 407–415. doi: 10.1016/j.scitotenv.2017.12.142
Norton, J. M., Klotz, M. G., Stein, L. Y., Arp, D. J., Bottomley, P. J., Chain, P. S., et al. (2008). Complete genome sequence of Nitrosospira multiformis, an ammonia-oxidizing bacterium from the soil environment. Appl. Environ. Microbiol. 74, 3559–3572. doi: 10.1128/AEM.02722-07
Oksanen, J., Blanchet, F., Friendly, M., Kindt, R., Legendre, P., McGlinn, D., et al. (2018). Vegan: Community ecology package. R package version 2.5-2. 2018. Vienna: R Core Team.
Olesen, J. M., Bascompte, J., Dupont, Y. L., and Jordano, P. (2007). The modularity of pollination networks. Proc. Natl. Acad. Sci. U. S. A. 104, 19891–19896. doi: 10.1073/pnas.0706375104
Ortiz, M., Legatzki, A., Neilson, J. W., Fryslie, B., Nelson, W. M., Wing, R. A., et al. (2014). Making a living while starving in the dark: metagenomic insights into the energy dynamics of a carbonate cave. ISME J. 8, 478–491. doi: 10.1038/ismej.2013.159
Pace, N. (1971). Caves and saltpeter: a novel hypothesis for saltpeter formation. Caving in the Rockies. 13, 7–9.
Palmer, K., Biasi, C., and Horn, M. A. (2012). Contrasting denitrifier communities relate to contrasting N2O emission patterns from acidic peat soils in arctic tundra. ISME J. 6, 1058–1077. doi: 10.1038/ismej.2011.172
Palmer, K., and Horn, M. A. (2012). Actinobacterial nitrate reducers and proteobacterial denitrifiers are abundant in N2O-metabolizing palsa peat. Appl. Environ. Microbiol. 78, 5584–5596. doi: 10.1128/AEM.00810-12
Pandey, C. B., Kumar, U., Kaviraj, M., Minick, K. J., Mishra, A. K., and Singh, J. S. (2020). DNRA: a short-circuit in biological N-cycling to conserve nitrogen in terrestrial ecosystems. Sci. Total Environ. 738:139710. doi: 10.1016/j.scitotenv.2020.139710
Parker, C., Wolf, J., Auler, A., Barton, H., and Senko, J. (2013). Microbial reducibility of Fe(III) phases associated with the genesis of iron ore caves in the iron quadrangle, Minas Gerais, Brazil. Minerals 3, 395–411. doi: 10.3390/min3040395
Parks, D. H., Tyson, G. W., Hugenholtz, P., and Beiko, R. G. (2014). STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics 30, 3123–3124. doi: 10.1093/bioinformatics/btu494
Portillo, M. C., Alloza, R., and Gonzalez, J. M. (2009). Three different phototrophic microbial communities colonizing a single natural shelter containing prehistoric paintings. Sci. Total Environ. 407, 4876–4881. doi: 10.1016/j.scitotenv.2009.05.038
Prakash, O., and Lal, R. (2006). Description of Sphingobium fuliginis sp. nov., a phenanthrene-degrading bacterium from a fly ash dumping site, and reclassification of Sphingomonas cloacae as Sphingobium cloacae comb. nov. Int. J. Syst. Evol. Microbiol. 56, 2147–2152. doi: 10.1099/ijs.0.64080-0
Pranamuda, H., Tokiwa, Y., and Tanaka, H. (1997). Polylactide degradation by an Amycolatopsis sp. Appl. Environ. Microbiol. 63, 1637–1640. doi: 10.1128/AEM.63.4.1637-1640.1997
R Core Team (2020). R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing.
Ramasamy, D., Kokcha, S., Lagier, J. C., Nguyen, T. T., Raoult, D., and Fournier, P. E. (2012). Genome sequence and description of Aeromicrobium massiliense sp. nov. Stand. Genomic Sci. 7, 246–257. doi: 10.4056/sigs.3306717
Raven, J. A. (2009). Contributions of anoxygenic and oxygenic phototrophy and chemolithotrophy to carbon and oxygen fluxes in aquatic environments. Aquat. Microb. Ecol. 56, 177–192. doi: 10.3354/ame01315
Raza, S., Miao, N., Wang, P., Ju, X., Chen, Z., Zhou, J., et al. (2020). Dramatic loss of inorganic carbon by nitrogen-induced soil acidification in Chinese croplands. Glob. Chang. Biol. 26, 3738–3751. doi: 10.1111/gcb.15101
Ridge, J. P., Lin, M., Larsen, E. I., Fegan, M., McEwan, A. G., and Sly, L. I. (2007). A multicopper oxidase is essential for manganese oxidation and laccase-like activity in Pedomicrobium sp. ACM 3067. Environ. Microbiol. 9, 944–953. doi: 10.1111/j.1462-2920.2006.01216.x
Rodriguez, V., Moskwa, L. M., Oses, R., Kuhn, P., Riveras-Munoz, N., Seguel, O., et al. (2022). Impact of climate and slope aspects on the composition of soil bacterial communities involved in pedogenetic processes along the Chilean coastal cordillera. Microorganisms 10, 1–20. doi: 10.3390/microorganisms10050847
Rütting, T., Boeckx, P., Müller, C., and Klemedtsson, L. (2011). Assessment of the importance of dissimilatory nitrate reduction to ammonium for the terrestrial nitrogen cycle. Biogeosciences 8, 1779–1791. doi: 10.5194/bg-8-1779-2011
Segata, N., Izard, J., Waldron, L., Gevers, D., Miropolsky, L., Garrett, W. S., et al. (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12:R60. doi: 10.1186/gb-2011-12-6-r60
Seki, T., Matsumoto, A., Shimada, R., Inahashi, Y., Omura, S., and Takahashi, Y. (2012). Conexibacter arvalis sp. nov., isolated from a cultivated field soil sample. Int. J. Syst. Evol. Microbiol. 62, 2400–2404. doi: 10.1099/ijs.0.036095-0
Selensky, M. J., Masterson, A. L., Blank, J. G., Lee, S. C., and Osburn, M. R. (2021). Stable carbon isotope depletions in lipid biomarkers suggest subsurface carbon fixation in lava caves. J. Geophys. Res. Biogeosci. 126:e2021JG006430. doi: 10.1029/2021jg006430
Shi, Y., Sheng, L., Wang, Z., Zhang, X., He, N., and Yu, Q. (2016). Responses of soil enzyme activity and microbial community compositions to nitrogen addition in bulk and microaggregate soil in the temperate steppe of Inner Mongolia. Eurasian Soil Sci. 49, 1149–1160. doi: 10.1134/s1064229316100124
Shu, X., Zhang, K., Zhang, Q., and Wang, W. (2019). Ecophysiological responses of Jatropha curcas L. seedlings to simulated acid rain under different soil types. Ecotoxicol. Environ. Saf. 185:109705. doi: 10.1016/j.ecoenv.2019.109705
Soons, M. B., Hefting, M. M., Dorland, E., Lamers, L. P. M., Versteeg, C., and Bobbink, R. (2017). Nitrogen effects on plant species richness in herbaceous communities are more widespread and stronger than those of phosphorus. Biol. Conserv. 212, 390–397. doi: 10.1016/j.biocon.2016.12.006
Spence, J., and Telmer, K. (2005). The role of sulfur in chemical weathering and atmospheric CO2 fluxes: evidence from major ions, δ13CDIC, and δ34SSO4 in rivers of the Canadian cordillera. Geochim. Cosmochim. Acta 69, 5441–5458. doi: 10.1016/j.gca.2005.07.011
Spilde, M. N., Northup, D. E., Boston, P. J., Schelble, R. T., Dano, K. E., Crossey, L. J., et al. (2005). Geomicrobiology of cave ferromanganese deposits: a field and laboratory investigation. Geomicrobiol J. 22, 99–116. doi: 10.1080/01490450590945889
Stephan, M. P., Oliveira, M., Teixeira, K. R. S., Martinez-Drets, G., and Döbereiner, J. (1997). Physiology and dinitrogen fixation of Acetobacter diazotrophicus. FEMS Microbiol. Lett. 77, 67–72. doi: 10.1111/j.1574-6968.1991.tb04323.x
Steven, B., Pollard, W. H., Greer, C. W., and Whyte, L. G. (2008). Microbial diversity and activity through a permafrost/ground ice core profile from the Canadian high Arctic. Environ. Microbiol. 10, 3388–3403. doi: 10.1111/j.1462-2920.2008.01746.x
Suzuki, K. I. (2015). “Rubrobacter,” in Bergey’s Manual of Systematics of Archaea and Bacteria. ed. M. Goodfellow (John Wiley & Sons, Inc.), 1–6. doi: 10.1002/9781118960608.gbm00224
Tian, D., and Niu, S. (2015). A global analysis of soil acidification caused by nitrogen addition. Environ. Res. Lett. 10:024019. doi: 10.1088/1748-9326/10/2/024019
Too, C. C., Ong, K. S., Lee, S. M., Yule, C. M., and Keller, A. (2018). Draft genome sequence of Dyella sp. strain C9, isolated from a Malaysian tropical peat swamp forest. Microbiol. Resour. Announc. 7:e01083-18. doi: 10.1128/MRA.01083-18
Van den Heuvel, R. N., Bakker, S. E., Jetten, M. S., and Hefting, M. M. (2011). Decreased N2O reduction by low soil pH causes high N2O emissions in a riparian ecosystem. Geobiology 9, 294–300. doi: 10.1111/j.1472-4669.2011.00276.x
van Kessel, M. A., Speth, D. R., Albertsen, M., Nielsen, P. H., Op den Camp, H. J., Kartal, B., et al. (2015). Complete nitrification by a single microorganism. Nature 528, 555–559. doi: 10.1038/nature16459
Wang, C., Liu, D., and Bai, E. (2018a). Decreasing soil microbial diversity is associated with decreasing microbial biomass under nitrogen addition. Soil Biol. Biochem. 120, 126–133. doi: 10.1016/j.soilbio.2018.02.003
Wang, C., Lu, X., Mori, T., Mao, Q., Zhou, K., Zhou, G., et al. (2018b). Responses of soil microbial community to continuous experimental nitrogen additions for 13 years in a nitrogen-rich tropical forest. Soil Biol. Biochem. 121, 103–112. doi: 10.1016/j.soilbio.2018.03.009
Wang, L., Qiu, S., Guo, J., and Ge, S. (2021). Light irradiation enables rapid start-up of nitritation through suppressing nxrB gene expression and stimulating ammonia-oxidizing bacteria. Environ. Sci. Technol. 55, 13297–13305. doi: 10.1021/acs.est.1c04174
Wang, J., Shi, X., Zheng, C., Suter, H., and Huang, Z. (2021). Different responses of soil bacterial and fungal communities to nitrogen deposition in a subtropical forest. Sci. Total Environ. 755:142449. doi: 10.1016/j.scitotenv.2020.142449
Wang, M., Zheng, F., Wang, T., Lyu, Y. M., Alteen, M. G., Cai, Z. P., et al. (2019). Characterization of stackebrandtia nassauensis GH 20 beta-hexosaminidase, a versatile biocatalyst for chitobiose degradation. Int. J. Mol. Sci. 20:1243. doi: 10.3390/ijms20051243
Ward, L. M., Johnston, D. T., and Shih, P. M. (2021). Phanerozoic radiation of ammonia oxidizing bacteria. Sci. Rep. 11:2070. doi: 10.1038/s41598-021-81718-2
Weber, K. A., Achenbach, L. A., and Coates, J. D. (2006). Microorganisms pumping iron: anaerobic microbial iron oxidation and reduction. Nat. Rev. Microbiol. 4, 752–764. doi: 10.1038/nrmicro1490
Webster, K. D., Schimmelmann, A., Drobniak, A., Mastalerz, M., Lagarde, L. R., Boston, P. J., et al. (2022). Diversity and composition of methanotroph communities in caves. Microbiol. Spectr. 10, e01566–e01521. doi: 10.1101/412213
Wemheuer, F., Taylor, J. A., Daniel, R., Johnston, E., Meinicke, P., Thomas, T., et al. (2020). Tax4Fun2: prediction of habitat-specific functional profiles and functional redundancy based on 16S rRNA gene sequences. Environ. Microbiome 15:11. doi: 10.1186/s40793-020-00358-7
Wickham, H. (2016). Package ‘ggplot2’: elegant graphics for data analysis. Springer Verlag New York.
Xu, M., Zhang, Q., Xia, C., Zhong, Y., Sun, G., Guo, J., et al. (2014). Elevated nitrate enriches microbial functional genes for potential bioremediation of complexly contaminated sediments. ISME J. 8, 1932–1944. doi: 10.1038/ismej.2014.42
Xuan, C., Xiaoran, S., Zhaoji, S., Jiaen, Z., Zhong, Q., Huimin, X., et al. (2021). Analysis of the Spatio-temporal changes in acid rain and their causes in China (1998–2018). J. Res. Ecol. 12, 593–599. doi: 10.5814/j.issn.1674-764x.2021.05.002
Yang, Z., Cheng, X., Wang, H., Zeng, Z., Liu, X., Cao, J., et al. (2021). Environmental driving mechanisms and community assembly process of bacterial communities in the Luohandu cave, Guilin,Guangxi Province, China. Acta Microbiol Sin. 12, 4118–4136. doi: 10.13343/j.cnki.wsxb.20210328
Yao, M., Rui, J., Li, J., Dai, Y., Bai, Y., Heděnec, P., et al. (2014). Rate-specific responses of prokaryotic diversity and structure to nitrogen deposition in the Leymus chinensis steppe. Soil Biol. Biochem. 79, 81–90. doi: 10.1016/j.soilbio.2014.09.009
Yu, Z., He, Z., Tao, X., Zhou, J., Yang, Y., Zhao, M., et al. (2016). The shifts of sediment microbial community phylogenetic and functional structures during chromium (VI) reduction. Ecotoxicology 25, 1759–1770. doi: 10.1007/s10646-016-1719-6
Yun, Y., Wang, H., Man, B., Xiang, X., Zhou, J., Qiu, X., et al. (2016a). The relationship between pH and bacterial communities in a single karst ecosystem and its implication for soil acidification. Front. Microbiol. 7:1955. doi: 10.3389/fmicb.2016.01955
Yun, Y., Xiang, X., Wang, H., Man, B., Gong, L., Liu, Q., et al. (2016b). Five-year monitoring of bacterial communities in dripping water from the Heshang cave in Central China: implication for paleoclimate reconstruction and ecological functions. Geomicrobiol J. 33, 1–11. doi: 10.1080/01490451.2015.1062062
Zeng, Z., Cheng, X., Wang, H., Cao, J., Yang, Z., Liu, X., et al. (2022). Niche specificity and potential functions of microbial communities in karst caves as exampled by the Panlong Cave in Guilian City, Guangxi province. Earth Sci. 1–24.
Zhang, X., Wei, H., Chen, Q., and Han, X. (2014). The counteractive effects of nitrogen addition and watering on soil bacterial communities in a steppe ecosystem. Soil Biol. Biochem. 72, 26–34. doi: 10.1016/j.soilbio.2014.01.034
Zhao, R., Summers, Z. M., Christman, G. D., Yoshimura, K. M., and Biddle, J. F. (2020). Metagenomic views of microbial dynamics influenced by hydrocarbon seepage in sediments of the Gulf of Mexico. Sci. Rep. 10:5772. doi: 10.1038/s41598-020-62840-z
Zhao, R., Wang, H., Cheng, X., Yun, Y., and Qiu, X. (2018). Upland soil cluster gamma dominates the methanotroph communities in the karst Heshang cave. FEMS Microbiol. Ecol. 94, 1–13. doi: 10.1093/femsec/fiy192
Zhao, R., Wang, H., Yang, H., Yun, Y., and Barton, H. A. (2016). Ammonia-oxidizing archaea dominate ammonia-oxidizing communities within alkaline cave sediments. Geomicrobiol J. 34, 511–523. doi: 10.1080/01490451.2016.1225861
Zhou, J., Deng, Y., Luo, F., He, Z., Tu, Q., and Zhi, X. (2010). Functional molecular ecological networks. MBio 1:e00169-10. doi: 10.1128/mBio.00169-10
Zhou, J., Guan, D., Zhou, B., Zhao, B., Ma, M., Qin, J., et al. (2015). Influence of 34-years of fertilization on bacterial communities in an intensively cultivated black soil in Northeast China. Soil Biol. Biochem. 90, 42–51. doi: 10.1016/j.soilbio.2015.07.005
Zhou, Z., Wang, C., Zheng, M., Jiang, L., and Luo, Y. (2017). Patterns and mechanisms of responses by soil microbial communities to nitrogen addition. Soil Biol. Biochem. 115, 433–441. doi: 10.1016/j.soilbio.2017.09.015
Zhu, J., He, N., Wang, Q., Yuan, G., Wen, D., Yu, G., et al. (2015). The composition, spatial patterns, and influencing factors of atmospheric wet nitrogen deposition in Chinese terrestrial ecosystems. Sci. Total Environ. 511, 777–785. doi: 10.1016/j.scitotenv.2014.12.038
Keywords: karst cave, subsurface biosphere, habitat specialization, nitrogen cycling, cooccurrence network, microbial function
Citation: Liu X, Wang H, Wang W, Cheng X, Wang Y, Li Q, Li L, Ma L, Lu X and Tuovinen OH (2023) Nitrate determines the bacterial habitat specialization and impacts microbial functions in a subsurface karst cave. Front. Microbiol. 14:1115449. doi: 10.3389/fmicb.2023.1115449
Edited by:
Yizhi Sheng, China University of Geosciences, Beijing, ChinaReviewed by:
Jianjun Wang, Nanjing Institute of Geography and Limnology (CAS), ChinaLiang Guo, Xi'an University of Science and Technology, China
Copyright © 2023 Liu, Wang, Wang, Cheng, Wang, Li, Li, Ma, Lu and Tuovinen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongmei Wang, ✉ d2FuZ2htZWkwNEAxNjMuY29t; ✉ aG13YW5nQGN1Zy5lZHUuY24=
 Xiaoyan Liu
Xiaoyan Liu Hongmei Wang
Hongmei Wang Weiqi Wang
Weiqi Wang Xiaoyu Cheng
Xiaoyu Cheng Yiheng Wang
Yiheng Wang Qing Li
Qing Li Lu Li1
Lu Li1 Liyuan Ma
Liyuan Ma Xiaolu Lu
Xiaolu Lu Olli H. Tuovinen
Olli H. Tuovinen